- 1Department of Gastroenterology, The Third Xiangya Hospital of Central South University, Changsha, China
- 2Hunan Key Laboratory of Non-Resolving Inflammation and Cancer, The Third Xiangya Hospital, Central South University, Changsha, China
- 3Center for Medical Genetics and Hunan Key Laboratory of Medical Genetics, School of Life Sciences, Central South University, Changsha, Hunan, China
- 4Xiangya School of Medicine, Central South University, Changsha, Hunan, China
Background: Ulcerative colitis (UC) is a lifelong inflammatory disease affecting the rectum and colon with numerous treatment options that require an individualized treatment plan. Histone modifications regulate chromosome structure and gene expression, resulting in effects on inflammatory and immune responses. However, the relationship between histone modification-related genes and UC remains unclear.
Methods: Transcriptomic data from GSE59071 and GSE66407 were obtained from the Gene Expression Omnibus (GEO), encompassing colonic biopsy expression profiles of UC patients in inflamed and non-inflamed status. Differentially expressed gene (DEG) analyses, functional enrichment analyses, weighted gene co-expression network analysis (WGCNA), and random forest were performed to identify histone modification-related core genes associated with UC inflammation. Features were screened through the least absolute shrinkage and selection operator (LASSO) and support vector machine‐recursive feature elimination (SVM‐RFE), establishing a molecular inflammatory predictive model using logistic regression. The model was validated in the GSE107499 dataset, and the performance of the features was assessed using receiver operating characteristic (ROC) and calibration curves. Immunohistochemistry (IHC) staining of colonic biopsy tissues from UC patients treated with infliximab was used to further confirm the clinical application value. Univariate logistic regression on GSE14580 highlighted features linked to infliximab response.
Results: A total of 253 histone modification-related DEGs were identified between inflammatory and non-inflammatory patients with UC. Seven key genes (IL-1β, MSL3, HDAC7, IRF4, CAMK2D, AUTS2, and PADI2) were selected using WGCNA and random forest. Through univariate logistic regression, three core genes (CAMK2D, AUTS2, and IL-1β) were further incorporated to construct the molecular inflammatory predictive model. The area under the curve (AUC) of the model was 0.943 in the independent validation dataset. A significant association between CAMK2D protein expression and infliximab response was observed, which was validated in another independent verification set of GSE14580 from the GEO database.
Conclusion: The molecular inflammatory predictive model based on CAMK2D, AUTS2, and IL-1β could reliably distinguish the mucosal inflammatory status of UC patients. We further revealed that CAMK2D was a predictive marker of infliximab response. These findings are expected to provide a new evidence base for personalized treatment and management strategies for UC patients.
Introduction
Ulcerative colitis (UC) is a subtype of inflammatory bowel disease (IBD) characterized by recurrent and remitting chronic superficial mucosal ulceration and inflammation of the colon and rectum. The specific etiology of UC remains incompletely understood, while genetic susceptibility, immunological factors, and multiple environmental factors seem to contribute to the development of UC (1). The conventional therapies for ulcerative colitis include aminosalicylates, corticosteroids, and immunomodulators. A series of advanced treatments such as biologics and small-molecule drugs have emerged along with increased knowledge of the pathogenesis underlying UC. Nevertheless, the above available treatments remain unsatisfactory, possibly due to individual variations in patients’ intestinal inflammation molecular expression. For both patients and clinicians, selecting from various treatment options frequently presents a clinical challenge. Prior studies indicate that intestinal inflammation of patients with UC is associated with long-term outcomes and drug response (2, 3). Disease molecular measures may augment current endpoints and provide a more profound understanding of the disease mechanism (4). Therefore, the exploration of a molecular predictive model for inflammation might facilitate the customization of patient treatments.
Histone modifications regulate chromatin states and gene transcription, including histone acetylation, methylation, and phosphorylation. Emerging evidence suggests that patients with UC exhibit aberrant histone modifications accompanying inflammatory activity (5). Compared to healthy individuals, histone modification levels have been altered in the inflamed colonic mucosa of UC patients (6). Furthermore, increased histone acetylation at the promoters of pro-inflammatory cytokine genes, such as tumor necrosis factor-alpha (TNF-α) and interleukin-1β (IL-1β), has been associated with their upregulation in UC (7, 8). Similarly, altered histone methylation patterns have been linked to dysregulated expression of genes involved in immune cell activation and tissue damage (9, 10). Hence, exploring potential molecular biomarkers could provide insights into disease activity, and histone modifications have emerged as a promising avenue in this regard.
Biologic agents are crucial therapeutic options for patients with moderate-to-severe UC, and infliximab ranked first with the highest evidence for efficacy in inducing endoscopic improvement (11, 12). However, some patients show primary non-response or secondary loss of response to infliximab (13). Numerous studies have demonstrated that therapeutic drug and anti-drug antibody concentrations, clinical characteristics, mucosal immune features, gut microbial dysbiosis, and genetic factors are predictors of infliximab response (14–19). Notably, the anti-inflammatory effect of butyrate primarily arises from the activity of histone deacetylase inhibitors (20). Infliximab alleviates inflammation by binding to TNF-α. Similarly, histone modification could affect the expression of TNF-α and regulate the function of immune cells and anti-inflammatory pathways. It implied a possible synergistic effect of histone modification in the efficacy of infliximab treatment, making it a promising therapeutic target for UC (21). However, limited research has investigated the significance of histone modifications in forecasting inflammation and infliximab response among individuals with ulcerative colitis.
To address this gap in knowledge, the gene expression profiles of inflamed and non-inflamed colonic biopsies were retrieved from UC patients using the Gene Expression Omnibus (GEO) database, and histone modification-associated differentially expressed genes (DEGs) were identified using Wilcox analysis. Weighted gene co-expression network analysis (WGCNA) was employed to identify co-expressed gene modules. The key module eigengenes were functionally annotated through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Key genes were then selected using the random forest and support vector machine‐recursive feature elimination (SVM‐RFE) algorithms. Finally, a logistic regression model was constructed using the key genes for prediction purposes. The understanding of the gene expression regulation and immune response influences of histone modifications is expected to develop new therapeutic strategies for personalized approaches to UC management.
Materials and methods
Data selection and processing
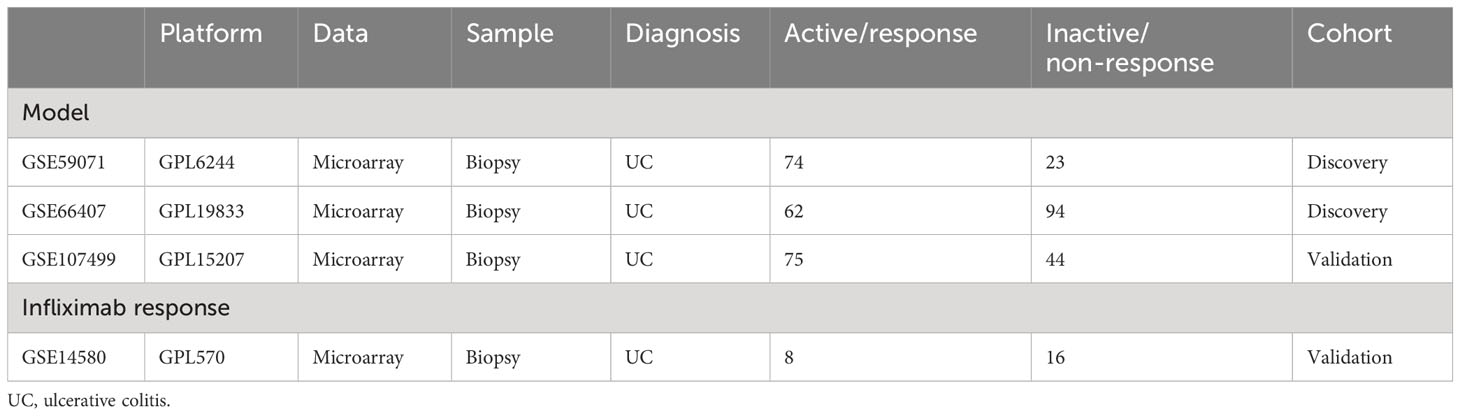
Public data and full clinical annotation were downloaded from the GEO database. Combining GSE59071 and GSE66407 as the training cohort, the data standardization process was carried out before analysis, including data alignment, missing value processing, and probe annotation. When multiple probes corresponded to the same gene, the average value was selected as the expression level of the gene. Inflamed and non-inflamed colonic biopsies from UC patients were selected for the following analysis. Using the “sva” package to reduce the batch effect in different independent cohorts, GSE107499 and GSE14580 were selected as the test cohorts for the molecular inflammatory predictive model and infliximab response validation, respectively (Table 1).
Screening for histone modification-related DEGs
In the training dataset, samples were categorized into UC_noninflammation (n = 117) and UC_inflammation (n = 136) groups. Histone modification-associated DEGs were identified using Wilcox analysis, setting an adjusted p-value threshold of <0.05. The significance criteria for DEGs were established accordingly. A volcano plot was generated using the R package “EnhancedVolcano”, while the heatmap of histone modification-related DEGs (22) was created using the R package “pheatmap”.
WGCNA and functional enrichment analysis
WGCNA identified co-expressed gene modules, exploring gene network–phenotype relationships and key network genes. Using the “WGCNA” R package, histone modification-related genes were analyzed. Gene correlations were calculated, and weighted connections were established. A hierarchical clustering tree was constructed using correlation coefficients, indicating diverse gene modules through colors. The crucial module linked to inflammation was pinpointed, and module eigengenes were extracted for further analysis.
GO and KEGG pathway functional enrichment analyses of the module eigengenes were performed using the “clusterProfiler”, “org.Hs.eg.db”, “enrichplot”, and “ggplot2” packages in R software. Terms with p < 0.05 and q < 0.05 were considered significantly enriched.
Select core histone modification-related DEGS
Random forest was used to screen key genes using the “randomforest” package. The error rate for 1–500 trees was calculated, and the lowest error rate and the best number of stable trees were determined. Further, the random forest method was used to screen the key genes, and the Gini coefficient method was used to calculate the dimensional significance value. Genes with a significant value greater than three were considered key genes. The intersection of the module eigengenes and the key genes were regarded as the core genes. Correlations between core genes were analyzed.
Analysis of immune cell infiltration
We assessed the proportions of different infiltrating immune cell types between the UC_inflammation group and UC_noninflammation group using the CIBERSORT algorithm. We used LM22 and CIBERSORT matrices to predict the proportion of 22 infiltrating immune cell subtypes in a single sample of the dataset (23). We identified differentially enriched immune cells between UC_inflammation and UC_noninflammation samples and analyzed their correlation with core genes.
Construction of a molecular inflammatory predictive model
Univariate logistic regression was used to screen core genes significantly associated with inflammation. To avoid model overfitting and filter out the most important events, the “glmnet” package in R was used to screen the most significant predictors using the least absolute shrinkage and selection operator (LASSO) regression (24). Meanwhile, an SVM‐RFE model was established using the “e1071” package in R, which was compared by the average misjudgment rates of their fivefold cross‐validations. Furthermore, a multivariate logistic regression model based on the most important predictors was constructed. For univariate and multivariate logistic regression analyses, p-value <0.05 was considered the cutoff for the significant correlation.
Nomogram plot and calibration curve
A nomogram was established for a better clinical application of the model (25). A calibration curve was constructed using the “rms” package in R to compare the difference between the model-predicted probability and the actual probability.
Verification using validation datasets
On the validation dataset (GSE107499), the multivariate logistic regression model was tested for effectiveness verification. The area under the curve (AUC) was calculated using the “ROCR” package in R. Univariate logistic regression was used on the validation dataset (GSE14580) to screen the core genes significantly associated with infliximab response.
Clinical specimens and immunohistochemistry
Clinical data and endoscopic colon biopsy paraffin sections from February 2020 to December 2022 from 40 UC patients in the Third Xiangya Hospital were retrospectively collected. The inflamed (n = 20) and non-inflamed (n = 20) lesions were respectively obtained from the endoscopic activity and remission patients evaluated using Mayo endoscopic subscore from colonoscopy reports (26). The 20 patients presenting with endoscopic inflammation at baseline underwent subsequent treatment with infliximab. They were followed up for 8 weeks, and the infliximab response was assessed based on the Mayo endoscopic subscore. Ethical approval for the current study was granted by the institutional review board (IRB) of the Third Xiangya Hospital, Central South University (No. E.23188). More available details of these patients are depicted in Supplementary Table S1.
Paraffin sections were dewaxed, rehydrated, and incubated with 3% hydrogen peroxide for 15 minutes to block endogenous peroxidase. Then, sections were placed in 0.01 mol/L sodium citrate buffer for antigen repair, followed by washing with phosphate-buffered saline (pH = 7.4) two times for 5 minutes each and blocking with normal goat serum for 30 minutes. Immunostaining was performed by incubating with anti-CAMK2D antibody (1:100, DF12015, Affinity Biosciences, Jiangsu, China) or anti-IL-1β antibody (1:80, AF06643, AiFang biological, Hunan, China) at 4°C overnight. Sections were then washed in phosphate-buffered saline with Tween three times for 5 minutes each and incubated with secondary antibody for 30 minutes at room temperature. Bound antibodies were detected using diaminobenzidine, and slides were counterstained with hematoxylin.
A semi-quantitative scoring system was used to assess the expression of CAMK2D and IL-1β (protein expression = expression intensity + expression area). Expression intensity was scored using a 4-point scale from 0 to 3. Expression area according to the percentage of positive-stained cells was scored using a 5-point scale from 0 to 4. Finally, the summation scores divided each slide into low expression (0–2) and high expression (3–7). These results were confirmed by two independent pathologists who were blinded to patient clinical data.
Statistical analysis
All the statistical analyses and visualization of the GEO set samples were performed using R 4.1.1. A two-sided p-value of <0.05 was considered statistically significant unless otherwise noted.
Results
Screening for histone modification-related DEGs
The flowchart of the dataset analysis is described in Figure 1A. We merged the GSE59071 and GSE66407 datasets and removed batch effects. To further explore DEGs between inflamed and non-inflamed tissues, we conducted a differential analysis of 253 patients diagnosed with ulcerative colitis and 431 histone modification-related genes. We identified a total of 253 histone modification-related DEGs, including 129 downregulated genes and 124 upregulated genes (Figures 1B–E; Supplementary Table S2).
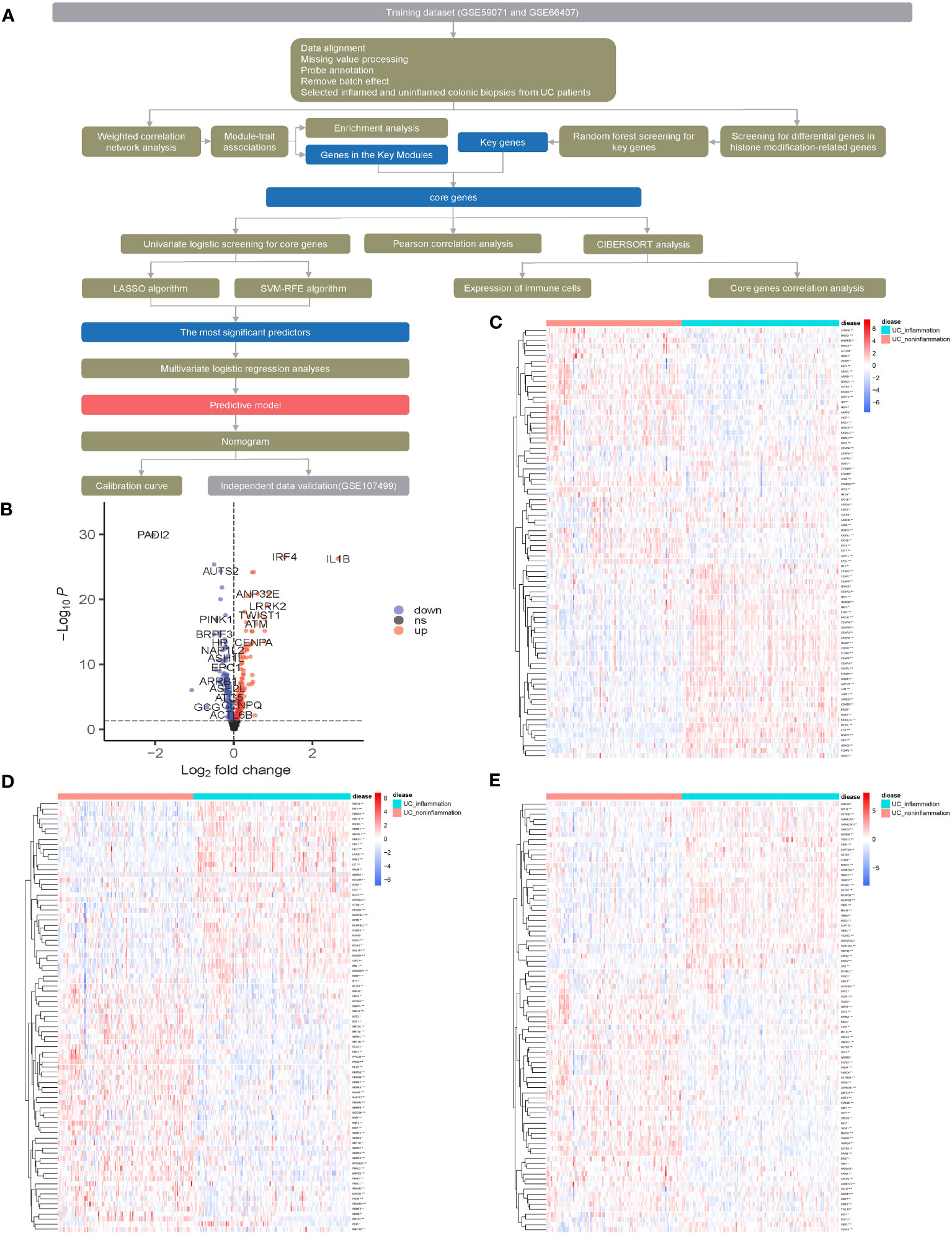
Figure 1 Screening for histone modification-related differentially expressed genes (DEGs) between inflamed and non-inflamed tissues of ulcerative colitis patients. (A) Flowchart of the dataset analysis. (B) Volcano plot and (C–E) heatmap of histone modification-related DEGs between inflamed and non-inflamed tissues.
Selecting key DEGs related to inflammation of UC
The above 253 histone modification-related DEGs were classified into 10 modules by hierarchical clustering tree (Figures 2A, B). The blue module was identified as having the most significant positive correlation with inflammation (0.64) related to ulcerative colitis. GO and KEGG functional enrichment analyses on the 46 genes contained in the blue module were performed. A total of 31 genes were identified to be significantly enriched in pathways related to biological processes, 11 genes in pathways related to cellular components, and 14 genes in pathways related to molecular function (p < 0.05 and q < 0.05). The top five biological processes, cellular components, and molecular function pathways are shown in Figures 2C–E, respectively. The KEGG annotation results indicated eight genes significantly enriched in Th17 cell differentiation and acute myeloid leukemia pathways (Figure 2F). The lowest error rate and the best number of stable trees were determined using random forest (Figure 3A). The most contributable 11 genes were intersected in the random forest (MeanDecreaseGini > 3) and the significant 46 module eigengenes in WGCNA (Figures 3B, C). Seven genes (IL-1β, MscS-Like 3 [MSL3], histone deacetylase 7 [HDAC7], interferon regulatory factor 4 [IRF4], calcium/calmodulin-dependent kinase II-delta [CAMK2D], autism susceptibility candidate 2 [AUTS2], and peptidyl arginine deiminase 2 [PADI2]) were assessed as the core genes. Higher expression of IL-1β, MSL3, HDAC7, and IRF4 and lower expression of CAMK2D, AUTS2, and PADI2 were correlated with inflammation in UC patients (Figure 3D). Further correlation analysis revealed a universal correlation between core histone modification-related genes in UC inflammation (Figure 3E).
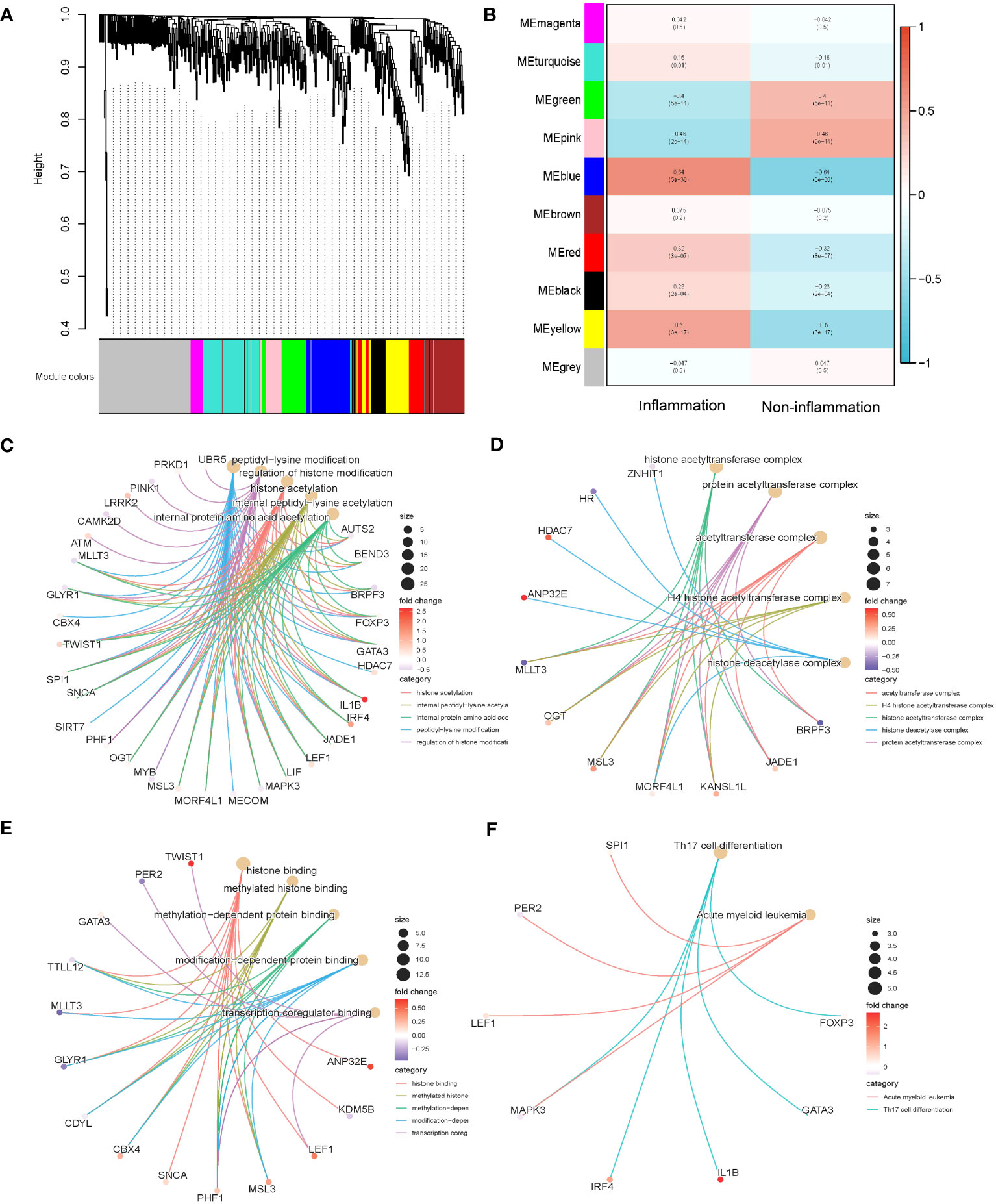
Figure 2 Co-expression and functional enrichment analysis of histone modification-related differentially expressed genes. (A) Gene dendrogram and module colors. (B) Correlation results between the 10 modules and two inflammatory states. (C) GO functional enrichment analysis of the intersecting genes with BP terms in the blue module. (D) GO functional enrichment analysis of the intersecting genes with CC terms in the blue module. (E) GO functional enrichment analysis of the intersecting genes with MF terms in the blue module. (F) KEGG annotation map of genes in the blue module. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes.
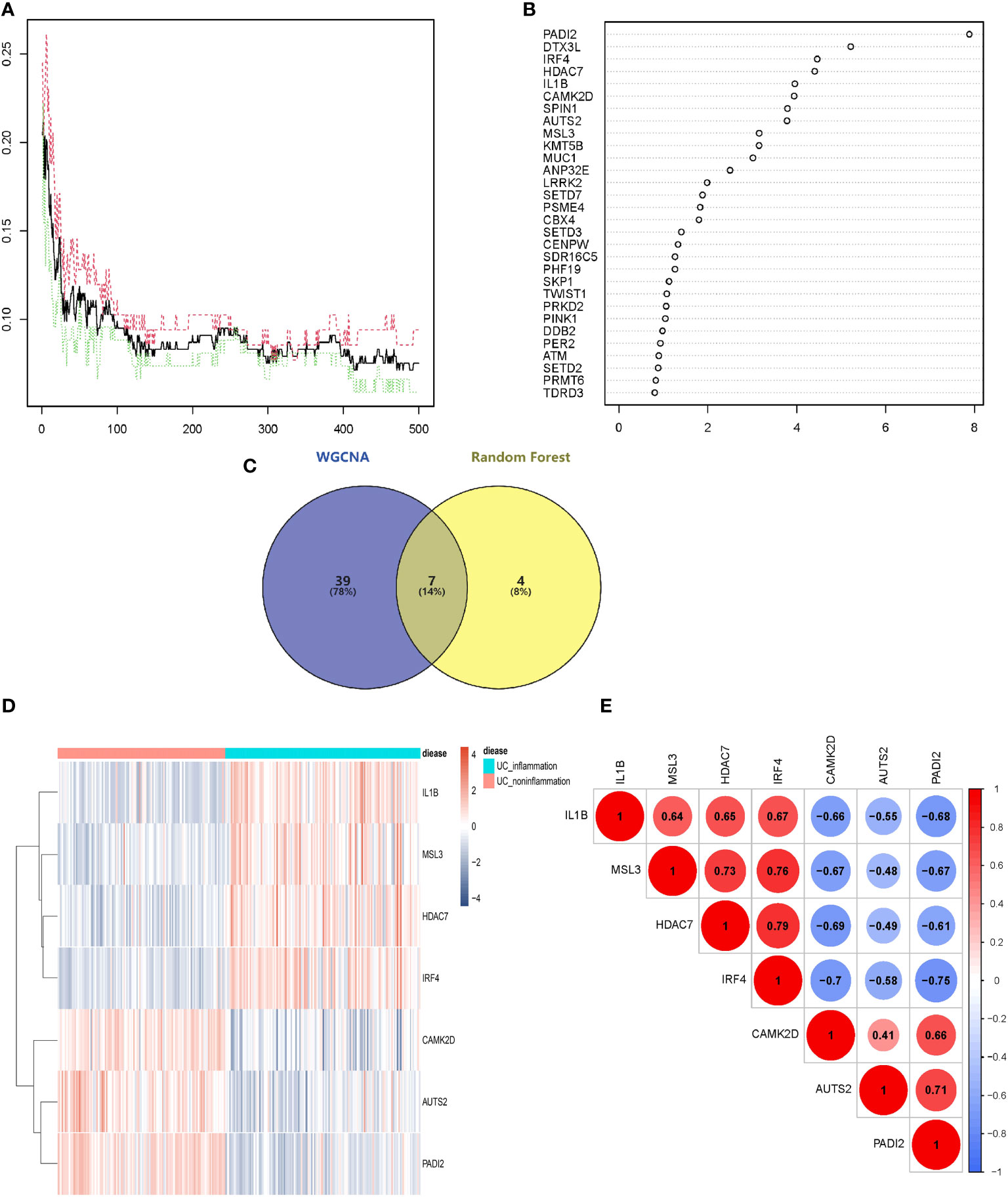
Figure 3 Selecting core histone modification-related differentially expressed genes. (A) The lowest error rate and the best number of stable trees were determined. (B) The top 11 significant genes were screened (seed = 213, MeanDecreaseGini > 3). (C) Venn diagram indicating intersections of leading genes in WGCNA and top significant genes in random forest. (D) Heatmap to explore the correlations between core genes and inflammatory status of UC. (E) Correlation analysis of seven core histone modification-related genes. UC, ulcerative colitis; WGCNA, weighted gene co-expression network analysis.
Analysis of immune cell infiltration
We evaluated the distribution and calculated the immune cell proportion of 22 immune cell types using CIBERSORT (Figures 4A, B). Significant differences in the proportion of immune cell features were exhibited between inflamed and non-inflamed UC patients, including plasma cells, CD8+ T cells, CD4+ memory resting T cells, CD4+ memory activated T cells, follicular helper T cells, regulatory T cells, γδ T cells, activated NK cells, M0 macrophages, M1 macrophages, M2 macrophages, activated dendritic cells, resting mast cells, activated mast cells, eosinophils, and neutrophils. The above immune cells may play an important role in the inflammatory process of UC.
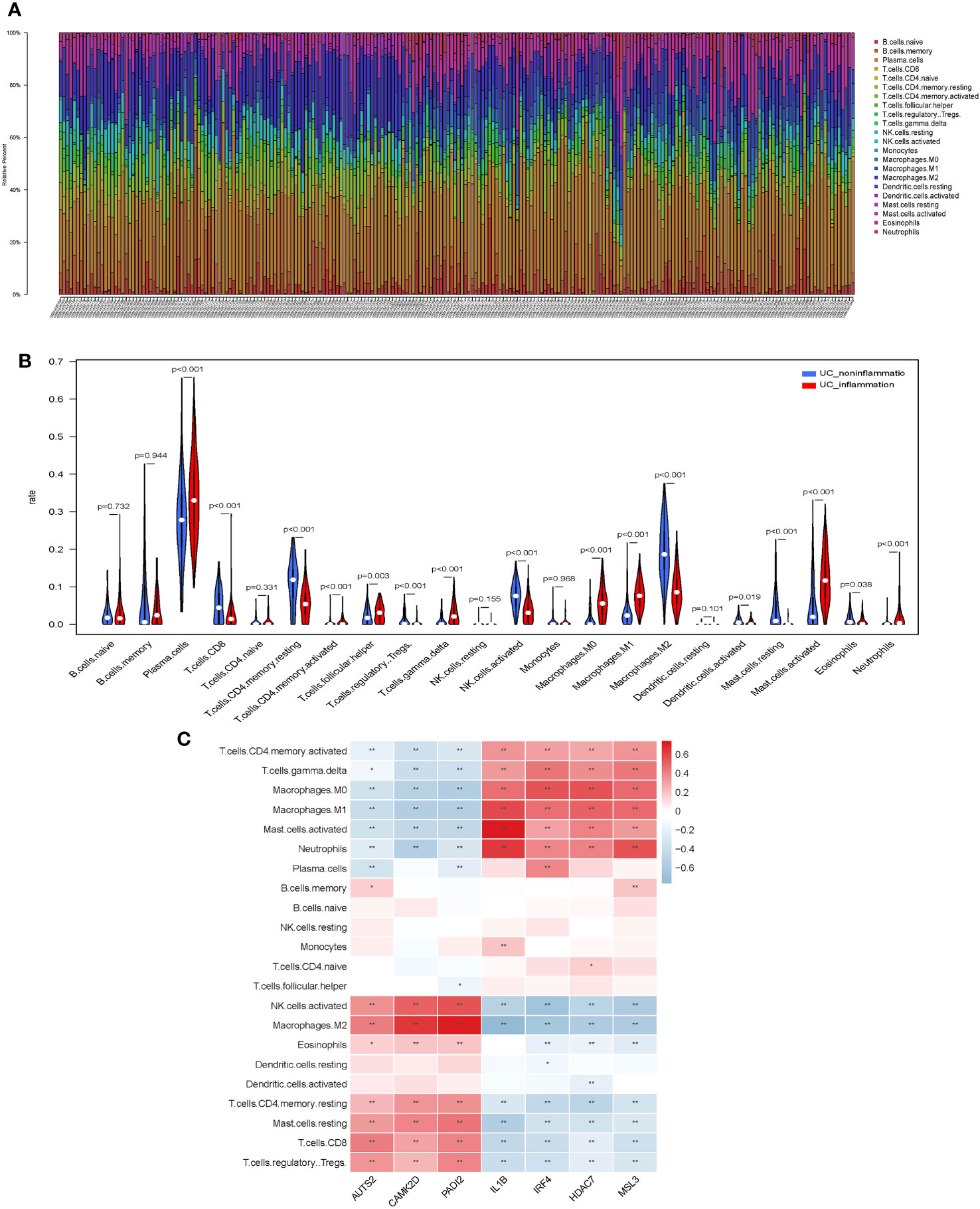
Figure 4 Immunological features of core histone modification-related genes in UC inflammation. (A) Bar plot of percentage infiltration of 22 immune cells in each sample. (B) Differential infiltration of the 22 immune cells in UC patients with endoscopic inflamed and non-inflamed conditions. (C) Correlation heatmap of 22 immune cell types with seven core genes. UC, ulcerative colitis. Asterisks represent levels of significance (*p < 0.05, **p < 0.01).
Subsequently, we assessed the correlation between core histone modification-related genes and immune cells in UC inflammation (Figure 4C). Activated NK cells, M2 macrophages, eosinophils, CD4+ memory resting T cells, resting mast cells, CD8+ T cells, and regulatory T cells were more abundant in the non-inflammatory UC patients and positively correlated with AUTS2, CAMK2D, and PADI2. On the contrary, CD4+ memory-activated T cells, γδ T cells, M0 macrophages, M1 macrophages, activated mast cells, and neutrophils were more abundant in the inflammatory UC patients and positively correlated with IL-1β, IRF4, HDAC7, and MSL3.
Constructing and validating the molecular inflammatory predictive model
Univariate logistic regression, LASSO regression, and the SVM‐RFE model were performed to screen potential inflammatory predictive markers from core histone modification-related genes in UC inflammation (Figures 5A–D). As shown in Figures 5E, F, three core genes (AUTS2, CAMK2D, and IL-1β) were further applied in multivariate logistic regression. A nomogram model was established (Figure 5G), and the C-index value of the model was 0.948. Calibration curves showed the accuracy of this model in predicting the inflammation rate of UC patients (Figure 5H). The inflammatory predictive model was validated on the GSE107499 dataset, and the AUC value was 0.943, which showed that the model had high accuracy in predicting the inflammatory status of UC patients (Figure 5I).
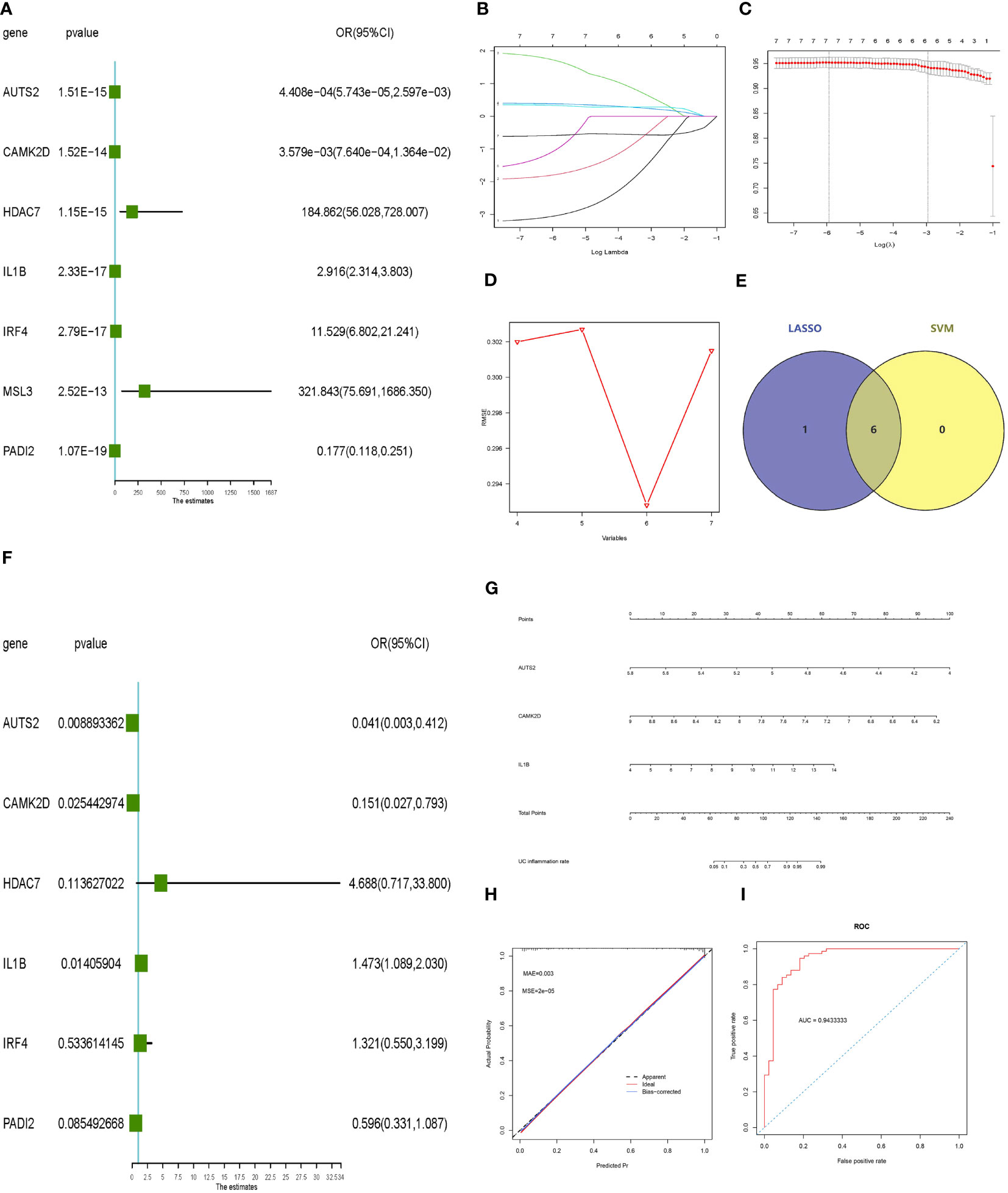
Figure 5 Constructing and validating the inflammatory predictive model. (A) Forest plot lines of significant core genes related to UC inflammation using univariate logistic regression. (B, C) LASSO regression and (D) SVM‐RFE model of the combined training dataset. (E) Venn diagram of core genes related to UC inflammation between LASSO analysis and SVM‐RFE model. (F) Forest plot lines of significant core genes related to UC inflammation using multivariate logistic regression. (G, H) Nomogram plot and calibration curve of the inflammatory predictive model. (I) Evaluation of the inflammatory predictive model in the validation dataset. UC, ulcerative colitis; LASSO, least absolute shrinkage and selection operator; SVM‐RFE, support vector machine‐recursive feature elimination.
Predictive gene validation in UC patients and infliximab response
We performed immunohistochemistry to detect the relationship between endoscopic inflammatory status and CAMK2D protein expression. We leveraged clinical data on erythrocyte sedimentation rate (ESR) from the electronic medical record system of the Third Xiangya Hospital, which is a common clinical inflammation indicator for disease diagnosis and management of UC patients. The level of ESR was lower in patients with higher CAMK2D expression (Supplementary Figure S1). A negative correlation between the histology severity of colon biopsies and CAMK2D protein expression level was found (p = 0.0024). To our knowledge, IL-1β is a traditional inflammatory cytokine and has been reported as highly expressed in UC patients. As shown in Figures 6A–D, CAMK2D protein expression was negatively correlated with endoscopic inflammation, whereas IL-1β protein expression was positively correlated with endoscopic inflammation. These results indicated that CAMK2D worked as an anti-inflammation molecule in UC.
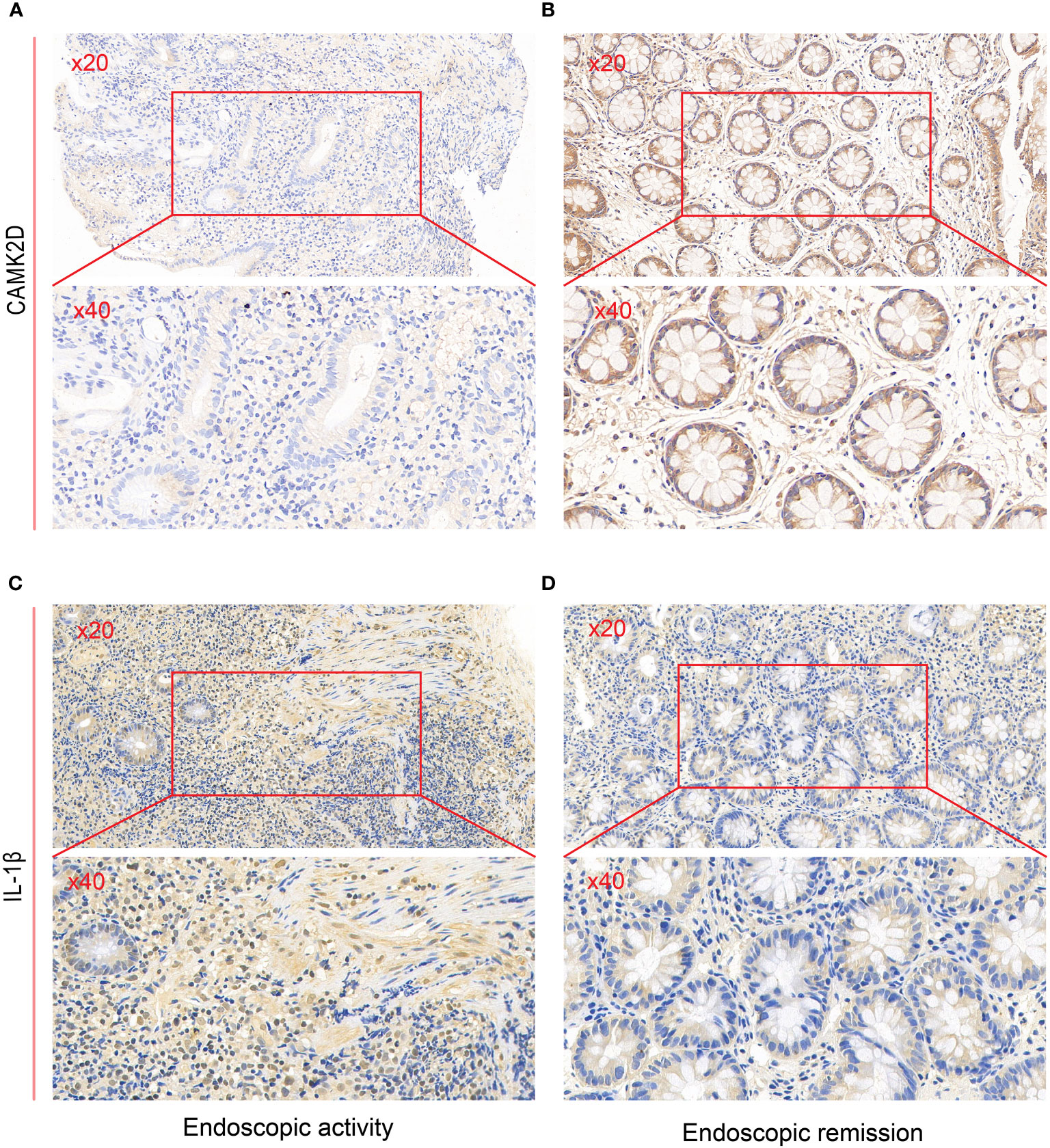
Figure 6 Validating predictive genes (IL-1β and CAMK2D) by IHC in UC patients. IHC staining of CAMK2D in (A) endoscopic activity and (B) endoscopic remission patients. IHC staining of IL-1β in (C) endoscopic activity and (D) endoscopic remission patients. UC, ulcerative colitis; IHC, immunohistochemistry.
We further followed up on 20 UC patients with endoscopic inflammation at baseline and initiated infliximab treatment for 8 weeks. Patients with high CAMK2D protein expression were associated with infliximab response (Supplementary Figure S2). We validated the result in GSE14580 (Supplementary Figure S3), which indicated a predictive role of CAMK2D in infliximab response.
Discussion
Over the past two decades, the goal in treating IBD has shifted from symptom relief to achieving consistent clinical and endoscopic remission to halt the natural disease progression and reduce the risk of complications and surgery; however, the rate is not encouraging (27). A meta-analysis reported that UC patients with endoscopic remission after initiation of therapy were associated with improved long-term outcomes such as corticosteroid-free clinical and endoscopic remission (28). Long-term and persistent colonic inflammation in UC increases the risk of relapse, steroid dependency, colectomy, and even colorectal cancer (29, 30). Therefore, it is important to focus on endoscopic inflammation status at baseline.
A range of inflammation biomarkers were upregulated in UC, which exacerbates chronic colon inflammation and mucosal damage, such as TNF-α, IL-1β, IL-6, and IFN-γ. Histone acetylation promotes these pro-inflammatory gene expressions and inflammasome activation (7, 8). Evidence showed that aberrant histone acetylation exists in UC patients (6) and colitis mice colon (31), while using histone deacetylase (HDAC) inhibitors could attenuate colitis by suppressing pro-inflammatory cytokines (32, 33), acetylating transcription of STAT1 and NF-κB (34, 35), increasing the formation of IL-35 via EBI3 (36), interacting with non-coding RNAs to fine-tune inflammatory responses (37), etc. Additionally, aberrant histone methylation patterns contribute to dysregulated inflammation (38, 39). Therefore, targets for histone modification may be potential candidate treatments for UC patients with endoscopic inflammation (21, 40). Machine learning algorithms are increasingly being used to create decision models that aid in disease diagnosis and treatment. In the current study, we identified histone modification-related inflammatory genes of UC within endoscopic activity versus endoscopic remission groups through integrated bioinformatics analysis, suggesting a key role of IL-1β, AUTS2, and CAMK2D in UC endoscopic inflammatory state. IL-1β is a classical pro-inflammatory cytokine regulated by histone modification that increases intestinal epithelial permeability, recruits neutrophils to inflamed colon tissue, and compromises the epithelial barrier (9, 10, 41, 42). Canakinumab treatment demonstrated significantly higher response rates in patients with very early onset IBD (43). Preclinical studies supported the effectiveness of anti-IL-1β monoclonal antibodies 7F IgG and FL-BsAb1/17 in mitigating colitis (44, 45), while anakinra showed efficacy in the UC model of primary anti-TNF non-responders (46). Targeting IL-1β is a potential therapeutic option for ameliorating UC. AUTS2, mainly expressed in the brain, is linked to various neurological diseases (47). A genome-wide association study reported a downregulated expression of AUTS2 in endoscopic pinch biopsies of Crohn’s disease (CD) patients compared to healthy individuals (48). CAMK2D has been implicated in the modulation of inflammatory processes. NF-κB regulates CAMK2D transcription, and CAMK2D protein modulates NF-κB signaling and leukocyte trans-endothelial migration to inflammatory sites (49–51). Therefore, histone modification may play an important role in UC, which needs to be explored further.
Emerging evidence suggests the innate and adaptive immune response plays a crucial role in inducing intestinal inflammation (52). We also identified significant differences in the proportions of some important immune cells such as plasma cells, T cells, NK cells, macrophages, neutrophils, and mast cells between inflamed and non-inflamed mucosa in UC patients using CIBERSORT. Mounting evidence reported the association between these immune cells and histone modification-related genes. Toll-like receptor (TLR) signaling, a key factor in the innate immune response, is characterized by histone modifications, which induce the expression of inflammation-related genes (53). N-Butyrate enhances histone acetylation in macrophages, downregulates pro-inflammatory mediators, and regulates B-lymphocyte differentiation (54). It helps maintain tolerance to commensal organisms by dampening colonic macrophage response (55). Moreover, monocytes differentiate into dendritic cells through CD14 and CD209 gene regulation, influenced by histone modifications, impacting IBD pathogenesis (56, 57). HDAC6 inhibitor may attenuate colonic inflammation by reducing the activation of neutrophils (58). Additionally, IBD patients exhibit hyperactive effector CD4+ T cells and an imbalance of regulatory T (Treg) and Th17 cells associated with histone deacetylase and methylation (59–62). EZH2, a histone methyltransferase, decreased in IBD patients (63), and the deficiency in Treg cells induces pro-inflammatory cytokines and spontaneous IBD (64). The function of Treg cells in IBD patients has been reported to correlate with treatment response and can be regulated by HDAC inhibitors (18, 65). Consistent with the content reported in the above literature, our data showed that the abundance of macrophages, monocytes, neutrophils, and T cells was associated with histone modification-related genes, such as IL-1β, AUTS2, and CAMK2D. These elements contribute to the importance of immune dysfunction and histone modifications in UC pathogenesis.
We conducted validation on two independent datasets to ensure the robustness of our results. First, we utilized the validation dataset GSE107499 to assess the accuracy of the predictive model that we developed. The model demonstrated excellent performance on this validation dataset as well (AUC > 0.9), indicating that AUTS2, CAMK2D, and IL-1β are indeed potential diagnostic markers capable of distinguishing between endoscopic inflammatory and non-inflammatory UC patients. Subsequently, we followed up 20 UC patients treated with infliximab and revealed an association between CAMK2D high expression and infliximab response, which was further validated in an external dataset. CAMK2D has been reported to be involved in several classical inflammatory pathways associated with UC, including NF-κB, STAT3, IL-6, and TNF-α (50, 66, 67). Dahlen et al. demonstrated lower mucosal IFN-γ, IL-1β, IL-6, and TNF-α mRNA expression in UC patients with anti-TNF response (68). A model based on these serum cytokines can classify UC patients as primary infliximab responders and non-responders (69). Notably, CAMK2D is an IFN-γ-induced gene calculated as a substitute indicator for IFN-γ level, emphasizing its potential in assessing immune responses (70). The findings suggested that CAMK2D may partially explain the poorer immune response in patients with endoscopic inflammatory states and may serve as a predictive marker of infliximab treatment response.
However, there were several limitations to our study. First, our study primarily relied on bioinformatics analysis and limited clinical validation. While we validated the results in different databases, potential bias from different microarray platforms, RNA extraction methods, statistical methods, and limited clinical samples could not be avoided. Thus, more experimental and clinical studies are needed to further prove the current findings. Moreover, we lacked detailed in vitro and in vivo experiments to elucidate the specific mechanisms underlying histone modification genes, particularly CAMK2D, in relation to infliximab response and endoscopic inflammatory status. Finally, due to the COVID-19 pandemic, declined enrollment (71), and limited colonic biopsy, we lacked participants treated with other biologic agents and small-molecule drugs. Therefore, further research is necessary to explore the predictive function of CAMK2D for other treatments.
To conclude, our study identified pivotal molecular mechanisms and potential therapeutic implications of targeting histone modifications for UC. We identified and validated three histone modification-DEGs related to endoscopic inflammatory status. Moreover, CAMK2D has been preliminarily identified as a key predictive biomarker of response to infliximab, potentially exerting its influence on progression through histone modifications. These findings are expected to provide a new evidence base for personalized treatment and management strategies for UC patients.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by The Ethics Committee of China’s Third Xiangya Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by-product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
SY: Conceptualization, Methodology, Formal analysis, Writing – original draft. YL: Conceptualization, Formal analysis, Methodology, Writing – original draft. LC: Methodology, Writing – review & editing. YW: Methodology, Writing – original draft. YH: Formal analysis, Writing – review & editing. QL: Writing – review & editing, Writing – original draft. LT: Writing – review & editing, Methodology. FL: Methodology, Writing – review & editing. XW: Writing – review & editing, Conceptualization, Formal analysis. FA: Conceptualization, Writing – review & editing, Methodology, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The cost of publication was covered by the Natural Science Foundation of Changsha City (kq2208350).
Acknowledgments
Colon specimens were provided by the Third Xiangya Hospital of Central South University with the approval of the Ethics Committee of China’s Third Xiangya Hospital.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1282136/full#supplementary-material
References
1. Du L, Ha C. Epidemiology and pathogenesis of ulcerative colitis. Gastroenterol Clin North Am (2020) 49(4):643–54. doi: 10.1016/j.gtc.2020.07.005
2. de Frias Gomes CG, de Almeida ASR, Mendes CCL, Ellul P, Burisch J, Buhagiar T, et al. Histological inflammation in the endoscopically uninflamed mucosa is associated with worse outcomes in limited ulcerative colitis. Inflammation Bowel Dis (2022) 28(3):350–7. doi: 10.1093/ibd/izab069
3. Cortesi PA, Fiorino G, Peyrin-Biroulet L, Mantovani LG, Jairath V, Paridaens K, et al. Non-invasive monitoring and treat-to-target approach are cost-effective in patients with mild-moderate ulcerative colitis. Aliment Pharmacol Ther (2023) 57(5):486–95. doi: 10.1111/apt.17261
4. Argmann C, Hou R, Ungaro RC, Irizar H, Al-Taie Z, Huang R, et al. Biopsy and blood-based molecular biomarker of inflammation in IBD. Gut (2023) 72(7):1271–87. doi: 10.1136/gutjnl-2021-326451
5. Lin Y, Qiu T, Wei G, Que Y, Wang W, Kong Y, et al. Role of histone post-translational modifications in inflammatory diseases. Front Immunol (2022) 13:852272. doi: 10.3389/fimmu.2022.852272
6. Mokry M, Middendorp S, Wiegerinck CL, Witte M, Teunissen H, Meddens CA, et al. Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology (2014) 146(4):1040–7. doi: 10.1053/j.gastro.2013.12.003
7. Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh , et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol (2013) 14(5):454–60. doi: 10.1038/ni.2550
8. Hwang I, Lee E, Jeon SA, Yu JW. Histone deacetylase 6 negatively regulates NLRP3 inflammasome activation. Biochem Biophys Res Commun (2015) 467(4):973–8. doi: 10.1016/j.bbrc.2015.10.033
9. Wang B, Chen J, Santiago FS, Janes M, Kavurma MM, Chong BH, et al. Phosphorylation and acetylation of histone H3 and autoregulation by early growth response 1 mediate interleukin 1beta induction of early growth response 1 transcription. Arterioscler Thromb Vasc Biol (2010) 30(3):536–45. doi: 10.1161/ATVBAHA.109.193821
10. Schaafsma W, Zhang X, van Zomeren KC, Jacobs S, Georgieva PB, Wolf SA, et al. Long-lasting pro-inflammatory suppression of microglia by LPS-preconditioning is mediated by RelB-dependent epigenetic silencing. Brain Behav Immun (2015) 48:205–21. doi: 10.1016/j.bbi.2015.03.013
11. Singh S, Murad MH, Fumery M, Dulai PS, Sandborn WJ. First- and second-line pharmacotherapies for patients with moderate to severely active ulcerative colitis: an updated network meta-analysis. Clin Gastroenterol Hepatol (2020) 18(10):2179–2191 e6. doi: 10.1016/j.cgh.2020.01.008
12. Burr NE, Gracie DJ, Black CJ, Ford AC. Efficacy of biological therapies and small molecules in moderate to severe ulcerative colitis: systematic review and network meta-analysis. Gut (2021) 71:1976–87. doi: 10.1136/gutjnl-2021-326390
13. Ben-Horin S, Chowers Y. Tailoring anti-TNF therapy in IBD: drug levels and disease activity. Nat Rev Gastroenterol Hepatol (2014) 11(4):243–55. doi: 10.1038/nrgastro.2013.253
14. Liefferinckx C, Minsart C, Toubeau JF, Cremer A, Amininejad L, Quertinmont E, et al. Infliximab trough levels at induction to predict treatment failure during maintenance. Inflammation Bowel Dis (2017) 23(8):1371–81. doi: 10.1097/MIB.0000000000001120
15. Kantasiripitak W, Wicha SG, Thomas D, Hoffman I, Ferrante M, Vermeire S, et al. A model-based tool for guiding infliximab induction dosing to maximize long-term deep remission in children with inflammatory bowel diseases. J Crohns Colitis (2023) 17(6):896–908. doi: 10.1093/ecco-jcc/jjad009
16. Zhou Y, Xu ZZ, He Y, Yang Y, Liu L, Lin Q, et al. Gut microbiota offers universal biomarkers across ethnicity in inflammatory bowel disease diagnosis and infliximab response prediction. mSystems (2018) 3(1). doi: 10.1128/mSystems.00188-17
17. Hoyhtya M, Korpela K, Saqib S, Junkkari S, Nissilä E, Nikkonen A, et al. Quantitative fecal microbiota profiles relate to therapy response during induction with tumor necrosis factor alpha antagonist infliximab in pediatric inflammatory bowel disease. Inflammation Bowel Dis (2023) 29(1):116–24. doi: 10.1093/ibd/izac182
18. Di Sabatino A, Biancheri P, Piconese S, Rosado MM, Ardizzone S, Rovedatti L, et al. Peripheral regulatory T cells and serum transforming growth factor-beta: relationship with clinical response to infliximab in Crohn’s disease. Inflammation Bowel Dis (2010) 16(11):1891–7. doi: 10.1002/ibd.21271
19. Noor NM, Verstockt B, Parkes M, Lee JC. Personalised medicine in Crohn’s disease. Lancet Gastroenterol Hepatol (2020) 5(1):80–92. doi: 10.1016/S2468-1253(19)30340-1
20. Couto MR, Gonçalves P, Magro F, Martel F. Microbiota-derived butyrate regulates intestinal inflammation: Focus on inflammatory bowel disease. Pharmacol Res (2020) 159:104947. doi: 10.1016/j.phrs.2020.104947
21. Felice C, Lewis A, Armuzzi A, Lindsay JO, Silver A. Review article: selective histone deacetylase isoforms as potential therapeutic targets in inflammatory bowel diseases. Aliment Pharmacol Ther (2015) 41(1):26–38. doi: 10.1111/apt.13008
22. Chen Y, Xu R, Ruze R, Yang J, Wang H, Song J, et al. Construction of a prognostic model with histone modification-related genes and identification of potential drugs in pancreatic cancer. Cancer Cell Int (2021) 21(1):291. doi: 10.1186/s12935-021-01928-6
23. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA, et al. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol (2018) 1711:243–59. doi: 10.1007/978-1-4939-7493-1_12
24. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw (2010) 33(1):1–22. doi: 10.18637/jss.v033.i01
25. Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol (2008) 26(8):1364–70. doi: 10.1200/JCO.2007.12.9791
26. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med (1987) 317(26):1625–9. doi: 10.1056/NEJM198712243172603
27. Le Berre C, Ricciuto A, Peyrin-Biroulet L, Turner D. Evolving short- and long-term goals of management of inflammatory bowel diseases: getting it right, making it last. Gastroenterology (2022) 162(5):1424–38. doi: 10.1053/j.gastro.2021.09.076
28. Shah SC, Colombel JF, Sands BE, Narula N. Mucosal healing is associated with improved long-term outcomes of patients with ulcerative colitis: A systematic review and meta-analysis. Clin Gastroenterol Hepatol (2016) 14(9):1245–1255 e8. doi: 10.1016/j.cgh.2016.01.015
29. Tursi A, Elisei W, Picchio M, Penna A, Lecca PG, Forti G, et al. Managing ambulatory ulcerative colitis patients with infliximab: a long term follow-up study in primary gastroenterology centers. Eur J Intern Med (2014) 25(8):757–61. doi: 10.1016/j.ejim.2014.07.007
30. Rutter M, Saunders B, Wilkinson K, Rumbles S, Schofield G, Kamm M, et al. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology (2004) 126(2):451–9. doi: 10.1053/j.gastro.2003.11.010
31. Wang JM, Lin SR, Zhu YB, Yuan J, Wang YM, Zhang Q, et al. Proteomic analysis of lysine acetylation reveals that metabolic enzymes and heat shock proteins may be potential targets for DSS-induced mice colitis. Int Immunopharmacol (2021) 101(Pt B):108336. doi: 10.1016/j.intimp.2021.108336
32. Glauben R, Batra A, Fedke I, Zeitz M, Lehr HA, Leoni F, et al. Histone hyperacetylation is associated with amelioration of experimental colitis in mice. J Immunol (2006) 176(8):5015–22. doi: 10.4049/jimmunol.176.8.5015
33. Glauben R, Batra A, Stroh T, Erben U, Fedke I, Lehr HA, et al. Histone deacetylases: novel targets for prevention of colitis-associated cancer in mice. Gut (2008) 57(5):613–22. doi: 10.1136/gut.2007.134650
34. Mudter J, Weigmann B, Bartsch B, Kiesslich R, Strand D, Galle PR, et al. Activation pattern of signal transducers and activators of transcription (STAT) factors in inflammatory bowel diseases. Am J Gastroenterol (2005) 100(1):64–72. doi: 10.1111/j.1572-0241.2005.40615.x
35. Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature (2007) 446(7135):557–61. doi: 10.1038/nature05698
36. Wetzel A, Scholtka B, Gerecke C, Kleuser B. Epigenetic histone modulation contributes to improvements in inflammatory bowel disease via EBI3. Cell Mol Life Sci (2020) 77(23):5017–30. doi: 10.1007/s00018-020-03451-9
37. Scott GK, Mattie MD, Berger CE, Benz SC, Benz CC. Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res (2006) 66(3):1277–81. doi: 10.1158/0008-5472.CAN-05-3632
38. Saccani S, Natoli G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev (2002) 16(17):2219–24. doi: 10.1101/gad.232502
39. Manni W, Jianxin X, Weiqi H, Siyuan C, Huashan S. JMJD family proteins in cancer and inflammation. Signal Transduct Target Ther (2022) 7(1):304. doi: 10.1038/s41392-022-01145-1
40. Ventham NT, Kennedy NA, Nimmo ER, Satsangi J. Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics. Gastroenterology (2013) 145(2):293–308. doi: 10.1053/j.gastro.2013.05.050
41. Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol (2014) 14(5):329–42. doi: 10.1038/nri3661
42. Mateer SW, Mathe A, Bruce J, Liu G, Maltby S, Fricker M, et al. IL-6 drives neutrophil-mediated pulmonary inflammation associated with bacteremia in murine models of colitis. Am J Pathol (2018) 188(7):1625–39. doi: 10.1016/j.ajpath.2018.03.016
43. Shaul E, Conrad MA, Dawany N, Patel T, Canavan MC, Baccarella A, et al. Canakinumab for the treatment of autoinflammatory very early onset- inflammatory bowel disease. Front Immunol (2022) 13:972114. doi: 10.3389/fimmu.2022.972114
44. Oh YS, Kwak MK, Kim K, Cho EH, Jang SE. Development and application of an antibody that binds to interleukin-1beta of various mammalian species for the treatment of inflammatory diseases. Biochem Biophys Res Commun (2020) 527(3):751–6. doi: 10.1016/j.bbrc.2020.04.073
45. Yin Q, Pi X, Jiang Y, Ren G, Liu Z, Liu H, et al. An immuno-blocking agent targeting IL-1beta and IL-17A reduces the lesion of DSS-induced ulcerative colitis in mice. Inflammation (2021) 44(5):1724–36. doi: 10.1007/s10753-021-01449-4
46. Liso M, Verna G, Cavalcanti E, De Santis S, Armentano R, Tafaro A, et al. Interleukin 1beta blockade reduces intestinal inflammation in a murine model of tumor necrosis factor-independent ulcerative colitis. Cell Mol Gastroenterol Hepatol (2022) 14(1):151–71. doi: 10.1016/j.jcmgh.2022.03.003
47. Oksenberg N, Ahituv N. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet (2013) 29(10):600–8. doi: 10.1016/j.tig.2013.08.001
48. Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, et al. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflammation Bowel Dis (2007) 13(7):807–21. doi: 10.1002/ibd.20110
49. Federman N, de la Fuente V, Zalcman G, Corbi N, Onori A, Passananti C, et al. Nuclear factor kappaB-dependent histone acetylation is specifically involved in persistent forms of memory. J Neurosci (2013) 33(17):7603–14. doi: 10.1523/JNEUROSCI.4181-12.2013
50. Martin TP, McCluskey C, Cunningham MR, Beattie J, Paul A, Currie S. CaMKIIdelta interacts directly with IKKbeta and modulates NF-kappaB signalling in adult cardiac fibroblasts. Cell Signal (2018) 51:166–75. doi: 10.1016/j.cellsig.2018.07.008
51. Dalal PJ, Sullivan DP, Weber EW, Sacks DB, Gunzer M, Grumbach IM, et al. Spatiotemporal restriction of endothelial cell calcium signaling is required during leukocyte transmigration. J Exp Med (2021) 218(1). doi: 10.1084/jem.20192378
52. Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev (2014) 13(1):3–10. doi: 10.1016/j.autrev.2013.06.004
53. Scarpa M, Stylianou E. Epigenetics: Concepts and relevance to IBD pathogenesis. Inflammation Bowel Dis (2012) 18(10):1982–96. doi: 10.1002/ibd.22934
54. Xu J, Xu HM, Yang MF, Liang YJ, Peng QZ, Zhang Y, et al. New insights into the epigenetic regulation of inflammatory bowel disease. Front Pharmacol (2022) 13:813659. doi: 10.3389/fphar.2022.813659
55. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U.S.A. (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111
56. Bullwinkel J, Lüdemann A, Debarry J, Singh PB. Epigenotype switching at the CD14 and CD209 genes during differentiation of human monocytes to dendritic cells. Epigenetics (2011) 6(1):45–51. doi: 10.4161/epi.6.1.13314
57. Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, et al. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest (2008) 118(6):2269–80. doi: 10.1172/JCI34610
58. Do A, Reid RC, Lohman RJ, Sweet MJ, Fairlie DP, Iyer A. An HDAC6 inhibitor confers protection and selectively inhibits B-cell infiltration in DSS-induced colitis in mice. J Pharmacol Exp Ther (2017) 360(1):140–51. doi: 10.1124/jpet.116.236711
59. Koenen HJ, Smeets RL, Vink PM, van Rijssen E, Boots AM, Joosten I, et al. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood (2008) 112(6):2340–52. doi: 10.1182/blood-2008-01-133967
60. Ray G, Longworth MS. Epigenetics, DNA organization, and inflammatory bowel disease. Inflammation Bowel Dis (2019) 25(2):235–47. doi: 10.1093/ibd/izy330
61. Renaude E, Kroemer M, Loyon R, Binda D, Borg C, Guittaut M, et al. The fate of Th17 cells is shaped by epigenetic modifications and remodeled by the tumor microenvironment. Int J Mol Sci (2020) 21(5). doi: 10.3390/ijms21051673
62. Nakase H, Sato N, Mizuno N, Ikawa Y. The influence of cytokines on the complex pathology of ulcerative colitis. Autoimmun Rev (2022) 21(3):103017. doi: 10.1016/j.autrev.2021.103017
63. Meddens CA, van der List ACJ, Nieuwenhuis EES, Mokry M. Non-coding DNA in IBD: from sequence variation in DNA regulatory elements to novel therapeutic potential. Gut (2019) 68(5):928–41. doi: 10.1136/gutjnl-2018-317516
64. Sarmento OF, Svingen PA, Xiong Y, Sun Z, Bamidele AO, Mathison AJ, et al. The role of the histone methyltransferase enhancer of Zeste homolog 2 (EZH2) in the pathobiological mechanisms underlying inflammatory bowel disease (IBD). J Biol Chem (2017) 292(2):706–22. doi: 10.1074/jbc.M116.749663
65. Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med (2007) 13(11):1299–307. doi: 10.1038/nm1652
66. O’Brien BJ, Singer HA, Adam AP, Ginnan RG. CaMKIIdelta is upregulated by pro-inflammatory cytokine IL-6 in a JAK/STAT3-dependent manner to promote angiogenesis. FASEB J (2021) 35(4):e21437. doi: 10.1096/fj.202002755R
67. Gray CB, Suetomi T, Xiang S, Mishra S, Blackwood EA, Glembotski CC, et al. CaMKIIdelta subtypes differentially regulate infarct formation following ex vivo myocardial ischemia/reperfusion through NF-kappaB and TNF-alpha. J Mol Cell Cardiol (2017) 103:48–55. doi: 10.1016/j.yjmcc.2017.01.002
68. Dahlen R, Magnusson MK, Bajor A, Lasson A, Ung KA, Strid H, et al. Global mucosal and serum cytokine profile in patients with ulcerative colitis undergoing anti-TNF therapy. Scand J Gastroenterol (2015) 50(9):1118–26. doi: 10.3109/00365521.2015.1031167
69. Obraztsov IV, Shirokikh KE, Obraztsova OI, Shapina MV, Wang MH, Khalif IL, et al. Multiple cytokine profiling: A new model to predict response to tumor necrosis factor antagonists in ulcerative colitis patients. Inflammation Bowel Dis (2019) 25(3):524–31. doi: 10.1093/ibd/izy358
70. Qian J, Wang C, Wang B, Yang J, Wang Y, Luo F, et al. The IFN-gamma/PD-L1 axis between T cells and tumor microenvironment: hints for glioma anti-PD-1/PD-L1 therapy. J Neuroinflamm (2018) 15(1):290. doi: 10.1186/s12974-018-1330-2
Keywords: histone modification, ulcerative colitis, molecular inflammatory predictive model, infliximab response, CAMK2D
Citation: Ye S, Lyu Y, Chen L, Wang Y, He Y, Li Q, Tian L, Liu F, Wang X and Ai F (2024) Construction of a molecular inflammatory predictive model with histone modification-related genes and identification of CAMK2D as a potential response signature to infliximab in ulcerative colitis. Front. Immunol. 14:1282136. doi: 10.3389/fimmu.2023.1282136
Received: 23 August 2023; Accepted: 19 December 2023;
Published: 11 January 2024.
Edited by:
Giuseppe Murdaca, University of Genoa, ItalyReviewed by:
Hiroshi Nakase, Sapporo Medical University, JapanAntonella Fazio, University Medical Center Hamburg-Eppendorf, Germany
Copyright © 2024 Ye, Lyu, Chen, Wang, He, Li, Tian, Liu, Wang and Ai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feiyan Ai, YWlmZWl5YW4xMTA1QGNzdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Shuyu Ye
Shuyu Ye Yongqing Lyu
Yongqing Lyu Libin Chen
Libin Chen Yiwei Wang1,4
Yiwei Wang1,4 Li Tian
Li Tian Xiaoyan Wang
Xiaoyan Wang Feiyan Ai
Feiyan Ai