
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 06 November 2024
Sec. Mucosal Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1415475
This article is part of the Research TopicImmune Responses at Barrier Tissues: Insights from Synthetic Biology in Therapeutics, Diagnostics, Mechanisms, and BeyondView all 6 articles
 Emily L. Webb1
Emily L. Webb1 Stefan Petkov2
Stefan Petkov2 Heejin Yun3
Heejin Yun3 Laura Else4
Laura Else4 Limakatso Lebina5
Limakatso Lebina5 Jennifer Serwanga6
Jennifer Serwanga6 Azure-Dee A. P. Pillay5
Azure-Dee A. P. Pillay5 Thabiso B. Seiphetlo5
Thabiso B. Seiphetlo5 Susan Mugaba6
Susan Mugaba6 Patricia Namubiru6
Patricia Namubiru6 Geoffrey Odoch6
Geoffrey Odoch6 Daniel Opoka6
Daniel Opoka6 Andrew S. Ssemata6
Andrew S. Ssemata6 Pontiano Kaleebu6
Pontiano Kaleebu6 Saye Khoo4
Saye Khoo4 Neil Martinson5
Neil Martinson5 Julie Fox7
Julie Fox7 Clive M. Gray8
Clive M. Gray8 Carolina Herrera3†‡
Carolina Herrera3†‡ Francesca Chiodi2*‡ on behalf of CHAPS consortium
Francesca Chiodi2*‡ on behalf of CHAPS consortiumIntroduction: Tight junctions (TJs) serve as permeability filters between the internal and external cellular environment. A large number of proteins have been identified to be localized at the TJs. Due to limitations in tissue collection, TJs in the male genital tract have been understudied.
Methods: We analysed the transcriptomics of 132 TJ genes in foreskin tissue of men requesting voluntary medical male circumcision (VMMC) and enrolled in the Combined HIV Adolescent Prevention Study (CHAPS) trial conducted in South Africa and Uganda (NCT03986970). The trial evaluated the dose requirements for event-driven HIV pre-exposure prophylaxis (PrEP) with emtricitabine-tenofovir (FTC-TDF) or emtricitabine-tenofovir alafenamide (FTC-TAF) during insertive sex. A total of 144 participants were randomized to either control arm or one of 8 PrEP arms (n=16/arm), receiving oral FTC-TDF or FTC-TAF over one or two days. Following in vivo oral PrEP dosing and VMMC, the expression level of three important TJ proteins (CLDN-1, OCN and ZO-1) was measured ex vivo in foreskin tissue by Western blot. The expression of cytokine genes implicated in TJ regulation was determined. Non-parametric Kruskal-Wallis tests were used to compare TJ gene expression and protein levels by type of PrEP received, and Spearman’s correlation coefficients were calculated to assess whether TJ gene expression levels were related to cytokine gene levels or to PrEP drug concentrations and their active intracellularly phosphorylated metabolites.
Results: A high level of expression in foreskin tissue was found for 118 (of 132) TJ genes analysed; this finding contributed to create a map of TJ components within the male genital tract. Importantly, PrEP regimens tested in the CHAPS trial did not affect the expression of TJ genes and the analysed proteins in the foreskin; thus, further supporting the safety of this prevention strategy against HIV-1 transmission during insertive sex. Additionally, we identified the level of several cytokines’ genes to be correlated to TJ gene expression: among them, IL-18, IL-33 and VEGF.
Discussion: TJs can limit viral entry into target cells; to affect this biological function viruses can reduce the expression of TJ proteins. Our study, on the expression and regulation of TJs in the foreskin, contribute important knowledge for PrEP safety and further design of HIV-1 prophylaxis.
Adhesion between epithelial cells is provided by desmosomes, gap junctions, adherens junctions and tight junctions (1). The latter are intercellular adhesion complexes constituting the continuous intercellular barrier between epithelial cells which serves as a permeability filter by dividing the internal and external cellular milieus, thus controlling the movement of solutes across the epithelium (2). Networks of paired, spatially organized, TJ strands form a ring surrounding the cell, which regulate the diffusion of solutes throughout the cellular sheet. Several proteins are localized at the TJs, including integral membrane proteins as well as cytosolic adaptor proteins. Transmembrane proteins include claudins (CLDNs), protein crumbs homologue 3 (CRB3), MARVEL domain proteins such as occludin (OCLN), blood vessel epicardial substance (BVES), junctional adhesion molecules (JAMS), and other immunoglobulin (Ig)-type adhesion proteins such as nectins and e-cadherin. CLDNs form strands of similar sizes that can result from the polymerization of different CLDNs. The channels established by CLDN strands regulate the paracellular ions and solutes movement between epithelial cells. Similarly, OCLN regulates the paracellular passage between cells; however, it polymerizes forming short strand fragments. Functionality of transmembrane proteins is interlinked with their interactions to cytosolic adaptors, such as zonula occludens (ZO) proteins, which connect TJs to the actin-cytoskeleton and adherens junctions [reviewed in (2)]. ZO-1 is involved in the initial steps of TJ formation between cells forming primordial junctions where they couple the assembly of transmembrane proteins to form mature TJs (3, 4). The protein composition of TJs may be dictated by the functional properties of the different epithelial cell types in tissues including cell polarity, signaling and vesicle trafficking [reviewed in (5)]. The number of proteins considered part of the TJs family has been progressively growing during the recent years; approximately 130 genes are currently listed in the gene ontology term “cell-cell junction assembly” (https://amigo.geneontology.org/amigo/term/GO:0007043). Multiple functions are included under this gene ontology class allowing gene classification under different major subfamilies (Supplementary Table 1). The formation of TJs involves a high degree of functional redundancy as shown in each family; for example, CLDN1 and OCLN, two transmembrane proteins, are part of the “bicellular tight junction assembly” class. CLDN1 and OCLN interact with the cytosolic adaptor ZO-1, encoded by gene TJP1 which belongs to the gene ontology family of “cell-cell junction assembly”.
The abundance of TJ proteins in tissue is regulated by different cytokines. As examples, CLDN1 expression is down-regulated by TGFβ1 (6) and IL-33 (7), whereas TNF-α increases the expression of CLDN 1, 4 and 7 in tubular cells (8). Inflammatory conditions may also be a dramatic driving force for changes in the expression of TJ proteins. In this respect, it was shown that local cytokine production in the labial salivary gland was impaired in Sjögren’s syndrome patients, with this alteration affecting the TJs’ integrity of epithelial cells, a condition which could be mimicked in vitro by TNF-α and IFN-γ (9).
In view of their role as physical barriers, TJs can block viral entry into host cells. Viruses, however, have been shown to impair epithelial barriers for the purpose of increasing infection rate of target cells [reviewed in (10)]; this can be achieved by affecting the expression of TJ proteins and function, including permeability (11). The list of viruses able to promote infection of target cells by affecting TJs is comprehensive and includes retroviruses (HIV), flaviviruses (HCV, ZIKV, DENV), rabies virus and respiratory viruses (IAV, RSV, SARS-CoV, rotavirus) (12); these families of viruses have developed their own unique molecular strategies to utilize TJs for the purpose of infecting target tissues. HIV-1 was shown to induce down-regulation of TJ expression, particularly CLDN 2, OCLN and ZO-1 at the vaginal barrier (11–13) likely leading to increased permeability of the tissue to HIV-1 (12); down-regulation of TJ expression induced by viruses can also alter the epithelial TJ barrier role as sensors of host innate immune system (14).
The difficulty of obtaining tissue specimens hampers the possibility of building a complete profile of TJs in the human male genital tract, as well as the characterization of factors that can modulate their expression. The results obtained from randomized controlled trials conducted in Sub Saharan Africa demonstrated that voluntary medical male circumcision (VMMC) provided over 50% protection from HIV infection (15–17). The mechanism for the protective effect of VMMC is not fully clarified; however, as the foreskin carries high densities of HIV target cells (CD4+ T cells and Langerhans cells), the most likely mechanism is removal of tissue containing HIV target cells (18–20). VMMC has represented an important source of foreskin tissue useful to dissect physiological and pathological events in the male genital tract. A decreased expression of CLDN-1 was detected in the foreskin of asymptomatic HSV-2 seropositive individuals suggesting a fragile epithelial barrier in the genital tract which could increase the risk of contracting HIV-1 infection (21). A study on the foreskin of men who have sex with men or transgender women revealed subclinical changes in the inner foreskin, symptomatic of an inflammatory state with the potential of modifying epidermal barriers and availability of target cells for HIV-1 infection (22). Furthermore, ex vivo pre-clinical studies with human tissues (23–28) and clinical trials (29–32) have shown that event-driven oral and topical PrEP induce changes at the proteomic and transcriptomic levels in the female and male genital tracts as well as in the colorectum. However, no studies have assessed in parallel the impact of PrEP at the proteomic and transcriptomic levels in foreskin tissue and specifically the potential effect on epithelial integrity.
A recent open label randomized controlled trial (NCT03986970) conducted by the CHAPS consortium in South Africa and Uganda evaluated the dose for event-driven HIV PrEP for insertive sex (33). The trial included HIV-1 negative males requesting VMMC who were randomized to a control arm or to receive emtricitabine-tenofovir (FTC-TDF) or emtricitabine-tenofovir alafenamide (FTC-TAF) over one or two days. The primary outcome was to determine ex vivo protection against HIV-1 challenge of foreskin tissue samples (obtained at VMMC) (33). The results of the trial showed that all regimens tested with either FTC-TDF or FTC-TAF given prior to VMMC provided protection against ex vivo HIV-1 challenge. Ex vivo analysis of foreskin tissue from the CHAPS trial participants revealed that in vivo short-course of event-driven oral PrEP in men induced changes at the transcriptomic level, including modulation of genes involved in inflammation, mitochondrial function and cell proliferation (34).
As the number of individuals receiving HIV PrEP continues to grow world-wide, it is of high interest to study whether PrEP induces changes in the biology of the male genital tract which could be beneficial, or detrimental, for HIV protection. Hence, we further analyzed the data collected from the CHAPS trial focusing on the genes coding for TJ proteins in foreskin. The study provides the expression profile of the foreskin TJs and cytokine genes influencing their expression in this tissue.
The CHAPS trial recruited HIV negative males aged 13-24 years from VMMC clinics at Chris Hani Baragwanath Academic Hospital, Soweto, South Africa and Entebbe General Hospital, Entebbe, Uganda. Participants were randomized to either a control arm (receiving no PrEP) or PrEP trial arms where they received orally either FTC-TDF or FTC-TAF over two days or one day, before undergoing VMMC. Following in vivo dosing, or not for the control arm, samples including foreskin tissue, peripheral blood mononuclear cells (PBMCs) and plasma were collected from all individuals at time of VMMC, for ex vivo analyses. Resected inner and outer foreskin tissues were processed in the laboratory and cut into explants of different sizes specific for each ex vivo assay. Comparative analysis of outer and inner foreskins was not in the scope of this trial, and for each analysis an explant of outer was mixed with a piece of inner tissue. Full details of the CHAPS trial design and results for primary and key secondary outcomes have been described previously (33). At the circumcision visit, all participants provided midstream urine for Chlamydia trachomatis and Neisseria gonorrhea testing via nucleic acid amplification testing (NAAT) prior to surgery.
Details on preparation of resected foreskin and RNA sequencing of this tissue, including RNA-seq data processing and analysis were published (34). Briefly, total RNA was isolated from homogenized foreskin tissue samples (RNeasy kit, Qiagen, Hilden, Germany) and cDNA libraries prepared for Illumina sequencing with an Illumina Novaseq 6000 S4 flowcell (Illumina, San Diego, CA, USA). Reads were aligned to the Ensembl GRCh38 reference genome using STAR (v2.6.1d). Counts for each gene were obtained using featureCounts (v1.5.1). The transcriptome data presented in the study are deposited in the GenBank Data Libraries repository, accession number PRJNA884284.
For each participant, an inner and an outer foreskin explant of approximately 10 mg each were cut, pooled, and dry frozen at -80°C until analysis. Explants were lysed with a solution of Tris-HCl (pH 7.4) and 1x complete protease inhibitor cocktail (MilliporeSigma, Darmstadt, Germany) in Lysing Matrix A (MP Bio-medicals, Santa Ana, CA, USA), and homogenized with a FastPrep® Ribolyser FP120 (Thermo Scientific Savant™, Waltham, MA, USA). Lysed tissues were spun down and supernatants transferred to Vivaspin® 500 tubes (Sartorius, Göttingen, Germany) to concentrate the lysates. Recovered protein concentrates were quantified with Pierce TM BCA Protein Assay kit (Thermo Scientific). Ten µg of protein/well were loaded in a 9% SDS-polyacrylamide gel and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). The blots were blocked in PBS-5% skimmed milk overnight and then probed with rabbit anti-OCLN Ab (1:500; ab167161 Abcam, Cambridge, UK), mouse anti-CLDN-1 Ab (1:125; 37-4900 Invitrogen, Waltham, MA, USA) and rabbit anti-ZO1 Ab (1:5,000; ab2272 Sigma-Aldrich, Burlington, MA, USA). Membranes were then incubated with secondary-horseradish peroxidase (HRP) antibodies (HRP anti-rabbit IgG: 1:2,000; NA 943 and HRP anti-mouse IgG: 1:2,000; NA 931 GE Healthcare, Chicago, IL, USA). Proteins bands were visualized with ECL ClarityTM Western ECL Substrate (Bio-Rad). Normalization of TJ protein signals was conducted via detection of β-actin levels (HRP mouse anti-β-actin Ab, 1:10,000; ab49900 Abcam). Western blot images were captured by Azure c300 Imaging System (Azure biosystem, Dublin, CA, USA) and analyzed with Image Studio Lite software Ver 5.2 (LI-COR Bioscience, Lincoln, NE, USA).
Concentrations of TFV, FTC, the pro-drug TAF, and the active phosphorylated intracellular metabolites, tenofovir-diphosphate (TFV-DP) and emtricitabine-triphosphate (FTC-TP), were determined in foreskin tissue as described previously (35). Briefly, for each participant, inner and outer foreskin explants of 10 mg each were cut and frozen at -80°C in ice-cold chelating solution, methanol and 20 mM EDTA-EGTA (70:30 V/V). Explants were homogenized and analyte quantification was performed using a SCIEX 4500 or 5500 triple quadrupole mass spectrometer (AB Sciex UK Limited; Warrington, UK). Drug and metabolite concentrations in foreskin were quantified using a ng/sample or pmol/sample calibration curve and values normalized to ng/g or pmol/g of tissue.
Ex vivo susceptibility to HIV-1 infection was evaluated as described previously (33). Briefly, foreskin tissues resected by VMMC from each trial participant, were cut into explants and transferred to 96-well U-bottom plates in a non-polarized system. Explants were challenged ex vivo with HIV-1BaL for 20 h in the absence of drug. Infectivity was measured during 15 days of culture by analysis of p24 concentration in culture supernatants (Innotest HIV antigen ELISA, Fujirebio Europe, Ghent, Belgium).
Ethical clearance to conduct the trial was obtained from the South African Health Products Regulatory Authority (20181004); the Uganda Virus Research Institute Research Ethics Committee (GC/127/18/12/680); Uganda National Council of Science and Technology (HS 2534); Uganda National Drug Authority (618/NDA/DPS/09/2019) and the London School of Hygiene and Tropical Medicine Research Ethics Committee (Ref:17403). Informed written consent was collected from all participants. The Swedish Ethics Review Authority approved the laboratory studies of the collected specimens at the Karolinska Institutet (2020-00941).
Participant characteristics were summarized with frequencies for categorical variables and means and standard deviations for continuous variables. Characteristics of participants were compared between South Africa and Uganda using Wilcoxon ranksum test to compare age (as non-normally distributed), t-test to compare BMI, and chi-squared tests to compare trial arm. The distributions of TJ gene expression levels, foreskin genes encoding cytokines and TJ proteins were visualized and summarized as medians with interquartile ranges. Since distributions of these measures were either skewed or bi-modal, and could not be normalized through transformation, non-parametric statistical methods were used for all analyses. Spearman’s correlation coefficients and corresponding p-values were calculated to determine associations between (1) each pair of TJ genes, (2) each pair of foreskin genes encoding cytokines, and (3) each possible pair of TJ genes and foreskin genes encoding cytokines. Since expression levels of many of the TJ genes were strongly positively correlated, principal component analysis (PCA) of TJ gene expression levels was conducted as a data reduction technique to generate a new variable (the first principal component) comprising a linear combination of the TJ gene expression levels which captured a high degree of their variability; a high value for this variable represents high expression levels for many of the TJ genes, while a low value for this variable represent low expression levels for many of the TJ genes.
The effect of oral PrEP (trial arm) was examined by comparing gene expression levels (and the first principal component representing their combination) and genes encoding cytokines between trial arms (FTC-TDF versus FTC-TAF versus no PrEP) using the Kruskal-Wallis test. These analyses were repeated, separately for each country (South Africa and Uganda). Spearman’s correlation coefficients and corresponding p-values were calculated to assess the relationship of TJ genes and genes encoding cytokines with foreskin drug levels. Levels of TJ proteins measured by western blot were compared between trial arms (FTC-TDF versus FTC-TAF versus no PrEP) using Kruskal-Wallis tests. All analyses were adjusted for multiple testing using the false discovery rate (FDR): FDR-adjusted p-values are reported throughout unless otherwise indicated.
Of the 144 participants enrolled in CHAPS, foreskin TJs and cytokine gene expression as well as TJ proteins abundance data were available from 139, comprising 68 of the 72 participants enrolled in South Africa, and 71 of the 72 participants enrolled in Uganda. Characteristics of participants with data available for this analysis are summarized in Table 1. Overall, the median age of participants was 19 years (interquartile range 16-21 years) and the distributions of age and other characteristics were comparable across the two countries. A minority of individuals included in the trial (6 out of 139; 2 individuals from South Africa and 4 from Uganda) tested positive for Chlamydia trachomatis and all individuals were negative for Neisseria gonorrhea (Table 1); these results were not considered in the analyses.
Of the 132 TJ genes evaluated, 14 were excluded from further analyses as they were expressed in fewer than 70% of participants overall (Supplementary Table 2). Figure 1 and Supplementary Figure 1 show the expression levels for each participant for the remaining 118 genes, with participants ordered by trial arm and country, respectively. Among all participants combined, the 10 most highly expressed genes were TRAF4, RAP2B, CCND1, NPHP4, TJAP1, ACTB, CRB3, PRKCZ, CDH5 and ATP7B; the function of proteins coded by these genes is shown in Table 2.
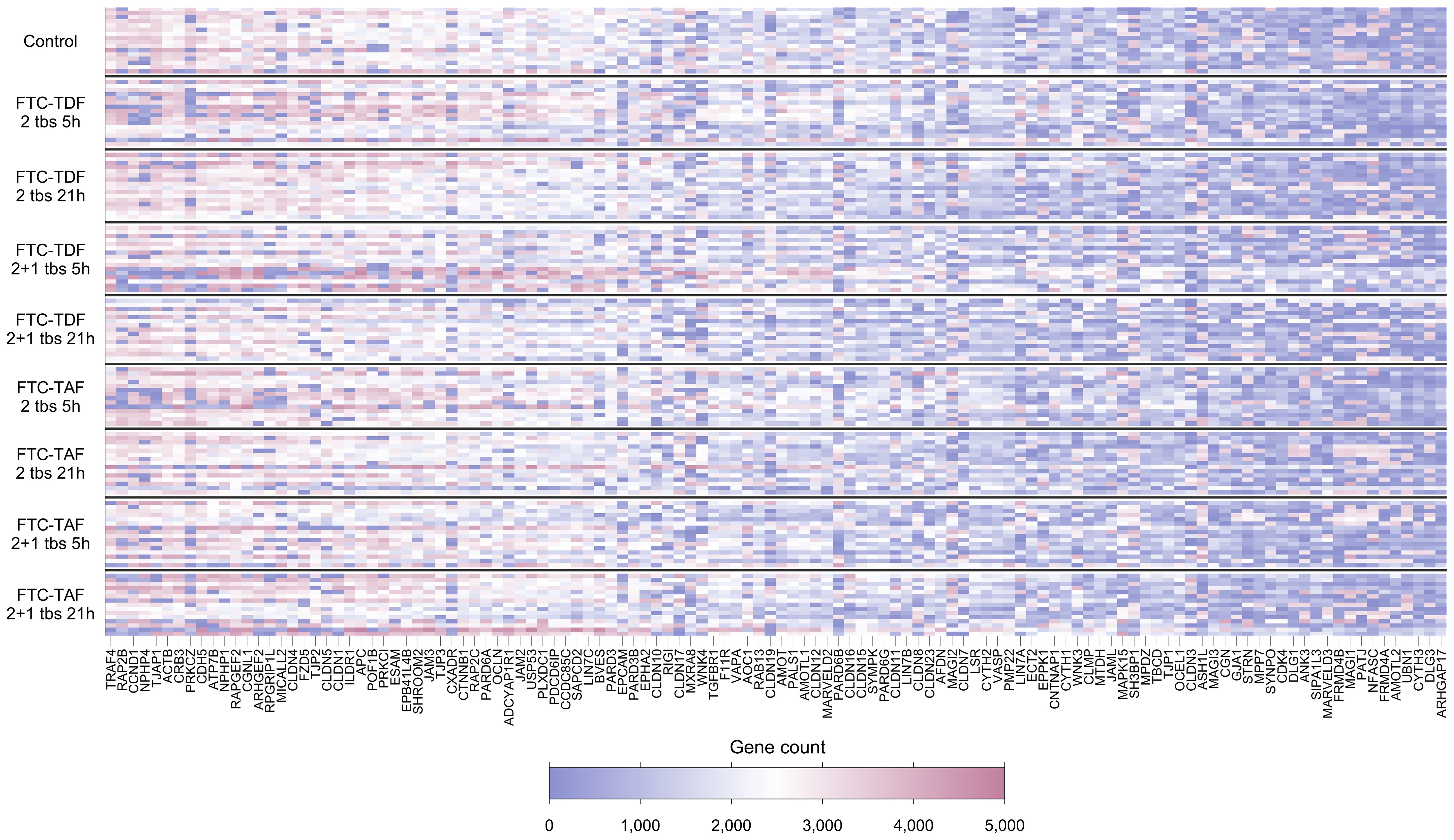
Figure 1. Heatmap showing expression levels (gene counts) of 118 TJ genes for 139 individuals included in the CHAPS study. Each row represents one individual, and each column represents one gene. Individuals (rows) are grouped according to their CHAPS trial arm. Genes (columns) are ordered by median gene expression level (high to low). Darker red values indicate higher gene counts while darker blue values indicate low gene counts, as shown in the legend.
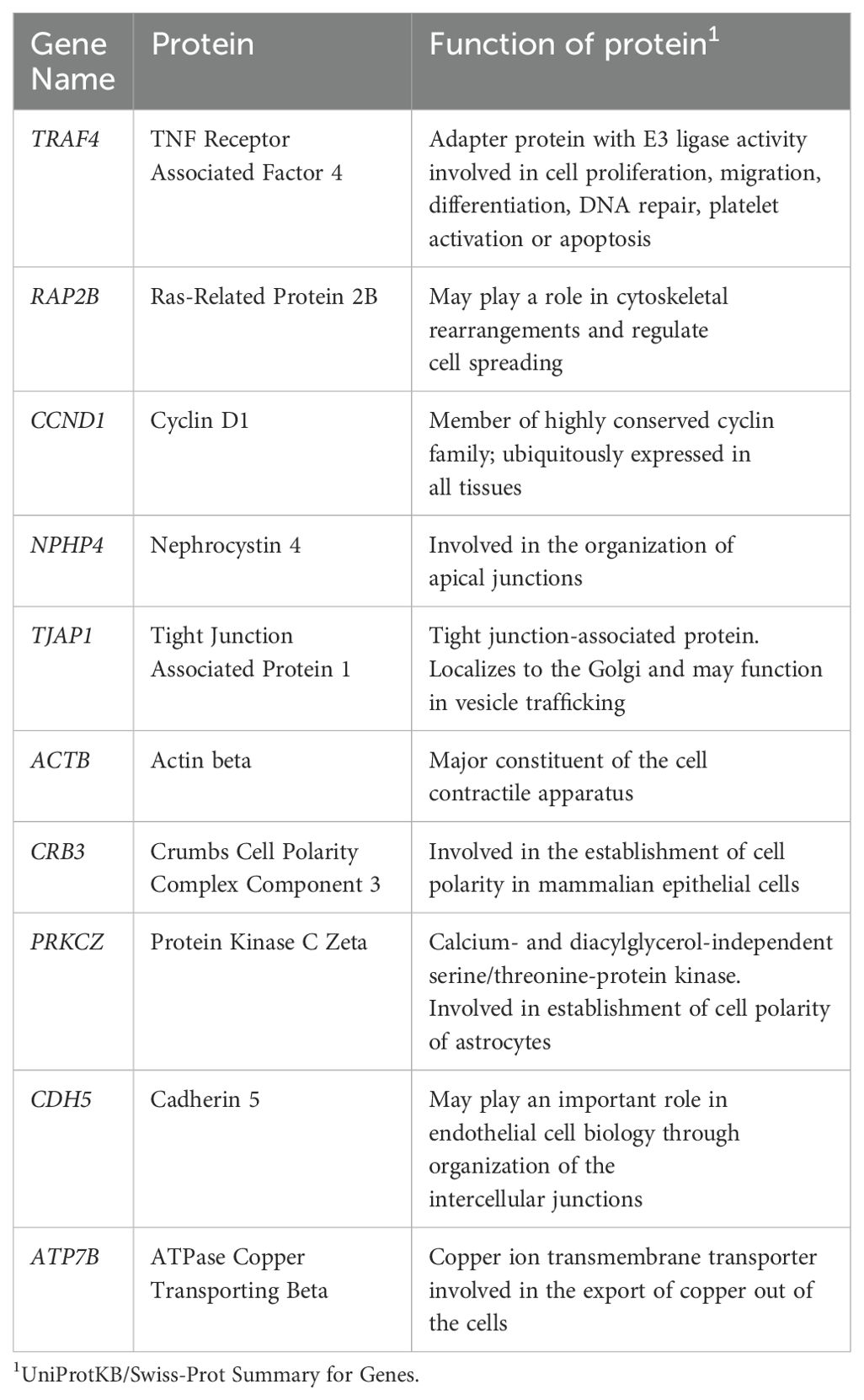
Table 2. Function of 10 most highly expressed tight junctions genes in foreskin samples from CHAPS participants.
Expression levels were strongly positively correlated between genes; of the 6,903 gene-gene pairs, 3,323 (48%) showed significant pairwise correlation after adjustment for multiple testing using FDR (all FDR-adjusted p-values <0.05; Supplementary Figure 2). In PCA analysis, the first component captured 31% of the variability in expression levels for all 118 TJ genes.
There were differences in gene expression levels between Ugandan and South African participants, with 25 of the 118 genes showing evidence of a difference (after FDR adjustment). For 22 of these 25 genes, expression was higher in samples from South African participants; the median expression ranged from 1.2 to 2-fold higher with corresponding p-values 0.04 to <0.0001 (Figure 2) whereas the expression of SH3BP1 (p<0.001), MICALL2 (p=0.006) and MSRA8 (p=0.04) was higher in individuals from Uganda.
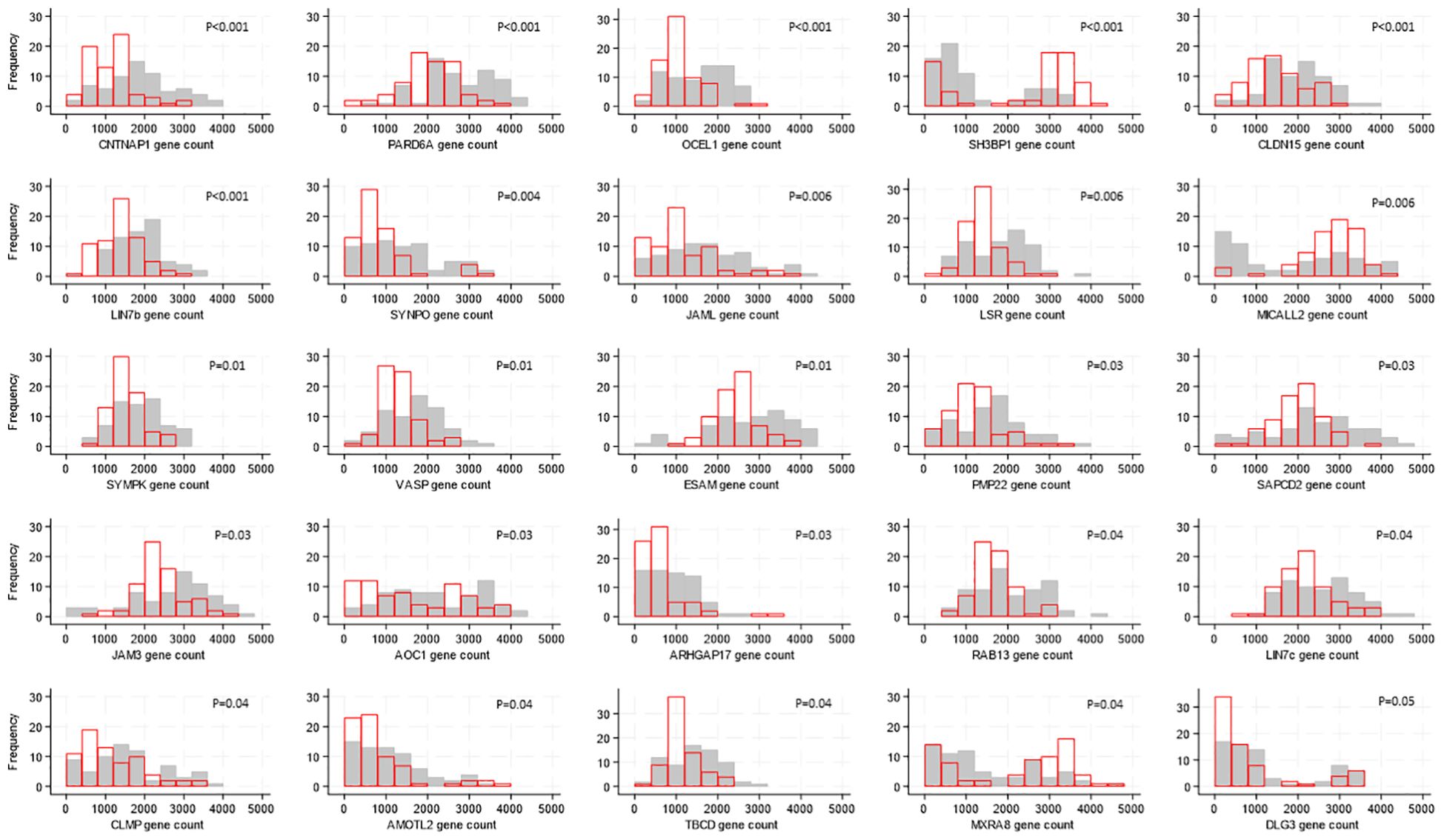
Figure 2. Distributions of tight junction gene expression levels by country, shown for the 25 genes whose expression levels differed by country. Gene counts are indicated by the values on the x-axis, categorized into groups of width 500, and the number of participants falling into each gene count group is shown on the y-axis. Grey bars show distribution of expression levels from South African participants, red bars show distribution of expression levels from Ugandan participants. P-values for differences in distribution of gene expression levels between countries were generated by Kruskal-Wallis tests and adjusted for multiple testing using the false discovery rate approach are indicated on each graph.
When examining the effect of oral PrEP through comparing gene expression levels by CHAPS trial arm, there was no impact of oral PrEP on foreskin TJ gene expression, either when comparing expression levels for each gene individually between trial arms (smallest FDR-adjusted p-value 0.14; Supplementary Table 3), or when comparing the score for the first principal component between trial arms (p=0.71; Supplementary Figure 3). This was also the case when participants from each country were analyzed separately (Supplementary Tables 4, 5).
Cytokines are known to influence and regulate the expression of TJ proteins (6–8); utilizing our transcriptomic data we evaluated whether the gene expression of cytokines known to affect TJ proteins correlated with the expression of TJ genes. Of the 21 genes encoding cytokines and evaluated in foreskin, eight were detectable in less than 70% of participants and were not analyzed further (Supplementary Table 6). Individuals’ levels for the remaining 13 are shown in Figure 3. The distribution of three genes differed between South African and Ugandan participants: TGFB1 was significantly higher in Ugandan participants (p<0.001), while VEGF (p<0.001) and MMP9 (p=0.03) were significantly increased in South African participants (Figure 4). There was no effect of oral PrEP on the distribution of these outcomes, either overall or when countries were analyzed separately (Supplementary Table 7).
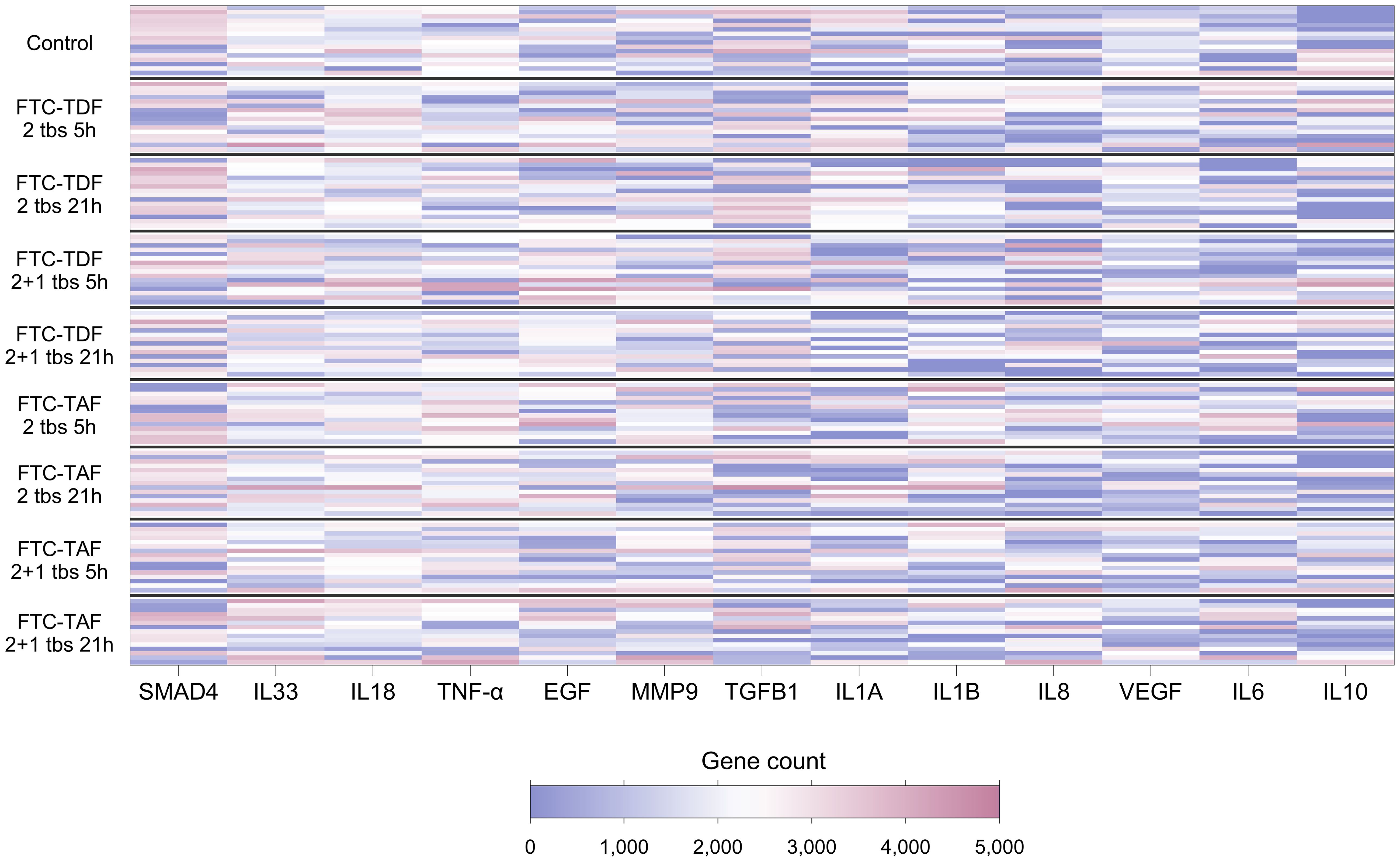
Figure 3. Heatmap showing expression levels of foreskin genes encoding cytokines, for 139 individuals included in the CHAPS study. Each row represents one individual, and each column represents one gene. Individuals (rows) are grouped according to their CHAPS trial arm. Genes encoding cytokines (columns) are ordered by median gene count (high to low). Darker red values indicate higher gene counts while darker blue values indicate lower gene counts, as shown in the legend.
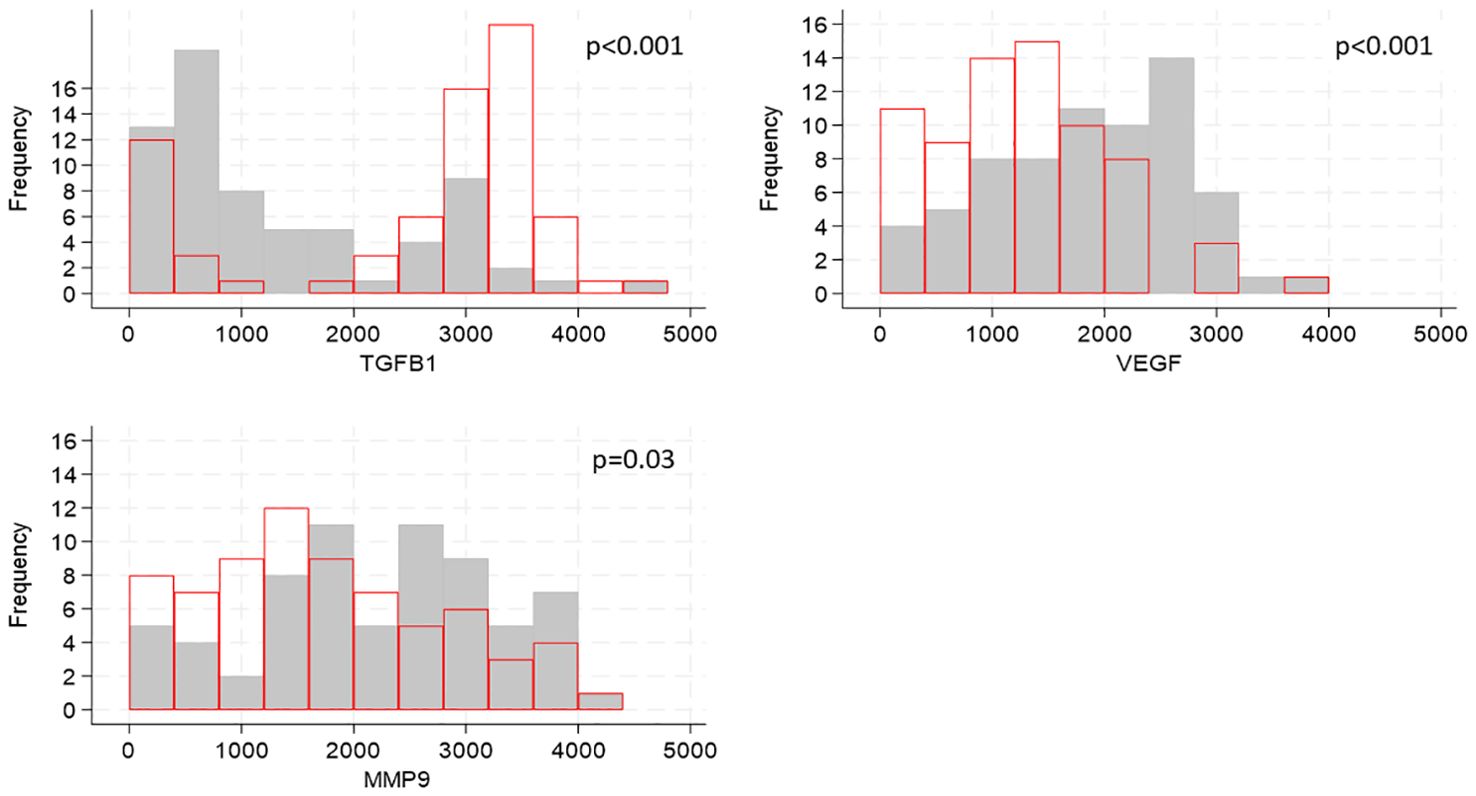
Figure 4. Distributions of cytokine gene expression by country, shown for genes whose distributions differed by country. Cytokine gene counts are indicated by the values on the x-axis, categorized into groups of width 400, and the number of participants falling into each group is shown on the y-axis. Grey bars show distribution of cytokine gene expression from South African participants, red bars show distribution of cytokine concentrations from Ugandan participants. P-values for differences in distributions between the two countries were generated by Kruskal-Walis tests and adjusted for multiple testing using the false discovery rate approach are indicated on each graph.
Foreskin genes encoding cytokines were strongly positively correlated with TJ gene expression levels. Of the 1534 gene-cytokine pairs, 481 (31%) showed significant pairwise correlation (Figure 5). The cytokine genes most strongly correlated with levels of TJ gene expression were IL-18, VEGF and IL-33. Supplementary Figure 4 displays the relationship of these cytokine gene counts with the score for the first principal component (which captures a large degree of the variability in expression levels for genes analyzed); correlation coefficients were 0.71, 0.56 and 0.45, respectively (all p<0.001).
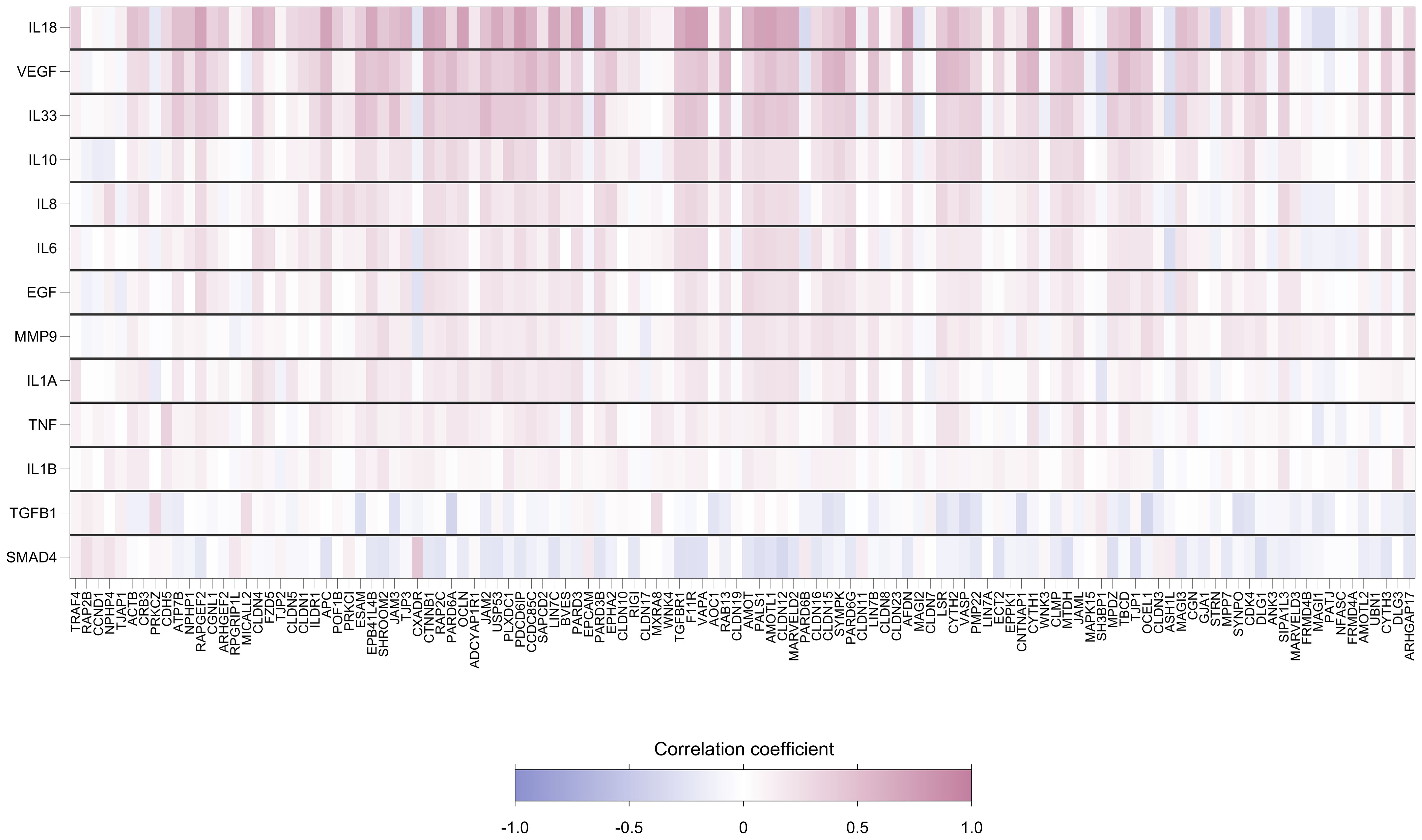
Figure 5. Heatmap showing pairwise correlation coefficients between expression levels of 118 foreskin tight junctions and 13 cytokine genes. Each row represents a gene encoding cytokine, and each column represents a TJ gene. Spearman’s correlation coefficient was calculated for each row-column pair. Darker red values indicate higher positive correlation while darker blue values indicate higher negative correlation, as shown in the legend.
We found no evidence that TJs and cytokine gene expression levels were associated with the concentrations of TFV-DP and FTC-TP measured in foreskin, either overall or in separate country-specific analyses (Supplementary Table 8). No evidence of correlation was found between p24 concentrations measured in foreskin tissue challenged ex vivo with HIV-1 and either TJs or cytokine gene levels (Supplementary Table 9). The pharmacodynamic and pharmacokinetic profiles of these drugs in foreskin have been extensively presented (33).
We then analyzed the level of expression for three TJ proteins, CLDN-1, OCLN and ZO-1, via western blot. Samples from 138 participants were available; one participant from South Africa included in the transcriptomic analysis did not have a sample available for proteomic analyses. When examining the effect of oral PrEP regimens tested in the CHAPS trial on these proteins, no differences between treatment arms were detected (Supplementary Table 10, Supplementary Figure 5).
Our study is the first to report the complete expression profile of TJ genes in the foreskin tissue of individuals receiving oral PrEP (both TDF-FTC and TAF-FTC) in two different sub-Saharan countries, South Africa and Uganda. The results showed that the expression of TJ genes is not altered in subjects receiving short-term PrEP and that there was no correlation of expressed TJ genes with the concentrations of TFV-DP and FTC-TP measured in foreskin of these subjects nor with the ex vivo HIV-1BaL infectivity levels (33).
Our consortium conducted a clinical trial comparing efficacy of FTC-TDF versus FTC-TAF dosing prior to VMMC on HIV-1BaL infection of foreskin using an ex vivo challenge model. This trial simulated the relative protection that could be attained for insertive sex by pre- and post-exposure regimens. Foreskin tissue was utilized to determine dosing schedules and duration of protection against ex vivo challenge (35); as well as concentrations of drug and active metabolites in foreskin tissue. The levels of drug metabolites TFV-DP and FTC-TP in foreskin were comparable to exposures previously reported in cervical and vaginal tissues (36, 37). It is of interest that, in men included in the CHAPS trial, timing and dose of F/TDF and F/TAF had no significant impact on CD4+CCR5+ cell numbers in foreskins and on CCR5 expression levels on CD4+ cells compared to the control arm, as shown by immunohistochemistry imaging (38); these results suggested that PrEP does not alter the immunological microenvironment of the foreskin and that PrEP induced protection from HIV infection is not the result of a diminished expression of HIV co-receptor CCR5 on CD4+ cells (38).
In our study, both FTC-TDF and FTC-TAF were well tolerated and highly effective against ex vivo challenge of foreskin tissue with a clade B R5-tropic laboratory adapted isolate, HIV-1BaL (33). It is relevant that in spite of the measurable levels of FTC-TDF and FTC-TAF in the foreskin tissue and the protection that these drug levels provided to HIV-1BaL ex vivo challenge, the expression of foreskin TJ genes studied by us did not change significantly when the control group was compared to groups receiving either FTC-TDF or FTC-TAF. That TJ expression remains unchanged in our study in individuals receiving PrEP is an encouraging information as a large number of individuals receive PrEP world-wide. The results obtained from the short time PrEP administration tested in the CHAPS trial are corroborated by the results presented in a previous study by Hladik et al. (29) where the effect of tenofovir 1% gel on rectal mucosa was studied in individuals treated with once-daily applications for seven consecutive days. In this study, Supplementary Materials listed genes which were up- or down regulated in biopsies obtained at day 7 of treatment in relation to biopsies obtained at enrolment. We assessed whether any of the 118 genes included in the gene ontology term “cell-cell junction assembly” studied in the present work were dysregulated in the supplementary results presented by Hladik and co-authors (29). Interestingly, only one gene MARVELD3 (involved in bicellular TJ assembly) was down-regulated after 7-days of treatment (29). Deletion of CLDN-1, an important TJs component, was demonstrated to be highly lethal in mice after birth because of defects in epidermal barrier function (39). We have previously performed immunohistochemistry analysis of CLDN-1 protein abundance in the foreskin tissues collected during CHAPS trial (38); the results showed that the geometric mean of percentage CLDN-1 levels was 34% higher in foreskin tissue from participants receiving PrEP, compared with the control arm, a result which was no longer significant, after allowing for multiple comparisons. In the present study, measurement of CLDN-1, OCN and ZO-1 by western blot revealed that the abundance level in foreskin tissue from the PrEP treated individuals did not differ from the control group. Considering the different body compartments studied by us and Hladik and co-authors (29), the different length of treatment and the multiple methods used for analyses we can anticipate that PrEP has a minimal impact, if any, on the expression of TJ genes.
The expression of all 118 TJ genes found in over 70% of individuals included in the present study was in general high, with read counts >500. Among the most highly expressed (>2600 copies), genes with a broad range of functions could be found; the mapping of these expressed genes to the foreskin may be useful in the context of mechanism of diseases and therapeutical approaches. It is interesting that the expression of 25 genes coding for TJ proteins differed between participants in South Africa and Uganda. Whether this difference in the expression of a group of genes reflects a specialized biological function remains to be studied. One hypothesis is that genetically determined differences, as previously reported by the African Genome Variation Project (40), or diet, could be at the basis for these differences; in this context, it was reported that the intestinal TJ barrier could be regulated by dietary factors in connection to different inflammatory disorders [reviewed in (41)]. There are 9654 genes present in the human gene database (https://www.genecards.org) which are associated to HIV; among those the expression of TJ genes CNTNAP1, PARD6A, LIN7B, SYNPO, LSR, SYMPK, JAM3, AOC1, RAB13, LIN7C, AMOTL2, TBCD, as detected in our study, was found to be higher in individuals from South Africa. Further studies could reveal if the increased expression of the 12 TJ genes associated with HIV in South African males render them less susceptible to HIV infection.
We found that the expression levels of the majority of TJ genes were strongly positively correlated. Correlations in gene expression can be used to deduce functional and regulatory relationships between genes; the optimal setting to study gene correlations is to use single cell transcriptomics (42). In our study, we used the whole foreskin tissue to study gene expression; considering that the studied genes all belong to the gene ontology TJs it is likely that the highly positive correlations between TJ genes further strengthen the concept that the selected genes represent a cluster which components together carry out complex biological functions.
It is of interest that foreskin genes encoding cytokines were strongly correlated with TJ gene expression levels. That expression of TJ proteins is influenced by cytokines has been reported previously in several contexts (6–9, 43, 44). In our analyses, IL-18, VEGF and IL-33 were the cytokine genes which most strongly correlated with levels of TJ gene expression. Dysregulated expression of these cytokines can be linked to several types of disorders in humans: induction of VEGF has been reported to be induced by several viruses associated with cancer (45); IL-18 has been linked to several inflammatory diseases including HIV-1 (46); IL-33 has been linked to HIV-associated neurological disorders (47). HIV-1 has been shown to induce down-regulation of TJs to facilitate infection of target cells (11–13).
Our study presents with limitations which should be taken in consideration in follow-up investigations. The function of TJ genes has been previously studied in detail in epithelial cells whereas the complete picture of TJ expression and function in other cells, including immune cells, remains incomplete. The expression of TJ genes in the present study was obtained for the whole foreskin tissue and individual cellular components were not separated. The foreskin tissue has, in fact, been shown to be rich in epithelial cells and Langerhans’ cells which are target cells for HIV-1 infection (48); in addition, dendritic cells, macrophages and T cells are also present in foreskin although they are predominantly dermal. Furthermore, a separate analysis for inner and outer foreskins was not performed. Considering the heterogeneity of HIV-target cells between inner and outer tissues (49, 50), further studies will be essential to determine any tissue-specific modulation of TJs by cytokines/chemokines and potential impact of PrEP regimes. Another important limitation in the present study is that PrEP administration in the CHAPS trial was limited to a maximum of two doses administered with a maximum interval of 24 hours; it is important for these studies to be repeated in individuals who have received PrEP for a prolonged period of time. The opportunity to conduct novel studies on the relevance of genital tissue barriers for protection from HIV or other sexually transmitted infections could be provided by trials focused on long-acting PrEP drugs. A phase 3 trial enrolling young women and adolescent girls in Uganda and South Africa (51) demonstrated that twice-yearly administration of subcutaneous Lenacapavir, a long-acting agent approved by the Food and Drug Administration (FDA) for HIV treatment, prevented HIV infection. The incidence of laboratory-diagnosed, sexually transmitted infections C. trachomatis, N. gonorrhoeae, or T. vaginalis infections was however high in the Lenacapavir group and comparable to groups of women receiving F/TAF or F/TDF in the same trial (51).
The mRNA gene expression has been reported to highly correlate (90%) with protein level (52); our study is mostly based on mRNA gene expression, and it would be interesting to study whether any difference can be found when methods aimed at determining protein levels in tissue will be applied to biological specimens from individuals receiving PrEP. Additional studies should verify the biological role of individual TJ proteins in the foreskin tissue. Another important aspect of the CHAPS trial is the focus on young males between the ages of 13 and 24-years old. Testosterone has been shown to modulate the expression level of TJs in the male genital tract. Expression of ZO-1 and CLDN-1 has been shown to be upregulated by testosterone (53, 54). The immune system is also affected by hormonal changes during adolescence (55), which could impact the formation of TJ in foreskin tissue. In this study we did not have statical power to identify potential changes in TJ expression levels and the potential impact of PrEP, between different age groups within the range of participants recruited.
The major findings of this study are: i) the expression map of TJ components in the foreskin tissue; ii) PrEP does not affect the expression of TJ genes and proteins in the foreskin; iii) identification of cytokines whose gene expression is linked to TJ gene expression.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical clearance to conduct the trial was obtained from the South African Health Products Regulatory Authority (20181004); the Uganda Virus Research Institute Research Ethics Committee (GC/127/18/12/680); Uganda National Council of Science and Technology (HS 2534); Uganda National Drug Authority (618/NDA/DPS/09/2019) and the London School of Hygiene and Tropical Medicine Research Ethics Committee (Ref: 17403). Informed written consent was collected from all participants. The Swedish Ethics Review Authority approved the laboratory studies of the collected specimens at the Karolinska Institutet (2020-00941). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
EW: Conceptualization, Formal Analysis, Writing – original draft, Writing – review & editing, Visualization, Validation. SP: Formal Analysis, Visualization, Writing – original draft, Writing – review & editing, Investigation, Validation. HY: Formal Analysis, Writing – review & editing, Investigation, Validation. LE: Formal Analysis, Writing – review & editing, Investigation, Validation. LL: Conceptualization, Writing – review & editing, Data curation. JS: Data curation, Writing – review & editing, Investigation. AP: Investigation, Writing – review & editing, Data curation. TS: Investigation, Writing – review & editing, Data curation. SM: Investigation, Writing – review & editing, Data curation. PN: Investigation, Writing – review & editing, Data curation. GO: Writing – review & editing, Investigation, Data curation. DO: Writing – review & editing, Investigation, Data curation. AS: Writing – review & editing, Data curation, Investigation. PK: Project administration, Resources, Writing – review & editing. SK: Resources, Writing – review & editing. NM: Writing – review & editing, Project administration, Resources. JF: Project administration, Writing – review & editing, Conceptualization, Funding acquisition. CG: Funding acquisition, Conceptualization, Writing – review & editing. CH: Conceptualization, Formal Analysis, Funding acquisition, Writing – original draft, Writing – review & editing, Supervision, Validation. FC: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing, Resources, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by EDCTP2 programme from the European Union (grant number RIA2016MC-1616-CHAPS), and from the Swedish Research Council (Francesca Chiodi and Carolina Herrera; Vetenskapsrådet 2019–04596).
We thank Dr Bethany Schneiderman, Imperial College London, for technical assistance. We would like to acknowledge all study participants and their parents for their involvement in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1415475/full#supplementary-material
1. Campbell HK, Maiers JL, DeMali KA. Interplay between tight junctions & adherens junctions. Exp Cell Res. (2017) 358:39–44. doi: 10.1016/j.yexcr.2017.03.061
2. Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. (2016) 17:564–80. doi: 10.1038/nrm.2016.80
3. Rajasekaran AK, Hojo M, Huima T, Rodriguez-Boulan E. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J Cell Biol. (1996) 132:451–63. doi: 10.1083/jcb.132.3.451
4. Maiers JL, Peng X, Fanning AS, DeMali KA. ZO-1 recruitment to α-catenin–a novel mechanism for coupling the assembly of tight junctions to adherens junctions. J Cell Sci. (2013) 126:3904–15. doi: 10.1242/jcs.126565
5. Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol. (2009) 1:a002584. doi: 10.1101/cshperspect.a002584
6. Wang K, Pascal LE, Li F, Chen W, Dhir R, Balasubramani GK, et al. Tight junction protein claudin-1 is downregulated by TGF-beta1 via MEK signaling in benign prostatic epithelial cells. Prostate. (2020) 80:1203–15. doi: 10.1002/pros.v80.14
7. Ryu WI, Lee H, Bae HC, Jeon J, Ryu HJ, Kim J, et al. IL-33 down-regulates CLDN1 expression through the ERK/STAT3 pathway in keratinocytes. J Dermatol Sci. (2018) 90:313–22. doi: 10.1016/j.jdermsci.2018.02.017
8. Amoozadeh Y, Dan Q, Anwer S, Huang HH, Barbieri V, Waheed F, et al. Tumor necrosis factor-alpha increases claudin-1, 4, and 7 expression in tubular cells: role in permeability changes. J Cell Physiol. (2017) 232:2210–20. doi: 10.1002/jcp.v232.8
9. Ewert P, Aguilera S, Alliende C, Kwon YJ, Albornoz A, Molina C, et al. Disruption of tight junction structure in salivary glands from Sjogren’s syndrome patients is linked to proinflammatory cytokine exposure. Arthritis Rheumatol. (2010) 62:1280–9. doi: 10.1002/art.27362
10. Bomsel M, Alfsen A. Entry of viruses through the epithelial barrier: pathogenic trickery. Nat Rev Mol Cell Biol. (2003) 4:57–68. doi: 10.1038/nrm1005
11. Ding G, Shao Q, Yu H, Liu J, Li Y, Wang B, et al. Tight junctions, the key factor in virus-related disease. Pathogens. (2022) 11. doi: 10.3390/pathogens11101200
12. Nazli A, Chan O, Dobson-Belaire WN, Ouellet M, Tremblay MJ, Gray-Owen SD, et al. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PloS Pathog. (2010) 6:e1000852. doi: 10.1371/journal.ppat.1000852
13. Passmore JA, Jaspan HB, Masson L. Genital inflammation, immune activation and risk of sexual HIV acquisition. Curr Opin HIV AIDS. (2016) 11:156–62. doi: 10.1097/COH.0000000000000232
14. Nazli A, Dizzell S, Zahoor MA, Ferreira VH, Kafka J, Woods MW, et al. Interferon-beta induced in female genital epithelium by HIV-1 glycoprotein 120 via Toll-like-receptor 2 pathway acts to protect the mucosal barrier. Cell Mol Immunol. (2019) 16:178–94. doi: 10.1038/cmi.2017.168
15. Auvert B, Taljaard D, Lagarde E, Sobngwi-Tambekou J, Sitta R, Puren A. Randomized, controlled intervention trial of male circumcision for reduction of HIV infection risk: the ANRS 1265 Trial. PloS Med. (2005) 2:e298. doi: 10.1371/journal.pmed.0020298
16. Gray RH, Kigozi G, Serwadda D, Makumbi F, Watya S, Nalugoda F, et al. Male circumcision for HIV prevention in men in Rakai, Uganda: a randomised trial. Lancet. (2007) 369:657–66. doi: 10.1016/S0140-6736(07)60313-4
17. Bailey RC, Moses S, Parker CB, Agot K, Maclean I, Krieger JN, et al. Male circumcision for HIV prevention in young men in Kisumu, Kenya: a randomised controlled trial. Lancet. (2007) 369:643–56. doi: 10.1016/S0140-6736(07)60312-2
18. Fahrbach KM, Barry SM, Anderson MR, Hope TJ. Enhanced cellular responses and environmental sampling within inner foreskin explants: implications for the foreskin’s role in HIV transmission. Mucosal Immunol. (2010) 3:410–8. doi: 10.1038/mi.2010.18
19. Zhou Z, Xu L, Sennepin A, Federici C, Ganor Y, Tudor D, et al. The HIV-1 viral synapse signals human foreskin keratinocytes to secrete thymic stromal lymphopoietin facilitating HIV-1 foreskin entry. Mucosal Immunol. (2018) 11:158–71. doi: 10.1038/mi.2017.23
20. Gray CM, O’Hagan KL, Lorenzo-Redondo R, Olivier AJ, Amu S, Chigorimbo-Murefuet N, et al. Impact of chemokine C-C ligand 27, foreskin anatomy and sexually transmitted infections on HIV-1 target cell availability in adolescent South African males. Mucosal Immunol. (2020) 13:118–27. doi: 10.1038/s41385-019-0209-6
21. Rohl M, Tjernlund A, Mehta SD, Pettersson P, Bailey RC, Broliden K. Comparable mRNA expression of inflammatory markers but lower claudin-1 mRNA levels in foreskin tissue of HSV-2 seropositive versus seronegative asymptomatic Kenyan young men. BMJ Open. (2015) 5:e006627. doi: 10.1136/bmjopen-2014-006627
22. Lemos MP, Lama JR, Karuna ST, Fong Y, Montano SM, Ganoza C, et al. The inner foreskin of healthy males at risk of HIV infection harbors epithelial CD4+ CCR5+ cells and has features of an inflamed epidermal barrier. PloS One. (2014) 9:e108954. doi: 10.1371/journal.pone.0108954
23. Fischetti L, Barry SM, Hope TJ, Shattock RJ. HIV-1 infection of human penile explant tissue and protection by candidate microbicides. AIDS. (2009) 23:319–28. doi: 10.1097/QAD.0b013e328321b778
24. Herrera C, McRaven MD, Laing KG, Dennis J, Hope TJ, Shattock RJ. Early colorectal responses to HIV-1 and modulation by antiretroviral drugs. Vaccines (Basel). (2021) 9. doi: 10.3390/vaccines9030231
25. Brand RM, Siegel A, Myerski A, Metter EJ, Engstrom J, Brand RE, et al. Ranpirnase reduces HIV-1 infection and associated inflammatory changes in a human colorectal explant model. AIDS Res Hum Retroviruses. (2018) 34:838–48. doi: 10.1089/aid.2017.0308
26. Crakes KR, Herrera C, Morgan JL, Olstad K, Hessell AJ, Ziprin P, et al. Efficacy of silk fibroin biomaterial vehicle for in vivo mucosal delivery of Griffithsin and protection against HIV and SHIV infection ex vivo. J Int AIDS Soc. (2020) 23:e25628. doi: 10.1002/jia2.v23.10
27. Biswas N, Rodriguez-Garcia M, Crist SG, Shen Z, Bodwell JE, Fahey JV, et al. Effect of tenofovir on nucleotidases and cytokines in HIV-1 target cells. PloS One. (2013) 8:e78814. doi: 10.1371/journal.pone.0078814
28. Biswas N, Rodriguez-Garcia M, Shen Z, Crist SG, Bodwell JE, Fahey JV, et al. Effects of tenofovir on cytokines and nucleotidases in HIV-1 target cells and the mucosal tissue environment in the female reproductive tract. Antimicrob Agents Chemother. (2014) 58:6444–53. doi: 10.1128/AAC.03270-14
29. Hladik F, Burgener A, Ballweber L, Gottardo R, Vojtech L, Fourati S, et al. Mucosal effects of tenofovir 1% gel. Elife. (2015) 4. doi: 10.7554/eLife.04525
30. Keller MJ, Wood L, Billingsley JM, Ray LL, Goymer J, Sinclair S, et al. Tenofovir disoproxil fumarate intravaginal ring for HIV pre-exposure prophylaxis in sexually active women: a phase 1, single-blind, randomised, controlled trial. Lancet HIV. (2019) 6:e498–508. doi: 10.1016/S2352-3018(19)30145-6
31. Thurman AR, Schwartz JL, Brache V, Clark MR, McCormick T, Chandra N, et al. Randomized, placebo controlled phase I trial of safety, pharmacokinetics, pharmacodynamics and acceptability of tenofovir and tenofovir plus levonorgestrel vaginal rings in women. PloS One. (2018) 13:e0199778. doi: 10.1371/journal.pone.0275794
32. McGowan I, Hoesley C, Cranston RD, Andrew P, Janocko L, Dai JY, et al. A phase 1 randomized, double blind, placebo controlled rectal safety and acceptability study of tenofovir 1% gel (MTN-007). PloS One. (2013) 8:e60147. doi: 10.1371/journal.pone.0060147
33. Herrera C, Serwanga J, Else L, Limakatso L, Opoka D, Ssemata AS, et al. Dose finding study for on-demand HIV pre-exposure prophylaxis for insertive sex in sub-Saharan Africa: results from the CHAPS open label randomised controlled trial. EBioMedicine. (2023) 93:104648. doi: 10.1016/j.ebiom.2023.104648
34. Petkov S, Herrera C, Else L, Lebina L, Opoka D, Seiphetlo TB, et al. Short-term oral pre-exposure prophylaxis against HIV-1 modulates the transcriptome of foreskin tissue in young men in Africa. Front Immunol. (2022) 13:1009978. doi: 10.3389/fimmu.2022.1009978
35. Else L, Penchala SD, Pillay AD, Seiphetlo TB, Lebina L, Callebaut C, et al. Pre-clinical evaluation of tenofovir and tenofovir alafenamide for HIV-1 pre-exposure prophylaxis in foreskin tissue. Pharmaceutics. (2022) 14. doi: 10.3390/pharmaceutics14061285
36. Cottrell ML, Garrett KL, Prince HMA, Sykes C, Schauer A, Emerson CW, et al. Single-dose pharmacokinetics of tenofovir alafenamide and its active metabolite in the mucosal tissues. J Antimicrob Chemother. (2017) 72:1731–40. doi: 10.1093/jac/dkx064
37. Thurman AR, Schwartz JL, Cottrell ML, Brache V, Chen BA, Cochon L, et al. Safety and pharmacokinetics of a tenofovir alafenamide fumarate-emtricitabine based oral antiretroviral regimen for prevention of HIV acquisition in women: A randomized controlled trial. EClinicalMedicine. (2021) 36:100893. doi: 10.1016/j.eclinm.2021.100893
38. Rametse CL, Webb EL, Herrera C, Alinde B, Besethi A, Motaung B, et al. A randomized clinical trial of on-demand oral pre-exposure prophylaxis does not modulate lymphoid/myeloid HIV target cell density in the foreskin. AIDS. (2023) 37:1651–9. doi: 10.1097/QAD.0000000000003619
39. Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol. (2002) 156:1099–111. doi: 10.1083/jcb.200110122
40. Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, et al. The African Genome Variation Project shapes medical genetics in Africa. Nature. (2015) 517:327–32. doi: 10.1038/nature13997
41. Suzuki T. Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim Sci J. (2020) 91:e13357. doi: 10.1111/asj.13357
42. Chapman AR, Lee DF, Cai W, Ma W, Li X, Sun W, et al. Correlated gene modules uncovered by high-precision single-cell transcriptomics. Proc Natl Acad Sci U S A. (2022) 119:e2206938119. doi: 10.1073/pnas.2206938119
43. Gruber R, Bornchen C, Rose K, Daubmann A, Volksdorf T, Wladykowski E, et al. Diverse regulation of claudin-1 and claudin-4 in atopic dermatitis. Am J Pathol. (2015) 185:2777–89. doi: 10.1016/j.ajpath.2015.06.021
44. Li X, Akhtar S, Choudhry MA. Alteration in intestine tight junction protein phosphorylation and apoptosis is associated with increase in IL-18 levels following alcohol intoxication and burn injury. Biochim Biophys Acta. (2012) 1822:196–203. doi: 10.1016/j.bbadis.2011.09.019
45. Tavakolian S, Tabaeian SP, Namazi A, Faghihloo E, Akbari A. Role of the VEGF in virus-associated cancers. Rev Med Virol. (2024) 34:e2493. doi: 10.1002/rmv.v34.1
46. Samarani S, Allam O, Sagala P, Aldabah Z, Jenabian MA, Mehraj V, et al. Imbalanced production of IL-18 and its antagonist in human diseases, and its implications for HIV-1 infection. Cytokine. (2016) 82:38–51. doi: 10.1016/j.cyto.2016.01.006
47. Gougeon ML. Alarmins and central nervous system inflammation in HIV-associated neurological disorders. J Intern Med. (2017) 281:433–47. doi: 10.1111/joim.2017.281.issue-5
48. McCoombe SG, Short RV. Potential HIV-1 target cells in the human penis. AIDS. (2006) 20:1491–5. doi: 10.1097/01.aids.0000237364.11123.98
49. Galiwango RM, Yegorov S, Joag V, Prodger J, Shahabi K, Huibner S, et al. Characterization of CD4(+) T cell subsets and HIV susceptibility in the inner and outer foreskin of Ugandan men. Am J Reprod Immunol. (2019) 82:e13143. doi: 10.1111/aji.2019.82.issue-1
50. Liu A, Yang Y, Liu L, Meng Z, Li L, Qiu C, et al. Differential compartmentalization of HIV-targeting immune cells in inner and outer foreskin tissue. PloS One. (2014) 9:e85176. doi: 10.1371/journal.pone.0085176
51. Bekker LG, Das M, Abdool Karim Q, Ahmed K, Batting J, Brumskine W, et al. Twice-yearly lenacapavir or daily F/TAF for HIV prevention in cisgender women. N Engl J Med. (2024). doi: 10.1056/NEJMoa2407001. ahead of print.
52. Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. (2015) 347:664–7. doi: 10.1126/science.1260793
53. Kabbesh H, Riaz MA, Jensen AD, Scheiner-Bobis G, Konrad L. Long-term maintenance of viable adult rat sertoli cells able to establish testis barrier components and function in response to androgens. Cells. (2021) 10:2405. doi: 10.3390/cells10092405
54. Gye MC. Expression of claudin-1 in mouse testis. Arch Androl. (2003) 49:271–9. doi: 10.1080/01485010390204913
Keywords: emtricitabine tenofovir, pre-exposure prophylaxis PrEP, foreskin, transcriptomes, tight junctions, cytokines
Citation: Webb EL, Petkov S, Yun H, Else L, Lebina L, Serwanga J, Pillay A-DAP, Seiphetlo TB, Mugaba S, Namubiru P, Odoch G, Opoka D, Ssemata AS, Kaleebu P, Khoo S, Martinson N, Fox J, Gray CM, Herrera C and Chiodi F (2024) Gene expression of tight junctions in foreskin is not affected by HIV pre-exposure prophylaxis. Front. Immunol. 15:1415475. doi: 10.3389/fimmu.2024.1415475
Received: 10 April 2024; Accepted: 16 October 2024;
Published: 06 November 2024.
Edited by:
Hernan Felipe Peñaloza, Pontificia Universidad Católica de Chile, ChileReviewed by:
Morgane Bomsel, Centre National de la Recherche Scientifique (CNRS), FranceCopyright © 2024 Webb, Petkov, Yun, Else, Lebina, Serwanga, Pillay, Seiphetlo, Mugaba, Namubiru, Odoch, Opoka, Ssemata, Kaleebu, Khoo, Martinson, Fox, Gray, Herrera and Chiodi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Chiodi, ZnJhbmNlc2NhLmNoaW9kaUBraS5zZQ==
†Present address: Carolina Herrera, CONRAD, Macon & Joan Brock Virginia Health Sciences at Old Dominion University, Norfolk, VA, United States
‡These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.