
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 16 July 2024
Sec. Mucosal Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1432651
This article is part of the Research Topic The Synthesis of Secretory Immunoglobulin A in Mucosal Tissue: Mucosal-associated Invariant T, T Follicular Helper, and B Cells View all 5 articles
 Mengge Gao1
Mengge Gao1 Xiaosu Zhao1,2,3*
Xiaosu Zhao1,2,3*Mucosa-associated invariant T (MAIT) cells are a subset of innate-like non-conventional T cells characterized by multifunctionality. In addition to their well-recognized antimicrobial activity, increasing attention is being drawn towards their roles in tissue homeostasis and repair. However, the precise mechanisms underlying these functions remain incompletely understood and are still subject to ongoing exploration. Currently, it appears that the tissue localization of MAIT cells and the nature of the diseases or stimuli, whether acute or chronic, may induce a dynamic interplay between their pro-inflammatory and anti-inflammatory, or pathogenic and reparative functions. Therefore, elucidating the conditions and mechanisms of MAIT cells’ reparative functions is crucial for fully maximizing their protective effects and advancing future MAIT-related therapies. In this review, we will comprehensively discuss the establishment and potential mechanisms of their tissue repair functions as well as the translational application prospects and current challenges in this field.
Over the past three decades, innate-like lymphocytes such as mucosa-associated invariant T (MAIT) cells, NKT cells, and γδ T cells have emerged as a unique class of immune cells that bridge innate and adaptive immunity. These cells acquire their effector functions during development and stably reside in peripheral tissues, ready to respond immediately upon recognizing primary antigens (1–5). MAIT cells (CD3+CD161hiVα7.2+) express a conserved TCRα chain Vα7.2-Jα33 (corresponding to mouse Vα19–Jα33) (4–6), primarily recognizing monomorphic MHC-1-related molecule (MR1). This recognition allows them to respond to small molecule antigens such as 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU) and 5-(2-oxoethylideneamino)-6-D-ribitylaminouracil (5-OE-RU), derived from the riboflavin metabolic pathway produced by the microbiota (7, 8). Consequently, MAIT cells are considered particularly dependent on the microbiota. In germ-free (GF) mice, the frequency of MAIT cells is significantly lower compared to those raised under specific-pathogen-free (SPF) conditions (2, 9, 10).
In recent years, transcriptomic technologies have greatly advanced the exploration of MAIT cell heterogeneity across various contexts. Beyond their well-documented anti-infective activities against bacteria and viruses (11–17), recent studies have highlighted the crucial role of MAIT cells in tissue homeostasis and repair (2, 18–22). Current research indicates that MAIT cells contribute to tissue repair primarily through homing and chemotactic migration. These cells interact with mucosal microbiota or other immune cells, leading to the inhibition of inflammatory responses or the secretion of repair mediators. This process maintains mucosal barrier function and facilitates tissue repair. The following discussion provides a detailed examination of the significant properties of MAIT cells in tissue repair and their potential future applications.
MAIT cells are enriched in human intestines (10%) (2, 23–27), lungs (3%) (28), liver (10%-40%) (29), skin (0.5%-2%) (8) (Above frequency as % of total T cells or αβT cells.), as well as urogenital tract (30–32). They constitute a crucial subset of immune cells within mucosal tissues. These tissues are also colonized by diverse microbial communities, which play a significant role in the development, differentiation, and activation of MAIT cells (2, 15–18, 33). MAIT cells are classified as MAIT1 and MAIT17 based on the expression of transcription factors T-bet and RORγt, respectively. However, in humans, MAIT cells exhibit a mixed gene expression pattern without distinct functional subsets (34, 35). In naive SPF mice, tissue-resident MAIT cells are predominantly RORγt+T-bet− (MAIT17), producing IL-17A, while a minor population of circulating MAIT cells is T-bet+RORγt− (MAIT1), producing IFN-γ. Following intranasal infection with pathogens such as Salmonella or Legionella, an increase in RORγt+T-bet+ lung MAIT cells is observed. This suggests that the diverse stimuli received by human MAIT cells may explain some interspecies differences (36, 37). Additionally, in the female reproductive tract and oral mucosa, barrier MAIT cells predominantly exhibit a mature CD4−CD8− MAIT17 phenotype with higher RORγt and lower T-bet expression (23, 38, 39). The transcriptional profile of mouse tissues reveals specific differences with lung tissue tending towards MAIT17 and liver tissue towards MAIT1 (34), likely due to microbial exposure or other environmental factors.
The enrichment of MAIT cells within tissues is primarily due to their high surface expression of various chemokine receptors and tissue residency markers, such as CD69 and CD103 (23, 24, 29, 40). The tissue homing process is driven by the master transcription factor promyelocytic leukemia zinc finger protein (PLZF), which decreases the expression of Klf2 and its target CD62L (7, 33, 40–45). Within the thymus, MAIT cells acquire distinctive tissue residency traits (33, 41–44) and exhibit high levels of CCR2, CCR8, and CXCR6 expression. Upon exiting the thymus, these cells are directed towards different non-lymphoid tissues, with tropism potentially varying among cell subsets (29, 33–35, 46). MAIT1 cells are primarily localized in the spleen, lymph nodes, and liver, whereas MAIT17 cells are enriched in barrier tissues such as the lungs, skin, and intestines (1, 33–35, 41–46), possibly driven by differentiation programs involving the expression of T-bet and RORγt and surface chemokine receptor profiles.
Single-cell sequencing data reveal that circulating MAIT cells express various chemokine receptor receptors, including CCR2 (associated with inflammation tissue infiltration), CCR5, CCR8 (potentially targeting skin and lung tissues), CXCR3 (homing to inflamed tissues), CCR6 (tropism towards skin, gut, and brain), and CXCR6 (tropism towards gut-liver interfaces) (23, 28, 29, 33, 47). CXCR3 is predominantly expressed in the MAIT1 subset, facilitating their migration towards sites of infection or inflammation (35, 45, 46, 48), while MAIT17 cells exhibit high expression of CCR6 and CXCR6 (35, 45, 46, 49), associated with mucosal migration to organs such as the liver (29, 50–52), intestines (53), and lungs (28). This diversity leads to heterogeneity in MAIT cell phenotypes and subsets between blood and tissues, as well as within tissues themselves. However, it is currently unclear whether tissue-resident mucosal MAIT cells can exit mucosal tissues and recirculate.
Under pathological conditions such as mucosal inflammation or tissue injury, MAIT cell numbers at the site of pathology rapidly increase through both in-situ proliferation and recruitment from circulation (Figure 1). For instance, in pulmonary infection with Francisella tularensis live vaccine strain (LVS), CXCR6 facilitates long-term retention of MAIT cells in affected tissues, where they proliferate locally rather than being recruited from secondary lymphoid tissues (28). Conversely, studies in mice have shown that increased MAIT cells at the skin wound recruit from the circulation system in a CXCR6-CXCL16-dependent manner and initiate wound repair functions (54). Our studies using NOD-SCID-IL-2Rg−/−(NSG) mouse transplant models simulated the dynamic distribution of MAIT cells and indicated that circulating MAIT cells may recruit to intestinal tissues via CXCR6 (55).
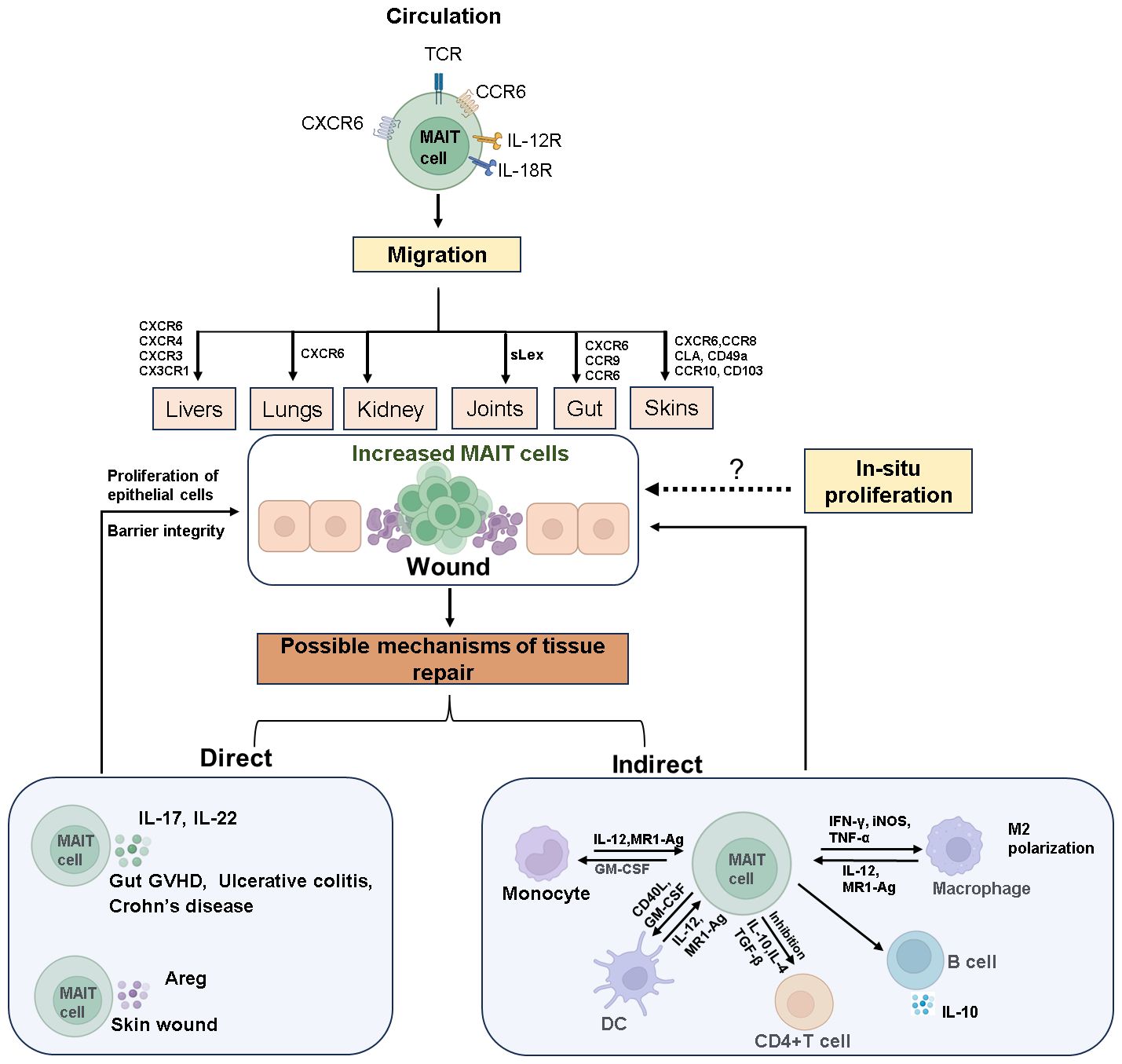
Figure 1 The mechanisms of MAIT cell accumulation at wound sites and their role in tissue repair. Circulating MAIT cells express various cytokine and chemokine receptors, enabling them to target and migrate to different tissues. Upon tissue damage, MAIT cells accumulate at the site of wound through chemotaxis or in-situ proliferation. The invasion of mucosal microbes or inflammatory stimuli rapidly activates MAIT cells, prompting them to secrete tissue repair factors such as IL-17, IL-22, and Areg. These cells also interact with other immune cells to facilitate the repair of damaged tissue. The dashed arrows in the Figure indicate that these processes require further experimental evidence for support. Created with Biorender.com.
MAIT cell migration and chemotaxis are prominent in clinical diseases. In immune-mediated diseases like primary Sjögren’s syndrome (pSS) (56), primary biliary cholangitis (PBC) (57), inflammatory bowel disease (IBD) (58–61), rheumatoid arthritis (RA) (62), and type 1 diabetes (T1D) (63–65), the chemokine and cytokine milieu in inflamed tissues drives MAIT cell via various chemokine receptors such as CCR9, CXCR5, CXCR4, CXCR3, CCR6, and CCR10 (Figure 1). For instance, in PBC, MAIT cells accumulate in the liver through CXCL12-CXCR4 chemotaxis (57), while in IBD, chemokines like CCL20, CXCL10, CXCL16, and CCL25 might guide MAIT cells to the inflamed gut (58). In newly diagnosed T1D children, the frequency of blood MAIT cells positively correlates with CCR6 expression, suggesting a role for CCR6 in their migration to inflamed tissues. CCR6-expressing cells recognize CCL20 and β-defensins, which are elevated in the pancreas and intestines of diabetic patients and mouse models (66, 67). Other homing receptors like CCR10, CD49a, CD103, and skin lymphocyte-associated antigen may direct MAIT cells to dermatitis herpetiform lesions (24, 38, 68, 69). Additionally, in crescentic glomerulonephritis (cGN), MAIT17 cells interact with pro-inflammatory myeloid cells in the kidney via the CXCR6–CXCL16 axis, suppressing their tissue-destructive capabilities (70).
In some diseases like systemic lupus erythematosus (SLE), Type 2 diabetes, and obesity, MAIT cells exhibit an activated and exhausted state (71–73). The decreased frequency of blood MAIT cells may reflect their migration to inflamed tissues or exhaustion upon activation. As inflammation progresses to chronic stages (74), other unidentified signals might sustain the accumulation and survival of MAIT cells in specific inflamed tissues. Although specific subsets of MAIT cells that preferentially traffic to different tissues have not been fully elucidated, their abundant expression of chemokine receptors and migratory capacity are crucial for their roles in tissue homeostasis and repair.
Similar to iNKT cells, MAIT cells can be activated via their TCR, recognizing microbial riboflavin metabolism intermediates like 5-OP-RU bound to MR1, or independently by pro-inflammatory cytokines (8, 18, 19, 75–77). These activation mechanisms differ different kinetics: TCR-mediated activation leads to rapid production of a broad array of pro-inflammatory cytokines and chemokines, including IL-1A, IL-1B, IL-2, IL-22, GM-CSF, CCL3, CCL4, and CCL20, with rapid IFN-γ release within 6 hours. In contrast, MAIT cells activation by IL-12 and IL-18 primarily induces IFN-γ production at 20-24 hours (18, 19, 75, 76).
Given their strategic location in mucosal tissues and activation by microbial metabolites, MAIT cells play crucial roles in maintaining tissue homeostasis. Previous studies indicate the response of unconventional H2-M3-restricted Tc17 cells to commensal S epidermidis, suggesting a role in regulating tissue homeostasis similar to other innate-like T cells involved in barrier surface homeostasis (36, 78, 79). Supported by a diverse gut microbiome, MAIT cells, along with Vδ2 unconventional T cells, are supported by a diverse gut microbiome and associated with favorable prognosis in patients post-allogeneic hematopoietic cell transplantation (HCT) (79). In gut graft-versus-host disease (GVHD) mouse models, Mr1-deficient mice lacking MAIT cells exhibit reduced gut microbial diversity, akin to IL-17A-deficient animals (22), highlighting the protective role of MAIT cells in intestinal inflammation. Recent findings by El Morr et al. demonstrated that during intestinal inflammation, MAIT cells detect microbiota-derived metabolites and promote tissue repair. Under normal conditions, these metabolites from aerotolerant bacteria in the colonic mucosa activate MAIT cells. During inflammation, increased production of these ligands crosses the intestinal barrier, activating MAIT cells to express repair genes and produce barrier-enhancing mediators, facilitating colitis resolution (80). Understanding these interactions between MAIT cells and the microbiome is crucial for elucidating their role in maintaining tissue homeostasis.
Transcriptional analyses in both mice and humans reveal a strong enrichment of tissue repair signatures in MAIT cells, particularly the MAIT17 subset [18,19,43,80]. In mucosal or wound tissues, the predominant phenotype of MAIT cells is MAIT17 (8), attributed to: ① Under steady-state conditions, the microbial environment of mucosal barriers promotes the differentiation of MAIT17 and the maintenance of its repair functional programs; ② Under stress, recruitment of MAIT17 cells from circulation to wounds becomes predominant. The functional effects of these transcriptional programs have been demonstrated both in vivo and in vitro. Using an in vitro wound healing assay with the Caco2 intestinal epithelial cell line, it was found that MAIT cells trigger inducible tissue repair programs in an MR1-dependent manner and accelerate wound closure in this system (19). This was demonstrated in mouse models, where Mr1-/- NOD mice exhibited increased intestinal permeability compared to Mr1+/+ NOD mice, indicating a protective role of MR1-mediated MAIT cells in maintaining intestinal homeostasis (60). Similarly, in vitro human cholangiocyte cell line H69 wound healing assay, MAIT cells showed wound healing characteristics dependent on MR1-antigen-TCR interaction conduction (81). Direct application of 5-OP-RU on injured skin was also sufficient to expand MAIT cells and accelerate tissue repair in mice (10). However, in a recent human-like mouse model of full-thickness skin excision, MAIT cells in the skin express tissue repair programs in a steady-state, but the recruitment and tissue repair functions of MAIT cells do not depend on MR1-mediated antigen presentation (54). The potential reasons for this contradictory result include, firstly, the need for additional signals to initiate and amplify tissue repair programs, apart from solely relying on MR1 molecules (18, 19, 82–86). Secondly, pro-inflammatory cytokines, along with TCR signaling, trigger robust and sustained effector functions of MAIT cells, which are not only reflected in the upregulation of repair factors but also in the release of various cytokines such as IFN-γ and TNF-α (18, 19, 82–86), affecting the inflammation response and immune cell activity, thereby helping to regulate the level of inflammation during tissue injury and repair processes. Lastly, classical TCR signaling through CD3/CD28 stimulation also increases the expression of mucosal protective factors such as IL-17A (84), although possibly to a lesser extent than activation via the MR1 pathway.
In summary, tissue repair is a complex process regulated by various factors. When MAIT cells are stimulated through TCR without pro-inflammatory cytokines, they secrete epithelial repair factors, contributing to the maintenance of homeostatic barriers. However, in the presence of cytokines accompanying wounds or inflammation, MAIT cells initiate additional anti-infective responses, aiding in emergency barrier repair and bolstering host defense mechanisms (19). This dual role underscores the versatility and adaptability of MAIT cells in maintaining tissue integrity and responding to different challenges.
Upon activation at mucosal or pathological sites, MAIT cells exert their effects through two pathways (Figure 1): direct action by secreting tissue repair factors to promote wound healing and indirect action by influencing other immune cell subsets through immunoregulation.
Transcriptomic studies of MAIT cells from human and mice have revealed shared expression of genes involved in tissue protection and repair. These genes include immune genes (TNF, PTGES2, TGFB1, CCL3, HMGB1), proteases (Furin, MMP25), growth factors (GM-CSF, M-CSF, PDGFB, LIF), and angiogenic genes (HIF1A, VEGFB) (10, 18–20). In the latest study by Sayaf. K et al., the MAIT cell stimulation leads to the production of growth factors, potentially mediating their critical role in tissue repair and regeneration following injury through the VEGF-VEGFR2 signaling pathway (87).
The roles of IL-17 (22, 88, 89) and amphiregulin (Areg) (54, 90) in tissue repair have been well studied. Areg is an epidermal growth factor-like molecule, mediates keratinocyte proliferation (90). During the skin injury repair, MAIT cells induce wound healing by secreting Areg (54). MAIT cells also play a crucial role in maintaining intestinal mucosal integrity by producing IL-17 and IL-22 (21, 22). In Type 1 Diabetes (T1D), a reduction in IL-17 and IL-22 production by MAIT cells leads to compromised mucosal barrier integrity and increased intestinal permeability (23, 63–65, 89, 91–93). This can trigger local intestinal inflammation and facilitate the translocation of bacterial compounds to the liver or pancreatic lymph nodes, exacerbating autoimmunity and disease progression (63, 64). In a mouse transplant model, residual intestinal MAIT cells from recipient mice secrete IL-17 to maintain intestinal integrity and suppress gut GVHD (22, 94). IL-22 promotes the survival and proliferation of epithelial cells, while IL-17 regulates tight junction proteins to prevent excessive barrier permeability during epithelial injury (89, 95, 96). Additionally, MAIT cell-deficient mice have been shown to develop intestinal leakage, though underlying mechanisms require further investigation (63).
MAIT cells also maintain tissue homeostasis at the meningeal barrier by expressing antioxidant molecules such as Selenop and Fth1, which enhance the expression of cell adhesion molecules like E-cadherin and Claudin11, contributing to meningeal barrier integrity (97). These findings highlight the crucial protective role of tissue repair factors secreted by MAIT cells in mucosal homeostasis and repair.
In addition to direct tissue repair, MAIT cells exert immunomodulatory effects indirectly. They trigger dendritic cells (DCs) maturation in a CD40L and GM-CSF-dependent manner, inducing anti-inflammatory macrophage polarization and promoting B cell differentiation and antibody production (62, 98–100). In experimental autoimmune encephalomyelitis (EAE), MAIT cells inhibit disease development by modulating pro-inflammatory molecules and promoting IL-10 production by B cells, reducing disease severity (6). Activated MAIT cells can induce differentiation of monocytes/macrophages into an M2 phenotype in vitro (50) and promote long-term survival of neutrophils and their differentiation into APC-like neutrophils via TNF, IFN-γ, and GM-CSF mediation (101–103). By modulating neutrophils and DCs, MAIT cells increase the number of effector and memory conventional CD4+ and CD8+ T cells at infection sites and new arrivals (62, 101). They may also recruit other tissue repair immune cells such as macrophages by producing chemotactic factors like CCL3 (19, 104, 105). Additionally, MAIT cells can inhibit CD4+ T cell proliferation in vitro (88–90). In an allogeneic reaction in vitro model, MAIT cells effectively control or delay the occurrence of GVHD through immunosuppressive effects (106–108), correlating with reduced infiltrating human T cell numbers, proliferation, and effector function in diseased mouse tissues, along with reduced circulating levels of IFN-γ and TNF-α and increased levels of IL-10 (108). In summary, MAIT cells can engage in repair or antimicrobial responses within mucosal tissues through cross-talk with other immune cells. However, detailed studies using mucosal tissue samples, such as those from the gastrointestinal tract, are necessary to better understand this phenomenon.
Recent evidence shows that MAIT cells play a significant role in the immune response to SARS-CoV-2. In COVID-19 patients, circulating MAIT cells significantly decrease and become enriched in the airways (109–114). The remaining blood MAIT cells are activated, with increased CD69, CD38, HLA-DR, CD56, and granzyme B expression, along with decreased CXCR3 expression. Airway MAIT produce IL-17A and TNF, associated with chemokines like CXCL10 and CX3CL1 (109, 112). Regarding clinical outcomes, current research shows inconsistent results. Parrot T et al. found that high CD69 and low CXCR3 expression on MAIT cells are associated with mortality (112). In contrast, research by Jouan Y et al. indicated that CD69 expression on MAIT and iNKT cells at admission is associated with improved oxygenation on day 7 and increased discharge from intensive care by day 15 (113), suggesting a beneficial role for MAIT cells in COVID-19. The data suggest MAIT cell dual role in contributing to inflammation and aiding in disease resolution. Additionally, during SARS-CoV-2 infection, SARS-CoV-2-infected macrophages can activate MAIT cell through MR1-dependent degranulation or the cytokine IL-18 (114). In severe COVID-19 patients, IL-10 suppresses monocyte HLA-DR expression, leading to MAIT cell dysfunction (115). Overall, the balance between protective and pro-inflammatory roles of MAIT cells, and their potential tissue repair function, remains unclear, but their distinct changes during SARS-CoV-2 infection underscore their significant role.
In summary, in chronic pathological conditions like viral infections and autoimmune, inflammatory, and metabolic diseases, MAIT cells can exert pathogenic effects through sustained inflammation and cytotoxicity. The balance between their pathogenic and protective roles may depend on factors such as activation status, tissue localization, cytokine profile, and disease chronicity (24). In conditions of chronic inflammation, tissue-resident MAIT cells may lose their homeostatic function, contributing to barrier integrity disruption.
MAIT cells’ significant roles in maintaining homeostasis and promoting tissue repair in the intestinal mucosa, meningeal barrier, and skin have garnered increasing attention. Beyond their well-established antimicrobial responses, there is a growing interest in their regulatory role in wound healing. From a translational medicine perspective, efforts are being made to utilize the unique homeostatic functions of MAIT cells to address chronic inflammation and restore tissue integrity. For example, chronic skin wounds such as leg ulcers, sacral pressure sores, or burns could potentially benefit from local reintroduction of symbiotic organisms capable of synthesizing riboflavin or the application of synthetic MAIT cell ligands (98).
Recent research indicates that MAIT cells lack alloreactive potential (116, 117), suggesting potential for developing universal MAIT cell adoptive therapy to overcome HLA disparities. Strict regulation of cell surface MR1 during allo-HSCT can minimize the risk of off-target effects of MAIT cells (75, 118). Additionally, due to their natural tropism for mucosal tissues, MAIT cells can effectively target mucosal tissues such as the gut and skin (118, 119). This highly conservative MR1 regulation makes allogeneic MAIT cell transplantation less likely to induce severe GVHD. Leveraging the effector or tissue repair/regulatory properties of MAIT cells in adoptive immunotherapy offers novel clinical strategies for treating GVHD. MAIT cell transplantation has been preliminarily validated in mouse models and could readily transition to clinical trials. Overall, the tissue repair and regulatory functions of MAIT cells open new avenues for clinical benefits. Furthermore, these characteristics make MAIT cells highly promising candidates for engineered chimeric antigen receptor (CAR)-MAIT therapy.
However, challenges remain, particularly in obtaining human mucosal tissue and developing ex vivo research models that accurately simulating in vivo disease environments. This limitation hinders comprehensive investigations into the reparative mechanisms of MAIT cells across different diseases or microenvironments. At present, MAIT cells can toggle between pro-inflammatory and anti-inflammatory, or pathogenic and reparative functions, depending on tissue localization and disease types (21, 57, 59, 60). Therefore, several potential obstacles must be addressed before successful clinical translation. Firstly, in-depth exploration of MAIT cell tissue localization and maintenance of repair functions, as well as the conditions or mechanisms triggering them, is crucial for developing targeted therapies for wounds. Secondly, studying the functions and tissue localization of different MAIT cell subtypes is necessary to determine the optimal cell type for transplantation. Finally, a thorough investigation into the antigens or stimuli present in healthy and different diseased tissue microenvironments is essential to understand how these conditions affect MAIT cell functional differentiation, thereby guiding the development of MAIT cell-related therapies conducive to disease recovery in various scenarios. It is noteworthy that understanding the degree of crosstalk between MAIT cells and other immune/non-immune cells and symbiotic microorganisms under normal conditions or when activated in disease microenvironments is also necessary.
To advance the study of MAIT cell reparative functions, it is imperative to develop novel ex vivo research models. For instance, fully utilizing current organoid culture techniques may be beneficial. Organoids, self-organizing, miniaturized organs derived from a series of stem cells, replicating key structural and functional features of their in vivo counterparts (109, 120–124). Constructing 3D models of human ex vivo skin, gastrointestinal tract, and other organoids, and exploring the interaction between co-culture systems and immune cells, can provide deeper insights into the functional aspects of MAIT cells in organoid models under different disease conditions. This includes migration, cytokine production, tissue repair, and antimicrobial activity (109). Additionally, these models can help describe the crosstalk between MAIT cells and resident mucosal immune cells and the consequent impacts on mucosal integrity and immunity. Recently, this technology has been applied to study MAIT cells in acute intestinal inflammation (125). In patient-derived appendiceal organoid (PDAO) models, circulating MAIT cells upregulated chemokine receptors and showed enhanced E. coli-pulsed PDAO infiltration in a CCR1-, CCR2-, and CCR4-dependent manner (125). This serves as an excellent preclinical model for investigating the roles of MAIT cells in mucosal organs. It is anticipated that this approach will be applied to other clinical disease models involving MAIT cells in the future, further advancing the clinical translation of MAIT cell research.
In conclusion, this review outlines the tissue repair effects of MAIT cells and their involvement in various diseases, summarizing current MAIT-related researches. We envisage using organoid models to enhance understanding the interactions between MAIT cells and different disease microenvironments. This approach provides a more robust theoretical basis and preclinical research means for the development of MAIT cell therapies.
GM: Methodology, Writing – original draft. ZX: Funding acquisition, Methodology, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was supported by the Beijing Municipal Natural Science Foundation (No. 7242155), the National Natural Science Foundation of China (No. 62331025), the National Key Research and Development Program of China (2021YFC2500300), the major Program of the National Natural Science Foundation of China (No. 82293630, 82293633) and Peking University Medicine Fund for world’s leading discipline or discipline cluster development (No. 71003Y3035).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Legoux F, Salou M, Lantz O. MAIT cell development and functions: the microbial connection. Immunity. (2020) 53:710–23. doi: 10.1016/j.immuni.2020.09.009
2. Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. (2003) 422:164–9. doi: 10.1038/nature01433
3. Koay HF, Godfrey DI, Pellicci DG. Development of mucosal-associated invariant T cells. Immunol Cell Biol. (2018) 96:598–606. doi: 10.1111/imcb.12039
4. Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. (1994) 180:1097–106. doi: 10.1084/jem.180.3.1097
5. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PloS Biol. (2009) 7:e54. doi: 10.1371/journal.pbio.1000054
6. Croxford JL, Miyake S, Huang YY, Shimamura M, Yamamura T. Invariant V(alpha)19i T cells regulate autoimmune inflammation. Nat Immunol. (2006) 7:987–94. doi: 10.1038/ni1370
7. Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J Immunol. (1998) 161:4066–77. doi: 10.4049/jimmunol.161.8.4066
8. Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. (2014) 509:361–5. doi: 10.1038/nature13160
9. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. (2016) 17:1300–11. doi: 10.1038/ni.3565
10. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. (2019) 366:eaax6624. doi: 10.1126/science.aax6624
11. Kawachi I, Maldonado J, Strader C, Gilfillan S. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J Immunol. (2006) 176:1618–27. doi: 10.4049/jimmunol.176.3.1618
12. Lantz O, Legoux F. MAIT cells: an historical and evolutionary perspective. Immunol Cell Biol. (2018) 96:564–72. doi: 10.1111/imcb.1034
13. Greene JM, Dash P, Roy S, McMurtrey C, Awad W, Reed JS, et al. MR1-restricted mucosal-associated invariant T (MAIT) cells respond to mycobacterial vaccination and infection in nonhuman primates. Mucosal Immunol. (2017) 10:802–13. doi: 10.1038/mi.2016.91
14. Chua WJ, Truscott SM, Eickhoff CS, Blazevic A, Hoft DF, Hansen TH. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect Immun. (2012) 80:3256–67. doi: 10.1128/IAI.00279-12
15. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PloS Biol. (2010) 8:e1000407. doi: 10.1371/journal.pbio.1000407
16. Meermeier EW, Harriff MJ, Karamooz E, Lewinsohn DM. MAIT cells and microbial immunity. Immunol Cell Biol. (2018) 96:607–17. doi: 10.1111/imcb.12022
17. Chengalroyen MD. Current perspectives and challenges of MAIT cell-directed therapy for tuberculosis infection. Pathogens. (2023) 12:1343. doi: 10.3390/pathogens12111343
18. Lamichhane R, Schneider M, de la Harpe SM, Harrop TWR, Hannaway RF, Dearden PK, et al. TCR- or cytokine-activated CD8+ Mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. (2019) 28:3061–3076.e5. doi: 10.1016/j.celrep.2019.08.054
19. Leng T, Akther HD, Hackstein CP, Powell K, King T, Friedrich M, et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. (2019) 28:3077–3091.e5. doi: 10.1016/j.celrep.2019.08.050
20. Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. (2019) 28:3249–3262.e5. doi: 10.1016/j.celrep.2019.07.039
21. Waterhölter A, Wunderlich M, Turner JE. MAIT cells in immune-mediated tissue injury and repair. Eur J Immunol. (2023) 53:e2350483. doi: 10.1002/eji.202350483
22. Varelias A, Bunting MD, Ormerod KL, Koyama M, Olver SD, Straube J, et al. Recipient mucosal-associated invariant T cells control GVHD within the colon. J Clin Invest. (2018) 128:1919–36. doi: 10.1172/JCI91646
23. Nel I, Bertrand L, Toubal A, Lehuen A. MAIT cells, guardians of skin and mucosa? Mucosal Immunol. (2021) 14:803–14. doi: 10.1038/s41385-021-00391-w
24. Toubal A, Nel I, Lotersztajn S, Lehuen A. Mucosal-associated invariant T cells and disease. Nat Rev Immunol. (2019) 19:643–57. doi: 10.1038/s41577-019-0191-y
25. Reantragoon R, Corbett AJ, Sakala IG, Gherardin NA, Furness JB, Chen Z, et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J Exp Med. (2013) 210:2305–20. doi: 10.1084/jem.20130958
26. Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. (2015) 212:1095–108. doi: 10.1084/jem.20142110
27. Booth JS, Salerno-Goncalves R, Blanchard TG, Patil SA, Kader HA, Safta AM, et al. Mucosal-associated invariant T cells in the human gastric mucosa and blood: role in helicobacter pylori infection. Front Immunol. (2015) 6:466. doi: 10.3389/fimmu.2015.00466
28. Yu H, Yang A, Liu L, Mak JYW, Fairlie DP, Cowley S. CXCL16 stimulates antigen-induced MAIT cell accumulation but trafficking during lung infection is CXCR6-independent. Front Immunol. (2020) 11:1773. doi: 10.3389/fimmu.2020.01773
29. Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. (2011) 117:1250–9. doi: 10.1182/blood-2010-08-303339
30. Gibbs A, Leeansyah E, Introini A, Paquin-Proulx D, Hasselrot K, Andersson E, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. (2017) 10:35–45. doi: 10.1038/mi.2016.30
31. Terpstra ML, Remmerswaal EBM, van der Bom-Baylon ND, Sinnige MJ, Kers J, van Aalderen MC, et al. Tissue-resident mucosal-associated invariant T (MAIT) cells in the human kidney represent a functionally distinct subset. Eur J Immunol. (2020) 50:1783–97. doi: 10.1002/eji.202048644
32. Law BMP, Wilkinson R, Wang X, Kildey K, Giuliani K, Beagley KW, et al. Human tissue-resident mucosal-associated invariant T (MAIT) cells in renal fibrosis and CKD. J Am Soc Nephrol. (2019) 30:1322–35. doi: 10.1681/ASN.2018101064
33. Bugaut H, El Morr Y, Mestdagh M, Darbois A, Paiva RA, Salou M, et al. A conserved transcriptional program for MAIT cells across mammalian evolution. J Exp Med. (2024) 221:e20231487. doi: 10.1084/jem.20231487
34. Chandra S, Ascui G, Riffelmacher T, Chawla A, Ramírez-Suástegui C, Castelan VC, et al. Transcriptomes and metabolism define mouse and human MAIT cell populations. Sci Immunol. (2023) 8:eabn8531. doi: 10.1126/sciimmunol.abn8531
35. Garner LC, Amini A, FitzPatrick MEB, Lett MJ, Hess GF, Filipowicz Sinnreich M, et al. Single-cell analysis of human MAIT cell transcriptional, functional and clonal diversity. Nat Immunol. (2023) 24:1565–78. doi: 10.1038/s41590-023-01575-1
36. Amini A, Pang D, Hackstein CP, Klenerman P. MAIT cells in barrier tissues: lessons from immediate neighbors. Front Immunol. (2020) 11:584521. doi: 10.3389/fimmu.2020.584521
37. Wang H, Kjer-Nielsen L, Shi M, D’Souza C, Pediongco TJ, Cao H, et al. IL-23 costimulates antigen-specific MAIT cell activation and enables vaccination against bacterial infection. Sci Immunol. (2019) 4:eaaw0402. doi: 10.1126/sciimmunol.aaw0402
38. Sobkowiak MJ, Davanian H, Heymann R, Gibbs A, Emgård J, Dias J, et al. Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. Eur J Immunol. (2019) 49:133–43. doi: 10.1002/eji.201847759
39. Dias J, Boulouis C, Gorin JB, van den Biggelaar RHGA, Lal KG, Gibbs A, et al. The CD4-CD8- MAIT cell subpopulation is a functionally distinct subset developmentally related to the main CD8+ MAIT cell pool. Proc Natl Acad Sci U S A. (2018) 115:E11513–22. doi: 10.1073/pnas.1812273115
40. Thomas SY, Scanlon ST, Griewank KG, Constantinides MG, Savage AK, Barr KA, et al. PLZF induces an intravascular surveillance program mediated by long-lived LFA-1-ICAM-1 interactions. J Exp Med. (2011) 208:1179–88. doi: 10.1084/jem.20102630
41. Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. (2019) 20:1110–28. doi: 10.1038/s41590-019-0444-8
42. Koay HF, Su S, Amann-Zalcenstein D, Daley SR, Comerford I, Miosge L, et al. A divergent transcriptional landscape underpins the development and functional branching of MAIT cells. Sci Immunol. (2019) 4:eaay6039. doi: 10.1126/sciimmunol.aay6039
43. Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. (2019) 366:494–9. doi: 10.1126/science.aaw2719
44. Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. (2019) 216:133–51. doi: 10.1084/jem.20181483
45. Legoux F, Gilet J, Procopio E, Echasserieau K, Bernardeau K, Lantz O. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat Immunol. (2019) 20:1244–55. doi: 10.1038/s41590-019-0465-3
46. Lee M, Lee E, Han SK, Choi YH, Kwon DI, Choi H, et al. Single-cell RNA sequencing identifies shared differentiation paths of mouse thymic innate T cells. Nat Commun. (2020) 11:4367. doi: 10.1038/s41467-020-18155-8
47. Lee CH, Zhang HH, Singh SP, Koo L, Kabat J, Tsang H, et al. C/EBPδ drives interactions between human MAIT cells and endothelial cells that are important for extravasation. Elife. (2018) 7:e32532. doi: 10.7554/eLife.32532
48. Oldham KA, Parsonage G, Bhatt RI, Wallace DM, Deshmukh N, Chaudhri S, et al. T lymphocyte recruitment into renal cell carcinoma tissue: a role for chemokine receptors CXCR3, CXCR6, CCR5, and CCR6. Eur Urol. (2012) 61:385–94. doi: 10.1016/j.eururo.2011.10.035
49. Singh SP, Zhang HH, Tsang H, Gardina PJ, Myers TG, Nagarajan V, et al. PLZF regulates CCR6 and is critical for the acquisition and maintenance of the Th17 phenotype in human cells. J Immunol. (2015) 194:4350–61. doi: 10.4049/jimmunol.1401093
50. Li Y, Huang B, Jiang X, Chen W, Zhang J, Wei Y, et al. Mucosal-associated invariant T cells improve nonalcoholic fatty liver disease through regulating macrophage polarization. Front Immunol. (2018) 9:1994. doi: 10.3389/fimmu.2018.01994
51. Billerbeck E, Kang YH, Walker L, Lockstone H, Grafmueller S, Fleming V, et al. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc Natl Acad Sci U S A. (2010) 107:3006–11. doi: 10.1073/pnas.0914839107
52. Sato T, Thorlacius H, Johnston B, Staton TL, Xiang W, Littman DR, et al. Role for CXCR6 in recruitment of activated CD8+ lymphocytes to inflamed liver. J Immunol. (2005) 174:277–83. doi: 10.4049/jimmunol.174.1.277
53. Mandai Y, Takahashi D, Hase K, Obata Y, Furusawa Y, Ebisawa M, et al. Distinct roles for CXCR6(+) and CXCR6(-) CD4(+) T cells in the pathogenesis of chronic colitis. PloS One. (2013) 8:e65488. doi: 10.1371/journal.pone.0065488
54. du Halgouet A, Darbois A, Alkobtawi M, Mestdagh M, Alphonse A, Premel V, et al. Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing. Immunity. (2023) 56:78–92.e6. doi: 10.1016/j.immuni.2022.12.004
55. Gao MG, Kong J, Sun YQ, Wang ZD, Han TT, Wang Y, et al. G-CSF induces MAIT cells to exert anti-intestinal gvhd effects dependent on CXCR6 mediated chemotaxis. Blood. (2023) 142:2048. doi: 10.1182/blood-2023-181445
56. Hinrichs AC, Kruize AA, Leavis HL, van Roon JAG. In patients with primary Sjögren's syndrome innate-like MAIT cells display upregulated IL-7R, IFN-γ, and IL-21 expression and have increased proportions of CCR9 and CXCR5-expressing cells. Front Immunol. (2022) 13:1017157. doi: 10.3389/fimmu.2022.1017157
57. Chen Z, Liu S, He C, Sun J, Wang L, Chen H, et al. CXCL12-CXCR4-mediated chemotaxis supports accumulation of mucosal-associated invariant T cells into the liver of patients with PBC. Front Immunol. (2021) 12:578548. doi: 10.3389/fimmu.2021.578548
58. Ju JK, Cho YN, Park KJ, Kwak HD, Jin HM, Park SY, et al. Activation, deficiency, and reduced IFN-γ Production of mucosal-associated invariant T cells in patients with inflammatory bowel disease. J Innate Immun. (2020) 12:422–34. doi: 10.1159/000507931
59. Serriari NE, Eoche M, Lamotte L, Lion J, Fumery M, Marcelo P, et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin Exp Immunol. (2014) 176:266–74. doi: 10.1111/cei.12277
60. Haga K, Chiba A, Shibuya T, Osada T, Ishikawa D, Kodani T, et al. MAIT cells are activated and accumulated in the inflamed mucosa of ulcerative colitis. J Gastroenterol Hepatol. (2016) 31:965–72. doi: 10.1111/jgh.13242
61. Hiejima E, Kawai T, Nakase H, Tsuruyama T, Morimoto T, Yasumi T, et al. Reduced numbers and proapoptotic features of mucosal-associated invariant T cells as a characteristic finding in patients with inflammatory bowel disease. Inflammation Bowel Dis. (2015) 21:1529–40. doi: 10.1097/MIB.0000000000000397
62. Kim M, Yoo SJ, Kang SW, Kwon J, Choi I, Lee CH. TNFα and IL-1β in the synovial fluid facilitate mucosal-associated invariant T (MAIT) cell migration. Cytokine. (2017) 99:91–8. doi: 10.1016/j.cyto.2017.07.007
63. Rouxel O, Da Silva J, Beaudoin L, Nel I, Tard C, Cagninacci L, et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol. (2017) 18:1321–31. doi: 10.1038/ni.3854
64. Rouland M, Beaudoin L, Rouxel O, Bertrand L, Cagninacci L, Saffarian A, et al. Gut mucosa alterations and loss of segmented filamentous bacteria in type 1 diabetes are associated with inflammation rather than hyperglycaemia. Gut. (2022) 71:296–308. doi: 10.1136/gutjnl-2020-323664
65. Gazali AM, Schroderus AM, Näntö-Salonen K, Rintamäki R, Pihlajamäki J, Knip M, et al. Mucosal-associated invariant T cell alterations during the development of human type 1 diabetes. Diabetologia. (2020) 63:2396–409. doi: 10.1007/s00125-020-05257-7
66. Ito T, Carson WF4, Cavassani KA, Connett JM, Kunkel SL. CCR6 as a mediator of immunity in the lung and gut. Exp Cell Res. (2011) 317:613–9. doi: 10.1016/j.yexcr.2010.12.018
67. Miani M, Le Naour J, Waeckel-Enée E, Verma SC, Straube M, Emond P, et al. Gut microbiota-stimulated innate lymphoid cells support β-defensin 14 expression in pancreatic endocrine cells, preventing autoimmune diabetes. Cell Metab. (2018) 28:557–572.e6. doi: 10.1016/j.cmet.2018.06.012
68. Teunissen MBM, Yeremenko NG, Baeten DLP, Chielie S, Spuls PI, de Rie MA, et al. The IL-17A-producing CD8+ T-cell population in psoriatic lesional skin comprises mucosa-associated invariant T cells and conventional T cells. J Invest Dermatol. (2014) 134:2898–907. doi: 10.1038/jid.2014.261
69. Li J, Reantragoon R, Kostenko L, Corbett AJ, Varigos G, Carbone FR. The frequency of mucosal-associated invariant T cells is selectively increased in dermatitis herpetiformis. Australas J Dermatol. (2017) 58:200–4. doi: 10.1111/ajd.12456
70. Gnirck AC, Philipp MS, Waterhölter A, Wunderlich M, Shaikh N, Adamiak V, et al. Mucosal-associated invariant T cells contribute to suppression of inflammatory myeloid cells in immune-mediated kidney disease. Nat Commun. (2023) 14:7372. doi: 10.1038/s41467-023-43269-0
71. Rouxel O, Lehuen A. Mucosal-associated invariant T cells in autoimmune and immune-mediated diseases. Immunol Cell Biol. (2018) 96:618–29. doi: 10.1111/imcb.12011
72. Magalhaes I, Pingris K, Poitou C, Bessoles S, Venteclef N, Kiaf B, et al. Mucosal-associated invariant T cell alterations in obese and type 2 diabetic patients. J Clin Invest. (2015) 125:1752–62. doi: 10.1172/JCI78941
73. Carolan E, Tobin LM, Mangan BA, Corrigan M, Gaoatswe G, Byrne G, et al. Altered distribution and increased IL-17 production by mucosal-associated invariant T cells in adult and childhood obesity. J Immunol. (2015) 194:5775–80. doi: 10.4049/jimmunol.1402945
74. Leeansyah E, Ganesh A, Quigley MF, Sönnerborg A, Andersson J, Hunt PW, et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. (2013) 121:1124–35. doi: 10.1182/blood-2012-07-445429
75. Ussher JE, van Wilgenburg B, Hannaway RF, Ruustal K, Phalora P, Kurioka A, et al. TLR signaling in human antigen-presenting cells regulates MR1-dependent activation of MAIT cells. Eur J Immunol. (2016) 46:1600–14. doi: 10.1002/eji.201545969
76. Klenerman P, Hinks TSC, Ussher JE. Biological functions of MAIT cells in tissues. Mol Immunol. (2021) 130:154–8. doi: 10.1016/j.molimm.2020.12.017
77. Sattler A, Dang-Heine C, Reinke P, Babel N. IL-15 dependent induction of IL-18 secretion as a feedback mechanism controlling human MAIT-cell effector functions. Eur J Immunol. (2015) 45:2286–98. doi: 10.1002/eji.201445313
78. Linehan JL, Harrison OJ, Han SJ, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. (2018) 172:784–796.e18. doi: 10.1016/j.cell.2017.12.033
79. Andrlová H, Miltiadous O, Kousa AI, Dai A, DeWolf S, Violante S, et al. MAIT and Vδ2 unconventional T cells are supported by a diverse intestinal microbiome and correlate with favorable patient outcome after allogeneic HCT. Sci Transl Med. (2022) 14:eabj2829. doi: 10.1126/scitranslmed.abj2829
80. El Morr Y, Fürstenheim M, Mestdagh M, Franciszkiewicz K, Salou M, Morvan C, et al. MAIT cells monitor intestinal dysbiosis and contribute to host protection during colitis. Sci Immunol. (2024) 9:eadi8954. doi: 10.1126/sciimmunol.adi8954
81. Xiao MH, Wu S, Liang P, Ma D, Zhang J, Chen H, et al. Mucosal-associated invariant T cells promote ductular reaction through amphiregulin in biliary atresia. EBioMedicine. (2024) 103:105138. doi: 10.1016/j.ebiom.2024.105138
82. Slichter CK, McDavid A, Miller HW, Finak G, Seymour BJ, McNevin JP, et al. Distinct activation thresholds of human conventional and innate-like memory T cells. JCI Insight. (2016) 1(8):e86292. doi: 10.1172/jci.insight.86292
83. Berkson JD, Prlic M. The MAIT conundrum - how human MAIT cells distinguish bacterial colonization from infection in mucosal barrier tissues. Immunol Lett. (2017) 192:7–11. doi: 10.1016/j.imlet.2017.09.013
84. Schubert K, Karkossa I, Schor J, Engelmann B, Steinheuer LM, Bruns T, et al. A multi-omics analysis of mucosal-associated-invariant T cells reveals key drivers of distinct modes of activation. Front Immunol. (2021) 12:616967. doi: 10.3389/fimmu.2021.616967
85. Azzout M, Dietrich C, Machavoine F, Gastineau P, Bottier A, Lezmi G, et al. IL-33 enhances IFNγ and TNFα Production by human MAIT cells: A new pro-th1 effect of IL-33. Int J Mol Sci. (2021) 22:10602. doi: 10.3390/ijms221910602
86. Dias J, Leeansyah E, Sandberg JK. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc Natl Acad Sci U S A. (2017) 114:E5434–43. doi: 10.1073/pnas.1705759114
87. Sayaf I K, Tasin HD, Akther LC, Garner N, Ramamurthy CP, Hackstein FP. et al, MAIT cells play a role in liver tissue repair via growth factor secretion. Digestive Liver Dis. (2024) 56, Supplement 1:S61. doi: 10.1016/j.dld.2024.01.106
88. Konieczny P, Xing Y, Sidhu I, Subudhi I, Mansfield KP, Hsieh B, et al. Interleukin-17 governs hypoxic adaptation of injured epithelium. Science. (2022) 377:eabg9302. doi: 10.1126/science.abg9302
89. Lee JS, Tato CM, Joyce-Shaikh B, Gulen MF, Cayatte C, Chen Y, et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity. (2015) 43:727–38. doi: 10.1016/j.immuni.2015.09.003
90. Zaiss DMW, Gause WC, Osborne LC, Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. (2015) 42:216–26. doi: 10.1016/j.immuni.2015.01.020
91. Nel I, Beaudoin L, Gouda Z, Rousseau C, Soulard P, Rouland M, et al. MAIT cell alterations in adults with recent-onset and long-term type 1 diabetes. Diabetologia. (2021) 64:2306–21. doi: 10.1007/s00125-021-05527-y
92. Maxwell JR, Zhang Y, Brown WA, Smith CL, Byrne FR, Fiorino M, et al. Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity. (2015) 43:739–50. doi: 10.1016/j.immuni.2015.08.019
93. Eyerich K, Dimartino V, Cavani A. IL-17 and IL-22 in immunity: Driving protection and pathology. Eur J Immunol. (2017) 47:607–14. doi: 10.1002/eji.201646723
94. Gao MG, Zhao XS. Mining the multifunction of mucosal-associated invariant T cells in hematological Malignancies and transplantation immunity: A promising hexagon soldier in immunomodulatory. Front Immunol. (2022) 13:931764. doi: 10.3389/fimmu.2022.931764
95. Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol. (2015) 33:747–85. doi: 10.1146/annurev-immunol-032414-112123
96. Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. (2008) 118:534–44. doi: 10.1172/JCI33194
97. Zhang Y, Bailey JT, Xu E, Singh K, Lavaert M, Link VM, et al. Mucosal-associated invariant T cells restrict reactive oxidative damage and preserve meningeal barrier integrity and cognitive function. Nat Immunol. (2022) 23:1714–25. doi: 10.1038/s41590-022-01349-1
98. Hinks TSC, Zhang XW. MAIT cell activation and functions. Front Immunol. (2020) 11:1014. doi: 10.3389/fimmu.2020.01014
99. Salio M, Gasser O, Gonzalez-Lopez C, Martens A, Veerapen N, Gileadi U, et al. Activation of human mucosal-associated invariant T cells induces CD40L-dependent maturation of monocyte-derived and primary dendritic cells. J Immunol. (2017) 199:2631–8. doi: 10.4049/jimmunol.1700615
100. Bennett MS, Trivedi S, Iyer AS, Hale JS, Leung DT. Human mucosal-associated invariant T (MAIT) cells possess capacity for B cell help. J Leukoc Biol. (2017) 102:1261–9. doi: 10.1189/jlb.4A0317-116R
101. Davey MS, Morgan MP, Liuzzi AR, Tyler CJ, Khan MWA, Szakmany T, et al. Microbe-specific unconventional T cells induce human neutrophil differentiation into antigen cross-presenting cells. J Immunol. (2014) 193:3704–16. doi: 10.4049/jimmunol.1401018
102. Schneider M, Hannaway RF, Lamichhane R, de la Harpe SM, Tyndall JDA, Vernall AJ, et al. Neutrophils suppress mucosal-associated invariant T cells in humans. Eur J Immunol. (2020) 50:643–55. doi: 10.1002/eji.201948394
103. Provine NM, Klenerman P. MAIT cells in health and disease. Annu Rev Immunol. (2020) 38:203–28. doi: 10.1146/annurev-immunol-080719-015428
104. DiPietro LA, Burdick M, Low QE, Kunkel SL, Strieter RM. MIP-1alpha as a critical macrophage chemoattractant in murine wound repair. J Clin Invest. (1998) 101:1693–8. doi: 10.1172/JCI1020
105. Li Z, Hodgkinson T, Gothard EJ, Boroumand S, Lamb R, Cummins I, et al. Epidermal Notch1 recruits RORγ(+) group 3 innate lymphoid cells to orchestrate normal skin repair. Nat Commun. (2016) 7:11394. doi: 10.1038/ncomms11394
106. Gao MG, Hong Y, Zhao XY, Pan XA, Sun YQ, Kong J, et al. The potential roles of mucosa-associated invariant T cells in the pathogenesis of gut graft-versus-host disease after hematopoietic stem cell transplantation. Front Immunol. (2021) 12:720354. doi: 10.3389/fimmu.2021.720354
107. Bhattacharyya A, Hanafi LA, Sheih A, Golob JL, Srinivasan S, Boeckh MJ, et al. Graft-derived reconstitution of mucosal-associated invariant T cells after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant. (2018) 24:242–51. doi: 10.1016/j.bbmt.2017.10.003
108. Talvard-Balland N, Lambert M, Chevalier MF, Minet N, Salou M, Tourret M, et al. Human MAIT cells inhibit alloreactive T cell responses and protect against acute graft-versus-host disease. JCI Insight. (2024) 9:e166310. doi: 10.1172/jci.insight.166310
109. Konecny AJ, Huang Y, Setty M, Prlic M. Signals that control MAIT cell function in healthy and inflamed human tissues. Immunol Rev. (2024) 323:138–49. doi: 10.1111/imr.13325
110. Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. (2020) 5:eabd7114. doi: 10.1126/sciimmunol.abd7114
111. Huang X, Kantonen J, Nowlan K, Nguyen NA, Jokiranta ST, Kuivanen S, et al. Mucosal-Associated Invariant T Cells are not susceptible in vitro to SARS-CoV-2 infection but accumulate into the lungs of COVID-19 patients. Virus Res. (2024) 341:199315. doi: 10.1016/j.virusres.2024.199315
112. Parrot T, Gorin JB, Ponzetta A, Maleki KT, Kammann T, Emgård J, et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci Immunol. (2020) 5:eabe1670. doi: 10.1126/sciimmunol.abe1670
113. Jouan Y, Guillon A, Gonzalez L, Perez Y, Boisseau C, Ehrmann S, et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J Exp Med. (2020) 217:e20200872. doi: 10.1084/jem.20200872
114. Flament H, Rouland M, Beaudoin L, Toubal A, Bertrand L, Lebourgeois S, et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat Immunol. (2021) 22:322–35. doi: 10.1038/s41590-021-00870-z
115. Yang Q, Wen Y, Qi F, Gao X, Chen W, Xu G, et al. Suppressive monocytes impair MAIT cells response via IL-10 in patients with severe COVID-19. J Immunol. (2021) 207:1848–56. doi: 10.4049/jimmunol.2100228
116. Ben Youssef G, Tourret M, Salou M, Ghazarian L, Houdouin V, Mondot S, et al. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J Exp Med. (2018) 215:459–79. doi: 10.1084/jem.20171739
117. Tourret M, Talvard-Balland N, Lambert M, Ben Youssef G, Chevalier MF, Bohineust A, et al. Human MAIT cells are devoid of alloreactive potential: prompting their use as universal cells for adoptive immune therapy. J Immunother Cancer. (2021) 9:e003123. doi: 10.1136/jitc-2021-003123
118. Cogswell DT, Gapin L, Tobin HM, McCarter MD, Tobin RP. MAIT cells: partners or enemies in cancer immunotherapy? Cancers (Basel). (2021) 13:1502. doi: 10.3390/cancers13071502
119. Parrot T, Healy K, Boulouis C, Sobkowiak MJ, Leeansyah E, Aleman S, et al. Expansion of donor-unrestricted MAIT cells with enhanced cytolytic function suitable for TCR redirection. JCI Insight. (2021) 6:e140074. doi: 10.1172/jci.insight.140074
120. Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. (2014) 345:1247125. doi: 10.1126/science.1247125
121. Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in immunological research. Nat Rev Immunol. (2020) 20:279–93. doi: 10.1038/s41577-019-0248-y
122. Günther C, Winner B, Neurath MF, Stappenbeck TS. Organoids in gastrointestinal diseases: from experimental models to clinical translation. Gut. (2022) 71:1892–908. doi: 10.1136/gutjnl-2021-326560
123. Puschhof J, Pleguezuelos-Manzano C, Martinez-Silgado A, Akkerman N, Saftien A, Boot C, et al. Intestinal organoid cocultures with microbes. Nat Protoc. (2021) 16:4633–49. doi: 10.1038/s41596-021-00589-z
124. Matthe DM, Dinkel M, Schmid B, Vogler T, Neurath MF, Poeck H, et al. Novel T cell/organoid culture system allows ex vivo modeling of intestinal graft-versus-host disease. Front Immunol. (2023) 14:1253514. doi: 10.3389/fimmu.2023.1253514
Keywords: mucosal-associated invariant T cell, tissue repair, chemotaxis, tissue phenotype, immunoregulatory effects
Citation: Gao M and Zhao X (2024) Insights into the tissue repair features of MAIT cells. Front. Immunol. 15:1432651. doi: 10.3389/fimmu.2024.1432651
Received: 14 May 2024; Accepted: 04 July 2024;
Published: 16 July 2024.
Edited by:
Rezwanul Wahid, University of Maryland, United StatesReviewed by:
Milton Maciel Jr., National Institute of Allergy and Infectious Diseases (NIH), United StatesCopyright © 2024 Gao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaosu Zhao, emhhby54aWFvc3VAb3V0bG9vay5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.