- 1Department of Immunobiology, Yale School of Medicine, New Haven, CT, United States
- 2Department of Biochemistry and Molecular Biology and Infection and Immunity Program, Biomedicine Discovery Institute, Monash University, Melbourne, VIC, Australia
- 3Yale Center for Immuno-Oncology, Yale Center for Systems and Engineering Immunology, Yale Center for Infection and Immunity, Yale School of Medicine, New Haven, CT, United States
Advances in immunotherapy rely on targeting novel cell surface antigens, including therapeutically relevant peptide fragments presented by HLA molecules, collectively known as the actionable immunopeptidome. Although the immunopeptidome of classical HLA molecules is extensively studied, exploration of the peptide repertoire presented by non-classical HLA-E remains limited. Growing evidence suggests that HLA-E molecules present pathogen-derived and tumor-associated peptides to CD8+ T cells, positioning them as promising targets for universal immunotherapies due to their minimal polymorphism. This mini-review highlights recent developments in mass spectrometry (MS) technologies for profiling the HLA-E immunopeptidome in various diseases. We discuss the unique features of HLA-E, its expression patterns, stability, and the potential for identifying new therapeutic targets. Understanding the broad repertoire of actionable peptides presented by HLA-E can lead to innovative treatments for viral and pathogen infections and cancer, leveraging its monomorphic nature for broad therapeutic efficacy.
Introduction
The development of checkpoint blockade immunotherapy, CAR T cells, bispecific T-cell antibodies, mRNA-based vaccines, and other T-cell-based immunotherapeutic strategies have emerged in the treatment landscape over the last decade (1–4). Advances in such immunotherapies often rely on identifying actionable T cell targets represented by relatively short peptide fragments presented by HLA class I or class II molecules. It is now established that those peptides presented by HLA molecules are collectively called the immunopeptidome, which can be interrogated using mass spectrometry (MS) technologies (5). Over the last thirty years, the science of investigating the immunopeptidome, i.e. MS-based immunopeptidomics, has been mostly applied to identify and quantify peptides presented by classical HLA molecules: HLA-A, -B, and -C for class I molecules, and HLA-DP, -DQ, -DR for class II molecules (6–10). Those genes are the most polymorphic across the entire human genome, with more than 27,300 alleles for class I and 11,600 alleles for class II (https://www.ebi.ac.uk/ipd/imgt/hla/about/statistics/), making the human immunopeptidome tremendously complex from a population perspective (11–13). In contrast, very little is known about the collection of peptides that can potentially be presented by non-classical HLA-I molecules (HLA-Ib) (14), including HLA-E, which is relatively non-polymorphic with only two known alleles: HLA-E*01:01 and HLA-E*01:03 (15). Thus, the HLA-E immunopeptidome should be much simpler and widely shared across humans.
Historically, HLA-E was first established as a key regulator of innate immunity (16, 17). Its primary function is to present canonical signal peptides derived from degraded HLA-A, -B, and -C proteins to the CD94/NKG2 receptors on NK cells, thereby inhibiting NK cell functions. However, the experimental detection of HLA-E-restricted CD8+ T cells within tumors and in infection profiles of cytomegalovirus (CMV), Salmonella enterica serovar Typhimurium, Mycobacterium tuberculosis (Mtb), human immunodeficiency virus (HIV), and Epstein-Barr virus strongly supports the notion that HLA-E also participates in adaptive immune responses (15). It does so by presenting pathogenic and tumor-associated peptides as ligands for the αβTCR (T-cell receptor) of CD8+ T cells (15, 18). The emerging role of HLA-E in CD8+ T cell activation has recently increased interest in developing universal vaccination and T-cell-based immunotherapy strategies targeting HLA-E-bound viral or tumor-associated epitopes that may be shared across all individuals.
Given the tremendous advances in MS technologies in recent years, revisiting the composition of the peptide repertoire presented by HLA-E in various disease conditions using state-of-the-art MS-immunopeptidomics methods may thus provide valuable information for new therapeutic approaches. This review will discuss the features of HLA-E, its immunological positioning, the potential impact of MS technologies in exploring the HLA-E immunopeptidome, and its role in developing universal anti-viral and anti-cancer immunotherapies that could serve as safe and effective alternatives to current treatment options.
HLA-E expression patterns and stability in human tissues and cell types
Non-classical HLA-E has significantly lower RNA expression levels than classical HLA-A, -B, and -C. Specifically, a study showed that HLA-E transcripts exhibit the highest median expression in the blood (275 RPKM), spleen (187 RPKM), lung (142 RPKM), and adipose tissue (141 RPKM) whereas classical HLA-I molecules show the highest median expression levels in the whole blood (1210 RPKM), spleen (850 RPKM), bone marrow (615 RPKM), lung (433 RPKM), small intestine (482 RPKM), and lymph node (422 RPKM) (19). The Human Protein Atlas (https://www.proteinatlas.org/ENSG00000204592-HLA-E/tissue) is also a valuable resource for exploring HLA-E expression at both RNA and protein levels, providing bulk and spatial information of its expression across a wide range of tissues and cell types (Figure 1) (20). According to the Human Protein Atlas data, HLA-E expression appears to be ubiquitous yet predominant in the respiratory system, kidney, urinary bladder, bone marrow, and lymphoid tissues (Figure 1A). Upon examination at the single-cell level in the lung, endothelial cells exhibit the highest expression of HLA-E, followed by infiltrating T cells and macrophages (Figure 1B). Thus, expression data suggest that the HLA-E immunopeptidome is present across the entire organism (20).
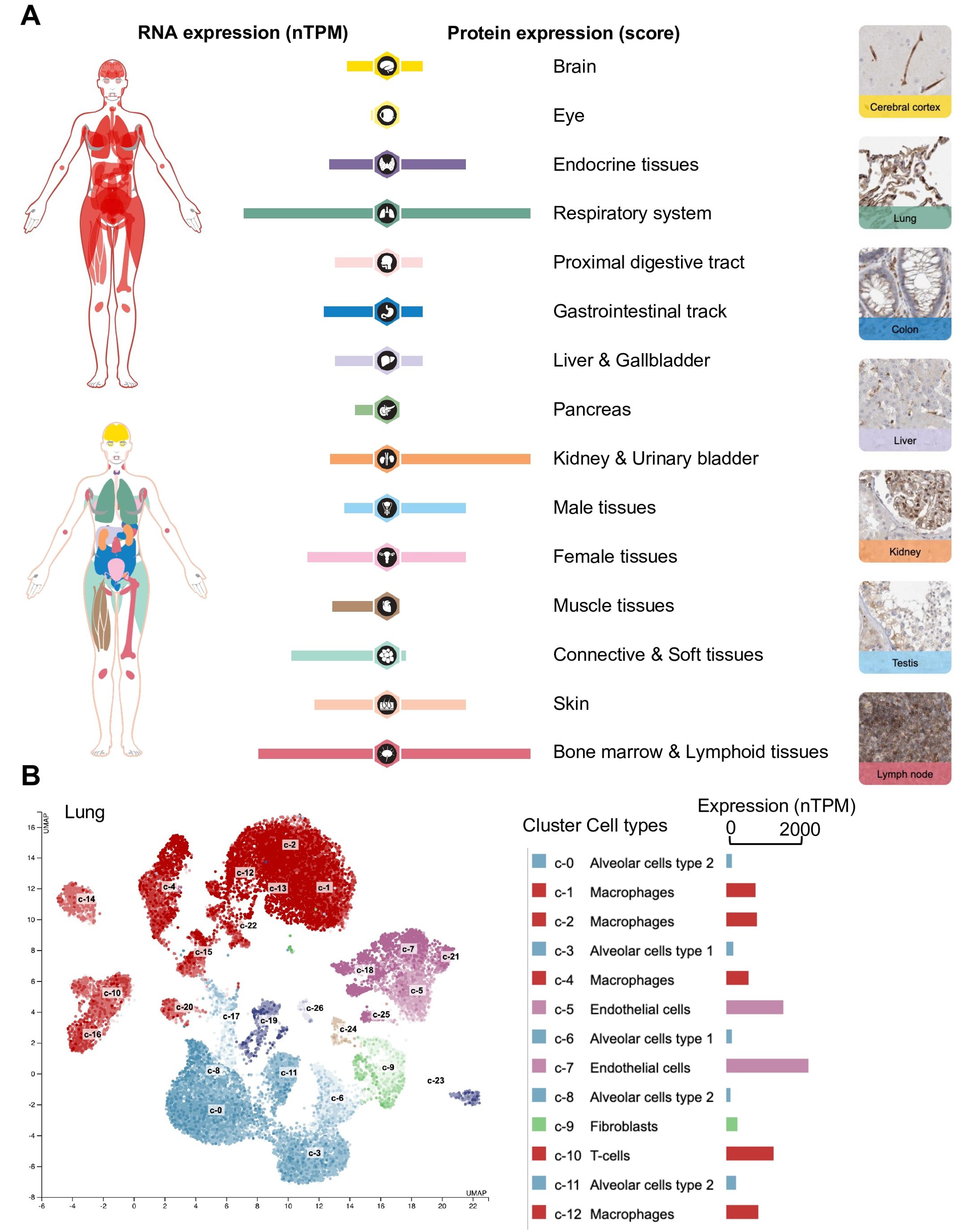
Figure 1. Exploration of HLA-E expression at the RNA and protein levels in human tissues and cell types using the Human Protein Atlas. (A) Expression profiles in human tissues of HLA-E both on the mRNA and protein level. The protein expression data from 44 normal human tissue types is derived from antibody-based protein profiling using conventional and multiplex immunohistochemistry. Each bar represents the highest expression score found in a particular group of tissues. Protein expression scores are based on a best estimate of the “true” protein expression from a knowledge-based annotation, described more in detail under Assays & annotation. The mRNA expression data is derived from RNA-seq from 40 different normal tissue types. RNA expression summary shows the consensus data based on normalized expression (nTPM) values from two different sources: internally generated Human Protein Atlas RNA-seq data and RNA-seq data from the Genotype-Tissue Expression (GTEx) project. (B) RNA expression of HLA-E at the single-cell level. The single-cell RNA sequencing (scRNAseq) data analysis was based on publicly available genome-wide expression data. Twelve out of twenty-six cluster cell types (right) are shown in the legend for simplicity. Use https://www.proteinatlas.org/ENSG00000204592-HLA-E for an in-depth exploration of HLA-E expression across human tissues and cell types. (B) was generated automatically using the Human Protein Atlas (https://www.proteinatlas.org/) (20).
Since >99% of the human population possesses either HLA-E*01:01 or HLA-E*01:03, HLA-E is relatively monomorphic. They differ by a single amino acid substitution: Arg (HLA-E*01:01) is replaced by Gly (HLA-E*01:03) at position 107, outside of the peptide-binding groove (21). The presence of Gly in HLA-E*01:03 provides increased thermal stability over Arg when bound to an equivalent peptide, resulting in a more stable expression of HLA-E*01:03 (15). Such increased stability potentially prolongs the interaction time between peptide-HLA-E (pHLA-E) complex and immune effector cells, which may generally improve the level of immunogenicity for antigenic HLA-bound peptides (22). Nevertheless, the stability of HLA-E molecules on cellular surfaces remains much lower compared to classical HLA-Ia molecules (23).
In this regard, thermostability (as a surrogate of stability) profiling of the immunopeptidome has recently been conducted for classical HLA-I-associated peptides (24). Still, it has rarely been performed for the HLA-E immunopeptidome. For instance, Walters et al. individually studied and compared the stability and structural features of the VL9- and non-VL9-HLA-E immunopeptides, demonstrating differences in peptide-binding motif and surface T cell recognition profiles (25). Thus, high-throughput profiling could provide valuable insights into predicting the duration of pHLA-E-TCR interactions, consequently improving the predictability of the likelihood of T-cell activation and peptide immunogenicity models.
Assessing the global thermostability of pHLA-E complexes could potentially yield crucial information for understanding diseases and developing universal T-cell-directed vaccines by shortlisting the potential targets. Key to this effort will be identifying the disease conditions that lead to the expansion of stable, disease-specific pHLA-E complexes recognized by CD8+ T cells. Indeed, the nomination of HLA-E-restricted T cell targets depends on establishing novel therapeutics by identifying new antigenic peptides that can stably bind to the HLA-E peptide clefts (pockets). This aim can be fulfilled with the tetramerization of peptide-HLA (pHLA) complexes to induce specific T cells. For example, Vaurs et al. produced photosensitive peptides to assemble HLA-E/pUV complexes and develop pHLA-E complexes through peptide exchange. They used ELISA to characterize the UV exchanges and utilized a new approach for peptide exchange detection by size exclusion chromatography (SEC) (26).
What is the scope of peptides exclusively presented by HLA-E alleles: a narrow range or a broad spectrum? Insights from immunopeptidomics studies
For many years, it was believed that HLA-E primarily bound a limited set of highly conserved nonameric 9-mer self-peptides derived from the leader sequences of classical HLA-Ia proteins, characterized by the consensus sequence VMAPRTL/VV/L/FL, and hence termed VL9 peptides (27). However, recent discoveries have shown that HLA-E ligands are not restricted to VL9 peptides and can originate from multiple host proteins and pathogenic sources (Table 1 and Supplementary Table S1). Additionally, various self-peptides presented by HLA-E molecules have been identified in MS-based immunopeptidomics studies (Table 1 and Supplementary Table S1). For instance, in 2016, Celik et al. conducted an immunopeptidomic study using human HLA-negative B-lymphoblastoid cell lines (LCL.721.221 and LCL.721.220) transduced with lentiviral vectors encoding soluble forms of HLA-E*01:01 or HLA-E*01:03. They identified 56 peptides of varying lengths (9-17 amino acids) in the absence of class I leader peptides, originating from diverse cellular proteins including myosin-9, histones H1.5, H2A 1-D, H2B 1-J or 1-L, and several 60S ribosomal proteins (23). However, further experimental validation is needed to confirm the HLA-E specificity of the peptides identified by MS, as antibodies that may not be specific to HLA-E, such as W6/32 (anti-HLA-ABC), are often used (Table 1).
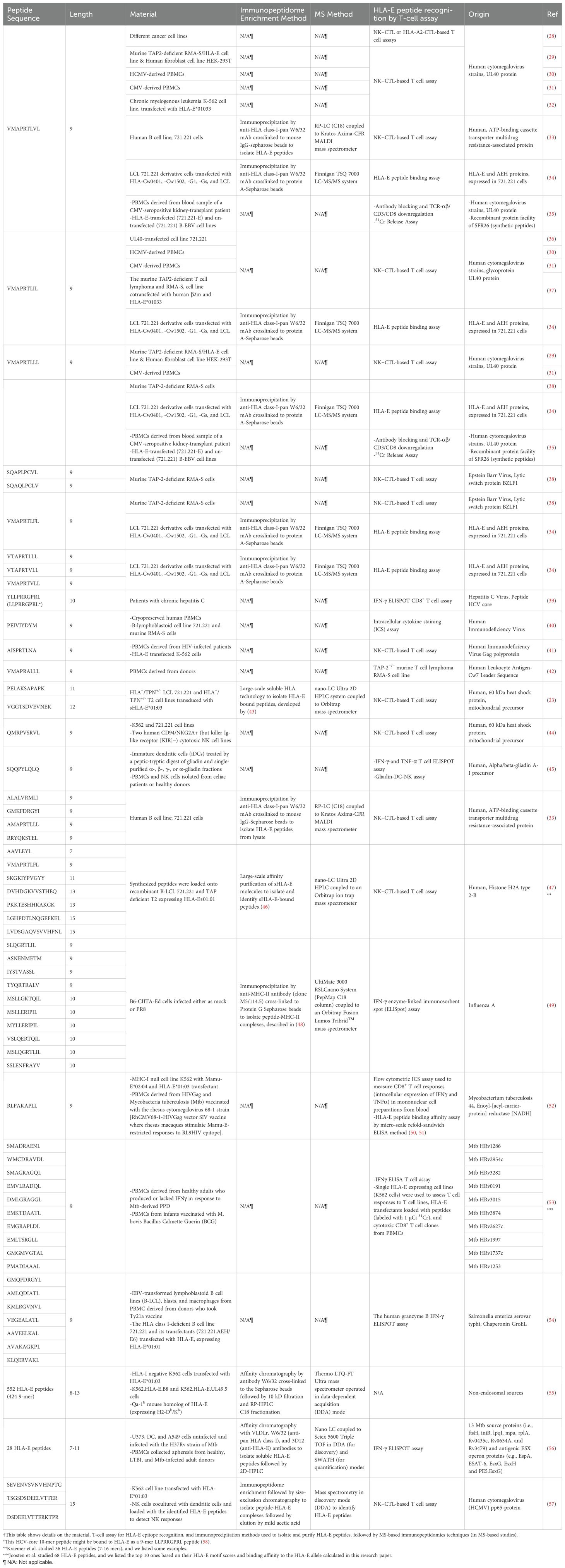
Table 1. A representative list of known HLA-E-associated epitopes originating from various protein sources †.
Proteasome-derived peptides must be bound to HLA class I molecules with enough binding affinity in the endoplasmic reticulum (ER) system to be presented on the cell surface for T cell immunosurveillance (59, 60). TAP (Transporter associated with Antigen Processing) is a key player in antigen processing, which transports peptide fragments from the cytosol into the ER system. Upon TAP impairment, new peptides have been reported to be presented by HLA-E. Specifically, in 2015, Kraemer et al. used TAP-deficient T2 cell lines transduced with soluble HLA-E*01:01. They found 36 HLA-E*01:01-bound peptides with 27 being longer than 10 amino acids (11-16 mers) and originating from various source proteins, including cell cycle regulatory proteins, matrix proteins, DNA repair proteins, and stress-induced proteins (47). However, those peptides need further experimental validation. In 2013, another study demonstrated that the repertoire of HLA-E-bound peptides retained the binding motif of HLA-A*02:01 upon TAP inhibition (55). Specifically, the authors used TAP-deficient K562 cells made by introducing the herpes virus evasion molecule UL49.5, which blocks TAP function and targets it for proteasomal degradation. Despite reduced peptide transport, surface HLA-E levels remained unchanged, indicating stabilization by an alternative, TAP-independent peptide repertoire (55). MS analysis of HLA-E-binding peptides from K562 cells revealed 552 unique peptide sequences in TAP-deficient cells (55) (Supplementary Table S1), with only eight overlapping with the TAP-proficient counterpart, indicating a distinct peptide repertoire in the absence of TAP function (55). Interestingly, the majority of the HLA-E peptide ligands were 9-mers, and few peptides were longer than 11 amino acids. The peptide sources were diverse, and many did not fit previously known processing mechanisms, suggesting other pathways, such as autophagy, might contribute to the TAP-independent peptide repertoire (55). Voogd et al. also described potential TAP-independent routes facilitating peptide processing and presentation onto HLA-E molecules (15). An alternative pathway involves furin, a protease in the trans-Golgi, which exhibits endoproteolytic activity by cleaving pro-proteins at designated C-terminal cleavage sites (15). Therefore, despite TAP impairment, HLA-E can present diverse arrays of peptides through alternative pathways. The lengths and identities of these peptides vary across different studies. In fact, a comprehensive analysis of HLA-E-bound peptides from multiple independent MS-based and non-MS studies reveals that these peptides are generally unique to each study, with minimal overlap (Figure 2 and Supplementary Table S1). Four peptides were found to be common between MS and non-MS studies (Figure 2A). All peptides were 9-mers in non-MS studies whereas peptide lengths were more variable in MS-based studies (Figure 2B). Predicted binding affinity to HLA-E using NetMHCpan 4.1 indicates a ~2-fold increase in the proportion of non-binders (NB) in MS-based studies compared to non-MS studies (Figure 2C). These predictions should be approached with caution since prediction algorithms for HLA-E ligands are still relatively sub-optimal in comparison with classical HLA-ABC, and more data are required to improve their performances. Additional comparisons between several studies show limited overlap in common HLA-E peptides (Figures 2D, E). Thus, our analysis underscores the necessity for further investigations to enhance our understanding of the HLA-E immunopeptidome under various conditions, including TAP perturbations, viral infections, and oncogenic processes.
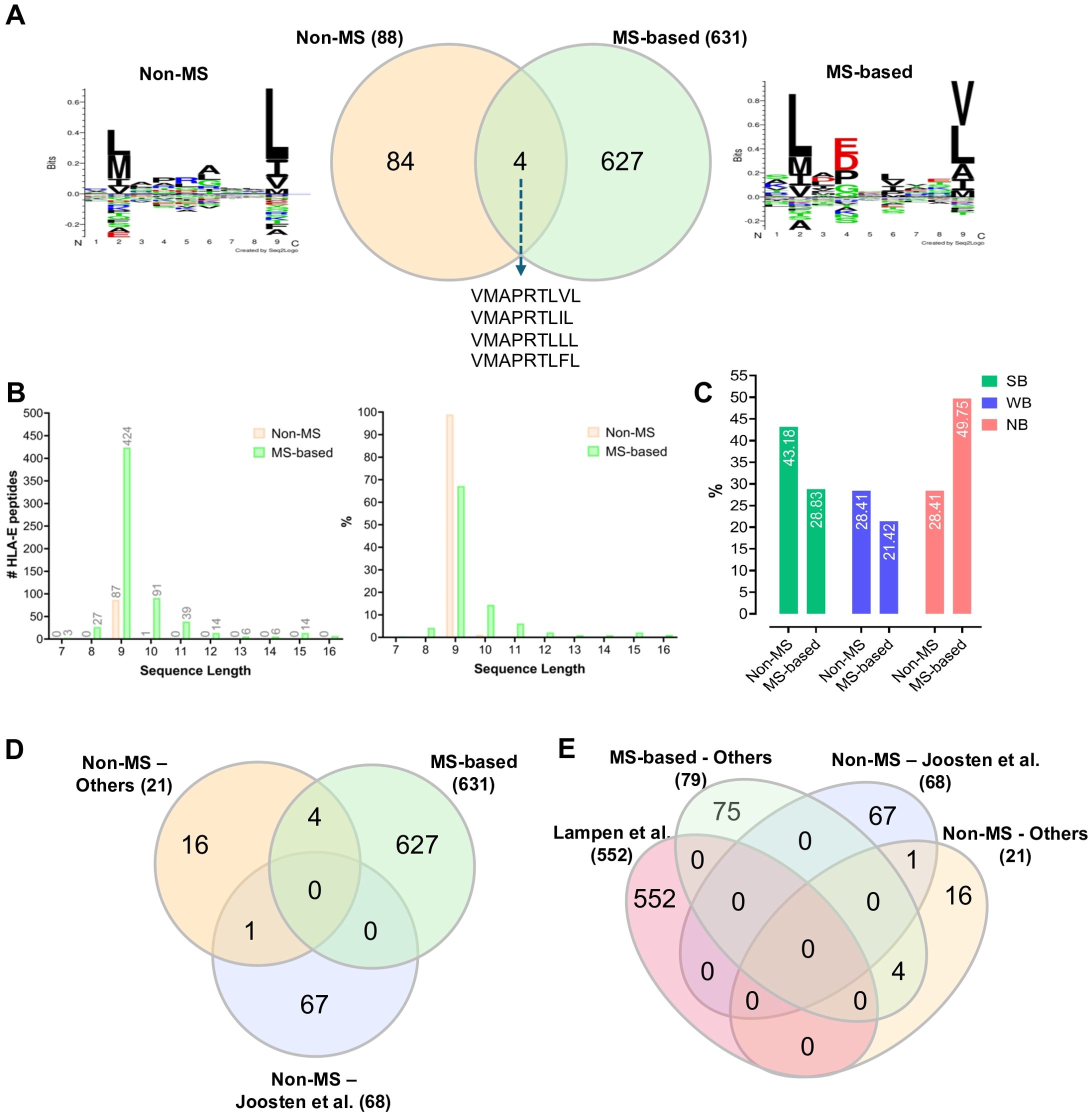
Figure 2. Comparative analysis of the previously reported HLA-E-bound peptides identified by non-MS versus MS-based immunopeptidomics techniques. (A) A two-way Venn diagram showing the overlap between the previously reported HLA-E-bound peptides and the corresponding sequence motif for each dataset. (B) Histogram showing length distribution of the previously reported HLA-E-bound peptides. (C) Histogram showing NetMHCpan 4.1 analysis of the previously reported HLA-E-bound peptides: percentages of predicted Strong Binders (SB); Weak Binders (WB) and Non-Binders (NB) are indicated (71). (D) A three-way Venn diagram comparing MS-identified HLA-E-bound peptides with findings from Joosten et al. (53), a notable non-MS study, and other non-MS studies. (E) A four-way Venn diagram illustrating the overlap among several studies that have characterized or examined higher numbers of HLA-E-bound peptides, including Lampen et al. (55) (MS-based), Joosten et al. (53) (non-MS), and other studies (see also Supplementary Table S1). Venn diagrams were generated by https://www.interactivenn.net/ (72). The sequence motifs were generated by Seq2Logo v. 2.0 (https://services.healthtech.dtu.dk/services/Seq2Logo-2.0/) (73) and GibbsCluster v. 2.0 (https://services.healthtech.dtu.dk/services/GibbsCluster-2.0/) (74) software tools. The bar graphs were generated by GraphPad Prism v. 9.3.1.
In this regard, tumor cells were shown to downregulate TAP subunits to avert T-cell clearance (61). In cancer, HLA-E peptides may take advantage of non-classical TAP-independent pathways if the classical ER-based, TAP-dependent route is unavailable. It has been found that murine TAP-/- T-cell lymphoma cells present immunogenic alternative peptides on the mouse HLA-E ortholog, Qa1, and CD8+ T-cell clone production was restricted (15). These alternative peptides are unique and have little overlap with their TAP-proficient counterparts (55). One alternative peptide was discovered from MS-based immunopeptidomic procedures: a 9-mer epitope named M-SL9 (SLQGRTLIL) associated with influenza A virus (IAV) Puerto Rico/8/1934 (PR8) within IAV-infected C57BL/6 mice (Table 1). This epitope elicits a co-immunodominant, unconventional cytolytic T-cell response, uniquely presented by Qa-1–the murine homolog of HLA-E– with cryptic origin (49). Notably, it represents the first viral CD8+ T-cell epitope sequenced to be restricted to Qa-1 and does not interact with NK cells. Researchers originally intended to investigate cryptic epitopes presented on MHC-II following IAV infection. Through this procedure, they could detect the M-SL9 epitope from a cryptic viral translation product restricted to Qa-1 that drives a large percentage of the CD8+ T-cell response (49). The preliminary success of this procedure demonstrates the potential of identifying other viral cryptic antigens presented by HLA-E. On a technical note, this group employed an anti-MHC-II antibody that actually cross-reacted against Qa-1, leading to the unexpected isolation and detection of the cryptic epitope. With this notable cross-reactivity in mind, it has been found that antibodies 3D12 and 4D12 recognize HLA-E without cross-reacting with classical HLA-Ia molecules, serving as potential antibodies for future HLA-E-specialized immunopeptidomic procedures (62). However, it is worth mentioning that these two antibodies have different recognition patterns that could impact immunopeptidome recovery.
Utilizing MHC-E-restricted CD8+ T cells in vaccines and immunotherapies against pathogens; viruses and cancer
MHC-E-restricted CD8+ T cells in T-cell-based vaccines and immune therapies offer distinct advantages over classically restricted CD8+ T cells. HLA-E molecules are ubiquitously expressed and have only two major allomorphs, which possess identical peptide-binding grooves. This characteristic enables the design of vaccines and therapies for the general population, eliminating the need for personalized approaches often required with MHC-Ia-restricted T cells (49). For instance, in the context of pathogen infections, a recent study described a TCR-based bispecific molecule that potently and selectively binds HLA-E in complex with a peptide encoded by the inhA gene of Mtb (63). The authors demonstrated the elimination of Mtb-infected cells and the reduction of intracellular Mtb growth. Moreover, the researchers revealed the biophysical and structural bases underpinning the potency and specificity of their TCR-based bispecific molecule, thereby providing a proof of principle for an innovative TCR-based therapeutic strategy capable of surmounting HLA polymorphism hurdles and thus promising broader applicability across patient populations.
Within the realm of vaccines, MHC-E-restricted CD8+ T cells have demonstrated distinctive protective abilities against HIV/SIV infections (64–66). HIV infection is generally known to downregulate classical HLA-Ia antigen expression via Nef proteins (67). In contrast, HLA-E expression is not affected by HIV infection and is continuously expressed to shield cells from NK-mediated lysis (68). The continuous expression of HLA-E upon infection has sparked interest in identifying and targeting MHC-E-associated epitopes for vaccine strategies against other viruses. To this end, an important aspect of consideration is the consistent and stable presentation of viral peptides within the HLA-E groove to ensure robust on-target T-cell activity and clinical effectiveness. High stability is particularly challenging for non-classical pHLA-E complexes, as they have been shown to be less stable than classical pHLA-Ia complexes (18). For instance, Wallace et al. recently employed immunopeptidomics in an effort to identify stable HLA-E-bound HIV-derived peptides that could be utilized to generate bispecific high-affinity T cell receptors (ImmTAX) (18). However, the instability of the predicted HIV-derived HLA peptide ligands posed a challenge in identifying HLA-E-bound peptides as potential therapeutic targets. As a result, Wallace et al. were unable to detect any HIV-derived HLA-E-bound peptides using MS-based immunopeptidomics techniques with the 3D12 antibody (18).
Recently, an mRNA vaccine was developed to trigger an unclassical CD8+ T-cell response against IAV within infected mice, demonstrating the feasibility of employing mRNA vaccines to investigate the regulative and protective functions of HLA-E-restricted CD8+ T cells (49). As aforementioned, we anticipate that systematic thermostability profiling studies of the HLA-E immunopeptidome after infection and/or vaccination will become critically important in the future to discover stable and immunogenic HLA-E peptides for the optimal design of universal HLA-E-restricted CD8+ T cell vaccines (24).
In the field of oncology, a recent study explored the potential of rhesus cytomegalovirus (RhCMV) vectors, genetically engineered to trigger an HLA-E-restricted CD8+ T cell response targeting tumor-associated antigens (TAAs), in generating an effective anti-tumor reaction against prostate cancer (69). Interestingly, the authors showed that their cancer vaccine could trigger an HLA-E-restricted CD8+ T cells response to non-canonical epitopes, including a peptide encoded by prostatic acidic phosphatase (PAP) (69). Their data indicate that T cell responses to TAAs were comparable to viral antigen-specific responses and, therefore, suggest that the HLA-E immunopeptidome can be exploited for CD8+ T cell-based immunotherapies against cancer.
Finally, the observed rapid internalization of HLA-E-peptide complexes into the cell could also represent an opportunity for therapeutic innovation. For instance, antibody-drug conjugates (ADCs) tailored to target precise TAAs bound to HLA-E could capitalize on rapid intracellular internalization, enabling highly effective delivery of potent cytotoxic agents directly into cancer cells, thereby acting as “biological missile” (70). The strategic screening of ADCs for optimal binding to HLA-E-TAA complexes and rapid internalization properties hold promise for significant therapeutic advancements in clinical applications.
Conclusion
In summary, peptides presented by HLA-E, a non-classical HLA-Ib molecule, hold promise as actionable targets for a wide patient population. MS-based immunopeptidomics stands as a valuable tool in identifying these clinically relevant targets. Despite limited studies to date, employing MS-based immunopeptidomics has successfully uncovered both canonical and non-canonical CD8+ T cell epitopes presented by HLA-E molecules. As antibody specificity for isolation of pHLA-E and MS instrumentation sensitivity continue to improve, there is considerable potential for high-throughput discovery and validation of HLA-E viral epitopes and tumor antigens. In the context of anti-cancer immunotherapeutics, tumors often manipulate the antigen processing and presentation pathway to evade the immune response, such as through TAP-deficiency in various tumor cells. As discussed earlier, the downregulation of TAP leads to an expansion of the HLA-E immunopeptidome, suggesting a plausible avenue for anti-cancer treatments centered around targeting TAP-independent HLA-E epitopes. By continuing to explore and exploit the potential of the HLA-E immunopeptidome in immunotherapy using MS technologies, we may unlock new avenues for improving patient outcomes across a diverse range of diseases.
Author contributions
MW: Writing – original draft. MS: Formal analysis, Writing – review & editing. SK: Writing – review & editing. EC: Supervision, Funding acquisition, Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This article was supported by a startup package provided by Yale University to establish the Caron laboratory.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1442783/full#supplementary-material
References
1. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. (2020) 20:651–68. doi: 10.1038/s41577-020-0306-5
2. Brudno JN, Kochenderfer JN. Current understanding and management of CAR T cell-associated toxicities. Nat Rev Clin Oncol. (2024) 21(7):501–21. doi: 10.1038/s41571-024-00903-0
3. Hsiue EH-C, Wright KM, Douglass J, Hwang MS, Mog BJ, Pearlman AH, et al. Targeting a neoantigen derived from a common TP53 mutation. Science. (2021) 371:eabc8697. doi: 10.1126/science.abc8697
4. Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. (2020) 586:594–9. doi: 10.1038/s41586-020-2814-7
5. Caron E, Kowalewski D, Chiek Koh C, Sturm T, Schuster H, Aebersold R. Analysis of major histocompatibility complex (MHC) immunopeptidomes using mass spectrometry*. Mol Cell Proteomics. (2015) 14:3105–17. doi: 10.1074/mcp.O115.052431
6. Abelin JG, Harjanto D, Malloy M, Suri P, Colson T, Goulding SP, et al. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity. (2019) 51(4):766–79.e17. doi: 10.1016/j.immuni.2019.08.012
7. Phulphagar KM, Ctortecka C, Jacome ASV, Klaeger S, Verzani EK, Hernandez GM, et al. Sensitive, high-throughput HLA-I and HLA-II immunopeptidomics using parallel accumulation-serial fragmentation mass spectrometry. Mol Cell Proteomics. (2023) 22(6):100563. doi: 10.1016/j.mcpro.2023.100563
8. Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, Sidney J, et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity. (2017) 46:315–26. doi: 10.1016/j.immuni.2017.02.007
9. Caron E, Vincent K, Fortier MH, Laverdure JP, Bramoullé A, Hardy MP, et al. The MHC I immunopeptidome conveys to the cell surface an integrative view of cellular regulation. Mol Syst Biol. (2011) 7:533–. doi: 10.1038/msb.2011.68
10. Fortier M-H, Caron É, Hardy M-P, Voisin G, Lemieux S, Perreault C, et al. The MHC class I peptide repertoire is molded by the transcriptome. J Exp Med. (2008) 205:595–610. doi: 10.1084/jem.20071985
11. Admon A, Bassani-Sternberg M. The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol Cell Proteomics. (2011) 10:O111.011833. doi: 10.1074/mcp.O111.011833
12. Caron E, Aebersold R, Banaei-Esfahani A, Chong C, Bassani-Sternberg M. A case for a human immuno-peptidome project consortium. Immunity. (2017) 47:203 8. doi: 10.1016/j.immuni.2017.07.010
13. Vizcaíno JA, Kubiniok P, Kovalchik KA, Ma Q, Duquette JD, Mongrain I, et al. The human immunopeptidome project: A roadmap to predict and treat immune diseases*. Mol Cell Proteomics. (2020) 19:31–49. doi: 10.1074/mcp.R119.001743
14. Dudek NL, Purcell AW. Repertoire of nonclassical MHC I (HLA-E, HLA-F, HLA-G, and orthologues). In: Ratcliffe MJH, editor. Encyclopedia of Immunobiology. Academic Press, Oxford (2016). p. 215–9. doi: 10.1016/j.it.2022.03.002
15. Voogd L, Ruibal P, Ottenhoff THM, Joosten SA. Antigen presentation by MHC-E: a putative target for vaccination? Trends Immunol. (2022) 43:355–65. doi: 10.1016/j.it.2022.03.002
16. Huisman BD, Guan N, Rückert T, Garner L, Singh NK, McMichael AJ, et al. High-throughput characterization of HLA-E-presented CD94/NKG2x ligands reveals peptides which modulate NK cell activation. Nat Commun. (2023) 14:4809. doi: 10.1038/s41467-023-40220-1
17. D’Souza MP, Adams E, Altman JD, Birnbaum ME, Boggiano C, Casorati G, et al. Casting a wider net: Immunosurveillance by nonclassical MHC molecules. PloS Pathogens. (2019) 15:e1007567. doi: 10.1371/journal.ppat.1007567
18. Wallace Z, Heunis T, Paterson RL, Suckling RJ, Grant T, Dembek M, et al. Instability of the HLA-E peptidome of HIV presents a major barrier to therapeutic targeting. Mol Ther. (2024) 32:678–88. doi: 10.1016/j.ymthe.2024.01.010
19. Boegel S, Löwer M, Bukur T, Sorn P, Castle JC, Sahin U. HLA and proteasome expression body map. BMC Med Genomics. (2018) 11:36. doi: 10.1186/s12920-018-0354-x
20. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. (2015) 347:1260419. doi: 10.1126/science.1260419
21. Barker DJ, Maccari G, Georgiou X, Cooper MA, Flicek P, Robinson J, et al. The IPD-IMGT/HLA database. Nucleic Acids Res. (2022) 51:D1053–D60. doi: 10.1093/nar/gkac1011
22. Blaha DT, Anderson SD, Yoakum DM, Hager MV, Zha Y, Gajewski TF, et al. High-throughput stability screening of neoantigen/HLA complexes improves immunogenicity predictions. Cancer Immunol Res. (2019) 7:50–61. doi: 10.1158/2326-6066.CIR-18-0395
23. Celik AA, Kraemer T, Huyton T, Blasczyk R, Bade-Döding C. The diversity of the HLA-E-restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics. (2016) 68:29–41. doi: 10.1007/s00251-015-0880-z
24. Jappe EC, Garde C, Ramarathinam SH, Passantino E, Illing PT, Mifsud NA, et al. Thermostability profiling of MHC-bound peptides: a new dimension in immunopeptidomics and aid for immunotherapy design. Nat Commun. (2020) 11:6305. doi: 10.1038/s41467-020-20166-4
25. Walters LC, Rozbesky D, Harlos K, Quastel M, Sun H, Springer S, et al. Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes. Cell Rep. (2022) 39:110959. doi: 10.1016/j.celrep.2022.110959
26. Vaurs J, Douchin G, Echasserieau K, Oger R, Jouand N, Fortun A, et al. A novel and efficient approach to high-throughput production of HLA-E/peptide monomer for T-cell epitope screening. Sci Rep. (2021) 11:17234. doi: 10.1038/s41598-021-96560-9
27. Braud VM, Allan DSJ, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. (1998) 8:1–10. doi: 10.1016/S0960-9822(98)70014-4
28. Sensi M, Pietra G, Molla A, Nicolini G, Vegetti C, Bersani I, et al. Peptides with dual binding specificity for HLA-A2 and HLA-E are encoded by alternatively spliced isoforms of the antioxidant enzyme peroxiredoxin 5. Int Immunol. (2009) 21:257–68. doi: 10.1093/intimm/dxn141
29. Pietra G, Romagnani C, Mazzarino P, Falco M, Millo E, Moretta A, et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci. (2003) 100:10896–901. doi: 10.1073/pnas.1834449100
30. Mazzarino P, Pietra G, Vacca P, Falco M, Colau D, Coulie P, et al. Identification of effector-memory CMV-specific T lymphocytes that kill CMV-infected target cells in an HLA-E-restricted fashion. Eur J Immunol. (2005) 35:3240–7. doi: 10.1002/(ISSN)1521-4141
31. Hoare HL, Sullivan LC, Pietra G, Clements CS, Lee EJ, Ely LK, et al. Structural basis for a major histocompatibility complex class Ib–restricted T cell response. Nat Immunol. (2006) 7:256–64. doi: 10.1038/ni1312
32. Ulbrecht M, Hofmeister V, Yüksekdag G, Ellwart JW, Hengel H, Momburg F, et al. HCMV glycoprotein US6 mediated inhibition of TAP does not affect HLA-E dependent protection of K-562 cells from NK cell lysis. Hum Immunol. (2003) 64:231–7. doi: 10.1016/S0198-8859(02)00788-7
33. Wooden SL, Kalb SR, Cotter RJ, Soloski MJ. Cutting edge: HLA-E binds a peptide derived from the ATP-binding cassette transporter multidrug resistance-associated protein 7 aSnd inhibits NK cell-mediated lysis1. J Immunol. (2005) 175:1383–7. doi: 10.4049/jimmunol.175.3.1383
34. Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences1. J Immunol. (1998) 160:4951–60. doi: 10.4049/jimmunol.160.10.4951
35. Allard M, Tonnerre P, Nedellec S, Oger R, Morice A, Guilloux Y, et al. HLA-E-restricted cross-recognition of allogeneic endothelial cells by CMV-associated CD8 T cells: A potential risk factor following transplantation. PloS One. (2012) 7:e50951. doi: 10.1371/journal.pone.0050951
36. Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. (2000) 287:1031–3. doi: 10.1126/science.287.5455.1031
37. Heatley SL, Pietra G, Lin J, Widjaja JML, Harpur CM, Lester S, et al. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells*. J Biol Chem. (2013) 288:8679–90. doi: 10.1074/jbc.M112.409672
38. García P, Llano M, Heredia A, Willberg CB, Caparrós E, Aparicio P, et al. Human T cell receptor-mediated recognition of HLA-E. Eur J Immunol. (2002) 32:936–44. doi: 10.1002/(ISSN)1521-4141
39. Schulte D, Vogel M, Langhans B, Krämer B, Körner C, Nischalke HD, et al. The HLA-ER/HLA-ER genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E–restricted recognition of an HCV-derived peptide by interferon-γ–secreting human CD8+ T cells. J Infect Diseases. (2009) 200:1397–401. doi: 10.1086/605889
40. Hannoun Z, Lin Z, Brackenridge S, Kuse N, Akahoshi T, Borthwick N, et al. Identification of novel HIV-1-derived HLA-E-binding peptides. Immunol Letters. (2018) 202:65–72. doi: 10.1016/j.imlet.2018.08.005
41. Nattermann J, Nischalke HD, Hofmeister V, Kupfer B, Ahlenstiel G, Feldmann G, et al. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antiviral Ther. (2004) 10:95–107. doi: 10.1177/135965350501000107
42. Romagnani C, Pietra G, Falco M, Millo E, Mazzarino P, Biassoni R, et al. Identification of HLA-E-specific alloreactive T lymphocytes: A cell subset that undergoes preferential expansion in mixed lymphocyte culture and displays a broad cytolytic activity against allogeneic cells. Proc Natl Acad Sci. (2002) 99:11328–33. doi: 10.1073/pnas.172369799
43. Kunze-Schumacher H, Blasczyk R, Bade-Doeding C. Soluble HLA technology as a strategy to evaluate the impact of HLA mismatches. J Immunol Res. (2014) 2014:246171. doi: 10.1155/2014/246171
44. Michaëlsson J, Matos C, Achour A, Lanier LL, Kärre K, Söderström K. A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J Exp Med. (2002) 196:1403–14. doi: 10.1084/jem.20020797
45. Terrazzano G, Sica M, Gianfrani C, Mazzarella G, Maurano F, Giulio BD, et al. Gliadin regulates the NK-dendritic cell cross-talk by HLA-E surface stabilization. J Immunol. (2007) 179:372–81. doi: 10.4049/jimmunol.179.1.372
46. Bade-Doeding C, Cano P, Huyton T, Badrinath S, Eiz-Vesper B, Hiller O, et al. Mismatches outside exons 2 and 3 do not alter the peptide motif of the allele group B*44:02P. Hum Immunol. (2011) 72:1039–44. doi: 10.1016/j.humimm.2011.08.004
47. Kraemer T, Celik AA, Huyton T, Kunze-Schumacher H, Blasczyk R, Bade-Döding C. HLA-E: presentation of a broader peptide repertoire impacts the cellular immune response—Implications on HSCT outcome. Stem Cells Int. (2015) 2015:346714. doi: 10.1155/2015/346714
48. Purcell AW, Ramarathinam SH, Ternette N. Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc. (2019) 14:1687–707. doi: 10.1038/s41596-019-0133-y
49. Hogan MJ, Maheshwari N, Begg BE, Nicastri A, Hedgepeth EJ, Muramatsu H, et al. Cryptic MHC-E epitope from influenza elicits a potent cytolytic T cell response. Nat Immunol. (2023) 24:1933–46. doi: 10.1038/s41590-023-01644-5
50. Strong RK, Holmes MA, Li P, Braun L, Lee N, Geraghty DE. HLA-E allelic variants: Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities*. J Biol Chem. (2003) 278:5082–90. doi: 10.1074/jbc.M208268200
51. Sylvester-Hvid C, Kristensen N, Blicher T, Ferré H, Lauemøller SL, Wolf XA, et al. Establishment of a quantitative ELISA capable of determining peptide – MHC class I interaction. Tissue Antigens. (2002) 59:251–8. doi: 10.1034/j.1399-0039.2002.590402.x
52. Walters LC, Harlos K, Brackenridge S, Rozbesky D, Barrett JR, Jain V, et al. Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat Commun. (2018) 9:3137. doi: 10.1038/s41467-018-05459-z
53. Joosten SA, Meijgaarden K, Weeren P, Kazi F, Geluk A, Savage NDL, et al. Mycobacterium tuberculosis Peptides Presented by HLA-E Molecules Are Targets for Human CD8+ T-Cells with Cytotoxic as well as Regulatory Activity. PloS Pathogens. (2010) 6:e1000782. doi: 10.1371/journal.ppat.1000782
54. Salerno-Gonçalves R, Fernandez-Viña M, Lewinsohn DM, Sztein MB. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with salmonella enterica serovar typhi strain ty21a typhoid vaccine. J Immunol. (2004) 173:5852–62. doi: 10.4049/jimmunol.173.9.5852
55. Lampen MH, Hassan C, Sluijter M, Geluk A, Dijkman K, Tjon JM, et al. Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol Immunol. (2013) 53:126–31. doi: 10.1016/j.molimm.2012.07.009
56. McMurtrey C, Harriff MJ, Swarbrick GM, Duncan A, Cansler M, Null M, et al. T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PloS One. (2017) 12:e0188288. doi: 10.1371/journal.pone.0188288
57. Martín Almazán N, Sala BM, Sandalova T, Sun Y, Resink T, Cichocki F, et al. Non-classical HLA-E restricted CMV 15-mer peptides are recognized by adaptive NK cells and induce memory responses. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1230718
58. Walters LC, McMichael AJ, Gillespie GM. Detailed and atypical HLA-E peptide binding motifs revealed by a novel peptide exchange binding assay. Eur J Immunol. (2020) 50:2075–91. doi: 10.1002/eji.202048719
59. Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. (2011) 11:823. doi: 10.1038/nri3084
60. Rock KL, Reits E, Neefjes J. Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. (2016) 37:724–37. doi: 10.1016/j.it.2016.08.010
61. Marijt KA, Doorduijn EM, Hall Tv. TEIPP antigens for T-cell based immunotherapy of immune-edited HLA class Ilow cancers. Mol Immunol. (2019) 113:43–9. doi: 10.1016/j.molimm.2018.03.029
62. Brackenridge S, John N, Früh K, Borrow P, McMichael AJ. The antibodies 3D12 and 4D12 recognise distinct epitopes and conformations of HLA-E. Front Immunol. (2024) 15:1329032. doi: 10.3389/fimmu.2024.1329032
63. Paterson RL, Manna MPL, Souza VAD, Walker A, Gibbs-Howe D, Kulkarni R, et al. An HLA-E-targeted TCR bispecific molecule redirects T cell immunity against Mycobacterium tuberculosis. Proc Natl Acad Sci United States America. (2024) 121:e2318003121. doi: 10.1073/pnas.2318003121
64. Hansen SG, Hancock MH, Malouli D, Marshall EE, Hughes CM, Randall KT, et al. Myeloid cell tropism enables MHC-E–restricted CD8+ T cell priming and vaccine efficacy by the RhCMV/SIV vaccine. Sci Immunol. (2022) 7:eabn9301. doi: 10.1126/sciimmunol.abn9301
65. Verweij MC, Hansen SG, Iyer R, John N, Malouli D, Morrow D, et al. Modulation of MHC-E transport by viral decoy ligands is required for RhCMV/SIV vaccine efficacy. Science. (2021) 372(6541):eabe9233. doi: 10.1126/science.abe9233
66. Hansen SG, Piatak M, Ventura AB, Hughes CM, Gilbride RM, Ford JC, et al. Immune clearance of highly pathogenic SIV infection. Nature. (2013) 502:100–4. doi: 10.1038/nature12519
67. Collins KL, Chen BK, Kalams SA, Walker BD, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. (1998) 391:397–401. doi: 10.1038/34929
68. Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. (1999) 10:661–71. doi: 10.1016/S1074-7613(00)80065-5
69. Iyer RF, Verweij MC, Nair SS, Morrow D, Mansouri M, Chakravarty D, et al. CD8+ T cell targeting of tumor antigens presented by HLA-E. Sci Adv (2024) 10(19):eadm7515. doi: 10.1126/sciadv.adm7515
70. Fu Z, Li S, Han S, Shi C, Zhang Y. Antibody drug conjugate: the “biological missile” for targeted cancer therapy. Signal Transduction Targeted Ther. (2022) 7:93. doi: 10.1038/s41392-022-00947-7
71. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. (2020) 48:W449–W54. doi: 10.1093/nar/gkaa379
72. Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. (2015) 16:169. doi: 10.1186/s12859-015-0611-3
73. Thomsen MCF, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. (2012) 40:W281–W7. doi: 10.1093/nar/gks469
Keywords: HLA-E peptides, immunopeptidome, mass spectrometry, vaccine design, viral infection, cancer immunotherapy
Citation: Weitzen M, Shahbazy M, Kapoor S and Caron E (2024) Deciphering the HLA-E immunopeptidome with mass spectrometry: an opportunity for universal mRNA vaccines and T-cell-directed immunotherapies. Front. Immunol. 15:1442783. doi: 10.3389/fimmu.2024.1442783
Received: 02 June 2024; Accepted: 15 August 2024;
Published: 05 September 2024.
Edited by:
Simone A. Joosten, Leiden University Medical Center (LUMC), NetherlandsReviewed by:
Huang Huang, Gilead, United StatesCopyright © 2024 Weitzen, Shahbazy, Kapoor and Caron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Etienne Caron, ZXRpZW5uZS5jYXJvbkB5YWxlLmVkdQ==
†These authors have contributed equally to this work
 Maya Weitzen
Maya Weitzen Mohammad Shahbazy
Mohammad Shahbazy Saketh Kapoor
Saketh Kapoor Etienne Caron
Etienne Caron