
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 18 September 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1269409
This article is part of the Research Topic Nanoparticle-Based Delivery Systems on Immunomodulation View all 4 articles
Porcine epidemic diarrhea virus (PEDV) causes acute watery diarrhea and high mortality in newborn piglets. Activation of intestinal mucosal immunity is crucial to anti-PEDV infection. To develop a vaccine capable of stimulating intestinal mucosal immunity, we prepared a bacterium (Lactococcus lactis)-like particle (BLP) vaccine (S1-BLPs) displaying the S1 protein, a domain of PEDV spike protein (S), based on gram-positive enhancer matrix (GEM) particle display technology. We further compared the effects of different vaccination routes on mucosal immune responses in mice induced by S1-BLPs. The specific IgG titer in serum of intramuscularly immunized mice with S1-BLPs was significantly higher than that of the intranasally administered. The specific IgA antibody was found in the serum and intestinal lavage fluid of mice vaccinated intranasally, but not intramuscularly. Moreover, the intranasally inoculated S1-BLPs induced higher levels of IFN-γ and IL-4 in serum than the intramuscularly inoculated. In addition, the ratio of serum IgG2a/IgG1 of mice inoculated intramuscularly was significantly higher with S1-BLPs compared to that of with S1 protein, suggesting that the immune responses induced by S1-BLPs was characterized by helper T (Th) cell type 1 immunity. The results indicated that S1-BLPs induced systemic and local immunity, and the immunization routes significantly affected the specific antibody classes and Th immune response types. The intranasally administered S1-BLPs could effectively stimulate intestinal mucosal specific secretory IgA response. S1-BLPs have the potential to be developed as PEDV mucosal vaccine.
Porcine epidemic diarrhea virus (PEDV), a highly contagious enterovirus, causes acute diarrhea, vomiting, dehydration and death in pigs, causing huge economic losses to the swine industry worldwide (1, 2). PEDV infects pigs of all ages, but neonatal piglets under 7 days old are more susceptible to PEDV infections, with a mortality of up to 100% (2, 3). In addition to direct and indirect fecal-oral routes, it has been confirmed that PEDV can be transmitted to the intestinal epithelium through the respiratory route (4). PEDV belongs to the genus Alphacoronavirus in the family Coronaviridae and is an enveloped single-stranded positive-sense RNA virus, with a genome size of about 28 k (5). Spike glycoprotein (S), composed of 1383-1386 amino acids (aa), is a type I membrane protein consisting of S1 and S2 subunits on the viral surface as a trimer. During viral infection, the N-terminal S1 subunit (1-789 aa) is responsible for receptor binding, and the C-terminal S2 subunit (790-1383 aa) is involved in the fusion of the viral envelope with the host cell membrane (6, 7). The S1 subunit is an important determinant of the virulence of PEDV and a major target of neutralizing antibodies (8–11). Therefore, subunit vaccines based on full-length or truncated S1 protein can effectively elicit protective antibody responses in vivo (12–15).
In general, although PEDV can also cause transient viremia in young piglets, it mainly causes localized intestinal infections. This phenomenon requires new vaccination strategies that focus on the induction of mucosal immunity to protect the intestinal mucosa. Moreover, due to the high susceptibility and immaturity of the immune system in neonatal piglets, passive lactogenic immunity to PEDV is critical for suckling piglets to obtain protection. IgA titers in colostrum are correlated with PEDV-neutralizing antibody titers (16, 17). Therefore, increasing the specific secretory IgA (sIgA) titers in colostrum via maternal immunity is the most effective strategy to protect newborn piglets against PEDV (18, 19). To date, attenuated or inactivated PEDV vaccines have been widely used (20, 21). However, existing vaccines are not so effective that some vaccinated sows or gilts do not develop protective lactogenic immunity for the neonatal. Meanwhile, there are some difficulties in the cultivation of PEDV, resulting in high production costs of attenuated or inactivated PEDV vaccines. Existing vaccines also have potential biosafety risks. Though vaccination via the traditional route such as intramuscular injection is effective in inducing systemic immune responses, it is difficult to elicit antigen-specific mucosal immune responses (22). Additionally, the triggering of immune responses is closely correlated with the nature of the antigen and vaccination routes. Even the same antigen with different vaccination routes causes different immune response types. Therefore, it is of practical significance to explore new vaccines and vaccination routes that can induce mucosal immune responses for the prevention and control of PEDV.
The heterologous display of proteins or peptides on the surface of microorganisms is an emerging and interesting technology with wide applications in various fields. Heat-killed non-recombinant lactic acid bacteria (LAB) or non-viable bacterium-like particles (BLPs) obtained by the pretreatment of whole bacteria in hot acid are designated as Gram-positive enhancer matrix (GEM) particles, which consisted mainly of bacterium-derived peptidoglycan spheres without other intact cell wall components and intracellular components (23). The GEM particles provide a suitable cell surface that can display various heterologous proteins through a peptidoglycan-binding domain, i.e., protein anchor (PA). The PA is derived from the Lactococcus lactis peptidoglycan hydrolase AcmA and contains three LysM motifs consisting of about 45 amino acids separated by spacer sequences, which can specifically bind to GEM particles and enable the display of heterologous proteins on their surface (24). Therefore, the GEM-PA is not only a safe, attractive and affordable antigenic surface display system but also a mucosal vaccine delivery system. BLPs can improve the systemic immune responses and local mucosal immune responses in animals through intranasal, oral and intramuscular injection immunization routes (25–27).
To develop a safe PEDV vaccine that can induce robust immune responses in the intestinal mucosa, S1-PA fusion protein was expressed by Escherichia coli (E. coli) in this study, and S1-BLPs displaying S1 protein was prepared using the strategy described in Figure 1. Mice were immunized with S1-BLPs by either intramuscular injection or intranasal administration, and we compared the differences in specific local mucosal and systemic immune responses induced by S1-BLPs between two immunization routes.
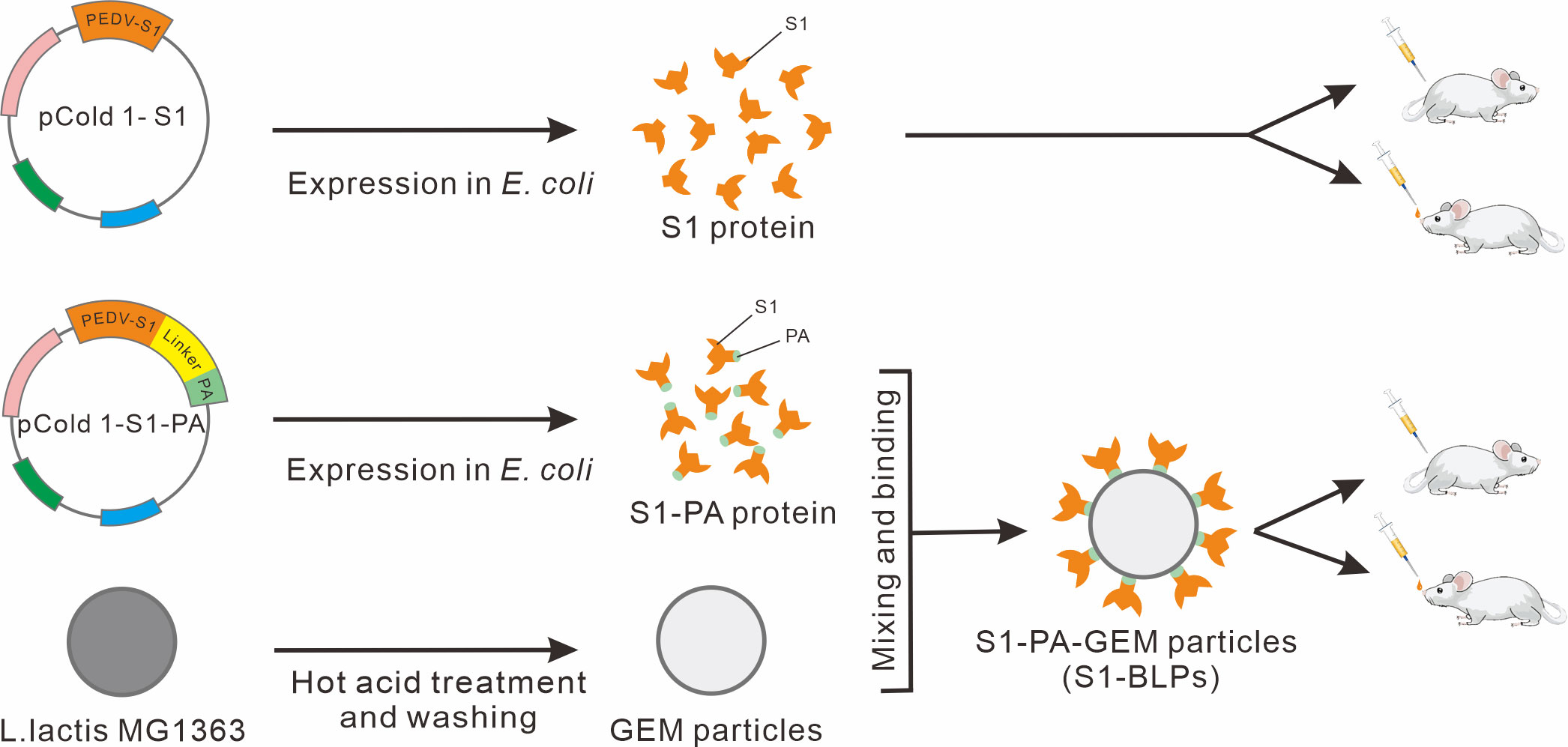
Figure 1 Schematic diagram of S1 protein, S1-PA-GEM particle preparation, and animal immunization. The pCold 1-S1 refers to S1 protein recombinant expression plasmids, and pCold 1-S1-PA refers to expression plasmids of recombinant protein (S1-PA) of S1 protein and the protein anchor (PA), containing a flexible linker sequence between S1 gene and PA gene sequences. The recombinant protein S1-PA and the gram-positive enhancer matrix (GEM) particles were used to prepare S1-BLPs for intramuscular or intranasal immunization of BALB/c mice.
The PEDV S1 gene was amplified from the genome of PEDV QY-2016 strain (GenBank ID: MH244927) preserved in our laboratory by PCR using primers S1-F and S1-R (Table 1). The DNA sequence of the PA gene based on the peptidoglycan hydrolase AcmA (GenBank ID: U17696) was synthesized by Sangon Biotech (Shanghai, China). The S1-PA fusion protein gene S1-PA was obtained using overlap extension PCR (OE-PCR) by splicing the segments of S1 gene and PA gene through a flexible linker (GGSG). All the primers in this study were synthesized by Sangon Biotech (Shanghai, China). To obtain the recombinant expression plasmid pCold1-S1 or pCold1-S1-PA, the gene S1 and the fusion protein gene S1-PA were digested with EcoR I and Sal I, respectively, and then cloned into the pCold 1 vector carrying a 6×his tag (Takara Bio, #3360) also digested with the above restriction endonucleases using T4 ligase. Finally, the single clone carrying S1 or S1-PA gene was verified by PCR and double digestion with EcoR I and Sal I, and then was sequenced by Sangon Biotech (Shanghai).
The recombinant plasmids pCold1-S1 and pCold1-S1-PA were transformed into competent E. coli BL21 (DE3) cells (Tiangen Biotech, #CB105) using the heat shock method. The transformed E. coli cells were inoculated on Luria Broth (Amp+/LB) Agar plates containing 50 μg/mL ampicillin and cultured at 37°C for 18 h. The single colony containing the target gene was selected and inoculated into Amp+/LB broth for culture at 37°C. When the OD600 nm value of the bacterial culture reached 0.6-0.8, the culture was moved to 4°C for 1 h. After adding 0.5 mmol/L isopropyl-β-d-thiogalactoside (IPTG), the bacteria were cultured at 16°C for 24 h to induce the expression of recombinant proteins. The bacterial cells were harvested and re-suspended in PBS (pH 7.4), followed by sonication in an ice bath, the supernatant was collected for SDS-PAGE analysis after centrifugation at 10,000 r/min for 10 min. The expressed proteins were purified using the HisTrap™ HP (GE company, #17524801) on the AKTA protein purification system (GE company, USA), and confirmed by Western blotting. The protein concentration was measured using the BCA protein assay kit (Takara Bio, #T9300A).
SDS-PAGE was used to analyze the expression and purification of S1 and S1-PA proteins, as well as the proteins bound to BLPs. S1 protein, S1-PA and BLPs-bound S1 protein were identified by Western blotting. Briefly, the recombinant protein was transferred to a polyvinylidene difluoride (PVDF) membrane following SDS-PAGE, and blocked overnight at 4°C. The membrane was incubated with anti-His-tagged mouse monoclonal antibody (1:5000; Cowin Biotech, #CW0285) or Rabbit anti-S1 protein polyclonal antibody (1:500; prepared and stored in our lab) at room temperature (RT) for 1 h. After washing, the membrane was incubated with 1:5000 diluted HRP-conjugated goat anti-mouse/-rabbit antibody (Solarbio Tech, #SE131, #SE134) at RT for 1 h. After washing, the membrane was incubated in the DAB chromogenic solution for color development, and the results were observed directly.
Lactococcus (L.) lactis MG1363 was cultured in GM17 broth (Hope Bio, #HB0391) at 30°C for 24 h with shaking. Bacterial cells were harvested by centrifugation at 8,000 r/min for 15 min, washed twice with sterile PBS, re-suspended in 25 mM sulfuric acid, and heated at 100°C for 30 min. The treated L. lactis (GEM) particles were centrifuged, and the pellet was washed three times with PBS. Finally, the GEM particles (GEMs) were re-suspended in PBS at a concentration of 2.5×109 particles/mL, which was referred to as 1 U (28). The GEM particles were directly used for the preparation of S1-BLPs or stored at -20°C.
To prepare S1-BLPs, the GEM particles (2.5×109 particles/mL) were mixed with 6.25 mg of the fusion protein S1-PA followed by incubation at RT for 30 min with shaking. The GEM particles bound to S1 protein via PA, designated S1-BLPs, were collected by centrifugation at 6,000 r/min for 5 min, washed 3 times with PBS, and re-suspended in PBS. To confirm the binding of the fusion protein S1-PA to the GEM particles, S1-BLPs was treated with 10% SDS at 100°C for 10 min to observe whether S1-PA was dissociated from the GEM particles. GEM particles control was set up at the same time. After centrifugation at 10,000 r/min for 2 min, the supernatant was collected and analyzed by SDS-PAGE to confirm the presence of S1-PA. Meanwhile, BCA protein assay kit was used to determine the total S1-PA protein concentration in the supernatant, and using the following formula to calculate the amount of protein bound by GEMs per unit (2.5×109 particles/mL). Amounts of GEMs binding S1-PA protein per unit (μg) = (Total protein amounts in per unit of S1-BLP supernatant) - (Total protein amounts in per unit of GEM supernatant).
The samples were dropped onto the copper grid, negatively stained with 2% phosphotungstic acid, and vacuum dried. The samples were examined on a transmission electron microscope (TEM) (JEM1400, JEOL, Tokyo, Japan), which was operated at 80 kV and equipped with An AMT Camera. The particle sizes were measured using Image J.
S1-BLPs was evenly spread on the polylysine coated slides, air-dried, and blocked with 3% BSA in PBS at RT for 30 min. After washing twice with PBS, the slides were incubated with anti-PEDV S1 protein rabbit polyclonal antibody (1:100) at RT for 60 min. Meanwhile, the anti-S1 protein antibody negative rabbit serum control was set up. After washing 3 times with PBS, the slides were incubated with FITC-labeled goat anti-rabbit IgG (1:200, Solarbio Tech, # SF134) for 60 min at RT in the dark. The slides were washed three times with PBS, and the fluorescence was observed under an Axio Observer D1 fluorescence microscope (ZEISS, Gottingen, Germany).
The S1 protein or S1-BLPs were mixed with an equal volume of Montanide™ IMS1313N VG (IMS1313 for short) water adjuvant (Seppic, Paris, France). Forty specific-pathogen-free (SPF) female BALB/c mice (Changsheng Biotech, Liaoning, China) aged 6-8 weeks were randomly divided into 5 groups (S1/IM; S1-BLPs/IM, S1/IN, S1-BLPs/IN, and Blank group) with 8 mice in each group. Each mouse in S1/IM and S1/IN groups was immunized three times by intramuscular (IM) and intranasal inoculation (IN) with a dose 80 μL contained 40 μg of S1 protein at 2-week intervals, respectively. And each one in BLPs IM and BLPs IN groups was immunized with S1-BLPs containing 40 μg S1 protein by the same routes as above, respectively. In the blank group, mice were not immunization. Blood samples were collected from the tail vein of mice weekly before and after immunization, and serum collection was conducted and stored at -20°C for subsequent tests. At 14 days after the second immunization and 21 days after the third immunization, 4 mice were randomly selected from each group and anesthetized by intraperitoneal injection of 10% chloral hydrate (0.1 mL/10g body weight), and then intestine and lung airway lavage fluid were collected for specific secretory IgA detection.
PEDV S1-specific IgG, IgA, IgG1, IgG2a and IgG2b antibodies in serum were measured by ELISA, and IgG titer was determined on day 21 after the third immunization. Moreover, the ratio of IgG2a/IgG1 in serum of S1/IM and S1-BLPs/IM groups was analyzed. Briefly, 96-well ELISA plates (Biofil, #FEP101896) were coated with PEDV S1 protein (2 μg/well) diluted with Coating buffer (0.1 M carbonate buffer, pH 9.0) at 37°C for 1 h and overnight at 4°C. The plates were blocked with Blocking buffer (5% skim milk powder in PBST) at 37°C for 1 h. After washing 3 times with PBST (0.05% Tween-20 in phosphate-buffered saline (PBS), pH 7.4), 100 μL of samples (serum, or intestine or lung airway lavage fluid), or 4-fold serially diluted (1:100 to 1:204 800) serum were added in the wells of the plate to incubate at 37°C for 60 min. At the same time, positive, negative, and blank controls were set up. After washing 3 times, 100 μL of HRP-conjugated goat anti-mouse IgG (1:15000, Biodragon Tech, #BF03001) or IgA/IgG1/IgG2a/IgG2b antibodies (1:1000, Biodragon Tech, #BF03007, #BF03050, #BF03051, #BF03052) were added at 37°C for 60 min. After washing 3 times, TMB single-component substrate solution (Solarbio Tech, #PR1200) was added, 100 μL/well, to incubate in the dark at RT for 15 min. Finally, 50 μL stop solution was added to each well to terminate the reaction, and the OD450 nm value was measured using a Multimode Microplate reader (Biotek Synergy HTX, USA). Three parallel repeated tests were performed for each sample.
The levels of IFN-γ, IL-2 and IL-4 in serum were detected using mouse cytokine ELISA kits (Shanghai Enzyme-linked Biotech, #m1002277, #ml063136, #ml063156) on day 21 after the third immunization. Briefly, 50 μL of serum diluted 1:3 and 100 μL of HRP-labeled specific antibody were added into each well followed by incubation at 37°C for 60 min. After washing 3 times with PBST, the substrate solution was added into each well to develop in the dark at 37°C for 15 min. The reaction was stopped immediately with the stop solution, 50 μL/well, and the OD values were measured at 450 nm using a Multimode Microplate reader (Biotek Synergy HTX, USA). Three parallel repeated tests were performed for each sample.
All data presented in this study were expressed as mean ± standard deviation (SD) and analyzed using GraphPad Prism 8.0. Statistical analysis was conducted using One-way ANOVA, followed by Duncan’s multiple comparisons. Differences among different groups were considered significant when P < 0.05 (*) or P < 0.01 (*), and the comparison without a statistics bar and asterisk were found to be non-significant (P > 0.05).
The constructed recombinant expression plasmids, pCold 1-S1 and pCold 1-S1-PA, were identified by PCR, the restriction endonuclease (EcoR I and Sal I) digestion, and sequencing and analysis, respectively. After agarose gel electrophoresis, target bands of the same size, 2244 bp and 2943 bp, as the expected S1 and the S1-PA recombinant protein was observed, respectively (Supplementary Figure 1). The sequencing results were consistent with the reference sequences.
SDS-PAGE analysis and Western blotting showed the recombinant proteins of S1 and S1-PA were successfully expressed in the E. coli BL21 (DE3) cells by IPTG-induced 24 h at 16°C (Figures 2A–C). The expressed proteins existed mainly in the supernatant of the bacterial lysate at 80 kDa and 104 kDa, respectively. Recombinant S1 protein was purified by metal affinity chromatography, and after desalination and concentration, the protein concentration was 2.4-3.2 mg/mL. S1-PA recombinant protein was directly used for the subsequent preparation of S1-BLPs.
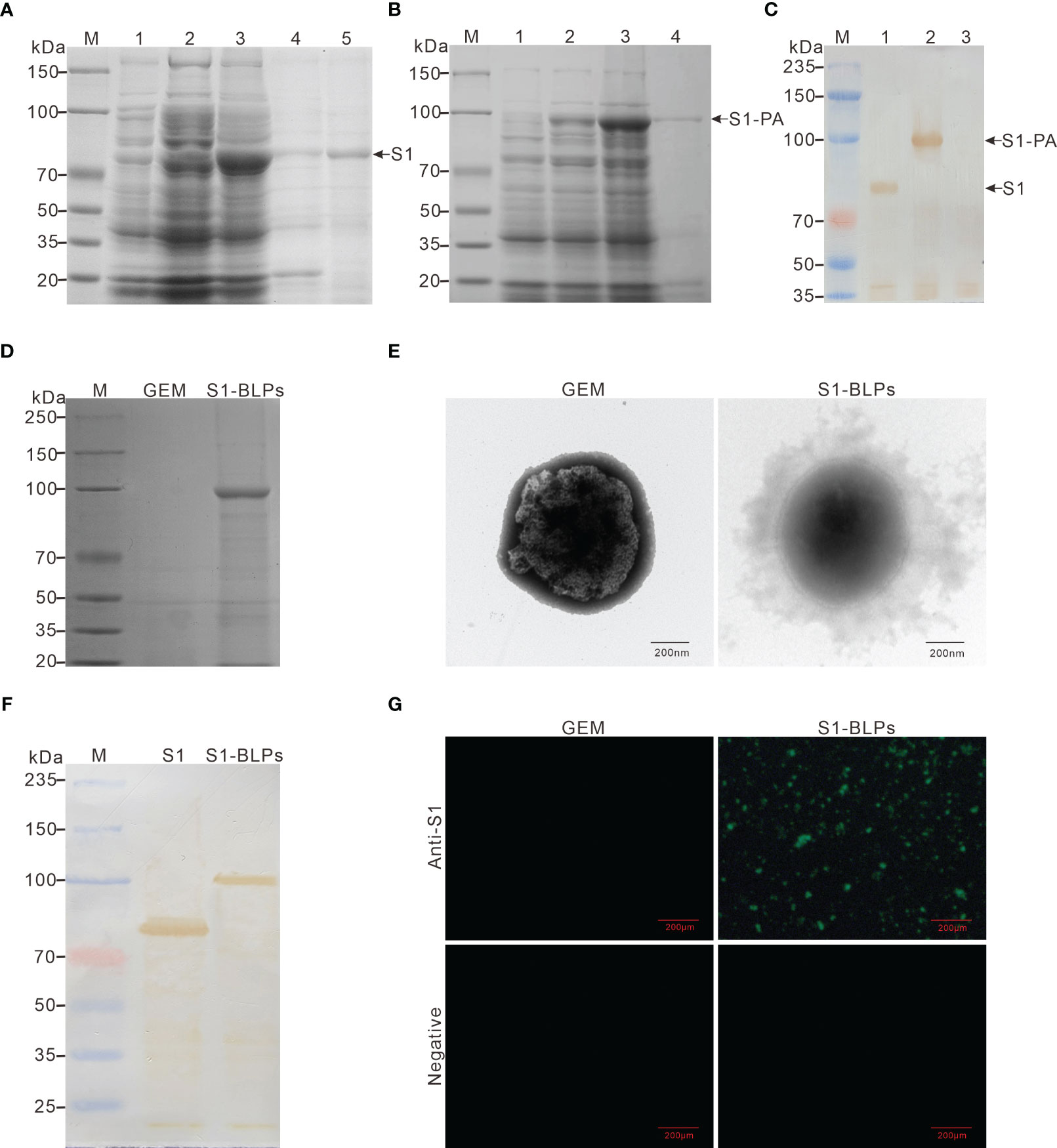
Figure 2 Expression of recombinant proteins and preparation of S1-BLPs. (A–C) Identification of expressed recombinant protein. SDS-PAGE analysis of recombinant proteins S1 (A) and S1-PA (B), respectively. M: Protein marker; 1 and 2: E. coli BL21 (DE3) before and after induction with IPTG, respectively; 3: Lysate supernatant of induced E. coli BL21 (DE3); 4: Lysate precipitate of induced E. coli BL21 (DE3); lane 5: Purified S1 protein. (C) Identification of recombinant proteins S1 and S1-PA by Western blotting. M: Protein marker; 1: Protein S1; 2: Protein S1-PA; 3: Negative control. (D) SDS-PAGE analysis of S1-PA recombinant protein on the surface of GEM. (E) Morphology of S1-BLPs under the transmission electron microscopy. (F) Identification of S1 protein on S1-BLPs by Western blotting. S1 (the control) or S1-BLPs reacted with Rabbit anti-S1 protein polyclonal antibody. (G) Indirect immunofluorescence assay. GEMs and S1-BLPs reacted with Rabbit anti-S1 protein polyclonal antibody or the anti-S1 protein antibody negative rabbit serum, respectively.
After incubation of GEM particles with the S1-PA protein, S1 antigen could be displayed on GEMs surface by anchoring protein PA. When S1-BLPs was subjected to SDS-PAGE electrophoresis, S1-PA recombinant protein bands were seen in the corresponding lanes (Figure 2D). BCA protein assay showed that each unit (2.5×109 particles/mL) of the GEMs could bind to 371-410 μg of S1-PA protein. Furthermore, the electron microscopy revealed a ring of flocculation around the GEMs in S1-BLPs with a diameter of about 800-1000 nm, but not around the GEMs that did not bind the S1 protein (Figure 2E). These results showed that S1 protein was conjugated with GEMs and displayed on the surface of GEMs.
The antigens in S1-BLPs were detected by Western blotting and immunofluorescence assay. As shown in Figure 2F, a clear brown band of around 104 kDa appeared on the PVDF membrane in the lane of S1-BLPs following Western blotting. As expected, S1-BLPs glowed green fluorescence after indirect immunofluorescence staining while GEMs control not (Figure 2G). The results demonstrated that S1 protein was conjugated with GEMs by the protein anchor.
In order to analyze the influences of immunization routes on immune responses and compare systematic specific IgG antibody induced by S1-BLPs and S1 protein, female BALB/c mice were immunized with S1-BLPs or S1 protein through the intramuscular and intranasal route, respectively, and serum was collected weekly (Figure 3A). After immunization, the specific IgG antibody levels in two intramuscular immunization groups (S1/IM and S1-BLPs/IM) were significantly higher than those in two intranasal inoculation groups (S1/IN and S1-BLPs/IN) (P<0.05) (Figure 3B). Although the IgG antibody in S1-BLPs intranasal immunization (S1-BLPs/IN) group remained at a relatively low level, it higher than that in S1/IN group. At 14 days after the primary immunization, the IgG level in S1-BLPs/IM group was significantly higher than that in S1/IM group (P<0.05), but there were no significant differences after the second and third immunizations (P>0.05). No specific IgG antibody was found in S1/IN and the blank group. The antibody titers in groups S1/IM and S1-BLPs/IM reached 1:102400 and 1:51200, respectively on day 21 after the third immunization, which were significantly higher than 1:1600 in the intranasal S1-BLPs group (Figure 3C), indicating that the intranasal immunization of S1-BLPs has limited ability to stimulate systemic specific IgG antibodies.
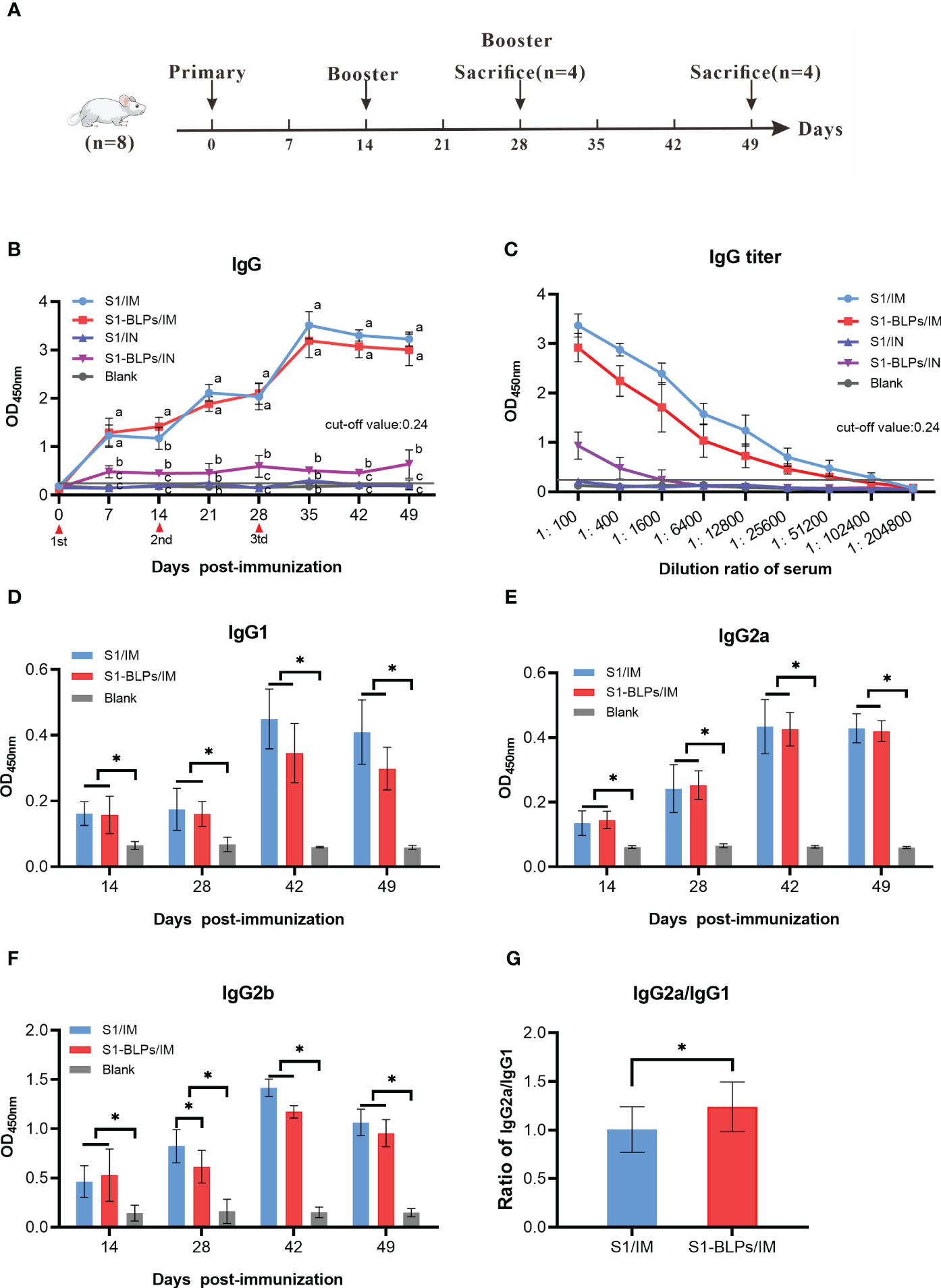
Figure 3 Timeline of the mouse immunization and specific IgG and IgG subclasses in serum. (A) Timeline of experimental treatment of mice. 40 female BALB/c mice aged 6-8 weeks were randomly divided into 5 groups with 8 mice in each group. Before and after immunization, blood samples were collected from the tail vein of mice weekly. At 14 days after the second immunization and 21 days after the third immunization, 4 mice were randomly selected from each group for the collection of intestine and lung airway lavage fluid, respectively. (B) Dynamics of specific IgG. (C) Specific IgG titers on day 21 after the third immunization. (D–F) Specific IgG1, IgG2a, IgG2b antibodies in serum. (G) Ratios of IgG2a to IgG1 at 14, 28, 42, and 49 days post immunization. The data came from three parallel replicates of each sample. Bars show means ± SD. Lowercase or *P<0.05.
The levels of specific IgG subclasses in serum were also tested by ELISA. Due to the low total IgG levels in the intranasal immunization groups, only IgG subclasses of the intramuscular immunization groups were measured. As shown in Figure 3D, the IgG1 antibody levels in the S1/IM group was slightly higher than that in the S1-BLPs/IM group at 14 and 28 days after the third immunization, but there was no significant difference between them (P>0.05). S1/IM group and S1-BLPs/IM group had similar IgG2a levels (P>0.05), and the former exhibited a higher IgG2b antibody level on day 14 after the second immunization (P<0.05) (Figures 3E, F). Moreover, S1-BLPs/IM group had a higher ratio of IgG2a/IgG1compared to the S1/IM group (P<0.05) (Figure 3G), indicating that the immune response induced by S1-BLPs was characterized by T helper (Th) cell type 1 (Th1) immunity.
To evaluate the effects of immunization routes on the specific IgA and sIgA responses, we compared the levels of IgA in serum and sIgA in the intestine and bronchoalveolar lavage fluid. From 7 days after immunization until the end of the experiment, only seroconversion of IgA presented in S1-BLPs/IN group (P<0.05). There was no significant difference between groups S1/IM, S1-BLPs/IM or S1/IN and the blank group (P>0.05) (Figure 4A). At 14 days after the second immunization and 21 days after the third immunization, the OD values of specific sIgA in intestine lavage fluid were 0.246 and 0.330 in S1/IN group, and 0.660 and 0.796 in S1-BLPs/IN group, respectively (Figure 4B). There was a significant difference between the two groups (P<0.01). No specific sIgA was found in the other groups (Figure 4B). In bronchoalveolar lavage fluid of all groups, specific sIgA was not detected (Figure 4C). The result showed that S1-BLPs could induce systemic and intestinal mucosal specific IgA responses by intranasal immunization.
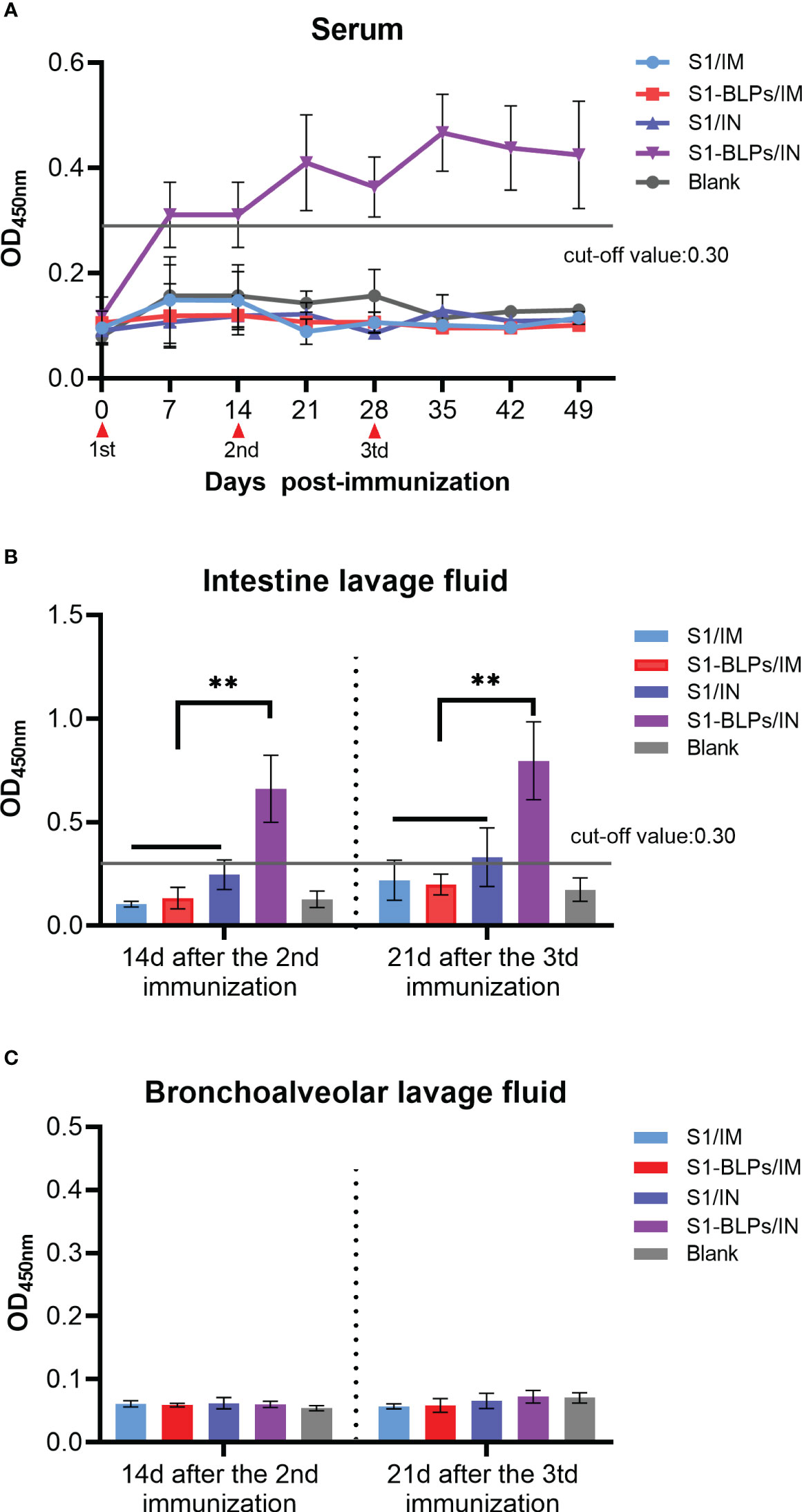
Figure 4 IgA in serum and secretory IgA in lavage fluid by ELISA. When the OD value is >0.3, the specific IgA is positive, and when it <0.3, the specific IgA is negative. (A) Dynamics of specific IgA in serum. (B) Specific secretory IgA in small intestinal lavage fluid. (C) Specific secretory IgA levels in bronchoalveolar lavage fluid. The data came from three replicates of each sample. Bars show mean ± SD. **P<0.01. The data came from three parallel replicates of each sample.
To further evaluate cytokine responses induced by S1-BLPs, the levels of IFN-γ, IL-2 and IL-4 in serum were analyzed by ELISA, and the results were shown in Figure 5. In addition to group S1/IN, serum IFN-γ levels in groups S1/IM, S1-BLPs/IM and S1-BLPs/IN were higher than that in the blank group. Furthermore, there was a stronger IFN-γ response in group S1-BLPs/IN compared to group S1-BLPs/IM and S1/IM (P<0.05), but there was no significant difference between groups S1/IN and S1/IM (Figure 5A). IL-2 levels were similar between the immunized groups and between the immunized and the blank group (Figure 5B). The level of IL-4 in group S1-BLPs/IN was higher than that in S1/IN and the blank group (P<0.05) (Figure 5C). The results indicated that S1-BLPs promoted IFN-γ response, and intranasal inoculation could significantly increase IFN-γ levels. Moreover, intranasal inoculation of S1-BLPs significantly induced IL-4 production.
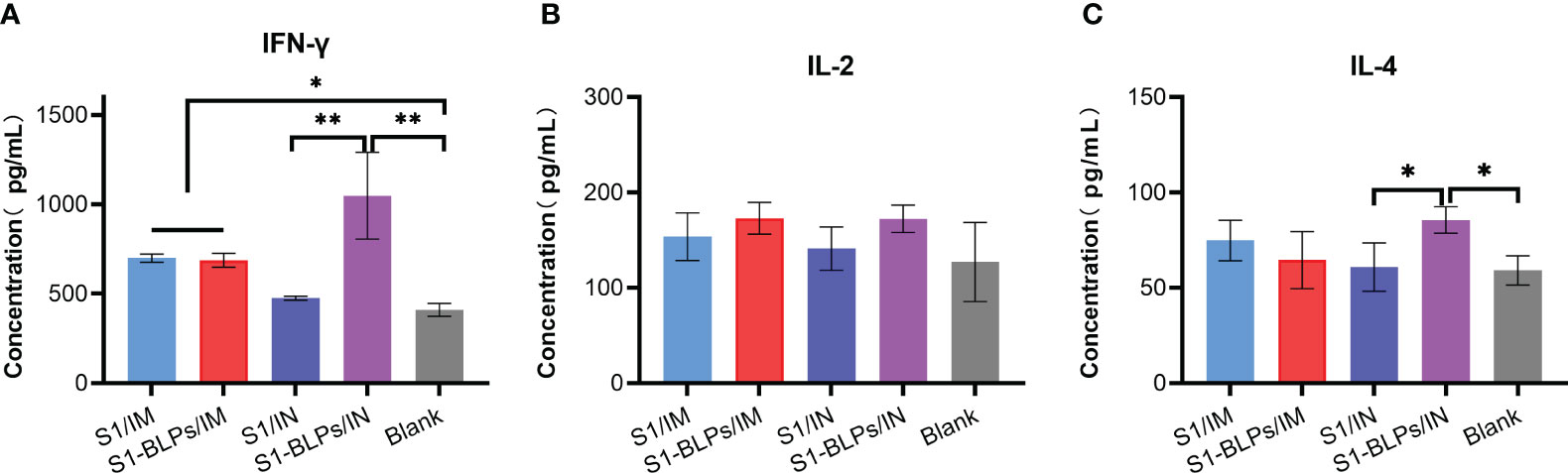
Figure 5 The levels of cytokines in serum by ELISA at 21 days post-immunization. (A–C) Concentration of IFN-γ, IL-2, IL-4 in serum, respectively. The data came from three parallel replicates of each sample. Bars show mean ± SD. **P<0.01; *P<0.05.
Since 2010, outbreaks of PEDV genogroup 2 (GII) have caused devastating losses in the global swine industry, especially the high mortality for newborn piglet (29–31). According to the genetic evolution analysis of the viral genome, PEDV strains around the world are divided into two groups (GI and GII). The GI consists of two subgroups (GIa and GIb), and the GII consists of three subgroups (GIIa, GIIb and GIIc) (32). The groups or subgroups of strains circulating in different countries are various, such as GIb and GIIb in the US, and GIIa, GIIb and GIIc in China (33–35). Cross-immune protection between different subgroups is low or variable (2), which brings challenges to prevent and control effectively PEDV. The S1 protein gene in this study was derived from PEDV QY2016 strain, which belongs to the GIIa subgroup and is a currently circulating strain. Therefore, the prepared S1-BLPs is expected to provide good immune protection against the current circulating strains. In addition, due to the pathogenic characteristics of PEDV, intestine mucosal immunity and the specific sIgA play a critical role in host resistance to the viral infection (36).
Because the mucosal immunity act as a crucial actor in the process of defense against pathogen infection, it is necessary to trigger mucosal immunity through various way (37). The intranasal route is one of the most direct and effective way of vaccination. Intranasal immunization can induce not only systemic specific but also specific local mucosal immune responses (38, 39). However, compared with subcutaneous and intramuscular injection of antigens, intranasal vaccination is less efficient in inducing systemic immunity (40, 41), which is consistent with the stronger serum IgG responses in the intramuscular vaccination (S1/IM and S1-BLPs/IM) groups than those in the intranasal vaccination (S1/IN and S1-BLPs/IN) groups in this study.
The routes of vaccination significantly affect mucosal immune response. In this study, intramuscular inoculation of S1 protein or S1-BLPs with adjuvant IMS1313 could induce high levels of the systemic specific IgG in mice, but could not trigger specific IgA response either systemic or mucosal immune responses. Consistent with other studies (41), our results demonstrated that the immunization route was particularly important for stimulating mucosal immunity. In addition, by comparing specific IgA levels in serum and intestinal lavage, it is found that S1-BLPs can stimulate mucosal immune response better than S1 protein. This result is similar to previous studies on influenza (42), respiratory syncytial virus (RSV) (43), and streptococcus pneumoniae bacterium-like particles (44). Moreover, Induction of mucosal immunity by S1-BLPs might be associated with the immunomodulatory effects of GEM particles as an adjuvant. According to the common mucosal immune system (CMIS), intranasal route is more practical to stimulate broad and disseminated antigen-specific mucosal and systemic immune responses (45). For the prevention and control of PEDV or other enteroviruses, the feasibility of intranasal vaccination needs further study. Most noteworthy, no specific sIgA was detected in bronchoalveolar lavage fluid of all groups, even by intranasal immunization in present study. This result is different from previous study reports on influenza mucosal BLPs vaccines (23, 42). We speculate that this result is related to the diversity of antigen characteristics and tissue tropism of PEDV. Studies have shown that the target organ of PEDV infection is the small intestine rather than the respiratory organ, and S1 protein is the key for PEDV to bind to the cell receptors (46). In addition, A combined immunization scheme (i.e., subcutaneous inoculation followed by intranasal inoculation) can induce stronger systemic and mucosal immune responses than one immunization route alone (40). Therefore, we will try different routes of combined immunization to enhance the mucosal immune effect in the future.
Furthermore, Th cell immune responses are divided into two types, Th1 response and Th2 response (47). The Th1-type immune response induces cellular immunity, while the Th2 immune response favors humoral immunity. Cytokines play a vital role in regulating immune response and maintaining the immune balance between Th1-type and Th2-type responses (48). IFN-γ, IL-2 and TNF-α/β, and IgG2a increase in Th1-type responses, while IL-4, IL-6 and IL-10, and IgG1 are elevated in Th2-type responses (47, 49). IgG2a response are associated with increased efficacy of vaccination, and more efficient at clearing virus infection (50). After intranasal administration, GEM particles, can effectively stimulate systemic and local immune responses, and enhance Th1-type immunity (25, 51). This study found that S1-BLPs significantly increase the levels of IFN-γ or IgG2a when intranasal and intramuscular administration, respectively, both of which are manifestation of Th1 type immunity. At the same time, it was found that S1-BLPs also increase the expression of IL-4 which is related to humoral immune response via intranasal inoculation. These scenarios indicate that S1-BLPs enhanced the cellular immune responses, but also stimulated humoral immune response, thereby maintaining the immune balance.
GEM particles derived from different species or strains of lactic acid bacteria display various adjuvant properties (52). Previous studies demonstrated that GEM particles from Lactobacillus rhamnosus CRL1505 shows stronger antiviral immune responses in porcine intestinal epithelial cells than those from Lactobacillus plantarum CRL2506 (53). Orally administered recombinant HEV capsid protein (ORF2) and GEMs from Lactobacillus rhamnosus strain IBLPS027 induced a more effective specific secretory IgA than that of strain CRL1505 in the gut of mice (54). Therefore, the diversity of immune-enhancing effects of lactic acid bacteria should be considered in the development of bacteria-like particle vaccines based on GEMs.
In conclusion, S1-BLPs prepared by GEM particle surface antigen display technology could induce specific systemic and intestinal mucosal immune responses in mice. Intramuscularly administered S1-BLPs significantly induced the serum specific IgG responses and increased the ratio of IgG2a/IgG1, but did not stimulate systemic and intestinal specific secretory IgA responses. Intranasally immunized S1-BLPs but not S1, could induce systemic and intestinal specific secretory IgA responses. Moreover, intranasally inoculated S1-BLPs significantly increased IFN-γ responses which is conducive to Th1-type immunity. S1-BLPs has the potential to be developed as PEDV mucosal vaccine. The next step will be to confirm whether oral inoculation of S1-BLPs can also induce the same immune effect, and to conduct in vivo experiments in pigs to systematically evaluate its clinical application effect.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was approved by Hebei Agricultural University (China) Animal Welfare and Ethical Review Board. The study was conducted in accordance with the local legislation and institutional requirements.
KS: Conceptualization, Data curation, Formal Analysis, Methodology, Visualization, Writing – original draft. YW: Data curation, Formal Analysis, Methodology, Visualization, Software, Writing – review & editing. CY: Data curation, Methodology, Visualization, Writing – review & editing. YZ: Data curation, Visualization, Writing – review & editing. YL: Data curation, Writing – review & editing. TL: Data curation, Software, Writing – review & editing. QS: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing, Software.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Center of Technology Innovation for Pigs (NCTIP-XD/B11) and the Science and Technology Project of the Department of Education of Hebei Province, China (ZD2022145).
We gratefully thank Dr. Zekun Yu for generously providing the lactic acid bacterium Lactococcus lactis subsp.cremoris MG1363 for this study. We would also like to thank coworkers Tiantian Ma, Huaining Yue, Yajuan Ma and Jing Ren for their enthusiastic help in animal experiments and sample collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1269409/full#supplementary-material
1. Chen P, Wang K, Hou Y, Li H, Li X, Yu L, et al. Genetic evolution analysis and pathogenicity assessment of porcine epidemic diarrhea virus strains circulating in part of China during 2011-2017. Infection Genet Evol (2019) 69(1):153–65. doi: 10.1016/j.meegid.2019.01.022
2. Jung K, Saif LJ, Wang Q. Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res (2020) 286(1):198045–63. doi: 10.1016/j.virusres.2020.198045
3. Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerging Infect Dis (2012) 18(1):161–3. doi: 10.3201/eid1801.111259
4. Li Y, Wu Q, Huang L, Yuan C, Wang J, Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat Commun (2018) 9(1):3811. doi: 10.1038/s41467-018-06056-w
5. Song D, Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes (2016). 44(2):167–75. doi: 10.1007/s11262-012-0713-1
6. Li W, van Kuppeveld FJM, He Q, Rottier PJM, Bosch BJ. Cellular entry of the porcine epidemic diarrhea virus. Virus Res (2016) 226(1):117–27. doi: 10.1016/j.virusres.2016.05.031
7. Lee C. Porcine epidemic Diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J (2015) 12(1):193–208. doi: 10.1186/s12985-015-0421-2
8. Chang CY, Cheng IC, Chang YC, Tsai PS, Lai SY, Huang YL, et al. Identification of neutralizing monoclonal antibodies targeting novel conformational epitopes of the porcine epidemic diarrhoea virus spike protein. Sci Rep (2019) 9(1):2529–40. doi: 10.1038/s41598-019-39844-5
9. Gong L, Gong Y, Lin Y, Qin J, Liu Y, Qian X, et al. Identification and characterization of a novel linear epitope in the spike protein of the porcine epidemic diarrhea virus. Acta virologica (2019) 63(1):88–95. doi: 10.4149/av_2019_111
10. Li C, Li W, Lucio de Esesarte E, Guo H, van den Elzen P, Aarts E, et al. Cell attachment domains of the porcine epidemic diarrhea virus spike protein are key targets of neutralizing antibodies. J Virol (2017) 91(12):273–85. doi: 10.1128/jvi.00273-17
11. Suzuki T, Terada Y, Enjuanes L, Ohashi S, Kamitani W. S1 subunit of spike protein from a current highly virulent porcine epidemic diarrhea virus is an important determinant of virulence in piglets. Viruses (2018) 10(9):467–78. doi: 10.3390/v10090467
12. Wang J, Huang L, Mou C, Zhang E, Wang Y, Cao Y, et al. Mucosal immune responses induced by oral administration recombinant bacillus subtilis expressing the Coe antigen of PEDV in newborn piglets. Bioscience Rep (2019) 39(3):2028–40. doi: 10.1042/bsr20182028
13. Chang YC, Chang CY, Tsai PS, Chiou HY, Jeng CR, Pang VF, et al. Efficacy of heat-labile enterotoxin B subunit-adjuvanted parenteral porcine epidemic diarrhea virus trimeric spike subunit vaccine in piglets. Appl Microbiol Biotechnol (2018) 102(17):7499–507. doi: 10.1007/s00253-018-9110-6
14. Li Q, Xu Z, Wu T, Peng O, Huang L, Zhang Y, et al. A flagellin-adjuvanted ped subunit vaccine improved protective efficiency against PEDV variant challenge in pigs. Vaccine (2018) 36(29):4228–35. doi: 10.1016/j.vaccine.2018.05.124
15. Kang KJ, Kim DH, Hong EJ, Shin HJ. The carboxy terminal region on spike protein of porcine epidemic diarrhea virus (PEDV) is important for evaluating neutralizing activity. Pathog (Basel Switzerland) (2021) 10(6):683–94. doi: 10.3390/pathogens10060683
16. Langel SN, Paim FC, Lager KM, Vlasova AN, Saif LJ. Lactogenic immunity and vaccines for porcine epidemic diarrhea virus (PEDV): historical and current concepts. Virus Res (2016) 226:93–107. doi: 10.1016/j.virusres.2016.05.016
17. Song Q, Stone S, Drebes D, Greiner LL, Dvorak CMT, Murtaugh MP. Characterization of anti-porcine epidemic diarrhea virus neutralizing activity in mammary secretions. Virus Res (2016) 226:85–92. doi: 10.1016/j.virusres.2016.06.002
18. Langel SN, Paim FC, Alhamo MA, Buckley A, Van Geelen A, Lager KM, et al. Stage of gestation at porcine epidemic diarrhea virus infection of pregnant swine impacts maternal immunity and lactogenic immune protection of neonatal suckling piglets. Front Immunol (2019) 10:727. doi: 10.3389/fimmu.2019.00727
19. Chattha KS, Roth JA, Saif LJ. Strategies for design and application of enteric viral vaccines. Annu Rev Anim Biosci (2015) 3:375–95. doi: 10.1146/annurev-animal-022114-111038
20. Hou Y, Wang Q. Emerging highly virulent porcine epidemic diarrhea virus: molecular mechanisms of attenuation and rational design of live attenuated vaccines. Int J Mol Sci (2019) 20(21):5478–92. doi: 10.3390/ijms20215478
21. Collin EA, Anbalagan S, Okda F, Batman R, Nelson E, Hause BM. An inactivated vaccine made from a U.S. Field isolate of porcine epidemic disease virus is immunogenic in pigs as demonstrated by a dose-titration. BMC veterinary Res (2015) 11(1):62–70. doi: 10.1186/s12917-015-0357-1
22. Lycke N. Recent progress in mucosal vaccine development: potential and limitations. Nat Rev Immunol (2012) 12(8):592–605. doi: 10.1038/nri3251
23. Van Braeckel-Budimir N, Haijema BJ, Leenhouts K. Bacterium-like particles for efficient immune stimulation of existing vaccines and new subunit vaccines in mucosal applications. Front Immunol (2013) 4:282(1). doi: 10.3389/fimmu.2013.00282
24. Bosma T, Kanninga R, Neef J, Audouy SA, van Roosmalen ML, Steen A, et al. Novel surface display system for proteins on non-genetically modified gram-positive bacteria. Appl Environ Microbiol (2006) 72(1):880–9. doi: 10.1128/aem.72.1.880-889.2006
25. Raya-Tonetti F, Müller M, Sacur J, Kitazawa H, Villena J, Vizoso-Pinto MG. Novel lysm motifs for antigen display on lactobacilli for mucosal immunization. Sci Rep (2021) 11(1):21691–705. doi: 10.1038/s41598-021-01087-8
26. Liu W, Tan Z, Liu H, Zeng Z, Luo S, Yang H, et al. Nongenetically modified lactococcus lactis-adjuvanted vaccination enhanced innate immunity against helicobacter pylori. Helicobacter (2017) 22(5):12426–36. doi: 10.1111/hel.12426
27. Jee PF, Tiong V, Shu MH, Khoo JJ, Wong WF, Abdul Rahim R, et al. Oral immunization of a non-recombinant lactococcus lactis surface displaying influenza hemagglutinin 1 (Ha1) induces mucosal immunity in mice. PloS One (2017) 12(11):187718–35. doi: 10.1371/journal.pone.0187718
28. Zhang S, Yan F, Liu D, Li E, Feng N, Xu S, et al. Bacterium-like particles displaying the rift valley fever virus Gn head protein induces efficacious immune responses in immunized mice. Front Microbiol (2022) 13:799942. doi: 10.3389/fmicb.2022.799942
29. Jung K, Wang Q, Scheuer KA, Lu Z, Zhang Y, Saif LJ. Pathology of us porcine epidemic diarrhea virus strain Pc21a in gnotobiotic pigs. Emerging Infect Dis (2014) 20(4):662–5. doi: 10.3201/eid2004.131685
30. Su M, Li C, Qi S, Yang D, Jiang N, Yin B, et al. A molecular epidemiological investigation of PEDV in China: characterization of co-infection and genetic diversity of S1-based genes. Transboundary emerging Dis (2020) 67(3):1129–40. doi: 10.1111/tbed.13439
31. Kikuti M, Drebes D, Robbins R, Dufresne L, Sanhueza JM, Corzo CA. Growing pig incidence rate, control and prevention of porcine epidemic diarrhea virus in a large pig production system in the United States. Porcine Health Manage (2022) 8(1):23–30. doi: 10.1186/s40813-022-00268-9
32. Zhang H, Zou C, Peng O, Ashraf U, Xu Q, Gong L, et al. Global dynamics of porcine enteric coronavirus PEDV epidemiology, evolution, and transmission. Mol Biol Evol (2023) 40(3):52–64. doi: 10.1093/molbev/msad052
33. Sun Y, Chen Y, Han X, Yu Z, Wei Y, Zhang G. Porcine epidemic diarrhea virus in asia: an alarming threat to the global pig industry. Infection Genet Evol (2019) 70(1):24–6. doi: 10.1016/j.meegid.2019.02.013
34. Chen X, Zhang XX, Li C, Wang H, Wang H, Meng XZ, et al. Epidemiology of porcine epidemic diarrhea virus among chinese pig populations: A meta-analysis. Microbial pathogenesis (2019) 129(1):43–9. doi: 10.1016/j.micpath.2019.01.017
35. Guo J, Fang L, Ye X, Chen J, Xu S, Zhu X, et al. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transboundary emerging Dis (2019) 66(1):111–8. doi: 10.1111/tbed.12991
36. Li Z, Ma Z, Li Y, Gao S, Xiao S. Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microbial pathogenesis (2020) 149(1):104553–65. doi: 10.1016/j.micpath.2020.104553
37. Lockhart A, Mucida D, Parsa R. Immunity to enteric viruses. Immunity (2022) 55(5):800–18. doi: 10.1016/j.immuni.2022.04.007
38. Bernasconi V, Norling K, Bally M, Höök F, Lycke NY. Mucosal vaccine development based on liposome technology. J Immunol Res (2016) 2016:5482087–101. doi: 10.1155/2016/5482087
39. Riese P, Sakthivel P, Trittel S, Guzmán CA. Intranasal formulations: promising strategy to deliver vaccines. Expert Opin Drug delivery (2014) 11(10):1619–34. doi: 10.1517/17425247.2014.931936
40. Zhang Y, Guo X, Guo M, Chen X, Li B, Yu J, et al. Combined prime-boost immunization with systemic and mucosal pneumococcal vaccines based on pneumococcal surface protein a to enhance protection against lethal pneumococcal infections. Immunologic Res (2019) 67(4-5):398–407. doi: 10.1007/s12026-019-09107-6
41. Bi J, Li F, Zhang M, Wang H, Lu J, Zhang Y, et al. An hiv-1 vaccine based on bacterium-like particles elicits Env-specific mucosal immune responses. Immunol Lett (2020) 222(1):29–39. doi: 10.1016/j.imlet.2020.03.002
42. de Haan A, Haijema BJ, Voorn P, Meijerhof T, van Roosmalen ML, Leenhouts K. Bacterium-like particles supplemented with inactivated influenza antigen induce cross-protective influenza-specific antibody responses through intranasal administration. Vaccine (2012) 30(32):4884–91. doi: 10.1016/j.vaccine.2012.04.032
43. Rigter A, Widjaja I, Versantvoort H, Coenjaerts FE, van Roosmalen M, Leenhouts K, et al. A protective and safe intranasal Rsv vaccine based on a recombinant prefusion-like form of the F protein bound to bacterium-like particles. PLoS One (2013) 8(8):71072–85. doi: 10.1371/journal.pone.0071072
44. Wang D, Lu J, Yu J, Hou H, Leenhouts K, Van Roosmalen ML, et al. A novel pspa protein vaccine intranasal delivered by bacterium-like particles provides broad protection against pneumococcal pneumonia in mice. Immunol investigations (2018) 47(4):403–15. doi: 10.1080/08820139.2018.1439505
45. Li M, Wang Y, Sun Y, Cui H, Zhu SJ, Qiu HJ. Mucosal vaccines: strategies and challenges. Immunol Lett (2020) 217(1):116–25. doi: 10.1016/j.imlet.2019.10.013
46. Jung K, Miyazaki A, Saif LJ. Immunohistochemical detection of the vomiting-inducing monoamine neurotransmitter serotonin and enterochromaffin cells in the intestines of conventional or gnotobiotic (Gn) pigs infected with porcine epidemic diarrhea virus (PEDV) and serum cytokine responses of Gn pigs to acute pedv infection. Res veterinary Sci (2018) 119(1):99–108. doi: 10.1016/j.rvsc.2018.06.009
47. Saravia J, Chapman NM, Chi H. Helper T cell differentiation. Cell Mol Immunol (2019) 16(7):634–43. doi: 10.1038/s41423-019-0220-6
48. Read KA, Powell MD, Sreekumar BK, Oestreich KJ. In vitro differentiation of effector Cd4(+) T helper cell subsets. Methods Mol Biol (Clifton NJ) (2019) 1960(1):75–84. doi: 10.1007/978-1-4939-9167-9_6
49. Kanai N, Min WP, Ichim TE, Wang H, Zhong R. Th1/th2 xenogenic antibody responses are associated with recipient dendritic cells. Microsurgery (2007) 27(4):234–9. doi: 10.1002/micr.20342
50. Mancardi DA, Iannascoli B, Hoos S, England P, Daëron M, Bruhns P. Fcgammariv is a mouse ige receptor that resembles macrophage fcepsilonri in humans and promotes Ige-induced lung inflammation. J Clin Invest (2008) 118(11):3738–50. doi: 10.1172/jci36452
51. Ramirez K, Ditamo Y, Rodriguez L, Picking WL, van Roosmalen ML, Leenhouts K, et al. Neonatal mucosal immunization with a non-living, non-genetically modified lactococcus lactis vaccine carrier induces systemic and local Th1-type immunity and protects against lethal bacterial infection. Mucosal Immunol (2010) 3(2):159–71. doi: 10.1038/mi.2009.131
52. Raya Tonetti F, Arce L, Salva S, Alvarez S, Takahashi H, Kitazawa H, et al. Immunomodulatory properties of bacterium-like particles obtained from immunobiotic lactobacilli: prospects for their use as mucosal adjuvants. Front Immunol (2020) 11:15(1). doi: 10.3389/fimmu.2020.00015
53. Albarracin L, Kobayashi H, Iida H, Sato N, Nochi T, Aso H, et al. Transcriptomic analysis of the innate antiviral immune response in porcine intestinal epithelial cells: influence of immunobiotic lactobacilli. Front Immunol (2017) 8:57(1). doi: 10.3389/fimmu.2017.00057
54. Arce LP, Raya Tonetti MF, Raimondo MP, Müller MF, Salva S, Álvarez S, et al. Oral vaccination with hepatitis E virus capsid protein and immunobiotic bacterium-like particles induce intestinal and systemic immunity in mice. Probiotics antimicrobial Proteins (2020) 12(3):961–72. doi: 10.1007/s12602-019-09598-7
Keywords: PEDV, S1 protein, bacterium-like particles, immunization routes, mucosal immunity
Citation: Su K, Wang Y, Yuan C, Zhang Y, Li Y, Li T and Song Q (2023) Intranasally inoculated bacterium-like particles displaying porcine epidemic diarrhea virus S1 protein induced intestinal mucosal immune response in mice. Front. Immunol. 14:1269409. doi: 10.3389/fimmu.2023.1269409
Received: 30 July 2023; Accepted: 31 August 2023;
Published: 18 September 2023.
Edited by:
Pedro Augusto Carvalho Costa, Federal University of Minas Gerais, BrazilReviewed by:
Jianzhong Wang, Jilin Agriculture University, ChinaCopyright © 2023 Su, Wang, Yuan, Zhang, Li, Li and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinye Song, c29uZ3FpbnllQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.