
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 24 October 2023
Sec. Molecular Innate Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1265038
This article is part of the Research Topic Tuberculosis: Host Immunity, Diagnostics and Therapeutics View all 5 articles
 Sajad A. Bhat1,2,3
Sajad A. Bhat1,2,3 Mahmoud Elnaggar3†
Mahmoud Elnaggar3† Thomas J. Hall1
Thomas J. Hall1 Gillian P. McHugo1
Gillian P. McHugo1 Cian Reid3†
Cian Reid3† David E. MacHugh1,2
David E. MacHugh1,2 Kieran G. Meade1,2,4*
Kieran G. Meade1,2,4*Bovine tuberculosis (bTB), caused by infection with Mycobacterium bovis, continues to cause significant issues for the global agriculture industry as well as for human health. An incomplete understanding of the host immune response contributes to the challenges of control and eradication of this zoonotic disease. In this study, high-throughput bulk RNA sequencing (RNA-seq) was used to characterise differential gene expression in γδ T cells – a subgroup of T cells that bridge innate and adaptive immunity and have known anti-mycobacterial response mechanisms. γδ T cell subsets are classified based on expression of a pathogen-recognition receptor known as Workshop Cluster 1 (WC1) and we hypothesised that bTB disease may alter the phenotype and function of specific γδ T cell subsets. Peripheral blood was collected from naturally M. bovis-infected (positive for single intradermal comparative tuberculin test (SICTT) and IFN-γ ELISA) and age- and sex-matched, non-infected control Holstein-Friesian cattle. γδ T subsets were isolated using fluorescence activated cell sorting (n = 10–12 per group) and high-quality RNA extracted from each purified lymphocyte subset (WC1.1+, WC1.2+, WC1- and γδ-) was used to generate transcriptomes using bulk RNA-seq (n = 6 per group, representing a total of 48 RNA-seq libraries). Relatively low numbers of differentially expressed genes (DEGs) were observed between most cell subsets; however, 189 genes were significantly differentially expressed in the M. bovis-infected compared to the control groups for the WC1.1+ γδ T cell compartment (absolute log2 FC ≥ 1.5 and FDR Padj. ≤ 0.1). The majority of these DEGs (168) were significantly increased in expression in cells from the bTB+ cattle and included genes encoding transcription factors (TBX21 and EOMES), chemokine receptors (CCR5 and CCR7), granzymes (GZMA, GZMM, and GZMH) and multiple killer cell immunoglobulin-like receptor (KIR) proteins indicating cytotoxic functions. Biological pathway overrepresentation analysis revealed enrichment of genes with multiple immune functions including cell activation, proliferation, chemotaxis, and cytotoxicity of lymphocytes. In conclusion, γδ T cells have important inflammatory and regulatory functions in cattle, and we provide evidence for preferential differential activation of the WC1.1+ specific subset in cattle naturally infected with M. bovis.
Bovine tuberculosis (bTB), caused by Mycobacterium bovis is endemic in many countries and has significant economic and animal welfare impacts across global agricultural systems (1). Additionally, M. bovis is a member of the Mycobacterium tuberculosis complex (MTBC), which can infect multiple species including humans and livestock making it a zoonotic pathogen and a significant threat to public health (2). The reasons for a failure to eradicate bTB are multifactorial (3) but scientific understanding of M. bovis persistence has been limited in part due to the complexity of host-pathogen interactions that occur under conditions of natural infection. The immune system in humans as well as in livestock is key to disease resistance (4) and suboptimal or dysregulated immunity is thought to contribute to bTB disease susceptibility, progression to clinical disease, and the failure of current generation diagnostics to detect all truly infected cattle. Addressing this important knowledge gap is critical toward achieving the ultimate goal of bTB eradication.
As an intracellular pathogen, immunological research on M. bovis has logically concentrated on the innate macrophage and cell-mediated arm of the immune response, and in particular on the CD4+ helper T cell subset (5). As major producers of the cytokine interferon-γ, which is not only associated with protection against infection but is the principal diagnostic measurement in the in vitro interferon-γ (IFN-γ) release assay (IGRA) used to detect bTB+ cattle, CD4+ T cells are considered major players in anti-mycobacterial immunity (6). While other T cell subsets have received less attention, it is likely that these too play an effector and immunomodulatory role during the course of M. bovis infection. For example, CD8+ cytotoxic T cells have been shown to express IFN-γ after experimental infection with M. bovis but negatively impact on protection to bTB following vaccination (7, 8).
Another T cell subset, γδ T cells have attracted significant recent attention due in part to their ability to span innate and adaptive arms of the immune response but also because the full extent of their development and polyfunctionality remains enigmatic (9). γδ T cells are a diverse group of T lymphocytes that express T-cell receptor (TCR) consisting of TCR-γ and TCR-δ chains. Like classical αβ T cells, γδ T cells are an evolutionary conserved (>430 million years) lymphocyte lineage found in the immune systems of all jawed vertebrates, including humans, mice and cattle (10). An interesting feature of the γδ T cell compartment in ruminants is that unlike in humans and mice where these cells represent less than 5% of the circulating peripheral lymphocyte pool, γδ T cells constitute up to 60% of the total blood lymphocyte population in young calves and approximately 30% in adult cattle (11).
In addition to γδ TCR expression, bovine γδ T cells, express workshop cluster (WC) 1 receptor, a transmembrane glycoprotein and pathogen recognition receptor similar to human CD163c-α (12). Based on WC1 expression, γδ T cells are broadly divided into two major subsets, WC1+ and WC1-. WC1-expressing γδ T cells are further classified into two main subpopulations, WC1.1+ or WC1.2+ (13). Murine and human studies have shown how various lineages of γδ T cells can have proinflammatory (IFN-γ expression) or anti-inflammatory function (IL-10 production) and therefore it is clear that their phenotypic plasticity and development of regulatory functions could have important consequences for the ability of cattle to mount an effective anti-mycobacterial immune response against M. bovis infection (14). γδ T cells are of particular interest to the study of mycobacterial immunity as they have been shown to recognize M. bovis antigens (15, 16), secrete sentinel cytokines including IFN-γ (17, 18), and also influence the activity of other key innate cells including dendritic cells and macrophages during mycobacterial infection (19–21). One study demonstrated that circulating bovine γδ T cells spontaneously secrete the anti-inflammatory cytokine IL-10 and can inhibit proliferation of both CD4+ and CD8+ T cells, thereby documenting a major regulatory role for these cells (22).
Multiple studies now show the importance of crosstalk between innate and adaptive cells in influencing cell subtype and function, including recent studies showing an additional influence of the host microbiome on γδ T cell function (23, 24). However, the number of studies that have comprehensively assessed bovine γδ T cells under natural infection conditions are limited. Therefore, we sought to use bulk RNA-sequencing (RNA-seq) of total lymphocyte and γδ T cell subset populations from naturally infected cattle to capture the true functional status of these critical cell subsets in response to M. bovis infection. Our previous work showed significantly higher number of circulating γδ T cells in M. bovis infected cattle (25), but their functions have not yet been elucidated. In this study we hypothesized that M. bovis infection induces a specific γδ T cell functional phenotype that may play a role in the progression to clinical disease and that may impact on the ability of the host to clear mycobacterial infection.
Male Holstein-Friesian (Bos taurus) cattle were used for this study (age range 18-30 months). For the infected group, animals (n = 12) were selected from a herd of animals naturally infected with M. bovis maintained at the Department of Agriculture, Food and the Marine research farm in Longtown, Co. Kildare, Ireland. Only naturally infected males were available for use in this study as to house females, facilities at the resource farm would require a milking parlour which makes management, logistics and cost of maintaining female cattle significantly higher. There are also additional advantages to using males for the assessment of bTB+ specific changes in that the considerable impact of lactation physiology on immunity (26) does not exist, enabling us to more clearly detect disease-associated immune response differences. The infected group animals had tested positive for bovine tuberculosis by single intradermal comparative tuberculin test (SICTT) and also by the whole blood IFN-γ release assay (IGRA, University College Dublin, Ireland). For the control group, non-infected (negative for both the SICTT and IFN-γ release assay), age and sex matched animals (n = 10) were selected from a herd with that were free from bTB for more than five years. The non-infected cattle had tested negative by SICTT within the previous 84-151 days prior to sampling and were confirmed negative by IGRA at time of sampling. Test Response and bTB classification information on all cattle is detailed in Supplementary Table S1. All animal procedures and experimental protocols in this study were approved by the Teagasc Animal Ethics Committee (TAEC217-2019) and carried out in accordance with the relevant institutional guidelines and under license from the Irish Health Products Regulatory Authority (HPRA no. AE19132/I019).
Peripheral blood from the jugular vein of M. bovis-infected and control animals was collected in 10 ml Vacutainer® tubes containing EDTA anticoagulant (BD Diagnostics, Oxford, UK). For cell separation, whole blood was diluted 1:1 with phosphate buffered saline (PBS) in 50-ml conical Falcon tubes (Corning, Inc., Kaiserslautern, Germany). Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coat fractions using Histopaque-1077 (Sigma-Aldrich Ireland Ltd., Wicklow, Ireland) and the Leucosep system (Greiner Bio-One Ltd., Stonehouse, UK) with gradient centrifugation at 1034 × g for 25 min. PBMCs were collected and washed in PBS to remove platelets. Contaminating red blood cells were removed using cell lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA).
PBMCs were washed and re-suspended in PBS containing 2% bovine serum albumin (BSA). For labelling, cells were mixed with anti-bovine γδ TCR (GB21A, IgG2b), WC1.1 (BAG25A, IgM) and WC1.2 (CACTB32A, IgG1) monoclonal antibodies (mAbs, 1 μg per 2 × 106 of each, Monoclonal Antibody Center, Pullman, WA, USA), incubated at 4°C for 20 min, washed and re-suspended in 2% BSA PBS. Cells were then incubated in the dark for 30-45 min on ice with goat anti-mouse fluorochrome conjugated isotype specific mAbs (Life Technologies Corporation, Invitrogen, USA), washed twice and re-suspended in 2% BSA PBS. The cells were collected, analysed and sorted using the FACSAria Fusion Sorter (BD Biosciences, Wokingham, UK). FACSDiva™ (BD Biosciences) and FCS Express™ (De Novo Dotmatics) software programs were used for the analysis, sorting and flow plot generation. Compensation was used to eliminate residual spectral overlaps between individual fluorochromes. Side and forward scatter area/forward scatter width parameters or characteristics were used for the identification of viable single lymphocytes and exclusion of doublets. Propidium iodide (Thermo Fisher Scientific) was used for the exclusion of dead cells. Sorted γδ T cells and subsets were identified as having viability greater than 95% and purity greater than 99%. The γδ T cells were centrifuged and cell pellets were snap frozen on dry ice first and then stored at -80°C. An unpaired Students t-test (GraphPad Prism v9) was used to assess significant differences using a P value cut off of <0.05 between both groups after assessment for normality.
Total RNA was extracted from the cell pellets using an AllPrep DNA/RNA Kit (Qiagen). RNA quantity, integrity and purity were assessed using a NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific) and an Agilent 2100 Bioanalyzer using an RNA 6000 Nano LabChip kit (Agilent Technologies Ltd., Cork, Ireland), according to the manufacturers’ instructions. Only RNA samples had RNA integrity number (RIN) values > 8 were subsequently used for transcriptomic analysis (n = 6 per group). RNA libraries were prepared from a starting quantity of 100 ng high quality RNA using the TruSeq RNA Library Prep Kit v2 (Illumina Inc., San Diego, CA, USA). RNA libraries were prepared from a starting quantity of 100 ng high quality RNA using the TruSeq RNA Library Prep Kit v2 (Illumina Inc., San Diego, CA, USA). Individually barcoded RNA-seq libraries were pooled in equimolar quantities and the quantity and quality of the final pooled libraries (two different pools in total) were assessed as described above. RNA-seq library sample pool construction was performed using an NEB Next® Ultra RNA Library Prep Kit for Illumina® (Cat No. 7530) and detailed in Supplementary Table S2. Cluster generation and high-throughput 150 bp paired-end sequencing of the pooled RNA-seq libraries were performed on a NovaSeq 6000 using an S4 flow cell prepared with the Illumina 300 cycle Reagent Kit (v1.5). All RNA-seq data generated for this study have been deposited in the ArrayExpress database under project accession number E-MTAB-13111.
Sequence quality and composition were checked using FastQC (version 0.11.8) software (27). Adapter sequence reads were removed and quality trimming was carried out using the fastP adapter removal software (28). RNA seq data was processed as described previously (29). Briefly, 50 million, 2 × 150 bp paired-end sequence reads were generated from each RNA sample. Quality filtering of RNA-seq read pairs yielded a mean of 69,161,585 reads per individual library (48 libraries in total). The filtered RNA seq paired-end reads were mapped to the most recently annotated version of the Bos taurus reference genome (ARS-UCD1.2, GenBank assembly accession: GCA_002263795.2) (30) using the STAR aligner (version 2.7) (31). A mean of 64,133,177 read pairs (92.63%) were uniquely mapped to locations in the ARS-UCD1.2 bovine genome assembly (Supplementary Table S2). Aligned reads were assigned to genomic features using featureCounts (32) and the resulting quantification files were annotated at gene level via biomaRt and GO.db (33). The annotated gene counts were then normalised and differential expression analysis performed with DESeq2 (version 1.20.0) (34). Lowly expressed reads and extreme count outliers were removed within DESeq2 using the Cook’s distance. P values were adjusted for multiple testing using the Benjamini-Hochberg (B-H) false discovery rate (FDR) method (35). The criteria for detection of significantly differentially expressed genes (DEGs) were an FDR-adjusted P-value less than 0.1 (Padj. < 0.1) and a |log2 FC| ≥ 1.5, which was incorporated into the statistical model in DESeq2.
The Ingenuity® Pathway Analysis (IPA) software package (36) with the Ingenuity® Knowledge Base (Qiagen, Redwood City, CA, USA; release date December 2022) was used to identify overrepresented (enriched) canonical pathways and interaction networks for the set of DEGs. IPA® Core Analysis was performed using the default settings with the user data set as the background, high predicted confidence and all nodes selected. The significance of the association of genes with each canonical pathway was determined using a B-H adjusted right-tailed Fisher’s exact test. Gene networks (graphical representations of the molecular interactions among genes) were generated based on their relevance and connectivity with the genes in the input data. Each network was assigned a score equivalent to the negative exponent of the right-tailed Fisher’s exact test for each network. The functions of the DEGs were evaluated using the Ingenuity® Knowledge Base, the GeneCards Suite (37), and the NCBI resources (38) and Uniprot (39) databases.
The proportions of γδ T subpopulations in the bTB+ (n = 12) and non-infected control (n = 10) cattle groups were assessed using flow cytometry on live lymphocytes in peripheral blood mononuclear cells (PBMCs). The gating strategy shows the identification of WC1.1+, WC1.2+ and WC1- subpopulations within the γδ TCR+ subpopulation and γδ- cells were identified by gating on the γδ TCR- cells (Figure 1). No significant difference in the total γδ+ T cells proportions (Figure 2A), or WC1.1+ (Figure 2B), WC1.2+ (Figure 2C) or WC1- (Figure 2D) subsets was detected in the M. bovis-infected animals compared to the non-infected controls. Significant inter-individual animal variation in all four subsets was apparent in both experimental groups (Figure 2E). A significantly higher proportion of WC1+ γδ T lymphocytes was detected in the bTB+ cattle in previous work by our group (25), discordance which may be explained by seasonal changes in circulating immune cell populations or age-related differences between cattle cohorts.
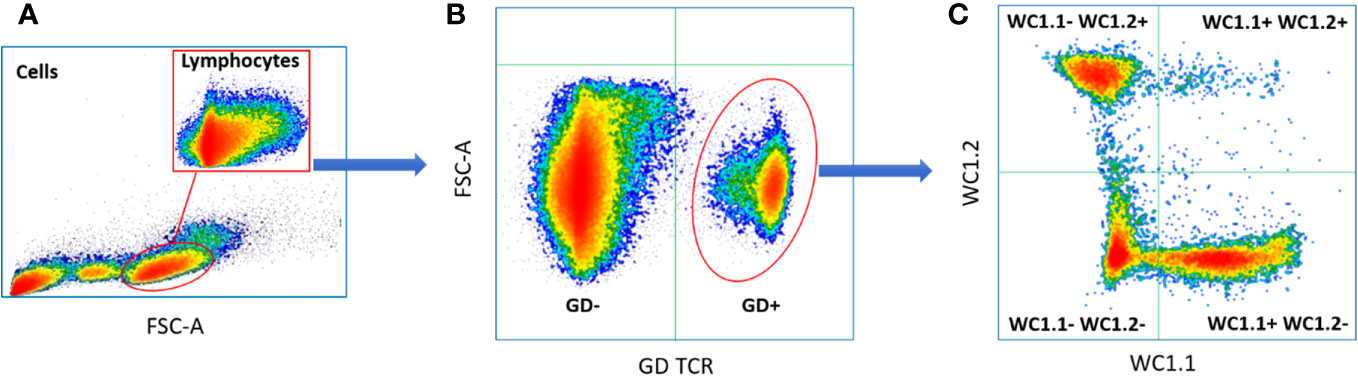
Figure 1 Typical gating strategy for flow cytometric γδ T cell analysis and sorting: (A) Forward and side scatter shows the gating on lymphocytes; (B) Lymphocyte separated into γδ TCR+ and γδ TCR- sub-populations; (C) γδ TCR+ T cells separated into three major sub compartments based on the expression of WC1.1+; WC1.2+ or WC1-.
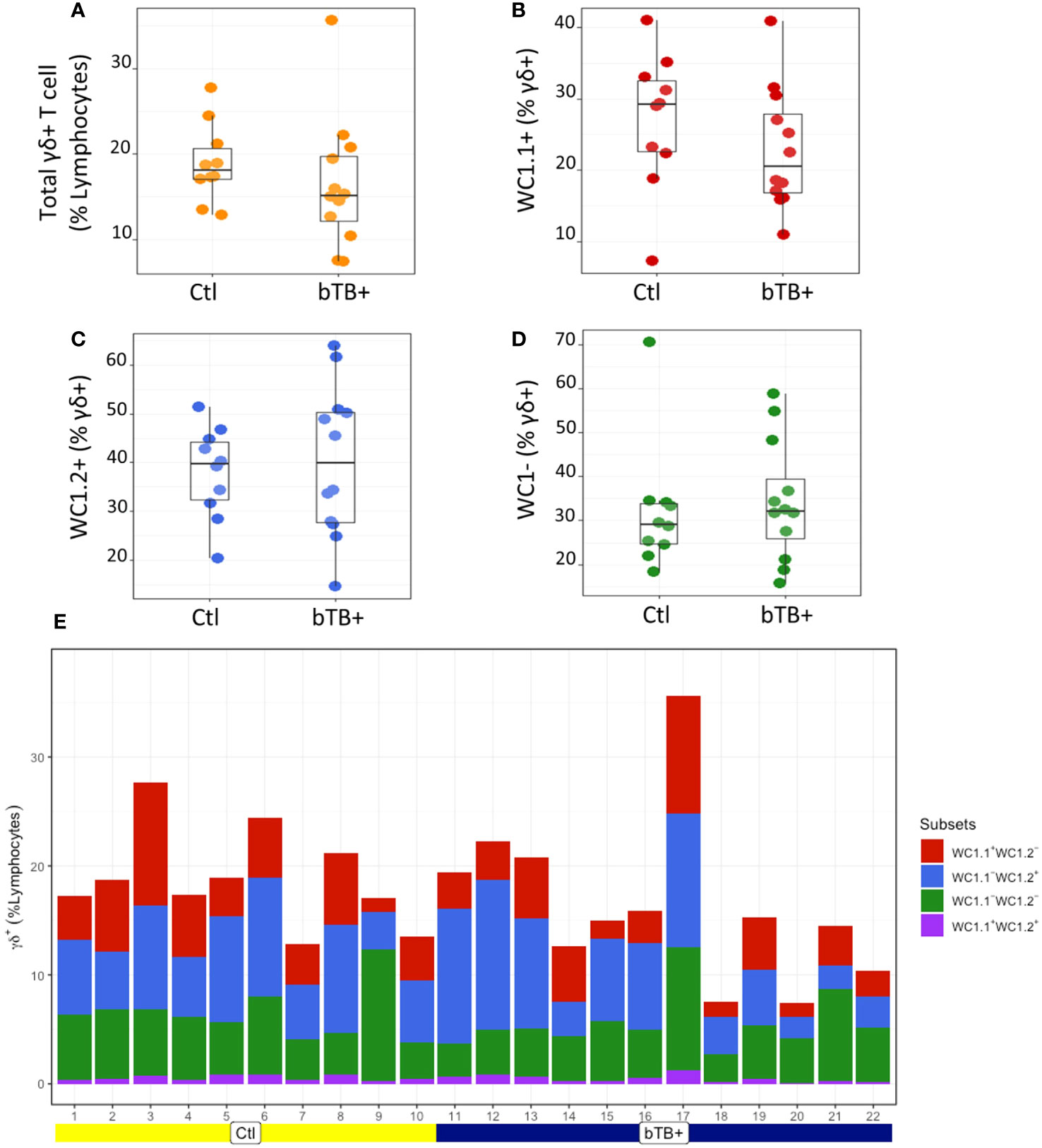
Figure 2 Assessment of (A) Total γδ TCR+; (B) WC1.1+; (C) WC1.2+ and (D) WC1- subsets in bTB+ (n = 12) and non-infected control (n = 10) cattle. Data presented as standard error mean (SEM) of percentages; (E) Individual animal data for γδ T cell subsets in control and bTB+ groups, expressed as a % of lymphocytes. An unpaired Students t-test (GraphPad Prism v9) was used to assess significant differences using a P value cut off of <0.05 between both groups after assessment for normality.
Statistical analysis demonstrated that differential gene expression was evident between the bTB+ and the non-infected control cattle groups, predominantly in the WC1.1+ γδ T cell compartment. A total of 189 genes were observed to be differentially expressed in WC1.1+ γδ T cells. A smaller number of genes were differentially expressed between groups in the WC1.2+ γδ T cell (19 genes) and WC1- γδ T cell (33 genes) compartments. Only seven genes were differentially expressed in the γδ- T cells. Supplementary Table S3 provides detailed information on the differential expression analysis results for each comparison. A Venn diagram for the significant DEGs across cell compartments between the bTB+ and non-infected control cattle groups is shown in Figure 3, and very minor overlap in DEGs is apparent between subsets. Most of the DE genes in the WC1.1+ γδ T cells were increased in expression in the bTB+ cattle (89%) and this asymmetric response is shown in the volcano plot in Figure 4. The range in log2 fold change for genes exhibiting significantly different expression in the bTB+ group ranged from -21.83 to 29.2 (Supplementary Table S3).
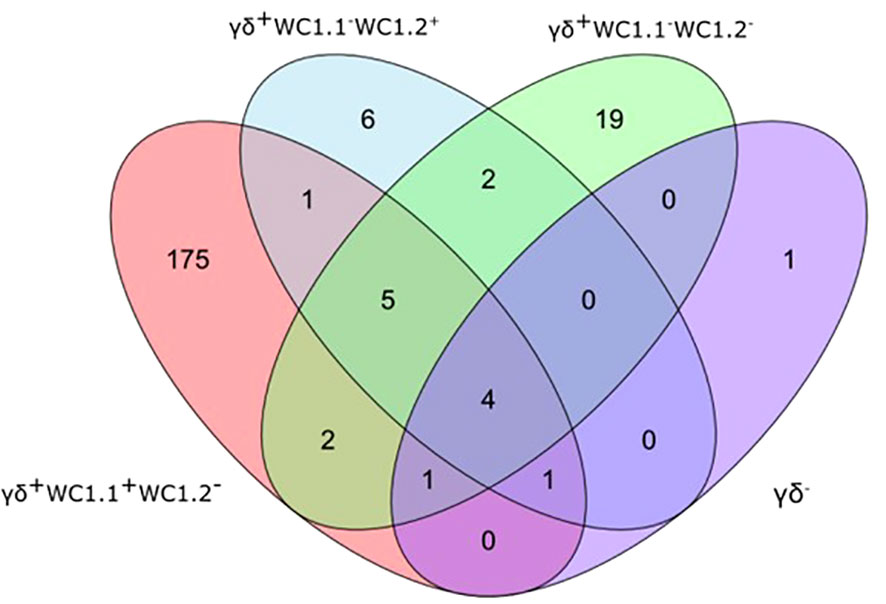
Figure 3 Venn Diagram showing the overlap of DEGs (FDR-Padj. < 0.10; |log2FC| ≥ 1.5) between γδ T cell subsets in the bTB+ group (n = 6) compared to the non-infected control group (n = 6).
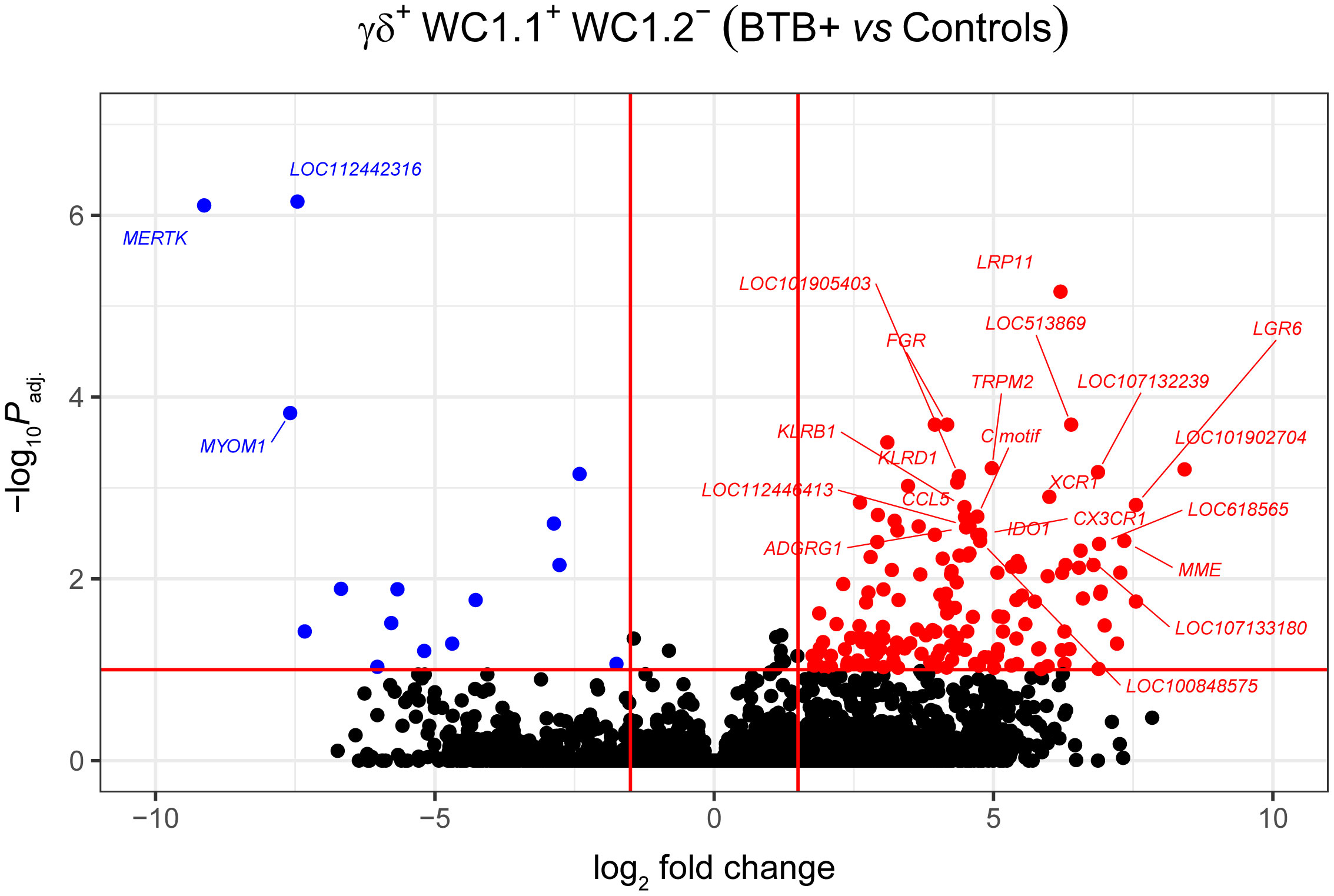
Figure 4 Volcano plot showing differential gene expression for the WC1.1+ γδ T cell subset in the bTB+ group (n = 6) compared to the non-infected control group (n = 6). The FDR-Padj. < 0.10 and |log2FC| ≥ 1.5 thresholds are shown as red lines.
The 168 significantly upregulated genes includes genes which encode proteins with well characterised immune roles evident from Cluster of Differentiation (CD) protein identifiers, including the cell adhesion marker CD2 (log2FC = 2.92; FDR-P = 0.0039), cell surface glycoproteins CD6 (log2FC = 2.75; FDR-P = 0.0455), CD8A (log2FC = 2.6; FDR-P = 0.0443), CD8B (log2FC = 2.6; FDR-P = 0.0330), and CD38 (log2FC = 3.47; FDR-P = 0.0010), an enzyme that functions in intracellular signalling and as a receptor with a role in endothelial adhesion (40). Interestingly, a new CD8A/B expressing subset of γδ T cells that can also express IFN-γ has been reported (41) but to-date, this subset has not been identified in domestic cattle. The CD86 gene encoding a co-stimulatory molecule involved in T cell activation (42) was also increased in expression (log2FC = 2.42; FDR-P = 0.0867). CD244, a gene encoding a signalling lymphocyte activation molecule (SLAM) family immunoregulatory receptor found on many immune cell types was also upregulated (log2FC = 2.38; FDR-P = 0.0901). In addition, CCL5, which encodes a potent cytotoxic T cell activator (C-C motif chemokine ligand 5) (43), exhibited increased expression (log2FC = 4.49; FDR-P = 0.0021). The CCR5 gene encoding a receptor for CCL5 was also upregulated (log2FC = 4.0; FDR-P = 0.0706), as was the CCR7 gene (log2FC = 2.45; FDR-P = 0.0447). The CCR7 protein has been shown to regulate T cell homing and dendritic cell maturation (44). The indoleamine-2,3-dioxygenase 1 gene (IDO1) involved in T regulatory cell development (45) and the interleukin 2 receptor gene (IL2RB), which regulates T cell-mediated immune responses, were also significantly increased in expression for the bTB+ group (log2FC = 4.71; FDR-P = 0.0033; and log2FC = 2.31; FDR-P = 0.0114, respectively). A microRNA (miRNA) gene (MIR2901) exhibited the largest expression fold-change (log2FC = 23.67; FDR-P = 8.53 × 10-56); however, no functional data exists for the mir-2901 miRNA in cattle. Finally, 42 of the upregulated genes had NCBI LOC symbols for which detailed functional information is also limited (Supplementary Table S3).
The 21 genes that exhibited decreased expression in the bTB+ group included the gamma-aminobutyric acid receptor subunit alpha-2 gene (GABRA2: log2FC = -14.14; FDR-P = 2.58 × 10-6) and the protocadherin 11 X gene (PCDH11X: log2FC = -11.09; FDR-P = 0.0006). MERTK also exhibited decreased expression (log2FC = -9.13; FDR-P = 7.78 ×10-7) and encodes a receptor tyrosine kinase, which is a key phagocytic receptor in the immune system that regulates many physiological processes including phagocytosis of apoptotic cells (46). Also downregulated was expression of the C-type lectin domain family 5 member A gene (CLEC5A), which has a role in the induction of many cytokines and chemokines thereby amplifying the innate immune response (log2FC = -5.67; FDR-P = 0.01302) (47). Interestingly, however, differential expression of cytokine genes was not a defining feature of the DEG data set. The 20 genes that exhibited the highest expression fold changes (up- and downregulated) are shown in Table 1.
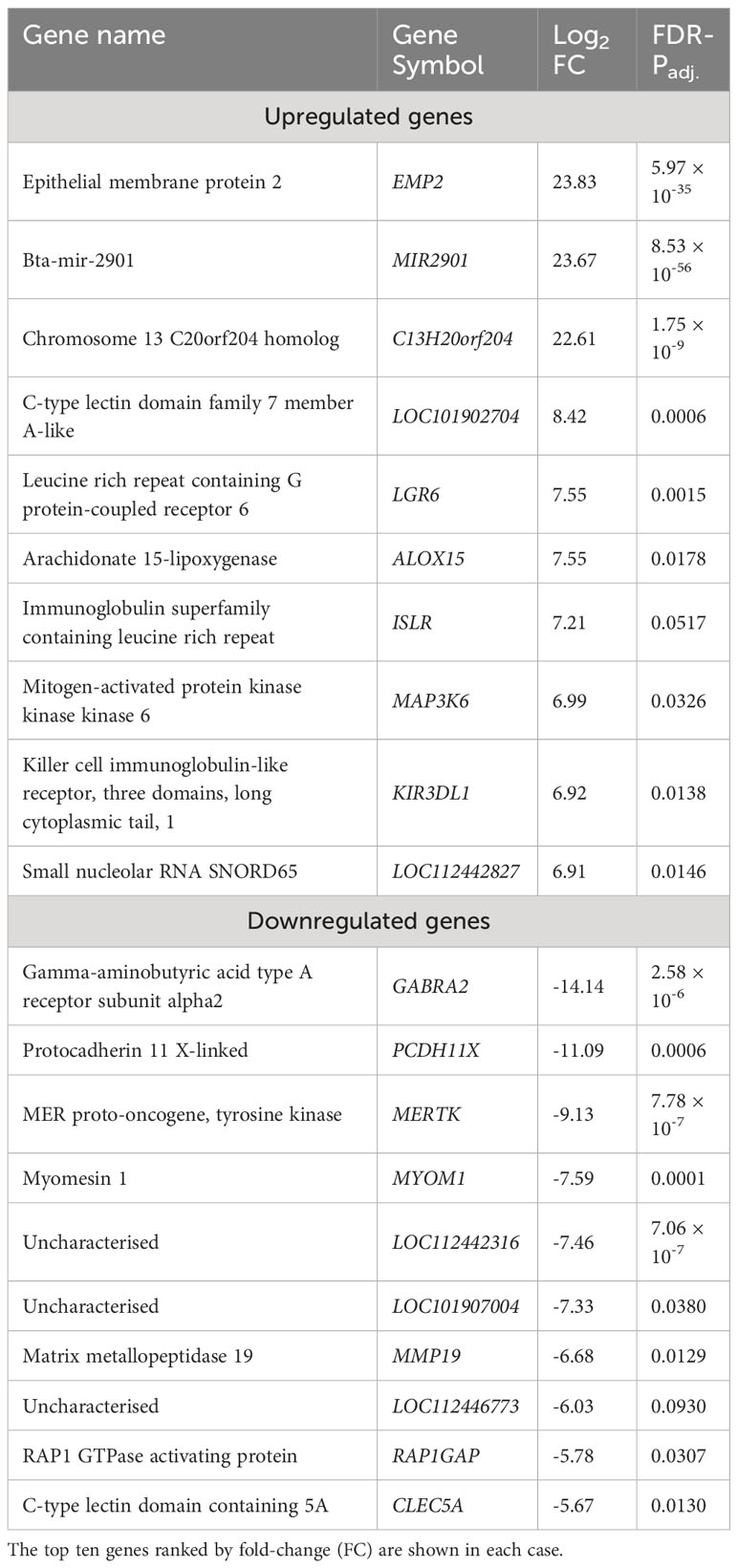
Table 1 Genes exhibiting statistically significant (FDR-Padj. < 0.10; |log2FC| ≥ 1.5) increased and decreased expression in WC1.1+ γδ T cells for the bTB+ group (n = 6) compared to the non-infected control group (n = 6).
The WC1.1+ γδ T cell DEG data set was further analysed using IPA® to identify enriched canonical pathways and biological interaction networks. Out of the 189 significant DEGs, 125 analysis-ready genes could be mapped to genes in the Ingenuity® Knowledge Base, of which 111 had increased expression and 14 had decreased expression, against a background set of 14,109 analysis-ready genes (7857 with increased expression, 5876 with decreased expression and 376 with no change in expression) from the 18,310 detectable genes that were mapped by IPA®. Canonical pathway analysis revealed that the DEGs were enriched in several notable signalling pathways (Figure 5A). The top biological pathways were Natural Killer Cell Signalling, Crosstalk between Dendritic Cells and Natural Killer Cells, Th1 and Th2 Activation Pathway, CTLA4 Signalling in Cytotoxic T Lymphocytes, and the CREB Signalling in Neurons represented by the DEGs shown in Figure 5B.
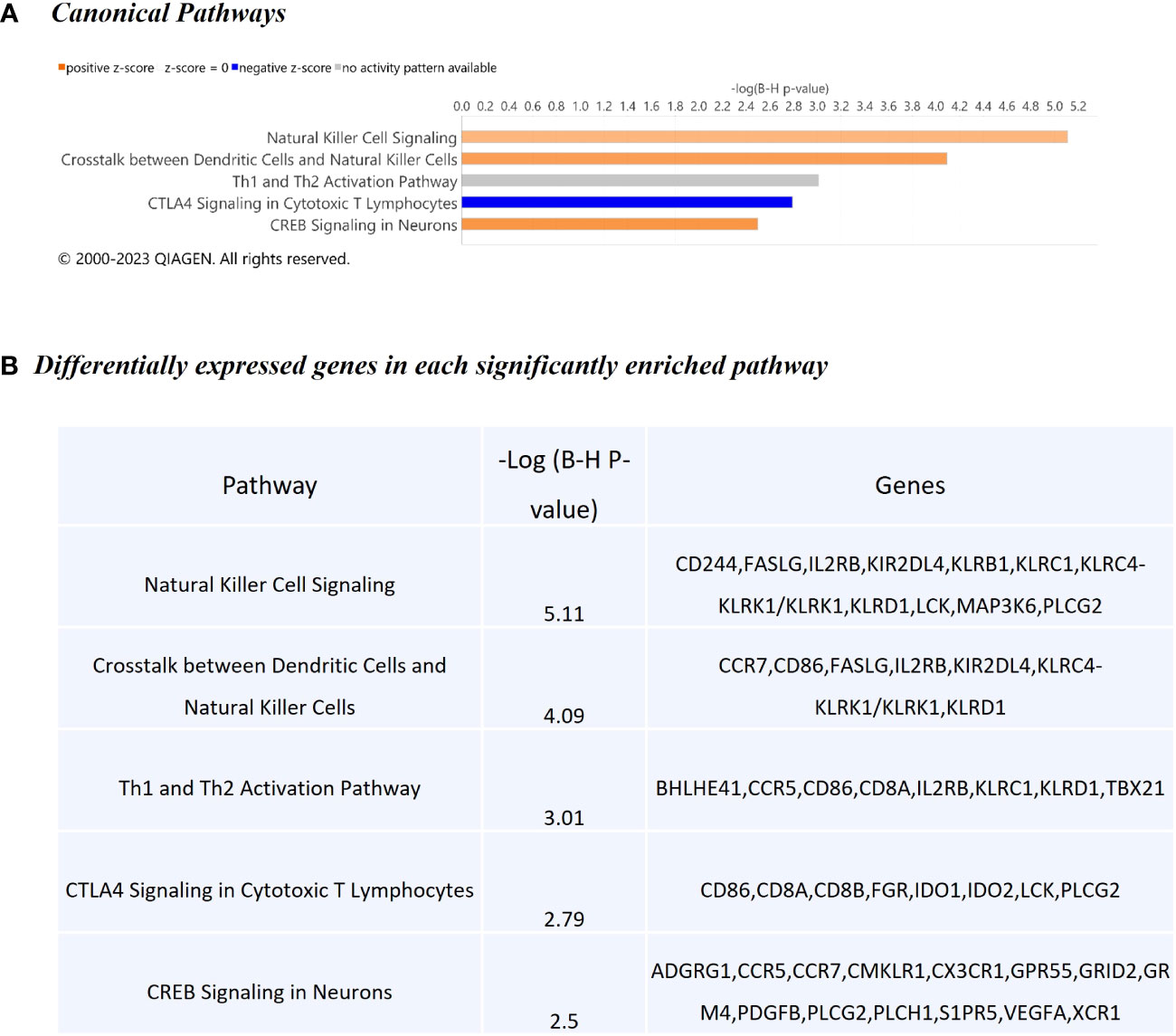
Figure 5 Canonical pathway enrichment results from IPA® showing (A) Top five biological pathways associated with M. bovis infection in cattle; (B) The DEGs present within each enriched pathway.
The top diseases and biological functions included Inflammatory Response, Organismal Injury and Abnormalities, Cell-to-Cell Signalling and Interaction, Cell Death and Survival, Haematological System Development and Function and Immune Cell Trafficking (see Tables 2A–C). The network analysis showed that the M. bovis infection in cattle affected diverse biological functions and cellular processes including cellular crosstalk and cytotoxic function (Supplementary Tables S4, S6). Eleven biological interaction networks were generated from the 125 DEGs set using the IPA® Knowledge Base (Supplementary Table S5). Figure 6 shows the highest-ranked biological network (with a network score of 33 and 17 focus molecules), which includes ACER2, ADGRG1, Calmodulin, CCR5, CCR7, CEMIP2, CMKLR1, CX3CR1, Erm, Fc gamma receptor, Focal adhesion kinase, G protein alpha i, Gpcr, GPR55, GRM4, Integrin, NFkB (complex), Nr1h, OSCAR, p85 (pik3r), Pka, PLC, PLC gamma, PLCG2, PLCH1, PPEF1, PPIF, PTCH1, Rac, RAS, S1PR5, Sfk, SRC (family), STAT and XCR1. This network yields interesting insight into the inflammatory response cascades present in WC1.1+ γδ T cells in bTB+ infected cattle, and as CCR5 has bene previously associated with mycobacterial immune subversion (48), this data contributes to our understanding of bTB pathogenesis.
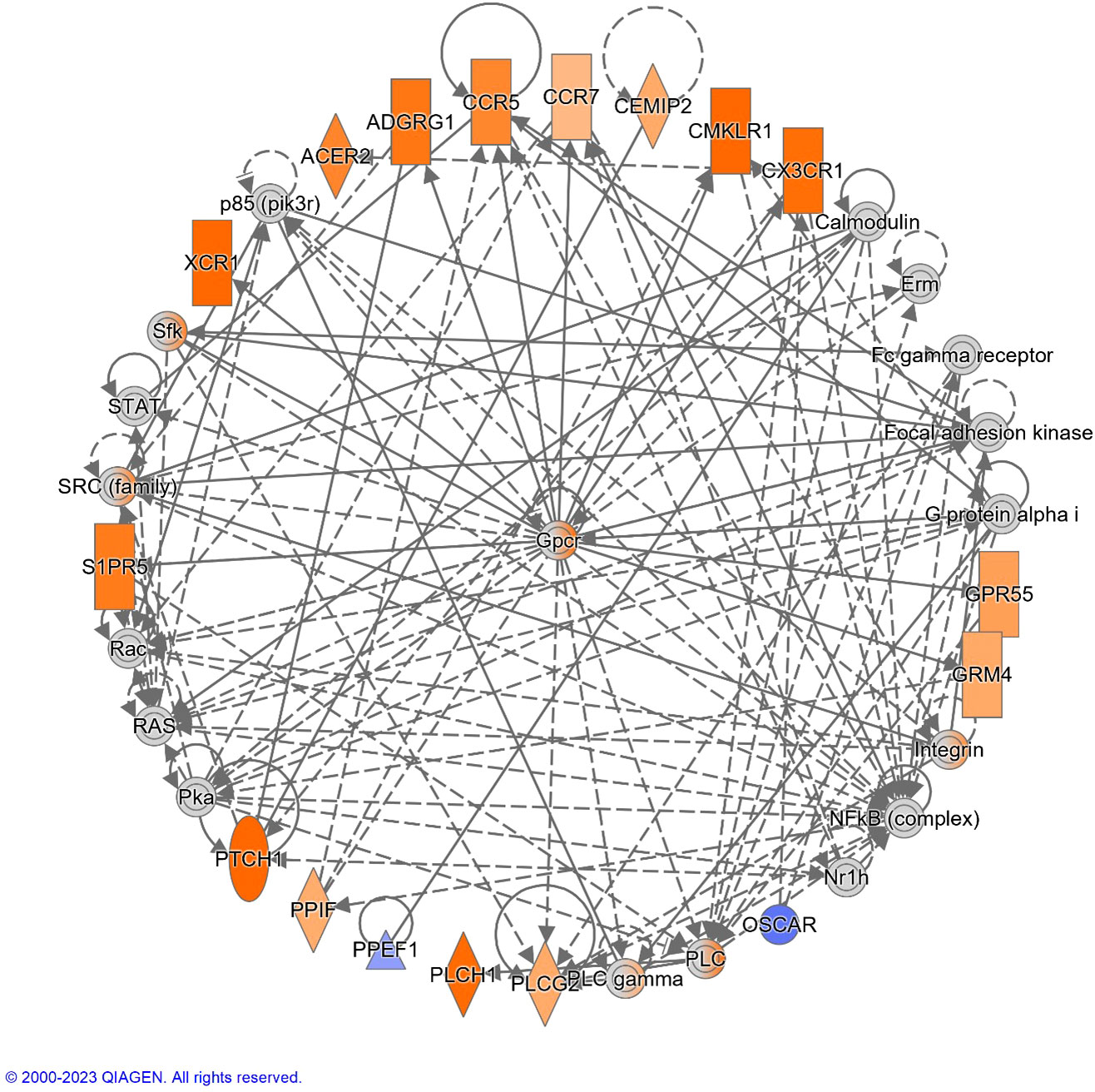
Figure 6 The top-ranked network generated using IPA®, which consisted of 17 focus genes (Network Score = 33). The top Diseases and Functions for this network are Cell-mediated Immune Response (P-value = 3.8 × 10-7), Cell-To-Cell Signaling and Interaction (P-value = 7.3 × 10-7) and Cellular Movement (P-value = 1.0 × 10-6). Increased and decreased gene expression is indicated with the orange and blue colour scales, respectively. Genes that were not detectable are indicated in grey. A full legend is available at: https://qiagen.my.salesforce-sites.com/KnowledgeBase/articles/Basic_Technical_Q_A/Legend.
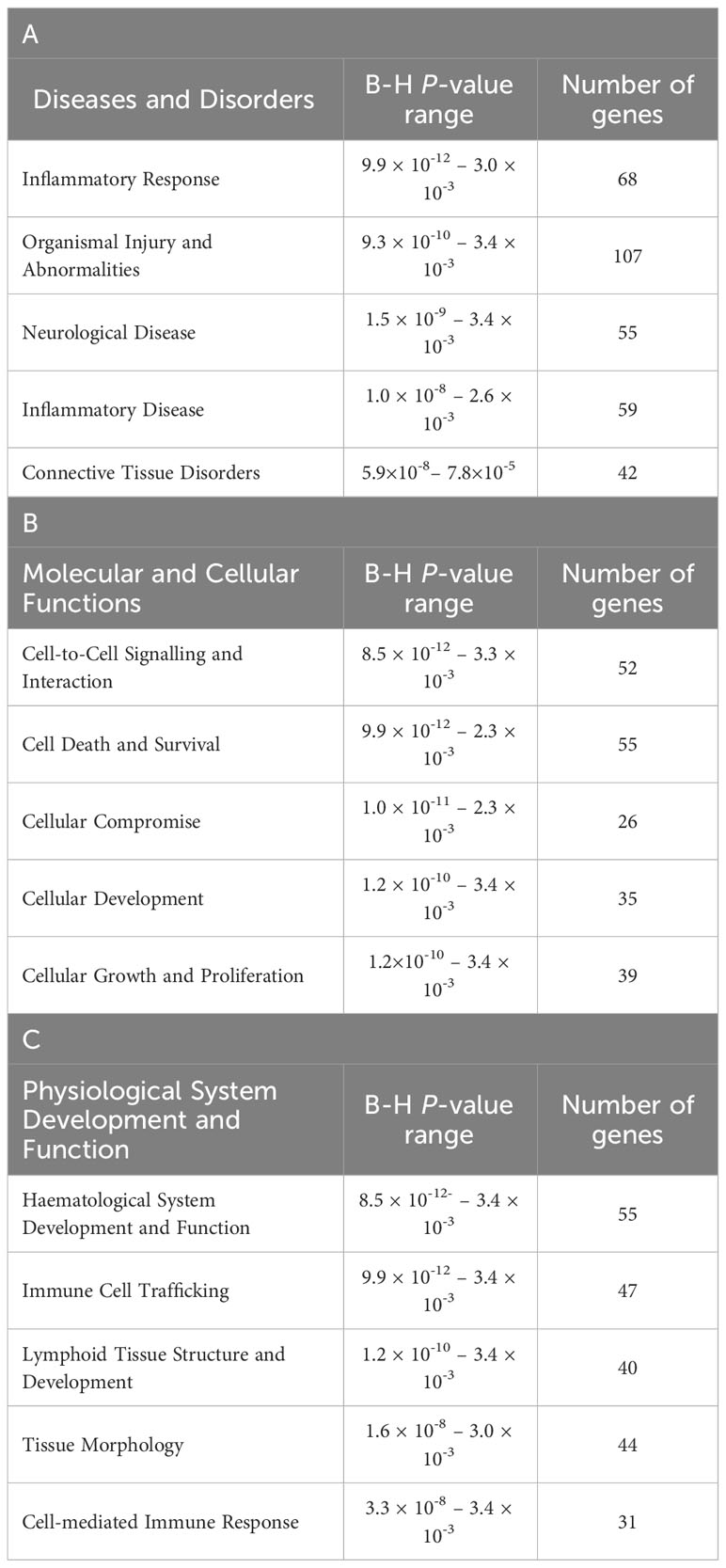
Table 2 The top-ranked Diseases and Disorders (A), Molecular and Cellular Functions (B) and (C) Physiological System Development and Function categories associated with M. bovis infection status identified using IPA®.
Despite no apparent difference in overall γδ T cell or subset numbers between the bTB+ and control groups, our results clearly show preferential activation of the WC1.1+ γδ T cell compartment in bTB+ infected cattle. It is surprising that of all the direct comparisons between cell types from the bTB+ and healthy, control cattle, numbers of DEG are low. This includes the examination of the γδ TCR- lymphocytes, suggesting that a differential response is only maintained in the WC1.1+ γδ T cell compartment, even in the absence of exogenous antigen stimulation. Other studies have found that this γδ T cell subset is more inflammatory (49) and it is the WC1.1+ subset that localize to the site of infection after BCG vaccination (50), thereby supporting the immunoreactive ability of these cells to mycobacterial antigens.
One of the defining features of our analysis was the overrepresentation of genes that encode markers more often associated with the function of natural killer (NK) cells. NK cells are cytotoxic lymphocytes endowed with multiple mechanisms for killing infected cells, including those infected with mycobacteria (51). Cytotoxic cells can directly kill intracellular bacteria through granulysin-mediated delivery of granzymes (52) and these molecules have documented antimycobacterial efficacy against M. tuberculosis (53).
In the DEG data set generated for the present study, CD244, encoding a cell surface receptor expressed on natural killer (NK) cells and other T cells is upregulated in bTB+ cattle (log2FC = 2.38; FDR-P = 0.0901). The CD244 protein modulates cellular cytotoxicity and plays an important role in regulating production of IFN-γ as well as both CD4+ and CD8+ T cell immunity during TB disease (54, 55). The natural killer cell granule protein 7 gene (NKG7), a regulator of lymphocyte granule exocytosis and inflammation, also exhibited increased expression (log2FC = 2.09; FDR-P = 0.0929). NK receptors also play a direct role in the regulation of γδ T-cell-mediated immune responses, likely reflecting important cellular crosstalk in this context, and their activation leads to the release of cytotoxic granules containing granzymes (56). Also of relevance is the differential expression of granzyme A and M genes (GZMA and GZMM), which have documented antimycobacterial activity (log2FC = 4.04; FDR-P = 0.0618 and log2FC = 4.25; FDR-P = 0.0082, respectively), have been previously reported to be expressed in the γδ T cells of other species (57, 58). In conjunction with the granzyme H gene (currently annotated to LOC788601; log2FC = 6.88; FDR-P = 0.0980), these genes all exhibit increased expression in bTB+ cattle, suggesting a role in the immune response to M. bovis infection.
The eomesodermin gene (EOMES) encodes a transcription factor with a crucial role in regulating cytotoxic function, development, and survival of a range of immune cells such as NK and CD8+ T cells (59–61). In addition, the T-box transcription factor 21 gene (TBX21) encodes a transcription factor that plays a central role in CD4+ Th1 lineage development (62, 63). Consequently, EOMES and TBX21 encode proteins that control the development and function of a repertoire of cells in the innate and adaptive compartments of the immune system (61). Both EOMES and TBX21 showed increased expression in bTB+ cattle (log2FC = 4.35; FDR-P = 0.0447 and log2FC = 3.95; FDR-P = 0.0002, respectively). The genes currently annotated to LOC104968634 and LOC112448791 were also upregulated (log2FC = 4.14; FDR-P = 0.0191 and log2FC = 4.57; FDR-P = 0.0052, respectively) and encode the NK2B and granulysin (GNLY) antimicrobial peptides, respectively. LOC104968634 has also been shown to be upregulated in bovine alveolar macrophages challenged in vitro with M. bovis (64). The differential gene expression for the DEGs with cytotoxic functions is shown in Figure 7.
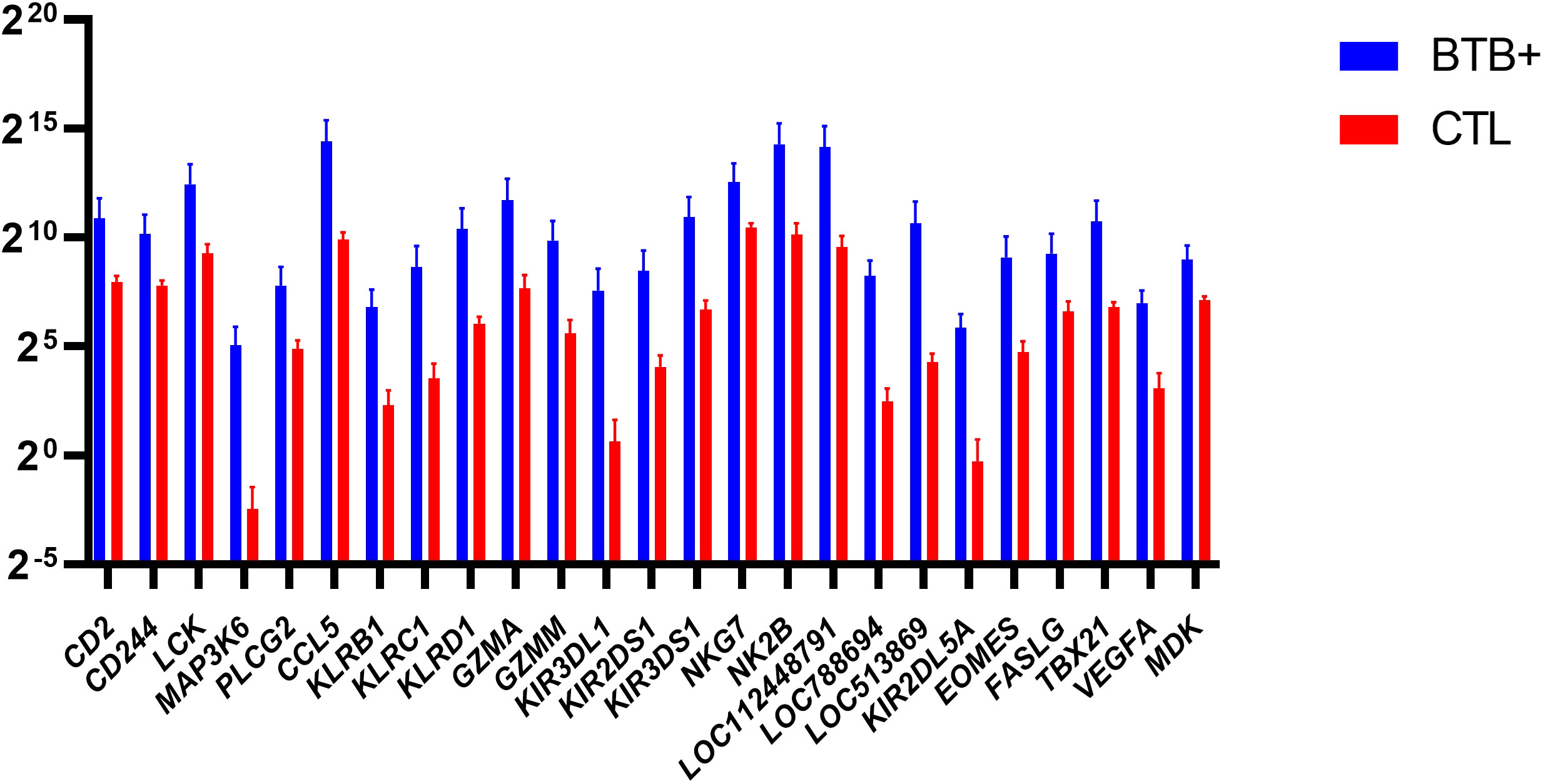
Figure 7 Natural Killer (NK) related genes identified as differentially expressed (FDR-Padj. < 0.10; |log2FC| ≥ 1.5) in WC1.1+ γδ T cells from the bTB+ group (n = 6) compared to the non-infected control group (n = 6). The log2 normalized read counts ± SEM are shown.
Other genes that exhibited increased expression in bTB+ cattle included four members of the killer cell lectin like receptor (KLR) family of immune inhibitory receptor genes (KLRB1, KLRC1, KLRD1, and KLRK1). KLRB1 (log2FC = 4.48; FDR-P = 0.0016) encodes a protein that has been reported to have both costimulatory and coinhibitory roles in T-cells and is attracting significant attention with other KLR proteins for their potent therapeutic potential (65, 66). KLRD1 (log2FC = 4.35; FDR-P = 0.0009) encodes a protein that has been shown to induce functional exhaustion of cytotoxic NK and CD8+ T cells (67–69). KLRK1 (log2FC = 3.3; FDR-P = 0.0171) encodes the natural killer group 2D protein (NKG2D), which has a documented roles in immunoprotection from infection via γδ T cell activation (70), and pulmonary clearance of bacteria (71). Multiple killer cell immunoglobulin like receptor (KIR) genes were also upregulated in the present study, including KIR2DL5A, KIR2DS1, KIR3DL1, and KIR3DS1. Individual KIR genes as well as specific haplotypes has been previously associated with resilience to TB disease in human populations (72, 73).
The association of some of these cytotoxic functions has only been superficially investigated in γδ T cells previously and has not been documented in cattle naturally infected with M. bovis indicating potential relevance to bTB disease. These findings are supported by related research in murine models. Whereas γδ TCR knockout mice support a protective role for γδ T cells in mycobacterial infection (74), control of mycobacterial infection was seriously impaired in NK cell knockout mice (75), suggesting that the adoption of these functions by γδ T cells is a critical disease control measure. It is of further relevance that depletion of the NK cell subset has been reported in human patients with TB (76) and the authors report that this subset is GZMB+, therefore indicating a potential loss of granzyme function, at least from the NK cell subset associated with the progression to clinical disease. It is therefore plausible that γδ T cell granzyme expression may partially compensate for this loss of anti-mycobacterial function. However, comprehensive characterization of the NK cell subset has not been performed in M. bovis-infected cattle to date but based on the results described here, this avenue warrants further investigation.
Smaller numbers of significant DEGs were detected between groups in the other cellular compartments. In the WC1.2+ γδ T cell, 15 DEGs were identified between the bTB+ and non-infected control cattle groups. Six genes were upregulated, and nine genes were downregulated in the bTB+ group, respectively. Genes encoding cytochrome 8B (COX8B) (log2FC = 18.9; FDR-P = 2.41 × 10-16), cytochrome P450 family 17 subfamily A member 1 (CYP17A1; annotated to LOC112444495) (log2FC = 28.38; FDR-P = 0.0025), and LDL receptor related protein 11 (LRP11) (log2FC = 5.31; FDR-P = 0.0012) were increased in expression. The myomesin 1 gene (MYOM1), the protocadherin 11 X-linked gene (PCDH11X), and a solute carrier family 22 member 9-like gene (annotated to LOC517475) were decreased in expression.
Of the 28 DEGs in the WC1- γδ T cell compartment, the majority (64%) are also decreased in expression in the bTB+ cattle. Some of the transcripts are differentially expressed in the same manner as the WC1.2+ subset. These include upregulation of the cytochrome P450 17A1 gene (LOC112444495; log2FC = 28.25; FDR-P = 0.0019) and the LDL receptor related protein 11 gene (LRP11; log2FC = 4.47; FDR-P = 0.0198), and downregulation of the myomesin 1 gene (MYOM1; log2FC = -12.65; FDR-P = 1.44 × 10-10) and the protocadherin 11 X-linked gene (PCDH11X; log2FC = -10.32; FDR-P = 0.0040) indicating differential expression of these genes is not specific to particular cell subsets. Downregulation of additional genes including the aldehyde oxidase gene (AOX1; log2FC = -20.04; FDR-P = 1.10 × 10-8) was also detected. Aldehyde oxidase catalyses the production of hydrogen peroxide and can also catalyse the formation of superoxide (77). which may impact production of ROS species in these cells.
Only six DEGs were detected in the γδ- T cells, all of which were decreased in expression in the bTB+ cattle group. Consistent decreased expression in PCDH11X (log2FC = -11.46; FDR-P = 0.0037) and MYOM1 (log2FC = -8.81; FDR-P value 3.56 × 10-5) was apparent in these lymphocytes. A full list of DEGs detected across the different biological contrasts is provided in Supplementary Table S3.
This comprehensive whole transcriptome dataset will be of value to the research community for insights into the cell subset specificity of effector molecule expression on γδ T cells in both healthy and bTB+ cattle. The cell-specific expression of KIR receptors in cattle, for example, remains incomplete and emerging research documents a broader expression than has been found in primates, including in bovine monocytes and in B cells (78). This work therefore contributes to the documentation of KIR expression in an additional cell subset, the bovine γδ T cell, which may contribute to more effective targeting of these cells to improve vaccination responses to BCG (79). Naturally infected cattle offer a very valuable model for the study of bTB, but this model is subject to certain caveats as the complete infection, environmental and management history of the cattle cannot be completely controlled. Delineating any potential effects of comorbidities including subclinical disease will be an important focus of future work to further refine the specificity of the bovine γδ T cell response with a specific focus on the cytotoxic functions of the WC1.1+ subset and their role in the development of protective immunity against mycobacteria.
The eradication of bTB is a major policy aim in countries where the disease is endemic as it has widespread negative impacts on the agri-food sector and poses a zoonotic risk to human health (3). A comprehensive understanding of all aspects of the host-pathogen relationship are required to better understand immune mechanisms associated with pathogenesis and to improve diagnostic sensitivity and vaccination performance (80).
Gamma-delta (γδ) T cells connect the innate and adaptive arms of the immune response with known anti-mycobacterial function but although expanded in cattle, knowledge of their functional capacity in livestock species remains limited. Here we document significant differential expression indicating specific activation of WC1.1+ γδ T cells in response to M. bovis infection. We have identified differential regulation of multiple genes regulating cytotoxic functions of anti-mycobacterial relevance and that shed new light on the functional capacity of bovine γδ T cells under natural infection conditions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ebi.ac.uk/arrayexpress/, E-MTAB-13111.
All animal procedures and experimental protocols in this study were approved by the Teagasc Animal Ethics Committee (TAEC217-2019) and carried out in accordance with the relevant institutional guidelines and under license from the Irish Health Products Regulatory Authority (HPRA no. AE19132/I019). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
SB: Investigation, Writing – original draft, Writing – review & editing, Formal Analysis. ME: Formal Analysis, Investigation, Writing – review & editing. TH: Formal Analysis, Writing – review & editing, Data curation, Methodology. GM: Formal Analysis, Methodology, Writing – review & editing. CR: Formal Analysis, Methodology, Writing – review & editing. DM: Methodology, Writing – review & editing. KM: Writing – review & editing, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was funded by Science Foundation Ireland (SFI) Grants 17/CDA/4717 and SFI/15/IA/3154.
The authors would like to express their gratitude to Edward Mulligan, Colm Brady and the staff at the Department of Agriculture, Food, and the Marine (DAFM) farm in Longtown, Kildare, Ireland for their help with sample collection. In addition, we would like to thank Barry Moran (Trinity College Dublin) for his expertise with flow cytometry and Dr Alia Parveen (University College Dublin) for her assistance with data file submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1265038/full#supplementary-material
Supplementary Table 1 | Single intradermal comparative tuberculin test (SICTT), IFN-γ release assay results, bTB classification and sampling information (Contained in Excel file – Supplementary Information File 1).
Supplementary Table 2 | Filtering and mapping statistics for 48 RNA-seq libraries from M. bovis-infected and non-infected control animals (Contained in Excel file – Supplementary Information File 2).
Supplementary Table 3 | Differentially expressed genes (DEGs) in the γδ T cell subsets from M. bovis-infected and non-infected control animals (statistically significant DEGs (included are those which had RNA seq reads ≥ 3 animals and showed ≥ ± 1.5-fold change in expression with FDR p adj < 0.1) shown. (Contained in Excel file – Supplementary Information File 3).
Supplementary Table 4 | Canonical pathways identified using IPA for RNA-seq DE gene results in the WC1.1+ γδ T cells from M. bovis-infected and non-infected control animals (statistically significant pathways (adjusted P ≤ 0.1) shown and ranked according to P-value [smallest to largest]). (Contained in Excel file – Supplementary Information File 4).
Supplementary Table 5 | Biological networks identified using IPA for RNA-seq DE gene results in the WC1.1+ γδ T cells from M. bovis-infected and non-infected control animals (statistically significant networks (adjusted P ≤ 0.1) shown and ranked or scored according to the connectivity and interactions between the genes in the dataset [largest to smallest]). (Contained in Excel file – Supplementary Information File 5).
Supplementary Table 6 | Diseases and biological (cellular and molecular) functions identified using IPA for RNA-seq DE gene results in the WC1.1+ γδ T cells from M. bovis-infected and non-infected control animals (statistically significant (adjusted P ≤ 0.1) shown and ranked according to P-value [smallest to largest]). (Contained in Excel file – Supplementary Information File 6).
1. Borham M, Oreiby A, El-Gedawy A, Hegazy Y, Khalifa HO, Al-Gaabary M, et al. Review on bovine tuberculosis: an emerging disease associated with multidrug-resistant mycobacterium species. Pathogens (2022) 11(7). doi: 10.3390/pathogens11070715
2. Quadri NS, Brihn A, Shah JA, Kirsch JD. Bovine tuberculosis: A re-emerging zoonotic infection. J Agromedicine (2021) 26(3):334–9. doi: 10.1080/1059924X.2020.1771497
3. Allen AR, Skuce RA, Byrne AW. Bovine tuberculosis in Britain and Ireland - A perfect storm? the confluence of potential ecological and epidemiological impediments to controlling a chronic infectious disease. Front Vet Sci (2018) 5:109. doi: 10.3389/fvets.2018.00109
4. Cassidy JP, Martineau AR. Innate resistance to tuberculosis in man, cattle and laboratory animal models: nipping disease in the bud? J Comp Pathol (2014) 151(4):291–308. doi: 10.1016/j.jcpa.2014.08.001
5. Hogan LH, Heninger E, Elsner RA, Vonderheid HA, Hulseberg P, Co D, et al. Requirements for CD4(+) T cell levels in acute Mycobacterium bovis strain bacille Calmette Guerin (BCG)-induced granulomas differ for optimal mycobacterial control versus granuloma formation. Int Immunol (2007) 19(5):627–33. doi: 10.1093/intimm/dxm028
6. Ngai P, McCormick S, Small C, Zhang X, Zganiacz A, Aoki N, et al. Gamma interferon responses of CD4 and CD8 T-cell subsets are quantitatively different and independent of each other during pulmonary Mycobacterium bovis BCG infection. Infect Immun (2007) 75(5):2244–52. doi: 10.1128/IAI.00024-07
7. Villarreal-Ramos B, McAulay M, Chance V, Martin M, Morgan J, Howard CJ. Investigation of the role of CD8+ T cells in bovine tuberculosis in vivo. Infect Immun (2003) 71(8):4297–303. doi: 10.1128/IAI.71.8.4297-4303.2003
8. Boer MC, van Meijgaarden KE, Joosten SA, Ottenhoff TH. CD8+ regulatory T cells, and not CD4+ T cells, dominate suppressive phenotype and function after in vitro live Mycobacterium bovis-BCG activation of human cells. PLoS One (2014) 9(4):e94192. doi: 10.1371/journal.pone.0094192
9. Fahl SP, Coffey F, Wiest DL. Origins of gammadelta T cell effector subsets: a riddle wrapped in an enigma. J Immunol (2014) 193(9):4289–94. doi: 10.4049/jimmunol.1401813
10. Holderness J, Hedges JF, Ramstead A, Jutila MA. Comparative biology of gammadelta T cell function in humans, mice, and domestic animals. Annu Rev Anim Biosci (2013) 1:99–124. doi: 10.1146/annurev-animal-031412-103639
11. Guerra-Maupome M, Slate JR, McGill JL. Gamma delta T cell function in ruminants. Vet Clin North Am Food Anim Pract (2019) 35(3):453–69. doi: 10.1016/j.cvfa.2019.08.001
12. Herzig CT, Waters RW, Baldwin CL, Telfer JC. Evolution of the CD163 family and its relationship to the bovine gamma delta T cell co-receptor WC1. BMC Evol Biol (2010) 10:181. doi: 10.1186/1471-2148-10-181
13. Chen C, Herzig CT, Telfer JC, Baldwin CL. Antigenic basis of diversity in the gammadelta T cell co-receptor WC1 family. Mol Immunol (2009) 46(13):2565–75. doi: 10.1016/j.molimm.2009.05.010
14. Boom WH. Gammadelta T cells and Mycobacterium tuberculosis. Microbes Infect (1999) 1(3):187–95. doi: 10.1016/S1286-4579(99)80033-1
15. Welsh MD, Kennedy HE, Smyth AJ, Girvin RM, Andersen P, Pollock JM. Responses of bovine WC1(+) gammadelta T cells to protein and nonprotein antigens of Mycobacterium bovis. Infect Immun (2002) 70(11):6114–20. doi: 10.1128/IAI.70.11.6114-6120.2002
16. McGill JL, Sacco RE, Baldwin CL, Telfer JC, Palmer MV, Waters WR. Specific recognition of mycobacterial protein and peptide antigens by gammadelta T cell subsets following infection with virulent Mycobacterium bovis. J Immunol (2014) 192(6):2756–69. doi: 10.4049/jimmunol.1302567
17. Vesosky B, Turner OC, Turner J, Orme IM. Gamma interferon production by bovine gamma delta T cells following stimulation with mycobacterial mycolylarabinogalactan peptidoglycan. Infect Immun (2004) 72(8):4612–8. doi: 10.1128/IAI.72.8.4612-4618.2004
18. Rusk RA, Palmer MV, Waters WR, McGill JL. Measuring bovine gammadelta T cell function at the site of Mycobacterium bovis infection. Vet Immunol Immunopathol (2017) 193-194:38–49. doi: 10.1016/j.vetimm.2017.10.004
19. Baquero MM, Plattner BL. Bovine WC1(+) and WC1(neg) gammadelta T Lymphocytes Influence Monocyte Differentiation and Monocyte-Derived Dendritic Cell Maturation during In Vitro Mycobacterium avium Subspecies paratuberculosis Infection. Front Immunol (2017) 8:534. doi: 10.3389/fimmu.2017.00534
20. Baquero MM, Plattner BL. Bovine peripheral blood WC1(+) and WC1(neg) gammadelta T lymphocytes modulate monocyte-derived macrophage effector functions during in vitro Mycobacterium avium subspecies paratuberculosis infection. Cell Immunol (2017) 315:34–44. doi: 10.1016/j.cellimm.2017.01.009
21. Baquero MM, Plattner BL. Bovine WC1(+) gammadelta T lymphocytes modify monocyte-derived macrophage responses during early Mycobacterium avium subspecies paratuberculosis infection. Vet Immunol Immunopathol (2016) 170:65–72. doi: 10.1016/j.vetimm.2015.12.002
22. Guzman E, Hope J, Taylor G, Smith AL, Cubillos-Zapata C, Charleston B. Bovine gammadelta T cells are a major regulatory T cell subset. J Immunol (2014) 193(1):208–22. doi: 10.4049/jimmunol.1303398
23. Eberl M, Roberts GW, Meuter S, Williams JD, Topley N, Moser B. A rapid crosstalk of human gammadelta T cells and monocytes drives the acute inflammation in bacterial infections. PLoS Pathog (2009) 5(2):e1000308. doi: 10.1371/journal.ppat.1000308
24. Papotto PH, Yilmaz B, Silva-Santos B. Crosstalk between gammadelta T cells and the microbiota. Nat Microbiol (2021) 6(9):1110–7. doi: 10.1038/s41564-021-00948-2
25. Doherty R, Whiston R, Cormican P, Finlay EK, Couldrey C, Brady C, et al. The CD4(+) T cell methylome contributes to a distinct CD4(+) T cell transcriptional signature in Mycobacterium bovis-infected cattle. Sci Rep (2016) 6:31014. doi: 10.1038/srep31014
26. Eder JM, Gorden PJ, Lippolis JD, Reinhardt TA, Sacco RE. Lactation stage impacts the glycolytic function of bovine CD4(+) T cells during ex vivo activation. Sci Rep (2020) 10(1):4045. doi: 10.1038/s41598-020-60691-2
27. Andrews S. FastQC: a quality control tool for high throughput sequence data. Cambridge, UK: Bioinformatics Group, Babraham Institute, Babraham Research Campus (2019).
28. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (2018) 34(17):i884–i90. doi: 10.1093/bioinformatics/bty560
29. Hall TJ, Mullen MP, McHugo GP, Killick KE, Ring SC, Berry DP, et al. Integrative genomics of the mammalian alveolar macrophage response to intracellular mycobacteria. BMC Genomics (2021) 22(1):343. doi: 10.1186/s12864-021-07643-w
30. Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience (2020) 9(3). doi: 10.1093/gigascience/giaa021
31. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (2013) 29(1):15–21. doi: 10.1093/bioinformatics/bts635
32. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (2014) 30(7):923–30. doi: 10.1093/bioinformatics/btt656
33. Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, et al. BioMart–biological queries made easy. BMC Genomics (2009) 10:22. doi: 10.1186/1471-2164-10-22
34. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15(12):550. doi: 10.1186/s13059-014-0550-8
35. Benjamini YH Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc (1995) 57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
36. Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics (2014) 30(4):523–30. doi: 10.1093/bioinformatics/btt703
37. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The geneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinf (2016) 54:1 30 1–1 3. doi: 10.1002/cpbi.5
38. Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res (2022) 50(D1):D20–D6. doi: 10.1093/nar/gkab1112
39. UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res (2021) 49(D1):D480–D9. doi: 10.1093/nar/gkaa1100
40. Piedra-Quintero ZL, Wilson Z, Nava P, Guerau-de-Arellano M. CD38: an immunomodulatory molecule in inflammation and autoimmunity. Front Immunol (2020) 11:597959. doi: 10.3389/fimmu.2020.597959
41. Kadivar M, Petersson J, Svensson L, Marsal J. CD8alphabeta+ gammadelta T cells: A novel T cell subset with a potential role in inflammatory bowel disease. J Immunol (2016) 197(12):4584–92. doi: 10.4049/jimmunol.1601146
42. Lim TS, Goh JK, Mortellaro A, Lim CT, Hammerling GJ, Ricciardi-Castagnoli P. CD80 and CD86 differentially regulate mechanical interactions of T-cells with antigen-presenting dendritic cells and B-cells. PLoS One (2012) 7(9):e45185. doi: 10.1371/journal.pone.0045185
43. Appay V, Dunbar PR, Cerundolo V, McMichael A, Czaplewski L, Rowland-Jones S. RANTES activates antigen-specific cytotoxic T lymphocytes in a mitogen-like manner through cell surface aggregation. Int Immunol (2000) 12(8):1173–82. doi: 10.1093/intimm/12.8.1173
44. Seth S, Oberdorfer L, Hyde R, Hoff K, Thies V, Worbs T, et al. CCR7 essentially contributes to the homing of plasmacytoid dendritic cells to lymph nodes under steady-state as well as inflammatory conditions. J Immunol (2011) 186(6):3364–72. doi: 10.4049/jimmunol.1002598
45. Merlo LMF, DuHadaway JB, Montgomery JD, Peng WD, Murray PJ, Prendergast GC, et al. Differential roles of IDO1 and IDO2 in T and B cell inflammatory immune responses. Front Immunol (2020) 11:1861. doi: 10.3389/fimmu.2020.01861
46. Zizzo G, Hilliard BA, Monestier M, Cohen PL. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol (2012) 189(7):3508–20. doi: 10.4049/jimmunol.1200662
47. Teng O, Chen ST, Hsu TL, Sia SF, Cole S, Valkenburg SA, et al. CLEC5A-mediated enhancement of the inflammatory response in myeloid cells contributes to influenza virus pathogenicity in vivo. J Virol (2017) 91(1). doi: 10.1128/JVI.01813-16
48. Das S, Banerjee S, Majumder S, Chowdhury BP, Goswami A, Halder K, et al. Immune subversion by Mycobacterium tuberculosis through CCR5 mediated signaling: involvement of IL-10. PLoS One (2014) 9(4):e92477. doi: 10.1371/journal.pone.0092477
49. Rogers AN, VanBuren DG, Hedblom E, Tilahun ME, Telfer JC, Baldwin CL. Function of ruminant gammadelta T cells is defined by WC1.1 or WC1.2 isoform expression. Vet Immunol Immunopathol (2005) 108(1-2):211–7. doi: 10.1016/j.vetimm.2005.08.008
50. Price S, Davies M, Villarreal-Ramos B, Hope J. Differential distribution of WC1(+) gammadelta TCR(+) T lymphocyte subsets within lymphoid tissues of the head and respiratory tract and effects of intranasal M. bovis BCG vaccination. Vet Immunol Immunopathol (2010) 136(1-2):133–7. doi: 10.1016/j.vetimm.2010.02.010
51. Garand M, Goodier M, Owolabi O, Donkor S, Kampmann B, Sutherland JS. Functional and Phenotypic Changes of Natural Killer Cells in Whole Blood during Mycobacterium tuberculosis Infection and Disease. Front Immunol (2018) 9:257. doi: 10.3389/fimmu.2018.00257
52. Walch M, Dotiwala F, Mulik S, Thiery J, Kirchhausen T, Clayberger C, et al. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell (2014) 157(6):1309–23. doi: 10.1016/j.cell.2014.03.062
53. Dieli F, Troye-Blomberg M, Ivanyi J, Fournie JJ, Krensky AM, Bonneville M, et al. Granulysin-dependent killing of intracellular and extracellular Mycobacterium tuberculosis by Vgamma9/Vdelta2 T lymphocytes. J Infect Dis (2001) 184(8):1082–5. doi: 10.1086/323600
54. Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A (2015) 112(29):E3883–92. doi: 10.1073/pnas.1501662112
55. Yang B, Wang X, Jiang J, Cheng X. Involvement of CD244 in regulating CD4+ T cell immunity in patients with active tuberculosis. PLoS One (2013) 8(4):e63261. doi: 10.1371/journal.pone.0063261
56. Ribeiro ST, Ribot JC, Silva-Santos B. Five layers of receptor signaling in gammadelta T-cell differentiation and activation. Front Immunol (2015) 6:15. doi: 10.3389/fimmu.2015.00015
57. Rasi V, Wood DC, Eickhoff CS, Xia M, Pozzi N, Edwards RL, et al. Granzyme A produced by gamma(9)delta(2) T cells activates ER stress responses and ATP production, and protects against intracellular mycobacterial replication independent of enzymatic activity. Front Immunol (2021) 12:712678. doi: 10.3389/fimmu.2021.712678
58. Krenacs L, Smyth MJ, Bagdi E, Krenacs T, Kopper L, Rudiger T, et al. The serine protease granzyme M is preferentially expressed in NK-cell, gamma delta T-cell, and intestinal T-cell lymphomas: evidence of origin from lymphocytes involved in innate immunity. Blood (2003) 101(9):3590–3. doi: 10.1182/blood-2002-09-2908
59. Lino CNR, Barros-Martins J, Oberdorfer L, Walzer T, Prinz I. Eomes expression reports the progressive differentiation of IFN-gamma-producing Th1-like gammadelta T cells. Eur J Immunol (2017) 47(6):970–81. doi: 10.1002/eji.201646753
60. Chen L, He W, Kim ST, Tao J, Gao Y, Chi H, et al. Epigenetic and transcriptional programs lead to default IFN-gamma production by gammadelta T cells. J Immunol (2007) 178(5):2730–6. doi: 10.4049/jimmunol.178.5.2730
61. Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol (2012) 12(11):749–61. doi: 10.1038/nri3307
62. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell (2000) 100(6):655–69. doi: 10.1016/S0092-8674(00)80702-3
63. Yang R, Mele F, Worley L, Langlais D, Rosain J, Benhsaien I, et al. Human T-bet governs innate and innate-like adaptive IFN-gamma immunity against mycobacteria. Cell (2020) 183(7):1826–47.e31. doi: 10.1016/j.cell.2020.10.046
64. Malone KM, Rue-Albrecht K, Magee DA, Conlon K, Schubert OT, Nalpas NC, et al. Comparative 'omics analyses differentiate Mycobacterium tuberculosis and Mycobacterium bovis and reveal distinct macrophage responses to infection with the human and bovine tubercle bacilli. Microb Genom (2018) 4(3). doi: 10.1099/mgen.0.000163
65. Konduri V, Oyewole-Said D, Vazquez-Perez J, Weldon SA, Halpert MM, Levitt JM, et al. CD8(+)CD161(+) T-cells: cytotoxic memory cells with high therapeutic potential. Front Immunol (2020) 11:613204. doi: 10.3389/fimmu.2020.613204
66. Spear P, Wu MR, Sentman ML, Sentman CL. NKG2D ligands as therapeutic targets. Cancer Immun (2013) 13:8.
67. Bortolotti D, Gentili V, Rizzo S, Rotola A, Rizzo R. SARS-CoV-2 spike 1 protein controls natural killer cell activation via the HLA-E/NKG2A pathway. Cells (2020) 9(9). doi: 10.3390/cells9091975
68. Andre P, Denis C, Soulas C, Bourbon-Caillet C, Lopez J, Arnoux T, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell (2018) 175(7):1731–43 e13. doi: 10.1016/j.cell.2018.10.014
69. Kamiya T, Seow SV, Wong D, Robinson M, Campana D. Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J Clin Invest (2019) 129(5):2094–106. doi: 10.1172/JCI123955
70. Rincon-Orozco B, Kunzmann V, Wrobel P, Kabelitz D, Steinle A, Herrmann T. Activation of V gamma 9V delta 2 T cells by NKG2D. J Immunol (2005) 175(4):2144–51. doi: 10.4049/jimmunol.175.4.2144
71. Borchers MT, Harris NL, Wesselkamper SC, Zhang S, Chen Y, Young L, et al. The NKG2D-activating receptor mediates pulmonary clearance of Pseudomonas aeruginosa. Infect Immun (2006) 74(5):2578–86. doi: 10.1128/IAI.74.5.2578-2586.2006
72. Pydi SS, Sunder SR, Venkatasubramanian S, Kovvali S, Jonnalagada S, Valluri VL. Killer cell immunoglobulin like receptor gene association with tuberculosis. Hum Immunol (2013) 74(1):85–92. doi: 10.1016/j.humimm.2012.10.006
73. Salie M, Daya M, Moller M, Hoal EG. Activating KIRs alter susceptibility to pulmonary tuberculosis in a South African population. Tuberculosis (Edinb) (2015) 95(6):817–21. doi: 10.1016/j.tube.2015.09.003
74. Ladel CH, Blum C, Dreher A, Reifenberg K, Kaufmann SH. Protective role of gamma/delta T cells and alpha/beta T cells in tuberculosis. Eur J Immunol (1995) 25(10):2877–81. doi: 10.1002/eji.1830251025
75. Feng CG, Kaviratne M, Rothfuchs AG, Cheever A, Hieny S, Young HA, et al. NK cell-derived IFN-gamma differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J Immunol (2006) 177(10):7086–93. doi: 10.4049/jimmunol.177.10.7086
76. Cai Y, Dai Y, Wang Y, Yang Q, Guo J, Wei C, et al. Single-cell transcriptomics of blood reveals a natural killer cell subset depletion in tuberculosis. EBioMedicine (2020) 53:102686. doi: 10.1016/j.ebiom.2020.102686
77. Kundu TK, Velayutham M, Zweier JL. Aldehyde oxidase functions as a superoxide generating NADH oxidase: an important redox regulated pathway of cellular oxygen radical formation. Biochemistry (2012) 51(13):2930–9. doi: 10.1021/bi3000879
78. Hay AL, Birch J, Ellis S, Burns D, Mansour S, Khakoo SI, et al. Cattle killer immunoglobulin-like receptor expression on leukocyte subsets suggests functional divergence compared to humans. Vet Immunol Immunopathol (2023) 263:110646. doi: 10.1016/j.vetimm.2023.110646
79. Guerra-Maupome M, McGill JL. Characterization of local and circulating bovine gammadelta T cell responses to respiratory BCG vaccination. Sci Rep (2019) 9(1):15996. doi: 10.1038/s41598-019-52565-z
Keywords: WC1+ γδ T cells, natural Mycobacterium bovis, bovine, Nk receptors, RNA-seq - RNA sequencing
Citation: Bhat SA, Elnaggar M, Hall TJ, McHugo GP, Reid C, MacHugh DE and Meade KG (2023) Preferential differential gene expression within the WC1.1+ γδ T cell compartment in cattle naturally infected with Mycobacterium bovis. Front. Immunol. 14:1265038. doi: 10.3389/fimmu.2023.1265038
Received: 21 July 2023; Accepted: 02 October 2023;
Published: 24 October 2023.
Edited by:
Amy Barczak, Massachusetts General Hospital and Harvard Medical School, United StatesReviewed by:
Berit Carow, Novavax, SwedenCopyright © 2023 Bhat, Elnaggar, Hall, McHugo, Reid, MacHugh and Meade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kieran G. Meade, a2llcmFuLm1lYWRlQHVjZC5pZQ==
†Present addresses: Mahmoud Elnaggar, Department of Veterinary Medicine, College of Applied and Health Sciences, A’Sharqiyah University, Ibra, Oman Department of Microbiology, Faculty of Veterinary Medicine, Alexandria University, Alexandria, Egypt
Cian Reid, School of Biochemistry and Immunology, Trinity College Dublin, Dublin, Ireland
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.