- Faculty of Biotechnology, Hanoi University of Pharmacy, Hanoi, Vietnam
The burgeoning field of pharmacomicrobiomics offers promising insights into the intricate interplay between the microbiome and cancer, shaping responses to diverse treatment modalities. This review aims to analyze the molecular mechanisms underlying interactions between distinct microbiota types and cancer, as well as their influence on treatment outcomes. We explore how the microbiome impacts antitumor immunity, and response to chemotherapy, immunotherapy, and radiation therapy, unveiling its multifaceted roles in cancer progression and therapy resistance. Moreover, we discuss the challenges hindering the development of microbiome-based interventions in cancer therapy, including standardization, validation, and clinical translation. By synthesizing clinical evidence, we underscore the transformative potential of harnessing pharmacomicrobiomics in guiding cancer treatment decisions, paving the way for improved patient outcomes in clinical practice.
1 Introduction
Precision medicine, or personalized therapy, is a rapidly evolving trend in modern healthcare, tailoring treatment to individual patients by selecting the right drug, at the right dose and at the right time based on their specific cellular, molecular, and genetic characteristics. This approach optimizes treatment effectiveness while minimizing the risk of adverse drug reactions (ADRs). Among therapeutic fields, cancer treatment has seen particularly swift advancements in precision medicine due to its typical characteristics such as the highly heterogeneous nature of tumors, substantial variability in drug response among individuals, and the significant side effects associated with cancer therapies (1). Additionally, the high costs of treatment and the invaluable cost of life opportunities necessitate precise treatment protocols without room for trial and error. Biomarkers used to predict treatment response and disease prognosis not only aid in selecting appropriate drugs for patients but also drive innovation in drug development, reducing time and costs for clinical trials and substantially increasing success rates (2).
To date, the most extensively utilized biomarker in personalized cancer therapy is the somatic mutations found in tumors. These mutations assist in identifying patients who are likely to respond favorably to targeted therapy and immunotherapy. Furthermore, the distinctive germline mutations present in individual patients significantly influence the pharmacokinetics and pharmacodynamics of chemotherapy, enabling the prediction of those at risk of experiencing severe ADRs (3, 4). As a result, pharmacogenomics has emerged as a rapidly advancing field, essential for integrating personalized cancer therapy into clinical practice. However, clinical studies and trials have indicated that pharmacogenomic biomarkers only partially account for the inter-individual variation in drug response (3, 5). Therefore, to fully leverage the benefits of personalized therapy for patients, additional research is necessary to investigate other types of biomarkers that can help elucidate the differences among individual patients with impacts on treatment response.
Although the role of human microbiome in health and various diseases, including cancer, has been known for a long time, the concept of Pharmacomicrobiomics only emerged around 2010 as an extension of Pharmacogenomics (6). The human microbiome refers to the community of microorganisms residing in specific body environments, with the gut microbiome being the first and most extensively studied (7). However, nowadays, increasing attention is being paid to the important role of microbiota in other body environments such as the skin microbiome, vaginal microbiome … and even more recently, microbiota discovered within the microenvironments of tumors (6). Pharmacomicrobiomics is a field that investigates the interaction between individual’s microbiomes and drug response to understand how the composition and activity of these microorganisms influence the pharmacokinetics and pharmacodynamics of various medications (8). Pharmacomicrobiomics explores how differences in the microbiome among individuals can affect drug metabolism, efficacy, and toxicity, ultimately influencing an individual’s response to treatment. This field has implications for personalized medicine, as it may help optimize drug therapy by considering an individual’s microbiome profile.
There have been numerous studies on the role of various human microbiomes in tumorigenesis and progression across different cancer types (9–14). However, only in recent years have scientists begun to explore the influence of the microbiome on cancer treatment response (15). With the increasing demand for the rapid adoption of personalized medicine in cancer therapy to prolong survival and improve the quality of life of patients, pharmacomicrobiomics may contribute vital biomarkers to enhance the translation of precision medicine into clinical practice. This review analyzes the molecular mechanisms of interaction between different microbiome types and tumors, as well as their response to various cancer treatment modalities. It also analyzes the challenges to consider in developing this application direction, proposes potential solutions to benefit patients, and ultimately provides clinical evidence for the advancement of pharmacomicrobiomics in practice.
2 The role of microbiome in cancer
2.1 The microbiota dysbiosis in cancer development and progression
There is compelling evidence indicating a correlation between various types of human microbiomes and different types of cancer (16). Among these, the gut microbiome has been the first and most extensively studied. It is well-documented that the gut microbiome influences systemic metabolic balance and immune function, thus playing a significant role in the tumorigenesis and progression of various cancer types, from gastrointestinal cancers to breast or lung cancer (Table 1). Additionally, changes in the bacterial ecosystem on the skin, which is the body’s largest organ, also have implications for breast health and the risk of developing breast cancer (10). Reduced alpha diversity within the oral microbiome could potentially correlate with an elevated likelihood of developing lung cancer, offering a potential indicator for predicting lung cancer risk (11). In particular, recent discoveries regarding the existence of intratumoral microbiota also underscore their significant influence on tumor development, invasion, and metastasis (12, 56).
Numerous studies have revealed that tumors can interact with various components within the body as well as metabolisms (57), such as platelets (58), circulating tumor cells (CTCs) (59), exosomes (60), and modify these components to serve the tumor’s growth. Similarly, an important mechanism explaining the causal relationship between the human microbiome and cancer development is the interplay between microbiota and tumor cells, leading to significant alterations in the composition and function of microbiomes in cancer patients. The dysbiosis in various microbiota has been observed in cancer patients with changed variability of ecosystems compared to healthy people. Fusobacterium nucleatum and Parvimonas micra were found to be more prevalent, while Clostridia and Bacteroidia decreased in the gut of colorectal cancer (CRC) patients (61). Additionally, Lactococcus and Fusobacteria exhibited higher abundance, whereas Pseudomonas and Escherichia-Shigella were downregulated in CRC tissues compared to adjacent non-cancerous ones (62, 63). Likewise, the gut microbiota of cervical cancer patients exhibited notable variations in the abundance of seven genera, namely Escherichia-Shigella, Roseburia, Succinivibrio, Lachnoclostridium, Lachnospiraceae_UCG-004, Dorea, and Pseudomonas (30).
Reprogrammed ecosystems were also identified in cancer patients beyond the gastrointestinal tract, involving other different microbiota. Cervical cancer exhibits the greatest diversity in vaginal microbiota, with the enrichment of Ralstonia,Lactobacillus, Gardnerella, Sneathia and Prevotella. Similarly, Gardnerella, Prevotella, and Sneathia exhibit higher prevalence within the HPV-positive cervical intraepithelial neoplasia (CIN) group. In contrast, Gardnerella and Prevotella are more prevalent in the HPV-positive non-CIN group (38).
The significance of microbiota variation in tumorigenesis is evident when comparing the microbiome across healthy tissue, pre-cancerous lesions, and malignant tissues. A transitional microbial dysbiosis is observed from healthy skin to pre-malignant actinic keratosis (AK) and further to squamous cell carcinoma (SCC), marked by an elevated presence of the pathobiont Staphylococcus aureus, surpassing the commensal Cutibacterium acnes in SCC (44). In a recent study, Bacteroides, Trueperella, Staphylococcus, Streptococcus, and Fusobacterium were found to be notably more abundant in the microbiome of melanoma tissue, while Corynebacterium 1, Clostridium sensu stricto 1, and Lactobacillus were identified as the genera exhibiting significantly higher levels in the microbiome of healthy skin (46). Furthermore, there is a significant differentiation in the microbial enrichment of microbiome between patients at various stages of cancer (33). The well-documented heterogeneity of tumor cells and the tumor microenvironment extends beyond diversity at the cellular and molecular levels to include microbial clusters forming micro niches with varying species composition and quantities within a tumor mass. This specific distribution of clusters has been reported with multiple types of tumors, from skin cancer to CRC or gastric cancer (48, 50).
2.2 Reprogrammed microbiota impact cancer hallmarks
Not only limited to changes in microbial composition, in the cancer state, there are significant alterations in the functions of microorganisms, especially in metabolism, generating metabolites that favor the hallmarks of cancer cells (Table 1). The toxin produced by enterotoxigenic Bacteroides fragilis (ETBF), known as B. fragilis toxin (BFT), is implicated in colitis and prompts a pro-cancerous inflammatory response. This inflammation, driven by Stat3 and T helper type 17 (T(H)17) cells, contributes to colonic hyperplasia and the development of tumors (64). Similarly, through the Stat3 pathway and interleukin-17-positive (IL-17+) γδ T-cells axis, the oral microbiota found in periodontitis, particularly with a dominant presence of Porphyromonas, has been implicated in the promotion of oral SCC (39). The homotrimeric form of Collagen Type 1, comprising three α1 chains, derived from pancreatic cancer cells, has been shown to facilitate oncogenic signaling via Discoidin domain receptor 1 and integrin α3β1. This signaling pathway promotes cancer cell proliferation and the formation of tumor organoids. Additionally, it leads to an increased abundance of Bacteroidales within the intratumoral microbiome (47).
Angiogenesis is another important hallmark, essentially contributing to the growth of tumors. There is ample evidence demonstrating the relationship between microbiomes at various locations in the body, from the gut to ocular microbiota, and the process of neovascularization (65, 66). The immortality of tumor cells is originated from the capability to resist apoptosis. On the other hand, apoptosis can be impacted by metabolites from certain taxa in various microbiota. P. anaerobius, found in abundance in CRC patients, secretes trans-3-indoleacrylic acid (IDA), which promotes CRC development by counteracting ferroptosis, a form of cell death characterized by uncontrolled lipid peroxidation and subsequent membrane damage. Inhibiting key mediators of IDA, such as Apoptosis-inducing factor 2, aldehyde dehydrogenase 1 family member A3, or Aryl Hydrocarbon Receptor, reversed this effect and suppressed tumor growth. On the other hand, feeding IDA or introducing P. anaerobius accelerated CRC development in mouse models (67). In recent years, advancements in next-generation sequencing (NGS) technology have facilitated the exploration of interactions between metagenomics across diverse microbiomes and the host genome. In a case-control study, a correlation was discovered between the colon microbiota and specific mutations and genome stability in CRC tumors. This study revealed that the presence of the Kirsten rat sarcoma virus (KRAS) mutant type was positively linked to Faecalibacterium while inversely associated with the presence of Bifidobacterium (18).
A characteristic of cancer cells leading to recurrence and treatment failure is their ability to metastasize and invade surrounding tissues. The role of gut microbiota in the dissemination, survival, and colonization of metastatic cancer cells has long been recognized through numerous studies across various cancer types (68). Recently, along with the discovery of microbiota residing within tumors, their significant role in tumor migration and metastasis has been unveiled. Research conducted on a murine model of spontaneous breast tumors revealed that during the process of metastatic colonization, bacteria residing within the tumor were transported by CTCs. These bacteria played a role in enhancing the survival of host cells by increasing their resistance to fluid shear stress during the metastasizing process through the reorganization of the actin cytoskeleton. Additionally, the direct administration of certain bacterial strains, such as Staphylococcus and Lactobacillus, isolated from the microbiota of breast tumors, promoted metastasis in experimental tumor models. Conversely, when breast intratumor bacteria were depleted, there was a significant reduction in lung metastasis (52). CRC cells infected with F. nucleatum exhibit enhanced invasiveness into their surrounding environment and attract myeloid cells to the bacterial niches. This process accelerates migration rates significantly by mediating various signaling pathways crucial for cancer metastasis, including extracellular matrix remodeling and modulation of cell adhesion and migration via ERK1 and ERK2 (48).
2.3 Reprogrammed microbiota modifies the host immune system
The characteristic changes in microbiota composition in cancer patients can result in alterations of the composition and functions of immune cells including T-cells, natural killer (NK) cells, dendritic cells (DCs), and macrophages, implicated in antitumor immunity. A comparative analysis of intraepithelial lymphocytes in CRC tissue versus healthy colonic tissue revealed a reduction in γδ T-cells and resident memory T-cells within cancerous tissue. These populations exhibited a regulatory CD39-expressing phenotype in the cancer microenvironment. Moreover, distinct patterns of T-cell proliferative responses to various commensal bacteria were observed in CRC patients, while B cell memory responses to certain bacteria/yeast were notably elevated. This increase in B cell memory responses was accompanied by higher proportions of circulating effector memory B cells, transitional B cells, and plasmablasts (69).
The influence of microbiota on lymphocyte populations is partially attributed to the modulation of antigen presentation cells through microbiota metabolites. Specifically, butyrate, a short-chain fatty acid (SCFA) metabolite produced by microbiota, has been shown to hinder DCs presentation of tumor-associated antigens. Consequently, this impediment affects the infiltration of T-CD8+ cells in an IFN-γ-dependent manner. Therefore, the depletion of butyrate-producing strains in the gut microbiota through vancomycin treatment has been observed to enhance the antitumor response to radiotherapy (70). Likewise, phytosphingosine, a metabolite derived from the gut microbiota, can increase the expression of HLA class I on cancer cells. This sensitizes the cells to targeted antigen-specific cytotoxic T lymphocyte destruction, both in vitro and within living organisms, thereby enhancing the efficacy of immuno checkpoint inhibitor (ICI) treatments (71).
Besides altering the antigen-presenting function of immune cells, microbiota also has the capability to influence the production of pro-inflammatory cytokines like interleukin-12 (IL-12) and IFN-γ. These cytokines play crucial roles in activating and enhancing the function of cytotoxic T-cells and NK cells (72, 73). Apart from the systemic immune modulation exerted by gut microbiota, the intratumoral microbiota plays a role in shaping the immune profile within the tumor microenvironment. In oral SCC, an enrichment of genera such as Capnocytophaga, Fusobacterium, and Treponema correlated with the presence of effector subsets of tumor-infiltrating lymphocytes and the associated gene expression involved in the recruitment of B cells and T-cells. This enrichment ultimately leads to immunosuppressive effects within the tumor microenvironment (74).
Fascinatingly, the microbiota not only can interact with tumors to modulate immune responses toward decreased tumor surveillance, but it can also interact with therapeutic drugs to alter the immune system in a synergistic antitumor direction. A recent study revealed that ICI induces the movement of specific native gut bacteria into secondary lymphoid organs and subcutaneous melanoma tumors. Specifically, ICI prompts the restructuring of lymph nodes and activation of DCs, facilitating the migration of a specific subgroup of gut bacteria to extraintestinal tissues. This migration promotes optimal antitumor T-cell responses in both the tumor-draining lymph nodes (TDLNs) and the primary tumor. Furthermore, antibiotic treatment leads to reduced translocation of gut microbiota into mesenteric lymph nodes (MLNs) and TDLNs, resulting in weakened DCs and effector CD8+ T-cell responses, as well as diminished responses to ICI. These findings offer opportunities for leveraging microbiota in a beneficial direction for treatment (75).
Immune checkpoint molecules, such as programmed cell death protein 1 (PD-1), Programmed Death Ligand 1 (PD-L1), and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), play pivotal roles in regulating T-cell responses. Research indicates that certain microbial species and their metabolites can influence the expression of these checkpoint molecules, thereby impacting T-cell activation and facilitating immune evasion by tumors. A study examining the urogenital microbial community in bladder cancer patients revealed an increased presence of Leptotrichia, Roseomonas, and Propionibacterium, along with a reduction in certain bacterial genera, such as Prevotella and Massilia, among those exhibiting PD-L1 expression on cancerous tissues compared to those who tested negative for PD-L1 expression (76). Similarly, Veillonella dispar was prevalent in the lung microbiome of lung cancer patients exhibiting high PD-L1 expression, whereas the abundance of Neisseria was notably elevated in patients with low PD-L1 expression. Consequently, V. dispar predominated in the group of patients showing a positive response, whereas Haemophilus influenzae and Neisseria perflava were prevalent in the non-responder group (77). In the context of CTLA4 blockade therapy, heightened concentrations of butyrate and propionate in the bloodstream correlate with treatment resistance and an increased proportion of Treg cells. Mouse studies reveal that butyrate impeded the CTLA-4-induced elevation of CD80/CD86 expression on DCs and ICOS expression on T-cells, along with the buildup of tumor-specific T-cells and memory T-cells. In patients, elevated blood butyrate levels mitigated the ipilimumab-induced accumulation of memory and ICOS + CD4 + T-cells and IL-2 production, suggesting that SCFA restricts the efficacy of anti-CTLA-4 therapies (78).
Overall, the molecular intricacies governing the interplay between the microbiome and antitumor immunity encompass complex interactions among microbial elements, host immune cells, and the tumor microenvironment. Grasping these mechanisms is imperative for devising microbiome-centered interventions aimed at bolstering antitumor immune responses and refining cancer treatment outcomes.
3 The impact of microbiota on cancer treatment
Given the crucial role in tumor formation, development, metastasis, and host immunity, microbiota can exert significant influences on patients’ responses to cancer treatment modalities, including both therapeutic efficacy and toxicity. Among these therapies, the most extensively investigated area with compelling evidence supporting the role of microbiota is the field of immunotherapy - the latest cancer treatment method that has made remarkable advances in clinical application. Additionally, microbiota also affect response to other cancer therapies, suggesting perspectives of interventions to achieve precision medicines (Figure 1).
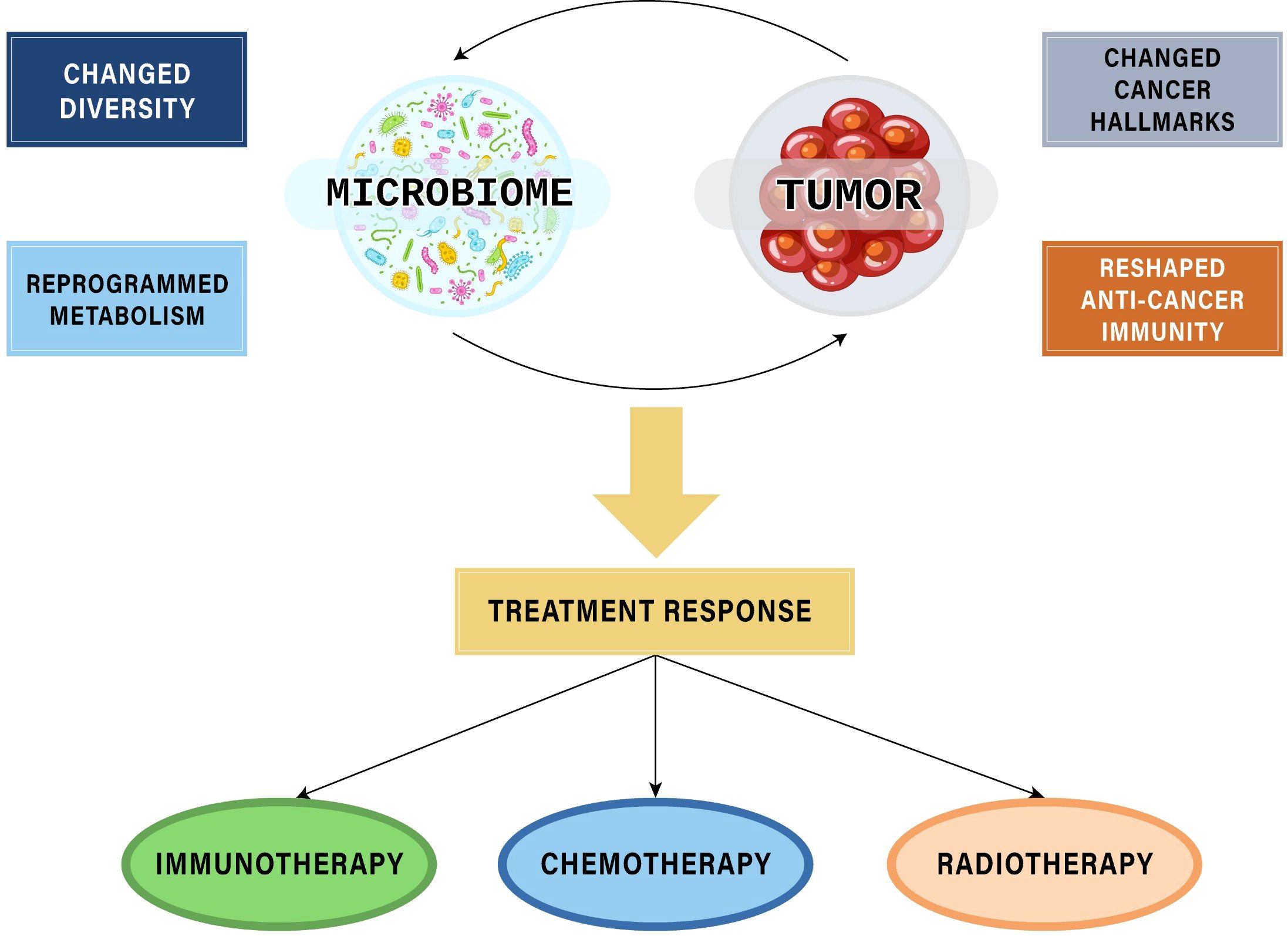
Figure 1. The interplays between microbiomes and tumors affecting responses to cancer treatment modalities.
3.1 Microbiota and immunotherapy response
In precision medicine, the most important aspect is to identify biomarkers for the stratification of patient groups to select appropriate drugs for each group. Although some molecular biomarkers have been applied in personalized medicine with immunotherapies, their true effectiveness in clinical practice remains controversial, requiring supplementation or support from other types of biomarkers. Significant variations in the composition of gut microbiota have been observed between patients who respond favorably to ICI and non-responders across a range of different cancer types. Responding melanoma patients exhibited notably higher alpha diversity, along with a relative abundance of Ruminococcaceae bacteria. Intriguingly, metagenomic analysis revealed an enrichment of amino acid biosynthesis in responders, thereby contributing to enhanced immunity characterized by increased infiltration of CD4+ and CD8+ T-cells (79). Likewise, an analysis of gut microbiota utilizing 16S ribosomal RNA sequencing revealed increased alpha diversity among responders to ICI and CTLA-4 inhibitors across various cancer types. The microbiota composition of responders resembled that of healthy individuals. Additionally, certain bacteria, including Prevotella copri and Faecalibacterium prausnitzii, were linked to a favorable treatment outcome (80). Together with the enhanced alpha diversity, the enrichment of g-Blautia has been suggested as a potential predictor of responders to ICI with longer progression-free survival (PFS) in patients with non-small cell lung cancer (NSCLC) (81). Interestingly, Sarfaty et al. not only identified cancer type-specific microbiome signatures to distinguish between favorable responders and non-responders but also observed certain similarities in the microbiome signatures of non-responders across three different cancer types including lung, urothelial, and melanoma. This suggests the potential utility of these signatures as common pharmacomicrobiomic biomarkers (82).
Despite the treatment efficacy advantages of such an advanced therapeutic modality, ICI, like other cancer treatment regimens, also have unintended effects, notably immune-related adverse events (irAEs), which can impact treatment response and patient adherence. Among melanoma patients, responders experiencing irAEs from grade 2 in the Common Terminology Criteria for Adverse Events (CTCAE) exhibited a predominance of Bacteroides plebeius and Bacteroides coprophilus in their gut microbiota, while those without irAEs showed an enrichment of Eubacterium siraeum (83). Apart from alterations in gut microbiota, there is also a distinction in the skin microbiome among melanoma patients experiencing cutaneous irAEs following ICI treatment. This is characterized by an increase in Staphylococci and Proteobacteria, whereas patients without irAEs exhibited enrichment in anaerobic Eubacteriales (84). Identifying predictors of irAEs can aid in mitigating severe ADRs and preventing patient suffering.
Beyond merely identifying differences in microbiota between patient groups responding and not responding to immunotherapies, scientists can actively modify treatment responses by intervening in the microbiota. In vivo experiments have shown that transplanting fecal material from patients who respond to treatment with an abundance of Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium into germ-free mice could result in enhanced tumor control, increased T-cell responses, and improved efficacy of anti-PD-L1 therapy (85). Conversely, mice that received fecal transplants from patients with poor responsiveness exhibited elevated frequencies of regulatory CD4+FoxP3+ T-cells and CD4+IL-17+ T-cells in the spleen. This indicates compromised host immune responses, ultimately resulting in the failure of ICI treatment (79). A thorough study on improving the gut microbiota to finetune cancer immunotherapy shows that certain bacteria in the gut are very important in determining the immune responses linked to CTLA-4 checkpoint blocking therapy. By regulating the gut flora, this understanding has the potential to enhance the therapeutic effectiveness of ICI and potentially reduce its immune-mediated toxicity (86).
In addition to impacting gut microbiota through fecal transplantation, skin bacteria can be genetically modified to induce changes in systemic anti-cancer immune responses. A recent study revealed that modifying the skin bacterium Staphylococcus epidermidis to express tumor antigens could enhance highly specific adaptive immune responses mediated by T-cells, leading to significant improvements in melanoma immunotherapy efficacy (87). Additionally, a more straightforward approach to modulate gut microbiota and thus modify the response to ICI treatment is through dietary interventions. Melanoma patients who consumed a diet rich in dietary fiber experienced significantly extended PFS while undergoing ICI treatment. Conversely, mice fed a low-fiber diet exhibited poor responsiveness to anti-PD1 therapy, characterized by a reduced frequency of interferon-γ–positive cytotoxic T-cells in the tumor microenvironment (88).
3.2 Microbiota and chemotherapy response
In addition to its impact on immunotherapy, the microbiota plays a significant role in influencing the effectiveness of other treatments such as chemotherapy, whose response relies heavily on pharmacokinetics and pharmacodynamics (Figure 2). Like many other therapeutic groups, the bioavailability of chemotherapy agents is determined by hepatic metabolic enzymes and transporters, including the cytochrome P450 superfamily, which is responsible for metabolizing a majority of medications. Research has shown that alterations in gut microbiota can affect the expression of key pharmacokinetic proteins like CYP3A1, UGT1A1, and P-glycoprotein (P-gp) in the liver. Specifically, changes in the composition of gut microbiota have been observed to impact the metabolism and bioavailability of drugs like cyclosporine. For instance, higher levels of Alloprevolleta and Oscillospiraceae UCG 005 have been associated with reduced bioavailability of cyclosporin, while increased levels of Parasutterella and the Eubacterium xylanophilum group have been linked to increased bioavailability (89). Previously, only the biotransformation processes of drugs by human hepatic enzymes were known. However, recent studies have shown that enzymes from the intestinal microbiota also participate in drug metabolism reactions, significantly affecting the plasma concentration of drugs and their metabolite as well as the drug’s elimination half-life (90, 91). Consequently, this impacts the efficacy and toxicity of the treatment. This is particularly significant for drugs with a narrow therapeutic window such as chemotherapies.
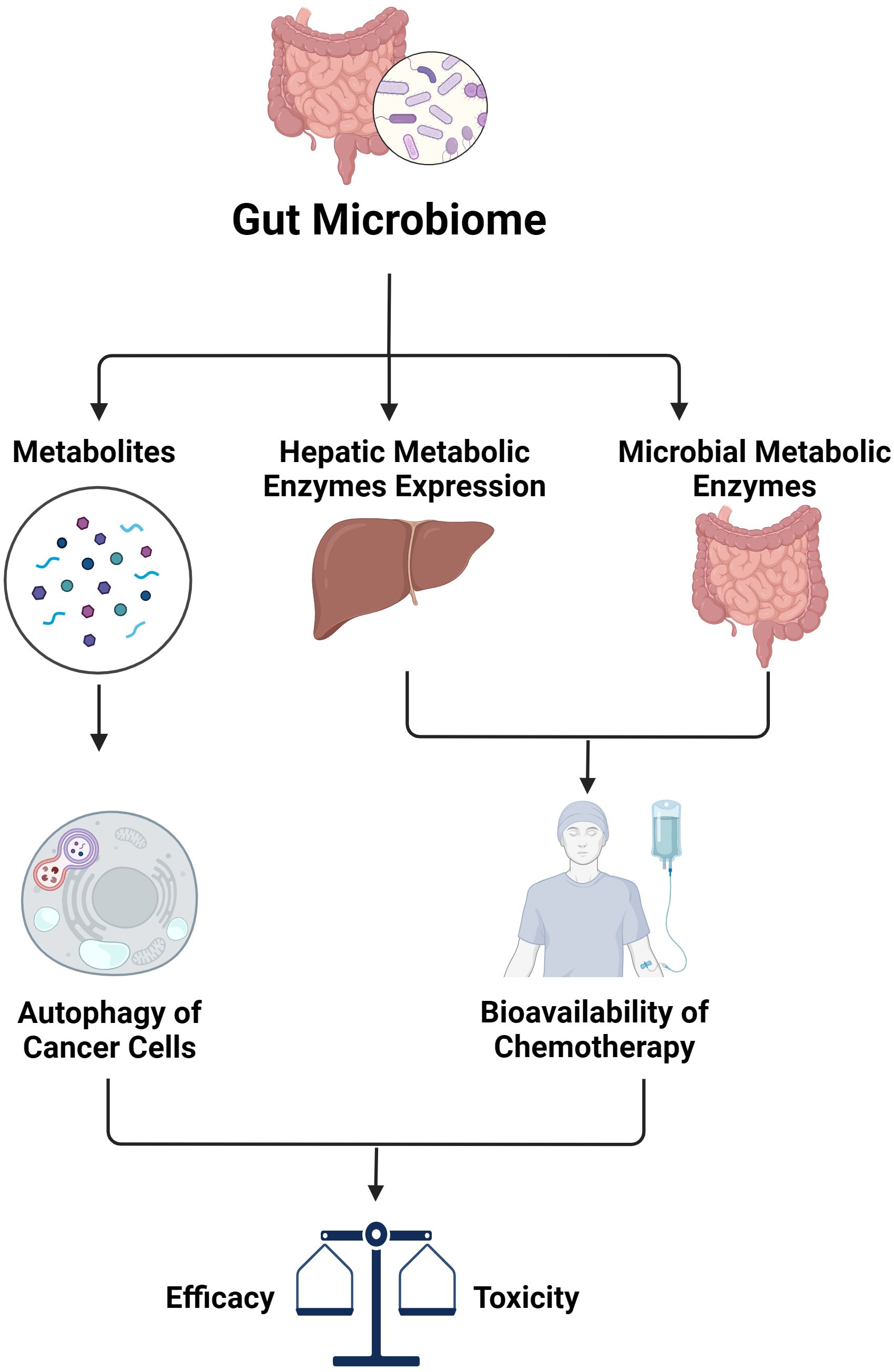
Figure 2. The impact of gut microbiome on efficacy and toxicity of chemotherapy (created with BioRender.com).
Similar to immunotherapies, there exists a significant disparity in both the composition and abundance of microorganisms within the gut microbiota between patients who respond favorably to chemotherapy and those who do not, across various types of cancer. This contrast has been observed in locally advanced rectal cancer patients, where intestinal microbes associated with butyrate production, such as Roseburia, Dorea, and Anaerostipes, were found to be more prevalent in responders to neoadjuvant chemotherapy, whereas Coriobacteriaceae and Fusobacterium were more prevalent in non-responders. A set of ten predictors, including Dorea, Anaerostipes, and Streptococcus, was identified for the stratification of responders, achieving an area under the curve value of up to 93.57% (92).
The favorable efficacy of chemotherapies is associated with some specific microbiota-derived metabolites, suggesting their utility as solutions to enhance the benefit of such a popular cancer therapy. The microbiota-derived tryptophan metabolite, indole-3-acetic acid (3-IAA) which plays a crucial role in the autophagy process of cancer cells was found in higher concentrations in pancreatic cancer patients who positively responded to treatment. Studies have shown that interventions such as fecal microbiota transplantation, short-term dietary adjustments focusing on tryptophan, and oral administration of 3-IAA enhance the effectiveness of chemotherapy in humanized murine models of pancreatic cancer. Furthermore, a significant correlation between the levels of 3-IAA and the chemotherapy’s efficacy has been observed in two separate cohorts of patients with pancreatic ductal adenocarcinoma (PDAC) (93). By utilizing machine learning models incorporating both drug response and microbiota data, Hermida et al. demonstrated that the microbiota profile emerges as a superior predictor of clinical outcomes when compared to clinical variables across seven distinct cancer types, including chemotherapy treatments for bladder urothelial carcinoma, docetaxel treatment for breast invasive carcinoma and sarcoma, as well as various treatments for stomach adenocarcinoma (94).
Not only can the microbiota profile predict the efficacy of chemotherapies, but it can also help anticipate specific ADRs caused by chemotherapy agents. In acute lymphoblastic leukemia (ALL), the initial gut microbiome composition, marked by an abundance of Proteobacteria, served as a predictive factor for febrile neutropenia following chemotherapy. Notably, a prevalence of Enterococcaceae was associated with a significantly higher likelihood of experiencing subsequent febrile neutropenia and diarrheal ADRs. Additionally, the dominance of Streptococcaceae predicted a remarkably increased risk of subsequent diarrheal adverse events (95). Similarly, the presence of Bacteroides and Blautia2 in the gut microbiota of rectal cancer patients could predict ADRs such as fatigue, sleep disturbance, or depression following chemotherapy with an accuracy of 74% (96). Also in rectal cancer cases, dynamic changes observed in the tumor microbiome throughout and following chemoradiation therapy were associated with drug-related toxicity. Specifically, patients who experienced heightened toxicity by week 5 displayed elevated relative counts of Clostridia, Actinobacteria, and Clostridiales at the outset of treatment (97).
3.3 Microbiota and radiotherapy response
Radiotherapy stands as a crucial cancer treatment modality with interindividual variations in therapeutic effectiveness and toxicity. Among its most significant antitumor mechanisms is the stimulation of both innate and adaptive immunity. Consequently, the microbiota, which exerts profound influences on the host immune system, can significantly influence therapeutic outcomes. The intricate relationship between intestinal microbiota and post-radiation immune responses in mouse models of breast cancer and melanoma has been uncovered. While the exclusion of gut fungi enhanced the anti-tumor effects of radiation, the use of antibiotics to deplete bacteria diminished responsiveness, leading to the proliferation of commensal fungi. Moreover, the expression level of intratumoral Dectin-1, a key innate fungal sensor was essential for the impact of commensal fungi in mice undergoing radiation therapy and could predict survival rates in breast cancer (98).
Butyrate, a common metabolite produced by intestinal bacteria, is renowned for its impact on immune function. Recent research using murine models has revealed a negative correlation between the abundance of butyrate-producing gut bacteria and anticancer responses to radiation. Butyrate inhibited STING-activated type I IFN expression in DCs by blocking TBK1 and IRF3 phosphorylation. This inhibition abolished radiation-induced tumor-specific cytotoxic T-cell immune responses, without directly shielding CRC and melanoma cells from radiation. These results underscore the potential of selectively targeting butyrate-producing microbiota as a novel therapeutic approach to enhance tumor radiation sensitivity (99). The contribution of gut microbiota via the STING pathway to antitumor immune responses has also been observed in both hepatocellular carcinoma (HCC) patients and experimental HCC models (100).
Radiation therapeutic response is not solely influenced by the gut microbiome. A recent investigation using CRC mouse models revealed that modifications in oral microbiota led to shifts in bacterial makeup within CRC tumors while leaving adjacent peritumor tissues unaffected. Notably, the migration of Fusobacterium nucleatum from the oral cavity to the CRC site was observed, hindering the effectiveness of radiotherapy and impacting prognosis. The administration of metronidazole successfully countered the detrimental effects of oral microbiome alterations on CRC radiotherapy outcomes. Furthermore, the oral microbiota was found to correlate with radiation-induced intestinal damage through its influence on intestinal microbial communities (101).
One drawback of radiotherapy is the emergence of undesired side effects affecting various organs in the body, such as toxicity to the digestive or nervous systems. Around 90% of cancer patients undergoing pelvic radiotherapy experience gastrointestinal (GI) toxicity, including symptoms like bloody diarrhea and gastritis, with many linked to gut dysbiosis. Hence, the gut microbiome, pivotal in regulating digestive function, significantly influences gastrointestinal ADRs to radiation. In a preclinical investigation, radiation-induced damage to intestinal villi height and mucosal thickness was observed, along with induced neural inflammation and cell death (102). Intriguingly, altering the gut microbiota effectively mitigated toxicity in both the gastrointestinal and neural systems, suggesting a key to the challenge of radiotherapy. A study conducted on gynecologic cancer patients yielded similar findings, demonstrating that modifying the vaginal microbiota resulted in changes to radiation-induced vaginal toxicities, including pain, dyspareunia, and sexual dysfunction (103).
4 Perspectives for microbiome-targeted solutions to improve cancer treatment outcomes
Understanding the significant influence of the various microbiota on the development, metastasis, and response to treatment of tumors not only allows the use of microbiome components as biomarkers for selecting appropriate treatment methods, achieving high efficacy with minimized toxicity but also opens perspectives for intervening to alter the microbiome to bring about favorable outcomes for cancer patients. The advantages of microbiome-targeted interventions lie in their high feasibility, as they do not require overly advanced, costly methods, are minimally invasive, have fewer long-term systemic side effects, and are non-irreversible for patients. Another particularly notable aspect is that interventions targeting the microbiome can be personalized according to the unique microbiome characteristics of each patient. Additionally, microbiome-targeted interventions can be combined with various therapeutic modalities in a personalized manner to maximize benefits for patients. The followings are primary strategies for microbiome interventions.
4.1 Diet and supplement-based interventions
Substantial evidence highlights significant differences in microbiota composition, beneficial/pathogenic microbe abundance, and metabolite profiles between healthy individuals and cancer patients, as well as among cancer patients with varied treatment responses (63). Consequently, modifying dietary habits to promote beneficial microbe growth and diminish harmful ones can positively influence treatment outcomes in cancer patients. Recognized as pivotal components of cancer precision medicine, diet, and supplement-based interventions target specific dietary factors and nutritional supplements to optimize treatment efficacy. For example, embracing a diet abundant in fiber, fruits, vegetables, and fermented foods fosters a diverse and healthy microbiota. Additionally, a proactive approach to microbiome influence involves supplementing beneficial microbes and their substrates through microbiome modulators such as probiotics, prebiotics, and postbiotics to elicit desired effects. This notion finds support in numerous clinical studies across diverse cancer types (Table 2).
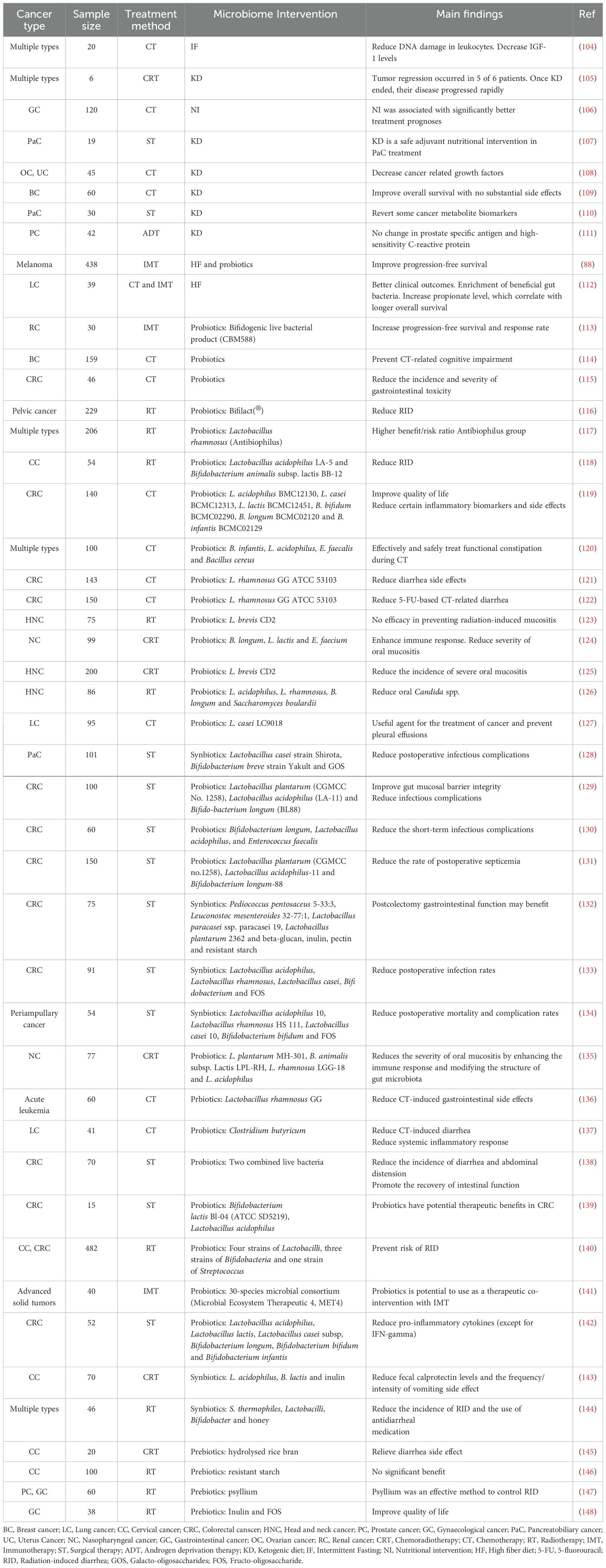
Table 2. Clinical evidence supporting diet and supplement-based interventions for better cancer treatment outcomes.
Despite increasing interest in these interventions, challenges persist regarding standardization, efficacy, and safety (149). Additionally, some contrary findings have been reported, in which, probiotics use compromised the efficacy of ICIs in cancer patients (150). Therefore, rigorous clinical trials are indispensable to assess the efficacy and safety of diet approaches, establish optimal dosages and formulations, and ascertain their compatibility with conventional cancer therapies. Furthermore, personalized strategies are imperative to tailor diet and supplement interventions to individual patient characteristics, including cancer type, stage, genetic profile, and lifestyle factors.
4.2 Fecal microbiota transplantation
Bacteriotherapy, which involves utilizing microbes or their byproducts to treat illnesses, encompasses various approaches. Alongside supplementing specific microbes through probiotics, Fecal Microbiota Transplantation (FMT) is emerging as a promising method to utilize the gut microbiome’s potential to modulate therapeutic responses and enhance patient outcomes. FMT entails transferring fecal material from a healthy or therapeutically responsive donor into the gastrointestinal tract of a patient to restore or manipulate the microbiota composition (151). According to a Europe-wide survey conducted in 2019, 31 FMT centers across 17 countries performed a total of 1,874 procedures. However, the sole officially approved indication for FMT remains Clostridioides difficile infection (152).
Despite accumulating evidence demonstrating the potential benefits of FMT in enhancing outcomes of cancer treatment in experimental models, its application in cancer patients is currently limited to research and clinical trials (Table 3). Often integrated with other therapeutic approaches, some of these trials have shown promising results, suggesting a potential avenue for effective cancer treatment. FMT has been demonstrated to enhance the efficacy of immunotherapy by bolstering anti-tumor immune responses in CRC and melanoma (156, 195). Additionally, FMT has proven effective in mitigating treatment-related toxicities, such as chemotherapy-induced gastrointestinal symptoms (194). In particular, several clinical studies have demonstrated the efficacy of FMT in treating Gastrointestinal Acute Graft-versus-Host Disease (GI-aGvHD), a severe and potentially life-threatening complication arising from Allogeneic Stem Cell Transplantation (allo-SCT), an advanced therapeutic approach utilized in the management of hematologic malignancies.
However, challenges and considerations accompany the implementation of FMT in cancer treatment. These include standardizing donor selection and screening procedures, optimizing FMT protocols, determining optimal timing and dosing, and managing potential risks such as infection transmission and immune-related adverse events. That is the reason why FMT has not been approved for use in clinical setting, except for Clostridioides difficile infection, in European countries. Despite these challenges, FMT represents a promising avenue for precision oncology, offering a personalized and microbiome-based approach to cancer therapy. Further research is necessary to elucidate the mechanisms of action, optimize treatment protocols, and identify patient subgroups most likely to benefit from FMT. Overall, FMT holds potential as an innovative strategy to complement existing cancer treatment modalities and improve outcomes for cancer patients.
4.3 Antibiotic-based interventions for microbiome
Antibiotics, traditionally used to combat bacterial infections, have garnered attention for their potential to influence cancer treatment responses through modulation of the microbiome through both preclinical and clinical research (196). Despite promising findings in preclinical research indicating potential benefits of antibiotics in enhancing treatment efficacy and reducing adverse reactions in cancer therapy, clinical studies across diverse cancer types consistently demonstrate that antibiotic usage before or during treatment is associated with worsened outcomes, notably in immunotherapy (Table 4). These observations suggest an additional strategy for regulating microbiota in cancer precision medicine through the selective use of antibiotics given the requirement for thorough research for the appropriate antibiotic. Targeting harmful microbes with antibiotics to manipulate microbial communities can optimize treatment responses and reduce adverse effects. However, careful consideration, especially in antibiotic dosage is necessary to avoid disturbing the beneficial microbiota, leading to clinical complications (209) and the danger of antibiotic resistance (210).
4.4 Modulation of other local microbiotas beyond the gut
Local interventions targeting microbiotas beyond the gut microbiota are emerging as promising strategies in cancer precision therapy, aiming to harness the influence of various microbial communities on tumor biology and treatment responses. While much attention has been focused on the gut microbiota, other microbiotas throughout the body, including those in the oral cavity, skin, respiratory tract, urogenital tract, and tumor microenvironment, also play significant roles in cancer development and treatment.
Research has revealed the intricate interactions between these microbiotas and cancer, highlighting their potential as therapeutic targets for precision therapy (15, 16). Local interventions seek to modulate the composition and function of these microbiotas to enhance treatment efficacy, reduce treatment-related toxicities, and improve patient outcomes. These interventions encompass a range of approaches, including probiotics, prebiotics, antibiotics, microbial metabolites, targeted therapies, and physical interventions.
In the oral cavity, interventions targeting the oral microbiota, such as mouthwash formulations containing probiotics or antimicrobial agents, hold promise for preventing oral mucositis and reducing the risk of secondary infections in patients undergoing radiation therapy or chemotherapy for head and neck cancers (101). Similarly, interventions targeting the skin microbiota may involve topical applications of probiotic formulations or antimicrobial agents to alleviate radiation-induced dermatitis and enhance wound healing in patients with skin cancers (52, 87).
In the respiratory tract, interventions may include inhalation therapies with probiotics or antimicrobial agents to improve treatment responses and reduce the risk of respiratory infections in patients with lung cancers (211). Likewise, interventions targeting the urogenital microbiota may involve the use of vaginal probiotics or antimicrobial agents to prevent urinary tract infections and enhance treatment tolerability in patients with genitourinary cancers (76, 212).
Moreover, interventions targeting the tumor microenvironment may encompass immunomodulatory therapies, such as ICI or adoptive cell therapies, aimed at modulating the local immune response and tumor growth in various cancer types. Additionally, microbial-based therapies, such as oncolytic viruses or bacteria engineered to target tumors, offer novel strategies for directly targeting tumor cells and modulating the tumor microenvironment (213).
Despite the potential benefits of local interventions on non-gut microbiotas for cancer precision therapy, challenges exist in their implementation and optimization. These include the need for further research to elucidate the complex interactions between microbiotas and cancer, as well as the development of targeted and personalized interventions tailored to individual patient characteristics and tumor biology.
5 Conclusion
The field of pharmacomicrobiomics holds immense promise for revolutionizing precision cancer therapy by leveraging the intricate interplay between the microbiome and drug response. From preclinical investigations elucidating molecular mechanisms to clinical trials evaluating patient outcomes, advancements in this field offer unprecedented opportunities to optimize treatment strategies tailored to individual patients. By harnessing the potential of pharmacomicrobiomics, we can enhance treatment efficacy, minimize adverse effects, and ultimately improve patient outcomes in the era of precision oncology. However, challenges such as standardization, validation, and clinical translation remain, underscoring the need for continued research and collaborative efforts across disciplines.
Author contributions
KL: Visualization, Writing – original draft, Writing – review & editing. TP: Visualization, Writing – original draft, Writing – review & editing. TN: Writing – review & editing. PH: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tsimberidou AM, Fountzilas E, Nikanjam M, Kurzrock R. Review of precision cancer medicine: evolution of the treatment paradigm. Cancer Treat Rev. (2020) 86:102019. doi: 10.1016/j.ctrv.2020.102019
2. Huang X, Zhu M, Chen R, Ni J, Zhao W, Li S, et al. Innovative drugs promote precision cancer therapy. Clin Cancer Bull. (2023) 2:1. doi: 10.1007/s44272-023-00002-8
3. Weinshilboum RM, Wang L. Pharmacogenomics: precision medicine and drug response. Mayo Clin Proc. (2017) 92:1711–22. doi: 10.1016/j.mayocp.2017.09.001
4. Saugstad AA, Petry N. Pharmacogenetic review: germline genetic variants possessing increased cancer risk with clinically actionable therapeutic relationships. Front Genet. (2022) 13:857120. doi: 10.3389/fgene.2022.857120
5. Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. (2015) 526:343–50. doi: 10.1038/nature15817
6. Doestzada M, Vila AV, Zhernakova A, Koonen DPY, Weersma RK, Touw DJ, et al. Pharmacomicrobiomics: A novel route towards personalized medicine? Protein Cell. (2018) 9:432–45. doi: 10.1007/s13238-018-0547-2
7. Hou K, Wu Z-X, Chen X-Y, Wang J-Q, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther. (2022) 7:(2022–04-23). doi: 10.1038/s41392-022-00974-4
8. Cai W, Liu Z, Miao L, Xiang X. Pharmacogenomics in precision medicine from a perspective of ethnic differences. 1st. ed. Singapore: Springer Singapore (2020).
9. Kandalai S, Li H, Zhang N, Peng H, Zheng Q. The human microbiome and cancer: A diagnostic and therapeutic perspective. Cancer Biol Ther. (2023) 24:2240084. doi: 10.1080/15384047.2023.2240084
10. Wang K, Nakano K, Naderi N, Bajaj-Elliott M, Mosahebi A. Is the skin microbiota a modifiable risk factor for breast disease?: A systematic review. Breast. (2021) 59:279–85. doi: 10.1016/j.breast.2021.07.014
11. Zhang K, He C, Qiu Y, Li X, Hu J, Fu B. Association of oral microbiota and periodontal disease with lung cancer: A systematic review and meta-analysis. J Evid Based Dent Pract. (2023) 23:101897. doi: 10.1016/j.jebdp.2023.101897
12. Zhou X, Ji L, Ma Y, Tian G, Lv K, Yang J. Intratumoral microbiota-host interactions shape the variability of lung adenocarcinoma and lung squamous cell carcinoma in recurrence and metastasis. Microbiol Spectr. (2023) 11:e0373822. doi: 10.1128/spectrum.03738-22
13. Cui W, Guo M, Liu D, Xiao P, Yang C, Huang H, et al. Gut microbial metabolite facilitates colorectal cancer development via ferroptosis inhibition. Nat Cell Biol. (2024) 26:124–37. doi: 10.1038/s41556-023-01314-6
14. Chen F, Dai X, Zhou CC, Li KX, Zhang YJ, Lou XY, et al. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut. (2022) 71:1315–25. doi: 10.1136/gutjnl-2020-323476
15. Blake SJ, Wolf Y, Boursi B, Lynn DJ. Role of the microbiota in response to and recovery from cancer therapy. Nat Rev Immunol. (2023) 24:308–25. doi: 10.1038/s41577-023-00951-0
16. Cullin N, Azevedo Antunes C, Straussman R, Stein-Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell. (2021) 39:1317–41. doi: 10.1016/j.ccell.2021.08.006
17. Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, et al. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. (2023) 72:1129–42. doi: 10.1136/gutjnl-2022-327156
18. Yuan D, Tao Y, Wang H, Wang J, Cao Y, Cao W, et al. A comprehensive analysis of the microbiota composition and host driver gene mutations in colorectal cancer. Invest New Drugs. (2022) 40:884–94. doi: 10.1007/s10637-022-01263-1
19. Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carra A, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. (2019) 363. doi: 10.1126/science.aar7785
20. Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. (2015) 60:208–15. doi: 10.1093/cid/ciu787
21. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. (2013) 14:207–15. doi: 10.1016/j.chom.2013.07.007
22. Kovacs T, Miko E, Vida A, Sebo E, Toth J, Csonka T, et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci Rep. (2019) 9:1300. doi: 10.1038/s41598-018-37664-7
23. Zhu J, Liao M, Yao Z, Liang W, Li Q, Liu J, et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome. (2018) 6:136. doi: 10.1186/s40168-018-0515-3
24. Goedert JJ, Hua X, Bielecka A, Okayasu I, Milne GL, Jones GS, et al. Postmenopausal breast cancer and oestrogen associations with the iga-coated and iga-noncoated faecal microbiota. Br J Cancer. (2018) 118:471–9. doi: 10.1038/bjc.2017.435
25. Miko E, Vida A, Kovacs T, Ujlaki G, Trencsenyi G, Marton J, et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim Biophys Acta Bioenerg. (2018) 1859:958–74. doi: 10.1016/j.bbabio.2018.04.002
26. Zhao F, An R, Wang L, Shan J, Wang X. Specific gut microbiome and serum metabolome changes in lung cancer patients. Front Cell Infect Microbiol. (2021) 11:725284. doi: 10.3389/fcimb.2021.725284
27. Zheng Y, Fang Z, Xue Y, Zhang J, Zhu J, Gao R, et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. (2020) 11:1030–42. doi: 10.1080/19490976.2020.1737487
28. Zhuang H, Cheng L, Wang Y, Zhang YK, Zhao MF, Liang GD, et al. Dysbiosis of the gut microbiome in lung cancer. Front Cell Infect Microbiol. (2019) 9:112. doi: 10.3389/fcimb.2019.00112
29. Zhang WQ, Zhao SK, Luo JW, Dong XP, Hao YT, Li H, et al. Alterations of fecal bacterial communities in patients with lung cancer. Am J Transl Res. (2018) 10:3171–85.
30. Wang Z, Wang Q, Zhao J, Gong L, Zhang Y, Wang X, et al. Altered diversity and composition of the gut microbiome in patients with cervical cancer. AMB Express. (2019) 9. doi: 10.1186/s13568-019-0763-z
31. Golombos DM, Ayangbesan A, O'Malley P, Lewicki P, Barlow L, Barbieri CE, et al. The role of gut microbiome in the pathogenesis of prostate cancer: A prospective, pilot study. Urology. (2018) 111:122–8. doi: 10.1016/j.urology.2017.08.039
32. Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. (2015) 5:8096. doi: 10.1038/srep08096
33. Witt RG, Cass SH, Tran T, Damania A, Nelson EE, Sirmans E, et al. Gut microbiome in patients with early-stage and late-stage melanoma. JAMA Dermatol. (2023) 159:1076–84. doi: 10.1001/jamadermatol.2023.2955
34. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via gammadelta T cells. Cell. (2019) 176:998–1013 e16. doi: 10.1016/j.cell.2018.12.040
35. Liu HX, Tao LL, Zhang J, Zhu YG, Zheng Y, Liu D, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer. (2018) 142:769–78. doi: 10.1002/ijc.31098
36. Cameron SJS, Lewis KE, Huws SA, Hegarty MJ, Lewis PD, Pachebat JA, et al. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PloS One. (2017) 12:e0177062. doi: 10.1371/journal.pone.0177062
37. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. (2016) 102:89–95. doi: 10.1016/j.lungcan.2016.10.016
38. Li X, Wu J, Wu Y, Duan Z, Luo M, Li L, et al. Imbalance of vaginal microbiota and immunity: two main accomplices of cervical cancer in chinese women. Int J Women's Health. (2023) 15:987–1002. doi: 10.2147/IJWH.S406596
39. Wei W, Li J, Shen X, Lyu J, Yan C, Tang B, et al. Oral microbiota from periodontitis promote oral squamous cell carcinoma development via gammadelta T cell activation. mSystems. (2022) 7:e0046922. doi: 10.1128/msystems.00469-22
40. Yang Y, Cai Q, Shu XO, Steinwandel MD, Blot WJ, Zheng W, et al. Prospective study of oral microbiome and colorectal cancer risk in low-income and african american populations. Int J Cancer. (2019) 144:2381–9. doi: 10.1002/ijc.31941
41. Komiya Y, Shimomura Y, Higurashi T, Sugi Y, Arimoto J, Umezawa S, et al. Patients with colorectal cancer have identical strains of fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. (2019) 68:1335–7. doi: 10.1136/gutjnl-2018-316661
42. Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, et al. Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. (2017) 77:6777–87. doi: 10.1158/0008-5472.CAN-17-1296
43. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut. (2018) 67:120–7. doi: 10.1136/gutjnl-2016-312580
44. Voigt AY, Emiola A, Johnson JS, Fleming ES, Nguyen H, Zhou W, et al. Skin microbiome variation with cancer progression in human cutaneous squamous cell carcinoma. J Invest Dermatol. (2022) 142:2773–82 e16. doi: 10.1016/j.jid.2022.03.017
45. Chiba A, Bawaneh A, Velazquez C, Clear KYJ, Wilson AS, Howard-McNatt M, et al. Neoadjuvant chemotherapy shifts breast tumor microbiota populations to regulate drug responsiveness and the development of metastasis. Mol Cancer Res. (2020) 18:130–9. doi: 10.1158/1541-7786.MCR-19-0451
46. Mekadim C, Skalnikova HK, Cizkova J, Cizkova V, Palanova A, Horak V, et al. Dysbiosis of skin microbiome and gut microbiome in melanoma progression. BMC Microbiol. (2022) 22. doi: 10.1186/s12866-022-02458-5
47. Chen Y, Yang S, Tavormina J, Tampe D, Zeisberg M, Wang H, et al. Oncogenic collagen I homotrimers from cancer cells bind to α3β1 integrin and impact tumor microbiome and immunity to promote pancreatic cancer. Cancer Cell. (2022) 40:818–34.e9. doi: 10.1016/j.ccell.2022.06.011
48. Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. (2022) 611:810–7. doi: 10.1038/s41586-022-05435-0
49. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappab, and up-regulating expression of microrna-21. Gastroenterology. (2017) 152:851–66 e24. doi: 10.1053/j.gastro.2016.11.018
50. Yang W, Zhao Y, Ge Q, Wang X, Jing Y, Zhao J, et al. Genetic mutation and tumor microbiota determine heterogenicity of tumor immune signature: evidence from gastric and colorectal synchronous cancers. Front Immunol. (2022) 13:947080. doi: 10.3389/fimmu.2022.947080
51. Dai D, Yang Y, Yu J, Dang T, Qin W, Teng L, et al. Interactions between gastric microbiota and metabolites in gastric cancer. Cell Death Dis. (2021) 12:1104. doi: 10.1038/s41419-021-04396-y
52. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–72 e26. doi: 10.1016/j.cell.2022.02.027
53. Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol. (2018) 9:951. doi: 10.3389/fmicb.2018.00951
54. Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. (2016) 82:5039–48. doi: 10.1128/AEM.01235-16
55. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
56. Wang M, Yu F, Li P. Intratumor microbiota in cancer pathogenesis and immunity: from mechanisms of action to therapeutic opportunities. Front Immunol. (2023) 14:1269054. doi: 10.3389/fimmu.2023.1269054
57. Huong PT, Lap NT, Anh DTM, Bac NX, Van Hien M, Ha PTT, et al. The essential role of glucose metabolism in chemoresistance of colorectal cancer-a mini review. Tạp chí Nghiên cứu Dược và Thông tin Thuốc. (2023) 14:63–71. doi: 10.59882/1859-364X/155
58. Huong PT, Nguyen LT, Nguyen XB, Lee SK, Bach DH. The role of platelets in the tumor-microenvironment and the drug resistance of cancer cells. Cancers (Basel). (2019) 11. doi: 10.3390/cancers11020240
59. Thanh Huong P, Gurshaney S, Thanh Binh N, Gia Pham A, Hoang Nguyen H, Thanh Nguyen X, et al. Emerging role of circulating tumor cells in gastric cancer. Cancers (Basel). (2020) 12. doi: 10.3390/cancers12030695
60. Paskeh MDA, Entezari M, Mirzaei S, Zabolian A, Saleki H, Naghdi MJ, et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol. (2022) 15:83. doi: 10.1186/s13045-022-01305-4
61. Zhao L, Cho WC, Nicolls MR. Colorectal cancer-associated microbiome patterns and signatures. Front Genet. (2021) 12:787176. doi: 10.3389/fgene.2021.787176
62. Gao Z, Guo B, Gao R, Qin H. Microbiota dysbiosis is associated with colorectal cancer. Front Microbiol. (2015) 6:20. doi: 10.3389/fmicb.2015.00020
63. Asseri AH, Bakhsh T, Ali S, Rather IA. The gut dysbiosis-cancer axis: illuminating novel insights and implications for clinical practice. Front Pharmacol. (2023) 14:1208044. doi: 10.3389/fphar.2023.1208044
64. Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. (2009) 15:1016–22. doi: 10.1038/nm.2015
65. Lee HJ, Yoon CH, Kim HJ, Ko JH, Ryu JS, Jo DH, et al. Ocular microbiota promotes pathological angiogenesis and inflammation in sterile injury-driven corneal neovascularization. Mucosal Immunol. (2022) 15:1350–62. doi: 10.1038/s41385-022-00555-2
66. Andriessen EMMA, Wilson AM, Mawambo G, Dejda A, Miloudi K, Sennlaub F, et al. Gut microbiota influences pathological angiogenesis in obesity-driven choroidal neovascularization. EMBO Mol Med. (2016) 8:1366–79-79. doi: 10.15252/emmm.201606531
67. Zhang R, Kang R, Tang D. Gut microbiome mediates ferroptosis resistance for colorectal cancer development. Cancer Res. (2024) 84(6):796–7. doi: 10.1158/0008-5472.Can-24-0275
68. Yu S, Wang S, Xiong B, Peng C. Gut microbiota: key facilitator in metastasis of colorectal cancer. Front Oncol. (2023) 13:1270991. doi: 10.3389/fonc.2023.1270991
69. Noble A, Pring ET, Durant L, Man R, Dilke SM, Hoyles L, et al. Altered immunity to microbiota, B cell activation and depleted Γδ/resident memory T cells in colorectal cancer. Cancer Immunol Immunother. (2022) 71:2619–29. doi: 10.1007/s00262-021-03135-8
70. Uribe-Herranz M, Rafail S, Beghi S, Gil-de-Gómez L, Verginadis I, Bittinger K, et al. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J Clin Invest. (2020) 130:466–79. doi: 10.1172/JCI124332
71. Ferrari V, Lo Cascio A, Melacarne A, Tanasković N, Mozzarelli AM, Tiraboschi L, et al. Sensitizing cancer cells to immune checkpoint inhibitors by microbiota-mediated upregulation of hla class I. Cancer Cell. (2023) 41:1717–30.e4. doi: 10.1016/j.ccell.2023.08.014
72. Mirlekar B, Pylayeva-Gupta Y. Il-12 family cytokines in cancer and immunotherapy. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13020167
73. Li Q, Li Y, Wang Y, Xu L, Guo Y, Wang Y, et al. Oral administration of bifidobacterium breve promotes antitumor efficacy via dendritic cells-derived interleukin 12. Oncoimmunology. (2021) 10:e1868122-1-15. doi: 10.1080/2162402x.2020.1868122
74. Pratap Singh R, Kumari N, Gupta S, Jaiswal R, Mehrotra D, Singh S, et al. Intratumoral microbiota changes with tumor stage and influences the immune signature of oral squamous cell carcinoma. Microbiol Spectr. (2023) 11:e04596–22. doi: 10.1128/spectrum.04596-22
75. Choi Y, Lichterman JN, Coughlin LA, Poulides N, Li W, Del Valle P, et al. Immune checkpoint blockade induces gut microbiota translocation that augments extraintestinal antitumor immunity. Sci Immunol. (2023) 8:eabo2003. doi: 10.1126/sciimmunol.abo2003
76. Chen C, Huang Z, Huang P, Li K, Zeng J, Wen Y, et al. Urogenital microbiota: potentially important determinant of pd-L1 expression in male patients with non-muscle invasive bladder cancer. BMC Microbiol. (2022) 22:7. doi: 10.1186/s12866-021-02407-8
77. Jang HJ, Choi JY, Kim K, Yong SH, Kim YW, Kim SY, et al. Relationship of the lung microbiome with pd-L1 expression and immunotherapy response in lung cancer. Respir Res. (2021) 22. doi: 10.1186/s12931-021-01919-1
78. Coutzac C, Jouniaux J-M, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic short chain fatty acids limit antitumor effect of ctla-4 blockade in hosts with cancer. Nat Commun. (2020) 11:2168. doi: 10.1038/s41467-020-16079-x
79. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti–pd-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
80. Chang JW-C, Hsieh J-J, Tsai C-Y, Chiu H-Y, Lin Y-F, Wu C-E, et al. Gut microbiota and clinical response to immune checkpoint inhibitor therapy in patients with advanced cancer. Biomed J. (2024), 100698. doi: 10.1016/j.bj.2024.100698
81. Shoji F, Yamaguchi M, Okamoto M, Takamori S, Yamazaki K, Okamoto T, et al. Gut microbiota diversity and specific composition during immunotherapy in responders with non-small cell lung cancer. Front Mol Biosci. (2022) 9:1040424. doi: 10.3389/fmolb.2022.1040424
82. Sarfaty M, Desjardins CA, Giardina P, Bardhan K, Pandian S, Halley K, et al. Assessment of cancer-specific microbiome signature of response to immune checkpoint inhibitors. J Clin Oncol. (2021) 39:2574. doi: 10.1200/JCO.2021.39.15_suppl.2574
83. Williams N, Wheeler CE, Husain M, Hoyd R, Meara AS, Lynn M, et al. De-correlating immune checkpoint inhibitor toxicity and response in melanoma via the microbiome. J Clin Oncol. (2023) 41:9569. doi: 10.1200/JCO.2023.41.16_suppl.9569
84. Kraehenbuehl L, Frame J, Taur Y, Chekalil S, Liu C, Goleva E, et al. Abstract 646: characteristics of skin microbiome in immune related cutaneous adverse events. Cancer Res. (2023) 83:646. doi: 10.1158/1538-7445.AM2023-646
85. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-pd-1 efficacy in metastatic melanoma patients. Science. (2018) 359:104–8. doi: 10.1126/science.aao3290
86. Pitt JM, Vetizou M, Waldschmitt N, Kroemer G, Chamaillard M, Boneca IG, et al. Fine-tuning cancer immunotherapy: optimizing the gut microbiome. Cancer Res. (2016) 76:4602–7. doi: 10.1158/0008-5472.CAN-16-0448
87. Chen YE, Bousbaine D, Veinbachs A, Atabakhsh K, Dimas A, Yu VK, et al. Engineered skin bacteria induce antitumor T cell responses against melanoma. Science. (2023) 380:203–10. doi: 10.1126/science.abp9563
88. Spencer CN, McQuade JL, Gopalakrishnan V, McCulloch JA, Vetizou M, Cogdill AP, et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science. (2021) 374:1632–40. doi: 10.1126/science.aaz7015
89. Zhou J, Zhang R, Guo P, Li P, Huang X, Wei Y, et al. Effects of intestinal microbiota on pharmacokinetics of cyclosporine a in rats. Front Microbiol. (2022) 13:1032290. doi: 10.3389/fmicb.2022.1032290
90. Yu H, Xu H, Yang X, Zhang Z, Hu J, Lu J, et al. Gut microbiota-based pharmacokinetic-pharmacodynamic study and molecular mechanism of specnuezhenide in the treatment of colorectal cancer targeting carboxylesterase. J Pharm Anal. (2023) 13:1024–40. doi: 10.1016/j.jpha.2023.06.012
91. Džidić-Krivić A, Kusturica J, Sher EK, Selak N, Osmančević N, Karahmet Farhat E, et al. Effects of intestinal flora on pharmacokinetics and pharmacodynamics of drugs. Drug Metab Rev. (2023) 55:126–39. doi: 10.1080/03602532.2023.2186313
92. Yi Y, Shen L, Shi W, Xia F, Zhang H, Wang Y, et al. Gut microbiome components predict response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer: A prospective, longitudinal study. Clin Cancer Res. (2021) 27:1329–40. doi: 10.1158/1078-0432.CCR-20-3445
93. Tintelnot J, Xu Y, Lesker TR, Schönlein M, Konczalla L, Giannou AD, et al. Microbiota-derived 3-iaa influences chemotherapy efficacy in pancreatic cancer. Nature. (2023) 615:168–74. doi: 10.1038/s41586-023-05728-y
94. Hermida LC, Gertz EM, Ruppin E. Abstract 2914: analyzing the tumor microbiome to predict cancer patient survival and drug response. Cancer Res. (2021) 81:2914. doi: 10.1158/1538-7445.AM2021-2914
95. Hakim H, Dallas R, Wolf J, Tang L, Schultz-Cherry S, Darling V, et al. Gut microbiome composition predicts infection risk during chemotherapy in children with acute lymphoblastic leukemia. Clin Infect Dis. (2018) 67:541–8. doi: 10.1093/cid/ciy153
96. González-Mercado VJ, Henderson WA, Sarkar A, Lim J, Saligan LN, Berk L, et al. Changes in gut microbiome associated with co-occurring symptoms development during chemo-radiation for rectal cancer: A proof of concept study. Biol Res Nurs. (2021) 23:31–41. doi: 10.1177/1099800420942830
97. Lin D, El Alam MB, Jaoude JA, Kouzy R, Phan JL, Elnaggar JH, et al. Microbiome dynamics during chemoradiation therapy for anal cancer. Int J Radiat Oncol Biol Phys. (2022) 113:974–84. doi: 10.1016/j.ijrobp.2022.04.037
98. Shiao SL, Kershaw KM, Limon JJ, You S, Yoon J, Ko EY, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. (2021) 39:1202–13.e6. doi: 10.1016/j.ccell.2021.07.002
99. Yang K, Hou Y, Zhang Y, Liang H, Sharma A, Zheng W, et al. Suppression of local type I interferon by gut microbiota–derived butyrate impairs antitumor effects of ionizing radiation. J Exp Med. (2021) 218:e20201915. doi: 10.1084/jem.20201915
100. Li Z, Zhang Y, Hong W, Wang B, Chen Y, Yang P, et al. Gut microbiota modulate radiotherapy-associated antitumor immune responses against hepatocellular carcinoma via sting signaling. Gut Microbes. (2022) 14:2119055. doi: 10.1080/19490976.2022.2119055
101. Dong J, Li Y, Xiao H, Zhang S, Wang B, Wang H, et al. Oral microbiota affects the efficacy and prognosis of radiotherapy for colorectal cancer in mouse models. Cell Rep. (2021) 37:109886. doi: 10.1016/j.celrep.2021.109886
102. Venkidesh BS, Shankar SR, Narasimhamurthy RK, Rao SBS, Mumbrekar KD. Radioprotective potential of probiotics against gastrointestinal and neuronal toxicity: A preclinical study. Clin Trans Oncol. (2023) 25:3165–73. doi: 10.1007/s12094-023-03184-8
103. Tsementzi D, Meador R, Eng T, Patel P, Shelton J, Arluck J, et al. Changes in the vaginal microbiome and associated toxicities following radiation therapy for gynecologic cancers. Front Cell Infect Microbiol. (2021) 11:680038. doi: 10.3389/fcimb.2021.680038
104. Dorff TB, Groshen S, Garcia A, Shah M, Tsao-Wei D, Pham H, et al. Safety and feasibility of fasting in combination with platinum-based chemotherapy. BMC Cancer. (2016) 16:1. doi: 10.1186/s12885-016-2370-6
105. Klement RJ, Sweeney RA, Klement RJ, Sweeney RA. Impact of a ketogenic diet intervention during radiotherapy on body composition: I. Initial clinical experience with six prospectively studied patients. BMC Res Notes. (2016) 9. doi: 10.1186/s13104-016-1959-9
106. Nguyen LT, Dang AK, Duong PT, Phan HBT, Pham CTT, Nguyen ATL, et al. Nutrition intervention is beneficial to the quality of life of patients with gastrointestinal cancer undergoing chemotherapy in Vietnam. Cancer Med. (2021) 10:1668–80. doi: 10.1002/cam4.3766
107. Ok JH, Lee H, Chung H-Y, Lee SH, Choi EJ, Kang CM, et al. The potential use of a ketogenic diet in pancreatobiliary cancer patients after pancreatectomy. Anticancer Res. (2018) 38:6519–27. doi: 10.21873/anticanres.13017
108. Cohen CW, Fontaine KR, Arend RC, Alvarez RD, III CAL, Huh WK, et al. A ketogenic diet reduces central obesity and serum insulin in women with ovarian or endometrial cancer. J Nutr. (2018) 148:1253–60. doi: 10.1093/jn/nxy119
109. Khodabakhshi A, Akbari ME, Mirzaei HR, Mehrad-Majd H, Kalamian M, Davoodi SH. Feasibility, safety, and beneficial effects of mct-based ketogenic diet for breast cancer treatment: A randomized controlled trial study. Nutr Cancer. (2020) 72:627–34. doi: 10.1080/01635581.2019.1650942
110. Kang CM, Yun B, Kim M, Song M, Y-h K, SH L, et al. Postoperative serum metabolites of patients on a low carbohydrate ketogenic diet after pancreatectomy for pancreatobiliary cancer: A nontargeted metabolomics pilot study. Sci Rep. (2019) 9. doi: 10.1038/s41598-019-53287-y
111. Freedland SJ, Howard L, Allen J, Smith J, Stout J, Aronson W, et al. A Lifestyle Intervention of Weight Loss Via a Low-Carbohydrate Diet Plus Walking to Reduce Metabolic Disturbances Caused by Androgen Deprivation Therapy among Prostate Cancer Patients: Carbohydrate and Prostate Study 1 (Caps1) randomized Controlled Trial. Prostate Cancer Prostatic Dis. (2019) 22:428–37. doi: 10.1038/s41391-019-0126-5
112. Richard C, Benlaifaoui M, Ouarzadi OE, Diop K, Desilets A, Malo J, et al. 679 high fiber diet modifies gut microbiome, propionate production, intratumor immune response and is associated with outcome in patients with lung cancer treated with immune checkpoint inhibitors. J ImmunoTher Cancer. (2020) 8. doi: 10.1136/jitc-2020-SITC2020.0679
113. Dizman N, Meza L, Bergerot P, Alcantara M, Dorff T, Lyou Y, et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: A randomized phase 1 trial. Nat Med. (2022) 28:704–12. doi: 10.1038/s41591-022-01694-6
114. Juan Z, Chen J, Ding B, Yongping L, Liu K, Wang L, et al. Probiotic supplement attenuates chemotherapy-related cognitive impairment in patients with breast cancer: A randomised, double-blind, and placebo-controlled trial. Eur J Cancer. (2022) 161:10–22. doi: 10.1016/j.ejca.2021.11.006
115. Mego M, Chovanec J, Vochyanova-Andrezalova I, Konkolovsky P, Mikulova M, Reckova M, et al. Prevention of irinotecan induced diarrhea by probiotics: A randomized double blind, placebo controlled pilot study. Complement Ther Med. (2015) 23:356–62. doi: 10.1016/j.ctim.2015.03.008
116. Demers M, Dagnault A, Desjardins J. A randomized double-blind controlled trial: impact of probiotics on diarrhea in patients treated with pelvic radiation. Clin Nutr. (2014) 33:761–7. doi: 10.1016/j.clnu.2013.10.015
117. Urbancsek H, Kazar T, Mezes I, Neumann K. Results of a double-blind, randomized study to evaluate the efficacy and safety of antibiophilus in patients with radiation-induced diarrhoea. Eur J Gastroenterol Hepatol. (2001) 13:391–6. doi: 10.1097/00042737-200104000-00015
118. Linn YH, Thu KK, Win NHH, Linn YH, Thu KK, Win NHH. Effect of probiotics for the prevention of acute radiation-induced diarrhoea among cervical cancer patients: A randomized double-blind placebo-controlled study. Probiotics Antimicrobial Proteins. (2018) 11:638–47. doi: 10.1007/s12602-018-9408-9
119. Golkhalkhali B, Rajandram R, Paliany AS, Ho GF, Ishak WZW, Johari CS, et al. Strain-specific probiotic (Microbial cell preparation) and omega-3 fatty acid in modulating quality of life and inflammatory markers in colorectal cancer patients: A randomized controlled trial. Asia-Pacific J Clin Oncol. (2018) 14:179–91. doi: 10.1111/ajco.12758
120. Liu J, Huang X-E. Efficacy of bifidobacterium tetragenous viable bacteria tablets for cancer patients with functional constipation. Asian Pacific J Cancer Prev. (2015) 15:10241–4. doi: 10.7314/APJCP.2014.15.23.10241
121. Holma R, Korpela R, Sairanen U, Blom M, Rautio M, Poussa T, et al. Colonic methane production modifies gastrointestinal toxicity associated with adjuvant 5-fluorouracil chemotherapy for colorectal cancer. J Clin Gastroenterol. (2013) 47:45–51. doi: 10.1097/MCG.0b013e3182680201
122. Österlund P, Ruotsalainen T, Korpela R, Saxelin M, Ollus A, Valta P, et al. Lactobacillus supplementation for diarrhoea related to chemotherapy of colorectal cancer: A randomised study. Br J Cancer. (2007) 97:1028–34. doi: 10.1038/sj.bjc.6603990
123. Sanctis VD, Belgioia L, Cante D, Porta MRL, Caspiani O, Guarnaccia R, et al. Lactobacillus brevis cd2 for prevention of oral mucositis in patients with head and neck tumors: A multicentric randomized study. Anticancer Res. (2019) 39:1935–42. doi: 10.21873/anticanres.13303
124. Jiang C, Wang H, Xia C, Dong Q, Chen E, Qiu Y, et al. A randomized, double-blind, placebo-controlled trial of probiotics to reduce the severity of oral mucositis induced by chemoradiotherapy for patients with nasopharyngeal carcinoma. Cancer. (2019) 125:1081–90. doi: 10.1002/cncr.31907
125. Sharma A, Rath GK, Chaudhary SP, Thakar A, Mohanti BK, Bahadur S. Lactobacillus brevis cd2 lozenges reduce radiation- and chemotherapy-induced mucositis in patients with head and neck cancer: A randomized double-blind placebo-controlled study. Eur J Cancer. (2012) 48:875–81. doi: 10.1016/j.ejca.2011.06.010
126. Doppalapudi R, Vundavalli S, Prabhat MPV. Effect of probiotic bacteria on oral candida in head- and neck-radiotherapy patients: A randomized clinical trial. J Cancer Res Ther. (2020) 16:470–7. doi: 10.4103/jcrt.JCRT_334_18
127. Masuno T, Kishimoto S, Ogura T, Honma T, Niitani H, Fukuoka M, et al. A comparative trial of lc9018 plus doxorubicin and doxorubicin alone for the treatment of Malignant pleural effusion secondary to lung cancer. Cancer. (1991) 68:1495–500. doi: 10.1002/1097-0142(19911001)68:7<1495::AID-CNCR2820680705>3.0.CO;2-6
128. Sugawara G, Nagino M, Nishio H, Ebata T, Takagi K, Asahara T, et al. Perioperative synbiotic treatment to prevent postoperative infectious complications in biliary cancer surgery: A randomized controlled trial. Ann Surg. (2006) 244:706–14. doi: 10.1097/01.sla.0000219039.20924.88
129. Liu Z, Qin H, Yang Z, Xia Y, Liu W, Yang J, et al. Randomised clinical trial: the effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery – a double-blind study. Alimentary Pharmacol Ther. (2011) 33:50–63. doi: 10.1111/j.1365-2036.2010.04492.x
130. Yang Y, Xia Y, Chen H, Hong L, Feng J, Yang J, et al. The effect of perioperative probiotics treatment for colorectal cancer: short-term outcomes of a randomized controlled trial. Oncotarget. (2016) 7:8432–40. doi: 10.18632/oncotarget.7045
131. Liu Z-H, Huang M-J, Zhang X-W, Wang L, Huang N-Q, Peng H, et al. The effects of perioperative probiotic treatment on serum zonulin concentration and subsequent postoperative infectious complications after colorectal cancer surgery: A double-center and double-blind randomized clinical trial. Am J Clin Nutr. (2013) 97:117–26. doi: 10.3945/ajcn.112.040949
132. Theodoropoulos GE, Memos NA, Peitsidou K, Karantanos T, Spyropoulos BG, Zografos G. Synbiotics and gastrointestinal function-related quality of life after elective colorectal cancer resection. Ann Gastroenterol. (2016) 29(1):56–62.
133. Flesch AT, Tonial ST, Contu PDC, Damin DC. Perioperative synbiotics administration decreases postoperative infections in patients with colorectal cancer: A randomized, double-blind clinical trial. Rev do Colégio Brasileiro Cirurgiões. (2017) 44:567–73. doi: 10.1590/0100-69912017006004
134. Sommacal HM, Bersch VP, Vitola SP, Osvaldt AB. Perioperative synbiotics decrease postoperative complications in periampullary neoplasms: A randomized, double-blind clinical trial. Nutr Cancer. (2015) 67:457–62. doi: 10.1080/01635581.2015.1004734
135. Xia C, Li W, Hong H, Wei H, Xin H, Chen T. A phase ii randomized clinical trial and mechanistic studies using improved probiotics to prevent oral mucositis induced by concurrent radiotherapy and chemotherapy in nasopharyngeal carcinoma. Front Immunol. (2021) 12:618150. doi: 10.3389/fimmu.2021.618150
136. Reyna-Figueroa J, Barrón-Calvillo E, García-Parra C, Galindo-Delgado P, Contreras-Ochoa C, Lagunas-Martínez A, et al. Probiotic supplementation decreases chemotherapy-induced gastrointestinal side effects in patients with acute leukemia. J Pediatr Hematol/Oncol. (2019) 41:468–72. doi: 10.1097/MPH.0000000000001497
137. Tian Y, Li M, Song W, Jiang R, Li YQ. Effects of probiotics on chemotherapy in patients with lung cancer. Oncol Lett. (2019) 17:2836–48. doi: 10.3892/ol.2019.9906
138. Chen H, Xia Y, Shi C, Liang Y, Yang Y, Qin H. Effects of perioperative probiotics administration on patients with colorectal cancer. Oncotarget. (2014) 7(7):8432–40. doi: 10.1002/central/CN-00993213
139. Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, et al. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. (2017) 4. doi: 10.1136/bmjgast-2017-000145
140. Delia P, Sansotta G, Donato V, Frosina P, Messina G, Renzis CD, et al. Use of probiotics for prevention of radiation-induced diarrhea. World J Gastroenterol. (2007) 13:912–5. doi: 10.3748/wjg.v13.i6.912
141. Spreafico A, Heirali AA, Araujo DV, Tan TJ, Oliva M, Schneeberger PHH, et al. First-in-class microbial ecosystem therapeutic 4 (Met4) in combination with immune checkpoint inhibitors in patients with advanced solid tumors (Met4-io trial). Ann Oncol. (2023) 34:520–30. doi: 10.1016/j.annonc.2023.02.011
142. Zaharuddin L, Mokhtar NM, Muhammad Nawawi KN, Raja Ali RA, Zaharuddin L, Mokhtar NM, et al. A randomized double-blind placebo-controlled trial of probiotics in post-surgical colorectal cancer. BMC Gastroenterol. (2019) 19. doi: 10.1186/s12876-019-1047-4
143. Rodríguez LHDL, Ortiz GG, Moragrega PR, Brizuela IEV, Gutiérrez JFS, Sánchez ARR, et al. Effect of symbiotic supplementation on fecal calprotectin levels and lactic acid bacteria, bifidobacteria, escherichia coli and salmonella DNA in patients with cervical cancer. Nutricion hospitalaria. (2018) 35:1394–400. doi: 10.20960/nh.1762
144. Mansouri-Tehrani HS, Rabbani Khorasgani M, Roayaei M. Effects of probiotics with or without honey on radiation-induced diarrhea. Int J Radiat Res. (2016) 14:206–13. doi: 10.18869/acadpub.ijrr.14.3.205
145. Itoh Y, Mizuno M, Ikeda M, Nakahara R, Kubota S, Ito J, et al. A randomized, double-blind pilot trial of hydrolyzed rice bran versus placebo for radioprotective effect on acute gastroenteritis secondary to chemoradiotherapy in patients with cervical cancer. Evid-Based Complement Altern Med. (2015) 2015:974390. doi: 10.1155/2015/974390
146. Sasidharan BK, Ramadass B, Viswanathan PN, Samuel P, Gowri M, Pugazhendhi S, et al. A phase 2 randomized controlled trial of oral resistant starch supplements in the prevention of acute radiation proctitis in patients treated for cervical cancer. J Cancer Res Ther. (2019) 15:1383–91. doi: 10.4103/jcrt.JCRT_152_19
147. Murphy J, Stacey D, Crook J, Thompson B, Panetta D. Testing control of radiation-induced diarrhea with a psyllium bulking agent: A pilot study. Can Oncol Nurs J. (2000) 10:96–100. doi: 10.5737/1181912x10396100
148. Garcia-Peris P, Velasco C, Hernandez M, Lozano MA, Paron L, de la Cuerda C, et al. Effect of inulin and fructo-oligosaccharide on the prevention of acute radiation enteritis in patients with gynecological cancer and impact on quality-of-life: A randomized, double-blind, placebo-controlled trial. Eur J Clin Nutr. (2015) 70:170–4. doi: 10.1038/ejcn.2015.192
149. Mercier BD, Tizpa E, Philip EJ, Feng Q, Huang Z, Thomas RM, et al. Dietary interventions in cancer treatment and response: A comprehensive review. Cancers (Basel). (2022) 14. doi: 10.3390/cancers14205149
150. Zitvogel L, Derosa L, Kroemer G. Modulation of cancer immunotherapy by dietary fibers and over-the-counter probiotics. Cell Metab. (2022) 34:350–2. doi: 10.1016/j.cmet.2022.02.004
151. Zhang J, Wu K, Shi C, Li G. Cancer immunotherapy: fecal microbiota transplantation brings light. Curr Treat Options Oncol. (2022) 23. doi: 10.1007/s11864-022-01027-2
152. Baunwall SMD, Terveer EM, Dahlerup JF, Erikstrup C, Arkkila P, Vehreschild MJ, et al. The use of faecal microbiota transplantation (Fmt) in europe: A europe-wide survey. Lancet Reg Health Eur. (2021) 9:100181. doi: 10.1016/j.lanepe.2021.100181
153. Zhao W, Lei J, Ke S, Chen Y, Xiao J, Tang Z, et al. Fecal microbiota transplantation plus tislelizumab and fruquintinib in refractory microsatellite stable metastatic colorectal cancer: an open-label, single-arm, phase ii trial (Renmin-215). eClinicalMedicine. (2023) 66. doi: 10.1016/j.eclinm.2023.102315
154. ClinicalTrials.gov. Fecal microbiota transplant and re-introduction of anti-pd-1 therapy (Pembrolizumab or nivolumab) for the treatment of metastatic colorectal cancer in anti-pd-1 non-responders. Available online at: https://clinicaltrials.gov/study/NCT04729322 (accessed March 19, 2024).
155. ClinicalTrials.gov. Fmt combined with immune checkpoint inhibitor and tki in the treatment of crc patients with advanced stage. Available online at: https://clinicaltrials.gov/study/NCT05279677 (accessed March 19, 2024).
156. Del Giglio A, Atui FC. Fecal transplantation in patient with metastatic melanoma refractory to immunotherapy: A case report. World J Clin cases. (2023) 11:5830–4. doi: 10.12998/wjcc.v11.i24.5830
157. Routy B, Lenehan JG, Miller WH, Jamal R, Messaoudene M, Daisley BA, et al. Fecal microbiota transplantation plus anti-pd-1 immunotherapy in advanced melanoma: A phase I trial. Nat Med. (2023) 29:2121–32. doi: 10.1038/s41591-023-02453-x
158. Borgers JSW, Burgers F, Terveer EM, van Leerdam M, Korse TM, Kessels R, et al. 120tip conversion of response to immune checkpoint inhibition by fecal microbiota transplantation in patients with metastatic melanoma: A randomized phase ib/iia trial. Immuno-Oncol Technol. (2022) 16. doi: 10.1016/j.iotech.2022.100224
159. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin JM, Morrison RM, et al. Fecal microbiota transplant overcomes resistance to anti-pd-1 therapy in melanoma patients. Science. (2021) 371:595–602. doi: 10.1126/science.abf3363
160. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science. (2021) 371:602–9. doi: 10.1126/science.abb5920
161. ClinicalTrials.gov. Assessing the tolerance and clinical benefit of fecal transplantation in patients with melanoma (Picasso). Available online at: https://clinicaltrials.gov/study/NCT04988841 (accessed March 19, 2024).
162. ClinicalTrials.gov. Fecal microbial transplantation non-small cell lung cancer and melanoma (Fmt-luminate). Available online at: https://clinicaltrials.gov/study/NCT04951583 (Accessed March 19 2024).
163. ClinicalTrials.gov. A phase ib trial to evaluate the safety and efficacy of fmt and nivolumab in subjects with metastatic or inoperable melanoma, msi-H, dmmr or nsclc. Available online at: https://clinicaltrials.gov/study/NCT04521075. (accessed March 19, 2024).
164. ClinicalTrials.gov. Microbiota transplant in advanced lung cancer treated with immunotherapy. Available online at: https://clinicaltrials.gov/study/NCT04924374 (accessed March 19, 2024).
165. ClinicalTrials.gov. Gut microbiota reconstruction for nsclc immunotherapy. Available online at: https://www.clinicaltrials.gov/study/NCT05008861 (accessed March 19, 2024).
166. Porcari S, Ciccarese C, Pinto F, Quaranta G, Giorgi SD, Rondinella D, et al. Fecal microbiota transplantation to improveefficacy of immune checkpoint inhibitors inrenal cell carcinoma (TACITO trial). J Clin Oncol. (2022). doi: 10.1200/JCO.2022.40.6_suppl.TPS407
167. Fernandes R, Parvathy SN, Ernst DS, Haeryfar M, Burton J, Silverman M, et al. Preventing adverse events in patients with renal cell carcinoma treated with doublet immunotherapy using fecal microbiota transplantation (Fmt): initial results from perform a phase I study. Nat Med. (2022) 40(16_suppl):4553–. doi: 10.1200/JCO.2022.40.16_suppl.4553
168. Park SR, Kim G, Kim Y, Cho B, Kim S-Y, Do E-J, et al. Fecal microbiota transplantation combined with anti-pd-1 inhibitor for unresectable or metastatic solid cancers refractory to anti-pd-1 inhibitor. Cell Host & Microbe. (2023) 41(16_suppl):105–. doi: 10.1200/JCO.2023.41.16_suppl.105
169. ClinicalTrials.gov. Fmt with nivolumab in patients with advanced solid cancers who have progressed during anti-pd-(L)1 therapy. Available online at: https://clinicaltrials.gov/study/NCT05533983 (accessed March 19, 2024).
170. Peng Z, Zhang X, Xie T, Cheng S, Han Z, Wang S, et al. Efficacy of fecal microbiota transplantation in patients with anti-pd-1–resistant/refractory gastrointestinal cancers. Gut Microbes. (2023) 41(4_suppl):389–. doi: 10.1200/JCO.2023.41.4_suppl.389
171. Fasanello MK, Robillard KT, Boland PM, Bain AJ, Kanehira K. Use of fecal microbial transplantation for immune checkpoint inhibitor colitis. ACG Case Rep J. (2020) 7:e00360. doi: 10.14309/crj.0000000000000360
172. Wang Y, Varatharajalu K, Shatila M, Campbell MT, Msaouel P, Kovitz CA. First-line treatment of fecal microbiota transplantation for immune-mediated colitis. J Clin Oncol. (2023) 41(16_suppl):2510–. doi: 10.1200/JCO.2023.41.16_suppl.2510
173. ClinicalTrials.gov. Fecal microbiota transplant and pembrolizumab for men with metastatic castration resistant prostate cancer. Available online at: https://clinicaltrials.gov/study/NCT04116775 (accessed March 19, 2024).
174. ClinicalTrials.gov. A single dose fmt infusion as an adjunct to keytruda for metastatic mesothelioma. Available online at: https://www.clinicaltrials.gov/study/NCT04056026 (accessed March 19, 2024).
175. Ali H, Ma W, Khurana S, Jiang Z-D, DuPont HL, Okhuysen P, et al. Safety and efficacy of fecal microbiota transplantation (Fmt) in the management of recurrent clostridioides difficile infection (Rcdi) in cancer patients. J Cancer. (2020) 38:e24048–e. doi: 10.1200/JCO.2020.38.15_suppl.e24048.
176. Amin R, Thomas AS, Wang Y. Fecal transplant sustained colitis remission on immunotherapy resumption. Ann Intern Med Clin Cases. (2022) 1:e220490. doi: 10.7326/aimcc.2022.0490.
177. Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. (2018) 24:1804–8. doi: 10.1038/s41591-018-0238-9
178. ClinicalTrials.gov. Fecal microbiota transplantation in patients with Malignancies not responding to cancer immunotherapy. Available online at: https://clinicaltrials.gov/study/NCT05273255 (accessed March 19, 2024).
179. Youngster I, Eshel A, Geva M, Danylesko I, Henig I, Zuckerman T, et al. Fecal microbiota transplantation in capsules for the treatment of steroid refractory and steroid dependent acute graft vs. Host disease: A pilot study. Bone marrow Transplant. (2024) 59:409–16. doi: 10.1038/s41409-024-02198-2
180. Malard F, Loschi M, Huynh A, Cluzeau T, Guenounou S, Legrand F, et al. Pooled allogeneic faecal microbiota maat013 for steroid-resistant gastrointestinal acute graft-versus-host disease: A single-arm, multicentre phase 2 trial. EClinicalMedicine. (2023) 62:102111. doi: 10.1016/j.eclinm.2023.102111
181. Rashidi A, Ebadi M, Rehman TU, Elhusseini H, Kazadi D, Halaweish H, et al. Randomized double-blind phase ii trial of fecal microbiota transplantation versus placebo in allogeneic hematopoietic cell transplantation and aml. J Clin Oncol: Off J Am Soc Clin Oncol. (2023) 41:5306–19. doi: 10.1200/jco.22.02366
182. Aurore D, Aurélie R, Alexandrine C, Aurélie C, Mathieu W, Bruno P, et al. Faecal microbiota transplantation to prevent complications after allogeneic stem cell transplantation for haematological Malignancies: A study protocol for a randomised controlled phase-ii trial (the fmt-allo study). BMJ Open. (2023) 13:e068480. doi: 10.1136/bmjopen-2022-068480
183. Liu Y, Zhao Y, Qi J, Ma X, Qi X, Wu D, et al. Fecal microbiota transplantation combined with ruxolitinib as a salvage treatment for intestinal steroid-refractory acute gvhd. Exp Hematol Oncol. (2022) 11:96. doi: 10.1186/s40164-022-00350-6
184. Zhao Y, Li X, Zhou Y, Gao J, Jiao Y, Zhu B, et al. Safety and efficacy of fecal microbiota transplantation for grade iv steroid refractory gi-gvhd patients: interim results from fmt2017002 trial. Front Immunol. (2021) 12:678476. doi: 10.3389/fimmu.2021.678476
185. Goeser F, Sifft B, Stein-Thoeringer C, Farowski F, Strassburg CP, Brossart P, et al. Fecal microbiota transfer for refractory intestinal graft-versus-host disease - experience from two german tertiary centers. Eur J Haematol. (2021) 107:229–45. doi: 10.1111/ejh.13642
186. van Lier YF, Davids M, Haverkate NJE, de Groot PF, Donker ML, Meijer E, et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci Trans Med. (2020) 12. doi: 10.1126/scitranslmed.aaz8926
187. Qi X, Li X, Zhao Y, Wu X, Chen F, Ma X, et al. Treating steroid refractory intestinal acute graft-vs.-host disease with fecal microbiota transplantation: A pilot study. Front Immunol. (2018) 9:2195. doi: 10.3389/fimmu.2018.02195
188. Spindelboeck W, Schulz E, Uhl B, Kashofer K, Aigelsreiter A, Zinke-Cerwenka W, et al. Repeated fecal microbiota transplantations attenuate diarrhea and lead to sustained changes in the fecal microbiota in acute, refractory gastrointestinal graft-versus-host-disease. Haematologica. (2017) 102:e210–e3. doi: 10.3324/haematol.2016.154351
189. Kakihana K, Fujioka Y, Suda W, Najima Y, Kuwata G, Sasajima S, et al. Fecal microbiota transplantation for patients with steroid-resistant acute graft-versus-host disease of the gut. Blood. (2016) 128:2083–8. doi: 10.1182/blood-2016-05-717652
190. ClinicalTrials.gov. Fecal microbiota transplantation for steroid resistant and steroid dependent gut acute graft versus host disease. Available online at: https://clinicaltrials.gov/study/NCT03214289 (accessed March 19, 2024).
191. ClinicalTrials.gov. Fecal microbiota transplantation for treatment of refractory graft versus host disease-a pilot study. Available online at: https://clinicaltrials.gov/study/NCT03549676 (accessed March 19, 2024).
192. ClinicalTrials.gov. Fecal Microbiota Transplantation (Fmt) in Recipients after Allogeneic Hematopoietic Cell Transplantation (Hct). Available online at: https://www.clinicaltrials.gov/study/NCT03720392 (accessed March 19, 2024).
193. Malard F, Vekhoff A, Lapusan S, Isnard F, D'Incan-Corda E, Rey J, et al. Gut microbiota diversity after autologous fecal microbiota transfer in acute myeloid leukemia patients. Nat Commun. (2021) 12:3084. doi: 10.1038/s41467-021-23376-6
194. Hefazi M, Patnaik MM, Hogan WJ, Litzow MR, Pardi DS, Khanna S. Safety and efficacy of fecal microbiota transplant for recurrent clostridium difficile infection in patients with cancer treated with cytotoxic chemotherapy: A single-institution retrospective case series. Mayo Clinic Proc. (2017) 92:1617–24. doi: 10.1016/j.mayocp.2017.08.016
195. Yu H, Li XX, Han X, Chen BX, Zhang XH, Gao S, et al. Fecal microbiota transplantation inhibits colorectal cancer progression: reversing intestinal microbial dysbiosis to enhance anti-cancer immune responses. Front Microbiol. (2023) 14:1126808. doi: 10.3389/fmicb.2023.1126808
196. Patel P, Poudel A, Kafle S, Thapa Magar M, Cancarevic I. Influence of microbiome and antibiotics on the efficacy of immune checkpoint inhibitors. Cureus. (2021) 13:e16829. doi: 10.7759/cureus.16829
197. Grant CV, Jordan K, Seng MM, Pyter LM. Antibiotic treatment inhibits paclitaxel chemotherapy-induced activity deficits in female mice. PloS One. (2023) 18:e0284365. doi: 10.1371/journal.pone.0284365
198. Pinato DJ, Li X, Mishra-Kalyani P, D'Alessio A, Fulgenzi CAM, Scheiner B, et al. Association between antibiotics and adverse oncological outcomes in patients receiving targeted or immune-based therapy for hepatocellular carcinoma. JHEP Rep. (2023) 5:100747. doi: 10.1016/j.jhepr.2023.100747
199. Kim CG, Koh JY, Shin SJ, Shin JH, Hong M, Chung HC, et al. Prior antibiotic administration disrupts anti-pd-1 responses in advanced gastric cancer by altering the gut microbiome and systemic immune response. Cell Rep Med. (2023) 4:101251. doi: 10.1016/j.xcrm.2023.101251
200. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. (2018) 29:1437–44. doi: 10.1093/annonc/mdy103
201. Thompson J, Szabo A, Arce-Lara C, Menon S. P1.07-008 microbiome & Immunotherapy: antibiotic use is associated with inferior survival for lung cancer patients receiving pd-1 inhibitors. J Thorac Oncol. (2017) 12:S1998. doi: 10.1016/j.jtho.2017.09.926
202. Eng L, Sutradhar R, Niu Y, Liu N, Liu Y, Kaliwal Y, et al. Impact of antibiotic exposure before immune checkpoint inhibitor treatment on overall survival in older adults with cancer: A population-based study. J Clin Oncol. (2023) 41:3122–34. doi: 10.1200/JCO.22.00074
203. Vihinen H, Jokinen A, Laajala TD, Wahid N, Peltola L, Kettunen T, et al. Antibiotic treatment is an independent poor risk factor in nsclc but not in melanoma patients who had received anti-pd-1/L1 monotherapy. Clin Lung Cancer. (2023) 24:295–304. doi: 10.1016/j.cllc.2023.01.004
204. Lu SSM, Mohammed Z, Häggström C, Myte R, Lindquist E, Gylfe Å, et al. Antibiotics use and subsequent risk of colorectal cancer: A swedish nationwide population-based study. JNCI: J Natl Cancer Institute. (2022) 114:38–46. doi: 10.1093/jnci/djab125
205. Wardill HR, van der Aa SAR, da Silva Ferreira AR, Havinga R, Tissing WJE, Harmsen HJM. Antibiotic-induced disruption of the microbiome exacerbates chemotherapy-induced diarrhoea and can be mitigated with autologous faecal microbiota transplantation. Eur J Cancer. (2021) 153:27–39. doi: 10.1016/j.ejca.2021.05.015
206. Fulop DJ, Zylberberg HM, Wu YL, Aronson A, Labiner AJ, Wisnivesky J, et al. Association of antibiotic receipt with survival among patients with metastatic pancreatic ductal adenocarcinoma receiving chemotherapy. JAMA Network Open. (2023) 6. doi: 10.1001/jamanetworkopen.2023.4254
207. Chambers LM, Kuznicki M, Yao M, Chichura A, Gruner M, Reizes O, et al. Impact of antibiotic treatment during platinum chemotherapy on survival and recurrence in women with advanced epithelial ovarian cancer. Gynecol Oncol. (2020) 159:699–705. doi: 10.1016/j.ygyno.2020.09.010
208. Hamada K, Yoshimura K, Hirasawa Y, Hosonuma M, Murayama M, Narikawa Y, et al. Antibiotic usage reduced overall survival by over 70% in non-small cell lung cancer patients on anti-pd-1 immunotherapy. Anticancer Res. (2021) 41:4985–93. doi: 10.21873/anticanres.15312
209. Elkrief A, Derosa L, Kroemer G, Zitvogel L, Routy B. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: A new independent prognostic factor? Ann Oncol. (2019) 30:1572–9. doi: 10.1093/annonc/mdz206
210. Fong W, Li Q, Yu J. Gut microbiota modulation: A novel strategy for prevention and treatment of colorectal cancer. Oncogene. (2020) 39:4925–43. doi: 10.1038/s41388-020-1341-1
211. Huynh M, Crane MJ, Jamieson AM. The lung, the niche, and the microbe: exploring the lung microbiome in cancer and immunity. Front Immunol. (2022) 13:1094110. doi: 10.3389/fimmu.2022.1094110
212. Zhang W, Yang F, Mao S, Wang R, Chen H, Ran Y, et al. Bladder cancer-associated microbiota: recent advances and future perspectives. Heliyon. (2023) 9:e13012. doi: 10.1016/j.heliyon.2023.e13012
Keywords: cancer therapy, microbiota, microbiome intervention, pharmacomicrobiome, precision medicine
Citation: Le Ngoc K, Pham TTH, Nguyen TK and Huong PT (2024) Pharmacomicrobiomics in precision cancer therapy: bench to bedside. Front. Immunol. 15:1428420. doi: 10.3389/fimmu.2024.1428420
Received: 08 May 2024; Accepted: 19 August 2024;
Published: 09 September 2024.
Edited by:
Matija Rijavec, University Clinic of Pulmonary and Allergic Diseases Golnik, SloveniaReviewed by:
Urska Janzic, University Clinic of Pulmonary and Allergic Diseases Golnik, SloveniaYuting Ke, Massachusetts Institute of Technology, United States
Copyright © 2024 Le Ngoc, Pham, Nguyen and Huong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Phung Thanh Huong, aHVvbmdwdEBodXAuZWR1LnZu
†These authors have contributed equally to this work and share first authorship
 Khanh Le Ngoc
Khanh Le Ngoc Tran Thu Ha Pham
Tran Thu Ha Pham Tiep Khac Nguyen
Tiep Khac Nguyen Phung Thanh Huong
Phung Thanh Huong