- 1Institute for Virology, University Hospital Essen, University Duisburg-Essen, Essen, Germany
- 2Virology and Microbiology, Center for Biomedical Education and Research (ZBAF), Faculty of Health, Witten/Herdecke University, Witten, Germany
For the development of new adenovirus (AdV)-based vectors, it is important to understand differences in immunogenicity. In a side-by-side in vitro analysis, we evaluated the effect of 40 AdV types covering human AdV (HAdV) species A through G on the expression of 11 activation markers and the secretion of 12 cytokines by AdV-transduced dendritic cells, and the effect on CD8+ T cell proliferation capacity. We found that the expression of activation markers and cytokines differed widely between the different HAdV types, and many types were able to significantly impair the proliferation capacity of CD8+ T cells. Univariate and multivariate regression analyses suggested an important role of type I interferons in mediating this suppression of CD8+ T cells, which we confirmed experimentally in a proliferation assay using a type I interferon receptor blocking antibody. Using Bayesian statistics, we calculated a prediction model that suggests HAdV types HAdV-C1, -D8, -B7, -F41, -D33, -C2, -A31, -B3 and -D65 as the most favorable candidates for vaccine vector development.
Introduction
The virus family Adenoviridae comprises a large number of animal and human adenoviruses (AdV), with the International Committee on Taxonomy of Viruses listing 54 human AdV (HAdV) types in the genus Mastadenovirus and 64 non-human primate AdV types (1), with the non-human primate AdV types belonging to the subgenus Simian Mastadenovirus or to the subgenus Human Mastadenovirus due to their close phylogenetic relationship. The Human Adenovirus Working Group, on the other hand, even assigns more than 100 HAdV types (2). HAdV are grouped into seven different species A through G, and can result in mainly respiratory, gastrointestinal, ocular or genitourinary infections.
AdVs are popular vectors for exogenous gene delivery due to their ability to infect both dividing and non-dividing cells, ease of purification to high titers, ability to accommodate long exogenous genes, and their episomal, non-integrating nature [reviewed in (3)]. While later generations of AdV-based vectors have been developed that lack more or all of the AdV genes, the first-generation vectors which are lacking E1 and often also E3 are the most commonly used vector type in the vaccine setting. While the deletion of the E1 region results in replication deficiency and creates space for the transgene-encoding cassette, the deletion of E3 prevents the downregulation of the major histocompatibility complex I on transduced cells (4), resulting in improved immunogenicity and allowing for insertion of larger transgenes.
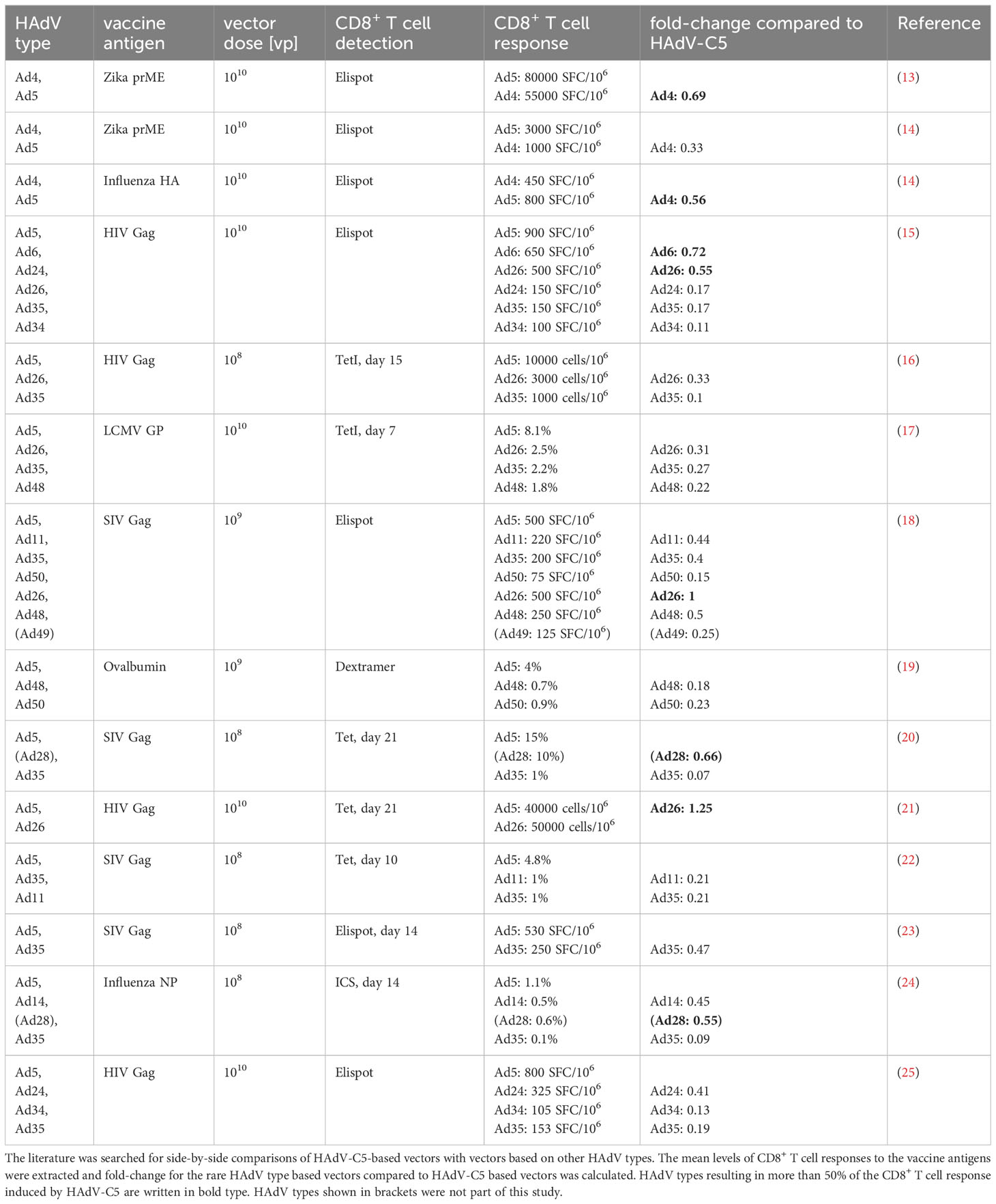
Originally, the species C HAdV types 2 and 5 were most often used for vector development (5–9), but their applicability had been questioned due to high levels of pre-existing immunity in many populations, especially in Africa and Asia (10, 11), and after high pre-existing immunity against HAdV-C5 was found to be linked to higher HIV-1 infection rates of male, non-circumcised vaccinees in the phase IIb STEP vaccine trial, the research focus shifted to so-called “rare” HAdV types with low seroprevalence for the development of new HAdV-based vectors. In addition to the seroprevalence, another important factor to consider when selecting AdV types for future vector development is their immunogenicity, which refers to the ability to trigger an immune response in the host, including the activation of innate immune mechanisms and of antigen presenting immune cells and the promotion of cellular immunity. AdV types with high immunogenicity can effectively activate the immune system and enhance the immune response to the antigens carried as transgenes by the vector, thus acting as adjuvants and improving the protective efficacy of the vaccine. HAdV types differ in their immunogenicity, resulting in differential activation of antigen-presenting cells and, consequently, of CD8+ T cells [reviewed in (12)]. A range of HAdV types other than HAdV-C5 have been evaluated as vectors for immunization, with some of them showing efficacy comparable to that of HAdV-C5 (Table 1). While many HAdV have been shown to be clearly inferior to HAdV-C5 with regard to their ability to induce transgene-specific CD8+ T cell responses, a few, most notably HAdV-D26, -D28, -C6 and -E4 showed good potential in immunization models. Subsequently, vectors based on HAdV-C5 and -D26 and the chimpanzee adenovirus Y25 based ChAdOx-1 vector have proven highly effective in the last few years in their widespread use for immunization against SARS-CoV-2 (26–29) and on a smaller scale against Ebola virus (30, 31). The very successful deployment of these vaccines has shown the great potential of AdV-based vectors.
The large-scale deployment also means, however, that a considerable part of the population now has some degree of immunity against these vector types. This vaccine-induced vector immunity may make these vector types less effective if applied again in a boost immunization or in the future as the vector for a different immunization. Thus, it is highly desirable to enlarge the repertoire of effective AdV-based vaccine vectors for future vaccine development and epidemic preparedness. Toward this goal, we have investigated in previous studies the seroprevalence of 39 HAdV types in a cohort of medical students (32) and in a cohort of patients with neuromuscular disease (33) and identified rare HAdV types with significantly lower seroprevalence compared to HAdV-C5. In the study presented here, we aimed to characterize the same broad range of 39 HAdV types with regard to their immunogenicity. Although many reports have constructed AdV-based vectors and evaluated their immunogenicity in vitro and in vivo, most studies have only focused on a few individual AdV types (Table 1), and there is currently a lack of such systematic studies on a wide range of AdV types. In the present study, we screened 40 AdV types from species A through G in vitro, including 39 HAdV types and the chimpanzee AdV Y25 derived vector ChAdOx-1, for their impact on the proliferation capacity of CD8+ T cells, as well as their influence on bone-marrow-derived dendritic cell (bmDC) activation. Ultimately, we identified the most promising types of HAdVs for future vector development.
Materials and methods
Viruses and viral vectors
In this study, a total of 39 types of HAdVs were employed, consisting of 34 wild-type HAdVs (HAdV-A12, -A18, -A31, -B3, -B7, -B11, -B14, -B21, -B34, -B35, -C1, -C2, -C5, -C6, -D8, -D9, -D10, -D13, -D17, -D20, -D24, -D25, -D26, -D27, -D33, -D37, -D70, -D73, -D74, -D75, -D80, -E4, -F41, and -G52) and five adenoviral vectors encoding eGFP and/or firefly luciferase (Ad16.GLN, Ad65.GLN, and Ad69.GLN (encoding eGFP and luciferase), Ad48.GFP and Ad50.Luc). Comprehensive details regarding the cultivation and purification of these HAdVs and HAdV-based vectors can be found in our previous study (32). In addition, the chimpanzee adenovirus-derived vector ChAdOx1-eGFP [kindly provided by Sarah Gilbert, University of Oxford (34)] was also included in this study.
For UV inactivation, AdV particles in 30 µl of PBS were irradiated with UV light (254 nm) for 30 minutes at 4°C.
Mice
C57BL/6 background human CD46-transgenic mice (B6-CD46tg) were obtained by backcrossing IFNAR-/-SLAM Ge CD46 Ge S1 mice (35) onto C57BL/6 background and selecting single transgenic mice, and were kindly provided by Matthias Tenbusch, University of Erlangen-Nürnberg, Gemany, with permission from Roberto Cattaneo, Mayo Clinic, Rochester, MN. T cell receptor-transgenic C57BL/6 mice express a CD8+ T cell receptor specific for the immunodominant Friend virus CD8+ T cell epitope GagL85-93 (36).
All mice were bred and maintained in the animal facility of the Institute for Virology at the University Hospital Essen. All procedures were carried out in compliance with national regulations and followed the institutional guidelines of the University Hospital Essen, Essen, Germany.
Isolation and culture of bmDCs
Bone marrow-derived dendritic cells (bmDCs) were cultured from the bone marrow of B6-CD46tg mice. The femurs of the mice were flushed to obtain the bone marrow, which was subsequently cultured in R10 medium (RPMI-1640 medium supplemented with 10% heat-inactivated FCS, 50 μg/ml gentamicin, 20 μg/ml ciprofloxacin, 2 mM L-glutamine, 50 μM β-mercaptoethanol, 10 mM HEPES and 1 mM sodium pyruvate) plus 20 ng/ml GM-CSF and 1.25 ng/ml IL-4, with an addition of an equal volume of fresh medium on day 4. The bone marrow cells were maintained at 37°C, 5% CO2 and 95% humidity, and loosely adherent cells were harvested on the seventh day as bmDCs for further experiments (37).
In vitro bmDC stimulation assay
The bmDC stimulation assay was performed in 96-well plates, wherein the bmDCs were transduced with various types of HAdV at a multiplicity of infection (MOI) of 1000. In addition, a negative control devoid of any stimulant and a positive control stimulated with 10 or 100 μg/ml LPS, were included. bmDCs were fixed with 2% PFA at 24 h post-transduction, and the cells were stained with anti-CD11c-BUV496 (clone HL3, BD Biosciences, Heidelberg, Germany), anti-MHC-I-AF647 (clone KH95, BioLegend, Fell, Germany), anti-MHC-II-PE/Dazzle594 (clone M5/115.14.2, BioLegend), anti-CD40-AF488 (clone HM40-3, BioLegend), anti-CD80-PE-Cy5 (clone 16-10A1, BioLegend), anti-CD86-BV510 (clone GL-1, BioLegend), anti-4-1BBL-PE (clone TKS-1, BioLegend), anti-CD252-PE-Cy7 (clone RM134L, BioLegend), anti-CD54-BUV737 (clone 3E2, BD Biosciences), anti-CD83-BV650 (clone Michel-19, BioLegend), anti-PD-L1-PerCP (clone MIH5, BD Biosciences), anti-PD-L2-BV421 (clone TY25, BioLegend) and Fixable Viability Dye eFluor 780 (eBioscience, Frankfurt, Germany). Data were acquired on a BD FACSymphony A5 flow cytometer (BD Biosciences) and analyzed using FlowJo software (TreeStar, Ashland, OR).
In vitro proliferation assay
Friend virus GagL85-93-specific CD8+ T cells were isolated from spleen cells of the T cell receptor transgenic mice by magnetic cell sorting using CD8 microbeads (Miltenyi, Bergisch-Gladbach, Germany). Subsequently, the isolated T cells were labeled with 1.5 μM carboxyfluorescein succinimidyl ester (CFSE) to facilitate tracking of their proliferation. The bmDCs were loaded with the peptide GagL85-93 and simultaneously transduced with HAdVs at a multiplicity of infection (MOI) of 1000 and then co-cultured with CFSE-labelled CD8+ T cells at an initial ratio of 1:2.5 for 3 days. CFSE intensity was analysed using flow cytometry after staining CD8+ T cells with BV421-anti-CD8 antibody (clone 53-6.7, BioLegend). Untreated bmDCs were used in the negative control coculture, while bmDCs loaded exclusively with the peptide without undergoing transduction by HAdVs were employed in the positive control stimulation to provide a baseline for comparison.
Additional proliferation assays were conducted in a transwell system, where HAdV-transduced bmDCs were placed in the upper transwell chamber, while GagL85-93-loaded bmDCs, as well as CD8+ T cells, were placed in the lower chamber. Following a three-day co-incubation period, the CD8+ T cells were subjected to staining and analysis, as described above.
Data were acquired on a BD FACSymphony A5 flow cytometer (BD Biosciences) and analyzed using FlowJo software (TreeStar, Ashland, OR). The division index of individual samples was calculated in FlowJo software as the total number of cell divisions divided by the total number of cells at the start of the culture of that individual sample as calculated from the division peaks.
Bead-based multiplex cytokine assay
For cytokine analysis, bmDCs were transduced with different HAdVs or left unstimulated (negative control) or stimulated with 10 or 100 μg/ml LPS (positive controls). The culture supernatants were harvested and subjected to analysis of mouse cytokines using the bead-based LEGENDplex multi-analyte flow assay kit (BioLegend), which encompassed a range of cytokines including interferons (IFN-α, -β, -γ), interleukins (IL-1β, -6, -10, -12), chemokines (CCL2, CCL5, CXCL1), TNF-α, and GM-CSF. Experiments were performed following the manufacturer’s instructions. Data were acquired by flow cytometry on a BD FACSymphony A5 flow cytometer (BD Biosciences) and analyzed by Legendplex V8.0 software (BioLegend).
Statistical analysis
Data were analyzed for statistically significant differences using GraphPad Prism software version 8, applying a one-way analysis of variance (ANOVA) for the comparison among multiple groups or an unpaired t test for pairwise comparisons. Univariate regression analysis was performed by Pearson correlation analysis in GraphPad Prism software. Multivariate regression analysis was performed by random forest analysis in R software using the packages randomForest and randomForestExplainer. Bayesian regression analysis was performed in R software using the package rstanarm using the mean fold-change of the division index of CD8+ T cells in the in vitro proliferation assay, the mean fold-change of bmDC surface marker and the mean fold-change of cytokine secretion levels, and the classification of HAdV-C5, -C6, -D26, -E4 and ChAdOx as “favorable vectors” and HAdV-B11, -B14, -B34, -B35, -B50, -D24 and -D48 as “unfavorable vectors” according to previously published data (Table 1). The results of the Bayesian regression analysis were visualized using the packages bayesplot, ggplot and ggforce.
Results
Reduction in CD8+ T cell proliferation after stimulation with AdV-transduced bmDCs
We observed in the past that vectors based on different HAdV types encoding the same transgene had strikingly different potential to induce transgene-specific CD8+ T cells in vivo and to stimulate the proliferation of transgene-specific CD8+ T cells in an in vitro proliferation assay (19). To inform future selection of HAdV types for the development of new HAdV-based vectors, we decided to screen a wide range of HAdV types in an in vitro proliferation assay and to characterize their influence on antigen-presenting cells.
To analyze the impact of different HAdV types on the proliferation capacity of transgene-specific CD8+ T cells, we used murine TCR transgenic CD8+ T cells that are specific for the Leader-Gag-derived epitope GagL85-93 of the Friend retrovirus, which is a model retrovirus infection that has been used by us and others in the past to study immune mechanisms and vaccine approaches targeting retrovirus infections [reviewed in (38)]. While the CD8+ T cells proliferated strongly in the in vitro proliferation assay when they were coincubated with bmDCs loaded with the GagL85-93 peptide alone, their proliferation activity varied greatly when the peptide-loaded bmDCs were transduced with different HAdV types (Figure 1A–C). While some HAdV types, such as HAdV-A31, -B50, -C2, -C5, -C6, -D37, -E4, -F41 and ‐G52, did not affect CD8+ T cell proliferation, many other HAdV types, notably almost all tested species D types but also HAdV-A18, some species B types and, to a moderate extent, HAdV-C1, led to significantly reduced CD8+ T cell proliferation. When we performed a complementary experiment with the most strongly inhibitory HAdV types, HAdV-D10, -D24 and -D80, we found that the inhibitory effect was abolished when the viruses were UV-inactivated (Figure 1D).
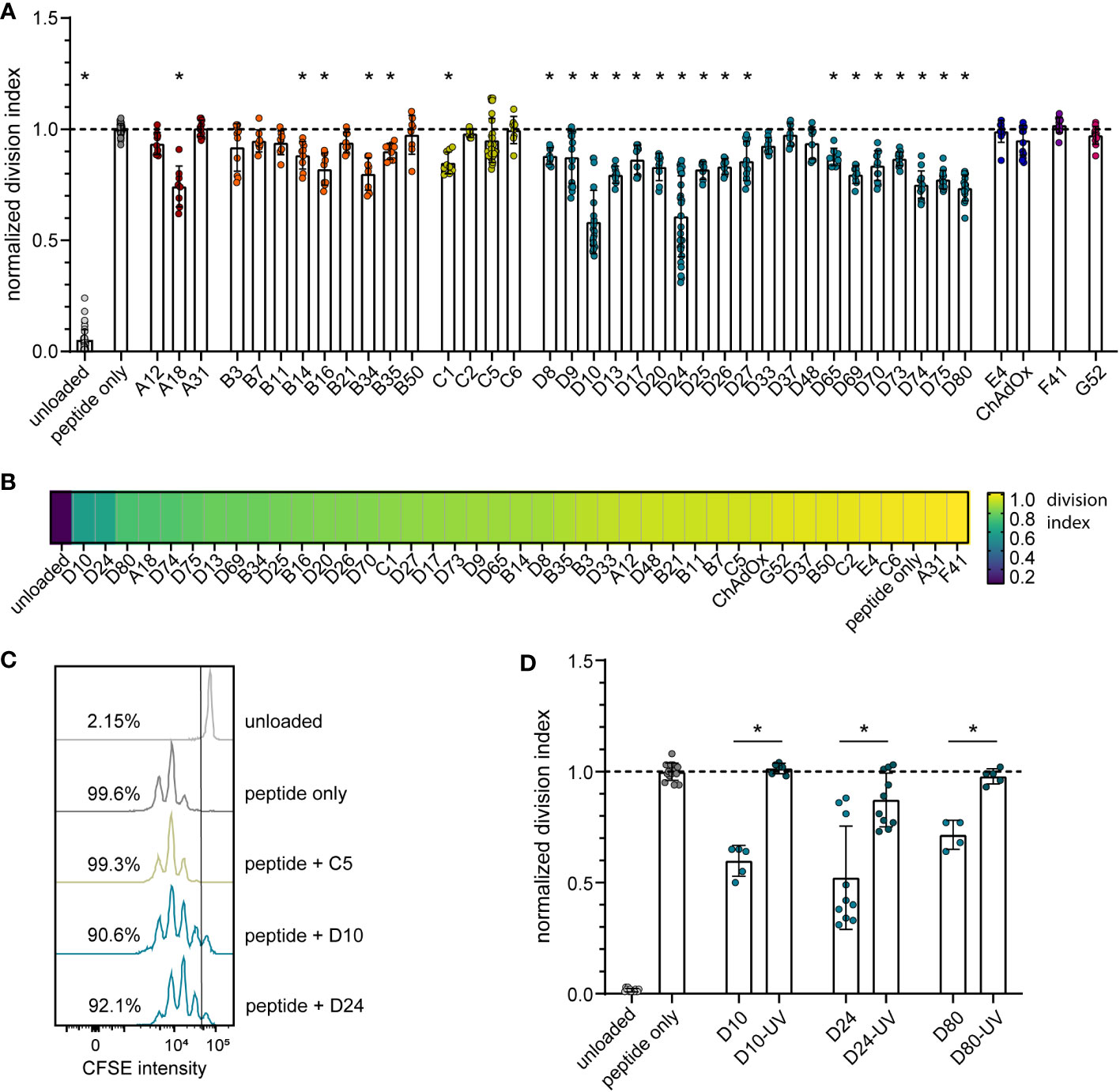
Figure 1 Reduction in CD8+ T cell proliferation after transduction of bmDCs with AdV. A CD8+ T cell proliferation assay was performed using FV GagL85-93 specific CD8+ T cells and bmDCs loaded with the GagL85-93 peptide with and without additional transduction with the indicated HAdV types or ChAdOx. (A) Proliferation of the CD8+ T cells was analyzed by dilution of CFSE staining intensity after 3 days of coculture, and the division index of the CD8+ T cells was calculated and normalized to the division index of the peptide-only stimulation. (B) A heatmap representation of the mean normalized division indices of CD8+ T cells stimulated with peptide and the indicated AdV types. (C) An overlay of representative histograms of different CD8+ T cell stimulations. (D) The influence on CD8+ T cell proliferation of bmDCs transduced with different untreated and UV-inactivated HAdV; the graph shows the division index normalized to the peptide-only control. Each dot indicates an individual sample, bars indicate mean values, and whiskers indicate the standard deviation. Data were acquired in at least two independent experiments (A, B, D). The dashed line indicates the normalized value of 1 (A, D), * indicates a statistically significant difference compared to the peptide-only control (A; P < 0.05, one-way ANOVA) or between the indicated groups (D; P < 0.05, unpaired t test).
Influence of AdV transduction on bmDC surface marker expression
To gain mechanistic insight into the differential effect of HAdV types on CD8+ T cell proliferation, we characterized the impact of HAdV transduction of bmDCs on the expression of stimulatory and inhibitory DC surface markers (Figure 2; Supplementary Figure S1) and on the secretion of cytokines by the bmDCs (Figure 3; Supplementary Figure S2). The bmDC surface molecules showed different patterns in response to transduction by different HAdV types. The surface expression of the co-stimulatory molecules 4‐1BBL, CD83 and CD86 was only slightly affected by most of the tested HAdV types and only significantly reduced by a few species B and D HAdV types (4-1BBL: HAdV-B34, ‐D33, ‐D37, -D48; CD86: HAdV-A18, -D17, -D20, -D69, -D74, -D75; Figure 2A). The expression of the co-stimulatory molecule CD40 was increased by a majority of the tested HAdV types, whereas the expression of CD54 and CD80 was increased to a lesser extent by some HAdV types and decreased by others, and the expression of CD252, on the other hand, was decreased by all tested HAdV types. The expression of the peptide-presenting molecules MHC-I (Figures 2A, B) and MHC-II was differentially and in many cases opposingly influenced by the different HAdV types, with a significant upregulation of MHC-I observed only for HAdV-B16, -B34, -B35, -D20, -D26, -D33, -D37 and -D65. The expression of the inhibitory molecule PD-L1 (Figure 2C) was significantly upregulated by most of the tested HAdV types, with the strongest induction (> 2.5-fold) observed for HAdV-B16, -B21, -B35, -D17, -D25, -D27, -D65, -D69 and -D74. Expression of the inhibitory molecule PD-L2, on the other hand, was not as strongly affected and was significantly upregulated only by HAdV-B7, -B34, -C1, and -D26, whereas the expression showed a strong trend toward a decrease after transduction with other HAdV types such as HAdV-A18, -B21, -B50, -D17 and -D74. Notably, the single animal-derived AdV type, ChAdOx, had limited impact on any of the markers, similar to HAdV types A31, C6, F41, and G52. In addition, comparisons at the species level showed that HAdVs of species B and D had a greater impact on the expression of bmDC surface molecules, while species F and G HAdVs exerted a relatively mediocre effect.
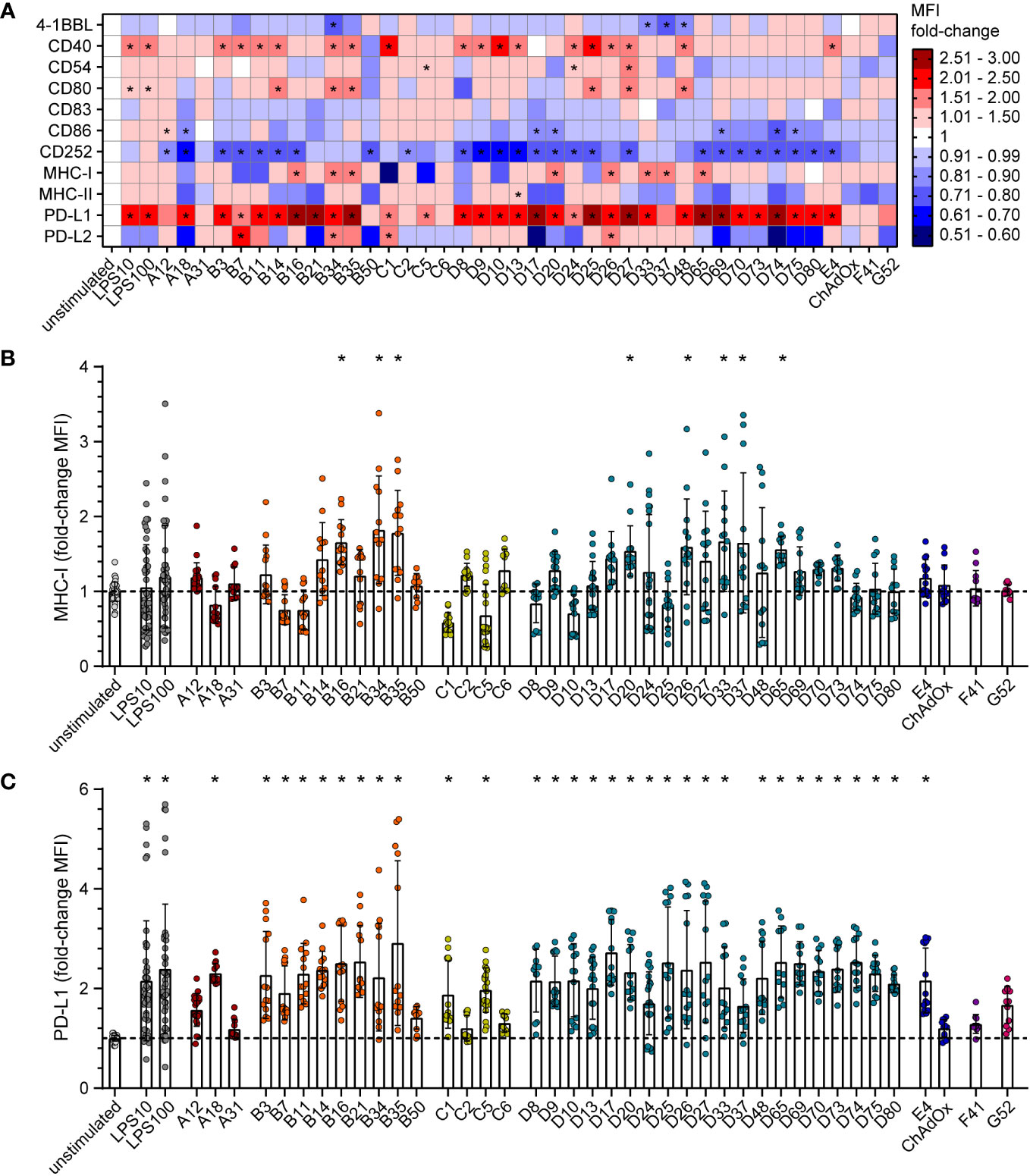
Figure 2 Differential expression of bmDC surface markers after transduction with AdV. bmDCs were transduced with the indicated AdV types or stimulated with 10 µg/ml or 100 µg/ml LPS. After 24 hours, the expression of surface molecules was analyzed by flow cytometry. The median fluorescence intensity (MFI) of the surface markers was determined, and fold-changes compared to the unstimulated bmDCs were calculated. (A) A heatmap representation of the mean fold-change of the MFI of the indicated surface markers after stimulation with the indicated AdV types. (B, C) Dot plot representations of the MFI fold-change for MHC-I (B) and PD-L1 (C). Each dot indicates an individual sample (B, C), bars indicate mean values, and whiskers indicate the standard deviation. Data were acquired in at least two independent experiments (A–C). The dashed line indicates the normalized value of 1 (B, C), * indicates a statistically significant difference compared to the unstimulated control (P < 0.05, one-way ANOVA).
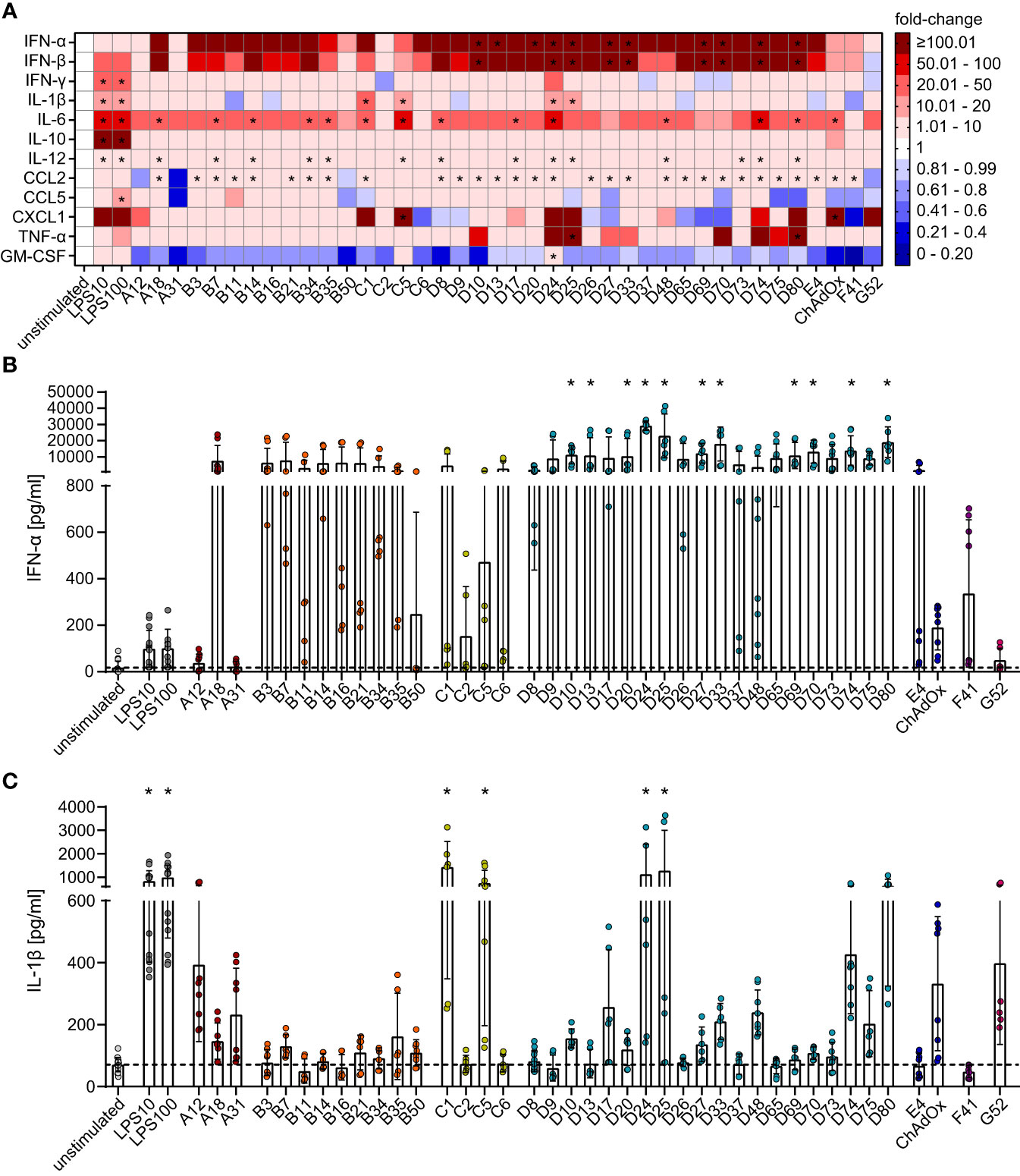
Figure 3 Differential secretion of cytokines by bmDCs after transduction with AdV. bmDCs were transduced with the indicated AdV types or stimulated with 10 µg/ml or 100 µg/ml LPS. After 24 hours, the supernatants were collected and analyzed for the presence of the indicated cytokines. The cytokine concentrations were determined and fold-changes compared to the unstimulated bmDCs were calculated. (A) A heatmap representation of the mean fold-change of the concentrations of the indicated cytokines after stimulation with the indicated AdV types. (B, C) Dot plot representations of the concentrations of IFN-α (B) and IL-1β (C) in the supernatant of bmDCs after the indicated stimulations. Each dot indicates an individual sample (B, C), bars indicate mean values, and whiskers indicate the standard deviation. Data were acquired in at least two independent experiments (A–C). The dashed line indicates the mean concentration of the indicated cytokine in the supernatant of unstimulated bmDCs (B, C), * indicates a statistically significant difference compared to the unstimulated control (P < 0.05, one-way ANOVA).
Influence of AdV transduction on bmDC cytokine secretion
The effect of bmDC HAdV transduction on cytokine production revealed a more homogenous picture for most of the analytes (Figure 3A). Compared to the unstimulated control, HAdV transduction resulted in a substantial increase in the concentrations of most cytokines, with the most pronounced effects observed for IFN-α (Figures 3A, B) and IFN-β (Figure 3A), which showed a clear trend toward strong induction by all HAdV types and statistically significant increases by most species D HAdV types. Concentrations of IL-6, IL-12, and CCL2 were also significantly upregulated, but to a lesser extent than IFN-α and IFN-β, and did not differ distinctly across HAdV species. On the other hand, the secretion of IL-1β (Figures 3A, C), CXCL-1, and TNF-α (Figure 3A) was not strongly affected by most HAdVs, and only a few specific HAdV types induced significant increases in their secretion (IL-1β: HAdV-C1, -C5, -D24 and -D25; CXCL1: HAdV-C5 and ChAdOx; TNF-α: HAdV-D25 and -D80), whereas there was a trend toward reduced secretion after transduction with other HAdV types. It is worth noting that the secretion of several cytokines by bmDCs, including IFN-γ, IL-10, and CCL5, was not significantly altered upon transduction with any type of HAdV tested in this study. Interestingly, the secretion of GM-CSF tended to be reduced under the influence of all tested AdV types except for HAdV-C5 and ‐D24.
Correlation of CD8+ T cell proliferation with bmDC surface marker expression and cytokine secretion
In a regression analysis, we analyzed the correlation of the impact of the different HAdV types on bmDC surface marker expression and cytokine secretion levels with the impact on CD8+ T cell proliferation capacity. In the analysis of the bmDC surface markers (Figure 4), we found a significant and strong positive correlation of CD8+ T cell proliferation with the expression levels of the co-stimulatory molecules CD86 (Figure 4F) and CD252 (Figure 4J) and a negative correlation with the expression level of the inhibitory molecule PD-L1 (Figure 4C). We also found significant negative correlations of CD8+ T cell proliferation with the secretion of INF-β, IFN-α, IL-12, CCL2, IFN-γ, GM-CSF, TNF-α and IL-6 (in descending order, Figure 5). To further investigate the contribution of the individual parameters, we performed a multivariate regression analysis by performing a random forest analysis. This analysis confirmed the results of the univariate analyses and predicted the level of IFN-β as the most important variable for the outcome of the CD8+ T cell proliferation rate, followed by IFN-α, CD86 and TNF-α (Figure 6).
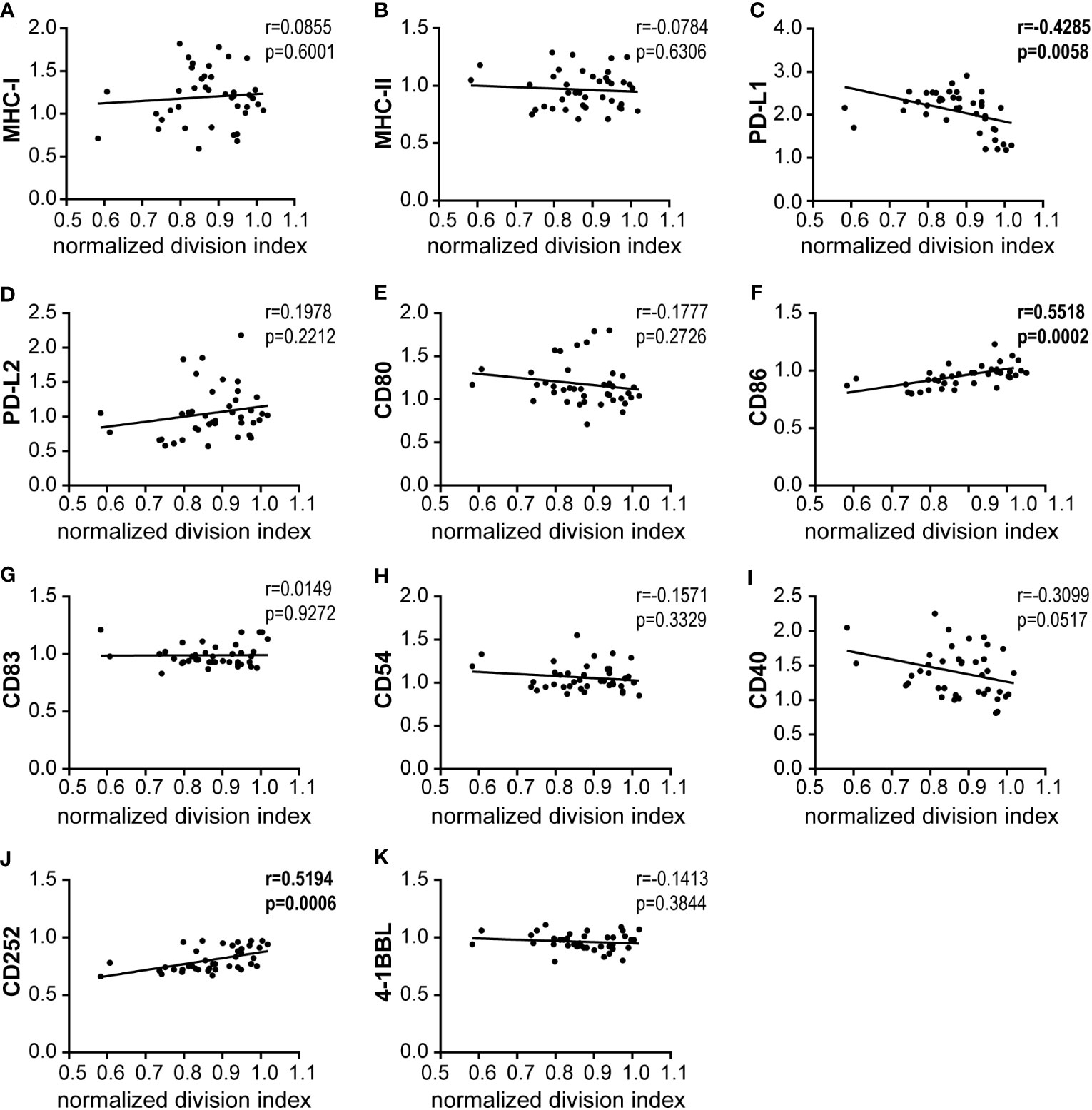
Figure 4 Correlation analysis of bmDC surface markers and CD8+ T cell proliferation. (A–K) Pairwise Pearson correlation analyses were performed using the mean CD8+ T cell division indices as shown in Figure 1 and the mean fold-change of MFI values for the indicated bmDC surface molecules as shown in Figure 2. r, Pearson correlation coefficient; statistically significant correlations are indicated by bold type (P < 0.05).
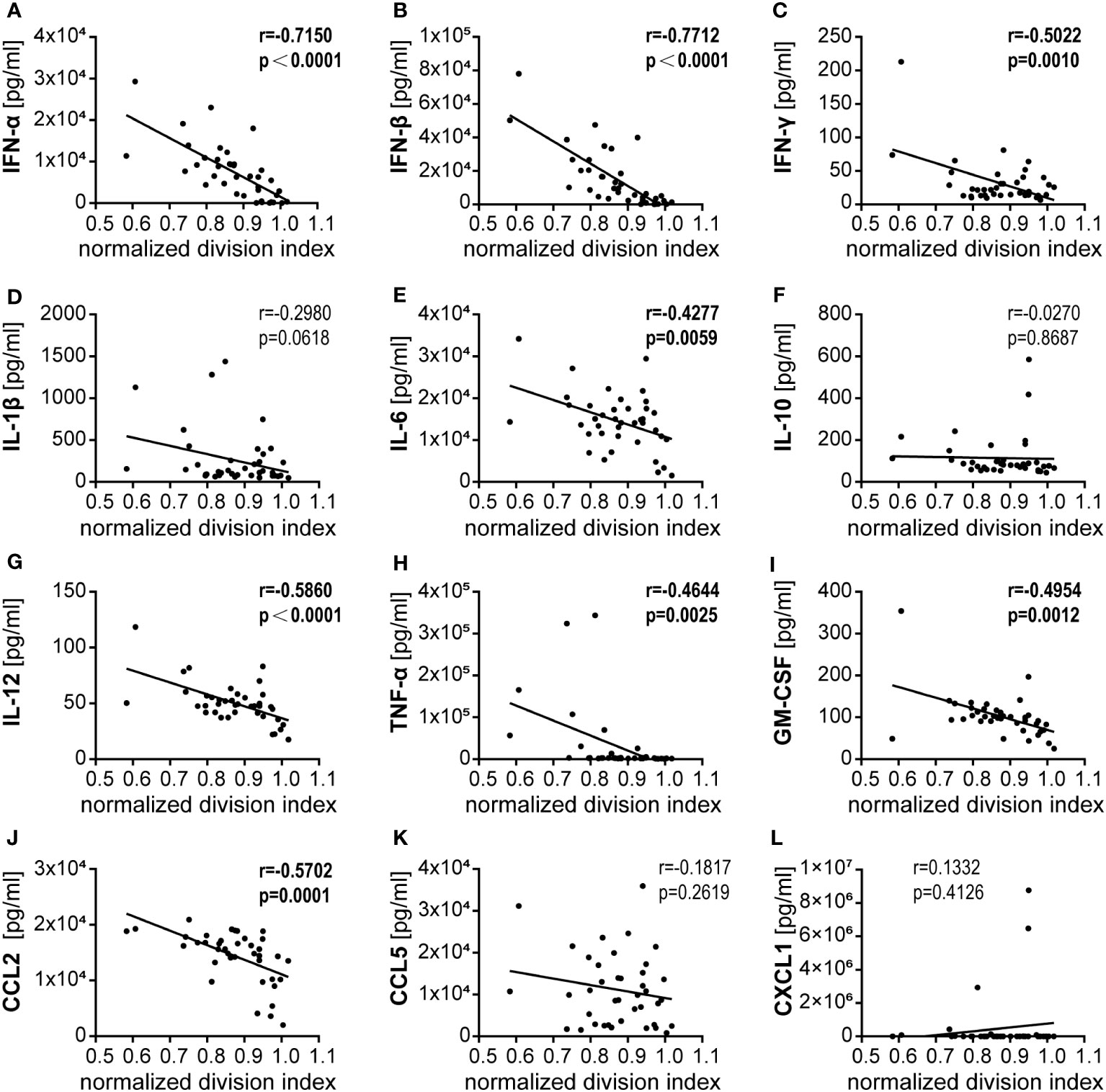
Figure 5 Correlation analysis of bmDC cytokine secretion and CD8+ T cell proliferation. (A–L) Pairwise Pearson correlation analyses were performed using the mean CD8+ T cell division indices as shown in Figure 1 and the mean concentrations of cytokines secreted by bmDCs as shown in Figure 3. r, Pearson correlation coefficient; statistically significant correlations are indicated by bold type (P < 0.05).
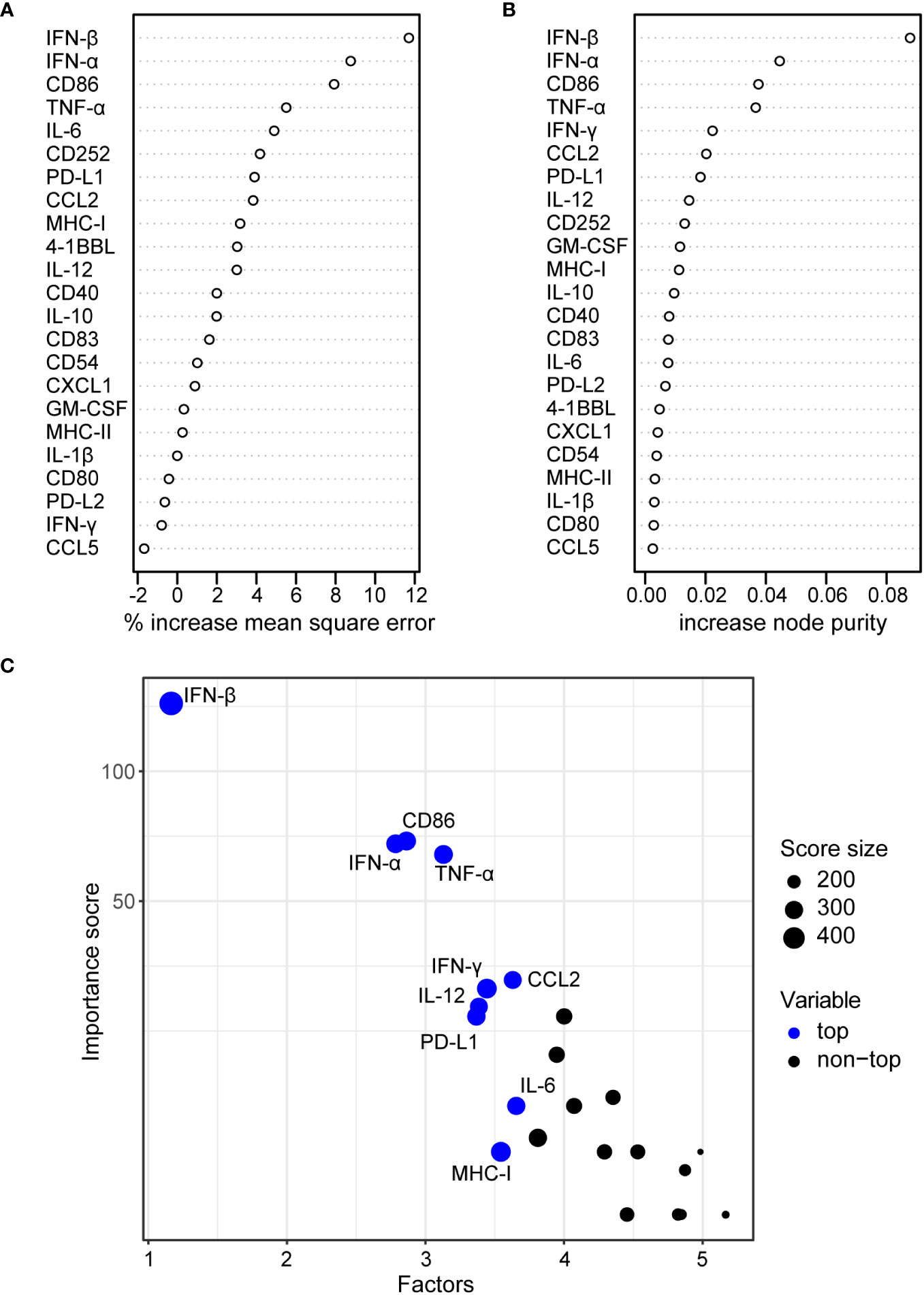
Figure 6 Random forest regression analysis of bmDC cytokine secretion and surface marker expression and CD8+ T cell proliferation. A random forest analysis was performed using the mean values for the CD8+ T cell division index as shown in Figure 1, and the bmDC surface marker expression measured by their MFI as shown in Figure 2 and the bmDC cytokine secretion as shown in Figure 3 as explanatory factors. Percent increase in mean square error (A) and increase in node purity (B) for the 23 tested explanatory factors are shown. The importance plot in (C) highlights the variables with the highest importance scores.
Inhibition of CD8+ T cell proliferation is mediated by high levels of type I interferons
To address these findings experimentally, we performed a CD8+ T cell proliferation assay in a transwell system, where the HAdV-transduced bmDCs were spatially separated from the peptide-loaded bmDCs and the CD8+ T cells. Here, we found that the inhibition of CD8+ T cell proliferation that we observed before when bmDCs were transduced with HAdV-D10 was not changed in the transwell system (Figure 7A), confirming that soluble factors mediated the suppressive effect. As the regression analyses ranked IFN-β and IFN-α most highly, we next performed a CD8+ T cell proliferation assay in the presence of an interferon α/β receptor (IFNAR) blocking antibody (Figure 7B). We selected six HAdV types for which we had observed strong induction of type I interferons (Figure 3), four of which had shown the strongest suppression of CD8+ T cell proliferation (Figure 1). In the control cultures without the addition of IFNAR-blocking antibody, we observed robust suppression of CD8+ T cell proliferation as before, which was completely abrogated in the presence of the IFNAR-blocking antibody, confirming the mechanistic role of the induction of type I interferon signaling in the suppression of CD8+ T cell proliferation by HAdV in this setting.
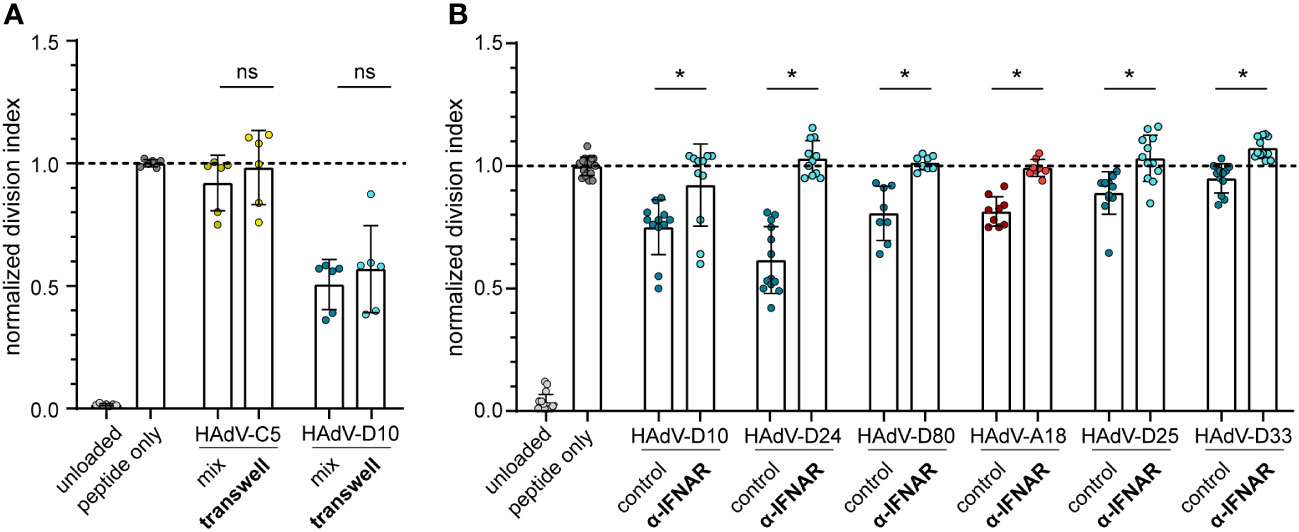
Figure 7 CD8+ T cell proliferation in transwell plates and in the presence of type I IFN receptor blocking antibodies. (A) A CD8+ T cell proliferation assay was performed using FV GagL85-93 specific CD8+ T cells and bmDCs loaded with GagL85-93 peptide, with HAdV-transduced bmDCs either in the same well (mix) or separated from the CD8+ T cells and peptide-loaded bmDCs in a transwell chamber (transwell). Proliferation of the CD8+ T cells was analyzed by dilution of CFSE staining intensity after 3 days of coculture, and the division index of the CD8+ T cells was calculated and normalized to the division index of the peptide-only stimulation. (B) A CD8+ T cell proliferation assay was performed using FV GagL85-93 specific CD8+ T cells and bmDCs loaded with GagL85-93 peptide and transduced with the indicated HAdV in the absence (control) or presence of an α-IFNAR antibody (α-IFNAR). Proliferation of the CD8+ T cells was analyzed by dilution of CFSE staining intensity after 3 days of coculture, and the division index of the CD8+ T cells was calculated and normalized to the division index of the peptide-only stimulation. Each dot indicates an individual sample, bars indicate mean values, and whiskers indicate the standard deviation. Data were acquired in at least two independent experiments. The dashed line indicates the normalized value of 1, * indicates a statistically significant difference between the indicated groups (P < 0.05, unpaired t test), ns, not significant.
Prediction of favorable HAdV types for vector development
For the selection of strong candidates for future HAdV-based vector development, it was of great interest to combine all observations into a statistical model to estimate the potential of the tested HAdV types. We therefore performed a Bayes regression analysis using the data of HAdV-B11, -B14, -B34, -B35, -B50, -C5, -C6, -D24, -D26, -D48 and-E4, which have already been evaluated side-by-side with HAdV-C5 for in vivo vaccination efficacy, and of ChAdOx, which has shown its high efficacy as a SARS-CoV-2 vaccine vector, for the calculation of the posteriors. We classified HAdV-C5, -C6, -D26, -E4 and ChAdOx as “favorable vectors” and HAdV-B11, -B14, -B34, -B35, -B50, -D24 and -D48 as “unfavorable vectors” based on the previously published efficacy data, as summarized in Table 1. It is noteworthy that in this prediction of good vector candidates, the individual factors are weighted quite differently than in the random forest analysis of the contribution to the repression of CD8+ T cell proliferation, and the highest posterior estimates were obtained for the division index of CD8+ T cells in the in vitro proliferation assay and the expression levels of the DC surface markers MHC-II, CD83, CD86 and CD252, but the secretion levels of some cytokines also tended to have positive posterior estimates (Figure 8A). Applying the obtained model to predict promising vector candidates, the HAdV types HAdV-C1, -D8, -B7, -F41, -D33, -C2, -A31, -B3 and -D65 were assigned prediction values higher than 0.5 (in descending order; Figure 8B).
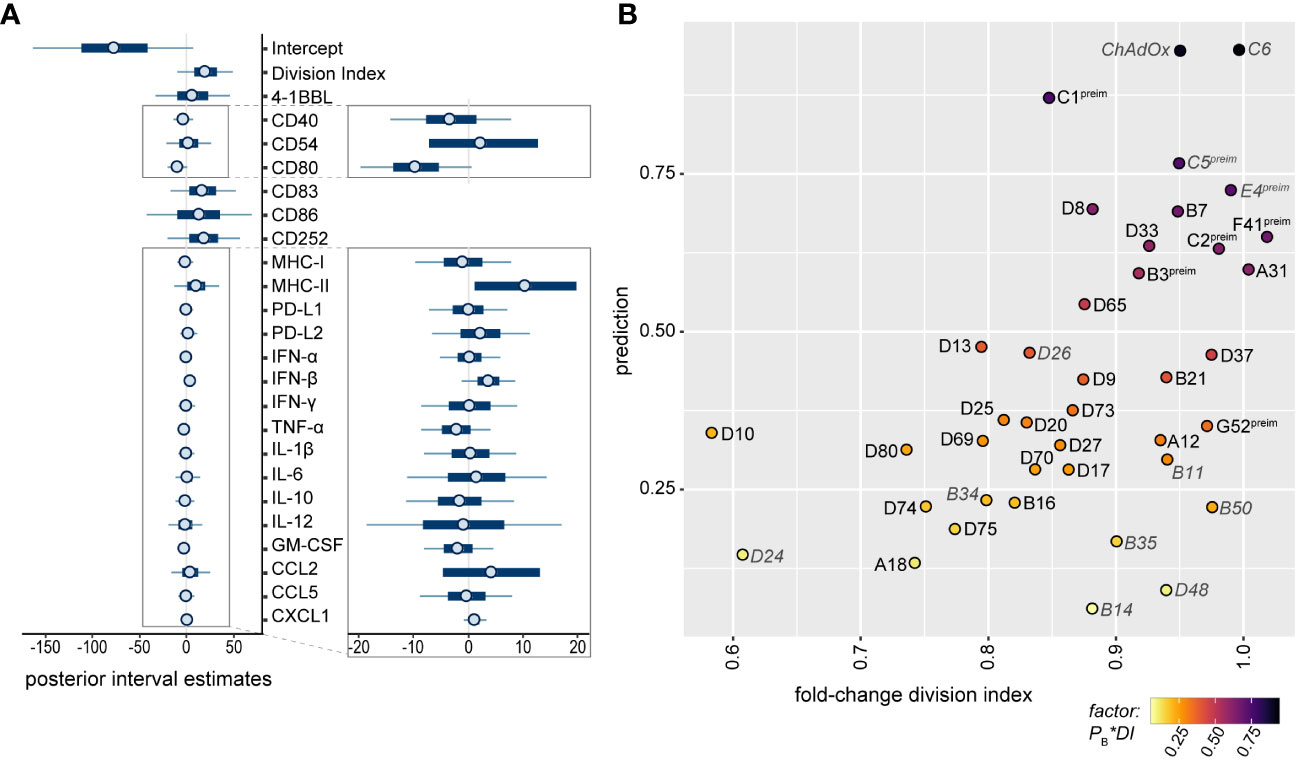
Figure 8 Bayesian regression analysis-based prediction model for the selection of HAdV types for vector development. A Bayesian regression analysis was performed using the mean fold-change of the division index of CD8+ T cells in the in vitro proliferation assay, the mean fold-change of all tested bmDC surface markers and the mean fold-change of the cytokine secretion levels as factors, and the classification of HAdV-C5, -C6, -D26, -E4 and ChAdOx as “favorable vectors” and HAdV-B11, -B14, -B34, -B35, -B50, -D24 and -D48 as “unfavorable vectors” according to previously published data (Table 1) to train the model. Posterior interval estimates are shown in (A), and the right-hand figure shows a zoomed-in graph for improved readability. (B) Prediction values for the individual AdV types were plotted against the fold-change division index, and the color of the dots indicates the factor of the Bayes prediction value (PB) and the fold-change division index (DI). AdV types that were used to train the model are labelled in grey italics. preim: high pre-existing immunity has been shown previously (32, 33).
Discussion
AdV-based vectors have been explored during the last decades as gene delivery vectors for gene therapy, oncolytic therapy and immunization, and the last few years have shown the great potential of AdV-based immunization, since AdV-based vaccines were important assets in the effort to control the SARS-CoV-2 pandemic and to curb Ebola virus outbreaks in recent years (39). However, the vast diversity of different HAdV types has not yet been explored for vector development, and systematic comparisons of the immunogenicity of a large number of viruses are currently lacking. In this detailed in vitro analysis, we analyzed the effect of 40 AdV types on DC activation and cytokine secretion and on CD8+ T cell proliferation capacity and identified HAdV-C1, -D8, -B7, -F41, -D33, -C2, -A31, -B3 and -D65 as having the most favorable immunogenicity profile for vaccine vector development.
In our in vitro analysis, we found that the capacity to induce type I interferons correlated with the potential to suppress the proliferative capacity of CD8+ T cells. It is recognized that AdV-based vectors are able to stimulate multiple innate immune pathways, whereby they can have an adjuvant effect on the immune response towards the transgene. Multiple innate immune factors influencing the immunogenicity of HAdV types have been described including Toll-like receptors and cGAS/STING [for comprehensive reviews, see (40) and (41)]. Differences in the activation of innate immune mechanisms have been attributed to several factors: in a first step, receptor and integrin binding result in activation of proteins involved in intracellular signaling pathways such as mitogen activated protein kinase by binding to CAR and phosphoinositide 3-kinase by integrin binding, leading to production of pro-inflammatory cytokines such as IL-1α, IL-6, IL-8 and TNF-α (42–45). Differences in receptor usage will result in differential activation of intracellular signaling [reviewed in (46)]. After cell entry, AdV particles have to escape the endosome, which the different AdV types do at different stages along the endosomal-lysosomal axis, resulting in membrane damage and differential release or exposure of endosomal components that induce activation of the NLRP3 inflammasome and of authophagy (47, 48). Finally, differences in the DNA sequence will influence the detection of viral DNA by TLR9 and cGAS/STING and the activation of the subsequent signaling, which results in activation of type I interferon expression (49–51).
Interestingly, it has been shown before that while a moderate ability to stimulate the innate immune system is advantageous for vector potency, excessive stimulation of type I interferons by chimpanzee AdV-based vectors in comparison to HAdV-C5-based vectors resulted in reduced transgene-specific antibody responses (52) and CD8+ T cell responses (53), which seemed to correlate with reduced transgene expression in the presence of type I interferons. Furthermore, it was shown that vectors based on HAdV-D28 and HAdV-B35 induced stronger IFN-α in vitro and in vivo, which resulted in a reduction in transgene expression, a reduced induction of transgene-specific CD8+ T cells but also a qualitatively changed CD8+ T cell response, with a more pronounced polyfunctional and memory phenotype (54), which the authors attributed to both autocrine and paracrine effects of the elevated IFN-α levels. Taken together, these findings support our in vitro data and suggest that the levels of type I interferons play an equally important role in the in vivo setting.
Perreau et al. suggested that the number of TLR9 agonist motifs in the HAdV genome correlates with immunogenicity, as they observed a positive correlation between TLR9 agonist motif genome content and the induction of CD86 expression and TNF-α secretion by DCs transduced with HAdV immune complexes (55). Our data also show a positive correlation of the ratio of stimulatory and inhibitory TLR9 motif genome content and the expression level of the DC surface marker CD86 and a negative correlation of the TLR9 motif ratio with the expression level of PD-L1 and the secretion of cytokines, which was most pronounced for type I interferons, CCL2, GM-CSF and IL-12 (Supplementary Figure 3). It can still be observed that HAdV types with very similar, low ratios of stimulatory and inhibitory TLR9 motifs induce quite different levels of cytokines, which is in agreement with the finding that multiple innate sensing pathways are involved in HAdV sensing and the induced innate immune response (40), as discussed above.
The model that we developed for the prediction of favorable vector candidates attributes the highest importance to the division index of CD8+ T cells in the in vitro proliferation assay and the expression levels of the DC surface markers MHC-II, CD83, CD86 and CD252, as well as the secretion levels of some cytokines. Data from in vivo studies where different HAdV-based vectors were compared side-by-side demonstrated widely varying potency, and limited induction of cellular immunity was observed for the HAdV types HAdV-B11, -B14, -B34, -B35, -B50, -D24 and -D48 [(15–20, 22–25); Table 1]. Interestingly, not all of them showed a strong impact on the CD8+ T cell proliferation capacity in our in vitro assay, highlighting the complex immune mechanisms underlying the induction of effective immune responses and the value of analyzing a wide range of immune parameters for the prediction of favorable HAdV types for vector development.
According to our Bayes statistic model, the HAdV types HAdV-C1, -D8, -B7, -F41, -D33, -C2, -A31, -B3 and -D65 are the most promising vector candidates. It has to be taken into account, however, that some of these HAdV types showed rather high seroprevalence in our previous studies: neutralizing antibody levels against HAdV-B3, -C1, -C2, and -F41 were similar to those against HAdV-C5, or even higher, in both cohorts (32, 33), precluding their widespread use.
Because of the widespread application of AdV-based vectors for SARS-CoV-2 immunization, it has been noted that in very rare cases, immunization would result in the severe adverse event of vaccine-induced thrombosis with thrombocytopenia syndrome (56, 57), which has been attributed to the induction of autoimmune antibodies directed against platelet factor 4 (PF4), leading to the activation of platelets and granulocytes (58–60). Complex formation of ChAdOx, HAdV-C5 and HAdV-D26 particles with PF4 could be demonstrated experimentally and was found to be in the nanomolar affinity range (61). Importantly, it was recently shown that different HAdV types have distinct affinities for PF4 binding, and HAdV-B11, -B34, -D13 and -D25 exhibited no or very low detectable PF4 binding (62). While the prediction based on our Bayesian statistic modelling is slightly below 0.5 for HAdV-D13, its favorable PF4-non-binding profile may make this an HAdV vector candidate not to be discarded quite yet.
One limitation of our study is that all experiments were performed using murine cells, but there are studies in which murine and human DCs were used side-by-side for AdV infection experiments that support the conclusion that similar results would be obtained using human cells: Johnson et al. performed infection experiments with murine bmDCs and human plasmacytoid DCs and observed the same differences in IFN-α induction by HAdV-C5, -D28 and -B35 in both cell types (54). In a study using only HAdV-C5 based vectors, Strack et al. also showed comparable effects of HAdV-C5 on murine bmDCs and human peripheral blood derived DCs (63). Quinn et al. have performed extensive analyses of innate immune responses using both human peripheral blood mononuclear cells (PBMCs) infected in vitro as well as mouse cells from in vivo vaccination, and compared the immune response against HAdV-C5 to that against, among others, HAdV-B35, and -D28 (53). They showed that while there was some discordance in up- and down-regulated genes in murine lymph nodes and human PBMCs, expression patterns in murine and human PBMCs were highly comparable. Importantly, they also showed in the in vivo model that immunization with HAdV-B35 and -D28 based vectors led to a significantly stronger expression of IFN-α in draining lymph nodes and higher IFN-α levels in the serum compared to HAdV-C5, which also confirms our in vitro data. Teigler et al. evaluated the innate immune responses against HAdV-C5, -B35, -D26 and -D48 in rhesus monkeys and in human PBMCs, and reported a strong induction of IFN-α2 in human PBMCs for HAdV‐B35, ‐D26 and -D48 and much less for HAdV-C5 (64), which is in accordance with our findings in murine DCs. Interestingly, Tran et al. have shown in in vitro experiments that HAdV-C5 infected human DCs and bystander DCs produced significantly higher levels of a range of cytokines if the HAdV-C5 was complexed with IgG (65), suggesting that cytokine responses in vivo may be higher in the presence of pre-existing immunity. Chen et al. analyzed cytokine levels in sera of HAdV-B3 and -B7 infected hospitalized children, and made similar observations of increased levels of CCL-2, IL-6, IL-10, IFN-α and -γ and TNF- α in the infected children in comparison to healthy controls (66). While the authors observed less pronounced IFN-α levels than could be expected from the in vitro studies, this may be due to the time point of serum collection. In accordance with the above-mentioned similarities, it has been noted in various reviews (67, 68) that human and mouse DCs share many similarities and that they behave very similarly if they have been prepared, cultured and activated in the same way. Finally, it should be noted that those AdV types that had been most effective in various mouse models have proven their high efficacy in recent years when they were applied as SARS-CoV-2 vaccines, lending validity to results obtained in mouse experiments. Overall, we conclude that the results obtained in our in vitro assays with murine cells are meaningful and valid also for possible translation into human use in the future, and that vectors derived from the identified favorable HAdV types can be expected to also have favorable in vivo immunogenicity.
Our data provide important insight into the different immunogenicity profiles of 40 AdV types. Taken together with our previously reported data on seroprevalence, we suggest that HAdV-D8, -B7, -D33, -A31 and -D65 are the most favorable HAdV types to be explored as vaccine vectors and evaluated in vivo in animal models in the future.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The animal study was approved by Internal Review Board of the Animal Facility of the University Hospital Essen. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XW: Data curation, Formal Analysis, Funding acquisition, Investigation, Visualization, Writing – original draft, Writing – review & editing. MH: Data curation, Formal Analysis, Investigation, Methodology, Writing – review & editing. WZ: Investigation, Methodology, Resources, Writing – review & editing. AE: Conceptualization, Investigation, Resources, Writing – review & editing. WB: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a stipend from the China Scholarship Council to XW and by a grant from the Deutsche Forschungsgemeinschaft to WB (BA 4432/1-4). We acknowledge support from the Open Access Publication Fund of the University of Duisburg-Essen.
Acknowledgments
The authors would like to thank Dr. Sarah Gilbert for kindly providing the ChAdOx-1 vector for this comparative study.
Conflict of interest
WZ and AE are mentioned as co-inventors on a patent with application number 554 PCT/EP2017/058306 relevant for the vectors Ad16.GLN, Ad65.GLN, and Ad69.GLN.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1286622/full#supplementary-material
References
1. Benko M, Aoki K, Arnberg N, Davison AJ, Echavarria M, Hess M, et al. Ictv virus taxonomy profile: adenoviridae 2022. J Gen Virol (2022) 103(3):001721. doi: 10.1099/jgv.0.001721
2. Seto D, Chodosh J, Brister JR, Jones MS, Members of the adenovirus research Community. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol (2011) 85(11):5701–2. doi: 10.1128/JVI.00354-11
3. Sallard E, Zhang W, Aydin M, Schröer K, Ehrhardt A. The adenovirus vector platform: novel insights into rational vector design and lessons learned from the COVID-19 vaccine. Viruses (2023) 15(1):204. doi: 10.3390/v15010204
4. Wold WSM, Tollefson AE, Hermiston TW. E3 transcription unit of adenovirus. The molecular repertoire of adenoviruses I. Curr Topics 199/I Microbiol Immunol (1995) . p:237–74. doi: 10.1007/978-3-642-79496-4_13
5. Saito I, Oya Y, Yamamoto K, Yuasa T, Shimojo H. Construction of nondefective adenovirus type 5 bearing a 2.8-kilobase hepatitis B virus DNA near the right end of its genome. J Virol (1985) 54(3):711–9. doi: 10.1128/JVI.54.3.711-719.1985
6. Yamada M, Lewis JA, Grodzicker T. Overproduction of the protein product of a nonselected foreign gene carried by an adenovirus vector. Proc Natl Acad Sci U.S.A. (1985) 82(11):3567–71. doi: 10.1073/pnas.82.11.3567
7. Haj-Ahmad Y, Graham FL. Development of a helper-independent human adenovirus vector and its use in the transfer of the herpes simplex virus thymidine kinase gene. J Virol (1986) 57(1):267–74. doi: 10.1128/JVI.57.1.267-274.1986
8. Massie B, Gluzman Y, Hassell JA. Construction of a helper-free recombinant adenovirus that expresses polyomavirus large T antigen. Mol Cell Biol (1986) 6(8):2872–83. doi: 10.1128/mcb.6.8.2872-2883.1986
9. Saito I, Oya Y, Shimojo H. Novel RNA family structure of hepatitis B virus expressed in human cells, using a helper-free adenovirus vector. J Virol (1986) 58(2):554–60. doi: 10.1128/JVI.58.2.554-560.1986
10. Mast TC, Kierstead L, Gupta SB, Nikas AA, Kallas EG, Novitsky V, et al. International epidemiology of human pre-existing adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies: correlates of high Ad5 titers and implications for potential HIV vaccine trials. Vaccine (2010) 28(4):950–7. doi: 10.1016/j.vaccine.2009.10.145
11. Vogels R, Zuijdgeest D, van Rijnsoever R, Hartkoorn E, Damen I, de Béthune M-P, et al. Replication-Deficient Human Adenovirus Type 35 Vectors for Gene Transfer and Vaccination: Efficient Human Cell Infection and Bypass of Preexisting Adenovirus Immunity. J Virol (2003) 77(15):8263–71. doi: 10.1128/jvi.77.15.8263-8271.2003
12. Marquez-Martinez S, Vijayan A, Khan S, Zahn R. Cell entry and innate sensing shape adaptive immune responses to adenovirus-based vaccines. Curr Opin Immunol (2023) 80:102282. doi: 10.1016/j.coi.2023.102282
13. Bullard BL, Corder BN, Gorman MJ, Diamond MS, Weaver EA. Efficacy of a T cell-biased adenovirus vector as a Zika virus vaccine. Sci Rep (2018) 8(1):18017. doi: 10.1038/s41598-018-35755-z
14. Bullard BL, Corder BN, Gordon DN, Pierson TC, Weaver EA. Characterization of a species E adenovirus vector as a Zika virus vaccine. Sci Rep (2020) 10(1):3613. doi: 10.1038/s41598-020-60238-5
15. Colloca S, Barnes E, Folgori A, Ammendola V, Capone S, Cirillo A, et al. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Trans Med (2012) 4:115ra2. doi: 10.1126/scitranslmed.3002925
16. Tan WG, Jin H-T, West EE, Penaloza-MacMaster P, Wieland A, Zilliox MJ, et al. Comparative analysis of simian immunodeficiency virus Gag-specific effector and memory CD8+ T cells induced by different adenovirus vectors. J Virol (2013) 87(3):1359–72. doi: 10.1128/jvi.02055-12
17. Penaloza-MacMaster P, Provine NM, Ra J, Borducchi EN, McNally A, Simmons NL, et al. Alternative serotype adenovirus vaccine vectors elicit memory T cells with enhanced anamnestic capacity compared to Ad5 vectors. J Virol (2013) 87(3):1373–84. doi: 10.1128/jvi.02058-12
18. Abbink P, Lemckert AAC, Ewald BA, Lynch DM, Denholtz M, Smits S, et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol (2007) 81(9):4654–63. doi: 10.1128/jvi.02696-06
19. Hrycak CP, Windmann S, Bayer W. Comparative evaluation of the vaccine efficacies of three adenovirus-based vector types in the Friend retrovirus infection model. J Virol (2019) 93(21):e01155-19. doi: 10.1128/jvi.01155-19
20. Quinn KM, Da Costa A, Yamamoto A, Berry D, Lindsay RWB, Darrah PA, et al. Comparative analysis of the magnitude, quality, phenotype, and protective capacity of simian immunodeficiency virus gag-specific CD8+ T cells following human-, simian-, and chimpanzee-derived recombinant adenoviral vector immunization. J Immunol (2013) 190(6):2720–35. doi: 10.4049/jimmunol.1202861
21. Chen H, Xiang ZQ, Li Y, Kurupati RK, Jia B, Bian A, et al. Adenovirus-based vaccines: comparison of vectors from three species of adenoviridae. J Virol (2010) 84(20):10522–32. doi: 10.1128/jvi.00450-10
22. Lemckert AAC, Sumida SM, Holterman L, Vogels R, Truitt DM, Lynch DM, et al. Immunogenicity of heterologous prime-boost regimens involving recombinant adenovirus serotype 11 (Ad11) and Ad35 vaccine vectors in the presence of anti-Ad5 immunity. J Virol (2005) 79(15):9694–701. doi: 10.1128/jvi.79.15.9694-9701.2005
23. Barouch DH, Pau MG, Custers JHHV, Koudstaal W, Kostense S, Havenga MJE, et al. Immunogenicity of recombinant adenovirus serotype 35 vaccine in the presence of pre-existing anti-Ad5 immunity. J Immunol (2004) 172(10):6290–7. doi: 10.4049/jimmunol.172.10.6290
24. Kahl CA, Bonnell J, Hiriyanna S, Fultz M, Nyberg-Hoffman C, Chen P, et al. Potent immune responses and in vitro pro-inflammatory cytokine suppression by a novel adenovirus vaccine vector based on rare human serotype 28. Vaccine (2010) 28(35):5691–702. doi: 10.1016/j.vaccine.2010.06.050
25. Shiver JW, Emini EA. Recent advances in the development of HIV-1 vaccines using replication-incompetent adenovirus vectors. Annu Rev Med (2004) 55(1):355–72. doi: 10.1146/annurev.med.55.091902.104344
26. Sadoff J, Gray G, Vandebosch A, Cardenas V, Shukarev G, Grinsztejn B, et al. Safety and efficacy of single-dose Ad26.Cov2.S vaccine against COVID-19. N Engl J Med (2021) 384(23):2187–201. doi: 10.1056/NEJMoa2101544
27. Zhu FC, Guan XH, Li YH, Huang JY, Jiang T, Hou LH, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (2020) 396(10249):479–88. doi: 10.1016/S0140-6736(20)31605-6
28. Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet (2021) 397(10275):671–81. doi: 10.1016/s0140-6736(21)00234-8
29. Ramasamy MN, Minassian AM, Ewer KJ, Flaxman AL, Folegatti PM, Owens DR, et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (Cov002): A single-blind, randomised, controlled, phase 2/3 trial. Lancet (2020) 396(10267):1979–93. doi: 10.1016/s0140-6736(20)32466-1
30. Milligan ID, Gibani MM, Sewell R, Clutterbuck EA, Campbell D, Plested E, et al. Safety and immunogenicity of novel adenovirus type 26- and modified vaccinia ankara-vectored Ebola vaccines: A randomized clinical trial. JAMA (2016) 315(15):1610–23. doi: 10.1001/jama.2016.4218
31. Zhu FC, Wurie AH, Hou LH, Liang Q, Li YH, Russell JB, et al. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: A single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (2017) 389(10069):621–8. doi: 10.1016/S0140-6736(16)32617-4
32. Wang X, Kerkmann L, Hetzel M, Windmann S, Trilling M, Zhang W, et al. Analysis of the prevalence of binding and neutralizing antibodies against 39 human adenovirus types in student cohorts reveals low-prevalence types and a decline in binding antibody levels during the Sars-Cov-2 pandemic. J Virol (2022) 96(22):e0113322. doi: 10.1128/jvi.01133-22
33. Klann PJ, Wang X, Elfert A, Zhang W, Köhler C, Güttsches A-K, et al. Seroprevalence of binding and neutralizing antibodies against 39 human adenovirus types in patients with neuromuscular disorders. Viruses (2022) 15(1):79. doi: 10.3390/v15010079
34. Dicks MDJ, Spencer AJ, Edwards NJ, Wadell G, Bojang K, Gilbert SC, et al. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PloS One (2012) 7(7):e40385. doi: 10.1371/journal.pone.0040385
35. Mrkic B, Pavlovic J, Rülicke T, Volpe P, Buchholz CJ, Hourcade D, et al. Measles virus spread and pathogenesis in genetically modified mice. J Virol (1998) 72(9):7420–7. doi: 10.1128/jvi.72.9.7420-7427.1998
36. Öhlén C, Kalos M, Cheng LE, Shur AC, Hong DJ, Carson BD, et al. CD8+ T cell tolerance to a tumor-associated antigen is maintained at the level of expansion rather than effector function. J Exp Med (2002) 195(11):1407–18. doi: 10.1084/jem.20011063
37. Balkow S, Krux F, Loser K, Becker JU, Grabbe S, Dittmer U. Friend retrovirus infection of myeloid dendritic cells impairs maturation, prolongs contact to naïve T cells, and favors expansion of regulatory T cells. Blood (2007) 110(12):3949–58. doi: 10.1182/blood-2007-05-092189
38. Dittmer U, Sutter K, Kassiotis G, Zelinskyy G, Bánki Z, Stoiber H, et al. Friend retrovirus studies reveal complex interactions between intrinsic, innate and adaptive immunity. FEMS Microbiol Rev (2019) 43(5):435–56. doi: 10.1093/femsre/fuz012
39. Jacob-Dolan C, Barouch DH. COVID-19 vaccines: adenoviral vectors. Annu Rev Med (2022) 73(1):41–54. doi: 10.1146/annurev-med-012621-102252
40. Coughlan L. Factors which contribute to the immunogenicity of non-replicating adenoviral vectored vaccines. Front Immunol (2020) 11:909. doi: 10.3389/fimmu.2020.00909
41. Daussy CF, Pied N, Wodrich H. Understanding post entry sorting of adenovirus capsids; a chance to change vaccine vector properties. Viruses (2021) 13(7):1221. doi: 10.3390/v13071221
42. Di Paolo NC, Miao EA, Iwakura Y, Murali-Krishna K, Aderem A, Flavell RA, et al. Virus binding to a plasma membrane receptor triggers interleukin-1α-mediated proinflammatory macrophage response in vivo. Immunity (2009) 31(1):110–21. doi: 10.1016/j.immuni.2009.04.015
43. Farmer C, Morton PE, Snippe M, Santis G, Parsons M. Coxsackie adenovirus receptor (Car) regulates integrin function through activation of p44/42 MAPK. Exp Cell Res (2009) 315(15):2637–47. doi: 10.1016/j.yexcr.2009.06.008
44. Philpott NJ, Nociari M, Elkon KB, Falck-Pedersen E. Adenovirus-induced maturation of dendritic cells through a Pi3 kinase-mediated Tnf-alpha induction pathway. Proc Natl Acad Sci U.S.A. (2004) 101(16):6200–5. doi: 10.1073/pnas.0308368101
45. Tamanini A, Nicolis E, Bonizzato A, Bezzerri V, Melotti P, Assael BM, et al. Interaction of adenovirus type 5 fiber with the coxsackievirus and adenovirus receptor activates inflammatory response in human respiratory cells. J Virol (2006) 80(22):11241–54. doi: 10.1128/jvi.00721-06
46. Nestić D, Božinović K, Pehar I, Wallace R, Parker AL, Majhen D. The revolving door of adenovirus cell entry: not all pathways are equal. Pharmaceutics (2021) 13(10):1585. doi: 10.3390/pharmaceutics13101585
47. Barlan AU, Griffin TM, McGuire KA, Wiethoff CM. Adenovirus membrane penetration activates the Nlrp3 inflammasome. J Virol (2011) 85(1):146–55. doi: 10.1128/jvi.01265-10
48. Montespan C, Marvin SA, Austin S, Burrage AM, Roger B, et al. Multi-layered control of galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPXY motif in the viral capsid. PloS Pathog (2017) 13(2):e1006217. doi: 10.1371/journal.ppat.1006217
49. Fejer G, Drechsel L, Liese J, Schleicher U, Ruzsics Z, et al. Key role of splenic myeloid Dcs in the Ifn-Aβ Response to adenoviruses in vivo. PloS Pathog (2008) 4(11):e1000208. doi: 10.1371/journal.ppat.1000208
50. Stein SC, Falck-Pedersen E. Sensing adenovirus infection: activation of interferon regulatory factor 3 in Raw 264.7 cells. J Virol (2012) 86(8):4527–37. doi: 10.1128/jvi.07071-11
51. Zhu J, Huang X, Yang Y. Innate immune response to adenoviral vectors is mediated by both toll-like receptor-dependent and -independent pathways. J Virol (2007) 81(7):3170–80. doi: 10.1128/jvi.02192-06
52. Hensley SE, Cun AS, Giles-Davis W, Li Y, Xiang Z, Lasaro MO, et al. Type I interferon inhibits antibody responses induced by a chimpanzee adenovirus vector. Mol Ther (2007) 15(2):393–403. doi: 10.1038/sj.mt.6300024
53. Quinn KM, Zak DE, Costa A, Yamamoto A, Kastenmuller K, Hill BJ, et al. Antigen expression determines adenoviral vaccine potency independent of IFN and STING signaling. J Clin Invest (2015) 125(3):1129–46. doi: 10.1172/jci78280
54. Johnson MJ, Petrovas C, Yamamoto T, Lindsay RWB, Loré K, Gall JGD, et al. Type I IFN induced by adenovirus serotypes 28 and 35 has multiple effects on T cell immunogenicity. J Immunol (2012) 188(12):6109–18. doi: 10.4049/jimmunol.1103717
55. Perreau M, Welles HC, Pellaton C, Gjoksi B, Potin L, Martin R, et al. The number of toll-like receptor 9-agonist motifs in the adenovirus genome correlates with induction of dendritic cell maturation by adenovirus immune complexes. J Virol (2012) 86(11):6279–85. doi: 10.1128/jvi.00123-12
56. Greinacher A, Thiele T, Warkentin TE, Weisser K, Kyrle PA, Eichinger S. Thrombotic thrombocytopenia after ChAdOx1 nCoV-19 vaccination. New Engl J Med (2021) 384(22):2092–101. doi: 10.1056/NEJMoa2104840
57. Elberry MH, Abdelgawad HAH, Hamdallah A, Abdella WS, Ahmed AS, Ghaith HS, et al. A systematic review of vaccine-induced thrombotic thrombocytopenia in individuals who received COVID-19 adenoviral-vector-based vaccines. J Thromb Thrombolysis (2022) 53(4):798–823. doi: 10.1007/s11239-021-02626-w
58. Greinacher A, Selleng K, Palankar R, Wesche J, Handtke S, Wolff M, et al. Insights in Chadox1 nCoV-19 vaccine-induced immune thrombotic thrombocytopenia. Blood (2021) 138(22):2256–68. doi: 10.1182/blood.2021013231
59. Buoninfante A, Andeweg A, Baker AT, Borad M, Crawford N, Dogné J-M, et al. Understanding thrombosis with thrombocytopenia syndrome after COVID-19 vaccination. NPJ Vaccines (2022) 7(1):141. doi: 10.1038/s41541-022-00569-8
60. Gresele P, Momi S, Marcucci R, Ramundo F, De Stefano V, Tripodi A. Interactions of adenoviruses with platelets and coagulation and the vaccine-induced immune thrombotic thrombocytopenia syndrome. Haematologica (2021) 106(12):3034–45. doi: 10.3324/haematol.2021.279289
61. Baker AT, Boyd RJ, Sarkar D, Teijeira-Crespo A, Chan CK, Bates E, et al. ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome. Sci Adv (2021) 7(49):eabl8213. doi: 10.1126/sciadv.abl8213
62. Sallard E, Pembaur D, Schröer K, Schellhorn S, Koukou G, Schmidt N, et al. Adenovirus type 34 and HVR1-deleted adenovirus type 5 do not bind to PF4: clearing the path towards vectors without thrombosis risk. bioRxiv (2022). doi: 10.1101/2022.11.07.515483
63. Strack A, Deinzer A, Thirion C, Schrödel S, Dörrie J, Sauerer T, et al. Breaking entry-and species barriers: lentiboost® plus polybrene enhances transduction efficacy of dendritic cells and monocytes by adenovirus 5. Viruses (2022) 14(1):92. doi: 10.3390/v14010092
64. Teigler JE, Iampietro MJ, Barouch DH. Vaccination with adenovirus serotypes 35, 26, and 48 elicits higher levels of innate cytokine responses than adenovirus serotype 5 in rhesus monkeys. J Virol (2012) 86(18):9590–8. doi: 10.1128/jvi.00740-12
65. Tran TTP, Eichholz K, Amelio P, Moyer C, Nemerow GR, Perreau M, et al. Humoral immune response to adenovirus induce tolerogenic bystander dendritic cells that promote generation of regulatory T cells. PloS Pathog (2018) 14(8):e1007127. doi: 10.1371/journal.ppat.1007127
66. Chen Q, Liu J, Liang W, Chen Y, Dou M, Liu Z, et al. Clinical features, replication competence, and innate immune responses of human adenovirus type 7 infection. J Infect Dis (2021) 223(8):1390–9. doi: 10.1093/infdis/jiaa524
67. Shortman K, Liu Y-J. Mouse and human dendritic cell subtypes. Nat Rev Immunol (2002) 2(3):151–61. doi: 10.1038/nri746
Keywords: human adenovirus (HAdV), adenoviral vector, adenovirus-based immunization, CD8 T cell response, CD8+ T cell response, T cell response, immunogenicity
Citation: Wang X, Hetzel M, Zhang W, Ehrhardt A and Bayer W (2023) Comparative analysis of the impact of 40 adenovirus types on dendritic cell activation and CD8+ T cell proliferation capacity for the identification of favorable immunization vector candidates. Front. Immunol. 14:1286622. doi: 10.3389/fimmu.2023.1286622
Received: 31 August 2023; Accepted: 04 October 2023;
Published: 17 October 2023.
Edited by:
Jason Upton, Auburn University, United StatesReviewed by:
Akihiko Sakamoto, Kanazawa University, JapanDmitry M. Shayakhmetov, Emory University, United States
Copyright © 2023 Wang, Hetzel, Zhang, Ehrhardt and Bayer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wibke Bayer, d2lia2UuYmF5ZXJAdWstZXNzZW4uZGU=
 Xiaoyan Wang
Xiaoyan Wang Mario Hetzel1
Mario Hetzel1 Wibke Bayer
Wibke Bayer