
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 21 November 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1305937
 Ekramy E. Sayedahmed1
Ekramy E. Sayedahmed1 Nelly O. Elshafie1
Nelly O. Elshafie1 GuangJun Zhang1
GuangJun Zhang1 Sulma I. Mohammed1
Sulma I. Mohammed1 Suryaprakash Sambhara2*
Suryaprakash Sambhara2* Suresh K. Mittal1*
Suresh K. Mittal1*Introduction: Nonhuman adenoviral (AdV) gene delivery platforms have significant value due to their ability to elude preexisting AdV vector immunity in most individuals. Previously, we have demonstrated that intranasal (IN) immunization of mice with BAd-H5HA, a bovine AdV type 3 (BAdV3) vector expressing H5N1 influenza virus hemagglutinin (HA), resulted in enhanced humoral and cell-mediated immune responses. The BAd-H5HA IN immunization resulted in complete protection following the challenge with an antigenically distinct H5N1 virus compared to the mouse group similarly immunized with HAd-H5HA, a human AdV type 5 (HAdV5) vector expressing HA.
Methods: Here, we attempted to determine the activation of innate immune responses in the lungs of mice inoculated intranasally with BAd-H5HA compared to the HAd-H5HA-inoculated group.
Results: RNA-Seq analyses of the lung tissues revealed differential expression (DE) of genes involved in innate and adaptive immunity in animals immunized with BAd-H5HA. The top ten enhanced genes were verified by RT-PCR. Consistently, there were transient increases in the levels of cytokines (IL-1α, IL-1β, IL-5, TNF- α, LIF, IL-17, G-CSF, MIP-1β, MCP-1, MIP-2, and GM-CSF) and toll-like receptors in the lungs of the group inoculated with BAdV vectors compared to that of the HAdV vector group.
Conclusion: These results demonstrate that the BAdV vectors induce enhanced innate and adaptive immunity-related factors compared to HAdV vectors in mice. Thus, the BAdV vector platform could be an excellent gene delivery system for recombinant vaccines and cancer immunotherapy.
Adenoviruses (AdV) are icosahedral non-enveloped viruses with 25-48 kb dsDNA genomes (1). AdV vector-based vaccines are capable of eliciting both humoral and cell-mediated immune (CMI) responses (2, 3) by stimulating innate immunity through both Toll-like receptor (TLR)-mediated and TLR-independent pathways (4, 5). Unlike subunit or inactivated virus vaccines, Adv vector-based vaccines do not require an adjuvant for their immunogenicity. AdV vector-based influenza vaccines have shown immense promise in eliciting protective immunity in animal models (6, 7) and human clinical trials (8, 9). Also, Adv vectors are excellent delivery vehicles for cancer gene therapy (10–12).
Due to the potential presence of more than 100 AdV types in humans, there is a high possibility of developing Adv-specific neutralizing antibodies, known as ‘preexisting vector immunity, in the general population (13–15). This vector immunity could adversely impact the efficacy of several human Ad (HAdV) vector-based delivery systems. Hence, several nonhuman Ads have been developed as gene delivery vectors to avoid vector immunity (16, 17). These nonhuman Ad vectors can be based on bovine AdV (BAdV), simian AdV, ovine AdV, canine AdV, porcine AdV, avian AdV, or murine AdV (16–18).
We have shown that the BAdV3 vector system can induce humoral and CMI responses against HA of an H5N1 influenza virus even in exceptionally high levels of HAdV vector immunity (19). Moreover, preexisting HAdV-neutralizing antibodies in humans do not cross-neutralize BAdV3 (20), and HAdV-specific CMI response does not cross-react with BAdV3 (5). BAdV3 internalization into the cells is independent of the HAdV5 receptors [Coxackievirus-adenovirus receptor (CAR) and αvβ3 or αvβ5 integrin] (21); however it utilizes α(2,3)-linked and α(2,6)-linked sialic acid-binding proteins as major receptors for internalization (22). BAdV3 efficiently transduces the heart, kidney, lung, liver, and spleen. The vector persists longer than a HAdV5 vector, especially in the heart, kidney, and lung in a mouse model (23). Sequential administration of HAdV5 and BAdV3 vectors overcomes vector immunity in an immunocompetent mouse model of breast cancer (20), and the persistence of the BAdV3 genome in human and nonhuman cell lines is similar to HAdV5 vectors (24). Therefore, BAdV3 vectors offer an attractive replacement to HAdV vectors for circumventing high levels of preexisting HAdV immunity with comparable safety profiles as HAdV vectors.
In an earlier study, we demonstrated that the BAd-H5HA, a BAdV vector expressing hemagglutinin (HA) of the H5N1 influenza virus, elicited significantly better immune responses compared to HAd-H5HA, even at a reduced dose (25). To understand the factors responsible for the antigen-specific enhanced immune responses with the BAdV vector-based platform, we employed transcriptome analyses of the lung tissues from BAd-H5HA-inoculated mouse groups and compared with HAd-H5HA-inoculated groups. The genes involved in innate and adaptive immune responses were highly expressed in BAd-H5HA-inoculated groups. The top ten of these enhanced genes involved in innate and adaptive immunity were validated by qRT-PCR analyses. In addition, the upregulation of Toll like receptor genes (TLR2, TLR3, TLR4, TLR7, and TLR9) in the lungs compared to the HAdV vector was also confirmed by qRT-PCR. Furthermore, some of these highly expressed genes related to innate and/or adaptive immune responses-related molecules or cytokines were verified by multiplex assay. Our results suggest that higher expression of factors associated with innate and adaptive immune responses could be critical in eliciting better immune responses in animals immunized with the BAdV vector platform compared to the HAdV vector-based delivery system.
Previously, we have shown that IN (intranasal) immunization of mice with BAd-H5HA induced enhanced humoral and CMI responses compared to the group vaccinated similarly with HAd-H5HA, even with a reduced vaccine dose (25). We hypothesized that enhanced expression of immune response-related genes might be a critical difference between BAd-H5HA and HAd-H5HA. To test this hypothesis, we collected lung tissues at 6, 12, 24, and 48 h post-infection (PI) from mouse groups infected with BAd-H5HA, HAd-H5HA, or PBS (Table 1). Total RNA extracted from the lung tissues was used for RNA-Seq analyses to uncover DE genes.
A heat map of RNA-Seq data is shown where the values of data points are represented to visualize gene expression levels (Figure 1). Each row represents a gene, each column represents a sample, and a color gradient indicates the expression level of each gene. The heat map shows the clusters of genes that are co-regulated and the samples with similar expression profiles. The PBS-treated groups clustered together at 6, 12, 24, or 48 h (basal group), indicating the homogeneity of mock samples. While both BAd-H5HA and HAd-H5HA treated groups at 6 and 12 h form separated clusters from the mock samples (Figure 1). More interestingly, at the later stages (24 and 48 h), BAd-H5HA and HAd-H5HA groups showed a distinct gene expression profile, where the BAd-H5HA groups clustered mainly with the early states (6 h and 12 h), but the HAd-H5HA groups were clustered with the PBS basal group annotated with arrows (Figure 1). This difference suggests that the HAd-H5HA-induced DE gene expression levels decline rapidly. In contrast, the BAd-H5HA-induced DE maintained its overexpression status for a longer period.
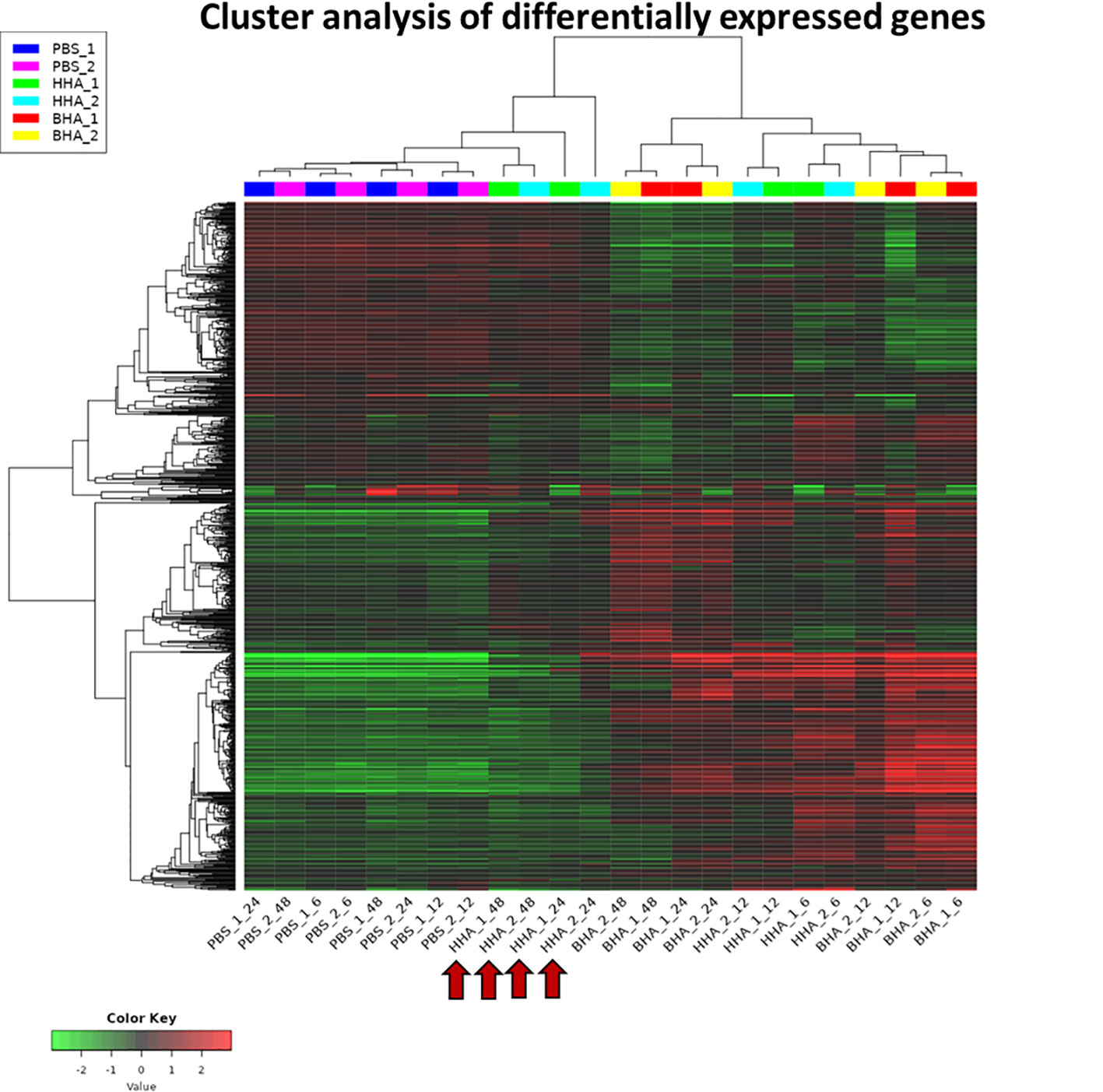
Figure 1 A heatmap uncovering the differential expressed (DE) genes in the lungs of mice at various times post-inoculation (PI) with BAd-H5HA (BHA), HAd-H5HA (HHA), or mock (PBS). Two independent samples for each group are shown, depicting an increase or decrease in expression levels, with red or green color, respectively. PBS_1_06, PBS_1_12, PBS_1_24, & PBS_1_48 represent 6, 12, 24, and 48 h PI samples from mouse #1 in mock-inoculated group. HHA_1_06, HHA_1_12, HHA_1_24, & HHA_1_48 represent 6, 12, 24, and 48 h PI samples from mouse #1 in HAd-H5HA-inoculated group. BHA_1_06, BHA_1_12, BHA_1_24, & BHA_1_48 represent 6, 12, 24, and 48 h PI samples from mouse #1 in BAd-H5HA-inoculated group. The same time points are repeated with mouse #2. The red arrows are showing the later stages (24 and 48 h) of HAd-H5HA groups gene expression clustering close to the PBS basal group.
To determine variability in the expression of DE genes in the BAd-H5HA or the HAd-H5HA group, a graphical representation by Venn diagram analysis was examined to infer the overall distribution of DE genes between the vaccine groups at 6, 12, 24, and 48 h PI (Figure 2). The average DE genes within the group was used to compare the gene expression patterns between samples. The overlapping region between the circles indicates the number of DE genes shared between the groups. Compared to the PBS group, the BAd-H5HA (BHA) group shows the highest number of DE genes at all times than the HAd-H5HA (HHA) group. The BAd-H5HA group shows variable numbers of DE genes compared to the HAd-H5HA group at all time points.
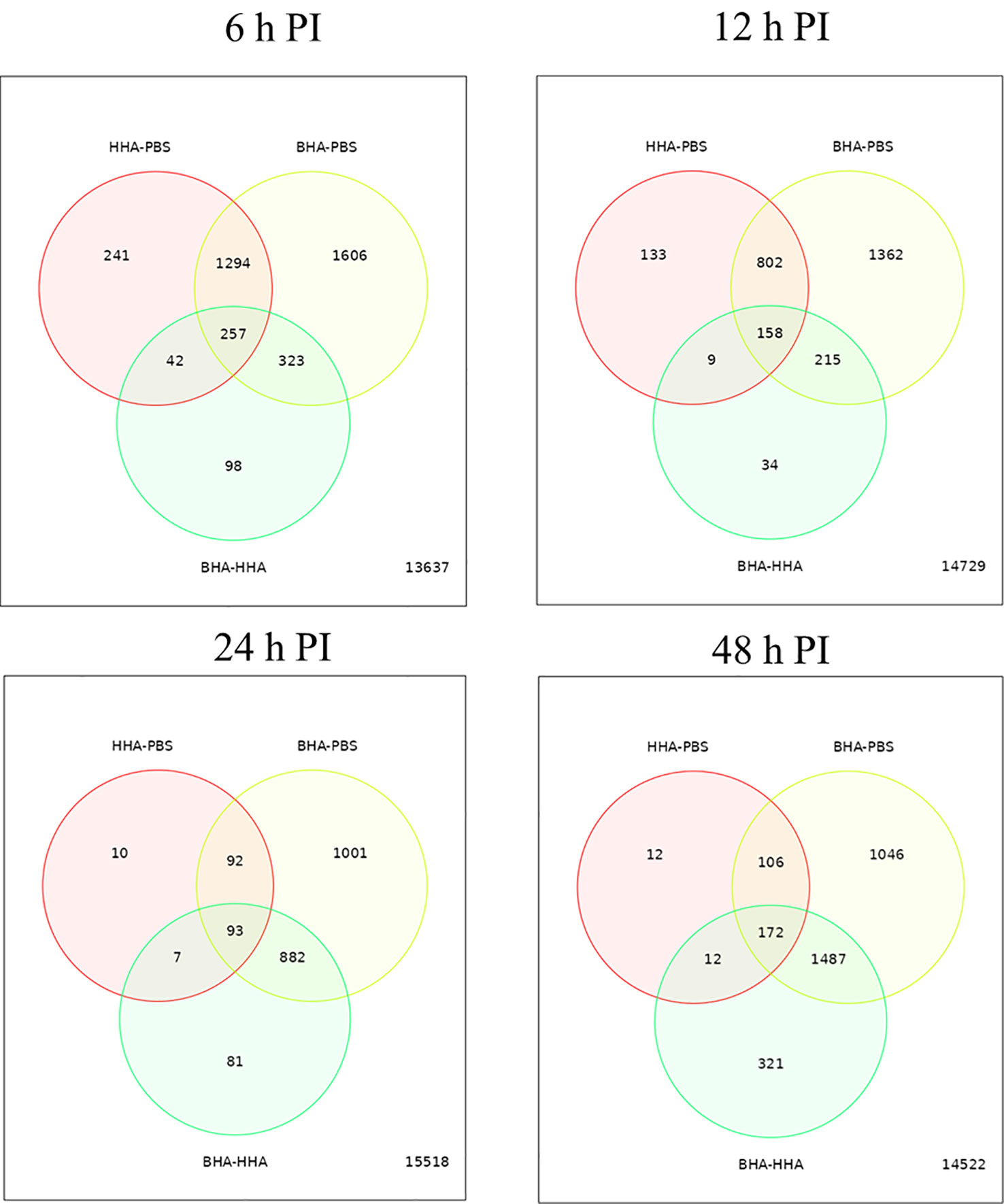
Figure 2 Venn diagram of differentially expressed (DE) genes in the lungs of mice at 6, 12, 24, and 48 h post-inoculation (PI) with BAd-H5HA (BHA), HAd-H5HA (HHA) or mock (PBS). The red circle represents the number of differentially expressed genes (DEGs) in the HAd-H5HA(HHA) compared to the PBS group. The yellow circle represents the number of DEGs in the BAd-H5HA (BHA) compared to the PBS group. The green circle represents the number of DEGs in the BAd-H5HA (BHA) compared to HAd-H5HA (HHA) group. The overlapping region between the circles indicates the number of DE genes shared between the groups.
Further in Volcano plot analysis, there were 664, 320, 901, and 1865 DE genes at 6, 12, 24, and 48 h PI, respectively, between BAd-H5HA and HAd-H5HA groups (Figure 3). The overall gene expression profiles are similar at 6 h and 12 h, while quite different at 24 h and 48 h time points. Our data suggest that host responses to BAd-H5HA and HAd-H5HA are somewhat similar at early stages (6 and 12 h PI) than the later stages (24 and 48 h PI). The top ten overexpressed genes in the BAd-H5HA group compared to the HAd-H5HA group at 6, 12, 24, and 48 h PI are listed (Supplementary Table 2).
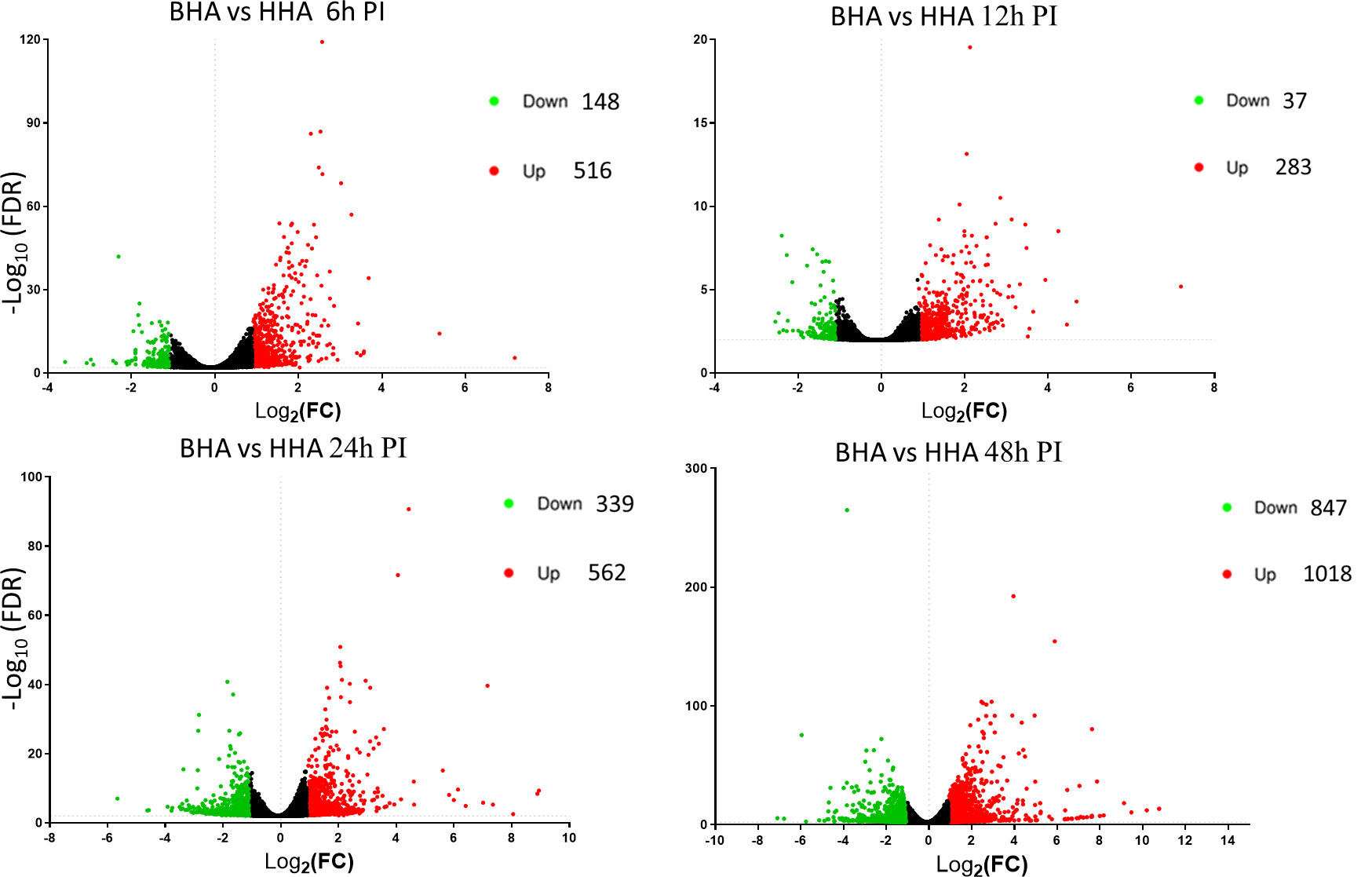
Figure 3 Volcano plots from the DESeq2 analysis of differentially expressed (DE) genes in the lungs of mice at 6, 12, 24, and 48 h post-inoculation (PI) with BAd-H5HA (BHA), HAd-H5HA (HHA), or mock (PBS). Log2 fold changes are plotted on the x-axis, and -log10 p-values on the y-axis. Each point represents a gene, the red color dots representing the upregulated genes, and the green color dots representing the downregulated genes. The cutoff of the log2 fold change is 2.5, and the cutoff of the -log10 (FDR<0.05) is 1.3.
To better understand the biological functions of the DE genes, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses were conducted for the BAd-H5HA group compared to the HAd-H5HA group at 6, 12, 24, and 48 h PI. The primary biological pathways relevant to the innate and adaptive immune responses that were highlighted include cytokine-cytokine receptor interaction (19 genes), IL-17 signaling pathways (8 genes), TLR signaling pathway (7 genes), NOD-like receptor signaling pathway (9 genes), and TNF signaling pathway (7 genes) at 6 h PI; cytokine-cytokine receptor interaction (35 genes), natural killer cell-mediated cytotoxicity (12 genes), IL-17 signaling pathways (11 genes), TLR signaling pathway (11 genes), and TNF signaling pathway (10 genes) at 12 h PI; cytokine-cytokine receptor interaction (33 genes), IL-17 signaling pathways (19 genes), TNF signaling pathway (19 genes), NOD-like receptor signaling pathway (18 genes), chemokine signaling pathway (16 genes), and TLR signaling pathway (11 genes) at 24 h PI; and cytokine-cytokine receptor interaction (39 genes), hematopoietic cell linkage (20 genes), IL-17 signaling pathways (19 genes), cell adhesion molecules (20 genes), TNF signaling pathway (16 genes), and chemokine signaling pathway (19 genes), at 48 h PI (Figure 4). Overall, our transcriptome study of DE genes suggests that the major signaling pathways that were upregulated included cytokine-cytokine receptor interaction, IL-17 signaling pathway, TLR signaling pathway, TNF signaling pathway, and NOD-like receptor signaling pathway. The top 10 pathways by KEGG database pathway analysis are listed (Table 2). The cytokine-cytokine receptor interaction pathway has the highest number of DE genes (228). This pathway has multiple chemokine and cytokine genes upregulated in the BAd-H5HA group than the HAd-H5HA group (Figure 5).
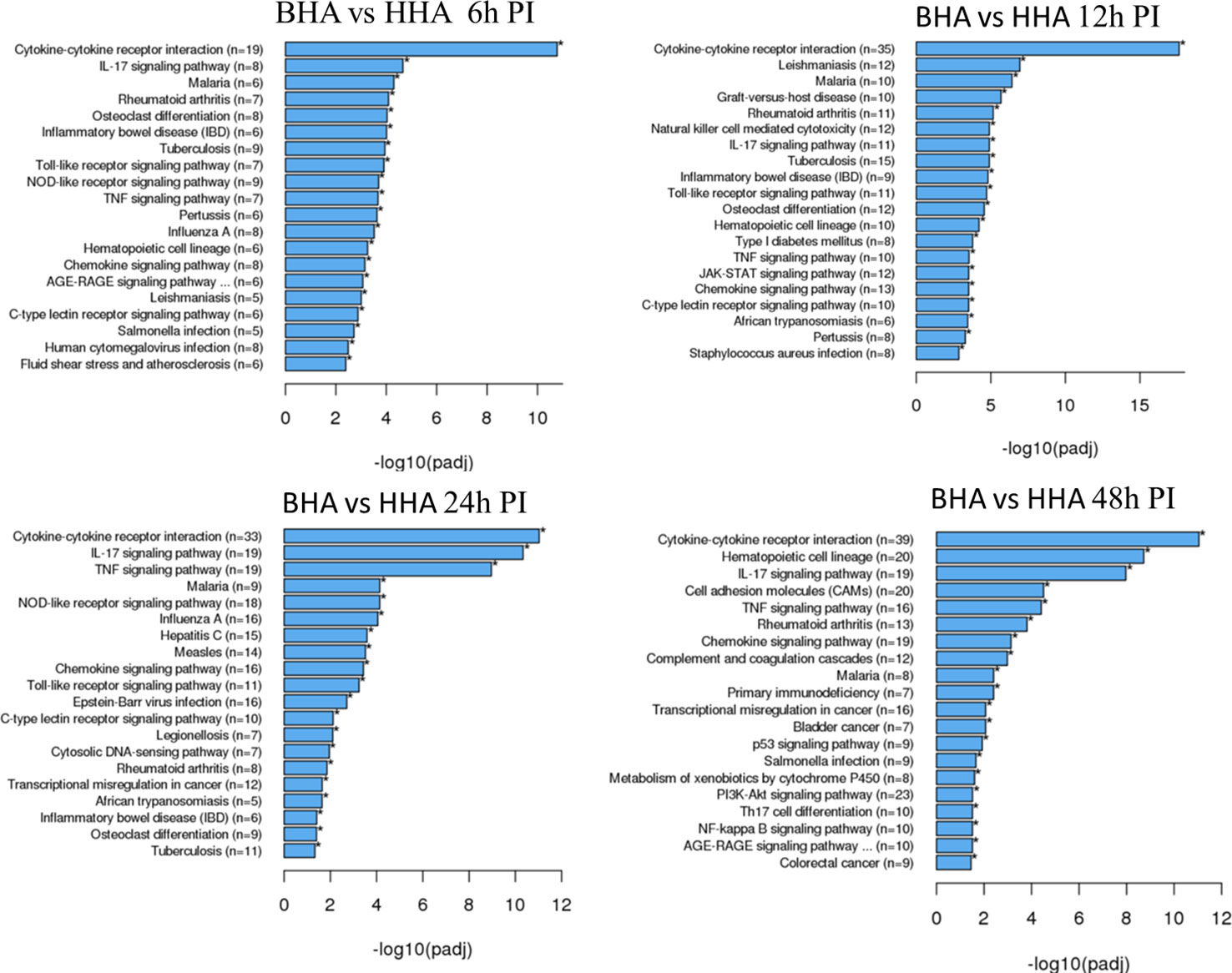
Figure 4 KEGG pathways highlighting differentially expressed (DE) groups of genes within the same biological pathway in the lungs of mice at 6, 12, 24, and 48 h post-inoculation (PI) with BAd-H5HA (BHA) compared to the HAd-H5HA (HHA) groups.
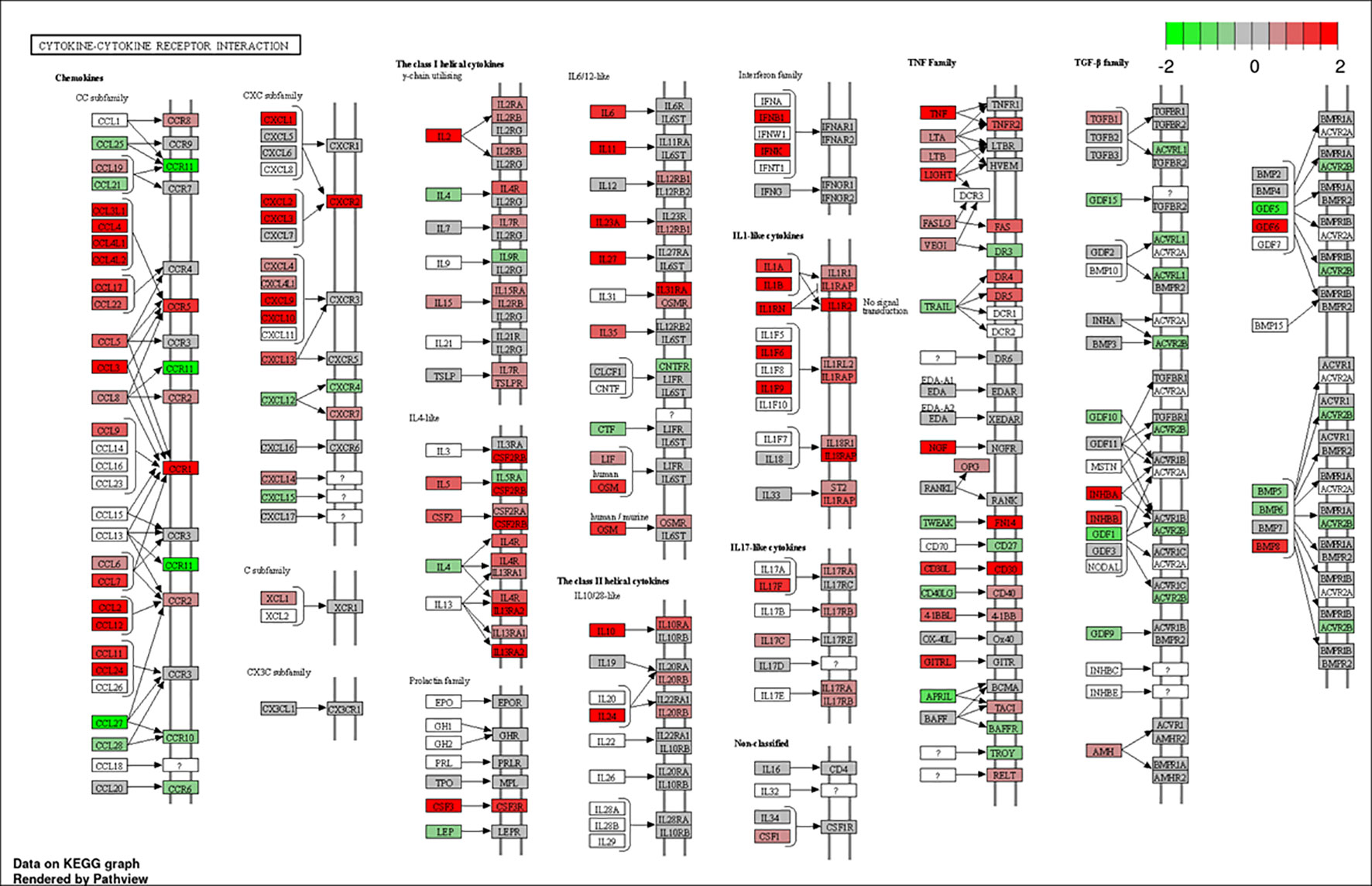
Figure 5 Cytokine-cytokine receptor interaction pathway. DE genes KEGG pathway analysis with significance cutoff (FDR) 0.1 was performed for BAd-H5HA versus HAd-H5HA. Red and green represent up-regulated and down-regulated genes, respectively. The KEGG database pathway analysis with a false discovery rate (FDR) 0.1 cutoff value for significance was used to show the top 10 pathways.
To further validate our identified DE genes by RNA-Seq, we examined the top ten upregulated genes (Supplementary Table 2) by examining RNA extracted from the lung tissues of BAd-H5HA- or HAd-H5HA-inoculated mice using qRT-PCR. Indeed, all the 10 genes were upregulated in BAd-H5HA groups compared to HAd-H5HA groups at 6, 12, 24, and 24 h PI (Figure S1A). The gene ontology (GO) biological processes of these DE genes showed highly enriched natural killer cell pathways (Figure S1B). The KEGG pathway analysis indicated the involvement of four genes in the cytokine-cytokine receptors interaction, two genes in the TLR process, and two genes in viral protein interaction with cytokine and cytokine receptor interaction beside other pathways (Figure S1C). A tree map (Figure S1D right) and network map (Figure S1D left) illustrated potential interactions of different KEGG pathways.
AdV plays a vital role in activating TLR-mediated innate immunity (4). To ascertain enhanced expression of TLR genes in the lungs of the BAd-H5HA-inoculated mouse group compared to the HAd-H5HA group, qRT-PCR analyses were performed. The qRT-PCR analyses showed that the BAdV groups at various time points have higher expression levels of TLR2, TLR3, TLR4, TLR7, and TLR9 than the HAdV groups (Figure S2).
To further verify whether there was enhanced induction of innate and adaptive immunity-related factors, cytokines, and chemokines in the lungs of mice inoculated with BAd-H5HA, the lung wash samples from mock-, HAd-ΔE1E3-, BAd-ΔE1E3-, HAd-H5HA-, or BAd-H5HA-inoculated groups were assayed using a 32-multiplex kit assay. There was a transient increase in the levels of IL-1α, IL-1β, IL-5, tumor necrosis factor-alpha (TNF-α), Leukemia inhibitory factor LIF, IL-17, G-CSF, and GM-CSF cytokines in the lung washes of the group inoculated with BAd-H5HA or BAd-ΔE1E3 compared to the group inoculated with HAd-H5HA or HAd-ΔE1E3 (Figure 6). There were transient increases in the levels of chemokines CCL2, CCL4, CCL8, CXCL1 and CXCL10 in the lung washes of the group inoculated with BAd-H5HA or BAd-ΔE1E3 compared to the group inoculated with HAd-H5HA or HAd-ΔE1E3 (Figure 7). These data suggest that the BAdV vector platform is a better gene delivery system than the HAdV vector in stimulating the innate immune responses in mice.
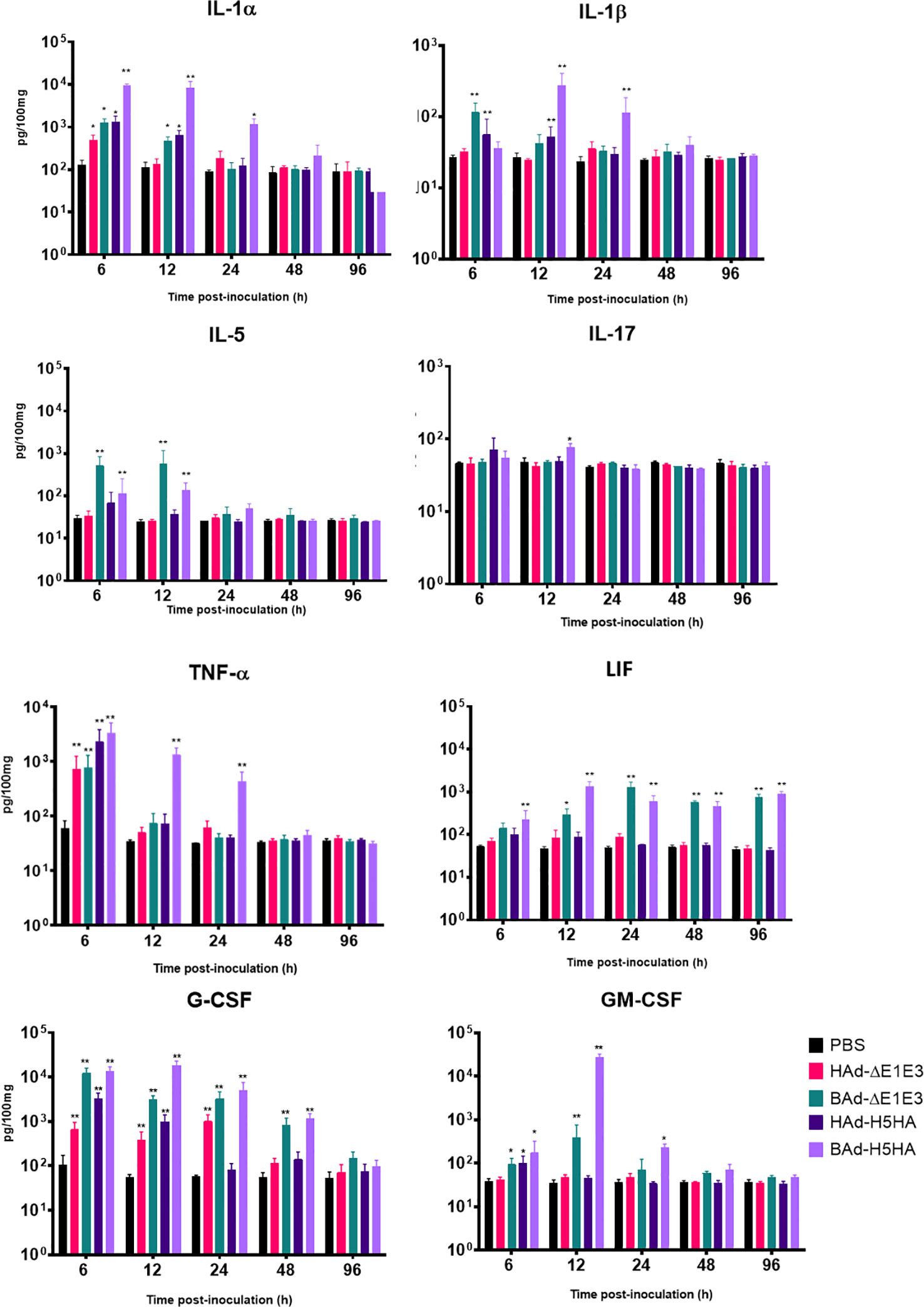
Figure 6 Innate cytokine multiplex assay showing the increase in cytokines levels in BAdV vectors inoculated groups. BALB/c mice (3 animals/group) were inoculated IN once with PBS or with 3 × 107 PFU per animal of HAd-ΔE1E3, BAd-ΔE1E3, HAd-H5HA or BAd-H5HA, and at 6, 12, 24, 48 and 96 h after inoculation and the animals were euthanized. The lung washes were collected and used to monitor levels of innate cytokines by multiplex assays using a 32-plex Kit from Millipore Sigma. *, significant at p<0.05; **, significant at p<0.01.
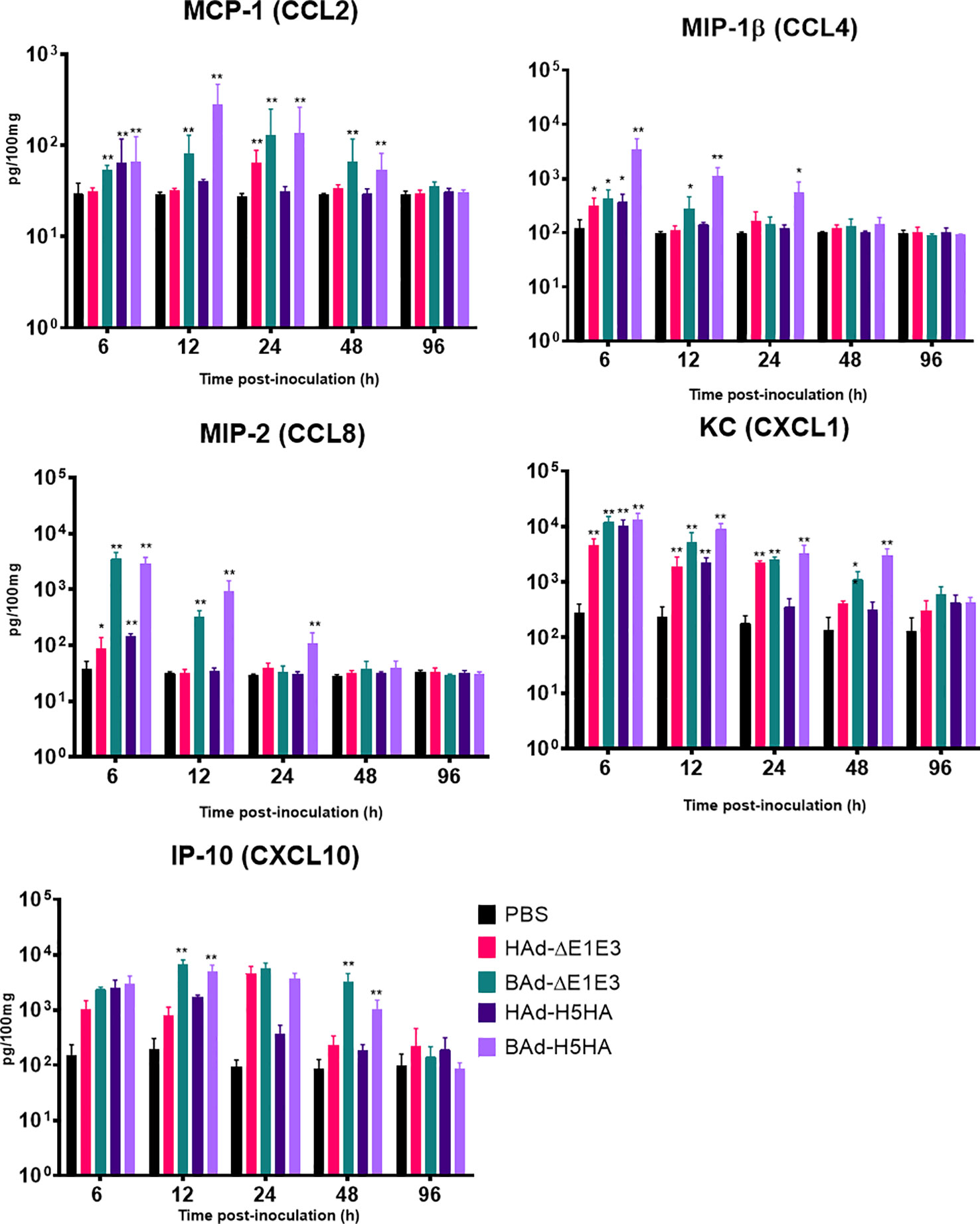
Figure 7 Innate chemokines multiplex assay showing the increase in chemokines levels in BAdV vectors inoculated groups. BALB/c mice (3 animals/group) were inoculated IN once with PBS or with 3 × 107 PFU per animal of HAd-ΔE1E3, BAd-ΔE1E3, HAd-H5HA or BAd-H5HA, and at 6, 12, 24, 48 and 96 h after inoculation and the animals were euthanized. The lung washes were collected to monitor levels of innate chemokines by multiplex assays using a 32-plex Kit from Millipore Sigma. *, significant at p<0.05; **, significant at p<0.01.
AdV vectors have enormous potential as a gene delivery platform for developing recombinant vaccines against infectious diseases and cancer immunotherapy (26–32). The concept of preexisting AdV vector immunity in humans (33) has created a niche for developing novel AdV vectors that can overcome the preexisting vector immunity barrier. Along this direction, we developed a BAdV vector platform (34) and demonstrated its utility in eluding an exceptionally high level of vector immunity in a mouse model (35). We have shown that IN immunization of mice with BAd-H5HA elicited significantly higher levels of humoral (including mucosal) and CMI responses at a lower vector dose, resulting in complete protection following challenge with an antigenically distinct influenza virus compared to the HAd-H5HA-immunized group (25). Enhanced immune responses with BAd-H5HA were also observed with the IM route of immunization (25); however, complete protection was conferred with a 30-fold vaccine dose compared to the vaccine dose used for the IN inoculation. These observations led us to pursue the potential factors responsible for high levels of humoral and CMI responses following IN immunization with BAd-H5HA.
Our RNA-Seq analyses of the lung samples from vector-inoculated mice revealed a distinct spectrum and longer duration of DE genes, including genes associated with innate and adaptive immunity in the BAd-H5HA-inoculated group compared to the HAd-H5HA-inoculated group. These results showed that the BAdV vector induced significantly higher levels of several innate and adaptive immunity-related host factors compared to the HAdV vector. Indirectly, these transcriptome analyses also implicate that the BAdV vaccine platform could serve as an excellent gene delivery vehicle for recombinant vaccines. Further studies are needed to investigate the roles of the vital DE genes and their cellular origin in the BAd-H5HA-inoculated group in inducing enhanced immune responses.
Enhanced induction of innate and adaptive immunity-related host factors, cytokines, and chemokines in the lungs of mice inoculated with BAd-H5HA broadly supports the outcomes of transcriptome analyses. It is not a surprise since previously we have demonstrated the enhanced expression of CCL2, CCL3, CCL4, CCL5, CXCL2, TNF-α, CXCL-10, interferon-gamma (IFN-γ), IL-6, TLR2, TLR-3, TLR-4, TLR-7 and TLR9 in the spleen at early time points in BAdV vector-inoculated groups compared to that of HAdV vector-inoculated groups by intravenous (IV) injection (36). It further emphasizes that the BAdV vector platform can induce higher levels of innate and adaptive immunity-related factors, cytokines, and chemokines that could lead to the enhancement of immune responses to BAdV vaccines. This process could significantly impact the development of transgene-specific immune responses.
The BAdV vector stimulated higher expression of TLR2, TLR3, TLR4, TLR7, and TLR9 genes in the lungs than the HAdV vector. These results are aligned with our previously described data (36) where the same TLRs were upregulated in the spleen of the BAdV vector group. TLR-mediated pathways are critical for the activation of transcription factors like interferon regulatory factors (IRFs) and nuclear factor κB (NF-κB), which determine the outcome of the innate immune responses (37). The qRT-PCR analyses of the top ten DE genes identified by RNA-Seq in the lungs of BAd-H5HA confirmed their upregulated state. These genes are involved in natural killer cell pathways, TLR pathways, cytokine-receptor interactions, and viral protein interaction with cytokine, suggesting the advantage of the BAdV vector as a vaccine vector might be explained through its better stimulation of the innate and adaptive immune factors.
In summary, there are enhanced innate and adaptive immunity-related factors in the lungs of mice in the BAdV vector group compared to the HAdV vector group, suggesting the preeminence of the BAdV vaccine platform for developing effective vaccines against emerging infectious diseases and cancer immunotherapy.
293 (human embryonic kidney cells expressing HAdV5 E1 proteins) (38), BHH3 (bovine-human hybrid clone 3) (39), and BHH2C (bovine-human hybrid clone 2C) (39) cells as monolayer cultures were grown using minimum essential medium (MEM) (Life Technologies, Thermo Fisher Scientific, Waltham, MA) supplemented with either 10% reconstituted fetal bovine serum or fetal calf serum (Hyclone, Thermo Fisher Scientific) and gentamycin (50 µg/ml).
The generation and characterization of BAd-ΔE1E3 (BAdV3 E1 and E3 deleted empty vector) (40), BAd-H5HA [BAd3 E1 and E3 deleted vector expressing HA of A/Hong Kong/156/97(H5N1) (HK/156)] (19), HAd-ΔE1E3 (HAdV5 E1 and E3 deleted empty vector) (41), HAd-H5HA [HAdV5 E1 and E3 deleted vector expressing HA of HK/156] (42), were described earlier. BAd-ΔE1E3 and BAd-H5HA were replicated and titrated in BHH3 cells as described elsewhere (19), whereas HAd-ΔE1E3 and HAd-H5HA were grown in 293 cells and titrated in BHH2C cells as described earlier (43). As described previously, all vectors were purified by cesium chloride density-gradient ultracentrifugation (44, 45).
All mouse studies were conducted with the approvals of the Institutional Animal Care and Use Committee (IACUC) and the Institutional Biosafety Committee (IBC). Six-to-eight-week-old BALB/c mice (Envigo RMS, Inc., Indianapolis, IN) were mock-inoculated with phosphate-buffered saline (PBS), pH 7.2 or inoculated IN with 3 × 107 PFU of BAd-ΔE1E3 [4.2 × 108 virus particles (VP)], BAd-H5HA (6.8 × 108 VP), HAd-H5HA (1.2 × 109 VP), or HAd-ΔE1E3 (8.9 × 108 VP). The plaque forming units and virus particle counts were calculated as previously described (46). At 6, 12, 24, 48, and 96 h PI, three animals/group were anesthetized with ketamine-xylazine solution, and the lung washes were prepared by homogenizing one lung from each animal in 1 mL of PBS as described (25, 47). The lung tissue samples were also collected in Invitrogen RNAlater Stabilization Solution (Thermo Fisher Scientific # AM7020) for RNA extraction.
The lung tissue samples in RNAlater were used to extract RNA using Monarch Total RNA Miniprep Kit (Thermo Fisher Scientific #50-152-7886), and the RNA quality was confirmed by 1% agarose gel electrophoresis and Agilent 2100 bioanalyzer 2100 Total RNA Nano Series II. RNA samples were sent for RNA-Seq using 250-300 bp insert cDNA library with Illumina NovaSeq platforms with paired-end 150 bp (PE 150) sequencing strategy using 20 million reads/sample and ≥ 6 G Raw Data/Sample depth (Novogene, Sacramento, CA). The data quality parameters are shown (Supplementary Table 1). The data were analyzed by a bioinformatics specialist using a combination of programs, including STAR, HTseq, Cufflink, and wrapped scripts. Alignments were parsed using the Tophat program, and differential expressions were determined through DESeq2/edgeR. GO and KEGG enrichments were implemented by ClusterProfiler. Gene fusion and the difference of alternative splicing events were detected by Star-fusion and rMATS software.
Sequences were quality-checked using FastQC for completeness, depth, and read quality. Sequences were aligned to the mm10 Mus musculus reference genome using a STAR aligner (48). Gene quantification was done using HTSeq-count (49).
DESeq2 determines DE genes between at least two experimental groups (50–52). Genes with low counts are filtered for subdued expression by their normalized mean counts, and raw p-values are adjusted for multiple testing using the Benjamini-Hochberg correction. For this experiment, genes considered significantly DE are those with adjusted p-values controlled at False Discovery Rates (FDR) < 0.01 specific threshold. Volcano plots were produced in the Enhanced Volcano R package (53) and showed the un-transformed log (2) fold change of each gene plotted against its adjusted p-value, both of which were calculated in DESEq2. Twenty genes with the highest mean-normalized counts were selected to generate the heatmap.
The extracted RNA samples from the lungs were converted to a single complementary DNA strand (cDNA) using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific) according to the manufacturing recommendation. 500 nanogram RNA from each sample was converted to cDNA and used in the downstream qPCR.
TaqMan Fast Universal PCR 2X Master Mix (Applied Biosystems, Thermo Fisher Scientific) was used to evaluate the differential gene expression between the groups. A real-time PCR was conducted with a 20 µL reaction volume with ROX dye as a passive internal reference to normalize non-PCR-related fluorescence fluctuations. The QuantStudio 3 real-time PCR System (Thermo Fisher, Fisher Scientific) was used to detect differences in each target quantity. TaqMan Gene Expression Assays (Applied Biosystems, Thermo Fisher Scientific) were used to evaluate the differentially expressed TLRs. The 18S ribosomal RNA gene was used as an internal housekeeping gene. The primers and probes of the top ten DE genes identified by RNA-Seq were obtained from TaqMan assays and arrays (Thermo Fisher Scientific) (Supplementary Table 3).
ShinyGo 0.77 (http://bioinformatics.sdstate.edu/go/), the web-based bioinformatics data analysis tool, was used to analyze gene/s functional pathways and process the enrichment analysis for the DE top ten genes at 6, 12, 24, and 48 h PI; Mus musculus was used as the input species. The DE genes were annotated and used in the functional enrichment analysis by the default settings for determining the significance of Gene Ontology (GO) Biological Processes and Kyoto Encyclopedia of Genes and Genomes KEGG pathways. The ShinyGO is another web-based data analysis tool used to graphically visualize the upregulated genes’ functions and enrichment results (54). The P-value cutoff set was set at a false discovery rate (FDR) = 0.05.
The lung washes were used to monitor levels of innate and adaptive immunity-related factors, cytokines, and chemokines by multiplex assays using Mouse Cytokine/Chemokine Magnetic Bead Panel - Premixed 32 Plex - Immunology Multiplex Assay (Millipore Sigma, St. Louis, MO as described (55). Briefly, the samples were centrifuged at 15,000 g for 10 min, and analyzed for interleukin (IL)-1 alpha, IL-1 beta, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17, interferon-gamma inducible protein (IP10), keratinocyte-derived chemokine (KC), eotaxin, monocyte chemoattractant protein 1 (MCP-1), monokine-induced by gamma interferon (MIG), macrophage inflammatory protein-2 alpha (MIP-2α), macrophage inflammatory protein-1 alpha (MIP-1α), macrophage inflammatory protein-1 beta (MIP-1β), IFN-γ, lipopolysaccharide-induced CXC chemokine (LIX), TNF-α, regulated upon activation normal T cell expressed and secreted (RANTES), leukemia inhibitory factor (LIF), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (M-CSF), vascular endothelial growth factor (VEGF). Samples were 2-fold diluted and incubated with the pre-mixed capture antibody-coupled beads in 96-well plates at 4°C overnight. The beads were washed and incubated with the biotinylated secondary antibodies for 2 h. Streptavidin–phycoerythrin was added and incubated for 30 min, and the beads were washed and resuspended in sheath fluid. The standard curve range was 3.2-10,000 pg/ml. The analysis was performed by a Bio-Plex 200 System with High Throughput Fluidics (HTF) Multiplex Array System (Indiana University Cancer Center Facility, Indianapolis, IN).
One and two-way ANOVA with Bonferroni post-test were performed to determine statistical significance for the multiplex assay. The p-value below 0.05 was considered statistically significant.
The datasets presented in this study are deposited in NCBI BioProject accession number: PRJNA1039157.
The animal study was approved by Institutional Animal Care and Use Committee (IACUC) and the Institutional Biosafety Committee (IBC). The study was conducted in accordance with the local legislation and institutional requirements.
ES: Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. NE: Formal Analysis, Investigation, Methodology, Software, Writing – review & editing. GZ: Conceptualization, Supervision, Validation, Writing – review & editing. SM: Investigation, Supervision, Writing – review & editing. SS: Conceptualization, Formal Analysis, Supervision, Writing – review & editing. SM: Conceptualization, Formal Analysis, Funding acquisition, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Public Health Service grants AI059374 and AI158177 from the National Institute of Allergy and Infectious Diseases and the Hatch funds.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention, U.S. Department of Health and Human Services.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1305937/full#supplementary-material
1. Benkő M, Aoki K, Arnberg N, Davison AJ, Echavarría M, Hess M, et al. Ictv report: ICTV virus taxonomy profile: adenoviridae 2022. J Gen Virol (2022) 103(3). doi: 10.1099/jgv.0.001721
2. Vemula SV, Mittal SK. Production of adenovirus vectors and their use as a delivery system for influenza vaccines. Expert Opin Biol Ther (2010) 10(10):1469–87. doi: 10.1517/14712598.2010.519332
3. Ahi YS, Bangari DS, Mittal SK. Adenoviral vector immunity: its implications and circumvention strategies. Curr Gene Ther (2011) 11(4):307–20. doi: 10.2174/156652311796150372
4. Zhu J, Huang X, Yang Y. Innate immune response to adenoviral vectors is mediated by both Toll-like receptor-dependent and -independent pathways. J Virol (2007) 81(7):3170–80. doi: 10.1128/jvi.02192-06
5. Sharma A, Tandon M, Ahi YS, Bangari DS, Vemulapalli R, Mittal SK. Evaluation of cross-reactive cell-mediated immune responses among human, bovine and porcine adenoviruses. Gene Ther (2010) 17(5):634–42. doi: 10.1038/gt.2010.1
6. Gao W, Soloff AC, Lu X, Montecalvo A, Nguyen DC, Matsuoka Y, et al. Protection of mice and poultry from lethal H5N1 avian influenza virus through adenovirus-based immunization. J Virol (2006) 80(4):1959–64. doi: 10.1128/jvi.80.4.1959-1964.2006
7. Hoelscher MA, Jayashankar L, Garg S, Veguilla V, Lu X, Singh N, et al. New pre-pandemic influenza vaccines: an egg- and adjuvant-independent human adenoviral vector strategy induces long-lasting protective immune responses in mice. Clin Pharmacol Ther (2007) 82(6):665–71. doi: 10.1038/sj.clpt.6100418
8. Van Kampen KR, Shi Z, Gao P, Zhang J, Foster KW, Chen DT, et al. Safety and immunogenicity of adenovirus-vectored nasal and epicutaneous influenza vaccines in humans. Vaccine (2005) 23(8):1029–36. doi: 10.1016/j.vaccine.2004.07.043
9. Gurwith M, Lock M, Taylor EM, Ishioka G, Alexander J, Mayall T, et al. Safety and immunogenicity of an oral, replicating adenovirus serotype 4 vector vaccine for H5N1 influenza: a randomised, double-blind, placebo-controlled, phase 1 study. Lancet Infect Dis (2013) 13(3):238–50. doi: 10.1016/s1473-3099(12)70345-6
10. Crystal RG. Adenovirus: the first effective in vivo gene delivery vector. Hum Gene Ther (2014) 25(1):3–11. doi: 10.1089/hum.2013.2527
11. Sharma A, Tandon M, Bangari DS, Mittal SK. Adenoviral vector-based strategies for cancer therapy. Curr Drug Ther (2009) 4(2):117–38. doi: 10.2174/157488509788185123
12. Zhang C, Zhou D. Adenoviral vector-based strategies against infectious disease and cancer. Hum Vaccin Immunother (2016) 12(8):2064–74. doi: 10.1080/21645515.2016.1165908
13. Bangari DS, Mittal SK. Porcine adenoviral vectors evade preexisting humoral immunity to adenoviruses and efficiently infect both human and murine cells in culture. Virus Res (2004) 105(2):127–36. doi: 10.1016/j.virusres.2004.05.003
14. Kostense S, Koudstaal W, Sprangers M, Weverling GJ, Penders G, Helmus N, et al. Adenovirus types 5 and 35 seroprevalence in AIDS risk groups supports type 35 as a vaccine vector. Aids (2004) 18(8):1213–6. doi: 10.1097/00002030-200405210-00019
15. Nwanegbo E, Vardas E, Gao W, Whittle H, Sun H, Rowe D, et al. Prevalence of neutralizing antibodies to adenoviral serotypes 5 and 35 in the adult populations of The Gambia, South Africa, and the United States. Clin Diagn Lab Immunol (2004) 11(2):351–7. doi: 10.1128/cdli.11.2.351-357.2004
16. Bangari DS, Mittal SK. Development of nonhuman adenoviruses as vaccine vectors. Vaccine (2006) 24(7):849–62. doi: 10.1016/j.vaccine.2005.08.101
17. Mittal SK, Ahi YS, Vemula SV. 19 - xenogenic adenoviral vectors A2 - curiel, david T. In: Adenoviral Vectors for Gene Therapy (Second Edition). San Diego: Academic Press (2016). doi: 10.1016/B978-0-12-800276-6.00019-X
18. Alhashimi M, Elkashif A, Sayedahmed EE. and Mittal SK: Nonhuman adenoviral vector-based platforms and their utility in designing next generation of vaccines for infectious diseases. Viruses (2021) 13(8):1493. doi: 10.3390/v13081493
19. Singh N, Pandey A, Jayashankar L, Mittal SK. Bovine adenoviral vector-based H5N1 influenza vaccine overcomes exceptionally high levels of pre-existing immunity against human adenovirus. Mol Ther (2008) 16(5):965–71. doi: 10.1038/mt.2008.12
20. Tandon M, Sharma A, Vemula SV, Bangari DS, Mittal SK. Sequential administration of bovine and human adenovirus vectors to overcome vector immunity in an immunocompetent mouse model of breast cancer. Virus Res (2012) 163(1):202–11. doi: 10.1016/j.virusres.2011.09.031
21. Bangari DS, Sharma A, Mittal SK. Bovine adenovirus type 3 internalization is independent of primary receptors of human adenovirus type 5 and porcine adenovirus type 3. Biochem Biophys Res Commun (2005) 331(4):1478–84. doi: 10.1016/j.bbrc.2005.04.058
22. Li X, Bangari DS, Sharma A, Mittal SK. Bovine adenovirus serotype 3 utilizes sialic acid as a cellular receptor for virus entry. Virology (2009) 392(2):162–8. doi: 10.1016/j.virol.2009.06.029
23. Sharma A, Bangari DS, Tandon M, Pandey A, HogenEsch H, Mittal SK. Comparative analysis of vector biodistribution, persistence and gene expression following intravenous delivery of bovine, porcine and human adenoviral vectors in a mouse model. Virology (2009) 386(1):44–54. doi: 10.1016/j.virol.2009.01.008
24. Sharma A, Bangari DS, Vemula SV, Mittal SK. Persistence and the state of bovine and porcine adenoviral vector genomes in human and nonhuman cell lines. Virus Res (2011) 161(2):181–7. doi: 10.1016/j.virusres.2011.08.002
25. Sayedahmed EE, Hassan AO, Kumari R, Cao W, Gangappa S, York I, et al. A bovine adenoviral vector-based H5N1 influenza -vaccine provides enhanced immunogenicity and protection at a significantly low dose. Mol Ther Methods Clin Dev (2018) 10:210–22. doi: 10.1016/j.omtm.2018.07.007
26. Sayedahmed EE, Araújo MV, Silva-Pereira TT, Chothe SK, Elkashif A, Alhashimi M, et al. Impact of an autophagy-inducing peptide on immunogenicity and protection efficacy of an adenovirus-vectored SARS-CoV-2 vaccine. Mol Ther Methods Clin Dev (2023) 30:194–207. doi: 10.1016/j.omtm.2023.06.009
27. Wang WC, Sayedahmed EE, Sambhara S, Mittal SK. Progress towards the development of a universal influenza vaccine. Viruses (2022) 14(8):1684. doi: 10.3390/v14081684
28. Elkashif A, Alhashimi M, Sayedahmed EE, Sambhara S, Mittal SK. Adenoviral vector-based platforms for developing effective vaccines to combat respiratory viral infections. Clin Transl Immunol (2021) 10(10):e1345. doi: 10.1002/cti2.1345
29. Khan A, Sayedahmed EE, Singh VK, Mishra A, Dorta-Estremera S, Nookala S, et al. A recombinant bovine adenoviral mucosal vaccine expressing mycobacterial antigen-85B generates robust protection against tuberculosis in mice. Cell Rep Med (2021) 2(8):100372. doi: 10.1016/j.xcrm.2021.100372
30. Su C. Adenovirus-based gene therapy for cancer. In: Xu DK, editor. Viral Gene Therapy. Rijeka, Croatia: InTech (2011). doi: 10.5772/19757
31. Matsunaga W, Gotoh A. Adenovirus as a vector and oncolytic virus. Curr Issues Mol Biol (2023) 45(6):4826–40. doi: 10.3390/cimb45060307
32. Wold WS, Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther (2013) 13(6):421–33. doi: 10.2174/1566523213666131125095046
33. Wang WC, Sayedahmed EE, Mittal SK. Significance of preexisting vector immunity and activation of innate responses for adenoviral vector-based therapy. Viruses (2022) 14(12):2727. doi: 10.3390/v14122727
34. Mittal SK, Prevec L, Graham FL, Babiuk LA. Development of a bovine adenovirus type 3-based expression vector. J Gen Virol (1995) 76(Pt 1):93–102. doi: 10.1006/viro.2000.0350
35. Moffatt S, Hays J, HogenEsch H, Mittal SK. Circumvention of vector-specific neutralizing antibody response by alternating use of human and non-human adenoviruses: implications in gene therapy. Virology (2000) 272(1):159–67. doi: 10.1006/viro.2000.0350
36. Sharma A, Bangari DS, Tandon M, Hogenesch H, Mittal SK. Evaluation of innate immunity and vector toxicity following inoculation of bovine, porcine or human adenoviral vectors in a mouse model. Virus Res (2010) 153(1):134–42. doi: 10.1016/j.virusres.2010.07.021
37. Rosenberger K, Derkow K, Dembny P, Krüger C, Schott E, Lehnardt S. The impact of single and pairwise Toll-like receptor activation on neuroinflammation and neurodegeneration. J Neuroinflamm (2014) 11:166. doi: 10.1186/s12974-014-0166-7
38. Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol (1977) 36(1):59–74. doi: 10.1099/0022-1317-36-1-59
39. van Olphen AL, Mittal SK. Development and characterization of bovine x human hybrid cell lines that efficiently support the replication of both wild-type bovine and human adenoviruses and those with E1 deleted. J Virol (2002) 76(12):5882–92. doi: 10.1128/jvi.76.12.5882-5892.2002
40. Bangari DS, Shukla S, Mittal SK. Comparative transduction efficiencies of human and nonhuman adenoviral vectors in human, murine, bovine, and porcine cells in culture. Biochem Biophys Res Commun (2005) 327(3):960–6. doi: 10.1016/j.bbrc.2004.12.099
41. Noblitt LW, Bangari DS, Shukla S, Knapp DW, Mohammed S, Kinch MS, et al. Decreased tumorigenic potential of EphA2-overexpressing breast cancer cells following treatment with adenoviral vectors that express EphrinA1. Cancer Gene Ther (2004) 11(11):757–66. doi: 10.1038/sj.cgt.7700761
42. Hoelscher MA, Garg S, Bangari DS, Belser JA, Lu X, Stephenson I, et al. Development of adenoviral-vector-based pandemic influenza vaccine against antigenically distinct human H5N1 strains in mice. Lancet (2006) 367(9509):475–81. doi: 10.1016/s0140-6736(06)68076-8
43. Vemula SV, Ahi YS, Swaim AM, Katz JM, Donis R, Sambhara S, et al. Broadly protective adenovirus-based multivalent vaccines against highly pathogenic avian influenza viruses for pandemic preparedness. PLoS One (2013) 8(4):e62496. doi: 10.1371/journal.pone.0062496
44. Pandey A, Singh N, Vemula SV, Couetil L, Katz JM, Donis R, et al. Impact of preexisting adenovirus vector immunity on immunogenicity and protection conferred with an adenovirus-based H5N1 influenza vaccine. PLoS One (2012) 7(3):e33428. doi: 10.1371/journal.pone.0033428
45. Sayedahmed EE, Kumari R, Mittal SK. Current use of adenovirus vectors and their production methods. In: Manfredsson FP, Benskey MJ, editors. Viral Vectors for Gene Therapy: Methods and Protocols. New York, NY: Springer New York (2019). doi: 10.1007/978-1-4939-9065-8_9
46. Sayedahmed EE, Mittal SK. A potential approach for assessing the quality of human and nonhuman adenoviral vector preparations. Can J Vet Res (2020) 84(4):314–8.
47. Papp Z, Middleton DM, Mittal SK, Babiuk LA, Baca-Estrada ME. Mucosal immunization with recombinant adenoviruses: induction of immunity and protection of cotton rats against respiratory bovine herpesvirus type 1 infection. J Gen Virol (1997) 78(Pt 11):2933–43. doi: 10.1099/0022-1317-78-11-2933
48. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (2013) 29(1):15–21. doi: 10.1093/bioinformatics/bts635
49. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics (2015) 31(2):166–9. doi: 10.1093/bioinformatics/btu638
50. Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinf (2010) 11:422. doi: 10.1186/1471-2105-11-422
51. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26(1):139–40. doi: 10.1093/bioinformatics/btp616
52. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15(12):550. doi: 10.1186/s13059-014-0550-8
53. Blighe K, Rana S, Lewis M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. (2019). Available at: https://bioconductor.org/packages/devel/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html.
54. Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics (2020) 36(8):2628–9. doi: 10.1093/bioinformatics/btz931
Keywords: innate immunity, mucosal immunity, mucosal immunization, bovine adenoviral vector, human adenoviral vector, TLR, cytokine, chemokine
Citation: Sayedahmed EE, Elshafie NO, Zhang G, Mohammed SI, Sambhara S and Mittal SK (2023) Enhancement of mucosal innate and adaptive immunity following intranasal immunization of mice with a bovine adenoviral vector. Front. Immunol. 14:1305937. doi: 10.3389/fimmu.2023.1305937
Received: 02 October 2023; Accepted: 01 November 2023;
Published: 21 November 2023.
Edited by:
Bingdong Zhu, Lanzhou University, ChinaReviewed by:
Eric Weaver, University of Nebraska-Lincoln, United StatesCopyright © 2023 Sayedahmed, Elshafie, Zhang, Mohammed, Sambhara and Mittal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suresh K. Mittal, bWl0dGFsQHB1cmR1ZS5lZHU=; Suryaprakash Sambhara, c3NhbWJoYXJhQGNkYy5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.