- 1Department of Neurosurgery, National Health Commission Key Laboratory of Cell Transplantation, The First Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang, China
- 2Department of Neurosurgery, Hainan Cancer Hospital, Haikou, Hainan, China
- 3School of Life Science, Northeast Agricultural University, Harbin, Heilongjiang, China
Background: The gut microbiota (GM) has been implicated in neurological disorders, but the relationship with hydrocephalus, especially the underlying mechanistic pathways, is unclear. Using Mendelian randomization (MR), we aim to discover the mediating role of inflammatory factors in the relationship between GM and hydrocephalus.
Methods: After removing confounders, univariable and multivariable MR analyses were performed using summary statistics to assess the causal relationships between GM, inflammatory factors (IL-17A and IL-27), and types of hydrocephalus. Meta-analyses were used to reconcile the differences in MR results between different hydrocephalus sources. Finally, mediator MR analyses were applied to determine the mediating effect of inflammatory factors. Various sensitivity analysis methods were employed to ensure the reliability and stability of the results.
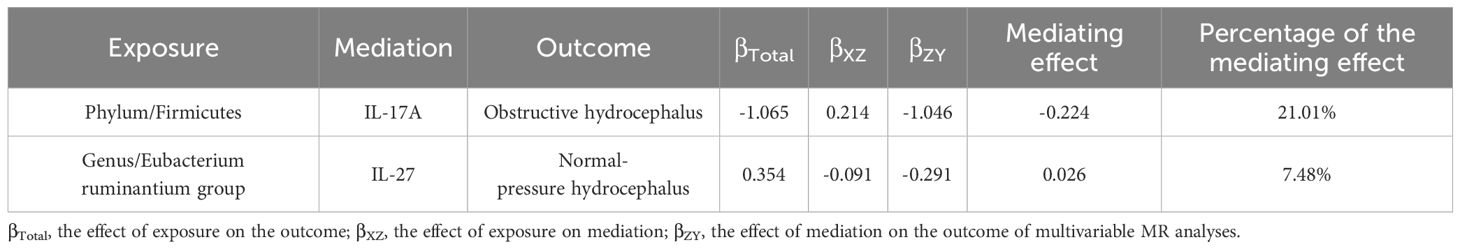
Results: After correction for P-values, Firmicutes (phylum) (OR, 0.34; 95%CI, 0.17–0.69; P = 2.71E-03, PFDR = 2.44E-02) significantly reduced the risk of obstructive hydrocephalus. The remaining 18 different taxa of GM had potential causal relationships for different types of hydrocephalus. In addition, Firmicutes (phylum) decreased the risk of obstructive hydrocephalus by increasing levels of IL-17A (mediating effect = 21.01%), while Eubacterium ruminantium group (genus) increased the risk of normal-pressure hydrocephalus by decreasing levels of IL-27 (mediating effect = 7.48%).
Conclusion: We reveal the connection between GM, inflammatory factors (IL-17A and IL-27), and hydrocephalus, which lays the foundation for unraveling the mechanism between GM and hydrocephalus.
1 Introduction
Hydrocephalus is an old category of disorder that has been documented since ancient times. It is characterized by enlargement of the ventricular system due to an imbalance between the production and absorption of cerebrospinal fluid (CSF) (1). The healthcare burden of hydrocephalus is enormous in both developing and developed countries. Although surgery can reduce CSF production or facilitate shunting, the failure rate of surgery and the long-term infection rate cannot be ignored (2). The processes leading to impaired circulation of CSF remain unknown. However, microscopic explorations beyond the primary lesion may offer insights into the development and aggravation of hydrocephalus. According to recent studies, choroidal arteries trigger cytokine storms that promote hydrocephalic developments in cases of posthemorrhagic hydrocephalus. Neuroinflammation is regarded as a crucial factor in the development of hydrocephalus (3).
The gut microbiota (GM) relies on the intestinal environment to carry out immune, metabolic, and endocrine functions. Emerging studies have indicated the GM’s crucial role in controlling T cell homeostasis and neuroinflammation in neurological disorders (4). The gut-brain axis linking the gut and nervous system is becoming increasingly acknowledged. Previous research demonstrates that the GM has a role in various neurological conditions, such as Alzheimer’s disease, ischemic stroke, epilepsy, and Parkinson’s disease (5, 6). Furthermore, a correlation between GM and neuroinflammation following a stroke has been established. It is not just altering the GM composition after stroke; transferring specific gut microbes into animal models after hemorrhage has also reduced neuroinflammation (4). Thus, a correlation between the gut and neurological inflammation-related disorders may exist.
Interleukins (IL) is a large family with protective immune and pro-inflammatory mediators among the many inflammatory factors. It has now been found that some of the interleukins may have bidirectional physiopathologic roles, such as IL-17A and IL-27. IL-17A was the first member of the IL-17 family to be identified. In addition to its ability to induce cellular activation to produce inflammatory mediators, IL-17A has been found to play a central role in protective immunity against Candida and other fungal pathogens (7). In addition, IL-17A drives epithelial hypoxia-Inducible factor 1-α to promote wound repair via glycolysis (8). IL-27, on the other hand, has been found to activate multiple signaling cascades that promote/inhibit different TH cell activation and thus produce opposite immune effects, respectively (9). Also, IL-27 is associated with neurological pathologies, including myasthenia gravis, Guillain-Barre syndrome, and ischemia-reperfusion, and may even play a neuroprotective role in reducing neuroinflammation (10–12).
The secretions of GM found so far can interfere with the expression of inflammatory factors and thus contact distant tissues (13). Mendelian randomization (MR) is a new way of exploring causality. Since any given allele has the same chance of being randomly inherited in a person, the causality of MR studies is plausible (14). However, we need to be aware of studies elaborating on the relationship between gut microbiome, inflammatory factors, and hydrocephalus. Therefore, we will perform multiple MR analyses with the help of Genome-Wide Association Studies (GWAS) to further analyze the causal relationship between GM, inflammatory factors, and hydrocephalus.
2 Materials and methods
2.1 Study design and ethics statement
In all analyses, we used single nucleotide polymorphisms (SNPs) as instrumental variables (IVs) and relied on three main assumptions: (1) IVs are strongly associated with exposure; (2) IVs are not associated with confounders; and (3) IVs can only influence outcome through exposure (15). The second and third assumptions, collectively referred to as independence of horizontal pleiotropy, were assessed using various statistical methods. Initially, we chose the SNPs based on a locus-wide significance threshold (P< 1 × 10-5) and conducted bidirectional MR analyses to investigate the association between GM and hydrocephalus. Secondly, we reselected SNPs based on a genome-wide statistical significance cutoff (P< 5 × 10-8) for GM to enhance the precision of the causal link between GM and hydrocephalus. Subsequently, we employed meta-analysis to coordinate differences in the results of MR analyses derived from hydrocephalus in different cohorts. Lastly, we conducted mediator MR analyses to elucidate whether GM impacts hydrocephalus through inflammatory factors (16) (Figure 1).
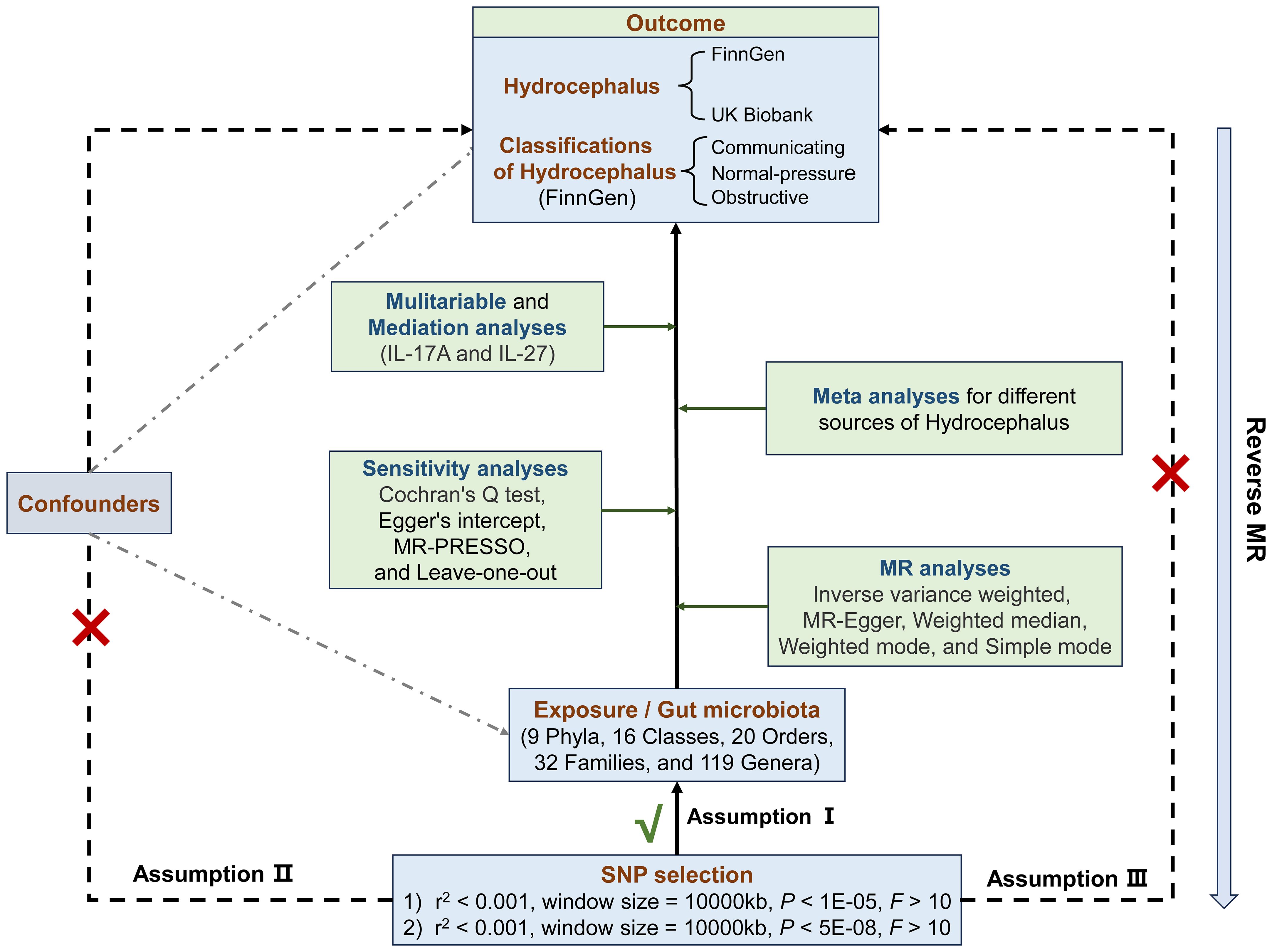
Figure 1 Flowchart of the Mendelian randomization and three key assumptions. MR, Mendelian randomization; SNP, single nucleotide polymorphism; MR-PRESSO, MR pleiotropy residual sum and outlier.
We did not require additional ethical approval since we used publicly available data, which had already received approval from the appropriate ethical and institutional review boards. This study is reported in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization guidelines (STROBE-MR) (17) (Supplementary Table S1).
2.2 Data source
We obtained the GWAS dataset of the human GM from the MiBioGen consortium. This multi-ethnic, genome-wide analysis focuses on human autosomal genetic variation and the GM to investigate host-genetics-microbiome associations (18). From this comprehensive study, we identified 211 bacterial taxa: 9 phyla, 16 classes, 20 orders, 35 families (3 unknown), and 131 genera (12 unknown), resulting in 196 bacteria representing gut species in our analyses (details in Supplementary Table S2).
We initially obtained hydrocephalus GWAS data based on ICD-10 diagnostic criteria (G91) from the FinnGen consortium. An additional data source on hydrocephalus came from a study of a generalized linear mixed-model association tool on samples from the UK Biobank. For detailed analysis, we acquired GWAS data from FinnGen for specific hydrocephalus types based on ICD-10 classifications: communicating hydrocephalus (G91.0), normal-pressure hydrocephalus (G91.2), and obstructive hydrocephalus (G91.1). IL-17A was derived from measurements of 2,994 plasma proteins from 3,301 individuals with a total SNP number of 10,534,735 (19), whereas IL-27 was derived from measurements of 90 proteins from 21,759 participants in 13 cohorts with a total SNP number of 13,102,608 (20). Notably, all populations in these datasets were of European descent (Supplementary Table S2).
2.3 Selection of SNPs
First, we used SNPs with P< 1 × 10-5 as the initial genetic IVs for all MR analyses, which was determined to be the optimal threshold for the MR studies associated with GM as well as the other exposures in this study to maximize the amount of genetic variation explained by genetic predictors and to increase the number of SNPs eligible for sensitivity analysis (21). Second, we selected IVs for GM with a genome-wide significance cutoff (P< 5 × 10-8) to understand the corresponding exposure (22, 23). Subsequently, we applied linkage disequilibrium (r2< 0.001, window size = 10,000 kb) to exclude non-compliant SNPs (24). Also, we used F-statistic values to represent the intensity of IVs:
with R2 indicating the degree of variance elucidated by the instruments, n signifying the sample size, and k representing the number of chosen IVs, with F-statistic values< 10 were excluded (25). We removed SNPs significantly associated with the outcome based on the criterion: P outcome< P exposure. Finally, we excluded SNPs with palindromic structures to ensure alignment of exposure and outcome alleles.
2.4 Statistical analysis
In the MR analyses, we employed five MR-related statistical methods: inverse variance weighted (IVW), MR-Egger regression, weighted median, weighted mode, and simple mode. The IVW method enables the estimation of the causal effect of exposure on the outcome by integrating ratio estimates for each SNP (26). It transforms MR estimates into a weighted regression of SNP-outcome effects against SNP-exposure effects. IVW provides the most precise estimates among the methods considered, assuming all SNPs are valid instruments. If the β values of other methods were consistent, a significant result (P< 0.05) was obtained by the IVW method, and there is no pleiotropy and heterogeneity can be considered positive, even if other methods did not yield significant results (27). MR-PRESSO was also used to augment the robustness of the results. If the causal estimate conforms with the beta value of the method above and the P-value is less than 0.05, it enhances the robustness of the results.
We applied a significance level of P< 0.05 to assess heterogeneity and horizontal pleiotropy. Initially, we used Cochran’s Q test to evaluate heterogeneity (24). Then, we conducted the MR Egger’s intercept test and the global test of MR-PRESSO to assess horizontal pleiotropy. MR-PRESSO was used to identify outliers in IVs. If any outliers were found, we re-ran the MR analysis after excluding them. Additionally, we performed leave-one-out analyses by systematically excluding each instrumental SNP and re-executing the IVW analyses (28). Finally, we would not consider the causal relationship valid if horizontal pleiotropy was present and we could not correct for outliers.
For each outcome, we corrected the P-values for the false discovery rate (FDR) based on different categories of GM and inflammatory factors, resulting in a corresponding P FDR. In our analysis, a significant causal relationship between exposure and outcome is considered when P< 0.05 and P FDR< 0.1, whereas a potential association is suggested when P< 0.05 and P FDR ≥ 0.1 (29, 30).
When types of hydrocephalus were used as outcomes for exposures causally related to hydrocephalus, we searched the PhenoScanner database for common confounders associated with hydrocephalus to minimize their interference (including hypertension, BMI, and cholesterol) (31). After removing these associated SNPs, we repeated the MR and sensitivity analyses.
2.5 Replication and meta-analysis for hydrocephalus
When GM was used as the exposure and hydrocephalus (G91) as the outcome, we used hydrocephalus data from the UK Biobank as additional replication analyses. We conducted meta-analyses to consolidate the results with causality from FinnGen and UK Biobank sets. The heterogeneity of meta-analyses was checked using I2 and corresponding P-values.
2.6 Mediation analysis
Regarding mediation, initially, we recorded the causality between GM and classifications of hydrocephalus as the total effects (βTotal). Next, we examined the causal relationships between inflammatory factors (IL-17A, IL-27) and various classifications of hydrocephalus using two-sample MR analyses. Building on the previous information, the first step in the two-step MR was to assess the causal relationships of GM on inflammatory factors (IL-17A, IL-27), denoted by βXZ. The second step was to evaluate the causal relationships of inflammatory factors on classifications of hydrocephalus by multivariable MR (MVMR), denoted by βZY. βXZ × βZY denotes the mediating effect, and (βXZ × βZY)/βTotal denotes the percentage of this mediating effect. Mediating effects are considered to exist if (βXZ × βZY) and βTotal share consistent directions (as shown in Figure 2A).
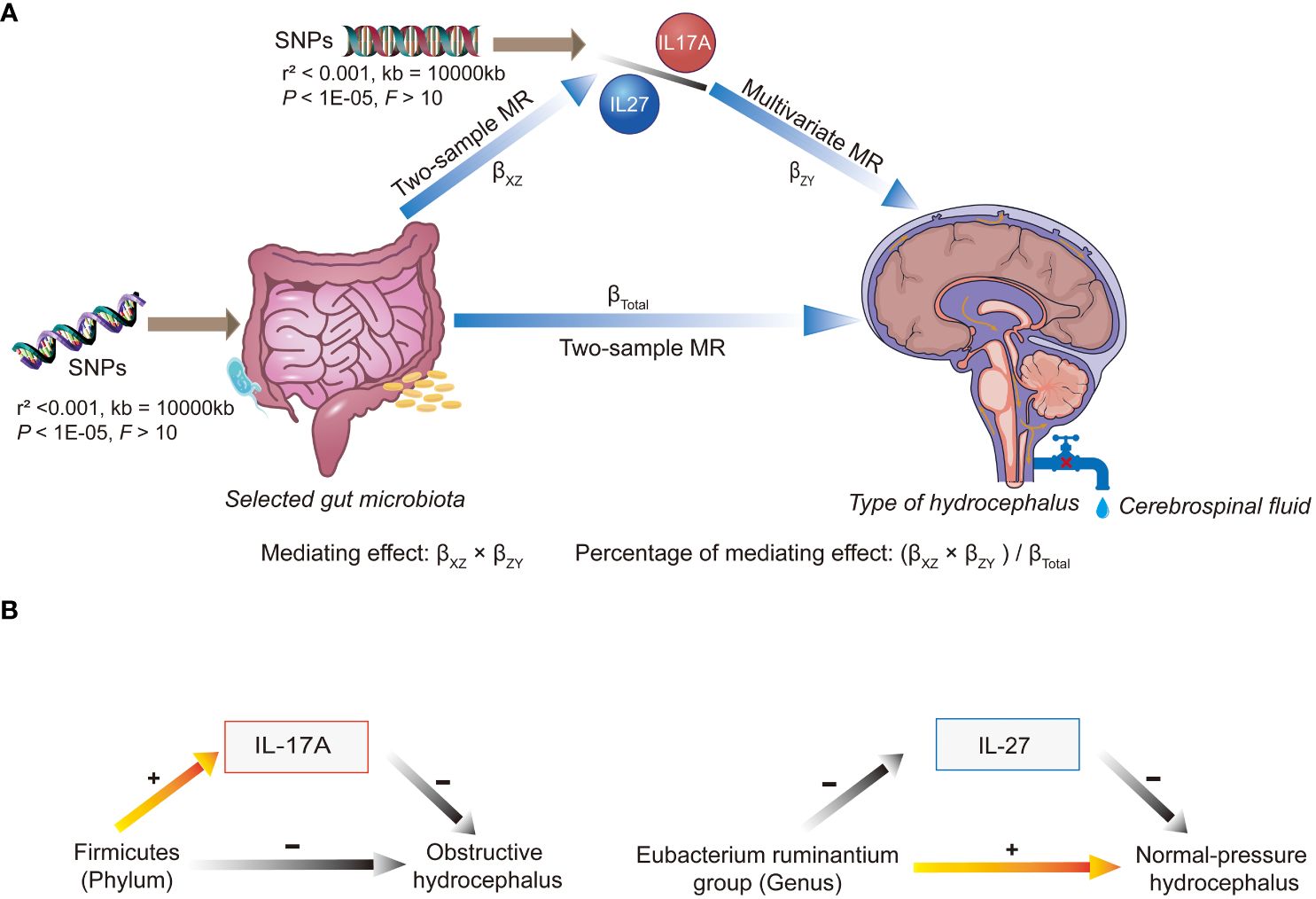
Figure 2 An overview chart of mediation analysis. SNP, single nucleotide polymorphism; MR, Mendelian randomization. (A) When multiple gut microbiota taxa were used as exposures, different microbiota affected the different types of hydrocephalus via IL-17A or IL-27. Mediating effects were calculated from beta values at multiple stages. (B) Elevated abundance of Firmicutes (phylum) reduced the risk of obstructive hydrocephalus by increasing levels of IL-17A. On the other hand, the Eubacterium ruminantium group (genus) increased the risk of normal-pressure hydrocephalus by decreasing levels of IL-27.
All statistical analyses were conducted with R (version 4.3.0), and MR analyses were executed using the TwosampleMR, Mendelian Randomization, MVMR, and MR-PRESSO R packages.
3 Results
3.1 Causal relationship between gut microbiota and hydrocephalus (P < 1 × 10-5)
We identified 192 distinct bacterial taxa for analyses following a rigorous IVs screening process. Then, we selected the corresponding SNPs for hydrocephalus (FinnGen) (n = 2029), communicating hydrocephalus (FinnGen) (n = 2042), normal-pressure hydrocephalus (FinnGen) (n = 2028), and obstructive hydrocephalus (FinnGen) (n = 2032), and hydrocephalus (UK Biobank) (n =2064). The F-values of all IVs were greater than 10, eliminating the bias of weak instruments. The results of MR analysis for GM and FinnGen-derived hydrocephalus are shown in Figures 3 and 4. After removing the confounders-related SNPs (Supplementary Tables S3, S4), we found that only Firmicutes (phylum) had a significant causal relationship with obstructive hydrocephalus (FinnGen) according to the criteria, indicating a protective effect (OR, 0.34; 95%CI, 0.17–0.69; P = 2.71E-03; PFDR = 2.44E-02) (Shown in Figure 5 and Supplementary Table S5).
Figure 3 Causal effects of the gut microbiota on hydrocephalus (A) and communicating hydrocephalus (B) based on MR analyses. The data source for hydrocephalus derived from FinnGen. From outside to inside, the beta values of simple mode, weighted mode, weighted median, MR- Egger, inverse variance weighted, and P -value of inverse variance weighted are represented, respectively. MR, Mendelian randomization; IVW, Inverse variance weighted.
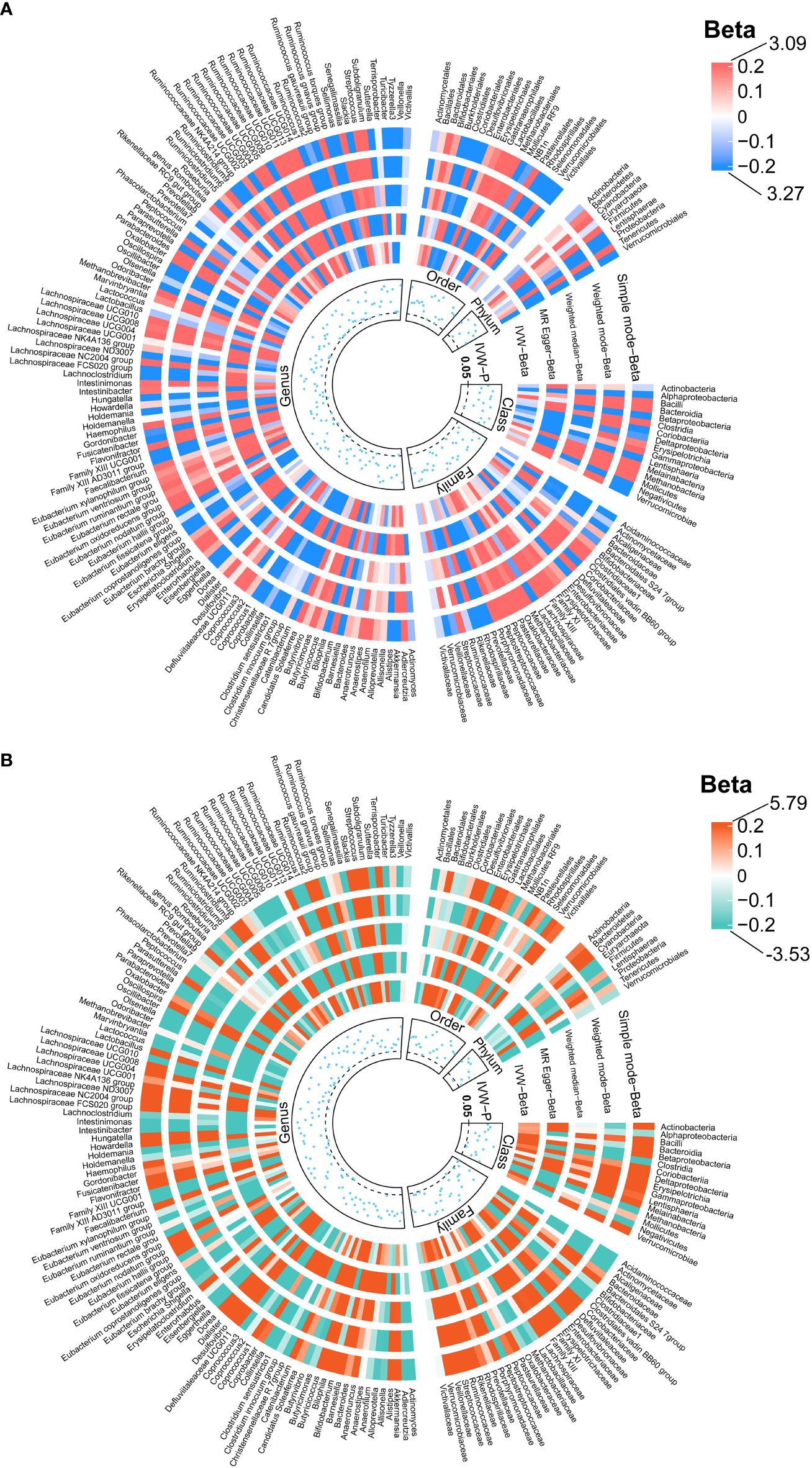
Figure 4 Causal effects of the gut microbiota on normal-pressure hydrocephalus (A) and obstructive hydrocephalus (B) based on MR analyses. The data source for hydrocephalus derived from FinnGen. From outside to inside, the beta values of simple mode, weighted mode, weighted median, MR- Egger, inverse variance weighted, and P -value of inverse variance weighted are represented, respectively. MR, Mendelian randomization; IVW, Inverse variance weighted.
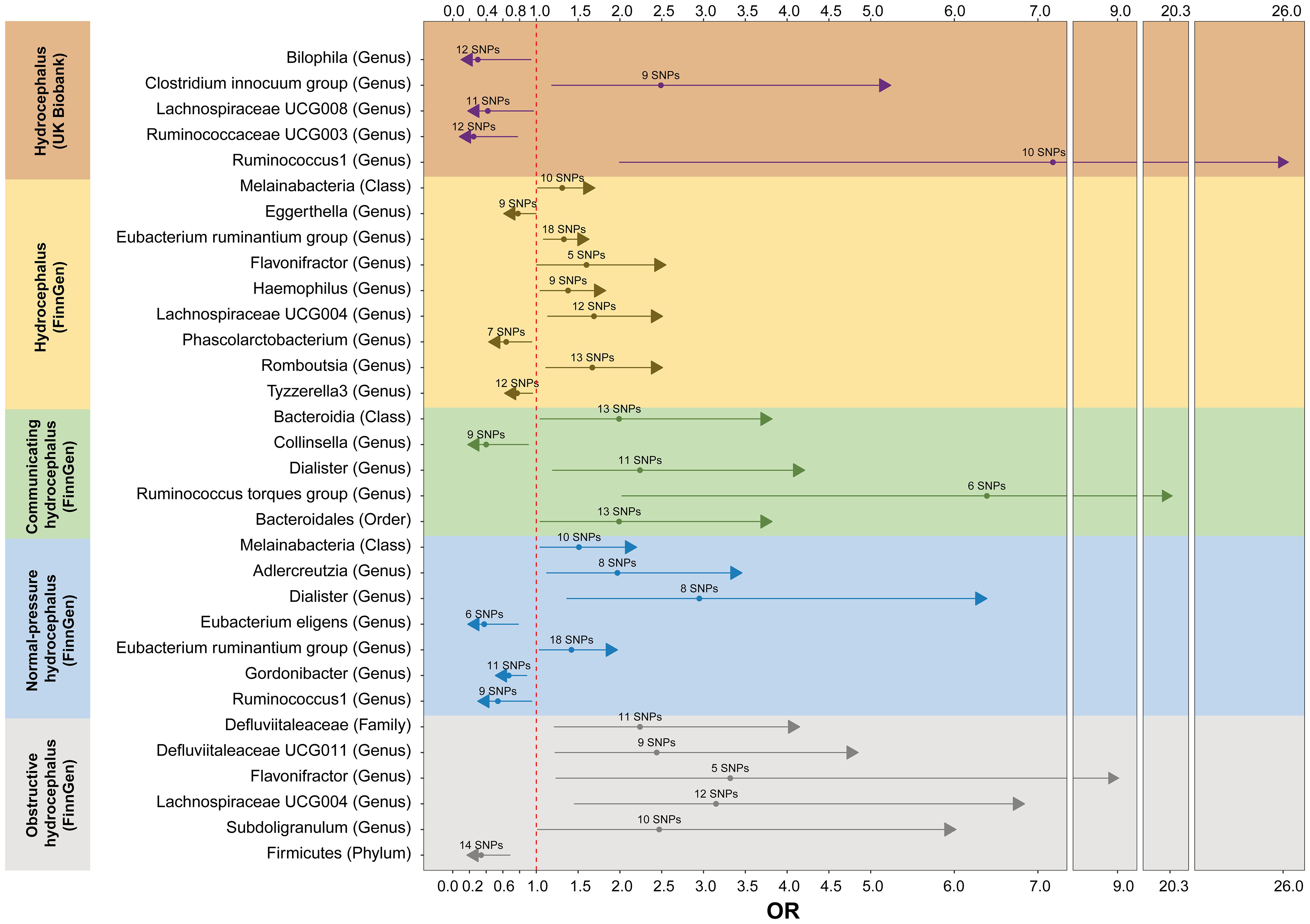
Figure 5 The forest plot of MR analyses between gut microbiota and hydrocephalus when selected instrumental variables with a locus-wide significance threshold (P< 1 × 10-5). Different color blocks represent different types of hydrocephalus as outcomes. The method used for MR analyses is inverse variance weighted. MR, Mendelian randomization; OR, Odds ratio.
For hydrocephalus (FinnGen), we identified several GMs with potential causal relationships. Specifically, Melainabacteria (class), Eubacterium ruminantium group (genus), Flavonifractor (genus), Haemophilus (genus), Lachnospiraceae UCG004 (genus), and Romboutsia (genus) potentially increased the risk of hydrocephalus. While Eggerthella (genus), Phascolarctobacterium (genus), and Tyzzerella3 (genus) may be protective against it (Figure 5 and Supplementary Table S5).
In assessing the causal effects of communicating hydrocephalus (FinnGen), several genetically predicted GMs were identified as potentially influential after removing the confounders-related SNPs (rs35866622 and rs2016057). Collinsella (genus) exhibited a potential protective effect against communicating hydrocephalus. In contrast, Bacteroidia (genus), Dialister (genus), Ruminococcus torques group (genus), and Bacteroidales (order) were associated with a potentially increased risk of communicating hydrocephalus (Figure 5 and Supplementary Table S5).
For normal-pressure hydrocephalus (FinnGen), after removing the confounders-related SNP (rs10167839), we found that Eubacterium eligens (genus), Gordonibacter (genus), and Ruminococcus1 (genus) exerted potentially protective effects. Conversely, Melainabacteria (class), Adlercreutzia (genus), Dialister (genus), and Eubacterium ruminantium group (genus) were associated with a potentially increased risk of normal-pressure hydrocephalus (Figure 5 and Supplementary Table S5).
Regarding obstructive hydrocephalus (FinnGen), besides the significant protective effect of Firmicutes (phylum), we found that Defluviitaleaceae (family), Defluviitaleaceae UCG011 (genus), Flavonifractor (genus), Lachnospiraceae UCG004 (genus), and Subdoligranulum (genus) potentially increased the risk of obstructive hydrocephalus (Figure 5 and Supplementary Table S5).
Finally, to perform replication analyses, we used UK Biobank-derived data on hydrocephalus as the outcome. The results suggest that the Clostridium innocuum group (genus) and Ruminococcus1 (genus) potentially increased the risk of hydrocephalus. In contrast, Bilophila (genus), Lachnospiraceae UCG008 (genus), and Ruminococcaceae UCG003 (genus) potentially decreased the risk of hydrocephalus (Figure 5 and Supplementary Table S5).
In the sensitivity analyses, the P-values of Cochran’s Q tests were greater than 0.05, indicating no significant heterogeneity (Supplementary Table S6). In the MR-PRESSO test for Dialister (genus) and normal-pressure hydrocephalus (FinnGen), we identified two outlier SNPs. After removing these outliers and re-running all analyses, the MR-PRESSO results were satisfactory, further enhancing the robustness of the results (Supplementary Table S7). Additionally, the leave-one-out analyses showed that none of the identified causal associations were influenced by any single IV (Supplementary Table S8).
3.2 Causal relationship of hydrocephalus on selected gut microbiota
We performed reverse MR analyses using different types of hydrocephalus as exposures, selecting corresponding IVs to explore their causal effects on the various microbiotas mentioned.
The results of the selected SNPs and reverse MR analyses are provided in Supplementary Tables S9 and Supplementary Tables S10. Based on the criteria, we found no significant results. That is, the abundance of selected GM did not change after hydrocephalus. In subsequent sensitivity analyses, despite no detected causality, we found heterogeneity in the causality assessment between the Eubacterium ruminantium group (genus) and hydrocephalus (FinnGen). Similar assessments were found for Dialister (genus) and normal-pressure hydrocephalus (FinnGen). Therefore, we used multiplicative random effects to represent the IVW results. We also identified a few outliers using MR-PRESSO and re-ran the MR analysis after excluding these abnormal SNPs. As shown in Supplementary Tables S11 and S12, the results indicate no significant horizontal pleiotropy and no noteworthy effects on individual IV results.
3.3 Causal relationship between gut microbiota and hydrocephalus (P < 5 × 10-8)
We selected SNPs meeting genome-wide statistical significance criteria for individual and total GMs, as shown in Supplementary Table S13. After removing confounder-associated SNPs (Supplementary Table S14), we found that Tyzzerella3 (genus) served as a potential protective factor against hydrocephalus (FinnGen). Bifidobacteriales (order) (OR, 8.56; 95%CI, 1.20–61.25; P = 2.75E-02; PFDR = 5.50E-02) significantly increased the risk of communicating hydrocephalus (FinnGen), while Bifidobacteriaceae (family), Bifidobacterium (genus), and Oxalobacter (genus) were identified as potential risk factors for communicating hydrocephalus (FinnGen). Additionally, when hydrocephalus (UK Biobank) served as an outcome, Tyzzerella3 (genus) significantly increased the risk of hydrocephalus (OR, 36.41; 95%CI, 5.42–244.42; P = 2.15E-04; PFDR = 2.37E-03), with Candidatus Soleaferrea (genus) and Oxalobacteraceae (genus) playing potential promoting roles (Figure 6 and Supplementary Table S15).
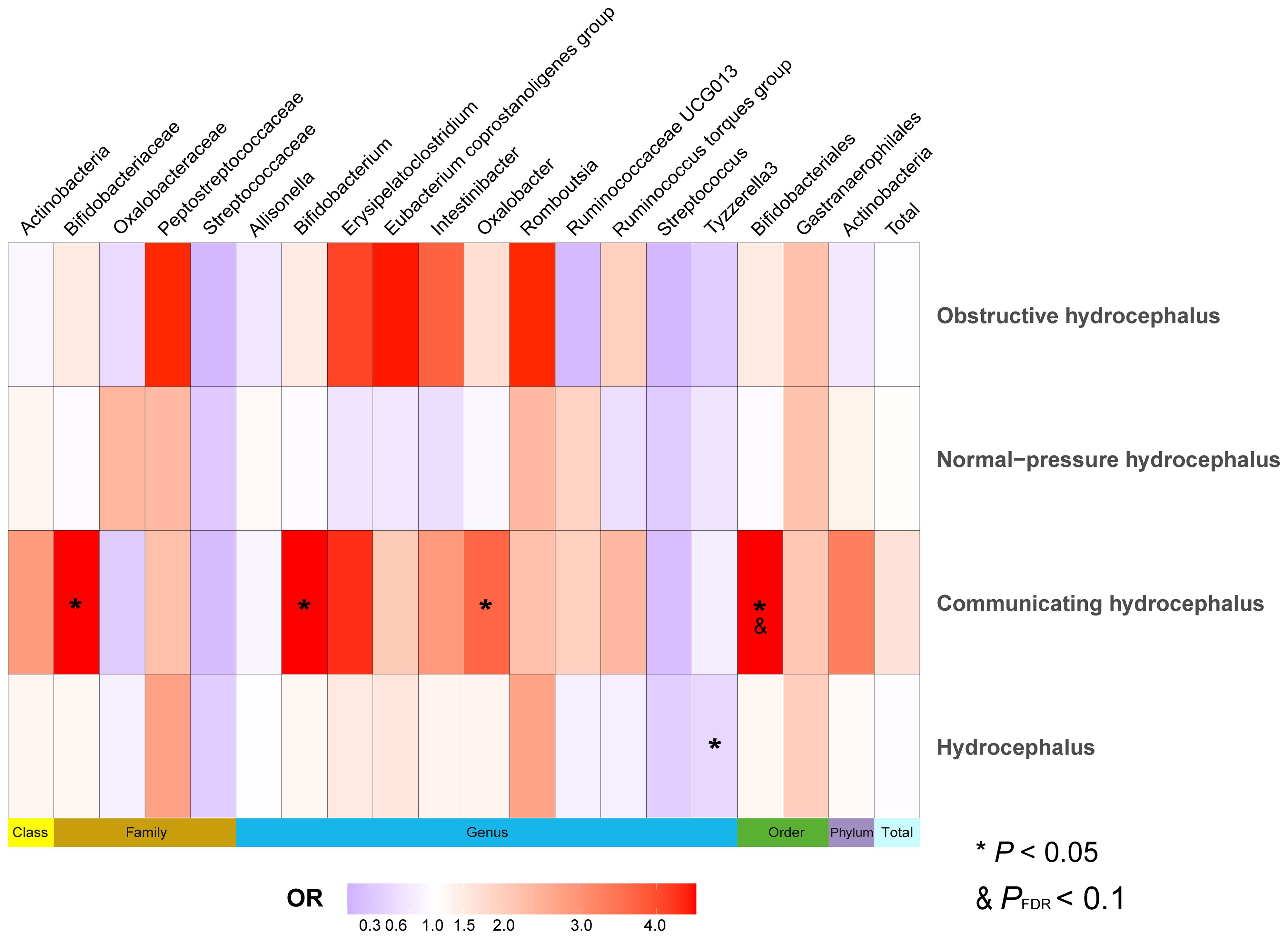
Figure 6 Heatmap of MR analysis between gut microbiota and hydrocephalus when selected instrumental variables with a genome-wide threshold of statistical significance (P< 5 × 10-8). Data on different types of hydrocephalus were obtained from FinnGen. Different color blocks represent different odds ratio values. The methods used for MR analyses include Wald ratio and Inverse variance weighted. “Total” indicates exposure of the entire gut microbiota. MR, Mendelian randomization; OR, Odds ratio.
When analyzing GM as an overall exposure, we found no causal relationship with any type of hydrocephalus, and sensitivity analyses revealed no significant outliers. Due to the limited number of SNPs, we could not perform further tests of heterogeneity and horizontal pleiotropy when analyzing GM separately as an exposure (Supplementary Tables S16 and S17). Therefore, these causal relationships could not be confirmed under more stringent SNP screening thresholds.
3.4 Meta-analyses for causal relationship of hydrocephalus
To reconcile potential discrepancies when GM was the exposure and hydrocephalus from different sources (FinnGen and UK Biobank) was the outcome, we used meta-analysis. As shown in Supplementary Table 18, When the threshold for targeting SNPs was P< 1 × 10-5, Melainabacteria (class) (OR, 1.32; 95%CI, 1.04–1.70; P = 2.38E-02), Eubacterium ruminantium group (genus) (OR, 1.31; 95%CI, 1.08–1.59; P = 6.30E-03), Haemophilus (genus) (OR, 1.14; 95%CI, 1.07–1.85; P =1.31E-02) and Lachnospiraceae UCG004 (genus) (OR, 1.66; 95%CI, 1.14–2.43; P = 8.10E-03) still retained the causal relationships and all served as potential risk factors for hydrocephalus. However, when the threshold was P< 5 × 10-8, Oxalobacteraceae (family) and Tyzzerella3 (genus) no longer hold causal relationships.
3.5 Causal relationship between inflammatory factors and hydrocephalus
We then evaluated the association between inflammatory factors and hydrocephalus using two-sample MR analysis. As shown in Supplementary Table S19, we used IL-17A and IL-27 as exposures and identified IVs that met the criteria. After removing confounder-associated SNPs (rs10774624) (Supplementary Table S20), our results indicated that IL-17A (OR, 0.66; 95%CI, 0.51–0.87; P = 2.63E-03; P FDR = 1.32E-02) and IL-27 (OR, 0.75; 95%CI, 0.59–0.95; P = 1.70E-02; P FDR = 8.50E-02) were significantly associated with lower risks of obstructive hydrocephalus and normal-pressure hydrocephalus, respectively (Figure 7 and Supplementary Table S21). In the MR results indicating causality, sensitivity analyses did not reveal significant heterogeneity, pleiotropy, or abnormal individual IVs (Supplementary Tables S22 and S23).
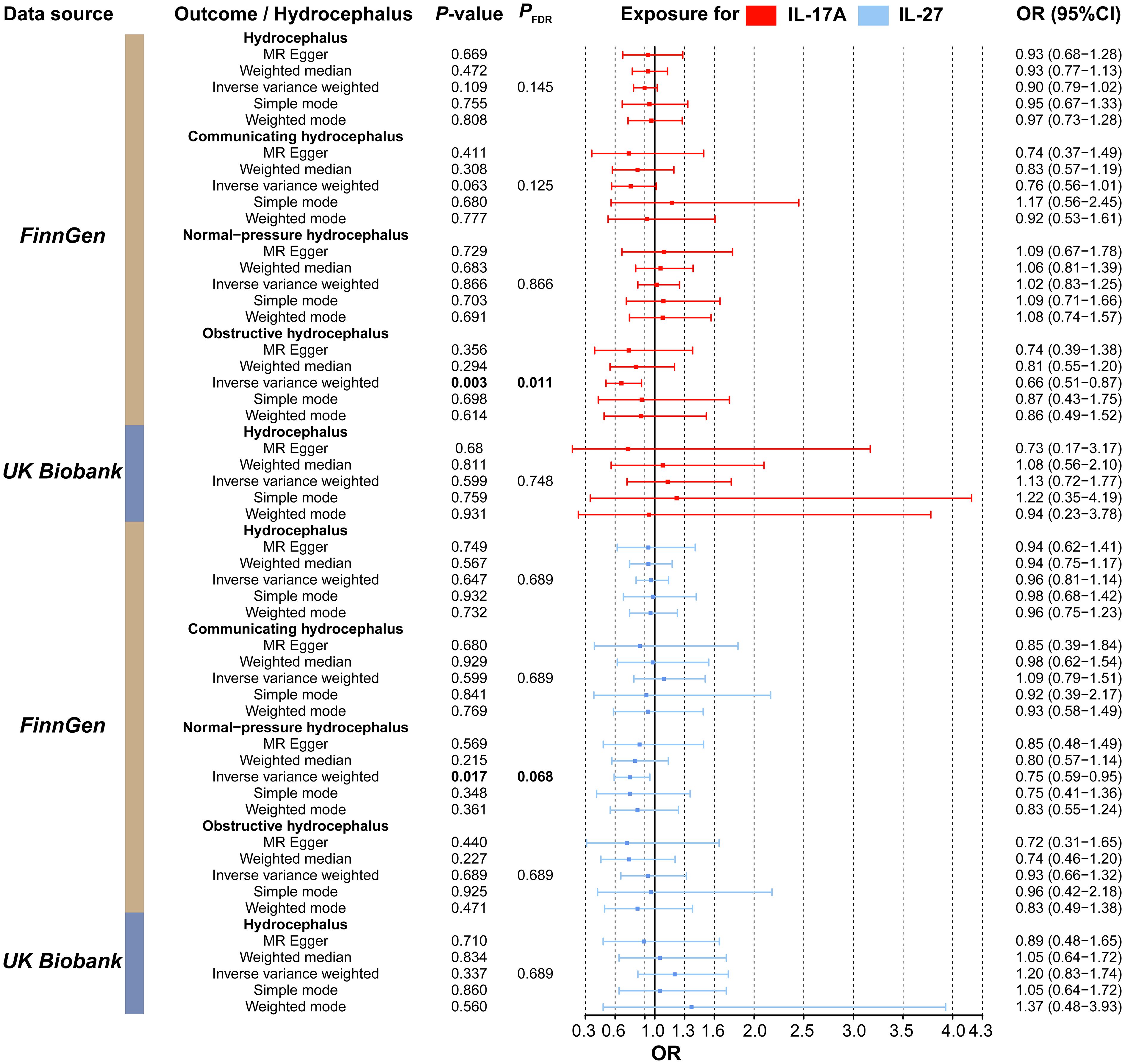
Figure 7 Forest plot of the association between the inflammatory factors (IL-17A and IL-27) and types of hydrocephalus. Red represents MR results when IL-17A was used as the exposure, while blue represents results when IL-27 was used as the exposure. MR, Mendelian randomization; CI, Confidence interval; OR, Odds ratio.
3.6 Causal relationship between selected gut microbiota and inflammatory factors
Next, we selected GMs causally associated with normal-pressure hydrocephalus and obstructive hydrocephalus, given that IL-17A and IL-27 are known to be associated with these conditions. In our evaluation of IL-17A, we found that a significantly increased abundance of Firmicutes (phylum) (β, 0.214; 95%CI, 0.002 ~ 0.426; P = 4.81E-02; P FDR = 4.81E-02) led to elevated IL-17A levels. Concerning IL-27, we observed that Melainabacteria (class) (β, 0.139; 95%CI, 0.064 ~ 0.213; P= 2.83E-04; PFDR = 2.83E-04) significantly increased IL-27 levels. Conversely, the Eubacterium ruminantium group (genus) (β, -0.091; 95%CI, -0.159~-0.023; P = 8.47E-03; P FDR = 8.47E-02) had the inverse effect, resulting in significantly decreased IL-27 levels (Figure 8, Supplementary Tables S24 and S25).
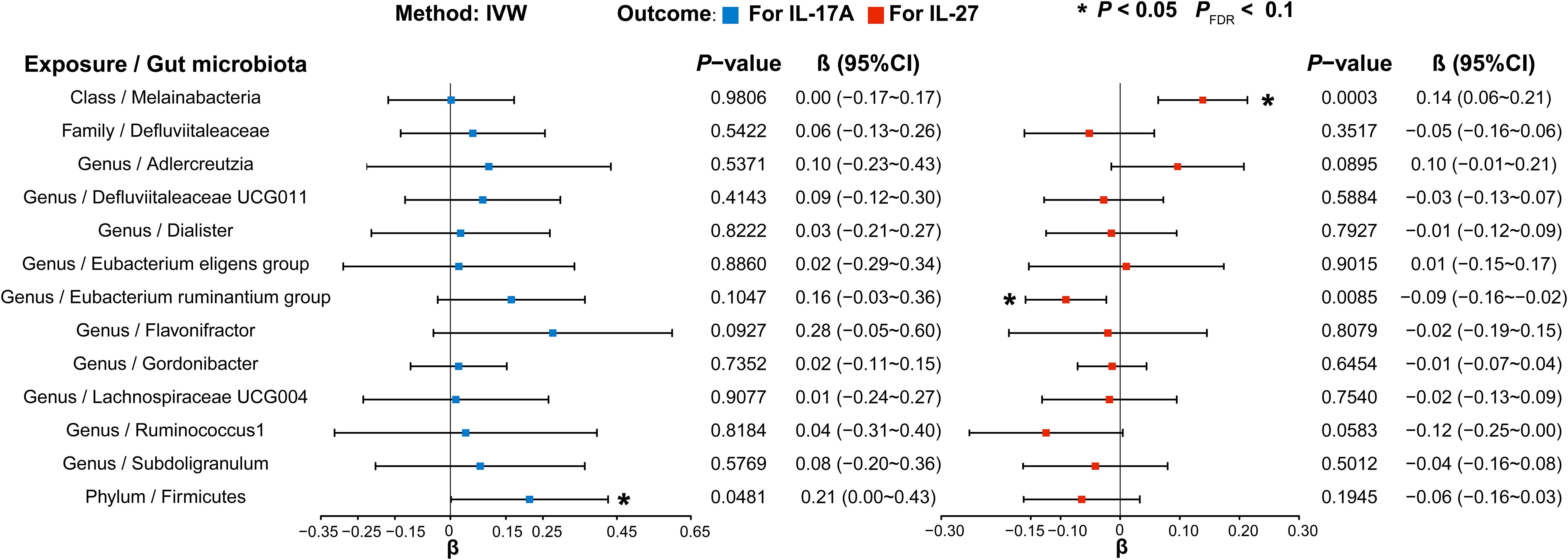
Figure 8 Forest plot illustrating the causal effects of gut microbiota on inflammatory factors using IVW methods. Blue (left) represents MR analysis results when IL-17A served as the outcome, and red (right) represents the results when IL-27 acted as the outcome. IVW, Inverse variance weighted.
In assessments of causal relationships, our sensitivity analysis revealed no significant heterogeneity and evidence of horizontal pleiotropy. Additionally, the leave-one-out analysis confirmed that no single instrumental variable significantly impacted the identified causal associations (Supplementary Tables S26 and S27).
3.7 Causal relationship between exposures and hydrocephalus using MVMR
To further evaluate the independent effects of inflammatory factors on hydrocephalus, we utilized MVMR analysis. After removing SNP (rs10774624) associated with confounders, results indicated that IL-17A (OR, 0.35; 95%CI, 0.18–0.68; P = 1.97E-03) still significantly reduced the risk of obstructive hydrocephalus, and IL-27 (OR, 0.75; 95%CI, 0.58–0.97; P = 2.96E-02) also remained a significantly protective factor against normal-pressure hydrocephalus (shown in Table 1). The absence of heterogeneity and horizontal pleiotropy further strengthened the reliability of the results (Supplementary Table S28).
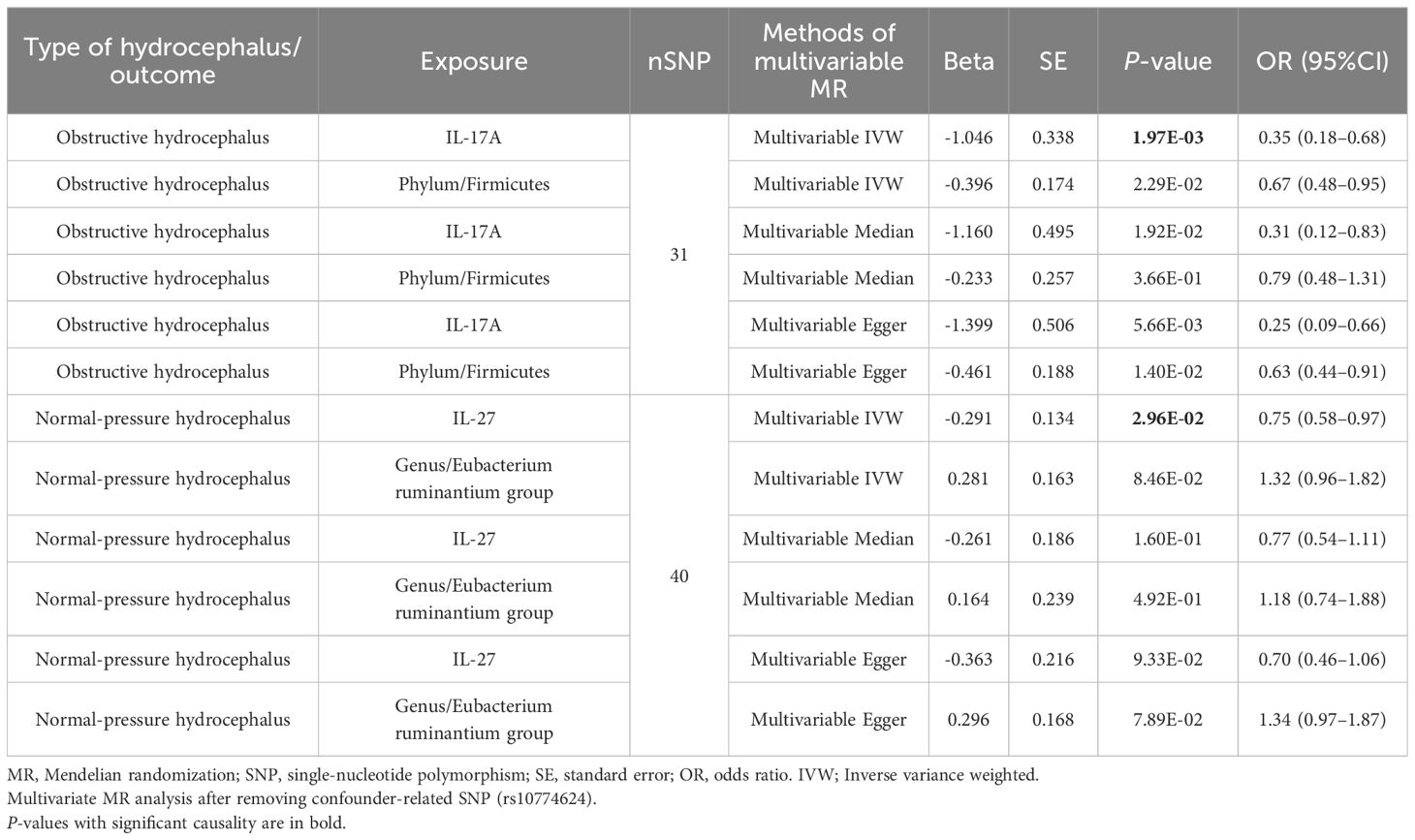
Table 1 Multivariable mendelian randomization analyses results between gut microbiota, inflammatory factors, and hydrocephalus.
3.8 Mediated MR analysis
When IL-27 was employed as a mediator, we observed a causal relationship between Melainabacteria (class) and IL-27. However, this relationship exhibited a different direction compared to the βTotal (Melainabacteria/class and hydrocephalus). Consequently, IL-27 could not mediate the causal association between Melainabacteria (class) and hydrocephalus.
Table 2 and Figure 2B displayed that IL-17A played a noteworthy role in the causal pathway from Firmicutes (phylum) to obstructive hydrocephalus (with 21.01% mediated effect). While there was only a potential causal relationship between Eubacterium ruminantium group (genus) and normal-pressure hydrocephalus, L-27 may also play an essential role in the causal pathway from Eubacterium ruminantium group (genus) to normal-pressure hydrocephalus (with 7.48% mediated effect).
4 Discussion
To the best of our knowledge, this is the initial study exploring causal relationships among human GM, inflammation, and hydrocephalus. A series of MR analyses enabled us to definitively identify 4 different GMs as potentially causally associated with hydrocephalus. When focusing on the classifications of hydrocephalus, we identified 17 different GMs that had potential causal relationships with different types of hydrocephalus. In addition, Firmicutes (phylum) demonstrated a significant protective effect against obstructive hydrocephalus. We did not observe a causal relationship between hydrocephalus and the GM. Furthermore, we found that IL-17A and IL-27 reduced the risk of obstructive hydrocephalus and normal-pressure hydrocephalus, respectively. Finally, mediation analyses provided insight into the fact that Firmicutes (phylum) reduced the risk of obstructive hydrocephalus by increasing the concentrations of IL-17A, whereas Eubacterium ruminantium group (genus) potentially increased the risk of normal-pressure hydrocephalus by decreasing the concentrations of IL-27. In conclusion, we have linked GM, inflammatory factors, and hydrocephalus, which provides a foundation for exploring the mechanisms of hydrocephalus and its prevention and treatment.
GM can connect to the brain in multiple mechanisms, including metabolites, extracellular vehicles (EVs), and inflammation. First, GM can produce various tryptophan metabolites, including indole compounds. Indoles can affect brain function and behavior when they enter the circulation. Administration of indole to the cecum of rats increased blinking frequency, c-Fos expression in the dorsal vagal complex, and anxiety behavior (32). The abundance of GM-producing short-chain fatty acids (SCFAs) was reduced in rat models of bilateral carotid artery occlusion. Behavioral abnormalities and hippocampal neuronal apoptosis in rats were significantly ameliorated by transplantation of the GM-producing SCFAs (33). In patients with bipolar disorder, various metabolites, including kynurenic acid, gamma-aminobutyric acid, and SCFAs, differed significantly from controls. These metabolites were influenced by the abundance of GM, which included Akkermansia muciniphila, Citrobacter freundii, Enterobacter cloacae, and Enterobacter kobei (34). Second, EVs have been implicated as potent markers in various disease processes (35, 36). GM-derived EVs act as novel communication modes that increase tau phosphorylation and lead to cognitive impairment (37). EVs from Pseudomonas aeruginosa contain lipopolysaccharides (LPS), which can induce a potent neuroinflammatory response (38). In addition, GM may also regulate hippocampal and axonal development by affecting miRNA expression (an important EV component) in the hippocampal region through kynurenine pathway enzyme (39, 40).
Inflammation also acts as a modulator of disease states, extensively linking GM and the nervous system. The abundance of Lactococcus and Ligilactobacillus was found to be strongly correlated with a high-fat diet. Following an inhibitory high-fat diet, a decrease in their abundance inhibited astrocyte activity in mouse neural tissue, attenuating neuroinflammation (41). Administering ampicillin increases the abundance of Klebsiella oxytoca in the mouse intestine, leading to the aggregation of microglia, monocytes, and dendritic cells in the hippocampus. At this time, nuclear factor kappa-B, IL-1β, and tumor necrosis factor (TNF)-α in the brain will be significantly increased, promoting the anxiety state of mice, and administering L. reuteri can significantly improve this condition (42). LPS acts as a receptor involved in various inflammatory responses in vivo. In Parkinson’s patients, the abundance of butyric acid-producing and anti-inflammatory genera (Blautia, Coprococcus, and Roseburia) is significantly lower, whereas the abundance of LPS-producing Oscillospira and Bacteroides is higher. Additionally, Bacteroides cell wall components contain LPS, which activates the immune system via Toll-like receptor 4 (TLR4), triggering autoimmune responses (43). These findings illustrate the complex relationship between GM, inflammation, and neurological disease (44). However, previous basic and clinical retrospective studies were limited by translation issues and the direction of causation, preventing an accurate link between GM and inflammation in hydrocephalus patients. Using Mendelian randomization (MR), this complex association can be initially explored. We found a link between GM, inflammation, and hydrocephalus. Specifically, the Firmicutes (phylum) could reduce the risk of obstructive hydrocephalus by increasing the levels of IL-17A, while the Eubacterium ruminantium group (genus) potentially increased the risk of normal-pressure hydrocephalus by decreasing the levels of IL-27.
Recent studies suggest that inflammatory activation plays a crucial role in the development of hydrocephalus. Firstly, in hemorrhage-induced hydrocephalus, large amounts of hemoglobin, its degradation products, and lipopolysaccharides act as inflammatory activators. This leads to the polarization of microglial cells (M2 type), activation of resident immune cells and infiltrating macrophages, promoting the release of cytokines and chemokines, leukocyte recruitment, increased vascular permeability, and cerebral edema formation, with potential direct neurotoxic effects (45). Additionally, the choroid plexus consists of an epithelial cell membrane that includes both immune and mesenchymal cells. As a site of CSF production, the choroid plexus is critical for collecting CSF and blood (46). At the choroid plexus-CSF interface, activated cytokines exacerbate hydrocephalus by activating the phosphorylation of TNF receptor-associated stress-activated protein kinases, which leads to increased CSF production by choroid epithelial cells (3). The expression of aquaporin is upregulated during inflammatory cascade activation, further exacerbating ventricular burden (47). Elevated levels of inflammatory markers chitinase-3-like protein 1 and soluble triggering receptor expressed on myeloid cells 2 have been found in the CSF of patients with idiopathic normal-pressure hydrocephalus (48). On the other hand, adenosine, known for its neuroprotective properties, significantly decreased in the peripheral blood of affected individuals (49). These findings provide further evidence for the involvement of inflammatory components in the development and progression of hydrocephalus.
To explore the pathophysiological mechanisms of inflammatory factors in hydrocephalus, we analyzed IL-17A and IL-27, key players in inflammatory activation and immunoprotection. Rigorous MR analysis revealed that IL-17A reduces the risk of obstructive hydrocephalus, while IL-27 reduces the risk of normal-pressure hydrocephalus. IL-17A is primarily derived from TH17 cells, with various factors affecting its release. A hallmark function of IL-17A is the induction of cytokine (50). IL-17A induces inflammatory factors such as C-X-C motif chemokine ligands 1 and 2, granulocyte colony-stimulating factor, and IL-6 in acute or chronic infections. When its regulatory capacity is exceeded, IL-17A leads to pathogenic inflammation (51). IL-17A plays a well-defined role in the pathology of psoriasis, psoriatic arthritis, and ankylosing spondylitis. Dysregulated IL-17A responses are considered a risk factor for these diseases (52, 53). However, IL-17A also induces the release of antimicrobial peptides such as β-defensins and S100A8. IL-17A protects the host from microbial infections and increases the barrier function of local epithelia, such as the intestinal epithelium (54). Colonization of commensal bacteria, including Staphylococcus epidermidis, induces non-classical MHC Ib-activated Tc17 cells to produce IL-17A locally in the skin without causing significant inflammation (55). IL-17A can also induce the production of the Hypoxia-Inducible Factor 1-alpha, a re-epithelialization factor controlled by RORγt + cells, through the extracellular signal-regulated kinase (ERK) 1/2-protein kinase b-mammalian target of rapamycin pathway (8). Thus, IL-17A plays an essential role in epithelial repair after wounding. In addition, IL-17A utilizes the ERK1/2 pathway to upregulate the expression of vascular endothelial growth factor, promoting the proliferation of lymphatic vessel endothelial cells and facilitating lymphatic vessel growth (56, 57). Numerous studies have demonstrated the importance of lymphatic drainage in the resorption of hydrocephalus (58). Therefore, enhancing infection barrier function and promoting lymphatic drainage may explain the protective effect of IL-17A on obstructive hydrocephalus. IL-27 is a pleiotropic cytokine released primarily by antigen-presenting cells, capable of exerting both anti-inflammatory and pro-inflammatory effects. It has been demonstrated that IL-27 not only antagonizes IL-2 production by T cells (59), but also assists IL-10 production by Th1, Th2, and Th17 cells, thereby limiting the response of helper cells during neuroinflammation (60). When administered directly to the brain in an inflammatory state, IL-27 may inhibit granulocyte-macrophage colony-stimulating factor in CD4+ T cells and induce PD-L1 mRNA in central inflammatory cells, ultimately inhibiting autoimmune encephalomyelitis development (61). Furthermore, An IL-27-producing innate B-1a cell (i27-Breg), which accumulates and is maintained in the CNS and lymphoid tissues during neuroinflammation, exerts a protective effect against CNS autoimmune diseases. Specifically, i27-Breg immunotherapy upregulates inhibitory receptors (Lag3, PD-1) and converts conventional B cells in the CNS into lymphocytes that secrete neuroprotective IL-10 and IL-35 (62). A range of inflammatory protective effects explains the reduced risk of normal-pressure hydrocephalus associated with IL-27.
We first confirmed that Firmicutes (phylum) reduces the risk of obstructive hydrocephalus by increasing the levels of IL-17A. Specifically, segmented filamentous bacilli (SFB) are widely distributed Gram-positive clostridia within the Firmicutes (phylum). Studies have shown that SFB can promote dendritic cell migration in mesenteric lymph nodes via its antigen, activating RORγt+ T cells and promoting IL-17A secretion (63). As mentioned earlier, IL-17A promotes barrier function to protect the host from harmful microorganisms and acts on intestinal epithelial cells to trigger antimicrobial peptide release, maintaining intestinal homeostasis (64). Meanwhile, IL-17A released by Firmicutes promotes the production of lymphatic endothelium. Additionally, Clostridium butyricum (a member of Firmicutes) activates the TLR-2/ERK-AP-1 kinase pathway and promotes the release of transforming growth factor-β (TGF-β) (65). whereas upregulation of TGF-β prevents neuroinflammatory infiltration and demyelination (66). Thus, Firmicutes (phylum) is inextricably linked to inflammation and may reduce the risk of obstructive hydrocephalus by enhancing the gut barrier and lymph angiogenesis via IL-17A. We also found that the Eubacterium ruminantium group (genus) potentially increases the risk of normal-pressure hydrocephalus by decreasing the levels of IL-27. Eubacterium groups play a broad regulatory role in host inflammation. Eubacterium is reported to be the main SCFA-producing group. SCFA promotes the activation of G protein-coupled receptor 109A, further activating macrophages and dendritic cells, which express free fatty acid receptor 2 and block IL-27 expression (67, 68). The relationship between Eubacterium ruminantium group (genus) and IL-27 remains unclear. Recent studies have found that Eubacterium ruminantium can reduce the expression of inflammatory factors, such as IL-1β and TNF-α (69). However, the Eubacterium ruminantium group (genus) also upregulates extracellular matrix protein 1 in bladder tissues, increasing bladder cancer risk by raising matrix metalloproteinase 9 expression through the ERK1/2 phosphorylation pathway (70). Thus, Eubacterium ruminantium group (genus) may act as a pleiotropic bacterium with a complex association with inflammatory factors. Its inhibition of certain inflammatory factors may also inhibit IL-27, which further may lead to the dysregulation of inflammatory factors, thereby promoting the development of normal-pressure hydrocephalus.
Our analyses still have some limitations. First, all data sources were from European populations, which limits the generalization of the results to other ethnic populations. Therefore, more refined cohort studies of different ancestries are needed to increase the generalizability of the results. Second, the MR analyses used summarized data, which did not allow us to perform nonlinear correlation analyses. Third, since the selection of SNPs with P< 5 × 10-8 was too limited for the GM data, we chose SNPs with P< 1 × 10-5 as the IVs of GM for analysis. Subsequently, we screened a series of SNPs to minimize the spurious associations introduced by the loose threshold. Finally, the minimal categorization of the dataset for GM limited our more microscopic assessment of causality. Nonetheless, we established a relationship between GM, inflammatory factors, and various types of hydrocephalus through multiple MR analyses, laying the foundation for further research.
5 Conclusion
We identified 4 different taxa of GM with potential causal effects on hydrocephalus and 16 taxa of GM as potentially causal for different classifications of hydrocephalus. Furthermore, Firmicutes (phylum) reduced the risk of obstructive hydrocephalus by increasing the concentrations of IL-17A, whereas the Eubacterium ruminantium group (genus) potentially increased the risk of normal-pressure hydrocephalus by decreasing the concentrations of IL-27. Therefore, modifying the abundance of GM could potentially alter the inflammatory state of the organism, thereby reducing the risk of hydrocephalus. Our study lays an essential foundation for explaining the mechanisms of the gut-brain axis. However, more refined basic experiments and clinical trials are still necessary.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YS: Conceptualization, Data curation, Formal analysis, Methodology, Software, Visualization, Writing – original draft. CL: Conceptualization, Software, Writing – review & editing. XZ: Data curation, Investigation, Visualization, Writing – review & editing. YW: Resources, Software, Visualization, Writing – review & editing. HZ: Conceptualization, Resources, Validation, Writing – review & editing. ZY: Software, Writing – review & editing. BG: Formal analysis, Validation, Writing – review & editing. RH: Investigation, Resources, Writing – review & editing. QL: Software, Visualization, Writing – review & editing. AG: Project administration, Supervision, Writing – review & editing. HL: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundations of China (No.82071368, 82271340) and the Outstanding Youth Funding of Harbin Medical University (No. HYD2020JQ0016).
Acknowledgments
We thank all the volunteers who participated in this study and expressed our gratitude to the participants and researchers in the MiBioGen, IEU open GWAS project, FinnGen and UK Biobank study groups.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1372051/full#supplementary-material
References
1. Kousi M, Katsanis N. The genetic basis of hydrocephalus. Annu Rev Neurosci. (2016) 39:409–35. doi: 10.1146/annurev-neuro-070815-014023
2. Hochstetler A, Raskin J, Blazer-Yost BL. Hydrocephalus: historical analysis and considerations for treatment. Eur J Med Res. (2022) 27:168. doi: 10.1186/s40001-022-00798-6
3. Robert SM, Reeves BC, Kiziltug E, Duy PQ, Karimy JK, Mansuri MS, et al. The choroid plexus links innate immunity to CSF dysregulation in hydrocephalus. Cell. (2023) 186:764–785.e21. doi: 10.1016/j.cell.2023.01.017
4. Yu X, Zhou G, Shao B, Zhou H, Xu C, Yan F, et al. Gut microbiota dysbiosis induced by intracerebral hemorrhage aggravates neuroinflammation in mice. Front Microbiol. (2021) 12:647304. doi: 10.3389/fmicb.2021.647304
5. Quigley EMM. Microbiota-brain-gut axis and neurodegenerative diseases. Curr Neurol Neurosci Rep. (2017) 17:94. doi: 10.1007/s11910-017-0802-6
6. Peh A, O'donnell JA, Broughton BRS, Marques FZ. Gut microbiota and their metabolites in stroke: A double-edged sword. Stroke. (2022) 53:1788–801. doi: 10.1161/STROKEAHA.121.036800
7. Davidson L, Van Den Reek J, Bruno M, Van Hunsel F, Herings RMC, Matzaraki V, et al. Risk of candidiasis associated with interleukin-17 inhibitors: A real-world observational study of multiple independent sources. Lancet Reg Health Eur. (2022) 13:100266. doi: 10.1016/j.lanepe.2021.100266
8. Konieczny P, Xing Y, Sidhu I, Subudhi I, Mansfield KP, Hsieh B, et al. Interleukin-17 governs hypoxic adaptation of injured epithelium. Science. (2022) 377:eabg9302. doi: 10.1126/science.abg9302
9. Meka RR, Venkatesha SH, Dudics S, Acharya B, Moudgil KD. IL-27-induced modulation of autoimmunity and its therapeutic potential. Autoimmun Rev. (2015) 14:1131–41. doi: 10.1016/j.autrev.2015.08.001
10. Peng J, Zhang H, Liu P, Chen M, Xue B, Wang R, et al. IL-23 and IL-27 levels in serum are associated with the process and the recovery of guillain-barré Syndrome. Sci Rep. (2018) 8:2824. doi: 10.1038/s41598-018-21025-5
11. Luo C, Li B, Chen L, Zhao L, Wei Y. IL-27 Protects the Brain from Ischemia-Reperfusion Injury via the gp130/STAT3 Signaling Pathway. J Mol Neurosci. (2021) 71:1838–48. doi: 10.1007/s12031-021-01802-0
12. Yi M, Zhang LJ, Liu XJ, Wang N, Huang CN, Liu MQ, et al. Increased serum IL-27 concentrations and IL-27-producing cells in MG patients with positive AChR-Ab. J Clin Neurosci. (2021) 86:289–93. doi: 10.1016/j.jocn.2020.11.017
13. Li G, Liu L, Lu T, Sui Y, Zhang C, Wang Y, et al. Gut microbiota aggravates neutrophil extracellular traps-induced pancreatic injury in hypertriglyceridemic pancreatitis. Nat Commun. (2023) 14:6179. doi: 10.1038/s41467-023-41950-y
14. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. Jama. (2017) 318:1925–6. doi: 10.1001/jama.2017.17219
15. Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. Mendelian randomization. Nat Rev Methods Primers. (2022) 2:6. doi: 10.1038/s43586-021-00092-5
16. Carter AR, Sanderson E, Hammerton G, Richmond RC, Davey Smith G, Heron J, et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. (2021) 36:465–78. doi: 10.1007/s10654-021-00757-1
17. Skrivankova VW, Richmond RC, Woolf B, Davies NM, Swanson SA, Vanderweele TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. Bmj. (2021) 375:n2233. doi: 10.1136/bmj.n2233
18. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. (2021) 53:156–65. doi: 10.1038/s41588-020-00763-1
19. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature. (2018) 558:73–9. doi: 10.1038/s41586-018-0175-2
20. Folkersen L, Gustafsson S, Wang Q, Hansen DH, Hedman Å K, Schork A, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. (2020) 2:1135–48. doi: 10.1038/s42255-020-00287-2
21. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PloS Genet. (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
22. Xiang K, Wang P, Xu Z, Hu YQ, He YS, Chen Y, et al. Causal effects of gut microbiome on systemic lupus erythematosus: A two-sample mendelian randomization study. Front Immunol. (2021) 12:667097. doi: 10.3389/fimmu.2021.667097
23. He Q, Wang W, Xiong Y, Tao C, Ma L, Ma J, et al. A causal effects of gut microbiota in the development of migraine. J Headache Pain. (2023) 24:90. doi: 10.1186/s10194-023-01609-x
24. Chen X, Hong X, Gao W, Luo S, Cai J, Liu G, et al. Causal relationship between physical activity, leisure sedentary behaviors and COVID-19 risk: a Mendelian randomization study. J Transl Med. (2022) 20:216. doi: 10.1186/s12967-022-03407-6
25. Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
26. Choi KW, Chen CY, Stein MB, Klimentidis YC, Wang MJ, Koenen KC, et al. Assessment of bidirectional relationships between physical activity and depression among adults: A 2-sample mendelian randomization study. JAMA Psychiatry. (2019) 76:399–408. doi: 10.1001/jamapsychiatry.2018.4175
27. Chen X, Kong J, Diao X, Cai J, Zheng J, Xie W, et al. Depression and prostate cancer risk: A Mendelian randomization study. Cancer Med. (2020) 9:9160–7. doi: 10.1002/cam4.3493
28. Yang J, He X, Qian L, Zhao B, Fan Y, Gao F, et al. Association between plasma proteome and childhood neurodevelopmental disorders: A two-sample Mendelian randomization analysis. EBioMedicine. (2022) 78:103948. doi: 10.1016/j.ebiom.2022.103948
29. Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U.S.A. (2003) 100:9440–5. doi: 10.1073/pnas.1530509100
30. Li P, Wang H, Guo L, Gou X, Chen G, Lin D, et al. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. (2022) 20:443. doi: 10.1186/s12916-022-02657-x
31. Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. (2019) 35:4851–3. doi: 10.1093/bioinformatics/btz469
32. Jaglin M, Rhimi M, Philippe C, Pons N, Bruneau A, Goustard B, et al. Indole, a signaling molecule produced by the gut microbiota, negatively impacts emotional behaviors in rats. Front Neurosci. (2018) 12:216. doi: 10.3389/fnins.2018.00216
33. Xiao W, Su J, Gao X, Yang H, Weng R, Ni W, et al. The microbiota-gut-brain axis participates in chronic cerebral hypoperfusion by disrupting the metabolism of short-chain fatty acids. Microbiome. (2022) 10:62. doi: 10.1186/s40168-022-01255-6
34. Li Z, Lai J, Zhang P, Ding J, Jiang J, Liu C, et al. Multi-omics analyses of serum metabolome, gut microbiome and brain function reveal dysregulated microbiota-gut-brain axis in bipolar depression. Mol Psychiatry. (2022) 27:4123–35. doi: 10.1038/s41380-022-01569-9
35. Li Y, Li Y, Yu S, Qian L, Chen K, Lai H, et al. Circulating EVs long RNA-based subtyping and deconvolution enable prediction of immunogenic signatures and clinical outcome for PDAC. Mol Ther Nucleic Acids. (2021) 26:488–501. doi: 10.1016/j.omtn.2021.08.017
36. Lai H, Li Y, Zhang H, Hu J, Liao J, Su Y, et al. exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. (2022) 50:D118–d128. doi: 10.1093/nar/gkab1085
37. Wei S, Peng W, Mai Y, Li K, Wei W, Hu L, et al. Outer membrane vesicles enhance tau phosphorylation and contribute to cognitive impairment. J Cell Physiol. (2020) 235:4843–55. doi: 10.1002/jcp.29362
38. Liu X, Shen L, Wan M, Xie H, Wang Z. Peripheral extracellular vesicles in neurodegeneration: pathogenic influencers and therapeutic vehicles. J Nanobiotechnology. (2024) 22:170. doi: 10.1186/s12951-024-02428-1
39. Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, et al. Extracellular Vesicles Long RNA Sequencing Reveals Abundant mRNA, circRNA, and lncRNA in Human Blood as Potential Biomarkers for Cancer Diagnosis. Clin Chem. (2019) 65:798–808. doi: 10.1373/clinchem.2018.301291
40. Zhao L, Ye Y, Gu L, Jian Z, Stary CM, Xiong X. Extracellular vesicle-derived miRNA as a novel regulatory system for bi-directional communication in gut-brain-microbiota axis. J Transl Med. (2021) 19:202. doi: 10.1186/s12967-021-02861-y
41. Huang Q, Liu L, Tan X, Wang S, Wang S, Luo J, et al. Empagliflozin alleviates neuroinflammation by inhibiting astrocyte activation in the brain and regulating gut microbiota of high-fat diet mice. J Affect Disord. (2024) 360:229–41. doi: 10.1016/j.jad.2024.05.150
42. Jang HM, Lee HJ, Jang SE, Han MJ, Kim DH. Evidence for interplay among antibacterial-induced gut microbiota disturbance, neuro-inflammation, and anxiety in mice. Mucosal Immunol. (2018) 11:1386–97. doi: 10.1038/s41385-018-0042-3
43. Yan Y, Zhou X, Guo K, Zhou F, Yang H. Chlorogenic acid protects against indomethacin-induced inflammation and mucosa damage by decreasing bacteroides-derived LPS. Front Immunol. (2020) 11:1125. doi: 10.3389/fimmu.2020.01125
44. Keshavarzian A, Green SJ, Engen PA, Voigt RM, Naqib A, Forsyth CB, et al. Colonic bacterial composition in Parkinson's disease. Mov Disord. (2015) 30:1351–60. doi: 10.1002/mds.26307
45. Holste KG, Xia F, Ye F, Keep RF, Xi G. Mechanisms of neuroinflammation in hydrocephalus after intraventricular hemorrhage: a review. Fluids Barriers CNS. (2022) 19:28. doi: 10.1186/s12987-022-00324-0
46. Dani N, Herbst RH, Mccabe C, Green GS, Kaiser K, Head JP, et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell. (2021) 184:3056–3074.e21. doi: 10.1016/j.cell.2021.04.003
47. Qing WG, Dong YQ, Ping TQ, Lai LG, Fang LD, Min HW, et al. Brain edema after intracerebral hemorrhage in rats: the role of iron overload and aquaporin 4. J Neurosurg. (2009) 110:462–8. doi: 10.3171/2008.4.JNS17512
48. Yang F, Yang L, Fang X, Deng Y, Mao R, Yan A, et al. Increased cerebrospinal fluid levels of soluble triggering receptor expressed on myeloid cells 2 and chitinase-3-like protein 1 in idiopathic normal-pressure hydrocephalus. J Alzheimers Dis. (2023) 93:1341–54. doi: 10.3233/JAD-221180
49. Casati M, Arosio B, Gussago C, Ferri E, Magni L, Assolari L, et al. Down-regulation of adenosine A1 and A2A receptors in peripheral cells from idiopathic normal-pressure hydrocephalus patients. J Neurol Sci. (2016) 361:196–9. doi: 10.1016/j.jns.2015.12.040
50. Mills KHG. IL-17 and IL-17-producing cells in protection versus pathology. Nat Rev Immunol. (2023) 23:38–54. doi: 10.1038/s41577-022-00746-9
51. Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. (2014) 14:585–600. doi: 10.1038/nri3707
52. Baeten D, Sieper J, Braun J, Baraliakos X, Dougados M, Emery P, et al. Secukinumab, an interleukin-17A inhibitor, in ankylosing spondylitis. N Engl J Med. (2015) 373:2534–48. doi: 10.1056/NEJMoa1505066
53. Ghoreschi K, Balato A, Enerbäck C, Sabat R. Therapeutics targeting the IL-23 and IL-17 pathway in psoriasis. Lancet. (2021) 397:754–66. doi: 10.1016/S0140-6736(21)00184-7
54. Li J, Casanova JL, Puel A. Mucocutaneous IL-17 immunity in mice and humans: host defense vs. excessive inflammation. Mucosal Immunol. (2018) 11:581–9. doi: 10.1038/mi.2017.97
55. Linehan JL, Harrison OJ, Han SJ, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. (2018) 172:784–796.e18. doi: 10.1016/j.cell.2017.12.033
56. Chen X, Xie Q, Cheng X, Diao X, Cheng Y, Liu J, et al. Role of interleukin-17 in lymphangiogenesis in non-small-cell lung cancer: Enhanced production of vascular endothelial growth factor C in non-small-cell lung carcinoma cells. Cancer Sci. (2010) 101:2384–90. doi: 10.1111/j.1349-7006.2010.01684.x
57. Chauhan SK, Jin Y, Goyal S, Lee HS, Fuchsluger TA, Lee HK, et al. A novel pro-lymphangiogenic function for Th17/IL-17. Blood. (2011) 118:4630–4. doi: 10.1182/blood-2011-01-332049
58. Rustenhoven J, Tanumihardja C, Kipnis J. Cerebrovascular anomalies: perspectives from immunology and cerebrospinal fluid flow. (2021) 129:174–94. doi: 10.1161/CIRCRESAHA.121.318173
59. Villarino AV, Stumhofer JS, Saris CJ, Kastelein RA, De Sauvage FJ, Hunter CA. IL-27 limits IL-2 production during Th1 differentiation. J Immunol. (2006) 176:237–47. doi: 10.4049/jimmunol.176.1.237
60. Tait Wojno ED, Hunter CA, Stumhofer JS. The immunobiology of the interleukin-12 family: room for discovery. Immunity. (2019) 50:851–70. doi: 10.1016/j.immuni.2019.03.011
61. Casella G, Finardi A, Descamps H, Colombo F, Maiorino C, Ruffini F, et al. IL-27, but not IL-35, inhibits neuroinflammation through modulating GM-CSF expression. Sci Rep. (2017) 7:16547. doi: 10.1038/s41598-017-16702-w
62. Choi JK, Yu CR, Bing SJ, Jittayasothorn Y, Mattapallil MJ, Kang M, et al. IL-27-producing B-1a cells suppress neuroinflammation and CNS autoimmune diseases. Proc Natl Acad Sci U.S.A. (2021) 118:e2109548118. doi: 10.1073/pnas.2109548118
63. Flannigan KL, Denning TL. Segmented filamentous bacteria-induced immune responses: a balancing act between host protection and autoimmunity. Immunology. (2018) 154:537–46. doi: 10.1111/imm.12950
64. Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol. (2019) 4:35–45. doi: 10.1038/s41564-018-0278-4
65. Kashiwagi I, Morita R, Schichita T, Komai K, Saeki K, Matsumoto M, et al. Smad2 and smad3 inversely regulate TGF-β Autoinduction in clostridium butyricum-activated dendritic cells. Immunity. (2015) 43:65–79. doi: 10.1016/j.immuni.2015.06.010
66. Wu Q, Miao X, Zhang J, Xiang L, Li X, Bao X, et al. Astrocytic YAP protects the optic nerve and retina in an experimental autoimmune encephalomyelitis model through TGF-β signaling. Theranostics. (2021) 11:8480–99. doi: 10.7150/thno.60031
67. Lavoie S, Chun E, Bae S, Brennan CA, Gallini Comeau CA, Lang JK, et al. Expression of free fatty acid receptor 2 by dendritic cells prevents their expression of interleukin 27 and is required for maintenance of mucosal barrier and immune response against colorectal tumors in mice. Gastroenterology. (2020) 158:1359–1372.e9. doi: 10.1053/j.gastro.2019.12.027
68. Mukherjee A, Lordan C, Ross RP, Cotter PD. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. (2020) 12:1802866. doi: 10.1080/19490976.2020.1802866
69. Hu S, Ma Y, Xiong K, Wang Y, Liu Y, Sun Y, et al. Ameliorating effects of vitamin K2 on dextran sulfate sodium-induced ulcerative colitis in mice. Int J Mol Sci. (2023) 24:2986. doi: 10.3390/ijms24032986
Keywords: gut microbiota, hydrocephalus, inflammation, mediator, Mendelian randomization
Citation: Shen Y, Li C, Zhang X, Wang Y, Zhang H, Yu Z, Gui B, Hu R, Li Q, Gao A and Liang H (2024) Gut microbiota linked to hydrocephalus through inflammatory factors: a Mendelian randomization study. Front. Immunol. 15:1372051. doi: 10.3389/fimmu.2024.1372051
Received: 17 January 2024; Accepted: 27 June 2024;
Published: 15 July 2024.
Edited by:
Abbas Yadegar, Shahid Beheshti University of Medical Sciences, IranReviewed by:
Shuming Li, Capital Medical University, ChinaJiachen Liu, Washington University in St. Louis, United States
Yuchen Li, Stanford University, United States
Copyright © 2024 Shen, Li, Zhang, Wang, Zhang, Yu, Gui, Hu, Li, Gao and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongsheng Liang, bGlhbmdob25nc2hlbmdAaHJibXUuZWR1LmNu; Aili Gao, Z2FvYWlsaTIwMDRAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Yingjie Shen
Yingjie Shen Changyu Li2†
Changyu Li2† Yaolou Wang
Yaolou Wang Zhao Yu
Zhao Yu Binbin Gui
Binbin Gui Qi Li
Qi Li Hongsheng Liang
Hongsheng Liang