- 1College of Agriculture and Biological Science, Dali University, Dali, China
- 2Team for Aquatic Ecology in Erhai Lake Watershed, Dali University, Dali, China
- 3Co-Innovation Center for Cangshan Mountain and Erhai Lake Integrated Protection and Green Development of Yunnan Province, Dali University, Dali, China
- 4Key Laboratory of Freshwater Aquatic Genetic Resources, Ministry of Agriculture, Shanghai Ocean University, Shanghai, China
- 5National Experimental Teaching Demonstration Center of Aquatic Science, Shanghai Ocean University, Shanghai, China
- 6Research and Development (R & D) Center, Qinghai Minze Longyangxia Ecological Hydroponics Co., Ltd, Hainan, China
Astaxanthin is an important aquatic feed additive that enhances the antioxidant capacity, and immune function of rainbow trout (Oncorhynchus mykiss); however, very limited information is available on its underlying molecular mechanisms. Haematococcus pluvialis powder, Phaffia rhodozyma powder, and synthetic astaxanthin were added to the commercial feed (no astaxanthin, NA) to prepare three experimental feeds, referred to as the HPA, PRA, and SA groups, respectively, and their actual astaxanthin contents were 31.25, 32.96, and 31.50 mg.kg-1, respectively. A 16-week feeding trial was conducted on the O. mykiss with an initial body weight of 669.88 ± 36.22 g. Serum and head kidney samples from commercial-sized O. mykiss were collected for metabolomics and transcriptomics analysis, respectively. Metabolomics analysis of the serum revealed a total of 85 differential metabolites between the astaxanthin-supplemented group and the control group. These metabolites were involved in more than 30 metabolic pathways, such as glycerophospholipid metabolism, fatty acid biosynthesis, linoleic acid metabolism, and arginine and proline metabolism. It is speculated that different sources of dietary astaxanthin may regulate antioxidant capacity and immunity mainly by affecting lipid metabolism and amino acid metabolism. Transcriptomic analysis of the head kidney revealed that the differentially expressed genes between the astaxanthin-supplemented group and the control group, such as integrin beta-1 (ITGB1), alpha-2-macroglobulin (A2M), diamine acetyltransferase 1 (SAT1), CCAAT/enhancer-binding protein beta (CEBPB) and DNA damage-inducible protein 45 alpha (GADD45A), which are involved in cell adhesion molecules, the FoxO signaling pathway, phagosomes, and arginine and proline metabolism and play regulatory roles in different stages of the antioxidant and immune response of O. mykiss.
1 Introduction
Rainbow trout (Oncorhynchus mykiss) is an important commercial aquaculture species that has been farmed extensively worldwide. It is popular among consumers because of its abundance of amino acids, minerals, vitamins, highly unsaturated fatty acids [such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA)], and other beneficial properties (1). Currently, rainbow trout are generally farmed in high-density or intensive ways, such as through cage culture and running water culture. However, these farming methods can increase fishery production but inevitably cause a series of problems. Previous studies have shown that infectious diseases caused by various bacterial and viral pathogens remain a major obstacle to rainbow trout farming and are often the leading cause of economic losses and restricting the development of aquaculture (2–4). Therefore, enhancing the immunity and disease resistance of rainbow trout is currently a significant concern for aquatic animal nutritionists.
Astaxanthin (3, 3’ - dihydroxy - β, β-carotene-4, 4’ - dione) is a keto-carotenoid that is widely found in microorganisms and marine organisms, such as shrimp, crab, salmon, trout, krill, microalgae, and yeast. It has strong antioxidant properties, as well as anti-inflammatory, anti-apoptotic, and immune-modulatory functions (5, 6). Therefore, it is generally used in food (7), biomedical science (8), aquatic feed (9), and other fields. At present, natural astaxanthin and synthetic astaxanthin are the two main sources of astaxanthin, of which natural astaxanthin sources mainly include green algae (Haematococcus pluvialis) (10), red yeast (Phaffia rhodozyma) (11), bacteria (Paracoccus carotinifaciens) (12) and Antarctic krill (Euphausia superba) (13). The price of astaxanthin from H. pluvialis is relatively high, while the price of astaxanthin from P. rhodozyma and P. carotinifaciens is moderate, and both have been used in the salmon aquaculture industry (12, 14).
Astaxanthin not only plays a role in regulating the color of salmon and trout, but also has important biological functions, such as antioxidant activity, immune response regulation and disease resistance in fish (6, 15, 16), and different physiological functions have been found among different sources of astaxanthin (11, 17–20). Previous studies on the effects of dietary supplementation with astaxanthin on the antioxidant and immune functions of salmon and trout have focused mainly on the activities of antioxidant and immune-related enzymes and their gene expression levels. These studies have shown that dietary P. rhodozyma astaxanthin could significantly reduce the activities of catalase (CAT), glutamic oxaloacetic transaminase (GOT), glutamic pyruvic transaminase (GPT) and lipid peroxide (LPO) in the plasma or serum of O. mykiss (21, 22), but increase the total carotenoid content, total antioxidant capacity (T-AOC) and superoxide dismutase (SOD) activity in plasma or serum (21, 23). Dietary supplementation with synthetic astaxanthin significantly increased the levels of antioxidants such as retinol, alpha-tocopherol, astaxanthin, and ascorbic acid in the muscle and liver of Atlantic salmon (Salmo salar) (24). In addition, supplementation with 100 mg.kg-1 synthetic astaxanthin in feed significantly enhanced the liver T-AOC of O. mykiss (25). However, relatively few studies have investigated the molecular regulatory effects of different sources (or different configurations) of astaxanthin on the antioxidant and immune functions of O. mykiss.
With the rapid development of next-generation sequencing (NGS) technology, it is possible to understand and characterize the molecular reactions of salmon and trout species in depth (26). Metabolomics is a new technology and discipline revealing the essence of metabolism; nuclear magnetic resonance (NMR), liquid chromatography-mass spectrometry (LC-MS), and gas chromatography-mass spectrometry (GC-MS) are generally used to qualitatively and quantitatively analyze small molecule metabolites in living organisms, and investigate the relationships between endogenous metabolites in response to internal or external factors (27, 28). Transcriptomics is a discipline that studies the transcription and regulation of genes in cells and is used to analyze the differences in the expression levels of certain genes in different tissues or different physiological states and to identify important functional genes or unknown genes with specific physiological functions (29, 30). At present, metabolomics and transcriptomics have been widely applied in studies of growth and development, disease resistance and immunity, phylogeny, and biotoxicological processes and related mechanisms in aquatic animals (31–34). For example, high-throughput sequencing technology has been used to systematically and comprehensively analyze fish immune response mechanisms (35), mine core immune-related gene clusters (36), and research immune response pathways (37).
This study was conducted to investigate the molecular regulatory effect of different sources of dietary astaxanthin on the antioxidant and immune functions of commercial-sized O. mykiss based on comprehensive metabolomics and transcriptomics analyses. These results could not only deepen our understanding of the molecular regulatory effects of different sources of astaxanthin on the antioxidant and immune functions of O. mykiss but also provide a reference for studies of antioxidant and immune functions in fish.
2 Materials and methods
2.1 Experimental diets
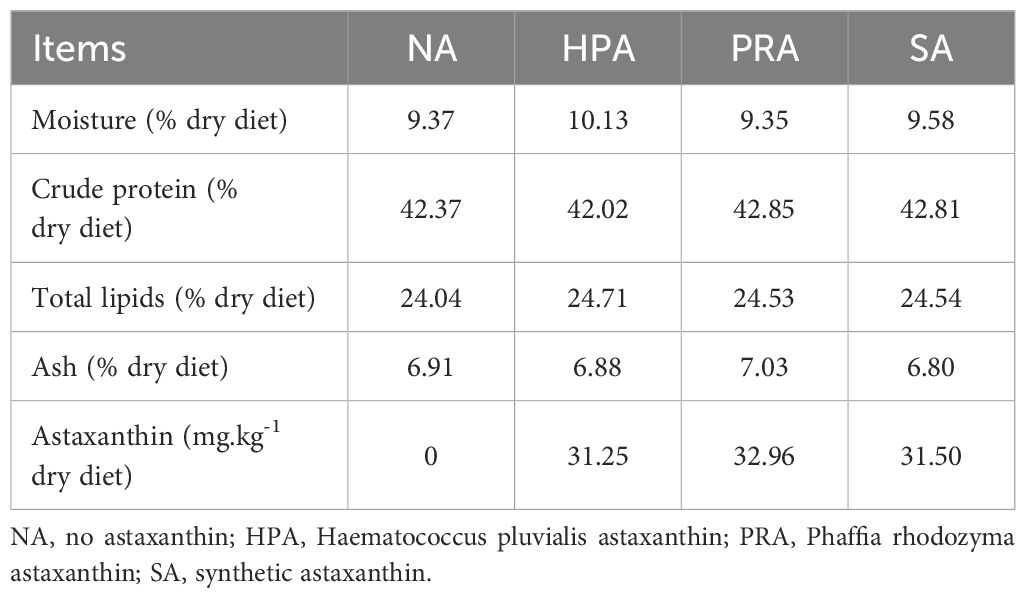
The commercial feed of rainbow trout was used as the basic diet (no astaxanthin, NA, crude protein: 42% dry diet; total lipids: 20% dry diet). Three different sources of astaxanthin (H. pluvialis powder, P. rhodozyma powder, and synthetic astaxanthin) were added to the basic diet to prepare three experimental diets; the theoretical content of dietary astaxanthin was 30 mg.kg-1, and the actual astaxanthin content was 31.25, 32.96, and 31.50 mg.kg-1, respectively (defined as the HPA, PRA, and SA groups, respectively). All experimental diets were processed by the Officially Aller Aqua (Qingdao) Co., Ltd., and sealed in a black bag, transported to the Qinghai Minze Longyangxia Ecological Aquatic Co., Ltd, Qinghai Province, China, and stored at -20°C for later use. The analysis of moisture, crude protein, and ash in experimental diets was conducted according to AOAC procedures (38). Total lipids were extracted with chloroform-methanol (2:1, v/v) according to the method described by Folch et al. (39). The nutrient compositions of the four experimental diets (NA, HPA, PRA, and SA) are shown in Table 1. The ingredients of experimental feeds mainly include fish meal, soybean meal, rapeseed meal, fish oil, soybean oil, soy lecithin, calcium dihydrogen phosphate, vitamin premix, and mineral premix. Since the feed formulations (the proportions of the ingredients) involved commercial confidentiality from the Officially Aller Aqua (Qingdao) Co., Ltd., this information could not be provided in this paper.
2.2 Experimental design and culture management
This experiment was conducted at Qinghai Minze Longyangxia Ecological Aquatic Co., Ltd, Qinghai Province, China. A total of 1800 healthy and active O. mykiss juveniles with a body weight of 669.88 ± 36.22 g were selected and randomly stocked into 12 net cages (length × width × height = 4 m × 4 m × 5 m). There were four diet groups (defined as NA, HPA, PRA, and SA), with each group having three replicate net cages (150 fish per net cage). Before the feeding trial, all the O. mykiss were acclimated to the culture conditions for 1 week and fed the NA diet (without astaxanthin). During the feeding trial, the water temperature ranged from 12-19°C, and the dissolved oxygen content ranged from 6-9 mg. L-1, and the water transparency was approximately 2.50 m; all the fish were fed to apparent satiation twice (at 8:00 and 16:00) daily for 4 months.
2.3 Sample collection
At the end of the feeding trial, the fish were fasted for 24 hours before sampling. Three fish were selected randomly and anesthetized with 30 mg. L-1 eugenol from each net cage; therefore, six fish were selected for each diet group. Approximately 10 mL of blood was drawn from the caudal vein with a syringe. The blood samples were centrifuged at 5, 000 × g for 10 min at 4°C, after which the supernatant (serum) was collected, divided into several aliquots, snap-frozen in liquid nitrogen, and stored at -80°C for subsequent metabolomics analysis. When metabolomics analysis was performed, a total of 6 fish serum samples were taken from each diet group. The rainbow trout were dissected, and the head kidney tissues were sampled and put into a 2 mL cryotube. Then 1.5 mL of RNAlater (Cat.R0901, Merck Company, Germany) was added, and the samples were stored at -80°C for subsequent transcriptomic analysis.
2.4 Serum metabolomics analysis
One hundred microliters of serum in a 1.5 mL Eppendorf tube was mixed with 300 μL of methanol-acetonitrile (2:1, v/v) and 10 μL of L-2-chlorophenylalanine (0.3 mg.mL-1 stock in dH2O, as an internal quantitative standard) and vortexed for 1 min by a vortex mixer (QL-901, Jiangsu Haimen Qilin Beier Instrument Manufacturing Co., Ltd., Haimen, China) before being placed in -20°C freezer for 30 min. The mixture was then centrifuged at 13000 r·min-1 and 4°C for 10 min (Eppendorf 5417R, Eppendorf Co., Hamburg, Germany); subsequently, 200 μL of the supernatant was collected into a 2 mL HPLC glass vial and blown dry with nitrogen, and 300 μL of methanol-water (1:4, v/v) was added to each sample and vortexed for 30 s and placed at 20°C for 2 h. The mixture was then centrifuged at 13000 r·min-1 at 4°C for 10 min, after which 150 μL of the supernatant was filtered with a syringe through a 0.22 μm organic phase pinhole filter membrane, and collected into a 2 mL HPLC glass vial for later analysis.
The metabolites in the serum were analyzed using a liquid-mass spectrometry system consisting of a Nexera UPLC ultra-high performance liquid chromatography-tandem QE high-resolution mass spectrometer. The chromatography was performed on an ACQUITY UPLC HSS T3 (100 mm × 2.1 mm, 1.8 μm). A stepwise elution program with two mobile phases, A-water (containing 0.1% formic acid), and B-acetonitrile (containing 0.1% formic acid), was used (Supplementary Table 1). The flow rate was 0.35 mL·min-1 and the column temperature was 45°C. The injection volume was 2 μL. An electrospray ion source (ESI) was used as the ion source. The positive and negative ion scanning modes were used to collect the sample quality spectrum signals. The mass spectrum parameters are shown in Supplementary Table 2.
2.5 Head kidney transcriptomics analysis
2.5.1 Total RNA extraction
Total RNA in the head kidney was extracted using TRIzol reagent (TaKaRa, Dalian, China), and the concentration and purity of total RNA were detected and assessed by Nanodrop 2000 (Thermo Scientific, USA) and agarose gel electrophoresis, respectively. RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Subsequently, cDNA libraries were constructed using TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Then, these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten), and 125 bp/150 bp paired-end reads were generated. The transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China).
2.5.2 De novo assembly and analysis of the transcriptome
The raw data (raw reads) in fastq format were first processed using Trimmomatic and the low-quality reads were removed to obtain the clean reads. The clean reads were mapped to the reference genome using HISAT2. Position information on the reference genome or gene, as well as sequence feature information unique to the sequenced sample, was obtained. The FPKM of each gene was calculated using Cufflinks, and the read counts of each gene were obtained by HTSeqcount. Differential expression analysis was performed using the DESeq (2012) R package. A P value < 0.05 and fold change > 2 or < 0.5 were set as the thresholds for significantly differential expression. Enrichment analysis was performed on differentially expressed genes (DEGs) to determine the biological functions or pathways affected by the DEGs.
2.5.3 Validation of the RNA-seq profiles by quantitative real-time PCR
Four randomly selected DEGs identified by transcriptome sequencing analysis were used for RNA-seq validation via qRT-PCR. First-strand cDNA synthesis was performed with a Prime-Script™ 1st Strand cDNA Synthesis Kit (RR036A, Takara Bio, Japan) following the manufacturer’s instructions. The qRT-PCR assays were performed with three replicates, and the β-actin gene was used as an internal control to normalize the expression level of the target genes. Specific primers were designed according to the unigene sequences using Primer 5.0 software (Supplementary Table 3). Gene expression levels were assessed using SYBR® Premix Ex Taq (TaKaRa) in a 20 μL reaction with a QuantStudio 3 real-time PCR instrument. The relative gene expression was calculated by 2−△△Ct method. The results were analyzed by Student’s t-test using SPSS 16.0 software, and the significance level was set at P < 0.05.
3 Results
3.1 Serum metabolomics analysis
3.1.1 Serum metabolic profile and comparison
The orthogonal partial least squares discriminant analysis (OPLS-DA) of the LC-MS metabolic profiles of the serum showed significantly separated clusters between the NA and the HPA groups, between the NA and the PRA groups, between the NA and SA groups, and between the HPA and the PRA and the SA groups, respectively, and all the samples from the four diet groups were within the 95% Hotelling’s T2 ellipse (Figure 1).
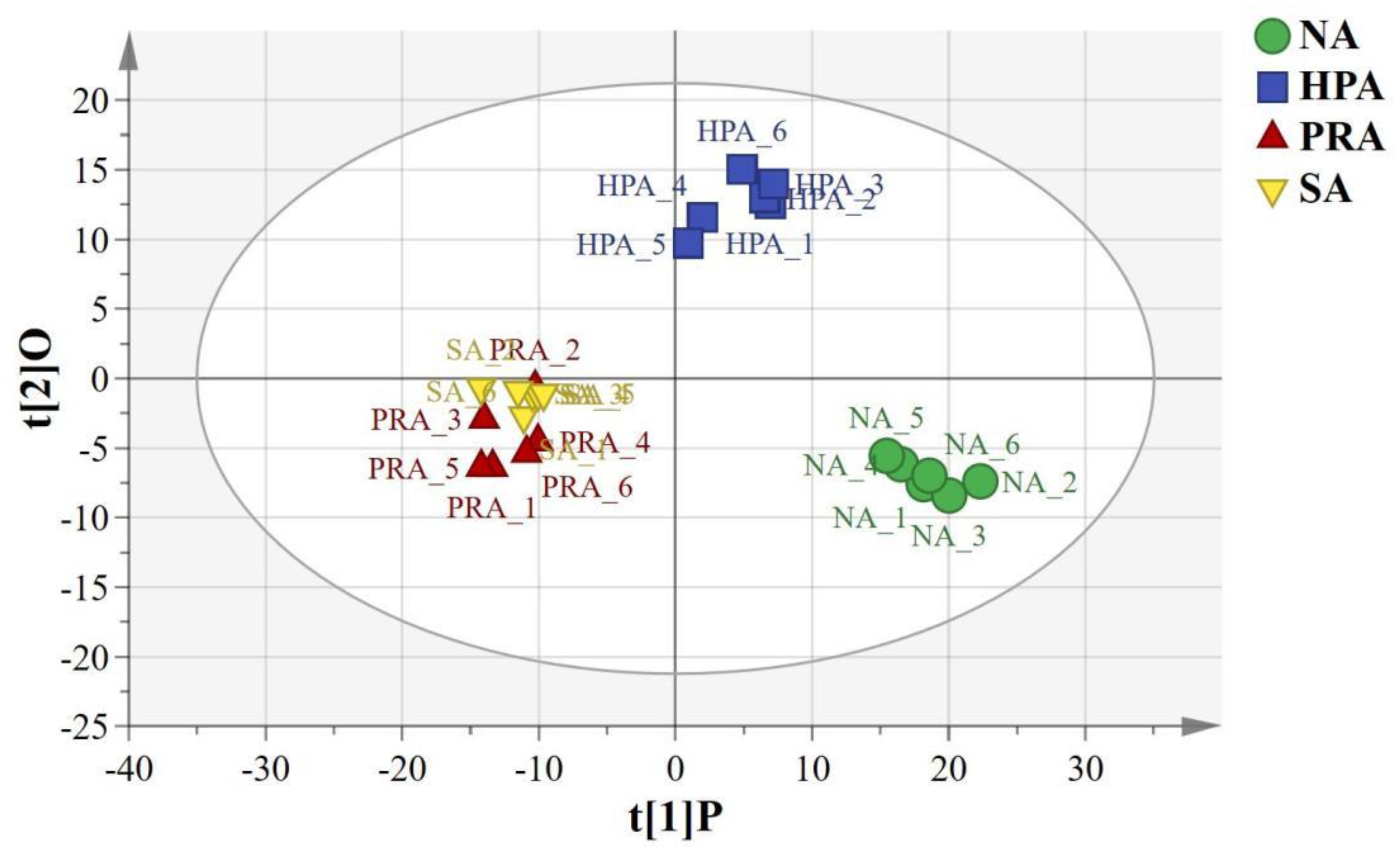
Figure 1 OPLS-DA score plots based on the LC-MS spectra of serum samples from O. mykiss among the different sources of dietary astaxanthin groups. “O” means “Orthogonal” and “P” means “Predictive” in OPLS-DA. R2X [1] =0.174, R2Y [2]= 0.0637, Ellipse: Hotelling’s T2 (95%). NA, no astaxanthin; HPA, Haematococcus pluvialis astaxanthin; PRA, Phaffia rhodozyma astaxanthin; SA, synthetic astaxanthin.
The total number of metabolites detected within the serum was 983. There were 11 significantly differential metabolites in the serum between the NA and HPA groups, among which 8 metabolites were up-regulated and 3 metabolites were down-regulated in the HPA group compared with the NA group (Supplementary Table 4). There were 15 differential metabolites in the serum of O. mykiss between the NA and PRA groups, among which 11 metabolites were up-regulated and 5 metabolites were down-regulated in the PRA group (Supplementary Table 5). There were 18 differential metabolites in the serum of O. mykiss between the NA and SA groups, among which 12 metabolites were up-regulated and 5 metabolites were down-regulated in the SA group (Supplementary Table 6). There were 21 differential metabolites in the serum of O. mykiss between the HPA and PRA groups, among which 11 metabolites were up-regulated and 10 metabolites were down-regulated in the PRA group (Supplementary Table 7). There were 18 differential metabolites in the serum of O. mykiss between the HPA and SA groups, among which 10 metabolites were up-regulated and 8 metabolites were down-regulated in the SA group (Supplementary Table 8). These differential metabolites among the four diet groups mainly included phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), LysoPC, palmitic acid and palmitoleic acid, aniline, glycyl-glycine, L-histidine, sphingosine, etc.
3.1.2 Serum metabolic pathway analysis
The differential metabolites identified in the serum between the NA and HPA groups were mainly involved in glycerophospholipid metabolism, fatty acid biosynthesis, autophagy-other, autophagy-animal, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, linoleic acid metabolism, fatty acid elongation, alpha-linolenic acid metabolism, galactose metabolism, histidine metabolism, and fatty acid degradation (Table 2). The differential metabolites identified in the serum between the NA and PRA groups were mainly involved in glycerophospholipid metabolism, alpha-linolenic acid metabolism, autophagy-other, autophagy-animal, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, linoleic acid metabolism, neuroactive ligand-receptor interaction, fatty acid biosynthesis, and arachidonic acid metabolism (Table 3). The differential metabolites identified in the serum between the NA and SA groups were mainly involved in histidine metabolism, glycerophospholipid metabolism, D-arginine and D-ornithine metabolism, linoleic acid metabolism, neuroactive ligand-receptor interaction, alpha-linolenic acid metabolism, fatty acid biosynthesis, phenylalanine metabolism, arachidonic acid metabolism, arginine and proline metabolism, and drug metabolism - cytochrome P450 (Table 4). The differential metabolites identified in the serum between the HPA and PRA groups were mainly involved in histidine metabolism, glycerophospholipid metabolism, apoptosis, necroptosis, D-arginine and D-ornithine metabolism, sphingolipid metabolism, linoleic acid metabolism, alpha-linolenic acid metabolism, galactose metabolism, phenylalanine metabolism, biosynthesis of unsaturated fatty acids, arachidonic acid metabolism, and arginine and proline metabolism (Table 5). The differential metabolites identified in the serum between the HPA and SA groups were mainly involved in the biosynthesis of unsaturated fatty acids, histidine metabolism, D-arginine and D-ornithine metabolism, D-glutamine and D-glutamate metabolism, nitrogen metabolism, arginine biosynthesis, alanine, aspartate and glutamate metabolism, vitamin B6 metabolism, fatty acid elongation, galactose metabolism, fatty acid degradation, glycine, serine and threonine metabolism, aminoacyl-tRNA biosynthesis, glycerophospholipid metabolism, drug metabolism-other enzymes, fatty acid biosynthesis, phenylalanine metabolism, glyoxylate and dicarboxylate metabolism, pyrimidine metabolism, arginine and proline metabolism, ABC transporters, purine metabolism, and steroid hormone biosynthesis (Table 6). These differential metabolites between the different diet groups were mainly related to fatty acid metabolism and amino acid metabolism.
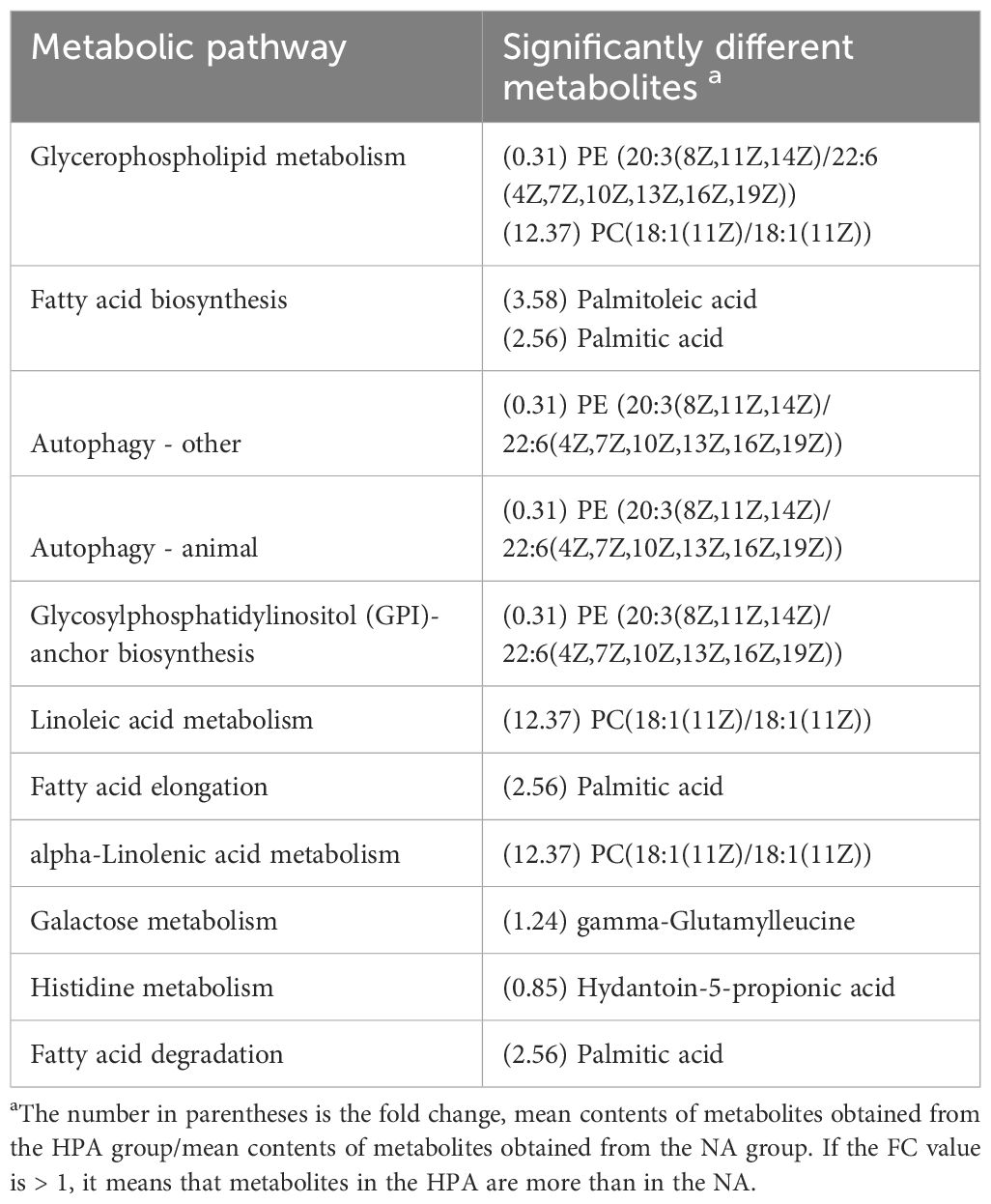
Table 2 Metabolic pathways identified from the significantly different metabolites in the serum of O. mykiss between the NA and HPA groups.
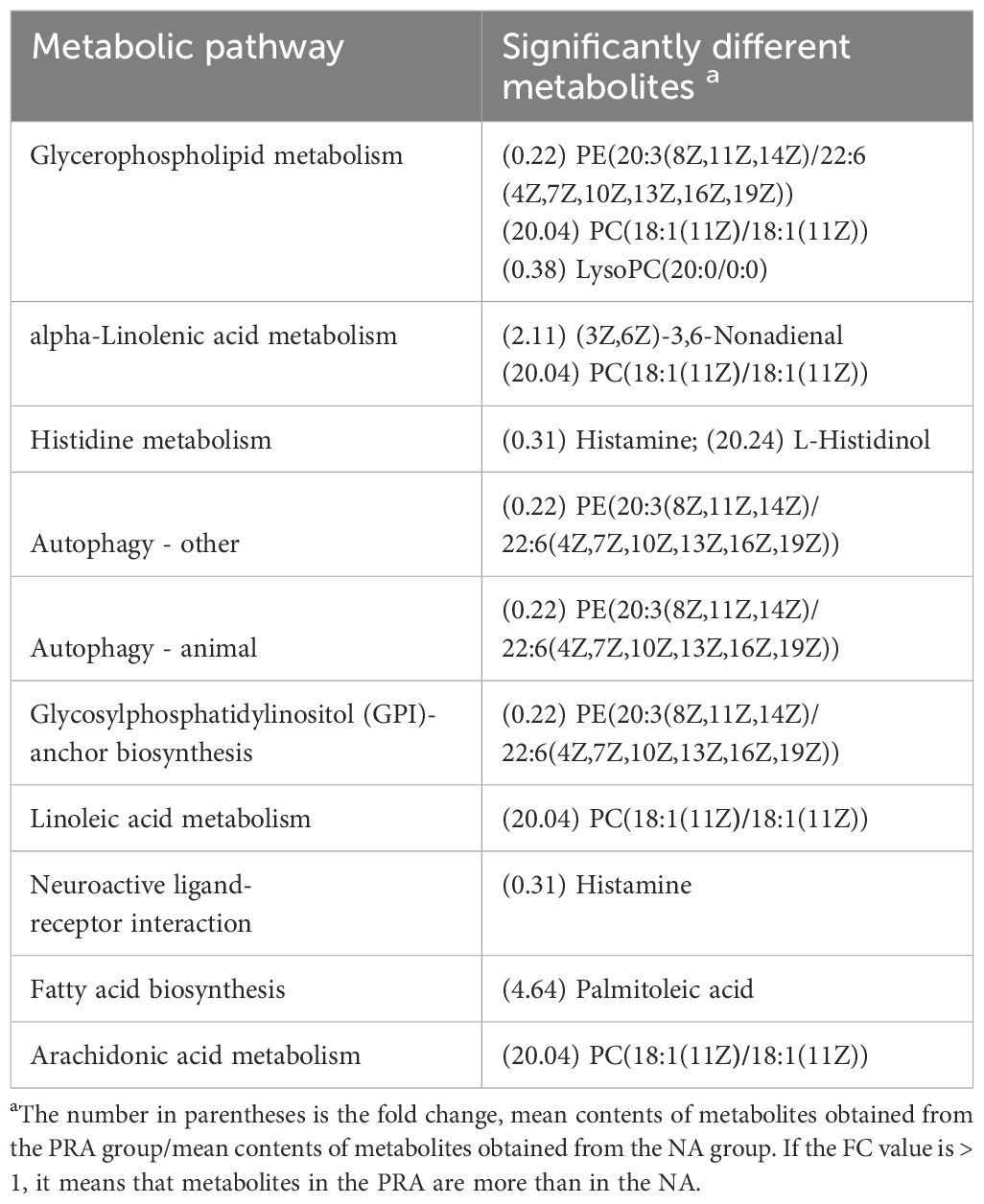
Table 3 Metabolic pathways identified from the significantly different metabolites in the serum of O. mykiss between the NA and PRA groups.

Table 4 Metabolic pathways identified from the significantly different metabolites in the serum of O. mykiss between the NA and SA groups.
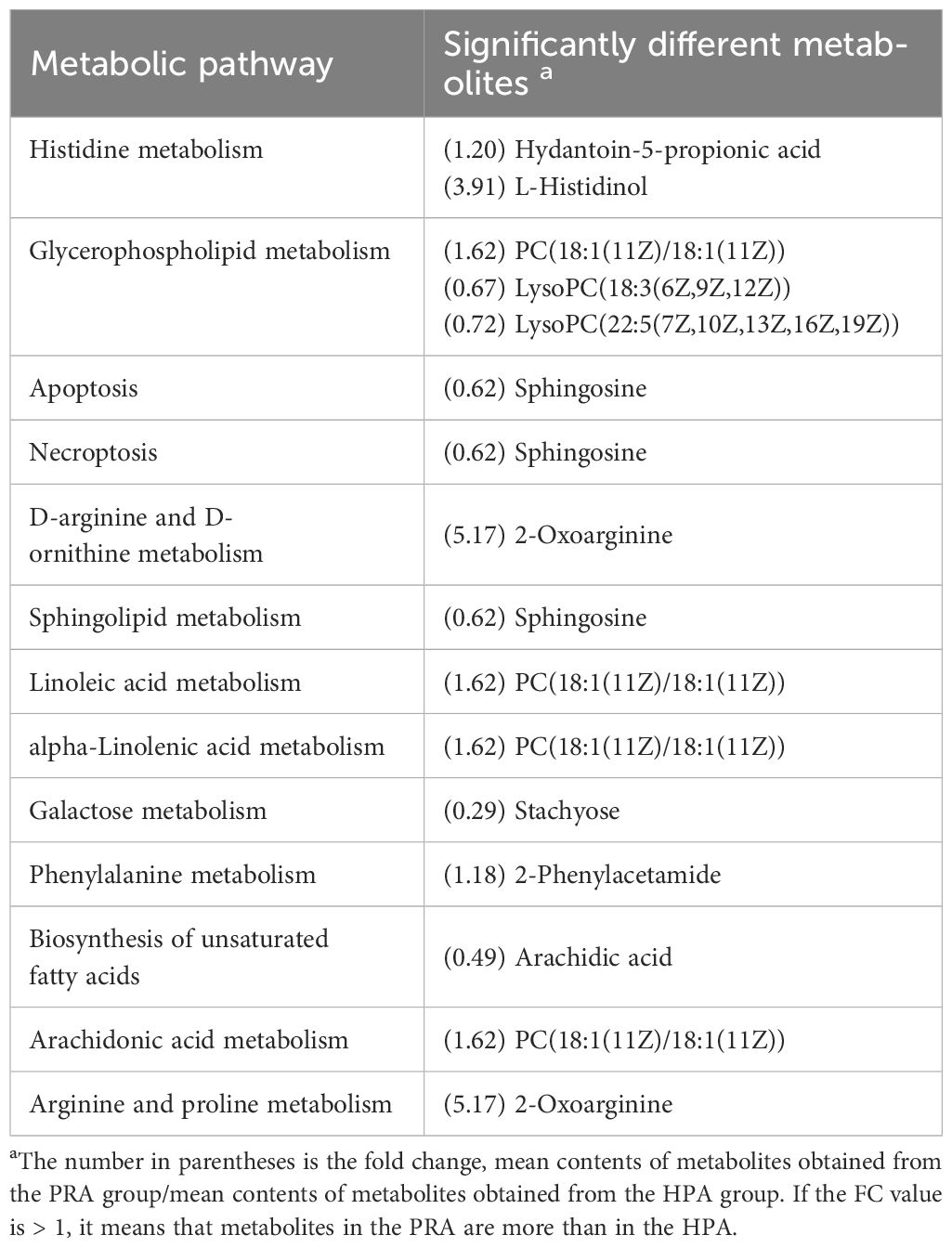
Table 5 Metabolic pathways identified from the significantly different metabolites in the serum of O. mykiss between the HPA and PRA groups.
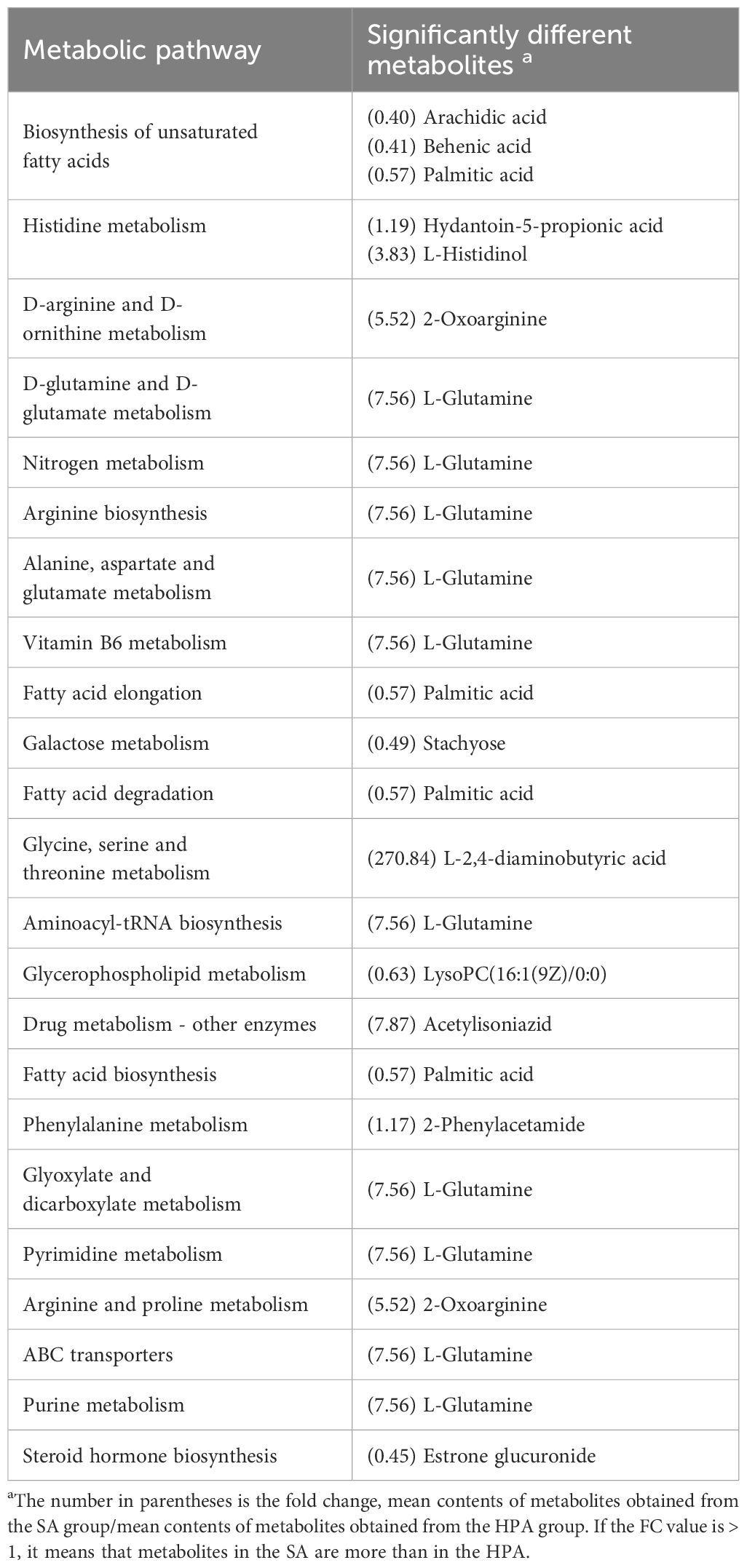
Table 6 Metabolic pathways identified from the significantly different metabolites in the serum of O. mykiss between the HPA and SA groups.
3.2 Head kidney transcriptomics analysis
3.2.1 Transcriptome sequence assessment and annotation
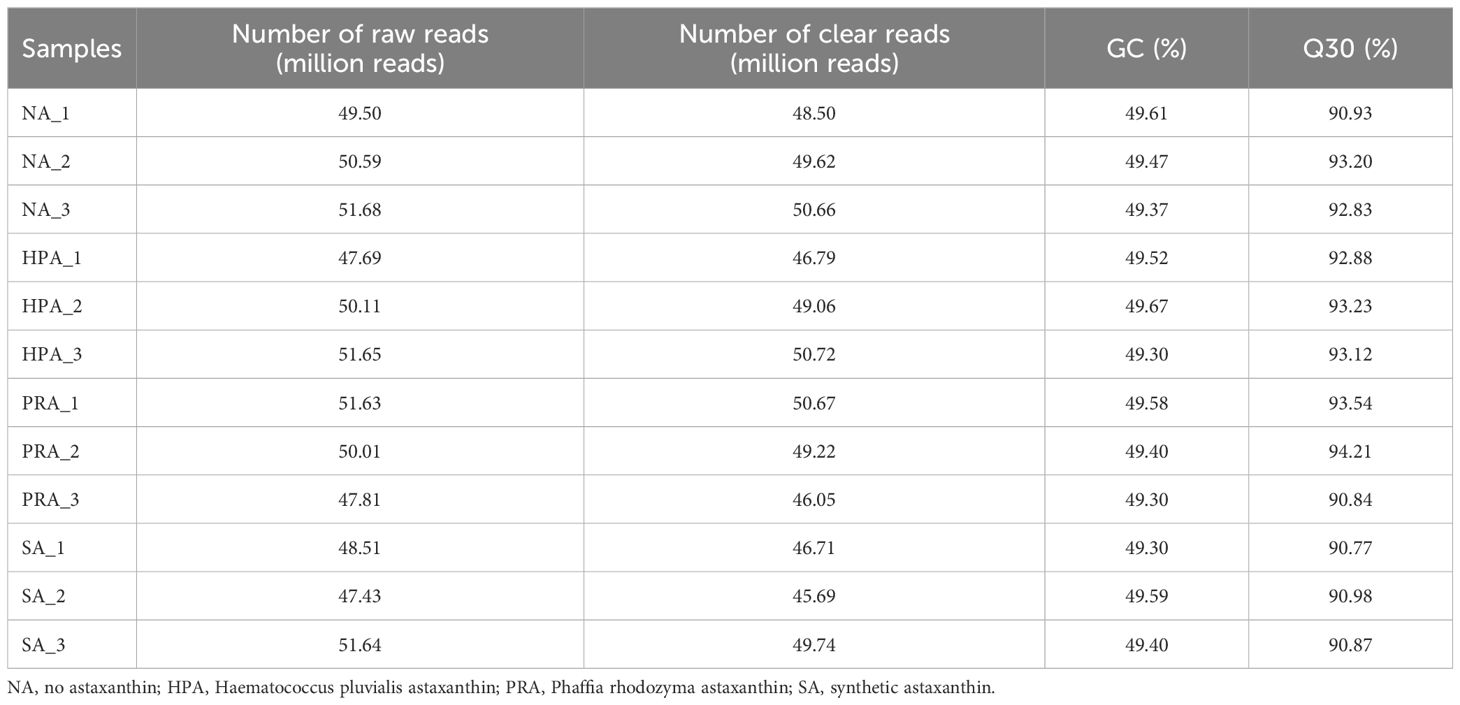
A total of 86.15 G of clean reads were obtained from the head kidney transcriptome sequencing of the four diet groups after removing adaptor sequences and low-quality reads (Table 7). The effective amount of each sample was 6.72-7.5 G, the Q30 base content was 90.77-94.21%, and the average GC content was 49.46%. Clean reads were compared to the reference genome of the species using HISAT2, and 88.61-90.14% of the genome was compared for each sample.
3.2.2 Analysis of differentially expressed genes
A total of 211 differentially expressed genes (DEGs) in the head kidney were identified among the four diet groups (Figure 2). There were 24 DEGs in the head kidney of O. mykiss between the NA and HPA groups, among which 13 DEGs were up-regulated and 11 DEGs were down-regulated in the HPA group. These DEGs mainly included interleukin-2 receptor subunit beta, hemoglobin subunit beta-1, immunoglobulin kappa variable 1-33, and perforin-1. There were 13 DEGs in the head kidney of O. mykiss between the NA and PRA groups, among which 6 DEGs were up-regulated and 7 DEGs were down-regulated in the PRA group. These DEGs included mainly alpha-2-macroglobulin and integrin beta-1. There were 50 DEGs in the head kidney of O. mykiss between the NA and SA groups, among which 31 DEGs were up-regulated and 19 DEGs were down-regulated in the SA group. These DEGs mainly included interferon-induced protein 44-like, CXC chemokine receptor type 2, major histocompatibility complex class I-related gene protein, and tumor necrosis factor alpha-induced protein 2.
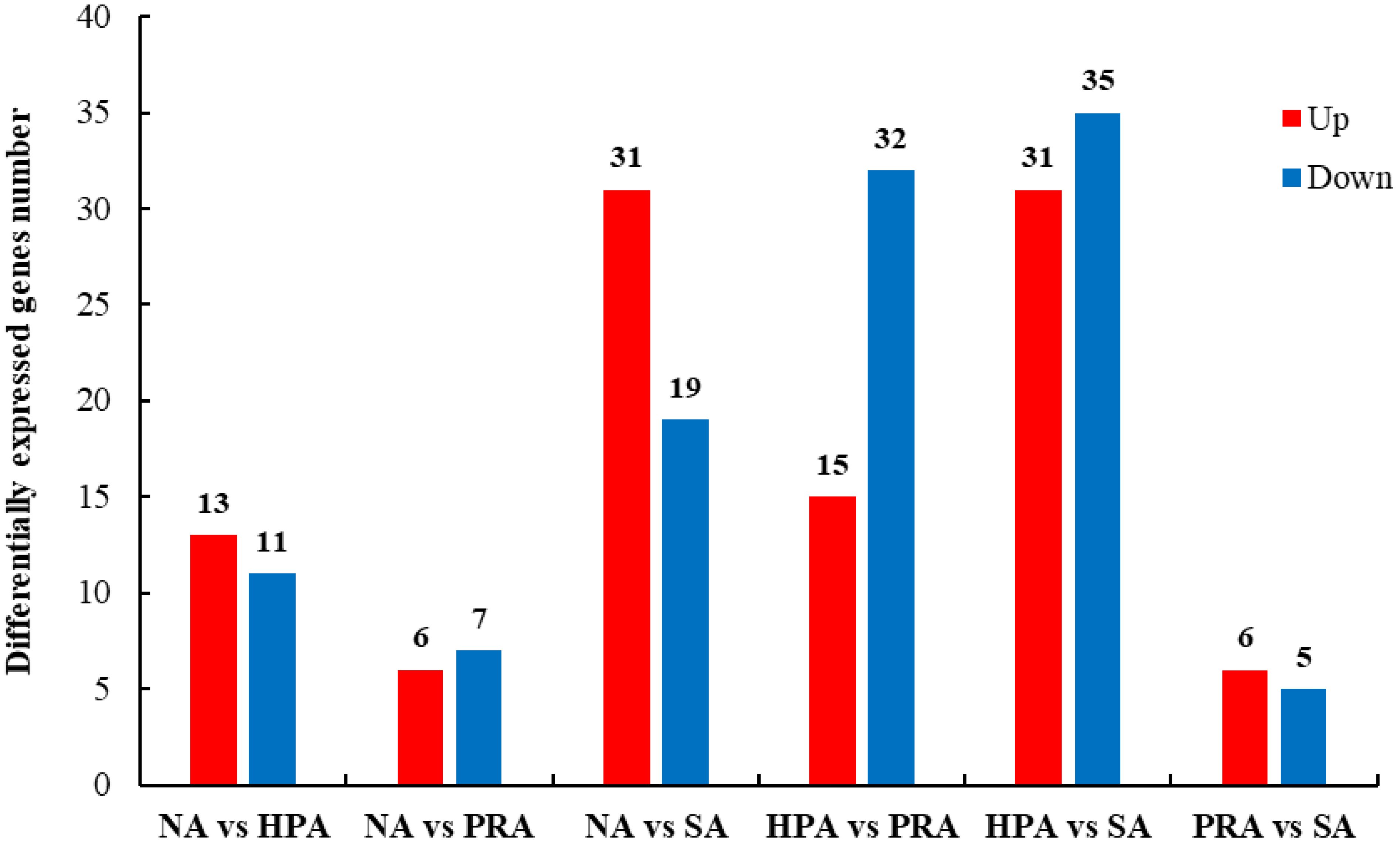
Figure 2 Summary of differentially expressed genes (DEGs) in the transcriptome of the head kidney of O. mykiss among different diet groups. NA, no astaxanthin; HPA, Haematococcus pluvialis astaxanthin; PRA, Phaffia rhodozyma astaxanthin; SA, synthetic astaxanthin.
3.2.3 GO functional annotation analysis of differentially expressed genes
To understand the functions of these DEGs, gene ontology (GO) assignments were conducted. A total of 211 DEGs were annotated to 1462 GO terms, among which 795 were annotated to biological processes, 369 were annotated to molecular functions, and 298 were annotated to cellular components. The DEGs between the NA and HPA groups (Figure 3A), the NA and PRA groups (Figure 3B), the NA and SA groups (Figure 3C), the HPA and PRA groups (Figure 3D), the HPA and SA groups (Figure 3E), the PRA and SA groups (Figure 3F) were annotated into 186, 107, 300, 376, 448, and 45 GO items, respectively. Among these, antioxidant and immune-related GO terms mainly included immune response (GO: 0006955), immunoglobulin production (GO: 0002377), positive regulation of the innate immune response (GO: 0045089), hemoglobin complex (GO: 0005833), and antigen binding (GO: 0003823).
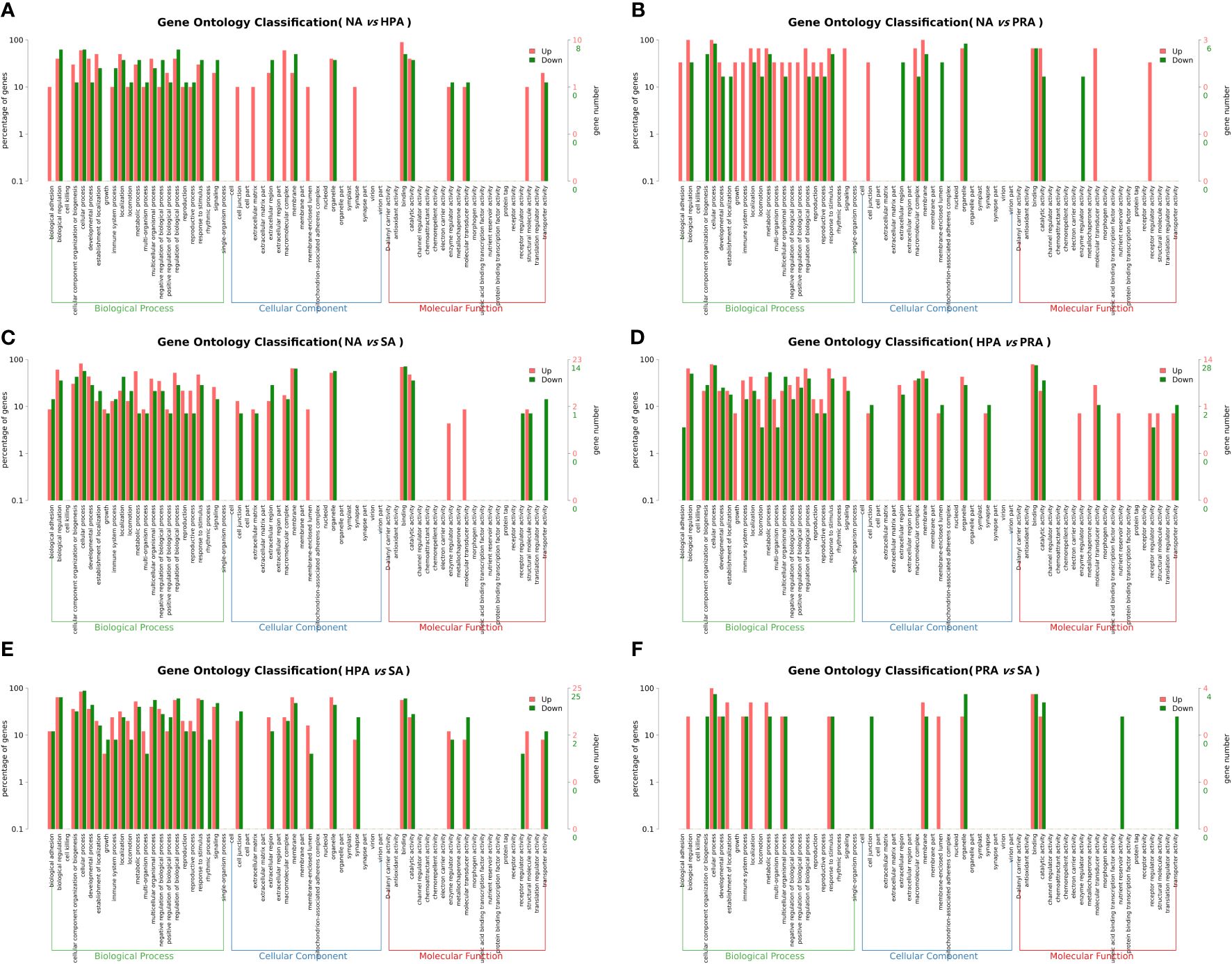
Figure 3 GO enrichment analyses of differentially expressed genes (DEGs) in the head kidney among different diet groups. (A) NA vs HPA; (B) NA vs PRA; (C) NA vs SA; (D) HPA vs PRA; (E) HPA vs SA; (F) PRA vs SA. NA, no astaxanthin; HPA, Haematococcus pluvialis astaxanthin; PRA, Phaffia rhodozyma astaxanthin; SA, synthetic astaxanthin.
3.2.4 KEGG enrichment analysis of differentially expressed genes
KEGG enrichment analysis revealed that the DEGs in the head kidney of rainbow trout among all diet groups were enriched in 191 specific KEGG metabolic pathways. The 29 significant enriched KEGG pathways included metabolism, organismal systems, genetic information processing, cellular processes, and environmental information processing, such as transport and catabolism, signaling molecules and interaction, signal transduction, translation, folding, sorting and degradation, immune system, lipid metabolism, energy metabolism, amino acid metabolism, carbohydrate metabolism, and endocrine system (Table 8).
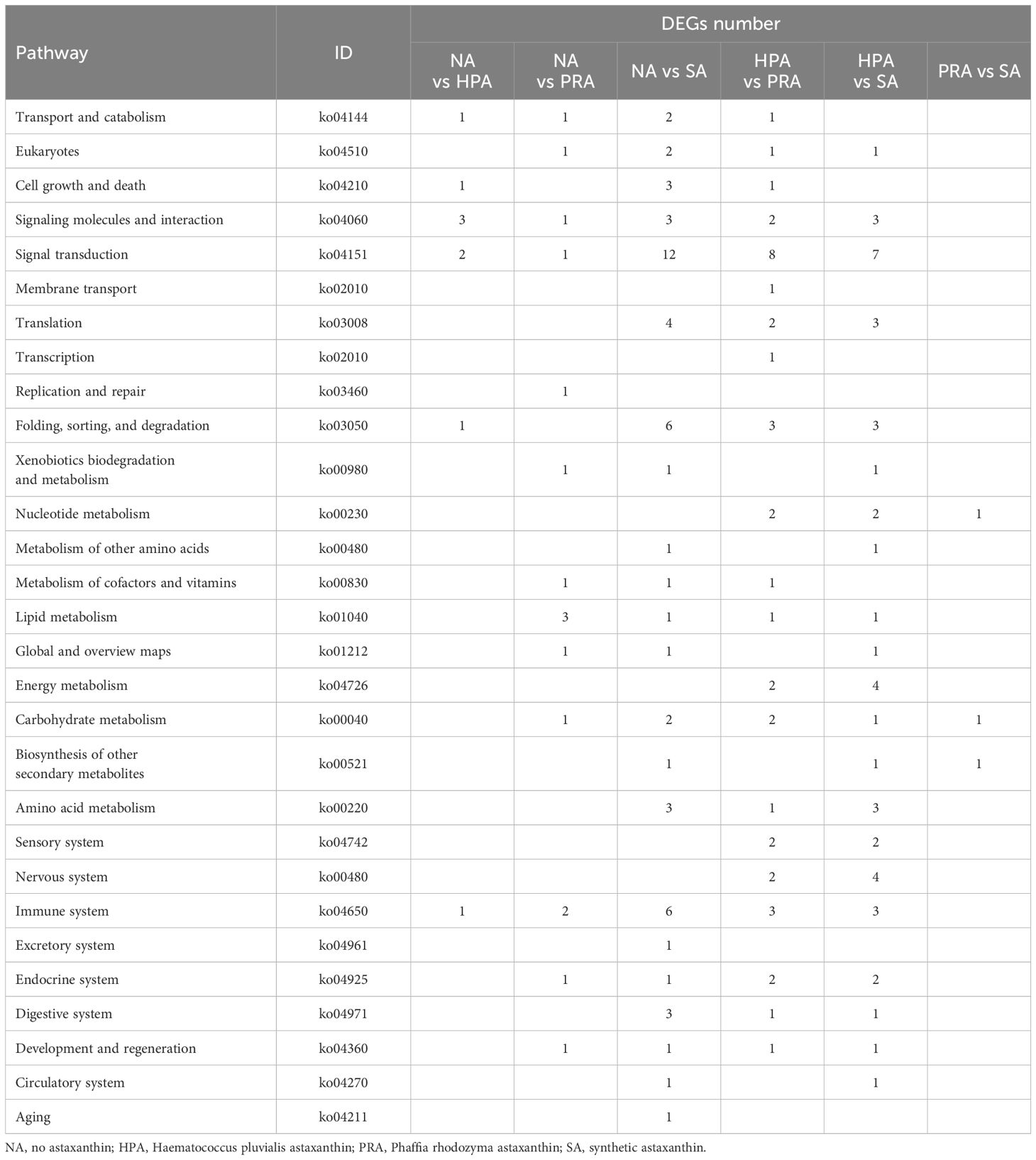
Table 8 Significantly altered pathways according to KEGG analysis of the genes whose expression significantly changed in the head kidney of O. mykiss fed diets with different sources of astaxanthin.
3.3 Validation of transcriptomic sequencing by quantitative PCR analysis
To validate the expression levels of the DEGs obtained from RNA-Seq data, four DEGs were selected for qRT-PCR analysis using the same RNA samples and the differences of fold changes were compared between RNA-Seq and qRT-PCR. The fold changes from qRT-PCR were consistent with the results from RNA-Seq data, indicating the reliability and accuracy of transcriptome data and quantified gene expression (Figure 4). Of the four DEGs selected for qRT-PCR, the expression level of nod like receptor C3 (NLRC3) was significantly down-regulated in the HPA group compared with the NA group (Figure 5A). The expression level of alpha-2-macroglobulin (A2M) was significantly down-regulated (Figure 5B), while the expression level of integrin beta-1 (ITGB1) was significantly up-regulated in the PRA group compared with the NA group (Figure 5C). The expression level of CXC chemokine receptor type 2 (CXCR2) was significantly up-regulated in the SA group compared with the NA group (Figure 5D).
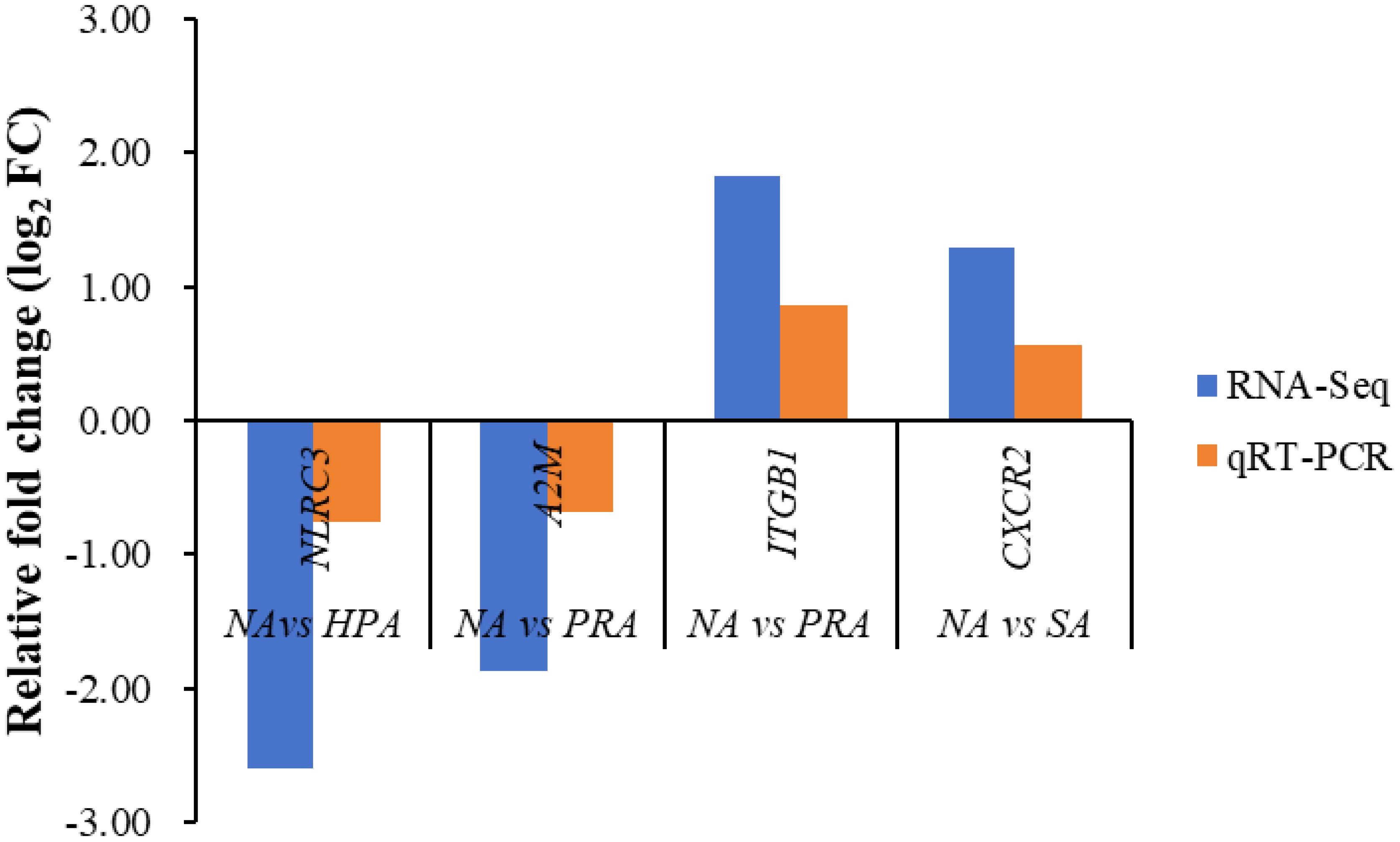
Figure 4 Validation results of RNA-seq profiles by qPCR. The expression of the following proteins was detected by RNA-seq and real-time qPCR. NLRC3, nod like receptor C3; A2M, alpha-2-macroglobulin; ITGB1, integrin beta-1; CXCR2, CXC chemokine receptor type 2.
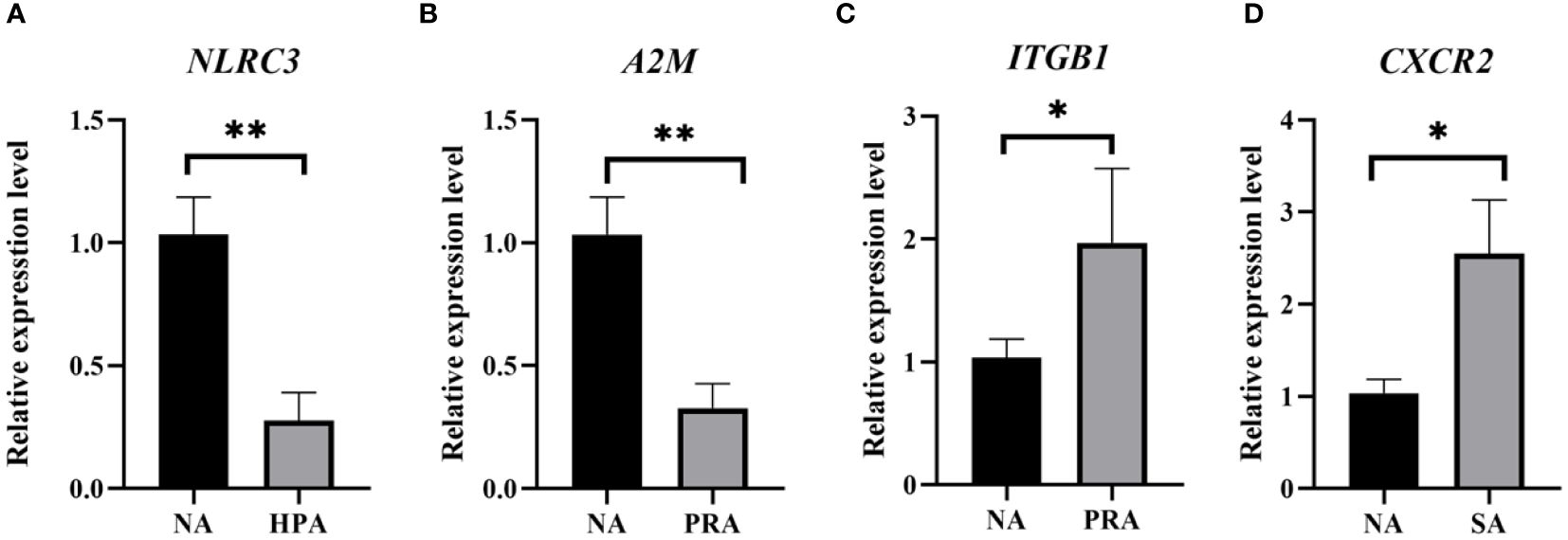
Figure 5 Validation of gene expression patterns in the head kidney of O. mykiss by qPCR among different diet groups. The significant differences between the two groups are represented by “*” (P < 0.05) and “**” (P < 0.01). NA: no astaxanthin; HPA: Haematococcus pluvialis astaxanthin; PRA, Phaffia rhodozyma astaxanthin; SA, synthetic astaxanthin. (A) NLRC3, nod like receptor C3; (B) A2M, alpha-2-macroglobulin; (C) ITGB1, integrin beta-1; (D) CXCR2, CXC chemokine receptor type 2.
4 Discussion
In this study, OPLS-DA revealed differences in the serum metabolite profiles between the NA and the HPA groups, the NA and the PRA groups, the NA and SA groups, and the HPA and the PRA and the SA groups which indicated that dietary supplementation with astaxanthin had significant effects on the physiological metabolism of commercial-sized rainbow trout. The differential metabolites among the different diet groups were enriched in glycerophospholipid metabolism, linoleic acid metabolism, alpha-linolenic acid metabolism, biosynthesis of unsaturated fatty acids, arginine and proline metabolism, glycine, serine and threonine metabolism and other pathways related to lipid and amino acid metabolism.
Glycerophospholipids are important components of biofilms and play important roles in ensuring the normal operation of membrane-binding proteins, ion channels, and receptors (40). Under the stress condition, the change in cell membrane composition and fluidity is associated with the immune response (41). Previous metabolomics analysis showed that largemouth bass (Micropterus salmoides) fed with oxidized fish oil reduced phospholipid unsaturation, which may impair the normal function of cells by affecting the function of receptors in the membrane. This may be one of the causes of increased oxidative stress and decreased immunity in fish (42). In this study, the serum metabolites, such as phosphatidylcholine (PC) and phosphatidyl inositol (PI), which contain multiple double bond structures involved in glycerophospholipid metabolism, were significantly up-regulated in the astaxanthin-supplemented diet groups, resulting in an increase in the unsaturation of phospholipids, thus contributing to the normal operation of receptors in the cell membrane and thus maintaining the normal function of cells.
As one of the important components of the cell membrane, phosphatidylcholine (PC) is a precursor of signaling molecules (43), and plays an important role in the regulation of cell membrane fluidity, permeability, and other functions (44). It can regulate serum lipid levels (such as reducing cholesterol levels), strengthen immunity, and promote growth (45). In this study, serum phosphatidylcholine involved in linoleic acid metabolism and α-linolenic acid metabolism was significantly up-regulated in the HPA, PRA, and SA groups compared with the NA group. These results indicate that the dietary supplementation of astaxanthin is conducive to the biosynthesis of linoleic acid and α-linolenic acid. In addition to reducing blood lipids and participating in the phospholipid composition of cell membranes, linoleic acid can also affect the production of intestinal microbiota and inflammatory mediators (46). Similarly, n-3 poly-unsaturated fatty acids (n-3 PUFAs) such as eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) are produced by the extension of the carbon chain under the action of fatty acid desaturase, which has anti-inflammatory effects and plays a key role in regulating body homeostasis (47). This study suggested that dietary supplementation with astaxanthin was beneficial to the accumulation of fatty acids in commercial-sized rainbow trout, which may be conducive to enhancing the polyunsaturated fatty acid content, increasing the nutritional value, and improving the antioxidant capacity.
The head kidney is an important immune and hematopoietic organ in teleost fish (48–50) and plays an important physiological role in the initial stage of the immune response (51). In this study, antioxidant and immune-related differentially expressed genes (DEGs), such as interleukin-2 receptor subunit beta (IL2RB) and the glycine receptor subunit alpha Z1 (GLRA1) were detected between the NA and HPA groups; alpha-2-macroglobulin (A2M) and integrin beta-1 (ITGB1), between the NA and PRA groups, and CXC chemokine receptor type 2 (CXCR2), CCAAT/enhancer-binding protein beta (CEBPB), dual specificity protein phosphatase 1 (DUSP1), growth arrest and DNA damage-inducible protein GADD45 alpha (GADD45A), ornithine decarboxylase 1 (ODC1A), and diamine acetyltransferase 1 (SAT1), between the NA and SA groups. These DEGs were mainly involved in signaling pathways related to immunity, anti-inflammation, and antioxidants, including cell adhesion molecules (CAMs), ubiquitin-mediated proteolysis, FoxO signaling pathway, phagosome, complement and coagulation cascades, natural killer cell-mediated cytotoxicity, RIG-I-like receptor signaling pathway, NF-kappa B signaling pathway, TNF signaling pathway, arginine and proline metabolism, glutathione metabolism, etc.
Cell adhesion molecules (CAMs) can promote the migration of immune cells during immune responses (52). The integrin family is one of the categories of cell adhesion molecules, and it is a heterodimeric transmembrane glycoprotein that includes α and β subunits and plays important roles in cell migration, differentiation, and survival (53). Previous studies have shown that integrin subunits were cellular receptors for white spot syndrome virus infection in Chinese shrimp (Fenneropenaeus chinensis) (54, 55) and Kuruma prawn (Marsupenaeus japonicas) (56). The antibacterial activity of integrins has been reported in Chinese mitten crab (Eriocheir sinensis) (53). Additionally, integrins are involved in precursor activation, phagocytosis, and immunomodulatory antioxidant systems in Pacific white shrimp (Litopenaeus vannamei) (57). In this study, the head kidney DEGs between the NA and PRA groups were enriched in integrin binding, integrin-mediated signaling pathway, integrin complex, and other related pathways. Significantly different signaling pathways, CAMs, and related DEGs (such as Integrin beta-1 (ITGB1) and Claudin-4 (CLDN4)) were found between the NA and PRA groups and between the HPA and SA groups. These results suggested that the addition of astaxanthin from different sources may enhance the mutual recognition and adhesion between cells and extracellular mediators by enhancing integrins, thus improving the immunity of fish.
The ubiquitin proteolytic system plays important roles in regulations of the cell cycle, and immune and inflammatory responses, as well as in the control of signal transduction pathways, development, and differentiation (58). In fish, studies on ubiquitin-mediated proteolysis have focused mainly on viral infection (59). The expression of ubiquitin-related proteins in rainbow trout is disrupted after bacterial infection (60), and ubiquitin degradation can promote MHC molecule synthesis, the endoplasmic reticulum pathway, antigen processing and delivery, etc., to assist the body’s immune system clearing exogenous antigens. In this study, the E3 ubiquitin/ISG15 ligase TRIM25 (TRIM25) was significantly up-regulated in the PRA group compared with the NA group. This result suggested that the addition of P. rhodozyma astaxanthin to the diet could maintain the normal translation of the protein to a certain extent, thus enhancing the immune response.
Chemokines are cytokines that mediate the migration of immune cells. When the body is injured or infected, chemokines will chemoattract immune-related cells to the site of injury or infection and bind to G protein-coupled receptors on target cells, leading tochemotaxis (61). In response to pathogenic exposure, chemokines not only promote leukocyte mobilization but also regulate immune responses and the differentiation of recruited cells to orchestrate the first steps of both innate and adaptive immunity (62, 63). Previous studies have shown that the expression of CXCR2 increased in the kidneys, spleen, and liver of large yellow croaker (Larimichthys crocea) after challenge with Vibrio anguillarum; likely, high expression of CXCR2 was also detected in the kidneys and spleen of rock bream (Oplegnathus fasciatus) (64). These studies suggested that CXCR2 was very important in resisting bacterial infection (65). A study by Hu et al. (2020) showed that CXCR2 was highly expressed in the spleen, intestine, liver, and head kidney of tilapia after infection with Streptococcus agalactiae and Aeromonas hydrophila, which indicates that CXCR2 plays a role in the immune response (62). In this study, the expression level of the head kidney gene CXCR2, which is involved in the chemokine signaling pathway, was significantly up-regulated in the SA group compared to the NA group. This result indicated that the addition of synthetic astaxanthin to the diet was beneficial in enhancing the immune function of rainbow trout, which may be related to the gene expression of cytokines, chemotaxis, and several immunity-related signaling molecules (66). These results will provide a basis for further research on the function of CXCR2.
The FoxO signaling pathway is related to a variety of biological processes, including cell metabolism, immunity, and antioxidant stress (67). In terms of oxidative stress, the FoxO signaling pathway can drive the expression of genes involved in fighting oxidative stress and accelerating cell cycle circulation, thereby protecting cell function (68) Gao et al. (2019) reported that FoxO plays an important role in the immune response to bacterial infection in Channel catfish (Ictalurus punctatus) (69). In this study, the head kidney gene, growth arrest, and DNA damage-inducible protein GADD45 alpha (GADD45A), which is involved in the FoxO signaling pathway, were significantly up-regulated in the SA group compared with the NA group, which may represent the host’s early defense against bacterial infection.
Phagocytosis is a central mechanism of tissue remodeling, inflammation, and defense against infection, and it is the process by which cells transport invading pathogens into cells for elimination (70). In addition to providing cells with specific immune functions, phagocytes are also an important part of the non-specific defense system and play an important role in all stages of foreign pathogen invasion in fish (71, 72). A study by Song et al. (2021) showed that the phagosome pathway was significantly enriched in the head kidney of red sea bream (Pagrus major) after infection with Vibrio anguillarum (9). Transcriptomics analysis results showed that phagosome pathways related to immunity were also enriched in rainbow trout after heat stress (73). In this study, head kidney genes, such as integrin beta-1 (ITGB1) and immunoglobulin heavy variable 3-43 (IGHV3-43), which are involved in phagosome pathways, were significantly up-regulated in the PRA group compared with the NA and HPA groups. These results indicated that the addition of astaxanthin could improve the body’s ability to fight inflammation and defend against infection, which may be related to the strong antioxidant and immune functions of astaxanthin.
The complement system, the earliest evolved immune response system, exists in the serum and can mediate a variety of functions (including phagocytosis, cytolysis, inflammation, dissolution of immune complexes, clearance of apoptotic cells, and promotion of humoral immune response), mainly by regulating antigens to assist antigen phagocytosis, cell lysis, and chemotaxis. The complement system not only plays an important role in innate immunity but also regulates the level of specific immune responses by assisting antigen phagocytosis (74–77). After invasion, complement binds to the surface of the pathogen, inducing phagocytes to recognize and engulf the pathogen (76, 78). In this study, the expression of head kidney genes involved in the complement and coagulation cascades, such as alpha-2-macroglobulin (A2M) and plasminogen activator inhibitor 1 (SERPINE1), was up-regulated in the PRA and SA groups compared to the NA group. A2M is a member of the alpha-macroglobulin family (α-macroglobulin, αMs), a class of macromolecular glycoproteins present in a variety of animal plasma, and has been shown to regulate host cell apoptosis and cancer cell metabolism, enhance prothrombin activation and thrombin potential, mediate T-cell proliferation, and induce macrophage proliferation and activation (79, 80). It has been reported that the A2M and complement components (C3 and C4) of L. vannamei are highly conserved during evolution and play a certain role in bacterial infection (81, 82). Variable splicing of the A2M gene also plays a role in immune diseases in carp and shrimp (83, 84). In pearl oysters (Pinctada fucata), A2M significantly reduced the in vivo phagocytosis of Vibrio alginolyticus by blood cells (85). In addition, in the swimming crab (Portunus trituberculatus), PtA2Ms may play a role in host defense by regulating phagocytosis and the expression of antimicrobial peptide genes (79). In general, these genes play an important role in the initial stage of the immune response, promoting innate immunity development, indicating that the complement and coagulation cascade systems are activated and work synergistically to eliminate bacteria, which is conducive to enhancing the immunity of rainbow trout.
In this study, the arginine and proline metabolism pathway exists in both metabolomics analysis and transcriptomic analysis results. Arginine and proline are two important fatty acid amides that play important roles in osmotic stress. Arginine can produce nitric oxide (NO), and participate in immune function regulation in fish (15). Some studies have shown that arginine affects the growth performance, immunity, and nutrient metabolism of fish (86, 87), and the metabolism of arginine and proline is closely related to the progression of oxidative stress (88, 89). Proline plays important roles in protein synthesis and structure, metabolism and nutrition, as well as wound healing, antioxidative reactions, and immune responses (90). In this study, the expression levels of diamine acetyltransferase 1 (SAT1) and ornithine decarboxylase 1 (ODC1A) which are involved in the proline and arginine metabolism pathways, were significantly up-regulated in the SA group. This result suggested that dietary supplementation with synthetic astaxanthin may contribute to the maintenance of osmotic pressure and resistance to oxidative stress.
In addition, both the transcriptome and metabolome pathway enrichment results showed the presence of lipid metabolism and amino acid metabolism pathways. Further analysis of the data reveled that PRA, compared to NA, screened out the gene HSD17B12A (very-long-chain 3-oxoacyl-CoA reductase-A), which is related to lipid metabolism and specifically involved in fatty acid metabolism and biosynthesis of unsaturated fatty acids. Additionally, the genes PLPP1 (phospholipid phosphatase 1), ARG2 (arginase-2 and mitochondrial), and ODC1-A (ornithine decarboxylase 1) were screened in both the SA and NA groups. PLPP1 is primarily involved in glycerophospholipid metabolism, ARG2 is involved in the biosynthesis of amino acids and arginine and proline metabolism, and ODC1-A is mainly involved in arginine and proline metabolism and glutathione metabolism.
PLPP1 is a multifunctional phospholipase that can catalyze the hydrolysis of various fatty acid phospholipids. Studies have shown that cholic acid has a hepatoprotective effect on CCL4-induced acute liver injury in mice by regulating the lipid metabolism pathway (especially the metabolism of glycerophospholipids, arachidonic acid, and linoleic acid) and altering the expression of Ptgr1 and PLPP1 genes (91). ARG2 is a mitochondrial enzyme involved in arginine metabolism that promotes the resolution of macrophage inflammation and is a target of microRNA-155 (miR-155). Mechanistically, the catalytic activity and presence of ARG2 in mitochondria are critical for oxidative phosphorylation. Moreover, ARG2 is essential for IL-10-mediated downregulation of the inflammatory mediators succinate, hypoxia-inducible factor 1α (HIF-1α), and IL-1β in vitro (92, 93). These results suggest that the addition of P. rhodozyme astaxanthin and synthetic astaxanthin can alter the body’s antioxidant and immune responses by up-regulating the expression of genes related to amino acid and lipid metabolism.
5 Conclusion
This study showed that dietary supplementation with astaxanthin was conducive to the biosynthesis of linoleic acid, α-linolenic acid, arachidonic acid and glycerophospholipids, significantly increased the content of astaxanthin in the serum of rainbow trout, and was conducive to improving the health status; the contents of phosphatidylcholine and phosphatidylinositol were significantly increased, which was helpful to regulate the stability of cell membrane function of rainbow trout. Dietary supplementation with astaxanthin could enhance the antioxidant and immune functions of rainbow trout by affecting the expression of integrin beta-1 (ITGB1), CXC chemokine receptor type 2 (CXCR2), and ornithine decarboxylase 1 (ODC1A) and regulating the FoxO signaling pathway, phagosome, complement and coagulation cascade, and arginine and proline metabolism pathways. These results provide some information for further understanding of the molecular mechanism related to astaxanthin regulation of antioxidant and immune functions in rainbow trout and provide a reference for the healthy and sustainable development of rainbow trout aquaculture.
Data availability statement
The datasets presented in this article are not readily available due to the deletion of the raw data by a third-party. Requests to access the datasets should be directed to the corresponding author.
Ethics statement
The animal study was approved by the Animal Ethics Committee of Dali University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LC: Conceptualization, Investigation, Methodology, Writing – original draft. LW: Formal analysis, Investigation, Methodology, Writing – original draft. YL: Formal Analysis, Investigation, Writing – review & editing. XW: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. XL: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Yunnan Fundamental Research Projects (No. 202101AT070028), the Yunnan Province "Xingdian Talent Support Plan" Young Talents Special Project (No. 230212524080), the Technology Transformation Project of Yunnan Alphy Biotech Co., Ltd. (No. 2019310031000118), and the Dali University Doctoral Research Startup Fee Project (No. KYBS2021064).
Acknowledgments
The authors would like to thank Yunnan Alphy Biotech Co., Ltd., for donating Haematococcus pluvialis powder for this research and the Officially Aller Aqua (Qingdao) Co., Ltd., for their help in the preparation of the experimental feed. Most sincere thanks are also given to Caidan Gongbao, Peng Zhao, and Wenyan Fan for their assistance with the fish culture and sampling.
Conflict of interest
Author YL was employed by the company R & D Center of Qinghai Minze Longyangxia Ecological Hydroponics Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1408168/full#supplementary-material
References
1. Rebolé S, Velasco S, Rodriguez ML, Treviño J, Alzueta C, Tejedor JL, et al. Nutrient content in the muscle and skin of fillets from farmed rainbow trout (Oncorhynchus mykiss). Food Chem. (2015) 174:614–20. doi: 10.1016/j.foodchem.2014.11.072
2. Bondad-Reantaso MG, Subasinghe RP, Arthur JR, Ogawa K, Chinabut S, Adlard R, et al. Disease and health management in Asian aquaculture. Vet Parasitol. (2005) 132:249–72. doi: 10.1016/j.vetpar.2005.07.005
3. Carbone D, Faggio C. Importance of prebiotics in aquaculture as immunostimulants. Effects on immune system of Sparus aurata and Dicentrarchus labrax. Fish Shellfish Immun. (2016) 54:172–8. doi: 10.1016/j.fsi.2016.04.011
4. Round JL, Stentiford GD, Sritunyalucksana K, Flegel TW, Williams BAP, Withyachumnarnkul B, et al. New paradigms to help solve the global aquaculture disease crisis. PloS Pathog. (2017) 13:e1006160. doi: 10.1371/journal.ppat.1006160
5. Raza SHA, Naqvi SRZ, Abdelnour SA, Schreurs N, Mohammedsaleh ZM, Khan I, et al. Beneficial effects and health benefits of astaxanthin molecules on animal production: A review. Res Vet Sci. (2021) 138:69–78. doi: 10.1016/j.rvsc.2021.05.023
6. Li M, Wu W, Zhou P, Xie F, Zhou Q, Mai K. Comparison effect of dietary astaxanthin and Haematococcus pluvialis on growth performance, antioxidant status and immune response of large yellow croaker Pseudosciaena crocea. Aquaculture. (2014) 434:227–32. doi: 10.1016/j.aquaculture.2014.08.022
7. Cao Y, Yang L, Qiao X, Xue C, Xu J. Dietary astaxanthin: an excellent carotenoid with multiple health benefits. Crit Rev Food Sci. (2021) 63:3019–45. doi: 10.1080/10408398.2021.1983766
8. Chang MX, Xiong F. Astaxanthin and its effects in inflammatory responses and inflammation-associated diseases: recent advances and future directions. Molecules. (2020) 25:5342. doi: 10.3390/molecules25225342
9. Song XL, Wang L, Li XQ, Chen ZZ, Liang GY, Leng XJ. Dietary astaxanthin improved the body pigmentation and antioxidant function, but not the growth of discus fish (Symphysodon spp). Aquac Res. (2017) 48:1359–67. doi: 10.1111/are.13200
10. Wu XG, Zhao L, Long XW, Liu JG, Su F, Cheng YX. Effects of dietary supplementation of Haematococcus pluvialis powder on gonadal development, coloration and antioxidant capacity of adult male Chinese mitten crab (Eriocheir sinensis). Aquac Res. (2017) 48:5214–23. doi: 10.1111/are.13333
11. Bjerkeng B, Peisker M, von Schwartzenberg K, Ytrestøyl T, Åsgård T. Digestibility and muscle retention of astaxanthin in Atlantic salmon, Salmo salar, fed diets with the red yeast Phaffia rhodozyma in comparison with synthetic formulated astaxanthin. Aquaculture. (2007) 269:476–89. doi: 10.1016/j.aquaculture.2007.04.070
12. Ishibashi T. Manufacture production of carotenoid from Paracoccus bacterium. Seibutsu-kogaku kaishi. (2015) 93:388–90.
13. Roncarati A, Sirri F, Felici A, Stocchi L, Melotti P, Meluzzi A. Effects of dietary supplementation with krill meal on pigmentation and quality of flesh of rainbow trout (Oncorhynchus mykiss). Ital J Anim Sci. (2016) 10:139–45. doi: 10.4081/ijas.2011.e27
14. European, Food, Safety Authority. Scientific Opinion of the Panel on Biological Hazards (BIOHAZ) - Monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic VTEC types. Efsa J. 5:579 (2007).
15. Sheikhzadeh N, Tayefi-Nasrabadi H, Oushani AK, Enferadi MH. Effects of Haematococcus pluvialis supplementation on antioxidant system and metabolism in rainbow trout (Oncorhynchus mykiss). Fish Physiol Biochem. (2012) 38:413–9. doi: 10.1007/s10695-011-9519-7
16. Elia AC, Prearo M, Dörr AJM, Pacini N, Magara G, Brizio P, et al. Effects of astaxanthin and canthaxanthin on oxidative stress biomarkers in rainbow trout. J Toxicol Env Heal A. (2019) 82:760–8. doi: 10.1080/15287394.2019.1648346
17. Ju ZY, Deng DF, Dominy WG, Forster IP. Pigmentation of pacific white shrimp, Litopenaeus vannamei, by dietary astaxanthin extracted from Haematococcus pluvialis. J World Aquacul Soc. (2011) 42:633–44. doi: 10.1111/j.1749-7345.2011.00511.x
18. Liu XJ, Luo QX, Rakariyatham K, Cao Y, Goulette T, Liu X, et al. Antioxidation and anti-ageing activities of different stereoisomeric astaxanthin in vitro and in vivo. J Funct Foods. (2016) 25:50–61. doi: 10.1016/j.jff.2016.05.009
19. Liu XJ, Luo QX, Cao Y, Goulette T, Liu X, Xiao H. Mechanism of different stereoisomeric astaxanthin in resistance to oxidative stress in Caenorhabditis elegans. J Food Sci. (2016) 81:H2280–7. doi: 10.1111/1750-3841.13417
20. Rao AR, Sindhuja HN, Dharmesh SM, Sankar KU, Sarada R, Ravishankar GA. Effective inhibition of skin cancer, tyrosinase, and antioxidative properties by astaxanthin and astaxanthin esters from the green alga Haematococcus pluvialis. J Agric Food Chem. (2013) 61:3842–51. doi: 10.1021/jf304609j
21. Choi J, Rahman MM, Le SY, Chang KH, Lee SM. Effects of dietary inclusion of fermented soybean meal with Phaffia rhodozyma on growth, muscle pigmentation, and antioxidant activity of juvenile rainbow trout (Oncorhynchus mykiss). Turk J Fish Aquat Sci. (2016) 16:91–101. doi: 10.4194/1303-2712-v16_1_10
22. Nakano T, Kanmuri T, Sato M, Takeuchi M. Effect of astaxanthin rich red yeast (Phaffia rhodozyma) on oxidative stress in rainbow trout. Bba-Bioenergetics. (1999) 1426:119–25. doi: 10.1016/S0304-4165(98)00145-7
23. Amar E, Kiron V, Satoh S, Watanabe T. Enhancement of innate immunity in rainbow trout (Oncorhynchus mykiss Walbaum) associated with dietary intake of carotenoids from natural products. Fish Shellfish Immun. (2004) 16:527–37. doi: 10.1016/j.fsi.2003.09.004
24. Christiansen R, Torrissen OJ. Growth and survival of Atlantic salmon, Salmo salar L. fed different dietary levels of astaxanthin. Juveniles Aquacult Nutr. (1996) 2:55–62. doi: 10.1111/j.1365-2095.1996.tb00008.x
25. Zhang JJ, Li XQ, Leng XJ, Zhang CL, Han ZY, Zhang FG. Effects of dietary astaxanthins on pigmentation of flesh and tissue antioxidation of rainbow trout (Oncorhynchus mykiss). Aquacul Int. (2013) 21:579–89. doi: 10.1007/s10499-012-9590-9
26. Valenzuela-Muñoz V, Valenzuela-Miranda D, Gallardo-Escárate C. Comparative analysis of long non-coding RNAs in Atlantic and Coho salmon reveals divergent transcriptome responses associated with immunity and tissue repair during sea lice infestation. Dev Com Immunol. (2018) 87:36–50. doi: 10.1016/j.dci.2018.05.016
27. Saghatelian A, Cravatt BF. Global strategies to integrate the proteome and metabolome. Curr Opin Chem Biol. (2005) 9:62–8. doi: 10.1016/j.cbpa.2004.12.004
28. Utpott M, Rodrigues E, Rios AD, Mercali GD, Flo SH. Metabolomics: An analytical technique for food processing evaluation. Food Chem. (2021) 366:130685. doi: 10.1016/j.foodchem.2021.130685
29. Konga TT, Lin SM, Ren X, Li SK, Gong Y. Transcriptome and metabolome integration analysis of mud crab Scylla paramamosain challenged to Vibrio parahaemolyticus infection. Fish Shellfish Immun. (2020) 103:430–7. doi: 10.1016/j.fsi.2020.05.069
30. Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. (2009) 48:249–57. doi: 10.1016/j.ymeth.2009.03.016
31. Alfaro AC, Young T. Showcasing metabolomic applications in aquaculture: a review. Rev Aquacult. (2018) 10:135–52. doi: 10.1111/raq.12152
32. Ma QQ, Chen Q, Shen ZH, Li DL, Han T, Qin JG, et al. The metabolomics responses of Chinese mitten-hand crab (Eriocheir sinensis) to different dietary oils. Aquaculture. (2017) 479:188–99. doi: 10.1016/j.aquaculture.2017.05.032
33. Patti GJ, Yanes O, Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Bio. (2012) 13:263–9. doi: 10.1038/nrm3314
34. Roques S, Deborde C, Richard N, Skiba-Cassy S, Moing A, Fauconneau B. Metabolomics and fish nutrition: a review in the context of sustainable feed development. Rev aquacult. (2018) 12:261–82. doi: 10.1111/raq.12316
35. Yang H, Gao X, Li X, Zhang HH, Chen N, Zhang YY, et al. Comparative transcriptome analysis of red swamp crayfish (Procambarus clarkia) hepatopancreas in response to WSSV and Aeromonas hydrophila infection. Fish Shellfish Immun. (2018) 83:397–405. doi: 10.1016/j.fsi.2018.09.051
36. Ning X, Sun L. Gene network analysis reveals a core set of genes involved in the immune response of Japanese flounder (Paralichthys olivaceus) against Vibrio Anguillarum infection. Fish Shellfish Immun. (2020) 98:800–9. doi: 10.1016/j.fsi.2019.11.033
37. Chu Q, Gao YH, Xu GL, Wu CW, Xu TJ. Transcriptome comparative analysis revealed poly(I:C) activated RIG-I/MDA5-mediated signaling pathway in miiuy croaker. Fish Shellfish Immun. (2015) 47:168–74. doi: 10.1016/j.fsi.2015.08.032
38. AOAC, Association of Official Analytical Chemists. Official methods of analysis. Gait Hersburg Maryland, USA: Association of Official Analytical Chemists (2000).
39. Folch J, Lees M, Stanley G. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. (1957) 226:497–509. doi: 10.1016/S0021-9258(18)64849-5
40. Fokina NN, Ruokolainen TR, Bakhmet IN, Nemova NN. Role of lipids in adaptation of mussels Mytilus edulis L. of the White Sea to rapid changes in temperature. Dokl Biochem Biophys. (2014) 457:155–7. doi: 10.1134/S1607672914040103
41. Arts MT, Kohler CC. Health and condition in fish: the influence of lipids on membrane competency and immune response. Lipids in Aquatic Ecosystems. (2009), 237–56. doi: 10.1007/978-0-387-89366-2_10
42. Xie SW, Yin P, Tian LX, Liu YJ, Niu J. Lipid metabolism and plasma metabolomics of juvenile largemouth bass Micropterus salmoides were affected by dietary oxidized fish oil. Aquaculture. (2020) 522:735158. doi: 10.1016/j.aquaculture.2020.735158
43. Cahu CL, Infante JZ, Barbosa V. Effect of dietary phospholipid level and phospholipid: neutral lipid value on the development of sea bass (Dicentrarchus labrax) larvae fed a compound diet. Brit J Nutr. (2003) 90:21–8. doi: 10.1079/BJN2003880
44. Robinson BS, Yao ZM, Baisted DJ, Vance DE. Lyso-phosphatidylcholine metabolism and lipoprotein secretion bycultured rat hepatocytes deficient in choline. Biochem J. (1989) 260:207–14. doi: 10.1042/bj2600207
45. Krogdahl Å, Hansen AKG, Kortner TM, Björkhem I, Krasnov A, Berge GM, et al. Choline and phosphatidylcholine, but not methionine, cysteine, taurine and taurocholate, eliminate excessive gut mucosal lipid accumulation in Atlantic salmon (Salmo salar L). Aquaculture. (2020) 528:735552. doi: 10.1016/j.aquaculture.2020.735552
46. Lou MS, Yu H, Shen SR. Association of dietary linoleic acid with gut microbiome and chronic metabolic diseases. J Mod Med Health. (2013) 39:3611–9. doi: 10.3969/j.issn.1009-5519.2023.21.003
47. Saini RK, Keum YS. Omega-3 and omega-6 polyunsaturated fatty acids: Dietary sources, metabolism, and significance: A review. Life Sci. (2018) 203:255–67. doi: 10.1016/j.lfs.2018.04.049
48. Zhong AH, Dai XX. Comparative transcriptome analysis of the head kidney and trunk kidney in adult yellow catfish (Pelteobagrus fulvidraco). Oceanologia Limnologia Sin. (2021) 52:1486–95. doi: 10.11693/hyhz20210500110
49. Jing HY, Zhang QR, Li S, Gao XJ. Pb exposure triggers MAPK-dependent inflammation by activating oxidative stress and miRNA-155 expression in carp head kidney. Fish Shellfish Immun. (2020) 106:219–27. doi: 10.1016/j.fsi.2020.08.015
50. Song LF, Dong XZ, Hu GB. Transcriptome analysis of red sea bream (Pagrus major) head kidney and spleen infected by Vibrio Anguillarum. Aquacult Rep. (2021) 21:100789. doi: 10.1016/j.aqrep.2021.100789
51. Castro R, Coll J, Blanco MM, Rodriguez-Bertos A. Spleen and head kidney differential gene expression patterns in trout infected with Lactococcus garvieae correlate with spleen granulomas. Vet Res. (2019) 50:32. doi: 10.1186/s13567-019-0649-8
52. Ge H, Lin KB, Zhou C, Lin Q, Zhang ZP, Wu JS, et al. A multi-omic analysis of orange-spotted grouper larvae infected with nervous necrosis virus identifies increased adhesion molecules and collagen synthesis in the persistent state. Fish Shellfish Immun. (2020) 98:595–604. doi: 10.1016/j.fsi.2020.01.056
53. Huang Y, Zhao LL, Feng JL, Zhu HX, Huang X, Ren Q, et al. A novel integrin function in innate immunity from Chinese mitten crab (Eriocheir sinensis). Dev Comp Immunol. (2015) 52:155–65. doi: 10.1016/j.dci.2015.05.005
54. Escobedo-Bonilla CM, Alday- Sanz V, Wille M, Sorgeloos P, Pensaert MB, Nauwynck HJ. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J Fish Dis. (2008) 31:1–18. doi: 10.1111/j.1365-2761.2007.00877.x
55. Tang X, Wang X, Zhan W. An integrin β subunit of Chinese shrimp Fenneropenaeus chinensis involved in WSSV infection. Aquaculture. (2012) 368:1–9. doi: 10.1016/j.aquaculture.2012.08.004
56. Li DF, Zhang MC, Yang HJ, Zhu YB, Xu X. beta-integrin mediates WSSV infection. Virology. (2007) 368:122–32. doi: 10.1016/j.virol.2007.06.027
57. Lin YC, Chen JC, Chen YY, Liu CH, Cheng W, Hsu CH, et al. Characterization of white shrimp Litopenaeus vannamei integrin beta and its role in immunomodulation by dsRNA-mediated gene silencing. Dev Comp Immunol. (2013) 40:167–79. doi: 10.1016/j.dci.2013.01.001
58. Tsukamoto S, Yokosawa H. Natural products inhibiting the ubiquitin-proteasome proteolytic pathway, a target for drug development. Curr Med Chem. (2006) 13:745–54. doi: 10.2174/092986706776055571
59. Huang RQ, Zhang J, Zhu GH, He JG, Xie JF. The core ubiquitin system of mandarin fish, Siniperca chuatsi, can be utilized by infectious spleen and kidney necrosis virus. Fish Shellfish Immun. (2017) 70:293–301. doi: 10.1016/j.fsi.2017.09.017
60. Rivas-Aravena A, Fuentes-Valenzuela M, Escobar-Aguirre S, Gallardo-Escarate C, Molina A, Valdés JA. Transcriptomic response of rainbow trout (Oncorhynchus mykiss) skeletal muscle to Flavobacterium psychrophilum. Comp Biochem Phys D. (2019) 31:100596. doi: 10.1016/j.cbd.2019.100596
61. Hu QM, Ao QW, Zhu JJ. Response of chemokine receptors CXCR2 and integrin β2 after Streptococcus agalactiae and Aeromonas hydrophila challenge in GIFT strain of Nile tilapia Oreochromis niloticus. Dev Comp Immunol. (2020) 115:103897. doi: 10.1016/j.dci.2020.103897
62. Bird S, Tafalla C. Teleost chemokines and their receptors. Biology. (2015) 4:756–84. doi: 10.3390/biology4040756
63. Luo K, Di J, Han PP, Zhang SH, Xia LH, Tian GM, et al. Transcriptome analysis of the critically endangered Dabry's sturgeon (Acipenser dabryanus) head kidney response to Aeromonas hydrophila. Fish Shellfish Immun. (2018) 83:249–61. doi: 10.1016/j.fsi.2018.09.044
64. Umasuthan N, Wan Q, Revathy KS, Whang I, Noh JK, Kim S, et al. Molecular aspects, genomic arrangement and immune responsive mRNA expression profiles of two CXC chemokine receptor homologs (CXCR1 and CXCR2) from rock bream, Oplegnathus fasciatus. Fish Shellfish Immun. (2014) 40:304–18. doi: 10.1016/j.fsi.2014.06.006
65. Liu XX, Kang LS, Liu W, Lou B, Wu CW, Jiang LH. Molecular characterization and expression analysis of the large yellow croaker (Larimichthys crocea) chemokine receptors CXCR2, CXCR3, and CXCR4 after bacterial and poly I:C challenge. Fish Shellfish Immun. (2017) 70:228–39. doi: 10.1016/j.fsi.2017.08.029
66. Paneru B, Al-Tobasei R, Palti Y, Wiens GD, Salem M. Differential expression of long non-coding RNAs in three genetic lines of rainbow trout in response to infection with Flavobacterium psychrophilum. Sci REP-UK. (2016) 6:36032. doi: 10.1038/srep36032
67. Lees J, Hay J, Moles MW, Michie AM. The discrete roles of individual FOXO transcription factor family members in B-cell Malignancies. Front Immunol. (2023) 14:1179101. doi: 10.3389/fimmu.2023.1179101
68. Gross DN, van den Heuvel APJ, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. (2008) 27:2320–36. doi: 10.1038/onc.2008.25
69. Gao L, Yuan ZH, Zhou T, Yang YJ, Gao DY, Dunham R, et al. FOXO genes in channel catfish and their response after bacterial infection. Dev Comp Immunol. (2019) 97:38–44. doi: 10.1016/j.dci.2019.03.010
70. Zhao LJ, Tu JG, Zhang YL, Wang JF, Yang L, Wang WM, et al. Transcriptomic analysis of the head kidney of Topmouth culter (Culter alburnus) infected with Flavobacterium columnare with an emphasis on phagosome pathway. Fish Shellfish Immun. (2016) 57:413–8. doi: 10.1016/j.fsi.2016.09.001
71. Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PloS Pathog. (2007) 3:e51. doi: 10.1371/journal.ppat.0030051
72. Rieger AM, Barreda DR. Antimicrobial mechanisms of fish leukocytes. Dev Comp Immunol. (2011) 35:1238–45. doi: 10.1016/j.dci.2011.03.009
73. Huang JQ, Li YJ, Liu Z, Kang YJ, Wang JF. Transcriptomic responses to heat stress in rainbow trout Oncorhynchus mykiss head kidney. Fish Shellfish Immunol. (2018) 82:32–40. doi: 10.1016/j.fsi.2018.08.002
74. Amara U, Rittirsch D, Flierl M, Bruckner U, Klos A, Gebhard F, et al. Interaction between the coagulation and complement system. Curr Top complement II. (2008) 632:71–9. doi: 10.1007/978-0-387-78952-1_6
75. Markiewski MM, Nilsson B, Ekdahl KN, Mollnes TE, Lambris JD. Complement and coagulation: strangers or partners in crime? Trends Immunol. (2007) 28:184–92. doi: 10.1016/j.it.2007.02.006
76. Nakao M, Tsujikura M, Ichiki S, Vo TK, Somamoto T. The complement system in teleost fish: progress of post-homolog-hunting researches. Dev Comp Immunol. (2011) 35:1296–308. doi: 10.1016/j.dci.2011.03.003
77. Wang LL, Zhang H, Wang LL, Zhang DX, Lv Z, Liu ZQ, et al. The RNA-seq analysis suggests a potential multi-component complement system in oyster Crassostrea gigas. Dev Comp Immunol. (2017) 76:209–19. doi: 10.1016/j.dci.2017.06.009
78. Chen YX, Xu KD, Li JJ, Wang XY, Ye YY, Qi PZ. Molecular characterization of complement component 3 (C3) in Mytilus coruscus improves our understanding of bivalve complement system. Fish Shellfish Immun. (2018) 76:41–7. doi: 10.1016/j.fsi.2018.02.044
79. Ning JH, Liu Y, Gao FT, Song CW, Cui ZX. Two alpha-2 macroglobulin from Portunus trituberculatus involved in the prophenoloxidase system, phagocytosis and regulation of antimicrobial peptides. Fish Shellfish Immun. (2019) 89:574–85. doi: 10.1016/j.fsi.2019.04.033
80. Pizzo SV. When is a proteinase inhibitor a hormone? The strange tale of α2-macroglobulin. J Nat Sci. (2015) 1:1–5.
81. Ho PY, Cheng CH, Cheng WT. Identification and cloning of the alpha2-macroglobulin of giant freshwater prawn Macrobrachium rosenbergii and its expression in relation with the molt stage and bacteria injection. Fish Shellfish Immun. (2009) 26:459–66. doi: 10.1016/j.fsi.2009.01.007
82. Lin YC, Vaseeharan B, Chen JC. Molecular cloning and phylogenetic analysis on α2-macroglobulin (α2-M) of white shrimp Litopenaeus vannamei. Dev Comp Immunol. (2008) 32:317–29. doi: 10.1016/j.dci.2007.07.002
83. Ma HM, Wang B, Zhang JQ, Li FH, Xiang JH. Multiple forms of alpha-2 macroglobulin in shrimp Fenneropenaeus chinesis and their transcriptional response to WSSV or Vibrio pathogen infection. Dev Comp Immunol. (2010) 34:677–84. doi: 10.1016/j.dci.2010.01.014
84. Onara DF, Forlenza M, Gonzalez SF, Rakus KL, Pilarczyk A, Irnazarow G, et al. Differential transcription of multiple forms of alpha-2-macroglobulin in carp (Cyprinus carpio) infected with parasites. Dev Comp Immunol. (2008) 32:339–47. doi: 10.1016/j.dci.2007.06.007
85. Wang ZL, Wang B, Chen G, Lu YS, Jian JC, Wu ZH. An alpha-2 macroglobulin in the pearl oyster Pinctada fucata: characterization and function in hemocyte phagocy-tosis of Vibrio alginolyticus. Fish Shellfish Immun. (2016) 55:585–94. doi: 10.1016/j.fsi.2016.06.037
86. Chen GF, Liu Y, Jiang J, Jiang WD, Kuang SY, Tang L, et al. Effect of dietary arginine on the immune response and gene expression in head kidney and spleen following infection of Jian carp with Aeromonas hydrophila. Fish Shellfish Immun. (2015) 44:195–202. doi: 10.1016/j.fsi.2015.02.027
87. Andersen SM, Waagbø R, Espe M. Functional amino acids in fish nutrition, health and welfare. Front Biosci. (2015) 8:143–69. doi: 10.2741/757
88. Zhang J, Liu XS, Hu CY, Chen XJ, Sun X, Xu NJ. Physiological and transcriptome analysis of exogenous l-arginine in the alleviation of high-temperature stress in Gracilariopsis lemaneiformis. Front Mar Sci. (2021) 8:784586. doi: 10.3389/fmars.2021.784586
89. Hoseini SM, Khan MA, Yousefi M, Costas B. Roles of arginine in fish nutrition and health: insights for future researches. Rev Aquacult. (2020) 12:2091–108. doi: 10.1111/raq.12424
90. Wu GY, Baze FW, Burghardt RC, Johnson GA, Kim SW, Knabe DA, et al. Proline and hydroxyproline metabolism: implications for animal and human nutrition. Amino Acids. (2011) 40:1053–63. doi: 10.1007/s00726-010-0715-z
91. Zhang Z, Sun Y, Zeng Y, Cui N, Li B, Zhang W, et al. Elucidating the hepatoprotective mechanisms of cholic acid against CCl4-Induced acute liver injury: A transcriptomic and metabolomic study. J Ethnopharmacol. (2024) 328:118052. doi: 10.1016/j.jep.2024.118052
92. De Santi C, Nally FK, Afzal R, Duffy CP, Fitzsimons S, Annett SL, et al. Enhancing arginase 2 expression using target site blockers as a strategy to modulate macrophage phenotype. Mol Ther Nucleic Acids. (2022) 29:643–55. doi: 10.1016/j.omtn.2022.08.004
Keywords: astaxanthin, Oncorhynchus mykiss, metabolomics, transcriptomics, antioxidant capacity, immunity
Citation: Chen L, Wang L, Li Y, Wu X and Long X (2024) Comprehensive metabolomics and transcriptomics analyses investigating the regulatory effects of different sources of dietary astaxanthin on the antioxidant and immune functions of commercial-sized rainbow trout. Front. Immunol. 15:1408168. doi: 10.3389/fimmu.2024.1408168
Received: 27 March 2024; Accepted: 26 June 2024;
Published: 26 September 2024.
Edited by:
Kartik Baruah, Swedish University of Agricultural Sciences, SwedenReviewed by:
Meimei Liu, Jiangsu Ocean University, ChinaMin Jin, Ningbo University, China
Tor Gjøen, University of Oslo, Norway
Copyright © 2024 Chen, Wang, Li, Wu and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xugan Wu, eGd3dUBzaG91LmVkdS5jbg==; Xiaowen Long, bG9uZ3hpYW93ZW4xMjNAc2luYS5jb20=
†These authors have contributed equally to this work
 Li Chen1,2,3†
Li Chen1,2,3† Xugan Wu
Xugan Wu Xiaowen Long
Xiaowen Long