- Department of Neurology, The First Affiliated Hospital of China Medical University, Shenyang, China
Myasthenia gravis (MG) stands as a perplexing autoimmune disorder affecting the neuromuscular junction, driven by a multitude of antibodies targeting postsynaptic elements. However, the mystery of MG pathogenesis has yet to be completely uncovered, and its heterogeneity also challenges diagnosis and treatment. Growing evidence shows the differential expression of non-coding RNAs (ncRNAs) in MG has played an essential role in the development of MG in recent years. Remarkably, these aberrantly expressed ncRNAs exhibit distinct profiles within diverse clinical subgroups and among patients harboring various antibody types. Furthermore, they have been implicated in orchestrating the production of inflammatory cytokines, perturbing the equilibrium of T helper 1 cells (Th1), T helper 17 cells (Th17), and regulatory T cells (Tregs), and inciting B cells to generate antibodies. Studies have elucidated that certain ncRNAs mirror the clinical severity of MG, while others may hold therapeutic significance, showcasing a propensity to return to normal levels following appropriate treatments or potentially foretelling the responsiveness to immunosuppressive therapies. Notably, the intricate interplay among these ncRNAs does not follow a linear trajectory but rather assembles into a complex network, with competing endogenous RNA (ceRNA) emerging as a prominent hub in some cases. This comprehensive review consolidates the landscape of dysregulated ncRNAs in MG, briefly delineating their pivotal role in MG pathogenesis. Furthermore, it explores their promise as prospective biomarkers, aiding in the elucidation of disease subtypes, assessment of disease severity, monitoring therapeutic responses, and as novel therapeutic targets.
1 Introduction
Myasthenia gravis (MG) is an autoimmune disorder affecting the neuromuscular junction, primarily instigated by the presence of antibodies targeting various postsynaptic components (1). Among these antibodies, anti-acetylcholine receptor antibodies (AChR-Ab) stand as the most prevalent, while antibodies against MuSK (MuSK-Ab), Lrp4, and agrin are comparatively less common (2). Additionally, biomarkers such as ColQ, Kv1.4, titin, and RyR are indeed valuable, albeit their specific pathogenic roles remain uncertain (3). The hallmark feature of MG is fatigable muscle weakness, accentuated post-exertion and relieved during rest (4). MG classification encompasses subgroups based on antibody profiles, clinical manifestations, age of onset, and thymus pathology (2). Notably, thymus involvement plays a pivotal role in AChR-Ab-positive MG (AChR-MG), with hyperplasia observed in early-onset MG (EOMG) and atrophy in late-onset MG (LOMG) (2). Thymoma-associated MG (TAMG) represents a distinct subgroup (2). The advent of high-throughput sequencing technology has redefined our perception of ncRNAs, once considered “junk DNA,” as critical cellular regulators (5). This vast class of genomic elements is typically categorized into two main groups: small or short non-coding RNAs and long non-coding RNAs (lncRNAs), based on whether their length exceeds 200 nucleotides (6). Additionally, circular RNAs (circRNAs), unique single-stranded RNA molecules formed by covalent closure at the 5’ and 3’ ends, have emerged as noteworthy constituents of ncRNAs (7). Intriguingly, ncRNAs have exhibited profound associations with diverse diseases, spanning cancer, cardiovascular and cerebrovascular conditions, as well as metabolic and autoimmune disorders (8–10). In systemic lupus erythematosus (SLE), some miRNAs, such as miR-125a, miR-125b, miR-21, miR-148a, miR,223, and miR-31, expressed abnormally, others, like miR-7, miR-155, miR-146, and miR-182 were reported to regulate B cells, T cells, or the formation of germinal centers (GCs) (11–13). While in rheumatoid arthritis (RA), miR-146a, miR-155, miR-22, and miR-10a-5p were related to the inflammatory environment (13). Let-7g-5p and NEAT1, a lncRNA, were proved to promote the proliferation of Th17 (14). With this backdrop, the investigation of ncRNAs on MG pathogenesis has become a compelling area of study. Despite significant advancements in MG management, several challenges persist (2). The etiological “triggers,” underlying molecular mechanisms, and regulatory factors at the molecular level remain elusive, confounding researchers (15). Furthermore, a subset of MG patients fails to achieve remission or substantial clinical improvement with current immunotherapies (16), a concern exacerbated by the drawbacks of long-term immunosuppressant usage, including intolerance, delayed therapeutic onset, and systemic toxicity (17). Our conviction lies in the potential of precision-targeted immunotherapy as a promising avenue to tackle these challenges. In this review, we delve into the role of ncRNAs in MG pathogenesis and explore the significance of ncRNAs in MG treatment and therapeutic monitoring.
2 miRNAs in MG
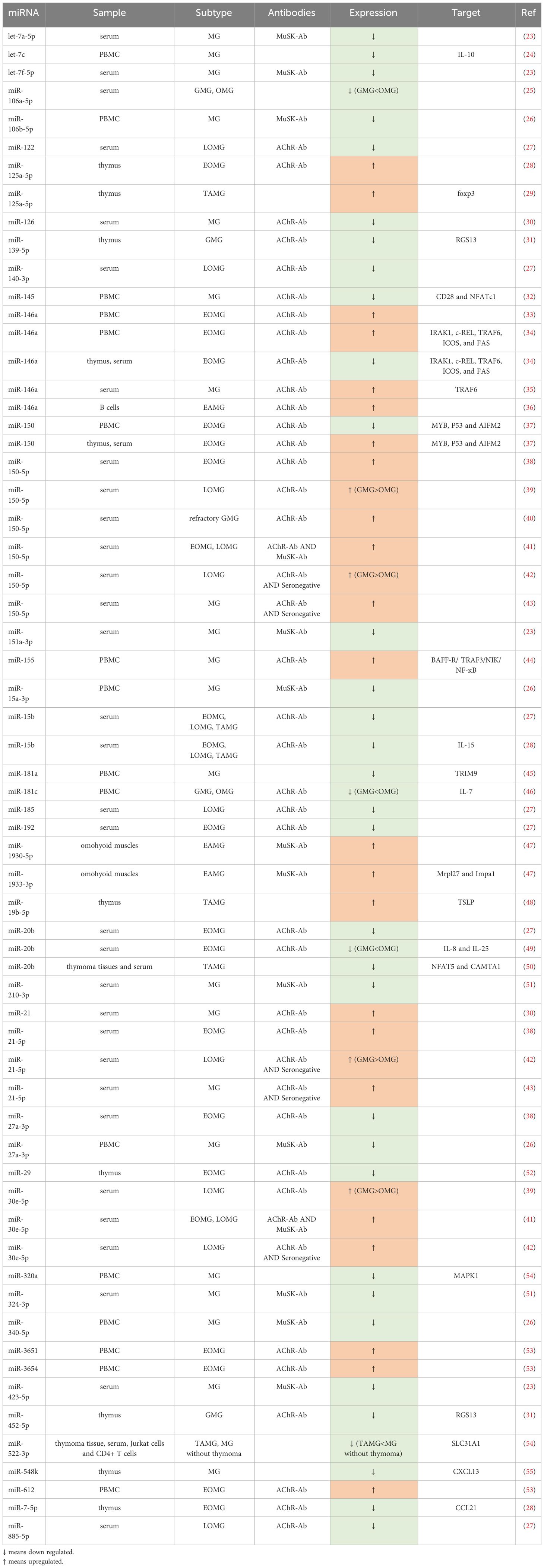
Among the diverse categories of small non-coding RNAs, microRNAs (miRNAs) supreme as one of the most prominent classes, distinguished by their sheer number and prevalence in research (18). These endogenous single-stranded molecules typically comprise approximately 22 nucleotides (19) and were initially discovered by Lee et al. in the early 1990s (20). In the realm of immune system modulation, miRNAs play an integral role, which explains why disruptions in their function have been associated with a range of autoimmune conditions, such as MG (21, 22) (Table 1).
2.1 miRNAs involved in MG pathogenesis
In patients with MG, a series of dysfunctions occur at the neuromuscular junction (NMJ), driven by T cells and mediated by B cells, resulting in a complex pathogenic process (24). This process involves an array of miRNAs and proinflammatory cytokines, potentially contributing to the development of MG (Figure 1).
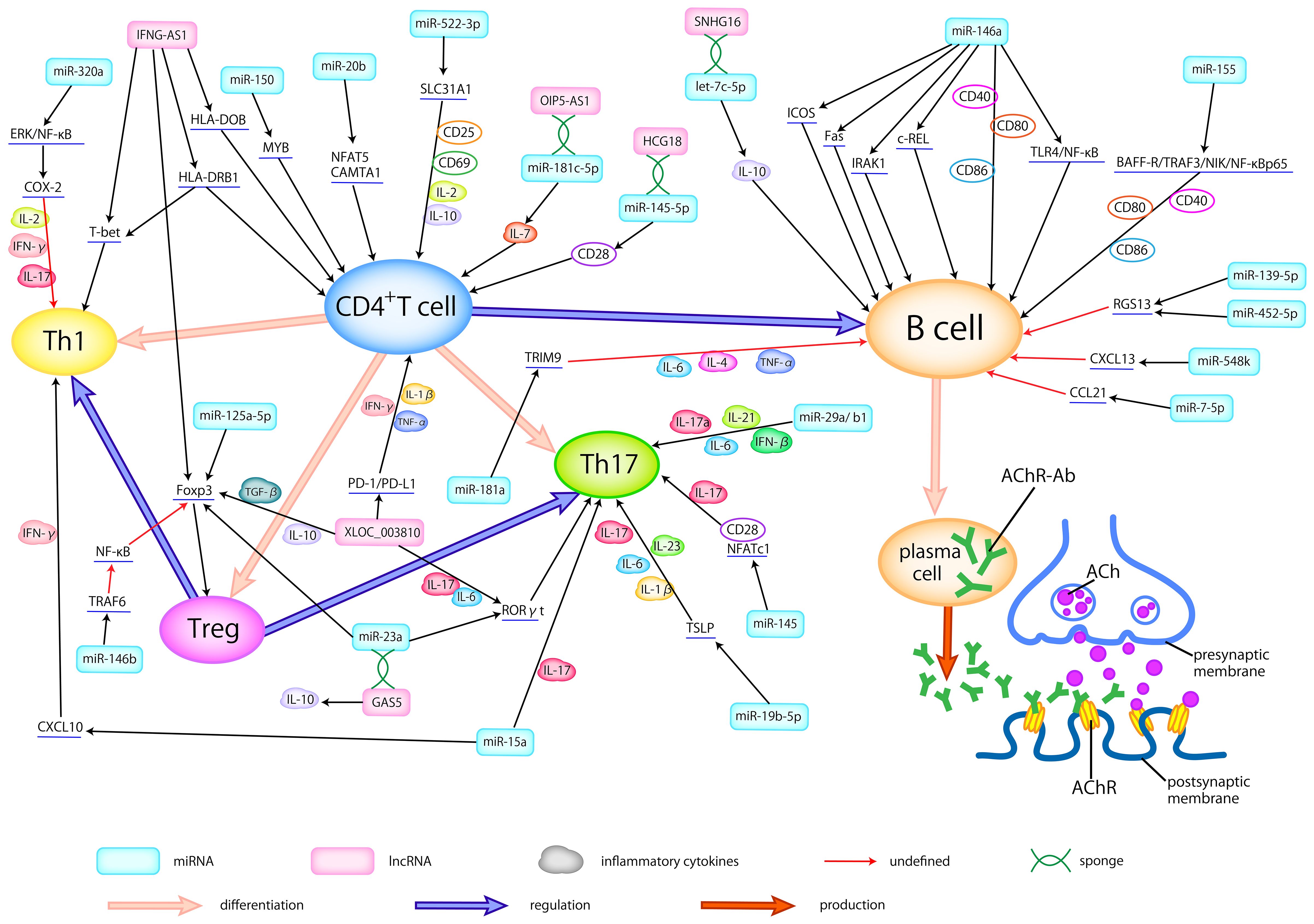
Figure 1 Dysregulated ncRNAs in MG pathogenesis. Abnormally expressed ncRNAs in different types of samples actively participate in the pathogenesis of AChR-MG by regulating the differentiation of T cells, broking the balance between helper T cells (Th1 and Th17) and Treg cells, activating the B cells, modulating the production of inflammatory cytokines, thus promoting the production of antibodies. Here, we delineate the regulatory relationship between abnormally expressed ncRNAs and immune cells, cytokines, and related signaling pathways.
Several studies have explored the role of dysregulated miRNAs in MG by compiling a catalog of genes associated with MG susceptibility based on existing literature (27, 28, 53, 56). Yang et al. (28) conducted an analysis of 93 miRNAs associated with MG risk pathways, shedding light on their regulatory role within these pathways. Their work underscored the potential significance of some specific miRNAs in MG pathogenesis, including miR-497, miR-15a, miR-15b, miR-16, and miR-195. Cao et al. (56) pinpointed miRNA-146a as an important contributor to the regulation of numerous MG risk pathways, underscoring its pivotal role in MG pathogenesis. Moreover, in the study of Bo et al. (53), 13 dysregulated miRNAs, including miR-29b-3p, miR-145-5p, and miR-451a, were believed to play a role in MG pathogenesis. Among these risk-related miRNAs, miR-145-5p was the sole miRNA found to exhibit differential expression, suggesting its potential pivotal function in MG. In a complementary study, Qian et al. (27) reported 30 abnormally expressed miRNAs, which may participate in MG development by modulating Tregs. Collectively, these identified miRNAs offer valuable insights and may serve as guiding principles for future research endeavors aimed at unraveling the intricate pathogenesis of MG.
2.1.1 Modulation of Tregs, Th17 and Th1
It is well-established that the initiation of MG critically hinges on the activation of autoreactive T cells (57). Consequently, the miRNAs exhibiting abnormal expression and responsibility for T cell proliferation and activation may represent a pivotal link in MG pathogenesis. Certain miRNAs have been identified to be involved in the regulation of T cells in MG. Cron et al. (58) discovered that miR-150 could target MYB, a proto-oncogene, thus affecting the survival of CD4+ and CD8+T cells, which may contribute to the maintenance of an immunologically activated state. Research showed that miR-20b would inhibit T cell proliferation and activation by suppressing the NFAT signaling pathway through downregulating NFAT5 and CAMTA1 (33). Moreover, decreased miR-522-3p can lead to the overexpression of CD25, CD69, IL-2, and IL-10 by targeting SLC31A1, consequently promoting the activation and proliferation of Jurkat cells, a T cell leukemia line (31). Dysregulation of key immune cell populations, including Tregs, Th17, and Th1, exerts a significant influence on MG pathogenesis. Studies have reported that the expression of Tregs is associated with disease severity, with observations of decreased Foxp3, a Treg-related cytokine, in peripheral blood mononuclear cells (PBMCs) of MG patients (39). In an AIRE knockout mouse model, reduced Tregs and increased Th17 cells in the thymus rendered the mice more susceptible to experimental autoimmune myasthenia gravis (EAMG) (42), mirroring the pathogenesis of LOMG and TAMG (15). Villegas et al. (46) noted that an imbalance between Tregs and Th17 cells is associated with chronic thymic inflammatory conditions in AChR-MG.
The lapses in Treg function result in the loss of their inhibitory effect on excessive inflammatory responses, leading to uncontrolled inflammatory cell activity in MG (25). This unchecked immune activity gives rise to chronic inflammation and thymic hyperplasia, further disrupting immune tolerance mechanisms. Recent research outcomes have unveiled a miR-146b-TRAF6-NF-κB-Foxp3 pathway, which plays a crucial role in suppressing Treg proliferation and its inhibitory function (49, 59). Intriguingly, Yan et al. (60) proposed that overexpressed miR-146a may contribute to the pathogenesis of MG by promoting TRAF6 expression, which could further disrupt Treg function. miR-125a-5p was confirmed to negatively regulate Foxp3, consequently inhibiting the generation of Treg with a great possibility (37). Additionally, there is a positive correlation between the reduced miR-126 and Foxp3 mRNA in MG, providing strong evidence for the downregulation of Treg activity (50). In summary, differentially expressed miRNAs in MG have been observed to inhibit Treg proliferation and weaken their inhibitory function. Consequently, these miRNA-mediated changes impair immunological tolerance mechanisms and play an important role in the pathogenesis of MG.
Compelling scholarly evidence has demonstrated that the activation of Th17 and Th1 cells, when losing the control of Tregs, can lead to the release of pro-inflammatory cytokines such as INF-γ and IL-17 (61), which may build the bridge between some dysregulated miRNAs and MG. For instance, Wang et al. (62) revealed that miR-145 could serve as a regulator that promotes T cell proliferation and differentiation into Th17 cells, by targeting CD28 and NFATc1. Similarly, Cron et al. (34) reported a reduction of the miR-29 family within thymic tissues of MG, potentially in connection with an increase of IFN-β. Among these, miR-29a/b1, part of a miR-29a genomic cluster, could possibly facilitate the flourishing of Th17 cells. Besides, miR-19b-5p in TAMG has been shown to post-transcriptionally inhibit thymic stromal lymphopoietin (TSLP), thereby regulating Th17 cells and their related cytokines (44). It’s reasonable to speculate that these abnormally expressed miRNAs may function as regulators, promoting the release of Th17-related cytokines (IL-6, TGF-β, IL-17, IL-1β, and IL-23) and the proliferation of Th17 cells. On the grounds of research by Cheng et al. (45), with the target of MAPK1, miR-320a may influence the Th1-associated cytokines, like IL-2, and IFN-γ, by adjusting COX-2 through the regulation of ERK/NF-κB pathways. In addition, several miRNAs contribute to the pathogenesis of MG by influencing both Th1 and Th17 responses concurrently. Liu et al. (63) reported that miR-15a could affect the CXCL10 gene by regulating Th1- and Th17-related cytokines to generate immune responses. In conclusion, the dysregulation of miRNAs can disrupt Treg function, promote the proliferation of Th1 and Th17 cells, and result in the overexpression of pro-inflammatory cytokines, thus modulating the pathogenesis of MG.
2.1.2 miRNAs as regulators of B cell activation
B cells, integral players in the pathogenesis of MG, participate in the formation of ectopic GCs, receive signals from antigen-presenting cells (APCs), and contribute to the production of autoimmune antibodies (38). Many dysregulated miRNAs can modulate chemokines and specific signaling pathways to promote the maturation of autoreactive B cells and the development of GCs.
miR-155, found to be elevated in B cells, engaged in the immune response through the BAFF-R/TRAF3/NIK/NF-κB p65 pathway, mediating the survival of activated B cells and increasing the production of AChR antibodies by regulating co-stimulatory molecules (48). The binding of BAFF to BAFF-R is necessary for B cell maturation and survival (43). And miR-155 can promote the expression of BAFF-R and TRAF3 to facilitate the phosphorylation of NIK, thus helping the NF-κB to translocate into the nuclear of B cells (48). At the same time, miR-155 can also modulate the co-stimulatory molecules, such as CD40, CD80, and CD86, to enhance the production of antibodies by B cells. Two independent studies mentioned an elevation of miR-146a in AchR-specific B cells and they also reported that miR-146a could affect B cell immunity (64, 65). Notably, one of these studies proposed that miR-146a may facilitate the activation of B cells and the production of antibodies by modulating the TLR4 and NF-κB pathways (64). In a study by Bortone et al. (66), the downregulated miR-146a showed a noteworthy negative correlation with IRAK1, c-REL, ICOS, and FAS in the follicular hyperplastic thymus of EOMG. The genes targeted by miR-146a appear to collectively contribute to the activation of B cells and the formation of GCs. Firstly, the downregulation of miR-146a allows IRAK1 to induce excessive inflammation and disrupt immune tolerance through the TLR signaling pathway. Secondly, the deficiency of miR-146a can enhance the activation of c-REL, thereby promoting the proliferation and differentiation of B cells and amplifying GCs formation. Furthermore, reduced miR-146a may fail to effectively restrict the aggregation of follicular helper T cells (Tfhs) and the proliferation of B cells in GCs by targeting ICOS. Lastly, miR-146a can downregulate FAS expression, promoting lymphoproliferation and GCs formation. Wang et al. (67) found that miR-181a may influence the levels of inflammatory cytokines, such as TNF-α, IL-4, and IL-6, affecting B cell proliferation by regulating TRIM9.
Additionally, miR-548k, reduced in the thymus of MG, can target CXCL13 (40), which plays a role in directing B cells and facilitating GCs formation. miR-452-5p and miR-139-5p were associated with the promotion of RGS13 expression, leading to B cell proliferation and GCs expansion (54). miR-150-5p has shown a positive correlation with CD19+ and CD27+ B cells, suggesting its involvement in B cell differentiation, memory B cell formation, and further possible immune responses, including B cell activation and antibody production (30). Additionally, the abnormally expressed miR-126 and miR-21 in PBMCs were proved to upregulate the expression of IL-6, thus promoting the proliferation and differentiation of B cells, consequently facilitating the production of antibodies (50). let-7c was reported to target IL-10, probably participating in MG pathogenesis by stimulating B cells (68). Besides, the reduced miR-7 in thymus was reported to upregulate the expression of CCL21, thus helping the formation of GCs (69).
2.2 miRNAs as clinical biomarkers and therapeutic targets
2.2.1 miRNAs engaged in AChR-MG
AChR-Ab is the most prevalent antibody in MG, accounting for approximately 85% of cases (70). AChR-MG can be further classified according to clinical features. Interestingly, there are some differences in thymus pathology among EOMG, LOMG and TAMG. EOMG refers to individuals under the age of 50, characterized by hyperplastic thymus pathology. In contrast, LOMG manifests in individuals over the age of 50, commonly associated with thymic atrophy. TAMG, on the other hand, represents a paraneoplastic syndrome of thymoma (4). A few of studies (54, 55, 58, 64, 66, 69, 71) revealed some differentially expressed miRNAs, such as miR-139-5p, miR-452-5p, miR-612, miR-3654, miR-365, miR-150, miR-20b, miR-192, miR-7, miR-125a-5p, and miR-146a, in the AChR antibody-positive EOMG (AChR-EOMG) patients. A decrease in miR-15b expression was observed in the serum of both early-onset and late-onset AChR-MG (71), which aligns with findings from an animal model of EAMG in mice injected with Torpedo AChR (72). In addition, in patients with AChR antibody-positive LOMG (AChR-LOMG), miR-122, miR-140-3p, miR-185, miR-885-5p, miR-106b-3p, miR-223-5p, miR-140-5p, miR-19b-3p, miR-30e-5p, and miR-150-5p were found to express abnormally (29, 32, 71). These findings suggest a nuanced expression profile of dysregulated miRNAs between EOMG and LOMG. Many miRNAs were reported to express aberrantly in TAMG, including miR-125a-5p, miR-19b-5p, miR-20b and miR-522-3p (25, 33, 50, 58). Additionally, a study by Shi et al. (72) indicated a decrease in miR-15b expression in TAMG, EOMG, and LOMG patients.
Several dysregulated miRNAs in AChR-MG, compared with ocular myasthenia gravis (OMG), have more severe abnormalities in generalized myasthenia gravis (GMG), some of which are correlated with quantitative myasthenia gravis scores (QMGs) or myasthenia gravis composite scores (MGCs), reflecting the severity of MG and further serving as potential biomarkers for disease surveillance with great possibility. Research findings indicated that both OMG and GMG patients showed elevated miR-150-5p, miR-21-5p, and miR-30e-5p, which had a positive correlation with MGCs and significantly more overexpressed in GMG than in OMG (29, 32). Conversely, the lower expressed miR-181a, miR-106a-5p, and miR-20b in MG, had a negative relationship between QMGs, with GMG displaying notably reduced levels compared to OMG (73–75). Additionally, several studies (51, 76) on curative effect indicated a parallel between the reduction of AChR-Ab titers and the improvement of symptoms. A fast decrease of AChR-Ab titers more than 50% also made a clinical sense (23). Interestingly, some studies showed that miR-126, and miR-145 were negatively correlated with AChR-Ab titer (50, 62), while miR-21 was positively related to AChR-Ab titer (50). We therefore speculate that specific ncRNAs may also be useful for efficacy detection.
MiR-150-5p, miR-30e-5p, and miR-146a levels in the serum of AChR-MG patients have shown promising responses to various treatments like thymectomy and immunosuppression, indicating a correlation between their levels and clinical improvement (26, 30, 32, 35, 52, 58, 66). These miRNAs could potentially serve as biomarkers for monitoring therapeutic efficacy and reflecting the disease severity. Furthermore, research by Zhang et al. (65) suggested that silencing miR-146a could reduce the expression of CD40, CD80, and CD86 on the surface of B cells, inhibit B cell differentiation, and consequently reduce the production of anti-AChR antibodies. By comparing the level of miR-146a and the expression of c-REL in the thymus of MG in corticosteroid-naïve patients and corticosteroid-treated patients, Bortone et al. (66) reported that after corticosteroid treatment, the immune response of c-REL, which was originally active in GCs and infiltrating B cells of untreated patients, was strongly inhibited, highlighting the miR-146a/c-REL axis as a potential therapeutic target for immunosuppressants. These discoveries suggests that miR-146a could serve as both a therapeutic target and a monitoring indicator for treatment efficacy. Moreover, Wang et al. (48) demonstrated that silencing miR-155 could inhibit the production of anti-T-AChR antibodies, making miR-155 another potential treatment target. Sengupta et al. (54) proposed that miR-139-5p and miR-452-5p might be considered for MG treatment, especially for EOMG, as their mimics inhibit B cell chemotaxis and GCs formation. Additionally, Cavalcante et al. reported that AChR-MG with abnormally expressed miRNAs such as miR-323b-3p, miR-409-3p, miR-485-3p, miR-181d-5p, and miR-340-3p are not sensitive to immunosuppressants (47), suggesting the presence of these miRNAs may result in poor treatment. In other words, these miRNAs may be used to forecast clinical response.
In essence, miRNAs exhibit subtle variations in expression across MG subgroups. Certain miRNAs with abnormal expression levels have the potential to facilitate disease monitoring, aid in targeted therapy, and assess treatment efficacy. Firstly, several miRNAs tend to normalize after appropriate treatment. Secondly, certain miRNAs may be implicated in antibody production, making them potential targets for MG treatment. Lastly, the presence of specific miRNAs may indicate the response to treatment. These miRNAs hold promise as specific biomarkers reflecting disease severity or treatment responses and may serve as novel targets for MG therapy. Consequently, there arises the prospect of precision therapy and personalized monitoring for individuals with MG.
2.2.2 miRNAs intertwined with MuSK-MG
MuSK antibodies are present in approximately 1-10% of MG cases, predominantly affecting young women under the age of 40 (77). Although research on MuSK-Ab-positive MG (MuSK-MG) has not been as extensive as that on AChR-MG, several studies have reported differentially expressed miRNAs in this subgroup. Sabre et al. (36) observed a significant decrease in miR-210-3p and miR-324-3p levels in the serum of MuSK-MG. Punga et al. (78) identified differential expression of let-7a-5p, let-7f-5p, miR-151a-3p, and miR-423-5p in the serum of MuSK-MG compared to healthy controls. An investigation of PBMCs in MuSK-MG revealed 5 overexpressed and 96 under expressed miRNAs, with marked decreases observed in miR-340-5p, miR-106b-5p, miR-27a-3p, and miR-15a-3p (79). Moreover, an animal study identified 13 abnormally expressed miRNAs in the omohyoid muscle of MuSK-Ab-positive EAMG mice, particularly highlighting the elevation of miR-1933-3p and miR-1930-5p (80). Additionally, in a therapeutic study, the serum level of miR-151a-3p in MuSK-MG patients decreased after treatment (81), indicating a possibility for miR-151a-3p as potential therapeutic target and monitor for treatment efficacy in MuSK-MG.
Unlike AChR and MuSK antibodies, Lrp4 and agrin antibodies are rare in MG, resulting in limited research on these subgroups. Although some miRNAs have been identified as potential regulators of Lrp4 or agrin (82–88), the precise relationship between miRNAs and these two antibodies remains undiscovered (89).
3 lncRNAs in MG
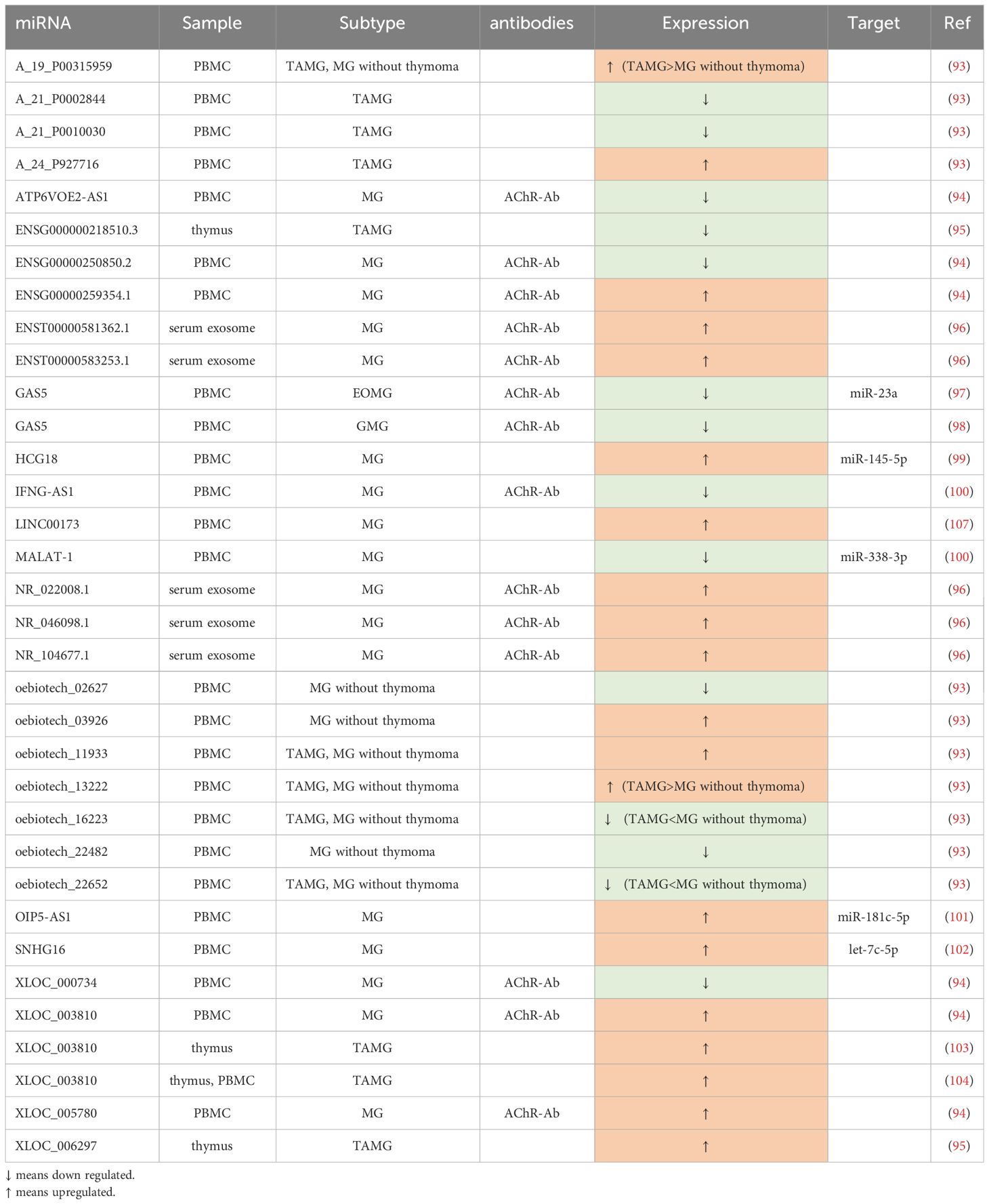
Being different from miRNAs, lncRNA genes exhibit low conservation across evolution (90). They play a pivotal role in modulating the immune system through various mechanisms, such as guiding the development of immune cell lineages, orchestrating dynamic transcriptional programs that activate immune cells (91), and regulating immune-related genes (92). As an autoimmune disease, MG has a close connection with lncRNAs (Table 2).
3.1 The role of lncRNAs in MG pathogenesis
LncRNAs have the capacity to modulate the balance between T cell subtypes and regulate proinflammatory cytokines, thereby participating in the pathogenesis of MG (Figure 1). Interestingly, some lncRNAs can serve as the ceRNA, certain RNA sequestering another RNA to influence its primary targets through microRNA response elements (MREs) (95) to regulate the development of MG.
Several studies have aimed to construct lncRNA-related networks to uncover potential relationships among genes, ncRNAs, and signaling pathways, thus shedding light on MG pathogenesis. For instance, Xu et al. (104) identified LINC00173, FAM13A-AS1, and OIP5-AS1 as closely associated with phosphatase and tensin homolog (PTEN), with LINC00173 showing promise as a potential MG biomarker. Another study by Lu et al. (97) suggested that lncRNAs such as NR_104677.1, NR_022008.1, and ENST00000581362.1 may play a role in MG progression by acting as miRNA sponges in specific tuples involving miRNAs like miR-15b-5p and miR-146a-5p. Hong et al. (93), in their research involving culturing human primary myoblast cells with AChR antibodies, established a co-expressed network of lncRNAs and protein-coding RNAs. They found that MEG3, RP11-184M15.1, and SNHG3 were co-expressed with several protein-coding RNAs, and MEG3, in particular, was linked to cellular homeostasis pathways. The dysregulation of MEG3 might contribute to the development of MG by disrupting cellular equilibrium.
3.1.1 Regulation of immune cells and cytokines
Just like miRNAs, dysregulated lncRNAs can also influence pro-inflammatory cytokines and associated signaling pathways, promoting the proliferation of CD4+ T cells and B cells, inhibiting the proliferation of Treg cells, and activating Th17 and Th1 cells to disrupt immune homeostasis in MG.
In the study by Xu et al. (98), GAS5 was found to decrease in CD4+T cells and directly negatively regulate the expression of miR-23a. Furthermore, overexpressed GAS5 was shown to disrupt the balance between Th17 and Treg cells, restraining Th17 differentiation by sponging miR-23a. Another related study (103) indicated that the upregulation of GAS5 was associated with increased levels of IL-10, coinciding with an improvement in MG symptoms. Two studies by Hu et al. (94, 102) demonstrated that XLOC_003810 was elevated in thymic CD4+T cells in MG. In one study, overexpression of XLOC_003810 in TAMG disrupted the balance between Treg and Th17 by favoring Th17 differentiation and increasing Th17-associated markers such as RORγt, IL-6, and IL-17, while reducing Treg-related markers like Foxp3, TGF-β1, and IL-10. Another study emphasized that XLOC_003810 promoted the expression of CD4+T cells and their inflammatory cytokines, such as IFN-γ, TNF-α, and IL-1β, highlighting its significant role in MG pathogenesis, especially in TAMG. Luo et al. (100) found lower levels of IFNG-AS1 in PBMCs of MG, which were negatively correlated with HLA-DOB and HLA-DRB1. They also demonstrated that increased IFNG-AS1 could suppress the proliferation of Th1 cells and promote the expansion of Treg cells, along with some of their transcription factors. IFNG-AS1 was considered to downregulate CD40L and T-bet in CD4+T cells of MG, partly dependent on HLA-DRB1, implying the involvement of IFNG-AS1 in CD4+ T-related immune responses. HCG18, an upregulated ceRNA in PBMCs, sponging miR-145-5p to modulate CD28, was proved to inhibit apoptosis and enhance the proliferation of Jurkat cells (99). Moreover, OIP5-AS1 was also verified to have a similar effect on Jurkat cells in another study (105), which can be achieved through regulating IL-7 by sponging miR-181c-5p. Additionally, Wang et al. (101) demonstrated that the upregulated SNHG16 can serve as a ceRNA, competitively binding with let-7c-5p in PBMCs of MG. This action not only facilitates Jurkat cell proliferation and inhibits their apoptosis but also influences the level of IL-10, potentially further promoting the activation of B cells.
3.2 Clinical prospects and therapeutic potential of lncRNAs
In AChR-MG, some of the abnormally expressed lncRNAs are closely related to QMGs and MG Impairment Index (MGII), making them indicative of disease severity, while others may aid in subgroup diagnosis, particularly for TAMG. Luo et al. (100) found that IFNG-AS1 in PBMCs exhibited significant negative correlations with QMGs. Besides, GAS5 was also observed to have strong associations with QMGs and MGII (103). In another expression profile, five lncRNAs, NR_104677.1, ENST00000583253.1, NR_046098.1, NR_022008.1, and ENST00000581362.1, were reported to remarkedly overexpress in MG exosome, wherein, NR_046098.1 was upregulated prominently with the severity of MG (97). Another study profiling lncRNA expression identified numerous dysregulated lncRNAs in PBMCs (96). Among them, ENSG00000250850.2, ATP6VOE2-AS1, and XLOC_000734 were the three most significantly downregulated lncRNAs, while XLOC_003810, XLOC_005780, and ENSG00000259354.1 were the top three highly upregulated lncRNAs when compared to healthy controls. In the context of TAMG, there were 3,699 upregulated lncRNAs and 661 downregulated lncRNAs identified. Notably, XLOC_006297 exhibited the highest expression, while ENSG000000218510.3 had the lowest expression levels among the identified lncRNAs (106). In a study involving 34 MG patients and 13 healthy controls, Luo et al. (107) identified significant dysregulation of lncRNAs in PBMCs. Their findings revealed distinct expression patterns when comparing different experimental and control groups. Some of their key observations relied on the fact that, compared with the control group, oebiotech_11933 and A_24_P927716 were the top two overexpressed lncRNAs, while A_21_P0010030 and A_21_P0002844 were the least expressed in TAMG patients. They have also verified that the expression of A_19_P00315959 and oebiotech_13222 in TAMG was much more elevated than that in non-thymoma MG, while the level of oebiotech_22652 and oebiotech_16223 in TAMG patients was much lower. Finally, oebiotech_11933 and oebiotech_03926 were significantly upregulated, whereas ebiotech_02627 and oebiotech_22482 were substantially reduced in MG patients without thymoma versus healthy controls.
In addition to serving as biomarkers, the dysregulated lncRNAs also have the potential to be targeted for treatment. Kong et al. (108) suggested that MALAT-1 may function as an endogenous sponge, competing with male-specific lethal 2 (MSL2) to bind miR-338-3p. This interaction could lead to the inhibition of T cells and potentially play a protective role, making the MALAT-1-miR-338-3p-MSL2 network a promising therapeutic target.
4 circRNAs in MG
circRNAs were first reported in viroids by Sanger et al. (109) in 1976. These molecules, characterized by their circular structure, can be categorized into four main types (110): exonic circRNAs (ecircRNAs), exon-intron circRNAs (EIciRNAs), intronic circRNAs (ciRNAs), and tRNA intronic circular RNAs (tricRNAs). While the functions of most circRNAs remain poorly understood, they are known to play crucial roles in immune regulation, including the adjustment of immune cells, handling immune responses, and modulating immune signaling pathways (111).
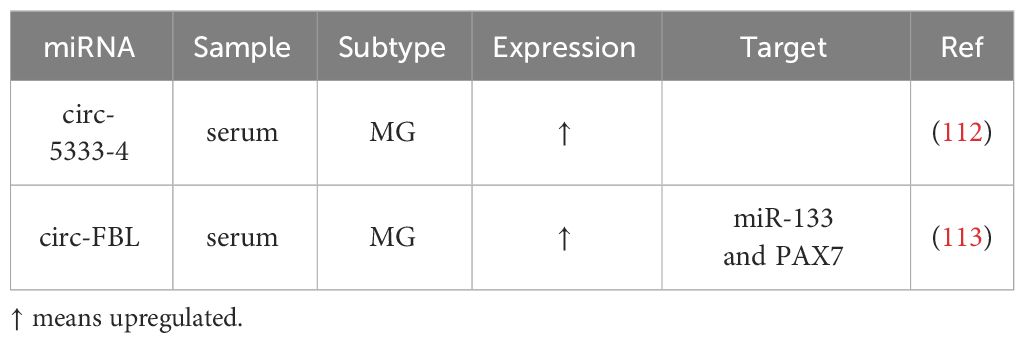
Recent research has begun to shed light on aberrant circRNAs in MG (Table 3), although only a limited number of studies have explored the connection between circRNAs and MG to date. These abnormally expressed circRNAs may contribute to the pathogenesis of MG, reflect disease severity, and present a potential as therapeutic targets.
Two published studies on circRNAs and MG have identified several circRNAs with altered expression patterns that may participate in MG pathogenesis and serve as potential biomarkers: First, four circRNAs, circ-5333-4, circ-0076490, circ-0047056, and circ-16293-1, were found to be dysregulated in the peripheral blood of MG patients. Among these, circ-5333-4 exhibited significant upregulation and was associated with QMGs, indicating a potential connection between its expression and MG severity (112). The specificity of circ-5333-4 for MG was highlighted by comparing MG and SLE cohorts. And circ-5333-4 was proposed to be part of a ceRNA regulation network involving miR-4310 and MORF4L2, warranting further investigation of potential for MG diagnosis and monitoring. Second, in a more recent study investigating circRNAs in MG serum, circ-FBL was identified as an overexpressed circRNA (113). It was suggested to function as a ceRNA by sponging miR-133, thereby promoting the expression of Pax7. This interaction was found to enhance myoblast proliferation, potentially compensating for MG-related muscle weakness, suggesting a therapeutic effect when Pax7 regulated by circ-FBL expressed to a great extent.
5 Conclusions and future directions
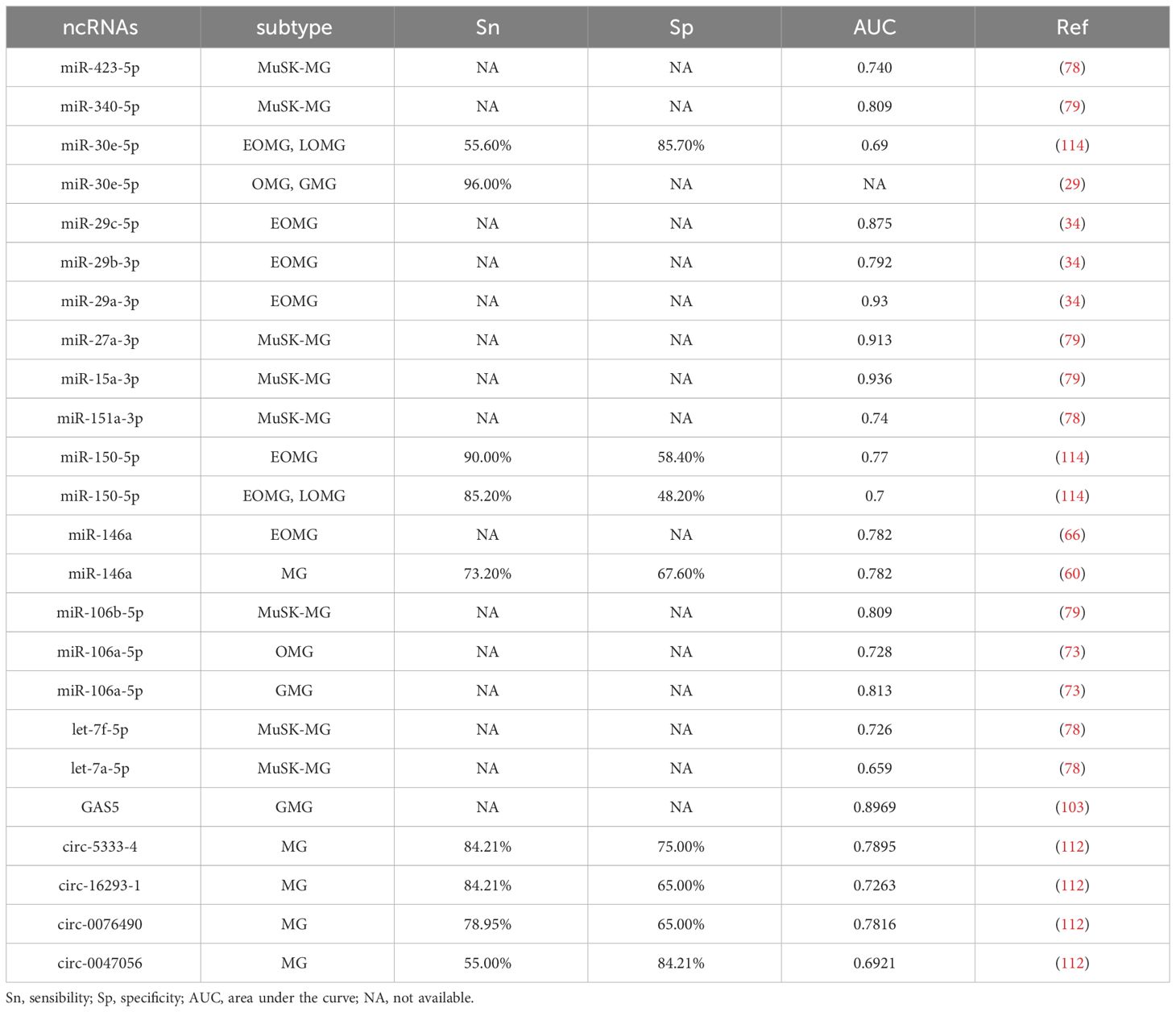
MG is characterized by complexity in diagnosis, treatment variability, and lacks curative options. ncRNAs have emerged as vital biomarkers for delineating MG subgroups, assessing severity, monitoring responses, and offering treatment targets. Research highlights aberrant ncRNAs in MG pathogenesis, suggesting their potential as biomarkers (Table 4). While immune dysregulation and antibody production are known MG drivers, precise disease initiators remain elusive. ncRNAs are integral in MG pathogenesis, potentially offering avenues for disease prevention. Some differentially expressed ncRNAs could modulate T cell differentiation, disrupt the delicate balance between helper T cells and regulatory T cells, activate B cells, regulate inflammatory cytokine production, and function as ceRNAs or not to construct intricate networks involving related pathways. Moreover, Losen et al. (115) reported that short hairpin RNA (shRNA) could disturb the neuromuscular transmission by reducing the level of rapsyn, a bridge protein between AChR and the cytoskeleton in the postsynaptic membrane (2). In addition, some ncRNAs were reported to protect the muscle endplate from the complement attack (116). The regulation of several miRNAs, such as miR-206, miR-127, and miR-29b, can affect the differentiation of satellite cells, the main muscle stem cells (117). A few of ncRNAs were found to serve as ceRNAs to modulate the muscle development (113, 117). It follows that ncRNAs may not only influence the production of specific antibodies, but also take part in the stabilization of postsynaptic membrane, the regulation of complement, and the regeneration of muscle.
Several drugs targeting T cells and B cells have sprung up to treat MG. In parallel, the burgeoning interest in the role of ncRNAs in modulating B cells and T cells presents a fertile ground for exploration. With the potential for regulating GCs formation, B cell differentiation, certain ncRNAs, such as miR-146a, miR-155, miR-139-5p, and miR-452-5p, exhibit potential as innovative therapeutic targets, while others may serve as prognostic indicators for clinical response. Furthermore, some ncRNAs with protective roles in MG may harbor therapeutic potential, like circ-FBL (113).
Beyond their therapeutic implications, ncRNAs can also serve as indicators for monitoring clinical therapeutic responsiveness, by changing in their levels before and after the treatment and effecting on antibody titers, thus facilitating individualized treatment approaches. Notably, the presence of some miRNAs, such as miR-323b-3p, miR-409-3p, miR-485-3p, miR-181d-5p and miR-340-3p, made patients insensitive to immunosuppressive therapies (47), suggesting the need for a reevaluation of treatment strategies.
The preceding findings underscore the significance of further exploration into the role of ncRNAs in MG. However, several critical areas remain uncharted. Firstly, a subset known as ‘seronegative MG’ (41) patients that lack detectable antibodies still present a diagnostic challenge, and the exploration of ncRNAs may offer insights into the diagnosis of them. Furthermore, we know less about the ncRNAs associated with uncommon antibodies at the neuromuscular junction, such as anti-LRP4 and Agrin antibodies. Secondly, many studies have focused solely on documenting the aberrant expression of ncRNAs without delving into their specific roles in MG pathogenesis. Lastly, there is a paucity of research on circRNAs in MG, despite their potential to uncover novel aspects of MG pathogenesis and open new avenues for treatment. In summary, the enigmatic pathogenesis of MG and its association with ncRNAs necessitate comprehensive and ongoing exploration to expand our understanding. This expanded knowledge will better equip us to navigate the complexities associated with the disease and develop more tailored and effective treatment approaches.
Author contributions
BW: Writing – review & editing, Writing – original draft. YZ: Writing – review & editing. DL: Writing – review & editing. CH: Writing – review & editing. RZ: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (81501006, 81400950). The design collection and analysis were supported by the funding.
Acknowledgments
We are deeply grateful to all participants of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
References
1. Huijbers MG, Marx A, Plomp JJ, Le Panse R, Phillips WD. Advances in the understanding of disease mechanisms of autoimmune neuromuscular junction disorders. Lancet Neurol. (2022) 21:163–75. doi: 10.1016/S1474-4422(21)00357-4
2. Gilhus NE, Tzartos S, Evoli A, Palace J, Burns TM, Verschuuren JJGM. Myasthenia gravis. Nat Rev Dis Primers. (2019) 5:30. doi: 10.1038/s41572-019-0079-y
3. Punga AR, Maddison P, Heckmann JM, Guptill JT, Evoli A. Epidemiology, diagnostics, and biomarkers of autoimmune neuromuscular junction disorders. Lancet Neurol. (2022) 21:176–88. doi: 10.1016/S1474-4422(21)00297-0
4. Dresser L, Wlodarski R, Rezania K, Soliven B. Myasthenia gravis: epidemiology, pathophysiology and clinical manifestations. J Clin Med. (2021) 10:2235. doi: 10.3390/jcm10112235
5. Alexander RP, Fang G, Rozowsky J, Snyder M, Gerstein MB. Annotating non-coding regions of the genome. Nat Rev Genet. (2010) 11:559–71. doi: 10.1038/nrg2814
6. Meller VH, Joshi SS, Deshpande N. Modulation of chromatin by noncoding RNA. Annu Rev Genet. (2015) 49:673–95. doi: 10.1146/annurev-genet-112414-055205
7. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
8. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. (2017) 127:761–71. doi: 10.1172/JCI84424
9. Mazzone R, Zwergel C, Artico M, Taurone S, Ralli M, Greco A, et al. The emerging role of epigenetics in human autoimmune disorders. Clin Epigenet. (2019) 11:34. doi: 10.1186/s13148-019-0632-2
10. Keijzers M, Nogales-Gadea G, de Baets M. Clinical and scientific aspects of acetylcholine receptor myasthenia gravis. Curr Opin Neurol. (2014) 27:552–7. doi: 10.1097/WCO.0000000000000125
11. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, et al. Circular RNAs in immune responses and immune diseases. Theranostics. (2019) 9:588–607. doi: 10.7150/thno.29678
12. Schell SL, Rahman ZSM. miRNA-mediated control of B cell responses in immunity and SLE. Front Immunol. (2021) 12:683710. doi: 10.3389/fimmu.2021.683710
13. Zhang L, Wu H, Zhao M, Chang C, Lu Q. Clinical significance of miRNAs in autoimmunity. J Autoimmun. (2020) 109:102438. doi: 10.1016/j.jaut.2020.102438
14. Ali SA, Peffers MJ, Ormseth MJ, Jurisica I, Kapoor M. The non-coding RNA interactome in joint health and disease. Nat Rev Rheumatol. (2021) 17:692–705. doi: 10.1038/s41584-021-00687-y
15. Melzer N, Ruck T, Fuhr P, Gold R, Hohlfeld R, Marx A, et al. Clinical features, pathogenesis, and treatment of myasthenia gravis: a supplement to the Guidelines of the German Neurological Society. J Neurol. (2016) 263:1473–94. doi: 10.1007/s00415-016-8045-z
16. Schneider-Gold C, Gilhus NE. Advances and challenges in the treatment of myasthenia gravis. Ther Adv Neurol Disord. (2021) 14:17562864211065406. doi: 10.1177/17562864211065406
17. Rodolico C, Nicocia G, Damato V, Antonini G, Liguori R, Evoli A. Benefit and danger from immunotherapy in myasthenia gravis. Neurol Sci. (2021) 42:1367–75. doi: 10.1007/s10072-021-05077-6
18. Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. (2016) 17:1712. doi: 10.3390/ijms17101712
19. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
20. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y
21. Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. (2016) 15:1171–80. doi: 10.1016/j.autrev.2016.09.003
22. Le Panse R, Berrih-Aknin S. Autoimmune myasthenia gravis: autoantibody mechanisms and new developments on immune regulation. Curr Opin Neurol. (2013) 26:569–76. doi: 10.1097/WCO.0b013e328364d6cd
23. Marcuse F, Brandts L, Moens D, Damoiseaux J, Hochstenbag M, Hoeijmakers JGJ, et al. The association between anti-acetylcholine receptor antibody level and clinical improvement in myasthenia gravis. Eur J Neurol. (2022) 29:1187–97. doi: 10.1111/ene.15238
24. Zhao R, Luo S, Zhao C. The role of innate immunity in myasthenia gravis. Autoimmun Rev. (2021) 20:102800. doi: 10.1016/j.autrev.2021.102800
25. Berrih-Aknin S, Le Panse R. Myasthenia gravis: a comprehensive review of immune dysregulation and etiological mechanisms. J Autoimmun. (2014) 52:90–100. doi: 10.1016/j.jaut.2013.12.011
26. Molin CJ, Sabre L, Weis CA, Punga T, Punga AR. Thymectomy lowers the myasthenia gravis biomarker miR-150-5p. Neurol Neuroimmunol Neuroinflamm. (2018) 5:e450. doi: 10.1212/NXI.0000000000000450
27. Qian K, Xu JX, Deng Y, Peng H, Peng J, Ou CM, et al. Signaling pathways of genetic variants and miRNAs in the pathogenesis of myasthenia gravis. Gland Surg. (2020) 9:1933–44. doi: 10.21037/gs
28. Yang L, Wang J, Sun X, Cao Y, Ning S, Zhang H, et al. Identifying a polymorphic ‘switch’ that influences miRNAs’ regulation of a myasthenia gravis risk pathway. PloS One. (2014) 9:e104827. doi: 10.1371/journal.pone.0104827
29. Sabre L, Maddison P, Wong SH, Sadalage G, Ambrose PA, Plant GT, et al. miR-30e-5p as predictor of generalization in ocular myasthenia gravis. Ann Clin Transl Neurol. (2019) 6:243–51. doi: 10.1002/acn3.692
30. Zhong H, Lu J, Jing S, Xi J, Yan C, Song J, et al. Low-dose rituximab lowers serum Exosomal miR-150-5p in AChR-positive refractory myasthenia gravis patients. J Neuroimmunol. (2020) 348:577383. doi: 10.1016/j.jneuroim.2020.577383
31. Lu H, Wang H, Sun P, Wang J, Li S, Xu T. MiR-522-3p inhibits proliferation and activation by regulating the expression of SLC31A1 in T cells. Cytotechnology. (2021) 73:483–96. doi: 10.1007/s10616-021-00472-5
32. Sabre L, Maddison P, Sadalage G, Ambrose PA, Punga AR. Circulating microRNA miR-21-5p, miR-150-5p and miR-30e-5p correlate with clinical status in late onset myasthenia gravis. J Neuroimmunol. (2018) 321:164–70. doi: 10.1016/j.jneuroim.2018.05.003
33. Xin Y, Cai H, Lu T, Zhang Y, Yang Y, Cui Y. miR-20b inhibits T cell proliferation and activation via NFAT signaling pathway in thymoma-associated myasthenia gravis. BioMed Res Int. (2016) 2016:9595718. doi: 10.1155/2016/9595718
34. Cron MA, Payet CA, Fayet OM, Maillard S, Truffault F, Fadel E, et al. Decreased expression of miR-29 family associated with autoimmune myasthenia gravis. J Neuroinflammat. (2020) 17:294. doi: 10.1186/s12974-020-01958-3
35. Punga T, Le Panse R, Andersson M, Truffault F, Berrih-Aknin S, Punga AR. Circulating miRNAs in myasthenia gravis: miR-150-5p as a new potential biomarker. Ann Clin Transl Neurol. (2014) 1:49–58. doi: 10.1002/acn3.24
36. Sabre L, Guptill JT, Russo M, Juel VC, Massey JM, Howard JF Jr, et al. Circulating microRNA plasma profile in MuSK+ myasthenia gravis. J Neuroimmunol. (2018) 325:87–91. doi: 10.1016/j.jneuroim.2018.10.003
37. Li J, Qiu D, Chen Z, Du W, Liu J, Mo X. Altered expression of miR-125a-5p in thymoma-associated myasthenia gravis and its down-regulation of foxp3 expression in Jurkat cells. Immunol Lett. (2016) 172:47–55. doi: 10.1016/j.imlet.2016.02.005
38. Zhang X, Liu S, Chang T, Xu J, Zhang C, Tian F, et al. Intrathymic tfh/B cells interaction leads to ectopic GCs formation and anti-AChR antibody production: central role in triggering MG occurrence. Mol Neurobiol. (2016) 53:120–31. doi: 10.1007/s12035-014-8985-1
39. Masuda M, Matsumoto M, Tanaka S, Nakajima K, Yamada N, Ido N, et al. Clinical implication of peripheral CD4+CD25+ regulatory T cells and Th17 cells in myasthenia gravis patients. J Neuroimmunol. (2010) 225:123–31. doi: 10.1016/j.jneuroim.2010.03.016
40. Li J, Qiu D, Chen Z, Du W, Liu J, Mo X. miR-548k regulates CXCL13 expression in myasthenia gravis patients with thymic hyperplasia and in Jurkat cells. J Neuroimmunol. (2018) 320:125–32. doi: 10.1016/j.jneuroim.2018.03.021
41. Lenti MV, Rossi CM, Melazzini F, Gastaldi M, Bugatti S, Rotondi M, et al. Seronegative autoimmune diseases: A challenging diagnosis. Autoimmun Rev. (2022) 21:103143. doi: 10.1016/j.autrev.2022.103143
42. Aricha R, Feferman T, Scott HS, Souroujon MC, Berrih-Aknin S, Fuchs S. The susceptibility of Aire (-/-) mice to experimental myasthenia gravis involves alterations in regulatory T cells. J Autoimmun. (2011) 36:16–24. doi: 10.1016/j.jaut.2010.09.007
43. McAllister E, Jellusova J. BAFF signaling in B cell metabolism. Curr Opin Immunol. (2021) 71:69–74. doi: 10.1016/j.coi.2021.05.011
44. Wang Z, Chen Y, Xu S, Yang Y, Wei D, Wang W, et al. Aberrant decrease of microRNA19b regulates TSLP expression and contributes to Th17 cells development in myasthenia gravis related thymomas. J Neuroimmunol. (2015) 288:34–9. doi: 10.1016/j.jneuroim.2015.08.013
45. Cheng Z, Qiu S, Jiang L, Zhang A, Bao W, Liu P, et al. MiR-320a is downregulated in patients with myasthenia gravis and modulates inflammatory cytokines production by targeting mitogen-activated protein kinase 1. J Clin Immunol. (2013) 33:567–76. doi: 10.1007/s10875-012-9834-5
46. Villegas JA, Van Wassenhove J, Le Panse R, Berrih-Aknin S, Dragin N. An imbalance between regulatory T cells and T helper 17 cells in acetylcholine receptor-positive myasthenia gravis patients. Ann N Y Acad Sci. (2018) 1413:154–62. doi: 10.1111/nyas.13591
47. Cavalcante P, Mizrachi T, Barzago C, Scandiffio L, Bortone F, Bonanno S, et al. MicroRNA signature associated with treatment response in myasthenia gravis: A further step towards precision medicine. Pharmacol Res. (2019) 148:104388. doi: 10.1016/j.phrs.2019.104388
48. Wang YZ, Tian FF, Yan M, Zhang JM, Liu Q, Lu JY, et al. Delivery of an miR155 inhibitor by anti-CD20 single-chain antibody into B cells reduces the acetylcholine receptor-specific autoantibodies and ameliorates experimental autoimmune myasthenia gravis. Clin Exp Immunol. (2014) 176:207–21. doi: 10.1111/cei.12265
49. Lu Y, Hippen KL, Lemire AL, Gu J, Wang W, Ni X, et al. miR-146b antagomir-treated human Tregs acquire increased GVHD inhibitory potency. Blood. (2016) 128:1424–35. doi: 10.1182/blood-2016-05-714535
50. Huang P, He XY, Xu M. Expression and significance of microRNA-126 and microRNA-21 in peripheral blood mononuclear cells in patients with myasthenia gravis. Neuroimmunomodulation. (2021) 3:1–8. doi: 10.1159/000510714
51. Howard JF Jr, Bril V, Burns TM, Mantegazza R, Bilinska M, Szczudlik A, et al. Efgartigimod MG Study Group. Randomized phase 2 study of FcRn antagonist efgartigimod in generalized myasthenia gravis. Neurology. (2019) 92:e2661–73. doi: 10.1212/WNL.0000000000007600
52. Punga AR, Andersson M, Alimohammadi M, Punga T. Disease specific signature of circulating miR-150-5p and miR-21-5p in myasthenia gravis patients. J Neurol Sci. (2015) 356:90–6. doi: 10.1016/j.jns.2015.06.019
53. Bo C, Zhang H, Cao Y, Lu X, Zhang C, Li S, et al. Construction of a TF-miRNA-gene feed-forward loop network predicts biomarkers and potential drugs for myasthenia gravis. Sci Rep. (2021) 11:2416. doi: 10.1038/s41598-021-81962-6
54. Sengupta M, Wang BD, Lee NH, Marx A, Kusner LL, Kaminski HJ. MicroRNA and mRNA expression associated with ectopic germinal centers in thymus of myasthenia gravis. PloS One. (2018) 13:e0205464. doi: 10.1371/journal.pone.0205464
55. Barzago C, Lum J, Cavalcante P, Srinivasan KG, Faggiani E, Camera G, et al. A novel infection- and inflammation-associated molecular signature in peripheral blood of myasthenia gravis patients. Immunobiology. (2016) 221:1227–36. doi: 10.1016/j.imbio.2016.06.012
56. Cao Y, Lu X, Wang J, Zhang H, Liu Z, Xu S, et al. Construction of an miRNA-regulated drug-pathway network reveals drug repurposing candidates for myasthenia gravis. Int J Mol Med. (2017) 39:268–78. doi: 10.3892/ijmm.2017.2853
57. Marx A, Yamada Y, Simon-Keller K, Schalke B, Willcox N, Ströbel P, et al. Thymus and autoimmunity. Semin Immunopathol. (2021) 43:45–64. doi: 10.1007/s00281-021-00842-3
58. Cron MA, Maillard S, Truffault F, Gualeni AV, Gloghini A, Fadel E, et al. Causes and consequences of miR-150-5p dysregulation in myasthenia gravis. Front Immunol. (2019) 10:539. doi: 10.3389/fimmu.2019.00539
59. Ni X, Kou W, Gu J, Wei P, Wu X, Peng H, et al. TRAF6 directs FOXP3 localization and facilitates regulatory T-cell function through K63-linked ubiquitination. EMBO J. (2019) 38:e99766. doi: 10.15252/embj.201899766
60. Yan M, Fu Y, Rao H, Zhou H, Liang X. Expression and clinical significance of miR-146a and tumor necrosis factor receptor-associated factor 6 (TRAF6) in myasthenia gravis patient serum. BioMed Res Int. (2021) 2021:5573469. doi: 10.1155/2021/5573469
61. Cavalcante P, Bernasconi P, Mantegazza R. Autoimmune mechanisms in myasthenia gravis. Curr Opin Neurol. (2012) 25:621–9. doi: 10.1097/WCO.0b013e328357a829
62. Wang J, Zheng S, Xin N, Dou C, Fu L, Zhang X, et al. Identification of novel MicroRNA signatures linked to experimental autoimmune myasthenia gravis pathogenesis: down-regulated miR-145 promotes pathogenetic Th17 cell response. J Neuroimmune Pharmacol. (2013) 8:1287–302. doi: 10.1007/s11481-013-9498-9
63. Liu XF, Wang RQ, Hu B, Luo MC, Zeng QM, Zhou H, et al. MiR-15a contributes abnormal immune response in myasthenia gravis by targeting CXCL10. Clin Immunol. (2016) 164:106–13. doi: 10.1016/j.clim.2015.12.009
64. Lu J, Yan M, Wang Y, Zhang J, Yang H, Tian FF, et al. Altered expression of miR-146a in myasthenia gravis. Neurosci Lett. (2013) 555:85–90. doi: 10.1016/j.neulet.2013.09.014
65. Zhang J, Jia G, Liu Q, Hu J, Yan M, Yang B, et al. Silencing miR-146a influences B cells and ameliorates experimental autoimmune myasthenia gravis. Immunology. (2015) 144:56–67. doi: 10.1111/imm.12347
66. Bortone F, Scandiffio L, Marcuzzo S, Bonanno S, Frangiamore R, Motta T, et al. miR-146a in myasthenia gravis thymus bridges innate immunity with autoimmunity and is linked to therapeutic effects of corticosteroids. Front Immunol. (2020) 11:142. doi: 10.3389/fimmu.2020.00142
67. Wang Q, Liu Y, Kuang S, Li R, Weng N, Zhou Z. miR-181a ameliorates the progression of myasthenia gravis by regulating TRIM9. Evid Based Complement Alternat Med. (2021) 2021:1303375. doi: 10.1155/2021/1303375
68. Jiang L, Cheng Z, Qiu S, Que Z, Bao W, Jiang C, et al. Altered let-7 expression in Myasthenia gravis and let-7c mediated regulation of IL-10 by directly targeting IL-10 in Jurkat cells. Int Immunopharmacol. (2012) 14:217–23. doi: 10.1016/j.intimp.2012.07.003
69. Cron MA, Maillard S, Delisle F, Samson N, Truffault F, Foti M, et al. Analysis of microRNA expression in the thymus of Myasthenia Gravis patients opens new research avenues. Autoimmun Rev. (2018) 17:588–600. doi: 10.1016/j.autrev.2018.01.008
70. Lazaridis K, Tzartos SJ. Autoantibody specificities in myasthenia gravis; implications for improved diagnostics and therapeutics. Front Immunol. (2020) 11:212. doi: 10.3389/fimmu.2020.00212
71. Nogales-Gadea G, Ramos-Fransi A, Suárez-Calvet X, Navas M, Rojas-García R, Mosquera JL, et al. Analysis of serum miRNA profiles of myasthenia gravis patients. PloS One. (2014) 9:e91927. doi: 10.1371/journal.pone.0091927
72. Shi L, Liu T, Zhang M, Guo Y, Song C, Song D, et al. miR-15b is downregulated in myasthenia gravis patients and directly regulates the expression of interleukin-15 (IL-15) in experimental myasthenia gravis mice. Med Sci Monit. (2015) 21:1774–80. doi: 10.12659/MSM.893458
73. Xu H, Bao Z, Liang D, Li M, Wei M, Ge X, et al. Plasma exosomal miR-106a-5p expression in myasthenia gravis. Muscle Nerve. (2020) 61:401–7. doi: 10.1002/mus.26785
74. Zhang Y, Guo M, Xin N, Shao Z, Zhang X, Zhang Y, et al. Decreased microRNA miR-181c expression in peripheral blood mononuclear cells correlates with elevated serum levels of IL-7 and IL-17 in patients with myasthenia gravis. Clin Exp Med. (2016) 16:413–21. doi: 10.1007/s10238-015-0358-1
75. Chunjie N, Huijuan N, Zhao Y, Jianzhao W, Xiaojian Z. Disease-specific signature of serum miR-20b and its targets IL-8 and IL-25, in myasthenia gravis patients. Eur Cytokine Netw. (2015) 26:61–6. doi: 10.1684/ecn.2015.0367
76. Howard JF Jr, Bril V, Vu T, Karam C, Peric S, Margania T, et al. Safety, efficacy, and tolerability of efgartigimod in patients with generalised myasthenia gravis (ADAPT): a multicentre, randomised, placebo-controlled, phase 3 trial. Lancet Neurol. (2021) 20:526–36. doi: 10.1212/WNL.96.15_supplement.4520
77. Evoli A, Tonali PA, Padua L, Monaco ML, Scuderi F, Batocchi AP, et al. Clinical correlates with anti-MuSK antibodies in generalized seronegative myasthenia gravis. Brain. (2003) 126:2304–11. doi: 10.1093/brain/awg223
78. Punga T, Bartoccioni E, Lewandowska M, Damato V, Evoli A, Punga AR. Disease specific enrichment of circulating let-7 family microRNA in MuSK+ myasthenia gravis. J Neuroimmunol. (2016) 292:21–6. doi: 10.1016/j.jneuroim.2016.01.003
79. Tan Y, Zhu L, Cui L, Guan Y. Differential expression of miRNA in the peripheral blood mononuclear cells in myasthenia gravis with muscle-specific receptor tyrosine kinase antibodies. Crit Rev Eukaryot Gene Expr. (2021) 31:1–15. doi: 10.1615/CritRevEukaryotGeneExpr.v31.i2
80. Bogatikov E, Lindblad I, Punga T, Punga AR. miR-1933-3p is upregulated in skeletal muscles of MuSK+ EAMG mice and affects Impa1 and Mrpl27. Neurosci Res. (2020) 151:46–52. doi: 10.1016/j.neures.2019.02.003
81. Zhou Y, Yan C, Gu X, Zhou L, Lu J, Zhu W, et al. Short-term effect of low-dose rituximab on myasthenia gravis with muscle-specific tyrosine kinase antibody. Muscle Nerve. (2021) 63:824–30. doi: 10.1002/mus.27233
82. Ma L, Pan L, Liu W, Liu Y, Xiang X, Pan Y, et al. Agrin Influences Botulinum Neurotoxin A-Induced Nerve Sprouting via miR-144-agrin-MuSK Signaling. Front Cell Dev Biol. (2020) 8:15. doi: 10.3389/fcell.2020.00015
83. Chaudhry N, Muhammad H, Seidl C, Downes D, Young DA, Hao Y, et al. Highly efficient CRISPR-Cas9-mediated editing identifies novel mechanosensitive microRNA-140 targets in primary human articular chondrocytes. Osteoarthritis Cartilage. (2022) 30:596–604. doi: 10.1016/j.joca.2022.01.005
84. Yu H, Zhang J, Liu X, Li Y. microRNA-136-5p from bone marrow mesenchymal stem cell-derived exosomes facilitates fracture healing by targeting LRP4 to activate the Wnt/β-catenin pathway. Bone Joint Res. (2021) 10:744–58. doi: 10.1302/2046-3758.1012.BJR-2020-0275.R2
85. Li SC, Shi H, Khan M, Caplin M, Meyer T, Öberg K, et al. Roles of miR-196a on gene regulation of neuroendocrine tumor cells. Mol Cell Endocrinol. (2015) 412:131–9. doi: 10.1016/j.mce.2015.06.003
86. Xie S, Li X, Qian L, Cai C, Xiao G, Jiang S, et al. An integrated analysis of mRNA and miRNA in skeletal muscle from myostatin-edited Meishan pigs. Genome. (2019) 62:305–15. doi: 10.1139/gen-2018-0110
87. Yu Z, Zhang M, Luo B, Jing H, Yu Y, Wang S, et al. Lrp4 in hippocampal astrocytes serves as a negative feedback factor in seizures. Cell Biosci. (2020) 10:135. doi: 10.1186/s13578-020-00498-w
88. Mao Z, Wang Z, Zhang S, Pu Y, Wang J, Zhang T, et al. LRP4 promotes migration and invasion of gastric cancer under the regulation of microRNA-140-5p. Cancer biomark. (2020) 29:245–53. doi: 10.3233/CBM-190571
89. Wang L, Zhang L. Emerging roles of dysregulated microRNAs in myasthenia gravis. Front Neurosci. (2020) 14:507. doi: 10.3389/fnins.2020.00507
90. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
91. Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA. (2018) 9:e1471. doi: 10.1002/wrna.1471
92. Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. (2017) 18:962–72. doi: 10.1038/ni.3771
93. Hong Y, Liang X, Gilhus NE. AChR antibodies show a complex interaction with human skeletal muscle cells in a transcriptomic study. Sci Rep. (2020) 10:11230. doi: 10.1038/s41598-020-68185-x
94. Hu B, Niu L, Jiang Z, Xu S, Hu Y, Cao K. LncRNA XLOC_003810 promotes T cell activation and inhibits PD-1/PD-L1 expression in patients with myasthenia gravis-related thymoma. Scand J Immunol. (2020) 92:e12886. doi: 10.1111/sji.12886
95. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
96. Zhang F, Liu G, Bu Y, Ma X, Hao J. Expression profile of long noncoding RNAs and mRNAs in peripheral blood mononuclear cells from myasthenia gravis patients. J Neuroimmunol. (2016) 299:124–9. doi: 10.1016/j.jneuroim.2016.09.005
97. Lu W, Lu Y, Wang CF, Chen TT. Expression profiling and bioinformatics analysis of exosomal long noncoding RNAs in patients with myasthenia gravis by RNA sequencing. J Clin Lab Anal. (2021) 35:e23722. doi: 10.1002/jcla.23722
98. Xu Y, Ouyang Y. Long non-coding RNA growth arrest specific 5 regulates the T helper 17/regulatory T balance by targeting miR-23a in myasthenia gravis. J Int Med Res. (2022) 50:3000605211053703. doi: 10.1177/03000605211053703
99. Li S, Wang X, Wang T, Zhang H, Lu X, Liu L, et al. Identification of the regulatory role of lncRNA HCG18 in myasthenia gravis by integrated bioinformatics and experimental analyses. J Transl Med. (2021) 19:468. doi: 10.1186/s12967-021-03138-0
100. Luo M, Liu X, Meng H, Xu L, Li Y, Li Z, et al. IFNA-AS1 regulates CD4+ T cell activation in myasthenia gravis though HLA-DRB1. Clin Immunol. (2017) 183:121–31. doi: 10.1016/j.clim.2017.08.008
101. Wang J, Cao Y, Lu X, Wang X, Kong X, Bo C, et al. Identification of the regulatory role of lncRNA SNHG16 in myasthenia gravis by constructing a competing endogenous RNA network. Mol Ther Nucleic Acids. (2020) 19:1123–33. doi: 10.1016/j.omtn.2020.01.005
102. Niu L, Jiang J, Yin Y, Hu B. LncRNA XLOC_003810 modulates thymic Th17/Treg balance in myasthenia gravis with thymoma. Clin Exp Pharmacol Physiol. (2020) 47:989–96. doi: 10.1111/1440-1681.13280
103. Peng S, Huang Y. LncRNA GAS5 positively regulates IL-10 expression in patients with generalized myasthenia gravis. Brain Behav. (2022) 12:e2457. doi: 10.1002/brb3.2457
104. Xu S, Wang T, Lu X, Zhang H, Liu L, Kong X, et al. Identification of LINC00173 in myasthenia gravis by integration analysis of aberrantly methylated- differentially expressed genes and ceRNA networks. Front Genet. (2021) 12:726751. doi: 10.3389/fgene.2021.726751
105. Wang X, Zhang H, Lu X, Li S, Kong X, Liu L, et al. LncRNA OIP5-AS1 modulates the proliferation and apoptosis of Jurkat cells by sponging miR-181c-5p to regulate IL-7 expression in myasthenia gravis. PeerJ. (2022) 10:e13454. doi: 10.7717/peerj.13454
106. Ke J, Du X, Cui J, Yu L, Li H. LncRNA and mRNA expression associated with myasthenia gravis in patients with thymoma. Thorac Canc. (2022) 13:15–23. doi: 10.1111/1759-7714.14201
107. Luo Z, Li Y, Liu X, Luo M, Xu L, Luo Y, et al. Systems biology of myasthenia gravis, integration of aberrant lncRNA and mRNA expression changes. BMC Med Genomics. (2015) 8:13. doi: 10.1186/s12920-015-0087-z
108. Kong X, Wang J, Cao Y, Zhang H, Lu X, Wang Y, et al. The long noncoding RNA MALAT-1 functions as a competing endogenous RNA to regulate MSL2 expression by sponging miR-338-3p in myasthenia gravis. J Cell Biochem. (2019) 120:5542–50. doi: 10.1002/jcb.27838
109. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
110. Chen B, Huang S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. (2018) 418:41–50. doi: 10.1016/j.canlet.2018.01.011
111. Xie R, Zhang Y, Zhang J, Li J, Zhou X. The role of circular RNAs in immune-related diseases. Front Immunol. (2020) 11:545. doi: 10.3389/fimmu.2020.00545
112. Lv J, Ren L, Han S, Zhang J, Zhao X, Zhang Y, et al. Peripheral blood hsa-circRNA5333-4: A novel biomarker for myasthenia gravis. Clin Immunol. (2021) 224:108676. doi: 10.1016/j.clim.2021.108676
113. Lai X, Bi Z, Yang X, Hu R, Wang L, Jin M, et al. Upregulation of circ-FBL promotes myogenic proliferation in myasthenia gravis by regulation of miR-133/PAX7. Cell Biol Int. (2021) 45:2287–93. doi: 10.1002/cbin.11676
114. Beretta F, Huang YF, Punga AR. Towards Personalized Medicine in Myasthenia Gravis: Role of Circulating microRNAs miR-30e-5p, miR-150-5p and miR-21-5p. Cells. (2022) 11:740. doi: 10.3390/cells11040740
115. Martínez-Martínez P, Phernambucq M, Steinbusch L, Schaeffer L, Berrih-Aknin S, Duimel H, et al. Silencing rapsyn in vivo decreases acetylcholine receptors and augments sodium channels and secondary postsynaptic membrane folding. Neurobiol Dis. (2009) 35:14–23. doi: 10.1016/j.nbd.2009.03.008
116. Kaminski HJ, Li Z, Richmonds C, Lin F, Medof ME. Complement regulators in extraocular muscle and experimental autoimmune myasthenia gravis. Exp Neurol. (2004) 189:333–42. doi: 10.1016/j.expneurol.2004.06.005
Keywords: myasthenia gravis, non-coding RNAs, pathogenesis, treatment, biomarker
Citation: Wang B, Zhu Y, Liu D, Hu C and Zhu R (2024) The intricate dance of non-coding RNAs in myasthenia gravis pathogenesis and treatment. Front. Immunol. 15:1342213. doi: 10.3389/fimmu.2024.1342213
Received: 21 November 2023; Accepted: 11 March 2024;
Published: 27 March 2024.
Edited by:
Amanda Piquet, University of Colorado Anschutz Medical Campus, United StatesCopyright © 2024 Wang, Zhu, Liu, Hu and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruixia Zhu, enJ4XzIwMDYyNjMxM0AxNjMuY29t
 Benqiao Wang
Benqiao Wang Ruixia Zhu
Ruixia Zhu