
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 28 November 2023
Sec. NK and Innate Lymphoid Cell Biology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1269614
This article is part of the Research Topic Killer Re-Design: Synthetic Biology Approaches for Natural Killer-Cell Therapy View all 8 articles
Natural killer (NK) cells integrate heterogeneous signals for activation and inhibition using germline-encoded receptors. These receptors are stochastically co-expressed, and their concurrent engagement and signaling can adjust the sensitivity of individual cells to putative targets. Against cancers, which mutate and evolve under therapeutic and immunologic pressure, the diversity for recognition provided by NK cells may be key to comprehensive cancer control. NK cells are already being trialled as adoptive cell therapy and targets for immunotherapeutic agents. However, strategies to leverage their naturally occurring diversity and agility have not yet been developed. In this review, we discuss the receptors and signaling pathways through which signals for activation or inhibition are generated in NK cells, focusing on their roles in cancer and potential as targets for immunotherapies. Finally, we consider the impacts of receptor co-expression and the potential to engage multiple pathways of NK cell reactivity to maximize the scope and strength of antitumor activities.
Natural killer (NK) cells are agile lymphocytes capable of immune polarization and rapid responsiveness to eliminate virally infected or malignant cells. Though they were initially described for their ability to discern “self” from “non-self” cells based on expression of class I major histocompatibility molecules (MHC) (1), NK cells’ function and underlying molecular toolkit is now understood to be much broader. Upon receiving signals from healthy, stressed, infected, or transformed cells, and in response to environmental signals, NK cells either perform cytotoxicity, stimulate subsequent immunity, regulate immunity, or do nothing at all (2, 3). This “polyvalency” to integrate signals for activation and inhibition creates a unique form of diversity in the immune system by using germline-encoded receptors, making NK cells attractive candidates for cell-based cancer immunotherapies (4, 5).
NK cells differ from other group one innate lymphoid cells (ILCs) by their relatively high expression of the transcription factor eomesodermin (EOMES), and the ability to mediate direct cytolysis of target cells via degranulation to release perforin and granzyme (6). In contrast to other lymphocytes, NK cells do not express antigen-specific clonotypic receptors (1, 7–10). Instead, NK cells use a constellation of germline-encoded activating and inhibitory receptors (Table 1). These receptors are differentially expressed, co-expressed, and armed among the NK cells that comprise an individual’s NK cell repertoire, and can range from relatively immature and inexperienced to adaptive and highly functional cells. This creates diversity to respond to the extensively variable phenotypes created by transformation, infection, and disease (7, 10, 13). Indeed, though HLA loss eliminates a dominant inhibitory signal to NK cells, other inhibitory mechanisms within the tumor microenvironment can also interfere with NK cell inhibition; these include metabolic dysregulation (14), inhibitory cytokine release (15), regulatory immune populations (16), and even biophysical properties of the cancer cells themselves (17).
Cells become targets for NK cell-mediated killing when they acquire DNA mutations, express ligands associated with uncontrolled proliferation, stress, transformation, or are bound by antibodies (4, 18–23). Clinical presentation of a tumor often occurs after transformed cells have evolved mechanisms to escape immune-mediated recognition, and the patient’s NK cells and other immune mechanisms no longer fully control tumor growth (4, 24). Sustained expression of inhibitory checkpoints, shed ligands which block activating receptors, and metabolic dysregulation can impair NK cell-mediated recognition and elimination of tumors (4, 25). Concurrently, tumors can recruit other immune cells that regulate and suppress anti-tumor immune responses, and promote tumor invasion and growth (26) and/or establish physical stromal barriers that can prevent cellular and drug penetration into the tumor’s core (27–30).
NK cells are key effectors against hematologic malignancies, and agents of antibody-dependent cellular cytotoxicity (ADCC) in response to monoclonal antibody therapies (31–33). The presence of NK cells in solid tumors is associated with improved overall survival in a variety of cancers (4, 34, 35) and NK cells contribute to the efficacy of cancer treatment, including an abscopal effect following low-dose radiation, rescue of activity in the presence of immune checkpoint blockade, or activation by chemotherapy (36, 37). Recognizing the anti-cancer potential of NK cells, current clinical trials aim to deliberately support NK cell activation or deliver NK cells expanded ex vivo as cellular immunotherapy, including those that employ additional engineering to support strong anti-cancer activity (5, 38–40). In each of these contexts, inflammatory signals, or mechanisms to otherwise strengthen activating signals received by NK cells may tip the balance toward immune-mediated cancer control.
NK cell receptors meet target cell ligands in clusters at an immunologic synapse, whose formation is catalyzed by adhesion molecules (primarily ICAM and LFA-1) working in a coordinated effort alongside the reorganization of cytoskeletal components (41, 42). Single receptor-ligand engagement is insufficient to activate the cell and often, concurrent signals for activation and inhibition occur. Synergy between multiple receptors is necessary for cross-phosphorylation of membrane-proximal kinases to initiate a signal cascade that can lead to NK cell activation and degranulation (43). Inside the cell, phosphatases and kinases compete to respond to incoming signals in a dynamic tug-of-war whose outcome can be impacted by environmental features, immunogenetic variation and concurrently-received signals (Figure 1) (44).
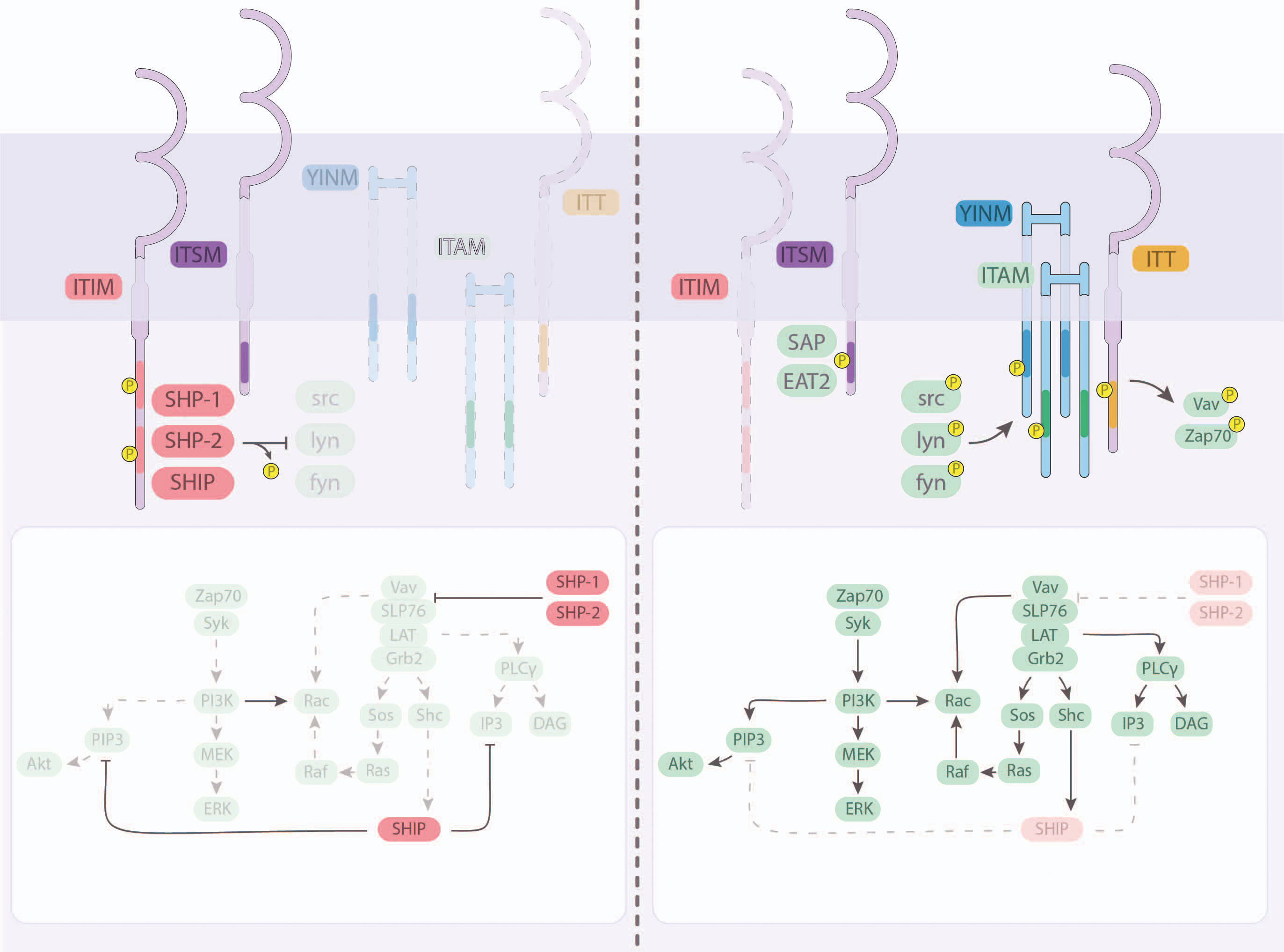
Figure 1 Intracellular signaling and integration downstream of the major NK cell receptors. Most NK cell receptors signal via transmembrane domains, including immunotyrosine-based inhibitory motifs (ITIM), immunoreceptor tyrosine-based switch motifs (ITSM), tyrosine-based signaling motif (YINM), immunotyrosine-based activation motifs (ITAM) and immunoglobulin tail tyrosine (ITT). (Left) Inhibitory signals received by NK cells are facilitated by the recruitment and activation of inhibitory SHP-1, SHP-2, and SHIP through ITIM and ITSM. (Right) Activating receptor clustering at the immunologic synapse facilitates the activation of intracellular domains by src-family kinases (src, lyn, fyn), or other kinases, including SAP and EAT2. Signals compete to activate or inhibit downstream intermediaries that can lead to transcription factor-mediated cytotoxicity and cytokine production.
Recognizing the anti-cancer potential of NK cells, current clinical trials aim to deliberately support NK cell activation or deliver NK cells expanded ex vivo as cellular immunotherapy, including those that employ additional engineering to support strong anti-cancer activity (5, 38–40, 45, 46). With engineering, local cytokine support, inflammatory signals, or mechanisms to otherwise strengthen the activating signals received by NK cells may tip the balance toward immune-mediated cancer control. In this review, we discuss the receptor-ligand pairs that govern NK cell interactions with transformed and cancerous cells, their pathways for intracellular signaling, and how alone or in combination, they present opportunities for cancer immunotherapy.
Most interactions between NK cells and neighbouring cells result in non-activation or inhibition, as NK cells survey neighboring, and not necessarily damaged, cells. Provision of inhibitory signals prevents the NK cell exhaustion that might otherwise result from persistent activation, and facilitates maintenance of NK cell education (47). Inhibitory signals are conveyed via engagement of classical and non-classical MHC I molecules, and immune checkpoints (7, 48). Major MHC I-binding receptors include members of the family of killer immunoglobulin-like receptors (KIRs) and the natural killer group 2 (NKG2) family member-A (NKG2A) (49, 50). Other inhibitory receptors may also control NK cell activation, including classical immune checkpoint receptors: TACTILE (CD96), PD-1, TIM-3, LAG-3 and TIGIT (51). Many of these inhibitory receptors convey signaling via immunoreceptor tyrosine-based inhibitory motifs (ITIMs), which, when phosphorylated, recruit phosphatases including the Src homology-containing tyrosine phosphatases (SHP)-1, SHP-2, and SH2 domain-containing inostitol-5-phosphatase (SHIP) (52–54). These phosphatases compete directly with activating signals received by Src-family kinases (SFKs), including Lck, Fyn and Syk (55, 56).
SFKs are key mediators of NK cell activation, dephosphorylated at rest and quickly phosphorylated upon receptor clustering to activate signal intermediaries (57). A subset of KIRs, the Fc receptor CD16a, and the natural cytotoxicity receptors (NCRs) convey activating signals via immunoreceptor tyrosine activating motifs (ITAMs), which activate SFKs, and other signaling intermediaries including ZAP-70 and Vav-1, then phospholipase C, PI3K, Rho-family GTPases (58, 59). Additional activating receptors include the NKG2 family members NKG2C and NKG2D, which signal via DAP12 and DAP10, respectively, and therefore bypass the need for SFKs, instead shunting directly to activating downstream mediators including Vav-1, PI3K, Rho-family GTPases and phospholipase C (60–63). Immunoglobulin tail tyrosine motifs (ITTs), used by DNAM-1, are phosphorylated by SFKs and similarly drive downstream activation (64, 65). Hence, although they are initiated separately, multiple activating pathways converge on SFKs, so their signaling can be additive.
Immunotyrosine-based switch motifs (ITSMs), encoded in SLAM-family receptors and PD-1, combine the features of ITAMs and ITIMs by associating with SH2 domain-containing proteins, including SHP-1, SHP-2, and SHIP-1, (which convey inhibitory signaling), and SLAM-associated protein (SAP) and Ewing’s sarcoma-activated transcript 2 (EAT2), which support activation (66). Activation signals proceed via SFKs. Inhibitory SH2 domain-containing proteins prevent these signals in two ways: directly, by occupying the docking site on the ITSM, and indirectly, by the phosphatase activity of SHIP, SHP-1 and SHP-2 (66). In this way, one ITSM-containing receptor can switch between signaling for inhibition or activation.
The outcome of concurrent activating and inhibitory signals result in adjustments to the levels of transcription factors, including NF-κB, which mediate NK cell activation-related genes (67). These genes regulate NK cell functions such as cytotoxicity, microtubule organization and granule polarization (67). In some cases, NK cells are induced to become “adaptive” or memory-like NK cells: a state driven by epigenetic changes where NK cells are noted for their long lifespans, ability to recall previous challenges, and rapid, potent responses (68–70).
The redundancy of the intracellular signaling cascades downstream of the major NK cell inhibitory and activating receptors enables crosstalk and integration of signals. Hence, with combined and simultaneous signalling from different receptors, each NK cell can balance and calibrate an appropriate response.
Human leukocyte antigens (HLA) are the most polymorphic gene in the human genome, and expressed on all healthy nucleated cells at variable densities that reflect cellular health and HLA allelic diversity (71, 72). HLA-A, HLA-B and HLA-C are together classified as “conventional HLA I”, or class Ia molecules, and represent the most polymorphic HLA I alleles (72). Unconventional HLA I molecules, classified as class Ib, include HLA-E, HLA-F, and HLA-G (71). These exhibit relatively low polymorphism, and have important roles in tolerance, inhibition, and pregnancy, with HLA-E remaining relatively conserved between human and primates (72–76). Each HLA I, except for HLA-F, is comprised of three alpha domains, and stabilized by β2 microglobulin and by peptides presented in the binding groove of its variable domain (57). HLA-F is less understood, but can associate with β2 microglobulin and exhibits at least two configurations: closed, where it seems to not present peptides, and open, where it presents uncommonly long peptides in an open-ended binding groove (77).
HLA I is co-evolving with the killer immunoglobulin-like receptors (KIR) and members of the NKG2 family (78). Their interactions can signal for activation or inhibition, depending on the specific receptors ligated, bound peptides, immunologic experience, and compound allelic partnerships (3, 79) (Figure 2A, B). Groups of conventional HLA I molecules display conserved public epitopes that enable groupings of HLA I molecules as “KIR ligands”: HLA-C molecules expressing Asp or Lys at position 80 define the HLA-C1 or C2 subgroups, respectively, and account for all HLA-C molecules (80, 81). A subset of HLA-A and B molecules each contain the HLA-Bw4 motif, and some specific alleles of HLA-A (HLA-A*03 and *11) can act as NK cell ligands, and the balance of HLA-A and -B alleles are not known to engage with any KIR (79, 82, 83).
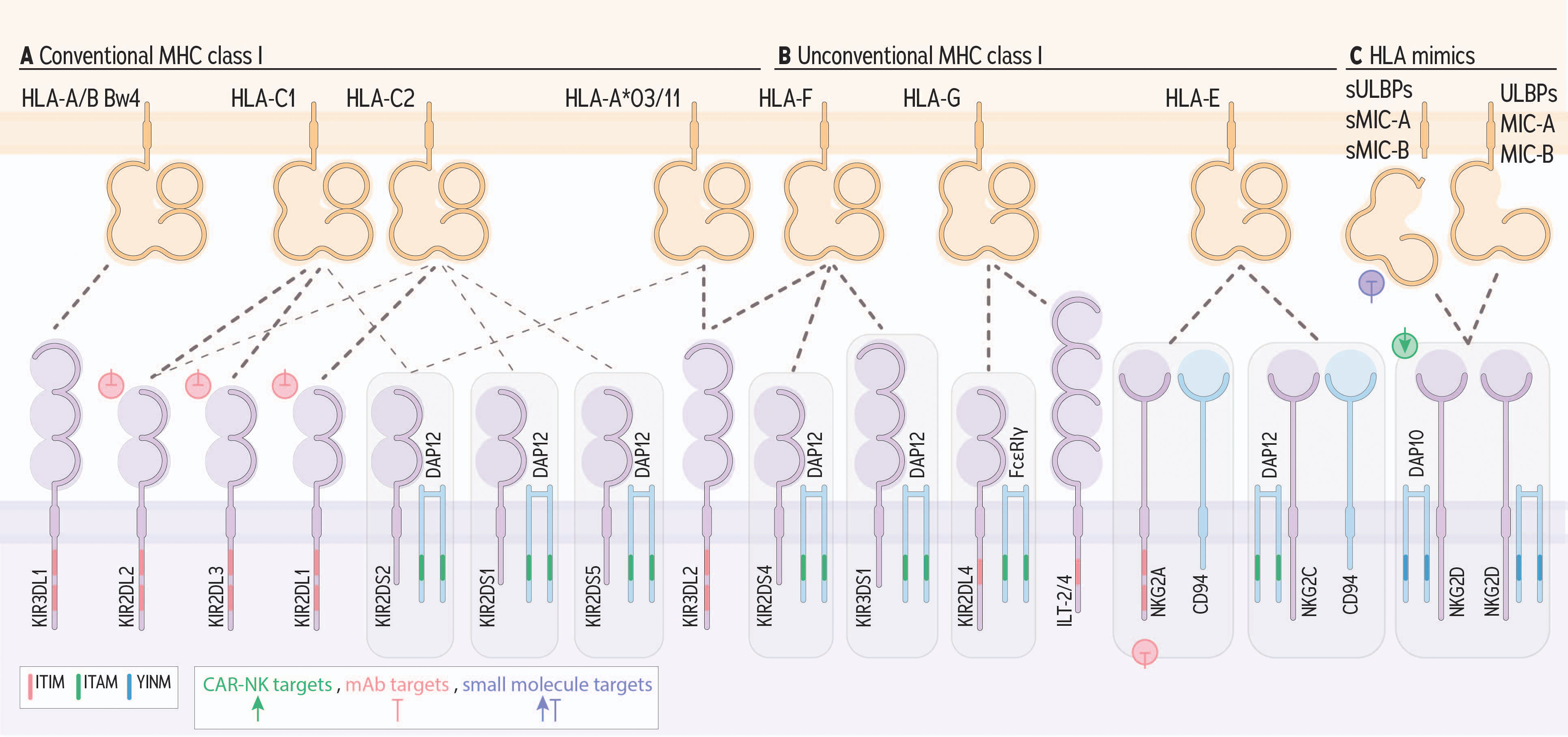
Figure 2 NK cell receptors engaging HLA I and conveying activating or inhibitory signals. KIRs engage conserved epitopes on groups of HLA I molecules. (A) KIR receptors with long (L) cytoplasmic tails generally convey signals for inhibition via an ITIM, except for KIR2DL4. (B) KIR receptors with short (S) cytoplasmic tails and KIR2DL4 convey signals for activation by engaging DAP12, or FcϵRIγ, respectively, which contain ITAMs. Beyond KIR, ILT-2 and 4, and NKG2A (heterodimerized with CD94) bind with MHC I and convey inhibitory signals via ITIMs. NKG2C (heterodimerized with CD94) and NKG2D engage with HLA-E or HLA orthologs which include ULBPs, MIC-A and MIC-B. (C) Finally, NKG2C associates with DAP12 and NKG2D signals via DAP10, a YNIM-containing receptor, each to signal for activation. Green, pink, and purple symbols indicate CAR-NK targets, mAb targets, and small molecule targets, respectively.
KIR molecules with long cytoplasmic tails signal for inhibition via ITIMs encoded in their transmembrane regions. HLA-C1 engages KIR2DL3 and HLA-C2 engages KIR2DL1 (84). KIR2DL2 preferentially binds with HLA-C1 molecules, but some alleles also exhibit binding affinity with HLA-C2 (85, 86). HLAs harbouring the Bw4 motif bind with KIR3DL1; the remainder of HLA-B molecules and most of the HLA-A molecules are not known to encode a ligand for KIR (87). KIR3DL2 binds to HLA-A*03 and A*11 molecules, but only when they present certain peptides, including exogenous CpG DNA and peptides derived from Epstein-Barr virus (82, 83, 88). Inhibitory KIR2DL4 and immunoglobulin-like transcript (ILT)-2 and -4 molecules on NK cells bind HLA-G and signal via ITIM (89). HLA-G is an unconventional HLA molecule whose expression is normally limited to immune privileged sites including the cornea (90) and placenta (91), but has also been found upregulated, and associated with poorer prognosis in several cancers, including colorectal cancer (92), pancreatic cancer (93), breast cancer (94), and ovarian cancer (95). Soluble HLA-G has been found to dampen NK cell function, including impairment of NK cell migration to inflamed tissues (96).
KIR molecules with a short cytoplasmic tail recruit the ITAM-containing molecule DAP12 to the immunologic synapse. Activating KIR, including KIR2DS1, KIR2DS2, and KIR3DS1 are each known to bind with HLA I molecules, albeit with lower affinity than their inhibitory counterparts (97, 98). Additional activating KIRs (KIR2DS3, KIR2DS5) also have ITAMs and may contribute to activation and may bind with specific alleles (99), though the impact of these ligands and their corresponding signals have not been well established.
KIR2DS1 binds HLA-C2 (Lys80) and, at high levels, individuals that are homozygous for HLA-C2 have KIR2DS1+ NK cells that are hyporesponsive (97, 100, 101). This has implications in hematopoietic cell transplantation, where HLA-matching is only beneficial in the absence of homozygous HLA-C2 (102, 103). Moreover, for the treatment of B-cell malignancies, addition of rituximab to chemotherapy did not improve survival outcomes in patients with KIR2DS1-HLA-C2/C2 (104), suggesting that KIR2DS1 and HLA-C status could be a predictive marker for efficacy of rituximab due to the potential of hyporesponsive NK cells (104).
Although KIR are not known to recognize HLA-presented peptides specifically, the biochemical features and structural/allosteric variations that they introduce can influence receptor binding and consequent signaling (57, 105). This is the case for KIR2DS2, which binds conditionally to HLA-C1 and HLA-A*11 alleles, when they present viral peptides, including those derived from viral helicases (3, 106), though their role in cancer is unclear. Similarly, KIR3DS1 is activated specifically by the open (peptide presenting) configuration of HLA-F (107).
The NKG2 family members engage with HLA-E, but in a manner distinct from conventional KIR-HLA interactions (108). NKG2A, an ITIM-containing receptor, and NKG2C, which engages ITAM-containing DAP12 for signaling, both bind with HLA-E and heterodimerize with CD94 (49, 50, 109–111). HLA-E molecules present the leader peptides of conventional HLA molecules but also present antigenic peptides, including neoantigens, to both T and NK cells (105). In healthy tissues, HLA-E expression is low (112), but high HLA-E expression is linked to poor patient outcomes in several cancer types, including gastric cancer (113, 114), colorectal cancer (113), pancreatic cancer (93), breast cancer (94), renal cell carcinoma (115), ovarian cancer (116), and glioma (111, 117), suggesting a dominant impact of NK inhibition on limiting anti-cancer activity. Indeed, high expression of NKG2A, both alone and with high HLA-E expression, is associated with poorer patient survival in liver cancer (118). Likewise, co-expression of the high-density HLA-E allele (HLA-E*01:03) and NKG2A associates with Epstein-Barr virus-associated lymphoma (119). Given the known impacts of HLA-E expression on cancer development, it is likely that these are exacerbated by NKG2A expression on NK cells in other cancer types, though this remains to be studied extensively.
In addition to signaling during target cell engagement, KIR-HLA and NKG2A:HLA-E interactions are central to a process called NK cell “education”, “tuning” or “licensing” (120–122). Each of KIR and NKG2A are expressed on only a subset of NK cells, their HLA ligands may or may not be available in the host, and the avidity of receptor-ligand binding is variable. Therefore, the extent to which each NK cell can be inhibited by “self” HLA varies. An individual’s NK cell repertoire therefore consists of both educated and uneducated cells (85, 123). Resultantly, the reactive thresholds of individual NK cells differ, and create a spectrum of responsiveness and anti-tumor effector function that varies within and between individuals (3, 122, 124).
NK cells that are most sensitive to inhibition by “self” HLA I exhibit a greater mobilization of activating receptors in the actin meshwork (8), greater DNAM-1 expression (125), a greater density of granzyme B (9) and lower levels of SHP-1 (53)– making them more easily activated in the absence of strong inhibition. Sensitivity to inhibition, however, is the Achille’s heel of educated NK cells in cancer therapy because HLA I on tumors often persists, or becomes upregulated in response to IFN-γ (126). Strategic selection of KIR and HLA allelic combinations and NK education status, or blocking signals for inhibition, will therefore be critically important for the success of NK cell-based immunotherapy (3, 124).
During cancer development, there is extensive pressure on HLA expression driven by immune activity: T cell-mediated recognition may select for clones lacking HLA expression, but this may create a target for NK cells by interrupting signals for inhibition. Downregulation of classical HLA I expression is reported for several solid cancers, including melanoma, cervical, breast, colorectal and lung cancers (127–130). HLA I loss may represent partial or complete losses of HLA gene loci (129, 131), or components of the HLA processing and presentation pathway (127, 131). Frequently, loss of heterozygosity for the HLA-ABC genes, and other genes involved in HLA I processing and presentation, is observed following acquired resistance to immune checkpoint inhibitors like anti-PD1/PD-L1 (127, 132, 133).
KIR is highly polymorphic; an individual’s KIR configuration has been associated with both the risk of developing cancer and their ability to respond to cancer therapies. KIR expression is not known to change as a function of cancer, but the presence of self-sensitive KIR can enable inhibition of otherwise-activated NK cells. Beyond strategic donor selection to optimize KIR/HLA genotype for potent NK cell alloreactivity, both KIR and NKG2A can be directly targeted through monoclonal antibodies. Lirilumab, an anti-KIR2DL1/2/3, has been tested in a phase I clinical trial against multiple myeloma, and supports alloreactivity of NK cells (134). Monalizumab (anti-NKG2A), in combination with Cetuximab, has shown anti-tumor effects in a clinical trial of patients with squamous cell carcinoma of head and neck with high expression of HLA-E and NK cell infiltration. Taken together, this implies the important role of selecting/regulating KIR/NKG2A in cancer treatment.
Perhaps the best known member of the NKG2 family, NKG2D, is unique in that it forms homodimers (135), and signals for activation via DAP10 (62). Ligands for NKG2D are structural homologs that co-evolved alongside HLA I molecules and include MHC class I chain-related molecule A and B (MICA/B), and UL16 binding proteins 1,2,3,4,5 and 6 (ULBP1-6) (Figure 2C) (136, 137). NKG2D ligands are typically expressed at low cell surface densities, but their density on the cell surface increases in response to DNA damage (138), oncogene activation (138), infection, excessive proliferation, and oxidative stress, earning them the title of “stress ligands” (138). The NKG2D receptor itself is responsive to cytokines associated with immune priming (ie. IL-2, IL-15), which increase basal phosphorylation of DAP10, priming the cell to deliver activating signals (139). Together, NKG2D and its ligands generate a robust system to detect hallmark features of stressed cells.
Presented on a target cell, NKG2D ligands signal for NK cell activation, and NKG2D is central in cancer immunosurveillance. In mouse models, antibody-mediated neutralization of NKG2D interrupted immunosurveillance that otherwise prevented carcinogen-induced tumors (140). Likewise, elimination of NKG2D using microRNA silencing rendered mice more susceptible to cancer growth (140–142). In humans, histological studies have confirmed high and co-expression of NKG2D ligands in cancers of the breast (142, 143), colon (144, 145), gastric system (146, 147), lung (148, 149), skin (150), ovary (151, 152), pancreas (153), prostate (148, 154), and kidney (148). For NKG2D, diminished receptor expression is known to occur in response to hypoxia and diminished STAT3 activity (155), and in response to shed soluble ligands released following protease-mediated cleavage, which block the receptors from signaling (156).
Existing and nascent approaches to cancer therapy alter the expression of NKG2D ligands and encourage NK cell-mediated tumor killing. For example, ionizing radiation (157, 158), histone deacetylase (HDAC) inhibitors (159), and chemotherapy (158, 160) each prompt increased levels of NKG2D ligands. Furthermore, agents that prevent ligand matrix metalloproteinase activity by small molecule inhibitors can potentiate NK cell response by preventing ligand shedding (161).
The NKG2D receptor is a logical target for immunotherapy, and it is now being incorporated into immunotherapeutic approaches including NKG2D-CAR-NK, NKG2D-CAR-T cells (162, 163), CAR-T cells with a DAP10 intracellular domain (164), and bi-specific killer-engagers (BiKEs) (165, 166). While these therapies have indeed enhanced NK cell mediated anti-tumor activity, complete tumor control will require combination therapies that extend beyond the targeting of NKG2D alone.
The tumor necrosis factor receptor superfamily (TNFRSF) is a group of proteins that primarily regulate cell activation, differentiation, and survival, either as membrane-bound factors or cleaved, soluble factors. Here, we focus our discussion on those studied for their roles in NK cell function and reactivity, including the death receptor ligands Fas and TNF-related apoptosis inducing ligand (TRAIL), and costimulatory members 4-1BB and CD40L (Figure 3). TNFRSFs engage TNF superfamily (TNFSF) ligands, which can often be in membrane bound and soluble forms (167). This family of receptors and ligands signal primarily through NF-κB to induce proinflammatory function, or induce apoptosis of the target cell (168). Notably, while other TNFSF members have been reported to be expressed by NK cells, or to impact their function [i.e. GITR (CD357) (169), LIGHT (CD258) (170) and CD70 (171)], relatively little is known on their roles and targetability in cancer; these remain open questions and opportunities.
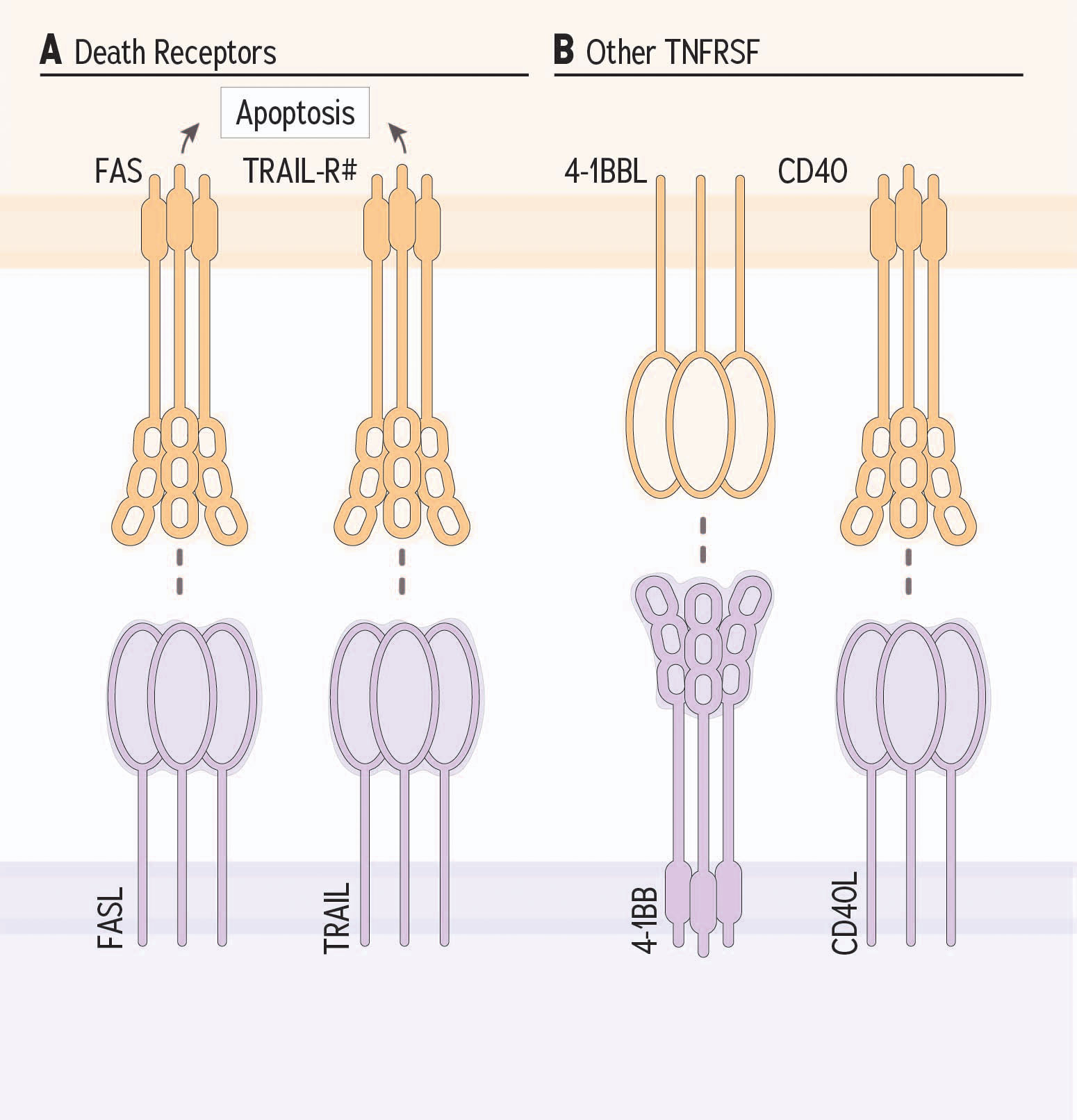
Figure 3 TNF receptor superfamily receptor family member receptor and ligand partnerships and NK cells. (A) Death receptors expressed by target cells are induced for apoptosis when their ligand is provided by NK and other cellular sources. (B) Costimulatory TNFRSF members, 4-1BB and CD40L potentiate NK cell response against target cells.
“Death receptors” include Fas and TNF-related apoptosis inducing ligand (TRAIL) receptor; they are present on putative target cells and facilitate the induction of apoptosis when bound by death receptor ligands (172–174) (Figure 3A). The typical role for these receptor-ligand partnerships is to facilitate normal cell turnover and immunoediting; for instance, to remove aged or damaged cells, or reduce an expanded population of effector cells once an infection is cleared (175). Each of FasL and (TRAIL) can be provided by NK cells, and exist as membrane-bound or soluble ligands (176). The role of soluble TRAIL is not well understood and only membrane-bound FasL, not soluble FasL, is capable of inducing target cell apoptosis (177).
FasL has a single known receptor: Fas, and TRAIL has multiple receptors: TRAIL-R1, TRAIL-R2, TRAIL-R3, TRAIL-R4 and the soluble decoy receptor osteoprotegrin (OPG) (178). Fas, TRAIL-R1, and TRAIL-R2 each contain a death domain that bind Fas-Associated Death Domain (FADD) adaptor, which recruits the Death Inducing Signaling Complex (DISC) to initiate caspase-8 activation and apoptosis (179, 180). Fas and TRAIL receptor signalling can also initiate activation of NF-κB which, in contrast to the pro-apoptotic signaling generated by the death receptors, can paradoxically contribute to target cell survival and proliferation (181, 182). For this reason, TRAIL-R3, TRAIL-R4 and OPG, which lack a death domain and have been classified as decoy receptors, may contribute to cancer cell survival and proliferation (183). Targeting apoptosis, but not cellular activation, will therefore be critical in therapies that aim to leverage death receptor signaling for cancer killing.
Agonists have been developed to target TRAIL receptors on cancer cells; they have been demonstrated as safe, but relatively ineffective as monotherapy [reviewed in Snajdauf et al., 2021 (184)]. For example, mapatumumab and conatumumab, TRAIL-R1 agonistic antibodies are well-tolerated, but ineffective as monotherapy to patients with solid tumors (185–188). However, stable disease was enabled in the presence of mapatuxumab combined with apoptosis-inducing chemotherapy, or the tyrosine kinase inhibitor sorafenib (184, 189), highlighting how the simultaneous targeting of multiple death pathways might prevent tumor escape. Similar agonistic approaches may be possible for targeting Fas, but preclinical models have revealed a potential unexpected benefit of blocking Fas: survival of tumor-infiltrating T cells, which may, in turn, enhance the efficacy of immunologic checkpoint blockade (190). Altogether, the available, but limited studies, reveal a potential role for death receptor signaling in cancer immunotherapy, but little is known specifically of the roles of NK cells in these pathways.
Costimulatory molecules of the TNFSF are best studied for their roles in T cells, and although many can also impact NK cell function, relatively few experiments have explored this. Here, we highlight two TNFSF members with known costimulatory function in NK cells, acknowledging that further research is required to classify whether additional TNFSF members are expressed, functional and relevant on NK cells (Figure 3B).
CD137 (4-1BB) is a transmembrane glycoprotein expressed on NK cells that engages 4-1BBL during NK cell activation to enhance cytotoxicity and expansion by increasing the expression of effector molecules including granzyme B, perforin and FasL (191–193). These functional enhancements are a result of increased MAPK signaling resulting in NF-κB activation within NK cells (193). The inclusion of 4-1BBL on feeder cells (in combination with IL-21) is used in expansion of NK cells for clinical trials (190). Addition of 4-1BBL promotes upregulation of activating receptors including the NCRs, CD16, and SLAM family members, alongside inhibitory KIR, all via STAT3 activation (194).
CD40 ligand (CD40L, CD154) is best understood as a ligand upregulated on T cells and macrophages, but can be upregulated on NK cells in response to IL-2 stimulation and during NK cell expansion (195–197). The significance of CD40L on NK cells remains to be investigated, but when CD40L is provided by T cells, the result is signals for activation and maturation in antigen presenting cells. Indeed, in mouse models, depletion of NK cells and blockade of CD40L had a similar effect on antigen presentation in the tumor draining lymph nodes: both resulted in lower cytotoxic T cell priming, but whether CD40L is obligately provided by NK cells was not studied (198). Nevertheless, these, and other co-stimulatory TNFSFR members expressed on NK cells could represent good candidates for immunotherapy.
The natural cytotoxicity triggering receptors (NCRs) are a group of HLA III genes that primarily generate activating signals in response to ligand binding through a transmembrane-encoded ITAM, or association with ITAM-containing adapter molecules (199) (Figure 4A). The NCRs do not themselves encode ITAMs, and instead recruit and signal through ITAM adapter molecules, including DAP12, CD3ζ and FcϵRIγ (199). Each exist as splice variants, with some isoforms conveying inhibitory signals via an ITIM-like sequence (200). Although there are others, three NCRs that have been studied in the context of cancer are NKp30, NKp44, and NKp46.
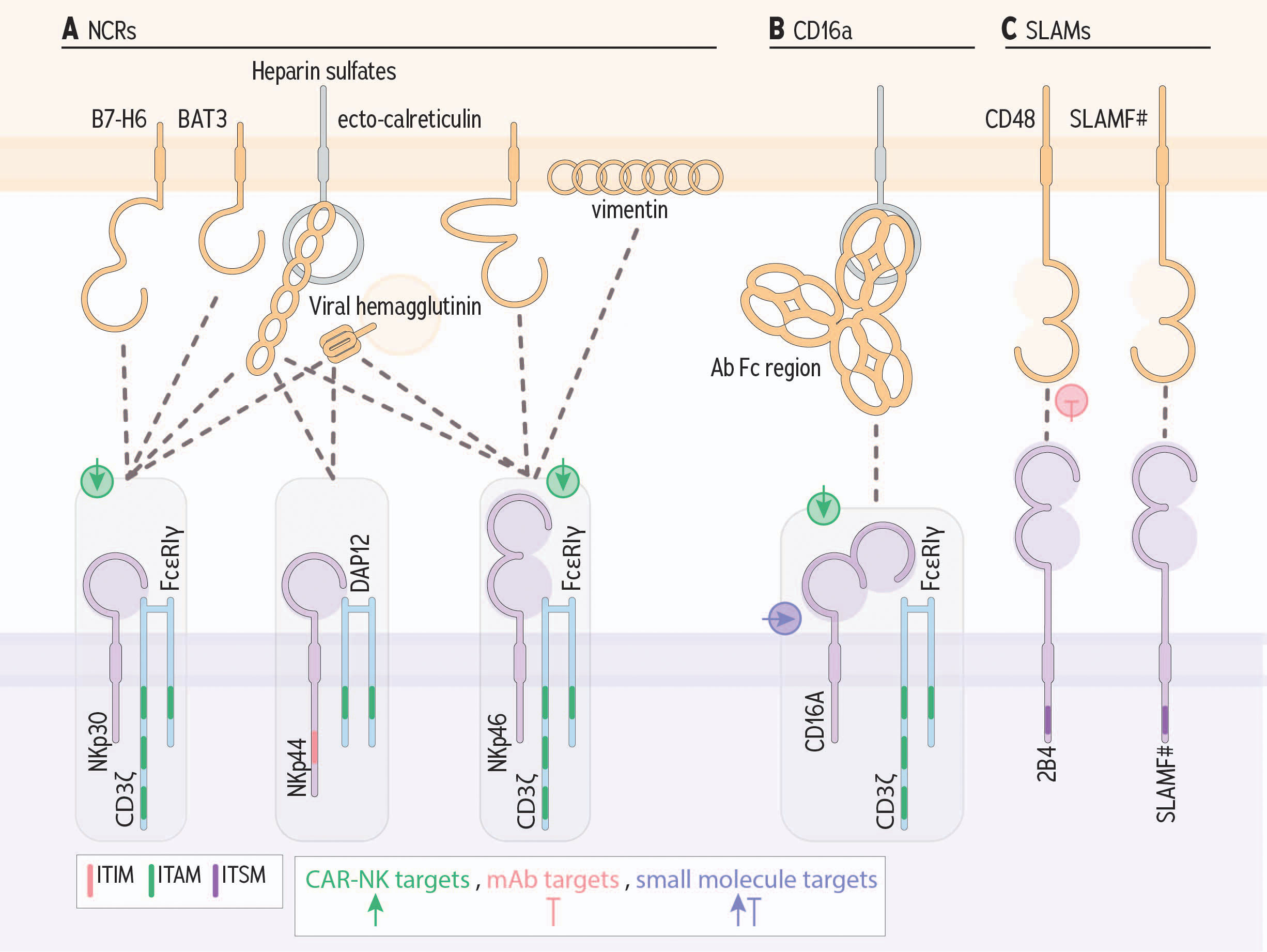
Figure 4 Germline encoded NK cell receptors and ligands that contribute to cancer cell killing. (A) Natural cytotoxicity receptors bind an array of ligands; shown are the most important to tumor cell recognition. (B) CD16a, binds the constant region/fragment crystallizable (Fc), portion of antibodies that, when bound to target cells, enable cross linking of receptors. (C) SLAM family members can signal for activation or inhibition via an immunotyrosine switch motif. Green, pink, and purple symbols indicate CAR-NK targets, mAb targets, and small molecule targets, respectively.
NCR ligands typically become available as a result of cellular stress, inflammation, and transformation (199). They are often expressed on tumor cells, or released during oncogenesis, and likely evolved to respond to ligands available in disease contexts. Cancer-derived ligands are not the exclusive binding partners for the NCRs, but for simplicity, we focus on those involved in cancer pathology here.
● B7-H6 is a B7 costimulatory family member that is overexpressed and associated with poorer outcomes in cancer (201, 202). B7-H6 and BAT-3 can be shed from the membrane and antagonize NCR binding to prevent NK cell activation (203).
● BAT-3 normally contributes to stabilizing p53 and contributing to tumor suppression (204). It may also be released in exosomes derived from cancer cells; in this configuration they can drive NK cell activation against cells that have lost normal p53 function (205).
● Heparin sulfates are components of the extracellular matrix that are exposed during cellular migration and tumor metastasis (206).
● Platelet-derived growth factor (PDGF) is involved in angiogenesis and cellular proliferation and is frequently expressed by tumor cells (207–209). There are at least five dimeric isoforms; among them, PDGF-DD is established as the ligand for NKp44 [reviewed in (199)].
● Proliferating cellular nuclear antigen (PCNA) is involved in DNA replication, repair, and remodelling (210). Among the ligands for NKp44, PCNA is the only one known to generate inhibitory signals. Evidence supports a protective immunoregulatory role in pregnancy, and the overexpression of PCNA by tumors may enable escape from NK-mediated immunosurveillance (211).
● Vimentins are structural components of mesenchymal cells, and they have been used as a biomarker of epithelial-to-mesenchymal transitioning, which is associated with metastasis, in an array of solid tumors (212, 213).
● Ecto-calreticulin is externalized calreticulin, which is typically released from the endoplasmic reticulum in response to stress. Most notably, this occurs in response to chemotherapy-induced cell death and senescence (18).
NKp30 binds B7-H6 (205), BAT-3 (205), and heparin sulfates (214, 215). NKp30’s transmembrane domain associates with the ITAM-containing adaptors CD3ζ and FcϵRIγ. There are at least six splice variants of NKp30, which differ in their tissue distribution, engagement with adapter molecules, and outcomes upon ligand binding, notably on production of IFN-γ and IL-10 (216). Variants a-c are the most common, with a and b favouring IFN-γ production and NKp30c associating with IL-10 production (216). B7-H6 has been proposed as a biomarker for development and progression in an array of cancers (217–219); by extension, leveraging this expression as a target for immunotherapy may be possible.
NKp44 signals for activation when bound by PDGF and heparan sulfates, and signals for inhibition upon binding to PCNA (199, 215, 220, 221). Three splice variants are described for NKp44, where the transmembrane domain in the NKp44b and NKp44c isoforms associate with DAP12 to signal for NK cell activation (222). NKp44a signals via an ITIM-like domain and binds PCNA, and at high surface densities, this splice isoform can inhibit NK cell function (200, 223). Dominant expression of this “inhibitory” NKp44 isoform is associated with poorer survival in patients with acute myelogenous leukemia (223), and PCNA is currently being explored as a biomarker and target for monoclonal antibodies (224).
NKp46 associates with CD3ζ and FcεRΙγ (225, 226). Key cancer associated ligands for NKp46 include heparan sulfates (215), vimentin (199, 225), and ecto-calreticulin (18). NKp46 expression is correlated with the degree of NK cell response generated (227) and IFN-γ produced via NKp46 signaling alters the deposition of fibronectin limiting metastasis in murine melanoma (228). Like NKp30, the ligands for NKp46 have been proposed as biomarkers for cancer severity and progression (229), endorsing these as potential targets for NK cell-based immunotherapies.
Targeting NCRs or their ligands for cancer immunotherapy is an area of active exploration that warrants investigation in NK cell-based cancer immunotherapy. Current studies are investigating the potential of using BiKEs or CARs that use the NCRs to enhance tumor recognition (230, 231). For example, a trifunctional natural killer cell engager (TriKEs) targeting the AML antigen, CD123, while simultaneously binding NKp46 and CD16a on NK cells has demonstrated efficacy in murine models and nonhuman primates (232) and is currently being tested in early clinical trials (NCT05086315). A similar strategy – coupling NKp46 engagement to anti-CD20, CD16 and the IL-2R beta chain has likewise generated promising preclinical results (233). NKp30 has been targeted on tri-specific engagers, coupling NKp30 engagers with Fab and anti-EGFR to create a potent pathway for tumor cell lysis and NK cell cytokine production (234). These strategies are in their infancy, but illustrate the promise of targeting such conserved NK cell ligands for immunotherapeutic purposes.
CD16a is expressed on NK cells, monocytes, and macrophages, and is the major receptor engaged for ADCC (Figure 4B) (235). CD16a encodes an ITAM motif, which recruits the signaling adapters FcϵRIγ, and CD3ζ to signal NK cells for proliferation, survival, cytokine production and degranulation (43, 236). Notably, CD16a activates NK cell degranulation, even in the absence of other activating signals (237).
Antibodies can function by neutralizing targets or opsonizing them for phagocytosis or killing. Target cell binding by antibodies enables cross-linking of CD16a; these antibodies can be produced endogenously by plasma cells or delivered as therapy (21). In NK cells, CD16a Fc receptor crosslinking triggers ADCC (238). NK cell-mediated ADCC is a central mechanism to the killing primed by anti-HER2 (trastuzumab) (239) anti-GD2, (dinutuximab) (240), and anti-CD20 (rituximab) (241). In mice deficient for CD16a, or in which CD16a engagement with antibodies is blocked, tumor growth is exacerbated (239). Noteworthy, CD16a is susceptible to cleavage by metalloproteases within the tumor environment, and these soluble fragments could block CD16a on NK cells and create an additional opportunity to evade ADCC (242). Strategies to strengthen antibody binding and Fc receptor signaling are being explored and include antibody Fc engineering to maximize binding and activation, non-cleavable CD16 molecules, bi-specific antibodies, and antibody-drug conjugates (37, 238, 243).
Signalling lymphocytic activating molecule (SLAM)-family receptors are a group of type I transmembrane receptors, expressed on hematopoietic cells (244), especially during and shortly after differentiation (Figure 4C) (245). SLAM-family receptors expressed on NK cells include SLAMF1, SLAMF2 (CD48), SLAMF3, SLAMF4 (2B4), SLAMF6, SLAMF7 (CRACC) and SLAMF8– with CD48, 2B4, and CRACC being the most prominent (246).
These SLAM-family receptors, excluding CD48 and SLAMF8, signal through ITSMs (247). SLAM-family receptors are homotypic, except for the partnership of 2B4 and CD48 (248, 249). CD48 and 2B4 also, uniquely, bind in both trans (ie. with other ligand-expressing cells) or in cis (ie. on the NK cell surface) (250, 251). Cis binding conveys baseline 2B4 ITSM phosphorylation, and a higher signalling threshold for activation due to competition with ligands for binding in trans (248, 250, 251).
SLAM-family receptors can be expressed on hematologic (252) and solid tumors (245, 249). Indeed, CD48-2B4 signalling has been linked to early NK cell activation by monocytes, followed by exhaustion (253). NK cell monocytes isolated from hepatocellular carinoma highly express CD48, and blocking this CD48-2B4 interaction leads to relatively decreased NK cell activation and sequential exhaustion (253). Monocytes isolated from hepatocellular carcinoma have high CD48 expression and blocking the 2B4-CD48 interaction decreased NK cell activation and exhaustion (253). This provides evidence that a multitarget approach might be necessary for checkpoint therapy, and that targeting SLAM-family receptors (whether inhibiting or activating these receptors) is not a one-size-fits all for each tumor. Additionally, both CD48 and CRACC are highly expressed on multiple myeloma, and these are now being investigated as targets for monoclonal antibody therapies (254). Monoclonal CD48 has shown promising pre-clinical results at decreading multiple myeloma tumor growth (252). Undoubtedly, the extensive expression of SLAM family members and their ability to contribute to NK cell activation will promote further investigation toward their clinical use.
The DNAX-accessory molecule-1 (DNAM-1) and TIGIT receptors are commonly expressed on NK cells and T cells, and to interact with the same ligands: CD155 and CD112 (64, 255, 256). DNAM-1 is an activating receptor that signals via ITT; TIGIT is an inhibitory immune checkpoint that encodes an ITIM (65, 257) (Figure 5). CD96 (TACTILE) also contains an ITIM, and binds to CD155, but not CD112 (257), and is thought to have secondary roles in cell-cell adhesion (257). Recently, KIR2DL5, an ITIM-containing KIR, has also been identified as a receptor for CD155 (258).
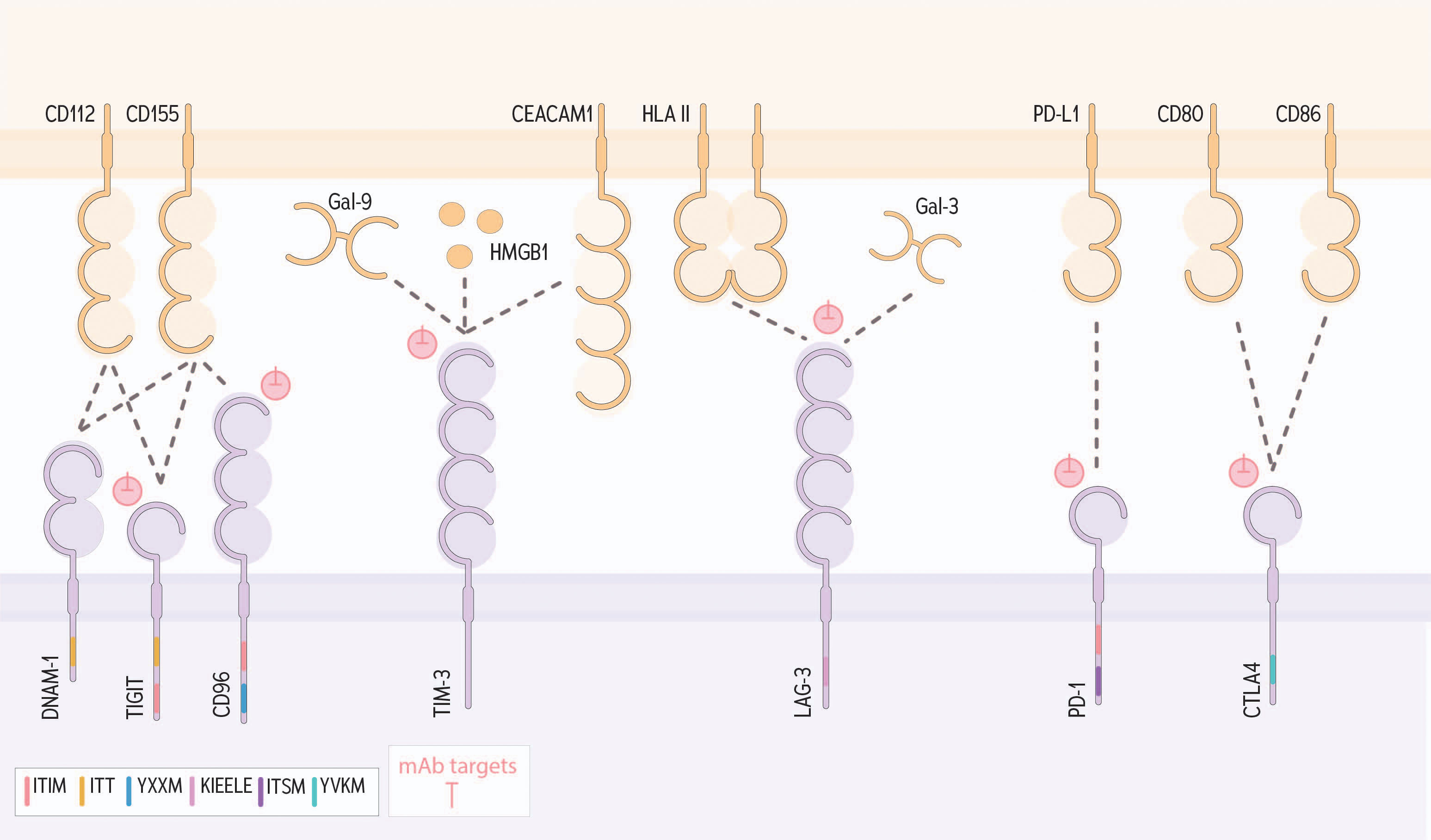
Figure 5 DNAM-1, TIGIT, CD96 and other immunologic checkpoints that contribute to NK cell regulation within a tumor. The adhesion molecule CD112 is recognized by both DNAM-1 and TIGIT. Engagement with DNAM-1 results in activation via ITT signaling, conversely binding to TIGIT results in net inhibition signed through an ITIM. CD155, another adhesion molecule, binds to DNAM-1, TIGIT and CD96; signaling through CD96 through the signaling motifs tyrosine-based sorting motif (YXXM) and ITIM. TIM-3 can bind a variety of ligands including Gal-9, HMGB1 and CEACM1 to signal for inhibition. LAG-3 is known to bind HLA II and Gal-3. PD-1 binds PD-L1 and CTLA4 binds to CD80/CD86 to signal for inhibition. Pink symbols indicate mAb targets.
Each of CD155 and CD112 bind TIGIT and DNAM-1, signaling for inhibition through TIGIT or activation through DNAM-1. CD155 and CD112 both bind TIGIT with a higher affinity that DNAM-1 (259, 260). CD155 and CD112 are expressed at low levels on healthy cells and are upregulated in response to inflammation, cellular stress, reactive oxygen and nitrogen species (261–263), and following chemotherapy (264). Overexpression of these ligands on the tumor is usually associated with poorer prognosis for patients: increased tumor expression of CD112 correlates with increased tumor size and stage, in cancers of the gallbladder (265), colon (266), ovary (267), and pancreas (268). Likewise, tumor overexpression of CD155 is associated with poor prognosis and progression of multiple cancers (269) including breast cancer (270), gastric cancer (271), non-small cell lung cancer (272), melanoma (273), colorectal cancer (274), and sarcoma (275).
TIGIT modulates DNAM-1 mediated activation through the competitive binding of their shared ligands (276). Further, TIGIT directly inhibits DNAM-1 activation through cis interactions that interfere with DNAM-1 homodimerization (277). TIGIT blockade enhances NK cell responsiveness both in vitro and in vivo, particularly when DNAM-1/CD155 interactions remain intact (277, 278). In mouse models, blockade of CD96/CD155 binding results in reduced metastatic spread in vivo– in part by enabling DNAM-1/CD155 binding through loss of CD96 competitive binding (279, 280).
Through alternative splicing, CD155 can be released in a soluble form (sCD155), which binds to DNAM-1 with greater affinity than TIGIT or membrane-bound CD155, and block binding and signaling for activation via DNAM-1 (281). High expression of sCD155 is associated with poor prognosis and increased cancer progression in lung, breast, liver, and gynecological cancers (282–284). sCD155 binding can also result in the endocytosis of DNAM-1, making it less available to generate signals for activation (285, 286).
The opposing roles these receptors play is evident in cancer progression; in acute myeloid leukemia, high expression of DNAM-1 is associated with longer progression-free survival and overall survival (287), while increased TIGIT expression was correlated with NK cell dysfunction and poorer patient outcomes (288). Further, CD96 expression was linked with an increased immunosuppressive immune signature and poor patient prognosis in gastric cancer (289).
Targeting TIGIT, and thereby blocking its inhibitory interaction with CD155 and/or CD112, has been the focus of recent research, with several monoclonal antibodies currently undergoing clinical trial. A phase II trial assessing tiragolumab in combination with atezolizumab demonstrated an objective response rate of 37%, with an objective response rate of 66% in patients with a high PD-L1 status. However, some adverse events were observed, with two treatment-related patient deaths (290). Phase III trials are currently under way for both small cell and non-small cell lung carcinoma (NCT04256421; NCT04294810), as well as trials of tiragolumab against other malignancies, such as melanoma (NCT05116202; NCT03554083), pancreatic cancer (NCT03193190), gastric cancer (NCT0493322; NCT05251948), and cervical cancer (NCT04300647). Of course, administration of these antibodies can impact other lymphocytes, and understanding the specific roles of NK cells compared with other lymphocytes are needed.
Immunologic checkpoints are best studied in the context of T cells in cancer but have similar functions in NK cells (51). Immunologic checkpoints known to impact T cells may also inhibit NK cells. These include T cell immunoglobulin and mucin-domain containing-3 (TIM-3), lymphocyte activation gene (LAG-3), programmed cell death protein-1 (PD-1) and cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) (Figure 5). Like the NCRs, these immunologic checkpoints have and share several ligands associated with cellular transformation and cancer. The ligands are discussed below, first alone, and then in the context of their receptors.
● Galectin-9 (Gal-9) is a C-type lectin that is bound by carbohydrate moieties found on membrane-bound proteins across several lymphocytes, including T cells and NK cells (291, 292). Gal-9 is expressed on the cell surface, and can be cleaved by metalloproteinases and secreted in soluble form (293). Gal-9 is involved in cell adhesion and migration (294, 295) and binds to TIM-3 on lymphocytes (296, 297).
● High mobility group box 1 (HMGB-1) is a nuclear protein that binds to and stabilizes DNA at steady state, but can translocate to the cytoplasm of stressed cells and act as a damage-associated molecular pattern (DAMP) that drives activation of innate immunity via toll-like receptors (298).
● Carcinoembryonic Antigen-Related Cell Adhesion Molecule 1 (CEACAM-1) is an immunoglobulin superfamily member most commonly found on epithelial cells, but also on immune cells with regulatory functions (299, 300). In cancer, expression of CEACAM-1 has been associated with both better and worse prognosis (301). In melanoma specifically, its expression is associated with aggressive metastasis (302) and immune exclusion (303).
● Phoshatidylserine is a component of the inner leaflet of healthy cell membranes, and contributes to maintenance of membrane integrity, cell signaling, adhesion and trafficking and apoptosis (304). When externalized – a consequence of apoptosis and cell death (305) – phosphatidylserine signals to phagocytose the damaged cell (304). Phosphatidylserine signaling generally promotes immune regulation, ostensibly to limit inflammation and prevent autoimmunity (306); in cancer, phosphatidylserine can interfere with lymphocyte-mediated tumor killing (307).
● HLA II presents exogenous antigens to helper (CD4+) T cells. HLA II is typically expressed on the surface of professional antigen presenting cells (308), and can be presented by cancer cells and antigen presenting cells in the tumor microenvironment (309).
● Lymph node sinusoidal endothelial cell C-type lectin (LSECtin) is an adhesion molecule that promotes tumor invasion and metastasis. LSECtin has a regulatory role, acting on cellular microRNAs to diminish immune cell activation (310).
● Programmed death ligand-1 and -2 (PD-L1/PD-L2) are type 1 transmembrane proteins of the immunoglobulin superfamily, commonly expressed on hematopoietic and non-hematopoietic cells such as endothelial cells, keratinocytes, and pancreatic islet cells (311). PD-L1 and PD-L2 expression is limited under homeostasis but upregulated in response to environmental stimuli, disease, and inflammatory cytokines including IFN-γ, TNF, and IL-6, likely to limit the extent of inflammation (312–314). PD-L1 overexpression has been observed in lung cancer, lymphoma, and pancreatic cancer (312, 315, 316) and associated with poorer outcomes, including worse overall survival and decreased progression free survival (317–319).
● CD80 and CD86 are members of the B7 receptor-ligand family and are expressed by antigen presenting cells and T regulatory cells (Tregs) (320, 321).
Reinvigoration of T cells has been the major goal of monoclonal antibodies against TIM-3, LAG-3 and PD-L1/L2, but NK cells may be also rescued by their inhibition in the cancer microenvironment. TIM-3 signals for inhibition via five conserved tyrosine residues, and Bat-3 is associated with its transmembrane domain at steady state (322, 323). Known ligands for TIM-3 include Gal-9, HMGB-1, CEACAM-1, and phosphatidylserine. NK cells expressing TIM-3 have suppressed cell-mediated cytotoxicity and can be rescued with TIM-3 blockade (324, 325).
The specific role for Gal-9’s interactions with TIM-3 on NK cells is unclear, with reports of both stimulation for IFN-γ production by the NK-92 cell line (296) and immunoregulation in viral infections and pregnancy (326, 327), suggesting that its role may be nuanced or influenced by microenvironmental features. In T cells, Gal-9/TIM-3 binding triggers release of Bat-3, which liberates an immunosuppressive signal (328). TIM-3 signaling results in inhibition of T cell proliferation and cytokine productions, potentially leading to T cell death (291, 292).
In cancer, the impacts of TIM3:Gal-9 interactions on NK cells are similarly controversial, with reports alternately ascribing pro- and anti-tumor roles. TIM-3:Gal-9 interactions are associated with NK cell exhaustion and decreased cytotoxicity in AML and gastrointestinal tumors (329, 330). Increased Gal-9 expression is associated with worse overall survival, decreased progression-free survival, and increased metastasis in cancers of the liver (331), kidney (332) and virus-associated cancers (333). Conversely, and consistent with an activating role for TIM-3:Gal-9 interactions, higher expression of Gal-9 is associated with less tumor dedifferentiation and metastasis in cervical cancer (334). Gal-9 drives apoptosis of melanoma, leukemia, and lymphoma cell lines (335, 336). Higher Gal-9 expression is associated with superior outcomes in patients with gastric cancer (337) and triple-negative breast cancer (338).
LAG-3 is a member of the immunoglobulin superfamily receptors, and binds MHC class II, and LSECtin (339–343). On T cells, LAG-3 is induced by cell activation, ostensibly to enable control of ongoing lymphocyte responses (344, 345), associates with the TCR and binds HLA II with higher affinity than CD4, to regulate signaling and inhibit proliferation (344–346). LAG-3 is known to be expressed by NK cells (347) with the highest expression being reported on activated, adaptive, and mature NK cells (348).
The expression of MHC II in the tumor microenvironment is typically associated with improved immune cell infiltration and patient outcomes (349, 350). Expression of LAG-3, on the other hand, is well established as a marker of increased tumor progression and aggressiveness across cancer types (351–353). As a result, therapies directly targeting LAG-3 are currently being explored, both as monotherapy and in combination with immune checkpoint blockade. While studies evaluating LAG-3 therapies rarely profile NK cells, one study evaluating the response of patients with melanoma to a combination of anti-LAG-3 and anti-PD-1 found that adaptive LAG-3+ NK cells were most prominent in those who responded to immunotherapy (348). This data supports the role for NK cells influencing response to checkpoint inhibitors. Interestingly, this has also been suggested by earlier work where NK cells isolated from a murine model with LAG-3 deficiency exhibited defects in NK cell mediated anti-tumor immunity (354). Additional research is needed to better understand the role and importance of LAG-3 in NK cell cancer killing.
PD-1 is expressed most prominently on T cells, and has recently been described to be present, but at lower densities on mouse and human NK cells (355, 356). Interference with PD-1/PD-L1/2 signaling with monoclonal antibodies on these mouse NK cells increases their target cell cytotoxicity, confirming a parallel function to that described for T cells (357, 358). PD-1 expression on human NK cells is not yet well understood and seems to be limited to the adaptive NK cell subset (359–361). Nonetheless, blockade of the PD-1/PD-L1 pathway improves NK cell responsiveness both in vitro and in vivo (362, 363). Blocking PD-L1+ NK cells with anti-PD-L1 improves degranulation and cytokine production, as well as control tumor in vivo, indicating that NK cells may contribute to the success of checkpoint inhibition independent of PD-1 expression (364).
Monoclonal antibodies that interfere with PD-L1/L2-PD-1 interactions have become part of treatment for an array of tumors. Durvalumab and atezolizumab (both anti-PD-L1 monoclonal antibodies) are approved as first-line treatments for non-small cell lung cancer (365). Further, durvalumab is approved for bladder cancer and atezolizumab is approved for treatment of triple-negative breast cancer and liver cancer (366–369). Pembrolizumab (anti-PD-1) is also approved for use in MSI-H/dMMR cancers (319). Whether and to what extent these impact NK cell function with impacts on cancer control in patients is unknown.
CTLA-4 is a member of the B7/CD28 family and is constitutively expressed on regulatory T cells, as well as upregulated on other T cell subsets upon activation (370). CTLA-4 inhibits CD28 signalling (371), and T cell activation, through the competitive binding of its ligands CD80/CD86 (372). Expression of CTLA-4 has been reported in both activated mouse and human NK cells (355, 373, 374). In healthy human donors, CTLA-4 expression on NK cell populations is low, and associated with decreased production of activating cytokines and an increase in IL-10 production (355). CTLA-4 expression was found on mediastinal lymph node-derived NK cells of NSCLC patients (375) and on tumor infiltrating NK cells in early-stage lung cancer (376).
The exact role of CTLA-4 expressing NK cells has not yet been fully elucidated. Nonetheless, NK cells have been shown to play a role in the success of anti-CTLA-4 therapies. In melanoma, patients’ response to ipilimumab correlated with an activated NK cell signature (377). Further, ipilimumab was shown to directly bind NK cells through immunofluorescence microscopy. Along with directly binding ipilimumab, NK cells further target CTLA-4 expressing Tregs in vivo and target them through ADCC (378, 379).
Monoclonal antibodies against CTLA-4 are currently in the clinic both alone and in combination with other checkpoint inhibitors. Ipilimumab was first approved as a single-therapy treatment in melanoma (380) and has since been approved as a combination treatment with nivolumab in several cancer types, including colorectal (381), liver (382), renal cell (381), and lung cancer (383). Tremelimumab has recently been approved in combination with durvalumab for the treatment of patients with unresectable liver cancer (384).
Altogether, the immunologic checkpoints that impact T cells may also inhibit NK cells (and be rescued by immune checkpoint blockade). Whether these classical checkpoints are effective targets, or other mechanisms of NK cell inhibition would enable superior anticancer activity remains to be defined.
Though most studies consider the role of NK cell receptor-ligand partnerships in isolation, individual NK cells express constellations of receptors, and their potential impacts should be considered as a composite response. For instance, even strong signals for activation can be outweighed by concurrent signals for inhibition via KIR-HLA interactions (33). Moreover, NK cell function, receptor expression and arming are impacted by local signals, and can change as the environment does (8, 385). Finally, strategies to intervene and manipulate NK cells must also consider the impacts on other cells; for instance, delivery of receptor agonists and cytokines may have impacts beyond their direct impacts on NK cells (386, 387).
Although immunotherapies have dramatically changed outcomes for people with cancer, there remains significant mortality from the disease, refractory cancers, and relapse of cancers with acquired therapeutic resistance (388–390). The tumor microenvironment is often highly immunosuppressive, and tumor cells themselves diversified, so effective treatment with a single-targeting agent is challenging. The variety of functional capabilities and mechanisms of effector:tumor engagement that direct NK cell function make them intriguing targets for immunotherapy, because they may provide a mechanism for agile and ongoing recognition of plastic cancer cells. Already, NK cells are recruited by standard of care treatments and immunotherapeutic strategies, including those that use monoclonal antibodies against tumor antigens and immunologic checkpoint blockade. Strategies exist to expand, engineer, and transfer allogeneic NK cells. Hence, it is feasible to recruit and use NK cells as immunotherapy; the next challenge will be to adequately direct them for cancer killing. We expect this will be achieved by targeting multiple NK cell features simultaneously.
Standard approaches to treatment may be combined with NK cell-targeting therapies to achieve more complete tumor control. Fas agonism, for example, is rendered less-toxic in the context of chemotherapy, so combining these agents may create a synergistic impact to allow greater tumor recognition and control (243). Recognizing that one of NK cells’ intrinsic functions is to respond to “stress” ligands, it is unsurprising that inflammation-inducing therapies can support NK cell activity. For example, radiation induces CXCL8 production in pancreatic tumors, attracting CD56dim NK cells; this associates with prolonged survival (35). Other studies have reported NK cell mediated responses to oncolytic virus infected cells (391), and chemotherapy (34). These treatments all induce inflammatory responses, which may serve to alter the immunosuppressive tumor microenvironment and attract and activate NK cells.
Creating a supportive environment for NK cell function may be facilitated by provision of immune modulating cytokines. Early cytokine therapies included systemic delivery of IL-2 (392), but the concentration of IL-2 needed to achieve meaningful clinical responses was associated with severe adverse toxicities (393). In addition to T cells, NK cells can be activated by IL-2, but high-dose IL-2 can ultimately deprive NK cells of IL-2 as it instead supports regulatory T cell activation and expansion (394). These initial findings led to the attempted use of lower dosing, less potent analogues, ex vivo cytokine treatment of NK cells, or more specific cytokines signals, including IL-15, which strongly supports NK cell proliferation and activation.
IL-15 is most potent in the context of its receptor, IL-15Rα. As therapy, IL-15 and its receptors have been engineered to enhance stability and efficacy. These constructs include the IL-15 super-agonist complex (ALT-803), which is a complex containing a mutated IL-15 (N7D) and IL-15Rα (395). ALT-803 increases NK cell proliferation in the ascites of ovarian cancer patients and increases healthy donor NK cell function against ovarian cancer cell lines (395). When combined with IL-12 and IL-18, IL-15 can induce the adaptive, memory-like NK cell features, including enhanced anti-cancer function (396). More recently, a heteromeric fusion protein complex (HFPC) platform combined IL-12, IL-15, and IL-18, that enhanced primary NK cell proliferation more efficiently compared to the cytokines administered (397).
Immune stimulating agents, including agonists of the stimulator of interferon genes (STING) pathway can likewise achieve an inflammatory and immune-permissive microenvironment (398). STING silencing is a mechanism used by tumors to quiet immune responses (399, 400), and activating STING can enhance NK-cell mediated immunotherapy (401, 402) and NK cell trafficking via CXCR3 (401, 402). IL-2 and STING-agonists together support T and NK cell activation against treatment-refractory mouse tumor models (401, 403). Potentially identifying a mechanism that contributes to these responses, a recent single-cell approach identified that STING mediates its anti-tumor immune stimulating impact, in part, through CXCR3 upregulation and the subsequential recruitment and activation of NK cells (402).
Beyond general signals for inflammation, NK cells can contribute to antigen-specific anti-cancer responses via ADCC driven by therapeutic monoclonal antibodies. More recently, these have been created with cytokines and other agents to drive activation and killing by NK cells simultaneously. One such fusion protein, for example, combines the ALT-803 backbone with rituximab (404). In mouse models, this compound, “N-803” induces NK cells for increased secretion of cytokines, chemokines and growth factors, cytotoxicity, and control of rituximab resistant bone lesions when compared to those cultured or treated with rituximab and ALT-803 while NK cells exhibited enhanced expression of NKG2D, CD16, NCRs, and enhanced cytotoxicity (405). Other fusion proteins focus on enhancing signaling through activating receptors on NK cells. For example, CD123-NKCE, is a TriKE recently developed to bind CD123 on acute myeloid leukemia, while simultaneously signalling through NKp46 and CD16a on NK cells (232). This engager prevented CD64-mediated ADCC inhibition which is a usual mechanism of evasion undertaken by acute myeloid leukemia cells. These strategies direct NK cell mediated ADCC while also focusing on enhancing the NK itself.
NK cells express the immune checkpoints, PD-1 and LAG-3, and immune checkpoint blockade may enable NK cell anti-tumor responses. In the blood of patients with metastatic melanoma treated with relatlimab (anti-LAG-3) and nivolumab (anti-PD-1) the “adaptive” NK cell subset exhibited the highest LAG-3 expression, and responding patient NK cells were activated with treatment (348). A pre-clinical approach leverages the extensive availability of PD-L1 in the tumor microenvironment and employs NK cells equipped with a chimeric switch receptor linking PD-1 to activating domains CD3ζ, DAP10, or DAP12 (406). These cells had superior cytotoxicity towards PD-L1+ target cells compared to wildtype NK cells (406). In pre-clinical studies, inhibition of other immune checkpoints TGFβ and CIS (an NK cell IL-15 signalling checkpoint encoded by CISH) simultaneously results in enhanced NK cell activation and decreased MC38 colorectal cancer tumor burden in mice (407). Similarly, CISH is another member of the suppressor of cytokine signaling (SOCS) family encoded by CISH that when targeted, improved NK cell effector functions (408).
A major advantage of NK cells is that they can be adoptively transferred across allogeneic barriers and expanded extensively ex vivo, opening the possibility of “off the shelf” cancer therapy. Strategies to select and/or engineer NK cells with the greatest anti-cancer potential are still in development and approaches that maximize NK cell activation against tumors will likely be the most effective against cancer. For example, recent work demonstrating that isolating single-KIR+NKG2C+ NK cells from donors harboring large adaptive NK cell subsets could be used to optimize response against HLA-mismatched acute myeloid leukemia (409). Other cellular sources for NK cells, including umbilical cord blood stem cells or induced pluripotent stem cells (iPSCs) may provide further flexibility and opportunities to tailor NK cells as adoptive cell therapy [reviewed recently by (410, 411)]. For example, NK cell metabolism could be reprogramed in iPSC derived NK cells through the deletion of CISH, which normally opposes IL-15 signaling, to enhance in vivo persistence and efficacy (412).
NK cell expansion protocols provide an ideal platform for modulating NK cell populations through pharmacologic interventions such as small molecule inhibitors. There are several small molecule inhibitors available that target proteins used by NK cells to regulate cell signalling. For example, glycogen synthetase kinase (GSK)3 inhibitors can halt GSK3 mediated NFκB inhibition, thereby promoting NK cell activation (413). Indeed, administering a GSK3 inhibitor, CHIR99021, to IL-15 expanded adaptive NK cells enhanced cytokine production, natural cytotoxicity, and antibody-dependent cytotoxicity (414). These small molecule inhibitors can also be harnessed to enhance NK cell resistance against tumor-mediated suppression. Canonical TGF-β signaling suppresses NK cell function and remains a barrier to intra-tumoral NK cell activation (415). TGF-β signaling is facilitated through SMAD3, which can be inhibited through small molecule inhibitor SIS3 (416). Indeed, the use of SIS3 in vitro and in vivo has demonstrated the ability to release E4BP4/NFIL3 NK cell differentiation and promote NK cell mediated lung tumor control (417).
Engineered chimeric antigen receptor (CAR)-NK cells allow durable, antigen-directed targets alongside the anticancer activity of NK cells and offer the combined advantages of NK cells and potent antigen targeting through CAR. CAR-NK have exhibited exceptional efficacy. In a clinical trial employing CD19-targeting CAR-NK cells in patients with lymphoid malignancies, CAR-NK cells were well-tolerated, and generated complete remission in 7/11 treated patients (38). Combined strategies to maximize key features of NK cells are in their infancy, but possible and may further enhance the efficacy of NK cellular therapy. For example, CAR-NK cells have been further modified to enhance ADCC can by inclusion of non-cleavable CD16 and a membrane-bound IL-15 fusion molecule (358). There are many combinations that are feasible, but following the biology of NK cells and the tumors against which they act may help to design rational, bespoke approaches to comprehensive tumor targeting.
NK cells are equipped with a toolkit of germline-encoded activating and inhibitory receptors, which act together to integrate incoming signals. Since the receptors on the NK cells that comprise a person’s repertoire are variable, they provide extensive diversity to recognize a variety of target cell phenotypes. NK cells are critical agents of immunosurveillance and participate in existing approaches to treat cancer.
The next generation of immunotherapies are multivalent: they simultaneously target more than one feature of tumors to prevent immune escape. Strategies to quantify the strength of signaling associations based on both NK cell receptors and the ligands present will be required to prioritize targets. Understood, these will enable development of multitargeted, precision NK cell-based cancer immunotherapies.
SN: Conceptualization, Validation, Visualization, Writing – original draft, Writing – review & editing. EC: Conceptualization, Writing – original draft, Writing – review & editing. SL: Conceptualization, Writing – original draft, Writing – review & editing. LW: Writing – original draft, Writing – review & editing. JB: Conceptualization, Funding acquisition, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. SN, EC and SL are trainee members of the Beatrice Hunter Cancer Research Institute. SN is supported by a Vanier Canada Graduate Scholarship from the Canadian Institutes of Health Research and a Killam Predoctoral Award and Nova Scotia Graduate Scholarship through Dalhousie University. EC has funds provided by the Dalhousie Medical Research Foundation’s Crease Endowment for Cancer Research. SL is supported by a Canada Graduate Scholarship from the Canadian Institutes of Health Research and a Nova Scotia Graduate Scholarship through Dalhousie University. This work is supported by a joint J.D. Irving/Canadian Cancer Society grant co-funded by the Canadian Institutes of Health Research and the Cancer Research Society’s operating grant to JB.
Dalhousie University and the Boudreau laboratory are in Mi’kma’ki, the traditional and unceded territories of the Mi’kmaq peoples. We acknowledge our responsibilities laid out in the Peace and Friendship treaties and are committed to understanding and reconciliation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kiessling R, Klein E, Wigzell H. “Natural” killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol (1975) 5(2):112–7. doi: 10.1002/eji.1830050208
2. Pfefferle A, Jacobs B, Netskar H, Ask EH, Lorenz S, Clancy T, et al. Intra-lineage plasticity and functional reprogramming maintain natural killer cell repertoire diversity. Cell Rep (2019) 29(8):2284–94 e4. doi: 10.1016/j.celrep.2019.10.058
3. Boudreau JE, Hsu KC. Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol (2018) 39(3):222–39. doi: 10.1016/j.it.2017.12.001
4. Huntington ND, Cursons J, Rautela J. The cancer-natural killer cell immunity cycle. Nat Rev Cancer. (2020) 20(8):437–54. doi: 10.1038/s41568-020-0272-z
5. Laskowski TJ, Biederstadt A, Rezvani K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat Rev Cancer. (2022) 22(10):557–75. doi: 10.1038/s41568-022-00491-0
6. Cherrier DE, Serafini N, Di Santo JP. Innate lymphoid cell development: A T cell perspective. Immunity (2018) 48(6):1091–103. doi: 10.1016/j.immuni.2018.05.010
7. Boudreau JE, Hsu KC. Natural killer cell education in human health and disease. Curr Opin Immunol (2018) 50:102–11. doi: 10.1016/j.coi.2017.11.003
8. Guia S, Jaeger BN, Piatek S, Mailfert S, Trombik T, Fenis A, et al. Confinement of activating receptors at the plasma membrane controls natural killer cell tolerance. Sci Signal (2011) 4(167):ra21. doi: 10.1126/scisignal.2001608
9. Goodridge JP, Jacobs B, Saetersmoen ML, Clement D, Hammer Q, Clancy T, et al. Remodeling of secretory lysosomes during education tunes functional potential in NK cells. Nat Commun (2019) 10(1):514. doi: 10.1038/s41467-019-08384-x
10. Lopes N, Galluso J, Escaliere B, Carpentier S, Kerdiles YM, Vivier E. Tissue-specific transcriptional profiles and heterogeneity of natural killer cells and group 1 innate lymphoid cells. Cell Rep Med (2022) 3(11):100812. doi: 10.1016/j.xcrm.2022.100812
11. Kim AMJ, Nemeth MR, Lim SO. 4-1BB: A promising target for cancer immunotherapy. Front Oncol (2022) 12:968360. doi: 10.3389/fonc.2022.968360
12. Strohm L, Ubbens H, Munzel T, Daiber A, Daub S. Role of CD40(L)-TRAF signaling in inflammation and resolution-a double-edged sword. Front Pharmacol (2022) 13:995061. doi: 10.3389/fphar.2022.995061
13. O’Brien KL, Finlay DK. Immunometabolism and natural killer cell responses. Nat Rev Immunol (2019) 19(5):282–90. doi: 10.1038/s41577-019-0139-2
14. Poznanski SM, Singh K, Ritchie TM, Aguiar JA, Fan IY, Portillo AL, et al. Metabolic flexibility determines human NK cell functional fate in the tumor microenvironment. Cell Metab (2021) 33(6):1205–20 e5. doi: 10.1016/j.cmet.2021.03.023
15. Crane CA, Han SJ, Barry JJ, Ahn BJ, Lanier LL, Parsa AT. TGF-beta downregulates the activating receptor NKG2D on NK cells and CD8+ T cells in glioma patients. Neuro Oncol (2010) 12(1):7–13. doi: 10.1093/neuonc/nop009
16. Li H, Han Y, Guo Q, Zhang M, Cao X. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J Immunol (2009) 182(1):240–9. doi: 10.4049/jimmunol.182.1.240
17. Zheng X, Hou Z, Qian Y, Zhang Y, Cui Q, Wang X, et al. Tumors evade immune cytotoxicity by altering the surface topology of NK cells. Nat Immunol (2023) 24(5):802–13. doi: 10.1038/s41590-023-01462-9
18. Sen Santara S, Lee DJ, Crespo A, Hu JJ, Walker C, Ma X, et al. The NK cell receptor NKp46 recognizes ecto-calreticulin on ER-stressed cells. Nature (2023) 616(7956):348–56. doi: 10.1038/s41586-023-05912-0
19. Hammer Q, Dunst J, Christ W, Picarazzi F, Wendorff M, Momayyezi P, et al. SARS-CoV-2 Nsp13 encodes for an HLA-E-stabilizing peptide that abrogates inhibition of NKG2A-expressing NK cells. Cell Rep (2022) 38(10):110503. doi: 10.1016/j.celrep.2022.110503
20. Beziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A, et al. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur J Immunol (2012) 42(2):447–57. doi: 10.1002/eji.201141826
21. Wang SY, Racila E, Taylor RP, Weiner GJ. NK-cell activation and antibody-dependent cellular cytotoxicity induced by rituximab-coated target cells is inhibited by the C3b component of complement. Blood (2008) 111(3):1456–63. doi: 10.1182/blood-2007-02-074716
22. Le Bert N, Gasser S. Advances in NKG2D ligand recognition and responses by NK cells. Immunol Cell Biol (2014) 92(3):230–6. doi: 10.1038/icb.2013.111
23. Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: the rheostat model. J Immunol (2009) 182(8):4572–80. doi: 10.4049/jimmunol.0803900
24. Lo HC, Xu Z, Kim IS, Pingel B, Aguirre S, Kodali S, et al. Resistance to natural killer cell immunosurveillance confers a selective advantage to polyclonal metastasis. Nat Cancer. (2020) 1(7):709–22. doi: 10.1038/s43018-020-0068-9
25. Seliger B, Koehl U. Underlying mechanisms of evasion from NK cells as rationale for improvement of NK cell-based immunotherapies. Front Immunol (2022) 13:910595. doi: 10.3389/fimmu.2022.910595
26. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol (2021) 21(8):485–98. doi: 10.1038/s41577-020-00490-y
27. Watt J, Kocher HM. The desmoplastic stroma of pancreatic cancer is a barrier to immune cell infiltration. Oncoimmunology (2013) 2(12):e26788. doi: 10.4161/onci.26788
28. Swamy K. Vascular normalization and immunotherapy: Spawning a virtuous cycle. Front Oncol (2022) 12:1002957. doi: 10.3389/fonc.2022.1002957
29. Vazquez-Garcia I, Uhlitz F, Ceglia N, Lim JLP, Wu M, Mohibullah N, et al. Ovarian cancer mutational processes drive site-specific immune evasion. Nature (2022) 612(7941):778–86. doi: 10.1038/s41586-022-05496-1
30. Mi H, Sivagnanam S, Betts CB, Liudahl SM, Jaffee EM, Coussens LM, et al. Quantitative spatial profiling of immune populations in pancreatic ductal adenocarcinoma reveals tumor microenvironment heterogeneity and prognostic biomarkers. Cancer Res (2022) 82(23):4359–72. doi: 10.1158/0008-5472.CAN-22-1190
31. Nersesian S, Schwartz SL, Grantham SR, MacLean LK, Lee SN, Pugh-Toole M, et al. NK cell infiltration is associated with improved overall survival in solid cancers: A systematic review and meta-analysis. Transl Oncol (2021) 14(1):100930. doi: 10.1016/j.tranon.2020.100930
32. Kannan GS, Aquino-Lopez A, Lee DA. Natural killer cells in Malignant hematology: A primer for the non-immunologist. Blood Rev (2017) 31(2):1–10. doi: 10.1016/j.blre.2016.08.007
33. Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest. (2012) 122(9):3260–70. doi: 10.1172/JCI62749
34. Jimenez-Sanchez A, Cybulska P, Mager KL, Koplev S, Cast O, Couturier DL, et al. Unraveling tumor-immune heterogeneity in advanced ovarian cancer uncovers immunogenic effect of chemotherapy. Nat Genet (2020) 52(6):582–93. doi: 10.1038/s41588-020-0630-5
35. Walle T, Kraske JA, Liao B, Lenoir B, Timke C, von Bohlen Und Halbach E, et al. Radiotherapy orchestrates natural killer cell dependent antitumor immune responses through CXCL8. Sci Adv (2022) 8(12):eabh4050. doi: 10.1126/sciadv.abh4050
36. Dixon KJ, Wu J, Walcheck B. Engineering anti-tumor monoclonal antibodies and fc receptors to enhance ADCC by human NK cells. Cancers (Basel). (2021) 13(2):1–13. doi: 10.3390/cancers13020312
37. Musolino A, Gradishar WJ, Rugo HS, Nordstrom JL, Rock EP, Arnaldez F, et al. Role of Fcgamma receptors in HER2-targeted breast cancer therapy. J Immunother Cancer (2022) 10(1):1–16. doi: 10.1136/jitc-2021-003171
38. Liu E, Marin D, Banerjee P, Macapinlac HA, Thompson P, Basar R, et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N Engl J Med (2020) 382(6):545–53. doi: 10.1056/NEJMoa1910607
39. Gleason MK, Ross JA, Warlick ED, Lund TC, Verneris MR, Wiernik A, et al. CD16xCD33 bispecific killer cell engager (BiKE) activates NK cells against primary MDS and MDSC CD33+ targets. Blood (2014) 123(19):3016–26. doi: 10.1182/blood-2013-10-533398
40. Felices M, Lenvik TR, Davis ZB, Miller JS, Vallera DA. Generation of biKEs and triKEs to improve NK cell-mediated targeting of tumor cells. Methods Mol Biol (2016) 1441:333–46. doi: 10.1007/978-1-4939-3684-7_28
41. Verron Q, Forslund E, Brandt L, Leino M, Frisk TW, Olofsson PE, et al. NK cells integrate signals over large areas when building immune synapses but require local stimuli for degranulation. Sci Signal (2021) 14(684):1–15. doi: 10.1126/scisignal.abe2740
42. Tomaz D, Pereira PM, Guerra N, Dyson J, Gould K, Henriques R. Nanoscale colocalization of NK cell activating and inhibitory receptors controls signal integration. Front Immunol (2022) 13:868496. doi: 10.3389/fimmu.2022.868496
43. Bryceson YT, March ME, Ljunggren HG, Long EO. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood (2006) 107(1):159–66. doi: 10.1182/blood-2005-04-1351
44. Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol (2013) 31:227–58. doi: 10.1146/annurev-immunol-020711-075005
45. Bodden M, Hacker A, Roder J, Kiefer A, Zhang C, Bhatti A, et al. Co-expression of an IL-15 superagonist facilitates self-enrichment of GD(2)-targeted CAR-NK cells and mediates potent cell killing in the absence of IL-2. Cancers (Basel) (2023) 15(17):1–21. doi: 10.3390/cancers15174310
46. Zhu H, Kaufman DS. Engineered human pluripotent stem cell-derived natural killer cells: the next frontier for cancer immunotherapy. Blood Sci (2019) 1(1):4–11. doi: 10.1097/BS9.0000000000000023
47. Boudreau JE, Liu XR, Zhao Z, Zhang A, Shultz LD, Greiner DL, et al. Cell-extrinsic MHC class I molecule engagement augments human NK cell education programmed by cell-intrinsic MHC class I. Immunity (2016) 45(2):280–91. doi: 10.1016/j.immuni.2016.07.005
48. Karre K. Natural killer cell recognition of missing self. Nat Immunol (2008) 9(5):477–80. doi: 10.1038/ni0508-477
49. Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics (2000) 51(4-5):268–80. doi: 10.1007/s002510050620
50. Colucci F. Uterine NK cells ace an “A” in education: NKG2A sets up crucial functions at the maternal-fetal interface. J Immunol (2022) 209(8):1421–5. doi: 10.4049/jimmunol.2200384
51. Poggi A, Zocchi MR. Natural killer cells and immune-checkpoint inhibitor therapy: Current knowledge and new challenges. Mol Ther Oncolytics. (2022) 24:26–42. doi: 10.1016/j.omto.2021.11.016
52. Le Drean E, Vely F, Olcese L, Cambiaggi A, Guia S, Krystal G, et al. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. Eur J Immunol (1998) 28(1):264–76. doi: 10.1002/(SICI)1521-4141(199801)28:01<264::AID-IMMU264>3.0.CO;2-O
53. Wu Z, Park S, Lau CM, Zhong Y, Sheppard S, Sun JC, et al. Dynamic variability in SHP-1 abundance determines natural killer cell responsiveness. Sci Signal (2021) 14(708):eabe5380. doi: 10.1126/scisignal.abe5380
54. Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C. Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244). Blood (2005) 105(12):4722–9. doi: 10.1182/blood-2004-09-3796
55. Yusa S, Catina TL, Campbell KS. SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol (2002) 168(10):5047–57. doi: 10.4049/jimmunol.168.10.5047
56. Fasbender F, Claus M, Wingert S, Sandusky M, Watzl C. Differential requirements for src-family kinases in SYK or ZAP70-mediated SLP-76 phosphorylation in lymphocytes. Front Immunol (2017) 8:789. doi: 10.3389/fimmu.2017.00789
57. Mbiribindi B, Mukherjee S, Wellington D, Das J, Khakoo SI. Spatial clustering of receptors and signaling molecules regulates NK cell response to peptide repertoire changes. Front Immunol (2019) 10:605. doi: 10.3389/fimmu.2019.00605
58. Tomasello E, Blery M, Vely F, Vivier E. Signaling pathways engaged by NK cell receptors: double concerto for activating receptors, inhibitory receptors and NK cells. Semin Immunol (2000) 12(2):139–47. doi: 10.1006/smim.2000.0216
59. Cecchetti S, Spadaro F, Lugini L, Podo F, Ramoni C. Functional role of phosphatidylcholine-specific phospholipase C in regulating CD16 membrane expression in natural killer cells. Eur J Immunol (2007) 37(10):2912–22. doi: 10.1002/eji.200737266
60. Grewal RK, Das J. Spatially resolved in silico modeling of NKG2D signaling kinetics suggests a key role of NKG2D and Vav1 Co-clustering in generating natural killer cell activation. PloS Comput Biol (2022) 18(5):e1010114. doi: 10.1371/journal.pcbi.1010114
61. Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity (1998) 8(6):693–701. doi: 10.1016/S1074-7613(00)80574-9
62. Billadeau DD, Upshaw JL, Schoon RA, Dick CJ, Leibson PJ. NKG2D-DAP10 triggers human NK cell-mediated killing via a Syk-independent regulatory pathway. Nat Immunol (2003) 4(6):557–64. doi: 10.1038/ni929
63. Segovis CM, Schoon RA, Dick CJ, Nacusi LP, Leibson PJ, Billadeau DD. PI3K links NKG2D signaling to a CrkL pathway involved in natural killer cell adhesion, polarity, and granule secretion. J Immunol (2009) 182(11):6933–42. doi: 10.4049/jimmunol.0803840
64. Andrade LF, Smyth MJ, Martinet L. DNAM-1 control of natural killer cells functions through nectin and nectin-like proteins. Immunol Cell Biol (2014) 92(3):237–44. doi: 10.1038/icb.2013.95
65. Zhang Z, Wu N, Lu Y, Davidson D, Colonna M, Veillette A. DNAM-1 controls NK cell activation via an ITT-like motif. J Exp Med (2015) 212(12):2165–82. doi: 10.1084/jem.20150792
66. Veillette A, Dong Z, Perez-Quintero LA, Zhong MC, Cruz-Munoz ME. Importance and mechanism of ‘switch’ function of SAP family adapters. Immunol Rev (2009) 232(1):229–39. doi: 10.1111/j.1600-065X.2009.00824.x
67. Vicioso Y, Wong DP, Roy NK, Das N, Zhang K, Ramakrishnan P, et al. NF-kappaB c-Rel Is Dispensable for the Development but Is Required for the Cytotoxic Function of NK Cells. Front Immunol (2021) 12:652786. doi: 10.3389/fimmu.2021.652786
68. Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity (2015) 42(3):443–56. doi: 10.1016/j.immuni.2015.02.008
69. Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity (2015) 42(3):431–42. doi: 10.1016/j.immuni.2015.02.013
70. Nikzad R, Angelo LS, Aviles-Padilla K, Le DT, Singh VK, Bimler L, et al. Human natural killer cells mediate adaptive immunity to viral antigens. Sci Immunol (2019) 4(35):1–13. doi: 10.1126/sciimmunol.aat8116
71. Yamamoto F, Suzuki S, Mizutani A, Shigenari A, Ito S, Kametani Y, et al. Capturing differential allele-level expression and genotypes of all classical HLA loci and haplotypes by a new capture RNA-seq method. Front Immunol (2020) 11:941. doi: 10.3389/fimmu.2020.00941
72. Gonzalez-Galarza FF, McCabe A, Santos E, Jones J, Takeshita L, Ortega-Rivera ND, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res (2020) 48(D1):D783–D8. doi: 10.1093/nar/gkz1029
73. Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev (2001) 183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x
74. Parga-Lozano C, Reguera R, Gomez-Prieto P, Arnaiz-Villena A. Evolution of major histocompatibility complex G and C and natural killer receptors in primates. Hum Immunol (2009) 70(12):1035–40. doi: 10.1016/j.humimm.2009.07.017
75. Hackmon R, Pinnaduwage L, Zhang J, Lye SJ, Geraghty DE, Dunk CE. Definitive class I human leukocyte antigen expression in gestational placentation: HLA-F, HLA-E, HLA-C, and HLA-G in extravillous trophoblast invasion on placentation, pregnancy, and parturition. Am J Reprod Immunol (2017) 77(6):1–11. doi: 10.1111/aji.12643
76. Burrows CK, Kosova G, Herman C, Patterson K, Hartmann KE, Velez Edwards DR, et al. Expression quantitative trait locus mapping studies in mid-secretory phase endometrial cells identifies HLA-F and TAP2 as fecundability-associated genes. PloS Genet (2016) 12(7):e1005858. doi: 10.1371/journal.pgen.1005858
77. Dulberger CL, McMurtrey CP, Holzemer A, Neu KE, Liu V, Steinbach AM, et al. Human leukocyte antigen F presents peptides and regulates immunity through interactions with NK cell receptors. Immunity (2017) 46(6):1018–29 e7. doi: 10.1016/j.immuni.2017.06.002
78. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci (2012) 367(1590):800–11. doi: 10.1098/rstb.2011.0266
79. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol (2013) 13(2):133–44. doi: 10.1038/nri3370
80. Boyington JC, Brooks AG, Sun PD. Structure of killer cell immunoglobulin-like receptors and their recognition of the class I MHC molecules. Immunol Rev (2001) 181:66–78. doi: 10.1034/j.1600-065X.2001.1810105.x
81. Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology (2011) 132(3):315–25. doi: 10.1111/j.1365-2567.2010.03398.x
82. Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol (2004) 34(6):1673–9. doi: 10.1002/eji.200425089
83. Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med (2009) 206(11):2557–72. doi: 10.1084/jem.20091010
84. Hilton HG, Parham P. Missing or altered self: human NK cell receptors that recognize HLA-C. Immunogenetics (2017) 69(8-9):567–79. doi: 10.1007/s00251-017-1001-y
85. Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol (2013) 190(12):6198–208. doi: 10.4049/jimmunol.1300464
86. Le Luduec JB, Kudva A, Boudreau JE, Hsu KC. Novel multiplex PCR-SSP method for centromeric KIR allele discrimination. Sci Rep (2018) 8(1):14853. doi: 10.1038/s41598-018-33135-1
87. Rajagopalan S, Long EO. Understanding how combinations of HLA and KIR genes influence disease. J Exp Med (2005) 201(7):1025–9. doi: 10.1084/jem.20050499
88. Pugh J, Guethlein L, Parham P. Abundant CpG-sequences in human genomes inhibit KIR3DL2-expressing NK cells. PeerJ (2021) 9:e12258. doi: 10.7717/peerj.12258
89. Lin A, Yan WH. HLA-G/ILTs targeted solid cancer immunotherapy: opportunities and challenges. Front Immunol (2021) 12:698677. doi: 10.3389/fimmu.2021.698677
90. Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in human cornea, an immune-privileged tissue. Hum Immunol (2003) 64(11):1039–44. doi: 10.1016/j.humimm.2003.08.346
91. Zhuang B, Shang J, Yao Y. HLA-G: an important mediator of maternal-fetal immune-tolerance. Front Immunol (2021) 12:744324. doi: 10.3389/fimmu.2021.744324
92. Guo ZY, Lv YG, Wang L, Shi SJ, Yang F, Zheng GX, et al. Predictive value of HLA-G and HLA-E in the prognosis of colorectal cancer patients. Cell Immunol (2015) 293(1):10–6. doi: 10.1016/j.cellimm.2014.10.003
93. Hiraoka N, Ino Y, Hori S, Yamazaki-Itoh R, Naito C, Shimasaki M, et al. Expression of classical human leukocyte antigen class I antigens, HLA-E and HLA-G, is adversely prognostic in pancreatic cancer patients. Cancer Sci (2020) 111(8):3057–70. doi: 10.1111/cas.14514
94. de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol (2010) 185(12):7452–9. doi: 10.4049/jimmunol.1002629
95. Menier C, Prevot S, Carosella ED, Rouas-Freiss N. Human leukocyte antigen-G is expressed in advanced-stage ovarian carcinoma of high-grade histology. Hum Immunol (2009) 70(12):1006–9. doi: 10.1016/j.humimm.2009.07.021
96. Morandi F, Ferretti E, Castriconi R, Dondero A, Petretto A, Bottino C, et al. Soluble HLA-G dampens CD94/NKG2A expression and function and differentially modulates chemotaxis and cytokine and chemokine secretion in CD56bright and CD56dim NK cells. Blood (2011) 118(22):5840–50. doi: 10.1182/blood-2011-05-352393
97. Morvan M, David G, Sebille V, Perrin A, Gagne K, Willem C, et al. Autologous and allogeneic HLA KIR ligand environments and activating KIR control KIR NK-cell functions. Eur J Immunol (2008) 38(12):3474–86. doi: 10.1002/eji.200838407
98. Liu J, Xiao Z, Ko HL, Shen M, Ren EC. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc Natl Acad Sci U S A. (2014) 111(7):2662–7. doi: 10.1073/pnas.1322052111
99. Blokhuis JH, Hilton HG, Guethlein LA, Norman PJ, Nemat-Gorgani N, Nakimuli A, et al. KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans. Immun Inflammation Dis (2017) 5(4):461–8. doi: 10.1002/iid3.178
100. Le Luduec JB, Boudreau JE, Freiberg JC, Hsu KC. Novel approach to cell surface discrimination between KIR2DL1 subtypes and KIR2DS1 identifies hierarchies in NK repertoire, education, and tolerance. Front Immunol (2019) 10:734. doi: 10.3389/fimmu.2019.00734
101. Pittari G, Liu XR, Selvakumar A, Zhao Z, Merino E, Huse M, et al. NK cell tolerance of self-specific activating receptor KIR2DS1 in individuals with cognate HLA-C2 ligand. J Immunol (2013) 190(9):4650–60. doi: 10.4049/jimmunol.1202120
102. Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M, et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med (2012) 367(9):805–16. doi: 10.1056/NEJMoa1200503
103. Boudreau JE, Giglio F, Gooley TA, Stevenson PA, Le Luduec JB, Shaffer BC, et al. KIR3DL1/HLA-B subtypes govern acute myelogenous leukemia relapse after hematopoietic cell transplantation. J Clin Oncol (2017) 35(20):2268–78. doi: 10.1200/JCO.2016.70.7059
104. Kaddu-Mulindwa D, Altmann B, Robrecht S, Ziepert M, Regitz E, Tausch E, et al. KIR2DS1-HLA-C status as a predictive marker for benefit from rituximab: a post-hoc analysis of the RICOVER-60 and CLL8 trials. Lancet Haematol (2022) 9(2):e133–e42. doi: 10.1016/S2352-3026(21)00369-0
105. Lin Z, Bashirova AA, Viard M, Garner L, Quastel M, Beiersdorfer M, et al. HLA class I signal peptide polymorphism determines the level of CD94/NKG2-HLA-E-mediated regulation of effector cell responses. Nat Immunol (2023) 24(7):1087–97. doi: 10.1038/s41590-023-01523-z
106. Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S, et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci Immunol (2017) 2(15):1–11. doi: 10.1126/sciimmunol.aal5296
107. Garcia-Beltran WF, Holzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR, et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat Immunol (2016) 17(9):1067–74. doi: 10.1038/ni.3513
108. Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature (1998) 391(6669):795–9. doi: 10.1038/35869
109. Barrett DM, Gustafson KS, Wang J, Wang SZ, Ginder GD. A GATA factor mediates cell type-restricted induction of HLA-E gene transcription by gamma interferon. Mol Cell Biol (2004) 24(14):6194–204. doi: 10.1128/MCB.24.14.6194-6204.2004
110. Gallegos CE, Michelin S, Dubner D, Carosella ED. Immunomodulation of classical and non-classical HLA molecules by ionizing radiation. Cell Immunol (2016) 303:16–23. doi: 10.1016/j.cellimm.2016.04.005
111. Hrbac T, Kopkova A, Siegl F, Vecera M, Ruckova M, Kazda T, et al. HLA-E and HLA-F are overexpressed in glioblastoma and HLA-E increased after exposure to ionizing radiation. Cancer Genomics Proteomics. (2022) 19(2):151–62. doi: 10.21873/cgp.20311
112. Boegel S, Lower M, Bukur T, Sorn P, Castle JC, Sahin U. HLA and proteasome expression body map. BMC Med Genomics (2018) 11(1):36. doi: 10.1186/s12920-018-0354-x
113. Zeestraten EC, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker JW, Liefers GJ, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br J Cancer. (2014) 110(2):459–68. doi: 10.1038/bjc.2013.696
114. Morinaga T, Iwatsuki M, Yamashita K, Matsumoto C, Harada K, Kurashige J, et al. Evaluation of HLA-E expression combined with natural killer cell status as a prognostic factor for advanced gastric cancer. Ann Surg Oncol (2022) 29(8):4951–60. doi: 10.1245/s10434-022-11665-3
115. Seliger B, Jasinski-Bergner S, Quandt D, Stoehr C, Bukur J, Wach S, et al. HLA-E expression and its clinical relevance in human renal cell carcinoma. Oncotarget (2016) 7(41):67360–72. doi: 10.18632/oncotarget.11744
116. Gooden M, Lampen M, Jordanova ES, Leffers N, Trimbos JB, van der Burg SH, et al. HLA-E expression by gynecological cancers restrains tumor-infiltrating CD8(+) T lymphocytes. Proc Natl Acad Sci U S A. (2011) 108(26):10656–61. doi: 10.1073/pnas.1100354108
117. Wu Z, Liang J, Wang Z, Li A, Fan X, Jiang T. HLA-E expression in diffuse glioma: relationship with clinicopathological features and patient survival. BMC Neurol (2020) 20(1):59. doi: 10.1186/s12883-020-01640-4
118. Sun C, Xu J, Huang Q, Huang M, Wen H, Zhang C, et al. High NKG2A expression contributes to NK cell exhaustion and predicts a poor prognosis of patients with liver cancer. Oncoimmunology (2017) 6(1):e1264562. doi: 10.1080/2162402X.2016.1264562
119. Vietzen H, Staber PB, Berger SM, Furlano PL, Kuhner LM, Lubowitzki S, et al. Inhibitory NKG2A(+) and absent activating NKG2C(+) NK cell responses are associated with the development of EBV(+) lymphomas. Front Immunol (2023) 14:1183788. doi: 10.3389/fimmu.2023.1183788
120. Ewen EM, Pahl JHW, Miller M, Watzl C, Cerwenka A. KIR downregulation by IL-12/15/18 unleashes human NK cells from KIR/HLA-I inhibition and enhances killing of tumor cells. Eur J Immunol (2018) 48(2):355–65. doi: 10.1002/eji.201747128
121. Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today (1990) 11(7):237–44. doi: 10.1016/0167-5699(90)90097-S
122. Pollock NR, Harrison GF, Norman PJ. Immunogenomics of killer cell immunoglobulin-like receptor (KIR) and HLA class I: coevolution and consequences for human health. J Allergy Clin Immunol Pract (2022) 10(7):1763–75. doi: 10.1016/j.jaip.2022.04.036
123. Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, Hsu KC. KIR3DL1 and HLA-B density and binding calibrate NK education and response to HIV. J Immunol (2016) 196(8):3398–410. doi: 10.4049/jimmunol.1502469
124. Mehta RS, Rezvani K. Can we make a better match or mismatch with KIR genotyping? Hematol Am Soc Hematol Educ Program (2016) 2016(1):106–18. doi: 10.1182/asheducation-2016.1.106
125. Shang QN, Yu XX, Xu ZL, Cao XH, Liu XF, Zhao XS, et al. Functional competence of NK cells via the KIR/MHC class I interaction correlates with DNAM-1 expression. J Immunol (2022) 208(2):492–500. doi: 10.4049/jimmunol.2100487
126. Zhang S, Kohli K, Black RG, Yao L, Spadinger SM, He Q, et al. Systemic interferon-gamma increases MHC class I expression and T-cell infiltration in cold tumors: results of a phase 0 clinical trial. Cancer Immunol Res (2019) 7(8):1237–43. doi: 10.1158/2326-6066.CIR-18-0940
127. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med (2016) 375(9):819–29. doi: 10.1056/NEJMoa1604958
128. Mehta AM, Jordanova ES, Kenter GG, Ferrone S, Fleuren GJ. Association of antigen processing machinery and HLA class I defects with clinicopathological outcome in cervical carcinoma. Cancer Immunol Immunother. (2008) 57(2):197–206. doi: 10.1007/s00262-007-0362-8
129. Cabrera T, Angustias Fernandez M, Sierra A, Garrido A, Herruzo A, Escobedo A, et al. High frequency of altered HLA class I phenotypes in invasive breast carcinomas. Hum Immunol (1996) 50(2):127–34. doi: 10.1016/0198-8859(96)00145-0
130. Menon AG, Morreau H, Tollenaar RA, Alphenaar E, Van Puijenbroek M, Putter H, et al. Down-regulation of HLA-A expression correlates with a better prognosis in colorectal cancer patients. Lab Invest. (2002) 82(12):1725–33. doi: 10.1097/01.LAB.0000043124.75633.ED
131. McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell (2017) 171(6):1259–71 e11. doi: 10.1016/j.cell.2017.10.001
132. Aptsiauri N, Garrido F. The challenges of HLA class I loss in cancer immunotherapy: facts and hopes. Clin Cancer Res (2022) 28(23):5021–9. doi: 10.1158/1078-0432.CCR-21-3501
133. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (2016) 351(6280):1463–9. doi: 10.1126/science.aaf1490
134. Benson DM Jr., Cohen AD, Jagannath S, Munshi NC, Spitzer G, Hofmeister CC, et al. A phase I trial of the anti-KIR antibody IPH2101 and lenalidomide in patients with relapsed/refractory multiple myeloma. Clin Cancer Res (2015) 21(18):4055–61. doi: 10.1158/1078-0432.CCR-15-0304
135. Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, et al. Pillars article: activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science (1999) 285:727–9. doi: 10.1126/science.285.5428.727
136. Choy MK, Phipps ME. MICA polymorphism: biology and importance in immunity and disease. Trends Mol Med (2010) 16(3):97–106. doi: 10.1016/j.molmed.2010.01.002
137. Vivier E, Tomasello E, Paul P. Lymphocyte activation via NKG2D: towards a new paradigm in immune recognition? Curr Opin Immunol (2002) 14(3):306–11. doi: 10.1016/S0952-7915(02)00337-0
138. Jung H, Hsiung B, Pestal K, Procyk E, Raulet DH. RAE-1 ligands for the NKG2D receptor are regulated by E2F transcription factors, which control cell cycle entry. J Exp Med (2012) 209(13):2409–22. doi: 10.1084/jem.20120565
139. Sutherland CL, Rabinovich B, Chalupny NJ, Brawand P, Miller R, Cosman D. ULBPs, human ligands of the NKG2D receptor, stimulate tumor immunity with enhancement by IL-15. Blood (2006) 108(4):1313–9. doi: 10.1182/blood-2005-11-011320
140. Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous Malignancy. Immunity (2008) 28(4):571–80. doi: 10.1016/j.immuni.2008.02.016
141. Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med (2005) 202(5):583–8. doi: 10.1084/jem.20050994
142. Shen J, Pan J, Du C, Si W, Yao M, Xu L, et al. Silencing NKG2D ligand-targeting miRNAs enhances natural killer cell-mediated cytotoxicity in breast cancer. Cell Death Dis (2017) 8(4):e2740. doi: 10.1038/cddis.2017.158
143. de Kruijf EM, Sajet A, van Nes JG, Putter H, Smit VT, Eagle RA, et al. NKG2D ligand tumor expression and association with clinical outcome in early breast cancer patients: an observational study. BMC Cancer. (2012) 12:24. doi: 10.1186/1471-2407-12-24
144. Watson NF, Spendlove I, Madjd Z, McGilvray R, Green AR, Ellis IO, et al. Expression of the stress-related MHC class I chain-related protein MICA is an indicator of good prognosis in colorectal cancer patients. Int J Cancer. (2006) 118(6):1445–52. doi: 10.1002/ijc.21510
145. McGilvray RW, Eagle RA, Watson NF, Al-Attar A, Ball G, Jafferji I, et al. NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin Cancer Res (2009) 15(22):6993–7002. doi: 10.1158/1078-0432.CCR-09-0991
146. Chen Y, Lin WS, Zhu WF, Lin J, Zhou ZF, Huang CZ, et al. Tumor MICA status predicts the efficacy of immunotherapy with cytokine-induced killer cells for patients with gastric cancer. Immunol Res (2016) 64(1):251–9. doi: 10.1007/s12026-015-8743-0
147. Ribeiro CH, Kramm K, Galvez-Jiron F, Pola V, Bustamante M, Contreras HR, et al. Clinical significance of tumor expression of major histocompatibility complex class I-related chains A and B (MICA/B) in gastric cancer patients. Oncol Rep (2016) 35(3):1309–17. doi: 10.3892/or.2015.4510
148. Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A. (1999) 96(12):6879–84. doi: 10.1073/pnas.96.12.6879
149. Okita R, Yukawa T, Nojima Y, Maeda A, Saisho S, Shimizu K, et al. MHC class I chain-related molecule A and B expression is upregulated by cisplatin and associated with good prognosis in patients with non-small cell lung cancer. Cancer Immunol Immunother. (2016) 65(5):499–509. doi: 10.1007/s00262-016-1814-9
150. Vetter CS, Groh V, thor Straten P, Spies T, Brocker EB, Becker JC. Expression of stress-induced MHC class I related chain molecules on human melanoma. J Invest Dermatol (2002) 118(4):600–5. doi: 10.1046/j.1523-1747.2002.01700.x
151. Li K, Mandai M, Hamanishi J, Matsumura N, Suzuki A, Yagi H, et al. Clinical significance of the NKG2D ligands, MICA/B and ULBP2 in ovarian cancer: high expression of ULBP2 is an indicator of poor prognosis. Cancer Immunol Immunother. (2009) 58(5):641–52. doi: 10.1007/s00262-008-0585-3
152. McGilvray RW, Eagle RA, Rolland P, Jafferji I, Trowsdale J, Durrant LG. ULBP2 and RAET1E NKG2D ligands are independent predictors of poor prognosis in ovarian cancer patients. Int J Cancer. (2010) 127(6):1412–20. doi: 10.1002/ijc.25156
153. Chen J, Zhu XX, Xu H, Fang HZ, Zhao JQ. Expression and prognostic significance of unique ULBPs in pancreatic cancer. Onco Targets Ther (2016) 9:5271–9. doi: 10.2147/OTT.S107771
154. Sakiyama MJ, Espinoza I, Reddy A, de Carlo F, Kumar A, Levenson AS, et al. Race-associated expression of MHC class I polypeptide-related sequence A (MICA) in prostate cancer. Exp Mol Pathol (2019) 108:173–82. doi: 10.1016/j.yexmp.2019.04.010
155. Solocinski K, Padget MR, Fabian KP, Wolfson B, Cecchi F, Hembrough T, et al. Overcoming hypoxia-induced functional suppression of NK cells. J Immunother Cancer (2020) 8(1):1–12. doi: 10.1136/jitc-2019-000246
156. Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol (2008) 180(11):7249–58. doi: 10.4049/jimmunol.180.11.7249
157. Lee YS, Heo W, Nam J, Jeung YH, Bae J. The combination of ionizing radiation and proteasomal inhibition by bortezomib enhances the expression of NKG2D ligands in multiple myeloma cells. J Radiat Res (2018) 59(3):245–52. doi: 10.1093/jrr/rry005
158. Weiss T, Schneider H, Silginer M, Steinle A, Pruschy M, Polic B, et al. NKG2D-dependent antitumor effects of chemotherapy and radiotherapy against glioblastoma. Clin Cancer Res (2018) 24(4):882–95. doi: 10.1158/1078-0432.CCR-17-1766
159. Skov S, Pedersen MT, Andresen L, Straten PT, Woetmann A, Odum N. Cancer cells become susceptible to natural killer cell killing after exposure to histone deacetylase inhibitors due to glycogen synthase kinase-3-dependent expression of MHC class I-related chain A and B. Cancer Res (2005) 65(23):11136–45. doi: 10.1158/0008-5472.CAN-05-0599
160. Jennings VA, Scott GB, Rose AMS, Scott KJ, Migneco G, Keller B, et al. Potentiating oncolytic virus-induced immune-mediated tumor cell killing using histone deacetylase inhibition. Mol Ther (2019) 27(6):1139–52. doi: 10.1016/j.ymthe.2019.04.008
161. Zhou Q, Gil-Krzewska A, Peruzzi G, Borrego F. Matrix metalloproteinases inhibition promotes the polyfunctionality of human natural killer cells in therapeutic antibody-based anti-tumour immunotherapy. Clin Exp Immunol (2013) 173(1):131–9. doi: 10.1111/cei.12095
162. Leivas A, Valeri A, Cordoba L, Garcia-Ortiz A, Ortiz A, Sanchez-Vega L, et al. NKG2D-CAR-transduced natural killer cells efficiently target multiple myeloma. Blood Cancer J (2021) 11(8):146. doi: 10.1038/s41408-021-00537-w
163. Chang YH, Connolly J, Shimasaki N, Mimura K, Kono K, Campana D. A chimeric receptor with NKG2D specificity enhances natural killer cell activation and killing of tumor cells. Cancer Res (2013) 73(6):1777–86. doi: 10.1158/0008-5472.CAN-12-3558
164. Zhang C, Roder J, Scherer A, Bodden M, Pfeifer Serrahima J, Bhatti A, et al. Bispecific antibody-mediated redirection of NKG2D-CAR natural killer cells facilitates dual targeting and enhances antitumor activity. J Immunother Cancer (2021) 9(10):1–15. doi: 10.1136/jitc-2021-002980
165. Prajapati K, Perez C, Rojas LBP, Burke B, Guevara-Patino JA. Functions of NKG2D in CD8(+) T cells: an opportunity for immunotherapy. Cell Mol Immunol (2018) 15(5):470–9. doi: 10.1038/cmi.2017.161
166. Baumeister SH, Murad J, Werner L, Daley H, Trebeden-Negre H, Gicobi JK, et al. Phase I trial of autologous CAR T cells targeting NKG2D ligands in patients with AML/MDS and multiple myeloma. Cancer Immunol Res (2019) 7(1):100–12. doi: 10.1158/2326-6066.CIR-18-0307
167. Kucka K, Wajant H. Receptor oligomerization and its relevance for signaling by receptors of the tumor necrosis factor receptor superfamily. Front Cell Dev Biol (2020) 8:615141. doi: 10.3389/fcell.2020.615141
168. Dostert C, Grusdat M, Letellier E, Brenner D. The TNF family of ligands and receptors: communication modules in the immune system and beyond. Physiol Rev (2019) 99(1):115–60. doi: 10.1152/physrev.00045.2017
169. Baltz KM, Krusch M, Bringmann A, Brossart P, Mayer F, Kloss M, et al. Cancer immunoediting by GITR (glucocorticoid-induced TNF-related protein) ligand in humans: NK cell/tumor cell interactions. FASEB J (2007) 21(10):2442–54. doi: 10.1096/fj.06-7724com
170. Fan Z, Yu P, Wang Y, Wang Y, Fu ML, Liu W, et al. NK-cell activation by LIGHT triggers tumor-specific CD8+ T-cell immunity to reject established tumors. Blood (2006) 107(4):1342–51. doi: 10.1182/blood-2005-08-3485
171. Al Sayed MF, Ruckstuhl CA, Hilmenyuk T, Claus C, Bourquin JP, Bornhauser BC, et al. CD70 reverse signaling enhances NK cell function and immunosurveillance in CD27-expressing B-cell Malignancies. Blood (2017) 130(3):297–309. doi: 10.1182/blood-2016-12-756585
172. Takeda K, Hayakawa Y, Smyth MJ, Kayagaki N, Yamaguchi N, Kakuta S, et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat Med (2001) 7(1):94–100. doi: 10.1038/83416
173. Halaas O, Vik R, Ashkenazi A, Espevik T. Lipopolysaccharide induces expression of APO2 ligand/TRAIL in human monocytes and macrophages. Scand J Immunol (2000) 51(3):244–50. doi: 10.1046/j.1365-3083.2000.00671.x
174. Janssen EM, Droin NM, Lemmens EE, Pinkoski MJ, Bensinger SJ, Ehst BD, et al. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature (2005) 434(7029):88–93. doi: 10.1038/nature03337
175. OR E, Tirincsi A, Logue SE, Szegezdi E. The janus face of death receptor signaling during tumor immunoediting. Front Immunol (2016) 7:446. doi: 10.3389/fimmu.2016.00446
176. Ehrlich S, Infante-Duarte C, Seeger B, Zipp F. Regulation of soluble and surface-bound TRAIL in human T cells, B cells, and monocytes. Cytokine (2003) 24(6):244–53. doi: 10.1016/S1043-4666(03)00094-2
177. OR LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, et al. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature (2009) 461(7264):659–63. doi: 10.1038/nature08402
178. Sordo-Bahamonde C, Lorenzo-Herrero S, Payer AR, Gonzalez S, Lopez-Soto A. Mechanisms of apoptosis resistance to NK cell-mediated cytotoxicity in cancer. Int J Mol Sci (2020) 21(10):1–27. doi: 10.3390/ijms21103726
179. Cardoso Alves L, Corazza N, Micheau O, Krebs P. The multifaceted role of TRAIL signaling in cancer and immunity. FEBS J (2021) 288(19):5530–54. doi: 10.1111/febs.15637
180. Levoin N, Jean M, Legembre P. CD95 structure, aggregation and cell signaling. Front Cell Dev Biol (2020) 8:314. doi: 10.3389/fcell.2020.00314
181. Kreuz S, Siegmund D, Rumpf JJ, Samel D, Leverkus M, Janssen O, et al. NFkappaB activation by Fas is mediated through FADD, caspase-8, and RIP and is inhibited by FLIP. J Cell Biol (2004) 166(3):369–80. doi: 10.1083/jcb.200401036
182. Braeuer SJ, Buneker C, Mohr A, Zwacka RM. Constitutively activated nuclear factor-kappaB, but not induced NF-kappaB, leads to TRAIL resistance by up-regulation of X-linked inhibitor of apoptosis protein in human cancer cells. Mol Cancer Res (2006) 4(10):715–28. doi: 10.1158/1541-7786.MCR-05-0231
183. O’Leary L, van der Sloot AM, Reis CR, Deegan S, Ryan AE, Dhami SP, et al. Decoy receptors block TRAIL sensitivity at a supracellular level: the role of stromal cells in controlling tumour TRAIL sensitivity. Oncogene (2016) 35(10):1261–70. doi: 10.1038/onc.2015.180
184. Snajdauf M, Havlova K, Vachtenheim J Jr., Ozaniak A, Lischke R, Bartunkova J, et al. The TRAIL in the treatment of human cancer: an update on clinical trials. Front Mol Biosci (2021) 8:628332. doi: 10.3389/fmolb.2021.628332
185. von Pawel J, Harvey JH, Spigel DR, Dediu M, Reck M, Cebotaru CL, et al. Phase II trial of mapatumumab, a fully human agonist monoclonal antibody to tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1), in combination with paclitaxel and carboplatin in patients with advanced non-small-cell lung cancer. Clin Lung Cancer. (2014) 15(3):188–96 e2. doi: 10.1016/j.cllc.2013.12.005
186. Trarbach T, Moehler M, Heinemann V, Kohne CH, Przyborek M, Schulz C, et al. Phase II trial of mapatumumab, a fully human agonistic monoclonal antibody that targets and activates the tumour necrosis factor apoptosis-inducing ligand receptor-1 (TRAIL-R1), in patients with refractory colorectal cancer. Br J Cancer. (2010) 102(3):506–12. doi: 10.1038/sj.bjc.6605507
187. Greco FA, Bonomi P, Crawford J, Kelly K, Oh Y, Halpern W, et al. Phase 2 study of mapatumumab, a fully human agonistic monoclonal antibody which targets and activates the TRAIL receptor-1, in patients with advanced non-small cell lung cancer. Lung Cancer. (2008) 61(1):82–90. doi: 10.1016/j.lungcan.2007.12.011
188. Fuchs CS, Fakih M, Schwartzberg L, Cohn AL, Yee L, Dreisbach L, et al. TRAIL receptor agonist conatumumab with modified FOLFOX6 plus bevacizumab for first-line treatment of metastatic colorectal cancer: A randomized phase 1b/2 trial. Cancer (2013) 119(24):4290–8. doi: 10.1002/cncr.28353
189. Ciuleanu T, Bazin I, Lungulescu D, Miron L, Bondarenko I, Deptala A, et al. A randomized, double-blind, placebo-controlled phase II study to assess the efficacy and safety of mapatumumab with sorafenib in patients with advanced hepatocellular carcinoma. Ann Oncol (2016) 27(4):680–7. doi: 10.1093/annonc/mdw004
190. Zhu J, Powis de Tenbossche CG, Cane S, Colau D, van Baren N, Lurquin C, et al. Resistance to cancer immunotherapy mediated by apoptosis of tumor-infiltrating lymphocytes. Nat Commun (2017) 8(1):1404. doi: 10.1038/s41467-017-00784-1
191. Navabi S, Doroudchi M, Tashnizi AH, Habibagahi M. Natural killer cell functional activity after 4-1BB costimulation. Inflammation (2015) 38(3):1181–90. doi: 10.1007/s10753-014-0082-0
192. Reithofer M, Rosskopf S, Leitner J, Battin C, Bohle B, Steinberger P, et al. 4-1BB costimulation promotes bystander activation of human CD8 T cells. Eur J Immunol (2021) 51(3):721–33. doi: 10.1002/eji.202048762
193. Vidard L, Dureuil C, Baudhuin J, Vescovi L, Durand L, Sierra V, et al. CD137 (4-1BB) engagement fine-tunes synergistic IL-15- and IL-21-driven NK cell proliferation. J Immunol (2019) 203(3):676–85. doi: 10.4049/jimmunol.1801137
194. Cubukcu HC, Mesutoglu PY, Seval GC, Beksac M. Ex vivo expansion of natural killer cells for hematological cancer immunotherapy: a systematic review and meta-analysis. Clin Exp Med (2023) 23(6):2503–33. doi: 10.1007/s10238-022-00923-z
195. Terrazzano G, Pisanti S, Grimaldi S, Sica M, Fontana S, Carbone E, et al. Interaction between natural killer and dendritic cells: the role of CD40, CD80 and major histocompatibility complex class i molecules in cytotoxicity induction and interferon-gamma production. Scand J Immunol (2004) 59(4):356–62. doi: 10.1111/j.0300-9475.2003.01387.x
196. Carbone E, Ruggiero G, Terrazzano G, Palomba C, Manzo C, Fontana S, et al. A new mechanism of NK cell cytotoxicity activation: the CD40-CD40 ligand interaction. J Exp Med (1997) 185(12):2053–60. doi: 10.1084/jem.185.12.2053
197. Jung D, Baek YS, Lee IJ, Kim KY, Jang H, Hwang S, et al. Ex vivo expanded allogeneic natural killer cells have potent cytolytic activity against cancer cells through different receptor-ligand interactions. J Exp Clin Cancer Res (2021) 40(1):333. doi: 10.1186/s13046-021-02089-0
198. Lindsay RS, Melssen MM, Stasiak K, Annis JL, Woods AN, Rodriguez AB, et al. NK cells reduce anergic T cell development in early-stage tumors by promoting myeloid cell maturation. Front Oncol (2022) 12:1058894. doi: 10.3389/fonc.2022.1058894
199. Barrow AD, Martin CJ, Colonna M. The natural cytotoxicity receptors in health and disease. Front Immunol (2019) 10:909. doi: 10.3389/fimmu.2019.00909
200. Shemesh A, Brusilovsky M, Kundu K, Ottolenghi A, Campbell KS, Porgador A. Splice variants of human natural cytotoxicity receptors: novel innate immune checkpoints. Cancer Immunol Immunother. (2018) 67(12):1871–83. doi: 10.1007/s00262-017-2104-x
201. Zhu Z, Teng KY, Zhou J, Xu Y, Zhang L, Zhao H, et al. B7H6 serves as a negative prognostic marker and an immune modulator in human pancreatic cancer. Front Oncol (2022) 12:814312. doi: 10.3389/fonc.2022.814312
202. Zhao L, Lian T, Li J, Wei S, Li H, Li C, et al. NCR3LG1 (B7-H6) is a potential prognostic factor for bladder cancer patients. Biomarkers (2021) 26(3):260–7. doi: 10.1080/1354750X.2021.1883110
203. Schlecker E, Fiegler N, Arnold A, Altevogt P, Rose-John S, Moldenhauer G, et al. Metalloprotease-mediated tumor cell shedding of B7-H6, the ligand of the natural killer cell-activating receptor NKp30. Cancer Res (2014) 74(13):3429–40. doi: 10.1158/0008-5472.CAN-13-3017
204. Zhang R, Cui D, Xue T, Lang Y, Zhang Y, Li L, et al. HLA-B-associated transcript 3 (Bat3) stabilizes and activates p53 in a HAUSP-dependent manner. J Mol Cell Biol (2020) 12(2):99–112. doi: 10.1093/jmcb/mjz102
205. Pogge von Strandmann E, Simhadri VR, von Tresckow B, Sasse S, Reiners KS, Hansen HP, et al. Human leukocyte antigen-B-associated transcript 3 is released from tumor cells and engages the NKp30 receptor on natural killer cells. Immunity (2007) 27(6):965–74. doi: 10.1016/j.immuni.2007.10.010
206. Elgundi Z, Papanicolaou M, Major G, Cox TR, Melrose J, Whitelock JM, et al. Cancer metastasis: the role of the extracellular matrix and the heparan sulfate proteoglycan perlecan. Front Oncol (2019) 9:1482. doi: 10.3389/fonc.2019.01482
207. Yan J, Xiao G, Yang C, Liu Q, Lv C, Yu X, et al. Cancer-associated fibroblasts promote lymphatic metastasis in cholangiocarcinoma via the PDGF-BB/PDGFR-beta mediated paracrine signaling network. Aging Dis (2023) 14:1–21. doi: 10.14336/AD.2023.0420
208. Brahmi M, Lesluyes T, Dufresne A, Toulmonde M, Italiano A, Mir O, et al. Expression and prognostic significance of PDGF ligands and receptors across soft tissue sarcomas. ESMO Open (2021) 6(1):100037. doi: 10.1016/j.esmoop.2020.100037
209. Turrell FK, Orha R, Guppy NJ, Gillespie A, Guelbert M, Starling C, et al. Age-associated microenvironmental changes highlight the role of PDGF-C in ER(+) breast cancer metastatic relapse. Nat Cancer. (2023) 4(4):468–84. doi: 10.1038/s43018-023-00525-y
210. Boehm EM, Gildenberg MS, Washington MT. The many roles of PCNA in eukaryotic DNA replication. Enzymes (2016) 39:231–54. doi: 10.1016/bs.enz.2016.03.003
211. Shemesh A, Kugel A, Steiner N, Yezersky M, Tirosh D, Edri A, et al. NKp44 and NKp30 splice variant profiles in decidua and tumor tissues: a comparative viewpoint. Oncotarget (2016) 7(43):70912–23. doi: 10.18632/oncotarget.12292
212. Paolillo M, Schinelli S. Extracellular matrix alterations in metastatic processes. Int J Mol Sci (2019) 20(19):1–18. doi: 10.3390/ijms20194947
213. Mimeault M, Batra SK. Molecular biomarkers of cancer stem/progenitor cells associated with progression, metastases, and treatment resistance of aggressive cancers. Cancer Epidemiol Biomarkers Prev (2014) 23(2):234–54. doi: 10.1158/1055-9965.EPI-13-0785
214. Bloushtain N, Qimron U, Bar-Ilan A, Hershkovitz O, Gazit R, Fima E, et al. Membrane-associated heparan sulfate proteoglycans are involved in the recognition of cellular targets by NKp30 and NKp46. J Immunol (2004) 173(4):2392–401. doi: 10.4049/jimmunol.173.4.2392
215. Hecht ML, Rosental B, Horlacher T, Hershkovitz O, De Paz JL, Noti C, et al. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res (2009) 8(2):712–20. doi: 10.1021/pr800747c
216. Kruse PH, Matta J, Ugolini S, Vivier E. Natural cytotoxicity receptors and their ligands. Immunol Cell Biol (2014) 92(3):221–9. doi: 10.1038/icb.2013.98
217. Liu X, Guo R, Xu Y. B7-H6 as a diagnostic biomarker for cervical squamous cell carcinoma. Genet Test Mol Biomarkers. (2021) 25(7):463–70. doi: 10.1089/gtmb.2020.0313
218. Cherif B, Triki H, Charfi S, Bouzidi L, Kridis WB, Khanfir A, et al. Immune checkpoint molecules B7-H6 and PD-L1 co-pattern the tumor inflammatory microenvironment in human breast cancer. Sci Rep (2021) 11(1):7550. doi: 10.1038/s41598-021-87216-9
219. Yuan L, Sun L, Yang S, Chen X, Wang J, Jing H, et al. B7-H6 is a new potential biomarker and therapeutic target of T-lymphoblastic lymphoma. Ann Transl Med (2021) 9(4):328. doi: 10.21037/atm-20-5308
220. Rosental B, Brusilovsky M, Hadad U, Oz D, Appel MY, Afergan F, et al. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J Immunol (2011) 187(11):5693–702. doi: 10.4049/jimmunol.1102267
221. Horton NC, Mathew SO, Mathew PA. Novel interaction between proliferating cell nuclear antigen and HLA I on the surface of tumor cells inhibits NK cell function through NKp44. PloS One (2013) 8(3):e59552. doi: 10.1371/journal.pone.0059552
222. Cantoni C, Bottino C, Vitale M, Pessino A, Augugliaro R, Malaspina A, et al. NKp44, a triggering receptor involved in tumor cell lysis by activated human natural killer cells, is a novel member of the immunoglobulin superfamily. J Exp Med (1999) 189(5):787–96. doi: 10.1084/jem.189.5.787
223. Shemesh A, Brusilovsky M, Hadad U, Teltsh O, Edri A, Rubin E, et al. Survival in acute myeloid leukemia is associated with NKp44 splice variants. Oncotarget (2016) 7(22):32933–45. doi: 10.18632/oncotarget.8782
224. Knaneh J, Hodak E, Fedida-Metula S, Edri A, Eren R, Yoffe Y, et al. mAb14, a monoclonal antibody against cell surface PCNA: A potential tool for sezary syndrome diagnosis and targeted immunotherapy. Cancers (Basel) (2023) 15(17):1–20. doi: 10.3390/cancers15174421
225. Westgaard IH, Berg SF, Vaage JT, Wang LL, Yokoyama WM, Dissen E, et al. Rat NKp46 activates natural killer cell cytotoxicity and is associated with FcepsilonRIgamma and CD3zeta. J Leukoc Biol (2004) 76(6):1200–6. doi: 10.1189/jlb.0903428
226. Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature (2001) 409(6823):1055–60. doi: 10.1038/35059110
227. Sivori S, Pende D, Bottino C, Marcenaro E, Pessino A, Biassoni R, et al. NKp46 is the major triggering receptor involved in the natural cytotoxicity of fresh or cultured human NK cells. Correlation between surface density of NKp46 and natural cytotoxicity against autologous, allogeneic or xenogeneic target cells. Eur J Immunol (1999) 29(5):1656–66. doi: 10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1
228. Glasner A, Levi A, Enk J, Isaacson B, Viukov S, Orlanski S, et al. NKp46 receptor-mediated interferon-gamma production by natural killer cells increases fibronectin 1 to alter tumor architecture and control metastasis. Immunity (2018) 48(2):396–8. doi: 10.1016/j.immuni.2018.01.010
229. Oto J, Le QK, Schafer SD, Kiesel L, Mari-Alexandre J, Gilabert-Estelles J, et al. Role of syndecans in ovarian cancer: new diagnostic and prognostic biomarkers and potential therapeutic targets. Cancers (Basel) (2023) 15(12):1–16. doi: 10.3390/cancers15123125
230. Wu MR, Zhang T, Gacerez AT, Coupet TA, DeMars LR, Sentman CL. B7H6-specific bispecific T cell engagers lead to tumor elimination and host antitumor immunity. J Immunol (2015) 194(11):5305–11. doi: 10.4049/jimmunol.1402517
231. Gauthier L, Morel A, Anceriz N, Rossi B, Blanchard-Alvarez A, Grondin G, et al. Multifunctional natural killer cell engagers targeting NKp46 trigger protective tumor immunity. Cell (2019) 177(7):1701–13 e16. doi: 10.1016/j.cell.2019.04.041
232. Gauthier L, Virone-Oddos A, Beninga J, Rossi B, Nicolazzi C, Amara C, et al. Control of acute myeloid leukemia by a trifunctional NKp46-CD16a-NK cell engager targeting CD123. Nat Biotechnol (2023) 41:1296–306. doi: 10.1038/s41587-022-01626-2
233. Demaria O, Gauthier L, Vetizou M, Blanchard Alvarez A, Vagne C, Habif G, et al. Antitumor immunity induced by antibody-based natural killer cell engager therapeutics armed with not-alpha IL-2 variant. Cell Rep Med (2022) 3(10):100783. doi: 10.1016/j.xcrm.2022.100783
234. Klausz K, Pekar L, Boje AS, Gehlert CL, Krohn S, Gupta T, et al. Multifunctional NK cell-engaging antibodies targeting EGFR and NKp30 elicit efficient tumor cell killing and proinflammatory cytokine release. J Immunol (2022) 209(9):1724–35. doi: 10.4049/jimmunol.2100970
235. Ravetch JV, PeRussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils. Cell type-specific expression of two genes that differ in single nucleotide substitutions. J Exp Med (1989) 170(2):481–97. doi: 10.1084/jem.170.2.481
236. Costello RT, Sivori S, Marcenaro E, Lafage-Pochitaloff M, Mozziconacci MJ, Reviron D, et al. Defective expression and function of natural killer cell-triggering receptors in patients with acute myeloid leukemia. Blood (2002) 99(10):3661–7. doi: 10.1182/blood.V99.10.3661
237. Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol Rev (2006) 214:73–91. doi: 10.1111/j.1600-065X.2006.00457.x
238. Pinto S, Pahl J, Schottelius A, Carter PJ, Koch J. Reimagining antibody-dependent cellular cytotoxicity in cancer: the potential of natural killer cell engagers. Trends Immunol (2022) 43(11):932–46. doi: 10.1016/j.it.2022.09.007
239. Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med (2000) 6(4):443–6. doi: 10.1038/74704
240. Liu XY, Chen YL, Liu GJ, Deng XN, Cui Y, Tan J, et al. Development of a variant of dinutuximab with enhanced antitumor efficacy and reduced induction of neuropathic pain. FEBS Open Bio. (2022) 12(9):1644–56. doi: 10.1002/2211-5463.13464
241. Yanaka S, Yogo R, Yagi H, Onitsuka M, Wakaizumi N, Yamaguchi Y, et al. Negative interference with antibody-dependent cellular cytotoxicity mediated by rituximab from its interactions with human serum proteins. Front Immunol (2023) 14:1090898. doi: 10.3389/fimmu.2023.1090898
242. Romee R, Foley B, Lenvik T, Wang Y, Zhang B, Ankarlo D, et al. NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood (2013) 121(18):3599–608. doi: 10.1182/blood-2012-04-425397
243. Liu H, Saxena A, Sidhu SS, Wu D. Fc engineering for developing therapeutic bispecific antibodies and novel scaffolds. Front Immunol (2017) 8:38. doi: 10.3389/fimmu.2017.00038
244. Veillette A. SLAM-family receptors: immune regulators with or without SAP-family adaptors. Cold Spring Harb Perspect Biol (2010) 2(3):a002469. doi: 10.1101/cshperspect.a002469
245. Chen S, Dong Z. NK cell recognition of hematopoietic cells by SLAM-SAP families. Cell Mol Immunol (2019) 16(5):452–9. doi: 10.1038/s41423-019-0222-4
246. Farhangnia P, Ghomi SM, Mollazadehghomi S, Nickho H, Akbarpour M, Delbandi AA. SLAM-family receptors come of age as a potential molecular target in cancer immunotherapy. Front Immunol (2023) 14:1174138. doi: 10.3389/fimmu.2023.1174138
247. Ishibashi M, Morita R, Tamura H. Immune functions of signaling lymphocytic activation molecule family molecules in multiple myeloma. Cancers (Basel). (2021) 13(2):1–15. doi: 10.3390/cancers13020279
248. Claus M, Urlaub D, Fasbender F, Watzl C. SLAM family receptors in natural killer cells - Mediators of adhesion, activation and inhibition via cis and trans interactions. Clin Immunol (2019) 204:37–42. doi: 10.1016/j.clim.2018.10.011
249. McArdel SL, Terhorst C, Sharpe AH. Roles of CD48 in regulating immunity and tolerance. Clin Immunol (2016) 164:10–20. doi: 10.1016/j.clim.2016.01.008
250. Claus M, Wingert S, Watzl C. Modulation of natural killer cell functions by interactions between 2B4 and CD48 in cis and in trans. Open Biol (2016) 6(5):1–11. doi: 10.1098/rsob.160010
251. Wu N, Zhong MC, Roncagalli R, Perez-Quintero LA, Guo H, Zhang Z, et al. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat Immunol (2016) 17(4):387–96. doi: 10.1038/ni.3369
252. Hosen N, Ichihara H, Mugitani A, Aoyama Y, Fukuda Y, Kishida S, et al. CD48 as a novel molecular target for antibody therapy in multiple myeloma. Br J Haematol (2012) 156(2):213–24. doi: 10.1111/j.1365-2141.2011.08941.x
253. Wu Y, Kuang DM, Pan WD, Wan YL, Lao XM, Wang D, et al. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology (2013) 57(3):1107–16. doi: 10.1002/hep.26192
254. Ashour R, Ri M, Aly SS, Yoshida T, Tachita T, Kanamori T, et al. Expression analysis of two SLAM family receptors, SLAMF2 and SLAMF7, in patients with multiple myeloma. Int J Hematol (2019) 110(1):69–76. doi: 10.1007/s12185-019-02649-3
255. Freed-Pastor WA, Lambert LJ, Ely ZA, Pattada NB, Bhutkar A, Eng G, et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing pancreatic cancer. Cancer Cell (2021) 39(10):1342–60 e14. doi: 10.1016/j.ccell.2021.07.007
256. Ziegler AE, Fittje P, Muller LM, Ahrenstorf AE, Hagemann K, Hagen SH, et al. The co-inhibitory receptor TIGIT regulates NK cell function and is upregulated in human intrahepatic CD56(bright) NK cells. Front Immunol (2023) 14:1117320. doi: 10.3389/fimmu.2023.1117320
257. Yu X, Harden K, Gonzalez LC, Francesco M, Chiang E, Irving B, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol (2009) 10(1):48–57. doi: 10.1038/ni.1674
258. Ren X, Peng M, Xing P, Wei Y, Galbo PM, Corrigan D, et al. Blockade of the immunosuppressive KIR2DL5/PVR pathway elicits potent human NK cell–mediated antitumor immunity. J Clin Invest (2022) 132(22):1–17. doi: 10.1172/JCI163620
259. Deuss FA, Gully BS, Rossjohn J, Berry R. Recognition of nectin-2 by the natural killer cell receptor T cell immunoglobulin and ITIM domain (TIGIT). J Biol Chem (2017) 292(27):11413–22. doi: 10.1074/jbc.M117.786483
260. Liu J, Qian X, Chen Z, Xu X, Gao F, Zhang S, et al. Crystal structure of cell adhesion molecule nectin-2/CD112 and its binding to immune receptor DNAM-1/CD226. J Immunol (2012) 188(11):5511–20. doi: 10.4049/jimmunol.1200324
261. Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat Rev Immunol (2015) 15(4):243–54. doi: 10.1038/nri3799
262. Fionda C, Abruzzese MP, Zingoni A, Soriani A, Ricci B, Molfetta R, et al. Nitric oxide donors increase PVR/CD155 DNAM-1 ligand expression in multiple myeloma cells: role of DNA damage response activation. BMC Cancer. (2015) 15(1):17. doi: 10.1186/s12885-015-1023-5
263. Ardolino M, Zingoni A, Cerboni C, Cecere F, Soriani A, Iannitto ML, et al. DNAM-1 ligand expression on Ag-stimulated T lymphocytes is mediated by ROS-dependent activation of DNA-damage response: relevance for NK–T cell interaction. Blood (2011) 117(18):4778–86. doi: 10.1182/blood-2010-08-300954
264. Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, et al. ATM-ATR–dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood (2009) 113(15):3503–11. doi: 10.1182/blood-2008-08-173914
265. Miao X, Yang Z, Xiong L, Zou Q, Yuan Y, Li J, et al. Nectin-2 and DDX3 are biomarkers for metastasis and poor prognosis of squamous cell/adenosquamous carcinomas and adenocarcinoma of gallbladder. Int J Clin Exp Pathol (2013) 6(2):179–90.
266. Karabulut M, Gunaldi M, Alis H, Afsar CU, Karabulut S, Serilmez M, et al. Serum nectin-2 levels are diagnostic and prognostic in patients with colorectal carcinoma. Clin Trans Oncol (2016) 18(2):160–71. doi: 10.1007/s12094-015-1348-1
267. Bekes I, Löb S, Holzheu I, Janni W, Baumann L, Wöckel A, et al. Nectin-2 in ovarian cancer: How is it expressed and what might be its functional role? Cancer Sci (2019) 110(6):1872–82. doi: 10.1111/cas.13992
268. Liang S, Yang Z, Li D, Miao X, Yang L, Zou Q, et al. The clinical and pathological significance of nectin-2 and DDX3 expression in pancreatic ductal adenocarcinomas. Dis Markers. (2015) 2015:1–8. doi: 10.1155/2015/379568
269. Zhang D, Liu J, Zheng M, Meng C, Liao J. Prognostic and clinicopathological significance of CD155 expression in cancer patients: a meta-analysis. World J Surg Oncol (2022) 20(1):1–13. doi: 10.1186/s12957-022-02813-w
270. Li Y-C, Zhou Q, Song Q-K, Wang R-B, Lyu S, Guan X, et al. Overexpression of an immune checkpoint (CD155) in breast cancer associated with prognostic significance and exhausted tumor-infiltrating lymphocytes: A cohort study. J Immunol Res (2020) 2020:1–9. doi: 10.1155/2020/3948928
271. Wang J-B, Li P, Liu X-L, Zheng Q-L, Ma Y-B, Zhao Y-J, et al. An immune checkpoint score system for prognostic evaluation and adjuvant chemotherapy selection in gastric cancer. Nat Commun (2020) 11(1):1–14. doi: 10.1038/s41467-020-20260-7
272. Oyama R, Kanayama M, Mori M, Matsumiya H, Taira A, Shinohara S, et al. CD155 expression and its clinical significance in non−small cell lung cancer. Oncol Lett (2022) 23(5):1–8. doi: 10.3892/ol.2022.13286
273. Bevelacqua V, Bevelacqua Y, Candido S, Skarmoutsou E, Amoroso A, Guarneri C, et al. Nectin like-5 overexpression correlates with the Malignant phenotype in cutaneous melanoma. Oncotarget (2012) 3(8):882–92. doi: 10.18632/oncotarget.594
274. Murakami D, Matsuda K, Iwamoto H, Mitani Y, Mizumoto Y, Nakamura Y, et al. Prognostic value of CD155/TIGIT expression in patients with colorectal cancer. PloS One (2022) 17(3):e0265908. doi: 10.1371/journal.pone.0265908
275. Atsumi S, Matsumine A, Toyoda H, Niimi R, Iino T, Sudo A. Prognostic significance of CD155 mRNA expression in soft tissue sarcomas. Oncol Letters. (2013) 5(6):1771–6. doi: 10.3892/ol.2013.1280
276. Zhang D, Liu Y, Ma J, Xu Z, Duan C, Wang Y, et al. Competitive binding of CD226/TIGIT with poliovirus receptor regulates macrophage polarization and is involved in vascularized skin graft rejection. Am J Transplant. (2023) 23(7):920–34. doi: 10.1016/j.ajt.2023.04.007
277. Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell (2014) 26(6):923–37. doi: 10.1016/j.ccell.2014.10.018
278. Wang F, Liu S, Liu F, Xu T, Ma J, Liang J, et al. TIGIT immune checkpoint blockade enhances immunity of human peripheral blood NK cells against castration-resistant prostate cancer. Cancer Lett (2023) 568:216300. doi: 10.1016/j.canlet.2023.216300
279. Chan CJ, Martinet L, Gilfillan S, Souza-Fonseca-Guimaraes F, Chow MT, Town L, et al. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol (2014) 15(5):431–8. doi: 10.1038/ni.2850
280. Blake SJ, Stannard K, Liu J, Allen S, Yong MC, Mittal D, et al. Suppression of metastases using a new lymphocyte checkpoint target for cancer immunotherapy. Cancer Discovery (2016) 6(4):446–59. doi: 10.1158/2159-8290.CD-15-0944
281. Okumura G, Iguchi-Manaka A, Murata R, Yamashita-Kanemaru Y, Shibuya A, Shibuya K. Tumor-derived soluble CD155 inhibits DNAM-1–mediated antitumor activity of natural killer cells. J Exp Med (2020) 217(4):1–9. doi: 10.1084/jem.20191290
282. Iguchi-Manaka A, Okumura G, Kojima H, Cho Y, Hirochika R, Bando H, et al. Increased soluble CD155 in the serum of cancer patients. PloS One (2016) 11(4):e0152982. doi: 10.1371/journal.pone.0152982
283. Iguchi-Manaka A, Okumura G, Ichioka E, Kiyomatsu H, Ikeda T, Bando H, et al. High expression of soluble CD155 in estrogen receptor-negative breast cancer. Breast Cancer. (2020) 27(1):92–9. doi: 10.1007/s12282-019-00999-8
284. Jin AL, Yang YH, Su X, Yang WJ, Liu T, Chen W, et al. High serum soluble CD155 level predicts poor prognosis and correlates with an immunosuppressive tumor microenvironment in hepatocellular carcinoma. J Clin Lab Anal (2022) 36(3):1–11. doi: 10.1002/jcla.24259
285. Pesce S, Tabellini G, Cantoni C, Patrizi O, Coltrini D, Rampinelli F, et al. B7-H6-mediated downregulation of NKp30 in NK cells contributes to ovarian carcinoma immune escape. OncoImmunology (2015) 4(4):e1001224. doi: 10.1080/2162402X.2014.1001224
286. Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature (2002) 419(6908):734–8. doi: 10.1038/nature01112
287. ChashChina A, Marklin M, Hinterleitner C, Salih HR, Heitmann JS, Klimovich B. DNAM-1/CD226 is functionally expressed on acute myeloid leukemia (AML) cells and is associated with favorable prognosis. Sci Rep (2021) 11(1):18012. doi: 10.1038/s41598-021-97400-6
288. Liu G, Zhang Q, Yang J, Li X, Xian L, Li W, et al. Increased TIGIT expressing NK cells with dysfunctional phenotype in AML patients correlated with poor prognosis. Cancer Immunol Immunother. (2022) 71(2):277–87. doi: 10.1007/s00262-021-02978-5
289. Xu C, Fang H, Gu Y, Yu K, Wang J, Lin C, et al. Impact of intratumoural CD96 expression on clinical outcome and therapeutic benefit in gastric cancer. Cancer Sci (2022) 113(12):4070–81. doi: 10.1111/cas.15537
290. Cho BC, Abreu DR, Hussein M, Cobo M, Patel AJ, Secen N, et al. Tiragolumab plus atezolizumab versus placebo plus atezolizumab as a first-line treatment for PD-L1-selected non-small-cell lung cancer (CITYSCAPE): primary and follow-up analyses of a randomised, double-blind, phase 2 study. Lancet Oncol (2022) 23(6):781–92. doi: 10.1016/S1470-2045(22)00226-1
291. Heusschen R, Griffioen AW, Thijssen VL. Galectin-9 in tumor biology: a jack of multiple trades. Biochim Biophys Acta (2013) 1836(1):177–85. doi: 10.1016/j.bbcan.2013.04.006
292. Chabot S, Kashio Y, Seki M, Shirato Y, Nakamura K, Nishi N, et al. Regulation of galectin-9 expression and release in Jurkat T cell line cells. Glycobiology (2002) 12(2):111–8. doi: 10.1093/glycob/12.2.111
293. John S, Mishra R. Galectin-9: From cell biology to complex disease dynamics. J Biosciences. (2016) 41(3):507–34. doi: 10.1007/s12038-016-9616-y
294. Mansour AA, Raucci F, Sevim M, Saviano A, Begum J, Zhi Z, et al. Galectin-9 supports primary T cell transendothelial migration in a glycan and integrin dependent manner. BioMed Pharmacother. (2022) 151:113171. doi: 10.1016/j.biopha.2022.113171
295. Iqbal AJ, Krautter F, Blacksell IA, Wright RD, Austin-Williams SN, Voisin MB, et al. Galectin-9 mediates neutrophil capture and adhesion in a CD44 and β2 integrin-dependent manner. FASEB J (2022) 36(1):1–18. doi: 10.1096/fj.202100832R
296. Gleason MK, Lenvik TR, McCullar V, Felices M, O’Brien MS, Cooley SA, et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood (2012) 119(13):3064–72. doi: 10.1182/blood-2011-06-360321
297. Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol (2005) 6(12):1245–52. doi: 10.1038/ni1271
298. Tang D, Lotze MT. Tumor immunity times out: TIM-3 and HMGB1. Nat Immunol (2012) 13(9):808–10. doi: 10.1038/ni.2396
299. Jeon SH, Kang M, Jeon M, Chung Y, Kim AR, Lee YJ, et al. CEACAM1 marks highly suppressive intratumoral regulatory T cells for targeted depletion therapy. Clin Cancer Res (2023) 29(9):1794–806. doi: 10.1158/1078-0432.CCR-22-1843
300. Yu S, Ren X, Meng F, Guo X, Tao J, Zhang W, et al. TIM3/CEACAM1 pathway involves in myeloid-derived suppressor cells induced CD8(+) T cells exhaustion and bone marrow inflammatory microenvironment in myelodysplastic syndrome. Immunology (2023) 168(2):273–89. doi: 10.1111/imm.13488
301. Xu J, Liu B, Ma S, Zhang J, Ji Y, Xu L, et al. Characterizing the tumor suppressor role of CEACAM1 in multiple myeloma. Cell Physiol Biochem (2018) 45(4):1631–40. doi: 10.1159/000487730
302. Azimi A, Patrick E, Teh R, Kim J, Fernandez-Penas P. Proteomic profiling of cutaneous melanoma explains the aggressiveness of distant organ metastasis. Exp Dermatol (2023) 32(7):1072–84. doi: 10.1111/exd.14814
303. Abou-Hamad J, Hodgins JJ, de Souza CT, Garland B, Labreche C, Marotel M, et al. CEACAM1 is a direct SOX10 target and inhibits melanoma immune infiltration and stemness. iScience (2022) 25(12):105524. doi: 10.1016/j.isci.2022.105524
304. Appelt U, Sheriff A, Gaipl US, Kalden JR, Voll RE, Herrmann M. Viable, apoptotic and necrotic monocytes expose phosphatidylserine: cooperative binding of the ligand Annexin V to dying but not viable cells and implications for PS-dependent clearance. Cell Death Differ (2005) 12(2):194–6. doi: 10.1038/sj.cdd.4401527
305. Kay JG, Koivusalo M, Ma X, Wohland T, Grinstein S. Phosphatidylserine dynamics in cellular membranes. Mol Biol Cell (2012) 23(11):2198–212. doi: 10.1091/mbc.e11-11-0936
306. Wang W, Wu S, Cen Z, Zhang Y, Chen Y, Huang Y, et al. Mobilizing phospholipids on tumor plasma membrane implicates phosphatidylserine externalization blockade for cancer immunotherapy. Cell Rep (2022) 41(5):111582. doi: 10.1016/j.celrep.2022.111582
307. Kelleher RJ Jr., Balu-Iyer S, Loyall J, Sacca AJ, Shenoy GN, Peng P, et al. Extracellular vesicles present in human ovarian tumor microenvironments induce a phosphatidylserine-dependent arrest in the T-cell signaling cascade. Cancer Immunol Res (2015) 3(11):1269–78. doi: 10.1158/2326-6066.CIR-15-0086
308. Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol (2015) 15(4):203–16. doi: 10.1038/nri3818
309. Yi R, Hong S, Zhang Y, Lin A, Ying H, Zou W, et al. MHC-II signature correlates with anti-tumor immunity and predicts anti-PD-L1 response of bladder cancer. Front Cell Dev Biol (2022) 10:757137. doi: 10.3389/fcell.2022.757137
310. Zhang Y, Feng Z, Xu Y, Jiang S, Zhang Q, Zhang Z, et al. Novel roles of LSECtin in gastric cancer cell adhesion, migration, invasion, and lymphatic metastasis. Cell Death Dis (2022) 13(7):593. doi: 10.1038/s41419-022-05026-x
311. Zhang Y, Zheng J. Functions of immune checkpoint molecules beyond immune evasion. In: Advances in Experimental Medicine and Biology. New York: Springer Singapore (2020). p. 201–26.
312. Lu C, Paschall AV, Shi H, Savage N, Waller JL, Sabbatini ME, et al. The MLL1-H3K4me3 axis-mediated PD-L1 expression and pancreatic cancer immune evasion. J Natl Cancer Inst (2017) 109(6):1–12. doi: 10.1093/jnci/djw283
313. Lamano JB, Lamano JB, Li YD, Didomenico JD, Choy W, Veliceasa D, et al. Glioblastoma-derived IL6 induces immunosuppressive peripheral myeloid cell PD-L1 and promotes tumor growth. Clin Cancer Res (2019) 25(12):3643–57. doi: 10.1158/1078-0432.CCR-18-2402
314. Wang X, Yang L, Huang F, Zhang Q, Liu S, Ma L, et al. Inflammatory cytokines IL-17 and TNF-α up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol Letters. (2017) 184:7–14. doi: 10.1016/j.imlet.2017.02.006
315. Roemer MGM, Advani RH, Ligon AH, Natkunam Y, Redd RA, Homer H, et al. PD-L1 and PD-L2 genetic alterations define classical hodgkin lymphoma and predict outcome. J Clin Oncol (2016) 34(23):2690–7. doi: 10.1200/JCO.2016.66.4482
316. George J, Saito M, Tsuta K, Iwakawa R, Shiraishi K, Scheel AH, et al. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin Cancer Res (2017) 23(5):1220–6. doi: 10.1158/1078-0432.CCR-16-1069
317. Yu J, Wang X, Teng F, Kong L. PD-L1 expression in human cancers and its association with clinical outcomes. OncoTargets Ther (2016) 9:5023–39. doi: 10.2147/OTT.S105862
318. Yi M, Zheng X, Niu M, Zhu S, Ge H, Wu K. Combination strategies with PD-1/PD-L1 blockade: current advances and future directions. Mol Cancer. (2022) 21(1):1–27. doi: 10.1186/s12943-021-01489-2
319. Zhang X, Wu T, Cai X, Dong J, Xia C, Zhou Y, et al. Neoadjuvant immunotherapy for MSI-H/dMMR locally advanced colorectal cancer: new strategies and unveiled opportunities. Front Immunol (2022) 13:795972. doi: 10.3389/fimmu.2022.795972
320. Hathcock KS, Laszlo G, Pucillo C, Linsley P, Hodes RJ. Comparative analysis of B7-1 and B7-2 costimulatory ligands: expression and function. J Exp Med (1994) 180(2):631–40. doi: 10.1084/jem.180.2.631
321. Trzupek D, Dunstan M, Cutler AJ, Lee M, Godfrey L, Jarvis L, et al. Discovery of CD80 and CD86 as recent activation markers on regulatory T cells by protein-RNA single-cell analysis. Genome Med (2020) 12(1):55. doi: 10.1186/s13073-020-00756-z
322. Yang J, Xiang J, Zhu M, Zhao Y, Zhou L, Hu B, et al. Down-regulation of HLA-B-associated transcript 3 impairs the tumoricidal effect of natural killer cells through promoting the T cell immunoglobulin and mucin domain-containing-3 signaling in a mouse head and neck squamous cell carcinoma model. Immunobiology (2022) 227(1):152127. doi: 10.1016/j.imbio.2021.152127
323. Lee J, Su EW, Zhu C, Hainline S, Phuah J, Moroco JA, et al. Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol Cell Biol (2011) 31(19):3963–74. doi: 10.1128/MCB.05297-11
324. Ndhlovu LC, Lopez-Verges S, Barbour JD, Jones RB, Jha AR, Long BR, et al. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood (2012) 119(16):3734–43. doi: 10.1182/blood-2011-11-392951
325. da Silva IP, Gallois A, Jimenez-Baranda S, Khan S, Anderson AC, Kuchroo VK, et al. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol Res (2014) 2(5):410–22. doi: 10.1158/2326-6066.CIR-13-0171
326. Hu XH, Tang MX, Mor G, Liao AH. Tim-3: Expression on immune cells and roles at the maternal-fetal interface. J Reprod Immunol (2016) 118:92–9. doi: 10.1016/j.jri.2016.10.113
327. Ju Y, Hou N, Meng J, Wang X, Zhang X, Zhao D, et al. T cell immunoglobulin- and mucin-domain-containing molecule-3 (Tim-3) mediates natural killer cell suppression in chronic hepatitis B. J Hepatology. (2010) 52:322–9. doi: 10.1016/j.jhep.2009.12.005
328. Rangachari M, Zhu C, Sakuishi K, Xiao S, Karman J, Chen A, et al. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3-mediated cell death and exhaustion. Nat Med (2012) 18(9):1394–400. doi: 10.1038/nm.2871
329. Komita H, Koido S, Hayashi K, Kan S, Ito M, Kamata Y, et al. Expression of immune checkpoint molecules of T cell immunoglobulin and mucin protein 3/galectin-9 for NK cell suppression in human gastrointestinal stromal tumors. Oncol Rep (2015) 34(4):2099–105. doi: 10.3892/or.2015.4149
330. Kikushige Y, Miyamoto T, Yuda J, Jabbarzadeh-Tabrizi S, Shima T, Takayanagi S-I, et al. A TIM-3/gal-9 autocrine stimulatory loop drives self-renewal of human myeloid leukemia stem cells and leukemic progression. Cell Stem Cell (2015) 17(3):341–52. doi: 10.1016/j.stem.2015.07.011
331. Jiao J, Jiao D, Yang F, Zhang J, Li Y, Han D, et al. Galectin-9 expression predicts poor prognosis in hepatitis B virus-associated hepatocellular carcinoma. Aging (2022) 14(4):1879–90. doi: 10.18632/aging.203909
332. Jikuya R, Kishida T, Sakaguchi M, Yokose T, Yasui M, Hashizume A, et al. Galectin-9 expression as a poor prognostic factor in patients with renal cell carcinoma. Cancer Immunology Immunother (2020) 69(10):2041–51. doi: 10.1007/s00262-020-02608-6
333. Okoye I, Xu L, Motamedi M, Parashar P, Walker JW, Elahi S. Galectin-9 expression defines exhausted T cells and impaired cytotoxic NK cells in patients with virus-associated solid tumors. J ImmunoTherapy Cancer. (2020) 8(2):e001849. doi: 10.1136/jitc-2020-001849
334. Liang M, Ueno M, Oomizu S, Arikawa T, Shinonaga R, Zhang S, et al. Galectin-9 expression links to Malignant potential of cervical squamous cell carcinoma. J Cancer Res Clin Oncol (2008) 134(8):899–907. doi: 10.1007/s00432-008-0352-z
335. Kuroda J, Yamamoto M, Nagoshi H, Kobayashi T, Sasaki N, Shimura Y, et al. Targeting activating transcription factor 3 by Galectin-9 induces apoptosis and overcomes various types of treatment resistance in chronic myelogenous leukemia. Mol Cancer Res (2010) 8(7):994–1001. doi: 10.1158/1541-7786.MCR-10-0040
336. Wiersma VR, De Bruyn M, Van Ginkel RJ, Sigar E, Hirashima M, Niki T, et al. The glycan-binding protein galectin-9 has direct apoptotic activity toward melanoma cells. J Invest Dermatol (2012) 132(9):2302–5. doi: 10.1038/jid.2012.133
337. Jiang J, Jin M-S, Kong F, Cao D, Ma H-X, Jia Z, et al. Decreased galectin-9 and increased tim-3 expression are related to poor prognosis in gastric cancer. PloS One (2013) 8(12):e81799. doi: 10.1371/journal.pone.0081799
338. Yoshikawa K, Ishida M, Yanai H, Tsuta K, Sekimoto M, Sugie T. Prognostic significance of the expression levels of T−cell immunoglobulin mucin−3 and its ligand galectin−9 for relapse−free survival in triple−negative breast cancer. Oncol Letters. (2022) 23(6):1–10. doi: 10.3892/ol.2022.13318
339. Xu F, Liu J, Liu D, Liu B, Wang M, Hu Z, et al. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res (2014) 74(13):3418–28. doi: 10.1158/0008-5472.CAN-13-2690
340. Wang J, Sanmamed MF, Datar I, Su TT, Ji L, Sun J, et al. Fibrinogen-like protein 1 is a major immune inhibitory ligand of LAG-3. Cell (2019) 176(1-2):334–47 e12. doi: 10.1016/j.cell.2018.11.010
341. Mao X, Ou MT, Karuppagounder SS, Kam TI, Yin X, Xiong Y, et al. Pathological alpha-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science (2016) 353(6307):1–13. doi: 10.1126/science.aah3374
342. Baixeras E, Huard B, Miossec C, Jitsukawa S, Martin M, Hercend T, et al. Characterization of the lymphocyte activation gene 3-encoded protein. A new ligand for human leukocyte antigen class II antigens. J Exp Med (1992) 176(2):327–37. doi: 10.1084/jem.176.2.327
343. Kouo T, Huang L, Pucsek AB, Cao M, Solt S, Armstrong T, et al. Galectin-3 shapes antitumor immune responses by suppressing CD8+ T cells via LAG-3 and inhibiting expansion of plasmacytoid dendritic cells. Cancer Immunol Res (2015) 3(4):412–23. doi: 10.1158/2326-6066.CIR-14-0150
344. Demeure CE, Wolfers J, Martiin-Garcia N, Gaulard P, Triebel F. T Lymphocytes infiltrating various tumour types express the MHC class II ligand lymphocyte activation gene-3 (LAG-3): role of LAG-3/MHC class II interactions in cell–cell contacts. Eur J Cancer. (2001) 37:1709–18. doi: 10.1016/S0959-8049(01)00184-8
345. Merino A, Zhang B, Dougherty P, Luo X, Wang J, Blazar BR, et al. Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J Clin Invest (2019) 129(9):3770–85. doi: 10.1172/JCI125916
346. Maruhashi T, Okazaki I-M, Sugiura D, Takahashi S, Maeda TK, Shimizu K, et al. LAG-3 inhibits the activation of CD4+ T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat Immunol (2018) 19(12):1415–26. doi: 10.1038/s41590-018-0217-9
347. Khan M, Arooj S, Wang H. NK cell-based immune checkpoint inhibition. Front Immunol (2020) 11:167. doi: 10.3389/fimmu.2020.00167
348. Huuhtanen J, Kasanen H, Peltola K, Lonnberg T, Glumoff V, Bruck O, et al. Single-cell characterization of anti-LAG-3 and anti-PD-1 combination treatment in patients with melanoma. J Clin Invest (2023) 133(6):1–18. doi: 10.1172/JCI164809
349. Park IA, Hwang S-H, Song IH, Heo S-H, Kim Y-A, Bang WS, et al. Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PloS One (2017) 12(8):e0182786. doi: 10.1371/journal.pone.0182786
350. He Y, Rozeboom L, Rivard CJ, Ellison K, Dziadziuszko R, Yu H, et al. MHC class II expression in lung cancer. Lung Cancer. (2017) 112:75–80. doi: 10.1016/j.lungcan.2017.07.030
351. Deng W-W, Mao L, Yu G-T, Bu L-L, Ma S-R, Liu B, et al. LAG-3 confers poor prognosis and its blockade reshapes antitumor response in head and neck squamous cell carcinoma. OncoImmunology (2016) 5(11):e1239005. doi: 10.1080/2162402X.2016.1239005
352. Marcq E, Waele JD, Audenaerde JV, Lion E, Santermans E, Hens N, et al. Abundant expression of TIM-3, LAG-3, PD-1 and PD-L1 as immunotherapy checkpoint targets in effusions of mesothelioma patients. Oncotarget (2017) 8(52):89722–35. doi: 10.18632/oncotarget.21113
353. Yang Z-Z, Kim HJ, Villasboas JC, Chen Y-P, Price-Troska T, Jalali S, et al. Expression of LAG-3 defines exhaustion of intratumoral PD-1+ T cells and correlates with poor outcome in follicular lymphoma. Oncotarget (2017) 8(37):61425–39. doi: 10.18632/oncotarget.18251
354. Miyazaki T, Dierich A, Benoist C, Mathis D. Independent modes of natural killing distinguished in mice lacking Lag3. Science (1996) 272(5260):405–8. doi: 10.1126/science.272.5260.405
355. Esen F, Deniz G, Aktas EC. PD-1, CTLA-4, LAG-3, and TIGIT: The roles of immune checkpoint receptors on the regulation of human NK cell phenotype and functions. Immunol Letters. (2021) 240:15–23. doi: 10.1016/j.imlet.2021.09.009
356. Hasim MS, Marotel M, Hodgins JJ, Vulpis E, Makinson OJ, Asif S, et al. When killers become thieves: Trogocytosed PD-1 inhibits NK cells in cancer. Sci Adv (2022) 8(15):1–15. doi: 10.1126/sciadv.abj3286
357. Makowska A, Meier S, Shen L, Busson P, Baloche V, Kontny U. Anti-PD-1 antibody increases NK cell cytotoxicity towards nasopharyngeal carcinoma cells in the context of chemotherapy-induced upregulation of PD-1 and PD-L1. Cancer Immunology Immunother (2021) 70(2):323–36. doi: 10.1007/s00262-020-02681-x
358. Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault MC, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. (2018) 128(10):4654–68. doi: 10.1172/JCI99317
359. Judge SJ, Dunai C, Aguilar EG, Vick SC, Sturgill IR, Khuat LT, et al. Minimal PD-1 expression in mouse and human NK cells under diverse conditions. J Clin Invest. (2020) 130(6):3051–68. doi: 10.1172/JCI133353
360. Pesce S, Greppi M, Tabellini G, Rampinelli F, Parolini S, Olive D, et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J Allergy Clin Immunol (2017) 139(1):335–46 e3. doi: 10.1016/j.jaci.2016.04.025
361. Pesini C, Hidalgo S, Arias MA, Santiago L, Calvo C, Ocariz-Diez M, et al. PD-1 is expressed in cytotoxic granules of NK cells and rapidly mobilized to the cell membrane following recognition of tumor cells. Oncoimmunology (2022) 11(1):2096359. doi: 10.1080/2162402X.2022.2096359
362. Concha-Benavente F, Kansy B, Moskovitz J, Moy J, Chandran U, Ferris RL. PD-L1 mediates dysfunction in activated PD-1(+) NK cells in head and neck cancer patients. Cancer Immunol Res (2018) 6(12):1548–60. doi: 10.1158/2326-6066.CIR-18-0062
363. Liu Y, Cheng Y, Xu Y, Wang Z, Du X, Li C, et al. Increased expression of programmed cell death protein 1 on NK cells inhibits NK-cell-mediated anti-tumor function and indicates poor prognosis in digestive cancers. Oncogene (2017) 36(44):6143–53. doi: 10.1038/onc.2017.209
364. Dong W, Wu X, Ma S, Wang Y, Nalin AP, Zhu Z, et al. The mechanism of anti-PD-L1 antibody efficacy against PD-L1-negative tumors identifies NK cells expressing PD-L1 as a cytolytic effector. Cancer Discovery (2019) 9(10):1422–37. doi: 10.1158/2159-8290.CD-18-1259
365. Mathieu L, Shah S, Pai-Scherf L, Larkins E, Vallejo J, Li X, et al. FDA approval summary: atezolizumab and durvalumab in combination with platinum-based chemotherapy in extensive stage small cell lung cancer. Oncologist (2021) 26(5):433–8. doi: 10.1002/onco.13752
366. Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med (2017) 377(20):1919–29. doi: 10.1056/NEJMoa1709937
367. Sonpavde GP, Sternberg CN, Loriot Y, Marabelle A, Lee JL, Flechon A, et al. Primary results of STRONG: An open-label, multicenter, phase 3b study of fixed-dose durvalumab monotherapy in previously treated patients with urinary tract carcinoma. Eur J Cancer. (2022) 163:55–65. doi: 10.1016/j.ejca.2021.12.012
368. Emens LA, Adams S, Barrios CH, Dieras V, Iwata H, Loi S, et al. Corrigendum to ‘First-line atezolizumab plus nab-paclitaxel for unresectable locally advanced or metastatic triple-negative breast cancer: IMpassion130 final overall survival analysis’: Annals of Oncology 2021; volume 32: 983-993. Ann Oncol (2021) 32(10):1308. doi: 10.1016/j.annonc.2021.10.002
369. Gordan JD, Kennedy EB, Abou-Alfa GK, Beg MS, Brower ST, Gade TP, et al. Systemic therapy for advanced hepatocellular carcinoma: ASCO guideline. J Clin Oncol (2020) 38(36):4317–45. doi: 10.1200/JCO.20.02672
370. Alegre ML, Shiels H, Thompson CB, Gajewski TF. Expression and function of CTLA-4 in Th1 and Th2 cells. J Immunol (1998) 161(7):3347–56. doi: 10.4049/jimmunol.161.7.3347
371. Rudd CE, Taylor A, Schneider H. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol Rev (2009) 229(1):12–26. doi: 10.1111/j.1600-065X.2009.00770.x
372. Komata T, Tanaka R, Yamamoto K, Oda T, Ono K, Yoshida S, et al. B7-1(CD80)-transfected human glioma cells and interleukin-12 directly stimulate allogeneic CD8+ T cells. J Immunother. (1997) 20(4):256–64. doi: 10.1097/00002371-199707000-00002
373. Stojanovic A, Fiegler N, Brunner-Weinzierl M, Cerwenka A. CTLA-4 is expressed by activated mouse NK cells and inhibits NK Cell IFN-gamma production in response to mature dendritic cells. J Immunol (2014) 192(9):4184–91. doi: 10.4049/jimmunol.1302091
374. Lougaris V, Tabellini G, Baronio M, Patrizi O, Gazzurelli L, Mitsuiki N, et al. CTLA-4 regulates human Natural Killer cell effector functions. Clin Immunol (2018) 194:43–5. doi: 10.1016/j.clim.2018.06.010
375. Cikman DI, Esen F, Engin A, Turna A, Agkoc M, Yilmaz A, et al. Mediastinal lymph node removal modulates natural killer cell exhaustion in patients with non-small cell lung cancer. Immunol Res (2023) 1–13. doi: 10.1007/s12026-023-09410-3
376. Mollavelioglu B, Cetin Aktas E, Cabioglu N, Abbasov A, Onder S, Emiroglu S, et al. High co-expression of immune checkpoint receptors PD-1, CTLA-4, LAG-3, TIM-3, and TIGIT on tumor-infiltrating lymphocytes in early-stage breast cancer. World J Surg Oncol (2022) 20(1):349. doi: 10.1186/s12957-022-02810-z
377. Davis-Marcisak EF, Fitzgerald AA, Kessler MD, Danilova L, Jaffee EM, Zaidi N, et al. Transfer learning between preclinical models and human tumors identifies a conserved NK cell activation signature in anti-CTLA-4 responsive tumors. Genome Med (2021) 13(1):129. doi: 10.1186/s13073-021-00944-5
378. Sanseviero E, O’Brien EM, Karras JR, Shabaneh TB, Aksoy BA, Xu W, et al. Anti-CTLA-4 activates intratumoral NK cells and combined with IL15/IL15Ralpha complexes enhances tumor control. Cancer Immunol Res (2019) 7(8):1371–80. doi: 10.1158/2326-6066.CIR-18-0386
379. Arce Vargas F, Furness AJS, Litchfield K, Joshi K, Rosenthal R, Ghorani E, et al. Fc effector function contributes to the activity of human anti-CTLA-4 antibodies. Cancer Cell (2018) 33(4):649–63 e4. doi: 10.1016/j.ccell.2018.02.010
380. Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med (2010) 363(8):711–23. doi: 10.1056/NEJMoa1003466
381. Motzer RJ, Escudier B, McDermott DF, Aren Frontera O, Melichar B, Powles T, et al. Survival outcomes and independent response assessment with nivolumab plus ipilimumab versus sunitinib in patients with advanced renal cell carcinoma: 42-month follow-up of a randomized phase 3 clinical trial. J Immunother Cancer (2020) 8(2):1–18. doi: 10.1136/jitc-2020-000891
382. Yau T, Kang YK, Kim TY, El-Khoueiry AB, Santoro A, Sangro B, et al. Efficacy and safety of nivolumab plus ipilimumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib: the checkMate 040 randomized clinical trial. JAMA Oncol (2020) 6(11):e204564. doi: 10.1001/jamaoncol.2020.4564
383. Paz-Ares L, Ciuleanu TE, Cobo M, Schenker M, Zurawski B, Menezes J, et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): an international, randomised, open-label, phase 3 trial. Lancet Oncol (2021) 22(2):198–211. doi: 10.1016/S1470-2045(20)30641-0
384. Kudo M. Durvalumab plus tremelimumab in unresectable hepatocellular carcinoma. Hepatobiliary Surg Nutr (2022) 11(4):592–6. doi: 10.21037/hbsn-22-143
385. Felices M, Lenvik AJ, McElmurry R, Chu S, Hinderlie P, Bendzick L, et al. Continuous treatment with IL-15 exhausts human NK cells via a metabolic defect. JCI Insight (2018) 3(3):1–15. doi: 10.1172/jci.insight.96219
386. Fromm G, de Silva S, Schreiber TH. Reconciling intrinsic properties of activating TNF receptors by native ligands versus synthetic agonists. Front Immunol (2023) 14:1236332. doi: 10.3389/fimmu.2023.1236332
387. Cichocki F, Bjordahl R, Goodridge JP, Mahmood S, Gaidarova S, Abujarour R, et al. Quadruple gene-engineered natural killer cells enable multi-antigen targeting for durable antitumor activity against multiple myeloma. Nat Commun (2022) 13(1):7341. doi: 10.1038/s41467-022-35127-2
388. Pantel K, Alix-Panabieres C. Crucial roles of circulating tumor cells in the metastatic cascade and tumor immune escape: biology and clinical translation. J Immunother Cancer. (2022) 10(12):1–9. doi: 10.1136/jitc-2022-005615
389. Ma F, Vayalil J, Lee G, Wang Y, Peng G. Emerging role of tumor-derived extracellular vesicles in T cell suppression and dysfunction in the tumor microenvironment. J Immunother Cancer. (2021) 9(10):1–15. doi: 10.1136/jitc-2021-003217
390. Haugh A, Daud A. Resistance to immune checkpoint blockade: IFNgamma or MHC-I? Cancer Immunol Res (2023) 11(7):864. doi: 10.1158/2326-6066.CIR-23-0373
391. Leung EYL, Ennis DP, Kennedy PR, Hansell C, Dowson S, Farquharson M, et al. NK cells augment oncolytic adenovirus cytotoxicity in ovarian cancer. Mol Ther Oncolytics. (2020) 16:289–301. doi: 10.1016/j.omto.2020.02.001
392. Dutcher JP, Schwartzentruber DJ, Kaufman HL, Agarwala SS, Tarhini AA, Lowder JN, et al. High dose interleukin-2 (Aldesleukin) - expert consensus on best management practices-2014. J Immunother Cancer. (2014) 2(1):26. doi: 10.1186/s40425-014-0026-0
393. Siegel JP, Puri RK. Interleukin-2 toxicity. J Clin Oncol (1991) 9(4):694–704. doi: 10.1200/JCO.1991.9.4.694
394. Sitrin J, Ring A, Garcia KC, Benoist C, Mathis D. Regulatory T cells control NK cells in an insulitic lesion by depriving them of IL-2. J Exp Med (2013) 210(6):1153–65. doi: 10.1084/jem.20122248
395. Felices M, Chu S, Kodal B, Bendzick L, Ryan C, Lenvik AJ, et al. IL-15 super-agonist (ALT-803) enhances natural killer (NK) cell function against ovarian cancer. Gynecol Oncol (2017) 145(3):453–61. doi: 10.1016/j.ygyno.2017.02.028
396. Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, et al. Cytokine activation induces human memory-like NK cells. Blood (2012) 120(24):4751–60. doi: 10.1182/blood-2012-04-419283
397. Cubitt CC, McClain E, Becker-Hapak M, Foltz JA, Wong P, Wagner JA, et al. A novel fusion protein scaffold 18/12/TxM activates the IL-12, IL-15, and IL-18 receptors to induce human memory-like natural killer cells. Mol Ther Oncolytics. (2022) 24:585–96. doi: 10.1016/j.omto.2022.02.009
398. Kabashima A, Matsuo Y, Ito S, Akiyama Y, Ishii T, Shimada S, et al. cGAS-STING signaling encourages immune cell overcoming of fibroblast barricades in pancreatic cancer. Sci Rep (2022) 12(1):10466. doi: 10.1038/s41598-022-14297-5
399. Hayman TJ, Baro M, MacNeil T, Phoomak C, Aung TN, Cui W, et al. STING enhances cell death through regulation of reactive oxygen species and DNA damage. Nat Commun (2021) 12(1):2327. doi: 10.1038/s41467-021-22572-8
400. Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, Sundararaman SK, et al. Suppression of STING associated with LKB1 loss in KRAS-driven lung cancer. Cancer Discovery (2019) 9(1):34–45. doi: 10.1158/2159-8290.CD-18-0689
401. Berger G, Knelson EH, Jimenez-Macias JL, Nowicki MO, Han S, Panagioti E, et al. STING activation promotes robust immune response and NK cell-mediated tumor regression in glioblastoma models. Proc Natl Acad Sci U S A. (2022) 119(28):e2111003119. doi: 10.1073/pnas.2111003119
402. Knelson EH, Ivanova EV, Tarannum M, Campisi M, Lizotte PH, Booker MA, et al. Activation of tumor-cell STING primes NK-cell therapy. Cancer Immunol Res (2022) 10(8):947–61. doi: 10.1158/2326-6066.CIR-22-0017
403. Wolf NK, Blaj C, Picton LK, Snyder G, Zhang L, Nicolai CJ, et al. Synergy of a STING agonist and an IL-2 superkine in cancer immunotherapy against MHC I-deficient and MHC I(+) tumors. Proc Natl Acad Sci U S A. (2022) 119(22):e2200568119. doi: 10.1073/pnas.2200568119
404. Foltz JA, Hess BT, Bachanova V, Bartlett NL, Berrien-Elliott MM, McClain E, et al. Phase I trial of N-803, an IL15 receptor agonist, with rituximab in patients with indolent non-hodgkin lymphoma. Clin Cancer Res (2021) 27(12):3339–50. doi: 10.1158/1078-0432.CCR-20-4575
405. Chu Y, Nayyar G, Kham Su N, Rosenblum JM, Soon-Shiong P, Lee J, et al. Novel cytokine-antibody fusion protein, N-820, to enhance the functions of ex vivo expanded natural killer cells against Burkitt lymphoma. J Immunother Cancer (2020) 8(2):1–14. doi: 10.1136/jitc-2020-001238
406. Susek KH, Schwietzer YA, Karvouni M, Gilljam M, Keszei M, Hussain A, et al. Generation of NK cells with chimeric-switch receptors to overcome PD1-mediated inhibition in cancer immunotherapy. Cancer Immunol Immunother. (2023) 72(5):1153–67. doi: 10.1007/s00262-022-03317-y
407. Souza-Fonseca-Guimaraes F, Rossi GR, Dagley LF, Foroutan M, McCulloch TR, Yousef J, et al. TGFbeta and CIS inhibition overcomes NK-cell suppression to restore antitumor immunity. Cancer Immunol Res (2022) 10(9):1047–54. doi: 10.1158/2326-6066.CIR-21-1052
408. Bernard PL, Delconte R, Pastor S, Laletin V, Costa Da Silva C, Goubard A, et al. Targeting CISH enhances natural cytotoxicity receptor signaling and reduces NK cell exhaustion to improve solid tumor immunity. J Immunother Cancer (2022) 10(5):1–13. doi: 10.1136/jitc-2021-004244
409. Haroun-Izquierdo A, Vincenti M, Netskar H, van Ooijen H, Zhang B, Bendzick L, et al. Adaptive single-KIR(+)NKG2C(+) NK cells expanded from select superdonors show potent missing-self reactivity and efficiently control HLA-mismatched acute myeloid leukemia. J Immunother Cancer (2022) 10(11):1–14. doi: 10.1136/jitc-2022-005577
410. Cichocki F, van der Stegen SJC, Miller JS. Engineered and banked iPSCs for advanced NK- and T-cell immunotherapies. Blood (2023) 141(8):846–55. doi: 10.1182/blood.2022016205
411. Lin C, Schwarzbach A, Sanz J, Montesinos P, Stiff P, Parikh S, et al. Multicenter long-term follow-up of allogeneic hematopoietic cell transplantation with omidubicel: A pooled analysis of five prospective clinical trials. Transplant Cell Ther (2023) 29(5):338 e1– e6. doi: 10.1016/j.jtct.2023.06.009
412. Zhu H, Blum RH, Bernareggi D, Ask EH, Wu Z, Hoel HJ, et al. Metabolic Reprograming via Deletion of CISH in Human iPSC-Derived NK Cells Promotes In Vivo Persistence and Enhances Anti-tumor Activity. Cell Stem Cell (2020) 27(2):224–37 e6. doi: 10.1016/j.stem.2020.05.008
413. Parameswaran R, Ramakrishnan P, Moreton SA, Xia Z, Hou Y, Lee DA, et al. Repression of GSK3 restores NK cell cytotoxicity in AML patients. Nat Commun (2016) 7:11154. doi: 10.1038/ncomms11154
414. Cichocki F, Valamehr B, Bjordahl R, Zhang B, Rezner B, Rogers P, et al. GSK3 inhibition drives maturation of NK cells and enhances their antitumor activity. Cancer Res (2017) 77(20):5664–75. doi: 10.1158/0008-5472.CAN-17-0799
415. Slattery K, Woods E, Zaiatz-Bittencourt V, Marks S, Chew S, Conroy M, et al. TGFbeta drives NK cell metabolic dysfunction in human metastatic breast cancer. J Immunother Cancer (2021) 9(2):1–14. doi: 10.1136/jitc-2020-002044
416. Trotta R, Dal Col J, Yu J, Ciarlariello D, Thomas B, Zhang X, et al. TGF-beta utilizes SMAD3 to inhibit CD16-mediated IFN-gamma production and antibody-dependent cellular cytotoxicity in human NK cells. J Immunol (2008) 181(6):3784–92. doi: 10.4049/jimmunol.181.6.3784
Keywords: natural killer cells, killer immunoglobulin-like receptors (KIR), signal integration, cancer immunotherapy, innate lymphoid cells
Citation: Nersesian S, Carter EB, Lee SN, Westhaver LP and Boudreau JE (2023) Killer instincts: natural killer cells as multifactorial cancer immunotherapy. Front. Immunol. 14:1269614. doi: 10.3389/fimmu.2023.1269614
Received: 30 July 2023; Accepted: 30 October 2023;
Published: 28 November 2023.
Edited by:
Scott McComb, National Research Council Canada (NRC), CanadaReviewed by:
Kamalakannan Rajasekaran, Genentech Inc., United StatesCopyright © 2023 Nersesian, Carter, Lee, Westhaver and Boudreau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeanette E. Boudreau, SmVhbmV0dGUuYm91ZHJlYXVAZGFsLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.