
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 24 July 2024
Sec. Nutritional Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1356414
This article is part of the Research Topic Gut Microbiota and Immunity in Health and Disease: dysbiosis and eubiosis's effects on the human body View all 12 articles
Background: The gut microbiota significantly influences the onset and progression of juvenile idiopathic arthritis (JIA) and associated uveitis (JIAU); however, the causality remains unclear. This study aims to establish a causal link between gut microbiota and JIA or JIAU.
Methods: Using publicly available genome-wide association studies (GAWS) summary data, we conducted a two-sample Mendelian randomisation (MR) analysis employing various methods, namely inverse variance weighted (IVW), simple mode, weighted mode, weighted median and MR-Egger regression methods, to assess the causal association between JIA or JIAU and gut microbiota. Sensitivity analyses, including Cochrane’s Q test, MR-Egger intercept test, leave-one-out analysis and MR-PRESSO, were performed to evaluate the robustness of the MR results. Subsequently, reverse MR analysis was conducted to determine causality between gene-predicted gut microbiota abundance and JIA or JIAU.
Results: The MR analysis revealed a causal association between gut microbiota abundance variations and JIA or JIAU risk. Specifically, the increased abundance of genus Ruminococcaceae UCG013 (OR: 0.055, 95%CI: 0.006–0.103, p = 0.026) and genus Ruminococcaceae UCG003 (β: 0.06, 95%CI: 0.003–0.117, p = 0.041) correlated with an increased risk of JIA, while genus Lachnospiraceae UCG001 (OR: 0.833, 95%CI: 0.699~0.993, p = 0.042) was associated with a reduced risk of JIA, among others. Sensitivity analysis confirmed MR analysis robustness.
Conclusions: This study provides substantial evidence supporting a causal association between genetically predicted gut microbiota and JIA or JIAU. It highlights the significant role of intestinal flora in JIA or JIAU development, suggesting their potential as novel biomarkers for diagnosis and prevention. These findings offer valuable insights to mitigate the impact of JIA or JIAU.
Juvenile idiopathic arthritis (JIA) is a heterogeneous condition characterised by arthritis of unknown origin manifesting before the age of 16 (1), often resulting in functional limitations and, in severe cases, disability (2). Globally, approximately three million children are affected by JIA (3, 4), making it the most prevalent chronic inflammatory rheumatic disease in childhood. Severe cases may exhibit persistent systemic symptoms, joint inflammation, severe drug side effects and macrophage activation syndrome (MAS) occurrence, which can pose life-threatening consequences, thereby placing a considerable burden on children’s health and socioeconomic systems (5). Juvenile idiopathic arthritis-associated uveitis (JIAU) is commonly acknowledged as a prevalent and severe extra-articular manifestation of JIA (6), characterised by chronic bilateral recurrent anterior uveitis, a leading cause of disability and visual impairment. The incidence of JIAU varies from 5.0% to 19.1% among JIA populations across different geographical regions (7). Although multiple factors, such as genetics, environment and immunity, are hypothesised to contribute to the pathogenesis and underlying mechanisms of JIA and JIAU (6, 8), their precise aetiology and pathogenesis remain unclear, necessitating further investigation to enhance diagnosis, treatment and reduce associated disease burden. The human intestinal microbiota, comprising approximately 100 trillion bacteria of 1000–1150 species, forms a symbiotic relationship with the host, playing critical roles in human metabolism, nutrient absorption, immune responses and other aspects (9). Additionally, the intestinal microbiota is intricately linked to the onset and progression of various human diseases. In the context of predictive, preventive and personalised medicine, systemic inflammation serves as an essential communication bridge between the human host and the gut microbiota (10). Recent evidence suggests a potential role of gut microbiota in immune-mediated diseases such as rheumatoid arthritis (RA), wherein dysbiosis of the gut microbiota disrupts intestinal barrier function, leading to increased permeability and immune imbalance. Consequently, this dysregulation allows immune cells to migrate to extraintestinal sites, including joints, thereby triggering localised inflammation (11, 12).
Recent case-control studies spanning three continents provide compelling evidence of the influence of gut microbiota dysbiosis on the onset and progression of JIA (13–16). A prospective study focussing on the gut microbiota in patients with JIA in Italy and the Netherlands reports the presence of gut microbial dysbiosis in this population. Moreover, significant differences in microbial diversity and composition exist between patients with JIA and healthy individuals (17). Another study conducted in China yielded similar findings, indicating that patients with JIA display a significantly lower abundance of Anaerostipes, Dorea, Lachnospira and Roseburia compared to the control group. The study also identified 12 genera that could potentially serve as biomarkers and predictive factors for JIA (18). Nevertheless, the causal relationship between JIA and the gut microbiota remains uncertain, necessitating further investigation. Furthermore, observational studies are prone to the impact of confounding variables and reverse causality, potentially biasing results (19).
To address these limitations, we employ Mendelian randomisation (MR) to investigate the genetic-level association between JIA or JIAU and gut microbiota. MR utilises single nucleotide polymorphisms (SNPs) as instrumental variables (IVs) to estimate potential causal links between exposure variables and health outcomes (20). Leveraging extensive genome-wide association studies (GWAS) data, we employ a bidirectional two-sample MR approach to explore the causal relationship between the gut microbiota and these diseases, offering novel perspectives into potential therapeutic strategies targeting the microbiota (21–23).
This study employs a bidirectional two-sample MR approach to evaluate the causal associations between the genetic prediction of JIA or JIAU and the genetic prediction of gut microbiota. Three key assumptions guide the selection of SNPs as IVs: 1) relevance, requiring a substantial correlation between the SNP and the exposure variable; 2) exclusion restriction, indicating that the SNP influences the outcome solely through the exposure variable and not by any alternative causal pathway; and 3) independence, implying that the SNP is independent of the outcome variable and potential confounding factors. In forward MR analysis, gut microbiota serves as the exposure variable, while JIA or JIAU is considered the outcome variable. Conversely, in reverse MR analysis, JIA or JIAU is regarded as the exposure variable, while gut microbiota is treated as the outcome variable. This approach aims to investigate potential causal links between JIA or JIAU and gut microbiota in both directions. Figure 1 illustrates the schematic representation of the MR causality research design, elucidating the underlying principles of MR studies. Figure 2 presents a flowchart outlining the step-by-step process involved in conducting such a study. This study adheres to the reporting guidelines outlined in STROBE-MR, supplemented with materials that include a checklist according to STROBE-MR and a checklist based on the Critical Appraisal Checklist for examining MR research (24, 25). The Supplementary Materials provide a detailed explanation of the checklist items.
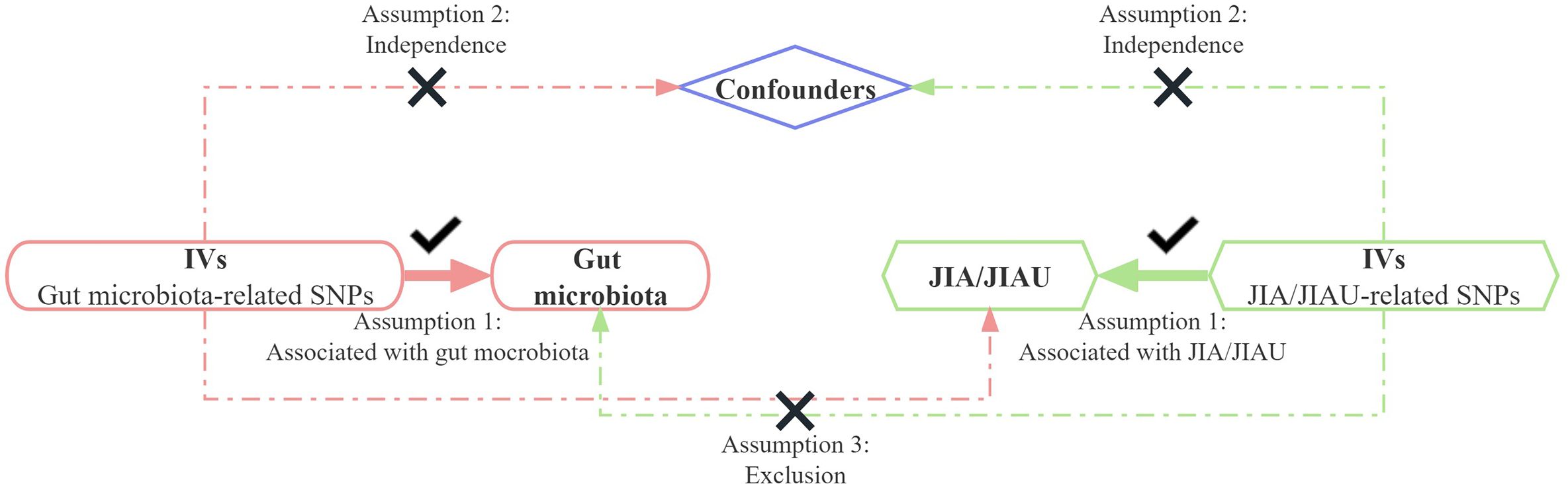
Figure 1 Bidirectional two-sample Mendelian randomisation between JIA/JIAU and gut microbiota abundance outcomes. Directed acyclic graph (DAG) of the causal association between JIA/JIAU and gut microbiota abundance. IVs, instrumental variables; SNPs, single nucleotide polymorphisms; JIA, juvenile idiopathic arthritis; JIAU, JIA-associated uveitis.
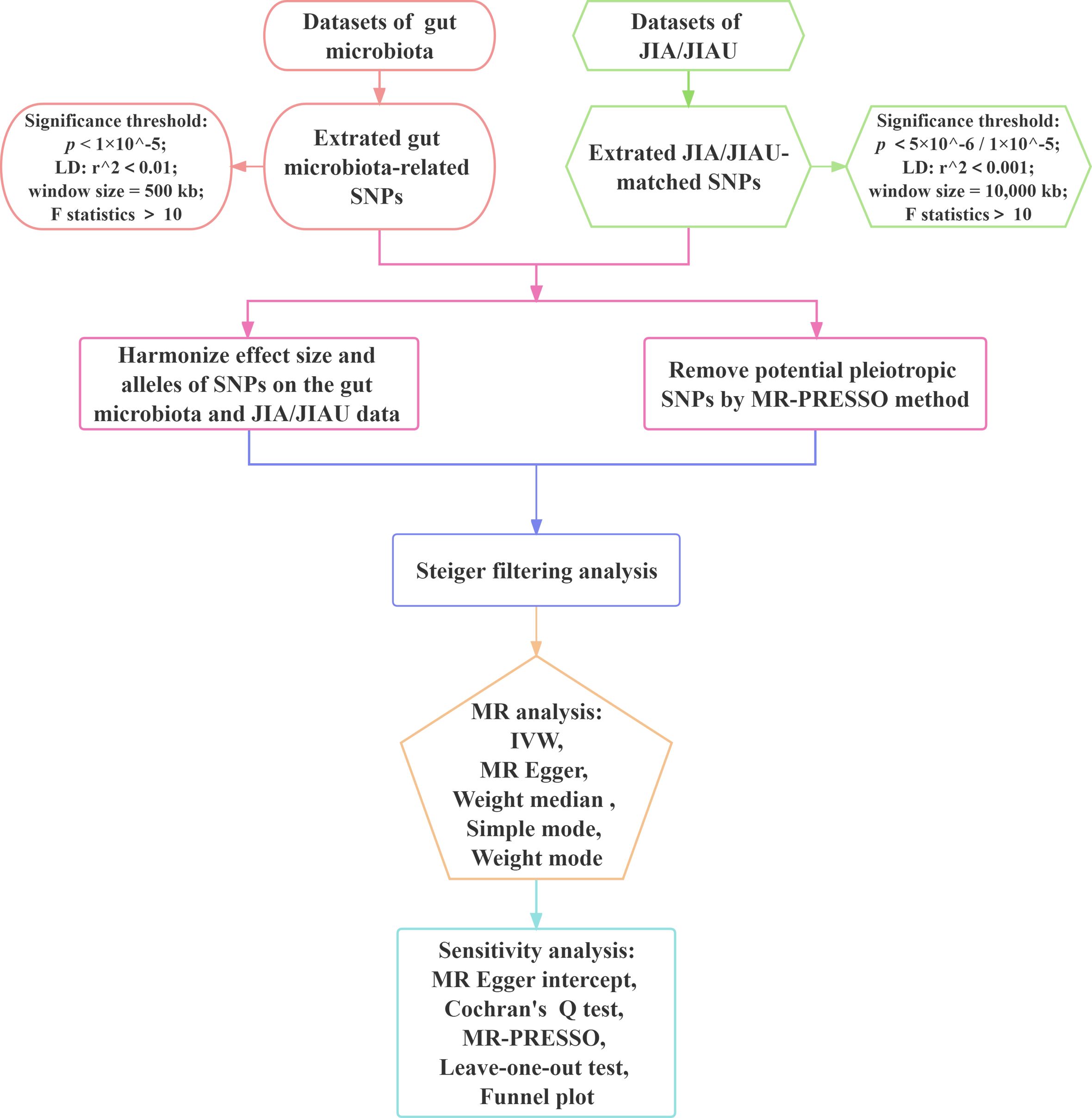
Figure 2 Workflow of Mendelian randomisation study revealing causality between gut microbiota abundance and JIA/JIAU risk. JIA, juvenile idiopathic arthritis; JIAU, JIA-associated uveitis; SNPs, single nucleotide polymorphisms; LD, linkage disequilibrium; IVW, inverse variance weighted; MR, Mendelian randomisation; MR-PRESSO, MR pleiotropy residual sum and outlier.
GWAS data for JIA are based on a recent meta-analysis comprising 6,056 patients with JIA and 25,086 European ancestry controls (26). The detailed research process is described in previous studies by Lopez-Isac E and McIntosh LA (26, 27). The diagnostic criteria for JIA follow international standards published by the International League of Associations for Rheumatology (ILAR), with the specificity and common susceptibility loci for the various JIA ILAR subtypes systematically examined using the JIA GWAS data. The GWAS data for JIAU were sourced from Haasnoot et al.’s study (28), encompassing 192 patients with JIAU of European ancestry and 330 JIA control patients without uveitis.
GWAS data related to gut microbiota were obtained from the Mi BioGen consortium, representing the most extensive dataset available (https://mibiogen.gcc.rug.nl/). The GWAS dataset includes 16S rRNA gene information from the faecal microbiomes of 18,340 individuals across 24 cohorts, alongside whole-genome genotyping data. Specifically, the study investigated the species composition of gut microbiota utilising the 16S rRNA gene regions V1-V2, V3-V4 and V4, representing three distinct variable regions. The dataset comprises 211 intestinal flora, categorised by kingdom, phylum, order, family, genus and species. These species exhibit relative abundance or genetic variations and can be further categorised into 9 phyla, 16 classes, 20 orders, 35 families and 131 genera. After excluding 12 unknown genera, we incorporated a total of 119 genera as classification units for the bidirectional MR study. Supplementary Table 1 presents the features of the data sources utilised in this investigation.
To ensure the validity of the causal relationship between gut microbiota composition and JIA or JIAU risk, rigorous quality control measures were applied in selecting appropriate genetic IVs.
Given the limited number of genetic variants associated with gut microbiota, we established a significance threshold of p< 1.0×10-5 based on the majority of MR studies on gut microbiota to ensure adequate candidate instrument numbers for forward MR analysis (29–31). Additionally, to ensure independence between IVs and avoid bias from linkage disequilibrium (LD) between SNPs, we applied LD clustering with a cutoff of r2< 0.01 and a clustering distance of 500 kb. Finally, to minimise instrument bias, IVs with an F statistic less than 10 were excluded, where , with N representing the sample size and R2 the squared correlation coefficient (32).
For reverse MR, the significance threshold was set at p< 5.0×10-6 to screen SNPs as IVs for JIA and p< 1.0×10-5 for JIAU, with LD set at r2< 0.001 and a clumping distance of 10,000 kb.
To determine the direction of influence of specific IVs on the outcome, Steiger filtering analysis was employed (33). IVs with a ‘TRUE’ result indicating the expected association direction were included in subsequent analysis, with those classified as ‘False’ were excluded.
MR analysis was conducted to explore the causality between JIA/JIAU and gut microbiota using R software version 4.2.2 and three specific R packages: ‘TwoSampleMR’ (v.0.5.6) (33), ‘MRPRESSO’ (v.1.0) (34) and ‘MendelianRandomization’ (35). A significance threshold of p< 0.05 was considered for causation. To account for multiple comparisons at every taxonomic level (phylum, class, order, family and genus), a significance threshold of 0.05/n was set, where n represents the number of distinct bacterial species at that particular taxonomic level.
Five statistical methodologies were employed: inverse variance weighted (IVW), simple mode, weight mode, weighted median and MR-Egger regression. IVW, considered the primary method, was used for outcome determination, supplemented by the other four methods. Different methods possess different underlying assumptions about horizontal pleiotropy. IVW employs the inverse of the squared standard error (SE) of the outcome as weights and does not include an intercept component in regression. Additionally, random effects were utilised for IVW modelling, with overall estimates derived from weighted linear regression of the Wald estimate for each SNP (36). The other four methods complement IVW by providing more robust estimates in broader scenarios, albeit with lower effect values (wider CI) (34). Moreover, the MR-Egger method offers a consistent causal effect test when SNPs directly linked with the outcome or exhibiting horizontal pleiotropy are excluded (37). Weighted median methodology can produce reliable estimates even when up to 50% of findings are from erroneous SNPs (38). Meanwhile, the simple mode provides an unweighted mode of estimating causal effects’ empirical density function (39). Supplementary Table 2 lists the advantages, disadvantages, efficacy and applications of these five methods.
Following MR analysis, sensitivity analysis was conducted to assess the robustness of the results. Firstly, the intercept of the MR-Egger regression was examined to detect the presence of horizontal pleiotropy, with p > 0.05 suggesting a weaker potential for pleiotropy in the causal analysis (34). Additionally, MR-PRESSO was employed to identify and address possible outliers by screening for heterogeneity and outliers, followed by a re-analysis of the MR. To assess heterogeneity in the IVW method, Cochran’s Q test (40) and funnel plots were used. Furthermore, to evaluate the impact of individual SNPs on the primary causal relationship, a leave-one-out analysis was conducted by sequentially excluding individual SNPs.
All GWAS data utilised in our analysis were obtained from publicly available datasets with ethical permission granted by the relevant ethical review boards. These datasets do not contain personal information, ensuring compliance with ethical standards.
In the initial steps, a total of 3036 SNPs relevant to gut microbiota at the phylum, class, order, family and genus levels were identified, each with an F statistic > 10, indicating minimal instrumental bias (Supplementary Table 2).
Additionally, Steiger filtering analysis revealed no SNPs with a reverse causal direction for gut microbiota on JIA (Supplementary Table 3), while 849 SNPs were eliminated for JIAU (Supplementary Table 5). For our analysis, a total of 131 genera, 35 families, 16 classes, 20 orders and 9 phyla were identified as IVs, with 5 to 26 IVs obtained from each classification.
To address multiple comparisons, significance thresholds for gut microbiota on JIA and JIAU were determined using the Bonferroni correction: For JIA, thresholds were set at phyla (p = 5.56 × 10-3), class (p = 3.13 × 10-3), order (p = 2.50 × 10-3), family (p = 1.35 × 10-3) and genus (p = 3.82× 10-4). Similarly, for JIAU, thresholds were set at phyla (p = 5.56 × 10-3), class (p = 3.13 × 10-3), order (p = 2.94 × 10-3), family (p = 1.43 × 10-3) and genus (p = 3.82× 10-4).
Utilising the IVW method, an association between JIA risk and gut microbiota at one phylum and four genera was identified (Figure 3). MR analysis revealed that family FamilyXI (OR: 1.148, 95%CI: 1.008~1.307, p = 0.037) increased the risk of JIA, while genera Rikenellaceae RC9 gut group (OR: 0.843, 95%CI: 0.74~0.96, p = 0.01), Lachnospiraceae UCG001 (OR: 0.833, 95%CI: 0.699~0.993, p = 0.042), Intestinimonas (OR: 0.840, 95%CI: 0.709~0.994, p = 0.0426) and Clostridium innocuum group (OR: 0.862, 95%CI: 0.73~1.0, p = 0.0495) decreased the risk of JIA. Notably, consistent effect directions were observed for family FamilyXI, genus Lachnospiraceae UCG001, genus Intestinimonas, and genus Clostridium innocuum group across multiple MR methods. However, for the genus Rikenellaceae RC9 gut group, the results from the MR-Egger method differed from the IVW method but remained consistent with the weighted median, simple mode, and weighted mode methods. A circular heat map visualised the outcomes of MR analysis conducted on two sample groups, illustrating the causal effects estimated by the five different MR methods, with gut microbiota abundance as the exposure variable and JIA as the outcome variable (Figure 4). Supplementary Figures 1, 2 display the scatter plot and forest plot, respectively.

Figure 3 Forest plots of Mendelian randomisation for two samples indicated the causal effects of five different Mendelian randomisation methods with gut microbiota abundance as exposure and juvenile idiopathic arthritis (JIA) as the outcome. The effect estimates are presented as the effect size (OR) and 95% confidence interval (CI). snp, single nucleotide polymorphism.
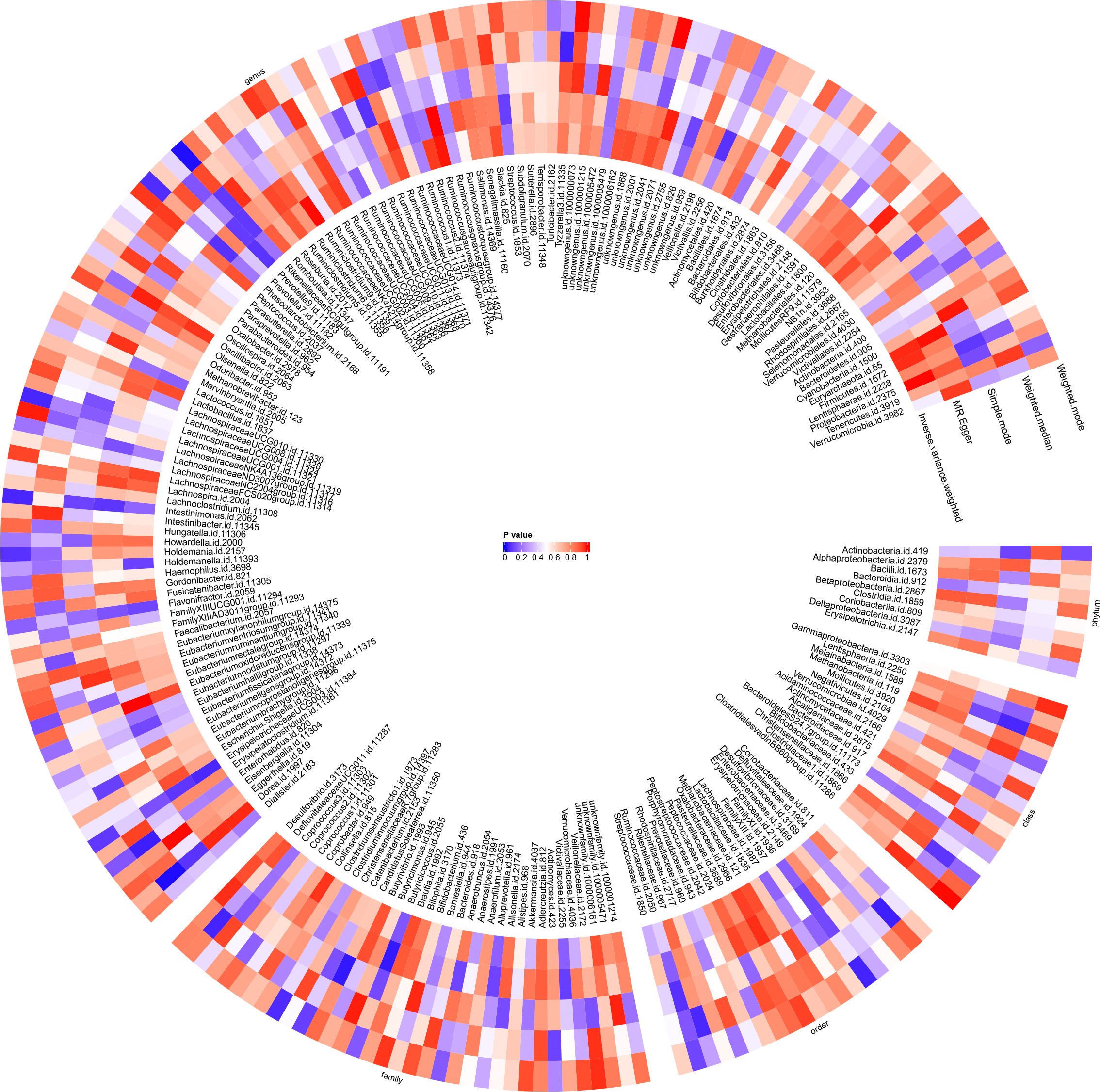
Figure 4 Circular heat map was generated to visualise the results of Mendelian randomisation analysis conducted on two sample groups, illustrating the causal effects estimated by five different Mendelian randomisation methods, with gut microbiota abundance as the exposure variable and juvenile idiopathic arthritis (JIA) as the outcome variable.
A causal relationship was found between one genus of gut microbiota and the risk of JIAU using the IVW method (Figure 5). MR analysis showed that genus Prevotella7 reduced the likelihood of JIAU (OR: 0.398, 95% CI: 0.191–0.83, p = 0.014). Additionally, the MR-Egger, weighted median, simple mode and weighted mode methods revealed consistent effect directions. A circular heat map provided insights into the relationship between gut microbiota abundance (exposure variable) and JIAU (outcome variable) (Figure 6), with scatter plots (Supplementary Figures 5) and forest plots (Supplementary Figure 6) further illustrating these relationships.
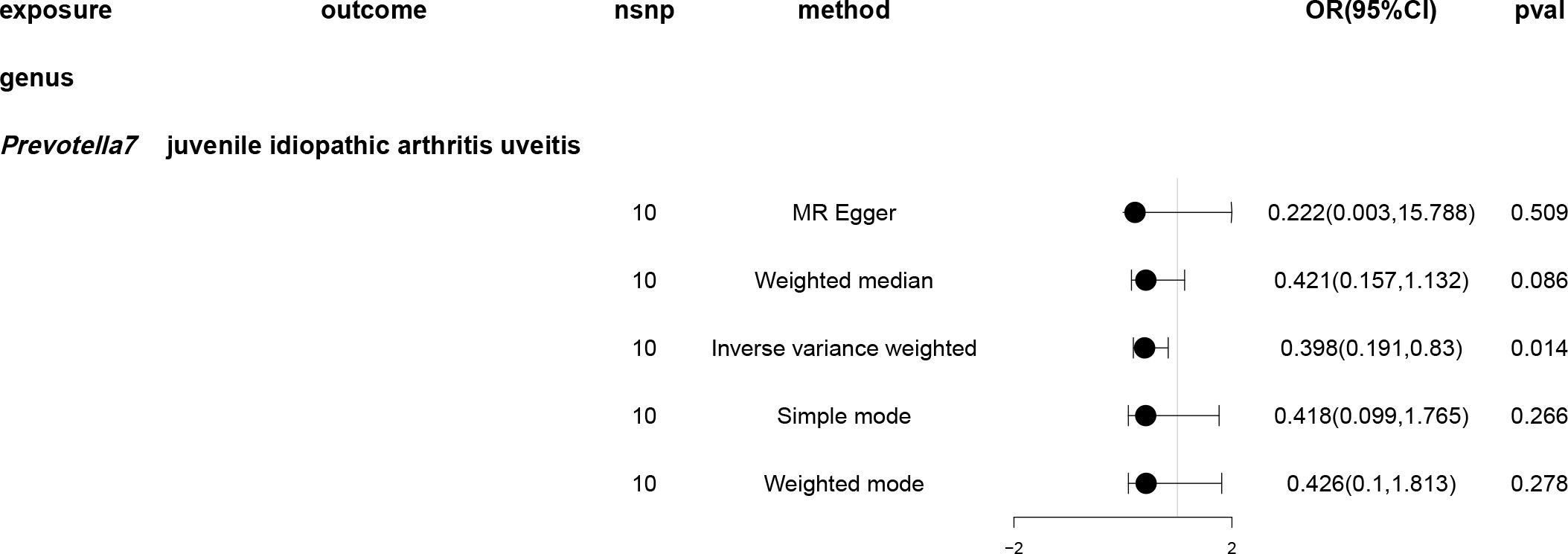
Figure 5 Forest plots of Mendelian randomisation for two samples demonstrated the causal effects of five different Mendelian randomisation methods with gut microbiota abundance as exposure and juvenile idiopathic arthritis associated uveitis (JIAU) as the outcome. The effect estimates are presented as the effect size (OR) and 95% confidence interval (CI). snp, single nucleotide polymorphism.
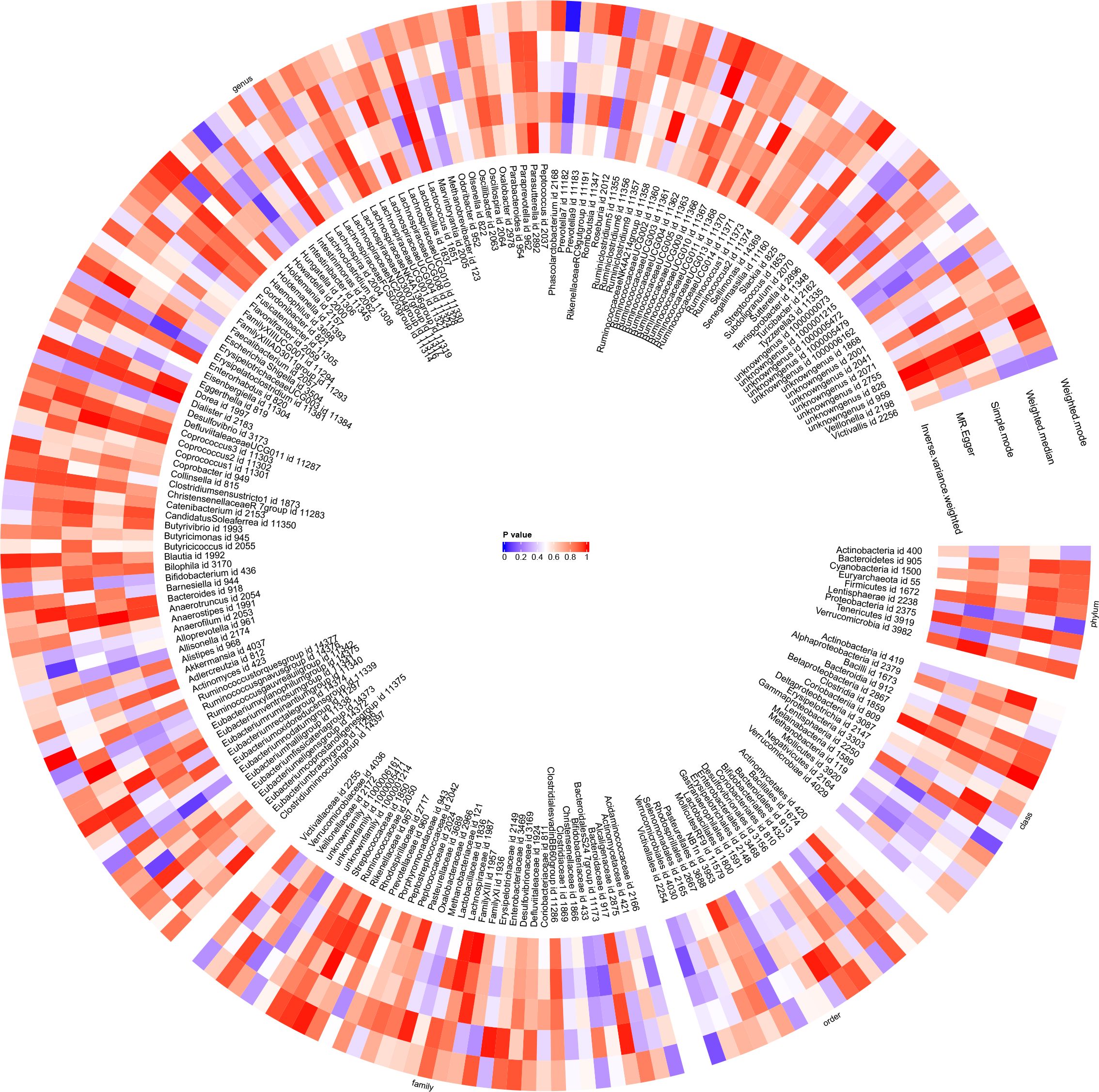
Figure 6 With gut microbiota abundance as the exposure variable and juvenile idiopathic arthritis associated uveitis (JIAU) as the outcome variable, a circular heat map was generated to visualise the findings of the Mendelian randomisation analysis performed on two sample groups. The heat map illustrated the causal effects estimated by five different Mendelian randomisation methods.
For JIA, 13 SNPs and for JIAU, 6 SNPs met the IV screening criteria, each with an F-statistics > 10, indicating minimal instrumental bias (Supplementary Tables 13, 19). Additionally, Steiger filtering analysis revealed no SNPs with opposing causal orientations for either condition (Supplementary Tables 14, 20).
The SNP findings summarised data from a total of 208 gut microbiota classifications, including 16 classes, 126 genera, 33 families, 20 orders and 9 phyla, used to study the connection between JIA and gut microbiota (Supplementary Table 15). Significance thresholds for multiple comparisons at distinct taxonomic classifications were set as follows: phylum (p = 5.56×10-3), class (p = 3.13×10-3), order (p = 2.50×10-3), family (p = 1.52×10-3) and genus (p = 3.97×10-4), and were adjusted using the Bonferroni correction method. Similarly, for JIAU and gut microbiota, 208 different gut microbiota classifications were included in the data summary for the SNP results (Supplementary Table 21). These classifications encompassed 16 classes, 128 genera, 34 families, 20 orders and 9 phyla. Significance thresholds for multiple comparisons at different taxonomic classifications were set as follows: phylum (p = 5.56×10-3), class (p = 3.13×10-3), order (p = 2.50×10-3), family (p = 1.47×10-3) and genus (p = 3.91×10-4), and were adjusted using the Bonferroni correction method.
Using the IVW method, causal associations between JIA and gut microbiota were identified at one phylum, one order, one family and eight genera (Figure 7). MR analysis revealed that JIA increased the abundance of class betaproteobacteria (β: 0.061, 95%CI: 0.014–0.109, p = 0.012), order Burkholderiales (β: 0.061, 95%CI: 0.013–0.109, p = 0.013), family Alcaligenaceae (β: 0.056, 95%CI: 0.008–0.104, p = 0.023), genus Ruminococcaceae UCG013 (β: 0.055, 95%CI: 0.006–0.103, p = 0.026), genus Roseburia (β: 0.051, 95%CI: 0.003–0.099, p = 0.036), genus Ruminococcaceae UCG003 (β: 0.06, 95%CI: 0.003–0.117, p = 0.041) and genus Anaerofilum (β: 0.097, 95%CI: 0.003–0.192, p = 0.044), while decreasing the abundance of genus Olsenella (β: -0.137, 95%CI: -0.24- -0.033, p = 0.01), genus Coprococcus2 (β: -0.065, 95%CI: -0.123- -0.006, p = 0.03), genus Romboutsia (β: -0.057, 95%CI: -0.11, -0.005, p = 0.033) and genus Eisenbergiella (β: -0.145, 95%CI: -0.285- -0.006, p = 0.041). For class beta proteobacteria, order Burkholderiales, family Alcaligenaceae, genus Ruminococcaceae UCG013, genus Roseburia, genus Ruminococcaceae UCG003, genus Olsenella, genus Romboutsia and genus Eisenbergiella, the MR-Egger, weighted median, mode-based estimator and weighted mode methods revealed directional effects that were consistent with the IVW method. However, for genus Anaerofilum and genus Coprococcus2, the results from the MR-Egger method differed from those of the IVW method, while the results from the weighted median, mode-based estimator and weighted mode methods aligned with the IVW method. A circular heat map visually represented the outcomes of MR analysis performed on the two sample groups, depicting the causal effects estimated by the five different MR methods and providing insights into the relationship between JIA (exposure variable) and gut microbiota abundance (outcome variable) (Figure 8). Scatter plots (Supplementary Figure 9) and forest plots Supplementary Figure 10) further illustrated these relationships.
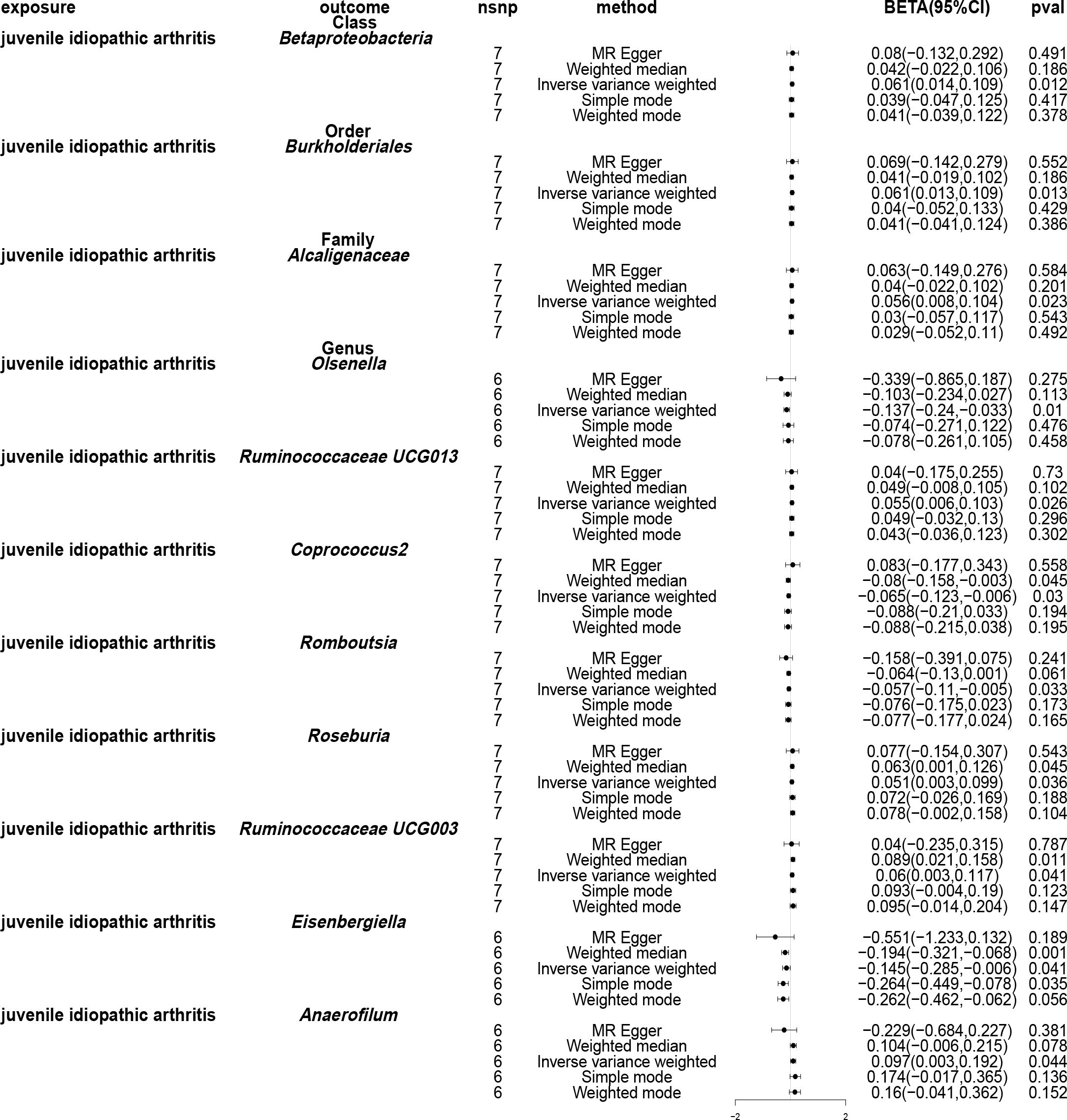
Figure 7 Forest plots of Mendelian randomisation for two samples, depicted the causal effects of five different Mendelian randomisation methods with juvenile idiopathic arthritis (JIA) as exposure and gut microbiota as the outcome. The effect estimates are presented as the effect size (BETA) and 95% confidence interval (CI). snp, single nucleotide polymorphism.
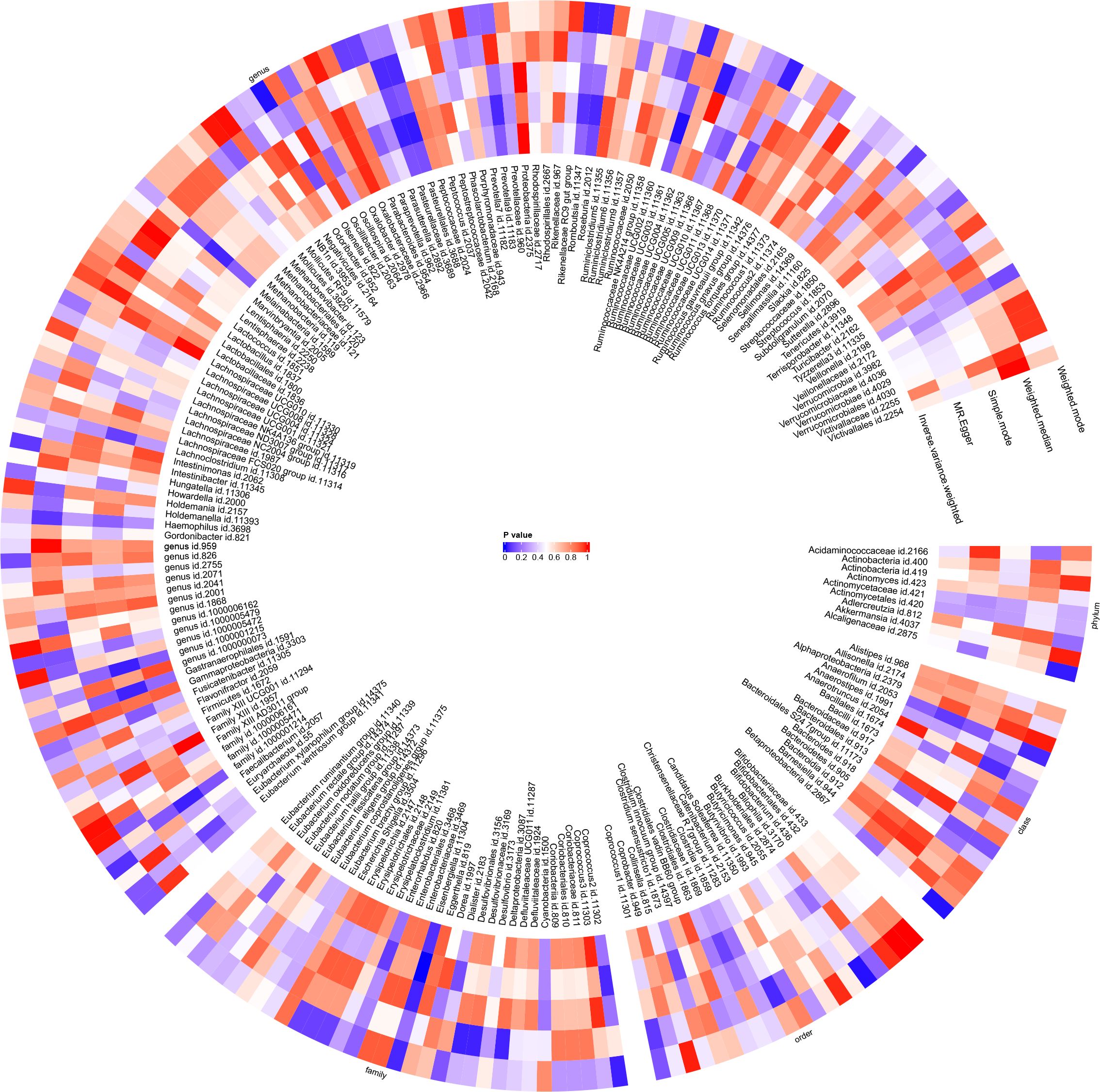
Figure 8 Circular heat map was generated to visually represent the outcomes of Mendelian randomisation analysis performed on two sample groups, depicting the causal effects estimated by five different Mendelian randomisation methods, with juvenile idiopathic arthritis (JIA) as the exposure variable and gut microbiota abundance as the outcome variable.
Employing the IVW approach, a causal association was found between JIAU and the gut microbiota at one phylum, one class, one order, two families and three genera (Figure 9). MR analysis demonstrated that JIAU increased the abundance of genus Phascolarctobacterium (β: 0.023, 95%CI: 0.001–0.044, p = 0.04) but decreased the abundance of phylum Lentisphaerae (β: -0.041, 95%CI: -0.074–0.007, p = 0.017), class Lentisphaeria (β: -0.043, 95%CI: -0.076- -0.009, p=0.012), order Victivallales (β: -0.043, 95%CI: -0.076- -0.009, p=0.012), family Peptostreptococcaceae (β: -0.019, 95%CI: -0.036- -0.001, p = 0.041), family Victivallaceae (β: -0.045, 95%CI: -0.082- -0.008, p = 0.016), genus Slackia (β: -0.04, 95%CI: -0.075- -0.005, p = 0.026) and genus Alloprevotella (β: -0.086, 95%CI: -0.168- -0.005, p = 0.038). For phylum Lentisphaerae, class Lentisphaeria, order Victivallales, family Victivallaceae, genus Slackia and genus Phascolarctobacterium, MR-Egger, weighted median, simple mode and weighted mode methods revealed effect directions consistent with the IVW results. However, for the family Peptostreptococcaceae, the results from the MR-Egger method differed from that of the IVW method; however, the weighted median, simple mode and weighted mode methods revealed results consistent with the IVW findings. A circular heat map was generated to visually represent the outcomes of MR analysis performed on the two sample groups, depicting the causal effects estimated by the five different MR methods, thereby providing insights into the relationship between the exposure (JIAU) and outcome (gut microbiota abundance) (Figure 10). Supplementary Figures 13, 14 present the scatter and forest plots, respectively. Supplementary Table 25 lists the top five microbial genera and species and their characterisation with potent effects associated with JIA or JIAU as derived from forward MR and reverse MR.
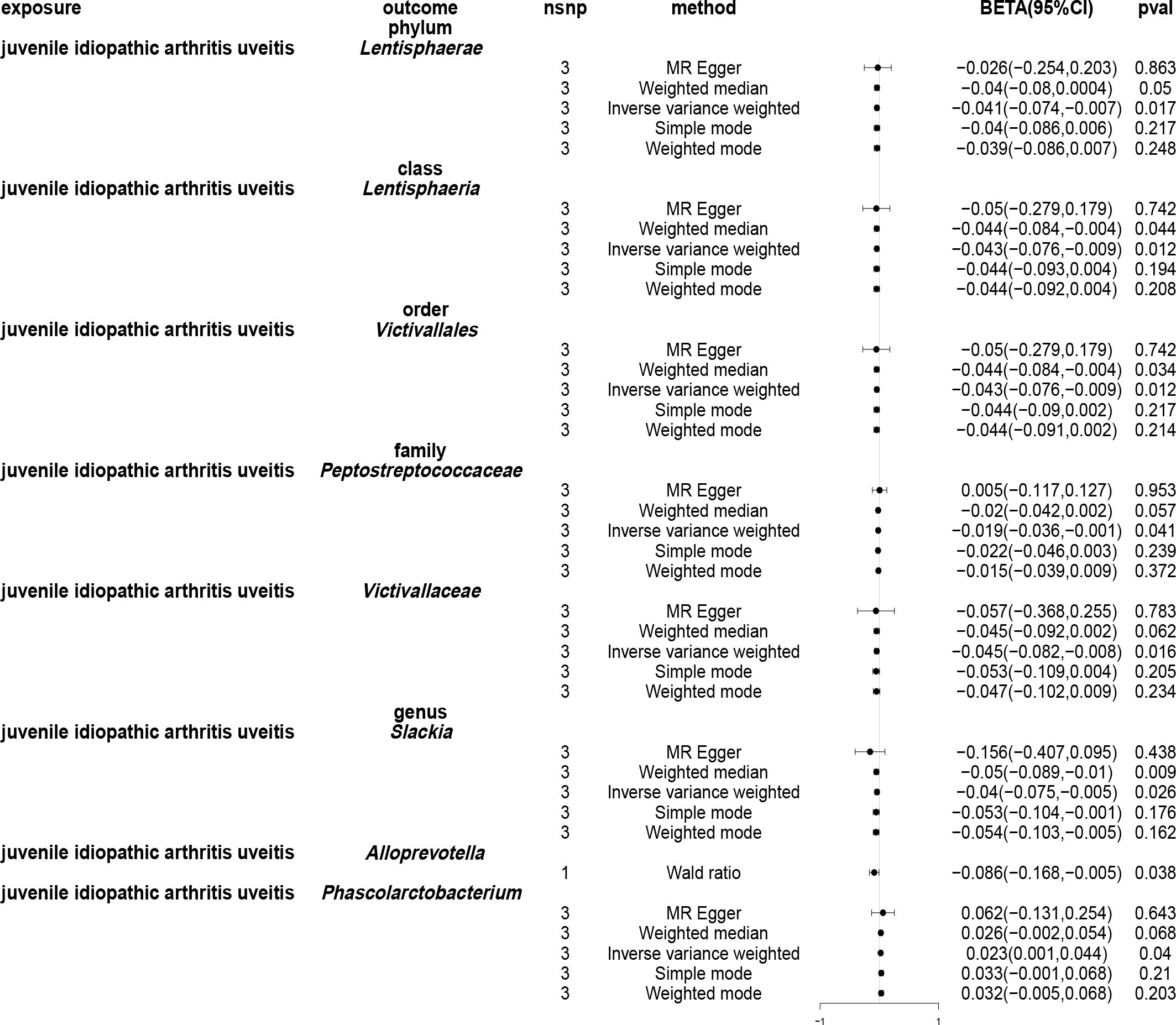
Figure 9 Forest plots of Mendelian randomisation for two samples illustrated the causal effects of five different Mendelian randomisation methods with juvenile idiopathic arthritis associated uveitis (JIAU) as exposure and gut microbiota as the outcome. The effect estimates are presented as the effect size (BETA) and 95% confidence interval (CI). snp, single nucleotide polymorphism.
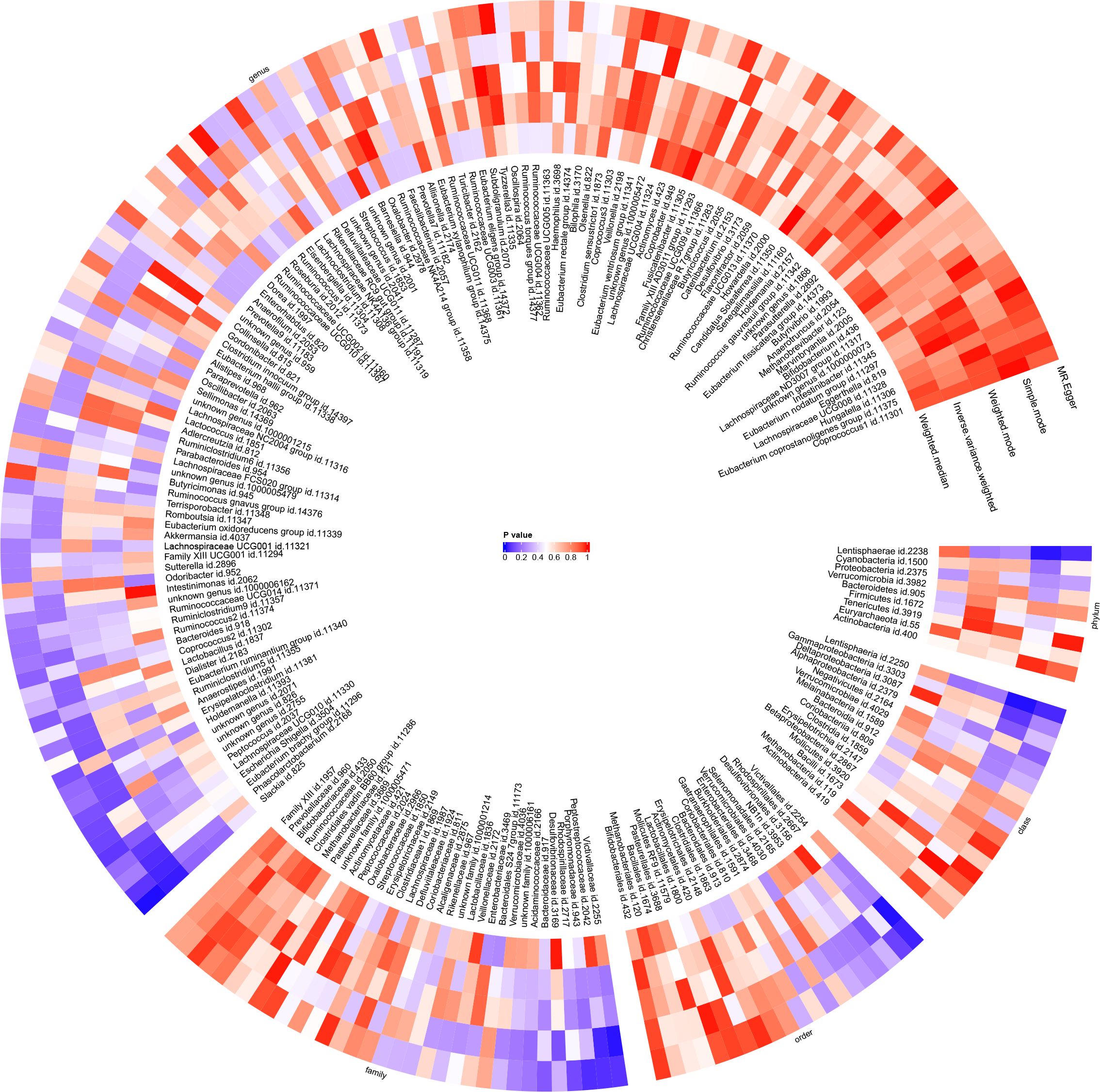
Figure 10 The results of Mendelian randomisation study on two sample groups were visualised via a circular heat map, which showed the causal effects estimated by five different Mendelian randomisation methods, with juvenile idiopathic arthritis associated uveitis (JIAU) as the variable of exposure and gut microbiota abundance as the variable of outcome.
When the variables of the gut microbiota or JIA or JIAU samples were analysed using the MR-Egger regression intercept method, no evidence of horizontal pleiotropy was detected (Supplementary Tables 8, 11, 17, 23). We conducted outlier screening and removal in the MR-PRESSO analysis, and the global p-value test of MR-PRESSO did not indicate horizontal pleiotropy for gut microbiota or JIA or JIAU (Supplementary Tables 9, 12, 18). Furthermore, the majority of Cochrane’s Q test results did not reveal significant heterogeneity (p > 0.05) (Supplementary Table 7, Supplementary Tables 10, 16, 22). In the presence of significant heterogeneity, we employed random effects models and the IVW model as the main analytical results. Sensitivity analyses using the leave-one-out method and funnel plots are depicted in Supplementary Figures 3, 4, 7, 8, 11, 12, 15, 16.
JIA is a common autoimmune rheumatic disease that poses a significant threat to the overall health and well-being of children, triggering joint deformities, functional impairments, stunted growth and osteoporosis, among other serious complications. Meanwhile, JIAU, a prevalent extra-articular complication of JIA, further complicates the clinical picture with chronic, non-granulomatous and non-infectious uveitis, often affecting the anterior segment of the eye. Approximately 10% to 14% of patients with JIA exhibit signs of uveitis before arthritis onset (41). Current treatment modalities for JIA and JIAU primarily focus on symptomatic relief, lacking definitive curative options (8). Previous studies have reported that siblings of patients with JIA have a higher risk of developing JIA than the general population, with the onset age, disease type and disease course of twin siblings exhibiting a similar trajectory (42). These findings indicate a genetic factor underlying JIA pathogenesis (43, 44). Moreover, subsequent studies have reported a genetic susceptibility locus for JIA, further confirming this speculation. However, as research advances, scientists have discovered that these genetic susceptibility factors can only explain 18% of the causes of JIA. Currently, the progression of JIA is speculated to be the outcome of the interaction between genetic susceptibility genes, environmental stimuli and immune dysregulation (45–47).
Emerging evidence underscores the microbiota’s involvement in immune-related disorders like RA and inflammatory bowel disease (IBD) (11). The intestinal microbiota predominately contributes to JIA pathogenesis by altering intestinal mucosal permeability and regulating the host immune system (48). Notably, altered microbial compositions, such as increased phyla Bacteroidetes and decreased Firmicutes, have been observed in paediatric patients with JIA compared to healthy individuals (15). This study suggests an increase in the abundance of class betaproteobacteria in patients with JIA, consistent with previous research findings (15).
In animal experiments, an extraperitoneal injection of intestinal bacterial cell wall components into mice has been demonstrated to induce arthritis in a conventional environment, but not in a sterile environment (49), suggesting a close relationship between gut microbiota alterations and arthritis development. Early investigations by Malin et al. (50) highlighted elevated bacterial urease activity in the stool samples of children with JIA compared to those of healthy children, suggesting an anaerobic dysbiosis of the intestinal flora. Interestingly, oral Lactobacillus administration to patients with JIA reduced urease activity. Results from a multicentre study conducted by van Dijkhuizen et al. (78 Italian children and 21 Dutch children) (17) revealed that Italian children with JIA exhibited an elevated abundance of Faecalibacterium prausnitzii, Erysipelotrichaceae, Enterococcus, Parabactteroides and Ruminococccaceae, but reduced levels of Allobaculum, Gemellaceae, Propionibacterium acnes and Turicibacter compared to the healthy control group. Similarly, our study also suggests an increase in the abundance of genera Ruminococcaceae UCG013 and Ruminococcaceae UCG003 in patients with JIA. Additionally, a significant decrease in the level of intestinal flora has been reported in samples from active and inactive states compared to healthy children (17). Kindgren et al.’s study on population queues (51) revealed a higher content of Acidaminococcales, Prevotella 9 and Veillonella parvula in JIA cases, while Coprococcus, Subdoligranulum, Phascolarctobacterium, Dialister spp, Bifidobacterium breve, Fusicatenibacter saccharivorans, Roseburia intestinalis and Akkermansia muciniphila exhibited reduced abundance. Notably, studies have linked the presence of Parabacteroides distasonis to an increased risk of subsequent JIA occurrence, alongside shorter breastfeeding duration and increased antibiotic exposure, particularly in genetically susceptible populations. Therefore, it can be concluded that environmental stimuli have a stronger effect on genetically predisposed infants and that microbial dysbiosis during infancy may initiate or accelerate the development of JIA. However, t our MR study findings indicate an increase in the abundance of genus Phascolarctobacterium in JIAU, contrasting previous results. This inconsistency may stem from JIAU’s nature as a complication of JIA, potentially exerting different effects on the gut microbiota. However, these findings support the notion that environmental stimuli exert a significant influence. In another cross-sectional investigation, Qian et al. reported that the JIA group exhibited a significant reduction in the relative abundance of four genera (Anaerostipes, didiister, Lachnospira and Roseburia) compared to the control group (18). These four genera are known to produce short-chain fatty acids (SCFAs), whose decrease is associated with severe clinical complications. Additionally, the study identified the genus Lachnospiraceae UCG001 as potentially reducing the risk of JIA. However, contrary to previous findings (18), an increase in the level of genus Roseburia in JIA was observed in our study.
Previous research consistently highlights differences in the gut microbiota composition between children with JIA and their healthy counterparts. However, the causal relationship between gut microbiota and the disease remains unclear. It is uncertain whether microbiota imbalance precedes JIA or arises as a consequence of long-term inflammation, abnormal metabolism or behavioural changes associated with JIA symptoms. Given the limitations of previous studies, including susceptibility to confounding factors and reverse causation effects, further investigation is warranted to elucidate the link between JIA or JIAU and the gut microbiota. In our study, leveraging GWAS data and MR analysis, we investigated the causality between JIA or JIAU and gut microbiota. In addition to our primary findings, we observed an elevated risk of FamilyXI, while genera Rikenellaceae RC9 gut group, Intestinimonas and Clostridium innocuum group were linked to decreased JIA risk. Conversely, a reduced abundance of genus Prevotella7 was associated with a decreased likelihood of JIAU. Furthermore, the abundance of order Burkholderiales, family Alcaligenaceae and genus Anaerofilum increased, while the abundance of genera Olsenella, Coprococcus2, Romboutsia and Eisenbergiella decreased in JIA cases. JIAU decreased the abundance of phylum Lentisphaerae, class Lentisphaeria, order Victivallales, family Peptostreptococcaceae, family Victivallaceae and genus Slackia. These novel findings contribute to existing knowledge by uncovering previously unexplored associations.
Given the pivotal role of gut microbiota in arthritis, targeted probiotics are emerging as a novel therapeutic avenue for rheumatic diseases. Since the mature and stable state of the microbiota once formed is difficult to change, and childhood is a critical period to acquire basic functions (such as immune tolerance to commensal microbiota), these findings present a unique opportunity for early intervention and potentially modifying the disease progression by targeting the microbiota (12). Clinical trials investigating prebiotics and probiotics are on the rise, with an expanding body of evidence reporting favourable tolerance and potential benefits for restoring infant microbiota to health (52, 53). However, a randomised controlled trial of probiotics by Shukl et al. demonstrated good tolerance in patients with Enthesitis-related arthritis (ERA) but did not show any favourable clinical or immunological effects compared to non-steroidal anti-inflammatory medication therapy (54). Therefore, to assess the efficacy and safety of probiotics for JIA, further clinical data are warranted to ensure better recommendations for clinical practice.
Our research offers several advantages. To the best of our knowledge, this is the first investigation to explore the causal relationship between JIA/JIAU and gut microbiota abundance using MR analyses. Relative to other research methods, MR analyses have several advantages in the study of the causal relationship between gut microbiota and JIA or JIAU. First, in terms of evidence-based medicine levels of evidence, MR has the third highest evidence-based rating after systematic reviews of randomised controlled trials (RCTs) and RCTs when RCTs are feasible, and the highest evidence-based rating when RCTs are not feasible, ranking first (55). Whereas, due to practical and ethical reasons, random assignment of specific gut flora cannot be done, RCT studies are not feasible when conducting causal studies of gut flora with JIA and JIAU. Observational study data are relatively more readily available and closer to real-world situations, but problems such as smaller sample sizes, confounding factors, and causal inversions often limit inferences about cause and effect (56), so appropriate causal modelling is needed to infer causal associations between exposure factors and disease outcomes. Animal studies have an even lower evidence-based rating. Therefore, for the study of the causal relationship of gut microbiota with JIA or JIAU, MR studies provide an effective way to solve the above problems. By utilising MR, which relies on the random allocation of allelic genes at conception, we established a temporal relationship (‘cause before effect’) between genetic variations and disease development, which is free from the influence of postnatal environmental factors and social behaviours. This approach minimised the risk of reverse causal linkages and allowed for more effective control of confounding variables. Moreover, to ensure the robustness of the auxiliary variables used in the MR analysis, we sourced GWAS data for gut microbiota, JIA and JIAU from the largest available database. Furthermore, to minimise the possible effects of weak IV bias, we adopted appropriate thresholds for the genomic instruments based on a threshold of p< 1.0×10-5 in this bidirectional study. This criterion was selected based on the availability of an adequate number of SNPs with suitable statistical power for the majority of gut microbiota to effectively prevent confounding.
While our study boasts several strengths, it also has certain limitations. Firstly, although our analysis yields statistically significant p-values, they may not be as robust as those corrected using the Bonferroni method for significance. Nevertheless, our research is hypothesis-driven and supported by substantial biological data and past studies that establish the epidemiological link between gut microbiota and JIA or JIAU. However, future research may need to include samples from a larger population of patients with JIA and JIAU to further strengthen these results. Secondly, adjusting p-values for multiple comparisons may increase the risk of false positives and potentially weaken the number and multi-level structure of microbial communities (abundance and correlation between microbial strains) as well as the correlation between JIA or JIAU. Therefore, cautious interpretation is warranted when considering unfavourable outcomes or potentially significant p-values. Furthermore, the two-sample MR method is a theoretical causal analysis method, and the conclusions of our study may contradict epidemiological studies suggesting the impact of environmental risk factors on JIA or JIAU. Additionally, as the specific mechanisms of the gut microbiome in the onset of JIA or JIAU remain unclear, further research, including clinical studies and other ex vivo and in vivo studies such as animal experiments, is needed to verify the results of this study. New research techniques and methods such as metagenomics and metabolomics technologies, where available, are needed to further validate the results of the present study and to understand the relationship between the observed intestinal flora and JIA, as well as the mechanisms behind the relationship of JIAU. Lastly, as our study primarily includes participants of European descent in the JIA group, generalising our results to other ethnic populations should be done with caution. When GWAS summary data on JIA, JIAU and gut microbiota from other races become available in the future, we also hope to analyse and study them to improve the applicability and generalisability of these findings.
This study presents compelling evidence supporting a causal association between genetically predicted gut microbiota and JIA or JIAU development. This study underscores the significant interactive influence of intestinal flora on these conditions, suggesting their potential as novel biomarkers for diagnosis and prevention. These findings offer valuable insights that can aid in addressing and mitigating the impact of JIA or JIAU.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
J-BH: Conceptualization, Writing – original draft, Writing – review & editing, Formal analysis, Methodology, Validation, Visualization. Y-XC: Formal analysis, Writing – original draft, Writing – review & editing, Methodology, Validation, Visualization. Z-YS: Formal analysis, Validation, Writing – original draft, Writing – review & editing. X-YC: Methodology, Validation, Writing – review & editing. Y-NL: Conceptualization, Validation, Writing – review & editing. J-HY: Conceptualization, Funding acquisition, Writing – review & editing, Validation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Department of Science and Technology of Guangdong Province, under the project ‘Efficacy and safety of the Jianer Jiedu Formula for the treatment of novel coronavirus infections in children-a real-world and randomised controlled study’ (No. 2023B1111020004); the State Administration of Traditional Chinese Medicine, under the project ‘a project for Chinese Medicine on Ying Lv’s Renowned Expert Inheritance Studio’ (No. E43729); Bureau of Science and Technology of Guangzhou Municipality, under the project ‘Mechanism Study on the Regulation of NLRP3-mediated Pyroptosis by Jianer Jiedu Formula for the Treatment of RSV Pneumonia in Children based on the Lingnan DampHeat Theory’ (No. 2024A03J0125); Guangdong Provincial Key Laboratory of Research on Emergency in TCM (No. 2023B1212060062); the Open Fund of Guangdong Provincial Key Laboratory of Research and Development in Traditional Chinese Medicine (No.KFKT02–006); and the 2024 Guangdong Renowned Traditional Chinese Medicine Practitioner Inheritance Studio Construction Project-Xu Youjia (No. 0102018005).
We extend our gratitude to the researchers for generously publishing their valuable data on JIA, JIAU and gut microbiota, which greatly contributed to this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1356414/full#supplementary-material
1. Martini A, Lovell DJ, Albani S, Brunner HI, Hyrich KL, Thompson SD, et al. Juvenile idiopathic arthritis. Nat Rev Dis Primers. (2022) 8(1):5. doi: 1038/s41572-021-00332-8
2. Prakken B, Albani S, Martini A. Juvenile idiopathic arthritis. Lancet. (2011) 377:2138–49. doi: 10.1016/S0140-6736(11)60244-4
3. Dave M, Rankin J, Pearce M, Foster HE. Global prevalence estimates of three chronic musculoskeletal conditions: Club foot, juvenile idiopathic arthritis and juvenile systemic lupus erythematosus. Pediatr Rheumatol Online J. (2020) 18:49. doi: 10.1186/s12969-020-00443-8
4. McHugh J. Global prevalence of jia, jsle and club foot. Nat Rev Rheumatol. (2020) 16:408. doi: 10.1038/s41584-020-0465-6
5. Al-Mayouf SM, Al Mutairi M, Bouayed K, Habjoka S, Hadef D, Lotfy HM, et al. Epidemiology and demographics of juvenile idiopathic arthritis in Africa and Middle East. Pediatr Rheumatol Online J. (2021) 19:166. doi: 10.1186/s12969-021-00650-x
6. Sen ES, Dick AD, Ramanan AV. Uveitis associated with juvenile idiopathic arthritis. Nat Rev Rheumatol. (2015) 11:338–48. doi: 10.1038/nrrheum.2015.20
7. Consolaro A, Giancane G, Alongi A, van Dijkhuizen EHP, Aggarwal A, Al-Mayouf SM, et al. Phenotypic variability and disparities in treatment and outcomes of childhood arthritis throughout the world: An observational cohort study. Lancet Child Adolesc Health. (2019) 3:255–63. doi: 10.1016/S2352-4642(19)30027-6
8. Bansal N, Pasricha C, Kumari P, Jangra S, Kaur R, Singh R. A comprehensive overview of juvenile idiopathic arthritis: From pathophysiology to management. Autoimmun Rev. (2023) 22:103337. doi: 10.1016/j.autrev.2023.103337
9. Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. (2019) 76:473–93. doi: 10.1007/s00018-018-2943-4
10. Behzadi P, Dodero VI, Golubnitschaja O. Systemic inflammation as the health-related communication tool between the human host and gut microbiota in the framework of predictive, preventive, and personalized medicine. In: Wang W, editor. All around Suboptimal Health : Advanced Approaches by Predictive, Preventive and Personalised Medicine for Healthy Populations. Springer Nature Switzerland, Cham (2024). p. 203–41.
11. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The Oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. (2015) 21:895–905. doi: 10.1038/nm.3914
12. De Filippo C, Di Paola M, Giani T, Tirelli F, Cimaz R. Gut microbiota in children and altered profiles in juvenile idiopathic arthritis. J Autoimmun. (2019) 98:1–12. doi: 10.1016/j.jaut.2019.01.001
13. Stoll ML, Kumar R, Morrow CD, Lefkowitz EJ, Cui X, Genin A, et al. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res Ther. (2014) 16:486. doi: 10.1186/s13075-014-0486-0
14. Di Paola M, Cavalieri D, Albanese D, Sordo M, Pindo M, Donati C, et al. Alteration of fecal microbiota profiles in juvenile idiopathic arthritis. Associations with hla-B27 allele and disease status. Front Microbiol. (2016) 7:1703. doi: 10.3389/fmicb.2016.01703
15. Tejesvi MV, Arvonen M, Kangas SM, Keskitalo PL, Pirttila AM, Karttunen TJ, et al. Faecal microbiome in new-onset juvenile idiopathic arthritis. Eur J Clin Microbiol Infect Dis. (2016) 35:363–70. doi: 10.1007/s10096-015-2548-x
16. Aggarwal A, Sarangi AN, Gaur P, Shukla A, Aggarwal R. Gut microbiome in children with enthesitis-related arthritis in a developing country and the effect of probiotic administration. Clin Exp Immunol. (2017) 187:480–9. doi: 10.1111/cei.12900
17. van Dijkhuizen EHP, Del Chierico F, Malattia C, Russo A, Pires Marafon D, Ter Haar NM, et al. Microbiome analytics of the gut microbiota in patients with juvenile idiopathic arthritis: a longitudinal observational cohort study. Arthritis Rheumatol. (2019) 71:1000–10. doi: 10.1002/art.40827
18. Qian X, Liu YX, Ye X, Zheng W, Lv S, Mo M, et al. Gut microbiota in children with juvenile idiopathic arthritis: Characteristics, biomarker identification, and usefulness in clinical prediction. BMC Genomics. (2020) 21:286. doi: 10.1186/s12864-020-6703-0
19. Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet. (2015) 16:327–50. doi: 10.1146/annurev-genom-090314-050016
20. Bae SC, Lee YH. Coffee consumption and the risk of rheumatoid arthritis and systemic lupus erythematosus: a mendelian randomization study. Clin Rheumatol. (2018) 37:2875–9. doi: 10.1007/s10067-018-4278-9
21. Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, et al. The effect of host genetics on the gut microbiome. Nat Genet. (2016) 48:1407–12. doi: 10.1038/ng.3663
22. Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic determinants of the gut microbiome in uk twins. Cell Host Microbe. (2016) 19:731–43. doi: 10.1016/j.chom.2016.04.017
23. Wang J, Thingholm LB, Skieceviciene J, Rausch P, Kummen M, Hov JR, et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. (2016) 48:1396–406. doi: 10.1038/ng.3695
24. Skrivankova VW, Richmond RC, Woolf BAR, Davies NM, Swanson SA, VanderWeele TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (Strobe-Mr): explanation and elaboration. BMJ (2021) 375:n2233. doi: 10.1136/bmj.n2233
25. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the Strobe-Mr statement. JAMA (2021) 326(16):1614–21. doi: 10.1001/jama.2021.18236
26. Lopez-Isac E, Smith SL, Marion MC, Wood A, Sudman M, Yarwood A, et al. Combined genetic analysis of juvenile idiopathic arthritis clinical subtypes identifies novel risk loci, target genes and key regulatory mechanisms. Ann Rheum Dis (2021) 80(3):321–8. doi: 10.1136/annrheumdis-2020-218481
27. McIntosh LA, Marion MC, Sudman M, Comeau ME, Becker ML, Bohnsack JF, et al. Genome-wide association meta-analysis reveals novel juvenile idiopathic arthritis susceptibility loci. Arthritis Rheumatol (2017) 69(11):2222–32. doi: 10.1002/art.40216
28. Haasnoot AJW, Schilham MW, Kamphuis S, Hissink Muller PCE, Heiligenhaus A, Foell D, et al. Identification of an amino acid motif in Hla-Drbeta1 that distinguishes uveitis in patients with juvenile idiopathic arthritis. Arthritis Rheumatol (2018) 70(7):1155–65. doi: 10.1002/art.40484
29. Li R, Guo Q, Zhao J, Kang W, Lu R, Long Z, et al. Assessing causal relationships between gut microbiota and asthma: Evidence from two sample mendelian randomization analysis. Front Immunol. (2023) 14:1148684. doi: 10.3389/fimmu.2023.1148684
30. Chen S, Han H, Sun X, Zhou G, Zhou Q, Li Z. Causal effects of specific gut microbiota on musculoskeletal diseases: a bidirectional two-sample mendelian randomization study. Front Microbiol. (2023) 14:1238800. doi: 10.3389/fmicb.2023.1238800
31. Wu J, Zhang B, Zhou S, Huang Z, Xu Y, Lu X, et al. Associations between gut microbiota and sleep: a two-sample, bidirectional mendelian randomization study. Front Microbiol. (2023) 14:1236847. doi: 10.3389/fmicb.2023.1236847
32. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for mendelian randomization studies using multiple genetic variants. Int J Epidemiol. (2011) 40:740–52. doi: 10.1093/ije/dyq151
33. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using gwas summary data. PLoS Genet. (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
34. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
35. OYJS MendelianRandomization: Mendelian Randomization Package R package version 070 (2023). Available online at: https://CRANR-projectorg/package=MendelianRandomization.
36. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. (2013) 37:658–65. doi: 10.1002/gepi.21758
37. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
38. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. (2017) 46:1985–98. doi: 10.1093/ije/dyx102
39. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The mr-base platform supports systematic causal inference across the human phenome. Elife. (2018) 7. doi: 10.7554/eLife.34408
40. International Genetics of Ankylosing Spondylitis C, Cortes A, Hadler J, Pointon JP, Robinson PC, Karaderi T, et al. Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat Genet. (2013) 45:730–8. doi: 10.1038/ng.2667
41. Vitale AT, Graham E, de Boer JH. Juvenile idiopathic arthritis-associated uveitis: clinical features and complications, risk factors for severe course, and visual outcome. Ocul Immunol Inflammation. (2013) 21:478–85. doi: 10.3109/09273948.2013.815785
42. Runstadler JA, Saila H, Savolainen A, Leirisalo-Repo M, Aho K, Tuomilehto-Wolf E, et al. Hla-drb1, tap2/tap1, and hla-dpb1 haplotypes in finnish juvenile idiopathic arthritis: more complexity within the mhc. Genes Immun. (2004) 5:562–71. doi: 10.1038/sj.gene.6364129
43. Ombrello MJ, Remmers EF, Tachmazidou I, Grom A, Foell D, Haas JP, et al. Hla-drb1*11 and variants of the mhc class ii locus are strong risk factors for systemic juvenile idiopathic arthritis. Proc Natl Acad Sci USA. (2015) 112:15970–5. doi: 10.1073/pnas.1520779112
44. De Benedetti F, Meazza C, Vivarelli M, Rossi F, Pistorio A, Lamb R, et al. Functional and prognostic relevance of the -173 polymorphism of the macrophage migration inhibitory factor gene in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. (2003) 48:1398–407. doi: 10.1002/art.10882
45. Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet. (2013) 45:664–9. doi: 10.1038/ng.2614
46. Scher JU. Intestinal dysbiosis and potential consequences of microbiome-altering antibiotic use in the pathogenesis of human rheumatic disease. J Rheumatol. (2015) 42:355–7. doi: 10.3899/jrheum.150036
47. Lin YT, Wang CT, Gershwin ME, Chiang BL. The pathogenesis of oligoarticular/polyarticular vs systemic juvenile idiopathic arthritis. Autoimmun Rev. (2011) 10:482–9. doi: 10.1016/j.autrev.2011.02.001
48. Xin L, He F, Li S, Zhou ZX, Ma XL. Intestinal microbiota and juvenile idiopathic arthritis: current understanding and future prospective. World J Pediatr. (2021) 17:40–51. doi: 10.1007/s12519-020-00371-3
49. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife. (2013) 2:e01202. doi: 10.7554/eLife.01202
50. Malin M, Verronen P, Mykkanen H, Salminen S, Isolauri E. Increased bacterial urease activity in faeces in juvenile chronic arthritis: evidence of altered intestinal microflora? Br J Rheumatol. (1996) 35:689–94. doi: 10.1093/rheumatology/35.7.689
51. Kindgren E, Ahrens AP, Triplett EW, Ludvigsson J. Infant gut microbiota and environment associate with juvenile idiopathic arthritis many years prior to disease onset, especially in genetically vulnerable children. EBioMedicine. (2023) 93:104654. doi: 10.1016/j.ebiom.2023.104654
52. Millar M, Seale J, Greenland M, Hardy P, Juszczak E, Wilks M, et al. The microbiome of infants recruited to a randomised placebo-controlled probiotic trial (Pips trial). EBioMedicine. (2017) 20:255–62. doi: 10.1016/j.ebiom.2017.05.019
53. Panigrahi P, Parida S, Nanda NC, Satpathy R, Pradhan L, Chandel DS, et al. A randomized synbiotic trial to prevent sepsis among infants in Rural India. Nature. (2017) 548:407–12. doi: 10.1038/nature23480
54. Shukla A, Gaur P, Aggarwal A. Effect of probiotics on clinical and immune parameters in enthesitis-related arthritis category of juvenile idiopathic arthritis. Clin Exp Immunol. (2016) 185:301–8. doi: 10.1111/cei.12818
55. Arsenault BJ. From the garden to the clinic: How mendelian randomization is shaping up atherosclerotic cardiovascular disease prevention strategies. Eur Heart J. (2022) 43:4447–9. doi: 10.1093/eurheartj/ehac394
Keywords: juvenile idiopathic arthritis, uveitis, gut microbiota, causality, bidirectional, Mendelian randomisation analysis
Citation: Hong J-b, Chen Y-x, Su Z-y, Chen X-y, Lai Y-n and Yang J-h (2024) Causal association of juvenile idiopathic arthritis or JIA-associated uveitis and gut microbiota: a bidirectional two-sample Mendelian randomisation study. Front. Immunol. 15:1356414. doi: 10.3389/fimmu.2024.1356414
Received: 15 December 2023; Accepted: 04 July 2024;
Published: 24 July 2024.
Edited by:
Servaas Antonie Morré, VU Medical Center, NetherlandsReviewed by:
Payam Behzadi, Islamic Azad University, ShahreQods, IranCopyright © 2024 Hong, Chen, Su, Chen, Lai and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-ni Lai, bGFpeWFubmkxMjA0QDE2My5jb20=; Jing-hua Yang, ZG91bWlhb21hbWFAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.