
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 10 June 2024
Sec. Viral Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1380982
This article is part of the Research TopicImmunogenetics in times of COVID-19 pandemicView all 20 articles
Introduction: Viral infections have been implicated as a risk factor for laryngeal cancer. Given the possible effects of Corona virus disease 2019 (COVID-19) on the laryngeal tissue, we investigated the causal link between COVID-19 and laryngeal cancer using a two-sample Mendelian randomization (MR) approach.
Methods: We utilized genetic data from the 5th Genome-wide association studies (GWAS) edition of the COVID-19 Host Genetics Initiative (published on January 18, 2021) and a large-scale laryngeal cancer GWAS comprising 180 cases and 218,612 controls of European ancestry. We applied inverse variance weighting, MR Egger, and weighted median methods to infer causality. We performed sensitivity analysis using the “leave-one-out” method to verify robustness.
Results: We found no evidence of a causal association between gene-predicted COVID-19 and laryngeal cancer [Odds ratio (OR)=0.24 (95% Confidence intervals (CI), 0.05–1.26), P=0.09]. However, we observed significant inverse associations between gene-predicted COVID-19 hospitalization [OR=0.51 (95% CI, 0.28–0.95), P=0.03] and severe patients [OR=0.62 (95% CI, 0.43–0.90), P=0.01] and laryngeal cancer. Notably, the study detected important genetic variants, such as rs13050728, that modulate the expression of interferon alpha receptor 2 (IFNAR2), indicating possible roles for immune response pathways in both COVID-19 and cancer.
Discussion: This study reveals a potential interaction between COVID-19 severity, genetic factors, and laryngeal cancer, underscoring the importance of investigating the immune response mechanisms in both conditions. These findings contribute to the understanding of the complex interactions between COVID-19 and laryngeal cancer and may guide future research on the role of immune response, particularly involving IFNAR2.
Corona virus disease 2019 (COVID-19), characterized by a range of febrile and respiratory symptoms with potentially lethal risks caused by SARS-CoV-2, has garnered widespread global attention since its initial report. After three years of a global pandemic, COVID-19 has resulted in over 700 million infections, leading to millions of deaths (1–3). While the vast majority of COVID-19 patients have recovered, some individuals still experience Post-COVID-19 symptoms, indicating its long-term effects on various organ functions (4).
Laryngeal cancer primarily manifests as squamous cell carcinoma within the larynx. The global annual incidence rate stands at 2.76 per 100,000 inhabitants, with higher prevalence among men than women. Its causes are linked to smoking, alcohol consumption, exposure to physical and chemical stimuli, and variations in hormone secretion levels (5, 6). As a malignant tumor localized in the larynx, laryngeal cancer obstructs patients’ upper respiratory tracts and esophagus, leading to difficulties in breathing and swallowing. Simultaneously, it affects patients’ voices, resulting in hoarseness and articulation challenges (7). Additionally, viral infections, particularly human papilloma virus (HPV), constitute significant risk factors for laryngeal cancer. Studies suggest an approximate 28% HPV infection rate among patients with laryngeal cancer. HPV, upon infecting the larynx, may induce gene mutations and disruptions in gene expression through various mechanisms, thereby causing cell cycle dysregulation, abnormal proliferation, and ultimately leading to the development of laryngeal cancer (8). Furthermore, several studies indicate that HPV-positive patients exhibit a more favorable prognosis than HPV-negative patients, underscoring the role of viral infections in oropharyngeal cancer development and treatment (9–11).
Given that COVID-19 is a viral infection closely associated with the respiratory system, there is curiosity regarding its potential impact on the occurrence of laryngeal cancer through analogous effects. Several case reports have indicated that COVID-19 can prompt acute laryngitis and epiglottitis, hinting at potential adverse effects on laryngeal function (12–14). Previous studies have also highlighted a notable correlation between COVID-19 and the incidence of diverse cancers (15, 16). This has prompted investigations into the potential correlation between COVID-19 and laryngeal cancer. However, laryngeal cancer is a condition with a multifaceted etiology, and conventional randomized controlled studies are susceptible to confounding variables and reverse causation. These studies are also time-consuming and resource-intensive. Hence, this study opts for Mendelian randomization (MR), leveraging genetic variation as an instrumental variable based on Mendel’s law of inheritance. MR helps circumvent confounding factors and reverse causation, allowing for the investigation of the relationship between COVID-19 and laryngeal cancer (17).
In this study, COVID-19 served as the exposure factor and was categorized based on severity: non-hospitalized patients, hospitalized patients, and those with severe respiratory symptoms. Laryngeal cancer was selected as the outcome factor and investigated using the MR methodology. MR studies hinge on three critical assumptions: association, independence, and exclusivity (18). Our study assumes: (i) a significant association between the instrumental variables (IV, i.e., Single Nucleotide Polymorphism, SNPs) and COVID-19 patients; (ii) independence of SNPs from all confounders of COVID-19 and laryngeal cancer; (iii) the instrumental variables solely impacting laryngeal cancer through their associations with COVID-19 (Figure 1).
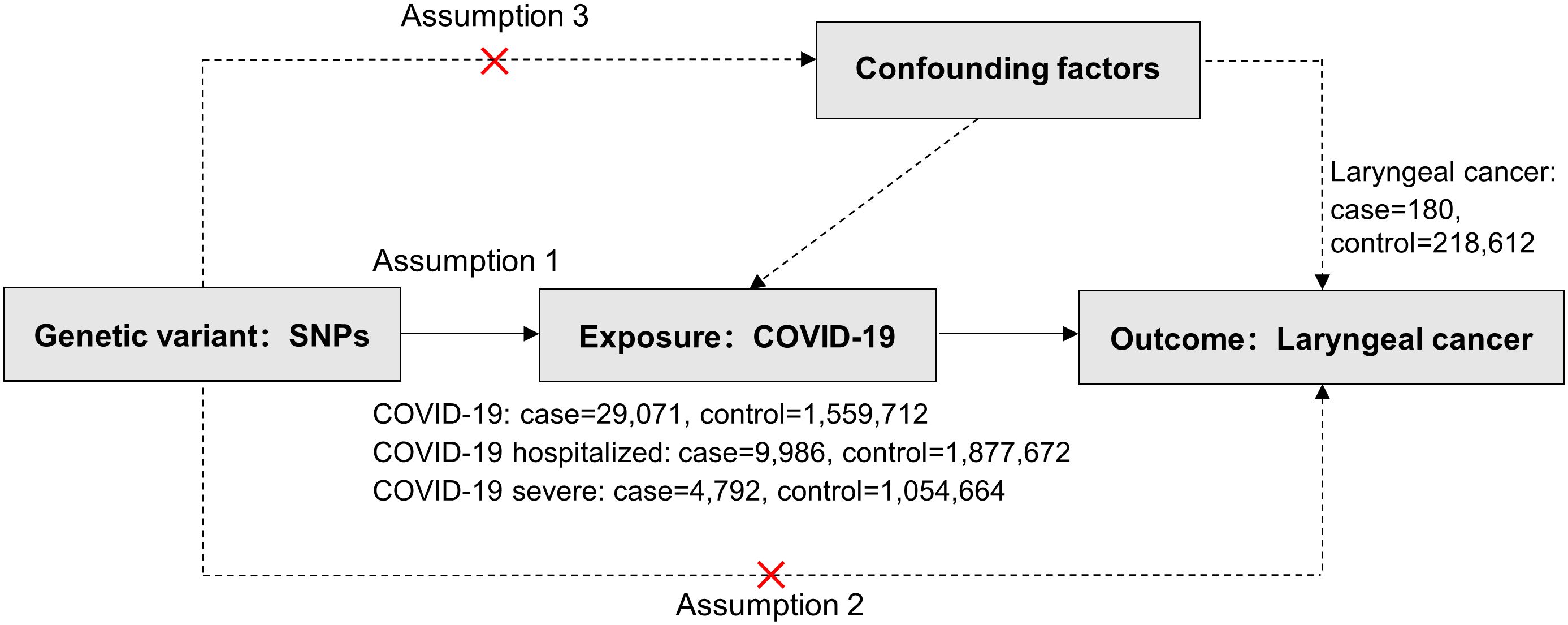
Figure 1 Three critical assumptions of COVID-19 on Laryngeal cancer via Mendelian randomization. Three distinct assumptions are illustrated by three paths. Assumption 1: The association of SNPs with COVID-19 (the exposure). Assumption 2: The SNPs influence laryngeal cancer solely through COVID-19 (as the exposure) and not through any alternative causal pathways. Assumption 3: The SNPs are entirely independent of potential confounding factors that might influence both COVID-19 and laryngeal cancer.
Genetic association summary statistics for COVID-19 risk were derived from COVID-19 Genome-wide association studies (GWAS) version 5, released on January 18, 2021, per the 2020 COVID-19 Host Genetics Initiative (19). This database included three phenotypes to explore correlations: non-hospitalized (29,071 patients, 1,559,712 healthy controls), hospitalized (9,986 patients, 1,877,672 healthy controls), and severe respiratory symptom patients (4,792 patients, 1,054,664 healthy controls) from European populations (Supplementary Table S1). Ethical approvals were obtained from the original study.
The laryngeal cancer GWAS dataset was sourced from the FinnGen Biobank’s 5th round of data collection in Finland (20), encompassing 180 patients and 218,612 healthy control European subjects (Supplementary Table S1). Ethical approvals were obtained from the original study.
Significant SNPs (P<5×10-8) related to COVID-19 from the entire genome of 1,000 individuals were compiled. The R2 threshold was set to 0.001, and kilobase pairs to 10,000 to eliminate linkage disequilibrium interference. Heterogeneity tests removed significantly divergent SNPs, yielding valid SNPs associated with COVID-19 as instrumental variables. An F value >10 indicated the absence of weak instrumental variable bias (, where N is the sample size, K is the number of SNPs, and R2 is the variance proportion explained by the SNPs) (21). R2 was calculated as , where EAF represents the frequency of the effect allele, β denotes the allele effect value, and SD stands for the standard deviation (22).
MR analysis, an effective epidemiological tool, mitigates confounding factors and reverse causality. This study primarily employed inverse variance-weighted (IVW), MR-Egger regression, and weighted median methods for analysis. Utilizing these approaches, the causal impact of the exposure on the outcome was computed by determining the ratio of the SNP associated with the exposures (Wald ratio estimate). IVW weighs each instrumental variable’s inverse variance, providing the standard MR analysis result (23). MR-Egger considers the intercept term in regression and uses inverse variance as weights (24). Weighted median estimator (WME) yields consistent causality estimates if at least half of the valid instruments are available (25). IVW results hold more significance than MR-Egger and WME methods. All analyses were carried out utilizing R version 4.3.1, and the MR analysis was performed using version 0.5.7 of the “TwoSampleMR” package.
The heterogeneity test assessed differences between individual IVs, highlighting significant differences as considerable heterogeneity. The pleiotropy test evaluated horizontal pleiotropy in multiple IVs, primarily using MR-Egger’s intercept term. A non-zero intercept suggests horizontal pleiotropy (26).
This study utilized the “Leave-one-out” method to assess result reliability. Eliminating each IV in turn and comparing the resulting total against the initial MR result determined whether an IV had a non-specific effect on estimation results (27).
In the present study, MR analyses of COVID-19 and laryngeal cancer were conducted using primarily three methods: MR-Egger, IVW, and WME. COVID-19 patients were categorized into three phenotypes based on severity: non-hospitalized, inpatients, and those with severe respiratory symptoms. Figure 2 illustrates the effect estimates of the MR analyses regarding the association between COVID-19 and laryngeal cancer. We utilized IVW, MR-Egger, simple mode, weighted median, and weighted mode-based estimations to assess the causal associations between genetically predicted COVID-19 severity and laryngeal cancer (Figure 3).

Figure 2 Mendelian randomization estimates from instrument variants for COVID-19 on laryngeal cancer. Depicts the causal relationships between COVID-19, hospitalized COVID-19 and severe COVID-19, and laryngeal cancer through the utilization of five distinct MR methods.
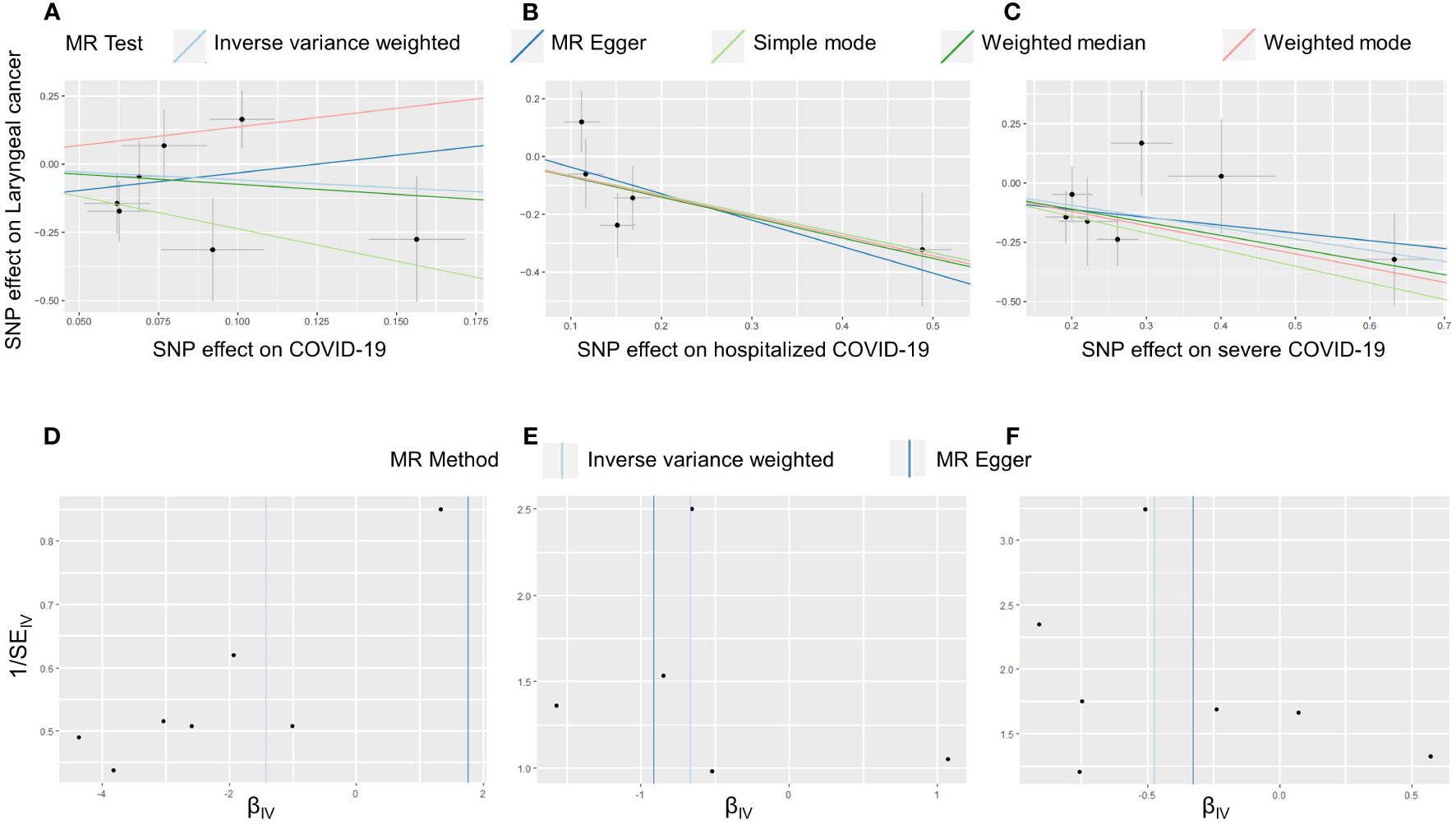
Figure 3 Causal effect of COVID-19 on laryngeal cancer. Scatter plots and funnel plots of Mendelian randomization analyses for the effect of COVID-19 (A, D), hospitalized COVID-19 (B, E) and severe COVID-19 (C, F) on laryngeal cancer. (A–C) The association between the SNP effect size estimate for COVID-19 (on the x-axis) and the corresponding effect size estimate for laryngeal cancer outcomes (on the y-axis) is illustrated. The x-axis trait signifies the exposure, the y-axis trait signifies the outcome, and each intersection point denotes an instrumental variant. The dots and brief lines traversing the dots indicate the effect sizes (B) and the 95% confidence intervals (CI) of COVID-19 concerning the outcomes of laryngeal cancer. (D–F) The funnel plots show the inverse variance weighted MR estimate of COVID-19 with Laryngeal cancer versus 1/standard error.
For the non-hospitalized COVID-19 patient group, 7 SNP loci were selected as instrumental variables (Supplementary Table S2). The IVW method’s results indicated an Odds ratio (OR) of 0.24 (95% Confidence intervals (CI), 0.05–1.26) with a P-value of 0.09, suggesting no correlation between non-hospitalized COVID-19 patients and laryngeal cancer (Figure 2; Table 1). The Cochran Q test showed no heterogeneity (P=0.13), and the MR-Egger intercept test demonstrated no horizontal pleiotropy (P=0.29) (Table 1).
In the COVID-19 inpatient group, 5 SNP loci were chosen as instrumental variables (Supplementary Table S2). The IVW method revealed an OR of 0.51 (95% CI, 0.28–0.95) with a P-value of 0.03, suggesting an inverse correlation between COVID-19 inpatients and laryngeal cancer (Figure 2; Table 1). The Cochran Q test showed no heterogeneity (P=0.29), and the MR-Egger intercept test indicated no horizontal multiplicity (P=0.70) (Table 1).
Within the COVID-19 patients exhibiting severe respiratory symptoms, 7 SNP loci were utilized as instrumental variables (Supplementary Table S2). The IVW method showed an OR of 0.62 (95% CI, 0.43–0.90) with a P-value of 0.01, suggesting a significant inverse correlation between COVID-19 patients with severe respiratory symptoms requiring hospitalization and laryngeal cancer (Figure 2; Table 1). The Cochran Q test indicated no heterogeneity (P=0.64), and the MR-Egger intercept test revealed no horizontal multiplicity (P=0.74) (Table 1).
The causal effect of each SNP associated with COVID-19 and laryngeal cancer was shown in Supplementary Figure S1. Leave-one-out sensitivity analysis was performed for each group of COVID-19. Results indicated that after individually removing each SNP, the remaining SNP analysis results closely mirrored the initial analysis, suggesting robust and unbiased data (Figure 4).
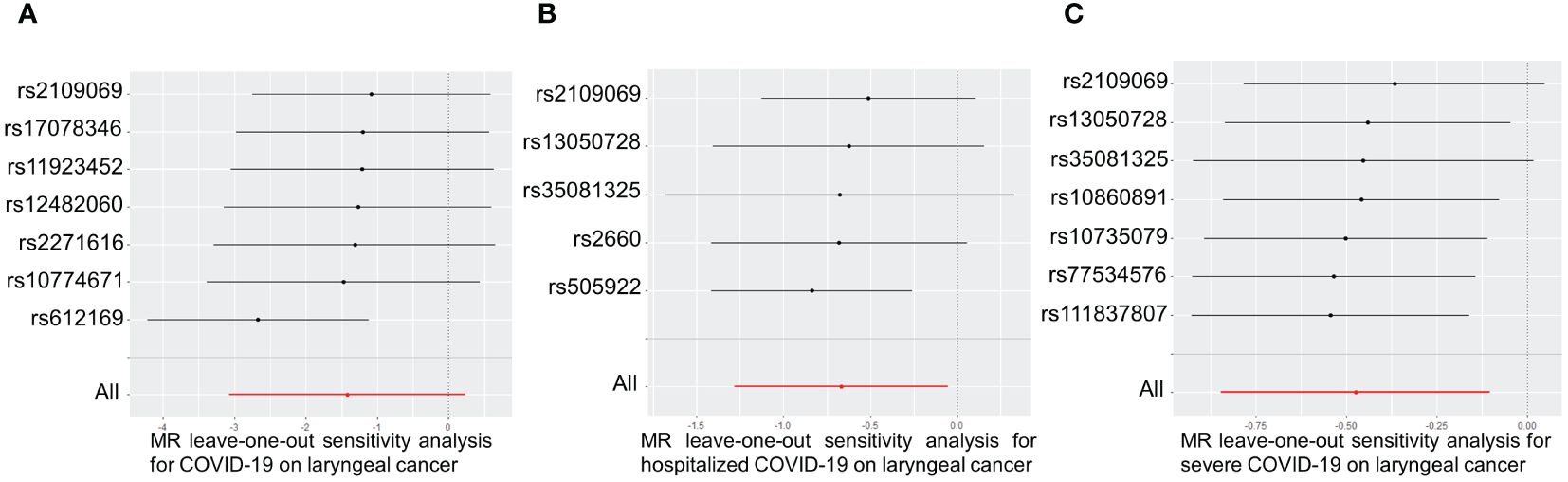
Figure 4 Leave-one-out of SNPs associated between COVID-19 with laryngeal cancer. Leave-one-out of SNPs associated between COVID-19 (A), hospitalized COVID-19 (B) and severe COVID-19 (C) with laryngeal cancer. Each black point represents the outcome of the Inverse Variance Weighted (IVW) MR method, estimating the causal effect by excluding a specific SNP from the analysis. Each red point represents the IVW estimate using all SNPs.
It has been demonstrated that COVID-19, as a viral disease, can cause acute inflammation of the larynx and epiglottis (12–14). This suggests that SARS-CoV-2 might have the potential to infect the larynx or pharynx similarly to HPV (8). Patients co-infected with HPV and afflicted with laryngeal or oropharyngeal cancer have also exhibited favorable responses to immunotherapy and chemotherapy (10, 11). HPV infection has been suggested as a prognostic indicator for laryngeal and oropharyngeal cancer. Moreover, as a global pandemic, COVID-19 has left many patients with various degrees of post-recovery sequelae, highlighting the potential long-term effects of COVID-19 infection on organ function (4). This has prompted our interest in investigating the correlation between the severity of COVID-19 infection and laryngeal cancer, with the aim of utilizing MR to explore whether COVID-19 infection can serve as a predictive factor for the onset and prognosis of laryngeal cancer. Few MR studies have explored the relationship between the severity of COVID-19 infection and the risk of various cancers, including laryngeal and oropharyngeal cancers (15, 16). However, these studies have not extensively investigated the specific association between COVID-19 and laryngeal cancer. Our research aims to fill this gap by comprehensively examining the correlation between COVID-19 and laryngeal cancer, while also analyzing potential pathways that may influence this relationship.
Despite HPV being a significant risk factor for oropharyngeal cancer, studies consistently show better outcomes among HPV-positive patients, highlighting viral infections’ pivotal role in oropharyngeal cancer’s development and treatment (9–11). Interestingly, we found similar effect of COVID-19 on laryngeal cancer. Our study reveals a significant correlation between both COVID-19 hospitalized patients (OR=0.51, 95% CI: 0.28–0.95, P=0.03) and patients with severe respiratory symptoms (OR=0.62, 95% CI: 0.43–0.90, P=0.01) and laryngeal cancer, indicating a possible protective role against laryngeal cancer. Such results raise the question of why a disease widely impacting the respiratory system could act as a protective factor for laryngeal cancer. Thus, we commenced our investigation with SNP loci associated with COVID-19. Specifically, five SNP loci related to COVID-19 hospitalized patients and seven SNP loci linked to COVID-19 patients with severe respiratory symptoms were chosen as instrumental variables, with three overlapping loci: rs13050728, rs2109069, rs35081325. These three SNPs are believed to elucidate the correlation between COVID-19 and laryngeal cancer.
Previous research (28) has highlighted the strong connection between a genetic marker, rs13050728, and severe COVID-19 cases, influencing the expression of a key immune gene called interferon alpha receptor 2 (IFNAR2). This gene’s role in both COVID-19 and cancer is being explored in this study. Specifically, the C allele of rs13050728, identified in this research, demonstrates a robust link with heightened IFNAR2 expression, potentially impacting immune response mechanisms (28).
Type I interferons (IFNs), pivotal in combating infections and tumors, have been extensively studied (29–32). In the context of COVID-19, delays in the body’s IFN response have been observed, leading to more severe symptoms in affected individuals (28, 33–36). Interestingly, individuals with severe COVID-19 cases and certain cancer patients display escalated levels of IFNAR2 (37, 38). Multiple studies have also confirmed that high expression of IFNAR2 has a significant impact on the good prognosis of immunotherapy, and has a positive impact on patient survival rate and survival time (39–41). This study underscores IFNAR2’s vital role in immune responses to infections and tumors in both diseases. We propose that interference with IFN α and IFN β hinders type I interferon response, impacting COVID-19 severity and increasing IFNAR2 expression. Maintaining high expression of IFNAR2 and restoring normal levels of type I interferon can effectively inhibit the proliferation of tumor cells, thereby reducing the risk of laryngeal cancer or improving the treatment effect of laryngeal cancer patients during IFN treatment.
Laryngeal cancer, often linked to viral infection, paradoxically exhibits improved prognoses in viral-infection-positive cases. A study assessing anti-PD-1 treatment in nasopharyngeal carcinoma patients who received the COVID-19 vaccine reported higher objective response and disease control rates than the non-vaccinated subgroup (42). Meanwhile, certain related studies have suggested that COVID-19 vaccine administration could enhance the therapeutic efficacy of PD-1 inhibitors in various cancers (43, 44). These studies also suggest that COVID-19 vaccination leads to an increase in CD4+ T cell levels in patients, potentially aiding in the activation of CD4+ T cells and reshaping the tumor microenvironment (43). HPV-positive laryngeal cancer patients display extended survival periods and enhanced sensitivity to treatments like immunotherapy and chemotherapy (45–48). This suggests viral infections both pose a risk for laryngeal cancer and positively impact its prognosis, aligning with our findings. We hypothesize that viral infections activate the immune system, potentially influencing a better prognosis.
Use of MR in our study, employing a European population and the largest COVID-19 GWAS, ensured robustness and minimized confounding factors (19). We validated our selected SNPs as instrumental variables, observing no significant heterogeneity or horizontal pleiotropy. However, limitations exist: (i) the novelty of the COVID-19 outbreak might not reflect short-term outcomes in laryngeal cancer, and high mortality rates early in the pandemic may introduce bias; (ii) while statistical methods addressed current issues, alternative pathways influenced by instrumental variables in laryngeal cancer cannot be entirely ruled out; (iii) due to the limited availability of publicly accessible GWAS data on COVID-19 patients and laryngeal cancer, it is currently not feasible to assess the impact of gender and age on their correlation. Further investigation into COVID-19-related genes and their impact on patients is crucial.
In summary, our research suggests that the exacerbation of COVID-19 infection may be an important factor in reducing the risk of laryngeal cancer, and the increase in IFNAR2 levels may play a significant role in this. This inference underscores the significant role of the type I interferon response in mitigating the risk of laryngeal cancer, thereby suggesting that interferon-related immunotherapy holds substantial promise for both the prevention and treatment of this condition. Furthermore, such immunotherapy interventions may offer considerable potential for reducing tumor staging preoperatively and prolonging survival postoperatively among patients with laryngeal cancer.
In conclusion, genes associated with COVID-19 development might hinder tumor development, potentially lowering laryngeal cancer risk or improving prognoses in severe COVID-19 cases post-immunotherapy. Detailed mechanisms warrant exploration through additional studies.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
Due to the utilization of published studies and publicly available databases, no additional ethical approval from an institutional review board was necessary.
HW: Data curation, Formal analysis, Methodology, Validation, Writing – original draft, Writing – review & editing. NF: Data curation, Methodology, Supervision, Validation, Writing – review & editing. PM: Writing – original draft, Writing – review & editing. RJ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. XW: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study received funding from the General Program of the National Natural Science Foundation of China (No. 82071031).
We are sincerely grateful to the original GWAS researchers and participants for their invaluable contributions. The extensive data collection and management were pivotal for this study, and we deeply appreciate the tremendous effort and dedication of all individuals involved in making these invaluable resources accessible for research purposes.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1380982/full#supplementary-material
COVID-19, Corona virus disease 2019; HPV, Human papilloma virus; MR, Mendelian randomization; IV, Instrumental variables; SNP, Single Nucleotide Polymorphism; GWAS, Genome-wide association studies; IVW, Inverse variance-weighted; WME, Weighted median estimator; OR, Odds ratio; CI, Confidence intervals; IFNAR2, Interferon alpha receptor 2; IFNs, Interferons.
1. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. (2020) 109:102433. doi: 10.1016/j.jaut.2020.102433
2. Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L, et al. Presumed asymptomatic carrier transmission of COVID-19. Jama. (2020) 323:1406–7. doi: 10.1001/jama.2020.2565
3. Wang H, Paulson KR, Pease SA, Watson S, Comfort H, Zheng P, et al. Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020–21. Lancet. (2022) 399:1513–36. doi: 10.1016/S0140-6736(21):02796-3
4. Kalia R, Kalia R, Musih J, Cubelo M, Popat J. Post-COVID-19 syndrome: A novel diagnosis. Cureus J Med Sci. (2022) 14:e28266. doi: 10.7759/cureus.28266
5. Nocini R, Molteni G, Mattiuzzi C, Lippi G. Updates on larynx cancer epidemiology. Chin J Cancer Res. (2020) 32:18–25. doi: 10.21147/j.issn.1000-9604.2020.01.03
6. Wunsch Filho V. The epidemiology of laryngeal cancer in Brazil. Sao Paulo Med J Rev Paulista Med. (2004) 122:188–94. doi: 10.1590/S1516-31802004000500002
7. Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA: A Cancer J Clin. (2016) 67:31–50. doi: 10.3322/caac.21386
8. Yang D, Shi Y, Tang Y, Yin H, Guo Y, Wen S, et al. Effect of HPV infection on the occurrence and development of laryngeal cancer: A review. J Cancer. (2019) 10:4455–62. doi: 10.7150/jca.34016
9. Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clinic Proc. (2016) 91:386–96. doi: 10.1016/j.mayocp.2015.12.017
10. Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal D, El-Naggar A, Garden AS, et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol. (2014) 32:3365–73. doi: 10.1200/JCO.2014.55.1937
11. Rieckmann T, Tribius S, Grob TJ, Meyer F, Busch C-J, Petersen C, et al. HNSCC cell lines positive for HPV and p16 possess higher cellular radiosensitivity due to an impaired DSB repair capacity. Radiother Oncol. (2013) 107:242–6. doi: 10.1016/j.radonc.2013.03.013
12. Kimura Y, Hirabayashi E, Yano M, Fujitani S, Shioiri S. COVID-19 Omicron variant-induced laryngitis. Auris Nasus Larynx. (2023) 50:637–40. doi: 10.1016/j.anl.2022.08.007
13. Iwamoto S, Sato MP, Hoshi Y, Otsuki N, Doi K. COVID-19 presenting as acute epiglottitis: A case report and literature review. Auris Nasus Larynx. (2023) 50:165–8. doi: 10.1016/j.anl.2021.12.007
14. Asaoka M, Chubachi S, Yamada Y, Fukunaga K. A rare case of COVID-19 infection with laryngeal involvement. BMJ Case Rep. (2021) 14:e242426. doi: 10.1136/bcr-2021-242426
15. Li J, Bai H, Qiao H, Du C, Yao P, Zhang Y, et al. Causal effects of COVID-19 on cancer risk: A Mendelian randomization study. J Med Virol. (2023) 95:e28722. doi: 10.1002/jmv.28722
16. Li Z, Wei Y, Zhu G, Wang M, Zhang L. Cancers and COVID-19 risk: A mendelian randomization study. Cancers (Basel). (2022) 14:2086. doi: 10.3390/cancers14092086
17. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization. Jama. (2021) 326:1614–21. doi: 10.1001/jama.2021.18236
18. Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol. (2015) 30:543–52. doi: 10.1007/s10654-015-0011-z
19. The COVID-19 Host Genetics Initiative. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. (2020) 28:715–8. doi: 10.1038/s41431-020-0636-6
20. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. (2023) 613:508–18. doi: 10.1038/s41586-023-05837-8
21. Zhou S, Zhu G, Xu Y, Gao R, Zhang M, Zeng Q, et al. Mendelian randomization study on the causal effect of chickenpox on dementia. J Med Virol. (2022) 95:e28420. doi: 10.1002/jmv.28420
22. Boehm FJ, Zhou X. Statistical methods for Mendelian randomization in genome-wide association studies: A review. Comput Struct Biotechnol J. (2022) 20:2338–51. doi: 10.1016/j.csbj.2022.05.015
23. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. (2013) 37:658–65. doi: 10.1002/gepi.21758
24. Slob EAW, Groenen PJF, Thurik AR, Rietveld CA. A note on the use of Egger regression in Mendelian randomization studies. Int J Epidemiol. (2017) 46:2094–7. doi: 10.1093/ije/dyx191
25. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
26. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
27. Gronau QF, Wagenmakers E-J. Limitations of bayesian leave-one-out cross-validation for model selection. Comput Brain Behav. (2018) 2:1–11. doi: 10.1007/s42113-018-0011-7
28. Gaziano L, Giambartolomei C, Pereira AC, Gaulton A, Posner DC, Swanson SA, et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med. (2021) 27:668–76. doi: 10.1038/s41591-021-01310-z
29. Sun Y, Wang G, Wang R, Ren L, Yuan Z, Liu Y, et al. Serum levels of type I interferon (IFN-I) is associated with the severity of COVID-19. J Med Microbiol. (2023) 72:001694. doi: 10.1099/jmm.0.001694
30. Schreiber G. The role of type I interferons in the pathogenesis and treatment of COVID-19. Front Immunol. (2020) 11:595739. doi: 10.3389/fimmu.2020.595739
31. Nagano. Synergistic antitumor effect of interferon-ß with gemcitabine in interferon-α-non-responsive pancreatic cancer cells. Int J Oncol. (2011) 38:1237–43. doi: 10.3892/ijo
32. Shyu RY, Su HL, Yu JC, Jiang SY. Direct growth suppressive activity of interferon-alpha and -gamma on human gastric cancer cells. J Surg Oncol. (2000) 75:122–30. doi: 10.1002/(ISSN)1096-9098
33. Pahl MC, Le Coz C, Su C, Sharma P, Thomas RM, Pippin JA, et al. Implicating effector genes at COVID-19 GWAS loci using promoter-focused Capture-C in disease-relevant immune cell types. Genome Biol. (2022) 23:125. doi: 10.1186/s13059-022-02691-1
34. Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID-19. Nature. (2020) 591:92–8. doi: 10.1038/s41586-020-03065-y
35. Salman AA, Waheed MH, Ali-Abdulsahib AA, Atwan ZW. Low type I interferon response in COVID-19 patients: Interferon response may be a potential treatment for COVID-19. BioMed Rep. (2021) 14:43. doi: 10.3892/br
36. Goncalves D, Mezidi M, Bastard P, Perret M, Saker K, Fabien N, et al. Antibodies against type I interferon: detection and association with severe clinical outcome in COVID-19 patients. Clin Transl Immunol. (2021) 10:e1327. doi: 10.1002/cti2.1327
37. Fricke-Galindo I, Martinez-Morales A, Chavez-Galan L, Ocana-Guzman R, Buendia-Roldan I, Perez-Rubio G, et al. IFNAR2 relevance in the clinical outcome of individuals with severe COVID-19. Front Immunol. (2022) 13:949413. doi: 10.3389/fimmu.2022.949413
38. Vitale G, van Eijck CHJ, van Koetsveld Ing PM, Erdmann JI, Speel EJM, van der Wansem Ing K, et al. Type I interferons in the treatment of pancreatic cancer. Ann Surgery. (2007) 246:259–68. doi: 10.1097/01.sla.0000261460.07110.f2
39. Furuya N, Kamai T, Shirataki H, Yanai Y, Fukuda T, Mizuno T, et al. Serum interferon alpha receptor 2 mRNA may predict efficacy of interferon alpha with/without low-dose sorafenib for metastatic clear cell renal cell carcinoma. Cancer Immunol Immunother. (2011) 60:793–808. doi: 10.1007/s00262-011-0989-3
40. Ota H, Nagano H, Doki Y, Sekimoto M, Kondo M, Wada H, et al. Expression of type I interferon receptor as a predictor of clinical response to interferon-α therapy of gastrointestinal cancers. Oncol Rep. (2006) 16:249–55. doi: 10.3892/or
41. Tomimaru Y, Eguchi H, Wada H, Tomokuni A, Kobayashi S, MArubashi S, et al. Synergistic antitumor effect of interferon-β with gemcitabine in interferon-α-non-responsive pancreatic cancer cells. Int J Oncol. (2011) 38:1237–43. doi: 10.3892/ijo.2011.954
42. Hua YJ, Liu YL, Wen K, Kurts C, Wu H, Mei Q, et al. Potentially improved response of COVID-19 vaccinated nasopharyngeal cancer patients to combination therapy with anti-PD-1 blockade and chemotherapy. Ann Oncol. (2023) 34:121–3. doi: 10.1016/j.annonc.2022.10.002
43. Qian Y, Zhu Z, Mo YY, Zhang Z. COVID-19 vaccination is associated with enhanced efficacy of anti-PD-(L)1 immunotherapy in advanced NSCLC patients: a real-world study. Infect Agent Cancer. (2023) 18:50. doi: 10.1186/s13027-023-00526-7
44. Mei Q, Hu G, Yang Y, Liu B, Yin J, Li M, et al. Impact of COVID-19 vaccination on the use of PD-1 inhibitor in treating patients with cancer: a real-world study. J Immunother Cancer. (2022) 10:e004157. doi: 10.1136/jitc-2021-004157
45. Bates JE, Morris CG, Hitchcock KE, Dziegielewski PT, Mendenhall WM, Amdur RJ. Locally advanced hypopharyngeal and laryngeal cancer: Influence of HPV status. Radiother Oncol. (2019) 140:6–9. doi: 10.1016/j.radonc.2019.05.013
46. Shaughnessy JN, Farghaly H, Wilson L, Redman R, Potts K, Bumpous J, et al. HPV: A factor in organ preservation for locally advanced larynx and hypopharynx cancer? Am J Otolaryngol. (2014) 35:19–24. doi: 10.1016/j.amjoto.2013.08.006
47. Tong F, Geng J, Yan B, Lou H, Chen X, Duan C, et al. Prevalence and prognostic significance of HPV in laryngeal squamous cell carcinoma in Northeast China. Cell Physiol Biochem. (2018) 49:206–16. doi: 10.1159/000492858
Keywords: COVID-19, laryngeal cancer, Mendelian randomization, IFNAR2, immune response, viral infections
Citation: Wang H, Fang N, Mozumder P, Jiang R and Wang X (2024) Exploring the protective association between COVID-19 infection and laryngeal cancer: insights from a Mendelian randomization study. Front. Immunol. 15:1380982. doi: 10.3389/fimmu.2024.1380982
Received: 02 February 2024; Accepted: 27 May 2024;
Published: 10 June 2024.
Edited by:
Aranzazu Mediero, Health Research Institute Foundation Jimenez Diaz (IIS-FJD), SpainReviewed by:
Jun-Feng Cao, Chengdu Medical College, ChinaCopyright © 2024 Wang, Fang, Mozumder, Jiang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richeng Jiang, anJjMTIyNEBqbHUuZWR1LmNu; Xin Wang, eHdhbmc5OUBqbHUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.