- 1Department of Ophthalmology, Shanghai Pudong New Area Gongli Hospital, Shanghai, China
- 2Department of Ophthalmology, Changzheng Hospital, Second Affiliated Hospital of Naval Medical University, Shanghai, China
- 3Department of Anesthesiology, Changzheng Hospital, Second Affiliated Hospital of Naval Medical University, Shanghai, China
Uveal melanoma (UM) is a highly aggressive and fatal tumor in the eye, and due the special biology of UM, immunotherapy showed little effect in UM patients. To improve the efficacy of immunotherapy for UM patients is of great clinical importance. Single-cell RNA sequencing(scRNA-seq) provides a critical perspective for deciphering the complexity of intratumor heterogeneity and tumor microenvironment(TME). Combing the bioinformatics analysis, scRNA-seq could help to find prognosis-related molecular indicators, develop new therapeutic targets especially for immunotherapy, and finally to guide the clinical treatment options.
Introduction
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults, accounting for 3%-5% of all melanoma (1). UM originates from melanocytes of the iris (3–5%), ciliary body (5–8%), or choroid (approximately 85%) (2). The average incidence of UM in the world is 0.0001%~0.0009%, with obvious regional and ethnic differences, with the highest incidence in white people, followed by yellow people, and less common in black people (3). The incidence is slightly higher in males than in females. The advances of diagnosis and treatment have improved the local control rate of UM, but the overall survival(OS) remains unchanged (4). About 50% of patients with UM eventually develop metastases, most involving the liver (5). And the median OS for metastatic UM(mUM) patients is shorter than 12 months (6). Improving the survival of UM patient is the main clinical goal at present.
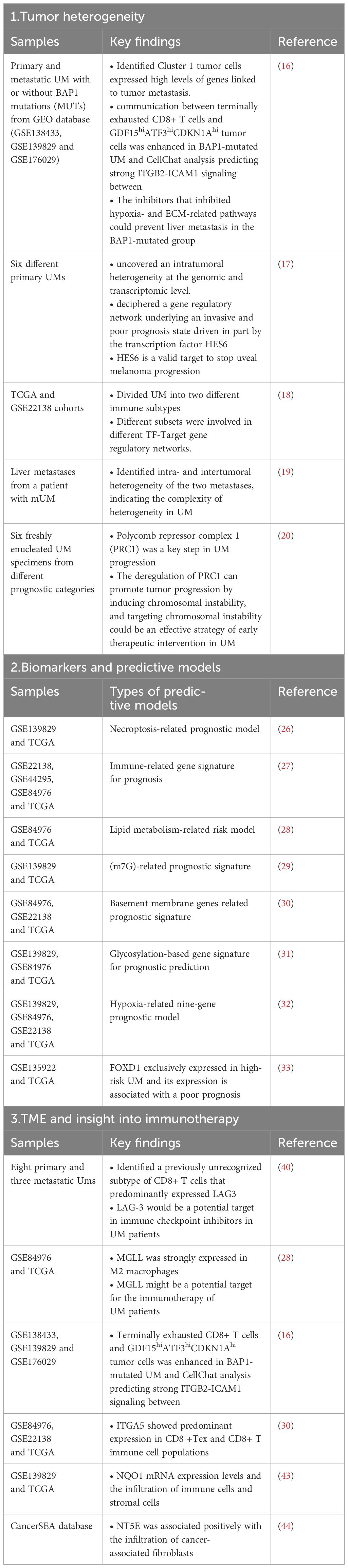
Single-cell RNA sequencing(scRNA-seq) is a cutting-edge technology that could provide in-depth information of tumor heterogeneity and tumor microenvironment(TME) (7, 8) and has yielded remarkable results towards the molecular biology of UM. Combined with bioinformatic analysis, scRNA-seq could provide detailed information regarding the occurrence, progression and metastasis of UM and help to develop new therapeutic targets and prognostic models (9). Immunotherapy has been widely used in anti-tumor therapy, but UM shows little response to immunotherapy as it has the lowest tumor mutation burden in all types of cancer (10, 11). Moreover, eyes are immune privileged and could protects the eye tissue from the immune system attack (12). As is known that the efficacy of immunotherapy is closely related the TME (13), using scRNA-seq to analyze the features of TME in UM patients could give insight to improve the efficiency of immunotherapy for UM patients. In this review, we are going to summarize the recent progresses on the application of scRNA-seq in UM and explore their insight to immunotherapy (Table 1).
Tumor heterogeneity
UM a highly heterogeneous and complex disease, and intertumoral heterogeneity has been observed with markedly different genetic backgrounds, histopathological features, and clinical behaviors. Apart from intertumoral heterogeneity, intratumoral heterogeneity (ITH) has only been recognized within the past decades with the advances in sequencing technology. ITH originates from the genetic heterogeneity of cancer cells and could lead to distinct populations present in the tumor (14, 15). Each clone has unique functional properties, such as the ability to form metastases or respond to specific therapies.
BAP1 inactivating mutations was a risk factor for metastasis in UM patients. To understand the mechanisms of BAP1 deficiency driving UM metastasis, Jiaoduan Li et al. analyzed scRNA-seq data that included primary and metastatic UM with or without BAP1 mutations (MUTs). The malignant cells were divided into five clusters, and malignant cells expressing high levels of GDF15, ATF3, and CDKN1A were linked to tumor metastasis (16). Charlotte Pandiani et al. use single-cell RNA sequencing of six different primary uveal melanomas and uncovered distinct intratumoral heterogeneity at the genomic and transcriptomic level. Further analysis identified a gene regulatory network underlying an invasive and poor prognosis state driven by the transcription factor HES6 (17). By integration of bulk RNA-seq and scRNA-seq, Guohong Gao et al. had divided UM patients into two different immune subtypes and each subtypes a had its unique characteristics in prognosis, immune-related molecules, immune score, and immune cell infiltration (18). Weitao Lin et al. utilized scRNA-seq of two metastases from a mUM patients who had received immunotherapy and the results demonstrated that two metastases were largely infiltrated by noncancerous cells with significant variability in cell types. Further single-nucleotide polymorphism(SNP) and copy-number variations (CNVs) uncovered both intra- and intertumoral heterogeneity of the two metastases, indicating the complexity of heterogeneity in UM (19). Mathieu F. Bakhoum et al. explored the relationship between chromosomal instability, epigenetics, tumor progression and metastasis by integrating information by scRNA-seq, and the results revealed that alterations in a series of molecules driven by the deletion of Polycomb repressor complex 1(PRC1) was a key step in UM progression. The deregulation of PRC1 can promote tumor progression by inducing chromosomal instability, and targeting chromosomal instability could be an effective strategy of early therapeutic intervention in UM (20).
These results above verified the existence of ITH within the same tumor, and each subset of heterogeneity had its unique characteristics. ScRNA-seq allows the researchers to evaluate the genetic heterogeneity at single-cell resolution and helps to reveal the complex biological diversity within the tumor and to make the individualized treatment plan.
Biomarkers and predictive models
Based on the 8th edition AJCC cancer staging, UM is classified by tumor size (diameter and thickness), anatomical extent (ciliary involvement, and extrascleral extension) (21). With the advances of genetics, the deletion of chromosome 3 and the increase of chromosome 8 have been found to be associated with poor prognosis (22). Therefore, Dogrusöz et al. and Bagger et al. added chromosomal information to the AJCC staging system to further improve the prognostic prediction (23). In 2004, Onken et al. used unsupervised clustering in primary UM transcriptome analysis and uncovered two GEP-based classifications, one with a better prognosis and the other with a poor prognosis (24). With the application of next-generation sequencing technology, the driver genes were discovered in UM development. The primary driver genes are GNAQ, GNA11, and secondary driver genes are BAP1, SF3B1, EIF1MX (25). About 90% of UM patients carry GNAQ and GNA11 gene mutations, which activate the tumorigenesis. BAP1 mutations have the worst clinical prognosis, followed by SF3B1 and EIF1MX (25). ScRNA-seq could provide in-depth information about mechanism of tumorigenesis and tumor progression and could avoid the ITH with the tumor brought by the bulk RNA-seq. Biomarkers and prediction model based on the scRNA-seq data might be more accurate than those based on the bulk RNA-seq.
Jiaheng Xie et al. used the online scRNA-seq data of UM, and identified a hub of necroptosis-related genes. By COX regression and Lasso regression, the authors constructed a necroptosis-related prognostic model that could distinguish different survival time (26). Wanpeng Wang et al. identified the immune infiltration pattern of UM and constructed an immune-related gene prognostic signature which had strong predictive ability for UM patients (27). Similarly, Yao Tan et al. investigated the expression patterns of lipid metabolism in UM patients and established a risk model based on the genes involved with lipid metabolism which could accurately predict survival in patients with UM (28). Moreover, researchers had constructed a m7G-related prognostic signature by integrating TCGA and GEO database and suggested PAG1 as biomarker for diagnosis and treatment of UM (29). Yunyue Li et al. also developed a prognostic risk model based on basement membrane protein-related genes (30), and glycosylation-based gene signature and hypoxia-related gene signature were described as well by integrating single-cell analysis and machine learning (31, 32). Re-analyzing publicly available single cell RNA sequencing, researchers found that FOXD1 exclusively expressed in high-risk UM and its expression is associated with a poor prognosis, suggesting FOXD1 as a new biomarker for the diagnosis of UM (33).
Base on the scRNA-seq data, especially those online public data, researchers have developed a series of prediction model and biomarkers with the intention to accurately predict the prognosis of UM patients. However, the clinical validation of those models and biomarkers is still uncertain. Apart from developing new biomarkers and models, the validation seems more practical.
TME and insight into immunotherapy
Although UM originated from melanocytes, the biological and clinical features of UM are quite different from cutaneous melanoma(CM). CM harbors higher tumor mutation burdens(TMB) and shows good response to immune checkpoint inhibitos(ICIs) (34). High TMB could lead to the continuous production of neoantigens which makes the tumors highly immunogenic and sensitive to ICIs (35). However, UM has the lowest TMB among all types of tumors, which makes it irresponsible to ICIs (36). Besides the TMB, the response to ICIs is closely related to TME. TME is mainly composed of tumor cells and surrounding tumor-associated immune cells, such as tumor-infiltrating lymphocytes (TILs), natural killer (NK) cells, tumor-associated non-immune cells (fibroblasts, lipid cells, etc.), and extracellular matrix(ECM), which plays a key role in the occurrence, progression, and metastasis of tumor. As previously described, constant interactions between tumor cells and their surrounding microenvironment was closely related with treatment response and prognosis of malignancies (37, 38). ScRNA-seq is a powerful tool to explore the complex TME, and exploring the mechanism of the interaction between TME and tumor cells may lead to a breakthrough in immunotherapy for UM.
Michael A Durante et al. interrogated the tumor microenvironment using scRNA-seq of 59,915 tumor and non-neoplastic cells from 8 primary and 3 metastatic samples. The tumor-infiltrating immune cells comprised a previously unrecognized subtype of CD8+ T cells that predominantly expressed the checkpoint marker LAG3, rather than PD-1 and CTLA-4, indicating that LAG-3 would be a potential target in immune checkpoint inhibitors in UM patients (39). LAG-3, like PD-1 and CTLA-4, is another ICI protein. LAG-3 is not expressed on naïve T cells, but can be induced on CD4+ and CD8+ T cells when stimulated by antigens to inhibit T cell function (40). It has been reported that anti-PD-1 and anti-CTLA-4 cannot improve the prognosis of mUM patients (41), and anti-LAG-3 might be a new direction to the immunotherapy of UM patients. Yao Tan et al. explored the expression patterns of lipid metabolism in 80 UM patients from the TCGA database and integrated the results with single-cell sequencing analysis on UM patients from the GEO data to characterize the lipid metabolism in TME. And the results showed that monoacylglycerol lipase (MGLL) was strongly expressed in macrophages, specifically M2 macrophages, which might function in the M2 polarization and M2 macrophage activation, suggesting that MGLL might be a potential target for the immunotherapy of UM patients (28). Macrophages that recruited in TME are mostly M2 phenotype, which plays a critical role in promoting tumor progression (42). Strategies targeting tumor-associated macrophages(TAMs) are reducing the number of TAMs or altering their functionality within the TME. And by uncovering the specific markers on TAMs, targeted antibodies towards those markers could be applied to either eliminate or disfunction those tumor -promoting TAMs. In BAP1-mutated UM, researchers found that ITGB2-ICAM1 signaling was enhanced between terminally exhausted CD8+ T cells and GDF15hiATF3hiCDKN1Ahi tumor cells, and inhibiting either ICAM1 or ITGB2 could prevent liver metastasis in the BAP1-mutated patients (16). In the study of Yunyue Li et al, ADAMTS10 and ITGA5 were expressed in various immune cell, and ITGA5 showed predominant expression in CD8 +Tex and CD8+ T immune cell populations. Further studies should be carried out to study the function of those basement membrane protein-related genes in TME and immunotherapy of UM (30). Liping Shen et al. conducted a pan cancer research of NQO1 and found that NQO1 was significantly upregulated in most cancer types. The scRNA-seq analysis revealed a potential relationship between the NQO1 mRNA expression levels and the infiltration of immune cells and stromal cells, and inhibition of NQO1 might be a promising strategy for cancer immunotherapy (43). Likewise, Xinmiao Xue et al. identified NT5E as a novel prognostic biomarker in a pan-cancer analysis. The overexpression of NT5E was related to worse overall survival in UM patients and NT5E was associated positively with the infiltration of cancer-associated fibroblasts (CAFs) through epithelial-mesenchymal transition(EMT) which provided the insight for immunotherapy by targeting EMT (44).
The existing anti-PD-1/PD-L1 and anti-CTLA-1 antibodies showed little response in UM patients. Although studies have demonstrated that LAG-3 might be a promising ICIs in UM, little is reported about the clinical application anti-LAG-3 antibody in UM patients. With the improvements of scRNA-seq, detailed analysis of TME could be carried out and more ICIs and other targeted therapies would be developed to improve the immunotherapy of UM.
Discussion
ScRNA-seq is promising technology that could provide thorough information of a single cell and could also address the complex TME. The adoption of scRNA-seq has provided us the deeper understanding of the gene expression, and cell type, sub-species type, state and development trajectory within the tumor. But there are still some limitations about scRNA-seq. As to apply the scRNA-seq, tissues must to be digested into single cells. During the digestion cell integrity and cell viability are often compromised and the spatial location of tumor cells is also lost which is a very important information in TME. The recently developed spatial transcriptomics(ST) could provide the gene expression profile within intact tissue (45), which would be perfect supplement to scRNA-seq. And by integrating ST and scRNA-seq should be more valuable to study the molecular mechanisms and TME. Another limitation is the high cost of scRNA-seq, which have greatly limited its application across the world. Reduced sequencing costs would facilitate the widespread use of scRNA-seq in tumor research.
As the most common primary intraocular malignancy in adults, UM patients can seldom benefit from immunotherapy due to the special immune characteristics of UM- low tumor mutation burden and low immune infiltration. Although there were no major breakthroughs in the immunotherapy in UM, the immune system still plays an important role in the development of UM. With the adoption of scRNA-seq in tumor research, hopes have been lit up upon the immunotherapy in UM patients. Traditional bulk RNA-seq is difficult to capture the heterogeneity within the tumor, but scRNA-seq could provide detailed information about the interaction between tumor cells and immune cells in TME, which would be the potential target for immunotherapy (46). New immune check point proteins and molecular mechanisms have been uncovered by illustrating the comprehensive picture of TME in UM by scRNA-seq. And clinical validations should be carried out to confirm the findings obtained in scRNA-seq. However, the overall results are still insufficient and a quite number of studies used the online public data. More detailed studies should be encouraged by scRNA-seq. With the development of scRNA-seq, more in-depth mechanisms of UM have been uncovered and by designing the specific drugs targeting the mechanisms, the treatment would be more effective and thus prolong the survival of UM patients.
Author contributions
ST: Conceptualization, Writing – original draft. YZ: Formal analysis, Investigation, Writing – review & editing. SH: Investigation, Writing – review & editing. TZ: Investigation, Writing – review & editing. XH: Conceptualization, Funding acquisition, Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funding by Shanghai Pudong New Area Health System Discipline leading training program (PWRd2023-13)
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Radivoyevitch T, Zabor EC, Singh AD. Uveal melanoma: Long-term survival. PloS One. (2021) 16:e0250939. doi: 10.1371/journal.pone.0250939
2. Shields CL, Furuta M, Thangappan A, Nagori S, Mashayekhi A, Lally DR, et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch Ophthalmol. (2009) 127:989–98. doi: 10.1001/archophthalmol.2009.208
3. Hu DN, Yu GP, McCormick SA, Schneider S, Finger PT. Population-based incidence of uveal melanoma in various races and ethnic groups. Am J Ophthalmol. (2005) 140:612–7. doi: 10.1016/j.ajo.2005.05.034
4. Aronow ME, Topham AK, Singh AD. Uveal melanoma: 5-year update on incidence, treatment, and survival (SEER 1973–2013). Ocul Oncol Pathol. (2018) 4:145–51. doi: 10.1159/000480640
5. Carvajal RD, Sacco JJ, Jager MJ, Eschelman DJ, Olofsson Bagge R, Harbour JW, et al. Advances in the clinical management of uveal melanoma. Nat Rev Clin Oncol. (2023) 20:99–115. doi: 10.1038/s41571-022-00714-1
6. Khoja L, Atenafu EG, Suciu S, Leyvraz S, Sato T, Marshall E, et al. Meta-analysis in metastatic uveal melanoma to determine progression free and overall survival benchmarks: an international rare cancers initiative (IRCI) ocular melanoma study. Ann Oncol. (2019) 30:1370–80. doi: 10.1093/annonc/mdz176
7. Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol. (2020) 17:807–21. doi: 10.1038/s41423-020-0488-6
8. Huang D, Ma N, Li X, Gou Y, Duan Y, Liu B, et al. Advances in single-cell RNA sequencing and its applications in cancer research. J Hematol Oncol. (2023) 16:98. doi: 10.1186/s13045-023-01494-6
9. Wang MM, Chen C, Lynn MN, Figueiredo CR, Tan WJ, Lim TS, et al. Applying single-cell technology in uveal melanomas: current trends and perspectives for improving uveal melanoma metastasis surveillance and tumor profiling. Front Mol Biosci. (2020) 7:611584. doi: 10.3389/fmolb.2020.611584
10. Finck A, Gill SI, June CH. Cancer immunotherapy comes of age and looks for maturity. Nat Commun. (2020) 11:3325. doi: 10.1038/s41467-020-17140-5
11. Masaoutis C, Kokkali S, Theocharis S. Immunotherapy in uveal melanoma: novel strategies and opportunities for personalized treatment. Expert Opin Investig Drugs. (2021) 30:555–69. doi: 10.1080/13543784.2021.1898587
12. Wu CS, Cioanca AV, Gelmi MC, Wen L, Di Girolamo N, Zhu L, et al. The multifunctional human ocular melanocortin system. Prog Retin Eye Res. (2023) 95:101187. doi: 10.1016/j.preteyeres.2023.101187
13. Ren X, Zhang L, Zhang Y, Li Z, Siemers N, Zhang Z. Insights gained from single-cell analysis of immune cells in the tumor microenvironment. Annu Rev Immunol. (2021) 39:583–609. doi: 10.1146/annurev-immunol-110519-071134
14. De Sousa EMF, Vermeulen L, Fessler E, Medema JP. Cancer heterogeneity–a multifaceted view. EMBO Rep. (2013) 14:686–95. doi: 10.1038/embor.2013.92
15. Vitale I, Shema E, Loi S, Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat Med. (2021) 27:212–24. doi: 10.1038/s41591-021-01233-9
16. Li J, Cao D, Jiang L, Zheng Y, Shao S, Zhuang A, et al. ITGB2-ICAM1 axis promotes liver metastasis in BAP1-mutated uveal melanoma with retained hypoxia and ECM signatures. Cell Oncol (Dordr). [ahead of print] (2023). doi: 10.1007/s13402-023-00908-4
17. Pandiani C, Strub T, Nottet N, Cheli Y, Gambi G, Bille K, et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ. (2021) 28:1990–2000. doi: 10.1038/s41418-020-00730-7
18. Gao G, Deng A, Liang S, Liu S, Fu X, Zhao X, et al. Integration of bulk RNA sequencing and single-cell RNA sequencing to reveal uveal melanoma tumor heterogeneity and cells related to survival. Front Immunol. (2022) 13:898925. doi: 10.3389/fimmu.2022.898925
19. Lin W, Beasley AB, Ardakani NM, Denisenko E, Calapre L, Jones M, et al. Intra- and intertumoral heterogeneity of liver metastases in a patient with uveal melanoma revealed by single-cell RNA sequencing. Cold Spring Harb Mol Case Stud. (2021) 7. doi: 10.1101/mcs.a006111
20. Bakhoum MF, Francis JH, Agustinus A, Earlie EM, Di Bona M, Abramson DH, et al. Loss of polycomb repressive complex 1 activity and chromosomal instability drive uveal melanoma progression. Nat Commun. (2021) 12:5402. doi: 10.1038/s41467-021-25529-z
21. Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. (2017) 67:93–9. doi: 10.3322/caac.21388
22. Ewens KG, Kanetsky PA, Richards-Yutz J, Al-Dahmash S, De Luca MC, Bianciotto CG, et al. Genomic profile of 320 uveal melanoma cases: chromosome 8p-loss and metastatic outcome. Invest Ophthalmol Vis Sci. (2013) 54:5721–9. doi: 10.1167/iovs.13-12195
23. Dogrusoz M, Bagger M, van Duinen SG, Kroes WG, Ruivenkamp CA, Bohringer S, et al. The prognostic value of AJCC staging in uveal melanoma is enhanced by adding chromosome 3 and 8q status. Invest Ophthalmol Vis Sci. (2017) 58:833–42. doi: 10.1167/iovs.16-20212
24. Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. (2004) 64:7205–9. doi: 10.1158/0008-5472.CAN-04-1750
25. Field MG, Durante MA, Anbunathan H, Cai LZ, Decatur CL, Bowcock AM, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Commun. (2018) 9:116. doi: 10.1038/s41467-017-02428-w
26. Xie J, Chen L, Tang Q, Wei W, Cao Y, Wu C, et al. A necroptosis-related prognostic model of uveal melanoma was constructed by single-cell sequencing analysis and weighted co-expression network analysis based on public databases. Front Immunol. (2022) 13:847624. doi: 10.3389/fimmu.2022.847624
27. Wang W, Zhao H, Wang S. Identification of a novel immune-related gene signature for prognosis and the tumor microenvironment in patients with uveal melanoma combining single-cell and bulk sequencing data. Front Immunol. (2023) 14:1099071. doi: 10.3389/fimmu.2023.1099071
28. Tan Y, Pan J, Deng Z, Chen T, Xia J, Liu Z, et al. Monoacylglycerol lipase regulates macrophage polarization and cancer progression in uveal melanoma and pan-cancer. Front Immunol. (2023) 14:1161960. doi: 10.3389/fimmu.2023.1161960
29. Xie J, Chen L, Cao Y, Ma C, Zhao W, Li J, et al. Single cell sequencing analysis constructed the N7-methylguanosine (m7G)-related prognostic signature in uveal melanoma. Aging (Albany NY). (2023) 15:2082–96. doi: 10.18632/aging.204592
30. Li Y, Cai H, Yang J, Xie X, Pei S, Wu Y, et al. Decoding tumor heterogeneity in uveal melanoma: basement membrane genes as novel biomarkers and therapeutic targets revealed by multi-omics approaches for cancer immunotherapy. Front Pharmacol. (2023) 14:1264345. doi: 10.3389/fphar.2023.1264345
31. Liu J, Zhang P, Yang F, Jiang K, Sun S, Xia Z, et al. Integrating single-cell analysis and machine learning to create glycosylation-based gene signature for prognostic prediction of uveal melanoma. Front Endocrinol (Lausanne). (2023) 14:1163046. doi: 10.3389/fendo.2023.1163046
32. Zhang X, Qiu J, Huang F, Han P, Shan K, Zhang C. Construction and verification of a hypoxia-related nine-gene prognostic model in uveal melanoma based on integrated single-cell and bulk RNA sequencing analyses. Exp Eye Res. (2022) 223:109214. doi: 10.1016/j.exer.2022.109214
33. van den Bosch QCC, Nguyen JQN, Brands T, van den Bosch TPP, Verdijk RM, Paridaens D, et al. FOXD1 is a transcription factor important for uveal melanocyte development and associated with high-risk uveal melanoma. Cancers (Basel). (2022) 14. doi: 10.3390/cancers14153668
34. Long GV, Swetter SM, Menzies AM, Gershenwald JE, Scolyer RA. Cutaneous melanoma. Lancet. (2023) 402:485–502. doi: 10.1016/S0140-6736(23)00821-8
35. Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. (2021) 39:154–73. doi: 10.1016/j.ccell.2020.10.001
36. Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. (2018) 33:151. doi: 10.1016/j.ccell.2017.12.013
37. Hui L, Chen Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. (2015) 368:7–13. doi: 10.1016/j.canlet.2015.07.039
38. Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. (2017) 387:61–8. doi: 10.1016/j.canlet.2016.01.043
39. Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun. (2020) 11:496. doi: 10.1038/s41467-019-14256-1
40. Aggarwal V, Workman CJ, Vignali DAA. LAG-3 as the third checkpoint inhibitor. Nat Immunol. (2023) 24:1415–22. doi: 10.1038/s41590-023-01569-z
41. Algazi AP, Tsai KK, Shoushtari AN, Munhoz RR, Eroglu Z, Piulats JM, et al. Clinical outcomes in metastatic uveal melanoma treated with PD-1 and PD-L1 antibodies. Cancer. (2016) 122:3344–53. doi: 10.1002/cncr.30258
42. Atri C, Guerfali FZ, Laouini D. Role of human macrophage polarization in inflammation during infectious diseases. Int J Mol Sci. (2018) 19. doi: 10.3390/ijms19061801
43. Shen L, Jiang S, Yang Y, Yang H, Fang Y, Tang M, et al. Pan-cancer and single-cell analysis reveal the prognostic value and immune response of NQO1. Front Cell Dev Biol. (2023) 11:1174535. doi: 10.3389/fcell.2023.1174535
44. Xue XM, Liu YY, Chen XM, Tao BY, Liu P, Zhou HW, et al. Pan-cancer analysis identifies NT5E as a novel prognostic biomarker on cancer-associated fibroblasts associated with unique tumor microenvironment. Front Pharmacol. (2022) 13:1064032. doi: 10.3389/fphar.2022.1064032
45. Wang Y, Liu B, Zhao G, Lee Y, Buzdin A, Mu X, et al. Spatial transcriptomics: Technologies, applications and experimental considerations. Genomics. (2023) 115:110671. doi: 10.1016/j.ygeno.2023.110671
Keywords: uveal melanoma, single-cell sequencing, tumor microenvironment, immunotherapy, prognostic model
Citation: Tang S, Zhang Y, Huang S, Zhu T and Huang X (2024) Single cell RNA-sequencing in uveal melanoma: advances in heterogeneity, tumor microenvironment and immunotherapy. Front. Immunol. 15:1427348. doi: 10.3389/fimmu.2024.1427348
Received: 03 May 2024; Accepted: 03 June 2024;
Published: 20 June 2024.
Edited by:
Haoran Feng, Ruijin Hospital, ChinaCopyright © 2024 Tang, Zhang, Huang, Zhu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojing Huang, MTM5MTcwMDAyMzVAMTYzLmNvbQ==; Tengfei Zhu, emh1dGYzMjFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Shiyi Tang
Shiyi Tang Yun Zhang
Yun Zhang Shengmei Huang1†
Shengmei Huang1† Xiaojing Huang
Xiaojing Huang