
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 04 July 2024
Sec. Cancer Immunity and Immunotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1407837
 Zheng Wang1†
Zheng Wang1† Anqi Li2†
Anqi Li2† Yujie Lu1†
Yujie Lu1† Mengyuan Han1†
Mengyuan Han1† Miao Ruan2
Miao Ruan2 Chaofu Wang2*
Chaofu Wang2* Xiaotian Zhang3
Xiaotian Zhang3 Changbin Zhu3
Changbin Zhu3 Kunwei Shen1*
Kunwei Shen1* Lei Dong2*
Lei Dong2* Xiaosong Chen1*
Xiaosong Chen1*The aim of this study was to evaluate the mutation spectrum of homologous recombination repair (HRR) genes and its association with tumor immune infiltration and prognosis in triple-negative breast cancer (TNBC). TNBC patients (434 patients from Ruijin cohort) were evaluated with targeted next-generating sequencing for mutations in HRR genes. The frequencies of mutations were compared with public reference cohorts (320 TNBC patients from METABRIC, 105 from TCGA, and 225 from MSKCC 2018). Associations between mutation status and tumor immune infiltration and prognosis were analyzed. HRR genes mutations were seen in 21.89% patients, with BRCA1/2 mutations significantly enriched in tumors with breast/ovarian cancer family history (P = 0.025) and high Ki-67 levels (P = 0.018). HRR genes mutations were not related with recurrence-free survival (RFS) (adjusted P = 0.070) and overall survival (OS) (adjusted P = 0.318) for TNBC patients, regardless of carboplatin treatment (P > 0.05). Moreover, tumor immune infiltration and PD-L1 expression was positively associated with HRR or BRCA1/2 mutation (all P < 0.001). Patients with both HRR mutation and high CD8+ T cell counts had the best RFS and OS, whereas patients with no HRR mutation and low CD8+ T cell counts had the worst outcomes (RFS P < 0.001, OS P = 0.019). High frequency of HRR gene mutations was found in early TNBC, with no prognostic significance. Immune infiltration and PD-L1 expression was positively associated with HRR mutation, and both HRR mutation and high CD8+ T cell infiltration levels were associated with superior disease outcome.
The homologous recombination repair (HRR) pathway is responsible for cell differentiation, DNA mismatch repair, transcriptional regulation, and cell apoptosis in order to maintain genomic stability and suppress oncogenesis (1). BRCA1/2 are key genes to the HRR process following DNA double strand breaks (DSBs) (2), and other genes involved in the HRR pathway include ATM, BARD1, PALB2 and RAD51C etc. (3–6) HRR gene mutation results in DNA repair failures and increases lifelong breast cancer risks by 60–70% (7). Germline and somatic BRCA1/2 mutations occur in 10–20% and 3–5% of all TNBC patients, respectively (1, 8), but the mutation pattern of the other HRR genes and the distribution of mutation sites is still uncovered.
Despite its role in oncogenesis, HRR gene mutation still has its silver lining. HRR gene mutation causes genomic loss of heterozygosity, telomeric allelic imbalance, and large-scale state transitions, named as homologous recombination deficiency (HRD) (9). Previous studies have demonstrated that BRCA mutations and HRD-positive status are associated with improved response rates to chemotherapy, but not necessarily with prolonged survival (10, 11). Moreover, BRCA mutation and HRD-positive status also acted as a potential predictor in platinum-containing chemotherapy efficacy (12–17). Yet, most of the findings on treatment response were based on neoadjuvant and metastatic settings, little was understood in the role of HRR gene mutations in early TNBC.
Furthermore, HRR gene mutation may have a potential role in guiding immunotherapy. Previous studies demonstrated that BRCA1-PALB2 interaction disruption was positively associated with programmed cell death-ligand 1 (PD-L1) expression and T-lymphocyte infiltration in hepatocellular carcinoma (HCC) (18), and in ovarian cancer, DNA damage response-deficient (DDRD) breast tumors were associated with attenuated T cell inflammation (19, 20). However, the correlation of HRR gene mutation with immune infiltration, and their impact on the prognosis in TNBC patients have not been well elucidated. With the development of immune checkpoint inhibitors (ICIs) in TNBC (21, 22), understanding the association between HRR gene mutation, immune infiltration and PD-L1 expression, as well as their effect on prognosis, is crucial for guiding treatment and predicting prognosis in early TNBC patients.
Therefore, in the present study, we aimed to identify the mutation spectrum of HRR genes, and explore the association between HRR gene mutation and immune infiltration, as well as its impact on patient prognosis in early TNBC patients.
For Ruijin Cohort 1, we retrospectively screened consecutive breast cancer patients treated at the Comprehensive Breast Health Center, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (RJBC-CBHC) from January 2012 to March 2019. Patients who met the following eligibility criteria were included: 1) invasive breast cancer, 2) pathologically diagnosed with TNBC, 3) available formalin fixed paraffin-embedded (FFPE) tissues. Exclusion criteria were as follows: 1) male breast cancer, 2) de novo stage IV, 3) incomplete immunohistochemistry (IHC) information.
For Ruijin Cohort 2, TNBC patients newly diagnosed and treated in RJBC-CBHC from June 2020 to January 2023 were prospectively recruited. Patients were eligible if they successfully received HRR genotyping. All tumor size, lymph node status, comorbidities, and adjuvant therapy strategies were permitted. Patients with metastatic or recurrent disease were excluded.
Archival FFPE blocks were selected from the biobank at the Department of Pathology, Shanghai Ruijin Hospital. Elaborate clinical data were retrieved from Shanghai Jiao Tong University Breast Cancer Database (SJTU-BCDB). All patients were given informed consent and our study was approved by the Ethical Committees of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (Clinical Trial Ethics Approval Number: 2016; 5–3). All procedures were in accordance with the 1964 Helsinki declaration and its later amendments.
At least two experienced pathologists (A. Li and M. Ruan) from the Department of Pathology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, contributed to the tumor histopathological analysis. The IHC testing was used to determine the status of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor-2 (HER-2) and proliferation index (Ki-67). ER and PR positivity were defined as no less than 1% stained nuclei, as was described in our previous publications (23, 24). Ki67 ≥ 30% was classified as high-expression. HER-2 status was classified as “HER-2 Low” if IHC HER-2 1+ or HER-2 2+/Fluorescence in situ hybridization (FISH) negative, and HER-2 0 patients were defined as “HER-2 negative”.
Patients from Ruijin Cohort 1 went through regular outpatient follow-ups or follow-up calls, once every three months within the first two year after surgery, once every six months through the third to the fifth year, and once every year ever since, in light of the American Society of Clinical Oncology Guidelines (ASCO guidelines) (25). Overall survival (OS) was defined as the time interval between surgery and death. Recurrence-free survival (RFS) was defined as the time interval between surgery and the event of local recurrence, distant metastasis or death. For patients with no events, OS and RFS were define as the time interval between surgery and the last follow-up date (Feb 12th 2023).
Genomic DNA from FFPE blocks was extracted, purified, and quantified using the MagPure FFPE DNA LQ Kit (Magen). A commercially available targeted AmoyDx HANDLE HRR NGS Panel covering 32 genes (17 HRR genes: ATM, ATR, BARD1, BRCA1, BRCA2, BRIP1, CHEK1, CHEK2, FANCA, FANCL, MRE11, NBN, PALB2, RAD51B, RAD51C, RAD51D, RAD54L; other cancer predisposition genes: AR, BRAF, CDH1, CDK12, ERBB2, ESR1, HDAC2, HOXB13, KRAS, NRAS, PIK3CA, PPP2R2A, PTEN, STK11, TP53) was used for next-generating sequencing analysis (Amoy Diagnostics, Xiamen, China). Prepared libraries were sequenced on the Illumina MiSeq system (Paired-End Reads, 2×150 cycles) and analyzed on the AmoyDx ANDAS Data Analyzer (Amoy Diagnostics, Xiamen, China) to accurately detect single nucleotide variants (SNVs) and short insertions/deletions (InDels) with detection sensitivity at variant allele frequency (VAF) ≥ 5%. The average coverage depth was 543X, and the results were further manually filtered to ensure that no false positives were reported.
The avidin-biotin-peroxidase complex method was adopted to perform immunohistochemistry (IHC) staining, as previously described (26, 27). After deparaffinization, rehydration and antigen retrieval, FFPE slides were incubated with CD8 (BD Pharmingen, US) and PD-L1 (Abcam, UK) primary antibodies overnight at 4°C. The secondary antibodies (Abcam, UK) were then applied and incubated at 37°C for an hour. After that, the slides were rinsed with phosphate buffer saline (PBS), stained with 3,3′-diaminobenzidine (DAB), and counterstained with hematoxylin. All specimens were independently and blindly evaluated by two expert pathologists (A. Li and M. Ruan). For CD8 staining, the number of positive cells was calculated in a 0.5 mm diameter cylinder and expressed as the mean value of the five fields per sample (cells per spot) (26). Individual values were used for correlation and survival analyses. Immunotype, as previously described (28), including “inflamed”, “immune excluded” and “immune desert”, was defined based on CD8+ T cell infiltration pattern at the tumor core and stroma. “Inflamed” represents diffuse CD8+ T cell infiltration in the tumor. “Immune excluded” is characterized by CD8+ T cell infiltration limited only to the invasive margin of the tumor. “Immune desert” stands for the absence of CD8+ T cell infiltration in the tumor. Tumor tissues with PD-L1+ tumor cells over 1% are considered PD-L1 positive. For combined survival analysis of HRR mutation and immune infiltration, patients were categorized into three groups: Group I, patients without HRR mutation and a low level of T cells; Group III, patients with HRR mutation and with a high level of T cells; Group II, the remaining patients (the average number of CD8+ T cells as cut-off value).
Data from the Cancer Genome Atlas (TCGA), the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), and the Memorial Sloan Kettering Cancer Center database 2018 (MSKCC 2018) was utilized to compare mutational frequencies. TCGA and MSKCC data was downloaded from the cBioPortal (29) (http://cbioportal.org/, files “TCGA, Firehose Legacy”, “MSK, Cancer cell 2018”). A total of 105 TNBC patients from the TCGA database and 225 TNBC MSK data were analyzed. METABRIC was retrieved as described previously (30), and 320 TNBC patients were used for analysis.
The data distribution was characterized by frequency tabulation and summary statistics. Differences in categorical data was assessed through the Chi-square test or Fisher’s exact test. Kaplan-Meier curves and the Log-rank test were used to compare unadjusted survival between study groups. Cox proportional hazards models were used to evaluate hazard ratios across subgroups and to adjust for patients’ clinicopathological and therapeutic parameters. Two-sided P < 0.05 was considered statistically significant. All statistical analyses were performed using Stata version 16.1 (StataCorp LP, College Station, Texas).
A total of 238 and 196 TNBC patients were included in the Ruijin Cohort 1 and Ruijin Cohort 2, respectively. Public reference cohorts retrieved from cancer genome databases consist of 320 TNBC patients from METABRIC, 105 from TCGA, and 225 from MSKCC. All cohorts were used for mutation rate analysis. The pooled cohort (n = 434) of Ruijin Cohort 1 and Ruijin Cohort 2 was used for clinicopathological characteristics analysis, from which a subset of 383 patients with follow-up data was used for survival and predictive value of carboplatin treatment efficacy analysis (Figure 1).
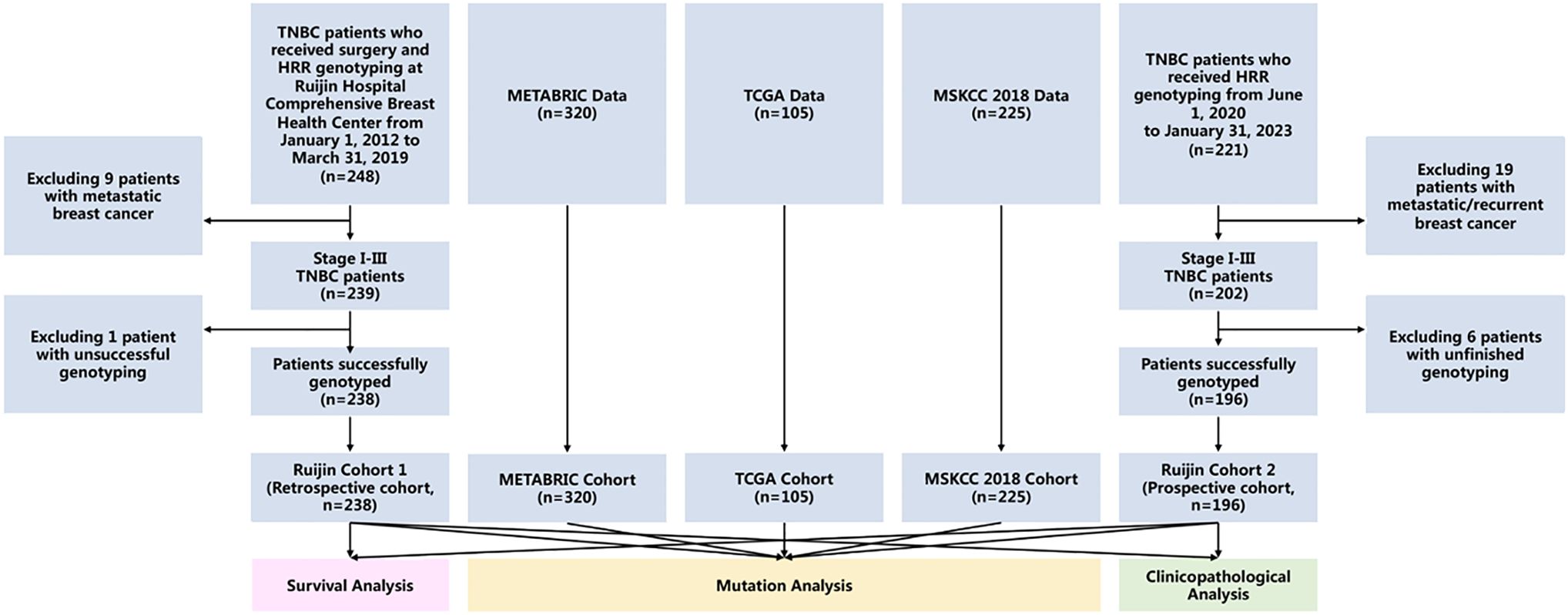
Figure 1 Study flowchart. Abbreviations: METABRIC, the Molecular Taxonomy of Breast Cancer International Consortium; MSKCC, the Memorial Sloan Kettering Cancer Center; TCGA, the Cancer Genome Atlas; TNBC, triple negative breast cancer.
Patient characteristics according to mutation status are shown in Table 1. Patients below 50 years accounted for 46.31% of the total cohort. Breast or ovarian cancer history was found in 63 (14.52%) patients, whereas 31 (7.14%) patients had a family history of other cancers. Twenty-three (5.30%) patients had at least one more primary malignancy other than breast cancer. Invasive ductal carcinoma (IDC) was observed in 84.56% of the study cohort, whereas the proportion of patients with a histological grade of III and a Ki-67 level above 30% were 67.74% and 76.04%, respectively. And HER-2 low status was seen in 55.07% of patients (Table 1).
Tumor mutations in 17 HRR genes were detected in 21.89% (95/434) patients (Figure 2A). In Ruijin Cohort 1, BRCA1 was the most frequently mutated HRR gene (10.08%), followed by BRCA2 (5.88%), ATR (2.52%), BARD1 (2.10%) and PALB2 (2.10%) (Table 2). As for Ruijin Cohort 2, BRCA1 was also the most frequently mutated HRR gene (11.22%), followed by ATM (2.04%) and BRCA2 (1.53%) (Table 2). Notably, BRCA1 was the most frequently mutated HRR gene across all cohorts (except MSKCC 2018), with a mutation rate ranging from 2.67% to 11.22% (Figures 2A–D). In MSKCC 2018, the most frequently mutated HRR gene was ATM (5.33%). The pathological variation (PV) prevalence of BRCA1 was similar across public controls (2.67% to 2.86%), but was much higher (10.60%) in the study cohort, whereas the PV prevalence of BRCA2 in our study (3.92%) and MSKCC 2018 (4.44%) greatly outnumbered that of TCGA (0.95%) and METABRIC (0.63%) (Figure 2E).
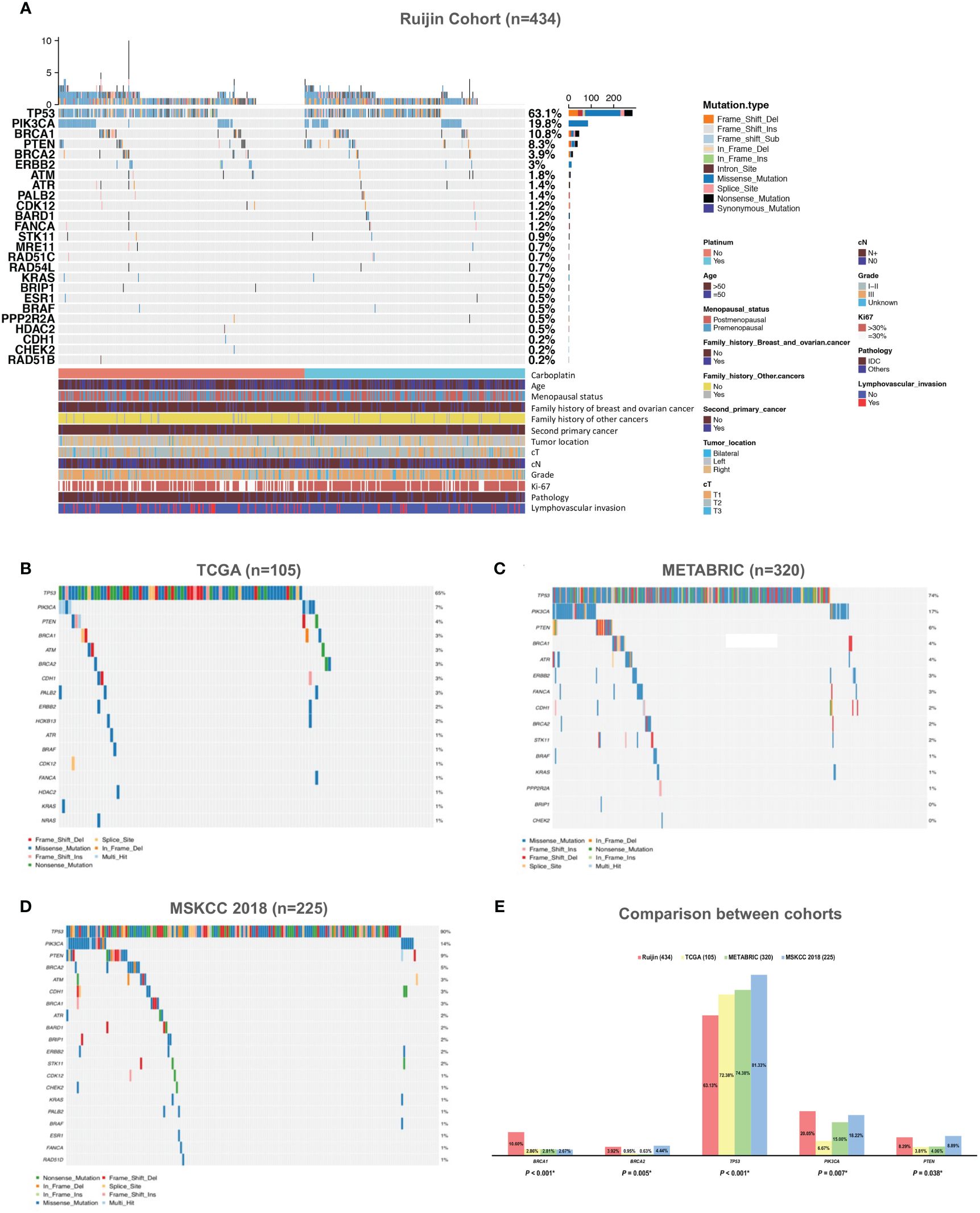
Figure 2 Mutation spectrum of HRR genes and other panel genes in TNBC. (A) Mutation spectrum of 434 TNBC patients from Ruijin Cohort. (B) Mutation spectrum of 105 TNBC patients from TCGA cohort. (C) Mutation spectrum of 320 TNBC patients from METABRIC cohort. (D) Mutation spectrum of 225 TNBC patients from MSKCC 2018 cohort. (E) Comparison between the prevalence of pathogenic variants of 17 homologous recombination repair genes across four cohorts. Abbreviations: HER-2, human epidermal growth factor receptor 2; HRR, homologous recombination repair; IDC, invasive ductal carcinoma; METABRIC, the Molecular Taxonomy of Breast Cancer International Consortium; MSKCC, the Memorial Sloan Kettering Cancer Center; PV, pathogenic variant; TCGA, the Cancer Genome Atlas; TNBC, triple negative breast cancer.
Mutations in other cancer predisposition genes were detected in 59.91% (260/434) of patients. Both in Ruijin Cohort 1 and 2, TP53 was the most frequently mutated panel gene (Ruijin Cohort 1: 61.76%; Ruijin Cohort 2: 64.80%), followed by PIK3CA (Ruijin Cohort 1: 23.53%; Ruijin Cohort 2: 15.82%), and PTEN (Ruijin Cohort 1: 7.98%; Ruijin Cohort 2: 8.67%) (Table 2). TP53, PIK3CA, and PTEN were also the most highly mutated gene across all cohorts, ranging from 55.88% to 81.33%, 6.67% to 23.53% and 3.81% to 8.89% of mutation rate in TCGA, METABRIC and MSKCC 2018, respectively (Figures 2A–E).
Locations of mutations and domains in proteins encoded by BRCA1, BRCA2, and other predisposition genes are shown by lollipop structures (Figure 3). The mutation hotspot of BRCA1 lay at protein domain p.I1824Dfs*3, which accounts for 6.52% (3/46) of BRCA1 mutations; and no mutation hotspot was detected for BRCA2. Mutations of TP53 mainly located at p.R273H/C/L (6.57%, 18/274). The most frequently mutated protein domain of PIK3CA was p.H1047R/L (70.11%, 61/87). PTEN mutations most frequently occurred at p.R130*/Q (5.56%, 2/36). And ERBB2 mutations mainly located on p.V777L (23.08%, 3/13). Further correlation analysis between HRR gene mutations indicated several co-mutation patterns, including PALB2 and RAD51B, NBN and RAD54L, ATR and RAD54B, ATR and RAD51L, FANCA and NBN, CHEK2 and PPP2R2A, and PPP2R2A and RAD54L (all odds ratios > 5.0, all P < 0.001) (Supplementary Figure 1A and Figure 1B).
In 434 patients from the pooled cohort of Ruijin Cohort 1 and Ruijin Cohort 2 with early TNBC, BRCA tumor mutation was associated with menopausal status (mutation rate 18.32% in premenopausal women versus 10.78% in postmenopausal women, P = 0.035), family history of breast/ovarian cancer (mutation rate 23.81% in women with family history versus 12.67% in women without family history, P = 0.016) and high Ki-67 levels (mutation rate 16.97% in high Ki-67 group versus 5.77% in low Ki-67 group, P = 0.005). TP53 mutation was associated with high Ki-67 levels (mutation rate 68.18% in high Ki-67 group versus 47.12% in low Ki-67 group, P < 0.001). A significantly higher PIK3CA mutation rate was found in patients above 50 years (mutation rate 24.89% in patients above 50 years versus 14.43% in patients below 50 years, P = 0.007) and postmenopausal women (mutation rate 26.29% in postmenopausal women versus 12.87% in premenopausal women, P < 0.001), and was also associated with low tumor grade (mutation rate = 24.49% in grade I-II tumors versus 17.01% in grade III tumors, P = 0.049), HER-2 low status (mutation rate = 26.78% in HER-2 low tumors versus 11.79% in HER-2 negative tumors, P < 0.001) and low proliferation indices (mutation rate = 29.81% in low Ki-67 group versus 16.97% in high Ki-67 group, P = 0.004). No association between tumor mutation and secondary primary tumor was found in the study cohort (Table 3). Multivariate analysis further showed that breast/ovarian cancer family history (OR = 2.15, 95%CI 1.10–4.19, P = 0.025) and high Ki-67 levels (OR = 2.91, 95%CI 1.20–7.07, P = 0.018) were two independent predictive factors for BRCA mutation. A high Ki-67 level is also an independent predictive factor for TP53 mutation (OR = 2.41, 95%CI 1.53–3.77, P < 0.001). Moreover, HER-2 low status was an independent predictive factor for PIK3CA mutation (OR = 2.37, 95%CI 1.37–4.08, P = 0.002) (Supplementary Table 1).
After a median follow-up of 44.73 months, eighty-six (22.45%) out of 383 patients with follow-up data experienced with locoregional recurrence and (or) distant metastasis, whereas a total of 46 (12.01%) patients died. Univariate analysis indicated that 5-year RFS (79.88% versus 70.32%, P = 0.030), but not OS (88.99% versus 82.46%, P = 0.230) was better in HRR gene mutated patients compared with their wildtype counterparts (Figure 4A; Supplementary Figure 2). Yet after adjusting for age, tumor location, tumor grade, cT stage, cN stage, Ki67 level, lymphovascular invasion status, and carboplatin administration status, no difference in RFS (Adjusted P = 0.070) and OS (Adjusted P = 0.318) was observed between HRR gene mutated and non-mutated patients (Supplementary Figures 3A, B).
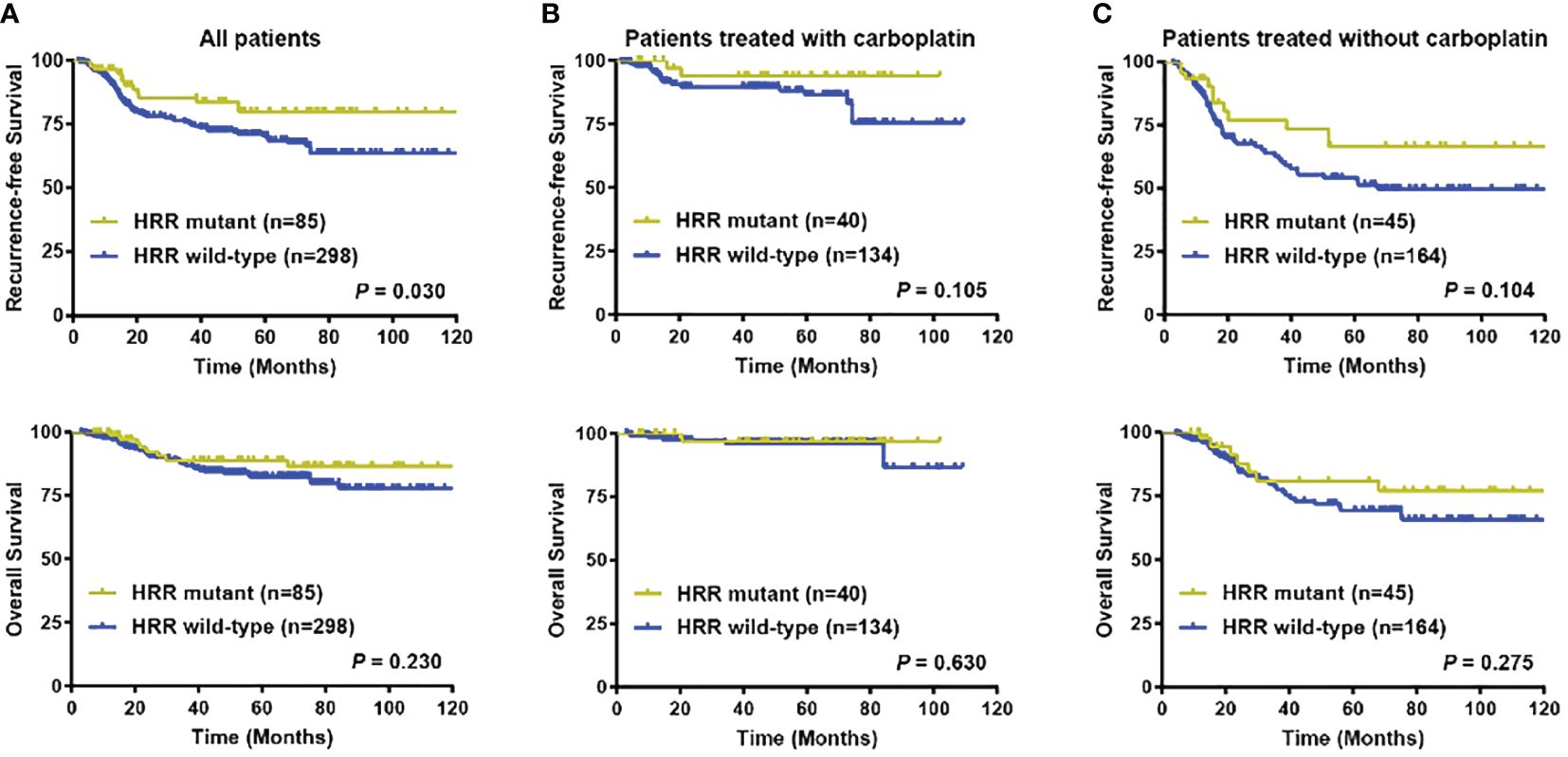
Figure 4 Prognostic significance of HRR gene mutation status. (A) Recurrence-free survival and overall survival in all patients according to HRR gene mutation status; (B) Recurrence-free survival and overall survival in patients treated with carboplatin chemotherapy according to HRR gene mutation status; (C) Recurrence-free survival and overall survival in patients treated without carboplatin chemotherapy according to HRR gene mutation status. Abbreviations: HRR, homologous recombination repair; OS, overall survival; RFS, recurrence-free survival.
A total of 174 (45.43%) patients received carboplatin-containing adjuvant chemotherapy. Within the carboplatin treatment group, RFS events were found in 19 (10.92%) patients, while death occurred in 6 (3.45%) patients. In the rest 209 (54.57%) patients treated without carboplatin, locoregional recurrence and (or) distant metastasis was observed in 67 (32.06%) patients, whereas death was seen in 40 (19.14%) patients. Subgroup analysis further indicated that in patients treated with carboplatin chemotherapy, no survival difference was seen between the HRR genes mutation and non-mutation groups in 5-year RFS (93.84% versus 86.34%, P = 0.105) and OS (96.88% versus 96.39%, P = 0.630) (Figure 4B). And in subgroup without carboplatin treatment, no significant difference was found in terms of 5-year RFS (66.44% versus 52.74%, P = 0.104) and OS (80.97% versus 69.29%, P = 0.275) between HRR gene mutation carriers and non-carriers (Figure 4C).
In the study cohort, a lower percentage of invasive ductal carcinoma (P = 0.026) and a higher frequency of unknown tumor grade (P = 0.003) was seen in the carboplatin treatment group than in the non-carboplatin treatment group, whereas Ki67 levels were higher in the carboplatin treatment group than in the non-carboplatin treatment group (P < 0.001). No difference was observed in other clinicopathological parameters (Supplementary Table 2). The 5-year RFS (88.17% versus 56.06%, P < 0.001) and OS (96.47% versus 72.15%, P < 0.001) in patients treated with carboplatin chemotherapy were significantly higher than those without (Supplementary Figure 4A). In HRR gene mutation carriers, superior 5-year RFS (93.84% versus 66.44%, P = 0.006) and OS (96.88% versus 80.97%, P = 0.030) were observed in the carboplatin treatment group compared with the non-carboplatin group (Supplementary Figure 4B). Patients without HRR gene mutations showed better 5-year RFS (86.34% versus 52.74%, P < 0.001) and OS (96.39% versus 69.29%, P < 0.001) when carboplatin was added to the treatment regimen (Supplementary Figure 4C). Furthermore, in multivariate analysis, carboplatin administration acted as an independent protective factor for RFS (HR = 0.23, 95%CI 0.13–0.38, P < 0.001) and OS (HR = 0.15, 95%CI 0.06–0.37, P < 0.001) in early TNBC (Supplementary Figures 3A, B).
Across TP53, PIK3CA, and PTEN-PIK3CA pathway mutation and non-mutation subgroups, RFS and OS for early TNBC were significantly higher in patients treated with carboplatin chemotherapy than without regardless of gene mutation status (All Interaction P > 0.05) (Figures 5A, B; Supplementary Figures 5C–F). Nevertheless, survival benefit from carboplatin chemotherapy was only seen in BRCA mutation non-carriers. In BRCA mutation carriers, survival benefit from carboplatin chemotherapy was only observed in RFS (P = 0.046), but not OS (P = 0.077) (Supplementary Figures 5A, B).
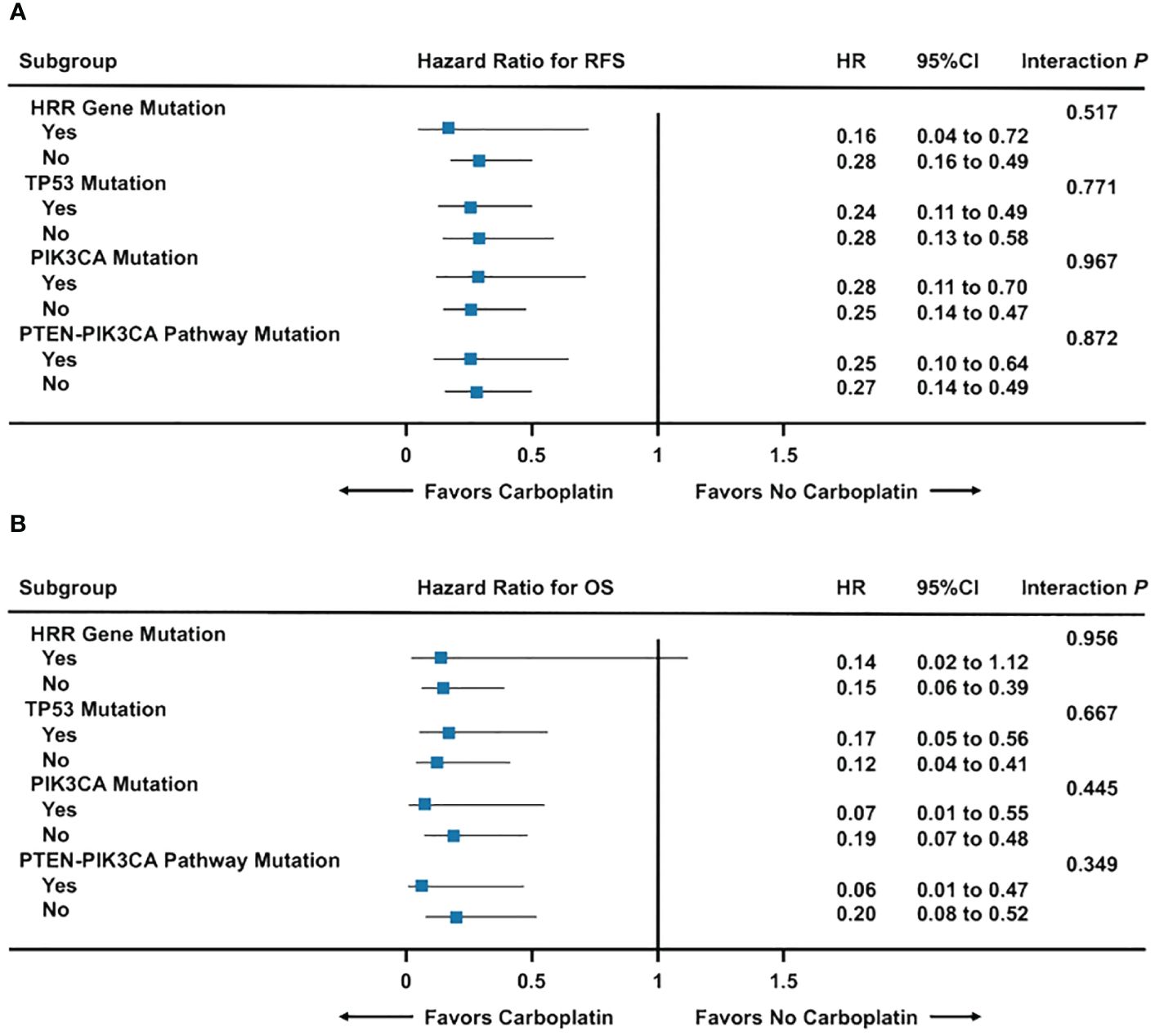
Figure 5 Forest plot of carboplatin chemotherapy efficacy among gene mutation subgroups. (A) Recurrence-free survival among gene mutation subgroups; (B) Overall survival among gene mutation subgroups. Abbreviations: HRR, homologous recombination repair; OS, overall survival; RFS, recurrence-free survival.
In Ruijin Cohort 1, tumor specimens from 162 patients successfully achieved CD8 and PD-L1 immune staining and evaluation (Figure 6A). Forty-six patients were immune inflamed, 39 were immune exclude, and 77 were immune desert, respectively. Of 31 HRR gene mutated patients, 21 (68%) were immune inflamed, 5 (16%) were immune excluded, and 5 (16%) were immune desert, while in 131 HRR gene wildtype patients, 25 (19%) were immune inflamed, 34 (26%) were immune excluded, and 72 (55%) were immune desert (P < 0.001) (Figure 6B). Twenty-one patients were BRCA mutation carriers, consisting of 14 (67%) immune inflamed, 2 (10%) immune excluded, and 5 (23%) immune desert immunotypes, whereas within 141 noncarriers, the distribution of inflamed, immune excluded and immune desert immunotypes were 32 (23%), 37 (26%) and 72 (51%), respectively (P < 0.001) (Figure 6C). The level of immune infiltration and CD8+ T cell count in HRR gene or BRCA mutation carriers were significantly higher than their wildtype counterparts (both P < 0.001) (Figures 6D, E). Eighty one percent of HRR gene mutated tumors expressed PD-L1, which was 53% in HRR gene wildtype tumors (P < 0.001) (Figure 6F). In BRCA mutation carriers and non-carriers, 76% and 56% tumors expressed PD-L1 (P = 0.003), respectively (Figure 6G). Kaplan-Meier curves found that patients with both HRR gene mutation and high CD8+ T cell counts (average count as cut-off value) had the best RFS and OS, whereas patients without HRR gene mutation and low CD8+ T cell counts had the worst RFS (P < 0.001) and OS (P = 0.019) (Figures 6H, I).
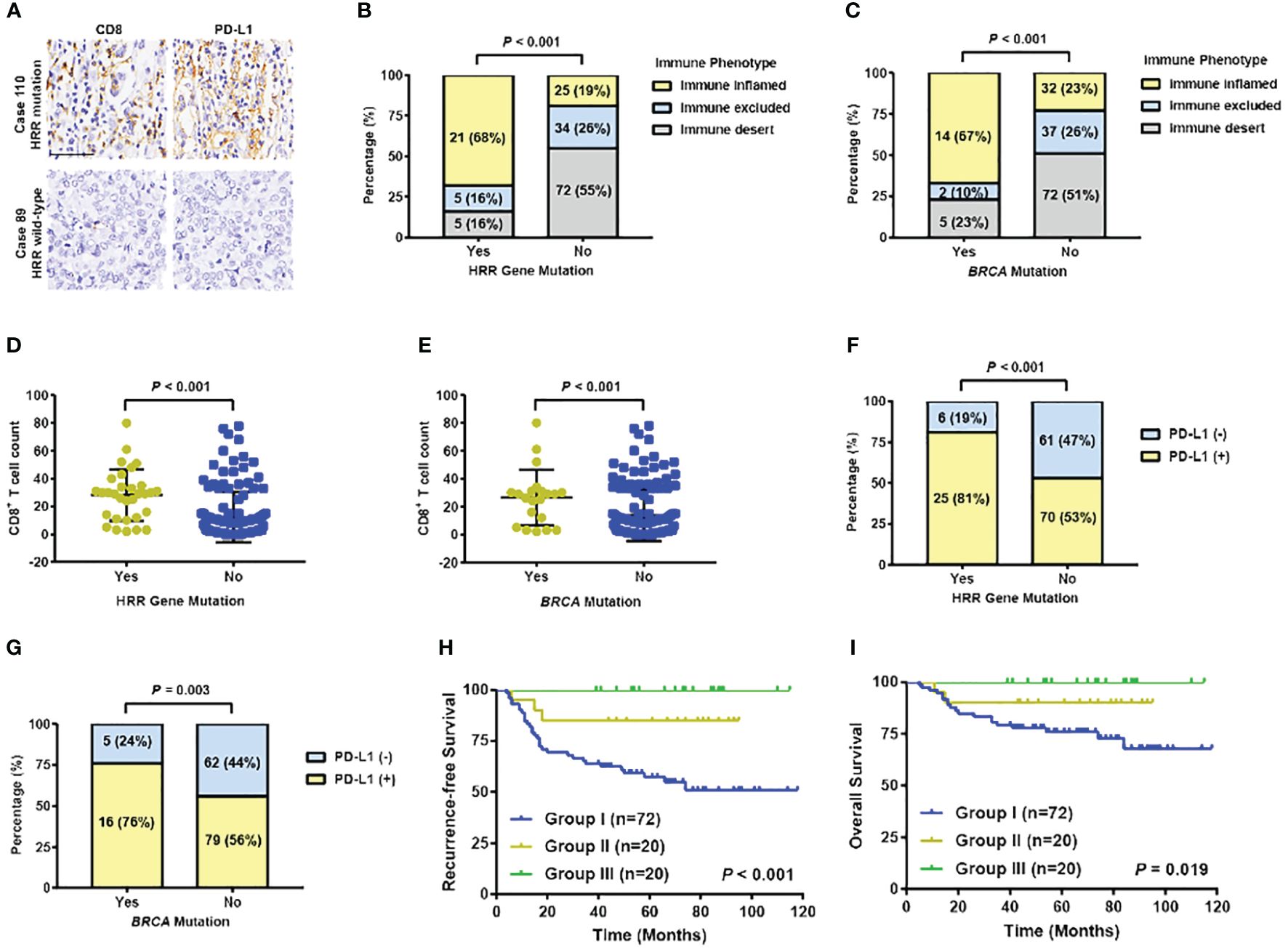
Figure 6 Association between HRR gene mutation and immune infiltration in TNBC patients. (A) Representative images of IHC staining of CD8 and PD-L1 in HRR-mutated and wildtype TNBC samples. (Up: high expression of CD8 and PD-L1; bottom: low expression of CD8 and PD-L1). Scale bar, 50 μm. (B) Distribution of immune phenotype by HRR gene mutation status. (C) Distribution of immune phenotype by BRCA mutation status. (D) Distribution of CD8+ T cell counts by HRR gene mutation status. (E) Distribution of CD8+ T cell counts by BRCA mutation status. (F) PD-L1 expression by HRR gene mutation status. (G) PD-L1 expression by BRCA mutation status. (H) Recurrence-free survival by HRR gene mutation status and CD8+ T cell counts (Group I: patients without HRR mutation and a low level of T cells; Group III: patients with HRR mutation and with a high level of T cells; Group II: the remaining patients). (I) Overall survival by HRR gene mutation status and CD8+ T cell counts (Group I: patients without HRR mutation and a low level of T cells; Group III: patients with HRR mutation and with a high level of T cells; Group II: the remaining patients). Abbreviations: HRR, homologous recombination repair; OS, overall survival; PD-L1, programmed cell death-ligand 1; RFS, recurrence-free survival; TNBC, triple-negative breast cancer.
In this large cohort of 434 Chinese patients with early TNBC, BRCA1, BRCA2, and ATM were the most frequently mutated HRR genes, and for other cancer predisposition genes, the top three ranking were TP53, PIK3CA, and PTEN, respectively. The mutation rate of BRCA1, FANCA, and RAD51C was higher in Ruijin Cohorts than in the TCGA, METABRIC and MSKCC 2018 cohorts. One reason lies in the difference of coverage depth. Deep coverage aids in differentiating sequencing errors from single nucleotide polymorphisms and increases accuracy in the final results. Based on previous studies, an average depth above 30X is required to detect all SNVs (31), and the depth of coverage in our study cohort, METABRIC (32), TCGA (33), and MSKCC 2018 (34) was 543X, 50X, 100X, and 771X, respectively, which was more than sufficient to achieve accurate results, especially in our study. Another reason was that data on public reference controls mainly report somatic mutations, while our study presented tumor mutations with next-generation target sequencing in FFPE samples, which may increase the mutation rate in some panel genes. The overall mutation rate of HRR genes was 21.89% in early TNBC, adding to a high clinical value and a great potential to carry out further research into HRR gene mutation’s association with clinicopathological parameters and survival prognosis.
Previous studies showed that the mutation hotspot of BRCA1 was p.IGQ1824–1826*, which was p.I1824Dfs*3 in our study, indicating different types of mutations at the same location. And similar to our study, BRCA2 also showed mutations on sporadic sites without a detected hotspot (35). The most frequently mutated domain of TP53 was p.R175H according to Kim Y et al (36), p.R248Q according to Jouali F et al (37), but p.R273H/C/L in our study. The mutation hotspot of PIK3CA was p.H1047R/L in our cohort, which is in line with Martínez-Sáez O et al’s and Jouali F et al’s studies (38, 39). PTEN mutations most frequently occurred at p.K60SfsX98 (40), which was p.R130*/Q in the Ruijin cohort. And ERBB2 mutation hotspot was located at p.V777L in our study, which was p.V842I according to Robichaux JP et al’s study (41). Collectively, these detected mutation hotspots in our study still require further exploration to verify their biological functions. In terms of the association between gene mutation status and clinicopathological factors, BRCA mutation was associated with high proliferation indices, probably due to the fact that BRCA mutated cancers are more likely to show basal-like phenotypes (42). PIK3CA mutations, on the contrary, shows a tendency towards low proliferation indices, indicating that the mutation of BRCA and PIK3CA might be mutually exclusive, which was in line with previous studies (43, 44). The mechanism under this mutually exclusiveness is still uncovered, and further investigations are still needed.
In survival prognosis analysis, tumor mutations in HRR genes didn’t have significant impact on RFS and OS, which was similar with Punie K et al. ‘s study, indicating that there was no significant survival difference between germline BRCA mutated and non-mutated breast cancer. Similar results with the GeparSixto trial, all the patients received chemotherapy in our cohort, and BRCA mutation didn’t significantly affect survival prognosis in early TNBC patients, probably due to the DNA damage chemotherapeutic agents (13). Although the prognosis of patients with HRR genes mutation carriers and non-carriers in early TNBC were similar, future studies are needed to identify certain gene combinations within the HRR pathway where mutation status could predict survival.
Moreover, carboplatin-containing chemotherapy could significantly improve response rates in TNBC patients, according to GeparSixto trial (13), indicating carboplatin is an effect agent in TNBC. However, it remains uncertain whether carboplatin could improve prognosis in early TNBC patients and patients with HRR gene mutation. In our study, carboplatin treatment significantly improved survival prognosis regardless of HRR gene, TP53 and PIK3CA mutation status and acted as an independent protective factor for RFS and OS. This result might derive from the fact that most of the patients received carboplatin chemotherapy sequenced with anthracyclines and cyclophosphamide, all of which were DNA damaging agents and could cast severe DNA damage to tumor cells when being used together (45). When the degree of damage exceeded the DNA repair capacity of the HRR pathway, cell death could occur regardless of HRR status, which might be the possible molecular mechanism in our study. Although no difference in carboplatin efficacy was observed between HRR gene mutation carriers and non-carriers, HRR gene mutation’s prognostic role in patients treated with other agents, such as PARP inhibitors (46), still requires further investigation.
Furthermore, based on our study, immune infiltration and PD-L1 expression was positively associated with HRR gene and BRCA mutation, which was in line with the results in liver and ovarian cancers (18, 19). According to previous researches, the underlying mechanism of this phenomenon could be listed as follows. On one hand, the stimulator of interferon genes (STING) pathway, which plays a crucial role in innate immune response to cancer, can be activated at DNA damage repair (DDR) deficiency and exposure to DNA-damaging agents (47), which stimulates interferon (IFN) production and in turn recruit T lymphocytes through the STING-interferon regulatory factor 3 (IRF3) pathway (48, 49), leading to T cell infiltration in HRR gene mutated tumors. On the other hand, the activated STING pathway induces PD-L1 expression through the STING-IRF3-signal transducer and activator of transcription 1 (STAT1) pathway, while DNA damage further upregulates PD-L1 mRNA expression, leading to increased PD-L1 expression in HRR mutation carriers (50–52). Additionally, patients with both HRR gene mutation and high immune infiltration has the superior disease outcome in our study. Potential mechanism might lie in that tumors with HRR gene mutation have deficiency in repairing DSBs caused by chemotherapy, whereas damaged tumor cells become more recognizable and vulnerable to immunologic cytotoxicity caused by infiltrating T lymphocytes (20). Future researches are still needed to further identify the underlying mechanisms.
There are potential limitations to this study. The cohort in which we analyzed survival prognosis is a single-center retrospective cohort, including a continuous cohort of triple negative breast cancer patients who underwent surgery, which might be susceptible to selection bias and lack of representativeness and requires further validation in further multicenter prospective studies. In addition, tumor-specific somatic mutations can be further distinguished from tumor mutations in FFPE samples by stroking out background germline mutations, which can be achieved through targeted-NGS in peripheral blood or normal breast tissues. Besides, new generation method in assessing immune infiltration status, such as multiple immunofluorescences, spatial transcriptomics, might help to provide more precise classification. Furthermore, though HRR mutation status could not predict carboplatin efficacy, there might be a better model, to name a few, HRD score (12), Neo-Family History Score (53), and ten HRR gene-model (17) to differentiate survival prognosis between platinum-containing treatment groups and their counterparts, which needs further biomarker studies in the future.
High frequency of tumor mutations in HRR genes was found in early TNBC patients, but showed no significant association with survival outcome, regardless of carboplatin treatment. Immune infiltration and PD-L1 expression was positively associated with HRR mutation, and patients with both HRR mutation and high CD8+ T cell infiltration levels had superior disease outcome. Further genetic testing implication and novel clinical trials with specific targeted HRR pathway therapies need to be investigated.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by Ethical Committees of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
ZW: Writing – review & editing, Writing – original draft, Project administration, Methodology, Investigation, Funding acquisition, Data curation. AL: Writing – review & editing, Writing – original draft, Validation, Resources, Methodology, Investigation, Formal analysis, Data curation. YL: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Data curation, Conceptualization. MH: Writing – review & editing, Visualization, Software. MR: Writing – review & editing, Resources, Methodology, Formal analysis. CW: Writing – review & editing, Validation, Supervision, Resources, Methodology. XZ: Writing – review & editing, Resources, Methodology, Formal analysis. CZ: Writing – review & editing, Validation, Software, Resources, Methodology, Formal analysis. KS: Writing – review & editing, Validation, Supervision, Resources, Project administration, Funding acquisition, Data curation, Conceptualization. LD: Writing – review & editing, Supervision, Project administration, Conceptualization. XC: Writing – review & editing, Validation, Supervision, Resources, Project administration, Methodology, Funding acquisition, Data curation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82072937 and 82072897), Innovative Research Team of High-Level Local Universities in Shanghai (SHSMU-ZDCX20212200), Natural Science Foundation of Shanghai Science and Technology Committee (23ZR1439500), and Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20172007).
XZ and CZ were employed by Amoy Diagnostics Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1407837/full#supplementary-material
1. Atchley DP, Albarracin CT, Lopez A, Valero V, Amos CI, Gonzalez-Angulo AM, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer, J. Clin Oncol. (2008) 26:4282–8. doi: 10.1200/JCO.2008.16.6231
2. Yun MH, Hiom K. CtIP−BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. (2009) 459:460–3. doi: 10.1038/nature07955
3. Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. (2010) 108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0
4. Tarsounas M, Sung P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication, Nat. Rev Mol Cell Biol. (2020) 21:284–99. doi: 10.1038/s41580-020-0218-z
5. Li A, Geyer FC, Blecua P, Lee JY, Selenica P, Brown DN, et al. Homologous recombination DNA repair defects in PALB2-associated breast cancers. NPJ Breast Cancer. (2019) 5:23. doi: 10.1038/s41523-019-0115-9
6. Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science (2001) 291:1284–9. doi: 10.1126/science.1056154
7. Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. (2003) 72:1117–30. doi: 10.1086/375033
8. Belli C, Duso BA, Ferraro E, Curigliano G. Homologous recombination deficiency in triple negative breast cancer. Breast. (2019) 45:15–21. doi: 10.1016/j.breast.2019.02.007
9. Marquard AM, Eklund AC, Joshi T, Krzystanek M, Favero F, Wang ZC, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. biomark Res. (2015) 3:9. doi: 10.1186/s40364-015-0033-4
10. Sharma P, Barlow WE, Godwin AK, Pathak H, Isakova K, Williams D, et al. Impact of homologous recombination deficiency biomarkers on outcomes in patients with triple-negative breast cancer treated with adjuvant doxorubicin and cyclophosphamide (SWOG S9313). Ann Oncol. (2018) 29:654–60. doi: 10.1093/annonc/mdx821
11. Copson ER, Maishman TC, Tapper WJ, Cutress RI, Greville-Heygate S, Altman DG, et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. Lancet Oncol. (2018) 19:169–80. doi: 10.1016/S1470-2045(17)30891-4
12. Loibl S, Weber KE, Timms KM, Elkin EP, Hahnen E, Fasching PA, et al. Survival analysis of carboplatin added to an anthracycline/taxane-based neoadjuvant chemotherapy and HRD score as predictor of response-final results from GeparSixto. Ann Oncol. (2018) 29:2341–7. doi: 10.1093/annonc/mdy460
13. Hahnen E, Lederer B, Hauke J, Loibl S, Kröber S, Schneeweiss A, et al. Germline mutation status, pathological complete response, and disease-free survival in triple-negative breast cancer: secondary analysis of the geparSixto randomized clinical trial. JAMA Oncol. (2017) 3:1378–85. doi: 10.1001/jamaoncol.2017.1007
14. Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer, Clin. Cancer Res. (2016) 22:3764–73. doi: 10.1158/1078-0432.CCR-15-2477
15. Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. (2018) 24:628–37. doi: 10.1038/s41591-018-0009-7
16. Martin M, Ramos-Medina R, Bernat R, García-Saenz JA, Del Monte-Millan M, Alvarez E, et al. Activity of docetaxel, carboplatin, and doxorubicin in patient-derived triple-negative breast cancer xenografts. Sci Rep. (2021) 11:7064. doi: 10.1038/s41598-021-85962-4
17. Huang L, Lang GT, Liu Q, Shi JX, Shao ZM, Cao AY. A predictor of pathological complete response to neoadjuvant chemotherapy in triple-negative breast cancer patients with the DNA repair genes. Ann Transl Med. (2021) 9:301. doi: 10.21037/atm-20–4852
18. Ma H, Kang Z, Foo TK, Shen Z, Xia B. Disrupted BRCA1-PALB2 interaction induces tumor immunosuppression and T-lymphocyte infiltration in HCC through cGAS-STING pathway. Hepatology. (2023) 77:33–47. doi: 10.1002/hep.32335
19. Bruand M, Barras D, Mina M, Ghisoni E, Morotti M, Lanitis E, et al. Cell-autonomous inflammation of BRCA1-deficient ovarian cancers drives both tumor-intrinsic immunoreactivity and immune resistance via STING. Cell Rep. (2021) 36:109412. doi: 10.1016/j.celrep.2021.109412
20. Jiang M, Jia K, Wang L, Li W, Chen B, Liu Y, et al. Alterations of DNA damage response pathway: Biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm Sin B. (2021) 11:2983–94. doi: 10.1016/j.apsb.2021.01.003
21. Schmid P, Cortes J, Dent R, Pusztai L, McArthur H, Kümmel S, et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N Engl J Med. (2022) 386:556–67. doi: 10.1056/NEJMoa2112651
22. Mittendorf EA, Zhang H, Barrios CH, Saji S, Jung KH, Hegg R, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet. (2020) 396:1090–100. doi: 10.1016/S0140-6736(20)31953-X
23. Ding S, Sun X, Lu S, Wang Z, Chen X, Shen K. Association of molecular subtype concordance and survival outcome in synchronous and metachronous bilateral breast cancer. Breast. (2021) 57:71–9. doi: 10.1016/j.breast.2021.03.005
24. Zhu S, Li Y, Chen W, Fei X, Shen K, Chen X. Molecular subtype may be more associated with prognosis and chemotherapy benefit than tumor size in T1N0 breast cancer patients: an analysis of 2,168 patients for possible de-escalation treatment. Front Oncol. (2021) 11:636266. doi: 10.3389/fonc.2021.636266
25. Khatcheressian JL, Wolff AC, Smith TJ, Grunfeld E, Muss HB, Vogel VG, et al. American Society of Clinical Oncology 2006 update of the breast cancer follow-up and management guidelines in the adjuvant setting. J Clin Oncol. (2006) 24:5091–7. doi: 10.1200/JCO.2006.08.8575
26. Wang Z, Wang F, Ding XY, Li TE, Wang HY, Gao YH, et al. Hippo/YAP signaling choreographs the tumor immune microenvironment to promote triple negative breast cancer progression via TAZ/IL-34 axis. Cancer Lett. (2022) 527:174–90. doi: 10.1016/j.canlet.2021.12.016
27. Tong Y, Huang J, Ren W, Yu J, Zhang X, Wang Z, et al. Association of tumor immune microenvironment profiling and 21-gene recurrence assay in early breast cancer patients. Eur J Med Res. (2022) 27:293. doi: 10.1186/s40001-022-00917-3
28. Li TE, Zhang Z, Wang Y, Xu D, Dong J, Zhu Y, et al. A novel immunotype-based risk stratification model predicts postoperative prognosis and adjuvant TACE benefit in chinese patients with hepatocellular carcinoma. J Cancer. (2021) 12:2866–76. doi: 10.7150/jca.54408
29. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
30. Hwang KT, Kim BH, Oh S, Park SY, Jung J, Kim J, et al. Prognostic role of KRAS mRNA expression in breast cancer. J Breast Cancer. (2019) 22:548–61. doi: 10.4048/jbc.2019.22.e55
31. Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. (2014) 15:121–32. doi: 10.1038/nrg3642
32. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. (2016) 7:11479. doi: 10.1038/ncomms11479
33. Wang VG, Kim H, Chuang JH. Whole-exome sequencing capture kit biases yield false negative mutation calls in TCGA cohorts. PloS One. (2018) 13:e0204912. doi: 10.1371/journal.pone.0204912
34. Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. (2018) 34:427–38. doi: 10.1016/j.ccell.2018.08.008
35. Ma D, Chen SY, Ren JX, Pei YC, Jiang CW, Zhao S, et al. Molecular features and functional implications of germline variants in triple-negative breast cancer. J Natl Cancer Inst. (2021) 113:884–92. doi: 10.1093/jnci/djaa175
36. Kim Y, Kim J, Lee HD, Jeong J, Lee W, Lee KA. Spectrum of EGFR gene copy number changes and KRAS gene mutation status in Korean triple negative breast cancer patients. PloS One. (2013) 8:e79014. doi: 10.1371/journal.pone.0079014
37. Jouali F, El Ansari FZ, Marchoudi N, Barakat A, Zmaimita H, Samlali H, et al. EGFR, BRCA1, BRCA2 and TP53 genetic profile in Moroccan triple negative breast cancer cases. Int J Mol Epidemiol Genet. (2020) 11:16–25.
38. Martínez-Sáez O, Chic N, Pascual T, Adamo B, Vidal M, González-Farré B, et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. (2020) 22:45. doi: 10.1186/s13058-020-01284-9
39. Jouali F, Marchoudi N, Talbi S, Bilal B, El Khasmi M, Rhaissi H, et al. Detection of PIK3/AKT pathway in Moroccan population with triple negative breast cancer. BMC Cancer (2018) 18:900. doi: 10.1186/s12885-018-4811-x
40. Yang J, Ren Y, Wang L, Li B, Chen Y, Zhao W, et al. PTEN mutation spectrum in breast cancers and breast hyperplasia. J Cancer Res Clin Oncol. (2010) 136:1303–11. doi: 10.1007/s00432-010-0781-3
41. Robichaux JP, Elamin YY, Vijayan RSK, Nilsson MB, Hu L, He J, et al. Pan-cancer landscape and analysis of ERBB2 mutations identifies poziotinib as a clinically active inhibitor and enhancer of T-DM1 activity. Cancer Cell. (2019) 36:444–57. doi: 10.1016/j.ccell.2019.09.001
42. Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. (2003) 95:1482–5. doi: 10.1093/jnci/djg050
43. Wu H, Wang W, Du J, Li H, Wang H, Huang L, et al. The distinct clinicopathological and prognostic implications of PIK3CA mutations in breast cancer patients from Central China. Cancer Manage Res. (2019) 11:1473–92. doi: 10.2147/CMAR.S195351
44. Severson TM, Peeters J, Majewski I, Michaut M, Bosma A, Schouten PC, et al. BRCA1-like signature in triple negative breast cancer: Molecular and clinical characterization reveals subgroups with therapeutic potential. Mol Oncol. (2015) 9:1528–38. doi: 10.1016/j.molonc.2015.04.011
45. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. (2016) 13:674–90. doi: 10.1038/nrclinonc.2016.66
46. Tutt ANJ, Garber JE, Kaufman B, Viale G, Fumagalli D, Rastogi P, et al. OlympiA clinical trial steering committee and investigators. Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N Engl J Med. (2021) 384:2394–405. doi: 10.1056/NEJMoa2105215
47. Brzostek-Racine S, Gordon C, Van Scoy S, Reich NC. The DNA damage response induces IFN. J Immunol. (2011) 187:5336–45. doi: 10.4049/jimmunol.1100040
48. Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. (2016) 17:1142–9. doi: 10.1038/ni.3558
49. Rivera Vargas T, Benoit-Lizon I, Apetoh L. Rationale for stimulator of interferon genes-targeted cancer immunotherapy. Eur J Cancer. (2017) 75:86–97. doi: 10.1016/j.ejca.2016.12.028
50. Permata TBM, Hagiwara Y, Sato H, Yasuhara T, Oike T, Gondhowiardjo S, et al. Base excision repair regulates PD-L1 expression in cancer cells. Oncogene. (2019) 38:4452–66. doi: 10.1038/s41388-019-0733-6
51. Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. (2017) 8:1751. doi: 10.1038/s41467-017-01883-9
52. Vendetti FP, Karukonda P, Clump DA, Teo T, Lalonde R, Nugent K, et al. ATR kinase inhibitor AZD6738 potentiates CD8+ T cell-dependent antitumor activity following radiation. J Clin Invest. (2018) 128:3926–40. doi: 10.1172/JCI96519
53. Xu Y, Lin Y, Wang Y, Zhou L, Xu S, Wu Y, et al. Association of Neo-Family History Score with pathological complete response, safety, and survival outcomes in patients with breast cancer receiving neoadjuvant platinum-based chemotherapy: An exploratory analysis of two prospective trials. EClinicalMedicine. (2021) 38:101031. doi: 10.1016/j.eclinm.2021.101031
Keywords: triple negative breast cancer, homologous recombination repair genes, mutation spectrum, clinicopathological factors, immune infiltration, prognosis
Citation: Wang Z, Li A, Lu Y, Han M, Ruan M, Wang C, Zhang X, Zhu C, Shen K, Dong L and Chen X (2024) Association of tumor immune infiltration and prognosis with homologous recombination repair genes mutations in early triple-negative breast cancer. Front. Immunol. 15:1407837. doi: 10.3389/fimmu.2024.1407837
Received: 27 March 2024; Accepted: 21 June 2024;
Published: 04 July 2024.
Edited by:
Lisheng Wang, University of Ottawa, CanadaCopyright © 2024 Wang, Li, Lu, Han, Ruan, Wang, Zhang, Zhu, Shen, Dong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaosong Chen, Y2hlbnhpYW9zb25nMDE1NkBob3RtYWlsLmNvbQ==; Lei Dong, ZGwxMTk2OEByamguY29tLmNu; Kunwei Shen, a3dzaGVuQG1lZG1haWwuY29tLmNu; Chaofu Wang, d2NmMTE5NTZAcmpoLmNvbS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.