- State Key Laboratory of Oncology in South China, Guangdong Provincial Clinical Research Center for Cancer, Sun Yat-sen University Cancer Center, Guangzhou, China
The tumor microenvironment (TME) is a heterogeneous ecosystem comprising cancer cells, immune cells, stromal cells, and various non-cellular components, all of which play critical roles in controlling tumor progression and response to immunotherapies. Methyltransferase-like 3 (METTL3), the core component of N6-methyladenosine (m6A) writer, is frequently associated with abnormalities in the m6A epitranscriptome in different cancer types, impacting both cancer cells and the surrounding TME. While the impact of METTL3 on cancer cells has been extensively reviewed, its roles in TME and anti-cancer immunity have not been comprehensively summarized. This review aims to systematically summarize the functions of METTL3 in TME, particularly its effects on tumor-infiltrating immune cells. We also elaborate on the underlying m6A-dependent mechanism. Additionally, we discuss ongoing endeavors towards developing METTL3 inhibitors, as well as the potential of targeting METTL3 to bolster the efficacy of immunotherapy.
1 Introduction
Tumor is a complicated ecosystem that is composed of cancer cells, various non-neoplastic cell components and extracellular matrix (1). The cellular and non-cellular components surrounding cancer cells form the tumor microenvironment (TME), which consists of (i) immune cells such as tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), natural killer cells (NKs), neutrophils, dendritic cells (DCs), T cells, and B cells; (ii) stromal cells such as cancer-associated fibroblasts (CAFs) and vascular endothelial cells (VECs); and (iii) extracellular matrix such as extracellular vesicles (EVs) (Figure 1). It is acknowledged that TME plays critical roles throughout the development and progression of tumors. Specifically, TME harbors dual potentials for suppressing or promoting cancer. During the early stages of tumor growth, infiltrating immune cells and associated matrix components are recruited and activated by tumor cells to establish an anti-tumor TME, preventing the formation and progression of tumors (2, 3). However, persistent immune stimulation causes depletion or remodeling of effector cells, ultimately leading to an immune suppressive TME (4, 5). Therefore, eliminating the suppressive effects and restoring the innate anti-tumor capability of the immune system represents a promising strategy for cancer therapy (6, 7).
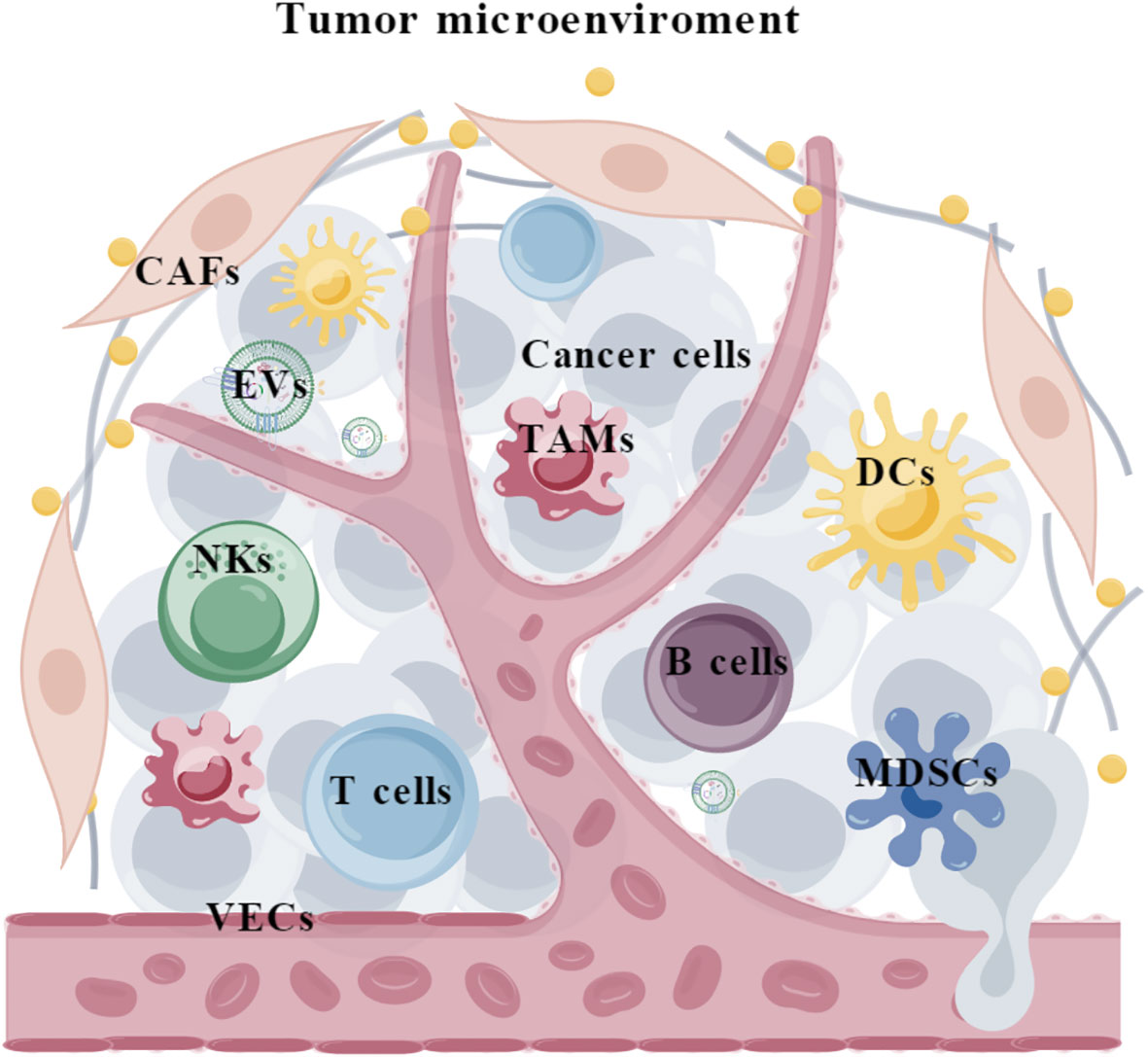
Figure 1 The composition of tumor microenvironment. TAMs, tumor-associated macrophages; MDSCs, myeloid-derived suppressor cells; NKs, natural killer cells; DCs, Dendritic cells; CAFs, cancer-associated fibroblasts; VECs, vascular endothelial cells; EVs, extracellular vesicles.
Recently, accumulating evidence has shown that abnormalities in epitranscriptomic are frequently associated with immune evasion in cancer and targeting deregulated RNA epigenetic machinery can reprogram TME to improve cancer immunotherapy (8). As one of the most abundant modifications in the epitranscriptome, N6-methyladenosine (m6A) has been extensively investigated in cancer. It is dynamically regulated by methyltransferases (also known as “writers”) that catalyze the transition of adenosine (A) to m6A, and demethylases (also known as “erasers”) that are in charge of removing methyl group. Through interacting with different m6A-binding proteins (“readers”), the m6A modification affects every fundamental aspect of RNA metabolism. For instance, YTHDF2 mediates RNA decay, YTHDF1 facilitates mRNA translation, YTHDC1 controls RNA exportation, IGF2BPs promotes RNA stabilization and translation, and hnRNPA2B1 mediates microRNA processing (9). m6A modification has been shown to affect nearly every aspect of RNA metabolism, and thus participates in various physiological and pathological processes, particularly in immune response and tumorigenesis (10–13).
Methyltransferase-like 3 (METTL3) is the catalytic component of the m6A methyltransferase complex (MTC), which mediates m6A deposition on RNA substrates (14). Structurally, METTL3 consists of a methyltransferase domain (MTD) and a zinc finger domain (ZFD) (15). The MTD binds to the methyl donor S-adenosylmethionine (SAM), while the ZFD specifically recognizes the RNA GGACU consensus and cooperates with the MTD of METTL3-METTL14 to catalyze (15, 16) (Figure 2). By catalyzing m6A on functional critical transcripts, METTL3 regulates gene expression and plays a significant oncogenic role in most types of cancers (17, 18), except for a tumor-suppressive function in renal cell carcinoma and endometrial tumors (19, 20). These findings strongly suggest that METTL3 plays multifaceted functions in cancer, which may be context-dependent. Therefore, investigations of the functions and mechanisms of METTL3 in cancer cells and TME might enhance our comprehension of the roles of METTL3 in cancer. While the impact of METTL3 on cancer cells has been extensively reviewed, its roles in TME and anti-cancer immunity have not been comprehensively summarized.
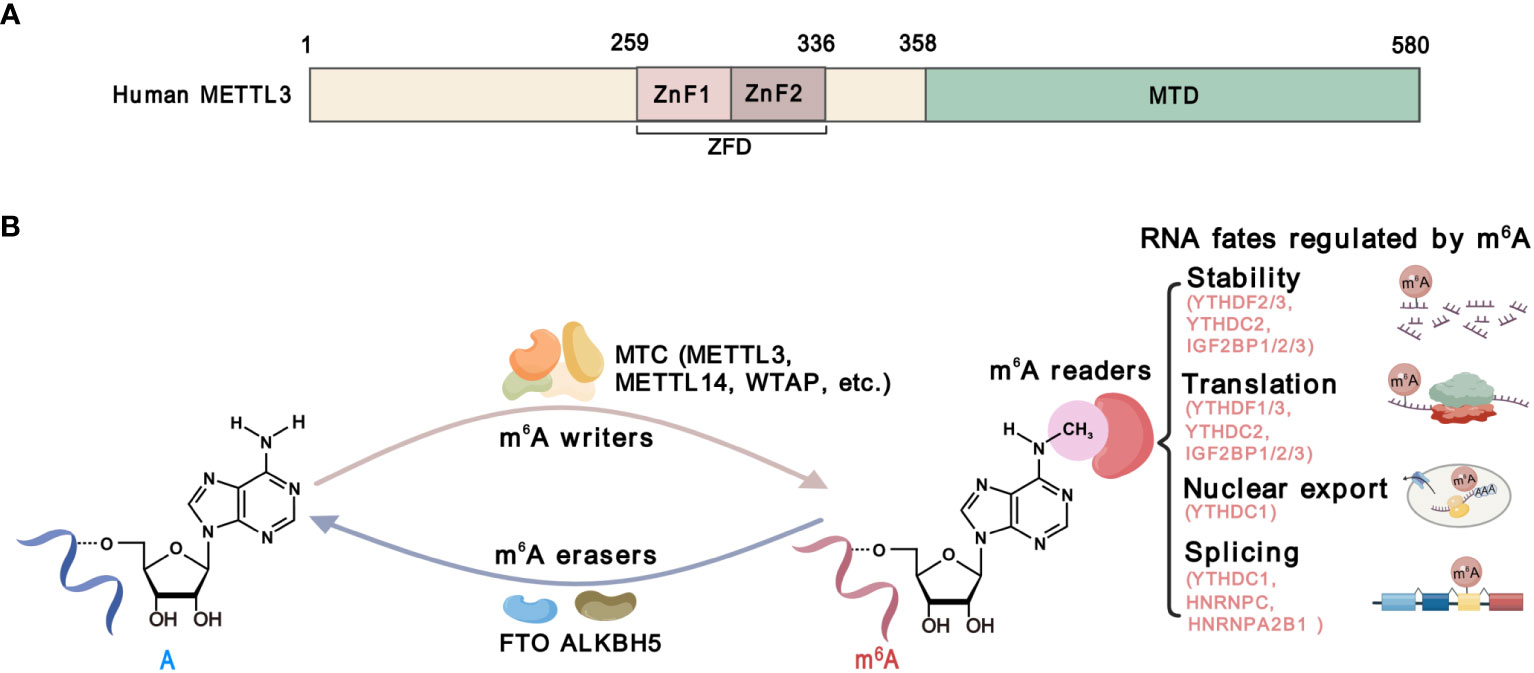
Figure 2 The structure and function of METTL3. (A) Schematic domain structure of METTL3. The zinc-finger domain (ZFD), from 259 to 336 amino acid, comprises two tandem CCC-H type Zn2+ fingers (ZnF1 and ZnF2). The methyltransferase domain (MTD) at the C-terminal of METTL3 is responsible for SAM binding and catalysis. (B) The m6A modification machinery. The m6A methylation is catalyzed by methyltransferase complexes (MTCs) including METTL13, METTL14, WTAP, and other accessory components, which is known as m6A “writers”. Demethylases, including FTO and ALKBH5, serve as m6A”erasers”. m6A-binding proteins, including YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3, HNRNPC and HNRNPA2B1, serve as m6A “readers” that determine the fate of m6A-modified RNAs.
In this review, we aim to provide a comprehensive overview of the roles and implications of METTL3 in TME. We systematically summarize the functions and mechanisms of METTL3-mediated m6A modification in innate immunity and adaptive immunity, with a discussion of recent advances in developing small molecular inhibitors of METTL3 and their potential in immunotherapy.
2 The roles of METTL3 in TME
Immunosuppression is a common characteristic of TME, which is characterized by an imbalance of distinct immune cells and stromal cells (21, 22). Emerging evidence shows that METTL3 and it-mediated m6A are involved in reshaping TME through affecting both innate immune (Figure 3) and adaptive immune (Figure 4) compartments (Table 1).
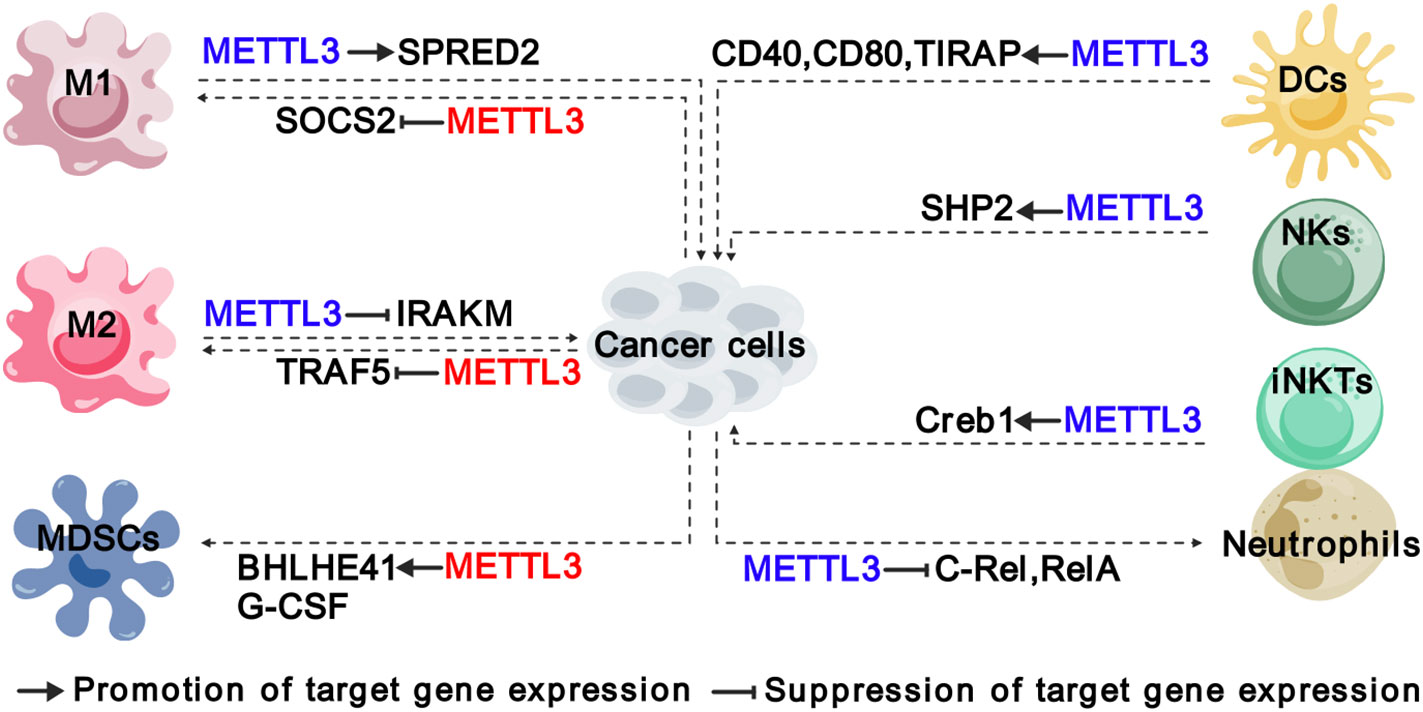
Figure 3 Functions and mechanisms of METTL3 in regulating innate immune cells. The colors of “METTL3” indicate its functions in cancer: red represents tumor promoting function, while blue represents tumor suppressive function. The solid arrows indicate that METTL3 promotes target gene expression, while the inhibition arrows indicate that METTL3 represses target gene expression. The dashed arrows between different cell types indicate the interaction between cancer cells and TME. M1, “classically” activated macrophage; M2, “alternatively” activated macrophage; MDSCs, myeloid-derived suppressor cells; DCs, Dendritic cells; NKs, natural killer cells; iNKTs, innate-like T cells.
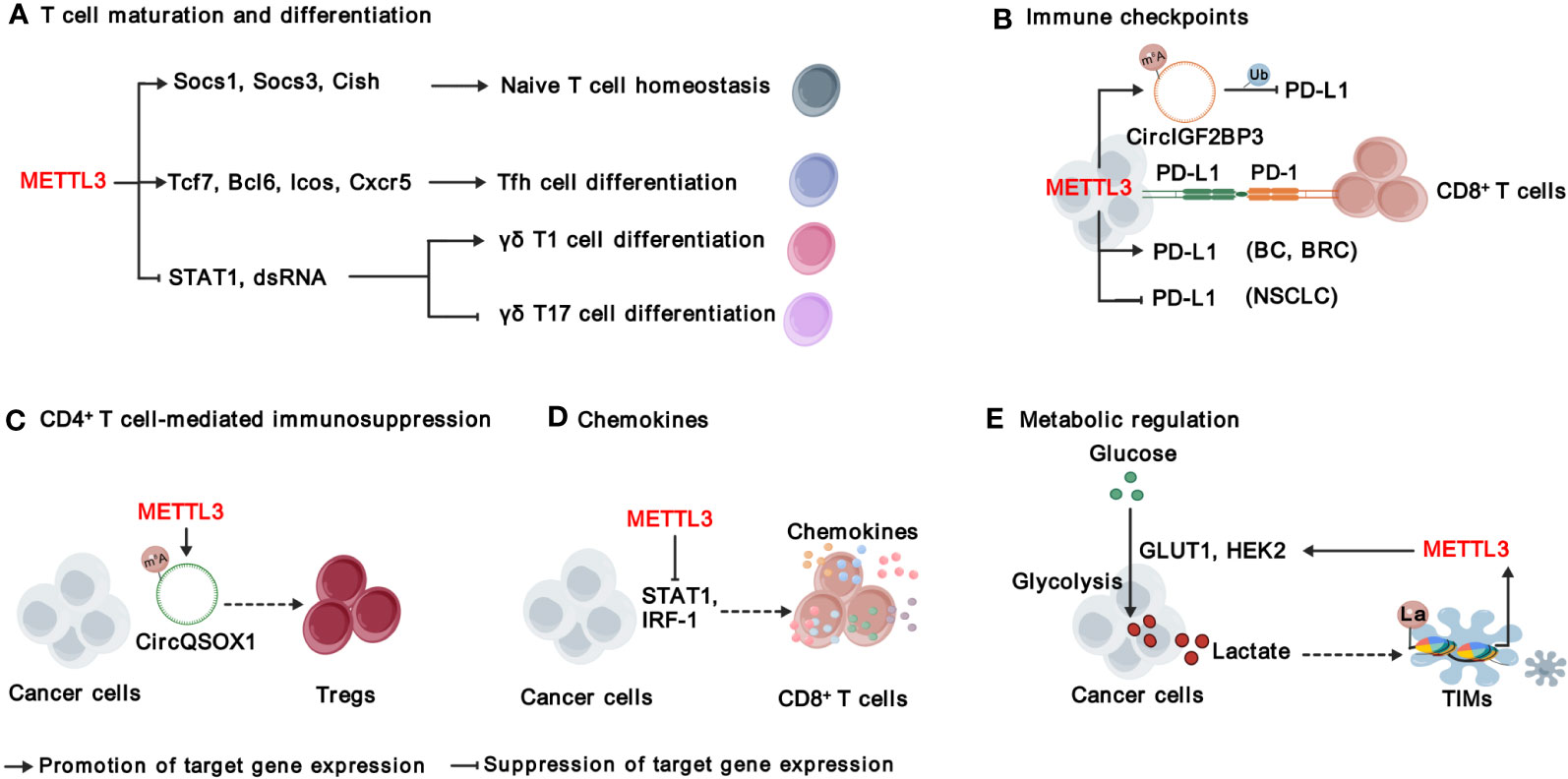
Figure 4 Functions and mechanisms of METTL3 in modulating adaptive immunity. METTL3 regulates T cell maturation and differentiation (A), immune checkpoints (B), CD4+ T cell-mediated immunosuppression (C), chemokines (D), and metabolic microenvironment (E). “METTL3” in red indicates that it plays tumor promoting function. The solid arrows indicate that METTL3 promotes target gene expression, while the inhibition arrows indicate that METTL3 represses target gene expression. The dashed arrows between different cell types indicate the interaction between cancer cells and TME. BC, bladder cancer; BRC, breast Cancer; NSCLC, non-small-cell lung cancer. La, lactylation; Ub, ubiquitination.
2.1 Roles of METTL3 in regulating innate immunity in TME
2.1.1 METTL3 regulates the polarization and function of TAMs
As evolutionarily ancient immune cells, macrophages are present in virtually all tissues and respond to many diseases such as cancer (43, 44). TAMs represent the most abundant innate immune population in TME, which are generally heterogeneous and can exert dual influence on cancer depending on their active state. There are commonly two distinct subsets of TAMs: the “classically” activated macrophage (M1), which generally exerts pro-inflammatory and anti-tumoral functions, and the “alternatively” activated macrophage (M2) with immunosuppressive and pro-tumoral functions (38, 45). Recently, the modification of m6A has been recognized as an important regulatory mechanism in macrophage activation. Tong et al. identified several components of m6A MTCs, including Mettl3, Mettl14 and Rbm15, were key regulators of macrophage activation based on their CRISPR screening experiments (38). In particular, METTL3 facilitated M1 polarization by regulating the expression of signal transducer and activator of transcription (STAT) proteins, a family of master transcription factors (TFs), that control the polarization of M1 and M2 macrophages (46). Therefore, depletion of Mettl3 in myeloid cells contributes to the formation of an immunosuppressive microenvironment, including increased infiltration of M1/M2-like TAMs and regulatory T cells, and promotes the growth and metastasis of Lewis lung carcinoma and melanoma in mice (40). Mechanistically, the loss of METTL3 impairs the m6A-YTHDF2-SPRED2 axis, which enhances the activation of NF-kB and STAT3 phosphorylation (40). METTL3 also exerts a similar function in glioma to inhibit M1 polarization by promoting m6A methylation of suppressor of cytokine signaling 2 (SOCS2) and reducing its expression, which is a negative regulator of the JAK/STAT pathway (47, 48).
Furthermore, METTL3 significantly influences the function of both M1 and M2 macrophages. Mettl3 deficiency in macrophages upregulates the expression of interleukin-1 receptor-associated kinase M (Irakm) through m6A, leading to a reduction in TNF-α production and attenuating their ability to fight against pathogens and eliminate tumors in vivo (38). Additionally, METTL3-mediated m6A mechanism contributes to promoting M2-associated therapeutic resistance, such as oxaliplatin resistance in colorectal cancer (CRC) (27) and immuno-resistance in lung adenocarcinoma (LUAD) (33). In CRC, the infiltration of CD206+CD163+ M2-TAMs is dramatically higher in oxaliplatin-resistant patients, which enables oxaliplatin resistance through METTL3-mediated m6A modification on TRAF5 mRNAs (27). Similarly, M2-TAMs are greatly increased in the tumor tissues of patients with immunoresistant LUAD and enhance the expression of METTL3 and total m6A RNA level, while interfering with METTL3 could significantly reverse immunoresistance (33). Cumulatively, these findings establish METTL3 as an m6A writer that governs the polarization and function of TAMs.
2.1.2 METTL3 induces MDSCs migration and accumulation in TME
MDSCs were initially reported in 2007 as a group of myeloid cells with immunosuppressive functions, which promoted the growth and advancement of tumors by suppressing immune responses, particularly the activity of T cells (49). Recent findings have revealed a significant positive association between the expression of METTL3 and the density of intratumoral CD33+ MDSCs in cervical cancer (50). Coincidentally, the expression of METTL3 in CRC cells activates the transcription of C-X-C motif chemokine ligand 1 (CXCL1) through upregulation of basic helix-loop-helix family member 41 (BHLHE41) in an m6A-dependent manner, and thereby enhances the migration of MDSCs to inhibit CD8+ T cells, forming an immunosuppressive TME (28). Whereas inhibition of METTL3 expression in bladder cancer (BC) by cisplatin decreases m6A modification on granulocyte colony-stimulating factors (G-CSF) mRNA and the production of G-CSF, resulting in a reduced the number of fibrocytic MDSCs (f-MDSCs) during intra-arterial infusion chemotherapy (23). Collectively, METTL3 plays a crucial role in promoting the migration and accumulation of MDSCs in TME.
2.1.3 METTL3 modulates the homeostasis and function of NK and invariant natural killer T cells
NK cells are cytotoxic lymphocytes of the innate immune system that can directly kill infected or cancerous cells without exposure to an antigen, constituting the first line of defense against the formation of cancerous cells (51). METTL3-mediated m6A methylation on SH2 domain-containing protein tyrosine phosphatase-2 (SHP-2) has been found to activate the AKT-mTOR and MAPK-ERK signaling pathways in NK cells in response to IL-15 stimulation (39). Downregulation of METTL3 in NK cells is highly associated with poor infiltration and function of NK cells within TME, which could accelerate tumor progression and shorten the survival of mice models with MC38 or B16-F10 syngeneic tumors (39). Therefore, METTL3 is considered essential for maintaining homeostasis and tumor immunosurveillance function in NK cells.
Invariant natural killer T (iNKT) cells are an innate-like T cell subset that expresses an invariant T cell receptor (TCR) α-chain and recognizes lipids presented on CD1d (52). iNKT cells are involved in bridging adaptive and innate immunity and are considered promising targets for immunotherapy. As METTL3 and METTL14 subunits are mutually required for protein stability, METTL14 knockdown in mature iNKT cells diminishes their cytokine production, correlating with decreased TCR signaling suggesting a potential function of METTL3 in iNKT cells (53). Indeed, You et al. found that conditional depletion of Mettl3 in CD4+ CD8+ double-positive thymocytes impaired iNKT cells proliferation, differentiation, and cytokine secretion through regulating the transcription factor Creb1, in turn facilitating the development of B16-F10 melanoma in mice (41).
2.1.4 METTL3 regulates the activation and function of neutrophils and DCs
TME is characterized by persistent chronic inflammation and infiltration of neutrophils, which leads to a dichotomous role in either promoting or impeding tumor growth (54–57). Research has shown that METTL3-mediated m6A modification on TLR4 plays a role in regulating neutrophil activation by modulating TLR4/Myd88/NF-κB signaling (58). In addition to TLR4, c-Rel and RelA have been identified as crucial downstream targets of METTL3 in tumor-associated neutrophils (TANs) (42). By regulating the mRNA homeostasis of c-Rel and RelA in an m6A-YTHDF2-dependent manner, METTL3 suppresses the production of IL-8 and IL8-induced recruitment of TANs, consequently restricting the progression of papillary thyroid cancer (PTC) (42). It remains unclear whether the suppressive function of METTL3 on TANs is PTC-specific or generally applicable to most cancer types, which requires further investigation.
DCs are specialized cells that present antigens and play a crucial role in bridging innate and adaptive immunity (59). The proper function of DCs is essential for both activating immune defense and maintaining immune tolerance (60). Recent research has shown that METTL3 and m6A modification in DCs are essential for cytokine production and the stimulation of T cell activation (61, 62). Specifically, the expression of METTL3 in DCs promotes the production of co-stimulatory molecules CD40, CD80, and cytokine IL-12, and stimulates T cell activation, which is mediated by m6A modification on CD40, CD80 and Toll/IL-1 receptor domain-containing adaptor protein (TIRAP) (61). However, the roles of METTL3 in tumor-infiltrating DCs are not yet clear. Further investigations into the relationship between METTL3 and tumor-infiltrating DCs, as well as other innate immune cells in TME, can significantly enhance our comprehension of the regulatory mechanisms governing innate immunity within TME and can pave the way for reshaping TME for cancer therapy.
2.2 Roles of METTL3 in modulating adaptive immunity
2.2.1 METTL3 regulates T cell maturation and differentiation
T cells are an essential part of the immune system and play a central role in immunotherapy (63). There are two main types of T cells: cytotoxic T-cells (also known as CD8+ T cells) that can directly destroy abnormal cells, and helper T-cells (Th, also known as CD4+ T cells) that assist other immune cells in their functions. CD4+ T cells can be further divided into T helper 1 (Th1), T helper 2 (Th2), regulatory T cells (Tregs) and T follicular helper (Tfh) cells (64). It is critical to maintain the homeostasis of T cells in immune system; however, Li et al. found that deficient of METTL3 impaired mouse T cell homeostasis and differentiation (65). Applying a lymphogenic mouse adoptive transfer model, they demonstrated that depletion of Mettl3 prevented naïve T cells from undergoing homeostatic expansion and remained in naïve state for months, through impairment of the timely degradation of Socs1, Scos3, and Cish mRNAs in response to IL-7 signaling (65). METTL3-mediated m6A is also important for the expression of Tfh signature genes, including Tcf7, Bcl6, Icos and Cxcr5, and thereby essentially for Tfh differentiation and germinal center responses (66). In addition, METTL3-mediated m6A modification orchestrates Stat1 stability and double- stranded RNA (dsRNA) contents to promote γδT17 but inhibits γδT1, and Mettl3 deficient γδT cells produces less IL-17 and less pathogenic (67). In sum, METTL3-mediated m6A modification is required for proper T cell homeostasis.
2.2.2 METTL3 regulates immune checkpoints
Signaling suppression by inhibitory immunoreceptors (also known as immune checkpoints) is an imperative mechanism to maintain self-tolerance, but is utilized by tumors to evade T cell-mediated anti-tumor immunity (68). Immune checkpoint molecules include, but are not limited to, programmed cell death protein 1 (PD-1), cytotoxic T lymphocyte-associated protein 4 (CTLA-4), Lymphocyte-activation gene 3 (LAG3), T-cell immunoglobulin and mucin domain-containing protein 3 (TIM3), T cell immunoreceptor with Ig and ITIM domains (TIGIT), and B and T lymphocyte attenuator (BTLA) (68). Recent studies focus on the roles of METTL3 in regulating PD-1 ligand, PD-L1. In bladder and breast cancer, METTL3 mediates PD-L1 mRNA m6A modification and upregulates its expression through IGF2BP3-dependent manner, resulting the exhaustion of T-cells and the suppression of T cell activation and infiltration (24, 26). On contrast, Yu et al., and Liu et al., have reported that METTL3 destabilizes PD-L1 mRNA in an m6A-dependent manner and mediates ubiquitin-dependent proteasomal degradation of PD-L1 proteins through the circIGF2BP3/PKP3 axis in non-small-cell lung cancer (NSCLC) (35, 36) Despite the effects of METTL3 on PD-L1 expression is controversial, all of the above research demonstrate that inhibiting METTL3 enhances the treatment efficacy of anti-PD-1 therapy, suggesting that unknown mechanisms warrant further investigation (24, 26, 35, 36). In addition, whether METTL3-mediated m6A modification regulates other immune checkpoint molecules warrants further investigation.
2.2.3 METTL3 regulates CD4+ T cell-mediated immunosuppression
CD4+ T cells exhibit multifaceted functions in anti-cancer immunity, contingent upon the functional specialization of different subtypes of CD4+ T cells. Tregs, an immunosuppressive subset of CD4+ T cells, hinder the protective immunosurveillance of neoplasia (69). Lineage-specific depletion of Mettl3 in CD4+ T cells leads to loss of Treg suppression function and severe autoimmune diseases by modulating m6A modification on Sertoli cell-only syndrome (SCOS) mRNA and IL-2/STAT5 signaling pathway (70). Interestingly, METTL3-mediated m6A modification enhances Tregs cell-driven immune evasion through regulating circQSOX1 in CRC cells, ultimately boosting CRC carcinogenesis (29). However, the understanding of METTL3’s functions in CD4+ T cells within the context of TME is still limited, and further investigation is required.
2.2.4 METTL3 regulates chemokines in TME
The infiltration and migration of immune cells in TME, as well as their interaction with other cell components, are controlled by their movement towards gradient chemokines. As mentioned above, METTL3 regulates the production of CXCL1, a key chemotactic factor for recruiting MDSCs in CRC (28). Furthermore, deletion of Mettl3 or Mettl14 promoted IFN-γ-Stat1-Irf1 signaling and increased the secretion of Ifn‐γ, Cxcl9 and Cxcl10 in TME, facilitating the infiltration of CD8+ T cells (30). In addition, METTL3 elevates the expression of pro-tumorigenic chemokines, including CXCL1, CXCL5, and CCL20 in NSCLC (35). These findings together suggest that targeting METTL3 reprograms a more inflamed TME, which can improve immune therapy.
2.2.5 Roles of METTL3 in metabolic regulation of immune response
The metabolic regulation of immune signals has emerged as a crucial regulation for cancer immunity (71). As a product of glucose metabolism, excessive lactate is secreted to TME, which is not just a waste product, but plays important roles in regulating immune responses via causing extracellular acidification and protein lactylation, a posttranslational modification initially reported by Zhao’s research group in 2019 (72). Recently, several studies have revealed the interplay between METTL3-mediated m6A modification and lactation/lactylation in CRC. On one hand, METTL3 regulates glycolysis and lactate production in CRC through regulating the expression of glucose transporter GLUT1 and Hexokinase 2 (HK2) in m6A-dependent manner (73, 74). On the other hand, lactate accumulated in the CRC TME potently induces the upregulation of METTL3 in tumor-infiltrating myeloid cells (TIMs) via the lactylation of histone H3 lysine 18 (H3K18) (31). Lactylation-driven METTL3 mediates the activation of JAK1-STAT3 signaling pathway, which further supports the sustained immunosuppressive activity of TIMs (31). Overall, METTL3-mediated m6A modification significantly shapes the immunosuppressive TME, highlighting targeting METTL3 as a promising strategy to improve immunotherapy.
2.3 Roles of METTL3 in regulating stromal cells
In addition to immune cells, cancer-associated fibroblasts (CAFs), vascular endothelial cells (VECs), and other stromal cells infiltrating in the tumor, forming a non-immune tumor microenvironment. CAFs play a crucial role in TME by promoting tumor development, angiogenesis, and chemoresistance through the production of cytokines, inflammatory ligands, extracellular vehicles. It has been unveiled that COL10A1, which is secreted by lung squamous cell carcinoma (LUSC)-derived CAFs, can accelerate LUSC tumor growth (34). METTL3 promotes the malignant behavior of LUSC cells through mediating m6A modification on COL10A1 mRNA and sustaining its high expression in CAFs (34). Moreover, it was discovered that CAFs-secreted vascular endothelial growth factor A (VEGFA) promoted the metastatic potential of NSCLC cells through METTL3-m6A-RAC3 mechanism (37).
Tumor angiogenesis, characterized by the recruitment of endothelial progenitor cells that subsequently differentiate into mature VECs in response to local microenvironmental stimuli, is a pivotal step in tumor development and metastasis (75). METTL3 was found to promote VECs viability, proliferation, and tube formation through the m6A modification of target transcripts, such as LDL Receptor Related Protein 6 (LRP6) and dishevelled 1 (DVL1) (76). Additionally, METTL3 could promote angiogenesis and metastasis in gastric cancer (GC) and bladder cancer through regulating m6A and mRNA stability of hepatoma-derived growth factor (HDGF), tyrosine kinase endothelial (TEK) and VEGFA, respectively (25, 32). While the expression of METTL3 itself is induced by P300-mediated histone H3 lysine 27 acetylation (H3K27ac) (25). Taken together, METTL3 plays a crucial role in regulating stromal cells and their interaction with cancer cells (Figure 5).
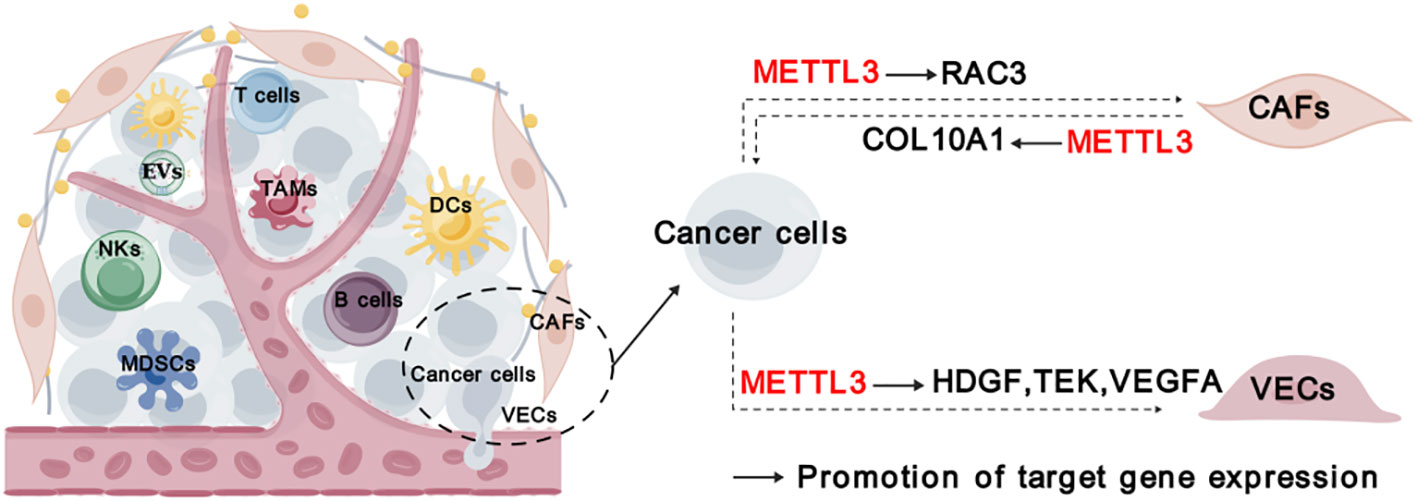
Figure 5 Roles of METTL3 in stromal cells. “METTL3” in red indicates that it plays tumor promoting function. The solid arrows indicate that METTL3 promotes target gene expression. The dashed arrows between different cell types indicate the interaction between cancer cells and TME. CAFs, cancer-associated fibroblasts; VECs, vascular endothelial cells.
3 The implications of METTL3 in cancer immunotherapy
To date, immune checkpoint blockades (ICBs) have emerged as one of the most prominent immunotherapies, fundamentally revolutionizing the paradigm of tumor treatment and achieving significant success (77). Several ICBs, including CTLA4 antibody (anti-CTLA4) and the PD-1/PD-L1 inhibitors (anti-PD-1/anti-PD-L1), have been approved to treat a variety of cancers, such as melanoma, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer (mCRC), NSCLC and hepatocellular carcinoma (HCC) etc. However, not all cancer patients respond to ICBs therapy. Compelling evidence indicates that METTL3 is negatively correlated with the efficacy of anti-PD-1 therapy in various tumor cells, including CRC, melanoma, and NSCLC (28, 30, 35). Furthermore, METTL3 inhibits the efficacy of anti-PD-L1 treatment in breast and bladder cancer, as well as the efficacy of anti-CTLA4 treatment in CRC (24, 26, 29). Targeting METTL3 using single guide RNAs (sgRNAs) has been shown to reprogram TME and enhance the effectiveness of anti-PD-1 treatment, providing proof-of-concept for targeting METTL3 to improve immunotherapy (28, 35).
In the past few years, a great effort has been devoted to discovering small molecular inhibitors targeting METTL3 and exploiting their potential for therapeutic applications, which leads to the discovery of several small molecular inhibitors as potential therapeutic agents. Due to the requirement of MTDs for METTL3’s function, the current strategy for inhibiting METTL3 mainly involves designing compounds that act as substrate (i.e. SAM) competitors of MTD (78). The discovery of the first nucleoside METTL3 inhibitor began with screening of analogues and derivatives of adenosine by high throughput docking into the SAM binding site of METTL3 (79). Two compounds showed good ligand efficiency, but their clinical efficacy still needs to be elucidated (79). Later, screening by in vitro enzyme assays identified Eltrombopag as an allosteric inhibitor of the METTL3-METTL14 complex (80). Eltrombopag shows anti-proliferative effects in acute myeloid leukemia (AML) cell lines by suppressing the m6A level of mRNA and displays synergistic effects when combined with current AML drugs, venetoclax and cytarabine (80).
Another non-nucleoside METTL3 inhibitor, UZH1a, has been discovered by a structure-based drug discovery approach (81). Treatment with UZH1a could significantly reduce mRNA m6A levels and inhibit proliferation and self-renewal in glioblastoma stem cells (GSCs) (81). On the basis of optimizing UZH1a analogues, a more potent inhibitor of METTL3, UZH2, was obtained (82). UZH2 shows target engagement in cells and is able to reduce the m6A/A level in acute myeloid leukemia and prostate cancer cell lines at a sub-micromolar level (82).
Studies have illustrated that METTL3-mediated m6A modification suppresses both the innate immune response and adoptive immunity through regulating double stranded RNA (dsRNA) formation, implying that the antitumor immune response may be enhanced by METTL3 inhibition (83). Indeed, STC-15, an orally bioavailable small molecule inhibitor of METTL3 in Phase 1 clinical study, can restrain the growth of AML cell lines and patient-derived AML samples with sub-micromolar to 1 micromolar IC50 values through activation of anti-cancer immune responses (84). Mechanistically, treatment with STC-15 in AML cell lines leads to prominent accumulation of dsRNAs and upregulation of genes associated with innate immunity, including type-I and type-III IFNs, along with many interferon-stimulated genes, which results in the activation of innate immune pathways and CD8+ T-cells to inhibit tumor growth (84). Inspiringly, the combination of SCT-15 with anti-PD1 therapy can synergistically enhance the anti-tumor properties resulting in tumor regression in MC38 CRC and A20 lymphoma syngeneic models, with mice remaining tumor-free long after treatment has ceased (84).
In addition, STM2457 is recently discovered to be a highly potent and selective METTL3 inhibitor, which can effectively impair leukemic stem cell function and prevent AML development (85). Daily treatment with STM2457 leads to impaired engraftment and prolonged survival in the MLL-AF9/Flt3-ITD mouse model or AML patient-derived xenografts (PDX) with NPM1c or MLL-AF6 PDX, suggesting that STM2457 might benefit the therapy of AML patients with MLL rearrangement or NPMc1 mutation (85). In addition to AML, STM2457 can also inhibit METTL3 expression in prostate cancer and cholangiocarcinoma cells with impaired cell proliferation and invasion (86, 87). Most recently, it has been reported that targeting METTL3 by STM2457 or the second-generation inhibitor STM3006 could activate dsRNA-mediated interferon signaling and anti-viral mimicry response in ovarian cancer cells, augmenting antigen-dependent T-cell killing (88). Strikingly, treatment with STM2457 in mice exhibited dramatically synergistic effects with anti-PD-1, providing a rationale for employing METTL3 inhibitors to promote antitumor immunity in the clinic (88). Although research on METTL3 inhibitors is still in its infancy, it is believed that targeting METTL3 represents a promising therapeutic strategy, especially when combined with immunotherapy (Figure 6).
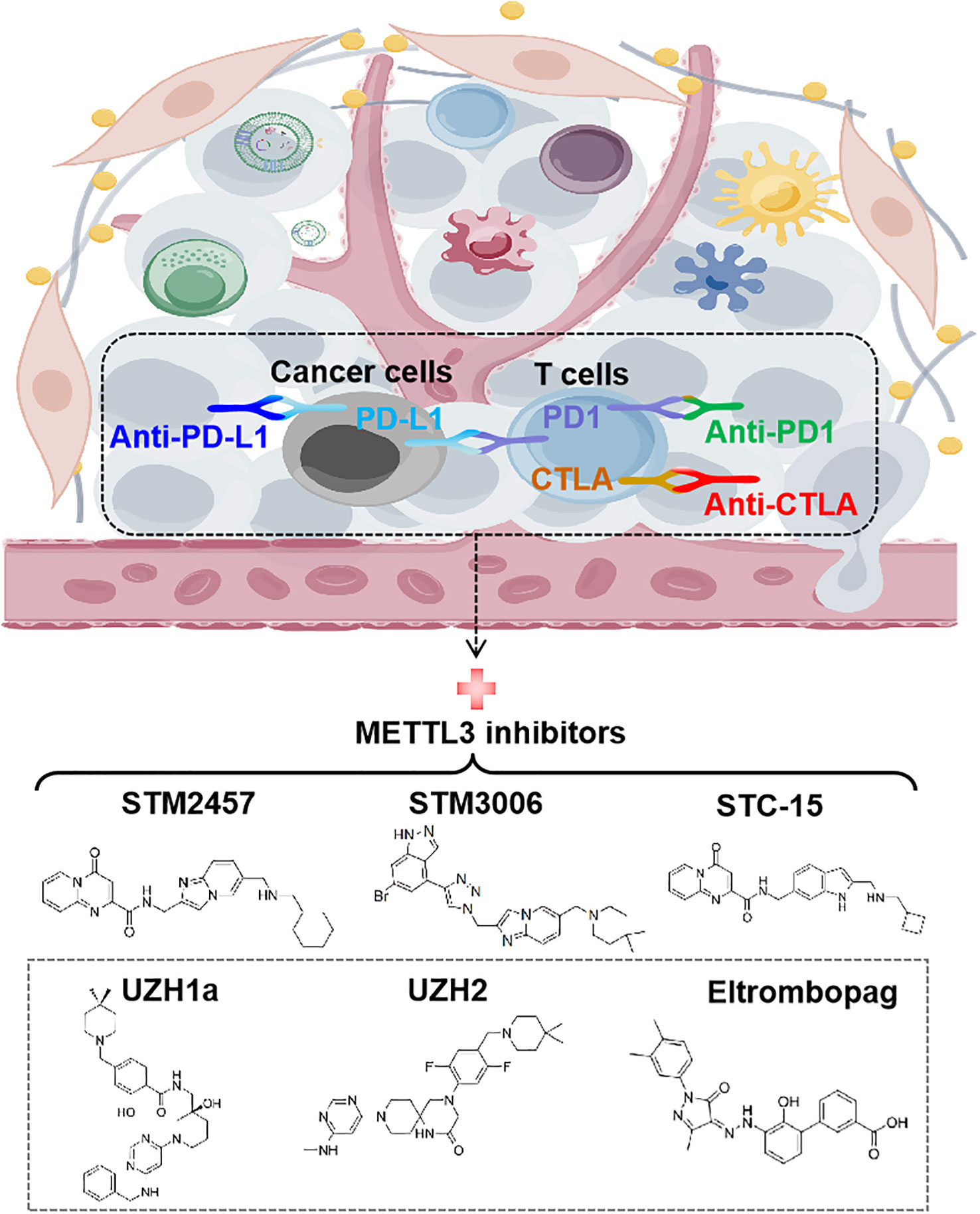
Figure 6 Targeting METTL3 for anti-cancer immunotherapy. The structures of recent discovered small-molecule inhibitors of METTL3 are shown. STM2457, STM3006, and STC-15 exhibit synergistic effects with anti-PD-1 in cancer therapy. PD-1, programmed cell death protein-1; PD-L1, programmed cell death ligand-1; CTLA-4, cytotoxic T lymphocyte-associated protein 4.
4 Discussion and perspective
Recent years have witnessed the rapid expansion of our knowledge of tumor immunity and its epigenetic regulation. Abundant evidence demonstrates that METTL3, as the primary m6A writer, plays imperative roles in various types of cancers, by affecting both cancer cells themselves and the surrounding immune cells and non-immune cells in TME. In summary, the roles of METTL3 in TME include: (i) affecting the differentiation and maturation of innate immune and adaptive immune cells; (ii) regulating dsRNA formation and activation of innate immune response; (iii) regulating immune checkpoints; (iv) affecting the extracellular biochemical microenvironment which mainly comprises metabolites, cytokines and inflammatory factors. It should be noted that recent research is focusing on the m6A-dependent function of METTL3 in TME, and key m6A-modified targets have been identified in these processes. However, emerging evidence also suggests that METTL3 plays catalytic activity-independent functions to promote the translation of target genes in certain types of cancers, such as LUAD and GC (89, 90). Therefore, it is possible that METTL3 might also exert m6A writer-independent functions in TME. Further studies are warranted to fully uncover the roles and mechanisms of METTL3 in TME.
Furthermore, research on METTL3 inhibitors is still in its infancy with many unanswered questions. For instance, recent studies are focusing on leukemia. It would be of great interest to know whether METTL3 inhibitors could effectively kill cancer cells or boost ICBs’ efficacy in solid tumors. It should also be noted that most studies of METTL3 inhibitors are still in the pre-clinical stage. Therefore, evidence from clinical research is urgently needed to support the translational application. In addition, screening or designing novel small molecular inhibitors utilizing artificial intelligence (AI) techniques and developing other targeting strategies such as targeted protein degradation may open new avenues for targeting METTL3 in cancer therapy.
Author contributions
WS: Writing – original draft, Writing – review & editing. LC: Funding acquisition, Writing – original draft, Writing – review & editing. WL: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. HH: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by National Natural Science Foundation of China 82173058 (to HH), 82173289 (to WL), 82372986 (to WL) and 82304180 (to LC), Guangdong Basic and Applied Basic Research Foundation 2021A1515010425 (to HH), China Postdoctoral Science Foundation 2022M713585 (to LC), Young Talents Program of Sun Yat-sen University Cancer Center YTP-SYSUCC-0044 (to HH) and the Fundamental Research Funds for the Central Universities, Sun Yat-sen University 23xkjc018 (to WL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. de Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell (2023) 41(3):374–403. doi: 10.1016/j.ccell.2023.02.016
2. Xie Y, Xie F, Zhang L, Zhou X, Huang J, Wang F, et al. Targeted anti-tumor immunotherapy using tumor infiltrating cells. Adv Sci (Weinh) (2021) 8(22):e2101672. doi: 10.1002/advs.202101672
3. Tay C, Tanaka A, Sakaguchi S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell (2023) 41(3):450–65. doi: 10.1016/j.ccell.2023.02.014
4. Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol (2015) 15(11):669–82. doi: 10.1038/nri3902
5. Tang T, Huang X, Zhang G, Hong Z, Bai X, Liang T. Advantages of targeting the tumor immune microenvironment over blocking immune checkpoint in cancer immunotherapy. Signal Transduction Targeted Ther (2021) 6(1):72. doi: 10.1038/s41392-020-00449-4
6. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (Time) for effective therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
7. Bilotta MT, Antignani A, Fitzgerald DJ. Managing the tme to improve the efficacy of cancer therapy. Front Immunol (2022) 13:954992. doi: 10.3389/fimmu.2022.954992
8. Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med (2019) 25(3):403–18. doi: 10.1038/s41591-019-0376-8
9. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol (2014) 15(5):313–26. doi: 10.1038/nrm3785
10. Lan Q, Liu PY, Bell JL, Wang JY, Hüttelmaier S, Zhang XD, et al. The emerging roles of rna M(6)a methylation and demethylation as critical regulators of tumorigenesis, drug sensitivity, and resistance. Cancer Res (2021) 81(13):3431–40. doi: 10.1158/0008-5472.Can-20-4107
11. Huang H, Weng H, Chen J. M(6)a modification in coding and non-coding rnas: roles and therapeutic implications in cancer. Cancer Cell (2020) 37(3):270–88. doi: 10.1016/j.ccell.2020.02.004
12. Deng X, Qing Y, Horne D, Huang H, Chen J. The roles and implications of rna M(6)a modification in cancer. Nat Rev Clin Oncol (2023) 20(8):507–26. doi: 10.1038/s41571-023-00774-x
13. Li X, Ma S, Deng Y, Yi P, Yu J. Targeting the rna M(6)a modification for cancer immunotherapy. Mol Cancer (2022) 21(1):76. doi: 10.1186/s12943-022-01558-0
14. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cdna cloning of the adomet-binding subunit of the human mrna (N6-adenosine)-methyltransferase. Rna (1997) 3(11):1233–47.
15. Huang J, Dong X, Gong Z, Qin LY, Yang S, Zhu YL, et al. Solution structure of the rna recognition domain of mettl3-mettl14 N(6)-methyladenosine methyltransferase. Protein Cell (2019) 10(4):272–84. doi: 10.1007/s13238-018-0518-7
16. Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of mettl3 and mettl14 methyltransferases. Mol Cell (2016) 63(2):306–17. doi: 10.1016/j.molcel.2016.05.041
17. Zeng C, Huang W, Li Y, Weng H. Roles of mettl3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol (2020) 13(1):117. doi: 10.1186/s13045-020-00951-w
18. Wang J, Yu H, Dong W, Zhang C, Hu M, Ma W, et al. N6-Methyladenosine-Mediated up-Regulation of Fzd10 Regulates Liver Cancer Stem Cells’ Properties and Lenvatinib Resistance through Wnt/B-Catenin and Hippo Signaling Pathways. Gastroenterology (2023) 164(6):990–1005. doi: 10.1053/j.gastro.2023.01.041
19. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. M(6)a mrna methylation regulates akt activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol (2018) 20(9):1074–83. doi: 10.1038/s41556-018-0174-4
20. Zhan Y, Liu Y, Yang R, Chen Q, Teng F, Huang Y, et al. Circpten suppresses human clear cell renal carcinoma progression and resistance to mtor inhibitors by targeting epigenetic modification. Drug Resist Update (2023) 71:101003. doi: 10.1016/j.drup.2023.101003
21. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
22. Li M, Zha X, Wang S. The role of N6-methyladenosine mrna in the tumor microenvironment. Biochim Biophys Acta Rev Cancer (2021) 1875(2):188522. doi: 10.1016/j.bbcan.2021.188522
23. Mu X, Wu K, Zhu Y, Zhu Y, Wang Y, Xiao L, et al. Intra-arterial infusion chemotherapy utilizing cisplatin inhibits bladder cancer by decreasing the fibrocytic myeloid-derived suppressor cells in an M6a-dependent manner. Mol Immunol (2021) 137:28–40. doi: 10.1016/j.molimm.2021.06.012
24. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. Mettl3/igf2bp3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of pd-L1 mrna in breast cancer. Mol Cancer (2022) 21(1):60. doi: 10.1186/s12943-021-01447-y
25. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. Mettl3-mediated M(6)a modification of hdgf mrna promotes gastric cancer progression and has prognostic significance. Gut (2020) 69(7):1193–205. doi: 10.1136/gutjnl-2019-319639
26. Ni Z, Sun P, Zheng J, Wu M, Yang C, Cheng M, et al. Jnk signaling promotes bladder cancer immune escape by regulating mettl3-mediated M6a modification of pd-L1 mrna. Cancer Res (2022) 82(9):1789–802. doi: 10.1158/0008-5472.Can-21-1323
27. Lan H, Liu Y, Liu J, Wang X, Guan Z, Du J, et al. Tumor-associated macrophages promote oxaliplatin resistance via mettl3-mediated M(6)a of traf5 and necroptosis in colorectal cancer. Mol Pharm (2021) 18(3):1026–37. doi: 10.1021/acs.molpharmaceut.0c00961
28. Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, et al. Mettl3 inhibits antitumor immunity by targeting M(6)a-bhlhe41-cxcl1/cxcr2 axis to promote colorectal cancer. Gastroenterology (2022) 163(4):891–907. doi: 10.1053/j.gastro.2022.06.024
29. Liu Z, Zheng N, Li J, Li C, Zheng D, Jiang X, et al. N6-methyladenosine-modified circular rna qsox1 promotes colorectal cancer resistance to anti-ctla-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Update (2022) 65:100886. doi: 10.1016/j.drup.2022.100886
30. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. M(6) a rna methyltransferases mettl3/14 regulate immune responses to anti-pd-1 therapy. EMBO J (2020) 39(20):e104514. doi: 10.15252/embj.2020104514
31. Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven mettl3-mediated rna M(6)a modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell (2022) 82(9):1660–77 e10. doi: 10.1016/j.molcel.2022.02.033
32. Wang G, Dai Y, Li K, Cheng M, Xiong G, Wang X, et al. Deficiency of mettl3 in bladder cancer stem cells inhibits bladder cancer progression and angiogenesis. Front Cell Dev Biol (2021) 9:627706. doi: 10.3389/fcell.2021.627706
33. Wu L, Cheng D, Yang X, Zhao W, Fang C, Chen R, et al. M2-tams promote immunoresistance in lung adenocarcinoma by enhancing mettl3-mediated M6a methylation. Ann Transl Med (2022) 10(24):1380. doi: 10.21037/atm-22-6104
34. Li Y, Li X, Deng M, Ye C, Peng Y, Lu Y. Cancer-associated fibroblasts hinder lung squamous cell carcinoma oxidative stress-induced apoptosis via mettl3 mediated M(6)a methylation of col10a1. Oxid Med Cell Longevity (2022) 2022:4320809. doi: 10.1155/2022/4320809
35. Yu H, Liu J, Bu X, Ma Z, Yao Y, Li J, et al. Targeting mettl3 reprograms the tumor microenvironment to improve cancer immunotherapy. Cell Chem Biol (2023) S2451-9456(23)00291-X. doi: 10.1016/j.chembiol.2023.09.001
36. Liu Z, Wang T, She Y, Wu K, Gu S, Li L, et al. N(6)-methyladenosine-modified circigf2bp3 inhibits cd8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of pd-L1 in non-small cell lung cancer. Mol Cancer (2021) 20(1):105. doi: 10.1186/s12943-021-01398-4
37. Chen M, Zhang Q, Zheng S, Guo X, Cao L, Ren Y, et al. Cancer-associated fibroblasts promote migration and invasion of non-small cell lung cancer cells via mettl3-mediated rac3 M(6)a modification. Int J Biol Sci (2023) 19(5):1616–32. doi: 10.7150/ijbs.79467
38. Tong J, Wang X, Liu Y, Ren X, Wang A, Chen Z, et al. Pooled crispr screening identifies M(6)a as a positive regulator of macrophage activation. Sci Adv (2021) 7(18):eabd4742. doi: 10.1126/sciadv.abd4742
39. Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. Mettl3-mediated M(6)a rna methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun (2021) 12(1):5522. doi: 10.1038/s41467-021-25803-0
40. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. Rna M6a methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun (2021) 12(1):1394. doi: 10.1038/s41467-021-21514-8
41. You M, Liu J, Li J, Ji C, Ni H, Guo W, et al. Mettl3-M(6)a-creb1 forms an intrinsic regulatory axis in maintaining inkt cell pool and functional differentiation. Cell Rep (2023) 42(6):112584. doi: 10.1016/j.celrep.2023.112584
42. He J, Zhou M, Yin J, Wan J, Chu J, Jia J, et al. Mettl3 restrains papillary thyroid cancer progression via M(6)a/C-rel/il-8-mediated neutrophil infiltration. Mol Ther (2021) 29(5):1821–37. doi: 10.1016/j.ymthe.2021.01.019
43. Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discovery (2022) 21(11):799–820. doi: 10.1038/s41573-022-00520-5
44. Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct Target Ther (2021) 6(1):75. doi: 10.1038/s41392-021-00484-9
45. Murray PJ. Macrophage polarization. Annu Rev Physiol (2017) 79:541–66. doi: 10.1146/annurev-physiol-022516-034339
46. Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N(6)-methyladenosine (M(6)a)-forming enzyme mettl3 facilitates M1 macrophage polarization through the methylation of stat1 mrna. Am J Physiol Cell Physiol (2019) 317(4):C762–c75. doi: 10.1152/ajpcell.00212.2019
47. Zhong C, Tao B, Yang F, Xia K, Yang X, Chen L, et al. Histone demethylase jmjd1c promotes the polarization of M1 macrophages to prevent glioma by upregulating mir-302a. Clin Transl Med (2021) 11(9):e424. doi: 10.1002/ctm2.424
48. Zhou D, Chen L, Yang K, Jiang H, Xu W, Luan J. Socs molecules: the growing players in macrophage polarization and function. Oncotarget (2017) 8(36):60710–22. doi: 10.18632/oncotarget.19940
49. Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, et al. The terminology issue for myeloid-derived suppressor cells. Cancer Res (2007) 67(1):425. doi: 10.1158/0008-5472.Can-06-3037
50. Ni HH, Zhang L, Huang H, Dai SQ, Li J. Connecting mettl3 and intratumoural cd33(+) mdscs in predicting clinical outcome in cervical cancer. J Transl Med (2020) 18(1):393. doi: 10.1186/s12967-020-02553-z
51. Myers JA, Miller JS. Exploring the nk cell platform for cancer immunotherapy. Nat Rev Clin Oncol (2021) 18(2):85–100. doi: 10.1038/s41571-020-0426-7
52. Crosby CM, Kronenberg M. Tissue-specific functions of invariant natural killer T cells. Nat Rev Immunol (2018) 18(9):559–74. doi: 10.1038/s41577-018-0034-2
53. Cao L, Morgun E, Genardi S, Visvabharathy L, Cui Y, Huang H, et al. Mettl14-dependent M(6)a modification controls inkt cell development and function. Cell Rep (2022) 40(5):111156. doi: 10.1016/j.celrep.2022.111156
54. Beauvillain C, Delneste Y, Scotet M, Peres A, Gascan H, Guermonprez P, et al. Neutrophils efficiently cross-prime naive T cells in vivo. Blood (2007) 110(8):2965–73. doi: 10.1182/blood-2006-12-063826
55. Mantovani A. The yin-yang of tumor-associated neutrophils. Cancer Cell (2009) 16(3):173–4. doi: 10.1016/j.ccr.2009.08.014
56. Dumitru CA, Lang S, Brandau S. Modulation of neutrophil granulocytes in the tumor microenvironment: mechanisms and consequences for tumor progression. Semin Cancer Biol (2013) 23(3):141–8. doi: 10.1016/j.semcancer.2013.02.005
57. Giese MA, Hind LE, Huttenlocher A. Neutrophil plasticity in the tumor microenvironment. Blood (2019) 133(20):2159–67. doi: 10.1182/blood-2018-11-844548
58. Luo S, Liao C, Zhang L, Ling C, Zhang X, Xie P, et al. Mettl3-mediated M6a mrna methylation regulates neutrophil activation through targeting tlr4 signaling. Cell Rep (2023) 42(3):112259. doi: 10.1016/j.celrep.2023.112259
59. Rowley DA, Fitch FW. The road to the discovery of dendritic cells, a tribute to ralph steinman. Cell Immunol (2012) 273(2):95–8. doi: 10.1016/j.cellimm.2012.01.002
60. Godoy-Tena G, Ballestar E. Epigenetics of dendritic cells in tumor immunology. Cancers (Basel) (2022) 14(5):1179. doi: 10.3390/cancers14051179
61. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mrna M(6)a methylation promotes dendritic cell activation. Nat Commun (2019) 10(1):1898. doi: 10.1038/s41467-019-09903-6
62. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mrna M(6)a methylation and ythdf1 in dendritic cells. Nature (2019) 566(7743):270–4. doi: 10.1038/s41586-019-0916-x
63. Lei K, Kurum A, Tang L. Mechanical immunoengineering of T cells for therapeutic applications. Accounts Chem Res (2020) 53(12):2777–90. doi: 10.1021/acs.accounts.0c00486
64. Chatzileontiadou DSM, Sloane H, Nguyen AT, Gras S, Grant EJ. The many faces of cd4(+) T cells: immunological and structural characteristics. Int J Mol Sci (2020) 22(1):73. doi: 10.3390/ijms22010073
65. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)a mrna methylation controls T cell homeostasis by targeting the il-7/stat5/socs pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
66. Yao YP, Yang Y, Guo WH, Xu LF, You MH, Zhang YC, et al. Mettl3-dependent M(6)a modification programs T follicular helper cell differentiation. Nat Commun (2021) 12(1):1333. doi: 10.1038/s41467-021-21594-6
67. Xiao Z, Wang S, Tian Y, Lv W, Sheng H, Zhan M, et al. Mettl3-mediated M6a methylation orchestrates mrna stability and dsrna contents to equilibrate Γδ T1 and Γδ T17 cells. Cell Rep (2023) 42(7):112684. doi: 10.1016/j.celrep.2023.112684
68. He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res (2020) 30(8):660–9. doi: 10.1038/s41422-020-0343-4
69. Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol (2019) 16(6):356–71. doi: 10.1038/s41571-019-0175-7
70. Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP, et al. M(6)a mrna methylation sustains treg suppressive functions. Cell Res (2018) 28(2):253–6. doi: 10.1038/cr.2018.7
71. Chapman NM, Boothby MR, Chi H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol (2020) 20(1):55–70. doi: 10.1038/s41577-019-0203-y
72. Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature (2019) 574(7779):575–80. doi: 10.1038/s41586-019-1678-1
73. Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, et al. M(6)a-dependent glycolysis enhances colorectal cancer progression. Mol Cancer (2020) 19(1):72. doi: 10.1186/s12943-020-01190-w
74. Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J, et al. Rna N(6)-methyladenosine methyltransferase mettl3 facilitates colorectal cancer by activating the M(6)a-glut1-mtorc1 axis and is a therapeutic target. Gastroenterology (2021) 160(4):1284–300.e16. doi: 10.1053/j.gastro.2020.11.013
75. Jiang X, Wang J, Deng X, Xiong F, Zhang S, Gong Z, et al. The role of microenvironment in tumor angiogenesis. J Exp Clin Cancer Res (2020) 39(1):204. doi: 10.1186/s13046-020-01709-5
76. Yao MD, Jiang Q, Ma Y, Liu C, Zhu CY, Sun YN, et al. Role of mettl3-dependent N(6)-methyladenosine mrna modification in the promotion of angiogenesis. Mol Ther (2020) 28(10):2191–202. doi: 10.1016/j.ymthe.2020.07.022
77. Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J Clin Invest (2015) 125(9):3335–7. doi: 10.1172/jci83871
78. Xu P, Ge R. Roles and drug development of mettl3 (Methyltransferase-like 3) in anti-tumor therapy. Eur J Medicinal Chem (2022) 230:114118. doi: 10.1016/j.ejmech.2022.114118
79. Bedi RK, Huang D, Eberle SA, Wiedmer L, Śledź P, Caflisch A. Small-molecule inhibitors of mettl3, the major human epitranscriptomic writer. ChemMedChem (2020) 15(9):744–8. doi: 10.1002/cmdc.202000011
80. Lee JH, Choi N, Kim S, Jin MS, Shen H, Kim YC. Eltrombopag as an allosteric inhibitor of the mettl3-14 complex affecting the M(6)a methylation of Rna in acute myeloid leukemia cells. Pharm (Basel) (2022) 15(4):440. doi: 10.3390/ph15040440
81. Moroz-Omori EV, Huang D, Kumar Bedi R, Cheriyamkunnel SJ, Bochenkova E, Dolbois A, et al. Mettl3 inhibitors for epitranscriptomic modulation of cellular processes. ChemMedChem (2021) 16(19):3035–43. doi: 10.1002/cmdc.202100291
82. Dolbois A, Bedi RK, Bochenkova E, Müller A, Moroz-Omori EV, Huang D, et al. 1,4,9-triazaspiro[5.5]Undecan-2-one derivatives as potent and selective mettl3 inhibitors. J Med Chem (2021) 64(17):12738–60. doi: 10.1021/acs.jmedchem.1c00773
83. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. M6a modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol (2019) 20(2):173–82. doi: 10.1038/s41590-018-0275-z
84. Ofir-Rosenfeld Y, Vasiliauskaite L, Saunders C, Sapetschnig A, Tsagkogeorga G, Albertella M, et al. Stc-15, an oral small molecule inhibitor of the rna methyltransferase mettl3, inhibits tumour growth through activation of anti-cancer immune responses associated with increased interferon signalling, and synergises with T cell checkpoint blockade. Eur J Cancer (2022) 174:S123. doi: 10.1016/S0959-8049(22)01128-5
85. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of mettl3 as a strategy against myeloid leukaemia. Nature (2021) 593(7860):597–601. doi: 10.1038/s41586-021-03536-w
86. Haigh DB, Woodcock CL, Lothion-Roy J, Harris AE, Metzler VM, Persson JL, et al. The mettl3 rna methyltransferase regulates transcriptional networks in prostate cancer. Cancers (Basel) (2022) 14(20):5148. doi: 10.3390/cancers14205148
87. Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ, Huang XT, et al. Mettl3 promotes intrahepatic cholangiocarcinoma progression by regulating ifit2 expression in an M(6)a-ythdf2-dependent manner. Oncogene (2022) 41(11):1622–33. doi: 10.1038/s41388-022-02185-1
88. Guirguis AA, Ofir-Rosenfeld Y, Knezevic K, Blackaby W, Hardick D, Chan YC, et al. Inhibition of mettl3 results in a cell-intrinsic interferon response that enhances antitumor immunity. Cancer Discovery (2023) 13(10):2228–47. doi: 10.1158/2159-8290.Cd-23-0007
89. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. Mrna circularization by mettl3-eif3h enhances translation and promotes oncogenesis. Nature (2018) 561(7724):556–60. doi: 10.1038/s41586-018-0538-8
Keywords: methyltransferase-like 3 (METTL3), N6-methyladenosine (m6A), tumor-infiltrating immune cells (TICs), small molecular inhibitors, cancer immunotherapy, immune checkpoint inhibitors (ICI)
Citation: Su W, Che L, Liao W and Huang H (2024) The RNA m6A writer METTL3 in tumor microenvironment: emerging roles and therapeutic implications. Front. Immunol. 15:1335774. doi: 10.3389/fimmu.2024.1335774
Received: 09 November 2023; Accepted: 04 January 2024;
Published: 22 January 2024.
Edited by:
Chengwu Zeng, Jinan University, ChinaReviewed by:
Di Chen, Zhejiang University, ChinaKai Song, University of California, Los Angeles, United States
Chen Xue, Zhejiang University, China
Copyright © 2024 Su, Che, Liao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenting Liao, bGlhb3d0QHN5c3VjYy5vcmcuY24=; Huilin Huang, aHVhbmdobDFAc3lzdWNjLm9yZy5jbg==
†These authors have contributed equally to this work
 Weiqi Su
Weiqi Su Lin Che
Lin Che Wenting Liao
Wenting Liao Huilin Huang
Huilin Huang