
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 12 December 2023
Sec. Comparative Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1289769
 Walter Conca1,2,3*†‡
Walter Conca1,2,3*†‡ Soad M. Saleh2†‡
Soad M. Saleh2†‡ Rana Al-Rabiah2‡
Rana Al-Rabiah2‡ Ranjit Singh Parhar2
Ranjit Singh Parhar2 Mahmoud Abd-Elnaeim4
Mahmoud Abd-Elnaeim4 Hussein Al-Hindas2‡
Hussein Al-Hindas2‡ Alexander Tinson5‡
Alexander Tinson5‡ Katharina Birgit Kroell6
Katharina Birgit Kroell6 Klaus Roman Liedl6‡
Klaus Roman Liedl6‡ Kate Collison2‡
Kate Collison2‡ Uday Kishore7‡
Uday Kishore7‡ Futwan Al-Mohanna2,3‡
Futwan Al-Mohanna2,3‡Introduction: The evolution of adaptive immunity in Camelidae resulted in the concurrent expression of classic heterotetrameric and unconventional homodimeric heavy chain-only IgG antibodies. Heavy chain-only IgG bears a single variable domain and lacks the constant heavy (CH) γ1 domain required for pairing with the light chain. It has not been reported whether this distinctive feature of IgG is also observed in the IgA isotype.
Methods: Gene-specific primers were used to generate an IgA heavy chain cDNA library derived from RNA extracted from the dromedary’s third eyelid where isolated lymphoid follicles and plasma cells abound at inductive and effector sites, respectively.
Results: Majority of the cDNA clones revealed hallmarks of heavy chain-only antibodies, i.e. camelid-specific amino acid substitutions in framework region 1 and 2, broad length distribution of complementarity determining region 3, and the absence of the CHα1 domain. In a few clones, however, the cDNA of the canonical IgA heavy chain was amplified which included the CHα1 domain, analogous to CHγ1 domain in IgG1 subclass. Moreover, we noticed a short, proline-rich hinge, and, at the N-terminal end of the CHα3 domain, a unique, camelid-specific pentapeptide of undetermined function, designated as the inter-α region. Immunoblots using rabbit anti-camel IgA antibodies raised against CHα2 and CHα3 domains as well as the inter-α region revealed the expression of a ~52 kDa and a ~60 kDa IgA species, corresponding to unconventional and canonical IgA heavy chain, respectively, in the third eyelid, trachea, small and large intestine. In contrast, the leporine anti-CHα1 antibody detected canonical, but not unconventional IgA heavy chain, in all the examined tissues, milk, and serum, in addition to another hitherto unexplored species of ~45 kDa in milk and serum. Immunohistology using anti-CHα domain antibodies confirmed the expression of both variants of IgA heavy chains in plasma cells in the third eyelid’s lacrimal gland, conjunctiva, tracheal and intestinal mucosa.
Conclusion: We found that in the dromedary, the IgA isotype has expanded the immunoglobulin repertoire by co-expressing unconventional and canonical IgA heavy chains, comparable to the IgG class, thus underscoring the crucial role of heavy chain-only antibodies not only in circulation but also at the mucosal frontiers.
Within the wider context of exploring mechanisms of adaptation of ocular immune defense to biohazards in the desert habitat, the present study was based on the fundamental knowledge that one-humped Arabian camels or dromedaries (Camelus dromedarius) share with other species of the Camelidae family, which includes Camelus ferus (two-humped wild or feral camel), Camelus bactrianus (two-humped Bactrian camel), Lama glama (llama), Lama guanicoe (guanaco), Vicugna vicugna (vicuña) and Vicugna pacos (alpaca), a distinguishing characteristic of circulating antibodies of the IgG isotype, namely the absence of the constant heavy (CH) γ1 domain that covalently pairs with the constant domain of the light chain (CL), resulting in the formation of homodimeric heavy chain-only (alias ‘only-heavy-chain’) antibodies where the paratope comprises the variable domain of the heavy chain variant, termed VHH/VHH/VHH (all three acronyms are in use) (1–3). These unconventional antibodies were also discovered in the nurse shark (Ginglymostoma cirratum) and labeled as neo or nurse shark antigen receptor (4, 5). Over the years, these discoveries were widely exploited and resulted in several medical innovations (6–8). Although the etiology and pathways which drove B cell ontogeny in Camelidae to produce a functional IgG antibody alternative remain enigmatic, it is important to bear in mind that unconventional heavy chain-only antibody subclasses IgG2 (IgG2a and IgG2c) and IgG3 circulate beside canonical heterotetrameric subclass IgG1 (IgG1a and IgG1b), inferring that either antibody variant is a key effector in humoral immune defense (9–14). Whether in the Arabian camel other immunoglobulin classes share this exceptional feature of antibody structure has so far not been reported.
Therefore, we embarked on studies in the dromedary and focused on IgA as the established protagonist of adaptive immune defense at the ocular surface, which is protected by a tear fluid film produced by lacrimal and sebaceous glands as well as goblet cells in the conjunctiva (15–18). Primers deduced from amino acid sequences in constant domains of IgA heavy chain of alpaca and two-humped Bactrian camel served to create a library of IgA heavy chain cDNA amplified by PCR from mRNA isolated from the third eyelid of the dromedary, which harbors the conjunctiva-associated lymphoid tissue (CALT), a large sebaceous gland and the gland of Harder or Harderian gland (HG) (19–23). We show here that most of the amplified IgA heavy chain transcripts were devoid of the CHα1 domain, the fingerprint of heavy chain-only antibodies, while few corresponded to conventional IgA heavy chain transcripts. We also provide protein data confirming the translation of both IgA subclasses not only in CALT, but also in the upper respiratory and the intestinal tract. These observations fill a gap in our knowledge about the composition of the dromedary’s mucosal IgA, and favor an evolutionary scenario in which exposure to microbial pathogens and commensal microflora (24), probably forced by environmental hazards in the desert habitat, such as unabated exposure to UV light, temperature and humidity extremes (25), as well as metabolic and osmotic derangements during starvation and dehydration (26), accounted for a distinct and likely “rescue” or “salvage” pathway of antibody synthesis capable of complementing or alternating with, yet not replacing, the highly complex, fine-tuned endoplasmic assembly of heavy and light chains (27–29).
Third eyelids in toto, tissue samples of the trachea, as well as the small and large intestine were harvested at necropsy from three apparently healthy, subadult (one to two years of age) male one-humped camels (C1, C2 and C3), crossbreeds of Sofor and Waddah (alias Wodeh), at the municipal abattoir in Riyadh, Central Region of Saudi Arabia, immediately fixed in 10% (v/v) neutral buffered formalin for >24 hours for histomorphology and immunohistochemistry, or snap frozen in liquid nitrogen for RNA and protein extraction. After fixation, tissues were paraffin-embedded and cut into 4-μm sections. Sequential sections were either stained with hematoxylin and eosin (H&E) or analyzed by immunohistology. Tissues of camel C3 were used for further analyses. Blood was drawn by venipuncture from the external jugular vein from healthy female and male dromedaries at a camel farm in Buraidah, Central Region of Saudia Arabia, allowed to clot and sera were stored at -20°C. Camel milk was collected at a local farm in the vicinity of Riyadh. Approval to carry out the sampling procedures was obtained from “The Animal Care and Use Committee” of the King Faisal Specialist Hospital & Research Center (RAC#2180-023).
Total RNA was isolated from the third eyelid of dromedary C3 with Zymo® according to the manufacturer’s instructions (Zymo Research, Irvine, CA, USA). RNA was quantified by NanoDrop™ spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and its integrity assessed by 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA, USA). We first generated a ~500 base pair (bp) product using primers derived from the constant domains of IgA heavy chain from alpaca and Bactrian camel. The cDNA sequences obtained showed ~97% identity with nucleotide sequences of constant domains of IgA heavy chains of these two camelids. Thereafter, we amplified further towards the N-terminal and C-terminal ends of the cDNA using the RACE technique. RACE is a PCR-based technique which facilitates the cloning of full-length cDNA sequences. RACE is widely used to amplify unknown cDNA sequences towards the 5’ and 3’ ends of mRNA using gene-specific primers (GSP) derived from a known short internal stretch of the mRNA of interest (30). RACE was performed with the Invitrogen™ GeneRacer™ Kit (Thermo Fisher Scientific, Waltham, MA, USA) to generate full-length cDNA as follows: 4μg of total RNA was dephosphorylated with calf intestine alkaline phosphatase to remove free 5’ phosphates from rRNA, fragmented RNA, tRNA and contaminating DNA. Full-length capped mRNA was treated with tobacco acid pyrophosphatase to remove the 5’ cap structure. This treatment left a phosphate at the 5’ end required for ligation to 44-base-long GeneRacer™ RNA Oligo (5’-CGACTGGAGCACGAGGACACTGACATGGACTGAAGAGTAGAAA-3’) using T4 RNA ligase, which provides a priming site for GeneRacer™ 5’ primer and 5’ nested primer. Ligated mRNA was reverse transcribed using Superscript™ III Reverse Transcription (RT) and the 60-base GeneRacer™ oligo (dT) primer (5’-GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)24-3’) containing a dT tail of 24 nucleotides to create RACE-ready first-strand cDNA with known priming sites at the 5’ and 3’ ends, and restriction sites for efficient cDNA synthesis for cloning and sequencing purposes after PCR amplification. The GeneRacer™ oligo (dT) primer sequence contains the priming sites for the GeneRacer™ 3’ and the GeneRacer™ 3’ nested primers. After comparing nucleotide sequences of constant domains of IgA heavy chains of other camelids, reverse (5’-AGCAGGTGGACCTGGGGCCGGAT-3’) and forward (5’- ATCCGGCCCCAGGTCCACCTGCT-3’) GSP were chosen from the CHα3 domain of IgA heavy chain of the Bactrian camel (GenBank #KP999944.1; nucleotides 643-665 of a partial coding sequence; N-terminus of CHα3 domain) using the online primer designing tool in NCBI (NCBI Primer Blast) to amplify the 5’ and 3’ ends of dromedary IgA heavy chain cDNA, respectively. As an internal control, β-actin mRNA was amplified. PCR amplifications of 1 μl RT template were performed in 50 μl aqueous reaction mixture consisting of High-Fidelity PCR buffer, 2 mM MgSO4, 200 nM of each dNTP, 200 nM forward or reverse GSP, 600 nM GeneRacer™ 5’ primer or 3’ primer, and 2.5 U Platinum® Taq DNA polymerase High Fidelity that generated blunt end products (Thermo Fisher Scientific, USA), using hot start and touchdown PCR, following the manufacturer’s recommendations. To obtain 5’ ends, first-strand cDNA was amplified using the reverse GSP and the GeneRacer™ 5’ primer and 5’ nested primer. Only mRNA that had the GeneRacer™ RNA Oligo ligated to the 5’ end and was completely reverse transcribed, was amplified by PCR. To obtain the 3’ ends, first-strand cDNA was amplified using the forward GSP and GeneRacer™ 3’ primer and GeneRacer™ 3’ nested primer. Only mRNA that had a poly(A) tail and was reverse transcribed was amplified by PCR. Three negative PCR controls (no template; no GSP; no GeneRacer™ 5’ or 3’ primers) were included to eliminate non-specific amplification. Cycling parameters were as follows: 1 cycle: 2 min at 94°C; 5 cycles: 30 sec at 94°C and 1min/1kb DNA at 72°C; 25 cycles: 30 sec at 94°C; 45 sec at 65°C and 2 min at 68°C; 1 cycle for 10 min at 70°C. After PCR, 10 μl of the amplified product was analyzed on a 1.2% (w/v) agarose gel.
PCR-amplified 5’ and 3’ cDNA fragments were separated on a 1.2% (w/v) agarose gel. Bands were eluted using QIAquick Gel Extraction Kit (Qiagen, Inc., Chatsworth, CA, USA), then ligated into Zero Blunt™ TOPO™ vector (Invitrogen, Carlsbad, CA, USA) using DNA Ligation Kit, Mighty Mix (Takara Bio USA, San Jose, CA, USA), and transformed into chemically competent TOP10 E. coli cells (Invitrogen, Carlsbad, CA, USA). Individual colonies were grown, and plasmids purified using a QIAprep Spin Miniprep Kit (Qiagen, Inc., Chatsworth, CA, USA). For analysis of the 5’ RACE products, those colonies with complete sequence of the GeneRacer™ RNA Oligo and reverse GSP were selected indicating that the intact 5’ RACE product was ligated to GeneRacer™ RNA Oligo. DNA was isolated from selected transformants and sequenced from both directions using M13 forward and M13 reverse sequencing primers with BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems™, Inc. Foster City, CA, USA) at the King Faisal Specialist Hospital & Research Center sequencing facility. Forward and reverse sequences were aligned using EMBOSS needle pairwise sequence alignment algorithms and DNASTAR™ program and merged using EMBOSS. The same strategy was used for 3’ RACE colonies obtained after amplification in the presence of GeneRacer™ oligo (dT) primer sequence and forward GSP. Overlapping 5’ and 3’ RACE sequences were then combined, and the complete cDNA sequence of IgA heavy chain obtained.
From conceptually translated amino acid sequences of the constant domains CHα1 (SPSVFPLGPSYDKASRQVALACLVHGFFPPAPLKVTWGLSGQNVSVMDFPAVQPASGVLYTMSSQLTTPVEQCPDSEIVTCQVQHLSSSSKTVNVPCK), CHα2 (CCNPSLALHPPALEDLLLGSNASLTCTLSGLRNPEGAQFTWTPSGGKVAVQQSPKSDPCGCFSVSSVLPGCAEQWNSKTNFSCSATHPESKNTLYATITK) and CHα3 (IRPQVHLLPPPSEELALNE MVTLTCVVRGFSPKDVLVRWLHGNQELPREKYLTWRPLPEPEQSITTYAVTSLLRVEAEAWKQGDNYSCMVGHEALPLAFTQKTIDRLSGKPTHVNVSVVMAEAEGVCY) the cDNA sequence was derived, His tagged and expressed in E. coli.
All proteins were custom-made by GenScript HK Limited, Hong Kong SAR (PolyExpress Premium antigen specific affinity purified pAb package). Freund’s adjuvant was added to the immunogen. Six New Zealand rabbits (two rabbits/recombinant protein) were immunized with three injections by conventional protocol. Seven days after the third immunization, the titre of antiserum was tested by ELISA to detect antibody response. Total IgG from pre-immune serum was used as a negative control. Rabbits were sacrificed according to animal welfare principles. The final bleed antiserum was purified by antigen affinity column. 0.02% (w/v) sodium azide was added as preservative, and antibody preparations were stored at -20°C.
The oligopeptide, ATITKSSEDAIRPQVC, spanning the inter-α region (IAR), was generated with a cysteine added at the C-terminus by 9-fluorenylmethyloxycarbonyl (Fmoc) solid phase peptide synthesis on resin and then removed from the resin by treatment with trifluoroacetic acid (31). Oligopeptides were conjugated with keyhole limpet hemocyanin, then purified by reverse-phase HPLC with acetonitrile and water, and verified by mass spectrometry. The purified product was used for immunization of two rabbits following the same protocol adopted for the immunization with CHα domains and the antibody was purified by antigen affinity column.
Serum proteins were solubilized in ice-cold RIPA lysis buffer (Sigma-Aldrich, St. Louis, MO, USA) containing cOmplete™ Mini protease inhibitors cocktail (Roche Diagnostics, Mannheim, Germany). Similarly, RIPA lysates were made from snap-frozen samples of various camel tissues. 10-20 µg of total protein were mixed with SDS-PAGE loading buffer, pH 7.0, containing 5% (v/v) β-mercaptoethanol, heated to 95°C for 5 min, and separated by SDS-PAGE at 20 mA using NuPAGE™ 10% (w/v), Bis-Tris, 1.5 mm, Mini Protein gels (Invitrogen, Carlsbad, CA, USA) in a Mini-PROTEAN Tetra vertical electrophoresis cell (Bio-Rad, Hercules, CA, USA). The gels were fixed and stained with 50% (v/v) methanol, 10% (v/v) acetic acid, 0.1% (w/v) Coomassie Brilliant Blue R-250, and de-stained with 40% (v/v) methanol and 10% (v/v) acetic acid. Proteins were then transferred to 0.2 µm nitrocellulose membranes (Bio-Rad, Hercules, CA, USA) using the Trans-Blot® Turbo™ semi-dry transfer system (BioRad, Hercules, CA, USA). Anti-CHα1 domain (1/2500), anti-CHα2 domain (1/2500), anti-CHα3 domain (1/2500) and anti-IAR (1/2500) primary antibodies, diluted in 3% (w/v) non-fat dry milk, were added in TBS-Tween buffer overnight at 4°C, followed by washing in TBS-Tween buffer. Thereafter, horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG antibody (1:10000) (Promega, Madison, WI, USA), diluted in 3% (w/v) non-fat dry milk, was added and the blot incubated in TBS-Tween buffer for 2 hours at room temperature, followed by three washes with PBS-Tween buffer. Proteins were detected using Pierce™ ECL Plus Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA, USA) and images were acquired via ChemiDoc Imaging System (Bio-Rad, Hercules, CA, USA). Densitometry analysis was performed using Image Lab software (Bio-Rad, Hercules, CA, USA).
ultraView® Universal DAB Detection Kit (Ventana Medical Systems, Oro Valley, AZ, USA) in combination with BenchMark ULTRA IHC/ISH System (Ventana Medical Systems, Oro Valley, AZ, USA) fully automated instrument, was used for the detection of IgA heavy chains in formalin-fixed paraffinized tissue sections of various organs using rabbit anti-CHα1, CHα2, and anti-IAR primary antibodies. The reagents provided with the kit do not contain biotin, and therefore, non-specific staining from endogenous biotin is eliminated. Deparaffinization was done at 72°C, followed by cell conditioning (heat-induced epitope retrieval) with ULTRA CC1 solution for 64 min at 95°C. Primary antibodies were applied manually, all other reagents automatically in a pre-diluted dispenser, to tissue sections placed on glass microscope slides and were then exposed to a “cocktail” of secondary antibodies, consisting of goat anti-rabbit IgG, goat anti-mouse IgG and IgM, conjugated to HRP by long-arm linkers (HRP multimer). Primary antibody incubation time was 20 min, followed by secondary antibody incubation time for 8 min at room temperature. The immune complex was then visualized with the chromogen 3,3’-diaminobenzidine tetrahydrochloride (DAB) and H2O2, which upon oxidation formed a brown precipitate. Secondary antibody alone was used as a negative control. Counterstaining was performed with hematoxylin first, followed by post-counterstaining with bluing reagent for 8 min. Microphotographs were taken with an Olympus BX53 digital camera (Olympus Corporation, Tokyo, Japan).
The histomorphologic correlates of the mucosal immune response in the third eyelid are shown in Figure 1. In Figure 1A, the bulbar part of the HG is adjacent to an agglomerate of isolated lymphoid follicles (ILF) lacking germinal centers, each one situated beneath the specialized follicle-associated epithelium (arrow) and the subepithelial dome (*), a complex functional unit that consists of a cluster of B cells built on stromal cells, surrounded by many dendritic cells and few T cells. Here, at the conjunctival cul-de-sac, antigen uptake via M cells and processing by antigen-presenting cells is known to take place, followed by IgA class switching in B cells, mostly independent of T cells. Therefore, ILFs represent an inductive site of conjunctival B cell maturation in response to airborne antigens at the ocular surface and resemble the architecture of lymphoid tissue in mucosa-associated lymphoid tissues (MALT) of other species (32–41). In Figure 1B, acini in the secretory endpieces of the tubulo-alveolar mucous HG are lined with columnar cells with basally situated nuclei and a narrow lumen (*). They are surrounded by stroma which harbors clusters of plasma cells, the effector cells of humoral immunity and principal source of lacrimal IgA, displaying their characteristic morphology, with eccentric nucleus, condensed chromatin resulting in “clock face” morphology and cytoplasmic “Hof” (arrows).
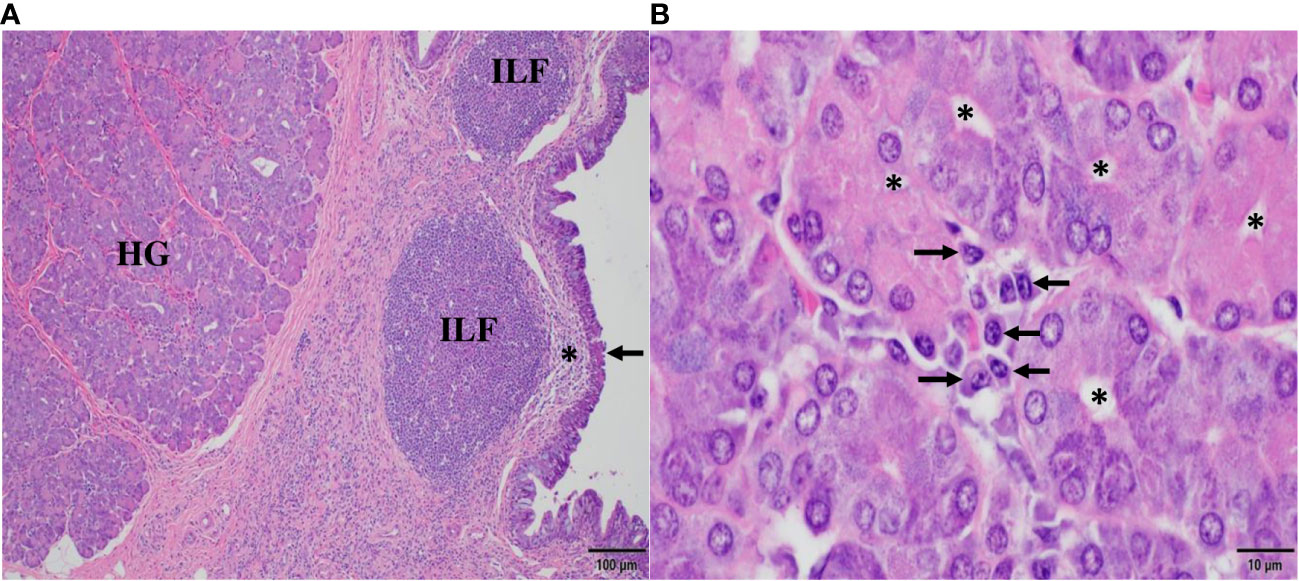
Figure 1 Histomorphology of conjunctiva-associated lymphoid tissue and the Harderian gland (HG) in the third eyelid of the dromedary. In (A), posterior (bulbar) to the HG, numerous isolated lymphoid follicles (ILF) beneath subepithelial domes (*) and the specialized follicle-associated epithelium (arrow) of the conjunctiva represent the inductive site of mucosal B cell maturation. In (B), numerous plasma cells (arrows) cluster between the acini of the HG and form the principal effector site of the mucosal humoral immune response. Asterisks (*) mark the lumen of acini. Magnifications and scale bar lengths are as follows: in (A) 10x and 100 μm, respectively; in (B), 100x and 10 μm, respectively. Sections were stained with H&E.
5’ RACE PCR products separated on 1.2% (w/v) agarose gels revealed two species: most were ~800 bp, a few ~1100 bp long (Figure 2A). In contrast, a single amplification product of ~450 bp length was obtained from 3’ RACE (Figure 2B). 5’ and 3’ PCR species were ligated into Zero Blunt™TOPO™ vector for sequencing. Out of 124 clones analyzed, two corresponded to the longer 5’ PCR products and included the CHα1 domain, whereas all other clones of the shorter 5’ PCR product lacked the CHα1 domain, which defines an unconventional IgA heavy chain. 5’ and 3’ RACE PCR products were joined together through their overlapping ends to generate two species of full-length cDNA of ~1.3 kbp and ~1.6 kbp, corresponding to unconventional and canonical IgA heavy chain, respectively. For comparative purposes, representative, conceptually translated complete protein sequences of classic IgA heavy chain (IgA classic; 485 amino acids; top) and unconventional IgA heavy chain (IgA HC; 388 amino acids; bottom) are shown in Figure 2C. While scrutinizing databases for dromedary IgA sequences we found a partial amino acid sequence (292 amino acids) of the constant domain of classic IgA heavy chain, starting with an internal methionine (UniProt #A0A5N4E0V1. A0A5N4E0V1_CAMDR; “IgA alpha-1 chain C region”) that was translated from the whole genome shotgun sequence “Camelus dromedarius/ breed African/ isolate Drom800/ chromosome 6/IgA alpha-1 chain C region” (GenBank #JWIN03000006.1; protein_id #KAB1277053.1) (45). Individual components of the variable domain of classic IgA heavy chain (VH) and unconventional IgA heavy chain (VHH), i.e. framework regions (FR) 1, 2, 3 and 4, and complementarity determining regions (CDR) 1, 2 and 3, as well as constant domains CHα1, CHα2 and CHα3, are indicated above the amino acid sequence, with normal and bold font letters alternating for a better illustration. Variable domains were defined according to IMGT® delimitations (http://www.imgt.org, IMGT Scientific chart>Numbering>IMGT unique numbering for V-DOMAIN and V-LIKE DOMAIN) (42). A 19-mer leader peptide precedes the constituents of the variable domain FR1, CDR1, FR2, CDR2, FR3, CDR3 and FR4. The multiple constant domains of classic IgA heavy chain are numbered from the N-terminal to the C-terminal end as follows: CHα1, CHα2 and CHα3. The proline-rich (VPPPPPP) hinge region is positioned between the CHα1 and the CHα2 domain in canonical IgA heavy chain, which like its unconventional counterpart ends in the tailpiece (Tp), an 18-mer oligopeptide extension critical for the formation of IgA polymers. These constant domains were specified according to the scheme of human myeloma IgA1 Bur and IgA2 But (43, 44). In contrast, in unconventional IgA heavy chain, the CHα1 domain spanning 102 amino acids is absent, as indicated by dashes, and therefore, FR4 of the VHH domain, with the canonical J motif W-G-X-G and the variable/constant switch region consisting of the tripeptide VSS, directly precedes the heptameric hinge, which was identical in all clones. Downstream of the hinge region, we observed 100% identity among the variants of dromedary IgA heavy chains. 101 amino acid sequences of heavy chain-only IgA were aligned in the Supplementary File 1. In Figure 2C, key substitutions with hydrophilic amino acids in FR1 (S for L) and FR2 (F for V, E for G, R for L, G for W) of heavy chain-only IgA are underlined as they are considered crucial for the conformation and solubility of the VHH domain (2). As expected, the highest variability in length and amino acid composition was observed in the V-D-J recombined CDR3, ranging from 14 to 26 amino acids in our library. Of note, we observed a distinct pentapeptide, consisting of S, E, D, A and I at the N-terminal end of the CHα3 domain, intriguingly different from other mammals at this site (Figure 3). We labeled the pentapeptide as the IAR and used it to immunize rabbits.
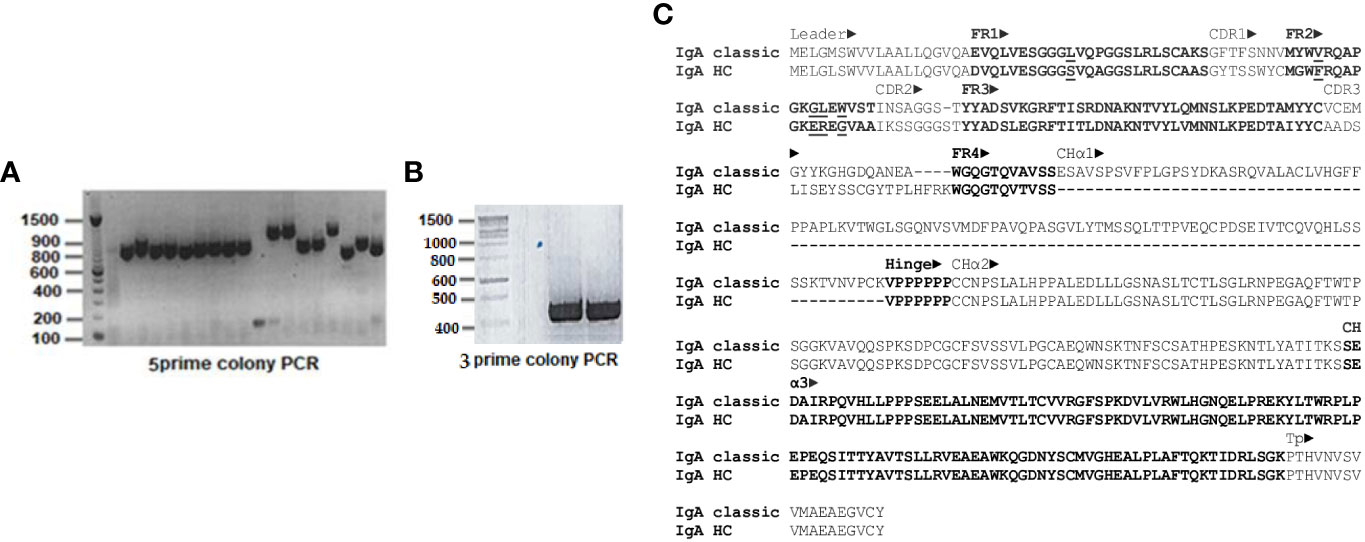
Figure 2 Rapid amplification of 5’ and 3’ cDNA ends (RACE) of dromedary IgA heavy chain and primary structure of classic and unconventional IgA heavy chain. In (A), PCR products of 5’ RACE, and in (B), 3’ RACE are shown after agarose gel electrophoresis, with a DNA ladder loaded in the first well. The majority of 5’ PCR species were ~800 bp, a few were ~1.1 kbp. In contrast, 3’ RACE PCR species consisted uniformly of ~450 bp. There were some minor variations in the length of the ~800 kbp 5’ PCR products, reflecting the heterogeneity of CDR in VHH domains. The difference of ~300 bp among the two 5’ PCR species, however, was explained by identification of the CHα1 domain in classic IgA heavy chain. In (C), two representative amino acid sequences of dromedary IgA heavy chains chosen from our library were aligned, with conventional IgA heavy chain (IgA classic) at the top and unconventional IgA heavy chain (IgA HC) at the bottom. Variable domains of both heavy chains (VH and VHH), preceded by a 19-mer leader peptide, comprised four framework regions (FR1, FR2, FR3 and FR4) and three complementarity determining regions (CDR1, CDR2 and CDR3), whereas constant domains of classic IgA heavy chain encompassed three (CHα1, CHα2 and CHα3), in contrast to only two domains (CHα2 and CHα3) in the unconventional format. A short hinge region, consisting of valine and six consecutive prolines, was located between CHα1 and CHα2 domains in conventional IgA heavy chain, in contrast to the hinge region in heavy chain-only IgA which separated FR4 of the VHH domain from the CHα2 domain. The tailpiece (Tp) represented the C-terminal end of all IgA heavy chains variants. Dashes indicate deficient single amino acid residues as well as lack of the entire CHα1 domain in heavy chain-only IgA. Definition of the constituents of the variable and constant domains (marked by alternating normal font and bold letters) was accomplished according to IMGT® delimitations (42) and the numbering scheme of human IgA1 and IgA2 chains, respectively (43, 44). Underlined residues in FR1 and FR2 indicate amino acid changes considered critical for conformation and solubility of the VHH domain of heavy chain-only IgA. Further downstream, a unique pentapeptide (SEDAI) marked the N-terminus of the CHα3 domain.

Figure 3 Amino acid sequences of the CHα2-CHα3 boundary in mammals. Amino acid sequences at the 3’ terminal end of CHα2 and the 5’ terminus of CHα3 domains of the dromedary, Bactrian camel and alpaca were compared with those of other mammals. Names in Latin are to the left. The gap between CHα2 and CHα3 domains is deliberate. A distinct pentapeptide (bold letters) consisting of serine (leucine in alpaca), glutamic acid, aspartic acid, alanine (proline in alpaca) and isoleucine was only found in camelids, upstream of a highly conserved arginine followed by an invariant proline at the N-terminus of the CHα3 domain. Glutamic acid at the 5’ end of CHα3 was also present in gelada baboon, horse, donkey, guinea pig, red squirrel, and California grey whale, whereas aspartic acid at this position was identified only in the greater bamboo lemur. In contrast, the most frequently encountered motif in the interdomain region of other mammals at the N-terminal end of the CHα3 domain was a tetrapeptide consisting of glycine, asparagine, threonine (or methionine, isoleucine, and valine) and phenylalanine.
Proteins of tissue lysates (Figure 4) of the small (lane 1) and large intestine (lane 2), trachea (lane 3), third eyelid (lane 4), and also from whey milk (lane 5), dromedary serum (female and male, lane 6 and lane 7, respectively) and human serum from a male donor (lane 8), were separated by SDS-PAGE electrophoresis under reducing conditions, transferred to nitrocellulose membranes and probed with four different polyclonal antibodies raised in rabbits against IgA CH domains: CHα1 (panel A), CHα2 (panel B), CHα3 (panel C) and the IAR (panel D). We predicted that the anti-CHα1 antibody would visualize canonical IgA heavy chain, but not the unconventional counterpart, resulting in a single band in immunoblots, whereas anti-CHα2, anti-CHα3 and anti-IAR antibodies were expected to detect two protein species of different Mr. As shown in Figure 4A, the anti-CHα1 antibody indeed revealed one protein species of estimated Mr of ~60 kDa that corresponds to the conventional IgA heavy chain in small (lane 1) and large intestine (lane 2), third eyelid (lane 4), whey milk (lane 5), serum of female (lane 6) and male Arabian camel (lane 7), as well as two bands of ~60 kDa and ~65 kDa in human serum, likely cross-reacting with the two known IgA subclasses IgA1 (higher Mr) and IgA2 (lower Mr) (lane 8). The signal intensity of canonical IgA heavy chain band was faint in the tissue extract from the trachea, reflecting a lower density of plasma cells in the mucosa of the upper respiratory tract (lane 3). Surprisingly, the anti-CHα1 antibody also detected a polypeptide species with an estimated Mr of ~45 kDa in milk and serum, which merits further investigations. In contrast, the expression patterns of IgA heavy chains observed using anti-CHα2 (Figure 4B), anti-CHα3 (Figure 4C) and anti-IAR (Figure 4D) antibodies, which detect canonical as well as unconventional IgA heavy chain, were similar: the higher Mr species of ~60 kDa corresponded to canonical IgA heavy chain, whereas the lower Mr species of ~52 kDa represented the unconventional heavy chain variant. Whereas in the tissues examined both IgA heavy chain variants were expressed at comparable levels (lanes 1-4), the ~60 kDa band was more prominent in milk (lane 5). In circulation, however, only canonical IgA heavy chain was found, favoring the absence of heavy chain-only IgA in the blood (lanes 6 and 7). Weak cross-reactivity was observed with a ~60 kDa species in human serum (lane 8). Expectedly, the anti-IAR antibody failed to visualize any human IgA heavy chain species as the oligopeptide used for immunization is unique for Camelidae. Of note, the band density observed with the anti-IAR antibody appeared identical to that generated by the anti-CHα2 antibody, indicating comparable immunogenicity of the 15-mer oligopeptide and the CHα2 or the CHα3 domains, both comprising ~100 amino acids.
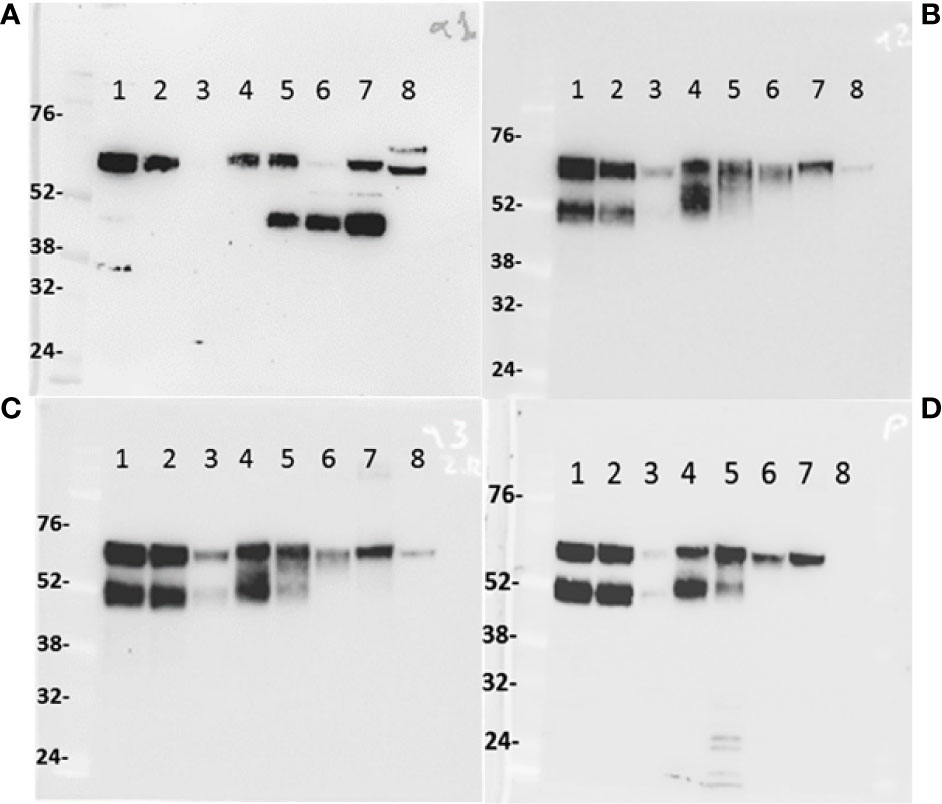
Figure 4 IgA heavy chain variants in the intestine, trachea, third eyelid, milk, and serum. One-humped camel proteins (~20 μg) from small (lane 1) and large intestine (lane 2), trachea (lane 3), third eyelid (lane 4), milk (lane 5), female (lane 6) and male (lane 7) serum and human male serum (lane 8) were separated under reducing conditions by 10% (w/v) SDS-PAGE, transferred to nitrocellulose membranes, probed with anti-CHα1 (A), anti-CHα2 (B), anti-CHα3 (C), and anti-IAR (D) antibodies, and visualized with HRP-conjugated anti-rabbit goat antibodies. In (A), the anti-CHα1 antibody reacted with a ~60 kDa species, indicating the expression of canonical IgA heavy chains, with cross-reactivity to two protein species of ~60 kDa and ~65 kDa in human serum (lane 8). In camel milk (lane 5) and sera (lanes 6 and 7), an additional prominent protein species of ~45 kDa was identified by the anti-CHα1, but not by other antibodies, most likely representing a proteolytic fragment of canonical IgA heavy chain. In (B–D), two protein bands of ~52 kDa and ~60 kDa of comparable intensity were identified in tissues (lanes 1, 2, 3 and 4), but not in milk (lane 5) nor serum (lanes 6 and lane 7) where the higher molecular weight species corresponding to canonical IgA heavy chains prevailed. Level of IgA expression in the trachea (lane 3) appeared low, likely reflecting lower plasma cell density in the mucosa of the upper respiratory tract. Cross-reactivity of circulating human IgA species was observed with all anti-CH domain antibodies, except the camel-specific anti-IAR antibody. Molecular weight markers in kDa are indicated to the left of each panel.
After delineating the histomorphologic features of CALT in the third eyelid, we studied by immunohistochemistry the expression of IgA heavy chains not only in the third eyelid but also in other MALT using primary antibodies raised in rabbits against different CHα domains (Figures 5–8). The aim was, first, to define the expression patterns of IgA heavy chains in MALT of the dromedary, and second, to determine the spatial distribution of IgA+ plasma cells within the tissues examined. We took advantage of purposely generated antibodies specific for CHα1, CHα2 and IAR, but refrained from using the anti-CHα3 antibody on tissue sections because the results obtained by immunoblotting with the anti-CHα3 antibody were identical to those observed with the anti-CHα2 and anti-IAR antibodies (Figure 4). Based on the results from Western blots we inferred that antibodies binding the CHα1 domain will detect classic, yet not unconventional IgA heavy chain, while antibodies binding the CHα2 domain and IAR would identify both variants of IgA heavy chain.
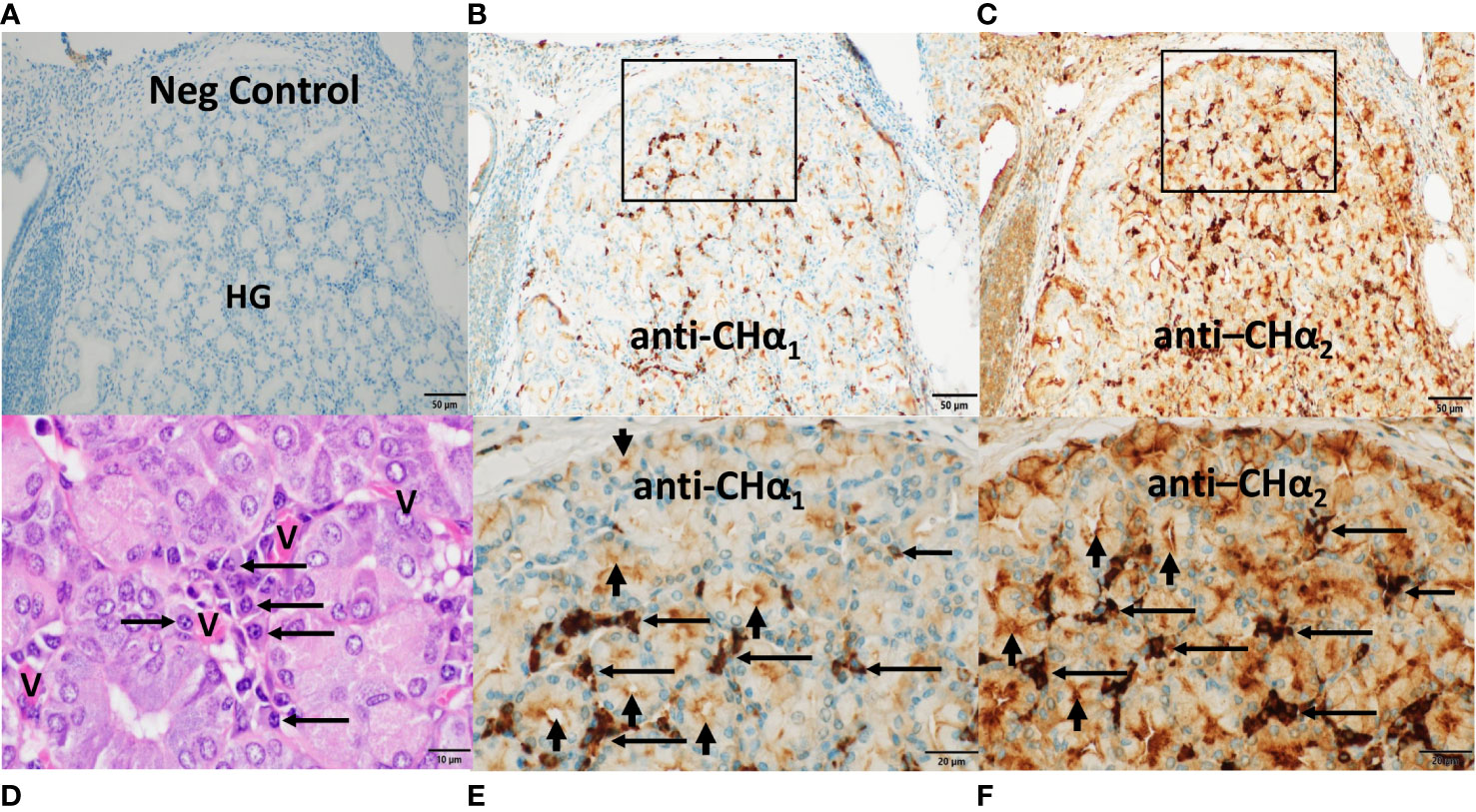
Figure 5 Expression of IgA heavy chain variants in the Harderian gland. Immunohistochemistry was performed on a section through the lacrimal (Harderian) gland (HG) of the third eyelid of the dromedary. Anti-CHα1 domain antibodies were used in (B, E) and anti-CHα2 domain antibodies in (C) and (F) and visualized by goat anti-rabbit HRP conjugated antibodies and hydrogen peroxide/DAB as substrates. The anti-CHα1 antibody detected canonical IgA heavy chains, whereas canonical and unconventional IgA heavy chains were both reactive to the anti-CHα2 antibody. In (A), no IgA was identified with secondary antibodies only. In (B, C), immunoreactivity was observed in the periacinar spaces and within epithelial cells throughout the lobule with anti-CHα1 and anti-CHα2 antibodies, respectively. In (E, F), two rectangular areas selected in (B, C) show, at higher power, intense immunostaining primarily in plasma cells in the periacinar spaces (horizontal arrows), and at the luminal apices of epithelial cells (short vertical arrows). In (D), an H&E-stained section demonstrates an agglomerate of typical plasma cells (horizontal arrows) and red blood cells within capillaries (V) adjacent to the basal membranes of acini. Magnifications and scale bar lengths were as follows: in (A–C), 20x and 50μm, respectively; in (D), 100x and 10 μm, respectively; in (E, F), 60x and 20μm, respectively.
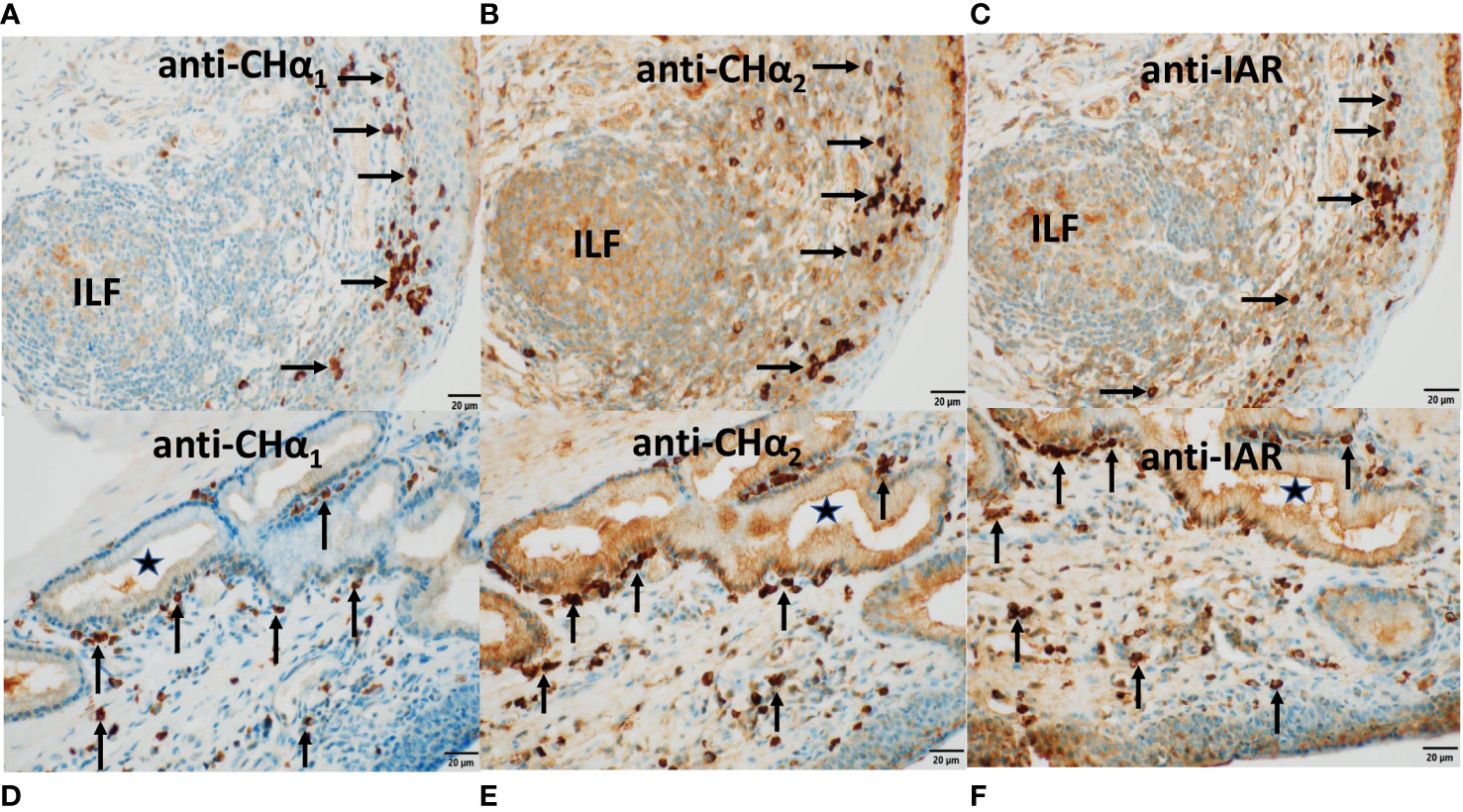
Figure 6 Expression of IgA heavy chain variants in MALT of the third eyelid and trachea. Immunostaining was performed on sections through the conjunctiva of the third eyelid (top panels) and on sections through the trachea (bottom panels) using antibodies raised against CHα1 domain in (A, D), CHα2 domain in (B, E), and IAR in (C, F), visualized by goat anti-rabbit HRP conjugated antibodies and hydrogen peroxide/DAB as substrates. The anti-CHα1 antibody detected canonical IgA heavy chain only. By contrast, both canonical and unconventional IgA heavy chains were identified by the anti-CHα2 as well as the anti-IAR antibody. In (A–C), IgA heavy chain immunoreactivity to all three antibodies was identified in plasma cells within the stratified epithelial layer of the conjunctiva and in the subconjunctival stroma (horizontal arrows) as well as within an isolated lymphoid follicle (ILF). IgA heavy chain immunoreactivity was also observed along the surface of the conjunctiva. In (D–F), IgA+ plasma cells (vertical arrows) were noticed alongside the basal membrane of a subepithelial mucous gland lined by columnar epithelium and in the subepithelial stroma of the pseudostratified tracheal epithelium. The lumen of the gland is marked by (*). Immunoreactivity was also present within the glandular epithelial layer, reflecting transcytosis. Of note, the brown color generated with the anti-CHα2 and the anti-IAR antibody was more intense. In this series of microphotographs, magnification and scale bar length was 40x and 20μm, respectively.
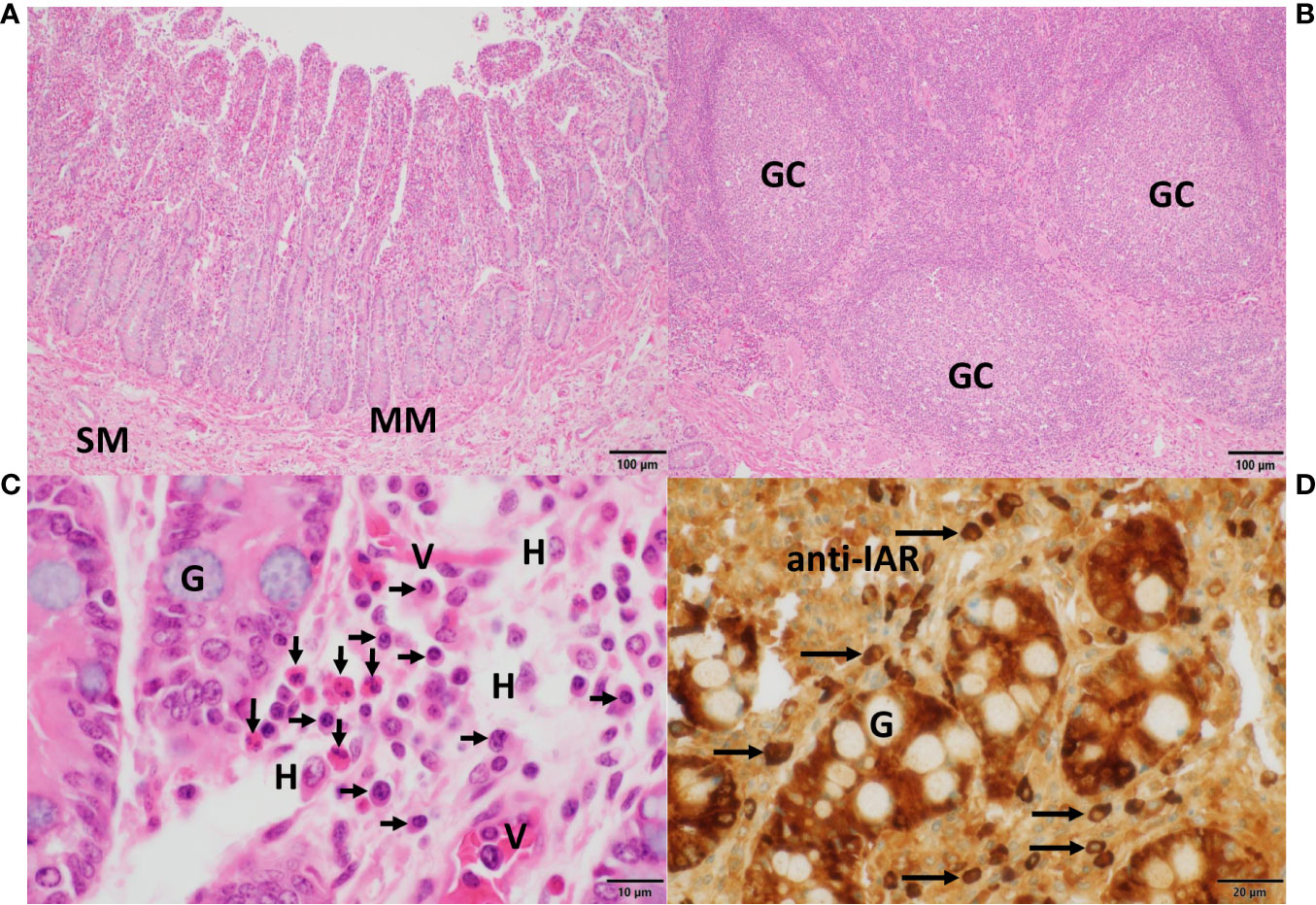
Figure 7 Histomorphology, expression of IgA heavy chains and tissue eosinophilia in the small intestine. In (A), villi and crypts characterized the mucosa of the small intestine, which was separated from the submucosa (SM) by the muscularis mucosae (MM). In (B), aggregates of submucosal lymphoid follicles with germinal centers (GC) surrounded by numerous lymphocytes, typical of Peyer’s patches, were present. In (C), details of crypts revealed interspersed goblet cells filled with mucin (G). In the adjacent lamina propria, numerous eosinophils (vertical arrows), plasma cells (horizontal arrows), histiocytes (H) and small vessels (V) were observed. Sections in (A–C) were stained with H&E. In (D), strong IgA heavy chain immunoreactivity to the anti-IAR antibody was detected in plasma cells (horizontal arrows) and within enterocytes while sparing mucin in goblet cells (G). Magnifications and scale bare lengths were as follows: in (A, B), 10x and 100μm, respectively; in (C), 100x and 10μm, respectively; in (D), 60x and 20μm, respectively.
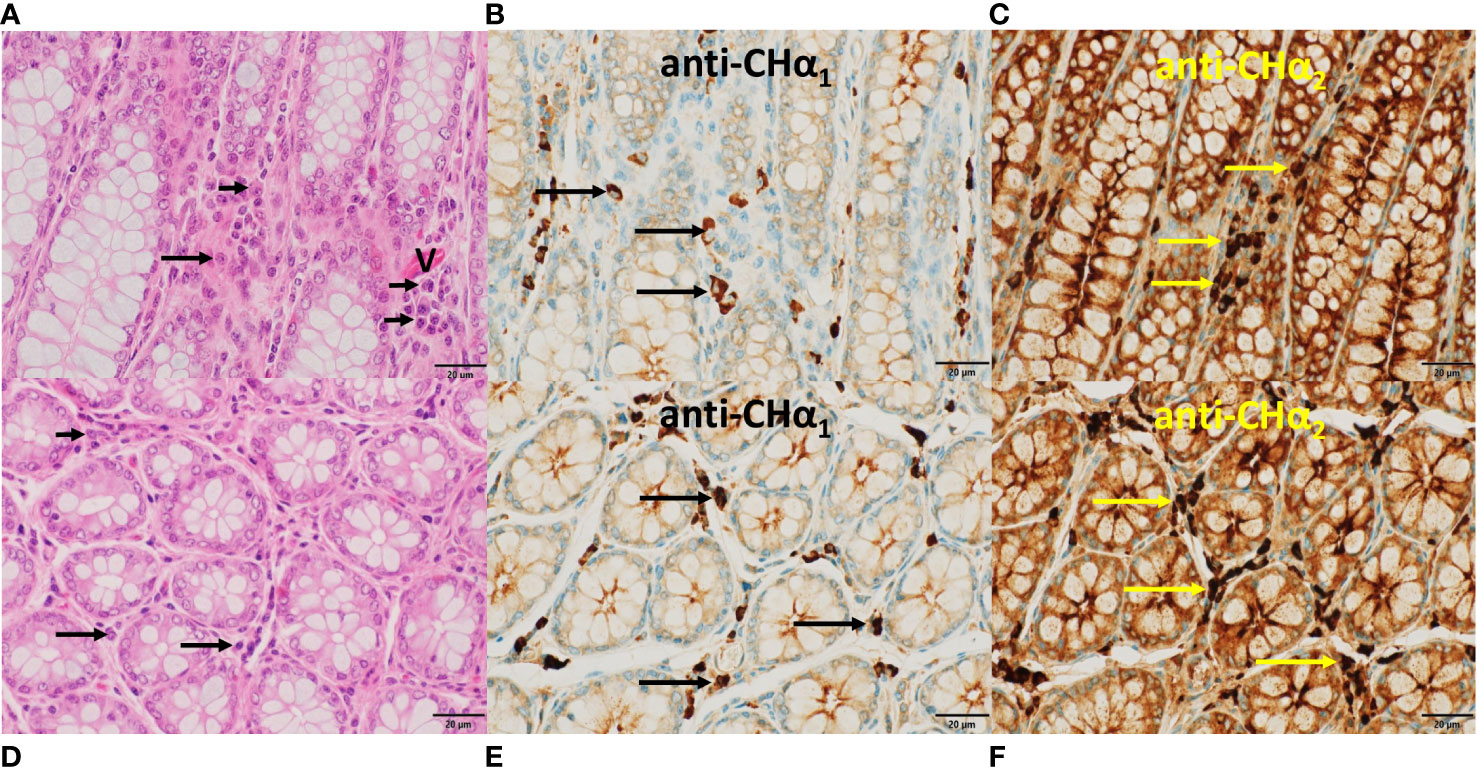
Figure 8 Histomorphology and expression of IgA heavy chain variants in the large intestine. In (A, D), the hallmarks of colon mucosa are shown in H&E-stained sections. Upper row panels depict longitudinal, lower row panels transverse sections. Crypts were lined primarily by mucin-enriched epithelial cells (goblet cells) and surrounded by lamina propria, where plasma cells (arrows) were found. In (B, E), plasma cells showed IgA heavy chain immunoreactivity to the anti-CHα1 (arrows), and in (C, F) to the anti-CHα2 antibody (yellow arrows). Immunoreactivity was also observed in the enterocytes within crypts, indicative of transcytosis. In (A), a capillary (V) was identified by the presence of red blood cells. In contrast to the lamina propria of the small intestine (Figure 7C) eosinophils were absent in this area of the colon. In all microphotographs magnification was 60x, and scale bar length was 20μm.
In sections through a lobule of the HG (Figure 5), we noticed that classic IgA heavy chain was expressed in plasma cells throughout the HG using the anti-CHα1 antibody, as shown in Figure 5B, and at higher magnification in Figure 5E. We surmise that heavy chains of both IgA subclasses were co-expressed in plasma cells using the anti-CHα2 antibody, as shown in Figure 5C and at higher magnification in Figure 5F. IgA+ plasma cells are indicated by horizontal arrows (Figures 5E, F). A negative control section is shown (Figure 5A), and, in an H&E-stained section (Figure 5D), a nest of plasma cells (horizontal arrows) resided in the periacinar stroma adjacent to the basal lamina of the columnar epithelial cells and next to capillaries (V) identified by intravascular red blood cells. The more intense brown color observed with both, the anti-CHα2 antibody in Figures 5, 6, 8, as well as the anti-IAR antibody in Figures 6, 7, was possibly related to non-specific background staining seen throughout the section. However, we hypothesize that an additive effect of immunoreactivity resulted in the apparent darker brown color as anti-CHα2 and anti-IAR antibodies react to both types of IgA heavy chains, whereas the anti-CHα1 antibody was selective for canonical IgA heavy chains. Immunostaining was also observed at the apical pole of acinar epithelial cells towards the luminal surface (Figures 5E, F; short vertical arrows), where IgA is transported via polymeric Ig receptor (pIgR)/secretory component (SC)-mediated transcytosis (46).
In CALT (Figure 6, top row), plasma cells (horizontal arrows) expressing IgA heavy chains detected by anti-CHα1 (Figure 6A), anti-CHα2 (Figure 6B) and anti-IAR (Figure 6C) antibodies are shown. They resided along the basal lamina of the stratified conjunctiva on the bulbar side of the third eyelid overlying an ILF, while some of them were found within the epithelium. The observation of intraepithelial plasma cells was also reported in other Artiodactyla, horse as well as the duck (22, 47). ILFs showed weak central immunoreactivity compatible with intrafollicular IgA class switching. Bottom panels in Figure 6 demonstrate IgA+ plasma cells (vertical arrows) reactive to anti-CHα1 (Figure 6D), anti-CHα2 (Figure 6E) and anti-IAR (Figure 6F) antibodies in the subepithelial stroma of the pseudostratified columnar epithelium of the trachea and in the vicinity of a subepithelial mucous gland lined by columnar epithelium (*). We attribute the immunostaining of epithelial cells, which was more intense with the anti-CHα2 and the anti-IAR antibody, to the transcytosis of IgA. Of note, the intensity of the color reaction with the anti-IAR domain antibody appeared identical to that generated by the anti-CHα2 antibody, suggesting comparable immunogenicity or avidity.
The histomorphology of the mucosa of the small intestine is shown in Figure 7. Villi and crypts are separated by the muscularis mucosae (MM) from the submucosa (SM) (Figure 7A). Aggregations of lymphoid follicles with germinal centers (GC), typical of Peyer’s patches, are shown in Figure 7B. Numerous eosinophilic neutrophils (vertical arrows), abundant plasma cells (horizontal arrows) along with scattered histiocytes (H) were the principal cellular components in the highly vascular (V) lamina propria (Figure 7C). A few goblet cells (G) were found between the enterocytes. In Figure 7D, IgA heavy chains expressed in plasma cells (horizontal arrows) and in enterocytes exhibited strong reactivity to the anti-IAR antibody, whereas the mucin in goblet cells (G) remained unstained.
In the mucosa of the large intestine (Figure 8), with crypts lined by numerous goblet cells, plasma cells (horizontal arrows) were found in the lamina propria between crypts as shown in longitudinal (upper row panels) and transverse sections (lower row panels). In Figures 8B, E, representative plasma cells (horizontal arrows) showed IgA immunoreactivity to the anti-CHα1 antibody, and in Figures 8C, F, to the anti-CHα2 antibody. Immunostaining was also associated with colonic enterocytes, indicative of IgA transcytosis, but not with mucin in goblet cells. In contrast to the small intestine, eosinophils were absent in this section of the large intestine.
Table 1 (A: immunoblots; B: immunohistology) summarizes a balanced expression profile of IgA heavy chain variants in MALT of the third eyelid, upper respiratory tract, small intestine and colon. In milk and serum, however, classic IgA heavy chains predominated.

Table 1 Expression profile of IgA variants in the third eyelid, trachea, small intestine, colon, milk, and serum of the dromedary.
We demonstrate that dromedary IgA, the protagonist immunoglobulin class at mucosal sites in mammals, maintains the distinctive dualistic structure of canonical and unconventional heavy chains, which was discovered in the IgG class of the one-humped camel and later in the nurse shark (1, 4). We also show that classic and unconventional IgA heavy chains are co-expressed in MALT of the Arabian camel.
The nictitating membrane was chosen as a relatively easily accessible source of IgA because it integrates inductive and effector components of mucosal immune defense against airborne antigens. The histomorphology of the third eyelid was extensively explored and compared in many quadrupeds, including dromedaries and alpacas (23, 48), and the sites of synthesis of IgA and SC determined (22). On the immune-inductive site, CALT consists of characteristic mucosal ILF lacking germinal centers located underneath the innermost (bulbar) aspect of the nictitating membrane, whereas at immune-effector sites, plasma cells sparse throughout the subconjunctiva or clustered in the periacinar interstitium of the HG express IgA heavy chains that engage via the J-chain the epithelial pIgR located at the basolateral surface of the glandular epithelial cell for transcytosis, eventually resulting in the release of sIgA, a multimeric protein complex that includes the SC, which is proteolytically generated from the pIgR (49, 50). Based on data obtained from the analyses of nasopharynx-associated lymphoid tissue situated in the tonsils and gut-associated lymphoid tissue, both representing inductive sites for B cell maturation and differentiation, it can be inferred that in the ILF of CALT, similar immunoregulatory events of B cell maturation, including IgA class-switching, take place (51–53). ILF located in the lamina propria below the epithelium of the third eyelid’s bulbar conjunctiva, constitutes an immune-inductive site, whereas IgA+ plasma cells residing in the lamina propria of the conjunctiva or grouped in the periacinar stroma of the HG constitute the effector arm of conjunctival humoral immunity. From inductive sites, IgA+ B cells are thought to migrate via the lymphatic system to reach regional lymph nodes, where they acquire redirecting homing molecules for regionalized tissue-specific localization in the lamina propria of the conjunctiva and in the periacinar stroma of the lacrimal gland. However, given the proximity of inductive and effector sites in the third eyelid, it is tempting to speculate on an alternative, T-cell independent pathway in which IgA+ B cells bypass further maturation in lymph nodes and directly access their terminal effector niches (54).
The RACE results showed heterogeneity in the length of the 5’ PCR products in contrast to a single 3’ PCR species. Sequencing of two 5’ PCR species running at ~1100 bp confirmed that they were conventional IgA heavy chains encompassing the ~300 bp CHα1 domain, whereas most clones with the ~800 bp 5’ PCR products were void of the CHα1 domain, demonstrating that plasma cells at the effector site of the third eyelid were capable of co-expressing both types of IgA heavy chains. 101 deduced amino acid sequences of VHH and constant domains from our IgA cDNA library are listed in the Supplementary File 1. The 19-mer leader oligopeptide, MELGLSWVVLAALLQGVQA, was earlier found associated with variable domains of classic (VH) as well as unconventional heavy chains (VHH) in a genomic library generated from dromedary liver DNA (55). The V domains following this particular leader sequence derive from a subgroup of IGHV3 genes (equivalent to human IGHV3 family clan III) (56). We found that the cardinal amino acid substitutions in the VHH domain of unconventional IgA heavy chain were identical to those reported for the VHH domain of IgG (2). These substitutions were as follows: in FR1, serine for leucine; in FR2, phenylalanine for valine, glutamic acid for glycine, arginine for leucine, and glycine for tryptophan. It was reported that these amino acids reshape the protein surface and increase the solubility of isolated VHH domains (57, 58). Regarding the length of CDR in the VHH domain, the rearranged CDR3 is more extended than in other species with a maximum of 26 amino acids in our IgA heavy chain cDNA library. However, there was considerable length variation among the CDR3, less so in CDR2, as shown in Supplementary File 1. Of interest was the presence of a short hinge region in all clones, which consisted of six consecutive prolines, preceded by a valine, almost identical to the proline pentamer of human IgA2 and chimpanzee IgA2 hinges, known to confer remarkable resistance to highly specific IgA1 bacterial proteases (59).
Co-expression of both IgA heavy chain variants in tissue extracts was not only observed in the third eyelid, but also in the trachea, as well as in the upper and lower intestinal tract, and inferred by immunohistochemistry in plasma cells of the HG, the conjunctiva, trachea, and upper as well as lower intestine. The findings were different in milk, where classic IgA heavy chain prevailed, and in serum, where unconventional heavy chain-only IgA was undetectable. The latter observation contrasted the original discovery of circulating heavy chain-only IgG in the serum of dromedaries which amount to ~3/4 of total IgG (1). However, the apparent absence of unconventional IgA heavy chain in serum could be accounted for by very low IgA levels, analogous to the situation in human blood where IgA levels reach ~1/5 of the concentration of IgG, and therefore very low levels of the heavy chain-only variant could escape detection. Of interest was the demonstration of the presence of a ~45 kDa protein species in circulation, which was observed with the anti-CHα1 antibody only. It may represent a cleaved fragment of classic IgA since serum and milk contain a variety of proteolytic systems that generate Fab fragments of IgA (60) and IgG class as well (61, 62). Bacterial IgA1 proteases exist that cleave proline-serine bonds specifically in the IgA1 hinge region (63), and two proline-serine bonds are found upstream and central in the CHα2 domain of the dromedary. If enzymatic cleavage at these sites with subsequent degradation of the C-terminal fragment took place, the non-reactivity of the anti-CHα2, anti-CHα3 and anti-IAR antibody would find an explanation. Cross-reactivity of the anti-CHα1 antibody with another immunoglobulin class circulating in serum or secreted in the milk offers an alternative, yet less likely, explanation. Further characterization of the observed protein species is needed.
At the N-terminus of the CHα3 domain, we noticed a novel pentapeptide, consisting of serine, glutamic acid, aspartic acid, alanine or sporadically threonine, and isoleucine. This sequence was identical in the Bactrian camel. At the N-terminus of the CHα3 domain in alpaca, leucine was observed instead of serine, and proline for alanine. A comparison with other mammals revealed that only camelids have a pair of acidic amino acids and isoleucine at this site of transition between the CHα2 and CHα3 domains, where immediately 3’ to the pentapeptide a highly conserved arginine followed by an invariant proline marks the N-terminus of the CHα3 domain (43, 44). However, two amino acid sequences (No 109 and No 111 in the Supplementary File 1) were an exception to this rule as the pentapeptide was found to contain not acidic but basic residues (arginine and histidine). By contrast, in other mammals a tetrapeptide consisting of glycine, asparagine, threonine, and phenylalanine represents the most common motif in the interdomain region, with the notable exception of the dog (threonine-glutamic acid-histidine-isoleucine), rabbit (glycine-serine-leucine-threonine), horse and donkey (glutamic acid-proline-leucine-phenylalanine), and greater bamboo lemur (glycine-aspartic acid-asparagine-phenylalanine). We surmise that in camelids, and possibly a few other species, negatively (or rarely positively) charged amino acids centered between basic residues, i.e. lysine upstream, arginine and histidine downstream, may affect structural and functional properties of the interdomain region likely accounting for novel protein-protein interactions with a putative receptor or a soluble protein.
Reports on the mucosal immune system in camelids in general and on the IgA system in particular are scarce. Recently, the topography of the pIgR was described in the lung of the Bactrian camel (64). Earlier, the small intestine was thoroughly examined in Bactrian camels where the concurrent expression of IgA and IgG was found and interpreted as a functional dual barrier of mucosal immune defense (65, 66). Predominant IgA over IgG expression was also described in a specific area of the three-chambered stomach of the dromedary (67). Yet, the probing antibodies used to locate these isotypes were not designed to differentiate between canonical and heavy chain-only antibodies and molecular data were not provided. Therefore, the question whether heavy chain-only antibodies existed in the gut of the Bactrian camel was not answered. Based on our observations an additional defense line of mucosal adaptive immunity in the dromedary must now be considered since both variants of IgA heavy chains were jointly and comparably expressed as judged by the density of the protein bands obtained by immunoblotting. In addition, IgA+ plasma cells were detected at effector sites in the lamina propria of the mucosa of the small as well as the large intestine by anti-CHα1, anti-CHα2 and anti-IAR antibodies, with the first antibody selective for conventional IgA heavy chains, while the other two antibodies were reactive to both IgA heavy chain variants. Although the darker brown color observed with the anti-CHα2 and anti-IAR antibodies compared to the staining intensity of the anti-CHα1 antibody suggested an additive effect, we cannot exclude that the differences in color intensities were caused by non-specific background staining. We were therefore unable to determine the ratio of IgA+ plasma cells producing classic heavy chains versus those expressing unconventional heavy chains. Yet, the results obtained from Western blots performed on tissue extracts suggest that they were in equilibrium at the time the tissue samples were obtained. Moreover, we assume that in the dromedary plasma cells have the potential to switch transcription between either IgA heavy chain variant, an unlikely scenario in the gut of the Bactrian camel or the alpaca which possess only one Cα gene (68, 69).
Another observation deserves attention, namely the proximity of eosinophils and plasma cells in the lamina propria of the small intestine, but not in a sample of the lower bowel, suggesting a functional relationship between terminally differentiated cells of innate and adaptive immunity. We do not know whether in the small intestine of the Bactrian camel similar conspicuous eosinophilic infiltrates exist in proximity of plasma cells. The role of eosinophils in providing key survival factors, such as APRIL (a proliferation-inducing ligand), IL-6, IL-4, IL-10 and TNF-α, to plasma cells has been described in the bone marrow, whereas in the digestive tract of mice, eosinophils maintain the function of IgA+ plasma cells and participate in T cell-independent class-switching via a TGF-β-dependent mechanism (70–73). Therefore, it is conceivable that eosinophils, beside their pivotal effector function in innate and FcϵR-mediated immune responses against gastrointestinal parasites (protozoa and helminths) modulate the synthesis of IgA heavy chains and promote survival of IgA+ plasma cells in niches of the intestinal lamina propria in the dromedary (74–77). Furthermore, interactions between eosinophils and IgA+ plasma cells may be reciprocal as IgA binds to and activates eosinophils via a highly glycosylated isoform of FcαRI (78).
The existence of heavy chain-only IgA in the dromedary is at odds with available data in other members of the Camelidae family, the closest species being the two-humped Bactrian camel and the more distantly related South American family member alpaca, which both miss a second Cα gene, a sine qua non for the transcription of unconventional Ig heavy chains. In the IgH locus of the alpaca, only one gene for Cα, Cδ, Cϵ and Cμ was identified in contrast to four Cγ genes, whereas Bactrian camels possess six Cγ genes and one gene for Cα, Cδ, Cϵ and Cμ (68, 69, 79). Moreover, partial protein sequences of IgA deduced from a genomic library in Bactrian camel (GenBank #EPY85672.1) (68) and alpaca (GenBank #CAO79580.1) (69) as well as from a spleen cDNA library of the Bactrian camel (GenBank #KP999944.1) also contain the CHα1 domain, indicating that IgA in these Camelidae is composed of a bona fide IgA heavy chain. While five Cγ genes were found in the dromedary, conclusive numbers on Cα genes were not revealed (10). Whether our findings of the co-expression of canonical and unconventional IgA heavy chains can be extrapolated to all dromedaries demands confirmation as phenotypically different breeds of dromedaries exist, 14 alone in Saudi Arabia (6, 80, 81). The dromedaries examined here were crossbreeds of the Sofor and Waddah types. Therefore, we cannot at present generalize these findings because the genetic makeup could differ among various breeds (82). Given the numerical variation of Cγ genes in Camelidae (i.e. five in dromedary, six in Bactrian camel and llama, four in alpaca), the presence of a second Cα gene in one-humped camels coding for an alternative IgA heavy chain is plausible. Two genes code for IgA subclasses, IgA1 and IgA2, in humans, chimpanzees and gorillas, and gibbons express two IgA subclasses as well, whereas most other species examined have just one Cα gene (83–85). Concerning the numbers of Cα genes, however, the absolute champion is the rabbit, with a remarkable number of probably more than 15 Cα genes (86, 87).
Considering the advances in characterizing the mucosal immune system in general and IgA in particular ((88–90) with its eminent role in natural and induced mucosal immune defense against newly emerged viral pathogens such as SARS-CoV-2 in humans (91–94) and MERS-CoV in the dromedary (95, 96), it was our aim to determine whether in the dromedary both subclasses of IgA antibodies co-exist at mucosal surfaces. Our study demonstrates that the IgA class maintains the typical, camelid-specific dualism of antibody structure which was acquired over millions of years throughout transcontinental migration granting the control of infections under the harshest climatic and extreme physiologic conditions (97). There is evidence that the affinity of heavy chain-only antibodies of the IgG class for their epitopes is superior to that of canonical heterotetrameric antibodies, providing a clear advantage in antigen recognition and neutralization, and the same could be translated to the IgA class at mucosal surfaces (11). The observations that camelids are less susceptible to some clinical infectious diseases in comparison to ruminants, such as foot-and-mouth disease and vesicular stomatitis (75), and that dromedaries are capable of mounting a specific and robust antibody response with mild or subclinical disease, e.g. in MERS-CoV infection (98), strongly support the key role of the dual antibody system in infection control. With the use of CH domain-specific antibodies, comparative studies on the kinetics of unconventional versus canonical IgG levels during seroconversion in a variety of infections and on IgA subclass responses to intestinal pathogens (77) could delineate the respective contribution of each IgG or IgA variant in the neutralization or elimination of the causative infectious agent. Although the advantages of doubling the IgG defense line in circulation and in tissues are obvious, a conclusive explanation why functional antibodies with a simpler structure emerged exclusively in two evolutionary remote, terrestrial (Camelidae) and aquatic creatures (nurse shark), remains so far elusive. Furthermore, the detection of canonical and unconventional IgA heavy chains in dromedaries adds another firewall of immune defense at the frontline of mucosal surfaces as other species of the Camelidae family do not possess a second Cα gene at the heavy chain locus with the specific G → A transition mutation in the canonical splicing site, i.e. GT, flanking the first exon, which by analogy with Cγ genes, is a requisite for transcribing unconventional heavy chain genes (3, 99, 100). Our findings expand the antimicrobial spectrum of single-domain heavy chain-only antibodies to the mucosal membranes of the eye, the upper respiratory tract, and the intestine. Genomic data will determine whether in the dromedary the expression of the unconventional heavy chain-only IgA variant is indeed the result of a gene duplication event combined with a splicing defect that occurred when one-humped camels emerged, and whether there are further IgA subclasses or allotypes.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was approved by King Faisal Specialist Hospital and Research Centre, Riyadh. The study was conducted in accordance with the local legislation and institutional requirements.
WC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft. SMS: Data curation, Methodology, Validation, Investigation, Writing – review & editing. RA-R: Data curation, Methodology, Visualization, Writing – review & editing. RSP: Methodology, Supervision, Investigation, Writing – review & editing. MA-E: Methodology, Data curation, Investigation, Writing – review & editing. HA-H: Data curation, Investigation, Methodology, Writing – review & editing. AT: Data curation, Investigation, Methodology, Writing – review & editing. KBK: Data curation, Formal analysis, Methodology, Software, Visualization, Writing – review & editing. KRL: Data curation, Formal analysis, Methodology, Software, Writing – review & editing. KC: Investigation, Supervision, Writing – review & editing. UK: Formal Analysis, Investigation, Supervision, Visualization, Writing – review & editing. FA-M: Funding acquisition, Investigation, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by grant Nr. 11-BIO2073-20 (The Long-Term Comprehensive National Science, Technology and Innovation Plan) and grant Nr. 42-1 (MERS Research Grant Program) from King Abdulaziz City for Science and Technology (KACST), Riyadh, Saudi Arabia. UK acknowledges funding by an UAEU grant (12F043).
We thank Dr. A. Alaiya, Dr. R. Al Hijailan, Mrs. A. Inglis, Mrs. A. Alanazi, Mrs. M. Kafaar and Mr. I. Keragala for technical assistance and Mrs. C. Touzinte and Mr. M. Noreldin for secretarial work. We also thank Dr. G. Wick for review and comments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1289769/full#supplementary-material
Amino acids, one-letter symbols; bp, base pair; CALT, conjunctiva-associated lymphoid tissue; CDR, complementarity determining region; C, constant; CH, constant heavy; DAB, 3,3’-diaminobenzidine tetrahydrochloride; FR, framework region; GSP, gene specific primer; HC, heavy chain; H&E, hematoxylin and eosin; HG, Harderian gland; HRP, horseradish peroxidase; IAR, inter-α region; Ig, immunoglobulin; ILF, isolated lymphoid follicle; MALT, mucosa-associated lymphoid tissue; PCR, polymerase chain reaction; pIgR, polymeric Ig receptor; RACE, rapid amplification of cDNA ends; SC, secretory component; sIgA, secretory IgA; Tp, tailpiece; V, variable; VH, variable heavy; VHH, variable heavy heavy.
1. Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature (1993) 363(6428):446–8. doi: 10.1038/363446a0
2. Muyldermans S, Atarhouch T, Saldanha J, Barbosa JA, Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng (1994) 7(9):1129–35. doi: 10.1093/protein/7.9.1129
3. Nguyen VK, Hamers R, Wyns L, Muyldermans S. Loss of splice consensus signal is responsible for the removal of the entire C(H)1 domain of the functional camel IGG2A heavy-chain antibodies. Mol Immunol (1999) 36(8):515–24. doi: 10.1016/s0161-5890(99)00067-x
4. Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature (1995) 374(6518):168–73. doi: 10.1038/374168a0
5. Roux KH, Greenberg AS, Greene L, Strelets L, Avila D, McKinney EC, et al. Structural analysis of the nurse shark (new) antigen receptor (NAR): molecular convergence of NAR and unusual mammalian immunoglobulins. Proc Natl Acad Sci U.S.A. (1998) 95(20):11804–9. doi: 10.1073/pnas.95.20.11804
6. Ali A, Baby B, Vijayan R. From desert to medicine: A review of camel genomics and therapeutic products. Front Genet (2019) 10:17. doi: 10.3389/fgene.2019.00017
7. Jovčevska I, Muyldermans S. The therapeutic potential of nanobodies. BioDrugs (2020) 34(1):11–26. doi: 10.1007/s40259-019-00392-z
8. Arbabi-Ghahroudi M. Camelid single-domain antibodies: promises and challenges as lifesaving treatments. Int J Mol Sci (2022) 23(9):5009. doi: 10.3390/ijms23095009
9. Nguyen VK, Desmyter A, Muyldermans S. Functional heavy-chain antibodies in Camelidae. Adv Immunol (2001) 79:261–96. doi: 10.1016/s0065-2776(01)79006-2
10. Conrath KE, Wernery U, Muyldermans S, Nguyen VK. Emergence and evolution of functional heavy-chain antibodies in Camelidae. Dev Comp Immunol (2003) 27(2):87–103. doi: 10.1016/s0145-305x(02)00071-x
11. Flajnik MF, Deschacht N, Muyldermans S. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PloS Biol (2011) 9(8):e1001120. doi: 10.1371/journal.pbio.1001120
12. Tillib SV, Vyatchanin AS, Muyldermans S. Molecular analysis of heavy chain-only antibodies of Camelus bactrianus. Biochem (Mosc) (2014) 79(12):1382–90. doi: 10.1134/S000629791412013X
13. Griffin LM, Snowden JR, Lawson AD, Wernery U, Kinne J, Baker TS. Analysis of heavy and light chain sequences of conventional camelid antibodies from Camelus dromedarius and Camelus bactrianus species. J Immunol Methods (2014) 405:35–46. doi: 10.1016/j.jim.2014.01.003
14. Ciccarese S, Burger PA, Ciani E, Castelli V, Linguiti G, Plasil M, et al. The camel adaptive immune receptors repertoire as a singular example of structural and functional genomics. Front Genet (2019) 10:997. doi: 10.3389/fgene.2019.00997
15. Gionfriddo JR, Melgarejo T, Morrison EA, Alinovi CA, Asem EK, Krohne SG. Comparison of tear proteins of llamas and cattle. Am J Vet Res (2000) 61(10):1289–93. doi: 10.2460/ajvr.2000.61.1289
16. Knop E, Knop N, Claus P. Local production of secretory IgA in the eye-associated lymphoid tissue (EALT) of the normal human ocular surface. Invest Ophthalmol Vis Sci (2008) 49(6):2322–9. doi: 10.1167/iovs.07-0691
17. Shamsi FA, Chen Z, Liang J, Li K, Al-Rajhi AA, Chaudhry IA, et al. Analysis and comparison of proteomic profiles of tear fluid from human, cow, sheep, and camel eyes. Invest Ophthalmol Vis Sci (2011) 52(12):9156–65. doi: 10.1167/iovs.11-8301
18. Abuagla IA, Ali HA, Ibrahim ZH. An anatomical study on the eye of the one-humped camel (Camelus dromedarius). Int J Vet Sci (2016) 5(3):137–41.
19. Bang BG, Bang FB. Localized lymphoid tissues and plasma cells in paraocular and paranasal organ systems in chickens. Am J Pathol (1968) 53(5):735–51.
20. Albini B, Wick G, Rose E, Orlans E. Immunoglobulin production in chicken Harderian glands. Int Arch Allergy Appl Immunol (1974) 47(1):23–34. doi: 10.1159/000231198
22. Schlegel T, Brehm H, Amselgruber WM. IgA and secretory component (SC) in the third eyelid of domestic animals: a comparative study. Vet Ophthalmol (2003) 6(2):157–61. doi: 10.1046/j.1463-5224.2003.00284.x
23. Al-Ramadan SY, Ali AM. Morphological studies on the third eyelid and its related structures in the one-humped camel (Camelus dromedarius). J Vet Anat (2012) 5(2):71–81. doi: 10.21608/JVA.2012.44875
24. Mantis NJ, Rol N, Corthésy B. Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol (2011) 4(6):603–11. doi: 10.1038/mi.2011.41
25. Conca W, Al-Nuaimi K, Nagelkerke N. The complexity of regional warming in the United Arab Emirates in the period 1982–2009. Int J Global Warming (2010) 2(3):225–33. doi: 10.1504/IJGW.2010.036134
26. Wu H, Guang X, Al-Fageeh MB, Cao J, Pan S, Zhou H, et al. Camelid genomes reveal evolution and adaptation to desert environments. Nat Commun (2014) 5:5188. doi: 10.1038/ncomms6188
27. Knittler MR, Haas IG. Interaction of BiP with newly synthesized immunoglobulin light chain molecules: cycles of sequential binding and release. EMBO J (1992) 11(4):1573–81. doi: 10.1002/j.1460-2075.1992.tb05202.x
28. Kaloff CR, Haas IG. Coordination of immunoglobulin chain folding and immunoglobulin chain assembly is essential for the formation of functional IgG. Immunity (1995) 2(6):629–37. doi: 10.1016/1074-7613(95)90007-1
29. Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, et al. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell (2009) 34(5):569–79. doi: 10.1016/j.molcel.2009.04.028
30. Yeku O, Frohman MA. Rapid amplification of cDNA ends (RACE). Methods Mol Biol (2011) 703:107–22. doi: 10.1007/978-1-59745-248-9_8
31. Chan WC, White PD. Fmoc solid phase peptide synthesis: A practical approach. Oxford: Oxford Univ Press (1999), 376. doi: 10.1093/oso/9780199637256.001.0001
32. Knop E, Knop N. Anatomy and immunology of the ocular surface. Chem Immunol Allergy (2007) 92:36–49. doi: 10.1159/000099252
33. Chodosh J, Nordquist RE, Kennedy RC. Anatomy of mammalian conjunctival lymphoepithelium. Adv Exp Med Biol (1998) 438:557–65. doi: 10.1007/978-1-4615-5359-5_79
34. Sakimoto T, Shoji J, Inada N, Saito K, Iwasaki Y, Sawa M. Histological study of conjunctiva-associated lymphoid tissue in mouse. Jpn J Ophthalmol (2002) 46(4):364–69. doi: 10.1016/s0021-5155(02)00503-8
35. Hamada H, Hiroi T, Nishiyama Y, Takahashi H, Masunaga Y, Hachimura S, et al. Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J Immunol (2002) 168(1):57–64. doi: 10.4049/jimmunol.168.1.57
36. Giuliano EA, Moore CP, Phillips TE. Morphological evidence of M cells in healthy canine conjunctiva-associated lymphoid tissue. Graefes Arch Clin Exp Ophthalmol (2002) 240(3):220–6. doi: 10.1007/s00417-002-0429-3
37. Astley RA, Kennedy RC, Chodosh J. Structural and cellular architecture of conjunctival lymphoid follicles in the baboon (Papio anubis). Exp Eye Res (2003) 76(6):685–94. doi: 10.1016/s0014-4835(03)00062-9
38. Brandtzaeg P, Pabst R. Let's go mucosal: communication on slippery ground. Trends Immunol (2004) 25(11):570–7. doi: 10.1016/j.it.2004.09.005
39. Knop N, Knop E. Ultrastructural anatomy of CALT follicles in the rabbit reveals characteristics of M-cells, germinal centres and high endothelial venules. J Anat (2005) 207(4):409–26. doi: 10.1111/j.1469-7580.2005.00470.x
40. Tsuji M, Suzuki K, Kinoshita K, Fagarasan S. Dynamic interactions between bacteria and immune cells leading to intestinal IgA synthesis. Semin Immunol (2008) 20(1):59–66. doi: 10.1016/j.smim.2007.12.003
41. Lin M, Du L, Brandtzaeg P, Pan-Hammarström Q. IgA subclass switch recombination in human mucosal and systemic immune compartments. Mucosal Immunol (2014) 7(3):511–20. doi: 10.1038/mi.2013.68
42. Lefranc MP, Lefranc G. IMGT® and 30 years of immunoinformatics insight in antibody V and C domain structure and function. Antibodies (2019) 8(2):29. doi: 10.3390/antib8020029
43. Toraño A, Putnam FW. Complete amino acid sequence of the alpha 2 heavy chain of a human IgA2 immunoglobulin of the A2m (2) allotype. Proc Natl Acad Sci U.S.A. (1978) 75(2):966–9. doi: 10.1073/pnas.75.2.966
44. Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J Biol Chem (1979) 254(8):2865–74.
45. Elbers JP, Rogers MF, Perelman PL, Proskuryakova AA, Serdyukova NA, Johnson WE, et al. Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary. Mol Ecol Resour (2019) 19(4):1015–26. doi: 10.1111/1755-0998.13020
46. Kaetzel CS, Bruno MEC. Epithelial transport of igA by the polymeric immunoglobulin receptor. In: Kaetzel CS, editor. Mucosal immune defense: immunoglobulin A. New York: Springer (2007). p. 43–89.
47. Oliveira CA, Telles LF, Oliveira AG, Kalapothakis E, Gonçalves-Dornelas H, Mahecha GA. Expression of different classes of immunoglobulin in intraepithelial plasma cells of the Harderian gland of domestic ducks Anas platyrhynchos. Vet Immunol Immunopathol (2006) 113(3-4):257–66. doi: 10.1016/j.vetimm.2006.05.008
48. Klećkowska-Nawrot J, Nowaczyk R, Goździewska-Harłajczuk K, Krasucki K, Janeczek M. Histological, histochemical and fine structure studies of the lacrimal gland and superficial gland of the third eyelid and their significance on the proper function of the eyeball in alpaca (Vicugna pacos). Folia Morphol (Warsz) (2015) 74(2):195–205. doi: 10.5603/FM.2015.0001
49. Brandtzaeg P. Mucosal and glandular distribution of immunoglobulin components: differential localization of free and bound SC in secretory epithelial cells. J Immunol (1974) 112(4):1553–9. doi: 10.4049/jimmunol.112.4.1553
50. Brandtzaeg P, Prydz H. Direct evidence for an integrated function of J chain and secretory component in epithelial transport of immunoglobulins. Nature (1984) 311(5981):71–3. doi: 10.1038/311071a0
51. Brandtzaeg P. Immune functions of nasopharyngeal lymphoid tissue. Adv Otorhinolaryngol (2011) 72:20–4. doi: 10.1159/000324588
52. Lycke NY, Bemark M. The regulation of gut mucosal IgA B-cell responses: recent developments. Mucosal Immunol (2017) 10(6):1361–74. doi: 10.1038/mi.2017.62
53. Fenton TM, Jørgensen PB, Niss K, Rubin SJS, Mörbe UM, Riis LB, et al. Immune profiling of human gut-associated lymphoid tissue identifies a role for isolated lymphoid follicles in priming of region-specific immunity. Immunity (2020) 52(3):557–570.e6. doi: 10.1016/j.immuni.2020.02.001
54. Allman D, Wilmore JR, Gaudette BT. The continuing story of T-cell independent antibodies. Immunol Rev (2019) 288(1):128–35. doi: 10.1111/imr.12754
55. Nguyen VK, Muyldermans S, Hamers R. The specific variable domain of camel heavy-chain antibodies is encoded in the germline. J Mol Biol (1998) 275(3):413–8. doi: 10.1006/jmbi.1997.1477
56. Deschacht N, De Groeve K, Vincke C, Raes G, De Baetselier P, Muyldermans S. A novel promiscuous class of camelid single-domain antibody contributes to the antigen-binding repertoire. J Immunol (2010) 184(10):5696–704. doi: 10.4049/jimmunol.0903722
57. Davies J, Riechmann L. 'Camelising' human antibody fragments: NMR studies on VH domains. FEBS Lett (1994) 339(3):285–90. doi: 10.1016/0014-5793(94)80432-x
58. Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol (1996) 3(9):803–11. doi: 10.1038/nsb0996-803
59. Mistry D, Stockley RA. IgA1 protease. Int J Biochem Cell Biol (2006) 38(8):1244–8. doi: 10.1016/j.biocel.2005.10.005
60. Mansa B, Kilian M. Retained antigen-binding activity of Fab alpha fragments of human monoclonal immunoglobulin A1 (IgA1) cleaved by IgA1 protease. Infect Immun (1986) 52(1):171–4. doi: 10.1128/iai.52.1.171-174.1986
61. Quan CP, Ruffet E, Arihiro K, Pires R, Bouvet JP. High affinity serum-derived Fab fragments as another source of antibodies in the gut lumen of both neonates and adults. Scand J Immunol (1996) 44(2):108–14. doi: 10.1046/j.1365-3083.1996.d01-288.x
62. Dallas DC, Murray NM, Gan J. Proteolytic systems in milk: perspectives on the evolutionary function within the mammary gland and the infant. J Mammary Gland Biol Neoplasia (2015) 20(3-4):133–47. doi: 10.1007/s10911-015-9334-3
63. Senior BW, Woof JM. Effect of mutations in the human immunoglobulin A1 (IgA1) hinge on its susceptibility to cleavage by diverse bacterial IgA1 proteases. Infect Immun (2005) 73(3):1515–22. doi: 10.1128/IAI.73.3.1515-1522.2005
64. He WH, Zhang WD, Cheng CC, Lu J, Liu L, Chen ZH, et al. Expression characteristics of polymeric immunoglobulin receptor in Bactrian camel (Camelus bactrianus) lungs. PloS One (2022) 17(3):e0264815. doi: 10.1371/journal.pone.0264815
65. Zhang WD, Wang WH, Jia S. Distribution of immunoglobulin G antibody secretory cells in small intestine of Bactrian camels (Camelus bactrianus). BMC Vet Res (2015) 11:222. doi: 10.1186/s12917-015-0538-y
66. Zhang WD, Wang WH, Jia S. The distribution of SIgA and igG antibody-secreting cells in the small intestine of bactrian camels (Camelus bactrianus) of different ages. PloS One (2016) 11(6):e0156635. doi: 10.1371/journal.pone.0156635
67. Hassan Omer ZI, Lu J, Cheng YJ, Li PX, Chen ZH, Wang WH. Age-dependent changes in the anatomical and histological characteristics of the aggregated lymphoid nodules in the stomach of Dromedary camels (Camelus Dromedarius). PloS One (2023) 18(3):e0279417. doi: 10.1371/journal.pone.0279417
68. Bactrian Camels Genome Sequencing and Analysis Consortium, Jirimutu, Wang Z, Ding G, Chen G, Sun Y, et al. Genome sequences of wild and domestic bactrian camels. Nat Commun (2012) 3:1202. doi: 10.1038/ncomms2192
69. Achour I, Cavelier P, Tichit M, Bouchier C, Lafaye P, Rougeon F. Tetrameric and homodimeric camelid IgGs originate from the same IgH locus. J Immunol (2008) 181(3):2001–9. doi: 10.4049/jimmunol.181.3.2001
70. Chu VT, Berek C. Immunization induces activation of bone marrow eosinophils required for plasma cell survival. Eur J Immunol (2012) 42(1):130–7. doi: 10.1002/eji.201141953
71. Chu VT, Beller A, Rausch S, Strandmark J, Zänker M, Arbach O, et al. Eosinophils promote generation and maintenance of immunoglobulin-A-expressing plasma cells and contribute to gut immune homeostasis. Immunity (2014) 40(4):582–93. doi: 10.1016/j.immuni.2014.02.014
72. Berek C. Eosinophils: important players in humoral immunity. Clin Exp Immunol (2016) 183(1):57–64. doi: 10.1111/cei.12695
73. Nguyen DC, Duan M, Ali M, Ley A, Sanz I, Lee FE. Plasma cell survival: The intrinsic drivers, migratory signals, and extrinsic regulators. Immunol Rev (2021) 303(1):138–53. doi: 10.1111/imr.13013
74. Capron M, Soussi Gounni A, Morita M, Truong MJ, Prin L, Kinet JP, et al. Eosinophils: from low- to high-affinity immunoglobulin E receptors. Allergy (1995) 50(25 Suppl):20–3. doi: 10.1111/j.1398-9995.1995.tb04270.x
75. Fowler ME. Camelids are not ruminants. In: Fowler ME, Miller RE, editors. Zoo and wild animal medicine, Sixth Edition. Philadelphia: Saunders (2008). p. 375–85. doi: 10.1016/B978-1-4160-4047-7.50049-X
76. Huang L, Appleton JA. Eosinophils in helminth infection: defenders and dupes. Trends Parasitol (2016) 32(10):798–807. doi: 10.1016/j.pt.2016.05.004
77. Bouragba M, Laatamna A, Cheddad FE, Baroudi D, Houali K, Hakem A. Gastrointestinal parasites of dromedary camel (Camelus dromedarius) in Algeria. Vet World (2020) 13(8):1635–40. doi: 10.14202/vetworld.2020.1635-1640
78. Decot V, Woerly G, Loyens M, Loiseau S, Quatannens B, Capron M, et al. Heterogeneity of expression of IgA receptors by human, mouse, and rat eosinophils. J Immunol (2005) 174(2):628–35. doi: 10.4049/jimmunol.174.2.628
79. Liang Z, Wang T, Sun Y, Yang W, Liu Z, Fei J, et al. A comprehensive analysis of immunoglobulin heavy chain genes in the Bactrian camel (Camelus bactrianus). Front Agr Sci Eng (2015) 2(3):249–59. doi: 10.15302/J-FASE-2015056
80. Abdallah HR, Faye B. Phenotypic classification of Saudi Arabian camel (Camelus dromedarius) by their body measurements. Emir J Food Agric (2012) 24(3):272–80.
81. Al-Atiyat RM, Suliman G, AlSuhaibani E, El-Waziry A, Al-Owaimer A, Basmaeil S. The differentiation of camel breeds based on meat measurements using discriminant analysis. Trop Anim Health Prod (2016) 48(5):871–8. doi: 10.1007/s11250-015-0990-5
82. Piro M. Aspects of molecular genetics in dromedary camel. Front Genet (2021) 12:723181. doi: 10.3389/fgene.2021.723181
83. Kawamura S, Saitou N, Ueda S. Concerted evolution of the primate immunoglobulin alpha-gene through gene conversion. J Biol Chem (1992) 267(11):7359–67. doi: 10.1016/S0021-9258(18)42525-2
84. Woof JM, Kerr MA. IgA function–variations on a theme. Immunology (2004) 113(2):175–7. doi: 10.1111/j.1365-2567.2004.01958.x
85. Steffen U, Koeleman CA, Sokolova MV, Bang H, Kleyer A, Rech J, et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat Commun (2020) 11(1):120. doi: 10.1038/s41467-019-13992-8
86. Pinheiro A, de Sousa-Pereira P, Strive T, Knight KL, Woof JM, Esteves PJ, et al. Identification of a new European rabbit IgA with a serine-rich hinge region. PloS One (2018) 13(8):e0201567. doi: 10.1371/journal.pone.0201567
87. Pinheiro A, de Sousa-Pereira P, Esteves PJ. The IgA of hares (Lepus sp.) and rabbit confirms that the leporids IgA explosion is old and reveals a new case of trans-species polymorphism. Front Immunol (2023) 14:1192460. doi: 10.3389/fimmu.2023.1192460
88. Brandtzaeg P. Secretory igA: designed for anti-microbial defense. Front Immunol (2013) 4:222. doi: 10.3389/fimmu.2013.00222
89. de Sousa-Pereira P, Woof JM. IgA: structure, function, and developability. Antibodies (Basel) (2019) 8(4):57. doi: 10.3390/antib8040057
90. Kumar N, Arthur CP, Ciferri C, Matsumoto ML. Structure of the secretory immunoglobulin A core. Science (2020) 367(6481):1008–14. doi: 10.1126/science.aaz5807
91. Russell MW, Moldoveanu Z, Ogra PL, Mestecky J. Mucosal immunity in COVID-19: A neglected but critical aspect of SARS-coV-2 infection. Front Immunol (2020) 11:611337. doi: 10.3389/fimmu.2020.611337
92. Chao YX, Rötzschke O, Tan EK. The role of igA in COVID-19. Brain Behav Immun (2020) 87:182–3. doi: 10.1016/j.bbi.2020.05.057
93. Quinti I, Mortari EP, Fernandez Salinas A, Milito C, Carsetti R. IgA antibodies and igA deficiency in SARS-coV-2 infection. Front Cell Infect Microbiol (2021) 11:655896. doi: 10.3389/fcimb.2021.655896
94. Sterlin D, Mathian A, Miyara M, Mohr A, Anna F, Claër L, et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med (2021) 13(577):eabd2223. doi: 10.1126/scitranslmed.abd2223
95. Stalin Raj V, Okba NMA, Gutierrez-Alvarez J, Drabek D, van Dieren B, Widagdo W, et al. Chimeric camel/human heavy-chain antibodies protect against MERS-CoV infection. Sci Adv (2018) 4(8):eaas9667. doi: 10.1126/sciadv.aas9667
96. Mubarak A, Alturaiki W, Hemida MG. Middle east respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J Immunol Res (2019) 2019:6491738. doi: 10.1155/2019/6491738
97. Burger PA. The history of Old World camelids in the light of molecular genetics. Trop Anim Health Prod (2016) 48(5):905–13. doi: 10.1007/s11250-016-1032-7
98. Wernery U, Corman VM, Wong EY, Tsang AK, Muth D, Lau SK, et al. Acute middle East respiratory syndrome coronavirus infection in livestock dromedaries, Dubai, 2014. Emerg Infect Dis (2015) 21(6):1019–22. doi: 10.3201/eid2106.150038
99. Ming L, Wang Z, Yi L, Batmunkh M, Liu T, Siren D, et al. Chromosome-level assembly of wild Bactrian camel genome reveals organization of immune gene loci. Mol Ecol Resour (2020) 20(3):770–80. doi: 10.1111/1755-0998.13141
100. Lado S, Elbers JP, Rogers MF, Melo-Ferreira J, Yadamsuren A, Corander J, et al. Nucleotide diversity of functionally different groups of immune response genes in Old World camels based on newly annotated and reference-guided assemblies. BMC Genomics (2020) 21(1):606. doi: 10.1186/s12864-020-06990-4
Keywords: Arabian camel immunoglobulin A, heavy chain-only IgA, unconventional IgA heavy chain, IgA variable heavy heavy (VHH) domain, third eyelid, mucosal immunity
Citation: Conca W, Saleh SM, Al-Rabiah R, Parhar RS, Abd-Elnaeim M, Al-Hindas H, Tinson A, Kroell KB, Liedl KR, Collison K, Kishore U and Al-Mohanna F (2023) The immunoglobulin A isotype of the Arabian camel (Camelus dromedarius) preserves the dualistic structure of unconventional single-domain and canonical heavy chains. Front. Immunol. 14:1289769. doi: 10.3389/fimmu.2023.1289769
Received: 06 September 2023; Accepted: 30 October 2023;
Published: 12 December 2023.
Edited by:
Pedro Jose Esteves, Centro de Investigacao em Biodiversidade e Recursos Geneticos (CIBIO-InBIO), PortugalReviewed by:
Xu-Jie Zhang, Huazhong Agricultural University, ChinaCopyright © 2023 Conca, Saleh, Al-Rabiah, Parhar, Abd-Elnaeim, Al-Hindas, Tinson, Kroell, Liedl, Collison, Kishore and Al-Mohanna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walter Conca, Y29uY2F3YWx0ZXI2QGdtYWlsLmNvbQ==
†These authors share first authorship
‡ORCID: Walter Conca, orcid.org/0000-0003-3427-2664
Souad M. Saleh, orcid.org/0009-0008-3368-3465
Rana Al-Rabiah, orcid.org/0009-0005-1968-7193
Hussein Al-Hindas, orcid.org/0000-0003-4189-5843
Alexander Tinson, orcid.org/0000-0002-6925-4223
Klaus Roman Liedl, orcid.org/0000-0002-0985-2299
Kate Collison, orcid.org/0000-0002-0691-6859
Uday Kishore, orcid.org/0000-0002-6033-6759
Futwan Al-Mohanna, orcid.org/0000-0001-5931-0837
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.