- 1Division of Infectious Diseases, Department of Medicine, University of British Columbia, Vancouver, BC, Canada
- 2Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, Canada
Leishmania donovani, an intracellular protozoan parasite, is the causative agent of visceral leishmaniasis, the most severe form of leishmaniasis in humans. It is becoming increasingly clear that several intracellular pathogens target host cell RNA interference (RNAi) pathways to promote their survival. Complexes of Argonaute proteins with small RNAs are core components of the RNAi. In this study, we investigated the potential role of host macrophage Argonautes in Leishmania pathogenesis. Using Western blot analysis of Leishmania donovani-infected macrophages, we show here that Leishmania infection selectively increased the abundance of host Argonaute 1 (Ago1). This increased abundance of Ago1 in infected cells also resulted in higher levels of Ago1 in active Ago-complexes, suggesting the preferred use of Ago1 in RNAi in Leishmania-infected cells. This analysis used a short trinucleotide repeat containing 6 (TNRC6)/glycine-tryptophan repeat protein (GW182) protein-derived peptide fused to Glutathione S-transferase as an affinity matrix to capture mature Ago-small RNAs complexes from the cytosol of non-infected and Leishmania-infected cells. Furthermore, Ago1 silencing significantly reduced intracellular survival of Leishmania, demonstrating that Ago1 is essential for Leishmania pathogenesis. To investigate the role of host Ago1 in Leishmania pathogenesis, a quantitative whole proteome approach was employed, which showed that expression of several previously reported Leishmania pathogenesis-related proteins was dependent on the level of macrophage Ago1. Together, these findings identify Ago1 as the preferred Argonaute of RNAi machinery in infected cells and a novel and essential virulence factor by proxy that promotes Leishmania survival.
1 Introduction
Infection with intracellular pathogens belonging to the genus Leishmania results in a broad spectrum of clinical manifestations, mainly divided into three distinct clinical syndromes: visceral, cutaneous, and mucocutaneous leishmaniasis (1). Leishmania donovani, the focus of this study, causes visceral leishmaniasis, which can be life-threatening if left untreated (2). Leishmania has a digenetic life cycle, alternating between the non-motile amastigote form in the vertebrate host and the motile promastigote form in the invertebrate vector, the phlebotomine sandfly (3). The infection is initiated when an infected female sandfly injects metacyclic promastigotes into a mammalian host during a regular blood meal. Parasites released are rapidly taken up by phagocytic cells, including macrophages and neutrophils (4). The macrophages are the final resident cells which support Leishmania growth and proliferation, establishing chronic infection in humans. After internalization, promastigotes settle in phagolysosomes of macrophages, where they differentiate into amastigote forms, which divide many times and ultimately rupture macrophages and infect other surrounding macrophages (1).
Leishmaniasis is responsible for severe morbidity and mortality in parts of the tropics, subtropics and southern Europe. Leishmaniasis is classified as a neglected tropical disease. Unfortunately, leishmaniasis is rising due to increased global travel and a lack of effective therapeutic and prophylactic methods. Available drugs are expensive, have severe toxicity, and are circumvented by emerging parasite resistance (5). Although tremendous effort has been made to develop Leishmania vaccines, only some have been commercially approved for canine leishmaniasis (6), while none have yet been licensed for human leishmaniasis (5). Expanding the knowledge on the biological interaction between the parasite and the host is crucial for developing a comprehensive strategy to fight the disease and improve leishmaniasis treatment. The pathogenesis of Leishmania involves the alteration of the macrophage phenotype, where Leishmania causes deactivation of the host by interfering with host-cell gene expression and signal transduction (7). One potential mechanism of interest of Leishmania pathogenesis is targeting host small non-coding RNAs (sncRNAs), which are involved in post-transcriptional gene expression regulation, known as RNA interference (RNAi).
It is becoming increasingly clear that non-coding RNAs (ncRNAs), which do not encode proteins but account for the majority of the human genome, play important roles in many biological processes and diseases (8–11). Among them are small ncRNAs, such as microRNAs (miRNAs), Piwi-interacting RNAs, and tRNA-derived RNA fragments (tRFs), which have been shown to play a critical role in cancer, stress response, metabolic abnormalities and viral infections (12–16). Out of all species of ncRNAs, sncRNAs represent the core of the regulatory mechanisms of gene expression in eukaryotic cells. The mechanisms of action of most sncRNAs require a group of effector proteins as part of an RNA-induced silencing complex (RISC). These effector proteins are necessary for the stabilization, transport and regulatory activity of ncRNAs on their target RNA transcripts. RNA-binding proteins Argonautes (Agos) are members of the Argonaute family that bind sncRNAs that act as guide RNA and direct them to specific RNA transcripts for silencing. In fact, these highly conserved proteins are central components of the RISC in RNAi (17, 18). There are eight Ago family members in humans consisting of four ubiquitously expressed Ago1-4 of the Ago clade and four of the PIWI clade (PIWIL1-4), which are mainly restricted to the germline (19). Deep sequencing of human Ago-associated sncRNAs revealed Ago association with RNA fragments of diverse origin, such as miRNAs, small nucleolar RNAs, vault RNAs, and tRFs, to carry out their gene silencing function (20).
Ago proteins are transiently held in open conformation by heat shock proteins (HSPs) to allow loading of sncRNA, resulting in the closed conformation Ago-sncRNA complex, also known as the mature RISC. The mature RISC further interacts with the scaffolding protein trinucleotide repeat containing 6 (TNRC6)/glycine-tryptophan repeat protein (GW182), which preferentially binds to mature RISCs over guide-free Ago proteins (21) and facilitates downstream silencing processes, possibly by recruiting other RNA binding proteins (RBPs) (22). Finally, the RISC interacts with and represses expression of messenger RNA (mRNA) targets whose sequences are partially or fully complementary to those of the sncRNAs in the RISC.
Small ncRNAs (sncRNAs), such as microRNAs (miRNAs) and tRNA-derived RNA fragments (tRFs), have been clearly demonstrated to play an important role in microbial infections (12–14, 23). Our recent study showed that Leishmania infection causes genome-wide attenuation of macrophage miRNAs by decreasing miRNAs gene transcription. miRNA downregulation was linked to the upregulation of macrophage transcription factor c-Myc. c-Myc silencing reversed the Leishmania-mediated miRNA suppression and also reduced the intracellular survival of Leishmania (24). We have recently discovered that sncRNAs like tRFs and ribosomal RNA-derived RNA fragments (rRFs) are highly enriched in exosomes secreted by L. donovani and L. braziliensis (25), and these secretory exosomes have been shown to modulate innate and adaptive immune responses through predominantly immunosuppressive effects, such as promoting IL-10 production and inhibiting TNF-alpha in human monocytes and monocyte-derived dendritic cells (26). Overall, the emerging role of sncRNAs (such as miRNAs) from both host and pathogen (such as tRFs) during Leishmania infection represents a novel virulence paradigm that invites further examination. Thus, it was of interest to investigate the possibility that Leishmania targets host macrophage Ago(s) to regulate host gene expression to facilitate its survival.
In this study, we show that L. donovani selectively increases the abundance of host macrophage Ago1. Strikingly, the enhanced level of Ago1 in infected cells correlated with an increased level of Ago1 containing-complexes isolated using a recently published biochemical isolation of Argonaute protein complexes by “Ago proteins Affinity Purification by Peptide” (Ago-APP). Deliberate silencing of host Ago1 significantly attenuated intracellular survival of Leishmania in human macrophages, demonstrating that Ago1 is essential for Leishmania pathogenesis. SILAC (stable isotope labeling using amino acids in cell culture)-based quantitative proteomic analysis revealed that the expression of several previously reported Leishmania pathogenesis-related proteins was dependent on the level of macrophage Ago1, suggesting the role of Ago1 in Leishmania pathogenesis. Together, these findings identify Ago1 as the preferred Argonaute of RNAi machinery in infected cells and a novel and essential virulence factor by proxy that promotes Leishmania survival.
2 Materials and methods
2.1 THP-1 cell culture and differentiation
THP-1 cells (human leukemia monocytic cells) were obtained from ATCC (TIB-202TM), and maintained at 37°C, 5% CO2 in RPM1 1640 media (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 10 mM HEPES (MilliporeSigma), 100 U/ml penicillin/streptomycin (MilliporeSigma) and 2 mM L-glutamine (MilliporeSigma). For differentiation, THP-1 cells were treated with 10 ng/ml Phorbol 12-myristate 13-acetate (PMA) overnight (16–18 h). Adhered differentiated THP-1 (dTHP-1) cells were washed with Hanks’ balanced salt solution (HBSS) (MilliporeSigma) and given fresh media without PMA. The cells were rested for 24 h prior to further treatment.
2.2 Parasite culture and in vitro infection
Leishmania donovani Sudan strain 2S was provided by Dr. Kwang Poo Chang (Rockefeller University, NY, USA), and continuously cultured in the lab. Virulence was maintained by regular passages via Syrian Golden hamster from which fresh amastigotes were purified and transformed in vitro into promastigotes (24). Promastigotes were cultured in M199 medium (MilliporeSigma) supplemented with 10% heat-inactivated fetal bovine serum, 10 mM HEPES, 10 µg/ml folic acid (MilliporeSigma), 3 µg/ml hemin (MilliporeSigma), 2 mM L-Glutamine, 100 U/ml penicillin/streptomycin and 100 mM adenosine (MilliporeSigma) at 26°C. The parasites were subcultured every three days for a maximum of 25-30 passages. For Leishmania infections, stationary phase promastigotes were used at an MOI of 20:1 unless otherwise stated.
2.3 Determination of infection rate
To determine the infection rate, Leishmania-infected cells were washed with warm PBS, then fixed with 2% paraformaldehyde in PBS for 20 min on ice, and mounted on glass slides using VECTASHIELD Antifade Mounting Medium with DAPI, which stains macrophage and parasite nuclei. Cells were viewed under fluorescence using the Axioplan II (Carl Zeiss Inc.) microscope. For each treatment, at least 10 images were taken at 40x magnification using the AxioCam MRm Camera and the AxioVision software Version 4.8.2 (Carl Zeiss Inc.). At least 100 cells were counted for each condition to determine the average number of parasites per macrophage and the percentage of macrophages infected.
2.4 Parasite rescue and transformation assay
PMA differentiated THP-1 cells (dTHP-1) were infected with stationary-phase L. donovani promastigotes at an MOI of 20:1 for 24 h. After the duration of infection, cells were thoroughly washed with HBSS to remove non-internalized Leishmania. Controlled lysing of infected cells was performed using 0.01% SDS as previously described (24, 27) to release intracellular amastigotes. The rescued Leishmania amastigotes were transformed to promastigotes in M199 media by incubating at 26°C for 72 h. Their growth was evaluated after 72 h of incubation by counting transformed promastigotes stained with trypan blue solution (0.4% w/v in PBS) on a hemocytometer.
2.5 Western blotting
dTHP-1 cells were washed with HBSS and lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 5 mM NaF, 1 mM Na3VO4, 1 mM EDTA), supplemented with 1 mM PMSF, 5 µg/ml aprotinin and 5 µg/ml leupeptin. Whole cell lysates were clarified by centrifugation and proteins in supernatant were separated by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad) using a semi-dry transfer apparatus. Transferred proteins were probed with the appropriate antibodies, according to the manufacturer’s instructions: Anti-Ago1 (Cell Signaling 5053), anti-Ago2 (Cell Signaling 2897), anti-GST (Santa Cruz sc-138), anti-GAPDH (Applied Biological Materials (Abm) G041), anti-Lamin A/C (Cell Signaling 2032), and anti-Actin (Santa Cruz SC-47778). Reactive protein signals were captured on Blue X-ray film (Carestream) using SuperSignal West Pico Plus or Femto Chemiluminescence Substrate (Thermo). Densitometric analysis was performed in ImageJ, and results were normalized to Actin.
2.6 THP-1 cell culture in SILAC media
For SILAC-based labeling, RPMI 1640 media without L-glutamine, L-arginine and L-lysine (Caisson labs) was supplemented with 10% heat-inactivated dialyzed fetal bovine serum (Gibco), 2 mM L-glutamine, 10 mM HEPES and 100 U/ml penicillin/streptomycin. The RPMI medium was then split into three: the “light” RPMI was supplemented with normal isotopic abundance arginine (21 mg/l) and lysine (36.5 mg/l), while the “medium” RPMI was supplemented with 13C6-arginine (20.25 mg/l) and 2H4-lysine (37.5 mg/l), and “heavy” RPMI was supplemented with 13C615N4-arginine (22.25 mg/l) and 13C615N2-lysine (38.5 mg/l).
THP-1 cells were spun down, one third of which were resuspended in the respective SILAC media, incubated at 37°C and 5% CO2, and passaged for a minimum of five times prior to use. Cells were differentiated using 10 ng/ml of PMA for 16-18 h and then washed with HBSS and rested for 24 h in fresh respective SILAC media prior to infection.
2.7 siRNA-mediated knockdown of Ago1 in THP-1 cells
Three siRNAs targeting human Ago1 (SR308857), as well as non-specific control (scrambled) siRNA (SR30004), were purchased from OriGene. Cells were transfected for 52 h with non-specific or Ago1 siRNA (50 pmol/well) using HiPerFect transfection reagent (Qiagen) according to the manufacturer’s instructions. We have previously used HiPerFect transfection reagent (Qiagen) to silence the expression of various proteins in THP-1 cells (28) and in human monocyte-derived macrophages (24) using siRNAs from OriGene, with transfection efficiency between 70-80%, as measured by fluorescent-labeled transfection control siRNA (OriGene). After the duration of transfection, THP-1 cells were differentiated with PMA for further use.
2.8 Expression and purification of recombinant proteins
For the expression and purification of required recombinant proteins we used a previously published protocol (29). Briefly, GST(glutathione S-transferase) and GST-T6B plasmid constructs were transformed in DH5 alpha E. coli and then expressed in BL21. Subsequently, 2 L isopropyl β-D-1-thio- galactopyranoside–induced culture (OD600 = 0.6) was grown overnight at 18°C. The cells were harvested by centrifugation at 4,400 × g, 30 min at 4°C and the resulting pellet was then resuspended in resuspension buffer (PBS containing 1 mM PMSF and 1 mM DTT) supplemented with 1 mg/ml lysozyme. After sonication for 4 × 30 sec, the lysate was cleared by centrifugation at 40,000 × g, 40 min at 4°C. Then the supernatant was incubated with 3 ml washed Glutathione-Agarose beads (MilliporeSigma) at 4°C for 3 h. Thereafter, the GST/GST-T6B-coupled beads were loaded onto a column and washed with 10 column volumes (cv) of resuspension buffer. GST/GST-T6B was eluted with 4 cv of elution buffer (PBS supplemented with 20 mM Tris-HCl, pH 8.0, and 10 mM reduced glutathione), pooled and then concentrated with Amicon Ultra-15 centrifugal filter units. The concentrated fractions were loaded onto a desalting chromatography column (Zeba™ Spin Desalting Columns) that was equilibrated in PBS with 5% (v/v) glycerol. Desalted fractions were adjusted to ∼2 mg/ml proteins, aliquoted and stored at −80°C. The purity of recombinant proteins was assessed by staining with Coomassie brilliant blue.
2.9 Preparation of cytosolic fraction
For the preparation of the cytosolic fraction, we followed a published protocol (30). Briefly, dTHP-1 cells were extensively washed and lysed in hypotonic lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.3% NP-40, 1 mM NaF, 1 mM Na3VO4) supplemented with 5 µg/ml aprotinin, 5 µg/ml leupeptin, and 1 mM PMSF, and passed ten times through a 22-gauge needle on ice to disrupt cells. Then disrupted cells were centrifuged at 2,300 × g for 20 min at 4°C. The supernatant was kept as the cytoplasmic fraction. The purity of cytoplasmic fractions was assessed using antibodies against specific markers for the cytoplasm (GAPDH) and nuclear fraction (Lamin A/C) in a Western blot assay. These cytoplasmic fractions were used for the purification of Agos and associated proteins using GST-T6B affinity beads.
2.10 GST-T6B affinity chromatography
To prepare the affinity beads, 50 µl of Glutathione-Agarose beads were incubated with approximately 100 µg of GST/GST-T6B recombinant proteins for 3 h at 4°C with end-over-end rotation. Non-bound peptides were removed by washing the beads three times with excess of ice-cold PBS. In order to remove non-specific proteins which might have interacted with GST alone or Glutathione-Agarose beads, the cell lysates of interest prepared in NET buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 10% glycerol, 1 mM NaF; supplemented with 0.5 mM DTT and 1 mM PMSF) were pre-incubated with 50 µl of Glutathione-Agarose beads loaded with 100 µg GST for 1 h at 4°C with end-over-end rotation. Non-bound materials were then incubated with the GST-T6B affinity beads for 3 h, at 4°C to capture interacting proteins. Beads were spun down, washed three times with NET buffer to remove non-bound material, and once with PreScission cleavage buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT). The affinity beads were eluted by PreScission enzyme (MilliporeSigma) cleavage at 4°C or by addition of 2x Laemmli sample loading buffer and incubation at 95°C for 7 min.
2.11 Liquid chromatography-tandem mass spectrometry and protein identification
For mass spectroscopy analysis of siRNA-mediated Ago1 downregulated macrophages, equal amounts of proteins from each treatment were mixed, reduced, alkylated (31) and run on 10% SDS-PAGE. Proteins in the entire lane were digested in gel with MS-grade trypsin (Promega) (32) and resulting peptides were cleaned on C-18 STop And Go Extraction (STAGE) tips (33) using 40% (v/v) acetonitrile in 0.1% (v/v) formic acid as the elution buffer. Peptides were analysed on NanoDrop One (Thermo Scientific - A205, scopes) to load approximately 200 ng of peptides on Orbitrap Exploris 480 coupled to easy nLC 1200 (Thermo Scientific) with ionopticks’ Aurora series 25 cm x 75 µm C18 1.6 µm analytical column heated to 40°C. Peptides were separated on a 62 minute gradient with a LC flow rate of 250 nl/min. Buffer A consisted of 2% (v/v) acetonitrile and 0.1% (v/v) formic acid; buffer B consisted of 80% (v/v) acetonitrile and 0.1% (v/v) formic acid. Spray voltage was set to 1,900 V and ion transfer tube temperature to 290°C. FAIMS was not enabled with data acquisition. Data dependent mode was set for 20 scans and the orbitrap resolution was set to 120,000 for full MS and 15,000 for fragment MS, with AGC target set to 100% at 20 ms for full MS and 50% for fragment MS. Intensity threshold was set at 8,000, isolation window at 2 m/z, with normalized collision energy at 28%. Dynamic exclusion was enabled to exclude after 1 time for 45 s.
Acquired data were searched on MaxQuant version 2.1.0.0 (34) against Uniprot’s human sequences (UP000005640), Leishmania donovani BPK282A1, and common contaminant sequences. SILAC labels of medium arginine (13C6), medium lysine (D4), heavy arginine (13C6, 15N4), and heavy lysine (13C6, 15N2) were set for quantitation enabling iBAQ, requantify, and match-between-runs options. The data were filtered for 1% false discovery at protein, peptide and PSM levels.
Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD037042. The reviewer account for the PRIDE data has the username reviewer_pxd037042@ebi.ac.uk and password o1Lv0bxR.
2.12 Bioinformatic analysis of LC-MS/MS data
All downstream analysis was done using R software (http://www.r-project.org). Of the total protein groups (hereafter referred to simply as ‘proteins’) detected by LC-MS/MS, we removed those that were commonly occurring contaminants, identified by peptides found to be part of a protein derived from the reversed part of the decoy database, or non-human, yielding a filtered high-confidence dataset of human proteins. Proteins were further filtered to only keep those with a minimum of two unique peptides per protein identification and which were identified in at least two of the three replicates. For all analyses, log2 transformed normalized ratios (fold changes) were used.
The one-sample T-test was performed to statistically compare log2 transformed normalized ratios to the hypothetical value of 0 (representing no change). A p-value of less than 0.05 was considered statistically significant. No cut-off was set as the minimum fold change for a protein to be considered modulated. Instead, where applicable, the significance of each protein modulation was ranked by distance from the origin measured along axes (Manhattan distance), when represented in a volcano plot of -log10(p-value) vs. log2(fold change). The higher the Manhattan score, the more significant the modulation of the protein was considered. For proteins that were both Leishmania-modulated and Ago1-dependent, the sum of their Manhattan scores was used to rank their combined significance.
We compared our dataset with the Pathways Commons collection of databases (https://www.pathwaycommons.org/), which collects biological pathway and interaction data from various partner databases—including, but not limited to, BioGRID, SwissProt, NCI Pathway Interaction Database: Pathway, and Innate DB.
For Gene Ontology (GO) annotation, Uniprot Princeton GO Mapper (https://go.princeton.edu/cgi-bin/GOTermMapper) was used with the following parameters: Organism (Annotation) set to “Homo sapiens (GOA @EBI + Ensembl)” and Ontology set to “Generic slim”. The proteins were annotated for each of three biological aspects: Biological process (“process”), Molecular function (“function”), and Cellular component (“component”). We omitted GO terms with 3 or fewer protein members or that were very broad and encompassed many subcategories (“Organelle”, with 68/71 proteins). Conversely, we combined similar highly specific GO terms (“Catalytic activity acting on a protein”, “Catalytic activity acting on RNA”, “activity acting on DNA”) into their corresponding umbrella term (“Catalytic activity”). Finally, for sets of two nearly identical GO terms, we retained only one (“Extracellular space” but not “Extracellular region”, and “Extracellular matrix” rather than “External encapsulating structure”).
3 Results
3.1 The host Ago1 is upregulated during Leishmania infection and downregulation of Ago1 attenuates Leishmania survival
Argonautes 1 and 2 are the most abundant in mammalian cells and have been extensively studied in various biological contexts such as regulation of the biogenesis of small RNA, mRNA translation, small RNA-guided host defense against exogenous nucleic acids, genome editing and even disease progression (17). Thus, we investigated the modulation of host macrophage Ago1 and Ago2 in response to L. donovani infection in PMA-differentiated THP-1 cells (dTHP-1). These cells have been extensively used as a model system to study human leishmaniasis (35–37). To examine Ago1 and Ago2 levels, dTHP-1 cells were infected with stationary-phase L. donovani promastigotes at MOI 20:1 for 24 h and 48 h. Ago1 and Ago2 levels were monitored by Western blot assay using their respective antibodies. As shown in Figures 1A, B, the expression level of Ago1 was selectively and strongly upregulated, while the Ago2 level remained unaltered in infected cells, both at 24 and 48 h post-infection. We checked the specificity of commercial Ago1 antibody from Cell signaling using whole cell lysate of dTHP-1 cells (positive control) and whole cell lysate of Leishmania promastigotes in a Western blot assay. Anti-Ago1 showed no detectable reactivity in the whole cell lysate of Leishmania promastigotes (data not shown), thus rejecting any potential contribution of Leishmania Ago1 in the observed upregulation of host Ago1 in infected cells. Furthermore, to rule out the possibility that Leishmania-induced Ago1 is due to the general phagocytosis of external particles, we incubated dTHP-1 cells with latex beads at different MOIs (5:1, 10:1 and 20:1) for 24 h. Western blot analysis of the bead-treated cells for Ago1 level revealed no significant induction of Ago1 compared to non-treated control cells (Figure S1). This strongly suggests that the increase in Ago1 abundance following Leishmania infection is not mediated by general phagocytotic processes.
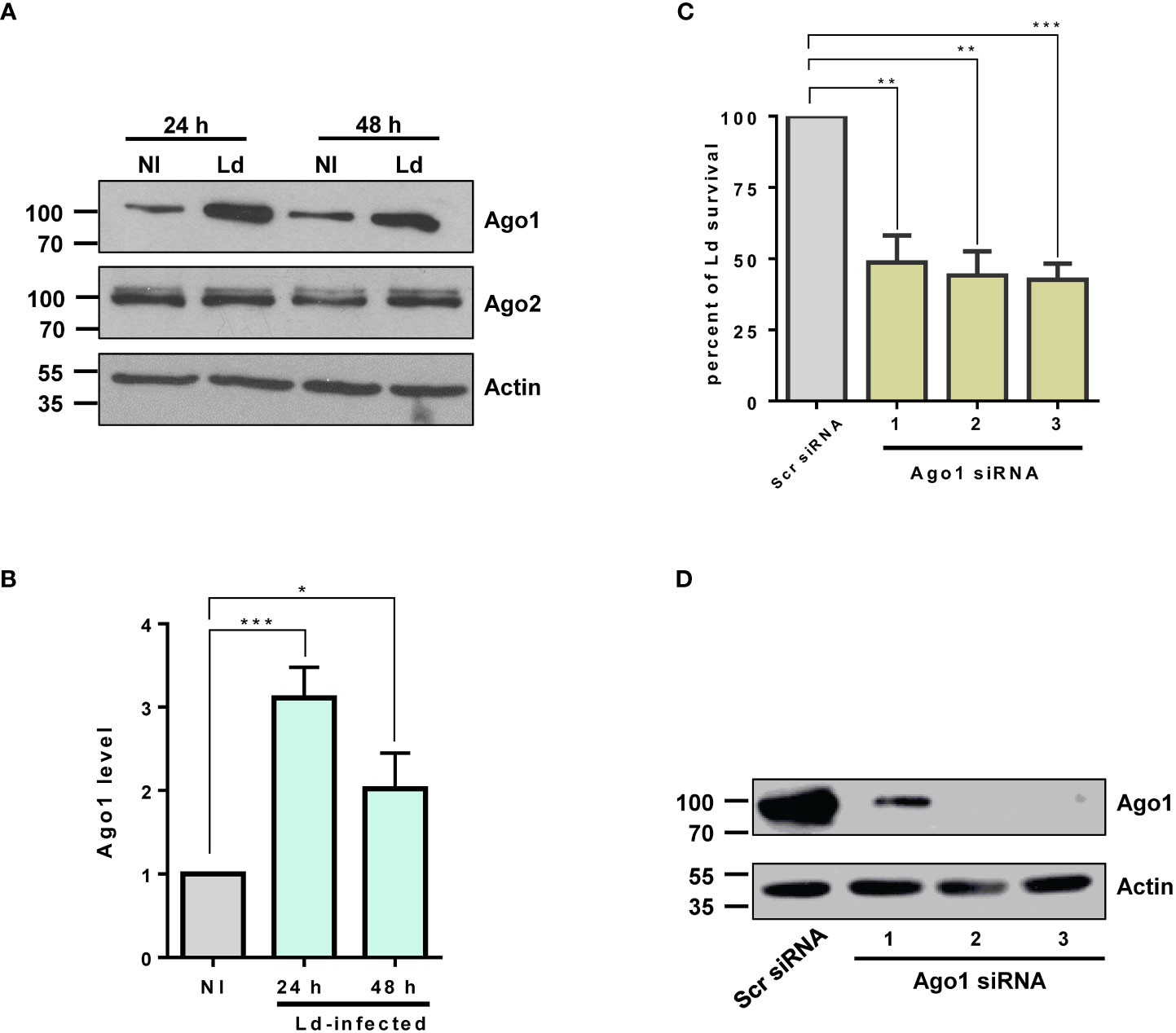
Figure 1 Ago1 is upregulated in infected macrophages. (A) dTHP-1 cells were infected with L. donovani (Ld) or not for 24 h and 48 h, whole cell lyates were from non-infected (NI) and Ld-infected macrophages were analysed by Western blotting with antibodies specific for Ago1, Ago2 and Actin (loading control). (B) Densitometric analysis was used to measure Ago1 and Ago2, normalized to Actin. The histogram shown is the mean ± S.D. of the densitometric analyses of three independent experiments. (C) Ago1 knockdown attenuates Leishmania survival. THP-1 cells were treated with control scrambled siRNAs or three different Ago1-specific siRNAs for 52 h Control and Ago1-knockdown cells were differentiated and subsequently incubated with Leishmania for 24 h At the end of the experiment, a parasite rescue assay as described in “Materials and methods” was performed. Shown is the mean ± S.D. of the densitometric analyses of three independent experiments (*p-value < 0.05, **p-value < 0.01, ***p-value < 0.001). (D) In parallel, the cells treated with siRNAs and differentiated for parasite rescue assay were analyzed by Western blotting for Ago1 and Actin levels.
Given that Ago1 binds a variety of sncRNAs, the central component of RISC in the RNA silencing pathway, it seemed likely that Ago1 upregulation would have pleiotropic and important effects on the macrophage phenotype. In addition, it seemed probable that this upregulation of Ago1 itself, independent of any impact on the RNAi pathway, would have consequences for macrophage biology. Thus, to address the biological relevance of Leishmania-mediated selective increase in host Ago1 level, we investigated the possibility that Ago1 may confer a pathogen survival advantage. We therefore assessed Leishmania intracellular survival in host macrophages treated with Ago1 siRNAs. To this end, we downregulated host Ago1 using three unique Ago1-targeting siRNAs. A non-specific scrambled siRNA was used as a negative control. Ago1-knockdown cells and control cells were infected with L. donovani for 24 h. At the end of the experiment, the infected cells were lysed for parasite rescue, as described in “Materials and methods.” The results presented in Figure 1C show that reduced Ago1 level correlated with decreased survival of Leishmania. In parallel, the downregulation of Ago1 was confirmed by immunoblotting using Ago1 antibody. The efficiency of siRNA-mediated Ago1 knockdown is shown in Figure 1D. All three unique siRNAs were able to downregulate Ago1. The transfection of siRNAs 2 and 3 resulted in nearly complete downregulation of Ago1. Together, these results show that host Ago1 is essential for the intracellular survival of Leishmania.
Results presented thus far show enhanced levels of Ago1 in infected cells. Deliberate downregulation of Ago1 resulted in decreased survival of Leishmania inside host cells. Given that Argonaute proteins participate in the RNA silencing pathway as the central component of RISC (17, 38), it was important to investigate the possibility that an enhanced level of Ago1 in infected cells also results in a higher level of Ago1-containing mature RISC complexes required for RNAi. Thus, we isolated Ago protein complexes from non-infected control and Leishmania-infected cells.
3.2 Preparation of GST-TNRC6 affinity beads and validation for its interaction with Argonautes
We used a recently published procedure, “Ago protein Affinity Purification by Peptides,” to isolate Ago-containing complexes from control and infected cells (29). This affinity column-based approach allows for the simultaneous isolation of all Argonautes and associated proteins from a variety of species and cell lines. This robust pull-down assay involves the (GST)-T6B peptide, a part of the TNRC6/GW182 protein family that efficiently interacts with all Ago proteins (29).
To prepare recombinant GST-T6B peptide and control affinity beads, purified GST-T6B proteins and GST alone were loaded respectively on Glutathione-Agarose beads as described in “Materials and methods.” The purity of concentrated and desalted recombinant proteins is shown in Figure S2A. Without antibodies against the T6B peptide, expected molecular mass and reactivity to anti-GST antibodies were used to correctly identify recombinant proteins (Figure S2B). To test that recombinant GST-T6B peptide is active and successfully isolates Ago proteins from macrophages, whole cell lysate was incubated with GST alone and GST-T6B affinity beads separately. Bound proteins were released by heating in the Laemmli sample buffer. The presence of Agos in the bound materials was investigated by immunoblotting using appropriate antibodies. As expected, Ago1 and Ago2 were detected in material eluted from GST-T6B beads but absent from material eluted from GST alone, thus validating the specificity and effectiveness of peptide affinity beads in the isolation of Ago proteins (Figures S2C, D).
3.3 Ago protein-containing complexes isolated from Leishmania-infected cells are enriched in Ago1
As described above, it was of interest to investigate whether this increase in Ago1 correlates with an increase in Ago1 being part of RISC, which is responsible for the RNAi process for gene regulation.
It is well established that most Ago complexes are restricted to the cytoplasm (22, 39) to participate in canonical RNAi. Thus, we used cytoplasmic fractions to isolate Ago protein-containing complexes using GST-T6B affinity beads for further study. GAPDH was used as the cytoplasm marker, and Lamin A/C was used as the nuclear marker. The results presented in Figure 2A show cytoplasmic fraction nearly devoid of nuclear content. Bound complexes from GST alone and GST-T6B affinity beads were released by heating in Laemmli sample buffer. The presence of Agos in the bound materials was investigated by immunoblotting using appropriate antibodies. As expected and shown in Figure 2B, Ago1 and Ago2 are enriched in cytoplasm and nearly completely absent in the nuclear fraction. After confirmation of the enrichment of Agos in the cytoplasm, cytoplasmic and nuclear fractions from control and infected cells were separately incubated with GST-T6B affinity beads to isolate Ago-containing complexes. As shown in Figure 2C, the amount of Ago1 was strikingly enriched in complexes isolated from cytoplasmic fractions of Leishmania-infected cells compared to non-infected cells. This finding of Ago1 enrichment in RISC isolated from infected cells positively correlates with the abundance of Ago1 in Leishmania-infected cells. Taken together, results obtained suggest that Leishmania infection promotes Ago1-containing mature complexes, which are essential for RNAi.
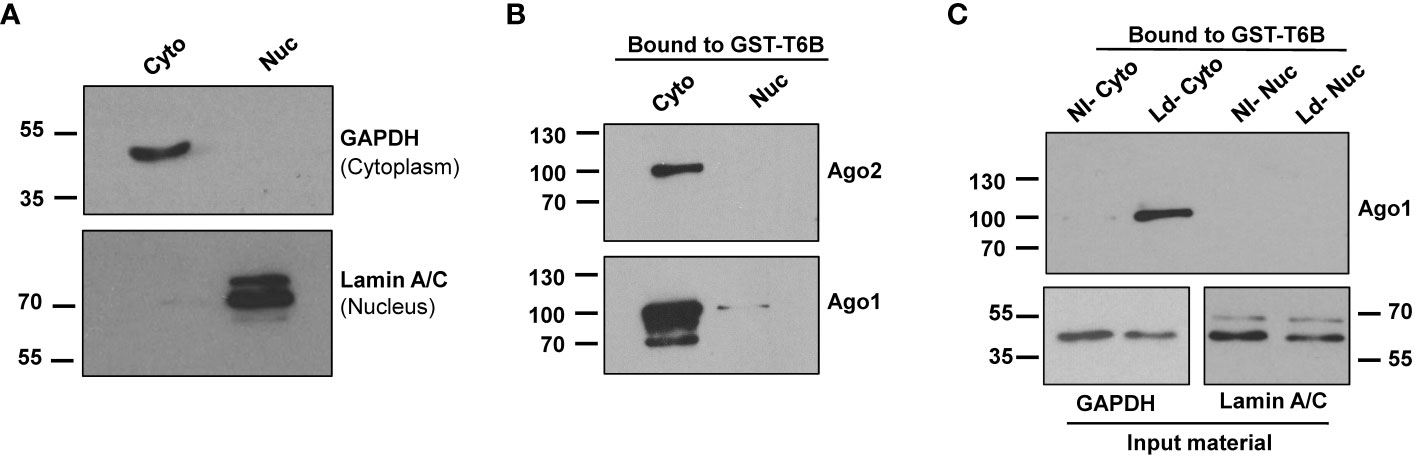
Figure 2 Analysis of cytoplasmic and nuclear fractions from non-infected and infected macrophages for the presence of Ago1. (A) dTHP-1 cells were subjected to cell fractionation for cytoplasmic and nuclear fractions as described in “Materials and methods” and the purity of fractions was assessed using indicated antibodies in a Western blot assay. (B) The cytoplasmic and nuclear extracts from dTHP-1 cells were incubated separately with GST-T6B affinity beads and bound proteins were subjected to Western blotting using indicated antibodies for the presence of Ago1 and Ago2. (C) Immunoblot comparing levels of Ago1 in bound materials eluted from GST-T6B affinity beads incubated with cytosolic or nuclear lysates of non-infected (NI) and Leishmania-infected (Ld) dTHP-1 cells. In parallel, aliquots from the cytoplasmic fraction from NI and Leishmania-infected cells were tested for GAPDH and Lamin A/C level. The data shown is one representative of three independent experiments.
The results thus far show that Ago complexes are enriched in Ago1-containing complexes, and host macrophage Ago1 is critical for optimal intracellular survival of Leishmania. Together these findings prompted us to further study the role of Ago1 in the pathogenesis of Leishmania infection. To this end, we used a comprehensive, unbiased SILAC-based quantitative proteomic analysis of Leishmania-infected cells in an Ago1-deficient condition.
3.4 SILAC-based quantitative proteomic analysis of Leishmania-infected macrophages in normal and Ago1-deficient condition
Our approach to comprehensively identify and characterize host macrophage proteins modulated by Leishmania and their dependence on Ago1 involves the gene silencing of host cell Ago1 by siRNAs unique to human Ago1, followed by SILAC-based quantitative LC-MS/MS analysis (outlined in Figure S3A). For SILAC-based quantitative analysis, THP-1 cells were cultured in corresponding SILAC media as shown in Figure S3A. The “light” (L) cell population was transfected with control siRNA (non-infected). The “medium” (M) cell population was transfected with control siRNA and infected with L. donovani. The “heavy” (H) cell population was transfected with Ago1-specific siRNAs and infected with L. donovani. Due to the restricted availability of SILAC isotopes, we elected to pool two unique siRNAs (#2 and #3, as shown above in Figure 1D). As expected and shown in Figure S3B, pooling of unique siRNAs was effective in downregulating host Ago1, and the use of control siRNA did not prevent Leishmania-mediated upregulation of Ago1. After 24 h incubation with Leishmania, cells were lysed, and equal amounts of proteins from “light,” “medium” and “heavy” populations were mixed in a 1:1:1 ratio. Proteins were subsequently identified and quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described in “Materials and methods”.
3.5 Global proteomic quantitation-based identification of Leishmania-modulated host proteins in infected macrophages
In order to identify Leishmania-modulated proteins which are dependent on Ago1 and which may have relevance to Leishmania pathogenesis, we first identified host macrophage proteins that were differentially expressed after Leishmania infection. As shown in Figure S4A, we filtered the initial 3379 proteins identified through LC-MS/MS to retain only high-confidence human proteins, resulting in 2151 proteins. Of these, we considered only those with ratios detected in at least two out of three replicates. We performed one-sample T-tests comparing the log2(M/L) ratios to zero, which represents no change in response to Leishmania infection. Of the total 1778 proteins identified, 331 were significantly modulated by Leishmania (Table S1). 212 of the differentially expressed proteins were downregulated, while 119 proteins were upregulated in response to Leishmania infection (Figure 3A). For the ease of readers, Leishmania-modulated proteins were also represented in a volcano plot, where the -log10(p-value) was plotted against the log2(M/L) ratio (Figure 3B).
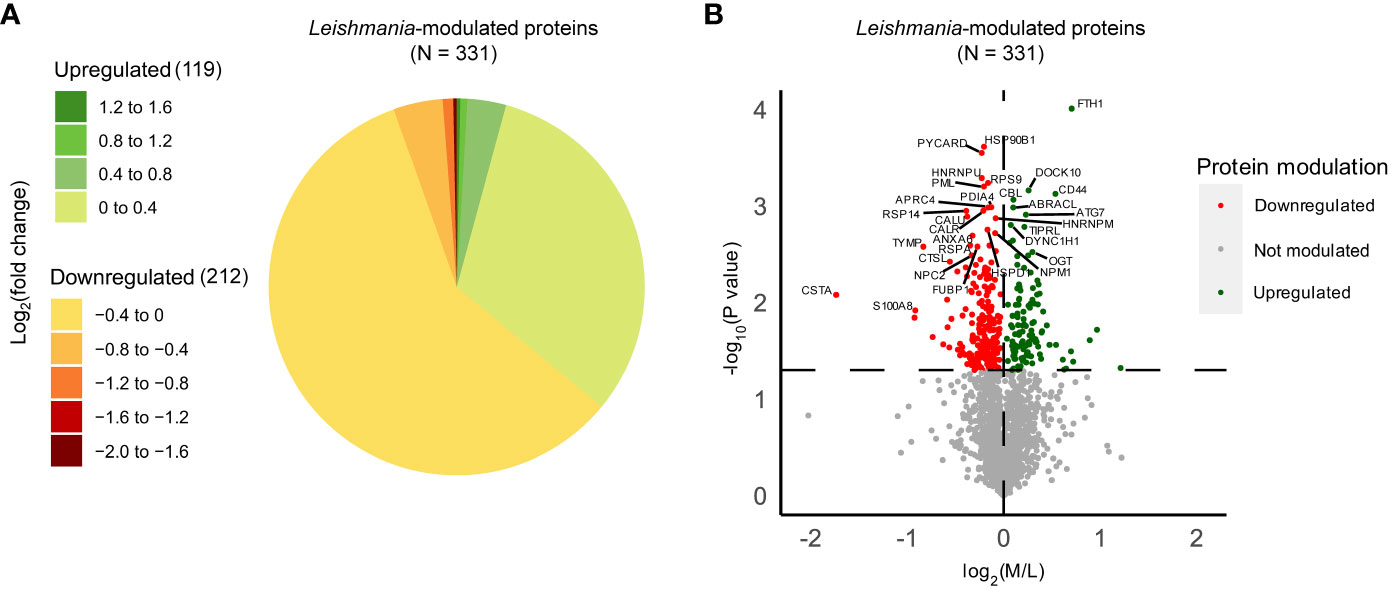
Figure 3 Quantitative proteomic analysis of non-infected and Leishmania-infected macrophages. THP-1 cells were cultured in SILAC media as described in “Materials and methods”. Labeled THP-1 cells were treated with an equal mix of two Ago1 siRNAs or with control scrambled siRNA. Control and Ago1-knockdown cells were differentiated and incubated with Leishmania for 24 h. Subsequently, control and infected cells were washed extensively and mass spectrometry was performed on a mix of three lysates allowing for a quantitative comparison between non-infected, infected cells and infected in Ago1 knockdown condition (three independent experiments). Shown is the comparison between non-infected (L) and Leishmania-infected (M) cells. (A) Pie chart of the log2(fold change) distribution of Leishmania-modulated proteins. M/L normalized ratios were log2 transformed and averaged for at least 2 replicates. P-values < 0.05 from one-sample T-tests were considered significant. Proteins with a negative log2(fold change) were downregulated in Leishmania-infected cells compared to non-infected cells, whereas proteins with a positive log2(fold change) were upregulated in Leishmania-infected cells compared to non-infected cells. (B) Volcano plot comparing protein expression levels in Leishmania-infected cells with those in non-infected cells, both treated with control siRNA. Proteins significantly upregulated by Leishmania infection are shown as green dots, and those that were downregulated, as red dots. The horizontal dashed line marks the p-value cut off of 0.05 or -log10(p-value) = 1.301. The vertical dashed line represents log2(M/L) = 0. L, Light (Scrambled siRNA-treated, Non-infected); M, Medium (Scrambled siRNA-treated, Ld-infected).
For in-depth analysis in terms of their biological relevance, we considered the top 30 host proteins ranked by Manhattan distance (as described in “Materials and methods”) that were significantly modulated by Leishmania (Table 1). Interestingly, 14/30 have been shown to be implicated in Leishmania infection-related studies, such as Ferritin heavy chain (FTH1), Cystatin-A, Apoptosis-associated speck-like protein containing a CARD (PYCARD), CD44 antigen (CD44) and Heterogeneous nuclear ribonucleoprotein U (HNRNPU). Together, our proteomic analysis of differentially expressed proteins in Leishmania-infected host cells exhibits significant overlap with those that have been previously shown to be implicated in Leishmania infection, thus providing added confidence to our quantitative proteomic analysis.
3.6 Global identification of Leishmania-modulated Ago1-dependent host proteins in infected macrophages
Next, we sought to determine which of the differentially expressed proteins in Leishmania-infected macrophages are dependent on host Ago1 by combining two sets of proteins as shown in Figure S4B: (i) proteins that are upregulated in response to Leishmania infection (log2M/L > 0), and whose levels revert back closer to non-infected control levels, i.e. are recovered, (log2H/M < 0) in response to Ago1 knockdown, and (ii) proteins that are downregulated by Leishmania infection (log2M/L < 0) and which are recovered by Ago1 knockdown (log2H/M > 0), where H (heavy) represents levels in L. donovani-infected, Ago1 siRNA-transfected cells and M (medium) represents levels in L. donovani-infected, scrambled siRNA-transfected cells. The abundances of 97/331 proteins were significantly affected following Ago1 knockdown of Leishmania-infected macrophages, compared to corresponding control Leishmania-infected macrophages with normal levels of Ago1. Of these 97 proteins, the levels of 71 proteins (21 upregulated, 50 downregulated) were recovered by Ago1 knockdown as shown in Figures 4A, S4B. The overall importance of these protein modulations and recoveries were ranked by combining the Manhattan scores of Leishmania modulation and Ago1 modulation (as described in “Materials and methods”) and listed in descending order in Table S2. The proteins were also classified by GO terms in three categories: biological process, cellular component, and molecular function (Figures 4B-D).
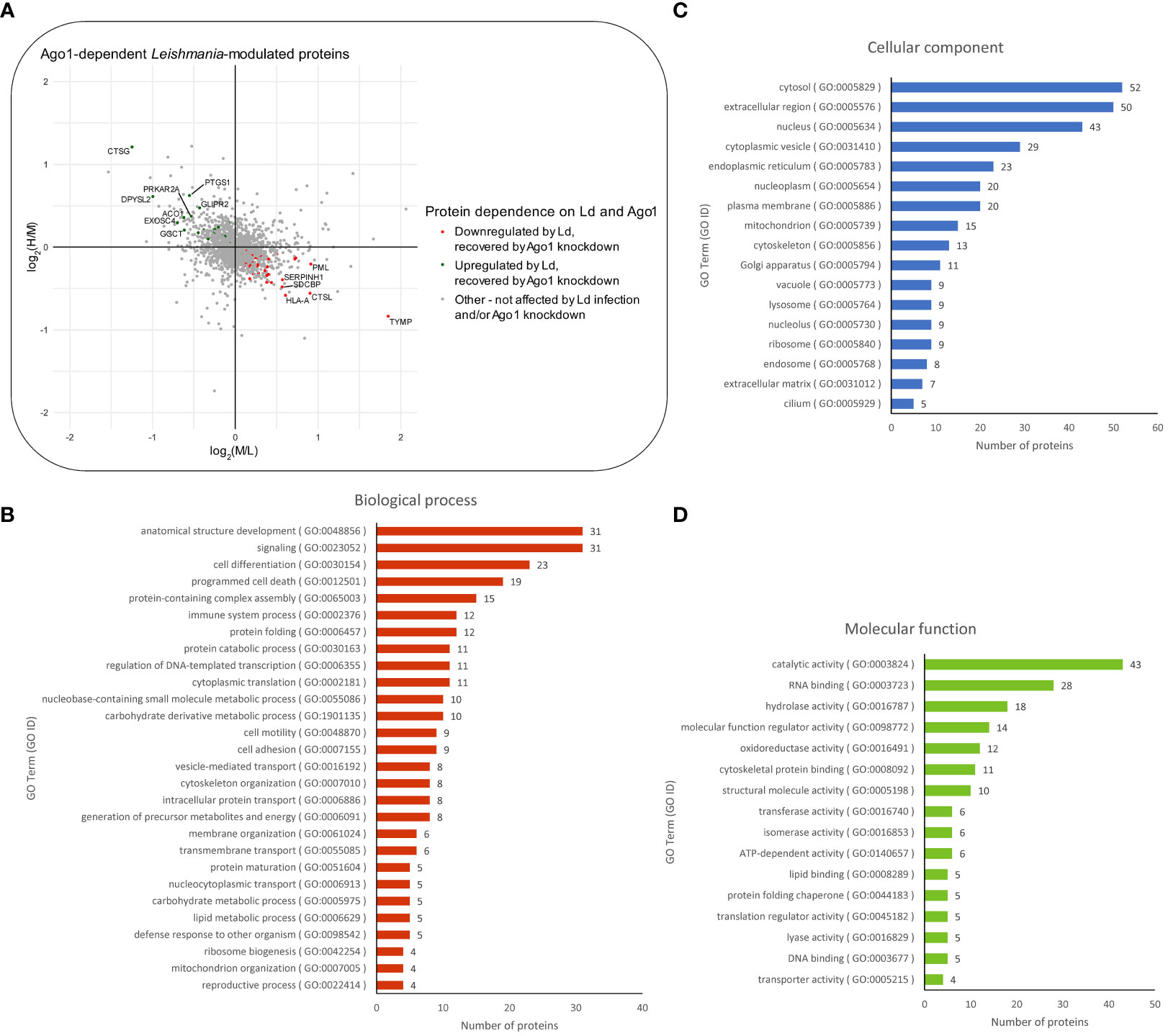
Figure 4 Proteomic analysis of Ago1-dependent Leishmania-modulated macrophage proteins. (A) Scatter plot of log2 transformed M/L ratios (y-axis) vs. log2 transformed H/M ratios (x-axis) of proteins. Proteins that were upregulated by Leishmania infection and recovered to any degree following Ago1 knockdown are shown in green. Proteins that were downregulated by Leishmania-infection and recovered to any degree following Ago1 knockdown are shown in red. The Ago1-dependent Leishmania-modulated proteins were also classified by GO terms in three categories: (B) biological process, (C) cellular component, and (D) molecular function. P-values < 0.05 from one-sample T-tests were considered significant for both log2 transformed M/L and H/M fold changes. L, Light (Scrambled siRNA-transfected, Non-infected); M, Medium (Scrambled siRNA-transfected, Ld-infected); H, Heavy (Ago1 siRNA-transfected, Ld-infected).
It was of interest to find out how many of the Ago1-dependent Leishmania-modulated proteins have relevance to Leishmania pathogenesis. For this, we searched the literature relevant to Leishmania-macrophage interactions, mainly using PubMed. Of the 71 Leishmania-modulated Ago1-dependent proteins, 20 have previously been reported in Leishmania infection-related studies. Of these 20 proteins, 5 proteins were upregulated by Leishmania (CTSG, PFKL, SEPT9, PTGS1 and ACO1) (Table 2) and 15 were downregulated by Leishmania (CTSL, EIF3A, ALDOA, NQO1, ANXA2, P4HB, HLA-A, HSP90AB1, PRDX1, PKM, HSPH1, LMNA, HSPA5, RPL13 and HSPA4) (Table 3).
Together, the quantitative proteomics analysis of Leishmania-modulated proteins in Ago1-deficient host cells reveals that a significant number of Ago1-dependent proteins have potential implications in Leishmania pathogenesis based on previously published studies. How many of the remaining Leishmania-modulated Ago1-dependent host proteins have a role to play in Leishmania-infection related activities remains to be investigated.
4 Discussion
It is well established that Leishmania parasites have evolved to acquire the ability to alter the cell biology of their host cells to favor their survival and pathogenesis. Since sncRNAs are involved in post-transcriptional regulation of gene expression via RNAi, these biomolecules can play critical roles in regulating host-pathogen interactions. In this context, several recent studies have clearly shown that sncRNAs play a crucial role in microbial infections (12–14, 23) including protozoan parasite infections such as Leishmania infection (163–165). There is growing evidence that sncRNAs are involved in the outcome of infections. Dysregulation of host miRNA has been associated with attenuated immune response and thus pathogen survival. On the other hand, sncRNAs can be employed by the host to counter parasite survival. Extracellular vesicles seem to facilitate this communication (164).
The current study was initiated based on our hypothesis that Leishmania regulates the host cell RNAi process to promote its survival in infected host cells. Since Argonautes are central components of the RNAi pathway, we began by investigating whether Leishmania regulates the two most abundant host cell Argonautes—Ago1 and Ago2—to its advantage. We found that Leishmania infection selectively enhanced macrophage Ago1 expression. Strikingly, enhanced level of Ago1 in infected cells correlated with higher levels of Ago1-containing complexes, suggesting preference for Ago1 to regulate RNAi-mediated host genes regulation. Our finding of upregulated expression of Ago1 and enrichment of Ago1 complexes in Leishmania-infected host cells prompted us to assess the potential involvement of Ago1 in Leishmania survival. For this study, we knocked down Ago1 using specific siRNAs followed by parasite rescue assay. Interestingly, downregulation of host macrophage Ago1 resulted in significant reduction of Leishmania survival in infected cells. Together, these results strongly demonstrate the involvement of Ago1 in promoting Leishmania survival in infected cells. At present, we do not know the mechanism of Leishmania-mediated upregulation of Ago1 in infected cells. This upregulation can occur at any point in the transcription-translation process. This includes an increase in the rate of transcription, stability of mRNA, rate of translation and stability of protein.
Upregulation of Ago1 during intracellular infection is not unique to Leishmania infection, and changes in host RISC composition have also been reported in other infections. For example, increased expressions of Ago1 and Dicer1 were detected upon slow bee paralysis virus (SBPV) infection in bumblebees and linked to 17 differentially expressed miRNAs upon infection (166). Also, depletion of Ago1 and Dicer1 in the mosquito Anopheles gambiae during Plasmodium berghei infection led to a two-fold increase in the number of oocysts (167). In another study, Ago1 and/or 3 were required for optimal resistance to flu virus (168).
It has recently been shown in Epstein-Barr virus (EBV)-infected mammalian cells that sncRNAs other than miRNAs were selectively loaded onto Ago1, but not Ago2 (169). Indeed, differences in the affinity of sncRNAs for Argonaute members have been observed in both lower organisms and mammals. In Drosophila, it has been shown that perfectly matched sncRNAs duplexes are loaded onto Ago2, whereas non-perfectly matched sncRNAs sorted to Ago1 (170, 171). Similarly, in the RNAi pathway of Caenorhabditis elegans, sncRNA duplexes that perfectly match their target mRNA are preferentially incorporated into Argonaute RNAi defective-1 (RDE-1) and non-perfectly matched sncRNA duplexes are loaded onto Argonaute family members ALG-1 or ALG-2 (172, 173). In contrast, sncRNA-mediated selectivity of Ago proteins in mammals is not well understood. However, there are reports suggesting the existence of Ago selectivity for certain distinct sncRNAs. For example, RNA-sequencing of Ago1-, Ago2- and Ago3-associated miRNAs revealed that some have a bias toward particular Ago proteins (20). Together, these findings strongly suggest the existence of selective Ago sorting mechanisms that direct distinct sncRNAs onto specific Ago-containing RISCs, and that Ago1 may be a preferred Ago for the loading of non-perfectly matching sncRNAs, including sncRNAs from pathogens.
To further investigate the role of Ago1 in Leishmania infection, we used SILAC-based quantitative LC-MS/MS proteomics analysis of L. donovani-infected Ago1-deficient cells, which revealed differentially expressed proteins in response to Leishmania infection that are dependent on Ago1. As expected, many of the modulated proteins that we identified have been previously shown to be implicated in Leishmania infection-related processes. For example, strongly modulated proteins Ferritin heavy chain (FTH1) and Cystatin A (CSTA) were upregulated and downregulated respectively, in agreement with previous studies (40, 41, 174). Apoptosis-associated speck-like protein containing a CARD (PYCARD, also known as ASC), which was downregulated upon Leishmania infection in our study, has previously been reported in multiple studies covering a range of Leishmania species and host cell types to be differentially expressed upon Leishmania infection (42–44).
From the validated set of 331 Leishmania-modulated proteins, a subset of 71 Ago1-dependent proteins was identified. Of these, 20 were found in previous studies relevant to leishmaniasis. Of interest, among the Leishmania-upregulated Ago1-dependent proteins, Cathepsin G (CTSG), has been shown to be significantly enriched at the late chronic phase of visceral leishmaniasis in mice (59). Another study reported a remarkable upregulation of CTSG in murine infection with L. donovani (60). Among the proteins downregulated by infection and recovered by Ago1-knockdown, Cathepsin L (CTSL) has been shown to play a crucial role for a Th1-type immune response during L. major infection by processing of soluble Leishmania antigen (SLA) for presentation on MHC class II molecules (51). Further, treatment of L. major-infected mice with CLIK148, a specific inhibitor of CTSL, exacerbated the disease by enhancing an SLA-specific Th2-type response (51).
In addition, it was of interest to analyze the Leishmania-modulated Ago1-dependent proteins in relation to other intracellular pathogen infections. Interestingly, 53 out of the 71 proteins have been shown to be related to the pathogenesis of other intracellular pathogens (Tables 2, 3). For example, host glyoxylate reductase/hydroxypyruvate reductase and mitochondrial very long-chain specific acyl-CoA dehydrogenase are both suppressed by the protozoan pathogen Trypanosoma cruzi (67). Protein promyelocytic leukemia protein/tripeptide motif protein (PML/TRIM19) has been reported to show activity against several viruses (80). Lassa virus and lymphocytic choriomeningitis virus replicate to higher levels in PML-deficient cells (81), whereas human foamy virus replication efficiency was found to be decreased upon overexpression of PML (82). Another protein of interest is calreticulin, which is known to be released onto the cell surface as an eat-me signal as a result of infection, such as in Mycobacterium tuberculosis and cytomegalovirus infections (90, 92). Furthermore, deliberate downregulation of calreticulin significantly increased the intracellular survival of M. tuberculosis (92).
Interestingly, based on the Pathways Commons collection of databases, 31 out of 71 Leishmania-modulated Ago1-knockdown-recovered proteins have reported interaction with Ago1, providing additional confidence in our Ago1-dependent proteins. We also note that 346 out of 417 proteins that were differentially expressed upon Ago1-knockdown were not modulated upon Leishmania-infection. Nevertheless, the data provided can act as a valuable resource for data mining by other investigators interested in Ago1 functions and its role in the RNAi pathway (Figure S5 and Table S1). We note that our data is restricted to proteomic analysis. It will be of interest to complement these findings with relevant biological assays and may be considered for future studies.
Given that Ago1 knockdown results in broad effects on the host cell proteomic profile, our results generate an attractive hypothesis that Leishmania-mediated upregulation of Ago1 is a strategy to regulate host cell gene expression in its favour. Since Ago1 interacts directly with sncRNAs, it is probable that Leishmania skews host RNAi by selectively uploading sncRNAs onto Ago1, including its own sncRNAs (Figure 5). This model also has support of the following published findings: [1] Leishmania exosomes delivered to the host cytosol contain Leishmania sncRNAs (25); [2] Leishmania infection results in a broad-based downregulation of host macrophage miRNA expression, thus freeing host Agos for exogenous sncRNA loading (24); [3] Ago1 does not require full complementarity of sncRNA to target mRNA, making it favorable for the loading of pathogen sncRNAs (170, 171); [4] Unloaded Ago proteins are not stable and are degraded by the proteasome degradation pathway (175, 176) and thus, increased Ago1 abundance is presumably complemented with increased loading of sncRNAs onto Ago1 for its increased stability (176). Together, this model adds to increasing evidence of cross-kingdom RNAi in which sncRNAs are transported bi-directionally between the host and their pathogens as a host defense mechanism or strategy of pathogens to regulate host cell biology in their favor (177).
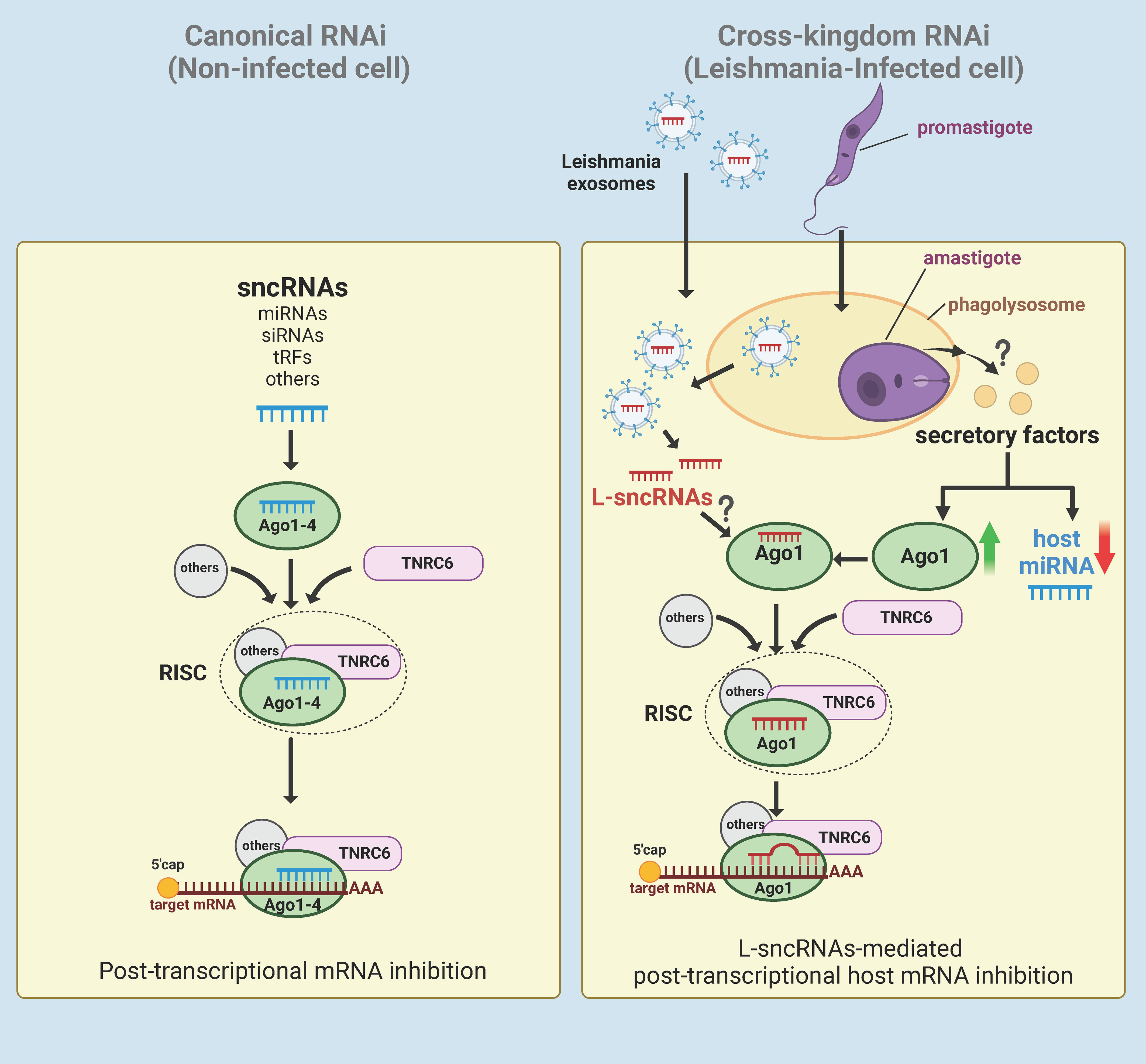
Figure 5 Hypothetical model of Leishmania-macrophage cross-kingdom RNAi. In canonical RNAi (left panel), endogenous sncRNAs interact with Ago1-4 proteins and then assembles with TNRC6 and other proteins to form the RISC. The RISC then regulates gene expression of the cell at post-transcriptional level by targeting the 3’UTR of mRNAs. In Leishmania-infected cells (right panel), Leishmania induces broad-based downregulation of host miRNAs and upregulation of Ago1 via an unknown mechanism, possibly by secretory factors. Leishmania exosomes containing ncRNAs secreted by amastigotes in the phagolysosomes of either the host cell or infected bystander cells are delivered to the cytosol. For RNAi, the delivered Leishmania exosomal sncRNAs (L-sncRNAs) are preferentially loaded onto host Ago1, which does not require full complementarity to target the mRNA. Created with BioRender.com.
To our knowledge, this is the first report showing the role of host macrophage Ago1 in the pathogenesis of an intracellular parasitic infection. The prior understanding of the role of RNA interference mechanisms in infection is mainly based on plants and insects. These findings represent a framework for further study of the role of RNAi in Leishmania pathogenesis in humans. A deep understanding of cross-kingdom RNAi in context of Leishmania infection could provide novel therapeutic strategies for managing and treating leishmaniasis and may have implications for other intracellular pathogens.
Data availability statement
The datasets presented in this study can be found in online repositories/Supplementary Material. The names of the repository/repositories and accession number(s) can be found in the article.
Author contributions
AM: Formal Analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. SC: Data curation, Formal Analysis, Methodology, Writing – original draft, Writing – review & editing. SK: Investigation, Writing – review & editing. K-MM: Formal Analysis, Methodology, Software, Writing – review & editing. LF: Formal Analysis, Funding acquisition, Writing – review & editing. NR: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. DN: Conceptualization, Investigation, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The funding for this research was provided by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-04991) and the Canadian Institute of Health Research (PJT-162191) awarded to NR, and Genome British Columbia (264PRO) awarded to LF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We appreciate Dr. Gunter Meister from the University of Regensburg for providing us the GST-T6B affinity construct. Portions of the findings from this manuscript were previously made available in an online format within a thesis submitted as a requirement for the completion of the Master’s program at the University of British Columbia. The thesis is titled: “Leishmania donovani upregulates host macrophage Argonaute 1 to persist inside infected host, and quantitative proteomic analysis of RNA-induced silencing complex isolated from infected host” and is archived in the University of British Columbia library (178).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1287539/full#supplementary-material
Supplementary Figure 1 | Macrophage Ago1 abundance is not affected in response to phagocytosis of latex beads. dTHP-1 cells were treated with latex beads at different multiplicities of infection (MOIs) or not for 24 h, whole cell lysates from non-treated and latex beads treated macrophages were analysed by Western blotting with antibodies specific for Ago1 and Actin (loading control). Densitometric analysis was used to measure Ago1 normalized to Actin. The histogram shown is the mean ± S.D. of the densitometric analyses of three independent experiments.
Supplementary Figure 2 | Preparation of recombinant proteins for GST-T6B affinity mediated pull-down assay for the isolation of Ago complexes. Recombinant GST-T6B peptide and GST alone were prepared as described in “Materials and methods”. (A) Purity assessment of recombinant proteins (stained with Coomassie Blue) used for the preparation of affinity beads to perform GST-T6B peptide mediated pull-down assay as described in “Materials and methods”. (B) The samples from purified and concentrated recombinant proteins were analyzed by Western blotting using anti-GST antibodies. (C) GST-T6B affinity based pull-down assay was performed to isolate Ago complexes from dTHP-1 whole cell lysates. The presence of Ago proteins was tested using indicated antibodies in a Western blot assay. GST alone affinity beads were used as a negative control for non-specific protein interaction. The result shown represents one of three independent experiments.
Supplementary Figure 3 | SiRNA-mediated Ago1 knockdown in macrophages for SILAC-based comparative proteomics analysis. (A) Schematic diagram of metabolic SILAC quantitative proteomic strategy. (B) Labeled THP-1 cells were treated with an equal mix of two human Ago1 siRNAs or with control scrambled siRNA. Control and Ago1-knockdown cells were differentiated and incubated with Leishmania for 24 h. Subsequently, control and infected cells were washed, and whole cell lysates were prepared for SILAC-based proteomic analysis. In parallel, samples were assessed by Western blotting to confirm the downregulation of Ago1 protein levels by Ago1 siRNAs and the preservation of Leishmania-mediated Ago1 induction in the presence of scrambled siRNA. The fold changes are normalized to scrambled siRNA-transfected non-infected cells. Shown is the mean ± S.D. of the densitometric analyses of three independent experiments. (**p-value < 0.01).
Supplementary Figure 4 | Schematic diagram detailing the data processing steps involved in the determination of (A) Leishmania infection-modulated host proteins; and (B) Ago1-dependent (i.e. recovered by Ago1 knockdown) Ld-modulated proteins.
Supplementary Figure 5 | Quantitative proteomic analysis of Ago1 knockdown-affected host macrophage proteins in the presence of infection. (A) Pie chart of the log2(fold change) distribution of Ago1 knockdown-affected proteins. H/M normalized ratios were log2 transformed and averaged for at least 2 replicates. P-values < 0.05 from one-sample T-tests were considered significant. Proteins with a negative log2(fold change) were downregulated in Leishmania-infected cells treated with Ago1 siRNA compared to Leishmania-infected cells transfected with scrambled siRNA, whereas proteins with a positive log2(fold change) were upregulated in Leishmania-infected cells treated with Ago1 siRNA compared to Leishmania-infected cells treated with scrambled siRNA. (B) Volcano plot comparing protein expression levels in Ago1-knockdown cells with those in cells transfected with control scrambled siRNA, both infected with Leishmania. Proteins significantly upregulated following Ago1 knockdown are shown as green dots, and those that were downregulated, as red dots. The top 30 modulated proteins based on Manhattan distance are labeled. The horizontal dashed line marks the p-value cut off of 0.05 or -log10(p-value) = 1.301. The vertical dashed line represents log2(H/M) = 0. M: Medium (Scrambled siRNA-treated, Ld-infected), H: Heavy (Ago siRNA-treated, Ld-infected).
References
1. Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: a review. F1000Research (2017) 6:750. doi: 10.12688/f1000research.11120.1
2. Martins-Melo FR, Lima Mda S, Ramos AN Jr, Alencar CH, Heukelbach J. Mortality and case fatality due to visceral leishmaniasis in brazil: a nationwide analysis of epidemiology, trends and spatial patterns. PloS One (2014) 9(4):e93770. doi: 10.1371/journal.pone.0093770
3. Serafim TD, Coutinho-Abreu IV, Dey R, Kissinger R, Valenzuela JG, Oliveira F, et al. Leishmaniasis: the act of transmission. Trends Parasitol (2021) 37(11):976–87. doi: 10.1016/j.pt.2021.07.003
4. de Menezes JP, Saraiva EM, da Rocha-Azevedo B. The site of the bite: Leishmania interaction with macrophages, neutrophils and the extracellular matrix in the dermis. Parasit Vectors. (2016) 9:264. doi: 10.1186/s13071-016-1540-3
5. Dinc R. Leishmania vaccines: the current situation with its promising aspect for the future. Korean J Parasitol (2022) 60(6):379–91. doi: 10.3347/kjp.2022.60.6.379
6. Velez R, Gállego M. Commercially approved vaccines for canine leishmaniosis: a review of available data on their safety and efficacy. Trop Med Int Health (2020) 25(5):540–57. doi: 10.1111/tmi.13382
7. Yasmin H, Adhikary A, Al-Ahdal MN, Roy S, Kishore U. Host–pathogen interaction in leishmaniasis: immune response and vaccination strategies. Immuno (2022) 2(1):218–54. doi: 10.3390/immuno2010015
8. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet (2006) 15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046
9. de Almeida RA, Fraczek MG, Parker S, Delneri D, O’Keefe RT. Non-coding RNAs and disease: the classical ncRNAs make a comeback. Biochem Soc Trans (2016) 44(4):1073–8. doi: 10.1042/BST20160089
10. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev (2016) 96(4):1297–325. doi: 10.1152/physrev.00041.2015
11. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet (2011) 12(12):861–74. doi: 10.1038/nrg3074
12. Guo Z, Li Y, Ding SW. Small RNA-based antimicrobial immunity. Nat Rev Immunol (2019) 19(1):31–44. doi: 10.1038/s41577-018-0071-x
13. Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet (2019) 20(2):89–108. doi: 10.1038/s41576-018-0073-3
14. Sullivan CS, Ganem D. MicroRNAs and viral infection. Mol Cell (2005) 20(1):3–7. doi: 10.1016/j.molcel.2005.09.012
15. Chahar HS, Corsello T, Kudlicki AS, Komaravelli N, Casola A. Respiratory syncytial virus infection changes cargo composition of exosome released from airway epithelial cells. Sci Rep (2018) 8(1):387. doi: 10.1038/s41598-017-18672-5
16. Choi EJ, Ren J, Zhang K, Wu W, Lee YS, Lee I, et al. The importance of AGO 1 and 4 in post-transcriptional gene regulatory function of tRF5-GluCTC, an respiratory syncytial virus-induced tRNA-derived RNA fragment. Int J Mol Sci (2020) 21(22):8766. doi: 10.3390/ijms21228766
17. Wu J, Yang J, Cho WC, Zheng Y. Argonaute proteins: Structural features, functions and emerging roles. J Adv Res. (2020) 24:317–24.
18. Azlan A, Dzaki N, Azzam G. Argonaute: The executor of small RNA function. J Genet Genomics Yi Chuan Xue Bao (2016) 43(8):481–94. doi: 10.1016/j.jgg.2016.06.002
19. Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell (2007) 26(5):611–23. doi: 10.1016/j.molcel.2007.05.001
20. Burroughs AM, Ando Y, de Hoon MJL, Tomaru Y, Suzuki H, Hayashizaki Y, et al. Deep-sequencing of human argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin. RNA Biol (2011) 8(1):158–77. doi: 10.4161/rna.8.1.14300
21. Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell (2015) 162(1):84–95. doi: 10.1016/j.cell.2015.06.029
22. Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet (2013) 14(7):447–59. doi: 10.1038/nrg3462
23. Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther J Am Soc Gene Ther (2013) 21(2):368–79. doi: 10.1038/mt.2012.237
24. Colineau L, Lambertz U, Fornes O, Wasserman WW, Reiner NE. C-myc is a novel leishmania virulence factor by proxy that targets the host miRNA system and is essential for survival in human macrophages. J Biol Chem (2018) 293(33):12805–19. doi: 10.1074/jbc.RA118.002462
25. Lambertz U, Oviedo Ovando ME, Vasconcelos EJR, Unrau PJ, Myler PJ, Reiner NE. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world leishmania providing evidence for conserved exosomal RNA packaging. BMC Genomics (2015) 16:151. doi: 10.1186/s12864-015-1260-7
26. Silverman JM, Clos J, Horakova E, Wang AY, Wiesgigl M, Kelly I, et al. Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J Immunol Baltim Md 1950 (2010) 185(9):5011–22. doi: 10.4049/jimmunol.1000541
27. Jain SK, Sahu R, Walker LA, Tekwani BL. A parasite rescue and transformation assay for antileishmanial screening against intracellular leishmania donovani amastigotes in THP1 human acute monocytic leukemia cell line. J Vis Exp JoVE (2012) 70:4054. doi: 10.3791/4054
28. Thomas SA, Nandan D, Kass J, Reiner NE. Countervailing, time-dependent effects on host autophagy promotes intracellular survival of leishmania. J Biol Chem (2018) 293(7):2617–30. doi: 10.1074/jbc.M117.808675
29. Hauptmann J, Schraivogel D, Bruckmann A, Manickavel S, Jakob L, Eichner N, et al. Biochemical isolation of Argonaute protein complexes by Ago-APP. Proc Natl Acad Sci U.S.A. (2015) 112(38):11841–5. doi: 10.1073/pnas.1506116112
30. Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep (2014) 6(1):211–21. doi: 10.1016/j.celrep.2013.12.013
31. Goodman JK, Zampronio CG, Jones AME, Hernandez-Fernaud JR. Updates of the in-gel digestion method for protein analysis by mass spectrometry. Proteomics (2018) 18(23):e1800236. doi: 10.1002/pmic.201800236
32. Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem (1996) 68(5):850–8. doi: 10.1021/ac950914h
33. Ishihama Y, Rappsilber J, Andersen JS, Mann M. Microcolumns with self-assembled particle frits for proteomics. J Chromatogr A (2002) 979(1–2):233–9. doi: 10.1016/S0021-9673(02)01402-4
34. Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc (2016) 11(12):2301–19. doi: 10.1038/nprot.2016.136
35. Perea-Martínez A, García-Hernández R, Manzano JI, Gamarro F. Transcriptomic analysis in human macrophages infected with therapeutic failure clinical isolates of leishmania infantum. ACS Infect Dis (2022) 8(4):800–10. doi: 10.1021/acsinfecdis.1c00513
36. Singh AK, Pandey RK, Siqueira-Neto JL, Kwon YJ, Freitas-Junior LH, Shaha C, et al. Proteomic-based approach to gain insight into reprogramming of THP-1 cells exposed to Leishmania donovani over an early temporal window. Infect Immun (2015) 83(5):1853–68. doi: 10.1128/IAI.02833-14
37. Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol (2014) 23(1):37–45. doi: 10.1016/j.intimp.2014.08.002
38. Müller M, Fazi F, Ciaudo C. Argonaute proteins: from structure to function in development and pathological cell fate determination. Front Cell Dev Biol (2019) 7:360. doi: 10.3389/fcell.2019.00360
39. Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb Perspect Biol (2011) 3(10):a003772. doi: 10.1101/cshperspect.a003772
40. Ben-Othman R, Flannery AR, Miguel DC, Ward DM, Kaplan J, Andrews NW. Leishmania-mediated inhibition of iron export promotes parasite replication in macrophages. PloS Pathog (2014) 10(1):e1003901. doi: 10.1371/journal.ppat.1003901
41. Kar S, Ukil A, Das PK. Cystatin cures visceral leishmaniasis by NF-κB-mediated proinflammatory response through co-ordination of TLR/MyD88 signaling with p105-Tpl2-ERK pathway. Eur J Immunol (2011) 41(1):116–27. doi: 10.1002/eji.201040533
42. Dillon LAL, Suresh R, Okrah K, Corrada Bravo H, Mosser DM, El-Sayed NM. Simultaneous transcriptional profiling of Leishmania major and its murine macrophage host cell reveals insights into host-pathogen interactions. BMC Genomics (2015) 16:1108. doi: 10.1186/s12864-015-2237-2
43. Esch KJ, Schaut RG, Lamb IM, Clay G, Morais Lima ÁL, do Nascimento PRP, et al. Activation of autophagy and nucleotide-binding domain leucine-rich repeat-containing-like receptor family, pyrin domain-containing 3 inflammasome during Leishmania infantum-associated glomerulonephritis. Am J Pathol (2015) 185(8):2105–17. doi: 10.1016/j.ajpath.2015.04.017
44. Veras PST, Bezerra de Menezes JP. Using proteomics to understand how leishmania parasites survive inside the host and establish infection. Int J Mol Sci (2016) 17(8):E1270. doi: 10.3390/ijms17081270
45. Holowka T, Castilho TM, Garcia AB, Sun T, McMahon-Pratt D, Bucala R. Leishmania-encoded orthologs of macrophage migration inhibitory factor regulate host immunity to promote parasite persistence. FASEB J Off Publ Fed Am Soc Exp Biol (2016) 30(6):2249–65. doi: 10.1096/fj.201500189R
46. Chaparro V, Graber TE, Alain T, Jaramillo M. Transcriptional profiling of macrophages reveals distinct parasite stage-driven signatures during early infection by Leishmania donovani. Sci Rep (2022) 12(1):6369. doi: 10.1038/s41598-022-10317-6
47. Alzahrani KJH, Ali JAM, Eze AA, Looi WL, Tagoe DNA, Creek DJ, et al. Functional and genetic evidence that nucleoside transport is highly conserved in Leishmania species: Implications for pyrimidine-based chemotherapy. Int J Parasitol Drugs Drug Resist (2017) 7(2):206–26. doi: 10.1016/j.ijpddr.2017.04.003
48. LaFon SW, Nelson DJ, Berens RL, Marr JJ. Purine and pyrimidine salvage pathways in Leishmania donovani. Biochem Pharmacol (1982) 31(2):231–8. doi: 10.1016/0006-2952(82)90216-7
49. Dogra N, Warburton C, McMaster WR. Leishmania major abrogates gamma interferon-induced gene expression in human macrophages from a global perspective. Infect Immun (2007) 75(7):3506–15. doi: 10.1128/IAI.00277-07
50. Negrão F, Giorgio S, Eberlin MN, Yates JR. Comparative Proteomic Analysis of Murine Cutaneous Lesions Induced by Leishmania amazonensis or Leishmania major. ACS Infect Dis (2019) 5(8):1295–305. doi: 10.1021/acsinfecdis.8b00370
51. Onishi K, Li Y, Ishii K, Hisaeda H, Tang L, Duan X, et al. Cathepsin L is crucial for a Th1-type immune response during Leishmania major infection. Microbes Infect (2004) 6(5):468–74. doi: 10.1016/j.micinf.2004.01.008
52. Zhang T, Maekawa Y, Sakai T, Nakano Y, Ishii K, Hisaeda H, et al. Treatment with cathepsin L inhibitor potentiates Th2-type immune response in Leishmania major-infected BALB/c mice. Int Immunol (2001) 13(8):975–82. doi: 10.1093/intimm/13.8.975
53. Isnard A, Christian JG, Kodiha M, Stochaj U, McMaster WR, Olivier M. Impact of Leishmania infection on host macrophage nuclear physiology and nucleopore complex integrity. PloS Pathog (2015) 11(3):e1004776. doi: 10.1371/journal.ppat.1004776
54. Muxel SM, Acuña SM, Aoki JI, Zampieri RA, Floeter-Winter LM. Toll-Like Receptor and miRNA-let-7e Expression Alter the Inflammatory Response in Leishmania amazonensis-Infected Macrophages. Front Immunol (2018) 9:2792. doi: 10.3389/fimmu.2018.02792
55. Vogl T, Eisenblätter M, Völler T, Zenker S, Hermann S, van Lent P, et al. Alarmin S100A8/S100A9 as a biomarker for molecular imaging of local inflammatory activity. Nat Commun (2014) 5:4593. doi: 10.1038/ncomms5593
56. Guerfali FZ, Laouini D, Guizani-Tabbane L, Ottones F, Ben-Aissa K, Benkahla A, et al. Simultaneous gene expression profiling in human macrophages infected with Leishmania major parasites using SAGE. BMC Genomics (2008) 9:238. doi: 10.1186/1471-2164-9-238
57. Prakash S, Saini S, Kumari S, Singh B, Kureel AK, Rai AK. Retinoic acid restores the levels of cellular cholesterol in Leishmania donovani infected macrophages by increasing npc1 and npc2 expressions. Biochimie (2022) 198:23–32. doi: 10.1016/j.biochi.2022.03.002
58. Losada-Barragán M, Umaña-Pérez A, Durães J, Cuervo-Escobar S, Rodríguez-Vega A, Ribeiro-Gomes FL, et al. Thymic microenvironment is modified by malnutrition and leishmania infantum infection. Front Cell Infect Microbiol (2019) 9:252. doi: 10.3389/fcimb.2019.00252
59. Agallou M, Athanasiou E, Kammona O, Tastsoglou S, Hatzigeorgiou AG, Kiparissides C, et al. Transcriptome analysis identifies immune markers related to visceral leishmaniasis establishment in the experimental model of BALB/c mice. Front Immunol (2019) 10:2749. doi: 10.3389/fimmu.2019.02749
60. Ashwin H, Seifert K, Forrester S, Brown N, MacDonald S, James S, et al. Tissue and host species-specific transcriptional changes in models of experimental visceral leishmaniasis. Wellcome Open Res (2018) 3:135. doi: 10.12688/wellcomeopenres.14867.1
61. Nde PN, Lima MF, Johnson CA, Pratap S, Villalta F. Regulation and use of the extracellular matrix by Trypanosoma cruzi during early infection. Front Immunol (2012) 3:337. doi: 10.3389/fimmu.2012.00337
62. Osorio y Fortéa J, de la Llave E, Regnault B, Coppée JY, Milon G, Lang T, et al. Transcriptional signatures of BALB/c mouse macrophages housing multiplying Leishmania amazonensis amastigotes. BMC Genomics (2009) 10:119. doi: 10.1186/1471-2164-10-119
63. Ohms M, Ferreira C, Busch H, Wohlers I, Guerra de Souza AC, Silvestre R, et al. Enhanced glycolysis is required for antileishmanial functions of neutrophils upon infection with leishmania donovani. Front Immunol (2021) 12:632512. doi: 10.3389/fimmu.2021.632512
64. Smirlis D, Dingli F, Sabatet V, Roth A, Knippschild U, Loew D, et al. Identification of the host substratome of leishmania-secreted casein kinase 1 using a SILAC-based quantitative mass spectrometry assay. Front Cell Dev Biol (2021) 9:800098. doi: 10.3389/fcell.2021.800098
65. Akil A, Peng J, Omrane M, Gondeau C, Desterke C, Marin M, et al. Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV. Nat Commun (2016) 7:12203. doi: 10.1038/ncomms12203
66. Boddy KC, Gao AD, Truong D, Kim MS, Froese CD, Trimble WS, et al. Septin-regulated actin dynamics promote Salmonella invasion of host cells. Cell Microbiol (2018) 20(10):e12866. doi: 10.1111/cmi.12866
67. Zhang S, Kim CC, Batra S, McKerrow JH, Loke P. Delineation of diverse macrophage activation programs in response to intracellular parasites and cytokines. PloS Negl Trop Dis (2010) 4(3):e648. doi: 10.1371/journal.pntd.0000648
68. Rabhi I, Rabhi S, Ben-Othman R, Rasche A, Daskalaki A, Trentin B, et al. Transcriptomic signature of Leishmania infected mice macrophages: a metabolic point of view. PloS Negl Trop Dis (2012) 6(8):e1763. doi: 10.1371/journal.pntd.0001763
69. O’Neill GP, Mancini JA, Kargman S, Yergey J, Kwan MY, Falgueyret JP, et al. Overexpression of human prostaglandin G/H synthase-1 and -2 by recombinant vaccinia virus: inhibition by nonsteroidal anti-inflammatory drugs and biosynthesis of 15-hydroxyeicosatetraenoic acid. Mol Pharmacol (1994) 45(2):245–54.
70. Aerbajinai W, Joo J, Yan M, Kumkhaek C, Rodgers GP. Glia maturation factor-gamma regulates the intracellular growth of salmonella via modulation of ferroportin in murine macrophages. Blood (2015) 126(23):2208–8. doi: 10.1182/blood.V126.23.2208.2208
71. Kobets T, Čepičková M, Volkova V, Sohrabi Y, Havelková H, Svobodová M, et al. Novel loci controlling parasite load in organs of mice infected with leishmania major, their interactions and sex influence. Front Immunol (2019) 10:1083. doi: 10.3389/fimmu.2019.01083
72. Pisu D, Huang L, Grenier JK, Russell DG. Dual RNA-seq of Mtb-infected macrophages in vivo reveals ontologically distinct host-pathogen interactions. Cell Rep (2020) 30(2):335–350.e4. doi: 10.1016/j.celrep.2019.12.033
73. Wu J, Gu J, Shen L, Fang D, Zou X, Cao Y, et al. Exosomal microRNA-155 inhibits enterovirus A71 infection by targeting PICALM. Int J Biol Sci (2019) 15(13):2925–35. doi: 10.7150/ijbs.36388
74. Diaz A, Ahlquist P. Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication. Curr Opin Microbiol (2012) 15(4):519–24. doi: 10.1016/j.mib.2012.04.007
75. Bruckert WM, Abu Kwaik Y. Complete and ubiquitinated proteome of the Legionella-containing vacuole within human macrophages. J Proteome Res (2015) 14(1):236–48. doi: 10.1021/pr500765x
76. Aslam Y, Williamson J, Romashova V, Elder E, Krishna B, Wills M, et al. Human cytomegalovirus upregulates expression of HCLS1 resulting in increased cell motility and transendothelial migration during latency. iScience (2019) 20:60–72. doi: 10.1016/j.isci.2019.09.016
77. Jamwal S, Midha MK, Verma HN, Basu A, Rao KVS, Manivel V. Characterizing virulence-specific perturbations in the mitochondrial function of macrophages infected with Mycobacterium tuberculosis. Sci Rep (2013) 3:1328. doi: 10.1038/srep01328
78. Badaoui B, Rutigliano T, Anselmo A, Vanhee M, Nauwynck H, Giuffra E, et al. RNA-sequence analysis of primary alveolar macrophages after in vitro infection with porcine reproductive and respiratory syndrome virus strains of differing virulence. PloS One (2014) 9(3):e91918. doi: 10.1371/journal.pone.0091918
79. Li W, Yue H. Thymidine phosphorylase is increased in COVID-19 patients in an acuity-dependent manner. Front Med (2021) 8:653773. doi: 10.3389/fmed.2021.653773
80. Nisole S, Stoye JP, Saïb A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol (2005) 3(10):799–808. doi: 10.1038/nrmicro1248
81. Asper M, Sternsdorf T, Hass M, Drosten C, Rhode A, Schmitz H, et al. Inhibition of different Lassa virus strains by alpha and gamma interferons and comparison with a less pathogenic arenavirus. J Virol (2004) 78(6):3162–9. doi: 10.1128/JVI.78.6.3162-3169.2004
82. Regad T, Saib A, Lallemand-Breitenbach V, Pandolfi PP, de Thé H, Chelbi-Alix MK. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J (2001) 20(13):3495–505. doi: 10.1093/emboj/20.13.3495
83. Kahle T, Volkmann B, Eissmann K, Herrmann A, Schmitt S, Wittmann S, et al. TRIM19/PML restricts HIV infection in a cell type-dependent manner. Viruses (2015) 8(1):E2. doi: 10.3390/v8010002
84. Lunardi A, Gaboli M, Giorgio M, Rivi R, Bygrave A, Antoniou M, et al. A role for PML in innate immunity. Genes Cancer (2011) 2(1):10–9. doi: 10.1177/1947601911402682
85. Ribet D, Lallemand-Breitenbach V, Ferhi O, Nahori MA, Varet H, de Thé H, et al. Promyelocytic leukemia protein (PML) controls listeria monocytogenes infection. mBio (2017) 8(1):e02179-16. doi: 10.1128/mBio.02179-16
86. Liu X, Huuskonen S, Laitinen T, Redchuk T, Bogacheva M, Salokas K, et al. SARS-CoV-2-host proteome interactions for antiviral drug discovery. Mol Syst Biol (2021) 17(11):e10396. doi: 10.15252/msb.202110396
87. Pires D, Marques J, Pombo JP, Carmo N, Bettencourt P, Neyrolles O, et al. Role of cathepsins in mycobacterium tuberculosis survival in human macrophages. Sci Rep (2016) 6:32247. doi: 10.1038/srep32247
88. Nepal RM, Mampe S, Shaffer B, Erickson AH, Bryant P. Cathepsin L maturation and activity is impaired in macrophages harboring M. avium and M. tuberculosis. Int Immunol (2006) 18(6):931–9. doi: 10.1093/intimm/dxl029
89. Xu X, Greenland J, Baluk P, Adams A, Bose O, McDonald DM, et al. Cathepsin L protects mice from mycoplasmal infection and is essential for airway lymphangiogenesis. Am J Respir Cell Mol Biol (2013) 49(3):437–44. doi: 10.1165/rcmb.2013-0016OC
90. Zhu J, Newkirk MM. Viral induction of the human autoantigen calreticulin. Clin Investig Med Med Clin Exp (1994) 17(3):196–205.
91. Birkle T, Brown GC. I’m infected, eat me! Innate immunity mediated by live, infected cells signaling to be phagocytosed. Infect Immun (2021) 89(5):e00476-20. doi: 10.1128/iai.00476-20
92. Jo SH, Choi JA, Lim YJ, Lee J, Cho SN, Oh SM, et al. Calreticulin modulates the intracellular survival of mycobacteria by regulating ER-stress-mediated apoptosis. Oncotarget (2017) 8(35):58686–98. doi: 10.18632/oncotarget.17419
93. Kim Y, Chang KO. Protein disulfide isomerases as potential therapeutic targets for influenza A and B viruses. Virus Res (2018) 247:26–33. doi: 10.1016/j.virusres.2018.01.010
94. Subramani C, Nair VP, Anang S, Mandal SD, Pareek M, Kaushik N, et al. Host-virus protein interaction network reveals the involvement of multiple host processes in the life cycle of hepatitis E virus. mSystems (2018) 3(1):e00135-17. doi: 10.1128/mSystems.00135-17
95. Frank B, Marcu A, de Oliveira Almeida Petersen AL, Weber H, Stigloher C, Mottram JC, et al. Autophagic digestion of Leishmania major by host macrophages is associated with differential expression of BNIP3, CTSE, and the miRNAs miR-101c, miR-129, and miR-210. Parasit Vectors. (2015) 8:404. doi: 10.1186/s13071-015-0974-3
96. Tien CF, Cheng SC, Ho YP, Chen YS, Hsu JH, Chang RY. Inhibition of aldolase A blocks biogenesis of ATP and attenuates Japanese encephalitis virus production. Biochem Biophys Res Commun (2014) 443(2):464–9. doi: 10.1016/j.bbrc.2013.11.128
97. Zhai Y, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, et al. Host transcriptional response to influenza and other acute respiratory viral infections–A prospective cohort study. PloS Pathog (2015) 11(6):e1004869. doi: 10.1371/journal.ppat.1004869
98. Reverte M, Eren RO, Jha B, Desponds C, Snäkä T, Prevel F, et al. The antioxidant response favors Leishmania parasites survival, limits inflammation and reprograms the host cell metabolism. PloS Pathog (2021) 17(3):e1009422. doi: 10.1371/journal.ppat.1009422
99. Lata S, Ali A, Sood V, Raja R, Banerjea AC. HIV-1 Rev downregulates Tat expression and viral replication via modulation of NAD(P)H:quinine oxidoreductase 1 (NQO1). Nat Commun (2015) 6:7244. doi: 10.1038/ncomms8244
100. Cheng ST, Hu JL, Ren JH, Yu HB, Zhong S, Wai Wong VK, et al. Dicoumarol, an NQO1 inhibitor, blocks cccDNA transcription by promoting degradation of HBx. J Hepatol (2021) 74(3):522–34. doi: 10.1016/j.jhep.2020.09.019
101. Sun G, Liu F, Lin TJ. Identification of Pseudomonas aeruginosa-induced genes in human mast cells using suppression subtractive hybridization: up-regulation of IL-8 and CCL4 production. Clin Exp Immunol (2005) 142(1):199–205. doi: 10.1111/j.1365-2249.2005.02909.x
102. Boon ACM, deBeauchamp J, Hollmann A, Luke J, Kotb M, Rowe S, et al. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J Virol (2009) 83(20):10417–26. doi: 10.1128/JVI.00514-09
103. Gao X, Hardwidge PR. Ribosomal protein s3: a multifunctional target of attaching/effacing bacterial pathogens. Front Microbiol (2011) 2:137. doi: 10.3389/fmicb.2011.00137
104. Janke R, Genzel Y, Wetzel M, Reichl U. Effect of influenza virus infection on key metabolic enzyme activities in MDCK cells. BMC Proc (2011) 5(Suppl 8):P129. doi: 10.1186/1753-6561-5-S8-P129
105. Hayward RJ, Marsh JW, Humphrys MS, Huston WM, Myers GSA. Chromatin accessibility dynamics of Chlamydia-infected epithelial cells. Epigenet Chromatin (2020) 13(1):45. doi: 10.1186/s13072-020-00368-2
106. Ma H, Kien F, Manière M, Zhang Y, Lagarde N, Tse KS, et al. Human annexin A6 interacts with influenza a virus protein M2 and negatively modulates infection. J Virol (2012) 86(3):1789–801. doi: 10.1128/JVI.06003-11
107. Musiol A, Gran S, Ehrhardt C, Ludwig S, Grewal T, Gerke V, et al. Annexin A6-balanced late endosomal cholesterol controls influenza A replication and propagation. mBio (2013) 4(6):e00608-00613. doi: 10.1128/mBio.00608-13
108. Hua Y, Wu J, Fu M, Liu J, Li X, Zhang B, et al. Enterohemorrhagic escherichia coli effector protein espF interacts with host protein ANXA6 and triggers myosin light chain kinase (MLCK)-dependent tight junction dysregulation. Front Cell Dev Biol (2020) 8:613061. doi: 10.3389/fcell.2020.613061
109. Thai M, Thaker SK, Feng J, Du Y, Hu H, Ting Wu T, et al. MYC-induced reprogramming of glutamine catabolism supports optimal virus replication. Nat Commun (2015) 6:8873. doi: 10.1038/ncomms9873
110. Derry MC, Sutherland MR, Restall CM, Waisman DM, Pryzdial ELG. Annexin 2-mediated enhancement of cytomegalovirus infection opposes inhibition by annexin 1 or annexin 5. J Gen Virol (2007) 88(Pt 1):19–27. doi: 10.1099/vir.0.82294-0
111. Dziduszko A, Ozbun MA. Annexin A2 and S100A10 regulate human papillomavirus type 16 entry and intracellular trafficking in human keratinocytes. J Virol (2013) 87(13):7502–15. doi: 10.1128/jvi.00519-13
112. Koga R, Kubota M, Hashiguchi T, Yanagi Y, Ohno S. Annexin A2 mediates the localization of measles virus matrix protein at the plasma membrane. J Virol (2018) 92(10):e00181-18. doi: 10.1128/jvi.00181-18
113. Santos CXC, Stolf BS, Takemoto PVA, Amanso AM, Lopes LR, Souza EB, et al. Protein disulfide isomerase (PDI) associates with NADPH oxidase and is required for phagocytosis of Leishmania chagasi promastigotes by macrophages. J Leukoc Biol (2009) 86(4):989–98. doi: 10.1189/jlb.0608354
114. Stolf BS, Smyrnias I, Lopes LR, Vendramin A, Goto H, Laurindo FRM, et al. Protein disulfide isomerase and host-pathogen interaction. ScientificWorldJournal (2011) 11:1749–61. doi: 10.1100/2011/289182
115. Parakh S, Atkin JD. Novel roles for protein disulphide isomerase in disease states: a double edged sword? Front Cell Dev Biol (2015) 3:30. doi: 10.3389/fcell.2015.00030
116. Auwerx J, Isacsson O, Söderlund J, Balzarini J, Johansson M, Lundberg M. Human glutaredoxin-1 catalyzes the reduction of HIV-1 gp120 and CD4 disulfides and its inhibition reduces HIV-1 replication. Int J Biochem Cell Biol (2009) 41(6):1269–75. doi: 10.1016/j.biocel.2008.10.031
117. Gilbert J, Ou W, Silver J, Benjamin T. Downregulation of protein disulfide isomerase inhibits infection by the mouse polyomavirus. J Virol (2006) 80(21):10868–70. doi: 10.1128/JVI.01117-06
118. Tsai B, Rapoport TA. Unfolded cholera toxin is transferred to the ER membrane and released from protein disulfide isomerase upon oxidation by Ero1. J Cell Biol (2002) 159(2):207–16. doi: 10.1083/jcb.200207120
119. Nyambura LW, Jarmalavicius S, Walden P. Impact of Leishmania donovani infection on the HLA I self peptide repertoire of human macrophages. PloS One (2018) 13(7):e0200297. doi: 10.1371/journal.pone.0200297
120. Reiner NE, Ng W, McMaster WR. Parasite-accessory cell interactions in murine leishmaniasis. II. Leishmania donovani suppresses macrophage expression of class I and class II major histocompatibility complex gene products. J Immunol Baltim Md 1950 (1987) 138(6):1926–32. doi: 10.4049/jimmunol.138.6.1926
121. Crux NB, Elahi S. Human leukocyte antigen (HLA) and immune regulation: how do classical and non-classical HLA alleles modulate immune response to human immunodeficiency virus and hepatitis C virus infections? Front Immunol (2017) 8:832. doi: 10.3389/fimmu.2017.00832
122. Vanderboom PM, Mun DG, Madugundu AK, Mangalaparthi KK, Saraswat M, Garapati K, et al. Proteomic signature of host response to SARS-coV-2 infection in the nasopharynx. Mol Cell Proteomics MCP (2021) 20:100134. doi: 10.1016/j.mcpro.2021.100134
123. Li D, Rawle DJ, Wu Z, Jin H, Lin MH, Lor M, et al. eEF1A demonstrates paralog specific effects on HIV-1 reverse transcription efficiency. Virology (2019) 530:65–74. doi: 10.1016/j.virol.2019.01.023
124. Wei T, Li D, Marcial D, Khan M, Lin MH, Snape N, et al. The eukaryotic elongation factor 1A is critical for genome replication of the paramyxovirus respiratory syncytial virus. PloS One (2014) 9(12):e114447. doi: 10.1371/journal.pone.0114447
125. Mahmood F, Xu R, Awan MUN, Song Y, Han Q, Xia X, et al. PDIA3: Structure, functions and its potential role in viral infections. BioMed Pharmacother Biomed Pharmacother (2021) 143:112110. doi: 10.1016/j.biopha.2021.112110
126. Bzdyl NM, Scott NE, Norville IH, Scott AE, Atkins T, Pang S, et al. Peptidyl-Prolyl Isomerase ppiB Is Essential for Proteome Homeostasis and Virulence in Burkholderia pseudomallei. Infect Immun (2019) 87(10):e00528-19. doi: 10.1128/IAI.00528-19
127. Joshi P, Sloan B, Torbett BE, Stoddart CA. Heat shock protein 90AB1 and hyperthermia rescue infectivity of HIV with defective cores. Virology (2013) 436(1):162–72. doi: 10.1016/j.virol.2012.11.005
128. Joshi P, Stoddart CA. Impaired infectivity of ritonavir-resistant HIV is rescued by heat shock protein 90AB1. J Biol Chem. (2013) 286(28):24581–92. doi: 10.1074/jbc.M111.248021
129. Matsumura K, Iwai H, Kato-Miyazawa M, Kirikae F, Zhao J, Yanagawa T, et al. Peroxiredoxin 1 Contributes to Host Defenses against Mycobacterium tuberculosis. J Immunol Baltim Md 1950 (2016) 197(8):3233–44. doi: 10.4049/jimmunol.1601010
130. Jamaluddin M, Wiktorowicz JE, Soman KV, Boldogh I, Forbus JD, Spratt H, et al. Role of peroxiredoxin 1 and peroxiredoxin 4 in protection of respiratory syncytial virus-induced cysteinyl oxidation of nuclear cytoskeletal proteins. J Virol (2010) 84(18):9533–45. doi: 10.1128/JVI.01005-10
131. Nchoutmboube JA, Viktorova EG, Scott AJ, Ford LA, Pei Z, Watkins PA, et al. Increased long chain acyl-Coa synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PloS Pathog (2013) 9(6):e1003401. doi: 10.1371/journal.ppat.1003401
132. Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MAR, Sheedy FJ, Gleeson LE, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab (2015) 21(1):65–80. doi: 10.1016/j.cmet.2014.12.005
133. Lang BJ, Gorrell RJ, Tafreshi M, Hatakeyama M, Kwok T, Price JT. The Helicobacter pylori cytotoxin CagA is essential for suppressing host heat shock protein expression. Cell Stress Chaperones (2016) 21(3):523–33. doi: 10.1007/s12192-016-0680-x
134. Wuerth JD, Habjan M, Kainulainen M, Berisha B, Bertheloot D, Superti-Furga G, et al. eIF2B as a target for viral evasion of PKR-mediated translation inhibition. mBio (2020) 11(4):e00976–20. doi: 10.1128/mBio.00976-20
135. Valiente-Echeverría F, Melnychuk L, Vyboh K, Ajamian L, Gallouzi IE, Bernard N, et al. eEF2 and Ras-GAP SH3 domain-binding protein (G3BP1) modulate stress granule assembly during HIV-1 infection. Nat Commun (2014) 5:4819. doi: 10.1038/ncomms5819
136. Toribio-Fernández R, Zorita V, Rocha-Perugini V, Iborra S, Martínez Del Hoyo G, Chevre R, et al. Lamin A/C augments Th1 differentiation and response against vaccinia virus and Leishmania major. Cell Death Dis (2018) 9(1):9. doi: 10.1038/s41419-017-0007-6
137. DiGiuseppe S, Rollins MG, Bartom ET, Walsh D. ZNF598 plays distinct roles in interferon-stimulated gene expression and poxvirus protein synthesis. Cell Rep (2018) 23(5):1249–58. doi: 10.1016/j.celrep.2018.03.132
138. Li L, Yu C, Gao H, Li Y. Argonaute proteins: potential biomarkers for human colon cancer. BMC Cancer (2010) 10:38. doi: 10.1186/1471-2407-10-38
139. Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. The Tudor SND1 protein is an m6A RNA reader essential for replication of Kaposi's sarcoma-associated herpesvirus. Elife (2019) 8:e47261. doi: 10.7554/eLife.47261
140. Wang X, Zhang C, Wang S, Rashu R, Thomas R, Yang J, et al. SND1 promotes Th1/17 immunity against chlamydial lung infection through enhancing dendritic cell function. PloS Pathog (2021) 17(2):e1009295. doi: 10.1371/journal.ppat.1009295
141. Galluzzi L, Diotallevi A, De Santi M, Ceccarelli M, Vitale F, Brandi G, et al. Leishmania infantum induces mild unfolded protein response in infected macrophages. PloS One (2016) 11(12):e0168339. doi: 10.1371/journal.pone.0168339
142. Reid SP, Shurtleff AC, Costantino JA, Tritsch SR, Retterer C, Spurgers KB, et al. HSPA5 is an essential host factor for Ebola virus infection. Antiviral Res (2014) 109:171–4. doi: 10.1016/j.antiviral.2014.07.004
143. Heikkilä O, Merilahti P, Hakanen M, Karelehto E, Alanko J, Sukki M, et al. Integrins are not essential for entry of coxsackievirus A9 into SW480 human colon adenocarcinoma cells. Virol J (2016) 13(1):171. doi: 10.1186/s12985-016-0619-y
144. Spooner R, Yilmaz Ö. Nucleoside-diphosphate-kinase: a pleiotropic effector in microbial colonization under interdisciplinary characterization. Microbes Infect (2012) 14(3):228–37. doi: 10.1016/j.micinf.2011.10.002
145. Bombaci G, Sarangdhar MA, Andina N, Tardivel A, Yu ECW, Mackie GM, et al. LRR-protein RNH1 dampens the inflammasome activation and is associated with COVID-19 severity. Life Sci Alliance (2022) 5(6):e202101226. doi: 10.26508/lsa.202101226
146. Zhang Z, Trypsteen W, Blaauw M, Chu X, Rutsaert S, Vandekerckhove L, et al. IRF7 and RNH1 are modifying factors of HIV-1 reservoirs: a genome-wide association analysis. BMC Med (2021) 19(1):282. doi: 10.1186/s12916-021-02156-5
147. Venugopal G, Bird JT, Washam CL, Roys H, Bowlin A, Byrum SD, et al. In vivo transcriptional analysis of mice infected with Leishmania major unveils cellular heterogeneity and altered transcriptomic profiling at single-cell resolution. PloS Negl Trop Dis (2022) 16(7):e0010518. doi: 10.1371/journal.pntd.0010518
148. Guan J, Han S, Wu J, Zhang Y, Bai M, Abdullah SW, et al. Ribosomal protein L13 participates in innate immune response induced by foot-and-mouth disease virus. Front Immunol (2021) 12:616402. doi: 10.3389/fimmu.2021.616402
149. Han S, Sun S, Li P, Liu Q, Zhang Z, Dong H, et al. Ribosomal protein L13 promotes IRES-driven translation of foot-and-mouth disease virus in a helicase DDX3-dependent manner. J Virol (2020) 94(2):e01679–19. doi: 10.1128/JVI.01679-19
150. Jiang J, Smith HN, Ren D, Dissanayak Mudiyanselage SD, Dawe AL, Wang L, et al. Potato spindle tuber viroid modulates its replication through a direct interaction with a splicing regulator. J Virol (2018) 92(20):e01004-18. doi: 10.1128/JVI.01004-18
151. Dissanayaka Mudiyanselage SD, Qu J, Tian N, Jiang J, Wang Y. Potato spindle tuber viroid RNA-templated transcription: factors and regulation. Viruses (2018) 10(9):E503. doi: 10.3390/v10090503
152. Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol Cell (2017) 67(1):71–83. doi: 10.1016/j.molcel.2017.05.021
153. Fichtenbauer D, Xu XM, Jackson D, Kragler F. The chaperonin CCT8 facilitates spread of tobamovirus infection. Plant Signal Behav (2012) 7(3):318–21. doi: 10.4161/psb.19152
154. Degrossoli A, Colhone MC, Arrais-Silva WW, Giorgio S. Hypoxia modulates expression of the 70-kD heat shock protein and reduces Leishmania infection in macrophages. J BioMed Sci (2004) 11(6):847–54. doi: 10.1159/000081832
155. Xu T, Lin Z, Wang C, Li Y, Xia Y, Zhao M, et al. Heat shock protein 70 as a supplementary receptor facilitates enterovirus 71 infections in vitro. Microb Pathog (2019) 128:106–11. doi: 10.1016/j.micpath.2018.12.032
156. Reyes-Del Valle J, Chávez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol (2005) 79(8):4557–67. doi: 10.1128/JVI.79.8.4557-4567.2005
157. Li G, Zhang J, Tong X, Liu W, Ye X. Heat shock protein 70 inhibits the activity of Influenza A virus ribonucleoprotein and blocks the replication of virus in vitro and in vivo. PloS One (2011) 6(2):e16546. doi: 10.1371/journal.pone.0016546
158. Zhang Z, Pan L, Ding Y, Lv J, Zhou P, Fang Y, et al. eEF1G interaction with foot-and-mouth disease virus nonstructural protein 2B: Identification by yeast two-hybrid system. Microb Pathog (2017) 112:111–6. doi: 10.1016/j.micpath.2017.09.039
159. Sammaibashi S, Yamayoshi S, Kawaoka Y. Strain-specific contribution of eukaryotic elongation factor 1 gamma to the translation of influenza A virus proteins. Front Microbiol (2018) 9:1446. doi: 10.3389/fmicb.2018.01446
160. Warren K, Wei T, Li D, Qin F, Warrilow D, Lin MH, et al. Eukaryotic elongation factor 1 complex subunits are critical HIV-1 reverse transcription cofactors. Proc Natl Acad Sci USA (2012) 109(24):9587–92. doi: 10.1073/pnas.1204673109
161. Liu XY, Li GQ, Ma Y, Zhao LJ. Topological centrality-based identification of hub genes and pathways associated with acute viral respiratory infection in infants. Genet Mol Res GMR (2015) 14(4):18334–43. doi: 10.4238/2015.December.23.21
162. Wei J, Kishton RJ, Angel M, Conn CS, Dalla-Venezia N, Marcel V, et al. Ribosomal proteins regulate MHC class I peptide generation for immunosurveillance. Mol Cell (2019) 73(6):1162–1173.e5. doi: 10.1016/j.molcel.2018.12.020
163. Rashidi S, Mansouri R, Ali-Hassanzadeh M, Ghani E, Barazesh A, Karimazar M, et al. Highlighting the interplay of microRNAs from Leishmania parasites and infected-host cells. Parasitology (2021) 148(12):1434–46. doi: 10.1017/S0031182021001177
164. Bayer-Santos E, Marini MM, da Silveira JF. Non-coding RNAs in host-pathogen interactions: subversion of mammalian cell functions by protozoan parasites. Front Microbiol (2017) 8:474. doi: 10.3389/fmicb.2017.00474
165. Afrin F, Khan I, Hemeg HA. Leishmania-host interactions-an epigenetic paradigm. Front Immunol (2019) 10:492. doi: 10.3389/fimmu.2019.00492
166. Niu J, Meeus I, De Coninck DI, Deforce D, Etebari K, Asgari S, et al. Infections of virulent and avirulent viruses differentially influenced the expression of dicer-1, ago-1, and microRNAs in Bombus terrestris. Sci Rep (2017) 7:45620. doi: 10.1038/srep45620
167. Winter F, Edaye S, Hüttenhofer A, Brunel C. Anopheles Gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res (2007) 35(20):6953–62. doi: 10.1093/nar/gkm686
168. Van Stry M, Oguin TH, Cheloufi S, Vogel P, Watanabe M, Pillai MR, et al. Enhanced susceptibility of Ago1/3 double-null mice to influenza A virus infection. J Virol (2012) 86(8):4151–7. doi: 10.1128/JVI.05303-11
169. Yamakawa N, Okuyama K, Ogata J, Kanai A, Helwak A, Takamatsu M, et al. Novel functional small RNAs are selectively loaded onto mammalian Ago1. Nucleic Acids Res (2014) 42(8):5289–301. doi: 10.1093/nar/gku137
170. Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA N Y N (2010) 16(1):43–56. doi: 10.1261/rna.1972910
171. Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell (2007) 130(2):287–97. doi: 10.1016/j.cell.2007.05.056
172. Jannot G, Boisvert MEL, Banville IH, Simard MJ. Two molecular features contribute to the Argonaute specificity for the microRNA and RNAi pathways in C. elegans. RNA (2008) 14(5):829–35. doi: 10.1261/rna.901908
173. Steiner FA, Hoogstrate SW, Okihara KL, Thijssen KL, Ketting RF, Plasterk RHA, et al. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol (2007) 14(10):927–33. doi: 10.1038/nsmb1308
174. Das L, Datta N, Bandyopadhyay S, Das PK. Successful therapy of lethal murine visceral leishmaniasis with cystatin involves up-regulation of nitric oxide and a favorable T cell response. J Immunol Baltim Md 1950 (2001) 166(6):4020–8. doi: 10.4049/jimmunol.166.6.4020
175. Smibert P, Yang JS, Azzam G, Liu JL, Lai EC. Homeostatic control of Argonaute stability by microRNA availability. Nat Struct Mol Biol (2013) 20(7):789–95. doi: 10.1038/nsmb.2606
176. Kobayashi H, Shoji K, Kiyokawa K, Negishi L, Tomari Y. Iruka eliminates dysfunctional argonaute by selective ubiquitination of its empty state. Mol Cell (2019) 73(1):119–29. doi: 10.1016/j.molcel.2018.10.033
177. Zeng J, Gupta VK, Jiang Y, Yang B, Gong L, Zhu H. Cross-kingdom small RNAs among animals, plants and microbes. Cells (2019) 8(4):E371. doi: 10.3390/cells8040371
Keywords: Leishmania, host-pathogen interactions, parasitic infection, RNAi, proteomics
Citation: Moradimotlagh A, Chen S, Koohbor S, Moon K-M, Foster LJ, Reiner N and Nandan D (2023) Leishmania infection upregulates and engages host macrophage Argonaute 1, and system-wide proteomics reveals Argonaute 1-dependent host response. Front. Immunol. 14:1287539. doi: 10.3389/fimmu.2023.1287539
Received: 01 September 2023; Accepted: 26 October 2023;
Published: 30 November 2023.
Edited by:
Abhay Satoskar, The Ohio State University, United StatesReviewed by:
Aislan Carvalho Vivarini, Federal University of Rio de Janeiro, BrazilLaila Gutierrez Kobeh, National Autonomous University of Mexico, Mexico
Copyright © 2023 Moradimotlagh, Chen, Koohbor, Moon, Foster, Reiner and Nandan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Devki Nandan, ZG5hbmRhbkBtYWlsLnViYy5jYQ==
†These authors have contributed equally to this work
 Atieh Moradimotlagh
Atieh Moradimotlagh Stella Chen
Stella Chen Sara Koohbor1
Sara Koohbor1 Kyung-Mee Moon
Kyung-Mee Moon Devki Nandan
Devki Nandan