- 1Department of Laboratory Medical Center, General Hospital of Northern Theater Command, Shenyang, China
- 2Bioinformatics of Department, Key laboratory of Cell Biology, School of Life Sciences, China Medical University, Shenyang, China
Objective: Recently, circulating donor-derive cell free DNA (dd-cfDNA) has gained growing attention in the field of solid organ transplantation. The aim of the study was to analyze circulating dd-cfDNA levels in graft rejection, ACR and AMR separately for each rejection type compared with non-rejection, and assessed the diagnostic potential of dd-cfDNA levels in predicting graft rejection after lung transplantation.
Methods: A systematic search for relevant articles was conducted on Medline, Web of Science, China National Knowledge Infrastructure (CNKI), and Wanfang databases without restriction of languages. The search date ended on June 1, 2023. STATA software was used to analyze the difference between graft rejection, ACR, AMR and stable controls, and evaluate the diagnostic performance of circulating dd-cfDNA in detecting graft rejection.
Results: The results indicated that circulating dd-cfDNA levels in graft rejection, ACR, and AMR were significantly higher than non-rejection (graft rejection: SMD=1.78, 95% CI: 1.31-2.25, I2 = 88.6%, P< 0.001; ACR: SMD=1.03, 95% CI: 0.47-1.59, I2 = 89.0%, P < 0.001; AMR: SMD= 1.78, 95% CI: 1.20-2.35, I2 = 89.8%, P < 0.001). Circulating dd-cfDNA levels distinguished graft rejection from non-rejection with a pooled sensitivity of 0.87 (95% CI: 0.80-0.92) and a pooled specificity of 0.82 (95% CI: 0.76-0.86). The corresponding SROC yield an AUROC of 0.90 (95% CI: 0.87-0.93).
Conclusion: Circulating dd-cfDNA could be used as a non-invasive biomarker to distinguish the patients with graft rejection from normal stable controls.
Systematic Review Registration: https://www.crd.york.ac.uk/prospero/, identifier CRD42023440467.
Introduction
Lung transplantation has become the live-saving treatment for patients with end-stage pulmonary disease. Dramatic advancements have been made in surgery, medications, and postoperative management, and increased the overall survival rate to 85% and 59% at the end of the first and five years of transplantation, respectively (1). However, graft rejection, including acute cellular rejection (ACR) and antibody-mediated rejection (AMR), remains a substantial cause of morbidity and mortality after lung transplantation. ACR, as one of the most common complications, is detected in 27% of lung transplant recipients (LTRs) within the first year, and significantly related to chronic allograft dysfunction (2, 3). Moreover, due to the absence of the standardized diagnostic and treatment approaches, it is estimated that the incidence rate of AMR ranges from 4% to >50% in LTRs (4). AMR, characterized by donor-specific antibodies (DSA), innate immune infiltration, and evidence of complement activation, has been consistently identified as a contributor to morbidity, chronic lung allograft dysfunction (CLAD), and graft failure (5, 6). To date, invasive biopsy is the golden standard for diagnosis of graft rejection, performed either when rejection is suspected or for surveillance (7). However, invasive graft biopsy is difficult to practice for most LTRs in clinical settings. Apart from the possibility of sampling error, high economic cost, and physician dependence, a potential risk of complications has prevented invasive biopsy from becoming a routine screening approach (8, 9). Thus, a non-invasive, relatively objective, and sensitive biomarker is urgently needed for rejection diagnosis after transplantation.
Donor-derive cell free DNA (dd-cfDNA) refers to the free DNA released by damaged cells of graft tissue after transplantation. Recently, circulating dd-cfDNA has gained growing attention in the field of solid organ transplantation. The dd-cfDNA examination takes advantage of sensitivity and specificity of genome sequencing and the wide genomic difference between donors and recipients to quantify the circulating dd-cfDNA (10). The assay methods of dd-cfDNA includes quantitative polymerase chain reaction, digital droplet PCR, shotgun sequencing, and HLA alleles-mismatches. Xiao H, et al. summarized that dd-cfDNA can be a helpful marker for the diagnosis of AMR in transplantation recipients suspected of renal dysfunction (11). Wijtvliet VPWM, et al. showed that dd-cfDNA may be a useful marker for AMR diagnosis, but probably not for ACR (12). Mounting evidence has suggested that dd-cfDNA was a potential non-invasive marker for allograft injury and rejection and could be used for graft rejection surveillance by the International Society for Heart and Lung Transplantation (ISHLT) (13–15). To the best our knowledge, no relevant systematic review and meta-analysis evaluated the circulating dd-cfDNA levels in graft rejection, ACR, and AMR, as well as the diagnostic value in graft rejection after lung transplantation.
To date, the clinical detection of circulating dd-cfDNA levels in lung transplantation has been documented in more than 10 studies, focusing on ACR, AMR or both. However, the data on the circulating dd-cfDNA levels in graft rejection after lung transplantation are conflicting. We therefore conducted a systematic review and meta-analysis to analyze circulating dd-cfDNA levels in graft rejection, ACR and AMR separately for each rejection type compared with non-rejection, and assessed the diagnostic potential of dd-cfDNA in predicting graft rejection after lung transplantation.
Materials and methods
This systematic review and meta-analysis was performed following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA 2020) guidelines. Quality assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool and Newcastle Ottawa scale (NOS) tool were used to assess the quality of included studies, respectively. The protocol was registered in the International Prospective Register of Systematic Reviews (PROSPERO; CRD 42023440467).
Literature search
A systematic search for relevant articles was conducted on Medline, Web of Science, China National Knowledge Infrastructure (CNKI), and Wanfang databases without restriction of languages. No time restrictions were applied regarding the start date of publication, and the search ended on June 1, 2023. The search strategy was conducted using the following keywords: (“donor” OR “tissue donors”) AND (“cell-free DNA” OR “cell-free nucleic acids”) AND (“rejection”) AND (“lung transplantation” OR “lung grafting” OR “pulmonary transplant” OR “pulmonary allograft”) OR “lung transplant” OR “lung allograft”). Two reviewers (LB AND LYH) screened the titles and abstracts of articles in the initial search to identify the relevant publications. Then, full texts were obtained and reviewed in detail.
Inclusion and exclusion criteria
Articles were considered eligible if they met all of the following inclusion and exclusion criteria. The inclusion criteria included: (1) case-control studies or cohort studies; (2) studies conducted on lung transplantation, including graft rejection or ACR or AMR cases; (3) studies assessing the circulating dd-cfDNA levels, or studies including sufficient data to calculate the number of true positive (TP), true negative (TN), false positive (FP), and false negative (FN) values. Moreover, studies that met the following criteria were excluded: (1) duplicate publications; (2) review articles, editorials, comments, and conference proceedings; (3) animal experiments; (4) essential data for pooled analysis or quality assessment were not retrievable.
Two reviewers (LB and LYH) independently selected and cross-checked the articles. Any discrepancies were resolved by discussion.
Data extraction and quality assessment
The following data were extracted from the selected articles: first author’s name, publication year, country, study design, sample number, rejection type, control type, assay method, dd-cfDNA levels, thresholds, the area under the receiver operating characteristic curve (AUROC), and its sensitivity, specificity, TP, FP, FN, and TN.
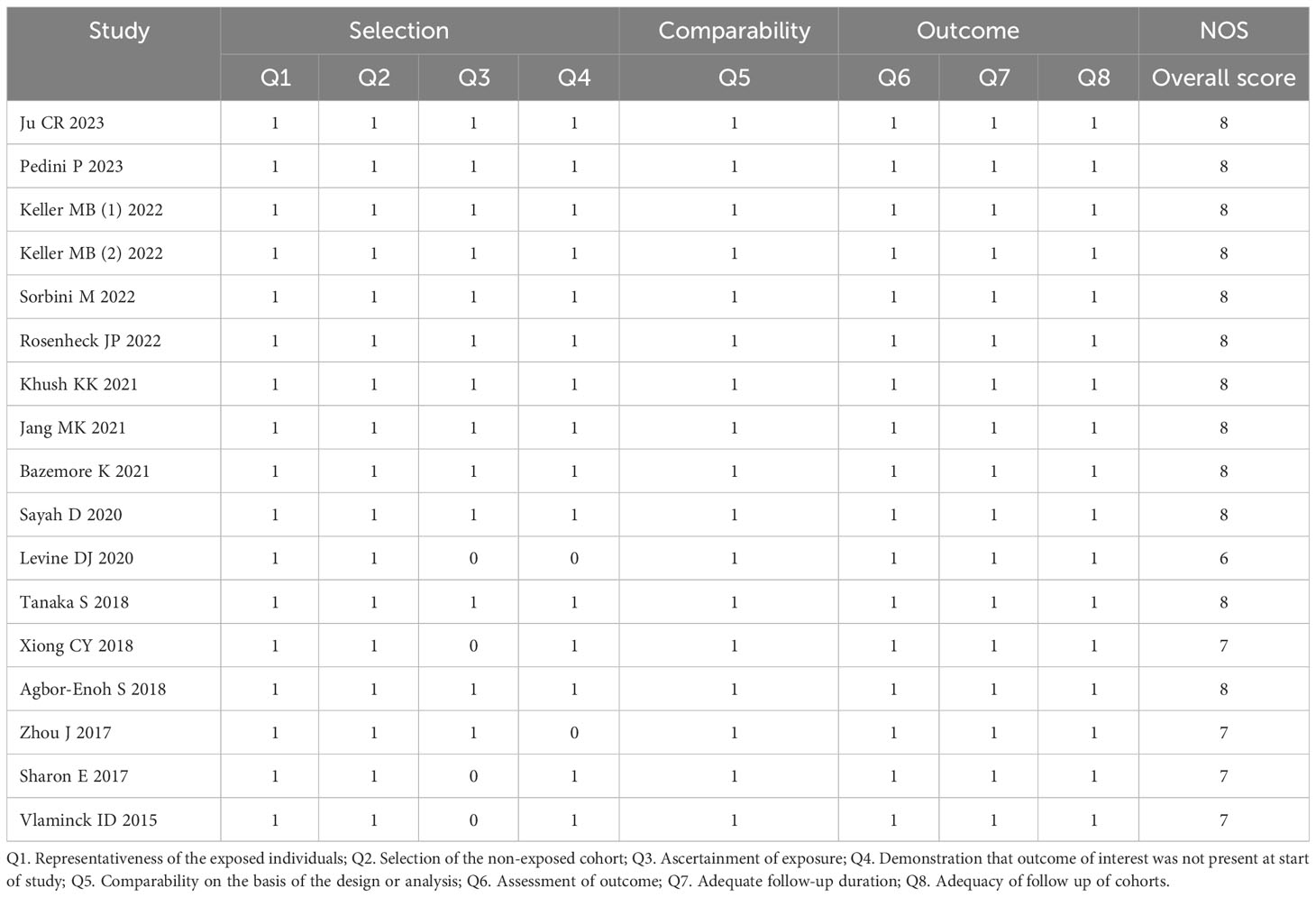
The case‐control study or cohort study was evaluated according to the NOS tool containing eight items that could be classified into three main categories (selection, comparability, and exposure/outcome evaluation) and assigned on a score of 0-9. In the NOS assessment, scores of 7-9, 4-6, and 0-3 were considered “high quality”, “moderate quality”, and “low quality”, respectively. The quality of diagnostic studies was evaluated using QUADAS‐2 tool, which consisted of seven items covering case selection, index test, golden standard, and flow and timing. The total score ≥4 (full score= 7) indicated that the quality of the study was high.
Statistical analysis
STATA software (version 15.0, Stata Corp) was used to analyze the original data. We performed the meta-analyses of circulating dd-cfDNA levels in patients with graft rejection, ACR, and AMR in comparison with normal stable cases. Due to different assay methods of dd-cfDNA levels, standard mean difference (SMD) along with 95% confidence interval (CI) was used to compare the difference. If the original articles only provide median and interquartile ranges (IQR), the methods suggested by Luo DH, et al. and Wan X, et al. were used to convert them into means and standard deviations (SDs) (16, 17). Given the variability in the sample characteristics, diagnostic criteria, and dd-cfDNA assay methods between studies, the chi-square Q test and I 2 statistics were used to determine the heterogeneity of eligible studies. If I 2 > 50% or P < 0.05, we considered heterogeneity to be significant and adopted a random-effects model, otherwise, a fixed-effects model was adopted. Publication bias was assessed through funnel plot asymmetry and Begg's test among the included studies. Sensitivity analysis was performed to assess the impact of single study on the overall result by serial removal of a specific study.
STATA software was used to estimate the diagnostic test accuracy of circulating dd-cfDNA in graft rejection by calculating the pooled ROC area of all studies. We performed pooled analysis of sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and area under the summary receiver operating characteristic curve (AUSROC), along with the corresponding 95% CI. Deek’s funnel plot was used to assess publication bias among studies. Fagan nomogram can show the correlation between prior probability, likelihood ratio and post-test probability. The more the difference between pre-test and post-test probability, the more important the biomarker is, which can provide a reference for clinical application evaluation (18). A P value < 0.05 was considered statistically significant.
Results
Search results
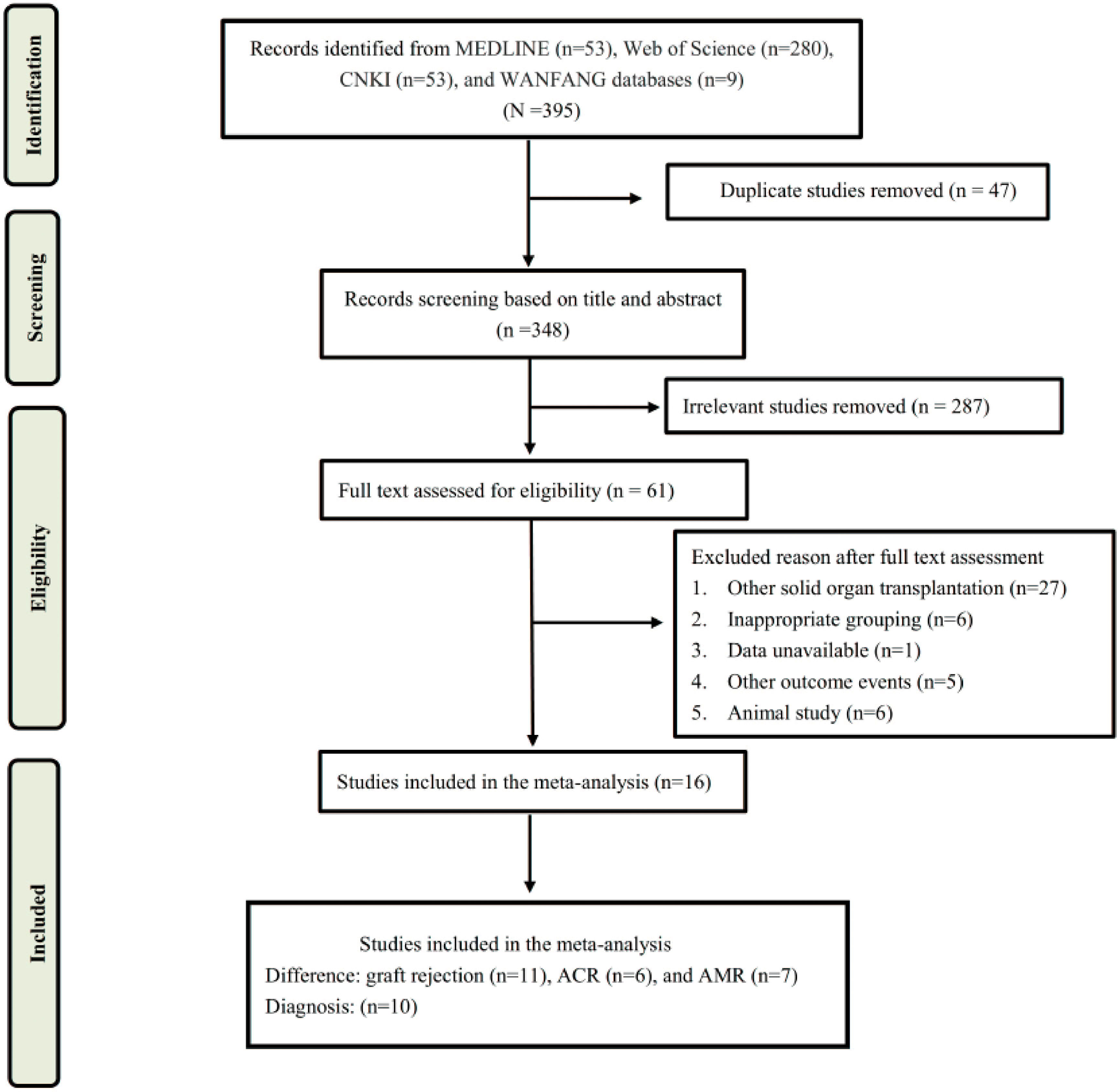
The literature search and selection process were shown in Figure 1. A total of 395 articles were screened, including 53 from Medline, 280 from Web of Science, 53 from CNKI, and 9 from WANFANG database. After a careful review of the titles and abstracts, 304 articles were excluded for duplicate and irrelevance, of which 61 articles were selected for additional screening based on the full text. Finally, a total of 16 studies were included in this systematic review and meta-analysis (19–34).
Study characteristics and quality assessment
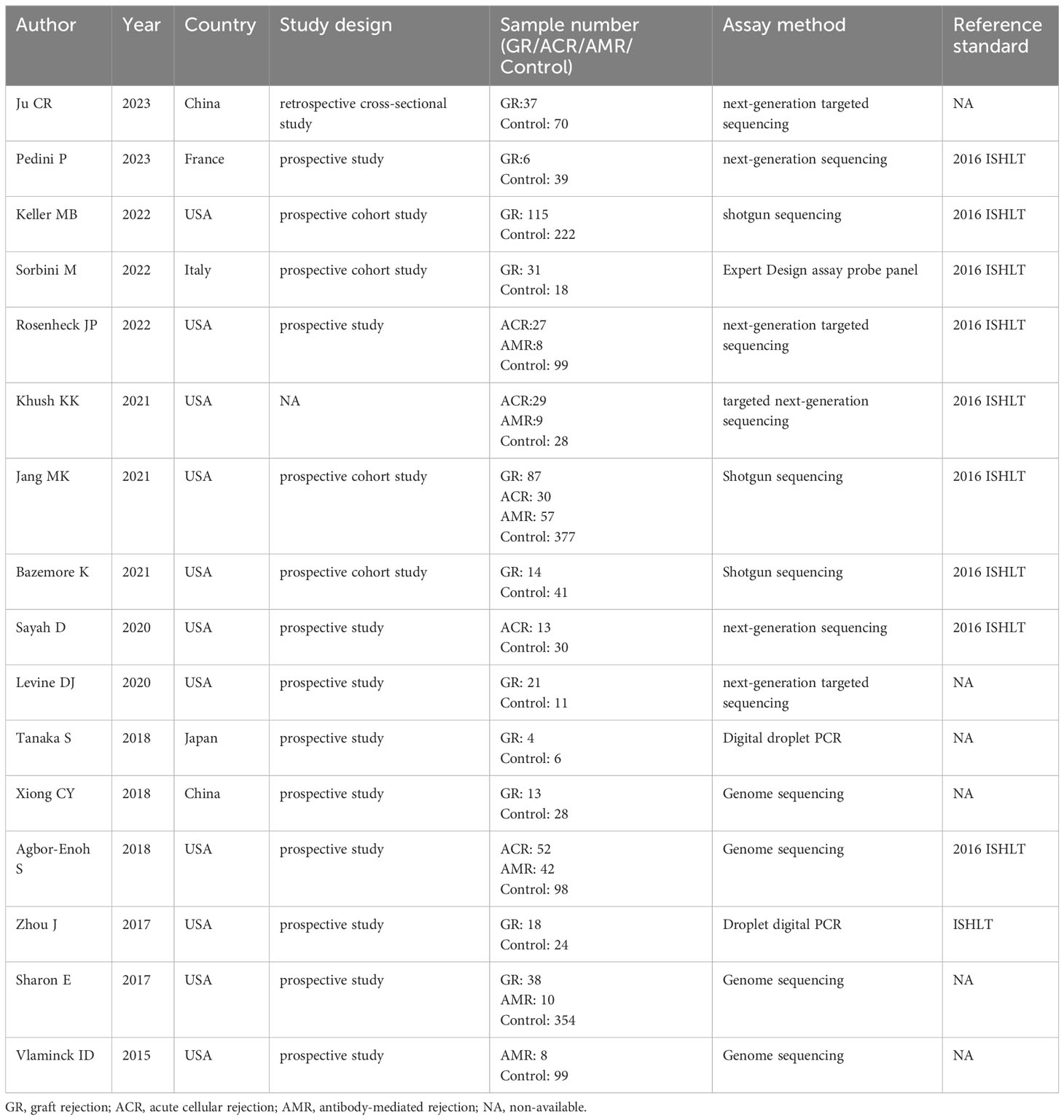
A summary of characteristics of the included studies was shown in Tables 1, Table 2. Eleven studies were from USA (21, 23–28, 31–36), 1 study from France (20), 1 study from Italy (22), and 1 study from Japan (29), and two studies from China (19, 30). In total, eleven studies measured dd-cfDNA levels in graft rejection (19, 21, 22, 25, 26, 28–30, 32, 34), 6 studies measured dd-cfDNA levels in ACR (23–25, 27, 31, 33), and 7 studies measured dd-cfDNA levels in AMR (21, 23–25, 31, 33, 34), respectively. Eleven studies evaluated the diagnostic accuracy of circulating dd-cfDNA in graft rejection (19, 21–25, 27, 28, 30, 33), providing AUROC, sensitivity and specificity. Except for one study was retrospective study (19), all studies were prospective studies. Table 3 and Figure 2 present the quality results of NOS tool and QUADAS-2 tool, respectively.

Figure 2 Quality assessment of the included literature. (A) QUADAS-2 summary plot of bias assessment. (B) QUADAS-2 bar plot of the individual risk of bias domains and applicability concerns.
Circulating dd-cfDNA levels in graft rejection, ACR and AMR
A total of 11 studies, including 384 graft rejection samples and 929 no-rejection samples, were included in the comparison of circulating dd-cfDNA levels in graft rejection and non-rejection groups. Due to high heterogeneity, the random-effects model was used. Meta-analysis indicted that circulating dd-cfDNA levels in graft rejection group were significantly higher than non-rejection group (SMD: 1.78, 95% CI: 1.31-2.25, I2 = 88.6%, P< 0.001, Figure 3A). Six studies including 178 samples with ACR and 731 samples without ACR showed that circulating dd-cfDNA levels in ACR group was significantly higher than non-ACR group (SMD: 1.03, 95% CI: 0.47-1.59, I2 = 89.0%, P < 0.001, Figure 3B). Moreover, seven studies including 213 samples with AMR and 1,277 samples without AMR, showed that circulating dd-cfDNA levels in AMR was significantly higher than non-AMR (SMD: 1.78, 95% CI: 1.20-2.35, I2 = 89.8%, P < 0.001, Figure 3C).
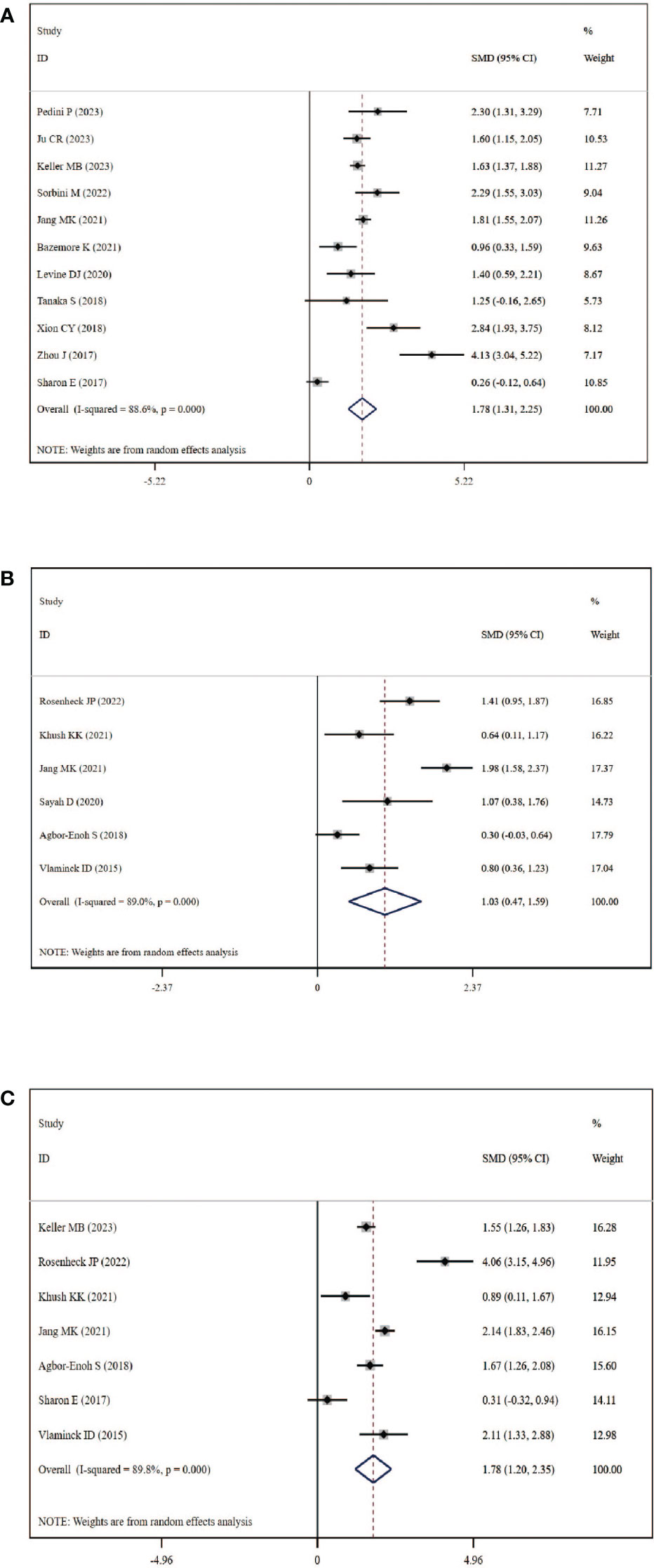
Figure 3 Forest plot of circulating dd-cfDNA levels in patients with rejection. (A) dd-cfDNA levels in graft rejection, (B) dd-cfDNA levels in ACR, (C) dd-cfDNA levels in AMR.
Begg’s test was used to explore the publication bias of the included studies in our meta-analysis, and a visual funnel plot was plotted to identify the publication bias (Figure S1). The funnel plots showed symmetrical features which indicated the absence of publication bias, identified by Begg’s tests (graft rejection: P=0.533; ACR: P=0.851; AMR: P=0.764). Moreover, sensitivity analysis which removed each study in turn showed that the overall result was stable and no study could influence the final result in this meta-analysis (Figure S2).
Diagnostic performance of circulating dd-cfDNA in graft rejection
Circulating dd-cfDNA levels distinguished graft rejection from non-rejection with a pooled sensitivity of 0.87 (95% CI: 0.80-0.92) and a pooled specificity of 0.82 (95% CI: 0.76-0.86) (Figure 4A). The combined PLR, NLR, and DOR were 4.7 (95% CI: 3.4-6.5), 0.16 (95% CI:0.10-0.26), and 29 (95% CI: 14-61), respectively (Figure S3). The corresponding SROC yield an AUROC of 0.90 (95% CI: 0.87-0.93, Figure 4B), indicating high accuracy in diagnostic performance of graft rejection. There was no significant publication bias identified by Deek’s funnel plot asymmetry test (P=0.41, Figure 4C).
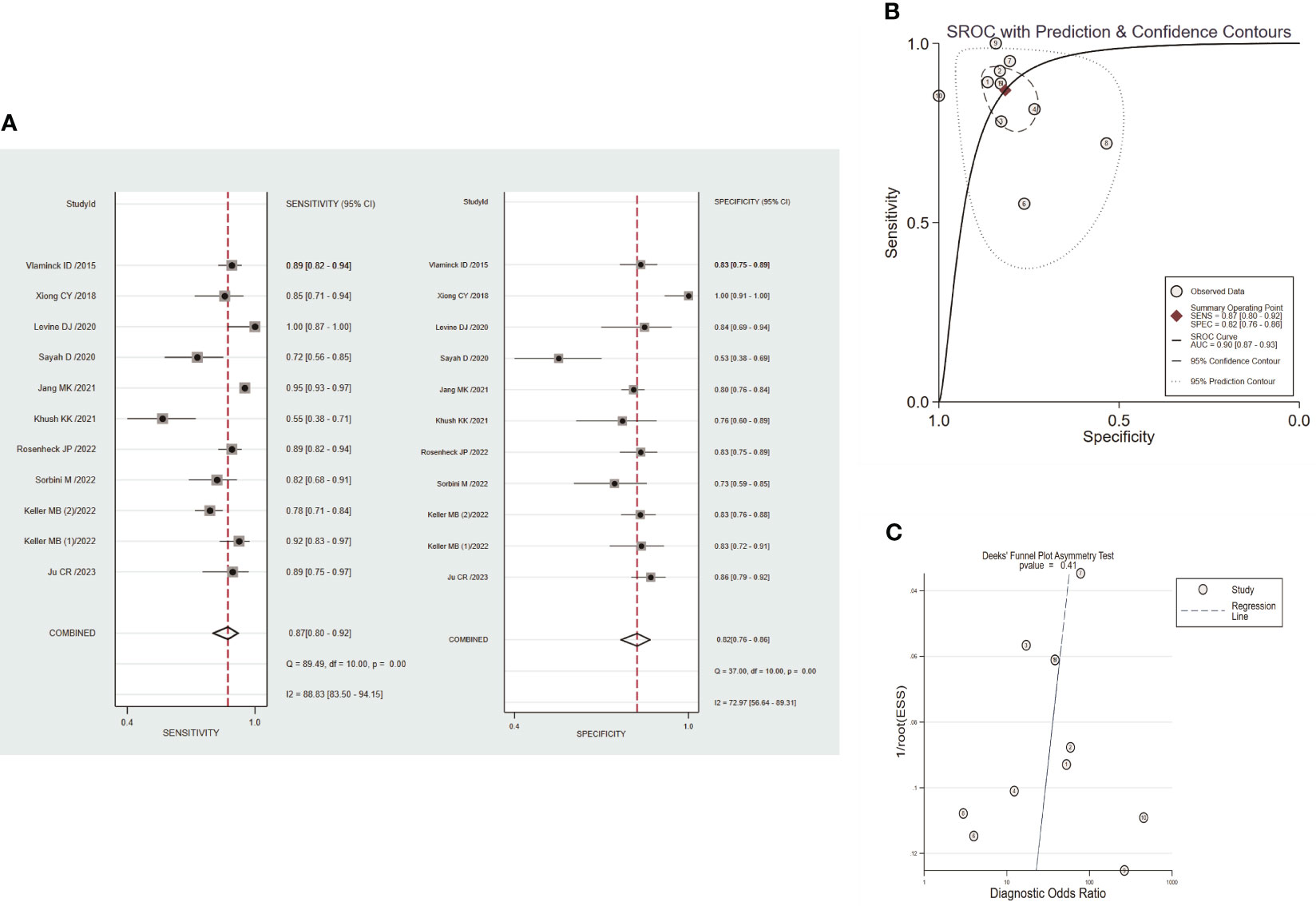
Figure 4 Performance of circulating dd-cfDNA in graft rejection detection. (A) forest plots of pooled diagnostic performance for sensitivity and specificity of circulating dd-cfDNA in graft rejection detection; (B) summary receiver operating characteristic (SROC) plot of circulating dd-cfDNA in graft rejection detection; (C) Deeks’ funnel plot for publication bias.
Fagan nomogram
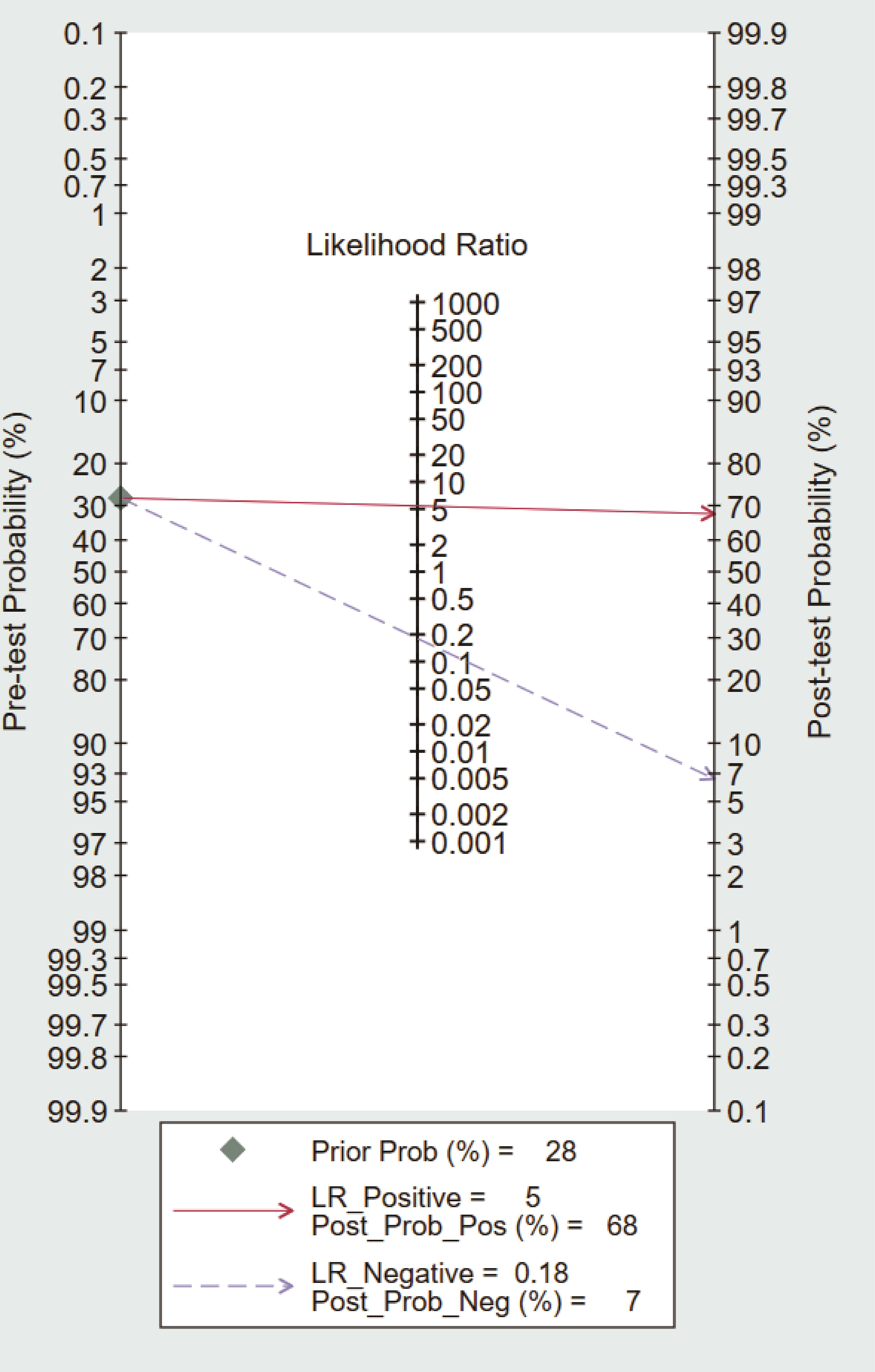
To further explore the potential value of circulating dd-cfDNA level for graft rejection in lung transplantation, we built a Fagan nomogram. ISHLT reported that 28% of LTRs experience at least one episode of treated ACR within the first year (37). Assuming a 28% pre-test probability of developing graft rejection, we used circulating dd-cfDNA levels to predict a 68% post-probability of developing graft rejection and a 7% risk of not developing graft rejection (Figure 5). The Fagan nomogram showed that the post-probability was increased by 40% in patients with positive pre-test and decreased by 21% in patients with negative pre-test. Our findings suggest that circulating dd-cfDNA was a clinically beneficial diagnostic marker in predicting graft rejection after lung transplantation.
Discussion
Rejection remains a significant problem following lung transplantation. In spite of advances in surgical techniques and medication, long term outcomes after lung transplantation remains worse compared with other solid organ transplantation (38). The clinical data indicated that 28% of adult recipients experienced at least one episode of acute rejection during the first year after lung transplantation (37). Currently, the biopsy with detailed pathology is regarded as the gold standard for making diagnosis based on histologic lesions associated with acute rejection. However, frequent coexistence of rejection and infection, disagreement between different pathologists, and invasive complication limited the accuracy, timeliness, and repeatability. The value of dd-cfDNA as a non-invasive biomarker for lung transplantation has been investigated in some studies. Moreover, most studies were published in the last three years, underlining the emerging interest in circulating dd-cfDNA detection as a monitor of graft rejection. However, does dd-cfDNA play a role in the current clinical paradigm for the monitoring of underlying graft rejection? In this review and meta-analysis, we found that circulating dd-cfDNA levels in lung transplantation patients with graft rejection, ACR, and AMR were higher than patients without rejection or stable patients, and circulating dd-cfDNA was a non-invasive potential marker in diagnosis of graft rejection after lung transplantatiopn.
The dd-cfDNA, detected in the blood of transplant recipients, provided huge potential as a sensitive, non-invasive, cost-effective biomarker for long-term monitoring of graft health, based on the fact that organ transplantations are also genome transplants. The plasma dd-cfDNA levels were generally low in stable transplant recipients, and dd-cfDNA levels increased due to tissue injury after solid organ transplantation (39). Snyder TM, et al. found that dd-cfDNA levels were less than 1% at stable patients, and increased to 3-4% during rejection episode after heart transplantation (40). In lung transplantation, similar results were found that dd-cfDNA had a strong concordance with clinical indicators of rejection (33). In liver transplantation, dd-cell free DNA was elevated significantly after engrafting, followed by a steady decrease to 10% of total cfDNA after 1 week (41). Moreover, dd-cfDNA accounts for <0.5% of total plasma cell free DNA in renal transplantation recipients without allograft injury, and normal dd-cfDNA could reduce the probability of detecting rejection (42). Early diagnosis of subclinical rejection might improve the clinical prognosis. Agbor-Rnoh S, et al. reported allograft injury detected by dd-cfDNA preceded clinical AMR diagnosis by a median of 2.8 months (31). The result from a total of 1,092 adult kidney transplant recipients across 7 transplant centers showed that the elevation of dd-cfDNA was significantly correlated with clinical and subclinical graft rejection (43). However, not all injurious processes may release substantial cell lysis or cause the dysregulation leading to an elevation in dd-cfDNA (44). Khush KK, et al. reported that dd-cfDNA level with AMR was no difference with stable patients after lung transplantation (24). Sayah D, et al. found that dd-cfDNA levels with ACR were not different with allograft infection (27). Moreover, dd-cfDNA values in different studies were presented as fraction (% ddcfDNA of total cfDNA) or absolute value (copies/ml plasma), which lead to varied thresholds among rejection types and clinical status. A recent study demonstrated that relative change in dd-cfDNA% has gained improved diagnostic performance compared with absolute values (45). In view of the conflicting results, we extracted the relative value of dd-cfDNA of included studies, and found that dd-cfDNA fraction levels were significantly higher in graft rejection, not only in ACR, but also in AMR in this meta-analysis.
The release of cell-free DNA into the blood is influenced by many physiological and pathological factors, such as physical and psychological stress, inflammation, and autoimmunity (46). Remarkably, dd-cfDNA was elevated in the presence of infection, in keeping with the notion that dd-cfDNA could predict graft damage. Compared with asymptomatic respiratory tract infections, symptomatic respiratory tract infections were also more likely to have high %ddcfDNA (36). A multicenter prospective cohort study of Genomic Research Alliance for Transplantation found that dd-cfDNA is a marker of pathogen associated allograft injury and may detect subclinical injury (26). In addition, very few patients underwent a prolonged decline in dd-cfDNA levels within the first 10 days after kidney transplantation, graft rejection might be suspected incorrectly due to elevated dd-cfDNA levels (47). Hence, we performed a synthesis of the currently available knowledge of circulating dd-cfDNA in relation to rejection diagnosis. The results indicated that circulating dd-cfDNA has a high accuracy in the diagnosis of graft rejection with pooled sensitivity and pooled specificity of 0.87 and 0.82, respectively. The PLR and NLR were 4.7 and 0.16, which indicated that probability of positive dd-cfDNA in graft rejection was 4.7 times higher than normal stable patients, and normal stable patients with negative results have about one-sixth chance of developing graft rejection. The DOR value was 29, indicating that circulating dd-cfDNA could effectively distinguish graft rejection from normal stable patients. Based on the Fagan’s nomogram, if the pre-test probability was 28%, the post-test probability of dd-cfDNA’s positive diagnostic result for graft rejection was 68%. In addition, the SROC curve showed that corresponding AUC was 0.90 (0.87-0.93), which was much higher than the common diagnostic criteria (>0.8). Above results indicated that circulating dd-cfDNA has a promising diagnostic performance in predicting graft rejection. However, the heterogeneity in these studies was considerable. The clinical characteristics of patients, methodological differences, cut-off values, and sample preparation protocols may lead to hidden bias in the pooled estimation. In view of no sufficient studies to perform subgroup analysis, in depth analysis of heterogeneity was not possible. With the advances in laboratory technology and knowledge, the subgroup analysis of circulating dd-cfDNA levels is needed in the future.
The present study has some limitations. First, the sample size was relatively small, and there are not many studies related to circulating dd-cfDNA levels in graft rejection in recent years. Second, the diagnostic criteria were not standardized in each study. Third, it was difficult to obtain the raw data for some included studies, which restricts us to investigate the diagnostic value of dd-cfDNA absolute quantification. Forth, owing to the different measurement methods, the circulating dd-cfDNA cut-off values used among the included studies were not completely consistent, and different cut-off values would affect the accuracy of the combined results to a certain extent. Most importantly, sources of heterogeneity in the results should still be considered carefully. Nonetheless, our study clearly showed that circulating dd-cfDNA was a promising marker in diagnosis of graft rejection, despite the fact that the meta-analysis has the limitations mentioned.
Conclusion
Generally, our meta-analysis suggested that circulating dd-cfDNA was higher in patients with graft rejection, ACR, and AMR. Circulating dd-cfDNA could be used as a non-invasive biomarker to distinguish the patients with graft rejection from normal stable controls. Further evidence is required to explore the diagnostic accuracy of circulating dd-cfDNA in each type of graft rejection, thus paving the way for clinical application of circulating dd-cfDNA detection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
BL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – original draft. YL: Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1263389/full#supplementary-material
Supplementary Figure 1 | Funnel plot of the included studies to evaluate the publication bias. (A) graft rejection (n=10 studies); (B) ACR (n=6 studies); (C) AMR (n=6 studies).
Supplementary Figure 2 | Sensitivity analysis of individual study. (A) graft rejection (n=10 studies); (B) ACR (n=6 studies); (C) AMR (n=6 studies).
Supplementary Figure 3 | Forest plots of pooled diagnostic performance for PLR, NLR, and DOR.
References
1. Bos S, Vos R, Van Raemdonck DE, Verleden GM. Survival in adult lung transplantation: where are we in 2020? Curr Opin Organ Transplant (2020) 25(3):268–73. doi: 10.1097/MOT.0000000000000753
2. Greer M, Werlein C, Jonigk D. Surveillance for acute cellular rejection after lung transplantation. Ann Transl Med (2020) 8(6):410. doi: 10.21037/atm.2020.02.127
3. Steinack C, Gaspert A, Gautschi F, Hage R, Vrugt B, Soltermann A, et al. Transbronchial cryobiopsy compared to forceps biopsy for diagnosis of acute cellular rejection in lung transplants: analysis of 63 consecutive procedures. Life (Basel) (2022) 12(6):898. doi: 10.3390/life12060898
4. Witt CA, Gaut JP, Yusen RD, Byers DE, Iuppa JA, Bennett Bain K, et al. Acute antibody-mediated rejection after lung transplantation. J Heart Lung Transplant (2013) 32(10):1034–40. doi: 10.1016/j.healun.2013.07.004
5. Calabrese DR, Chong T, Singer JP, Rajalingam R, Hays SR, Kukreja J, et al. CD16(+) natural killer cells in bronchoalveolar lavage are associated with antibody-mediated rejection and chronic lung allograft dysfunction. Am J Transplant (2023) 23(1):37–44. doi: 10.1016/j.ajt.2022.10.006
6. Halverson LP, Hachem RR. Antibody-mediated rejection and lung transplantation. Semin Respir Crit Care Med (2021) 42(3):428–35. doi: 10.1055/s-0041-1728796
7. McWilliams TJ, Williams TJ, Whitford HM, Snell GI. Surveillance bronchoscopy in lung transplant recipients: risk versus benefit. J Heart Lung Transplant (2008) 27(11):1203–9. doi: 10.1016/j.healun.2008.08.004
8. Park CH, Paik HC, Haam SJ, Lim BJ, Byun MK, Shin JA, et al. HRCT features of acute rejection in patients with bilateral lung transplantation: the usefulness of lesion distribution. Transplant Proc (2014) 46(5):1511–6. doi: 10.1016/j.transproceed.2013.12.060
9. Hamour IM, Burke MM, Bell AD, Panicker MG, Banerjee R, Banner NR. Limited utility of endomyocardial biopsy in the first year after heart transplantation. Transplantation (2008) 85(7):969–74. doi: 10.1097/TP.0b013e318168d571
10. Agbor-Enoh S, Wang Y, Tunc I, Jang MK, Davis A, De Vlaminck I, et al. Donor-derived cell-free DNA predicts allograft failure and mortality after lung transplantation. EBioMedicine (2019) 40:541–53. doi: 10.1016/j.ebiom.2018.12.029
11. Xiao H, Gao F, Pang Q, Xia Q, Zeng X, Peng J, et al. Diagnostic accuracy of donor-derived cell-free DNA in renal-allograft rejection: A meta-analysis. Transplantation (2021) 105(6):1303–10. doi: 10.1097/TP.0000000000003443
12. Wijtvliet V, Plaeke P, Abrams S, Hens N, Gielis EM, Hellemans R, et al. Donor-derived cell-free DNA as a biomarker for rejection after kidney transplantation: a systematic review and meta-analysis. Transpl Int (2020) 33(12):1626–42. doi: 10.1111/tri.13753
13. Velleca A, Shullo MA, Dhital K, Azeka E, Colvin M, DePasquale E, et al. The International Society for Heart and Lung Transplantation (ISHLT) guidelines for the care of heart transplant recipients. J Heart Lung Transplant (2023) 42(5):e1–e141. doi: 10.1016/j.healun.2022.10.015
14. Rodgers N, Gerding B, Cusi V, Vaida F, Tada Y, Morris GP, et al. Comparison of two donor-derived cell-free DNA tests and a blood gene-expression profile test in heart transplantation. Clin Transplant (2023) 37(6):e14984. doi: 10.1111/ctr.14984
15. Oellerich M, Budde K, Osmanodja B, Bornemann-Kolatzki K, Beck J, Schutz E, et al. Donor-derived cell-free DNA as a diagnostic tool in transplantation. Front Genet (2022) 13:1031894. doi: 10.3389/fgene.2022.1031894
16. Luo D, Wan X, Liu J, Tong T. Optimally estimating the sample mean from the sample size, median, mid-range, and/or mid-quartile range. Stat Methods Med Res (2018) 27(6):1785–805. doi: 10.1177/0962280216669183
17. Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol (2014) 14:135. doi: 10.1186/1471-2288-14-135
18. Luo H, Yin L. Diagnostic value of superb microvascular imaging and color doppler for thyroid nodules: A meta-analysis. Front Oncol (2023) 13:1029936. doi: 10.3389/fonc.2023.1029936
19. Ju C, Xu X, Zhang J, Chen A, Lian Q, Liu F, et al. Application of plasma donor-derived cell free DNA for lung allograft rejection diagnosis in lung transplant recipients. BMC Pulm Med (2023) 23(1):37. doi: 10.1186/s12890-022-02229-y
20. Pedini P, Coiffard B, Cherouat N, Casas S, Fina F, Boutonnet A, et al. Clinical relevance of cell-free DNA quantification and qualification during the first month after lung transplantation. Front Immunol (2023) 14:1183949. doi: 10.3389/fimmu.2023.1183949
21. Keller MB, Meda R, Fu S, Yu K, Jang MK, Charya A, et al. Comparison of donor-derived cell-free DNA between single versus double lung transplant recipients. Am J Transplant (2022) 22(10):2451–7. doi: 10.1111/ajt.17039
22. Sorbini M, Togliatto G, Mioli F, Simonato E, Marro M, Cappuccio M, et al. Validation of a simple, rapid, and cost-effective method for acute rejection monitoring in lung transplant recipients. Transpl Int (2022) 35:10546. doi: 10.3389/ti.2022.10546
23. Rosenheck JP, Ross DJ, Botros M, Wong A, Sternberg J, Chen YA, et al. Clinical validation of a plasma donor-derived cell-free DNA assay to detect allograft rejection and injury in lung transplant. Transplant Direct (2022) 8(4):e1317. doi: 10.1097/TXD.0000000000001317
24. Khush KK, De Vlaminck I, Luikart H, Ross DJ, Nicolls MR. Donor-derived, cell-free DNA levels by next-generation targeted sequencing are elevated in allograft rejection after lung transplantation. ERJ Open Res (2021) 7(1):00462–2020. doi: 10.1183/23120541.00462-2020
25. Jang MK, Tunc I, Berry GJ, Marboe C, Kong H, Keller MB, et al. Donor-derived cell-free DNA accurately detects acute rejection in lung transplant patients, a multicenter cohort study. J Heart Lung Transplant (2021) 40(8):822–30. doi: 10.1016/j.healun.2021.04.009
26. Bazemore K, Rohly M, Permpalung N, Yu K, Timofte I, Brown AW, et al. Donor derived cell free DNA% is elevated with pathogens that are risk factors for acute and chronic lung allograft injury. J Heart Lung Transplant (2021) 40(11):1454–62. doi: 10.1016/j.healun.2021.05.012
27. Sayah D, Weigt SS, Ramsey A, Ardehali A, Golden J, Ross DJ. Plasma donor-derived cell-free DNA levels are increased during acute cellular rejection after lung transplant: pilot data. Transplant Direct (2020) 6(10):e608. doi: 10.1097/TXD.0000000000001063
28. Levine DJ, Ross DJ, Sako E. Single center “Snapshot” Experience with donor-derived cell-free DNA after lung transplantation. biomark Insights (2020) 15:1177271920958704. doi: 10.1177/1177271920958704
29. Tanaka S, Sugimoto S, Kurosaki T, Miyoshi K, Otani S, Suzawa K, et al. Donor-derived cell-free DNA is associated with acute rejection and decreased oxygenation in primary graft dysfunction after living donor-lobar lung transplantation. Sci Rep (2018) 8(1):15366. doi: 10.1038/s41598-018-33848-3
30. Xiong CY. The application of Noninvasive Diagnosis of Cell-Free DNA in the monitoring of Lung Transplant Rejection. (2018), MA. thesis. South China University of Technology.
31. Agbor-Enoh S, Jackson AM, Tunc I, Berry GJ, Cochrane A, Grimm D, et al. Late manifestation of alloantibody-associated injury and clinical pulmonary antibody-mediated rejection: Evidence from cell-free DNA analysis. J Heart Lung Transplant (2018) 37(7):925–32. doi: 10.1016/j.healun.2018.01.1305
32. Zou J, Duffy B, Slade M, Young AL, Steward N, Hachem R, et al. Rapid detection of donor cell free DNA in lung transplant recipients with rejections using donor-recipient HLA mismatch. Hum Immunol (2017) 78(4):342–9. doi: 10.1016/j.humimm.2017.03.002
33. De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A (2015) 112(43):13336–41. doi: 10.1073/pnas.1517494112
34. Sharon E, Shi H, Kharbanda S, Koh W, Martin LR, Khush KK, et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PloS Comput Biol (2017) 13(8):e1005629. doi: 10.1371/journal.pcbi.1005629
35. Charya AV, Ponor IL, Cochrane A, Levine D, Philogene M, Fu YP, et al. Clinical features and allograft failure rates of pulmonary antibody-mediated rejection categories. J Heart Lung Transplant (2023) 42(2):226–35. doi: 10.1016/j.healun.2022.09.012
36. Bazemore K, Permpalung N, Mathew J, Lemma M, Haile B, Avery R, et al. Elevated cell-free DNA in respiratory viral infection and associated lung allograft dysfunction. Am J Transplant (2022) 22(11):2560–70. doi: 10.1111/ajt.17125
37. Chambers DC, Yusen RD, Cherikh WS, Goldfarb SB, Kucheryavaya AY, Khusch K, et al. The Registry of the International Society for Heart and Lung Transplantation: Thirty-fourth Adult Lung And Heart-Lung Transplantation Report-2017; Focus Theme: Allograft ischemic time. J Heart Lung Transplant (2017) 36(10):1047–59. doi: 10.1016/j.healun.2017.07.016
38. Parulekar AD, Kao CC. Detection, classification, and management of rejection after lung transplantation. J Thorac Dis (2019) 11(Suppl 14):S1732–S9. doi: 10.21037/jtd.2019.03.83
39. Keller M, Agbor-Enoh S. Cell-free DNA in lung transplantation: research tool or clinical workhorse? Curr Opin Organ Transplant (2022) 27(3):177–83. doi: 10.1097/MOT.0000000000000979
40. Snyder TM, Khush KK, Valantine HA, Quake SR. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci (2011) 108(15):6229–34. doi: 10.1073/pnas.1013924108
41. Singal A, Schütz E, Fischer A, Beck J, Harden M, Koch M, et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: A prospective, observational, multicenter cohort study. PloS Med (2017) 14(4):e1002286. doi: 10.1371/journal.pmed.1002286
42. Graver AS, Lee D, Power DA, Whitlam JB. Understanding donor-derived cell-free DNA in kidney transplantation: an overview and case-based guide for clinicians. Transplantation (2022) 107(8):1675–1686. doi: 10.1097/TP.0000000000004482
43. Bu L, Gupta G, Pai A, Anand S, Stites E, Moinuddin I, et al. Clinical outcomes from the Assessing Donor-derived cell-free DNA Monitoring Insights of kidney Allografts with Longitudinal surveillance (ADMIRAL) study. Kidney Int (2022) 101(4):793–803. doi: 10.1016/j.kint.2021.11.034
44. Grabuschnig S, Bronkhorst AJ, Holdenrieder S, Rosales Rodriguez I, Schliep KP, Schwendenwein D, et al. Putative origins of cell-free DNA in humans: A review of active and passive nucleic acid release mechanisms. Int J Mol Sci (2020) 21(21):8062. doi: 10.3390/ijms21218062
45. Trindade AJ, Chapin KC, Gray JN, Furuya Y, Mullican A, Hoy H, et al. Relative change in donor-derived cell-free DNA is superior to absolute values for diagnosis of acute lung allograft dysfunction. Transplant Direct (2023) 9(6):e1487. doi: 10.1097/TXD.0000000000001487
46. Kustanovich A, Schwartz R, Peretz T, Grinshpun A. Life and death of circulating cell-free DNA. Cancer Biol Ther (2019) 20(8):1057–67. doi: 10.1080/15384047.2019.1598759
Keywords: donor-derived cell-free DNA, lung transplantation, graft rejection, acute cellular rejection, antibody-mediated rejection, meta-analysis
Citation: Li Y and Liang B (2023) Circulating donor-derived cell-free DNA as a marker for rejection after lung transplantation. Front. Immunol. 14:1263389. doi: 10.3389/fimmu.2023.1263389
Received: 19 July 2023; Accepted: 26 September 2023;
Published: 11 October 2023.
Edited by:
Ilias Doxiadis, University Hospital Leipzig, GermanyReviewed by:
Philippe Lemaitre, Columbia University, United StatesRoss Simon Francis, Princess Alexandra Hospital, Australia
Copyright © 2023 Li and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Liang, YmxpYW5nQGNtdS5lZHUuY24=
 Yunhui Li
Yunhui Li Bin Liang
Bin Liang