- 1Department of Laboratory Medicine, Linkou Chang Gung Memorial Hospital, Taoyuan, Taiwan
- 2Department of Medical Biotechnology and Laboratory Science, College of Medicine, Chang Gung University, Taoyuan, Taiwan
- 3Division of Rheumatology, Allergy, and Immunology, Linkou Chang Gung University and Memorial Hospital, Taoyuan, Taiwan
Introduction: Autoimmune diseases result from the loss of immune tolerance, and they exhibit complex pathogenic mechanisms that remain challenging to effectively treat. It has been reported that the altered expression levels of co-stimulatory/inhibitory molecules will affect the level of T/B cell activation and lead to the loss of immune tolerance.
Methods: In this study, we evaluated the gene polymorphisms of the ligand genes corresponding co-stimulatory system that were expressed on antigen-presenting cells (CD80, CD86, ICOSLG, and PDL1) from 60 systemic lupus erythematosus (SLE) patients and 60 healthy controls.
Results: The results showed that rs16829984 and rs57271503 of the CD80 gene and rs4143815 of the PDL1 gene were associated with SLE, in which the G-allele of rs16829984 (p=0.022), the A-allele of rs57271503 (p=0.029), and the GG and GC genotype of rs4143815 (p=0.039) may be risk polymorphisms for SLE.
Discussion: These SNPs are in the promoter and 3’UTR of the genes, so they may affect the transcription and translation activity of the genes, thereby regulating immune function and contributing to the development of SLE.
Introduction
Autoimmune diseases are one of the top ten major injuries in Taiwan, their pathogenesis remains unclear up till the present moment. The widely accepted notion is that these diseases are attributed to the over-activation of autoreactive immune cells and the loss of immune tolerance (1), leading to immune responses that target and attack one’s tissues (2).
Due to the advancement of medical technology, numerous single nucleotide polymorphisms (SNPs) have been discovered to be associated with autoimmune diseases (3). In the previous study, we investigated the association between systemic lupus erythematosus (SLE) and the SNPs of the genes that were involved in regulating T-cell activation, including cytotoxic T-lymphocyte-associated protein 4 (CTLA4), CD28, programmed cell death protein 1 (PDCD1), and inducible T cell costimulator (ICOS) (4). The aforementioned genes encode co-stimulatory/inhibitory molecules expressed on the surface of T cells, which provide the necessary second signal for T-cell activation when it binds with its ligand on antigen-presenting cells (APCs). However, the activation of immune cells requires interaction between receptors and ligands to exert their effects. Therefore, further understanding is needed to determine whether the gene polymorphisms of the ligands are indeed associated with autoimmune diseases.
The CD80 (B7-1) and CD86 (B7-2) genes, located on human chromosome 3, are ligand genes for CTLA4 and CD28. They are primarily expressed on the surface of APCs, such as dendritic cells, macrophages, and B cells, and they play an important role in the immune system by interacting with CD28 and CTLA4 (5). It has been shown that increased expression of CD80 and CD86 may be involved in the presentation of autoantigens and hyperactivation of T cells, in which the immune system attacks normal tissues and cells in the body, resulting in an exaggerated immune response in autoimmune diseases (6). In recent years, significant progress has been made in the research of receptor genes CD28 and CTLA4. However, it is still a poor understanding of their ligands genes, CD80 and CD86. In certain autoimmune diseases, such as rheumatoid arthritis (RA), autoimmune thyroid disease, Graves’ ophthalmopathy (GO), and autoimmune diabetes, studies have shown that excessive expression of CD80 and CD86 may lead to overactivation of T cells and enhanced inflammatory responses, thereby promoting the development of autoimmune pathologies (7–11).
Programmed death-ligand 1 (PD-L1) usually binds with its receptor, programmed cell death protein-1 (PD-1), to form the PD-L1/PD-1 signaling pathway, which plays an important role in immune tolerance and immune regulation (12). Studies have indicated that excessive expression of PD-L1 may be involved in the occurrence and progression of autoimmune diseases, such as RA, SLE, and autoimmune diabetes (13–15). Thus, the increased expression of PD-L1 may help immune cells escape immune surveillance and inhibit T-cell activation, thereby promoting the development of autoimmune diseases. Similarly, blocking the PD-L1/PD-1 signaling pathway is an important strategy in immunotherapy for treating autoimmune diseases. By suppressing the effect of PD-L1, the attack of T cells on pathological tissues can be restored, thus inhibiting autoimmune reactions (16).
The inducible T-cell co-stimulator ligand (ICOSL) gene (ICOSLG), located on chromosome 21, is a co-stimulatory molecule, that combines with ICOS on T cells, providing a second signal to activate and regulate the function of T cells. This is important for appropriate immune response and immune tolerance. Therefore, ICOSL may be an option for the treatment of autoimmune diseases. Studies have shown that the polymorphisms of ICOSLG were associated with an increased risk of RA, autoimmune thyroid diseases, and SLE (17–19).
Materials and methods
The aim of this study
In our previous study, we found that the gene polymorphisms of co-stimulatory molecules expression on the surface of T cells had an association with SLE. Here, we tried to find out the association between the gene polymorphisms of the ligands located on the APC that correspond to those co-stimulatory molecules and SLE.
The characteristics of participants
The Institutional Review Board of Chang Gung Memorial Hospital has reviewed and approved the study. The approval ID was 202002097B0. All study subjects signed informed consent and performed following relevant guidelines and regulations. In this case-control study, we evaluated 60 patients with SLE who were diagnosed based on the diagnostic criteria established by the American College of Rheumatology and 60 healthy controls without any immune abnormalities and not taking any immune-related drugs for analysis. The onset age of the SLE patients was 32.85 ± 1.76 years old, consisting of 52 females and 8 males. The average age of the SLE patients was 36.05 ± 0.87 years old, consisting of 48 females and 12 males.
Sample collection, DNA extraction, and sequencing
The remaining samples will be extracted with the QIAamp DNA Mini kit and the QIAamp RNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany) to extract DNA and RNA from the subjects. The purity and concentration of the DNA and RNA samples were evaluated by using a spectrophotometer to measure the absorbance at 260nm and 280nm. The PCR mixture contained 50ng of DNA, 7.5µl of Hotstar Taq DNA Polymerase (Qiagen GmbH, Hilden, Germany) or 2X Tag polymerase, each 1µl of forward and reverse primer (10μΜ), and 14.5µl of ddH2O. Then, the Big Dye Terminator Cycle Sequencing kit (Thermo Fisher, Waltham, Massachusetts, USA) and the ABI PRISM genetic analyzer (Thermo Fisher, Waltham, Massachusetts, USA) were used for direct sequencing according to the manufacturer’s instructions.
Primer design and candidate SNP selection
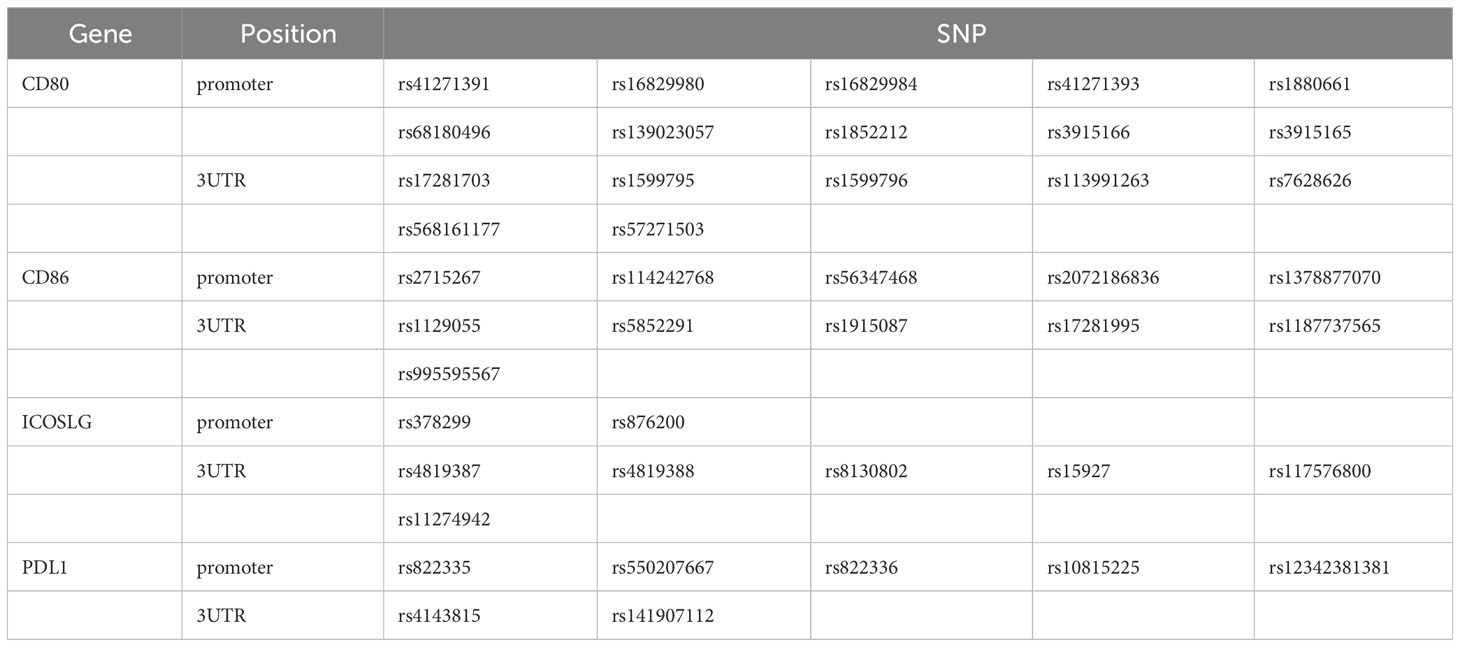
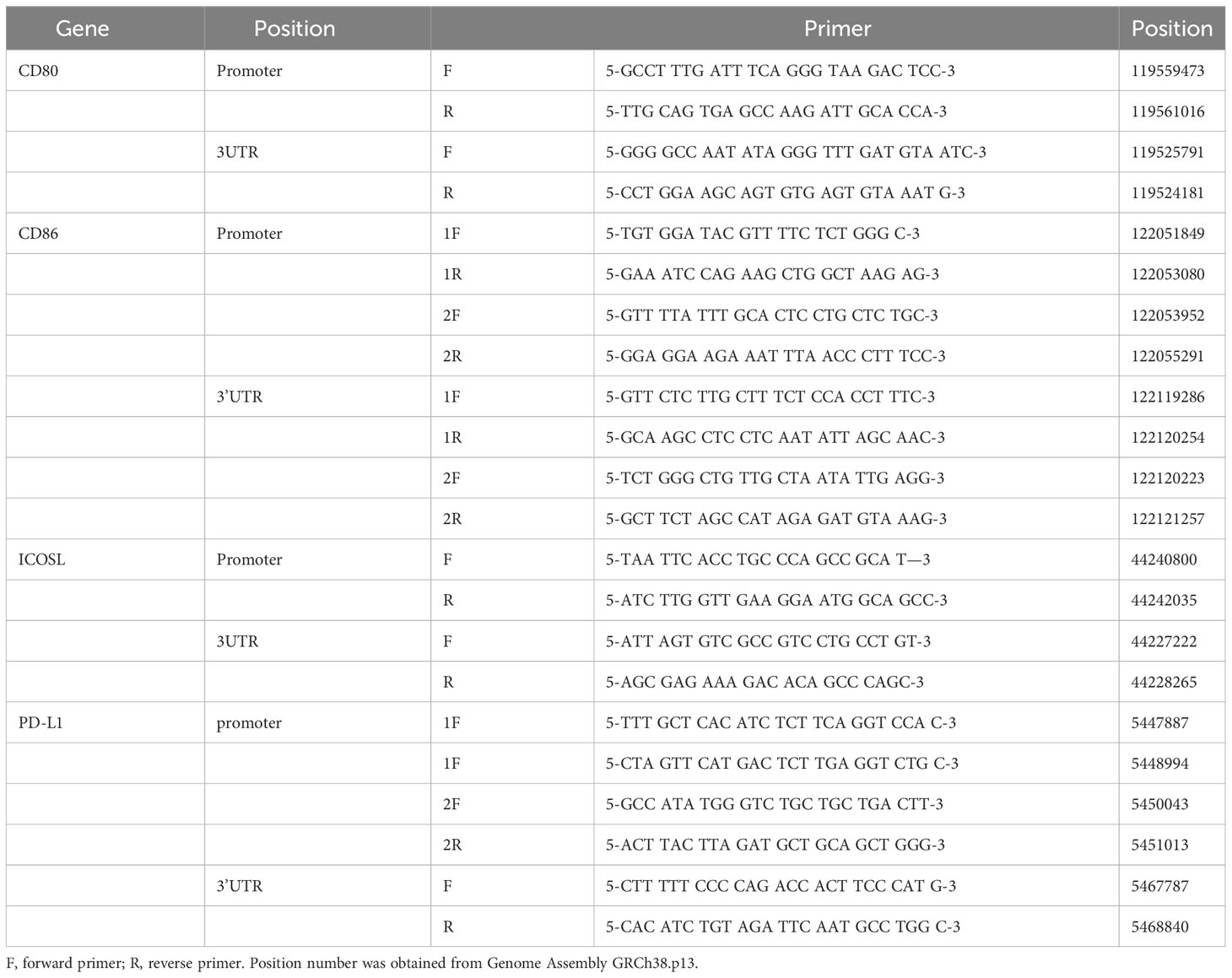
Primers were designed for the SNPs located in the promoter and 3’UTR of genes that have been reported to be associated with immune-related diseases. In this experiment, we focused on the CD80/CD86, PDL1, and ICOSLG, which were the corresponding ligands for CD28/CTLA4, PD1, and ICOS, respectively. There were 43 SNPs (Table 1) obtained by searching the literature from NCBI. The 11 pairs of primers (Table 2) were designed to amplify the genomic DNA fragments covering these 43 SNPs.
Statistical analysis
Statistical analysis was performed using the SPSS 17.0 software, employing chi-square tests or Fisher’s exact test. In the genetic model, the genotypes (AA, Aa, and aa) were evaluated, where the allele with a higher frequency was referred to as the major allele “A,” and the other allele was referred to as the minor allele “a”. All data were analyzed using the homozygous model (AA vs. aa), heterozygous model (AA vs. Aa), dominant model (AA vs. Aa + aa), recessive model (AA + Aa vs. aa), and additive model (AA vs. Aa vs. aa) to identify significant SNPs. The allele frequencies for each SNP in the control group were in Hardy-Weinberg equilibrium (HWE). The analysis of linkage disequilibrium (LD) is performed using Haploview 4.2 software (Broad Institute, Cambridge, MA, USA; http://www.broad.mit.edu/mpg/haploview) to assess the presence of linkage relationships and conduct haplotype analysis to identify the correct pathogenic genes. For multiple comparisons, the false discovery rate (FDR) Q values were calculated to evaluate the expected proportion of Type I errors.
Results
The analysis of genotype frequency
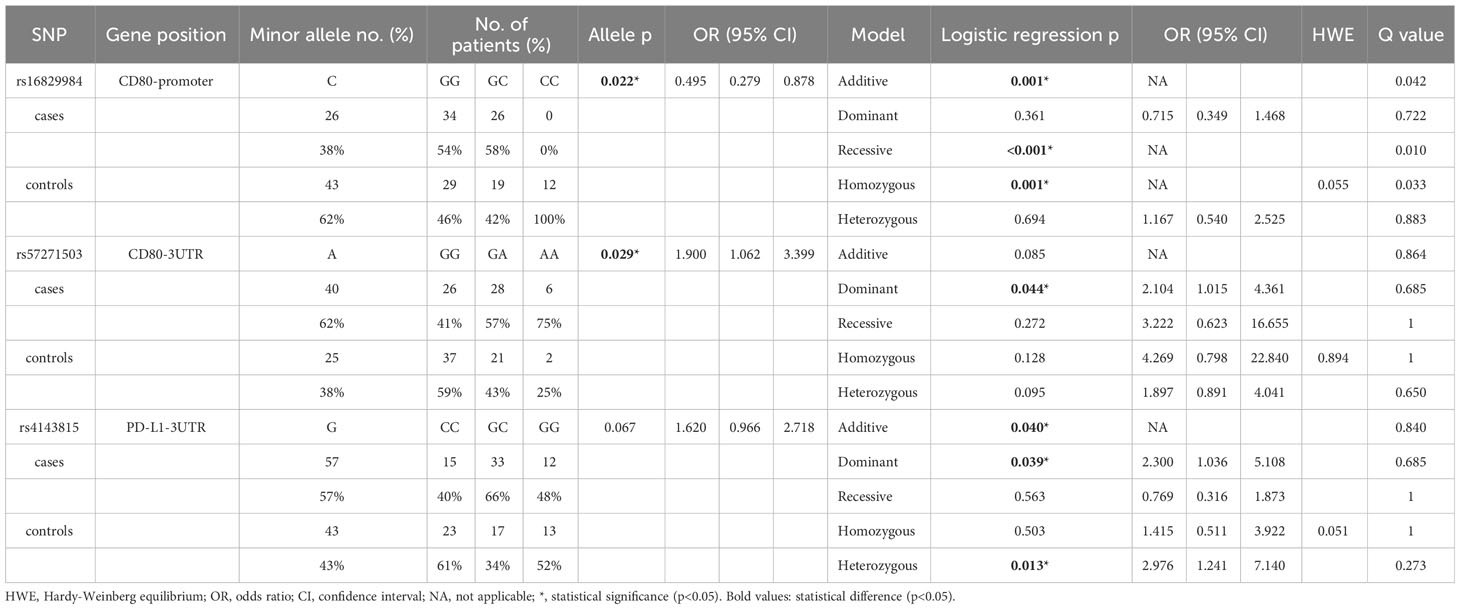
The SNP analysis revealed significant findings for two SNPs of CD80: rs16829984 located in the promoter region and rs57271503 located in the 3’UTR (Table 3). The genotype frequency of rs16829984 had a significant difference between SLE patients and healthy controls based on the additive model (GG vs. GC vs. CC, p=0.001), recessive model (GG + GC vs. CC, p<0.001), and homozygous model (GG vs. CC, p=0.001). Additionally, the allele frequency of rs16829984 was significantly different between SLE patients and healthy controls (G vs. C, p=0.022, OR=0.495, 95% CI. = 0.279-0.878), which indicated that the C-allele of rs16829984 had a decreased risk for SLE. On the other hand, the G-allele of rs16829984 was the risk allele for SLE. The polymorphisms of rs57271503 were associated with SLE based on allele frequency (G vs. A, p=0.029, OR=1.900, 95% CI. = 1.062-3.399) and dominant model (GG vs.GA + AA, p=0.044, OR=2.104, 95% CI. = 1.015-4.361), which was suggested individuals with at least one A-allele at rs57271503 would have a higher odds of SLE.
In addition, one SNPs of PDL1 had statistical significance: rs4143815 in the 3’UTR (Table 3). The genotype frequency of rs4143815 had a significant difference between SLE patients and healthy controls based on the additive model (CC vs. GC vs. GG, p=0.040), dominant model (CC vs. GC + GG, p=0.039, OR=2.300, 95% CI. = 1.036-5.108), and heterozygous model (CC vs. GC, p=0.013, OR=2.976, 95% CI. = 1.241-7.140), which was shown that individuals with GG and GC genotypes would have higher odds of SLE compared to CC genotype. The complete data is shown in Supplementary Table 1.
The analysis of linkage disequilibrium
Linkage disequilibrium (LD) analysis of each 4 genes was shown in Figures 1A–D. There were two haplotype blocks in the CD80 gene, in which one was composed of rs1599795 and rs1599796, and the other was composed of rs41271391, rs16829980, rs16829984, rs41271393, rs1880661, rs68180496, rs1852212, rs3915166, and rs3915165 (Figure 1), and there was one haplotype block in the ICOSLG, which was composed of rs4819388 and rs15927 (Figure 1C), while there was no haplotype in the CD86 and PDL1 gene.
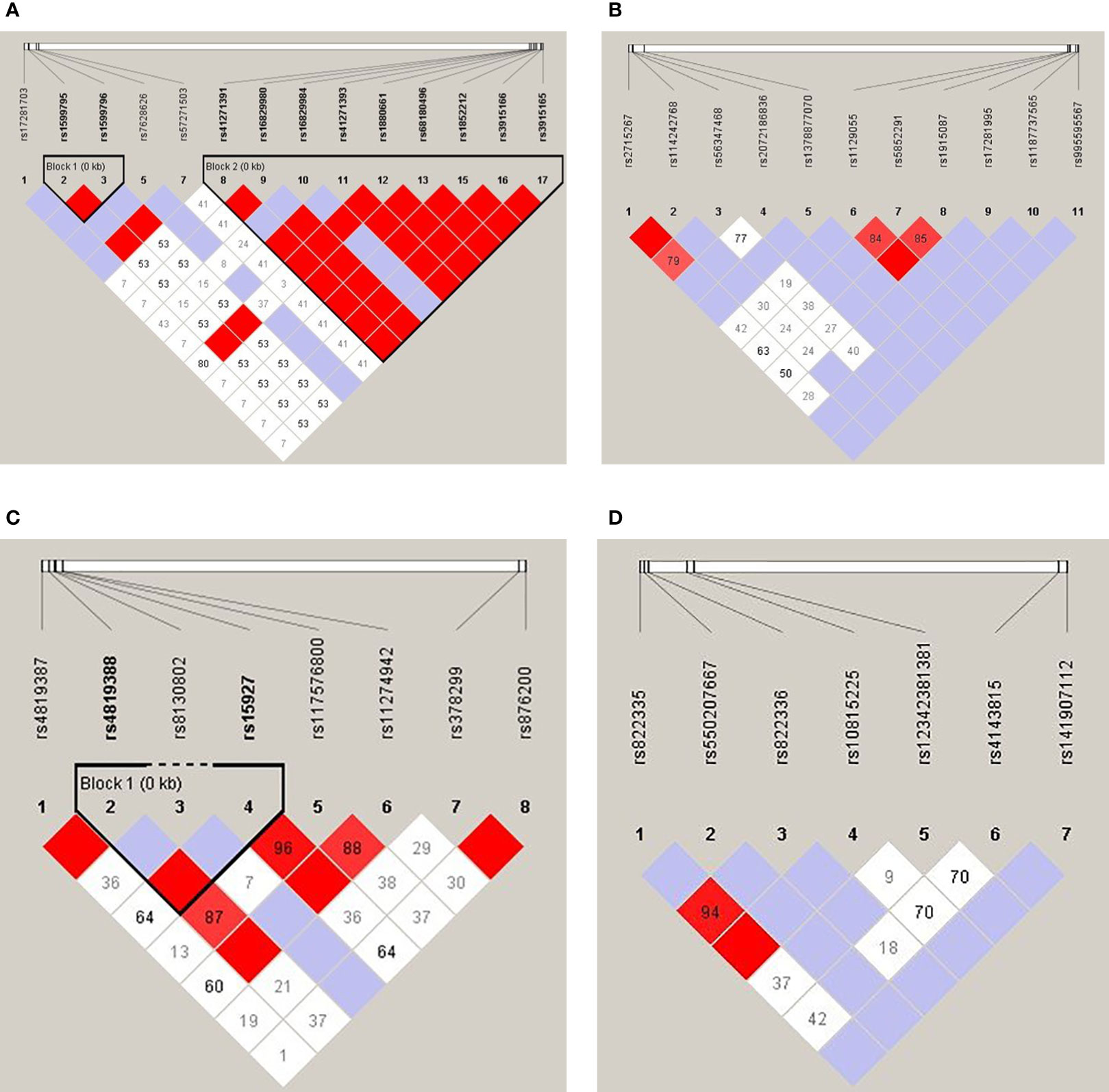
Figure 1 Linkage disequilibrium analysis of the CD80 (A), CD86 (B), ICOSLG (C), and PDL1 (D). The red color in the box indicates that two SNPs have strong linkage, whereas the less the linkage the closer to the white color of the box, and the light purple indicates no linkage.
Discussion
In this study, we compared the SNPs in the immune-related ligand genes between 60 patients with SLE and 60 healthy controls. Because the expression level of costimulatory molecules may affect the T-cell activation level (20) and genetic variations in the promoter and 3’ UTR of genes can influence gene expression (21, 22), this project focused on investigating the promoter region and 3’ UTR of the target genes. The analysis revealed significant findings for three SNPs: rs16829984 located in the promoter region of the CD80 gene, rs57271503 located in the 3’UTR of the CD80 gene, and rs4143815 in the 3’UTR of the PDL1 gene.
The CD80 gene is an important immunoregulatory gene that plays a crucial role in the human immune system. The protein encoded by the CD80 gene (CD80 molecule) is a co-stimulatory molecule, also known as B7.1, which is mainly expressed on the surface of antigen-presenting cells such as dendritic cells, macrophages, and B cells. After binding to CD28, the CD28-CD80 pathway provides an important costimulatory signal to activate and regulate the immune response of T cells. This signal is essential for T-cell activation, leading to T-cell proliferation, differentiation, and the release of immune factors to combat infection or pathological conditions. Thus, abnormal expression levels and functional alterations of CD80 are closely associated with various immune-related disorders, including autoimmune diseases, allergic reactions, and tumor immune evasion (23). Studies have shown that modulating the CD80 signaling pathway may hold significant importance in regulating immune system balance, treating certain diseases, and developing immunotherapies (24). This study found that the G-allele of rs16829984 and the A-allele of rs57271503 may be risk alleles for SLE. Additionally, the related studies have shown that rs16829984 was statistically associated with cervical cancer, while rs57271503 is statistically associated with rheumatoid arthritis (25, 26). Consequently, these genetic variations have susceptibility to diseases that may be due to the influence of gene transcription or translation. However, there is limited literature available on these two SNPs at present and the bio-function of these SNPs needs to be further explored. In addition, our published findings showed that several SNPs in the promoter region of CTLA4 were associated with SLE, while there was no susceptibility SNP in the CD28 gene (4). Integrating with the results of this project, it was suggested that the CD80/CTLA4 pathway, namely the inhibiting signal, may be more important for the development of SLE than the active pathway.
The PDL1 gene also plays a crucial role in the human immune system. PD-L1 is an immune checkpoint molecule that primarily functions to regulate the balance of immune responses and maintain the self-regulatory mechanisms of the immune system. The primary ligand for PD-L1 is the PD-1 receptor, which is typically expressed on activated T cells, B cells, and other immune cells. When PD-L1 binds to PD-1, it inhibits the activation and effector functions of T cells, which is crucial for maintaining immune tolerance and preventing excessive immune responses (27). In certain disease conditions, such as tumors and infections, tumor cells or pathogens may overexpress PD-L1. This overexpression can lead to the formation of immune evasion mechanisms, allowing the tumor or pathogen to evade attacks from the immune system (28–30). This study found that the rs4143815 variant located in the 3’UTR of the PD-L1 gene is statistically associated with SLE. Individuals with the GG or GC genotypes have a higher odds ratio of developing SLE. The polymorphism of this SNP was also associated with various diseases. For example, GG genotype was associated with poor prognosis in lung adenocarcinoma and squamous cell carcinoma (31), the CC genotype was associated with a two times higher risk of developing squamous cell lung cancer (32), G-allele was associated with poor outcomes in the early stage non-small cell lung cancer (33), the CC genotype has significantly reduced the risk of breast cancer (34), individuals carrying the C-allele of rs4143815 had less risk of type 1 diabetes (35), and so on. In addition, a functional study has already demonstrated that the rs4143815 C>G variant reduced transcriptional activity of the PDL1 gene (33), which indicated that the statistical association between rs4143815 and the diseases may be due to the gene expression level alteration caused by SNP variation, which in turn contributes to the development of the diseases.
Although the SNPs in non-coding gene regions do not directly affect protein function and structure, they play a crucial role in regulating gene expression (36). Gene variations in the promoter region may affect the initiation and activity of transcription through interactions with transcription factors, DNA methylation, or serving as targets for miRNA and other epigenetic mechanisms (21), and the sequence of the 3’UTR plays a crucial role in processes such as mRNA stability, localization, translational activity, pre-mRNA processing, and subsequent protein synthesis (22), leading to a significant association between these non-coding gene SNPs and numerous complex diseases. However, many SNP variations are neutral, namely they have no impact on biological function. Therefore, the functional effect of these disease susceptibility SNPs needs to be further explored in the future and it can be assisted by using silico analysis to predict transcription factor binding sites, performing expression quantitative trait loci analysis to understand the association between genetic variants and gene expression levels, and so on. At the gene level, it can only be used to predict the odds of disease. Moreover, the mRNA levels altered by non-coding gene SNPs are not a reliable indicator of corresponding protein abundance (37). To accurately reflect the actual protein expression, the use of flow cytometry to detect protein expression is also necessary.
According to current research literature, autoimmune diseases are usually complex and multifactorial, making it difficult to accurately determine their course and effectively treat them. Clinical experience has shown that the same medication may be effective for some individuals while ineffective for others, leading to time-consuming trial-and-error processes. Therefore, it is hoped that through this project, key mechanisms of autoimmune diseases can be identified, and personalized medicine can be achieved by selecting the most suitable medication based on genetic markers specific to each patient. Alternatively, the research findings from this project can provide new directions for the treatment of autoimmune diseases. It is worth noting that the mechanism of autoimmune diseases is a complex disease process that involves the interaction of numerous genes and immune regulatory factors. The role of the genes encoding co-stimulatory molecules and their ligands may only be the tip of the iceberg, and further research is still required to fully understand its specific role in autoimmune diseases.
In conclusion, the findings of this study revealed the association between gene polymorphisms of co-stimulatory ligand genes and SLE. However, these disease-associated SNP findings can only be used for statistical correlation to aid in clinical predictions and cannot explain direct causal relationships between SNPs and diseases. Therefore, in the future, it will be necessary to provide evidence of the correlation between gene variations and protein expression to elucidate the mechanisms by which the SNP affects the disease.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SS2137544506 and SS6403964567 - SS6403964612 (dbSNP; (https://www.ncbi.nlm.nih.gov/SNP/snp_viewBatch.cgi?sbid=1063574).
Ethics statement
The studies involving humans were approved by The Institutional Review Board of Chang Gung Memorial Hospital has reviewed and approved the study. The approval ID was 202002097B0 and 202102018B0C601. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
D-PC: Conceptualization, Funding acquisition, Writing – review & editing. W-TL: Data curation, Formal analysis, Methodology, Writing – original draft. F-PH: Data curation, Formal analysis, Methodology, Writing – original draft. K-HY: Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants to D-PC from the Chang Gung Memorial Hospital (CMRPG3N0021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1331796/full#supplementary-material
References
1. Bolon B. Cellular and molecular mechanisms of autoimmune disease. Toxicol Pathol (2012) 40(2):216–29. doi: 10.1177/0192623311428481
2. Lu P, Wang YL, Linsley PS. Regulation of self-tolerance by CD80/CD86 interactions. Curr Opin Immunol (1997) 9(6):858–62. doi: 10.1016/s0952-7915(97)80190-2
3. Wong M. What has happened in the last 50 years in immunology. J Paediatr Child Health (2015) 51(2):135–9. doi: 10.1111/jpc.12834
4. Chen DP, Lin WT, Yu KH. Investigation of the association between the genetic polymorphisms of the co-stimulatory system and systemic lupus erythematosus. Front Immunol (2022) 13:946456. doi: 10.3389/fimmu.2022.946456
5. Saito T. Molecular dynamics of co-signal molecules in T-cell activation. Adv Exp Med Biol (2019) 1189:135–52. doi: 10.1007/978-981-32-9717-3_5
6. Goronzy JJ, Weyand CM. T-cell co-stimulatory pathways in autoimmunity. Arthritis Res Ther (2008) 10 Suppl 1(Suppl 1):S3. doi: 10.1186/ar2414
7. Sfikakis PP, Via CS. Expression of CD28, CTLA4, CD80, and CD86 molecules in patients with autoimmune rheumatic diseases: implications for immunotherapy. Clin Immunol Immunopathol (1997) 83(3):195–8. doi: 10.1006/clin.1997.4368
8. Lorenzetti R, Janowska I, Smulski CR, Frede N, Henneberger N, Walter L, et al. Abatacept modulates CD80 and CD86 expression and memory formation in human B-cells. J Autoimmun (2019) 101:145–52. doi: 10.1016/j.jaut.2019.04.016
9. Watanabe A, Inoue N, Watanabe M, Yamamoto M, Ozaki H, Hidaka Y, et al. Increases of CD80 and CD86 expression on peripheral blood cells and their gene polymorphisms in autoimmune thyroid disease. Immunol Invest (2020) 49(1-2):191–203. doi: 10.1080/08820139.2019.1688343
10. Liao WL, Chen RH, Lin HJ, Liu YH, Chen WC, Tsai Y, et al. The association between polymorphisms of B7 molecules (CD80 and CD86) and Graves' ophthalmopathy in a Taiwanese population. Ophthalmology (2011) 118(3):553–7. doi: 10.1016/j.ophtha.2010.07.021
11. Zhang P, Yang CL, Du T, Liu YD, Ge MR, Li H, et al. Diabetes mellitus exacerbates experimental autoimmune myasthenia gravis via modulating both adaptive and innate immunity. J Neuroinflamm (2021) 18(1):244. doi: 10.1186/s12974-021-02298-6
12. Yi M, Zheng X, Niu M, Zhu S, Ge H, Wu K. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer (2022) 21(1):28. doi: 10.1186/s12943-021-01489-2
13. Hellbacher E, Sundström C, Molin D, Baecklund E, Hollander P. Expression of PD-1, PD-L1, and PD-L2 in lymphomas in patients with pre-existing rheumatic diseases-a possible association with high rheumatoid arthritis disease activity. Cancers (Basel) (2022) 14(6):1509. doi: 10.3390/cancers14061509
14. Liao W, Zheng H, Wu S, Zhang Y, Wang W, Zhang Z, et al. The systemic activation of programmed death 1-PD-L1 axis protects systemic lupus erythematosus model from nephritis. Am J Nephrol (2017) 46(5):371–9. doi: 10.1159/000480641
15. Quandt Z, Young A, Anderson M. Immune checkpoint inhibitor diabetes mellitus: a novel form of autoimmune diabetes. Clin Exp Immunol (2020) 200(2):131–40. doi: 10.1111/cei.13424
16. Hosseinzadeh R, Feizisani F, Shomali N, Abdelbasset WK, Hemmatzadeh M, Gholizadeh Navashenaq. J, et al. PD-1/PD-L1 blockade: Prospectives for immunotherapy in cancer and autoimmunity. IUBMB Life (2021) 73(11):1293–306. doi: 10.1002/iub.2558
17. Nanki T, Takada K, Komano Y, Morio T, Kanegane H, Nakajima A, et al. Chemokine receptor expression and functional effects of chemokines on B cells: implication in the pathogenesis of rheumatoid arthritis. Arthritis Res Ther (2009) 11(5):R149. doi: 10.1186/ar2823
18. Yoshie N, Watanabe M, Inoue N, Kawaguchi H, Hidaka Y, Iwatani Y. Association of polymorphisms in the ICOS and ICOSL genes with the pathogenesis of autoimmune thyroid diseases. Endocr J (2016) 63(1):61–8. doi: 10.1507/endocrj.EJ15-0435
19. Her M, Kim D, Oh M, Jeong H, Choi I. Increased expression of soluble inducible costimulator ligand (ICOSL) in patients with systemic lupus erythematosus. Lupus (2009) 18(6):501–7. doi: 10.1177/0961203308099176
20. Podojil JR, Miller SD. Molecular mechanisms of T-cell receptor and costimulatory molecule ligation/blockade in autoimmune disease therapy. Immunol Rev (2009) 229(1):337–55. doi: 10.1111/j.1600-065X.2009.00773.x
21. Vohra M, Sharma AR, Prabhu BN, Rai PS. SNPs in sites for dna methylation, transcription factor binding, and miRNA Targets leading to allele-specific gene expression and contributing to complex disease risk: A systematic review. Public Health Genomics (2020) 23(5-6):155–70. doi: 10.1159/000510253
22. Mayr C. What are 3' UTRs doing? Cold Spring Harb Perspect Biol (2019) 11(10):a034728. doi: 10.1101/cshperspect.a034728
23. Manzoor AM. Chapter 1 - Introduction to costimulation and costimulatory molecules. In: Manzoor AM, editor. developing costimulatory molecules for immunotherapy of diseases. Elsevier Publishers USA: Academic Press (2015). p. 1–43. doi: 10.1016/B978-0-12-802585-7.00001-7
24. Manzoor AM. Chapter 3 - Costimulation immunotherapy in infectious diseases. In: Manzoor AM, editor. developing costimulatory molecules for immunotherapy of diseases. Elsevier Publishers USA: Academic Press (2015). p. 83–129. doi: 10.1016/B978-0-12-802585-7.00003-0
25. Li CY, Yan ZL, Shi L, Yang HY, Hong C, Yu JK, et al. The association between the SNPs of CD80 gene and the initiation and development of cervical cancer in a Yunnan Han population. J Immunol (2017) 1:46–53.
26. Marquez Pete N, Maldonado Montoro MDM, Pérez Ramírez C, Sánchez Martín A, Martínez de la Plata JE, Martínez Martínez F, et al. Impact of single-nucleotide polymorphisms of CTLA-4, CD80 and CD86 on the effectiveness of abatacept in patients with rheumatoid arthritis. J Pers Med (2020) 10(4):220. doi: 10.3390/jpm10040220
27. Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev (2010) 236:219–42. doi: 10.1111/j.1600-065X.2010.00923.x
28. Zou J, Wu D, Li T, Wang X, Liu Y, Tan S. Association of PD-L1 gene rs4143815 C>G polymorphism and human cancer susceptibility: A systematic review and meta-analysis. Pathol Res Pract (2019) 215(2):229–34. doi: 10.1016/j.prp.2018.12.002
29. Hashemi M, Karami S, Sarabandi S, Moazeni-Roodi A, Małecki A, Ghavami S, et al. Association between PD-1 and PD-L1 polymorphisms and the risk of cancer: A meta-analysis of case-control studies. Cancers (Basel) (2019) 11(8):1150. doi: 10.3390/cancers11081150
30. Zhang W, Song Y, Zhang X. Relationship of programmed death-1 (PD-1) and programmed death ligand-1 (PD-L1) polymorphisms with overall cancer susceptibility: An updated meta-analysis of 28 studies with 60 612 subjects. Med Sci Monit (2021) 27:e932146. doi: 10.12659/MSM.932146
31. Yeo MK, Choi SY, Seong IO, Suh KS, Kim JM, Kim KH. Association of PD-L1 expression and PD-L1 gene polymorphism with poor prognosis in lung adenocarcinoma and squamous cell carcinoma. Hum Pathol (2017) 68:103–11. doi: 10.1016/j.humpath.2017.08.016
32. Moksud N, Wagner M, Pawełczyk K, Porębska I, Muszczyńska-Bernhard B, Kowal A, et al. Common inherited variants of PDCD1, CD274 and HAVCR2 genes differentially modulate the risk and prognosis of adenocarcinoma and squamous cell carcinoma. J Cancer Res Clin Oncol (2023) 149(9):6381–90. doi: 10.1007/s00432-023-04602-8
33. Lee SY, Jung DK, Choi JE, Jin CC, Hong MJ, Do SK, et al. Functional polymorphisms in PD-L1 gene are associated with the prognosis of patients with early stage non-small cell lung cancer. Gene (2017) 599:28–35. doi: 10.1016/j.gene.2016.11.007
34. Karami S, Sattarifard H, Kiumarsi M, Sarabandi S, Taheri M, Hashemi M, et al. Evaluating the possible association between PD-1 (rs11568821, rs2227981, rs2227982) and PD-L1 (rs4143815, rs2890658) polymorphisms and susceptibility to breast cancer in a sample of southeast Iranian women. Asian Pac J Cancer Prev (2020) 21(10):3115–23. doi: 10.31557/APJCP.2020.21.10.3115
35. Qian C, Guo H, Chen X, Shi A, Li S, Wang X, et al. Association of PD-1 and PD-L1 genetic polymorphyisms with type 1 diabetes susceptibility. J Diabetes Res (2018) 2018:1614683. doi: 10.1155/2018/1614683
36. Yang W, Zhang T, Song X, Dong G, Xu L, Jiang F. SNP-target genes interaction perturbing the cancer risk in the post-GWAS. Cancers (Basel) (2022) 14(22):5636. doi: 10.3390/cancers14225636
Keywords: systemic lupus erythematosus, promoter, 3 prime untranslated region, costimulatory molecules, immune regulatory genes
Citation: Chen D-P, Lin W-T, Hsu F-P and Yu K-H (2024) The susceptibility of single nucleotide polymorphisms located within co-stimulatory pathways to systemic lupus erythematosus. Front. Immunol. 14:1331796. doi: 10.3389/fimmu.2023.1331796
Received: 01 November 2023; Accepted: 30 November 2023;
Published: 01 February 2024.
Edited by:
Chris Wincup, King’s College Hospital NHS Foundation Trust, United KingdomReviewed by:
Xueyin Zhou, Wenzhou Medical University, ChinaLamjed Mansour, Carthage University, Tunisia
Copyright © 2024 Chen, Lin, Hsu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuang-Hui Yu, Z291dEBhZG0uY2dtaC5vcmcudHc=
 Ding-Ping Chen
Ding-Ping Chen Wei-Tzu Lin
Wei-Tzu Lin Fang-Ping Hsu
Fang-Ping Hsu Kuang-Hui Yu
Kuang-Hui Yu