
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Immunol. , 29 May 2024
Sec. Comparative Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1401626
Zinc finger Asp-His-His-Cys motif-containing (zDHHC) proteins, known for their palmitoyltransferase (PAT) activity, play crucial roles in diverse cellular processes, including immune regulation. However, their non-palmitoyltransferase immunomodulatory functions and involvement in teleost immune responses remain underexplored. In this study, we systematically characterized the zDHHC family in the large yellow croaker (Larimichthys crocea), identifying 22 members. Phylogenetic analysis unveiled that each of the 22 LczDHHCs formed distinct clusters with their orthologues from other teleost species. Furthermore, all LczDHHCs exhibited a highly conserved DHHC domain, as confirmed by tertiary structure prediction. Notably, LczDHHC23 exhibited the most pronounced upregulation following Pseudomonas plecoglossicida (P. plecoglossicida) infection of macrophage/monocyte cells (MO/MΦ). Silencing LczDHHC23 led to heightened pro-inflammatory cytokine expression and diminished anti-inflammatory cytokine levels in MO/MΦ during infection, indicating its anti-inflammatory role. Functionally, LczDHHC23 facilitated M2-type macrophage polarization, as evidenced by a significant skewing of MO/MΦ towards the pro-inflammatory M1 phenotype upon LczDHHC23 knockdown, along with the inhibition of MO/MΦ necroptosis induced by P. plecoglossicida infection. These findings highlight the non-PAT immunomodulatory function of LczDHHC23 in teleost immune regulation, broadening our understanding of zDHHC proteins in host-pathogen interactions, suggesting LczDHHC23 as a potential therapeutic target for immune modulation in aquatic species.
The zinc finger Asp-His-His-Cys motif-containing (zDHHC) proteins constitute a family of palmitoyltransferases (PATs) primarily tasked with catalyzing the addition of palmitate moieties onto protein substrates, a process known as S-palmitoylation (1). To date, 24 zDHHC members have been identified in mammalian species, with 23 in humans (zDHHC1–9, 11–24) and 24 in mice (zDHHC1–9, 11–25) (1). All of these members are characterized by the presence of a 50-residue long DHHC (Asp-His-His-Cys) domain, which serves as the active center for PATs (2). S-palmitoylation, the reversible attachment of palmitate to cysteine residues via thioester bonds, plays crucial roles in regulating protein membrane localization, trafficking, stability, and function (3). As integral players in protein S-palmitoylation, zDHHCs hence contribute significantly to the modulation of diverse cellular processes, including signal transduction, membrane dynamics, and protein-protein interactions (4). Recently, research has focused on the role of zDHHCs in modulating immune responses. Molecules involved in innate immunity, such as the pattern recognition receptor TLR2 (Toll-like receptor 2), the cytoplasmic DNA receptor cGAS (cyclic GMP-AMP synthase), the cytoplasmic bacterial peptidoglycan receptor NOD1 and NOD2 (nucleotide-binding oligomerization domain-containing protein 1/2), the type I interferon receptor IFNAR (interferon alpha/beta receptor), the transcription factor STAT3 (signal transducer and activator of transcription 3), and the cytokine TNF-α, have all been reported to undergo palmitoylation by various zDHHCs (5–9). Moreover, zDHHCs participate in the palmitoylation of adaptive immune molecules, such as CD4, CD8, and LAT (the inker for activation of T cells), which are associated with T cell function (10–12). Additionally, molecules involved in B cell function, like CD81, LAB (the linker for activation of B cells), were also palmitoylated by zDHHCs (13, 14). Despite primarily functioning as PAT for immune-related molecules, the non-PAT roles of zDHHCs in immunity are largely unexplored. Only zDHHC1 and zDHHC11 has been identified as a positive regulator of DNA virus-triggered signaling via STING interaction, independent of their PAT activity (15, 16). Therefore, investigating the non-PAT functions of the zDHHC family is a highly valuable area for further research.
To date, our understanding of the composition and function of the zDHHC family in lower species, including teleosts, remains limited. Systematic studies are needed to determine the number of zDHHCs present in teleosts and their respective functions, as well as to investigate the evolutionary clues of this family. Only five zDHHCs (zDHHC1, 13, 15, 16, and 17) have been reported and characterized for their functions in teleosts, with studies focusing mainly on zebrafish (zDHHC13, 15, 16, and 17) (17–20). Independently of their PAT activity, zebrafish zDHHC13 regulates embryonic differentiation through interaction with Smad6, zDHHC15b is implicated in the differentiation of dopamine (DA) neurons, and the anchor protein domain of zDHHC17 influences neuronal axon growth by regulating the formation of the TrkA tubulin complex (17, 19, 20). Only zDHHC16 promotes telencephalic neural stem cell proliferation by modulating the FGF/ERK signaling pathway through its PAT activity (18). Research on the immune regulatory roles of zDHHCs in teleosts is further limited. Only zDHHC1, identified in grass carp (Ctenopharyngodon idella) and Chinese perch (Siniperca chuatsi), has been reported to possess antiviral functions (21, 22).
In the current study, we systematically screened the large yellow croaker genome for members of the zDHHC family, identifying 22 zDHHC proteins (LczDHHC1–9, 11–18, 20–24). Subsequently, we cloned all 22 zDHHCs and conducted analyses on their evolutionary relationships, protein structures, and importantly, their potential association with Pseudomonas plecoglossicida (P. plecoglossicida) infection, a significant pathogenic bacterium affecting large yellow croaker (23). Among them, LczDHHC23 exhibited the highest upregulation in P. plecoglossicida infected large yellow croaker macrophage/monocyte cells (MO/MΦ). However, the immune regulatory functions of zDHHC23 have not been previously explored, even in mammals. In mammals, zDHHC23 primarily palmitoylates molecules implicated in mTOR signaling and tumorigenesis (24, 25). Therefore, investigating the mechanisms underlying the notable upregulation of LczDHHC23 expression post-infection holds significance. By silencing LczDHHC23 expression, we observed an upregulation of pro-inflammatory cytokines and a downregulation of anti-inflammatory cytokines during P. plecoglossicida infection, indicating the anti-inflammatory role of LczDHHC23. Further investigation revealed that LczDHHC23 predominantly promotes M2-type macrophage polarization while inhibiting M1-type polarization, as also evidenced by enhanced phagocytic activity in LczDHHC23-knockdown MO/MΦ. Additionally, LczDHHC23 was found to facilitate the necrosis of P. plecoglossicida-infected MO/MΦ, as evidenced by delayed and reduced phosphorylation of necrosis markers receptor−interacting serine/threonine kinase (RIP)1, RIP3, and mixed lineage kinase domain-like (MLKL) upon compromised LczDHHC23 expression. To our knowledge, our investigation represents the first study to elucidate the role of zDHHC23 in immune cells during infection, potentially laying the groundwork for a deeper understanding of zDHHC23’s function in modulating anti-pathogen immune responses.
Healthy large yellow croakers (100–120 g) were purchased from Xiangshan county farm (NingBo, China). All fish were temporarily kept in a recirculating seawater system maintained at 18–20°C for at least two weeks. All animal experiments were approved by the Institutional Animal Care and Use Committee of Ningbo University and conducted in accordance with the Guide for the Care and Use of Laboratory Animals issued by the National Institutes of Health. P. plecoglossicida strain (NZBD9) was cultivated at 18°C in Luria–Bertani (LB) broth with shaking and collected until reaching the logarithmic growth stage. The bacteria were washed with sterile phosphate-buffered solution (PBS), and then diluted to a final 5 ×104 colony forming units (CFU)/100g fish in 100 μL PBS for the in vivo challenge test. The large yellow croakers were intraperitoneally injected with P. plecoglossicida, while the control group received an equivalent volume of PBS. The tissue samples in each group (gill, head kidney, intestine, liver, and spleen) were collected at 0, 6, 12, 24, 48 and 72 hours after infection (hpi), and promptly snap-frozen in liquid nitrogen and stored at -80°C for subsequent Real-time quantitative polymerase chain reaction (RT-qPCR) analysis. The primers used are listed in Supplementary Table 1.
The 22 LczDHHC genes were cloned via reverse transcription PCR (RT-PCR), using sequences obtained from the NCBI database. Subsequently, the PCR products were sequenced for validation. The primers used are listed in Supplementary Table 1. Phylogenetic analysis of the whole family was conducted using the neighbor-joining method, supported by 1000 bootstrap repetitions in MEGA 7.0 software. The phylogenetic tree was modified using iTOL programs. Transmembrane (TM) domain analysis of LczDHHC23 was predicted by the TMHMM Server v. 2.0 program (http://www.cbs.dtu.dk/services/TMHMM-2.0/). Multiple sequence alignment was generated using ClustalW (http://clustalw.ddbj.nig.ac.jp/). The domain information of LczDHHC23 was predicted using SMART (http://smart.embl-heidelberg.de/), and the three-dimensional structures of all LczDHHCs were predicted using SWISS-MODEL (https://swissmodel.expasy.org/), followed by visualization of the PDB files using PyMOL software version 3.1. Sequences used in this study are listed in Supplementary Table 2.
Large yellow croaker head-kidney-derived MO/MΦ were isolated as previously described (26, 27). Leukocyte-enriched fractions were obtained by applying dissociated head-kidney to a Ficoll density gradient (1.077 g/mL; #17144002 GE Healthcare, Chicago, IL, USA). The cells were then seeded in 6 well plates at a density of 2×107/mL and cultured overnight at 26°C under 5% CO2. After washing away the nonadherent cells, attached cells were incubated with complete DMEM medium (10% fetal bovine serum, 100 U/mL penicillin, and 100μg/mL streptomycin) and cultured under the same conditions. Live P. plecoglossicida diluted in PBS at a multiplicity of infection (MOI) of 2 or PBS alone were added into cell culture medium. Cells were collected at 4, 8, 12, and 24 hpi for RNA extraction, and RT-qPCR was conducted as described. The primers used are listed in Supplementary Table 1.
Total RNA of both large yellow croaker tissues and MO/MΦ were isolated using RNAiso (#9108/9109, TaKaRa, Dalian, China), treated with DNase I (#2270A, TaKaRa), and reversed transcribed into first-strand cDNA using AMV reverse transcriptase (#2621, TaKaRa) according to manufacturer protocol. RT-qPCR was conducted on an ABI StepOne real-time PCR system (Applied Biosystems, Foster City, CA, USA) using SYBR premix Ex Taq II (#RR82WR, TaKaRa). The thermal cycling conditions were as follows: initial denaturation at 95°C for 10 s, followed by 40 cycles of amplification (95°C for 50 s and 60°C for 20 s), and final melting curve analysis (95°C for 60 s, 55°C for 30 s, and 95°C for 30 s). Relative gene expression was calculated using the 2−ΔΔCT method and the data were normalized against Lc18S rRNA. The primers used are listed in Supplementary Table 1. Each PCR trial was performed in triplicate and repeated at least three times.
The full-length open reading frame (ORF) of LczDHHC23 was amplified by PCR using PrimeSTAR GXL DNA polymerase (#R050A, Takara). The PCR product was ligated with pcDNA-HA vector to generate the HA-tagged LczDHHC23 plasmid using the pEASY®-Basic Seamless Cloning and Assembly Kit (#CU201–02, TransGen Biotech). The plasmid was subsequently transformed into competent Escherichia coli (E. coli) cells. The bacterial solution was spread onto LB nutrient agar plates, and single colonies were selected from overnight cultures for sequencing.
Hela cells were seeded at a density of 1×105/mL and transfected with HA-tagged LczDHHC23 plasmids. After 36 h of transfection, the cells were fixed with 4% paraformaldehyde in PBS (pH=7.4) and permeabilized with 0.5% saponin. After blocking, the cells were incubated with appropriate primary and secondary antibodies. Confocal images were obtained using Zeiss LSM 880 confocal microscope (Carl Zeiss AG, Oberkochen, Germany) and analyzed using the ZEN Blue software.
LczDHHC23-specific small interfering RNA (RiboBio, Guangzhou, China) was transfected into large yellow croaker MO/MΦ using Lipofectamine™ RNAiMAX Transfection Reagent (#13778150, Invitrogen/Life Technologies, Carlsbad, CA, USA) according to the previous studies for 24 and 48 h to evaluate the knock down efficiency (21, 28–30). The scrambled siRNA was used as the control. To evaluate the role of LczDHHC23 on MO/MΦ function following P. plecoglossicida stimulation, the isolated MO/MФ were transfected with 30 pmol LczDHHC23 siRNA, or scrambled siRNA for 24h before infected with P.plecoglossicida.
Isolated MO/MΦ were transfected with LczDHHC23 siRNA, or corresponding controls, for 24 h before being infected with live P. plecoglossicida at an MOI of 4 (30, 31). The phagocytosis of bacteria was proceeded for 30 minutes at 26°C under 5% CO2. The remaining bacteria attached to cell surface were killed using gentamicin (50 μg/mL), and then washed with sterile PBS. Each set of interfered MO/MФ were divided into two groups. One group (the uptake group) was lysed immediately with 1% Triton X-100 solution and plated onto solid LB agar medium to assess bacterial uptake. The other group (the kill group) was incubated for an additional 1.5 h before being lysed and plated on LB agar medium. After incubation at 28°C for 24 h, the CFUs of the plates were calculated. Bacterial survival was determined by dividing the CFUs in the kill group by those in the uptake group. Three independent experiments were performed.
To investigate the impact of LczDHHC23 on MO/MФ polarization, LPS-induced M1-type and cAMP-induced M2-type MO/MФ were prepared according to previous reported (32). MO/MФ cells were transfected with LczDHHC23 siRNA, or corresponding controls for 24 h before treatment with LPS (50 μg/mL; #L4391, Sigma Aldrich) or a cAMP analog (dibutyryl cAMP; 0.5 mg/mL, #28745-M, Sigma-Aldrich) for 18 h. Expression levels of the pro-inflammatory cytokines (IL-1β and IL-6) and the anti-inflammatory cytokines (IL-10 and TGF-β) were determined. Moreover, the markers of M1-type MO/MФ (C-X-C motif chemokine ligand 9 (CXCL9) and induced nitric oxide synthase (iNOS)) and the markers of M2-type MO/MФ (secreted phosphoprotein 1 (SPP1) and arginase) were also evaluated (33). In addition, the iNOS (M1-type) and arginase activities (M2-type) were assessed. iNOS activity was measured using a nitric oxide synthase assay kit (fluorescence probe method; #S0024, Beyotime, Shanghai, China) following the manufacturer’s instructions. The relative iNOS activity of each group was expressed as fold change relative to the value of the control group. Arginase activity was measured using an arginase activity assay kit (#MAK112, Sigma-Aldrich) following the manufacturer’s protocol. Absorbance was read at 430 nm, and arginase activity (U/L) was calculated according to comparison with urea-standard data.
Large yellow croaker MO/MΦ were transfected with LczDHHC23 siRNA, or corresponding controls, for 24 h prior to infection with P. plecoglossicida at an MOI of 10 (30, 31). Cell samples were collected at 1, 2, 4 and 8 hpi. The collected cells were washed and then labeled with Annexin V-FITC and propidium iodide (PI) for 15 minutes using a FITC Annexin V apoptosis detection kit I (#556547, BD Pharmingen, San Diego, CA, USA). Apoptosis was evaluated by flow cytometry using the MACSQuant Analyzer 10 (Miltenyi Biotec) within 15 min of staining, and data were analyzed using MACSQuant analysis software (Miltenyi Biotec). Three independent experiments were performed.
Large yellow croaker MO/MΦ cells were seeded in 6 well plates at a density of 2×107/mL the day before the experiment. Cells were washed twice with PBS and infected with P.plecoglossicida at an MOI of 10. Subsequently, cells were collected at 0, 1, 2, 4 and 8 hpi and lyzed by lysis buffer (20 mM Tris-HCl [pH=8.0], 2 mM EDTA, 120 mM NaCl, 1% NP-40) containing phosphatase inhibitor (#A32963, Thermo fisher). The soluble protein concentration was measured using the Bradford method. Proteins were separated on 10% or 15% SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes (#IPVH00010, EMD Millipore). The membranes were blocked with 5% non-fat milk for 1 h at room temperature, then incubated overnight at 4°C with apoptosis/necroptosis antibodies (#92570, Cell Signaling Technology), followed by incubation with appropriate HRP-conjugated secondary antibodies for 1 h at room temperature. The blots were subsequently visualized using a chemiluminescent detection system with ECL western blotting detection reagents (#32106, Thermo Fisher Scientific).
All data are presented as the means ± SEM. Statistical analysis was performed using one-way analysis of variance (ANOVA) with the Prism 8.02 software (GraphPad Software, San Diego, CA, USA). The p values *p < 0.05 were considered statistically significant.
Twenty-two LczDHHCs have been cloned from the large yellow croaker (LczDHHC1–9, 11–18, 20–24). Unlike observed in mammals, the zDHHC family in the large yellow croaker does not include zDHHC19 and zDHHC25 (1). Phylogenetic analysis revealed three major clades grouping the 22 subfamilies, with each of these 22 LczDHHCs forming clusters with their respective orthologues from other teleost species (Figure 1A). The tertiary structures of the LczDHHCs were predicted using SWISS-MODEL, revealing a highly conserved DHHC domain and 4 to 7 transmembrane (TM) domains in all members (Figures 1B–D; Supplementary Figure 1). Additionally, LczDHHC6 contains an SH3 domain, while LczDHHC13 and LczDHHC17 feature an ankyrin repeat domain. The TM helices of these LczDHHCs form pocket-like structures, and the DHHC domain contains 2 or more β-hairpin structures, except for LczDHHC13. This arrangement is reminiscent of the crystal structure of zDHHC proteins reported in humans and zebrafish (34). Typically, the DHHC domain of most LczDHHC proteins (excluding LczDHHC13, 17, and 23) is situated between the second and third TM domains (Figures 1B–D; Supplementary Figure 1).
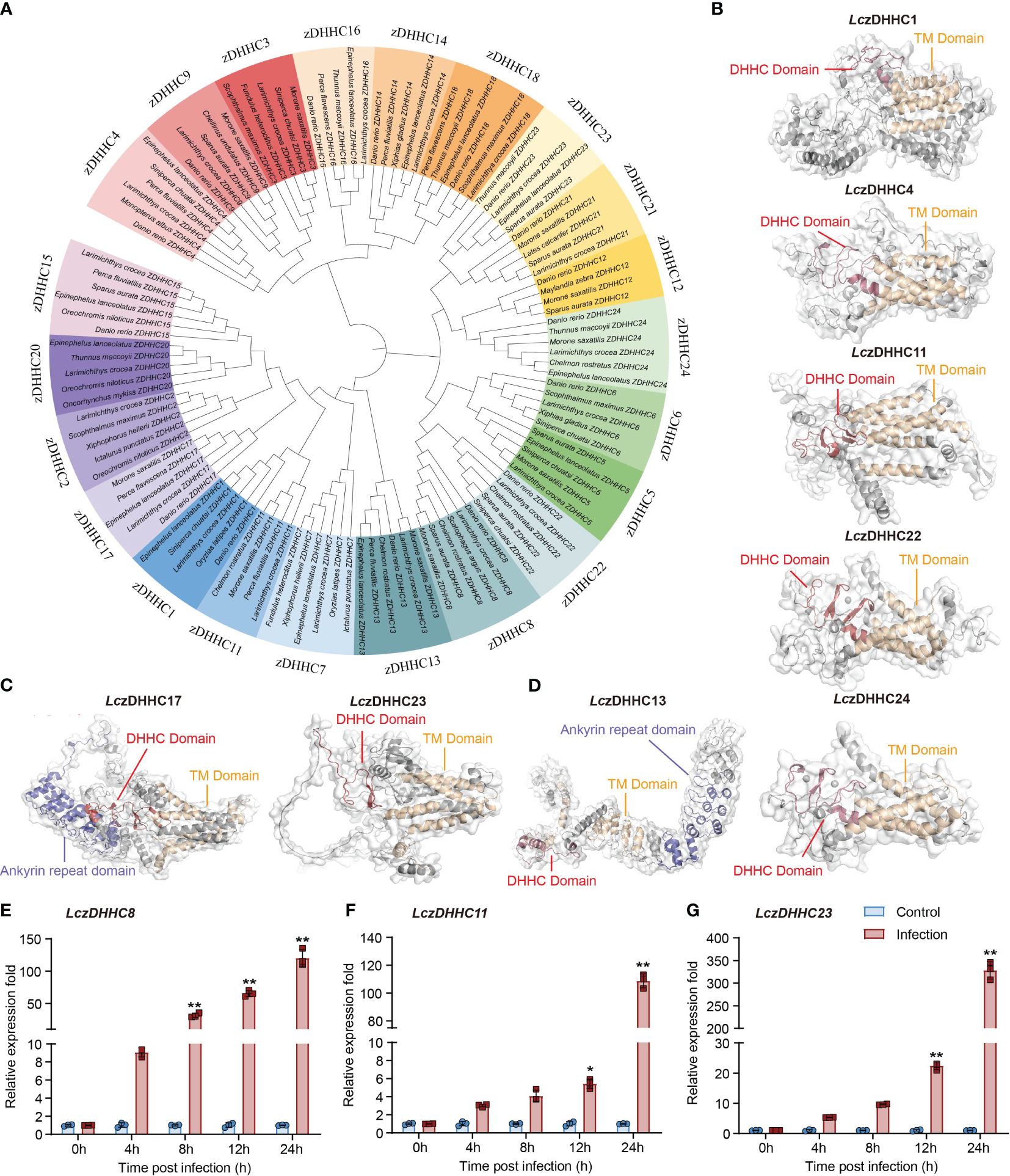
Figure 1 Bioinformatic analysis and expression of LczDHHC family in response to P. plecoglossicida infection in head kidney-derived MO/MΦ. (A) Phylogenetic tree showing the divergence of zDHHC proteins among teleost species, generated using the neighbor-joining method in MEGA 7.0 with 1000 bootstrap replications and modified using iTOL. (B–D) Predicted tertiary structures of LczDHHC family proteins with five (B), six (C), and seven (D) transmembrane (TM) domains. Structures were modeled using SWISS-MODEL and visualized using PyMOL software version 3.1. LczDHHC 1, 4, 11, 22, and 24 have five TM domains (B), LczDHHC17 and LczDHHC 23 have six TM domains (C), and LczDHHC13 has seven TM domains (D). Additionally, LczDHHC13 and 17 feature an ankyrin repeat domain. TM domains are depicted in yellow, DHHC domains in red, and ankyrin repeat domains in deep blue. (E–G) Quantitative analysis of significantly upregulated expression of LczDHHCs in response to P. plecoglossicida infection, including LczDHHC8 (E), LczDHHC11 (F), and LczDHHC23 (G). MO/MΦ from large yellow croakers were infected with P. plecoglossicida at an MOI of 2, with PBS-treated cells as controls. Samples were collected at 0, 4, 8, 12, and 24 hpi. Expression levels of each LczDHHC mRNA were normalized to Lc18S rRNA and then to the 0 h PBS control using the 2-ΔΔCT method. Data represent the means ± SEM of three replicates. *p < 0.05 and **p < 0.01.
We screened the 22 LczDHHCs for changes in expression during P. plecoglossicida infection in large yellow croaker head kidney-derived MO/MΦ. Among them, LczDHHC8, LczDHHC11, and LczDHHC23 showed significant induction, with LczDHHC23 exhibiting the highest induction, reaching over 300-fold at 24 hpi (Figures 1E–G). The expression changes of other LczDHHCs were minor, with induction or suppression within 20-fold (Supplementary Figures 2, 3). Consequently, we focused on studying the role of LczDHHC23 in regulating the immune response induced by P. plecoglossicida infection in the subsequent investigation.
The LczDHHC23 sequence spans 2355 nucleotides (nt), with an open reading frame (ORF) of 1178 base pairs (bp), encoding a protein of 392 amino acids (aa). A phylogenetic tree was constructed based on the amino acid sequences of zDHHC23 proteins from teleosts and other species. The analysis revealed that teleost zDHHC23s formed a distinct cluster, with LczDHHC23 showing the closest relationship to that of Nibea albiflora (Supplementary Figure 4A). Multiple sequence alignment demonstrated a high degree of conservation among zDHHC23 orthologs, comprising six transmembrane (TM) domains and a highly conserved DHHC domain (Supplementary Figure 4B). We further compared the tertiary structure of LczDHHC23 with that of human orthologue (HszDHHC23), and found that they both possess six TM helices, forming a pocket-like structure similar to that of most members of the zDHHC family. Additionally, they feature a highly conserved DHHC domain positioned between the fourth and fifth TM domains, contrasting with most zDHHC family proteins where it situated between the second and third TM domains. As depicted in Figure 2A, the DHHC domain of LczDHHC23 contains two β-hairpin structures, whereas the DHHC domain of HszDHHC23 has more. This suggests that the DHHC domain of HszDHHC23 has a more complex structure compared to that of the LczDHHC23.
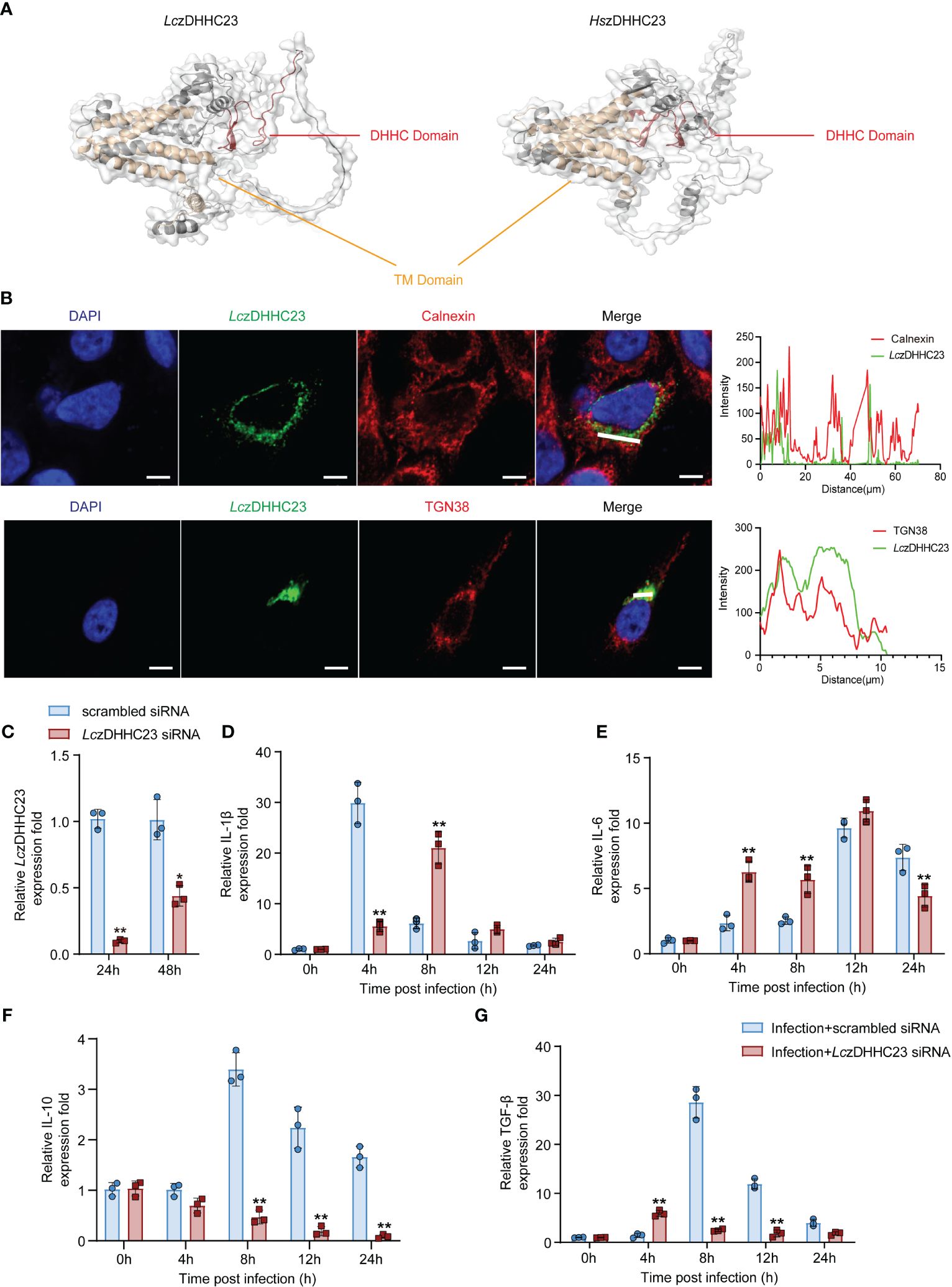
Figure 2 Molecular characterization of LczDHHC23 and its impact on cytokine expression of P. plecoglossicida-infected MO/MΦ. (A) Comparison of the tertiary structures of LczDHHC23 and Homo sapiens zDHHC23 (HszDHHC23). The tertiary structures were modeled using SWISS-MODEL and visualized using PyMOL software version 3.1. TM domains are represented in yellow, and DHHC domain is depicted in red. (B) Immunofluorescence images showing the subcellular localization of LczDHHC23 with the endoplasmic reticulum marker Calnexin and Golgi apparatus marker TGN38. HeLa cells were seeded at a density of 1 × 105/mL and transfected with HA-tagged LczDHHC23 plasmid. After 36 h of transfection, the cells were fixed and examined using a confocal microscope. The white line indicates the color pickup line, with the fluorescence intensity curve shown in green and red on the right side, analyzed with ZEN software. Scale bars, 5 μm. Results are representative of three independent experiments. (C–G) Impact of LczDHHC23 on the cytokine expression in P. plecoglossicida infected MO/MΦ. (C) RT-qPCR assesses the interference efficiency of LczDHHC23 siRNA. Transcript levels of LczDHHC23 were normalized against 18S rRNA and then to the 24 h scrambled siRNA control group using the 2-ΔΔCT method. *p < 0.05 and **p < 0.01. (D–G) RT-qPCR analysis evaluates the effects of LczDHHC23 interference on cytokine mRNA expression in P. plecoglossicida-infected MO/MΦ. Large yellow croaker MO/MΦ were transfected with LczDHHC23 siRNA for 24 h before P. plecoglossicida infection at an MOI of 2. Samples were collected at 0, 4, 8, 12, and 24 hpi. Expressions levels of IL-1β, IL-6, IL-10, and TGF-β were detected using RT-qPCR. mRNA expression was normalized to that of 18S rRNA and then to the respective 0 h control using the 2-ΔΔCT method. Data represent the means ± SEM of three replicates. *p < 0.05 and **p < 0.01.
We further evaluated the molecular characteristics of LczDHHC23. Previous studies have indicated that members of the zDHHC family primarily localize to the endoplasmic reticulum, the Golgi apparatus, or both (35). Our subcellular localization analysis via immunofluorescence revealed that LczDHHC23 predominantly co-localizes with the Golgi apparatus marker TGN38, rather than the endoplasmic reticulum marker Calnexin (Figure 2B), indicating a difference from observations in mammals (36). The tissue expression pattern of LczDHHC23 in major immune tissues (intestine, head kidney, liver, gill, and spleen) of healthy large yellow croaker revealed that the spleen exhibited the highest levels of LczDHHC23 transcripts, followed by the liver and head kidney (Supplementary Figure 5A). Following P. plecoglossicida injection, the expression of LczDHHC23 significantly increased in all examined tissues except for the liver, which showed a decrease at 12 hpi. The highest expression levels of LczDHHC23 in the head kidney and gill were observed at 12 hpi, followed by a decrease. In the intestine, LczDHHC23 expression initially decreased significantly and then sharply increased. LczDHHC23 levels in the spleen increased at 12 hours and remained stable until 72 hpi (Supplementary Figures 5B–F).
To assess the impact of LczDHHC23 on immune responses following P. plecoglossicida infection, we initially synthesized LczDHHC23 siRNA (5’-CACCACTGCATCTGGATAA-3’) to downregulate its expression in large yellow croaker MO/MΦ. The expression of LczDHHC23 was notably reduced after 24 and 48 hours of interference compared to the control group, reaching 9.8% and 43.8%, respectively (Figure 2C). After confirming the efficacy of LczDHHC23 siRNA knockdown, we investigated the impact of LczDHHC23 expression interference on the expression of pro-inflammatory (IL-1β and IL-6) and anti-inflammatory (IL-10 and TGF-β) cytokines following P. plecoglossicida infection. The results revealed that inhibiting LczDHHC23 significantly upregulated the expression of pro-inflammatory cytokines (IL-1β and IL-6; Figures 2D, E) and downregulated the expression of anti-inflammatory cytokines (IL-10 and TGF-β; Figures 2F, G), compared to the control group. These findings suggest that LczDHHC23 may indeed play an anti-inflammatory role in MO/MΦ during P. plecoglossicida infection in large yellow croaker.
We further evaluated whether LczDHHC23 exert its anti-inflammatory roles by affecting the polarization of large yellow croaker MO/MΦ. Fish MO/MФ can differentiate into two main types: the M1-type, characterized by the expression of pro-inflammatory cytokines and the production of reactive oxygen species (ROS) and NO, and the M2-type, characterized by the expression of anti-inflammatory cytokines and increased arginase activity (33, 37). We induced M1-type and M2-type polarization of isolated large yellow croaker MO/MΦ using lipopolysaccharide (LPS) and cyclic adenosine monophosphate (cAMP), respectively, as previously reported (32). As shown in Figure 3, knockdown of LczDHHC23 further promoted the induced expression of pro-inflammatory cytokines (IL-1β and IL-6) and M1-type markers (CXCL9 and iNOS), as well as the iNOS activity in M1-type MO/MΦ (Figures 3A, B, I, J, M). Conversely, LczDHHC23 silencing further decreased the already low expression of anti-inflammatory cytokines (IL-10 and TGF-β) in M1-type MO/MΦ, indicating an inhibiting role of LczDHHC23 on the M1-type function (Figures 3E, F). Moreover, LczDHHC23 silencing significantly upregulated the expression of IL-1β and IL-6 in cAMP-induced M2-type MO/MΦ, while downregulating the expression of IL-10, TGF-β, as well as the M2 marker SSP1, and the arginase activity (Figures 3C, D, G, H, K, L, N). These findings suggest that LczDHHC23 may promote M2-type MO/MΦ polarization to exert its anti-inflammatory function. In support of this, we observed a significant augmentation in bacterial killing activity in LczDHHC23-silenced MO/MΦ. Specifically, the bacterial killing activity decreased from 22.84% to 6.07% in LczDHHC23-deficient cells compared to control cells (Supplementary Figure 6).
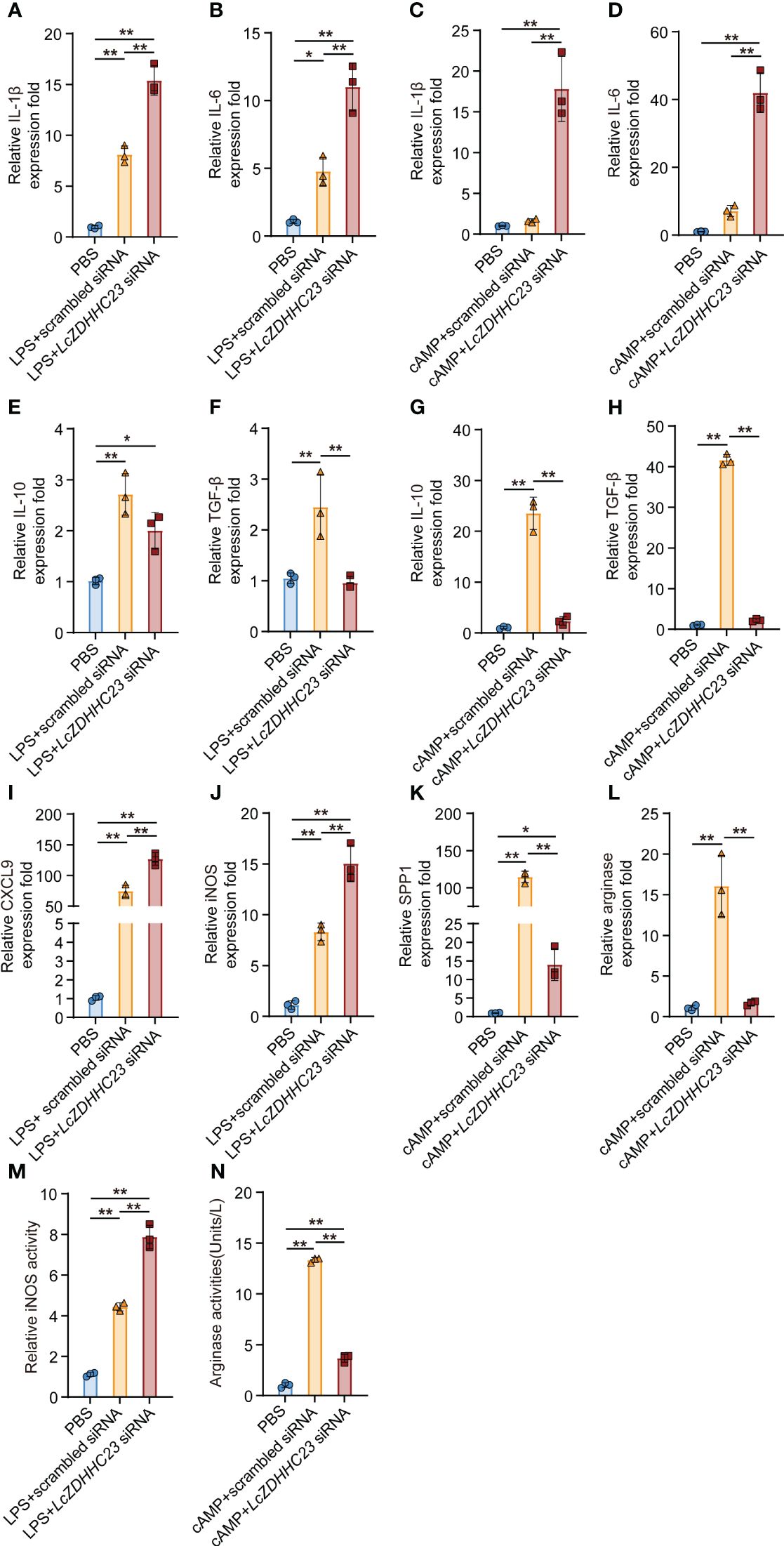
Figure 3 Effect of LczDHHC23 deficiency on the polarization of large yellow croaker MO/MФ. Large yellow croaker MO/MΦ were transfected with LczDHHC23 siRNA for 24 h before stimulation with LPS or cAMP for 18 h. (A, B, E, F) Knockdown of LczDHHC23 promotes M1-type polarization of MO/MΦ. Silencing LczDHHC23 significantly induced the expression of pro-inflammatory cytokines IL-1β (A) and IL-6 (B), while decreased the expression of anti-inflammatory cytokines IL-10 (E) and TGF-β (F) following LPS stimulation. (I, J, M) Expression of M1-type MO/MФ markers CXCL9 (E), iNOS (F), as well as the iNOS activity (G) were increased with LczDHHC23 knockdown. (C, D, G, H) LczDHHC23 knockdown inhibits the M2-type polarization of large yellow croaker MO/MΦ. Knockdown of LczDHHC23 significantly increased the expression of pro-inflammatory cytokines IL-1β (C) and IL-6 (D), while decreasing the expression of anti-inflammatory cytokines IL-10 (G) and TGF-β (H) following cAMP stimulation. (K, L, N) Expression of M2-type MO/MФ markers SPP1 (K), arginase (L), as well as the arginase activity (N) were also decreased with LczDHHC23 inhibition. Data represent the means ± SEM of three replicates. *p < 0.05 and **p < 0.01.
Given LczDHHC23’s role in promoting M2-type polarization and anti-inflammatory responses in large yellow croaker MO/MΦ, and considering the impact of inflammatory responses on cell states during pathogen infection (38), we further assessed the involvement of LczDHHC23 in the apoptosis/necroptosis of P. plecoglossicida-infected large yellow croaker MO/MΦ. The findings indicate that P. plecoglossicida infection predominantly triggers necroptosis in MO/MΦ, with the proportion of necroptotic cells gradually increasing during infection, reaching 58.43 ± 8.54% at 8 hpi. However, upon silencing of LczDHHC23, the induction of MO/MΦ necroptosis by P. plecoglossicida infection was significantly reduced, with only 26.03 ± 1.25% of cells undergoing necroptosis at 8 hpi (Figure 4A). We further validated the impact of LczDHHC23 on P. plecoglossicida infection-induced MO/MΦ necroptosis by examining necroptosis markers, including phosphorylated RIP3 (p-RIP3), RIP1 (p-RIP1), and MLKL (p-MLKL), as well as assessing Caspase-3/8 activity inhibition (39). Results demonstrated a significant increase in the phosphorylation levels of these proteins at 2 hpi, with further elevation observed as the infection progressed. Concurrently, cleaved Caspase-3 and Caspase-8 remained at very low levels, with minimal observation during the late stage of infection when signs of apoptosis were also noted (Figures 4A, B). Upon silencing of LczDHHC23, the heightened phosphorylation of RIP1, RIP3, and MLKL induced by P. plecoglossicida infection was mitigated or delayed (Figure 4B), indicating the involvement of LczDHHC23 in promoting immune cell necroptosis during pathogen infection.
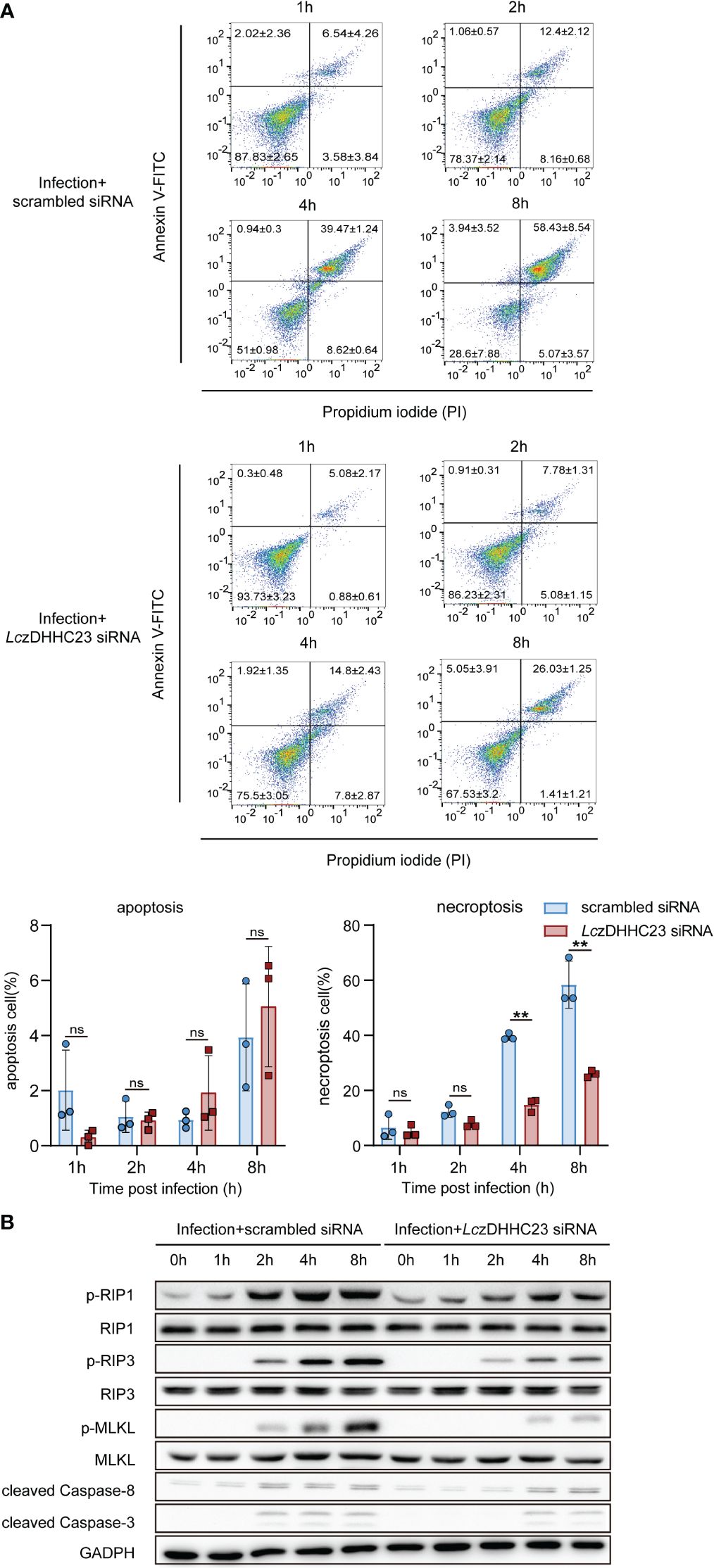
Figure 4 Impact of LczDHHC23 deficiency on P. plecoglossicida-induced necroptosis of large yellow croaker MO/MФ. (A) MO/MΦ were transfected with LczDHHC23 siRNA for 24 h prior to P. plecoglossicida infection at an MOI of 10. After 1, 2, 4 and 8 h of infection, cells were harvested and stained with Annexin V-FITC and PI for flow cytometry analysis. Live cells, are negative for both PI and Annexin V-FITC (lower left quadrant); dead cells are positive for PI but negative for Annexin V-FITC (lower right quadrant); necroptotic cells are positive for both PI and Annexin V-FITC (upper right quadrant), while early apoptotic cells are positive for Annexin V-FITC but negative for PI (upper left quadrant). Histograms represent the percentage of apoptosis and necroptosis. Data represent the means ± SEM of three replicates. **p < 0.01, ns represent not significant. (B) Cells collected from the above experiment were also collected and lysed for Western blot analysis to evaluate the expression of necroptosis-related proteins (RIP1, RIP3, MLKL, p-RIP1, p-RIP3, and p-MLKL) and apoptosis-related proteins (cleaved Caspase-3 and cleaved Caspase-8). GAPDH was used as an internal control.
Previous studies have underestimated the non-PAT immunomodulatory functions of zDHHC family proteins, but primarily focused on the palmitoylation modification of immune-related molecules mediated by the zDHHC members (11, 15, 16). Also, research on the immune regulatory roles of zDHHCs in teleosts remains limited, with only zDHHC1 molecules investigated for their antiviral functions (21, 22). Herein, we identified 22 zDHHC proteins in the large yellow croaker genome, excluding zDHHC10 and 19, and analyzed their evolutionary relationships. Interestingly, we observed the absence of the zDHHC19 gene in teleosts, contrasting with its presence in mammals (1). Among the identified zDHHC proteins, LczDHHC23 stood out due to its significant upregulation following P. plecoglloscida infection in MO/MΦ, prompting further investigation into its potential immune regulatory role. Our choice to focus on LczDHHC23 was also influenced by previous studies implicating zDHHC23 in immune-related processes, particularly its involvement in mTOR signaling and tumorigenesis (24, 25). Thus, we aimed to elucidate the specific immune functions of LczDHHC23 in the context of pathogen infection induced immune responses in teleosts.
Our bioinformatics analyses provided insights into the functional domains and evolutionary patterns of LczDHHC23. Phylogenetic analyses revealed its clustering with zDHHC23 from teleosts, particularly Nibea albiflora, suggesting evolutionary conservation. Furthermore, structural predictions indicated similarities between LczDHHC23 and HszDHHC23 proteins, particularly in their DHHC domains and transmembrane regions. Despite these similarities, we noted differences in the subcellular localization of LczDHHC23, predominantly within the Golgi apparatus in contrast to mammalian studies (36).
In examining the immune function of LczDHHC23, we observed its widespread expression in immune-related tissues of healthy large yellow croaker, with significant upregulation following P. plecoglloscida infection, except in the liver. The expression pattern of LczDHHC23 initially increased, followed by a decrease, in both the liver and gills after P. plecoglossicida infection. This suggests that the immune system might boost immune cell activity and inflammatory signaling pathways by upregulating the expression of the zDHHC23 gene to combat pathogen invasion, while its gradual decrease in expression aims to alleviate excessive inflammation and immune-related damage (40). These findings underscore the involvement of LczDHHC23 in the immune response against bacterial infection (41, 42). Silencing LczDHHC23 expression in MO/MΦ resulted in heightened pro-inflammatory cytokine expression and reduced anti-inflammatory cytokine levels during P. plecoglossicida infection, indicating its anti-inflammatory role. Meanwhile, we observed a significant increase in the expression of TGF-β at 4 h, whereas the expression of IL-1β and IL-6 significantly decreased at 4 and 24 h, respectively. Typically, MO/MΦ release inflammatory cytokines rapidly upon pathogen infection. However, to mitigate excessive inflammation and immune damage, the expression of inhibitory inflammatory cytokines may gradually rise (43). This result could be attributed to the regulatory role of zDHHC23, a non-PAT, in the cytokine signaling pathway. Currently, there is insufficient literature on the non-PAT functions of the zDHHC family and their involvement in cytokine signaling pathways. Further research is warranted to elucidate specific targets and underlying mechanisms.
Moreover, our study revealed the impact of LczDHHC23 deficiency on MO/MΦ polarization, shifting the balance towards the pro-inflammatory M1 phenotype. These findings further support the anti-inflammatory role of LczDHHC23 in MO/MΦ and its role in promoting M2-typ MO/MΦ polarization (32, 44). Further, silencing LczDHHC23 reduced P. plecoglloscida infection induced necroptosis of MO/MΦ, which was also supported by decreased phosphorylation levels of necroptosis markers, suggesting a protective role against cell death with LczDHHC23 deficiency (45, 46). Meanwhile, we also observed a slight decrease in the expression of cleaved Caspase-3 and 8 in the LczDHHC23 knockout group 2 hpi (Figure 4B). However, no further significant differences were observed with prolonged infection. Furthermore, upon calculating the relative grey intensity of the Cleaved Caspase-3 band to that of GAPDH (data not shown), we found that the minor decrease of the 2 hpi band were not statistically significant. We thought that the weak cleavage of Caspase-3 may not be significant enough to indicate a notable effect. Additionally, our statistical analysis of flow cytometry data representing apoptosis revealed no significant difference between the LczDHHC23 knockdown and control groups (Figure 4A). Therefore, we thought the influence of LczDHHC23 on cell death induced by infection primarily pertains to necroptosis. Given the emerging role of necroptosis in host defense against pathogens, elucidating the precise mechanisms underlying LczDHHC23-mediated regulation of necroptosis could provide valuable insights into host-pathogen interactions in teleosts.
In summary, our study elucidates the immune regulatory functions of LczDHHC23 in large yellow croaker, highlighting its anti-inflammatory properties in MO/MΦ. By modulating cytokine expression, polarization, and necroptosis, LczDHHC23 plays a crucial anti-inflammatory role in the immune response against bacterial infection. However, further investigations are warranted to unravel the precise mechanisms underlying LczDHHC23-mediated immune regulation and its potential as a therapeutic target for immune modulation in teleosts.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The animal study was approved by Institutional Animal Care and Use Committee of Ningbo University. The study was conducted in accordance with the local legislation and institutional requirements.
TD: Investigation, Methodology, Visualization, Writing – original draft. ZZ: Methodology, Writing – review & editing, Investigation, Visualization. TZ: Investigation, Methodology, Visualization, Writing – review & editing. CF: Investigation, Visualization, Writing – review & editing. LN: Methodology, Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. JC: Writing – review & editing, Conceptualization, Project administration, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Zhejiang Province (LY23C190002), and the Natural Science Foundation of Ningbo City (202003N4011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1401626/full#supplementary-material
1. Lemonidis K, Werno MW, Greaves J, Diez-Ardanuy C, Sanchez-Perez MC, Salaun C, et al. The zDHHC family of S-acyltransferases. Biochem Soc Trans. (2015) 43:217–21. doi: 10.1042/BST20140270
2. Salaun C, Locatelli C, Zmuda F, Cabrera Gonzalez J, Chamberlain LH. Accessory proteins of the zDHHC family of S-acylation enzymes. J Cell Sci. (2020) 133(22):jcs251819. doi: 10.1242/jcs.251819
3. Chamberlain LH, Shipston MJ. The physiology of protein S-acylation. Physiol Rev. (2015) 95:341–76. doi: 10.1152/physrev.00032.2014
4. Zmuda F, Chamberlain LH. Regulatory effects of post-translational modifications on zDHHC S-acyltransferases. J Biol Chem. (2020) 295:14640–52. doi: 10.1074/jbc.REV120.014717
5. Lu Y, Zheng Y, Coyaud É, Zhang C, Selvabaskaran A, Yu Y, et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science. (2019) 366(6464):460–7. doi: 10.1126/science.aau6391
6. Kim YC, Lee SE, Kim SK, Jang HD, Hwang I, Jin S, et al. Toll-like receptor mediated inflammation requires FASN-dependent MYD88 palmitoylation. Nat Chem Biol. (2019) 15:907–16. doi: 10.1038/s41589–019-0344–0
7. Shi C, Yang X, Liu Y, Li H, Chu H, Li G, et al. ZDHHC18 negatively regulates cGAS-mediated innate immunity through palmitoylation. EMBO J. (2022) 41:e109272. doi: 10.15252/embj.2021109272
8. Collura KM, Niu J, Sanders SS, Montersino A, Holland SM, Thomas GM. The palmitoyl acyltransferases ZDHHC5 and ZDHHC8 are uniquely present in DRG axons and control retrograde signaling via the Gp130/JAK/STAT3 pathway. J Biol Chem. (2020) 295:15427–37. doi: 10.1074/jbc.RA120.013815
9. Claudinon J, Gonnord P, Beslard E, Marchetti M, Mitchell K, Boularan C, et al. Palmitoylation of interferon-alpha (IFN-alpha) receptor subunit IFNAR1 is required for the activation of Stat1 and Stat2 by IFN-alpha. J Biol Chem. (2009) 284:24328–40. doi: 10.1074/jbc.M109.021915
10. Bieerkehazhi S, Fan Y, West SJ, Tewari R, Ko J, Mills T, et al. Ca2+-dependent protein acyltransferase DHHC21 controls activation of CD4+ T cells. J Cell Sci. (2022) 135(5):jcs258186. doi: 10.1242/jcs.258186
11. Lin H. Protein cysteine palmitoylation in immunity and inflammation. FEBS J. (2021) 288:7043–59. doi: 10.1111/febs.15728
12. Ladygina N, Martin BR, Altman A. Dynamic palmitoylation and the role of DHHC proteins in T cell activation and anergy. Adv Immunol. (2011) 109:1–44. doi: 10.1016/B978–0-12–387664–5.00001–7
13. Delandre C, Penabaz TR, Passarelli AL, Chapes SK, Clem RJ. Mutation of juxtamembrane cysteines in the tetraspanin CD81 affects palmitoylation and alters interaction with other proteins at the cell surface. Exp Cell Res. (2009) 315:1953–63. doi: 10.1016/j.yexcr.2009.03.013
14. Hundt M, Harada Y, De Giorgio L, Tanimura N, Zhang W, Altman A. Palmitoylation-dependent plasma membrane transport but lipid raft-independent signaling by linker for activation of T cells. J Immunol. (2009) 183:1685–94. doi: 10.4049/jimmunol.0803921
15. Liu Y, Zhou Q, Zhong L, Lin H, Hu MM, Zhou Y, et al. ZDHHC11 modulates innate immune response to DNA virus by mediating MITA-IRF3 association. Cell Mol Immunol. (2018) 15:907–16. doi: 10.1038/cmi.2017.146
16. Zhou Q, Lin H, Wang S, Wang S, Ran Y, Liu Y, et al. The ER-associated protein ZDHHC1 is a positive regulator of DNA virus-triggered, MITA/STING-dependent innate immune signaling. Cell Host Microbe. (2014) 16:450–61. doi: 10.1016/j.chom.2014.09.006
17. Chen X, Shi W, Wang F, Du Z, Yang Y, Gao M, et al. Zinc finger DHHC-type containing 13 regulates fate specification of ectoderm and mesoderm cell lineages by modulating Smad6 activity. Stem Cells Dev. (2014) 23:1899–909. doi: 10.1089/scd.2014.0068
18. Shi W, Chen X, Wang F, Gao M, Yang Y, Du Z, et al. ZDHHC16 modulates FGF/ERK dependent proliferation of neural stem/progenitor cells in the zebrafish telencephalon. Dev Neurobiol. (2016) 76:1014–28. doi: 10.1002/dneu.22372
19. Shi W, Wang F, Gao M, Yang Y, Du Z, Wang C, et al. ZDHHC17 promotes axon outgrowth by regulating TrkA-tubulin complex formation. Mol Cell Neurosci. (2015) 68:194–202. doi: 10.1016/j.mcn.2015.07.005
20. Wang F, Chen X, Shi W, Yao L, Gao M, Yang Y, et al. Zdhhc15b regulates differentiation of diencephalic dopaminergic neurons in zebrafish. J Cell Biochem. (2015) 116:2980–91. doi: 10.1002/jcb.25256
21. Xu X, Li M, Wu Z, Wang H, Wang L, Huang K, et al. Endoplasmic reticulum transmembrane proteins ZDHHC1 and STING both act as direct adaptors for IRF3 activation in teleost. J Immunol. (2017) 199:3623–33. doi: 10.4049/jimmunol.1700750
22. Chen SN, Zhang S, Li L, Laghari ZA, Nie P. Molecular and functional characterization of zinc finger aspartate-histidine-histidine-cysteine (DHHC)-type containing 1, ZDHHC1 in Chinese perch Siniperca chuatsi. Fish Shellfish Immunol. (2022) 130:215–22. doi: 10.1016/j.fsi.2022.09.023
23. Tang Y, Xin G, Zhao LM, Huang LX, Qin YX, Su YQ, et al. Novel insights into host-pathogen interactions of large yellow croakers ( Larimichthys crocea) and pathogenic bacterium Pseudomonas plecoglossicida using time-resolved dual RNA-seq of infected spleens. Zool Res. (2020) 41:314–27. doi: 10.24272/j.issn.2095–8137.2020.035
24. Sadhukhan T, Bagh MB, Appu AP, Mondal A, Zhang W, Liu A, et al. In a mouse model of INCL reduced S-palmitoylation of cytosolic thioesterase APT1 contributes to microglia proliferation and neuroinflammation. J Inherit Metab Dis. (2021) 44:1051–69. doi: 10.1002/jimd.12379
25. Jeong DW, Park JW, Kim KS, Kim J, Huh J, Seo J, et al. Palmitoylation-driven PHF2 ubiquitination remodels lipid metabolism through the SREBP1c axis in hepatocellular carcinoma. Nat Commun. (2023) 14:6370. doi: 10.1038/s41467–023-42170–0
26. Li CH, Chen J, Nie L, Chen J. MOSPD2 is a receptor mediating the LEAP-2 effect on monocytes/macrophages in a teleost, Boleophthalmus pectinirostris. Zool Res. (2020) 41:644–55. doi: 10.24272/j.issn.2095–8137.2020.211
27. Wu XY, Nie L, Lu XJ, Fei CJ, Chen J. Molecular characterization, expression and functional analysis of large yellow croaker (Larimichthys crocea) peroxisome proliferator-activated receptor gamma. Fish Shellfish Immunol. (2022) 123:50–60. doi: 10.1016/j.fsi.2022.02.021
28. Chen Q, Chen J, Chen J, Lu XJ. Molecular and functional characterization of liver X receptor in ayu, Plecoglossus altivelis: Regulator of inflammation and efferocytosis. Dev Comp Immunol. (2016) 65:358–68. doi: 10.1016/j.dci.2016.08.007
29. Shen HX, Lu XJ, Lu JF, Chen J. Beta-adrenergic receptor stimulation influences the function of monocytes/macrophages in ayu (Plecoglossus altivelis). Dev Comp Immunol. (2020) 103:103513. doi: 10.1016/j.dci.2019.103513
30. Ning YJ, Lu XJ, Chen J. Molecular characterization of a tissue factor gene from ayu: A pro-inflammatory mediator via regulating monocytes/macrophages. Dev Comp Immunol. (2018) 84:37–47. doi: 10.1016/j.dci.2018.02.002
31. Nie L, Cai SY, Sun J, Chen J. MicroRNA-155 promotes pro-inflammatory functions and augments apoptosis of monocytes/macrophages during Vibrio Anguillarum infection in ayu, Plecoglossus altivelis. Fish Shellfish Immunol. (2019) 86:70–81. doi: 10.1016/j.fsi.2018.11.030
32. Joerink M, Ribeiro CM, Stet RJ, Hermsen T, Savelkoul HF, Wiegertjes GF. Head kidney-derived macrophages of common carp (Cyprinus carpio L.) show plasticity and functional polarization upon differential stimulation. J Immunol. (2006) 177:61–9. doi: 10.4049/jimmunol.177.1.61
33. Bill R, Wirapati P, Messemaker M, Roh W, Zitti B, Duval F, et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science. (2023) 381:515–24. doi: 10.1126/science.ade2292
34. Rana MS, Kumar P, Lee CJ, Verardi R, Rajashankar KR, Banerjee A. Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science. (2018) 359(6372):eaao6326. doi: 10.1126/science.aao6326
35. Philippe JM, Jenkins PM. Spatial organization of palmitoyl acyl transferases governs substrate localization and function. Mol Membr Biol. (2019) 35:60–75. doi: 10.1080/09687688.2019.1710274
36. Korycka J, Lach A, Heger E, Boguslawska DM, Wolny M, Toporkiewicz M, et al. Human DHHC proteins: a spotlight on the hidden player of palmitoylation. Eur J Cell Biol. (2012) 91:107–17. doi: 10.1016/j.ejcb.2011.09.013
37. Joerink M, Savelkoul HF, Wiegertjes GF. Evolutionary conservation of alternative activation of macrophages: structural and functional characterization of arginase 1 and 2 in carp (Cyprinus carpio L). Mol Immunol. (2006) 43:1116–28. doi: 10.1016/j.molimm.2005.07.022
38. Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. (2009) 22:240–73. doi: 10.1128/CMR.00046–08
39. Tong X, Tang R, Xiao M, Xu J, Wang W, Zhang B, et al. Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research. J Hematol Oncol. (2022) 15:174. doi: 10.1186/s13045–022-01392–3
40. Sayyaf Dezfuli B, Lorenzoni M, Carosi A, Giari L, Bosi G. Teleost innate immunity, an intricate game between immune cells and parasites of fish organs: who wins, who loses. Front Immunol. (2023) 14:1250835. doi: 10.3389/fimmu.2023.1250835
41. Wu N, Song YL, Wang B, Zhang XY, Zhang XJ, Wang YL, et al. Fish gut-liver immunity during homeostasis or inflammation revealed by integrative transcriptome and proteome studies. Sci Rep. (2016) 6:36048. doi: 10.1038/srep36048
42. Secombes CJ, Wang T. The innate and adaptive immune system of fish. Infect Dis Aquac. (2012), 3–68. doi: 10.1533/9780857095732.1.3
43. Guo H, Dixon B. Understanding acute stress-mediated immunity in teleost fish. Fish Shellfish Immunol Rep. (2021) 2:100010. doi: 10.1016/j.fsirep.2021.100010
44. Huang X, Li Y, Fu M, Xin HB. Polarizing macrophages in vitro. Methods Mol Biol. (2018) 1784:119–26. doi: 10.1007/978–1-4939–7837-3_12
45. Zhao H, Jaffer T, Eguchi S, Wang Z, Linkermann A, Ma D. Role of necroptosis in the pathogenesis of solid organ injury. Cell Death Dis. (2015) 6:e1975. doi: 10.1038/cddis.2015.316
Keywords: LczDHHC23, immune regulation, macrophage polarization, necroptosis, large yellow croaker
Citation: Dai T, Zhao Z, Zhu T, Fei C, Nie L and Chen J (2024) The anti-inflammatory role of zDHHC23 through the promotion of macrophage M2 polarization and macrophage necroptosis in large yellow croaker (Larimichthys crocea). Front. Immunol. 15:1401626. doi: 10.3389/fimmu.2024.1401626
Received: 15 March 2024; Accepted: 06 May 2024;
Published: 29 May 2024.
Edited by:
Jia Cai, Guangdong Ocean University, ChinaCopyright © 2024 Dai, Zhao, Zhu, Fei, Nie and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Nie, bmllbGlAbmJ1LmVkdS5jbg==; Jiong Chen, Y2hlbmppb25nQG5idS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.