
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Immunol. , 03 April 2024
Sec. Inflammation
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1381340
This article is part of the Research Topic The Role of Inflammation and Immune Control in Digestive Disease and Therapeutic Approaches View all 20 articles
 Jiawen Huang1†
Jiawen Huang1† Jiayu Li1†
Jiayu Li1† Yuan Peng2†
Yuan Peng2† Tianqi Cui1
Tianqi Cui1 Jingyi Guo1
Jingyi Guo1 Siwei Duan1
Siwei Duan1 Kaili Zhou1
Kaili Zhou1 Shangyi Huang1
Shangyi Huang1 Jiabing Chen1
Jiabing Chen1 Qincheng Yi1
Qincheng Yi1 Min Qiu1
Min Qiu1 Tingting Chen1
Tingting Chen1 Xiaoqin Wu1
Xiaoqin Wu1 Chenlu Ma1
Chenlu Ma1 Ziyi Zhang1
Ziyi Zhang1 Yi Zheng1
Yi Zheng1 Xi Tang1
Xi Tang1 Yanqing Pang3
Yanqing Pang3 Lei Zhang1,4*
Lei Zhang1,4* Chong Zhong5*
Chong Zhong5* Yong Gao1*
Yong Gao1*Background: In addition to abnormal liver inflammation, the main symptoms of non-alcoholic steatohepatitis (NASH) are often accompanied by gastrointestinal digestive dysfunction, consistent with the concept of spleen deficiency (SD) in traditional Chinese medicine. As an important metabolic sensor, whether peroxisome proliferator-activated receptor alpha (PPARα) participates in regulating the occurrence and development of NASH with SD (NASH-SD) remains to be explored.
Methods: Clinical liver samples were collected for RNA-seq analysis. C57BL/6J mice induced by folium sennae (SE) were used as an SD model. qPCR analysis was conducted to evaluate the inflammation and metabolic levels of mice. PPARα knockout mice (PPARαko) were subjected to SE and methionine–choline-deficient (MCD) diet to establish the NASH-SD model. The phenotype of NASH and the inflammatory indicators were measured using histopathologic analysis and qPCR as well.
Results: The abnormal expression of PPARα signaling, coupled with metabolism and inflammation, was found in the results of RNA-seq analysis from clinical samples. SD mice showed a more severe inflammatory response in the liver evidenced by the increases in macrophage biomarkers, inflammatory factors, and fibrotic indicators in the liver. qPCR results also showed differences in PPARα between SD mice and control mice. In PPARαko mice, further evidence was found that the lack of PPARα exacerbated the inflammatory response phenotype as well as the lipid metabolism disorder in NASH-SD mice.
Conclusion: The abnormal NR signaling accelerated the vicious cycle between lipotoxicity and inflammatory response in NAFLD with SD. Our results provide new evidence for nuclear receptors as potential therapeutic targets for NAFLD with spleen deficiency.
Non-alcoholic fatty liver disease (NAFLD) is the most common chronic liver disease worldwide, affecting 12.5% of the global population (1). Non-alcoholic steatohepatitis (NASH) is the advanced stage of NAFLD, which typically increases the risk of developing into cirrhosis and hepatocellular carcinoma (HCC), ultimately leading to HCC (2). So far, NASH has become the fastest-developing cause of HCC. In clinical practice, changing lifestyle and drug therapy are the commonly used coping strategies mainly because there is currently no specific treatment method for NASH and the specific cause of NASH is not yet clear (3).
NASH is a chronic liver disease closely related to metabolic disorders, characterized by hepatocyte steatosis, inflammation, and fibrosis (4). Among them, inflammation is an important feature of NASH, which is a complex process caused by multiple factors. In brief, immune cells such as macrophages and T lymphocytes are activated by fatty acids and oxidative stress, releasing pro-inflammatory cytokines, leading to the progression of inflammation and fibrosis (5, 6). Secondly, abnormal fatty acid metabolism is one of the key factors in NASH inflammation. Once liver fatty acid metabolism is imbalanced, fatty acids accumulate excessively in hepatocytes, leading to the formation and enlargement of lipid droplets, thereby triggering an inflammatory response in the liver (7). Moreover, when exposed to excessive fatty acids and free radicals, hepatocytes can also result in increased oxidative stress, further increasing the inflammatory response and exacerbating the progress of NASH (8). In traditional Chinese medicine (TCM), spleen deficiency (SD) is the basis of NASH, which can be manifested as chest and rib swelling and pain, sighing, depression or irritability, lack of appetite, bloating, and loose stools (9). Mice gavaged with folium sennae were usually used as the SD model, exhibiting diarrhea, imbalance of short-chain fatty acid metabolism, and severe inflammation (10). However, it is not clear whether this severe inflammation will cause an inflammatory response in the liver and ultimately lead to metabolic disorders in the liver.
Peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors of the nuclear hormone receptor superfamily comprised of three subtypes, namely, PPARα, PPARβ/δ, and PPARγ, and they play a crucial role in glucose and lipid metabolism and inflammation (11). Among them, PPARα can promote the uptake and oxidation of fatty acids, inhibit adipocyte differentiation, and also promote the reverse transport of cholesterol by regulating the expression of related genes (12, 13). Although abnormal PPARα signaling is often associated with abnormal liver lipid metabolism, its abnormal expression is also accompanied by inflammatory reactions in the liver (14). Recent studies have begun to consider the crosstalk between hepatocytes and immunocytes. It is known that lipotoxicity caused by lipid metabolism disorders in hepatocytes can induce inflammatory reactions as shown in our previous study (15). The administration of the PPARα/γ dual agonist tesaglitazar had anti-inflammatory effects in ob/ob mice by reducing the number of pro-inflammatory adipose tissue macrophages (16). The current study investigated the hypothesis that SD-induced liver inflammation leads to abnormalities in the PPARα pathway, thereby accelerating susceptibility to NASH.
It is believed that once the inflammatory response intensifies, it will further induce lipid metabolism disorders to form a vicious cycle, ultimately exacerbating the development of the disease (17). Similar to this point of view, our previous study proved that hepatic Zbtb18 protein transcriptionally activates FAO and CLTC expression, which inhibits NLRP3 inflammasome’s activity, alleviating inflammatory stress and insulin resistance. In this study, WT mice and PPARα knockout (PPARαko) mice gavaged with folium sennae (SE) were used as the SD model, a methionine–choline-deficient (MCD) diet was used as an incentive for NASH, and the molecular biology techniques and transcriptomics were utilized for detecting changes in inflammation, lipid metabolism, and PPARα expression.
Six- to 8-week-old C57BL/6J mice were housed in an SPF-level breeding environment (24°C ± 2°C, a 12/12-h light/dark cycle) and had free access to water and food supervised by the Ethics Committee of Guangzhou University of Chinese Medicine. Mice were divided into the CON group and the SD group. SD mice were gavaged with 0.4 ml of 10% SE (w/v), while mice in the CON group were given the same equivalent normal saline. Seven days later, mice were executed, and the liver of each mouse was collected for further examination.
Six- to 8-week-old WT mice and PPARα knockout (PPARαko) mice were fed with chow diet (CD) or MCD diet with SE gavage for 7 days. The serum and the liver of each mouse were collected for further detection.
For qPCR, the total mRNA of each liver sample was extracted, followed by reverse transcription into cDNA, and then quantitative PCR was performed using Master Mix (ABclonal Co., Wuhan, China). The specific primers are shown in Supplementary Table S1.
For histopathologic analysis, liver samples were sliced after embedding and fixation. For immunohistochemistry, hematoxylin and eosin (H&E) staining kit, Oil Red O staining kit, and Sirius Red staining kit were used following the instructions. For immunohistochemistry (IHC), after dewaxing, hydration, and antigen repair, slices were blocked and incubated with antibodies overnight at 4°C, followed by incubation with the secondary antibody for 1 h, and pictures were observed using a microscope after being sealed.
The liver samples of NAFLD and NAFLD-SD patients were collected from the First Affiliated Hospital of Guangzhou University of Chinese Medicine in accordance with the guidelines of the Declaration of Helsinki, and written informed consent was obtained from each patient supervised by the First Affiliated Hospital of Guangzhou University of Chinese Medicine Institutional Review Board and Ethics Committee.
For RNA-seq analysis, total RNA was extracted and RNA purity, quantification, and integrity were evaluated. Sequencing was conducted and the libraries were constructed according to the manufacturer’s instructions. The RNA-seq analysis was conducted by Berry Genomics Corporation (Beijing, China).
Data were shown as means ± SEM, and the significant differences were evaluated using Student’s t-test. A p-value <0.05 was considered statistically significant.
To gain insight into the underlying molecular mechanisms of NAFLD and NAFLD-SD, we performed RNA-seq analysis of clinical liver samples from patients with NAFLD and NAFLD-SD (Figure 1A). The results showed that 213 genes were differently expressed with 106 upregulated and 107 downregulated differentially expressed genes (DEGs) (Figure 1B). RNA-seq data showed a significant difference between NAFLD-SD and NAFLD (Figure 1C), and the KEGG pathway analysis showed that the main differential pathways included metabolism and inflammation, with the PPAR signaling pathway closely associated with them (Figure 1D). In line with the KEGG analysis, gene set enrichment analysis (GSEA) showed that the DEGs (NAFLD-SD vs. NAFLD) were negatively correlated with the “PPAR signaling pathway” categories and were positively correlated with inflammation, suggesting that these changes could regulate the nuclear receptor and inflammation (Figures 1E, F). Pearson’s correlation analysis of the RNA-seq analysis data was performed to further explore the correlation between PPAR and genes involved in inflammation from the NAFLD-SD vs. NAFLD. The results showed that PPARα and PPARr mRNA levels were negatively correlated with the expression of the gene related to inflammation (Figure 1G).
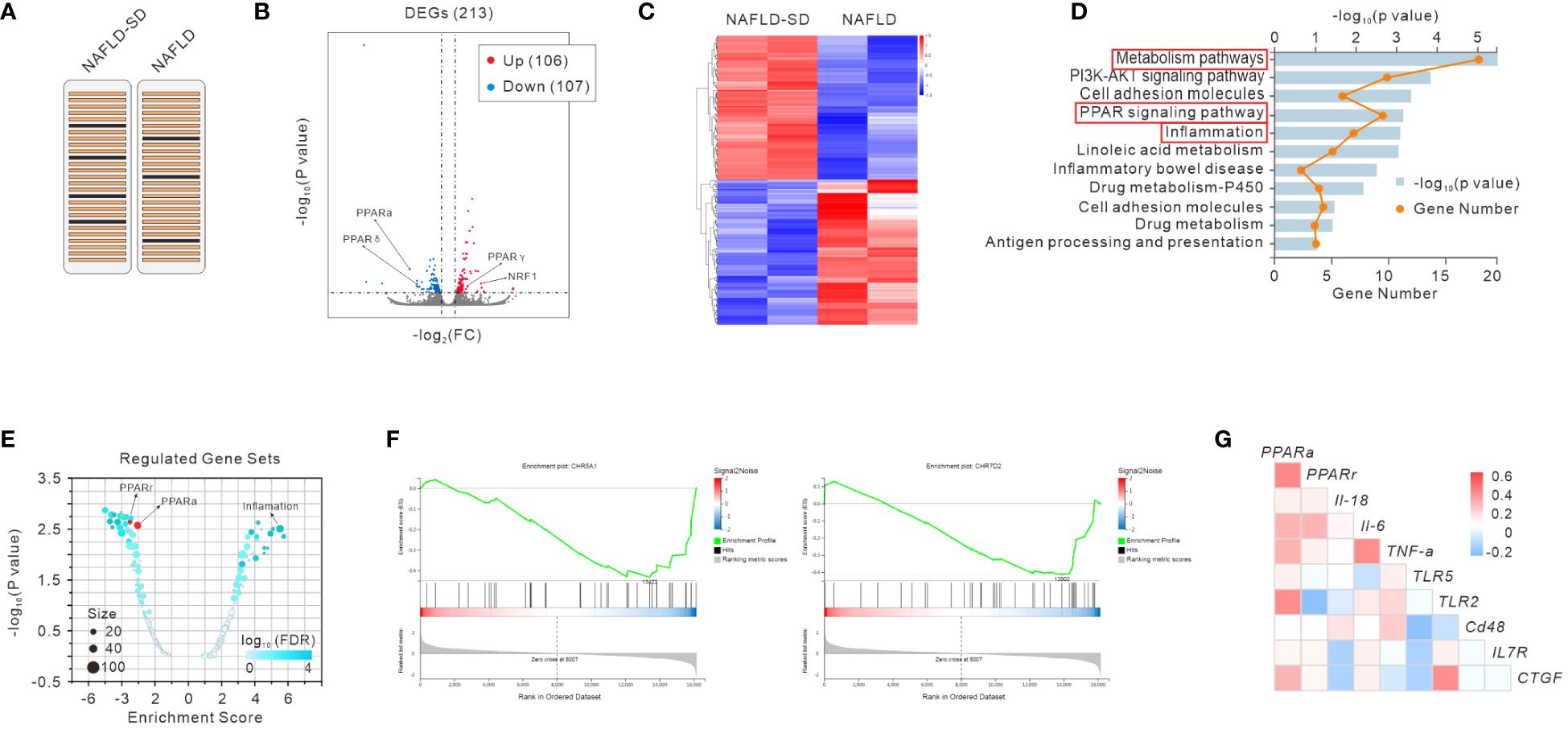
Figure 1 RNA-seq analysis of liver samples of NAFLD and NAFLD-SD patients (A). Scatter plot (B), heatmap (C), and KEGG analysis (D) of RNA-seq data. Regulated gene sets (E), gene set enrichment analysis (GSEA) (F), and gene correlation analysis (G) of RNA-seq data.
In order to explore the changes caused by SE-induced SD in the liver, qPCR analysis was conducted. The results showed that inflammatory genes such as Il6, Tnfα, Il1β, and nfkb were highly expressed in the SD group (Figure 2A). The macrophage infiltration marker genes such as cd86 and iNos were also significantly increased in the liver of SD mice (Figure 2B). PPARα signaling-related genes were also detected, and it was found that PPARα was downregulated while Pgc1α was upregulated by SD intervention (Figure 2C). Moreover, the mRNA expression levels of asma, Tgfβ, Col2a1, and Col5a1 were increased in the liver of SD mice, suggesting an increased tendency to liver fibrosis (Figure 2D).
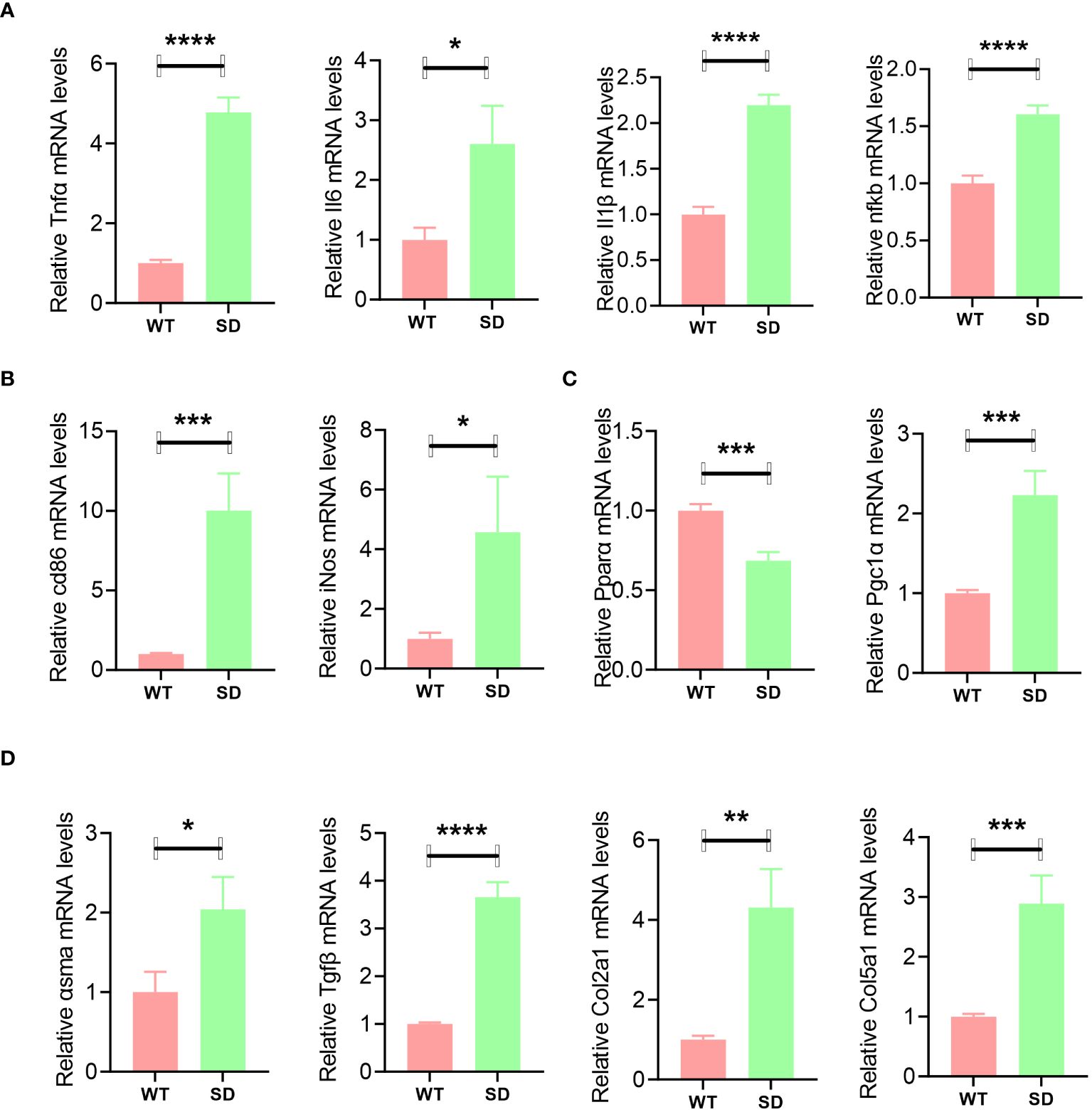
Figure 2 qPCR analysis of liver samples of WT and SD mice, including inflammatory genes such as Il6, Tnfα, Il1β, and nfkb (A); macrophage marker genes cd86 and iNos (B); PPARα signaling-related genes such as PPARα and Pgc1α (C); and fibrosis-related genes such as asma, Tgfβ, Col2a1, and Col5a1 (D). Data are shown as the mean ± SEM, n = 6. *p < 0.05, **p < 0.01, ***p < 0.005, and ****p < 0.001.
PPARαko mice fed with MCD diet as well as gavaged with SE (MCD+SD) were used for further exploration. When fed with CD, there was barely any difference between WT and PPARαko mice in liver histopathological analysis, while when subjected to MCD+SD intervention, more severe pathological manifestations of lipid deposition and fibrosis were observed in the liver of PPARαko mice compared with WT mice (Figure 3A). Mice in the MCD+SD group exhibited high mRNA expression levels of TGFβ1 and Col1a1 in the qPCR results, with PPARαko mice exhibiting higher expression compared with WT mice (Figure 3B).
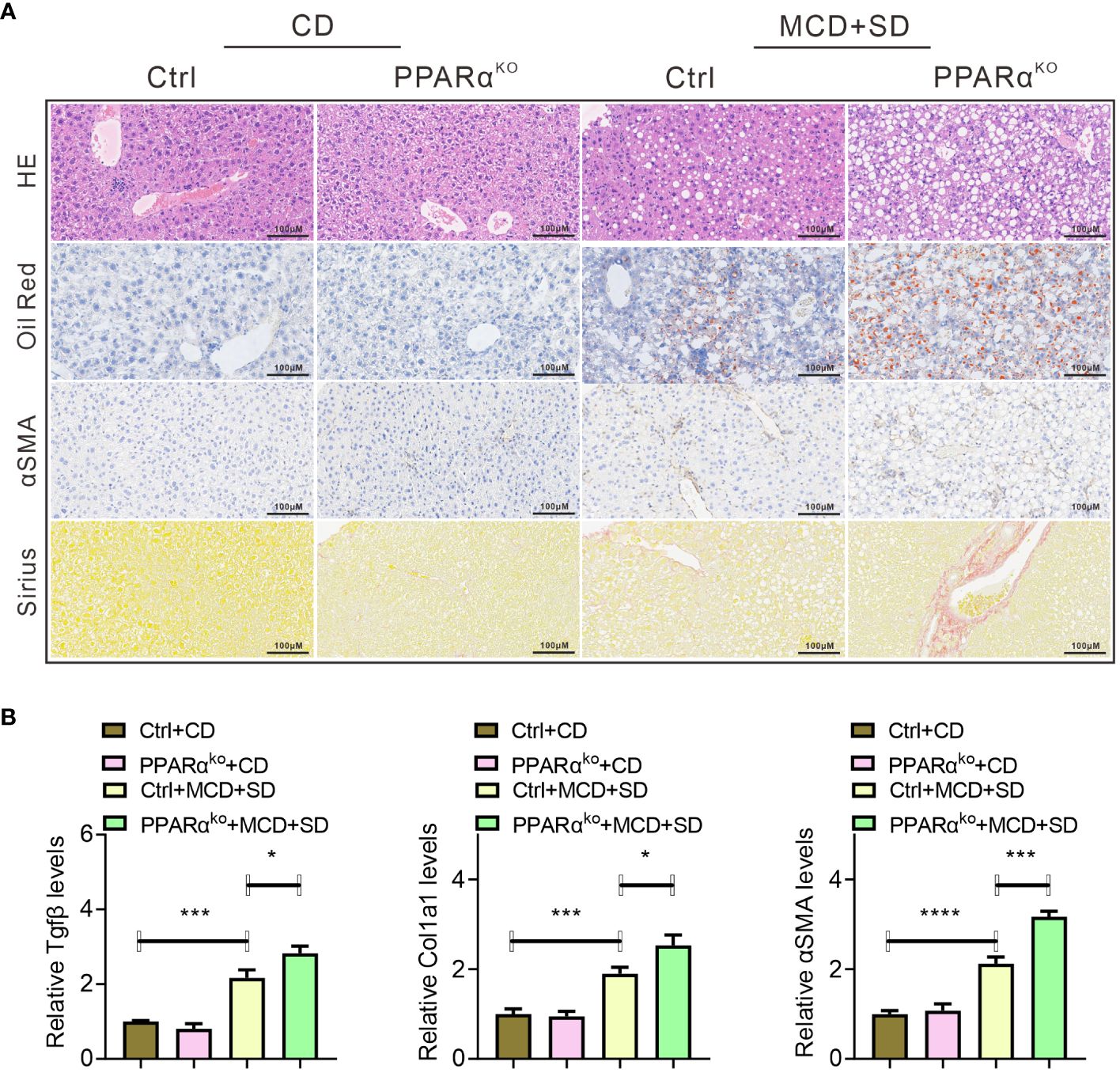
Figure 3 Histopathological analysis of liver samples of WT and PPARαko mice intervened with CD or MCD+SD, including H&E staining, Oil Red O staining, IHC of αSMA, and Sirius Red staining (A). qPCR analysis of liver samples of those mice, including fibrosis-related genes such as Tgfβ, Col1a1, and αSMA (B). Data are shown as the mean ± SEM, n = 6. *p < 0.05, ***p < 0.005, and ****p < 0.001.
Further experiments focused on the liver inflammation of those mice. When fed with CD, there was no significant difference in the expression levels of inflammatory genes including Il6, Tnfα, and Il1β as well as F4/80 in the liver of both WT and PPARαko mice, while when intervened with MCD+SD, those genes in PPARαko mice were significantly higher than those in WT mice (Figure 4).
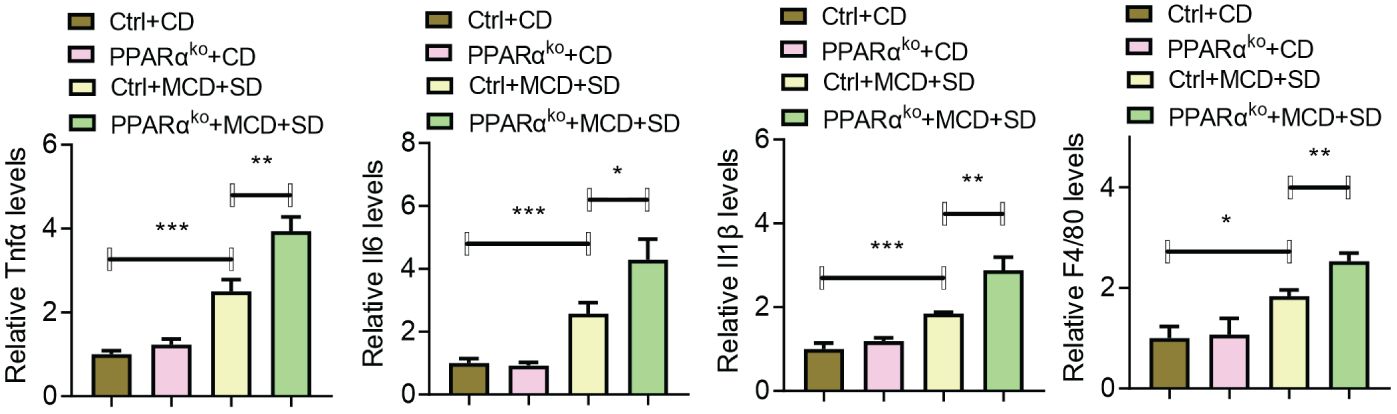
Figure 4 qPCR analysis of liver samples of WT and PPARαko mice intervened with CD or MCD+SD, including inflammatory genes such as Il6, Tnfα, Il1β, and f4/80. Data are shown as the mean ± SEM, n = 6. *p < 0.05, **p < 0.01, and ***p < 0.005.
The pandemic of NASH has forced us to consider countermeasures, and identifying susceptible populations of NASH and preventing their process in NASH are two of the feasible ways. TCM believes that SD, which is involved in the abnormal function of the spleen and liver as well as organs of the digestive system, is the key to the occurrence and development of NASH (18). An individual with SD is more likely to exhibit symptoms of digestive and metabolic abnormalities, which may trigger or accelerate NASH (19). Also, TCM research on NASH has always focused on SD (20). SE is a commonly used inducer for establishing SD models, and mice treated with its intervention may experience metabolic abnormalities, diarrhea, and other symptoms, which are typical manifestations of SD (21). This project outlines the increased susceptibility of SD individuals to NASH and points out one of its potential therapeutic targets—PPARα.
Inflammation is one of the important triggers in the progression of NAFLD to NASH (22). In this study, there are significant differences in metabolic and inflammatory pathways in the RNA-seq analysis of liver samples from NASH-SD and NASH patients, suggesting that metabolism and inflammation may be triggers for the development of SD toward NASH, as demonstrated by SD mice in our study. This is consistent with a recent study on increased inflammation and metabolic disorders in SD mice (10). However, SD-induced severe diarrhea in mice may also be associated with intestinal barrier dysfunction and dysbiosis of gut microbiota (23). For example, when transferred to sterile mice, specific microbial characteristics may be reshaped, and the progression of NASH can be delayed, indicating that mitochondrial dysfunction promotes the progression of NASH by exacerbating the gut–liver status as well as inflammation (24). Overall, the changes in the microenvironment of liver inflammation caused by different reasons will drive the activation of hepatic stellate cells and thereby accelerate the development of NASH, which is consistent with our research (25).
In NASH patients, not only severe inflammation and lipid deposition but also a tendency for liver fibrosis was observed (26). Similar results were observed in SD mice, indicating that individuals with SD are more susceptible to NASH. Meanwhile, NASH treatment strategies either seek to alleviate metabolic disorders and cell damage or directly target the accompanying inflammation and fibrosis, with targets for reducing the activation of inflammatory cascade reactions including nuclear receptor agonists (27). In this study, we observed that the liver of SD mice exhibited not only elevated levels of inflammation and a tendency toward fibrosis but also abnormal PPARα signaling, in which PPARα is one of the nuclear receptors. When entering the nucleus, PPARα can bind to promoters that are associated with β-oxidation-related factors such as ACADM and CPT1A and also enhance their transcriptional activity and ultimately drive the process of fatty acid β-oxidation (28). Also, activated PGC-1α can regulate fatty acid β-oxidation through synergism with transcription factors such as PPARα, indicating the crucial role that PPARα/PGC1α signaling plays in liver fatty acid metabolism homeostasis (29). PPARα is downregulated in many liver diseases such as NAFLD and NASH (30). In this study, we found for the first time that PPARα is downregulated in the liver of SD mice. Thus, the abnormal expression levels of PPARα and PGC1α occurred simultaneously with fibrosis and elevated inflammatory response in SD mice, indicating that this signaling may be the reason for the increased susceptibility of SD mice to NASH.
In further research, we investigated whether PPARα deficiency increased the susceptibility to NASH, using PPARαko mice and WT mice with MCD+SD intervention as the study subjects and conducting histopathological observations. H&E staining and Oil Red staining are commonly used to observe fat deposition in mouse liver, while Sirius staining and immunohistochemistry for detecting αSMA expression are commonly used to observe the severity of liver fibrosis (31). In our study, PPARαko mice treated with MCD+SD intervention showed more severe fat deposition and fibrosis in the above liver pathological analysis compared with WT mice, but there was no difference between the two in the CD diet. It is known that PPARα is highly expressed in the liver and participates in regulating fatty acid metabolism (32). After binding to fatty acid ligands, PPARα stimulates the transcription of genes containing PPARα response elements in its enhancer, with the most significant being those genes involved in lipid metabolism and energy homeostasis (33). Previously, Li’s research showed that there was no significant difference in the liver between normally fed PPARko mice and WT mice, which is consistent with our research results (34). The absence of PPARα exacerbates the NASH performance caused by MCD+SD intervention, which is related to the functions of PPARα in regulating lipid metabolism homeostasis and inflammatory response. Meanwhile, our results also indirectly reflect that the absence of PPARα increases the susceptibility to NASH. In addition, only a preliminary exploration of the mechanism has been conducted, and the number of clinical samples used was limited, which cannot be representative of all situations. Further studies are needed in the future.
Overall, the lack of PPARα accelerated the vicious cycle between lipid metabolism and inflammatory response in NAFLD with SD. Similar results have been found in sepsis in which the metabolic and inflammatory responses to bacterial infection were impaired in the absence of PPARα, which leads to an enhancement in mortality due to bacterial sepsis (35). Our previous studies also exhibited that the ingredients of spleen-tonifying drugs can improve NAFLD or NASH by regulating PPAR signaling, which indirectly confirms the conclusion of this topic (36, 37). Unfortunately, there was no separate comparison between the MCD group and the MCD+SD group when setting up the groups, which will also be our next research direction. In addition, only a preliminary exploration of the mechanism has been conducted, and the number of clinical samples used was limited, which cannot be representative of all situations. Further studies are needed in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1061326.
The studies involving humans were approved by the First Affiliated Hospital of Guangzhou University of Chinese Medicine Institutional Review Board and Ethics Committee affiliated to Guangzhou University of Chinese Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by the Ethics Committee of Guangzhou University of Chinese Medicine affiliated to Guangzhou University of Chinese Medicine. The study was conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
JH: Writing – review & editing, Writing – original draft, Investigation, Funding acquisition, Formal analysis. JL: Writing – review & editing, Writing – original draft, Investigation, Formal analysis. YP: Writing – original draft, Writing – review & editing, Formal analysis. TQC: Writing – original draft, Visualization, Investigation. JG: Writing – original draft, Visualization, Investigation. SD: Writing – original draft, Software, Investigation. KZ: Writing – original draft, Software, Investigation. SH: Writing – original draft, Investigation, Data curation. JC: Writing – original draft, Investigation, Data curation. QY: Writing – original draft, Investigation. MQ: Writing – original draft, Investigation. TTC: Writing – original draft, Investigation. XW: Writing – original draft, Investigation. CM: Writing – original draft, Investigation. ZZ: Writing – original draft, Investigation. YZ: Writing – original draft, Investigation. XT: Writing – original draft, Investigation. YQP: Writing – review & editing, Conceptualization, Funding acquisition. LZ: Writing – review & editing, Writing – original draft, Resources, Formal analysis. CZ: Writing – review & editing, Writing – original draft, Resources, Formal analysis. YG: Writing – review & editing, Writing – original draft, Resources, Funding acquisition, Formal analysis, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (82160891, 82304826), Key Projects of Guangdong Provincial Department of Education (2021ZDZX2010), Basic and Applied Basic Research Project of Guangzhou (202201011501, 2022010101340, 202201011240), Guangdong Basic and Applied Basic Research Foundation (2020A1515110732), and Science and Technology Planning Project of Guangzhou (202201020349).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1381340/full#supplementary-material
1. Powell EE, Wong VW, Rinella M. Non-alcoholic fatty liver disease. Lancet. (2021) 397:2212–24. doi: 10.1016/S0140-6736(20)32511-3
2. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. (2016) 64:73–84. doi: 10.1002/Hep.28431
3. Kakleas K, Christodouli F, Karavanaki K. Nonalcoholic fatty liver disease, insulin resistance, and sweeteners: A literature review. Expert Rev Endocrinol Metab. (2020) 15:83–93. doi: 10.1080/17446651.2020.1740588
4. Cano L, Desquilles L, Ghukasyan G, Angenard G, Landreau C, Corlu A, et al. Sars-Cov-2 receptor ace2 is upregulated by fatty acids in human mash. Jhep Rep. (2024) 6:100936. doi: 10.1016/J.Jhepr.2023.100936
5. Zhou Y, Zhang H, Yao Y, Zhang X, Guan Y, Zheng F. Cd4(+) T cell activation and inflammation in Nash-related fibrosis. Front Immunol. (2022) 13:967410. doi: 10.3389/Fimmu.2022.967410
6. Barreby E, Chen P, Aouadi M. Macrophage functional diversity in nafld - more than inflammation. Nat Rev Endocrinol. (2022) 18:461–72. doi: 10.1038/S41574-022-00675-6
7. Zhao L, Zhang C, Luo X, Wang P, Zhou W, Zhong S, et al. Cd36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis. J Hepatol. (2018) 69:705–17. doi: 10.1016/J.Jhep.2018.04.006
8. Zhou J, Pang J, Tripathi M, Ho JP, Widjaja AA, Shekeran SG, et al. Spermidine-mediated hypusination of translation factor eif5a improves mitochondrial fatty acid oxidation and prevents non-alcoholic steatohepatitis progression. Nat Commun. (2022) 13:5202. doi: 10.1038/S41467-022-32788-X
9. Dai L, Xu JJ, Zhou WJ, Lu AP, Ji G. Appraisal of treatment outcomes in integrative medicine using metabonomics: taking non-alcoholic fatty liver disease with spleen deficiency syndrome as an example. J Integr Med. (2022) 20:524–33. doi: 10.1016/J.Joim.2022.08.002
10. Liu C, Song C, Wang Y, Xiao Y, Zhou Z, Cao G, et al. Deep-fried atractylodes lancea rhizome alleviates spleen deficiency diarrhea-induced short-chain fatty acid metabolic disorder in mice by remodeling the intestinal flora. J Ethnopharmacol. (2023) 303:115967. doi: 10.1016/J.Jep.2022.115967
11. Wagner N, Wagner KD. The role of Ppars in disease. Cells. (2020) 9(11):2367. doi: 10.3390/Cells9112367
12. Montaigne D, Butruille L, Staels B. Ppar control of metabolism and cardiovascular functions. Nat Rev Cardiol. (2021) 18:809–23. doi: 10.1038/S41569-021-00569-6
13. Gross B, Pawlak M, Lefebvre P, Staels B. Ppars in obesity-induced T2dm, dyslipidaemia and nafld. Nat Rev Endocrinol. (2017) 13:36–49. doi: 10.1038/Nrendo.2016.135
14. Li H, Zheng J, Xu Q, Yang Y, Zhou J, Guo X, et al. Hepatocyte adenosine kinase promotes excessive fat deposition and liver inflammation. Gastroenterology. (2023) 164:134–46. doi: 10.1053/J.Gastro.2022.09.027
15. Zhang L, Chen J, Yang X, Shen C, Huang J, Zhang D, et al. Hepatic zbtb18 (Zinc finger and btb domain containing 18) alleviates hepatic steatohepatitis via fxr (Farnesoid X receptor). Signal Transduct Target Ther. (2024) 9:20. doi: 10.1038/S41392-023-01727-7
16. Osinski V, Bauknight DK, Dasa S, Harms MJ, Kroon T, Marshall MA, et al. In vivo liposomal delivery of Pparalpha/gamma dual agonist tesaglitazar in A model of obesity enriches macrophage targeting and limits liver and kidney drug effects. Theranostics. (2020) 10:585–601. doi: 10.7150/Thno.36572
17. Minami Y, Hoshino A, Higuchi Y, Hamaguchi M, Kaneko Y, Kirita Y, et al. Liver lipophagy ameliorates nonalcoholic steatohepatitis through extracellular lipid secretion. Nat Commun. (2023) 14:4084. doi: 10.1038/S41467-023-39404-6
18. Hao W, Wang R, Li X, Chen J. Commentary on biological mechanism of liver qi depression and spleen deficiency syndrome based on the correlation of prescription and syndrome. China J of Traditional Chin Med And Pharm. (2023) 38:3005–11. doi: 10.14148/j.issn.1672-0482.2023.0384
19. Zheng XF, Tian JS, Liu P, Xing J, Qin XM. Analysis of the restorative effect of Bu-Zhong-Yi-Qi-Tang in the spleen-Qi deficiency rat model using (1)H-nmr-based metabonomics. J Ethnopharmacol. (2014) 151:912–20. doi: 10.1016/J.Jep.2013.12.001
20. Liang X, Lin S, Wang Y, Xu J, Liu Y, Xie C. Research progress on the mechanism of traditional Chinese medicine in the intervention of nonalcoholic steatohepatitis. J of Nanjing Univ of Traditional Chin Med. (2023) 39:384–92. doi: 10.14148/j.issn.1672-0482.2023.0384
21. Yu H, Meng Y, Wang E, Yuan J, Peng Y, Li X. Buzhong Yiqi decoction ameliorates spleen deficiency syndrome by regulating gut microbiota. China J of Chin Materia Med. (2023) 49:1028–43. doi: 10.19540/j.cnki.cjcmm.20231013.701
22. Schuster S, Cabrera D, Arrese M, Feldstein AE. Triggering and resolution of inflammation in Nash. Nat Rev Gastroenterol Hepatol. (2018) 15:349–64. doi: 10.1038/S41575-018-0009-6
23. Xu J, Liu C, Shi K, Sun X, Song C, Xu K, et al. Atractyloside-A ameliorates spleen deficiency diarrhea by interfering with tlr4/myd88/nf-kappab signaling activation and regulating intestinal flora homeostasis. Int Immunopharmacol. (2022) 107:108679. doi: 10.1016/J.Intimp.2022.108679
24. Juarez-Fernandez M, Goikoetxea-Usandizaga N, Porras D, Garcia-Mediavilla MV, Bravo M, Serrano-Macia M, et al. Enhanced mitochondrial activity reshapes A gut microbiota profile that delays Nash progression. Hepatology. (2023) 77:1654–69. doi: 10.1002/Hep.32705
25. Hammerich L, Tacke F. Hepatic inflammatory responses in liver fibrosis. Nat Rev Gastroenterol Hepatol. (2023) 20:633–46. doi: 10.1038/S41575-023-00807-X
26. Tripathi M, Singh BK, Zhou J, Tikno K, Widjaja A, Sandireddy R, et al. Vitamin B(12) and folate decrease inflammation and fibrosis in Nash by preventing syntaxin 17 homocysteinylation. J Hepatol. (2022) 77:1246–55. doi: 10.1016/J.Jhep.2022.06.033
27. Tacke F, Puengel T, Loomba R, Friedman SL. An integrated view of anti-inflammatory and antifibrotic targets for the treatment of Nash. J Hepatol. (2023) 79:552–66. doi: 10.1016/J.Jhep.2023.03.038
28. Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular actions of Pparalpha in lipid metabolism and inflammation. Endocr Rev. (2018) 39:760–802. doi: 10.1210/Er.2018-00064
29. Vandenbeek R, Khan NP, Estall JL. Linking metabolic disease with the pgc-1alpha gly482ser polymorphism. Endocrinology. (2018) 159:853–65. doi: 10.1210/En.2017-00872
30. Zhou S, You H, Qiu S, Yu D, Bai Y, He J, et al. A new perspective on nafld: focusing on the crosstalk between peroxisome proliferator-activated receptor alpha (Pparalpha) and farnesoid X receptor (Fxr). BioMed Pharmacother. (2022) 154:113577. doi: 10.1016/J.Biopha.2022.113577
31. Ye Q, Liu Y, Zhang G, Deng H, Wang X, Tuo L, et al. Deficiency of gluconeogenic enzyme pck1 promotes metabolic-associated fatty liver disease through pi3k/akt/pdgf axis activation in male mice. Nat Commun. (2023) 14:1402. doi: 10.1038/S41467-023-37142-3
32. Yan T, Luo Y, Yan N, Hamada K, Zhao N, Xia Y, et al. Intestinal peroxisome proliferator-activated receptor alpha-fatty acid-binding protein 1 axis modulates nonalcoholic steatohepatitis. Hepatology. (2023) 77:239–55. doi: 10.1002/Hep.32538
33. De Souza AT, Dai X, Spencer AG, Reppen T, Menzie A, Roesch PL, et al. Transcriptional and phenotypic comparisons of Ppara knockout and sirna knockdown mice. Nucleic Acids Res. (2006) 34:4486–94. doi: 10.1093/Nar/Gkl609
34. Li G, Brocker CN, Xie C, Yan T, Noguchi A, Krausz KW, et al. Hepatic peroxisome proliferator-activated receptor alpha mediates the major metabolic effects of wy-14643. J Gastroenterol Hepatol. (2018) 33:1138–45. doi: 10.1111/Jgh.14046
35. Paumelle R, Haas JT, Hennuyer N, Bauge E, Deleye Y, Mesotten D, et al. Hepatic Pparalpha is critical in the metabolic adaptation to sepsis. J Hepatol. (2019) 70:963–73. doi: 10.1016/J.Jhep.2018.12.037
36. Yang Z, Yu Y, Sun N, Zhou L, Zhang D, Chen H, et al. Ginsenosides rc, as A novel sirt6 activator, protects mice against high fat diet induced nafld. J Ginseng Res. (2023) 47:376–84. doi: 10.1016/J.Jgr.2020.07.005
Keywords: nuclear receptor, lipid metabolism, non-alcoholic steatohepatitis, inflammatory response, PPARα
Citation: Huang J, Li J, Peng Y, Cui T, Guo J, Duan S, Zhou K, Huang S, Chen J, Yi Q, Qiu M, Chen T, Wu X, Ma C, Zhang Z, Zheng Y, Tang X, Pang Y, Zhang L, Zhong C and Gao Y (2024) The lack of PPARα exacerbated the progression of non-alcoholic steatohepatitis in mice with spleen deficiency syndrome by triggering an inflammatory response. Front. Immunol. 15:1381340. doi: 10.3389/fimmu.2024.1381340
Received: 03 February 2024; Accepted: 14 March 2024;
Published: 03 April 2024.
Edited by:
Zhenhua Li, Jinan University, ChinaReviewed by:
Xinchen Wang, Zhaoqing University, ChinaCopyright © 2024 Huang, Li, Peng, Cui, Guo, Duan, Zhou, Huang, Chen, Yi, Qiu, Chen, Wu, Ma, Zhang, Zheng, Tang, Pang, Zhang, Zhong and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Gao, Z2FveW9uZ0BnenVjbS5lZHUuY24=; Chong Zhong, WmhvbmdjaG9uZ0BnenVjbS5lZHUuY24=; Lei Zhang, c3RlZmFuaWUwMzA0QHNpbmEuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.