
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 29 May 2024
Sec. Molecular Innate Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1424385
The nuclear-encoded mitochondrial protein Tu translation elongation factor, mitochondrial (TUFM) is well-known for its role in mitochondrial protein translation. Originally discovered in yeast, TUFM demonstrates significant evolutionary conservation from prokaryotes to eukaryotes. Dysregulation of TUFM has been associated with mitochondrial disorders. Although early hypothesis suggests that TUFM is localized within mitochondria, recent studies identify its presence in the cytoplasm, with this subcellular distribution being linked to distinct functions of TUFM. Significantly, in addition to its established function in mitochondrial protein quality control, recent research indicates a broader involvement of TUFM in the regulation of programmed cell death processes (e.g., autophagy, apoptosis, necroptosis, and pyroptosis) and its diverse roles in viral infection, cancer, and other disease conditions. This review seeks to offer a current summary of TUFM’s biological functions and its complex regulatory mechanisms in human health and disease. Insight into these intricate pathways controlled by TUFM may lead to the potential development of targeted therapies for a range of human diseases.
Tu translation elongation factor, mitochondrial (TUFM, also known as P43; EF-TU; EFTU; COXPD4; EF-Tu MT) is a nuclear-encoded mitochondrial protein participating in mitochondrial polypeptide biosynthesis. Of note, TUFM is one of the most abundant proteins in the mitochondria and the bacterial cell (1–3). The initial exploration of TUFM rooted in the study of Yeast Saccharomyces cerevisiae in 1970 (4). In 1990s, the subsequent investigation involved the examination of TUFM expression pattern in both Arabidopsis thaliana and Homo sapiens (5–7). TUFM proteins are conserved across a spectrum ranging from bacterial elongation factor Tu (EF-Tu) to eukaryotic TUFM homologs, and exhibit a higher degree of conservation in terms of structurally and functionally crucial amino acids (Figures 1A, B). S. cerevisiae TUFM and Escherichia coli EF-Tu are functionally interchangeable, showing notable nucleotide and amino acid homology, with percentages reaching 60% and 66%, respectively (4, 10, 11). In addition, distinctive structural variations are discernible among bacterial EF-Tu to eukaryotic TUFM homologs. The absence of the N terminus in bacterial EF-Tu distinguishes it from its eukaryotic counterparts TUFM, and bacterial EF-Tu possesses a distinctive KxKFxR motif (Figures 1A, B), serving as a pathogen-associated molecular pattern (PAMP) that elicits immune activation (12–15). A plausible scenario emerges wherein bacterial EF-Tu may be assimilated into host cells and undergo gradual evolution to assume the identity of TUFM. This evolutionary trajectory is marked by a functional shift from immune activation to immune suppression (14). Simultaneously, TUFM exhibits a significantly greater level of sequence divergence compared to its cytoplasmic counterparts, eukaryotic translation elongation factor 1 alpha (eEF1A) (Figure 1A), which is responsible for promoting recruitment of aminoacyl tRNAs to the ribosome (16, 17).
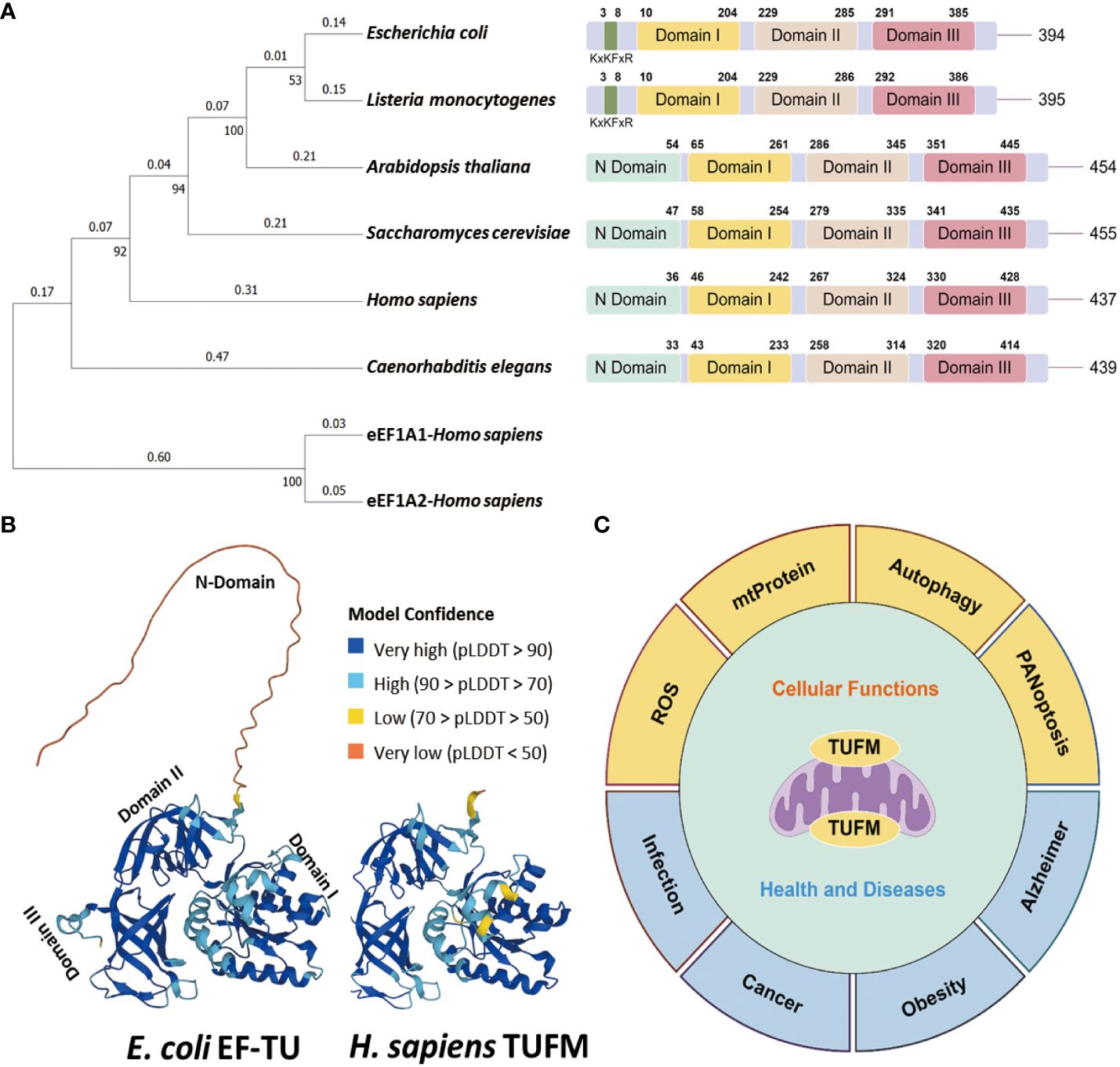
Figure 1 Conservation of TUFM protein. (A) Neighbor joining phylogenetic trees depicts the relationships among TUFM homologs (Left), and protein domain patterns of TUFM homologs juxtaposed with eEF1A (Right). (B) The AlphaFold 3D viewer showcases the structural comparison of TUFM proteins originated from E. coli (UniProt ID: P0CE47) and H. sapiens (UniProt ID: P49411) TUFM (8, 9). (C) Illustration of the diversified functions of TUFM, highlighting its multifaceted roles beyond mitochondrial protein translation and elongation.
The TUFM gene is located on chromosome 16p11.2, accompanied by a putative pseudogene or variant TUFML in close proximity to the centromere of chromosome 17. TUFM encompasses three distinct protein domains including N-terminal GTP-binding domain (domain I), domain II, and a C-terminal domain (domain III) (Figures 1A, B). Notably, domain I incorporates a mitochondrion targeting sequence (MTS). Functionally, TUFM forms a ternary complex with GTP and mitochondrial aminoacyl-tRNAs (aa-tRNA), delivers the aa-tRNA to the A-site of the ribosome, and plays a critical role in mediating translation of mitochondrial genes, which has been covered in detail in many other articles (18–21). In bacteria, EF-Tu is predominantly phosphorylated at serine/threonine residues, and phosphorylation at these universally conserved residues inhibits protein synthesis by preventing ternary complex formation (22–24). In eukaryotic cells, TUFM has been identified as a potential target of Src Family Kinases Fyn and c-Src (25–27). TUFM-tyrosin-266 is one of the major c-Src phosphorylation targets, and phosphorylation at this site, as well as its phosphomimic mutation to a glutamate residue, inhibits ternary complex formation (27).
Beyond its fundamental role in mitochondrial protein translation and elongation (28, 29), TUFM has garnered increasing attention for its involvement in various physiological and pathological processes (Figure 1C). This paper provides a comprehensive review of the diverse roles of TUFM in both health and disease, with a specific emphasis on its contributions to viral infection and oncogenesis.
Mitophagy is a selective type of macro-autophagy by which dysfunctional or excessive mitochondria are sequestered by autophagic vesicles for degradation. In contrast to type I interferon (IFN) production, which normally restricts viral infection, the function of autophagy could be either the antiviral or proviral role depending on the viruses and types of host cells. Typically, mitophagy is an active process that protects the host against viral infection via degrading invading viral particles, enhancing antigen presentation, or boosting inflammatory and non-inflammatory responses (30–32). Conversely, mitophagy can sometimes be exploited for viral replication, and virus-induced mitophagy has been demonstrated to attenuate IFN responses. At present, the majority of studies on TUFM in mitophagy tend to concentrate on the subject of viruses (Table 1, Figure 2).
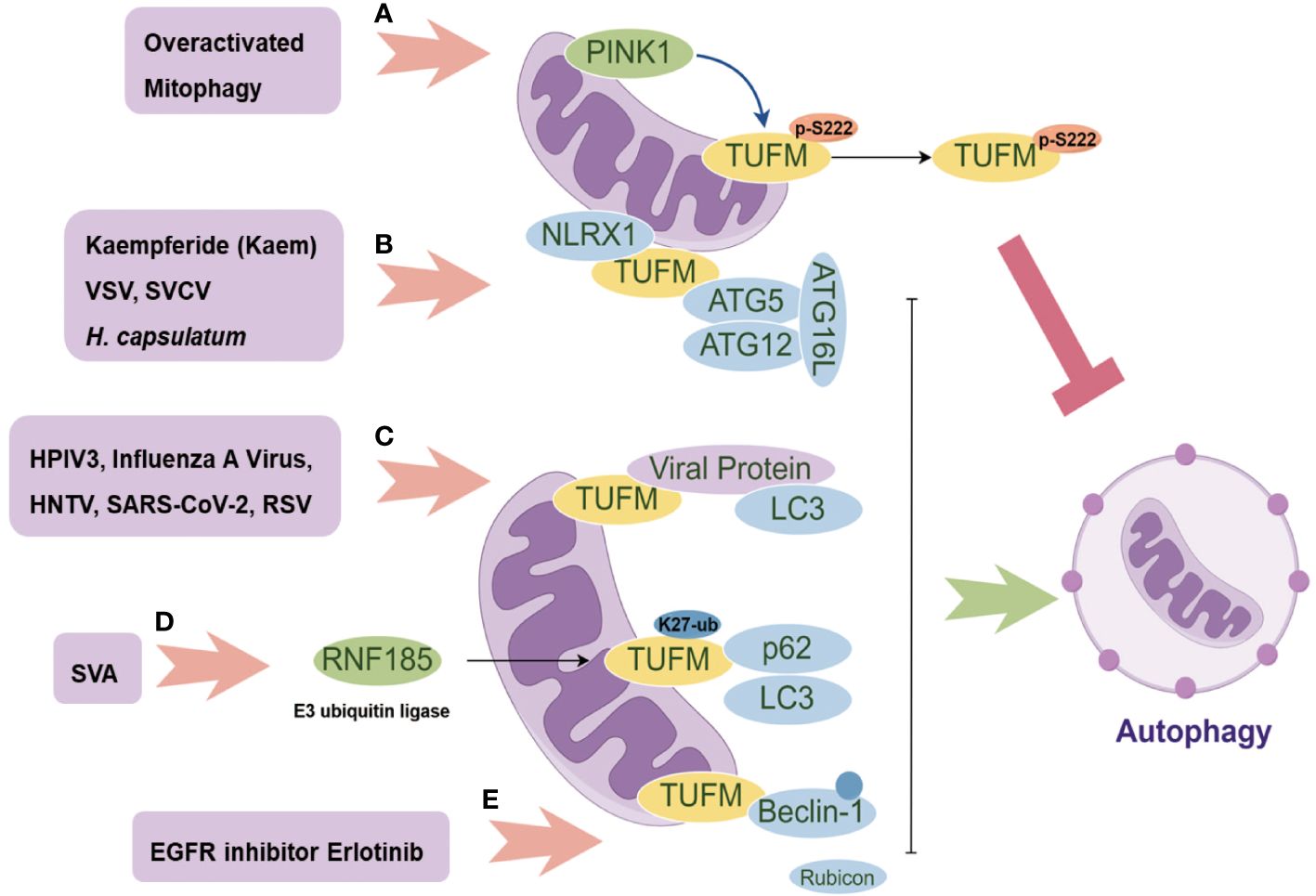
Figure 2 The Multifaceted Role of TUFM in Autophagy Modulation. PINK1 promotes the phosphorylation of TUFM at Serine222, leading to its delocalization from mitochondria to the cytoplasm and consequently releasing overactivated mitophagy (A); The mitochondrial proteins NLRX1 and TUFM form a complex that recruits the autophagy E3-like complex ATG5-ATG12-ATG16L1, thereby inducing autophagy (B); Several viral proteins can serve as autophagy adaptors, facilitating the interaction between TUFM and LC3 for autophagy induction (C); SVA promotes the K27-linked ubiquitination of TUFM, which in turn enhances its interaction with p62 and LC3, mediating mitophagy (D); Erlotinib promotes TUFM recruitment of Beclin-1 and its polyubiquitination, consequently disrupting its interaction with Rubicon for further autophagy induction (E).
TUFM plays different roles in modulating mitophagy. On the one hand, TUFM interacts with autophagy E3-like complex ATG12-ATG5 and ATG16L1, thus serving as a tether recruiting NLR Family Member X1 (NLRX1) and promoting mitophagy (14, 33). Additionally, TUFM and NLRX1 are comparable in inhibiting Retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) activation and promoting virus-mediated mitophagy that promotes vesicular stomatitis virus (VSV) infection (Figure 2B). Importantly, neither TUFM nor NLRX1 affect stimulator of interferon genes (STING, also known as MITA) pathway. TUFM proteins are conserved from fish to human (Figure 1), and mRNA levels of black carp TUFM (bcTUFM) are increased after spring viremia of carp virus (SVCV) infection (38). Similarly, bcTUFM cooperates with bcNLRX1 to inhibit mitochondrial antiviral-signaling protein (MAVS)-mediated antiviral signaling during SVCV infection (38) (Figure 2B). On the other hand, recent research has emphasized the precise modulation mechanism of TUFM in mitophagy, revealing that TUFM protein is a substrate of PINK1, which phosphorylates TUFM to suppress overactivated mitophagy (46). A PTEN-induced kinase 1 (PINK1)-dependent TUFM phosphoswitch at Serine222 determines conversion from activating to suppressing mitophagy. Mechanically, p-S222-TUFM is restricted predominantly to the cytosol, where it inhibits mitophagy by impeding ATG12-ATG5 formation (Figure 2A). Importantly, PINK1/TUFM is an evolutionarily conserved Parkin-independent pathway toward mitophagy.
Certain viral proteins induce complete mitophagy in the absence of Parkin, indicating the importance of TUFM in virus-induced mitophagy (Figure 2C), including influenza A virus PB1-F2, human parainfluenza virus (HPIV) M protein, Hantaan virus (HNTV) Gn protein, and respiratory syncytial virus (RSV) NS1 protein (34, 36, 39, 42). Substitution of glutamic acid (Glu, E) for lysine (Lys, K) at residue 627 of influenza A virus polymerase basic protein 2 (PB2) is regarded as overcoming host restriction and facilitate human infectivity (47). TUFM interacts with PB2627E as opposed to PB2627K (35, 48). By TUFM-dependent autophagy degradation for PB2627E, TUFM selectively restricts the replication of rWSN (a recombinant influenza A virus) PB2627E, while rWSN PB2627E induces TUFM-dependent autophagy. Conversely, TUFM is not involved in the replication of rWSN PB2627K or the autophagy that rWSN PB2627K induced (35). However, a recent study shows that TUFM benefits the replication of influenza A virus (39). Mechanically, influenza A virus PR8 PB1-F2 protein harbor a typical LIR motif, WxxL, at its C-terminal region and a TUFM interacting motif at its C terminal. The translocation of PB1-F2 to mitochondria is mediated by TUFM, and PB1-F2 in turn stimulates the interaction between TUFM and LC3, which mediates the induction of complete mitophagy. PB1-F2-induced mitophagy is critical for MAVS degradation and further type I IFN suppression.
Apart from influenza A virus, HPIV M protein (34), HTNV Gn protein (36), and RSV NS1 protein (42) translocate to mitochondria by interacting with TUFM, and then recruit LC3 to mitochondria for further TUFM and LC3 interaction (Figure 2C). Importantly, these interactions are required for mitophagy induction and further type I IFN inhibition. In addition, SARS-CoV-2 M protein interacts with TUFM, and autophagy is beneficial for SARS-CoV-2 replication (37) (Figure 2C), with unknown mechanism remained to be discovered. Notably, senecavirus A (SVA) 2C protein does not directly interact with LC3 under mitophagy induction status (41). SVA 2C protein promotes the K27-linked ubiquitination of TUFM catalyzed by the E3 ubiquitin ligase RNF185. Then, the ubiquitinated TUFM is recognized and bound by p62 (also called SQSTM1), which in turn interacts with LC3, thereby mediating complete mitophagy (Figure 2D). Of note, TUFM is discovered to interact directly with Beclin-1 and indirectly with the ATG12-ATG5 conjugate involved in mitophagosome formation (41).
African swine fever virus (ASFV) D1133 L and Japanese encephalitis virus (JEV) NS3 are both shown to interact with TUFM, while ASFV replication is considerably reduced by ectopic TUFM overexpression (40, 43). In addition, TUFM is involved in reactive oxygen species (ROS) inhibition (49, 50). Through a mechanism independent of ROS, NLRX1 facilitates LC3-Associated Phagocytosis (LAP) by interacting with TUFM which associates with autophagic proteins ATG5-ATG12 for LAPosome formation under fungus histoplasma capsulatum infection (Figure 2B). Of note, TUFM-mediated LAP induces MAPKs-AP-1 signal pathway activation for inflammatory cytokine response (51). In addition, Human Herpesvirus 8 (HHV-8) vIRF-1 is localized to mitochondria and stimulates TUFM dimerization for mitophagy induction which is associated with the inhibition of caspase-8-mediated apoptosis (44, 45) (Table 1).
Upregulation of TUFM has been reported in numerous types of tumors, including lung cancer (50), esophageal cancer (52), gastric cancer (53), intestinal cancer (54), bile duct cancer (55, 56), ovarian cancer (57, 58), pancreatic cancer (59), and others. Dysregulation of TUFM has been associated with various aspects of cancer progression, including tumor growth, metastasis, and therapeutic resistance (Figure 3).
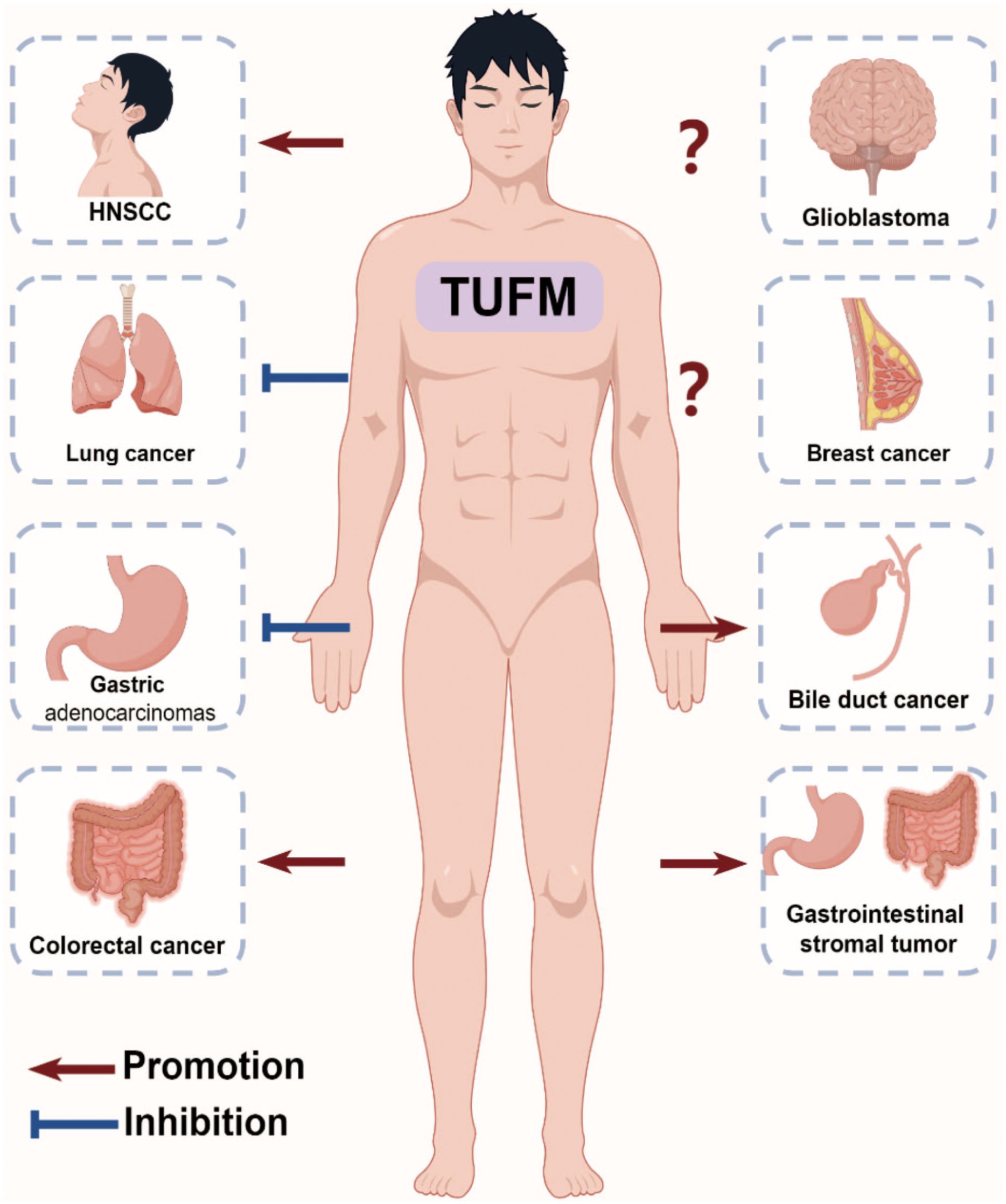
Figure 3 Role of TUFM in Cancer Progression. The multifunctional role of TUFM in various cancer progressions is evident, with overexpression in colorectal cancer leading to poorer clinical outcomes, promotion of proliferation and migration in gastrointestinal stromal tumors, negative correlations with serosal invasion and node involvement in gastric adenocarcinomas, promotion of autophagy and resistance to EGFR inhibitors in head and neck squamous cell carcinoma, inhibition of epithelial-mesenchymal transition in lung cancer, and controversial roles in glioblastoma and breast cancer.
Gastrointestinal cancer ranks top for incidence and second for mortality globally, constituting roughly 15.4% of all cancer cases (60). It has been well established that the majority of colorectal cancer (CRC) cases originate from adenomas (61, 62). Immunohistochemistry staining findings indicate a gradual increase in the expression of TUFM and tumor suppressor gene p53 from normal mucosa through adenoma to carcinoma tissues, correlating with the degree of dysplasia (63). Moreover, TUFM exhibits overexpression in CRC compared to matched normal colon mucosa, with minimal or weak expression in the stroma (54, 64). Increased TUFM expression in CRC correlates with poorer clinical outcomes, and serves as an independent prognostic factor for CRC patients. Specifically, TUFM expression significantly correlates with a higher 5-year recurrence rate in CRC patient, showing no significant association with additional clinical information, such as tumor node metastasis stage (54). TUFM is highly increased upon overexpression of microRNA-451a, a novel colorectal cancer-related gene (65). In addition, doxorubicin (DOX) and Berberine (BBR) have been shown to significantly suppress TUFM expression in CRC cells (66, 67). Mechanistically, TUFM is identified as a substrate of ubiquitin specific peptidase 5 (USP5). USP5 deubiquitinates TUFM at K48-linked chain and increases its stability, thereby promoting CRC cell growth, and TUFM, in turn, enhances the expression of Cyclin D1 (66). Consistent with the finding, both USP5 and TUFM expression are increased in CRC and are correlated with the prognosis of CRC patients.
Another in vitro analysis reveals that TUFM-knockdown results in decreased proliferation, as well as reduces lateral and vertical migration capacity of gastrointestinal stromal tumors (GISTs)-T1 and imatinib mesylate-resistant GIST-IR cells. In addition, there is an increase in the proportion of cells in the pre-G1 stage, with no alteration observed in the proportions of cells in the G1, S and G2/M stages in TUFM-knockdown GIST cells (68). In 5contrast to the reports mentioned above, high TUFM expression serves as a significant independent prognostic factor in gastric adenocarcinomas. TUFM is detected in both gastric adenocarcinoma and corresponding normal tissues, with high expression of TUFM also observed in gastric lymphocytes, vascular endothelial cells, and smooth muscle. Additionally, low expression of TUFM expression shows correlations with depth of serosal invasion and node involvement, and is associated with progression of gastric adenocarcinomas (53). Furthermore, TUFM expression varies among different gastric cancer cell lines, with high levels observed in SGC-7901, MKN-45, KatoIII, and AGS, while low levels are detected in MKN-28 and MKN-74 (53, 69).
Glioblastoma is the most common malignant primary brain tumor in adults, with the worst prognosis and overall survival approximately 15 months (70). TUFM has been proposed as a potential universal biomarker for glioblastoma (71–74). The protein levels of TUFM are found to be higher and comparable in glioblastoma tissue, neural stem cells (NSCs), glioblastoma stem cells (GSCs), and glioblastoma cell lines U251MG and U87MG, compared with normal brain tissue. Interestingly, the mRNA levels of TUFM are higher in GSCs, U251MG, and U87MG compared with normal brain tissue, glioblastoma tissue, and NSCs, of which TUFM mRNA expression does not exhibit significant differences between glioblastoma tissue and normal brain tissue (71). Significantly, the anti-TUFM nanobody Nb206 exhibits substantial cytotoxic effects, inducing apoptosis and necrosis, on the proliferation of GSCs and U87MG in particular, and to a lesser extent on U251MG (71). This finding is consistent with that genetic inhibition of TUFM-dependent mitochondrial translation suppresses GSCs proliferation (75). However, no significant reduction in cell growth is observed upon Nb206 treatment in NSCs, astrocytes, and HaCaT cells (71). These findings suggest that TUFM could play a significant role in promoting the progression and prognosis of glioblastoma. Although mRNA levels do not differ significantly between GBM and normal brain tissue, TUFM protein levels are higher in GBM tissue, indicating that post-transcriptional regulation of TUFM RNA could be a critical factor influencing its cancer-modulating function. Further studies are required to confirm these observations definitively.
Lung cancer stands as a leading cause of cancer death worldwide, with approximately 2 million new cases and 1.7 million deaths annually (60). Recent research shows that TUFM protein levels exhibits a progressive decrease with the advancement of lung adenocarcinoma (50). Intriguingly, despite TUFM knockdown reducing tumor growth in vivo, it induces Epithelial-to-mesenchymal transition (EMT), as evidenced by decreased levels of epithelial adhesion marker proteins ZO-1 and E-cadherin, and increased protein levels of mesenchymal marker fibronectin. This finding aligns with the observation that the protein levels of TUFM are found to be decreased during TGF-β1-induced EMT and apoptosis (50, 76). Furthermore, TUFM knockdown diminishes mitochondrial respiratory chain activity, escalates glycolysis and ROS production, and enhances migration, invasion, and metastasis of lung cancer. Mechanistically, TUFM knockdown induces EMT through activation of the AMPK-GSK3β/β-catenin pathway (50). Notably, TUFM knockdown also promotes autophagy, as indicated by the conversion of LC3 in lung cancer (50), which contrasts with the classical role of TUFM in autophagy modulation (14).
Breast cancer stands as the most commonly diagnosed cancer, with an estimated 2.3 million new cases and 6.8 million deaths annually (60). The current understanding of TUFM in breast cancer appears to be controversial. On one hand, TUFM knockdown induces EMT in MCF7 breast cancer cells, as evidenced by decreased levels of E-cadherin and γ-catenin (50). On the other hand, CBFB is found to interact with TUFM and enhances the binding of mitochondrial mRNAs to TUFM, thereby promoting breast cancer progression (77, 78). Interestingly, a novel resveratrol analog, HS-1793, exhibits anti-tumor activity in breast cancer by inhibiting the expression of mitochondrial translational protein TUFM and disrupting mitochondrial homeostasis, leading to sensitization of tumor cells to cell death specifically in MCF-7 cells (79). Similarly, ClpP agonists ONC201 and TR‐107 reduces the protein levels of TUFM and inhibits breast cancer progression (80–83).
Current studies show that blocking epidermal growth factor receptor (EGFR) inhibits HNSCC initiation and maintenance (84), with the inhibition of autophagy sensitizing HNSCC to EGFR blockade (85). Regarding TUFM, a reduction in its expression is associated with a poor response to EGFR inhibitor cetuximab or gefitinib treatment (86). Mechanistically, the deficiency of either NLRX1 or TUFM results in impaired autophagy when treated with EGFR inhibitors in HNSCC cells. The NLRX1/TUFM complex has been identified as a novel anchorage site for autophagy, facilitating the recruitment of Beclin-1 to mitochondria, enhancing its polyubiquitination, and disrupting its interaction with Rubicon (Figure 2E). Although no direct interaction between NLRX1 and Beclin-1 is observed, it is noteworthy that this protein complex also plays a crucial role in facilitating unfolded protein response (UPR) activation, potentially serving as an additional mechanism to enhance autophagy (86).
Recent studies have shown a reduction in TUFM protein levels in the brains of individuals with Alzheimer’s disease, as well as in mouse models of such disease (87–89). The downregulation of TUFM has been associated with decreased levels of β-amyloid converting enzyme 1 (BACE1) and its catalytic product, β-amyloid protein (87, 90). Silencing TUFM results in enhanced RNA stability of BACE1 through ROS but not NF-κB-dependent translation modulation. Furthermore, silencing TUFM is linked to apoptosis induction and tau phosphorylation, both of which are mitigated by the mitochondria-targeted antioxidant TEMPO. In addition, Kaempferide (Kaem), a natural flavonoid compound, is shown to facilitate the autophagic clearance of microtubule-associated protein tau by targeting TUFM (91). Taken together, these findings indicate that targeting TUFM may hold promise as a therapeutic strategy for ameliorating Alzheimer’s disease.
The relationship between TUFM-dependent autophagy and obesity, as well as its impact on the insulin cascade, has been extensively documented in the literature (92–96). In rats subjected to high-fat diet, TUFM expression is found to be increased in adipose tissue, liver and extensor digitorum longus muscle (97). Furthermore, pediatric adipose tissue samples from individuals with obesity also exhibits a significant upregulation of TUFM (98). Mouse models have further elucidated the effects of diet and physical activity on TUFM expression, showing that a western diet and sedentary lifestyle can lead to decreased TUFM expression in muscles compared to a normal chow diet and voluntary wheel running (99). Kaem exhibits a high binding affinity to the TUFM G172 and K256 residues, facilitating its interaction with the ATG12-ATG5 complex to induce autophagy and promote lipid degradation (49) (Figure 2B). These results offer valuable insights into the potential therapeutic implications of targeting TUFM in the management of metabolic syndrome.
TUFM is significantly upregulated in individuals diagnosed with pulmonary arterial hypertension (PAH) (100, 101), with predominant expression observed in pulmonary arterial smooth muscle cells (PASMCs) and minimal to absent expression in endothelial cells in a rat model of monocrotaline-induced PAH. Silencing of TUFM is shown to mitigate monocrotaline-induced PAH by enhancing apoptosis and reducing mitophagy (101). Mechanically, TUFM is found to promote PASMC proliferation and is induced under hypoxic conditions in PASMCs (101, 102). Additionally, TUFM silencing alleviates mitophagy and enhances apoptosis in PASMCs exposed to hypoxia, which is associated with AMPK/mTOR Pathway (101).
TUFM is widely expressed in the heart (103). In AC-16 cardiomyocytes, FUNDC1 recruits TUFM directly and facilitates its translocation to the mitochondria. This interaction between FUNDC1 and TUFM plays a crucial role in maintaining mitochondrial DNA stability and preventing PANoptosis-associated cell death when exposed to DOX (104). Additionally, the depletion of TUFM results in similar effects on the release of mitochondrial DNA and the upregulation of PANoptosis. It is important to note that the administration of DOX leads to a decrease in TUFM expression in heart tissues, a phenomenon that is not influenced by FUNDC1. In mouse models of diabetes and diabetic cardiomyopathy, mitochondrial argonaute 2 (ago2) is found to directly interact with TUFM to facilitate the translation of electron transport chain subunit, consequently leading to a decrease in ROS and providing protection (52). Of note, Ago2 is observed to enhance the binding of TUFM to mitochondrial mRNAs, while TUFM does not impact the interaction between Ago2 and mitochondrial RNAs. Furthermore, the increase in mitochondrial-encoded subunits, specifically cytochrome b (CYTB), targed by miRNAs miR-133a-3p, miR-143-3p, and miR-21, is lost in Ago2 or TUFM deficient myocytes.
A mitochondrial Mortality factor 4-like protein 1 (MRG15)-TUFM axis is tightly involved in the progression from simple steatosis to NASH (105). Reduced levels of TUFM are observed in the livers of patients with NASH or mouse models of NASH, with a negative correlation with MRG15 expression. Intriguingly, no significant changes are found in TUFM mRNA levels in liver tissues or primary hepatocytes. Mechanistically, MRG15 interacts with and deacetylates TUFM, specifically at the K82 and K91 sites within mitochondria. This interaction facilitates TUFM degradation through the mitochondrial ClpXP protease system, ultimately resulting in impaired mitophagy, increased ROS, and activated NLRP3 inflammasome.
TUFM, initially identified as a nuclear-encoded mitochondrial protein in yeast, exhibits high conservation from prokaryotes to eukaryotes (4–7), yet the evolutionary functions and origins of TUFM remain largely elusive. Although recognized for its involvement in mitochondrial protein translation, recent studies have unveiled its multifaceted functions beyond this realm. TUFM not only regulates programmed cell death but also shows potential as a drug target for treating virus- and cancer-related diseases. Mutations in TUFM have been linked to an early fatal outcome and a range of diseases, such as multiple sclerosis (106), dysplastic leukoencephalopathy (107), and premature ovarian insufficiency (108).
It is of significance to note that TUFM displays a higher susceptibility to proteinase K digestion in comparison to mitochondrial inner and matrix proteins, yet is similar to outer membrane proteins, indicating its steady-state presence on the mitochondrial outer membrane (44, 46). This finding may provide insight into the potential interaction between TUFM and cytosolic viral or host proteins for further mitophagy induction. Furthermore, prior research has indicated that Influenza A PB1F2 localizes to the mitochondrial inner membrane space, potentially facilitating its interaction with TUFM (39, 109). Based on our knowledge, TUFM is a dual-localized protein with a mitochondrial targeting sequence (46). It translocates to the matrix side of mitochondria through the outer and inner mitochondrial membrane channel under resting states. However, when overactivated mitophagy occurs, phosphorylation of TUFM at Ser222 by PINK1 switches its role from activating to suppressing mitophagy (46). This phosphorylated form of TUFM is mainly found in the cytosol, potentially protecting mitochondria from excessive degradation. The regulation of TUFM activation appears to be tightly controlled at spatially and temporally levels, although the specific mechanisms remain incompletely understood.
Recent research has elucidated the involvement of TUFM in non-canonical mitophagy during viral infection, where viral proteins exploit TUFM for mitophagy induction and viral replication. An intriguing area for further investigation lies in elucidating the specific mechanisms underlying TUFM-dependent autophagy. Additionally, in the context of cancer, increased expression of TUFM may play a role in promoting tumor metastasis and progression. Exploring the molecular pathways through which TUFM contributes to the initiation, progression, and resistance to therapy in cancer within the tumor microenvironment may reveal novel biomarkers and therapeutic targets for the treatment of cancer. Additionally, the evaluation of small molecule compounds or anti-TUFM antibody-drug conjugates could be a viable strategy to modulate TUFM activity for the management of diverse diseases.
In summary, continued investigation into the diverse functions of TUFM in both health and disease is imperative for elucidating its therapeutic efficacy and devising precise interventions for a range of human diseases.
NL: Writing – original draft. BP: Writing – original draft. LK: Writing – review & editing. DL: Writing – review & editing. XJ: Supervision, Writing – review & editing. C-MZ: Conceptualization, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82302523, 82203623), the Natural Science Foundation of Hebei (H2023206290), and the Hebei Province Program for Introducing Returned Overseas Researcher (C20230353).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Parmeggiani A, Nissen P. Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action. FEBS Lett. (2006) 580:4576–81. doi: 10.1016/j.febslet.2006.07.039
2. Furano AV. Content of elongation factor Tu in Escherichia coli. Proc Natl Acad Sci U.S.A. (1975) 72:4780–4. doi: 10.1073/pnas.72.12.4780
3. Kavaliauskas D, Nissen P, Knudsen CR. The busiest of all ribosomal assistants: elongation factor Tu. Biochemistry. (2012) 51:2642–51. doi: 10.1021/bi300077s
4. Richter D, Lipmann F. Separation of mitochondrial and cytoplasmic peptide chain elongation factors from yeast. Biochemistry. (1970) 9:5065–70. doi: 10.1021/bi00828a004
5. Kuhlman P, Palmer JD. Isolation, expression, and evolution of the gene encoding mitochondrial elongation factor Tu in Arabidopsis thaliana. Plant Mol Biol. (1995) 29:1057–70. doi: 10.1007/BF00014977
6. Wells J, Henkler F, Leversha M, Koshy R. A mitochondrial elongation factor-like protein is over-expressed in tumours and differentially expressed in normal tissues. FEBS Lett. (1995) 358:119–25. doi: 10.1016/0014-5793(94)01403-N
7. Koch I, Hofschneider PH, Lottspeich F, Eckerskorn C, Koshy R. Tumour-related expression of a translation-elongation factor-like protein. Oncogene. (1990) 5:839–43.
8. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. (2021) 596:583–9. doi: 10.1038/s41586-021-03819-2
9. Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. (2022) 50:D439–44. doi: 10.1093/nar/gkab1061
10. Nagata S, Tsunetsugu-Yokota Y, Naito A, Kaziro Y. Molecular cloning and sequence determination of the nuclear gene coding for mitochondrial elongation factor Tu of Saccharomyces cerevisiae. Proc Natl Acad Sci U.S.A. (1983) 80:6192–6. doi: 10.1073/pnas.80.20.6192
11. Piechulla B, Kuntzel H. Mitochondrial polypeptide elongation factor EF-Tu of Saccharomyces cerevisiae. Functional and structural homologies to Escherichia coli EF-Tu. Eur J Biochem. (1983) 132:235–40. doi: 10.1111/j.1432-1033.1983.tb07353.x
12. Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. (2004) 16:3496–507. doi: 10.1105/tpc.104.026765
13. Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. (2006) 125:749–60. doi: 10.1016/j.cell.2006.03.037
14. Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. (2012) 36:933–46. doi: 10.1016/j.immuni.2012.03.025
15. Harvey KL, Jarocki VM, Charles IG, Djordjevic SP. The diverse functional roles of elongation factor tu (EF-tu) in microbial pathogenesis. Front Microbiol. (2019) 10:2351. doi: 10.3389/fmicb.2019.02351
16. Andersen GR, Valente L, Pedersen L, Kinzy TG, Nyborg J. Crystal structures of nucleotide exchange intermediates in the eEF1A-eEF1Balpha complex. Nat Struct Biol. (2001) 8:531–4. doi: 10.1038/88598
17. Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, et al. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol Cell. (2000) 6:1261–6. doi: 10.1016/S1097-2765(00)00122-2
18. Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy F, Weir JR, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. (2009) 326:688–94. doi: 10.1126/science.1179700
19. Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. (2012) 1819:1035–54. doi: 10.1016/j.bbagrm.2011.11.009
20. Li Q, Yang H, Stroup EK, Wang H, Ji Z. Low-input RNase footprinting for simultaneous quantification of cytosolic and mitochondrial translation. Genome Res. (2022) 32:545–57. doi: 10.1101/gr.276139.121
21. Wang F, Zhang D, Zhang D, Li P, Gao Y. Mitochondrial protein translation: emerging roles and clinical significance in disease. Front Cell Dev Biol. (2021) 9:675465. doi: 10.3389/fcell.2021.675465
22. Alexander C, Bilgin N, Lindschau C, Mesters JR, Kraal B, Hilgenfeld R, et al. Phosphorylation of elongation factor Tu prevents ternary complex formation. J Biol Chem. (1995) 270:14541–7. doi: 10.1074/jbc.270.24.14541
23. Lippmann C, Lindschau C, Vijgenboom E, Schroder W, Bosch L, Erdmann VA. Prokaryotic elongation factor Tu is phosphorylated in vivo. J Biol Chem. (1993) 268:601–7. doi: 10.1016/S0021-9258(18)54193-4
24. Talavera A, Hendrix J, Versees W, Jurenas D, Van Nerom K, Vandenberk N, et al. Phosphorylation decelerates conformational dynamics in bacterial translation elongation factors. Sci Adv. (2018) 4:eaap9714. doi: 10.1126/sciadv.aap9714
25. Guedouari H, Ould Amer Y, Pichaud N, Hebert-Chatelain E. Characterization of the interactome of c-Src within the mitochondrial matrix by proximity-dependent biotin identification. Mitochondrion. (2021) 57:257–69. doi: 10.1016/j.mito.2020.12.012
26. Koc EC, Miller-Lee JL, Koc H. Fyn kinase regulates translation in mammalian mitochondria. Biochim Biophys Acta Gen Subj. (2017) 1861:533–40. doi: 10.1016/j.bbagen.2016.12.004
27. Koc EC, Hunter CA, Koc H. Phosphorylation of mammalian mitochondrial EF-Tu by Fyn and c-Src kinases. Cell Signal. (2023) 101:110524. doi: 10.1016/j.cellsig.2022.110524
28. Zhang T, Du W, Wilson AF, Namekawa SH, Andreassen PR, Meetei AR, et al. Fancd2 in vivo interaction network reveals a non-canonical role in mitochondrial function. Sci Rep. (2017) 7:45626. doi: 10.1038/srep45626
29. Chatla S, Du W, Wilson AF, Meetei AR, Pang Q. Fancd2-deficient hematopoietic stem and progenitor cells depend on augmented mitochondrial translation for survival and proliferation. Stem Cell Res. (2019) 40:101550. doi: 10.1016/j.scr.2019.101550
30. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. (2007) 315:1398–401. doi: 10.1126/science.1136880
31. Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. (2013) 494:201–6. doi: 10.1038/nature11866
32. Orvedahl A, MacPherson S, Sumpter R Jr., Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. (2010) 7:115–27. doi: 10.1016/j.chom.2010.01.007
33. Lei Y, Wen H, Ting JP. The NLR protein, NLRX1, and its partner, TUFM, reduce type I interferon, and enhance autophagy. Autophagy. (2013) 9:432–3. doi: 10.4161/auto.23026
34. Ding B, Zhang L, Li Z, Zhong Y, Tang Q, Qin Y, et al. The matrix protein of human parainfluenza virus type 3 induces mitophagy that suppresses interferon responses. Cell Host Microbe. (2017) 21:538–547 e534. doi: 10.1016/j.chom.2017.03.004
35. Kuo SM, Chen CJ, Chang SC, Liu TJ, Chen YH, Huang SY, et al. Inhibition of avian influenza A virus replication in human cells by host restriction factor TUFM is correlated with autophagy. mBio. (2017) 8(3):e00481-17. doi: 10.1128/mBio.00481-17
36. Wang K, Ma H, Liu H, Ye W, Li Z, Cheng L, et al. The glycoprotein and nucleocapsid protein of hantaviruses manipulate autophagy flux to restrain host innate immune responses. Cell Rep. (2019) 27:2075–2091 e2075. doi: 10.1016/j.celrep.2019.04.061
37. Hui X, Zhang L, Cao L, Huang K, Zhao Y, Zhang Y, et al. SARS-CoV-2 promote autophagy to suppress type I interferon response. Signal Transduct Target Ther. (2021) 6:180. doi: 10.1038/s41392-021-00574-8
38. Cao Y, Chen Z, Huang J, Wu H, Zou J, Feng H. Black carp TUFM collaborates with NLRX1 to inhibit MAVS-mediated antiviral signaling pathway. Dev Comp Immunol. (2021) 122:104134. doi: 10.1016/j.dci.2021.104134
39. Wang R, Zhu Y, Ren C, Yang S, Tian S, Chen H, et al. Influenza A virus protein PB1-F2 impairs innate immunity by inducing mitophagy. Autophagy. (2021) 17:496–511. doi: 10.1080/15548627.2020.1725375
40. Hao Y, Yang J, Yang B, Zhang T, Shi X, Yang X, et al. Identification and analysis of the interaction network of African swine fever virus D1133L with host proteins. Front Microbiol. (2022) 13:1037346. doi: 10.3389/fmicb.2022.1037346
41. Chen M, Zhang X, Kong F, Gao P, Ge X, Zhou L, et al. Senecavirus a induces mitophagy to promote self-replication through direct interaction of 2C protein with K27-linked ubiquitinated TUFM catalyzed by RNF185. Autophagy. (2023) 12:1–28. doi: 10.1080/15548627.2023.2293442
42. Cheng J, Wang Y, Yin L, Liang W, Zhang J, Ma C, et al. The nonstructural protein 1 of respiratory syncytial virus hijacks host mitophagy as a novel mitophagy receptor to evade the type I IFN response in HEp-2 cells. mBio. (2023) 14(6):e0148023. doi: 10.1128/mbio.01480-23
43. Qin L, Rao T, Li X, Chen H, Qian P. DnaJA2 interacts with Japanese encephalitis virus NS3 via its C-terminal to promote viral infection. Virus Res. (2023) 336:199210. doi: 10.1016/j.virusres.2023.199210
44. Choi CY, Vo MT, Nicholas J, Choi YB. Autophagy-competent mitochondrial translation elongation factor TUFM inhibits caspase-8-mediated apoptosis. Cell Death Differ. (2022) 29:451–64. doi: 10.1038/s41418-021-00868-y
45. Vo MT, Choi CY, Choi YB. The mitophagy receptor NIX induces vIRF-1 oligomerization and interaction with GABARAPL1 for the promotion of HHV-8 reactivation-induced mitophagy. PloS Pathog. (2023) 19:e1011548. doi: 10.1371/journal.ppat.1011548
46. Lin J, Chen K, Chen W, Yao Y, Ni S, Ye M, et al. Paradoxical mitophagy regulation by PINK1 and TUFm. Mol Cell. (2020) 80:607–620 e612. doi: 10.1016/j.molcel.2020.10.007
47. Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. (1993) 67:1761–4. doi: 10.1128/jvi.67.4.1761-1764.1993
48. Qi W, Tian J, Su S, Huang L, Li H, Liao M. Identification of potential virulence determinants associated H9N2 avian influenza virus PB2 E627K mutation by comparative proteomics. Proteomics. (2015) 15:1512–24. doi: 10.1002/pmic.v15.9
49. Kim D, Hwang HY, Ji ES, Kim JY, Yoo JS, Kwon HJ. Activation of mitochondrial TUFM ameliorates metabolic dysregulation through coordinating autophagy induction. Commun Biol. (2021) 4:1. doi: 10.1038/s42003-020-01566-0
50. He K, Guo X, Liu Y, Li J, Hu Y, Wang D, et al. TUFM downregulation induces epithelial-mesenchymal transition and invasion in lung cancer cells via a mechanism involving AMPK-GSK3beta signaling. Cell Mol Life Sci. (2016) 73:2105–21. doi: 10.1007/s00018-015-2122-9
51. Huang JH, Liu CY, Wu SY, Chen WY, Chang TH, Kan HW, et al. NLRX1 facilitates histoplasma capsulatum-induced LC3-associated phagocytosis for cytokine production in macrophages. Front Immunol. (2018) 9:2761. doi: 10.3389/fimmu.2018.02761
52. Zhan J, Jin K, Xie R, Fan J, Tang Y, Chen C, et al. Ago2 protects against diabetic cardiomyopathy by activating mitochondrial gene translation. Circulation. (2024) 149(14):1102–20. doi: 10.1161/CIRCULATIONAHA.123.065546
53. Xu C, Wang J, Li J, Fang R. Expression of elongation factor (EF)-Tu is correlated with prognosis of gastric adenocarcinomas. Int J Mol Sci. (2011) 12:6645–55. doi: 10.3390/ijms12106645
54. Shi H, Hayes M, Kirana C, Miller R, Keating J, Macartney-Coxson D, et al. TUFM is a potential new prognostic indicator for colorectal carcinoma. Pathology. (2012) 44:506–12. doi: 10.1097/PAT.0b013e3283559cbe
55. Srisomsap C, Sawangareetrakul P, Subhasitanont P, Panichakul T, Keeratichamroen S, Lirdprapamongkol K, et al. Proteomic analysis of cholangiocarcinoma cell line. Proteomics. (2004) 4:1135–44. doi: 10.1002/pmic.200300651
56. Tamai K, Nakamura-Shima M, Shibuya-Takahashi R, Kanno SI, Yasui A, Mochizuki M, et al. BEX2 suppresses mitochondrial activity and is required for dormant cancer stem cell maintenance in intrahepatic cholangiocarcinoma. Sci Rep. (2020) 10:21592. doi: 10.1038/s41598-020-78539-0
57. Koc ZC, Sollars VE, Bou Zgheib N, Rankin GO, Koc EC. Evaluation of mitochondrial biogenesis and ROS generation in high-grade serous ovarian cancer. Front Oncol. (2023) 13:1129352. doi: 10.3389/fonc.2023.1129352
58. Permuth-Wey J, Chen YA, Tsai YY, Chen Z, Qu X, Lancaster JM, et al. Inherited variants in mitochondrial biogenesis genes may influence epithelial ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. (2011) 20:1131–45. doi: 10.1158/1055-9965.EPI-10-1224
59. Grant AG, Flomen RM, Tizard ML, Grant DA. Differential screening of a human pancreatic adenocarcinoma lambda gt11 expression library has identified increased transcription of elongation factor EF-1 alpha in tumour cells. Int J Cancer. (1992) 50:740–5. doi: 10.1002/ijc.2910500513
60. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
61. He X, Wu K, Ogino S, Giovannucci EL, Chan AT, Song M. Association between risk factors for colorectal cancer and risk of serrated polyps and conventional adenomas. Gastroenterology. (2018) 155:355–373 e318. doi: 10.1053/j.gastro.2018.04.019
62. Becker WR, Nevins SA, Chen DC, Chiu R, Horning AM, Guha TK, et al. Single-cell analyses define a continuum of cell state and composition changes in the Malignant transformation of polyps to colorectal cancer. Nat Genet. (2022) 54:985–95. doi: 10.1038/s41588-022-01088-x
63. Xi HQ, Zhang KC, Li JY, Cui JX, Zhao P, Chen L. Expression and clinicopathologic significance of TUFM and p53 for the normal-adenoma-carcinoma sequence in colorectal epithelia. World J Surg Oncol. (2017) 15:90. doi: 10.1186/s12957-017-1111-x
64. Shi H, Hood KA, Hayes MT, Stubbs RS. Proteomic analysis of advanced colorectal cancer by laser capture microdissection and two-dimensional difference gel electrophoresis. J Proteomics. (2011) 75:339–51. doi: 10.1016/j.jprot.2011.07.025
65. Xu K, Zhang YY, Han B, Bai Y, Xiong Y, Song Y, et al. Suppression subtractive hybridization identified differentially expressed genes in colorectal cancer: microRNA-451a as a novel colorectal cancer-related gene. Tumour Biol. (2017) 39:1010428317705504. doi: 10.1177/1010428317705504
66. Xu X, Huang A, Cui X, Han K, Hou X, Wang Q, et al. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics. (2019) 9:4208–20. doi: 10.7150/thno.33803
67. Tong M, Liu H, Hao J, Fan D. Comparative pharmacoproteomics reveals potential targets for berberine, a promising therapy for colorectal cancer. Biochem Biophys Res Commun. (2020) 19:S0006-291X(20)30320-X. doi: 10.1016/j.bbrc.2020.02.052
68. Weng X, Zheng S, Shui H, Lin G, Zhou Y. TUFM-knockdown inhibits the migration and proliferation of gastrointestinal stromal tumor cells. Oncol Lett. (2020) 20:250. doi: 10.3892/ol
69. Kim HK, Park WS, Kang SH, Warda M, Kim N, Ko JH, et al. Mitochondrial alterations in human gastric carcinoma cell line. Am J Physiol Cell Physiol. (2007) 293:C761–771. doi: 10.1152/ajpcell.00043.2007
70. Schaff LR, Mellinghoff IK. Glioblastoma and other primary brain Malignancies in adults: A review. JAMA. (2023) 329:574–87. doi: 10.1001/jama.2023.0023
71. Zottel A, Jovcevska I, Samec N, Mlakar J, Sribar J, Krizaj I, et al. Anti-vimentin, anti-TUFM, anti-NAP1L1 and anti-DPYSL2 nanobodies display cytotoxic effect and reduce glioblastoma cell migration. Ther Adv Med Oncol. (2020) 12:1758835920915302. doi: 10.1177/1758835920915302
72. Samec N, Jovcevska I, Stojan J, Zottel A, Liovic M, Myers MP, et al. Glioblastoma-specific anti-TUFM nanobody for in-vitro immunoimaging and cancer stem cell targeting. Oncotarget. (2018) 9:17282–99. doi: 10.18632/oncotarget.v9i25
73. Jovcevska I, Zupanec N, Urlep Z, Vranic A, Matos B, Stokin CL, et al. Differentially expressed proteins in glioblastoma multiforme identified with a nanobody-based anti-proteome approach and confirmed by OncoFinder as possible tumor-class predictive biomarker candidates. Oncotarget. (2017) 8:44141–58. doi: 10.18632/oncotarget.v8i27
74. Lah TT, Majc B, Novak M, Susnik A, Breznik B, Porcnik A, et al. The cytotoxic effects of cannabidiol and cannabigerol on glioblastoma stem cells may mostly involve GPR55 and TRPV1 signalling. Cancers (Basel). (2022) 14(23):5918. doi: 10.3390/cancers14235918
75. Sighel D, Notarangelo M, Aibara S, Re A, Ricci G, Guida M, et al. Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth. Cell Rep. (2021) 35:109024. doi: 10.1016/j.celrep.2021.109024
76. Zhang KH, Tian HY, Gao X, Lei WW, Hu Y, Wang DM, et al. Ferritin heavy chain-mediated iron homeostasis and subsequent increased reactive oxygen species production are essential for epithelial-mesenchymal transition. Cancer Res. (2009) 69:5340–8. doi: 10.1158/0008-5472.CAN-09-0112
77. Malik N, Kim YI, Yan H, Tseng YC, du Bois W, Ayaz G, et al. Dysregulation of mitochondrial translation caused by CBFB deficiency cooperates with mutant PIK3CA and is a vulnerability in breast cancer. Cancer Res. (2023) 83:1280–98. doi: 10.1158/0008-5472.CAN-22-2525
78. Malik N, Yan H, Kim YI, Ayaz G, Wang S, Mondal P, et al. Autophagy inhibition as a potential therapeutic strategy for breast cancer with mitochondrial translation defect caused by CBFB-deficiency. Autophagy. (2023) 19:3026–8. doi: 10.1080/15548627.2023.2208481
79. Jeong SH, Song IS, Kim HK, Lee SR, Song S, Suh H, et al. An analogue of resveratrol HS-1793 exhibits anticancer activity against MCF-7 cells via inhibition of mitochondrial biogenesis gene expression. Mol Cells. (2012) 34:357–65. doi: 10.1007/s10059-012-0081-7
80. Mishukov A, Odinokova I, Mndlyan E, Kobyakova M, Abdullaev S, Zhalimov V, et al. ONC201-induced mitochondrial dysfunction, senescence-like phenotype, and sensitization of cultured BT474 human breast cancer cells to TRAIL. Int J Mol Sci. (2022) 23(24):15551. doi: 10.3390/ijms232415551
81. Greer YE, Hernandez L, Fennell EMJ, Kundu M, Voeller D, Chari R, et al. Mitochondrial matrix protease clpP agonists inhibit cancer stem cell function in breast cancer cells by disrupting mitochondrial homeostasis. Cancer Res Commun. (2022) 2:1144–61. doi: 10.1158/2767-9764.CRC-22-0142
82. Fennell EMJ, Aponte-Collazo LJ, Wynn JD, Drizyte-Miller K, Leung E, Greer YE, et al. Characterization of TR-107, a novel chemical activator of the human mitochondrial protease ClpP. Pharmacol Res Perspect. (2022) 10:e00993. doi: 10.1002/prp2.993
83. Greer YE, Porat-Shliom N, Nagashima K, Stuelten C, Crooks D, Koparde VN, et al. ONC201 kills breast cancer cells in vitro by targeting mitochondria. Oncotarget. (2018) 9:18454–79. doi: 10.18632/oncotarget.v9i26
84. Nie D, Wang X, Sun M, Feng Z, Pei F, Liu W, et al. The primary site of head and neck squamous cell carcinoma predicts survival benefits of EGFR inhibitors: A systematic review and meta-analysis. Radiother Oncol. (2021) 158:13–20. doi: 10.1016/j.radonc.2021.02.001
85. Bashiri H, Tabatabaeian H. Autophagy: A potential therapeutic target to tackle drug resistance in multiple myeloma. Int J Mol Sci. (2023) 24(7):6019. doi: 10.3390/ijms24076019
86. Lei Y, Kansy BA, Li J, Cong L, Liu Y, Trivedi S, et al. EGFR-targeted mAb therapy modulates autophagy in head and neck squamous cell carcinoma through NLRX1-TUFM protein complex. Oncogene. (2016) 35:4698–707. doi: 10.1038/onc.2016.11
87. Zhong BR, Zhou GF, Song L, Wen QX, Deng XJ, Ma YL, et al. TUFM is involved in Alzheimer's disease-like pathologies that are associated with ROS. FASEB J. (2021) 35:e21445. doi: 10.1096/fj.202002461R
88. Johnson ECB, Dammer EB, Duong DM, Ping L, Zhou M, Yin L, et al. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med. (2020) 26:769–80. doi: 10.1038/s41591-020-0815-6
89. Yang J, Long Y, Xu DM, Zhu BL, Deng XJ, Yan Z, et al. Age- and nicotine-associated gene expression changes in the hippocampus of APP/PS1 mice. J Mol Neurosci. (2019) 69:608–22. doi: 10.1007/s12031-019-01389-7
90. Gao J, Guo X, Yan C, Gong X, Ma P, Gu S, et al. Transcriptome-wide association study by different approaches reveals candidate causal genes for cannabis use disorder. Gene. (2023) 851:147048. doi: 10.1016/j.gene.2022.147048
91. Kim D, Hwang HY, Kwon HJ. A natural small molecule induces MAPT clearance via mTOR-independent autophagy. Biochem Biophys Res Commun. (2021) 568:30–6. doi: 10.1016/j.bbrc.2021.06.060
92. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. (2010) 42:937–48. doi: 10.1038/ng.686
93. Voisin S, Almen MS, Zheleznyakova GY, Lundberg L, Zarei S, Castillo S, et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. (2015) 7:103. doi: 10.1186/s13073-015-0225-4
94. Mercader JM, Puiggros M, Segre AV, Planet E, Sorianello E, Sebastian D, et al. Identification of novel type 2 diabetes candidate genes involved in the crosstalk between the mitochondrial and the insulin signaling systems. PloS Genet. (2012) 8:e1003046. doi: 10.1371/journal.pgen.1003046
95. Du X, Yan Y, Yu J, Zhu T, Huang CC, Zhang L, et al. SH2B1 tunes hippocampal ERK signaling to influence fluid intelligence in humans and mice. Res (Wash D C). (2023) 6:0269. doi: 10.34133/research.0269
96. Liu L, Fan Q, Zhang F, Guo X, Liang X, Du Y, et al. A genomewide integrative analysis of GWAS and eQTLs data identifies multiple genes and gene sets associated with obesity. BioMed Res Int. (2018) 2018:3848560. doi: 10.1155/2018/3848560
97. Gutierrez-Aguilar R, Kim DH, Woods SC, Seeley RJ. Expression of new loci associated with obesity in diet-induced obese rats: from genetics to physiology. Obes (Silver Spring). (2012) 20:306–12. doi: 10.1038/oby.2011.236
98. Mao K, Zhang M, Cao J, Zhao X, Gao L, Fu L, et al. Coding variants are relevant to the expression of obesity-related genes for pediatric adiposity. Obes (Silver Spring). (2021) 29:194–203. doi: 10.1002/oby.23046
99. Lee DE, Brown JL, Rosa ME, Brown LA, Perry RA, Washington TA, et al. Translational machinery of mitochondrial mRNA is promoted by physical activity in Western diet-induced obese mice. Acta Physiol (Oxf). (2016) 218:167–77. doi: 10.1111/apha.12687
100. Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, et al. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol. (2010) 298:H1235–1248. doi: 10.1152/ajpheart.00254.2009
101. Wei R, Lv X, Fang C, Liu C, Ma Z, Liu K. Silencing TUFM inhibits development of monocrotaline-induced pulmonary hypertension by regulating mitochondrial autophagy via AMPK/mTOR signal pathway. Oxid Med Cell Longev. (2022) 2022:4931611. doi: 10.1155/2022/4931611
102. Ma C, Wang X, He S, Zhang L, Bai J, Qu L, et al. Ubiquitinated AIF is a major mediator of hypoxia-induced mitochondrial dysfunction and pulmonary artery smooth muscle cell proliferation. Cell Biosci. (2022) 12:9. doi: 10.1186/s13578-022-00744-3
103. Kim HK, Kang YG, Jeong SH, Park N, Marquez J, Ko KS, et al. Cyclic stretch increases mitochondrial biogenesis in a cardiac cell line. Biochem Biophys Res Commun. (2018) 505:768–74. doi: 10.1016/j.bbrc.2018.10.003
104. Bi Y, Xu H, Wang X, Zhu H, Ge J, Ren J, et al. FUNDC1 protects against doxorubicin-induced cardiomyocyte PANoptosis through stabilizing mtDNA via interaction with TUFM. Cell Death Dis. (2022) 13:1020. doi: 10.1038/s41419-022-05460-x
105. Tian C, Min X, Zhao Y, Wang Y, Wu X, Liu S, et al. MRG15 aggravates non-alcoholic steatohepatitis progression by regulating the mitochondrial proteolytic degradation of TUFM. J Hepatol. (2022) 77:1491–503. doi: 10.1016/j.jhep.2022.07.017
106. Chen S, Mitchell GA, Soucy JF, Gauthier J, Brais B, La Piana R. TUFM variants lead to white matter abnormalities mimicking multiple sclerosis. Eur J Neurol. (2023) 30:3400–3. doi: 10.1111/ene.15982
107. Di Nottia M, Montanari A, Verrigni D, Oliva R, Torraco A, Fernandez-Vizarra E, et al. Novel mutation in mitochondrial Elongation Factor EF-Tu associated to dysplastic leukoencephalopathy and defective mitochondrial DNA translation. Biochim Biophys Acta Mol Basis Dis. (2017) 1863:961–7. doi: 10.1016/j.bbadis.2017.01.022
108. Zhang J, Zhou XY, Wang A, Lai YH, Zhang XF, Liu XT, et al. Novel Tu translation elongation factor, mitochondrial (TUFM) homozygous variant in a consanguineous family with premature ovarian insufficiency. Clin Genet. (2023) 104:516–27. doi: 10.1111/cge.14403
Keywords: TUFM, mitochondria, virus, autophagy, cancer
Citation: Liu N, Pang B, Kang L, Li D, Jiang X and Zhou C-m (2024) TUFM in health and disease: exploring its multifaceted roles. Front. Immunol. 15:1424385. doi: 10.3389/fimmu.2024.1424385
Received: 28 April 2024; Accepted: 20 May 2024;
Published: 29 May 2024.
Edited by:
Takumi Koshiba, Fukuoka University, JapanReviewed by:
Atsuko Kasahara, Tohoku University, JapanCopyright © 2024 Liu, Pang, Kang, Li, Jiang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia Jiang, amlhbmd4aWEwOTI1QGhlYm11LmVkdS5jbg==; Chuan-min Zhou, emhvdWNodWFubWluQHdodS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.