- Department of Orthopedics, Second Affiliated Hospital, Chongqing Medical University, Chongqing, China
Background: Ankylosing spondylitis (AS) is an autoimmune disease that affects millions of individuals. Immune cells have been recognized as having a crucial role in the pathogenesis of AS. However, their relationship has not been fully explored.
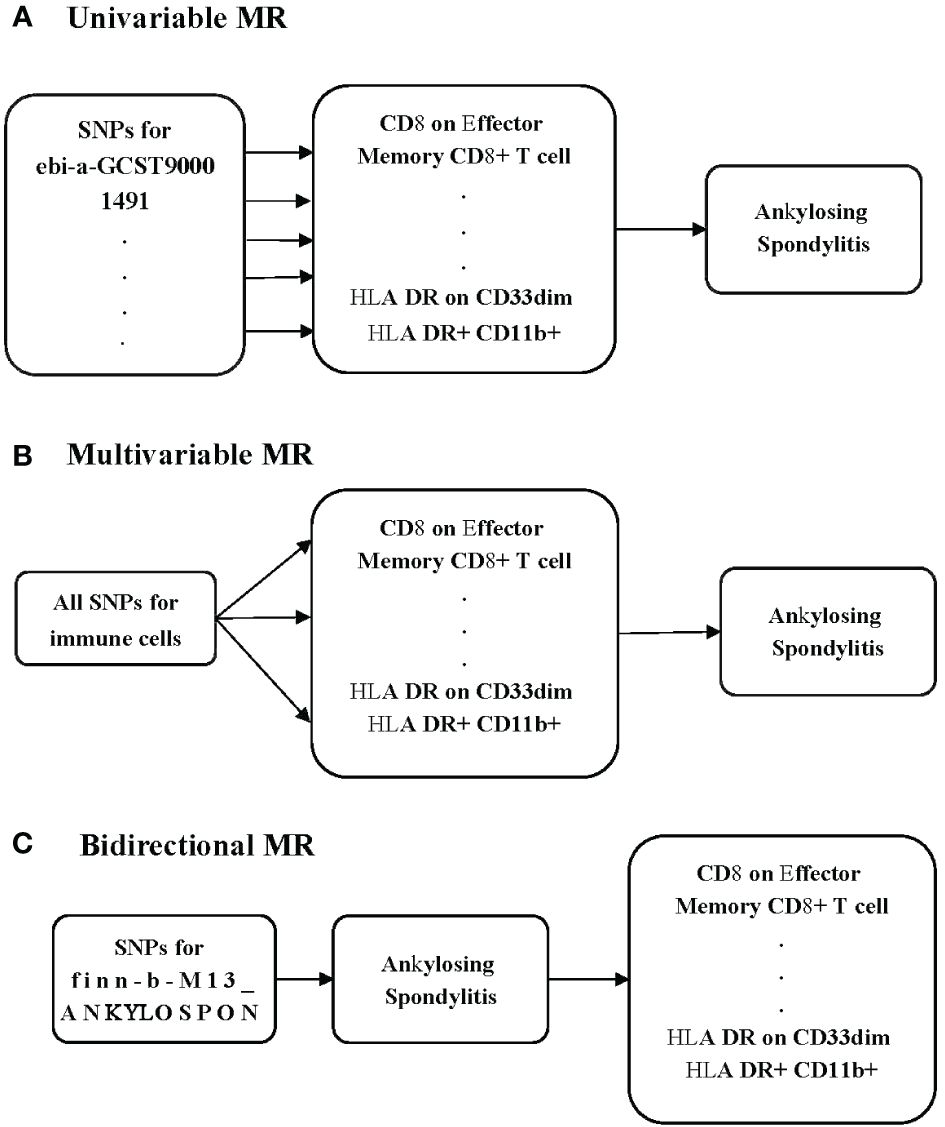
Methods: We chose to employ Mendelian randomization (MR) to investigate the potential correlation between immune cells and AS. We sourced the data on immune cells from the latest genome-wide association studies (GWASs). We obtained data on AS from the FinnGen consortium. Our comprehensive univariable MR analysis covered 731 immune cells to explore its potential causal relationship with AS. The primary analysis method was inverse-variance weighted (IVW). Additionally, we used Cochran’s Q test and the MR-Egger intercept test to assess the presence of pleiotropy and heterogeneity. We examined whether our results could be influenced by individual single-nucleotide polymorphisms (SNPs) using the leave-one-out test. We conducted a bidirectional MR to investigate the reverse relationship. We also applied multivariable MR to decrease the potential influence between the immune cells.
Results: Overall, our univariable MR analysis revealed eight immune cells associated with AS. Among these, four immune cells contributed to an increased risk of AS, while four immune cells were identified as protective factors for AS. However, the Bonferroni test confirmed only one risk factor and one protective factor with a significance level of p < 6.84E−05. CD8 on effector memory CD8+ T cell could increase the risk of AS (p: 1.2302E−05, OR: 2.9871, 95%CI: 1.8289–4.8786). HLA DR on CD33dim HLA DR+ CD11b+ could decrease the risk of AS (p: 1.2301E−06, OR: 0.5446, 95%CI: 0.4260–0.6962). We also identified a bidirectional relationship between CD4 on CD39+ activated CD4 regulatory T cells and AS utilizing the bidirectional MR. To address potential confounding among immune cells, we employed multivariable MR analysis, which revealed that only one immune cell had an independent effect on AS. HLA DR on CD33dim HLA DR+ CD11b+ could decrease the risk of AS (p: 2.113E−06, OR: 0.0.5423, 95%CI: 0.4210–0.6983). Our findings were consistently stable and reliable.
Conclusions: Our findings indicated a potential link between immune cells and AS, which could provide a new idea for future research. Nevertheless, the specific underlying mechanisms require further exploration.
1 Introduction
Ankylosing spondylitis (AS) is a chronic inflammatory disease with an incidence ranging from 1‰ to 3‰ in the general population (1). It is characterized by vertebral fusion, reduced mobility, and the potential for long-term disability in advanced stages (2, 3). Factors such as infection, environmental influences, and immune dysregulation have been suggested as potential triggers for AS (4–6). In recent years, the relationship between immunity and AS has garnered attention (3). Multiple immune cells are thought to be involved in the development of AS (7, 8). Regulatory T cells are lower in AS than in healthy individuals, leading to dysregulation of the immunity of the AS patient and thus to a decrease in the negative regulation of immunity in AS patients (9). However, the causal relationship between immunization and AS remains unclear and requires further investigation.
Mendelian randomization (MR) is a statistical method that uses genotype information as an instrumental variable (IV) to assess the causal relationship between exposure and outcome (10, 11). MR uses the Mendelian independent distribution law as a theoretical basis to explore the etiology of disease. Therefore, MR can effectively overcome the bias caused by confounding and reverse causation problems (10, 11). We plan to conduct an MR to investigate the relationship between immune cells and AS. This study mainly investigated the potential causal connection between immune cells and AS at a genetic level, utilizing univariable, bidirectional, and multivariable MR analyses.
2 Materials and methods
2.1 Study design
To investigate the potential causal relationship between immune cells and AS, we conducted a univariable MR analysis in our study (Figure 1A). All genetic variations used as IVs adhere to the three fundamental assumptions: 1) the chosen IVs had a clear association with the exposure, 2) the selected IVs had no relationship with any confounding factors, and 3) the IVs could influence the outcomes solely through their impact on the exposure (12). To minimize the potential for reverse connection, we conducted a bidirectional MR analysis to explore the relationship between immune cells and AS (Figure 1B). Recognizing that various immune cells may interact and affect each other due to genetic pleiotropy, potentially introducing confounding effects, we subsequently carried out a multivariable MR analysis to assess the direct influence of immune cells on AS (Figure 1C).
2.2 GWAS data source
The genome-wide association study (GWAS) data on immune cells were acquired from the latest study involving 3,757 individuals of Sardinian descent within the European population (13). This study included 3,757 cases and 3,027 controls, with a gender distribution of 43% male and 57% female. The ages of the participants ranged from 18 to 102 years (13). A total of 731 immunophenotypes were included, including 118 absolute cell (AC) counts, 389 median fluorescence intensities (MFIs) reflecting surface antigen levels, 32 morphological parameters (MPs), and 192 relative cell (RC) counts (13). The MFI, AC, and RC features encompass B cells, CDCs, mature stages of T cells, monocytes, myeloid cells, TBNK (T cells, B cells, and natural killer cells), and Treg panels, while the MP feature encompasses CDC and TBNK panels (13). This study measured 22 million genetic variations (13).
The data related to AS were sourced from the FinnGen database (https://www.finngen.fi/fi). This dataset encompasses 1,462 cases, 164,682 controls, and 16,380,022 single-nucleotide polymorphisms (SNPs). Diagnoses of AS were made in accordance with the International Classification of Diseases, specifically ICD-10 (M45), ICD-9 (7200), and ICD-8 (7124) coding standards. All populations included in the database were of European origin. Additional relevant information can be obtained from the FinnGen website (Risteys · Home (finregistry.fi)).
2.3 Selection of instrumental variables
We conducted various strict quality controls to choose IVs to satisfy the three core assumptions of MR analysis and ensure the robustness and reliability of MR analysis. First, we selected the SNPs of immune cells at a genome-wide significance threshold (p < 5E−08) (10). Second, we addressed the issue of linkage disequilibrium (LD) between SNPs by removing strongly linked variants (r2 = 0.001, clumping distance of 10,000 kb). This step aimed to mitigate any potential bias in the results caused by LD (10). Then, we utilized the PhenoScanner database to exclude the effect of confounder factors. Finally, we computed the F-statistics for all the selected SNPs. We excluded SNPs with F-statistics less than 10 to ensure that all remaining SNPs were strongly associated with the exposure (14). We calculated F-statistics using the formula F = beta2/standard error2(SE) (15, 16) (Supplementary Table 1).
2.4 Statistical analysis
In our MR analysis, we employed the inverse-variance weighted (IVW) method as our primary analytical approach. We established statistical significance by setting the threshold for p-values at 6.84E−05, which was adjusted using the Bonferroni method (0.05/731). Additionally, we considered p-values falling between 6.84E−05 and 0.05 suggestive of significance. Since the IVW method assumes the absence of an intercept term, we conducted the MR-Egger test to assess the presence of the intercept (17). Additionally, to enhance the robustness of our results, we employed the MR-Egger, weighted median, weighted mode, and simple mode methods.
pWe performed Cochran’s Q test and MR-Egger intercept analysis to ensure the absence of heterogeneity and pleiotropy. Heterogeneity was deemed present if the Q–p-value was less than 0.05, leading to the utilization of a random effects model for the analysis (18). MR-Egger intercept p-value exceeding 0.05 indicated the absence of pleiotropy (19). Finally, we employed leave-one-out analysis to investigate the potential impact of single SNP points on the causal relationship between immune cells and AS.
We conducted all statistical analyses using the “TwoSampleMR” (version 0.5.7) packages within the R statistical software (version 4.3.1).
3 Result
3.1 Univariable MR
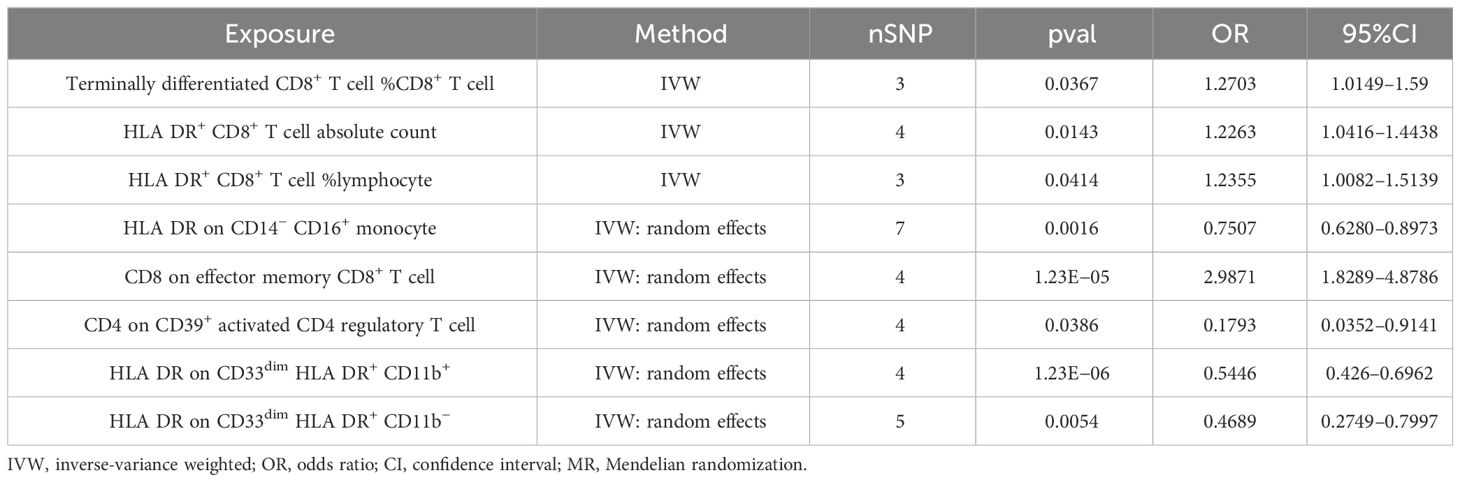
In our study, we investigated the association between 731 immune cells and AS (Supplementary Table 2). Our analysis revealed that 39 of these immune cells were significantly associated with AS, comprising 22 risk factors and 17 protective factors (Supplementary Table 3). However, we excluded 31 factors with fewer than three SNPs each due to limitations in the number of available SNPs. Consequently, we identified four potential pathogenic factors and four potential protective factors. We calculated terminally differentiated CD8+ T cell %CD8+ T cell, HLA DR+ CD8+ T cell absolute count, and HLA DR+ CD8+ T cell %lymphocyte using the fixed effects model (Table 1). For HLA DR on CD14− CD16+ monocyte, CD8 on effector memory CD8+ T cell, CD4 on CD39+ activated CD4 regulatory T cell, HLA DR on CD33dim HLA DR+ CD11b+, and HLA DR on CD33dim HLA DR+ CD11b−, we employed a random effects model for calculation (Table 1).
Following the application of the Bonferroni test, we identified one risk factor and one protective factor associated with AS (Table 1; Figures 2, 3). CD8 on effector memory CD8+ T cell could significantly increase the risk of AS (p: 1.2302E−05, OR: 2.9871, 95%CI: 1.8289–4.8786). HLA DR on CD33dim HLA DR+ CD11b+ could significantly decrease the risk of AS (p: 1.2301E−06, OR: 0.5446, 95%CI: 0.4260–0.6962).
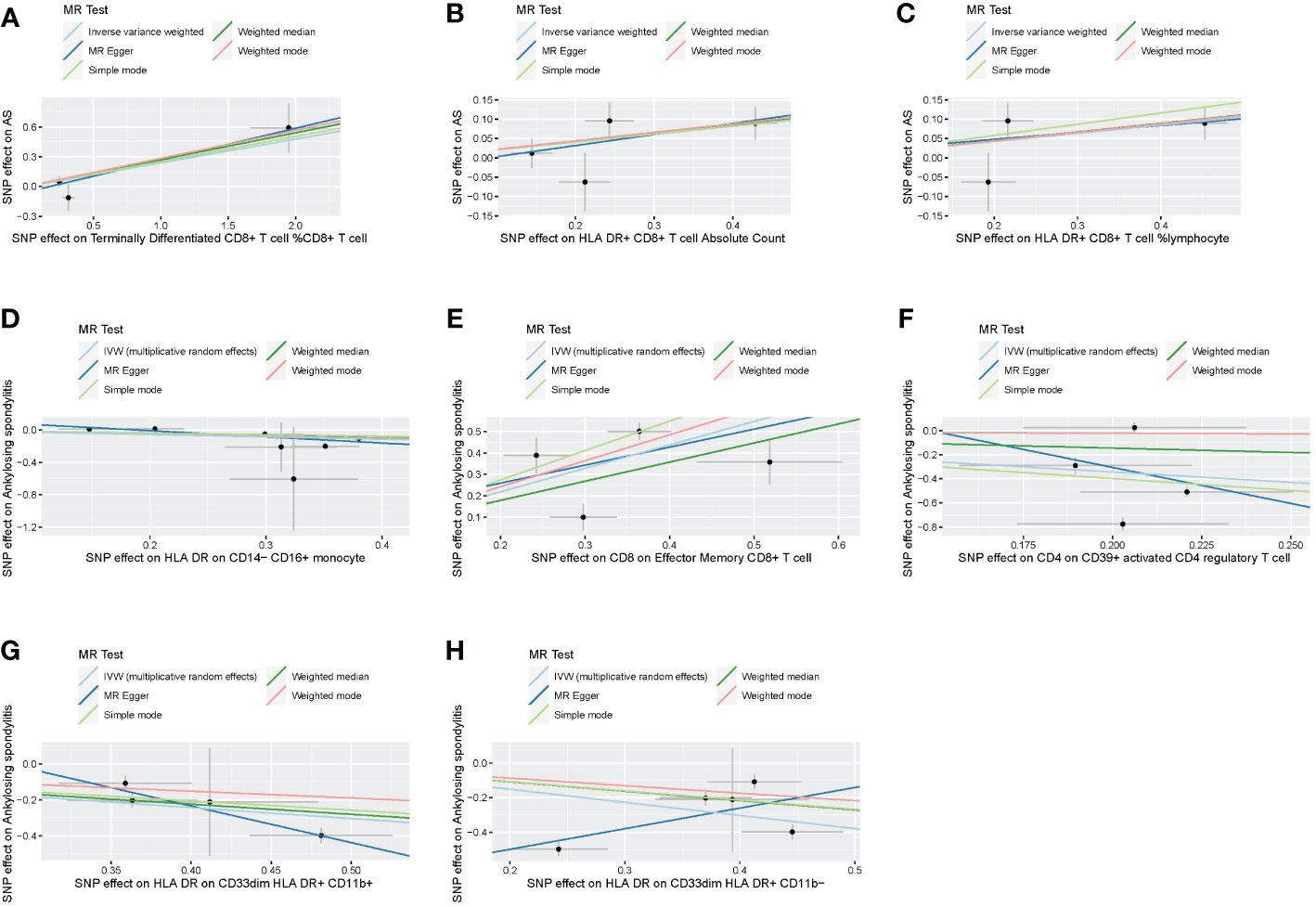
Figure 2 Scatter plots of effects of immune cells on the AS using univariable MR. (A) Terminally differentiated CD8+ T cell %CD8+ T cell. (B) HLA DR+ CD8+ T cell absolute count. (C) HLA DR+ CD8+ T cell %lymphocyte. (D) HLA DR on CD14− CD16+ monocyte. (E) CD8 on effector memory CD8+ T cell. (F) CD4 on CD39+ activated CD4 regulatory T cell. (G) HLA DR on CD33dim HLA DR+ CD11b+. (H) HLA DR on CD33dim HLA DR+ CD11b−. AS, ankylosing spondylitis; MR, Mendelian randomization.
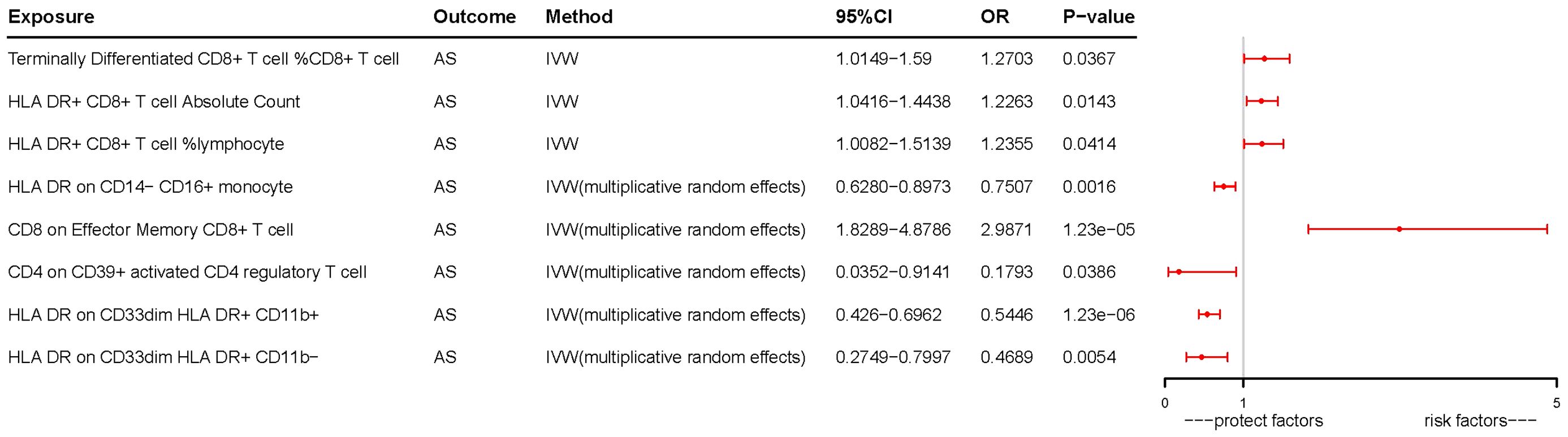
Figure 3 Forest plots of effects of immune cells on the AS using univariable MR. AS, ankylosing spondylitis; IVW, inverse-variance weighted; OR, odds ratio; CI, confidence interval; MR, Mendelian randomization.
3.2 Bidirectional MR
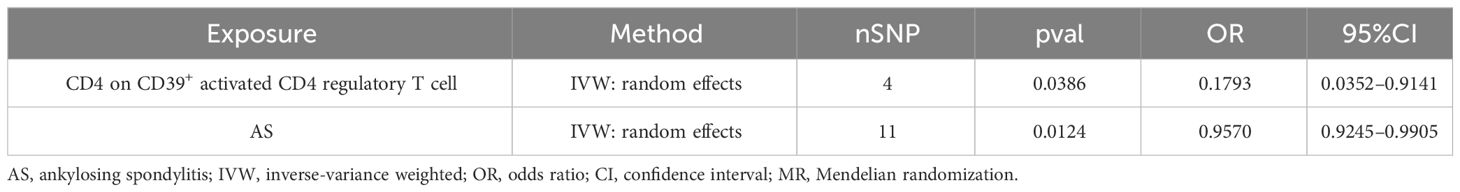
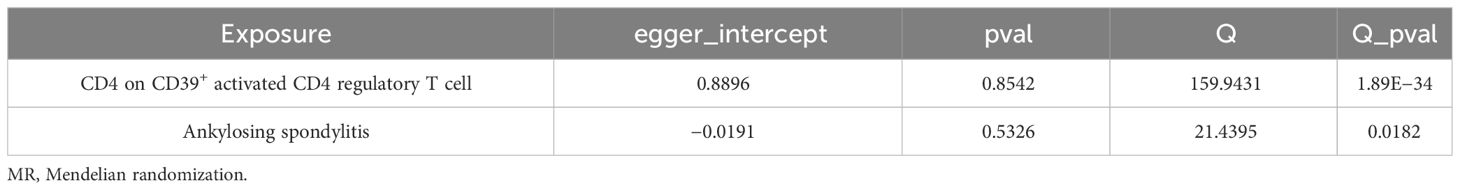
A bidirectional MR analysis was performed, and evidence of a reverse relationship was identified, particularly between CD4 on CD39+ activated CD4 regulatory T cells and AS. The bidirectional MR was analyzed by the random effects model. It was observed that AS could lead to a decrease in CD4 on CD39+ activated CD4 regulatory T cell (p: 0.0124, OR: 0.9570, 95%CI: 0.9245–0.9905) (Table 2; Figures 4, 5A). The potential causal relationship between them appears to be bidirectional.
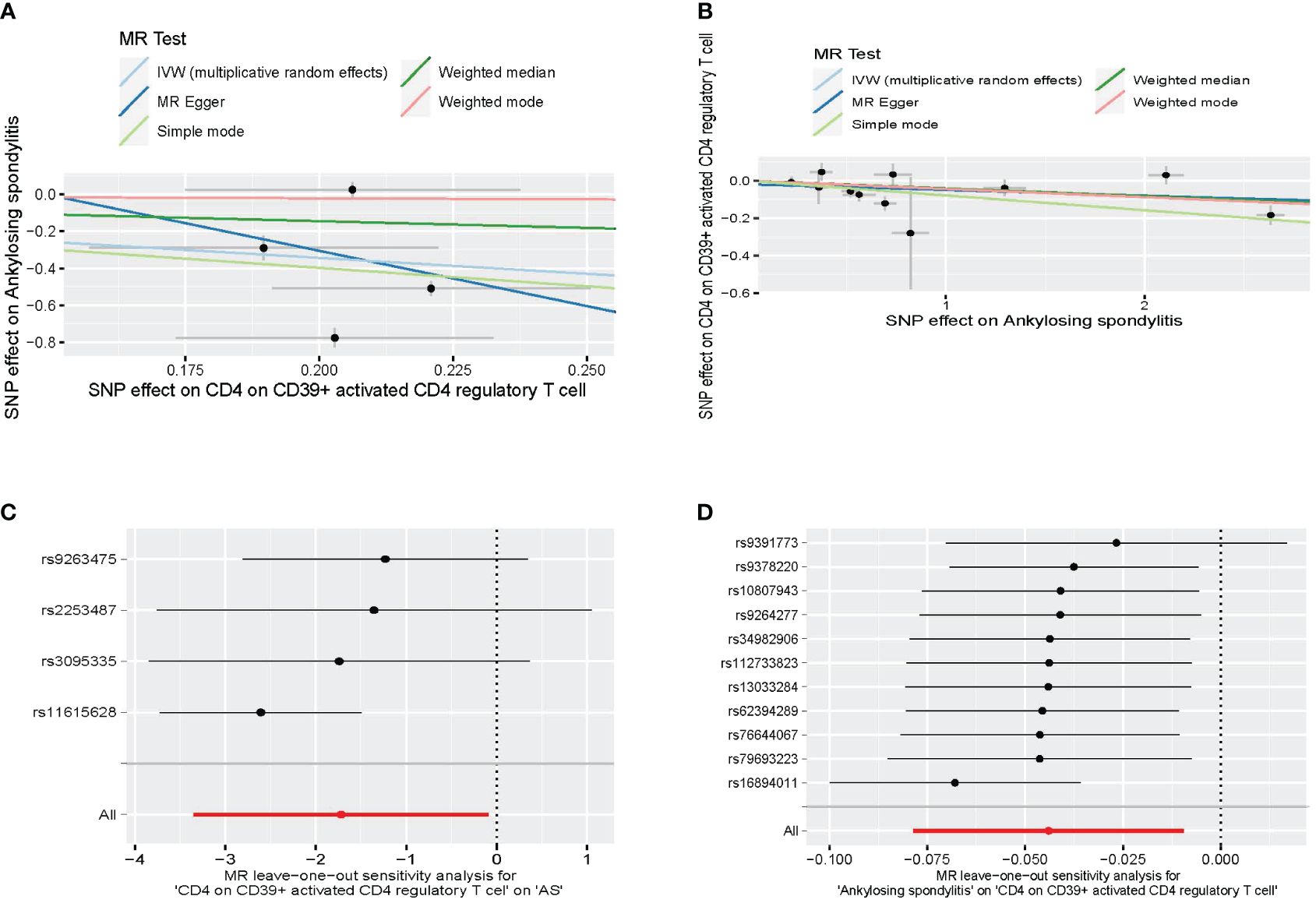
Figure 4 The result of bidirectional MR between CD4 on CD39+ activated CD4 regulatory T cell and AS. (A) The scatter plot of CD4 on CD39+ activated CD4 regulatory T cell on AS. (B) The scatter plot of AS on CD4 on CD39+ activated CD4 regulatory T cell. (C) The leave-one-out plot of CD4 on CD39+ activated CD4 regulatory T cell on AS. (D) The leave-one-out plot of AS on CD4 on CD39+ activated CD4 regulatory T cell. MR, Mendelian randomization; AS, ankylosing spondylitis.
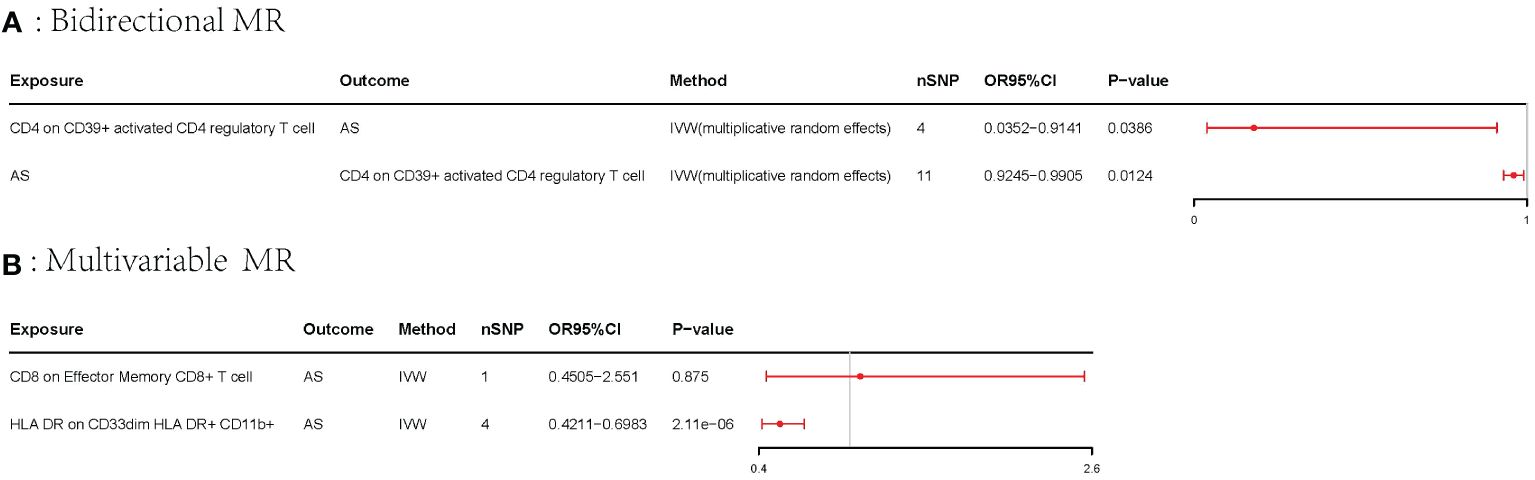
Figure 5 Forest plots of the bidirectional MR and multivariable MR. AS, ankylosing spondylitis; IVW, inverse-variance weighted; OR, odds ratio; CI, confidence interval; MR, Mendelian randomization. (A) Forest plots of bidirectional MR. (B) Forest plots of the multivariable MR.
3.3 Multivariable MR
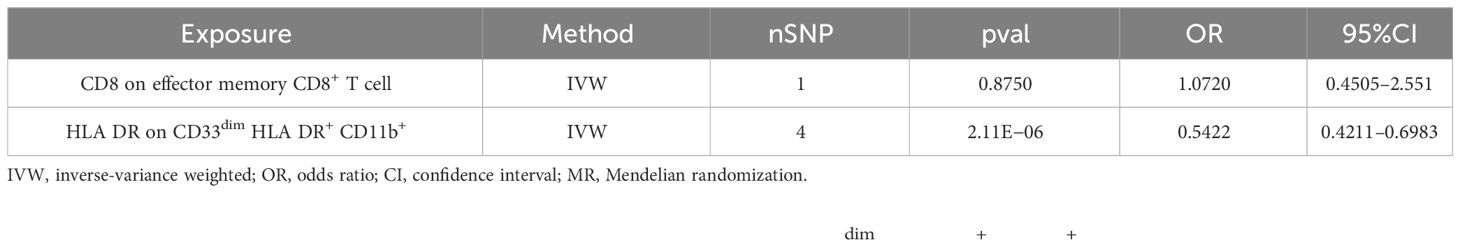
The multivariable MR analysis included HLA DR on CD33dim HLA DR+ CD11b+ and CD8 on effector memory CD8+ T cell. HLA DR on CD33dim HLA DR+ CD11b+ could decrease the risk of AS (p: 2.113E−06, OR: 0.0.5423, 95%CI: 0.4210–0.6983) (Table 3; Figure 5B).
3.4 Sensitivity analyses
3.4.1 Univariable MR
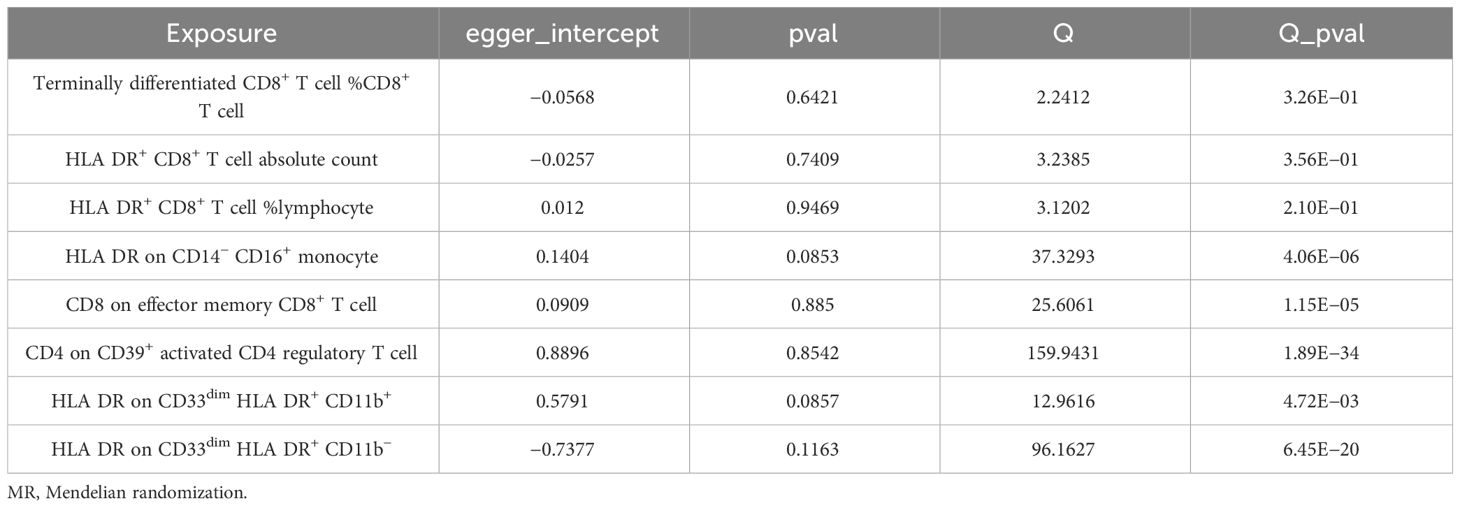
We conducted Cochran’s Q test and MR-Egger intercept test to assess the robustness of our results. Notably, we observed that only the MR-Egger intercept test p-value for HLA DR on CD14− CD16+ monocyte was less than 0.05, indicating the presence of horizontal pleiotropy (Table 4). During the leave-one-out test, we identified an anomaly with the SNP (rs6917212) (Figure 6). As a result, we chose to remove this SNP to ensure the stability of our results. Through this adjustment, horizontal pleiotropy for this exposure was no longer present, reinforcing the validity of our decision (Table 4). We found no evidence of horizontal pleiotropy in other analyses of immune cells (Table 5). Heterogeneity was not found among terminally differentiated CD8+ T cell %CD8+ T cell, HLA DR+ CD8+ T cell absolute count, and HLA DR+ CD8+ T cell %lymphocyte (Table 5). However, the p-values of Cochran’s Q test were less than 0.05 for HLA DR on CD14− CD16+ monocyte, CD8 on effector memory CD8+ T, CD4 on CD39+ activated CD4 regulatory T cell, HLA DR on CD33dim HLA DR+ CD11b+, and HLA DR on CD33dim HLA DR+ CD11b−. These results indicated the presence of heterogeneity (Table 5). Therefore, we employed the random effects model to analyze the effects on AS (Table 1, Figure 2). Furthermore, the leave-one-out test revealed that the association between immune cells and AS remained unaffected by the exclusion of individual SNP (Figure 7). In summary, our results have been validated and are considered reliable and acceptable.
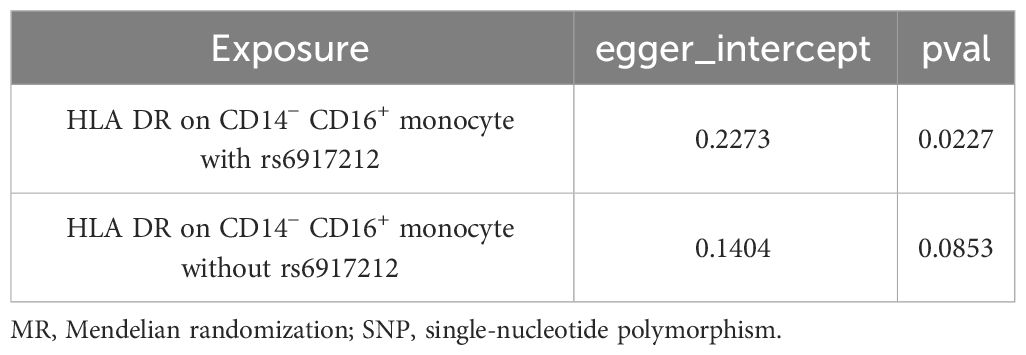
Table 4 The result of MR-Egger intercept test of HLA DR on CD14− CD16+ monocyte with or without the SNP of rs6917212.
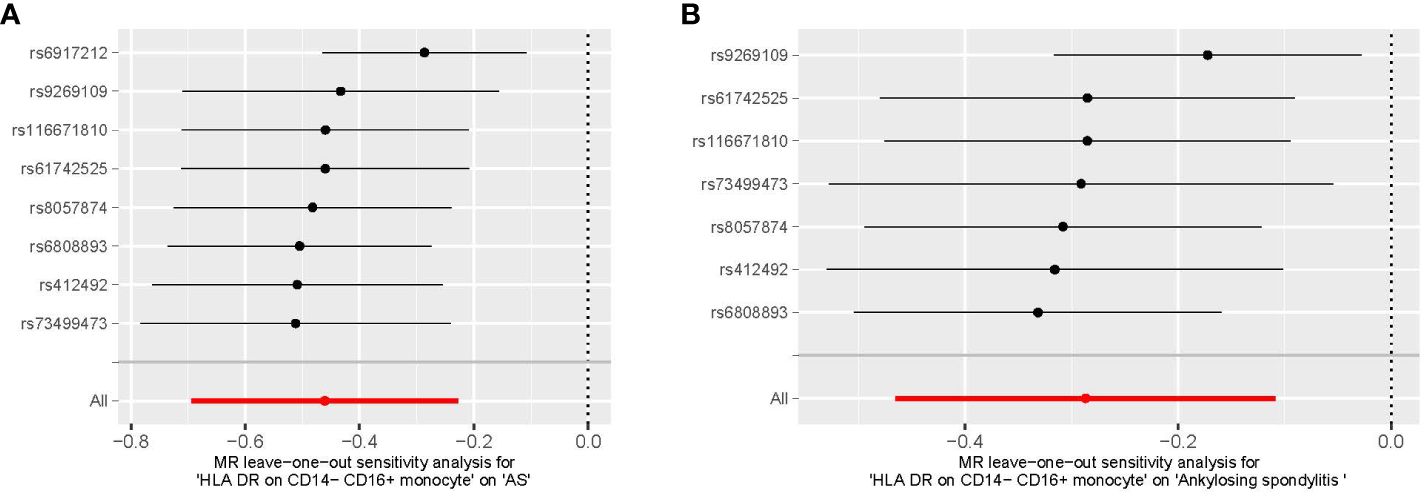
Figure 6 The leave-one-out plot of HLA DR on CD14− CD16+ monocyte on AS. (A) The leave-one-out plot of HLA DR on CD14− CD16+ monocyte on AS with the rs6917212. (B) The leave-one-out plot of HLA DR on CD14− CD16+ monocyte on AS without the rs6917212. AS, ankylosing spondylitis.
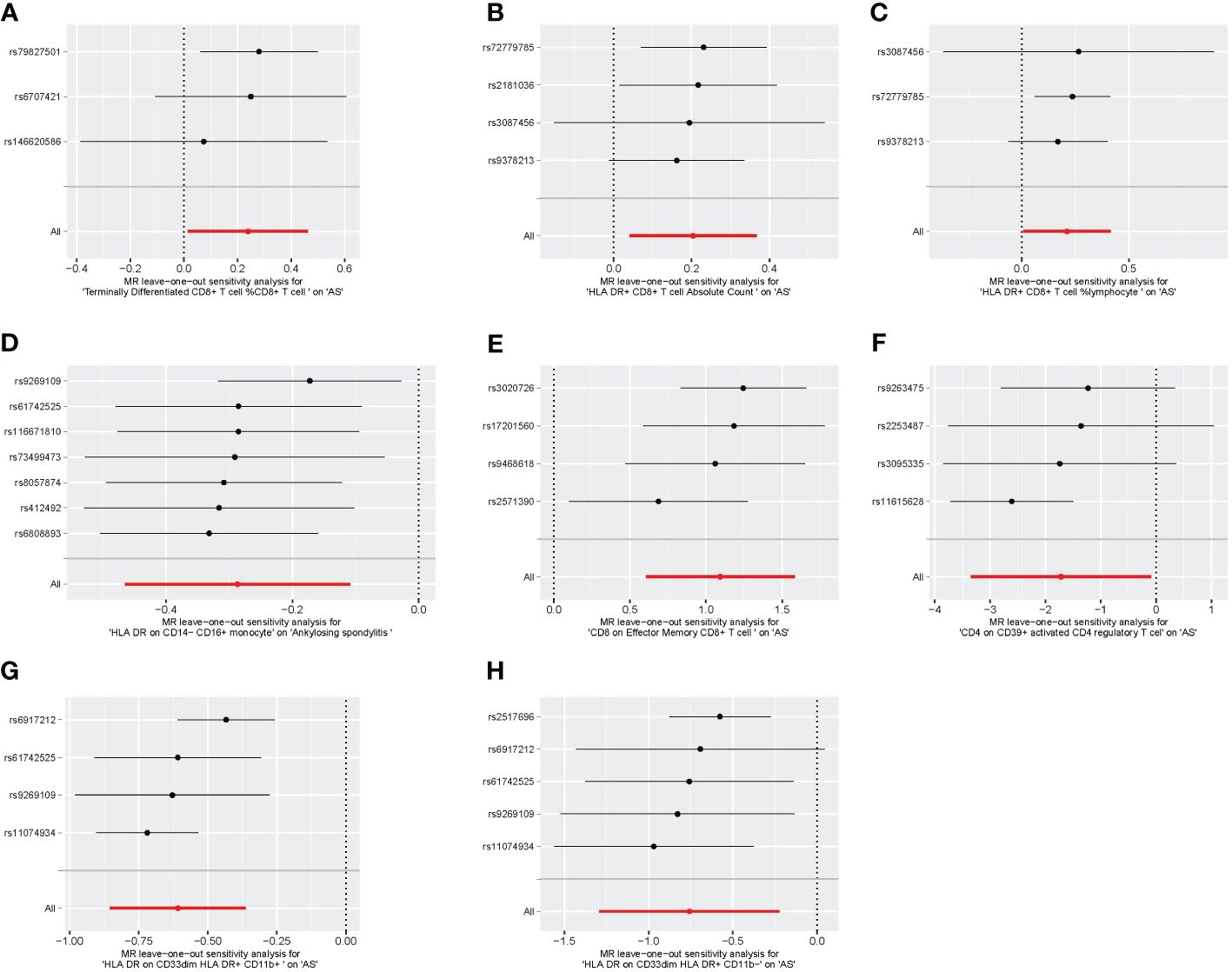
Figure 7 The leave-one-out plot of univariable MR. (A) Terminally differentiated CD8+ T cell %CD8+ T cell. (B) HLA DR+ CD8+ T cell absolute count. (C) HLA DR+ CD8+ T cell %lymphocyte. (D) HLA DR on CD14− CD16+ monocyte. (E) CD8 on effector memory CD8+ T cell. (F) CD4 on CD39+ activated CD4 regulatory T cell. (G) HLA DR on CD33dim HLA DR+ CD11b+. (H) HLA DR on CD33dim HLA DR+ CD11b−. MR, Mendelian randomization.
3.4.2 Bidirectional MR
We found heterogeneity in the bidirectional MR (Table 6). Therefore, we decided to use the random effects model (Table 2, Figure 4). The horizontal pleiotropy was absent in the bidirectional MR (Table 6). Meanwhile, the leave-one-out test indicated that the results were not decided by a single SNP (Figure 4).
4 Discussion
In this study, we employed univariable, bidirectional, and multivariable MR to explore the association between 731 immune cells and AS. Using the extensive public GWAS summary data, we uncovered the complex relationship between immune cells and AS. Our findings were confirmed by sensitivity analysis. Additionally, we used the Bonferroni test to further support the association between immune cells and AS. Through univariable MR coupled with the Bonferroni test, we validated one risk factor and one protective factor. Finally, we have conclusively demonstrated that HLA-DR expression on CD33dim HLA-DR+ CD11b+ cells decreases the risk of ankylosing spondylitis through multivariable MR, after controlling for the effect between immune cells. Employing bidirectional MR, we revealed a bidirectional link between immune cells and AS.
4.1 Univariable MR
Our findings indicated that HLA-DR exhibits both protective and promotive effects on AS across various immune cells. It is hypothesized that the presence of different HLA-DR subtypes on various immune cells determines their respective roles. Kchir reported elevated expression levels of HLA-DRB1*11 in AS patients, and HLA-DRB1*11 did not have a direct effect on AS but showed dependence on HLA-B27 (20). Meanwhile, their findings also indicated that HLA-DRB1*13 played a protective role in AS (20). These findings support our results on the role of HLA-DR in AS.
Our study demonstrated that CD8+ T cells can contribute to the development of AS. Terminally differentiated CD8+ T cell %CD8+ T cell, HLA DR+ CD8+ T cell absolute count, HLA DR+ CD8+ T cell %lymphocyte, and CD8 on effector memory CD8+ T cell could increase the risk of AS. Previous studies have shown a significant increase in IL-6 levels in CD8+ T cells in the peripheral blood of patients with AS (9). IL-6, a pro-inflammatory cytokine, may enhance the body’s inflammatory response and promote the progression of AS (9, 21). The role of CD8+ cells in the pathogenesis of AS is dependent on the presentation of antigens through HLA-B27, and it is widely acknowledged that HLA-B27 plays a significant role in promoting the development of AS (22–25). Moreover, cytotoxic cells restricted by HLA-B27 can be found in the synovial fluids of AS patients (26).
In this study, we also found that HLA DR on CD14− CD16+ monocytes was associated with a reduced risk of AS. Wright proposed that CD14− CD16+ mononuclear cells may contribute to AS by promoting T helper 17 cells (Th17) responses, which can produce the IL-17 to accelerate the pathogenesis of AS (24, 27–29). However, our findings differed from those reported by Wright. It is proposed that the protective influence exerted by HLA-DR offsets the promotional impact of CD14− CD16+ on AS. The protective influence of HLA-DR has been reported by a previous study (20).
4.2 Bidirectional MR
CD4 on CD39+ activated CD4 regulatory T cell has the capacity to decrease the risk of AS. Conversely, AS can diminish the population of CD4 on CD39+ activated CD4 regulatory T cells. However, CD4 cell activation leads to increased secretion of IL-10, which exerts an anti-inflammatory effect and diminishes the risk of AS (9, 21). However, the body experiences immune dysregulation in AS patients, impairing immune suppression function and potentially resulting in reduced IL-10 secretion by CD4 cells, thereby inhibiting their function (9). The collaborative action of CD39 and CD73 results in the conversion of ATP to ADP and cAMP, ultimately generating adenosine (30, 31). Adenosine can interact with multiple receptors, including A1, A2A, A2B, and A3, leading to various immune responses (31, 32). Adenosine stimulates immune responses through A1 and A3 receptors, while it exerts immunosuppressive effects when engaging with A2A and A2B receptors (32–34). In the context of AS pathogenesis, we hypothesize that CD39 utilizes the latter pathway to participate in AS.
4.3 Multivariable MR
First, we identified potential causative factors of AS through univariable MR. The use of the Bonferroni test reduced the probability of a type I error and increased the stability of our results. Furthermore, to eliminate mutual confounding between immune cells, we employed multivariate MR. These methods have significantly reduced the influence of confounding factors and greatly increased result confidence. It has been confirmed that the development of AS can be inhibited by HLA-DR on CD33dim HLA DR+ CD11b+.
It is hypothesized that HLA-DR reduces the risk of AS through its subtype HLA-DRB113. Previous studies have indicated decreased expression of HLA-DRB113 in AS patients (20). Further investigations are necessary to determine if other isoforms may also contribute. Furthermore, AS patients with low activity were found to have decreased CD11b expression compared to normal controls (35). This may be due to CD11b’s ability to inhibit the inflammatory response, thereby reducing the occurrence of AS. In mice, CD11b promotes neutrophil apoptosis to inhibit inflammation (36). It is suspected that a similar effect of reducing inflammation may occur in the human body. Additionally, CD33 inhibits the expression of pro-inflammatory cytokines, including IL-1β and TNF-α (37, 38). Consequently, CD33 may decrease the incidence of AS by mitigating the body’s inflammatory response. However, further research is required to elucidate the specific mechanisms.
Our MR analysis has several strengths. First, we used a comprehensive approach, including univariable, bidirectional, and multivariable MR, to address potential confounding factors and reverse causality. Second, we conducted multiple sensitivity analyses to validate our hypotheses and minimize bias. Third, our research was limited to the European population to reduce population bias. Finally, we used the Bonferroni test to confirm the causative role of AS.
However, it is important to note that our study has limitations. The conclusions drawn from our data could not be immediately generalized to other populations, as all of our data originated from European sources. Additionally, the relatively small sample size may introduce bias, highlighting the need for larger samples to ensure more robust results.
5 Conclusion
Our extensive MR analysis has unveiled the intricate relationship between immune cells and AS. These immune cells can serve as both contributing and protective factors, opening new perspectives for the treatment and prevention of AS. However, the specific mechanisms behind this phenomenon have yet to be fully explored. Therefore, further experiments are necessary to elucidate these underlying mechanisms, and our study can serve as a guiding foundation for future research.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://gwas.mrcieu.ac.uk/.
Author contributions
CQ: Data curation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing, Software. QY: Conceptualization, Methodology, Writing – review & editing, Supervision. ZD: Writing – review & editing, Supervision, Project administration, Methodology. YZ: Conceptualization, Methodology, Writing – review & editing, Writing – original draft, Data curation. MC: Conceptualization, Methodology, Supervision, Writing – review & editing. XW: Writing – review & editing. TH: Writing – review & editing. BL: Writing – review & editing. ZY: Conceptualization, Supervision, Writing – review & editing. SC: Writing – review & editing, Validation, Supervision, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Chongqing Medical Scientific Research Project (Grant No. 2024WSJK057).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1345416/full#supplementary-material
References
1. Wang R, Ward MM. Epidemiology of axial spondyloarthritis: an update. Curr Opin Rheumatol. (2018) 30:137–43. doi: 10.1097/BOR.0000000000000475
2. McKenna SP, Doward L. Functional disability and quality of life in patients with ankylosing spondylitis. Rheumatol Int. (2004) 24:57–8. doi: 10.1007/s00296-003-0376-2
3. Mauro D, Thomas R, Guggino G, et al. Ankylosing spondylitis: an autoimmune or autoinflammatory disease? Nat Rev Rheumatol. (2021) 17:387–404. doi: 10.1038/s41584-021-00625-y
4. Smith JA. Update on ankylosing spondylitis: current concepts in pathogenesis. Curr Allergy Asthma Rep. (2015) 15:489. doi: 10.1007/s11882-014-0489-6
5. Liu L, Yuan Y, Zhang S, Xu J, Zou J. Osteoimmunological insights into the pathogenesis of ankylosing spondylitis. J Cell Physiol. (2021) 236:6090–100. doi: 10.1002/jcp.30313
6. Walker J. Ankylosing spondylitis. Nurs Stand. (2006) 20:48–52. doi: 10.7748/ns2006.07.20.46.48.c4472
7. Zambrano-Zaragoza JF, Agraz-Cibrian JM, González-Reyes C, de Jesús Durán-Avelar M, Vibanco-Pérez N. Ankylosing spondylitis: from cells to genes. Int J Inflammation. (2013) 2013:501653. doi: 10.1155/2013/501653
8. Ciccia F, Bombardieri M, Rizzo A, Principato A, Giardina AR, Raiata F, et al. Over-expression of paneth cell-derived anti-microbial peptides in the gut of patients with ankylosing spondylitis and subclinical intestinal inflammation. Rheumatol Oxf Engl. (2010) 49:2076–83. doi: 10.1093/rheumatology/keq239
9. Duan Z, Gui Y, Li C, Lin J, Gober H-J, Qin J, et al. The immune dysfunction in ankylosing spondylitis patients. Biosci Trends. (2017) 11:69–76. doi: 10.5582/bst.2016.01171
10. Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing mendelian randomization investigations: update for summer 2023. Wellcome Open Res. (2019) 4:186. doi: 10.12688/wellcomeopenres.15555.3
11. Davies NM, Holmes MV, Davey Smith G. Reading mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ. (2018) 362:k601. doi: 10.1136/bmj.k601
12. Widding-Havneraas T, Zachrisson HD. A gentle introduction to instrumental variables. J Clin Epidemiol. (2022) 149:203–5. doi: 10.1016/j.jclinepi.2022.06.022
13. Orrù V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. (2020) 52:1036–45. doi: 10.1038/s41588-020-0684-4
14. Burgess S, Thompson SG. CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
15. Zeng R, Wang J, Jiang R, Yang J, Zheng C, Wu H, et al. Investigating causality and shared genetic architecture between neurodegenerative disorders and inflammatory bowel disease. Aging Dis. (2023) 14:1349–59. doi: 10.14336/AD.2022.12209
16. Higbee DH, Granell R, Hemani G, Davey Smith G, Dodd JW. Lung function, COPD and cognitive function: a multivariable and two sample Mendelian randomization study. BMC Pulm Med. (2021) 21:246. doi: 10.1186/s12890-021-01611-6
17. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. (2016) 45(6):1961–74. doi: 10.1093/ije/dyw220
18. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. (2018) 7:e34408. doi: 10.7554/eLife.34408
19. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
20. Kchir MM, Hamdi W, Laadhar L, Kochbati S, Kaffel D, Saadellaoui K, et al. HLA-B, DR and DQ antigens polymorphism in Tunisian patients with ankylosing spondylitis (a case-control study). Rheumatol Int. (2010) 30:933–9. doi: 10.1007/s00296-009-1079-0
21. Hwang MC, Ridley L, Reveille JD. Ankylosing spondylitis risk factors: a systematic literature review. Clin Rheumatol. (2021) 40:3079–93. doi: 10.1007/s10067-021-05679-7
22. Brewerton DA, Hart FD, Nicholls A, Caffrey M, James DC, Sturrock RD, et al. Ankylosing spondylitis and HL-A 27. Lancet Lond Engl. (1973) 1:904–7. doi: 10.1016/s0140-6736(73)91360-3
23. Schlosstein L, Terasaki PI, Bluestone R, Pearson CM. High association of an HL-A antigen, W27, with ankylosing spondylitis. N Engl J Med. (1973) 288:704–6. doi: 10.1056/NEJM197304052881403
24. Ranganathan V, Gracey E, Brown MA, Inman RD, Haroon N. Pathogenesis of ankylosing spondylitis - recent advances and future directions. Nat Rev Rheumatol. (2017) 13:359–67. doi: 10.1038/nrrheum.2017.56
25. Garrido-Mesa J, Brown MA. T cell Repertoire Profiling and the Mechanism by which HLA-B27 Causes Ankylosing Spondylitis. Curr Rheumatol Rep. (2022) 24:398–410. doi: 10.1007/s11926-022-01090-6
26. Hermann E, Yu DT, Meyer zum Büschenfelde KH, Fleischer B. HLA-B27-restricted CD8 T cells derived from synovial fluids of patients with reactive arthritis and ankylosing spondylitis. Lancet Lond Engl. (1993) 342:646–50. doi: 10.1016/0140-6736(93)91760-j
27. Wright PB, McEntegart A, McCarey D, McInnes IB, Siebert S, Milling SWF, et al. Ankylosing spondylitis patients display altered dendritic cell and T cell populations that implicate pathogenic roles for the IL-23 cytokine axis and intestinal inflammation. Rheumatol Oxf Engl. (2016) 55:120–32. doi: 10.1093/rheumatology/kev245
28. Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discovery. (2012) 11:763–76. doi: 10.1038/nrd3794
29. Baeten D, Baraliakos X, Braun J, Sieper J, Emery P, van der Heijde D, et al. Anti-interleukin-17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: a randomised, double-blind, placebo-controlled trial. Lancet Lond Engl. (2013) 382:1705–13. doi: 10.1016/S0140-6736(13)61134-4
30. Ohta A, Sitkovsky M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature. (2001) 414:916–20. doi: 10.1038/414916a
31. Akhtari M, Zargar SJ, Mahmoudi M, Vojdanian M, Rezaeimanesh A, Jamshidi A, et al. Ankylosing spondylitis monocyte-derived macrophages express increased level of A2A adenosine receptor and decreased level of ectonucleoside triphosphate diphosphohydrolase-1 (CD39), A1 and A2B adenosine receptors. Clin Rheumatol. (2018) 37:1589–95. doi: 10.1007/s10067-018-4055-9
32. Moesta AK, Li X-Y, Smyth MJ. Targeting CD39 in cancer. Nat Rev Immunol. (2020) 20:739–55. doi: 10.1038/s41577-020-0376-4
33. Mittal D, Sinha D, Barkauskas D, Young A, Kalimutho M, Stannard K, et al. Adenosine 2B receptor expression on cancer cells promotes metastasis. Cancer Res. (2016) 76:4372–82. doi: 10.1158/0008-5472.CAN-16-0544
34. Huang S, Apasov S, Koshiba M, Sitkovsky M. Role of A2a extracellular adenosine receptor-mediated signaling in adenosine-mediated inhibition of T-cell activation and expansion. Blood. (1997) 90:1600–10. doi: 10.1182/blood.V90.4.1600
35. Surdacki A, Sulicka J, Korkosz M, Mikolajczyk T, Telesinska-Jasiówka D, Klimek E, et al. Blood monocyte heterogeneity and markers of endothelial activation in ankylosing spondylitis. J Rheumatol. (2014) 41:481–9. doi: 10.3899/jrheum.130803
36. Coxon A, Rieu P, Barkalow FJ, Askari S, Sharpe AH, von Andrian UH, et al. A novel role for the beta 2 integrin CD11b/CD18 in neutrophil apoptosis: a homeostatic mechanism in inflammation. Immunity. (1996) 5:653–66. doi: 10.1016/s1074-7613(00)80278-2
37. Lajaunias F, Dayer J-M, Chizzolini C. Constitutive repressor activity of CD33 on human monocytes requires sialic acid recognition and phosphoinositide 3-kinase-mediated intracellular signaling. Eur J Immunol. (2005) 35:243–51. doi: 10.1002/eji.200425273
Keywords: immune cells, ankylosing spondylitis, multivariable Mendelian randomization, univariable Mendelian randomization, bidirectional Mendelian randomization
Citation: Qin C, Yu Q, Deng Z, Zhang Y, Chen M, Wang X, Hu T, Lei B, Yan Z and Cheng S (2024) Causal relationship between the immune cells and ankylosing spondylitis: univariable, bidirectional, and multivariable Mendelian randomization. Front. Immunol. 15:1345416. doi: 10.3389/fimmu.2024.1345416
Received: 27 November 2023; Accepted: 25 March 2024;
Published: 09 April 2024.
Edited by:
Mark Hwang, University of Texas Health Science Center at Houston, United StatesReviewed by:
Cecilia Contreras-Cubas, National Institute of Genomic Medicine (INMEGEN), MexicoFederico Diaz-Gonzalez, University of La Laguna, Spain
Copyright © 2024 Qin, Yu, Deng, Zhang, Chen, Wang, Hu, Lei, Yan and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengjian Yan, eWFuempAaG9zcGl0YWwuY3FtdS5lZHUuY24=; Si Cheng, MzA0MjM4QGNxbXUuZWR1LmNu
†These authors contributed equally to this work and share first authorship
 Chaofan Qin†
Chaofan Qin† Qingshuai Yu
Qingshuai Yu Si Cheng
Si Cheng