- 1Engineering Research Center of Southwest Animal Disease Prevention and Control Technology, Ministry of Education of the People’s Republic of China, Chengdu, China
- 2Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
- 3International Joint Research Center for Animal Disease Prevention and Control of Sichuan Province, Chengdu, China
- 4Institute of Veterinary Medicine and Immunology, Sichuan Agricultural University, Chengdu, China
- 5Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
3D polymerase, also known as RNA-dependent RNA polymerase, is encoded by all known picornaviruses, and their structures are highly conserved. In the process of picornavirus replication, 3D polymerase facilitates the assembly of replication complexes and directly catalyzes the synthesis of viral RNA. The nuclear localization signal carried by picornavirus 3D polymerase, combined with its ability to interact with other viral proteins, viral RNA and cellular proteins, indicate that its noncatalytic role is equally important in viral infections. Recent studies have shown that 3D polymerase has multiple effects on host cell biological functions, including inducing cell cycle arrest, regulating host cell translation, inducing autophagy, evading immune responses, and triggering inflammasome formation. Thus, 3D polymerase would be a very valuable target for the development of antiviral therapies. This review summarizes current studies on the structure of 3D polymerase and its regulation of host cell responses, thereby improving the understanding of picornavirus-mediated pathogenesis caused by 3D polymerase.
1 Introduction
Picornaviruses represent one of the largest virus groups and include several important human and animal pathogens, such as poliovirus (PV), coxsackievirus (CV), enterovirus (EV), rhinovirus (RV), encephalomyocarditis virus (EMCV), and foot-and-mouth disease virus (FMDV) (1). To date, the family Picornaviridae consists of 158 species grouped into 68 genera (as of March 2022), such as Enterovirus, Hepatovirus, Cardiovirus and Aphthovirus (2, 3).
The members of the Picornaviridae family are small, nonenveloped RNA viruses. The picornavirus virion has a symmetrical icosahedral spherical structure with an approximate diameter of 20-40 nm (4–6). The viral genome is a single-stranded, positive RNA strand approximately 6.7-10.1 kb in length that consists of an open reading frame (ORF), a highly structured 5′ untranslated region (5′ UTR), and a 3′ untranslated region (3′ UTR) with a [poly(A)] tail (Figure 1) (9). The viral genome-linked protein 3B (also known as VPg) is covalently bound to the 5′ end of the positive-sense RNA (10). The 5′ UTR harbors an internal ribosomal entry site (IRES) that recruits ribosomes and other host factors and mediates cap-independent translation (11, 12). The ORF initially encodes a single polyprotein that is co and posttranslationally cleaved by viral proteases to release the capsid proteins VP0, VP1, and VP3 and nonstructural proteins (2A, 2B, 2C, 3A, 3B, 3C, 3D), as well as some stable precursors, such as 3AB or 3CD, that are essential for the replication of viral RNA (13–15). Recently, a second ORF termed the upstream ORF (uORF) was identified in enteroviruses (16). Some genera of picornaviruses, such as Aphthoviruses and Cardioviruses, also have a leading conductor (L) protein at the N-terminus of the polyprotein (17). This review mainly focuses on 3D polymerase (3Dpol).
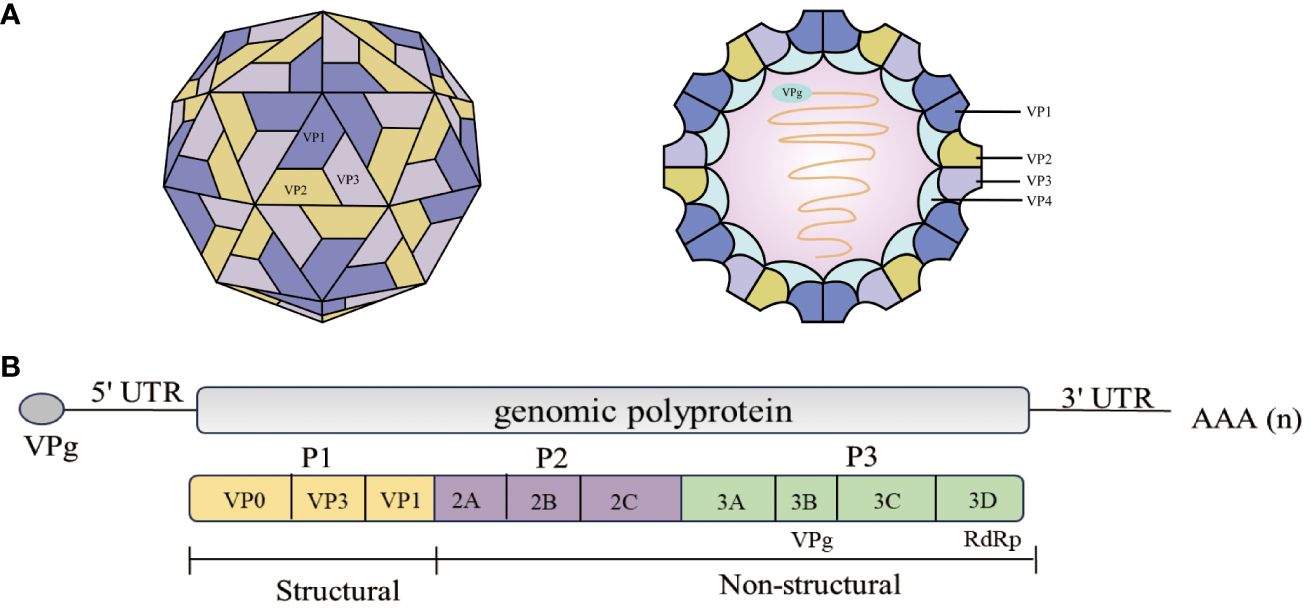
Figure 1 Schematic representation of the EV71 structure and genome structure of the virion. (A) Structure of the EV71 (6). EV71 is a small (circumference around 30 nm), non-enveloped, icosahedral particle that contains a single-stranded, positive-sense, polyadenylated virus RNA of approximately 7.4 kb (6). (B) The diagram demonstrates the EV71 genome structure. All the structural proteins are encoded by the P1 region (yellow) of the genome. The P2 (purple) and P3 (green) regions encode seven non-structural proteins—2A–2C and 3A–3D. The last part of the polyprotein is 3Dpol, the RNA-dependent RNA polymerase that is active only upon cleavage of the 3Cpro–3Dpol junction (7, 8).
The picornavirus 3Dpol, also known as RNA-dependent RNA polymerase (RdRp), is responsible for genome synthesis (18, 19). 3Dpol becomes active upon cleavage of the precursor 3CD protease (3CDpro) (7, 20). Previous studies have yielded a very good understanding of the 3Dpol structure and fundamental molecular mechanism for catalysis (21). The N-terminal region of 3Dpol acts as a nuclear localization signal (NLS), which is involved in nucleotide recognition and affects the incorporation of nucleotide analogs, suggesting the multifunctionality of the picornavirus polymerase domains (22). In addition, recent studies have revealed novel mechanisms for picornavirus invasion of host cells involving multiple previously undiscovered functions of 3Dpol that differ from its traditional role in viral replication. For example, EV71 3Dpol can enter the cellular nucleus through the NLS to associate with the core splicing factor pre-mRNA processing factor 8 (Prp8), affecting the normal function of Prp8 during the second catalytic splicing step, leading to the inhibition of pre-mRNA splicing, the accumulation of the lariat form, and a decrease in the resulting mRNA; or it can facilitate viral and host translation by forming complexes with small and large subunits of ribosomes (23, 24) (Figure 2B); 3Dpol also functions as an antagonist against the host innate immune response (25–27). In this review, we summarize the general structural features and functions of 3Dpol and discuss the role of 3Dpol in regulating virus−host interactions to promote viral replication.
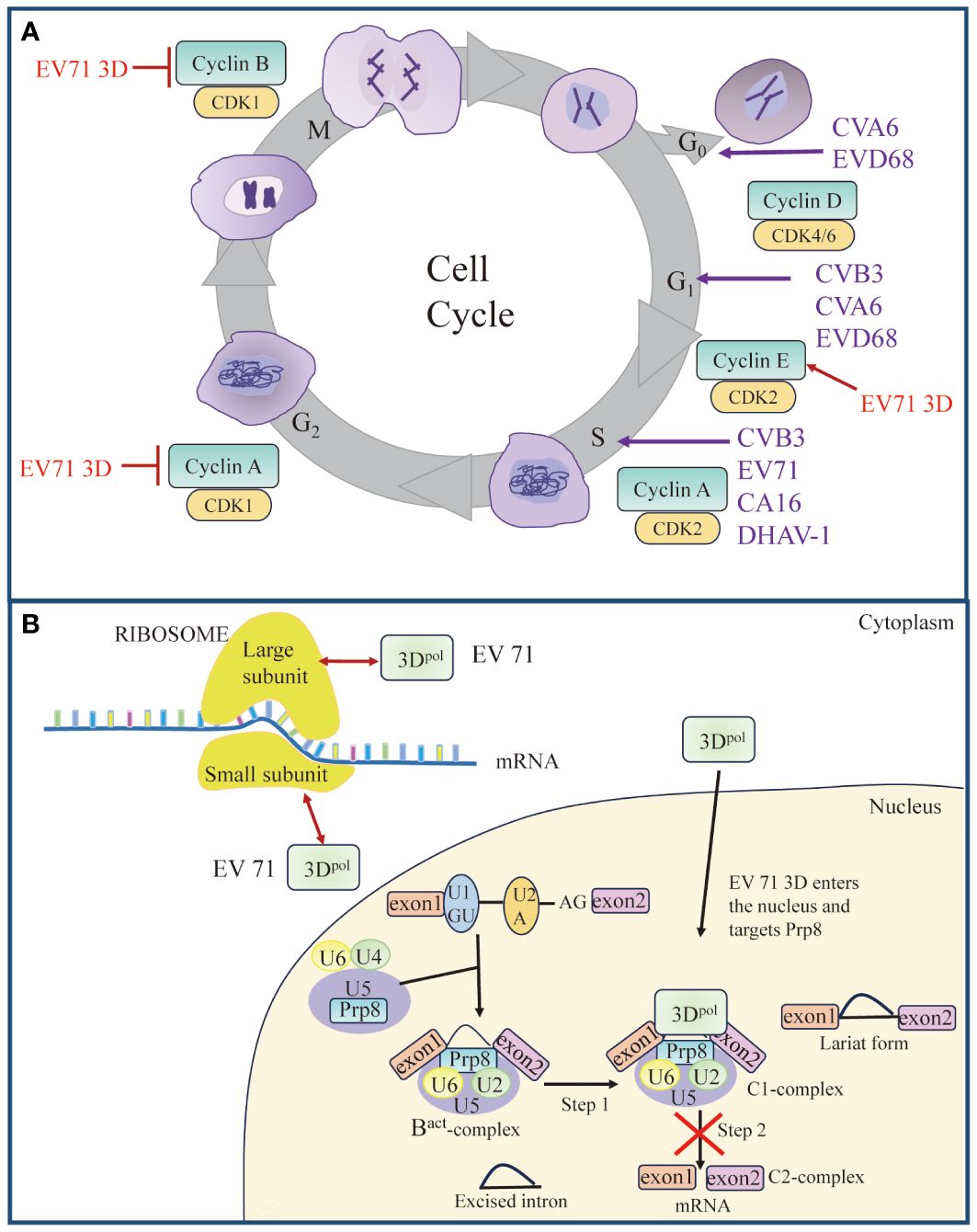
Figure 2 Roles of 3Dpol in cell cycle and cell translation. (A) Picornavirus 3Dpol induces cell cycle arrest. Red arrow represents “Upregulate”, and red vertical symbols represent “Inhibit”. (B) Schematic model of 3Dpol-mediated effects on cellular translation. EV 71 3Dpol could enhance EV-A71 IRES-dependent translation as well as cap-dependent translation by interacting with small and large subunits of ribosomes. Partially, 3Dpol also enters the nucleus and interacts with the core splicing factor Prp8, which interferes with the function of Prp8 in the C1-complex. The interference of the Prp8 function inhibits the second step of the splicing process and results in the accumulation of the lariat form and a reduction in mRNA synthesis. Red bidirectional arrows represent “Interact”. The figure was modified from (24).
2 Structural features of the picornavirus 3Dpol
2.1 Overall 3Dpol structure
Following the first report of the complete crystal structure of PV 3Dpol in 2004 (8), crystal structures of 3Dpol from HRV (28) and FMDV (29) were reported in succession. To date, there are several viral RdRp structures in the Protein Data Bank (PDB, www.wwpdb.org) related to different picornaviruses, including PV, CVB3, EV71, HRV, EMCV, and FMDV (21). Like other DNA and RNA polymerases, the crystal structure of the picornavirus 3Dpol resembles a cupped right hand, with three defined subdomains, termed the thumb, fingers and palm (Figures 3A, B) (8, 28, 29, 31–33). The finger domain can be further divided into distinct substructures that are sometimes referred to by the anatomical analogy of the index, ring, middle, and pinky domains. The thumb domain interacts with the finger domain to “close” the hand and envelops the active site, forming an NTP entry channel behind the RdRp (8, 28, 29, 31–33). The palm subdomain, consisting of two α spiral and five β-barrel domains (29), is the catalytic region of 3Dpol with a GDD-3 amino acid active site shared by all RdRps, and can bind Mg2+ and locate NTP substrates (34). In addition, 3Dpol contains seven conserved motifs (A to G) that play key roles in rNTP substrate recognition, template/primer binding and catalysis (35). Currently, available data provides high-resolution pictures for a range of conformational states associated to template and primer recognition, VPg uridylylation, rNTP recognition and binding, catalysis and chain translocation (36). These structural information provide insights into both initiation of RNA synthesis and the replication elongation processes in picornavirus (37, 38). The increased understanding of polymerase structure could help explore possible ways of vaccine development.
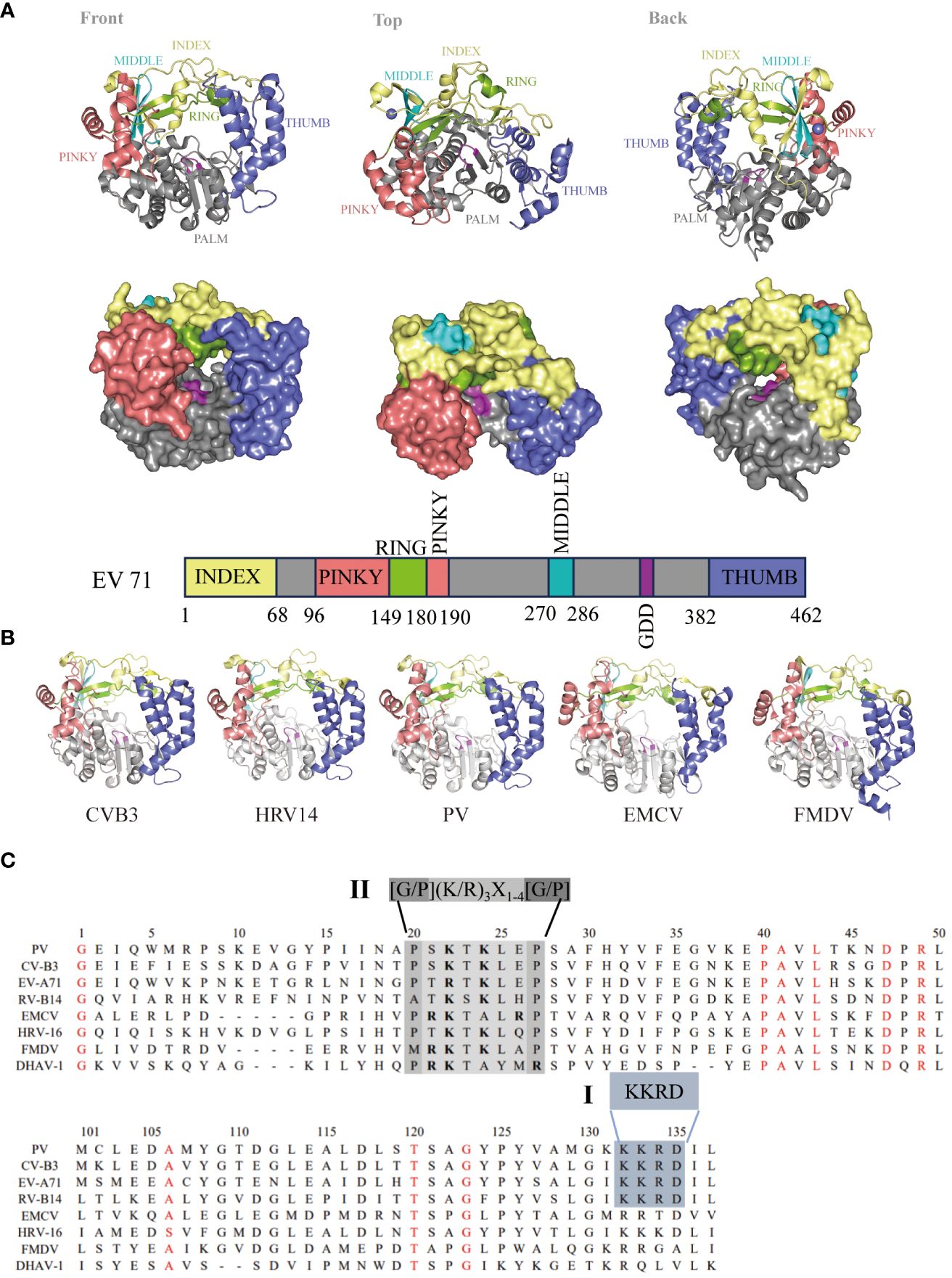
Figure 3 Genome and structure of picornavirus 3Dpol. (A) Cartoon and surface representations of EV71 3Dpol (Protein Data Bank: 3N6L) in three different orientations. The structure resembles a cupped right hand composed of palm, fingers, and thumb domains; an index finger (residues 1-68) in yellow; a middle finger (270–286) in canyon; a ring finger (150–179) in green; a pinky finger (96–149, 180–190) in pink; a palm (191–269, 287–381) in gray; a thumb (382–462) in purple; and a GDD active site (328–320) in magenta. Bar representation of the 3Dpol sequence colored according to the structural elements are shown in (A). (B) 3Dpol structures of CVB3 (PDB: 3DDK), HRV14 (PDB: 1XR5), PV (PDB: 2ILY), EMCV (PDB: 4NYZ) and FMDV (PDB: 1U09) are shown, and all of them exhibit a very high degree of structural homology. (C) Localization of the putative nuclear localization sequence (NLS) on picornavirus 3D sequences. Alignment of the amino acid sequences corresponding to the 3D amino termini from PV, CV, EV, RV, EMCV and DHAV-1. The single basic NLS KKRD (I) and the consensus NLS found in several yeast ribosomal proteins, G/P(KR)3X1-4[G/P] (II), are indicated (30).
2.2 Nuclear localization signal
The picornavirus 3Dpol primarily replicates in the host cytoplasm, but 3Dpol/3CDpro can enter the nucleus in virus-infected cells (39–41). Previous studies have shown that PV 3Dpol and 3CDpro enter the nucleus through a single basic type of nuclear localization signal (NLS), KKKRD, which spans 125–129 amino acids (aa) within 3Dpol (40, 41). The putative NLS is partially contained within the KKRD sequence (126–129 aa), which is typical among all known picornaviral 3Dpol (40, 42). However, these motifs are not completely reiterated in the other members of this family. In contrast, a NLS (15PRKTALRP22 in EMCV), similar to that in many yeast ribosomal proteins (43), was identified near the N-terminus of the EMCV 3Dpol sequence (44). An NLS similar to that of EMCV was also found in the 3Dpol of HRV16, FMDV and duck hepatitis A virus type 1 (DHAV-1) (30, 45, 46). By comparing the 3Dpol amino acid sequences, we found that NLSs within 3Dpol were mainly divided into two types: single basic NLSs, KKRDs, and other NLSs, which are similar to those found in several yeast ribosomal proteins, G/P(KR)3X1-4[G/P] (Figure 3C). Because studies have shown that the virus-encoded 3C protease (3Cpro) cleaves transcription factors at glutamine–glycine sites and is directly responsible for host cell transcription shut-off, it is likely that 3Cpro must enter the nucleus of infected cells as is or in the form of a precursor (47). These data suggest that the NLS present within 3Dpol plays a role in the nuclear entry of precursor 3CD or 3BCD. This mechanism may be a common feature of picornavirus infections (48–50).
NLS sequences have functions other than facilitating the entry of viral proteins into the nucleus. The 16MRKTKLAPT24 sequence in 3Dpol of FMDV was identified as an NLS, and substitutions at the K18 or K20 residues resulted in two conformational changes that reduced 3Dpol binding to RNA (22); moreover, K18 and K20 were demonstrated to be essential for virus proliferation (45). In addition, the T19 and L21 residues are important for maintaining the fidelity of FMDV RdRps and ensuring faithful replication of the FMDV genome (51). Thus, the role of this class of NLS motifs in picornavirus viral polymerases needs to be revisited.
2.3 RNA structure in the 3Dpol-coding region
The genomes of RNA viruses often contain RNA structures that are crucial for translation and RNA replication and may play additional roles during the viral replication cycle (52–55). For picornaviruses, within the ORF, several RNA structures have been identified. The cis-acting replication element in the 2C coding region (2C-CRE), which acts as a template for uridylylation of the VPg (3B) protein (56, 57), and an RNA structure carried in the 3Cpro ORF that potently inhibits the endonuclease activity of RNase L (an antiviral endoribonuclease) have been identified (58, 59). In addition, in the PV genome, two stem loops (referred to as loops α and β) within the coding region of 3Dpol that are important for proper RNA synthesis during viral infection have been identified (54, 60). Previous studies have further shown the existence of a novel functional interaction between these RNA structures in the 3Dpol-coding region and the viral protein (s) 3Cpro and/or its precursor 3CDpro (54). Three of the RNA structures (ORF-SL51, ORF-SL52, and ORF-SL53) within the coding region of FMDV 3Dpol have also been identified, and they are critical for efficient replication of the FMDV replicon (61). Thus, the RNA structures formed by those genomic regions may play a functional role in the picornavirus replication cycle.
3 Posttranslational modifications of 3Dpol
Ubiquitination and SUMOylation are widely studied posttranslational modifications (PTMs) that play critical roles in diverse biological processes (62, 63). The ubiquitin–proteasome system (UPS) also plays an important role in the different steps of the viral life cycle (64–66). The mechanisms by which the UPS regulates viral infection include the degradation of intracellular proteins or excess viral proteins and the modulation of viral protein function through ubiquitin-mediated modification or direct encoding of ubiquitin-related enzymes (67). An increasing number of studies have suggested that various viruses evolve different mechanisms to utilize or manipulate the host UPS for their own benefit (68–71). For picornaviruses, studies have shown that the UPS may regulate CVB3 replication through ubiquitinating viral 3Dpol, which is essential for initiating viral RNA replication (72). In addition, Senecavirus A (SVA) 3Dpol is ubiquitinated by UBE2L6, an E2 ubiquitin-conjugating enzyme, and this ubiquitination serves to inhibit the degradation of 3Dpol, thereby facilitating SVA infection (73) (Figure 4). Normally, the interplay between SUMOylation and ubiquitination often involves the stability of the target protein (76–78). EV71 3Dpol was modified by small ubiquitin-like modifier 1 (SUMO-1) both during infection and in vitro, and 3Dpol was ubiquitinated in a SUMO-dependent manner to enhance the stability of the viral polymerase (74). Moreover, residues K159 and L150/D151/L152 were found to be responsible for 3Dpol SUMOylation, and mutation of SUMOylation sites impaired 3Dpol activity and virus replication. Similarly, Hao et al. reported that the m6A methyltransferase METTL3 interacts with EV71 3Dpol and induces SUMOylation and ubiquitination of 3Dpol, which boosts viral replication (75) (Figure 4). SUMOylation and ubiquitination of viral polymerases have been reported not only in picornaviruses but also in other viral families, such as nonstructural protein 5 (NS5) of dengue virus (79) and polymerase basic protein 1 (PB1) of influenza virus (80). Recent studies have characterized the alterations in UPS-dependent protein homeostasis during infection with CVB3 and demonstrated that the activity of the proteasome is exploited for the processing of viral precursor proteins (81). Furthermore, both the viral 3Cpro and the viral 3Dpol have been reported to be subjected to UPS-dependent proteolysis. This may be a strategy used by picornaviruses to maintain the proper balance of the expression levels of these two viral proteins to prevent premature cell death and ensure effective viral replication.
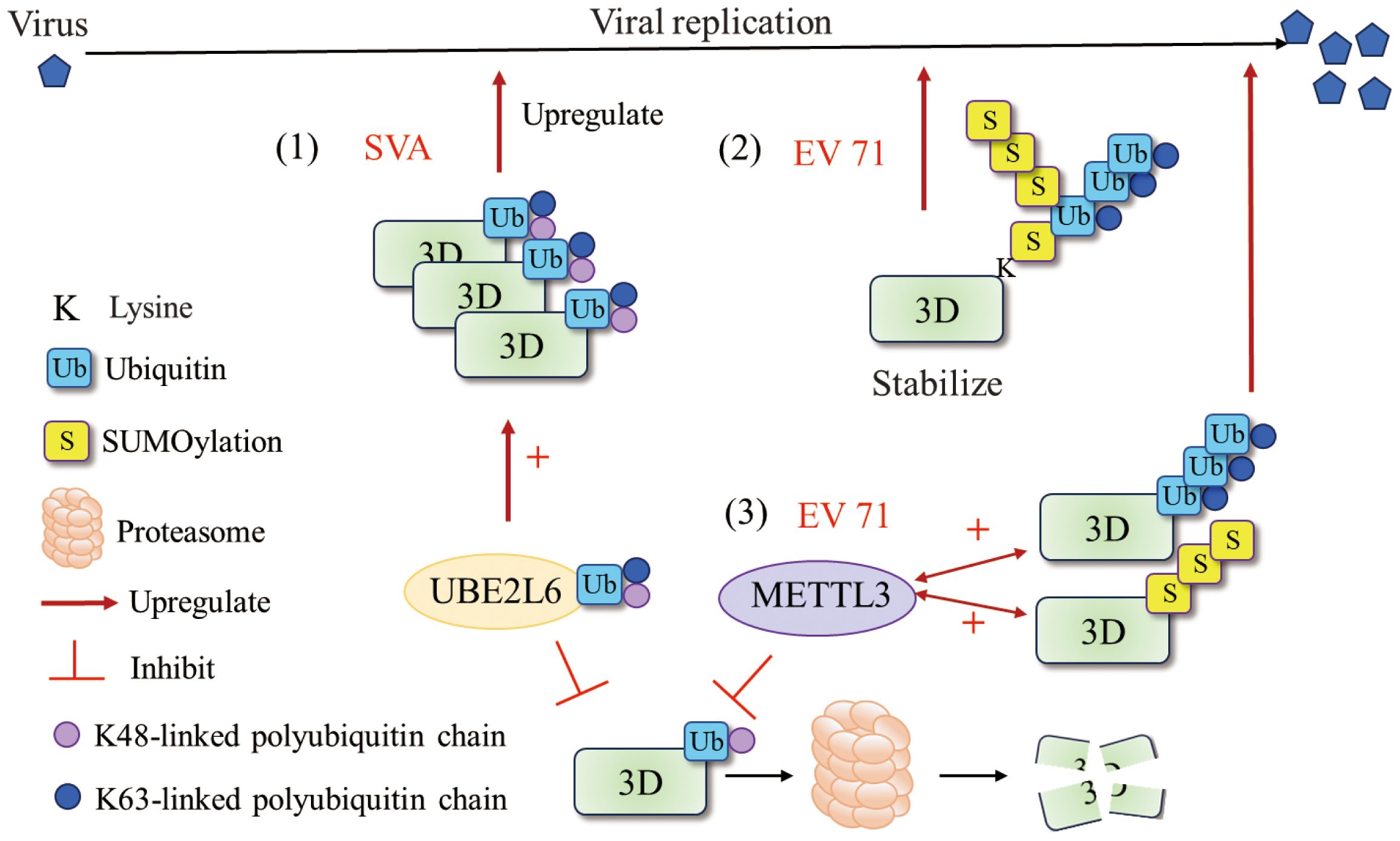
Figure 4 Ubiquitination and SUMOylation of 3Dpol. (1) UBE2L6 interacted with SVA 3Dpol and mediated K48/K63 chains to improve the stability of 3Dpol (73). (2) EV71 exploit the cross talk of SUMOylation and ubiquitination to stabilize the 3Dpol and enhance viral replication. SUMOylation and ubiquitination may share the same lysine residues and that 3Dpol was ubiquitinated in a SUMO-dependent manner (74). (3) METTL3 interacted with EV 71 3Dpol and increased K63-linked ubiquitination and SUMOylation of the 3Dpol that boosted viral replication (75). Figure adapted from (73).
Studies have proposed that SUMOylation and ubiquitination at specific 3Dpol sites contribute to maintaining the cellular level of 3Dpol and that corresponding deSUMOylation and deubiquitination may be necessary for 3Dpol to restore polymerase activity since, when 3Dpol is responsible for RNA genome replication, it is free of SUMOylation (74). These findings imply that the 3Dpol of picornaviruses exploits host cell modifications for efficient replication, revealing potential targets for antiviral therapy.
4 Roles of 3Dpol in picornavirus replication
The picornavirus 3Dpol plays a critical role in viral genome replication by catalyzing different steps of viral genomic RNA replication, from primer synthesis (VPg-uridylylation) to viral RNA synthesis and polyadenylation of progeny genomic RNA. The first step in picornavirus genome replication is uridylylation of VPg. In this process, 3Dpol catalyzes the covalent attachment of two uridine monophosphate (UMP) molecules to the hydroxyl group of tyrosine 3 (Y3) of VPg and generates VPg-pUpU-OH, which serves as a primer to initiate the replication process (82–84). This process has been extensively studied in different members of the Picornaviridae family (37, 85–90), and detailed information can be found in this Review (90). Subsequently, 3Dpol catalyzes the synthesis of viral negative- and positive-sense RNA within the replication complex (RC) (91). In addition, to ensure genome integrity, a variable poly(A) tail is regenerated on the 3′ UTR end of newly synthesized viral RNAs during each round of viral replication (92–94). The length of poly(A) affects viral mRNA translation and RNA replication (95, 96). Previous studies have shown that virus replication can be severely impaired when the poly(A) tail is curtailed to 14 or 12 adenines or less (96–98). Alanine mutations in PV 3Dpol change the sizes of poly(A) tails in virion RNA, suggesting that 3Dpol is primarily responsible for the sizes of poly(A) tails (99–101).
Picornavirus infection induces the redistribution and rearrangement of cytoplasmic organelles to form membrane-bound structures that contribute to viral RNA replication; these structures are known as replicating organelles (ROs) (102–106). ROs may originate from Golgi membranes or the endoplasmic reticulum (ER) and contain host factors such as the lipid kinase PI4KB (also called PI4K IIIβ) as well as viral proteins, including 3A and 3Dpol (104, 107–109), which are thought to protect viral RNAs from RNase degradation or cellular RNA sensor detection (110–112). Increasing evidence suggests that proteins of picornaviruses hijack host factors involved in membrane trafficking and biosynthesis pathways to promote efficient viral genome replication (102, 113). Membrane-associated protein 3A (114) recruits PI4KB to the replication site through interaction with acyl-CoA binding domain containing 3 (ACBD3) (107, 108). PI4KB then catalyzes the formation of a phosphatidylinositol 4-phosphate (PI4P)-rich microenvironment that facilitates the recruitment of 3Dpol (102, 108).
Viral nonstructural proteins and their precursors, such as the 3A and 2BC proteins, contain hydrophobic regions that interact extensively with cell membranes and assemble to form RCs with cellular proteins and viral RNAs on the RO surface (112, 115). However, since picornavirus 3Dpol is a soluble protein with no obvious membrane-binding region, 3Dpol can be recruited to complex only by protein–protein or protein–RNA interactions (87). As shown in Figure 5, four methods for recruiting 3Dpol to the RO surface have been described in existing studies: i) 3AB, a small basic protein with biochemical properties similar to those of membrane proteins (117), interacts with 3Dpol through its VPg domain and recruits 3Dpol to the RC (117–121); ii) the PI4P lipid-rich microenvironment promotes the recruitment and stabilization of the 3Dpol membrane (102, 122); iii) negatively charged lipids cooperate with membrane-anchored 3B to recruit the 3Dpol enzyme (116); and (iv) host proteins recruit 3Dpol by interacting directly with 3Dpol. Annexin A2 (ANXA2), which is localized on ROs, interacts with PI4KB, promotes the interaction of EV71 3Dpol with PI4KB and forms a higher-order protein complex with 3Dpol and PI4KB located in ROs (122). EV71 3Dpol interacts with host UDP-glucose glycoprotein glucosyltransferase 1 (UGGT1), a key ER protein involved in the unfolded protein response (UPR), to promote the formation of RCs on cellular membranes that enhance viral RNA synthesis (123). In addition, upon infection, the lysosomal tethered Ragulator-Rag complex promotes EV71/CVA16 replication by recruiting viral 3Dpol to the lysosomal surface through the interaction between 3Dpol and RagB (124). 3Dpol, as part of a replication complex of 3A and several other viral proteins, subsequently initiates RNA synthesis at these membranes.
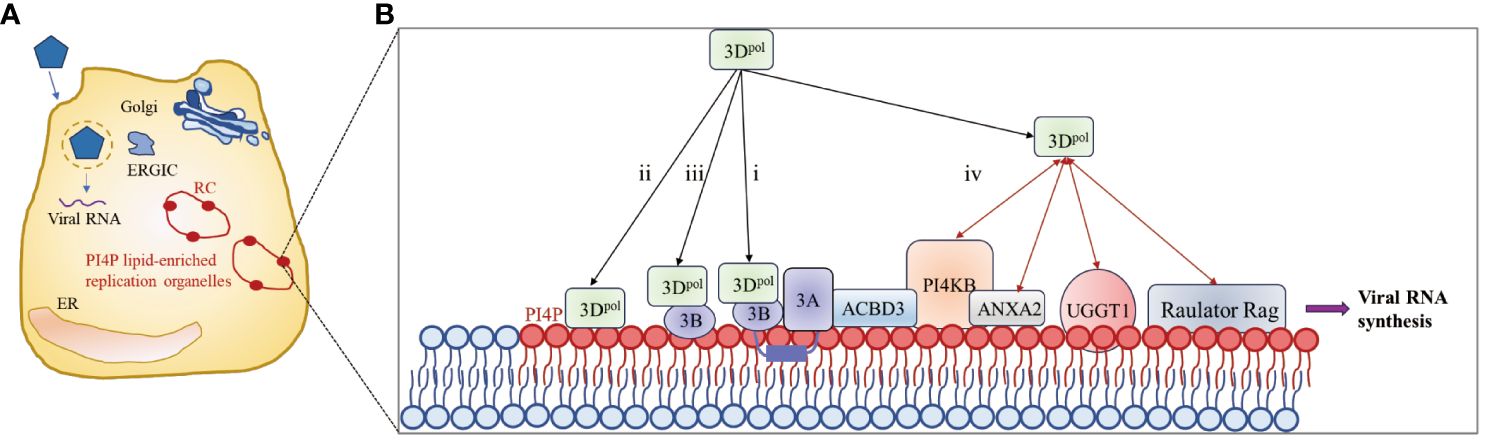
Figure 5 3Dpol is recruited to the surface of the lipid bilayer to promote RC formation. (A) Upon infection, synthesis of the 3A protein leads to remodeling of the Golgi/TGN and the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) into replication organelles (ROs). PI4P lipids are indicated by red ovals. The enrichment of PI4P in these ROs promotes the binding of 3Dpol or 3CD to the membrane, which in turn facilitates the assembly of replication complexes (RCs) and the synthesis of viral RNA. (B) Four methods for recruiting 3Dpol to the surface of the lipid bilayer. The red bidirectional arrows represent interactions between viral and host proteins. The figure was modified from (105, 116).
Interestingly, recent studies have shown that the CCT8, DBN1, IQGAP1 and ELMO2 proteins are involved in the regulation of cytoskeleton assembly and interact with EV71 3Dpol, suggesting that viral 3Dpol may also play a role in cytoskeletal rearrangement during infection (23).
5 Regulation of host cell responses by 3Dpol
Viruses have developed sophisticated mechanisms to manipulate host cellular pathways to facilitate viral replication and evade host defenses. In recent years, an increasing number of researchers have focused on the functions of 3Dpol (other than that of a RdRp) during viral infections. 3Dpol acts on host cells through interactions with host proteins and plays an important role in inducing cell cycle arrest (Figure 2A), regulating host cell translation (Figure 2B), inducing apoptosis and autophagy, evading immune responses (Figure 6), and activating the NLRP3 inflammasome (Figure 7). 3Dpol promotes the replication and proliferation of these viruses by regulating these responses.
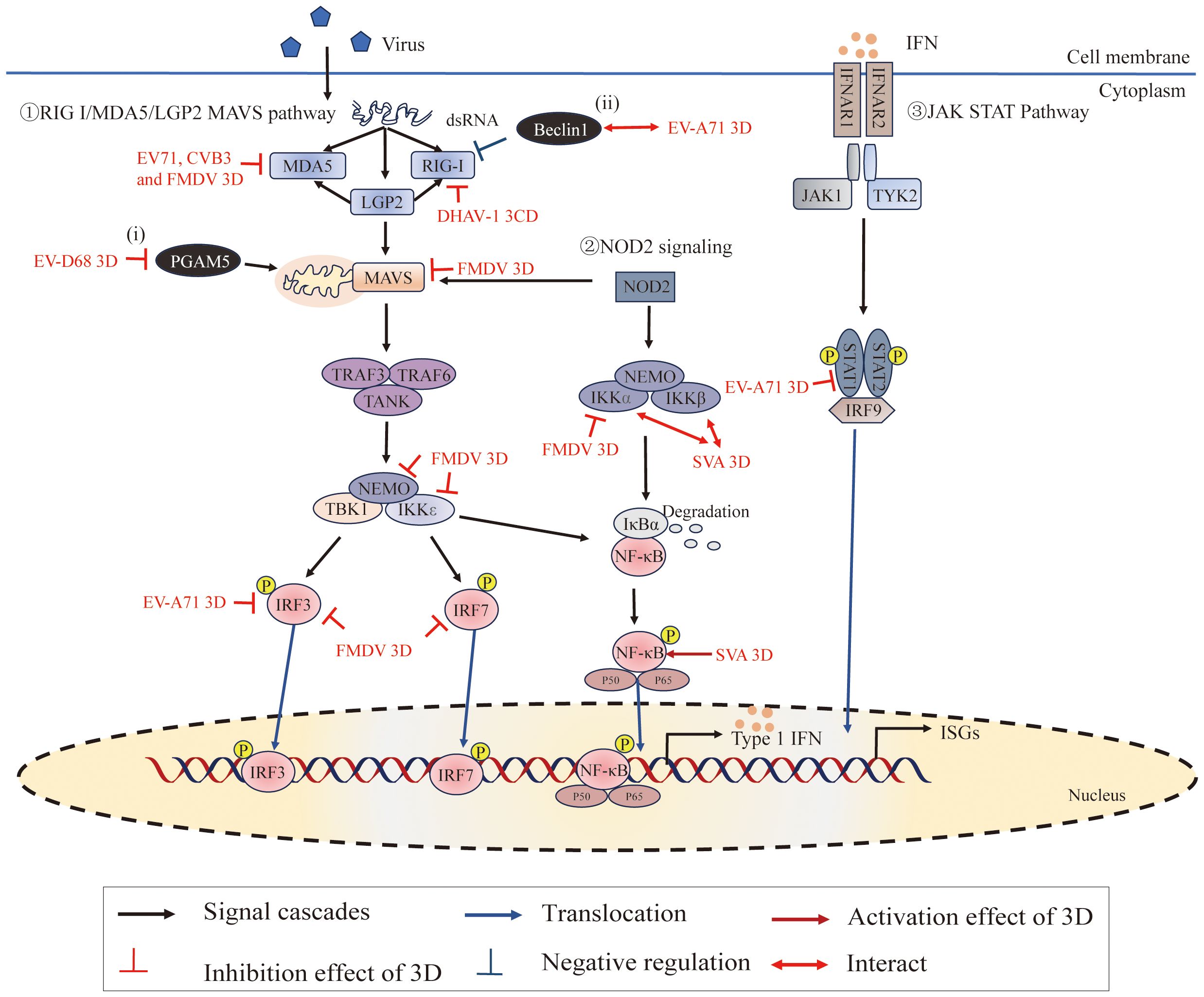
Figure 6 Roles of 3Dpol in subverting host innate immunity. ①The RIG-I/MDA5/LGP2-MAVS pathway recruits downstream adaptors, including tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3), TRAF6, and the TRAF family member-associated NF-κB activator (TANK), to directly induce the TBK1-IKKϵ-NEMO complex. These signaling cascades lead to the phosphorylation of interferon regulatory factors (IRFs) and NF-κB in the nucleus, where they promote the expression of interferons (IFNs), interferon-stimulated genes (ISGs) and proinflammatory cytokines (125). ②NOD2 signaling induces the activation of MAVS and the IKKα-IKKβ-NEMO complex. ③IFNs bind to IFN-α/β receptors (IFNARs), activating the Janus kinase-signal transducer and activator of transcription (JAK/STAT) pathway to amplify IFN production (126). Moreover, (i) PGAM5 affects mitochondrial morphology and affects the expression of MFN2, and MFN2 binds to MAVs to inhibit the RIG-I-like signaling pathway; (ii) Beclin-1 is a negative regulator of the RIG-I-MAVS-mediated IFN response. Figure adapted from (127).
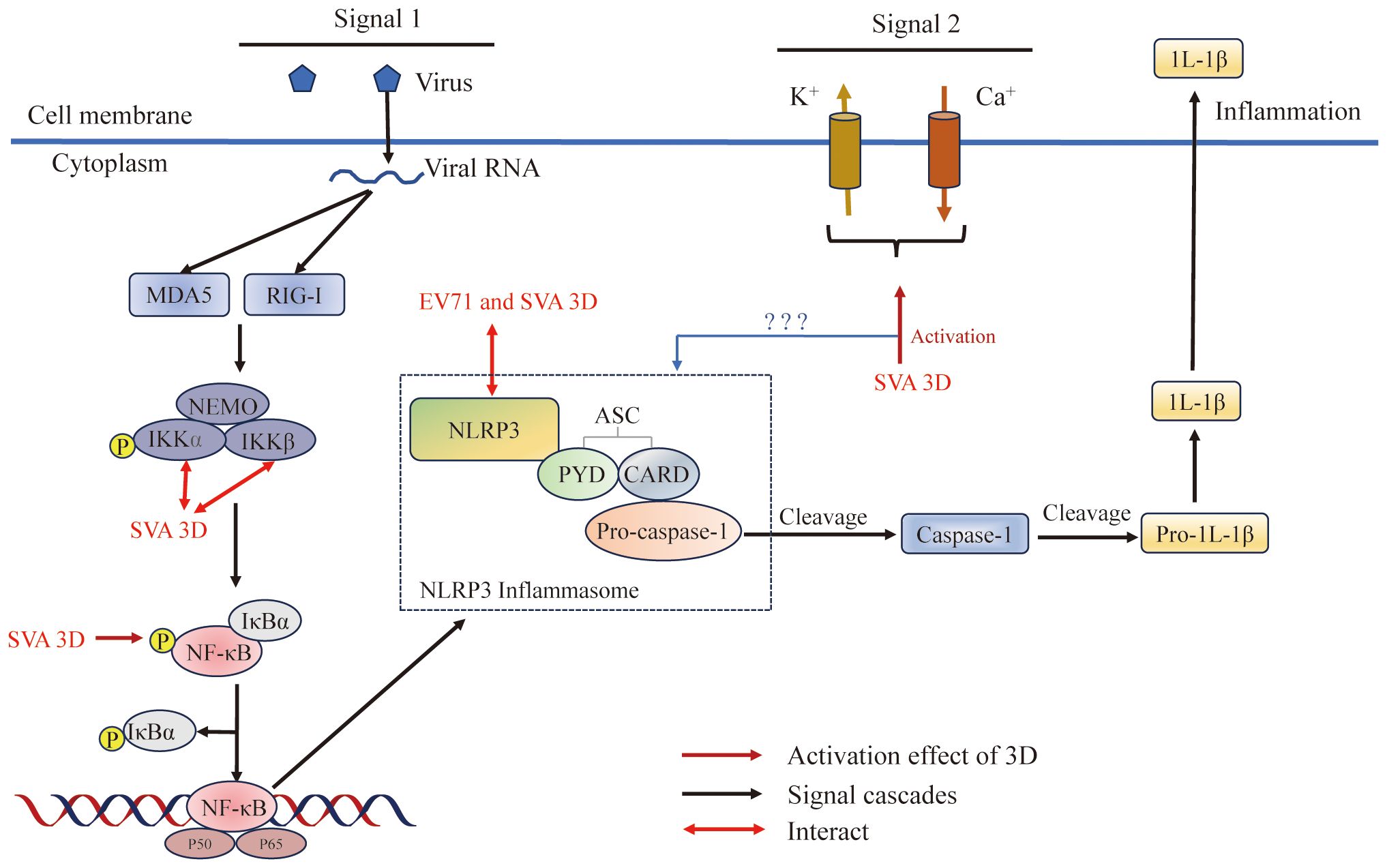
Figure 7 Picornavirus 3Dpol regulates inflammasome activation. The NLRP3 inflammasome is an oligomeric complex composed of the NOD-like receptor NLRP3, the adaptor protein ASC, and the effector protein pro-caspase-1 (128). 3D regulates inflammasome activation by inducing NF-κB activation, interacting with NLRP3 to facilitate NLRP3-ASC assembly or inducing calcium influx and potassium efflux. Figure adapted from (129).
5.1 Induction of cell cycle arrest and regulation of cellular translation
As part of their pathogenic mechanism, many viruses create a favorable environment for viral replication by manipulating the host cell cycle (130–133). The cell cycle is divided into a stationary G0 phase, interphase (G1, S and G2 phases), and a mitotic phase (M phase) (Figure 2A). Typically, the cell cycle is controlled by the binding of cyclin-dependent kinases (CDKs) to the corresponding cyclin regulatory subunits (134, 135). Cyclin E/CDK 2 is responsible for regulating cellular S-phase entry from G1 (136), cyclin A/CDK 2 regulates S-phase progression by replacing cyclin E (137, 138), and cyclin B/CDK 1 is involved in the mitotic process (139). Previous studies have indicated that the cell cycle affects picornavirus replication (140, 141). Studies using cell cycle inhibitors have revealed that cell cycle arrest at the G1 or G1/S phase could promote viral replication of CVB3 (142) (Figure 2A). In contrast, protein expression and virus yield were significantly reduced after cell cycle arrest in the G0 and G2/M phases (142). The expression of EV71 3Dpol increased the expression of cyclin E and phosphorylated CDK2 T160, which promoted S-phase entry, thereby facilitating viral production (143). In addition, coxsackievirus A16 (CA16)-induced S-phase arrest of the host cell cycle was also observed (143). However, the expression of CVA6 3Dpol induced cell cycle arrest in the G0/G1 phase, which promoted CVA6 replication and viral production (144). Surprisingly, EV71, CA16, and CVA6 all belong to the family Picornaviridae and cause hand, foot, and mouth disease; however, there are significant differences. Perhaps these viruses employ different strategies to promote their replication, which leads them to have different characteristics, such as clinical symptoms and epidemiological scopes (144). In addition, EV-D68 3Dpol was found to induce cell cycle arrest at the G0/G1 phase (145), DHAV-1 infection-induced cell cycle arrest in duck embryo fibroblasts (DEFs) in the S phase, and both the S phase and G0/G1 phase synchronization facilitated the replication of DHAV-1 (146). These results suggest that inducing cell cycle arrest in the S or G0/G1 phase and promoting viral replication are common strategies for picornaviruses.
Translation of most eukaryotic mRNAs is facilitated by a 5′ cap, a structure absent from picornavirus mRNA, which instead contains an IRES. Cap-dependent translation of cellular proteins is most robust during the G1 phase but is impaired at mitosis (142). Picornavirus IRESs evolved to operate in the G1 phase, a time at which cap-dependent translation is dominant. Upon infection, picornavirus caused inhibition of the cap-dependent translation machinery and utilized host translation machinery for cap-independent translation of viral proteins mediated by its IRES element within the 5′-UTR (142, 147, 148). Studies have shown that 2Apro and 3Cpro cleave eukaryotic initiation factor 4G (eIF4G) (149–151), eukaryotic initiation factor 4A (eIF4A) (152), and eukaryotic initiation factor 5B (eIF5B) (153), leading to host cell translation shutdown. However, it has been reported that EV71 3Dpol can enter the cellular nucleus through the NLS to associate with the core splicing factor Prp8 (24) (Figure 2B). 3Dpol affects the normal function of Prp8 during the second catalytic splicing step, resulting in the inhibition of pre-mRNA splicing and a decrease in the amount of resulting mRNA (24). In contrast to viral proteases blocking host transcription and translation mechanisms, picornaviruses utilize their polymerases to alter cellular gene expression by hijacking the splicing machinery, which potentially providing another advantage for virus replication. Interestingly, another study showed that EV71 3Dpol directly increases EV71 IRES-dependent translation as well as cap-dependent translation. 3Dpol, encoded by EV71, can interact with ribosomal proteins to form complexes with small and large subunits of ribosomes and activate viral and host translation (23) (Figure 2B). Since cellular factors known as ITAFs may regulate IRES-mediated translation initiation, 3Dpol increases the expression of these cellular proteins by facilitating host translation, favoring viral replication (130, 154). It is conceivable, therefore, that virus-induced cell cycle block may create an environment favorable for viral replication, which could then maximize virus production by manipulating the host cell translation devices.
5.2 Regulation of autophagy
Viruses have been shown to employ autophagic machinery to replicate and survive during the infection process (155–157). Recent studies have revealed a distinct mechanism by which EV71 induces apoptosis and autophagy in neural cells (158). EV71 3Dpol interacts with the peroxisomal protein acyl-CoA oxidase 1 (ACOX1), attenuates ACOX1 production, and enhances reactive oxygen species (ROS), thereby inducing apoptosis and autophagy in neuronal cells (158). In addition, EMCV 3Dpol induces autophagy in BHK-21 cells by activating the ER stress pathway, which ultimately benefits viral replication (159). Furthermore, EMCV-3Dpol has been demonstrated to regulate proteins associated with the PERK and ATF6α pathways. Other picornaviruses with similar structures/sequences to EV71 or EMCV 3Dpol may also have similar functions; however, further research is needed.
5.3 Regulation of the host cellular immune response
The innate immune system is the first line of defense against invading pathogens (160). Upon pathogenic microbial infection, they are recognized by pattern recognition receptors (PRRs), leading to the activation of signaling cascades to generate immune responses (161). Picornaviruses have evolved strategies to evade the innate immune response, and studies have focused mainly on the 2A (26, 162, 163), 2B (164–166), and 3Cpro (26, 167). To date, 3Dpol, essentially known for its significant role in viral genome RNA replication as a polymerase, has been the subject of very few studies concerning its action against the antiviral response. However, previous studies have shown that RdRp can also be involved in regulating innate immune responses (168, 169). The regulatory effect of 3Dpol on the host cell immune response mainly manifests as antagonistic effects (Figure 6).
5.3.1 3Dpol affects RNA sensors
Two cytoplasmic pathogen recognition receptors, melanoma differentiation-associated gene 5 (MDA5) and retinoic acid-inducible gene I (RIG-I), have been identified as sensors for recognizing RNA viruses and stimulating type I IFN expression (170–172). RIG-I recognizes cytoplasmic 5′ triphosphate single-stranded RNA with poly (U/A) motifs and short dsRNA, while MDA5 primarily recognizes long double-stranded RNAs (172–174). LGP2, the smallest member of the RIG-I-like receptors family, is pivotal in regulating the signaling pathway through positive and negative regulation of MDA5 and RIG-I, respectively (175–179). A recent study has shown that cleavage of MDA5 by the 3Cpro from Theilovirus leads to dysfunction of MDA5 as an innate immune RNA sensor for IFN induction (180). In addition, FMDV 3Cpro inhibits MDA5 protein expression as a mechanism to evade the innate immune response during FMDV infection (181).
RIG-I and MDA5 can sense viral RNA through their C-terminal domains (CTDs), and their caspase activation and recruitment domains (CARDs) can interact with CARDs of the downstream adaptor MAVS to transduce signals (125, 182). Recent studies have shown that EV71 3Dpol interacts with CARDs of MDA5 and plays a role in the inhibition of MDA5-mediated beta interferon (IFN-β) promoter activation and mRNA expression (25). This inhibition was also detected by using the RdRp activity knockout mutant (D330A) of EV71 3Dpol, which demonstrated that EV71 3Dpol inhibits IFN-β promoter activity without interfering with viral RNA replication. This study also has shown that CVB3 interacts with MDA5 and downregulates the antiviral signaling initiated by MDA5 (25). In addition, Sarry, Morgan et, al. found that FMDV 3Dpol interacts with MDA5 and IFN pathway proteins (IKKα, IKKϵ, IRF3, IRF7, NEMO, and MAVS), which may be responsible for the inhibitory effect on the IFN pathway induction phase by FMDV (127). Moreover, studies have shown that DHAV-1 3CD interacts with RIG-I, interferes with the interaction between RIG-I and MAVS, and degrades RIG-I protein through the proteasomal degradation pathway, thereby inhibiting its mediated antiviral innate immunity to promote DHAV-1 replication (183).
5.3.2 Interference with IFN-mediated signaling
Interferons are cytokines that play a crucial role in regulating and activating the host innate immune response to viral infection and limiting viral replication (126, 184). Upon the production and release of IFNs, the interferon α receptor (IFNAR) is ligated, which subsequently activates Janus-associated kinase 1/2 (Jak1/2) and recruits signal transducers and activators of transcription 1 (STAT1), ultimately leading to the expression of antiviral effector molecules (185–188). Experimental results have shown that EV71 3Dpol attenuates IFN-γ-induced tyrosine phosphorylation of STAT1 accompanied by a STAT1 decrease (189); either restoring STAT1 or inhibiting 3Dpol activity effectively reversed IFN-γ-induced IRF1 transactivation. However, it is still unknown how the 3Dpol regulates STAT1 activation and expression. The specific causes of the decrease in STAT1 transcriptional and/or posttranslational levels by the 3Dpol require further investigation.
5.3.3 3Dpol targets other proteins associated with the innate immunity response
PGAM family member 5 (PGAM5) can affect the fission/fusion process of mitochondria and inhibit the mitochondrial autophagy pathway (190–193). During EV-D68 replication, the 3Dpol, via its interaction with PGAM5, can affect the mitochondrial dynamics and suppress the expression of IFN-β by impacting the RIG-I-like receptor signal pathway (27) (Figure 2). In addition, extensive studies have shown that the innate immune response and autophagy constitute a mutually coordinated system (194, 195). The autophagy pathway is tightly controlled by numerous autophagy-related genes (ATG) (196–198). Among these, Beclin1 (which encodes BECN1, also called ATG6) is not only a critical regulator in both the early and late steps of autophagy but is also antagonistic to innate immune responses (199–201). It has been reported that EV71 possibly propels 3Dpol to interact with Beclin1 in order to regulate the process of autophagy to promote viral replication (202). Further, EV71 3Dpol makes use of the interaction with Beclin1 to suppress the type I IFN signaling pathway due to Beclin1 acting as a negative regulator of RIG-I-MAVS mediated IFN response (202, 203). In addition, recent studies have shown that inhibition of IKBKE expression by SERPINB1 induced autophagy to decrease type I interferon signaling, and ultimately promoted SVA proliferation (204). These studies imply the reciprocal coordination between autophagy and innate immunity. However, the mechanism of innate immunity and autophagy regulating viral proliferation and the interaction between these classical pathways remain unclear.
5.4 Regulation of the activation of the NLRP3 inflammasome
Inflammasome formation is an innate immune response induced in host cells in response to stimulation by microbial invasion that triggers the maturation of the proinflammatory cytokine interleukin-1β (IL-1β) (205). IL-1β causes the production of cytokines such as IL-6 and TNF-α, and plays a critical role in modulating the immune response during both acute and chronic viral infections (206, 207). IL-1β production is tightly regulated by the NLRP3 inflammasome complex, which consists of the NOD-like receptor NLRP3 and the adaptor protein ASC to recognize danger signals to promote cleavage of the effector protein pro-caspase-1 (128, 208–210). NLRP3 inflammasome activation requires NF-κB activation (priming signal) and assembly of NLRP3-ASC (second signal) (208, 210–212). First, PRRs (such as RIG-I or MDA5) induce a priming signal, which recognize viral nucleic acid and other molecular patterns and then induce NF-κB activation; NF-κB activation acts as a priming signal to initiate the transcription of pro-IL-1β and NLRP3 (210). The second signal is NLRP3-ASC inflammasome assembly, and there are three models for its induction: (i) the ion channel model (213); (ii) the lysosomal rupture model (214); and (iii) the reactive oxygen species (ROS) model (215). The ion channel model, which regulates the concentration of K+ or Ca2+ in the cells, ultimately helps pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) to enter into the cytosol or cause mitochondrial dysfunction to activate the NLRP3 inflammasome (129), the lysosomal rupture model, which causes the release of cathepsin B after lysosomal damage, leads to NLRP3 activation (216, 217), and the ROS model, which invigorates the circulation of K+ and induces NLRP3 inflammasome activation (218, 219).
As reported, SVA can induce IL-1β production (129). SVA has a +ssRNA genome, and it can be recognized by the RIG-I-like receptor of RIG-I/MDA5 and then induce the activation of NF-κB, which leads to the upregulation of NLRP3 and pro-IL-1β transcription (220) (Figure 7). Meanwhile, SVA 3Dpol promotes the activation of NF-κB by interacting IKKα and IKKβ, which upregulates the NLRP3 and pro-IL-1β transcription (129). These results suggested that the effects of SVA RNA and 3Dpol induction of NF-κB activation are superimposed. This study also proved that SVA 3Dpol directly interacts with the NATCH domain of NLRP3 through the N-terminus (amino acids 1 to 154) to facilitate NLRP3-ASC assembly, which induces IL-1β production (129). At the same time, 3Dpol also affects the production of IL-1β through ion channels. 3Dpol induces calcium influx and potassium efflux to activate the NLRP3 inflammasome at the second signaling step (129, 221). In addition, other studies revealed a novel mechanism by which EV71 stimulates the activation of NLRP3 inflammasome by the virus-encoded 3Dpol. 3Dpol interacts directly with NLRP3 to facilitate the assembly of NLRP3 inflammasome complex by forming a “3D-NLRP3-ASC” ring-like structure (222). These studies revealed a new role of picornavirus 3Dpol as an important regulator of inflammatory responses and provided new insights into the development of drugs for the treatment and prevention of virus-associated inflammation and diseases.
6 Conclusions
The past decade has been fruitful for the viral RdRp structure field, and providing insights into the initiation of RNA synthesis and the replication elongation processes in picornavirus (21, 36, 92). However, the NLS sequence carried by picornavirus 3Dpol, combined with its ability to interact with other viral proteins, viral RNA and cellular proteins, indicate that the noncatalytic role of picornavirus 3Dpol could be underestimated. In addition to its traditional role in replication, 3Dpol can interact with several host proteins, which participate in a variety of biological processes in host cells, such as cell cycle progression, protein synthesis, apoptosis and autophagy, and these interactions may result in multiple consequences that benefit the viruses in different lifecycle stages. Interactome analysis has been widely applied to explore virus–host interactions. Yeast-two-hybrid assays and proteomic approaches based on MALDI-TOF mass spectrometry have been used to screen host factors that may interact with viral proteins in infected cells (24, 123, 223, 224). Advanced approaches using immunoprecipitation coupled with liquid chromatography−tandem mass spectrometry (LC−MS/MS) can be practical to broadly detect cellular proteins that associate with viral proteins (23, 225). Further development of these related technologies and methods may help to identify and validate novel host proteins that interact with 3Dpol and provide a better understanding of how 3Dpol regulates and usurps host processes, while also helping to uncover the mechanisms underlying pathogenesis.
Importantly, there are currently only limited therapies for the treatment of picornavirus infection. The key role of 3Dpol in viral replication and its structural and sequence conservation make it a promising target for specific antiviral therapeutics (19, 31, 226). Several compounds that bind to 3Dpol active sites to block viral replication have been identified, which markedly reduce the synthesis of viral RNA by interacting with or occupying the 3Dpol active sites to inhibit enzyme function (227–229). Therefore, further elucidating the structures and molecular functions of 3Dpol is valuable and could be useful for future antiviral treatment of picornaviruses.
Author contributions
CX: Formal analysis, Investigation, Writing – original draft. MW: Formal analysis, Supervision, Writing – review & editing. AC: Funding acquisition, Supervision, Writing – review & editing. QY: Formal analysis, Supervision, Writing – review & editing. JH: Formal analysis, Supervision, Writing – review & editing. XO: Formal analysis, Supervision, Writing – review & editing. DS: Formal analysis, Supervision, Writing – review & editing. YH: Formal analysis, Supervision, Writing – review & editing. ZW: Formal analysis, Supervision, Writing – review & editing. YW: Formal analysis, Supervision, Writing – review & editing. SZ: Formal analysis, Supervision, Writing – review & editing. BT: Formal analysis, Supervision, Writing – review & editing. XZ: Formal analysis, Supervision, Writing – review & editing. ML: Formal analysis, Supervision, Writing – review & editing. DZ: Formal analysis, Supervision, Writing – review & editing. RJ: Formal analysis, Supervision, Writing – review & editing. SC: Formal analysis, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from China Agriculture Research System of MOF and MARA(CARS-42-17) and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tapparel C, Siegrist F, Petty TJ, Kaiser L. Picornavirus and enterovirus diversity with associated human diseases. Infect Genet Evol. (2013) 14:282–93. doi: 10.1016/j.meegid.2012.10.016
2. Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, et al. Changes to virus taxonomy and to the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2021). Arch Virol. (2021) 166:2633–48. doi: 10.1007/s00705-021-05156-1
3. Zell R. Picornaviridae-the ever-growing virus family. Arch Virol. (2018) 163:299–317. doi: 10.1007/s00705-017-3614-8
4. Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 a resolution. Science. (1985) 229:1358–65. doi: 10.1126/science.2994218
5. Rossmann MG, Arnold E, Erickson JW, Frankenberger EA, Griffith JP, Hecht HJ, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. (1985) 317:145–53. doi: 10.1038/317145a0
6. Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis. (2010) 10:778–90. doi: 10.1016/s1473-3099(10)70194-8
7. Marcotte LL, Wass AB, Gohara DW, Pathak HB, Arnold JJ, Filman DJ, et al. Crystal structure of poliovirus 3cd protein: virally encoded protease and precursor to the Rna-dependent Rna polymerase. J Virol. (2007) 81:3583–96. doi: 10.1128/jvi.02306-06
8. Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus Rna-dependent Rna polymerase. EMBO J. (2004) 23:3462–71. doi: 10.1038/sj.emboj.7600357
9. Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. (1993) 27:353–436. doi: 10.1146/annurev.ge.27.120193.002033
10. Lee YF, Nomoto A, Detjen BM, Wimmer E. A protein covalently linked to poliovirus genome Rna. Proc Natl Acad Sci U S A. (1977) 74:59–63. doi: 10.1073/pnas.74.1.59
11. Fitzgerald KD, Semler BL. Bridging Ires elements in Mrnas to the eukaryotic translation apparatus. Biochim Biophys Acta. (2009) 1789:518–28. doi: 10.1016/j.bbagrm.2009.07.004
12. Pelletier J, Kaplan G, Racaniello VR, Sonenberg N. Cap-independent translation of poliovirus Mrna is conferred by sequence elements within the 5' Noncoding region. Mol Cell Biol. (1988) 8:1103–12. doi: 10.1128/mcb.8.3.1103-1112.1988
13. Palmenberg AC. Proteolytic processing of Picornaviral polyprotein. Annu Rev Microbiol. (1990) 44:603–23. doi: 10.1146/annurev.mi.44.100190.003131
14. Jacobson MF, Baltimore D. Polypeptide cleavages in the formation of poliovirus proteins. Proc Natl Acad Sci U SA. (1968) 61:77–84. doi: 10.1073/pnas.61.1.77
15. Campagnola G, Peersen O. Co-folding and Rna activation of poliovirus 3c(Pro) polyprotein precursors. J Biol Chem. (2023) 299:105258. doi: 10.1016/j.jbc.2023.105258
16. Lulla V, Dinan AM, Hosmillo M, Chaudhry Y, Sherry L, Irigoyen N, et al. An upstream protein-coding region in enteroviruses modulates virus infection in gut epithelial cells. Nat Microbiol. (2019) 4:280–92. doi: 10.1038/s41564-018-0297-1
17. Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. Picornavirus morphogenesis. Microbiol Mol Biol Rev. (2014) 78:418–37. doi: 10.1128/mmbr.00012-14
18. Cameron CE, Oh HS, Moustafa IM. Expanding knowledge of P3 proteins in the poliovirus lifecycle. Future Microbiol. (2010) 5:867–81. doi: 10.2217/fmb.10.40
19. Wu Y, Lou Z, Miao Y, Yu Y, Dong H, Peng W, et al. Structures of Ev71 Rna-dependent Rna polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China. Protein Cell. (2010) 1:491–500. doi: 10.1007/s13238-010-0061-7
20. Andino R, Rieckhof GE, Achacoso PL, Baltimore D. Poliovirus Rna synthesis utilizes an Rnp complex formed around the 5'-end of viral Rna. EMBO J. (1993) 12:3587–98. doi: 10.1002/embj.1993.12.issue-9
21. Peersen OB. Picornaviral polymerase structure, function, and fidelity modulation. Virus Res. (2017) 234:4–20. doi: 10.1016/j.virusres.2017.01.026
22. Ferrer-Orta C, de la Higuera I, Caridi F, Sánchez-Aparicio MT, Moreno E, Perales C, et al. Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition. J Virol. (2015) 89:6848–59. doi: 10.1128/jvi.03283-14
23. Lee KM, Wu CC, Wu SE, Lin YH, Wang LT, Chang CR, et al. The Rna-dependent Rna polymerase of enterovirus A71 associates with ribosomal proteins and positively regulates protein translation. RNA Biol. (2020) 17:608–22. doi: 10.1080/15476286.2020.1722448
24. Liu YC, Kuo RL, Lin JY, Huang PN, Huang Y, Liu H, et al. Cytoplasmic viral Rna-dependent Rna polymerase disrupts the intracellular splicing machinery by entering the nucleus and interfering with Prp8. PLoS Pathog. (2014) 10:e1004199. doi: 10.1371/journal.ppat.1004199
25. Kuo RL, Chen CJ, Wang RYL, Huang HI, Lin YH, Tam EH, et al. Role of enteroviral Rna-dependent Rna polymerase in regulation of Mda5-mediated beta interferon activation. J Virol. (2019) 93:e00132–19. doi: 10.1128/jvi.00132-19
26. Feng Q, Langereis MA, Lork M, Nguyen M, Hato SV, Lanke K, et al. Enterovirus 2apro targets Mda5 and Mavs in infected cells. J Virol. (2014) 88:3369–78. doi: 10.1128/jvi.02712-13
27. Yang Z, Zheng H, Li H, Chen Y, Hou D, Fan Q, et al. The expression of Ifn-B Is suppressed by the viral 3d polymerase via its impact on Pgam5 expression during Enterovirus D68 infection. Virus Res. (2021) 304:198549. doi: 10.1016/j.virusres.2021.198549
28. Love RA, Maegley KA, Yu X, Ferre RA, Lingardo LK, Diehl W, et al. The crystal structure of the Rna-dependent Rna polymerase from human rhinovirus: A dual function target for common cold antiviral therapy. Structure. (2004) 12:1533–44. doi: 10.1016/j.str.2004.05.024
29. Ferrer-Orta C, Arias A, Perez-Luque R, Escarmís C, Domingo E, Verdaguer N. Structure of foot-and-mouth disease virus Rna-dependent Rna polymerase and its complex with a template-primer Rna. J Biol Chem. (2004) 279:47212–21. doi: 10.1074/jbc.M405465200
30. Amineva SP, Aminev AG, Palmenberg AC, Gern JE. Rhinovirus 3c protease precursors 3cd and 3cd' Localize to the nuclei of infected cells. J Gen Virol. (2004) 85:2969–79. doi: 10.1099/vir.0.80164-0
31. Campagnola G, Weygandt M, Scoggin K, Peersen O. Crystal structure of coxsackievirus B3 3dpol highlights the functional importance of residue 5 in picornavirus polymerases. J Virol. (2008) 82:9458–64. doi: 10.1128/jvi.00647-08
32. Thompson AA, Albertini RA, Peersen OB. Stabilization of poliovirus polymerase by Ntp binding and fingers-thumb interactions. J Mol Biol. (2007) 366:1459–74. doi: 10.1016/j.jmb.2006.11.070
33. Vives-Adrian L, Lujan C, Oliva B, van der Linden L, Selisko B, Coutard B, et al. The crystal structure of a Cardiovirus Rna-dependent Rna polymerase reveals an unusual conformation of the polymerase active site. J Virol. (2014) 88:5595–607. doi: 10.1128/jvi.03502-13
34. Yap TL, Xu T, Chen YL, Malet H, Egloff MP, Canard B, et al. Crystal structure of the dengue virus Rna-dependent Rna polymerase catalytic domain at 1.85-angstrom resolution. J Virol. (2007) 81:4753–65. doi: 10.1128/jvi.02283-06
35. Černý J, Černá Bolfíková B, Valdés JJ, Grubhoffer L, Růžek D. Evolution of tertiary structure of viral Rna dependent polymerases. PloS One. (2014) 9:e96070. doi: 10.1371/journal.pone.0096070
36. Ferrer-Orta C, Ferrero D, Verdaguer N. Rna-dependent Rna polymerases of Picornaviruses: from the structure to regulatory mechanisms. Viruses. (2015) 7:4438–60. doi: 10.3390/v7082829
37. Ferrer-Orta C, Agudo R, Domingo E, Verdaguer N. Structural insights into replication initiation and elongation processes by the Fmdv Rna-dependent Rna polymerase. Curr Opin Struct Biol. (2009) 19:752–8. doi: 10.1016/j.sbi.2009.10.016
38. Ferrer-Orta C, Arias A, Agudo R, Pérez-Luque R, Escarmís C, Domingo E, et al. The structure of a protein primer-polymerase complex in the initiation of genome replication. EMBO J. (2006) 25:880–8. doi: 10.1038/sj.emboj.7600971
39. Fernández-Tomás C. The presence of viral-induced proteins in nuclei from poliovirus-infected Hela cells. Virology. (1982) 116:629–34. doi: 10.1016/0042-6822(82)90154-4
40. Weidman MK, Sharma R, Raychaudhuri S, Kundu P, Tsai W, Dasgupta A. The interaction of cytoplasmic Rna viruses with the nucleus. Virus Res. (2003) 95:75–85. doi: 10.1016/s0168-1702(03)00164-3
41. Sharma R, Raychaudhuri S, Dasgupta A. Nuclear entry of poliovirus protease-polymerase precursor 3cd: implications for host cell transcription shut-off. Virology. (2004) 320:195–205. doi: 10.1016/j.virol.2003.10.020
42. Hansen JL, Long AM, Schultz SC. Structure of the Rna-dependent Rna polymerase of poliovirus. Structure. (1997) 5:1109–22. doi: 10.1016/S0969-2126(97)00261-X
43. Stugel R, Timmers ACJ, Raue HA, van’n Riet J. Nuclear import of ribosomal proteins: evidence for a novel type of nucleolar localization signal. In: The Ribosome Structure, Function, Antibiotics and Cellular Interaction. Washington: ASM Press (2000). p. 205–17.
44. Aminev AG, Amineva SP, Palmenberg AC. Encephalomyocarditis virus (Emcv) proteins 2a and 3bcd localize to nuclei and inhibit cellular Mrna transcription but not Rrna transcription. Virus Res. (2003) 95:59–73. doi: 10.1016/s0168-1702(03)00163-1
45. Sanchez-Aparicio MT, Rosas MF, Sobrino F. Characterization of a nuclear localization signal in the foot-and-mouth disease virus polymerase. Virology. (2013) 444:203–10. doi: 10.1016/j.virol.2013.06.011
46. Chen JH, Zhang RH, Lin SL, Li PF, Lan JJ, Gao JM, et al. Identification of a functional nuclear localization signal in 3d(Pol)/3cd of duck hepatitis a virus 1. Virus Res. (2019) 270:197670. doi: 10.1016/j.virusres.2019.197670
47. Clark ME, Lieberman PM, Berk AJ, Dasgupta A. Direct cleavage of human tata-binding protein by poliovirus protease 3c in vivo and in vitro. Mol Cell Biol. (1993) 13:1232–7. doi: 10.1128/mcb.13.2.1232-1237.1993
48. Belov GA, Evstafieva AG, Rubtsov YP, Mikitas OV, Vartapetian AB, Agol VI. Early alteration of nucleocytoplasmic traffic induced by some Rna viruses. Virology. (2000) 275:244–8. doi: 10.1006/viro.2000.0427
49. Gustin KE, Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. (2001) 20:240–9. doi: 10.1093/emboj/20.1.240
50. Gustin KE, Sarnow P. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J Virol. (2002) 76:8787–96. doi: 10.1128/jvi.76.17.8787-8796.2002
51. Kloc A, Rai DK, Gladue DP, Schafer E, Kenney M, Rieder E. Residues within the foot-and-mouth disease virus 3d(Pol) nuclear localization signal affect polymerase fidelity. J Virol. (2020) 94(17):e00833–20. doi: 10.1128/jvi.00833-20
52. Witwer C, Rauscher S, Hofacker IL, Stadler PF. Conserved Rna secondary structures in Picornaviridae genomes. Nucleic Acids Res. (2001) 29:5079–89. doi: 10.1093/nar/29.24.5079
53. McKnight KL, Lemon SM. The rhinovirus type 14 genome contains an internally located Rna structure that is required for viral replication. Rna. (1998) 4:1569–84. doi: 10.1017/s1355838298981006
54. Burrill CP, Westesson O, Schulte MB, Strings VR, Segal M, Andino R. Global Rna structure analysis of poliovirus identifies a conserved Rna structure involved in viral replication and infectivity. J Virol. (2013) 87:11670–83. doi: 10.1128/jvi.01560-13
55. Kloc A, Rai DK, Rieder E. The roles of Picornavirus untranslated regions in infection and innate immunity. Front Microbiol. (2018) 9:485. doi: 10.3389/fmicb.2018.00485
56. Rieder E, Paul AV, Kim DW, van Boom JH, Wimmer E. Genetic and biochemical studies of poliovirus Cis-acting replication element Cre in relation to Vpg uridylylation. J Virol. (2000) 74:10371–80. doi: 10.1128/jvi.74.22.10371-10380.2000
57. Ferhadian D, Contrant M, Printz-Schweigert A, Smyth RP, Paillart JC, Marquet R. Structural and functional motifs in influenza virus Rnas. Front Microbiol. (2018) 9:559. doi: 10.3389/fmicb.2018.00559
58. Han JQ, Townsend HL, Jha BK, Paranjape JM, Silverman RH, Barton DJ. A phylogenetically conserved Rna structure in the poliovirus open reading frame inhibits the antiviral Endoribonuclease Rnase L. J Virol. (2007) 81:5561–72. doi: 10.1128/jvi.01857-06
59. Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. A viral Rna competitively inhibits the antiviral endoribonuclease domain of Rnase L. Rna. (2008) 14:1026–36. doi: 10.1261/rna.958908
60. Song Y, Liu Y, Ward CB, Mueller S, Futcher B, Skiena S, et al. Identification of two functionally redundant Rna elements in the coding sequence of poliovirus using computer-generated design. Proc Natl Acad Sci U.S.A. (2012) 109:14301–7. doi: 10.1073/pnas.1211484109
61. Lasecka-Dykes L, Tulloch F, Simmonds P, Luke GA, Ribeca P, Gold S, et al. Mutagenesis mapping of Rna structures within the foot-and-mouth disease virus genome reveals functional elements localized in the polymerase (3d(Pol))-encoding region. mSphere. (2021) 6:e0001521. doi: 10.1128/mSphere.00015-21
63. Cappadocia L, Lima CD. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem Rev. (2018) 118:889–918. doi: 10.1021/acs.chemrev.6b00737
64. Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. (2004) 20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350
65. Shackelford J, Pagano JS. Tumor viruses and cell signaling pathways: deubiquitination versus ubiquitination. Mol Cell Biol. (2004) 24:5089–93. doi: 10.1128/mcb.24.12.5089-5093.2004
66. Furman MH, Ploegh HL. Lessons from viral manipulation of protein disposal pathways. J Clin Invest. (2002) 110:875–9. doi: 10.1172/jci16831
67. Fan Z, Zhuo Y, Tan X, Zhou Z, Yuan J, Qiang B, et al. Sars-Cov nucleocapsid protein binds to Hubc9, a ubiquitin conjugating enzyme of the sumoylation system. J Med Virol. (2006) 78:1365–73. doi: 10.1002/jmv.20707
68. Luo H, Zhang J, Dastvan F, Yanagawa B, Reidy MA, Zhang HM, et al. Ubiquitin-dependent proteolysis of Cyclin D1 is associated with coxsackievirus-induced cell growth arrest. J Virol. (2003) 77:1–9. doi: 10.1128/jvi.77.1.1-9.2003
69. Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, et al. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of Hiv-1 and Hiv-2. Proc Natl Acad Sci U S A. (2000) 97:13057–62. doi: 10.1073/pnas.97.24.13057
70. Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci U S A. (2000) 97:13063–8. doi: 10.1073/pnas.97.24.13063
71. Lin L, Qin Y, Wu H, Chen Y, Wu S, Si X, et al. Pyrrolidine dithiocarbamate inhibits enterovirus 71 replication by down-regulating ubiquitin-proteasome system. Virus Res. (2015) 195:207–16. doi: 10.1016/j.virusres.2014.10.012
72. Si X, Gao G, Wong J, Wang Y, Zhang J, Luo H. Ubiquitination is required for effective replication of Coxsackievirus B3. PLoS One. (2008) 3:e2585. doi: 10.1371/journal.pone.0002585
73. Li L, Bai J, Fan H, Yan J, Li S, Jiang P. E2 ubiquitin-conjugating enzyme Ube2l6 promotes Senecavirus a proliferation by stabilizing the viral Rna polymerase. PLoS Pathog. (2020) 16:e1008970. doi: 10.1371/journal.ppat.1008970
74. Liu Y, Zheng Z, Shu B, Meng J, Zhang Y, Zheng C, et al. Sumo modification stabilizes Enterovirus 71 polymerase 3d to facilitate viral replication. J Virol. (2016) 90:10472–85. doi: 10.1128/jvi.01756-16
75. Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, et al. N6-methyladenosine modification and mettl3 modulate Enterovirus 71 replication. Nucleic Acids Res. (2019) 47:362–74. doi: 10.1093/nar/gky1007
76. Miteva M, Keusekotten K, Hofmann K, Praefcke GJ, Dohmen RJ. Sumoylation as a signal for polyubiquitylation and proteasomal degradation. Subcell Biochem. (2010) 54:195–214. doi: 10.1007/978-1-4419-6676-6_16
77. Praefcke GJ, Hofmann K, Dohmen RJ. Sumo playing tag with ubiquitin. Trends Biochem Sci. (2012) 37:23–31. doi: 10.1016/j.tibs.2011.09.002
78. Denuc A, Marfany G. Sumo and ubiquitin paths converge. Biochem Soc Trans. (2010) 38:34–9. doi: 10.1042/bst0380034
79. Su CI, Tseng CH, Yu CY, Lai MMC. Sumo modification stabilizes dengue virus nonstructural protein 5 to support virus replication. J Virol. (2016) 90:4308–19. doi: 10.1128/jvi.00223-16
80. Han Q, Chang C, Li L, Klenk C, Cheng J, Chen Y, et al. Sumoylation of influenza a virus nucleoprotein is essential for intracellular trafficking and virus growth. J Virol. (2014) 88:9379–90. doi: 10.1128/jvi.00509-14
81. Voss M, Braun V, Bredow C, Kloetzel PM, Beling A. Coxsackievirus B3 exploits the ubiquitin-proteasome system to facilitate viral replication. Viruses. (2021) 13(7):1360. doi: 10.3390/v13071360
82. Paul AV, Yin J, Mugavero J, Rieder E, Liu Y, Wimmer E. A "Slide-back" Mechanism for the initiation of protein-primed Rna synthesis by the Rna polymerase of poliovirus. J Biol Chem. (2003) 278:43951–60. doi: 10.1074/jbc.M307441200
83. Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed Rna synthesis by purified poliovirus Rna polymerase. Nature. (1998) 393:280–4. doi: 10.1038/30529
84. Paul AV, Peters J, Mugavero J, Yin J, van Boom JH, Wimmer E. Biochemical and genetic studies of the Vpg uridylylation reaction catalyzed by the Rna polymerase of poliovirus. J Virol. (2003) 77:891–904. doi: 10.1128/jvi.77.2.891-904.2003
85. Gruez A, Selisko B, Roberts M, Bricogne G, Bussetta C, Jabafi I, et al. The crystal structure of coxsackievirus B3 Rna-dependent Rna polymerase in complex with its protein primer Vpg confirms the existence of a second Vpg binding site on picornaviridae polymerases. J Virol. (2008) 82:9577–90. doi: 10.1128/jvi.00631-08
86. Appleby TC, Luecke H, Shim JH, Wu JZ, Cheney IW, Zhong W, et al. Crystal structure of complete rhinovirus Rna polymerase suggests front loading of protein primer. J Virol. (2005) 79:277–88. doi: 10.1128/jvi.79.1.277-288.2005
87. Lyle JM, Clewell A, Richmond K, Richards OC, Hope DA, Schultz SC, et al. Similar structural basis for membrane localization and protein priming by an Rna-dependent Rna polymerase. J Biol Chem. (2002) 277:16324–31. doi: 10.1074/jbc.M112429200
88. Chen C, Wang Y, Shan C, Sun Y, Xu P, Zhou H, et al. Crystal structure of Enterovirus 71 Rna-dependent Rna polymerase complexed with its protein primer Vpg: implication for a trans mechanism of Vpg uridylylation. J Virol. (2013) 87:5755–68. doi: 10.1128/jvi.02733-12
89. Richards OC, Spagnolo JF, Lyle JM, Vleck SE, Kuchta RD, Kirkegaard K. Intramolecular and intermolecular uridylylation by poliovirus Rna-dependent Rna polymerase. J Virol. (2006) 80:7405–15. doi: 10.1128/jvi.02533-05
90. Eruera AR, McSweeney AM, McKenzie-Goldsmith GM, Ward VK. Protein nucleotidylylation in +Ssrna viruses. Viruses. (2021) 13(8):1549. doi: 10.3390/v13081549
91. Flanegan JB, Baltimore D. Poliovirus-specific primer-dependent Rna polymerase able to copy poly(a). Proc Natl Acad Sci U S A. (1977) 74:3677–80. doi: 10.1073/pnas.74.9.3677
92. Kempf BJ, Barton DJ. Picornavirus Rna polyadenylation by 3d(Pol), the viral Rna-dependent Rna polymerase. Virus Res. (2015) 206:3–11. doi: 10.1016/j.virusres.2014.12.030
93. Sarnow P. Role of 3'-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. (1989) 63:467–70. doi: 10.1128/jvi.63.1.467-470.1989
94. Spector DH, Baltimore D. Requirement of 3'-terminal poly(Adenylic acid) for the infectivity of poliovirus Rna. Proc Natl Acad Sci U S A. (1974) 71:2983–7. doi: 10.1073/pnas.71.8.2983
95. Bergamini G, Preiss T, Hentze MW. Picornavirus ireses and the poly(a) tail jointly promote cap-independent translation in a mammalian cell-free system. Rna. (2000) 6:1781–90. doi: 10.1017/s1355838200001679
96. Silvestri LS, Parilla JM, Morasco BJ, Ogram SA, Flanegan JB. Relationship between poliovirus negative-strand Rna synthesis and the length of the 3' Poly(a) tail. Virology. (2006) 345:509–19. doi: 10.1016/j.virol.2005.10.019
97. Zhao D, Li Y, Li Z, Zhu L, Sang Y, Zhang H, et al. Only fourteen 3'-end poly(a)S sufficient for rescuing Senecavirus a from its Cdna clone, but inadequate to meet requirement of viral replication. Virus Res. (2023) 328:199076. doi: 10.1016/j.virusres.2023.199076
98. Chen JH, Zhang RH, Lin SL, Li PF, Lan JJ, Song SS, et al. The functional role of the 3' Untranslated region and poly(a) tail of duck hepatitis a virus type 1 in viral replication and regulation of ires-mediated translation. Front Microbiol. (2018) 9:2250. doi: 10.3389/fmicb.2018.02250
99. Kempf BJ, Kelly MM, Springer CL, Peersen OB, Barton DJ. Structural features of a picornavirus polymerase involved in the polyadenylation of viral Rna. J Virol. (2013) 87:5629–44. doi: 10.1128/jvi.02590-12
100. Steil BP, Kempf BJ, Barton DJ. Poly(a) at the 3' End of Positive-Strand Rna and Vpg-Linked Poly(U) at the 5' End of Negative-Strand Rna Are Reciprocal Templates During Replication of Poliovirus Rna. J Virol. (2010) 84:2843–58. doi: 10.1128/jvi.02620-08
101. Meredith JM, Rohll JB, Almond JW, Evans DJ. Similar interactions of the poliovirus and rhinovirus 3d polymerases with the 3' Untranslated region of rhinovirus 14. J Virol. (1999) 73:9952–8. doi: 10.1128/jvi.73.12.9952-9958.1999
102. Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, et al. Viral reorganization of the secretory pathway generates distinct organelles for Rna replication. Cell. (2010) 141:799–811. doi: 10.1016/j.cell.2010.03.050
103. Schlegel A, Giddings TH Jr., Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. (1996) 70:6576–88. doi: 10.1128/jvi.70.10.6576-6588.1996
104. Baggen J, Thibaut HJ, Strating J, van Kuppeveld FJM. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol. (2018) 16:368–81. doi: 10.1038/s41579-018-0005-4
105. Sasvari Z, Nagy PD. Making of viral replication organelles by remodeling interior membranes. Viruses. (2010) 2:2436–42. doi: 10.3390/v2112436
106. Strating JR, van Kuppeveld FJ. Viral rewiring of cellular lipid metabolism to create membranous replication compartments. Curr Opin Cell Biol. (2017) 47:24–33. doi: 10.1016/j.ceb.2017.02.005
107. Lyoo H, van der Schaar HM, Dorobantu CM, Rabouw HH, Strating J, van Kuppeveld FJM. Acbd3 is an essential pan-enterovirus host factor that mediates the interaction between viral 3a protein and cellular protein Pi4kb. mBio. (2019) 10(1):e02742–18. doi: 10.1128/mBio.02742-18
108. Xiao X, Lei X, Zhang Z, Ma Y, Qi J, Wu C, et al. Enterovirus 3a facilitates viral replication by promoting phosphatidylinositol 4-kinase iiiβ-acbd3 interaction. J Virol. (2017) 91(19):e00791–17. doi: 10.1128/jvi.00791-17
109. Laufman O, Perrino J, Andino R. Viral generated inter-organelle contacts redirect lipid flux for genome replication. Cell. (2019) 178:275–89.e16. doi: 10.1016/j.cell.2019.05.030
110. Limpens RW, van der Schaar HM, Kumar D, Koster AJ, Snijder EJ, van Kuppeveld FJ, et al. The transformation of enterovirus replication structures: A three-dimensional study of single- and double-membrane compartments. mBio. (2011) 2(5):e00166–11. doi: 10.1128/mBio.00166-11
111. van der Schaar HM, Dorobantu CM, Albulescu L, Strating J, van Kuppeveld FJM. Fat(Al) attraction: Picornaviruses usurp lipid transfer at membrane contact sites to create replication organelles. Trends Microbiol. (2016) 24:535–46. doi: 10.1016/j.tim.2016.02.017
112. Suhy DA, Giddings TH Jr., Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. (2000) 74:8953–65. doi: 10.1128/jvi.74.19.8953-8965.2000
113. McPhail JA, Lyoo H, Pemberton JG, Hoffmann RM, van Elst W, Strating J, et al. Characterization of the C10orf76-pi4kb complex and its necessity for Golgi Pi4p levels and enterovirus replication. EMBO Rep. (2020) 21:e48441. doi: 10.15252/embr.201948441
114. Lama J, Carrasco L. Mutations in the hydrophobic domain of poliovirus protein 3ab abrogate its permeabilizing activity. FEBS Lett. (1995) 367:5–11. doi: 10.1016/0014-5793(95)00523-C
115. Barco A, Carrasco L. A human virus protein, poliovirus protein 2bc, induces membrane proliferation and blocks the exocytic pathway in the yeast saccharomyces cerevisiae. EMBO J. (1995) 14:3349–64. doi: 10.1002/embj.1995.14.issue-14
116. Dubankova A, Humpolickova J, Klima M, Boura E. Negative charge and membrane-tethered viral 3b cooperate to recruit viral Rna dependent Rna polymerase 3d (Pol). Sci Rep. (2017) 7:17309. doi: 10.1038/s41598-017-17621-6
117. Towner JS, Ho TV, Semler BL. Determinants of membrane association for poliovirus protein 3ab. J Biol Chem. (1996) 271:26810–8. doi: 10.1074/jbc.271.43.26810
118. Strauss DM, Wuttke DS. Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3ab and Vpg binding to the Rna-dependent Rna polymerase. J Virol. (2007) 81:6369–78. doi: 10.1128/jvi.02252-06
119. Ferrer-Orta C, Ferrero DS, Verdaguer N. Dual role of the foot-and-mouth disease virus 3b1 protein in the replication complex: as protein primer and as an essential component to recruit 3dpol to membranes. PLoS Pathog. (2023) 19:e1011373. doi: 10.1371/journal.ppat.1011373
120. Richards OC, Ehrenfeld E. Effects of poliovirus 3ab protein on 3d polymerase-catalyzed reaction. J Biol Chem. (1998) 273:12832–40. doi: 10.1074/jbc.273.21.12832
121. Rodriguez-Wells V, Plotch SJ, DeStefano JJ. Primer-dependent synthesis by poliovirus Rna-dependent Rna polymerase (3d(Pol)). Nucleic Acids Res. (2001) 29:2715–24. doi: 10.1093/nar/29.13.2715
122. Zhang Q, Li S, Lei P, Li Z, Chen F, Chen Q, et al. Anxa2 facilitates enterovirus 71 infection by interacting with 3d polymerase and Pi4kb to assist the assembly of replication organelles. Virol Sin. (2021) 36:1387–99. doi: 10.1007/s12250-021-00417-4
123. Huang PN, Jheng JR, Arnold JJ, Wang JR, Cameron CE, Shih SR. Uggt1 enhances enterovirus 71 pathogenicity by promoting viral Rna synthesis and viral replication. PloS Pathog. (2017) 13:e1006375. doi: 10.1371/journal.ppat.1006375
124. Wang X, Hu Z, Zhang W, Wu S, Hao Y, Xiao X, et al. Inhibition of lysosome-tethered ragulator-rag-3d complex restricts the replication of enterovirus 71 and coxsackie A16. J Cell Biol. (2023) 222(12):e20230108. doi: 10.1083/jcb.202303108
125. Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. (2009) 227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x
126. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. (2014) 14:36–49. doi: 10.1038/nri3581
127. Sarry M, Caignard G, Dupré J, Zientara S, Vitour D, Bakkali Kassimi L, et al. Host-specific interplay between foot-and-mouth disease virus 3d polymerase and the type-I interferon pathway. Viruses. (2023) 15(3):666. doi: 10.3390/v15030666
128. Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. (2016) 16:407–20. doi: 10.1038/nri.2016.58
129. Choudhury SM, Ma X, Zeng Z, Luo Z, Li Y, Nian X, et al. Senecavirus a 3d interacts with Nlrp3 to induce Il-1β Production by activating Nf-Kb and ion channel signals. Microbiol Spectr. (2022) 10:e0209721. doi: 10.1128/spectrum.02097-21
130. Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. The mechanism of translation initiation on type 1 Picornavirus iress. EMBO J. (2014) 33:76–92. doi: 10.1002/embj.201386124
131. Chen CJ, Makino S. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J Virol. (2004) 78:5658–69. doi: 10.1128/jvi.78.11.5658-5669.2004
132. Helt AM, Harris E. S-phase-dependent enhancement of dengue virus 2 replication in mosquito cells, but not in human cells. J Virol. (2005) 79:13218–30. doi: 10.1128/jvi.79.21.13218-13230.2005
133. He Y, Xu K, Keiner B, Zhou J, Czudai V, Li T, et al. Influenza a virus replication induces cell cycle arrest in G0/G1 phase. J Virol. (2010) 84:12832–40. doi: 10.1128/jvi.01216-10
134. Basu S, Greenwood J, Jones AW, Nurse P. Core control principles of the eukaryotic cell cycle. Nature. (2022) 607:381–6. doi: 10.1038/s41586-022-04798-8
135. Gavet O, Pines J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol. (2010) 189:247–59. doi: 10.1083/jcb.200909144
136. Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. (1992) 70:993–1006. doi: 10.1016/0092-8674(92)90249-C
137. Yam CH, Fung TK, Poon RY. Cyclin a in cell cycle control and cancer. Cell Mol Life Sci. (2002) 59:1317–26. doi: 10.1007/s00018-002-8510-y
138. Coverley D, Laman H, Laskey RA. Distinct roles for cyclins E and a during DNA replication complex assembly and activation. Nat Cell Biol. (2002) 4:523–8. doi: 10.1038/ncb813
139. Adeyemi RO, Pintel DJ. Parvovirus-induced depletion of cyclin B1 prevents mitotic entry of infected cells. PLoS Pathog. (2014) 10:e1003891. doi: 10.1371/journal.ppat.1003891
140. Eremenko T, Benedetto A, Volpe P. Virus infection as a function of the host cell life cycle: replication of Poliovirus Rna. J Gen Virol. (1972) 16:61–8. doi: 10.1099/0022-1317-16-1-61
141. Suarez M, Contreras G, Fridlender B. Multiplication of Coxsackie B1 virus in synchronized hela cells. J Virol. (1975) 16:1337–9. doi: 10.1128/jvi.16.5.1337-1339.1975
142. Feuer R, Mena I, Pagarigan R, Slifka MK, Whitton JL. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J Virol. (2002) 76:4430–40. doi: 10.1128/jvi.76.9.4430-4440.2002
143. Yu J, Zhang L, Ren P, Zhong T, Li Z, Wang Z, et al. Enterovirus 71 mediates cell cycle arrest in S phase through non-structural protein 3d. Cell Cycle. (2015) 14:425–36. doi: 10.4161/15384101.2014.980631
144. Wang Z, Wang Y, Wang S, Meng X, Song F, Huo W, et al. Coxsackievirus A6 induces cell cycle arrest in G0/G1 phase for viral production. Front Cell Infect Microbiol. (2018) 8:279. doi: 10.3389/fcimb.2018.00279
145. Wang ZY, Zhong T, Wang Y, Song FM, Yu XF, Xing LP, et al. Human enterovirus 68 interferes with the host cell cycle to facilitate viral production. Front Cell Infect Microbiol. (2017) 7:29. doi: 10.3389/fcimb.2017.00029
146. Liu Y, Li Y, Wang M, Cheng A, Ou X, Mao S, et al. Duck hepatitis a virus type 1 mediates cell cycle arrest in the S phase. Virol J. (2022) 19:111. doi: 10.1186/s12985-022-01839-6
147. Lai MC, Chen HH, Xu P, Wang RYL. Translation control of enterovirus A71 gene expression. J BioMed Sci. (2020) 27:22. doi: 10.1186/s12929-019-0607-9
148. Abedeera SM, Davila-Calderon J, Haddad C, Henry B, King J, Penumutchu S, et al. The repurposing of cellular proteins during enterovirus A71 infection. Viruses. (2023) 16(1):75. doi: 10.3390/v16010075
149. Sommergruber W, Ahorn H, Klump H, Seipelt J, Zoephel A, Fessl F, et al. 2a proteinases of coxsackie- and rhinovirus cleave peptides derived from Eif-4 gamma via a common recognition motif. Virology. (1994) 198:741–5. doi: 10.1006/viro.1994.1089
150. Gradi A, Svitkin YV, Imataka H, Sonenberg N. Proteolysis of Human Eukaryotic Translation Initiation Factor Eif4gii, but Not Eif4gi, Coincides with the Shutoff of Host Protein Synthesis after Poliovirus Infection. Proc Natl Acad Sci U.S.A. (1998) 95:11089–94. doi: 10.1073/pnas.95.19.11089
151. Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eif4g-eif4e complex is the target for direct cleavage by the rhinovirus 2a proteinase. J Virol. (1996) 70:8444–50. doi: 10.1128/jvi.70.12.8444-8450.1996
152. Li W, Ross-Smith N, Proud CG, Belsham GJ. Cleavage of translation initiation factor 4ai (Eif4ai) but not eif4aii by foot-and-mouth disease virus 3c protease: identification of the eif4ai cleavage site. FEBS Lett. (2001) 507:1–5. doi: 10.1016/S0014-5793(01)02885-X
153. de Breyne S, Bonderoff JM, Chumakov KM, Lloyd RE, Hellen CU. Cleavage of eukaryotic initiation factor eif5b by enterovirus 3c proteases. Virology. (2008) 378:118–22. doi: 10.1016/j.virol.2008.05.019
154. Shih SR, Stollar V, Li ML. Host factors in enterovirus 71 replication. J Virol. (2011) 85:9658–66. doi: 10.1128/jvi.05063-11
155. Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. (2007) 81:12128–34. doi: 10.1128/jvi.01356-07
156. Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. (2009) 19:359–78. doi: 10.1002/rmv.630
157. Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: A unique autophagy-related protein. Cell Res. (2007) 17:839–49. doi: 10.1038/cr.2007.78
158. You L, Chen J, Liu W, Xiang Q, Luo Z, Wang W, et al. Enterovirus 71 induces neural cell apoptosis and autophagy through promoting acox1 downregulation and Ros generation. Virulence. (2020) 11:537–53. doi: 10.1080/21505594.2020.1766790
159. Hou L, Ge X, Xin L, Zhou L, Guo X, Yang H. Nonstructural proteins 2c and 3d are involved in autophagy as induced by the encephalomyocarditis virus. Virol J. (2014) 11:156. doi: 10.1186/1743-422x-11-156
160. Chaplin DD. Overview of the immune response. J Allergy Clin Immunol. (2010) 125:S3–23. doi: 10.1016/j.jaci.2009.12.980
161. Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural basis for the activation of innate immune pattern-recognition receptor rig-I by viral Rna. Cell. (2011) 147:423–35. doi: 10.1016/j.cell.2011.09.039
162. Kuo RL, Kao LT, Lin SJ, Wang RY, Shih SR. Mda5 plays a crucial role in enterovirus 71 Rna-mediated Irf3 activation. PloS One. (2013) 8:e63431. doi: 10.1371/journal.pone.0063431
163. Wang B, Xi X, Lei X, Zhang X, Cui S, Wang J, et al. Enterovirus 71 protease 2apro targets mavs to inhibit anti-viral type I interferon responses. PloS Pathog. (2013) 9:e1003231. doi: 10.1371/journal.ppat.1003231
164. Zhu Z, Wang G, Yang F, Cao W, Mao R, Du X, et al. Foot-and-mouth disease virus viroporin 2b antagonizes rig-I-mediated antiviral effects by inhibition of its protein expression. J Virol. (2016) 90:11106–21. doi: 10.1128/jvi.01310-16
165. Li M, Xin T, Gao X, Wu J, Wang X, Fang L, et al. Foot-and-mouth disease virus non-structural protein 2b negatively regulates the rlr-mediated Ifn-B Induction. Biochem Biophys Res Commun. (2018) 504:238–44. doi: 10.1016/j.bbrc.2018.08.161
166. Xia X, Cheng A, Wang M, Ou X, Sun D, Mao S, et al. Functions of viroporins in the viral life cycle and their regulation of host cell responses. Front Immunol. (2022) 13:890549. doi: 10.3389/fimmu.2022.890549
167. Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, et al. The viral Rna recognition sensor rig-I is degraded during encephalomyocarditis virus (Emcv) infection. Virology. (2009) 393:311–8. doi: 10.1016/j.virol.2009.08.009
168. Li SX, Barrett BS, Heilman KJ, Messer RJ, Liberatore RA, Bieniasz PD, et al. Tetherin promotes the innate and adaptive cell-mediated immune response against retrovirus infection in vivo. J Immunol. (2014) 193:306–16. doi: 10.4049/jimmunol.1400490
169. Miller CM, Barrett BS, Chen J, Morrison JH, Radomile C, Santiago ML, et al. Systemic expression of a viral Rdrp protects against retrovirus infection and disease. J Virol. (2020) 94(9):e00071–20. doi: 10.1128/jvi.00071-20
170. Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. Mda-5: an interferon-inducible putative Rna helicase with double-stranded Rna-dependent Atpase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci U S A. (2002) 99:637–42. doi: 10.1073/pnas.022637199
171. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The Rna helicase Rig-I has an essential function in double-stranded Rna-induced innate antiviral responses. Nat Immunol. (2004) 5:730–7. doi: 10.1038/ni1087
172. Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of Mda5 and rig-I helicases in the recognition of Rna viruses. Nature. (2006) 441:101–5. doi: 10.1038/nature04734
173. Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, et al. Rig-I-mediated antiviral responses to single-stranded Rna bearing 5'-phosphates. Science. (2006) 314:997–1001. doi: 10.1126/science.1132998
174. Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M Jr. Innate immunity induced by composition-dependent Rig-I recognition of hepatitis C virus Rna. Nature. (2008) 454:523–7. doi: 10.1038/nature07106
175. Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor lgp2 activates antiviral signaling by regulating mda5-Rna interaction and filament assembly. Mol Cell. (2014) 55:771–81. doi: 10.1016/j.molcel.2014.07.003
176. Bruns AM, Pollpeter D, Hadizadeh N, Myong S, Marko JF, Horvath CM. Atp hydrolysis enhances Rna recognition and antiviral signal transduction by the innate immune sensor, laboratory of genetics and physiology 2 (Lgp2). J Biol Chem. (2013) 288:938–46. doi: 10.1074/jbc.M112.424416
177. Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, et al. The Rna helicase Lgp2 inhibits Tlr-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. (2005) 175:5260–8. doi: 10.4049/jimmunol.175.8.5260
178. Murali A, Li X, Ranjith-Kumar CT, Bhardwaj K, Holzenburg A, Li P, et al. Structure and function of lgp2, a dex(D/H) helicase that regulates the innate immunity response. J Biol Chem. (2008) 283:15825–33. doi: 10.1074/jbc.M800542200
179. Lee KY, Craig C, Patel SS. Unraveling blunt-end Rna binding and atpase-driven translocation activities of the rig-I family helicase lgp2. Nucleic Acids Res. (2024) 52:355–69. doi: 10.1093/nar/gkad1106
180. Miyamoto M, Himeda T, Ishihara K, Okuwa T, Kobayashi D, Nameta M, et al. Theilovirus 3c protease cleaves the C-terminal domain of the innate immune Rna sensor, melanoma differentiation-associated gene 5, and impairs double-stranded Rna-mediated Ifn response. J Immunol. (2023) 210:335–47. doi: 10.4049/jimmunol.2200565
181. Kim H, Kim AY, Choi J, Park SY, Park SH, Kim JS, et al. Foot-and-mouth disease virus evades innate immune response by 3c-targeting of Mda5. Cells. (2021) 10(2):271. doi: 10.3390/cells10020271
182. Yi J, Peng J, Yang W, Zhu G, Ren J, Li D, et al. Picornavirus 3c - a protease ensuring virus replication and subverting host responses. J Cell Sci. (2021) 134(5):jcs253237. doi: 10.1242/jcs.253237
183. Xia X, Cheng A, Wang M, Ou X, Sun D, Zhang S, et al. Dhav 3cd targets Irf7 and rig-I Proteins to block the type I interferon upstream signaling pathway. Vet Res. (2023) 54:5. doi: 10.1186/s13567-023-01134-4
184. Platanias LC. Mechanisms of type-I- and type-ii-interferon-mediated signalling. Nat Rev Immunol. (2005) 5:375–86. doi: 10.1038/nri1604
185. Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. (2004) 75:163–89. doi: 10.1189/jlb.0603252
186. Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. (2007) 96:41–101. doi: 10.1016/s0065-2776(07)96002-2
187. Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. (2008) 8:559–68. doi: 10.1038/nri2314
188. Biron CA. Interferons alpha and beta as immune regulators–a new look. Immunity. (2001) 14:661–4. doi: 10.1016/s1074-7613(01)00154-6
189. Wang LC, Chen SO, Chang SP, Lee YP, Yu CK, Chen CL, et al. Enterovirus 71 proteins 2a and 3d antagonize the antiviral activity of gamma interferon via signaling attenuation. J Virol. (2015) 89:7028–37. doi: 10.1128/jvi.00205-15
190. Ganzleben I, He GW, Günther C, Prigge ES, Richter K, Rieker RJ, et al. Pgam5 is a key driver of mitochondrial dysfunction in experimental lung fibrosis. Cell Mol Life Sci. (2019) 76:4783–94. doi: 10.1007/s00018-019-03133-1
191. Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, et al. The loss of pgam5 suppresses the mitochondrial degeneration caused by inactivation of pink1 in drosophila. PloS Genet. (2010) 6:e1001229. doi: 10.1371/journal.pgen.1001229
192. Yan C, Gong L, Chen L, Xu M, Abou-Hamdan H, Tang M, et al. Phb2 (Prohibitin 2) promotes pink1-prkn/parkin-dependent mitophagy by the parl-pgam5-pink1 axis. Autophagy. (2020) 16:419–34. doi: 10.1080/15548627.2019.1628520
193. Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in drosophila. Proc Natl Acad Sci U.S.A. (2008) 105:14503–8. doi: 10.1073/pnas.0803998105
194. Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. (2007) 27:135–44. doi: 10.1016/j.immuni.2007.05.022
195. Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, et al. Crosstalk between the Cgas DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. (2014) 15:228–38. doi: 10.1016/j.chom.2014.01.009
196. Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. (2008) 9:1004–10. doi: 10.1038/nrm2529
197. Barth S, Glick D, Macleod KF. Autophagy: assays and artifacts. J Pathol. (2010) 221:117–24. doi: 10.1002/path.2694
198. Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. (2012) 30:611–46. doi: 10.1146/annurev-immunol-020711-074948
199. Lee NR, Ban J, Lee NJ, Yi CM, Choi JY, Kim H, et al. Activation of rig-I-mediated antiviral signaling triggers autophagy through the mavs-traf6-beclin-1 signaling axis. Front Immunol. (2018) 9:2096. doi: 10.3389/fimmu.2018.02096
200. Chiok K, Pokharel SM, Mohanty I, Miller LG, Gao SJ, Haas AL, et al. Human respiratory syncytial virus Ns2 protein induces autophagy by modulating beclin1 protein stabilization and isgylation. mBio. (2022) 13:e0352821. doi: 10.1128/mbio.03528-21
201. Xu D, Zhang T, Xiao J, Zhu K, Wei R, Wu Z, et al. Modification of becn1 by isg15 plays a crucial role in autophagy regulation by type I Ifn/interferon. Autophagy. (2015) 11:617–28. doi: 10.1080/15548627.2015.1023982
202. Xiang Q, Wan P, Yang G, Huang S, Qin M, Yang H, et al. Beclin1 binds to enterovirus 71 3d protein to promote the virus replication. Viruses. (2020) 12(7):756. doi: 10.3390/v12070756
203. Jin S, Tian S, Chen Y, Zhang C, Xie W, Xia X, et al. Usp19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. (2016) 35:866–80. doi: 10.15252/embj.201593596
204. Yan J, Gao Y, Bai J, Li J, Li M, Liu X, et al. Serpinb1 promotes senecavirus a replication by degrading ikbke and regulating the Ifn pathway via autophagy. J Virol. (2023) 97:e0104523. doi: 10.1128/jvi.01045-23
205. Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. (2009) 27:229–65. doi: 10.1146/annurev.immunol.021908.132715
206. Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. (2019) 20(23):6008. doi: 10.3390/ijms20236008
207. Ma WT, Yao XT, Peng Q, Chen DK. The protective and pathogenic roles of Il-17 in viral infections: friend or foe? Open Biol. (2019) 9:190109. doi: 10.1098/rsob.190109
208. Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. (2009) 10:241–7. doi: 10.1038/ni.1703
209. Zheng D, Liwinski T, Elinav E. Inflammasome activation and regulation: toward a better understanding of complex mechanisms. Cell Discovery. (2020) 6:36. doi: 10.1038/s41421-020-0167-x
210. Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, et al. Cutting edge: Nf-Kappab activating pattern recognition and cytokine receptors license Nlrp3 inflammasome activation by regulating nlrp3 expression. J Immunol. (2009) 183:787–91. doi: 10.4049/jimmunol.0901363
211. de Zoete MR, Palm NW, Zhu S, Flavell RA. Inflammasomes. Cold Spring Harb Perspect Biol. (2014) 6:a016287. doi: 10.1101/cshperspect.a016287
212. Kelley N, Jeltema D, Duan Y, He Y. The nlrp3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci. (2019) 20(13):3328. doi: 10.3390/ijms20133328
213. Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, et al. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded Rna. J Biol Chem. (2006) 281:36560–8. doi: 10.1074/jbc.M607594200
214. Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, et al. The Nalp3 inflammasome is involved in the innate immune response to amyloid-Beta. Nat Immunol. (2008) 9:857–65. doi: 10.1038/ni.1636
215. Cassel SL, Eisenbarth SC, Iyer SS, Sadler JJ, Colegio OR, Tephly LA, et al. The nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U.S.A. (2008) 105:9035–40. doi: 10.1073/pnas.0803933105
216. Amaral EP, Riteau N, Moayeri M, Maier N, Mayer-Barber KD, Pereira RM, et al. Lysosomal cathepsin release is required for Nlrp3-inflammasome activation by mycobacterium tuberculosis in infected macrophages. Front Immunol. (2018) 9:1427. doi: 10.3389/fimmu.2018.01427
217. Jia C, Zhang J, Chen H, Zhuge Y, Chen H, Qian F, et al. Endothelial cell pyroptosis plays an important role in Kawasaki disease via Hmgb1/rage/cathespin B signaling pathway and Nlrp3 inflammasome activation. Cell Death Dis. (2019) 10:778. doi: 10.1038/s41419-019-2021-3
218. Abais JM, Xia M, Zhang Y, Boini KM, Li PL. Redox regulation of nlrp3 inflammasomes: ros as trigger or effector? Antioxid Redox Signal. (2015) 22:1111–29. doi: 10.1089/ars.2014.5994
219. Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Núñez G. K+ Efflux is the common trigger of nlrp3 inflammasome activation by bacterial toxins and particulate matter. Immunity. (2013) 38:1142–53. doi: 10.1016/j.immuni.2013.05.016
220. Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. (2012) 4(3):a006049. doi: 10.1101/cshperspect.a006049
221. Bagur R, Hajnóczky G. Intracellular Ca(2+) sensing: its role in calcium homeostasis and signaling. Mol Cell. (2017) 66:780–8. doi: 10.1016/j.molcel.2017.05.028
222. Wang W, Xiao F, Wan P, Pan P, Zhang Y, Liu F, et al. Ev71 3d protein binds with Nlrp3 and enhances the assembly of inflammasome complex. PloS Pathog. (2017) 13:e1006123. doi: 10.1371/journal.ppat.1006123
223. Teterina NL, Levenson E, Rinaudo MS, Egger D, Bienz K, Gorbalenya AE, et al. Evidence for functional protein interactions required for poliovirus Rna replication. J Virol. (2006) 80:5327–37. doi: 10.1128/jvi.02684-05
224. Yin J, Liu Y, Wimmer E, Paul AV. Complete protein linkage map between the P2 and P3 non-structural proteins of poliovirus. J Gen Virol. (2007) 88:2259–67. doi: 10.1099/vir.0.82795-0
225. Kuo RL, Chen CJ, Tam EH, Huang CG, Li LH, Li ZH, et al. Interactome analysis of ns1 protein encoded by influenza a H7n9 virus reveals an inhibitory role of Ns1 in host Mrna maturation. J Proteome Res. (2018) 17:1474–84. doi: 10.1021/acs.jproteome.7b00815
226. Chen TC, Chang HY, Lin PF, Chern JH, Hsu JT, Chang CY, et al. Novel antiviral agent Dtrip-22 targets Rna-dependent Rna polymerase of Enterovirus 71. Antimicrob Agents Chemother. (2009) 53:2740–7. doi: 10.1128/aac.00101-09
227. Li L, Wang M, Chen Y, Hu T, Yang Y, Zhang Y, et al. Structure of the enterovirus D68 Rna-dependent Rna polymerase in complex with nadph implicates an inhibitor binding site in the Rna template tunnel. J Struct Biol. (2020) 211:107510. doi: 10.1016/j.jsb.2020.107510
228. Theerawatanasirikul S, Semkum P, Lueangaramkul V, Chankeeree P, Thangthamniyom N, Lekcharoensuk P. Non-nucleoside inhibitors decrease foot-and-mouth disease virus replication by blocking the viral 3d(Pol). Viruses. (2022) 15(1):124. doi: 10.3390/v15010124
Keywords: picornavirus, 3D polymerase, virus replication, nuclear localization signal, interactions, innate immunity
Citation: Xu C, Wang M, Cheng A, Yang Q, Huang J, Ou X, Sun D, He Y, Wu Z, Wu Y, Zhang S, Tian B, Zhao X, Liu M, Zhu D, Jia R and Chen S (2024) Multiple functions of the nonstructural protein 3D in picornavirus infection. Front. Immunol. 15:1365521. doi: 10.3389/fimmu.2024.1365521
Received: 04 January 2024; Accepted: 21 March 2024;
Published: 02 April 2024.
Edited by:
Prof. Pei-Hui Wang, Shandong University, ChinaReviewed by:
Qiong Liu, Nanchang University, ChinaAkihiko Komuro, Niigata University of Pharmacy and Applied Life Sciences, Japan
Junfa Yuan, Huazhong Agricultural University, China
Pu Chen, University of Alberta, Canada
Copyright © 2024 Xu, Wang, Cheng, Yang, Huang, Ou, Sun, He, Wu, Wu, Zhang, Tian, Zhao, Liu, Zhu, Jia and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
 Chenxia Xu
Chenxia Xu Mingshu Wang
Mingshu Wang Anchun Cheng
Anchun Cheng Qiao Yang
Qiao Yang Juan Huang
Juan Huang Xumin Ou
Xumin Ou Di Sun
Di Sun Yu He1,2,3,4,5
Yu He1,2,3,4,5 Shaqiu Zhang
Shaqiu Zhang Bin Tian
Bin Tian Mafeng Liu
Mafeng Liu Dekang Zhu
Dekang Zhu Shun Chen
Shun Chen