- Department of Biochemistry, University of Hyderabad, Hyderabad, India
Mycobacterium tuberculosis (Mtb) and HIV are known to mutually support each other during co-infection by multiple mechanisms. This synergistic influence could be either by direct interactions or indirectly through secreted host or pathogen factors that work in trans. Mtb secretes several virulence factors to modulate the host cellular environment for its persistence and escaping cell-intrinsic immune responses. We hypothesized that secreted Mtb transcription factors that target the host nucleus can directly interact with host DNA element(s) or HIV LTR during co-infection, thereby modulating immune gene expression, or driving HIV transcription, helping the synergistic existence of Mtb and HIV. Here, we show that the Mtb-secreted protein, EspR, a transcription regulator, increased mycobacterial persistence and HIV propagation during co-infection. Mechanistically, EspR localizes to the nucleus of the host cells during infection, binds to its putative cognate motif on the promoter region of the host IL-4 gene, activating IL-4 gene expression, causing high IL-4 titers that induce a Th2-type microenvironment, shifting the macrophage polarization to an M2 state as evident from CD206 dominant population over CD64. This compromised the clearance of the intracellular mycobacteria and enhanced HIV propagation. It was interesting to note that EspR did not bind to HIV LTR, although its transient expression increased viral propagation. This is the first report of an Mtb transcription factor directly regulating a host cytokine gene. This augments our understanding of the evolution of Mtb immune evasion strategies and unveils how Mtb aggravates comorbidities, such as HIV co-infection, by modulating the immune microenvironment.
Introduction
HIV and tuberculosis (TB) are the two most devastating infectious diseases that are emerging as a syndemic. There is no cure for HIV and the anti-retroviral therapy only prolongs the life expectancy with severe side effects. The co-infection of HIV and TB spirals the complications leading to a fatal outcome. While Mycobacterium tuberculosis (Mtb) primarily infects and replicates in host macrophages and dendritic cells, HIV has a wider host tropism (1). HIV infection gradually declines the CD4+ T cells leading to a sharp decline in immune responses (2). The decline in CD4+ T cells is a conducive state for the activation of TB (3), the most common secondary infection in HIV-infected patients. The influence of each other during co-infection could be mediated either by direct interaction of host and pathogen factors or indirectly through secreted host or pathogen factors that work in trans. Many Mtb factors have been associated with augmenting HIV titers (4, 5), however, the molecular mechanisms behind the impact are not clear for all the factors.
Mtb secretes several proteins towards adaptation to hostile microbicidal stress conditions. Some of these proteins also positively regulate HIV LTR and therefore viral replication (6, 7). One of the secreted proteins of the Mtb ESX-1 secretory system, EspR, is a transcription factor that controls its virulence (8). Besides this, EspR also regulates the ESX-1 operon by binding to the upstream espACD locus (9) and is secreted out by its transcriptional targets, making it unavailable for further activation, and exhibiting a negative feedback mechanism (10). Raw 264.7 (a murine macrophage cell line) stably expressing EspR was shown to inhibit BCG-induced apoptosis by intercepting the MyD88 pathway, indicating the role of EspR in counteracting host-induced innate defense (11). Although the role of EspR in Mtb gene regulation is well understood, its role in host-pathogen interaction, when secreted in the host milieu, is limited.
Many mycobacterial surface components and secreted proteins interact with host-pathogen recognition receptors, such as TLR2, polarizing the host macrophages to the M1 state. The cytokines released by the M1 polarized macrophages are pro-inflammatory, which brings about a Th1-type immune response, responsible for cell-mediated immunity (12). The clearance of Mtb and HIV-infected cells requires a Th1-dominant environment that activates effector T cells and cytotoxic T cells, the failure of which has been associated with both HIV and TB disease progression (13, 14). In response to the hostile intracellular conditions, virulent mycobacteria secrete several proteins shifting the polarization of macrophages to an alternate or compromised repair state M2 (15). The cytokines released by macrophages in the M2 state favor a Th2-type immune response, promoting bacterial persistence and HIV replication by dampening the host defenses (16, 17). We and others have reported that TB patients and those with HIV co-infection have mixed Th1/Th2 responses with elevated levels of both the kind of cytokines (18–20). The molecular mechanisms and participation of mycobacterial factors behind such mixed responses are still elusive.
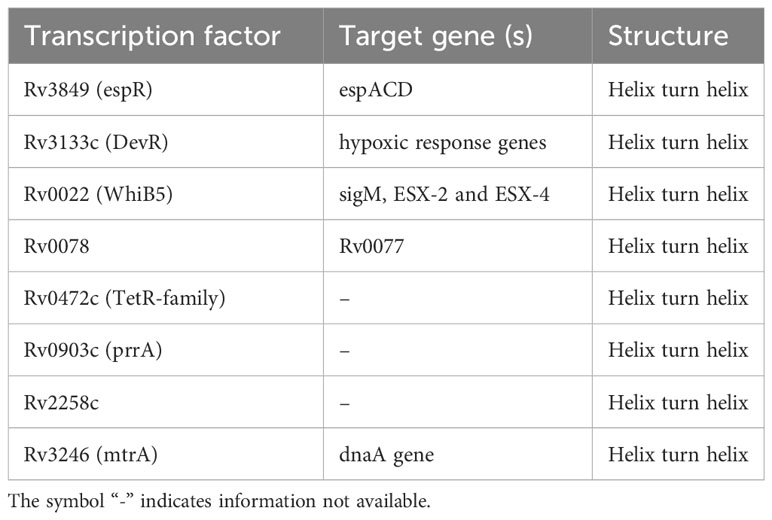
One of the ongoing studies of our laboratory is directed towards understanding the role of secreted Mtb transcription factors in mycobacterial pathogenesis and HIV-TB co-infection. In the present study, we shortlisted the transcription factors (Table 1) from the list of experimentally verified Mtb culture filtrate proteins (21). Among the shortlisted transcription factors, we focused on EspR and explored its functional role in the host macrophages. Here, we hypothesized that since EspR is a transcription factor, and if it gains access to the host nucleus, it could modulate host gene expression. In the present study, we found that EspR enters the host nucleus and commandeers the IL-4 gene expression by binding to its upstream promoter, leading to a Th2-type immune response and thereby increasing mycobacterial survival. We also observed that transiently expressing EspR or infection with EspR over-expressing mycobacteria in the background of HIV infection supported increased viral propagation, thereby adding new information to the role of mycobacterial factors in co-infection biology.
Methodology
The experiments were performed as per the approved protocols by the institutional biosafety committee of the University of Hyderabad, Hyderabad, Telangana, India.
Mycobacterial culture and HIV-1 strains
Mtb H37Rv and Mycobacterium smegmatis were grown either in 7H9 medium supplied with OADC (standard growth condition) or in Sauton’s minimal medium with acidic stress (pH 5.5), or oxidative stress (10 mM H2O2), or hypoxia (22–24) or nutrient starvation stress (grown in PBS) (25). Any possible contamination in Mtb culture was checked by Zeil-Neilson staining kit (HIMEDIA, India). HIV-1 strains used in the study include NL4.3 (X4 virus) or NL-ADA8 (R5 virus) (26–29).
Cell lines and antibodies
HEK293T cell line was maintained in DMEM (HIMEDIA, India) and THP-1 cell line (30) in RPMI-1640 (HIMEDIA, India) media, supplemented with 10% Fetal Bovine Serum (Gibco, USA), 100 U/ml of penicillin, and 100 ug/ml of streptomycin (HIMEDIA, India) at 37°C with 5% CO2 (31).
Different antibodies used to monitor the expression and localization of various proteins include anti-Myc antibody (Santacruz biotechnology (sc40); GAPDH (Santacruz biotechnology (sc47724); GFP antibody (Santacruz biotechnology, sc-9996 and ABclonal, AE011); anti-His antibody (Santacruz biotechnology, sc-8036), CD206 antibody (Santacruz biotechnology, sc-58986 PE), CD64 antibody (Santacruz biotechnology, sc-1184 FITC) and secondary antibodies include anti-mouse HRP (cell signaling, 08/2017), anti-rabbit HRP (Santacruz biotechnology, sc-2357) or anti-mouse Alexa 647 (Ab150115). The quantification of HIV for infection assays was performed using HIV-1 p24 Capture ELISA Assay Kit (ABL Inc, USA) and the released HIV antigens were measured using Microlisa HIV Ag & Ab assay kit (detects gp41, gp120, and p24) (JMitra &Co Pvt Ltd, India).
Transfection
THP-1 monocytes were differentiated into macrophages by treatment with PMA (Sigma, USA) (100 ng/ml) and incubated for 24 h. Subsequently, the cells were incubated in fresh media until macrophage morphology was observed (~24 h). For transfection, 6-8 x 105 HEK 293T cells or THP-1 macrophages were seeded in a 6-well plate 12-16 h before transfection. Fresh media was replaced before transfection with DNA complexes made using either Lipofectamine 2000 (Invitrogen, USA) (for THP-1 cells) or calcium phosphate (SIGMA, USA)(for HEK293T) (32) for about 6 h and used for experimental analyses 48 h post-transfection.
Plasmid constructs and recombinant EspR purification
The EspR ORF was amplified from Mtb H37Rv genomic DNA and cloned in pCDNA3.1 using BamH1 and XhoI sites (pC-EspR). pET-EspR was generated by cloning EspR ORF in pET28a using BamH1 and XhoI. pMSP12::mCherry-EspR was generated by cloning EspR ORF in pMSP12::mCherry (procured from the Addgene #30169) using KpnI and HindIII sites. pET-EspR transformed E. coli BL-21λDE3 (Agilent technologies, USA) cells were used for purification of recombinant MtbEspR protein (carrying N and C terminal 6X histidine tag) upon induction with 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Fermentas, USA) for 4 h at 37°C under native conditions using Talon-affinity resin (Clontech, USA) according to the manufacturer’s protocol and eluted with 200 mM imidazole containing PBS. Subsequently, rMtbEspR was dialyzed against PBS supplemented with 50 mM NaCl, and 10% glycerol. The protein concentration was measured using 1X Bradford Reagent (Bio-Rad, USA) according to the manufacturer’s instructions and the integrity of the protein was checked on 15% SDS PAGE (Supplementary Figures S1A, B).
Anti-EspR antibody generation
Anti-EspR antibody was raised as per protocol approved by the Institutional Animal Ethics Committee (UH/IAEC/SB/2022/45), by injecting 500 µg purified rMtbEspR diluted in a mixture of 0.5 ml PBS and 0.5 ml complete Freund’s adjuvant into a healthy rabbit as a prime dose. A booster dose was scheduled every two weeks with a gradual decrease (by half) of rMtbEspR protein and emulsified with incomplete Freund’s adjuvant and the cycle was repeated five times. At the end of the schedule, approximately 10 ml of blood was collected, and serum was isolated by centrifugation at 2000 x g for 10 min and confirmed by western blotting with purified protein including a non-specific protein as a negative control (Supplementary Figure S1C).
Electrophoretic mobility shift assay
EMSA was carried out as described with minor modifications (33). Briefly, the chemically synthesized EspR cognate sequences (FL, ΔS1, ΔS2) (IDT/Eurofins, Germany) were annealed and radiolabeled with γP32 using T4 PNK (NEB, England). The binding assays of recombinant EspR to different probes were performed in NEB 2.1 buffer supplemented with 50 ng/ml pdIdC, 10% glycerol, and in the presence or absence of unlabeled molar excess of cognate DNA (cold/homologous competition) for 30 min at ambient temperature (25-30°C). The DNA-protein complexes were resolved on a 5% native acrylamide gel (39: 1) at 200 V in 0.5 X TBE. Subsequently, the gels were dried and exposed to a phosphor imaging screen overnight (16-20 h) and imaged using a Typhoon scanner (BIO-RAD, USA) with Quant One software.
Infection and persistence
For infection, THP-1 macrophages were incubated with wildtype or transformed M. smegmatis at an MOI of 20 to 25 for 4 h. Subsequently, the cells were washed with PBS and replaced with fresh media. For bacterial persistence, the infected THP-1 cells were lysed in 100 ul water (per 104 cells) for 30 min at 37°C in 50 ul duplicates. Each aliquot was either processed by the Alamar blue method or plated on 7H10 plates and incubated over 4-5 days. The resulting colonies were counted and the cfu was calculated using the formula cfu/ml= no of colonies * dilution factor/volume of the culture plated. 100 ng/ml of p24 equivalent of HIV-1 (produced from pNL4.3 or NL-ADA8) was used for infection assays in the presence or absence of MtbEspR as described in the results section. The comparative release of viral antigens was quantified after 48 h and expressed as a relative percentage compared to the control experiment.
Immunofluorescence microscopy
HEK293T cells were washed twice with PBS (HIMEDIA, India) and fixed with 4% formaldehyde (Sigma, USA) for 20 min. The fixed cells were again washed with PBS to remove formaldehyde and subsequently permeabilized by using 0.5% Triton-X-100 (Qualigens, India) for 15 min followed by washing the cells twice with PBS and then blocking with 3% BSA (SRL, India) for 30 min. The cells were washed once with PBS and incubated in the appropriate primary antibody (in 1% BSA) for 2 h, followed by three washes with PBS. The cells were further incubated with the corresponding secondary antibody tagged with Alexa 647 (Ab150115) for 1 h. Subsequently, the cells were washed with PBS and the nuclei were stained with DAPI (Abcam, USA). Imaging was done by Leica confocal microscopy and visualized by Las X software. Plot profiles were generated by using Image J and Huygens software.
Live cell imaging
Around 0.3 million infected THP-1 cells (with M.smeg::pMSP12 or M.smeg::MtbEspR) were seeded in a live cell imaging dish (Ibidi GmbH, Germany). The cells were treated with Hoechst dye (Thermo Scientific, USA) for 15 min. Later, the cells were washed twice with PBS and finally replaced with fresh media and subjected to live cell imaging (Leica confocal microscope). The images were collected for 30 min to 1 h for every 30 s and visualized by Las X software.
Western blotting
Mtb H37Rv cultures grown under different stress conditions were used to harvest both cells and supernatant. The cells were resuspended in PBS and lysed using a bead beater for 10-20 cycles of 1 min pulse and 2 min pause. The culture supernatant was passed through a 0.2 μm filter. The protein concentrations (of both cell lysates and culture filtrate) were quantified by Bradford assay. Subsequently, the samples were fractionated on SDS-PAGE and transferred onto a nitrocellulose membrane (PALL, BioTrace™ NT nitrocellulose, USA) and confirmed by ponceau (Fluka Analytical, USA) staining. The blots were blocked with 5% skimmed milk (HIMEDIA, India) for 1 h at room temperature, followed by 3 washes with PBS and then incubated with primary antibody overnight at 4°C. The next day, the blot was washed thrice with PBS and further incubated with appropriate secondary antibody for 2 h and developed using a WesternBright™ ECL kit (Advansta, K-12045-D20) in a Biorad ChemiDoc instrument. The secreted levels of EspR were quantified by densitometry using Image J and represented in terms of fold change normalized to the total protein loaded in each condition.
RNA extraction and qRT-PCR
To measure the mRNA levels of IL-4 in THP-1 cells, the RNA was isolated by using a GeneJET RNA purification kit (Thermo Scientific, USA) according to the manufacturer’s protocol. For mycobacterial RNA, Mtb H37Rv cultures grown under different conditions were harvested and resuspended in Trizol (Invitrogen, USA) and lysed using a bead beater for 15 cycles of 1 min pulse and 2 min pause. The lysate was centrifuged at high speed for 10 min at 4°C and processed for RNA isolation following the manufacturer’s protocol. Further, 10 µg of isolated RNA was treated with DNase I (NEB, England) to remove any genomic DNA contamination. 1ug of DNase I treated RNA was converted to cDNA by using the iScript cDNA synthesis kit (BIO-RAD, USA) according to the manufacturer’s protocol. The diluted cDNA was used as the template for the qRT-PCR. The qRT-PCR was performed using the iTaq Universal SYBR green Supermix (BIO-RAD, USA) following the manufacturer’s protocol, in a 96-well plate (Applied Biosystems, USA) and analyzed using QuantStudio3 software.
Luciferase assay
HEK 293T or THP-1 cells transfected with appropriate plasmids were maintained as described for 48 h. The cells were washed with PBS and harvested by trypsin treatment (0.25 X for 1 min at 37°C) and again washed with PBS. The cells were lysed and processed for luciferase assay (Promega Luciferase Assay system, USA). Briefly, the samples were incubated at room temperature for 20 min followed by a brief vortex. The samples were frozen at -80°C for 1 h, freeze-thawed for 2 more cycles, and centrifuged at 11,000 rpm for 5 min at 4°C. The supernatant was collected in a fresh microfuge tube and the luminescence was recorded using the modulus single-tube multimode reader (Promega) according to the manufacturer’s protocol.
TZM-bl assay
TZM-bl cells, a cell line with a β-galactosidase reporter gene driven by HIV LTR (34), were infected with equal volume of culture supernatants collected from HEK293T cells transfected with either pCDNA or pC-EspR along with NL-ADA8 as described (35). Briefly, after 24 h of infection, the cells were washed with cold PBS (HIMEDIA, India) and harvested upon trypsin treatment. Cells were centrifuged at 1000 rpm for 5 min and washed once again with PBS and processed as described above.
Flow cytometry
HEK 293T or THP-1 cells transfected with appropriate plasmids were harvested by trypsin treatment and washed with PBS thrice. The cells were passed several times through a blunt 20-gauge needle fitted to a sterile syringe, to make a single-cell suspension (without any visual clumps). These cells were processed for immunostaining and acquired on an S3e cell sorter (FACS) to evaluate the cell surface markers for M1 and M2 polarization states and the data was analyzed using FACS Express 6 software.
Statistical analysis
Plots were generated and the statistical significance was determined by a two-tailed unpaired student’s t-test in GraphPad Prism. A p-value <0.05 was considered statistically significant. All the experiments were performed at least thrice.
Results
MtbEspR is differentially expressed and is secreted during microbicidal stress conditions
As it is known that MtbEspR is secreted, we confirmed the same in our experimental conditions. We investigated if it is differentially regulated under conditions mimicking intracellular microbicidal stresses. Towards this, Mtb was grown in well-established in vitro growth conditions including acidic stress, hypoxia, oxidative stress, and nutrient starvation conditions besides standard growth conditions. Total RNA was extracted from the cultures of each condition and the expression of espR RNA was analyzed by qRT-PCR. We observed an increase in the transcript levels of espR in acidic (~2-fold), oxidative (~3-fold), and hypoxic (~1.5-fold) conditions as compared to standard growth conditions (Figure 1A). Besides, we also investigated the protein levels of EspR, both in the cell lysates and culture filtrates, under these conditions (Figures 1B, C). We observed that EspR was secreted in all conditions but was elevated significantly in the culture filtrates of acidic stress as compared to standard growth conditions (Figures 1C, D). The levels of EspR, both at transcript and protein levels, were relatively low during nutrient starvation. Overall, these observations confirmed that EspR is secreted both during standard growth conditions and during abiotic stresses.
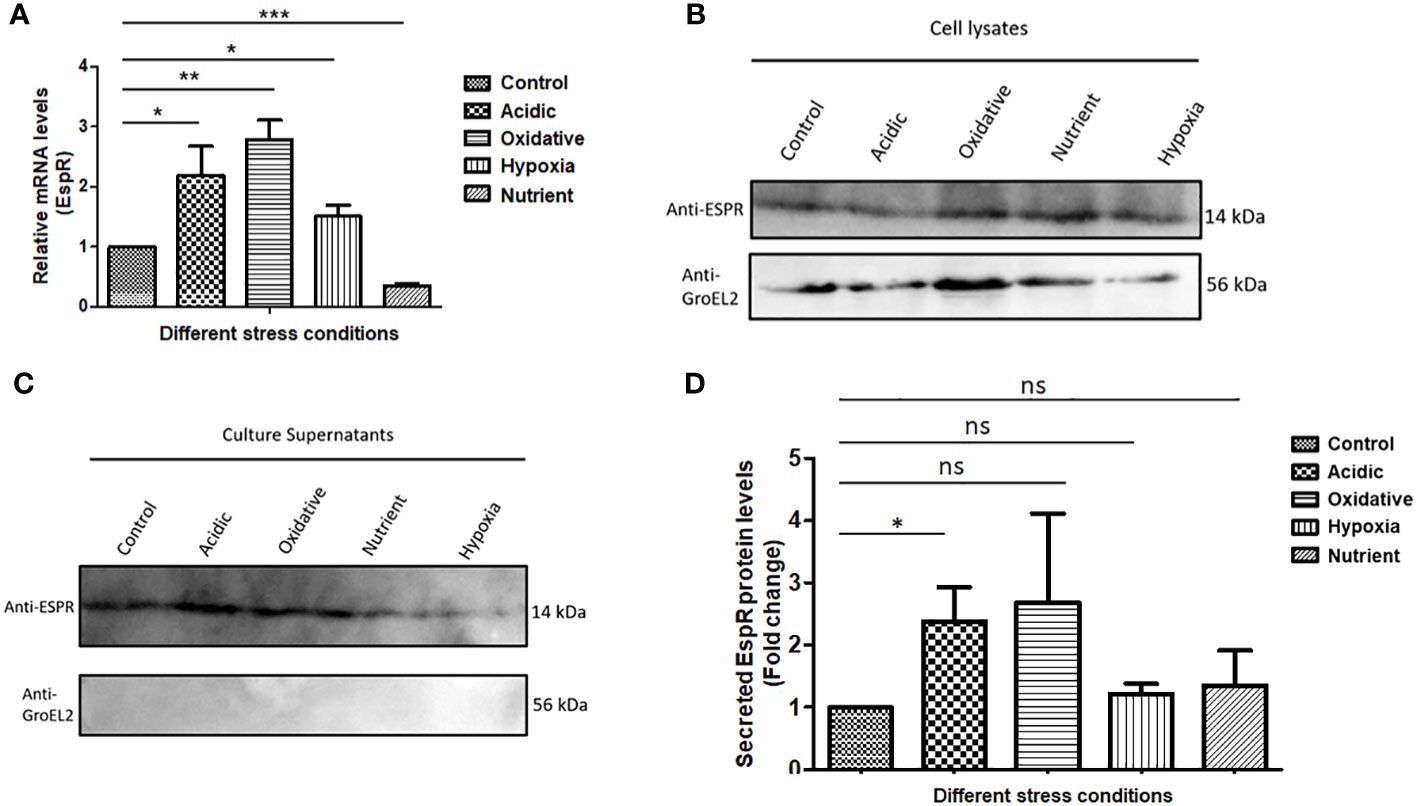
Figure 1 MtbEspR is differentially expressed and secreted during abiotic stress conditions (A) The levels of EspR RNA from Mtb H37Rv were measured in microbicidal stress conditions acidic, oxidative, hypoxia, and nutrient starvation as compared to standard growth conditions by qRT-PCR. (B) Immunoblots showing EspR protein levels in the microbicidal stress conditions in Mtb H37Rv cell lysates. GroEL2 was used as a lysis control. An equal amount (as measured by Bradford assay) of lysates was loaded in each well (C) Immunoblots showing the EspR protein levels in the respective culture supernatants. (D) Bar graph showing immunoblot-based quantification of EspR secretion for experiments represented in (C) The quantification for each condition was normalized to the total protein loaded. All the experiments were done at least 3 times. The significance was determined using an unpaired student’s t-test. The p-values are denoted as ***p <= 0.001; **p <= 0.01; and *p <= 0.05, while non-significant values are denoted by n.s.
MtbEspR supports the survival of mycobacteria and HIV propagation during co-infection
With increased expression and secretion of EspR under stress conditions, we investigated if over-expression of this protein provides an intracellular survival advantage. To prove the same, MtbEspR was ectopically expressed in M. smegmatis, an Mtb surrogate system, which has limited persistence inside macrophages and is generally cleared by macrophages within 4-9 h during in vitro experimental conditions (36). We generated two recombinant strains M. smegmatis::pMSP12 (vector control, from now onwards M.smeg::pMSP12) and M. smegmatis::MtbEspR (M.smeg::MtbEspR). The expression and secretion of MtbEspR, which is tagged with mCherry, were verified both in the cell lysates and culture supernatants by western blotting (Supplementary Figures S2A, B). The two strains showed no difference during in vitro growth kinetics in 7H9 media (Supplementary Figure S2C). After confirming secretion of MtbEspR from the recombinant M. smegmatis strains under in vitro growth conditions, THP-1 cells were infected with M.smeg::pMSP12 and M.smeg::MtbEspR, wherein we observed a 24% increase in the levels of M.smeg::MtbEspR than M.smeg::pMSP12, suggesting a higher survival of M. smegmatis over-expressing MtbEspR (Figure 2A). With numerous clinical reports showing the synergistic impact of secondary mycobacterial infections on HIV titers in co-infected patients (18), we further evaluated the effect of MtbEspR on mycobacterial survival and viral propagation during co-infection. THP-1 macrophages infected with HIV NL4.3 were subsequently infected with either M.smeg::pMSP12 or M.smeg::MtbEspR. As reported by us and others, we observed that mycobacterial survival was enhanced in HIV infection background. However, the persistence of M.smeg::MtbEspR increased over M.smeg::pMSP12, pointing to the role of MtbEspR in mycobacterial survival during co-infection (Figure 2B). Similarly, we also evaluated the release of viral proteins in culture supernatants during co-infection, wherein we observed an increase in viral proteins in the culture supernatant upon M.smeg::MtbEspR co-infection compared to M.smeg::pMSP12 (Figure 2C). To confirm that MtbEspR indeed influences HIV propagation, we transiently expressed MtbEspR in HEK293T cells. The released viral proteins were measured in the culture supernatant 48 h post-transfection by ELISA. We observed a ~24% increase in released viral proteins upon transient expression of MtbEspR (Figure 2D) as compared to vector control. Similarly, we performed experiments with the M-tropic (R5) virus of NL-ADA8 to verify the effect of MtbEspR on R5 viruses. We observed a similar increase in the released NL-ADA8 as we had observed for NL4.3 in the presence of MtbEspR (Supplementary Figures S2D, E). With these experiments, we confirmed that mCherry-tagged MtbEspR protein is secreted out from M.smeg::MtbEspR and that MtbEspR increases both mycobacterial survival and HIV propagation in the host cells.
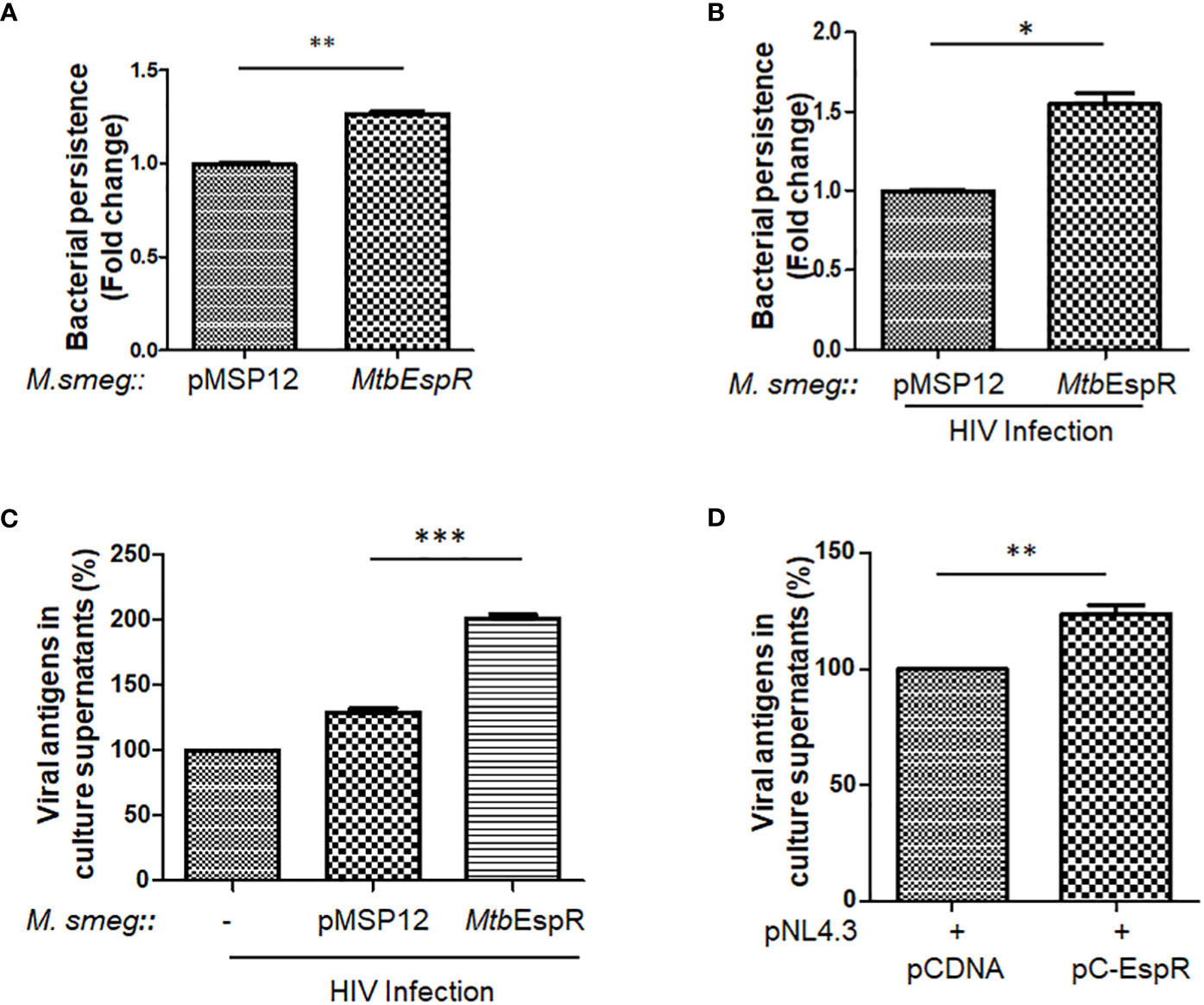
Figure 2 MtbEspR supports the survival of mycobacteria and HIV propagation in host macrophages (A) Fold change in the persistence of M.smeg::pMSP12 and M.smeg::MtbEspR strains as analyzed in THP-1 cells by Alamar Blue assay (B) Fold change in the persistence of M.smeg::pMSP12 and M.smeg::MtbEspR strains as analyzed in HIV infected THP-1 cells by CFU (C) Relative change in release of viral antigens in the culture supernatant as determined by ELISA in NL4.3, NL4.3+M.smeg::pMSP12; and NL4.3+M.smeg::MtbEspR infection of THP-1 cells, expressed in percentage (D) Relative change in release of viral antigens in the culture supernatant as determined by ELISA in pCDNA, pC-EspR transfected HEK293T cells (expressed in percentage). All the experiments were done at least 3 times. The significance was determined using an unpaired student’s t-test. The p-values are denoted as ***p<= 0.001; **p<= 0.01 and *p <= 0.05.
The secreted MtbEspR localizes to the host nucleus during infection
To comprehend the mechanism of action of MtbEspR and considering that it is a secreted transcription factor, we traced the localization of MtbEspR inside mammalian cells by transient expression of MtbEspR with a GFP tag (pEGFP-EspR) (Supplementary Figure S3A) in HEK293T cells and M.smeg::MtbEspR infected THP-1 cells using confocal microscopy by both fixed and live cell imaging. Towards this, we first transfected HEK293T cells with pEGFP-EspR and pC-EspR to identify its subcellular localization by confocal microscopy. We found that MtbEspR localized to the host nucleus as reflected by the cyan color in the merged confocal image (Figure 3A; Supplementary Figure S3B). The mCherry tagged MtbEspR in M.smeg::MtbEspR infected THP-1 also localized to the nuclei as observed in the merged confocal image and the line plots of representative single cells (Figure 3B; Supplementary Video S3C). The infection experiments confirmed that MtbEspR is released by M.smeg::MtbEspR and enters the host nucleus. With this, we concluded that MtbEspR enters the host nucleus both upon transient expression and during infection of THP-1 cells.
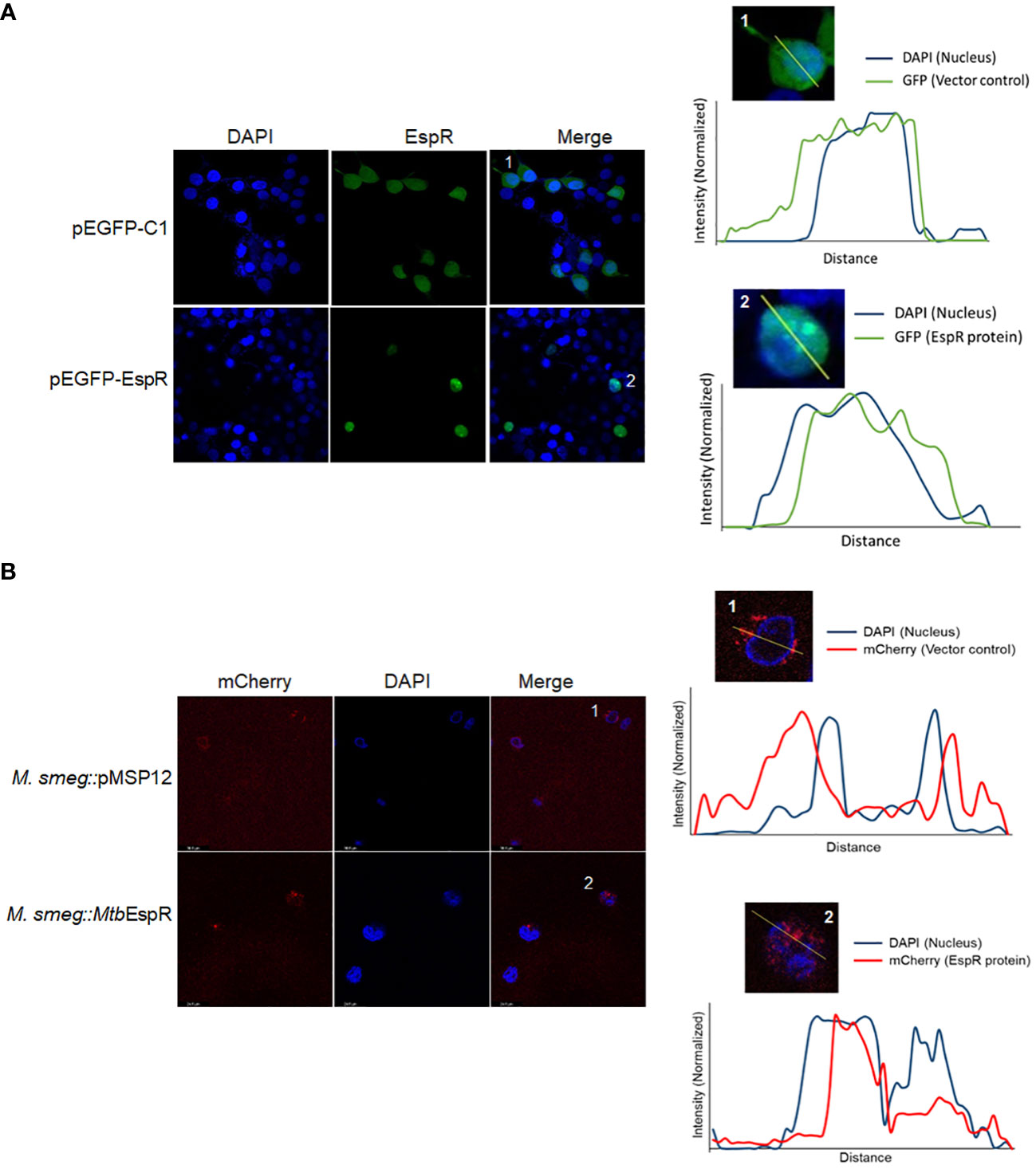
Figure 3 MtbEspR localizes to the host nucleus (A) Confocal microscopy of MtbEspR subcellular localization in HEK293T cells upon transfection with pEGFPC1 or pEGFP-EspR, along with plot profiles of representative single-cell images of the insets 1 and 2 were generated using ImageJ software (B) Confocal microscopy of MtbEspR localization in THP-1 cells upon infection with M.smeg::pMSP12, M.smeg::MtbEspR. The plot profiles of representative single-cell images of insets 1 and 2 were generated using ImageJ software. All experiments were done at least 3 times.
MtbEspR binds to the DNA elements in the upstream promoter region of the human IL-4 gene and positively regulates the IL-4 promoter
As mentioned earlier, MtbEspR is a mycobacterial transcription factor that binds to its cognate DNA element for transcriptional regulation (8). With the evidence that it indeed reaches the nucleus of the infected cell and also enhances mycobacterial survival and HIV propagation during co-infection, we next investigated if MtbEspR regulates any of the host gene expressions or HIV LTR. We manually screened for EspR consensus sequences in the promoters of the host innate immune genes as well as HIV LTR. Among the different innate immune gene regulators, we identified the EspR consensus sequence in IL-4 and IFN-γ promoters. We also found an EspR consensus binding site in HIV LTR (Figure 4A). Upon analysis of the EspR binding sites, we noted that human IL-4 and IFN-γ upstream regions have two adjacent EspR binding consensus sequences each, but HIV LTR had a single binding site. However, the orientation of the two binding sites is different for IL-4 and IFN-γ, with tandem orientation in the IL-4 promoter region and reverse in the IFN-γ promoter (Figure 4A). To confirm the occupancy of these sites by EspR, we performed EMSA using rMtbEspR with the probes of IL-4, IFN-γ promoters, and HIV LTR, wherein we observed that purified recombinant MtbEspR binds to the IL-4 promoter probe (Figure 4B), but not to that of IFN-γ or HIV LTR (Supplementary Figure S4).
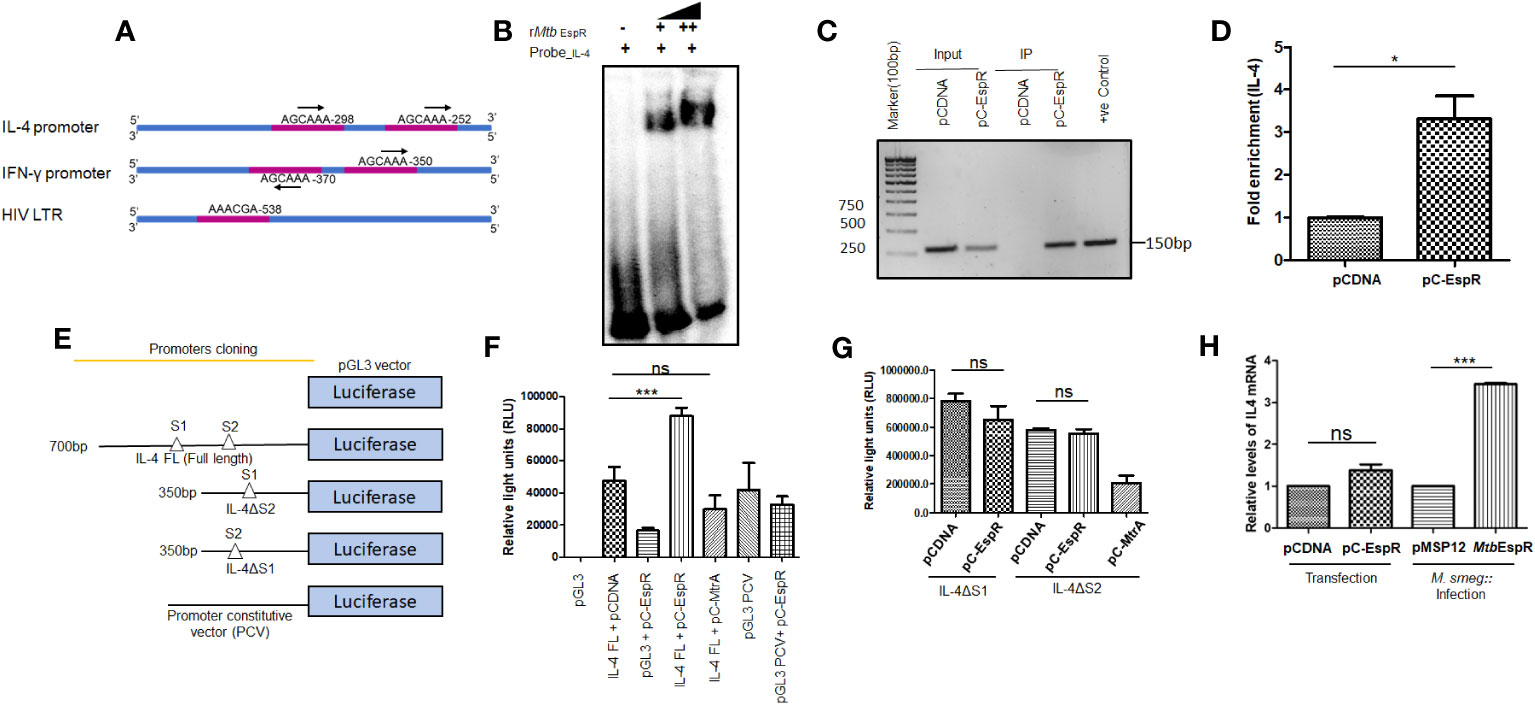
Figure 4 MtbEspR binds and positively regulates the IL-4 promoter (A) Schematic representation of the different EspR binding sites in the promoter regions of IL-4, IFN-γ and HIV LTR (B) Electrophoretic mobility shift assay depicting the binding of MtbEspR to the IL-4 promoter with increasing concentration of purified rMtb EspR protein (C) ChIP assay was performed using an anti-Myc antibody after transfecting with pCDNA or pC-EspR along with pGL3-IL-4 FL reporter followed by PCR using IL-4 promoter-specific primers (D) qRT-PCR was performed using IL-4 promoter-specific primers to quantify the occupancy of EspR on the IL-4 promoter region (E) Schematic illustration of the various IL-4 promoter constructs used for the luciferase reporter assay (F) HEK293T cells were transfected with full-length IL-4 reporter construct either in the presence or absence of MtbEspR (pC-EspR), followed by measurement of luciferase activity 48 h post-transfection. (G) The luciferase assay was performed with deletion constructs of IL-4 binding sites (IL-4ΔS1 and IL-4ΔS2 promoters) upon transient expression of MtbEspR (pC-EspR). Mtb MtrA was used as a non-specific Mtb protein (H) qRT-PCR was performed to quantify IL-4 mRNA in the presence and absence of MtbEspR protein either by transient transfection (pC-EspR) or during infection (M.smeg::MtbEspR). All experiments were done at least 3 times. The significance was determined using an unpaired student’s t-test. The p-values are denoted as ***p <=0.001 and *p <=0.05, while non-significant values are denoted by n.s.
To confirm the protein-DNA interaction of MtbEspR and IL-4 promoter in vivo, we performed chromatin immunoprecipitation assay (ChIP)-PCR in HEK293T cells transiently transfected with pC-EspR using anti-Myc antibody and IL-4 specific upstream primers. The ChIP analysis showed an enrichment of the IL-4 promoter in pC-EspR transfected HEK293T cells but not in its vector control cells as visualized by semi-quantitative (Figure 4C) and quantitative PCR (Figure 4D). Overall, these experiments suggested that MtbEspR binds to the IL-4 promoter both in vitro and in vivo.
After establishing that MtbEspR binds to the IL-4 promoter, we further investigated if MtbEspR indeed transcriptionally regulates the host IL-4 gene. Towards this, we cloned IL-4 full-length (FL, 700 bp upstream to IL-4 gene TSS), IL-4 ΔS1 (~350 bp upstream to IL-4 gene TSS), and IL-4 ΔS2 promoters (~350 bp upstream to IL-4 gene TSS) in the MCS of pGL3 basic vector (Figure 4E). These reporter constructs were transfected in HEK293T cells followed by luciferase assays. To evaluate the effect of MtbEspR, the reporter constructs, pGL3-IL-4 FL or pGL3 PCV (having a constitutive SV40 promoter as non-specific promoter control) were co-transfected with either pC-EspR or its vector control (pCDNA). An increase in luciferase activity (RLU) was observed upon co-transfection of pGL3-IL-4 FL and pC-EspR but not with the control pGL3 PCV signifying a positive regulation of IL-4 in the presence of MtbEspR (Figure 4F). The mutant reporter constructs with either S1 or S2 deletions did not influence luciferase activity as compared to respective vector controls, suggesting that both EspR consensus binding sites are required for EspR binding and regulation of the target gene expression (Figure 4G). With this, we concluded that MtbEspR is an activator of IL-4 gene expression.
To validate these observations in vivo, we monitored IL-4 expression at mRNA level in THP-1 macrophages both upon infection with M.smeg::MtbEspR, and transfection with pC-EspR by qRT-PCR. The results showed an increase in IL-4 mRNA levels both upon infection and transfection as compared to their respective vector controls (Figure 4H). Transfection of THP-1 cells with the pC-EspR could only marginally increase IL-4 transcript levels, but a 3-fold increase was observed during infection. These experiments suggested that MtbEspR increased IL-4 gene expression by binding to the upstream of the IL-4 promoter.
MtbEspR alters the host cytokine environment and macrophage polarization supporting mycobacterial persistence
Having observed that MtbEspR positively regulates the IL-4 promoter and knowing that IL-4 is critical for the shift towards Th2-type immune response, we next investigated the cytokines that are released from THP-1 upon infection with M.smeg::MtbEspR and compared that with M.smeg::pMSP12 infection. We scored for IFN-γ, IL-12 p70, IL-1 β, TNF-α, IL-10, and IL-4 in their culture supernatants upon infection. The analysis clearly showed that there were no differences in levels of the proinflammatory cytokines, TNF-α, IFN-γ, IL-12 p70, and IL-1 β upon either M.smeg::pMSP12 or M.smeg::MtbEspR infections. However, significant differences were noticed in the levels of regulatory cytokines IL-10 and IL-4 (Figures 5A–F), wherein M.smeg::MtbEspR infection showed higher cytokine titers, clearly pointing to an inclination towards Th2 immune response. We next checked the polarization of THP-1 macrophages upon infection with either M.smeg::pMSP12 or M.smeg::MtbEspR. The polarization of THP-1 macrophages was evaluated by FACS using CD64 and CD206 as M1 and M2 polarization surface markers respectively. THP-1 macrophages stimulated with either IFN-γ or IL-4 were used as positive controls for M1 and M2 polarization (Supplementary Figures S5A–C). We observed that transfection of pC-EspR or infection with M.smeg::MtbEspR shifted the macrophage population to the M2 state (Supplementary Figures S5A–C). To complement these experiments and delineate that the M2 polarization is indeed due to MtbEspR, we repeated the above experiment by transient transfection with pC-EspR. As observed in infection, we saw a similar trend in the levels of secreted cytokines upon transfection (Figures 5A–F) as in THP-1 polarization (Supplementary Figures S5A–D). To reconfirm that divergent polarized macrophage populations showed differential clearance of M.smeg::MtbEspR, we treated THP-1 cells with either IFN-γ or IL-4 before infecting them with M.smeg::pMSP12 and M.smeg::MtbEspR (Figure 5G). As evident from the plots in Figure 5G, we observed an increased load of M.smeg::MtbEspR as compared to M.smeg::pMSP12 upon IL-4 treatment. We also observed that M.smeg::MtbEspR marginally survived better even in IFN-γ treated THP-1, but the experiments were not statistically significant. The control experiments with M.smeg::pMSP12 infection alone are shown in Supplementary Figure S5E. With these experiments, we concluded that MtbEspR-mediated alterations in cytokine microenvironment to Th2-type promoted the increased survival of M.smeg::MtbEspR.
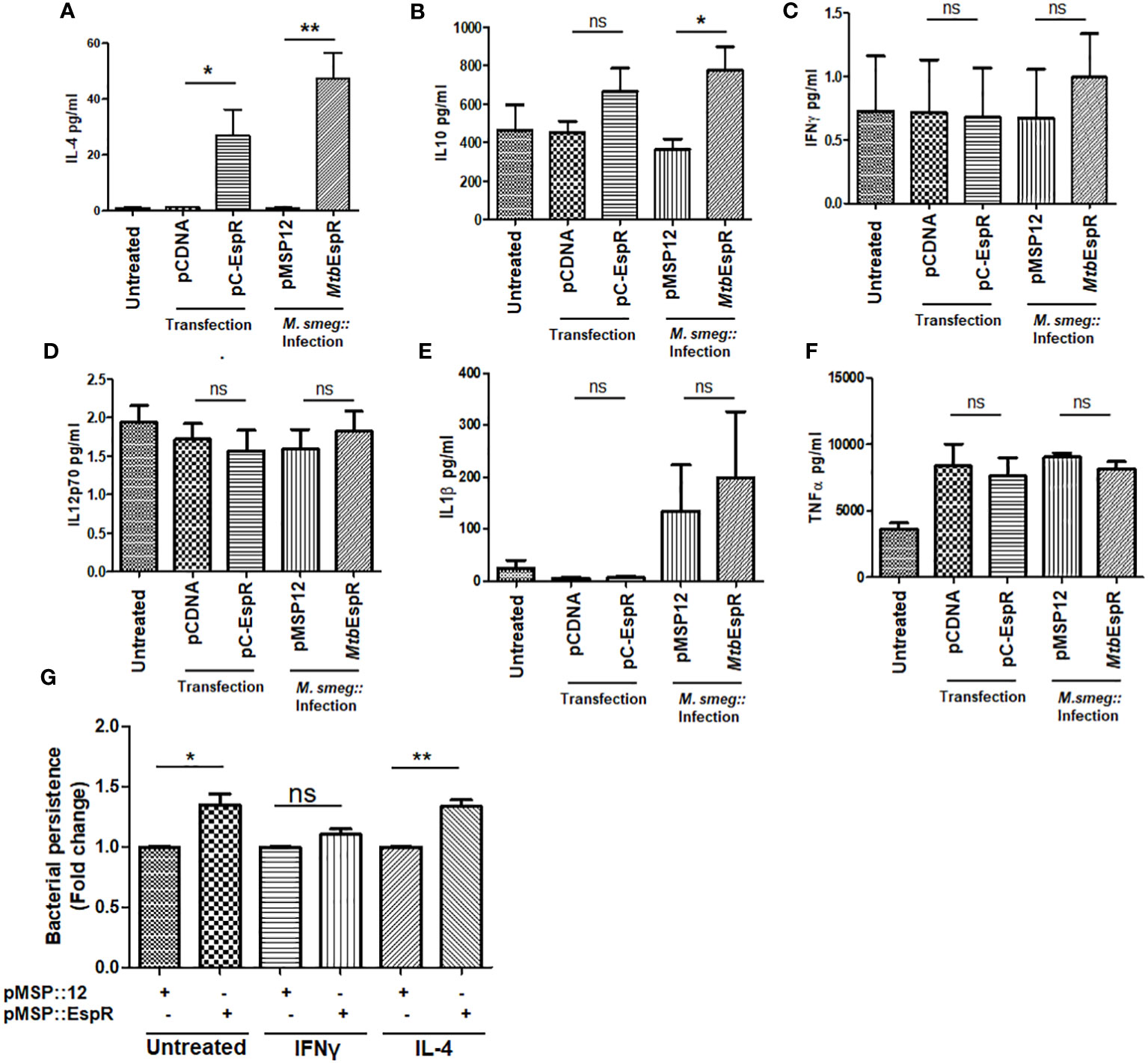
Figure 5 MtbEspR orients the macrophage immune response to Th2-type PMA-treated THP-1 macrophages either transiently expressing MtbEspR (pC-EspR) or infected with M.smeg::MtbEspR were evaluated for pro-inflammatory or anti-inflammatory cytokines by ELISA: (A) IL-4, (B) IL-10, (C) IFN-γ, (D) IL-12 p70, (E) IL-1β, (F) TNF-α. (G) Bacterial persistence using Alamar Blue assay was analyzed in IFN-γ, and IL-4 treated THP-1 macrophages, after infecting with M.smeg::pMSP12, M.smeg::MtbEspR. All experiments were done at least 3 times. The significance was determined using an unpaired student’s t-test. The p-values are denoted as **p <=0.01; and *p <=0.05, while non-significant values are denoted by n.s.
Conclusively, this study shows that the Mtb factor EspR is secreted during both standard growth conditions as well as microbicidal stresses and infection, localizes to the nucleus of the infected host cell, and binds to the IL-4 promoter, augmenting its expression, and thereby enhancing secreted IL-4 levels (Figure 6). In summary, this shifts the cytokine microenvironment to a Th2-type immune response enhancing mycobacterial persistence and HIV propagation both during single infection and during HIV co-infection.
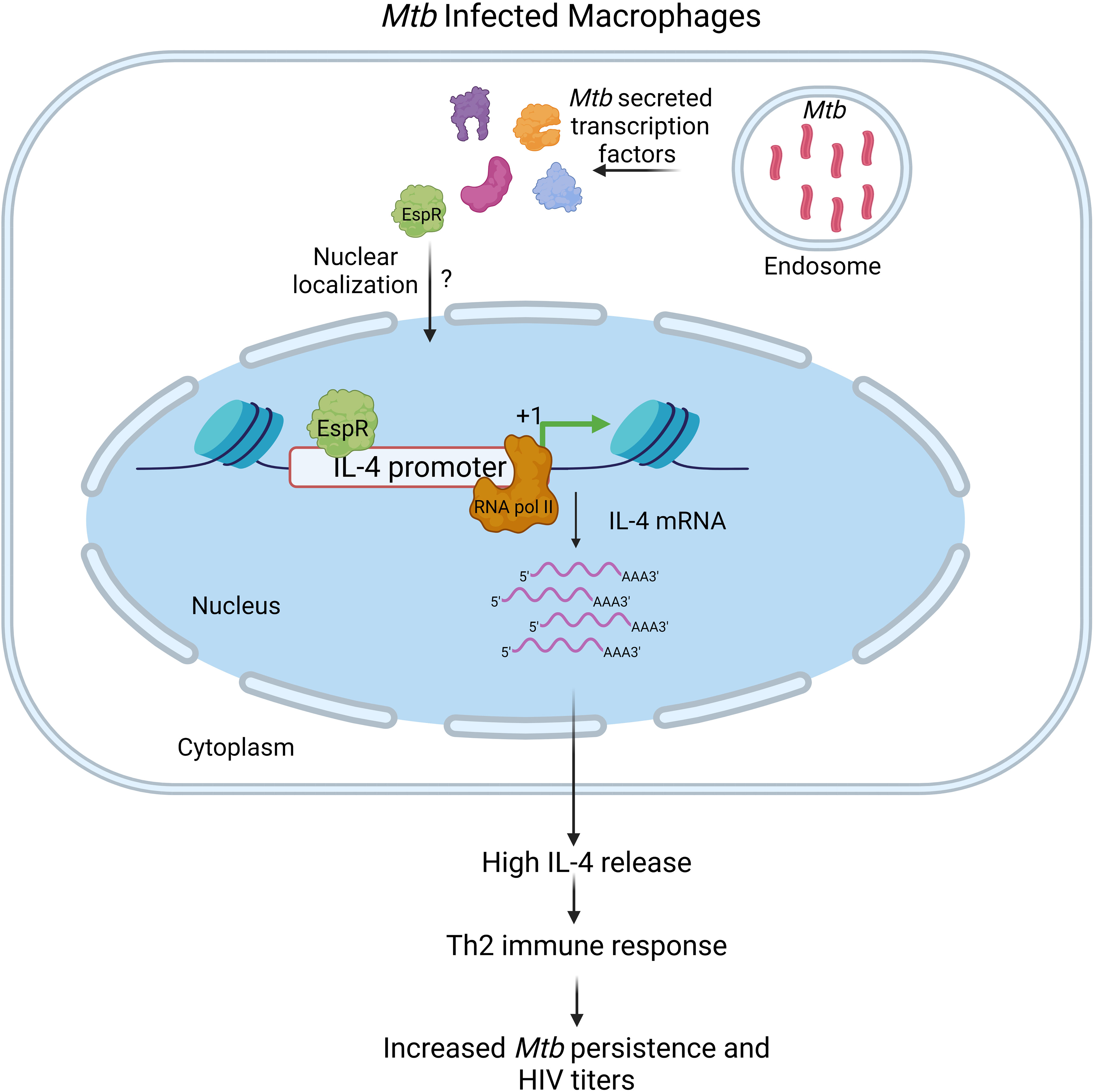
Figure 6 Schematic representation of the role of MtbEspR in Mycobacterium tuberculosis persistence Mycobacterium tuberculosis infects macrophages, where they primarily multiply and persist. Although the host exerts several innate defense mechanisms, they are thwarted by various mycobacterial proteins, either surface localized or secreted that have a major role in host-pathogen interaction. MtbEspR is one of the secreted proteins in response to microbicidal stress conditions, that enters the host nucleus, wherein it interacts with host DNA elements such as IL-4 and activates the host IL-4 gene expression and ultimately increasing IL-4 cytokine levels, shifting the immune response of the host from Th1- to Th2-type that is conducive for mycobacterial as well as viral multiplication. Created with Biorender.com.
Discussion
Mtb is a remarkably successful pathogen as it effectively thwarts host defenses through a multitude of mechanisms owing to its secreted proteins and metabolites. In the present study, we have revealed three new facets of MtbEspR in host-pathogen interaction (1). MtbEspR gains access to the host nucleus and directly regulates the host innate immune genes. (2) Mechanistic details of MtbEspR mediated shift in the Th1 and Th2 microenvironment. (3) MtbEspR positively regulates HIV propagation during co-infection.
Secretion of transcriptional regulator proteins from pathogens and their binding to host DNA elements is extremely rare in biology, with a few examples from viruses. For example, HIV Tat, a transcription factor, released from an infected cell can reach the nucleus of a bystander host cell and interact with DNA (37, 38). MtbEspR from this study represents a transcription factor from pathogenic mycobacteria that showed the capacity to alter the expression of a host gene by directly binding to the host DNA elements. While there is a study that reported that EspR is not secreted by Mtb (9), this study and others have shown that EspR is secreted (10, 39). It was also intriguing to note that as expected out of a transcription factor, MtbEspR indeed entered the host nucleus. Most proteins that are targeted to the nucleus possess a consensus nuclear localization signal (NLS) for nuclear entry. MtbEspR, however, lacks a conventional NLS and hence can be speculated to be using specific host nuclear import machinery to gain entry into the nucleus during infection. MtbEspR is a small protein and hence it may also gain access to the nucleus through nuclear pore complexes by simple diffusion. This may be addressed by exploring the host protein interaction network of MtbEspR during infection.
With the availability of the EspR binding sequence (8), we scanned the promoter regions of many human cytokine genes. Among them, IL-4 and IFN-γ gene upstream sequences showed two consensus EspR binding sites. Our earlier reports have shown that these two cytokines are concurrently upregulated in HIV-TB co-infected patients (18). Hence, we also scanned the HIV LTR sequence and observed the presence of a single EspR binding site. The orientation of EspR binding sites in the promoters of IL-4 and IFN-γ are different. While the repeats are tandem in IL-4, the orientation of the two EspR binding sequences are in reverse orientation in IFN-γ. With EMSA and ChIP data clearly showing that EspR binds to IL-4, but not to IFN-γ or HIV LTR sites, one can speculate that two tandem binding sequences are important for EspR-DNA interactions, although in-depth biochemistry and characterization of the DNA binding property of EspR is required. The reporter assays showed that EspR is an activator of the IL-4 promoter. We also noted that EspR needs two consensus-binding motifs for the regulation of transcription. When we deleted either of the consensus sequence from the IL-4 promoter and performed the reporter assays, we could not observe any significant enhancement in luciferase activity upon EspR expression, suggesting that EspR needs both the consensus sequences for the transcriptional regulation of the IL-4 promoter.
IL-4 is an anti-inflammatory cytokine and activates the Th2-type immune response and it also reciprocally regulates the Th1-type immune response. However, Mtb infection in macrophages causes a biphasic polarization with an early activation to the M1 polarization state (Th1-type immune response) and is soon shifted to an alternately polarized and compromised repair state M2 (Th2-type immune response) (40). With MtbEspR upregulating the expression of IL-4, one would expect that it will orient the response to the Th2-type. In our analysis of cytokine profile in the presence of MtbEspR, Th2 cytokines were dominant. Such observations have been made for many Mtb proteins, such as ZMP-1 (22), HupB (41), and PPE34 (42). These studies, although have indicated that many Mtb proteins skew the host responses to Th2-type, no mycobacterial factor has been shown to directly regulate the expression of Th2 cytokines by binding to host DNA elements.
Most intracellular infections require dominant Th1-type cytokine-induced cell-mediated responses to clear the pathogen. When we observed that MtbEspR is orienting the immune environment to the Th2-type, we speculated that it will also compromise the host cell to clear other intracellular pathogens associated with Mtb co-infections. As Mtb-HIV is one of the most common co-infections, we studied the impact of MtbEspR on the survival of both mycobacteria and HIV during co-infection. HIV is a known risk factor for developing into active TB, which compromises the immune system by depleting the CD4+T cells and accelerating the progression of TB (43). Simultaneously, HIV titers also increase during co-infection with TB (44, 45). Mtb can also make the cellular environment more conducive to HIV propagation by increasing the expression of host factors that are positive regulators of HIV LTR (7, 46). However, MtbEspR could not directly interact with HIV LTR, but the viral propagation increased in the presence of EspR over-expressing mycobacterial strain. With our study clearly showing that EspR does not directly bind to HIV LTR to influence HIV replication, we speculate that the bystander effect of EspR is through its impact on increased secretion of IL-4 by infected macrophages, which in turn generates a conducive environment for HIV replication. IL-4-mediated effect on HIV can be either stimulatory or inhibitory and is context-dependent. Such as treatment with IL-4 has been experimentally shown to stimulate HIV production by infected monocytes and macrophages (47). Further, Mikovits et al, reported that pre-treatment of peripheral monocytes for 48-72 h with IL-4 increased acute HIV infection, and similar effects were seen when the U1 and R-THP-1 monocytoid cell lines (the cell lines with restricted HIV expression) were treated with IL-4 (48). Another study showed that IL-4 stimulation enhanced transcription that is mediated by IL-4 stimulated NF-κB nuclear translocation. Further, they also state that IL-4 and IL-13 inhibited HIV replication at the transcriptional level in monocyte-derived macrophages after 3 to 5 days of adherence to the surface of plastic culture plates (49). Interestingly, it has been observed that IL-4 favored the replication of T-tropic (X4) virus as compared to M-tropic (R5) replication (50), favoring X4 virus generation. One may note that secondary infections like Mtb or other infections, are more frequent at chronic stages. It is known that during chronic stages of HIV infection, the M-tropic viruses (R5) are low and T-tropic viruses (X4), and dual tropic viruses (R5/X4) are predominant (51). Further, it is also indicated that Mtb induces the shift of the R5 virus to the X4 virus by increasing the expression of CXCR4 receptor (52). This suggests that mycobacteria-induced selection of X4 virus may also be mediated via altering IL-4 levels. In addition, that MtbEspR over-expression in HEK293T cells augmented propagation of both NL4.3 and NL-ADA8, supported the notion that increase in viral propagation by MtbEspR does not solely depend upon IL-4 secretion. It is reported that MtbEspR inhibits apoptosis in the macrophage cells (11). Such phenomena (inhibition of apoptosis) are supportive for active HIV replication (53). Therefore, we speculate that EspR enhances the HIV replication by several direct or indirect mechanisms, such as inhibiting apoptosis and promoting Th2-type microenvironment. MtbEspR interactome in host macrophages should be investigated to discover additional mechanisms.
In summary, in this study, we found that secreted mycobacterial protein EspR regulates host IL-4 cytokine levels, which skews the immune response to Th2-type leading to immune evasion. Our study shows for the first time that a bacterial transcription factor directly regulates the host gene transcription. This data provides new molecular insights into the mycobacterial protein-mediated regulation of the host genome and HIV propagation, adding to our understanding of mycobacterial infection and co-infection biology that can be used for elucidating new targets for therapeutic interventions aimed at controlling TB or HIV-TB co-infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SY: Formal Analysis, Writing – original draft, Writing – review & editing, Data curation, Investigation, Methodology, Validation, Visualization. AA: Formal Analysis, Investigation, Methodology, Writing – review & editing. AC: Formal Analysis, Investigation, Methodology, Writing – review & editing. SS: Formal Analysis, Investigation, Methodology, Writing – review & editing. KM: Formal Analysis, Investigation, Methodology, Writing – review & editing. SB: Formal Analysis, Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare financial support was received for this research from the Science and Research Engineering Board, India (SERB) Research grant (SPR/2021/000137) under the scheme SUPRA, and the Department of Biotechnology (DBT) (BT/HRD/NWBA/38/09/2018) to SB.
Acknowledgments
We also acknowledge the SERB POWER (SPG/2021/002627) grant to SB, UGC JRF-SRF to SY, DST-Inspire fellowship to AA, PMRF to SS, and NBRC core funds to AC. We thank Prof. Anand Kumar Kondapi, UoH, for permitting us to use their cell culture facility and providing pNL4.3. We thank Dr. Jayanta Bhattacharya, THSTI, Delhi for providing NL-ADA8. H37Rv and M. smegmatis were procured from JALMA, Agra, and LEPRA, India, Hyderabad respectively. We thank Dr. Kota Arun Kumar and his group for their help in EspR antibody generation. We thank Mr. Omkar Zade for his help in ELISA. We also thank Ms. Deepti for helping with confocal microscopy imaging at the Imaging facility, SLS, UoH. DST-FIST to the Department of Biochemistry, the DBT-BUILDER grant to the School of Life Sciences (BT/INF/22/SP41176/2020), and the Institution of Eminence supported projects RC1-20-017 and RC4-21-012 to SB and IoE to the University of Hyderabad MHRD (F11/9/2019- U3(A) towards infrastructure support to the Department of Biochemistry, School of Life Sciences and University common facilities are acknowledged. We also thank the UoH-NIAB BSL3/ABSL3 facility. BioRender.com was used to generate Figure 6.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1276817/full#supplementary-material
References
1. Yandrapally S, Mohareer K, Arekuti G, Vadankula GR, Banerjee S. HIV co-receptor-tropism: cellular and molecular events behind the enigmatic co-receptor switching. Crit Rev Microbiol (2021) 0:1–18. doi: 10.1080/1040841X.2021.1902941
2. Cloyd MW, Chen JJ, Adeqboyega P, Wang L. How does HIV cause depletion of CD4 lymphocytes? A mechanism involving virus signaling through its cellular receptors. Curr Mol Med (2001) 1:545–50. doi: 10.2174/1566524013363320
3. Scanga CA, Mohan VP, Yu K, Joseph H, Tanaka K, Chan J, et al. Depletion of CD4(+) T cells causes reactivation of murine persistent tuberculosis despite continued expression of interferon gamma and nitric oxide synthase 2. J Exp Med (2000) 192:347–58. doi: 10.1084/jem.192.3.347
4. Ranjbar S, Jasenosky LD, Chow N, Goldfeld AE. Regulation of mycobacterium tuberculosis-dependent HIV-1 transcription reveals a new role for NFAT5 in the toll-like receptor pathway. PloS Pathog (2012) 8:e1002620. doi: 10.1371/journal.ppat.1002620
5. Nakata K, Rom WN, Honda Y, Condos R, Kanegasaki S, Cao Y, et al. Mycobacterium tuberculosis enhances human immunodeficiency virus-1 replication in the lung. Am J Respir Crit Care Med (1997) 155:996–1003. doi: 10.1164/ajrccm.155.3.9117038
6. Toossi Z, Xia L, Wu M, Salvekar A. Transcriptional activation of HIV by Mycobacterium tuberculosis in human monocytes. Clin Exp Immunol (1999) 117:324–30. doi: 10.1046/j.1365-2249.1999.00952.x
7. Bhat KH, Chaitanya CK, Parveen N, Varman R, Ghosh S, Mukhopadhyay S. Proline-proline-glutamic acid (PPE) protein rv1168c of mycobacterium tuberculosis augments transcription from HIV-1 long terminal repeat promoter*. J Biol Chem (2012) 287:16930–46. doi: 10.1074/jbc.M111.327825
8. Rosenberg OS, Dovey C, Tempesta M, Robbins RA, Finer-Moore JS, Stroud RM, et al. EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins. Proc Natl Acad Sci USA (2011) 108:13450–5. doi: 10.1073/pnas.1110242108
9. Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, et al. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PloS Pathog (2012) 8:e1002621. doi: 10.1371/journal.ppat.1002621
10. Raghavan S, Manzanillo P, Chan K, Dovey C, Cox JS. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature (2008) 454:717–21. doi: 10.1038/nature07219
11. Jin C, Wu X, Dong C, Li F, Fan L, Xiong S, et al. EspR promotes mycobacteria survival in macrophages by inhibiting MyD88 mediated inflammation and apoptosis. Tuberculosis (2019) 116:22–31. doi: 10.1016/j.tube.2019.03.010
12. Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage M1–M2 polarization balance. Front Immunol (2014) 5:614. doi: 10.3389/fimmu.2014.00614
13. Vyas SP, Goswami R. Striking the right immunological balance prevents progression of tuberculosis. Inflammation Res (2017) 66:1031–56. doi: 10.1007/s00011-017-1081-z
14. Roff SR, Noon-Song EN, Yamamoto JK. The significance of interferon-γ in HIV-1 pathogenesis, therapy, and prophylaxis. Front Immunol (2014) 4:498. doi: 10.3389/fimmu.2013.00498
15. Mohareer K, Medikonda J, Vadankula GRGR, Banerjee S. Mycobacterial control of host mitochondria: bioenergetic and metabolic changes shaping cell fate and infection outcome. Front Cell Infect Microbiol (2020) 10:457. doi: 10.3389/fcimb.2020.00457
16. Rajavelu P, Das SD. Th2-type immune response observed in healthy individuals to sonicate antigen prepared from the most prevalent Mycobacterium tuberculosis strain with single copy of IS6110. FEMS Immunol Med Microbiol (2005) 45:95–102. doi: 10.1016/j.femsim.2005.02.011
17. Tiwari BM, Kannan N, Vemu L, Raghunand TR. The mycobacterium tuberculosis PE proteins rv0285 and rv1386 modulate innate immunity and mediate bacillary survival in macrophages. PloS One (2012) 7(12):e51686. doi: 10.1371/journal.pone.0051686
18. Benjamin R, Banerjee A, Sunder SR, Gaddam S, Valluri VL, Banerjee S. Discordance in CD4+T-cell levels and viral loads with co-occurrence of elevated peripheral TNF-α and IL-4 in newly diagnosed HIV-TB co-infected cases. PloS One (2013) 8:e70250. doi: 10.1371/journal.pone.0070250
19. Amelio P, Portevin D, Reither K, Mhimbira F, Mpina M, Tumbo A, et al. Mixed Th1 and Th2 Mycobacterium tuberculosis-specific CD4 T cell responses in patients with active pulmonary tuberculosis from Tanzania. PloS Negl Trop Dis (2017) 11:e0005817. doi: 10.1371/journal.pntd.0005817
20. Magombedze G, Eda S, Ganusov VV. Competition for Antigen between Th1 and Th2 Responses Determines the Timing of the Immune Response Switch during Mycobaterium avium Subspecies paratuberulosis Infection in Ruminants. PloS Comput Biol (2014) 10:e1003414. doi: 10.1371/journal.pcbi.1003414
21. Stamm CE, Pasko BL, Chaisavaneeyakorn S, Franco LH, Nair VR, Weigele BA, et al. Screening mycobacterium tuberculosis secreted proteins identifies mpt64 as a eukaryotic membrane-binding bacterial effector. mSphere (2019) 4(3):e00354-19. doi: 10.1128/mSphere.00354-19
22. Vemula MH, Ganji R, Sivangala R, Jakkala K, Gaddam S, Penmetsa S, et al. Mycobacterium tuberculosis zinc metalloprotease-1 elicits tuberculosis-specific humoral immune response independent of mycobacterial load in pulmonary and extra-pulmonary tuberculosis patients. Front Microbiol (2016) 7:418. doi: 10.3389/fmicb.2016.00418
23. Balakrishnan K, Mohareer K, Banerjee S. Mycobacterium tuberculosis Rv1474c is a TetR-like transcriptional repressor that regulates aconitase, an essential enzyme and RNA-binding protein, in an iron-responsive manner. Tuberculosis (2017) 103:71–82. doi: 10.1016/j.tube.2017.01.003
24. Rizvi A, Shankar A, Chatterjee A, More TH, Bose T, Gomez-casati DF, et al. Rewiring of metabolic network in mycobacterium tuberculosis during adaptation to different stresses. Front Microbiol (2019) 10:2417. doi: 10.3389/fmicb.2019.02417
25. Loebel RO, Shorr E, Richardson HB. The influence of adverse conditions upon the respiratory metabolism and growth of human tubercle bacilli. J Bacteriol (1933) 26:167–200. doi: 10.1128/jb.26.2.167-200.1933
26. Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, et al. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J Exp Med (1988) 167:1428–41. doi: 10.1084/jem.167.4.1428
27. Arendt V, Amand M, Iserentant G, Lemaire M, Masquelier C, Ndayisaba GF, et al. Predominance of the heterozygous CCR5 delta-24 deletion in African individuals resistant to HIV infection might be related to a defect in CCR5 addressing at the cell surface. J Int AIDS Soc (2019) 22:e25384. doi: 10.1002/jia2.25384
28. Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol (1986) 59:284–91. doi: 10.1128/JVI.59.2.284-291.1986
29. Bagnarelli P, Fiorelli L, Vecchi M, Monachetti A, Menzo S, Clementi M. Analysis of the functional relationship between V3 loop and gp120 context with regard to human immunodeficiency virus coreceptor usage using naturally selected sequences and different viral backbones. Virology (2003) 307(2):328–40. doi: 10.1016/S0042-6822(02)00077-6
30. Santos S, Obukhov Y, Nekhai S, Bukrinsky M, Iordanskiy S. Virus-producing cells determine the host protein profiles of HIV-1 virion cores. Retrovirology (2012) 9:65. doi: 10.1186/1742-4690-9-65
31. Banerjee A, Benjamin R, Balakrishnan K, Ghosh P, Banerjee S. Human protein Staufen-2 promotes HIV-1 proliferation by positively regulating RNA export activity of viral protein Rev. Retrovirology (2014) 11:18. doi: 10.1186/1742-4690-11-18
32. Kutner RH, Zhang X-Y, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc (2009) 4:495–505. doi: 10.1038/nprot.2009.22
33. Blasco B, Stenta M, Alonso-Sarduy L, Dietler G, Peraro MD, Cole ST, et al. Atypical DNA recognition mechanism used by the EspR virulence regulator of Mycobacterium tuberculosis. Mol Microbiol (2011) 82:251–64. doi: 10.1111/j.1365-2958.2011.07813.x
34. Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin C, Bilska M, Greene KM, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods (2014) 409:131–46. doi: 10.1016/j.jim.2013.11.022
35. Hotter D, Bosso M, Jønsson KL, Krapp C, Stürzel CM, Das A, et al. IFI16 targets the transcription factor sp1 to suppress HIV-1 transcription and latency reactivation. Cell Host Microbe (2019) 25:858–872.e13. doi: 10.1016/j.chom.2019.05.002
36. Anes E, Peyron P, Staali L, Jordao L, Gutierrez MG, Kress H, et al. Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell Microbiol (2006) 8:939–60. doi: 10.1111/j.1462-5822.2005.00675.x
37. Ruiz AP, Ajasin DO, Ramasamy S, DesMarais V, Eugenin EA, Prasad VR. A naturally occurring polymorphism in the HIV-1 tat basic domain inhibits uptake by bystander cells and leads to reduced neuroinflammation. Sci Rep (2019) 9:3308. doi: 10.1038/s41598-019-39531-5
38. Clark E, Nava B, Caputi M. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget (2017) 8(16):27569–81. doi: 10.18632/oncotarget.15174
39. Anil Kumar V, Goyal R, Bansal R, Singh N, Sevalkar RR, Kumar A, et al. EspR-dependent ESAT-6 protein secretion of mycobacterium tuberculosis requires the presence of virulence regulator phoP. J Biol Chem (2016) 291:19018–30. doi: 10.1074/jbc.M116.746289
40. Chatterjee S, Yabaji SM, Rukhlenko OS, Bhattacharya B, Waligurski E, Vallavoju N, et al. Channeling macrophage polarization by rocaglates increases macrophage resistance to Mycobacterium tuberculosis. iScience (2021) 24:102845. doi: 10.1016/j.isci.2021.102845
41. Choudhury M, Virivinti J, Kandi S, Sritharan V, Sritharan M. Th2 immune response by the iron-regulated protein HupB of Mycobacterium tuberculosis. Indian J Tuberc (2022) 69:90–9. doi: 10.1016/j.ijtb.2021.04.011
42. Bansal K, Sinha AY, Ghorpade DS, Togarsimalemath SK, Patil SA, Kaveri SV, et al. Src homology 3-interacting domain of rv1917c of mycobacterium tuberculosis induces selective maturation of human dendritic cells by regulating PI3K-MAPK-NF-κB signaling and drives th2 immune responses*. J Biol Chem (2010) 285:36511–22. doi: 10.1074/jbc.M110.158055
43. Bruchfeld J, Correia-Neves M, Kallenius G. Tuberculosis and HIV coinfection. Cold Spring Harb Perspect Med (2015) 5(7):a017871. doi: 10.1101/cshperspect.a017871
44. Ganji R, Dhali S, Rizvi A, Sankati S, Vemula MH, Mahajan G, et al. Proteomics approach to understand reduced clearance of mycobacteria and high viral titers during HIV-mycobacteria co-infection. Cell Microbiol (2016) 18(3):355–68. doi: 10.1111/cmi.12516
45. Ganji R, Dhali S, Rizvi A, Rapole S, Banerjee S. Understanding HIV-Mycobacteria synergism through comparative proteomics of intra-phagosomal mycobacteria during mono- and HIV co-infection. Sci Rep (2016) 6:22060. doi: 10.1038/srep22060
46. Benjamin R, Banerjee A, Balakrishnan K, Sivangala R, Gaddam S, Banerjee S. Mycobacterial and HIV infections up-regulated human zinc finger protein 134, a novel positive regulator of HIV-1 LTR activity and viral propagation. PloS One (2014) 9:1–11. doi: 10.1371/journal.pone.0104908
47. Kazazi F, Mathijs JM, Chang J, Malafiej P, Lopez A, Dowton D, et al. Recombinant interleukin 4 stimulates human immunodeficiency virus production by infected monocytes and macrophages. J Gen Virol (1992) 73(Pt 4):941–9. doi: 10.1099/0022-1317-73-4-941
48. Mikovits JA, Meyers AM, Ortaldo JR, Minty A, Caput D, Ferrara P, et al. IL-4 and IL-13 have overlapping but distinct effects on HIV production in monocytes. J Leukoc Biol (1994) 56:340–6. doi: 10.1002/jlb.56.3.340
49. Naif HM, Li S, Ho-Shon M, Mathijs JM, Williamson P, Cunningham AL. The state of maturation of monocytes into macrophages determines the effects of IL-4 and IL-13 on HIV replication. J Immunol (1997) 158:501–11. doi: 10.4049/jimmunol.158.1.501
50. Valentin A, Lu W, Rosati M, Schneider R, Albert J, Karlsson A, et al. Dual effect of interleukin 4 on HIV-1 expression: implications for viral phenotypic switch and disease progression. Proc Natl Acad Sci USA (1998) 95:8886–91. doi: 10.1073/pnas.95.15.8886
51. Parisi SG, Andreoni C, Sarmati L, Boldrin C, Buonomini AR, Andreis S, et al. HIV coreceptor tropism in paired plasma, peripheral blood mononuclear cell, and cerebrospinal fluid isolates from antiretroviral-naïve subjects. J Clin Microbiol (2011) 49(4):1441–5. doi: 10.1128/JCM.02564-10
52. Hoshino Y, Tse DB, Rochford G, Prabhakar S, Hoshino S, Chitkara N, et al. Mycobacterium tuberculosis-induced CXCR4 and chemokine expression leads to preferential X4 HIV-1 replication in human macrophages. J Immunol (2004) 172:6251–8. doi: 10.4049/jimmunol.172.10.6251
Keywords: Mycobacterium tuberculosis, EspR, Th1 immune response, Th2 immune response, cytokines, gene regulation
Citation: Yandrapally S, Agarwal A, Chatterjee A, Sarkar S, Mohareer K and Banerjee S (2023) Mycobacterium tuberculosis EspR modulates Th1-Th2 shift by transcriptionally regulating IL-4, steering increased mycobacterial persistence and HIV propagation during co-infection. Front. Immunol. 14:1276817. doi: 10.3389/fimmu.2023.1276817
Received: 13 August 2023; Accepted: 06 October 2023;
Published: 19 October 2023.
Edited by:
Elsa Anes, University of Lisbon, PortugalReviewed by:
Bibhuti Bhusan Mishra, Albany Medical College, United StatesJosé Miguel Azevedo-Pereira, University of Lisbon, Portugal
Copyright © 2023 Yandrapally, Agarwal, Chatterjee, Sarkar, Mohareer and Banerjee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sharmistha Banerjee, sbsl@uohyd.ac.in
†Present address: Archismita Chatterjee, Department of Cellular and Molecular Neuroscience, National Brain Research Centre (NBRC) Manesar, Gurgaon, India
‡These authors have contributed equally to this work
§These authors have contributed equally to this work
 Sriram Yandrapally
Sriram Yandrapally