
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Immunol. , 05 July 2024
Sec. T Cell Biology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1426620
This article is part of the Research Topic Metabolite-induced Metabolic Reprogramming in the Immune System View all articles
The nucleotide-binding and oligomerization domain-like receptors (NLRs) NLR family CARD domain-containing protein 5 (NLRC5) and Class II Major Histocompatibility Complex Transactivator (CIITA) are transcriptional regulators of major histocompatibility complex (MHC) class I and class II genes, respectively. MHC molecules are central players in our immune system, allowing the detection of hazardous ‘non-self’ antigens and, thus, the recognition and elimination of infected or transformed cells from the organism. Recently, CIITA and NLRC5 have emerged as regulators of selected genes of the butyrophilin (BTN) family that interestingly are located in the extended MHC locus. BTNs are transmembrane proteins exhibiting structural similarities to B7 family co-modulatory molecules. The family member BTN2A2, which indeed contributes to the control of T cell activation, was found to be transcriptionally regulated by CIITA. NLRC5 emerged instead as an important regulator of the BTN3A1, BTN3A2, and BTN3A3 genes. Together with BTN2A1, BTN3As regulate non-conventional Vγ9Vδ2 T cell responses triggered by selected metabolites of microbial origin or accumulating in hematologic cancer cells. Even if endogenous metabolites conform to the canonical definition of ‘self’, metabolically abnormal cells can represent a danger for the organism and should be recognized and controlled by immune system cells. Collectively, new data on the role of NLRC5 in the expression of BTN3As link the mechanisms regulating canonical ‘non-self’ presentation and those marking cells with abnormal metabolic configurations for immune recognition, an evolutionary parallel that we discuss in this perspective review.
Nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs) are best known for their role in the detection of specific danger-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs) that has been thoroughly discussed elsewhere (1, 2). In this perspective review, we instead focus on the transcriptional regulatory functions of NLR family CARD domain-containing protein 5 (NLRC5) and Class II Major Histocompatibility Complex Transactivator (CIITA). Through a molecular platform called ‘MHC-enhanceosome’, NLRC5 and CIITA regulate the constitutive and the interferon-induced transcription of major histocompatibility complex class I (MHCI) and II (MHCII) genes, respectively (3, 4). MHC molecules are the core of vertebrates’ immune system, enabling the distinction between ‘self’ and ‘non-self’ and as such are central to the function of different immune subsets, promoting immune activation or tolerance.
Intriguingly, recent findings reveal unappreciated parallels between the presentation to immune cells of ‘non-self’ molecules and metabolites derived from deregulated cell metabolism. Metabolism, the ensemble of processes that fulfill cellular bioenergetic and biosynthetic needs (5), is central to cellular function and can be adapted to respond to the cellular needs. In some cases, however, it can also represent a threat to the organism as alterations in metabolic configurations underlie – for example – malignant transformation (6, 7). It is conceivable that immunity evolved to identify metabolically abnormal cells, despite conforming to the classic definition of ‘self’. In this review, we will discuss transcriptional NLRs, with particular attention on the emerging ability of NLRC5 to regulate the transcription of butyrophilins (BTNs) that are key to reveal an altered mevalonate metabolism to immune cells (7–9).
CIITA, the master regulator of MHCII genes, can be found in different forms according to the cell type-specific promoters used for its transcription. The form I features an N-terminal CARD effector domain followed by all characteristic NLR domains; the nucleotide binding domain (NBD), the helix domain 1 (HD1), the winged helix domain (WHD), the helix domain 2 (HD2), and a short stretch of leucine-rich repeats (LRRs). Instead, forms III and IV, the latter induced by interferon, lack the CARD (3, 10), while maintaining all the other domains.
NLRC5, the transactivator of MHCI genes, shows the typical domain organization of the other NLRs and is the largest member of the NLR family (4, 11). At the N-terminus it exhibits an ‘untypical’ CARD (uCARD), while the C-terminus is characterized by a long stretch of LRRs, which is the longest among NLRs. NLRC5 is largely regulated by interferons, both type I and II; accordingly, two signal transducer and activator of transcription (STAT) binding sites have been identified in its promoter (12, 13). Furthermore, expression of NLRC5 via peroxisome proliferator-activated receptor γ isoform 1 (PPARγ1), the master regulator of adipogenesis and lipid metabolism, has been reported (14).
While CIITA is almost exclusively found in antigen-presenting cells (APCs) and appears to be equally distributed between the cytosol and the nucleus, NLRC5 is broadly expressed and resides mostly in the cytoplasm with a small fraction shuttling to the nucleus, (13, 15–19). In the nucleus, both NLRC5 and CIITA are recruited to the promoter of MHC and related genes through the MHC-enhanceosome (11, 18, 20, 21). Interestingly, both NLRC5 and CIITA do not show DNA binding motifs in their sequence, corroborating the importance of the interaction with MHC-enhanceosome components for their function (18, 22). Once occupying the promoter, these NLRs promote the transcription of target genes.
T lymphocytes constitute an essential arm of adaptive immunity. CD4+ T cells, which exert modulatory functions, express a T cell receptor (TCR) recognizing the cognate peptide in the context of MHCII molecules. MHCII glycoproteins are expressed on APCs and display peptides derived from phagocytosis or autophagy (20, 23). Instead, CD8+ T lymphocytes kill target cells upon detection of the cognate antigen presented onto classical MHCI molecules (Figure 1). Classical MHCI proteins are ubiquitously expressed and present proteasome-derived peptides, including pathogen-derived or tumor antigens in infected or transformed cells, respectively (20, 23). In fact, T cells are essentially selected to react toward ‘non-self’, while recognition of ‘self’-peptides is limited to the regulatory subsets (24). Further, ‘unconventional’ T cells are restricted to non-classical MHCI molecules, which bind to peptides or specific biochemical structures of microbial or host-derived origin, eliciting effector or regulatory responses by unconventional T cells (25–27). Therefore, MHC molecules are central to our immune system, allowing to discriminate between ‘self’ and ‘non-self’.
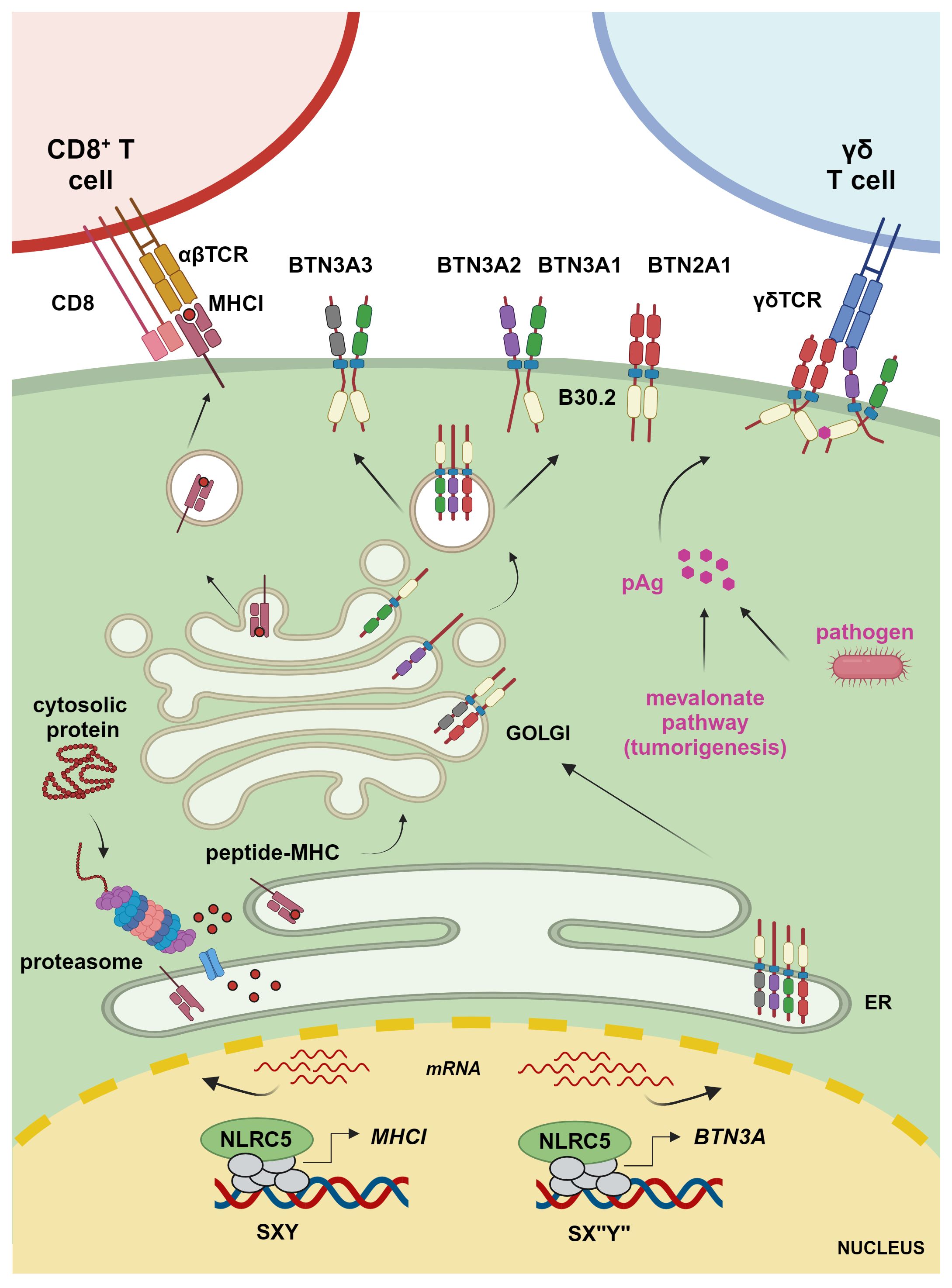
Figure 1 Transcriptional regulation of MHCI and BTN3A1–3 molecules. The transcriptional regulator NLRC5 translocates to the nucleus, where it binds to the SXY module of MHCI and BTN3A gene promoters along with a complex known as MHC-enhanceosome. NLRC5 recruits then chromatin modifiers, transcriptional elongation, and initiation factors to promote transcription of these target genes. MHCI molecules present proteasome-derived endogenous peptides to CD8+ T cells, whereas BTN3A1 binds – through the intracellular B30.2 domain – to pAgs accumulating due to infection or a hyperactive mevalonate pathway. This results in a conformational change inducing the interaction between the intracellular domain of BTN3A1, present as BTN3A1 homomer or heteromer with BTN3A molecules, and BTN2A1 and enabling the interaction between BTN2A1 and the TCR to mediate γδ T cell activation (figure was created by www.biorender.com).
CIITA regulates the expression of MHCII, which is composed of an alpha chain and a beta chain. In humans, three distinct MHCII molecules known as HLA-DR, HLA-DP, and HLA-DQ are encoded within the ‘MHC locus’, found on the short arm of chromosome 6. In mice instead, the ‘MHC locus’ located on chromosome 17 comprises two MHCII molecules, H2-A and H2-E. Moreover, CIITA regulates the expression of genes involved in MHCII loading, such as HLA-DM, HLA-DO, and CD74 (28–30). CIITA is therefore a key player in coordinating immunity, as shown by the effects of inactivating CIITA mutations that lead to an immunosuppressed condition known as bare lymphocyte syndrome (BLS) (31).
The expression of classical MHCI glycoproteins, which are composed of an alpha chain and the invariant β-2-microglobulin (β2m), is largely regulated by NLRC5 (13, 18, 32, 33). Classical MHCI genes in humans are HLA-A, HLA-B, and HLA-C, whereas the murine genes are H2-K, H2-D, and H2-L and are located within the ‘MHC locus’. NLRC5 also regulates the human non-classical MHCI genes HLA-E, HLA-F, and HLA-G and murine H2-M, H2-T, and H2-Q family members as well as the expression of genes involved in the MHCI pathway, such as β2m, selected proteasome subunits, and transporter associated with antigen processing 1 (TAP1), an essential molecule for antigen loading onto MHCI (34). Given its important function in the MHCI pathway, NLRC5 has been increasingly appreciated for its major role in regulating cytotoxic T cells toward cancerous cells (18, 33, 35–42).
Of note, while overexpression of NLRC5 does not induce MHCII expression, CIITA overexpression has been shown to induce MHCI transcription (43–46). However, in vivo, endogenous NLRC5 and CIITA show non-redundant functions (33, 34). This supports the idea that, due to the similar elements of the MHCI and MHCII promoters, overexpression of CIITA can lead to misleading results. In the following section, we will discuss in detail similarities and differences of MHC genes’ promoters and introduce in parallel the composition of the MHC-enhanceosome.
The promoter region of both MHCI and MHCII genes contains the SXY module, which consists of the S box, the X box further divided into X1 and X2, and the Y box, altogether responsible for the binding of the enhanceosome and the recruitment of the matching NLR (Figure 2) (21, 34, 48–50). Interestingly, both sequence and stereo-specific position of the boxes forming the SXY motif are conserved and crucial for the constitutive and induced expression of MHCI and MHCII genes (34, 51, 52). The X1 and Y boxes are bound by regulatory factor X (RFX) and nuclear factor Y (NF-Y) complexes, respectively (49, 53–55). The X2 box is bound by CREB/ATF1(cAMP response element-binding protein) family members (44, 56, 57). Interestingly, the S-box is important for both CIITA- and NLRC5-mediated transactivation and its sequence dictates the specificity of NLRC5 for MHCI (11, 34, 49). Despite this knowledge, the S-box binding factor(s) is still unknown.
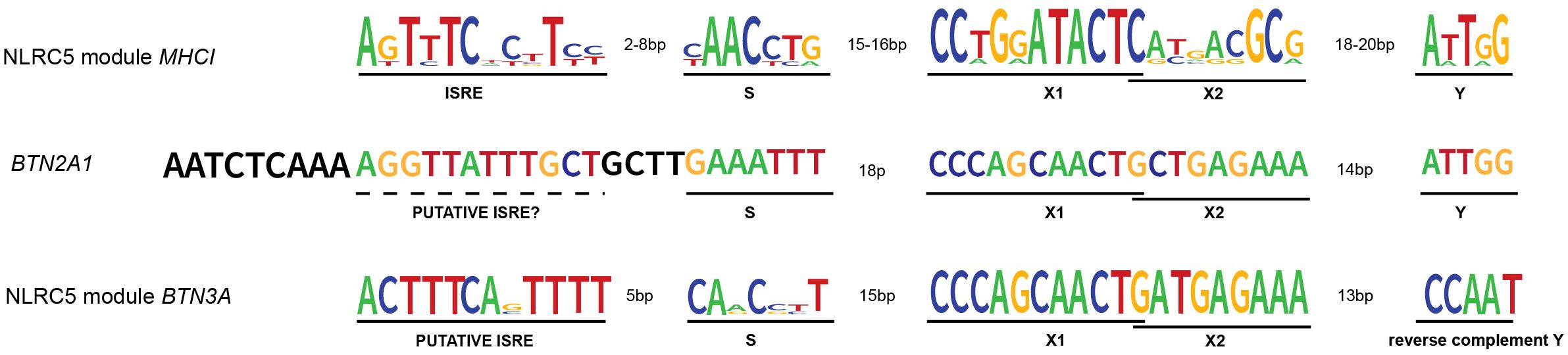
Figure 2 NLRC5’s transcription modules and BTN2A1 promoter. NLRC5 module from MHCI genes’ promoters: MHCI genes’ promoter presents the typical SXY module crucial for the binding of the enhanceosome and the recruitment of NLRC5. The X box is then further divided into X1 and X2. Upstream of the SXY motif, there is the interferon-stimulated regulatory element (ISRE), which mediates the binding of IRF1. NLRC5 module from BTN3A genes’ promoters: as for the MHCI genes, the promoter of BTN3A genes shows the presence of an SXY module with the peculiarity of a reverse complement Y motif. Interestingly, the alignment performed using SnapGene software (www.snapgene.com), shows the presence of a putative ISRE at the distance and orientation expected for MHCI genes’ promoter. BTN2A1 gene shows an SXY module with a Y motif in the same orientation as found in MHCI genes. However, our alignment did not show the presence of a highly conserved ISRE motif upstream of the SXY module. The logo for the consensus sequence of MHCI genes was created using http://weblogo.berkeley.edu (47) giving as input the respective ISRE, S, X, and Y sequences of HLA-A, B, C, E, F, and G genes. The same tool was used to generate the consensus for BTN3A genes giving as input the ISRE, S, X, and Y sequences of BTN3A1–3 genes. All the genomic sequences were retrieved from the genome browser (genome.ucsc.edu).
Upstream of the SXY box, the promoter of MHCI genes contains two additional regulatory sequences; the enhancer A and the interferon-stimulated responsive element (ISRE) bound by NF-κB and IRF1, respectively (Figure 2) (44, 54, 58, 59). The latter contributes to the interferon (IFN) type I- and type II-induced expression of MHCI genes.
The expression of MHCI and MHCII genes is tightly regulated, and epigenetics plays a crucial role. Indeed, different levels of histone modifications at the promoter region of MHCI and MHCII genes have been observed upon transcription activation, suggesting that histone modifiers might be associated with the MHC-enhanceosome (60–62). CIITA possesses per se a histone acetyltransferase (HAT) activity (63, 64) and has been shown to interact with chromatin remodeling factors such as histone lysine acetyltransferases p300/CREB-binding protein (CREBBP) and general control nonderepressible 5 (GCN5) (56, 57, 65). These two factors were also shown to enhance the transactivation activity of overexpressed NLRC5 (19, 49). Moreover, GCN5 has been recently reported to interact with and regulate NLRC5 nuclear retention (19). Yet, the mechanisms underlying the activity of NLRC5 and CIITA at the MHC-enhanceosome are most likely manifold and awaiting future investigation.
The best-studied activity of NLRC5 and CIITA is to control MHC genes’ expression and canonical antigen presentation pathways; however, additional targets have been identified that are regulated by CIITA or NLRC5 and a bona fide enhanceosome complex. Among these, BTN genes are of particular interest. Human BTN genes map to the telomeric end of the extended MHC locus, already suggesting a connection with MHC genes (66, 67). BTNs are transmembrane proteins exhibiting structural similarities to the B7 family of co-stimulatory molecules, which include the crucial immunomodulatory ligands PD-L1 and CD80 (67). The family member BTN2A2 indeed contributes to the control of T cell activation and proliferation and – accordingly – Btn2a2-knockout mice are more susceptible to T cell-mediated autoimmunity (68–72).
In 2016, Sarter et al. reported that CIITA transcriptionally regulates BTN2A2 gene expression both in the human and the mouse system (69). An SXY consensus was identified at the BTN2A2 promoter and the S-box sequence was compatible with CIITA-mediated regulation (8, 69).
The human BTN2A1, BTN3A1, BTN3A2, and BTN3A3 are emerging as key regulators of non-conventional Vγ9Vδ2 T cells, which are important effectors against selected microbial infections and cancers and have been shown to correlate with improved patient outcome when infiltrating tumoral tissue (73–85). Vγ9Vδ2 T cells sense and expand in response to so-called phosphoantigens (pAgs), metabolites of the isoprenoid synthesis pathway that accumulate in infected or transformed cells (Figure 1) (85). These metabolites, such as HMB-PP ((E)-4-hydroxy-3-methyl-but-2-enyl diphosphate), can be produced by parasitic and bacterial pathogens, including Mycobacterium tuberculosis or Listeria monocytogenes (86, 87). Isopentenyl pyrophosphate (IPP) is instead an endogenous form of pAg that accumulates in certain tumor cells (88). This is due to a dysfunctional mevalonate pathway, which is important for the biosynthesis of isoprenoid (terpenoid) molecules used in processes as diverse as cell membrane maintenance, protein modification, and synthesis of steroid hormones, coenzyme Q10, etc. (89). pAgs bind to the intracellular B30.2 domain, a multifunctional domain found in many innate immune sensors, of BTN3A1 (77, 90). This binding leads to a structural change in the complex formed by BTN3A1 and BTN2A1, which is required for the engagement of the Vγ9Vδ2 TCR, T cell proliferation, and cytotoxic attack of the abnormal cells (Figure 1) (83, 84, 91–95). In this context, BTN3A2 and BTN3A3 have been shown not only to increase the trafficking of BTN3A1 to the plasma membrane but also to be directly involved in the activation of Vγ9Vδ2 T cells (82, 94). Moreover, recent data hint at a crucial role for BTN3A3, which also possesses an intracellular B30.2 domain, in the activation of Vγ9Vδ2 T cells upon live Listeria monocytogenes infection (96). While the relevance of these BTN molecules has started to emerge over the last years, their exact role and dynamics await further elucidation (97).
Although BTNs and the associated Vγ9Vδ2 T cells are known to work in an MHC-independent manner, Dang et al. found a correlation between the MHC-regulating NLRC5 and BTN3A1–3 expression in cancerous cells and cells from healthy and M. tuberculosis-infected donors (8). Overexpressed NLRC5 was able to occupy and transactivate an atypical SXY module containing a reverse complement of the Y box, found in the promoter region of BTN3A1–3 genes leading to enhanced Vγ9Vδ2 T cell-mediated elimination of tumoral cells (Figures 1, 2). Recently, these data were confirmed and extended through a genome-wide CRISPR screen demonstrating the importance of endogenous NLRC5 for the expression of BTN3A1–3, and for cancer cells’ recognition and killing by Vγ9Vδ2 T cells (9, 98). This genome-wide screen also revealed a role for the histone acetylase CREBBP, an enzyme already found to promote the effects of NLRC5 and CIITA (49, 56). As CIITA is not found to play a major role in the activation of Vγ9Vδ2 T cells, these results support the idea that CREBBP is involved in NLRC5-dependent chromatin relaxation. This genetic screen also demonstrated the relevance of factors implicated in NLRC5/MHCI regulation, such as IRF1 and the enhanceosome factor RFXAP, for BTN3A1–3 expression (9). This is in line with our research group data, obtained from cell lines bearing mutations in RFX factors (8), that indicate the involvement of an enhanceosome complex in the regulation of BTN3A gene transcription. Together, these findings underline the association between antigen and pAg presentation.
While the data discussed above clearly imply enhanceosome factors in the regulation of BTN3A gene transcription, the presence of a reverse complement Y box raises questions on the reciprocal orientation of the complex subunits. This entails a degree of flexibility that might be fulfilled either by changes in the architecture of the complex or by DNA looping, aspects that to date remain unexplored.
Moreover, the recently reported importance of IRF1 for BTN3A1–3 gene expression and data indicating IRF1 binding proximal to the BTN3A1–3 transcription start site led us to verify the presence of an ISRE sequence in their promoters (9). Indeed, an ISRE was found at the close distance and orientation relative to the SXY typical of the previously reported promoters regulated by NLRC5 (Figure 2) (44, 59). These notions suggest that, not only from a biological but also from a promoter docking point of view, combining IRF1 and NLRC5 might be important, strengthening the parallels between MHCI and BTN3A transcriptional control.
To better appreciate the regulation of Vγ9Vδ2 T cells’ activity, another key question concerns the mechanisms controlling BTN2A1 transcription. Data from BLS-derived B cell lines indicate that BTN2A1 expression is largely dependent on the RFX complex but not on CIITA (8). With regard to NLRC5, we did not witness its role in the transcriptional induction of endogenous BTN2A1 and found just a weak correlation between NLRC5 and BTN2A1 transcript abundance in M. tuberculosis patients and ‘The Cancer Genome Atlas’ cancer samples (8). Interestingly, Liu and colleagues observed NLRC5-mediated transactivation of endogenous BTN2A1 only upon EBV-reactivation, which also leads to IFNγ upregulation in nasopharyngeal carcinoma cells (98). These differences might imply a complex transcriptional regulation of BTN2A1 at endogenous level, requiring the concomitant presence of NLRC5 together with additional transcriptional regulators, which – we might speculate – are stimulated by interferon. Indeed, an SXY module is present in the promoter of the BTN2A1 gene and weak IRF1 binding upstream of the transcription start site has been recently reported (9), even if at the canonical ISRE position a strong consensus is not observed (Figure 2).
Questions arise also on the possible contribution by NLRC5 and CIITA in controlling the transcription of other members of the BTN/BTN-like (BTNL) family. Next to BTN2A and BTN3A genes, BTN1A1, myelin oligodendrocyte glycoprotein (MOG), erythroid membrane-associated protein (ERMAP), BTNL2, 3, 8, and 9 are found in the human genome (66, 99). Interestingly, BTN1A1, MOG, and BTNL2 are located in the extended MHC locus on chromosome 6. Both BTN1A1 and BTNL2 have been reported to exert immunomodulatory functions and foster immune evasion when expressed on cancer cells (68, 100, 101) and BTNL2 has been associated with immunological disorders such as sarcoidosis (102). Furthermore, recent work showed how BTNL3 as part of BTNL3/8 heterodimers binds to Vγ4 γδ TCR via germline-encoded regions, suggesting an interaction modality important to maintain homeostasis of this lymphocyte subset (91).
The high expression of BTN1A1 and BTNL3/8 molecules by non-immune cells – largely mammary and intestinal epithelial cells (66) – does not match the expected patterns of NLRC5- or CIITA-driven expression and recent results indicate that hepatocyte nuclear factor 4 and caudal type homeobox factors are crucial for the transcription of these BTNL genes, underlying the importance of tissue-specific transcription factors in their regulation (103). Moreover, available data did not reveal the presence of BTN/BTNL genes among the top targets regulated by NLRC5 or CIITA (8, 18, 34, 104, 105), with the exception of MOG – coding for a protein highly expressed in the brain – which has been identified as a putative CIITA and RFX5 target by chromatin immunoprecipitation (105). However, these results are limited to the analyzed tissue/cell type and promoter scanning for SXY motifs, which can be functional even when partly degenerated, remains challenging unless accompanied by experimental approaches. Therefore, these observations encourage further investigations on the transcriptional regulation of BTN/BTNL genes.
MHC molecules are the core of vertebrates’ immune system, enabling the distinction between ‘self’ and ‘non-self’ and crucially promoting rejection or tolerance. NLRC5 regulates the transcription of MHCI, thus regulating cytotoxic T cell-mediated killing of infected and transformed cells. Intriguingly, recent results from others and us demonstrate that NLRC5 also controls the transcription of BTN3A1–3 molecules, which – together with BTN2A1 – are key for engaging Vγ9Vδ2 T cell-mediated killing of cells with a deregulated mevalonate pathway (Figure 1) (8, 9, 83, 84, 91–95). This reveals an unanticipated connection between the molecular circuits rendering cells presenting ‘non-self’ antigens and those with abnormal metabolic configurations detectable by the immune system.
Such a link is evolutionarily plausible as also an altered metabolism, albeit conforming to the definition of ‘self’, can represent a threat to the organism. Indeed, altered metabolism is central to malignant transformation. Along these lines, another target of NLRC5 is H2-M3 (32, 34, 35), a murine non-classical MHC gene presenting N-formylated peptides that can either originate from prokaryotic pathogens or mitochondria (25, 27). The NLRC5-H2-M3 axis might thus support T cell immune responses against bacteria as well as toward cells with perturbed oxidative metabolism. Moreover, multiple studies described the connection between NLRC5 and metabolic conditions including obesity, but also an interaction of NLRC5 with PPARγ1, a key regulator of lipid metabolism and beyond (106), fostering the transcriptional activity of the latter (14, 107–110). Recent work also revealed that the acetyltransferase GCN5 interacts with NLRC5, promoting NLRC5 nuclear localization and activity (19). Notably, GCN5 acetylates PPARγ coactivator-1 (PGC-1), thereby suppressing its transcriptional activity, which is crucial for PPARγ-dependent transcription as well as for mitochondrial biogenesis (111). These notions support the hypothesis that the interaction of NLRC5 with GCN5 might represent a second mechanism modulating PPARγ activity, for example by indirectly favoring PGC-1 de-acetylation. Finally, an elegant study demonstrating a role for NLRC5 in inflammatory cell death upon heme sensing found that NLRC5 levels were primed by depletion of oxidized nicotinamide adenine dinucleotide (NAD+), a crucial cofactor in multiple cellular metabolic reactions (112). Based on these data, we can speculate that conditions characterized by reduced NAD+ levels, such as ageing or obesity (113), might increase NLRC5 levels to influence cellular metabolism or license cell death. How these conditions relate to the transcriptional role of NLRC5 remains an open and exciting question to explore. Altogether these observations add a layer of complexity to the already novel node represented by NLRC5 at the intersection between ‘non-self’, ‘deregulated-self’, and immunity and raise therefore NLRC5 to a multilevel modulator of metabolism, anti-microbial, and anti-cancer immune responses.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author/s.
GG: Writing – original draft, Writing – review & editing. FB: Writing – original draft, Writing – review & editing. SN: Writing – original draft, Writing – review & editing. JG: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Swiss National Science Foundation (PP00P3_165833, 310030_197771 and CRSK-3_220949), the Novartis Foundation (#23C200), Basel, the Swiss Cancer Research Foundation (KFS 5141-08-2020), to GG and by the Fondazione IRB (Promising Pilot Projects) to JG. This study received funding from Novartis Foundation, Basel. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
We thank Caroline Junqueira, Hanif Javanmard Khameneh, and Irene Buzzago, IRB, Bellinzona, for critical reading of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Unrelated projects in GG laboratory are supported by OM Pharma.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. (2016) 16:407–20. doi: 10.1038/nri.2016.58
2. Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, Girardin SE. NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys. (2019) 670:69–81. doi: 10.1016/j.abb.2018.12.022
3. Reith W, Leibundgut-Landmann S, Waldburger JM. Regulation of MHC class II gene expression by the class II transactivator. Nat Rev Immunol. (2005) 5:793–806. doi: 10.1038/nri1708
4. Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nat Rev Immunol. (2012) 12:813–20. doi: 10.1038/nri3339
5. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. (2017) 168:960–76. doi: 10.1016/j.cell.2017.02.004
6. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
7. Gruenbacher G, Thurnher M. Mevalonate metabolism in immuno-oncology. Front Immunol. (2017) 8:1714. doi: 10.3389/fimmu.2017.01714
8. Dang AT, Strietz J, Zenobi A, Khameneh HJ, Brandl SM, Lozza L, et al. NLRC5 promotes transcription of BTN3A1-3 genes and Vgamma9Vdelta2 T cell-mediated killing. iScience. (2021) 24:101900. doi: 10.1016/j.isci.2020.101900
9. Mamedov MR, Vedova S, Freimer JW, Sahu AD, Ramesh A, Arce MM, et al. CRISPR screens decode cancer cell pathways that trigger gammadelta T cell detection. Nature. (2023) 621:188–95. doi: 10.1038/s41586-023-06482-x
10. Nickerson K, Sisk TJ, Inohara N, Yee CS, Kennell J, Cho MC, et al. Dendritic cell-specific MHC class II transactivator contains a caspase recruitment domain that confers potent transactivation activity. J Biol Chem. (2001) 276:19089–93. doi: 10.1074/jbc.M101295200
11. Chelbi ST, Dang AT, Guarda G. Emerging major histocompatibility complex class I-related functions of NLRC5. Adv Immunol. (2017) 133:89–119. doi: 10.1016/bs.ai.2016.11.003
12. Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent antiviral immune responses. J Immunol. (2010) 184:1990–2000. doi: 10.4049/jimmunol.0900557
13. Staehli F, Ludigs K, Heinz LX, Seguin-Estevez Q, Ferrero I, Braun M, et al. NLRC5 deficiency selectively impairs MHC class I- dependent lymphocyte killing by cytotoxic T cells. J Immunol. (2012) 188:3820–8. doi: 10.4049/jimmunol.1102671
14. Luan P, Jian W, Xu X, Kou W, Yu Q, Hu H, et al. NLRC5 inhibits neointima formation following vascular injury and directly interacts with PPARgamma. Nat Commun. (2019) 10:2882. doi: 10.1038/s41467-019-10784-y
15. Cressman DE, Chin KC, Taxman DJ, Ting JP. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. (1999) 10:163–71. doi: 10.1016/S1074-7613(00)80017-5
16. Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. GTP binding by class II transactivator: role in nuclear import. Science. (1999) 285:1402–5. doi: 10.1126/science.285.5432.1402
17. Hake SB, Masternak K, Kammerbauer C, Janzen C, Reith W, Steimle V. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol Cell Biol. (2000) 20:7716–25. doi: 10.1128/MCB.20.20.7716-7725.2000
18. Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U.S.A. (2010) 107:13794–9. doi: 10.1073/pnas.1008684107
19. Zhu B, Ouda R, An N, Tanaka T, Kobayashi KS. The balance between nuclear import and export of NLRC5 regulates MHC class I transactivation. J Biol Chem. (2024) 300:107205. doi: 10.1016/j.jbc.2024.107205
20. van den Elsen PJ, Holling TM, Kuipers HF, Van Der Stoep N. Transcriptional regulation of antigen presentation. Curr Opin Immunol. (2004) 16:67–75. doi: 10.1016/j.coi.2003.11.015
21. Neerincx A, Rodriguez GM, Steimle V, Kufer TA. NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J Immunol. (2012) 188:4940–50. doi: 10.4049/jimmunol.1103136
22. Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell. (1993) 75:135–46. doi: 10.1016/S0092-8674(05)80090-X
23. Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. (2011) 11:823–36. doi: 10.1038/nri3084
24. Pohar J, Simon Q, Fillatreau S. Antigen-specificity in the thymic development and peripheral activity of CD4(+)FOXP3(+) T regulatory cells. Front Immunol. (2018) 9:1701. doi: 10.3389/fimmu.2018.01701
25. Rodgers JR, Cook RG. MHC class Ib molecules bridge innate and acquired immunity. Nat Rev Immunol. (2005) 5:459–71. doi: 10.1038/nri1635
26. Godfrey DI, Uldrich AP, Mccluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. (2015) 16:1114–23. doi: 10.1038/ni.3298
27. Anderson CK, Brossay L. The role of MHC class Ib-restricted T cells during infection. Immunogenetics. (2016) 68:677–91. doi: 10.1007/s00251-016-0932-z
28. Doyle C, Ford PJ, Ponath PD, Spies T, Strominger JL. Regulation of the class II-associated invariant chain gene in normal and mutant B lymphocytes. Proc Natl Acad Sci U.S.A. (1990) 87:4590–4. doi: 10.1073/pnas.87.12.4590
29. Zhu L, Jones PP. Transcriptional control of the invariant chain gene involves promoter and enhancer elements common to and distinct from major histocompatibility complex class II genes. Mol Cell Biol. (1990) 10:3906–16. doi: 10.1128/mcb.10.8.3906-3916.1990
30. Brown AM, Barr CL, Ting JP. Sequences homologous to class II MHC W, X, and Y elements mediate constitutive and IFN-gamma-induced expression of human class II-associated invariant chain gene. J Immunol. (1991) 146:3183–9. doi: 10.4049/jimmunol.146.9.3183
31. Leon MaChado JA, Steimle V. The MHC class II transactivator CIITA: not (Quite) the odd-one-out anymore among NLR proteins. Int J Mol Sci. (2021) 22. doi: 10.3390/ijms22031074
32. Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting edge: impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J Immunol. (2012) 189:516–20. doi: 10.4049/jimmunol.1200064
33. Robbins GR, Truax AD, Davis BK, Zhang L, Brickey WJ, Ting JP. Regulation of class I major histocompatibility complex (MHC) by nucleotide-binding domain, leucine-rich repeat-containing (NLR) proteins. J Biol Chem. (2012) 287:24294–303. doi: 10.1074/jbc.M112.364604
34. Ludigs K, Seguin-Estevez Q, Lemeille S, Ferrero I, Rota G, Chelbi S, et al. NLRC5 exclusively transactivates MHC class I and related genes through a distinctive SXY module. PloS Genet. (2015) 11:e1005088. doi: 10.1371/journal.pgen.1005088
35. Yao Y, Wang Y, Chen F, Huang Y, Zhu S, Leng Q, et al. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. (2012) 22:836–47. doi: 10.1038/cr.2012.56
36. Chelbi ST, Guarda G. NLRC5, a promising new entry in tumor immunology. J Immunother Cancer. (2016) 4:39. doi: 10.1186/s40425-016-0143-z
37. Rodriguez Galaxia M, Bobbala D, Serrano D, Mayhue M, Champagne A, Saucier C, et al. NLRC5 elicits antitumor immunity by enhancing processing and presentation of tumor antigens to CD8+ T lymphocytes. OncoImmunology. (2016) 5. doi: 10.1080/2162402X.2016.1151593
38. Rota G, Ludigs K, Siegert S, Tardivel A, Morgado L, Reith W, et al. T cell priming by activated nlrc5-deficient dendritic cells is unaffected despite partially reduced MHC class I levels. J Immunol. (2016) 196. doi: 10.4049/jimmunol.1502084
39. Yoshihama S, Roszik J, Downs I, Meissner TB, Vijayan S, Chapuy B, et al. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci U S A. (2016) 113. doi: 10.1073/pnas.1602069113
40. Yoshihama S, Vijayan S, Sidiq T, Kobayashi KS. NLRC5/CITA: A key player in cancer immune surveillance. Trends Cancer. (2017) 3:28–38. doi: 10.1016/j.trecan.2016.12.003
41. Catalano C, Da Silva Filho MI, Frank C, Jiraskova K, Vymetalkova V, Levy M, et al. Investigation of single and synergic effects of NLRC5 and PD-L1 variants on the risk of colorectal cancer. PloS One. (2018) 13:e0192385. doi: 10.1371/journal.pone.0192385
42. Catalano C, Da Silva Filho MI, Jiraskova K, Vymetalkova V, Levy M, Liska V, et al. Short article: Influence of regulatory NLRC5 variants on colorectal cancer survival and 5-fluorouracil-based chemotherapy. Eur J Gastroenterol Hepatol. (2018) 30:838–42. doi: 10.1097/MEG.0000000000001154
43. Chang CH, Guerder S, Hong SC, Van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. (1996) 4:167–78. doi: 10.1016/S1074-7613(00)80681-0
44. Gobin SJ, Peijnenburg A, Keijsers V, Van Den Elsen PJ. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. (1997) 6:601–11. doi: 10.1016/S1074-7613(00)80348-9
45. Martin BK, Chin KC, Olsen JC, Skinner CA, Dey A, Ozato K, et al. Induction of MHC class I expression by the MHC class II transactivator CIITA. Immunity. (1997) 6:591–600. doi: 10.1016/S1074-7613(00)80347-7
46. Williams GS, Malin M, Vremec D, Chang CH, Boyd R, Benoist C, et al. Mice lacking the transcription factor CIITA–a second look. Int Immunol. (1998) 10:1957–67. doi: 10.1093/intimm/10.12.1957
47. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. (2004) 14:1188–90. doi: 10.1101/gr.849004
48. Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat Immunol. (2003) 4:132–7. doi: 10.1038/ni883
49. Meissner TB, Liu YJ, Lee KH, Li A, Biswas A, Van Eggermond MC, et al. NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J Immunol. (2012) 188:4951–8. doi: 10.4049/jimmunol.1103160
50. Anderson DA 3rd, Grajales-Reyes GE, Satpathy AT, Vasquez Hueichucura CE, Murphy TL, Murphy KM. Revisiting the specificity of the MHC class II transactivator CIITA in classical murine dendritic cells in vivo. Eur J Immunol. (2017) 47:1317–23. doi: 10.1002/eji.201747050
51. Gobin SJ, Van Zutphen M, Westerheide SD, Boss JM, Van Den Elsen PJ. The MHC-specific enhanceosome and its role in MHC class I and beta(2)-microglobulin gene transactivation. J Immunol. (2001) 167:5175–84. doi: 10.4049/jimmunol.167.9.5175
52. Scharer CD, Choi NM, Barwick BG, Majumder P, Lohsen S, Boss JM. Genome-wide CIITA-binding profile identifies sequence preferences that dictate function versus recruitment. Nucleic Acids Res. (2015) 43:3128–42. doi: 10.1093/nar/gkv182
53. Mantovani R, Pessara U, Tronche F, Li XY, Knapp AM, Pasquali JL, et al. Monoclonal antibodies to NF-Y define its function in MHC class II and albumin gene transcription. EMBO J. (1992) 11:3315–22. doi: 10.1002/embj.1992.11.issue-9
54. Gobin SJ, Peijnenburg A, Van Eggermond M, Van Zutphen M, Van Den Berg R, Van Den Elsen PJ. The RFX complex is crucial for the constitutive and CIITA-mediated transactivation of MHC class I and beta2-microglobulin genes. Immunity. (1998) 9:531–41. doi: 10.1016/S1074-7613(00)80636-6
55. Jabrane-Ferrat N, Nekrep N, Tosi G, Esserman L, Peterlin BM. MHC class II enhanceosome: how is the class II transactivator recruited to DNA-bound activators? Int Immunol. (2003) 15:467–75. doi: 10.1093/intimm/dxg048
56. Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. (1998) 18:6777–83. doi: 10.1128/MCB.18.11.6777
57. Fontes JD, Kanazawa S, Jean D, Peterlin BM. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. (1999) 19:941–7. doi: 10.1128/MCB.19.1.941
58. Giuliani C, Saji M, Napolitano G, Palmer LA, Taniguchi SI, Shong M, et al. Hormonal modulation of major histocompatibility complex class I gene expression involves an enhancer A-binding complex consisting of Fra-2 and the p50 subunit of NF-kappa B. J Biol Chem. (1995) 270:11453–62. doi: 10.1074/jbc.270.19.11453
59. Gobin SJ, Van Zutphen M, Woltman AM, Van Den Elsen PJ. Transactivation of classical and nonclassical HLA class I genes through the IFN-stimulated response element. J Immunol. (1999) 163:1428–34. doi: 10.4049/jimmunol.163.3.1428
60. Spilianakis C, Papamatheakis J, Kretsovali A. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol Cell Biol. (2000) 20:8489–98. doi: 10.1128/MCB.20.22.8489-8498.2000
61. Osborne A, Zhang H, Yang WM, Seto E, Blanck G. Histone deacetylase activity represses gamma interferon-inducible HLA-DR gene expression following the establishment of a DNase I-hypersensitive chromatin conformation. Mol Cell Biol. (2001) 21:6495–506. doi: 10.1128/MCB.21.19.6495-6506.2001
62. Wright KL, Ting JP. Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol. (2006) 27:405–12. doi: 10.1016/j.it.2006.07.007
63. Raval A, Howcroft TK, Weissman JD, Kirshner S, Zhu XS, Yokoyama K, et al. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol Cell. (2001) 7:105–15. doi: 10.1016/S1097-2765(01)00159-9
64. Soe KC, Devaiah BN, Singer DS. Transcriptional coactivator CIITA, a functional homolog of TAF1, has kinase activity. Biochim Biophys Acta. (2013) 1829:1184–90. doi: 10.1016/j.bbagrm.2013.09.001
65. Spilianakis C, Kretsovali A, Agalioti T, Makatounakis T, Thanos D, Papamatheakis J. CIITA regulates transcription onset viaSer5-phosphorylation of RNA Pol II. EMBO J. (2003) 22:5125–36. doi: 10.1093/emboj/cdg496
66. Abeler-Dorner L, Swamy M, Williams G, Hayday AC, Bas A. Butyrophilins: an emerging family of immune regulators. Trends Immunol. (2012) 33:34–41. doi: 10.1016/j.it.2011.09.007
67. Arnett HA, Viney JL. Immune modulation by butyrophilins. Nat Rev Immunol. (2014) 14:559–69. doi: 10.1038/nri3715
68. Smith IA, Knezevic BR, Ammann JU, Rhodes DA, Aw D, Palmer DB, et al. BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J Immunol. (2010) 184:3514–25. doi: 10.4049/jimmunol.0900416
69. Sarter K, Leimgruber E, Gobet F, Agrawal V, Dunand-Sauthier I, Barras E, et al. Btn2a2, a T cell immunomodulatory molecule coregulated with MHC class II genes. J Exp Med. (2016) 213:177–87. doi: 10.1084/jem.20150435
70. He X, Hu R, Luo P, Gao J, Yang W, Li J, et al. BTN2A2 protein negatively regulates T cells to ameliorate collagen-induced arthritis in mice. Sci Rep. (2021) 11:19375. doi: 10.1038/s41598-021-98443-5
71. Frech M, Danzer H, Uchil P, Azizov V, Schmid E, Schalter F, et al. Butyrophilin 2a2 (Btn2a2) expression on thymic epithelial cells promotes central T cell tolerance and prevents autoimmune disease. J Autoimmun. (2023) 139:103071. doi: 10.1016/j.jaut.2023.103071
72. Huang Y, Han F, Li J, Li Y, Gao J, Lai L, et al. BTN2A2-Ig protein inhibits the differentiation of pathogenic Th17 cells and attenuates EAE in mice. Immunol Lett. (2023) 260:58–67. doi: 10.1016/j.imlet.2023.06.009
73. Peedicayil A, Vierkant RA, Hartmann LC, Fridley BL, Fredericksen ZS, White KL, et al. Risk of ovarian cancer and inherited variants in relapse-associated genes. PloS One. (2010) 5:e8884. doi: 10.1371/journal.pone.0008884
74. Le Page C, Marineau A, Bonza PK, Rahimi K, Cyr L, Labouba I, et al. BTN3A2 expression in epithelial ovarian cancer is associated with higher tumor infiltrating T cells and a better prognosis. PloS One. (2012) 7:e38541. doi: 10.1371/journal.pone.0038541
75. Vavassori S, Kumar A, Wan GS, Ramanjaneyulu GS, Cavallari M, El Daker S, et al. Butyrophilin 3A1 binds phosphorylated antigens and stimulates human gammadelta T cells. Nat Immunol. (2013) 14:908–16. doi: 10.1038/ni.2665
76. Riano F, Karunakaran MM, Starick L, Li J, Scholz CJ, Kunzmann V, et al. Vgamma9Vdelta2 TCR-activation by phosphorylated antigens requires butyrophilin 3 A1 (BTN3A1) and additional genes on human chromosome 6. Eur J Immunol. (2014) 44:2571–6. doi: 10.1002/eji.201444712
77. Sandstrom A, Peigne CM, Leger A, Crooks JE, Konczak F, Gesnel MC, et al. The intracellular B30.2 domain of butyrophilin 3A1 binds phosphoantigens to mediate activation of human Vgamma9Vdelta2 T cells. Immunity. (2014) 40:490–500. doi: 10.1016/j.immuni.2014.03.003
78. Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. (2015) 21:938–45. doi: 10.1038/nm.3909
79. Benyamine A, Le Roy A, Mamessier E, Gertner-Dardenne J, Castanier C, Orlanducci F, et al. BTN3A molecules considerably improve Vgamma9Vdelta2T cells-based immunotherapy in acute myeloid leukemia. Oncoimmunology. (2016) 5:e1146843. doi: 10.1080/2162402X.2016.1146843
80. Benyamine A, Loncle C, Foucher E, Blazquez JL, Castanier C, Chretien AS, et al. BTN3A is a prognosis marker and a promising target for Vgamma9Vdelta2 T cells based-immunotherapy in pancreatic ductal adenocarcinoma (PDAC). Oncoimmunology. (2017) 7:e1372080. doi: 10.1080/2162402X.2017.1372080
81. Zocchi MR, Costa D, Vene R, Tosetti F, Ferrari N, Minghelli S, et al. Zoledronate can induce colorectal cancer microenvironment expressing BTN3A1 to stimulate effector gammadelta T cells with antitumor activity. Oncoimmunology. (2017) 6:e1278099. doi: 10.1080/2162402X.2016.1278099
82. Vantourout P, Laing A, Woodward MJ, Zlatareva I, Apolonia L, Jones AW, et al. Heteromeric interactions regulate butyrophilin (BTN) and BTN-like molecules governing gammadelta T cell biology. Proc Natl Acad Sci U.S.A. (2018) 115:1039–44. doi: 10.1073/pnas.1701237115
83. Karunakaran MM, Willcox CR, Salim M, Paletta D, Fichtner AS, Noll A, et al. Butyrophilin-2A1 directly binds germline-encoded regions of the vgamma9Vdelta2 TCR and is essential for phosphoantigen sensing. Immunity. (2020) 52:487–498 e6. doi: 10.1016/j.immuni.2020.02.014
84. Rigau M, Ostrouska S, Fulford TS, Johnson DN, Woods K, Ruan Z, et al. Butyrophilin 2A1 is essential for phosphoantigen reactivity by gammadelta T cells. Science. (2020) 367. doi: 10.1126/science.aay5516
85. Kumari R, Hosseini ES, Warrington KE, Milonas T, Payne KK. Butyrophilins: dynamic regulators of protective T cell immunity in cancer. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms24108722
86. Tanaka Y, Morita CT, Tanaka Y, Nieves E, Brenner MB, Bloom BR. Natural and synthetic non-peptide antigens recognized by human gamma delta T cells. Nature. (1995) 375:155–8. doi: 10.1038/375155a0
87. Chen ZW, Letvin NL. Vgamma2Vdelta2+ T cells and anti-microbial immune responses. Microbes Infect. (2003) 5:491–8. doi: 10.1016/S1286-4579(03)00074-1
88. Gober HJ, Kistowska M, Angman L, Jeno P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med. (2003) 197:163–8. doi: 10.1084/jem.20021500
89. Mullen PJ, Yu R, Longo J, Archer MC, Penn LZ. The interplay between cell signalling and the mevalonate pathway in cancer. Nat Rev Cancer. (2016) 16:718–31. doi: 10.1038/nrc.2016.76
90. D’Cruz AA, Babon JJ, Norton RS, Nicola NA, Nicholson SE. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. (2013) 22:1–10. doi: 10.1002/pro.2185
91. Willcox CR, Vantourout P, Salim M, Zlatareva I, Melandri D, Zanardo L, et al. Butyrophilin-like 3 directly binds a human vgamma4(+) T cell receptor using a modality distinct from clonally-restricted antigen. Immunity. (2019) 51:813–825 e4. doi: 10.1016/j.immuni.2019.09.006
92. Yang Y, Li L, Yuan L, Zhou X, Duan J, Xiao H, et al. A structural change in butyrophilin upon phosphoantigen binding underlies phosphoantigen-mediated vgamma9Vdelta2 T cell activation. Immunity. (2019) 50:1043–1053 e5. doi: 10.1016/j.immuni.2019.02.016
93. Hsiao CC, Nguyen K, Jin Y, Vinogradova O, Wiemer AJ. Ligand-induced interactions between butyrophilin 2A1 and 3A1 internal domains in the HMBPP receptor complex. Cell Chem Biol. (2022) 29:985–995.e5. doi: 10.1016/j.chembiol.2022.01.004
94. Karunakaran MM, Subramanian H, Jin Y, Mohammed F, Kimmel B, Juraske C, et al. A distinct topology of BTN3A IgV and B30.2 domains controlled by juxtamembrane regions favors optimal human γδ T cell phosphoantigen sensing. Nat Commun. (2023) 14:7617. doi: 10.1038/s41467-023-41938-8
95. Yuan L, Ma X, Yang Y, Qu Y, Li X, Zhu X, et al. Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate Vgamma9Vdelta2 T cells. Nature. (2023) 621:840–8. doi: 10.1038/s41586-023-06525-3
96. Fischer K, Bradlerova M, Decker T, Supper V. Vgamma9+Vdelta2+ T cell control of Listeria monocytogenes growth in infected epithelial cells requires butyrophilin 3A genes. Sci Rep. (2023) 13:18651. doi: 10.1038/s41598-023-45587-1
97. Cano CE, Pasero C, De Gassart A, Kerneur C, Gabriac M, Fullana M, et al. BTN2A1, an immune checkpoint targeting Vgamma9Vdelta2 T cell cytotoxicity against Malignant cells. Cell Rep. (2021) 36:109359. doi: 10.1016/j.celrep.2021.109359
98. Liu Y, Lui KS, Ye Z, Fung TY, Chen L, Sit PY, et al. EBV latent membrane protein 1 augments gammadelta T cell cytotoxicity against nasopharyngeal carcinoma by induction of butyrophilin molecules. Theranostics. (2023) 13:458–71. doi: 10.7150/thno.78395
99. Afrache H, Gouret P, Ainouche S, Pontarotti P, Olive D. The butyrophilin (BTN) gene family: from milk fat to the regulation of the immune response. Immunogenetics. (2012) 64:781–94. doi: 10.1007/s00251-012-0619-z
100. Du Y, Peng Q, Cheng D, Pan T, Sun W, Wang H, et al. Cancer cell-expressed BTNL2 facilitates tumour immune escape via engagement with IL-17A-producing gammadelta T cells. Nat Commun. (2022) 13:231. doi: 10.1038/s41467-021-27936-8
101. Kim YS, Lee SH, Park AH, Wu C, Hong BK, Jung H, et al. BTN1A1 is a novel immune checkpoint mutually exclusive to PD-L1. J Immunother Cancer. (2024) 12. doi: 10.1136/jitc-2023-008303
102. Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. (2005) 37:357–64. doi: 10.1038/ng1519
103. Suzuki T, Kilbey A, Casa-Rodriguez N, Lawlor A, Georgakopoulou A, Hayman H, et al. beta-catenin drives butyrophilin-like molecule loss and gammadelta T-cell exclusion in colon cancer. Cancer Immunol Res. (2023) 11:1137–55. doi: 10.1158/2326-6066.CIR-22-0644
104. Krawczyk M, Seguin-Estevez Q, Leimgruber E, Sperisen P, Schmid C, Bucher P, et al. Identification of CIITA regulated genetic module dedicated for antigen presentation. PloS Genet. (2008) 4:e1000058. doi: 10.1371/journal.pgen.1000058
105. Wong D, Lee W, Humburg P, Makino S, Lau E, Naranbhai V, et al. Genomic mapping of the MHC transactivator CIITA using an integrated ChIP-seq and genetical genomics approach. Genome Biol. (2014) 15:494. doi: 10.1186/s13059-014-0494-z
106. Hernandez-Quiles M, Broekema MF, Kalkhoven E. PPARgamma in metabolism, immunity, and cancer: unified and diverse mechanisms of action. Front Endocrinol (Lausanne). (2021) 12:624112. doi: 10.3389/fendo.2021.624112
107. Charlesworth JC, Peralta JM, Drigalenko E, Goring HH, Almasy L, Dyer TD, et al. Toward the identification of causal genes in complex diseases: a gene-centric joint test of significance combining genomic and transcriptomic data. BMC Proc. (2009) 3 Suppl 7:S92. doi: 10.1186/1753-6561-3-S7-S92
108. Lin X, Peng C, Greenbaum J, Li ZF, Wu KH, Ao ZX, et al. Identifying potentially common genes between dyslipidemia and osteoporosis using novel analytical approaches. Mol Genet Genomics. (2018) 293:711–23. doi: 10.1007/s00438-017-1414-1
109. Hosseinzadeh N, Mehrabi Y, Daneshpour MS, Zayeri F, Guity K, Azizi F. Identifying new associated pleiotropic SNPs with lipids by simultaneous test of multiple longitudinal traits: An Iranian family-based study. Gene. (2019) 692:156–69. doi: 10.1016/j.gene.2019.01.007
110. Bauer S, Aeissen V, Bubeck AM, Kienes I, Ellwanger K, Scheurenbrand M, et al. NLRC5 affects diet-induced adiposity in female mice and co-regulates peroxisome proliferator-activated receptor PPARgamma target genes. iScience. (2023) 26:106313. doi: 10.1016/j.isci.2023.106313
111. Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. (2006) 3:429–38. doi: 10.1016/j.cmet.2006.04.013
112. Sundaram B, Pandian N, Kim HJ, Abdelaal HM, Mall R, Indari O, et al. NLRC5 senses NAD(+) depletion, forming a PANoptosome and driving PANoptosis and inflammation. Cell. (2024) 187. doi: 10.1016/j.cell.2024.05.034
Keywords: NLRC5, CIITA, MHC, butyrophilins, Vγ9Vδ2 T cells, phosphoantigens
Citation: Brunschwiler F, Nakka S, Guerra J and Guarda G (2024) A Ménage à trois: NLRC5, immunity, and metabolism. Front. Immunol. 15:1426620. doi: 10.3389/fimmu.2024.1426620
Received: 01 May 2024; Accepted: 19 June 2024;
Published: 05 July 2024.
Edited by:
Thomas Herrmann, Julius Maximilian University of Würzburg, GermanyReviewed by:
Pierre Vantourout, King’s College London, United KingdomCopyright © 2024 Brunschwiler, Nakka, Guerra and Guarda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Greta Guarda, Z3JldGEuZ3VhcmRhQGlyYi51c2kuY2g=; Jessica Guerra, amVzc2ljYS5ndWVycmFAaXJiLnVzaS5jaA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.