
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 25 April 2024
Sec. Multiple Sclerosis and Neuroimmunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1347415
Objective: Emerging evidence has shown that gut diseases can regulate the development and function of the immune, metabolic, and nervous systems through dynamic bidirectional communication on the brain-gut axis. However, the specific mechanism of intestinal diseases and vascular dementia (VD) remains unclear. We designed this study especially, to further clarify the connection between VD and inflammatory bowel disease (IBD) from bioinformatics analyses.
Methods: We downloaded Gene expression profiles for VD (GSE122063) and IBD (GSE47908, GSE179285) from the Gene Expression Omnibus (GEO) database. Then individual Gene Set Enrichment Analysis (GSEA) was used to confirm the connection between the two diseases respectively. The common differentially expressed genes (coDEGs) were identified, and the STRING database together with Cytoscape software were used to construct protein-protein interaction (PPI) network and core functional modules. We identified the hub genes by using the Cytohubba plugin. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were applied to identify pathways of coDEGs and hub genes. Subsequently, receiver operating characteristic (ROC) analysis was used to identify the diagnostic ability of these hub genes, and a training dataset was used to verify the expression levels of the hub genes. An alternative single-sample gene set enrichment (ssGSEA) algorithm was used to analyze immune cell infiltration between coDEGs and immune cells. Finally, the correlation between hub genes and immune cells was analyzed.
Results: We screened 167 coDEGs. The main articles of coDEGs enrichment analysis focused on immune function. 8 shared hub genes were identified, including PTPRC, ITGB2, CYBB, IL1B, TLR2, CASP1, IL10RA, and BTK. The functional categories of hub genes enrichment analysis were mainly involved in the regulation of immune function and neuroinflammatory response. Compared to the healthy controls, abnormal infiltration of immune cells was found in VD and IBD. We also found the correlation between 8 shared hub genes and immune cells.
Conclusions: This study suggests that IBD may be a new risk factor for VD. The 8 hub genes may predict the IBD complicated with VD. Immune-related coDEGS may be related to their association, which requires further research to prove.
Vascular dementia (VD), the second major category of dementia, also known as vascular cognitive impairment (VCI), is mainly manifested as a progressive and fluctuating decline in memory, language, computation, visuospatial ability, attention, and power of execution (1). VD is caused by multiple factors, which can lead to neurovascular injury, further reducing cerebral blood flow, and cognitive dysfunction. Its main pathogenesis includes a neuroinflammatory response, oxidative stress, destruction of the blood-brain barrier (BBB), uncoupling of neurovascular units, destruction of mitochondrial morphology and function, demyelination and myelin injury, neurotoxicity, etc (2). The absence of well-established and universally accepted neuropathological criteria, along with the diversity and overlapping definitions of cerebrovascular lesions, pose challenges in determining the true prevalence of pure VCI in autopsy studies (3). However, the evidence has shown that the cognitive impairment of VD is more severe in comparison to other neurodegenerative diseases (4). An epidemiological investigation showed that there was significantly higher rate in hospitalization and mortality of VCI patients than those of non-VCI patients, similar to that of Alzheimer’s disease (AD) patients (5).
The evidence that intestinal diseases can affect nervous system function, such as depression, anxiety, dementia, Parkinson’s disease (PD), through the brain-gut axis (6), has been verified, and this relationship may be bidirectional (7–11). Inflammatory bowel disease (IBD), including crohn’s disease (CD) and ulcerative colitis (UC), is a chronic, relapsing, immune-mediated intestinal disease (6, 7). It is characterized by the overlapping effects of genetic mutations (12), environmental factors (13), intestinal microbiota disorders (14), and immune factors (15). The connection between IBD and the nervous system has been extensively studied. For example, a clinical research showed that the prevalence of anxiety and depression in IBD patients is 32.1% and 25.2% respectively (16). Similarly, patients with depression are more commonly diagnosed with IBD, and psychiatric symptoms are significantly associated with adverse disease activity-related outcomes in IBD (17). Furthermore, the relationship between IBD and PD is also bidirectional due to genetic overlap, abnormal transfer of α-synuclein from the gastrointestinal tract to the central nervous system (CNS), and neuroinflammatory responses. This connection has been corroborated by multiple studies and experiments (18–20). In addition, studies on dementia from the direction of the brain-gut axis involve many aspects, such as epidemiology, pathogenesis, and prognosis (21). For example, an epidemiological study showed that IBD might not only be a risk factor for dementia, but also lead to an earlier age of dementia. This was consistent with another study, revealing a positive association between IBD and dementia risk, particularly in patients with UC (8). In addition, a meta-study also uncovered an increased long-term risk of dementia in IBD patients (22). However, the association mechanism between IBD and dementia may be multifactor, which may be related to chronic inflammation, accelerated atherosclerosis, and hypercoagulability (23), especially thromboembolic events, thrombocytosis, hyperlipidemia, and hyperhomocysteinemia. All these factors may further aggravate the occurrence and development of VD (24). On the one hand, a study had shown that the gut microbes might affect neuroinflammatory responses through microglia (25). On the other hand, gut microbes could affect the generation ability of hippocampal neurons through metabolism, endocrine signaling, and nervous system driving mechanisms (26–28), thus affecting cognitive function. These findings uncover a fact that gut microbes can influence the development and impairment of cognitive function.
At present, the brain-gut axis not only connects the brain to the gut itself, but also represents the interaction between the brain-gut axis system and the central nervous system (CNS), endocrine chemical signaling system, immune regulation, microbiome, metabolism, and brain-gut barrier function (29) (Figure 1). Although extensive evidence has shown that gut diseases are closely associated with dementia, the underlying mechanisms have not been fully elucidated. In particular, limited research has been done to discover the relationship between VD and intestinal disease. Based on bioinformatics technology, this study explores the internal relationship between VD and IBD by means of searching for common differentially expressed genes (coDEGs) in VD brain tissue and UC colon tissue. The shared gene signatures identified are expected to elaborate the disease-related biological mechanism. The exploration of the common biological pathways can provide a new perspective for further study and treatment.
The overall design of the study is illustrated in Figure 2.
Gene expressions of VD and UC were downloaded from the GEO (http://www.ncbi.nlm.nih.gov/geo/) database with the type of microarray. The independent datasets of GSE122063 for VD, including 36 tissues from 8 VD patients and 44 tissues from 11 healthy persons from hippocampal and frontal lobes, and GSE47908 for UC, including 19 UC tissues and 15 healthy tissues from human colonic biopsies were download for better investigating the mechanistic relationship between VD and UC. The UC validation datasets GSE179285 contained 70 inflammatory colon tissues from IBD, 153 non-inflammatory colon tissues from IBD, and 31 colon tissues from healthy controls. After obtaining the gene expression matrix, the platform information was used for gene annotation.
Based on the bioinformatics analysis tools in Xiantao Academic website (https://www.xiantaozi.com/), we used the ClusterProfiler package in R software to implement Gene Set Enrichment Analysis (GSEA) analysis. The two data sets were enriched with reference to gene set c2.cp. kegg.v2022.1.Hs.symbols.Gmt (KEGG Pathway Database) to determine the GSEA KEGG pathways of VD and UC. The ggplot2 package was constructed to perform the classical visualization of GSEA graphs for the enrichment analysis results.
Based on the bioinformatics analysis tools in Xiantao Academic website, we used the package “limma” by implementing the R software to normalize the data obtained from GEO. Statistically differentially expressed genes (DEGs) were defined with adjusted p-value < 0.05 (P.adj < 0.05) and |log2 (fold change)| > 0.58 using limma package. The up-regulated and down-regulated DEGs of VD and UC were identified respectively using the limma package in R. The DEGs were visualized by volcano plots and heatmaps respectively. The ggplot2 package were used to generate the volcano plots of DEGs for VD and UC. Heatmaps were depicted using the ComplexHeatmap packages. Genes that were up-regulated or down-regulated in both VD and UC were respectively considered as up-coDEGs and down-coDEGs. The overlapped DEGs in the two datasets were detected using the VennDiagram R package.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for the selected coDEGs was carried out by using the R ClusterProfiler package. After ID conversion of the input molecule list, enrichment analysis was performed using the ClusterProfiler package, and the top 10 main pathways of biological processes (BP), cellular components (CC) and KEGG were visualized. We defined the threshold set for the coDEGs with FDR (q value) < 0.01 and P.adj < 0.05 as statistically significant.
The STRING database (https://string-db.org/) was used to search for molecular interactions and predict protein interactions. The PPI network of 167 coDEGs in VD and UC was constructed, and the reference value was set to confidence score > 0.4. Cytoscape (version 3.9.1) software was used to further visualize the PPI network diagram. The molecular complex detection (MCODE) plugin in Cytoscope was used to pick out PPI interacting core gene clusters in coDEGs. The screening criteria for key modules were MCODE score > 5, degree cut-off = 2, node score cut-off = 0.2, Max depth = 100, k-score = 2. By using the CytoHubba plugin of Cytoscape software, the top 10 key genes were selected from 167 coDEGs by applying the maximum cross correlation (MCC) algorithm, maximum neighborhood component (MNC) algorithm, Degree algorithm, and edge permeability component (EPC) algorithm. Then, the intersection of the VennDiagram R package was used to screen out the hub genes.
The GO and KEGG enrichment analysis of hub genes was performed using the ClusterProfiler package, and the results were visualized using the ggplot2 package. We used the qROC package in R software to draw a receiver operating characteristic (ROC) curve to evaluate the diagnostic value of the hub genes. We used the area under the curve (AUC) with 95% confidence intervals (CI) to assess the levels of hub genes respectively on VD and UC. In addition, the validation datasets GSE179285 were used for external verification of the shared hub genes, and the Mann-Whitney U test (Wilcoxon rank sum test) test was performed on hub genes according to the expression profile data characteristics to further verify the expression level of diagnostic biomarker genes.
The immune infiltration analysis was performed by single sample gene set enrichment analysis (ssGSEA) in the GSVA package for further verifying the results of bioinformatics analysis and exploring the potential relationship between the two Diseases. The background gene sets of 782 marker genes in 28 kinds of immune cells were constructed. The infiltration levels of VD and UC gene sets in 28 kinds of immune cells were calculated, and the infiltration levels of 16 kinds of immune cells were visualized. Combined with the data analysis of immune cells and immune functions, the heatmap software package was applied to draw the clustering heatmap of data sets and immune cells to show the infiltration degree of different immune cells in different samples. The ggplot2 package in R was used to draw a group box plot to compare the expression of different types of immune cells in the samples of the disease group and the control group. Meanwhile, the T test or Wilcox rank sum test was used to compare the differences between the groups, and there was a statistical difference when P < 0.05.
Finally, we extracted the expression matrix of the hub gene from the gene expression matrix, combined it with our immune cell abundance matrix, calculated the correlation coefficient and p-value, defined P < 0.05 as statistically significant differences, drew the correlation heatmap of immune cells and hub gene, and used the heatmap software package in R software for visual analysis.
The gene set of GSEA KEGG pathway reference MSigDB KEGG database (https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp), P. adj < 0.05 and FDR < 0.25 was considered statistically significant. In VD, 40 pathways were enriched, while 46 pathways were in UC, and there were 18 intersection pathways between the two diseases. Interestingly, among these intersecting KEGG categories, immunization items accounted for the largest proportion 44.44%, including immune system pathways at 33.33 and human immune diseases at 11.11. Among the enriched pathways of human diseases, the common enrichment pathways included immune diseases with systemic lupus erythematosus and organ rejection disease, neurological degeneration with PD, and other diseases, including viral myocarditis, leishmaniasis, and tumor pathways. The pathways enriched in biological organic systems include IgA-producing intestinal immune network, chemokine signaling pathway, toll-like receptor signaling pathway, nod-like receptor signaling pathway, complement and coagulation cascade, as well as natural killer cell-mediated cytotoxicity, all belonging to the immune system. Pathways enriched for environmental information processing include cytokine-cytokine receptor interactions, cell adhesion molecules, and JAK-STAT signaling pathways. In metabolism, we also enriched three common significant pathways, including oxidative phosphorylation, drug metabolism-cytochrome P450, and degradation of valine, leucine, and isoleucine (Figures 3A–F).
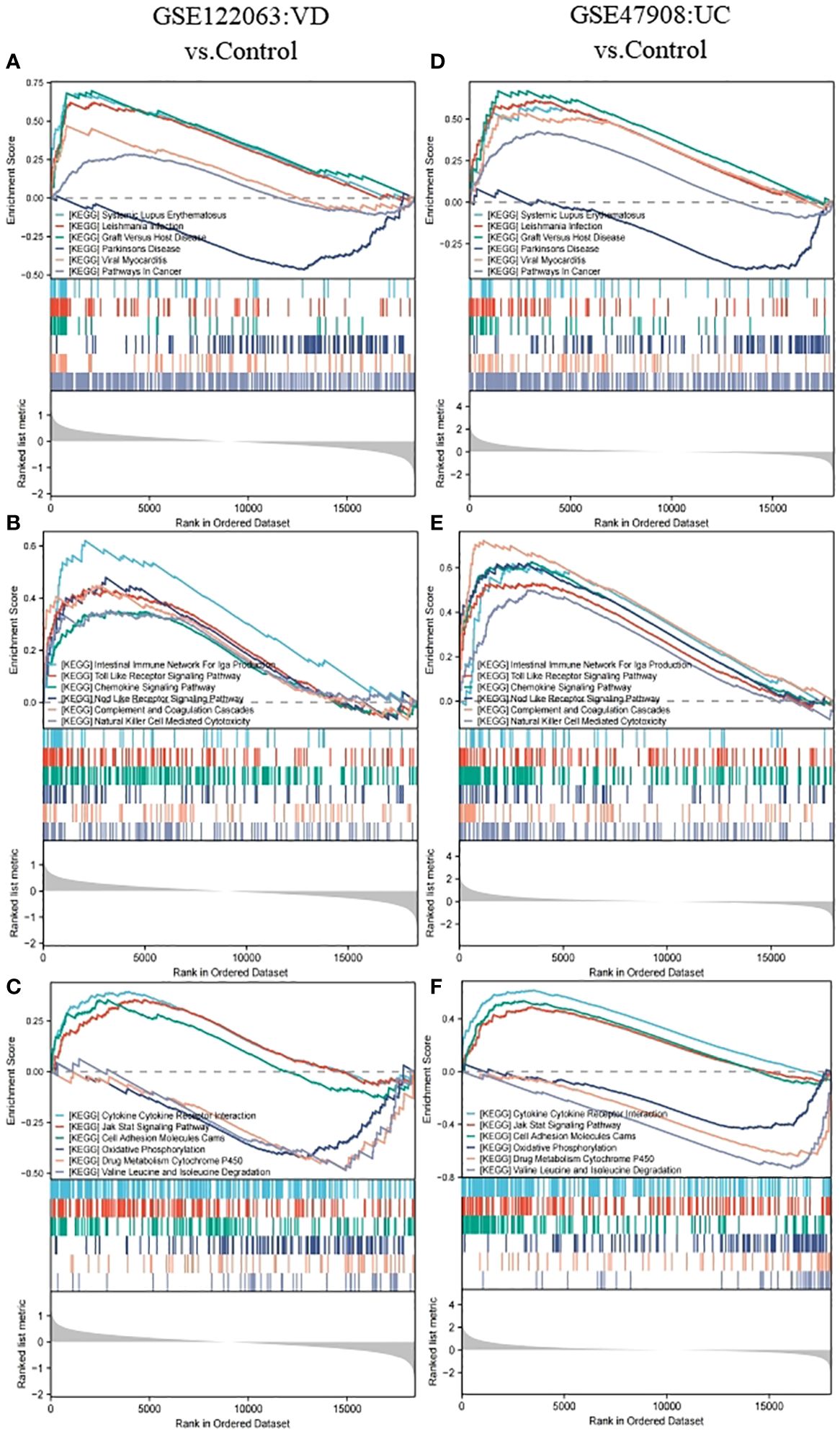
Figure 3 GSEA analysis for GSE122063 (VD) and GSE47908 (UC). (A, D) The genes of VD and UC were both enriched in terms of human diseases. (B, E) The genes of VD and UC were both enriched in terms of biological organic systems. (C, F) The genes of VD and UC were both enriched in terms of metabolism and environmental information processing.
Based on the GSE122063 datasets, 2446 DEGs were picked out between VD patients and healthy controls (1010 up-regulated DEGs, 1436 down-regulated DEGs) (Figures 4A, C). Based on the GSE47908 datasets, 2715 DEGs were distinguished from UC patients and healthy controls (1914 up-regulated DEGs, 801 down-regulated DEGs) (Figures 4B, D). Finally, we identified 167 co-DEGs in VD and UC, including 126 up-regulated coDEGs and 41 down-regulated coDEGs (Figure 4E). This suggests that there may be molecular mechanistic similarities between VD and UC.
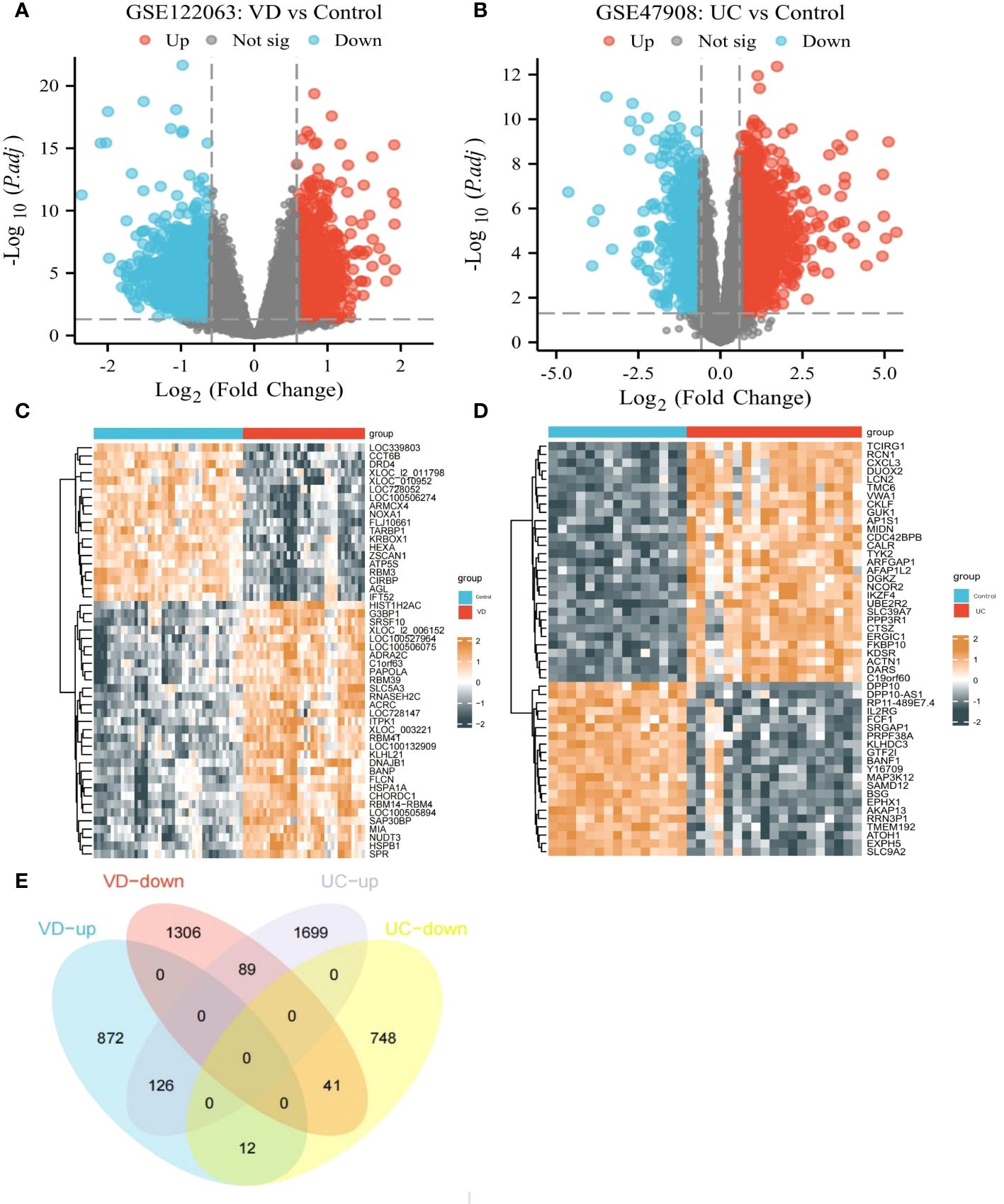
Figure 4 CoDEGs in VD and UC vs Control (healthy persons). (A) Volcano plots of DEGs from GSE122063. (B) Volcano plots of DEGs from GSE47908. (C) Heatmap of DEGs from GSE122063, including 36 tissues from VD and 44 tissues from healthy persons. (D) Heatmap of DEGs from GSE47908, including 19 UC tissues and 15 healthy tissues. (E) Venn diagram of co-DEGs extracted from DEGs of GSE122063 and GSE47908.
In order to better explain the biological functions of the shared genes, GO and KEGG pathway annotations were used to describe and analyze the 167 coDEGs screened in the above steps. GO was used to identify the top 10 significantly different BP, CC, and MF entries. KEGG pathway annotation identified the top 10 significantly enriched pathways of our interest.
Many of the GO/KEGG entries we enriched were related to immunity. In the BP category, genes were mainly concentrated in positive regulation of cytokine production, positive regulation of response to external stimulus, mononuclear cell differentiation, leukocyte cell-cell adhesion, regulation of leukocyte differentiation, regulation of leukocyte cell-cell adhesion, leukocyte mediated immunity, lymphocyte-mediated immunity, regulation of hemopoiesis and regulation of lymphocyte proliferation (Figure 5A). For the CC ontology, they were mainly located in the secretory granular membrane, cell-dissolved particles, and the outer part of the plasma membrane, and immune receptor activity was mainly identified in MF analysis (Figure 5B). KEGG genes were enriched in human immune system diseases such as inflammatory bowel disease and rheumatoid arthritis, as well as infectious diseases such as leishmaniasis, legionellosis, and malaria, and in the organic immune system, neutrophil extracellular trap formation was enriched. Phagosomes in cellular processes were also identified (Figure 5C).
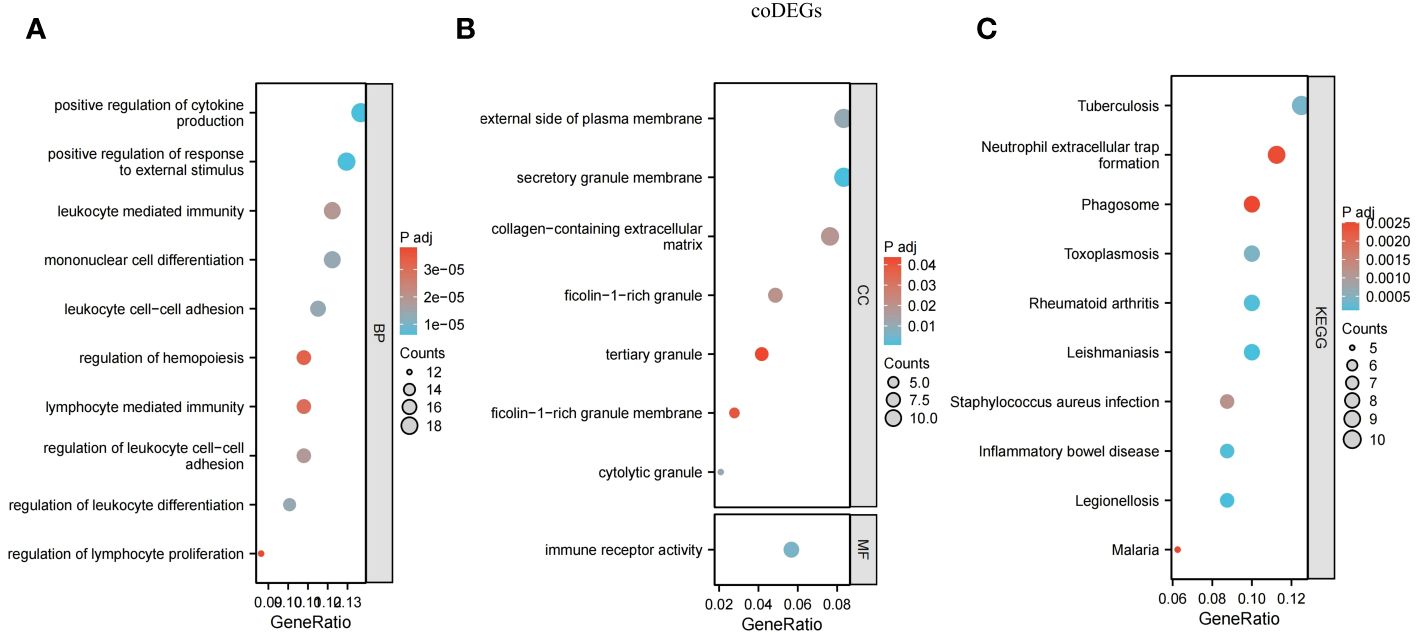
Figure 5 Functional annotation for coDEGs. (A) Biological process. (B) Cellular component and molecular function. (C) KEGG analysis. The size of the circle represents the number of genes enriched, and the larger the circle, the more genes enriched.
The 167 coDEGs analyzed in the above steps were located in the PPI interaction network to further explore their internal interactions in STRING database, with 157 nodes and 419 edges in total. 104 out of 167 genes were included in the network, including 85 out of 126 up-regulated genes and 19 out of 41 down-regulated genes. The functions of the rest of 63 genes have not yet been reported. Among them, purple nodes were up-regulated coDEGs, and blue nodes were down-regulated coDEGs. Node size was adjusted according to the value of connectivity. The higher the connectivity of nodes, the greater the weight of nodes in the PPI network diagram. As shown in Figure 6A, the top 10 gene with CASP1, ITGAL, IL18, IL10RA, CYBB, TLR2, BTK, ITGB2, IL1B, and PTPRC were picked out. Their weights were ≥20, all of which were up-regulated coDEGs. MCODE plug-in was used to screen and analyze the key modules of PPI. A total of 5 submodules were screened, and enrichment analysis was carried out in GO. Submodule 1 had 10 nodes and 41 edges (Figure 6B), belonging to NADPH oxidase complex, and its functions in GO enrichment analysis were as follows: positive regulation of helper T cell 1 factor, activation of the oxide synthesis process, active regulation of neuroinflammatory response, positive regulation of T-helper 1 cell cytokine production and reactive oxygen species biosynthetic process, positive regulation of granulocyte-macrophage colony-stimulating factor production). Submodule 2 had 9 nodes and 19 edges (Figure 6C), and the related functions were not enriched. Submodule 3 had 16 nodes and 31 edges (Figure 6D), which were related to the regulation of immune processes in GO enrichment analysis, including T cell activation, regulation of phagocytosis, and positive regulation of lymphocyte-mediated immunity. Submodule 4 had 5 nodes, and 8 edges (Figure 6E), GO enrichment analysis was related to respiratory electron transport chain, oxidative phosphorylation, and cellular respiration. Submodule 5 had 3 nodes and 3 edges (Figure 6F) and functions in the WikiPathway were related to osteoblast differentiation and related diseases.
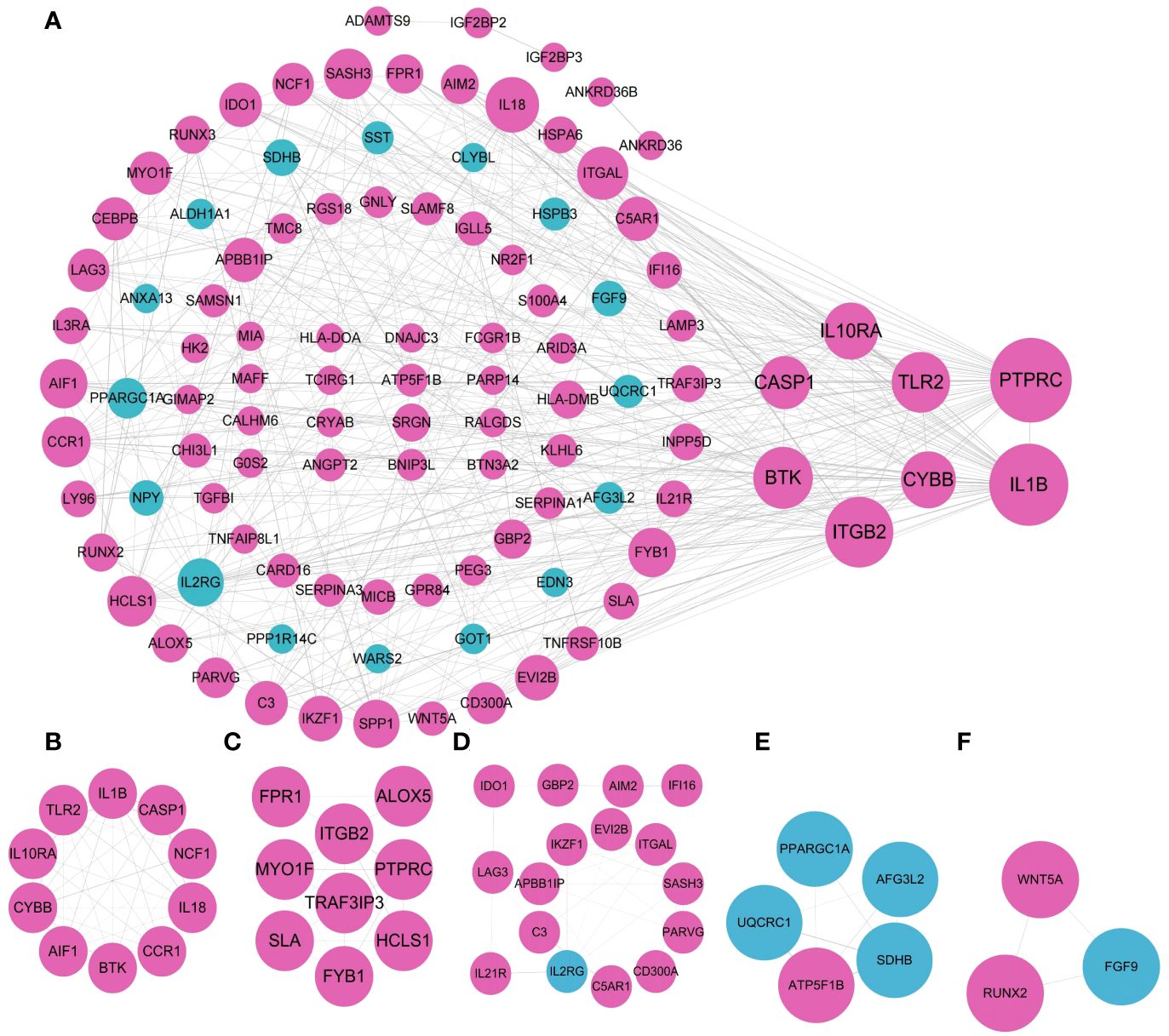
Figure 6 Visualization of the PPI network and the important modules. (A) PPI network. (B–F) submodule 1-5.
The 8 hub genes were obtained via the CytoHubba plugin of Cytoscape software: PTPRC, ITGB2, CYBB, IL1B, TLR2, CASP1, IL10RA, BTK (Figures 7A, B). GO/KEGG enrichment analysis of hub genes suggested that the biological processes (BP) mainly focused on the immune and inflammatory responses mediated by leukocytes, neuroinflammatory response, glial cell activation and regulation of inflammatory response, bone marrow leukocyte activation, and regulation of immune effectors. The pathways enriched in KEGG mainly included leishmaniosis, lipid and atherosclerosis, rheumatoid arthritis in the human disease group, and the obvious immune system pathways enriched in neutrophil extracellular trap formation, NOD-like receptor signaling pathway, and primary immunodeficiency disease (Figure 7C).
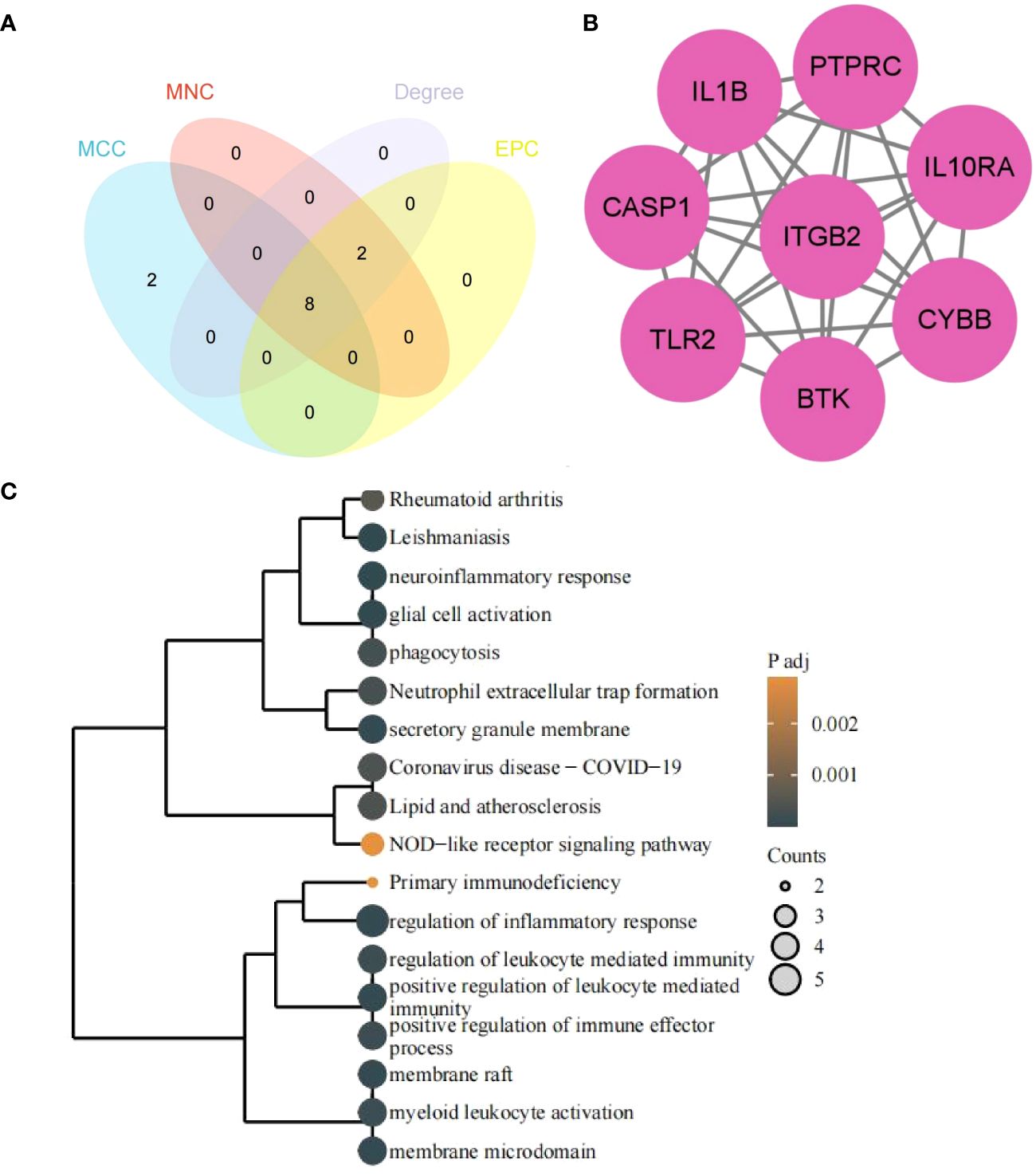
Figure 7 Hub genes and functional enrichment analysis. (A) Hub genes form four algorithms. (B) PPI network of hub genes. (C) GO/KEGG enrichment analysis of hub genes. The size of the circle represents the number of genes enriched, and the larger the circle, the more genes enriched.
ROC analyses were performed for VD and UC respectively to evaluate the ability of diagnosis of the 8 hub genes. In the area under the curve (AUC) of 8 hub genes used to discriminate patients from healthy controls in VD datasets, the AUC area was greater than 0.674 (Figure 8A). But in UC datasets, they were greater than 0.768 (Figure 8B). The ROC curves recommended that the 8 hub genes had the ability to predict the risk of UC with VD. The results provide a basis for the development of novel targeted therapies for VD and UC diseases with these hub genes.
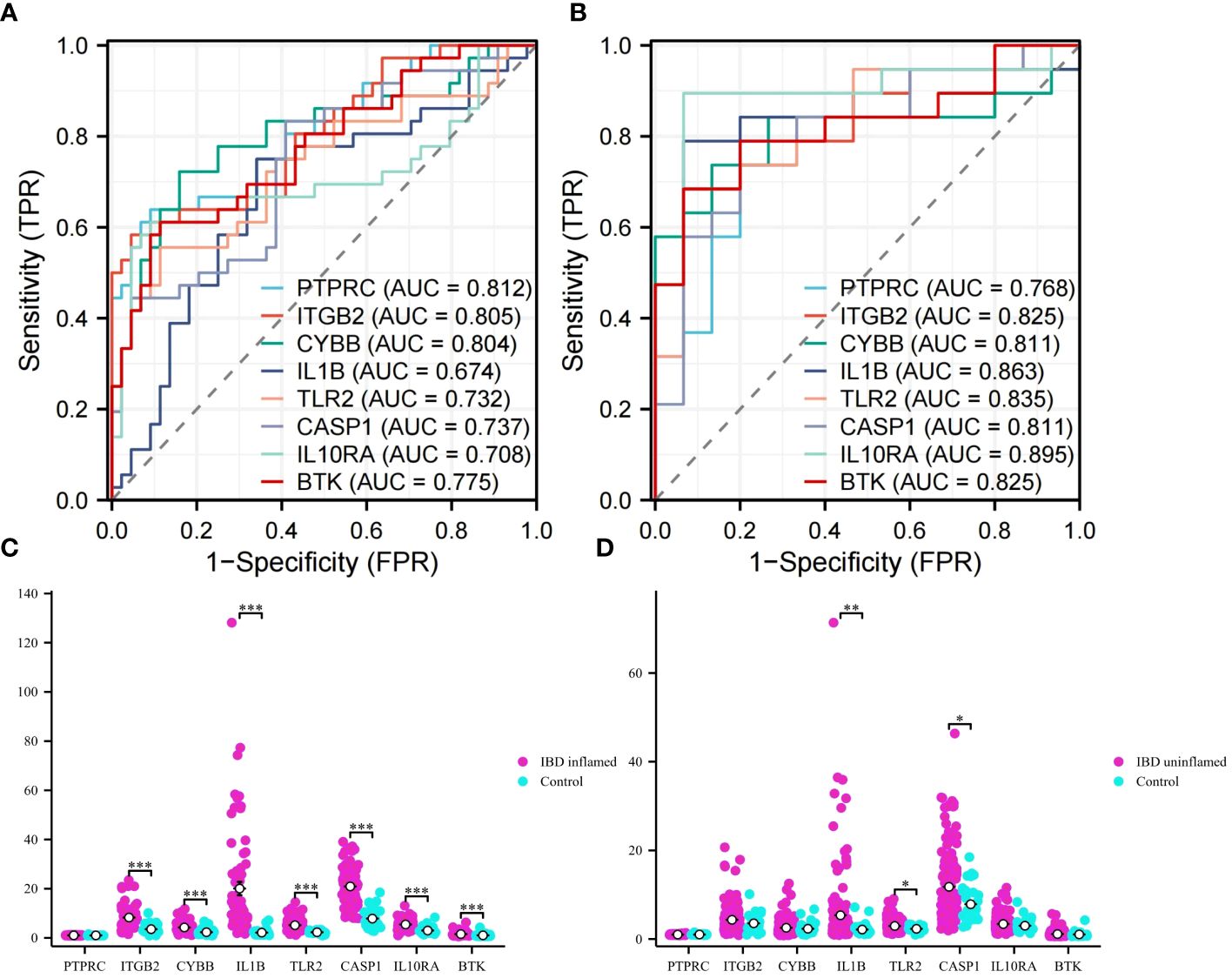
Figure 8 ROC analysis and validation. (A) ROC curves of hub genes in VD. (B) ROC curves of hub genes in UC. (C) Validation of expression levels in IBD inflammation datasets. (D) Validation of expression levels in IBM no inflammation dataset. This is a statistically significant marker that *P<0.05, **P<0.01, and ***P<0.001.
To obtain more accurate and reliable results, the UC external validation datasets were applied to verify the expression levels of 8 hub genes. It was found that the expression levels of genes ITGB2, CYBB, IL1B, TLR2, CASP1, IL10RA, and BTK in the samples of the IBD inflammation datasets were significantly higher than those in the healthy control (Figure 8C, P < 0.01). The results showed that these hub genes might have high diagnostic ability as characteristic biomarkers to predict VD in IBD patients. In addition, we verified the unaffected colon tissues of IBD patients in the validation datasets, and the results showed that IL1B, TLR2, and CASP1 genes had early diagnostic effects (Figure 8D, P < 0.05). Due to the lack of training datasets for VD, we had to abandon the validation on the VD datasets.
The algorithm of ssGSEA was used to quantify the distribution (Figures 9A, B) and relative proportions (Figures 10A, B) of the relative infiltration levels of 16 immune cells picked out from 28 immune cells in the GSE122063 and GSE47908 datasets. The correlation between the 8 hub genes and immune cell infiltration was analyzed (Figures 11A, B). The results could help us to better evaluate the connection about the immune pathways for the brain-gut axis between diseases and healthy controls.
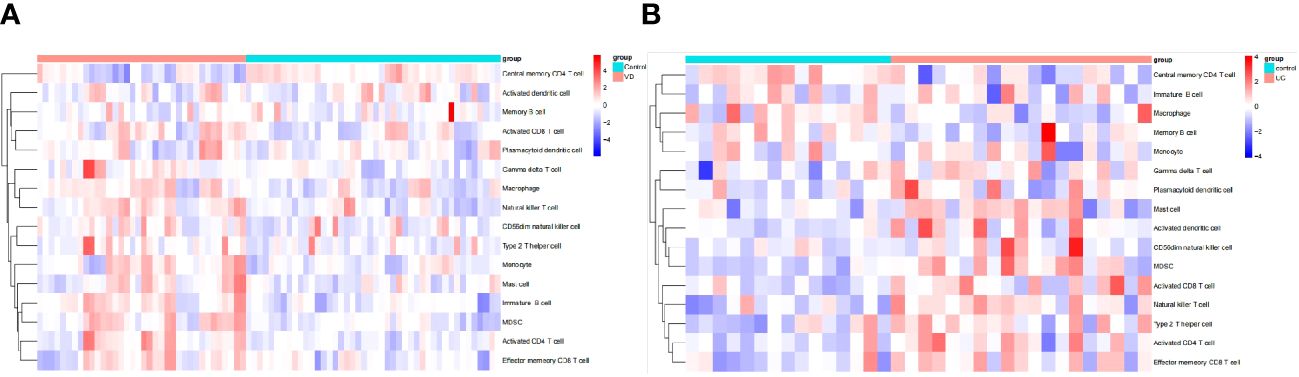
Figure 9 Immune cell infiltration analysis in VD and UC. (A) Hierarchical clustering of the distribution of the 16 immune cells in VD. (B) Hierarchical clustering of the distribution of the 16 immune cells in UC.
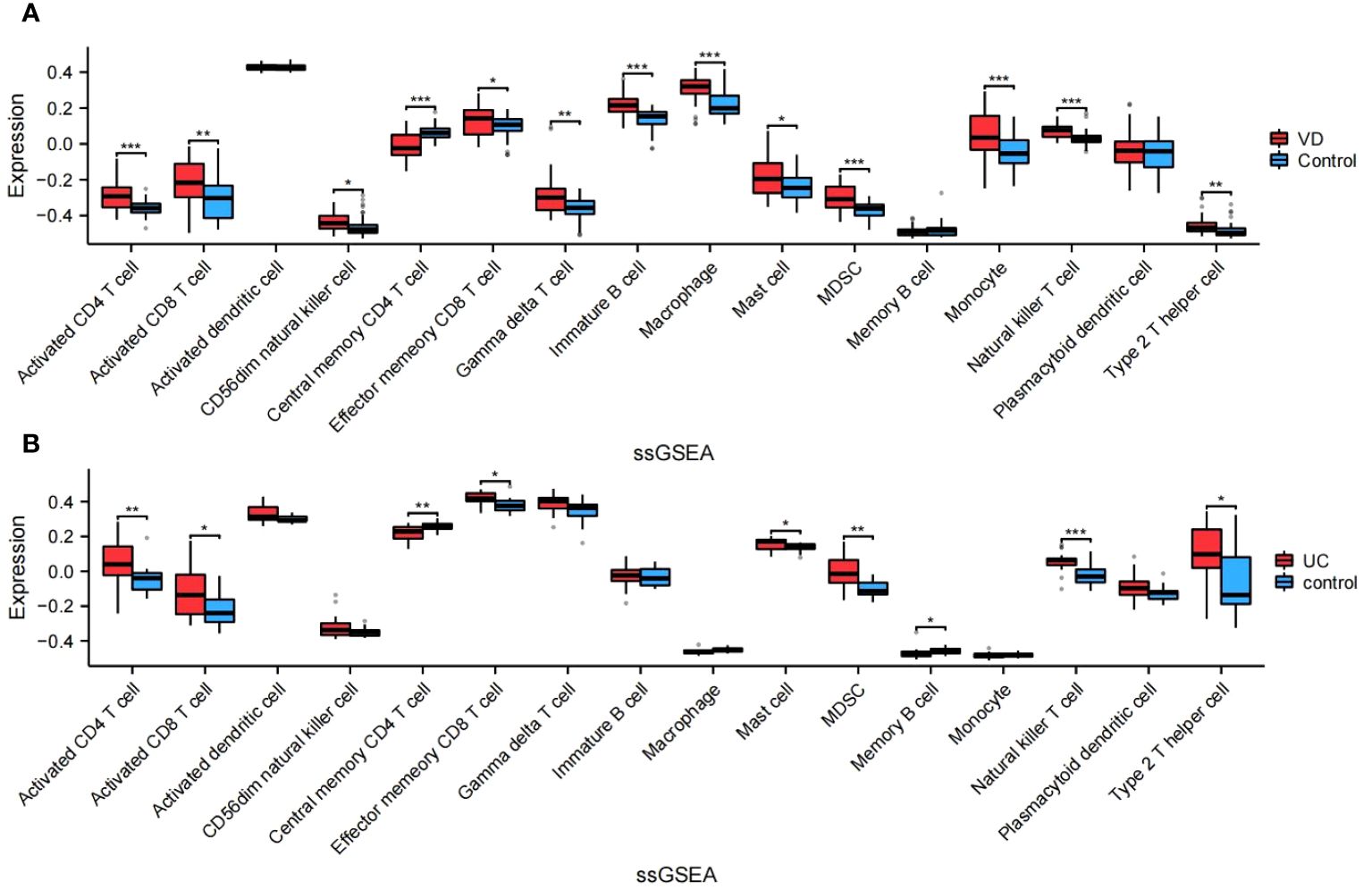
Figure 10 Relative proportions of the relative infiltration levels in VD and UC. (A) Relative proportions in VD. (B) Relative proportions in UC. This is a statistically significant marker that *P<0.05, **P<0.01, and ***P<0.001.
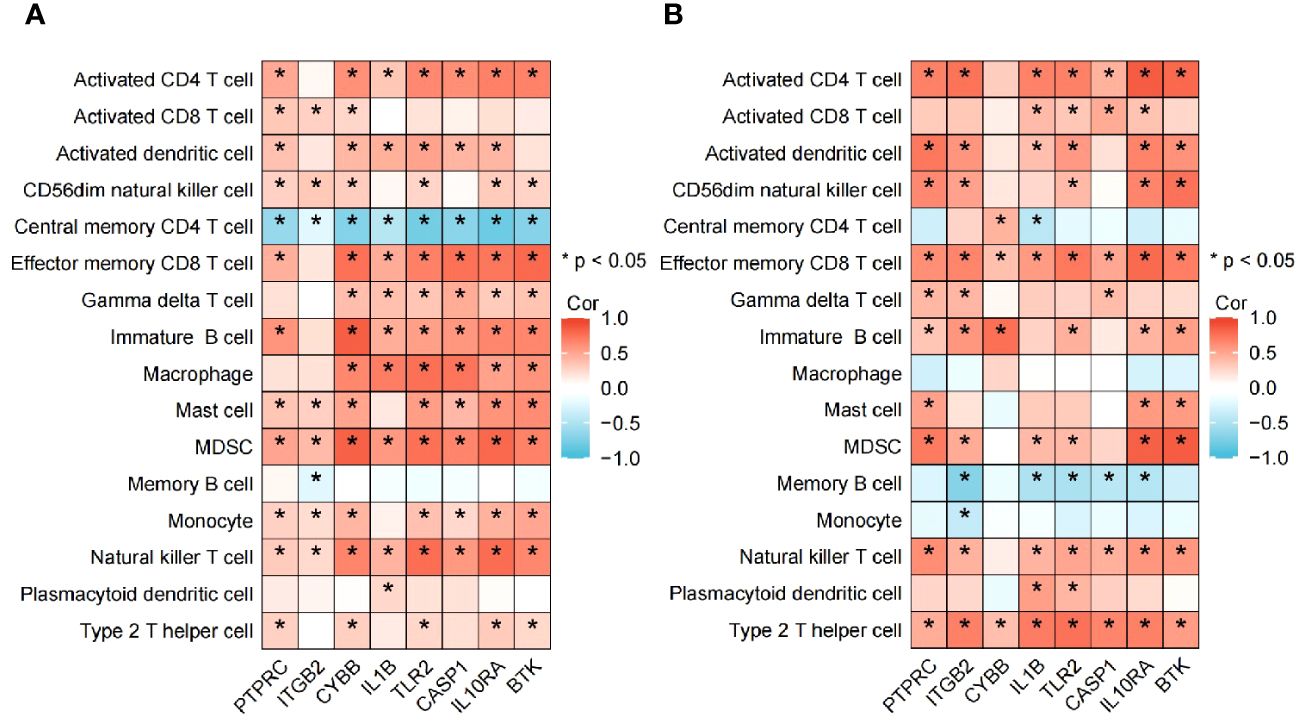
Figure 11 Association between the hub genes and immune cell infiltrationin VD and UC. (A) Heatmap of correlation analysis in VD. (B) Heatmap of correlation analysis in UC.
There has a significant difference in the distribution and proportion of multiple immune cells in VD and UC patients and healthy controls. Including activated CD4 T cell, activated CD8 T cell, CD56dim natural killer cell, effector memory CD8 T cell, gamma delta T cell, immature B cell, macrophage, mast cell, MDSC, monocyte, natural killer T cell, and type 2 T helper cell were up-regulated in VD. Central memory CD4 T cell was down-regulated in VD. There was no significant difference between activated dendritic cells, memory B cells, and plasmacytoid dendritic cells. The up-regulated immune cells in UC were activated CD4 T cell, effector memory CD8 T cell, activated CD8 T cell, mast cell, MDSC, memory B cell, natural killer (NK) T cell, type 2 T helper cell. The down-regulated immune cells in UC included central memory CD4 T cells, which were statistically different. There was no statistically significant in activated dendritic cells, CD56dim NK cells, gamma delta T cells, immature B cells, macrophages, Monocytes, and plasmacytoid dendritic cells.
Biomarkers of 8 hub genes and infiltration analysis of 28 immune cells suggested that in VD gene concentration, central memory CD4 T cells are significantly negatively correlated with all hub genes (P < 0.05), while MDSC and NK T cells are positively correlated with all hub genes (P < 0.05), indicating that hub genes have a good level of infiltration with immune cells in VD.
In UC gene concentration, all hub genes were significantly negatively correlated with memory B cell, particularly in ITGB2, IL1B, TLR2, CASP1, and IL10RA (P < 0.05), while positively correlated with effector memory CD8 T cell and type 2 T helper cell (P < 0.05). These results provide key immune pathway evidence for the link between IBD and VD, and further demonstrate the feasibility of studying the co-pathogenesis and drug intervention targets of the two from the direction of the brain-gut axis.
Vascular dementia is considered to be a disease that can be prevented and controlled. However, improvements in cognition have not been satisfactory in clinical trials, so medical practitioners have been focusing on innovation in vascular dementia mechanisms, risk factors, and new treatment options. Gut microbiota was considered to be an important influencing factor for stroke, and the bidirectional interaction between intestinal diseases and neurological diseases was still not clear. Integrative bioinformatics analyses are becoming more common in the exploration of new genes, potential diagnostic and prognostic biomarkers, underlying mechanisms, and potential therapeutic targets using big data. This approach can provide valuable insights into various diseases (30, 31).
The brain-gut axis is a bidirectional communication pathway. And all the central, intestinal, autonomic nervous systems, as well as the hypo-pituitary-adrenal (HPA) axis constitute a complete brain-gut axis system. The vagus nerve, immune and neuroendocrine systems, neurotransmitters and metabolites, and gut microbiota are the key nodes that play their respective mechanism effects in the brain-gut axis pathway (32–34). After decades of research by scientists, they have demonstrated a strong connection between the gut and the brain in mechanisms such as neurons, neurotransmitters, hormones, and immune-mediated regulation (35, 36). Whole-gene expression data may give us a hand to better understand the specific pathological links and provide molecular basis for brain-gut axis system. Firstly, 18 common pathways between GSE122063 (VD) and GSE47908 (UC) were shown by using he independent GSEA. Interestingly, the common pathways of both were highly clustered, such as mainly immune diseases and neurodegenerative diseases involved in human diseases, the common biological and organic systems belong to the immune system, and a considerable proportion of environmental information processing pathways, and metabolic pathways, suggesting that there might be a pathogenic correlation between VD and IBD. Next, GO and KEGG enrichment analyses were performed on 167 selected coDEGs. The main pathways of GO: BP function was mainly concentrated in the immune-mediated by leukocyte cells, monocytes, and lymphocytes, and the KEGG function enrichment was mainly concentrated in immune diseases, infectious disease, the immune system, and environmental information processing pathways. This suggested that the mechanism of connection between the two diseases might be related to immune and inflammatory regulation and environmental information processing. Interestingly, in a basic study (37, 38), scientists build a new microbiota-gut-brain axis, indicating that effector T cells migrate from the gut to the brain, specifically localizing in the leptomeninges. This migration enhances ischemic neuroinflammation by secreting interleukin-17 (IL-17), which in turn increases chemokine production in the brain parenchyma and leads to the infiltration of cytotoxic immune cells such as neutrophils. Furthermore, it had been well documented that inflammatory factors can lead to neuronal death resulting in cognitive and emotional dysfunction. In addition, in the study on the impact of ischemic stroke on the gut, cerebral ischemia could quickly cause intestinal ischemia, leading to a disruption in intestinal flora through free radical reactions, and enhance systemic inflammation and aggravate cerebral infarction. Transplantation of primary Intestinal Epithelial Stem Cells (IESCs) from healthy donors into post-stroke models could prevent depression-like behavior and cognitive impairment (39).
To further detect the coDEGs, 8 shared hub genes were selected, and GO/KEGG enrichment analysis was performed on the 8 shared genes. The GO: BP pathway mainly focused on the activation of glial cells, neuroinflammatory response and immune cell regulation, which was consistent with the results of previous studies (38). After verifying the functions of hub genes, we verified them in the IBD validation datasets, which showed the important role of 7 shared hub genes (ITGB2, CYBB, IL1B, TLR2, CASP1, IL10RA, and BTK) in IBD patients with VD and genes IL1B, TLR2, and CASP1 might play an early diagnostic role in IBD complicated with VD. Because gut microbes can directly affect the characteristics of the immune system, immune activation might be the pathway through which gut microbes act on the CNS. Finally, 167 coDEGs were analyzed for immune infiltration and immune correlation, and 8 hub genes were analyzed for immune correlation. The results further confirmed the association between VD and IBD between coDEGs and hub genes and the existence of 16 immune cells in immune activities. This suggests that inflammatory diseases of the gut may affect central nervous system function through a specific immune system, especially in relation to vascular dementia, but of course, there are other factors involved, which need to be further studied.
ITGB2 is predominantly expressed in immune cells and plays a crucial role in various metabolic pathways and immune functions, including leukocyte extravasation (40). ITGB2 plays a critical role in T-cell development and function. Research has also associated ITGB2 with chronic colitis (41, 42). In addition, a study suggested that Itgb2+ microglia were found to function as a time-specific multifunctional immunomodulatory subcluster during ischemia reperfusion injury (CIRI). It has been proved that ITGB2 plays a role in the pathological changes of IBD (43, 44), but the specific mechanism remains to be further explored. TLR2 plays a fundamental role in pathogen recognition and activation of innate immunity. Knockout of TLR2 has been shown to prevent impaired cerebral blood flow in early diabetes and VCI by counteracting overperfusion in long-term diabetes (45). The expression of TLR2 is crucial in enhancing barrier function during injury and preserving tight junctions (46, 47), and are crucial in IBD (48–50). Furthermore, neutrophil extracellular traps (NETs) activated by TLR2 and TLR4 have been shown to worsen colon tissue damage and promote thrombotic events with active inflammatory bowel disease (IBD) (50), resulting in stroke and VD. BTK plays a crucial role in B-cell development and functions as a key player in both innate and adaptive immunity (51). In AD brain tissue, BTK may improve cognition in AD patients by preventing microglia activation and synaptic loss. BTK deficiency has been shown in various experiments to lead to severe colitis. This mechanism includes the activation of the NLRP3 inflammasome due to BTK defects (52), as well as its important role in preserving gut immune balance and preventing inflammation through the regulation of T-cell polarization (53). However, the mechanism of BTK in VD remains to be further explored. CYBB, also known as NADPHoxidase 2 (NOX2), is involved in the generation of reactive oxygen species (ROS) during ischemic stroke (IS) and is found to be significantly increased (54, 55). As has been demonstrated in numerous studies, oxidative stress is an important mechanism of vascular dementia (56, 57). In addition, the inadequate ROS production results in an upregulation of nuclear factor kappa-B (NF-κB)-regulated inflammatory genes, leading to the over activation of NF-κB and inflammasome in phagocytes. This sustained activation triggers the continuous release of pro-inflammatory cytokines and contributes to inflammatory conditions, such as IBD (58). CASP1 plays a pivotal role in cellular immunity as an initiator of the inflammatory response. CASP1 initiates a pro-inflammatory reaction by cleaving the inflammatory cytokines IL1B and IL18, thus participating in various inflammatory processes (59–61). Additionally, CASP1 triggers pyroptosis (62). IL1B (Interleukin 1 Beta) is a member of the interleukin 1 cytokine family, known as a pro-inflammatory cytokine that plays a critical role in inflammatory disorders (63) and is typically regulated by the NLRP3-caspase1 axis (64). A number of studies have shown that the NLRP3/CASP1/IL1B pathway can participate in neuronal apoptosis and autophagy by mediating oxidative stress and neuroinflammatory response (65–67) in VD, leading to impairment and aggravation of cognitive function. Moreover, in IBD, numerous studies have demonstrated the activation of the NLRP3/CASP1/IL1B axis, which plays a crucial role in immune response, hyperinflammatory response, and pyroptosis, contributing to the pathogenesis and progression of the disease (68–71). IL10RA is cell surface receptor for the cytokine IL10 that participates in IL10-mediated anti-inflammatory functions, limiting excessive tissue disruption caused by inflammation. IL10RA modulates the miR15a/16-1/IL10RA/AKT3 axis to reduce brain damage and improve cognition in VD models (72). Intestinal microbial dysbiosis caused by IL10RA dysfunction can lead to early-onset IBD (73, 74) and refractory IBD (75). PTPRC requires for T-cell activation through the antigen receptor. Although there is substantial evidence for the role of T cells in VD and IBD (76, 77), the mechanism of action of PTPRC in both remains unclear. Despite the identification of shared genes using bioinformatics methods, further research is required to elucidate the interactions and relationships between these molecular markers through the brain-gut axis. It is evident that these biological pathways of molecular markers are primarily focused on immunity, inflammation, oxidative stress, microglia activation, among others. However, these pathways do not fully capture the signal transduction mechanisms between them, highlighting the necessity for additional pathway studies from the perspective of the brain-gut axis.
Neuroinflammatory response is mainly mediated by cytokines, chemokines, reactive oxygen species free radical transmitters, which are mainly secreted by microglia, astrocytes, endothelial cells (ECs) and peripheral derived immune cells. This response can be can be triggered by various damaging events, including hypoxia, ischemia, and infection (78). In normal organisms, neuroinflammation can maintain the stability of the immune microenvironment. However, it may also damage CNS due to excessive activation of the inflammatory response (79, 80), leading to cell injury, blood-brain barrier destruction, and other pathological changes, inducing or aggravating the occurrence and development of VD. There has been increasing evidence that the immune system plays a key role in brain-gut signal transduction (81). For example, studies in germ-free mice and mice treated with broad-spectrum antibiotics showed that the gut microbiota was involved in bacteria-related intestinal immunity, which also reflected the microbiota’s ability to locally regulate innate and adaptive immunity in the gastrointestinal tract (GI) and throughout the body (82). As we know, neurons and glia in the enteric nervous system are involved in intestinal immunity (83). Innate immune cells in the gut include dendritic cells, M cells, macrophages, mast cells, NK cells, and type 1, 2, and 3 innate lymphoid cells (ILCs), serving as the first line of defense in the intestinal barrier. Adaptive immune cells are divided into different cell subtypes (84). Cytotoxic CD8+T cells and CD4 effector cells (TH1 cells, TH2 cells, and Th17 cells) and regulatory T cells (Tregcells) are not only involved in action locally, but also migrate to other organs by immune migration, including the brain (85). Under normal conditions, the CNS is isolated from the periphery by the BBB, which is molecularly and cellularly similar to glial cells, and local immune functions are mainly supported by glial cells. Animal experiments had also demonstrated (86, 87) that when the BBB was damaged, its permeability increased, leading to microglia activation. Microglia are the main resident immune cells in the central nervous system, which have the ability of antigen presentation and bind to lymphocytes to limit the invasion of pathogens (88). In addition to cytokines, microglia can recognize neurotransmitters and neuromodulators, which trigger immune responses (88). As the most abundant glial cells in the nervous system, astrocytes play a key role in promoting the integrity of the BBB and participating in the immune response (89).
Blood ECs have long been known to modulate inflammation by regulating immune cell trafficking, activation status and function (90). ECs play a crucial role in BBB vascular biology by upholding permeability, homeostasis, vessel wall integrity, and preventing thrombosis (91), and are involved in peripheral immunity and neuroinflammation (92). In our study, we found that the functions of coDEGs are mainly concentrated in immune regulation and inflammatory response, and the dysfunction of these functions plays an important role in the permeability and function of ECs. For example, during sepsis, ECs enhance the immune response and activate the clotting system, thus shifting from an anticoagulant state to a pro-coagulant state (93, 94). They are both a target and source of inflammation, acting as a link between local and systemic immune responses. Inflammatory mediators also inhibited cAMP/Rac1signalling to increase endothelial cell permeability (95, 96). Severe inflammation can also result in parenchymal infiltration of peripheral immune cells and activation, causing structural damage to the BBB (97). When the BBB is damaged due to various reasons, the leaky BBB is involved in neuroinflammation, oxidative stress and excitotoxicity, leading to perivascular injury, leukocyte infiltration and brain parenchymal changes, leading to VD (98, 99). In addition, ECs can crosstalk with a variety of cells leading to cognitive impairment, for example, loss of endothelial-pericyte crosstalk is a major driving force in dementia pathology (100), and endothelium-macrophage crosstalk mediates BBB dysfunction in hypertension (101). Impaired BBB can cause a variety of neurological dysfunctions, which further leads to the imbalance of peripheral system homeostasis, leading to multiple intestinal complications, such as intestinal flora imbalance (GMD), IBD, necrotizing enterocolitis (NEC), and even colorectal cancer (CRC). In turn, these complications can worsen brain function and, in patients with IS, it can even disrupt the brain’s immune system (102).
Combined with the results of immune infiltration results of 8 hub genes, it can be inferred that intestinal diseases may migrate to the central nervous system through acquired immune cells such as Cytotoxic CD8+T cells, TH2 cells, and regulatory T cells to activate neural immunity. Of course, innate immune cells, especially NK cells, and mast cells are also directly involved in the formation of vascular dementia, which needs further experimental proof. Central memory CD4 T cells in VD and memory B cells in IBD can suppress this abnormal immune response and may provide targets for further intervention. Previous studies have found that intestinal immune cells themselves can also directly regulate neuroimmune homeostasis and brain responses to inflammation. Intestinal antigens stimulate B cells to differentiate into immunoglobulin A (IgA), secreting plasma cells to control the gut microbial community. Autoimmune diseases of the nervous system induce massive migration of intestinal IgA+ plasma cells to the brain and spinal cord (85). These findings are consistent with our results and may open new avenues for the treatment of nervous system disorders by using intestinal antigens to promote IgA+B cell production and promote neuroimmune suppression (103), and thus may provide potential immunotherapy targets for patients with IBD complicated with VD. In addition, evidence from animal studies suggested a potential role for short-chain fatty acids (SCFAs) in communication with the BBB and the brain-gut axis that supported BBB integrity (104), which could link butyric acid to memory, cognition, mood, and metabolism (105). Benakis et al. found that the intestinal flora of mice treated with antibiotics was changed, and the intestinal dysbiosis could increase regulatory T cells and decrease the number of IL-17+γδT cells by changing the activity of dendritic cells, and finally play a brain-protective role by limiting neuroinflammatory response (37). After middle cerebral artery occlusion, γδT cells would quickly infiltrate the lesion area and aggravate the BBB injury by releasing IL-17A.
Many scientists believe that there is a close link between microbial diversity and healthy aging. Studies in mice have shown that fecal microbiota grafts could correct age-related immune deficits (82), and that transplanting gut microbes from older mice into younger mice adversely affects critical CNS functions (106, 107). Neurological studies have shown that the microbiome also plays a role in neurodegeneration. Studies of fecal microbiota transplantation in transgenic mouse models indicated a causal relationship between gut microbiota, protein aggregation, and cognitive problems (108, 109). Animal and human clinical trials had explored dietary additives such as prebiotics, biostime, omega-3 polyunsaturated fatty acids, or phytochemicals to intervene in the intestinal microenvironment (110). For example, a high-fiber diet (which promoted SCFA production in the gut microbiota) was a promising intervention. It could overcome the cognitive and social dysfunction caused by maternal obesity (111). Fecal microbiota transplantation (FMT) was another potential treatment that had been shown to affect high-fat dietary intake in mice and reduce the risk of causing cognitive problems (112). Therefore, intestinal microbiome intervention is another promising target for improving cognitive impairment, which is consistent with the evidence of IBD and VD provided in this study.
This study has certain limitations. Initially, the data utilized in our research was sourced from the GEO database, which did not provide specific clinical details like gender, age, complications, etc. for each sample, resulting in their exclusion from our study. Additionally, the sample size derived from the database was limited, which might lead to false positive results. Moreover, the analysis of genes and pathways has certain limitations and cannot fully reflect the unknown mechanism of the disease. Consequently, further research with larger sample sizes is necessary to elucidate the precise relationship between VD and IBD and its various phenotypes. Finally, due to the difficulty in obtaining samples, we did not conduct in vitro experiments, particularly with regard to how these genes act on immune cells through which pathways and how they affect the occurrence and process of diseases.
This study elucidates the common pathogenesis of VD and IBD and the connection between the two through the brain-gut axis pathway at the molecular level, suggesting that IBD and VD are closely related, and IBD may be an independent risk factor for VD. In addition, the immunoinfiltration analysis of the coDEGs suggests that the two can interact through the innate and adaptive immune system, and the inflammatory response, which provides a new perspective for basic research and clinical research. The identification of 8 hub genes helps us to better search for new targets for the treatment of both, and is of great value for the prediction of IBD-combined VD.
The original data used in this study are all publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/ under the accession numbers GSE122063, GSE47908, and GSE479285.
Ethical approval was not required for the studies involving humans because All data in this paper are from public databases. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from gifted from another research group. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
YW: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. DX: Conceptualization, Funding acquisition, Writing – review & editing. SM: Data curation, Methodology, Validation, Writing – review & editing. NS: Project administration, Resources, Writing – review & editing. XZ: Project administration, Software, Writing – review & editing. XW: Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (No. 8187140739).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1347415/full#supplementary-material
1. Impellizzeri D, D’Amico R, Fusco R, Genovese T, Peritore AF, Gugliandolo E, et al. Açai berry mitigates vascular dementia-induced neuropathological alterations modulating nrf-2/beclin1 pathways. Cells. (2022) 11. doi: 10.3390/cells11162616
2. Tian Z, Ji X, Liu J. Neuroinflammation in vascular cognitive impairment and dementia: current evidence, advances, and prospects. Int J Mol Sci. (2022) 23. doi: 10.3390/ijms23116224
3. Medina-Rioja R, Patwardhan A, Mercado-Pompa A, Masellis M, Black SE. Ten things to remember (and not forget) about vascular cognitive impairment. Stroke. (2024) 55:e29–32. doi: 10.1161/strokeaha.123.042756
4. Gorelick PB, Scuteri A, Black SE, Decarli C, Greenberg SM, Iadecola C, et al. Vascular contributions to cognitive impairment and dementia: A statement for healthcare professionals from the American heart association/American stroke association. Stroke. (2011) 42:2672–713. doi: 10.1161/STR.0b013e3182299496
5. Rockwood K, Wentzel C, Hachinski V, Hogan DB, MacKnight C, McDowell I. Prevalence and outcomes of vascular cognitive impairment. Vascular cognitive impairment investigators of the Canadian study of health and aging. Neurology. (2000) 54:447–51. doi: 10.1212/wnl.54.2.447
6. Adolph TE, Meyer M, Schwärzler J, Mayr L, Grabherr F, Tilg H. The metabolic nature of inflammatory bowel diseases. Nat Rev Gastroenterol Hepatol. (2022) 19:753–67. doi: 10.1038/s41575-022-00658-y
7. Bisgaard TH, Allin KH, Keefer L, Ananthakrishnan AN, Jess T. Depression and anxiety in inflammatory bowel disease: epidemiology, mechanisms and treatment. Nat Rev Gastroenterol Hepatol. (2022) 19:717–26. doi: 10.1038/s41575-022-00634-6
8. Zingel R, Bohlken J, Kostev K. Association between inflammatory bowel disease and dementia: A retrospective cohort study. J Alzheimers Dis. (2021) 80:1471–8. doi: 10.3233/jad-210103
9. Zhang B, Wang HE, Bai YM, Tsai SJ, Su TP, Chen TJ, et al. Inflammatory bowel disease is associated with higher dementia risk: A nationwide longitudinal study. Gut. (2021) 70:85–91. doi: 10.1136/gutjnl-2020-320789
10. Villumsen M, Aznar S, Pakkenberg B, Jess T, Brudek T. Inflammatory bowel disease increases the risk of Parkinson’s disease: A Danish nationwide cohort study 1977-2014. Gut. (2019) 68:18–24. doi: 10.1136/gutjnl-2017-315666
11. Peter I, Dubinsky M, Bressman S, Park A, Lu C, Chen N, et al. Anti-tumor necrosis factor therapy and incidence of Parkinson disease among patients with inflammatory bowel disease. JAMA Neurol. (2018) 75:939–46. doi: 10.1001/jamaneurol.2018.0605
12. Liu TC, Stappenbeck TS. Genetics and pathogenesis of inflammatory bowel disease. Annu Rev Pathol. (2016) 11:127–48. doi: 10.1146/annurev-pathol-012615-044152
13. Singh N, Bernstein CN. Environmental risk factors for inflammatory bowel disease. United Eur Gastroenterol J. (2022) 10:1047–53. doi: 10.1002/ueg2.12319
14. Liu S, Zhao W, Lan P, Mou X. The microbiome in inflammatory bowel diseases: from pathogenesis to therapy. Protein Cell. (2021) 12:331–45. doi: 10.1007/s13238-020-00745-3
15. Holleran G, Lopetuso L, Petito V, Graziani C, Ianiro G, McNamara D, et al. The innate and adaptive immune system as targets for biologic therapies in inflammatory bowel disease. Int J Mol Sci. (2017) 18. doi: 10.3390/ijms18102020
16. Barberio B, Zamani M, Black CJ, Savarino EV, Ford AC. Prevalence of symptoms of anxiety and depression in patients with inflammatory bowel disease: A systematic review and meta-analysis. Lancet Gastroenterol Hepatol. (2021) 6:359–70. doi: 10.1016/s2468-1253(21)00014-5
17. Fairbrass KM, Lovatt J, Barberio B, Yuan Y, Gracie DJ, Ford AC. Bidirectional brain-gut axis effects influence mood and prognosis in ibd: A systematic review and meta-analysis. Gut. (2022) 71:1773–80. doi: 10.1136/gutjnl-2021-325985
18. Hui KY, Fernandez-Hernandez H, Hu J, Schaffner A, Pankratz N, Hsu NY, et al. Functional variants in the lrrk2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci Transl Med. (2018) 10. doi: 10.1126/scitranslmed.aai7795
19. Kim S, Kwon SH, Kam TI, Panicker N, Karuppagounder SS, Lee S, et al. Transneuronal propagation of pathologic α-synuclein from the gut to the brain models Parkinson’s disease. Neuron. (2019) 103:627–41.e7. doi: 10.1016/j.neuron.2019.05.035
20. Liu B, Fang F, Pedersen NL, Tillander A, Ludvigsson JF, Ekbom A, et al. Vagotomy and Parkinson disease: A Swedish register-based matched-cohort study. Neurology. (2017) 88:1996–2002. doi: 10.1212/wnl.0000000000003961
21. Kim JS, Chen MH, Wang HE, Lu CL, Wang YP, Zhang B. Inflammatory bowel disease and neurodegenerative diseases. Gut Liver. (2023) 17:495–504. doi: 10.5009/gnl220523
22. Zuin M, De Giorgio R, Capatti E, Boschetti E, Zuliani G. Inflammatory bowel disease as a new risk factor for dementia. Aging Clin Exp Res. (2022) 34:1725–8. doi: 10.1007/s40520-022-02076-1
23. Fousekis FS, Katsanos AH, Kourtis G, Saridi M, Albani E, Katsanos KH, et al. Inflammatory bowel disease and patients with mental disorders: what do we know? J Clin Med Res. (2021) 13:466–73. doi: 10.14740/jocmr4593
24. Katsanos AH, Kosmidou M, Giannopoulos S, Katsanos KH, Tsivgoulis G, Kyritsis AP, et al. Cerebral arterial infarction in inflammatory bowel diseases. Eur J Intern Med. (2014) 25:37–44. doi: 10.1016/j.ejim.2013.08.702
25. Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host microbiota constantly control maturation and function of microglia in the cns. Nat Neurosci. (2015) 18:965–77. doi: 10.1038/nn.4030
26. Fernández-Fernández L, Comes G, Bolea I, Valente T, Ruiz J, Murtra P, et al. Lmn diet, rich in polyphenols and polyunsaturated fatty acids, improves mouse cognitive decline associated with aging and alzheimer’s disease. Behav Brain Res. (2012) 228:261–71. doi: 10.1016/j.bbr.2011.11.014
27. Moreno-Jiménez EP, Flor-García M, Terreros-Roncal J, Rábano A, Cafini F, Pallas-Bazarra N, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with alzheimer’s disease. Nat Med. (2019) 25:554–60. doi: 10.1038/s41591-019-0375-9
28. Möhle L, Mattei D, Heimesaat MM, Bereswill S, Fischer A, Alutis M, et al. Ly6c(Hi) monocytes provide a link between antibiotic-induced changes in gut microbiota and adult hippocampal neurogenesis. Cell Rep. (2016) 15:1945–56. doi: 10.1016/j.celrep.2016.04.074
29. Chang L, Wei Y, Hashimoto K. Brain-gut-microbiota axis in depression: A historical overview and future directions. Brain Res Bull. (2022) 182:44–56. doi: 10.1016/j.brainresbull.2022.02.004
30. Zhu E, Shu X, Xu Z, Peng Y, Xiang Y, Liu Y, et al. Screening of immune-related secretory proteins linking chronic kidney disease with calcific aortic valve disease based on comprehensive bioinformatics analysis and machine learning. J Transl Med. (2023) 21:359. doi: 10.1186/s12967-023-04171-x
31. Dai S, Cao T, Shen H, Zong X, Gu W, Li H, et al. Landscape of molecular crosstalk between sars-cov-2 infection and cardiovascular diseases: emphasis on mitochondrial dysfunction and immune-inflammation. J Transl Med. (2023) 21:915. doi: 10.1186/s12967-023-04787-z
32. Cryan JF, O’Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, et al. The microbiota-gut-brain axis. Physiol Rev. (2019) 99:1877–2013. doi: 10.1152/physrev.00018.2018
33. Morais LH, Schreiber HL4, Mazmanian SK. The gut microbiota-brain axis in behavior and brain disorders. Nat Rev Microbiol. (2021) 19:241–55. doi: 10.1038/s41579-020-00460-0
34. Liang S, Wu X, Jin F. Gut-brain psychology: rethinking psychology from the microbiota-gut-brain axis. Front Integr Neurosci. (2018) 12:33. doi: 10.3389/fnint.2018.00033
35. Farzi A, Fröhlich EE, Holzer P. Gut microbiota and the neuroendocrine system. Neurotherapeutics. (2018) 15:5–22. doi: 10.1007/s13311-017-0600-5
36. Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. (2018) 1693:128–33. doi: 10.1016/j.brainres.2018.03.015
37. Benakis C, Brea D, Caballero S, Faraco G, Moore J, Murphy M, et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal Γδ T cells. Nat Med. (2016) 22:516–23. doi: 10.1038/nm.4068
38. Lee EC, Hong DY, Lee DH, Park SW, Lee JY, Jeong JH, et al. Inflammation and rho-associated protein kinase-induced brain changes in vascular dementia. Biomedicines. (2022) 10. doi: 10.3390/biomedicines10020446
39. Mani KK, El-Hakim Y, Branyan TE, Samiya N, Pandey S, Grimaldo MT, et al. Intestinal epithelial stem cell transplants as a novel therapy for cerebrovascular stroke. Brain Behav Immun. (2023) 107:345–60. doi: 10.1016/j.bbi.2022.10.015
40. Zhang X, Dong Y, Zhao M, Ding L, Yang X, Jing Y, et al. Itgb2-mediated metabolic switch in cafs promotes oscc proliferation by oxidation of nadh in mitochondrial oxidative phosphorylation system. Theranostics. (2020) 10:12044–59. doi: 10.7150/thno.47901
41. Moore SW, Sidler D, Zaahl MG. The itgb2 immunomodulatory gene (Cd18), enterocolitis, and hirschsprung’s disease. J Pediatr Surg. (2008) 43:1439–44. doi: 10.1016/j.jpedsurg.2007.12.057
42. George L, Menden H, Xia S, Yu W, Holmes A, Johnston J, et al. Itgb2 (Integrin β2) immunomodulatory gene variants in premature infants with necrotizing enterocolitis. J Pediatr Gastroenterol Nutr. (2021) 72:e37–41. doi: 10.1097/mpg.0000000000002941
43. Krabbendam L, Heesters BA, Kradolfer CMA, Haverkate NJE, Becker MAJ, Buskens CJ, et al. Cd127+ Cd94+ Innate lymphoid cells expressing granulysin and perforin are expanded in patients with crohn’s disease. Nat Commun. (2021) 12:5841. doi: 10.1038/s41467-021-26187-x
44. Goebel S, Huang M, Davis WC, Jennings M, Siahaan TJ, Alexander JS, et al. Vegf-a stimulation of leukocyte adhesion to colonic microvascular endothelium: implications for inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. (2006) 290:G648–54. doi: 10.1152/ajpgi.00466.2005
45. Hardigan T, Hernandez C, Ward R, Hoda MN, Ergul A. Tlr2 knockout protects against diabetes-mediated changes in cerebral perfusion and cognitive deficits. Am J Physiol Regul Integr Comp Physiol. (2017) 312:R927–r37. doi: 10.1152/ajpregu.00482.2016
46. Cari L, Rosati L, Leoncini G, Lusenti E, Gentili M, Nocentini G, et al. Association of gilz with muc2, tlr2, and tlr4 in inflammatory bowel disease. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms24032235
47. Ricci E, Roselletti E, Gentili M, Sabbatini S, Perito S, Riccardi C, et al. Glucocorticoid-induced leucine zipper-mediated tlr2 downregulation accounts for reduced neutrophil activity following acute dex treatment. Cells. (2021) 10. doi: 10.3390/cells10092228
48. Armstrong HK, Bording-Jorgensen M, Santer DM, Zhang Z, Valcheva R, Rieger AM, et al. Unfermented β-fructan fibers fuel inflammation in select inflammatory bowel disease patients. Gastroenterology. (2023) 164:228–40. doi: 10.1053/j.gastro.2022.09.034
49. Jia DJ, Wang QW, Hu YY, He JM, Ge QW, Qi YD, et al. Lactobacillus johnsonii alleviates colitis by tlr1/2-stat3 mediated cd206(+) macrophages(Il-10) activation. Gut Microbes. (2022) 14:2145843. doi: 10.1080/19490976.2022.2145843
50. Li T, Wang C, Liu Y, Li B, Zhang W, Wang L, et al. Neutrophil extracellular traps induce intestinal damage and thrombotic tendency in inflammatory bowel disease. J Crohns Colitis. (2020) 14:240–53. doi: 10.1093/ecco-jcc/jjz132
51. Horwood NJ, Page TH, McDaid JP, Palmer CD, Campbell J, Mahon T, et al. Bruton’s tyrosine kinase is required for tlr2 and tlr4-induced tnf, but not il-6, production. J Immunol. (2006) 176:3635–41. doi: 10.4049/jimmunol.176.6.3635
52. Mao L, Kitani A, Hiejima E, Montgomery-Recht K, Zhou W, Fuss I, et al. Bruton tyrosine kinase deficiency augments nlrp3 inflammasome activation and causes il-1β-mediated colitis. J Clin Invest. (2020) 130:1793–807. doi: 10.1172/jci128322
53. Guan D, Wang Z, Huo J, Xu S, Lam KP. Bruton’s tyrosine kinase regulates gut immune homeostasis through attenuating th1 response. Cell Death Dis. (2021) 12:431. doi: 10.1038/s41419-021-03702-y
54. Zeng X, Ren H, Zhu Y, Zhang R, Xue X, Tao T, et al. Gp91phox (Nox2) in activated microglia exacerbates neuronal damage induced by oxygen glucose deprivation and hyperglycemia in an in vitro model. Cell Physiol Biochem. (2018) 50:783–97. doi: 10.1159/000494243
55. Liu C, Li Z, Xi H. Bioinformatics analysis and in vivo validation of ferroptosis-related genes in ischemic stroke. Front Pharmacol. (2022) 13:940260. doi: 10.3389/fphar.2022.940260
56. Zhao X, Liu J, Yang S, Song D, Wang C, Chen C, et al. Ling-yang-gou-teng-decoction prevents vascular dementia through inhibiting oxidative stress induced neurovascular coupling dysfunction. J Ethnopharmacol. (2018) 222:229–38. doi: 10.1016/j.jep.2018.03.015
57. Zhu ML, Zhang J, Guo LJ, Yue RZ, Li SS, Cui BY, et al. Amorphous selenium inhibits oxidative stress injury of neurons in vascular dementia rats by activating nmdar pathway. Eur J Pharmacol. (2023) 955:175874. doi: 10.1016/j.ejphar.2023.175874
58. Trevelin SC, Shah AM, Lombardi G. Beyond bacterial killing: nadph oxidase 2 is an immunomodulator. Immunol Lett. (2020) 221:39–48. doi: 10.1016/j.imlet.2020.02.009
59. Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. (1992) 356:768–74. doi: 10.1038/356768a0
60. Alnemri ES, Fernandes-Alnemri T, Litwack G. Cloning and expression of four novel isoforms of human interleukin-1 beta converting enzyme with different apoptotic activities. J Biol Chem. (1995) 270:4312–7. doi: 10.1074/jbc.270.9.4312
61. Feng Q, Li P, Leung PC, Auersperg N. Caspase-1zeta, a new splice variant of the caspase-1 gene. Genomics. (2004) 84:587–91. doi: 10.1016/j.ygeno.2004.06.005
62. Kapplusch F, Schulze F, Rabe-Matschewsky S, Russ S, Herbig M, Heymann MC, et al. Casp1 variants influence subcellular caspase-1 localization, pyroptosome formation, pro-inflammatory cell death and macrophage deformability. Clin Immunol. (2019) 208:108232. doi: 10.1016/j.clim.2019.06.008
63. Church LD, Cook GP, McDermott MF. Primer: inflammasomes and interleukin 1beta in inflammatory disorders. Nat Clin Pract Rheumatol. (2008) 4:34–42. doi: 10.1038/ncprheum0681
64. Rathinam VA, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell. (2016) 165:792–800. doi: 10.1016/j.cell.2016.03.046
65. Du SQ, Wang XR, Zhu W, Ye Y, Yang JW, Ma SM, et al. Acupuncture inhibits txnip-associated oxidative stress and inflammation to attenuate cognitive impairment in vascular dementia rats. CNS Neurosci Ther. (2018) 24:39–46. doi: 10.1111/cns.12773
66. Wang R, Yin YX, Mahmood Q, Wang XJ, Gao YP, Gou GJ, et al. Calmodulin inhibitor ameliorates cognitive dysfunction via inhibiting nitrosative stress and nlrp3 signaling in mice with bilateral carotid artery stenosis. CNS Neurosci Ther. (2017) 23:818–26. doi: 10.1111/cns.12726
67. Meng N, Dong Y, Huo T, Song M, Jiang X, Xiao Y, et al. Past exposure to cigarette smoke aggravates cognitive impairment in a rat model of vascular dementia via neuroinflammation. Cell Mol Neurobiol. (2022) 42:1021–34. doi: 10.1007/s10571-020-00992-2
68. Chen X, Gao Y, Xie J, Hua H, Pan C, Huang J, et al. Identification of fcn1 as a novel macrophage infiltration-associated biomarker for diagnosis of pediatric inflammatory bowel diseases. J Transl Med. (2023) 21:203. doi: 10.1186/s12967-023-04038-1
69. Liu X, Zhou M, Dai Z, Luo S, Shi Y, He Z, et al. Salidroside alleviates ulcerative colitis via inhibiting macrophage pyroptosis and repairing the dysbacteriosis-associated th17/treg imbalance. Phytother Res. (2023) 37:367–82. doi: 10.1002/ptr.7636
70. Ning L, Ye N, Ye B, Miao Z, Cao T, Lu W, et al. Qingre xingyu recipe exerts inhibiting effects on ulcerative colitis development by inhibiting tnfα/nlrp3/caspase-1/il-1β Pathway and macrophage M1 polarization. Cell Death Discov. (2023) 9:84. doi: 10.1038/s41420-023-01361-w
71. Kulhari U, Kundu S, Mugale MN, Sahu BD. Nuciferine alleviates intestinal inflammation by inhibiting mapk/nf-κb and nlrp3/caspase 1 pathways in vivo and in vitro. Int Immunopharmacol. (2023) 115:109613. doi: 10.1016/j.intimp.2022.109613
72. Zhou C, Sun P, Xu Y, Chen Y, Huang Y, Hamblin MH, et al. Genetic deficiency of microrna-15a/16-1 confers resistance to neuropathological damage and cognitive dysfunction in experimental vascular cognitive impairment and dementia. Adv Sci (Weinh). (2022) 9:e2104986. doi: 10.1002/advs.202104986
73. Xue AJ, Miao SJ, Sun H, Qiu XX, Wang SN, Wang L, et al. Intestinal dysbiosis in pediatric crohn’s disease patients with il10ra mutations. World J Gastroenterol. (2020) 26:3098–109. doi: 10.3748/wjg.v26.i22.3098
74. Peng X, Lu Y, Wu B, Dong X, Li W, Wang H, et al. A novel 333 bp deletion of il10ra in Chinese patients with neonatal-onset inflammatory bowel disease. J Clin Immunol. (2021) 41:1095–8. doi: 10.1007/s10875-021-00973-0
75. Oh SH, Baek J, Liany H, Foo JN, Kim KM, Yang SC, et al. A synonymous variant in il10ra affects rna splicing in pediatric patients with refractory inflammatory bowel disease. J Crohns Colitis. (2016) 10:1366–71. doi: 10.1093/ecco-jcc/jjw102
76. Kaiser D, Weise G, Möller K, Scheibe J, Pösel C, Baasch S, et al. Spontaneous white matter damage, cognitive decline and neuroinflammation in middle-aged hypertensive rats: an animal model of early-stage cerebral small vessel disease. Acta Neuropathol Commun. (2014) 2:169. doi: 10.1186/s40478-014-0169-8
77. Krause P, Zahner SP, Kim G, Shaikh RB, Steinberg MW, Kronenberg M. The tumor necrosis factor family member tnfsf14 (Light) is required for resolution of intestinal inflammation in mice. Gastroenterology. (2014) 146:1752–62.e4. doi: 10.1053/j.gastro.2014.02.010
78. Sha S, Tan J, Miao Y, Zhang Q. The role of autophagy in hypoxia-induced neuroinflammation. DNA Cell Biol. (2021) 40:733–9. doi: 10.1089/dna.2020.6186
79. Dhapola R, Hota SS, Sarma P, Bhattacharyya A, Medhi B, Reddy DH. Recent advances in molecular pathways and therapeutic implications targeting neuroinflammation for alzheimer’s disease. Inflammopharmacology. (2021) 29:1669–81. doi: 10.1007/s10787-021-00889-6
80. Uddin MS, Kabir MT, Mamun AA, Barreto GE, Rashid M, Perveen A, et al. Pharmacological approaches to mitigate neuroinflammation in alzheimer’s disease. Int Immunopharmacol. (2020) 84:106479. doi: 10.1016/j.intimp.2020.106479
81. Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. (2017) 20:145–55. doi: 10.1038/nn.4476
82. Fung TC. The microbiota-immune axis as a central mediator of gut-brain communication. Neurobiol Dis. (2020) 136:104714. doi: 10.1016/j.nbd.2019.104714
83. Huh JR, Veiga-Fernandes H. Neuroimmune circuits in inter-organ communication. Nat Rev Immunol. (2020) 20:217–28. doi: 10.1038/s41577-019-0247-z
84. Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nat Rev Immunol. (2003) 3:331–41. doi: 10.1038/nri1057
85. Rojas OL, Pröbstel AK, Porfilio EA, Wang AA, Charabati M, Sun T, et al. Recirculating intestinal iga-producing cells regulate neuroinflammation via il-10. Cell. (2019) 176:610–24.e18. doi: 10.1016/j.cell.2018.11.035
86. Ju F, Ran Y, Zhu L, Cheng X, Gao H, Xi X, et al. Increased bbb permeability enhances activation of microglia and exacerbates loss of dendritic spines after transient global cerebral ischemia. Front Cell Neurosci. (2018) 12:236. doi: 10.3389/fncel.2018.00236
87. Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. (2005) 308:1314–8. doi: 10.1126/science.1110647
88. Deczkowska A, Amit I, Schwartz M. Microglial immune checkpoint mechanisms. Nat Neurosci. (2018) 21:779–86. doi: 10.1038/s41593-018-0145-x
89. Gyoneva S, Traynelis SF. Norepinephrine modulates the motility of resting and activated microglia via different adrenergic receptors. J Biol Chem. (2013) 288:15291–302. doi: 10.1074/jbc.M113.458901
90. Amersfoort J, Eelen G, Carmeliet P. Immunomodulation by endothelial cells - partnering up with the immune system? Nat Rev Immunol. (2022) 22:576–88. doi: 10.1038/s41577-022-00694-4
91. Huang X, Hussain B, Chang J. Peripheral inflammation and blood-brain barrier disruption: effects and mechanisms. CNS Neurosci Ther. (2021) 27:36–47. doi: 10.1111/cns.13569
92. Galea I. The blood-brain barrier in systemic infection and inflammation. Cell Mol Immunol. (2021) 18:2489–501. doi: 10.1038/s41423-021-00757-x
93. Dolmatova EV, Wang K, Mandavilli R, Griendling KK. The effects of sepsis on endothelium and clinical implications. Cardiovasc Res. (2021) 117:60–73. doi: 10.1093/cvr/cvaa070
94. Iba T, Levy JH. Inflammation and thrombosis: roles of neutrophils, platelets and endothelial cells and their interactions in thrombus formation during sepsis. J Thromb Haemost. (2018) 16:231–41. doi: 10.1111/jth.13911
95. Allingham MJ, van Buul JD, Burridge K. Icam-1-mediated, src- and pyk2-dependent vascular endothelial cadherin tyrosine phosphorylation is required for leukocyte transendothelial migration. J Immunol. (2007) 179:4053–64. doi: 10.4049/jimmunol.179.6.4053
96. Radeva MY, Waschke J. Mind the gap: mechanisms regulating the endothelial barrier. Acta Physiol (Oxf). (2018) 222. doi: 10.1111/apha.12860
97. Inoue Y, Shue F, Bu G, Kanekiyo T. Pathophysiology and probable etiology of cerebral small vessel disease in vascular dementia and alzheimer’s disease. Mol Neurodegener. (2023) 18:46. doi: 10.1186/s13024-023-00640-5
98. Díaz-Castro B, Robel S, Mishra A. Astrocyte endfeet in brain function and pathology: open questions. Annu Rev Neurosci. (2023) 46:101–21. doi: 10.1146/annurev-neuro-091922-031205
99. Rajeev V, Fann DY, Dinh QN, Kim HA, De Silva TM, Lai MKP, et al. Pathophysiology of blood brain barrier dysfunction during chronic cerebral hypoperfusion in vascular cognitive impairment. Theranostics. (2022) 12:1639–58. doi: 10.7150/thno.68304
100. Procter TV, Williams A, Montagne A. Interplay between brain pericytes and endothelial cells in dementia. Am J Pathol. (2021) 191:1917–31. doi: 10.1016/j.ajpath.2021.07.003
101. Santisteban MM, Ahn SJ, Lane D, Faraco G, Garcia-Bonilla L, Racchumi G, et al. Endothelium-macrophage crosstalk mediates blood-brain barrier dysfunction in hypertension. Hypertension. (2020) 76:795–807. doi: 10.1161/hypertensionaha.120.15581
102. Cooper J, Pastorello Y, Slevin M. A meta-analysis investigating the relationship between inflammation in autoimmune disease, elevated crp, and the risk of dementia. Front Immunol. (2023) 14:1087571. doi: 10.3389/fimmu.2023.1087571
103. Fitzpatrick Z, Frazer G, Ferro A, Clare S, Bouladoux N, Ferdinand J, et al. Gut-educated iga plasma cells defend the meningeal venous sinuses. Nature. (2020) 587:472–6. doi: 10.1038/s41586-020-2886-4
104. Hoyles L, Snelling T, Umlai UK, Nicholson JK, Carding SR, Glen RC, et al. Microbiome-host systems interactions: protective effects of propionate upon the blood-brain barrier. Microbiome. (2018) 6:55. doi: 10.1186/s40168-018-0439-y
105. Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne). (2020) 11:25. doi: 10.3389/fendo.2020.00025
106. D’Amato A, Di Cesare Mannelli L, Lucarini E, Man AL, Le Gall G, Branca JJV, et al. Fecal microbiota transplant from aged donor mice affects spatial learning and memory via modulating hippocampal synaptic plasticity- and neurotransmission-related proteins in young recipients. Microbiome. (2020) 8:140. doi: 10.1186/s40168-020-00914-w
107. Kundu P, Lee HU, Garcia-Perez I, Tay EXY, Kim H, Faylon LE, et al. Neurogenesis and prolongevity signaling in young germ-free mice transplanted with the gut microbiota of old mice. Sci Transl Med. (2019) 11. doi: 10.1126/scitranslmed.aau4760
108. Lach G, Fülling C, Bastiaanssen TFS, Fouhy F, Donovan ANO, Ventura-Silva AP, et al. Enduring neurobehavioral effects induced by microbiota depletion during the adolescent period. Transl Psychiatry. (2020) 10:382. doi: 10.1038/s41398-020-01073-0
109. Boehme M, Guzzetta KE, Bastiaanssen TFS, van de Wouw M, Moloney GM, Gual-Grau A, et al. Microbiota from young mice counteracts selective age-associated behavioral deficits. Nat Aging. (2021) 1:666–76. doi: 10.1038/s43587-021-00093-9
110. Codagnone MG, Spichak S, O’Mahony SM, O’Leary OF, Clarke G, Stanton C, et al. Programming bugs: microbiota and the developmental origins of brain health and disease. Biol Psychiatry. (2019) 85:150–63. doi: 10.1016/j.biopsych.2018.06.014
111. Liu X, Li X, Xia B, Jin X, Zou Q, Zeng Z, et al. High-fiber diet mitigates maternal obesity-induced cognitive and social dysfunction in the offspring via gut-brain axis. Cell Metab. (2021) 33:923–38.e6. doi: 10.1016/j.cmet.2021.02.002
Keywords: vascular dementia, inflammatory bowel disease, the brain-gut axis, pathogenesis, biomarkers, immune cells infiltration
Citation: Wang Y, Xie D, Ma S, Shao N, Zhang X and Wang X (2024) Exploring the common mechanism of vascular dementia and inflammatory bowel disease: a bioinformatics-based study. Front. Immunol. 15:1347415. doi: 10.3389/fimmu.2024.1347415
Received: 30 November 2023; Accepted: 15 April 2024;
Published: 25 April 2024.
Edited by:
Enzo Emanuele, 2E Science, ItalyReviewed by:
Joaquim Carreras, Tokai University, JapanCopyright © 2024 Wang, Xie, Ma, Shao, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daojun Xie, ZGFvanVueGllQGFodGNtLmVkdS5jbi9kYW9qdW54aWUxODdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.