- Institute of Pathogenic Biology, Hunan Provincial Key Laboratory for Special Pathogens Prevention and Control, School of Nursing, Hengyang Medical College, University of South China, Hengyang, China
Chlamydia trachomatis, is a kind of obligate intracellular pathogen. The removal of C. trachomatis relies primarily on specific cellular immunity. It is currently considered that CD4+ Th1 cytokine responses are the major protective immunity against C. trachomatis infection and reinfection rather than CD8+ T cells. The non-specific immunity (innate immunity) also plays an important role in the infection process. To survive inside the cells, the first process that C. trachomatis faces is the innate immune response. As the “sentry” of the body, mast cells attempt to engulf and remove C. trachomatis. Dendritic cells present antigen of C. trachomatis to the “commanders” (T cells) through MHC-I and MHC-II. IFN-γ produced by activated T cells and natural killer cells (NK) further activates macrophages. They form the body’s “combat troops” and produce immunity against C. trachomatis in the tissues and blood. In addition, the role of eosinophils, basophils, innate lymphoid cells (ILCs), natural killer T (NKT) cells, γδT cells and B-1 cells should not be underestimated in the infection of C. trachomatis. The protective role of innate immunity is insufficient, and sexually transmitted diseases (STDs) caused by C. trachomatis infections tend to be insidious and recalcitrant. As a consequence, C. trachomatis has developed a unique evasion mechanism that triggers inflammatory immunopathology and acts as a bridge to protective to pathological adaptive immunity. This review focuses on the recent advances in how C. trachomatis evades various innate immune cells, which contributes to vaccine development and our understanding of the pathophysiologic consequences of C. trachomatis infection.
1 Introduction
C. trachomatis, a specific intracellular pathogen, is closely associated with human epidemic diseases. According to the World Health Organization, in 2020, an estimated 129 million people would be infected with C. trachomatis (1). There are 19 serovars of C. trachomatis. In addition to trachoma, inclusion conjunctivitis, and infantile pneumonia(serovars A to C), it also causes more serious genitourinary tract infections(serovars D to K) and lymphogranuloma venereum(serovars L). The former is highly curable and can be treated with large-scale azithromycin (2), while the latter is insidious and chronic both in men and women (3). Although sensitive to antibiotics, their therapeutic benefit is limited mainly because of silent (asymptomatic) and recurrent infections attributable to immune evasion and the development of partial immunity from these infections. In the long term, these recurrent infections are thought to be contributing to the development of pelvic inflammatory disease (4), tubal infertility (5), cervical cancer (6), adverse pregnancy, miscarriage (7) in women, urethritis, epididymitis, orchitis, prostatitis, proctitis (8) or reactive arthritis (9) in men. These outcomes are thought to be culminating either alone or as a complex combination of Chlamydial pathogenesis, deficient or inferior immunological memory- immune escape and immunopathology. Chlamydia lives in membrane-coated vacuoles, called inclusion, which protect it from the humoral immune response. It has been found that Chlamydia obtains nutrients (amino acids, lipids (10), iron (11)) mainly from the host cell, and acquires ATP (12) entirely from the host. Chlamydia takes self-serving measures to deal with nutritional crisis, one of which is to limit the detection of innate immunity (13). NF-κB plays an important role in the inflammatory response. Surprisingly, no obvious signs of NF-κB activation were detected in Chlamydia-infected cells (14). C. trachomatis blocks NF-κB signaling through effector ChlaDUB1 reversal of IκB-α ubiquitination (15) and CPAF-mediated p65/RelA degradation (16). However, the in vivo role of CPAF in NF-κB signaling has not been proven. Inflammatory damage caused by chlamydial infection is largely due to both innate and adaptive immunity. Chlamydial infection stimulates host cells to produce interleukins, interferons, and tumor necrosis factor, which play a dual role in infection (17, 18). Consequent to infection of the upper genital tract, infiltration of neutrophils and monocytes that are responsible for potentially deleterious inflammation along with the bystander T cells both with the potential to cause immunopathology (19). Highly potent antibodies may also cause corresponding immunopathology through mechanisms such as activation of complement (20), ADCC (antibody-dependent cellular cytotoxicity) (21), and the emergence of a type IV hypersensitivity reaction (22). In addition, immune escape of Chlamydia is also an important reason for chronic recurrent infections, which will be described primarily in this review.
Recent studies indicate that C. trachomatis uses effector molecules like GarD that helps it to escape immune surveillance via avoiding ubiquitination and proteosomal degradation (23, 24). Recent findings indicate that Chlamydial lipopolysaccharide (LPS) (25) and lipooligosaccharide (LOS) (26) do not readily trigger innate inflammatory pathways, thus avoiding early innate recognition and promoting its survival and multiplication.
In summary, the key to maintaining the intracellular survival and persistence of C. trachomatis is to escape from the host’s innate immune cells. In recent years, it has been found that properly functioning immune cells have the potential to treat disease, and immune responses are often associated with the course of disease. When the body is in the midst of a persistent infection or the immune cells are not functioning properly, the ability of the immune system to clear the infection is declining. Then a critical point is reached where the immune cells are unable to clear the infected cells, and the disease occurs. The development of a novel vaccine against C. trachomatis infection benefits greatly from research on cancer immunotherapy (27) and experimental vaccinations against intracellular diseases. In fact, after H Su et al. immunized mice intravenously with bone marrow-derived dendritic cells stimulated in vitro by dead chlamydia, DC was able to efficiently perform its functions of bacterial phagocytosis and antigen presentation. The results showed that this method of immunization produced protective immunity against Chlamydia infection in the female genital tract comparable to that following in vivo infection (28). Karunakaran et al. have employed DCs pulsed with chlamydial immunopeptides in immunizations by adoptive transfer. Immunized mice developed Th1 protective immunity and partially resisted chlamydial lung and genital infections (29). However, there is a long way to go in the development of immune cell-targeted biologics in chlamydial immunity and immunopathology for prevention or treatment (30, 31).
2 Pathogenesis and immunity
Unlike other bacteria, C. trachomatis has a unique biphasic developmental cycle. In the initial step, non-replicating elementary bodies (EBs) bind to the host acetyl heparan sulfate proteoglycan (HSPG) and primarily the receptor tyrosine kinase (RTK), injecting various effector proteins called C. trachomatis secretory proteins (CtSPs) into the cytoplasm via the type III secretion system (T3SS) or other secretory mechanisms (32, 33). After entry, they differentiate into non-infectious replicating reticulate bodies (RBs) within parietal vesicles called inclusions, and RB-secreted inclusions membrane proteins (Incs) are incorporated into the membranes of the compartment (34). The interaction of C. trachomatis proteins with host proteins involves altering vesicular transport in extracellular vesicles, regulating cell survival pathways, and suppressing the innate host immune response. The Chlamydial effector proteins interfere with the host’s innate immune response, such as INCs, TepP (35), CPAF, Pgp3 (36), Pgp4 (37) and 60 other proteins (38), which promote intracellular survival of Chlamydia and limit the host response to infection. RBs continue to differentiate in inclusions and, at later stages, asynchronously undergo secondary differentiation to create new EBs. Studies have shown that C. trachomatis divides by a polarized budding mechanism, rather than binary fission (39). In the final process, intracellular EBs are release by two pathways described so far: lysis cell destruction or exit by extrusion formation. The particular form of cyclic propagation from EB to RB to EB occurs repeatedly in neighboring cells of the host, which takes approximately 36-48h to complete a developmental cycle.
The first line of defense in the human immune system is the skin and mucous membranes, and the second line is the phagocytes and bactericidal substances in the body fluids, which together constitute innate immunity. C. trachomatis genital serovars have a tropism for columnar epithelial cells located on mucosal surfaces (40, 41). On the one hand, host epithelial cells recognize the invasion of C. trachomatis antigens by surface receptors, endosomal receptors, and innate immune factors. These antigens are first blocked by the mucosal barrier and neutralized by mucosal antibodies. Microbiota namely lactobacillus along with lactoferrin confer an important mucosal defense in the cervicovaginal region (42). Upon breach of this barrier, the infected cells release cytokines and chemokines (43) that serve to recruit cells like neutrophils and monocytes as well as others that serve to curtail the infection and limit its spread (44). Macrophages engulf the bacteria and produce pro-inflammatory factors (45); IFN-γ secreted by NK cells not only kills infected host cells but also induces an immune response to Th1 (46, 47); When infection leads to the development of antigen-specific immunity, CD4+ T cells along with B cells produce immunity where chlamydia-specific Th1 CD4+ T cells and antibodies are considered protective (48, 49), whereas CD8+ T cell response is considered non-essential or even pathogenic (50, 51). These cells interact and collaborate to clear C. trachomatis.
On the other hand, this infection stimulates the establishment of immunogenicity is necessary to generate a good protective immune response. Chlamydia has evolved to evade immunity as well as actively subvert it by inhibiting the cytokines and chemotactic proteins produced by epithelial cells (52, 53). It interferes with the antigen-presenting function of antigen-presenting cells (APCs) (downregulation of MHC class I and II molecules) (54, 55), regulating specific cytokines with multiple effects (IL-18, IFN-γ, TNF-α), and anti-apoptosis (increased cell survival signaling and CPAF release) (56–58). In addition, recent studies have shown that intracellular RBs can enter uninfected neighboring cells via tunneling nanotubes (TNT) (59). This allows Chlamydia to remain unexposed to body fluids, thus evading to some extent the pursuit of various cytokines in body fluids.
3 Evasion of classical innate immune cells
3.1 Mφ
Tissue macrophages (Mφ) generally engulf bacteria early in the infection when they are outside the host cells. It is known that C. trachomatis entry into the monocyte macrophages is less efficient when compared to epithelial cells. Moreover, ROS-mediated microbicidal activity is unable to kill the engulfed bacteria, and only IFN-γ stimulated, highly activated phagocytes with sufficient effector molecules can effectively kill bacteria (17, 60) (Figure 1). Experiments by Chen, et al. in 2017 showed that both LPS/ATP and murine chlamydial infection of mouse bone marrow-derived macrophages (BMDM) could lead to caspase-1 cleavage and IL-1β released. However, unlike LPS/ATP, the inflammatory molecular switch RIP3 was not involved in the activation of NLRP3 inflammatory vesicles in murine Chlamydia-stimulated BMDM, suggesting that chlamydial infection leads to caspase-1 cleavage and IL-1β release by a different mechanism than LPS/ATP (61). Two years later, further experiments showed that lipopolysaccharide (LPS) of C. trachomatis in BMDM, unlike other Gram-negative bacteria, blocked downstream signaling of TRIF and MyD88 classical inflammatory pathways and failed to activate non-classical inflammatory pathways mediated through caspase-11 to evade innate immunity. This study explored the mechanism in more depth: trachoma LPS, while effectively binding CD14, cannot effectively induce TLR4/MD-2 complex dimerization or endocytosis, implying that the classical pathway is blocked at its source. It is speculated that this phenomenon may be related to the fact that lipid A of chlamydial LPS is penta-acylated, rather than hexa-acylated (25).
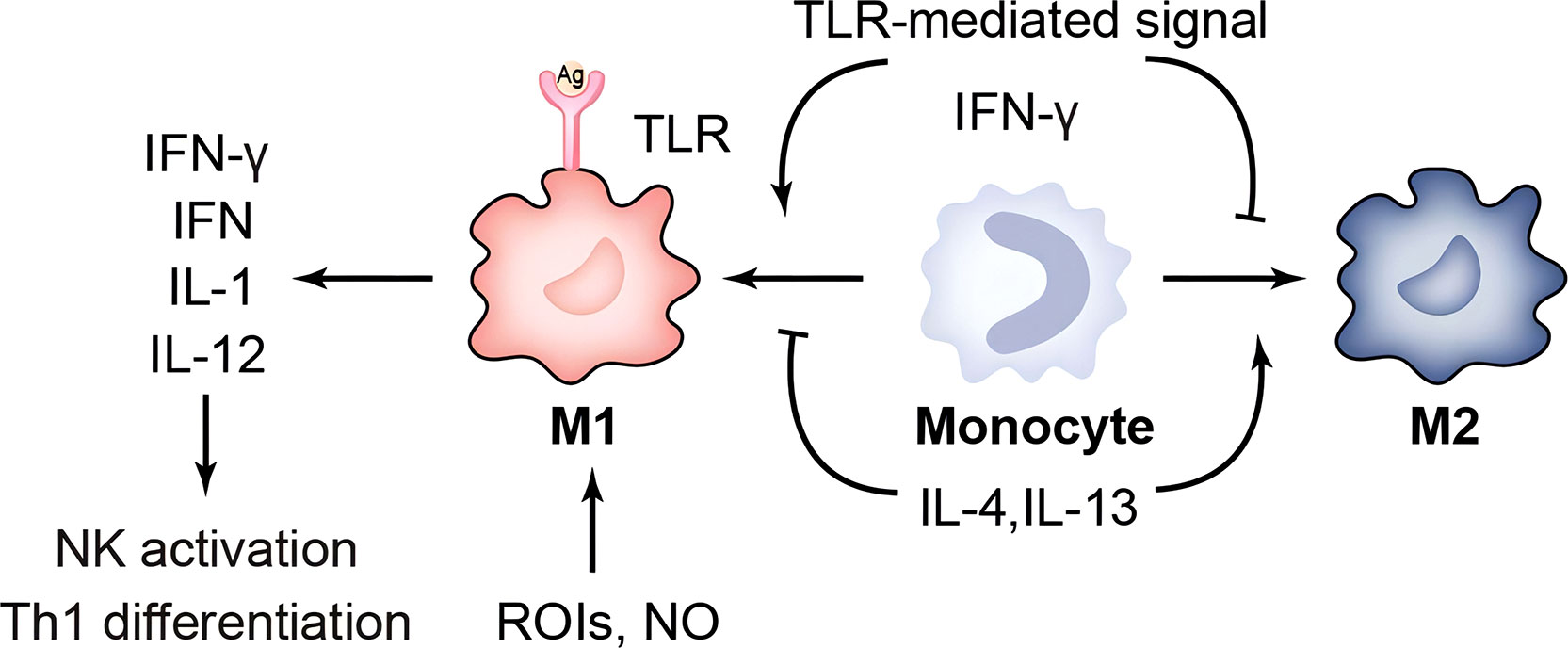
Figure 1 Immune response of macrophages. TLR, toll-like receptor; ROIs, reactive oxygen intermediates; IFN-γ, Interferon gamma; IL-1, interleukin-1.
Macrophages are divided into two phenotypes, M1 and M2: IFN-γ/LPS-induced M1 and selectively activated, IL-4-induced M2. M1 has a strong capacity to engulf and kill bacteria that triggers an inflammatory response by releasing chemokines and pro-inflammatory cytokines. And M2 cells inhibit inflammatory responses or participate in tissue repair and fibrosis. Buchacher T et al. showed that positive Chlamydia pneumoniae (Cpn) lipopolysaccharide staining and quantification of 16 S rDNA were significantly higher in M2 than in M1. A large number of intact perinuclear inclusions were found in M2-type macrophages, whereas rupture of inclusion bodies occurred in M1-type macrophages (62). The Chlamydia muridarum and murine bone-marrow derived macrophages showed that although M1 could mediate IFN-γ to control infection, it could not eliminate intracellular chlamydia (63). This suggests that M2-type macrophages provide a better environment for Cpn to survive. It has been shown that Nutlin-3 inhibits Chlamydia abortus growth and affects TNF-α secretion in M1 in a dose-dependent manner (64). In addition to improving the survival environment within macrophages, another important escape mechanism is the regulation of apoptosis. Utilizing CRISPR/Cas9 technology, Amy T.Y. Yeung and his colleagues hypothesized that deletion of the IL-10/IL-10R signaling pathway may resist macrophage apoptosis and thus sustain chlamydial growth (65). In addition, Chlamydia is expelled from the host cell by extrusion release, forming an inclusion-like structure called extrusions. Extrusions retard macrophage killing and eventually release infectious EB from macrophages, which facilitates the spread of EB to more distant sites. Extrusions are also able to survive longer in the extracellular environment than free C. trachomatis EB (66). This chlamydia-specific cell exit is likely a self-interested ability shaped by thousands of years of host-pathogen interactions. In summary, chlamydial survival in macrophages is suitable but limited. The restricted growth pattern is often associated with lysosomal trafficking, perforin-2 release, and nutrient starvation. Further work is needed to investigate the mechanisms of chlamydia-macrophage interactions, as well as to confirm them in vivo. This may, in some ways, increase the clearance susceptibility of the macrophage and decrease its ability to assist in chlamydial transmission.
3.2 NE
Neutrophil (NE) is the most abundant innate immune cells and the first leukocytes recruited to infected tissues. Neutrophils kill microorganisms primarily by phagocytosis and activation of the NADPH oxidase machinery (67). When neutrophils encounter pathogens, they activate their bactericidal reservoir to produce reactive oxygen species (ROS) and neutrophil extracellular traps (NETs). NETs are DNA structures decorated with cytoplasm, granules and nucleoproteins, and they can capture microorganisms including bacteria, viruses and 50 species of fungi. A variety of microorganisms are known to activate neutrophils and induce the formation of NETs. However, in order to evade or survive in neutrophils, some pathogens (68), such as C. trachomatis, have evolved mechanisms to degrade extracellular chromatin traps by secreting effector nucleases and proteases. It has been shown that chlamydial protease-like activating factor (CPAF) directly affects PMN survival and that formyl peptide receptor 2 (FPR2) is a key target of CPAF. In addition, C. trachomatis is able to reverse the short-lived neutrophils and delay apoptosis by activating ERK1/2 and PI3K/Akt survival signaling pathways (69); a key protein in human intracellular defense against Chlamydia is the tryptophan-degrading enzyme indoleamine 2,3-dioxygenase (IDO), but its activation does not inhibit chlamydial survival in neutrophils (70).
In pelvic inflammatory disease caused by C. trachomatis, interleukin1α(IL-1α) and type I interferon released upon the death of infected inflammatory cells favored the pathogen over the host. Persistent infection and chronic antigenic stimulation impair the ability of T cells to produce IFN-γ, thereby attenuating the protective response of Th1 and Th2 (4). In addition, Chlamydia-infected neutrophils exhibit high levels of extracellular ATP, which can act as a damage-associated molecular pattern (DAMP) to activate submucosal macrophage NLRP3 inflammatory vesicles, thereby driving damaging immunopathology (71). To some extent, these immune evasion mechanisms suggest that C. trachomatis infection can be overcome by the use of CPAF inhibitors, vaccines with Th1-inducing adjuvants providing antigens, etc.
3.3 DC
Classical dendritic cell (cDC) is the bridge between innate and adaptive immunity, and it is currently recognized as the most powerful specialized antigen-presenting cell in the body. From immature DC (iDC) to mature DC, it becomes progressively less capable of antigen uptake and processing (phagocytosis) and progressively more capable of antigen presentation. DCs are the sole initiators of initial T-cell activation (72). C. trachomatis, as an exogenous antigen, is internalized and processed by the APC, then processed by the proteasome in the cytoplasm, and finally activates CD4+T lymphocytes mainly through MHC-II molecules on the surface of DC. DC cells with the efficient presentation of cross-antigens activate CD8+T lymphocytes via surface-expressed MHC class I molecules in response to innate antigens in cells already infected with C. trachomatis. Moreover, CD8α+ dendritic cells were found to present antigen to CD8+T cells and induce Th1 production in mice at a better level than CD8α- using the adoptive transfer method (73). It has been shown that C. trachomatis hijacks the DC endocytic recycling system by recruiting Rab proteins associated with the recycling pathway around the inclusions, which leads to adverse changes in MHC-I intracellular transport (54).
As previously described, Chlamydia exits infected host cells via extrusion formation, which confers an infectious advantage to Chlamydia and promotes its survival within macrophages (74). It was found that the extrusion formation also prolonged bacterial survival within dendritic cells and altered the initial innate immune signaling of dendritic cells. Protective immunity against Chlamydia is primarily driven by IFN-γ producing T cells. Unlike macrophages, phagocytosis of extrudates leads to rapid apoptosis of DCs through a caspase 3/7-dependent mechanism, whereas exposure to free Chlamydia does not undergo apoptosis, directly preventing the initiation of adaptive immune responses (75). In 2017, Khamia Ryans et al. demonstrated that alpha enolase (ENO1) deficiency affects DC survival, maturation, and antigen presentation characteristics in vivo and in vitro experiments. Thus, modulation of ENO1 facilitates the enhancement of DC function, which could be used as an immunotherapeutic strategy to generate long-term immunity against Chlamydia-induced tubal lesions (76). In fact, using a combination of affinity chromatography and tandem mass spectrometry, it has previously been shown that DCs transferred with the chlamydial antigenic peptide Ags: PmpG125-500, RplF, or PmpE/F-225-575 overtly induced significant protective immunity against pulmonary and genital tract infections. Among them, PmpG125-500 is the most immunoprotective and could be a candidate for T cell protein-based subunit vaccines (77).
3.4 Other granulocytes
In addition to neutrophils, granulocytes include eosinophils, basophils and mast cells (Figure 2), which are rare in the study of C. trachomatis now. Eosinophils have unique chemokine receptors on their surface, and Vicetti Miguel et al. revealed in mouse experiments that IL-4-producing eosinophils promoted endometrial stromal cell (ESC) proliferation during primary Chlamydia infection, thereby repairing endometrial tissue induced by genital pathogens (78). In addition, a recent pathological study on endometriosis and endometritis revealed a significant increase in not only eosinophils but also basophils (79). And statistically, histological analysis of sexually transmitted infections caused by C. trachomatis in gay men presenting with proctitis revealed a scarcity of eosinophils (80). Mast cells are less frequent in studies of C. trachomatis infections and more frequent in allergic diseases. In a recent study of Chlamydia pneumoniae (C. pneumoniae), Norika Chiba et al. constructed mast cell-deficient mice (Wsh) and then found that the lungs of Wsh mice were more efficient at clearing Chlamydia pathology than those of WT mice. This suggests that the presence of mast cells exacerbates the inflammatory response and increases mortality, because it promotes the infiltration of immune cells into the air and provides a more favorable environment for C. pneumoniae to multiply (81).
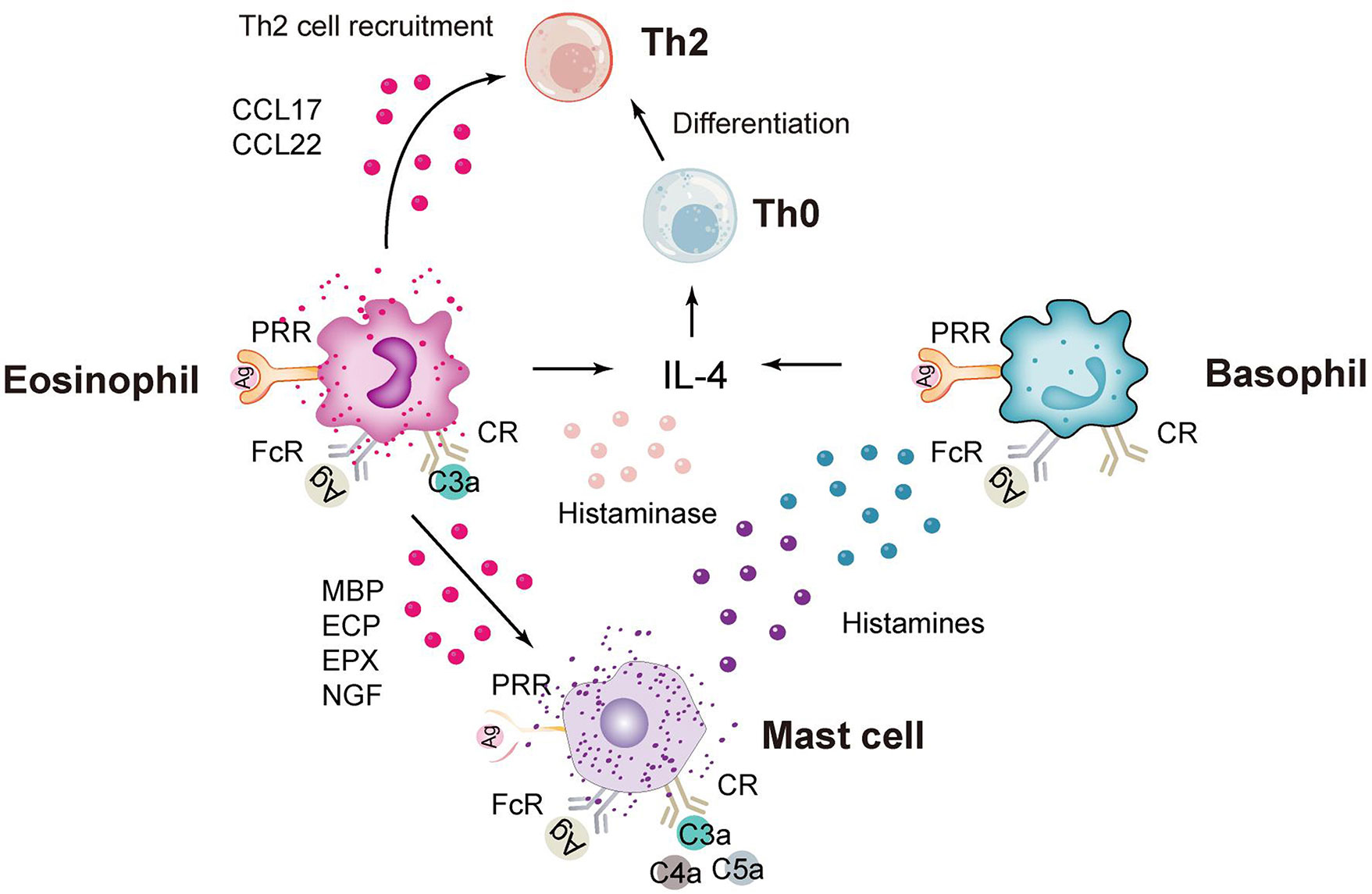
Figure 2 Immune response of eosinophils, basophils and mast cells. CCL17, chemokine (C-C motif) ligand 17; PRR, pattern recognition receptor; FcR, Fc receptor; CR, complement receptor; MBP, major basic protein; ECP, eosinophilic cationic protein; ECP, eosinophilic peroxidase; NGF, nerve growth factor.
4 Evasion of innate lymphoid cells [ILCs]
4.1 NKs and ILCs
The predominant innate lymphoid-like cells are natural killer cells (NKs), which do not have specific antigen recognition receptors like TCR and BCR or pattern recognition receptors like Mφ and DC. In the case of infection, killer activated receptors (KAR) and killer inhibitory receptors (KIR) on the surface of NK cells bind to target cell surface ligands (MHC-I), resulting in the deletion or downregulation of MHC-I molecules and abnormal expression or up-regulation of non-MHC-I molecules. At this point, the inhibitory signal is absent and the activating signal predominates, thereby initiating killing (82). MHC-I is down-regulated, and MICA (MHC class I-related protein A) is up-regulated on cells infected by C. trachomatis, which can be recognized by NK cells. Moreover, specific combinations of NKG2D (NK cell-activated receptor) and MICA alleles may promote escape of C. trachomatis from NK cell-mediated immune responses more effectively than others in different individuals, which may be a factor contributing to individual prognostic differences in female genital tract infections (83).
In addition to NK, other innate lymphoid cells that do not express specific antigen receptors and whose activation does not depend on recognition of antibodies are named ILCs, which are classified as ILC-1, ILC-2, and ILC-3. ILCs have no direct killing capacity, and their response depends on the activation of cytokines released by other cells, as they have only cytokine receptors. Activated ILCs also release corresponding cytokines to amplify the intensity of the previous immune response to perform an immunological function (84). Both ILC-1 and ILC-3 can secrete IFN-γ. IFN-γ produced by a variety of cells is known to be effective in clearing different sites or species of Chlamydia. IFN-γ+ILC-3 plays an important role in regulating colonization of the colon by Chlamydia muridarum and inhibiting endometrial chlamydial infection. IFN-γ produced by circulating cells such as NK cells and NKT cells prevents the spread of Chlamydia (85). Hong Xu and colleagues demonstrated that mouse ILCs significantly promote endometrial innate immunity in adaptive immunodeficient mice through interferon-dependent mechanisms, and that the role of ILC-3 is more important (86) in mice with C. trachomatis inoculation.
In addition to the frequently studied models of reproductive tract infection, gastrointestinal C. trachomatis is currently considered a seed bank for recurrent reproductive tract infections. Koprivsek et al. suggested in a murine model of Chlamydia muridarum-induced gastrointestinal infection that Chlamydia could evade IFN-γ from ILC-3 but not NK and maintain its long-term colonization in the colon (87). Furthermore, IFN-γ induced downregulation of c-Myc, a key regulator of host cell metabolism, in a STAT1-dependent manner, suggesting that c-Myc expression rescued the survival of C. trachomatis. IFN-γ caused the persistence of epithelial Chlamydia through infiltrative secretion by T cells and NK cells, as found in a pathological study of trachoma scarring (88, 89). Concerning therapy, it has been documented that NK cells are potential therapeutic targets in inflammatory bowel disease (IBD), atherosclerosis (AS), pulmonary arterial hypertension (PAH), and other inflammatory diseases (31). “NK therapies” are known to be widely used in cancer treatment. A nanoparticles (NP) therapy against the C. trachomatis antigen: Nanoparticles composed of poly (D,L-lactide-co-glycolide) (PLGA) have attracted much attention because of their biodegradability, biocompatibility and good colloidal stability. Co-delivery of C. trachomatis MOMP and immunostimulant (IS) with PLGA particles can prevent systemic adverse effects of immune boosters and activate dendritic cells and natural killer cells, thus enhancing the therapeutic effect of antigen-loaded PLGA particles (90).
5 Evasion of innate-like lymphocytes [ILLs]
5.1 NKT and γδT
Both Natural killer T cells (NKT cells) and γδ T cells belong to the atypical T cell family, and antigen recognition by these atypical T cells is not restricted by MHC class I and class II molecules (91). NKT cells are lymphocytes that express both the NK cell surface marker CD56 (NK1.1 in mice) and the T cell surface marker TCRαβ-CD3 complex. NKT cells are usually divided into type I and type II. Type I NKT cells are called semi-invariant NKT cells (iNKT) and recognize glycolipid and lipid antigens presented to them by the CD1d molecule, which respond rapidly to danger signals and pro-inflammatory cytokines (92). However, C. trachomatis infection can downregulate the CD1d molecule in human penile urothelial cells, which is associated with the CPAF protein. Activated type I NKT can promote the maturation and differentiation of DCs into CD8+/CD103+ DCs, which can further activate T cell differentiation into functional Th1 or Tc1-like peptide antigen-specific T cells. Activated antigen-specific CD4+Th1 and CD8+Tc1 can suppress bacterial infection (93, 94). However, CD1d-restricted NKT cells can regulate the immune response to chlamydial infection and cause immunopathological damage. Recent studies have shown that wild-type (WT) female mice have a significantly higher chlamydial burden than CD1d-/-(NKT-deficient) mice, suggesting that NKT cells delay chlamydial clearance and exacerbate immunopathology such as tubal effusion and obstruction. In contrast, there is no significant difference in the severity or incidence of tubal effusion in Jα18-/-(iNKT-deficient) mice compared with WT controls. Thus, non-invariant NKT cells have an immunopathogenic role in urogenital tract chlamydial infection (95). This partly explains a seemingly contradictory earlier study that NK T cells triggered pathological Th2 responses during chlamydial infection (96). It has also been shown that the activation of NKT triggers a pathology also associated with disruption of the CXCL13-CXCR5 axis. In this process, activated NKT cells increase chronic inflammation in the upper genital tract of mice by secreting cytokines or chemokines to recruit neutrophils and dendritic cells (97). These results suggest that NK T cells show protective Th1 immunity and pathological Th2 immunity in their role against Chlamydia.
γδT cells are known to bridge the gap between innate and adaptive immunity. γδT cells represent a small proportion of the population and are mainly distributed in mucosal tissues such as the peritoneal cavity, intestine and lung. They are the first to be recruited for mucosal infections. Experiments have shown that IL-17A plays a protective role against Chlamydia pulmonary infection, and it is produced rapidly but transiently by γδT cells in the early stages and mainly by Th17 in the later stages. Although the depletion of γδT cells led to a decrease in CD80 expression and an increase in IL-10 production by DCs, it had little effect on IL-12 production. There was no effect on type 1 T-cell responses after γδT cells depletion. In contrast, the decrease in IL-1α was more pronounced, suggesting that γδT cells could play a supportive but non-essential role in host defense against Chlamydia pulmonary infection (98, 99).
5.2 B-1
B cells are divided into B-1 cells, which mediate the innate immune response, and B-2 cells, which mediate the adaptive immune response. Depending on the presence or absence of surface CD5 molecules, B-1 cells can be further subdivided into B-1a (CD5+) and B-1b (CD5-). B-1 is mainly found in the peritoneal cavity, pleural cavity and lamina propria of the intestine. Most of the B cells are restricted to B-1 cells, which can be activated by TI-Ag (bacterial LPS, etc.) and produce antibodies with low specificity and do not produce memory cells. Mouse B-1 cells are thought to be the primary producers of natural antibodies to IgM (100, 101), which are independent of foreign antigen stimulation and mediate mucosal immunity below the mucosal lamina propria. Furthermore, in response to antigenic stimulation, it is estimated that 50% of serum IgA and IgG3 are also derived from B-1a cells (101). Lack of BCR diversity on the surface of B-1 cells and reconstitution of IgM-BCR complexes may explain the antigen-specific responses of self-reactive B-1 cells in response to infection by various pathogens, including bacteria, viruses, fungi and parasites (102). At the onset of infection, B-1a cells also spontaneously secrete IL-10 stimulated by lipopolysaccharide (LPS), GM-CSF and IL-3 (103). These natural antibodies and cytokines can protect the host from infection or reduce bacterial burden. In addition, B-1a cells are efficient antigen-presenting cells that provide effective signals to T cells through the co-stimulatory molecule CD80/CD86, which is constitutively expressed on B-1a cells (104). B-1 cells play an important role in assisting M1-type macrophages in the killing of Encephalitozoon cuniculi and in reducing its immune escape mechanism (105). In conclusion, the study of trends in B-1 cells and inflammation may lead to a paradigm shift toward sustainable treatment of various inflammatory diseases (101). C. trachomatis infection is an inflammatory disease and it is known that Chlamydia can colonize the gastrointestinal tract for long periods of time (106). Although B-1 cells have been little studied in C. trachomatis, this may, provide new ideas for C. trachomatis research.
6 Conclusions and perspectives
Currently, the interaction between various innate immune cells and C. trachomatis is still a challenging research topic, and both compete and promote each other in the course of a long-term battle (Figure 3). The complex set of mechanisms involved in the killing of innate immune cells against Chlamydia infection and Chlamydia immune escape from host cells can be broadly classified as follows:
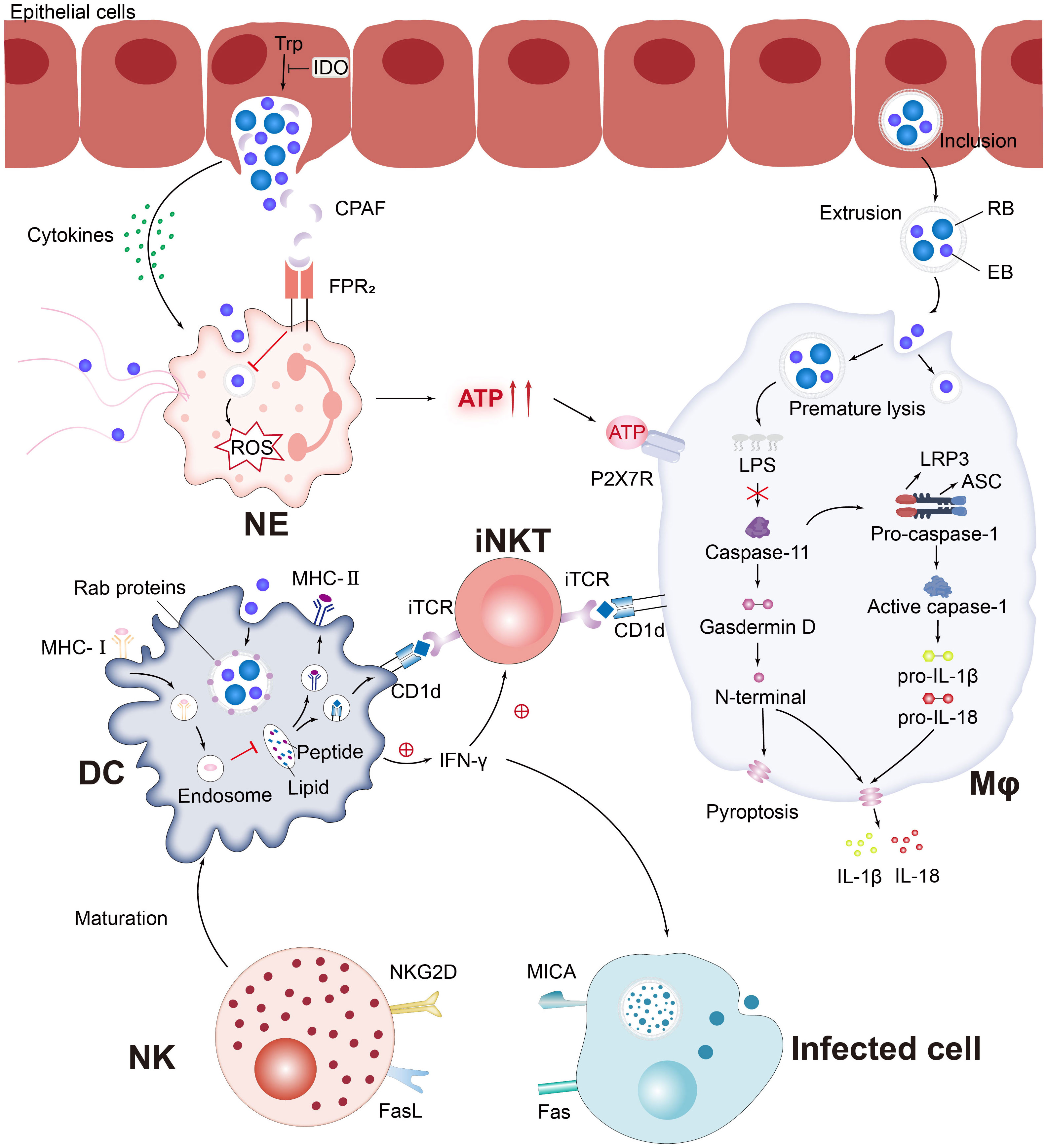
Figure 3 C. trachomatis evades the pursuit of innate immune cells. Pro-inflammatory cytokines secreted by C. trachomatis-infected cervical epithelial cells attract innate immune cells to the site of infection. Chlamydia protease-like activating factor (CPAF), which targets the cleaved NE surface receptor formyl peptide receptor 2 (FPR2), blocks the formation of neutrophil extracellular traps (NETs) and inhibits downstream reactive oxygen species (ROS) production, which paralyzes murine polymorphonuclear neutrophil (PMNs) activation. Chlamydia-infected NE produces elevated levels of extracellular ATP, adenosine triphosphate (ATP) that binds to P2X purinocreceptor 7 (P2X7R) and activates the NLRP3 inflammasome, thereby contributing to macrophage-associated immunopathology. Chlamydia is released from epithelial cells by extrusion and then forms extrusions that are taken up by Mφ. Interferon-induced GTPases are known to promote inclusions ubiquitination, leading to premature inclusion lysis. Bacterial lipid antigens are presented to iNKT cells via CD1d molecules on the surface of Mφ and DC. Activated NKT and NK promote DC maturation through the release of IFN-γ and positive feedback from cell-to-cell interactions. Rab proteins involved in the DC endocytic cycle are recruited around the inclusions and impede MHC-I intracellular trafficking. Notably, MICA upregulation occurs in parallel with MHC class I downregulation, affecting the sensitivity of C. trachomatis-infected cells to NK cell activity.
Firstly, resisting the phagocytic bactericidal effect of the phagocyte. C. trachomatis resists phagocytes through secreting specific effector proteins that evade the capture of NETs and prevent the activation of Mφ. In addition, the ability of nascent inclusions to evade fusion with phagocytic lysosomes is also related to these effector proteins, for example, IncE disrupts retromer and lysosome function by binding SNXs 5 and 6 (sorting nexin). However, the mechanism is not clear (38).
Secondly, blocking the activation of the lymphocytes. The persistent presence of C. trachomatis in innate immune cells may due to the pressure exerted by T lymphocyte-mediated immunity, which is the primary defense mechanism of the host against C. trachomatis infection. This prompts C. trachomatis to interfere with host antigen presentation by downregulating MHC molecules on the surface of target cells, including downregulation of MHC class I molecules that impede CD8+ CTL activation and downregulation of IFN-γ-induced MHC class II molecules that impede CD4+ T lymphocyte activation.
Thirdly, reproducing in immune cells (anti-apoptosis). In vitro experiments have shown that the surface structures of EB, like LPS and certain proteins, promote endocytosis of Chlamydia by susceptible cells (25). For example, the pore protein OmpA of the C. trachomatis outer membrane and the plasmid-encoded Pgp3 respectively inhibit apoptosis by targeting the pro-apoptotic proteins Bax and Bak (107), or block the activation of the apoptotic signaling pathway (108), which facilitates the pathogen to use the host cells for nutrition to multiply in and survival. In addition, C. trachomatis converts from RBs to AB (aberrant body) by changing the expression of HSP60, outer membrane proteins and LPS when it enters a crypt-infected state under the influence of external stresses (antibiotic use, iron deficiency or co-infection). This process is convenient for C. trachomatis to escape the anti-infective immune response of the host. ABs can be converted back to RBs in an active state, then RBs transformed into infectious EBs and released from the target cells when the external pressure is reduced or removed. This release mechanism is associated with the CPAF protein (32, 109).
Fourth, inducing the immune cells to apoptosis or directly killing immune cells. Host exposure to Chlamydia infection is known to exhibit high levels of metabolism, including sugar metabolism, nucleotide metabolism, etc., which is attributed to the reproduction-dependent nature of the bacteria. Moreover, Chlamydia infection causes excessive production of reactive oxygen species (ROS), causing oxidative DNA damage, resulting in single-strand breaks and even double-strand breaks, which can severely damage host cells. For example, Chlamydia not only causes macrophage foam (110), but also stimulates macrophages to produce TNF-α and induce apoptosis in neighboring T cells (111, 112). Hydrogen sulfide (H2S)-mammalian endogenous signaling gas transmitter is reported to exert protective effects on various innate immune cells against damage from ROS, immune or inflammatory hyperactivation, and also to control differentiation, maturation or polarization of immune cells (e.g. M2 polarization of macrophages) (113).
Nowadays, there are more studies on the interaction between C. trachomatis and classical innate immune cells, such as Mφ, NE and DC, but few studies on ILCs and ILLs. And the detailed aspects of how C. trachomatis evades innate immune cell pursuit need to be explored in depth by further techniques. Cellular immunotherapies have been reported to be emerging in the field of cancer (114), but this has rarely been studied in pathogenic infections. Therefore, an in-depth understanding of the interaction between Chlamydia and innate immune cells will provide further therapeutic interventions to combat this intractable epidemic.
Author contributions
XW: Writing – review & editing, Writing – original draft. HW: Writing – review & editing. CF: Writing – review & editing. ZL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 82272383, and 32070189), Hunan Provincial Natural Science Foundation of China (No.2021JJ30594), Scientific Research Project of Hunan Provincial Health Commission (No. C202304127239).
Acknowledgments
We thank the participation and attention to this research topics of all authors and readers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rodrigues R, Sousa C, Vale N. Chlamydia trachomatis as a current health problem: challenges and opportunities. Diagnostics (Basel Switzerland) (2022) 12(8):1795. doi: 10.3390/diagnostics12081795
2. Wolle MA, West SK. Ocular Chlamydia trachomatis infection: elimination with mass drug administration. Expert Rev anti-infective Ther (2019) 17(3):189–200. doi: 10.1080/14787210.2019.1577136
3. Mackern-Oberti JP, Motrich RD, Damiani MT, Saka HA, Quintero CA, Sánchez LR, et al. Male genital tract immune response against Chlamydia trachomatis infection. Reproduction (2017) 154(4):R99–r110. doi: 10.1530/rep-16-0561
4. Darville T. Pelvic inflammatory disease due to Neisseria gonorrhoeae and Chlamydia trachomatis: immune evasion mechanisms and pathogenic disease pathways. J Infect Dis (2021) 224(12 Suppl 2):S39–s46. doi: 10.1093/infdis/jiab031
5. Ling H, Luo L, Dai X, Chen H. Fallopian tubal infertility: the result of Chlamydia trachomatis-induced fallopian tubal fibrosis. Mol Cell Biochem (2022) 477(1):205–12. doi: 10.1007/s11010-021-04270-7
6. Arcia Franchini AP, Iskander B, Anwer F, Oliveri F, Fotios K, Panday P, et al. The role of Chlamydia trachomatis in the pathogenesis of cervical cancer. Cureus (2022) 14(1):e21331. doi: 10.7759/cureus.21331
7. Horne AW, Wheelhouse N, Horner PJ, Duncan WC. Association of past chlamydia trachomatis infection with miscarriage. JAMA network Open (2020) 3(10):e2018799. doi: 10.1001/jamanetworkopen.2020.18799
8. Zhong G. Chlamydia overcomes multiple gastrointestinal barriers to achieve long-lasting colonization. Trends Microbiol (2021) 29(11):1004–12. doi: 10.1016/j.tim.2021.03.011
9. Goulart ACX, Farnezi HCM, França J, Santos AD, Ramos MG, Penna MLF. Hiv, Hpv and Chlamydia trachomatis: impacts on male fertility. JBRA Assist Reprod (2020) 24(4):492–7. doi: 10.5935/1518-0557.20200020
10. Walpole GFW, Grinstein S, Westman J. The role of lipids in host-pathogen interactions. IUBMB Life (2018) 70(5):384–92. doi: 10.1002/iub.1737
11. Luo Z, Neville SL, Campbell R, Morey JR, Menon S, Thomas M, et al. Structure and metal binding properties of Chlamydia trachomatis Ytga. J bacteriology (2019) 202(1):E00580-19. doi: 10.1128/jb.00580-19
12. N'Gadjaga MD, Perrinet S, Connor MG, Bertolin G, Millot GA, Subtil A. Chlamydia trachomatis development requires both host glycolysis and oxidative phosphorylation but has only minor effects on these pathways. J Biol Chem (2022) 298(9):102338. doi: 10.1016/j.jbc.2022.102338
13. Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. Chlamydial intracellular survival strategies. Cold Spring Harbor Perspect Med (2013) 3(5):a010256. doi: 10.1101/cshperspect.a010256
14. Di Pietro M, Filardo S, Alfano V, Pelloni M, Splendiani E, Po A, et al. Chlamydia trachomatis elicits Tlr3 expression but disrupts the inflammatory signaling down-modulating Nfκb and Irf3 transcription factors in human Sertoli cells. J Biol regulators homeostatic Agents (2020) 34(3):977–86. doi: 10.23812/20-80-a-29
15. Le Negrate G, Krieg A, Faustin B, Loeffler M, Godzik A, Krajewski S, et al. Chladub1 of Chlamydia trachomatis suppresses Nf-Kappab activation and inhibits Ikappabalpha ubiquitination and degradation. Cell Microbiol (2008) 10(9):1879–92. doi: 10.1111/j.1462-5822.2008.01178.x
16. Christian J, Vier J, Paschen SA, Häcker G. Cleavage of the Nf-Kb family protein P65/rela by the chlamydial protease-like activity factor (Cpaf) impairs proinflammatory signaling in cells infected with Chlamydiae. J Biol Chem (2010) 285(53):41320–7. doi: 10.1074/jbc.M110.152280
17. Min S, He P, Zhou Q, Chen H. The dual role of cytokine responses to Chlamydia trachomatis infection in host pathogen crosstalk. Microbial pathogenesis (2022) 173(Pt A):105812. doi: 10.1016/j.micpath.2022.105812
18. Redgrove KA, McLaughlin EA. The role of the immune response in Chlamydia trachomatis infection of the male genital tract: A double-edged sword. Front Immunol (2014) 5:534. doi: 10.3389/fimmu.2014.00534
19. Lijek RS, Helble JD, Olive AJ, Seiger KW, Starnbach MN. Pathology after chlamydia trachomatis infection is driven by nonprotective immune cells that are distinct from protective populations. Proc Natl Acad Sci U S A (2018) 115(9):2216–21. doi: 10.1073/pnas.1711356115
20. Lausen M, Thomsen ME, Christiansen G, Karred N, Stensballe A, Bennike TB, et al. Analysis of complement deposition and processing on Chlamydia trachomatis. Med Microbiol Immunol (2021) 210(1):13–32. doi: 10.1007/s00430-020-00695-x
21. Moore T, Ananaba GA, Bolier J, Bowers S, Belay T, Eko FO, et al. Fc receptor regulation of protective immunity against Chlamydia trachomatis. Immunology (2002) 105(2):213–21. doi: 10.1046/j.0019-2805.2001.01354.x
22. Choroszy-Król IC, Frej-Mądrzak M, Jama-Kmiecik A, Bober T, Jolanta Sarowska J. Characteristics of the chlamydia trachomatis species - immunopathology and infections. Adv Clin Exp Med (2012) 21(6):799–808.
23. Haldar AK, Piro AS, Finethy R, Espenschied ST, Brown HE, Giebel AM, et al. Chlamydia trachomatis is resistant to inclusion Ubiquitination and associated host defense in gamma interferon-primed human epithelial cells. mBio (2016) 7(6):E01417-16. doi: 10.1128/mBio.01417-16
24. Walsh SC, Reitano JR, Dickinson MS, Kutsch M, Hernandez D, Barnes AB, et al. The bacterial effector gard shields Chlamydia trachomatis inclusions from Rnf213-mediated ubiquitylation and destruction. Cell Host Microbe (2022) 30(12):1671–84.e9. doi: 10.1016/j.chom.2022.08.008
25. Yang C, Briones M, Chiou J, Lei L, Patton MJ, Ma L, et al. Chlamydia trachomatis lipopolysaccharide evades the canonical and noncanonical inflammatory pathways to subvert innate immunity. mBio (2019) 10(2):E00595-19. doi: 10.1128/mBio.00595-19
26. Wang X, Rockey DD, Dolan BP. Chlamydia lipooligosaccharide has varied direct and indirect roles in evading both innate and adaptive host immune responses. Infection Immun (2020) 88(8):E00198-20. doi: 10.1128/iai.00198-20
27. Khatwani N, Romee R, Pillai AB. Editorial: innate immune cell therapy of cancer. Front Immunol (2022) 13:1004415. doi: 10.3389/fimmu.2022.1004415
28. Su H, Messer R, Whitmire W, Fischer E, Portis JC, Caldwell HD. Vaccination against Chlamydial Genital Tract Infection after Immunization with Dendritic Cells Pulsed Ex Vivo with Nonviable Chlamydiae. J Exp Med (1998) 188(5):809–18. doi: 10.1084/jem.188.5.809
29. Karunakaran KP, Rey-Ladino J, Stoynov N, Berg K, Shen C, Jiang X, et al. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol (Baltimore Md 1950) (2008) 180(4):2459–65. doi: 10.4049/jimmunol.180.4.2459
30. Rey-Ladino J, Ross AG, Cripps AW. Immunity, immunopathology, and human vaccine development against sexually transmitted Chlamydia trachomatis. Hum Vaccines immunotherapeutics (2014) 10(9):2664–73. doi: 10.4161/hv.29683
31. Xiao Q, Li X, Li Y, Wu Z, Xu C, Chen Z, et al. Biological drug and drug delivery-mediated immunotherapy. Acta Pharm Sin B (2021) 11(4):941–60. doi: 10.1016/j.apsb.2020.12.018
32. Elwell C, Mirrashidi K, Engel J. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol (2016) 14(6):385–400. doi: 10.1038/nrmicro.2016.30
33. Zhang JP, Stephens RS. Mechanism of C. Trachomatis attachment to eukaryotic host cells. Cell (1992) 69(5):861–9. doi: 10.1016/0092-8674(92)90296-o
34. Stelzner K, Vollmuth N, Rudel T. Intracellular lifestyle of Chlamydia trachomatis and host-pathogen interactions. Nat Rev Microbiol (2023) 21(7):448–62. doi: 10.1038/s41579-023-00860-y
35. Dolat L, Carpenter VK, Chen YS, Suzuki M, Smith EP, Kuddar O, et al. Chlamydia repurposes the actin-binding protein Eps8 to disassemble epithelial tight junctions and promote infection. Cell Host Microbe (2022) 30(12):1685–700.e10. doi: 10.1016/j.chom.2022.10.013
36. Cheong HC, Cheok YY, Chan YT, Tang TF, Sulaiman S, Looi CY, et al. Chlamydia trachomatis plasmid-encoding Pgp3 protein induces secretion of distinct inflammatory signatures from Hela cervical epithelial cells. BMC Microbiol (2023) 23(1):58. doi: 10.1186/s12866-023-02802-3
37. Lei L, Yang C, Patton MJ, Smelkinson M, Dorward D, Ma L, et al. A Chlamydial plasmid-dependent secretion system for the delivery of virulence factors to the host cytosol. mBio (2021) 12(3):e0117921. doi: 10.1128/mBio.01179-21
38. Bugalhão JN, Mota LJ. The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the Iceberg. Microbial Cell (Graz Austria) (2019) 6(9):414–49. doi: 10.15698/mic2019.09.691
39. Abdelrahman Y, Ouellette SP, Belland RJ, Cox JV. Polarized cell division of chlamydia trachomatis. PLoS Pathog (2016) 12(8):e1005822. doi: 10.1371/journal.ppat.1005822
40. Faris R, Andersen SE, McCullough A, Gourronc F, Klingelhutz AJ, Weber MM. Chlamydia trachomatis serovars drive differential production of proinflammatory cytokines and chemokines depending on the type of cell infected. Front Cell infection Microbiol (2019) 9:399. doi: 10.3389/fcimb.2019.00399
41. Mukura LR, Hickey DK, Rodriguez-Garcia M, Fahey JV, Wira CR. Chlamydia trachomatis regulates innate immune barrier integrity and mediates cytokine and antimicrobial responses in human uterine Ecc-1 epithelial cells. Am J Reprod Immunol (New York NY 1989) (2017) 78(6). doi: 10.1111/aji.12764
42. Valenti P, Rosa L, Capobianco D, Lepanto MS, Schiavi E, Cutone A, et al. Role of lactobacilli and lactoferrin in the mucosal cervicovaginal defense. Front Immunol (2018) 9:376. doi: 10.3389/fimmu.2018.00376
43. Deruaz M, Luster AD. Chemokine-mediated immune responses in the female genital tract mucosa. Immunol Cell Biol (2015) 93(4):347–54. doi: 10.1038/icb.2015.20
44. Barteneva N, Theodor I, Peterson EM, de la Maza LM. Role of neutrophils in controlling early stages of a Chlamydia trachomatis infection. Infection Immun (1996) 64(11):4830–3. doi: 10.1128/iai.64.11.4830-4833.1996
45. Tietzel I, Quayle AJ, Carabeo RA. Alternatively activated macrophages are host cells for Chlamydia trachomatis and reverse anti-chlamydial classically activated macrophages. Front Microbiol (2019) 10:919. doi: 10.3389/fmicb.2019.00919
46. Hook CE, Matyszak MK, Gaston JS. Infection of epithelial and dendritic cells by chlamydia trachomatis results in Il-18 and Il-12 production, leading to interferon-gamma production by human natural killer cells. FEMS Immunol Med Microbiol (2005) 45(2):113–20. doi: 10.1016/j.femsim.2005.02.010
47. Tseng CT, Rank RG. Role of Nk cells in early host response to chlamydial genital infection. Infection Immun (1998) 66(12):5867–75. doi: 10.1128/iai.66.12.5867-5875.1998
48. Jordan SJ, Gupta K, Ogendi BMO, Bakshi RK, Kapil R, Press CG, et al. The predominant Cd4(+) Th1 cytokine elicited to Chlamydia trachomatis infection in women is tumor necrosis factor alpha and not interferon gamma. Clin Vaccine Immunol CVI (2017) 24(4):E00010-17. doi: 10.1128/cvi.00010-17
49. Farris CM, Morrison SG, Morrison RP. Cd4+ T cells and antibody are required for optimal major outer membrane protein vaccine-induced immunity to Chlamydia muridarum genital infection. Infection Immun (2010) 78(10):4374–83. doi: 10.1128/iai.00622-10
50. Murthy AK, Li W, Chaganty BK, Kamalakaran S, Guentzel MN, Seshu J, et al. Tumor necrosis factor alpha production from Cd8+ T cells mediates oviduct pathological sequelae following primary genital chlamydia muridarum infection. Infection Immun (2011) 79(7):2928–35. doi: 10.1128/iai.05022-11
51. Vlcek KR, Li W, Manam S, Zanotti B, Nicholson BJ, Ramsey KH, et al. The contribution of chlamydia-specific Cd8+ T cells to upper genital tract pathology. Immunol Cell Biol (2016) 94(2):208–12. doi: 10.1038/icb.2015.74
52. Dong X, Zhang W, Hou J, Ma M, Zhu C, Wang H, et al. Chlamydial-secreted protease chlamydia high temperature requirement protein a (Chtra) degrades human cathelicidin ll-37 and suppresses its anti-chlamydial activity. Med Sci monitor (2020) 26:e923909. doi: 10.12659/msm.923909
53. Buckner LR, Lewis ME, Greene SJ, Foster TP, Quayle AJ. Chlamydia trachomatis infection results in a modest pro-inflammatory cytokine response and a decrease in T cell chemokine secretion in human polarized endocervical epithelial cells. Cytokine (2013) 63(2):151–65. doi: 10.1016/j.cyto.2013.04.022
54. Del Balzo D, Capmany A, Cebrian I, Damiani MT. Chlamydia trachomatis infection impairs Mhc-I intracellular trafficking and antigen cross-presentation by dendritic cells. Front Immunol (2021) 12:662096. doi: 10.3389/fimmu.2021.662096
55. Zhong G, Fan T, Liu L. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med (1999) 189(12):1931–8. doi: 10.1084/jem.189.12.1931
56. Rödel J, Grosse C, Yu H, Wolf K, Otto GP, Liebler-Tenorio E, et al. Persistent chlamydia trachomatis infection of hela cells mediates apoptosis resistance through a chlamydia protease-like activity factor-independent mechanism and induces high mobility group box 1 release. Infection Immun (2012) 80(1):195–205. doi: 10.1128/iai.05619-11
57. Luo F, Wen Y, Zhao L, Su S, Lei W, Chen L, et al. Lncrna Zeb1-As1/Mir-1224-5p / Map4k4 axis regulates mitochondria-mediated Hela cell apoptosis in persistent Chlamydia trachomatis infection. Virulence (2022) 13(1):444–57. doi: 10.1080/21505594.2022.2044666
58. Shu M, Bu J, Lei W, Chen L, Zhou Z, Lu C, et al. Pgp3 protein of chlamydia trachomatis inhibits apoptosis via Ho-1 upregulation mediated by Pi3k/Akt activation. Microbial pathogenesis (2023) 178:106056. doi: 10.1016/j.micpath.2023.106056
59. Jahnke R, Matthiesen S, Zaeck LM, Finke S, Knittler MR. Chlamydia trachomatis cell-to-cell spread through tunneling nanotubes. Microbiol Spectr (2022) 10(6):e0281722. doi: 10.1128/spectrum.02817-22
60. Lausen M, Christiansen G, Bouet Guldbæk Poulsen T, Birkelund S. Immunobiology of monocytes and macrophages during Chlamydia trachomatis infection. Microbes infection (2019) 21(2):73–84. doi: 10.1016/j.micinf.2018.10.007
61. Chen L, Liu X, Yu X, Ren R, Wang C, Zhao R, et al. Chlamydia muridarum infection of macrophages stimulates Il-1β Secretion and cell death via activation of caspase-1 in an Rip3-independent manner. BioMed Res Int (2017) 2017:1592365. doi: 10.1155/2017/1592365
62. Buchacher T, Ohradanova-Repic A, Stockinger H, Fischer MB, Weber V. M2 polarization of human macrophages favors survival of the intracellular pathogen chlamydia pneumoniae. PLoS One (2015) 10(11):e0143593. doi: 10.1371/journal.pone.0143593
63. Gracey E, Lin A, Akram A, Chiu B, Inman RD. Intracellular survival and persistence of Chlamydia muridarum is determined by macrophage polarization. PLoS One (2013) 8(8):e69421. doi: 10.1371/journal.pone.0069421
64. Diensthuber D, Simnacher U, Peters S, Walther P, Essig A, Hagemann JB. Clearing Chlamydia abortus infection in epithelial cells and primary human macrophages by use of antibiotics and the Mdm2-P53-inhibitor nutlin-3. Diagn Microbiol Infect Dis (2022) 103(3):115715. doi: 10.1016/j.diagmicrobio.2022.115715
65. Yeung ATY, Hale C, Lee AH, Gill EE, Bushell W, Parry-Smith D, et al. Exploiting induced pluripotent stem cell-derived macrophages to unravel host factors influencing Chlamydia trachomatis pathogenesis. Nat Commun (2017) 8:15013. doi: 10.1038/ncomms15013
66. Zuck M, Ellis T, Venida A, Hybiske K. Extrusions are phagocytosed and promote chlamydia survival within macrophages. Cell Microbiol (2017) 19(4). doi: 10.1111/cmi.12683
67. Kobayashi SD, DeLeo FR, Quinn MT. Microbes and the fate of neutrophils. Immunol Rev (2023) 314(1):210–28. doi: 10.1111/imr.13163
68. Kobayashi SD, Malachowa N, DeLeo FR. Neutrophils and bacterial immune evasion. J innate Immun (2018) 10(5-6):432–41. doi: 10.1159/000487756
69. Rajeeve K, Das S, Prusty BK, Rudel T. Chlamydia trachomatis paralyses neutrophils to evade the host innate immune response. Nat Microbiol (2018) 3(7):824–35. doi: 10.1038/s41564-018-0182-y
70. Virok DP, Tömösi F, Keller-Pintér A, Szabó K, Bogdanov A, Poliska S, et al. Indoleamine 2,3-dioxygenase cannot inhibit Chlamydia trachomatis growth in hl-60 human neutrophil granulocytes. Front Immunol (2021) 12:717311. doi: 10.3389/fimmu.2021.717311
71. Yang C, Lei L, Collins JWM, Briones M, Ma L, Sturdevant GL, et al. Chlamydia evasion of neutrophil host defense results in Nlrp3 dependent myeloid-mediated sterile inflammation through the purinergic P2x7 receptor. Nat Commun (2021) 12(1):5454. doi: 10.1038/s41467-021-25749-3
72. Vasilevsky S, Greub G, Nardelli-Haefliger D, Baud D. Genital Chlamydia trachomatis: understanding the roles of innate and adaptive immunity in vaccine research. Clin Microbiol Rev (2014) 27(2):346–70. doi: 10.1128/cmr.00105-13
73. Bilenki L, Wang S, Yang J, Fan Y, Jiao L, Joyee AG, et al. Adoptive transfer of Cd8alpha+ Dendritic cells (Dc) isolated from mice infected with Chlamydia muridarum are more potent in inducing protective immunity than Cd8alpha- Dc. J Immunol (Baltimore Md 1950) (2006) 177(10):7067–75. doi: 10.4049/jimmunol.177.10.7067
74. Zuck M, Sherrid A, Suchland R, Ellis T, Hybiske K. Conservation of extrusion as an exit mechanism for Chlamydia. Pathog Dis (2016) 74(7):FTW093. doi: 10.1093/femspd/ftw093
75. Sherrid AM, Hybiske K. Chlamydia trachomatis cellular exit alters interactions with host dendritic cells. Infection Immun (2017) 85(5):E00046-17. doi: 10.1128/iai.00046-17
76. Ryans K, Omosun Y, McKeithen DN, Simoneaux T, Mills CC, Bowen N, et al. The immunoregulatory role of alpha enolase in dendritic cell function during chlamydia infection. BMC Immunol (2017) 18(1):27. doi: 10.1186/s12865-017-0212-1
77. Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J Immunol (Baltimore Md 1950) (2009) 182(3):1602–8. doi: 10.4049/jimmunol.182.3.1602
78. Vicetti Miguel RD, Quispe Calla NE, Dixon D, Foster RA, Gambotto A, Pavelko SD, et al. Il-4-secreting eosinophils promote endometrial stromal cell proliferation and prevent chlamydia-induced upper genital tract damage. Proc Natl Acad Sci U S A (2017) 114(33):E6892–e901. doi: 10.1073/pnas.1621253114
79. Poli-Neto OB, Carlos D, Favaretto AJ, Rosa ESJC, Meola J, Tiezzi D. Eutopic endometrium from women with endometriosis and Chlamydial endometritis share immunological cell types and DNA repair imbalance: A transcriptome meta-analytical perspective. J Reprod Immunol (2021) 145:103307. doi: 10.1016/j.jri.2021.103307
80. Arévalo F, Rayme S, Zurita F, Ramírez R, Franco D, Montes P, et al. Immunohistochemical detection of Chlamydia trachomatis in sexually transmitted infectious proctitis. BMC Gastroenterol (2022) 22(1):171. doi: 10.1186/s12876-022-02233-w
81. Chiba N, Shimada K, Chen S, Jones HD, Alsabeh R, Slepenkin AV, et al. Mast cells play an important role in Chlamydia pneumoniae lung infection by facilitating immune cell recruitment into the airway. J Immunol (Baltimore Md 1950) (2015) 194(8):3840–51. doi: 10.4049/jimmunol.1402685
82. Zajonc DM. Unconventional peptide presentation by classical Mhc class I and implications for T and Nk cell activation. Int J Mol Sci (2020) 21(20):7561. doi: 10.3390/ijms21207561
83. Ibana JA, Aiyar A, Quayle AJ, Schust DJ. Modulation of mica on the surface of Chlamydia trachomatis-infected endocervical epithelial cells promotes Nk cell-mediated killing. FEMS Immunol Med Microbiol (2012) 65(1):32–42. doi: 10.1111/j.1574-695X.2012.00930.x
84. Ghaedi M, Takei F. Innate lymphoid cell development. J Allergy Clin Immunol (2021) 147(5):1549–60. doi: 10.1016/j.jaci.2021.03.009
85. Winner H, Friesenhahn A, Wang Y, Stanbury N, Wang J, He C, et al. Regulation of Chlamydial colonization by Ifnγ Delivered via distinct cells. Trends Microbiol (2023) 31(3):270–9. doi: 10.1016/j.tim.2022.09.002
86. Xu H, Su X, Zhao Y, Tang L, Chen J, Zhong G. Innate lymphoid cells are required for endometrial resistance to Chlamydia trachomatis infection. Infection Immun (2020) 88(7):E00152-20. doi: 10.1128/iai.00152-20
87. Koprivsek JJ, He Y, Song C, Zhang N, Tumanov A, Zhong G. Evasion of innate lymphoid cell-regulated gamma interferon responses by Chlamydia muridarum to achieve long-lasting colonization in mouse colon. Infection Immun (2020) 88(3):E00798-19. doi: 10.1128/iai.00798-19
88. Hu VH, Luthert PJ, Derrick T, Pullin J, Weiss HA, Massae P, et al. Immunohistochemical analysis of scarring trachoma indicates infiltration by natural killer and Undefined Cd45 negative cells. PLoS Negl Trop Dis (2016) 10(5):e0004734. doi: 10.1371/journal.pntd.0004734
89. Vollmuth N, Schlicker L, Guo Y, Hovhannisyan P, Janaki-Raman S, Kurmasheva N, et al. C-myc plays a key role in Ifn-Γ-induced persistence of Chlamydia trachomatis. eLife (2022) 11:e76721. doi: 10.7554/eLife.76721
90. Allahyari M, Mohit E. Peptide/protein vaccine delivery system based on plga particles. Hum Vaccines immunotherapeutics (2016) 12(3):806–28. doi: 10.1080/21645515.2015.1102804
91. Hayday AC. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity (2009) 31(2):184–96. doi: 10.1016/j.immuni.2009.08.006
92. Vogt S, Mattner J. Nkt cells contribute to the control of microbial infections. Front Cell infection Microbiol (2021) 11:718350. doi: 10.3389/fcimb.2021.718350
93. Shekhar S, Joyee AG, Yang X. Dynamics of Nkt-cell responses to chlamydial infection. Front Immunol (2015) 6:233. doi: 10.3389/fimmu.2015.00233
94. Zhao L, Yang X. Cross talk between natural killer T and dendritic cells and its impact on T cell responses in infections. Front Immunol (2022) 13:837767. doi: 10.3389/fimmu.2022.837767
95. Armitage CW, Carey AJ, Bryan ER, Kollipara A, Trim LK, Beagley KW. Pathogenic Nkt cells attenuate urogenital Chlamydial clearance and enhance infertility. Scandinavian J Immunol (2023) 97(5):e13263. doi: 10.1111/sji.13263
96. Bilenki L, Wang S, Yang J, Fan Y, Joyee AG, Yang X. Nk T cell activation promotes Chlamydia trachomatis infection in vivo. J Immunol (Baltimore Md 1950) (2005) 175(5):3197–206. doi: 10.4049/jimmunol.175.5.3197
97. Jiang J, Karimi O, Ouburg S, Champion CI, Khurana A, Liu G, et al. Interruption of cxcl13-cxcr5 axis increases upper genital tract pathology and activation of Nkt cells following chlamydial genital infection. PloS One (2012) 7(11):e47487. doi: 10.1371/journal.pone.0047487
98. Bai H, Gao X, Zhao L, Peng Y, Yang J, Qiao S, et al. Respective Il-17a production by Γδ T and Th17 cells and its implication in host defense against chlamydial lung infection. Cell Mol Immunol (2017) 14(10):850–61. doi: 10.1038/cmi.2016.53
99. Cong LH, Li T, Wang H, Wu YN, Wang SP, Zhao YY, et al. Il-17a-producing T cells exacerbate fine particulate matter-induced lung inflammation and fibrosis by inhibiting Pi3k/Akt/Mtor-mediated autophagy. J Cell Mol Med (2020) 24(15):8532–44. doi: 10.1111/jcmm.15475
100. Baumgarth N. B-1 cell heterogeneity and the regulation of natural and antigen-induced Igm production. Front Immunol (2016) 7:324. doi: 10.3389/fimmu.2016.00324
101. Aziz M, Holodick NE, Rothstein TL, Wang P. The role of B-1 cells in inflammation. Immunologic Res (2015) 63(1-3):153–66. doi: 10.1007/s12026-015-8708-3
102. Smith FL, Baumgarth N. B-1 cell responses to infections. Curr Opin Immunol (2019) 57:23–31. doi: 10.1016/j.coi.2018.12.001
103. Chin SS, Chorro L, Chan J, Lauvau G. Splenic innate B1 B cell plasmablasts produce sustained granulocyte-macrophage colony-stimulating factor and interleukin-3 cytokines during murine malaria infections. Infection Immun (2019) 87(12):E00482-19. doi: 10.1128/iai.00482-19
104. Rothstein TL, Griffin DO, Holodick NE, Quach TD, Kaku H. Human B-1 cells take the stage. Ann New York Acad Sci (2013) 1285:97–114. doi: 10.1111/nyas.12137
105. Pereira A, Alvares-Saraiva AM, Konno FTC, Spadacci-Morena DD, Perez EC, Mariano M, et al. B-1 cell-mediated modulation of M1 macrophage profile ameliorates microbicidal functions and disrupt the evasion mechanisms of Encephalitozoon cuniculi. PLoS Negl Trop Dis (2019) 13(9):e0007674. doi: 10.1371/journal.pntd.0007674
106. Kogut MH, Lee A, Santin E. Microbiome and pathogen interaction with the immune system. Poultry Sci (2020) 99(4):1906–13. doi: 10.1016/j.psj.2019.12.011
107. Waguia Kontchou C, Gentle IE, Weber A, Schoeniger A, Edlich F, Häcker G. Chlamydia trachomatis inhibits apoptosis in infected cells by targeting the pro-apoptotic proteins Bax and Bak. Cell Death differentiation (2022) 29(10):2046–59. doi: 10.1038/s41418-022-00995-0
108. Zou Y, Lei W, Su S, Bu J, Zhu S, Huang Q, et al. Chlamydia trachomatis plasmid-encoded protein Pgp3 inhibits apoptosis via the Pi3k-Akt-mediated Mdm2-P53 axis. Mol Cell Biochem (2019) 452(1-2):167–76. doi: 10.1007/s11010-018-3422-9
109. Yang Z, Tang L, Sun X, Chai J, Zhong G. Characterization of Cpaf critical residues and secretion during Chlamydia trachomatis infection. Infection Immun (2015) 83(6):2234–41. doi: 10.1128/iai.00275-15
110. Tumurkhuu G, Dagvadorj J, Porritt RA, Crother TR, Shimada K, Tarling EJ, et al. Chlamydia Pneumoniae hijacks a host autoregulatory Il-1β Loop to drive foam cell formation and accelerate atherosclerosis. Cell Metab (2018) 28(3):432–48.e4. doi: 10.1016/j.cmet.2018.05.027
111. Di Pietro M, Filardo S, De Santis F, Mastromarino P, Sessa R. Chlamydia pneumoniae and oxidative stress in cardiovascular disease: state of the art and prevention strategies. Int J Mol Sci (2014) 16(1):724–35. doi: 10.3390/ijms16010724
112. Rother M, Teixeira da Costa AR, Zietlow R, Meyer TF, Rudel T. Modulation of host cell metabolism by Chlamydia trachomatis. Microbiol Spectr (2019) 7(3). doi: 10.1128/microbiolspec.BAI-0012-2019
113. Dilek N, Papapetropoulos A, Toliver-Kinsky T, Szabo C. Hydrogen sulfide: an endogenous regulator of the immune system. Pharmacol Res (2020) 161:105119. doi: 10.1016/j.phrs.2020.105119
Keywords: Chlamydia trachomatis, innate immune cells, innate immunity, immune evasion, survival and growth
Citation: Wang X, Wu H, Fang C and Li Z (2024) Insights into innate immune cell evasion by Chlamydia trachomatis. Front. Immunol. 15:1289644. doi: 10.3389/fimmu.2024.1289644
Received: 06 September 2023; Accepted: 11 January 2024;
Published: 25 January 2024.
Edited by:
Lei Song, First Affiliated Hospital of Jilin University, ChinaReviewed by:
Karthika Rajeeve, Rajiv Gandhi Centre for Biotechnology, IndiaRoshan Pais, Rajiv Gandhi University of Health Sciences, India
Copyright © 2024 Wang, Wu, Fang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongyu Li, bHpoeTEwMjNAdXNjLmVkdS5jbg==
 Xinglv Wang
Xinglv Wang Hongrong Wu
Hongrong Wu