- Department of Interventional Vascular Surgery, Affiliated Hospital of Hebei University, Baoding, China
RNA 5-methylcytosine (m5C) methylation plays a crucial role in hepatocellular carcinoma (HCC). As reported, aberrant m5C methylation is closely associated with the progression, therapeutic efficacy, and prognosis of HCC. The innate immune system functions as the primary defense mechanism in the body against pathogenic infections and tumors since it can activate innate immune pathways through pattern recognition receptors to exert anti-infection and anti-tumor effects. Recently, m5C methylation has been demonstrated to affect the activation of innate immune pathways including TLR, cGAS-STING, and RIG-I pathways by modulating RNA function, unveiling new mechanisms underlying the regulation of innate immune responses by tumor cells. However, research on m5C methylation and its interplay with innate immune pathways is still in its infancy. Therefore, this review details the biological significance of RNA m5C methylation in HCC and discusses its potential regulatory relationship with TLR, cGAS-STING, and RIG-I pathways, thereby providing fresh insights into the role of RNA methylation in the innate immune mechanisms and treatment of HCC.
1 Introduction
Hepatocellular carcinoma (HCC) is renowned as the “Monarch of Malignancies” due to its high malignancy degree and dismal prognosis. Additionally, the clinical diagnosis and management of HCC are riddled with formidable challenges, such as difficult early detection, high recurrence and metastasis rates, lack of targeted therapies, and limited innovative drugs and treatment modalities (1, 2). Hence, it is pressing and pivotal to comprehensively investigate the molecular mechanisms of HCC.
As the frontline defense in the body, the innate immune system encompasses a diverse array of myeloid lineage cells, including dendritic cells (DCs), monocytes, and macrophages. Because these cells can function as professional antigen-presenting cells (APCs) or innate lymphocytes such as natural killer (NK) cells, they can swiftly detect microbial proteins or nucleic acid molecules on tumor cells by relying on pattern recognition receptors (PRRs) and other cell surface receptors, therefore orchestrating downstream immune responses (3, 4). For immune surveillance in HCC, various PRRs, such as cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS)-stimulator of interferon genes (STING) (5), retinoic acid-inducible gene I (RIG-I) (6), and Toll-like receptors (TLRs) (7), can favor the transcription of pro-inflammatory genes via pathways involving interferon regulatory factor 3 (IRF-3) and nuclear factor kappa B (NF-κB), inducing the production of interferons (IFNs), cytokines, and chemokines and ultimately potentiating cytotoxic anti-tumor responses mediated by effector T and NK cells. Nevertheless, little is known regarding mechanisms underlying the evasion of tumor cells from innate immune surveillance.
RNA methylation plays a critical role in modulating immune responses, among which 5-methylcytosine (m5C) methylation is an emerging RNA modification mechanism with the function of governing RNA stability, translational regulation, and various cellular functions such as proliferation and differentiation (8, 9). The constant expansion of the m5C methylase family has unveiled the critical involvement of m5C methylation in diverse diseases, prominently in cancers (10). m5C methylases have become a focal point in research on disease markers and potential therapeutic targets since their aberrant expression has been intricately associated with the development, progression, and treatment responses of tumors (11–13).
Several studies uncovered that m5C methylation affected the growth and immune evasion of tumors (14–16). Yet, it is still in the nascent stage to investigate the role of RNA m5C methylation in innate immunity against HCC. Accordingly, this review comprehensively analyzes the involvement of m5C methylation in the onset and progression of HCC and its mechanisms in the innate immune responses of HCC, thus furnishing a novel theoretical framework for the treatment of HCC.
2 Overview of RNA m5C methylation
2.1 Detection methods of m5C methylation sites
As a crucial reversible epigenetic modification, m5C methylation is extensively present in various RNA types, encompassing mRNA, rRNA, tRNA, long non-coding RNA, circular RNA, microRNA, and vtRNA. Methylation at diverse sites assumes a pivotal role in mediating the fate of RNA, including the stability, extranuclear transport, and transcription of RNA (13, 17). Intriguingly, m5C methylation sites on RNA can be detected and localized more precisely with the advent of cutting-edge high-throughput sequencing technologies, such as RNA bisulfite sequencing (RNA-BisSeq) (18, 19), 5-azacytidine-mediated RNA immunoprecipitation sequencing (AZA-IP-seq) (20), methylation individual nucleotide resolution crosslinking (miCLIP-seq) (21, 22), and immunoprecipitation sequencing (m5C-RIP-seq) (18, 23). For instance, m5C-RIP-seq can enrich m5C-containing RNA fragments for sequencing via antibodies specifically targeting m5C methylation sites15, (23) (Figure 1A). Although m5C-RIP-seq can effectively identify highly methylated RNA fragments, it has limited resolution of peaks and much lower rates and fails to obtain the precise location and absolute level of m5C in the transcriptome compared with other sequencing methods with single-nucleotide resolution since the RNA fragments enriched by antibodies are about 100−150 nt in length. In addition, m5C-RIP-seq cannot identify methylation on low-abundance mRNAs because the statistical analysis is relatively insensitive to low-coverage regions (24). miCLIP-seq can capture NOL1/NOP2/SUN domain 2 (NSUN2)-targeted RNA fragments through antibodies against NSUN2. Hussain et al. utilized miCLIP-seq technology to localize the m5C methylation site. Specifically, through ultraviolet light, overexpressed mutant NSUN2 (C271A) was cross-linked with RNA fragments to form RNA-protein complexes containing robust covalent bonds, and truncation or mutations were induced during reverse transcription polymerase chain reaction (RT-PCR) and then identified with sequencing data to map the m5C methylation site (21, 22) (Figure 1B). However, it has been also reported that because NSUN2 catalyzes m5C methylation in only a subset of RNAs, a comprehensive transcriptomic profile cannot be obtained with antibodies against a single RNA m5C methyltransferase. Furthermore, miCLIP-seq requires overexpression of NSUN2 with mutations in the m5C release domain, which creates an abnormal intracellular environment and limits the application of this technology to cells cultured in vitro (25). Furthermore, AZA-IP-seq can detect m5C sites in RNA by replacing cytosine with 5-azacytidine (20) (Figure 1C). Similar to miCLIP-seq, AZA-IP-seq also can only detect m5C sites in a subset of RNAs that are catalyzed by a single RNA m5C methyltransferase, with an incomplete transcriptomic map. Additionally, the detection results of AZA-IP-seq depend on the efficiency of 5-azaC introduction, which results in reduced sensitivity to m5C sites in low-abundance RNA (20). Although the use of AZA-IP-seq is limited to the detection of toxic and specific RNA subpopulations, this technology remains valuable for research on m5C methylation. RNA-BisSeq, additionally, is a widely applied detection method for RNA m5C methylation (18, 19) (Figure 1D), which converts cytosine to uracil through bisulfite treatment, followed by PCR detection of unconverted cytosine to determine the m5C site. Despite the advantages of single-nucleotide resolution and methylation level analysis, which avoids some of the drawbacks of other techniques, RNA-BisSeq fails to distinguish between unconverted cytosines derived from m5C and 5-hydroxymethylcytosine (hm5C) and may encounter recognition errors owing to the double-stranded structure. Conclusively, the evolution of these technologies tremendously advances the understanding of the pivotal role of RNA modifications in gene expression regulation, disease progression, and cellular functions.
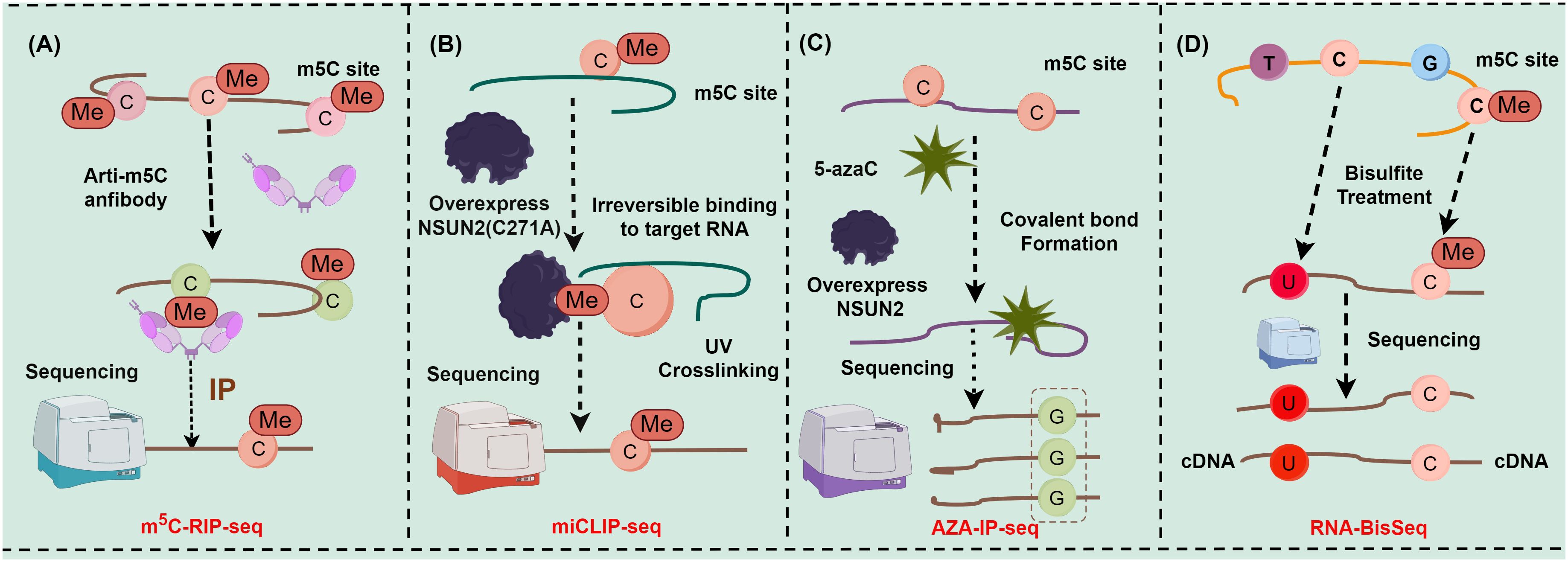
Figure 1 Major detection methods of m5C methylation sites (figures by Figdraw). (A) Detection steps for m5C-RIP-seq; (B) Detection steps for miCLIP-seq; (C) Detection steps for AZA-IP-seq; (D) Detection steps for RNA-BisSeq.
2.2 Molecular mechanisms underlying m5C methylation
First discovered in 1925, m5C methylation is currently known to be present on various RNA molecules in multiple organelles, including nuclei (Figure 2A), mitochondria (Figure 2B), and ribosomes (Figure 2C) (26–28). There are over 90,000 m5C sites detected in the human genome, and m5C methylation is preferentially deposited at the translation start site, near the 3’-untranslated region, and near the Argonaute binding region of mRNAs (29).
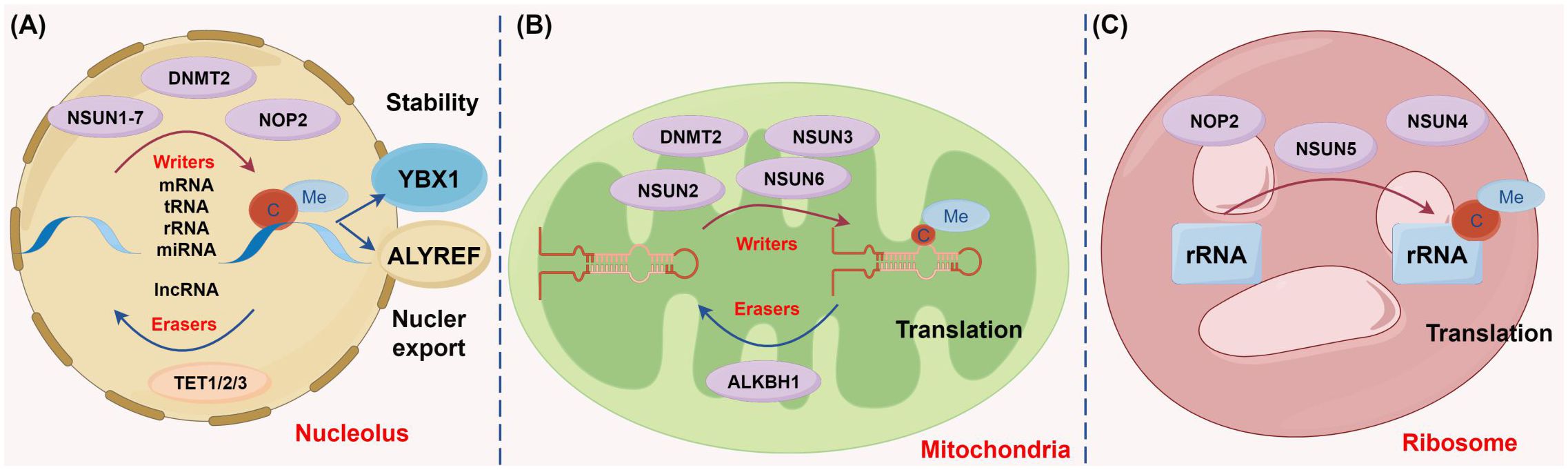
Figure 2 Diverse biological functions of m5C methylation regulators across different cellular compartments (figure by Figdraw). (A) In the nucleus, genes such as NSUN1-7, DNMT2, and NOP2 function as the catalysis enzymes of m5C methylation to modulate methylation modifications on multiple RNA types including mRNA, rRNA, tRNA, miRNA, and lncRNA. Conversely, the demethylases TET1, TET2, and TET3 erase m5C methylation of these RNAs. The m5C methylation recognition protein YBX1 stabilizes RNA, whereas ALYREF facilitates the nuclear export of RNA. (B) In mitochondria, m5C methylases, including NSUN2, NSUN3, NSUN6, and DNMT2, and the demethylase ALKBH1 collaboratively regulate mitochondrial tRNA stability and participate in protein translation. (C) In ribosomes, the methylases NOP2, NSUN4, and NSUN5 promote rRNA stability and are involved in protein translation.
Mechanistically, RNA m5C methylation primarily involves three categories of effectors, namely methyltransferases, demethyltransferases, and m5C readers.
(1) Methyltransferases: m5C methylation is typically catalyzed by methyltransferases, such as NSUN family members (NSUN1-7) and DNA methyltransferase homolog 2/tRNA-aspartic acid methyltransferase 1 (DNMT2/TRDMT1). During m5C methylation, the cysteine residue in the methyltransferase forms a covalent intermediate with the cytosine in the target RNA, which enables the C atom at position C5 to become nucleophilic and to bind to the methyl group of S-adenosyl methionine, contributing to the transfer of the methyl group (27). (2) Demethyltransferases: m5C demethylation is primarily achieved by the ten-eleven translocation (TET) family (TET1-3) and ALKBH1. Specifically, TET enzymes, as α-ketoglutarate- and Fe2+-dependent dioxygenases, are responsible for catalyzing m5C demethylation to generate hm5C. Recent studies have unveiled that TET overexpression substantially enhances RNA hm5C levels, highlighting that members in the TET family may also function as RNA demethylases to remove m5C methylation. Unlike TETs, ALKBH1 initially converts m5C to the intermediate hm5C and further oxidizes hm5C to produce 5-formylcytosine, thus catalyzing m5C demethylation (30). (3) m5C readers: m5C-modified RNA within cells is predominately recognized and regulated by ALYREF and YBX1. ALYREF encourages the nuclear export of mRNAs, whilst YBX1 stabilizes mRNAs by binding to m5C-modified mRNAs via its cold shock domain. Overall, the m5C regulatory network entails sophisticated molecular mechanisms and is essential for the regulation of cell functions and gene expression (28, 31).
3 Role of RNA m5C methylation in HCC
Aberrant expression of m5C correlates to the onset, prognosis, and immune microenvironment of various cancers, such as lung cancer (32), head and neck squamous cell carcinoma (33), HCC (34), and gastrointestinal cancer (35). Prior studies reported the marked up-regulation of m5C methylation levels and related regulatory factors, including NSUN2-7, TRDMT1, TET1-3, and ALYREF in HCC tissues (36–41). Particularly, high NSUN2 expression has been identified as an independent risk factor for HCC as it has been tightly associated with immune cell infiltration, malignant progression, and poor prognosis in HCC (34, 42, 43). Prior research has highlighted that genes upregulated in m5C methylation are primarily involved in phosphokinase pathways such as RAS and PI3K-AKT pathways. Additionally, NSUN2 expression shares a positive correlation with the expression of numerous genes, such as GRB2, RNF115, AATF, ADAM15, RTN3, and HDGF. Real-time PCR in a former study revealed that NSUN2 down-regulation significantly diminished the mRNA expression of these related genes and regulated the RAS pathway, causing cell cycle arrest and then sensitizing HCC cells to sorafenib (36). Several studies showed that NSUN2 overexpression increased HCC cell proliferation by facilitating the interaction between H19 and G3BP1 and enhancing FZR1 mRNA stability (43, 44). Additionally, another study exhibited that NOP2 overexpression suppressed HCC cell proliferation, migration, and invasion by elevating m5C methylation of XPD (45). Importantly, the latest study of Gu et al. demonstrated that NSUN5 accelerated HCC cell proliferation through a ZBED3-dependent mechanism, providing a new therapeutic target for HCC (46). Accumulating studies also reported the role of m5C readers in HCC. For instance, Wang et al. observed that ALYREF knockdown altered the biological phenotypes of HCC cells, influenced the level of immune cell infiltration, and was correlated with the overall survival of patients (47). Xue et al. found an association of ALYREF overexpression and eukaryotic translation initiation factor 4A3 (eIF4A3) upregulation with poor prognosis in HCC (48). In addition, Ru et al. noted that YBX1 up-regulation contributed to immune evasion by up-regulating PD-L1, whilst YBX1 knockout reversed therapy resistance, implicating that targeting m5C regulators may enhance the efficacy of immunotherapy (49). Collectively, aberrant expression of m5C methylation and alterations in the related regulators impact the onset and progression of HCC at various levels, offering crucial clues for the understanding of molecular mechanisms underlying HCC and providing guidance for future research and treatment of HCC.
4 Potential regulatory role of RNA m5C methylation in innate immune pathways of HCC
The innate immune system is one of the initial lines of defense in the body and comprises various cells, proteins, and molecules that collaborate to combat various external threats. The innate immune system not only combats pathogens but also exerts an anti-tumor role (3, 50). Recent research has underscored the significance of m5C methylation, a pivotal post-transcriptional modification, in the mediation of RNA structure and function (8, 51). Specifically, m5C methylation modulates the stability and translation of specific RNAs, thereby activating innate immune pathways (14–16). Within the innate immune system, specialized receptors, such as TLRs (16) and cytosolic nucleic acid sensors like cGAS-STING (14) and RIG-I-like receptors (RLRs) (15), can recognize abnormal tumor nucleic acids. When activated, these receptors initiate immune responses, including inflammation and immune cell activation, to bolster the defense against tumors. A deeper comprehension of the interplay between m5C methylation and TLRs, cGAS-STING, and RLRs offers novel insights into mechanisms underlying the anti-tumor actions of the immune system.
4.1 The TLR pathway
TLRs, as key PRRs within the innate immune system, are responsible for recognizing invading pathogens, double-stranded RNA, single-stranded RNA, and CpG-DNA. Upon recognition, TLRs stimulate immune responses by activating intracellular pathways and transcription factors such as IRF7 and NF-κB via adapter proteins including MyD88 (7). There are 10 types of TLRs in the human liver, which are distributed on the cell surface and within intracellular compartments (52). Given the constant exposure of the liver to gut microbes, TLRs assume crucial roles in repressing inappropriate pro-inflammatory responses. On the contrary, prolonged TLR activation may elicit chronic liver damage and elevate the risk of HCC (53). Notably, diverse biological functions vary across TLRs. For instance, TLR4 activation can obviously promote the development of HCC by up-regulating pro-inflammatory factors and malignant tumor-related molecules and mediating immune cell infiltration (54–59). In contrast, TLR3 activation can protect against HCC cell growth by inducing reactive oxygen species generation and activating apoptotic pathways (60–62). Additionally, TLR2 (63), TLR5 (64), TLR7 (65, 66), TLR8 (66), and TLR9 (67) are also implicated in the development of HCC, whose activities may affect various processes such as cell proliferation, migration, and apoptosis.
Through comparisons between m5C-modified RNA and unmodified RNA, Kariko et al. observed that m5C-modified RNA inhibited the expression of cytokines and activation markers in DCs and also suppressed the signaling activity activated by TLR3, TLR7, and TLR8, thereby affecting the immune activity of DC cells (16) (Figure 3). Furthermore, because base modification in mRNAs changes their immune activity, researchers used m5C in mRNA therapy alone or in combination with other modifications, which successfully inhibited the immune effects of TLR-mediated mRNAs (16). This breakthrough signifies a major advancement in the realms of RNA and immunotherapy. The research by Andries et al. further highlighted m5C-modified mRNAs as a novel mRNA therapy, partly because of their ability to boost evasion from the innate immune response by inhibiting endosomal TLR3 (68). In summary, m5C methylation may exert complex and critical effects on TLRs and the pathogenesis of HCC, offering essential insights into the immunological mechanisms of HCC and the development of treatment strategies for HCC.
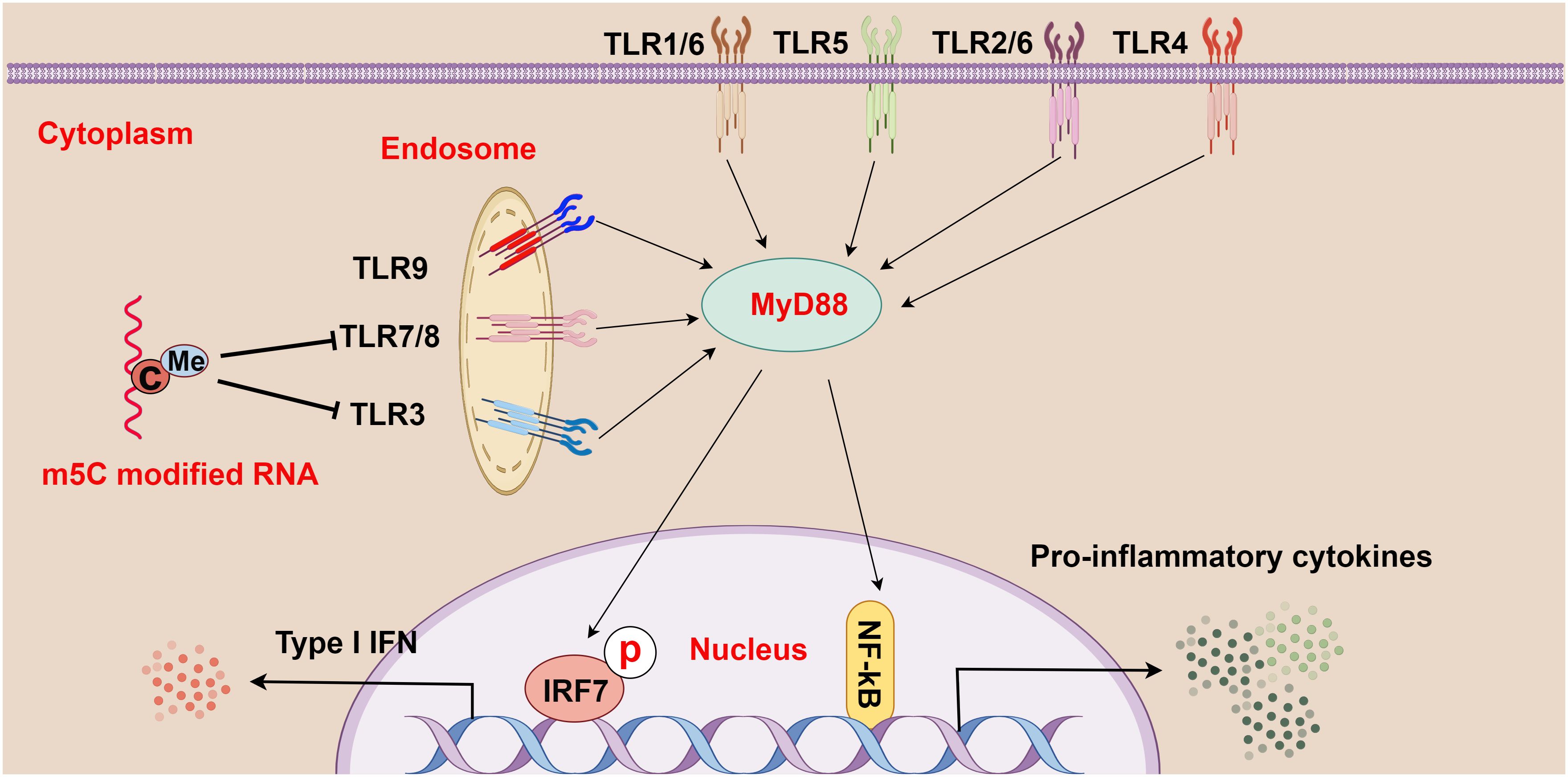
Figure 3 The relationship between m5C methylation and the Toll-like receptor (TLR) pathway (figure by Figdraw). TLR1-9 are ubiquitously distributed in cells, both on the cell surface and within internal organelles. Upon recognition of various pathogens and foreign molecules such as double-stranded RNA, single-stranded RNA, and CpG-DNA, these genes activate intracellular pathways and transcription factors (such as IRF7 and NF-κB), and these pathways signal through adapter proteins, such as MyD88. Moreover, m5C-modified RNA plays a crucial role in regulating this process by abrogating the signaling activity of TLR3, TLR7, and TLR8, thereby modulating the strength and scope of immune responses.
4.2 The cGAS-STING pathway
As a cytoplasmic PRR and a DNA sensor, cGAS activates the IFN pathway by recognizing cytoplasmic DNA, including DNA released from viruses, bacteria, mitochondria, tumor cells, or dead cells. The C-terminus of cGAS contains a conserved zinc ion-binding region, which favors DNA binding and dimerization of cGAS. DNA binding changes the structure of cGAS, leading to cGAS activation. STING, a small protein in the endoplasmic reticulum, is retained in the endoplasmic reticulum through interactions with the Ca2+ sensor STIM1. When binding to DNA, cGAS is activated by ATP and GTP, catalyzing the formation of cGAMP, which further activates STING. Activated STING is palmitoylated and phosphorylated to recruit TBK1 and IKK, among which TBK1 triggers the release of IFN (a pivotal pathway in cancers, particularly HCC) by activating IRF3, while IKK induces inflammation by activating NF-κB (69, 70).
The involvement of the cGAS-STING pathway in HCC has been increasingly reported. For instance, a prior study revealed that olaparib activated STING chemokine signaling to enhance radiation-induced systemic anti-tumor effects (71). Another study showed that the cGAS-STING pathway was involved in the promoting effects of oncolytic influenza viruses combined with PD-L1 antibodies on CD8+ T cell activation (72). Su et al. found that TAK1 deficiency modulated ferroptosis and macrophage cGAS-STING signaling to foster liver injury and tumorigenesis (73). A study by Li et al. demonstrated that hyperbaric oxygen boosted teniposide-induced cGAS-STING activation, therefore increasing the anti-tumor efficacy of PD-1 antibodies in HCC (74). Gut microbiota was reported to modulate radiotherapy-induced anti-tumor immune responses in HCC via STING signaling (75). Sorafenib combined with STAT3 knockdown was revealed to accelerate HCC cell apoptosis and encourage cGAS-STING-mediated anti-tumor immunity (76). The disruption of the BRCA1-PALB2 interaction was demonstrated to induce tumor immunosuppression and T lymphocyte infiltration in HCC through the cGAS-STING pathway (77). According to the results of Du et al., radiotherapy induced PD-L1 up-regulation via cGAS-STING activation to facilitate immune evasion in HCC (78). Zhao et al. observed that hypoxia-induced RNASEH2A constrained cGAS-STING signaling activation in HCC and might portend the poor prognosis of patients (79). Of note, another study displayed that STING deficiency accelerated tumor growth and reduced autophagy and apoptosis in mice with HCC, which could be nullified by STING agonists.
Furthermore, the research of Chen et al. demonstrated that NSUN5-mediated m5C methylation of GPX4 contributed to cGAS-STING signaling activation, which fostered anti-cancer immune responses in colon adenocarcinoma (80). Another study revealed that NSUN2 activation maintained global m5C methylation of RNA, including TREX2, in tumor cells and stabilized TREX2 expression, consequently impeding the accumulation of cytoplasmic double-stranded DNA and blocking the activation of the cGAS-STING pathway, which facilitated tumorigenesis and PD-L1 immunotherapy resistance (14). Additionally, NSUN2-mediated m5C methylation of IRF3 mRNA may negatively affect type I IFN responses in various viral infections (81). These findings underscore the important effect of m5C methylation on the cGAS-STING pathway in HCC and highlight its potential as a hot spot for future research on HCC (Figure 4).
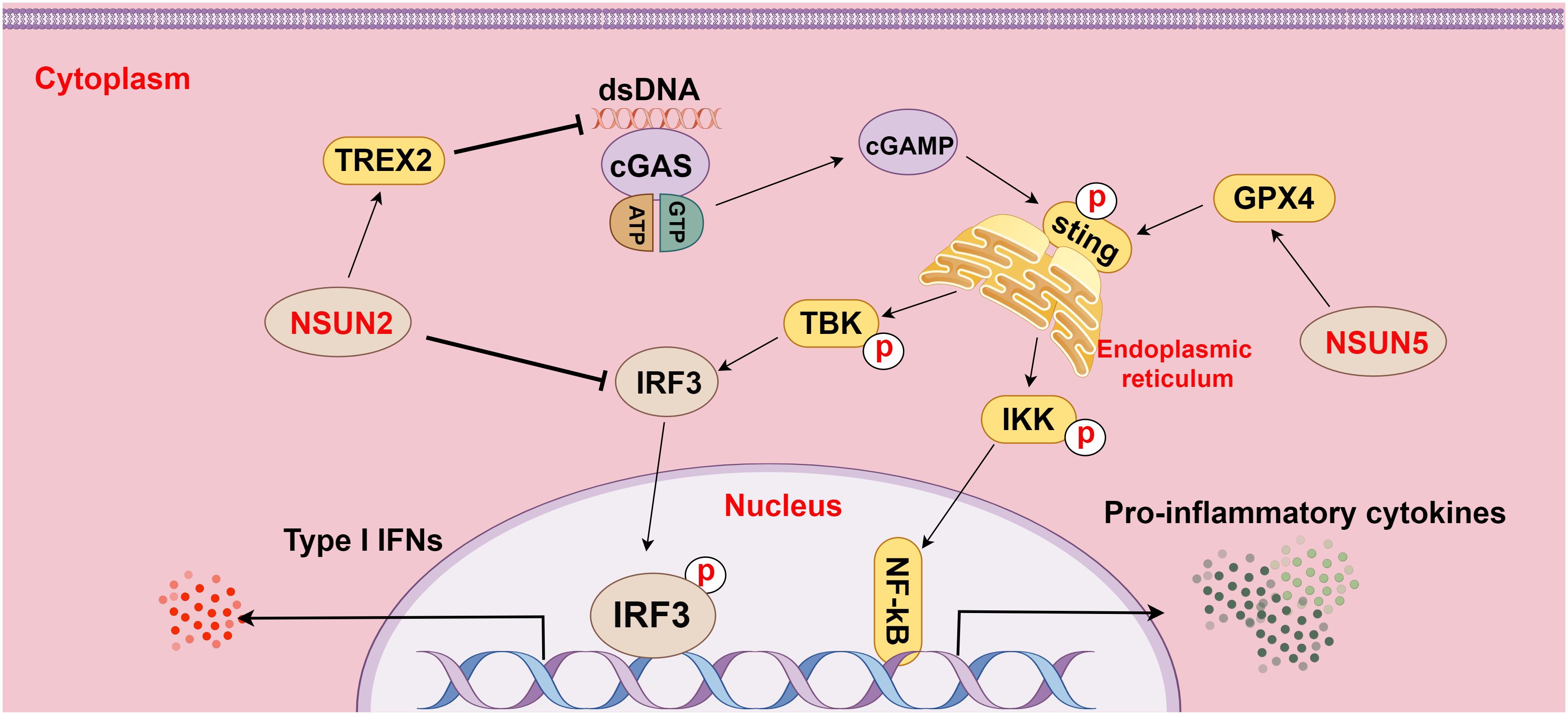
Figure 4 The relationship between m5C methylation and the cGAS-STING pathway (figure by Figdraw). cGAS is a cytoplasmic pattern recognition receptor and a DNA sensor, which activates the IFN pathway by recognizing double-stranded DNA (dsDNA) in the cytoplasm. NSUN2 inhibits dsDNA aggregation by methylating TREX2 mRNA and increasing TREX2 expression, thereby restraining the activation of the cGAS-STING pathway. Additionally, NSUN2 methylates IRF3 mRNA to facilitate IRF3 mRNA degradation and reduce IRF3 production, subsequently modulating the cGAS-STING pathway. Conversely, NSUN5 activates the cGAS-STING pathway by methylating GPX4 mRNA, thereby elevating the activity of the pathway.
4.3 The RIG-I pathway
RLRs, a class of intracellular PRRs, are primarily tasked with the surveillance of RNA viruses, DNA viruses, and pathogenic RNA stemming from certain bacterial infections. Upon detection, RLRs lead to the activation of IRF3, a transcription factor, transcriptionally up-regulating cytokines such as IFNs and then bolstering immune responses. Mounting evidence supports the indispensable role of intact IFN signaling in the efficacy of numerous traditional chemotherapeutic drugs and targeted anti-cancer drugs. Accordingly, activation of RIG-I or MDA5 signaling has emerged as a promising therapeutic strategy for cancers (82, 83).
A prior study exhibited that hepatocyte-specific RIG-I deficiency facilitated diethylnitrosamine-induced hepatocarcinogenesis but mitigated non-alcoholic fatty liver disease-induced hepatocarcinogenesis. This study also demonstrated that interleukin-6 (IL-6) diminished RIG-I expression in HCC progenitor cells (HcPCs), driving IL-6 effector signaling and the development of HCC from HcPCs (84). NFATc3 was found to impede HCC and hepatitis B virus replication by positively regulating RIG-I-mediated IFN transcription, illustrating that NFATc3 may exert an effect on the RIG-I pathway, which is vital for maintaining liver health and repressing viral replication (85). In stem cells, RIG-I deficiency led to TGF-β/AKT/Smad2 signaling activation and precipitated the formation of tolerogenic DCs, underscoring the significance of RIG-I in immune regulation (86). Additionally, RIG-I has been identified not only as an anti-oncogene but also as a biomarker for the efficacy of IFN-α treatment for HCC, emphasizing its critical role in modulating the growth and treatment responses of tumors (6, 87).
Moreover, a prior study displayed that deletion of the m5C methyltransferase NSUN2 boosted type I IFN responses and substantially depressed the replication of various RNA and DNA viruses and that NSUN2 loss diminished the RNA m5C methyl group in the host and elevated the polymerase III transcription of non-coding RNAs recognized by RIG-I, particularly RPPH1 and 7SL RNA, consequently enhancing type I IFN signaling. In addition, this study also showed that this m5C-mediated antiviral innate immunity was also conserved in mouse models (15). Overall, this study elucidates the role of m5C in regulating innate immunity and illustrates m5C as a promising target for the development of broad-spectrum antiviral and anti-tumor therapeutics (Figure 5).
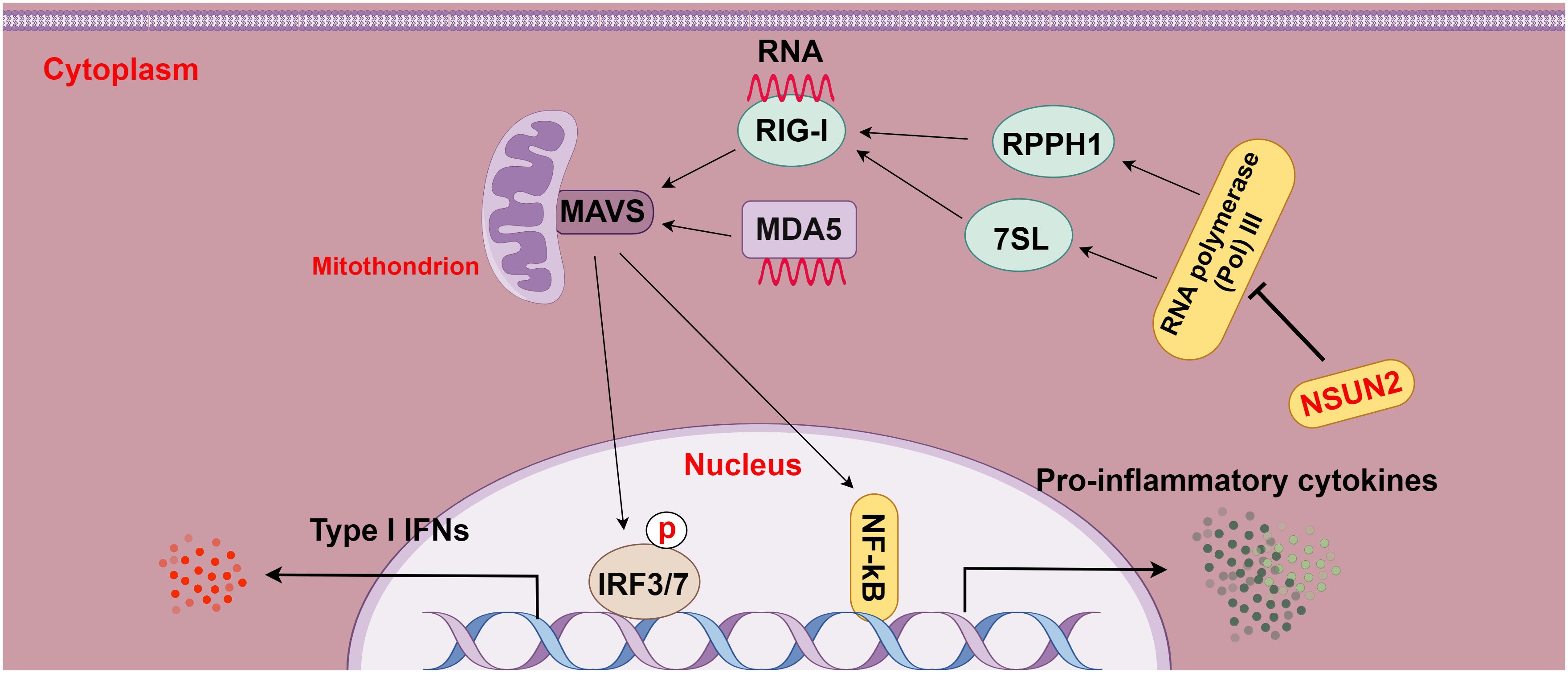
Figure 5 The relationship between m5C methylation and the RIG-I pathway (figure by Figdraw). RLRs, including RIG-I and MDA5, act as intracellular pattern recognition receptors (PRRs) that surveil RNA viruses, DNA viruses, and pathogenic RNA derived from tumors. Upon detection, RLRs activate the transcription factors IRF3/7, leading to the transcriptional up-regulation of interferons and other cytokines and then enhancing immune responses. NSUN2 mediates the RIG-I pathway by decreasing the transcription of non-coding RNAs regulated by host RNA polymerase III, particularly RPPH1 and 7SL RNA, ultimately suppressing RIG-I-mediated interferon responses, with no effect on MDA5 signaling.
5 Challenges and prospects
Although great strides have been made in understanding the regulatory effect of m5C methylation on innate immune pathways, the role and mechanism of m5C methylation in different tumors remain poorly identified because tumor cells are highly heterogeneous. Various immune cells, including macrophages, DCs, NK cells, and neutrophils, can recognize distinct tumor-related antigen modifications which may involve alterations in cell metabolism and tissue anatomy. These alterations potentially serve as markers for tumor identification (3, 50, 88). Chemotherapy and radiotherapy have been observed to induce immunogenic cell death in tumors, resulting in the release of damage-associated molecular patterns such as cell surface calreticulin and heat shock proteins (3, 89), which can favor antigen presentation and secretion of immune stimulatory factors to activate innate immune cells. Additionally, cytokines including CXCL9, CXCL10, type I IFN, IL-2, and IL-15 can enhance the activity of immune cells, such as CD8 T and NK cells (90). Therefore, further studies are warranted to delve into the mechanisms by which m5C methylation influences the recognition of tumors by innate immunity, which is essential for advancing the understanding of tumor-immune interactions and the development of novel therapeutic strategies.
Current research on the role of m5C methylation in the innate immune pathways of HCC largely relies on RNA methylation sequencing technologies and bioinformatics analysis (28). Therefore, it is imperative to develop more precise high-throughput detection technologies, such as Nanopore Sequencing (91), Chemical Labeling Coupled with Mass Spectrometry (92), Single-Molecule Real-Time Sequencing (93), and Selective Chemical Labeling Followed by Sequencing (94), for accurately quantifying and localizing m5C methylation. With the ongoing advancements in nanomedicine technology (95, 96), we can anticipate the application prospects of nanocarriers in the regulation of immune pathways by m5C methylation. These nanocarriers may serve as vehicles for delivering RNA methylation-related drugs or RNA (96), enabling precise regulation of the immune microenvironment in HCC and thereby enhancing the efficacy of tumor immunotherapy. Conclusively, more studies are required to further explore the role of m5C methylation in the innate immune pathways of HCC, thus providing a novel theoretical foundation and clinical direction for the advancement and application of immunotherapy for HCC.
Author contributions
SM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. BJ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. WH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. LM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. ZL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing. LS: Writing – original draft, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. (2018) 391:1301–14. doi: 10.1016/s0140-6736(18)30010-2
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
3. Li X, Dai H, Wang H, Han W. Exploring innate immunity in cancer immunotherapy: opportunities and challenges. Cell Mol Immunol. (2021) 18:1607–9. doi: 10.1038/s41423-021-00679-8
4. Paludan SR, Pradeu T, Masters SL, Mogensen TH. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat Rev Immunol. (2021) 21:137–50. doi: 10.1038/s41577-020-0391-5
5. Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol. (2015) 15:760–70. doi: 10.1038/nri3921
6. Hou J, Zhou Y, Zheng Y, Fan J, Zhou W, Ng IO, et al. Hepatic RIG-I predicts survival and interferon-α therapeutic response in hepatocellular carcinoma. Cancer Cell. (2014) 25:49–63. doi: 10.1016/j.ccr.2013.11.011
7. Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. (2009) 9:57–63. doi: 10.1038/nrc2541
8. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. (2017) 18:31–42. doi: 10.1038/nrm.2016.132
9. Xue C, Chu Q, Zheng Q, Jiang S, Bao Z, Su Y, et al. Role of main RNA modifications in cancer: N(6)-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct Target Ther. (2022) 7:142. doi: 10.1038/s41392-022-01003-0
10. Zhang Q, Liu F, Chen W, Miao H, Liang H, Liao Z, et al. The role of RNA m(5)C modification in cancer metastasis. Int J Biol Sci. (2021) 17:3369–80. doi: 10.7150/ijbs.61439
11. Chellamuthu A, Gray SG. The RNA methyltransferase NSUN2 and its potential roles in cancer. Cells. (2020) 9(8):1758. doi: 10.3390/cells9081758
12. Li M, Tao Z, Zhao Y, Li L, Zheng J, Li Z, et al. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J Transl Med. (2022) 20:214. doi: 10.1186/s12967-022-03427-2
13. He Y, Shi Q, Zhang Y, Yuan X, Yu Z. Transcriptome-wide 5-methylcytosine functional profiling of long non-coding RNA in hepatocellular carcinoma. Cancer Manag Res. (2020) 12:6877–85. doi: 10.2147/cmar.S262450
14. Chen T, Xu ZG, Luo J, Manne RK, Wang Z, Hsu CC, et al. NSUN2 is a glucose sensor suppressing cGAS/STING to maintain tumorigenesis and immunotherapy resistance. Cell Metab. (2023) 35:1782–1798.e8. doi: 10.1016/j.cmet.2023.07.009
15. Zhang Y, Zhang LS, Dai Q, Chen P, Lu M, Kairis EL, et al. 5-methylcytosine (m(5)C) RNA modification controls the innate immune response to virus infection by regulating type I interferons. Proc Natl Acad Sci U.S.A. (2022) 119:e2123338119. doi: 10.1073/pnas.2123338119
16. Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
17. He Y, Zhang Q, Zheng Q, Yu X, Guo W. Distinct 5-methylcytosine profiles of circular RNA in human hepatocellular carcinoma. Am J Transl Res. (2020) 12:5719–29.
18. Burrows CJ, Fleming AM. Bisulfite and nanopore sequencing for pseudouridine in RNA. Acc Chem Res. (2023) 56:2740–51. doi: 10.1021/acs.accounts.3c00458
19. Trixl L, Rieder D, Amort T, Lusser A. Bisulfite sequencing of RNA for transcriptome-wide detection of 5-methylcytosine. Methods Mol Biol. (2019) 1870:1–21. doi: 10.1007/978-1-4939-8808-2_1
20. Khoddami V, Cairns BR. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. (2013) 31:458–64. doi: 10.1038/nbt.2566
21. George H, Ule J, Hussain S. Illustrating the epitranscriptome at nucleotide resolution using methylation-iCLIP (miCLIP). Methods Mol Biol. (2017) 1562:91–106. doi: 10.1007/978-1-4939-6807-7_7
22. Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. (2021) 49:1006–22. doi: 10.1093/nar/gkaa1193
23. Gu X, Liang Z. Transcriptome-wide mapping 5-methylcytosine by m(5)C RNA immunoprecipitation followed by deep sequencing in plant. Methods Mol Biol. (2019) 1933:389–94. doi: 10.1007/978-1-4939-9045-0_24
24. Cui X, Liang Z, Shen L, Zhang Q, Bao S, Geng Y, et al. 5-methylcytosine RNA methylation in arabidopsis thaliana. Mol Plant. (2017) 10:1387–99. doi: 10.1016/j.molp.2017.09.013
25. Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip Rev RNA. (2019) 10:e1510. doi: 10.1002/wrna.1510
26. Qiu L, Jing Q, Li Y, Han J. RNA modification: mechanisms and therapeutic targets. Mol BioMed. (2023) 4:25. doi: 10.1186/s43556-023-00139-x
27. Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (Basel). (2019) 10(2):102. doi: 10.3390/genes10020102
28. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H, et al. Biological roles of RNA m5C modification and its implications in Cancer immunotherapy. biomark Res. (2022) 10:15. doi: 10.1186/s40364-022-00362-8
29. Huang Z, Pan J, Wang H, Du X, Xu Y, Wang Z, et al. Prognostic significance and tumor immune microenvironment heterogenicity of m5C RNA methylation regulators in triple-negative breast cancer. Front Cell Dev Biol. (2021) 9:657547. doi: 10.3389/fcell.2021.657547
30. Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. (2012) 139:1895–902. doi: 10.1242/dev.070771
31. Xu R, Wang Y, Kuang Y. Multi-omic analyses of m5C readers reveal their characteristics and immunotherapeutic proficiency. Sci Rep. (2024) 14:1651. doi: 10.1038/s41598-024-52110-7
32. Wang Y, Wei J, Feng L, Li O, Huang L, Zhou S, et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol Cancer. (2023) 22:81. doi: 10.1186/s12943-023-01780-4
33. Jin S, Li J, Shen Y, Wu Y, Zhang Z, Ma H. RNA 5-Methylcytosine Regulator NSUN3 promotes tumor progression through regulating immune infiltration in head and neck squamous cell carcinoma. Oral Dis. (2024) 30(2)313–28. doi: 10.1111/odi.14357
34. Gu X, Zhou H, Chu Q, Zheng Q, Wang J, Zhu H. Uncovering the association between m(5)C regulator-mediated methylation modification patterns and tumour microenvironment infiltration characteristics in hepatocellular carcinoma. Front Cell Dev Biol. (2021) 9:727935. doi: 10.3389/fcell.2021.727935
35. Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B, et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. (2021) 12:842. doi: 10.1038/s41419-021-04127-3
36. Song D, An K, Zhai W, Feng L, Xu Y, Sun R, et al. NSUN2-mediated mRNA m(5)C Modification Regulates the Progression of Hepatocellular Carcinoma. Genomics Proteomics Bioinf. (2023) 21:823–33. doi: 10.1016/j.gpb.2022.09.007
37. Zhang H, Zhai X, Liu Y, Xia Z, Xia T, Du G, et al. NOP2-mediated m5C Modification of c-Myc in an EIF3A-Dependent Manner to Reprogram Glucose Metabolism and Promote Hepatocellular Carcinoma Progression. Res (Wash D C). (2023) 6:184. doi: 10.34133/research.0184
38. He Y, Yu X, Li J, Zhang Q, Zheng Q, Guo W. Role of m(5)C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am J Transl Res. (2020) 12:912–22.
39. Zhang Q, Zheng Q, Yu X, He Y, Guo W. Overview of distinct 5-methylcytosine profiles of messenger RNA in human hepatocellular carcinoma and paired adjacent non-tumor tissues. J Transl Med. (2020) 18:245. doi: 10.1186/s12967-020-02417-6
40. Cui M, Qu F, Wang L, Liu X, Yu J, Tang Z, et al. m5C RNA methyltransferase-related gene NSUN4 stimulates Malignant progression of hepatocellular carcinoma and can be a prognostic marker. Cancer biomark. (2022) 33:389–400. doi: 10.3233/cbm-210154
41. Zhang XW, Wu LY, Liu HR, Huang Y, Qi Q, Zhong R, et al. NSUN5 promotes progression and predicts poor prognosis in hepatocellular carcinoma. Oncol Lett. (2022) 24:439. doi: 10.3892/ol.2022.13559
42. Yang X, Yang F, Lan L, Wen N, Li H, Sun X. Diagnostic and prognostic value of m5C regulatory genes in hepatocellular carcinoma. Front Genet. (2022) 13:972043. doi: 10.3389/fgene.2022.972043
43. Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S, et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. (2020) 39:6906–19. doi: 10.1038/s41388-020-01475-w
44. Zhai CT, Tian YC, Tang ZX, Shao LJ. RNA methyltransferase NSUN2 promotes growth of hepatocellular carcinoma cells by regulating fizzy-related-1 in vitro and in vivo. Kaohsiung J Med Sci. (2021) 37:991–9. doi: 10.1002/kjm2.12430
45. Sun GF, Ding H. NOP2-mediated m5C methylation of XPD is associated with hepatocellular carcinoma progression. Neoplasma. (2023) 70:340–9. doi: 10.4149/neo_2023_230110N17
46. Gu X, Li P, Gao X, Ru Y, Xue C, Zhang S, et al. RNA 5-methylcytosine writer NSUN5 promotes hepatocellular carcinoma cell proliferation via a ZBED3-dependent mechanism. Oncogene. (2024) 43:624–35. doi: 10.1038/s41388-023-02931-z
47. Wang ZZ, Meng T, Yang MY, Wang W, Zhang Y, Liu Y, et al. ALYREF associated with immune infiltration is a prognostic biomarker in hepatocellular carcinoma. Transl Oncol. (2022) 21:101441. doi: 10.1016/j.tranon.2022.101441
48. Xue C, Zhao Y, Li G, Li L. Multi-omic analyses of the m(5)C regulator ALYREF reveal its essential roles in hepatocellular carcinoma. Front Oncol. (2021) 11:633415. doi: 10.3389/fonc.2021.633415
49. Ru J, Lu J, Ge J, Ding B, Su R, Jiang Y, et al. IRGM is a novel regulator of PD-L1 via promoting S6K1-mediated phosphorylation of YBX1 in hepatocellular carcinoma. Cancer Lett. (2024) 581:216495. doi: 10.1016/j.canlet.2023.216495
50. Demaria O, Cornen S, Daëron M, Morel Y, Medzhitov R, Vivier E. Harnessing innate immunity in cancer therapy. Nature. (2019) 574:45–56. doi: 10.1038/s41586-019-1593-5
51. Song P, Tayier S, Cai Z, Jia G. RNA methylation in mammalian development and cancer. Cell Biol Toxicol. (2021) 37:811–31. doi: 10.1007/s10565-021-09627-8
52. Seki E, Park E, Fujimoto J. Toll-like receptor signaling in liver regeneration, fibrosis and carcinogenesis. Hepatol Res. (2011) 41:597–610. doi: 10.1111/j.1872-034X.2011.00822.x
53. Kiziltas S. Toll-like receptors in pathophysiology of liver diseases. World J Hepatol. (2016) 8:1354–69. doi: 10.4254/wjh.v8.i32.1354
54. Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. (2012) 21:504–16. doi: 10.1016/j.ccr.2012.02.007
55. Todoric J, Di Caro G, Reibe S, Henstridge DC, Green CR, Vrbanac A, et al. Fructose stimulated de novo lipogenesis is promoted by inflammation. Nat Metab. (2020) 2:1034–45. doi: 10.1038/s42255-020-0261-2
56. Yang LY, Luo Q, Lu L, Zhu WW, Sun HT, Wei R, et al. Increased neutrophil extracellular traps promote metastasis potential of hepatocellular carcinoma via provoking tumorous inflammatory response. J Hematol Oncol. (2020) 13:3. doi: 10.1186/s13045-019-0836-0
57. Lin Y, Yu LX, Yan HX, Yang W, Tang L, Zhang HL, et al. Gut-derived lipopolysaccharide promotes T-cell-mediated hepatitis in mice through Toll-like receptor 4. Cancer Prev Res (Phila). (2012) 5:1090–102. doi: 10.1158/1940-6207.Capr-11-0364
58. Pan C, Wu Q, Wang S, Mei Z, Zhang L, Gao X, et al. Combination with Toll-like receptor 4 (TLR4) agonist reverses GITR agonism mediated M2 polarization of macrophage in Hepatocellular carcinoma. Oncoimmunology. (2022) 11:2073010. doi: 10.1080/2162402x.2022.2073010
59. Yao RR, Li JH, Zhang R, Chen RX, Wang YH. M2-polarized tumor-associated macrophages facilitated migration and epithelial-mesenchymal transition of HCC cells via the TLR4/STAT3 signaling pathway. World J Surg Oncol. (2018) 16:9. doi: 10.1186/s12957-018-1312-y
60. Chen L, Xu YY, Zhou JM, Wu YY, Q. E and YY. Zhu: TLR3 dsRNA agonist inhibits growth and invasion of HepG2.2.15 HCC cells. Oncol Rep. (2012) 28:200–6. doi: 10.3892/or.2012.1791
61. Shen P, Jiang T, Lu H, Han H, Luo R. Combination of Poly I:C and arsenic trioxide triggers apoptosis synergistically via activation of TLR3 and mitochondrial pathways in hepatocellular carcinoma cells. Cell Biol Int. (2011) 35:803–10. doi: 10.1042/cbi20100739
62. Yuan MM, Xu YY, Chen L, Li XY, Qin J, Shen Y. TLR3 expression correlates with apoptosis, proliferation and angiogenesis in hepatocellular carcinoma and predicts prognosis. BMC Cancer. (2015) 15:245. doi: 10.1186/s12885-015-1262-5
63. Li S, Li F, Xu L, Liu X, Zhu X, Gao W, et al. TLR2 agonist promotes myeloid-derived suppressor cell polarization via Runx1 in hepatocellular carcinoma. Int Immunopharmacol. (2022) 111:109168. doi: 10.1016/j.intimp.2022.109168
64. Kairaluoma V, Kemi N, Huhta H, Pohjanen VM, Helminen O. Toll-like receptor 5 and 8 in hepatocellular carcinoma. Apmis. (2021) 129:470–9. doi: 10.1111/apm.13142
65. Ren X, Wang F, Ji B, Gao C. TLR7 agonist induced repression of hepatocellular carcinoma via the TLR7-IKK-NF-κB-IL6 signaling pathway. Oncol Lett. (2016) 11:2965–70. doi: 10.3892/ol.2016.4329
66. Zhou Z, Yu X, Zhang J, Tian Z, Zhang C. TLR7/8 agonists promote NK-DC cross-talk to enhance NK cell anti-tumor effects in hepatocellular carcinoma. Cancer Lett. (2015) 369:298–306. doi: 10.1016/j.canlet.2015.09.017
67. Zhou B, Yan J, Guo L, Zhang B, Liu S, Yu M, et al. Hepatoma cell-intrinsic TLR9 activation induces immune escape through PD-L1 upregulation in hepatocellular carcinoma. Theranostics. (2020) 10:6530–43. doi: 10.7150/thno.44417
68. Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release. (2015) 217:337–44. doi: 10.1016/j.jconrel.2015.08.051
69. Samson N, Ablasser A. The cGAS–STING pathway and cancer. Nat Cancer. (2022) 3:1452–63. doi: 10.1038/s43018-022-00468-w
70. Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. (2021) 21:548–69. doi: 10.1038/s41577-021-00524-z
71. Chen G, Zheng D, Zhou Y, Du S, Zeng Z. Olaparib enhances radiation-induced systemic anti-tumor effects via activating STING-chemokine signaling in hepatocellular carcinoma. Cancer Lett. (2024) 582:216507. doi: 10.1016/j.canlet.2023.216507
72. Sun F, Xu Y, Deng Z, Yang P. A recombinant oncolytic influenza virus expressing a PD-L1 antibody induces CD8(+) T-cell activation via the cGas-STING pathway in mice with hepatocellular carcinoma. Int Immunopharmacol. (2023) 120:110323. doi: 10.1016/j.intimp.2023.110323
73. Su W, Gao W, Zhang R, Wang Q, Li L, Bu Q, et al. TAK1 deficiency promotes liver injury and tumorigenesis via ferroptosis and macrophage cGAS-STING signalling. JHEP Rep. (2023) 5:100695. doi: 10.1016/j.jhepr.2023.100695
74. Li K, Gong Y, Qiu D, Tang H, Zhang J, Yuan Z, et al. Hyperbaric oxygen facilitates teniposide-induced cGAS-STING activation to enhance the antitumor efficacy of PD-1 antibody in HCC. J Immunother Cancer. (2022) 10(8):e004006. doi: 10.1136/jitc-2021-004006
75. Li Z, Zhang Y, Hong W, Wang B, Chen Y, Yang P, et al. Gut microbiota modulate radiotherapy-associated antitumor immune responses against hepatocellular carcinoma Via STING signaling. Gut Microbes. (2022) 14:2119055. doi: 10.1080/19490976.2022.2119055
76. Wang X, Hu R, Song Z, Zhao H, Pan Z, Feng Y, et al. Sorafenib combined with STAT3 knockdown triggers ER stress-induced HCC apoptosis and cGAS-STING-mediated anti-tumor immunity. Cancer Lett. (2022) 547:215880. doi: 10.1016/j.canlet.2022.215880
77. Ma H, Kang Z, Foo TK, Shen Z, Xia B. Disrupted BRCA1-PALB2 interaction induces tumor immunosuppression and T-lymphocyte infiltration in HCC through cGAS-STING pathway. Hepatology. (2023) 77:33–47. doi: 10.1002/hep.32335
78. Du SS, Chen GW, Yang P, Chen YX, Hu Y, Zhao QQ, et al. Radiation Therapy Promotes Hepatocellular Carcinoma Immune Cloaking via PD-L1 Upregulation Induced by cGAS-STING Activation. Int J Radiat Oncol Biol Phys. (2022) 112:1243–55. doi: 10.1016/j.ijrobp.2021.12.162
79. Zhao F, Liu A, Gong X, Chen H, Wei J, Chen B, et al. Hypoxia-induced RNASEH2A limits activation of cGAS-STING signaling in HCC and predicts poor prognosis. Tumori. (2022) 108:63–76. doi: 10.1177/03008916211026019
80. Chen B, Hong Y, Zhai X, Deng Y, Hu H, Tian S, et al. m6A and m5C modification of GPX4 facilitates anticancer immunity via STING activation. Cell Death Dis. (2023) 14:809. doi: 10.1038/s41419-023-06241-w
81. Wang H, Feng J, Zeng C, Liu J, Fu Z, Wang D, et al. NSUN2-mediated M(5)c methylation of IRF3 mRNA negatively regulates type I interferon responses during various viral infections. Emerg Microbes Infect. (2023) 12:2178238. doi: 10.1080/22221751.2023.2178238
82. Chen M, Hu S, Li Y, Jiang TT, Jin H, Feng L. Targeting nuclear acid-mediated immunity in cancer immune checkpoint inhibitor therapies. Signal Transduct Targeted Ther. (2020) 5:270. doi: 10.1038/s41392-020-00347-9
83. Jiang Y, Zhang H, Wang J, Chen J, Guo Z, Liu Y, et al. Exploiting RIG-I-like receptor pathway for cancer immunotherapy. J Hematol Oncol. (2023) 16:8. doi: 10.1186/s13045-023-01405-9
84. Li Z, Zhou Y, Jia K, Yang Y, Zhang L, Wang S, et al. JMJD4-demethylated RIG-I prevents hepatic steatosis and carcinogenesis. J Hematol Oncol. (2022) 15:161. doi: 10.1186/s13045-022-01381-6
85. Zao X, Cheng J, Shen C, Guan G, Feng X, Zou J, et al. NFATc3 inhibits hepatocarcinogenesis and HBV replication via positively regulating RIG-I-mediated interferon transcription. Oncoimmunology. (2021) 10:1869388. doi: 10.1080/2162402x.2020.1869388
86. Zhong M, Zhong C, Cui W, Wang G, Zheng G, Li L, et al. Induction of tolerogenic dendritic cells by activated TGF-β/Akt/Smad2 signaling in RIG-I-deficient stemness-high human liver cancer cells. BMC Cancer. (2019) 19:439. doi: 10.1186/s12885-019-5670-9
87. RIG-I is a tumor suppressor and biomarker of IFN-α efficacy in HCC. Cancer Discovery. (2014) 4:Of12. doi: 10.1158/2159-8290.Cd-rw2014-009
88. Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. (2013) 14:1014–22. doi: 10.1038/ni.2703
89. Hernandez C, Huebener P, Schwabe RF. Damage-associated molecular patterns in cancer: a double-edged sword. Oncogene. (2016) 35:5931–41. doi: 10.1038/onc.2016.104
90. Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. Innate immune signaling and regulation in cancer immunotherapy. Cell Res. (2017) 27:96–108. doi: 10.1038/cr.2016.149
91. Anreiter I, Mir Q, Simpson JT, Janga SC, Soller M. New twists in detecting mRNA modification dynamics. Trends Biotechnol. (2021) 39:72–89. doi: 10.1016/j.tibtech.2020.06.002
92. Wang J, Chew BLA, Lai Y, Dong H, Xu L, Liu Y, et al. A systems-level mass spectrometry-based technique for accurate and sensitive quantification of the RNA cap epitranscriptome. Nat Protoc. (2023) 18:2671–98. doi: 10.1038/s41596-023-00857-0
93. Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. (2012) 30:1232–9. doi: 10.1038/nbt.2432
94. Ding JH, Li G, Xiong J, Liu FL, Xie NB, Ji TT, et al. Whole-genome mapping of epigenetic modification of 5-formylcytosine at single-base resolution by chemical labeling enrichment and deamination sequencing. Anal Chem. (2024) 96:4726–35. doi: 10.1021/acs.analchem.4c00425
95. Kim BY, Rutka JT, Chan WC. Nanomedicine. N Engl J Med. (2010) 363:2434–43. doi: 10.1056/NEJMra0912273
Keywords: RNA m5C methylation, TLR, cGAS-STING, RIG-I, hepatocellular carcinoma
Citation: Meng S, Jiangtao B, Haisong W, Mei L, Long Z and Shanfeng L (2024) RNA m5C methylation: a potential modulator of innate immune pathways in hepatocellular carcinoma. Front. Immunol. 15:1362159. doi: 10.3389/fimmu.2024.1362159
Received: 27 December 2023; Accepted: 26 April 2024;
Published: 13 May 2024.
Edited by:
Wen Tan, Lanzhou University, ChinaReviewed by:
Leonard Maggi, Washington University in St. Louis, United StatesDaniele Vergara, University of Salento, Italy
Junjun Jia, Zhejiang Chinese Medical University, China
Copyright © 2024 Meng, Jiangtao, Haisong, Mei, Long and Shanfeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Shanfeng, c2ZlbmdsQGhidS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Sun Meng
Sun Meng Bai Jiangtao†
Bai Jiangtao†