
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Immunol. , 12 February 2024
Sec. Inflammation
Volume 15 - 2024 | https://doi.org/10.3389/fimmu.2024.1290523
This article is part of the Research Topic Long COVID and Brain Inflammation: Unravelling Mechanisms and Potential Therapies View all 10 articles
Severe COVID-19 leads to widespread transcriptomic changes in the human brain, mimicking diminished cognitive performance. As long noncoding RNAs (lncRNAs) play crucial roles in the regulation of gene expression, identification of the lncRNAs differentially expressed upon COVID-19 may nominate key regulatory nodes underpinning cognitive changes. Here we identify hundreds of lncRNAs differentially expressed in the brains of COVID-19 patients relative to uninfected age/sex-matched controls, many of which are associated with decreased cognitive performance and inflammatory cytokine response. Our analyses reveal pervasive transcriptomic changes in lncRNA expression upon severe COVID-19, which may serve as key regulators of neurocognitive changes in the brain.
Neurological symptoms including cognitive decline have been reported in individuals with COVID-19 (1–4). We and others have shown that severe COVID-19 induces widespread changes in protein-coding gene expression in the human frontal cortex (5, 6). However, the brain-related effects of COVID-19 on other RNA species such as long noncoding RNAs (lncRNAs), which may have widespread regulatory roles on transcriptional states despite lacking protein-coding potential (7), remain unclear. LncRNAs, which range from 200 base pairs to hundreds of kilobases, are a relatively understudied class of transcriptional regulators, often acting as scaffolds to recruit transcription factors and effectors to their target genes (8). Their target genes may reside near its gene locus (regulation in cis) or across the genome (regulation in trans) to regulate transcription (9–11). LncRNAs are expressed at different levels across brain areas and have been linked to synaptic plasticity, memory, and multiple brain disorders (7, 10, 12–17). Due to their potential roles in transcriptional regulation, we sought to determine the breadth of lncRNA changes upon COVID-19.
We analyzed our previously described total RNA-seq datasets (5), comprising of frontal cortex from 22 individuals with COVID-19 (23-84 years old) and 22 uninfected age- and sex-matched controls (± 2 years) (Figure 1A), to annotate both protein-coding and noncoding RNA genes (Figure 1B). Differential expression analysis revealed significantly increased (557) and decreased (269) expression levels of noncoding RNAs (ncRNAs) including numerous lncRNAs (long intergenic ncRNAs, antisense RNAs, and processed pseudogenes) associated with COVID-19 (Figures 1B, C; Supplementary Table). Clustering analysis using transcript abundances of significant differentially-expressed (DE) ncRNAs yielded a separation of COVID-19 cases from controls (Figure 1D). Interestingly, the top downregulated lncRNA, LINC01007, and one of the top upregulated lncRNAs LINC01094 upon COVID-19 were previously reported to follow a similar trend as in the brains of aged individuals and Alzheimer’s Disease (AD) patients (19, 20). Additional lncRNAs previously linked to brain aging and AD such as NEAT1, LINC00643, LINC00507, and MALAT1, were also identified (18, 19).
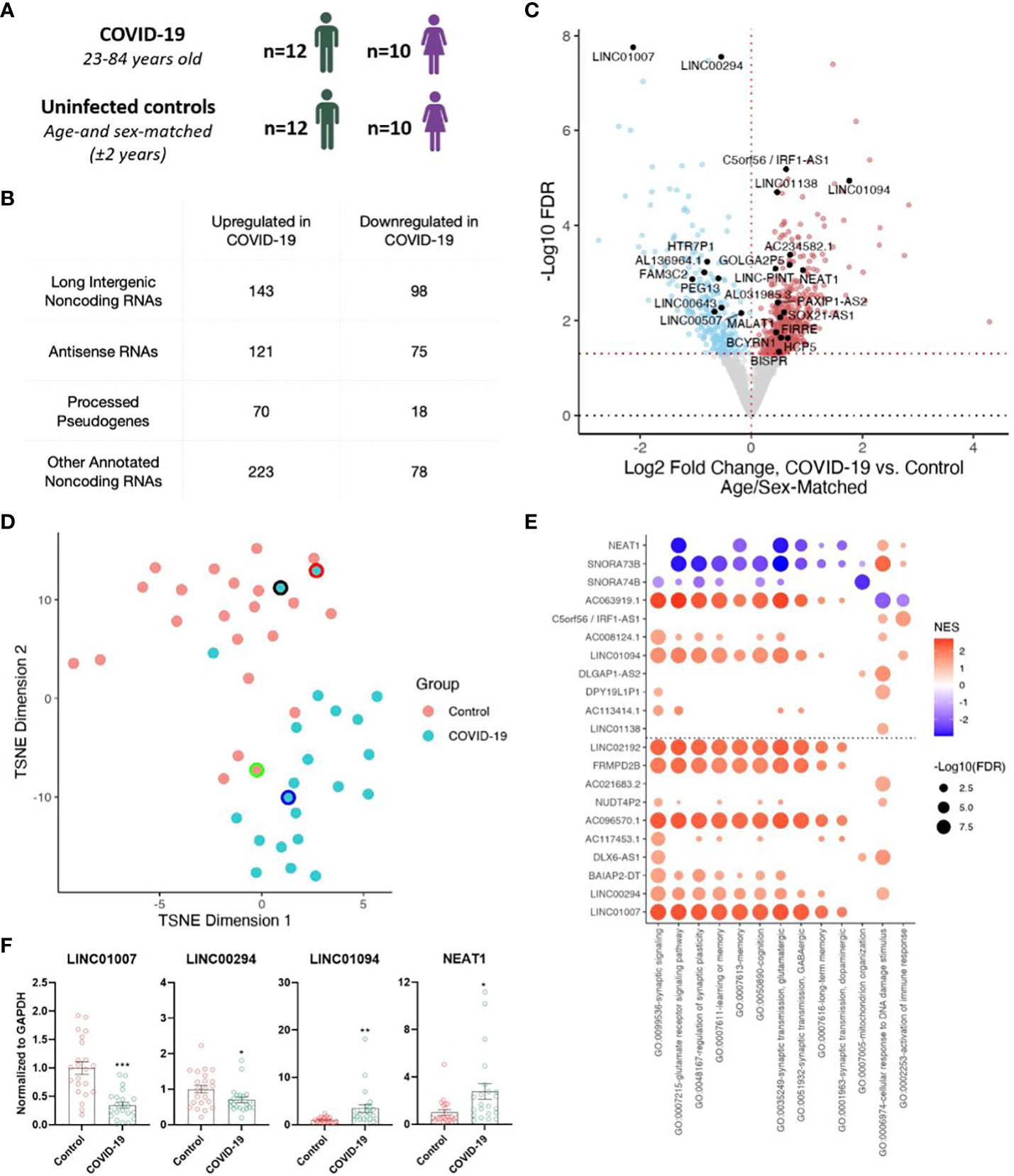
Figure 1 Severe COVID-19 changes the expression of long non-coding RNAs (lncRNAs) in the human frontal cortex. (A) Age and sex of individuals with COVID-19 and uninfected age/sex-matched control ( ± 2 years) groups (n=22/group) analyzed in this cohort; for further details see Mavrikaki et al. (5). (B) Tabulation of differentially expressed RNA species identified in our sequencing study. (C) Volcano plot showing the differentially expressed non-coding genes in the frontal cortex of COVID-19 cases versus age/sex-matched controls (n=22/group). Red points, significantly upregulated genes among COVID-19 cases (false discovery rate/FDR < 0.05). Blue points, significantly downregulated genes among COVID-19 cases. Black points, highlighted significant genes with corresponding gene symbols. (D) T-distributed stochastic neighbor embedding (TSNE) analysis of frontal cortex of COVID-19 cases and uninfected age/sex-matched controls, using significant differentially expressed noncoding RNA (ncRNA) expression levels as features. Black border, 23-year-old asymptomatic COVID-19 male. Red border, 62-year-old COVID-19 female individual with comorbid epilepsy. Blue border, 84-year-old COVID-19 female individual with comorbid Alzheimer’s Disease (AD). Green border, uninfected age/sex-matched control for the COVID case with comorbid AD. n=22/group. (E) Guilt-by-association-based Gene Ontology (GO) biological pathway analysis of top differentially expressed ncRNAs and NEAT1, a lncRNA involved in cognitive processes (18). (F) Validation of sequencing data using qRT-PCR. n=22/group. Two tailed unpaired t-test, *p<0.05, **p<0.01, ***p<0.001. LINC01007 t(42)= 5.377, p=0.000003, LINC00294 t(42)= 2.224, p=0.0316, LINC01094 t(42)= 2.844, p=0.0069, NEAT1 t(42)= 2.583, p=0.0134.
To better understand the roles of the differentially expressed noncoding RNAs in COVID-19, many of which have no known functional roles, we performed guilt-by-association pathway analysis for the top and bottom 10 COVID-19-regulated ncRNAs as well as NEAT1, a well-studied lncRNA involved in brain aging (19) and cognitive function (18). For each ncRNA we ranked the coexpression of protein-coding genes across the transcriptome-profiled samples from The Cancer Genome Atlas (TCGA), spanning multiple tissue samples and genetic backgrounds, and tested for pathway enrichment using these protein-coding gene rankings (21, 22). This analysis implicated many of these lncRNAs in pathways associated with cognitive function (e.g., memory and learning) (23–30) (Figure 1E). Finally, we validated the decreased expression of LINC01007 and LINC00294 and increased expression of LINC01094 and NEAT1 by qRT-PCR (Figure 1F; Supplementary Figure 1). We selected these genes because (1) LINC01007, LINC00294, and LINC01094 are among the top 10 up/down COVID-regulated genes, with LINC01007 and LINC00294 as the two most significantly downregulated lncRNAs, (2) LINC01007 and LINC01094 have been previously associated with aging (19), and (3) NEAT1, also a significantly upregulated lncRNA, is well-established as a regulator of cognitive function (18). Critically, overexpression of NEAT1 impairs cognitive function, whereas knockdown of NEAT1 improves memory in mice (18), in support of a functional role for NEAT1 upregulation in COVID-19-associated cognitive decline.
Next, we sought to evaluate whether the differential expression of these COVID-19-regulated ncRNAs was also associated with poor cognitive performance in humans. We utilized previously published cognitive and transcriptomic data, obtained from the same individuals, in the context of the ROSMAP cohort (31, 32). After splitting those cases (n=633: 406 females and 227 males) by median Mini-Mental State Examination (MMSE) score (high cognitive performance: ≥25, 207 females and 129 males, total 336; low cognitive performance: <25, 199 females and 98 males, total 297) and performing gene expression analysis, we found 1,307 downregulated ncRNAs and 1,322 upregulated ncRNAs in individuals with low cognitive performance (Figure 2A; Supplementary Table). The larger sample size of the ROSMAP cohort, in comparison to the COVID-19 cohort, likely contributes to increased statistical power and a greater number of significant differentially expressed ncRNAs in the ROSMAP cohort. By Gene Set Enrichment Analysis (GSEA) analysis, we found that ncRNAs associated with severe COVID-19 were also associated with low cognitive performance (Figures 2B, C). Moreover, the similarities in ncRNA expression profiles due to COVID-19 and poor cognitive performance are maintained in COVID-19 relative to control cases with history of intensive care unit or ventilator (ICU/VENT) treatment (n=9) (5), in support of potential roles for ncRNAs in COVID-19-induced cognitive changes independent of ICU/VENT-associated treatment (Figure 2D).
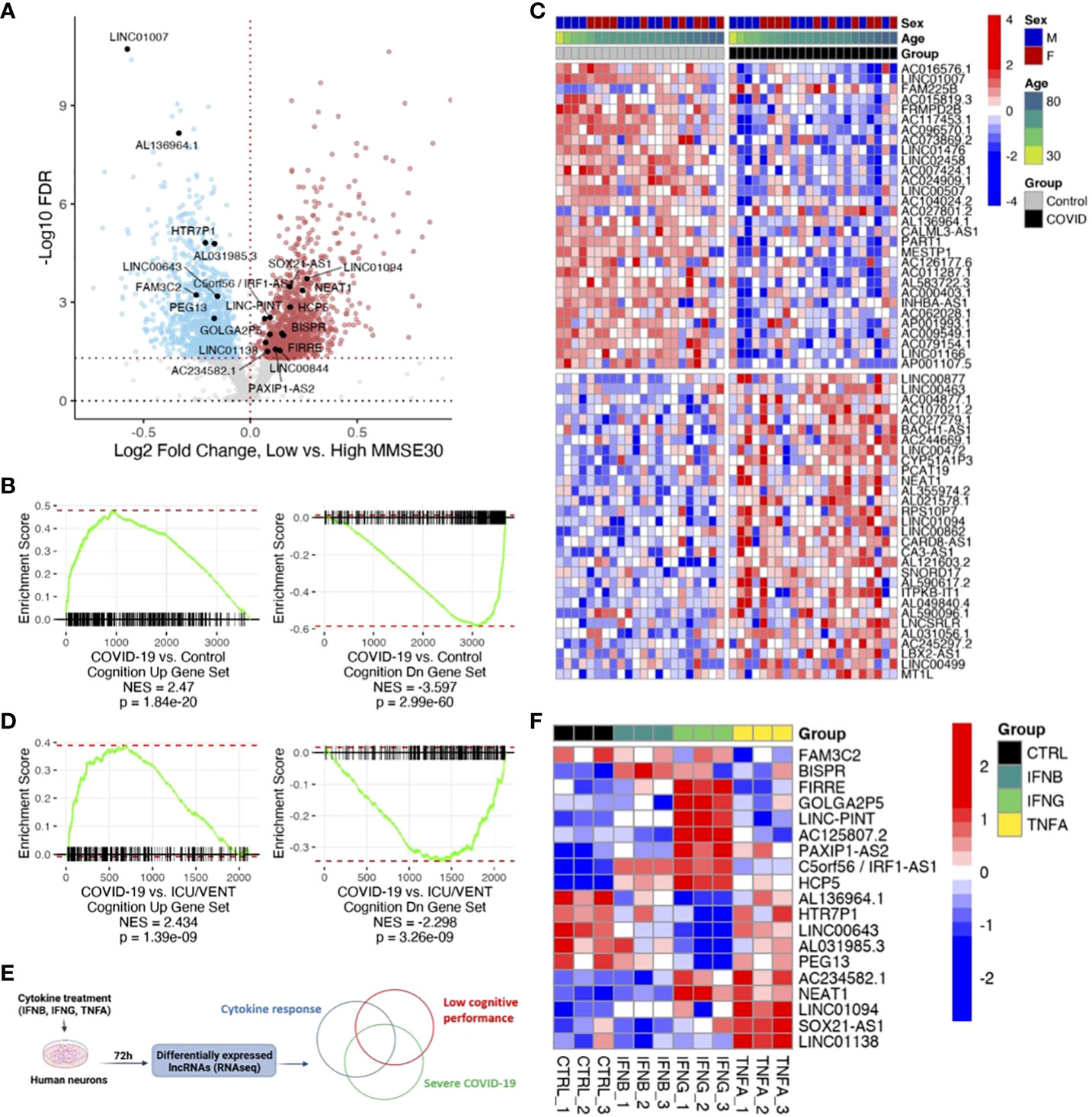
Figure 2 COVID-19 is associated with low cognitive performance-related noncoding RNAs (ncRNAs). (A) Volcano plot showing low cognitive performance-related ncRNAs identified in the ROSMAP cohort. Red points, significantly upregulated genes among individuals with low cognitive performance; MMSE scores <25 (false discovery rate/FDR < 0.05). Blue points, significantly downregulated genes with low cognitive performance; MMSE scores <25. Black points, highlighted significant genes with corresponding gene symbols (High MMSE 207 females and 129 males, total 336; Low MMSE 199 females and 98 males, total 297). (B) Gene set enrichment analysis (GSEA) of COVID-19-regulated ncRNAs using low cognitive performance-associated ncRNAs as gene sets. DEG ranks were assigned by signed -log10 FDR from frontal cortex transcriptome of COVID-19 versus transcriptome of age/sex-matched control (n=22/group). NES, normalized enrichment score. p, two-tailed GSEA p-value. (C) Heatmap of expression values (COVID-19 cohort) of top 30 upregulated ncRNAs and top 30 downregulated ncRNAs overlapping between COVID-19 and low cognitive performance-related ncRNAs. Red represents increased expression; Blue represents decreased expression. (D) GSEA of COVID-19-regulated ncRNAs using low cognitive performance-associated ncRNAs as gene sets. DEG ranks were assigned by signed -log10 FDR from frontal cortex transcriptomes of COVID-19 (n=22) versus transcriptomes of uninfected controls with ICU/VENT history (n=9). NES, normalized enrichment score. p, two-tailed GSEA p-value. (E) Schematic of in vitro cytokine treatment experiment and analytical approach. (F) Heatmap of expression values (in vitro human neurons) of significant ncRNAs overlapping between COVID-19, cognition, and cytokine response. IFNB: 1ng/ml-1; IFNG: 1μg/ml-1; TNFA: 100ng/ml-1. Red represents increased expression; Blue represents decreased expression.
Finally, as circulating inflammatory factors have been suggested to affect neurological states in COVID-19 (33), we utilized previously published total RNA sequencing data and assessed ncRNA expression changes in primary human neurons upon cytokine treatment (Figure 2E; Supplementary Table). We found 19 ncRNAs differentially expressed by at least one of IFNB, IFNG, or TNFA that are also differentially expressed in both severe COVID-19 and poor cognition (Figure 2F). Of these overlapping genes, LINC01094, NEAT1, and LINC00643 have been previously linked to brain aging and AD (19). Interestingly, loss of NEAT1 not only improves cognitive function (18), but also reduces inflammatory response (34). To obtain further insights into the effects of lncRNAs on protein-coding gene expression, we assessed whether the cognate sense genes (IRF1, PAXIP1, SOX21) of the three significant antisense lncRNAs (C5orf56/IRF1-AS1, PAXIP1-AS2, SOX21-AS1), which often transcriptionally regulate their corresponding sense gene (11), are also significant following cytokine treatment. We found that all three protein-coding genes follow similar expression patterns as the lncRNAs in our in vitro neuron datasets (Supplementary Figure 2). Of note, IRF1 is also significantly differentially expressed in both COVID-19 and ROSMAP comparisons. This gene is well-implicated in interferon regulation (35–37) and COVID-19 response (37, 38), in support of a role for IRF1-AS1 in the disease. Our analyses highlight the potential for lncRNAs as therapeutic targets to modulate neuroinflammation and mitigate associated cognitive deficits (15).
Given the cognitive decline reported in patients with milder COVID-19 (2), it is tempting to speculate that similar lncRNA expression changes might be found in milder COVID-19 cases. We note, however that our analysis is limited primarily to severe COVID-19 cases due to the availability of relevant specimens. Although we are not statistically powered to make comparisons in milder cases or in asymptomatic individuals with COVID-19, we have included one individual with asymptomatic COVID-19 in our analysis. We found that the ncRNA expression profile from this individual is more representative of control individuals rather than those with severe disease (Figures 1D, 2C).
In summary, we have identified widespread expression changes of numerous lncRNAs in the brain due to severe COVID-19 that are also associated with poor cognition. We link a number of these lncRNAs to transcriptomic changes in neurons upon inflammatory cytokine stimulation. As COVID-19 is associated with cognitive decline (2, 3), our findings suggest key roles for lncRNAs in cognitive decline in individuals with severe COVID-19 and support the idea that inflammation-associated lncRNAs may be targeted to alleviate cognitive deficits observed in COVID-19.
In this study, we analyzed our previously described total RNA-seq datasets (5). In that cohort, frozen COVID-19 frontal cortex specimens were collected following a protocol for waived consent for the use of excess tissue, approved by the Mass General Brigham Institutional Review Board. Frozen control frontal cortex specimens were obtained from the NIH NeuroBiobank and the NIH HBCC. Clinical features of the COVID-19 cohort have been previously described in Mavrikaki et al. and included 22 cases with pre-mortem or peri-mortem positive testing for SARS-CoV-2 by nasopharyngeal swab qPCR (COVID-19 group) with mean age 61.91 ± 3.1 years (12 males and 10 females), age/sex-matched (± 2 years) uninfected controls without any known psychiatric or neurological disease with mean age 61.86 ± 3.1 years, and an independent group of 9 uninfected cases with history of ICU or ventilator treatment (ICU/VENT) with mean age 57 ± 6.98 years (6 males and 3 females) (5). Total RNA from those samples was extracted using Trizol and phase separation. 450 ng of RNA for the frontal cortex specimens and 80 ng of RNA for the human primary neurons was used for library preparation (5). Libraries were prepared using the KAPA RNA HyperPrep kit with RiboErase (HMR; Roche; #08098131702) following the manufacturer’s recommendations, pooled together (4 runs), and processed for sequencing using NovaSeq 6000 (5). Total RNA-seq data for the COVID-19 cohort and in vitro neuron experiment are available at the Gene Expression Omnibus (GEO) with accession number GSE188847.
Raw. fastq sequencing files were aligned to Ensembl v104 using salmon v1.4.0, combining both protein-coding (cdna.all.fa) and noncoding RNA (ncrna.fa) sequences. Annotated gene biotypes were obtained from the Ensembl v96 release (April 2019), as distinction between antisense, processed pseudogene, and long intergenic noncoding RNA were not included in further Ensembl updates. Gene-level abundances were determined using tximport v1.18.0, and differential expression analysis was performed with DESeq2 v1.30.1 using lfcShrink to stabilize variance. Preprocessed ROSMAP gene abundances from n=633 (High MMSE 207 females and 129 males; Low MMSE 199 females and 98 males) and corresponding MMSE cognitive data were obtained from https://www.synapse.org/#!Synapse:syn8691134 and https://www.synapse.org/Portal.html#!Synapse:syn3157322, respectively (39), and differential expression analysis was performed with DESeq2 v1.30.1.
Guilt-by-association pathway analysis of lncRNAs was performed as follows. First, Pearson correlations between the expression levels (log2 transcripts per million + 1) of candidate lncRNAs and those of all protein-coding genes were determined across 9,830 patient transcriptome samples generated as part of The Cancer Genome Atlas Research Network: https://www.cancer.gov/tcga. Protein-coding genes ranked by correlation with each tested lncRNA were used as input for gene set enrichment analysis (fgsea v1.16.0), using gene sets of previously identified Gene Ontology pathways (5). In addition to NEAT1, the top and bottom 10 differentially expressed lncRNAs as ranked by FDR in the COVID-19 cohort and detected in the TCGA dataset were tested for pathway analysis (one snoRNA and one lncRNA were not detected).
Association testing between COVID-19 and ROSMAP cohorts was performed as follows. Signed -log10 FDRs from COVID-19 vs. Control or COVID-19 vs. ICU/VENT comparisons were used to rank ncRNA genes for gene set enrichment analysis via fgsea v1.16.0, filtering out genes with an FDR < 0.5. Cognition-associated gene sets were collated from ROSMAP Poor vs. Normal MMSE comparisons, using significant (FDR < 0.05) ncRNAs. Ensembl gene IDs were used for gene matching in this analysis.
R scripts, reference files, and realigned RNA-seq files used for these analyses are available at https://github.com/jonathandlee12/covid19-brain-lnc. All other reference datasets are either publicly available or will be provided upon reasonable request to the corresponding authors.
A total of 400 ng RNA from each sample was used for cDNA synthesis, and qRT-PCR for orthogonal validation was performed and analyzed as previously described in Mavrikaki et al. (5). GAPDH (Qiagen; QuantiTect primer assay: QT00079247) and RPS18 (Qiagen; QuantiTect primer assay: QT00248682) were used for normalization. Primers for LINC01007 (#qhsaLED0063333), LINC01094 (#qhsaLED0101136), and NEAT1 (#qhsaLED0134812) were purchased from Bio-Rad. Primers for LINC00294 were TGTGTTGTCCTCCAGAATCG (forward) and CCAACCAAGAGCCAACAAAG (reverse) (40), and were synthesized by IDT. Data were analyzed according to the 2-ΔΔCt method (41).
We reanalyzed previously published total RNA-seq data of primary human neurons (ScienCell Research Laboratories, 1520-5) treated with different cytokines (5) which are available on the GEO with accession number GSE188847. Primary neurons were treated with IFN-β (1 ng ml−1), IFN-γ (1 µg ml−1), TNF (100 ng ml−1) or nuclease-free water (control) for 72 h, and RNA was extracted using Trizol/phase separation, and 80ng of RNA was processed for total RNA-seq.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GSE188847 (GEO).
The studies involving human specimens were approved by Mass General Brigham Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements. Controls were obtained from the NIH Neurobiobank and the NIH HBCC as de-identified samples.
JL: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. IS: Resources, Writing – review & editing. FS: Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. MM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH award 1R01AG082093 to FS and 1R01AG079799 to MM.
We would like to thank the NIH Neurobiobank (the Harvard Brain Tissue Resource Center, the Miami Brain Endowment Bank, and the University of Maryland Brain and Tissue Bank) and the Human Brain Collection Core (HBCC) at NIH for providing control brain tissues. We thank the participants and study staff of the Religious Orders Study and the Rush Memory and Aging Project. The results published here are in part based on data obtained from the AD Knowledge Portal (https://adknowledgeportal.org).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1290523/full#supplementary-material
1. Solomon IH, Normandin E, Bhattacharyya S, Mukerji SS, Keller K, Ali AS, et al. Neuropathological features of covid-19. N Engl J Med (2020) 383:989–92. doi: 10.1056/NEJMc2019373
2. Douaud G, Lee S, Alfaro-Almagro F, Arthofer C, Wang C, McCarthy P, et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature (2022) 604:697–707. doi: 10.1038/s41586-022-04569-5
3. Hampshire A, Trender W, Chamberlain SR, Jolly AE, Grant JE, Patrick F, et al. Cognitive deficits in people who have recovered from COVID-19. EClinicalMedicine (2021) 39:101044. doi: 10.1016/j.eclinm.2021.101044
4. Kas A, Soret M, Pyatigoskaya N, Habert M. O, Hesters A, Le Guennec L, et al. The cerebral network of COVID-19-related encephalopathy: a longitudinal voxel-based 18F-FDG-PET study. Eur J Nucl Med Mol Imaging (2021) 48:2543–57. doi: 10.1007/s00259-020-05178-y
5. Mavrikaki M, Lee JD, Solomon IH, Slack FJ. Severe COVID-19 is associated with molecular signatures of aging in the human brain. Nat Aging (2022) 2:1130–7. doi: 10.1038/s43587-022-00321-w
6. Yang AC, Kern F, Losada PM, Agam MR, Maat CA, Schmartz GP, et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature (2021) 595:565–71. doi: 10.1038/s41586-021-03710-0
7. Liu Y, Chen X, Che Y, Li H, Zhang Z, Peng W, et al. LncRNAs as the regulators of brain function and therapeutic targets for alzheimer's disease. Aging Dis (2022) 13:837–51. doi: 10.14336/AD.2021.1119
8. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
9. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
10. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol (2023) 24:430–47. doi: 10.1038/s41580-022-00566-8
11. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature (2012) 491:454–7. doi: 10.1038/nature11508
12. Yang S, Lim KH, Kim SH, Joo JY. Molecular landscape of long noncoding RNAs in brain disorders. Mol Psychiatry (2021) 26:1060–74. doi: 10.1038/s41380-020-00947-5
13. Ni YQ, Xu H, Liu YS. Roles of long non-coding RNAs in the development of aging-related neurodegenerative diseases. Front Mol Neurosci (2022) 15:844193. doi: 10.3389/fnmol.2022.844193
14. Grinman E, Espadas I, Puthanveettil SV. Emerging roles for long noncoding RNAs in learning, memory and associated disorders. Neurobiol Learn Mem (2019) 163:107034. doi: 10.1016/j.nlm.2019.107034
15. Ahmad MA, Kareem O, Khushtar M, Akbar M, Haque MR, Iqubal A, et al. Neuroinflammation: a potential risk for dementia. Int J Mol Sci (2022) 23. doi: 10.3390/ijms23020616
16. Wang F, Wang Q, Liu B, Mei L, Ma S, Wang S, et al. The long noncoding RNA Synage regulates synapse stability and neuronal function in the cerebellum. Cell Death Differ (2021) 28:2634–50. doi: 10.1038/s41418-021-00774-3
17. Wei W, Zhao Q, Wang Z, Liau WS, Basic D, Ren H, et al. ADRAM is an experience-dependent long noncoding RNA that drives fear extinction through a direct interaction with the chaperone protein 14-3-3. Cell Rep (2022) 38:110546. doi: 10.1016/j.celrep.2022.110546
18. Butler AA, Johnston DR, Kaur S, Lubin FD. Long noncoding RNA NEAT1 mediates neuronal histone methylation and age-related memory impairment. Sci Signal (2019) 12(588):eaaw9277. doi: 10.1126/scisignal.aaw9277
19. Cao M, Li H, Zhao J, Cui J, Hu G. Identification of age- and gender-associated long noncoding RNAs in the human brain with Alzheimer's disease. Neurobiol Aging (2019) 81:116–26. doi: 10.1016/j.neurobiolaging.2019.05.023
20. Espuny-Camacho I, Arranz AM, Fiers M, Snellinx A, Ando K, Munck S, et al. Hallmarks of alzheimer's disease in stem-cell-derived human neurons transplanted into mouse brain. Neuron (2017) 93:1066–1081e1068. doi: 10.1016/j.neuron.2017.02.001
21. Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell (2018) 173:649–664e620. doi: 10.1016/j.cell.2018.03.052
22. Cancer Genome Atlas Research, N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet (2013) 45:1113–20. doi: 10.1038/ng.2764
23. Sudhof TC. Towards an understanding of synapse formation. Neuron (2018) 100:276–93. doi: 10.1016/j.neuron.2018.09.040
24. Chakraborty P, Dey A, Gopalakrishnan AV, Swati K, Ojha S, Prakash A, et al. Glutamatergic neurotransmission: A potential pharmacotherapeutic target for the treatment of cognitive disorders. Ageing Res Rev (2023) 85:101838. doi: 10.1016/j.arr.2022.101838
25. Prevot T, Sibille E. Altered GABA-mediated information processing and cognitive dysfunctions in depression and other brain disorders. Mol Psychiatry (2021) 26:151–67. doi: 10.1038/s41380-020-0727-3
26. Yan Z, Rein B. Mechanisms of synaptic transmission dysregulation in the prefrontal cortex: pathophysiological implications. Mol Psychiatry (2022) 27:445–65. doi: 10.1038/s41380-021-01092-3
27. Khacho M, Harris R, Slack RS. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat Rev Neurosci (2019) 20:34–48. doi: 10.1038/s41583-018-0091-3
28. Konopka A, Atkin JD. The role of DNA damage in neural plasticity in physiology and neurodegeneration. Front Cell Neurosci (2022) 16:836885. doi: 10.3389/fncel.2022.836885
29. Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature (2010) 464:529–35. doi: 10.1038/nature08983
30. Zhao F, Li B, Yang W, Ge T, Cui R. Brain-immune interaction mechanisms: Implications for cognitive dysfunction in psychiatric disorders. Cell Prolif (2022) 55:e13295. doi: 10.1111/cpr.13295
31. Bennett DA, Schneider JA, Arvanitakis Z, Wilson RS. Overview and findings from the religious orders study. Curr Alzheimer Res (2012) 9:628–45. doi: 10.2174/156720512801322573
32. Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson R Overview and findings from the rush Memory and Aging Project. Curr Alzheimer Res (2012) 9:646–63. doi: 10.2174/156720512801322663
33. Fernandez-Castaneda A, Lu P, Geraghty AC, Song E, Lee MH, Wood J, et al. Mild respiratory COVID can cause multi-lineage neural cell and myelin dysregulation. Cell (2022) 185:2452–2468e2416. doi: 10.1016/j.cell.2022.06.008
34. Zhang P, Cao L, Zhou R, Yang X, Wu M. The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat Commun (2019) 10:1495. doi: 10.1038/s41467-019-09482-6
35. Bonelli M, Dalwigk K, Platzer A, Olmos Calvo I, Hayer S, Niederreiter B, et al. IRF1 is critical for the TNF-driven interferon response in rheumatoid fibroblast-like synoviocytes : JAKinibs suppress the interferon response in RA-FLSs. Exp Mol Med (2019) 51:1–11. doi: 10.1038/s12276-019-0267-6
36. Carlin AF, Plummer EM, Vizcarra EA, Sheets N, Joo Y, Tang W, et al. An IRF-3-, IRF-5-, and IRF-7-independent pathway of dengue viral resistance utilizes IRF-1 to stimulate type I and II interferon responses. Cell Rep (2017) 21:1600–12. doi: 10.1016/j.celrep.2017.10.054
37. Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-coV-2 infection and cytokine shock syndromes. Cell (2021) 184:149–168e117. doi: 10.1016/j.cell.2020.11.025
38. Shin J, Toyoda S, Nishitani S, Onodera T, Fukuda S, Kita S, et al. SARS-CoV-2 infection impairs the insulin/IGF signaling pathway in the lung, liver, adipose tissue, and pancreatic cells. via IRF1. Metab (2022) 133:155236. doi: 10.1016/j.metabol.2022.155236
39. De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D, et al. A multi-omic atlas of the human frontal cortex for aging and Alzheimer's disease research. Sci Data (2018) 5:180142. doi: 10.1038/sdata.2018.142
40. Qiu J, Zhou S, Cheng W, Luo C. LINC00294 induced by GRP78 promotes cervical cancer development by promoting cell cycle transition. Oncol Lett (2020) 20:262. doi: 10.3892/ol.2020.12125
Keywords: noncoding RNAs, lncRNAs, cognitive decline, COVID-19, frontal cortex
Citation: Lee JD, Solomon IH, Slack FJ and Mavrikaki M (2024) Cognition-associated long noncoding RNAs are dysregulated upon severe COVID-19. Front. Immunol. 15:1290523. doi: 10.3389/fimmu.2024.1290523
Received: 07 September 2023; Accepted: 23 January 2024;
Published: 12 February 2024.
Edited by:
Jacob Raber, Oregon Health and Science University, United StatesReviewed by:
George Calin, University of Texas MD Anderson Cancer Center, United StatesCopyright © 2024 Lee, Solomon, Slack and Mavrikaki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Mavrikaki, bW1hdnJpa2FAYmlkbWMuaGFydmFyZC5lZHU=; Frank J. Slack, ZnNsYWNrQGJpZG1jLmhhcnZhcmQuZWR1
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.