
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 08 April 2024
Sec. Molecular Innate Immunity
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1380089
A correction has been applied to this article in:
Corrigendum: Immunomodulatory effects of a probiotic combination treatment to improve the survival of Pacific oyster (Crassostrea gigas) larvae against infection by Vibrio coralliilyticus
Introduction: The culture of Pacific oysters (Crassostrea gigas) is of significant socio-economic importance in the U.S. Pacific Northwest and other temperate regions worldwide, with disease outbreaks acting as significant bottlenecks to the successful production of healthy seed larvae. Therefore, the current study aims to describe the mechanisms of a probiotic combination in improving the survival of C. gigas larvae. Specifically, we investigate changes in C. gigas larval gene expression in response to V. coralliilyticus infection with or without a pre-treatment of a novel probiotic combination.
Methods: Treatment groups consisted of replicates of Pacific oyster larvae exposed to a) a combination of four probiotic bacteria at a total concentration of 3.0 x 105 CFU/mL at 18 hours post-fertilization (hpf), b) pathogenic V. coralliilyticus RE22 at a concentration of 6.0 x 103 CFU/mL at 48 hpf, and c) the probiotic combination at 18 hpf and V. coralliilyticus RE22 at 48 hpf. RNA was extracted from washed larvae after 72 hpf, and transcriptome sequencing was used to identify significant differentially expressed genes (DEGs) within each treatment.
Results: Larvae challenged with V. coralliilyticus showed enhanced expression of genes responsible for inhibiting immune signaling (i.e., TNFAIP3, PSMD10) and inducing apoptosis (i.e., CDIP53). However, when pre-treated with the probiotic combination, these genes were no longer differentially expressed relative to untreated control larvae. Additionally, pre-treatment with the probiotic combination increased expression of immune signaling proteins and immune effectors (i.e., IL-17, MyD88). Apparent immunomodulation in response to probiotic treatment corresponds to an increase in the survival of C. gigas larvae infected with V. coralliilyticus by up to 82%.
Discussion: These results indicate that infection with V. coralliilyticus can suppress the larval immune response while also prompting cell death. Furthermore, the results suggest that the probiotic combination treatment negates the deleterious effects of V. coralliilyticus on larval gene expression while stimulating the expression of genes involved in infection defense mechanisms.
Today’s aquaculture production satisfies half of the global seafood demand, and this proportion is increasing as harvests of wild fish and shellfish populations reach maximum sustainable yields (1). In 2021, commercial oyster production in the U.S. totalled 23.9 million pounds, valued at 222.5 million USD (2). Hatcheries that rear oyster larvae and supply oyster farmers with seed periodically face disease outbreaks caused by pathogenic bacteria. Infections by pathogenic Vibrio spp. (vibriosis) are primarily responsible for mass mortalities, which can cause significant bottlenecks in oyster larval production and subsequent economic losses (3).
Several Vibrio species have been reported to cause disease outbreaks in bivalve aquaculture, including V. aestuarianus, V. coralliilyticus, V. splendidus, V. tapetis, V. tasmaniensis, and V. tubiashii (4). Vibrio coralliilyticus is one of the most prominent pathogens in this industry, being linked to numerous mortality events and showing virulence towards a range of host species, including the Pacific oyster (Crassostrea gigas), Eastern oyster (Crassostrea virginica), Kumamoto oyster (Crassostrea sikamea), and Geoduck clams (Panope abrupta) (3, 5–8). Symptoms of vibriosis in larvae are often characterized by a rapid decline in larval motility, detachment of the velum, and soft tissue necrosis (9, 10). Although clinical symptoms have been observed, the oyster larvae’s immune and inflammatory response following pathogenic infection are not well understood.
As invertebrates, oysters deploy an innate immune response to protect themselves from pathogenic bacteria, viruses, and parasites (11). Pattern recognition receptors (PRRs) bind to pathogen-associated molecular patterns (PAMPs), stimulating downstream signalling pathways. Several immune signalling pathways may be employed to produce immune effector molecules, cytokines, and antimicrobial peptides (12). Unfortunately, even with a defence response, oyster larvae are still found to be highly susceptible to pathogens (13).
Methods for preventing bacterial infections in oyster larvae typically include maintaining good water quality (i.e., frequent water changes, ozonisation, ultra-violet radiation treatment) and, occasionally, adding antibiotics (14). Unfortunately, water treatments are usually not effective enough to reduce the impacts of pathogenic bacteria, and antibiotic treatments are often prohibited due to environmental concerns (15, 16). Thus, researching alternative disease control methods, such as probiotics, is a priority. In recent years, several putative probiotics have been investigated to protect oyster larvae from pathogenic bacteria (17–21). In addition, a new probiotic combination treatment introduced by Madison et al. (22) improved the relative percent survival of C. gigas larvae by 65.95% when challenged with V. coralliilyticus. Additionally, exposure to the probiotic combination significantly improved size and settlement rates of C. gigas larvae (22). The authors suggested a form of immune priming as one potential mechanism responsible for these changes. This study builds on this preliminary work and aims to characterize the defence response of C. gigas larvae exposed to V. coralliilyticus, either with or without pre-exposure to a combination of these novel probiotics.
Three of the four probiotic strains used in this study (B11, DM14, and D16) were previously isolated and evaluated as a single combination treatment (22). The DM14 and D16 isolates were identified as different Pseudoalteromonas spp., and B11 as an Epibacterium sp (22). The fourth strain (ASW1) was later isolated from larvae cultured in autoclaved seawater using methods outlined previously (22). Genetic identification of ASW1 was completed using 16s rRNA gene sequencing and the NCBI’s BLAST suite (23). The 16S rRNA sequence was submitted to GenBank (Accession OQ595186). ASW1, in combination with B11, DM14, and D16, was evaluated for efficiency in improving the survival of C. gigas larvae challenged with Vibrio coralliilyticus using well-plate assays as described by Madison et al. (22). To obtain isolates for larval assays, cells from glycerol stocks stored at -80°C were streaked on plates with seawater-based Luria-Bertani agar (LBSw; 10 g of tryptone, 5 g of yeast extract, and 15 g of agar, per liter of 10 µm filtered seawater) and incubated at 25°C for 48 hours. Colonies were grown in 5 mL of LBSw broth (10 g of tryptone, 5 g of yeast extract per liter of filtered seawater) at 25°C and agitated at 40 RPM on a roller drum (New Brunswick TC-7; New Brunswick Scientific, Enfield, CT, USA) for 24 hours. Cultures were then washed twice with autoclaved seawater by centrifuging at 3900 x g for 5 min and resuspended in autoclaved seawater. The optical density was measured at 600 nm (OD600) (Beckman DU 530, Beckman Coulter, Brea, CA, USA). Assuming that an OD600 measurement of 1.0 equalled approximately 8.0 x 108 CFU/mL, bacterial cultures were appropriately diluted to obtain target concentrations of 6.0 x 103 CFU/mL for V. coralliilyticus and 7.5 x 104 CFU/mL for each probiotic strain of the combination treatment used in larval challenge experiments.
Adult C. gigas were provided by the Molluscan Broodstock Program (MBP) at Oregon State University’s Hatfield Marine Science Centre. Oysters from genetically unrelated families were chosen for spawning. Oysters were held for two to four weeks in a conditioning system at 20°C to induce gametogenesis and continuously fed on a 50/50 (by cell concentration) mixed algal diet of Isochrysis galbana and Chaetoceros neogracile (total concentration of 10,000 cells/mL). When sexually mature, oysters were strip-spawned, and resulting gametes were fertilized following aseptic techniques (24). Larvae were hatched in sterile 250 mL Erlenmeyer flasks containing autoclaved seawater at 25°C, 32 ± 2 ppt salinity, and a pH of 8.2 ± 0.1. The seawater used to hatch larvae was pumped from the Yaquina Bay, Newport, passed through sand filters followed by 10-µm bag filters, and aerated with carbon-dioxide-stripped air overnight to adjust the pH before autoclaving. After the seawater was autoclaved, it was agitated and aerated on a shaker table (Benchmark, Sayreville, NJ, USA) at 80 RPM for 16 to 24 hours before adding fertilized eggs.
Our previous survival assays evaluated the effects of various probiotic combinations and concentrations in protecting developing C. gigas larvae against Vibrio coralliilyticus (22 and unpublished data). In this present study, the effects of the timing of probiotic additions on larval survival were evaluated by dosing larvae with the probiotics at different times post-fertilization of the eggs.
Following egg fertilization, embryos were transferred from a concentrated egg suspension in a single sterile 1 L beaker to sterile 250 mL flasks containing autoclaved seawater at a final concentration of 50 embryos per mL. This concentration allowed for the rearing of sufficient numbers of healthy normal larvae for the assays while considering handling constraints. The flasks were agitated and aerated on a shaker table at 50 RPM throughout the culturing the of larvae. Treatment groups were designated by the timing of probiotic addition. Probiotics were provided to larvae at 2 (2 hr PB Only), 6 (6 hr PB Only), 12 (12 hr PB Only), 18 (18 hr PB Only), or 24 hpf (24 hr PB Only). The four probiotics (ASW1, B11, DM14, D16) were added to larval cultures as a combination treatment at a concentration of 7.5 x 104 CFU/mL per probiotic strain (22). Each of these five probiotic treatment groups was cultured in triplicate flasks. Twenty-four hours post-fertilization (hpf), D-larvae from each flask were poured onto a sterile 40 µm sieve, rinsed with autoclaved seawater, and resuspended in probiotic-free autoclaved seawater, except for treatment groups receiving probiotics at 24 hpf. This 24 hpf treatment represented a “standard” method used in our earlier studies (22), whereby the probiotics were added to the larval cultures at 24 hpf, but the larvae were not resuspended in probiotic-free autoclaved seawater before the addition of V. coralliilyticus. Therefore, by adding the probiotic combination at time points before 24 hpf, we investigated if the probiotic additions could influence the larvae at different points early in larval development. The transfer of larvae into probiotic-free autoclaved seawater also limits potential interactions between the probiotics and pathogen within the culture environment and focuses on interactions within the gut of the host where infection begins.
After transferring larvae from the 2, 6, 12, and 18 hpf treatments to probiotic-free autoclaved sea water, the larvae were resuspended at a concentration of 35 larvae/mL. Then one mL of larvae suspension was transferred to each well of a 24-well plate (Figure 1A). Larval concentrations were reduced in the well plates to better compensate for the addition of bacteria and subsequent influence on abiotic factors (22). Once transferred into well plates, the 24 hpf probiotic treatment groups received the probiotic addition.
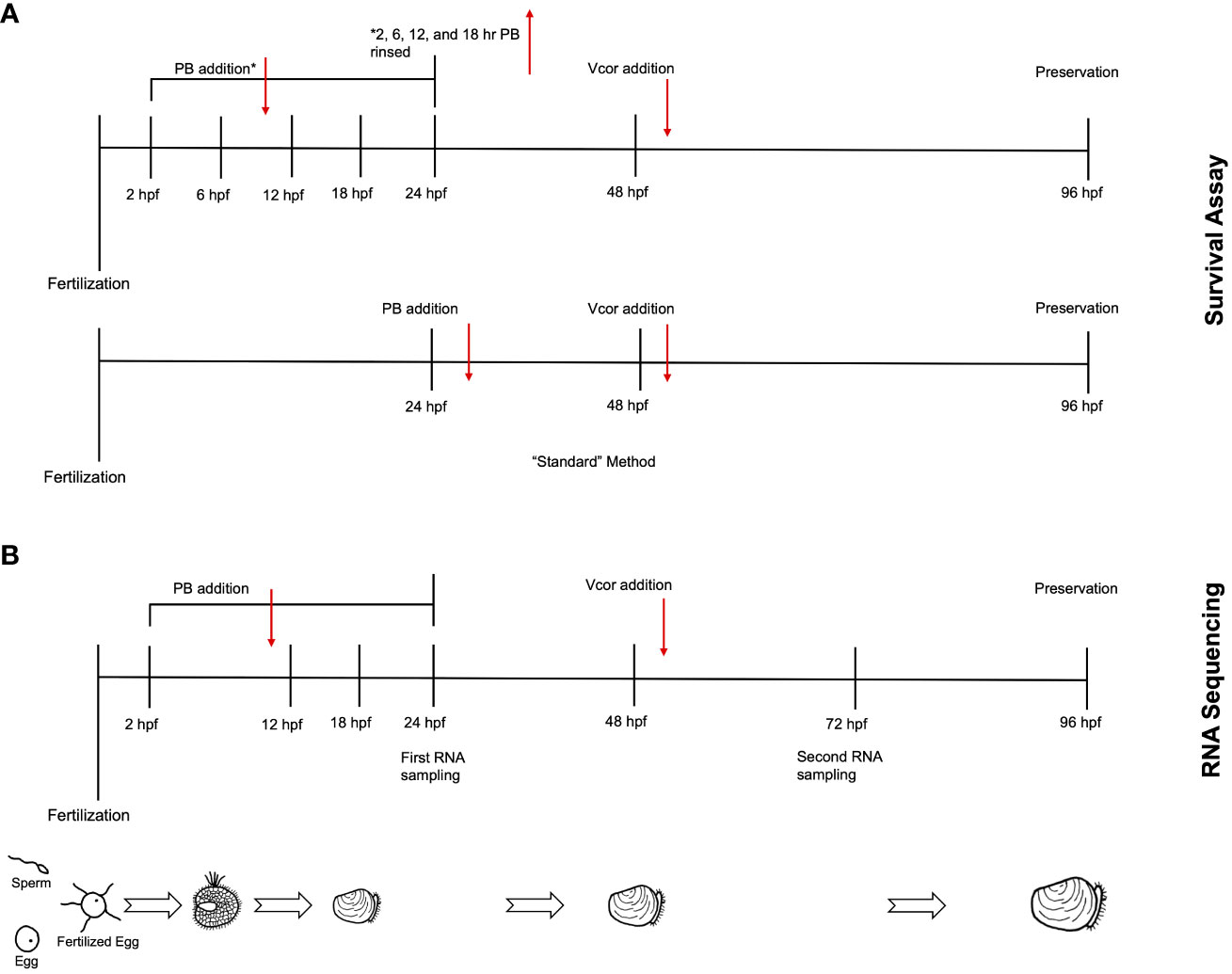
Figure 1 Experimental timeline for larval assays. (A) In the survival assay, probiotic treatment groups were designated by the timing of probiotic addition. Following fertilization, probiotics were added at either 2 (2 hr PB Only), 6 (6 hr PB Only), 12 (12 hr PB Only), or 18 hpf (18 hr PB Only). At 48 hpf, Vibrio coralliilyticus was added to probiotic-treated larvae (“2 hr PB + Vcor”, “6 hr PB + Vcor”, “12 hr PB + Vcor”, “18 hr PB + Vcor”, and “24 hr PB + Vcor”). A positive control of larvae receiving only V. coralliilyticus was inoculated at 48 hpf. An additional treatment group (24 hr PB Only) received probiotics at 24 hpf, which remained within the larvae culture until 96 hpf. (see methods). (B) For RNA sequencing, larvae were sampled from a similar yet separate experimental setup from that shown in (A) – see details in section 2.4. Larvae used for RNA extractions were collected from the “Larvae Only” control, the “Vcor Only” control, the “2 hr PB Only”, “12 hr PB Only”, “18 hr PB Only”, and the “2 hr PB + Vcor”, “12 hr PB + Vcor”, and “18 hr PB + Vcor” treatment groups at 72 hpf.
At 48 hpf, probiotic-treated larvae were challenged with 6.0 x 103 CFU/mL of V. coralliilyticus strain RE22 and incubated for 48 hours (these treatment groups were designated “2 hr PB + Vcor”, “6 hr PB + Vcor”, “12 hr PB + Vcor”, “18 hr PB + Vcor”, and “24 h PB + Vcor”). A positive control consisted of larvae infected with V. coralliilyticus alone without exposure to probiotics (Vcor Only). In addition, a negative control of larvae that received neither the probiotic treatment nor the pathogen was included (Larvae Only). Each treatment group consisted of six replicate wells (Table 1). At 96 hpf, larvae were preserved with 10 µL of 10% phosphate-buffered formalin at a pH of 8. Well-plates were observed under a 40X objective inverted light microscope. Larvae were counted as either live or dead, with larvae classified as dead, having less than 90% of their tissue remaining in their shells (22).
One liter flasks containing 500 mL autoclaved seawater were stocked with 1 x 104 embryos. This concentration was chosen to ensure that adequate amounts of live larvae were collected for RNA analysis (4). The flasks were agitated and aerated on a shaker table at 50 RPM throughout the culturing of the larvae. The experiment was conducted with four replicates per treatment group. The probiotic combination of ASW1, B11, DM14, and D16 was added to the embryos at 18 hpf (18 hr PB Only) at a total concentration of 3.0 x 105 CFU/mL. Unlike in the survival assay (Figure 1A), the larvae cultures were not rinsed and added to probiotic-free autoclaved seawater to minimize experimental effects on transcriptional changes and potential contamination and to increase the timing accuracy of bacterial exposures across all treatment groups and replicates. Vibrio coralliilyticus was then added to probiotic-treated larvae at 48 hpf (18 hr PB + Vcor). The larvae of the positive control received no probiotics, but V. coralliilyticus was added at 48 hpf (Vcor Only) (Figure 1B). A “no exogenous bacteria” negative control of larvae receiving no probiotic additions or V. coralliilyticus (Larvae Only) was maintained throughout this experiment.
All treatment and control groups were sampled at 72 hpf to ensure sufficient numbers of live larvae for RNA analysis after exposure to V. coralliilyticus. Five thousand larvae per treatment group were rinsed onto a sterile 40-µm sieve with autoclaved seawater and then rinsed with 2 mL of RNAlater (Thermo Scientific, Waltham, MA, USA). The larvae were then transferred to a microcentrifuge tube containing 1.5 mL RNAlater. In addition, samples of up to 100 larvae were collected from each of the four replicates into 10 mL shell vials (Thermo Scientific, Waltham, MA, USA) and preserved with 10% phosphate-buffered formalin (pH of 8) to evaluate survival, as described above.
Total RNA was extracted from each sample and DNase treated using the Zymo Direct-Zol RNA Microprep kit (Zymo Research, Irvine, CA, USA) with the addition of Trizol reagent (Thermo Scientific, Waltham, MA, USA). Total RNA extracted from each sample was quantified using the Qubit Broad Range RNA kit (Thermo Scientific, Waltham, MA, USA). The quality of total RNA was assessed using Agilent 2100 BioAnalyzer (Agilent, Santa Clara, CA, USA) with the Agilent RNA 6000 Nano kit (Agilent, Santa Clara, CA, USA). Typically, one of the four replicates within each treatment group had poor quality or quantity of total RNA; therefore, the three replicates from each treatment group with the highest quality and quantity of RNA were identified and prepared for sequencing.
The first step in sequence library preparation was the isolation of mRNA from the total RNA using the NEBNext Poly(A) Magnetic Isolation Module (New England BioLabs, Ipswich, MA, USA). Subsequently, complementary DNA (cDNA) libraries were constructed using the NEBNext Ultra II RNA Library Prep kit for Illumina (New England BioLabs, Ipswich, MA, USA), per the manufacturer’s instructions. Complementary DNA library quantification, normalization, and sequencing were then completed by Oregon State University’s Center for Quantitative Life Sciences (Corvallis, OR). Libraries were sequenced as 100-bp single-end reads with the NextSeq 2000. The P2 flow cell was utilized, allowing for 400 million reads to be sequenced per run. Four sequencing runs were completed, resulting in a sequencing depth of 32 million to 44 million reads per sample (Supplementary Table 1).
Raw sequencing reads were analysed for quality using FastQC 0.11.9 (25). Any identified poor-quality bases (quality scores lower than 20) and adapter sequences were removed using fastp (26). Processed reads were aligned to a C. gigas reference genome (version 1.0, Genbank GCA_902806645.1) via Hisat2 2.1.0 (27). Transcriptome assembly was performed using Stringtie (28) using default parameters. The overall alignment rate to the C. gigas reference genome using HISAT2 ranged from 45-80% for transcriptomes (Supplementary Table 1). Differential gene expression analysis was performed by comparing transcript counts between the treatment groups (“18 hr PB Only”, “18 hr PB + Vcor”, and “Vcor Only”) and the control (Larvae Only) using DESeq2, which was used also to normalize libraries to control for differences in sequence depth and remove outliers (29). Transcripts displaying a log fold change of ≥ 2 or ≤ −2 and a Benjamini-Hochberg adjusted p-value <.05 were considered significantly differentially expressed. A principal component analysis (PCA) was performed to observe the level of variation between treatment groups due to their level of differentially expressed genes. For significance testing of the PCA, a PERMANOVA was completed using the adonis2 function (30). Differentially expressed genes were annotated using KEGG’s GhostKOALA (KEGG Orthology and Links Annotation) program (31). Defence-related genes were annotated using Uniprot (32).
The “Larvae Only” negative control was used to normalize larval mortalities that were unrelated to experimental treatments. Relative percent survival (RPS) was calculated as RPS = [1-(percent mortality of treatment group/percent mortality of untreated control group)] x 100. Relative percent survival values were arcsine square root transformed before analysis. Statistical analyses were conducted using R statistical software (Version 4.0.3, R Project for Statistical Computing). Normality was assessed using the Shapiro-Wilk test and Q-Q plots. The homogeneity of variance was assessed using Levene’s test.
Nonparametric methods were used if violations of normality and variance assumptions were observed. For multiple comparisons of treatment groups, the Kruskal-Wallis one-way ANOVA was conducted. When significant differences (P <.05) were found, Dunn’s test with the Benjamini-Hochberg correction was used for pairwise comparisons. Comparisons between the two treatment groups were conducted using the Mann-Whitney U test.
In evaluating ASW1 as a potential addition to the probiotic combination treatment, ASW1 did not result in any mortalities when added to larvae at 24 hpf (Supplementary Table 2). Larvae that received ASW1 at 24 hpf and V. coralliilyticus at 48 hpf had an average relative percent survival of 76.9 ± 38.9%, which was 56.1% higher than that of larvae exposed to V. coralliilyticus alone (Supplementary Table 2). A BLASTN suite search identified ASW1 as an Alteromonas sp. based on 16S rRNA sequencing. Specifically, Alteromonas oceani was identified as the best match with a 99% query coverage and 98.71% percent identity.
Additions of the probiotic combination treatment at 2, 6, 12, 18, and 24 hpf were evaluated for the ability to reduce larval mortalities due to V. coralliilyticus. The negative control of larvae (Larvae Only) that received no bacterial additions and was used to calculate the relative percent survival yielded a survival rate of 99.7%. The positive control receiving only V. coralliilyticus (Vcor Only) resulted in an average relative percent survival of 8.7 ± 26.7% (Figure 2). When probiotics were added before pathogen exposure at 2, 6, 12, 18, or 24 hpf (PB + Vcor), the average relative percent survival significantly increased to 87.5 ± 18.9%, 89.3 ± 11.4%, 91.1 ± 12.9%, 85.9 ± 23.3%, and 83.6 ± 18.1%, respectively (P <.05) (Figure 2; Supplementary Table 3). There was no significant difference in survival among the different timings of probiotic additions (Supplementary Table 3); therefore, the 18-hour probiotic addition time was chosen as the focus for the RNA sequencing results to limit the possibility for changes in bacterial composition prior to the larvae being transferred into culture 24-well plates at 24 hpf.

Figure 2 Relative percent survival of 4-day-old D-larvae of C. gigas challenged with Vibrio coralliilyticus strain RE22 with or without probiotic supplementation. “Vcor Only” was the positive control for survival and did not receive any probiotics. A negative control (Larvae Only) did not receive any probiotics or the pathogen. Filled circles represent the average relative percent survival of replicate wells (n=18). The boxes indicate the upper and lower quartiles and the bar represents the median or middle quartile. The ends of the whiskers represent the most extreme values within the 1.5x interquartile range (IQR), and the empty circles indicate outliers. *** indicates statistical differences from “Vcor Only” at P ≤.001.
Larvae infected with V. coralliilyticus without the probiotic supplementation (Vcor Only; positive control for disease) had 267 differentially expressed genes (DEGs) compared to larvae that received no bacterial additions (Larvae Only; negative control) (Figure 3A). One hundred and twenty-nine DEGs were annotated. When the probiotics were added to larvae at 18 hpf without any pathogen (18 hr PB Only), 535 DEGs were identified, including 227 annotated DEGs (Figure 3A). The same number, though not an identical set, of DEGs were identified when V. coralliilyticus was added in addition to the probiotics (18 hr PB + Vcor) compared to the negative control (Larvae Only) (Figure 3A) (Supplementary Table 4). The “Vcor Only” treatment group had 125 total unique DEGs, 72 being annotated. Comparatively, the “18 hr PB Only” and “18 hr PB + Vcor” each had two unique annotated DEGs and two unique uncharacterized DEGs. Between all three treatment groups, a total of 141 DEGs were shared, 57 of them being annotated genes.
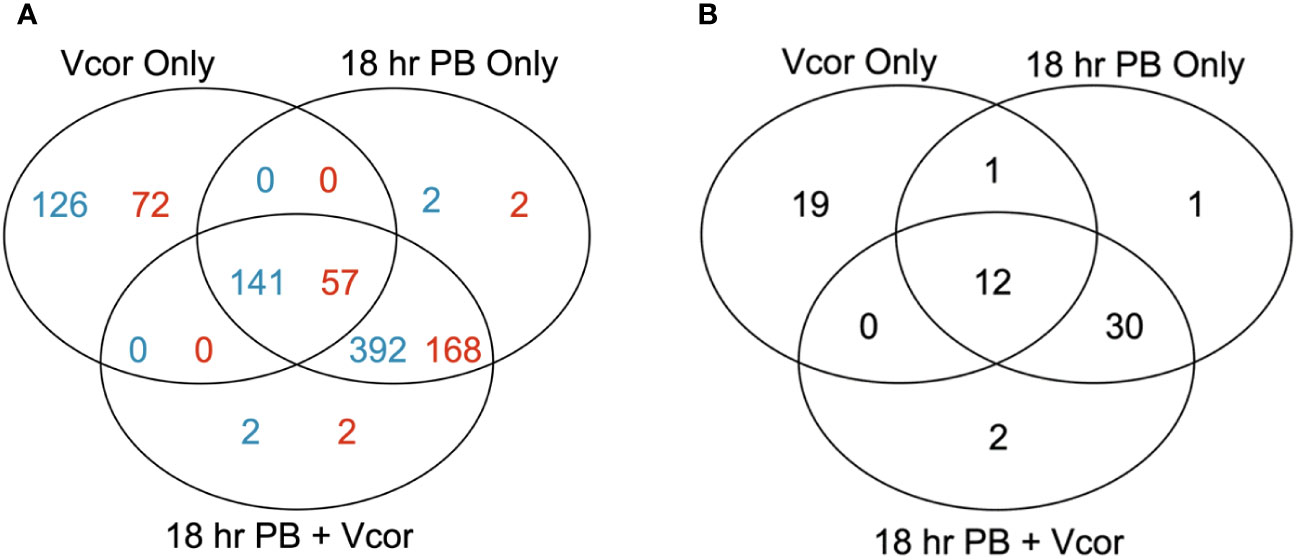
Figure 3 Numbers of differentially expressed genes in infection challenge and probiotic supplementation experiments. Venn diagrams represent the number of significant differentially expressed genes (DEGs) in each treatment group (“18 hr PB”, “18 hr PB + Vcor”, and “Vcor Only”) when compared to the “Larvae only” control. (A) The total numbers of all DEGs from each treatment, with the number of annotated genes listed in blue and the number of uncharacterized genes listed in red. (B) The total numbers of defence-related DEGs for each treatment compared to “Larvae only” controls.
When only defence-related DEGs were assessed, 32 DEGs were identified in the “Vcor Only” control compared to the “Larvae Only” control (Figure 3B). In comparison, there were 44 defence-related DEGs in both the “18 hr PB Only” and “18 hr PB + Vcor” treatment groups, 30 of which were identical (shared between both of the probiotic supplementation treatments). Conversely, the “Vcor Only” treatment group had 19 defence-related DEGs that were uniquely expressed in that treatment (Figure 3B). Twelve defence-related DEGs were shared between all three treatment groups (Supplementary Table 4).
Defence-related genes that were identified as significantly differentially expressed were organized into four categories: pattern recognition receptors, immune signalling, immune effectors, and other immune or inflammatory genes. These groups describe various components of the larva’s innate immune system that recognize foreign material and respond accordingly. For instance, recognition of bacteria is accomplished by pattern recognition receptors, which include both extra- and intracellular protein receptors. Subsequently, the immune signalling category includes genes coding for proteins that contribute to relaying a signal throughout various immune pathways involving toll-like signalling, nuclear factor- kappa B, mitogen-activated protein kinase, complement cascade, and others. Genes identified as immune effectors respond to upstream signalling and contribute to the defence response. Finally, the genes grouped into the “other” category may contribute to the defence response through other routes, such as apoptosis or phagocytosis.
Two-day-old larvae previously exposed to bacterial additions (probiotic, pathogen, or both) had an increased expression of a gene that encodes the leucine-rich repeat-containing G-protein coupled receptor 4 (LGR4), a protein that behaves as a negative regulator of toll-like receptors (Supplementary Table 5). In contrast, the toll-like receptor 6 gene (TLR6) was found to be differentially decreased in expression in response to all three bacterial exposure treatments (“Vcor Only”, “18 hr PB + Vcor”, and “18 hrs + PB Only”) (Figure 4).
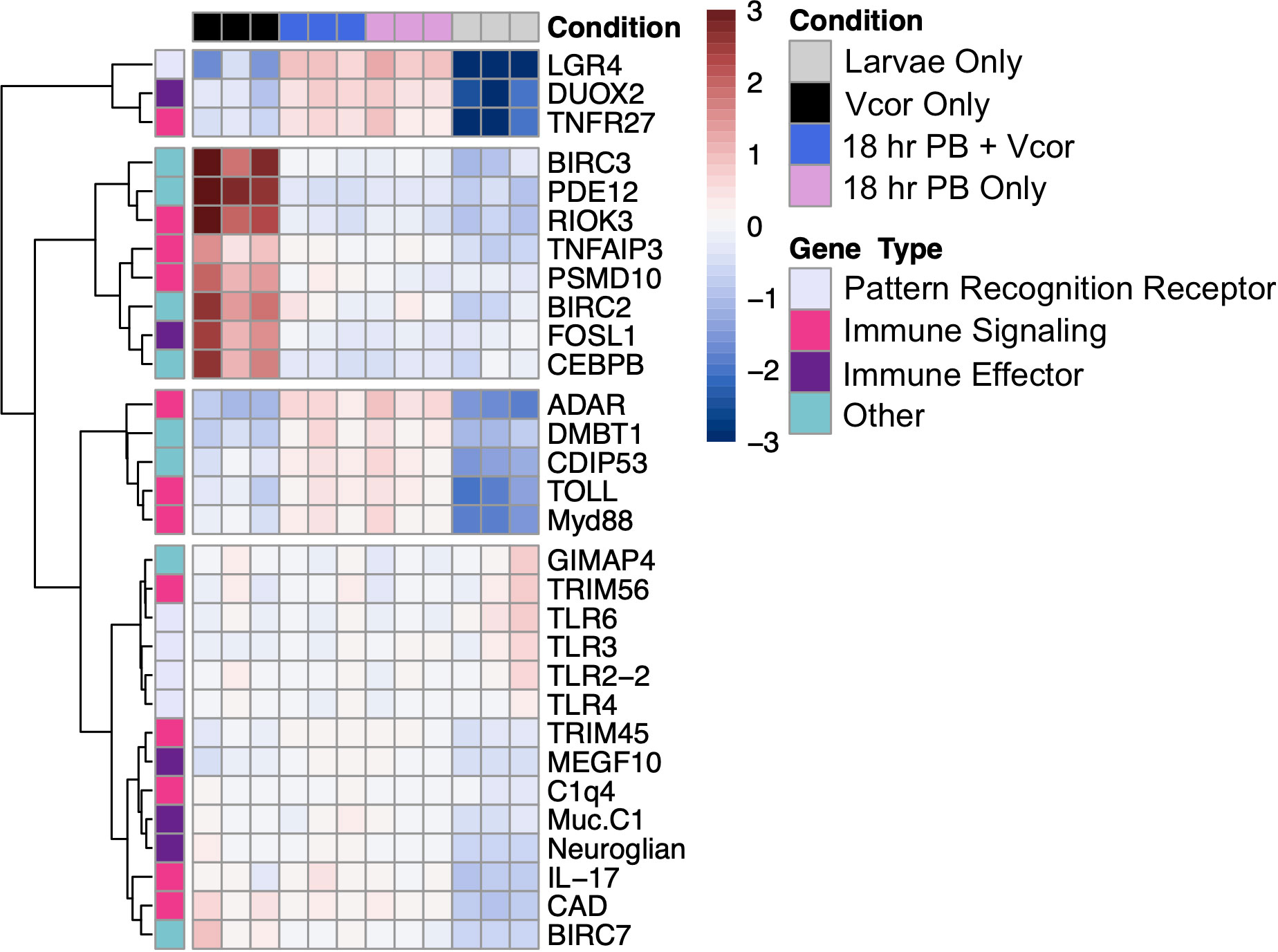
Figure 4 Differential gene expression of defence-related DEGs between the treatment groups (“Vcor Only”, “18 hr PB + Vcor”, and “18 hr PB Only”) and the negative control (Larvae Only). The treatment type is indicated at the top of the heatmap, with black signifying the “Vcor Only” infection control, blue the “18 hr PB + Vcor” treatment, pink the “18 hr PB Only” treatment, and grey representing the “Larvae Only” negative control. All samples were taken at 72 hpf, 24 hours after the addition of V. coralliilyticus to the “Vcor Only” and “18 h PB + Vcor” treatment groups. Heatmap and hierarchical clustering of selected genes is based on normalized read counts. Each column represents a single sample, with each row indicating the expression level of each gene. The colours represent the individual read count data are normalized to its average expression across all samples, with blue indicating lower than the genes’ average (decreased gene expression) and red indicating higher than the average (increased gene expression) (Gene names with corresponding abbreviations are found in Supplementary Table 6).
However, larvae that received the addition of only the probiotic combination (18 hr PB Only) showed a decreased and increased expression of the genes encoding the toll-like receptors 2 and 4 (TLR2-2, TLR4), respectively (Figure 4). In comparison, larvae exposed to V. coralliilyticus alone (Vcor Only) instead showed a decrease in the expression of the toll-like receptor 3 gene (TLR3) (Supplementary Table 5). Lastly, larvae exposed to the probiotics in addition to V. coralliilyticus (18 hr PB + Vcor) showed a similar gene expression pattern to the “18 hr PB Only” treatment group, presenting a decreased expression of the TLR2-2 gene in addition to an increased expression of the TLR4 gene (Figure 4).
The transcripts coding for interleukin-17-like protein (IL-17) and protein toll (TOLL) had elevated expression levels in all three bacterial treatment groups (“Vcor Only”, “18 hr PB Only”, “18 hr PB + Vcor”) compared to the “Larvae Only” group (Supplementary Table 5). Genes responsible for producing other immune signalling molecules showed higher expression levels in all three treatment groups, including tumour necrosis factor receptor superfamily member 27 (TNFR27) and Cis-aconitate decarboxylase (CAD), indicating the activation and regulation of NF-kB and MAPK pathways (Figure 4).
In larvae treated with the probiotic combination (18 hr PB Only), transcripts for the myeloid differentiation primary response protein (MyD88) were found with increased expression levels compared to the “Larvae Only” control. Despite the increased expression of positive regulators of the NF-kB signalling pathway, the tripartite motif-containing protein 45-like (TRIM45) gene, a repressor of the NF-kB pathway, showed higher expression levels in the “18 hr PB Only” treatment group relative to the “Larvae Only” group (Supplementary Table 5). Alternatively, the E3 ubiquitin-protein ligase TRIM56-like (TRIM56) gene showed decreased expression in the probiotic-treated larvae (18 hr PB Only) (Figure 4). Additionally, the genes encoding the double-stranded RNA-specific adenosine deaminase (ADAR) protein and the complement C1q-like protein 4 (C1q4), involved in the cytosolic DNA-sensing and complement pathways, respectively, were found with higher expression in probiotic-treated larvae (18 hr PB Only) compared to the control group (Larvae Only) (Figure 4).
Larvae exposed to V. coralliilyticus (Vcor Only) had increased expression of the toll-like signalling regulator, tumour necrosis factor alpha-induced protein 3-like protein (TNFAIP3) relative to the “Larvae Only” control (Figure 4). Furthermore, other genes involved in signal regulating or contributing to signal transduction were differentially expressed in the “Vcor Only” control. For example, the 26S proteasome non-ATPase regulatory subunit 10 (PSMD10) and the serine/threonine-protein kinase RIO3 (RIOK3) both had an increased expression in V. coralliilyticus-infected larvae, compared to the “Larvae Only” control (Figure 4).
Larvae receiving both probiotic and pathogenic bacteria (18 hr PB + Vcor) had similar immune signalling gene expression profiles to the “18 hr PB Only” treatment group. For example, the TRIM56 gene exhibited decreased expression while the genes encoding MyD88, TRIM45, ADAR, and C1q4 all showed differentially increased expression in the “18 hr PB + Vcor” treatment group compared to the “Larvae Only” control (Figure 4).
Bacterial exposure influenced the gene expression of multiple immune effectors in all three treatment groups. For example, transcripts for the cell-surface mucin, integumentary mucin C.1 protein (Muc.C1) were elevated in expression in all treatment groups (“Vcor Only”, “18 hr PB Only”, and “18 hr PB + Vcor”) when compared to the “Larvae Only” control (Figure 4; Supplementary Table 5). Similarly, several transcripts coding for the multiple epidermal growth factor-like domains protein 10 (MEGF10) and the dual oxidase 2 protein (DUOX2) had elevated expression in all treatment groups (Supplementary Table 4).
In contrast, larvae treated with probiotics alone (18 hr PB Only) exhibited differential expressions of transcripts coding for the mucin 2 (Muc2) and mucin 5AC (Muc5AC) proteins. Additionally, the serine protease inhibitor Cvsi-2 gene (Cvsi-2) showed an elevated expression in the probiotic-treated larvae compared to the “Larvae Only” control group (Supplementary Table 4).
The “Vcor Only” treatment group resulted in the increased expression of the fos-related antigen-1 (FOSL1) gene when compared to the “Larvae Only” control. In contrast, the “18 hr PB + Vcor” treatment group did not result in differential expression of the FOSL1 gene (Supplementary Table 4).
Larvae receiving bacterial additions (“Vcor Only”, “18hr PB Only”, and “18 hr PB + Vcor”) resulted in the increased expression of the cell-death inducing p53 (CDIP53) and the neuroglian genes when compared to the “Larvae Only” control (Supplementary Table 5). However, other genes involved in the regulation of apoptosis, including cell-death abnormality protein 1 (CDAP1) and the GTPase IMAP family members 4 and 7-like proteins (GIMAP4, GIMAP 7), had a decreased gene expression in the “18hr PB Only” treatment group when compared to the “Larvae Only” control (Supplementary Table 4). Additionally, the malignant brain tumours 1 protein gene (DMBT1) had an increased expression in the “18hr PB Only” treatment group compared to the “Larvae Only” control (Figure 4).
The differential expression of several inflammatory proteins was found to be unique to the “Vcor Only” treatment group. This included increased gene expression of the baculoviral IAP repeat-containing proteins 2, 3, and 7 (BIRC2, BIRC3, and BIRC7), the 2’,5’-phosphodiesterase 12 (PDE12), and the CCAAT/enhancer-binding protein beta (CEBPB) in pathogen-challenged larvae compared to the “Larvae Only” negative control (Figure 4).
Larvae exposed to both the probiotic and pathogenic bacteria (18 hr PB + Vcor) exhibited gene expressions comparable to that of the “18 hr PB Only” treatment group. For example, the CDAP1, GIMAP4, and GIMAP7 genes showed decreased expression levels, while the DMBT1 gene showed increased expression levels compared to the “Larvae Only” control (Figure 4).
This study describes gene expression changes and survival of three- and four-day-old C. gigas larvae after exposure to probiotic and/or pathogenic bacteria. Larvae infected with V. coralliilyticus survive better when pre-treated with the probiotic combination (22). The current study supports this even when the pre-treated larvae were rinsed and transferred into probiotic-free autoclaved seawater. Hence, these results suggest a mechanism of action aside from direct inhibition of the pathogen by competitive exclusion. However, the results from the RNA sequencing were produced from larvae that were not rinsed and returned to probiotic-free seawater before infection. Therefore, inhibition and exclusion of the pathogen via direct interaction with the probiotics should not be discounted from interpretation.
Our data are compatible with mechanisms whereby probiotics may mitigate the effects of pathogenic infection of the larvae through immune stimulation, enhanced cellular barrier function, and reduced inflammation. We hypothesize that immune priming may be the primary mechanism responsible for the beneficial effects of the probiotic treatment observed here. Our treatments resulted in differential gene expression similar to those found in other studies that have exposed adult oysters to poly(I:C), which mimics viral double-stranded DNA to study immune priming (33, 34). Genes with similar expression patterns include the double-stranded RNA-specific adenosine deaminase (ADAR), tripartite motif-containing proteins (TRIM45, TRIM56), toll-like receptors (TLRs), tumour necrosis factor receptor (TNFR27), and various lectins. Overall, our results suggest that the specific probiotic combination treatment used here induces pathogen-defence mechanisms in otherwise vulnerable oyster larvae.
The addition of ASW1 to a previously identified probiotic combination treatment of three bacterial isolates (22) further improved the relative percent survival of V. coralliilyticus-challenged C. gigas larvae compared to an infection control with no probiotics. While the relative percent survival between the three-strain and the four-strain combination was not statistically different (Supplementary Table 3), the latter increased the survival of infected larvae after an exposure of only six hours.
The probiotic treatment may directly or indirectly affect immune priming, with the latter possibly resulting from the probiotics influencing the early development of the oyster gut microbiome. It has previously been observed that C. gigas larvae begin ingesting small particles around 22 hpf (unpublished data), which allows the larvae two hours to ingest the probiotic bacteria before the larvae are rinsed with autoclaved seawater.
Ultimately, more work is needed to better understand the effects of probiotic additions on the possible development of the oyster gut microbiome and to determine the role of these probiotic bacteria in stimulating immune processes. Regardless, this work highlights a potentially beneficial and practical tool for promoting the health of larvae in oyster hatcheries and provides evidence for potential explanatory mechanisms that can be experimentally tested.
The current study’s results reveal that there is a generalized response to all exogenous additions of bacteria, regardless of whether they involved probiotic or pathogenic microbes. In addition to this generalized response to Gram-negative bacteria, we observed specific gene expression responses to each different bacterial treatment (i.e., “Vcor only”, “PB only”, and “PB + Vcor”).
It is generally understood that the first step of the bivalve innate immune response to foreign materials is for pattern recognition receptors to recognize and bind bacterial cell wall components, including lipopolysaccharides and peptidoglycans (12). Accordingly, the expression of multiple pattern recognition receptors involved in toll-like signalling was influenced by bacterial exposure in the following ways: 1) larvae exposed to any exogenous bacteria decreased expression of TLR6; 2) larvae that received probiotics at 18 hpf, regardless of whether they were exposed to V. coralliilyticus or not, demonstrated an additional decreased expression of TLR2-2 and TLR4. The coordinated expression of these receptors is supported by previous reports that TLR2 cooperates with TLR6 (35) and heterodimerizes with TLR4 in response to Gram-negative bacterial exposure (36); however, our results contradict previous studies, which reported upregulation of said toll-like receptors when C. gigas were exposed to live bacterial components, including peptidoglycan and lipopolysaccharides (37–39). Furthermore, larvae exposed to pathogenic V. coralliilyticus without any probiotic addition (Vcor Only) decreased expression of TLR3. In contrast to TLR2-2, TLR4, and TLR6, TLR3 proteins are found intracellularly, localized within the endosome organelle where they recognize viral double-stranded DNA, signalling for it to be internalized and then transported to the lysosome (40). The reasons for decreased expressions of the TLR genes are unclear; however, possible explanations include the increased expression of genes that code for proteins responsible for the negative regulation of TLRs. For instance, the leucine-rich repeat-containing G-protein coupled receptor 4 (LGR4) protein is hypothesized to be a negative regulator of toll-like signalling and was found to have elevated expression in all three probiotic treatment groups (“18 hr PB Only”, “18 hr PB + Vcor”, and “Vcor Only”) (Figure 5). In addition, larvae infected with V. coralliilyticus that did not receive any probiotic addition (Vcor Only) experienced an upregulation of the toll-like signalling negative regulator, tumour necrosis factor alpha-induced protein 3-like (TNFAIP3), compared to the larvae that did not receive any bacterial additions (Larvae Only). The differential expression of this protein seems unique to the larval response to V. coralliilyticus, as it was not differentially expressed in response to the probiotics. In contrast, both probiotic and pathogen exposure increased the expression of CAD, another negative regulator of toll-like receptors. The CAD protein works as a negative regulator of toll-like receptors by stimulating the expression of TNFAIP3 via CAD-dependent production of reactive oxygen species (ROS), leading to suppressed expression of toll-like receptors (Figure 5). Previously, itaconic acid, a by-product of cis-aconitate decarboxylase, has been found to inhibit ROS produced by phagocytes, therefore, regulating the innate immune response (41, 42). However, whether itaconic acid was increased in response to bacterial additions in this study is unknown. Van Nguyen & Alfaroo (43) found that adult mussels infected with a V. coralliilyticus/neptunius-like isolate experienced significantly higher ROS levels than non-infected individuals 6, 18, and 60 hours post-infection but that itaconic acid levels did not increase until 60 hours post-infection. Therefore, it is possible that increased expression of itaconic acid could occur in infected larvae, but not before 60 hours after exposure to V. coralliilyticus (108 hours post-fertilization).
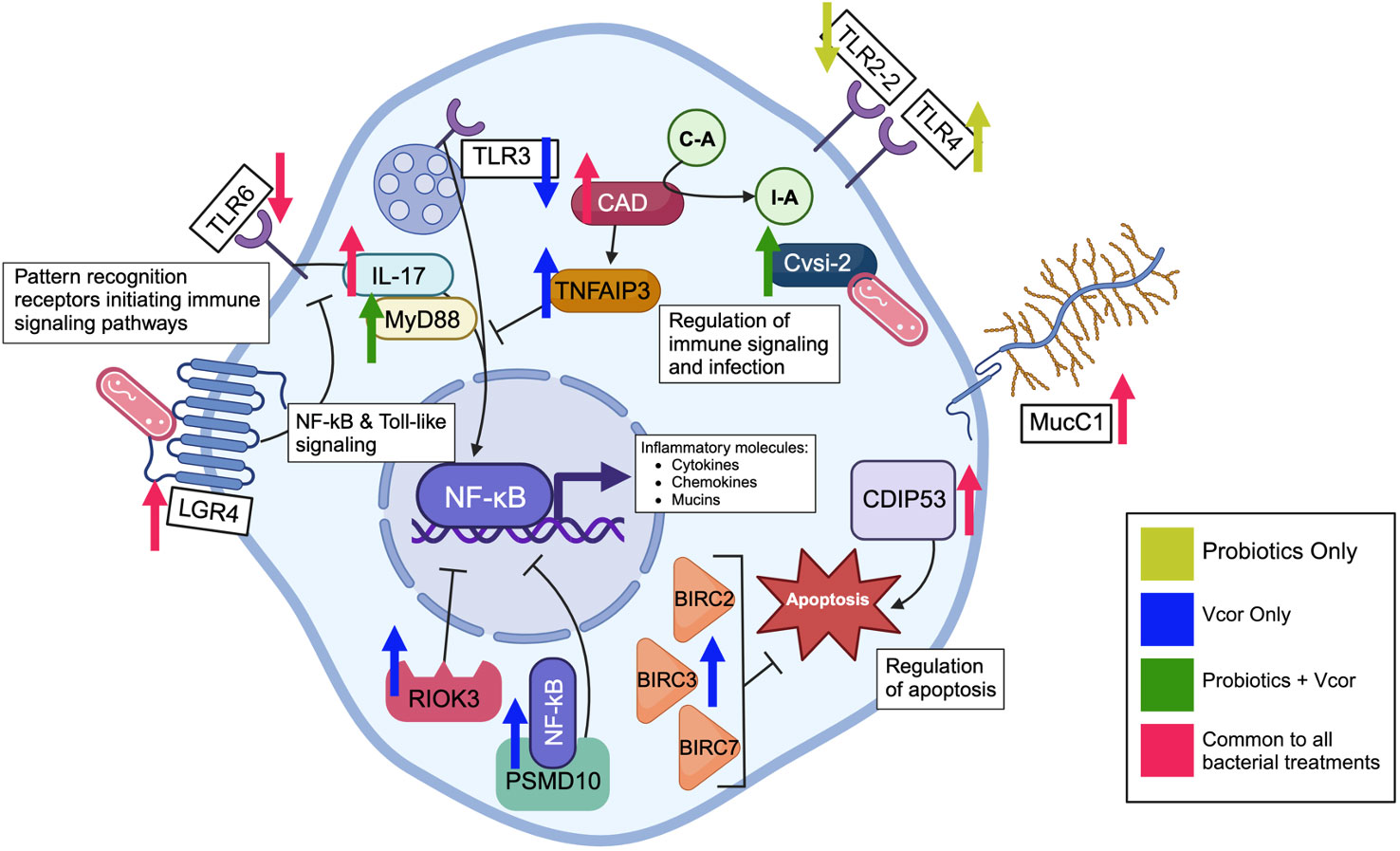
Figure 5 Defence-related genes in various cellular locations are differentially expressed in response to infection by Vibrio coralliilyticus and pre-treatment of the probiotic combination at 18 hours post-fertilization in C. gigas larvae. The arrows represent either an increase or decrease in gene expression caused by the exposure to the probiotic combination alone, V. coralliilyticus alone, V. coralliilyticus in addition to the probiotic treatment, or all three bacterial treatment groups. Pattern recognition receptors bind to the bacteria, sending an inflammatory signal through the NF-kB and toll-like signalling cascades. The activated NF-kB transcription factor leads to the production of cytokines, antimicrobial peptides, and immune effectors. Meanwhile, the tumour necrosis factor alpha-induced protein 3-like prevents activation of NF-kB transcription by inhibiting upstream signalling. The 26S proteasome non-ATPase regulatory subunit 10 binds to the NF-kB transcription factor and retains it in the cytosol as a negative regulator of the NF-kB signalling pathway. Additionally, the serine/threonine-protein kinase RIO3 inhibits NF-kB transcription, regulating inflammatory signalling. The immune effector serine protease inhibitor Cvsi2 directly interacts with endocytosed bacteria. Cell-surface mucins are produced and used as an inflammatory barrier of the cell. Apoptosis is enabled by the cell death-inducing p53-target protein 1 but inhibited by the baculoviral IAP repeat-containing proteins 2, 3, and 7.
Ultimately, even though multiple toll-like receptors had a lower expression level in the bacteria-treated larvae, downstream signalling molecules whose activation is typically induced via toll-like receptors, including interleukin-17 (IL-17), had significantly increased expression. IL-17 transcripts are expressed in the gills of Mytilus galloprovincialis in response to infection by Vibrio splendidus, suggesting a contribution to mucosal immunity (44). Additionally, IL-17 is known to be directly responsive to bacterial LPS and contributes to the activation of both myeloid differentiation primary response 88 (MyD88) and the NF-kB pathway (Figure 5). MyD88, found with elevated expression in larvae exposed to the probiotics, is directly stimulated by most toll-like receptors and plays a central role in the innate immune response, activating IL-1R associated kinases (IRAK) and subsequently the NF-kB and mitogen-activated protein kinase (MAPK) pathways (45). Therefore, MyD88 might be stimulated by long-term bacterial exposure as opposed to toll-like receptors whose expression may decline with continuous bacterial exposure (46).
In the absence of probiotics, infection by V. coralliilyticus caused an increase in expression of the 26S proteasome non-ATPase regulatory subunit 10 protein (PSMD10) and serine/threonine-protein kinase RIO3 (RIOK3), both of which have been found to inhibit NF-kB activation (Figure 5). For example, PSMD10 has been hypothesized to retain NF-kB in the cytoplasm of cells, subsequently inhibiting NF-kB activity, whereas RIOK3 has been shown to inhibit TNF-alpha and caspase-10 induced activation of the NF-kB pathway (47, 48). However, when larvae were exposed to both the probiotics and V. coralliilyticus, neither the RIOK3 nor PSMD10 genes were found to be differentially expressed. Therefore, this result may be due to the probiotics directly inhibiting V. coralliilyticus growth, virulence, or both, consequently preventing the initial upregulation of these two genes. Alternatively, some of the numerous uncharacterized DEGs influenced by the probiotics may function as regulators of RIOK3 and PSMD10. Further progress in the characterization of the C. gigas genome will be required to evaluate this possibility.
In addition to immune signalling pathways, treatment with both probiotic and pathogenic bacteria influenced the expression of various inflammation and effector molecules, including mucins. Elevated Mucin C1 (Muc.C1) expression was seen in all larvae exposed to bacteria; however, larvae treated with probiotics had particularly elevated levels of Mucin 2 (Muc2) and Mucin 5AC (Muc5AC) expression (Figure 5). These results correspond with previously identified localizations and proposed functions of each mucin. For example, Muc.C1 has been recognized as a transmembrane protein in many cell types, including human immune cells (49). Furthermore, Muc.C1 has been observed to behave as a binding site for bacterial pathogens, such as Pseudomonas aeruginosa. Following the binding of bacteria, Muc.C1 contributes to an anti-inflammatory response characterized by the inhibition of toll-like signalling (50). The resulting decreased expression of toll-like receptors identified in this study agrees with Muc.C1’s role within the anti-inflammatory process. In contrast, Muc2 and Muc5AC are mainly localized to the digestive tract and contribute to maintaining a mucosal barrier that will protect tissue surfaces and aid in removing unwanted material, including bacteria (49, 51, 52).
Aside from immune effectors, several proteins involved in apoptosis experienced differential expression due to bacterial exposure (Figure 5). For example, the cell-death-inducing p53 (CDIP53) protein had elevated expression in all bacteria-exposed larvae; this protein induces cellular apoptosis through the intrinsic pathway (53). In contrast, probiotic exposure seems to prevent apoptosis as there was a reduced expression of GTPase immune-associated proteins 4 and 7 (GIMAP4, GIMAP7) (apoptosis accelerators) and cell-death abnormality protein 1-like (a protein responsible for enabling phagocytes to engulf apoptotic cells) (54–57). Reduced apoptosis regulators suggest a reduced need for cell death as a defence mechanism. These results align with previous studies that evaluated the impacts of probiotic bacteria on bivalve larvae, including the Eastern oyster, Crassostrea virginica (4). Additionally, larvae infected with V. coralliilyticus displayed increased expression of various baculoviral IAP repeat-containing proteins (BIRCs), which are negative regulators of apoptosis. BIRCs present anti-apoptotic features by mediating multiple caspases, including caspase-3, 7, and 9 (58). The lack of self-induced apoptosis by V. coralliilyticus-infected cells might allow the pathogen to replicate and overwhelm this defence mechanism.
The current study describes the immune response to infection of C. gigas larvae by V. coralliilyticus and the modulation of this response by the application of a combination of probiotics that improved the survival of the Vibrio-challenged larvae. When larvae were pre-treated with the probiotics at a total concentration of 3.0 x 105 CFU/mL, gene expression patterns related to V. coralliilyticus exposure were suppressed, and the probiotic treatment stimulated inflammatory molecules supportive of an immune response that likely reduced the detrimental effects of V. coralliilyticus infection on larval survival. Further research should focus on the mechanics of beneficial bacteria that protect C. gigas larvae and other cultured bivalve species against microbial pathogens and improve the effectiveness of these and similar probiotic treatments in bivalve hatcheries.
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
The manuscript presents research on animals that do not require ethical approval for their study.
JH: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. RM: Conceptualization, Methodology, Supervision, Writing – review & editing, Funding acquisition. CL: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing. CS: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing, Project administration.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NOAA National Sea Grants (NA22OAR4170158 and NA18OAR4170346) awarded to CS, CL, and RM, a Pacific States Marine Fisheries Commission Award (NA18NMF4720007) awarded to CL, and an Oregon State University Agricultural Research Foundation award (ARF#9271A) awarded to CS. In addition, JH was awarded several scholarships by the Oregon State University’s Hatfield Marine Science Center in contribution to her Master’s thesis.
The authors thank MacKenna Hainey, Spencer Lunda, and the MBP staff for their support in rearing larvae, Dave Jacobson and Cristin Fitzpatrick for their help with the library preparation, MK English for the 16S rRNA gene sequencing of ASW1, Rachel Aitchison for her artistic contribution to the first figure, and Lynette Hawthorne for her administrative support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1380089/full#supplementary-material
1. Botta R, Asche F, Borsum JS, Camp EV. A review of global oyster aquaculture production and consumption. Mar Policy. (2020) 117:103952. doi: 10.1016/j.marpol.2020.103952
2. NOAA Fisheries. Landings (2024). Available online at: https://www.fisheries.noaa.gov/foss/f?p=215:200:25623307448388.
3. Dubert J, Barja JL, Romalde JL. New insights into pathogenic Vibrios affecting bivalves in hatcheries: present and future prospects. Front Microbiol. (2017) 8:762. doi: 10.3389/fmicb.2017.00762
4. Modak TH, Gomez-Chiarri M. Contrasting immunomodulatory effects of probiotic and pathogenic bacteria on eastern oyster, Crassostrea virginica, larvae. Vaccines. (2020) 4):588. doi: 10.3390/vaccines8040588
5. Elston RA, Hasegawa H, Humphrey KL, Polyak IK, Häse CC. Re-emergence of Vibrio tubiashii in bivalve shellfish aquaculture: severity, environmental drivers, geographic extent and management. Dis Aquat Organ. (2008) 82:119–34. doi: 10.3354/dao01982
6. Estes RM, Friedman CS, Elston RA, Herwig RP. Pathogenicity testing of shellfish hatchery bacterial isolates on pacific oyster Crassostrea gigas larvae. Dis Aquat Organ. (2004) 58:223–30. doi: 10.3354/dao058223
7. Gómez-León J, Villamil L, Salger SA, Sallum RH, Remacha-Triviño A, Leavitt DF, et al. Survival of eastern oysters Crassostrea virginica from three lines following experimental challenge with bacterial pathogens. Dis Aquat Organ. (2008) 79:95–105. doi: 10.3354/dao01902
8. Tubiash HS, Chanley PE, Leifson E. Bacillary necrosis, a disease of larval and juvenile bivalve mollusks I. etiology and epizootiology. J Bacteriol. (1965) 90:1036–44. doi: 10.1128/jb.90.4.1036-1044.1965
9. Ushijima B, Richards GP, Watson MA, Schubiger CB, Häse CC. Factors affecting infection of corals and larval oysters by Vibrio coralliilyticus. PLoS One. (2018) 13:e0199475. doi: 10.1371/journal.pone.0199475
10. Wang D, Loor A, Bels LD, Stappen GV, den Broeck WV, Nevejan N. Dynamic immune response to vibriosis in pacific oyster Crassostrea gigas larvae during the infection process as supported by accurate positioning of GFP-tagged Vibrio strains. Microorganisms. (2021) 9:1523. doi: 10.3390/microorganisms9071523
11. Wang L, Song X, Song L. The oyster immunity. Dev Comp Immunol. (2018) 80:99–118. doi: 10.1016/j.dci.2017.05.025
12. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
13. Song X, Wang H, Xin L, Xu J, Jia Z, Wang L, et al. The immunological capacity in the larvae of pacific oyster Crassostrea gigas. Fish Shellfish Immunol. (2016) 49:461–9. doi: 10.1016/j.fsi.2016.01.009
14. Prado S, Romalde JL, Barja JL. Review of probiotics for use in bivalve hatcheries. Vet Microbiol. (2010) 145:187–97. doi: 10.1016/j.vetmic.2010.08.021
15. Brown C. A study of two shellfish-pathogenic Vibrio strains isolated from a Long Island hatchery during a recent outbreak of disease. J Shellfish Res. (1981) 1:83–7.
16. Kang CH, Shin Y, Jang S, Yu H, Kim S, An S, et al. Characterization of Vibrio parahaemolyticus isolated from oysters in Korea: resistance to various antibiotics and prevalence of virulence genes. Mar pollut Bull. (2017) 118:261–6. doi: 10.1016/j.marpolbul.2017.02.070
17. Gibson LF, Woodworth J, George AM. Probiotic activity of Aeromonas media on the pacific oyster, Crassostrea gigas, when challenged with Vibrio tubiashii. Aquaculture. (1998) 169:111–20. doi: 10.1016/S0044-8486(98)00369-X
18. Karim M, Zhao W, Rowley D, Nelson D, Gomez-Chiarri M. Probiotic strains for shellfish aquaculture: protection of eastern oyster, Crassostrea virginica, larvae and juveniles against bacterial challenge. J Shellfish Res. (2013) 32:401–8. doi: 10.2983/035.032.0220
19. Kesarcodi-Watson A, Miner P, Nicolas JL, Robert R. Protective effect of four potential probiotics against pathogen-challenge of the larvae of three bivalves: pacific oyster (Crassostrea gigas), flat oyster (Ostrea edulis) and scallop (Pecten maximus). Aquaculture. (2012) 344–349:29–34. doi: 10.1016/j.aquaculture.2012.02.029
20. Khouadja S, Haddaji N, Hanchi M, Bakhrouf A. Selection of lactic acid bacteria as candidate probiotics for Vibrio parahaemolyticus depuration in pacific oysters (Crassostrea gigas). Aquac Res. (2017) 48:1885–94. doi: 10.1111/are.13026
21. Lim HJ, Kapareiko D, Schott EJ, Hanif A, Wikfors GH. Isolation and evaluation of new probiotic bacteria for use in shellfish hatcheries: I. isolation and screening for bioactivity. J Shellfish Res. (2011) 30:609–15. doi: 10.2983/035.030.0303
22. Madison D, Schubiger C, Lunda S, Mueller RS, Langdon C. A marine probiotic treatment against the bacterial pathogen Vibrio coralliilyticus to improve the performance of pacific (Crassostrea gigas) and kumamoto (C. sikamea) oyster larvae. Aquaculture. (2022) 560:738611. doi: 10.1016/j.aquaculture.2022.738611
23. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
24. Langdon CJ. Growth studies with bacteria-free oyster (Crassostrea gigas) larvae fed on semi-defined artificial diets. Biol Bull. (1983) 164:227–35. doi: 10.2307/1541141
25. Babraham Bioinformatics. FastQC A Quality Control tool for High Throughput Sequence Data (2024). Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
26. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinforma Oxf Engl. (2018) 34:i884–90. doi: 10.1093/bioinformatics/bty560
27. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
28. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. (2016) 11:1650–67. doi: 10.1038/nprot.2016.095
29. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
30. Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. (2001) 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x
31. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. (2017) 45:D353–61. doi: 10.1093/nar/gkw1092
32. UniProt (2024). Available online at: https://www.uniprot.org/.
33. Green TJ, Speck P. Antiviral defense and innate immune memory in the oyster. Viruses. (2018) 10:133. doi: 10.3390/v10030133
34. Lafont M, Vergnes A, Vidal-Dupiol J, de Lorgeril J, Gueguen Y, Haffner P, et al. A sustained immune response supports long-term antiviral immune priming in the pacific oyster, Crassostrea gigas. mBio. (2020) 11:e02777–19. doi: 10.1128/mbio.02777-19
35. Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. (2003) 21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126
36. Lorenz E. TLR2 and TLR4 expression during bacterial infections. Curr Pharm Des. (2006) 12:4185–93. doi: 10.2174/138161206778743547
37. Wang W, Zhang T, Wang L, Xu J, Li M, Zhang A, et al. A new non-phagocytic TLR6 with broad recognition ligands from pacific oyster Crassostrea gigas. Dev Comp Immunol. (2016) 65:182–90. doi: 10.1016/j.dci.2016.07.010
38. Wu Y, Liang H, Wang Z, Lei Q, Xia L. A novel toll-like receptor from the pearl oyster Pinctada fucata martensii is induced in response to stress. Comp Biochem Physiol B Biochem Mol Biol. (2017) 214:19–26. doi: 10.1016/j.cbpb.2017.08.006
39. Wang L, Zhang H, Wang M, Zhou Z, Wang W, Liu R, et al. The transcriptomic expression of pattern recognition receptors: Insight into molecular recognition of various invading pathogens in oyster Crassostrea gigas. Dev Comp Immunol. (2019) 91:1–7. doi: 10.1016/j.dci.2018.09.021
40. Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. (2014) 5:461. doi: 10.3389/fimmu.2014.00461
41. Nair S, Huynh JP, Lampropoulou V, Loginicheva E, Esaulova E, Gounder AP, et al. Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J Exp Med. (2018) 215:1035–45. doi: 10.1084/jem.20180118
42. Naujoks J, Tabeling C, Dill BD, Hoffmann C, Brown AS, Kunze M, et al. IFNs modify the proteome of Legionella-containing vacuoles and restrict infection via IRG1-derived itaconic acid. PLoS Pathog. (2016) 12:e1005408. doi: 10.1371/journal.ppat.1005408
43. Van Nguyen T, Alfaro AC. Targeted metabolomics to investigate antimicrobial activity of itaconic acid in marine molluscs. Metabolomics. (2019) 15:97. doi: 10.1007/s11306-019-1556-8
44. Saco A, Rey-Campos M, Rosani U, Novoa B, Figueras A. The evolution and diversity of interleukin-17 highlight an expansion in marine invertebrates and its conserved role in mucosal immunity. Front Immunol. (2021) 12:692997. doi: 10.3389/fimmu.2021.692997
45. Deguine J, Barton GM. MyD88: a central player in innate immune signaling. F1000Prime Rep. (2014) 6:97. doi: 10.12703/P6-97
46. Chen J, Lin J, Yu F, Zhong Z, Liang Q, Pang H, et al. Transcriptome analysis reveals the function of TLR4-MyD88 pathway in immune response of Crassostrea hongkongensis against Vibrio Parahemolyticus. Aquac Rep. (2022) 25:101253. doi: 10.1016/j.aqrep.2022.101253
47. Shan J, Wang P, Zhou J, Wu D, Shi H, Huo K. RIOK3 interacts with caspase-10 and negatively regulates the NF-κB signaling pathway. Mol Cell Biochem. (2009) 332:113–20. doi: 10.1007/s11010-009-0180-8
48. Chen Y, Li HH, Fu J, Wang XF, Ren YB, Dong LW, et al. Oncoprotein p28 GANK binds to RelA and retains NF-kappaB in the cytoplasm through nuclear export. Cell Res. (2007) 17:1020–9. doi: 10.1038/cr.2007.99
49. Hansson GC. Mucins and the microbiome. Annu Rev Biochem. (2020) 89:769. doi: 10.1146/annurev-biochem-011520-105053
50. Kato K, Lillehoj EP, Lu W, Kim KC. MUC1: the first respiratory mucin with an anti-inflammatory function. J Clin Med. (2017) 6:110. doi: 10.3390/jcm6120110
51. Gum JR, Hicks JW, Toribara NW, Siddiki B, Kim YS. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von willebrand factor. J Biol Chem. (1994) 269:2440–6. doi: 10.1016/S0021-9258(17)41965-X
52. Klomp LW, Van Rens L, Strous GJ. Cloning and analysis of human gastric mucin cDNA reveals two types of conserved cysteine-rich domains. Biochem J. (1995) 308:831–8. doi: 10.1042/bj3080831
53. Pietsch EC, Sykes SM, McMahon SB, Murphy ME. The p53 family and programmed cell death. Oncogene. (2008) 27:6507–21. doi: 10.1038/onc.2008.315
54. Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. (2005) 434:93–9. doi: 10.1038/nature03263
55. Nitta T, Takahama Y. The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends Immunol. (2007) 28:58–65. doi: 10.1016/j.it.2006.12.002
56. Ranjan A, Iwakuma T. Non-canonical cell death induced by p53. Int J Mol Sci. (2016) 17:2068. doi: 10.3390/ijms17122068
57. Jiang S, Jia Z, Xin L, Sun Y, Zhang R, Wang W, et al. The cytochemical and ultrastructural characteristics of phagocytes in the pacific oyster Crassostrea gigas. Fish Shellfish Immunol. (2016) 55:490–8. doi: 10.1016/j.fsi.2016.06.024
Keywords: Pacific oyster larvae, probiotics, immune response, Vibrio coralliilyticus, aquaculture
Citation: Hesser J, Mueller RS, Langdon C and Schubiger CB (2024) Immunomodulatory effects of a probiotic combination treatment to improve the survival of Pacific oyster (Crassostrea gigas) larvae against infection by Vibrio coralliilyticus. Front. Immunol. 15:1380089. doi: 10.3389/fimmu.2024.1380089
Received: 01 February 2024; Accepted: 18 March 2024;
Published: 08 April 2024.
Edited by:
Hany M. R. Abdel-Latif, Alexandria University, EgyptReviewed by:
Akram Shehata, Alexandria University, EgyptCopyright © 2024 Hesser, Mueller, Langdon and Schubiger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer Hesser, amVubmlmZXJoZXNzZXI5N0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.