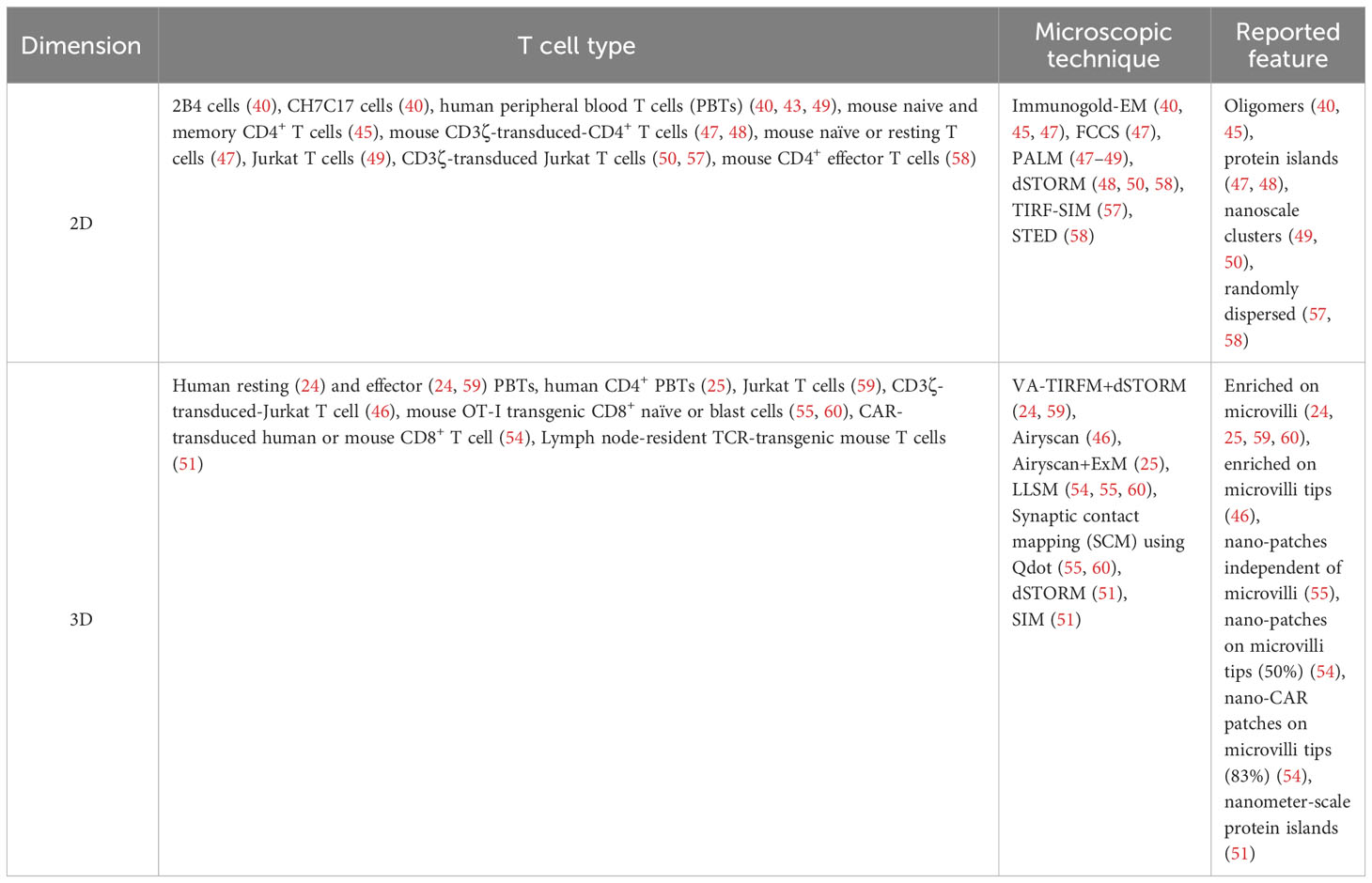
- 1Department of Nano-Biomedical Engineering, Advanced Science Institute, Yonsei University, Seoul, Republic of Korea
- 2Center for Nanomedicine, Institute for Basic Science, Seoul, Republic of Korea
T cell activation is initiated by the recognition of specific antigenic peptides and subsequently accomplished by complex signaling cascades. These aspects have been extensively studied for decades as pivotal factors in the establishment of adaptive immunity. However, how receptors or signaling molecules are organized in the resting state prior to encountering antigens has received less attention. Recent advancements in super-resolution microscopy techniques have revealed topographically controlled pre-formed organization of key molecules involved in antigen recognition and signal transduction on microvillar projections of T cells before activation and substantial effort has been dedicated to characterizing the topological structure of resting T cells over the past decade. This review will summarize our current understanding of how key surface receptors are pre-organized on the T-cell plasma membrane and discuss the potential role of these receptors, which are preassembled prior to ligand binding in the early activation events of T cells.
Introduction
T cells play a major role in establishing and orchestrating adaptive immune responses that defend the host against viral infections, cancer, and autoimmune diseases. Triggering of the antigen-specific T cell receptor (TCR) and subsequent signaling cascades have been extensively reviewed elsewhere (1, 2). This review focuses on the pre-organization of the T cell plasma membrane in the quiescent state, i.e., prior to antigen engagement, in order to provide a more comprehensive view of the molecular organization involved in the optimization of T cell activity. Pre-organized receptors are often reported to form nanoscale clusters. Although investigations using immunogold electron microscopy (EM) have revealed some pre-clustered receptors, many pre-organized proteins have remained unresolved due to diffraction limit until the advent of super-resolution imaging. Super-resolution imaging enables to resolve sub-diffraction limit by utilizing single-molecule localization precision (photoactivated localization microscopy (PALM) (3, 4), stochastic optical reconstruction microscopy (STORM) (5), points accumulation for imaging in nanoscale topography (PAINT) (6), etc.), or by shaping or modulating the illumination (stimulated emission depletion (STED) (7), structured-illumination microscopy (SIM) (8), lattice light sheet microscopy (LLSM (9), etc.). Another approach for super-resolution imaging, expansion microscopy (ExM) technique, improves resolution by physical expansion of hydrogel-embedded specimens (10). The detailed technical aspects of the super-resolution imaging techniques that are used for studying pre-organized clusters are reviewed elsewhere (11–13).
Receptor pre-organization is established and maintained in the naïve or resting T cell state and is, therefore, more likely to be controlled by the intrinsic physical properties of the plasma membrane or protein itself, rather than by the outcome of specific signaling events triggered by receptor-ligand interactions. For instance, the heterogeneous lipid composition, especially the presence of cholesterol or sphingolipids, leads to the compartmentalization of lipid domains known as lipid rafts within the plasma membrane. Lipid rafts are small (10–200) and dynamic assemblies capable of being clustered into microscopic domains (>300 nm) by protein–protein or protein–lipid interactions (14–16). These domains are commonly characterized by their liquid-ordered (Lo) phase (17, 18). The preferential associations between cholesterol and saturated lipids drive the formation of denser and thicker membrane domains that selectively recruit certain lipids and proteins, thereby contributing to receptor clustering, protein sorting, and downstream signaling events (15, 19, 20). Multimeric interactions between receptors can drive the formation of nanodomains in the plasma membrane (21, 22). Topographical structures, such as protrusions, can also cause the segregation of receptors (23–25). The ability of membrane proteins to associate with cortical cytoskeleton may also affect the localization and clustering of receptors (24, 26, 27). Presumably, the combination of these properties influences the arrangement of a pre-organized landscape on the T-cell plasma membrane, making it ready for sensitive, selective, and effective triggering and activation when they encounter cognate antigens.
Pre-existing clusters of T cell receptors
The TCR complex is composed of a disulfide-linked dimers consisting of TCRαβ, CD3γϵ and CD3δϵ, and a CD3ζζ homodimer (28). The extracellular domain of the TCRαβ dimer binds to specific antigens presented on MHC molecules on antigen presenting cells (APCs), leading to initiation of TCR signaling. This signaling is initiated by phosphorylation of multiple immune receptor tyrosine-based activation motifs (ITAMs) located in the intracellular domains of the CD3 subunits (1, 29, 30). Rapid changes in TCR localization have been observed upon activation using conventional microscopy. When TCRs bind to cognate antigens, they accumulate in the central region of the interface between the T cell and APC, called the immunological synapse (IS), forming clusters 1-5 µm in diameter observed by confocal microscopy (31, 32). Total internal reflection fluorescence microscopy (TIRFM) has been used to visualize TCR clusters of 300-800 nm in diameter along with other membrane associated signaling molecules, which formed within seconds after contact with antigen-functionalized glass surfaces or synthetic planar lipid bilayers (33–36). With the use of these conventional microscopies, TCRs appeared to be randomly distributed before activation. However, some studies have suggested that at least a fraction of TCRs forms dimers or multimers on unstimulated T cells (37–40), while other studies have suggested that TCRs are predominantly monovalent (41–44).
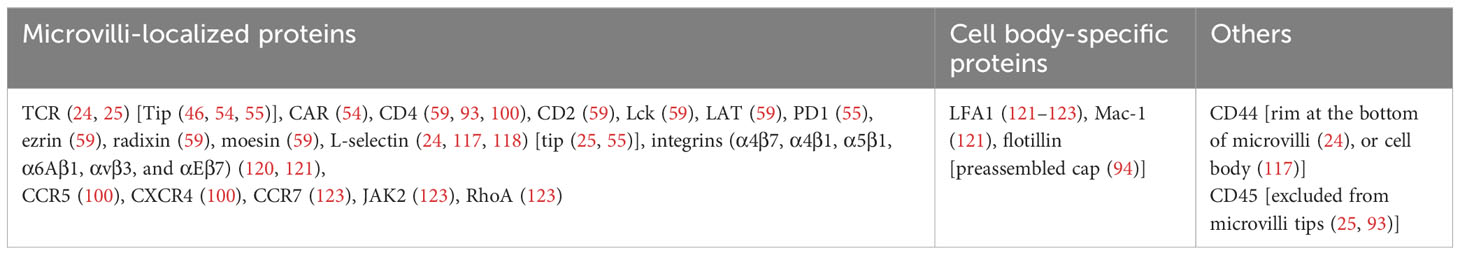
Immunogold-EM has been used to directly visualize multimeric TCRs on unstimulated cells (40, 45) (Figure 1A). After the introduction of fluorescence super-resolution imaging, Davis et al. reported the existence of pre-formed TCR nanoclusters on the plasma membrane of resting T cells using fluorescence super-resolution microscopic technique, PALM or direct STORM (dSTORM) (47, 48). TCR pre-clusters (protein islands) with a radius of 35–70 nm are observed in quiescent mouse T cells bound to both non-activating and activating surfaces. These pre-clusters then come together to form microclusters upon activation of the T cells.
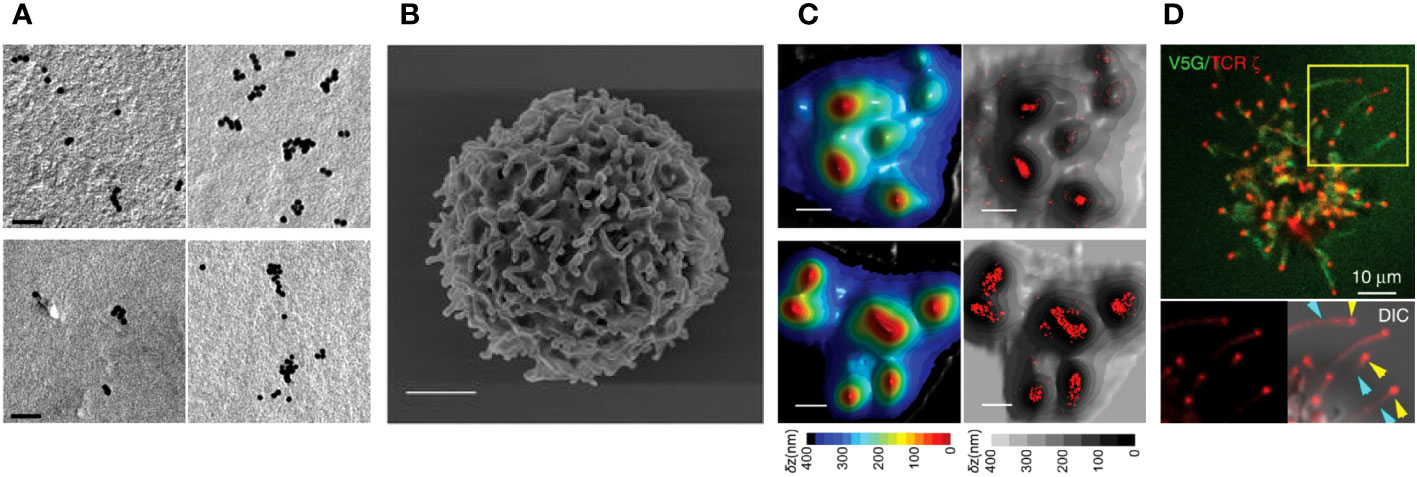
Figure 1 TCR pre-clusters of on human T cells. (A) Pre-clusters of TCRs on 2D surface of T cell membrane. Immunogold-EM images showing the distribution of gold particles of anti-CD3ϵ in human resting (upper-left) and effector (upper-right) T cells and anti-TCRβ in human resting (lower-left) and effector (lower right) T cells. (Scale bars: 50 nm). Reproduced from Kumar et al. (45) with permission from the Journal. (B) A scanning electron microscopy image of a human resting T cells isolated from peripheral blood mononuclear cells showing microvilli protrusions on the surface (Scale bar: 1 µm) (24). (C) High accumulation of TCRs on microvilli of human T cells. 3D surface topography reconstruction maps obtained from VA-TIRFM measurements of resting (upper-left) and effector (lower-left) T cells and STORM images of αβTCR (upper-right) and CD3ϵ molecules (lower-right) of the cells overlaid with the 3D surface reconstruction map presented on the left. Color bars indicate the relative distance (δz) from the surface. (Scale bars: 500 nm). (B, C) are reproduced from Jung et al. (24) with Copyright 2016 National Academy of Sciences. (D) TCRs localized at the tips of microvilli of Jurkat T cells. Airyscan confocal image of activated Jurkat T cell expressing V5G-GFP (green) and TCRζ-ptdTomato (red). Reproduced from Kim et al. (46), licensed under Creative Commons (CC BY 4.0).
The presence of TCR pre-clusters was confirmed by transmission EM (TEM) imaging, where they appeared as clusters having a 40-300 nm radius on resting T cells. Smaller pre-existing TCR nanoclusters have also been reported on T cells spread on non-activating surface using PALM or dSTORM (49, 50). In addition, nano-sized TCR clusters have been observed on lymph node-resident T cells in vivo using dSTORM and SIM (51). These studies revealed the existence of nanoscale pre-clusters prior to activation, but information on the three-dimensional (3D) distribution of TCRs on resting T cells was missing as the T cell plasma membrane is altered when the cells are spread on the two-dimensional (2D) surface.
In 2016, we first reported the localization of TCRs in relation to native surface protrusions using a combination of variable angle of TIRF microscopy (VA-TIRFM) and dSTORM (24). Our study revealed that CD3ϵ and TCRαβ are highly enriched on the microvilli of freshly isolated or effector human T cells in their quiescent state (Figures 1B, C). Microvilli are thin (~70-200 nm in diameter) and short (0.3-0.5µm in median length) actin-based finger-like membrane protrusions that are a dominant feature of the T cell surface (2-4 microvilli/μm2) (52, 53) (Figure 1B). A study using ExM in combination with Airyscan confocal imaging, which enables 3D super-resolution imaging of TCRs on the entire cell surface, has also shown significant accumulation of TCRs on microvilli (25). Visualization of microvilli on Jurkat T cells expressing a fluorescent protein-conjugated TCRζ by Airyscan confocal microscopy revealed high concentrations of TCRs, which specifically accumulated at the microvilli tips (Figure 1D) (46). Using LLSM, accumulation of endogenous TCRs and chimeric antigen receptors (CARs) on CAR- transduced human CD8+ T cell blasts were also observed in 50% and 87% of microvilli tips, respectively (54). However, in ovalbumin-specific TCR transgenic OT-I mice, TCR nano-clusters on CD8+ T cells were not restricted to microvilli, but increased microvilli occupancy was observed upon activation (55). Considering that microvilli are dynamic actin-based structures that undergo continuous transitions between growth, steady-state and retraction cycles (56), it is not surprising that some TCR clusters are not strictly localized in microvilli. It is also possible that the extent of TCR accumulation on microvilli may differ between human and mouse T cells or among different T cell subpopulations, and further studies may be needed to characterize these differences. Interestingly, the presence of high concentrations of B cell receptors (BCRs), which serve equivalent role to that of T cell TCRs, on the microvilli of human B cells has also been observed using super-resolution imaging (25). Although a couple of studies have reported a random distribution of TCRs on resting T cells (57, 58), the cells in these studies were spread on surfaces containing anti-CD45 antibodies or ICAM1, which can cause physical alterations or induce non-canonical signaling. This may explain the discrepancies between their results and other observations of pre-clustered TCRs. The reported features of TCR nanostructures studied in various cell types using different microscopic techniques are summarized in Table 1.
It has been proposed that TCRs in nanoclusters are more efficient for TCR triggering because 80% of the phosphorylated TCRζ is present in clusters containing 30% of the total TCRζ (50). The mechanisms underlying the trafficking of TCRs to microvilli are largely unknown. Actin filaments or specific motor proteins may play a role in transporting or maintaining TCRs on microvilli, as there is evidence that destabilization of the actin cytoskeleton by latrunculin-A leads to dispersion of TCR pre-clusters located on microvilli (24). In addition, microvilli were reported to collapse upon chemokine-induced Rac1 activation (61). These findings also raise interesting questions, such as whether the degree or form of TCR pre-organization depends on the T cell type, whether it changes during T cell development or differentiation, and how it affects T cell immunity.
CD45 pre-exclusion from the tips of microvilli
The tyrosine phosphatase CD45 is ubiquitously expressed in hematopoietic cells and has multiple isoforms produced by alternative splicing (62, 63). CD45 is known to play both positive and negative regulatory roles in T cell activation. CD45 can dephosphorylate both negative and positive regulatory tyrosine residues on lymphocyte-specific protein tyrosine kinase (Lck) (64–66), and CD45 depletion abolishes TCR-mediated signaling, supporting a positive effect on T cell activation (67–69). Other studies support a negative role of CD45, suggesting that its abundant phosphatase activity serves to prevent non-specific or undesirable activation of immune cells (64, 70). CD45 suppresses T cell hyperactivation in a wild-type context (64, 71), and CD45 directly inhibits TCR-induced phosphorylation of CD3 ITAMs, a hallmark of TCR activation (72). In addition, recent studies have reinforced the negative regulatory role of CD45 in inhibiting T cell activation in the absence of high affinity antigens (70, 73).
Upon activation, CD45 is observed to be largely excluded from the central region of the IS, where TCRs are highly accumulated (74, 75). This segregation of CD45 and TCRs during T cell activation has been proposed as a mechanism of TCR triggering (2). The phosphatase activity of TCR-proximal CD45 is negated by its physical separation from the IS, resulting in a shift in equilibrium to phosphorylation of the ITAMs on TCRs, and leading to T cell activation. Two major models have been proposed for the molecular mechanisms of CD45 exclusion. According to the kinetic segregation model, T cells form close contacts with APCs, which then drives small scale segregation of CD45 from the contact regions, resulting in initiation of TCR triggering (76, 77). In this model, the long extracellular domain of CD45 (~25-50 nm in length) causes its exclusion from the IS center due to the narrow gap (~13 nm) between the T cell and APC in this area, which is too short to accommodate the CD45 extracellular domain (77–79). Another model for CD45 exclusion proposes segregation by lipid rafts (80, 81). Approximately 95% of CD45 is found in the non-raft fraction of lipids, while a small fraction (<5%) of CD45 is detected in the raft fraction (80–85). Lipid raft crosslinking with cholera toxin B subunit (CTB) or TCR crosslinking have been shown to exclude CD45 from the aggregation of lipid rafts (81, 86, 87). Altered localization of CD45 in lipid rafts has been reported in patients with systemic lupus erythematosus (SLE) (88–90).
Using ExM combined with Airyscan confocal microscopy and 3D STORM, we recently reported the novel organization of pre-formed of CD45 clusters in resting T cells, where CD45 is excluded from the tips of microvilli before contact with APCs (25) (Figures 2A, B). This feature of CD45 pre-exclusion from microvilli tips is common to human or mouse CD4+, CD8+, and regulatory T cells, as well as B cells. This CD45 pre-exclusion is distinct from the close contact-driven exclusion of CD45 from the microvilli tips reported by other groups (91, 92). We proposed that this pre-exclusion is caused by the diffusion barrier formed by the slightly thicker plasma membranes at the tips of microvilli. This is due to the local enrichment of cholesterol (lipid rafts), which blocks the free diffusion of CD45 whose transmembrane integration limit (MIL) is too short to integrate into the thicker membrane (Figure 2C) (25). We demonstrated that extraction of cholesterol using methyl-β-cyclodextrin (MβCD) significantly reduced the pre-exclusion of CD45 from microvilli tips. Mutant CD45 with increased MIL length displayed reduced pre-exclusion, whereas mutant CD45 lacking the extracellular domain maintained the pre-exclusion at the tips of microvilli, suggesting that CD45 pre-exclusion is mediated by the transmembrane domain of CD45 rather than the extracellular domain (25). This study supports the proposed lipid raft-driven CD45 mechanism, at least when T cells are in a quiescent state. The exclusion of CD45 from the tips of protrusions of unstimulated Jurkat T cells has also been reported by another group (93). In another recent study, we demonstrated that large-scale CD45 exclusion is directly induced by synthetic lipid rafts formed by DNA origami containing multiple cholesterols in the absence of ligation or close contact formation (94). This indicates that local cholesterol concentration plays a direct role in the formation of lipid rafts at the plasma membrane of live T cells as well as the exclusion of CD45 from them.
Figure 2 Pre-exclusion of CD45 from the tips of microvilli. (A) A 4x-ExM-Airyscan image a human resting T cell labeled with anti-CD45 antibodies (green) showing depletion of CD45 from microvilli tips (yellow arrows). (B) Magnified 4x-ExM-Airyscan images of microvilli on human CD4+ T cells labeled with anti-CD45 (green) and FM 4-64FX membrane dye (red), reproduced from Jung et al. (25), licensed under Creative Commons (CC BY 4.0). (C) Schematic illustrating a model of pre-exclusion of CD45 from the microvillus tip of T cell. High accumulation of cholesterol at the microvillus tip thickens the lipid bilayer at the microvillus tip, creating a diffusion barrier for CD45, which has a short membrane integration limit (MIL). Unlike CD45, TCR complex has long MILs, which allow TCRs to localize to the tip of microvilli; thus, TCRs localized at the microvilli tips are free from the phosphatase activity of neighboring CD45 (25).
Given that the native form of the CD45 extracellular domain is physically excluded from residing at tight T cell-APC junctions and that TCR triggering induces the accumulation of lipid rafts at the interface, centric CD45 exclusion during IS formation is probably mediated by a combination of both mechanisms. The relative contribution of the extracellular CD45 domain versus the lipid raft-mediated CD45 exclusion during IS formation is not yet clear. However, our study (25) demonstrated that the pre-formed exclusion of CD45 from the tips of microvilli is directly mediated by the lipid composition of the membrane.
Pre-clustering of coreceptors and proximal TCR signaling molecules
The coreceptors CD4 and CD8 bind to MHC-II and MHC-I class molecules, respectively. Other costimulatory receptors, CD2 and CD28, bind to LFA3 (CD58) and CD80 or CD86, respectively. These receptors play a key role in enhancing adhesion, sensitivity in antigen recognition, and T cell signal augmentation (95–99). Immunogold EM revealed that CD4 is concentrated in microclusters on microvilli of human T cells along with chemokine receptors CCR5 and CXCR4 (100). Roh et al. demonstrated that CD4 is localized in nanoclusters separated from the TCR or Lck at the resting state (48). Another study showed that CD4 fused to the photo-switchable protein mEos2 forms nanoclusters with an average diameter of 100 nm on the surface of resting Jurkat T cells, and that the CD4 nanoclusters are regulated by the extracellular domain and receptor palmitoylation (101). Another study using super-resolution optical fluctuation imaging (SOFI) (102) also reported high-density CD4 regions on the surface of Jurkat T cells (101). Haran’s group also demonstrated that more than 90% of CD4 and CD2 accumulate on the microvilli of human effector T cells and Jurkat T cells (59). In a 3D super-resolution imaging study, CD4 was also reported to accumulate at the microvilli tips, while CD45 was excluded from the tips in unstimulated Jurkat T cells (93). Cai et al. reported that CD8 on mouse OT-I T cells exists as pre-clusters located close to the TCR clusters before activation (55). They also reported that CD28 and the inhibitory receptor PD-1 (103) also form clusters, which are separated from TCR clusters in the quiescent state. These studies indicate that major coreceptors that modulate TCR-induced T cell activation are also not randomly distributed but, rather, exist as pre-clusters even before activation.
Similarly, TCR-proximal signaling molecules that propagate TCR triggering intracellularly, such as Lck kinase and the adaptor protein linker for activation of T cells (LAT) (104), are highly enriched on the microvilli of resting human effectors and Jurkat T cells (59). The microvilli-specific fractions of Lck and LAT were lower than that of TCR, CD4, and CD2, but still quite high at 76% and 68%, respectively. The size of these clusters ranges from 80 to 170 nm, which is similar to the diameter of a microvillus. Previously, Lillemeier et al. reported that LAT is found in clusters distinct from TCR clusters in the plasma membrane of T cells that were flattened on artificial surfaces in the absence of stimulation. In contrast, Sherman et al. reported pre-existing LAT nanoscale clusters that partially overlap with TCR clusters on T cells spread on non-stimulating surfaces (49). The authors also reported that in the absence of stimulation, LAT nanoclusters either partially co-cluster or not with ζ chain-associated protein kinase 70 (Zap70) (105) or with phospholipase C-gamma1 (PLC-γ1) clusters (106, 107), the latter two signaling molecules known to be recruited to the TCRs or LAT, respectively, upon TCR stimulation,. Interestingly, another study also reported that the adaptor protein SH2-domain-containing leukocyte protein of 76 kDa (SLP-76), a cytosolic adaptor known to be recruited to LAT upon T cell activation (108), is recruited to the rim of the LAT clusters on the activating artificial surfaces and, furthermore, that the nano-organization of LAT and SLP-76 is dependent on actin polymerization. These observations may reflect alterations of microvilli as they spread over a 2D surface. It remains unclear whether these signaling molecules are individually present in small clusters on microvilli, but these findings suggest that pre-organized proximal signaling molecules, as well as coreceptors required for early TCR signaling, may be co-localized on a single microvillus. Although the pre-cluster formation of LAT is known to be regulated by protein-protein or protein-lipid interactions (49), the molecular mechanism of the formation of these pre-formed coreceptors and signaling molecules in microclusters is mostly unknown. Together, the high accumulation of these coreceptors, costimulatory receptors, and proximal signaling molecules on microvilli allows for their physical proximity to the TCRs, which favors efficient transmission and amplification of early TCR signals.
ERM proteins in microvilli
The ERM proteins, ezrin, radixin, and moesin, which belong to the band 4.1 superfamily, play an important role in cross-linking the cytoskeleton and plasma membrane (109, 110). Ezrin was first discovered at the intestinal brush border and is known to be highly expressed in intestinal microvilli (111). Radixin was originally purified as a barbed end-capping actin regulatory protein from adherent junctions isolated from rat liver (112), while moesin was isolated as a from bovine uterus heparin-binding protein (113). These three proteins are highly homologous, and binding to actin filaments via their C-terminal domains is induced by their phosphorylation. Once phosphorylated, each ERM protein can cross-link the actin cytoskeleton to specific plasma membrane proteins via their conserved N-terminal FERM domain (110, 114, 115) (Figure 3A). Although ERM proteins have been reported to be concentrated in microvilli at the intestinal brush border (114, 115), their distribution in relation to T cell microvilli was revealed in a recent study using combinatory super-resolution microscopic techniques (59). This study showed high colocalization of phosphorylated ERM proteins with TCRs and actin filaments in microvilli, suggesting that ERM proteins may stabilize TCR assemblies within microvilli prior to TCR ligand binding and T cell activation (Figure 3A). It was also shown that activation of the TCRs induces dephosphorylation of ERM proteins within minutes, leading to rapid dissociation of the actin cytoskeleton from the plasma membrane (116) (Figure 3B). These results imply that the preassembly of ERM proteins may limit the release of microvilli-localized TCRs from the actin cytoskeleton prior to initial TCR activation. However, further studies are needed in order to determine whether the interaction between the TCR and ERM proteins is direct or not, and whether the collapse of ERM protein-stabilized microvilli by actin depolymerization upon T cell activation affects the positioning of TCRs on microvilli.
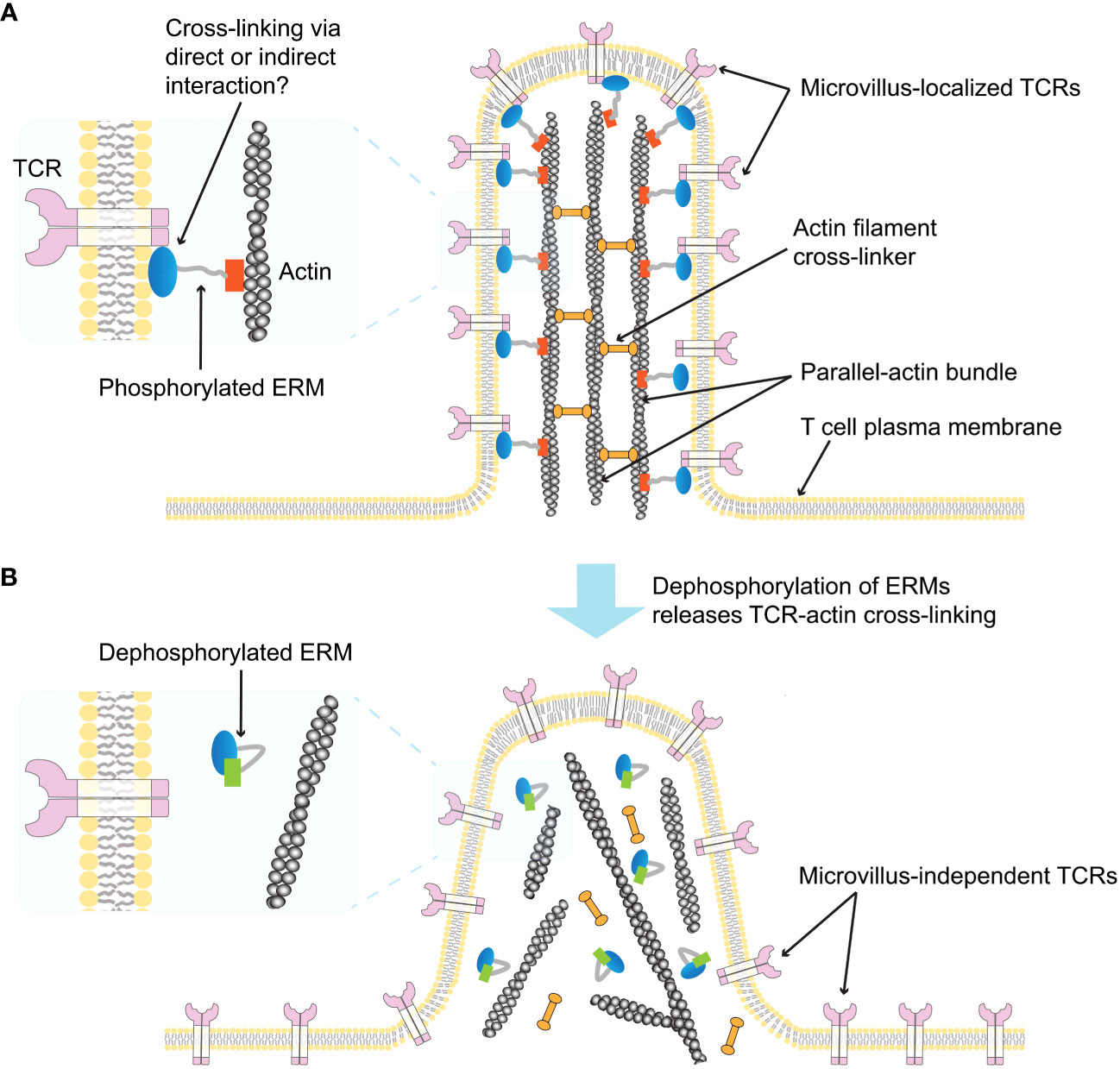
Figure 3 Schematic illustration of ERM-mediated TCR localization on microvilli (59). (A) A single T cell microvillus containing an actin bundle connected by actin-filament crosslinking proteins is depicted. The phosphorylated form of ERM crosslinks parallel actin filaments to the microvillus membrane, and the membrane-bound ERM links the TCR to the microvillus membrane (enlarged left). It is unclear whether the interaction between ERM proteins and the TCR is direct or indirect. (B) Dephosphorylation of ERM proteins releases TCR-actin crosslinks (enlarged left), abolishing the TCR confinement on the microvillus and inducing microvillus collapse.
Adhesion and chemokine receptors
T cells physically interact with their surrounding environment or other cells through a variety of adhesion molecules or chemokine receptors while migrating, homing, arresting, and rolling on the endothelium, or attaching to high endothelial venules in peripheral lymph nodes. Investigations of the localization of these proteins on 3D cell surfaces have relied heavily in the past on immunogold-EM. These studies have shown that L-selectin (117–119), integrins α4β7, α4β1, α5β1, α6Aβ1, αvβ3, and αEβ7 (120, 121), and the chemokine receptors CCR5 and CXCR4 (100) are preferentially localized to the microvilli tips of T cells, whereas CD44 (117) and integrins LFA-1 (CD11a/CD18 or αLβ2) (121, 122) and Mac-1 (CD11b/CD18 or αMβ2) (121) are mainly localized on the flat cell body. Modern super-resolution imaging techniques have also been applied to map the nanoscale distribution of these proteins in T cell plasma membrane. High accumulation of L-selectin (CD62L) on resting T cell microvilli has been demonstrated using several super-resolution imaging techniques (24, 25, 55). In contrast to L-selectin, CD44 has been reported to be localized on the cell body (117, 119). Lately, super-resolution imaging of resting effector human T cells has shown that CD44 accumulates around the base of the microvilli projections but not on the microvilli (24). Recently, the nanoscale topographical distribution of the chemokine receptor CCR7 (a member of G-protein coupled receptor family) and LFA1 has been reported. CCR7 clusters at the tips of microvilli, and 5% of LFA1 is found within a distance of ~20 nm from the CCR7 clusters, while the majority of the LFA1 is located on the cell body in the resting state (123). In addition, RhoA, a key GTPase involved in LFA-1 activation by CCR7-binding chemokines, is highly colocalized with CCR7 at the tips of microvilli. The tyrosine kinase JAK2, which is also involved in rapid CCR7-mediated LFA-1 activation, is also highly accumulated at the microvilli tips of human T cells (123). These results indicate that microvilli may function as signaling hubs ready to respond extremely rapidly to chemokine signals that play an important role in the arrest of T cells that roll on specialized vascular endothelial cells (124). Although it is unclear which membrane or cytoskeletal effector molecules control this exceptional segregation of chemokine receptors and adhesion receptors, the localization of pre-organized adhesive and signaling assemblies on or outside microvilli may also influence the mechanical forces experienced by T cells interacting with endothelial ligands under shear flow (125).
Preassembly of Flot1-rich domain on T cells
Flotillin-1 (Flot1 or Reggie 2) and flotillin-2 (Flot2 or Reggie 1) are ubiquitously expressed scaffold proteins known to localize to lipid rafts in the of plasma membrane of mammalian cells (126–129). Flot1 and Flot2 are oligomerized and are linked to the inner leaflet of the plasma membrane via palmitoylation or myristylation, respectively (130, 131). Interestingly, in T cells, flotillins are known to form polarized cap-like preassembled clusters on the plasma membrane in the resting state (132, 133) and these micron-sized preassembled flotillins are highly stable (94, 133). This preassembly of flotillins is a common feature of hematopoietic cells (132). The accumulation of flotillins in pre-clustered caps is greatly enhanced by cross-linking of glycosylphosphatidylinositol (GPI)-anchored proteins (134, 135), cholera toxins (132, 135), or by anti-CD3/anti-CD28 antibodies (136), which recruit TCRs and TCR-proximal signaling proteins (e.g., Fyn, Lck, or LAT) to the flotillin-rich caps (132–136). In addition, chemokine stimulation causes a high accumulation of flotillin in the T cell uropod, where the adhesion molecule PSGL-1 and phosphorylated ERMs are localized to cause morphological polarization and induce cell motility (137).
The mechanism of flotillins preassembly is unclear. Although a correlation between cytokinesis and polarized flotillins has been reported (132), the molecular mechanisms underlying the formation of the polarized cap of flotillins remain elusive (132). Deletion of the flotillin C-terminus domain results in failure to form preassembled flotillin caps, and cotransfection with a mutant Flot2 lacking its N-terminal domain impairs preassembly formation of Flot2 (133). Although this preassembly defect does not affect the TCR-induced ERK1/2 kinase activation, Ca2+ signaling, or phosphorylation of Zap70, impaired assembly of Flot2 using the N-terminal deletion mutants caused changes in the intracellular localization of Vav, a guanine nucleotide exchange factor that affects cytoskeletal remodeling and cell spreading (133, 138).
The presence of this unique flotillin-rich micron-sized compartment in the plasma membrane of resting T cells is interesting as it is distinct from the mostly reported small-sized lipid rafts (~10-200 nm) present in the plasma membrane (14, 81). Unlike known lipid rafts, the stability of flotillin-rich microdomains is resistant to cholesterol depletion (132, 133). In a recent study, we utilized DNA origami constructs of cholesterol nano-patches (CNPs) to create synthetic lipid rafts on the resting T cell plasma membrane and found that the flotillin-rich domain highly colocalized with the synthetic lipid rafts (94). We also observed that CNP binding induces/enhances the polarization of Flot1. These results indicate that cholesterol can influence the formation and polarization of flotillin assemblies (94). We also found that polarized Flot1 was localized to the cell body and not to microvilli in Jurkat T cells (94).
Little is known about the function of flotillin in T cells. A recent study reported that flotillin is involved in sorting of TCRs during internalization upon anti-CD3/anti-CD28 costimulation, whereas TCR recycling back to the IS is flotillin-independent (139). Depletion of Flot2 demonstrated its essential role in CD44 expression, but its absence affects the cytoskeleton, polarity, and homing defects, as well as the progression of chronic myeloid leukemia (CML), while acute myeloid leukemia (AML) remains unaffected (140). Flot1 deletion in CD8+ T cells resulted in altered morphology and motility in vivo, but had only minor effects on clonal expansion, homing, and migration (141).
The cause(s) of flotillins preassembly on the plasma membrane of resting T cells and its physiological role remains to be elucidated. It is also unclear whether the preassembled flotillins cause the segregation of lipid domains in the plasma membrane or other membrane proteins, or whether this preexisting domain differs from traditional lipid rafts. Similarly, it remains to be determined whether and how preassembled flotillins recruit lipid rafts-associated proteins, GPI-linked proteins, or cholera toxin, as well as IS-forming components, such as TCR, Lck, LAT upon TCR stimulation (142). Further studies will hopefully resolve these questions and provide a better understanding of the role of preassembled Flot1-rich domains in T cell function.
Implications and conclusions
Overall, many of the key receptors involved in T cell activation are not randomly distributed on the T cell plasma membrane but, rather, are present as nano-sized preclusters on the membrane prior to activation. In many cases, such as the TCR, Lck, LAT, CD4, or PD-1, the size of the cluster increases upon ligand binding and initiation of signal transduction (47–51, 55). In addition, significant fractions of many, if not all, of the pre-organized receptors are selectively associated with plasma membrane topography, specifically the microvilli structures, suggesting that microvilli play an important role in early T cell activation. The 3D topographical distribution of the major surface receptors or other proteins in relation to the microvilli structure discussed in this review is summarized in Figure 4A and Table 2. Upon activation, these molecules undergo active redistribution, and the dynamics and spatial arrangement of these receptors in IS formation are highly influenced by the strength and duration of antigenic stimulus, and the stage of activation. The structure of the IS has been described as classical synapses (centralized and symmetric configuration) (Figure 4B), multifocal synapses containing multiple foci of molecular clusters, and motility and asymmetric kinases, extensively reviewed elsewhere (143, 144). The spatial arrangement of receptors in fully developed classical synapses is depicted in Figure 4B. Nevertheless, there is still a lack of understanding how the three-dimensional molecular distribution of these elements evolves in the context of microvilli topographical alterations during the early stages.
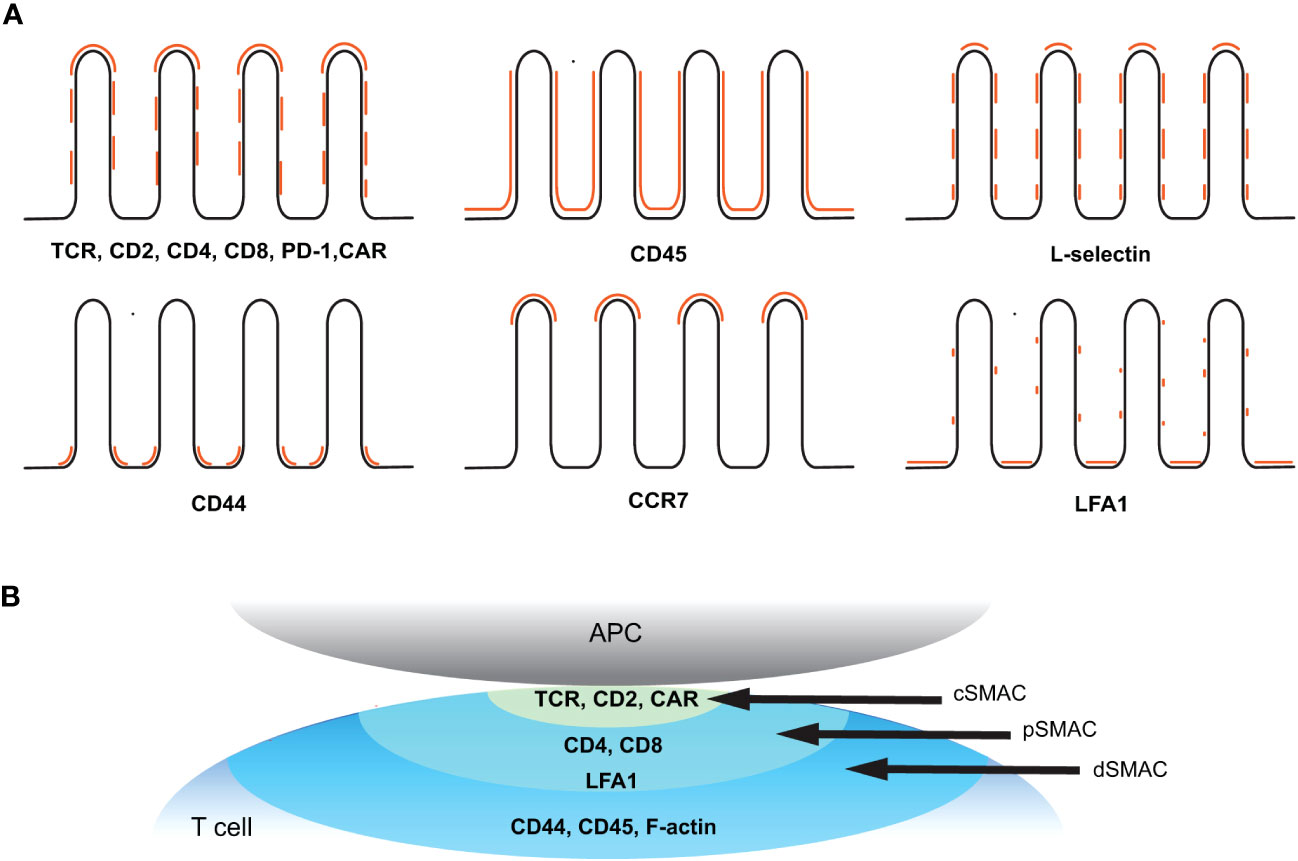
Figure 4 Spatial organization of key receptors before and after T cell activation. (A) Topographical distribution of key receptors on resting T cell membrane. Positions of the indicated receptors in relation to the microvilli (black line) are marked in orange. (B) Molecular distribution of key receptors within a classical immunological synapse after activation. The supramolecular activation cluster (SMAC) structure refers to the spatial organization of molecules within the IS. The central SMAC (cSMAC) contains TCRs and associated signaling molecules, the peripheral SMAC (pSMAC) surrounds the cSMAC and consists of costimulatory molecules and adhesion molecules, and the distal SMAC (dSMAC) at the outer edge of the synapse contains CD44, CD45, and F-actin.
Because T cells use their highly dynamic microvilli to efficiently scan the opposing surface of APCs within minutes (60), it is likely that TCRs, coreceptors, costimulatory receptors, and adhesion receptors preassembled on microvilli are the first molecules interacting with their microenvironment, including antigens presented on APCs. Furthermore, the exclusion of CD45 from the tips of microvilli may allow TCRs to efficiently and very rapidly recognize antigen and initiate contact and triggering during the microvilli-mediated scanning process (25). This implies that microvilli can serve as effective sensors for antigen recognition and signaling hubs for early stage T cell activation by TCR as well as by GPCR signaling (12, 24, 145).
Our understanding of the underlying mechanisms and functional consequences of microvilli in T cell immunity has recently expanded (125, 146–149). It has been demonstrated that microvilli-mediated contacts induce initial calcium triggering (25, 150–152) and that stabilization of the microvilli-mediated close contacts affects effector functions (54, 152). A study using nonporous membranes with varying pore depths, sizes, and interpore distances showed that microvilli confined in 200-nm pores altered gene expression and segregation of signaling molecules that lead to T cell activation and proliferation (153). A theoretical model study suggested that the size and dwell time of TCRs in CD45-depleted T cell-APC close contact regions (likely the microvilli tips), which are constrained by surface topography, are determinants of TCR triggering and ligand discrimination (154). Another recent study demonstrated that microvilli on T cells breach the highly charged glycocalyx barrier on target cells by formation of multiple close contacts via CD2-CD58 interactions while excluding CD45, thereby enabling sensitive antigen discrimination (152). Computational simulation of microvilli dynamics showed that microvilli motility optimizes the balance between scanning speed and antigen sensitivity, promoting positive signaling outcomes while moderately affecting antigen recognition (155).
In addition, the interplay between mechano-transduction and plasma membrane topography has been proposed to play a regulatory role in T cell activation (125, 148, 156, 157). The thin and flexible nature of microvilli influences mechanical forces, such as bending, tension, traction, shear, or the direction of forces exerted on TCR-pMHC binding. A theoretical study suggested that mechanical forces generated by the active motion of scanning microvilli stabilize the catch-bond (stimulatory bond) between TCR and pMHC, resulting in microvilli immobilization to promote further receptor engagement (158). It has also been reported that actin-based protrusions on cytotoxic T cells enhance their killing potency (159) and that actin-rich invadosome-like protrusions on memory and effector T cells can more easily penetrate glycocalyx-coated cells, especially cells of mesenchymal origin (e.g., endothelial and epithelial cells), to facilitate antigen recognition and calcium influx (160).
The unveiling of pre-organized receptor configurations on the T cell plasma membrane has shed light on the conundrum of how resting T cells are ready to rapidly and effectively respond to antigenic challenges. Before being activated, T cells are equipped with these pre-organized receptors on their surface, giving them an advantage in initiating and modulating selective and sensitive responses. The mechanisms underlying the formation and maintenance of these pre-organized receptors are largely unknown, and their physiological functions remain to be further elucidated. A better understanding of the topographical distribution of key surface receptors and the mechanisms that pre-organized these surface receptors on the T cell plasma membrane will help elucidate how T cells generate highly selective and sensitive responses to rare signals (e.g., binding of very few peptide-MHC complexes to the TCR). This will also expand our understanding of subsequent T cell activation, and such knowledge will potentially be highly beneficial for designing T cell-based therapeutics and vaccines.
Author contributions
YJ: Conceptualization, Data curation, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Research Foundation of Korea (NRF-2022R1I1A1A01070462).
Acknowledgments
I thank Prof. Gilad Haran and Prof. Ronen Alon for their kind comments and advice on the manuscript, and Prof. Amnon Altman for help with editing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Courtney AH, Lo W-L, Weiss A. TCR signaling: Mechanisms of initiation and propagation. Trends Biochem Sci (2018) 43(2):108–23. doi: 10.1016/j.tibs.2017.11.008
2. van der Merwe PA, Dushek O. Mechanisms for t cell receptor triggering. Nat Rev Immunol (2011) 11(1):47–55. doi: 10.1038/nri2887
3. Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science (2006) 313(5793):1642–5. doi: 10.1126/science.1127344
4. Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J (2006) 91(11):4258–72. doi: 10.1529/biophysj.106.091116
5. Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods (2006) 3(10):793–5. doi: 10.1038/nmeth929
6. Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci USA (2006) 103(50):18911–6. doi: 10.1073/pnas.0609643104
7. Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated-emission - stimulated-Emission-Depletion fluorescence microscopy. Optics Lett (1994) 19(11):780–2. doi: 10.1364/Ol.19.000780
8. Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci U.S.A. (2005) 102(37):13081–6. doi: 10.1073/pnas.0406877102
9. Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science (2014) 346(6208):1257998. doi: 10.1126/science.1257998
10. Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science (2015) 347(6221):543–8. doi: 10.1126/science.1260088
11. Schermelleh L, Ferrand A, Huser T, Eggeling C, Sauer M, Biehlmaier O, et al. Super-resolution microscopy demystified. Nat Cell Biol (2019) 21(1):72–84. doi: 10.1038/s41556-018-0251-8
12. Pettmann J, Santos AM, Dushek O, Davis SJ. Membrane ultrastructure and t cell activation. Front Immunology 9 (2018). doi: 10.3389/fimmu.2018.02152
13. Brameshuber M, Klotzsch E, Ponjavic A, Sezgin E. Understanding immune signaling using advanced imaging techniques. Biochem Soc Trans (2022) 50(2):853–66. doi: 10.1042/BST20210479
14. Pike LJ. Rafts defined: a report on the keystone symposium on lipid rafts and cell function. J Lipid Res (2006) 47(7):1597–8. doi: 10.1194/jlr.E600002-JLR200
15. Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science (2010) 327(5961):46–50. doi: 10.1126/science.1174621
16. Levental I, Levental KR, Heberle FA. Lipid rafts: Controversies resolved, mysteries remain. Trends Cell Biol (2020) 30(5):341–53. doi: 10.1016/j.tcb.2020.01.009
17. McConnell HM, Vrljic M. Liquid-liquid immiscibility in membranes. Annu Rev Biophysics Biomolecular Structure (2003) 32(1):469–92. doi: 10.1146/annurev.biophys.32.110601.141704
18. Sych T, Gurdap CO, Wedemann L, Sezgin E. How does liquid-liquid phase separation in model membranes reflect cell membrane heterogeneity? Membranes (2021) 11(5):323. doi: 10.3390/membranes11050323
19. Diaz-Rohrer BB, Levental KR, Simons K, Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc Natl Acad Sci (2014) 111(23):8500–5. doi: 10.1073/pnas.1404582111
20. Dinic J, Riehl A, Adler J, Parmryd I. The t cell receptor resides in ordered plasma membrane nanodomains that aggregate upon patching of the receptor. Sci Rep (2015) 5:10082. doi: 10.1038/srep10082
21. Castro M, van Santen HM, Férez M, Alarcón B, Lythe G, Molina-París C. Receptor pre-clustering and t cell responses: Insights into molecular mechanisms. Front Immunol (2014) 5:132. doi: 10.3389/fimmu.2014.00132
22. Woolf PJ, Linderman JJ. Self organization of membrane proteins via dimerization. Biophys Chem (2003) 104(1):217–27. doi: 10.1016/s0301-4622(02)00369-1
23. Ravid Y, Penič S, Mimori-Kiyosue Y, Suetsugu S, A. Iglič and NS. Gov: Theoretical model of membrane protrusions driven by curved active proteins. Front Mol Biosci (2023), 10. doi: 10.3389/fmolb.2023.1153420
24. Jung Y, Riven I, Feigelson SW, Kartvelishvily E, Tohya K, Miyasaka M, et al. Three-dimensional localization of t-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci (2016) 113(40):E5916–24. doi: 10.1073/pnas.1605399113
25. Jung Y, Wen L, Altman A, Ley K. CD45 pre-exclusion from the tips of t cell microvilli prior to antigen recognition. Nat Commun (2021) 12(1):3872.
26. Chichili GR, Rodgers W. Clustering of membrane raft proteins by the actin cytoskeleton*. J Biol Chem (2007) 282(50):36682–91. doi: 10.1074/jbc.M702959200
27. Mattila PK, Batista FD, Treanor B. Dynamics of the actin cytoskeleton mediates receptor cross talk: An emerging concept in tuning receptor signaling. J Cell Biol (2016) 212(3):267–80. doi: 10.1083/jcb.201504137
28. Arechaga I, Swamy M, Abia D, Schamel WA, Alarcón B, Valpuesta JM. Structural characterization of the TCR complex by electron microscopy. Int Immunol (2010) 22(11):897–903. doi: 10.1093/intimm/dxq443
29. Love PE, Hayes SM. ITAM-mediated signaling by the t-cell antigen receptor. Cold Spring Harb Perspect Biol (2010) 2(6):a002485. doi: 10.1101/cshperspect.a002485
30. Davis MM, Boniface JJ, Reich Z, Lyons D, Hampl J, Arden B, et al. Ligand recognition by αβ t cell receptors. Annu Rev Immunol (1998) 16(1):523–44. doi: 10.1146/annurev.immunol.16.1.523
31. Monks CRF, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in t cells. Nature (1998) 395(6697):82–6. doi: 10.1038/25764
32. Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, et al. The immunological synapse: A molecular machine controlling t cell activation. Science (1999) 285(5425):221–7. doi: 10.1126/science.285.5425.221
33. Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol (2002) 158(7):1263–75. doi: 10.1083/jcb.200203043
34. Yokosuka T, Sakata-Sogawa K, Kobayashi W, Hiroshima M, Hashimoto-Tane A, Tokunaga M, et al. Newly generated t cell receptor microclusters initiate and sustain t cell activation by recruitment of Zap70 and SLP-76. Nat Immunol (2005) 6(12):1253–62. doi: 10.1038/ni1272
35. Campi G, Varma R, Dustin ML. Actin and agonist MHC-peptide complex-dependent t cell receptor microclusters as scaffolds for signaling. J Exp Med (2005) 202(8):1031–6. doi: 10.1084/jem.20051182
36. Hashimoto-Tane A, Sakuma M, Ike H, Yokosuka T, Kimura Y, Ohara O, et al. Micro–adhesion rings surrounding TCR microclusters are essential for t cell activation. J Exp Med (2016) 213(8):1609–25. doi: 10.1084/jem.20151088
37. Exley M, Wileman T, Mueller B, Terhorst C. Evidence for multivalent structure of t-cell antigen receptor complex. Mol Immunol (1995) 32(11):829–39. doi: 10.1016/0161-5890(95)00046-H
38. Fernández-Miguel G, Alarcón B, Iglesias A, Bluethmann H, Alvarez-Mon M, Sanz E, et al. Multivalent structure of an αβT cell receptor. Proc Natl Acad Sci (1999) 96(4):1547–52. doi: 10.1073/pnas.96.4.1547
39. Niedergang F, Dautry-Varsat A, Alcover A. Peptide antigen or superantigen-induced down-regulation of TCRs involves both stimulated and unstimulated receptors. J Immunol (1997) 159(4):1703–10. doi: 10.4049/jimmunol.159.4.1703
40. Schamel WWA, Arechaga I, Risueño RM, van Santen HM, Cabezas P, Risco C, et al. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J Exp Med (2005) 202(4):493–503. doi: 10.1084/jem.20042155
41. James JR, White SS, Clarke RW, Johansen AM, Dunne PD, Sleep DL, et al. Single-molecule level analysis of the subunit composition of the t cell receptor on live t cells. Proc Natl Acad Sci (2007) 104(45):17662–7. doi: 10.1073/pnas.0700411104
42. Dunne PD, Fernandes RA, McColl J, Yoon JW, James JR, Davis SJ, et al. DySCo: Quantitating associations of membrane proteins using two-color single-molecule tracking. Biophys J (2009) 97(4):L5–7. doi: 10.1016/j.bpj.2009.05.046
43. James JR, McColl J, Oliveira MI, Dunne PD, Huang E, Jansson A, et al. The t cell receptor triggering apparatus is composed of monovalent or monomeric proteins. J Biol Chem (2011) 286(37):31993–2001. doi: 10.1074/jbc.M111.219212
44. Brameshuber M, Kellner F, Rossboth BK, Ta H, Alge K, Sevcsik E, et al. Monomeric TCRs drive t cell antigen recognition. Nat Immunol (2018) 19(5):487–96. doi: 10.1038/s41590-018-0092-4
45. Kumar R, Ferez M, Swamy M, Arechaga I, Rejas María T, Valpuesta Jose M, et al. Increased sensitivity of antigen-experienced t cells through the enrichment of oligomeric t cell receptor complexes. Immunity (2011) 35(3):375–87. doi: 10.1016/j.immuni.2011.08.010
46. Kim H-R, Mun Y, Lee K-S, Park Y-J, Park J-S, Park J-H, et al. T cell microvilli constitute immunological synaptosomes that carry messages to antigen-presenting cells. Nat Commun (2018) 9(1):3630. doi: 10.1038/s41467-018-06090-8
47. Lillemeier BF, Mörtelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and lat are expressed on separate protein islands on t cell membranes and concatenate during activation. Nat Immunol (2010) 11(1):90–6. doi: 10.1038/ni.1832
48. Roh KH, Lillemeier BF, Wang F, Davis MM. The coreceptor CD4 is expressed in distinct nanoclusters and does not colocalize with t-cell receptor and active protein tyrosine kinase p56lck. Proc Natl Acad Sci USA (2015) 112(13):E1604–13. doi: 10.1073/pnas.1503532112
49. Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, Akpan I, et al. Functional nanoscale organization of signaling molecules downstream of the t cell antigen receptor. Immunity (2011) 35(5):705–20. doi: 10.1016/j.immuni.2011.10.004
50. Pageon SV, Tabarin T, Yamamoto Y, Ma Y, Nicovich PR, Bridgeman JS, et al. Functional role of t-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc Natl Acad Sci USA (2016) 113(37):E5454–63. doi: 10.1073/pnas.1607436113
51. Hu YS, Cang H, Lillemeier BF. Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of t-cell receptors in lymph nodes. Proc Natl Acad Sci (2016) 113(26):7201–6. doi: 10.1073/pnas.1512331113
52. Polliack A, Froimovici M, Pozzoli E, Lambertenghi-Deliliers G. Acute lymphoblastic leukemia: A study of 25 cases by scanning electron microscopy. Blut (1976) 33(6):359–66. doi: 10.1007/BF00996568
53. Majstoravich S, Zhang J, Nicholson-Dykstra S, Linder S, Friedrich W, Siminovitch KA, et al. Lymphocyte microvilli are dynamic, actin-dependent structures that do not require wiskott-aldrich syndrome protein (WASp) for their morphology. Blood (2004) 104(5):1396–403. doi: 10.1182/blood-2004-02-0437
54. Beppler C, Eichorst J, Marchuk K, Cai E, Castellanos CA, Sriram V, et al. Hyperstabilization of t cell microvilli contacts by chimeric antigen receptors. J Cell Biol (2023) 222(3):e202205118. doi: 10.1083/jcb.202205118
55. Cai E, Beppler C, Eichorst J, Marchuk K, Eastman SW, Krummel MF. T cells use distinct topographical and membrane receptor scanning strategies that individually coalesce during receptor recognition. Proc Natl Acad Sci (2022) 119(32):e2203247119. doi: 10.1073/pnas.2203247119
56. Gorelik J, Shevchuk AI, Frolenkov GI, Diakonov IA, Lab MJ, Kros CJ, et al. Dynamic assembly of surface structures in living cells. Proc Natl Acad Sci (2003) 100(10):5819–22. doi: 10.1073/pnas.1030502100
57. Yi J, Balagopalan L, Nguyen T, McIntire KM, Samelson LE. TCR microclusters form spatially segregated domains and sequentially assemble in calcium-dependent kinetic steps. Nat Commun (2019) 10(1):277. doi: 10.1038/s41467-018-08064-2
58. Rossboth B, Arnold AM, Ta H, Platzer R, Kellner F, Huppa JB, et al. TCRs are randomly distributed on the plasma membrane of resting antigen-experienced t cells. Nat Immunol (2018) 19(8):821–7. doi: 10.1038/s41590-018-0162-7
59. Ghosh S, Di Bartolo V, Tubul L, Shimoni E, Kartvelishvily E, Dadosh T, et al. ERM-dependent assembly of t cell receptor signaling and co-stimulatory molecules on microvilli prior to activation. Cell Rep (2020) 30(10):3434–3447.e6. doi: 10.1016/j.ceLclrep.2020.02.069
60. Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by t cells. Science (2017) 356(6338):eaal3118. doi: 10.1126/science.aal3118
61. Nijhara R, van Hennik PB, Gignac ML, Kruhlak MJ, Hordijk PL, Delon J, et al. Rac1 mediates collapse of microvilli on chemokine-activated t lymphocytes. J Immunol (2004) 173(8):4985–93. doi: 10.4049/jimmunol.173.8.4985
62. Beverley PCL, Daser A, Michie CA, Wallace DL. Functional subsets of t cells defined by isoforms of CD45. Biochem Soc Trans (1992) 20(1):184–7. doi: 10.1042/bst0200184
63. Tonks NK, Charbonneau H, Diltz CD, Fischer EH, Walsh KA. Demonstration that the leukocyte common antigen CD45 is a protein tyrosine phosphatase. Biochemistry (1988) 27(24):8695–701. doi: 10.1021/bi00424a001
64. McNeill L, Salmond RJ, Cooper JC, Carret CK, Cassady-Cain RL, Roche-Molina M, et al. The differential regulation of lck kinase phosphorylation sites by CD45 is critical for t cell receptor signaling responses. Immunity (2007) 27(3):425–37. doi: 10.1016/j.immuni.2007.07.015
65. Alexander DR. The CD45 tyrosine phosphatase: a positive and negative regulator of immune cell function. Semin Immunol (2000) 12(4):349–59. doi: 10.1006/smim.2000.0218
66. Furlan G, Minowa T, Hanagata N, Kataoka-Hamai C, Kaizuka Y. Phosphatase CD45 both positively and negatively regulates t cell receptor phosphorylation in reconstituted membrane protein clusters. J Biol Chem (2014) 289(41):28514–25. doi: 10.1074/jbc.M114.574319
67. Pingel JT, Thomas ML. Evidence that the leukocyte-common antigen is required for antigen-induced t lymphocyte proliferation. Cell (1989) 58(6):1055–65. doi: 10.1016/0092-8674(89)90504-7
68. Koretzky GA, Picus J, Thomas ML, Weiss A. Tyrosine phosphatase CD45 is essential for coupling t-cell antigen receptor to the phosphatidyl inositol pathway. Nature (1990) 346(6279):66–8. doi: 10.1038/346066a0
69. Stone JD, Conroy LA, Byth KF, Hederer RA, Howlett S, Takemoto Y, et al. Aberrant TCR-mediated signaling in CD45-null thymocytes involves dysfunctional regulation of lck, fyn, TCR-zeta, and ZAP-70. J Immunol (1997) 158(12):5773–82. doi: 10.4049/jimmunol.158.12.5773
70. Courtney AH, Shvets AA, Lu W, Griffante G, Mollenauer M, Horkova V, et al. CD45 functions as a signaling gatekeeper in t cells. Sci Signaling (2019) 12(604):eaaw8151. doi: 10.1126/scisignal.aaw8151
71. D’Oro U, Ashwell JD. Cutting edge: The CD45 tyrosine phosphatase is an inhibitor of lck activity in thymocytes. J Immunol (1999) 162(4):1879–83. doi: 10.4049/jimmunol.162.4.1879
72. Furukawa T, Itoh M, Krueger NX, Streuli M, Saito H. Specific interaction of the CD45 protein-tyrosine phosphatase with tyrosine-phosphorylated CD3 zeta chain. Proc Natl Acad Sci (1994) 91(23):10928–32. doi: 10.1073/pnas.91.23.10928
73. Sibener LV, Fernandes RA, Kolawole EM, Carbone CB, Liu F, McAffee D, et al. Isolation of a structural mechanism for uncoupling t cell receptor signaling from peptide-MHC binding. Cell (2018) 174(3):672–687.e27. doi: 10.1016/j.cell.2018.06.017
74. Johnson KG, Bromley SK, Dustin ML, Thomas ML. A supramolecular basis for CD45 tyrosine phosphatase regulation in sustained t cell activation. Proc Natl Acad Sci (2000) 97(18):10138–43. doi: 10.1073/pnas.97.18.10138
75. Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity (2006) 25(1):117–27. doi: 10.1016/j.immuni.2006.04.010
76. Davis SJ, van der Merwe PA. The kinetic-segregation model: TCR triggering and beyond. Nat Immunol (2006) 7(8):803–9. doi: 10.1038/ni1369
77. Davis SJ, van der Merwe PA. The structure and ligand interactions of CD2: implications for t-cell function. Immunol Today (1996) 17(4):177–87. doi: 10.1016/0167-5699(96)80617-7
78. Wild MK, Cambiaggi A, Brown MH, Davies EA, Ohno H, Saito T, et al. Dependence of t cell antigen recognition on the dimensions of an accessory receptor–ligand complex. J Exp Med (1999) 190(1):31–42. doi: 10.1084/jem.190.1.31
79. Shaw AS, Dustin ML. Making the t cell receptor go the distance: a topological view of t cell activation. Immunity (1997) 6(4):361–9. doi: 10.1016/s1074-7613(00)80279-4
80. Hořejší V. Lipid rafts and their roles in t-cell activation. Microbes Infection (2005) 7(2):310–6. doi: 10.1016/j.micinf.2004.12.004
81. Janes PW, Ley SC, Magee AI, Kabouridis PS. The role of lipid rafts in t cell antigen receptor (TCR) signalling. Semin Immunol (2000) 12(1):23–34. doi: 10.1006/smim.2000.0204
82. Rodgers W, Rose JK. Exclusion of CD45 inhibits activity of p56lck associated with glycolipid-enriched membrane domains. J Cell Biol (1996) 135(6):1515–23. doi: 10.1083/jcb.135.6.1515
83. Zhang M, Moran M, Round J, Low TA, Patel VP, Tomassian T, et al. CD45 signals outside of lipid rafts to promote ERK activation, synaptic raft clustering, and IL-2 Production1. J Immunol (2005) 174(3):1479–90. doi: 10.4049/jimmunol.174.3.1479
84. Edmonds SD, Ostergaard HL. Dynamic association of CD45 with detergent-insoluble microdomains in t lymphocytes. J Immunol (2002) 169(9):5036–42. doi: 10.4049/jimmunol.169.9.5036
85. Irles C, Symons A, Michel F, Bakker TR, van der Merwe PA, Acuto O. CD45 ectodomain controls interaction with GEMs and lck activity for optimal TCR signaling. Nat Immunol (2003) 4(2):189–97. doi: 10.1038/ni877
86. Janes PW, Ley SC, Magee AI. Aggregation of lipid rafts accompanies signaling via the t cell antigen receptor. J Cell Biol (1999) 147(2):447–61. doi: 10.1083/jcb.147.2.447
87. Magee T, Pirinen N, Adler J, Pagakis SN, Parmryd I. Lipid rafts: cell surface platforms for t cell signaling. Biol Res (2002) 35(2):127–31. doi: 10.4067/s0716-97602002000200003
88. Jury EC, Kabouridis PS, Flores-Borja F, Mageed RA, Isenberg DA. Altered lipid raft-associated signaling and ganglioside expression in t lymphocytes from patients with systemic lupus erythematosus. J Clin Invest (2004) 113(8):1176–87. doi: 10.1172/jci20345
89. Krishnan S, Nambiar MP, Warke VG, Fisher CU, Mitchell J, Delaney N, et al. Alterations in lipid raft composition and dynamics contribute to abnormal t cell responses in systemic lupus Erythematosus12. J Immunol (2004) 172(12):7821–31. doi: 10.4049/jimmunol.172.12.7821
90. Jury EC, Kabouridis PS. T-lymphocyte signalling in systemic lupus erythematosus: a lipid raft perspective. Lupus (2004) 13(6):413–22. doi: 10.1191/0961203304lu1045rr
91. Razvag Y, Neve-Oz Y, Sajman J, Reches M, Sherman E. Nanoscale kinetic segregation of TCR and CD45 in engaged microvilli facilitates early t cell activation. Nat Commun (2018) 9(1):732. doi: 10.1038/s41467-018-03127-w
92. Chang VT, Fernandes RA, Ganzinger KA, Lee SF, Siebold C, McColl J, et al. Initiation of t cell signaling by CD45 segregation at 'close contacts'. Nat Immunol (2016) 17(5):574–82. doi: 10.1038/ni.3392
93. Franke C, Chum T, Kvíčalová Z, Glatzová D, Gentsch GJ, Rodriguez A, et al. Approach to map nanotopography of cell surface receptors. Commun Biol (2022) 5(1):218. doi: 10.1038/s42003-022-03152-y
94. Jung Y, Kim Y-J, Lee S, Nam J, Ha S, Cheon J, et al. Synthetic lipid rafts formed by cholesterol nano-patch induce t cell activation. bioRxiv (2023), 2023.02.22.529596. doi: 10.1101/2023.02.22.529596
95. Miceli MC, Parnes JR. The roles of CD4 and CD8 in t cell activation. Semin Immunol (1991) 3(3):133–41.
96. Lustgarten J, Waks T, Eshhar Z. CD4 and CD8 accessory molecules function through interactions with major histocompatibility complex molecules which are not directly associated with the t cell receptor-antigen complex. Eur J Immunol (1991) 21(10):2507–15. doi: 10.1002/eji.1830211030
97. Krensky AM, Sanchez-Madrid F, Robbins E, Nagy JA, Springer TA, Burakoff SJ. The functional significance, distribution, and structure of LFA-1, LFA-2, and LFA-3: cell surface antigens associated with CTL-target interactions. J Immunol (1983) 131(2):611–6. doi: 10.4049/jimmunol.131.2.611
98. Springer TA, Dustin ML, Kishimoto TK, Marlin SD. The lymphocyte function associated LFA-1, CD2, and LFA-3 molecules: Cell adhesion receptors of the immune system. Annu Rev Immunol (1987) 5(1):223–52. doi: 10.1146/annurev.iy.05.040187.001255
99. Esensten JH, Helou YA, Chopra G, Weiss A, Bluestone JA. CD28 costimulation: From mechanism to therapy. Immunity (2016) 44(5):973–88. doi: 10.1016/j.immuni.2016.04.020
100. Singer Irwin I, Scott S, Kawka Douglas W, Chin J, Daugherty Bruce L, DeMartino Julie A, et al. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and t cells. J Virol (2001) 75(8):3779–90. doi: 10.1128/JVI.75.8.3779-3790.2001
101. Lukeš T, Glatzová D, Kvíčalová Z, Levet F, Benda A, Letschert S, et al. Quantifying protein densities on cell membranes using super-resolution optical fluctuation imaging. Nat Commun (2017) 8(1):1731. doi: 10.1038/s41467-017-01857-x
102. Dertinger T, Colyer R, Iyer G, Weiss S, Enderlein J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI). Proc Natl Acad Sci (2009) 106(52):22287–92. doi: 10.1073/pnas.0907866106
103. Sharpe AH, Pauken KE. The diverse functions of the PD1 inhibitory pathway. Nat Rev Immunol (2018) 18(3):153–67. doi: 10.1038/nri.2017.108
104. Shah K, Al-Haidari A, Sun J, Kazi JU. T cell receptor (TCR) signaling in health and disease. Signal Transduction Targeted Ther (2021) 6(1):412. doi: 10.1038/s41392-021-00823-w
105. Au-Yeung BB, Shah NH, Shen L, Weiss A. ZAP-70 in signaling, biology, and disease. Annu Rev Immunol (2018) 36(1):127–56. doi: 10.1146/annurev-immunol-042617-053335
106. Braiman A, Barda-Saad M, Sommers CL, Samelson LE. Recruitment and activation of PLCγ1 in t cells: a new insight into old domains. EMBO J (2006) 25(4):774–84.
107. Sommers CL, Park C-S, Lee J, Feng C, Fuller CL, Grinberg A, et al. A LAT mutation that inhibits t cell development yet induces lymphoproliferation. Science (2002) 296(5575):2040–3.
108. Bunnell SC, Singer AL, Hong DI, Jacque BH, Jordan MS, Seminario M-C, et al. Persistence of cooperatively stabilized signaling clusters drives t-cell activation. Mol Cell Biol (2006) 26(19):7155–66.
109. Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol (2010) 11(4):276–87. doi: 10.1038/nrm2866
110. Ivetic A, Ridley AJ. Ezrin/radixin/moesin proteins and rho GTPase signalling in leucocytes. Immunology (2004) 112(2):165–76. doi: 10.1111/j.1365-2567.2004.01882.x
111. Bretscher A. Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J Cell Biol (1983) 97(2):425–32. doi: 10.1083/jcb.97.2.425
112. Tsukita S, Hieda Y, Tsukita S. A new 82-kD barbed end-capping protein (radixin) localized in the cell-to-cell adherens junction: purification and characterization. J Cell Biol (1989) 108(6):2369–82. doi: 10.1083/jcb.108.6.2369
113. Lankes W, Griesmacher A, Grünwald J, Schwartz-Albiez R, Keller R. A heparin-binding protein involved in inhibition of smooth-muscle cell proliferation. Biochem J (1988) 251(3):831–42. doi: 10.1042/bj2510831
114. Sato N, Funayama N, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. A gene family consisting of ezrin, radixin and moesin. its specific localization at actin filament/plasma membrane association sites. J Cell Sci (1992) 103(Pt 1):131–43. doi: 10.1242/jcs.103.1.131
115. Nakamura F, Amieva MR, Furthmayr H. Phosphorylation of threonine 558 in the carboxyl-terminal actin-binding domain of moesin by thrombin activation of human platelets. J Biol Chem (1995) 270(52):31377–85. doi: 10.1074/jbc.270.52.31377
116. Faure S, Salazar-Fontana LI, Semichon M, Tybulewicz VLJ, Bismuth G, Trautmann A, et al. ERM proteins regulate cytoskeleton relaxation promoting t cell–APC conjugation. Nat Immunol (2004) 5(3):272–9. doi: 10.1038/ni1039
117. von Andrian UH, Hasslen SR, Nelson RD, Erlandsen SL, Butcher EC. A central role for microvillous receptor presentation in leukocyte adhesion under flow. Cell (1995) 82(6):989–99.
118. Bruehl RE, Springer TA, Bainton DF. Quantitation of l-selectin distribution on human leukocyte microvilli by immunogold labeling and electron microscopy. J Histochem Cytochem (1996) 44(8):835–44. doi: 10.1177/44.8.8756756
119. Buscher K, Riese SB, Shakibaei M, Reich C, Dernedde J, Tauber R, et al. The transmembrane domains of l-selectin and CD44 regulate receptor cell surface positioning and leukocyte adhesion under flow*. J Biol Chem (2010) 285(18):13490–7. doi: 10.1074/jbc.M110.102640
120. Berlin C, Bargatze RF, Campbell JJ, von Andrian UH, Szabo MC, Hasslen SR, et al. Alpha 4 integrins mediate lymphocyte attachment and rolling under physiologic flow. Cell (1995) 80(3):413–22.
121. Abitorabi MA, Pachynski RK, Ferrando RE, Tidswell M, Erle DJ. Presentation of integrins on leukocyte microvilli: a role for the extracellular domain in determining membrane localization. J Cell Biol (1997) 139(2):563–71.
122. Tohya K, Kimura M. Ultrastructural evidence of distinctive behavior of l-selectin and LFA-1 (αLβ2 integrin) on lymphocytes adhering to the endothelial surface of high endothelial venules in peripheral lymph nodes. Histochem Cell Biol (1998) 110(4):407–16. doi: 10.1007/s004180050301
123. Ghosh S, Feigelson SW, Montresor A, Shimoni E, Roncato F, Legler DF, et al. CCR7 signalosomes are preassembled on tips of lymphocyte microvilli in proximity to LFA-1. Biophys J (2021) 120(18):4002–12. doi: 10.1016/j.bpj.2021.08.014
124. Shamri R, Grabovsky V, Gauguet J-M, Feigelson S, Manevich E, Kolanus W, et al. Lymphocyte arrest requires instantaneous induction of an extended LFA-1 conformation mediated by endothelium-bound chemokines. Nat Immunol (2005) 6(5):497–506. doi: 10.1038/ni1194
125. Al-Aghbar MA, Jainarayanan AK, Dustin ML, Roffler SR. The interplay between membrane topology and mechanical forces in regulating t cell receptor activity. Commun Biol (2022) 5(1):40. doi: 10.1038/s42003-021-02995-1
126. Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, et al. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J Biol Chem (2001) 276(21):18507–12. doi: 10.1074/jbc.M101113200
127. Otto GP, Nichols BJ. The roles of flotillin microdomains – endocytosis and beyond. J Cell Sci (2011) 124(23):3933–40. doi: 10.1242/jcs.092015
128. Ficht X, Ruef N, Stolp B, Samson GPB, Moalli F, Page N, et al. In vivo function of the lipid raft protein flotillin-1 during CD8+ t cell–mediated host surveillance. J Immunol (2019) 203(9):2377–87. doi: 10.4049/jimmunol.1900075
129. Langhorst MF, Reuter A, Stuermer CA. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol Life Sci (2005) 62(19-20):2228–40. doi: 10.1007/s00018-005-5166-4
130. Morrow IC, Rea S, Martin S, Prior IA, Prohaska R, Hancock JF, et al. Flotillin-1/reggie-2 traffics to surface raft domains via a novel golgi-independent pathway. identification of a novel membrane targeting domain and a role for palmitoylation. J Biol Chem (2002) 277(50):48834–41. doi: 10.1074/jbc.M209082200
131. Neumann-Giesen C, Falkenbach B, Beicht P, Claasen S, Lüers G, Stuermer CA, et al. Membrane and raft association of reggie-1/flotillin-2: role of myristoylation, palmitoylation and oligomerization and induction of filopodia by overexpression. Biochem J (2004) 378(Pt 2):509–18. doi: 10.1042/bj20031100
132. Rajendran L, Masilamani M, Solomon S, Tikkanen R, Stuermer CAO, Plattner H, et al. Asymmetric localization of flotillins/reggies in preassembled platforms confers inherent polarity to hematopoietic cells. Proc Natl Acad Sci (2003) 100(14):8241–6. doi: 10.1073/pnas.1331629100
133. Langhorst MF, Reuter A, Luxenhofer G, Boneberg E-M, Legler DF, Plattner H, et al. Preformed reggie/flotillin caps: stable priming platforms for macrodomain assembly in t cells. FASEB J (2006) 20(6):711–3. doi: 10.1096/fj.05-4760fje
134. Stuermer CAO, Langhorst MF, Wiechers MF, Legler DF, Hanwehr SHv, Guse AH, et al. PrPc capping in t cells promotes its association with the lipid raft proteins reggie-1 and reggie-2 and leads to signal transduction. FASEB J (2004) 18(14):1731–3. doi: 10.1096/fj.04-2150fje
135. Stuermer CAO, Lang DM, Kirsch F, Wiechers M, Deininger S-O, Plattner H. Glycosylphosphatidyl inositol-anchored proteins and fyn kinase assemble in noncaveolar plasma membrane microdomains defined by reggie-1 and -2. Mol Biol Cell (2001) 12(10):3031–45. doi: 10.1091/mbc.12.10.3031
136. Slaughter N, Laux I, Tu X, Whitelegge J, Zhu X, Effros R, et al. The flotillins are integral membrane proteins in lipid rafts that contain TCR-associated signaling components: implications for t-cell activation. Clin Immunol (2003) 108(2):138–51. doi: 10.1016/S1521-6616(03)00097-4
137. Affentranger S, Martinelli S, Hahn J, Rossy J, Niggli V. Dynamic reorganization of flotillins in chemokine-stimulated human t-lymphocytes. BMC Cell Biol (2011) 12(1):28. doi: 10.1186/1471-2121-12-28
138. Holsinger LJ, Graef IA, Swat W, Chi T, Bautista DM, Davidson L, et al. Defects in actin-cap formation in vav-deficient mice implicate an actin requirement for lymphocyte signal transduction. Curr Biol (1998) 8(10):563–72. doi: 10.1016/s0960-9822(98)70225-8
139. Redpath GMI, Ecker M, Kapoor-Kaushik N, Vartoukian H, Carnell M, Kempe D, et al. Flotillins promote t cell receptor sorting through a fast Rab5–Rab11 endocytic recycling axis. Nat Commun (2019) 10(1):4392. doi: 10.1038/s41467-019-12352-w
140. Kumar R, Pereira RS, Niemann J, Azimpour AI, Zanetti C, Karantanou C, et al. The differential role of the lipid raft-associated protein flotillin 2 for progression of myeloid leukemia. Blood Adv (2022) 6(12):3611–24. doi: 10.1182/bloodadvances.2021005992
141. Ficht X, Ruef N, Stolp B, Samson GPB, Moalli F, Page N, et al. In vivo function of the lipid raft protein flotillin-1 during CD8(+) t cell-mediated host surveillance. J Immunol (2019) 203(9):2377–87. doi: 10.4049/jimmunol.1900075
142. Rajendran L, Beckmann J, Magenau A, Boneberg E-M, Gaus K, Viola A, et al. Flotillins are involved in the polarization of primitive and mature hematopoietic cells. PloS One (2009) 4(12):e8290. doi: 10.1371/journal.pone.0008290
143. Thauland TJ, Parker DC. Diversity in immunological synapse structure. Immunology (2010) 131(4):466–72. doi: 10.1111/j.1365-2567.2010.03366.x
144. Dustin ML. T-cell activation through immunological synapses and kinapses. Immunol Rev (2008) 221:77–89. doi: 10.1111/j.1600-065X.2008.00589.x
145. Yi JC, Samelson LE. Microvilli set the stage for t-cell activation. Proc Natl Acad Sci (2016) 113(40):11061–2. doi: 10.1073/pnas.1613832113
146. Orbach R, Su X. Surfing on membrane waves: Microvilli, curved membranes, and immune signaling. Front Immunol (2020) 11:2187. doi: 10.3389/fimmu.2020.02187
147. Farrell MV, Webster S, Gaus K, Goyette J. T cell membrane heterogeneity aids antigen recognition and t cell activation. Front Cell Dev Biol (2020) 8:609. doi: 10.3389/fcell.2020.00609
148. Göhring J, Schrangl L, Schütz GJ, Huppa JB. Mechanosurveillance: Tiptoeing t cells. Front Immunol (2022) 13:886328. doi: 10.3389/fimmu.2022.886328
149. Kim HR, Jun CD. T cell microvilli: Sensors or senders? Front Immunol (2019) 10:1753. doi: 10.3389/fimmu.2019.01753
150. Fritzsche M, Fernandes RA, Chang VT, Colin-York H, Clausen MP, Felce JH, et al. Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci Adv (2017) 3(6):e1603032. doi: 10.1126/sciadv.1603032
151. Aramesh M, Mergenthal S, Issler M, Plochberger B, Weber F, Qin X-H, et al. Functionalized bead assay to measure three-dimensional traction forces during t-cell activation. Nano Lett (2021) 21(1):507–14. doi: 10.1021/acs.nanolett.0c03964
152. Jenkins E, Körbel M, O’Brien-Ball C, McColl J, Chen KY, Kotowski M, et al. Antigen discrimination by t cells relies on size-constrained microvillar contact. Nat Commun (2023) 14(1):1611. doi: 10.1038/s41467-023-36855-9
153. Aramesh M, Stoycheva D, Sandu I, Ihle SJ, Zünd T, Shiu J-Y, et al. Nanoconfinement of microvilli alters gene expression and boosts t cell activation. Proc Natl Acad Sci (2021) 118(40):e2107535118. doi: 10.1073/pnas.2107535118
154. Fernandes RA, Ganzinger KA, Tzou JC, Jönsson P, Lee SF, Palayret M, et al. A cell topography-based mechanism for ligand discrimination by the t cell receptor. Proc Natl Acad Sci (2019) 116(28):14002–10. doi: 10.1073/pnas.1817255116
155. Morgan J, Pettmann J, Dushek O, Lindsay AE. T cell microvilli simulations show operation near packing limit and impact on antigen recognition. Biophys J (2022) 121(21):4128–36. doi: 10.1016/j.bpj.2022.09.030
156. Puech P-H, Bongrand P. Mechanotransduction as a major driver of cell behaviour: mechanisms, and relevance to cell organization and future research. Open Biol (2021) 11(11):210256. doi: 10.1098/rsob.210256
157. Zhu C, Chen W, Lou J, Rittase W, Li K. Mechanosensing through immunoreceptors. Nat Immunol (2019) 20(10):1269–78. doi: 10.1038/s41590-019-0491-1
158. Pullen RH, Abel SM. Mechanical feedback enables catch bonds to selectively stabilize scanning microvilli at t-cell surfaces. Mol Biol Cell (2019) 30(16):2087–95. doi: 10.1091/mbc.E19-01-0048
159. Tamzalit F, Wang MS, Jin W, Tello-Lafoz M, Boyko V, Heddleston JM, et al. Interfacial actin protrusions mechanically enhance killing by cytotoxic t cells. Sci Immunol (2019) 4(33):eaav5445. doi: 10.1126/sciimmunol.aav5445
Keywords: pre-organization, pre-cluster, preassembly, T cell membrane, microvilli, T cell receptor, CD45
Citation: Jung Y (2023) Pre-organized landscape of T cell surface. Front. Immunol. 14:1264721. doi: 10.3389/fimmu.2023.1264721
Received: 24 July 2023; Accepted: 28 August 2023;
Published: 13 September 2023.
Edited by:
Anil K. Bamezai, Villanova University, United StatesReviewed by:
Jorge Ibanez-Vega, St. Jude Children’s Research Hospital, United StatesAlexandre M. Carmo, Universidade do Porto, Portugal
Copyright © 2023 Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunmin Jung, eXVubWluLmpAeW9uc2VpLmFjLmty
 Yunmin Jung
Yunmin Jung