
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 22 January 2024
Sec. B Cell Biology
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1320689
During lymphocyte development, a diverse repertoire of lymphocyte antigen receptors is produced to battle against pathogens, which is the basis of adaptive immunity. The diversity of the lymphocyte antigen receptors arises primarily from recombination-activated gene (RAG) protein-mediated V(D)J rearrangement in early lymphocytes. Furthermore, transcription factors (TFs), such as early B cell factor 1 (EBF1), paired box gene 5 (PAX5), and proto-oncogene myelocytomatosis oncogene (MYC), play critical roles in regulating recombination and maintaining normal B cell development. Therefore, the aberrant expression of these TFs may lead to hematologic neoplasms.
In early lymphocytes, RAG1/2-mediated V(D)J rearrangement of antigen receptor variable region genes results in the diversity of mammalian B cell receptors (BCRs) and T cell receptors (TCRs), which is the base of the adaptive immune response. In the process of recombination, RAG1/2 recognizes and scissors recombination signal sequences (RSSs) that flank the V, D, and J gene segments to generate double-strand breaks (DSBs) (1–4). Then, the broken DNA is repaired by non-homologous end joining (NHEJ) (5).
In the bone marrow (BM), B cells are derived from pluripotent stem cells that sequentially differentiate into lymphoid progenitors, progenitor-B (pro-B, including early and late pro-B), precursor-B (pre-B, including large and small pre-B), and immature B cells migrating to the spleen to further differentiate into mature B cells. Additionally, the V(D)J rearrangement of BCR heavy and light chain genes occurs at different stages (6–8). Following light chain gene (VL to JL) recombination at the small pre-B stage, heavy chain gene D to J first rearranges at the early pro-B stage, followed by V to DJ rearrangement at the late pro-B stage (9) (Figure 1).
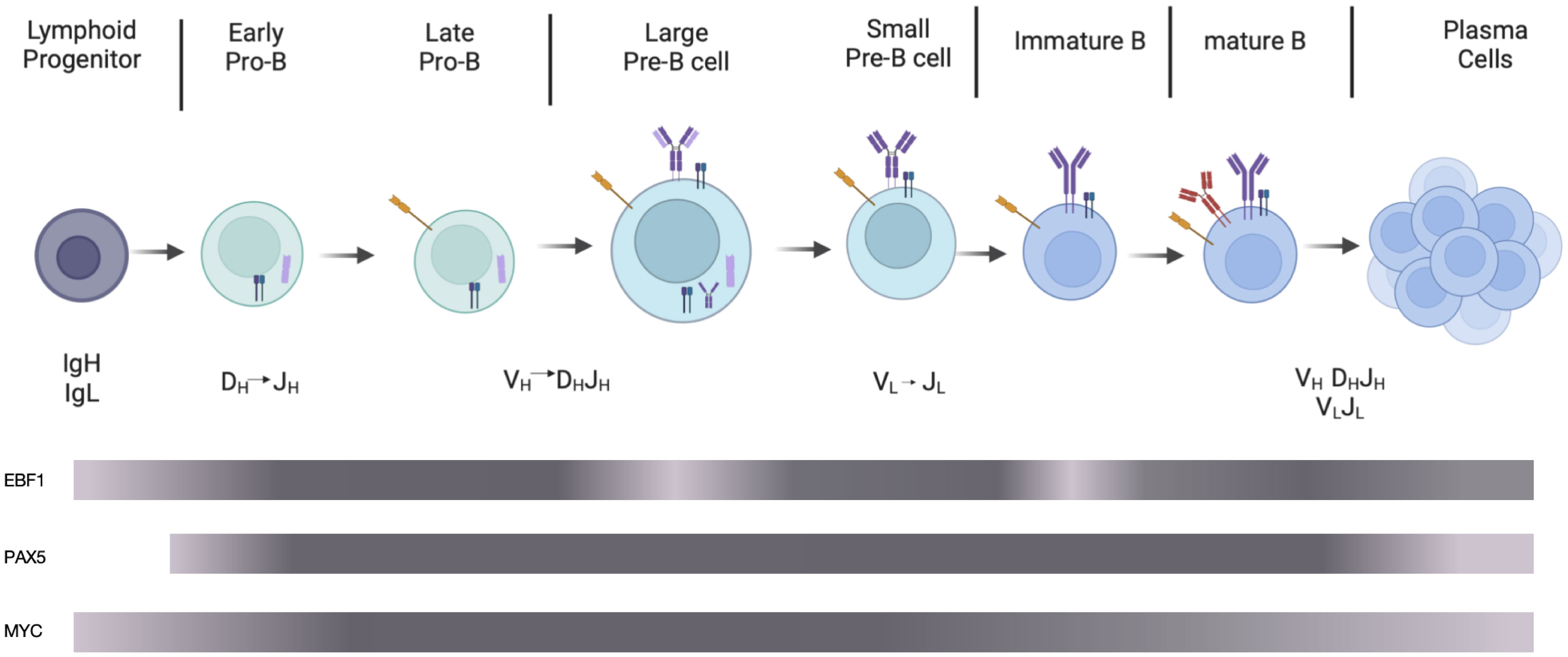
Figure 1 Schematic diagram of B cell development. In the bone marrow, partial lymphoid progenitors differentiate into early pro-B cells performing DH to JH rearrangement, followed by late pro-B cells in which VH to DHJH rearrangement occurs. After large pre-B cells expressing pre-BCR composed of heavy chains, VpreB1, and λ5, small pre-B cells perform light chain rearrangement. Immature B cells, preceding maturation in the periphery, express mature BCRs composed of heavy and light chains. Activated by antigens, mature B cells differentiate into plasma cells, which produce antibodies. The grey bars represent the expression levels of EBF1, PAX5, and MYC. The darker the color is, the higher the protein expression level is. The image was generated using the online platform https://www.biorender.com.
Accumulative evidence has proven that V(D)J rearrangement in B cells is regulated by various TFs, such as PU.1, Ikaros, E2A, EBF1, PAX5, and MYC (10–14). Forming complex networks, these TFs orderly activate B lineage-specific genes (15), ensuring normal lymphocyte development and avoiding B-cell malignancy (4). This review mainly focuses on the effects of EBF1, PAX5, and MYC on B cell development and their association with hematologic neoplasms.
EBF1 consists of four domains, including the conserved N-terminal DNA binding, the Ig/plexins/TF-like (IPT), the helix-loop-helix, and the C-terminal domain (CTD) (16). So far, only IPT and CTD domain functions have been studied in depth. The EBF1 CTD domain was found to bind to inaccessible genomic regions prior to detection of chromatin accessibility and recruit Brahma-related gene 1 to stabilize its binding to these regions (17, 18). Since a prion-like structure in the CTD domain promotes liquid-liquid phase separation via interaction with RNA-binding protein FUS, EBF1 enhances chromatin opening and facilitates the transition from lymphoid progenitor chromatin to B-lineage one (16, 18, 19). Similarly, Boller et al. found that the EBF1 CTD domain induces CpG demethylation to promote chromatin accessibility and, therefore, determines B cell commitment by regulating specific gene expression (17). The EBF1 IPT domain has been reported to bind to transporter protein 3 to facilitate nuclear import of EBF1 (20).
During stepwise development of B cells, the expression of EBF1 protein varies dynamically. EBF1 protein expression level reaches the highest at the heavy and light chain recombination stage, reaches moderate levels during cytokine-mediated proliferation phase (such as pre-BCR independent proliferation phase at large pre-B cell stage) and lowest at the pre-BCR and BCR signal transduction stage (21) (Figure 1). At the common lymphoid precursor (CLP) stage, E2A and FOXO1 activate the EBF1 gene and enhance its expression. In turn, EBF1 promotes FOXO1 protein expression (21–23). FOXO1 enhances RAG protein expression via direct binding to the RAG locus throughout early B cell development (24). At the highly-proliferating early B cell stage, increased EBF1 represses FOXO1 protein expression and its association with the RAG locus, negatively regulating RAG protein expression (25, 26). Barberi et al. also found that TF C/EBPɑ upregulates EBF1 protein expression at the CLP stage (27).
In previous studies, EBF1 has been identified to play critical roles in B cell development (28, 29). It was demonstrated that EBF1 is necessary not only for the survival, proliferation, and signaling of pro-B cells and peripheral B-cell subsets (such as B1 cells and marginal zone B cells) but also for germinal center formation and class switch recombination (30). Ikaros and PU.1 contribute to lymphoid progenitor cell differentiation into pro-B cells, and this process is blocked when these two TFs are deficient (31–34). The overexpression of EBF1 induces Ikaros-KO Lin-Sca1hic-Kithi cells to differentiate into pro-B cells (35). Further studies showed that EBF1 and E2A form a complex that binds to the cis-acting element of the Igh locus and activates DH-JH rearrangement (36, 37). Like E2A knockout mice, EBF1-deficient mice show blocked DH-JH rearrangement and arrested B-cell development (38). Using the diffuse large B cell lymphoma KIS-1 cell line, Bullerwell et al. found that exogenous expression of EBF1 in KIS-1 cells promote the expression of B cell-specific genes, such as CD19 and CD79b, partly because of enabling PAX5 to interact with mixed lineage leukemia H3K4 methyltransferase complexes (39). The results suggested that EBF1 is able to work synergistically with PAX5 to regulate B cell-specific gene expression. When the EBF1 N-terminus fused with the JAK2 C-terminus to form EBF1-JAK2 (E-J), the transcriptional activity of PAX5 was suppressed, which depends on PAX5 interaction with the fusion protein and JAK2-induced phosphorylation. But the detailed mechanism of inhibition remains elusive (40). The fusion protein was only found in 0.6% of adult BCR-ABL1-negative B-acute lymphoblastic leukemia (ALL) (41). Thus, these findings demonstrate that EBF1 plays essential roles in B cell-specific gene expression and B cell differentiation.
PAX5, also known as B cell specific activating protein (BSAP), contains the N-terminal paired domain responsible for binding DNA, C-terminal regulatory module controlling transcription, and middle conserved octapeptide and partial homeodomain (42–44).
PAX5 is restrictedly expressed in the B-cell lineage in the hematopoietic system, particularly high at the pro-B to mature B-cell stage (45–47) (Figure 1). It’s been identified that EBF1 binds directly to the PAX5 promoter to positively regulate its expression, which IL7-activated-STAT5 may improve at the early B cell stages (48–50). In addition, PU.1, interferon regulatory factor 4 (IRF4; functions as a transcriptional repressor), IRF8, and NF-kB are also important regulators of PAX5 (51). For example, activation of pre-BCR-induced-class IA phosphoinositide-3 kinase (PI3K) signaling promotes FOXO1 degradation, which upregulates PAX5 protein expression via repressing IRF4 protein expression (52).
It’s well-known that PAX5 plays essential roles in regulating B cell development and activating B cell-specific signaling proteins, such as CD19 and B-cell linker protein (BLNK), which determines the commitment and differentiation of B cells (52). In PAX5-deficient mice, B cells are severely blocked at the pro-B stage (47). Furthermore, PAX5-deleted mature B cells purified from the lymph nodes of CD19-cre PAX5fl/– Eμ-BCL2 mice dedifferentiated into functional T cells after transplantation into RAG2 KO mice (53).
How does PAX5 play regulatory roles in B cell development? David et al. found that in PAX5-deficient mice, the rearrangement of distal rather than proximal genes of Igh V is significantly impaired in pro-B cells (54). When PAX5 is reintroduced in PAX5 KO pro-B cells, large-scale contraction of Igh loci and rearrangement of distal VH-DHJH are induced, suggesting that PAX5 is essential for distal VH gene rearrangement in pro-B cells (55). Further studies revealed that due to chromatin activation and antisense transcription induction at the PAX5-activated intergenic repeat (PAIR) elements in the distal VH gene cluster, PAX5 promotes VH to DHJH recombination (56).
Histone modification may regulate the rearrangement of Igh V genes. For example, trimethylation of Lys-27 on histone H3 (H3K27me3) and dimethylation of Lys-36 on histone H3 (H3K36me2) is associated with repressive and active genes, respectively (57). Xu et al. also used CHIP and CHIP-on-CHIP techniques to find that H3K36me2 is enriched on distal VH genes, while H3K27me3 is exclusively on proximal VH genes. Via cooperation with IL-7 that promotes H3K36me2, PAX5 induces H3K27me3 to balance the proximal and distal VH gene recombination (58).
Recent studies also showed that cohesion complex–mediated chromatin loop extrusion results in Igh locus contraction that is helpful for RAG scanning (59–62). Recruiting polycomb-repressive complex 2 (PRC2), PAX5 downregulates the expression of Wings apart-like protein homolog (WAPL) that releases cohesion from chromatin and inhibits Igh loop extraction, enhancing VH gene rearrangement (59, 63, 64).
Moreover, Zhang et al. found that not only does PAX5 bind to the coding regions of VH gene segments, but it also interacts with the RAG1/2 complex to promote VH-to-DHJH recombination (65, 66).
Taken together, these studies demonstrate that Pax5 is crucial to VH (especial distal VH) to DHJH recombination and B cell commitment.
MYC, as a proto-oncogene, belongs to the basic region/helix-loop-helix/leucine zipper (bHLHZip) transcription factor family, containing the N-terminal transactivating domain (TAD) and the C-terminal DNA-binding domain. The TAD domain is an intrinsically disordered region responsible for the transcriptional activation of MYC. Dependent on the C-terminal domain with bHLHZip fragment, MYC forms a heterodimer with Max, enabling it to bind to DNA (67, 68). In addition to acting as a global gene expression amplifier, MYC regulates cell proliferation, differentiation, apoptosis, and homeostasis (69–72). Notably, MYC mutations or overexpression occur in many tumors (73–75).
In an early study, pre-B cells were found to express MYC in response to IL-7 stimulation, and splenic B cells expressed it due to lipopolysaccharide (LPS) stimulation (76). Moreover, MYC protein is continuously expressed from hematopoietic stem cells to lymphocytes, reaching highest in proliferating B cells, especially at the pro- to pre-B cell stage, and its expression decreases at the mature B cell stage (77, 78) (Figure 1).
Accumulated evidence showed that the coordinated action of factors regulates MYC expression. MYC is one of the EBF1-directly-targeted genes, and EBF1 promotes its expression in pro-B cells; in lack of EBF1 in pro-B cells, over-expression of PAX5 results in decreased MYC expression, which suggests EBF1 and PAX5 may play opposite roles in regulating MYC expression (79). However, Ramamoorthy et al. identified the EBF1+/−PAX5+/− mouse model for B-ALL and found that MYC is upregulated (80). The results imply that EBF1 and PAX5 may cooperate to limit MYC expression, partially avoiding transformation to B-ALL. Besides, Beer et al. found that EBF1 interacts with Epstein-Barr virus (EBV) nuclear antigen 2 (EBNA2) to stimulate MYC expression in human adenoid primary B cells (81). In pre-B cells, pre-BCR signaling-induced Ikaros and Aiolos are reported to negatively regulate MYC expression via binding its promoter (82, 83).
It was reported that deletion of MYC affects B cell development (84, 85). N-mycfl/fl c-mycfl/fl CD19cre+ mice exhibit blockage at the pro- to pre-B cell stage; MYC can elevate Ca2+ level in early B cells and activate genes targeted by MYC and Ca2+, thereby promoting the proliferation and differentiation of early B cells (86). Additionally, MYC has been reported to regulate cell cycle via activating cyclin-dependent protein kinase 1 (CDK1) to phosphorylate p27 (87), inducing cyclin D1 and D2, and sequestrating p27 and p21 (88). When MYC is suppressed in pre-B cells, the cell proliferation is inhibited due to induction of p27 and downregulation of cyclin D3 (83).
Additionally, MYC plays essential roles in further developing B cells in the germinal center (GC) where immune response occurs (89). Studies showed that in the GC response, B-cell lymphoma 6 (BCL6) is a direct inhibitor of MYC; most GC cells express either BCL6 or MYC (90). In the dark region of the GC, antigenic stimulation activates naïve B cells to express MYC and repress BCL6 transcription, leading to clonal expansion (91). After entering the light zone of GC, about 10% of B cells simultaneously express MYC and BCL6 to promote the cells to enter cell cycles (77, 91, 92).
In general, these studies suggest MYC is able to regulate not only the proliferation and differentiation of early B-cells but also B cell development in GC.
Since EBF1, PAX5, and MYC are essential for B cell development, their ectopic expression or mutants may lead to abnormal B cell development and hematologic neoplasms, such as B-lymphoid leukemia and lymphoma (93–100).
Accumulative evidence showed that EBF1 deletion results in the pathogenesis, drug resistance, and relapse of B-progenitor ALL (101, 102). In addition, EBF1 fused with platelet-derived growth factor receptor beta (PDGFRB) has been reported in Ph-like ALL. The fused gene contributes to EBF1 function deficiency and differentiation block (103–105). Moreover, a recent study showed that EBF1 is necessary for propagating mantle cell lymphoma, a lethal mature B cell lymphoma (106).
Depending on advanced assays such as whole genome sequencing, whole transcriptome sequencing, and single nucleotide polymorphism array, PAX5 alterations were identified in human B-ALL. These alterations mainly include rearrangements with partner genes (such as structural proteins, TFs, and signal transducers), mutation (such as R38H, P80R, and G183S), deletions (usually affecting one allele), and intragenic amplification (such as amplification of exons 2 to 5) (93, 102, 107–114). In addition to B-ALL, PAX5 mutations and rearrangements (such as PAX5 t (9, 14) (p13;q32)) were identified in Burkitt’s lymphoma, follicular lymphoma as well as diffuse large B cell lymphoma (DLBCL) (115–122).
Unlike other proto-oncogenes possibly activated by genetic mutations, aberrant MYC expression in transformed cells is caused by chromosomal translocation, gene amplification, and aberrant regulation of its expression (77). In B lymphoblastic leukemia with t (9, 22) BCR-ABL1, t (v;11) MLL, and t (12, 21) ETV6/RUNX1 rearrangement, triggered MYC overexpression was more common, while the frequency of MYC gene translocation and mutation was low (123–131). In contrast, Ig/MYC rearrangements induce MYC overexpression in B cell lymphomas, such as Burkitt’s lymphoma and plasmablastic lymphomas (132–134). In double-hit lymphoma, besides BCL6 rearrangement, MYC translocation was also observed (135, 136).
In short, as pivotal TFs in B cell development, the abnormalities of EBF1, PAX5, and MYC result in hematologic tumors.
The development of B cells is a complex process that is comprehensively regulated by multiple factors, such as TF EBF1, PAX5, and MYC. EBF1 is involved in the whole process of B cell development. At the early pro-B cell stage, EBF1 promotes the rearrangement of DH to JH. When DHJH recombination succeeds, EBF1 targets PAX5 to activate its expression and subsequently blocks the activation of non-B-lineage genes. PAX5 is critical to VH-DHJH recombination, especially distal VH gene rearrangement. Besides, EBF1 cooperates with PAX5 to regulate MYC expression, which may balance the proliferation and differentiation of early B cells. Therefore, EBF1, PAX5, and MYC lesions may drive B-cell malignancy and induce B-cell lymphomas.
In recent years, advances have been made in the regulatory mechanisms of EBF1, PAX5, and MYC in B cell development and maturation because of the application of DNA loop extrusion and phase separation, which still need in-depth research. Employing advanced test methods such as single-cell sequencing, more regulators and/or targeted genes of these TFs may be identified, which is helpful to optimize the regulatory mechanism of B cell development and maturation. Given that the aberrant expression and function of EBF1, PAX5, and MYC contribute to hematologic neoplasms, therapies targeting them may shed light on these diseases.
LL: Writing – original draft. DZ: Writing – review & editing. XC: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (81801551) and Central Government Funds of Guiding Local Scientific and Technological Development for Sichuan Province (2021ZYD0076).
The authors thank AiMi Academic Services (www.aimieditor.com) for English language editing and review services.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Krangel MS. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat Immunol (2003) 4(7):624–30. doi: 10.1038/ni0703-624
2. Ru H, Zhang P, Wu H. Structural gymnastics of rag-mediated DNA cleavage in V(D)J recombination. Curr Opin Struct Biol (2018) 53:178–86. doi: 10.1016/j.sbi.2018.11.001
3. Shetty K, Schatz DG. Recruitment of Rag1 and Rag2 to chromatinized DNA during V(D)J recombination. Mol Cell Biol (2015) 35(21):3701–13. doi: 10.1128/MCB.00219-15
4. Chi X, Li Y, Qiu X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: mechanism and regulation. Immunology (2020) 160(3):233–47. doi: 10.1111/imm.13176
5. Wang XS, Lee BJ, Zha S. The recent advances in non-homologous end-joining through the lens of lymphocyte development. DNA Repair (Amst) (2020) 94:102874. doi: 10.1016/j.dnarep.2020.102874
6. McLean KC, Mandal M. It takes three receptors to raise a B cell. Trends Immunol (2020) 41(7):629–42. doi: 10.1016/j.it.2020.05.003
7. Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med (1991) 173(5):1213–25. doi: 10.1084/jem.173.5.1213
8. Hughes AM, Kuek V, Kotecha RS, Cheung LC. The bone marrow microenvironment in B-cell development and Malignancy. Cancers (Basel) (2022) 14(9):2089. doi: 10.3390/cancers14092089
9. Wang Y, Liu J, Burrows PD, Wang JY. B cell development and maturation. Adv Exp Med Biol (2020) 1254:1–22. doi: 10.1007/978-981-15-3532-1_1
10. Revilla IDR, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J (2012) 31(14):3130–46. doi: 10.1038/emboj.2012.155
11. Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. A global network of transcription factors, involving E2a, Ebf1 and Foxo1, that orchestrates B cell fate. Nat Immunol (2010) 11(7):635–43. doi: 10.1038/ni.1891
12. Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol (2006) 18(2):127–34. doi: 10.1016/j.coi.2006.01.007
13. Kleiman E, Loguercio S, Feeney AJ. Epigenetic enhancer marks and transcription factor binding influence vkappa gene rearrangement in pre-B cells and pro-B cells. Front Immunol (2018) 9:2074. doi: 10.3389/fimmu.2018.02074
14. Rogers CH, Mielczarek O, Corcoran AE. Dynamic 3d locus organization and its drivers underpin immunoglobulin recombination. Front Immunol (2020) 11:633705. doi: 10.3389/fimmu.2020.633705
15. Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr Opin Immunol (2010) 22(2):161–7. doi: 10.1016/j.coi.2010.01.010
16. Kee BL. It's a phase that Ebf1 is going through. Immunity (2020) 53(6):1123–5. doi: 10.1016/j.immuni.2020.11.013
17. Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, et al. Pioneering activity of the C-terminal domain of ebf1 shapes the chromatin landscape for B cell programming. Immunity (2016) 44(3):527–41. doi: 10.1016/j.immuni.2016.02.021
18. Wang Y, Zolotarev N, Yang CY, Rambold A, Mittler G, Grosschedl R. A prion-like domain in transcription factor Ebf1 promotes phase separation and enables B cell programming of progenitor chromatin. Immunity (2020) 53(6):1151–67.e6. doi: 10.1016/j.immuni.2020.10.009
19. Schmiedel D, Hezroni H, Hamburg A, Shulman Z. Brg1 supports B cell proliferation and germinal center formation through enhancer activation. Front Immunol (2021) 12:705848. doi: 10.3389/fimmu.2021.705848
20. Bayer M, Boller S, Ramamoothy S, Zolotarev N, Cauchy P, Iwanami N, et al. Tnpo3 enables Ebf1 function in conditions of antagonistic notch signaling. Genes Dev (2022) 36(15-16):901–15. doi: 10.1101/gad.349696.122
21. Lee RD, Munro SA, Knutson TP, LaRue RS, Heltemes-Harris LM, Farrar MA. Single-cell analysis identifies dynamic gene expression networks that govern B cell development and transformation. Nat Commun (2021) 12(1):6843. doi: 10.1038/s41467-021-27232-5
22. Tsapogas P, Zandi S, Ahsberg J, Zetterblad J, Welinder E, Jonsson JI, et al. Il-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors. Blood (2011) 118(5):1283–90. doi: 10.1182/blood-2011-01-332189
23. Mansson R, Welinder E, Ahsberg J, Lin YC, Benner C, Glass CK, et al. Positive intergenic feedback circuitry, involving Ebf1 and Foxo1, orchestrates B-cell fate. Proc Natl Acad Sci U.S.A. (2012) 109(51):21028–33. doi: 10.1073/pnas.1211427109
24. Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol (2008) 9(6):613–22. doi: 10.1038/ni.1612
25. Timblin GA, Schlissel MS. Ebf1 and C-myb repress rag transcription downstream of stat5 during early B cell development. J Immunol (2013) 191(9):4676–87. doi: 10.4049/jimmunol.1301675
26. Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. Ebf1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol (2008) 181(5):3364–72. doi: 10.4049/jimmunol.181.5.3364
27. Barberi T, Cui C, Friedman AD. C/ebpalpha induces Ebf1 gene expression in common lymphoid progenitors. PloS One (2020) 15(12):e0244161. doi: 10.1371/journal.pone.0244161
28. Boller S, Li R, Grosschedl R. Defining B cell chromatin: lessons from Ebf1. Trends Genet (2018) 34(4):257–69. doi: 10.1016/j.tig.2017.12.014
29. Mercer EM, Lin YC, Benner C, Jhunjhunwala S, Dutkowski J, Flores M, et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity (2011) 35(3):413–25. doi: 10.1016/j.immuni.2011.06.013
30. Gyory I, Boller S, Nechanitzky R, Mandel E, Pott S, Liu E, et al. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev (2012) 26(7):668–82. doi: 10.1101/gad.187328.112
31. Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, et al. The ikaros gene is required for the development of all lymphoid lineages. Cell (1994) 79(1):143–56. doi: 10.1016/0092-8674(94)90407-3
32. Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol (2006) 7(4):382–91. doi: 10.1038/ni1314
33. Hayashi R, Takemori T, Kodama M, Suzuki M, Tsuboi A, Nagawa F, et al. The pu.1 binding site is a cis-element that regulates pro-B/pre-B specificity of Vkappa-jkappa joining. J Immunol (1997) 159(9):4145–9.
34. Le Coz C, Nguyen DN, Su C, Nolan BE, Albrecht AV, Xhani S, et al. Constrained chromatin accessibility in Pu.1-mutated agammaglobulinemia patients. J Exp Med (2021) 218(7):e20201750. doi: 10.1084/jem.20201750
35. Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by ikaros. Nat Immunol (2008) 9(8):927–36. doi: 10.1038/ni.1626
36. Goebel P, Janney N, Valenzuela JR, Romanow WJ, Murre C, Feeney AJ. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2a and early B cell factor in nonlymphoid cells. J Exp Med (2001) 194(5):645–56. doi: 10.1084/jem.194.5.645
37. Hesslein DG, Schatz DG. Factors and forces controlling V(D)J recombination. Adv Immunol (2001) 78:169–232. doi: 10.1016/s0065-2776(01)78004-2
38. Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor Ebf. Nature (1995) 376(6537):263–7. doi: 10.1038/376263a0
39. Bullerwell CE, Robichaud PP, Deprez PML, Joy AP, Wajnberg G, D'Souza D, et al. Ebf1 drives hallmark B cell gene expression by enabling the interaction of pax5 with the mll H3k4 methyltransferase complex. Sci Rep (2021) 11(1):1537. doi: 10.1038/s41598-021-81000-5
40. Kojima Y, Kawashima F, Yasuda T, Odaira K, Inagaki Y, Yamada C, et al. Ebf1-jak2 inhibits the pax5 function through physical interaction with pax5 and kinase activity. Int J Hematol (2023) 118(1):65–74. doi: 10.1007/s12185-023-03585-z
41. Yasuda T, Sanada M, Kawazu M, Kojima S, Tsuzuki S, Ueno H, et al. Two novel high-risk adult B-cell acute lymphoblastic leukemia subtypes with high expression of Cdx2 and Idh1/2 mutations. Blood (2022) 139(12):1850–62. doi: 10.1182/blood.2021011921
42. Eberhard D, Busslinger M. The partial homeodomain of the transcription factor Pax-5 (Bsap) is an interaction motif for the retinoblastoma and tata-binding proteins. Cancer Res (1999) 59(7 Suppl):1716s–24s; discussion 24s-25s.
43. Perez-Borrajero C, Okon M, McIntosh LP. Structural and dynamics studies of pax5 reveal asymmetry in stability and DNA binding by the paired domain. J Mol Biol (2016) 428(11):2372–91. doi: 10.1016/j.jmb.2016.04.004
44. Perez-Borrajero C, Heinkel F, Gsponer J, McIntosh LP. Conformational plasticity and DNA-binding specificity of the eukaryotic transcription factor Pax5. Biochemistry (2021) 60(2):104–17. doi: 10.1021/acs.biochem.0c00737
45. Shahjahani M, Norozi F, Ahmadzadeh A, Shahrabi S, Tavakoli F, Asnafi AA, et al. The role of Pax5 in leukemia: diagnosis and prognosis significance. Med Oncol (2015) 32(1):360. doi: 10.1007/s12032-014-0360-6
46. Dong Y, Bai H, Dong F, Zhang XB, Ema H. Gene knockout in highly purified mouse hematopoietic stem cells by Crispr/Cas9 technology. J Immunol Methods (2021) 495:113070. doi: 10.1016/j.jim.2021.113070
47. Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol (2007) 8(5):463–70. doi: 10.1038/ni1454
48. Hirokawa S, Sato H, Kato I, Kudo A. Ebf-regulating pax5 transcription is enhanced by Stat5 in the early stage of B cells. Eur J Immunol (2003) 33(7):1824–9. doi: 10.1002/eji.200323974
49. Goetz CA, Harmon IR, O'Neil JJ, Burchill MA, Johanns TM, Farrar MA. Restricted stat5 activation dictates appropriate thymic B versus T cell lineage commitment. J Immunol (2005) 174(12):7753–63. doi: 10.4049/jimmunol.174.12.7753
50. Kikuchi K, Lai AY, Hsu CL, Kondo M. Il-7 Receptor Signaling Is Necessary for Stage Transition in Adult B Cell Development through up-Regulation of Ebf. J Exp Med (2005) 201(8):1197–203. doi: 10.1084/jem.20050158
51. Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity (2009) 30(4):508–20. doi: 10.1016/j.immuni.2009.01.012
52. Abdelrasoul H, Werner M, Setz CS, Okkenhaug K, Jumaa H. Pi3k induces B-cell development and regulates B cell identity. Sci Rep (2018) 8(1):1327. doi: 10.1038/s41598-018-19460-5
53. Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature (2007) 449(7161):473–7. doi: 10.1038/nature06159
54. Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5' Igh V gene segments. Genes Dev (2003) 17(1):37–42. doi: 10.1101/gad.1031403
55. Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-dj rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev (2004) 18(4):411–22. doi: 10.1101/gad.291504
56. Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, et al. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity (2011) 34(2):175–87. doi: 10.1016/j.immuni.2011.02.005
57. Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol (2005) 6(11):838–49. doi: 10.1038/nrm1761
58. Xu CR, Schaffer L, Head SR, Feeney AJ. Reciprocal patterns of methylation of H3k36 and H3k27 on proximal vs. Distal igvh genes are modulated by Il-7 and Pax5. Proc Natl Acad Sci U.S.A. (2008) 105(25):8685–90. doi: 10.1073/pnas.0711758105
59. Dai HQ, Hu H, Lou J, Ye AY, Ba Z, Zhang X, et al. Loop extrusion mediates physiological igh locus contraction for rag scanning. Nature (2021) 590(7845):338–43. doi: 10.1038/s41586-020-03121-7
60. Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, et al. Cohesin loss eliminates all loop domains. Cell (2017) 171(2):305–20 e24. doi: 10.1016/j.cell.2017.09.026
61. Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature (2017) 551(7678):51–6. doi: 10.1038/nature24281
62. Wutz G, Varnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by Ctcf, Wapl, and Pds5 proteins. EMBO J (2017) 36(24):3573–99. doi: 10.15252/embj.201798004
63. Hill L, Ebert A, Jaritz M, Wutz G, Nagasaka K, Tagoh H, et al. Wapl repression by pax5 promotes V gene recombination by Igh loop extrusion. Nature (2020) 584(7819):142–7. doi: 10.1038/s41586-020-2454-y
64. Haarhuis JHI, van der Weide RH, Blomen VA, Yanez-Cuna JO, Amendola M, van Ruiten MS, et al. The cohesin release factor Wapl restricts chromatin loop extension. Cell (2017) 169(4):693–707.e14. doi: 10.1016/j.cell.2017.04.013
65. Zhang Z, Espinoza CR, Yu Z, Stephan R, He T, Williams GS, et al. Transcription factor Pax5 (Bsap) transactivates the rag-mediated V(H)-to-dj(H) rearrangement of immunoglobulin genes. Nat Immunol (2006) 7(6):616–24. doi: 10.1038/ni1339
66. Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science (2002) 296(5565):158–62. doi: 10.1126/science.1068768
67. Dai Y, Zhang J, Wang Y, Liu L, Gao J. Computational insights into the differentiated binding affinities of Myc, Max, and Omomyc Dimers to the E-boxes of DNA. J Mol Model (2022) 28(10):329. doi: 10.1007/s00894-022-05261-1
68. Sammak S, Hamdani N, Gorrec F, Allen MD, Freund SMV, Bycroft M, et al. Crystal structures and nuclear magnetic resonance studies of the apo form of the C-myc:Max bhlhzip complex reveal a helical basic region in the absence of DNA. Biochemistry (2019) 58(29):3144–54. doi: 10.1021/acs.biochem.9b00296
69. Carroll PA, Freie BW, Mathsyaraja H, Eisenman RN. The myc transcription factor network: balancing metabolism, proliferation and oncogenesis. Front Med (2018) 12(4):412–25. doi: 10.1007/s11684-018-0650-z
70. Tesi A, de Pretis S, Furlan M, Filipuzzi M, Morelli MJ, Andronache A, et al. An early myc-dependent transcriptional program orchestrates cell growth during B-cell activation. EMBO Rep (2019) 20(9):e47987. doi: 10.15252/embr.201947987
71. Eertink JJ, Zwezerijnen GJC, Wiegers SE, Pieplenbosch S, Chamuleau MED, Lugtenburg PJ, et al. Baseline radiomics features and myc rearrangement status predict progression in aggressive B-cell lymphoma. Blood Adv (2023) 7(2):214–23. doi: 10.1182/bloodadvances.2022008629
72. Hartl M, Bister K. Myc analysis in cancer and evolution. Methods Mol Biol (2021) 2318:87–117. doi: 10.1007/978-1-0716-1476-1_6
73. Lutz W, Leon J, Eilers M. Contributions of myc to tumorigenesis. Biochim Biophys Acta (2002) 1602(1):61–71. doi: 10.1016/s0304-419x(02)00036-7
74. Gurel B, Iwata T, Koh CM, Jenkins RB, Lan F, Van Dang C, et al. Nuclear myc protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol (2008) 21(9):1156–67. doi: 10.1038/modpathol.2008.111
75. Shroff EH, Eberlin LS, Dang VM, Gouw AM, Gabay M, Adam SJ, et al. Myc oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc Natl Acad Sci U.S.A. (2015) 112(21):6539–44. doi: 10.1073/pnas.1507228112
76. Morrow MA, Lee G, Gillis S, Yancopoulos GD, Alt FW. Interleukin-7 induces N-myc and C-myc expression in normal precursor B lymphocytes. Genes Dev (1992) 6(1):61–70. doi: 10.1101/gad.6.1.61
77. de Barrios O, Meler A, Parra M. Myc's fine line between B cell development and Malignancy. Cells (2020) 9(2):523. doi: 10.3390/cells9020523
78. Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of C-myc proto-oncogene expression during lymphocyte development revealed by a Gfp-C-myc knock-in mouse. Eur J Immunol (2008) 38(2):342–9. doi: 10.1002/eji.200737972
79. Somasundaram R, Jensen CT, Tingvall-Gustafsson J, Ahsberg J, Okuyama K, Prasad M, et al. Ebf1 and Pax5 control pro-B cell expansion via opposing regulation of the myc gene. Blood (2021) 137(22):3037–49. doi: 10.1182/blood.2020009564
80. Ramamoorthy S, Kometani K, Herman JS, Bayer M, Boller S, Edwards-Hicks J, et al. Ebf1 and Pax5 safeguard leukemic transformation by limiting Il-7 signaling, Myc expression, and folate metabolism. Genes Dev (2020) 34(21-22):1503–19. doi: 10.1101/gad.340216.120
81. Beer S, Wange LE, Zhang X, Kuklik-Roos C, Enard W, Hammerschmidt W, et al. Ebna2-Ebf1 complexes promote myc expression and metabolic processes driving S-phase progression of epstein-barr virus-infected B cells. Proc Natl Acad Sci U.S.A. (2022) 119(30):e2200512119. doi: 10.1073/pnas.2200512119
82. Ferreiros-Vidal I, Carroll T, Zhang T, Lagani V, Ramirez RN, Ing-Simmons E, et al. Feedforward regulation of myc coordinates lineage-specific with housekeeping gene expression during B cell progenitor cell differentiation. PloS Biol (2019) 17(4):e2006506. doi: 10.1371/journal.pbio.2006506
83. Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and aiolos inhibit pre-B-cell proliferation by directly suppressing C-myc expression. Mol Cell Biol (2010) 30(17):4149–58. doi: 10.1128/MCB.00224-10
84. Malynn BA, Demengeot J, Stewart V, Charron J, Alt FW. Generation of normal lymphocytes derived from N-myc-deficient embryonic stem cells. Int Immunol (1995) 7(10):1637–47. doi: 10.1093/intimm/7.10.1637
85. Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, et al. Hematopoietic stem cell function and survival depend on C-myc and N-myc activity. Cell Stem Cell (2008) 3(6):611–24. doi: 10.1016/j.stem.2008.09.005
86. Habib T, Park H, Tsang M, de Alboran IM, Nicks A, Wilson L, et al. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J Cell Biol (2007) 179(4):717–31. doi: 10.1083/jcb.200704173
87. Garcia-Gutierrez L, Bretones G, Molina E, Arechaga I, Symonds C, Acosta JC, et al. Myc stimulates cell cycle progression through the activation of cdk1 and phosphorylation of P27. Sci Rep (2019) 9(1):18693. doi: 10.1038/s41598-019-54917-1
88. Perez-Roger I, Kim SH, Griffiths B, Sewing A, Land H. Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of P27(Kip1) and P21(Cip1). EMBO J (1999) 18(19):5310–20. doi: 10.1093/emboj/18.19.5310
89. Elsner RA, Shlomchik MJ. Germinal center and extrafollicular B cell responses in vaccination, immunity, and autoimmunity. Immunity (2020) 53(6):1136–50. doi: 10.1016/j.immuni.2020.11.006
90. Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, et al. The proto-oncogene myc is required for selection in the germinal center and cyclic reentry. Nat Immunol (2012) 13(11):1083–91. doi: 10.1038/ni.2428
91. Luo W, Weisel F, Shlomchik MJ. B cell receptor and cd40 signaling are rewired for synergistic induction of the C-myc transcription factor in germinal center B cells. Immunity (2018) 48(2):313–26.e5. doi: 10.1016/j.immuni.2018.01.008
92. Ersching J, Efeyan A, Mesin L, Jacobsen JT, Pasqual G, Grabiner BC, et al. Germinal center selection and affinity maturation require dynamic regulation of mtorc1 kinase. Immunity (2017) 46(6):1045–58 e6. doi: 10.1016/j.immuni.2017.06.005
93. Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. Pax5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet (2019) 51(2):296–307. doi: 10.1038/s41588-018-0315-5
94. Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: A master regulator of B cell development and leukemogenesis. Adv Immunol (2011) 111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2
95. Pang SH, Carotta S, Nutt SL. Transcriptional control of pre-B cell development and leukemia prevention. Curr Top Microbiol Immunol (2014) 381:189–213. doi: 10.1007/82_2014_377
96. Somasundaram R, Prasad MA, Ungerback J, Sigvardsson M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood (2015) 126(2):144–52. doi: 10.1182/blood-2014-12-575688
97. Okuyama K, Strid T, Kuruvilla J, Somasundaram R, Cristobal S, Smith E, et al. Pax5 is part of a functional transcription factor network targeted in lymphoid leukemia. PloS Genet (2019) 15(8):e1008280. doi: 10.1371/journal.pgen.1008280
98. Prasad MA, Ungerback J, Ahsberg J, Somasundaram R, Strid T, Larsson M, et al. Ebf1 heterozygosity results in increased DNA damage in pro-B cells and their synergistic transformation by Pax5 haploinsufficiency. Blood (2015) 125(26):4052–9. doi: 10.1182/blood-2014-12-617282
99. Rosenthal A, Younes A. High grade B-cell lymphoma with rearrangements of myc and Bcl2 and/or Bcl6: double hit and triple hit lymphomas and double expressing lymphoma. Blood Rev (2017) 31(2):37–42. doi: 10.1016/j.blre.2016.09.004
100. Ganapathi KA, Brown LE, Prakash S, Bhargava P. New developments in non-hodgkin lymphoid Malignancies. Pathology (2021) 53(3):349–66. doi: 10.1016/j.pathol.2021.01.002
101. Liao D. Emerging roles of the ebf family of transcription factors in tumor suppression. Mol Cancer Res (2009) 7(12):1893–901. doi: 10.1158/1541-7786.MCR-09-0229
102. Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature (2007) 446(7137):758–64. doi: 10.1038/nature05690
103. Welsh SJ, Churchman ML, Togni M, Mullighan CG, Hagman J. Deregulation of kinase signaling and lymphoid development in Ebf1-Pdgfrb all leukemogenesis. Leukemia (2018) 32(1):38–48. doi: 10.1038/leu.2017.166
104. Chen YQ, Zheng YZ, Li J, Hua XL, Le SH. Acute lymphoblastic leukemia with Ebf1-pdgfrb fusion gene: two cases report and literature review. Zhonghua Xue Ye Xue Za Zhi (2023) 44(1):69–71. doi: 10.3760/cma.j.issn.0253-2727.2023.01.013
105. Ishihara T, Watakabe M, Ochi S, Akisada N, Nogami K. Early diagnosis of Ebf1-pdgfrb-positive acute lymphoblastic leukemia. Pediatr Int (2022) 64(1):e14955. doi: 10.1111/ped.14955
106. Jang JY, Hwang I, Pan H, Yao J, Alinari L, Imada E, et al. A foxo1-dependent transcription network is a targetable vulnerability of mantle cell lymphomas. J Clin Invest (2022) 132(24):e160767. doi: 10.1172/JCI160767
107. Familiades J, Bousquet M, Lafage-Pochitaloff M, Bene MC, Beldjord K, De Vos J, et al. Pax5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and Pax5 haploinsufficiency is associated with Bcr-abl1 and Tcf3-pbx1 fusion genes: A graall study. Leukemia (2009) 23(11):1989–98. doi: 10.1038/leu.2009.135
108. Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, et al. Incidence and diversity of Pax5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia (2009) 23(1):134–43. doi: 10.1038/leu.2008.306
109. Coyaud E, Struski S, Prade N, Familiades J, Eichner R, Quelen C, et al. Wide diversity of Pax5 alterations in B-all: A groupe francophone de cytogenetique hematologique study. Blood (2010) 115(15):3089–97. doi: 10.1182/blood-2009-07-234229
110. Iacobucci I, Lonetti A, Paoloni F, Papayannidis C, Ferrari A, Storlazzi CT, et al. The pax5 gene is frequently rearranged in bcr-abl1-positive acute lymphoblastic leukemia but is not associated with outcome. A report on behalf of the gimema acute leukemia working party. Haematologica (2010) 95(10):1683–90. doi: 10.3324/haematol.2009.020792
111. Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, et al. A recurrent germline Pax5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet (2013) 45(10):1226–31. doi: 10.1038/ng.2754
112. Ofverholm I, Tran AN, Heyman M, Zachariadis V, Nordenskjold M, Nordgren A, et al. Impact of Ikzf1 deletions and pax5 amplifications in pediatric B-cell precursor all treated according to nopho protocols. Leukemia (2013) 27(9):1936–9. doi: 10.1038/leu.2013.92
113. Auer F, Ruschendorf F, Gombert M, Husemann P, Ginzel S, Izraeli S, et al. Inherited susceptibility to pre B-all caused by germline transmission of Pax5 C.547g>A. Leukemia (2014) 28(5):1136–8. doi: 10.1038/leu.2013.363
114. Jia Z, Gu Z. Pax5 alterations in B-cell acute lymphoblastic leukemia. Front Oncol (2022) 12:1023606. doi: 10.3389/fonc.2022.1023606
115. Busslinger M, Klix N, Pfeffer P, Graninger PG, Kozmik Z. Deregulation of pax-5 by translocation of the emu enhancer of the igh locus adjacent to two alternative pax-5 promoters in a diffuse large-cell lymphoma. Proc Natl Acad Sci U.S.A. (1996) 93(12):6129–34. doi: 10.1073/pnas.93.12.6129
116. Poppe B, De Paepe P, Michaux L, Dastugue N, Bastard C, Herens C, et al. Pax5/igh rearrangement is a recurrent finding in a subset of aggressive B-nhl with complex chromosomal rearrangements. Genes Chromosomes Cancer (2005) 44(2):218–23. doi: 10.1002/gcc.20214
117. Krenacs L, Himmelmann AW, Quintanilla-Martinez L, Fest T, Riva A, Wellmann A, et al. Transcription factor B-cell-specific activator protein (Bsap) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood (1998) 92(4):1308–16. doi: 10.1182/blood.V92.4.1308
118. Arthur SE, Jiang A, Grande BM, Alcaide M, Cojocaru R, Rushton CK, et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat Commun (2018) 9(1):4001. doi: 10.1038/s41467-018-06354-3
119. Grande BM, Gerhard DS, Jiang A, Griner NB, Abramson JS, Alexander TB, et al. Genome-wide discovery of somatic coding and noncoding mutations in pediatric endemic and sporadic burkitt lymphoma. Blood (2019) 133(12):1313–24. doi: 10.1182/blood-2018-09-871418
120. Iida S, Rao PH, Nallasivam P, Hibshoosh H, Butler M, Louie DC, et al. The T(9;14)(P13;Q32) chromosomal translocation associated with lymphoplasmacytoid lymphoma involves the pax-5 gene. Blood (1996) 88(11):4110–7. doi: 10.1182/blood.V88.11.4110.4110
121. Ohno H, Takeoka K, Kishimori C, Nakagawa M, Fukutsuka K, Maekawa F, et al. T(9;14)(P13;Q32)/pax5-igh translocation as a secondary cytogenetic abnormality in diffuse large B-cell lymphoma. J Clin Exp Hematop (2021) 61(4):216–20. doi: 10.3960/jslrt.21025
122. Ohno H, Nakagawa M, Kishimori C, Fukutsuka K, Maekawa F, Takeoka K, et al. Diffuse large B-cell lymphoma carrying T(9;14)(P13;Q32)/pax5-immunoglobulin heavy chain gene is characterized by nuclear positivity of mum1 and pax5 by immunohistochemistry. Hematol Oncol (2020) 38(2):171–80. doi: 10.1002/hon.2716
123. Sawyers CL, Callahan W, Witte ON. Dominant negative myc blocks transformation by abl oncogenes. Cell (1992) 70(6):901–10. doi: 10.1016/0092-8674(92)90241-4
124. Afar DE, Goga A, McLaughlin J, Witte ON, Sawyers CL. Differential complementation of bcr-abl point mutants with C-myc. Science (1994) 264(5157):424–6. doi: 10.1126/science.8153630
125. Hyrenius-Wittsten A, Pilheden M, Sturesson H, Hansson J, Walsh MP, Song G, et al. De novo activating mutations drive clonal evolution and enhance clonal fitness in Kmt2a-rearranged leukemia. Nat Commun (2018) 9(1):1770. doi: 10.1038/s41467-018-04180-1
126. Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, et al. Polycomb repressive complex 2 is required for mll-af9 leukemia. Proc Natl Acad Sci U.S.A. (2012) 109(13):5028–33. doi: 10.1073/pnas.1202258109
127. Schreiner S, Birke M, Garcia-Cuellar MP, Zilles O, Greil J, Slany RK. Mll-enl causes a reversible and myc-dependent block of myelomonocytic cell differentiation. Cancer Res (2001) 61(17):6480–6. doi: 10.2307/3343410
128. Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, et al. Blockade of mir-150 maturation by Mll-fusion/Myc/Lin-28 is required for mll-associated leukemia. Cancer Cell (2012) 22(4):524–35. doi: 10.1016/j.ccr.2012.08.028
129. Chae H, Kim M, Lim J, Kim Y, Han K, Lee S. B lymphoblastic leukemia with etv6 amplification. Cancer Genet Cytogenet (2010) 203(2):284–7. doi: 10.1016/j.cancergencyto.2010.08.004
130. Bokemeyer A, Eckert C, Meyr F, Koerner G, von Stackelberg A, Ullmann R, et al. Copy number genome alterations are associated with treatment response and outcome in relapsed childhood Etv6/Runx1-positive acute lymphoblastic leukemia. Haematologica (2014) 99(4):706–14. doi: 10.3324/haematol.2012.072470
131. Cancelas JA, Williams DA. Rho gtpases in hematopoietic stem cell functions. Curr Opin Hematol (2009) 16(4):249–54. doi: 10.1097/MOH.0b013e32832c4b80
132. Valera A, Balague O, Colomo L, Martinez A, Delabie J, Taddesse-Heath L, et al. Ig/myc rearrangements are the main cytogenetic alteration in plasmablastic lymphomas. Am J Surg Pathol (2010) 34(11):1686–94. doi: 10.1097/PAS.0b013e3181f3e29f
133. Bemark M, Neuberger MS. The C-myc allele that is translocated into the Igh locus undergoes constitutive hypermutation in a Burkitt's lymphoma line. Oncogene (2000) 19(30):3404–10. doi: 10.1038/sj.onc.1203686
134. Seegmiller AC, Garcia R, Huang R, Maleki A, Karandikar NJ, Chen W. Simple karyotype and Bcl-6 expression predict a diagnosis of Burkitt lymphoma and better survival in Ig-Myc rearranged high-grade B-cell lymphomas. Mod Pathol (2010) 23(7):909–20. doi: 10.1038/modpathol.2010.76
135. Li S, Desai P, Lin P, Yin CC, Tang G, Wang XJ, et al. Myc/Bcl6 double-hit lymphoma (Dhl): A tumour associated with an aggressive clinical course and poor prognosis. Histopathology (2016) 68(7):1090–8. doi: 10.1111/his.12884
Keywords: EBF1, PAX5, MYC, V(D)J rearrangement, B cell development, hematologic neoplasms
Citation: Li L, Zhang D and Cao X (2024) EBF1, PAX5, and MYC: regulation on B cell development and association with hematologic neoplasms. Front. Immunol. 15:1320689. doi: 10.3389/fimmu.2024.1320689
Received: 12 October 2023; Accepted: 08 January 2024;
Published: 22 January 2024.
Edited by:
Christian Kosan, Friedrich Schiller University Jena, GermanyReviewed by:
René Winkler, Josep Carreras Leukaemia Research Institute (IJC), SpainCopyright © 2024 Li, Zhang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinmei Cao, bHpjeG1Ac3dtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.