
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 31 January 2024
Sec. Comparative Immunology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1329098
This article is part of the Research Topic The role of bats and their immune system in structuring host–pathogen interactions View all 10 articles
Background: GBPs (guanylate binding proteins), an evolutionary ancient protein family, play a key role in the host’s innate immune response against bacterial, parasitic and viral infections. In Humans, seven GBP genes have been described (GBP1-7). Despite the interest these proteins have received over the last years, evolutionary studies have only been performed in primates, Tupaia and rodents. These have shown a pattern of gene gain and loss in each family, indicative of the birth-and-death evolution process.
Results: In this study, we analysed the evolution of this gene cluster in several bat species, belonging to the Yangochiroptera and Yinpterochiroptera sub-orders. Detailed analysis shows a conserved synteny and a gene expansion and loss history. Phylogenetic analysis showed that bats have GBPs 1,2 and 4-6. GBP2 has been lost in several bat families, being present only in Hipposideidae and Pteropodidae. GBPs1, 4 and 5 are present mostly as single-copy genes in all families but have suffered duplication events, particularly in Myotis myotis and Eptesicus fuscus. Most interestingly, we demonstrate that GBP6 duplicated in a Chiroptera ancestor species originating two genes, which we named GBP6a and GBP6b, with different subsequent evolutionary histories. GBP6a underwent several duplication events in all families while GBP6b is present as a single copy gene and has been lost in Pteropodidae, Miniopteridae and Desmodus rotundus, a Phyllostomidae. With 14 and 15 GBP genes, Myotis myotis and Eptesicus fuscus stand out as having far more copies than all other studied bat species. Antagonistically, Pteropodidae have the lowest number of GBP genes in bats.
Conclusion: Bats are important reservoirs of viruses, many of which have become zoonotic diseases in the last decades. Further functional studies on bats GBPs will help elucidate their function, evolutionary history, and the role of bats as virus reservoirs.
Guanylate-binding proteins (GBPs) are a family of evolutionary ancient, conserved proteins that have vital roles in host defence against intracellular pathogens, ranging from viruses to bacteria (1, 2). These proteins belong to the large dynamin guanosine triphosphatases (GTPases) superfamily and the IFN-inducible guanosine triphosphatases (3) and share structural and biochemical similarities (3–5). The structure of these proteins comprises a globular N-terminal large GTPase (LG) domain connected by a hinge region to the middle domain (MD) and the GTPase effector domain (GED) at the C-terminus. The LG domain is involved in GTPase and GDPase activity and Mg2+ cofactor finding [reviewed in (3, 6)]. GBP expression is triggered by inflammatory signals, the most potent of which are interferons (IFN), but also interleukins (IL) and tumour necrosis factor (TNF) [reviewed in (3)]. As such, they are part of the cell-autonomous innate immune response and have been considered major players in the host’s innate immunity.
In mammals, GBPs are usually organized in tandem in one chromosome (5, 7) but in some rodents, like Mus musculus and Rattus norvegicus, GBPs are organized in two different gene clusters (8). Surprisingly, this gene family was only studied in primates, Tupaia and rodents (8–10). The evolutionary history of GBPs is complex, with duplications, deletions and neofunctionalization of genes as expected in gene families following the birth-and-death model of evolution (11). The number of GBP genes varies among species; not all GBP orthologs are present in every species and some are limited to a specific mammalian group. Seven GBPs have been described in humans, GBPs1-7 (1, 5). Of these, GBP3 seems to have emerged through a duplication of GBP1 in Simiiformes gaining a new function, regulation of caspase-4 activation (12). GBP7 most likely emerged from a duplication of GBP4 in primates being specific to this group (9). Muroids (Rodentia) share GBP2, GBP5 and GBP6 orthologs with primates and furthermore have four exclusive GBPs, GBPa-d. Each of the seven Muroid GBPs has its own pattern of duplications and deletions (8). In Tupaia, five GBPs have been described: GBP1, GBP2, GBP4, GBP5 and GBP7, all seemingly orthologs to primates’ GBPs except for GBP7 (10).
Bats belong to the order Chiroptera, the second largest mammalian order after Rodentia and have adapted to diverse ecological niches across the planet (13). The Chiroptera radiation occurred approximately 60 million years ago (mya) (14–16). Based on molecular genetics data, the order Chiroptera is subdivided into two suborders, Yangochiroptera (composed exclusively of microbat families) and Yinpterochiroptera (composed of five microbat families and all megabat families) (14). Bats are known reservoirs of many viruses in the animal kingdom, and have been assigned as the source of many human viral diseases in modern times, including SARS-CoV (China, 2002/2003) (17), Marburg virus (Africa, 2005) (18), MERS-CoV (Middle East, 2012) (19, 20), Ebola virus (West Africa, 2013) (21, 22) and the recent SARS-CoV2 (China, 2019) (23, 24). With shrinking habitats, caused by the Human population expansion, wild populations are co-existing in closer and closer proximity to humans leading to an increased threat of these events.
Studying the immune system of these species is key to assessing their resilience in a changing environment as well as what makes these unique mammals such good virus reservoirs. Still, the bats’ immune system is poorly understood. The innate immunity system (IIS) is the body’s first line of defence against pathogens. In this work, we analysed the GBP evolution in bats. We found a complex history of gene gain and loss with very different genetic repertoires between bat families.
Complete coding sequences of GBPs were obtained from publicly available databases. A total of 183 nucleotide sequences were collected from bats (129), primates (41), Tupaia glis (5) and Loxodonta africana (8). The accession numbers of these sequences can be found in Supplementary Material: Table 1. The L. africana GBP sequences were used as an outgroup. For bats, GBP sequences were collected only for species for which good-quality genomes are available at GenBank and Ensembl. Annotated GBP sequences were obtained through BLASTn searches using Human GBP sequences as queries. Searches were conducted in the NCBI’s GenBank (http://www.ncbi.nlm.nih.gov/genbank/) and Ensembl (https://www.ensembl.org/index.html) genome databases. Further BLASTn searches using Bat GBP sequences were conducted in both databases to ensure all bat GBP sequences were identified. In total, GBP sequences were obtained for 19 different species of bats belonging to eight families, Vespertilionidae, Miniopteridae, Phyllostomidae, Hipposideridae, Pteropodidae, Rhinolophidae, Mormoopidae and Molossidae.
Rodent GBP sequences, although available, were not used in this study. Muroid rodents GBP genes have a complex and seemingly specific pattern of evolution with four GBP genes that appear to be exclusive to Muroids (8). This diversity will add noise to our phylogenetic analysis causing a loss in resolution and correct identification of Bats GBPs.
Sequences were aligned with Clustal W (25) as implemented in BioEdit v7.2.5 (26), followed by visual inspection and necessary manual corrections. This dataset was screened for gene conversion using GARD (27); no recombination breakpoints were identified. The final nucleotide sequence alignment is given in Supplementary Material: Data 1.
The phylogenetic relationships between GBP nucleotide sequences were inferred using MEGA version 11 software (28) under a Maximum likelihood (ML) framework. The phylogenetic tree was constructed using the GTR+G+I model of nucleotide substitution, determined to be the best fitting model to our dataset by the Model Selection option in MEGA 11 (28). Node support was determined from 1000 bootstrap replicate trees.
The relative syntenic positions and transcription orientation of bats GBPs were assessed using NCBI (https://www.ncbi.nlm.nih.gov/genome/gdv/).
Genetic distances between the groups established based on the ML tree (see Figure 1) were calculated using MEGA 11 (28). The net between group mean distances function was used to obtain the genetic distances between bat and primate GBP groups. This option accounts for variance due to differences within groups. These were calculated in MEGA 11 (28) software using the p-distance method, uniform rates among sites, homogeneous rates among lineages and pairwise deletion of gaps options.
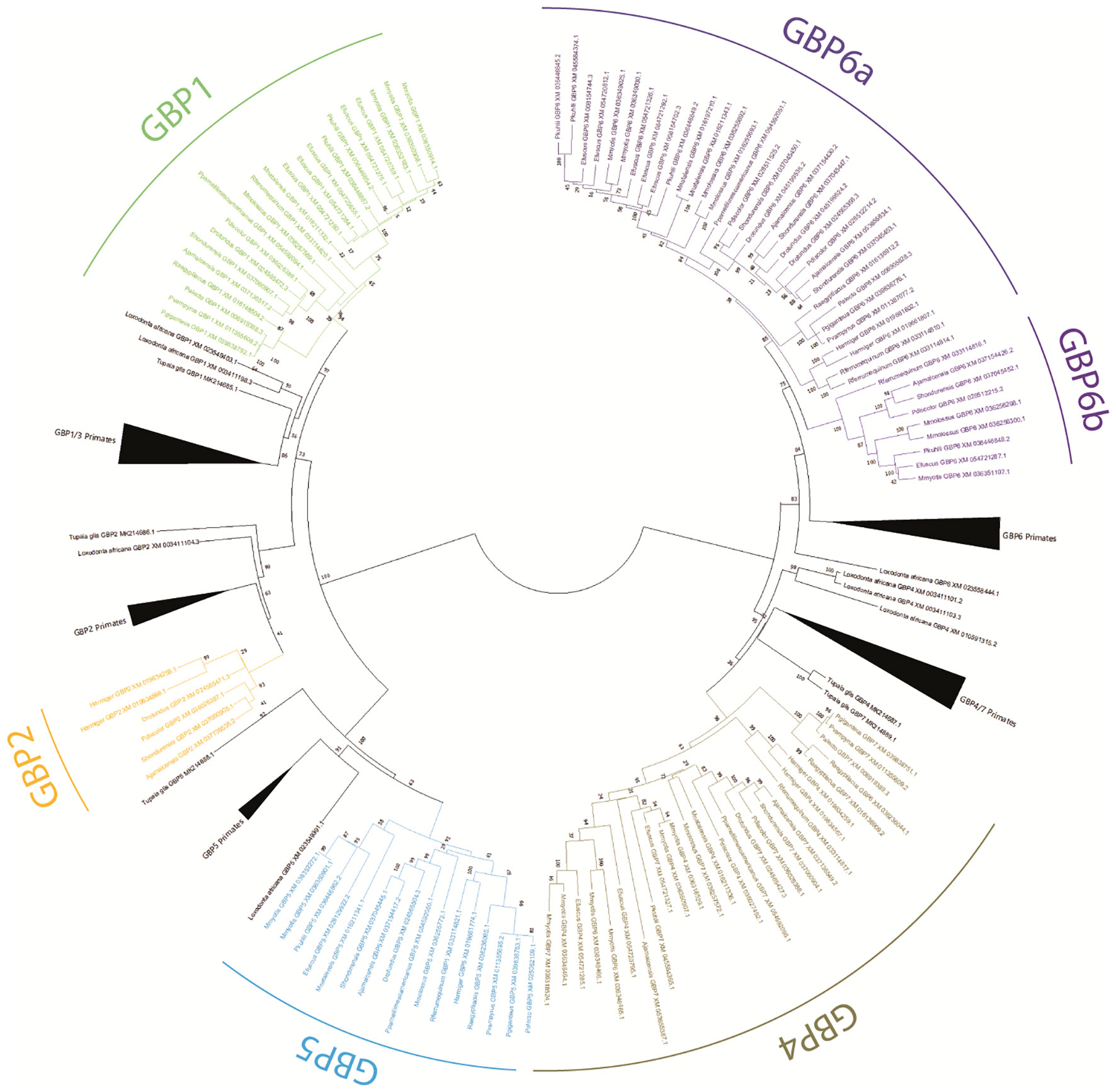
Figure 1 Phylogenetic tree of GBP genes in Chiroptera. A Maximum likelihood (ML) method and the GTR+G+I model of nucleotide substitution were used to obtain the GBP gene family phylogenetic tree. Bootstrap values are indicated near the most relevant branches.
The nucleotide substitution rate variation among the Chiroptera GBP6 genes was estimated in DnaSp version 6.12 (33). Sliding window analysis was performed with a window length of 250 nucleotides and a step size of 12 nucleotides along the nucleotide sequence alignment and plotting the differences as averages. Sites with alignment gaps were not counted.
The obtained ML phylogenetic tree shows bats, primates, Tupaia and L. africana GBP sequences grouped according to gene with good bootstrap support (Figure 1). Sequences for bats GBP1, GBP2, GBP4, GBP5, and GBP6 were identified. Within the GBP4 cluster, several sequences annotated as GBP6 and GBP7 appear. These seem misannotated and need to be reclassified (see Supplementary Material: Table 1).
Bat GBP1 is present in all species except Hipposideros armiger. An incomplete sequence resembling GBP1 was found for this species, located where GBP1 would be expected, but it is not possible to confirm whether it is GBP1 or not. Most species have a single copy of GBP1. However, Eptesicus fuscus has five GBP1 copies, Myotis myotis three GBP1 genes and Pippistrelus khuli has two copies, showing that duplication events have occurred in Vespertilionidae bats (Figures 1, 2).
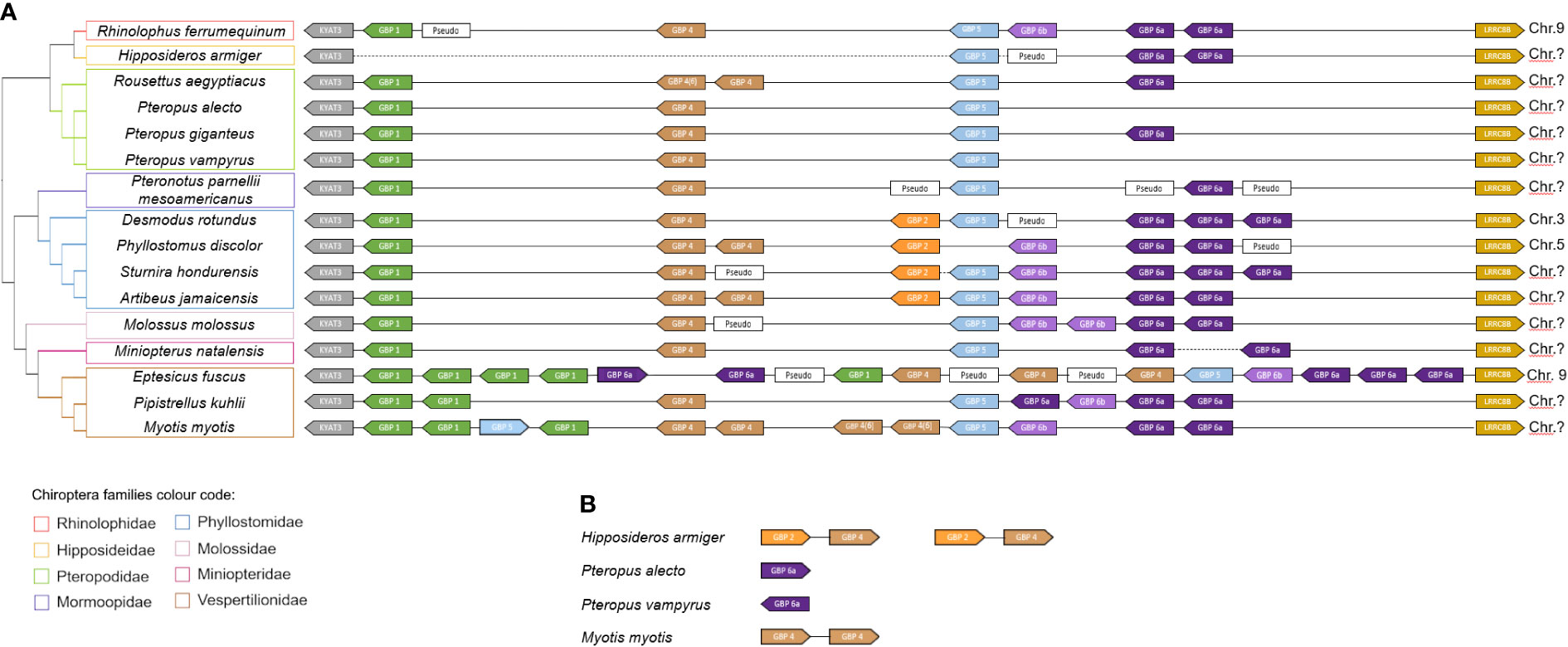
Figure 2 GBP gene family synteny in Chiroptera. (A) Organization of the GBP gene family in the studied species according to genomes available in NCBI (www.ncbi.nlm.nih.org). (B) Unplaced GBP in the bat genomes. The diagram is not drawn to scale. Arrows represent transcription orientation. Dashed lines represent gaps in the genome. Sequences excluded from the analysis for including stop codons or frameshifting indels are indicated as Pseudo. Chromosomes are indicated when information is available. Colour scheme:  GBP1,
GBP1,  GBP2,
GBP2,  GBP4,
GBP4,  GBP5,
GBP5,  GBP6a and
GBP6a and  GBP6b.
GBP6b.
Bat GBP2 was identified only for species of Phyllostomidae and Hipposideidae bats. Considering that these sequences are clustering with the primate sequences and are present in both Yangochiroptera (Phyllostomidae) and Yinpterochiroptera (Hipposideidae) suborders (Figures 1, 2), it suggests that GBP2 was present in the Chiroptera ancestor and was lost independently in several lineages.
Bat GBP4 and GBP5 are present in all studied species, the exception being Phyllostomus discolor which has lost GBP5. For these two genes, M. myotis has suffered duplications, being the only studied bat species with more than one GBP5 and having six GBP4 genes, contrasting with most other species which have one or two GBP4 copies (Figure 1, Table 1).
Within the GBP6 cluster, there are two well-supported subgroups (100 bootstrap for GBP6b and 85 bootstrap for GBP6a; Figure 1): a larger cluster containing sequences of the eight analysed bat families and a smaller cluster encompassing sequences for four of the analysed bat families. Within the larger cluster, most species have at least two copies of this GBP6 gene, except for the species belonging to the Pteropodidae family which carry only one copy (Figure 1, Table 1). This is in contrast with the smaller cluster for which most species have only one copy of the gene, the exception being Molossus molossus with two copies (Figure 1, Table 1). This pattern suggests that the GBP6 has duplicated in a Chiroptera ancestor originating two genes that have since followed distinct evolutionary patterns. Hereon, we shall designate the larger cluster as GBP6a and the smaller cluster as GBP6b (Figure 1).
It thus seems that the different bat GBPs have independently suffered deletions and/or duplications and are, thus, evolving under distinct evolutionary pressures. Table 1 shows a summary of the number and repertoire found for each studied bat species. Pteropodidae species, having lost GBP2 and GBP6b, have the least diversity of GBP genes and also the fewest copies since these have only one copy of each of the four other GBP genes present. The exception is Rousettus aegyptiacus, which has two copies of GBP4, resulting in five copies of GBP genes in R. aegyptiacus and four copies in the remaining Pteropodidae species (Table 1). In contrast, Vespertilionidae bats, in particular M. Myotis and E. fuscus, have an expansion of GBP genes, with a total of 14 and 15 GBP genes each, respectively (Table 1).
The bats’ GBP synteny is quite conserved (Figure 2). Despite the variability in the number of genes, bats’ GBPs are organized in tandem and flanked by KYAT3 and LRRC8B. For H. armiger, Pteropus alecto, Pteropus vampyrus and M. myotis, unplaced GBP genes were found. For H. armiger a genome gap exists between KYAT3 and the GBP genes, not being possible to determine the full GBP locus organization for this species. The unplaced GBP2 and GBP4 genes may be located in the canonical GBP locus, between KYAT3 and GBP5, where these would be expected to be according to the other bats’ synteny. The M. natalensis and S. hondurensis assemblies also have one genome gap in each one. The P. alecto, P. vampyrus and M. myotis unplaced genes are most likely retrotransposons inserted in other genomic locations.
The genetic distances confirm that bat GBPs are orthologs to primate GBPs, with low divergence between bat GBPs and their primate counterpart (7%-10.2%; Table 2) and high divergence between bats’ GBPs (6.8-28.6%; Table 2). Considering that primates GBP1 and GBP3 are considered two genes with a divergence as low as 4%, the divergence of 6.8% between the two subgroups of bats’ GBP6, GBP6a and GBP6b (Table 2), supports their classification as different genes that arose from a duplication of GBP6 in a bat ancestor. Furthermore, several amino acid residues that differentiate GBP6a from GBP6b are observable (Figure 3; see Supplementary Material: Data 3) and the amino acidic distance between these genes is high (11%). All the obtained results support the classification of GBP6a and GBP6b as different genes. This is considering 1) the good bootstrap support of the two bat GBP6 groups in the ML phylogenetic tree, 2) their genetic distance being higher than that between pGBP1 and pGBP3 and 3) the existence of amino acid characteristic positions between GBP6a and GBP6b.

Figure 3 Bats Gbp6a and Gbp6b amino acid sequences diversity comparison. Alignments of the 33 Gbp6a and 9 Gbp6b sequences were used to create the sequence’s logo graphical representations, using the WebLogo program (29). Only amino acid variable positions are depicted (see Supplementary Material: Data 3). Amino acid residues that differentiate GBP6a from GBP6b are in red boxes; the position in the alignment for these residues is indicated below the boxes.
The analysis of the nucleotide diversity along the two GBP6 genes shows that, overall, this parameter is higher in the 3’ end of the LG, MD and GED domains. Focusing only on non-synonymous sites shows that a high proportion of the nucleotide substitutions between GBP6a and GBP6b are non-synonymous and confirms that the two genes’ amino acid sequence is very divergent, particularly in the effector regions of the genes (Figure 4; see Supplementary Material: Data 2, 3).
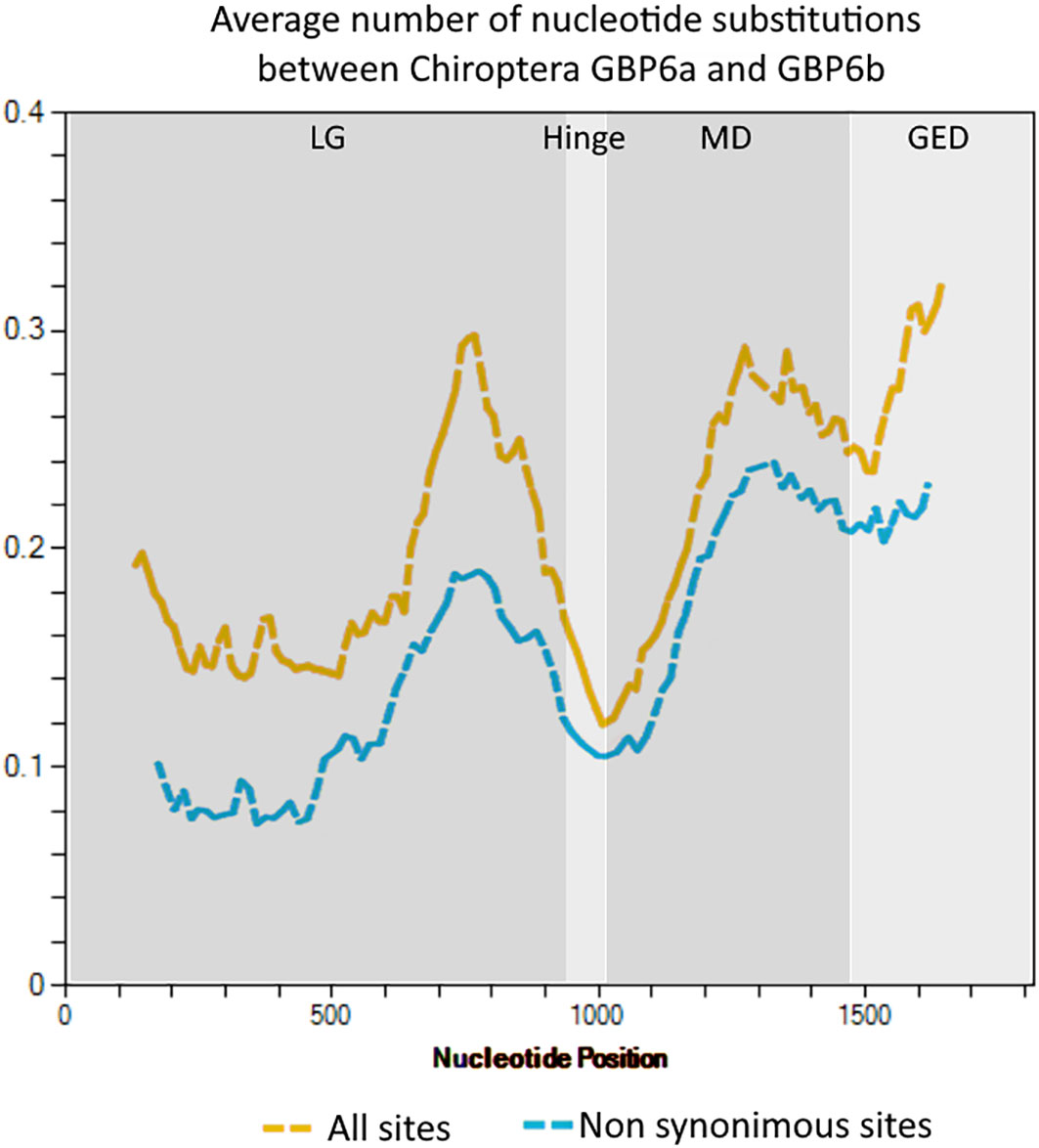
Figure 4 Sliding window analysis showing the nucleotide diversity along the GBP6a and GBP6b genes. The analysis was performed in DnaSP version 6.12 with a window length of 250 nucleotides and a step size of 12 nucleotides. The GBP domains are indicated.
Bats have been identified as a major reservoir for zoonotic viruses and have played roles in outbreaks of several emerging zoonotic viruses including SARS-CoV (China, 2002/2003) (17), Marburg virus (Africa, 2005) (18), MERS-CoV (Middle East, 2012) (19, 20), Ebola virus (West Africa, 2013) (21, 22) and the recent SARS-CoV2 (China, 2019) (23, 24). Viral reservoir species have an organized immunological response to the virus but show no overt clinical signs of disease. This means that the host usually carries a low viral load and is able to tolerate some viral replication. Studies have shown that bats have unique immunological approaches to enable coexistence with viral infections, causing no disease, while allowing enough viral replication for transmission (30–32). Screening the virome for over 4000 healthy bats from 40 different species of both Yangochiroptera and Yinpterochiroptera suborders, revealed an array of viruses belonging to diverse families, the most prevalent being Herpesviridae, Papillomaviridae, Retroviridae, Adenoviridae and Astroviridae, but also Coronaviridae, Caliciviridae, Polyomaviridae, Rhabdoviridae, among others (33), confirming bats as reservoirs of various virus. The unique ability of bats to act as reservoirs is thought to be mediated by a dampening of pro-inflammatory responses (30, 32, 34) as well as an increased resistance to infection mediated by special features of the antiviral type I interferon (IFN) system [reviewed in (35)]. GBPs are IFN-induced GTPases that have been shown to have vital roles in host immunity to infection and inflammation.
In this study, we screened available bat genomes for GBP genes. Sequences for bats GBP1, GBP2, GBP4, GBP5, and GBP6 were identified, indicating that these could be orthologs to their human counterparts. It would be of interest to perform functional assays to confirm if functions remain conserved, for example, both human and mouse GBP5 have been implicated in the NLRP3 activation upon bacterial infection (36). However, considering the obtained phylogenetic results, it is worth noting that some of the genes are poorly identified in the public databases. For example, some of the genes that are identified as GBP4 or GBP7 correspond, according to the presented phylogenetic tree, to GBP6 genes. Furthermore, many genes in the GBP4 group are annotated as GBP7. GBP7, however, has been shown to have emerged in primates as a duplication of pGBP4 (9), our phylogenetic tree does not show a distinction between bats GBP4 and GBP7 and these are also phylogenetically close to Tupaia glis and Loxodonta africana GBP4. Accordingly, these genes should be classified as GBP4. For these reasons, we propose a review of the nomenclature of these genes in the aforementioned databases and greater attention in subsequent studies aiming to sequence genomes of bat species to avoid this type of error (see Supplementary Material: Table 1). Gene misannotation in public databases compromises not only evolutionary studies but also, and maybe more relevantly, the biological and functional understanding of the gene (37), leading to erroneous conclusions.
Interestingly, our analysis shows the existence of two GBP6 genes in bats. The phylogenetic tree clustering (Figure 1), the calculated genetic distances (Table 2) and the existence of specific amino acids to both GBP6a and GBP6b, all support that these should be considered two independent genes. Furthermore, GBP6a and GBP6b seem to be evolving under different evolutionary pressures. The duplication of GBP6 occurred in a Chiroptera ancestor, circa 62 million years ago (38) given that both GBP6a and GBP6b are present in Yinpterochiroptera and Yangochiroptera suborders. GBP6a has persisted in all studied species and suffered duplications in all but the Pteropodidae family species, Pteropus sp and R. aegyptiacus. GBP6b, on the contrary, has been lost independently in several lineages and is present mostly as a single-copy gene. These different evolutionary patterns and the existence of characteristic amino acids in each gene suggest that GBP6a and GBP6b have evolved or are evolving to perform different functions. All sequences present the four conserved elements for nucleotide binding and hydrolysis (see alignment in Supplementary Material: Data 2); G1-G4, where G1 (45GxxxxGKS/T52), G2 present a threonine (75T), G3 97DxxG100 and G4 with 179T/AVRD183 (for GBP4, 6 and 7) (39). Despite this, the amino acid differences between GBP6a and GBP6b are numerous. As such, functional studies should be performed to understand how these differences influence the biological roles of these two genes (expression, localization and role against pathogens).
Of worth noticing, is also the discrepancy in the number and diversity of GBP genes between bat families. Pteropodidae species, having lost GBP2 and GBP6b, have the least diversity of GBP genes and also the fewest copies. On the opposite, Vespertilionidae bats, in particular M. Myotis and E. fuscus, have an expansion of GBP genes, each with a total of 15 GBP genes (Table 1). It is tempting to speculate that the loss of GBP2 led to the expansion of GBP4 in M. Myotis and E. fuscus since, in humans, upon Salmonella infection, GBP1 requires GBP2 and GBP4 to recruit caspase-4 to the surface of the bacteria (2, 40). However, in P. kuhlii or Pteropodidae bats, this trend is not observed. The resulting patterns in the number of GBPs seem to be species-specific and could have been caused by host-pathogen co-evolution since bats can be reservoirs for several viruses. Gene expansions have been described for other immune system genes in Yangochiroptera bats, such as the IFITM locus (41), and more specifically in the genus Myotis, for which an unusual expansion of the S100A7 genes occurred (42).
Although bat genomes are smaller than the genomes of other groups of mammals (13, 43), Pteropodidae (megabat) genomes tend to be smaller than the genomes of other bats (44). One of the reasons given is the fact that they have lost functions in an important line of long interspersed retrotransposable elements (LINES) known as LINE-1 (45). LINE-1 retrotransposons are the most abundant in mammals; in humans, for example, they account for around 15-20% of the genome (46). That said, it is possible that this and other similar events contributed to the notably reduced size of the megabat genome and more specific studies are needed to understand the reason for this reduction, as they are also the most distinct members of the chiropteran order, given their size, distribution, food, among others. The greater expansion of immune system genes in the genera Myotis and Eptesicus may be related to the success of these genera in colonizing new habitats (47).
This study further demonstrated that GBP3 and GBP7 are not present in bats, genes which are also absent in rodents (8). These results are congruent with the previous description of the emergence of these genes in primates, GBP3 as a duplication of GBP1 and GBP7 as a duplication of GBP4 (9). Several sequences of GBP pseudogenes were identified in the bat genomes (see synteny; Figure 2A), several genes were lost in different bat species and many GBP genes were duplicated. These data strongly support the birth-and-death model of evolution, which postulates that during evolution the genes from multigene families suffer duplications; some of these genes can be maintained in the genome, some can become pseudogenised and others can acquire a new function (3, 8, 9).
The results of this study show that several evolutionary processes occurred in the bats’ GBP gene family, such as gene deletions and duplications. These data are in accordance with the birth-and-death model of evolution, already attributed to members of this multigene family. An expansion of this gene family was also demonstrated in M. myotis and E. fuscus, and a reduction in it in members of the Pteropodidae family. A duplication of the GBP6 gene was identified, which gave rise to two new genes, here named GBP6a and GBP6b. These genes present several different amino acids between the two genes, changes which may affect function; therefore, it is suggested that specific studies on the functions of these new genes should be carried out. Here, we also propose a review of the nomenclature of this gene family in order Chiroptera, since our results demonstrate that GBP genes in bats were poorly annotated. Additionally, each bat species presents a specific GBP evolution, possibly due to host-pathogen co-evolution. More evolutionary studies should be carried out to fully understand the complex evolution of GBPs and provide more insights into their function. Additionally, it will be important to sequence more bat species and improve the quality of some currently available genomes, so that more complete evolutionary studies can be carried out.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
AP: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. JB: Formal analysis, Writing – original draft, Writing – review & editing. JC-R: Formal analysis, Writing – review & editing. PE: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by National Funds through Fundação para a Ciência e a Tecnologia (FCT) in the scope of the project UIDP/50027/2020). FCT also supported the JC-R PhD fellowship (DFA/BD/4965/2020) and the Investigator grant of PE (CEECIND/CP1601/CT0005).11.
BIOPOLIS – Enhancing the transference of scientific and technological knowledge through a new Centre of Excellence in Environmental Biology, Ecosystems and AgroBiodiversity (NORTE-01-0246-FEDER-000063), financed by Programa Operacional Regional do Norte (NORTE2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1329098/full#supplementary-material
1. Tretina K, Park ES, Maminska A, MacMicking JD. Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. J Exp Med (2019) 216(3):482–500. doi: 10.1084/jem.20182031
2. Kutsch M, Coers J. Human guanylate binding proteins: nanomachines orchestrating host defence. FEBS J (2021) 288(20):5826–49. doi: 10.1111/febs.15662
3. Schelle L, Corte-Real JV, Esteves PJ, Abrantes J, Baldauf HM. Functional cross-species conservation of guanylate-binding proteins in innate immunity. Med Microbiol Immunol (2023) 212(2):141–52. doi: 10.1007/s00430-022-00736-7
4. Praefcke GJK. Regulation of innate immune functions by guanylate-binding proteins. Int J Med Microbiol (2018) 308(1):237–45. doi: 10.1016/j.ijmm.2017.10.013
5. Li G, Zhang J, Sun Y, Wang H, Wang Y. The evolutionarily dynamic IFN-inducible GTPase proteins play conserved immune functions in vertebrates and cephalochordates. Mol Biol Evol (2009) 26(7):1619–30. doi: 10.1093/molbev/msp074
6. Zhang R, Li Z, Tang YD, Su C, Zheng C. When human guanylate-binding proteins meet viral infections. J BioMed Sci (2021) 28(1):17. doi: 10.1186/s12929-021-00716-8
7. Olszewski MA, Gray J, Vestal DJ. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J Interferon Cytokine Res (2006) 26(5):328–52. doi: 10.1089/jir.2006.26.328
8. Corte-Real JV, Baldauf HM, Melo-Ferreira J, Abrantes J, Esteves PJ. Evolution of guanylate binding protein (GBP) genes in muroid rodents (Muridae and cricetidae) reveals an outstanding pattern of gain and loss. Front Immunol (2022) 13:752186. doi: 10.3389/fimmu.2022.752186
9. Corte-Real JV, Baldauf HM, Abrantes J, Esteves PJ. Evolution of the guanylate binding protein (GBP) genes: Emergence of GBP7 genes in primates and further acquisition of a unique GBP3 gene in simians. Mol Immunol (2021) 132:79–81. doi: 10.1016/j.molimm.2021.01.025
10. Gu T, Yu D, Fan Y, Wu Y, Yao YL, Xu L, et al. Molecular identification and antiviral function of the guanylate-binding protein (GBP) genes in the chinese tree shrew (Tupaia belangeri chinesis). Dev Comp Immunol (2019) 96:27–36. doi: 10.1016/j.dci.2019.02.014
11. Nei M, Gu X, Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA (1997) 94(15):7799–806. doi: 10.1073/pnas.94.15.7799
12. Wandel MP, Kim BH, Park ES, Boyle KB, Nayak K, Lagrange B, et al. Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat Immunol (2020) 21(8):880–91. doi: 10.1038/s41590-020-0697-2
13. Teeling EC, Vernes SC, Davalos LM, Ray DA, Gilbert MTP, Myers E, et al. Bat biology, genomes, and the Bat1K project: To generate chromosome-level genomes for all living bat species. Annu Rev Anim Biosci (2018) 6:23–46. doi: 10.1146/annurev-animal-022516-022811
14. Amador LI, Moyers Arévalo RL, Almeida FC, Catalano SA, Giannini NP. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J Mamm Evolution. (2018) 25(1):37–70. doi: 10.1007/s10914-016-9363-8
15. Shi JJ, Rabosky DL. Speciation dynamics during the global radiation of extant bats. Evolution (2015) 69(6):1528–45. doi: 10.1111/evo.12681
16. Agnarsson I, Zambrana-Torrelio CM, Flores-Saldana NP, May-Collado LJ. A time-calibrated species-level phylogeny of bats (Chiroptera, mammalia). PloS Curr (2011) 3:RRN1212. doi: 10.1371/currents.RRN1212
17. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science (2005) 310(5748):676–9. doi: 10.1126/science.1118391
18. Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, et al. Seasonal pulses of marburg virus circulation in juvenile rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PloS Pathog (2012) 8(10):e1002877. doi: 10.1371/journal.ppat.1002877
19. Lau SKP, Fan RYY, Zhu L, Li KSM, Wong ACP, Luk HKH, et al. Isolation of MERS-related coronavirus from lesser bamboo bats that uses DPP4 and infects human-DPP4-transgenic mice. Nat Commun (2021) 12(1):216. doi: 10.1038/s41467-020-20458-9
20. Yang L, Wu Z, Ren X, Yang F, Zhang J, He G, et al. MERS-related betacoronavirus in vespertilio superans bats, china. Emerg Infect Dis (2014) 20(7):1260–2. doi: 10.3201/eid2007.140318
21. Mari Saez A, Weiss S, Nowak K, Lapeyre V, Zimmermann F, Dux A, et al. Investigating the zoonotic origin of the west african ebola epidemic. EMBO Mol Med (2015) 7(1):17–23. doi: 10.15252/emmm.201404792
22. Leroy EM, Epelboin A, Mondonge V, Pourrut X, Gonzalez JP, Muyembe-Tamfum JJ, et al. Human ebola outbreak resulting from direct exposure to fruit bats in luebo, democratic republic of congo, 2007. Vector Borne Zoonotic Dis (2009) 9(6):723–8. doi: 10.1089/vbz.2008.0167
23. Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in southeast asia. Nat Commun (2021) 12(1):972. doi: 10.1038/s41467-021-21240-1
24. Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature (2022) 604(7905):330–6. doi: 10.1038/s41586-022-04532-4
25. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL w: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res (1994) 22(22):4673–80. doi: 10.1093/nar/22.22.4673
26. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series. (1999) 41:3.
27. Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol (2006) 23(10):1891–901. doi: 10.1093/molbev/msl051
28. Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evolution. (2021) 38(7):3022–7. doi: 10.1093/molbev/msab120
29. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res (2004) 14(6):1188–90. doi: 10.1101/gr.849004
30. Xie J, Li Y, Shen X, Goh G, Zhu Y, Cui J, et al. Dampened STING-dependent interferon activation in bats. Cell Host Microbe (2018) 23(3):297–301 e4. doi: 10.1016/j.chom.2018.01.006
31. Wiederkehr MA, Qi W, Schoenbaechler K, Fraefel C, Kubacki J. Virus diversity, abundance, and evolution in three different bat colonies in switzerland. Viruses (2022) 14(9):1911. doi: 10.3390/v14091911
32. Prescott J, Guito JC, Spengler JR, Arnold CE, Schuh AJ, Amman BR, et al. Rousette bat dendritic cells overcome marburg virus-mediated antiviral responses by upregulation of interferon-related genes while downregulating proinflammatory disease mediators. mSphere (2019) 4(6):e00728–19. doi: 10.1128/mSphere.00728-19
33. Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J (2016) 10(3):609–20. doi: 10.1038/ismej.2015.138
34. Ahn M, Anderson DE, Zhang Q, Tan CW, Lim BL, Luko K, et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat Microbiol (2019) 4(5):789–99. doi: 10.1038/s41564-019-0371-3
35. Zhang D, Irving AT. Antiviral effects of interferon-stimulated genes in bats. Front Cell Infect Microbiol (2023) 13:1224532. doi: 10.3389/fcimb.2023.1224532
36. Shenoy AR, Wellington DA, Kumar P, Kassa H, Booth CJ, Cresswell P, et al. GBP5 promotes NLRP3 inflammasome assembly and immunity in mammals. Science (2012) 336(6080):481–5. doi: 10.1126/science.1217141
37. Klimke W, O'Donovan C, White O, Brister JR, Clark K, Fedorov B, et al. Solving the problem: Genome annotation standards before the data deluge. Stand Genomic Sci (2011) 5(1):168–93. doi: 10.4056/sigs.2084864
38. Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: A resource for timelines, timetrees, and divergence times. Mol Biol Evol (2017) 34(7):1812–9. doi: 10.1093/molbev/msx116
39. Jimah JR, Hinshaw JE. Structural insights into the mechanism of dynamin superfamily proteins. Trends Cell Biol (2019) 29(3):257–73. doi: 10.1016/j.tcb.2018.11.003
40. Santos JC, Boucher D, Schneider LK, Demarco B, Dilucca M, Shkarina K, et al. Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria. Nat Commun (2020) 11(1):3276. doi: 10.1038/s41467-020-16889-z
41. Scheben A, Mendivil Ramos O, Kramer M, Goodwin S, Oppenheim S, Becker DJ, et al. Long-read sequencing reveals rapid evolution of immunity- and cancer-related genes in bats. Genome Biol Evol (2023) 15(9):evad148. doi: 10.1093/gbe/evad148
42. Agueda-Pinto A, Castro LFC, Esteves PJ. The evolution of S100A7: an unusual gene expansion in myotis bats. BMC Evol Biol (2019) 19(1):102. doi: 10.1186/s12862-019-1433-0
43. Kapusta A, Suh A, Feschotte C. Dynamics of genome size evolution in birds and mammals. Proc Natl Acad Sci USA (2017) 114(8):E1460–E9. doi: 10.1073/pnas.1616702114
44. Smith JD, Gregory TR. The genome sizes of megabats (Chiroptera: Pteropodidae) are remarkably constrained. Biol Lett (2009) 5(3):347–51. doi: 10.1098/rsbl.2009.0016
45. Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. Loss of LINE-1 activity in the megabats. Genetics (2008) 178(1):393–404. doi: 10.1534/genetics.107.080275
46. Deininger PL, Moran JV, Batzer MA, Kazazian HH Jr. Mobile elements and mammalian genome evolution. Curr Opin Genet Dev (2003) 13(6):651–8. doi: 10.1016/j.gde.2003.10.013
Keywords: GBP, innate immunity, evolution, bats, genome, birth and death model of evolution
Citation: Pinheiro A, Borges JR, Côrte-Real JV and Esteves PJ (2024) Evolution of guanylate binding protein genes shows a remarkable variability within bats (Chiroptera). Front. Immunol. 15:1329098. doi: 10.3389/fimmu.2024.1329098
Received: 27 October 2023; Accepted: 16 January 2024;
Published: 31 January 2024.
Edited by:
Daniel Becker, University of Oklahoma, United StatesReviewed by:
Emmanuel Atangana Maze, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), FranceCopyright © 2024 Pinheiro, Borges, Côrte-Real and Esteves. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pedro J. Esteves, cGplc3RldmVzQGNpYmlvLnVwLnB0
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.