
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Immunol., 25 September 2023
Sec. Cancer Immunity and Immunotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1284124
This article is part of the Research TopicEpigenetic, Metabolic, and Transcriptional Regulation of Immune Cell Plasticity and Functions in Cancer and Non-Cancer DiseasesView all 11 articles
Editorial on the Research Topic
Epigenetic, metabolic, and transcriptional regulation of immune cell plasticity and functions in cancer and non-cancer diseases
High plasticity represents an essential characteristic of innate and adaptive immune cells, which allows their multi-directional transition into diverse phenotypes with different (even opposite) functions (1, 2). The significance of this property is further highlighted by an increasing number of recently emerging new phenotypes of immune cells, particularly involving many malignant and non-malignant disorders (3). Mechanistically, the phenotypic transition occurs via reprogramming of gene expression at the transcriptional level primarily driven by complex and interactive mechanisms involving microenvironment, intracellular signaling, transcription factors, epigenetic remodeling, metabolic rewiring, and post-translational modification (PTM) (1, 4–9).
Transcriptional reprogramming for the phenotypic transition of immune cells is primarily governed via various epigenetic mechanisms, including DNA methylation, histone PTMs, non-coding RNA, RNA modification, etc. (1, 10, 11). DNA methylation usually functions to silence the expression of tumor suppressors in most types of cancer (12). In contrast, many oncogenes are often hypomethylated to promote their expression in tumor cells. Interestingly, multiple differential methylation sites (DMSs) have been found in either the gene itself or the promoter of telomerase reverse transcriptase (TERT), a critical enzyme that controls the length of telomere and is thus considered an oncogene (Lin et al.). Moreover, hypermethylation of these DMSs significantly correlates with TERT expression, infiltration of immune cells [e.g., T cells, T helper 2 (Th2) cells, Treg, CD56dim natural killer (NK) cells, activated dendritic cells (DCs), and B cells], and immune checkpoints (e.g., LAG-3) in triple-negative breast cancer (TNBC) (Lin et al.). Of note, hypermethylation of at least some DMSs is associated with poor overall survival of patients with TNBC (Lin et al.). These findings suggest that TERT promoter hypermethylation may play a role in tumor microenvironment (TME), although the underlying mechanism remains to be explored.
5-methylcytosine (m5C) represents one of the important forms of RNA modification, which occurs on virtually all types of RNA (e.g., mRNA, tRNA, rRNA, lncRNA, and other RNAs) and plays diverse roles in RNA transcription, transportation, and translation (13). Thus, there is no doubt that m5C and its regulatory elements (including “writer”, “eraser”, and “reader”) account for malignant behaviors of both tumor cells and TME involving numerous immune cells (e.g., T cells, B/plasma cells, macrophages, granulocytes, NK cells, DCs, and mast cells) in numerous types of cancer, including solid tumor (e.g., liver, stomach, bladder, prostate, head and neck, breast, pancreas, kidney, and colon and rectum cancer) and hematologic malignancies (e.g., leukemia) (Gu et al.).
Recently, many novel forms of histone and non-histone protein PTMs have been discovered in physiological and/or pathological scenarios (14–17). Among them, lactylation occurs primarily at lysine residues of histones and in some circumstances, also non-histone proteins. Since lactylation is induced by lactate (the end product of glycolysis) (18), it thus represents a key linker between metabolic rewiring (paradigm shift from oxidative phosphorylation to glycolysis or vice versa) and epigenetic remodeling (19, 20). This new PTM may be particularly important in cancer, considering aerobic glycolysis as a metabolic hallmark of cancer known as the Warburg effect (21, 22). Indeed, fast-emerging evidence supports the functional role of lactylation in both tumor cells and TME involving tumor-infiltrating myeloid cells, including tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), and tumor-associated neutrophils (TANs), especially the communication between them (Su et al.). Thus, more reliable targets will become available for anti-tumor epigenetic therapy and immunotherapy soon.
As another protein PTM, ubiquitination is essential for protein turnover via the ubiquitin-proteasome system (UPS) or autophagy, among many other functions, in both normal and malignant cells (23). Consequently, agents targeting UPS (e.g., proteasome inhibitors such as bortezomib) have been extremely successful in the treatment of plasma cell neoplasms like multiple myeloma that relies on a highly-efficient protein turnover machinery to remove abundant useless but harmful immunoglobulin in tumor cells (24). Notably, many E3 ubiquitin ligases (e.g., WWP1/2, SMURF1/2, ITCH, FBXW7, FBXO3/6/21, HECTD1, and ULF1) deubiquitinases (e.g., USP3/5/7/13/14/15/49) that reciprocally regulate ubiquitination are associated with osteoarthritis (Zheng C. et al.), suggesting that this non-inflammatory degenerative joint arthritis may be another disorder caused by deficient protein turnover. In addition to a number of (de)ubiquitination-targeted agents undergoing development (Zheng C. et al.), it is interesting to see whether the FDA-approved proteasome inhibitors would be effective against this type of (de)ubiquitination-deficient disorders like osteoarthritis.
Among various immunotherapies, immune checkpoint inhibition represents a major therapeutic strategy. The most representative one is anti-PD1/PD-L1 monoclonal antibodies (mAbs), which have been approved for the treatment of many types of solid tumors, including lung cancer (25). However, although anti-PD1/PD-L1 mAbs display a striking efficacy, a considerable number of patients with lung cancer have not been benefited (26). To this end, numerous biomarkers have been discovered to pre-select patients who are most likely to benefit from this immunotherapy, including tumor-related markers (e.g., PD-L1 expression, tumor mutation burden, dMMR/MSI, and many tumor-specific genes), peripheral blood-based markers (e.g., ctDNA, immune cells/T cell receptor, and exosomal PD-L1/cytokines) and gut microbiome (Wang et al.). Exhaustion of T cells represents a major hurdle for immunotherapy (27), which may at least in part explain the unsatisfied efficacy of anti-PD1 mAb in most hematologic malignancies like acute myeloid leukemia (AML). In AML patients, accumulation of a subset of severely exhausted T cells (CD28−/PD-1+/TIGHT+) correlates with the presence of minimal residue disease, poor therapeutic response, and short disease-free survival (Huang et al.). Thus, a strategy combining agents targeting such exhausted T cells may improve the efficacy of PD-1 blockade in AML and probably other hematologic malignancies as well. Another approach to enhance the efficacy of immunotherapy is to combine it with radiotherapy, based on a potential mechanism of action involving oxidative stress (e.g., ROS) induced by radiation (Zheng Z. et al.). ROS promotes the release of tumor-associated antigens, which promote infiltration and differentiation of immune cells, modulate the expression of immune checkpoints, and remodel TME (Zheng Z. et al.). Interestingly, expression of certain cell cycle-related genes such as CENPE also correlates with the infiltration of immune cells (e.g., DCs, B cells, T cells, CD4+ or CD8+ memory T cells, macrophages, and mast cells) at least in some types of cancer (e.g., medulloblastoma) (Fang et al.). However, its functional role in TME, other than tumor cells themselves, and immunotherapy remains to be investigated.
In addition, many other immune checkpoints could also serve as potential targets to fill in the gap left behind. One example is CD47/SIRPα (known as a “don’t eat me” signal for phagocytosis by macrophages) (28). While this checkpoint has been well investigated, its targeted therapy has however not been as successful as anti-PD1/PD-L1 mAbs thus far, largely due to severe adverse events (e.g., anemia, because red blood cells highly express CD47) (29). In this circumstance, many alternative approaches, rather than mAbs, have been attempted to avoid this dark side (30). Targeting glutaminyl-peptide cyclotransferase-like protein (QPCTL) that catalyzes CD47 pyroglutamylation crucial for the binding between CD47 and SIRPα may be a promising approach for the treatment of glioma, in which QPCTL is highly expressed due to DNA hypomethylation and associated with poor outcomes (Liu et al.).
Last, unlike the rapid advance in immunotherapy for cancer, its application in non-cancer diseases lags way behind (31). Among many obstacles, the most important one may be the lack of defined target immune cells in autoimmune or inflammatory diseases, primarily due to the complexity of immune cells with diverse, plastic phenotypes. To this end, recently developed single-cell-analyzing techniques have been doing a really good job of identifying immune cells with different functions (3). For example, single-cell RNA sequencing (scRNAseq) and high-dimensional mass cytometry (CyTOF) have defined a hyper-inflammatory signature in peripheral blood mononuclear cells of patients with Prader-Willi syndrome, with a marked increase in CD16+ monocytes that likely drive hyper-inflammatory status of this disease and therefore represent a potential target for immunotherapy (Xu et al.).
Together, further basic, translational, and clinical research with the development and utilization of modern technology would provide much deeper insights into the mechanisms underlying immune cell plasticity (Figure 1), leading to the discovery of useful biomarkers or targets for precision medicine against cancer and non-cancer diseases.
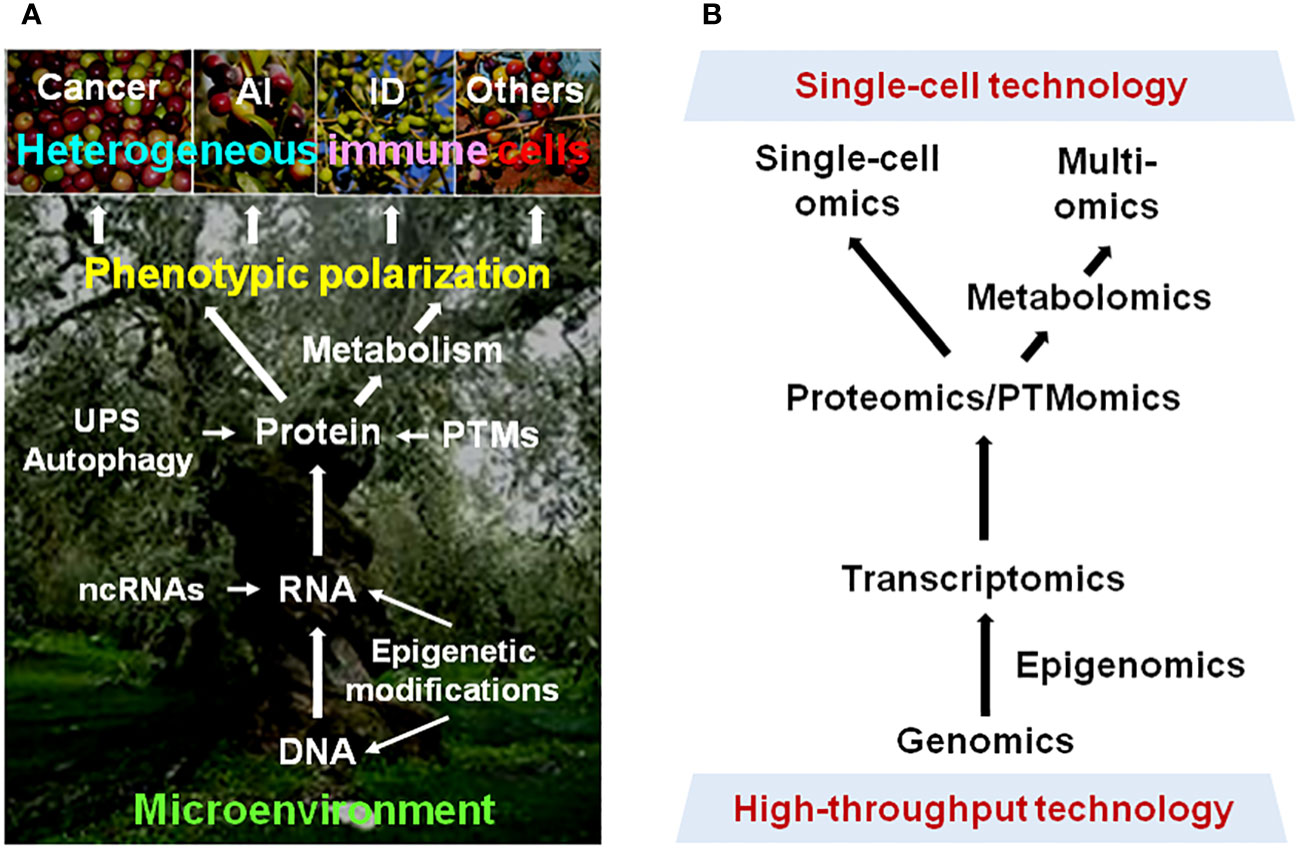
Figure 1 Phenotypic plasticity of immune cells in cancer and non-cancer diseases. (A) Multiple dimensions involving the mechanisms underlying the polarization of immune cells into heterogeneous phenotypes with diverse (even opposite) functions in different pathological scenarios. (B) Modern approaches for resolving the highly complex mechanisms for plasticity of immune cells in a disease-specific manner, thereby leading to immune microenvironment-targeted therapy for cancer and non-cancer diseases. AI, autoimmune disease; ID, inflammatory disease; UPS, ubiquitin-proteasome system; ncRNA, non-coding RNA; PTM, post-translational modification.
YD: Conceptualization, Funding acquisition, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. DR: Conceptualization, Writing – review & editing. YH: Conceptualization, Writing – review & editing. HY: Conceptualization, Resources, Writing – review & editing.
This study was funded by the National Natural Science Foundation of China (Nos. 81670189, 81870160, and 82270207), the Science and Technology Development Program of the Jilin Province (No. 20190201163JC).
We thank all authors who contributed their valuable work to this Research Topic.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Piccolo V, Curina A, Genua M, Ghisletti S, Simonatto M, Sabò A, et al. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross talks. Nat Immunol (2017) 18(5):530–40. doi: 10.1038/ni.3710
2. Munoz-Rojas AR, Kelsey I, Pappalardo JL, Chen M, Miller-Jensen K. Co-stimulation with opposing macrophage polarization cues leads to orthogonal secretion programs in individual cells. Nat Commun (2021) 12(1):301. doi: 10.1038/s41467-020-20540-2
3. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol (2018) 18(1):35–45. doi: 10.1038/nri.2017.76
4. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science (2014) 345(6204):1251086. doi: 10.1126/science.1251086
5. Chen L, Ge B, Casale FP, Vasquez L, Kwan T, Garrido-Martín D, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell (2016) 167(5):1398–414. doi: 10.1016/j.cell.2016.10.026
6. Stienstra R, Netea-Maier RT, Riksen NP, Joosten LAB, Netea MG. Specific and complex reprogramming of cellular metabolism in myeloid cells during innate immune responses. Cell Metab (2017) 26(1):142–56. doi: 10.1016/j.cmet.2017.06.001
7. Bekkering S, Arts RJW, Novakovic B, Kourtzelis I, van der Heijden CDCC, Li Y, et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell (2018) 172(1-2):135–46. doi: 10.1016/j.cell.2017.11.025
8. Schito L, Rey S. Cell-autonomous metabolic reprogramming in hypoxia. Trends Cell Biol (2018) 28(2):128–42. doi: 10.1016/j.tcb.2017.10.006
9. Zhang Q, Cao X. Epigenetic remodeling in innate immunity and inflammation. Annu Rev Immunol (2021) 39:279–311. doi: 10.1146/annurev-immunol-093019-123619
10. Stricker SH, Koferle A, Beck S. From profiles to function in epigenomics. Nat Rev Genet (2017) 18(1):51–66. doi: 10.1038/nrg.2016.138
11. Jin F, Li J, Guo J, Doeppner TR, Hermann DM, Yao G, et al. Targeting epigenetic modifiers to reprogramme macrophages in non-resolving inflammation-driven atherosclerosis. Eur Heart J Open (2021) 1(2):oeab022. doi: 10.1093/ehjopen/oeab022
12. Yang T, Liu X, Kumar SK, Jin F, Dai Y. Decoding DNA methylation in epigenetics of multiple myeloma. Blood Rev (2022) 51:100872. doi: 10.1016/j.blre.2021.100872
13. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol (2019) 20(10):608–24. doi: 10.1038/s41580-019-0168-5
14. Feinberg AP. The key role of epigenetics in human disease prevention and mitigation. N Engl J Med (2018) 378(14):1323–34. doi: 10.1056/NEJMra1402513
15. Stillman B. Histone modifications: insights into their influence on gene expression. Cell (2018) 175(1):6–9. doi: 10.1016/j.cell.2018.08.032
16. Narita T, Weinert BT, Choudhary C. Functions and mechanisms of nonhistone protein acetylation. Nat Rev Mol Cell Biol (2019) 20(3):156–74. doi: 10.1038/s41580-018-0081-3
17. Yang H, Sun Y, Li Q, Jin F, Dai Y. Diverse epigenetic regulations of macrophages in atherosclerosis. Front Cardiovasc Med (2022) 9:868788. doi: 10.3389/fcvm.2022.868788
18. Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature (2019) 574(7779):575–80. doi: 10.1038/s41586-019-1678-1
19. Izzo LT, Wellen KE. Histone lactylation links metabolism and gene regulation. Nature (2019) 574(7779):492–3. doi: 10.1038/d41586-019-03122-1
20. Ivashkiv LB. The hypoxia-lactate axis tempers inflammation. Nat Rev Immunol (2020) 20(2):85–6. doi: 10.1038/s41577-019-0259-8
21. Mehla K, Singh PK. Metabolic regulation of macrophage polarization in cancer. Trends Cancer (2019) 5(12):822–34. doi: 10.1016/j.trecan.2019.10.007
22. Murphy MP. Rerouting metabolism to activate macrophages. Nat Immunol (2019) 20(9):1097–9. doi: 10.1038/s41590-019-0455-5
23. Damgaard RB. The ubiquitin system: from cell signalling to disease biology and new therapeutic opportunities. Cell Death Differ (2021) 28(2):423–6. doi: 10.1038/s41418-020-00703-w
24. Wang W, Sun Y, Liu X, Kumar SK, Jin F, Dai Y. Dual-targeted therapy circumvents non-genetic drug resistance to targeted therapy. Front Oncol (2022) 12:859455. doi: 10.3389/fonc.2022.859455
25. Chang E, Pelosof L, Lemery S, Gong Y, Goldberg KB, Farrell AT, et al. Systematic review of PD-1/PD-L1 inhibitors in oncology: from personalized medicine to public health. Oncologist (2021) 26(10):e1786–99. doi: 10.1002/onco.13887
26. Kluger HM, Tawbi H, Feltquate D, LaVallee T, Rizvi NA, Sharon E, et al. Society for Immunotherapy of Cancer (SITC) checkpoint inhibitor resistance definitions: efforts to harmonize terminology and accelerate immuno-oncology drug development. J Immunother Cancer (2023) 11(7):e007309. doi: 10.1136/jitc-2023-007309
27. Chow A, Perica K, Klebanoff CA, Wolchok JD. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat Rev Clin Oncol (2022) 19(12):775–90. doi: 10.1038/s41571-022-00689-z
28. Kojima Y, Volkmer JP, McKenna K, Civelek M, Lusis AJ, Miller C, et al. CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature (2016) 536(7614):86–90. doi: 10.1038/nature18935
29. Wang F, Liu Y-H, Zhang T, Gao J, Xu Y, Xie G-Y, et al. Aging-associated changes in CD47 arrangement and interaction with thrombospondin-1 on red blood cells visualized by super-resolution imaging. Aging Cell (2020) 19(10):e13224. doi: 10.1111/acel.13224
30. Han S, Bao X, Zou Y, Wang L, Li Y, Yang L, et al. D-lactate modulates M2 tumor-associated macrophages and remodels immunosuppressive tumor microenvironment for hepatocellular carcinoma. Sci Adv (2023) 9(29):eadg2697. doi: 10.1126/sciadv.adg2697
Keywords: immune cells, epigenetic remodeling, transcriptional reprogramming, cancer, non-cancer disease
Citation: Dai Y, Ren D, He Y and Yi H (2023) Editorial: Epigenetic, metabolic, and transcriptional regulation of immune cell plasticity and functions in cancer and non-cancer diseases. Front. Immunol. 14:1284124. doi: 10.3389/fimmu.2023.1284124
Received: 28 August 2023; Accepted: 18 September 2023;
Published: 25 September 2023.
Edited and Reviewed by:
Lin Qi, Central South University, ChinaCopyright © 2023 Dai, Ren, He and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Dai, ZGFpeXVuQGpsdS5lZHUuY24=; Dong Ren, ZHJlbjNAaHMudWNpLmVkdQ==; Yafeng He, aGV5OUBtYWlsLm5paC5nb3Y=; Huanfa Yi, eWlodWFuZmFAamx1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.