
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 29 April 2024
Sec. Multiple Sclerosis and Neuroimmunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1370276
This article is part of the Research TopicNeuroinflammation, Neurodegeneration and Metabolic Disease: From Molecular Mechanisms to Therapeutic InnovationView all 11 articles
Background: Extensive observational studies have reported an association between inflammatory factors and autism spectrum disorder (ASD), but their causal relationships remain unclear. This study aims to offer deeper insight into causal relationships between circulating inflammatory factors and ASD.
Methods: Two-sample bidirectional Mendelian randomization (MR) analysis method was used in this study. The genetic variation of 91 circulating inflammatory factors was obtained from the genome-wide association study (GWAS) database of European ancestry. The germline GWAS summary data for ASD were also obtained (18,381 ASD cases and 27,969 controls). Single nucleotide polymorphisms robustly associated with the 91 inflammatory factors were used as instrumental variables. The random-effects inverse-variance weighted method was used as the primary analysis, and the Bonferroni correction for multiple comparisons was applied. Sensitivity tests were carried out to assess the validity of the causal relationship.
Results: The forward MR analysis results suggest that levels of sulfotransferase 1A1, natural killer cell receptor 2B4, T-cell surface glycoprotein CD5, Fms-related tyrosine kinase 3 ligand, and tumor necrosis factor-related apoptosis-inducing ligand are positively associated with the occurrence of ASD, while levels of interleukin-7, interleukin-2 receptor subunit beta, and interleukin-2 are inversely associated with the occurrence of ASD. In addition, matrix metalloproteinase-10, caspase 8, tumor necrosis factor-related activation-induced cytokine, and C-C motif chemokine 19 were considered downstream consequences of ASD.
Conclusion: This MR study identified additional inflammatory factors in patients with ASD relative to previous studies, and raised a possibility of ASD-caused immune abnormalities. These identified inflammatory factors may be potential biomarkers of immunologic dysfunction in ASD.
Autism spectrum disorders (ASD) are defined as a group of neurodevelopmental conditions of childhood with environmental causes that are still not fully understood. Most environmental factors during the perinatal stage appear to converge into a series of inflammatory conditions, such as bacterial and viral infections and inflammatory bowel disease (1, 2), suggesting that inflammatory responses could be an underlying factor in the etiology of ASD. Large population-based epidemiological studies have linked ASD with autoimmune disease and abnormal blood levels of various inflammatory cytokines and immunological biomarkers (3, 4). For instance, previous studies on inflammatory biomarkers have found increased concentrations of pro-inflammatory factors IL-1β, IL-4, IL-6, IL-8, and TNF-α (5–8), as well as decreased concentrations of anti-inflammatory factors IL-10 and IL-1Ra (9) in the peripheral blood of patients with ASD. These abnormal inflammatory cytokine levels are linked to greater impairments in language function and social interaction in children with ASD (6, 8). Therefore, it is necessary to further identify inflammatory biomarkers in ASD and uncover the causality between ASD and changes in the levels of inflammatory factors.
Observational studies are often susceptible to confounding, reverse causation, and multiple biases, which can lead to unreliable findings regarding the causal effects of exposures on outcomes. The Mendelian randomization (MR) method provides an alternative approach to investigate causality in epidemiological research, by utilizing genetic variants as instrumental variables to determine whether a risk factor has a causal effect on a health outcome. As an individual’s genotype is determined at conception and cannot be altered, this method avoids the reverse causality between genotype and outcome. In general, MR analysis rests on 3 assumptions: (1) genetic variants are associated with the risk factor; (2) genetic variants are not associated with confounders; and (3) genetic variants affect the outcome only through the risk factor (10). The advent of large-scale genome-wide association studies (GWAS) increases the accessibility of single-nucleotide polymorphisms (SNPs) as instrumental variables to infer causality in MR studies.
Based on the GWAS summary statistics, previous studies have examined causal relationships between 41 circulating inflammatory factors (11) and various complex diseases, including depression (12), epilepsy (13), and Alzheimer’s disease (14). Recently, Zhao et al. (15) extended previous works by conducting a genome-wide protein quantitative trait locus study which identified the genetic architecture of 91 circulating inflammatory factors in 14,824 European-ancestry participants. Several recent studies have included these 91 inflammatory factors for MR analysis, which further extends the use of inflammatory factors in MR studies (16, 17). Although previous observational studies have found a strong association between changes in levels of some circulating inflammatory proteins and ASD (5–9), their causal relationships remain undefined. The number of inflammatory factors analyzed in these studies is also relatively limited. Based on the knowledge above, we conducted the first bidirectional two-sample MR analysis (SNPs associated with the exposure and outcome are individually obtained from two independent samples) to determine the causal relationship between 91 inflammatory factors and ASD. The inflammatory biomarkers identified in this work may provide the basis for an objective test for early and accurate diagnosis of ASD and may shed light on the etiology and pathogenesis of ASD.
Figure 1 displays a schematic presentation of the study design. The data were obtained from the GWAS database and all included subjects had provided written informed consent in original research. The GWAS data sets for 91 circulating inflammatory factors are available in the GWAS Catalog (accession numbers from GCST90274758 to GCST90274848). These results come from a recent genome-wide protein quantitative trait locus study of 91 inflammation-related plasma factors in 14,824 European-ancestry participants (15). The genetic data on ASD were obtained from a genome-wide association meta-analysis of 18,381 ASD cases and 27,969 controls by Grove et al. (18) (OpenGWAS: ieu-a-1185). All participants were children born in Denmark between 1981 and 2005, diagnosed with ASD according to the 10th Revision of the International Classification of Diseases before 2014.
The SNPs strongly associated with inflammatory factors (p < 5e–06) were selected as instrumental variables. The linkage disequilibrium of these SNPs was removed using the clumping procedure in PLINK software (version v1.90), with the linkage disequilibrium parameter (r2) < 0.001 and a distance threshold of 10,000 kilobases. The r2 was calculated based on the 1000 Genomes Projects reference panel (Genomes Project C et al., 2012) (19). Additionally, we excluded palindrome SNPs and weak instrumental SNPs (F-statistics < 10). The F statistic was calculated by the following equation: F = R2 × (N−k−1)/k × (1 − R2), where N is the sample size of the exposure factor, K is the number of instrumental variables, and R2 is the proportion of variance explained by each instrumental variable. An F-statistic >10 typically indicates a strong correlation between instrumental variables and exposure factors (20).
Both statistical and sensitive analyses were conducted using the R software (version 4.2.1) and the “TwoSampleMR” R package (21, 22). The random-effects inverse variance weighted (IVW) method was employed as the main MR analysis to estimate causal effects, complemented by the weighted-median (WM) and MR-Egger methods to investigate potential pleiotropic effects. The IVW method can analyze individual Wald-type ratios of causal effects for each SNP, which provides the most accurate and unbiased causal estimates in the absence of horizontal pleiotropy (23). In the presence of horizontal pleiotropy, the WM method provides a consistent estimate even though half of the genetic variants are invalid instrumental variables (24). The estimation of causal effects of modifiable phenotypes on an outcome relies on the assumption of no pleiotropy, wherein genes solely influence the outcome via the given phenotype. If the genetic variants have a pleiotropic effect on the outcome, then the causal estimates may be biased. The MR-Egger regression intercept test was used to assess residual horizontal pleiotropy – intercepts around the zero indicate that SNPs do not have a direct effect on the outcome via the exposure (25). The Cochran’s Q statistic was utilized to evaluate the heterogeneity of SNPs in both the IVW and MR Egger methods (21). The core assumption of MR is not contradicted even if there is significant heterogeneity in the instrumental variables. A p-value of less than 0.05 suggests the presence of horizontal pleiotropy and heterogeneity. The association of individual SNPs was determined using leave-one-out sensitivity analysis to investigate whether the results were driven by any single SNP (26). For binary exposures, causal estimates were presented as odds ratio (OR) and 95% confidence interval (CI) per log-odds increment of genetic exposure risk. To account for multiple testing, a p-value of 0.00055 (0.05/91) was considered robust significance after the Bonferroni correction. A p-value between 0.00055 and 0.05 was deemed as suggestive evidence of potential causation. All statistical analyses were two-sided.
After the selection of SNPs based on the criteria, a total of 1,815 SNPs related to 91 circulating inflammatory factors extracted from the GWAS database were used as instrumental variables (Supplementary Table S1). The basic information on 91 circulating inflammatory factors is summarized in Supplementary Table S2. Due to the large number of inflammatory factors and associated SNPs, in this section, we focus on the presentation of positive results in MR analysis. SNPs used in the positive results of both forward and reverse MR analysis are shown in Supplementary Tables S3, S4. This compilation includes information on chromosome location, effect allele, and effect allele frequency. Moreover, all SNPs had F statistics greater than 10, indicating that they were free of weak instrumental bias.
The analysis results from IVW showed a statistically significant negative correlation between levels of interleukin-7 (IL-7) and ASD (OR = 0.858, 95% CI = 0.796 to 0.925, p = 6.69e-05). The results also showed possible positive associations between elevated levels of sulfotransferase 1A1 (SULT1A1) (OR = 1.109, 95% CI = 1.0423 to 1.181, p = 0.001), natural killer cell receptor 2B4 (CD244) (OR = 1.144, 95% CI = 1.040 to 1.259, p = 0.006), T-cell surface glycoprotein CD5 (CD5) (OR = 1.126, 95% CI = 1.028 to 1.233, p = 0.011), Fms-related tyrosine kinase 3 ligand (FLT3LG) (OR = 1.120, 95% CI = 1.013 to 1.238, p = 0.027), and tumor necrosis factor-related apoptosis-inducing ligand (TNFSF10) (OR = 1.093 95% CI = 1.009 to 1.184, p = 0.029) and an increased occurrence of ASD. Levels of interleukin-2 receptor subunit beta (IL2Rβ) (OR = 0.838, 95% CI = 0.749 to 0.936, p = 0.002), and interleukin-2 (IL-2) (OR = 0.874, 95% CI = 0.785 to 0.972, p = 0.013) were inversely associated with the risk of ASD. The results of the WM analysis for CD244 (OR = 1.150, 95% CI = 1.026 to 1.289, p = 0.016) and CD5 (OR = 1.181, 95% CI = 1.028 to 1.355, p = 0.018) also indicate a causal relationship with ASD, consistent with the trend observed in the IVW method. MR Egger analysis of all the inflammatory factors did not find any significant causal relationship with ASD. Figure 2 provide the results of the IVW, WM, and MR Egger analysis. None of the intercepts in the MR-Egger regression analysis significantly deviated from the zero (p > 0.05), suggesting no horizontal pleiotropy. Heterogeneity was observed only in TNFSF10 with a Cochran’s Q-derived p <0.05, but the causal estimate was acceptable when utilizing the random-effects IVW method (Table 1). The MR leave-one-out sensitivity analysis indicated that sequentially excluding individual SNP did not significantly influence the results, and all the estimates of the error lines were on the same side (Figure 3). The results of the IVW, WM, and MR Egger mode for all circulating inflammatory factors on ASD are displayed in Supplementary Table S5.
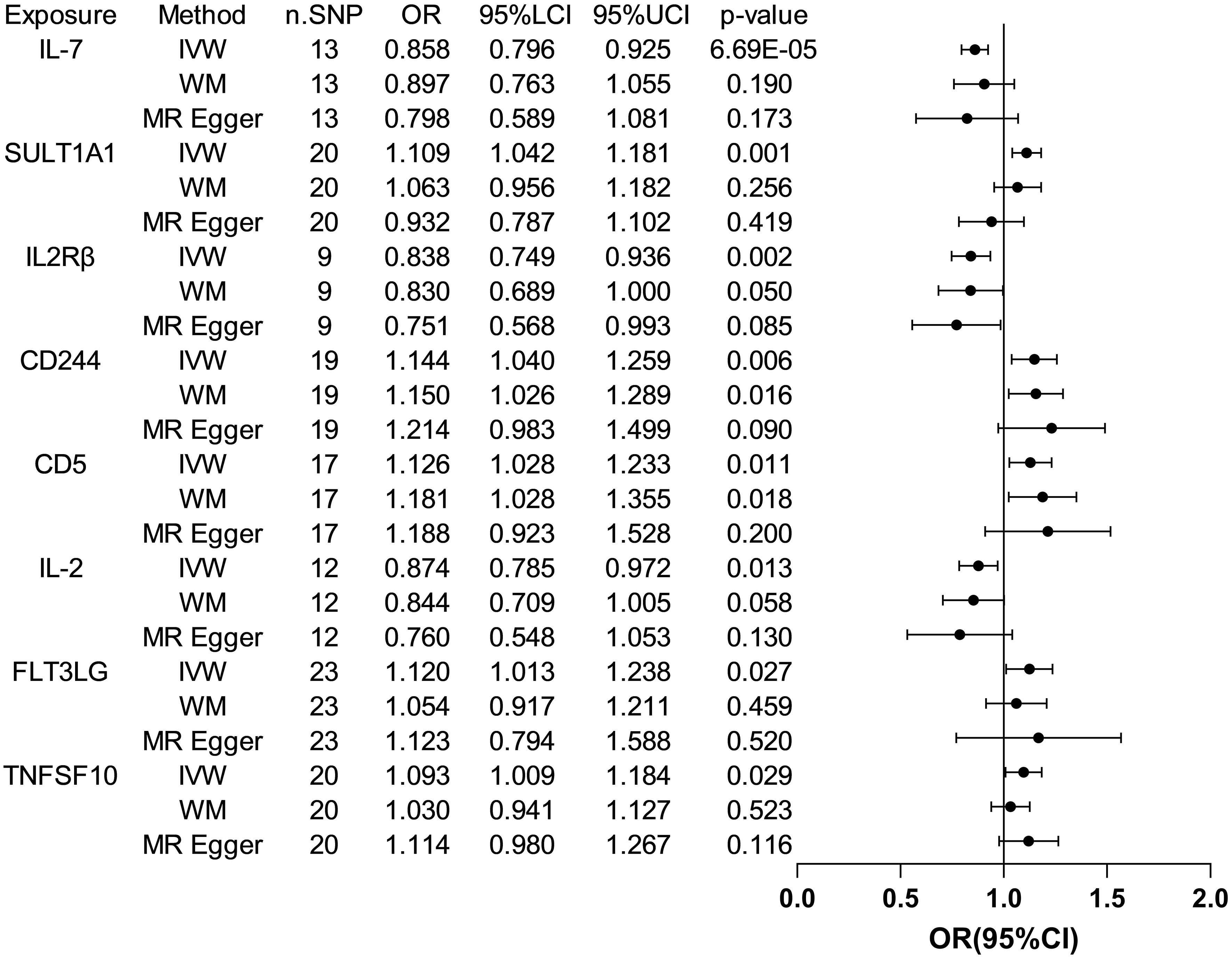
Figure 2 Forest plots of the pooled OR results between 8 inflammatory factors and ASD in the forward MR analysis.
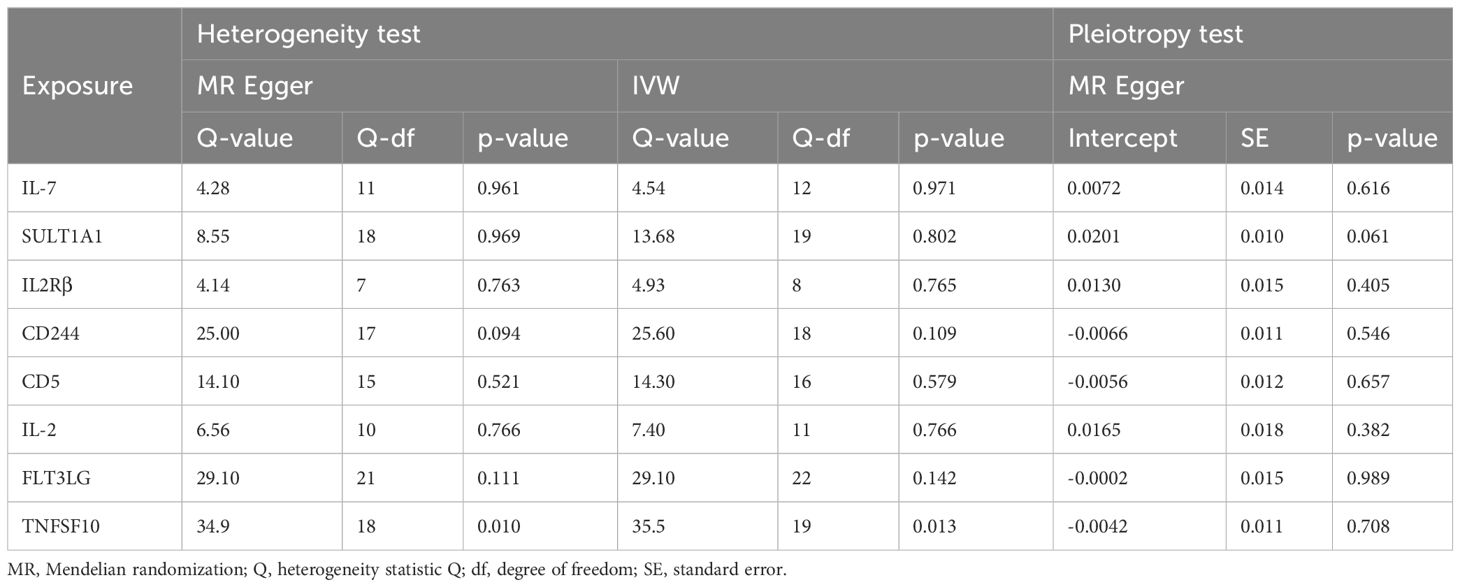
Table 1 The heterogeneity and horizontal pleiotropy results of the 8 inflammatory factors and ASD in the forward MR analysis.

Figure 3 Leave-one-out of SNPs results between 8 inflammatory factors and ASD in the forward MR analysis. Each black point indicates the MR result of all remaining SNPs after removing the SNP in this line. The MR results estimated by all SNPs are depicted by the red point. No single SNP strongly drives the overall causal effect in these sensitivity analyses.
When considering functional outcomes of ASD as exposures and the 91 circulating inflammatory factors as outcomes, the IVW results indicate that adverse functional outcomes following ASD may lead to increased levels of matrix metalloproteinase-10 (MMP10) and caspase 8 (CASP8) (OR = 1.067, 95% CI = 1.006 to 1.131, p = 0.032; OR = 1.064, 95% CI = 1.003-1.128, p = 0.040), as well as decreased levels of tumor necrosis factor-related activation-induced cytokine (TNFSF11) and C-C motif chemokine 19 (CCL19) (OR = 0.942, 95% CI = 0.888 to 0.998, p = 0.044; OR = 0.942 95% CI = 0.888 to 0.999, p = 0.047). The analysis results from both WM and MR Egger did not reveal any significant causality between ASD and the four inflammatory factors. Figure 4 provide the results of the IVW, WM, and MR Egger analysis. Cochran’s Q-test results showed no evidence of heterogeneity in the causal relationship between these SNPs. The p-value of the MR-Egger intercept was greater than 0.05, indicating that horizontal pleiotropy was not possible for these four associations (Table 2). Furthermore, the sensitivity analysis proved the robustness of these observed causal associations (Figure 5). The results of the IVW, WM, and MR Egger mode for ASD on all circulating inflammatory factors are displayed in Supplementary Table S6.
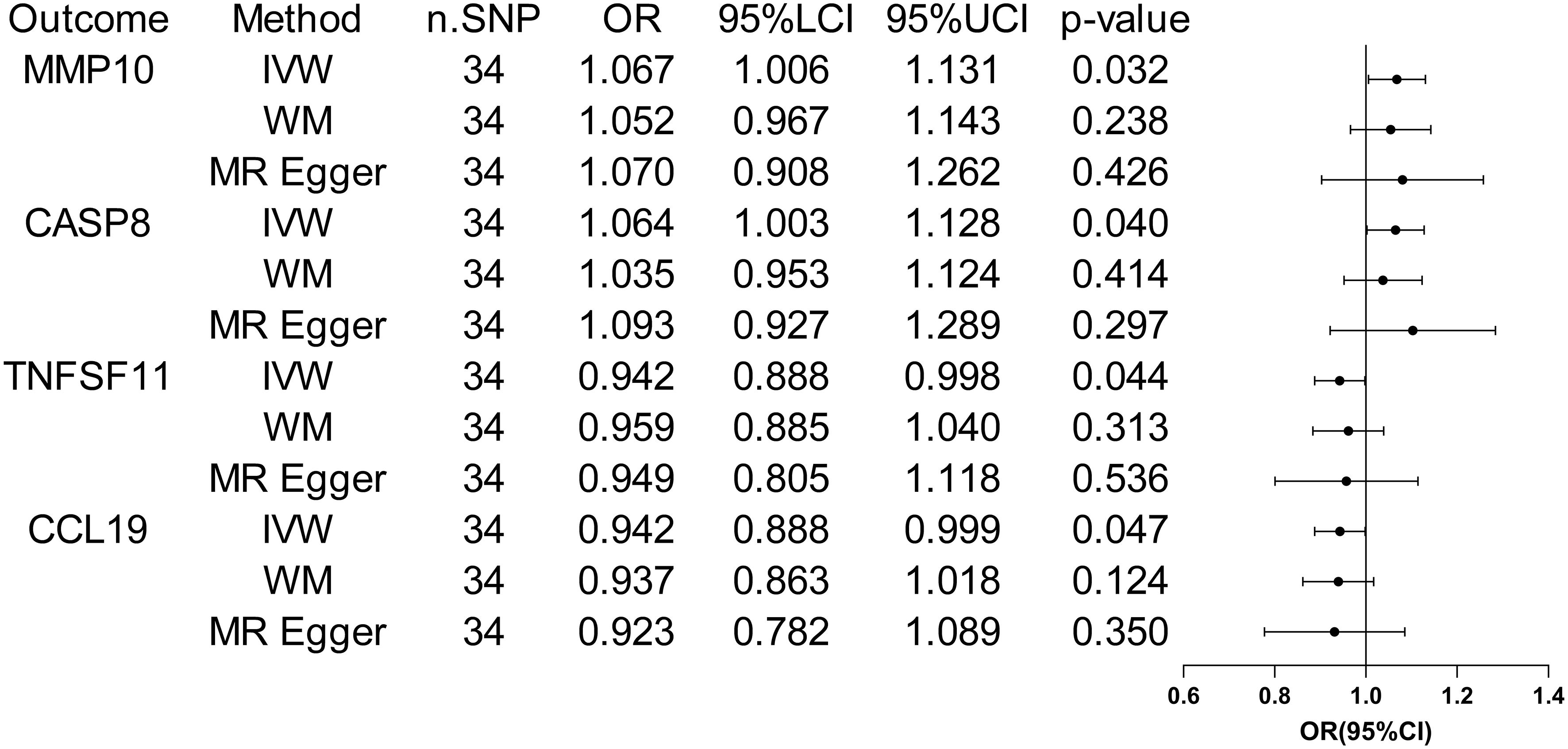
Figure 4 Forest plots of the pooled OR results between ASD and 4 inflammatory factors in the reverse MR analysis.
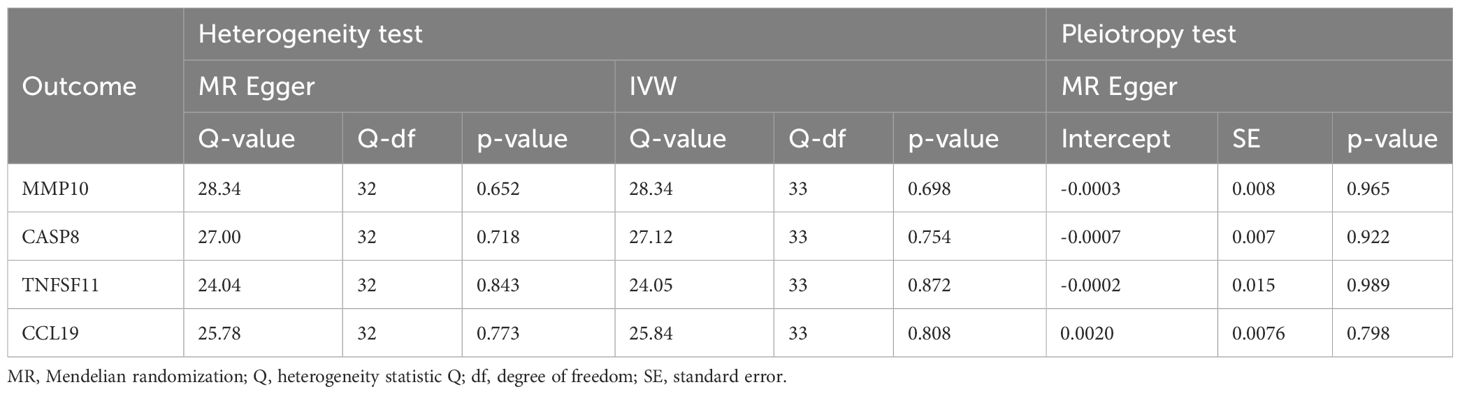
Table 2 The heterogeneity and horizontal pleiotropy results of the ASD and 4 inflammatory factors in the reverse MR analysis.
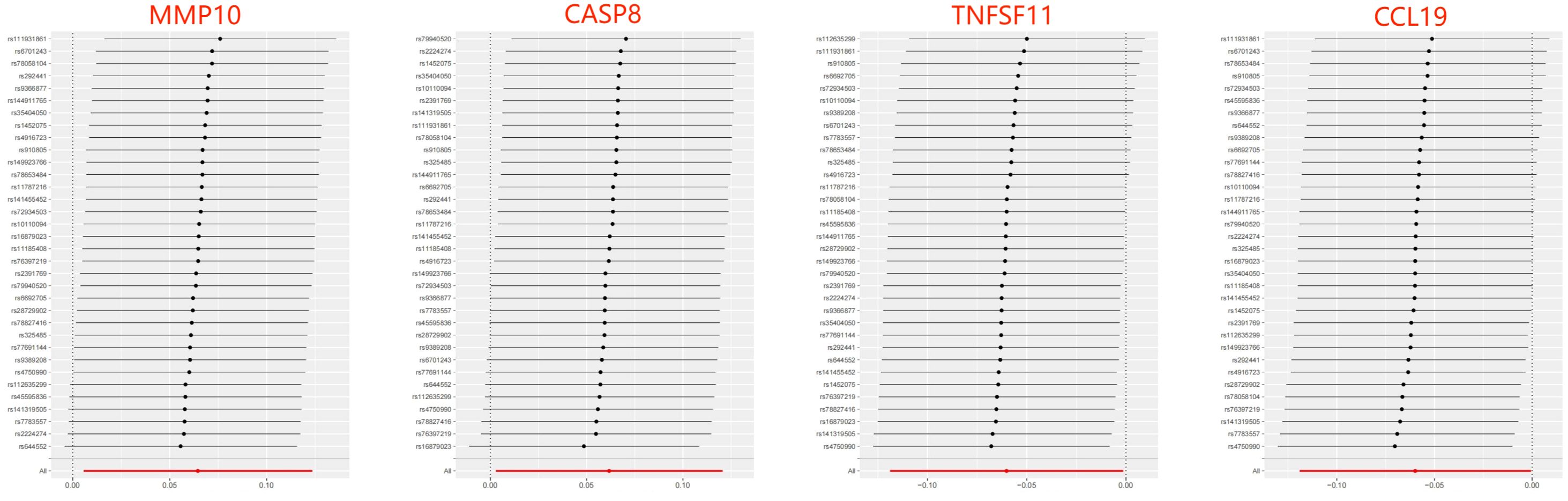
Figure 5 Leave-one-out of SNPs results between ASD and 4 inflammatory factors in the reverse MR analysis. Each black point indicates the MR result of all remaining SNPs after removing the SNP in this line. The MR results estimated by all SNPs are depicted by the red point. No single SNP strongly drives the overall causal effect in these sensitivity analyses.
Although recent observational studies have suggested an association between inflammatory factors and ASD (27–29), it is important to note that these results may be influenced by confounding factors, and the causal relationship between the two remains inconclusive. Using novel data and approaches, this bidirectional MR study offers a genetic insight into the potential causal relationship between circulating inflammatory factors and ASD. The findings of this study suggest that levels of SULT1A1, CD244, CD5, FLT3LG, and TNFSF10 are positively associated with the risk of ASD, while levels of IL-7, IL2Rβ, and IL-2 are inversely associated with the risk of ASD. Furthermore, the genetic susceptibility to ASD exhibited suggestive evidence of increased levels of MMP10 and CASP8, and decreased levels of TNFSF11 and CCL19. Importantly, sensitivity analyses supported the robustness of these results.
Individuals with ASD show alterations in circulating inflammatory factors along with abnormal peripheral blood levels of lymphocytes and macrophages across the lifespan (30). Inflammatory factor profiling can reveal the progression and status of immune system dysregulation in ASD, offering therapeutic targets for improving core autistic symptoms. Current results in the forward MR analysis demonstrated a strong causal association between a decreased level of circulating IL-7 and a higher liability of ASD based on the Bonferroni correction. IL-7 is mainly produced by stromal cells in lymphoid tissues and has pleiotropic effects on the development of T and B cells, as well as T-cell homeostasis. The administration or neutralization of IL-7 may enable the modulation of immune function in individuals with lymphocyte depletion or autoimmunity (31). Data from several studies have shown that plasma levels of IL-7 were higher in children with ASD than those observed in typically developing controls, but these differences did not reach statistical significance after correction for multiple comparisons (32–34). By contrast, Napolioni and colleagues (35) found that plasma levels of IL-7 were inversely correlated with full intelligence quotient in children with ASD using Spearman’s rank correlation analysis. Similarly, a large observational study on the risk of psychopathology found that decreased IL-7 levels in cord serum were linked to emotional symptoms and abnormal pro-social behavior in 5-year-old children (36). Overall, this MR analysis based on large populations replicates and extends these findings, highlighting a causal protective role of genetically encoded higher IL-7 levels against ASD.
Other results did not show robust causality after Bonferroni correction, which can only be regarded as suggestive evidence of potential causality. IL-2 functions as an essential immunoregulatory factor produced primarily by T cells, exerting its effects via binding to the high-affinity IL-2R comprising of α (IL-2Rα), β (IL-2Rβ), and γ (IL-2Rγ) subunits. Both IL-2 and IL-2Rβ have been implicated in clonal expansion and functional differentiation of T cells and natural killer cells (37, 38). Vojdani et al. have found that children with ASD appeared to suffer from decreased blood natural killer cell activity due to their low intracellular IL-2 levels (39). In patients with ASD, the proportion of DR+ (activated) T lymphocytes is abnormally increased, whereas the proportion of IL-2 receptor+ lymphocytes remains unchanged or even decreases. This is inversely proportional to the severity of autistic symptoms and similar to that seen in autoimmune diseases (40–42). In addition, previous studies have observed lower mRNA expression levels of IL-2 and percentages of IL-2 synthesizing CD4+ and CD8+ T cells in the peripheral blood of ASD children as compared to controls (43, 44). To sum up, our findings further support these observations and provide evidence of immune dysfunction and autoimmunity in patients with ASD.
Preliminary results suggest that there may be a positive association between levels of SULT1A1, CD244, CD5, FLT3LG, and TNFSF10 and risk of ASD. However, much of the research up to now has not dealt with the relationship between these inflammatory factors and ASD. SULT1A1 is responsible for the sulfonation of xenobiotics and has been implicated in several cancers by activating carcinogens (45). During autoimmune neuroinflammation, SULT1A is highly expressed in astrocytes, hindering the anti-inflammatory activity of endogenous estrogens (46). CD244 is an immune regulation receptor presented in all NK cells, which can stimulate NK cell cytotoxicity and IFN-γ production by interacting with CD48 on neighboring lymphocytes (47, 48). It has previously been observed that children with ASD had significantly higher serum and plasma levels of CD5 than those of normal controls, which is positively correlated with Childhood Autism Rating Scale score (49, 50). As a pan T cell marker, CD5 is highly expressed in a variety of autoimmune diseases, and this MR study provides new evidence that elevated levels of circulating CD5 may directly promote the development of ASD. In premature infants following respiratory viral infections, hyperoxia-induced high FLT3LG expression can lead to expansion and activation of lung CD103+ dendritic cells. The FLT3LG level is positively correlated with the level of proinflammatory cytokines (51). In addition, chronic HIV-1 patients also displayed significantly high levels of FLT3LG expression (52). TNFSF10 a proapoptotic member of the tumor necrosis factor family, has been shown to be highly up-regulated in patients with inflammatory bowel disease (53) and neurodegenerative diseases (54). The previous findings suggest a positive outcome for our MR results, indicating the need for further research on the current topic.
The occurrence of ASD may increase the levels of MMP10 and CASP8 and decrease the levels of TNFSF11 and CCL19 in the results of reverse MR analyses, suggesting that they may act as downstream factors in ASD. A recent proteomics analysis on plasma inflammation-related protein changes found that MMP-10 expression was significantly up-regulated in the ASD group compared with healthy children (29). Mild cognitive impairment individuals with elevated cerebrospinal fluid levels of MMP-10 had a higher likelihood of progression to Alzheimer’s type dementia and faster cognitive decline (55). CASP8 is a protease with both pro-death and pro-survival functions by mediating extrinsic apoptosis and suppressing necroptosis. Postmortem analysis results showed that the apoptosis was increased in the prefrontal cortex, hippocampus, and cerebellum of the autistic brain, as characterized by significantly increased levels of cleaved CASP8 (56). TNFSF11 exerts essential roles in lymph node organogenesis, cellular immunity, and osteoclastogenesis. For example, TNFSF11 can signal the augmentation of IFN-γ secretion and inhibit apoptosis of human monocyte-derived dendritic cells (57, 58). CCL19 has shown significant potential in the regulation of adaptive immune responses by coordinating dendritic cell migration and increasing interactions between dendritic cells, T cells, and B cells in secondary lymphoid tissues (59, 60). Hence, ASD-induced decreases in peripheral blood CCL19 and TNFSF11 levels may further exacerbate immune system disorders.
This MR study employed a large sample size and instrumental variables obtained from the GWAS database, ensuring statistical robustness in estimating causal associations and enhancing the credibility of results. By addressing the bias introduced by confounding factors and reverse causality through MR analysis, this study provides stronger evidence for assessing the causal relationship between inflammatory factor levels and the risk of ASD compared to traditional observational studies. Nevertheless, several limitations of the present study should be considered. Firstly, the present MR study can only provide statistical evidence for the causal association between circulating inflammatory factors and ASD, and further research is needed to investigate the potential mechanisms involved. Secondly, although current sensitivity analyses did not reveal any significant pleiotropy between SNPs, the effect of pleiotropy on the MR results cannot be completely ruled out. Thirdly, this MR study utilized pooled data from the GWAS database and did not analyze stratified risk factors related to ASD duration, severity, treatment, and comorbidities. Finally, the genetic data were mainly collected from individuals of European descent, and it is uncertain whether these findings are applicable to individuals of other ancestries.
Overall, this study provide novel ideas that IL-7, SULT1A1, IL2Rβ, CD244, CD5, IL-2, FLT3LG, and TNFSF10 may be upstream factors in the pathogenesis of ASD, while levels of MMP10, CASP8, TNFSF11, and CCL19 may act as downstream factors in ASD. These inflammatory factors could potentially serve as biomarkers for early diagnosis and treatment of ASD.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
JL: Writing – review & editing, Writing – original draft, Resources, Methodology, Investigation, Formal Analysis, Data curation, Conceptualization. HD: Writing – review & editing, Writing – original draft, Visualization, Software, Investigation, Formal Analysis. WS: Writing – review & editing, Visualization, Software, Methodology, Investigation. MM: Writing – review & editing, Validation, Resources, Methodology. HZ: Writing – review & editing, Writing – original draft, Supervision, Project administration, Investigation, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Key Research and Development Program of China (2018YFC2001703), China Disabled Persons Federation (CDPF2023KF00001), and Changping laboratory (2021B-01-01-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1370276/full#supplementary-material
1. Careaga M, Murai T, Bauman MD. Maternal immune activation and autism spectrum disorder: from rodents to nonhuman and human primates. Biol Psychiatry. (2017) 81:391–401. doi: 10.1016/j.biopsych.2016.10.020
2. Sadik A, Dardani C, Pagoni P, Havdahl A, Stergiakouli E, iPSYCH Autism Spectrum Disorder Working Group, et al. Parental inflammatory bowel disease and autism in children. Nat Med. (2022) 28:1406–11. doi: 10.1038/s41591-022-01845-9
3. Gesundheit B, Rosenzweig JP, Naor D, Lerer B, Zachor DA, Procházka V, et al. Immunological and autoimmune considerations of Autism Spectrum Disorders. J Autoimmun. (2013) 44:1–7. doi: 10.1016/j.jaut.2013.05.005
4. Hughes HK, Mills Ko E, Rose D, Ashwood P. Immune dysfunction and autoimmunity as pathological mechanisms in autism spectrum disorders. Front Cell Neurosci. (2018) 12:405. doi: 10.3389/fncel.2018.00405
5. Kordulewska NK, Kostyra E, Piskorz-Ogórek K, Moszyńska M, Cieślińska A, Fiedorowicz E, et al. Serum cytokine levels in children with spectrum autism disorder: Differences in pro- and anti-inflammatory balance. J Neuroimmunol. (2019) 337:577066. doi: 10.1016/j.jneuroim.2019.577066
6. Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah I, Van de Water J. Elevated plasma cytokines in autism spectrum disorders provide evidence of immune dysfunction and are associated with impaired behavioral outcome. Brain Behav Immun. (2011) 25:40–5. doi: 10.1016/j.bbi.2010.08.003
7. Zhao H, Zhang H, Liu S, Luo W, Jiang Y, Gao J. Association of peripheral blood levels of cytokines with autism spectrum disorder: A meta-analysis. Front Psychiatry. (2021) 12:670200. doi: 10.3389/fpsyt.2021.670200
8. Krakowiak P, Goines PE, Tancredi DJ, Ashwood P, Hansen RL, Hertz-Picciotto I, et al. Neonatal cytokine profiles associated with autism spectrum disorder. Biol Psychiatry. (2017) 81:442–51. doi: 10.1016/j.biopsych.2015.08.007
9. Saghazadeh A, Ataeinia B, Keynejad K, Abdolalizadeh A, Hirbod-Mobarakeh A, Rezaei N. Anti-inflammatory cytokines in autism spectrum disorders: A systematic review and meta-analysis. Cytokine. (2019) 123:154740. doi: 10.1016/j.cyto.2019.154740
10. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. (2017) 318:1925–6. doi: 10.1001/jama.2017.17219
11. Ahola-Olli AV, Würtz P, Havulinna AS, Aalto K, Pitkänen N, Lehtimäki T, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. (2017) 100:40–50. doi: 10.1016/j.ajhg.2016.11.007
12. Wang M, Jin G, Cheng Y, Guan SY, Zheng J, Zhang SX. Genetically predicted circulating levels of cytokines and the risk of depression: a bidirectional Mendelian-randomization study. Front Genet. (2023) 14:1242614. doi: 10.3389/fgene.2023.1242614
13. Sun H, Ma D, Hou S, Zhang W, Li J, Zhao W, et al. Exploring causal correlations between systemic inflammatory cytokines and epilepsy: A bidirectional Mendelian randomization study. Seizure. (2023) 114:44–9. doi: 10.1016/j.seizure.2023.11.006
14. Yeung CHC, Schooling CM. Systemic inflammatory regulators and risk of Alzheimer’s disease: a bidirectional Mendelian-randomization study. Int J Epidemiol. (2021) 50:829–40. doi: 10.1093/ije/dyaa241
15. Zhao JH, Stacey D, Eriksson N, Macdonald-Dunlop E, Hedman ÅK, Kalnapenkis A, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. (2023) 24:1540–51. doi: 10.1038/s41590-023-01588-w
16. Huang H, Fu Z, Yang M, Hu H, Wu C, Tan L. Levels of 91 circulating inflammatory proteins and risk of lumbar spine and pelvic fractures and peripheral ligament injuries: a two-sample mendelian randomization study. J Orthop Surg Res. (2024) 19:161. doi: 10.1186/s13018-024-04637-8
17. Shi W, Xu Y, Zhang A, Jia X, Liu S, Hu Z. Inflammatory cytokines and their potential role in Sjogren’s syndrome risk: insights from a mendelian randomization study. Adv Rheumatol. (2024) 64:14. doi: 10.1186/s42358-024-00354-2
18. Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H, et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. (2019) 51:431–44. doi: 10.1038/s41588-019-0344-8
19. 1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. (2012) 491:56–65. doi: 10.1038/nature11632
20. Burgess S, Thompson SG, CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
21. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. (2018) 7:e34408. doi: 10.7554/eLife.34408
22. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. (2017) 46:1734–9. doi: 10.1093/ije/dyx034
23. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. (2017) 26:2333–55. doi: 10.1177/0962280215597579
24. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
25. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
26. Liang Z, Zhao L, Lou Y, Liu S. Causal effects of circulating lipids and lipid-lowering drugs on the risk of epilepsy: a two-sample Mendelian randomization study. QJM. (2023) 116:421–8. doi: 10.1093/qjmed/hcad048
27. Gesundheit B, Zisman PD, Hochbaum L, Posen Y, Steinberg A, Friedman G, et al. Autism spectrum disorder diagnosis using a new panel of immune- and inflammatory-related serum biomarkers: A case-control multicenter study. Front Pediatr. (2023) 11:967954. doi: 10.3389/fped.2023.967954
28. Than UTT, Nguyen LT, Nguyen PH, Nguyen XH, Trinh DP, Hoang DH, et al. Inflammatory mediators drive neuroinflammation in autism spectrum disorder and cerebral palsy. Sci Rep. (2023) 13:22587. doi: 10.1038/s41598-023-49902-8
29. Bao XH, Chen BF, Liu J, Tan YH, Chen S, Zhang F, et al. Olink proteomics profiling platform reveals non-invasive inflammatory related protein biomarkers in autism spectrum disorder. Front Mol Neurosci. (2023) 16:1185021. doi: 10.3389/fnmol.2023.1185021
30. Arteaga-Henríquez G, Gisbert L, Ramos-Quiroga JA. Immunoregulatory and/or anti-inflammatory agents for the management of core and associated symptoms in individuals with autism spectrum disorder: A narrative review of randomized, placebo-controlled trials. CNS Drugs. (2023) 37:215–29. doi: 10.1007/s40263-023-00993-x
31. Fry TJ, Mackall CL. Interleukin-7: from bench to clinic. Blood. (2002) 99:3892–904. doi: 10.1182/blood.v99.11.3892
32. Inga Jácome MC, Morales Chacòn LM, Vera Cuesta H, Maragoto Rizo C, Whilby Santiesteban M, Ramos Hernandez L, et al. Peripheral inflammatory markers contributing to comorbidities in autism. Behav Sci. (2016) 6:29. doi: 10.3390/bs6040029
33. Suzuki K, Matsuzaki H, Iwata K, Kameno Y, Shimmura C, Kawai S, et al. Plasma cytokine profiles in subjects with high-functioning autism spectrum disorders. PloS One. (2011) 6:e20470. doi: 10.1371/journal.pone.0020470
34. Pecorelli A, Cervellati F, Belmonte G, Montagner G, Waldon P, Hayek J, et al. Cytokines profile and peripheral blood mononuclear cells morphology in Rett and autistic patients. Cytokine. (2016) 77:180–8. doi: 10.1016/j.cyto.2015.10.002
35. Napolioni V, Ober-Reynolds B, Szelinger S, Corneveaux JJ, Pawlowski T, Ober-Reynolds S, et al. Plasma cytokine profiling in sibling pairs discordant for autism spectrum disorder. J Neuroinflamm. (2013) 10:38. doi: 10.1186/1742-2094-10-38
36. Barbosa S, Khalfallah O, Forhan A, Galera C, Heude B, Glaichenhaus N, et al. Immune activity at birth and later psychopathology in childhood. Brain Behav Immun Health. (2020) 8:100141. doi: 10.1016/j.bbih.2020.100141
37. Fernandez IZ, Baxter RM, Garcia-Perez JE, Vendrame E, Ranganath T, Kong DS, et al. A novel human IL2RB mutation results in T and NK cell-driven immune dysregulation. J Exp Med. (2019) 216:1255–67. doi: 10.1084/jem.20182015
38. Henney CS, Kuribayashi K, Kern DE, Gillis S. Interleukin-2 augments natural killer cell activity. Nature. (1981) 291:335–8. doi: 10.1038/291335a0
39. Vojdani A, Mumper E, Granpeesheh D, Mielke L, Traver D, Bock K, et al. Low natural killer cell cytotoxic activity in autism: the role of glutathione, IL-2 and IL-15. J Neuroimmunol. (2008) 205:148–54. doi: 10.1016/j.jneuroim.2008.09.005
40. Denney DR, Frei BW, Gaffney GR. Lymphocyte subsets and interleukin-2 receptors in autistic children. J Autism Dev Disord. (1996) 26:87–97. doi: 10.1007/BF02276236
41. Plioplys AV, Greaves A, Kazemi K, Silverman E. Lymphocyte function in autism and Rett syndrome. Neuropsychobiology. (1994) 29:12–6. doi: 10.1159/000119056
42. Warren RP, Yonk J, Burger RW, Odell D, Warren WL. DR-positive T cells in autism: association with decreased plasma levels of the complement C4B protein. Neuropsychobiology. (1995) 31:53–7. doi: 10.1159/000119172
43. Eftekharian MM, Ghafouri-Fard S, Noroozi R, Omrani MD, Arsang-Jang S, Ganji M, et al. Cytokine profile in autistic patients. Cytokine. (2018) 108:120–6. doi: 10.1016/j.cyto.2018.03.034
44. Gupta S, Aggarwal S, Rashanravan B, Lee T. Th1- and Th2-like cytokines in CD4+ and CD8+ T cells in autism. J Neuroimmunol. (1998) 85:106–9. doi: 10.1016/s0165-5728(98)00021-6
45. Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, et al. Structural and kinetic implications of substrate inhibition. J Biol Chem. (2003) 278:7655–62. doi: 10.1074/jbc.M207246200
46. Guillot F, Garcia A, Salou M, Brouard S, Laplaud DA, Nicot AB. Transcript analysis of laser capture microdissected white matter astrocytes and higher phenol sulfotransferase 1A1 expression during autoimmune neuroinflammation. J Neuroinflamm. (2015) 12:130. doi: 10.1186/s12974-015-0348-y
47. Sun L, Gang X, Li Z, Zhao X, Zhou T, Zhang S, et al. Advances in understanding the roles of CD244 (SLAMF4) in immune regulation and associated diseases. Front Immunol. (2021) 12:648182. doi: 10.3389/fimmu.2021.648182
48. Messmer B, Eissmann P, Stark S, Watzl C. CD48 stimulation by 2B4 (CD244)-expressing targets activates human NK cells. J Immunol. (2006) 176:4646–50. doi: 10.4049/jimmunol.176.8.4646
49. Desoky T, Hassan MH, Fayed HM, Sakhr HM. Biochemical assessments of thyroid profile, serum 25-hydroxycholecalciferol and cluster of differentiation 5 expression levels among children with autism. Neuropsychiatr Dis Treat. (2017) 13:2397–403. doi: 10.2147/NDT.S146152
50. Halepoto DM, Alhowikan AM, Ayadhi LA. Cluster of differentiation 5 (CD5) levels in the plasma of children with autism spectrum disorder (ASD). J Coll Physicians Surg Pak. (2017) 27:149–52.
51. Cui TX, Brady AE, Zhang YJ, Fulton CT, Goldsmith AM, Popova AP. Early-life hyperoxia-induced Flt3L drives neonatal lung dendritic cell expansion and proinflammatory responses. Front Immunol. (2023) 14:1116675. doi: 10.3389/fimmu.2023.1116675
52. Ling L, Tang X, Huang X, Li J, Wang H, Chen Z. AAV-vectored fms-related tyrosine kinase 3 ligand inhibits CD34+ Progenitor cell engraftment in humanized mice. J Neuroimmune Pharmacol. (2018) 13:541–50. doi: 10.1007/s11481-018-9819-0
53. Begue B, Wajant H, Bambou JC, Dubuquoy L, Siegmund D, Beaulieu JF, et al. Implication of TNF-related apoptosis-inducing ligand in inflammatory intestinal epithelial lesions. Gastroenterology. (2006) 130:1962–74. doi: 10.1053/j.gastro.2006.03.022
54. Huang Y, Erdmann N, Peng H, Zhao Y, Zheng J. The role of TNF related apoptosis-inducing ligand in neurodegenerative diseases. Cell Mol Immunol. (2005) 2:113–22.
55. Martino Adami PV, Orellana A, García P, Kleineidam L, Alarcón-Martín E, Montrreal L, et al. Matrix metalloproteinase 10 is linked to the risk of progression to dementia of the Alzheimer’s type. Brain. (2022) 145:2507–17. doi: 10.1093/brain/awac024
56. Dong D, Zielke HR, Yeh D, Yang P. Cellular stress and apoptosis contribute to the pathogenesis of autism spectrum disorder. Autism Res. (2018) 11:1076–90. doi: 10.1002/aur.1966
57. Chen NJ, Huang MW, Hsieh SL. Enhanced secretion of IFN-gamma by activated Th1 cells occurs via reverse signaling through TNF-related activation-induced cytokine. J Immunol. (2001) 166:270–6. doi: 10.4049/jimmunol.166.1.270
58. Wong BR, Josien R, Lee SY, Sauter B, Li HL, Steinman RM, et al. TRANCE (tumor necrosis factor [TNF]-related activation-induced cytokine), a new TNF family member predominantly expressed in T cells, is a dendritic cell-specific survival factor. J Exp Med. (1997) 186:2075–80. doi: 10.1084/jem.186.12.2075
59. Marsland BJ, Bättig P, Bauer M, Ruedl C, Lässing U, Beerli RR, et al. CCL19 and CCL21 induce a potent proinflammatory differentiation program in licensed dendritic cells. Immunity. (2005) 22:493–505. doi: 10.1016/j.immuni.2005.02.010
Keywords: autism spectrum disorder, inflammatory factors, inflammation, Mendelian randomization, single nucleotide polymorphisms, genome-wide association study
Citation: Long J, Dang H, Su W, Moneruzzaman M and Zhang H (2024) Interactions between circulating inflammatory factors and autism spectrum disorder: a bidirectional Mendelian randomization study in European population. Front. Immunol. 15:1370276. doi: 10.3389/fimmu.2024.1370276
Received: 14 January 2024; Accepted: 16 April 2024;
Published: 29 April 2024.
Edited by:
Fawaz Alzaid, Sorbonne Universités, FranceReviewed by:
Xiaokuang Ma, The University of Arizona College of Medicine – Phoenix, United StatesCopyright © 2024 Long, Dang, Su, Moneruzzaman and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Zhang, Y3JyY3poMjAyMEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.