
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Immunol., 26 October 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1264705
This article is part of the Research TopicInteraction between the Gut Flora and Immunity in Intestinal DiseasesView all 13 articles
Background: Inflammatory bowel disease (IBD) has caused severe health concerns worldwide. Studies on gut microbiota have provided new targets for preventing and treating IBD. Therefore, it is essential to have a comprehensive understanding of the current status and evolution of gut microbiota and IBD studies.
Methods: A bibliometric analysis was performed on documents during 2003-2022 retrieved from the Scopus database, including bibliographical profiles, citation patterns, and collaboration details. Software programs of VOSviewer, CiteSpace, and the Bibliometrix R package visually displayed the mass data presented in the scientific landscapes and networks.
Results: 10479 publications were retrieved, showing a steadily growing tendency in interest. Xavier Ramnik J. group led the total number of publications (73 papers) and 19787 citations, whose productive work aroused widespread concern. Among the 1977 academic journals, the most prolific ones were Inflammatory Bowel Diseases, Frontiers in Immunology, and Nutrients. Research outputs from the United States (US, 9196 publications), China (5587), and Italy (2305) were highly ranked.
Conclusion: Our bibliometric study revealed that the role of gut microbiota has become a hot topic of IBD research worldwide. These findings are expected to improve understanding of research characteristics and to guide future directions in this field.
Inflammatory bowel disease (IBD) comprises a heterogeneous group of inflammatory disorders that are immune-mediated and primarily affect the gastrointestinal tract, with Crohn’s disease (CD) and ulcerative colitis (UC) being the two main subtypes (1, 2). IBD significantly impacts daily life and is a significant risk factor for the development of gastrointestinal cancers (3). The rapid evolution of social norms, lifestyles, diets, and the environment resulting from contemporary human behavior may instigate or contribute to the escalating prevalence of IBD, rendering it an emerging global concern (4). Increasing evidence suggests that the gut microbiome plays a crucial role in the development of IBD (5). IBD is associated with alterations in the gut microbiome, characterized by a consistent reduction in bacterial diversity (6). Meanwhile, fecal microbiota transplantation has been shown to restore intestinal microecological balance and treat IBD effectively (7).
Microbiota in the human digestive tract make up a complex ecological system. To date, over 3000 species have been detected in human feces; only 30% of this bacterial population is the typical core microflora shared between different individuals (8, 9). Investigations have indicated that gut microbiota is crucial in the maintenance of intestinal physiological function (10). The dynamic composition of the microbiota is influenced and regulated by a combination of endogenous and exogenous factors (11). Diet, hormones, medication, and health conditions of the host may affect the numbers and diversity of microflora in the gastrointestinal tract (10, 11). Dramatic perturbations like these may result in dysbiosis characterized by an altered composition and reduced stability (12). Moreover, microbiota dysbiosis could induce various human diseases like IBD in the pathological processes (13, 14).
Bibliometric analysis is an approach to evaluate the trends and characteristics of published literature in a particular domain over time. It provides an easy and direct way for scientists and researchers to access the field’s developing trends and research interests. The academic influence of leading publications and literature distributions from different origins is clearly present (15, 16). The conventional classification and summarization of literature heavily rely on the subjective judgment of authors, making it challenging to analyze a large volume of literature comprehensively and accurately. To address this issue, scientific cartography based on bibliometric quantitative analysis can be employed to examine the structure and development of research fields. This method facilitates the summarization and analysis of applied literature while uncovering key application areas and enables topic clustering using CiteSpace or VOSviewer software (17). Currently, bibliometric analysis has garnered increasing attention due to its distinctive advantages that enable investigators to delve into specific fields of study through the visualized analysis of citations, co-citations, geographic distribution, and term frequency, yielding highly valuable insights (18).
In this study, on the hotspot of gut microbiota and IBD, we conducted a bibliometric analysis of publications in the Scopus database during the past two decades to capture its research state and trends. Using software programs VOSviewer, CiteSpace, and Bibliometrix, we mapped the literature landscape and distribution layouts of active authors, journals, institutions, and countries. We also visualized the patterns of cooperation and citation. This study presents an overview and summary of the evolution of gut microbiota and IBD studies, and analyzes the current research state and future trends, aiming to assist researchers and policymakers gain a comprehensive understanding of the study on this topic and better grasp future directions.
A bibliometric search of research output on the gut microbiome and IBD, published from 2003 to 2022, was performed on July 10, 2023, using the Scopus database. Scopus by Elsevier is known to be the most comprehensive data source for detailed bibliometric evaluation from a quantitative and qualitative point of view (19–21). With the comprehensive coverage of scientific journals and the powerful performance of analytical tools, Scopus was selected as the literature source to retrieve abstracts, citations, and other bibliometric data at the initial stage.
Aiming to ensure reliable and accurate records, our primary keywords used in the literature search focused on gut microbiota and inflammation, along with the relevant synonyms based on Medical Subject Headings (MeSH) in MEDLINE (22). The terms “gastrointestinal microbiomes,” “gut microflora,” “gut microbiota,” “gastrointestinal flora,” “gut flora,” “gastrointestinal microbiota,” “gut microbiome,” “gastrointestinal microflora,” “intestinal microbiome,” “intestinal microbiota,” “intestinal microflora,” “intestinal flora” and “enteric bacteria” were used as the keywords of gut microbiota; the primary keywords of IBD were “inflammatory bowel disease” and “IBD” Meanwhile also includes “ulcer colitis”, “UC”, “Crohn disease” and “CD”. The two sets of keywords with the AND logic were searched in the field of “Article title/Abstract/Keywords.” The search was conducted in Scopus using the following terms: (TITLE-ABS-KEY (gastrointestinal AND microbiomes) OR TITLE-ABS-KEY (gut AND microflora) OR TITLE-ABS-KEY (gut AND microbiota) OR TITLE-ABS-KEY (gastrointestinal AND flora) OR TITLE-ABS-KEY (gut AND flora) OR TITLE-ABS-KEY (gastrointestinal AND microbiota) OR TITLE-ABS-KEY (gut AND microbiome) OR TITLE-ABS-KEY (gastrointestinal AND microflora) OR TITLE-ABS-KEY (intestinal AND microbiome) OR TITLE-ABS-KEY (intestinal AND microbiota) OR TITLE-ABS-KEY (intestinal AND microflora) OR TITLE-ABS-KEY (intestinal AND flora) OR TITLE-ABS-KEY (enteric AND bacteria) AND PUBYEAR > 2002 AND PUBYEAR < 2023) AND (TITLE-ABS-KEY (inflammatory bowel disease) OR TITLE-ABS-KEY (ulcer colitis) OR TITLE-ABS-KEY (crohn disease) OR TITLE-ABS-KEY (IBD) OR TITLE-ABS-KEY (UC) OR TITLE-ABS-KEY (CD) AND PUBYEAR > 2002 AND PUBYEAR < 2023).
The search outcomes from the Scopus database were exported into CSV format for further analysis, including bibliographical profiles, citation patterns, collaboration details, and other retrieved publications. We established the inclusion criteria as follows: 1. The literature pertains to topics of IBD and inflammation; 2. Articles published within the past two decades (2003-2022). Exclusions encompassed: 1. Incomplete or duplicated literature; 2. Non-academic documents such as conference proceedings, calls for papers, news reports, patent achievements, and newspaper abstracts.
Microsoft Excel and GraphPad Prism (Version 9.5.0, San Diego, CA, US) were applied to conduct statistical procedures, generating frequency distribution, sum, and average data. Further investigations were performed to determine the top, most prolific authors, journals, countries, institutions, and the most cited papers according to the standard competition ranking (SCR, also known as the 1-2-2-4 rule). We calculated the H-index to assess the number and level of academic output of researchers. In addition, we also calculate the G index and M-index as a supplement to the H index. The calculation method of G-index is as follows: the papers are sorted in descending order according to the number of citations, and the number of citations is superimposed according to the serial number. When the cumulative number of citations is equal to the square of the serial number, the serial number value is the G-index (23). The M-index is derived from the H-index of academic tenure, calculated by dividing the H-index by the number of years since an author’s initial publication (24).
Visualization analysis was applied for presenting a mass of data to display scientific landscapes and networks using software programs of VOSviewer (v.1.6.17) (25), CiteSpace (v.5.8.R2) (26), and the R package of Bibliometrix (27). VOSviewer conducts a visual analysis of country, institutional, author, and collaboration distribution, as well as keyword collaboration networks. Clustering is automatically completed using the similarity matrix and mapping techniques of VOSviewer, with corresponding labels added by the authors based on content. CiteSpace was utilized to analyze the distribution and collaboration among countries, institutions, keyword timelines, and reference data. Additionally, we employ R studio Desktop Software (v.2023.6.1.0) linked to the R Software (v.4.3.1) and converted into an R data frame. The Bibliometrix R package, which provides a web interface, was used for statistical analysis of the number of publications, references, and other data and visual analysis of national distribution and cooperation (27). The flow diagram for the searching and sorting process of related articles is shown in Figure 1. All raw data utilized in this study were sourced from publicly available databases, thus exempting the need for ethical review.
The 20-year period 2003-2022 saw the publication of 10479 articles in gut microbiota and IBD research. Global trends in the number of annual and accumulated publications related to this topic are shown in Figure 2A. During the first four years, 2003-2006, the number of annual articles ranged from117 to 156 and was relatively stable, implying that this crossover domain was not so attractive to scientists at that moment. Moreover, within a span of only two years (2007-2008), two important programs of the Human Microbiome Project (HMP) were launched by the United States National Institutes of Health (NIH) in 2007 (28), and Metagenomics of the Human Intestinal Tract (MetaHIT) by European Union in 2008 (29), and the number of annual articles increased to 209. Since then, the number has continued to rise, reaching 1813 publications in 2022 (or 906.5% of the articles in 2008). The active interest and intensive efforts from worldwide research communities in the past ten years have led to the enormous growth of this field, which is supported by a significant increase in the number of related publications. The number of articles published on this topic in the recent decade is more than 5.93 times in the first ten years since 2003. Furthermore, it is noted that the percentage of annual intestinal microbiota-related publications in the domain of inflammation research was increasing gradually, starting from 2009, when an increase from 0.9%-0.11% was observed (Figure 2B). Therefore, the combination of gut microbiota and inflammation as an immediate area of the research focus has attracted significant attention from scientists worldwide.
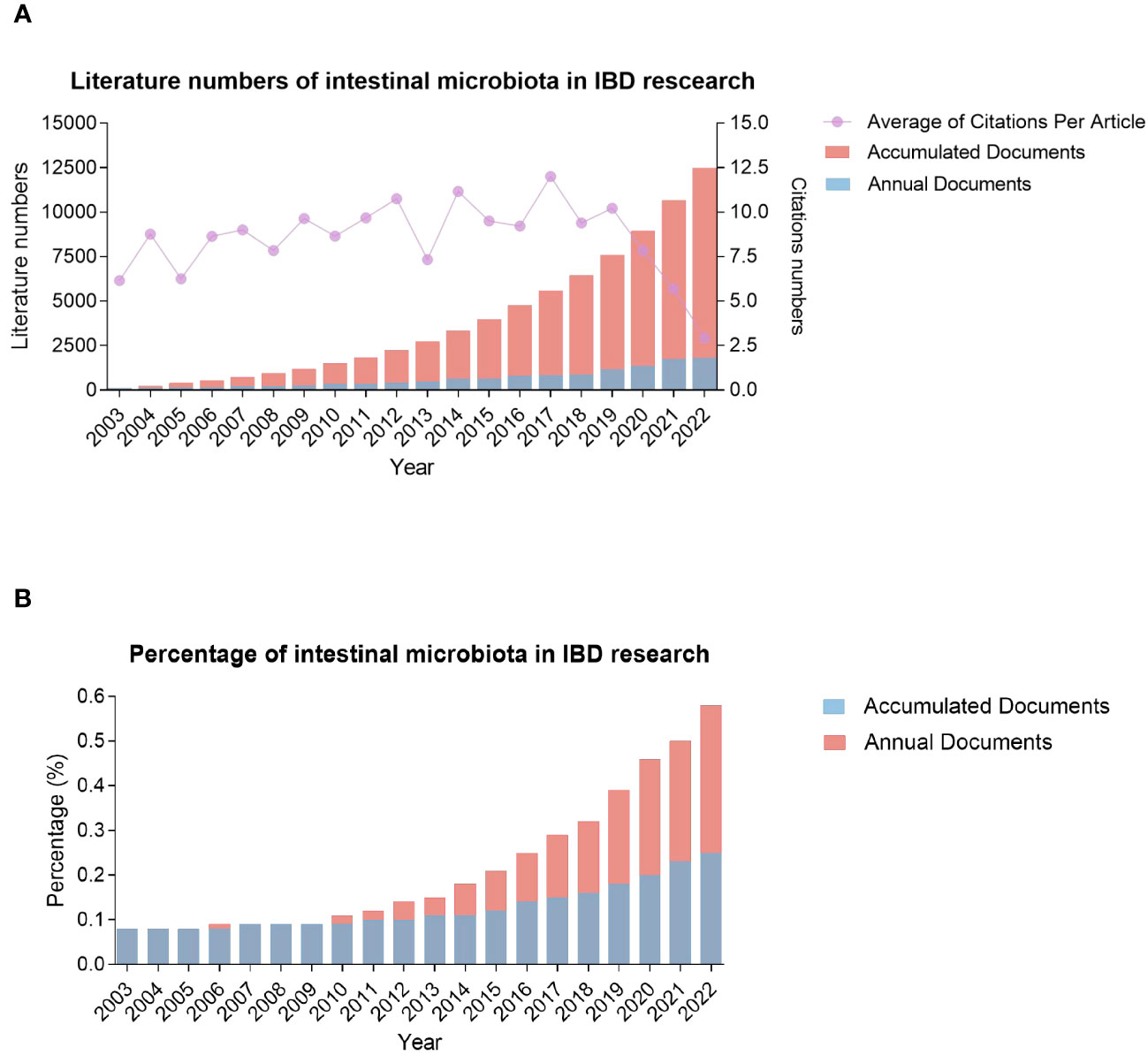
Figure 2 Global trends in the number of published articles related to gut microbiota and IBD over the past two decades from 2003 through 2022. (A) annual and accumulated publications of intestinal microbiota and IBD; (B) the percentage of intestinal microbiota-related publications in the IBD research.
Among the retrieved documents, the research articles (6544, 52.36%) and reviews (4629, 37.04%) made up the majority, while the others (10.60%) were conference papers, book chapters, short surveys, notes, and editorials, etc. (Figure 3A). Journals were the primary source of documents, accounting for 95.40% of all the publications (Figure 3B). In terms of subject distribution (Figure 3C), 8334 documents (66.69%) were related to Medicine, 2936 (23,49%) to Biochemistry, Genetics, and Molecular Biology, 2956 (23.65%) to Immunology and Microbiology, and 1204 (9.63%) to Agricultural and Biological Sciences. Of the 26 languages published, English was predominant (11857, 94.88%), followed by Chinese (236, 1.89%). Other languages, like German, Russian, French, etc., only covered less than 1% of publications (Figure 3D).
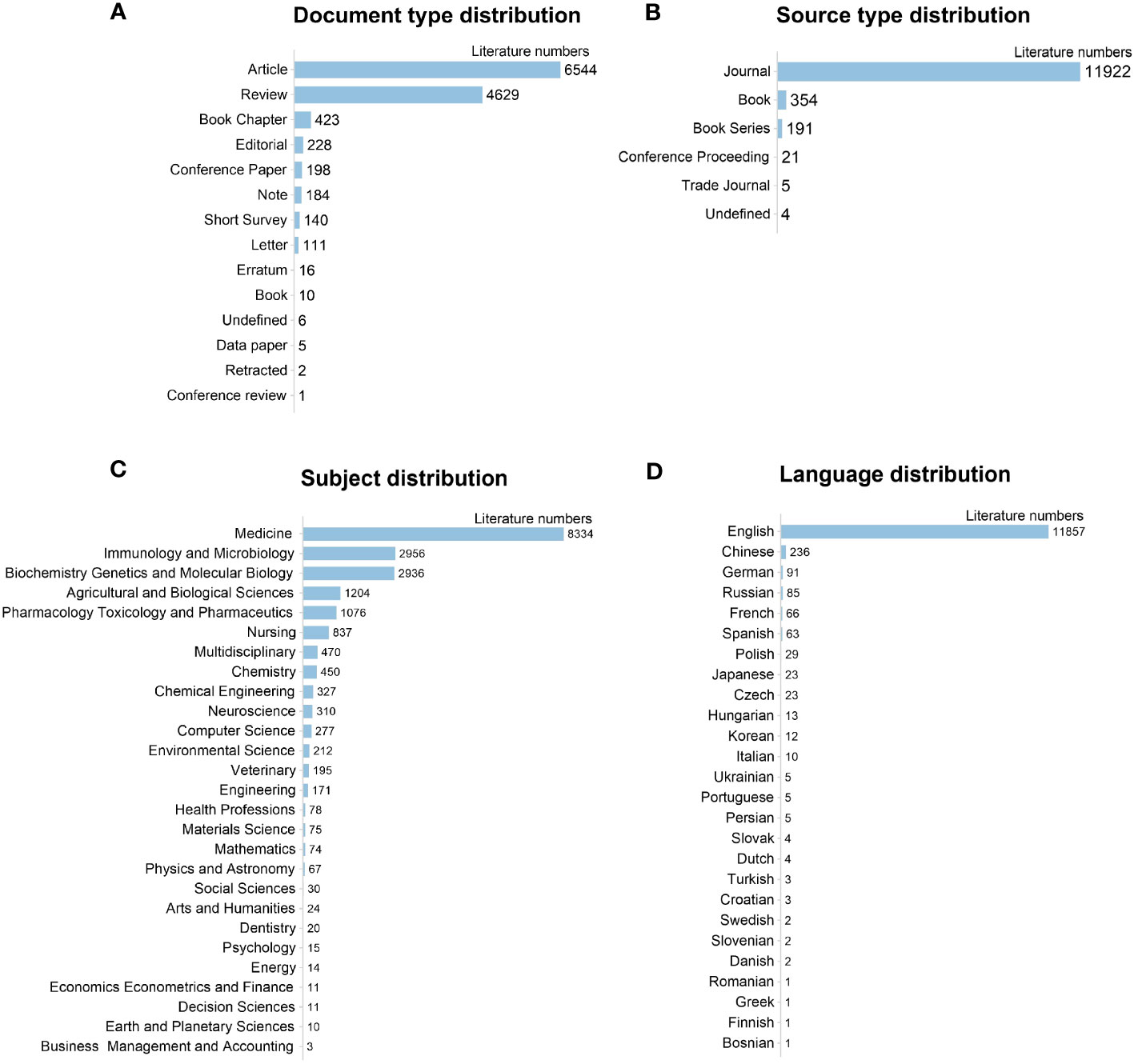
Figure 3 General information of retrieved 10479 publications on gut microbiota and IBD. (A) Document type distribution; (B) Source type distribution; (C) Subject distribution; (D) Language distribution.
Due to the source type and language heterogeneity among the retrieved documents, we set the inclusion criteria to limit publications to only research articles written in English to perform further analyses. Thus, papers of document types besides research articles and papers written in other languages were excluded (Figure 1). As a result, a total of 10479 English papers were included for the following analyses.
Although many factors may influence the citation impact of publications, it is widely regarded as a vital evaluation index for scientific documents. Table 1 presents the 20 most commonly cited papers between 2003 and 2022 (30–49). “Metabolic endotoxemia initiates obesity and insulin resistance,” published in Nature by David L.A. et al., was the most frequently cited article (5975 times) (30). In addition, the top journals were represented among the most cited articles in this field. Of the 20 most cited papers, four were published in Nature, and three were published in Proceedings of the National Academy of Sciences of the United States of America (PNAS), two from NEJM and two from Cell, respectively.
A total of 36335 different authors contributed to the 10479 papers included in this study. Further analysis revealed that the 722 most prolific authors who published ten articles or more accounted for 2.0% of all contributors. We created a historical map of the related publications and authors in the format of a bubble chart. This chart demonstrated the 20 most productive authors by year, as shown in Figure 4. Since 2003, Sartor R. Balfour, Colombel Jean Frederick, and Shanahan Ferguson have been pioneers in exploring the field of gut microbiota and inflammation, but at that time there were relatively few related papers on this topic. After a 5-year significant increase from 2007 to 2012, this emerging field witnessed explosive publication growth from 2013. More specifically, Xavier Ramnik J. group led in the total number of publications (73 papers), followed by Sokol Harry (67), Sartor R. Balfour (50), Colombel Jean-Frederic (49), and Huttenhower Curtis (42).
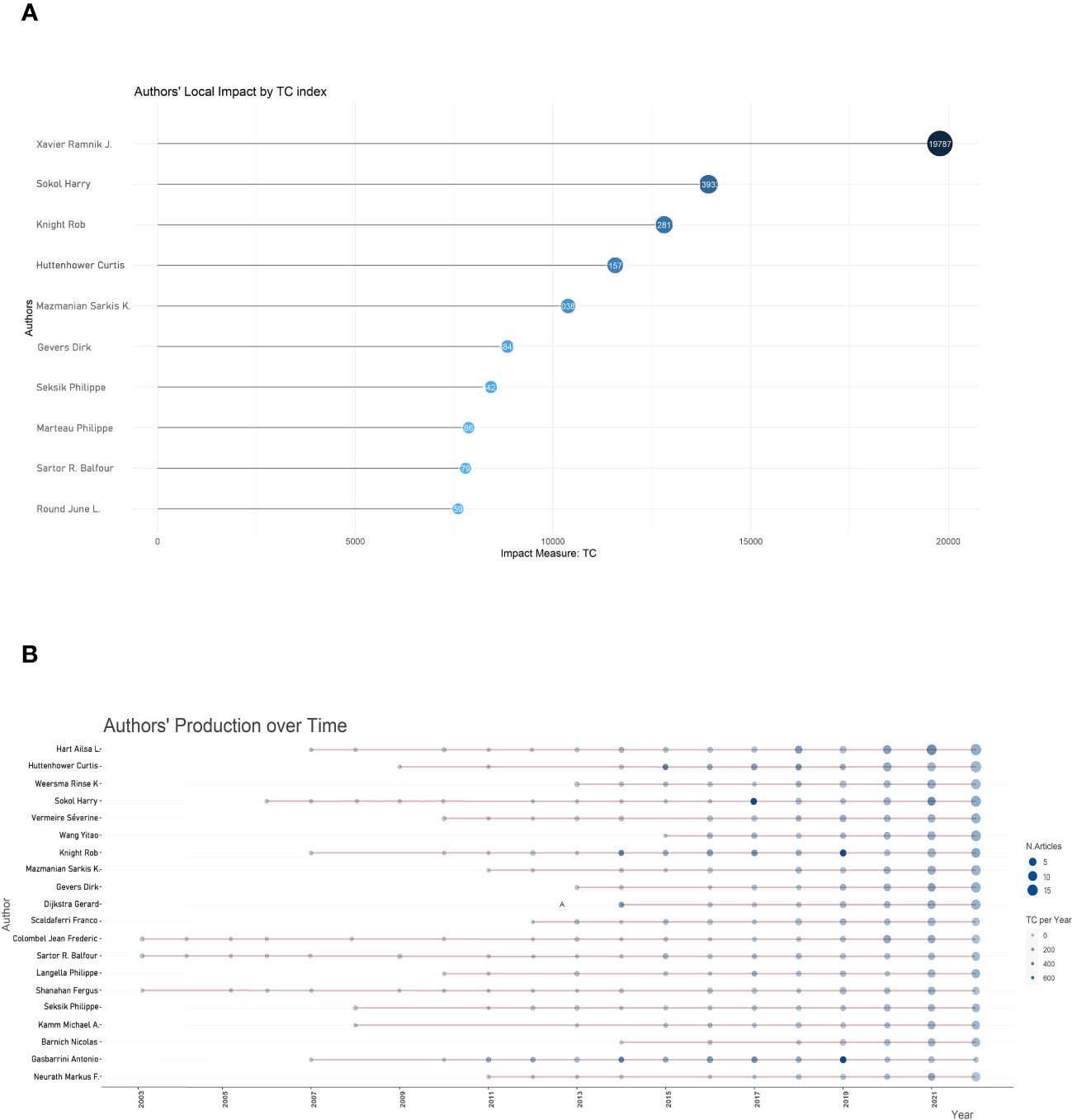
Figure 4 Bubble diagram depicting the influence of authors. (A) Top 10 authors with the highest citations; (B) Annual publication count and citation frequency. Larger shapes display more publications, and darker blue displays more citations.
The citation of the 234 most prolific authors with at least twenty publications was quantified and subjected to analysis, while the top 10 authors with the most published papers and the top 10 most cited authors on gut microbiota and IBD from 2003 to 2022 were presented in Table 2. Xavier, Ramnik J. led in the first place (19787 cited), followed by Sokol, Harry (13933) and Huttenhower, Curtis (11575). In addition, Xavier, Ramnik J. had the most citations (19787) among the top 20 most cited authors, followed by Sokol, Harry (13933) and Knight, Rob (12811). The 234 most prolific authors were also integrated into collaborative networks, as shown in Figure S1. The link thickness between any two authors indicates the extent of co-authorships (collaboration). The clusters in Figure S1 revealed a strong correlation between the number of publications produced by an author and co-authorship. In other words, the more muscular the total link strength of scientific collaboration, the more authored publications.
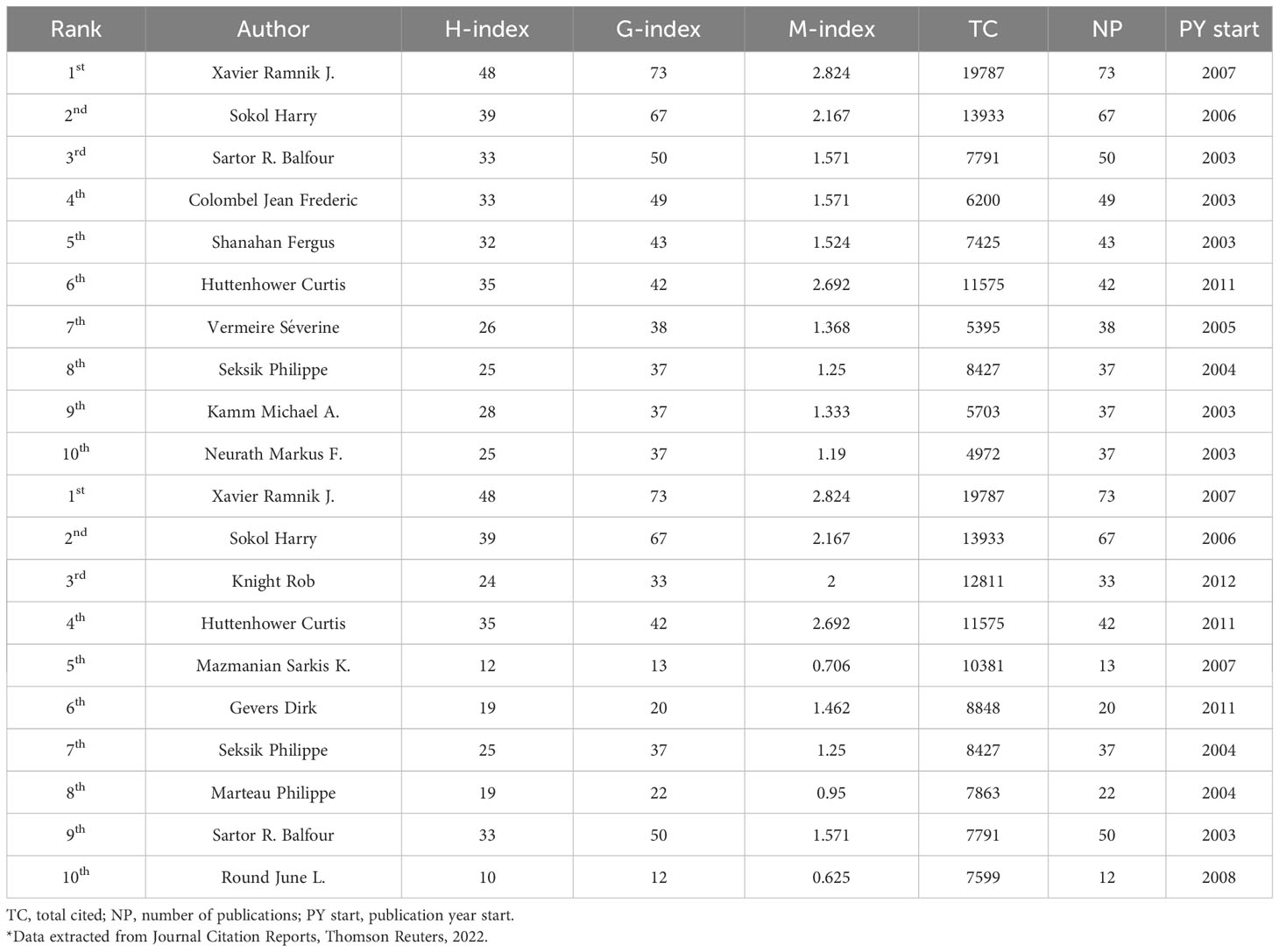
Table 2 Top 10 prolific authors and top 10 most cited authors on gut microbiota and IBD from 2003 to 2022.
The retrieved articles were published in 1977 different academic journals. Table 3 presents the top 20 active journals publishing articles on intestinal microflora and IBD, which produced 3269 articles (31.1%). Among them, Inflammatory Bowel Diseases took the leading position with 324 papers, followed by Frontiers in Immunology (301) and Nutrients (259). These three journals issued 8.5% of the total publications. According to IF, Nature Reviews Gastroenterology and Hepatology held the top position by an overwhelming high value of 65.1, owing to its excellent specialization in this field. In addition, Bradford’s law of scattering was applied here to reveal the distribution of the scientific literature in the research on gut microbiota and inflammation. Bradford zones acted as concentric zones of publication productivity with decreasing correlation, while each zone involved a similar number of articles. As shown in Table 4 and Figure S4, a total of 1977 journals were distributed in 3 Bradford’s zones in the field of gut microbiota and IBD. The average number of articles in each zone was 3493.
The number of citations also indicates the power and authority of journals in the field. Citation analysis of 380 journals with minimum productivity of 5 publications was presented in Figure 5A. Articles on gut microbiota and IBD published in Gut received the highest number of citations (28430), while those published in Nature (25407) and Inflammatory bowel disease (23871) ranked second and third, respectively. The dot size is proportionate to the number of citations, with yellow indicating higher average citations and blue indicating lower average citations (Figure 5A).

Figure 5 Citation analysis of journals. (A) Density map of 380 journals with a minimum productivity of 5 publications in this field. Journals with a higher number of citations have a yellower spot. (B) Heatmap illustrated the publication volume distributions per year of the top 20 most cited journals. The intensity of the blue hue is inversely proportional to the quantity of published papers, while the intensity of the red hue is directly proportional to the quantity of published papers.
Analyzing the top 20 most cited journals by year provided further insight into the level of journals directed to topic interest in Figure 5B. It was apparent that the journal citations have increased enormously, based on the great concentration in this area from 2008, which was consistent with patterns shown in Figure 1. Among the top 20 most cited journals, the quantity of publications regarding intestinal microflora and IBD has exhibited a consistent upward trend from 2003 to 2022. Frontiers in Immunology, Nutrients, and Frontiers in Microbiology experienced the most significant surge, demonstrating the most published research topics in intestinal microflora and IBD between 2003-2022.
The geographical distribution of research productivity from 217 countries/regions on six continents was presented in Figure 6. In the map, the darker blue color represented countries/regions with the higher productivity of gut microflora and IBD articles. The intensity of the color is directly proportional to the quantity of publications. Among the most productive countries, the United States (US) contributed most to the research productivity (9196 publications), followed by China (5587), Italy (2305), the United Kingdom (1806), and France (1740). International collaboration of active countries/regions was also assessed and presented in a network visualization map (Figures 6, S2). The thickness of the link between any two countries/regions indicated the strength of collaboration, while the density of the threads assigned for that country/region indicated the extent of international collaboration. The network visualization map shows that the most vigorous collaboration was between the US and China. With the densest line, the US implied the greatest extent of international collaboration with 80 countries/regions due to the wide range of its publications. From the cluster analysis in the map, countries/regions such as the US and Canada were observed in a close cluster, while Germany, Sweden, and Denmark were found in another close cluster. The ranking of production and national collaborations between the corresponding author’s countries are shown in Table S1. Citations analysis for countries in Table S2 showed that the US had been the most highly cited globally.
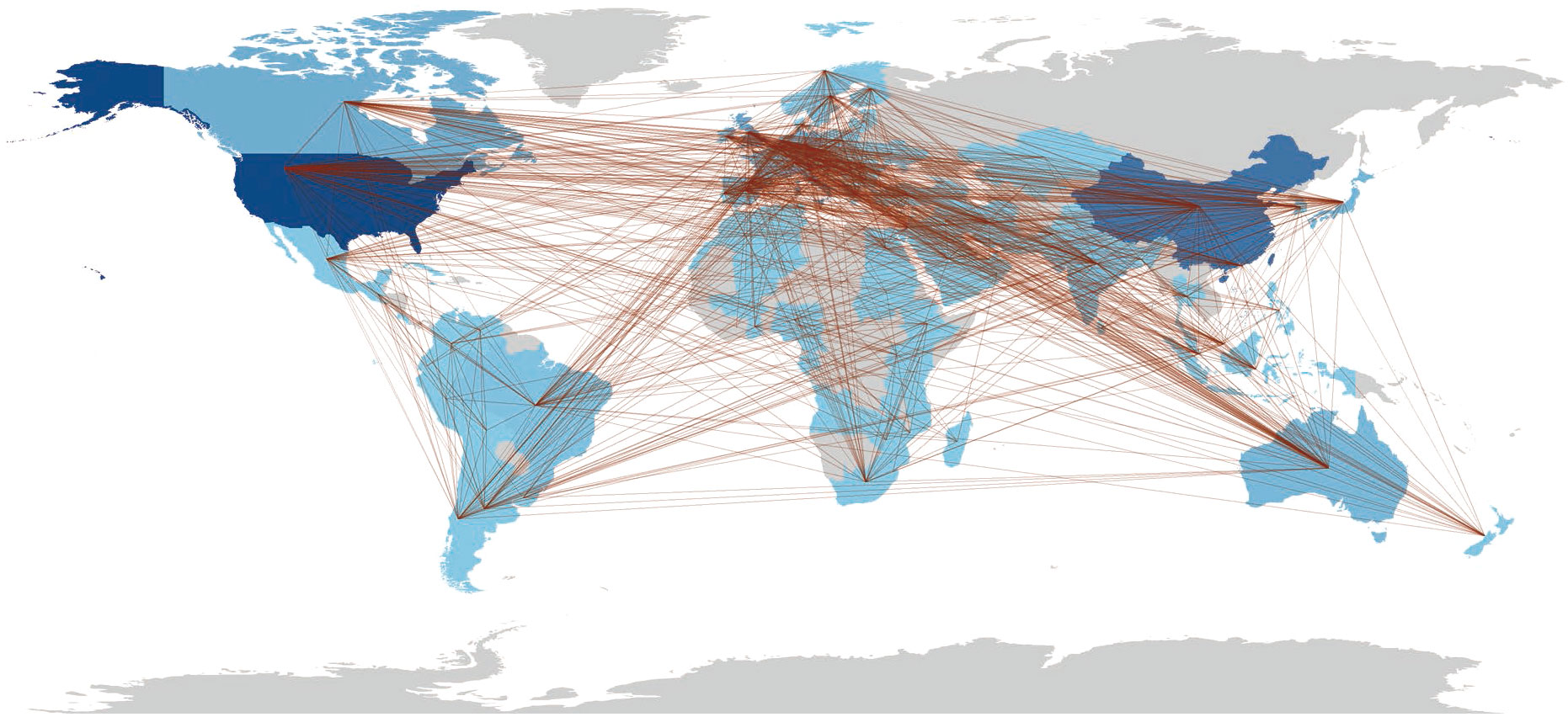
Figure 6 Trends in the publications on intestinal microbiota in IBD research involved 217 countries/regions over six continents. The interconnection among nations signifies collaborative efforts.
Table 5 lists the top 20 prolific institutions publishing papers on gut flora and IBD. Harvard Medical School ranked first in productivity with 336 scientific publications, followed by the University of California (274) and University College Cork (208). The 20 most active institutions are primarily in North America, with 10 in the US and 4 in Canada. The other six institutions are widely distributed in Europe (Ireland, Belgium, Denmark), Oceania (Australia), and Asia (China). The analysis of 277 organizations with high citations of more than 1000 times is shown in Figure S3. The Division of Biology, California Institute of Technology, Pasadena, CA, United States received the highest citation number (7833 citations), while the FAS Center for Systems Biology, Harvard University, Cambridge, MA, in the United States (6599) and Laboratory of Microbiology, Wageningen University Wageningen in Netherlands (6272) were in the second and third place, respectively.
This thematic analysis was performed on the terms that appeared in the information sources of retrieved publications from 2003-2022. The terms were mainly from the title, abstract, and keyword fields of academic literature, representing the authors’ main concepts and research interests for communication. A density visualization map is used to display which terms occur more often and how the terms interconnect (Figure 7). The larger the character fonts, the more frequently the terms are applied. A total of 525 terms that occurred more than 100 times were presented and divided into three groups with their interconnections. As a result, terms such as inflammatory bowel disease(IBD) were by far the most prevalent (8168 times), followed by patient (3202), gut microbiota (2651), microbiota (2346), and inflammation (2210).
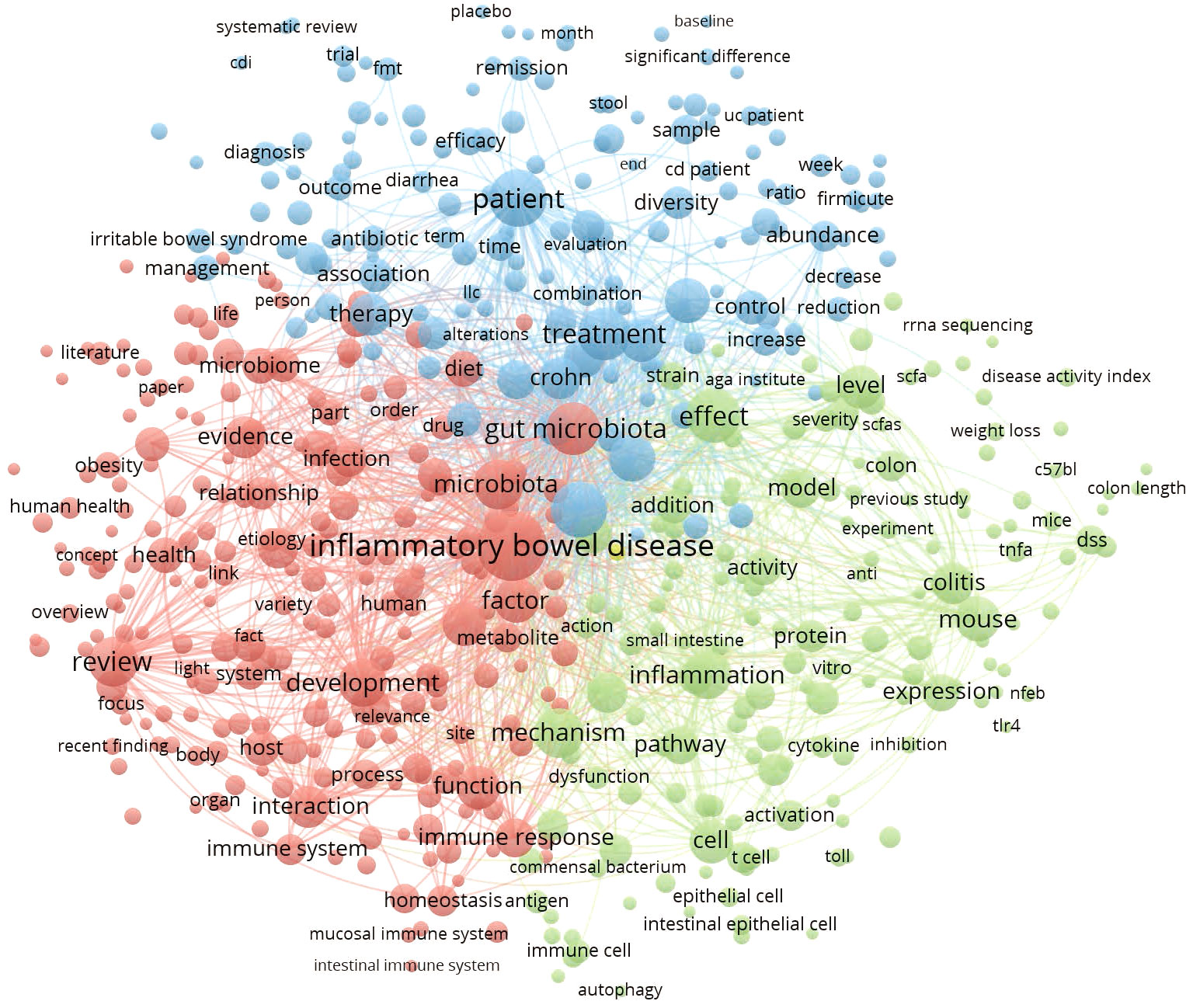
Figure 7 Density map of most frequently encountered terms extracted from the titles and abstracts of retrieved publications. A number of 525 terms met the threshold with a minimum number of occurrences of 100. The larger circle size or font size indicates higher occurrence.
IBD significantly impacts daily life and poses a significant risk for the development of gastrointestinal malignancies (3). Due to its clinical refractory nature, treating IBD has become a prominent topic in healthcare. However, the in-depth pathogenesis that accounts for IBD has been largely debated. The role of commensal microbiota in the onset and development of IBD has attracted increasing attention (6). Aberrant microbiota community structure and dysbiosis of the host’s microbiota may affect the gut’s immunological function and immune homeostasis (50). As metabolites of gut microbiota, short-chain fatty acid (SCFA) and bile acid have been shown to work on immune homeostasis through cell signaling receptors or epigenetic regulations (50, 51). The dysfunction of the intestinal barrier leading to pathological bacterial translocation also contributes to IBD (1).
The perspective of intestinal microbiota may shed new light on the investigation of IBD. Lifestyle and environmental factors could modulate the gut microbiota composition and bacterial diversity (52). Unhealthy dietary patterns result in microbiota dysbiosis, which becomes the facilitator of IBD (4). Gut microbiota-related approaches may have the potential to ameliorate IBD (7). Fecal microbiota transplantation has been shown to have beneficial effects, including alleviating colonic inflammation and promoting the restoration of intestinal homeostasis through multiple immune-mediated pathways (53). However, more in-depth investigations are needed to reach a thorough understanding of the interplay between intestinal microbiota and IBD.
Bibliometric analysis is a quantitative study of bibliographic information, including authors, institutions, publication types, source countries, funding and citation information, etc. (54). This approach can assess the academic performance of journals, authors, or countries and provide a comprehensive overview of a particular research domain. This study aimed to present a complete picture of intestinal microbiota and IBD research during the past two decades. The significance of gut microbiota in IBD-related research was investigated using a bibliometric method for the first time, and the scientific production and global trends of this field were also evaluated.
As revealed in the results, since 2003, Sartor R. Balfour, Colombel Jean Frederick, and Shanahan Ferguson have pioneered the field of gut microbiota and inflammation study, which has demonstrated sustained and exponential growth over the ensuing two decades. Especially in the last few years, the annual number of documents has been soaring since 2007, which coincides with the launch of the HMP and MetaHIT projects (55, 56). In recent years, gut microbiota research has been drawing increased attention to provide a new perspective on many complicated issues, it also accelerates the understanding of the origin and mechanism of IBD. The steady growth in the percentage of intestinal microbiota-related publications on IBD research indicates a promising future in this domain.
Through the utilization of keyword cluster analysis and timeline view, the current research hotspots pertaining to IBD and intestinal flora can be primarily categorized into two aspects: the microscopic mechanisms underlying IBD and its clinical treatment, with a particular emphasis on exploring the inflammatory pathways mediated by intestinal flora. The intestinal barrier and mucosal immunity play crucial roles in connecting gut microbiota with IBD. The TNF-α, TLR4, and other signaling pathways, as well as the metabolic mechanisms of intestinal flora such as SCFA, have garnered increasing attention in recent years. In studies of gut microbes associated with IBD, proteobacteria, and parabacteroides were the most heavily investigated in the past two decades, which have been reported in 287 and 154 articles, respectively, revealing the research hotspots in this field. Human-centered clinical study is experiencing steady growth and is poised to become a prominent topic in this field.
This study solely focused on publications within the Scopus database and did not encompass other databases, such as PubMed and Web of Science, which may yield marginally distinct outcomes. Despite being the largest peer-reviewed abstracts and citation database globally, it is plausible that several papers on this topic may have been published in journals not incorporated in Scopus. The other limitation of our study, which is inherent to any bibliometric approach, is that we did not examine individual article records beyond the random sample used for verifying index accuracy. Instead, we relied on MEDLINE indexes for classification purposes. Nevertheless, the manually verified 5% sample had a high level of accuracy; thus, we can confidently assert that the results obtained through bibliometric analysis are valid.
This extensive bibliometric study provides researchers with a global overview of academic trends, geographical distribution, and collaboration patterns in the field of gut microbiota and IBD research over the past 20 years. The comprehensive analysis and structured data presented in this study benefit scientists in screening academic interests and informing policymakers in developing policies related to this topic. Furthermore, the results reveal the significant role of gut microbiota in IBD and lay the groundwork for further explorations in the future.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
AZ: Writing – original draft, Writing – review & editing. FW: Writing – review & editing. DL: Writing – review & editing. C-ZW: Writing – review & editing. HY: Writing – original draft, Writing – review & editing. J-YW: Writing – review & editing, Writing – original draft. C-SY: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by the National Natural Science Foundation of China (82374324, 82274379); Fundamental Research Funds for the Central Universities (2022-JYB-JBZR-009); Beijing University of Chinese Medicine High-level Talent Start-up Research Project (2021-XJ-KYQD-004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1264705/full#supplementary-material
1. Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol (2015) 37:47–55. doi: 10.1007/s00281-014-0454-4
2. Gros B, Kaplan GG. Ulcerative colitis in adults: A review. JAMA (2023) 330:951–65. doi: 10.1001/jama.2023.15389
3. Axelrad JE, Lichtiger S, Yajnik V. Inflammatory bowel disease and cancer: The role of inflammation, immunosuppression, and cancer treatment. World J Gastroenterol (2016) 22:4794–801. doi: 10.3748/wjg.v22.i20.4794
4. Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome-searching the crime scene for clues. Gastroenterology (2021) 160:524–37. doi: 10.1053/j.gastro.2020.09.056
5. Kudelka MR, Stowell SR, Cummings RD, Neish AS. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat Rev Gastroenterol Hepatol (2020) 17:597–617. doi: 10.1038/s41575-020-0331-7
6. Wu R, Xiong R, Li Y, Chen J, Yan R. Gut microbiome, metabolome, host immunity associated with inflammatory bowel disease and intervention of fecal microbiota transplantation. J Autoimmun (2023) 2023:103062. doi: 10.1016/j.jaut.2023.103062
7. Allegretti JR, Kelly CR, Grinspan A, Mullish BH, Kassam Z, Fischer M. Outcomes of fecal microbiota transplantation in patients with inflammatory bowel diseases and recurrent clostridioides difficile infection. Gastroenterology (2020) 159:1982–84. doi: 10.1053/j.gastro.2020.07.045
8. Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science (2013) 341:1237439. doi: 10.1126/science.1237439
9. Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol (2014) 32:834–41. doi: 10.1038/nbt.2942
10. Hou K, Wu Z-X, Chen X-Y, Wang J-Q, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther (2022) 7:135. doi: 10.1038/s41392-022-00974-4
11. Li Y, Xia S, Jiang X, Feng C, Gong S, Ma J, et al. Gut microbiota and diarrhea: an updated review. Front Cell Infect Microbiol (2021) 11:625210. doi: 10.3389/fcimb.2021.625210
12. Peterson CT, Sharma V, Elmen L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol (2015) 179:363–77. doi: 10.1111/cei.12474
13. Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol (2017) 14:573–84. doi: 10.1038/nrgastro.2017.88
14. Federici S, Kredo-Russo S, Valdés-Mas R, Kviatcovsky D, Weinstock E, Matiuhin Y, et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell (2022) 185:2879–98.e24. doi: 10.1016/j.cell.2022.07.003
15. Cheng T, Pan Y, Hao M, Wang Y, Bryant SH. PubChem applications in drug discovery: a bibliometric analysis. Drug Discovery Today (2014) 19:1751–56. doi: 10.1016/j.drudis.2014.08.008
16. Dubner R. A bibliometric analysis of the Pain journal as a representation of progress and trends in the field. Pain (2009) 142:9–10. doi: 10.1016/j.pain.2009.01.003
17. Yu D, Hong X. A theme evolution and knowledge trajectory study in AHP using science mapping and main path analysis. Expert Syst Applications (2022) 205:117675. doi: 10.1016/j.eswa.2022.117675
18. Yu D, Xu Z, Wang W. Bibliometric analysis of fuzzy theory research in China: A 30-year perspective. Knowledge-Based Systems (2018) 141:188–99. doi: 10.1016/j.knosys.2017.11.018
19. Falagas ME, Pitsouni EI, Malietzis GA, Pappas G. Comparison of PubMed, Scopus, Web of Science, and Google Scholar: strengths and weaknesses. FASEB J (2008) 22:338–42. doi: 10.1096/fj.07-9492LSF
20. Choudhri AF, Siddiqui A, Khan NR, Cohen HL. Understanding bibliometric parameters and analysis. Radiographics (2015) 35:736–46. doi: 10.1148/rg.2015140036
21. Kulkarni AV, Aziz B, Shams I, Busse JW. Comparisons of citations in Web of Science, Scopus, and Google Scholar for articles published in general medical journals. Jama (2009) 302:1092–96. doi: 10.1001/jama.2009.1307
22. Humphrey SM. File maintenance of MeSH headings in Medline. J Am Soc Inf Sci (1984) 35:34–44. doi: 10.1002/asi.4630350106
23. Egghe L. Theory and practise of the g-index. Scientometrics [Internet] (2006) 69(1):131–52. doi: 10.1007/s11192-006-0144-7
24. Ball R. An introduction to bibliometrics: New development and trends: Chandos Publishing. (Cambridge, MA: Chandos Publishing, an imprint of Elsevier). (2017).
25. van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics (2010) 84:523–38. doi: 10.1007/s11192-009-0146-3
26. Chen C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. JASIST (2006) 57:359–77. doi: 10.1002/asi.20317
27. Aria M, Cuccurullo C. bibliometrix: An R-tool for comprehensive science mapping analysis. J Informetr (2017) 11:959–75. doi: 10.1016/j.joi.2017.08.007
28. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature (2007) 449:804–10. doi: 10.1038/nature06244
29. Blanco-Miguez A, Gutierrez-Jacome A, Fdez-Riverola F, Lourenco A, Sanchez B. MAHMI database: a comprehensive MetaHit-based resource for the study of the mechanism of action of the human microbiota. Database-Oxford (2017) 2017:baw157. doi: 10.1093/database/baw157
30. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature (2014) 505:559–63. doi: 10.1038/nature12820
31. Kaper JB, Nataro JP, Mobley HL. Pathogenic escherichia coli. Nat Rev Microbiol (2004) 2:123–40. doi: 10.1038/nrmicro818
32. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol (2009) 9:313–23. doi: 10.1038/nri2515
33. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature (2012) 489:220–30. doi: 10.1038/nature11550
34. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA (2008) 105:16731–36. doi: 10.1073/pnas.0804812105
35. O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep (2006) 7:688–93. doi: 10.1038/sj.embor.7400731
36. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell (2012) 148:1258–70. doi: 10.1016/j.cell.2012.01.035
37. Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet (2012) 13:260–70. doi: 10.1038/nrg3182
38. Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med (2009) 361:2066–78. doi: 10.1056/NEJMra0804647
39. Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe (2014) 15:382–92. doi: 10.1016/j.chom.2014.02.005
40. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med (2016) 375:2369–79. doi: 10.1056/NEJMra1600266
41. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol (2012) 13:R79. doi: 10.1186/gb-2012-13-9-r79
42. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature (2011) 474:307–17. doi: 10.1038/nature10209
43. Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature (2008) 453:620–25. doi: 10.1038/nature07008
44. Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut (2006) 55:205–11. doi: 10.1136/gut.2005.073817
45. Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA (2010) 107:12204–09. doi: 10.1073/pnas.0909122107
46. Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr (2010) 104 Suppl 2:S1–63. doi: 10.1017/S0007114510003363
47. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell (2011) 145:745–57. doi: 10.1016/j.cell.2011.04.022
48. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA (2007) 104:13780–85. doi: 10.1073/pnas.0706625104
49. Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet (2003) 361:512–19. doi: 10.1016/S0140-6736(03)12489-0
50. Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol (2020) 38:23–48. doi: 10.1146/annurev-immunol-070119-115104
51. Ikeda T, Nishida A, Yamano M, Kimura I. Short-chain fatty acid receptors and gut microbiota as therapeutic targets in metabolic, immune, and neurological diseases. Pharmacol Ther (2022) 239:108273. doi: 10.1016/j.pharmthera.2022.108273
52. Strasser B, Wolters M, Weyh C, Krüger K, Ticinesi A. The effects of lifestyle and diet on gut microbiota composition, inflammation and muscle performance in our aging society. Nutrients (2021) 13(6):2045. doi: 10.3390/nu13062045
53. Burrello C, Garavaglia F, Cribiu FM, Ercoli G, Lopez G, Troisi J, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun (2018) 9:5184. doi: 10.1038/s41467-018-07359-8
54. Cooper ID. Bibliometrics basics. J Med Libr Assoc (2015) 103:217–8. doi: 10.3163/1536-5050.103.4.013
55. Consortium IHiRN. The integrative human microbiome project. Nature (2019) 569:641–48. doi: 10.1038/s41586-019-1238-8
Keywords: intestinal microbiota, inflammatory bowel disease (IBD), bibliometric analysis, citation, research trends
Citation: Zhang A, Wang F, Li D, Wang C-Z, Yao H, Wan J-Y and Yuan C-S (2023) Emerging insights into inflammatory bowel disease from the intestinal microbiota perspective: a bibliometric analysis. Front. Immunol. 14:1264705. doi: 10.3389/fimmu.2023.1264705
Received: 21 July 2023; Accepted: 12 October 2023;
Published: 26 October 2023.
Edited by:
Ding Shi, Zhejiang University, ChinaReviewed by:
Yisheng He, The Chinese University of Hong Kong, Shenzhen, ChinaCopyright © 2023 Zhang, Wang, Li, Wang, Yao, Wan and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiqiang Yao, aGFpcWlhbmd5YW9Ab3V0bG9vay5jb20=; Jin-Yi Wan, d2FuamlueWkxMTI4QDE2My5jb20=
†ORCID: Haiqiang Yao, orcid.org/0000-0001-9716-0171
Jin-Yi Wan, orcid.org/0000-0001-8830-2035
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.