
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 06 October 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1269054
Microbial species that dwell human bodies have profound effects on overall health and multiple pathological conditions. The tumor microenvironment (TME) is characterized by disordered vasculature, hypoxia, excessive nutrition and immunosuppression. Thus, it is a favorable niche for microbial survival and growth. Multiple lines of evidence support the existence of microorganisms within diverse types of cancers. Like gut microbiota, intratumoral microbes have been tightly associated with cancer pathogenesis. Intratumoral microbiota can affect cancer development through various mechanisms, including induction of host genetic mutation, remodeling of the immune landscape and regulation of cancer metabolism and oncogenic pathways. Tumor-associated microbes modulate the efficacy of anticancer therapies, suggesting their potential utility as novel targets for future intervention. In addition, a growing body of evidence has manifested the diagnostic, prognostic, and therapeutic potential of intratumoral microorganisms in cancer. Nevertheless, our knowledge of the diversity and biological function of intratumoral microbiota is still incomplete. A deeper appreciation of tumor microbiome will be crucial to delineate the key pathological mechanisms underlying cancer progression and hasten the development of personalized treatment approaches. Herein, we summarize the most recent progress of the research into the emerging roles of intratumoral microbiota in cancer and towards clarifying the sophisticated mechanisms involved. Moreover, we discuss the effect of intratumoral microbiota on cancer treatment response and highlight its potential clinical implications in cancer.
The presence of bacteria within human tumors was first recognized in the 19th century (1). Due to difficulties in profiling the low biomass of microorganisms within tumors, our understanding of the link between tumor microbiota and cancer pathogenesis has made little progress during the first 50 years of the 20th century (2). In 1983, Warren et al. (3) observed that Helicobacter pylori, a causative agent of gastric cancer (GC), colonized the gastric epithelium. In 2006, bacteria-induced DNA damage was reported (4). Now, the advance in detection techniques is presenting unprecedented opportunities to investigate the diversity and functional features of intratumoral microbiota. Importantly, recent work has shed light on the perplexing roles of intratumoral microorganisms in cancer progression and their intercommunications with the immune system (5–7). These microbial residents can alter response to anticancer therapy and may be used as new biomarkers and therapeutic targets for cancer. Targeted manipulation of tumor microbiota will accelerate the pace to personalized medicine in cancer. However, the precise mechanisms that underlie the action of intratumoral microbiota in cancer development and treatment remain poorly understood. Gaining greater insight into the characteristics and biological functions of tumor-specific microbes would bring a new revolution in cancer management. In this review, we present the available data on the relationship between intratumoral microbes and cancer pathogenesis, and highlight the relevance of intratumoral microbiota in tumor immunity and therapeutic responses. We discuss the possibility of applying intratumoral microbiota as novel biomarkers and therapeutic targets for cancer and propose significant challenges for the development of tumor microbiota-targeted therapeutics.
The bacterial composition and richness of different cancer types are highly heterogeneous, as verified by a comprehensive study of the tumor microbiome by Ravid Straussman’s team (8). They profiled the microbial compositions across seven human tumor types, including glioblastoma multiforme (GBM), bone, brain, breast, lung, ovary and pancreatic tumors. Each tumor type possessed distinctive microbiota signatures (Table 1). The microbial communities within breast cancer were more abundant and diverse than those in other tumor types. The bacterial load and richness were also higher in breast tumors than their adjacent normal tissues. Particularly, the composition of microbial species was different across subtypes of the same tumor type. Bacteria belonging to the phyla Firmicutes and Proteobacteria dominated the microbiota of all cancer types, while members of the phylum Proteobacteria were the most abundant bacteria in pancreatic tumors. Bacteria from the families Corynebacteriaceae and Micrococcaceae were abundant in nongastrointestinal tumors (e.g., bone and breast cancer). At the species level, Fusobacterium nucleatum was enriched in breast and pancreatic tumors. Intratumoral bacteria were located inside both cancer and immune cells, especially macrophages. Live bacteria from the phyla Actinobacteria, Firmicutes and Proteobacteria were found inside breast tumors. Metagenomic analyses proved that fungi including Blastomyces gilchristii, Candida albicans, Malassezia globosa, Malassezia restricta and Saccharomyces cerevisiae were present in various human cancer types (breast, bone, brain, colon, lung, melanoma, ovary and pancreas) (9). All tumor types possessed increased fungal loads compared with normal controls. Like bacteria, fungi mainly localized to tumor cells and macrophages. Different cancers also displayed cancer type-specific mycobiome profiles. For instance, colon cancer had a high abundance of Saccharomycetes, while the relative abundance of Malasseziomycetes was higher in melanoma. Viral infection has been intimately intertwined with solid malignancies including breast, cervical, colon, esophageal, gastric, hepatocellular, lung and oral cancers (23–27). Several viruses such as Epstein-Barr virus (EBV), hepatitis B virus (HBV), hepatitis C virus (HCV), human papilloma virus (HPV), human T-cell lymphotropic virus (HTLV) and Kaposi sarcoma herpes virus (KSHV) are identified as risk factors for carcinogenesis (28). Particularly, the causal effect of HPV on the initiation of bladder cancer, cervical cancer, esophageal squamous cell carcinoma (ESCC) and head and neck cancers has been documented (10–13). In addition to HPV, EBV and polyomavirus infections can also lead to ESCC (13). HBV and HCV act as contributing factors to the oncogenesis of liver cancer and cholangiocarcinoma (14). Remarkably, the prevalence of Vibrio- and Streptococcus-inhabiting bacteriophage communities was previously verified in the gut of patients with colorectal cancer (CRC) (29). Collectively, the widespread existence of microorganisms in cancer cells underscores their implication in cancer pathogenesis. An in-depth investigation of the composition and function of tumor microbiota will open up new therapeutic opportunities for cancer. To gain a better understanding of heterogeneity in intratumoral microecology, cancer type-specific microbial profiles will be described below.
Dumont-Leblond et al. (15) profiled the lung microbiota of twenty-nine patients with non-small cell lung cancer (NSCLC) using a 16S rRNA sequencing approach. Diaphorobacter, Micrococcus, Paracoccus, Phascolarctobacterium and Ralstonia were overabundant in both healthy and cancerous tissues in NSCLC patients. Enteric, inflammatory or pathogenic bacteria, including Alloprevotella, Brevundimonas, Escherichia-Shigella, Faecalibacterium, Pseudomonas and unclassified Enterobacteriaceae, were only detected in cancerous tissues. Another study also revealed that pathogenic microbes, such as Acinetobacter Jungii, Enterococcus, Haemophilus haemolyticus, Haemophilus parainfluenzae and Streptococcus pneumoniae, were prevalent in lung tumor tissues (16). The common pathogenic microbes may cause lung infections due to the compromised immune system. It is intriguing whether these bacteria lead to pulmonary carcinogenesis or whether their enrichment is a result from the alteration of the pulmonary environment in cancer patients. Intratumoral pathogens could cause direct damage or induce chronic inflammation, contributing to lung cancer pathogenesis. Future work should concentrate on unraveling the involvement of intratumoral microbiota in lung carcinogenesis. The effect of NSCLC microbiota on clinical outcomes necessitates thorough exploration. Pathogenic microbes may also shape the tumor immune microenvironment. The crosstalk between intratumoral microbiota and host immune system is worthy of further research. In addition, there was an association between intratumoral microbiota and the microbial component in the bronchoalveolar lavage fluid (BALF) (16). Actinomyces Neesii, Haemophilus, H. haemolyticus, Haemophilus influenzae, human herpes virus type 7 (HHV-7), Neisseria lactose, Prevotella II, Streptococcus constellatus, Streptococcus crista, Streptococcus gordonii and S. pneumoniae were commonly present in the tumor tissue and BALF of NSCLC patients. It is reasonable to infer that exploration of microbial diversity and abundance in the BALF could provide hints regarding the NSCLC tumor condition.
The bacterial phyla Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria were abundant in tumor tissues of patients with primary liver cancer (PLC) (17). The relative abundance of Agrobacterium and Rhizobiaceae was dramatically increased in tumor tissues, while that of Pseudomonas was significantly reduced. Different histopathological subtypes (combined hepatocellular-cholangiocarcinoma (cHCC-CCA), intrahepatic cholangiocarcinoma (ICC) and hepatocellular carcinoma (HCC)) of PLC possessed their respective intratumoral microbial signatures. The HCC group exhibited special bacterial biomarkers (e.g., Enterobacteriaceae). The microbial biomarkers may provide a potential tool for the subtype stratification of PLC. Nevertheless, more clinical studies are warranted to track intratumoral microbiota dynamics in PLC patients. Upper and lower gastrointestinal tumors had different microbiota signatures (18). The five phyla Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria dominated the microbial composition in upper gastrointestinal tumors including esophageal carcinoma (ESCA) and stomach adenocarcinoma (STAD). The level of Bacteroidetes was higher while that of Firmicutes was lower in lower gastrointestinal tumors including colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) than upper gastrointestinal tumors. The Bacteroidetes/Firmicutes ratio was higher in lower gastrointestinal tumors than that in upper gastrointestinal tumors. The genera Capnocytophaga and Helicobacter were more abundant in upper gastrointestinal tumors than lower gastrointestinal tumors. Faecalibacterium and Porphyromonas were enriched in READ. The high levels of intratumoral Alistipes and Blautia correlated with better survival probability in COAD patients. The high abundances of Pasteurellales and Porphyromonas were associated with the tumor stage in COAD and READ, respectively. Profiling tumor-resident microbiota can be helpful in discovering prospective microbial biomarkers for cancer progression and prognosis. A great deal of further work needs to be done to elucidate the contribution of dysregulated microbiota in the pathogenesis of gastrointestinal cancer. Members of the genus Bifidobacterium were detected in CRC tissues (19). The amount of intratumoral Bifidobacteria DNA was associated with the extent of signet ring cells, alluding to a possible role of Bifidobacteria in regulating tumor microenvironment (TME) and tumor differentiation during CRC progression. Bifidobacteria, a natural proportion of the intestinal flora, are producers of lactic acid and acetate (30). The impairment of intestinal barrier function might lead to the entry of Bifidobacteria into CRC tissues (31). In the gut, Bifidobacteria prevented colorectal carcinogenesis (32). Nevertheless, Bifidobacteria seemed to behave oppositely in the TME. It was likely that Bifidobacteria-produced lactic acid and acetate served as an energy source for CRC cells, allowing for cancer growth and immune escape. The divergent roles of Bifidobacteria during CRC development remain to be explored in the future.
Intratumor pancreatic microbiome was characterized through large-scale sequencing data from The Cancer Genome Atlas (TCGA) (20). Members of the phylum Proteobacteria represented the dominant bacteria in pancreatic adenocarcinoma (PAAD) and were associated with tumor metastasis. The abundance of Citrobacter freundii, uncultured Pseudomonadales bacterium HF0500_12O04, Acidovorax ebreus TPSY, Shigella sonnei Ss046, the primary endosymbiont of Sitophilus zeamais, Toxypothrix sp. PCC 7601, Mycoplasma hyopneumoniae and uncultured bacterium HF0500_10F10 exhibited an association with the upregulation of oncogenic pathways, the downregulation of tumor-suppressive pathways, and immunosuppression. The high abundance of A. ebreus was linked with decreased levels of M2 macrophages, memory T cells and CD8+ T cells. Altogether, these pancreatic microbes might be involved in PAAD occurrence and development by reprograming the TME. It is still elusive how microbes colonize the pancreas. Permeability of intestinal barrier may be a cause of bacterial dissemination into the pancreatic duct, which requires further corroboration. Intratumoral microbes could affect tumor susceptibility to chemotherapy and patient outcome. Gammaproteobacteria acted to induce gemcitabine resistance via the bacterial enzyme cytidine deaminase (33). Oppositely, Bifidobacterium potentiated the efficacy of immune checkpoint inhibitor (ICI) (34). It is plausible to propose that antibiotic/probiotic administration in a tumor type-specific manner could sensitize cancer cells to anticancer treatments. More extensive studies will be required to evaluate the therapeutic benefits of microbial manipulation combined with conventional treatments in cancer patients.
The compositions of bacterial communities differed between renal cell carcinoma (RCC) tissues and adjacent normal tissues (21). The species diversity was reduced in RCC tissues compared to normal tissues. The phyla Proteobacteria (64.97%) and Firmicutes (14.09%) dominated the microbiota composition of RCC tissues. The levels of Burkholderiales and Comamonadaceae were lower in RCC tissues than those in normal tissues. The relative abundance of Actinomyces, Amycolatopsis, Brevundimonas, Deinococcus, Gordonia, Microlunatus, Nitriliruptor, Phyllobacterium, Pseudoclavibacter and Weissella was increased in RCC tissues compared with normal tissues. Deinococcus was considered as a factor contributing to GC development (35). The role of Deinococcus in RCC is worthy of more detailed investigation. Moreover, reduced levels of Chloroplast, Klebsiella and Streptophyta could differentiate RCC tissues from normal tissues with high sensitivity and specificity. The functional analysis of RCC-associated microbiota showed that pathways responsible for cell growth and death, membrane transport and transcription were enriched in RCC tissues. Dysregulated microbes might promote RCC development by regulating these intracellular pathways, which needs to be further deciphered. Substantial research efforts should be direct toward illustrating the genuine association between intratumor microbiota and RCC pathology.
The microbes belonging to the families Bacillaceae, Halobacteriaceae and Prevotellaceae were abundant in cervical cancer tissues from 121 patients (22). Cervical cancer tissues had increased levels of Fusobacterium, Peptoniphilus and Prevotella and decreased levels of Lactobacillus relative to healthy uterine cervix and vagina. Co-incubation experiments showed that Prevotella bivia increased the expression of three immunoregulatory proteins lysosomal associated membrane protein 3 (LAMP3), signal transducer and activator of transcription 1 (STAT1) and antigen peptide transporter 1 (TAP1) in cervical cancer cells under hypoxic conditions. LAMP3 acted as a tumor promoter contributing to cervical cancer metastasis (36). It is assumed that P. bivia could infiltrate cervical cancer tissues and propel cervical carcinogenesis through regulation of immunological pathways. This study suggested an indirect communication between cervical microbiota and cancer cells. It is intriguing whether intratumoral microbiota can affect the functionality of infiltrating immune cells. However, additional studies are necessary to decipher the host-microbiota interaction in cervical cancer.
Despite intratumoral microorganisms have attracted a great deal of attention, their origins remain a mystery. It is conceived that intratumoral microbes come from oral and intestinal microbiota, adjacent normal tissues, the circulatory system and mucosal sites (Figure 1). Compelling evidence has indicated that intestinal microbiota could be a main source of intratumoral microbes (37). Intestinal microbes can be detected in digestive system tumors (e.g., CRC and liver cancer), lung cancer and cervical cancer. All these organs have an externally exposed cavity, providing conditions propitious for microbial colonization. Microorganisms residing in the intestine may invade these tumor tissues owing to the permeability of intestinal barrier during carcinogenesis. A bacterial driver-passenger model was previously proposed (31). Specifically, intestinal bacteria such as Bacteroides fragilis and H. pylori acted as drivers of carcinogenesis and contributed to the creation of an appropriate condition that enabled the transfer of “passenger” bacteria, especially opportunistic bacteria, to the TME. Intestinal microbes are likely to play an important role in microbial intrusion into the TME. Considering the similar microbial composition between tumor tissues and their adjacent normal tissues, it is presumed that adjacent normal tissues are a potential source of intratumoral microbiota (37). The hypoxic and immunosuppressive TME favors microbial growth, which may be an explanation for microbial migration from noncancerous tissues to cancerous tissues. More research is required to clarify how microbes transfer from adjacent normal tissues to the tumor site. It is worth noting that the source of microbes within normal tissues remains ambiguous. Conversely, microorganisms inhabiting the tumor tissues may enter the normal tissues. This hypothesis requires further validation.
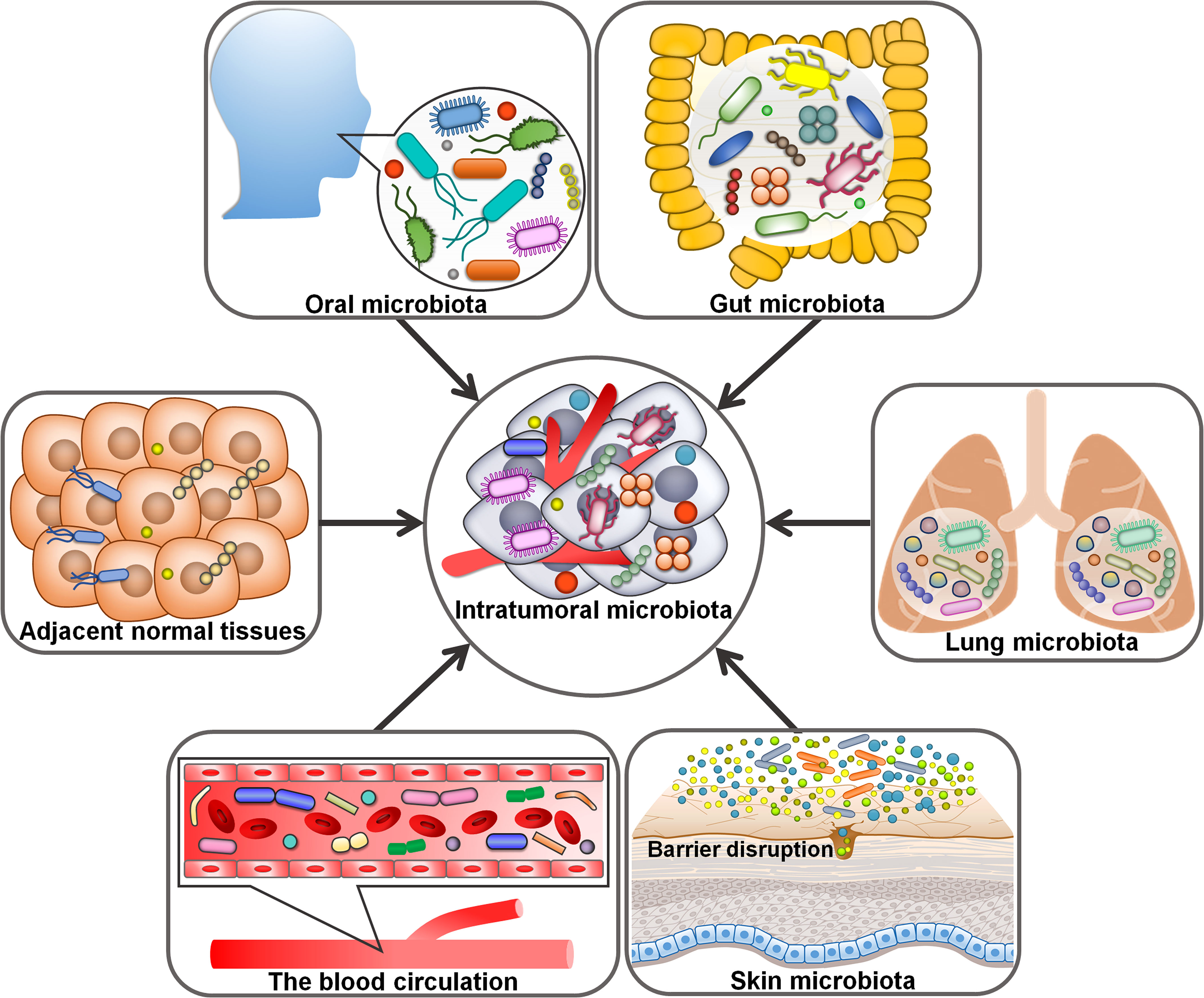
Figure 1 Potential origins of intratumoral microbiota. Oral and intestinal microbiota may be potential sources of intratumoral microbiota. The hypoxic and immunosuppressive tumor microenvironment could facilitate microbial migration from adjacent normal tissues to tumor tissues. For microorganisms intruding into the blood circulation from different loci, the chemotactic gradient or cellular debris released from necrotic tumor cells may support their transfer to the tumor microenvironment. Given abundant microbes exist in the mucosal organs (e.g., skin and lung), disruption in mucosal barriers may contribute to the entrance of intratumoral microorganisms.
Given the abundant and aberrant vascular architecture in tumors, oral and intestinal microorganisms could travel through the circulation system and enter distant tumor tissues via damaged blood vessels (38). For instance, the oral bacterium F. nucleatum could gain access to CRC tissues via the hematogenous route and colonize there (39). Escherichia coli impaired intestinal vascular barriers and migrated to the liver via the blood circulation, where it promoted the creation of a pre-metastatic microniche (40). It is not excluded that microbial species in the circulation system may directly invade the tumor tissues. Within the tumor tissue, microbes may be directly uptaken by tumor cells (41). For microbes intruding into the blood circulation from diverse loci, the chemotactic gradient or necrotic cell-released debris in tumors may be the cause of their transfer to the TME. Massive microbes exist in the mucosal organs including the skin and lung, and disruption in mucosal barriers may give rise to the entrance of intratumoral microbes (42). That is to say, intratumoral microbiota may originate from mucosal sites. Tumor-associated microorganisms are predominantly present in cancer cells and immune cells, implying that microbes may be delivered to the tumor site in the form of fractions or intact cells through cell migration (9, 43). Erythrocytes were considered as potential carriers for live bacteria into the tumors (44). Altogether, intratumoral microorganisms have various sources and show a close relationship with oral and intestinal microbiota. The mechanisms by which microorganisms enter the tumors are multitudinous and varied. Future work should be directed toward corroborating the diverse origins of intratumoral microbiota and elucidating the mechanisms regulating microbial translocation into the tumor site. A deeper understanding of the sources of intratumor microbes would provide new perspectives and theoretical basis for cancer prevention and treatment.
Intratumoral microbes have been identified as key regulators of cancer progression (Table 2). Liu et al. (45) characterized the microbial communities within CRC or precancerous adenoma. The levels of CRC-associated genera Bacteroides, Fusobacterium, Parvimonas and Prevotella were markedly different between CRC and adenoma tissues. The abundance of these genera varied across different sites of a single neoplasia, indicating the heterogeneity of microbial communities within a single CRC or nonmalignant adenoma. The amount of intra-neoplasia microbes with high variation such as Proteobacteria diminished along the adenoma-carcinoma sequence, suggesting the progressive alteration in abundance and diversity of microbes in the course of colorectal carcinogenesis. Firmicutes species (e.g., Clostridium, Parvimonas and Peptostreptococcus) were the most abundant members that displayed strong association with Kirsten rat sarcoma viral oncogene homologue (KRAS) mutation in CRC tumors and adenomas. Genera belonging to Proteobacteria (e.g., Dechloromonas and Gallionella) were the most abundant microbes that positively correlated with microsatellite instability (MSI) in CRC patients. The majority of intratumoral microbes that were related to CRC-associated genetic markers involving KRAS mutation and MSI showed high variation in abundance. The interplay between gut microbiota and host genetic mutation could accelerate cancer pathogenesis (68) (Figure 2). Host genetic factors in turn alter the microbial composition (69). More attention should be paid to the complex interrelation between gut microbiota and host genetic factors. Intratumoral microbiota heterogeneity may affect the metabolism of precancerous or cancer cells. Additional studies are suggested to elucidate the capricious compositional and functional profiles of intra-neoplasia microbiota as well as its contribution to colorectal carcinogenesis.
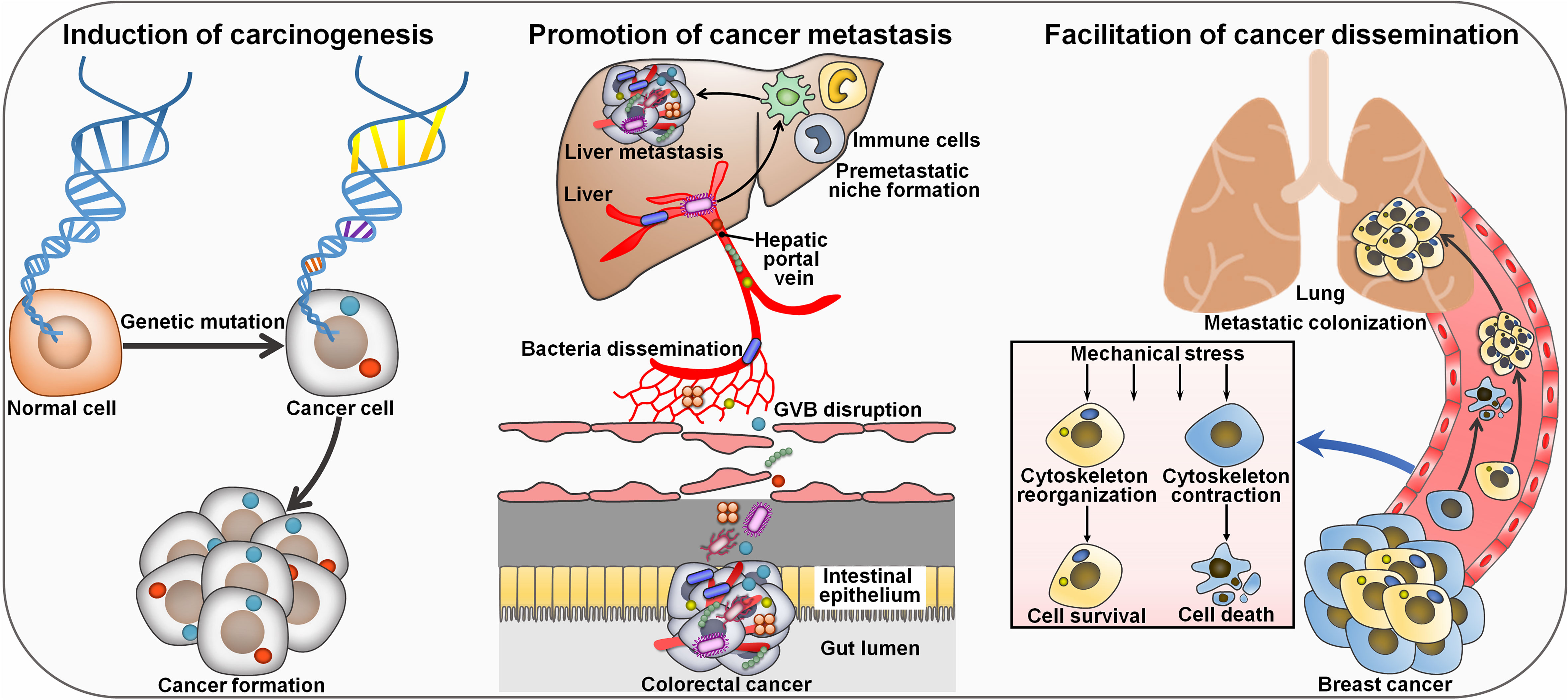
Figure 2 Mechanisms of intratumor microbiota affecting cancer pathogenesis. Intratumoral microorganisms act as a tumor inducer by causing genetic mutation in host cells. Colorectal cancer-resident microbes can impair the GVB and disseminate to the liver where they recruit immune cells (e.g., macrophages, neutrophils and inflammatory monocytes). These events lead to the formation of a pre-metastatic microenvironment that promotes cancer metastasis to the liver. Intratumoral microbes reorganize actin cytoskeleton in circulating breast cancer cells and foster their survival against mechanical stress in the circulation, culminating in enhanced lung metastasis. GVB, gut vascular barrier.
The liver is physiologically connected to the gut via the hepatic portal vein and is considered as the predominant organ site of primary CRC metastasis. However, gut vascular barrier (GVB) forms a critical barrier restricting intestinal bacteria dissemination. The high level of plasmalemma vesicle-associated protein-1 (PV-1) in primary tumors, a marker of disrupted GVB, was found to correlate with distant liver metastasis and poor prognosis in CRC patients (40). Importantly, CRC patients with high PV-1 expression in primary tumors possessed an increased amount of bacteria in metastatic liver lesions compared with paired healthy hepatic parenchyma, suggesting potential microbiota translocation through impaired GVB. Mechanistic investigation indicated that CRC-resident bacteria E. coli C17 directly broke down the GVB and caused PV-1 upregulation through a type III secretion system (TTSS) virulence factor (Virf)-dependent mechanism. Once GVB dysfunction, intestinal bacteria could disseminate to the liver where they fostered the recruitment of immune cells including macrophages, neutrophils and inflammatory monocytes. These events resulted in the creation of a pre-metastatic microenvironment that drove cancer metastasis to the liver. These findings revealed that intestinal bacteria targeted vascular gatekeepers to migrate into the secondary organ, contributing to the establishment of a favorable immune niche that supports the growth of disseminated cancer cells. PV-1 might act as a prognostic biomarker of gut vascular impairment and CRC distant recurrence, leading to liver metastasis formation. The clinical relevance of E. coli C17 for CRC progression should be further addressed. Furthermore, some beneficial bacteria (e.g., Lactobacillus paracasei) had the ability to restore the GVB. Therefore, there is a necessity of further research to dissect the functional link between intestinal bacteria and the metastatic process in CRC.
Spontaneous murine breast tumor displayed an expansion of commensal microorganisms that included Enterococcus, Lactobacillus, Staphylococcus and Streptococcus (46). The intratumoral microbes mainly colonized in tumor cell cytoplasm. Intracellular bacteria (e.g., Staphylococcus xylosus) together with host tumor cells, could travel through the circulation system and reside in distal organs. The invasion of various bacteria (e.g., Lactobacillus animalis, Streptococcus cuniculi and S. xylosus) promoted lung metastasis without affecting primary tumor growth. The intracellular bacteria reorganized actin cytoskeleton in circulating tumor cells and promoted host tumor cell survival by enhancing resistance to mechanical stress in the circulation system during metastatic colonization. Depletion of tumor-resident microbes suppressed the metastasis of breast tumor cells in vivo. Aerobic bacteria were increased in the lung metastasis, while the facultative anaerobes were decreased, implying a dynamic oxygen microenvironment in the tumor. The human and murine breast tumors possessed a similar microbial community profile, as characterized by high amounts of Enterococcus and Streptococcus. It was plausible to infer that intratumoral microbiota played an important role in the pathogenesis of human breast cancer. Intratumoral microbes may be an integral constituent of the tumor and act as pivotal drivers of tumor progression. Intratumoral microbiota could be a prospective target for better breast cancer management. It remains ambiguous how the bacteria enter tumor cells. The role of intratumor bacteria in tumor cell intravasation, extravasation and dissemination needs to be adequately delineated. Whether intratumoral microbiota, gut microbiota and the immune system cooperatively act to regulate cancer progression is an important question that emerges.
Increasing evidence indicates that intratumoral microbiota has a role in remodeling tumor immune microenvironment. Intratumoral microecology contributes to the recruitment and activation of tumor-supportive immune cells. Intratumoral microbiota induced interleukin-17 (IL-17) production to support the infiltration of B cells into the tumor tissues, culminating in colon cancer progression (70) (Figure 3). Polymorphonuclear neutrophils (PMNs), highly abundant immune cells in colon cancer, could reverse microbial dysbiosis in colon cancer tissues by reducing the amount of tumor-associated Akkermansia and increasing the amount of Proteobacteria. On the contrary, the absence of PMNs facilitated the outgrowth of colon microbiota and tumor-associated DNA damage. Depletion of colon microbiota or IL-17 inhibition reversed the pro-tumor effect of PMN deficiency. Collectively, PMNs limited colon cancer development by restraining the expansion of colon microbiota and reducing B cell infiltration through IL-17. It was possible that tumor-infiltrating PMNs evolved an immunosuppressive phenotype throughout colon cancer development (71). The perplexing contribution of PMNs in colon cancer progression deserves special attention. The mechanisms through which PMNs restrict colon microbiota expansion need to be further revealed. The high load of F. nucleatum inside MSI-high CRCs was markedly associated with tumor growth and invasion (47). F. nucleatum-enriched subset of MSI-high CRCs had decreased forkhead box P3 (FoxP3)+ T cells at both invasive margin and center of tumor areas. High load of F. nucleatum showed a positive relationship with an elevated ratio of CD163+ macrophages to CD68+ macrophages in MSI-high CRCs, alluding to increased population of M2-polarized macrophages in the tumor center. Thus, F. nucleatum might be connected with pro-tumoral immunity in MSI-high CRCs. Reducing F. nucleatum level could be effective in inhibiting CRC progression, which necessitates further study.
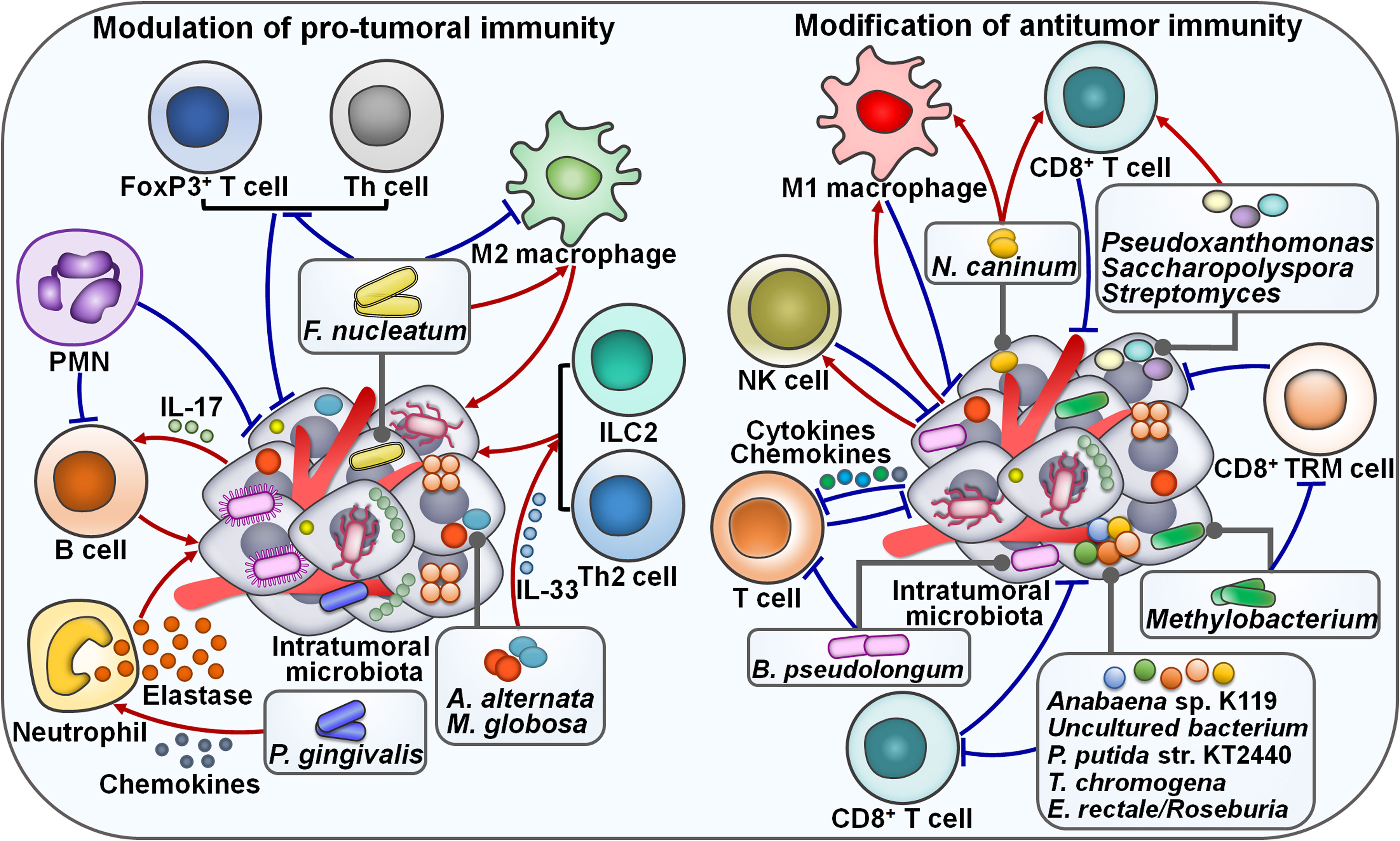
Figure 3 Emerging roles of intratumoral microbiota in tumor immunity. On the one hand, tumor-derived microbes are able to regulate pro-tumoral immunity. For instance, the F nucleatum load is associated with the population of M2 macrophages. The effects of F nucleatum on M2 macrophage activation vary depending on the tumor type. Intratumoral microbiota enhances IL-17 production to promote the infiltration of B cells into the tumor tissues, leading to cancer progression. PMNs restrict the expansion of tumor microbiota and suppress B cell infiltration through regulatory actions on IL-17. Intratumoral fungi A alternata and M. globosa recruit type 2 immune cells (e.g., ILC2s and Th2 cells) into the tumor microenvironment by increasing IL-33 release. Intratumoral P. gingivalis attracts tumor-associated neutrophils by increasing the secretion of neutrophilic chemokines. P. gingivalis also stimulates the secretion of neutrophil elastase from tumor-associated neutrophils, hence promoting cancer progression. On the other hand, tumor-derived microbiota plays an important role in modulating antitumor immune cell function. N. caninum correlates with the infiltration levels of macrophages and CD8+ T cells. Likewise, Pseudoxanthomonas, Saccharopolyspora and Streptomyces are related to the increased level of CD8+ T cells. The abundance of intratumoral microbiota shows a positive relationship with NK cell infiltration. Intratumor Methylobacterium reduces the frequency of CD8+ TRM cells within the tumor microenvironment. Intratumoral microbiota causes T cell exclusion by enhancing the secretion of specific chemokines and interleukins into the surrounding milieu. Particularly, B pseudolongum is shown to restrain T cell immunity. Anabaena sp. K119, uncultured bacterium, P. putida str. KT2440, T. chromogena and E rectale/Roseburia have a negative relationship with CD8+ T cells. FoxP3, forkhead box P3; Th cell, T helper cell; PMN, polymorphonuclear neutrophil; IL-17, interlekin-17; ILC2, type 2 innate lymphoid cell; IL-33, interlekin-33; NK cell, natural killer cell; TRM cell, tissue-resident memory T cell.
Porphyromonas gingivalis, a pathogen of periodontitis, was more abundant in pancreatic cancer (PC) tissues than in normal adjacent tissues, implying its momentous role in PC pathogenesis (48). Gavage with P. gingivalis accelerated tumor growth in PC-bearing mice. Remarkably, P. gingivalis was enriched in the tumor tissues of PC mice. P. gingivalis-treated PC tissues displayed a neutrophil-mediated proinflammatory TME. Mechanistically, intratumoral P. gingivalis recruited tumor-associated neutrophils by increasing the secretion of neutrophilic chemokines (C-X-C motif chemokine ligand 1 (CXCL1), CXCL2, C-X-C motif chemokine receptor 2 (CXCR2), IL-17F, S100a8 and S100a9). After that, P. gingivalis fostered the secretion of neutrophil elastase from tumor-associated neutrophils to drive PC development. The mechanism by which P. gingivalis enhances the secretion of neutrophil extracellular trap (NET)-associated proteases needs to be elucidated. P. gingivalis was commonly present in both the oral and intratumoral microbiota, implying the potentially bacterial migration from the oral cavity to the pancreas. The existence of P. gingivalis in the faeces of PC mice provided evidence of an oral-gut-pancreas translocation route. Inhibition of P. gingivalis infection may be beneficial for PC intervention. Future follow-up work is recommended to explore the mechanistic link between P. gingivalis colonization and the proinflammatory microenvironment in PC. It will be important to clarify whether other intratumoral pathogens can exert the same effect. The impact of intratumoral microbes on tumor immunity extends beyond bacterial species. Mycobiota has also been associated with cancer progression. Pancreatic ductal adenocarcinoma (PDAC) TME was infiltrated with type 2 immunocytes including T helper 2 (Th2) cells and type 2 innate lymphoid cells (ILC2s) (49). Notably, PDAC tumors contained a higher abundance of fungal communities than the normal pancreas. Intratumoral fungi Alternaria alternata and M. globosa actuated the dectin-1 signaling pathway in cancer cells that promoted KrasG12D-mediated IL-33 release. The released IL-33 recruited and induced type 2 immune cells into the tumor environment and facilitated PDAC development. As expected, in vivo experiment indicated that anti-fungal treatment or IL-33 deficiency decreased the infiltrating level of Th2 cells and ILC2s, leading to tumor regression and increased survival in PDAC-bearing mice. Retrograde transfer via the opening of the sphincter of Oddi might be a route for microbial migration from the duodenum to the pancreas. It is necessary to determine when the retrograde transfer of fungi occurs during the process of PDAC development. Continued research efforts are required to investigate whether intratumoral bacteria act synergistically with fungal communities to regulate host immune response and promote PDAC growth. ILC2s play divergent roles at different stages of cancer development. The inducing effects of intratumoral fungi on the IL-33/ILC2 axis potentially have variable consequences according to the cancer type, location and the state of the TME. The complex roles of intratumoral fungal communities in the occurrence and development of cancer should be a research priority in the future.
On the contrary, intratumoral microbiota also acts to counteract pro-tumoral immune responses. Reportedly, patients with F. nucleatum-positive oral squamous cell carcinoma (OSCC) had a lower recurrence rate, less frequent lymph node invasion and metastatic relapse than patients with F. nucleatum-negative tumors (50). Moreover, F. nucleatum-positive cases showed a trend toward longer overall survival (OS), better relapse-free survival (RFS) and metastasis-free survival (MFS) than F. nucleatum-negative cases. These findings demonstrated the prognostic significance of F. nucleatum in OSCC. The F. nucleatum load inversely correlated with the markers of B lymphocytes, fibroblasts, M2 macrophages and Th lymphocytes. Meanwhile, high F. nucleatum level was correlated with the decreased expression of OX40 ligand (tumor necrosis factor superfamily member 4 (TNFSF4)) and Toll-like receptor 4 (TLR4). The expression level of TNFSF9 receptor (TNFRSF9) was reduced while that of TNFSF9 and IL-1β was increased in OSCC tissues with high F. nucleatum load. F. nucleatum exerted both proinflammatory and immunosuppressive effects (72, 73). It was likely that OSCC-associated F. nucleatum remodeled the TME and rendered it insensitive to proinflammatory signals, as shown by the downregulation of TLR4 and M2 macrophages, thus leading to favorable clinical outcomes. Randomized clinical studies should be carried out to substantiate the interplay between F. nucleatum and tumor immune microenvironment. It will be equally important to examine the effects of F. nucleatum on OSCC pathogenesis and response to immunotherapy.
Intratumoral microbes can retard cancer development by augmenting antitumor immunity. Gram-negative bacteria were detected in the cytoplasm of osteosarcoma cells and tumor-associated macrophages (TAMs) (74). Osteosarcoma tissues from patients with local diseases were enriched in Gram-negative bacteria compared with those from patients with metastatic status. Moreover, the amount of antitumor M1 macrophages was also increased in osteosarcoma patients with local diseases. It was likely that the abundance of Gram-negative bacteria was associated with the expansion of M1-polarized macrophages. This study provided new insight into the mechanisms underlying osteosarcoma progression as well as therapeutic approaches against cancer. The possible functional linkage between intratumor microbiota and macrophage-mediated immune surveillance needs to be explored in further studies. Reportedly, the TLR4 signaling coordinated macrophage polarization towards M1 phenotype (75). It is therefore questioned whether the TLR4 signaling pathway mediates the effect of intratumor bacteria on macrophage activation in osteosarcoma. Soft tissue sarcoma harbored bacterial (e.g., Bacteroidetes, Firmicutes and Proteobacteria) and viral species (51). Respirovirus was strikingly enriched in patients without metastases compared with those with metastases. The abundance of intratumoral viruses was positively associated with natural killer (NK) cell infiltration. NK cells with an antiviral phenotype were related to improved clinical outcomes in patients with soft tissue sarcoma, underscoring an interrelation between the intratumoral virome, NK cell infiltration and favorable patient outcomes. However, the role and clinical significance of intratumoral microbiota in soft tissue sarcoma warrant more detailed investigation.
Increasing evidence indicates that tumor-associated microbiota functions as the focal point of local immune activation. Intratumoral inoculation of Neospora caninum induced melanoma cell death and noticeably suppressed tumor growth in mice (52). Mechanistic investigation demonstrated that N. caninum administration enhanced the production of Th1 cytokines including interferon-γ (IFN-γ), IL-2, IL-10, IL-12, programmed death-ligand 1 (PD-L1) and tumor necrosis factor-α (TNF-α) in the TME of melanoma-bearing mice, resulting in extensive tumor cell death. The infiltration levels of CD8+ T cells and macrophages were increased in N. caninum-treated mice. N. caninum also reversed intestinal microbiota dysbiosis by elevating the relative abundance of Adlercreutzia, Lachnospiraceae, Lactobacillus and Prevotellaceae. It seemed that N. caninum exerted a direct tumorolytic effect, but the death pathway of cancer cells involved remains to be ascertained in future studies. It is also intriguing whether the role of N. caninum in intensifying antitumor immunity can be partially ascribed to its effects on gut microbiota. Albeit these unsolved questions, the efficacy, safety, stability and facile culture properties of N. caninum make it an appropriate biopharmaceutical drug for clinical cancer treatment. Tumor microbiota diversity and three genera (Pseudoxanthomonas, Saccharopolyspora and Streptomyces) were correlated with CD8+ T cell densities in PDAC, hinting that tumor microbiota might induce antitumor immunity through attraction and activation of CD8+ T cells (53). Human-to-mice faecal microbiota transplant (FMT) experiments showed that gut microbiota could modify tumor microbiota by directly migrating into PDAC tissues or altering intratumoral microbial composition. Importantly, FMT from long-term survival (LTS) donors could restrict PDAC growth in recipient mice. Antibiotic administration or CD8+ T cell depletion attenuated the antitumor effect induced by LTS FMT. Collectively, gut microbiota could translocate to pancreatic tumors and regulate tumor progression by manipulating tumor microbiome and shaping host immunity. More studies are required to illuminate how gut microbiota alters tumor microbiota and induces immune activation. Modulation of tumor-associated microbiota could be an effective therapeutic strategy for PDAC intervention. The efficacy of antitumoral microbiota therapies in PDAC must be validated in clinical studies.
In contrast, some intratumoral microorganisms exert an inhibitory effect on antitumor immune cells. A retrospective cohort study including 802 patients with nasopharyngeal carcinoma (NPC) showed that Corynebacterium and Staphylococcus dominated the composition of tumor microbiota (76). The intratumoral bacterial load inversely correlated with disease-free survival (DFS), distant MFS and OS. These observations implied that intratumoral bacterial load was a potential prognostic indicator in NPC. The bacterial load had a negative relationship with T lymphocyte infiltration, suggesting that NPC microbiota fostered tumor progression by dampening antitumor immunity. The detailed mechanisms underlying this association deserve in-depth investigation. Anabaena sp. K119 and uncultured bacterium were the most abundant microbes in lung adenocarcinoma (LUAD), while Pseudomonas putida str. KT2440 and Thermostaphylospora chromogena were enriched in lung squamous cell carcinoma (LUSC) (54). These four bacteria had a negative relationship with CD8+ T cells and macrophages and a positive association with monocytes and neutrophils. The high abundance of P. putida str. KT2440 in LUSC was linked with reduced infiltration levels of both activated dendritic cells (DCs) and naïve B cells. T. chromogena was connected with reduced infiltration of naïve B cells, mast cells, resting NK cells and CD4+ T cells. The decreased amount of E. coli str. K-12 substr. W3110 and the increased amount of Staphylococcus aureus in LUAD were significantly correlated with patient survival. E. coli str. K-12 substr. W3110 might display a pro-tumor property. On the contrary, S. aureus seemed to function like a tumor suppressor. However, ongoing studies are still needed to reveal the role of dysregulated microbes in the pathogenesis of lung cancer.
Intratumoral microbiota was highly organized in the microniches with epithelial and immune cell functions across human tumors including CRC and OSCC (55). Intratumoral microbiota enhanced the secretion of specific chemokines (e.g., C-C motif chemokine ligand 2 (CCL2), CCL4, CXCL1 and CXCL10) and interleukins (e.g., IL-1β, IL-6 and IL-10) into the surrounding milieu, leading to T cell exclusion and tumor growth. Furthermore, CRC-derived F. nucleatum recruited myeloid cells to initiate an inflammatory response through the Janus kinase (JAK)/STAT signaling pathway and promoted transcriptional alterations in CRC epithelial cells that fostered invasion to the surrounding environment. Collectively, intratumor microbiota formed an essential component of the TME that could influence the biology of various cellular compartments, contributing to the suppression of antitumor immunity and migration of cancer epithelial cells. Noguti et al. (56) revealed that the high abundance of Eubacteria rectale/Roseburia in colon cancer tissues was correlated with a decreased level of CD8+ T cells and an increased risk of tumor recurrence. The intratumoral microbiota might promote colon cancer progression by limiting antitumor immunity. More clinical studies are required to corroborate these results. The causal link between E. rectale/Roseburia and colon cancer development is worthy of future investigation. Concerted research efforts should be encouraged to better understand the role of specific microorganisms in modulating antitumor immune responses. Intratumoral Methylobacterium strikingly correlated with poor prognosis in GC patients (57). It showed a negative relationship with the level of transforming growth factor-β (TGF-β) as well as the frequency of CD8+ tissue-resident memory T (TRM) cells within the TME. The in vivo experiment verified that Methylobacterium could reduce TGF-β expression and CD8+ TRM cells in gastric tumor tissues. Collectively, intratumoral Methylobacterium functioned as a driver of gastric carcinogenesis. The underlying mechanism through which Methylobacterium induces the exhaustion of CD8+ TRM cells during GC development needs to be revealed.
The microbial abundance was significantly increased in human PDAC tissues compared with normal pancreas (7). Intra-pancreatic microbiota in PDAC-bearing patients was enriched in bacteria belonging to the phyla Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria and the genera Elizabethkingia and Pseudomonas. B. pseudolongum was the most abundant Bifidobacterium species in KC mice, which developed spontaneous pancreatic tumors. Oral antibiotic administration reduced the abundance of the pancreatic microbiota and suppressed pancreatic carcinogenesis. Therefore, the gut and PDAC microbiome facilitated cancer progression, raising the possibility that regulation of gut microbiota via probiotics could reduce PDAC risk. Oppositely, microbial ablation diminished the intratumoral infiltration of myeloid-derived suppressor cells (MDSCs), enhanced T cell infiltration and skewed macrophages polarization towards a tumor-suppressive M1 phenotype. Depletion of gut microbiota also promoted Th1 polarization of CD4+ T cells and potentiated the cytotoxic activity of CD8+ T cells, as shown by high expression levels of IFN-γ, T-bet and TNF-α. In addition, microbial ablation significantly elevated expression of programmed cell death protein-1 (PD-1) on effector T cells, implying that oral antibiotics combined with checkpoint-centered immunotherapy may be a promising therapeutic option for PDAC treatment. Conversely, re-population using faeces derived from PDAC-bearing KC mice or B. pseudolongum supplementation supported pancreatic oncogenesis in germ-free KC mice, however, this tumor-promoting effect was abolished in the absence of the TLR signaling. Further study indicated that cell free extracts from B. pseudolongum could reprogram TAMs to increase the levels of tolerogenic cytokines (e.g., IL-10) by activating the TLR signaling cascade. It was previously reported that macrophage polarization affected effector T cell function in PDAC (77). Consistently, macrophages entrained by cell free extracts from B. pseudolongum resulted in the inhibition of T cell immunity. Lipopolysaccharides and flagellins derived from intra-pancreatic bacteria (e.g., Proteobacteria) could be potential TLR activators, which remains to be verified in the future. PDAC microbiota might play a critical role in mediating immunosuppression by modifying the cytokine milieu. The pancreatic duct might act as the conduit for bacterial translocation from the intestinal tract to the pancreas. Successive research efforts are necessary to characterize the mechanisms regulating the migration of specific intestinal bacteria. Altogether, manipulation of PDAC microbiota may be helpful in enhancing the efficacy of cancer immunotherapy.
Cancer metabolism is key and intimately linked to cancer progression. In recent years, the role of intratumoral microbiota in reprogramming of cancer metabolism has been increasingly recognized (Table 2). Bacillales, Burkholderiales, Clostridiales, Pseudomonadales, Sphingomonadales and Xanthomonadales were abundant in ICC tissues (58). Klebsiella pneumoniae, Paraburkholderia fungorum, Pseudomonas azotoformans and Staphylococcus capitis were verified to be present in ICC tissues. Intratumoral bacteria mainly resided in malignant cells of cancerous tissues, while they also localized to T cells, hinting that intratumor microbiota might be involved in tumor immunity. The amount of P. fungorum was noticeably higher in paracancerous tissues and had an inverse association with the carbohydrate antigen 199 (CA199) level, alluding to the potential antitumor potency of this bacterium. As expected, P. fungorum could repress the growth of human cholangiocarcinoma cells in vivo by regulating the metabolism of alanine, aspartate and glutamate (Figure 4). The hepatic-intestinal circulation may contribute to the crosstalk between intestinal bacteria and intratumoral microbes. The impact of intratumoral microbes on cancer metabolism should be an important subject for future investigation. Akkermansia muciniphila exhibited an inhibitory activity against lung cancer growth in vivo (59). This intestinal commensal bacterium could translocate to lung cancer tissues via systemic circulation. A. muciniphila altered the intratumoral metabolic pathways including amino acid metabolism, glycolysis/gluconeogenesis and fatty acid biosynthesis. Tumor-associated A. muciniphila also modified the composition of intratumor microbiota by increasing the abundance of Acidobacteriae, Akkermansia, Bacteroides, Bifidobacterium, Gammaproteobacteria, Lactobacillus, Sphingomonadales and Staphylococcus. A. muciniphila-influenced symbiotic microecology showed an association with the metabolic network in tumor tissues. Continual efforts should be made to clarify intratumoral microbe-mediated complicated cancer metabolic reprogramming. The specific intracellular microorganisms that are involved in the antitumor action of A. muciniphila should be identified.
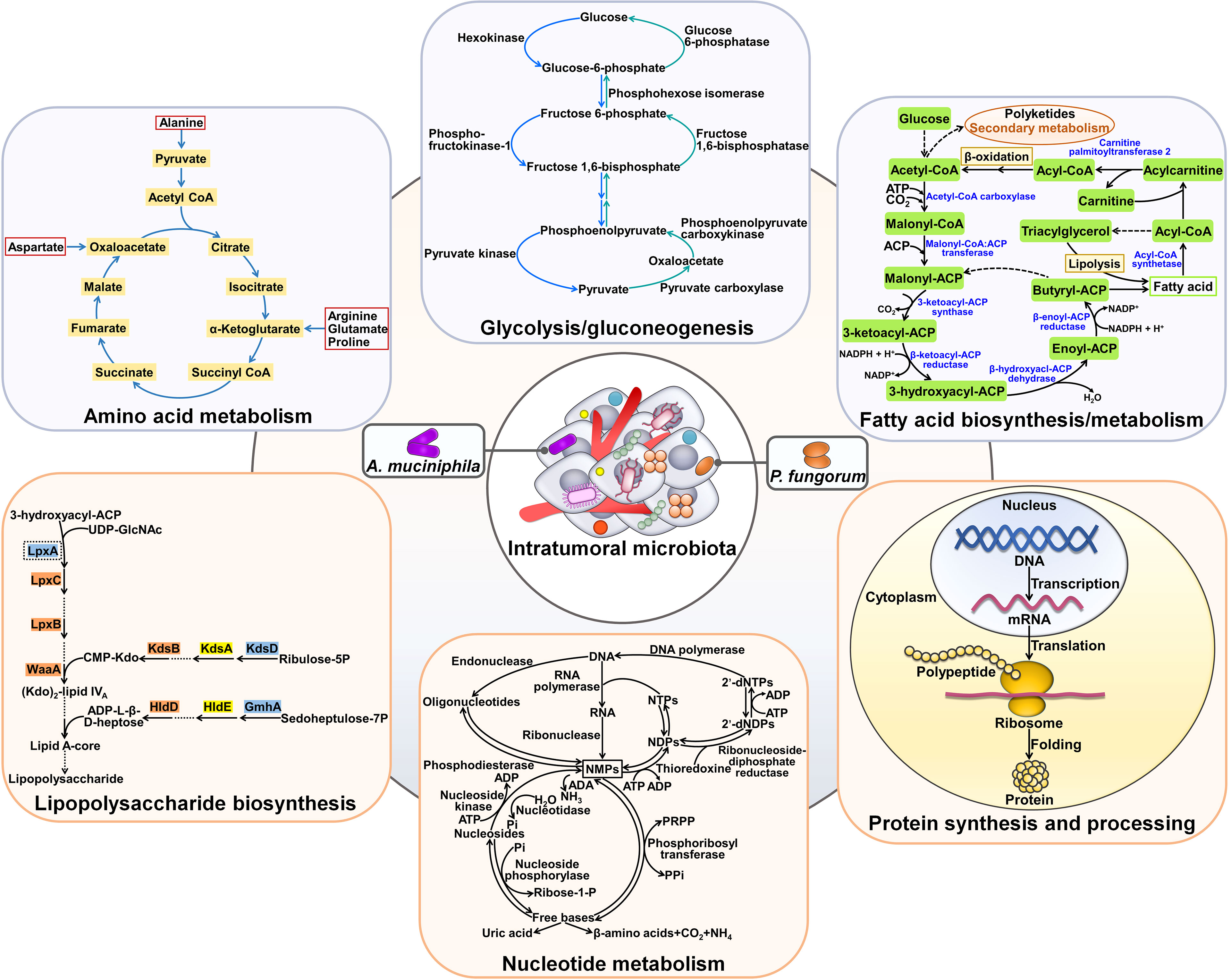
Figure 4 Effects of intratumoral microbiota on cancer metabolism. Tumor-resident microbes including A. muciniphila and P. fungorum can regulate amino acid metabolism. Moreover, intratumoral bacteria (e.g., A. muciniphila) affect glycolysis/gluconeogenesis and fatty acid biosynthesis and metabolism. In addition, intratumor microbial communities exert regulatory effects on lipopolysaccharide biosynthesis, nucleotide metabolism and protein synthesis and processing.
Bacterial communities were present in HCC cells and immune cells (78). Microbial diversity and richness were dramatically increased in HCC tissues compared with paracancerous tissues. The levels of commensal Enterobacteriaceae, Fusobacterium and Neisseria were higher in HCC tissues than in paracancerous tissues, whereas Agathobacter, Chryseobacterium, Dietzia, Faecalibacterium, Hydrogenophaga, Megamonas and Pseudomonas exhibited the opposite trend. It was reported that HCC-enriched Enterobacteriaceae utilized inflammatory byproducts within the microniches as energy sources, which eventually led to inflammation (79, 80). Likewise, the proinflammatory property of Fusobacterium was also documented (81). Since inflammation has been acknowledged as a risk factor for carcinogenesis, these bacteria may play a role in HCC formation (82). The high infiltration of pro-tumorigenic bacteria (e.g., Neisseria) and the reduced abundance of the tumor-suppressive bacteria (e.g., Pseudomonas) potentially propel HCC development (83, 84). Importantly, intratumoral microbes could alter cancer metabolic pathways. Fatty acid and lipopolysaccharide biosynthesis, and bacteria and disease pathways were remarkably enriched in HCC tissues. The resultant products of microbial metabolic activities may be a key source of fatty acids and lipids for cancer cells, thereby facilitating HCC cell proliferation and invasion. Intestinal permeability may be an explanation for bacterial translocation into HCC, calling for further verification. The mechanisms of action of intratumoral microbes in HCC carcinogenesis should be further explored.
Tumor microbiota communities were markedly different between PDAC patients with short-term survival (STS) and those with LTS (53). The STS tumors showed a predominance of Bacteroidea and Clostridia, while PDAC LTS cases were dominated by Alphaproteobacteria, Flavobacteria and Sphingobacteria. Actinobacteria (Saccharopolyspora and Streptomyces) and Proteobacteria (Pseudoxanthomonas) were more abundant in LTS patients versus STS patients. The heterogeneous tumor microbiota signatures contributed to different enrichment of metabolic functional pathways between STS and LTS patients. For instance, the STS cases were enriched in the pathways related to nucleotide metabolism, replication and repair, protein synthesis and processing, while the LTS cases demonstrated enrichment in the pathways associated with metabolism of amino acid, lipids and polyketides. The divergence in metabolic pathways may be an explanation for different clinical outcomes in PDAC patients, which necessitates additional research.
Intratumoral microbiota functions as key players in cancer-associated signaling cascades. The pathogens Acinetobacter, Actinomycetales, Ochrobacterium, Pseudomonas and Streptococcus were enriched in epithelial ovarian cancer (EOC) tissues (60). Intratumoral inoculation of Propionibacterium acnes propelled tumor growth and shortened the survival rate of EOC-bearing mice. The P. acnes strain could enhanced the expression of TNF-α and IL-1β, suggesting the proinflammatory potency of this bacterium. These two proinflammatory cytokines that were excreted by necrotic tumor cells could enhance Hedgehog (Hh) transcription (85). As expected, the Hh signaling pathway was abnormally motivated in P. acnes-treated mice, as evidenced by increased expression of glioma-associated oncogene 1 (Gli1), Gli2, patched homolog 1 (PTCH1), sonic hedgehog homolog (Shh) and smoothened homolog (SMO). Conversely, blockade of the Hh signaling prevented EOC progression caused by P. acnes. The proinflammatory P. acnes facilitated EOC pathogenesis by activating the Hh signaling cascade (Figure 5). Complementary in vitro and in vivo studies are warranted to delve into how intratumoral microbes interact with intracellular signaling pathways, providing important clues for improving cancer management.
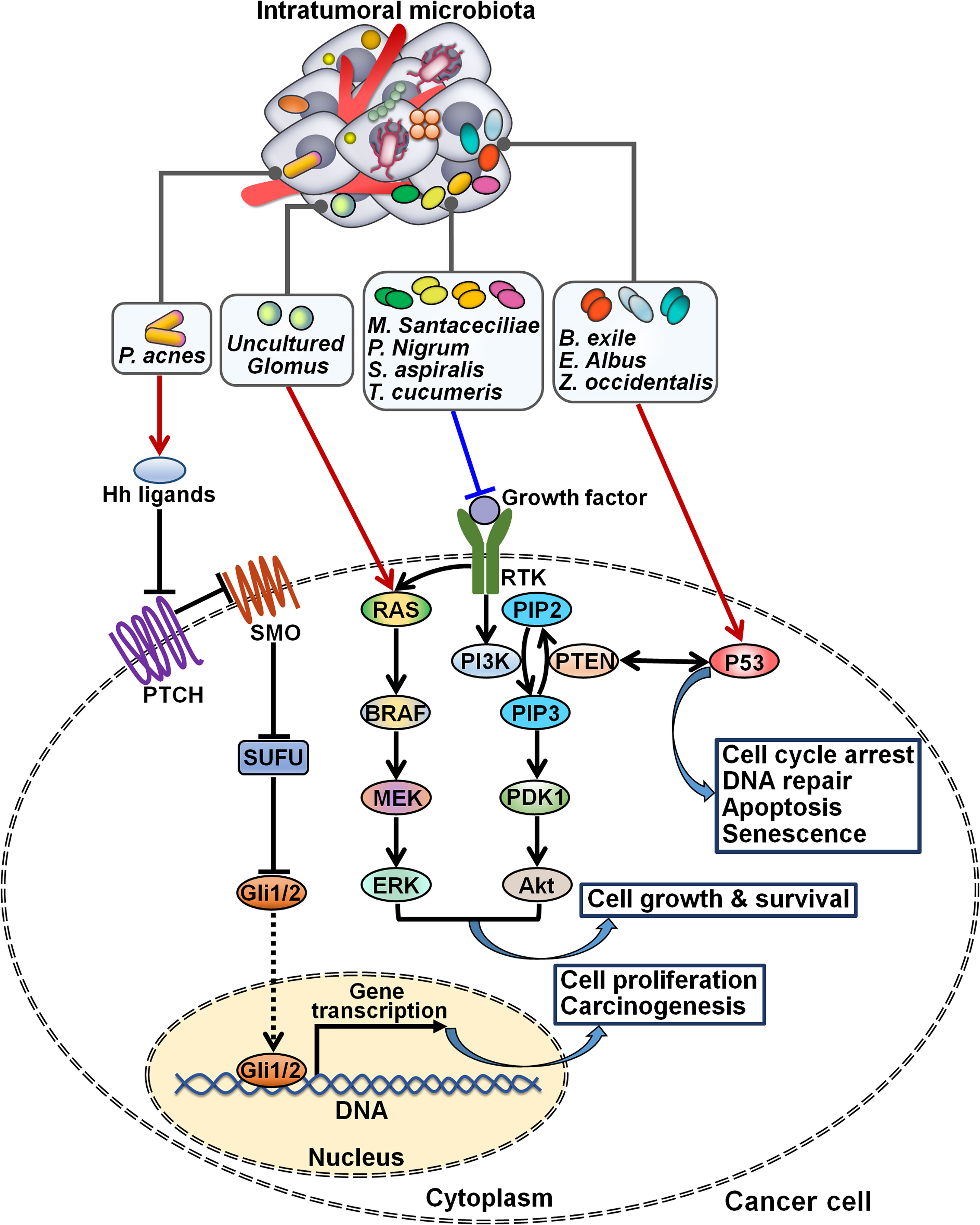
Figure 5 Regulation of intracellular signaling pathways by intratumoral microbiota. Intratumoral microbiota regulates the occurrence and development of cancer by coordinating diverse signal transduction cascades. The proinflammatory P. acnes induces carcinogenesis by activating the Hh signaling cascade. Uncultured Glomus is associated with the enhancement of BRAF kinase activity and the activation of the MAPK signaling cascade, while M. santaceciliae, P. nigrum, S. aspiralis and T. cucumeris are capable of inhibiting the PI3K/Akt pathway. B exile, E albus and Z. occidentalis act to motivate the p53 signaling cascade. Hh, Hedgehog; PTCH, patched homolog; SMO, smoothened homolog; SUFU, suppressor of fused; Gli, glioma-associated oncogene; RAS, rat sarcoma viral oncogene homolog; BRAF, B-Raf proto-oncogene serine/threonine kinase; MEK, mitogen-activated extracellular signal-regulated kinase; ERK, extracellular signal-regulated kinase; RTK, receptor tyrosine kinase; PI3K, phosphatidylinositol 3-kinase; PIP2, phosphatidylinositol-4,5-bisphosphate; PTEN, phosphatase and tensin homolog; PIP3, phosphatidylinositol-3,4,5-triphosphate; PDK1, 3-phosphoinositide-dependent kinase 1; Akt, protein kinase B.
The fungal species including Metarhizium acridum CQMa 102, Phaffia rhodozyma and S. cerevisiae YJM1338 were enriched in tumor tissues of patients with papillary thyroid carcinoma (PTC) (61). Three PTC subtypes, Classical (CPTC), Follicular Variant (FVPTC) and Tall Cell (TCPTC), showed a similar microbial composition and richness. For instance, Botrytis cinerea, Pichia cephalocereana and Trematosphaeria pertusa were overabundant in these subtypes. Rhizopus arrhizus and uncultured Uromyces were underabundant in both CPTC and TCPTC. The level of Chaetomium globosum CBS 148.51 was related to increasing pathologic stage in PTC patients. C. Albicans, Eremascus albus and Thanatephorus cucumeris were positively associated with a higher pathologic M stage, while Spiromyces aspiralis, uncultured Cryptomycota and Wickerhamiella pararugosa had a linkage with a higher pathological N stage. Further studies are required to characterize the influence of these fungi on PTC tumor metastasis. Importantly, the relative abundance of fungal microbes was related to PTC-specific oncogenic pathways. For instance, Metschnikowia santaceciliae, Pacynthium nigrum, Spriromyces aspiralis and T. cucumeris were associated with the downregulation of the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt) pathway in CPTC. Brevicellicium exile, E. albus and Zoophthora occidentalis correlated with the upregulation of the p53 signaling in TCPTC. Uncultured Glomus was linked with increased activity of BRAF kinase and the activation of the mitogen-activated protein kinase (MAPK) signaling in FVPTC. Additional investigation is needed to identify intracellular signaling pathways that mediate the tumor-promoting effects of intratumoral fungal species. The intricate signaling network underlying PTC development merits further study.
There is a growing appreciation of the effect of intratumoral microbiota on the response to anticancer therapies (Table 2). F. nucleatum, a predominant bacterium within CRC microbiota, was identified as a tumor-supportive bacterial species that was correlated with inferior patient prognosis (86). The first-line CRC chemotherapeutic drug, 5-fluorouracil (5-FU), exhibited an antagonistic activity against F. nucleatum (62). The intratumoral bacteria E. coli was resistant to 5-FU and could attenuate 5-FU toxicity toward F. nucleatum and human CRC epithelial cells (Figure 6). Mechanistically, E. coli contained a functional homolog of human dihydropyrimidine dehydrogenase (DPD) that converted 5-FU to nontoxic dihydrofluorouracil (DHFU). Thus, intratumoral microbiota had a significant impact on cancer cell sensitivity to chemotherapy. CRC patient-derived ex vivo tumor microbiota underwent community disruption upon exposure to 5-FU, characterized by an expansion of 5-FU-resistant bacteria (e.g., E. coli). The altered microbiota caused the depletion of 5-FU and thus blunted local chemotherapeutic efficacy. An improved understanding of the effect of intratumoral microbes on patient response to chemotherapy would assist in stratifying patients for combined treatment of chemotherapy with targeted antimicrobials. Further investigation into the detailed mechanisms behind intratumoral bacteria-mediated chemotherapeutic resistance are warranted.
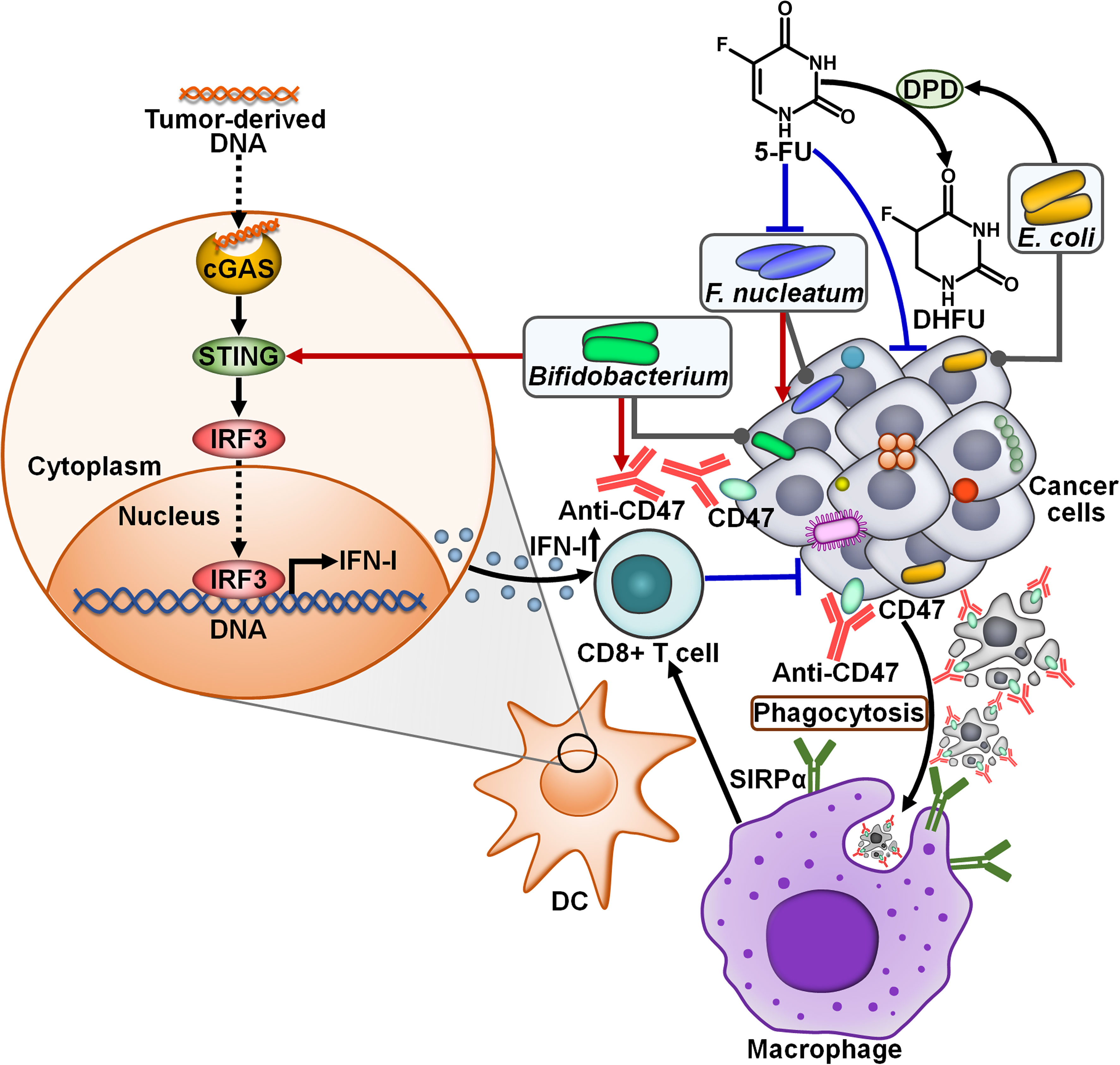
Figure 6 Impacts of intratumoral microbiota on the antitumor efficacy of cancer treatment. CD47, a surface membrane protein expressed by some cancer cells, binds to SIRPα on the surface of macrophages, contributing to impairment of phagocytic capacity. Blocking CD47 with anti-CD47 promotes phagocytosis of cancer cells by macrophages, which further initiates cytotoxic CD8+ T cell responses. Bifidobacterium can augment the antitumor effects of CD47-based therapy. In term of mechanisms, Bifidobacterium can actuate the STING signaling to enhance IFN-I production, resulting in the activation of CD8+ T cell-mediated immune responses. The intratumoral bacteria E. coli dampens the toxicity of 5-FU toward tumor-supportive F. nucleatum and cancer cells by transforming 5-FU into nontoxic DHFU. cGAS, cyclic guanosine monophosphate-adenosine monophosphate synthase; STING, stimulator of interferon genes; IRF3, interferon regulatory factor 3; IFN-I, type I interferon; DC, dendritic cell; 5-FU, 5-fluorouracil; DPD, dihydropyrimidine dehydrogenase; DHFU, dihydrofluorouracil; SIRPα, signal regulatory protein α.
Two mice models (Jax and Tac) had different gut microbiota profiles that led to distinct immune signatures (65). CRC-bearing Jax mice responded to anti-CD47 immunotherapy, while tumor-bearing Tac mice exhibited low responsiveness to CD47 blockade (63). Cohousing of tumor-bearing Jax mice with Tac mice restored the response of mice nonresponders (Tac mice) to CD47-based immunotherapy. Intriguingly, intratumoral inoculation of the antibiotic cocktail reduced the efficacy of anti-CD47 treatment in mice responders, hinting that intratumoral microbiota affected the efficacy of anti-CD47 immunotherapy. It was proposed that hypoxic environment within tumor tissues was beneficial for the colonization and growth of anaerobic commensals. Systemic or intratumoral administration of the Bifidobacterium cocktail (B. bifidum, B. breve, B. lactis and B. longum) rescued the capability of tumor inhibition by CD47 blockade in mice nonresponders. Intratumoral administration of the antibiotic cocktail blunted the therapeutic potency of Bifidobacterium-facilitated CD47 blockade. It could be concluded that Bifidobacterium had tumor-targeting ability and was essential for anti-CD47-mediated tumor inhibition effect. Moreover, specific inhibition of type I IFN (IFN-I) signaling in DCs within the TME dampened the antitumor efficacy of CD47-based therapy in mice nonresponders administrated with Bifidobacterium. The expression level of IFN-β was higher in tumor DCs derived from mice nonresponders treated with Bifidobacterium and CD47 blockade than those from mice only administrated with anti-CD47. The antibiotic cocktail-mediated removal of Bifidobacterium inside the TME attenuated IFN-β expression in tumor DCs, and also inhibited their cross-priming ability. Bifidobacterium administration failed to rescue the antitumor efficacy of CD47 blockade in T cell-deficient mice as well as in stimulator of interferon genes (STING)-knockout mice. Specific depletion of STING inside DCs also blocked the antitumor function of Bifidobacterium. Oppositely, the STING agonist enhanced the antitumor capacity of CD47 blockade in nonresponding mice. These results suggested that Bifidobacterium strengthened the antitumor effects of CD47-based therapy in a STING signaling- and T cell-dependent manner. It is likely that other anaerobic commensal microbes also have tumor-targeting potential and induce antitumor immunity. It is imperative to investigate whether these microorganisms accumulate in the TME and influence the antitumor effect of immunotherapies. Manipulation of the microbiota inside TME may represent an effective strategy to improve immunotherapeutic effectiveness. However, the pathogenicity of tumor-targeting bacteria must be adequately determined. Bifidobacterium is an anaerobic commensal bacterium with low toxicity and low possibility of residing in normal tissues, which make it a suitable tumor-targeting bacterium for clinical cancer intervention. Active bacteria can generate secondary metabolites to activate the STING signaling inside DCs. The mechanisms through which Bifidobacterium motivates the STING pathway remain to be determined. Special attention should be paid to the role of Bifidobacterium in antitumor immunity.
The core purpose of cancer immunotherapy is the persistent activation of tumor-specific T cells (87). Immune checkpoint-directed therapy can induce durable antitumor immune responses in cancer and has attracted increasing scientific and clinical interests in recent years. Intratumoral microbiota can affect the anticancer effect of immune checkpoint therapy. For instance, depletion of PDAC microbiota enhanced the efficacy of PD-1-targeted immunotherapy by increasing PD-1 expression (7). Introduction of Megasphaera into the breast cancer microenvironment yielded a better inhibitory effect on tumor growth when combined with anti-PD-1 therapy in vivo (64). Commensal Bifidobacterium promoted CD8+ T cell accumulation in the TME (65). The combination of Bifidobacterium administration and anti-PD-L1 treatment exerted a synergistic antitumor effect on melanoma. Short chain fatty acid (SCFA)-producing bacteria, including Eubacterium, Lactobacillus and Streptococcus, could improve the efficacy of anti-PD-1/PD-L1 treatment in gastrointestinal cancer (66). This effect might be attributed to the immunoregulatory function of SCFAs, which necessitates further study. In addition, B. fragilis elicited Th1 immune responses and favored the maturation of intratumoral DCs, which potentiated the therapeutic efficacy of cytotoxic T lymphocyte-associated protein 4 (CTLA-4) blockade in fibrosarcoma-bearing mice (67). Despite various studies indicating the relationship between intratumoral microbiota and immunotherapy effect, the underlying mechanisms remain equivocal. A deeper understanding of the influence of intratumoral microbiota on immunotherapy efficacy will provide a new direction for the clinical treatment of cancer.
The momentous role of intratumoral microbiota in cancer development and tumor immunity highlights its potential clinical implications in oncology. The diagnostic values of intratumoral microbiota have been explored. Wang et al. (88) revealed that head and neck squamous cell carcinoma (HNSCC)-depleted Actinobacteria/Actinomycetales/Actinomyces, Firmicutes, Lactobacillales, Rothia, Streptococcus and Veillonellales had solid predictive ability in differentiating tumor tissues from normal tissues. This intratumoral microbiota signature was associated with the clinicopathological characteristics such as age, gender, tumor stage and neoplasm histological grade. A microbial signature consisting of HCC-depleted Acidobacteriae and Parcubacteria as well as HCC-enriched Bacilli, Gammaproteobacteria and Saccharimonadia exhibited great discriminative accuracy and performance in HCC prediction (44). HCC-enriched Actinobacteriota, Firmicutes, Gammaproteobacteria, Proteobacteria and Saccharimonadia correlated with clinicopathological characteristics of HCC patients including tumor volume, cirrhosis grading (inflammation activity) and histological severity. These microbes might contribute to the onset and development of HCC. Intratumoral microbial signatures may be a valuable resource for unearthing prospective biomarkers for cancer diagnosis (Figure 7). The potential utility of specific intratumoral microbes in clinical settings should be illuminated in the future. Further study on the association between specific microbes and clinicopathological features is required to support the aforementioned results.
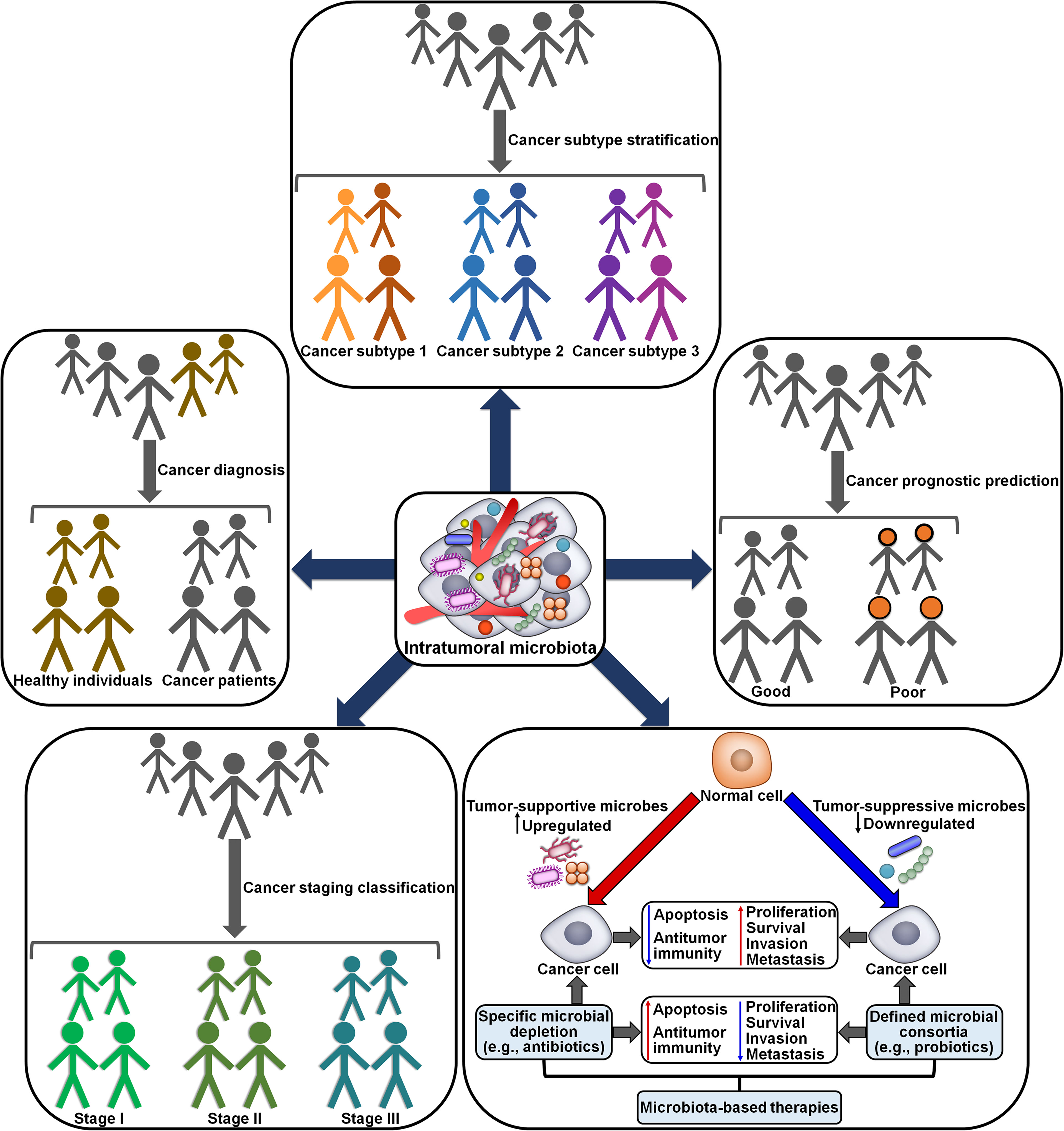
Figure 7 Potential clinical implications of intratumoral microbiota in cancer. Specific tumor-associated microbiota signatures are capable of discriminating cancer patients from healthy individuals. The microbial biomarkers also differentiate between and within subtypes and stages of cancers. Tumor-resident microbes may provide a valuable tool for cancer diagnosis, subtype stratification and staging classification. The abundance and diversity of intratumoral microbes are associated with clinical outcomes in cancer patients. Thus, intratumoral microbiota may have a promising role in cancer prognosis. Considering its important role in cancer pathogenesis, intratumoral microbiota may represent an attractive therapeutic target for cancer treatment. Microbiota modulation, such as depletion of tumor-supportive microbes and introduction of tumor-suppressive microbes, shows great potential in improving cancer treatment outcomes.
A microbial signature comprising Actinobacteria, Bacilli, Bacillales, Epsilonproteobacteria, Fusobacteria, Fusobacteriaceae, Lactobacillaceae, Negativicutes, Pasteurellales and Streptococcaceae could be a potential biomarker for subdividing different ESCA subtypes (ESCC and esophageal adenocarcinoma (EAD)) (89). ESCA-enriched Fusobacteriales positively correlated, while the microbial abundance of Lactobacillales inversely correlated with the tumor stage. Fusobacteria, present in multiple cancer types, promoted the establishment of a proinflammatory niche that supported cancer development (90, 91). Therapeutic manipulation of Fusobacteria could be a promising treatment option in cancer patients. High levels of Lactobacillus, Negativicutes and Proteobacteria reflected better prognosis, while the enrichment of Clostridiales and Fusobacteriales was associated with worse prognosis. Clostridia, Clostridiales, Fusobacteriia and Fusobacteriales might be independent prognostic biomarkers for ESCA. Orally derived Fusobacterium species including Fusobacterium animalis, F. nucleatum, Fusobacterium polymorphum and Fusobacterium vincentii were prevalent in tumor tissues of CRC patients (92). The Fusobacterium species were enriched in right-sided, microsatellite instability-high (MSI-H) and BRAF-mutant tumors. Fusobacterium might have prominent specificity for the inflamed CRC subtype. F. animalis was likely to correlate with disease progression and reduced survival in consensus molecular subtype 4 (CMS4) CRC patients, highlighting that intratumoral microbes might represent promising biomarkers for CRC subtype stratification and prognosis. GC tissues had a lower diversity of microbiota than adjacent nonmalignant tissues (93). MSI-H subtype might have a tendency toward an increased microbial diversity relative to other subtypes (diffuse and intestinal GC). The diversity of intratumoral microbiota may be a potential biomarker for the classification of GC subtypes. Further investigation of intratumoral microbial signatures will help to better understand key pathological processes and biological mechanisms during cancer pathogenesis, as well as facilitate the identification of new microbial biomarkers for cancer.
The high levels of Fusobacterium and Haemophilus correlated with the T3/T4 stage and lymphatic metastasis in patients with oral cancer (94). Bacillales and Gemella were more abundant in T1/T2 stage patients and the non-lymphatic metastasis cases. These results demonstrated that intratumoral microbiota could be connected with oral cancer progression and metastasis. Future work should concentrate on delineating the characteristics of the microbiota in metastatic lymph nodes and the mechanisms through which intratumoral microbes affect lymphatic metastasis. Acinetobacter, Pseudomonas, Ralstonia, Rhodococcus and Sphingomonas were the dominant bacteria in PTC tissues (95). PTC patients with advanced lesions had a higher microbiota diversity than patients with mild lesions. The abundance of Pseudomonas, Rhodococcus and Sphingomonas was high in mild lesion cases. Granulicatella and Streptococcus were more abundant in patients with advanced lesions than those with mild lesions. The eight-genera microbiota signature that was composed of g_norank_F_norank_o_Coriobacteriales, g_unclassified_o_Rhizobiales, Granulicatella, Haemophilus, Pseudomonas, Rhodococcus, Sphingomonas and Streptococcus had the potential to discriminate PTC invasion status. Intratumoral microbes potentially interacted with thyroid hormones and autoimmune antibodies, which remodeled the TME, contributing to PTC invasion and progression. The mechanism by which tumor-resident microbiota affects PTC development and outcome necessitates substantial attention. The increased abundance of Blumeria graminis f. sp. Hordei was strikingly associated with early cancer pathological stages in HNSCC, while the decreased number of Aspergillus flavus, Coccidioides immitis RS and Gaeumannomyces tritici R3-111a-1 correlated with the absence of perineural invasion (96). The high level of Podospora anserina S mat+ and S. cerevisiae EC1118 was linked with tumor neoplasm presence and advanced pathological N-stage. These altered microbes might give rise to cancer metastasis and progression. Moreover, Inosperma fulvum, Phlyctochytrium arcticum and uncultured Cryptomycota had a relationship with clinical characteristics of patients with HPV+ HNSCC. Punctularia strigosozonata HHB-11173 SS5 was linked to the upregulation of oncogenic pathways and poorer patient prognosis. These fungal microbes might be clinically relevant to patient outcome, but their genuine roles in HNSCC development needs further study.
Pathogenic oral microbes, including Firmicutes, Fusobacteriia, Fusobacteriales, Fusobacteriaceae and Fusobacterium, exhibited great predictive efficacy in OSCC diagnosis and prognosis (97). These microorganisms correlated with clinical characteristics of OSCC, including histological grade and tumor stage. Intratumoral microorganisms could be used as diagnostic and prognostic biomarkers for OSCC. The association between oral microbiome and clinical parameters of OSCC necessitates further clinical studies. H. parainfluenzae might be negatively associated with the first-line treatment outcomes in NSCLC patients (98). Staphylococcus crista and Staphylococcus haemolyticus were related to longer progression-free survival (PFS). Serratia marcescens was associated with better OS while Corynebacterium jergeri and H. parainfluenzae were associated with poorer OS. It is still obscure how intratumoral microbiota influences the treatment and prognosis of lung cancer. A microbiome analysis on biopsy samples from different sites of patients with distinct primary tumors (breast, CRC and lung) showed that Acinetobacter, Burkholderia, Corynebacterium, Cutibacterium, Flavobacterium, Pelomonas, Rheinheimera, Sphingobium, Staphylococcus and Streptococcus were the ten most represented genera in metastatic tumor tissues (99). The richness of intratumoral microbiota was significantly related to OS and PFS in cancer patients. Further experiments to determine the clinical value of intratumoral microbiota are required. The relative abundance of intratumoral Pseudomonas was significantly higher in PLC patients with LTS than in those with STS (17). The level of Pseudomonas was positively correlated with clinical prognosis in PLC patients. This bacterium might represent a prognostic biomarker for PLC. Tumor microbial diversity was positively correlated with OS in patients with resected PDAC (53). The increased number of Pseudoxanthomonas, Saccharopolyspora and Streptomyces was associated with better clinical outcomes in PDAC patients. Another study showed that the composition and diversity of intratumoral microbiota were apparently distinct between long-term and short-term PDAC survivors (64). The high abundance of Megasphaera and Sphingomonas positively correlated with the prolonged OS. The high level of opportunistic pathogen Clostridium was related to shortened survival time. These dysregulated microbes might be promising prognostic biomarkers for PDAC. The high load of intratumoral F. nucleatum was markedly related to better OS, DFS and MFS in patients with anal squamous cell carcinoma (ASCC) (100). In addition, the association between intratumoral fungi and patient survival and therapy response in different cancer types was also documented (9). The oncogenic fungus, M. globosa, was correlated with shorter OS in breast cancer patients (49). The Phaeosphaeria genus was dramatically linked with shorter PFS in patients with ovarian cancer. The high amount of Cladosporium might be negatively associated with immunotherapy response in patients with metastatic melanoma. Thus, intratumoral fungi hold the promise as potential biomarkers and therapeutic targets in cancer. The clinical implication of intratumoral fungi remains to be detected in larger cohorts. Given a paucity of publicly-available fungal genomes, a great deal of investigative work is required to better understand the fungal functional repertoires in cancer.
In light of recent advances in the field of intratumor microbiome, microbiota-based therapies are considered to hold great application potential (Figure 7). Many clinical trials exploring the therapeutic benefits of wild-type/modified microorganisms in cancer patients are now underway (Table 3). A randomized, placebo-controlled, phase I dose-escalation trial in 26 patients with advanced PC indicated the safety, immunogenicity and tolerability of an oral T cell vaccine VXM01, which was composed of live attenuated Salmonella typhi carrying a eukaryotic expression plasmid encoding vascular endothelial growth factor receptor 2 (VEGFR2) (101). Another phase I clinical trial demonstrated that the VXM01 vaccine was efficacious in inducing VEGFR2-specific effector T cell responses and attenuating tumor perfusion in 30 patients with advanced PC (102).
CRS-207 is a live attenuated Listeria monocytogenes expressing mesothelin (103). In a multicenter, open-label phase Ib study, CRS-207 infusion in combined with chemotherapy decreased tumor size in 35 patients with malignant pleural mesothelioma (MPM). CRS-207 treatment induced marked changes in the local TME, as evidenced by elevated infiltration levels of T cells, DCs and NK cells, an increased ratio of CD8+ T cells to regulatory T cells (Tregs), and the transformation of M2-like macrophages into M1-like macrophages (103). Accordingly, CRS-207 combined with chemotherapy resulted in enhanced antitumor immunity and induced objective tumor responses in MPM patients. A multicenter, randomized, phase II trial involving 90 PC patients indicated that administration of CRS-207 plus cyclophosphamide (Cy)/the cancer vaccine GVAX exhibited favorable safety profile (104). The combined treatment promoted mesothelin-specific CD8 T cell responses and prolonged survival of PC patients. However, a randomized, controlled phase IIb clinical trial in patients with advanced PC showed that CRS-207 in combination with Cy/GVAX did not improve OS over standard chemotherapy (105). The engineered Listeria strain ADXS11-001, which expresses the HPV16-E7 fusion protein to target HPV-positive tumors, could activate antigen-specific T cell responses and lead to the increased serum level of CCL22 in patients with HPV-associated oropharyngeal cancer (106). The combination of ADXS11-001 with standard chemoradiation was proven to be well tolerable in patients with anal cancer (107). All nine patients showed complete clinical response, and eight patients (89%) exhibited no evidence of disease at a median follow-up of 42 months. ADXS31-142 is an attenuated L. monocytogenes-based immunotherapy targeting prostate-specific antigen (PSA) (108). An open-label phase I/II KEYNOTE-046 study including 50 patients with metastatic castration-resistant prostate cancer (mCRPC) preliminarily verified the safety and tolerability of ADXS31-142 combined with pembrolizumab. This combination treatment might improve OS in mCRPC patients with visceral metastasis, which warrants additional evaluation in future studies. ADXS31-164, a highly attenuated, recombinant L. monocytogenes expressing a chimeric human HER2/neu fusion protein, induced robust T cell immune responses, retarded tumor growth and extended OS in animal models (109, 111). ADXS31-164 might have significant translational relevance for patients with HER2/neu-positive cancers. An open-label, multicenter, phase I study demonstrated that a single intratumoral inoculation of nontoxic Clostridium novyi (C. novyi-NT) reduced the size of the injected tumor in nine patients with injectable, treatment-refractory solid tumors (41%) (110). C. novyi-NT administration induced a temporary systemic cytokine response and potentiated tumor-specific T cell responses. However, C. novyi-NT treatment might raise safety concerns including limb abscess, pathological fracture, rash and respiratory insufficiency. Thus, it is of great importance to assess the safety and therapeutic value of C. novyi-NT. Future studies with larger cohorts will be required to verify the protective effect of these antitumor microbiota therapies. An in-depth investigation of the immune alterations in the TME will contribute to uncovering the mechanisms of action of microbiota-centered therapies.
Intratumoral microbiota is an emerging topic that has been suggested as an important component of the tumor ecosystem. Unlike its intestinal counterpart, the field of intratumoral microbiota is still in its nascency. Compelling evidence has verified the universal presence of intratumoral microbiota in distinct types of cancers. The origin and colonization mechanisms of intratumoral microbiota are poorly understood. Intratumoral microbes may originate from oral and intestinal microbiota. Adjacent normal tissues, mucosal organs and the circulation system are also deemed as potential sources of intratumoral microbiota. Nevertheless, how intratumoral microbes enter tumor cells and the TME is an important question that emerges. The Gal-GalNAc-binding lectin Fap2 of F. nucleatum may mediate its entry into Gal-GalNAc-overexpressing cancer cells (39). Whether the interaction between microbes and cancer cells forms the basis for their invasion needs further validation. The comparison of microbial compositions in potential sources (e.g., oral cavity, intestine and mucosal organs) and tumors will be conducive to ascertaining the genuine origin of tumor microbiota in different tumors. In vivo tracking of live microbes may assist researches in uncovering the dynamics of their migration process. The integrated 3D quantitative in situ intratumoral microbiota imaging method provided the direct evidence of the presence of bacteria within human glioma samples (112). This approach helped to visualize bacteria in their native context in host with single-cell resolution. The 3D in situ quantitative imaging technology may be a practical approach to examine the complex interplay among intratumoral microbiota, cancer cells, and the TME. Due to the relatively low microbial biomass within tumors, characterizing tumor microbiota remains a challenge. Undoubtedly, the advances in detection and functional analysis methods (e.g., targeted microbial imaging and multi-omics techniques) will accelerate the discovery of the compositional and functional profiles of intratumor microbial communities. Environmental contamination critically hinders the study of intratumor microbiota. Both experimental and bioinformatic approaches must be developed to exclude inaccurate data. It is intriguing whether the existence of multitudinous microbes within the tumor is a mechanism facilitating their own survival. In addition, the potential impacts of factors such as dietary patterns, drug use, sleep and stress on the composition and local diversity of tumor microbiota are worthy of further study.
The effect of intratumoral microbiota on cancer pathogenesis is an appealing aspect of the host-microbiota interaction. Several significant mechanisms underpinning the involvement of intratumoral microbiota in cancer development have been revealed. Intratumor microbiota can coordinate host genetic mutation and facilitate cancer cell dissemination and metastasis. Specific commensal microbes enhance antitumor immunity through modulation of tumor-associated neutrophils, antitumor M1 macrophages, NK cells and effector T cells, while certain microbes are capable of impelling immunosuppression through diverse mechanisms that affect the function of immunosuppressive cells. Tumor-associated microorganisms dominate cancer cell metabolism and oncogenic signal transduction pathways. Alternative mechanisms mediating the crosstalk between intratumor microbiota and cancer must exist and have yet to be determined. A great deal of work remains to be done to adequately unveil the complex mechanisms through which tumor microbiota affects various aspects of cancer biology. Tumor microbiota-derived metabolites, such as acetate, butyrate and deoxycholic acid, exerted tumor-promoting effects (113–115), which may constitute a crucial mechanism responsible for cancer pathogenesis. Microbe-derived peptides were found to be presented on the surface of melanoma cells, enabling them to be detected by T cells (116). It is inferred that intratumoral microbial peptides widely exist within the TME and thus actuate robust local and systemic immune responses. The role of microbial products in cancer development is therefore a research priority. The interplay between tumor microbiota and the TME has been an important research topic in oncoimmunology. As distinct intratumoral microbes polarize conflictingly different types of the immune responses that either drive or impede cancer progression, tumor microbiota may act in a tumor type- and context-dependent manner as a tumor promoter or suppressor. It is critical to define in which direction the subtle balance between intratumoral microbiota and host immune system is genuinely tipped for specific cancer types. Collectively, more in vivo studies will be mandatory to comprehensively characterize the reciprocal interaction between tumor microbiota and the host immune system, which will provide a basis and guidance for further research on this field.
Therapeutic resistance has been a principal obstacle to the success of cancer treatment (117). For instance, a substantial proportion of patients with melanoma and NSCLC demonstrated primary resistance to ICI (118–121). Hence, it is necessary to unearth novel strategies to improve treatment response in cancer patients. Tumor microbiota has the ability to alter therapeutic responsiveness in cancer. On the one hand, intratumoral microorganisms may synergize with anticancer therapeutic, especially immunotherapy, as intratumoral microbiota contributes to increased foreignness of the tumor, strengthening antitumor immune responses (122). On the other hand, intratumoral microorganisms may attenuate the response to anticancer therapies by directly acting on therapeutic agents or lessening antitumor immunity. Harnessing intratumoral microbiota may be helpful in reinforcing the therapeutic effect of existing anticancer regimen. Currently, the mechanisms through which tumor commensal microbes affect the antitumor efficacy of cancer treatment are yet to be fully elucidated. It is of great importance to find out how tumor microbiota protects cancer cells against tumor-killing effects by conventional therapies including chemotherapy, radiotherapy and immunotherapy.
Manipulation of tumor microbiota via targeted reconstitution or improvement of current therapies with supplementation of microbe-derived products may present key elements of future intervention. Several interventional approaches, such as FMT, defined microbial consortium, dietary intervention, antibiotic, prebiotic and probiotic-based approaches, may have the potential to change the composition of gut and tumor microbiota. Although previous clinical trials have positive results, the clinical translation of microbial treatment approaches still faces a myriad of challenges due to the scarcity of ample evidence. It must be noted that the terminal therapeutic effect of microbiota-centered treatments mainly lies on the functionality of tumor microbiota and its impact on the local/systemic immunity. Accordingly, exploring tumor microbiota and its multifaceted modes of action in cancer will offer opportunities for the discovery of new prevention and treatment modalities. Further investigation of microbiota-based treatment strategies aimed at regulating patient tumor microbes in larger patient cohorts represents the ideal next step. The identification and verification of common treatment-associated intratumor microbial signatures in different cancers will greatly advance development of off-the-shelf therapeutic modalities that enable broad applicability across diverse cancer types. Given the heterogeneity of intratumoral microbiota, personalized cancer treatments will be beneficial owing to their high therapeutic efficacy and precise targeting potential. Microbiota modulation has exhibited potential of reinforcing the efficacy of existing cancer treatments and reducing side effects. It is proposed that normalizing the microbiota within the tumor and introducing specific microbes into the TME represent feasible and attractive approaches to optimize the outcome of anticancer therapies (37). Notably, the effects of microbiota-based therapies may vary depending on tumor type, availability of other treatments and the immune status of cancer patients. More clinical studies are needed to determine their real value in assisting cancer intervention. Due to the crosstalk between tumor and gut microbiota, manipulating gut microbiota to treat cancer has been attempted. Nevertheless, it is arduous to disclose the causal mechanisms of microbiota-modulating treatments in complex in vivo environments. Further research efforts should be devoted to exploiting microbiota-related knowledge toward the development of improved cancer treatments.
In summary, intratumor microbiota, an integral component of the TME, has emerged as key players in carcinogenesis and cancer development. The potential clinical utility of intratumor microbiota in cancer management has drawn extensive attention in recent years. Thus, intratumor microbiota has become a new frontier in cancer research and a valuable resource of promising targets for personalized cancer care. However, there are still many challenges to be addressed in this field. Progress on all fronts is essential to move intratumor microbiota-related discoveries to the forefront of medical practice.
MW: Conceptualization, Supervision, Visualization, Writing – original draft. FY: Investigation, Resources, Writing – review & editing. PL: Conceptualization, Supervision, Writing – review & editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the grant from the Natural Science Foundation of Shandong Province, China (No. ZR2021MH018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Cogdill AP, Gaudreau PO, Arora R, Gopalakrishnan V, Wargo JA. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol (2018) 39(11):900–20. doi: 10.1016/j.it.2018.09.007
3. Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet (1983) 1(8336):1273–5. doi: 10.1016/S0140-6736(83)92719-8
4. Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science (2006) 313(5788):848–51. doi: 10.1126/science.1127059
5. Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, et al. Lack of commensal flora in helicobacter pylori-infected ins-gas mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology (2011) 140(1):210–20. doi: 10.1053/j.gastro.2010.09.048
6. Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: microbes inflame tumors. Cell (2014) 157(4):776–83. doi: 10.1016/j.cell.2014.03.006
7. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discovery (2018) 8(4):403–16. doi: 10.1158/2159-8290.CD-17-1134
8. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (2020) 368(6494):973–80. doi: 10.1126/science.aay9189
9. Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell (2022) 185(20):3789–806 e17. doi: 10.1016/j.cell.2022.09.005
10. Ohadian Moghadam S, Mansori K, Nowroozi MR, Afshar D, Abbasi B, Nowroozi A. Association of human papilloma virus (Hpv) infection with oncological outcomes in urothelial bladder cancer. Infect Agents Cancer (2020) 15:52. doi: 10.1186/s13027-020-00318-3
11. Laniewski P, Barnes D, Goulder A, Cui H, Roe DJ, Chase DM, et al. Linking cervicovaginal immune signatures, hpv and microbiota composition in cervical carcinogenesis in non-hispanic and hispanic women. Sci Rep (2018) 8(1):7593. doi: 10.1038/s41598-018-25879-7
12. Cantalupo PG, Katz JP, Pipas JM. Viral sequences in human cancer. Virology (2018) 513:208–16. doi: 10.1016/j.virol.2017.10.017
13. El-Zimaity H, Di Pilato V, Novella Ringressi M, Brcic I, Rajendra S, Langer R, et al. Risk factors for esophageal cancer: emphasis on infectious agents. Ann New York Acad Sci (2018) 1434(1):319–32. doi: 10.1111/nyas.13858
14. Wang Y, Yuan Y, Gu D. Hepatitis B and C virus infections and the risk of biliary tract cancers: A meta-analysis of observational studies. Infect Agents Cancer (2022) 17(1):45. doi: 10.1186/s13027-022-00457-9
15. Dumont-Leblond N, Veillette M, Racine C, Joubert P, Duchaine C. Non-small cell lung cancer microbiota characterization: prevalence of enteric and potentially pathogenic bacteria in cancer tissues. PloS One (2021) 16(4):e0249832. doi: 10.1371/journal.pone.0249832
16. Zhang M, Zhang Y, Han Y, Zhao X, Sun Y. Lung microbiota features of stage iii and iv non-small cell lung cancer patients without lung infection. Trans Cancer Res (2022) 11(2):426–34. doi: 10.21037/tcr-22-92
17. Qu D, Wang Y, Xia Q, Chang J, Jiang X, Zhang H. Intratumoral microbiome of human primary liver cancer. Hepatol Commun (2022) 6(7):1741–52. doi: 10.1002/hep4.1908
18. Wang J, Wang Y, Li Z, Gao X, Huang D. Global analysis of microbiota signatures in four major types of gastrointestinal cancer. Front Oncol (2021) 11:685641. doi: 10.3389/fonc.2021.685641
19. Kosumi K, Hamada T, Koh H, Borowsky J, Bullman S, Twombly TS, et al. The amount of bifidobacterium genus in colorectal carcinoma tissue in relation to tumor characteristics and clinical outcome. Am J Pathol (2018) 188(12):2839–52. doi: 10.1016/j.ajpath.2018.08.015
20. Chakladar J, Kuo SZ, Castaneda G, Li WT, Gnanasekar A, Yu MA, et al. The pancreatic microbiome is associated with carcinogenesis and worse prognosis in males and smokers. Cancers (2020) 12(9):2672. doi: 10.3390/cancers12092672
21. Wang J, Li X, Wu X, Wang Z, Zhang C, Cao G, et al. Uncovering the microbiota in renal cell carcinoma tissue using 16s rrna gene sequencing. J Cancer Res Clin Oncol (2021) 147(2):481–91. doi: 10.1007/s00432-020-03462-w
22. Lam KC, Vyshenska D, Hu J, Rodrigues RR, Nilsen A, Zielke RA, et al. Transkingdom network reveals bacterial players associated with cervical cancer gene expression program. PeerJ (2018) 6:e5590. doi: 10.7717/peerj.5590
23. Cai HZ, Zhang H, Yang J, Zeng J, Wang H. Preliminary assessment of viral metagenome from cancer tissue and blood from patients with lung adenocarcinoma. J Med Virol (2021) 93(8):5126–33. doi: 10.1002/jmv.26887
24. Mollerup S, Asplund M, Friis-Nielsen J, Kjartansdottir KR, Fridholm H, Hansen TA, et al. High-throughput sequencing-based investigation of viruses in human cancers by multienrichment approach. J Infect Dis (2019) 220(8):1312–24. doi: 10.1093/infdis/jiz318
25. An J, Kim D, Oh B, Oh YJ, Song J, Park N, et al. Comprehensive characterization of viral integrations and genomic aberrations in hbv-infected intrahepatic cholangiocarcinomas. Hepatology (2022) 75(4):997–1011. doi: 10.1002/hep.32135
26. Enokida T, Moreira A, Bhardwaj N. Vaccines for immunoprevention of cancer. J Clin Invest (2021) 131(9):e146956. doi: 10.1172/JCI146956
27. Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of helicobacter pylori caga into gastric epithelial cells by type iv secretion. Science (2000) 287(5457):1497–500. doi: 10.1126/science.287.5457.1497
28. Bai R, Lv Z, Xu D, Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. biomark Res (2020) 8:34. doi: 10.1186/s40364-020-00209-0
29. Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology (2018) 155(2):529–41 e5. doi: 10.1053/j.gastro.2018.04.018
30. Wang IK, Lai HC, Yu CJ, Liang CC, Chang CT, Kuo HL, et al. Real-time pcr analysis of the intestinal microbiotas in peritoneal dialysis patients. Appl Environ Microbiol (2012) 78(4):1107–12. doi: 10.1128/AEM.05605-11
31. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol (2012) 10(8):575–82. doi: 10.1038/nrmicro2819
32. O’Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol (2016) 7:925. doi: 10.3389/fmicb.2016.00925
33. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science (2017) 357(6356):1156–60. doi: 10.1126/science.aah5043
34. Longhi G, van Sinderen D, Ventura M, Turroni F. Microbiota and cancer: the emerging beneficial role of bifidobacteria in cancer immunotherapy. Front Microbiol (2020) 11:575072. doi: 10.3389/fmicb.2020.575072
35. Thorell K, Bengtsson-Palme J, Liu OH, Palacios Gonzales RV, Nookaew I, Rabeneck L, et al. In vivo analysis of the viable microbiota and helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infection Immun (2017) 85(10):e00031–17. doi: 10.1128/IAI.00031-17
36. Kanao H, Enomoto T, Kimura T, Fujita M, Nakashima R, Ueda Y, et al. Overexpression of lamp3/tsc403/dc-lamp promotes metastasis in uterine cervical cancer. Cancer Res (2005) 65(19):8640–5. doi: 10.1158/0008-5472.CAN-04-4112
37. Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, et al. Microbiota in tumors: from understanding to application. Advanced Sci (2022) 9(21):e2200470. doi: 10.1002/advs.202200470
38. Yang L, Li A, Wang Y, Zhang Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal transduction targeted Ther (2023) 8(1):35. doi: 10.1038/s41392-022-01304-4
39. Abed J, Emgard JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galnac. Cell Host Microbe (2016) 20(2):215–25. doi: 10.1016/j.chom.2016.07.006
40. Bertocchi A, Carloni S, Ravenda PS, Bertalot G, Spadoni I, Lo Cascio A, et al. Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell (2021) 39(5):708–24 e11. doi: 10.1016/j.ccell.2021.03.004
41. Baban CK, Cronin M, O’Hanlon D, O’Sullivan GC, Tangney M. Bacteria as vectors for gene therapy of cancer. Bioengineered bugs (2010) 1(6):385–94. doi: 10.4161/bbug.1.6.13146
42. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
43. Chen Y, Liu B, Wei Y, Kuang DM. Influence of gut and intratumoral microbiota on the immune microenvironment and anti-cancer therapy. Pharmacol Res (2021) 174:105966. doi: 10.1016/j.phrs.2021.105966
44. Huang JH, Wang J, Chai XQ, Li ZC, Jiang YH, Li J, et al. The intratumoral bacterial metataxonomic signature of hepatocellular carcinoma. Microbiol Spectr (2022) 10(5):e0098322. doi: 10.1128/spectrum.00983-22
45. Liu W, Zhang X, Xu H, Li S, Lau HC, Chen Q, et al. Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterology (2021) 160(7):2395–408. doi: 10.1053/j.gastro.2021.02.020
46. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell (2022) 185(8):1356–72 e26. doi: 10.1016/j.cell.2022.02.027
47. Lee JA, Yoo SY, Oh HJ, Jeong S, Cho NY, Kang GH, et al. Differential immune microenvironmental features of microsatellite-unstable colorectal cancers according to fusobacterium nucleatum status. Cancer immunology immunother (2021) 70(1):47–59. doi: 10.1007/s00262-020-02657-x
48. Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, et al. Periodontitis pathogen porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes (2022) 14(1):2073785. doi: 10.1080/19490976.2022.2073785
49. Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives il-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell (2022) 40(2):153–67 e11. doi: 10.1016/j.ccell.2022.01.003
50. Neuzillet C, Marchais M, Vacher S, Hilmi M, Schnitzler A, Meseure D, et al. Prognostic value of intratumoral fusobacterium nucleatum and association with immune-related gene expression in oral squamous cell carcinoma patients. Sci Rep (2021) 11(1):7870. doi: 10.1038/s41598-021-86816-9
51. Perry LM, Cruz SM, Kleber KT, Judge SJ, Darrow MA, Jones LB, et al. Human soft tissue sarcomas harbor an intratumoral viral microbiome which is linked with natural killer cell infiltrate and prognosis. J immunother Cancer (2023) 11(1):e004285. doi: 10.1136/jitc-2021-004285
52. Li X, Qi M, He K, Liu H, Yan W, Zhao L, et al. Neospora caninum inhibits tumor development by activating the immune response and destroying tumor cells in a B16f10 melanoma model. Parasites Vectors (2022) 15(1):332. doi: 10.1186/s13071-022-05456-8
53. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell (2019) 178(4):795–806 e12. doi: 10.1016/j.cell.2019.07.008
54. Wong LM, Shende N, Li WT, Castaneda G, Apostol L, Chang EY, et al. Comparative analysis of age- and gender-associated microbiome in lung adenocarcinoma and lung squamous cell carcinoma. Cancers (2020) 12(6):1447. doi: 10.3390/cancers12061447
55. Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature (2022) 611(7937):810–7. doi: 10.1038/s41586-022-05435-0
56. Noguti J, Chan AA, Bandera B, Brislawn CJ, Protic M, Sim MS, et al. Both the intratumoral immune and microbial microenvironment are linked to recurrence in human colon cancer: results from a prospective, multicenter nodal ultrastaging trial. Oncotarget (2018) 9(34):23564–76. doi: 10.18632/oncotarget.25276
57. Peng R, Liu S, You W, Huang Y, Hu C, Gao Y, et al. Gastric microbiome alterations are associated with decreased cd8+ Tissue-resident memory T cells in the tumor microenvironment of gastric cancer. Cancer Immunol Res (2022) 10(10):1224–40. doi: 10.1158/2326-6066.CIR-22-0107
58. Chai X, Wang J, Li H, Gao C, Li S, Wei C, et al. Intratumor microbiome features reveal antitumor potentials of intrahepatic cholangiocarcinoma. Gut Microbes (2023) 15(1):2156255. doi: 10.1080/19490976.2022.2156255
59. Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y, et al. Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes (2023) 15(1):2166700. doi: 10.1080/19490976.2023.2166700
60. Huang Q, Wei X, Li W, Ma Y, Chen G, Zhao L, et al. Endogenous propionibacterium acnes promotes ovarian cancer progression via regulating hedgehog signalling pathway. Cancers (2022) 14(21):5178. doi: 10.3390/cancers14215178
61. John D, Yalamarty R, Barakchi A, Chen T, Chakladar J, Li WT, et al. Transcriptomic analysis reveals dysregulation of the mycobiome and archaeome and distinct oncogenic characteristics according to subtype and gender in papillary thyroid carcinoma. Int J Mol Sci (2023) 24(4):3148. doi: 10.3390/ijms24043148
62. LaCourse KD, Zepeda-Rivera M, Kempchinsky AG, Baryiames A, Minot SS, Johnston CD, et al. The cancer chemotherapeutic 5-fluorouracil is a potent fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep (2022) 41(7):111625. doi: 10.1016/j.celrep.2022.111625
63. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates cd47-based immunotherapy via sting signaling. J Exp Med (2020) 217(5):e20192282. doi: 10.1084/jem.20192282
64. Huang Y, Zhu N, Zheng X, Liu Y, Lu H, Yin X, et al. Intratumor microbiome analysis identifies positive association between megasphaera and survival of chinese patients with pancreatic ductal adenocarcinomas. Front Immunol (2022) 13:785422. doi: 10.3389/fimmu.2022.785422
65. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-pd-L1 efficacy. Science (2015) 350(6264):1084–9. doi: 10.1126/science.aac4255
66. Peng Z, Cheng S, Kou Y, Wang Z, Jin R, Hu H, et al. The gut microbiome is associated with clinical response to anti-pd-1/pd-L1 immunotherapy in gastrointestinal cancer. Cancer Immunol Res (2020) 8(10):1251–61. doi: 10.1158/2326-6066.CIR-19-1014
67. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by ctla-4 blockade relies on the gut microbiota. Science (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
68. Kadosh E, Snir-Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, et al. The gut microbiome switches mutant P53 from tumour-suppressive to oncogenic. Nature (2020) 586(7827):133–8. doi: 10.1038/s41586-020-2541-0
69. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet (2021) 53(2):156–65. doi: 10.1038/s41588-020-00763-1
70. Triner D, Devenport SN, Ramakrishnan SK, Ma X, Frieler RA, Greenson JK, et al. Neutrophils restrict tumor-associated microbiota to reduce growth and invasion of colon tumors in mice. Gastroenterology (2019) 156(5):1467–82. doi: 10.1053/j.gastro.2018.12.003
71. Eruslanov EB, Bhojnagarwala PS, Quatromoni JG, Stephen TL, Ranganathan A, Deshpande C, et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J Clin Invest (2014) 124(12):5466–80. doi: 10.1172/JCI77053
72. Chen T, Li Q, Wu J, Wu Y, Peng W, Li H, et al. Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a tlr4-dependent mechanism. Cancer immunology immunother (2018) 67(10):1635–46. doi: 10.1007/s00262-018-2233-x
73. Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κb, and up-regulating expression of microrna-21. Gastroenterology (2017) 152(4):851–66.e24. doi: 10.1053/j.gastro.2016.11.018
74. Heymann CJF, Bobin-Dubigeon C, Muñoz-Garcia J, Cochonneau D, Ollivier E, Heymann MF, et al. Lipopolysaccharide-binding protein expression is associated to the metastatic status of osteosarcoma patients. J Bone Oncol (2022) 36:100451. doi: 10.1016/j.jbo.2022.100451
75. Zhou Y, Zhang T, Wang X, Wei X, Chen Y, Guo L, et al. Curcumin modulates macrophage polarization through the inhibition of the toll-like receptor 4 expression and its signaling pathways. Cell Physiol Biochem (2015) 36(2):631–41. doi: 10.1159/000430126
76. Qiao H, Tan XR, Li H, Li JY, Chen XZ, Li YQ, et al. Association of intratumoral microbiota with prognosis in patients with nasopharyngeal carcinoma from 2 hospitals in China. JAMA Oncol (2022) 8(9):1301–9. doi: 10.1001/jamaoncol.2022.2810
77. Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, et al. The necrosome promotes pancreatic oncogenesis via cxcl1 and mincle-induced immune suppression. Nature (2016) 532(7598):245–9. doi: 10.1038/nature17403
78. He Y, Zhang Q, Yu X, Zhang S, Guo W. Overview of microbial profiles in human hepatocellular carcinoma and adjacent nontumor tissues. J Trans Med (2023) 21(1):68. doi: 10.1186/s12967-023-03938-6
79. Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science (2012) 338(6103):120–3. doi: 10.1126/science.1224820
80. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-derived nitrate boosts growth of E. Coli Inflamed Gut. Sci (2013) 339(6120):708–11. doi: 10.1126/science.1232467
81. Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates muc2 mucin and tumor necrosis factor alpha. Infection Immun (2011) 79(7):2597–607. doi: 10.1128/IAI.05118-11
82. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer (2013) 13(11):759–71. doi: 10.1038/nrc3611
83. Xie X, Yang M, Ding Y, Chen J. Microbial infection, inflammation and epithelial ovarian cancer. Oncol Lett (2017) 14(2):1911–9. doi: 10.3892/ol.2017.6388
84. Chang L, Xiao W, Yang Y, Li H, Xia D, Yu G, et al. Pseudomonas aeruginosa-mannose-sensitive hemagglutinin inhibits epidermal growth factor receptor signaling pathway activation and induces apoptosis in bladder cancer cells in vitro and in vivo. Urologic Oncol (2014) 32(1):36 e11–8. doi: 10.1016/j.urolonc.2013.02.013
85. Kim JE, Phan TX, Nguyen VH, Dinh-Vu HV, Zheng JH, Yun M, et al. Salmonella typhimurium suppresses tumor growth via the pro-inflammatory cytokine interleukin-1beta. Theranostics (2015) 5(12):1328–42. doi: 10.7150/thno.11432
86. Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell (2017) 170(3):548–63 e16. doi: 10.1016/j.cell.2017.07.008
87. Li SY, Liu Y, Xu CF, Shen S, Sun R, Du XJ, et al. Restoring anti-tumor functions of T cells via nanoparticle-mediated immune checkpoint modulation. J Controlled release (2016) 231:17–28. doi: 10.1016/j.jconrel.2016.01.044
88. Wang Y, Wang Y, Wang J. A comprehensive analysis of intratumor microbiome in head and neck squamous cell carcinoma. Eur Arch oto-rhino-laryngology (2022) 279(8):4127–36. doi: 10.1007/s00405-022-07284-z
89. Wang Y, Guo H, Gao X, Wang J. The intratumor microbiota signatures associate with subtype, tumor stage, and survival status of esophageal carcinoma. Front Oncol (2021) 11:754788. doi: 10.3389/fonc.2021.754788
90. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe (2013) 14(2):207–15. doi: 10.1016/j.chom.2013.07.007
91. Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. Breast cancer colonization by fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun (2020) 11(1):3259. doi: 10.1038/s41467-020-16967-2
92. Younginger BS, Mayba O, Reeder J, Nagarkar DR, Modrusan Z, Albert ML, et al. Enrichment of oral-derived bacteria in inflamed colorectal tumors and distinct associations of fusobacterium in the mesenchymal subtype. Cell Rep Med (2023) 4(2):100920. doi: 10.1016/j.xcrm.2023.100920
93. Abate M, Vos E, Gonen M, Janjigian YY, Schattner M, Laszkowska M, et al. A novel microbiome signature in gastric cancer: A two independent cohort retrospective analysis. Ann Surg (2022) 276(4):605–15. doi: 10.1097/SLA.0000000000005587
94. Zeng B, Tan J, Guo G, Li Z, Yang L, Lao X, et al. The oral cancer microbiome contains tumor space-specific and clinicopathology-specific bacteria. Front Cell infection Microbiol (2022) 12:942328. doi: 10.3389/fcimb.2022.942328
95. Yuan L, Yang P, Wei G, Hu X, Chen S, Lu J, et al. Tumor microbiome diversity influences papillary thyroid cancer invasion. Commun Biol (2022) 5(1):864. doi: 10.1038/s42003-022-03814-x
96. Chakladar J, John D, Magesh S, Uzelac M, Li WT, Dereschuk K, et al. The intratumor bacterial and fungal microbiome is characterized by hpv, smoking, and alcohol consumption in head and neck squamous cell carcinoma. Int J Mol Sci (2022) 23(21):13250. doi: 10.3390/ijms232113250
97. Li Z, Fu R, Wen X, Wang Q, Huang X, Zhang L. The significant clinical correlation of the intratumor oral microbiome in oral squamous cell carcinoma based on tissue-derived sequencing. Front Physiol (2022) 13:1089539. doi: 10.3389/fphys.2022.1089539
98. Zhang M, Zhang Y, Sun Y, Wang S, Liang H, Han Y. Intratumoral microbiota impacts the first-line treatment efficacy and survival in non-small cell lung cancer patients free of lung infection. J healthcare Eng (2022) 2022:5466853. doi: 10.1155/2022/5466853
99. Hilmi M, Kamal M, Vacher S, Dupain C, Ibadioune S, Halladjian M, et al. Intratumoral microbiome is driven by metastatic site and associated with immune histopathological parameters: an ancillary study of the Shiva clinical trial. Eur J Cancer (2023) 183:152–61. doi: 10.1016/j.ejca.2023.01.024
100. Hilmi M, Neuzillet C, Lefevre JH, Svrcek M, Vacher S, Benhaim L, et al. Prognostic value of fusobacterium nucleatum after abdominoperineal resection for anal squamous cell carcinoma. Cancers (2022) 14(7):1606. doi: 10.3390/cancers14071606
101. Schmitz-Winnenthal FH, Hohmann N, Schmidt T, Podola L, Friedrich T, Lubenau H, et al. A phase 1 trial extension to assess immunologic efficacy and safety of prime-boost vaccination with vxm01, an oral T cell vaccine against vegfr2, in patients with advanced pancreatic cancer. Oncoimmunology (2018) 7(4):e1303584. doi: 10.1080/2162402X.2017.1303584
102. Schmitz-Winnenthal FH, Hohmann N, Niethammer AG, Friedrich T, Lubenau H, Springer M, et al. Anti-angiogenic activity of vxm01, an oral T-cell vaccine against vegf receptor 2, in patients with advanced pancreatic cancer: A randomized, placebo-controlled, phase 1 trial. Oncoimmunology (2015) 4(4):e1001217. doi: 10.1080/2162402X.2014.1001217
103. Hassan R, Alley E, Kindler H, Antonia S, Jahan T, Honarmand S, et al. Clinical response of live-attenuated, listeria monocytogenes expressing mesothelin (Crs-207) with chemotherapy in patients with Malignant pleural mesothelioma. Clin Cancer Res (2019) 25(19):5787–98. doi: 10.1158/1078-0432.CCR-19-0070
104. Le DT, Wang-Gillam A, Picozzi V, Greten TF, Crocenzi T, Springett G, et al. Safety and survival with gvax pancreas prime and listeria monocytogenes-expressing mesothelin (Crs-207) boost vaccines for metastatic pancreatic cancer. J Clin Oncol (2015) 33(12):1325–33. doi: 10.1200/JCO.2014.57.4244
105. Le DT, Picozzi VJ, Ko AH, Wainberg ZA, Kindler H, Wang-Gillam A, et al. Results from a phase iib, randomized, multicenter study of gvax pancreas and crs-207 compared with chemotherapy in adults with previously treated metastatic pancreatic adenocarcinoma (Eclipse study). Clin Cancer Res (2019) 25(18):5493–502. doi: 10.1158/1078-0432.CCR-18-2992
106. Krupar R, Imai N, Miles BA, Genden EM, Misiukiewicz KJ, Saenger YM, et al. Abstract lb-095: hpv E7 antigen-expressing listeria-based immunotherapy (Adxs11-001) prior to robotic surgery for hpv-positive oropharyngeal cancer enhances hpv-specific T cell immunity. Cancer Res (2016) 76(14_Supplement):LB–095. doi: 10.1158/1538-7445.AM2016-LB-095
107. Safran H, Leonard KL, Perez K, Vrees M, Klipfel A, Schechter S, et al. Tolerability of adxs11-001 lm-llo listeria-based immunotherapy with mitomycin, fluorouracil, and radiation for anal cancer. Int J Radiat oncol biol Phys (2018) 100(5):1175–8. doi: 10.1016/j.ijrobp.2018.01.004
108. Stein MN, Fong L, Tutrone R, Mega A, Lam ET, Parsi M, et al. Adxs31142 immunotherapy +/- pembrolizumab treatment for metastatic castration-resistant prostate cancer: open-label phase I/ii keynote-046 study. oncologist (2022) 27(6):453–61. doi: 10.1093/oncolo/oyac048
109. Mason NJ, Gnanandarajah JS, Engiles JB, Gray F, Laughlin D, Gaurnier-Hausser A, et al. Immunotherapy with a her2-targeting listeria induces her2-specific immunity and demonstrates potential therapeutic effects in a phase I trial in canine osteosarcoma. Clin Cancer Res (2016) 22(17):4380–90. doi: 10.1158/1078-0432.CCR-16-0088
110. Janku F, Zhang HH, Pezeshki A, Goel S, Murthy R, Wang-Gillam A, et al. Intratumoral injection of clostridium novyi-nt spores in patients with treatment-refractory advanced solid tumors. Clin Cancer Res (2021) 27(1):96–106. doi: 10.1158/1078-0432.CCR-20-2065
111. Shahabi V, Seavey MM, Maciag PC, Rivera S, Wallecha A. Development of a live and highly attenuated listeria monocytogenes-based vaccine for the treatment of her2/neu-overexpressing cancers in human. Cancer Gene Ther (2011) 18(1):53–62. doi: 10.1038/cgt.2010.48
112. Zhao J, He D, Lai HM, Xu Y, Luo Y, Li T, et al. Comprehensive histological imaging of native microbiota in human glioma. J biophotonics (2022) 15(4):e202100351. doi: 10.1002/jbio.202100351
113. Yao L, Jiang L, Zhang F, Li M, Yang B, Zhang F, et al. Acetate promotes snai1 expression by acss2-mediated histone acetylation under glucose limitation in renal cell carcinoma cell. Bioscience Rep (2020) 40(6):BSR20200382. doi: 10.1042/BSR20200382
114. Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, et al. Gut microbial metabolism drives transformation of msh2-deficient colon epithelial cells. Cell (2014) 158(2):288–99. doi: 10.1016/j.cell.2014.04.051
115. Flynn C, Montrose DC, Swank DL, Nakanishi M, Ilsley JN, Rosenberg DW. Deoxycholic acid promotes the growth of colonic aberrant crypt foci. Mol carcinogenesis (2007) 46(1):60–70. doi: 10.1002/mc.20253
116. Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria-derived hla-bound peptides in melanoma. Nature (2021) 592(7852):138–43. doi: 10.1038/s41586-021-03368-8
117. Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis (2013) 4(10):e838. doi: 10.1038/cddis.2013.350
118. Gide TN, Wilmott JS, Scolyer RA, Long GV. Primary and acquired resistance to immune checkpoint inhibitors in metastatic melanoma. Clin Cancer Res (2018) 24(6):1260–70. doi: 10.1158/1078-0432.CCR-17-2267
119. Brahmer JR, Govindan R, Anders RA, Antonia SJ, Sagorsky S, Davies MJ, et al. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of non-small cell lung cancer (Nsclc). J immunother Cancer (2018) 6(1):75. doi: 10.1186/s40425-018-0382-2
120. Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. New Engl J Med (2018) 378(22):2078–92. doi: 10.1056/NEJMoa1801005
121. Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (Poplar): A multicentre, open-label, phase 2 randomised controlled trial. Lancet (2016) 387(10030):1837–46. doi: 10.1016/S0140-6736(16)00587-0
Keywords: tumor microenvironment, intratumoral microbiota, cancer pathogenesis, tumor immunity, cancer metabolism, microbiota-targeted therapy
Citation: Wang M, Yu F and Li P (2023) Intratumor microbiota in cancer pathogenesis and immunity: from mechanisms of action to therapeutic opportunities. Front. Immunol. 14:1269054. doi: 10.3389/fimmu.2023.1269054
Received: 29 July 2023; Accepted: 26 September 2023;
Published: 06 October 2023.
Edited by:
Hiroaki Inaba, Okayama University, JapanReviewed by:
Masakazu Hamada, Osaka University, JapanCopyright © 2023 Wang, Yu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Man Wang, d2FuZ21hbkBxZHUuZWR1LmNu; Peifeng Li, cGVpZmxpQHFkdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.