- 1Abel Salazar Institute of Biomedical Sciences (ICBAS), University of Porto, Porto, Portugal
- 2Interdisciplinary Centre of Marine and Environmental Research (CIIMAR), University of Porto, Porto, Portugal
- 3Fish Immunology and Vaccinology Group, IBMC-Instituto de Biologia Molecular e Celular, Universidade do Porto, Porto, Portugal
- 4i3S - Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Porto, Portugal
- 5Departamento de Anatomía, Produción Animal e Ciencias Clínicas Veterinarias, Facultade de Veterinaria, Universidade de Santiago de Compostela, Lugo, Spain
Introduction: The marine aquaculture industry has been witnessing a worldwide emergence of tenacibaculosis, a poorly understood bacterial disease caused by Tenacibaculum maritimum that affects commercially important fish. So far, knowledge on the T. maritimum virulence mechanisms is scarce and the pathogen-host interaction operating in tenacibaculosis remain to be disclosed. This study aimed at contributing to a better understanding of this disease, by evaluating the early innate immune response triggered in European sea bass (Dicentrarchus labrax) by a bath-challenge with T. maritimum.
Methods: Groups of sea bass were bath-challenged with T. maritimum (challenged fish) or mock-challenged. Undisturbed fish were used as controls (time 0). Samples of blood, liver and mucosal organs (skin, gills and posterior-intestine) were collected at 0 h (control) and at 6, 24, 48 and 72 h post-challenge (n=12). Mucosal organs were used for analyzing the expression of immune-related genes by RT-qPCR, as well as blood samples for assessing haematological and innate humoral parameters and liver for oxidative stress assessment.
Results: An increased expression of il-1β, il8, mmp9 and hamp1 was detected in all mucosal organs of infected fish when compared with control and mock-challenged fish, suggesting a pro-inflammatory response against T. maritimum transversal to all organs. The faster induction of these pro-inflammatory genes was observed in the gills. Regarding the systemic response, challenged fish presented neutrophilia, monocytosis, signs of anemia, and a decrease of bactericidal and lysozyme activities in plasma. Almost no variations were observed regarding hepatic oxidative stress.
Discussion/Conclusions: The present study suggests that T. maritimum induces a local innate immune response upon bath infection not only in the skin of European sea bass, but also in the gills and posterior-intestine, likely triggered by the T. maritimum’s capacity to adhere, colonize and damage these organs that can function as entry ways to bacteria, leading ultimately to the seen host’s systemic response.
Highlights
● Bath-challenge with T. maritimum induces a pro-inflammatory response in fish mucosal organs;
● The response was faster in the gills than in the skin and posterior-intestine;
● Hemato-immunological parameters of challenged fish suggest a systemic response;
Introduction
Aquaculture is regarded as one of the fastest growing food production sectors, and, therefore, has the potential to fulfil the future demand for animal protein. This need is reflected in the tendency that aquaculture has to develop towards intensification (1), which in turn could enhance the susceptibility of the farmed aquatic organisms to disease outbreaks. The introduction and translocation of fish stocks between aquaculture facilities can also lead to the spread of diseases (2), which in association with the high stocking densities used in the aquaculture settings allow the thriving of several pathogens (3, 4).
In the last decades, the marine aquaculture sector has been witnessing a worldwide emergence of tenacibaculosis (formerly known as marine flexibacteriosis), a relatively unknown pathology that affects several commercially important species (5–7).
This disease has been responsible for countless losses, since it was first reported as a gliding bacterial infection affecting black seabream fry (Acanthopagrus schlegeli) reared in floating net cages in Japan (8). Since then, this pathogen was able to spread between aquaculture sites, reaching Europe in the French Mediterranean Coast, where it affected European sea bass (Dicentrarchus labrax) rearing facilities (9). Later on, cases of tenacibaculosis in cultured European sea bass were diagnosed in Italy, Greece and Turkey (10–12), increasing the concern regarding this disease.
Tenacibaculum maritimum is the etiological agent of tenacibaculosis, and has been described as a Gram-negative filamentous bacterium able to induce small lesions, upraised spots, scale loss and some disintegration of the epidermis in the host’s body surface, namely in the head, skin or fins (13–15). These lesions can establish a portal of entry for other opportunistic and frank pathogens, leading to mixed infections, which can ultimately lead to the host’s death (15–17).
In order to cause such detrimental symptomatology, T. maritimum presents a plethora of virulence mechanisms that allows a successful adhesion and colonization of its hosts. These bacteria rely on the production of exopolysaccharides, various adhesins and proteins with lectin or carbohydrate-binding motifs to strongly adhere to fish mucus, where they gather and accumulate the nutrients necessary for growth and proliferation (16, 18, 19). T. maritimum has also been described as a pathogen able to agglutinate erythrocytes from a wide range of species (20) and to directly compete with the host’s iron-binding proteins. In a study developed by (21), it was demonstrated that different T. maritimum strains have at least two different iron-uptake mechanisms, one related to the synthesis of siderophores and other involving the utilization of heme groups as iron sources (21). The proteolytic activity of several extracellular products (ECPs) has also been described and shown to include the ability to degrade gelatin, amylase, casein and nucleases (20). Furthermore, the genome analysis of T. maritimum revealed several proteins homologous to proteins that in other bacteria are known to act as toxins and virulence factors, such as sphingomyelinase and ceramidase (19). Despite these studies, knowledge regarding T. maritimum pathogenesis is scarce, and very few studies have approached the interactions between this pathogen and the host.
Guardiola et al. (22) focused on Senegalese sole (Solea senegalensis) mucosal and systemic immune responses following bath challenge with a sub-lethal dose of T. maritimum and further suggested the rudimentary systemic and the delayed host’s mucosal responses (22). Plasma’s antiprotease and bactericidal activities were mainly increased in challenged fish in the end of the trial, at 14-days post-challenge, and the same tendency was recorded for the haemolytic complement, lysozyme and peroxidase activities in skin mucus (22). This suggests that Senegalese sole immune response can be prolonged at least 14 days after being exposed to T. maritimum.
In a study developed by Faílde et al. (23), the haematological profile of turbot (Scophthalmus maximus) challenged subcutaneously with T. maritimum showed some alterations, including granulocytosis, lymphopenia and thrombocytopenia as well as mild decrease of haematocrit values. Due to the seen distribution of immunoglobulin positive cells in spleen, kidney, thymus, skin and intestine, it is suggested that tenacibaculosis is able to induce a humoral immune response in turbot, through the synthesis of specific antibodies in the spleen that later on migrate to lesion areas in the skin (23).
The present study aimed to bring more insights on the host responses against this fastidious bacterial pathogen by evaluating parameters of the short-term mucosal and systemic innate immune response in European sea bass (Dicentrarchus labrax) after bath-challenge with T. maritimum. To the best our knowledge, this is the first study approaching the host’s molecular immune response with focus on the three main mucosal organs (gills, skin and posterior-intestine).
Material and methods
Bacterial culture and inoculum preparation
The T. maritimum strain (ACC13.1) used in this study was isolated from Senegalese sole and belongs to the serotype O3 (24). The strain was kindly provided by Professor Alicia E. Toranzo (Departamento de Microbiología y Parasitología, Facultad de Biología, University of Santiago de Compostela, Spain) and stocks were kept frozen at −80°C until use. Recovery from frozen stocks was achieved using marine agar (MA; Laboratories CONDA, Spain) at 25°C for 48 h.
For inoculum preparation, bacteria were inoculated in 50 mL of marine broth (MB; Laboratories CONDA, Spain) in a 500 mL Erlenmeyer and grown at 25°C, with continuous shaking (180 rpm) for 48 h. Turbidity was measured at 600 nm (Spectrophotometer, UV-1600PC, VWR) and exponentially growing bacteria (OD=0.886) were collected by centrifugation at 3,000 x g for 10 min and resuspended in MB at a concentration of 5 x 105 CFU mL-1. The bacterial concentration was adjusted with the predetermined growth curve for this specific strain: y = 2 x 108x + 4 x 107 (25).
Fish husbandry and experimental design
The current study was conducted under the supervision of accredited researchers in laboratory animal science by the Portuguese Veterinary Authority following FELASA category C recommendations and in agreement with the guidelines for protection of animal used for scientific purposes according to European Union directive (2010/63/EU) (reviewed and approved by 0421/000/000/2020).
For this trial, European sea bass juveniles (45.45 ± 8.1 g) with no record of previous tenacibaculosis outbreaks were obtained from a commercial fish farm (Portugal) and were maintained in quarantine for 4 weeks at CIIMAR fish holding facilities in a recirculating aerated seawater system at 21.8 ± 0.4°C, salinity of 34.2 ± 0.4 ‰, 8.2 ± 0.2 mg mL-1 dissolved oxygen and a 12 h light/12 h dark photoperiod. Water quality was maintained with mechanical and biological filtration, and fish were fed daily with a commercial diet (Aquasoja, Portugal) at 2% of body weight, distributed by two meals a day. Ammonia and nitrite levels were measured daily using commercial kits. For screening purposes and to assess the health status of the stock fish, ten randomly selected individuals were sampled for histopathological assessment. Before the bacterial challenge, fish were randomly distributed into two closed recirculating seawater systems (10 kg m-3 stocking density, n= 25 fish per tank, 0.11 m3), one for the mock-challenged fish and another for the challenged fish, each with four aquaria (4 replicates for each treatment) for sampling purposes and two aquaria (two replicates for each treatment) to follow cumulative mortality, and acclimated for one week.
At the challenge, water temperature was increased to 25°C, to mimic temperature conditions at which tenacibaculosis outbreaks occur (25, 26). Fish, previously fasted for 24 h, were bath challenged for 2 h with T. maritimum (ACC13.1), prepared as described in the previous section (25), at a concentration of 5 x 105 CFU mL-1 (according to a pre-challenge to determine the LD30 for this strain). Challenge was performed in 50 L tanks with strong aeration at a stocking density of 25 kg m-3. Mock-challenged fish were submitted to the same treatment, but MB was used instead of bacterial inoculum. After challenge, fish were returned to the recirculating system where they were acclimated. Bacteria was re-isolated from aseptically collected blood from randomly selected challenge fish at 24 h post-challenged and identified as T. maritimum as described elsewhere (27).
Sampling
Fish were not fed during the trial period. Samples were collected post-mortem after euthanizing the fish with an overdose of anaesthetic, 0.7 mL/L (2-phenoxyethanol; Merck, ref. 807291, Germany). Sampling was performed before starting the bath challenge (n=12) (time 0, control) and at 6, 24, 48 and 72 h post-challenge. At each sampling time, three fish were removed from each tank (n=12 per treatment) and blood was collected from the caudal vein with heparinized 1 mL syringes and placed in heparinized 1.5 mL tubes. An aliquot was removed for haematological analysis, while the remaining blood was centrifuged for 10 min at 10,000 x g at 4°C for plasma collection and storage at ‐80°C. Skin (collected across the midline of the fish, beneath the dorsal fin, without any muscle), gills (portion of the second arch) and posterior intestine were also sampled and stored in RNA later (at a proportion of 1/10 w/v) at 4 °C for the first 24 h, and then stored at -80 °C for molecular biology analysis. Liver was collected and immediately frozen in liquid nitrogen, followed by storage at -80 °C. Samples of skin (across the midline of the fish, beneath the dorsal fin, not previously sampled for mucus, including 1 cm of subjacent muscle), gills (portion of the second arch) and posterior intestine were also collected for histological analyses.
Haematological parameters
The haematological profile was conducted according to Machado et al. (28). Total white (WBC) and red (RBC) blood cells were counted using a Neubauer chamber and haematocrit (Ht) and haemoglobin (Hb; SPINREACT kit, ref. 1001230, Spain) were also assessed, as previously described (28). The mean corpuscular volume (MCV), mean corpuscular haemoglobin (MCH) and mean corpuscular haemoglobin concentration (MCHC) were calculated (28).
Blood smears were done with 3 µL of gently homogenized blood, air dried and fixed for 1 min in formol-ethanol (10% of 37% formaldehyde in absolute ethanol). For identifying neutrophils, the peroxidase detection method described by Afonso et al. (29) was used (29). Blood smears were then stained with Wright’s stain (Haemacolor; Merck). Slides were examined under oil immersion (1,000 x) and 200 leucocytes were counted and categorized, based on their morphology, as thrombocytes, lymphocytes, monocytes and neutrophils. The percentage of each cell population was calculated and multiplied by total number of WBC in order to determine the number of cells per mL.
Innate immune parameters
Antiprotease and protease activities
The antiprotease activity was determined as described by Ellis (30) adapted for 96-well microplates. Shortly, 10 µL of plasma were incubated in microtubes with 10 µL of trypsin solution (5 mg mL−1 in 0.5% NaHCO3, pH 8.3) (Sigma, USA) for 10 min at 22°C. After incubation, 100 µL of phosphate buffer (115 mM NaH2PO4, pH 7.0) plus 125 µL of azocasein (20 mg mL−1 in 0.5% NaHCO3, pH 8.3) were added and incubated again for 1 h at 22°C in the dark, with agitation. Then, 250 µL of 10% cold trichloroacetic acid (TCA) were added and incubated for 30 min at 22°C, followed by centrifugation at 10,000 x g for 5 min at room temperature (RT). Finally, 100 µL were transferred, in duplicate, to a 96-well plate containing 100 µL of 1N NaOH per well and the OD (optical density) read at 450 nm in a Synergy HT microplate reader. Phosphate buffered saline was used as positive control, instead of plasma, and the percentage of trypsin activity was calculated as follows: 100 − ((sample absorbance/reference absorbance) × 100).
To assess protease activity, the same protocol was followed, but the initial incubation of the plasma with trypsin was omitted and the incubation with azocasein and phosphate buffer was maintained for 24 h instead of 1h, in constant agitation. Plasma was replaced by trypsin (5mg ml−1, Sigma) as a positive control or by PBS as negative control. The percentage of trypsin activity compared to the positive control was calculated as follows: (sample absorbance/positive reference) × 100.
Peroxidase
Peroxidase activity was determined in plasma as described by Quade and Roth (31). Briefly, in triplicates, 15 µL of plasma were placed into flat-bottomed 96-well plates and diluted in 135 µL of HBSS without Ca+2 and Mg+2 (Cytiva, USA). Then, 50 µL of 20 mM 3,3’,5,5’- tetramethylbenzidine hydrochloride (TMB; Sigma, USA) were added to each well. After 2 min the reaction was stopped by adding 50 µL of 2 M sulphuric acid and the absorbance was measured at 450 nm (Synergy HT microplate reader). Peroxidase activity (units mL-1 plasma) was calculated by defining one unit of peroxidase as the amount needed to produce an absorbance change of 1 OD.
Lysozyme activity
Lysozyme activity was assessed as described by Costas et al. (32). Firstly, Micrococcus lysodeikticus solution (0.5 mg mL−1 in 0.05 M sodium phosphate buffer, pH 6.2) was prepared. Then, 15 µL of plasma were added, in triplicates, to a microplate plus 250 µL of the Micrococcus lysodeikticus solution, for a final volume of 265 µL. After incubation at 25°C, the absorbance (450 nm) was measured after 0.5 and 20 min in a Synergy HT microplate reader. Lyophilized hen egg white lysozyme (Sigma) was successively diluted in sodium phosphate buffer (0.05 M, pH 6.2) to obtain a standard curve. The amount of lysozyme in the sample was calculated using the standard curve.
Bactericidal activity
The bactericidal activity assay was performed using T. maritimum ACC13.1 strain. Bacteria were grown on MA at 25°C for 24 h and resuspended in MB at a concentration of 1.6 x 108 CFUs mL-1, by measuring the turbidity at 600 nm (Synergy HT microplate reader) and using the previously mentioned growth curve. Plasma bactericidal activity was then determined following the method described by Graham et al. (33) with some modifications (28, 33). In a U-shaped 96-well plate, 20 µL of plasma were added in duplicates, and as positive control, MB was added to the wells instead of plasma. In each well, 20 µL of bacteria were added to the plate followed by an incubation for 2.5 h at 25°C. Afterwards, 25 µL of 3-(4, 5 dimethyl-2-yl)-2,5-diphenyltetrazolium bromide (MTT, 1 mg mL−1; Sigma) were added to each well and the plate was incubated for 10 min at 25°C. Plates were centrifuged at 2,000 × g for 10 min and formazan precipitate was dissolved with 200 µL of dimethyl sulfoxide (Sigma). The absorbance of the dissolved precipitate was measured at 560 nm (Synergy HT microplate reader). In this method, the difference between the formazan present in samples and in the positive controls (100%) enables to calculate the viable bacteria in each sample and, consequently, the percentage of non-viable bacteria.
Nitrite concentration
To indirectly access the nitric oxide (NO) concentration in plasma, a Nitrite/Nitrate colorimetric kit (Roche, 11746081001, Germany) was used according to the manufacturer’s instructions. Since nitrite and nitrate are endogenously produced as oxidative metabolites of the messenger molecule NO, these compounds are considered as indicative of NO production (34). To measure nitrite/nitrate, the samples were previously diluted 1:10 in distilled H2O in microtubes and the concentrations were expressed as µM.
Oxidative stress biomarkers
Liver tissue were homogenized 1/10 (w/v) in potassium phosphate buffer (0.2 M, pH 7.4). From the homogenized mixture, 200 µL were transferred to a microtube with 4 µL of 4% BHT (2,6-Di-tert-butyl-4-methylphenol) in methanol for lipid peroxidation (LPO) assessment.
For determining superoxide dismutase, catalase and glutathione-S-transferase activities, for each volume of tissue homogenate, a volume of potassium phosphate buffer (0.2 M, pH 7.4) was added followed by a centrifugation at 10,000 x g for 20 minutes at 4°C. The supernatants were collected and kept at −80°C. Protein concentration was measured using Pierce™ BCA Protein Assay kit, with bovine serum albumin as standard, according to the manufacturer’s instructions. For superoxide dismutase and catalase activity homogenates were diluted to achieve a final protein concentration of 0.3 and for total glutathione-S-transferase a concentration of 0.7 mg/mL.
LPO was determined using the protocol described by Bird and Draper (35) with some modifications (36). A volume of 100 µL of 100% TCA was added to the previously mentioned 204 µL of liver homogenate, and afterwards, 1 mL of 0.73% thiobarbituric acid solution (in Tris–HCl 60 mM, pH 7.4 with DTPA 0.1 mM). Samples were incubated for 1 h at 100°C in a kiln and then microtubes were centrifuged for 5 minutes at 15,000 x g. A volume of 200 µL of supernatant was transferred to a 96-well plate in triplicates and the absorbance was measured at 535 nm. The LPO was expressed as nmol of thiobarbituric acid reactive substances (TBARS) formed per g of wet tissue.
Catalase activity was quantified measuring the decrease in absorbance, through the consumption of H2O2, as described by Claiborne (37) but adapting the protocol to microplates as described by Rodrigues et al. (38). A sample of 10 µL was transferred to a UV light microplate in triplicates with 140 µL of potassium phosphate (0.05 M, pH 7.0) plus 150 µL of 30% H2O2. The absorbance was measured at 240 nm for 2 min. The catalase activity was quantified using H2O2 molar extinction coefficient at 240 nm of 40 M cm-1, expressed in U per mg of protein.
Superoxide dismutase (SOD) activity was assessed following the protocol describe by Almeida et al. (39), utilizing the cytochrome C method with xanthine/xanthine oxidase (39). A volume of 50 µL of each sample was transferred to a microplate in triplicates. Then, 200 µL of a reaction solution containing 50 mM potassium phosphate buffer (pH 7.8) containing 1 mM Na-EDTA, 0.7 mM xanthine and 0.03 mM cytochrome C were added. Promptly, 50 µL of 0.03 U mL-1 xanthine oxidase with 0.1 mM Na-EDTA were also added to the microplate. Absorbance was measured at 550 nm (Synergy HT microplate reader) at 20 s intervals for 3 min. Activity is described as units of SOD per mg of protein. One unit of activity was defined as the quantity of enzyme necessary to produce a 50% inhibition of the cytochrome C reduction rate.
Glutathione-S-transferase (GST) activity was accessed following the method of (40) adapted to microplate by Frasco and Guilhermino (41). Briefly, a 250 µL of a reaction solution containing 0.2 M potassium phosphate buffer (pH 6.5), 10 mM reduced glutathione (GSH) and 60 mM 1-chloro-2,4-dinitrobenzene (CDNB) was added to 50 µL of liver homogenate in triplicates. Absorbance was recorded at 340 nm for 5 min with 20 s intervals in microplate. GST activity was expressed as mU per mg of protein, using the molar extinction coefficient at 340 nm of 9.6 × 106 M/cm.
The reduced (GSH):oxidized (GSSG) glutathione ratio was determined using the microplate assay for GSH/GSSG commercial kit (Oxford Biomedical Research, UK) as previously described by Hamre et al. (42). This method relies on the quantitative determination at 412 nm of the total amount of glutathione (GSH + GSSG) and GSSG (43). Briefly, the determination of GSSG is obtained by adding a thiol scavenger (N-ethylmaleimide pyridine derivative solution, Oxford Biomedical Research, UK), which reacts with GSH to form a stable complex, therefore removing the GSH prior to the quantification of GSSG, without inhibiting GR activity. Through the addition of glutathione reductase, the available GSSG is reduced to GSH which reacts with 5,5’-dithiobis-2-nitrobenzoic acid (DTNB) allowing the measurement of pre-existent GSSG. The rate of the reaction is proportional to the GSH and GSSG concentration. The GSH/GSSG Ratio is calculated as follows: (GSHt – 2GSSG)/GSSG.
Histology and immunohistochemistry
At each sampling point, 12 fish per group (control, mock-challenged and challenged) were sampled. Tissue fragments from gills, skin and intestine were fixed with 4% buffered formaldehyde for 24-48 h, dehydrated and embedded in paraffin wax. Sections of 2-3 mm thickness were obtained and collected on silane coated slides, followed by drying overnight, dewaxing, hydration. Sections were then stained with haematoxylin and eosin (H&E) or used for immunohistochemistry (IHC). Regarding IHC, incubations were performed at RT in a humidified chamber and washing was performed by immersion for 5 min in phosphate-buffered saline (PBS; 8 mM Na2HPO4 3 mM NaH2PO4, 150 mM NaCl, pH 7.4) containing 0.5% (v/v) Tween 20. Endogenous peroxidase activity was quenched by incubation with peroxidase blocking buffer (Vector Labs, Burlingame, CA) for 1 h. The sections were washed once and blocked for 20 min in 2.5% normal horse serum (Vector Labs, Burlingame, CA), followed by incubation with 1:1000 (concentrations of mg mL-1) working dilution of rabbit anti-T. maritimum LL01.8.3.8 immunoadsorbed antibody (anti-Tm) for 1.5 h, according to Faílde et al. (23).
After washing again, the sections were incubated with ImmPRESS®-VR Horse Anti-Rabbit IgG Polymer-HRP (Vector Labs, Burlingame, CA) for 30 min, rinsed, and color development achieved with Vector® VIP Substrate Kit, Peroxidase (HRP) (Vector Labs, Burlingame, CA), as the chromogen. After a final wash, the slides were counterstained with haematoxylin, dehydrated and mounted.
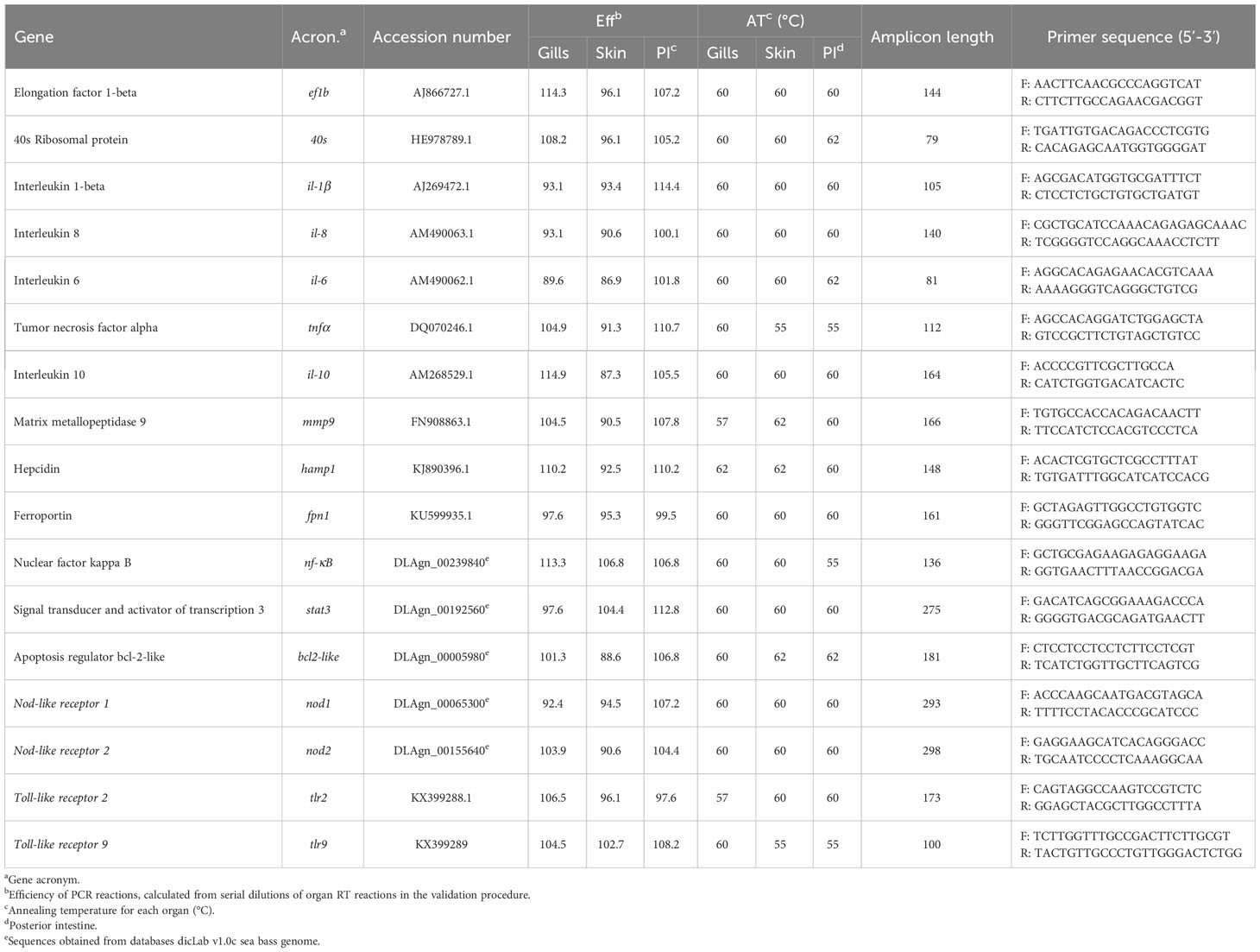
Gene expression analysis
Target organs (gills, skin and posterior-intestine) were weighted (up to 300 mg of organ), placed in 500 µL of Trizol (NZYTech, Lisbon, Portugal) and homogenized in a Precellys Evolution homogenizer at 6000 x g (2 x 20 s, 4°C). After this step, 150 µL of chloroform were added at 4°C and the samples were vortexed, followed by a centrifugation at 12,000 x g for 15 min at 4°C. The aqueous phase was transferred to a clean tube with 300 µL of 70% ethanol, mixed, and placed in NZYSpin Binding columns. After this step, the total RNA isolation was conducted with NZY Total RNA Isolation kit (NZYTech, Lisbon, Portugal) according to the manufacturer’s specifications. RNA samples were quantified and purity was assessed by spectrophotometry using DeNovix DS-11 FX (Wilmington, DE, USA) with absorbance ratios at 260 nm/280nm of 1.9–2.1. First-strand cDNA was synthesized and samples were standardized with NZY First-Strand cDNA Synthesis Kit (NZYTech, Lisbon, Portugal) with further storage at -80°C. For reverse transcriptase, a Veriti DX 96-well Thermal Cycler (Applied Biosystems, Foster City, CA, USA) was used. Real-time Quantitative PCR (qPCR) was performed with CFX384 Touch Real-Time PCR Detection System (Biorad, Hercules, CA, USA) using 4.4 µL of diluted cDNA mixed with 5 µL of iTaq Universal SYBR Green Supermix® (Biorad, Hercules, CA, USA) and 0.3 µL (10 µM) of each primer in a final volume of 10 µL. Primers were designed with NCBI Primer Blast Tool and IDT OligoAnalyzer ToolTM to amplify genes related with innate immune response in European sea bass. The known qPCR requirements (amplicon size, Tm difference between primers, GC content, and self-dimer or cross dimer formation) were respected. The template sequences used for the primer’s design were obtained from both NCBI and the databases dicLab v1.0c sea bass genome (44). The efficiency of each primer pair was determined by calculating the slope of the regression line of the cycle thresholds (Ct) vs. the relative concentration of cDNA, using serial 2-fold dilutions of cDNA. In order to ensure no amplification of primer dimers, melting curves were analyzed. The standard cycling conditions were 95 °C initial denaturation for 10 min, followed by 40 cycles of two steps (95 °C denaturation for 15 s followed by primer annealing temperature for 1 min), 95 °C for 1 min followed by 35 s at the annealing temperature, and finally, 95 °C for 15 s. The reactions were run in duplicates and target gene expression was normalized using the geometric mean of elongation factor 1β (ef1β) and ribosome 40s subunit (40s) and calculated according to the Pfaffl method (45).
Accession numbers, primer efficiencies and annealing temperatures for each organ, amplicon length and primer sequences are detailed in Table 1.
Statistical analysis
Mean and standard error of the mean (mean ± SEM) were calculated for all parameters. Data were analyzed for normality and homogeneity of variance, when necessary outliers were removed and gene expression data was Log-transformed before being statistically analyzed.
When all the assumptions were fulfilled, a T-student test or a One-Way ANOVA (Tukey post hoc test) was used under SPSS 27 program for WINDOWS. When the assumptions were not verified a Welch ANOVA (Games-Howell post hoc test) or a Kruskal-Wallis was performed. The level of significance used for all statistical tests was p ≤ 0.05.
Results
Bacterial challenge
Bath-challenge with 5 x 105 CFU mL-1 T. maritimum ACC13.1 resulted in 32.1% cumulative mortality, whereas, as expected, no mortality was recorded in mock-challenged fish (Figure 1; n=30 fish per treatment, X2<0.0008 for comparisons between treatments). Moreover, the mortalities in challenged fish occurred between days 3 and 4 after challenge (Figure 1).
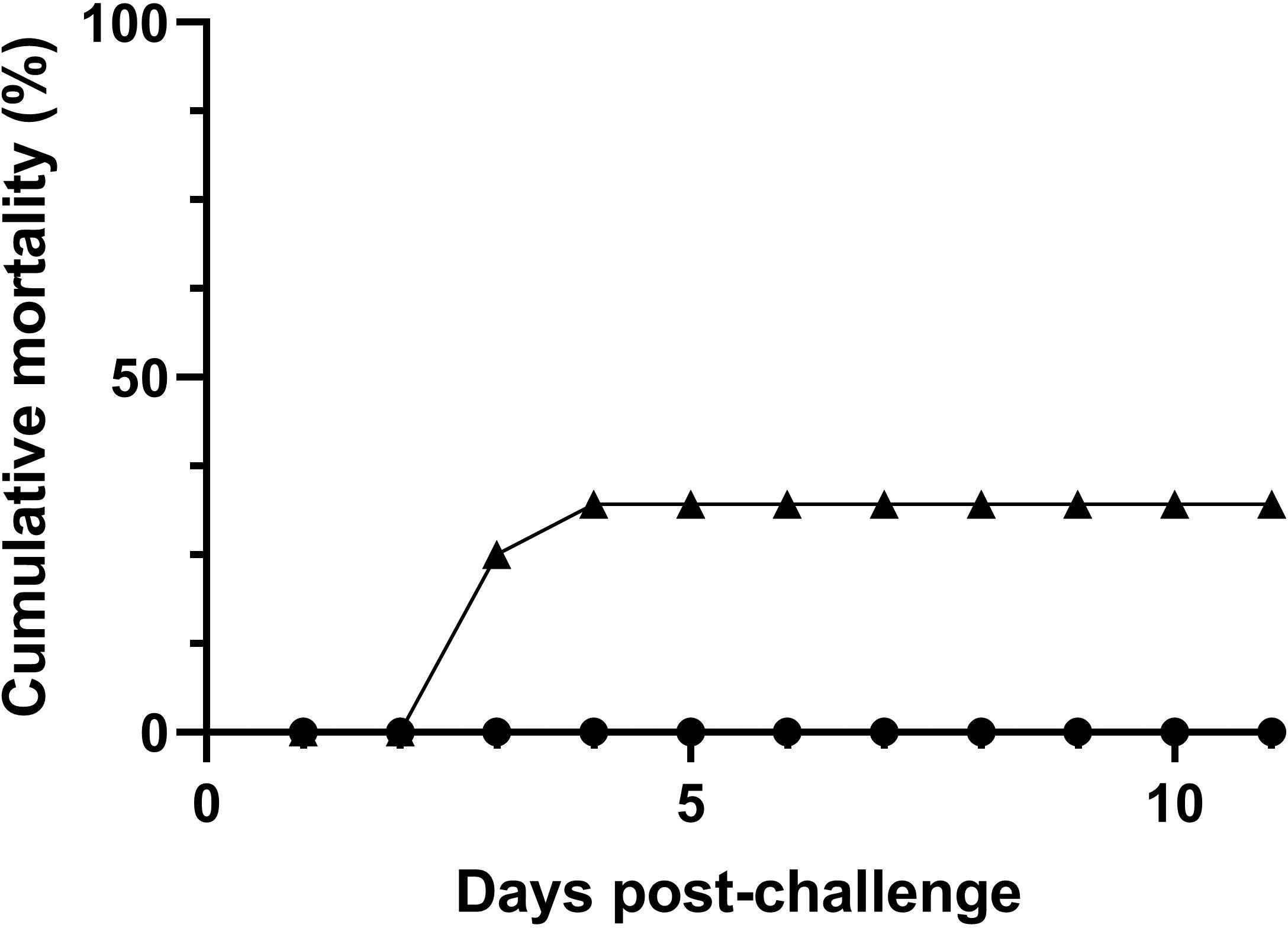
Figure 1 Mortality of European sea bass (Dicentrarchus labrax) after bath-challenge with 5 x 105 CFU mL-1 T. maritimum (▲) or with marine broth MB (●) (n=30 fish per treatment).
Haematological analysis
The concentration of red blood cells suffered a decrease in infected fish at 6, 24 and 48 h post-challenge, returning to a value similar to the control fish (0 h) at 72 h (Supplementary Table 1). In contrast, in mock-challenged fish, a slight decrease in red blood cells was only observed at 48 h post-challenge. Furthermore, the concentrations of red blood cells at 6, 24 and 48 h in challenged fish were lower than in mock-challenged animals (Supplementary Table 1). In agreement with this, haematocrit also decreased at 24 and 48 h post-challenged in infected fish, when compared to control value (0 h), and was lower in infected fish than in mock-challenged fish at all-time points analyzed (Supplementary Table 1). However, haemoglobin did not show any significant difference for the challenged fish (Supplementary Table 1). The mean corpuscular volume was increased at 6 h in infected fish, when compared to controls (0h) and to mock-challenged fish (Supplementary Table 1), whereas for mean corpuscular haemoglobin, an increase was observed in infected fish from 6 to 48 h post-challenge when compared to controls and mock-challenged animals (Supplementary Table 1). Regarding mean corpuscular haemoglobin concentration, an increase at 24 and 48 h post-challenge was observed in infected fish, but the values for the remaining time points were similar between mock and challenged fish (Supplementary Table 1).
The white blood cells’ counts in infected fish at 6, 24 and 48 h did not differ from the control value (0 h), but at 72 h, an increase in white blood cells number was observed (Supplementary Table 1). Likewise, an increase in white blood cells at 72 h was registered in the mock-challenged group (Supplementary Table 1). The differential counts showed that the number of circulating neutrophils increased at 6, 48 and 72 h post-challenge in infected fish, compared to controls (Supplementary Table 2). Infected fish also presented monocytosis at 48 and 72 h, compared to controls and mock-challenged fish (Supplementary Table 2). Lymphocytes’ concentration did not significant differences between groups (despite a wave-like variation on its values), except at 72 h, where an increase was observed for both mock and challenged fish, compared to controls (Supplementary Table 2). Regarding thrombocytes, a decrease was recorded at 6 h in infected fish compared to control and mock-challenged fish (Supplementary Table 2).
Innate humoral parameters
An increase in plasma antiprotease was only observe in infected fish at 48 and 72 h post-challenge (Supplementary Table 3). For plasma protease activity, a peak was reached at 24 h followed by a decrease at 48 h post-challenge for infected sea bass, returning to values similar to those from controls after 72 h (Supplementary Table 3). Although not significant, a similar pattern was observed for mock-challenge fish at 24 h post-challenge (Supplementary Table 3). Plasma peroxidase activity also increased over time reaching a peak at 48 h post-challenged for both mock-challenged and infected fish (Supplementary Table 3), with both groups showing similar patterns of activity. A strong decrease in lysozyme activity was observed in infected fish from 24-72 h post-challenge, with a minimum at 48 h (Supplementary Table 3). A similar tendency to decrease was seen for mock-challenged fish, although the values in infected fish at 24, 48 and 72 h were much lower than in mock-challenged animals. Plasma bactericidal activity decreased in infected sea bass from 6 to 48 h post-challenge, with values lower than the ones obtained for mock-challenged fish (Supplementary Table 3). For plasma NO levels, no significant differences were found between mock and challenged groups, despite an increase was observed at 48 h post-challenge for both groups when compared with control values (Supplementary Table 3).
Oxidative stress biomarkers
Hepatic catalase activity decreased over time until the end of the trial for both mock-challenged and challenged fish, with the two groups presenting similar values for each time point (Supplementary Table 4). Superoxide dismutase activity in liver was significantly higher for infected sea bass compared to control and mock-challenged fish at 6 h returning to basal values at the end of the trial (Supplementary Table 4). For mock-challenged specimens, a peak of activity was reached at 48 h post-challenge (Supplementary Table 4). No differences between time points were observed regarding lipid peroxidation, however, values were significantly lower for infected fish at 6 and 48 h post-challenge compared to mock-challenged group (Supplementary Table 4). Hepatic glutathione-S-transferase levels decreased slightly in challenged fish until the end of the trial, reaching its minimum value at 72 h post-challenge (Supplementary Table 4). Although a general tendency to decrease was also seen for mock-challenged fish, the values the values did not differ much from the basal ones (Supplementary Table 4).
Hepatic reduced glutathione decreased significantly between the control fish and both mock-challenged and infected fish after 72 h (Supplementary Table 4). Oxidized glutathione decreased until 48 h post-challenged in the liver of mock-challenged fish, and returned to basal values at 72 h. No differences were recorded for challenged fish (Supplementary Table 4). Regarding reduced:oxidized glutathione ratio in liver, no differences were recorded for challenged fish, but it was possible to distinguish a wave-like variation with a peak at 48 h post-challenge. The same was seen in the mock-challenge group, with a significantly higher value at that sampling time point, when compared to controls (0h) (Supplementary Table 4).
Histology and immunohistochemistry analyses
The fish sampled before the trial for screening purposes did not display any histopathological changes and the same was recorded for the individuals from the control and mock-challenge groups.
No histopathological changes were observed in the analyzed mucosal organs at 6 h post-challenge for bacteria-challenged fish. Instead, infected fish started to display typical tenacibaculosis symptoms at 24 h post-challenge, with ulcers in different areas of the skin and frayed fins. At 24 h the lesions in the skin of bath-challenged fish showed similar degrees of severity, presenting a considerable number of scattered inflammatory cells in the dermis and hypodermis, with severe necrosis of the dermis and detachment or loss of the epidermis (Figures 2A, B). In the samples of the remaining organs, no evidence of histopathological changes was observed for any of the specimens analyzed through sampling time points. Immunoreactivity was detected only in the skin of infected fish at 24 h and 48 h post-challenge, being mainly distributed across the dermis, revealing an extensive and fast progression of the bacteria; along with necrosis and vacuolization, it was possible to observe the recruitment of inflammatory cells to adjacent areas at 24 h (Figures 2C–F). No immunoreactivity was detected in the remaining organs for any specimens through sampling time points.
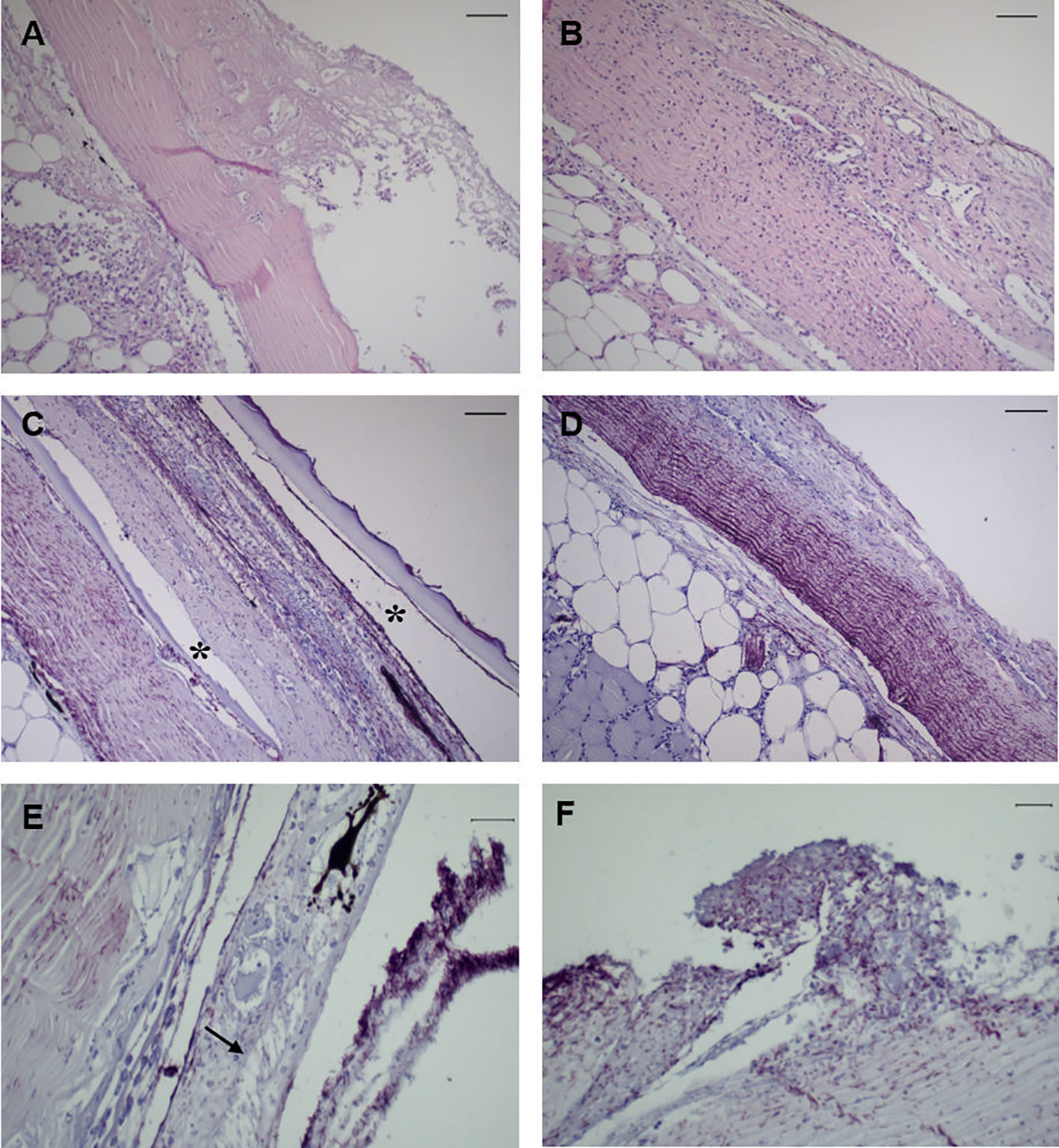
Figure 2 Representative images of skin tissue from European sea bass (Dicentrarchus labrax) bath-challenged with 5 x 105 CFU mL-1 T. maritimum. (A) Heavy infiltration of inflammatory cells in the dermis of challenge fish at 24 h. H–E. Bar 50 μm. (B) Extensive necrosis of the dermis associated with infiltration of inflammatory cells in the hypodermis of challenge fish at 24 h. H–E. Bar 50 μm. (C) Immunohistochemistry against T. maritimum antigen, revealing extensive proliferation of T. maritimum in the dermis of challenged fish at 24 h, with agglomerates of bacteria in the epidermis and scale pockets (*) Bar 50 μm. (D) Necrosis and agglomerates of bacteria in the dermis with infiltration of inflammatory cells in the hypodermis. Bar 50 μm. (E) Vacuolization of epithelial cells from the epidermis of challegend fish at 24 h (arrrow), with agglomerates of T. maritimum in the same area. Bar 50 μm. (F) Proliferation of these bacteria in challenged fish at 48 h post-challenge. Bar 20 μm. Section subjected to immunocytochemistry against T. maritimum antigen.
Gene expression analyses
Gills
Infected sea bass displayed a greater than 23-fold increase in the expression of the pro-inflammatory cytokine il-1β in the gills at 6 h post-challenge. At 24 h the expression was 9-fold higher than the expression in mock-challenged fish and returned to basal values after that sampling point (Figure 3A). A very similar pattern was also seen for il8 and mmp9 transcripts (Figures 3B, C). A high increase of hamp1 expression was observed at 6, 24 and 48 h post-challenge in infected sea bass compared to control and mock-challenged fish, with a 30-fold peak at 24 h (Figure 3D). On the contrary, a slight, albeit significant decrease in fpn expression was noticed in infected specimens at all sampling points compared to control and mock-challenged fish (Figure 3E). The expression of the anti-inflammatory cytokine il-10 did not change in the mock-challenged fish, but was increased at 6 h post-challenge for the infected ones (Figure 3F).
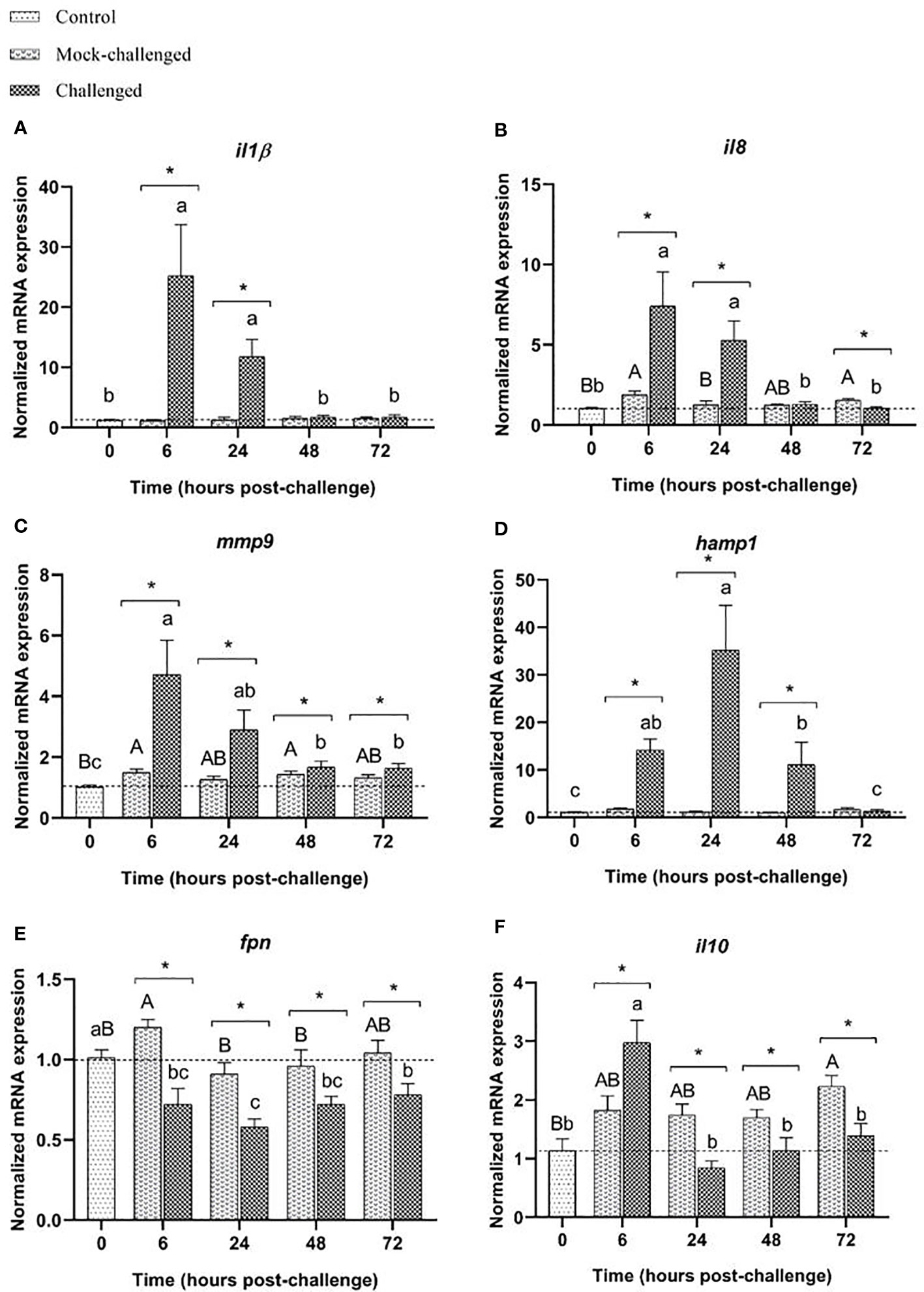
Figure 3 Expression of (A) il-1β, (B) il8, (C) mmp9, (D) hamp1, (E) fpn and (F) il10 in gills of European sea bass (Dicentrarchus labrax) after bacterial bath-challenge with 5 x 105 CFU mL-1 T. maritimum. Data are expressed as mean ± SEM (n=12 per treatment). Different capital letters indicate differences between control and mock-challenge and lower case letters indicate significant differences between control and challenged groups, while (*) represents statistical differences between mock and challenged fish at each sampling point (One-way ANOVA or Kruskal-Wallis; p ≤ 0.05).
The expression of the tlr2, tlr9, nod1 and nod2 receptors in the gills did not change significantly after bacterial exposure when compared to controls (0 h), although a tendency to decreased tlr2 expression was seen at 6 and 24 h post-infection (Supplementary Table 5). The mock-challenged fish showed higher tlr2 expression than the control or challenged fish at all-time points (Supplementary Table 5). Regarding tlr9 transcripts, the different treatment groups presented a similar pattern, with a downregulation at 24 h for infected fish (Supplementary Table 5). For the intracellular receptor nod1, an upregulation was observed at 6 h for challenged fish, returning afterwards to basal expression values (Supplementary Table 5). On the other hand, nod2 suffered a significant downregulation in infected fish compared to control and mock-challenged groups, with the lower expression registered at 24 h post-challenge. Both transcription factors, nf-κB and stat3, presented higher expression values at 6 h post-challenge for infected fish followed by a decrease in the remaining time points (Supplementary Table 5). In the mock-challenged fish, no changes in the nf-κB expression were observed (Supplementary Table 5). The same pattern of expression was seen for stat3 (Supplementary Table 5). Regarding bcl2-like, the expression in mock-challenged fish did not differ from the expression in control fish, but infected sea bass showed decreased expression throughout all sampling points (Supplementary Table 5). Expression of il-6 was downregulated in infected fish at 48 and 72 h post-challenge compared to control and mock-challenged animals (Supplementary Table 5). The mock-challenged fish did not present any major differences for il-6 expression. For the pro-inflammatory cytokine tnfα, the expression values for mock-challenged sea bass increased until the end of the time-course trial reaching its maximum at 72 h post-challenge, whereas a tendency to decrease was observed in the infected fish from 24 h onwards (Supplementary Table 5).
Skin
In what concerns the expression of pro-inflammatory mediators, the skin responded similarly to the gills. An upregulation of il-1β was observed at 6 h post-challenge in infected sea bass, with the higher expression (22-fold increase relative to control) recorded at 24 h (Figure 4A). A similar response was seen for il8 and mmp9 transcripts (Figures 4B, C). The antimicrobial peptide hamp1 registered a significant increase in mRNA levels at 6, 24 and 48 h post-challenge for infected fish, reaching its maximum expression value at 24 h post-challenge with a 90-fold increase compared to mock-challenged fish (Figure 4D). Regarding fpn mRNA expression an opposite pattern was recorded, with a significant downregulation at 6, 24 and 48 h post-challenge for the infected group, returning to basal expression values after 72 h (Figure 4E). The expression of the anti-inflammatory cytokine il-10 showed a significant, albeit moderate increase at 6 h post-challenge for both mock-challenged and infected groups, followed by a decrease at later time points to values similar to the basal level (Figure 4F).
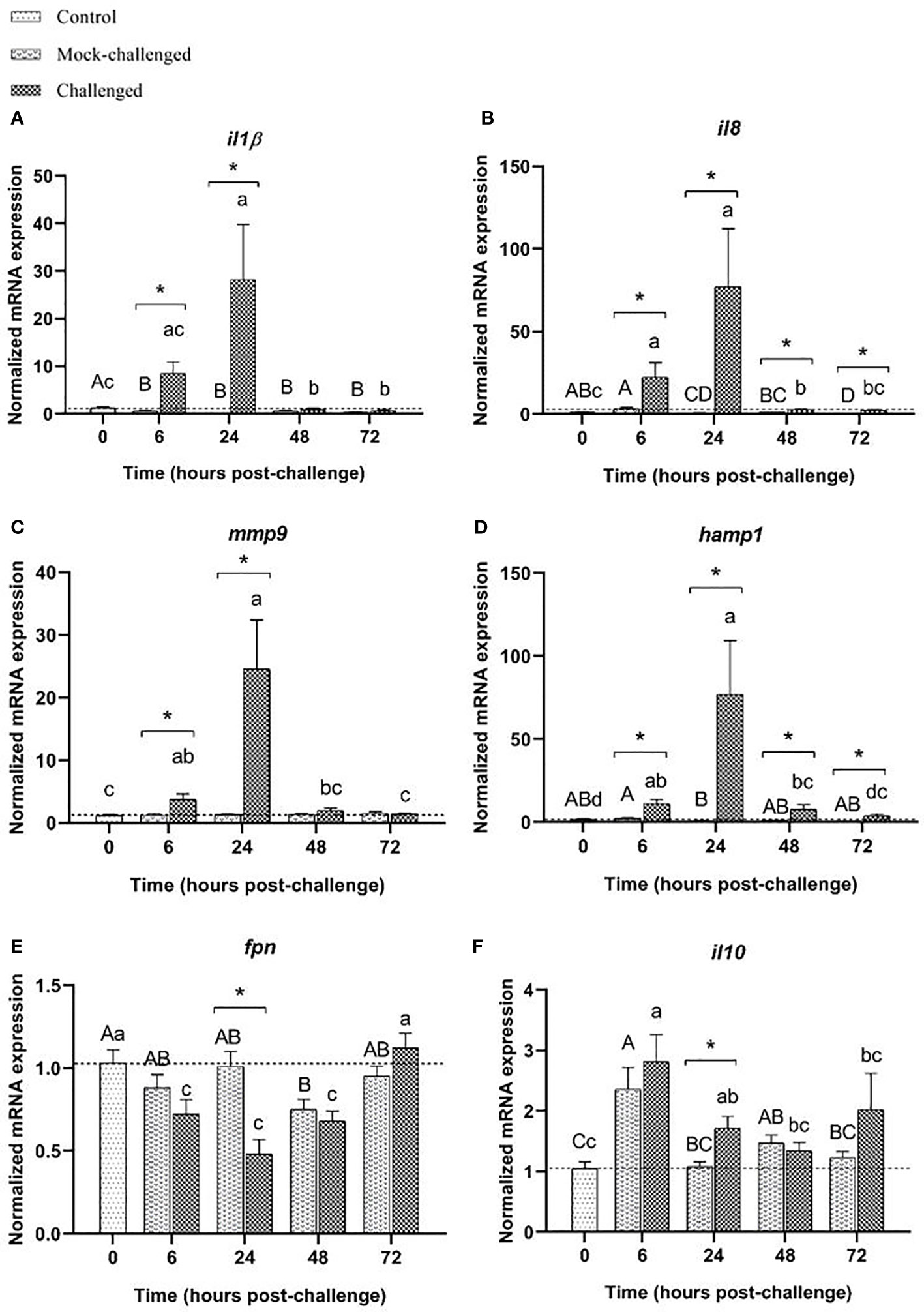
Figure 4 Expression of (A) il-1β, (B) il8, (C) mmp9, (D) hamp1, (E) fpn and (F) il10 in the skin of European sea bass (Dicentrarchus labrax) after bath-challenge with 5 x 105 CFU mL-1 T. maritimum. Data are expressed as mean ± SEM (n=12 per treatment). Different capital letters indicate differences between control and mock-challenge and lower case letters indicate significant differences between control and challenged groups, while (*) represents statistical differences between mock and challenged fish at each sampling point (One-way ANOVA or Kruskal-Wallis; p ≤ 0.05).
As in the gills, no major differences in the expression of the studied cell receptors were observed in the skin of sea bass bath exposed to T. maritimum. Infected fish showed a decrease in tlr2 transcripts at 6 and 24 h compared to the control group, and a higher expression at 72 h (Supplementary Table 6), whereas a tendency to increase was observed in the mock-challenged fish. No differences in the expression of the intracellular cell receptor tlr9 were recorded (Supplementary Table 6). The nod1 expression was slightly down-regulated in the mock-challenge group from 24 onwards. For the bath-challenged fish, nod1 expression was decreased at 48 h and 72 h post-challenge (Supplementary Table 6). Regarding nod2 expression levels, a wave pattern was observed in infected fish, with a significant decrease at 24 h followed by an upregulation at 48 and 72 h when compare to basal levels. In the case of mock-challenged fish, an upregulation of nod2 was seen from 24 h onwards (Supplementary Table 6). The nf-κB mRNA levels were increased at 6 and 24 h infected fish, but decreased at 48 and 72 h to levels lower than the control ones. The mock-challenged fish had lower transcripts throughout the time-course study compared to the controls (Supplementary Table 6). The same mRNA expression pattern was observed for stat3 (Supplementary Table 6). In the case of bcl-2like, a downregulation was seen throughout the time-course, especially at 24 h post-challenge for the infected fish compared to both control and mock-challenged groups. No differences were detected for mock-challenged ones (Supplementary Table 6). Mock-challenged and infected groups presented a similar pattern of il-6 expression, with a significant upregulation at 6 h post-challenge, followed by a decrease in later time-points (Supplementary Table 6). No significant differences in tnfα were recorded in mock-challenged and infected fish, despite a tendency to increase in the infected group (Supplementary Table 6).
Posterior-intestine
A clear increase in the levels of il-1β transcripts was recorded at 6 and 24 h in infected sea bass (46-fold and 126-fold increase, respectively compared to mock-challenged fish), similar to what was observed in the gills and skin (Figure 5A). The same patter occurred for il8 (Figure 5B), and mmp9 (Figure 5C), with an upregulation in infected sea bass at all-time points compared to control and mock-challenged groups. The antimicrobial peptide hamp1 showed marked increase in expression in challenged fish at 6 h (20-fold increase relative to mock fish), slowly decreasing after this time point, despite infected sea bass presented much higher values than the control or mock-challenged groups at all-time points (Figure 5D). While fpn transcripts presented a tendency to be downregulated, especially for infected sea bass at 6 h post-challenge, no significant differences were found (Figure 5E). The cytokine il-10 expression demonstrates a slightly different expression pattern to what was observed for the gills and skin response, with a significant increase at 24 h post-challenge (20-fold increase relative to mock-challenge). At 72 h post-challenge, values from infected fish returned to basal levels, similar to control ones (Figure 5F). Similarly, to what was seen in the gills and skin, no major differences in the expression of the studied cell receptors were observed in the posterior-intestine. Expression of tlr2 presents was slightly decreased in the challenged fish at 6 and 24 h, returning to basal values a 72 h. No differences were seen for mock-challenged group (Supplementary Table 7). Regarding tlr9 expression, no changes were detected in infected specimens when compared to controls, and a minor increase was recorded for mock-challenged fish at all time-points (Supplementary Table 7). For both nod1 and nod2, no major differences in expression were noticed in infected animals. In mock-challenged fish, an increased nod2 expression level was seen at 24 h post-challenge compared to control fish (Supplementary Table 7). Expression of nf-κB decreased from 24 h onwards in infected fish, and was also decreased in mock-challenged fish at all-time points (Supplementary Table 7). No differences in stat3 expression were noticed in challenged and mock-challenged groups, when compared to controls (Supplementary Table 7). The expression of bcl2-like in mock-challenged and infected groups was slightly lower than in control fish at all-time points (Supplementary Table 7). For il-6 mRNA levels no major differences were record (Supplementary Table 7). Regarding tnfα, no differences were recorded for mock-challenged fish, but a decreased expression was observed at 24 h post-challenge for infected fish, compared to control and mock-challenged groups (Supplementary Table 7).
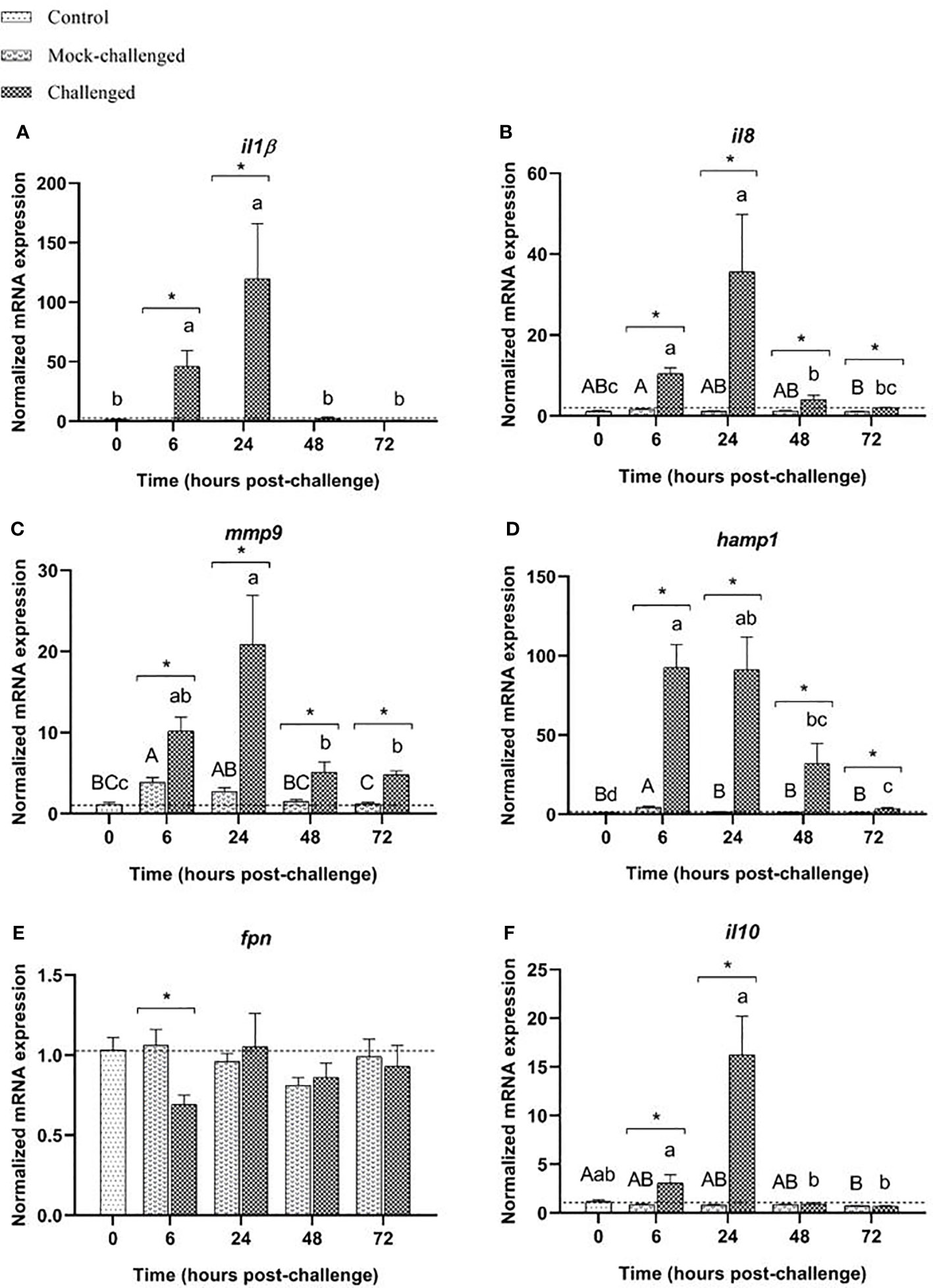
Figure 5 Expression of (A) il-1β, (B) il8, (C) mmp9, (D) hamp1, (E) fpn and (F) il10 in posterior-intestine of European sea bass (Dicentrarchus labrax) after bacterial bath-challenge with 5 x 105 CFU mL-1 T. maritimum. Data are expressed as mean ± SEM (n=12 per treatment). Different capital letters indicate differences between control and mock-challenge and lower case letters indicate significant differences between control and challenged groups, while (*) represents statistical differences between mock and challenged fish at each sampling point (One-way ANOVA or Kruskal-Wallis; p ≤ 0.05).
Discussion
With its ubiquitous distribution in marine environments, T. maritimum restrains the rearing of numerous fish species (46) and is considered one of the most threating bacterial infections (47) for aquaculture. However, available knowledge regarding T. maritimum pathogenesis is very limited. The present study evaluated cellular, humoral, oxidative and molecular short-term responses of European sea bass following T. maritimum bath challenge, providing an insight of the host’s responses against this particular bacterium.
In 1990s a few pathogenicity studies were developed with commercial fish species, which included European sea bass (48), Atlantic salmon (49), rainbow trout, and greenback flounder (Rhombosolea tapirina) (50), resulting in distinctive rates of cumulative mortality. Initially, prolonged immersion challenges of 18 h proved to be effective in reproducing tenacibaculosis in turbot (51). More recently, Mabrok et al. (25) also used a prolonged bath of 24 h at 23°C to successfully challenge Senegalese sole with different strains of T. maritimum (ACC6.1, ACC13.1 and ACC20.1), resulting in cumulative mortalities ranging from 50% to 100%. In this same study, the ACC13.1 T. maritimum strain (the same used in the present study) (9.6 × 105 cells mL−1) lead to cumulative mortalities of approximately 50% in Senegalese sole. In the last years, immersion challenge has been frequently used as a reliable method to experimentally reproduce tenacibaculosis in fish (52–57). In the present study, a mortality rate of 32.1% was obtained for the bath-challenged fish, while displaying tenacibaculosis clinical signs (e.g. ulcerative lesions in the skin and caudal fins, with haemorrhages, loose scales and abrasions). Since this challenge model was also able to successfully induce tenacibaculosis clinical signs and mortality in the bacteria exposed fish, it is suggested that immersion challenge (with a 2 h period of bacterial exposure) is an effective method to experimentally reproduce this disease in European sea bass. Similar mortality traits were also observed in previous studies following the same immersion challenge procedure (56, 57).
Fish bath-challenged with T. maritimum presented a moderate decrease in total RBCs counts and a decreased haematocrit, suggesting the occurrence of anemia in response to infection. Further studies are required to elucidate if the observed anemia results from bacterial-induced destruction of RBCs or to an insufficient supply of healthy RBCs. The complete genome sequence of T. maritimum was able to offer some insights about virulence/associated genes which encode the biosynthesis of hemolysins (19). The secretion of these hemolysins may be a possible explanation for the decrease of both RBCs and haematocrit in fish exposed to T. maritimum. However, these haematological parameters can also suggest anemia of inflammation, a host’s off-target strategy, where erythrocyte lifespan is shortened by activating macrophages allowing the sequestration of iron from serum by these cells (58, 59).
Analysis of the MCH showed that it increased after bacterial challenge, revealing a higher amount of hemoglobin inside the RBCs, also supported by a slight increase in MCHC. As previously mentioned, the presence of T. maritimum hemolysins may lead to RBCs lysis, which can lead to increased RBC production secondary to peripheral blood cell destruction, with the formed cells carrying more hemoglobin than normal-sized cells (60). Moreover, these incompletely processed RBCs, are slightly larger than the average RBC, increasing theses red cell indices (61).
Neutrophils are responsible to assemble an early and potent antimicrobial response against invading pathogens, being the first leukocytes to be recruited to inflammatory sites (62, 63). Even though blood total leucocyte numbers did not change in response to bath-challenge with T. maritimum, there was an increase in neutrophils at 48 and 72 h, suggesting that they are involved in the response to T. maritimum infection. These data are in agreement with the observed increased expression of the pro-inflammatory cytokines at the mucosal organs already at 6 h following infection. IL-8 has a potent chemotropic activity for neutrophils, monocytes, basophils and other immune cells (64), and several studies have demonstrated that increased expression of il-8 is related to acute inflammatory responses in teleosts upon infection with different bacterial species (65–68). Therefore, it is likely that the upregulation of il-8 expression in the mucosal organs of infected fish is related with the infiltration of inflammatory cells in the skin lesions, detected at 24 h post-challenge. The results obtained in the current study are in agreement with previous results obtained by Guardiola et al. (22) in Senegalese sole, which revealed a significant increase of neutrophils at 48 and 72 h after bath-challenge with T. maritimum (22), as well as with results reported in other studies for several bacterial fish pathogens (69–71).
Moreover, besides being a key mediator of the immune system, neutrophils are also the main responsible for the production of myeloperoxidase in the plasma (72). Therefore, the increase of peroxidase in the plasma of infected specimens from the present study can be mostly explained by the neutrophilia observed in bath-challenged fish.
During inflammation, circulating monocytes migrate to infection sites, following conditioning by pro-inflammatory cytokines, microbial products and local growth factors, differentiating afterwards into macrophages and dendritic cells (73). The increased number of monocytes at 72 h post-challenge may indicate a host’s attempt to increase the number of monocytes and their macrophage and dendritic-cell progeny, to fight against invading pathogens through phagocytosis, assist in the repair/regenerate of the damaged tissue, resolve the inflammation as well as to stablish the link with adaptive immunity by antigen presentation (74–76).
Studies have demonstrated that thrombocytes are involved in innate immune and inflammatory responses in fish, participating in phagocytic activities and in the killing of internalized bacteria (77–79). In the present study, the successive increase in circulating thrombocytes in bath-challenged fish up to 24 h following infection, allows to hypothesized that these cells are also migrating to infection sites. Since the teleost adaptive immune system implicates slow proliferation and maturation of lymphocytes (80, 81), their role in this time frame may not be as relevant as the other immune cells, as seen by the lack of variation during the first 72 h post-challenge.
Plasma lysozyme activity decreased in infected fish, whereas bactericidal activity in the plasma only started to increase upon 48 h following bacterial bath challenge, which can be related with the late influx of phagocytes (e.g. neutrophils and monocytes) (82) at the end of the time-course study. Many Gram-negative bacteria are able to produce lysozyme inhibitors that can significantly inhibited/decrease lysozyme activity in the host serum beginning from the early stages of host infection (83). Therefore, more studies would be needed to understand if T. maritimum could have similar evading mechanisms.
Pathogenic bacteria must be able to adapt to unpredictable environments and to cope with diverse stress-inducing factors, such as reactive oxygen species (ROS) produced by the host’s macrophages (19). T. maritimum’s genome encodes three different superoxide dismutases (SodA, SodB, and SodC) and two catalases/peroxidase (KatA and KatG), which may imply that these bacteria use a complex mechanism to fight oxidative stress (19). Therefore, it is tempting to speculate that T. maritimum is able to use these enzymes to cope with and to modulate the host’s immune response, resulting in the lack of changes regarding the analyzed oxidative stress parameters.
In order to recognize bacteria, the host’s immune system rely on pattern recognition receptors (PRRs), which are able to bind and recognize different pathogen-associated molecular patterns (PAMPs) and activate immune cells (84, 85).
Although no major changes in the expression of the studied PRRs and transcription factors in response to T. maritimum infection were detected in the present study, an immune response was indeed developed in bath-challenged fish, as a clear pro-inflammatory response was observed across all mucosal organs analyzed.
Several studies revealed that, when challenged with pathogenic bacteria, teleosts upregulate the expression of il-1β as initiation of the non-specific inflammatory response (66, 86, 87), with similar expression kinetics to the present study. The same upregulation is seen for mmp9, which increased expression was already linked to immune response against Listeria monocytogenes in infected zebrafish (88), Flavobacterium psychrophilum in rainbow trout (Oncorhynchus mykiss) (89), Aeromonas hydrophila in yellow catfish (Pelteobagrus fulvidraco) (90), and in peritoneal and peripheral blood leucocytes of stimulated common carp (Cyprinus carpio) (91). The skin and posterior-intestine of challenged fish responded quite similarly, with il-1β, il8 and mmp9 as the most highly induced genes, which provides evidence that an inflammatory response is activated upon infection with T. maritimum. The increase in il-1β expression in these mucosal organs may result in increased mucus secretion (92, 93), which could be advantageous during a T. maritimum’s infection. The increased expression of il8 and mmp9 can also be intertwined with il-1β expression, since this cytokine is able to promote the release of other cytokines and activate macrophages and other immune cells (82, 94). Not only T. maritimum was able to trigger a pro-inflammatory response in the host, but also modulated the expression of genes related to iron metabolism regulation. The response of mucosal organs may suggest that one of the mechanisms employed by the host to withstand T. maritimum is associated with hepcidin, a small antimicrobial peptide that is involved in iron metabolism regulation in mammals (95–97). The iron control in the extracellular environment is a known innate immune strategy developed to deprive pathogens of iron, an essential nutrient for bacterial growth, replication, and metabolic processes (98). This strategy, as a response to inflammatory stimuli, leads to high circulating levels of hepcidin, which in turn, negatively regulate the iron concentration in plasma (99) through occlusion of the open-outward conformation (100) or by internalization and degradation of ferroportin, an iron exporter (101, 102). Although there was no clear activation of the pro-inflammatory cytokine IL-6, the main hepcidin inducer (103), other experiments already revealed that hepcidin can also be induced by IL-1β (104, 105). Therefore, it is possible that in this study, hepcidin regulation was due to the inflammatory signals conducted by IL-1β, with the activation of NF-κB and JNK signaling pathways (105), leading to the seen transcriptional hamp induction and to the downregulation of fpn. The previously described mild anaemic condition of challenged fish, that can be referred to as anemia of inflammation, could have some repercussions in the expression levels of hepcidin, since in an anaemic situation, hepcidin synthesis is suppressed (97, 106, 107). However, this type of response was also recorded by 108, where Photobacterium damselae spp. piscicida challenged fish demonstrated that hepcidin responds to infection by increasing its expression levels in sea bass liver, despite the anaemia demonstrated by the infected fish. This hepcidin dynamic was already described for other fish species (109–112). These results are in accordance with other studies that also demonstrated the regulation of hepcidin and ferroportin by inflammatory signals induced by a pathogen (108, 113–116).
Although it is known that IL-1β is typically activated in situations where TNF-α is produced, no changes were detected in its expression after challenging sea bass with T. maritimum (117, 118). In a study developed by Nascimento et al. (119), a TNF-α up-regulation was briefly observed at 12 h post-infection with Photobacterium damselae subsp. piscicida in the head-kidney of European sea bass (119). Another study with Streptococcus iniae, presented the same results as the previous one for the head-kidney of European sea bass (120). Also, tnfα was significantly increased from 6 to 9 h post-infection in the head-kidney of European sea bass when intramuscularly infected with Betanodavirus (RGNNV), showing an early response of this gene (121).
Since tnf-α and il-6 did not show any major differences in their expression, an earlier sampling time point could be valuable to add information about their possible upregulation in a response against T. maritimum.
Due to its ubiquitous distribution and lack of host specificity, T. maritimum continuously inflicts significant losses among cultured marine species, as confirmed by the re-emerging nature of tenacibaculosis outbreaks in salmon farms globally (122). Although some progress has been made in the last decade regarding its pathogenic mechanisms, defining the host-pathogen relationship has proved to be very difficult to achieve. The present study offers a new insight regarding the mucosal innate immune response upon a pathogen inoculation pathway that mimics natural infection dynamics. In summary, the kinetics of the expression of molecular immune markers in gill, skin and posterior intestine of bath-challenged fish together with the findings observed for peripheral leucocytes demonstrate the occurrence of a pro-inflammatory response against T. maritimum in the studied mucosal organs, with a faster kinetic in the gills, which may suggest that this pathogen can use gill mucosa as a route of entry into the fish. The analysis of the humoral parameters suggests that the local response at the mucosal organs is followed by a response at systemic level.
Data availability statement
The data presented in the study are deposited in the “Figshare” repository, DOI 10.6084/m9.figshare.23998017.
Ethics statement
The animal study was reviewed and approved by 0421/000/000/2020. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
IF: Formal Analysis, Methodology, Writing – original draft. DP: Methodology, Writing – review & editing. AL: Methodology, Writing – review & editing. MQ: Supervision, Writing – review & editing. AV: Conceptualization, Supervision, Writing – review & editing. BC: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is supported by national funds through FCT - Foundation for Science and Technology - within the scope of UIDB/04423/2020 and UIDP/04423/2020. IF, DP, AdV, and BC benefited from grants by FCT (SFRH/BD/147750/2019, UI/BD/150900/2021, L57/2016/CP1355/CT0010 and 2020.00290.CEECIND). AdV was funded by Portuguese national funds through the FCT–Fundação para a Ciência e a Tecnologia, I.P., and, when eligible, by COMPETE 2020 FEDER funds, under the Scientific Employment Stimulus–Individual Call 2021.02251.CEECIND/CP1663/CT0016. This work also received funding from the European Union’s Horizon Europe research and innovation program under grant agreement No. 101084651 (project IGNITION).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
Views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union. The European Union cannot be held responsible for them.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1254677/full#supplementary-material
References
1. Ahmed N, Thompson S. The blue dimensions of aquaculture: A global synthesis. Sci Total Environ (2019) 652:851–61. doi: 10.1016/J.SCITOTENV.2018.10.163
2. Peeler EJ, Oidtmann BC, Midtlyng PJ, Miossec L, Gozlan RE. Non-native aquatic animals introductions have driven disease emergence in Europe. Biol Invasions (2011) 13(6):1291–303. doi: 10.1007/S10530-010-9890-9
3. Krkošek M. Host density thresholds and disease control for fisheries and aquaculture. Aquaculture Environ Interact (2010) 1(1):21–32. doi: 10.3354/AEI0004
4. Salama NKG, Murray AG. Farm size as a factor in hydrodynamic transmission of pathogens in aquaculture fish production. Aquaculture Environ Interact (2011) 2(1):61–74. doi: 10.3354/AEI00030
5. Fernández-Álvarez C, Santos Y. Identification and typing of fish pathogenic species of the genus Tenacibaculum. Appl Microbiol Biotechnol (2018) 102(23):9973–89. doi: 10.1007/S00253-018-9370-1
6. Avendaño-Herrera R, Collarte C, Saldarriaga-Córdoba M, Irgang R. New salmonid hosts for Tenacibaculum species: Expansion of tenacibaculosis in Chilean aquaculture. J Fish Dis (2020) 43(9):1077–85. doi: 10.1111/JFD.13213
7. Flores-Kossack C, Montero R, Köllner B, Maisey K. Chilean aquaculture and the new challenges: Pathogens, immune response, vaccination and fish diversification. Fish Shellfish Immunol (2020) 98:52–67. doi: 10.1016/J.FSI.2019.12.093
8. Masumura K, Wakabayashi H. An Outbreak of Gliding Bacterial Disease in Hatchery-born Red Seabream (Pagrus major) and Gilthead (Acanthopagrus schlegeli) Fry in Hiroshima. Fish Pathol (1977) 12(3):171–7. doi: 10.3147/JSFP.12.171
9. Pepin J-F, Emery E. Marine cytophaga-like bacteria (CLB) isolated from diseased reared sea bass (Dicentrarchus labrax L.) from French mediterranean coast. Bull Eur Assoc Fish Pathologists (1993) 13(5):165–7. Available at: https://archimer.ifremer.fr/doc/00271/38252/.
10. Salati F, Cubadda C, Viale I, Kusuda R. Immune response of sea bass Dicentrarchus labrax to Tenacibaculum maritimum antigens. Fisheries Sci (2005) 71(3):563–7. doi: 10.1111/J.1444-2906.2005.01000.X
11. Kolygas MN, Gourzioti E, Vatsos IN, Athanassopoulou F. Identification of Tenacibaculum maritimum strains from marine farmed fish in Greece. Veterinary Rec (2012) 170(24):623. doi: 10.1136/VR.100778
12. Yardımcı ER, Timur G. Isolation and identification of Tenacibaculum maritimum, the causative agent of tenacibaculosis in farmed sea bass (Dicentrarchus labrax) on the aegean sea coast of Turkey. Israeli J Aquaculture - Bamidgeh (2015) 67. doi: 10.46989/001C.20708
13. Van Gelderen R, Carson J, Nowak B. Experimentally induced marine flexibacteriosis in Atlantic salmon smolts Salmo salar. II. Pathology. Dis Aquat Organisms (2011) 95(2):125–35. doi: 10.3354/DAO02329
14. Haridy M, Hasheim M, Abdelgalil Ma, Sakai H, Yanai T. Pathological Findings of Tenacibaculum maritimus Infection in Black Damselfish, Neoglyphieodon melas and Picasso Triggerfish, Rhinecanthus assasi in Red Sea, Egypt. J Veterinary Sci Technol (2014) 06(02). doi: 10.4172/2157-7579.1000214
15. Lopez P, Saulnier D, Swarup-Gaucher S, David R, Lau C, Taputuarai R, et al. First isolation of virulent Tenacibaculum maritimum strains from diseased orbicular batfish (Platax orbicularis) farmed in Tahiti Island. BioRxiv (2021) 11:131. doi: 10.1101/2021.03.15.435441
16. Avendaño-Herrera R, Toranzo AE, Magariños B. Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Dis Aquat Organisms (2006) 71(3):255–66. doi: 10.3354/DAO071255
17. Handlinger J, Soltani M, Percival S. The pathology of Flexibacter maritimus in aquaculture species in Tasmania, Australia. J Fish Dis (1997) 20(3):159–68. doi: 10.1046/J.1365-2761.1997.00288.X
18. Magarinos B, Pazos F, Santos Y, Romalde JL, Toranzo AE. Response of Pasteurella piscicida and Flexibacter maritimus to skinmucus of marine fish. Dis Aquat Organisms (1995) 21(2):103–8. doi: 10.3354/DAO021103
19. Pérez-Pascual D, Lunazzi A, Magdelenat G, Rouy Z, Roulet A, Lopez-Roques C, et al. The complete genome sequence of the fish pathogen Tenacibaculum maritimum provides insights into virulence mechanisms. Front Microbiol (2017) 8:1542/BIBTEX. doi: 10.3389/FMICB.2017.01542/BIBTEX
20. Pazos F. Flexibacter Maritimus: Estudio Fenotípico, Inmunológico y Molecular. University of Santiago de Compostela, Santiago (1997). Ph.D. thesis.
21. Avendaño-Herrera R, Toranzo AE, Romalde JL, Lemos ML, Magariños B. Iron uptake mechanisms in the fish pathogen Tenacibaculum maritimum. Appl Environ Microbiol (2005) 71(11):6947–53. doi: 10.1128/AEM.71.11.6947-6953.2005/ASSET/E6233A18-7B60-4272-80AA-6B3A844A8CD7/ASSETS/GRAPHIC/ZAM0110560100004.JPEG
22. Guardiola FA, Mabrok M, MaChado M, Azeredo R, Afonso A, Esteban MA, et al. Mucosal and systemic immune responses in Senegalese sole (Solea Senegalensis Kaup) bath challenged with Tenacibaculum maritimum: A time-course study. Fish Shellfish Immunol (2019) 87:744–54. doi: 10.1016/J.FSI.2019.02.015
23. Faílde LD, Losada AP, Bermúdez R, Santos Y, Quiroga MI. Evaluation of immune response in turbot (Psetta maxima L.) tenacibaculosis: Haematological and immunohistochemical studies. Microbial Pathogenesis (2014) 76:1–9. doi: 10.1016/J.MICPATH.2014.08.008
24. Avendaño-Herrera R, Magariños B, Moriñigo MA, Romalde JL, Toranzo AE. A novel O-serotype in Tenacibaculum maritimum strains isolated from cultured sole (Solea Senegalensis). Bull Eur Ass. Fish Pathol (2005) 25(2):70.
25. Mabrok A. The host/pathogen interaction during experimental infection of Senegalese sole (Solea Senegalensis) by Tenacibaculum maritimum. J Fish Diseases (2016). doi: 10.1111/jfd.12483
26. Yamamoto T, Kawai K, Oshima S. Evaluation of an Experimental Immersion Infection Method with Tenacibaculum maritimum in Japanese Flounder Paralichthys olivaceus. Aquaculture Sci (2010) 58(4):481–9. doi: 10.11233/AQUACULTURESCI.58.481
27. Avendaño-Herrera R, Núnez S, Magarinos B, Toranzo AE. A non-destructive method for rapid detection of Tenacibaculum maritimum in farmed fish using nested PCR amplification. Bull Eur Assoc Fish Pathologists (2004) 24(6):280–6.
28. MaChado M, Azeredo R, Díaz-Rosales P, Afonso A, Peres H, Oliva-Teles A, et al. Dietary tryptophan and methionine as modulators of European seabass (Dicentrarchus labrax) immune status and inflammatory response. Fish Shellfish Immunol (2015) 42(2):353–62. doi: 10.1016/J.FSI.2014.11.024
29. Afonso A, Silva J, Lousada S, Ellis AE, Silva MT. Uptake of neutrophils and neutrophilic components by macrophages in the inflamed peritoneal cavity of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol (1998) 8(5):319–38. doi: 10.1006/FSIM.1998.0139
30. Ellis AE. Serum antiproteases in fish. In: Stolen W. B. V. M. F. T., Anderson DP, Roberson BS, editors. Techniques in Fish Immunology. Fair Haven, NJ: SOS Publications (1990).
31. Quade MJ, Roth JA. A rapid, direct assay to measure degranulation of bovine neutrophil primary granules. Veterinary Immunol Immunopathology (1997) 3–4(58):239–48. doi: 10.1016/s0165-2427(97)00048-2
32. Costas B, Conceição LEC, Dias J, Novoa B, Figueras A, Afonso A. Dietary arginine and repeated handling increase disease resistance and modulate innate immune mechanisms of Senegalese sole (Solea Senegalensis Kaup 1858). Fish Shellfish Immunol (2011) 31(6):838–47. doi: 10.1016/J.FSI.2011.07.024
33. Graham S, JEFFRIES AH, SECOMBES CJ. A novel assay to detect macrophage bactericidal activity in fish: factors influencing the killing of Aeromonas salmonicida. J Fish Dis (1988) 11(5):389–96. doi: 10.1111/J.1365-2761.1988.TB00734.X
34. Saeij JPJ, Verburg-Van Kemenade LBM, Van Muiswinkel WB, Wiegertjes GF. Daily handling stress reduces resistance of carp to Trypanoplasma borreli: In vitro modulatory effects of cortisol on leukocyte function and apoptosis. Dev Comp Immunol (2003) 27(3):233–45. doi: 10.1016/S0145-305X(02)00093-9
35. Bird RP, Draper HH. [35] comparative studies on different methods of malonaldehyde determination. Methods Enzymology (1984) 105(C):299–305. doi: 10.1016/S0076-6879(84)05038-2
36. Peixoto D, Pinto W, Gonçalves AT, MaChado M, Reis B, Silva J, et al. Microalgal biomasses have potential as ingredients in microdiets for Senegalese sole (Solea Senegalensis) post-larvae. J Appl Phycology (2021) 33(4):2241–50. doi: 10.1007/S10811-021-02431-1/FIGURES/2
37. Claiborne A. (1985). Catalase activity. In: Greenwald R. A., Ed., CRC Handbook of Methods for Oxygen Radical Research, CRC Press, Boca Raton, 283–4. Available at: https://www.scirp.org/(S(351jmbntvnsjt1aadkposzje))/reference/ReferencesPapers.aspx?ReferenceID=796054.
38. Rodrigues ACM, Gravato C, Quintaneiro C, Bordalo MD, Barata C, Soares AMVM, et al. Energetic costs and biochemical biomarkers associated with esfenvalerate exposure in Sericostoma vittatum. Chemosphere (2017) 189:445–53. doi: 10.1016/J.CHEMOSPHERE.2017.09.057
39. Almeida JR, Oliveira C, Gravato C, Guilhermino L. Linking behavioural alterations with biomarkers responses in the european seabass Dicentrarchus labrax l. exposed to the organophosphate pesticide fenitrothion. Ecotoxicology (2010) 19(8):1369–81. doi: 10.1007/S10646-010-0523-Y
40. Habig WH, Pabst MJ, Jakoby WB. Glutathione S-Transferases,the first enzymatic step in mercapturic acid formation. J Biol Chem (1974) 249(22):7130–9. doi: 10.1016/S0021-9258(19)42083-8
41. Frasco MF, Guilhermino L. Effects of dimethoate and beta-naphthoflavone on selected biomarkers of Poecilia reticulata. Fish Physiol Biochem (2002) 26(2):149–56. doi: 10.1023/A:1025457831923
42. Hamre K, Penglase SJ, Rasinger JD, Skjærven KH, Olsvik PA. Ontogeny of redox regulation in Atlantic cod (Gadus morhua) larvae. Free Radical Biol Med (2014) 73:337–48. doi: 10.1016/J.FREERADBIOMED.2014.05.017
43. Tietze F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: Applications to mamMalian blood and other tissues. Analytical Biochem (1969) 27(3):502–22. doi: 10.1016/0003-2697(69)90064-5
44. Kuhl H, Beck A, Wozniak G, Canario AVM, Volckaert FAM, Reinhardt R. The European sea bass Dicentrarchus labrax genome puzzle: Comparative BAC-mapping and low coverage shotgun sequencing. BMC Genomics (2010) 11(1):1–13. doi: 10.1186/1471-2164-11-68/FIGURES/5
45. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res (2001) 29(9):E45. doi: 10.1093/NAR/29.9.E45
46. Toranzo AE, Magariños B, Romalde JL. A review of the main bacterial fish diseases in mariculture systems. Aquaculture (2005) 246(1–4):37–61. doi: 10.1016/J.AQUACULTURE.2005.01.002
47. Cascarano MC, Stavrakidis-Zachou O, Mladineo I, Thompson KD, Papandroulakis N, Katharios P. Mediterranean aquaculture in a changing climate: Temperature effects on pathogens and diseases of three farmed fish species. Pathogens (2021) 10(9). doi: 10.3390/PATHOGENS10091205
48. Bernardet JF, Kerouault B, Michel C. Comparative Study on Flexibacter maritimus Strains Isolated from Farmed Sea Bass (Dicentrarchus labrax) in France. Fish Pathol (1994) 29(2):105–11. doi: 10.3147/JSFP.29.105
49. Powell M, Carson J, Van Gelderen R. Experimental induction of gill disease in Atlantic salmon Salmo salar smolts with Tenacibaculum maritimum. Dis Aquat Organisms (2004) 61(3):179–85. doi: 10.3354/DAO061179
50. Soltani M, Munday BL, Burke CM. The relative susceptibility of fish to infections by Flexibacter columnaris and Flexibacter maritimus. Aquaculture (1996) 140(3):259–64. doi: 10.1016/0044-8486(95)01157-9
51. Avendaño-Herrera R, Toranzo AE, Magariños B. A challenge model for Tenacibaculum maritimum infection in turbot, Scophthalmus maximus (L.). J Fish Dis (2006) 29(6):371–4. doi: 10.1111/J.1365-2761.2006.00712.X
52. López JR, Núñez S, Magariños B, Castro N, Navas JI, de la Herran R, et al. First isolation of Tenacibaculum maritimum from wedge sole, Dicologoglossa cuneata (moreau). J Fish Dis (2009) 32(7):603–10. doi: 10.1111/J.1365-2761.2009.01029.X
53. Nishioka T, Watanabe KI, Sano M. A bath challenge method with Tenacibaculum maritimum for Japanese flounder Paralichthys olivaceus. Fish Pathol (2009) 44(4):178–81. doi: 10.3147/JSFP.44.178
54. Frisch K, Småge SB, Vallestad C, Duesund H, Brevik J, Klevan A, et al. Experimental induction of mouthrot in Atlantic salmon smolts using Tenacibaculum maritimum from Western Canada. J Fish Dis (2018) 41(8):1247–58. doi: 10.1111/JFD.12818
55. Valdes S, Irgang R, Barros MC, Ilardi P, Saldarriaga-Córdoba M, Rivera–Bohle J, et al. First report and characterization of Tenacibaculum maritimum isolates recovered from rainbow trout (Oncorhynchus mykiss) farmed in Chile. J Fish Dis (2021) 44(10):1481–90. doi: 10.1111/JFD.13466
56. Resende D, Costas B, Sá T, Golfetto U, MaChado M, Pereira M, et al. Innovative swine blood hydrolysates as promising ingredients for European seabass diets: Impact on growth performance and resistance to Tenacibaculum maritimum infection. Aquaculture (2022) 561:738657. doi: 10.1016/J.AQUACULTURE.2022.738657
57. Ferreira M, MaChado M, Mota CSC, Abreu H, Silva J, Maia MRG, et al. Micro- and macroalgae blend modulates the mucosal and systemic immune responses of European seabass (Dicentrarchus labrax) upon infection with Tenacibaculum maritimum. Aquaculture (2023) 566:739222. doi: 10.1016/J.AQUACULTURE.2022.739222
58. Ganz T. Iron and infection. Int J Hematol (2018) 107(1):7–15. doi: 10.1007/S12185-017-2366-2/FIGURES/3
59. Díaz R, Troncoso J, Jakob E, Skugor S. Limiting access to iron decreases infection of Atlantic salmon SHK-1 cells with bacterium Piscirickettsia salmonis. BMC Veterinary Res (2021) 17(1). doi: 10.1186/S12917-021-02853-6
61. Borges MD, Sesti-Costa R. Macrophages: key players in erythrocyte turnover. Hematology Transfusion Cell Ther (2022) 44(4):574–81. doi: 10.1016/J.HTCT.2022.07.002
62. Havixbeck JJ, Barreda DR. Neutrophil development, migration, and function in teleost fish. Biology (2015) 4(4):715–34. doi: 10.3390/BIOLOGY4040715
63. Havixbeck JJ, Rieger AM, Wong ME, Hodgkinson JW, Barreda DR. Neutrophil contributions to the induction and regulation of the acute inflammatory response in teleost fish. J Leukocyte Biol (2016) 99(2):241–52. doi: 10.1189/JLB.3HI0215-064R
64. Remick DG. Interleukin-8. Critical Care Medicine (2005) 33(12 Suppl), S466–S467. doi: 10.1097/01.ccm.0000186783.34908.18
65. Chen N, Jiang J, Gao X, Li X, Zhang Y, Liu X, et al. Histopathological analysis and the immune related gene expression profiles of mandarin fish (Siniperca chuatsi) infected with Aeromonas hydrophila. Fish & Shellfish Immunol (2018) 83:410–415. doi: 10.1016/j.fsi.2018.09.023
66. Mohanty BR, Sahoo PK. Immune responses and expression profiles of some immune-related genes in Indian major carp, Labeo rohita to Edwardsiella tarda infection. Fish Shellfish Immunol (2010) 28(4):613–21. doi: 10.1016/J.FSI.2009.12.025
67. Nguyen TTT, Nguyen HT, Wang PC, Chen SC. Identification and expression analysis of two pro-inflammatory cytokines, TNF-α and IL-8, in cobia (Rachycentron canadum L.) in response to Streptococcus dysgalactiae infection. Fish & shellfish Immunol (2017) 67:159–171. doi: 10.1016/j.fsi.2017.06.014
68. Wang GL, Wang MC, Zhang XW, Chang MX, Xie HX, Nie P. (2017). Molecular cloning, biological effect, and tissue distribution of interleukin-8 protein in mandarin fish (Siniperca chuasti) upon Flavobacterium columnare infection. Fish & Shellfish Immunol 66:112–119. doi: 10.1016/j.fsi.2017.05.016
69. Wang Z, Lin L, Chen W, Zheng X, Zhang Y, Liu Q, et al. Neutrophil plays critical role during Edwardsiella piscicida immersion infection in zebrafish larvae. Fish Shellfish Immunol (2019) 87:565–72. doi: 10.1016/J.FSI.2019.02.008
70. Chen H, Yuan G, Su J, Liu X. Hematological and immune genes responses in yellow catfish (Pelteobagrus fulvidraco) with septicemia induced by Edwardsiella ictaluri. Fish Shellfish Immunol (2020) 97:531–9. doi: 10.1016/J.FSI.2019.11.071
71. Elbahnaswy S, Elshopakey GE. Differential gene expression and immune response of Nile tilapia (Oreochromis niloticus) challenged intraperitoneally with Photobacterium damselae and Aeromonas hydrophila demonstrating immunosuppression. Aquaculture (2020) 526. doi: 10.1016/J.AQUACULTURE.2020.735364
72. Arnhold J. The dual role of myeloperoxidase in immune response. Internt J of Mol Sci (2020) 21(21):8057. doi: 10.3390/ijms21218057
73. Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol (2011) 11(11):762. doi: 10.1038/NRI3070
74. Lichtnekert J, Kawakami T, Parks WC, Duffield JS. Changes in macrophage phenotype as the immune response evolves. Curr Opin Pharmacol (2013) 13(4):555–64. doi: 10.1016/J.COPH.2013.05.013
75. Oishi Y, Manabe I. Macrophages in inflammation, repair and regeneration. Int Immunol (2018) 30(11):511–28. doi: 10.1093/INTIMM/DXY054
76. Germic N, Frangez Z, Yousefi S, Simon HU. Regulation of the innate immune system by autophagy: monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differentiation (2019) 26(4):715–27. doi: 10.1038/S41418-019-0297-6
77. Stosik M, Deptuła W, Trávniček M, Baldy-Chudzik K. Phagocytic and bactericidal activity of blood thrombocytes in carps (Cyprinus carpio). Veterinarni Medicina (2002) 47(1):21–5. doi: 10.17221/5798-VETMED
78. Nagasawa T, Nakayasu C, Rieger AM, Barreda DR, Somamoto T, Nakao M. Phagocytosis by thrombocytes is a conserved innate immune mechanism in lower vertebrates. Front Immunol (2014) 5:445/ABSTRACT. doi: 10.3389/FIMMU.2014.00445/ABSTRACT
79. Mirhaj M, Labbaf S, Tavakoli M, Seifalian AM. (2022). Emerging treatment strategies in wound care. Int Wound J 19(7):1934–54. doi: 10.1111/iwj.13786
80. Whyte SK. The innate immune response of finfish - A review of current knowledge. Fish Shellfish Immunol (2007) 23(6):1127–51. doi: 10.1016/J.FSI.2007.06.005
81. Zhu W, Su J. Immune functions of phagocytic blood cells in teleost. Rev Aquaculture (2022) 14(2):630–46. doi: 10.1111/RAQ.12616
82. Ellis AE. Innate host defense mechanisms of fish against viruses and bacteria. Dev Comp Immunol (2001) 25(8–9):827–39. doi: 10.1016/S0145-305X(01)00038-6
83. Nishihara A, Morimoto N, Sumiyoshi T, Yasumoto S, Kondo M, Kono T, et al. Inhibition of lysozyme lytic activity by Ivy derived from Photobacterium damselae subsp. piscicida. Fish Shellfish Immunol (2022) 124:280–8. doi: 10.1016/J.FSI.2022.04.012
84. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol (2010) 11(5):373–84. doi: 10.1038/NI.1863
85. Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduction Targeted Ther (2021) 6(1). doi: 10.1038/S41392-021-00687-0
86. Pressley ME, Phelan PE, Eckhard Witten P, Mellon MT, Kim CH. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev Comp Immunol (2004) 29(6):501–13. doi: 10.1016/J.DCI.2004.10.007
87. Rojo I, de Ilárduya Ó.M, Estonba A, Pardo MÁ. Innate immune gene expression in individual zebrafish after Listonella Anguillarum inoculation. Fish Shellfish Immunol (2007) 23(6):1285–93. doi: 10.1016/J.FSI.2007.07.002
88. Shan Y, Zhang Y, Zhuo X, Li X, Peng J, Fang W. Matrix metalloproteinase-9 plays a role in protecting zebrafish from lethal infection with Listeria monocytogenes by enhancing macrophage migration. Fish Shellfish Immunol (2016) 54:179–87. doi: 10.1016/J.FSI.2016.04.003
89. Langevin C, Blanco M, Martin SAM, Jouneau L, Bernardet JF, Houel A, et al. Transcriptional responses of resistant and susceptible fish clones to the bacterial pathogen Flavobacterium psychrophilum. PloS One (2012) 7(6). doi: 10.1371/JOURNAL.PONE.0039126
90. Ke F, Wang Y, Hong J, Xu C, Chen H, Zhou SB. Characterization of MMP-9 gene from a norMalized cDNA library of kidney tissue of yellow catfish (Pelteobagrus fulvidraco). Fish Shellfish Immunol (2015) 45(2):260–7. doi: 10.1016/J.FSI.2015.04.012
91. Chadzinska M, Baginski P, Kolaczkowska E, Savelkoul HFJ, Lidy Verburg-Van Kemenade BM. Expression profiles of matrix metalloproteinase 9 in teleost fish provide evidence for its active role in initiation and resolution of inflammation. Immunology (2008) 125(4):601. doi: 10.1111/J.1365-2567.2008.02874.X
92. Lindenstrøm T, Secombes CJ, Buchmann K. Expression of immune response genes in rainbow trout skin induced by Gyrodactylus derjavini infections. Veterinary Immunol Immunopathology (2004) 97(3–4):137–48. doi: 10.1016/J.VETIMM.2003.08.016
93. Lindenstrøm T, Sigh J, Dalgaard MB, Buchmann K. Skin expression of IL-1β in East Atlantic salmon, Salmo salar L., highly susceptible to Gyrodactylus salaris infection is enhanced compared to a low susceptibility Baltic stock. J Fish Dis (2006) 29(2):123–8. doi: 10.1111/J.1365-2761.2006.00696.X
94. Secombes CJ, Wang T, Hong S, Peddie S, Crampe M, Laing KJ, et al. Cytokines and innate immunity of fish. Dev Comp Immunol (2001) 25(8–9):713–23. doi: 10.1016/S0145-305X(01)00032-5
95. Nicolas G, Viatte L, Bennoun M, Beaumont C, Kahn A, Vaulont S. Hepcidin, A new iron regulatory peptide. Blood Cells Molecules Dis (2002) 3(29):327–35. doi: 10.1006/BCMD.2002.0573
96. Anderson GJ, Vulpe CD. MamMalian iron transport. Cell Mol Life Sci (2009) 20(66):3241–61. doi: 10.1007/S00018-009-0051-1
97. Viatte L, Vaulont S. Hepcidin, the iron watcher. Biochimie (2009) 91(10):1223–8. doi: 10.1016/J.BIOCHI.2009.06.012
98. Page MGP. The role of iron and siderophores in infection, and the development of siderophore antibiotics. Clin Infect Diseases: Off Publ Infect Dis Soc America 69(Suppl (2019) 7):S529–37. doi: 10.1093/CID/CIZ825
99. Nairz M, Schroll A, Sonnweber T, Weiss G. The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol (2010) 12(12):1691–702. doi: 10.1111/J.1462-5822.2010.01529.X
100. Aschemeyer S, Qiao B, Stefanova D, Valore EV, Sek AC, Alex Ruwe T, et al. Structure-function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood (2018) 131(8):899–910. doi: 10.1182/BLOOD-2017-05-786590
101. Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe (2013) 13(5):509. doi: 10.1016/J.CHOM.2013.04.010
102. Nemeth E, Tuttle MS, Powelson J, Vaughn MD, Donovan A, Ward DMV, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science (2004) 306(5704):2090–3. doi: 10.1126/SCIENCE.1104742
103. Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, et al. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest (2004) 113(9):1271–6. doi: 10.1172/JCI20945
104. Lee P, Peng H, Gelbart T, Wang L, Beutler E. Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc Natl Acad Sci (2005) 102(6):1906–10. doi: 10.1073/PNAS.0409808102
105. Katsarou A, Pantopoulos K. Basics and principles of cellular and systemic iron homeostasis. Mol Aspects Med (2020) 75:100866–6. doi: 10.1016/J.MAM.2020.100866
106. Roy CN, Andrews NC. Anemia of inflammation: the hepcidin link. Curr Opin Hematol (2005) 12(2):107–11. doi: 10.1097/00062752-200503000-00001
107. Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr (2006) 26:323–42. doi: 10.1146/ANNUREV.NUTR.26.061505.111303
108. Neves JV, Caldas C, Wilson JM, Rodrigues PNS. Molecular mechanisms of hepcidin regulation in sea bass (Dicentrarchus labrax). Fish Shellfish Immunol (2011) 31(6):1154–61. doi: 10.1016/J.FSI.2011.10.006
109. Shike H, Lauth X, Westerman ME, Ostland VE, Carlberg JM, Van Olst JC, et al. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur J Biochem (2002) 269(8):2232–7. doi: 10.1046/J.1432-1033.2002.02881.X
110. Douglas SE, Gallant JW, Liebscher RS, Dacanay A, Tsoi SCM. Identification and expression analysis of hepcidin-like antimicrobial peptides in bony fish. Dev Comp Immunol (2003) 27(6–7):589–601. doi: 10.1016/S0145-305X(03)00036-3
111. Bao B, Peatman E, Li P, He C, Liu Z. Catfish hepcidin gene is expressed in a wide range of tissues and exhibits tissue-specific upregulation after bacterial infection. Dev Comp Immunol (2005) 29(11):939–50. doi: 10.1016/J.DCI.2005.03.006
112. Shen Y, Zhao Z, Zhao J, Chen X, Cao M, Wu M. Expression and functional analysis of hepcidin from mandarin fish (Siniperca chuatsi). Int J Mol Sci (2019) 20(22):5602. doi: 10.3390/IJMS20225602
113. Rodrigues PNS, Vázquez-Dorado S, Neves JV, Wilson JM. Dual function of fish hepcidin: Response to experimental iron overload and bacterial infection in sea bass (Dicentrarchus labrax). Dev Comp Immunol (2006) 12(30):1156–67. doi: 10.1016/J.DCI.2006.02.005
114. Pulgar R, Hödar C, Travisany D, Zuñiga A, Domínguez C, Maass A, et al. Transcriptional response of Atlantic salmon families to Piscirickettsia salmonis infection highlights the relevance of the iron-deprivation defence system. BMC Genomics (2015) 16(1):1–21. doi: 10.1186/S12864-015-1716-9/FIGURES/7
115. Agoro R, Mura C. Inflammation-induced up-regulation of hepcidin and down-regulation of ferroportin transcription are dependent on macrophage polarization. Blood Cells Molecules Dis (2016) 61:16–25. doi: 10.1016/J.BCMD.2016.07.006
116. Ma Y, Lee CJ, Kim SS, Kim DNJ, Nam BH, Kim YO, et al. Role of hepcidins from black rockfish (Sebastes schlegelii) in iron-metabolic function and bacterial defense. J Mar Sci Eng (2020) 8(7):493. doi: 10.3390/JMSE8070493
117. Saperstein S, Chen L, Oakes D, Pryhuber G, Finkelstein J. IL-1β Augments TNF-α–mediated inflammatory responses from lung epithelial cells. J Interferon Cytokine Res (2009) 29(5):273. doi: 10.1089/JIR.2008.0076
118. Umare V, Pradhan V, Nadkar M, Rajadhyaksha A, Patwardhan M, Ghosh KK, et al. Effect of proinflammatory cytokines (IL-6, TNF-α, and IL-1β) on clinical manifestations in Indian SLE patients. Mediators Inflammation (2014) 2014. doi: 10.1155/2014/385297
119. Nascimento DS, Pereira PJB, Reis MIR, do Vale A, Zou J, Silva MT, et al. Molecular cloning and expression analysis of sea bass (Dicentrarchus labrax L.) tumor necrosis factor-alpha (TNF-alpha). Fish Shellfish Immunol (2007) 23(3):701–10. doi: 10.1016/J.FSI.2007.02.003
120. El Aamri F, Real F, Acosta F, Bravo J, Román L, Déniz S, et al. Differential innate immune response of European seabass (Dicentrarchus labrax) against Streptococcus iniae. Fish Shellfish Immunol (2015) 46(2):436–41. doi: 10.1016/J.FSI.2015.05.054
121. Vaz M, Pires D, Pires P, Simões M, Pombo A, Santos P, et al. Early immune modulation in european seabass (Dicentrarchus labrax) juveniles in response to betanodavirus infection. Fishes (2022) 7(2):63. doi: 10.3390/FISHES7020063/S1
Keywords: tenacibaculosis, aquaculture, mucosal immunity, innate immunity, bacterial infection
Citation: Ferreira IA, Peixoto D, Losada AP, Quiroga MI, Vale Ad and Costas B (2023) Early innate immune responses in European sea bass (Dicentrarchus labrax L.) following Tenacibaculum maritimum infection. Front. Immunol. 14:1254677. doi: 10.3389/fimmu.2023.1254677
Received: 24 July 2023; Accepted: 15 August 2023;
Published: 04 September 2023.
Edited by:
Tor Gjøen, University of Oslo, NorwayReviewed by:
Carlo C. Lazado, Norwegian Institute of Food, Fisheries and Aquaculture Research (Nofima), NorwayMahmoud Tanekhy, Alexandria University, Egypt
Copyright © 2023 Ferreira, Peixoto, Losada, Quiroga, Vale and Costas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inês A. Ferreira, aW5lcy5mZXJyZWlyYUBjaWltYXIudXAucHQ=
 Inês A. Ferreira
Inês A. Ferreira Diogo Peixoto
Diogo Peixoto Ana Paula Losada
Ana Paula Losada María Isabel Quiroga5
María Isabel Quiroga5 Ana do Vale
Ana do Vale Benjamín Costas
Benjamín Costas