
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 04 January 2024
Sec. Autoimmune and Autoinflammatory Disorders : Autoimmune Disorders
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1286387
This article is part of the Research Topic Challenges Associated with Identifying Preclinical Animal Models for the Development of Immune-based Therapies View all 17 articles
 Suhee Kim1†
Suhee Kim1† Sung Hak Chun1†
Sung Hak Chun1† Yun-Hong Cheon1
Yun-Hong Cheon1 Mingyo Kim1
Mingyo Kim1 Hyun-Ok Kim1,2
Hyun-Ok Kim1,2 Hanna Lee1,2
Hanna Lee1,2 Seong-Tshool Hong3
Seong-Tshool Hong3 Sang-Jun Park4
Sang-Jun Park4 Myeong Soo Park4
Myeong Soo Park4 Young Sun Suh1,2*
Young Sun Suh1,2* Sang-Il Lee1*
Sang-Il Lee1*Introduction: The intricate connection between gut microbiota and rheumatoid arthritis (RA) pathogenesis has gained prominence, although the specific microbial species contributing to RA development remain largely unknown. Recent studies have sought to comprehensively explore alterations in the human microbiome, focusing on identifying disease-related microbial species through blood analysis. Consequently, this study aimed to identify RA-associated microbial species using a serum microbial array system and to investigate the efficacy and underlying mechanisms of potential microbial species for RA treatment.
Methods: Serum immunoglobulin M levels against 384 intestinal microbial species were assessed using a microbial microarray in patients with RA and healthy individuals. We investigated the therapeutic potential of the identified microbial candidate regarding arthritis development, immune responses, gut barrier function, and gut microbiome using a collagen-induced arthritis (CIA) mouse model.
Results: Our findings revealed significant alterations in antibody levels against 36 microbial species in patients with RA compared to healthy individuals. Notably, the antibody levels against Peptoniphilus gorbachii (PG) were decreased in patients with RA and exhibited an inverse correlation with RA disease activity. In vitro experiments demonstrated that PG produced acetate and butyrate, while exhibiting anti-inflammatory properties. In CIA mice, PG administration suppressed arthritis symptoms, reduced the accumulation of inflammatory monocytes in the mesenteric lymph nodes, and downregulated gene expression of pro-inflammatory cytokines in the ileum. Additionally, PG supplementation restored intestinal barrier integrity and partially resolved gut microbial dysbiosis in CIA mice. The fecal microbiota in PG-treated mice corresponded to improved intestinal barrier integrity and reduced inflammatory responses.
Conclusion: This study highlights the potential of serum-based detection of anti-microbial antibodies to identify microbial targets at the species level for RA treatment. Moreover, our findings suggest that PG, identified through the microbial microarray analysis, holds therapeutic potential for RA by restoring intestinal barrier integrity and suppressing the immunologic response associated with RA.
Microbiota has emerged as a crucial factor in human health. Accumulating evidence demonstrates that dysbiosis, defined as imbalanced microbiota composition, is evident in patients with rheumatoid arthritis (RA) (1–5). A balanced microbiota and commensal microbe-derived metabolites are pivotal for maintaining symbiotic relationships within the intestine by reinforcing the epithelial barrier and regulating the mucosal immune response (6). However, microbial dysbiosis can compromise epithelial barrier function, resulting in an influx of bacteria and their metabolites into the bloodstream, leading to increased exposure of immune cells to bacterial antigens and subsequent pro-inflammatory responses (7). Furthermore, the enterotoxin zonulin, secreted by intestinal epithelial cells following microbial stimulation, reduces the expression of intestinal tight junction proteins. This, in turn, induces damage to the gut barrier, T cell-mediated mucosal inflammation, and the transmigration of immune cells from the gut to the joints (8). Therefore, restoring microbiota homeostasis and intestinal barrier function emerges as a promising strategy for preventing or treating RA (9).
Several microorganisms, including Porphyromonas gingivalis and Prevotella copri, have been implicated in RA pathogenesis. P. gingivalis triggers and exacerbates RA by activating Th17 immune responses and encoding peptidyl arginine deiminase, which facilitates the generation of RA-related autoantigen citrullinated peptides (10, 11). Prevotella copri has been discovered in both the preclinical stage and in new-onset stages of untreated RA patients and has been shown to contribute to the development of the disease (12, 13). Lactobacillus salivarius is consistently enriched in patients with RA, particularly those with highly active RA (14). Conversely, another Prevotella species, Prevotella histicola, suppresses arthritis via mucosal immune regulation (15). Additionally, Lactobacillus casei significantly suppresses the induction of arthritis, restores gut microbiota, and reduces oxidative stress (16, 17). These findings underscore that even within the same genus, distinct species can exert different effects on RA. Therefore, identifying RA-related microorganisms at the species level is pivotal for discovering effective targets for RA treatment.
While 16S rRNA gene sequencing and shotgun metagenomic sequencing of fecal samples are commonly used for identifying RA-associated microorganisms (18), they present limitations. Experimental conditions, such as sequencing errors and genomic repeats, can impact the outcome of these methods (19, 20). Additionally, the fecal microbiome only partially represents the entire gastrointestinal tract microbiota and fails to represent the mouth and lungs microbiota, both implicated in RA pathogenesis (21, 22). Furthermore, the translocation of transient gut microbiota could elicit persistent systemic responses, detectable through a serum microbial antibody array but frequently missed by metagenomic sequencing of fecal samples (23).
As a solution to sequencing limitations, serological testing emerges as a viable alternative for identifying disease-related microorganisms. Clinical practice often employs serological tests to identify causative microorganisms in pneumonia, scrub typhus, and syphilis. Similarly, serological tests have recently been conducted to investigate RA-related microbiota using blood samples. For example, antibodies against P. gingivalis were highly observed in patients with RA compared to healthy controls and associated with the presence of anti-cyclic citrullinated peptide antibodies (ACPA) (24–27). Elevated serum antibody levels against Prevotella copri were also observed in individuals at risk of or diagnosed with RA (28). Another recent study, using blood samples, demonstrated altered microbial small RNA composition in plasma of patients with RA compared to controls (29). Such blood-based investigations offer the advantage of reflecting changes in microorganisms across various organs, including the mouth, lungs, and intestine, with minimal sample requirements (30).
This study aimed to identify species-level microbial candidates associated with RA through a serum microbial antibody microarray and to demonstrate the therapeutic effects and underlying mechanisms of these candidate RA-related microbial species in collagen-induced arthritis (CIA) mice.
The study included 81 patients with RA and 50 healthy controls (HC) matched for age, sex, and race. Human serum samples were provided by the Gyeongsang National University Hospital (GNUH)-Korea Biobank, sampled in 2017 and 2018. This study was approved by the Institutional Review Board (permit No: GNUH 2017-08-015). RA disease activity was assessed using the Disease Activity Score in 28 joints (DAS28) with measurements of erythrocyte sedimentation rate (ESR) and C-Reactive Protein (CRP) (31, 32). Patient information is summarized in Supplementary Table 1.
Immunoglobulin M (IgM) antibody levels targeting specific intestinal microbial species were evaluated in serum samples (33, 34). Specifically, 384 species of intestinal microbes obtained from the Gut Microbiota Bank (https://www.gutmicrobiotabank.com) were grown in yeast casitone fatty acid (YCFA) medium (Supplementary Table 2) and homogenized using an automill disruptor (Cosmo Bio, Japan). Microbial lysates were arrayed onto a nylon membrane using a microarray spotter (CapitalBio, China) and fixed by air drying, followed by a subsequent incubation at 80°C for 30 min. The membrane was then blocked in Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat dry milk at 37 °C for 30 min. Patient serum was added to the chips, followed by a 30-min incubation at 37°C. Subsequently, the chips were incubated with Alexa Fluor 647-conjugated goat anti-human IgM secondary antibodies (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C for 30 min. The fluorescence intensity was quantified using a Luxscan (CapitalBio, China). Each experiment involved at least three technical replicates and the average intensities of each sample were normalized using global normalization and Z-transform methods. Microbial richness and evenness were assessed based on species richness and Shannon index (α-diversity of species). Principal coordinate analysis (PCoA) was performed to identify microbial compositional diversity (β-diversity of species). Linear discriminant analysis (LDA) effect size (LEfSe) was performed using the Galaxy framework (https://huttenhower.sph.harvard.edu/galaxy/) to identify bacterial taxa enriched in the serum of healthy individuals and patients with RA (p < 0.01, LDA score > 3.0). A heatmap of bacterial species with differential relative abundances was generated (p < 0.01, Z-ratio > 1.5). Furthermore, a co-occurrence network was constructed using the Cytoscape v.3.10.0 plugin CoNet (p < 0.05 adjusted using the Benjamini-Hochberg correction, correlation coefficient > 0.5 based on four correlation measures including Spearman, Pearson, Bray-Curtis, and Kullback-Leibler).
Peptoniphilus gorbachii (PG) strain WAL 10418 (JBP9-00201) and P. gingivalis (JBP11-00401) obtained from the Gut Microbiota Bank were cultured anaerobically at 37°C in YCFA medium. Live PG pellets and culture supernatants were prepared by centrifugation of bacterial culture at 12,000 × g for 10 min. Additionally, live PG pellets were suspended in 10% skim milk in YCFA and freeze-dried. Heat-killed (HK)-PG was generated by incubating the bacterial culture at 60°C for 1 h.
PG and P. gingivalis were grown separately in YCFA medium until reaching an OD600 of 0.1. They were then mixed at a 1:1 ratio. Pure cultures of PG and P. gingivalis were also prepared as controls to assess competition. Co-cultures and single cultures were serially diluted 10-fold eight times after 0, 8, and 16 h of culture. These dilutions were then spread onto agar plates and incubated at 37°C for 48 h. All single colonies visualized on the agar plates were identified by light microscopy following Gram-staining.
SCFAs in PG-cultured supernatants were quantified using gas chromatography/mass spectrometry (GC-2010 Plus, GCMS-TQ 8030, Shimazu, Tokyo, Japan).
RAW 264.7, a murine macrophage cell line, was maintained in Dulbecco’s modified eagle’s medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin (GIBCO, Carlsbad, CA, USA). RAW 264.7 cells were seeded in a 24-well plate (1 × 105 cells/well) and cultured for 24 h. Subsequently, cells were exposed to HK-PG (1 × 105 cells/well) or PG supernatant (0.5%) for 20 h, followed by stimulation with LPS (1 µg/mL; O55:B5, Sigma-Aldrich, St. Louis, MO, USA) for 4 h. Total RNA was extracted from the cells to evaluate pro-inflammatory gene expression.
Mouse small intestinal lamina propria (siLP) cells were isolated as previously described (35). Single-cell suspensions of mesenteric lymph nodes (MLN) were prepared through mechanical grinding and filtering using a nylon mesh. Single-cell suspensions from siLP and MLN were seeded into a 96-well plate (5 × 105 cells/well) and stimulated with monoclonal anti-CD3e (1 µg/mL; clone 145-2C11, BD Biosciences, San Jose, CA, USA) and anti-CD28 (1 µg/mL; clone 37.51, BD Biosciences) antibodies, with or without HK-PG (5 × 103 cells/well) or PG supernatant (0.5%) for 24 h. Cell culture supernatants were used for the measurement of IFN-γ and IL-17A levels.
Five-week-old male DBA1/J mice were purchased from Central Laboratory Animal Inc. (Seoul, Korea). All mice were acclimatized for two weeks under specific pathogen-free conditions and fed standard rodent chow and sterile water. Animal experiments were conducted in accordance with the guidelines of Gyeongsang National University (GNU) and approved by the Institutional Animal Care and Use Committee (IACUC approval ID: GNU-200724-M0046) in Korea.
CIA was induced as previously described (36). Briefly, mice were immunized intradermally at the base of the tail with 100 μg bovine type II collagen (CII, Chondrex, Redmond, WA, USA) emulsified with complete Freund’s adjuvant (CFA, Sigma-Aldrich) on day 0. Three weeks later (day 21), mice were boosted with CII emulsified with incomplete Freund’s adjuvant (IFA, Chondrex). Mice were divided into four groups according to the treatment scheme: (1) naïve mice without treatment and immunization (n = 4), (2) CII-immunized mice without treatment (CIA, n = 8), (3) PG-treated CIA mice (CIA/PG, n = 9), and (4) antibiotic-treated CIA mice (CIA/AB, n = 8). PG treatment was initiated four weeks before the first CII injection and continued for 9 weeks until the end of the experiment (day 35). A total of 1 × 109 colony-forming units (CFUs) of live PG suspended in 200 µL of PBS were orally administered to mice three times a week. For antibiotic treatment, ampicillin (1 g/L), vancomycin (0.5 g/L), neomycin (1 g/L), and metronidazole (1 g/L) were administered to mice in the drinking water for a total period of 3 weeks, beginning on day 14 after the first CII injection.
Clinical arthritis scores were evaluated on a scale of 0–4 for each limb three times a week, starting after the second CII injection (day 21), as previously described (36). Mice were sacrificed five weeks post-immunization (day 35) for sample collection.
Ankles were formalin-fixed, decalcified with a decalcifying solution (Sigma-Aldrich), and embedded in paraffin. Ankle joint sections (5 μm) were subjected to hematoxylin and eosin (H&E) and safranin O staining. Slides were imaged using a Nikon microscope imaging system and NIS-Elements software (Nikon Instruments, Tokyo, Japan). The assessment of synovial inflammation, bone erosion, and cartilage damage was conducted as previously described (36).
Freshly collected mouse fecal pellets were smashed in 1 mL of YCFA medium, followed by a 10-fold serial dilution six times. One hundred microliters of the serially diluted fecal suspension were spread onto YCFA agar medium, and the agar plates were incubated at 37°C for 16 h in anaerobic conditions. Following visualization of single colonies on the agar plate, approximately 100 single colonies were selected from the plates. The 16S rDNA sequences, encompassing regions V1-V9, were amplified using colony PCR and universal primers (27F 5’-AGAGTTTGATCCTGGCTCAG-3’ and 1492R 5’-GGTTACCTTGTTACGACTT-3’). These amplified DNA fragments were sequenced using Sanger sequencing. Full-length 16S rRNA sequences were analyzed in a DNAbaser4.36 analyzer, and contigs of each individual DNA were uploaded to NCBI BLAST for bacterial species identification.
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) and reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). DNA was subjected to qPCR amplification using a SYBR Green PCR master mix (Thermo Fisher Scientific) on a ViiA 7 Real-time PCR Detection System (Applied Biosystems, Waltham, MA, USA). The expression of target genes relative to glyceraldehyde 3-phosphate dehydrogenase was calculated using the 2-ΔΔCt comparative method. The primer pairs used are listed in Supplementary Table 3.
To measure IFN-γ and IL-17A levels in cell culture supernatants, 96-well plates (Greiner Bio-One GmbH, Frickenhausen, Germany) were coated with IFN-γ (eBioscience, San Diego, CA, USA) or IL-17A monoclonal antibodies (eBioscience) and blocked with BD OptEIA™ Assay diluent (BD Biosciences). Samples were added to the wells and incubated overnight at 4°C. Biotinylated IFN-γ (eBioscience) or IL-17A (eBioscience) antibodies were added, followed by the addition of AKP streptavidin (BD Biosciences). Color was developed by adding 5 mM phosphatase substrate (Sigma-Aldrich), and absorbance was measured at 405 nm using a Versamax microplate reader with SoftMax Pro 6.5.1 software (Molecular Devices, Wokingham, UK).
Serum samples were prepared by centrifuging whole blood collected from mice at 2,000 × g for 10 min at 4°C. Subsequently, the serum samples were subjected to dilutions of 100,000-fold, 4-fold, and 1000-fold for the detection of anti-CII IgG antibody (Chondrex), anti-CCP antibody (MyBioSource Inc., San Diego, CA, USA), and zonulin (MyBioSource Inc.), respectively. The levels of serum autoantibodies and zonulin (MyBioSource Inc.) were measured in accordance with the manufacturer’s instructions.
Mouse colonic lamina propria (cLP) cells were isolated as previously described (35), and MLN and ILN cell suspensions were prepared through mechanical grinding. Cell surface and intracellular cytokine staining were conducted as previously described (35). Cells stained with fluorochrome-conjugated antibodies (Supplementary Table 4) were analyzed using a LSRFortessaTM X-20 flow cytometer (BD Biosciences). Data analysis was carried out using FlowJo (Tree Star Inc., Ashland, OR, USA).
Total bacterial genomic DNA from intracecal samples was extracted using the MagMAX™ Microbiome Ultra Nucleic Acid Isolation Kit on a KingFisher Flex automated DNA/RNA isolation system (Thermo Fisher Scientific), according to the manufacturer’s instructions. DNA samples were analyzed using a Nano-300 Micro-spectrophotometer (ALLSHENG, Hangzhou, China) and Qubit 3.0 fluorometer (Thermo Fisher Scientific) to assess concentration and purity of fecal DNA.
Libraries were constructed using the 16S Metagenomic Sequencing Library Preparation guide protocol (Illumina, San Diego, CA, USA). Variable regions 3 and 4 of the bacterial 16S rRNA gene were amplified using Kapa HiFi Hotstart Ready PCR Mix (Kapa Biosystems, Wilmington, MA, USA), with primers targeting MiSeq 341F and 805R. DNA indexing and second PCR were performed using Nextera XT index kits (Illumina). Each PCR product was purified using an Agencourt AMPure XP purification system (Beckman Coulter, Brea, CA, USA). Quantification and size estimation of the library were performed using the QIAxcel Advanced system and QIAxcel DNA High Resolution Kit (Qiagen, Hilden, Germany). Amplicons were pooled for sequencing using an Illumina MiSeq System (2 × 300 bp paired-end reads).
Sequence assembly and quality filtering were processed using QIIME2 software. Raw sequence reads were demultiplexed using the q2-demux plugin, followed by denoising using the DADA2 plugin to eliminate low-quality scores. All amplicon sequence variants (ASVs) were classified using the SILVA 138 99% database. QIIME2’s diversity analyses (q2-diversity plugin) and R software were used to present microbial richness, evenness diversity indices, and PCoA. The LEfSe algorithm was implemented using the Galaxy computational tool.
Data visualization and analysis were performed using GraphPad Prism (GraphPad, San Diego, CA, USA) or IBM SPSS (SPSS Inc., Chicago, IL, USA). Statistical significance was evaluated using a t-test or one- or two-way analysis of variance (ANOVA) followed by Fisher’s LSD post hoc test, or a Mann-Whitney test, Wilcoxon test or Kruskal-Wallis test followed by Dunn’s post hoc test, depending on the normal distribution of the data. Pearson’s correlation was used to assess the correlation between relative bacterial abundances and variables. Permutational multivariate analysis of variance (PERMANOVA) was applied using the QIIME2 plugin to evaluate differences in microbial community structure. Unless otherwise indicated, data are presented as the mean ± standard error of the mean (SEM), and a p value lower than 0.05 was considered significant.
To investigate microbial alterations in patients with RA based on serum antibody profiling, we conducted microbial species-specific IgM microarrays. The α-diversity of serum anti-microbial antibodies revealed a significant decrease in evenness in patients with RA compared to HC, while the richness remained similar between the two groups (Figure 1A). This diversity was further evident in the β-diversity assessment, where PCoA highlighted distinct clustering patterns representing the varying composition of serum anti-microbial antibodies in RA patients and HC (Figure 1B).
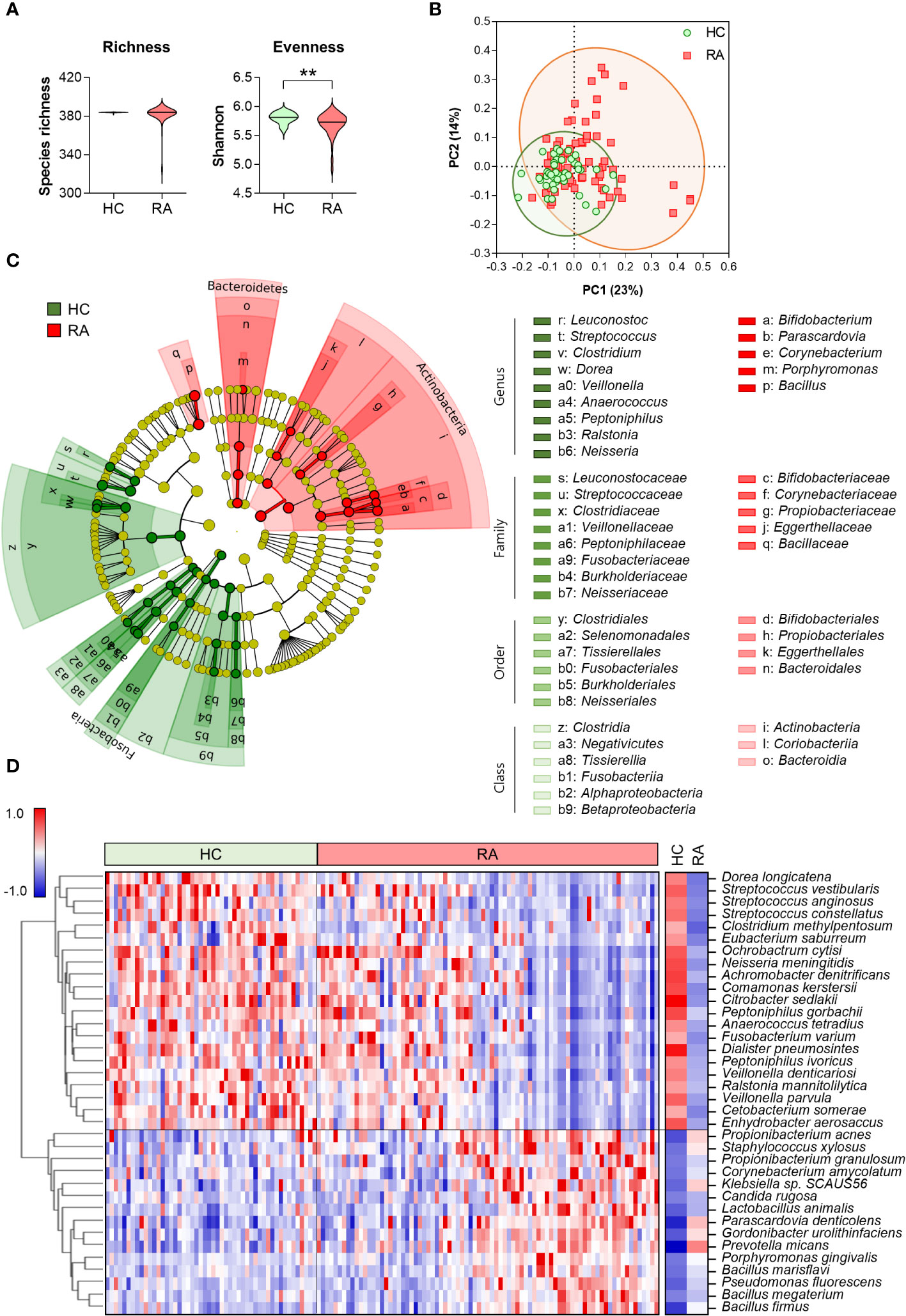
Figure 1 Serum anti-microbial antibody profile in healthy control (n = 50) and patients with RA (n = 81). (A) α-diversity assessed through microbial species-specific antibody abundance richness and evenness (Mann-Whitney test, **p < 0.01). (B) β-diversity analyzed using Bray-Curtis PCoA of microbial species-specific antibody communities (PERMANOVA, p < 0.001 vs HC). Symbols represent each individual. (C) Cladogram generated following a LEfSe analysis at the phylum to genus level (pairwise Wilcoxon test, p < 0.01, LDA score > 3.0). Green: taxa enriched in HC, Red: taxa enriched in RA. (D) Heatmap depicting differential anti-microbial antibody abundances between HC and patients with RA (t- and z-tests, p < 0.01, Z-ratio > 1.5). Blue: low abundance, Red: high abundance. HC: Healthy control, RA: Rheumatoid arthritis, PCoA: Principal coordinates analysis, LDA: Linear discriminant analysis, LEfSe: LDA effect size.
We identified discriminative core microbes employing a bacterial taxonomic rank based on the relative abundance of anti-microbial antibodies in the serum from HC and RA patients using an LEfSe analysis (Figure 1C). At the phylum level, Fusobacteria was significantly enriched in HC, whereas Actinobacteria and Bacteroidetes were enriched in patients with RA. At low taxonomic ranks, 6 and 3 classes, 6 and 4 orders, and 8 and 5 families were enriched in HC and patients with RA, respectively. The 9 genera including Streptococcus, Clostridium, and Peptoniphilus, were significantly enriched in HC, whereas 5 genera, including Corynebacterium, Prophyromonas, and Bacillus, were significantly enriched in patients with RA.
The heatmap depicting serum microbial species-specific antibody abundances highlighted significant alterations in antibody abundance against 36 microbial species in patients with RA compared to HC (Figure 1D). Specifically, antibodies against 21 microbial species were decreased, while antibodies against 15 microbial species were increased in patients with RA compared to HC.
Our study identified significant alterations in 36 anti-microbial antibodies in the comparison of patients with RA and HC (Figure 1D). Correlation analysis was conducted to investigate the association between the abundances of these significant anti-microbial antibodies and RA disease activity, determining associations with disease severity. Notably, anti-microbial antibodies decreased in patients with RA were inversely correlated with disease activity, whereas those elevated in patients were positively correlated with disease activity (Figure 2A). Specifically, the antibodies against Ochrobactrum cytisi (O. cytisi), P. gorbachii, Dialister pneumosintes (D. pneumosintes), and Veillonella denticariosi (V. denticariosi) were associated with lower disease severity (Figure 2B). Conversely, antibodies against Gordonibacter urolithinfaciens (G. urolithinfaciens) and Pseudomonas fluorescens (P. fluorescens) were associated with higher disease severity, as assessed by DAS28-CRP in patients with RA.
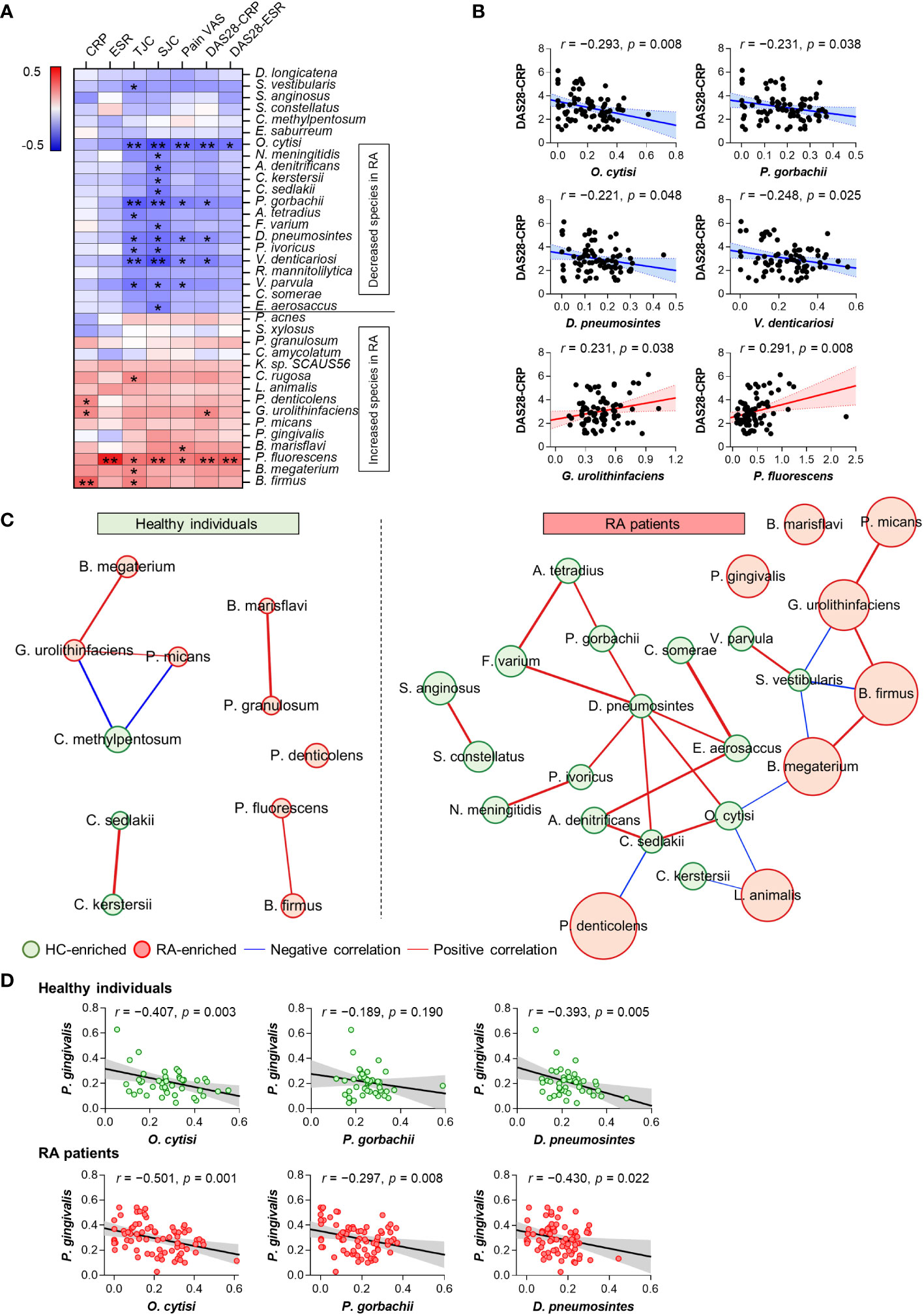
Figure 2 Association between altered microbial species-specific antibodies and disease activity in patients with RA (n = 81). (A) Correlations between differential anti-microbial antibody abundances and clinical features in patients with RA (Pearson’s correlation, *p <0.05, **p < 0.01). Blue: inverse correlation, Red: positive correlation. (B) The linear relationships between differential anti-microbial antibody abundances and DAS28 score in patients with RA (Pearson’s correlation). Solid lines represent the mean linear regression, while dotted lines indicate 95% confidence intervals. Symbols represent individual RA patients. (C) Co-occurrence network of differential anti-microbial antibodies in HC (n = 50) and patients with RA (n = 81). The size of each circle (node) represents the abundance of anti-microbial antibodies enriched in either HC (green node) or RA (red node) patients (Benjamini-Hochberg’s correction, p < 0.01). Lines connecting two nodes (edges) indicate the correlation efficiency between anti-microbial antibodies (chosen by Spearman, Pearson, Bray-Curtis, and Kullback-Leibler measures, r > 0.5, p < 0.05). Blue edges: negative correlation, Red edges: positive correlation. (D) The linear relationships between antibodies against P. gingivalis and disease activity-inversely correlated microbial species in HC and patients with RA (Pearson’s correlation). Solid lines represent the mean linear regression, while gray-filled areas indicate 95% confidence intervals. Symbols represent each individual. ESR: Erythrocyte Sedimentation Rate, CRP: C-Reactive Protein, TJC: Tender Joint Count, SJC: Swollen Joint Count, VAS: Visual Analog Scale, DAS: Disease Activity Score.
The CoNet approach was employed to infer association patterns among significant 36 anti-microbial antibodies in patients with RA and HC, revealing a higher number of networks in patients with RA compared to HC (Figure 2C), which is suggestive of increased co-occurrences of anti-microbial antibodies in patients with RA. In the RA patient group, most of the networks exhibited direct or indirect connections to one another. Specifically, antibodies against O. cytisi, P. gorbachii, and D. pneumosintes, which were enriched in HC and inversely associated with disease activity (Figure 2B), demonstrated positive connections either directly or indirectly within the RA patient network (Figure 2C). Additionally, the anti-G. urolithinfaciens antibody, enriched in RA and positively associated with disease activity (Figure 2B), showed positive connections with RA-enriched antibodies such as P. micans and B. firmus, while displaying an inverse connection with the HC-enriched S. vestibularis antibody. Overall, the altered anti-microbial antibodies exhibited associations with disease severity and interconnected relationships (networks) among themselves in patients with RA.
The analysis of microbial arrays revealed an increased abundance of antibodies targeting P. gingivalis in patients with RA, a putative pathogen implicated in RA through protein citrullination (Figure 1D). To further identify specific microbial candidates for RA therapy, we investigated the relationship between antibodies against P. gingivalis and three microbial species: O. cytisi, P. gorbachii, and D. pneumosintes. These species were selected based on their association with disease severity and their co-occurrence network (Figures 2B, C). The abundance of anti-O. cytisi and anti-D. pneumosintes antibodies exhibited an inverse correlation with the abundance of anti-P. gingivalis antibody in both patients with RA and HC (Figure 2D). Notably, the abundance of anti-P. gorbachii antibody showed an inverse correlation with anti-P. gingivalis antibody solely among patients with RA (Figure 2D). Importantly, P. gorbachii (PG) has been reported to inhibit infection (37) and produce SCFAs which were associated with RA suppression (38). Therefore, based on our bacterial microarray findings and the reported evidence (Supplementary Table 5), we selected PG as a potential specific target for RA treatment and further explored its involvement in RA.
To investigate the potential mechanisms underlying the therapeutic effects of PG in RA, we examined the competitive potential of PG against P. gingivalis, an RA-associated pathogen. Our in-vitro interspecies competition assay revealed an inhibitory impact of P. gingivalis on PG (Figure 3A). Despite similar growth rates in single cultures of both species, the growth of PG was strongly suppressed after 16 h of co-culture with P. gingivalis, indicating interference in bacterial growth between the two species.
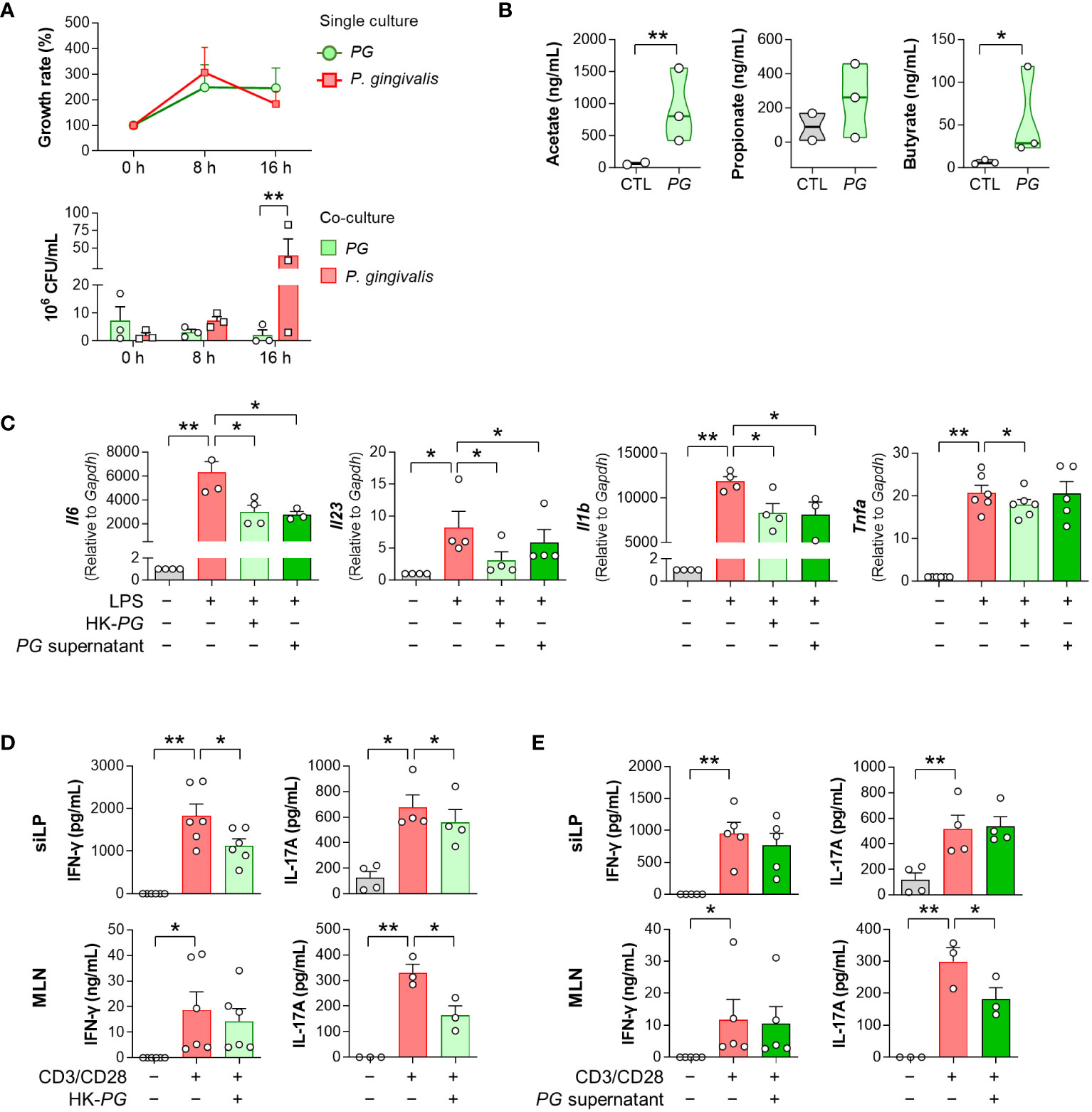
Figure 3 In vitro physiological and immunological properties of P. gorbachii (PG). (A) In vitro competition assay between PG and P. gingivalis. PG and P. gingivalis cultures were prepared each as a control (single culture) and then mixed (co-culture). After 0, 8, and 16 h of single and co-cultures, PG and P. gingivalis colonies were counted. n = 3 for each group and time (Two-way ANOVA, Fisher’s LSD post hoc test, **p < 0.01). (B) Measurement of short-chain fatty acids (SCFAs) in PG-cultured supernatants. n = 2–3 per group (t-test, *p < 0.05, **p < 0.01). Median and interquartile range (1st – 3rd quartile). (C) Pro-inflammatory gene expression of RAW 264.7 cells cultured in the absence or presence of heat-killed (HK)-PG or PG supernatant under LPS stimulation. n = 3–6 per group (One-way ANOVA, Fisher’s LSD post hoc test, *p < 0.05, **p < 0.01 vs LPS+ control). (D, E) IFN-γ and IL-17A concentrations in mouse siLP and MLN cell supernatants cultured in the absence or presence of HK-PG (D) or PG supernatant (E) under anti-CD3e and anti-CD28 stimulation. n = 3–6 per group (One-way ANOVA, Fisher’s LSD post hoc test, *p < 0.05, **p < 0.01 vs CD3/CD28+ control). Mean ± SEM. siLP: Small intestinal lamina propria, MLN: Mesenteric lymph node.
To validate the ability of PG to produce SCFAs, which have therapeutic implications in RA, we performed GC-MS analysis of PG-cultured media. PG produced SCFAs, including acetate and butyrate (Figure 3B), which have been associated with the alleviation of RA symptoms. Subsequently, we assessed the ability of PG to suppress immune responses through evaluation of pro-inflammatory cytokine profiles in RAW 264.7 macrophages, siLP, and MLN cells treated with HK-PG or PG supernatants under cell-stimulating conditions. Both HK-PG and PG supernatant downregulated the gene expression of LPS-induced Il6, Il23, Il1b, and Tnfa in RAW 264.7 cells (Figure 3C; Supplementary Figure 1). Moreover, HK-PG significantly reduced CD3/CD28-induced IFN-γ and IL-17A production in siLP cells, as well as IL-17A production in MLN cells (Figure 3D). However, PG supernatant only reduced CD3/CD28-induced IL-17A production in MLN cells (Figure 3E), indicating a specific anti-inflammatory suppression of immune cells. In an additional in-vitro experiment involving P. fluorescens, P. gingivalis, and P. micans, bacteria identified as elevated in the serum bacterial microarray analysis of patients with RA, treatment with P. fluorescens and P. gingivalis independently led to the upregulation of pro-inflammatory cytokine genes in RAW 264.7 cells in the absence of LPS (Supplementary Figure 2A). Meanwhile, the treatment of P. gingivalis and P. micans did not impact the induction of pro-inflammatory cytokine genes under LPS stimulated condition (Supplementary Figure 2B). Overall, PG demonstrated an outstanding effect in inhibiting the upregulation and secretion of pro-inflammatory cytokines associated with activated macrophages and CD4+ T cells. These findings underscore the critical role of PG in the modulating both innate and adaptive immune responses, involving key immune cell types such as macrophages and CD4+ T cells, which play a central role in the pathogenesis of RA.
To determine whether PG can prevent RA development in vivo, we evaluated the effect of PG treatment on arthritis in a CIA mouse model. First, we compared fecal CFUs of PG between CIA and PG-fed CIA (CIA/PG) mice to determine colonization of the intestine following oral administration. The fecal CFUs of PG were significantly higher in CIA/PG mice than in CIA mice, indicating stable intestinal colonization (Figure 4A).
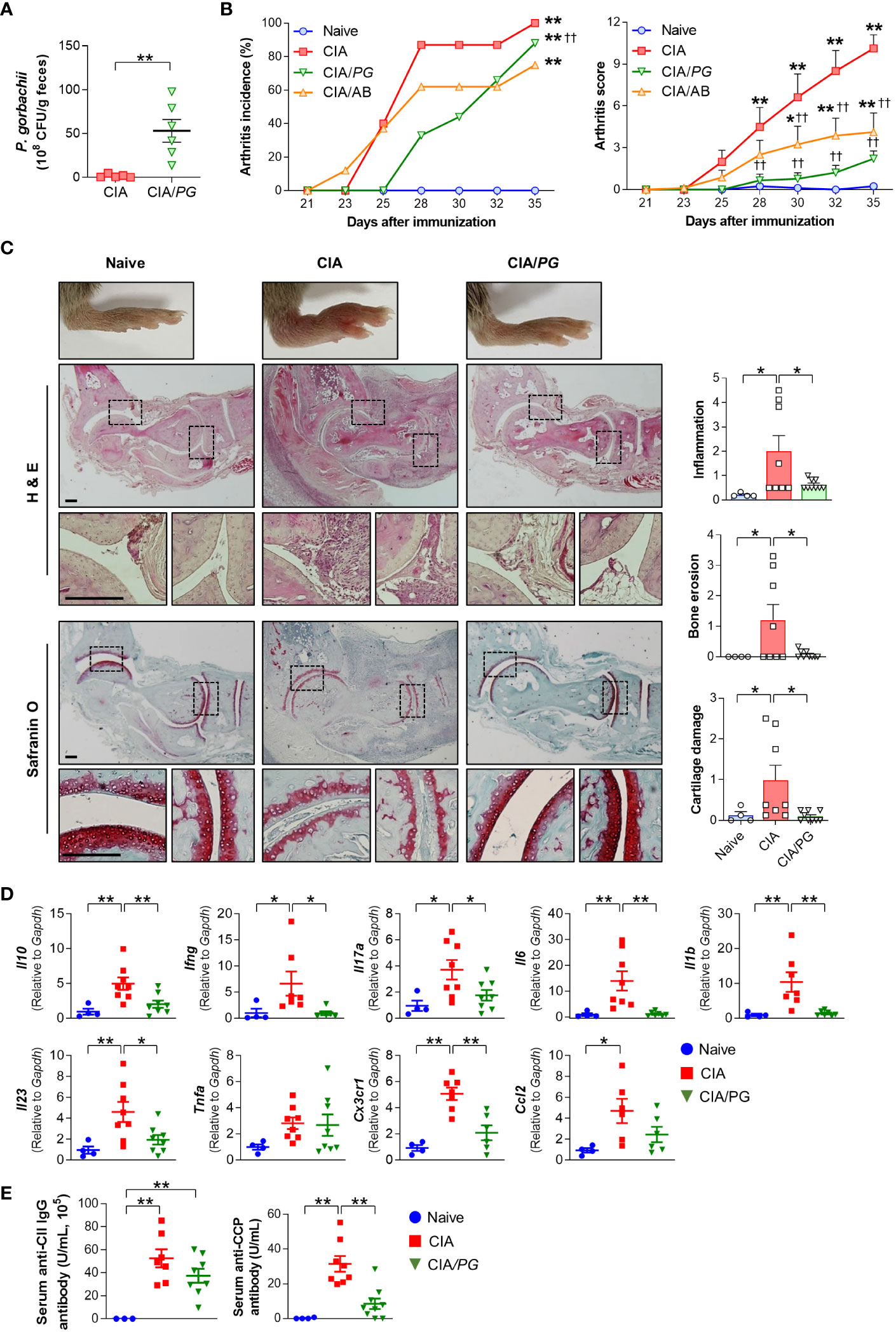
Figure 4 Effect of PG on arthritis in CIA mice. CIA was induced by intra-dermal injection of CII into mice. PG was orally administered for 4 weeks before the first CII injection (day 0) and continued until euthanasia at day 35. Samples were collected from naïve (n = 4), CIA (n = 8), and CIA/PG (n = 9) mice at day 35. (A) Colonization of PG in feces from CIA and CIA/PG mice (t-test, **p < 0.01). (B) Arthritis incidence and scores in the experimental groups (Log-rank test and Two-way ANOVA, Fisher’s LSD post hoc test, *p < 0.05, **p < 0.01 vs naïve; ††p < 0.01 vs CIA). (C) Representative histopathological images and scores of ankle joints stained with H&E and safranin O (One-way ANOVA, Fisher’s LSD post hoc test, *p < 0.05). Scale bar: 200 μm. (D) Expression of anti- and pro-inflammatory cytokine genes in ankle samples (One-way ANOVA, Fisher’s LSD post hoc test, *p < 0.05, **p < 0.01). (E) Serum anti-CII IgG and anti-CCP antibody levels (One-way ANOVA, Fisher’s LSD post hoc test, **p < 0.01). Mean ± SEM.
To clarify the effect of PG on arthritis, we treated a group of mice with antibiotics, as a broad spectrum of antibiotics has been reported to reduce arthritis severity in CIA mice (39). Both antibiotic- (CIA/AB) and PG-treated (CIA/PG) CIA mice exhibited reduced arthritis scores compared to CIA mice (Figure 4B). Furthermore, CIA/PG mice showed a clear prevention in arthritis development, with a lower incidence of arthritis than that of CIA mice (Figure 4B). Histopathological analysis using H&E staining revealed decreased inflammatory cell infiltration and bone erosion in CIA/PG mice (Figure 4C). Additionally, PG treatment led to the recovery of cartilage thinning in the joints of CIA mice.
To further confirm the suppressive effect of PG on arthritis, we investigated the gene expression of pro-inflammatory cytokines and chemokines in the ankles of the experimental groups. CIA/PG mice exhibited downregulated gene expression of pro-inflammatory cytokines (Ifng, Il17a, Il6, Il1b, and Il23) and a chemokine receptor (Cx3cr1), which were upregulated in CIA mice (Figure 4D; Supplementary Figure 3A). The gene expression of the anti-inflammatory cytokine Il10 was also upregulated in the ankles of CIA mice and normalized by PG treatment (Figure 4D).
Next, we assessed whether PG treatment influenced antigen-specific systemic autoimmunity by measuring serum anti-CII IgG and anti-CCP antibody levels. We observed no significant difference in anti-CII IgG antibody levels between CIA and CIA/PG mice; however, CIA/PG mice exhibited a trend for lower levels of anti-CII IgG antibody compared to CIA mice (Figure 4E). Furthermore, the elevated serum anti-CCP antibody levels in CIA mice were significantly decreased with PG treatment (Figure 4E). These findings suggest that PG not only attenuates arthritis severity but also reduces the production of antigen-specific autoantibodies, reflecting its potential therapeutic role in RA.
In our study, we explored PG’s role in regulating immune responses in the gut-joint axis, as prior research has suggested that intestinal inflammation can trigger the onset of RA (40). We conducted an analysis of T cell subsets in the ILN, MLN, and cLP of naïve, CIA, and CIA/PG mice using flow cytometry. The detailed gating strategies can be found in Supplementary Figure 4A.
Notably, we observed a significant decrease in the percentage of Th17 (CD4+IL-17A+) cells in the ILN of CIA/PG mice compared with that in CIA mice (Figure 5A). Furthermore, PG treatment significantly diminished the numbers of Th1 (CD4+IFN-γ+) and Th17 cells that had accumulated in the ILN of CIA mice. Interestingly, Treg (CD4+Foxp3+) cells, well-known for their immunosuppressive properties, were also increased in CIA mice relative to naïve and CIA/PG mice. However, the Th1/Treg and Th17/Treg ratios were higher in CIA mice than in naïve mice, suggesting a preferential differentiation or proliferation toward Th1 and Th17 cells in CIA. Importantly, PG treatment effectively restored the Th17/Treg imbalance observed in CIA mice by decreasing the Th17/Treg ratio in CIA/PG mice compared to CIA mice.
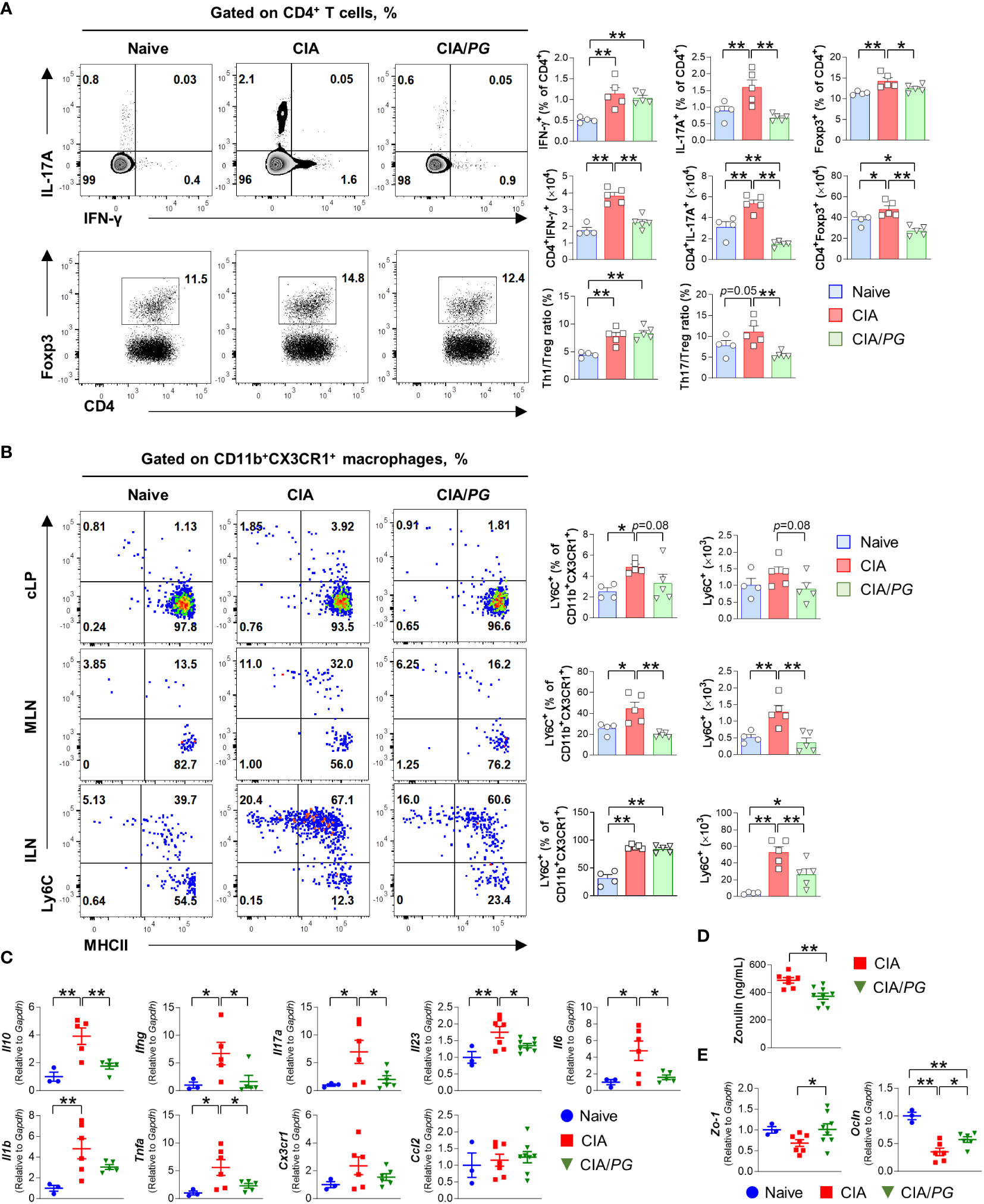
Figure 5 Role of PG in CIA-characteristic inflammation and intestinal barrier function. Samples were collected from naïve, CIA, and CIA/PG mice at 35 days after CII immunization. (A, B) Flow cytometry analysis of T cell subsets in the ILN (A) and macrophage lineage cells (B) from the cLP, MLN, and ILN of naïve (n = 4), CIA (n = 5), and CIA/PG (n = 5) mice. (C–E) Gene expression of pro-inflammatory cytokines in ileum (C), zonulin levels in serum (D), and gene expression of Zo-1 and Ocln in ileum (E) of naïve (n = 3), CIA (n = 6–7), and CIA/PG (n = 6–9) mice. Mean ± SEM. One-way ANOVA, Fisher’s LSD post hoc test (A–C, E) and t-test (D), *p < 0.05, **p < 0.01. cLP: Colonic intestinal lamina propria.
Although no difference was observed in Th1 and Th17 cells of cLP and MLN between naïve, CIA, and CIA/PG mice (Supplementary Figure 4B), PG treatment significantly reduced the accumulation of inflammatory monocytes (CD11b+CX3CR1+Ly6C+) in the MLN and ILN of CIA mice (Figure 5B). Furthermore, our study encompassed an analysis of conventional dendritic cells (cDCs), with a specific focus on the CD103– cDC2 subset, known for its role in promoting T cell differentiation into Th17 cells. However, our investigation did not reveal significant differences in CD103– cDC2 subset within the cLP and MLN when comparing non-treated CIA and PG-treated CIA mouse groups (Supplementary Figure 4C). Although PG treatment led to a reduction in both the number and percentage of CD103– cDC2 cells in the ILN of CIA mice, it did not restore normalcy to the population of all cDC subsets within cLP and MLN (Supplementary Figure 4C). Collectively, our findings highlight the potent inhibitory effect of PG on Th17 cells in ILN and the marked reduction in the accumulation of inflammatory monocytes in both ILN and gut-associated MLN.
To further understand the intestinal effect of PG on RA pathogenesis, we investigated the gene expression of pro-inflammatory cytokines in the ileum of naïve, CIA, and CIA/PG mice. The gene expression of Ifng, Il17a, Il23, Il6, and Tnfa was decreased in the ileal tissues of CIA/PG mice compared to those in CIA mice (Figure 5C). Moreover, we evaluated gut barrier integrity using zonulin, which is a protein secreted from gut epithelial cells in response to dietary, gut microbial, or inflammatory stimuli (8). Serum zonulin levels were decreased in CIA/PG mice compared to CIA mice, indicating improved gut barrier integrity (Figure 5D). Furthermore, genes encoding tight-junction proteins, zonula occludens-1 (Zo-1) and occludin (Ocln), were upregulated in the ileum of CIA/PG mice compared to those in CIA mice (Figure 5E; Supplementary Figure 3B). These data suggest a protective capacity of PG against arthritis through inhibition of CIA intestinal inflammation, reflected in the prevention of increased gut permeability and the disassembly of tight junctions.
To investigate the effects of PG treatment on the gut microbiome, we conducted 16S rRNA gene sequencing analysis of cecal samples from naïve, CIA, and CIA/PG mice. Although no significant differences were observed in α-diversity using Chao1 and Shannon indices among cecal microbiota from naïve, CIA, and CIA/PG mice (Figure 6A), CIA promoted a change in microbial communities with a clear separation in PCoA plots from the naïve and CIA/PG groups (Figure 6B). The cladogram showed discriminative bacterial clades by comparison of different mouse groups, where Bacteroidetes, Patescibacteria, and Firmicutes were the microbial phyla with differential abundances in the cecum of naïve, CIA, and CIA/PG mice, respectively (Figure 6C). At lower taxonomic ranks, bacterial taxa enriched in the cecum of naïve mice were Bacteroidaceae, Prevotellaceae, Staphylococcaceae, Eubacterium coprostanoligenes (E. coprostanoligenes) group, and Peptococcaceae families, as well as eight genera including Bacteroides, Alloprevotella, Staphylococcus, Lachnospiraceae_UCG-006, Butyricicoccaceae_UCG-009, and E. coprostanoligenes group. Bacterial taxa enriched in the cecum of CIA mice were the Saccharimonadaceae family, as well as the Desulfovibrio, Acetatifactor, and Candidatus saccharimonas genera. Bacterial taxa enriched in the cecum of CIA/PG mice were Lactobacillaceae, Streptococcaceae, Bacilli_unidentified, and Lachnospiraceae families, as well as six genera including Lactobacillus, Streptococcus, and Lachnospriaceae NK4A136 group.
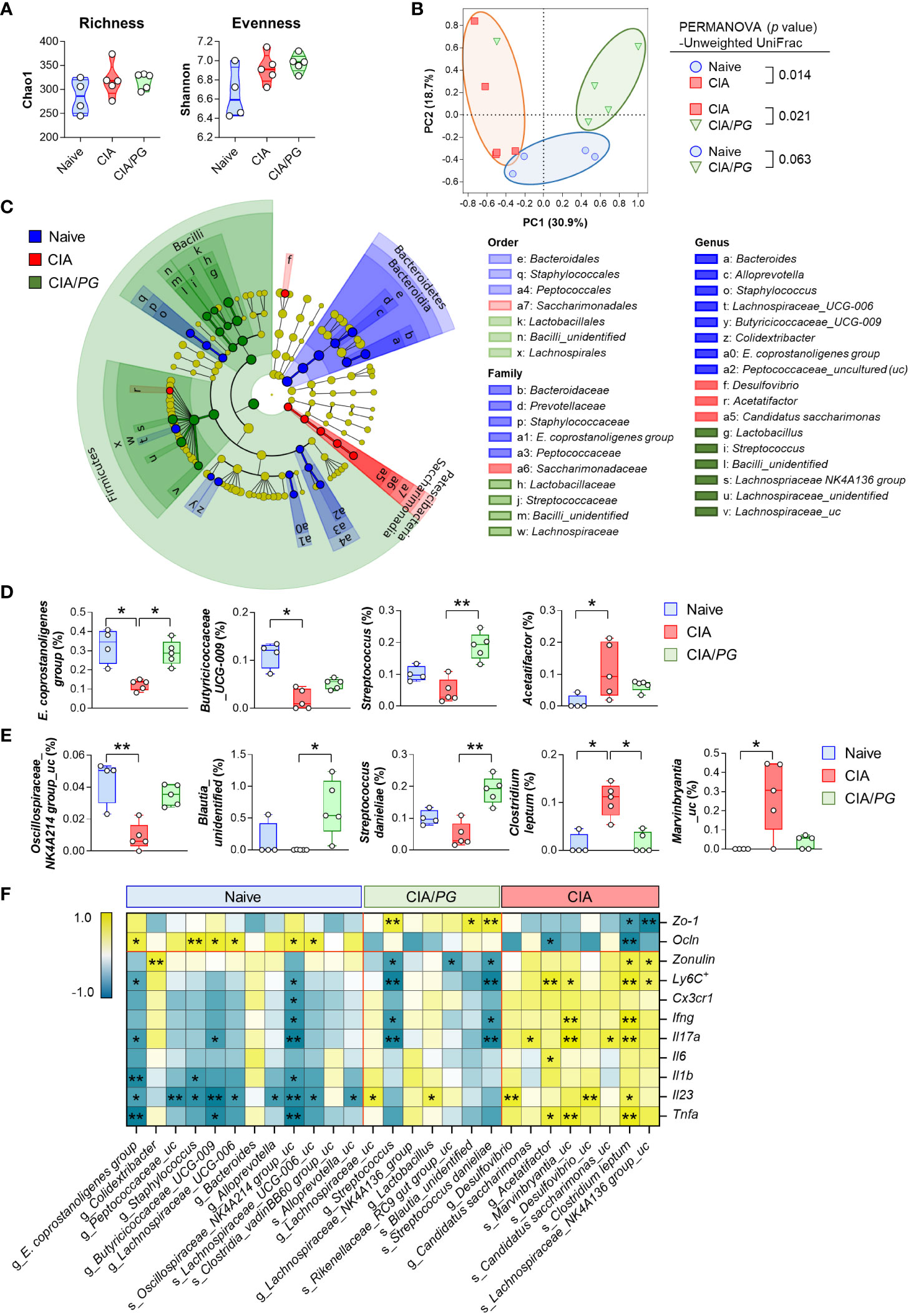
Figure 6 Role of PG in gut microbiome in CIA. Cecal samples were collected from naïve (n = 4), CIA (n = 5), and CIA/PG (n = 5) mice at 35 days after CII immunization and 16S rRNA gene sequencing was conducted. (A) α-diversity measured based on the Chao1 and Shannon indices (One-way ANOVA, Fisher’s LSD post hoc test, p > 0.05). Median and interquartile range (1st – 3rd quartile). (B) β-diversity assessed using PCoA based on unweighted UniFrac distance matrix. (C) Cladogram constructed based on the LEfSe analysis. Colors indicate bacterial taxa discriminately enriched in cecal microbiome of naive (blue), CIA (red), and CIA/PG (green) mice (Kruskal-Wallis, pairwise Wilcoxon test, p < 0.05, LDA score > 2.0). (D, E) Relative abundance of significantly different genera (D) and species (E) between groups (Kruskal-Wallis, Dunn’s post hoc test). Median ± min to max. (F) Relationship between gut-associated inflammatory profiles and cecal microbial taxa discriminately enriched in each experimental group (Pearson’s correlation, *p < 0.05, **p < 0.01). Colors indicate negative (turquoise) and positive (yellow) correlations.
PG treatment restored certain microbial genera and species to levels similar to those of naïve mice (Figures 6D, E). Notably, the relative abundance of the E. coprostanoligenes group and Clostridium leptum (C. leptum) was significantly normalized in the cecum of CIA/PG mice. In addition, PG treatment also promoted a significant increase in Blautia_unidentified and Streptococcus danieliae (S. danieliae) compared to CIA mice (Figure 6E). Overall, PG treatment partially resolved CIA-induced gut microbial dysbiosis, bring the gut microbial profile closer to that of naïve mice. To explore whether these changes in gut microbiota induced by PG treatment are linked to the production of SCFAs, we analyzed SCFA levels in cecal samples obtained from naive, CIA, and CIA/PG mice. Our analysis revealed a significant reduction in acetate and propionate levels in CIA mice; however, PG administration did not restore the decreased levels of acetate and propionate induced by CIA (Supplementary Figure 5). No significant changes in butyrate levels were observed among the three experimental mouse groups.
The correlation analysis between pathobiological molecules and bacterial taxa revealed that significantly abundant bacterial taxa in the cecum of naïve mice tended to be positively correlated with ileal Ocln gene expression and negatively correlated with an inflammatory response (Figure 6F). Conversely, significantly abundant bacterial taxa in the cecum of CIA mice showed a tendency for a negative correlation with intestinal epithelial barrier function and a positive correlation with an inflammatory response (Figure 6F). The genus Streptococcus and species S. danieliae were discriminately enriched in the cecum of CIA/PG mice and were associated with improved intestinal barrier integrity, showing a positive correlation with ileal Zo-1 gene expression and an inverse correlation with serum zonulin level (Figure 6F). Moreover, Streptococcus and S. danieliae were inversely correlated with the number of MLN Ly6C+ inflammatory monocytes and the gene expression of pro-inflammatory cytokines, such as Ifng and Il17a. Collectively, these findings demonstrated that the enriched microorganisms in the cecum of CIA/PG mice were closely linked to the amelioration of elevated gut permeability and inflammation.
In this study, we identified altered antibodies against 36 microbial species in patients with RA compared to healthy controls using a serum IgM antibody microarray against 384 microbial species. Among them, PG was selected as a candidate microorganism likely to have a therapeutic effect among the bacterial species that were decreased in RA patients and displayed a negative correlation with disease activity. PG showed therapeutic potential due to its competitive growth with P. gingivalis, its secretion of immunomodulatory SCFAs, and its suppression of inflammatory cytokines associated with macrophage and T cell activation. In a mouse model of CIA, PG administration exerted a strong inhibitory effect on Th17 cells in ILN and markedly inhibited inflammatory monocytes in MLN and ILN. Moreover, PG improved gut permeability and tight junction disassembly, inhibiting intestinal inflammation in CIA mice.
The most common method currently used for gut microbiota identification is 16S rRNA sequencing of fecal samples. As previously highlighted, some studies have attempted to overcome the limitations of sequencing methods using feces by focusing on serum-based approaches. Continuous exposure to microbes can trigger an antibody response against these microorganisms, offering a potential metric for assessing microbial exposure (41). Although IgA has traditionally been associated with immune responses against gut microbiota, recent research has highlighted the significance of IgM and IgG in host-microbiota interactions (42). A recent study revealed the presence of numerous IgM+ B cells in human intestine, which produce secretory IgM and intestinal IgM reacting to a variety of microbiota (43). IgM, the primary antibody produced by B cells upon antigen exposure before class switching to IgG or IgA (44), is characterized by its large pentameric structure, enabling it to bind to a broader range of bacterial antigens compared to IgG and IgA (45). Consequently, IgM-associated alterations are likely to signify diverse and robust systemic immune responses during the initial stages of disease. This may effectively mirror recent or ongoing shifts in microbiota, serving as a first responder against microbial antigens.
Several studies have explored the potential role of IgM in the immunopathogenesis of various diseases, often mediated through altered gut microbiota. For instance, individuals with obesity have shown elevated concentrations of plasma IgM (46) along with an increase in IgM-bound gut microbiota (47). Patients with RA exhibited higher IgM and IgA levels compared to control (48), while most patients with poor prognosis displayed elevated levels of IgM antibodies specifically binding to IgG (49). Moreover, RA patients exhibited significantly higher levels of multiple antimicrobial response factors such as EndoCAb IgM and EndoCAb IgA, which are released into circulation in response to microbial exposure when compared to healthy individuals (48). Elevated levels of IgM specific to certain bacterial species were also observed in RA patients (50). Notably, sera from rheumatoid factor (RF)-positive individuals showed significantly higher levels of IgM antibodies, but not IgG and to a lesser extent IgA, against Proteus mirabilis, Escherichia coli, and Klebsiella pneumoniae, which have been linked with RA (51).
This study was conducted to analyze the microbiota of patients with RA using a serum microbial array designed to detect IgM antibodies against gut microbiota in the serum, building upon the rationale supported by prior research. Our results using this novel method revealed a reduction in the evenness of anti-microbial antibody abundances in patients with RA compared to controls, as well as distinct compositions in these antibody levels between the two groups. Antibodies against a total of 36 microbial species were differentially expressed, with 21 being decreased and 15 being increased in patients with RA compared to HC patients. Notably, we found a significant correlation between the anti-PG antibody and RA disease activity, suggesting the potential effectiveness of the RA-specific bacteria, PG, as a treatment avenue for RA.
Preliminary data and the literature accentuate the plausibility of employing PG for RA treatment. First, PG belongs to the microbial taxa associated with disease-suppressive soils (37). Second, PG produces butyrate and acetate, immunomodulatory metabolites derived from microbiota (38). Butyrate has been associated with improved proliferation of intestinal epithelial cells, mucin synthesis, and maintenance of mucosal immune homeostasis (52). Acetate is a major SCFA that can be converted to butyrate by gut microbial enzymes (53, 54). Similarly, we showed that PG produces butyrate and acetate in-vitro, indicating its potential to restore commensal bacterial composition. In addition, the growth of PG was inhibited in the presence of P. gingivalis, a recognized pathogenic player in RA. Although the specific bacteria involved in competitive interactions with P. gingivalis are unknown, PG appears to play a role in such interactions. Notably, PG administration partially restored certain genera and species, contributing to the partial recovery of gut microbiota homeostasis. This study revealed that PG could potentially improve CIA by reinstating eubiosis.
This study has limitations in determining whether PG exerts its arthritis-suppressing effects through SCFA production. While our in-vitro experiment confirmed PG’s capacity to generate SCFAs, we observed no significant differences in cecal SCFA levels between non-treated CIA and PG-treated CIA mice. A prior study demonstrated differences in SCFA levels between gut and serum, suggesting the potential systemic transport of SCFAs within the body (55). Their research revealed an increase in serum butyrate levels upon administering Faeclibacterium prausnitzii, a representative butyrate-producing bacterium, in CIA mice. However, they reported no distinctions in cecal butyrate levels between non-treated CIA and F. prausnitzii-treated CIA mice. Although we were unable to analyze serum SCFAs in our present study, future investigations may be necessary to explore the influence of PG on the redistribution of SCFAs between the gut and the circulatory system in- vivo.
Pathophysiological immune mechanisms between gut microbiota and RA pathogenesis are potentially multifactorial. Some of these potential interactions might be underlined by the activation of antigen-presenting cells (APCs) via toll-like receptors, enzymatic peptide citrullination induction, antigenic mimicry, control of the host immune system (triggering T cell differentiation), and increased Th17-mediated mucosal inflammation (56). P gingivalis contributes to RA pathogenesis by activating Th17 cell-mediated pro-inflammatory responses and encoding peptidyl arginine deiminase, which facilitates the generation of ACPA (10, 11). Additionally, P. copri activates APCs through toll-like receptors and stimulates Th17 cell responses and ACPA production by B cells in patients with RA (56). Conversely, P. histicola can suppress arthritis development by modulating the immune response (regulation of DCs and generation of Treg cells), resulting in the suppression of Th17 responses and reduction of inflammatory cytokines (15). PG suppressed Th17 responses, as well as pro-inflammatory cytokines, inflammatory monocytes, and ACPA production. Our study further elucidates how PG contributes to CIA alleviation by curbing Th17 responses and inflammatory monocyte recruitment.
Lymphatic capillaries located within intestinal villi play a vital role in both nutrient absorption and immune cell trafficking to the MLN (57). Gut injuries can result in the release of pro-inflammatory factors that disrupt the integrity of the intestinal barrier (58). Recent evidence underscores the mesenteric lymph as a conduit for gut-derived harmful agents, ultimately contributing to systemic inflammation. These agents, upon absorption into the mesenteric lymphatic duct, possess the potential to activate and transport immune cells including monocytes/macrophages and DCs, thereby propagating systemic inflammation. During tissue injury or infection, monocytes rapidly infiltrate the affected tissues, acquiring pro-inflammatory characteristics, often classified as inflammatory monocytes (59). In inflamed colons, the recruitment of inflammatory monocytes to the LP is facilitated through CCR2, followed by their subsequent migration to the MLN, indicating colon inflammation (60).
In our study, although we did not observe a significant reduction in the population of inflammatory monocytes within the cLP of PG-treated CIA mice, we did find compelling evidence of PG’s inhibitory effect on the activation/recruitment of inflammatory monocytes within the MLN, implying its potential protective role against gut inflammation and injury. Furthermore, our study demonstrated a down-regulation of pro-inflammatory cytokine genes in the intestines of PG-treated CIA mice, accompanied by reduced serum zonulin levels, suggesting that PG’s impact on arthritis may be mediated through the modulation of the intestinal microenvironment. We also noted a trend toward reduced CCL2 gene expression in the ankles of PG-treated CIA mice, indicating a potential role for PG in inhibiting the migration of inflammatory monocytes to the synovium. Monocyte recruitment to the synovium relies on the interaction between chemokine receptors CCR2 and CX3CR1 on the monocyte surface and their ligands CCL2 and CX3CL1, which are produced by fibroblast-like synoviocytes (61). This interaction governs the egress of circulating monocytes and their subsequent recruitment into the RA synovium (62, 63).
Recent studies have highlighted the role of monocytes as APCs capable of activating T cells. In particular, Ly6C+ monocytes play a crucial role in transporting and presenting antigens to cognate T cells in LNs (59, 64). In RA, cytokines produced by monocytes/macrophages, such as IL-1β, IL-6, and IL-23, are instrumental in driving the polarization of Th17 cells and enhancing IL-17 production in CD4+ T cells (65). Inflammatory monocytes are likely the predominant monocyte subset involved in modulating the Th17 cells response, contributing to the activation and expansion of Th17 cells in inflamed synovial tissue (66, 67). Based on the findings from previous research, it is reasonable to speculate that PG contributes to the reduction of inflammatory monocyte recruitment/infiltration in the MLN by mitigating gut inflammation and barrier disruption. Concurrently, PG may hinder the trafficking of inflammatory monocytes to the synovium and its draining ILN by regulating CCL2, thereby influencing the induction of Th17 cells in the synovium and preventing the dissemination of systemic inflammation.
Nonetheless, recent studies have unveiled the intricate role of monocytes, including their actions within circulation and migration into tissues and lymphoid organs (59). Monocytes precursors originating from the bone marrow (BM) migrate to diverse organs, including lymphoid tissues like LNs and spleen, as well as nonlymphoid tissues such as the intestines, lung, and skin through blood circulation (68). CCL2 also plays a critical role in the egress of Ly6Chi monocytes from the BM (64). Activated monocytes migrate from the bloodstream into inflamed tissues through chemotaxis mediated CCL2 and CX3CL1, contributing to proinflammatory cytokine production. Monocytes can also traffic to LNs through tissues (69). Hence, it is conceivable that during arthritis, monocytes originating from the BM and circulating in the bloodstream may migrate to inflamed synovium or intestine, subsequently emigrating to their respective draining LNs. In the exploration of PG’s mechanisms for mitigating arthritis, unresolved questions remain regarding monocyte migration to tissues and draining LNs, as well as the interactions occurring within this lymphoid network.
Our findings indicate that PG influences intestinal permeability, suggesting a potential therapeutic role for PG in arthritis by modulating the intestinal environment. Zonulin, a central factor in this process, disrupts intestinal tight-junction proteins, enhances intestinal permeability, and promotes Th1 and Th17 infiltration in the lamina propria before the onset of arthritis (40). A recent study demonstrated that oral treatment with the zonulin antagonist larazotide before the onset of arthritis prevented increased intestinal barrier permeability and attenuated arthritis symptoms in CIA mice (40). ZO-1, an intracellular adaptor protein that binds to numerous transmembrane and cytoplasmic proteins, is required for the assembly of both adherens and tight junctions (70). Our study demonstrated that PG supplementation restored intestinal barrier integrity, decreased serum levels of zonulin, and increased gene expression of intestinal tight junction molecules (Zo-1 and Ocln), while also suppressing pro-inflammatory cytokines in the intestine. Thus, PG has the potential to ameliorate CIA by restoring intestinal barrier function. However, further mechanistic study is needed to determine which components of PG, including its structure or derived metabolites, contribute to the enhancement of intestinal function in vivo.
In the current investigation, certain limitations warrant acknowledgment. We did not compare the serum microbial array results with those obtained through fecal 16S RNA sequencing, the most common method for microbiota identification. Furthermore, since the serum microbial arrays examined only 384 microbial strains, the interpretation of the results could be somewhat limited. Lastly, the potential influence of anti-rheumatic drugs on microbiota was not factored into our analysis.
In conclusion, this study reiterates the utility of serum anti-microbial antibody arrays in uncovering therapeutic microbial targets for RA. Furthermore, we demonstrate the therapeutic effect of PG in restoring the intestinal barrier integrity and suppressing the immune response against RA. Therefore, PG may represent a promising microbiota-targeted therapy for individuals with RA.
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) repository, accession number PRJNA1048658 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1048658).
The studies involving humans were approved by GNUH Institutional Review Board in Korea (permit No: GNUH 2017-08-015). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by the Institutional Animal Care and Use Committee of GNU in Korea (IACUC approval ID: GNU-200724-M0046). The study was conducted in accordance with the local legislation and institutional requirements.
SK: Data curation, Formal analysis, Funding acquisition, Methodology, Visualization, Writing – original draft, Writing – review & editing. SC: Data curation, Formal analysis, Methodology, Visualization, Writing – original draft, Writing – review & editing. YC: Writing – review & editing. MK: Writing – review & editing. HK: Writing – review & editing. HL: Data curation, Formal analysis, Writing – review & editing. SH: Data curation, Formal analysis, Methodology, Resources, Writing – original draft, Conceptualization, Writing – review & editing. SP: Methodology, Resources, Writing – review & editing. MP: Methodology, Resources, Writing – review & editing. YS: Data curation, Formal analysis, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing, Conceptualization. SL: Data curation, Formal analysis, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (NRF-2018R1D1A1B07046903, NRF-2020R1I1A3071922, NRF-2022R1F1A1071114, http://www.nrf.re.kr/index) and the Biomedical Research Institute Fund (GNUHBRIF-2022-0001) from the Gyeongsang National University Hospital.
Authors S-JP and MP are employed by Bifido Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1286387/full#supplementary-material
1. Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep (2016) 6:28484. doi: 10.1038/srep28484
2. Higuchi BS, Rodrigues N, Gonzaga MI, Paiolo JCC, Stefanutto N, Omori WP, et al. Intestinal dysbiosis in autoimmune diabetes is correlated with poor glycemic control and increased interleukin-6: A pilot study. Front Immunol (2018) 9:1689. doi: 10.3389/fimmu.2018.01689
3. Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol (2013) 13(5):321–35. doi: 10.1038/nri3430
4. Sun Y, Chen Q, Lin P, Xu R, He D, Ji W, et al. Characteristics of gut microbiota in patients with rheumatoid arthritis in shanghai, China. Front Cell Infect Microbiol (2019) 9:369. doi: 10.3389/fcimb.2019.00369
5. van der Meulen TA, Harmsen HJM, Vila AV, Kurilshikov A, Liefers SC, Zhernakova A, et al. Shared gut, but distinct oral microbiota composition in primary sjogren’s syndrome and systemic lupus erythematosus. J Autoimmun (2019) 97:77–87. doi: 10.1016/j.jaut.2018.10.009
6. Kinashi Y. Hase K. Partners in leaky gut syndrome: Intestinal dysbiosis and autoimmunity. Front Immunol (2021) 12:673708. doi: 10.3389/fimmu.2021.673708
7. Akuzum B, Lee JY. Context-dependent regulation of type17 immunity by microbiota at the intestinal barrier. Immune Netw (2022) 22(6):e46. doi: 10.4110/in.2022.22.e46
8. Barbara G, Barbaro MR, Fuschi D, Palombo M, Falangone F, Cremon C, et al. Inflammatory and microbiota-related regulation of the intestinal epithelial barrier. Front Nutr (2021) 8:718356. doi: 10.3389/fnut.2021.718356
9. Rosenbaum JT, Asquith MJ. The microbiome: A revolution in treatment for rheumatic diseases? Curr Rheumatol Rep (2016) 18(10):62. doi: 10.1007/s11926-016-0614-8
10. Marchesan JT, Gerow EA, Schaff R, Taut AD, Shin SY, Sugai J, et al. Porphyromonas gingivalis oral infection exacerbates the development and severity of collagen-induced arthritis. Arthritis Res Ther (2013) 15(6):R186. doi: 10.1186/ar4376
11. Maresz KJ, Hellvard A, Sroka A, Adamowicz K, Bielecka E, Koziel J, et al. Porphyromonas gingivalis facilitates the development and progression of destructive arthritis through its unique bacterial peptidylarginine deiminase (pad). PloS Pathog (2013) 9(9):e1003627. doi: 10.1371/journal.ppat.1003627
12. Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis (2019) 78(5):590–3. doi: 10.1136/annrheumdis-2018-214514
13. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife (2013) 2:e01202. doi: 10.7554/eLife.01202
14. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med (2015) 21(8):895–905. doi: 10.1038/nm.3914
15. Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R, et al. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol (2016) 68(12):2878–88. doi: 10.1002/art.39785
16. Amdekar S, Singh V, Singh R, Sharma P, Keshav P, Kumar A. Lactobacillus casei reduces the inflammatory joint damage associated with collagen-induced arthritis (cia) by reducing the pro-inflammatory cytokines: Lactobacillus casei: Cox-2 inhibitor. J Clin Immunol (2011) 31(2):147–54. doi: 10.1007/s10875-010-9457-7
17. Pan H, Guo R, Ju Y, Wang Q, Zhu J, Xie Y, et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome (2019) 7(1):107. doi: 10.1186/s40168-019-0719-1
18. Gotschlich EC, Colbert RA, Gill T. Methods in microbiome research: Past, present, and future. Best Pract Res Clin Rheumatol (2019) 33(6):101498. doi: 10.1016/j.berh.2020.101498
19. Lindgreen S, Adair KL, Gardner PP. An evaluation of the accuracy and speed of metagenome analysis tools. Sci Rep (2016) 6:19233. doi: 10.1038/srep19233
20. Schirmer M, Ijaz UZ, D’Amore R, Hall N, Sloan WT, Quince C. Insight into biases and sequencing errors for amplicon sequencing with the illumina miseq platform. Nucleic Acids Res (2015) 43(6):e37. doi: 10.1093/nar/gku1341
21. Lkhagva E, Chung HJ, Hong J, Tang WHW, Lee SI, Hong ST, et al. The regional diversity of gut microbiome along the gi tract of male c57bl/6 mice. BMC Microbiol (2021) 21(1):44. doi: 10.1186/s12866-021-02099-0
22. Temoin S, Chakaki A, Askari A, El-Halaby A, Fitzgerald S, Marcus RE, et al. Identification of oral bacterial DNA in synovial fluid of patients with arthritis with native and failed prosthetic joints. J Clin Rheumatol (2012) 18(3):117–21. doi: 10.1097/RHU.0b013e3182500c95
23. Vogl T, Klompus S, Leviatan S, Kalka IN, Weinberger A, Wijmenga C, et al. Population-wide diversity and stability of serum antibody epitope repertoires against human microbiota. Nat Med (2021) 27(8):1442–50. doi: 10.1038/s41591-021-01409-3
24. Hitchon CA, Chandad F, Ferucci ED, Willemze A, Ioan-Facsinay A, van der Woude D, et al. Antibodies to porphyromonas gingivalis are associated with anticitrullinated protein antibodies in patients with rheumatoid arthritis and their relatives. J Rheumatol (2010) 37(6):1105–12. doi: 10.3899/jrheum.091323
25. Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC, et al. Antibody responses to porphyromonas gingivalis (p. Gingivalis) in subjects with rheumatoid arthritis and periodontitis. Int Immunopharmacol (2009) 9(1):38–42. doi: 10.1016/j.intimp.2008.09.008
26. Ogrendik M, Kokino S, Ozdemir F, Bird PS, Hamlet S. Serum antibodies to oral anaerobic bacteria in patients with rheumatoid arthritis. MedGenMed (2005) 7(2):2.
27. Okada M, Kobayashi T, Ito S, Yokoyama T, Komatsu Y, Abe A, et al. Antibody responses to periodontopathic bacteria in relation to rheumatoid arthritis in Japanese adults. J Periodontol (2011) 82(10):1433–41. doi: 10.1902/jop.2011.110020
28. Seifert JA, Bemis EA, Ramsden K, Lowell C, Polinski K, Feser M, et al. Association of antibodies to prevotella copri in anti-cyclic citrullinated peptide-positive individuals at risk of developing rheumatoid arthritis and in patients with early or established rheumatoid arthritis. Arthritis Rheumatol (2023) 75(4):507–16. doi: 10.1002/art.42370
29. Ormseth MJ, Wu Q, Zhao S, Allen RM, Solus J, Sheng Q, et al. Circulating microbial small rnas are altered in patients with rheumatoid arthritis. Ann Rheum Dis (2020) 79(12):1557–64. doi: 10.1136/annrheumdis-2020-217589
30. Castillo DJ, Rifkin RF, Cowan DA, Potgieter M. The healthy human blood microbiome: Fact or fiction? Front Cell Infect Microbiol (2019) 9:148. doi: 10.3389/fcimb.2019.00148
31. van Riel PL. The development of the disease activity score (das) and the disease activity score using 28 joint counts (das28). Clin Exp Rheumatol (2014) 32(5 Suppl 85):S–65-74.
32. van Riel PL, Schumacher HR Jr. How does one assess early rheumatoid arthritis in daily clinical practice? Best Pract Res Clin Rheumatol (2001) 15(1):67–76. doi: 10.1053/berh.2000.0126
33. Campanero-Rhodes MA, Lacoma A, Prat C, Garcia E, Solis D. Development and evaluation of a microarray platform for detection of serum antibodies against streptococcus pneumoniae capsular polysaccharides. Anal Chem (2020) 92(11):7437–43. doi: 10.1021/acs.analchem.0c01009
34. Li J, Zhai X, Ding C, Liu Y, Dong Q, Xu D, et al. Development of a bacterial macroarray for the rapid screening of targeted antibody-secreted hybridomas. SLAS Discovery (2019) 24(2):190–8. doi: 10.1177/2472555218804990
35. Kim M, Galan C, Hill AA, Wu WJ, Fehlner-Peach H, Song HW, et al. Critical role for the microbiota in cx3cr1(+) intestinal mononuclear phagocyte regulation of intestinal t cell responses. Immunity (2018) 49(1):151–63 e5. doi: 10.1016/j.immuni.2018.05.009
36. Kim M, Choe Y, Lee H, Jeon MG, Park JH, Noh HS, et al. Blockade of translationally controlled tumor protein attenuated the aggressiveness of fibroblast-like synoviocytes and ameliorated collagen-induced arthritis. Exp Mol Med (2021) 53(1):67–80. doi: 10.1038/s12276-020-00546-y
37. Gomez Exposito R, de Bruijn I, Postma J, Raaijmakers JM. Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front Microbiol (2017) 8:2529. doi: 10.3389/fmicb.2017.02529
38. Murphy EC, Frick IM. Gram-positive anaerobic cocci–commensals and opportunistic pathogens. FEMS Microbiol Rev (2013) 37(4):520–53. doi: 10.1111/1574-6976.12005
39. Jubair WK, Hendrickson JD, Severs EL, Schulz HM, Adhikari S, Ir D, et al. Modulation of inflammatory arthritis in mice by gut microbiota through mucosal inflammation and autoantibody generation. Arthritis Rheumatol (2018) 70(8):1220–33. doi: 10.1002/art.40490
40. Tajik N, Frech M, Schulz O, Schalter F, Lucas S, Azizov V, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun (2020) 11(1):1995. doi: 10.1038/s41467-020-15831-7
41. Zhang P, Minardi LM, Kuenstner JT, Zekan SM, Kruzelock R. Anti-microbial antibodies, host immunity, and autoimmune disease. Front Med (Lausanne) (2018) 5:153. doi: 10.3389/fmed.2018.00153
42. Sterlin D, Fadlallah J, Slack E, Gorochov G. The antibody/microbiota interface in health and disease. Mucosal Immunol (2020) 13(1):3–11. doi: 10.1038/s41385-019-0192-y
43. Magri G, Comerma L, Pybus M, Sintes J, Llige D, Segura-Garzon D, et al. Human secretory igm emerges from plasma cells clonally related to gut memory b cells and targets highly diverse commensals. Immunity (2017) 47(1):118–34 e8. doi: 10.1016/j.immuni.2017.06.013
44. Ouchida R, Mori H, Hase K, Takatsu H, Kurosaki T, Tokuhisa T, et al. Critical role of the igm fc receptor in igm homeostasis, b-cell survival, and humoral immune responses. Proc Natl Acad Sci U.S.A. (2012) 109(40):E2699–706. doi: 10.1073/pnas.1210706109
45. Johansen FE, Braathen R, Brandtzaeg P. Role of j chain in secretory immunoglobulin formation. Scand J Immunol (2000) 52(3):240–8. doi: 10.1046/j.1365-3083.2000.00790.x
46. Kosaraju R, Guesdon W, Crouch MJ, Teague HL, Sullivan EM, Karlsson EA, et al. B cell activity is impaired in human and mouse obesity and is responsive to an essential fatty acid upon murine influenza infection. J Immunol (2017) 198(12):4738–52. doi: 10.4049/jimmunol.1601031
47. Pearson JA, Ding H, Hu C, Peng J, Galuppo B, Wong FS, et al. Igm-associated gut bacteria in obesity and type 2 diabetes in c57bl/6 mice and humans. Diabetologia (2022) 65(8):1398–411. doi: 10.1007/s00125-022-05711-8
48. Ayyappan P, Harms RZ, Seifert JA, Bemis EA, Feser ML, Deane KD, et al. Heightened levels of antimicrobial response factors in patients with rheumatoid arthritis. Front Immunol (2020) 11:427. doi: 10.3389/fimmu.2020.00427
49. Nicolo A, Amendt T, El Ayoubi O, Young M, Finzel S, Senel M, et al. Rheumatoid factor igm autoantibodies control igg homeostasis. Front Immunol (2022) 13:1016263. doi: 10.3389/fimmu.2022.1016263
50. Newkirk MM, Goldbach-Mansky R, Senior BW, Klippel J, Schumacher HR Jr., El-Gabalawy HS. Elevated levels of igm and iga antibodies to proteus mirabilis and igm antibodies to escherichia coli are associated with early rheumatoid factor (rf)-positive rheumatoid arthritis. Rheumatol (Oxford) (2005) 44(11):1433–41. doi: 10.1093/rheumatology/kei036
51. Senior BW, McBride PD, Morley KD, Kerr MA. The detection of raised levels of igm to proteus mirabilis in sera from patients with rheumatoid arthritis. J Med Microbiol (1995) 43(3):176–84. doi: 10.1099/00222615-43-3-176
52. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U.S.A. (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111
53. Herrmann E, Young W, Rosendale D, Reichert-Grimm V, Riedel CU, Conrad R, et al. Rna-based stable isotope probing suggests allobaculum spp. As particularly active glucose assimilators in a complex murine microbiota cultured in vitro. BioMed Res Int (2017) 2017:1829685. doi: 10.1155/2017/1829685
54. Rios-Covian D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilan CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol (2016) 7:185. doi: 10.3389/fmicb.2016.00185
55. Moon J, Lee AR, Kim H, Jhun J, Lee SY, Choi JW, et al. Faecalibacterium prausnitzii alleviates inflammatory arthritis and regulates il-17 production, short chain fatty acids, and the intestinal microbial flora in experimental mouse model for rheumatoid arthritis. Arthritis Res Ther (2023) 25(1):130. doi: 10.1186/s13075-023-03118-3
56. Horta-Baas G, Romero-Figueroa MDS, Montiel-Jarquin AJ, Pizano-Zarate ML, Garcia-Mena J, Ramirez-Duran N. Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J Immunol Res (2017) 2017:4835189. doi: 10.1155/2017/4835189
57. Kim KW, Song JH. Emerging roles of lymphatic vasculature in immunity. Immune Netw (2017) 17(1):68–76. doi: 10.4110/in.2017.17.1.68
58. Ma Y, Yang X, Chatterjee V, Wu MH, Yuan SY. The gut-lung axis in systemic inflammation. Role of mesenteric lymph as a conduit. Am J Respir Cell Mol Biol (2021) 64(1):19–28. doi: 10.1165/rcmb.2020-0196TR
59. Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol (2017) 17(6):349–62. doi: 10.1038/nri.2017.28
60. Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, Friedlander G, et al. Ly6c hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity (2012) 37(6):1076–90. doi: 10.1016/j.immuni.2012.08.026
61. Ancuta P, Liu KY, Misra V, Wacleche VS, Gosselin A, Zhou X, et al. Transcriptional profiling reveals developmental relationship and distinct biological functions of cd16+ and cd16- monocyte subsets. BMC Genomics (2009) 10:403. doi: 10.1186/1471-2164-10-403
62. Rana AK, Li Y, Dang Q, Yang F. Monocytes in rheumatoid arthritis: Circulating precursors of macrophages and osteoclasts and, their heterogeneity and plasticity role in ra pathogenesis. Int Immunopharmacol (2018) 65:348–59. doi: 10.1016/j.intimp.2018.10.016
63. Sierra-Filardi E, Nieto C, Dominguez-Soto A, Barroso R, Sanchez-Mateos P, Puig-Kroger A, et al. Ccl2 shapes macrophage polarization by gm-csf and m-csf: Identification of ccl2/ccr2-dependent gene expression profile. J Immunol (2014) 192(8):3858–67. doi: 10.4049/jimmunol.1302821
64. Kratofil RM, Kubes P, Deniset JF. Monocyte conversion during inflammation and injury. Arterioscler Thromb Vasc Biol (2017) 37(1):35–42. doi: 10.1161/ATVBAHA.116.308198
65. Yoon BR, Yoo SJ, Choi Y, Chung YH, Kim J, Yoo IS, et al. Functional phenotype of synovial monocytes modulating inflammatory t-cell responses in rheumatoid arthritis (ra). PloS One (2014) 9(10):e109775. doi: 10.1371/journal.pone.0109775
66. Rossol M, Kraus S, Pierer M, Baerwald C, Wagner U. The cd14(bright) cd16+ monocyte subset is expanded in rheumatoid arthritis and promotes expansion of the th17 cell population. Arthritis Rheum (2012) 64(3):671–7. doi: 10.1002/art.33418
67. Winchester R, Giles JT, Nativ S, Downer K, Zhang HZ, Bag-Ozbek A, et al. Association of elevations of specific t cell and monocyte subpopulations in rheumatoid arthritis with subclinical coronary artery atherosclerosis. Arthritis Rheumatol (2016) 68(1):92–102. doi: 10.1002/art.39419
68. Smiljanovic B, Radzikowska A, Kuca-Warnawin E, Kurowska W, Grun JR, Stuhlmuller B, et al. Monocyte alterations in rheumatoid arthritis are dominated by preterm release from bone marrow and prominent triggering in the joint. Ann Rheum Dis (2018) 77(2):300–8. doi: 10.1136/annrheumdis-2017-211649
69. Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity (2013) 39(3):599–610. doi: 10.1016/j.immuni.2013.08.007
Keywords: rheumatoid arthritis, microbiota, Peptoniphilus gorbachii, intestinal barrier integrity, inflammatory reaction
Citation: Kim S, Chun SH, Cheon Y-H, Kim M, Kim H-O, Lee H, Hong S-T, Park S-J, Park MS, Suh YS and Lee S-I (2024) Peptoniphilus gorbachii alleviates collagen-induced arthritis in mice by improving intestinal homeostasis and immune regulation. Front. Immunol. 14:1286387. doi: 10.3389/fimmu.2023.1286387
Received: 31 August 2023; Accepted: 12 December 2023;
Published: 04 January 2024.
Edited by:
Rajan P. Adhikari, Integrated BioTherapeutics, Inc., United StatesCopyright © 2024 Kim, Chun, Cheon, Kim, Kim, Lee, Hong, Park, Park, Suh and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Young Sun Suh, dGF0YWJveDEyM0BoYW5tYWlsLm5ldA==; Sang-Il Lee, Z29sZGd1QGdudS5hYy5rcg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.