
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 02 September 2024
Sec. Microbial Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1418085
 Zhuhua Wu1,2†
Zhuhua Wu1,2† Qiuchan Tan3†
Qiuchan Tan3† Xiaojuan Jia4
Xiaojuan Jia4 Huizhong Wu2
Huizhong Wu2 Jing Liang5
Jing Liang5 Wenpei Wen2
Wenpei Wen2 Xuezhi Wang6
Xuezhi Wang6 Chenchen Zhang2
Chenchen Zhang2 Yuchuan Zhao2
Yuchuan Zhao2 Yuhui Chen2
Yuhui Chen2 Tingrong Luo1,7*
Tingrong Luo1,7* Wenjun Liu1,4*
Wenjun Liu1,4* Xunxun Chen2*
Xunxun Chen2*Background: microRNAs (miRNAs) were recognized as a promising source of diagnostic biomarker. Herein, we aim to evaluate the performance of an ultrasensitive method for detecting serum miRNAs using single molecule arrays (Simoa).
Methods: In this study, candidate miRNAs were trained and tested by RT-qPCR in a cohort of PTB patients. Besides that, ultrasensitive serum miRNA detection were developed using the Single Molecule Array (Simoa) platform. In this ultra-sensitive sandwich assay, two target-specific LNA-modified oligonucleotide probes can be simply designed to be complementary to the half-sequence of the target miRNA respectively. We characterized its analytical performance and measured miRNAs in the serum of patients with pulmonary tuberculosis and healthy individuals.
Results: We identified a five signature including three upregulated (miR-101, miR-196b, miR-29a) and two downregulated (miR-320b, miR-99b) miRNAs for distinguishing PTB patients from HCs, and validated in our 104 PTB patients. On the basis of Simoa technology, we developed a novel, fully automated digital analyser, which can be used to directly detect miRNAs in serum samples without pre-amplification. We successfully detected miRNAs at femtomolar concentrations (with limits of detection [LODs] ranging from 0.449 to 1.889 fM). Simoa-determined serum miR-29a and miR-99b concentrations in patients with PTB ((median 6.06 fM [range 0.00–75.22]), (median 2.53 fM [range 0.00–24.95]), respectively) were significantly higher than those in HCs ((median 2.42 fM [range 0.00–28.64]) (P < 0.05), (median 0.54 fM [range 0.00–9.12] (P < 0.0001), respectively). Serum levels of miR-320b were significantly reduced in patients with PTB (median 2.11 fM [range 0.00–39.30]) compared with those in the HCs (median 4.76 fM [range 0.00–25.10]) (P < 0.001). A combination of three miRNAs (miR-29a, miR-99b, and miR-320b) exhibited a good capacity to distinguish PTB from HCs, with an area under the curve (AUC) of 0.818 (sensitivity: 83.9%; specificity: 79.7%).
Conclusions: This study benchmarks the role of Simoa as a promising tool for monitoring miRNAs in serum and offers considerable potential as a non-invasive platform for the early diagnosis of PTB.
Tuberculosis (TB) remains a major public health problem worldwide. According to the WHO Global TB Report 2023, an estimated 10.6 million people fell ill with TB and about 1.3 million people died from it in 2022 (1). Early, rapid, and accurate diagnosis has been recognised as a pillar to achieving an end TB strategy (2). Therefore, it is crucial to identify novel biomarkers for TB to develop a practical method for diagnosis.
MicroRNAs (miRNAs) are highly conserved, short, non-coding RNAs that guide post-transcriptional gene regulation and involved in shaping immunity (3). Beyond their expression in specific tissue, miRNAs are also present in different body fluids (4). With their relatively stable, reproducible during freezing thawing and extended storage, substantial levels of sensitivity, miRNAs are considered to be promising diagnostic biomarkers (5). By 2024, numerous studies reported blood miRNA expression profiles in patients with pulmonary tuberculosis (PTB) and healthy individuals, indicating that miRNAs play a vital role in post-transcriptional regulation in the pathogenesis of Mycobacterium tuberculosis (Mtb) and have the potential to be diagnostic and prognostic non-invasive biomarkers (6, 7). However, there are inconsistencies in the data reported across all studies and overlapping expression patterns were rarely found. Whether this is due to the sequencing platform, small sample size and the screening criteria for candidates remains to be clarified.
Despite miRNAs have been scrutinized as prospective biomarkers, conventional miRNAs detection methods have not yet been performed in clinical routine due to their own limitations. Serum, plasma or whole peripheral blood is routinely collected and relatively non-invasive to sample, can be used as starting material. However, in many cases, isolation of miRNAs requires additional extraction and purification steps. In addition, due to extremely low levels of miRNAs, it is necessary to develop new detection technology with high sensitivity for low content miRNAs in complex biological clinical samples. Currently, conventional miRNA detection methods, including quantitative real-time polymerase chain reaction (RT-qPCR), next-generation sequencing (NGS), northern blot-based platforms, in situ hybridisation, and microarray, are far from ideal diagnostic tools for routine clinical use (8, 9). Recent studies reported various novel miRNA detection methods based on signal amplification and discrimination strategies (10). However, the performance of these new methods needs to be evaluated and optimised.
Single molecule array (Simoa) is an ultrasensitive technique capable of detecting single biomolecules (11). By utilizing Simoa, protein biomarkers as well as DNA with very low expression levels could be detected in many diseases, but few studies have been reported on miRNA detection (12–14). Simoa was first used for miRNA detection by Cohen et al. in 2017 (15). Simoa has the advantages of high sensitivity (with LODs ranging from 1 to 30 fM) and minimal processing steps (no additional extraction and purification steps required) and allows quantitative measurement of miRNAs without pre-amplification; it can be used to detect various disease-related miRNAs in real-time and has potential value in clinical practice.
In this study, we focused on assessing miRNA expression in the serum of patients with PTB in comparison to that in healthy controls (HCs) using Simoa. We performed an integrated miRNA-TB expression profiling analysis to screen out the miRNAs reported in at least three studies, these candidate miRNAs were further verified by RT-qPCR in a cohort of PTB patients. Besides that, we directly detects miRNAs without pre-amplification using Simoa, and apply it to the detection of miRNAs in serum samples from PTB and healthy individuals, and a potential diagnostic model was established by logistic regression.
A systematic literature review was performed to identify miRNA expression profiling datasets comparing patients with PTB and HCs. We conducted a systematic search of the PubMed databases, using the MESH search headings: (“Tuberculosis”[Mesh]) AND “MicroRNAs”[Mesh]. Reviewers independently screened titles and abstracts, relevant articles were retrieved and assessed independently, and extracted the following data including: first author, year of publication, country, platform, no. of samples, miRs validated, No. of qRT-PCR validated sample (TB/HC) and the list of up- and down-regulated miRNAs features, cut-off criteria and their relative fold change. Vote-counting rank was used to rank potential molecular biomarkers according to their importance as follows: (a) number of comparisons reported; (b) total number of samples for comparison in agreement; (c) average fold-changes reported for comparisons in agreement. Total number of samples was considered more important than average fold-change because many studies did not report a fold-change. For more information on the studies and dataset selection, see the Supplementary Material.
This study was approved by the Center for Tuberculosis Control of Guangdong Province Ethics Committee, Guangzhou, China. Written informed consent was obtained from all participants prior to their enrolment in the study.
Patients were recruited and enrolled at the outpatient department and health examination centres of the Center for Tuberculosis Control of Guangdong Province, Guangzhou, China. The demographic and clinical characteristics of the PTB and HC groups are summarised in Table 1. PTB patients diagnosed based on clinical signs, chest radiography, and smear microscopy. Participants who had another coexisting disease were excluded. Five millilitres of peripheral venous blood was extracted using a sterile polyolefin resin tube without anticoagulant. Serum samples were stored at -80°C until RNA was extracted.
Total miRNAs were isolated from serum using the miRNeasy Serum/Plasma Kit (Qiagen, Germany) according to the manufacturer’s instructions. The miRNAs were reverse-transcribed to cDNA using a TaqMan™ MicroRNA Reverse Transcription Kit (Applied Biosystem, USA). Thereafter, the quantification of miRNAs was performed using the HieffTM qPCR SYBR Green Master Mix (Vazyme, China) on a ViiA7 instrument. Ce_miR-39 was used as the reference gene. Relative fold changes of miRNA expression were calculated using the ΔΔCT method. RT-specific stem-loop primers and the miRNA-specific forward primer sequences used in this study are listed in Supplementary Table S1.
Locked nucleic acid (LNA) capture and detection probes were designed as previously described (16). LNA-modified capture and detection probes with sequences complementary to 11 or 12 bases of target miRNAs were used to increase hybridization specificity. The placement of bases within each LNA-modified capture
probes and detection probes were then adjusted according to the following criteria: (1) consistent melting temperature; (2) strong predicted binding between each probe and the target sequence; (3) low cross-reactivity. Specially, the capture probe was modified using 5’-Amino-Modifier C12 and detection probe was biotinylated on the 3’ end. The capture and detection probes used in this study are listed in Supplementary Table S2.
Affinity measurements was conducted as described previously (15). Capture probes, detection probes, and synthetic target miRNAs were synthesized from TaKaRa Biotechnology (Dalian, China). Affinity measurements were performed using the NeoSPR-M100 System (Neoline, China) to explore the potential interactions between the probes and target miRNAs.
miRNA Simoa assays were performed according to previously published procedures with some modifications (15). Briefly, the microbeads were diluted in hybridisation buffer to a concentration of 2 × 107 beads/mL after covalent coupling of the capture probes to paramagnetic microbeads. Samples and capture beads were placed in a 96-well conical bottom microtiter plate (Quanterix, USA) and incubated at 30°C for 60 min. Biotinylated detection probes were added after cleaning the capture beads. After incubation with specific biotinylated detection probes, streptavidin-β-galactosidase was added and incubated at 30°C with shaking for 20 min. Resorufin β-D-galactopyranoside and Simoa Sealing Oil were loaded onto the Simoa HD-1 Analyser (Quanterix, USA) according to the manufacturer’s instructions. The software on the HD-1 Analyser was used to determine the average number of enzymes per bead (AEB). The software was also used to interpolate the concentration of miRNAs in the samples from a calibration curve.
Calibration curves were fitted to a 4-parameter nonlinear regression model with a 1/Y2 weighting factor. SPSS software (version 16.0) was used for statistical analyses. Data from each group are presented as the mean ± standard deviation (SD). The Mann–Whitney U test (nonparametric) was used to compare the differences in serum miRNA concentrations among different groups. Statistical significance was set at P <0.05.
A total of 362 participants, including 184 patients with PTB and 178 HCs, were enrolled in the study. Patients with PTB and the HCs were divided into three phases: Screening set (n = 36), Validation set (n = 205), and Simoa assay phase (n = 121). The demographics and clinical parameters of the PTB and HC samples are listed in Table 1. No significant differences (P >0.05) were found in the distribution of sex and age among the patients with PTB and the HC samples.
A flowchart of the experimental design strategy is shown in Figure 1. Fifteen TB miRNA expression profiles were identified in this integrated miRNA-PTB expression profiling analysis (Supplementary Figure S1). The characteristics of each dataset are presented in Supplementary Table S3. Sixty-three differentially expressed miRNAs were identified in the fifteen miRNA expression profile datasets, of which fifty-two were upregulated and eleven were downregulated in at least three studies (Supplementary Table S4). Heatmap analysis showed that some miRNAs were inconsistently upregulated or downregulated (Supplementary Figure S2A). Following profiling, 12 of the 15 studies performed RT-qPCR validation of the selected 80 miRNAs. Among those 80 miRNAs, 26 were significantly (P <0.05) upregulated, and 4 were significantly (P <0.05) downregulated (Supplementary Figure S2B, Supplementary Table S5).
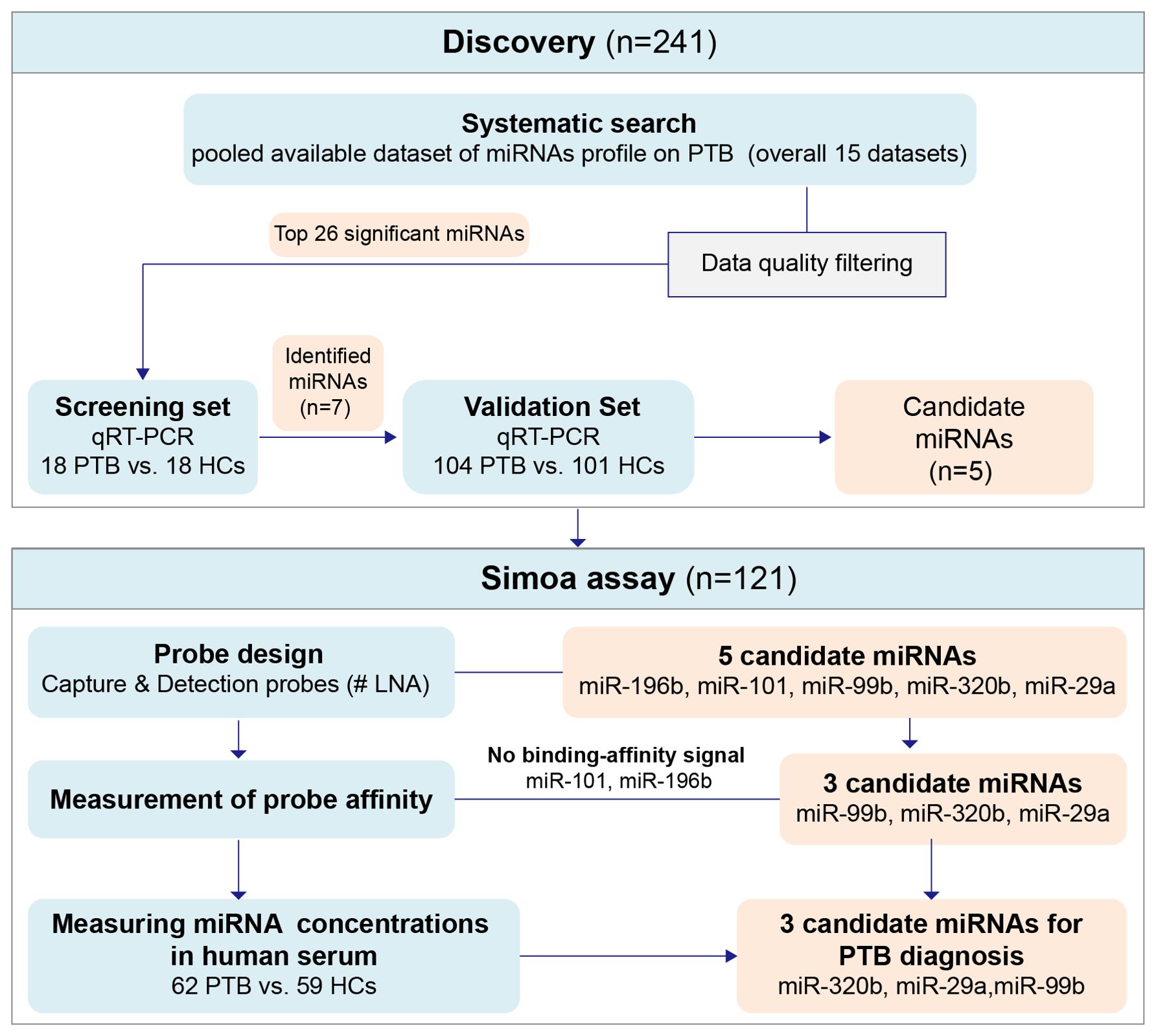
Figure 1. The flowchart of the experiment design strategy. Discovery phase: to discover and preliminarily validate the serum miRNAs differentially expressed between PTB patients and HCs, the signatures screened in the miRNAs expression profile were tested in the testing set and then applied in the external testing cohort. Simoa assay phase: to further validate the selected differentially expressed candidate miRNAs. HCs, Healthy controls; PTB, pulmonary tuberculosis.
Based on the miRNA-PTB expression profiling analysis of the miRNA screening results, we selected the top 26 significant miRNAs for further examination of PTB-associated miRNA expression using RT-qPCR. Seven miRNAs were differentially expressed in the screening set (fold change >2, P <0.05) (Supplementary Table S6) and were subsequently validated in the validation set, which included 104 patients with PTB and 101 HCs. The data showed that five miRNAs (miR-101, miR-320b, miR-99b, miR-196b, and miR-29a) were significantly differentially expressed between the PTB and HC groups (P <0.05) (Figure 2).
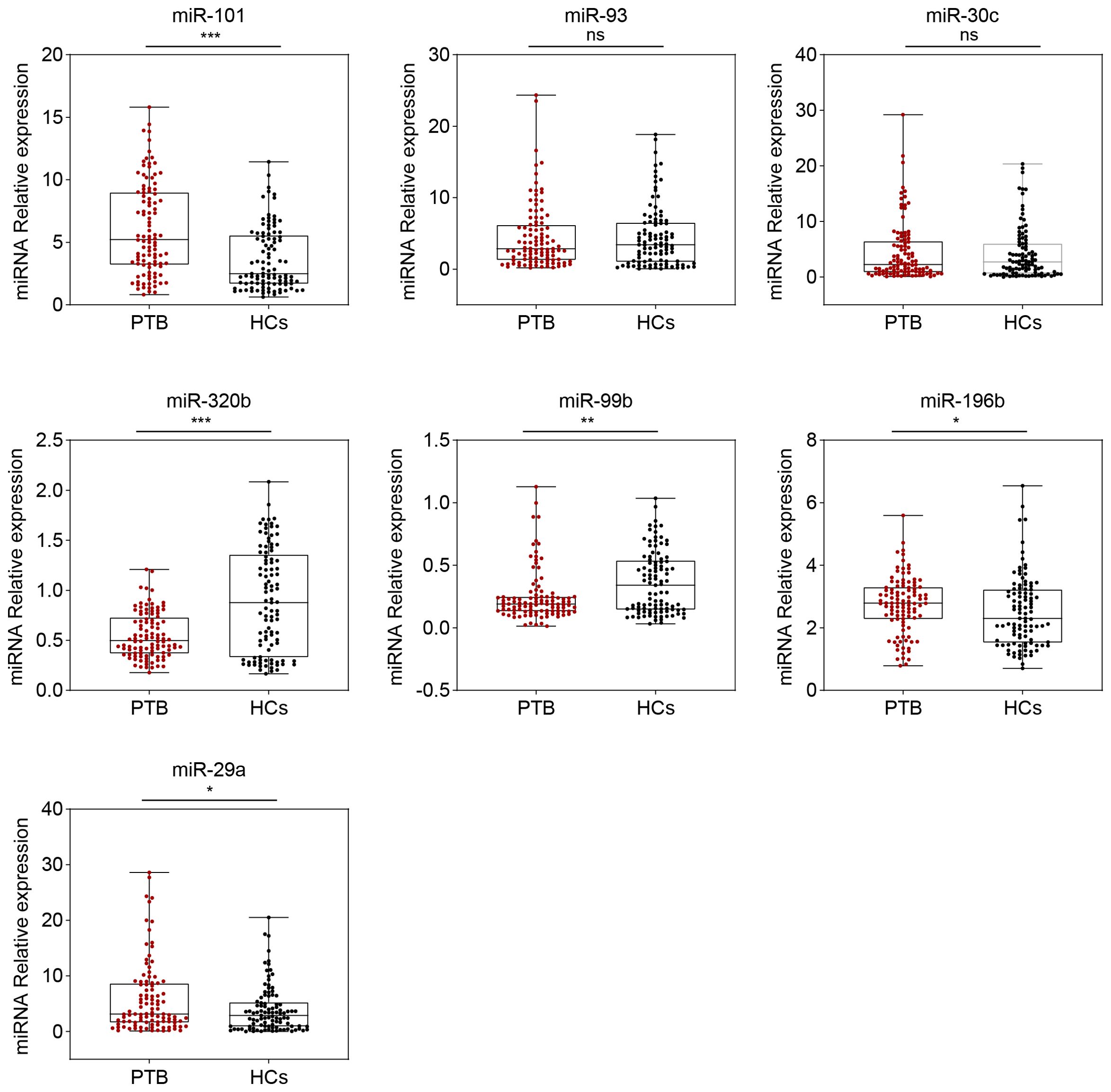
Figure 2. Seven serum miRNA levels in patients with PTB (n = 104) and HCs (n = 101) were selected for verification using the RT-qPCR (in the validation set). The relative abundance of the miRNAs is represented in boxplots. ns, no significance, *P <0.05, **P <0.01, ***P <0.001.
The objective of this study was to develop a Simoa assay to detect miRNAs directly in serum without nucleic acid purification and pre-amplification. Serum samples are incubated with paramagnetic microbeads coated with capture probes. The bead-miRNAs complexes are incubated sequentially with the biotinylated detection probes, SBG and loaded to the Simoa disc array. Figure 3 shows a schematic of the detection of miRNA using Simoa by developing a bead-based sandwich protocol.
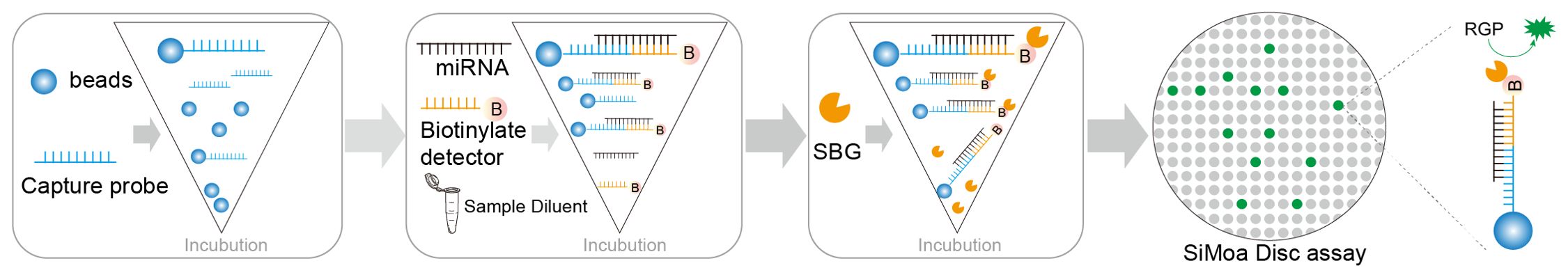
Figure 3. Overview of Sioma detection and probe affinity analysis. Schematic of the assay for miRNA detection using Simoa.
To examine the hybridisation efficiency, we examined the affinity of capture and detection probes for target miRNAs by the NeoSPR-M100 System. After the raw data were analysed, the KD values of capture and detection probes for miR-29a were 8.32 ×10-8 and 8.18×10-8 M, respectively (Figure 4A); The KD values of capture and detection probes for miR-99b were 1.07 ×10-7 and 3.00×10-8 M, respectively (Figure 4B); The KD values of capture and detection probes for miR-320b were 2.32 ×10-8 and 1.84×10-8 M, respectively (Figure 4C). Values of KD shown in Figure 4 indicate that the affinity of the capture/detection probes for the target miRNAs increased with increasing concentration. No binding-affinity signal was detected for miR-101 and miR-196b.
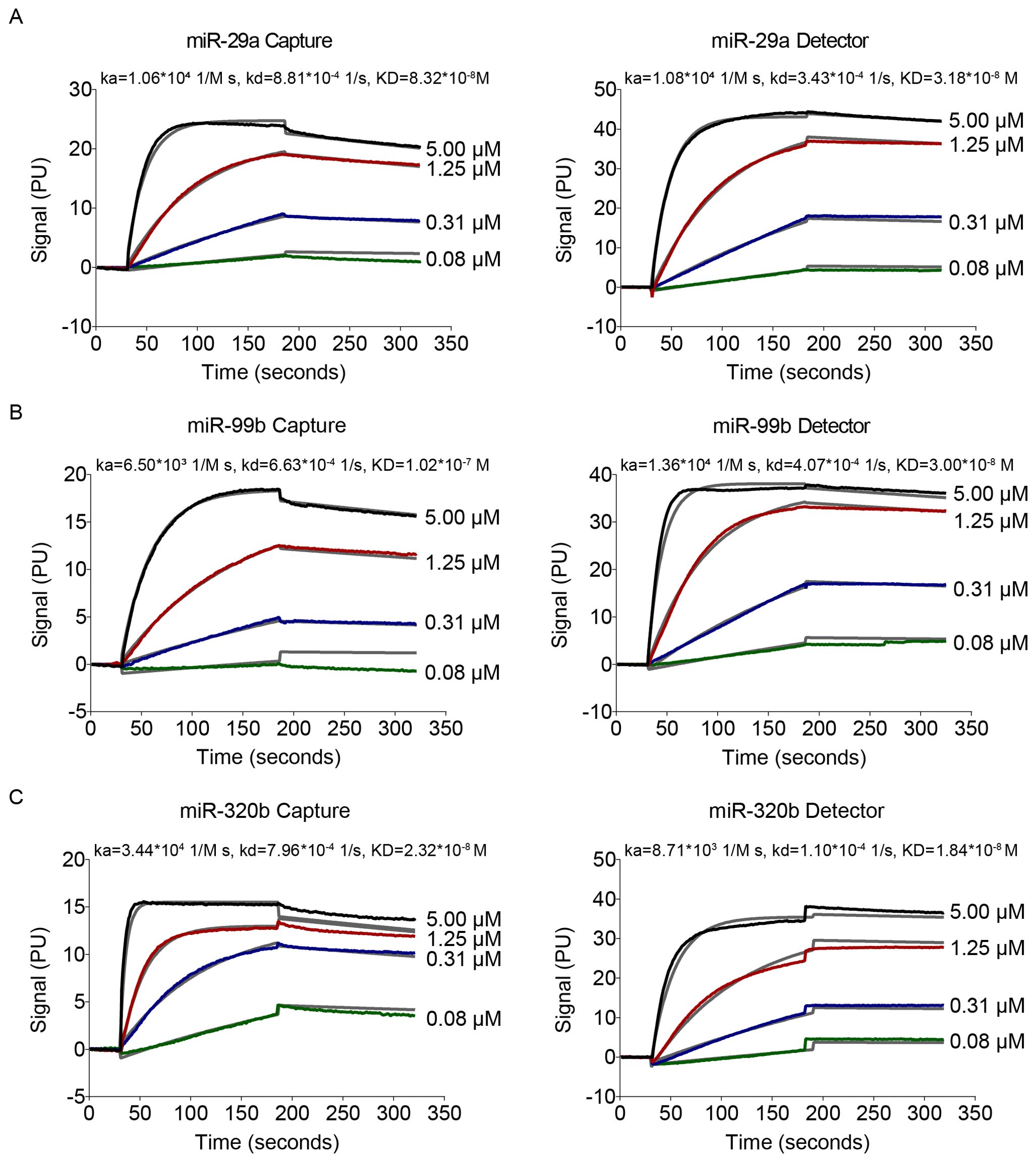
Figure 4. Data of affinity interactions between capture probes to target miRNA or detection probes to target miRNA. The binding affinity was determined using 4-fold serial dilutions at concentrations of 0.08–5 μM.
Figure 5 shows the AEB values for buffered solutions, into which known concentrations of a synthetic calibrator for each target miRNA were spiked, ranging from 0 to 10000 fM. All assays yielded similar results, with LOD of miR-99b, miR-320b, and miR-29a at 0.449, 0.462, and 1.889 fM, respectively (Figure 5). To demonstrate the potential use of the Simoa assay for clinical testing, we developed assays for the detection of miRNAs in human serum without a separate isolation step.
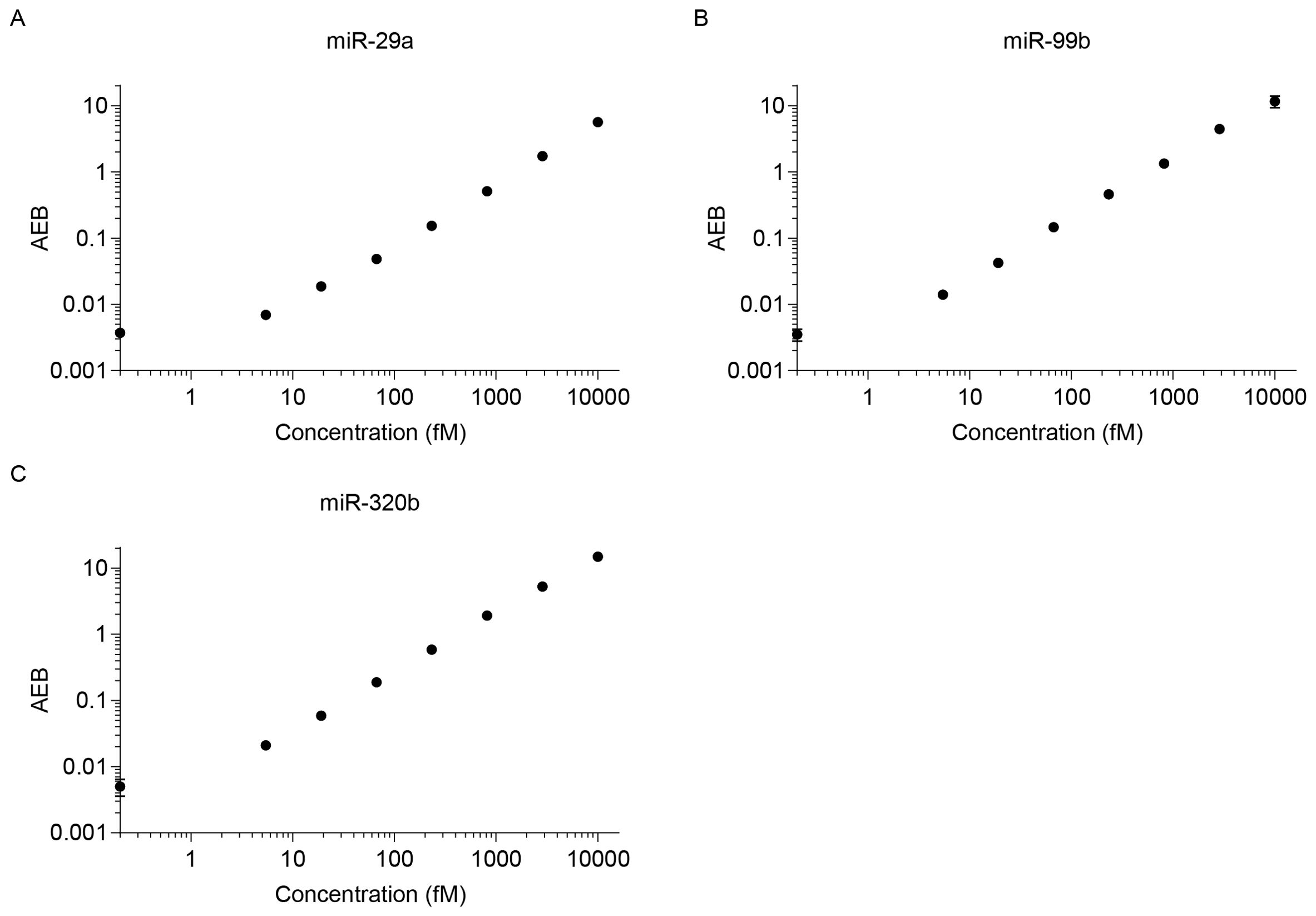
Figure 5. Calibration curves for various miRNAs. LODs were calculated as three standard deviations: (A) miR-29a (LOD 1.889 fM), (B) miR-99b (LOD 0.449 fM), and (C) miR-320b (LOD 0.462 fM).
Simoa-determined serum miR-29a concentrations in patients with PTB ((median 6.06 fM [range 0.00–75.22]) were significantly higher than those in HCs (median 2.42 fM [range 0.00–28.64]) (P < 0.05) (Figure 6A). The miR-99b concentrations also significantly differed between patients with PTB (median 2.53 fM [range 0.00–24.95]) and the HCs (median 0.54 fM [range 0.00–9.12] (P < 0.0001) (Figure 6B). Serum levels of miR-320b were significantly reduced in patients with PTB (median 2.11 fM [range 0.00–39.30]) compared with those in the HCs (median 4.76 fM [range 0.00–25.10]) (P < 0.001) (Figure 6C).
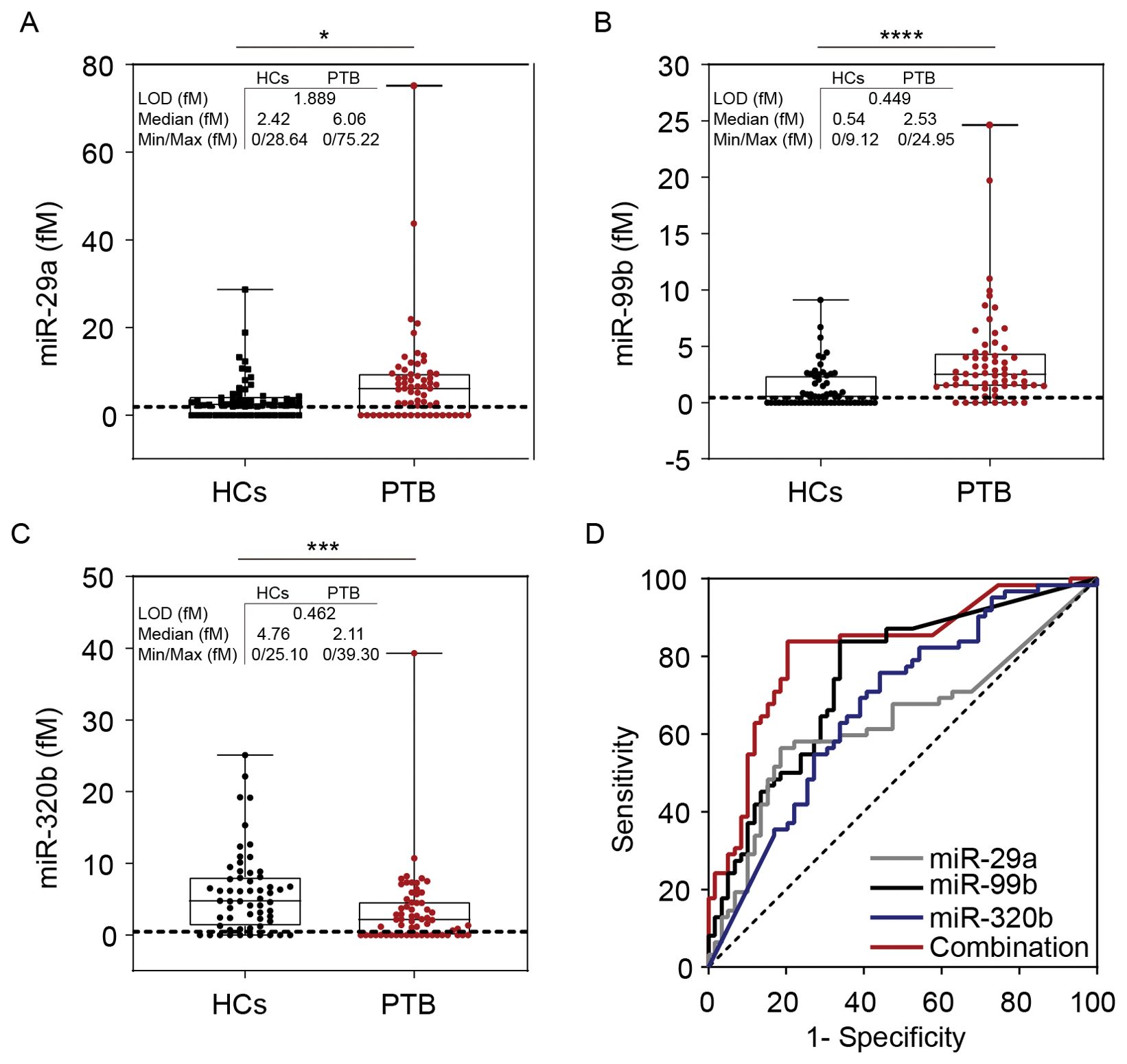
Figure 6. Simoa assay for miR-29a, miR-99b, and miR-320b. Direct detection of (A) miR-99b, (B) miR-29a, and (C) miR-320b concentrations in the serum of patients with PTB (n = 62) and HCs (n = 59). Dashed lines indicate LOD values. The concentration of the miRNAs is represented in boxplots. (D) The ROC curve of miR-29a, miR-99b, miR-320b, and the combined model were analysed. LOD, limit of detection. *P <0.05, ***P <0.001, ****P <0.0001.
Receiver operating characteristic curve (ROC) curve analyses were conducted to assess the diagnostic value for discriminating between patients with PTB and HCs, yielding the following AUCs: miR-29a, 0.633 (P <0.05, 95%CI: 0.531–0.734); miR-99b, 0.748 (P <0.0001, 95%CI: 0.661–0.834); miR-320b, 0.675 (P <0.01, 95%CI: 0.579–0.771). We performed a risk score analysis on the combined cohorts and found that the 3-miRNA panel had a greater separating capacity than one particular miRNA, as evidenced by an AUC of 0.818 (P <0.0001, 95% CI: 0.741–0.896). The sensitivity was 83.9% and the specificity was 79.7%, which reflected reasonable separation between the two groups (Figure 6D).
In the present study, we performed an integrated miRNA-PTB expression profiling analysis to screen out miRNAs reported in at least three studies that were trained and tested using RT-qPCR in a cohort of patients with PTB. We also evaluated the diagnostic accuracy of these candidate miRNAs as PTB biomarkers. In addition, we developed an ultrasensitive and non-invasive serum miRNA detection assay using capture and detection probes with fully automated Simoa technology. Our work systematically evaluated candidate miRNAs as diagnostic markers and indicated that Simoa shows promise for the ultrasensitive detection of miRNAs in human serum samples. This assay offers high-sensitivity, flexibility and affordability. The platform has a high degree of automation, all the measurements and exercise protocols can be performed in approximately 5 hours. Furthermore, Simoa technology allows for multiplexed detection, and from a reagent standpoint, the cost/test for the miRNAs assay would be expected to be comparable to conventional TB immunoassays due to our use of low cost bead-based reagents and commercially available probes.
Monumental studies have reported blood miRNA expression profiles in patients with PTB. However, there is a lack of consensus on miRNA panels for detecting PTB development. Summary and integration analysis of raw expression datasets is the preferred method for miRNA expression analysis; however, this rigorous approach is difficult to achieve because of the unavailability of raw data. Of the reported studies, 11 out of 15 conducted a preliminary analysis to evaluate miRNA expression in their cohorts using microarray or sequencing. Only Maertzdorf’s (17) and Zhang’s (18) published raw data on public data sources. Considering the inconsistent annotation and discovery of new miRNAs, we attempted to find a simple and effective way to improve the statistical power of microarray and sequencing data by integrating the results of each study. Hence, we performed an integrated miRNA-PTB expression profiling analysis to identify miRNAs reported in at least three studies. Our integrated miRNA-PTB expression profiling analysis distinguished significantly upregulated and downregulated miRNAs in patients with PTB compared with those in HCs; however, there were inconsistencies in the reported data, and overlapping expression patterns were rarely found.
For example, Zhang et al. (19) and Tu et al. (20) reported that miR-320b was downregulated in serum samples from patients with PTB. In contrast, miR-320b was upregulated in a study by Wang et al. (21, 22). According to our RT-qPCR validation, miR-320b was downregulated in patients with PTB compared with that in HCs. A systematic review of miRNAs as biomarkers in TB revealed that miR-320b was downregulated in patients with PTB compared with that in HCs, which is consistent with our RT-qPCR verification and Simoa detection results (23). In addition, four studies showed inconsistent results; two studies reported that miR-99b was upregulated in serum samples from patients with PTB (21, 22), whereas Wang et al. (18, 24) found that miR-99b expression was downregulated. Similarly, the RT-qPCR and Simoa assays conducted in this study reported inconsistent expression trends, and the RT-qPCR results did not coincide well with the Simoa measurements for both relative and absolute quantification. Previous studies have demonstrated that miR-99b is highly expressed in Mtb-infected dendritic cells, suggesting its participation in immune evasion (25) and suppression of inflammation (26); however, neutrophil-associated miR-99b transcript levels are significantly low in TB cases (27). Despite the heterogeneity of the results, some miRNAs have been identified as candidate biomarkers for TB in more than one study. A study on plasma used miRNA PCR panels to obtain a short-list of candidates that were validated via RT-qPCR. Notably, two miRNAs (miR-99b and miR-29a) with high expression in plasma were upregulated in patients with PTB compared with those in the controls (28). The remaining two miRNAs (miR-29a and miR-196b) showed consistent expression trends in serum samples (17, 18, 24, 29, 30). miR-29a and miR-196b levels were elevated in the serum and plasma samples of patients with TB, which is consistent with our RT-qPCR results. According to a new study, there was a statistically significant decrease in the expression of miR-454-3p, miR-15a-5p, miR-590-5p, miR-381, and miR-449a in the Pulmonary TB group compared with those in the healthy control group (31).
At present, northern blotting, microarray and qRT-PCR are conventional methods for miRNAs detection, however, they are not capable of detecting miRNAs in complex biological clinical samples and do not provide good performance in ultra-sensitive miRNAs detection (9). The Simoa technique has been previously utilized for ultra-sensitive detection of proteins (32), since the platform allows detection of sufficiently small particles, it has also been used to detect DNA (33) or miRNAs (12). Thus, we have developed an ultrasensitive and non-invasive serum miRNAs detection assay using capture and detector probes with fully automated Simoa technology. In the present study, we focused on the development and optimisation of efficient methods for miRNA detection in serum samples using the Simoa platform. In this process, it is crucial to design LNA-modified capture and detection probes. Both these probes must bind to the target miRNAs and generate signals. The results indicated that the target miRNAs had strong binding abilities with the capture/detection probes. Unfortunately, miR-101 and miR-196b was excluded from further Simoa detection because a binding affinity signal was not detected for it. We used the Simoa platform to measure miRNAs in the serum without a separate separation step. For any potential future clinical application, it is notable that all serum samples were well in the measurable range of the Simoa assay. We found that the Simoa platform exhibits limits of detection in the range of 0.449-1.889 fM. The WHO has included the development of diagnostic biomarkers in its high-priority target product profile (TPP), (non-sputum-based diagnostic: >90% sensitivity and >70% specificity) (34). For active case finding, Simoa detection did not meet TPP benchmarks for a triage test. However, our data demonstrates the moderate sensitivity and specificity offered by a combination of three miRNAs detection in distinguishing PTB from HCs. We are confident that the Simoa assay can
serve as a valuable tool for TB screening. There is a growing tendency to integrate multiple miRNAs into a biomarker panel, which can enhance both sensitivity and specificity. The composite expression profile of several miRNAs can furnish a more refined biomarker signature, enhancing the discriminative capability between PTB and HCs. Moreover, miRNA expression can vary with disease progression, suggesting the value of dynamic monitoring rather than relying solely on static levels. As the protocol for the Simoa assay is still under development, there are some limitations to our study. The detection sensitivity needs to be further optimised. The percentage of all serum samples for whom the miRNAs were consistently >LOD in any measurement was 69.4% (88/121) for miR-29a, 70.2% (85/121) for miR-99b, and 74.4% (90/121) for miR-320b. Secondly, because these miRNAs can be similarly expressed in other infections of the lungs, such as bacterial pneumonia or NTM infection. Although statistically significant differences in miRNA expression have been identified, translating these findings into clinical practice requires rigorous validation. To enhance the reliability, quality assurance, and validity of these biomarkers, it is essential to establish standardized procedures for the preparation of various sample types. Furthermore, the development of better methods to assess the quality of miRNA markers is critical. Therefore, in subsequent studies, we will optimise the sensitivity of Simoa assays by maximising the capture of target molecules and subsequently use the minimum concentrations of labelling reagents to yield the highest signal-to-background ratio. In addition, expanding the patients cohort for further validation is necessary in order to test fully the utility of these miRNA biomarker candidates.
Our results support the feasibility of quantifying miRNAs in serum samples as differentiators for PTB. More importantly, Simoa has great potential for miRNA detection owing to its high sensitivity and because it does not require pre-labelling, reverse transcription, or amplification. Simoa technology warrants further research in a large, well-characterised longitudinal patient cohort.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by Center for Tuberculosis Control of Guangdong Province Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
ZW: Funding acquisition, Validation, Writing – original draft, Writing – review & editing. QT: Data curation, Writing – original draft. XJ: Writing – review & editing, Funding acquisition. HW: Data curation, Writing – original draft. JL: Investigation, Writing – original draft. WW: Writing – original draft. XW: Writing – original draft. CZ: Writing – original draft. YZ: Writing – original draft. YC: Writing – original draft. TL: Supervision, Writing – review & editing. WL: Project administration, Writing – review & editing. XC: Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2021YFC2600200), the Science and Technology Program of Guangzhou, China (No. 202201011764), Guanedone Provincial Clinical Research Center for Tuberculosis (No. 202081111170014), the Science and Technology Planning Proiect of Guangdong Province, China (No. 202181212030003), and the Infectious Disease Prevention and Control of the National Science and Technique Major Project (No. 2018ZX10715004002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1418085/full#supplementary-material
1. Bagcchi S. WHO's global tuberculosis report 2022. Lancet Microbe. (2023) 4:e20. doi: 10.1016/S2666-5247(22)00359-7
2. Garcia JI, Allue-Guardia A, Tampi RP, Restrepo BI, Torrelles JB. New developments and insights in the improvement of mycobacterium tuberculosis vaccines and diagnostics within the end TB strategy. Curr Epidemiol Rep. (2021) 8:33–45. doi: 10.1007/s40471-021-00269-2
3. Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. (2013) 14:475–88. doi: 10.1038/nrm3611
4. Teoh SL, Das S. MicroRNAs in various body fluids and their importance in forensic medicine. Mini Rev Med Chem. (2022) 22:2332–43. doi: 10.2174/1389557522666220303141558
5. Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells-Basel. (2020) 9(2):276. doi: 10.3390/cells9020276
6. Daniel EA, Sathiyamani B, Thiruvengadam K, Vivekanandan S, Vembuli H, Hanna LE. MicroRNAs as diagnostic biomarkers for Tuberculosis: A systematic review and meta- analysis. Front Immunol. (2022) 13:954396. doi: 10.3389/fimmu.2022.954396
7. Agrawal P, Upadhyay A, Kumar A. microRNA as biomarkers in tuberculosis: a new emerging molecular diagnostic solution. Diagn Microbiol Infect Dis. (2024) 108:116082. doi: 10.1016/j.diagmicrobio.2023.116082
8. Dave VP, Ngo TA, Pernestig AK, Tilevik D, Kant K, Nguyen T, et al. MicroRNA amplification and detection technologies: opportunities and challenges for point of care diagnostics. Lab Invest. (2019) 99:452–69. doi: 10.1038/s41374-018-0143-3
9. Jet T, Gines G, Rondelez Y, Taly V. Advances in multiplexed techniques for the detection and quantification of microRNAs. Chem Soc Rev. (2021) 50:4141–61. doi: 10.1039/D0CS00609B
10. Chen YX, Huang KJ, Niu KX. Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens Bioelectron. (2018) 99:612–24. doi: 10.1016/j.bios.2017.08.036
11. Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. (2010) 28:595–9. doi: 10.1038/nbt.1641
12. Rissin DM, Lopez-Longarela B, Pernagallo S, Ilyine H, Vliegenthart A, Dear JW, et al. Polymerase-free measurement of microRNA-122 with single base specificity using single molecule arrays: Detection of drug-induced liver injury. PLoS One. (2017) 12:e179669. doi: 10.1371/journal.pone.0179669
13. Wei P, Wu F, Kang B, Sun X, Heskia F, Pachot A, et al. Plasma extracellular vesicles detected by Single Molecule array technology as a liquid biopsy for colorectal cancer. J Extracell Vesicles. (2020) 9:1809765. doi: 10.1080/20013078.2020.1809765
14. Preische O, Schultz SA, Apel A, Kuhle J, Kaeser SA, Barro C, et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer's disease. Nat Med. (2019) 25:277–83. doi: 10.1038/s41591-018-0304-3
15. Cohen L, Hartman MR, Amardey-Wellington A, Walt DR. Digital direct detection of microRNAs using single molecule arrays. Nucleic Acids Res. (2017) 45:e137. doi: 10.1093/nar/gkx542
16. Owczarzy R, Tataurov AV, Wu Y, Manthey JA, McQuisten KA, Almabrazi HG, et al. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. (2008) 36:W163–9. doi: 10.1093/nar/gkn198
17. Maertzdorf J, Weiner JR, Mollenkopf HJ, Bauer T, Prasse A, Muller-Quernheim J, et al. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U.S.A. (2012) 109:7853–8. doi: 10.1073/pnas.1121072109
18. Zhang H, Sun Z, Wei W, Liu Z, Fleming J, Zhang S, et al. Identification of serum microRNA biomarkers for tuberculosis using RNA-seq. PLoS One. (2014) 9:e88909. doi: 10.1371/journal.pone.0088909
19. Zhang X, Guo J, Fan S, Li Y, Wei L, Yang X, et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS One. (2013) 8:e81076. doi: 10.1371/journal.pone.0081076
20. Tu H, Yang S, Jiang T, Wei L, Shi L, Liu C, et al. Elevated pulmonary tuberculosis biomarker miR-423-5p plays critical role in the occurrence of active TB by inhibiting autophagosome-lysosome fusion. Emerg Microbes Infect. (2019) 8:448–60. doi: 10.1080/22221751.2019.1590129
21. Wang C, Liu CM, Wei LL, Shi LY, Pan ZF, Mao LG, et al. A group of novel serum diagnostic biomarkers for multidrug-resistant tuberculosis by iTRAQ-2D LC-MS/MS and solexa sequencing. Int J Biol Sci. (2016) 12:246–56. doi: 10.7150/ijbs.13805
22. Wang C, Yang S, Liu CM, Jiang TT, Chen ZL, Tu HH, et al. Screening and identification of four serum miRNAs as novel potential biomarkers for cured pulmonary tuberculosis. Tuberculosis (Edinb). (2018) 108:26–34. doi: 10.1016/j.tube.2017.08.010
23. Wang L, Xiong Y, Fu B, Guo D, Zaky MY, Lin X, et al. MicroRNAs as immune regulators and biomarkers in tuberculosis. Front Immunol. (2022) 13:1027472. doi: 10.3389/fimmu.2022.1027472
24. Qi Y, Cui L, Ge Y, Shi Z, Zhao K, Guo X, et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis. (2012) 12:384. doi: 10.1186/1471-2334-12-384
25. Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, et al. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem. (2013) 288:5056–61. doi: 10.1074/jbc.C112.439778
26. Sinigaglia A, Peta E, Riccetti S, Venkateswaran S, Manganelli R, Barzon L. Tuberculosis-associated microRNAs: from pathogenesis to disease biomarkers. Cells-Basel. (2020) 9. doi: 10.3390/cells9102160
27. van Rensburg IC, du Toit L, Walzl G, du Plessis N, Loxton AG. Decreased neutrophil-associated miRNA and increased B-cell associated miRNA expression during tuberculosis. Gene. (2018) 655:35–41. doi: 10.1016/j.gene.2018.02.052
28. Barry SE, Ellis M, Yang Y, Guan G, Wang X, Britton WJ, et al. Identification of a plasma microRNA profile in untreated pulmonary tuberculosis patients that is modulated by anti-mycobacterial therapy. J Infect. (2018) 77:341–8. doi: 10.1016/j.jinf.2018.03.006
29. Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. (2011) 49:4246–51. doi: 10.1128/JCM.05459-11
30. Tu Y, Cai Q, Zhu X, Xu M. Down-regulation of HCP5 inhibits cell proliferation, migration, and invasion through regulating EPHA7 by competitively binding miR-101 in osteosarcoma. Braz J Med Biol Res. (2021) 54:e9161. doi: 10.1590/1414-431x20209161
31. Cavusoglu C, Cogulu O, Durmaz A, Cengisiz Z, Yilmaz FF, Tasbakan MS, et al. Investigation of miRNA and cytokine expressions in latent tuberculosis infection and active tuberculosis. Turk J Med Sci. (2022) 52:649–57. doi: 10.55730/1300-0144.5357
32. Andersson E, Schultz N, Saito T, Saido TC, Blennow K, Gouras GK, et al. Cerebral Abeta deposition precedes reduced cerebrospinal fluid and serum Abeta42/Abeta40 ratios in the App(NL-F/NL-F) knock-in mouse model of Alzheimer's disease. Alzheimers Res Ther. (2023) 15:64. doi: 10.1186/s13195-023-01196-8
33. Song L, Shan D, Zhao M, Pink BA, Minnehan KA, York L, et al. Direct detection of bacterial genomic DNA at sub-femtomolar concentrations using single molecule arrays. Anal Chem. (2013) 85:1932–9. doi: 10.1021/ac303426b
Keywords: tuberculosis, microRNAs, single molecule array, biomarker, Serum
Citation: Wu Z, Tan Q, Jia X, Wu H, Liang J, Wen W, Wang X, Zhang C, Zhao Y, Chen Y, Luo T, Liu W and Chen X (2024) Single molecule measurements of microRNAs in the serum of patients with pulmonary tuberculosis. Front. Immunol. 15:1418085. doi: 10.3389/fimmu.2024.1418085
Received: 16 April 2024; Accepted: 16 August 2024;
Published: 02 September 2024.
Edited by:
Subash Babu, International Centers for Excellence in Research (ICER), IndiaReviewed by:
Kathirvel Maruthai, Johns Hopkins University, United StatesCopyright © 2024 Wu, Tan, Jia, Wu, Liang, Wen, Wang, Zhang, Zhao, Chen, Luo, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xunxun Chen, Z3JhY2VfY2hlbjUxNEAxNjMuY29t; Wenjun Liu, bGl1d2pAaW0gYWMuY24=; Tingrong Luo, dGluZ3JvbmdsdW9AZ3h1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.