
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 11 October 2023
Sec. Nutritional Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1277281
This article is part of the Research TopicMetabolic Regulation in Immune Processes against Microbial InfectionsView all 9 articles
Introduction: Metabolic reprogramming potentiates host protection against antibiotic-sensitive or -resistant bacteria. However, it remains unclear whether a single reprogramming metabolite is effective enough to combat both antibiotic-sensitive and -resistant bacteria. This knowledge is key for implementing an antibiotic-free approach.
Methods: The reprogramming metabolome approach was adopted to characterize the metabolic state of zebrafish infected with tetracycline-sensitive and -resistant Edwardsiella tarda and to identify overlapping depressed metabolite in dying zebrafish as a reprogramming metabolite.
Results: Aspartate was identify overlapping depressed metabolite in dying zebrafish as a reprogramming metabolite. Exogenous aspartate protects zebrafish against infection caused by tetracycline-sensitive and -resistant E. tarda. Mechanistically, exogenous aspartate promotes nitric oxide (NO) biosynthesis. NO is a well-documented factor of promoting innate immunity against bacteria, but whether it can play a role in eliminating both tetracycline-sensitive and -resistant E. tarda is unknown. Thus, in this study, aspartate was replaced with sodium nitroprusside to provide NO, which led to similar aspartate-induced protection against tetracycline-sensitive and -resistant E. tarda.
Discussion: These findings support the conclusion that aspartate plays an important protective role through NO against both types of E. tarda. Importantly, we found that tetracycline-sensitive and -resistant E. tarda are sensitive to NO. Therefore, aspartate is an effective reprogramming metabolite that allows implementation of an antibiotic-free approach against bacterial pathogens.
Edwardsiella tarda is a Gram-negative bacterium that causes infection in aquatic animals and humans, posing a threat to aquaculture industry and public health (1, 2). Antibiotics are effective against bacteria, but the extensive use of these drugs leads to the development of antibiotic resistance due to the consequent selective pressure. Bacterial strains that are resistant to antibiotics are difficult to treat and the increasing emergence of these strains limits sustainable development of aquaculture. Given that overuse of antibiotics triggers the development of antibiotic-resistant bacteria (3), antibiotic-free approaches are especially recommended to eliminate bacterial pathogens (4).
Reprogramming metabolomics is a new useful approach for controlling bacterial infection by using low doses of antibiotics or no antibiotics at all (5–11). This method requires the use of comparative metabolomics to identify crucial biomarkers, which are then used to induce metabolome-reprogramming for elevating bacterial sensitivity to antibiotics or improving host protection against infection (12–16). This leads to subsequent host protection against infections caused by antibiotic-sensitive or –resistant Vibrio alginolyticus and Edwardsiella tarda (17–21). However, the question of whether a single reprogramming metabolite can be used to combat both antibiotic-sensitive and –resistant bacteria has not been well defined. Importantly, antibiotic-free therapy is required to effectively combat both sensitive and resistant bacteria. This success will help in the development of antibiotic-free methods to eliminate bacterial pathogens.
Among antibiotic classes used in aquaculture, tetracyclines, including tetracycline and oxytetracycline, are the first-line agents used for treatment. As a result, there have been increasing incidences of tetracycline-resistant E. tarda. Lo et al., showed that 21.3% (20/94) of isolates were resistant to oxytetracycline (22). Further, Yu et al., isolated E. tarda CK41 from Japanese flounder diagnosed with edwardsiellosis, which is highly resistant to multiple antibiotics, including tetracycline (23). Lee and Wendy showed that out of 300 E. tarda and Aeromonas hydrophila strains, 58% were tetracycline-resistant (24). However, the reprogramming metabolomics approach has not yet been utilized to combat tetracycline-resistant bacteria. Importantly, for successful implementation of an antibiotic-free approach, reprogramming of a single metabolite must be effective for both antibiotic-sensitive and -resistant bacteria, but this has not yet been reported in the literature. Therefore, we sought to use tetracycline-resistant and -sensitive E. tarda may as representative bacterial strains to identify one reprogramming metabolite to eliminate both strains.
Herein, we used gas chromatography-mass spectrometry (GC-MS) based metabolomics to characterize differential metabolic profiles of zebrafish infected with a tetracycline-resistant strain (LTB4-RTET) and -sensitive strain (LTB4-S). All data were compared to uninfected control animals. In this way, we sought to identify an overlapping biomarker between LTB4-RTET and LTB4-S that could potentially be exploited as a reprogramming metabolite to improve host protection against both antibiotic-sensitive and –resistant E. tarda. Following data analysis, we identified aspartate as a crucial biomarker and subsequently used it to reverse the infective phenotype in zebrafish, thereby increasing survival.
The study was approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University, Guangzhou, China.
Zebrafish (Danio rerio), approximately 3 months old (body length, 3 ± 0.2 cm, body weight, 0.2 ± 0.05 g), were purchased from Guangdong Zebrafish Breeding Farm (Guangzhou, China). Zebrafish were acclimated for 2 weeks in 540 L water tanks with Closed Recirculating Aquaculture Systems. In the meantime, fish were fed twice a day, and water tanks were cleaned by siphoning the food debris and feces once every two days. Before experiments, all fish were tested to ensure they were not infected with E. tarda. E. tarda LTB4-S and LTB4-RTET were preserved in our laboratory. After a single colony was cultured for 24 h at 30°C, with shaking at 200 rpm in fresh 50 mL TSB medium, the bacterial suspensions were diluted 1:100 in fresh TSB medium cultured and grown to an optical density at 600 nm (OD600) of 1.0 at 30°C.
Exogenous metabolites were supplemented as previously described (25). Zebrafish were challenged with 1 × 105 CFU/fish. Then each zebrafish were intraperitoneally injected with 5 μL of a 12 μg/μL suspension of aspartate (60 μg total dose) or 0.75 ~ 6 μg sodium nitroprusside dissolved in saline for 3 days, once a day. Control fish were injected with 5 μL saline solution. After 30 h, humoral fluid was collected for GC-MS analysis and spleen samples were collected for qRT-PCR, nitric oxide synthase (NOS) activity, and NO content detection.
Next, qPCR was adopted to quantitate E. tarda in zebrafish as described previously (26). In brief, bacteria (103, 104, 105, 106, 107, and 108 CFU) were used to extract bacterial genomes. PCRs with primers for the gyrB gene were used to assess bacterial load along a standard curve. The same numbers of LTB4-S and LTB4-RTET (5 × 103 CFU) were intraperitoneally injected into zebrafish. The zebrafish that survived for 6–144 h were collected. The HiPure Tissue DNA Mini Kit (Magen Biotechnology, Guangzhou, China) was then used to extract genomic DNA and bacterial DNA was subsequently measured by qPCR. The amount of bacterial DNA was then calculated based on the aforementioned standard curve.
MICs of LTB4-S and LTB4-RTET were measured by antimicrobial susceptibility testing as described in Clinical & Laboratory Standards Institute guidelines (27). The bacterial growth curve was determined according to conventional procedures.
Samples were prepared as previously described (28). In brief, LTB4-S and LTB4-RTET were cultured in LB medium and collected by centrifuged to infect zebrafish using 1 × 105 CFU/fish. Dying zebrafish were collected, rinsed and wiped. These zebrafish were cut into six pieces, weighted, and added appropriate volume of saline (100 μL/100 mg) for humoral. After centrifugation, 50 μL supernatant was collected for metabolite extraction using precooled methanol. Following centrifugation, supernatant was collected and then dried by vacuum centrifuge device for GC-MS analysis.
Analysis was performed according to known methods (29). In brief, XCalibur software was used to analyze the mass fragmentation spectrum. The National Institute of Standards and Technology (NIST) library and NIST MS search 2.0 program were adopted to match the data to identify compounds. Software IBM SPSS Statistics 19 was used to analyze significant difference of the standardized data, when the differences were defined at P value < 0.05. SIMCA-P + 12.0 software were used to perform principal-component analysis (PCA) and S-plot analysis. iPath3.0 (https://pathways.embl.de/) was used to carry out interactive Pathways (iPath) analysis.
Next, qRT-PCR was performed according to previously published methods (30). Gene-specific primers that were used here are shown in Table 1. The β-actin gene served as the internal control.
A series of experiments were performed according to kit instructions (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The spleens from ten zebrafish were pooled into one biological sample and three biological replicates were measured per group.
LTB4-RTET and LTB4-S were obtained through sequential propagation of LTB4 in medium with and without tetracycline, respectively. This led to the revelation that LTB4-RTET has a 20-fold higher MIC for tetracycline compared to that of LTB4-S, which showed no change in the MIC during propagation (Figure 1A). Growth curves showed that LTB4-RTET proliferated more slowly than LTB4-S (Figure 1B). When zebrafish were infected with LTB4-S or LTB4-RTET, the two strains differentially persisted in the zebrafish. Specifically, more LTB4-S were detected in the fish during the 6–48 h time points compared to LTB4-RTET. Further, these bacteria were measured again at the 72, 120, and 144 h time points and we identified reduced numbers of LTB4-RTET compared to LTB4-S at the latter (Figure 1C). Similar survival rates were determined for LTB4-RTET and LTB4-S infected zebrafish (Figure 1D). Therefore, we concluded that both LTB4-RTET and LTB4-S infection causes differential growth rates and resistance phenotypes, but similar survival rates.
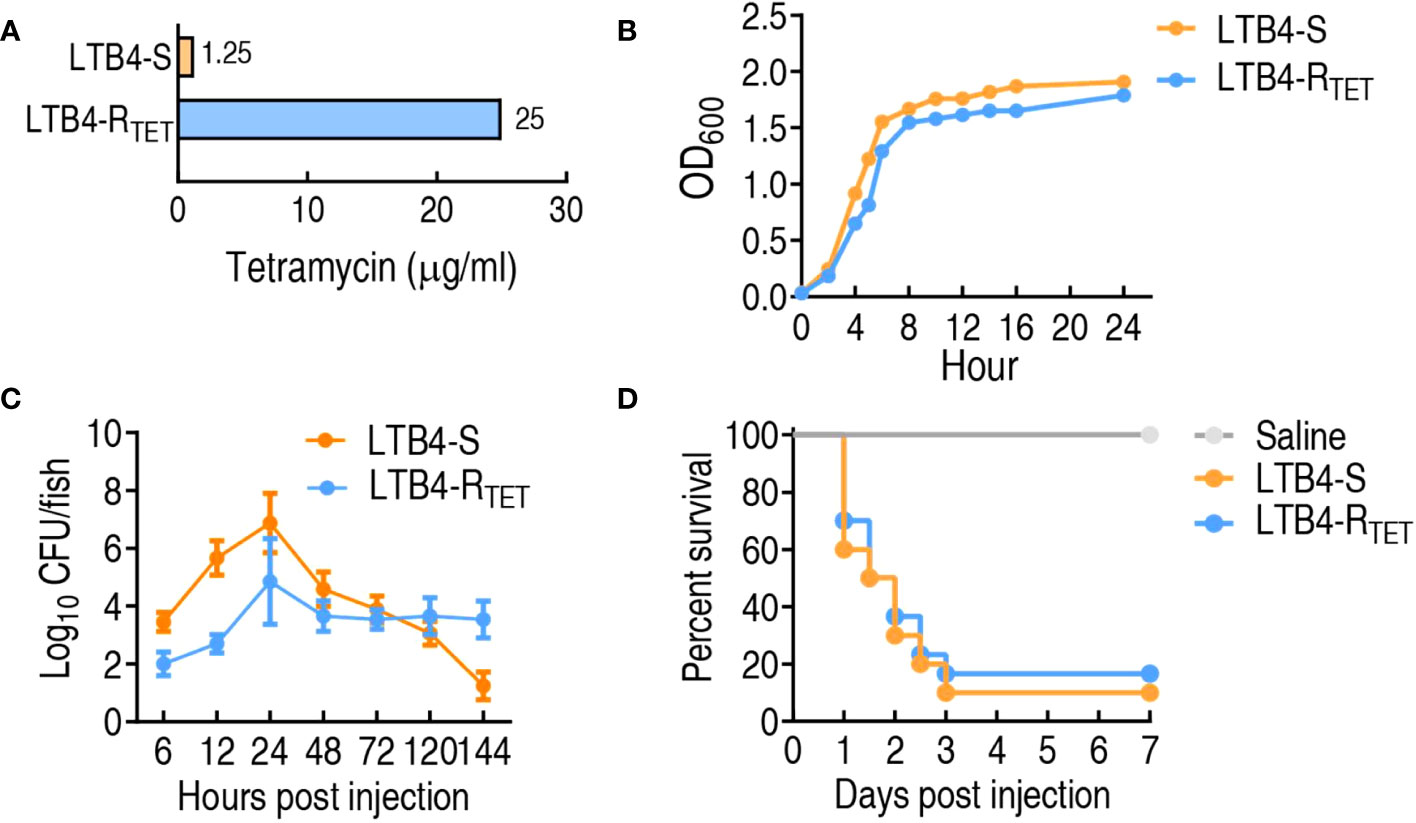
Figure 1 Resistance and growth phenotypes of LTB4-S and LTB4-RTET. (A) Minimum inhibitory concentration of LTB4-S and LTB4-RTET to tetracycline. (B) LTB4-S and LTB4-RTET growth curve. (C) Dynamic changes in bacterial number in zebrafish infected with LTB4-S and LTB4-RTET. (D) Survival of zebrafish infected with LTB4-S and LTB4-RTET.
Host metabolic state is correlated with susceptibility or resistance to bacterial pathogens (29, 31). Thus, the metabolic profiles of zebrafish infected with LTB4-RTET and LTB4-S were compared and uninfected control fish. To do this, a GC-MS-based metabolomics approach was utilized to characterize the metabolic profiles of dying animals in the two experimental groups and in the control group. Ten biological samples with two technical replicates were adopted for each group, yielding a total of 60 data sets. A total of 230 aligned individual peaks were obtained from each sample, where 77 metabolites were determined (Figure 2A). The correlation coefficient for technical replicates varied between 0.994 and 0.999 (Figure 2B), suggesting good repeatability of the data. Metabolic profiles of these 77 metabolites are displayed as a heatmap for each group, which shows that for each group, LTB4-S and LTB4-RTET infected and uninfected zebrafish clustered independently (Figure 2C). According to the KEGG annotation, the identified metabolites were classified as 37% amino acids, 32% carbohydrate, 13% nucleotide, 10% lipid, and 8% as other (Figure 2D). These data suggest that this metabolic platform provides reliable data for further analysis.
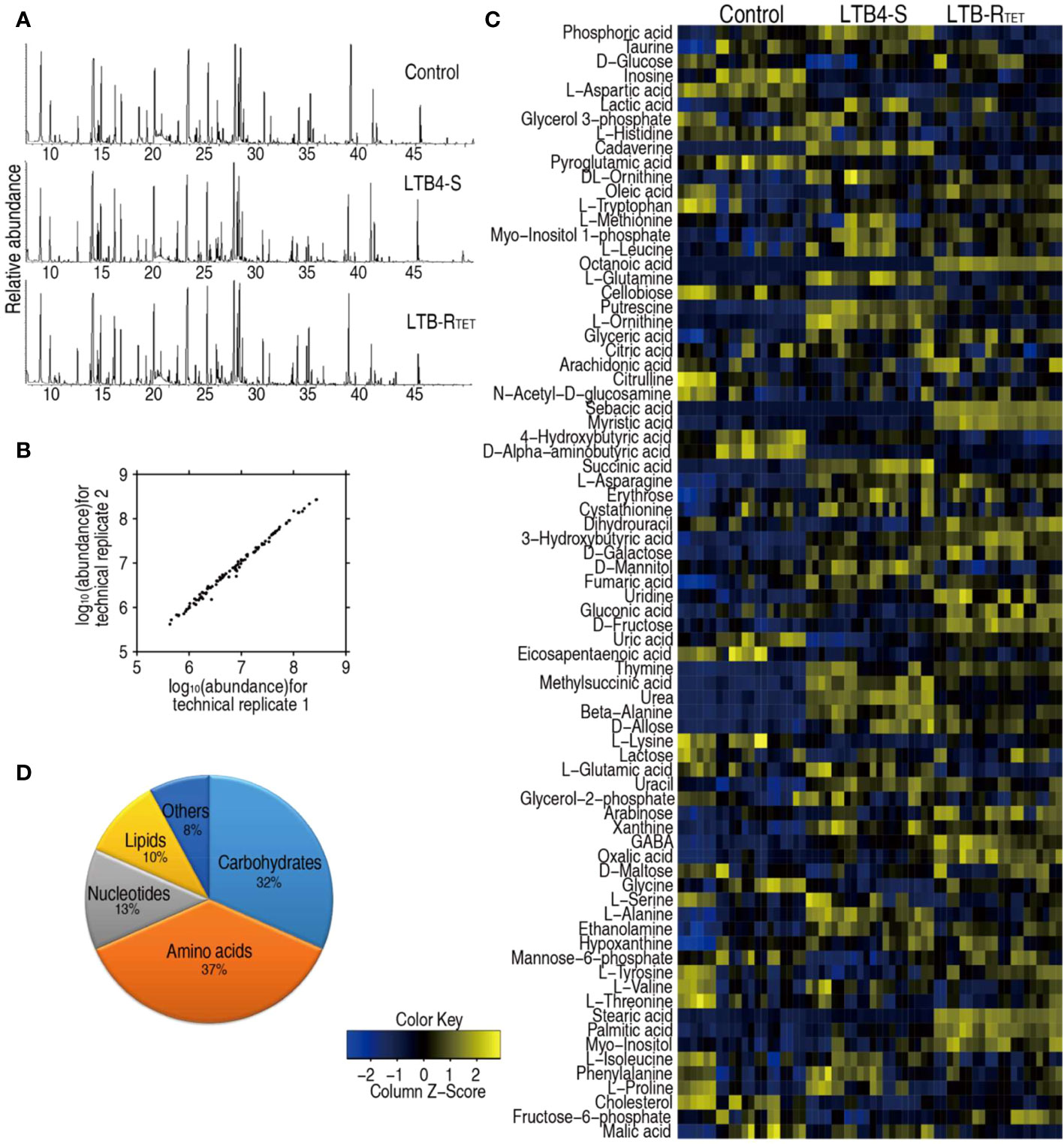
Figure 2 Metabolic profiles of LTB4-S and LTB4-R. (A) Samples of total ion current chromatogram separately from control uninfected animals and LTB4-S and LTB4-RTET-infected animals. (B) Evaluation for reproducibility of the metabolomic profiling platform. Pearson correlation coefficient of metabolite abundance between technical replicates varies between 0.994 and 0.999. (C) Heat map showing unsupervised hierarchical clustering by using metabolites (row). Blue, downregulated; yellow, upregulated (see color scale). (D) Categories of all metabolites by searching against KEGG.
To explore LTB4-RTET- and LTB4-S-induced metabolic features, a two-sided Mann–Whitney U test was used to identify differential abundance of metabolites in the two groups compared to control animals. We identified a total of 65 metabolites showing differential abundance (Figure 3A). Specifically, LTB4-S- and LTB4-RTET-infected fish had 57 and 52 differentially expressed metabolites, respectively. A Z-score calculation was used to display deviations between a value and the mean (Figure 3B). Among the 57 differentially expressed metabolites in LTB4-S-infected fish, 37 were upregulated while 20 were downregulated. Similarly, in LTB4-RTET- infected zebrafish, out of the 52 differentially abundant metabolites, 37 were also upregulated while 15 were downregulated. The top five downregulated metabolites in both groups overlapped, with ranking from the lowest to highest abundance of these five metabolites being: aspartate < histidine < aminobutyric acid < cellobiose < uric acid (Figure 3B). In total, we identified 44 overlapping metabolites between the two infection models as well as 13 (5 up- and 8 downregulated) and 8 (6 up- and 2 downregulated) unique changes in LTB4-S and LTB4-RTET infected fish, respectively. Among the 44 overlapping metabolites, 41 displayed the same change with 31 upregulations and 10 downregulations (Figure 3C). An increased number of upregulated metabolites were identified compared to downregulated metabolites in carbohydrate, amino acid, lipid and nucleotide groups of both infections (Figure 3D). These data suggest that a similar metabolic shift is characterized between infections caused by LTB4-S and LTB4-RTET.
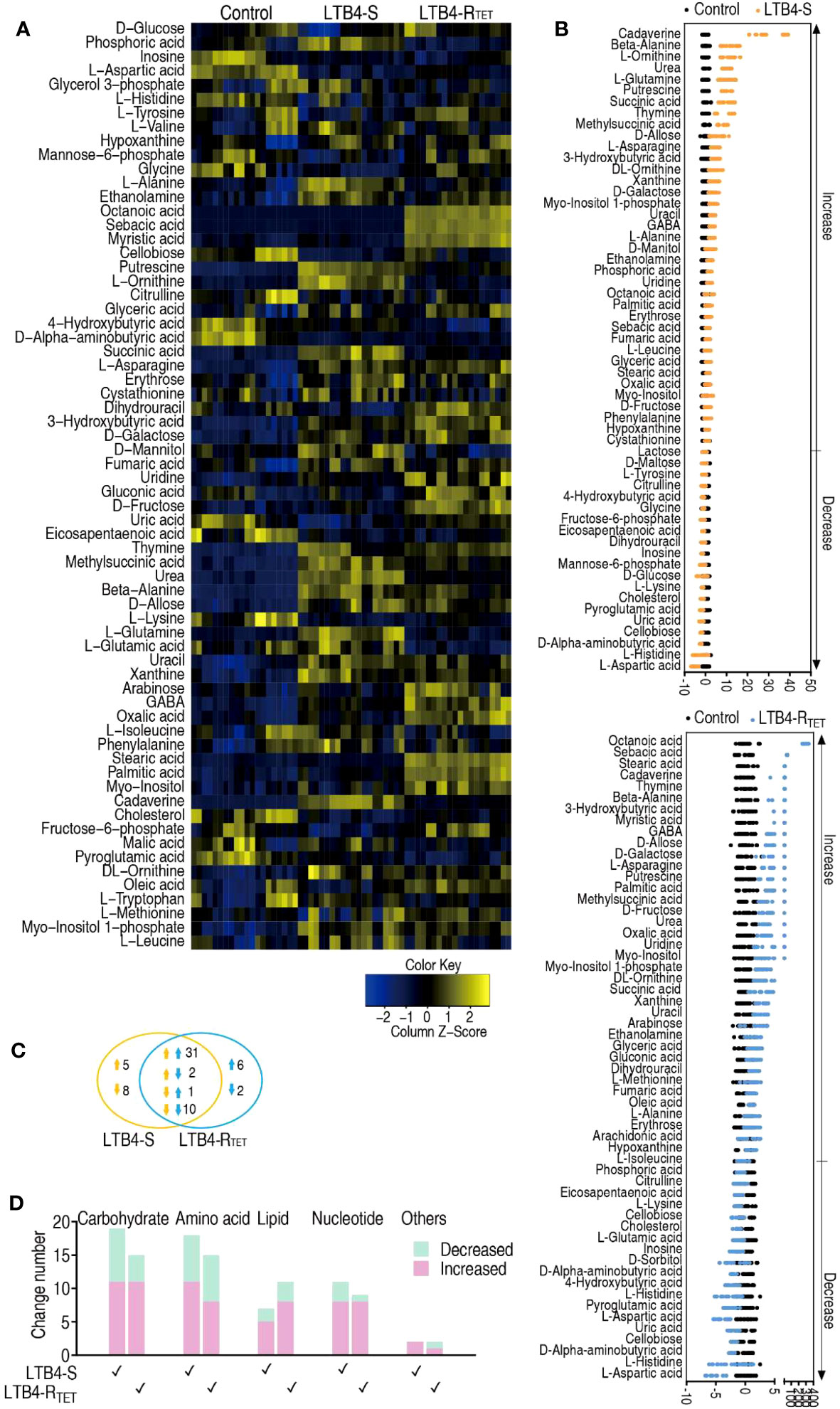
Figure 3 Metabolic profiles of differential metabolites. (A) Heat maps showing differentially expressed metabolites (row). Blue, downregulated; yellow, upregulated (see color scale). (B) Z-score plot showing the deviations of differential metabolites. (C) Venn diagram for the overlapping and unique metabolites with differential abundances between the two strains. Downward facing arrow, decreased metabolites; Upward facing arrow, increased metabolites. (D) Categories of metabolites with differential expression in the two strains.
Having data on metabolic pathway alterations provides information that aids our understanding of key changes in the metabolic state. Thus, differential metabolite expression was analyzed, leading to data showing the enrichment of 13 metabolic pathways. Among them, phenylalanine, tyrosine, and tryptophan; D-glutamine and D-glutamate metabolism; valine, leucine, and isoleucine biosynthesis; alanine, aspartate, and glutamate metabolism; beta-alanine metabolism, and arginine and proline metabolism were ranked as the top six enriched metabolic pathways (Figure 4A). With the exception of histidine metabolism, where the abundance of all metabolites (aspartate, histidine, and glutamic acid) was reduced, the other metabolic pathways exhibited both increased and decreased metabolite expression. We hypothesized that the downregulated metabolites were associated with the dying animals. Therefore, we became interested in identifying a key depressed metabolite that overlapped between LTB4-S and LTB4-RTET-infected animals as a potential reprogramming metabolite. A total of five metabolites overlapped among metabolites in these enriched metabolic pathways, including aspartate, histidine, pyroglutamic acid, lysine, and citrulline (Figure 4B).
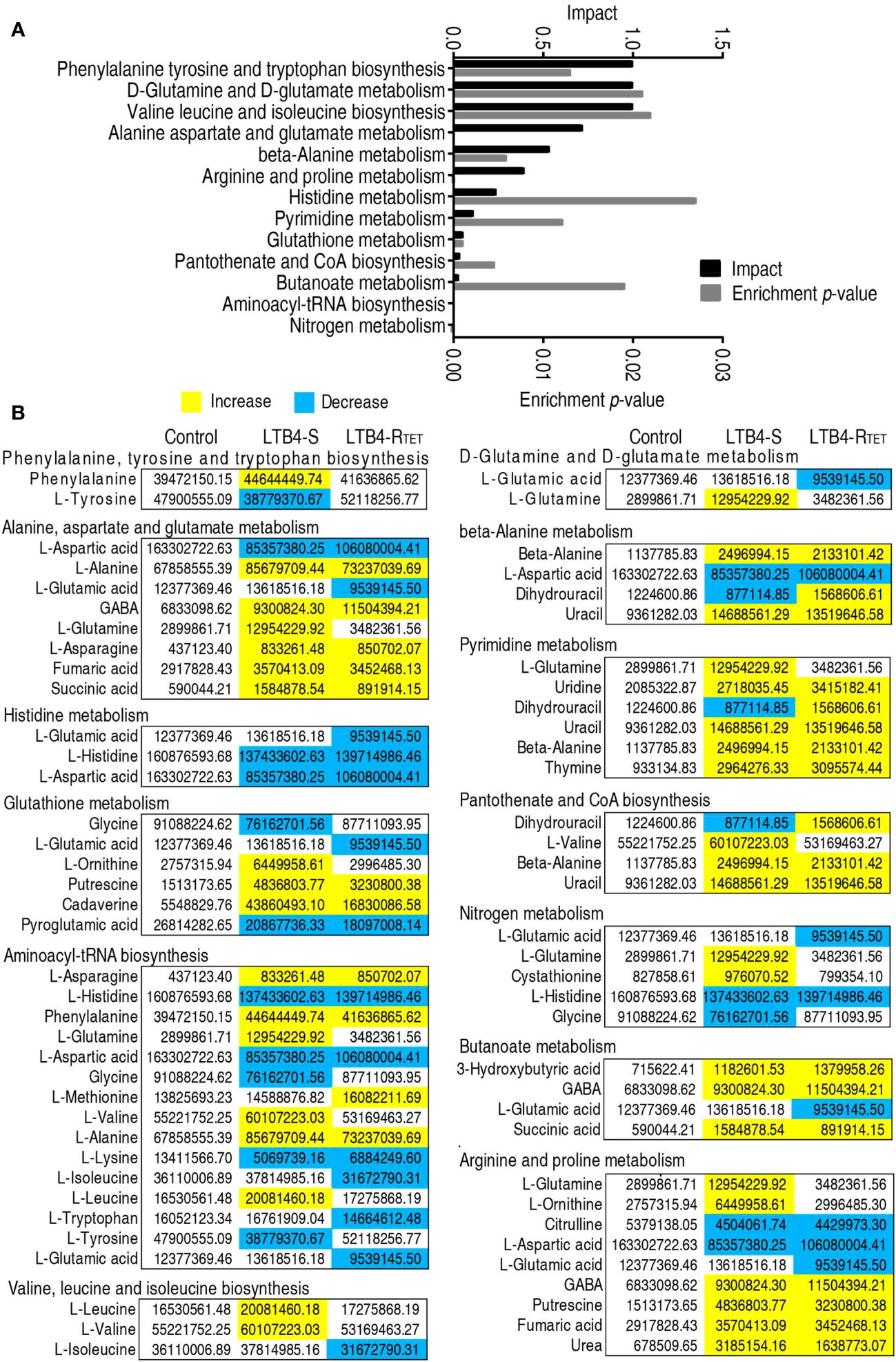
Figure 4 Metabolic pathway enrichment. (A) Enrichment of metabolic pathway by the metabolites with differential abundances. (B) Integrative analysis of the metabolites in enriched pathways. Yellow, upregulated metabolites; Blue, downregulated metabolites.
Pattern recognition is an efficient method to identify biomarkers in metabolomics analyses. Thus, orthogonal partial least square discriminant analysis (OPLS-DA) was employed to recognize the sample patterns of differential metabolomes. Here, t[1] was separated control and LTB4-RTET animals from the LTB4-S group, while t[2] differentiated control animals from the LTB4-RTET group and deviation of LTB4-S (Figure 5A). An S-plot was used to identify discriminating variables with cutoff values of ≤0.05 and ≥0.5 for the absolute value of covariance p and correlation p(corr), respectively, as biomarkers. The data showed that t[1] identified downregulated aspartate, glucose, tyrosine, glycine, cholesterol, mannose-6-phosphate, and upregulated ethanolamine, alanine, glutamine, cadaverine, phosphoric acid as biomarkers. In contrast, t[2] determined downregulated aspartate, inosine, pyroglutamic acid, and upregulated stearic acid, octanoic acid, palmitic acid, stearic acid, myo-inositol, myo-inositol 1 phosphate, ethanolamine, ornithinem, oleid acid as biomarkers (Figure 5B). Among them, downregulated aspartate was found to overlap (Figure 5C), thereby highlighting aspartate as the most promising biomarker related to zebrafish death in the two infection models.
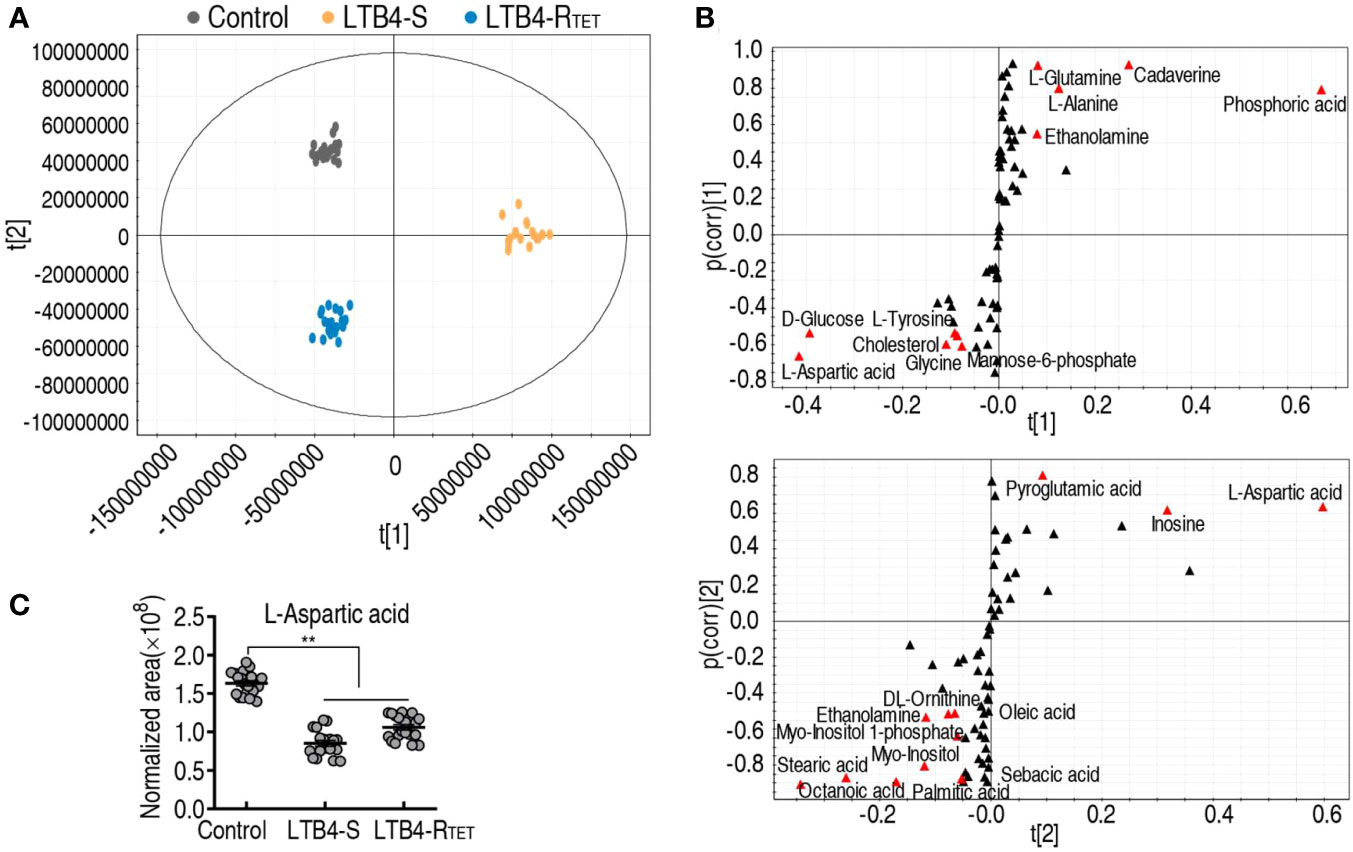
Figure 5 Identification of biomarkers. (A) PCA of uninfected control animals, LTB4-S, and LTB4-RTET. The technical replicates of samples are showed by dots in the plot. (B) S-plot is generated from OPLS-DA. Uninfected control animals and LTB4-RTET are differentiated from LTB4-S by using predictive component t[1] and correlation p(corr)[1]. Uninfected control animals and the deviations of LTB4-S are separated from LTB4-RTET by using predictive component t[2] and correlation p(corr)[2]. Ttriangle, metabolites; marked in red, candidate biomarkers. (C) Scatter diagram of crucial biomarkers in data (B). **, P, 0.01.
The above finding that the downregulated aspartate is related to zebrafish death suggested that the upregulation of aspartate may protect zebrafish against both LTB4-S and LTB4-RTET infections. To test this idea, aspartate was complemented in zebrafish which were then infected with LTB4-S and LTB4-RTET. Aspartate improved zebrafish survival from 10% and 16.7% to 50% and 36.7%, respectively (Figures 6A, B). Meanwhile, bacterial load was also used as an important index to evaluate the role of aspartate. Similar to the observed changes in survival, exogenous aspartate promoted the elimination of LTB4-S and LTB4-RTET. Specifically, exogenous aspartate reduced LTB4-S and LTB4-RTET by at least 2- and 4-fold after 24 h, respectively (Figures 6C, D). Therefore, aspartate is effective at protecting zebrafish from LTB4-S and LTB4-RTET infection.
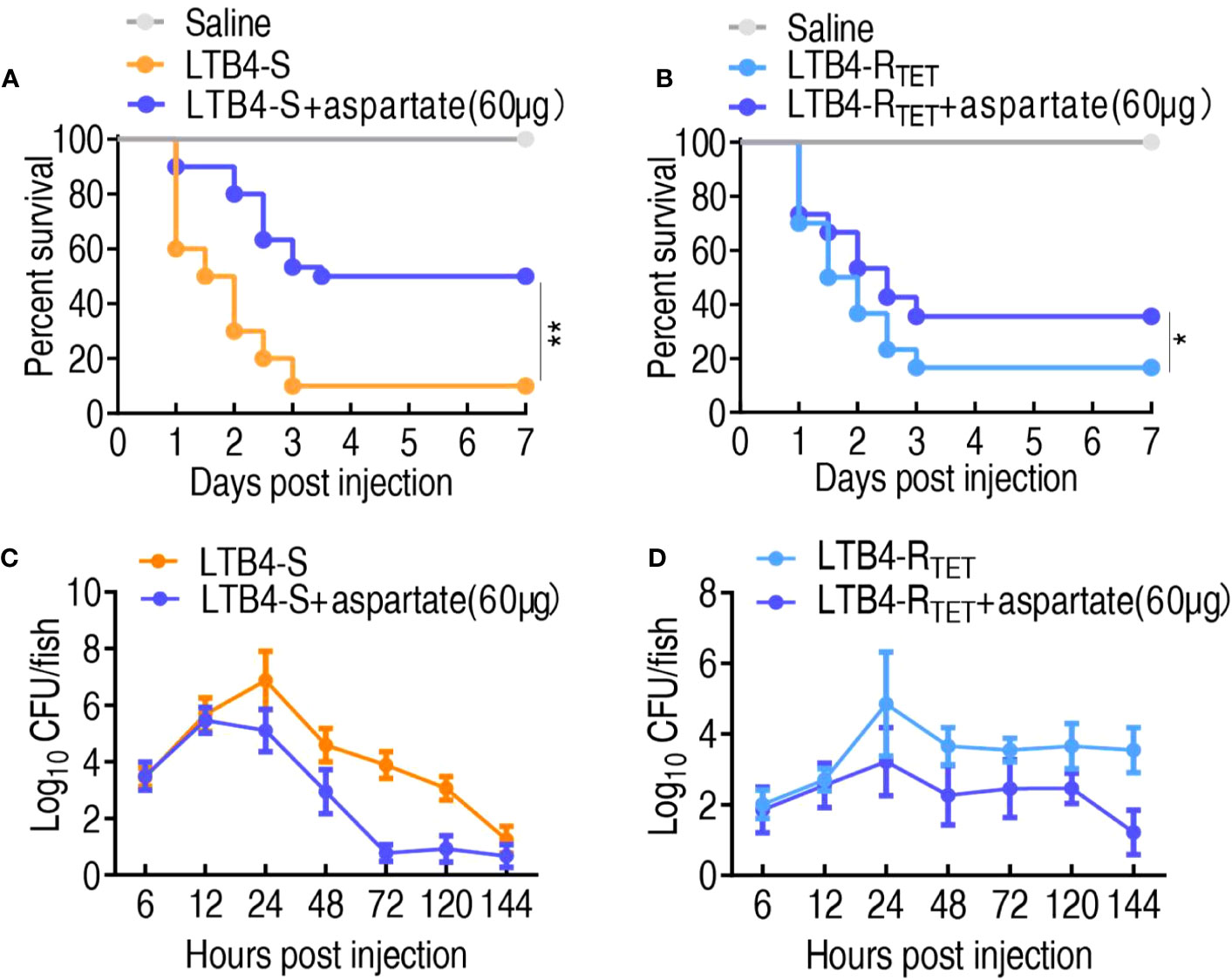
Figure 6 Role of aspartate in a zebrafish model. (A, B) Survival of zebrafish infected with and without aspartate and then infected with LTB4-S (A) or LTB4-RTET (B). (C, D) Bacterial load in the internal organs of zebrafish injected with and without aspartate and then infected with LTB4-S (C) or LTB4-RTET (D). *p < 0.05, **p < 0.01.
Aspartate has multiple metabolic pathways. To identify the affected pathway in LTB4-S and LTB4-RTET-infected zebrafish, iPath was employed to analyze differential abundances of metabolites (Figure 7A). LTB4-S and LTB4-RTET infections had their own specific differential pathways, but overlapping pathways were also identified. We were interested in the overlapping metabolic pathways that were involved in aspartate regulation. Our data showed that aspartate is involved in four overlapping metabolic pathways, where the metabolic flux from aspartate to the urea cycle is fully affected (Figure 7A). Aspartate is a source for the urea cycle, where NO is synthesized from an L-arginine by NOS (Figure 7B). Next, we performed qRT-PCR to measure expression of genes encoding the cycle in surviving and dying LTB4-S- and LTB4-RTET-infected zebrafish. Decreased and increased expression of ass1, as1, nos2a, and nos2b was measured in dying and surviving animals, respectively, compared to control animals. Interestingly, arg2 expression remained unchanged in the LTB4-S infection model. Notably, expression of the four genes was higher in animals infected with LTB4-S compared to LTB4-RTET (Figure 7C). When aspartate was complemented, higher expression of the four genes was measured (Figure 7D). Further, the activity of NOS and NO was lower in the dying animals but higher in the surviving animals compared to the control group (close to significant difference in NOS of the dying fish caused by LTB4-RTET) (Figures 7E, F). However, aspartate complement promoted NOS activity and NO level even if zebrafish were challenged by LTB4-S and LTB4-RTET (Figures 7G, H). Taken together, these data confirm that aspartate promotes NO production.
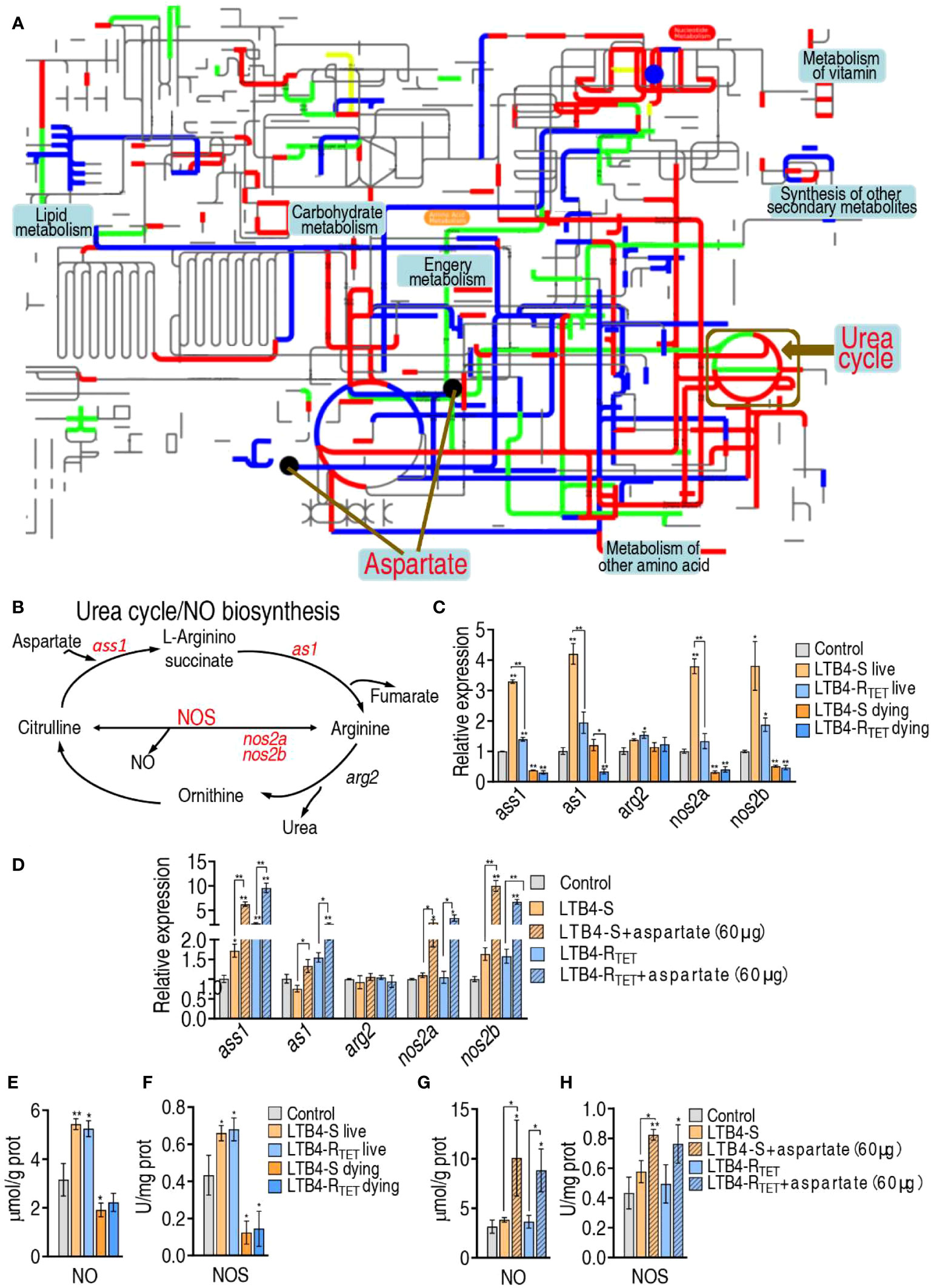
Figure 7 Role of NO in the aspartate potentiation. (A) iPath analysis for global metabolic changes. (B) Urea cycle and NO biosynthesis. (C, D) qRT-PCR for NO generation genes expression in control, LTB4-S, and LTB4-RTET (C) or plus aspartate (D). (E) NO level in control, LTB4-S, and LTB4-RTET animal groups. (F) NOS activity in control, LTB4-S, and LTB4-RTET animal groups. (G) NO level in control, LTB4-S, and LTB4-RTET animal groups in the presence of aspartate. (H) NOS activity in control, LTB4-S, and LTB4-RTET animal groups in the presence of aspartate. *, P , 0.05; **, P , 0.01.
To demonstrate that NO plays a role in the elimination of LTB4-S and LTB4-RTET, NO donor sodium nitroprusside was used to test protection against infection caused by LTB4-S and LTB4-RTET. NO was elevated during treatment with 3–6 μg sodium nitroprusside (Figures 8A, B). Sodium nitroprusside increased zebrafish survival in both infection models in a dose-dependent manner leading to survival rates of 55.5% and 48%, respectively, following treatment with 6 μg sodium nitroprusside (Figures 8C, D). In addition, we found that sodium nitroprusside consistently caused a reduction in bacterial load in both experimental groups. Specifically, sodium nitroprusside reduced LTB4-S and LTB4-RTET by at least 2- and 3-fold after 24 h, respectively (Figures 8E, F). These results indicate that NO plays an important role in protecting zebrafish from LTB4-S and LTB4-RTET infection.
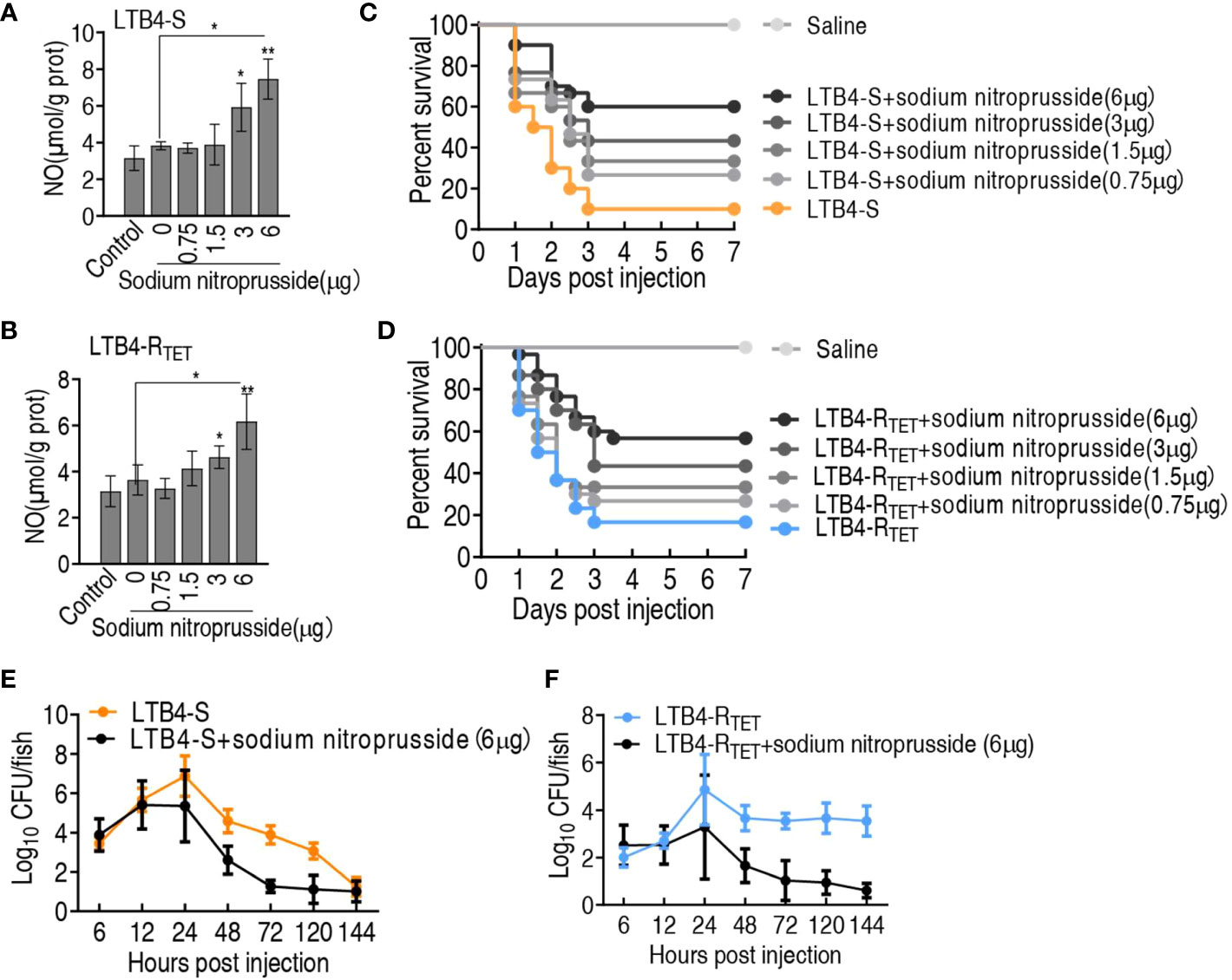
Figure 8 Role of sodium nitroprusside in the aspartate potentiation. (A, B) NO level of zebrafish in the indicated sodium nitroprusside plus LTB4-S (A) or LTB4-RTET (B). (C, D) Survival of zebrafish infected with LTB4-S (C) or LTB4-RTET (D) and with and without the indicated sodium nitroprusside. (E, F) Bacterial load in internal organs of zebrafish infected with LTB4-S (E) or LTB4-RTET (F) and with and without sodium nitroprusside. *p < 0.05, **p < 0.01.
Antibiotic-free therapy against bacterial infection is recommended as this strategy avoids antibiotic-related shortcomings (32, 33). Among the antibiotic-free therapy, reprogramming of the metabolome/metabolic state is an effective approach to restore host protection from microbes. In the present study, we used this approach to explore the potential of using a single reprogramming metabolite to combat both antibiotic-sensitive and -resistant E. tarda. In this way, we have identified aspartate as the ideal reprogramming metabolite and we have shown that it is downregulated in both LTB4-S- and LTB4-RTET-infected dying zebrafish. Moreover, exogenous aspartate protects zebrafish to eliminate LTB4-S and LTB4-RTET, thereby improving survival. Mechanistically, exogenous aspartate-mediated metabolic flux promotes NO biosynthesis to induce its protective effect, which is further validated by NO donor sodium nitroprusside that also promotes the elimination of both LTB4-S and LTB4-RTET in zebrafish to increase the survival rate of these infected animals as exogenous aspartate does. These results suggest that aspartate-mediated NO may be an effective approach for combating both antibiotic-sensitive and -resistant E. tarda, providing a previously unknown antibiotic-free therapeutic modality to eliminate E. tarda with both antibiotic sensitivities.
Reprogramming of the metabolome/metabolic state has previously been adopted to promote antibiotic killing efficacy (34–37), as well as to protect hosts against bacterial infection (20, 38–40). In the latter, reprogramming metabolites are identified from the comparison between control and surviving infected animals (41, 42). In a change from this, the present study explores whether reprogramming metabolites may be identified from the comparison between control and dying animals in E. tarda infection models. Our robust proof of concept study indicates that biomarkers identified from dying animals can be used as reprogramming metabolites. These data will be helpful in utilizing dying instead of surviving animals to identify reprogramming metabolites for low-antibiotic and antibiotic-free strategies against bacterial pathogens, which is especially important for animals with high economic value.
Thus, we have identified aspartate as the most promising biomarker, and demonstrated its downregulation in zebrafish infected with LTB4-S or LTB4-RTET compared to control fish. Exogenous aspartate was also shown to be an ideal reprogramming metabolite to combat both LTB4-S and LTB4-RTET infection. Based on aspartate metabolism, it is logical to hypothesize that high doses of aspartate promote NO generation, given aspartate acts as a source for its biosynthesis. On the other hand, NO is a key gas messenger in the pathogenesis of inflammation, where it links innate and adaptive immunity (43, 44). NO-mediated elimination of bacterial pathogens has previously been documented (45, 46), but there are no specific reports of NO-mediated elimination to E. tarda. Therefore, the present study focused on answering two questions: does exogenous aspartate promotes NO biosynthesis and does NO combat antibiotic-sensitive and -resistant E. tarda? Our results show that high doses of aspartate elevate NO biosynthesis and that sodium nitroprusside-induced elevation of NO also protects zebrafish against LTB4-S and LTB4-RTET infection. Therefore, downregulation of NO due to bacterial infection is a cause of zebrafish failure to eliminate LTB4-S and LTB4-RTET pathogens. Therefore, indirect complementation of NO by aspartate metabolic reprogramming or directly by sodium nitroprusside restores protection against LTB4-S and LTB4-RTET infection.
Herein, we show that aspartate is depressed in dying zebrafish infected with LTB4-S and LTB4-RTET and we have validated its role as a reprogramming metabolite. Exogenous aspartate restores zebrafish protection against LTB4-S and LTB4-RTET infection by increasing NO synthesis. These results suggest that reprogramming metabolites can be identified from both surviving and dying animals. Promoting NO production is an important method for promoting aspartate-mediated elimination of pathogenic bacteria. Because metabolites have multiple metabolic pathways to produce different effect products, only by identifying the metabolic pathways that play a role can we reveal underlying mechanisms and develop reversal strategies.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The animal study was approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (Approval No. SYSU-IACUC- 2020-B1267). The study was conducted in accordance with the local legislation and institutional requirements.
HL: Conceptualization, Writing – original draft, Writing – review & editing. JX: Data curation, Investigation, Methodology, Writing – original draft. M-YL: Investigation, Methodology, Writing – original draft.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by grants from the National Natural Science Foundation of China (31770045), the Innovation Group Project of Southern Marine Science, and the Engineering Guangdong Laboratory (Zhuhai) (No. 311020006).
The authors acknowledge Dr. Xuan-xian Peng for his supervise.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor CZ declared a shared affiliation with the authors at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Miwa S, Mano N. Infection with Edwardsiella tarda causes hypertrophy of liver cells in the Japanese flounder Paralichthys olivaceus. Dis Aquat Organ (2000) 42(3):227–31. doi: 10.3354/dao042227
2. Armwood AR, Griffin MJ, Richardson BM, Wise DJ, Ware C, Camus AC. Pathology and virulence of Edwardsiella tarda, Edwardsiella piscicida, and Edwardsiella Anguillarum in channel (Ictalurus punctatus), blue (Ictalurus furcatus), and channel × blue hybrid catfish. J Fish Dis (2022) 45(11):1683–98. doi: 10.1111/jfd.13691
3. Jiang M, Su YB, Ye JZ, Li H, Kuang SF, Wu JH, et al. Ampicillin-controlled glucose metabolism manipulates the transition from tolerance to resistance in bacteria. Sci Adv (2023) 9(10):eade8582. doi: 10.1126/sciadv.ade8582
4. Raza S, Matula K, Karoń S, Paczesny J. Resistance and adaptation of bacteria to non-antibiotic antibacterial agents: physical stressors, nanoparticles, and bacteriophages. Antibiotics (Basel) (2021) 10(4):435. doi: 10.3390/antibiotics10040435
5. Peng B, Su YB, Li H, Han Y, Guo C, Tian YM, et al. Exogenous alanine or/and glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab (2015) 21:249–61. doi: 10.1016/j.cmet.2015.01.008
6. Peng B, Li H, Peng XX. Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell (2015) 6:628–37. doi: 10.1007/s13238-015-0185-x
7. Cheng ZX, Guo C, Chen ZG, Yang TC, Zhang JY, Wang J, et al. Glycine, serine and threonine metabolism confounds efficacy of complement-mediated killing. Nat Commun (2019) 10:3325. doi: 10.1038/s41467-019-11129-5
8. Kou TS, Wu JH, Chen XW, Chen ZG, Zheng J, Peng B. Exogenous glycine promotes oxidation of glutathione and restores sensitivity of bacterial pathogens to serum-induced cell death. Redox Biol (2022) 58:102512. doi: 10.1016/j.redox.2022.102512
9. Wang Z, Li MY, Peng B, Cheng ZX, Li H, Peng XX. GC-MS-based metabolome and metabolite regulation in serum-resistant Streptococcus agalactiae. J Proteome Res (2016) 15(7):2246–53. doi: 10.1021/acs.jproteome.6b00215
10. Cao YC, Kou TS, Peng LT, Munang’andu HM, Peng B. Fructose promotes crucian carp survival against Aeromonas hydrophila infection. Front Immunol (2022) 3:865560. doi: 10.3389/fimmu.2022.865560
11. Peng B, Li H, Peng XX. Call for next-generation drugs that remove the uptake barrier to combat antibiotic resistance. Drug Discov Today (2023) 28:103753. doi: 10.1016/j.drudis.2023.103753
12. Zhao XL, Chen ZG, Yang TC, Jiang M, Wang J, Cheng ZX, et al. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci Transl Med (2021) 13:eabj0716. doi: 10.1126/scitranslmed.abj0716
13. Li L, Su YB, Peng B, Peng XX, Li H. Metabolic mechanism of colistin resistance and its reverting in Vibrio alginolyticus. Environ Microbiol (2020) 22:4295–313. doi: 10.1111/1462-2920.15021
14. Jiang M, Li X, Xie CL, Chen P, Luo W, Lin CX, et al. Fructose-enabled killing of antibiotic-resistant Salmonella enteritidis by gentamicin: Insight from reprogramming metabolomics. Int J Antimicrob Agents (2023) 62(3):106907. doi: 10.1016/j.ijantimicag.2023.106907
15. Peng LT, Li DL, Yang DX, Peng B. Taurine promotes Oreochromis niloticus survival against Edwardsiella tarda infection. Fish Shellfish Immunol (2022) 129:137–44. doi: 10.1016/j.fsi.2022.08.065
16. Chen XH, Zhang BW, Li H, Peng XX. Myo-inositol improves the host's ability to eliminate balofloxacin-resistant Escherichia coli. Sci Rep (2015) 5:10720. doi: 10.1038/srep10720
17. Zeng ZH, Du CC, Liu SR, Li H, Peng XX, Peng B. Glucose enhances tilapia against Edwardsiella tarda infection through metabolome reprogramming. Fish Shellfish Immunol (2017) 61:34–43. doi: 10.1016/j.fsi.2016.12.010
18. Yang MJ, Xu D, Yang DX, Li L, Peng XX, Chen ZG, et al. Malate enhances survival of zebrafish against Vibrio alginolyticus infection in the same manner as taurine. Virulence (2020) 11(1):349–64. doi: 10.1080/21505594.2020.1750123
19. Gong QY, Yang MJ, Yang LF, Chen ZG, Jiang M, Peng B. Metabolic modulation of redox state confounds fish survival against Vibrio alginolyticus infection. Microb Biotechnol (2020) 13(3):796–812. doi: 10.1111/1751-7915.13553
20. Gong Q, Yang D, Jiang M, Zheng J, Peng B. L-aspartic acid promotes fish survival against Vibrio alginolyticus infection through nitric oxide-induced phagocytosis. Fish Shellfish Immunol (2019) 97:359–66. doi: 10.1016/j.fsi.2019.12.061
21. Sun B, Sun B, Zhang B, Sun L. Temperature induces metabolic reprogramming in fish during bacterial infection. Front Immunol (2022) 13:1010948. doi: 10.3389/fimmu.2022.1010948
22. Lo DY, Lee YJ, Wang JH, Kuo HC. Antimicrobial susceptibility and genetic characterisation of oxytetracycline-resistant Edwardsiella tarda isolated from diseased eels. Vet Rec (2014) 175(8):203. doi: 10.1136/vr.101580
23. Yu JE, Cho MY, Kim JW, Kang HY. Large antibiotic-resistance plasmid of Edwardsiella tarda contributes to virulence in fish. Microb Pathog (2012) 52(5):259–66. doi: 10.1016/j.micpath.2012.01.006
24. Lee SW, Wendy W. Antibiotic and heavy metal resistance of Aeromonas hydrophila and Edwardsiella tarda isolated from red hybrid tilapia (Oreochromis spp.) coinfected with motile aeromonas septicemia and edwardsiellosis. Vet World (2017) 10(7):803–7. doi: 10.14202/vetworld.2017.803-807
25. Cheng ZX, Ma YM, Li H, Peng XX. N-acetylglucosamine enhances survival ability of tilapias infected by Streptococcus iniae. Fish Shellfish Immunol (2014) 40:524–30. doi: 10.1016/j.fsi.2014.08.008
26. Jiang M, Gong QY, Lai SS, Cheng ZX, Chen ZG, Zheng J, et al. Phenylalanine enhances innate immune response to clear ceftazidime-resistant Vibrio alginolyticus in Danio rerio. Fish Shellfish Immunol (2019) 84:912–9. doi: 10.1016/j.fsi.2018.10.071
27. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 32th informational supplement (M100). Wayne, PA: Clinical and Laboratory Standards Institute (2022). America. Available at: https://clsi.org/standards/products/microbiology/documents/m100/.
28. Zhao XL, Wu CW, Peng XX, Li H. Interferon-α2b against microbes through promoting biosynthesis of unsaturated fatty acids. J Proteome Res (2014) 13:4155–63. doi: 10.1021/pr500592x
29. Jiang M, Chen ZG, Li H, Zhang TT, Yang MJ, Peng XX, et al. Succinate and inosine coordinate innate immune response to bacterial infection. PloS Pathog (2022) 18(8):e1010796. doi: 10.1371/journal.ppat.1010796
30. Wang Z, Li MY, Peng B, Cheng ZX, Li H, Peng XX. GC–MS-based metabolome and metabolite regulation in serum-resistant Streptococcus agalactiae. J Proteome Res (2016) 15:2246–53. doi: 10.1021/acs.jproteome.6b00215
31. Yang DX, Yang MJ, Yin Y, Kou TS, Peng LT, Chen ZG, et al. Serine metabolism tunes immune responses to promote Oreochromis niloticus survival upon Edwardsiella tarda infection. mSystems (2021) 6(4):e0042621. doi: 10.1128/mSystems.00426-21
32. Wang Y, Yang Y, Shi Y, Song H, Yu C. Antibiotic-free antibacterial strategies enabled by nanomaterials: Progress and perspectives. Adv Mater (2020) 32(18):e1904106. doi: 10.1002/adma.201904106
33. Olson EG, Micciche AC, Rothrock MJ Jr, Yang Y, Ricke SC. Application of bacteriophages to limit Campylobacter in poultry production. Front Microbiol (2022) 12:458721. doi: 10.3389/fmicb.2021.458721
34. Su YB, Peng B, Li H, Cheng ZX, Zhang TT, Zhu JX, et al. The pyruvate cycle increases aminoglycosides efficacy and provides respiratory energy in bacteria. Proc Natl Acad Sci USA (2018) 115(7):E1578–87. doi: 10.1073/pnas.1714645115
35. Su YB, Kuang SF, Ye JZ, Tao JJ, Li H, Peng XX, et al. Enhanced biosynthesis of fatty acids is associated with the acquisition of ciprofloxacin resistance in Edwardsiella tarda. mSystems (2021) 6:e0069421. doi: 10.1128/mSystems.00694-21
36. Kuang SF, Chen YT, Chen JJ, Peng XX, Chen ZG, Li H. Synergy of alanine and gentamicin to reduce nitric oxide for elevating killing efficacy to antibiotic-resistant Vibrio alginolyticus. Virulence (2021) 12:1737–53. doi: 10.1080/21505594.2021.1947447
37. Kuang SF, Feng DY, Chen ZG, Liang ZZ, Xiang JJ, Li H, et al. Inactivation of nitrite-dependent nitric oxide biosynthesis is responsible for overlapped antibiotic resistance between naturally and artificially evolved Pseudomonas aeruginosa. mSystems (2021) 6:e00732–21. doi: 10.1128/mSystems.00732-21
38. Xu D, Wang J, Guo C, Peng XX, Li H. Elevated biosynthesis of palmitic acid is required for zebrafish against Edwardsiella tarda infection. Fish Shellfish Immunol (2019) 92:508–18. doi: 10.1016/j.fsi.2019.06.041
39. Kou TS, Wu JH, Chen XW, Peng B. Functional proteomics identify mannitol metabolism in serum resistance and therapeutic implications in Vibrio alginolyticus. Front Immunol (2022) 13:1010526. doi: 10.3389/fimmu.2022.1010526
40. Yang DX, Yang H, Cao YC, Jiang M, Zheng J, Peng B. Succinate promotes phagocytosis of monocytes/macrophages in teleost fish. Front Mol Biosci (2021) 8:644957. doi: 10.3389/fmolb.2021.644957
41. Chen XH, Liu SR, Peng B, Li D, Cheng ZX, Zhu JX, et al. Exogenous L-valine promotes phagocytosis to kill multidrug-resistant bacterial pathogens. Front Immunol (2017) 8:207. doi: 10.3389/fimmu.2017.00207
42. Jiang M, Yang LF, Chen ZG, Lai SS, Zheng J, Peng B. Exogenous maltose enhances Zebrafish immunity to levofloxacin-resistant Vibrio alginolyticus. Microb Biotechnol (2020) 13:1213–27. doi: 10.1111/1751-7915.13582
43. García-Ortiz A, Serrador JM. Nitric oxide signaling in T cell-mediated immunity. Trends Mol Med (2018) 24(4):412–27. doi: 10.1016/j.molmed.2018.02.002
44. Rasoul M, Rokhsareh M, Mohammad SM, Sajad K, Ahmadreza M. The Human immune system against Staphylococcus epidermidis. Crit Rev Immunol (2019) 39(3):151–63. doi: 10.1615/CritRevImmunol.2019031282
45. Kim JS, Kim YR, Yang CS. Host-directed therapy in tuberculosis: targeting host metabolism. Front Immunol (2020) 11:1790. doi: 10.3389/fimmu.2020.01790
Keywords: Edwardsiella tarda, aspartate, nitric oxide, reprogramming metabolome, antibiotic-free approach, zebrafish, sodium nitroprusside
Citation: Xiang J, Li M-y and Li H (2023) Aspartate metabolic flux promotes nitric oxide to eliminate both antibiotic-sensitive and -resistant Edwardsiella tarda in zebrafish. Front. Immunol. 14:1277281. doi: 10.3389/fimmu.2023.1277281
Received: 14 August 2023; Accepted: 15 September 2023;
Published: 11 October 2023.
Edited by:
Cheng Zhixue, Third Affiliated Hospital of Sun Yat-sen University, ChinaCopyright © 2023 Xiang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Li, bGlodWkzMkBzeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.