- 1State Key Laboratory of Cell Differentiation and Regulation, College of Life Science, Pingyuan Laboratory, Henan Normal University, Xinxiang, China
- 2National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, National Institute of Parasitic Diseases at Chinese Center for Disease Control and Prevention, Chinese Center for Tropical Diseases Research, Shanghai, China
- 3Key Laboratory of Parasite and Vector Biology, National Health Commission of the People’s Republic of China, Shanghai, China
- 4World Health Organization Collaborating Centre for Tropical Diseases, Shanghai, China
- 5The School of Global Health, Chinese Center for Tropical Diseases Research, Shanghai Jiao Tong University School of Medicine, Shanghai, China
Background: Schistosomiasis is a zoonotic parasitic disorder induced by the infestation of schistosomes, a genus of trematodes. MicroRNAs (miRNAs) in egg-derived exosomes are crucial for modulating the host’s immune responses and orchestrating the pathophysiological mechanisms. Although the exosomes secreted by S. japonicum contain abundant miRNAs, the specific roles of these miRNAs in the pathogenesis of schistosomiasis-induced hepatic fibrosis are yet to be comprehensively elucidated. The egg exosomes of S. japonicum secrete miRNA-30, a novel miRNA.
Methods: In vitro, the effect of miRNA-30 was evaluated by transfecting HSCs with miRNA mimics. The target gene biosignature for miRNA-30 was predicted using the miRDB software. The effect of miRNA-30 in hepatic fibrosis was evaluated by either elevating its expression in healthy mice or by inhibiting its activity in infected mice by administration of recombinant adeno-associated virus serotype eight vectors expressing miRNA-30 or miRNA sponges.
Results: This novel miRNA can activate hepatic stellate cells (HSCs), the prinary effector cells of hepatic fibrosis, in vitro, i.e., it significantly increases the fibrogenic factors Col1(α1), Col3(α1), and α-SMA at both mRNA and protein levels. In addition, miRNA-30 may activate HSCs by targeting the host RORA gene. In addition, in vivo experiments were conducted by administering a recombinant adeno-associated viral vector to modulate the expression levels of miRNA-30. The overexpression of miRNA-30 in healthy mice significantly elevated the expression of Col1(α1), Col3(α1), and α-SMA at both the transcriptomic and proteomic scales. This overexpression was coupled with a pronounced augmentation in the hepatic hydroxyproline content. Conversely, the in vivo silencing of miRNA-30 in infected mice induced a considerable reduction in the size of hepatic granulomas and areas of collagen deposition. Hence, in vivo, modulation of miRNA-30 expression may play a pivotal role in ameliorating the severity of hepatic fibrosis in mice afflicted with S. japonica.
Conclusions: The study results suggest that miRNA-30 may augment schistosomiasis-induced hepatic fibrosis through a probable interaction with the host RORA. Our study may improve the current theoretical framework regarding cross-species regulation by miRNAs of hepatic fibrosis in schistosomiasis.
Introduction
Schistosomiasis, a zoonotic infection, is caused by the parasitic flatworm Schistosoma spp (1). It has been reported from across 78 tropical and subtropical regions (2). It is a neglected tropical disease, even though it impacts an estimated 230 million people globally (3). The parasite lays its eggs within host tissues or the portal venous system, which triggers various immunopathological reactions in the host, such as the inflammation and blockage of the urinary system, multiple digestive issues such as diarrhea and bloody stools, as well as the inflammation of the liver and spleen, including liver fibrogenesis (notably in infections by Schistosoma mansoni and S. japonicum) (4, 5). The S. japonicum is reported to be historically pervasive in China. Although China has made substantial progress in the mitigation of schistosomiasis over the past decade, 112 counties (inclusive of cities and districts) of the country are still affected by the parasite. Presently, schistosomiasis transmission is primarily managed by using praziquantel (PZQ), an anthelmintic that is mainly effective against adult schistosomal parasites. It is ineffective against other developmental stages of the parasite (6). Therefore, the therapeutic resolution of schistosomiasis remains a formidable global health challenge (7).
Hepatic fibrosis is characterized by fibrous scars produced through the aggregation of extracellular matrix (ECM) proteins, which predominantly comprise cross-linked collagen types I and III. In a fibrotic liver, these fibrous scars substitute the damaged normal hepatic tissue (8, 9). Myofibroblasts are the principal components of the ECM in fibrotic livers. Hence, mitigating efforts for fibrosis must target myofibroblasts (10, 11). S. japonicum larvae infect their human hosts by direct penetration through the skin. Once the larvae reach the adult stage, they lay eggs, eliciting an intensified immune response in the host against the soluble egg antigen (SEA) (12, 13). This intense immune reaction induces the proliferation of hepatic stellate cells (HSCs), which subsequently transform into myofibroblasts (9, 14). This conversion is accompanied by an extensive synthesis of ECM, resulting in secondary hepatic fibrosis and cirrhosis (15, 16). Upon stimulation by the SEA, HSCs up-regulate the expression of α-SMA. Activated HSCs migrate to the injury site and secrete ECM, resulting in fibrous scarring (17, 18). Several cytokines directly stimulate Col1(α1) transcription in activated HSCs. Transforming growth factor-beta (TGF-β) is the most potent pro-fibrotic cytokine and drives smad2/smad3-dependent HSC activation (18, 19).
MicroRNAs (miRNAs) are short endogenous non-coding RNAs (18–25 nucleotides) that modulate post-transcriptional gene expression by binding to the 3′-untranslated region (3′-UTR) or open reading frame of target gene mRNAs (20, 21). Note that miRNAs are involved in a myriad of biological processes, including cell proliferation, differentiation, migration, and disease initiation and progression (21–25). They are attracting increasing research interest because of their role in extracellular vesicles, particularly exosomes, which are critical to intercellular communication (26, 27). miRNAs delicately regulate gene expression involved in fibrogenesis (28). Recent investigations have highlighted the critical role of host miRNAs in schistosomiasis-induced hepatic fibrosis (29, 30). During the course of the disease, S. japonicum continues to secrete exosomes that carry numerous worm-derived miRNAs (31, 32). These exosomes, upon release, deliver the worm-derived miRNAs to host cells, thereby influencing host immunity and pathology (32).
We have conducted the sequencing of S. japonicum egg exosomes in a previous study. We have also reported various novel miRNAs potentially involved in the regulation of hepatic fibrosis in the host. Building upon the previous report, we herein identify miRNA-30 as a particularly promising candidate for further investigation.
Materials and methods
Ethics statement
All animal experiments were performed in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals (approved by the State Council of the People’s Republic of China). Efforts were made to minimize suffering. The study was approved by the Laboratory Animal Welfare & Ethics Committee (LAWEC) of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention (Chinese Center for Tropical Diseases Research; approval ID: IPD 2019–12).
Cell culture and treatment
Human HSCs of the LX-2 line were acquired from Wuhan Servicebio Technology Co. Ltd. (China). Mouse HSCs (mHSCs) were procured from Zhejiang Meisen Cell Technology Co. Ltd. (China). For the propagation of LX-2 cells, a high-sugar Dulbecco’s Modified Eagle Medium (DMEM, 11995073, Gibco, USA) augmented with 10% fetal bovine serum (FBS, 10091148, Gibco, USA) and 1% penicillin-streptomycin (PS, 15070063, Gibco, USA) was employed. The mHSCs were cultivated in commercially obtained Dulbecco’s/F-12 medium (D/F-12, CTCC-002-035, Meisen, China). Both LX-2 and mHSCs were maintained under physiological conditions conducive to cellular growth, specifically within an incubator at 37°C and in an atmosphere comprising 5% CO2.
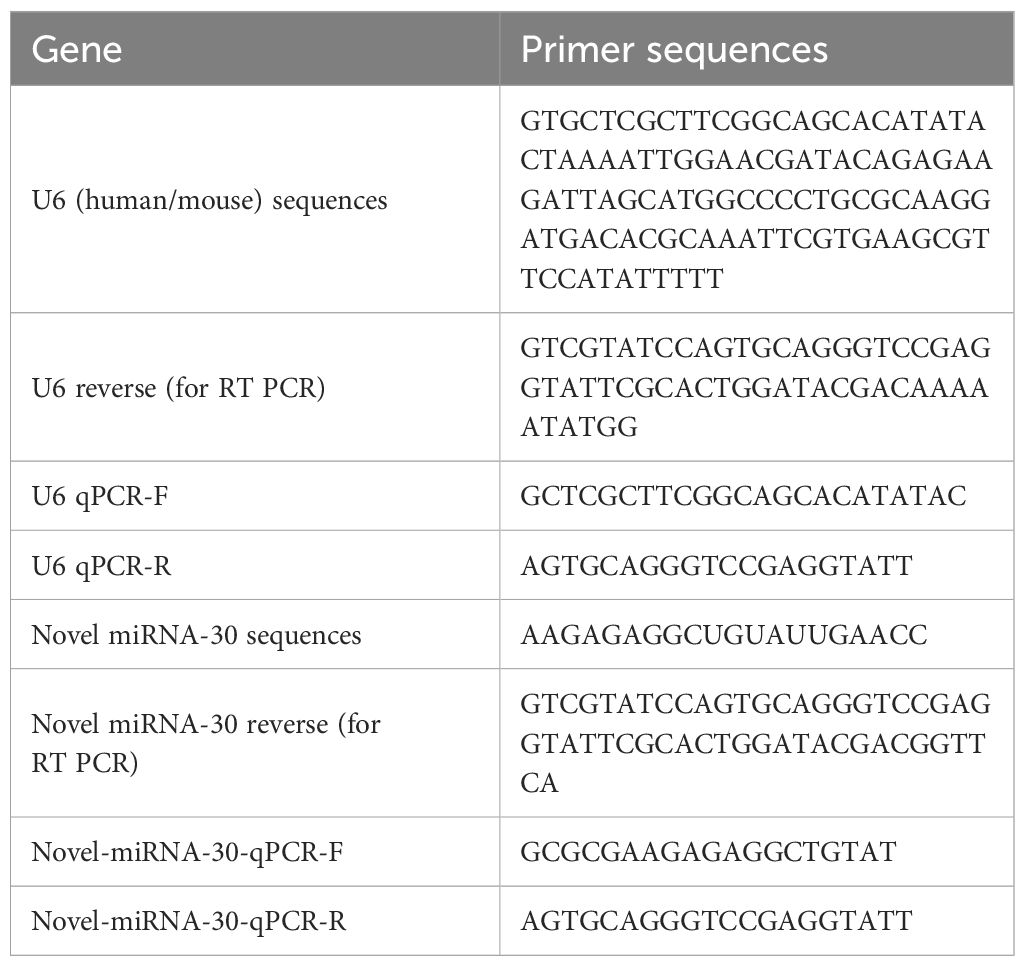
A novel mimic of miRNA-30, synthesized by RiboBio (Guangzhou, China), was utilized for the transfection of both LX-2 and mHSC lines. The transfection was initiated when 30–50% of cellular confluence was achieved. Manufacturer-provided protocols were adhered to during the transfection process, wherein the cells were exposed to either a negative control (NC) mimic or miR-30 mimic at concentrations determined optimal for each cell type (LX-2: 100 and 150 nM; mHSC: 50 nM) and subsequently cultured for 48 h. The detailed miRNA sequences and primer sequences are shown in Table 1.
Animals and parasite infections
All experimental animals were purchased from Shanghai Jihui Laboratory Animal Care Co., Ltd. (China) and were raised under specific pathogen-free conditions. All animal surgeries were performed under pentobarbital sodium-induced anesthesia to minimize pain and distress. The S. japonicum cercariae were supplied by the Vector Unit of the National Institute of Parasitic Diseases, which is part of the Chinese Center for Disease Control and Prevention under the National Research Centre for Tropical Diseases.
For the mass acquisition of S. japonicum eggs, three female New Zealand white rabbits were percutaneously exposed to 800 ± 20 S. japonicum cercariae. The rabbits were raised for 6 weeks and subsequently livers were harvested for the isolation of S. japonicum eggs. Fresh eggs were cultured for 1 week, the supernatant was collected every day, and the exosomes were separated by ultracentrifugation.
Fifty-four 6-week-old female C57BL/6 mice were randomly and equally divided into three groups. These mice were injected with 150 μL of PBS, rAAV8-SCR (blank vector), and rAAV8-miR-30 (overexpression vector) into the tail vein, with a viral titer of 1×1012 vector genomes per mouse. Then, on days 42, 56, and 70 post-injection, the mice were euthanized and liver samples were collected.
Fifty-four 6-week-old female C57BL/6 mice were randomly and equally allocated into three distinct groups and injected, via the tail vein, with 150 µL of PBS, rAAV8-SCR (blank vector), or rAAV8-anti-miR-30 (silencing vector), each at a viral load of 1×1012 vector genomes per mouse. At three weeks post-injection, the mice were percutaneously exposed to 20 ± 1 S. japonicum cercariae. On days 42 and 56 post-infection, these mice were euthanized, and the liver samples were extracted.
rAAV8 design and production
This study employed rAAV8-SCR, rAAV8-miR-30, and rAAV8-anti-miR-30 that were designed, generated, purified, and titered by Vigene Biosciences (Shandong, China).
Dual-luciferase reporter gene assay
The target gene biosignature for miRNA-30 was predicted using the miRDB software. Wild-type (WT) and 3′-UTR mutant reporter vectors for the NT5E and RORA genes were synthesized, and the dual-luciferase assay was executed.
rAAV8 design and production
This study employed rAAV8-SCR, rAAV8-miR-30, and rAAV8-anti-miR-30 that were designed, generated, purified, and titered by Vigene Biosciences (Shandong, China).
Western blot
Cellular and tissue specimens were lysed using the RIPA buffer with protease inhibitors. The protein concentrations were determined using the BCA Protein Assay Kit (PC0020, Solarbio, China) after conducting a thorough cell disruption. The samples were then heated for 10 min with 6 × Protein Loading Buffer. Next, 20–30 μg of protein was added per lane and the proteins were separated on SurePAGE™ (M00657, Genscript, China). After their separation, the proteins were transferred to a PVDF membrane, blocked with protein-free fast-blocking solution (PS108, Epizyme, China) for 30 min to prevent non-specific binding, and washed three times with 1 × Tris-buffered saline containing Tween 20 (1 × TBST). The membrane was then incubated with rabbit anti-GAPDH (5174S, CST, USA), anti-Col1(α1) (bs-10423R, Bioss Antibodies, USA), anti-Col3(α1) (bs-0548R, Bioss Antibodies), or anti-α-SMA (19245S, CST, USA) antibodies overnight at 4°C, followed by three 1×TBST washes. Then, the membrane was incubated with secondary antibodies (S0B4002, Starter, China) at room temperature for 1.5 h, followed by three 1 × TBST washes. Immunoreactive bands were detected using a ChemiDoc MP Imaging System (Bio-Rad, USA) and visualized with Tanon High-sig ECL Western Blotting Substrate (SQ201, Epizyme, China). The quantification and analysis were performed using ImageJ software to assess the protein expression levels. Antibody information is listed in Table 2.
Quantitative polymerase chain reaction
Total RNA from the cell and tissue samples was extracted using the trizol method. After measuring the total RNA concentration using NanoDrop™ 2000 software, reverse transcription was conducted according to the HyperScript™ III RT SuperMix for qPCR with gDNA Remover (R202-02, EnzyArtisan, China). Reverse transcription of miR-30 was performed according to the HyperScript™ III miRNA 1st Strand cDNA Synthesis Kit (by stem-loop) (R601, EnzyArtisan, China).
All the cDNA was analyzed by quantitative polymerase chain reaction (qPCR) using a 2 × S6 Universal SYBR qPCR Mix kit (Q204-05, EnzyArtisan, China). The endogenous control was U6 snRNA or GAPDH. The 2-△△t method was used to calculate the fold change in the expression of all mRNAs and miRNAs. The primers used in this study are listed in Table 3.
Determination of hydroxyproline content
We accurately weighed 30–100 mg of liver tissue using the hydroxyproline assay kit (A030-2-1, Nanjing Jiancheng Bioengineering Institute, China).
Statistical analysis
Statistical analysis was conducted using GraphPad Prism 9.0. Student’s t-tests were applied to assess differences between two groups. One-way analysis of variance (ANOVA), followed by Tukey’s post-tests, was used for three or more groups. Data were given as mean ± standard deviation and were shown as error bars for all experiments. Differences were considered significant at P < 0.05. The reported data were obtained from at least three biological replicates.
Results
Activation of HSCs by miRNA-30 derived from S. japonicum egg exosomes
As shown in (Figure 1A), we first identified egg-derived exosomes using transmission electron microscopy (TEM) and then extracted them through overspeed centrifugation. Next, we performed high-throughput sequencing and obtained a large number of miRNA sequences. From this pool of miRNA sequences, we identified six sequences associated with S. japonicum infection. We synthesized miRNA mimics for enhanced cellular miRNA enrichment in subsequent experimentations. These mimics were transfected into the LX-2 cell line. After 48 h, the total RNA was extracted to assess changes in the α-SMA levels—a hallmark of hepatic fibrosis. Based on the results obtained, we identified three types of novel miRNAs, including miRNA-33 (33) previously reported to promote HSC activation, capable of significantly up-regulating the α-SMA transcriptomic scales (Figure 1B). In contrast, other miRNAs, such as miRNA-15, miRNA-3, and miRNA-124-3p, had negligible effects on LX-2 cells. The other two novel miRNAs, miRNA-30 and miRNA-68, increased the transcription of α-SMA in LX-2 cells, suggesting their potential involvement in S. japonicum infection-induced hepatic fibrosis. In the subsequent investigations, we probed the regulatory role of miRNA-30 in S. japonicum-induced hepatic fibrosis.
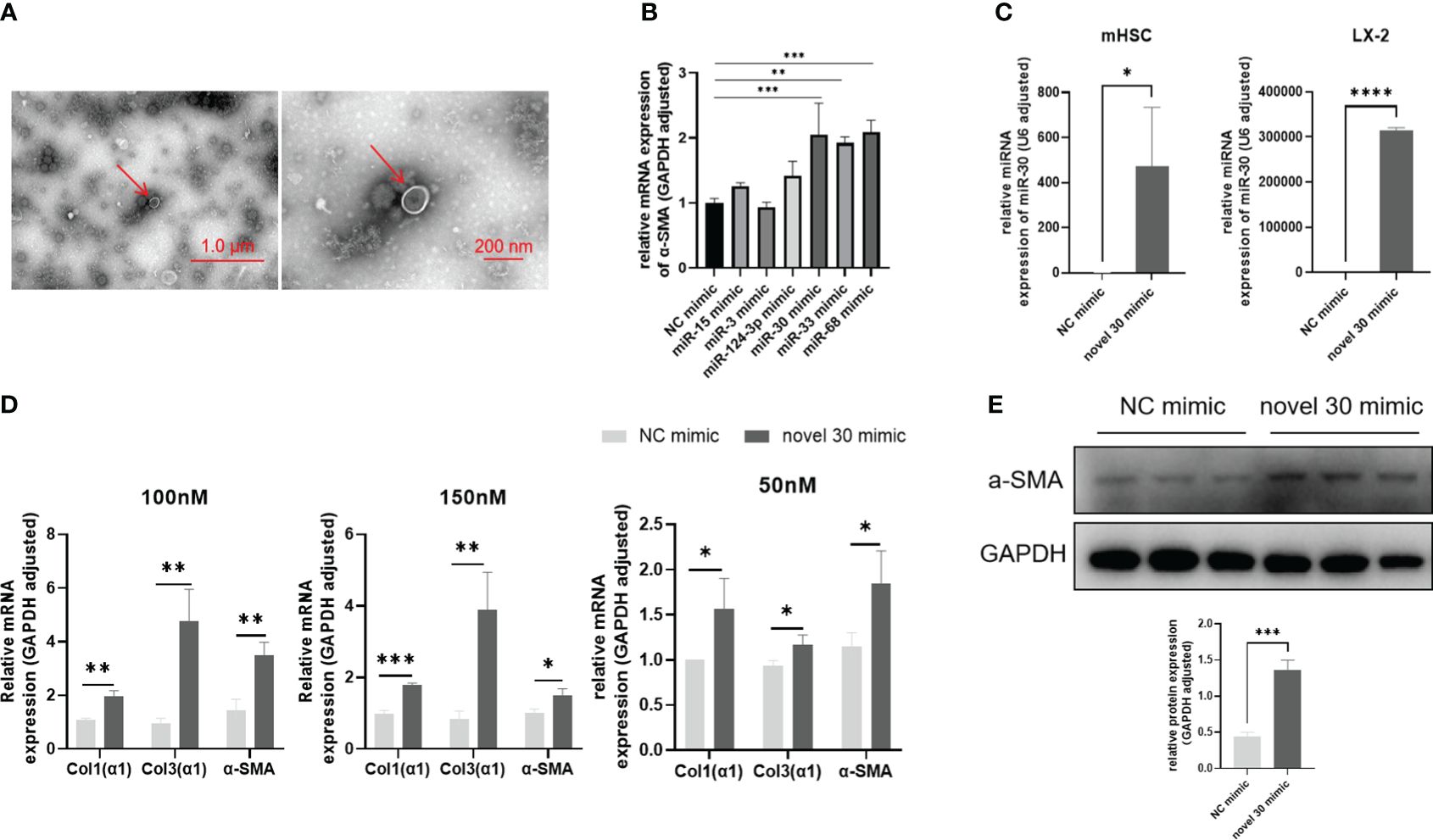
Figure 1 miRNA-30 derived from S. japonicum egg exosomes activates HSCs. (A) Exosomes were observed under transmission electron microscopy (TEM). (B) LX-2 cells were transfected with miRNA mimics for 48 h, followed by qPCR analysis to measure the mRNA levels of α-SMA. (C) LX-2 and mHSC cells were transfected with 50 nM of both NC mimic and miRNA-30 mimic, for 24 (h) Relative transcript levels of miRNA-30 were determined by qPCR. (D) LX-2 and mHSC cells were transfected with varying doses of NC mimic and miR-30 mimic for 48 h, followed by qPCR to quantify the mRNA levels of α-SMA, Col1(α1), and Col3(α1). (E) LX-2 cells were exposed to a 100 nM of both NC mimic and miRNA-30 mimic for 48 (h) The LX-2 cells were then subjected to Western blot analysis to detect their α-SMA protein expression levels. Both mRNA and protein levels of α-SMA, Col1(α1), and Col3(α1) were normalized against GAPDH. GAPDH, 37 kDa; α-SMA, 43 kDa. NC, negative control; 30 mimic, miRNA-30 mimic. The results are averaged from three independent experiments. Student’s t-tests were carried out to assess differences between the two groups (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
We initially assessed the transfection efficiency and cellular tolerance of miRNA-30 mimic. First, the LX-2 cells and mHSCs were transfected with an NC mimic and miRNA-30 mimic, for 24 h. Following the transfection, total RNA was extracted from both cell lines, and miRNA-30 levels were quantified via qPCR using the stem-loop method. The results demonstrated successful transfection and sustained presence of miRNA mimics within the HSCs of different species (human and mouse) (Figure 1C). Subsequently, we determined the optimal concentration of miRNA-30 mimic in both cell types. The LX-2 cells tolerated up to 150 nM of the miRNA mimics (Figure 1D), whereas >50 nM of miRNA mimics induced cell death in mHSCs (Figure 1D). Hence, we employed 50 nM of the miRNA-30 mimic for subsequent experimentations with mHSCs.
To further elucidate the activating influence of miRNA-30 on HSCs, we re-transfected the LX-2 cells with 100 and 150 nM of the NC mimic and miRNA-30 mimic. Total RNA was extracted after 48 h of transfection, and alterations in the hepatic fibrotic factor levels were assessed through qPCR. Our findings revealed that both 100 and 150 nM concentrations of miRNA-30 mimic led to an up-regulation of Col1(α1), Col3(α1), and α-SMA at the mRNA level (Figure 1D). Next, we conducted a Western blot analysis of the total protein extracted from the LX-2 cells transfected with 100 nM of both the NC mimic and miRNA-30 mimic to ascertain the upregulation of α-SMA at the protein level (Figure 1E). After establishing the ability of miRNA-30 to promote LX-2 activation, we extended our investigation to mHSCs. Remarkably, treatment with 50 nM of miRNA-30 mimic also resulted in the upregulation of Col1(α1), Col3(α1), and α-SMA at the mRNA level (Figure 1D).
Validation of the target genes of miRNA-30
MiRNA-30 target genes were predicted using the MicroRNA Target Prediction Database (miRDB), which revealed numerous potential target genes in both human and mouse models. The results showed 624 genes scored above 50 in humans and 651 in mice. Common to human and mouse, the top-scoring 42 target genes were validated by qPCR following the transfection of LX-2 cells with the NC and miRNA-30 mimics for 48 h in vitro. Among these, the mRNA levels of nine genes, including USB1, NT5E, RORA, SDK1, and IL10, were significantly up-regulated (Figure 2A), suggesting their involvement in the miRNA-30-mediated regulation of host hepatic fibrosis.
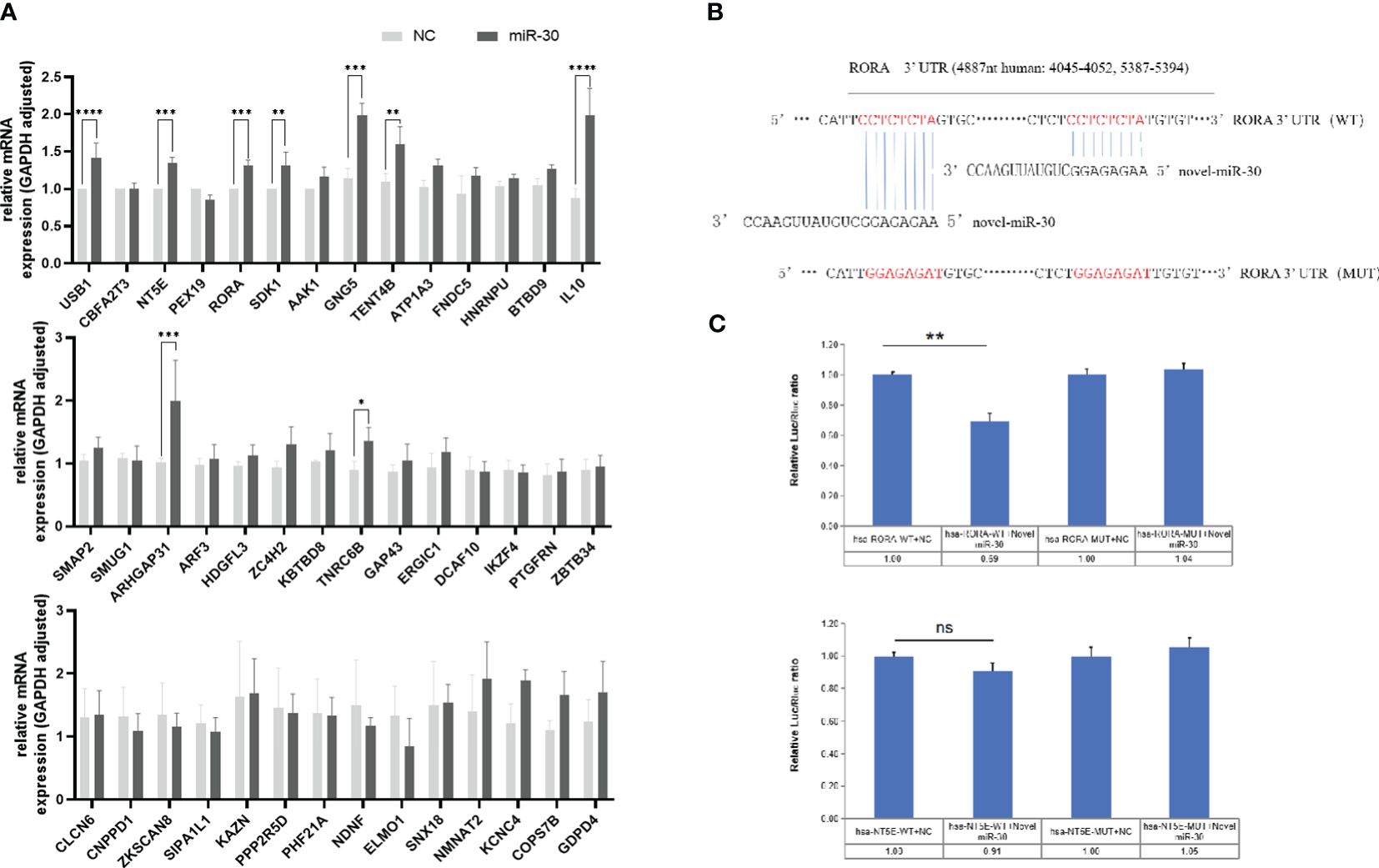
Figure 2 MiRNA-30 of S. japonicum egg exosome promotes the progression of hepatic fibrosis by targeting the host RORA gene in HSCs. (A) LX-2 cells were transfected with 150 nM of NC and miR-30 mimics for 48 (h) The mRNA expression of the first 42 candidate genes was measured by qPCR. (B) Sequence alignment was performed between miR-30 and the 3’-UTR of candidate gene RORA, which revealed two binding sites. (C) Validation of genes RORA and NT5E dual-luciferase target. The mRNA levels of 42 potential targets were normalized to GAPDH. NC, negative control; 30 mimic, miRNA-30 mimic. The results are averaged from three independent experiments. Student’s t-tests were employed to assess differences between the two groups (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001) ns, no significance.
Based on the prediction software scores, the retinoic acid receptor (RAR)-related orphan receptor alpha (RORA) and 5’-nucleotidase ecto (NT5E) genes were selected for further investigations. To verify whether RORA and NT5E are the direct target genes of miRNA-30, we cloned the 2 target genes 3’-UTR-indicated sequence into the pmiR-GLO dual luciferase reporter vector, respectively. As shown in (Figure 2B), two RORA gene sites are complementary to eight bases of miRNA-30 sequence. Co-transfection of tool cells (293T cells) with vectors constructed from both RORA and NT5E genes and miRNA-30 mimics or NC significantly reduced the luciferase activity in the cells co-transfected with the RORA WT vector and miRNA-30 mimics compared to that of the NC-treated cells (Figure 2C). Conversely, mutation of the seed-binding sequence in the 3’-UTR of the RORA gene removed this inhibition (Figure 2C). While binding of the NT5E gene to miRNA-30 decreased the luciferase activity, it did not reach significance (Figure 2C), suggesting an interaction between miRNA-30 and the 3’-UTR of LX-2 target gene RORA mRNA.
Promotion of hepatic fibrosis in mice by S. japonicum egg exosome-derived miRNA-30
To further investigate whether miRNA-30 could also contribute to the progression of hepatic fibrosis in mice, we conducted in vivo overexpression experiments by injecting rAAV8-miR-30 into the tail veins of healthy female C57BL/6 mice. Subsequently, we monitored dynamic changes in the hepatic fibrosis-related indicators.
Upon entry into the cell, recombinant adeno-associated virus (rAAV) can persist as additional DNA, exhibiting a stable presence within the cell. Typically, the gene expression peaks around weeks 2–3 post-entry and can endure for weeks to months. We injected the rAAV8-miR-30, rAAV8-SCR NC, and PBS blank control into the tail vein of C57BL/6 mice at a dosage of 5×1011 viral genomes per mouse. We obtained the liver and serum samples from the mice three weeks after the injection. Stem-loop reverse transcription of miRNA-30 followed by qPCR analysis showed that the sera of all the three groups (rAAV8-miR-30, rAAV8-SCR NC, and PBS blank control) did not exhibit overexpression of miRNA-30 (Figure 3A). Conversely, a marked upregulation of miRNA-30 was observed in the livers of the mice injected with rAAV8-miR-30 (Figure 3A), affirming the accurate targeting of the liver and processing of miRNA-30 within the livers of the mice. Additionally, the livers of the mice injected with the rAAV8-SCR NC exhibited no discernible variance compared to that shown by the PBS group, indicating that the recombinant adeno-associated viral empty vector did not exert any additional influence on the processing and maturation of miRNA-30 (Figure 3A). Consequently, the rAAV8-miR-30 overexpression group and the rAAV8-SCR NC were employed for the subsequent experiments.
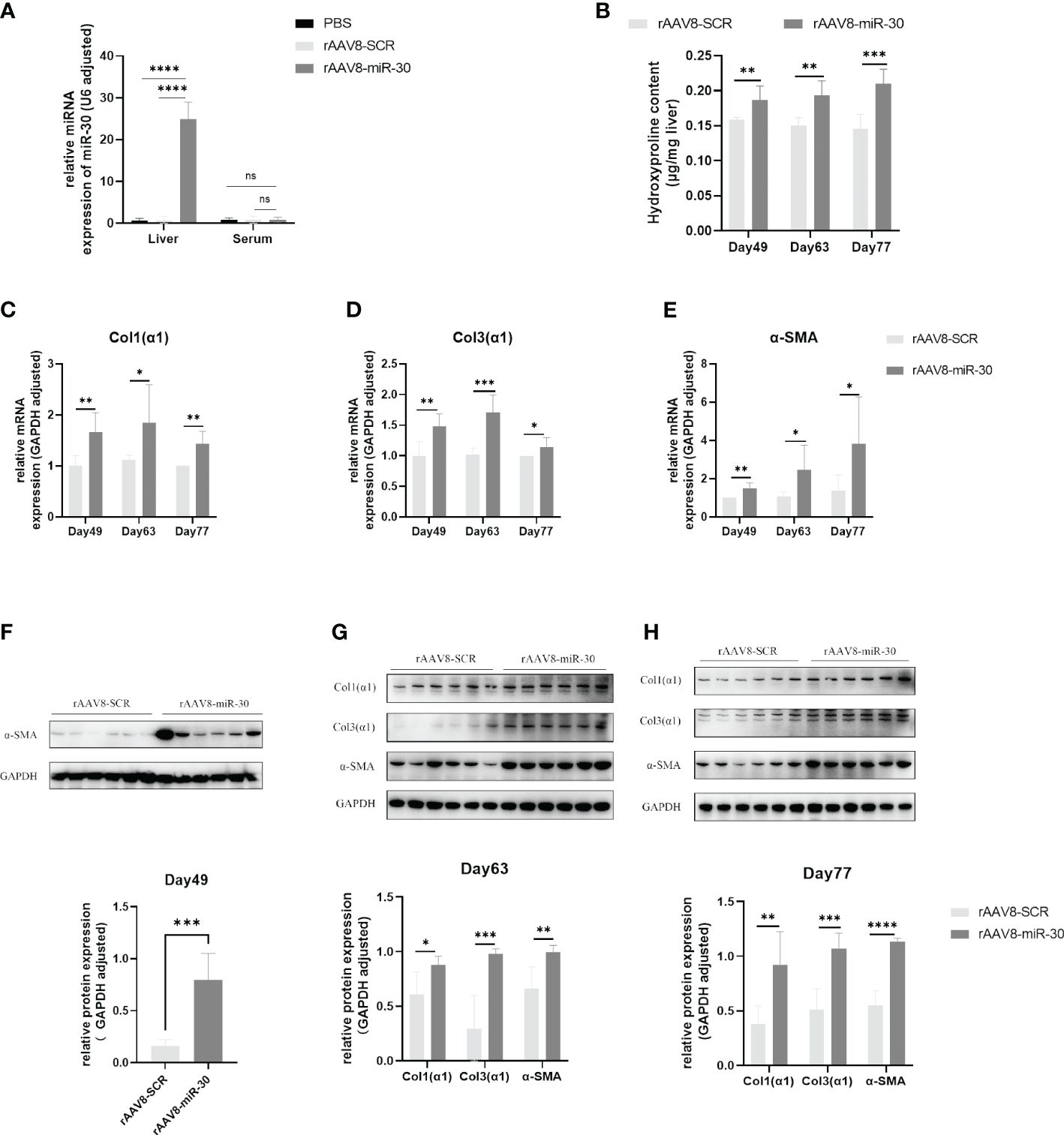
Figure 3 Overexpression of miRNA-30 induces hepatic fibrosis in naive mice. Healthy six-week-old female C57BL/6 mice were injected with of PBS, rAAV8-SCR (blank vector), and rAAV8-miR-30 (overexpression vector) into the tail vein, with a viral titer of 5 × 10 (11) or 1 × 10 (12) vector genomes per mouse. (A) Liver and serum samples were collected three weeks post-injection to measure the miRNA-30 expression through qPCR, with normalization to U6 RNA (n = 6). (B) Liver tissues harvested on days 49, 63, and 77 post-injection were analyzed for hydroxyproline content (n = 6). (C–E) qPCR to quantify the mRNA levels of α-SMA, Col1(α1), and Col3(α1) (n = 6). (F–H) Western blot to quantify the protein levels of α-SMA, Col1(α1), and Col3(α1) (n = 6). Both mRNA and protein levels of α-SMA, Col1(α1), and Col3(α1) were normalized against GAPDH. GAPDH, 37 kDa; α-SMA, 43 kDa; Col1(α1), 130 kDa; Col3(α1), 117 kDa. rAAV8, recombinant adeno-associated virus serotype 8; SCR, scrambled. Statistical significance between two or more groups was evaluated using Student’s t-test and one-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
We increased the viral titer to 1×1012 viral genomes per mouse and injected it into the tail vein of the C57BL/6 mice. The mice were euthanized and their liver tissue was collected on days 49, 63, and 77 post-injection to assess hepatic fibrosis-related markers. Our findings revealed a significant increase in the hydroxyproline content in the rAAV8-miR-30 overexpression group compared to that in the rAAV8-SCR NC group at all three time points (Figure 3B). Furthermore, the qPCR analysis of Col1(α1), Col3(α1), and α-SMA mRNA levels in the rAAV8-miR-30-induced mouse liver tissues demonstrated a substantial exacerbation of fibrosis at all three time points compared to that observed in the rAAV8-SCR NC group (Figures 3C–E). The Western blot results also corroborated this trend (Figures 3F–H). A comprehensive analysis of the hydroxyproline content and hepatic fibrosis factors (Col1(α1), Col3(α1), and α-SMA) at both the mRNA and protein levels over time revealed a progressive aggravation of hepatic fibrosis in mice following the in vivo overexpression of miRNA-30.
Inhibition of miRNA-30 obtained from S. japonicum egg-derived exosomes delays the progression of schistosomiasis-induced hepatic fibrosis in mice
After confirming the pro-fibrotic role of miRNA-30 in the WT female C57BL/6 mice, we hypothesized that inhibiting miRNA-30 could be a therapeutically promising option for schistosomiasis-induced hepatic fibrosis. To delve deeper into its involvement in S. japonicum-induced hepatic fibrosis, we administered rAAV8-anti-miR-30 to female C57BL/6 mice infected with S. japonicum. This in vivo approach aimed to silence miRNA-30 and assess its impact on the regulatory and therapeutic aspects of hepatic fibrosis in schistosomiasis. We employed the rAAV8-anti-miR-30 sponge (sponge vector) as the inhibitor. This sponge vector binds to miR-30 and inhibits its function. The potency and specificity of the sponge vector were validated in 293T cell lines.
We injected rAAV8-anti-miR-30 and rAAV8-SCR into the tail vein of 6-week-old healthy female C57BL/6 mice, while simultaneously exposing them percutaneously to 20 ± 1 S. japonicum cercariae. The liver tissues of these infected mice were then collected for immunohistochemistry, Western blot, and Masson trichrome staining analyses on days 42 and 56 post-infection. Masson trichrome staining demonstrated a notable decrease in the hepatic collagen areas and granuloma size in the mice injected with the rAAV8-anti-miR-30 sponge vector compared to those injected with the SCR control vector (Figure 4A). Western blotting results showed the down-regulation of α-SMA at the protein level (Figure 4B), while the number of S. japonicum eggs was not statistically different between the two groups (Figure 4C). Immunohistochemical analysis revealed significant reductions in the areas of Col1(α1), Col3(α1), and α-SMA on days 42 and 56 post-infection in the rAAV8-anti-miR-30-treated group compared to that in both the NC and PBS groups (Figures 4D, E). Moreover, qPCR results also showed the same trend (Figure 4F).
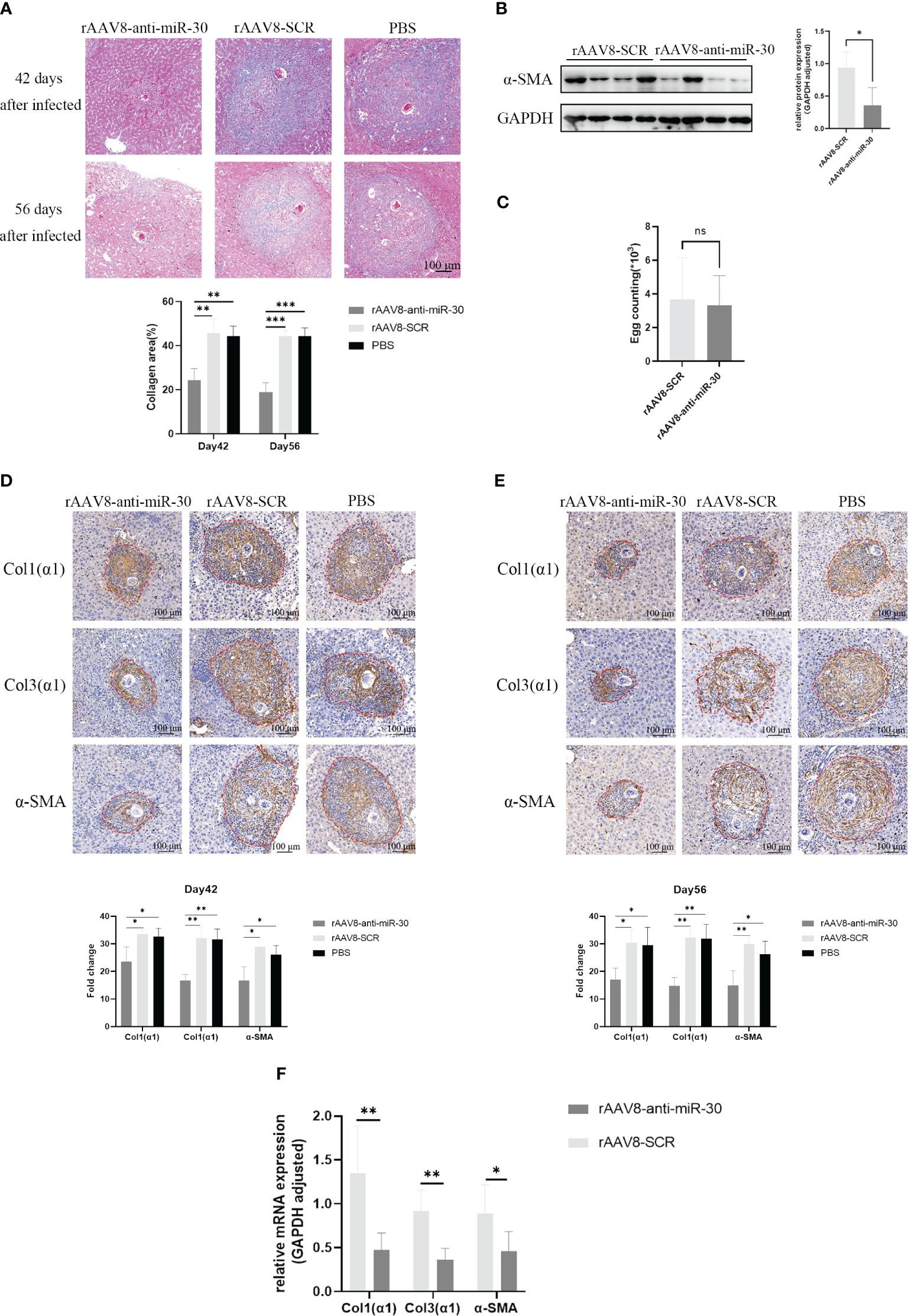
Figure 4 Inhibition of miRNA-30 from S. japonicum egg-derived exosomes delays the progression of hepatic fibrosis in schistosomiasis in mice. Mice received an injection of rAAV8-SCR or rAAV8-anti-miR-30-sponge vectors at a dose of 1 × 10 (12) virus genomes or PBS via the tail vein. After 3 weeks, these mice were dermally exposed to 20 ± 1 S. japonicum cercariae. Liver samples were collected at days 42 and 56 post-infection. (A) Granulomas and collagen deposition were assessed through Masson’s trichrome staining. (B) Expression of α-SMA at the protein level was examined on day 42 post-infection through Western blot analysis. (C) Egg count in mouse liver tissue showing there was no statistical difference between the two groups (n = 6). (D, E) Immunohistochemical analysis on days 42 and 56 post-S. japonicum infection revealed positively stained areas for Col1(α1), Col3(α1), and α-SMA. (F) qPCR to quantify the mRNA levels of α-SMA, Col1(α1), and Col3(α1) (n = 6). All images were magnified 10-fold. Quantification of α-SMA protein levels was normalized to GAPDH. GAPDH, 37 kDa; α-SMA, 43 kDa. rAAV8, recombinant adeno-associated virus serotype 8; SCR, scrambled. Statistical significance between two or more groups was evaluated using Student’s t-test and one-way ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001. ns, no significance.
Discussion
HSCs, constituting approximately 8–10% of total liver cells, are the precursor of principal fibroblasts in the liver (34). Their activation is crucial for initiating and advancing hepatic fibrosis, as they are responsible for the reduced degradation of the ECM and pathological deposition (35). Therefore, it is imperative to understand HSC activation. Various stimuli trigger HSC activation under pathological conditions, such as physical, chemical, viral, or biological insults, causing their transition into myofibroblasts characterized by α-SMA expression and ECM synthesis (36–38). Exogenous agents, such as exosomes or miRNAs, can also induce HSC activation. In our previous work, we have discussed the critical regulatory role of miRNAs in hepatic fibrosis (39). Herein, we investigate the activating effect of miRNA-30 mimics on HSCs via transfection. While higher doses of miRNA mimics (>50 nM) induced cell death in mHSCs, possibly because of the more demanding isolation and culture requirements of primary mHSCs, LX-2 cells could tolerate increased transfection doses. The subsequent analysis revealed significant upregulation of Col1(α1), Col3(α1), and α-SMA at both the mRNA and protein levels, indicating enhanced expression of fibrosis-associated factors in response to miRNA-30. These findings demonstrate that miRNA-30 derived from S. japonicum exosomes activates both human and mouse HSCs, promoting the expression of fibrosis-associated factors. Hence, we hypothesize that miRNA-30 exhibits cross-species activation potential.
In contrast to the conventional negative regulation of target genes by miRNAs (40), our study revealed that the S. japonicum egg exosome-derived miRNA-30 can trigger the activation of host HSCs by up-regulating its target gene, RORA, within host cells, which accelerates the fibrotic process. While miRNA-30 does not participate in the degradation or repression of target genes, its role in enhancing the stability of target genes or their translational processes cannot be overlooked. This phenomenon can be attributed to miRNAs regulating target genes or cell cycle arrest, where microRNPs toggle between the repression and activation of translation (41). In addition, miRNAs have been documented to boost the transcriptional and translational stability of mRNAs in viruses and eukaryotes alike (42). Thus, our findings offer insights into the potential positive regulation of host target genes by miRNAs across species.
RORA, an orphan receptor, is known to have a role in the regulation of various physiological processes, including metabolism, cellular immunity, inflammatory responses, and circadian rhythms (43, 44). This receptor operates across several human organs, such as the brain, heart, liver, testis, and skin (45). The RORA gene yields four isoforms through selective promoter and exon splicing: RORa1, RORa2, RORa3, and RORa4 (44). While these isoforms share common DNA and putative ligand-binding domains, they differ in their amino-terminal sequences, leading to slight variations in DNA-binding preferences (43, 46). Notably, RORa4 is the predominant isoform of hepatic nuclear receptor RORA. Its transcriptional activity can be modulated by various external stimuli in the HepG2 human hepatocellular carcinoma cells, thereby influencing disease progression (47). We propose that miRNA-30 derived from S. japonicum egg exosomes may enhance the transcriptional stability of RORa4 by targeting it in host HSCs, thereby promoting hepatic fibrosis under S. japonica infection. However, further investigation is warranted to elucidate the specific pro-fibrotic mechanisms involved.
The progression of hepatic pathology under S. japonicum infection is a multifaceted process, primarily characterized by the formation of granuloma and subsequent hepatic fibrosis. Apart from SEA, exosomes, their secreted proteins, and RNAs also play critical roles in hepatic fibrosis process (31, 48). All these events involve an intricate dysregulation of gene expression, including miRNAs, within the infected liver tissues. In a previous study, we showed that the miRNA-30 originating from S. japonicum egg exosomes can activate HSCs, the principal effector cells in hepatic fibrosis, in both human and murine models in vitro. Herein, our study validates the capability of S. japonicum egg exosomes to activate HSCs, which in turn influences hepatic fibrosis. The findings reveal that the overexpression of miRNA-30, which is exacerbated over time, substantially increases the number of fibrosis markers in the livers of healthy mice. Hence, the miRNA-30 derived from S. japonicum egg exosomes can induce hepatic fibrosis in mice. Furthermore, inhibition of miRNA-30 in infected mice using the rAAV8-anti-miR-30 sponge vector led to a reduction in both the granuloma area and the collagenous area surrounding liver granulomas, which delayed the progression of hepatic fibrosis. We have previously shown that, similar to miRNA-33, miRNA-30 also promotes hepatic fibrosis across species (39). In addition, miRNA-68 (Section 3.1) is capable of activating HSCs in vitro. Hence, the role of miRNA-68 in schistosomiasis-induced hepatic fibrosis should be further investigated.
Understanding the mechanisms underlying hepatic fibrosis has long been a primary focus of schistosomiasis research. Hepatic fibrosis in schistosomiasis is characterized as an immunopathologic condition. SEA, a key regulator in the progression of schistosomiasis, can induce a type-2 immune response. Furthermore, pro-fibrotic cytokines, such as IL-13 and TGF-β1, released during the immune response, can activate HSCs and drive hepatic fibrosis via the SMAD pathway (9). The study findings indicate that miRNA-30, specific to S. japonicum egg exosomes, promotes hepatic fibrosis in schistosomiasis across species.
To conclude, this study showed that in parasitic infections such as those by Schistosoma spp., certain miRNAs exhibit pro-fibrotic properties, while others act as inhibitors, regardless of whether they originate from the host or the parasite. The involvement of miRNAs in regulating hepatic fibrosis, coupled with their stable and easily detectable nature, offers novel avenues for monitoring and treating fibrotic liver diseases. This study sheds light on the pro-fibrotic role of miRNA-30 in schistosomiasis and enhances our understanding of how parasite-derived miRNAs regulate host pathology. It identifies potential targets that intervene in the treatment of schistosomiasis-induced hepatic fibrosis. In addition, it offers novel insights into the pathomechanisms responsible for hepatic fibrosis resulting from S. japonicum infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Laboratory Animal Welfare & Ethics Committee (LAWEC) of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention (Chinese Center for Tropical Diseases Research). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YC: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. YH: Formal analysis, Methodology, Writing – review & editing. HZ: Formal analysis, Methodology, Writing – original draft. NJ: Methodology, Writing – original draft. YW: Methodology, Writing – original draft. JZ: Methodology, Writing – original draft. YS: Formal analysis, Supervision, Writing – review & editing. GY: Supervision, Writing – review & editing. JC: Conceptualization, Formal analysis, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Nature Science Foundation of China (Nos. 82272369 and 81971969 to JC). The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We are grateful to Professor Zhongying Yuan, Mr. Quan Chen, and Mr. Xiaocheng Zhang from the National Institute of Parasitic Diseases at the Chinese Center for Disease Control and Prevention (Chinese Center for Tropical Diseases Research) for their assistance with the experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1425384/full#supplementary-material
Supplementary Data Sheet 1 | Excel spreadsheet containing, in separate sheets, the numerical data for Figures 1B–E, 2A, 3A–H, 4B, C, F.
Supplementary Data Sheet 2 | Sourced data for the immunoblotting, TEM, masson staining, immunofluorescence, and sequence alignment are shown in Figures 1A, E, 2B, C, 3F–H, 4A, B, D–F.
References
1. Hu W, Brindley PJ, McManus DP, Feng Z, Han ZG. Schistosome transcriptomes: new insights into the parasite and schistosomiasis. Trends Mol Med. (2004) 10:217–25. doi: 10.1016/j.molmed.2004.03.002
2. Agatsuma T. Origin and evolution of Schistosoma japonicum. Parasitol Int. (2003) 52:335–40. doi: 10.1016/S1383-5769(03)00049-7
3. Colley DG, Secor WE. Immunology of human schistosomiasis. Parasite Immunol. (2014) 36:347–57. doi: 10.1111/pim.12087
4. Deol AK, Fleming FM, Calvo-Urbano B, Walker M, Bucumi V, Gnandou I, et al. Schistosomiasis - Assessing progress toward the 2020 and 2025 global goals. N Engl J Med. (2019) 381:2519–28. doi: 10.1056/NEJMoa1812165
5. Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. (2006) 368:1106–18. doi: 10.1016/S0140-6736(06)69440-3
6. Cai P, Mu Y, Olveda RM, Ross AG, Olveda DU, McManus DP. Circulating miRNAs as footprints for liver fibrosis grading in schistosomiasis. EBioMedicine. (2018) 37:334–43. doi: 10.1016/j.ebiom.2018.10.048
7. Gardner JMF, Mansour NR, Bell AS, Helmby H, Bickle Q. The discovery of a novel series of compounds with single-dose efficacy against juvenile and adult Schistosoma species. PloS Negl Trop Dis. (2021) 15:e0009490. doi: 10.1371/journal.pntd.0009490
8. Friedman SL. Liver fibrosis – from bench to bedside. J Hepatol. (2003) 38 Suppl 1:S38–53. doi: 10.1016/S0168-8278(02)00429-4
9. Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. (2008) 134:1655–69. doi: 10.1053/j.gastro.2008.03.003
10. Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. (2011) 6:425–56. doi: 10.1146/annurev-pathol-011110-130246
11. Schmitt-Gräff A, Krüger S, Bochard F, Gabbiani G, Denk H. Modulation of alpha smooth muscle actin and desmin expression in perisinusoidal cells of normal and diseased human livers. Am J Pathol. (1991) 138:1233–42.
12. LoVerde PT. Schistosomiasis. Adv Exp Med Biol. (2019) 1154:45–70. doi: 10.1007/978-3-030-18616-6_3
13. Liu J, Zhou B, Chen D, Zhou C, Shi Q, Zheng C, et al. Transjugular intrahepatic portosystemic shunt placement in patients with schistosomiasis-induced liver fibrosis. Cardiovasc Intervent Radiol. (2019) 42:1760–70. doi: 10.1007/s00270-019-02295-6
14. Gomperts BN, Strieter RM. Fibrocytes in lung disease. J Leukoc Biol. (2007) 82:449–56. doi: 10.1189/jlb.0906587
15. Fallowfield JA, Mizuno M, Kendall TJ, Constandinou CM, Benyon RC, Duffield JS, et al. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J Immunol. (2007) 178:5288–95. doi: 10.4049/jimmunol.178.8.5288
16. Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. (2021) 18:151–66. doi: 10.1038/s41575-020-00372-7
17. Iwaisako K, Jiang C, Zhang M, Cong M, Moore-Morris TJ, Park TJ, et al. Origin of myofibroblasts in the fibrotic liver in mice. Proc Natl Acad Sci U S A. (2014) 111:E3297–305. doi: 10.1073/pnas.1400062111
18. Koyama Y, Wang P, Liang S, Iwaisako K, Liu X, Xu J, et al. Mesothelin/mucin 16 signaling in activated portal fibroblasts regulates cholestatic liver fibrosis. J Clin Invest. (2017) 127:1254–70. doi: 10.1172/JCI88845
19. Xu F, Liu C, Zhou D, Zhang L. TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J Histochem Cytochem. (2016) 64:157–67. doi: 10.1369/0022155415627681
20. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
21. Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. (2011) 481:190–4. doi: 10.1038/nature10661
22. Gee HE, Camps C, Buffa FM, Colella S, Sheldon H, Gleadle JM, et al. MicroRNA-10b and breast cancer metastasis. Nature. (2008) 455:E8–9. doi: 10.1038/nature07362
23. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. (2008) 455:1124–8. doi: 10.1038/nature07299
24. Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. (2009) 137:1005–17. doi: 10.1016/j.cell.2009.04.021
25. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. (2007) 449:682–8. doi: 10.1038/nature06174
26. Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol. (2009) 21:575–81. doi: 10.1016/j.ceb.2009.03.007
27. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. (2010) 73:1907–20. doi: 10.1016/j.jprot.2010.06.006
28. He X, Xie J, Zhang D, Su Q, Sai X, Bai R, et al. Recombinant adeno-associated virus-mediated inhibition of microRNA-21 protects mice against the lethal schistosome infection by repressing both IL-13 and transforming growth factor beta 1 pathways. Hepatology. (2015) 61:2008–17. doi: 10.1002/hep.27671
29. Cabantous S, Hou X, Louis L, He H, Mariani O, Sastre X, et al. Evidence for an important role of host microRNAs in regulating hepatic fibrosis in humans infected with Schistosoma japonicum. Int J Parasitol. (2017) 47:823–30. doi: 10.1016/j.ijpara.2017.05.007
30. Zhao Y, Dang Z, Chong S. Mmu-miR-92a-2-5p targets TLR2 to relieve Schistosoma japonicum-induced liver fibrosis. Int Immunopharmacol. (2019) 69:126–35. doi: 10.1016/j.intimp.2019.01.007
31. He X, Wang Y, Fan X, Lei N, Tian Y, Zhang D, et al. A schistosome miRNA promotes host hepatic fibrosis by targeting transforming growth factor beta receptor III. J Hepatol. (2020) 72:519–27. doi: 10.1016/j.jhep.2019.10.029
32. Liu J, Zhu L, Wang J, Qiu L, Chen Y, Davis RE, et al. Schistosoma japonicum extracellular vesicle miRNA cargo regulates host macrophage functions facilitating parasitism. PloS Pathog. (2019) 15:e1007817. doi: 10.1371/journal.ppat.1007817
33. Wang Y, Fan X, Lei N, He X, Wang X, Luo X, et al. A MicroRNA derived from Schistosoma japonicum promotes schistosomiasis hepatic fibrosis by targeting host secreted frizzled-related protein 1. Front Cell Infect Microbiol. (2020) 10:101. doi: 10.3389/fcimb.2020.00101
34. Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Delivery Rev. (2017) 121:27–42. doi: 10.1016/j.addr.2017.05.007
35. Trivedi P, Wang S, Friedman SL. The power of plasticity-metabolic regulation of hepatic stellate cells. Cell Metab. (2021) 33:242–57. doi: 10.1016/j.cmet.2020.10.026
36. Kisseleva T, Brenner DA. Hepatic stellate cells and the reversal of fibrosis. J Gastroenterol Hepatol. (2006) 21 Suppl 3:S84–7. doi: 10.1111/j.1440-1746.2006.04584.x
37. Kordes C, Sawitza I, Götze S, Herebian D, Häussinger D. Hepatic stellate cells contribute to progenitor cells and liver regeneration. J Clin Invest. (2014) 124:5503–15. doi: 10.1172/JCI74119
38. Puche JE, Saiman Y, Friedman SL. Hepatic stellate cells and liver fibrosis. Compr Physiol. (2013) 3:1473–92. doi: 10.1002/cphy.c120035
39. Wang Y, Gong W, Zhou H, Hu Y, Wang L, Shen Y, et al. A novel miRNA from egg-derived exosomes of Schistosoma japonicum promotes liver fibrosis in murine schistosomiasis. Front Immunol. (2022) 13:860807. doi: 10.3389/fimmu.2022.860807
40. Liu M, Cho WC, Flynn RJ, Jin X, Song H, Zheng Y. microRNAs in parasite-induced liver fibrosis: from mechanisms to diagnostics and therapeutics. Trends Parasitol. (2023) 39:859–72. doi: 10.1016/j.pt.2023.07.001
41. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. (2007) 318:1931–4. doi: 10.1126/science.1149460
42. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. (2007) 23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406
43. Giguère V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. (1994) 8:538–53. doi: 10.1101/gad.8.5.538
44. Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-André M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. (1994) 8:757–70. doi: 10.1210/mend.8.6.7935491
45. Giguère V, McBroom LD, Flock G. Determinants of target gene specificity for ROR alpha 1: monomeric DNA binding by an orphan nuclear receptor. Mol Cell Biol. (1995) 15:2517–26. doi: 10.1128/MCB.15.5.2517
46. Matysiak-Scholze U, Nehls M. The structural integrity of ROR alpha isoforms is mutated in staggerer mice: cerebellar coexpression of ROR alpha1 and ROR alpha4. Genomics. (1997) 43:78–84. doi: 10.1006/geno.1997.4757
47. Chauvet C, Bois-Joyeux B, Danan JL. Retinoic acid receptor-related orphan receptor (ROR) alpha4 is the predominant isoform of the nuclear receptor RORalpha in the liver and is up-regulated by hypoxia in HepG2 human hepatoma cells. Biochem J. (2002) 364:449–56. doi: 10.1042/bj20011558
Keywords: schistosomiasis, Schistosoma japonicum, exosomes, microRNA-30, hepatic fibrosis
Citation: Chen Y, Hu Y, Zhou H, Jiang N, Wang Y, Zhang J, Shen Y, Yu G and Cao J (2024) Induction of hepatic fibrosis in mice with schistosomiasis by extracellular microRNA-30 derived from Schistosoma japonicum eggs. Front. Immunol. 15:1425384. doi: 10.3389/fimmu.2024.1425384
Received: 29 April 2024; Accepted: 24 June 2024;
Published: 30 July 2024.
Edited by:
Romulo Dias Novaes, Federal University of Alfenas, BrazilReviewed by:
Celio Geraldo Freire-de-Lima, Federal University of Rio de Janeiro, BrazilMartin Roderfeld, University of Giessen, Germany
Copyright © 2024 Chen, Hu, Zhou, Jiang, Wang, Zhang, Shen, Yu and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianping Cao, Y2FvanBAY2hpbmFjZGMuY24=; Y2FvanBjZGNAMTYzLmNvbQ==; Guoying Yu, Z3VveWluZ3l1QGh0dS5lZHUuY24=
 Yang Chen
Yang Chen Yuan Hu
Yuan Hu Hao Zhou1,2,3,4
Hao Zhou1,2,3,4 Yiluo Wang
Yiluo Wang Yujuan Shen
Yujuan Shen Guoying Yu
Guoying Yu Jianping Cao
Jianping Cao