
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 30 August 2023
Sec. Comparative Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1247199
This article is part of the Research Topic Mucosal Barrier in Teleost Fish: Physical, Biochemical, and Immune Nature View all 7 articles
 Ignasi Sanahuja1*
Ignasi Sanahuja1* Laura Fernandez-Alacid2
Laura Fernandez-Alacid2 Silvia Torrecillas1
Silvia Torrecillas1 Alberto Ruiz1
Alberto Ruiz1 Eva Vallejos-Vidal3
Eva Vallejos-Vidal3 Joana P. Firmino1
Joana P. Firmino1 Felipe E. Reyes-Lopez4
Felipe E. Reyes-Lopez4 Lluis Tort5
Lluis Tort5 Dariel Tovar-Ramirez6
Dariel Tovar-Ramirez6 Antoni Ibarz2
Antoni Ibarz2 Enric Gisbert1
Enric Gisbert1The present study explores the effects of two supplementation levels of Debaryomyces hansenii (1.1% and 2.2%) as a probiotic in a reference low fish meal-based diet on the skin mucosal tissue in Sparus aurata. This study includes the evaluation of fish performance coupled with a holistic study of the skin mucosa: i) a transcriptomic study of the skin tissue, and ii) the evaluation of its secreted mucus both in terms of skin mucosal-associated biomarkers and its defensive capacity by means of co-culture analysis with two pathogenic bacteria. Results showed that after 70 days of diet administration, fish fed the diet supplemented with D. hansenii at 1.1% presented increased somatic growth and a better feed conversion ratio, compared to fish fed the control diet. In contrast, fish fed the diet including 2.2% of the probiotic presented intermediate values. Regarding gene regulation, the probiotic administration at 1.1% resulted in 712 differentially expressed genes (DEGs), among which 53.4% and 46.6% were up- and down-regulated, respectively. In particular, D. hansenii modulated some skin biological processes related to immunity and metabolism. Specifically, D. hansenii administration induced a strong modulation of some immune biological-related processes (61 DEGs), mainly involved in B- and T-cell regulatory pathways. Furthermore, dietary D. hansenii promoted the skin barrier function by the upregulation of anchoring junction genes (23 DEGs), which reinforces the physical defense against potential skin damage. In contrast, the skin showed modulated genes related to extracellular exosome and membrane organization (50 DEGs). This modulated functioning is of great interest, particularly in relation to the increased skin mucus defensive capacity observed in the bacterial co-culture in vitro trials, which could be related to the increased modulation and exudation of the innate immune components from the skin cells into the mucus. In summary, the modulation of innate immune parameters coupled with increased skin barrier function and cell trafficking potentiates the skin’s physical barrier and mucus defensive capacity, while maintaining the skin mucosa’s homeostatic immune and metabolic status. These findings confirmed the advantages of D. hansenii supplementation in low fish meal-based diets, demonstrating the probiotic benefits on cultured marine species.
The primary function of the epidermis is to separate the animals’ internal medium from the external environment, and this function has been modulated through species by physical and chemical adaptations (1). In fish, mucosal structures are considered mucosal-associated lymphoid tissues (MALT), which means that these specialized structures are capable of sensing and responding to environmental and biotic challenges (2). For this purpose, the mucosal immune system in fish has acquired throughout evolution a wide variety of interconnected cells with innate and adaptive components (3). Moreover, mucosae are dynamic and semipermeable barriers with several physiological capacities, among which are osmoregulation, respiration, nutrition, or locomotion (4–6). These characteristics could be modulated either by: (i) the improvement of husbandry and farming practices or (ii) providing a balanced diet. On this basis, adequate nutrition could avoid health deficiencies, meanwhile maintaining fish performance and protecting animals from biotic and abiotic stressors (7). In addition, nutritional strategies could be used to enhance fish health while promoting a sustainable practice by reducing the dependence on traditional marine raw materials (8, 9). However, balancing nutrients in aquafeeds with alternative resources to the fish-derived products is complex and usually a bottleneck to fish development and health (10).
One step further in the search for quality nutrition is supplementation to enhance certain aspects of fish biology or to counteract the deficiencies of sustainable non-animal-based aquafeeds. Among other supplements, prebiotics and probiotics confer benefits to the host by enhancing the gastrointestinal milieu, optimizing nutrient digestion and absorption, improving growth performance, and modulating the fish’s immune system (11, 12). These products could also possess mechanisms to modulate microbial communities, either by maintaining native microbiota or by introducing beneficial organisms that establish a synergistic symbiosis relevant to the host (13). One of these feed additives that globally encompass all the above-mentioned benefits is yeasts (14). Yeasts are single-celled organisms that contain wall-related bioactive compounds, which function as stimulants, favoring growth and the immune system and contributing to the overall animal condition and health (15). Debaryomyces hansenii, a yeast that belongs to the natural microbiota of carnivorous fish, offers different benefits in terms of health and performance, which in the aquaculture industry is of great interest. D. hansenii has been reported to enhance larval survival and development in several marine fish species (16–18), whereas in juveniles, it is reputed for promoting the immune system (17, 19–21). Several authors have attributed the beneficial effects of yeast to their β-glucan and polyamine wall-related contents (22, 23); however, its potential benefits and mode of action at the level of mucosal-associated lymphoid tissues have been poorly studied.
Focusing on MALT, available literature on D. hansenii primarily addresses its effects on gut-associated lymphoid tissue (GALT). Research has found that D. hansenii improves intestinal function and immunity, meanwhile modulating gut microbiota (19, 20, 23–25), which are effects that may be associated with its positive impacts on fish growth and performance (21). However, little is known about the effects of a D. hansenii dietary supplementation on other MALT, and especially the effects on the principal mucosa barrier against the environment, the skin. The skin and its exuded mucus present a similar multifactorial behavior against the environment (26), and due to their direct contact with the aquatic environment, play a principal role in protection (27). An example of this involvement is the production of a specialized mucus layer with a wide range of physical and biochemical defensive components (28, 29), which may be modulated through several factors, including dietary supplementation (30–32).
The present study aimed to determine the effects of a low fish meal-based diet (7%), supplemented with D. hansenii, in the skin mucosa of gilthead seabream (Sparus aurata). To provide insight into the effects of this probiotic on gilthead seabream skin, we combined a skin transcriptomic analysis, with the modification of the skin mucus-associated biomarkers (SMABs), in addition to the study of its antibacterial capacity through a co-culture with two pathogenic bacteria for this fish species.
Three experimental diets were designed to evaluate the effects of D. hansenii (CBS 8339) on fish performance and feed utilization and evaluate the effects of its supplementation on the skin through transcriptomic analysis. A 7% fish meal-based basal diet was formulated and considered a control diet (Table 1). Subsequently, the control diet was used as a basal diet to create both experimental yeast diets, which only differed on D. hansenii content proportion (Yeast1.1% diet with 1.1%, 17.2 x 105 cfu; and Yeast2.2% diet 2.2%, 35.7 x 105 cfu) as shown in Table 1. These percentages have been determined in previous studies (17–19, 21). Diets were isoproteic (48.4%), isolipidic (17.2%), and isoenergetic (21.7 MJ/Kg feed). Each of the diets (pellet size: 2mm) was manufactured by SPAROS Lda (Portugal) under the conditions detailed by Gisbert et al. (33) and stored at 4°C throughout the experiment to prevent feed deterioration. Diet manufacture conditions, packaging, and storage did not compromise the viability of the probiotic (18, 20, 21). D. hansenii was provided by CIBNOR (La Paz, Mexico) and cultured as described by Tovar-Ramírez et al. (18).
A total of 500 S. aurata juveniles (BW = 14.5 ± 1.2 g) were obtained from a commercial farm (PISCIMAR SL; AVRAMAR, Burriana, Spain) and transported and acclimated to IRTA-La Ràpita research facilities (La Ràpita, Spain). During acclimatization, fish were fed a commercial diet (Optibream, Skretting) and maintained in a 2,000 L tank connected to IRTAmar® recirculating system under natural conditions and constant temperature (23.0 ± 1.0°C).
Prior to the experiment, 300 fish were slightly anesthetized (50 mg/L tricaine methanesulfonate, MS-222, Sigma-Aldrich, Madrid, Spain) and randomly distributed in four replicate tanks per diet (N = 25 fish per tank). The experiment lasted for 70 days, and diets were distributed in four meals at 08:00, 11:00, 13:00, and 16:00 h (the corresponding feed ratio was distributed during 1 h), and uneaten pellets were collected and weighed to calculate daily feed ingesta values. The trial was run under natural photoperiod (14h light/8h dark), salinity (35-36 ‰), constant temperature (23.3 ± 1.3°C), dissolved oxygen (5.7 ± 0.2 mg/L) (OXI330, Crison Instruments, Spain), and pH (8.2 ± 0.1) (pHmeter 507, Crison Instruments). Ammonia and nitrite levels (0.13 ± 0.1 mg /L and 0.18 ± 0.1 mg /L, respectively) were continuously controlled using HACH DR 900 Colorimeter (Hach Company, Spain).
Growth performance and feed utilization indicators were calculated using the following formulae:
- Weight gain rate (WGR) % = 100 x [(BWf – BWi)/BWi]; where BWf and BWi are the final and initial mean BW of fish.
- Specific growth rate (SGR) % BW/day = 100 x [(ln BWf – ln BWi)/days]
- Survival rate (SR) % = 100 x (Final number of fish/Initial number of fish).
- Feed conversion ratio (FCR) = Feed intake (g)/Increase in fish biomass (g).
At the end of the experiment, all fish were anesthetized (100 mg/L tricaine methanesulfonate, MS222) and measured individually for the calculation of key performance indicators as previously described and to sample skin mucus following the method described by Fernandez-Alacid et al. (34). According to the results obtained from the growth and skin mucus metabolites, only control and Yeast1.1% diets were considered for the skin transcriptomics and skin mucus defensive capacity analyses.
In addition, three randomly selected fish per replicate tank (N = 12) were euthanized (300 mg/L MS222) for transcriptional analysis purposes. A ca. 1 cm2 of skin section from the mid-region of the fish body (over the lateral line, right side of each fish) was dissected and the muscle tissue attached to it was discarded. Samples were incubated overnight (4°C) in RNAlater™ and stored at -80°C for further RNA extraction.
At final sampling, 36 animals were randomly selected (12 fish for each dietary condition; three to four fish per tank) and their skin mucus was collected following the method described by Fernández-Alacid et al. (34). Glucose and lactate concentrations on skin mucus were determined by an enzymatic colorimetric test (LO-POD glucose, SPINREACT®, St. Esteve de Bas, Spain) and (LO-POD lactate, SPINREACT®, Barcelona, Spain), respectively, following the manufacturer’s instructions, but with slight modifications for fish skin mucus as described by Fernandez-Alacid et al. (34). Glucose and lactate values were expressed as μg/mL of skin mucus. Protein concentration (mg/mL of skin mucus) was determined using the Bradford assay (35), using bovine serum albumin as standard (BSA; Sigma Aldrich, Madrid, Spain). Cortisol levels were measured using an ELISA kit (IBL International, Tecan Group, Switzerland) following the manufacturer’s instructions for saliva determinations but with slight modifications for fish skin mucus (34). Cortisol values were expressed as ng/mL of skin mucus. The Ferric Antioxidant Power (FRAP) was measured by means of an enzymatic colorimetric test (Invitrogen, Thermo Fisher Scientific, Spain), following the manufacturer’s instructions for plasma, with minor modifications; values were expressed as µmol/mL of mucus. Mucus ratios referring to protein (glucose/protein, lactate/protein, cortisol/protein, and FRAP/protein) were calculated in order to avoid the dilution derived from mucus sampling. The glucose/lactate ratio was also calculated as a metabolic response indicator (34).
To study the skin mucus antibacterial capacity, three different bacteria were tested: two pathogenic bacteria, Vibrio anguillarum (CECT522T) and Pseudomonas anguilliseptica (CECT899T), and a non-pathogenic bacterium, Escherichia coli (DSMZ423). The pathogenic V. anguillarum and P. anguilliseptica strains were grown in Marine Broth culture media (MB, Difco Laboratories, Detroit) and the non-pathogenic E. coli strain was grown in Tryptic Soy Broth culture media (TSB, Conda, Spain). The skin mucus antibacterial capacity (from control and Yeast1.1% diets) on three strains’ viability was evaluated by monitoring the absorbance of co-culture grown at 400nm (per triplicate) for 14 hours in 96-well plates, as described by Fernandez-Alacid et al. (36).
Total RNA was extracted from the skin of 12 randomly selected fish per feeding treatment using the RNeasy® Mini Kit (Qiagen, Germany). Total RNA was eluted in a final volume of 35 µL nuclease-free and processed with DNAse (DNA-free™ DNA removal Kit; Invitrogen, Lithuania). Total RNA concentration and purity were measured using a Nanodrop-2000® spectrophotometer (Thermo Scientific, USA) and stored at -80°C for further analysis. Prior to microarray hybridization, the RNA samples were diluted to a concentration of 133.33 ng/µL and tested for RNA integrity (Agilent 2100 Bioanalyzer; Agilent Technologies, Spain). The samples were selected according to the criteria for RIN value > 8.5. Three different pools for each diet regimen were selected (N = four fish per pool).
Skin transcriptional analysis from both experimental groups was carried out using the Aquagenomics Sparus aurata oligonucleotide microarray v2.0 (4 × 44 K) (SAQ) platform. Detailed data and transcriptomic raw data are in the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus (GEO) public repository under accession numbers GPL13442 and GSE162504, respectively. Sampling labeling, hybridization, washing, and scanning were performed as described in (37). Briefly, a one-color RNA labeling (Agilent One-Color RNA Spike-In kit; Agilent Technologies, USA) was used. RNA from each pool (200 ng) was reverse-transcribed using spike-in. Total RNA was used as a template for Cyanine-3 (Cy3)-labeled cRNA synthesis and amplified using a Quick Amp Labeling kit (Agilent Technologies). cRNA samples were purified using an RNeasy® micro kit (Qiagen). Dye incorporation and cRNA yield were checked (NanoDrop ND-2000® spectrophotometer). Then, Cy3-labeled cRNA (1.5 mg) with >6.0 pmol Cy3/mg specific activity was then fragmented at 60°C for 30 min and hybridized at 65°C with the array in the presence of hybridization buffer (Gene expression hybridization kit, Agilent Technologies) for 17 h. For washes, microarrays were incubated with Gene expression wash buffers, and stabilization and drying solutions were according to the manufacturer’s instructions (Agilent Technologies). Microarray slides were scanned (Agilent G2505B Microarray Scanner System), and spot intensities and other quality control features were extracted (Agilent Feature Extraction software version 10.4.0.0).
The Search Tool for the Retrieval of Interacting Genes (STRING) public repository version 11.0 (https://string-db.org) was used to construct skin transcripteractome comparing fish fed with the control and Yeast1.1% diets. Protein-Protein interaction (PPI) Networks Functional Enrichment Analysis for all the differentially expressed genes (DEGs) was conducted with a medium-confidence interaction score (0.4) using Homo sapiens as the model organism (37, 38). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of all the DEGs were also evaluated through STRING (p < 0.05). A human orthology identification based on gene/protein name was accessed through the Genecards (www.genecards.org) (39) and Uniprot (www.uniprot.org) databases to confirm the match of gene acronyms between both H. sapiens and S. aurata. Additionally, protein-protein BLAST (BLASTp) was run (E < 10−7; query cover > 95%). The cluster aggrupation of nodes classified according to their biological processes was represented with the application ClueGO (v2.5.9) and CluePedia (v1.5.9) using Cytoscape (v3.9.1) software.
Equality of variances and normality were checked using Levene’s test. Differences among the key performance parameters and SMABs were performed using a one-way ANOVA test, followed by post-hoc Bonferroni’s test (if equal variances were assumed) or Dunnett’s test (if variances among groups were unbalanced) to test the effects of yeast supplementation levels (p < 0.05). In addition, curvilinear estimation of regression analysis was performed to a deep comprehension of KPI and SMAB differences (p < 0.05). Student’s t-test, assuming data homoscedasticity, was used to compare bacterial growth differences among the Yeast1.1% diet and control diet (p < 0.05). Data on KPI and SMABs were expressed as mean values ± standard deviations using SPSS Statistics for Windows (v.25; IBM Corp., Armonk, NY, USA), and bacterial growth values were expressed as mean ± standard error, with GraphPad PRISM 9.
Results on fish growth performance and key performance indicators are represented and summarized in Table 2. Briefly, fish fed the Yeast1.1% diet performed better than the rest of the dietary groups. Values of WGR, SGR, and FCR were improved in fish fed the Yeast1.1% diet compared to those from both control and Yeast2.2% diets. Furthermore, the feed conversion ratio (FCR) value was lower (p < 0.05) in fish fed the Yeast1.1% diet when compared to fish fed the reference diet, whereas fish fed the Yeast2.2% diet showed an intermediate value. However, no significant differences in feed intake (FI) were found among the experimental groups (Table 2; p > 0.05).
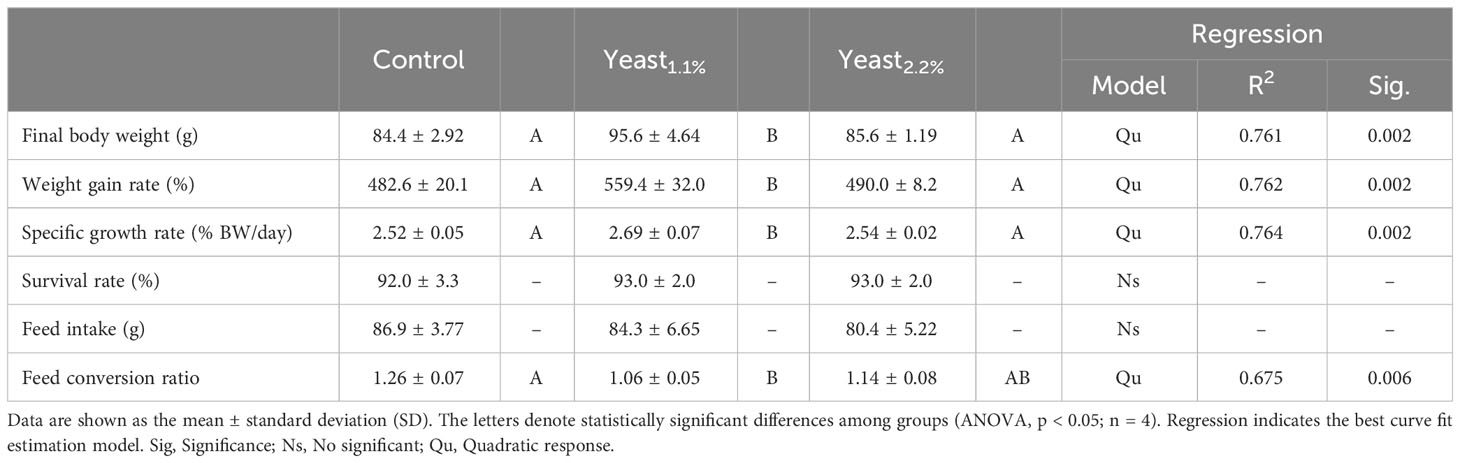
Table 2 Growth and feed performance indicators in gilthead sea bream (Sparus aurata) fed the experimental diets containing Debaryomyces hansenii (1.1%, and 2.2%) or devoid of the probiotic (Control diet).
Values of SMABs and their ratios, as well as the FRAP from skin mucus samples, are summarized in Table 3. No differences between the control and Yeast1.1% dietary groups were observed in terms of skin mucus biomarkers nor when they were normalized by protein values (p > 0.05). However, higher yeast supplementation (2.2% yeast diet) increased glucose levels in fish skin mucus, which was maintained even with protein normalization (p < 0.05).
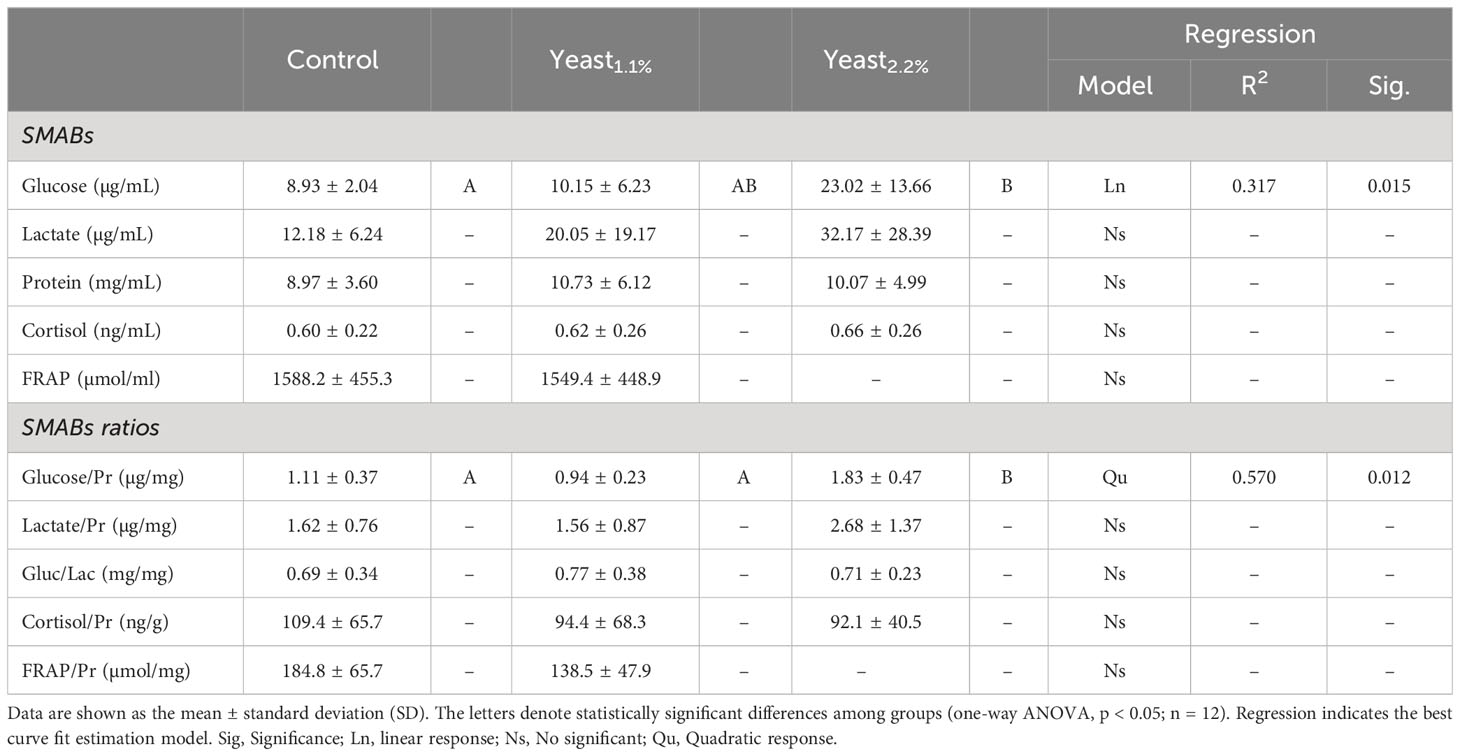
Table 3 Skin mucus-associated biomarkers (SMABs) of gilthead seabream (Sparus aurata) fed an experimental diet supplemented with 1.1% (Yeast1.1%) and 2.2% (Yeast2.2%) of D. hansenii, and the control diet devoid of the feed additive.
We evaluated the skin mucus’s defensive capacity to know whether the diet supplemented with yeast implies a functional protective mechanism against pathogenic bacterial growth on skin mucus. Taking into account results from KPI related to growth and feed utilization, only skin mucus samples from fish fed the control and the Yeast1.1% diets were analyzed.
The S. aurata skin mucus from both nutritional groups (Control and Yeast1.1%) showed a decrease in E. coli (Figure 1), V. anguillarum (Figure 2), and P. anguilliseptica (Figure 3) growth during all the co-culture periods. When cultured with the skin mucus from fish fed the Yeast1.1% diet, a reduction in the growth of the non-pathogenic bacteria E. coli was observed (p < 0.05; Figure 1A). Growth decrease was recorded between 4 and 14 h of bacterial culture. The most accentuated decrease in growth values compared with fish fed the control diet was found in the logarithmic phase (approximately 15% between 4h and 6h, 12% at 8h, and 10% until 14h) (Figure 1B). Regarding V. anguillarum, a decrease in bacterial growth was observed in the co-culture with the skin mucus from fish fed the supplemented diet (Figure 2A), being the most remarkable growth decrease between 8h-14h. In addition, the mucus of the fish fed with the Yeast1.1% diet had a substantial growth inhibiting capacity, being 20-30% greater than fish fed the control diet (Figure 2B). However, the most accentuated growth decline was observed in P. anguilliseptica (Figure 3), registering a maximum decrease in bacterial growth between 6h and 14h (percentage of inhibition over 20% compared with the control diet, p < 0.05) associated with the skin mucus of fish fed the yeast-supplemented diet.
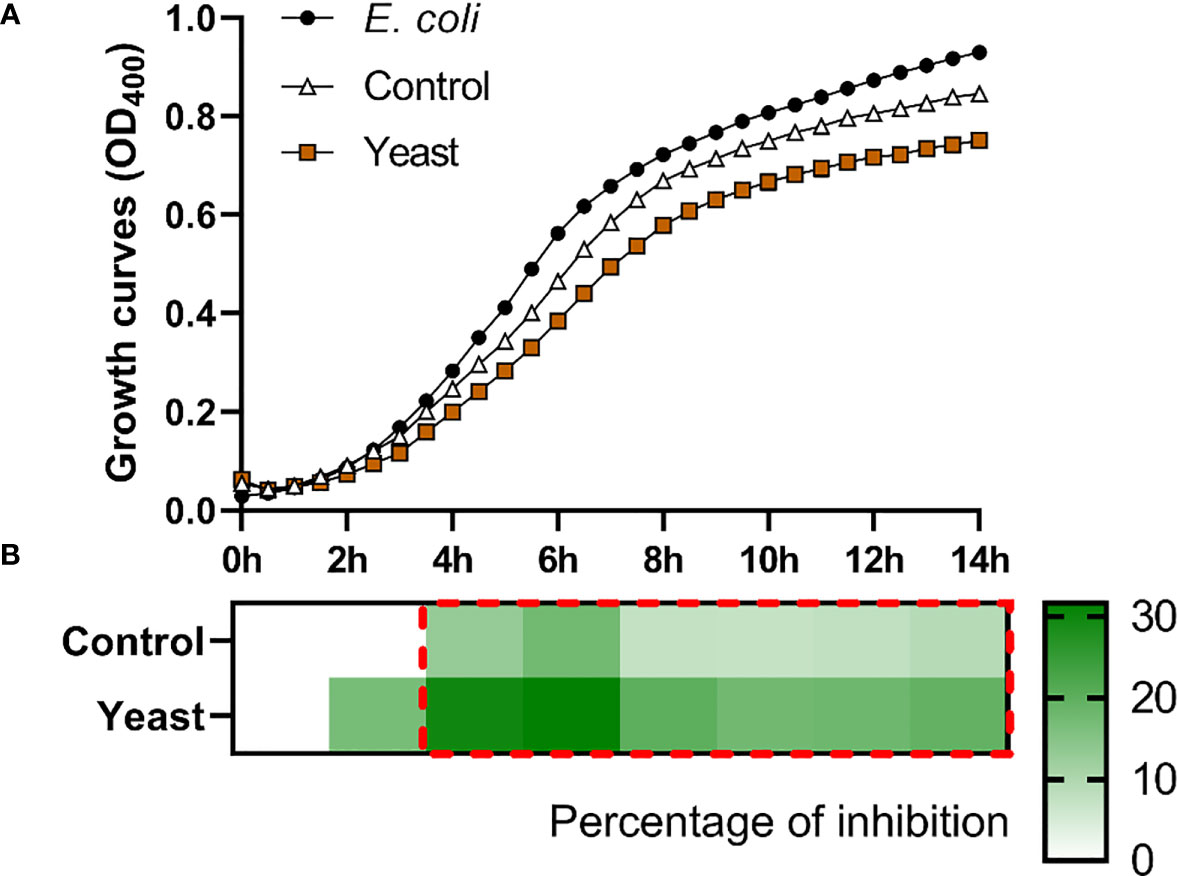
Figure 1 Co-culture growth of E. coli on the skin mucus of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast diet) or devoid of the probiotic (Control diet). Data from growth curves (A) correspond to the mean ± SEM of triplicate samples. Black circles correspond to E. coli growth in a medium devoid of mucus; white triangles correspond to E. coli growth in the mucus of fish fed the Control diet; brown squares correspond to E. coli growth in the mucus of fish fed the Yeast1.1% diet. The heat map shows the percentage of inhibition (B) of both diets in a red (negative) to green (positive) gradient. The dashed red line square indicates significant differences in bacterial growth between dietary groups (Student’s t-test, p < 0.05).
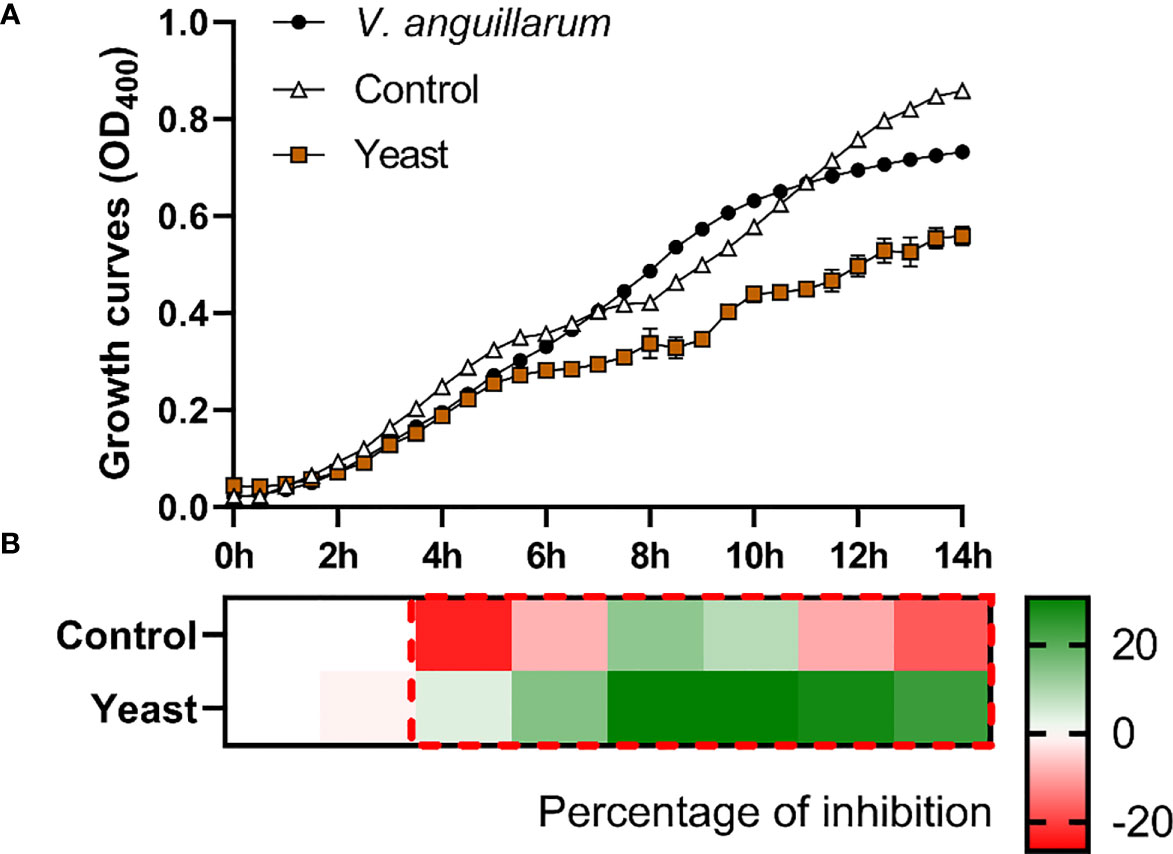
Figure 2 Co-culture growth of V. anguillarum on the skin mucus of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast diet) or devoid of the probiotic (Control diet). Data from growth curves (A) correspond to the mean ± SEM of triplicate samples. Black circles correspond to V. anguillarum growth in a medium devoid of mucus; white triangles correspond to V. anguillarum growth in the mucus of fish fed the Control diet; and brown squares correspond to V. anguillarum growth in the mucus of fish fed the Yeast1.1% diet. The heat map shows the percentage of inhibition (B) of both diets in a red (negative) to green (positive) gradient. The dashed red line square indicates significant differences in bacterial growth between dietary groups (Student’s t-test, p < 0.05).
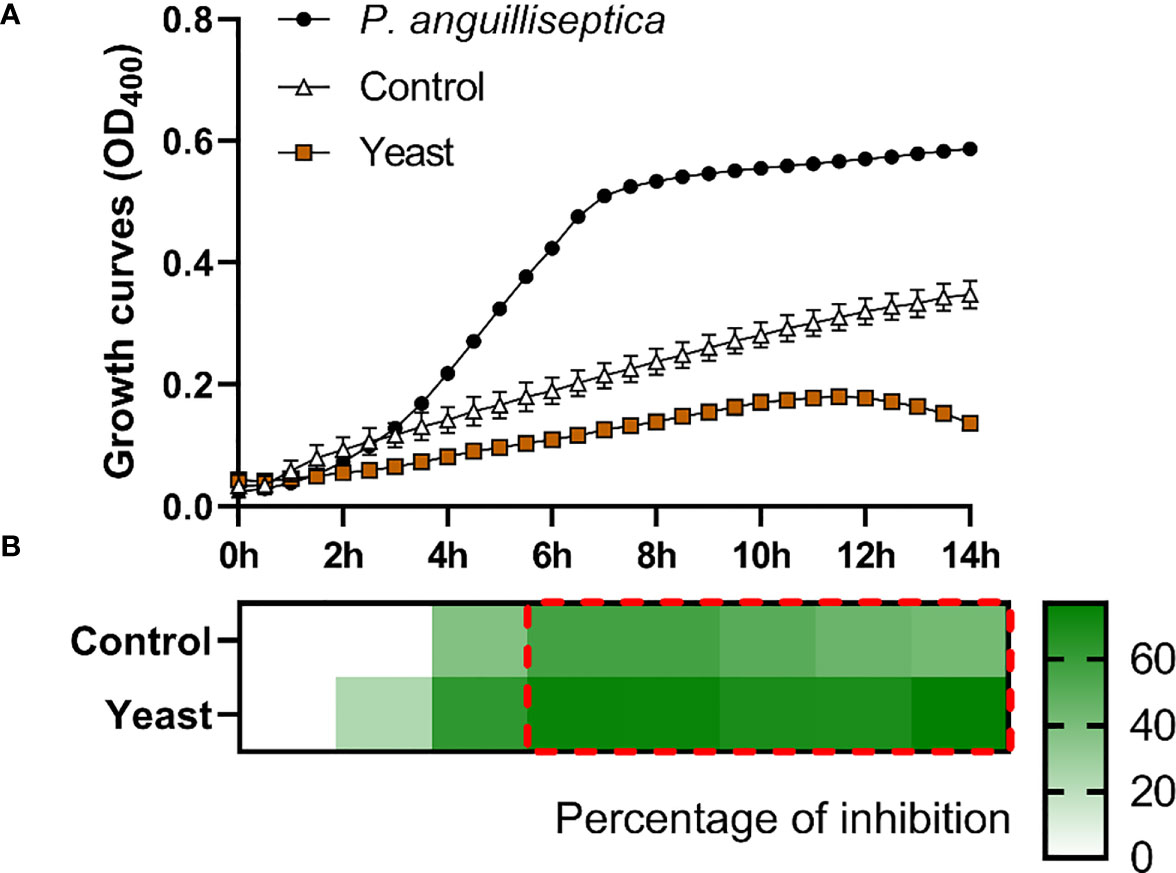
Figure 3 P. anguilliseptica co-culture growth on the skin mucus of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast diet) or devoid of the probiotic (Control diet). Data from growth curves (A) correspond to the mean ± SEM of triplicate samples. Black circles correspond to P. anguilliseptica growth in a medium devoid of mucus; white triangles correspond to P. anguilliseptica growth in the mucus of fish fed the Control diet; and brown squares correspond to P. anguilliseptica growth in the mucus of fish fed the Yeast1.1% diet. The heat map shows the percentage of inhibition (B) of both diets in a red (negative) to green (positive) gradient. The dashed red line square indicates significant differences in bacterial growth between dietary groups (Student’s t-test, p < 0.05).
In order to determine the effects of dietary supplementation of the yeast D. hansenii upon the S. aurata skin transcriptome, we conducted a microarray-based transcriptomic analysis. As results from fish condition neither indicated statistical differences between the Control and Yeast2.2% groups nor linear regression determined a dose-response effect, skin transcriptomics was only analyzed from fish fed the Yeast1.1% diet that showed the best results in terms of BWf and FCR values. A total of 712 differentially expressed genes (DEGs) were found in the skin of S. aurata fed the Yeast1.1% diet (Figure 4 and Supplementary Table 1). The DEGs were classified as up-regulated (UR-DEGs; N = 380) or down-regulated (DR-DEGs; N = 332) and ordered by their fold-change (FC) as shown in Figure 4A. The FC values obtained showed similar modulations at high-expressions ([FC ≥ 2], UR-DEGs = 11.3% and DR-DEGs = 15.7%), but at mid- and low-expressions the modulation percentages were remarkable ([1.5 ≤ FC < 2], UR-DEGs = 9.7% and DR-DEGs = 23.2%; [1 ≤ FC < 1.5], UR-DEGs = 79% and DR-DEGs = 61.1%). When comparing both inter- and intra-specific profiles from both experimental groups using a hierarchical clustering heatmap, a common aggregation among pool samples and an inverted response based on dietary administration was observed (Figure 4C). These results were supported by the PCA analysis showing differential profiles among dietary treatments in a Euclidean 3D space (Figure 4B).
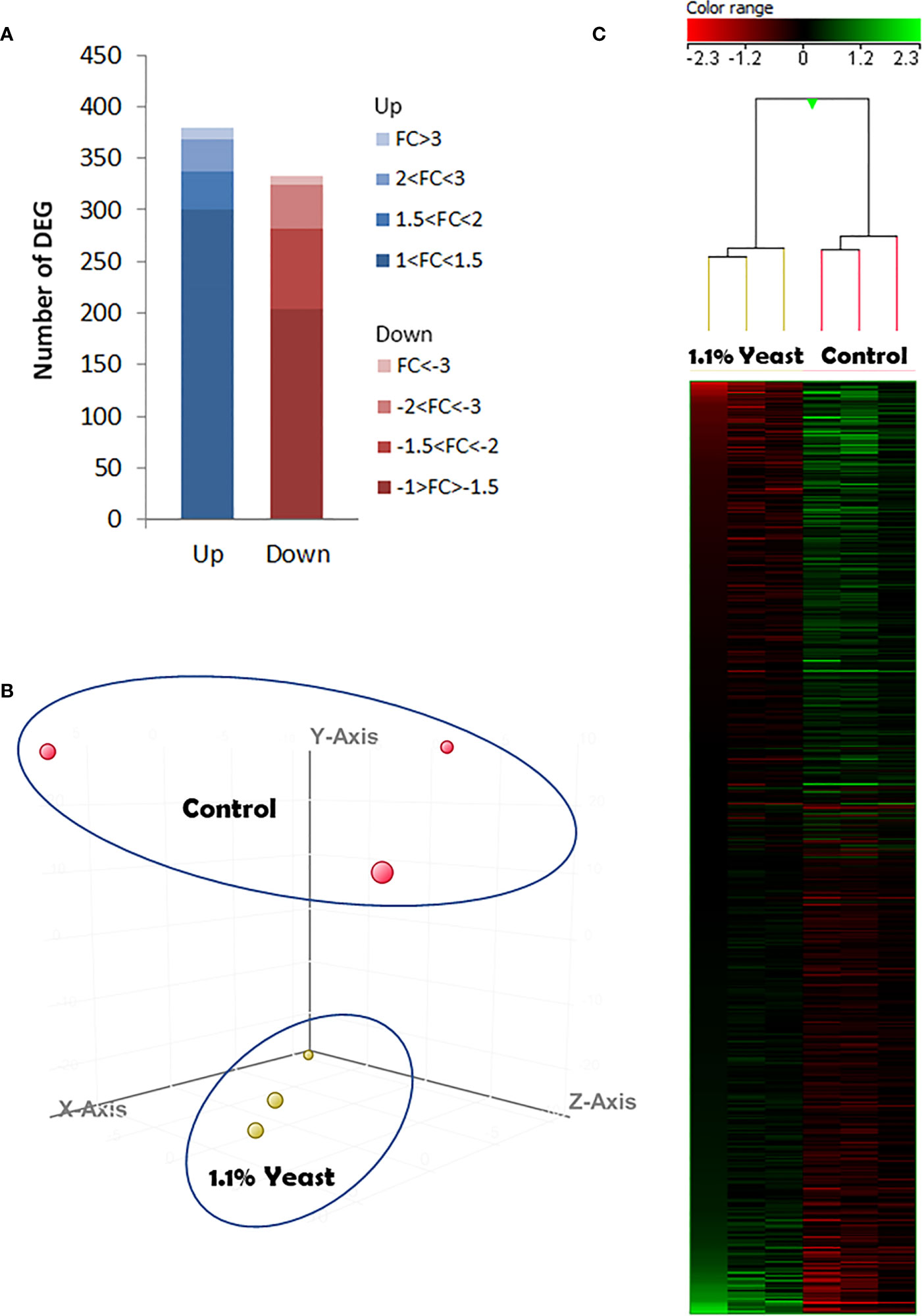
Figure 4 Skin microarrays-based transcriptomic analysis for the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). (A) The number of total differentially expressed genes (DEGs). The blue (up-regulated) and red (down-regulated) color schemes indicate the gene modulation according to its magnitude interval (fold-change). (B) 3D Principal component analysis (PCA). Control (red spheres) and D. hansenii groups (yellow spheres) are represented (each array included in the study is represented by one sphere). (C) Hierarchical clustering heatmap representing the 712 DEGs. The results of each microarray analyzed for the control and D. hansenii group are shown. The green (up-regulation) and red (down-regulation) color schemes indicate the gene normalized intensity values according to its magnitude interval.
Considering the complete list of DEGs, a transcriptomic-based biological network (transcripteractome) was generated containing 312 nodes (DEGs) which resulted in 478 interactions (edges) (Figure 5). The remaining DEGs (N = 388) were annotated as unknown genes and, therefore, these were excluded from the current analysis.
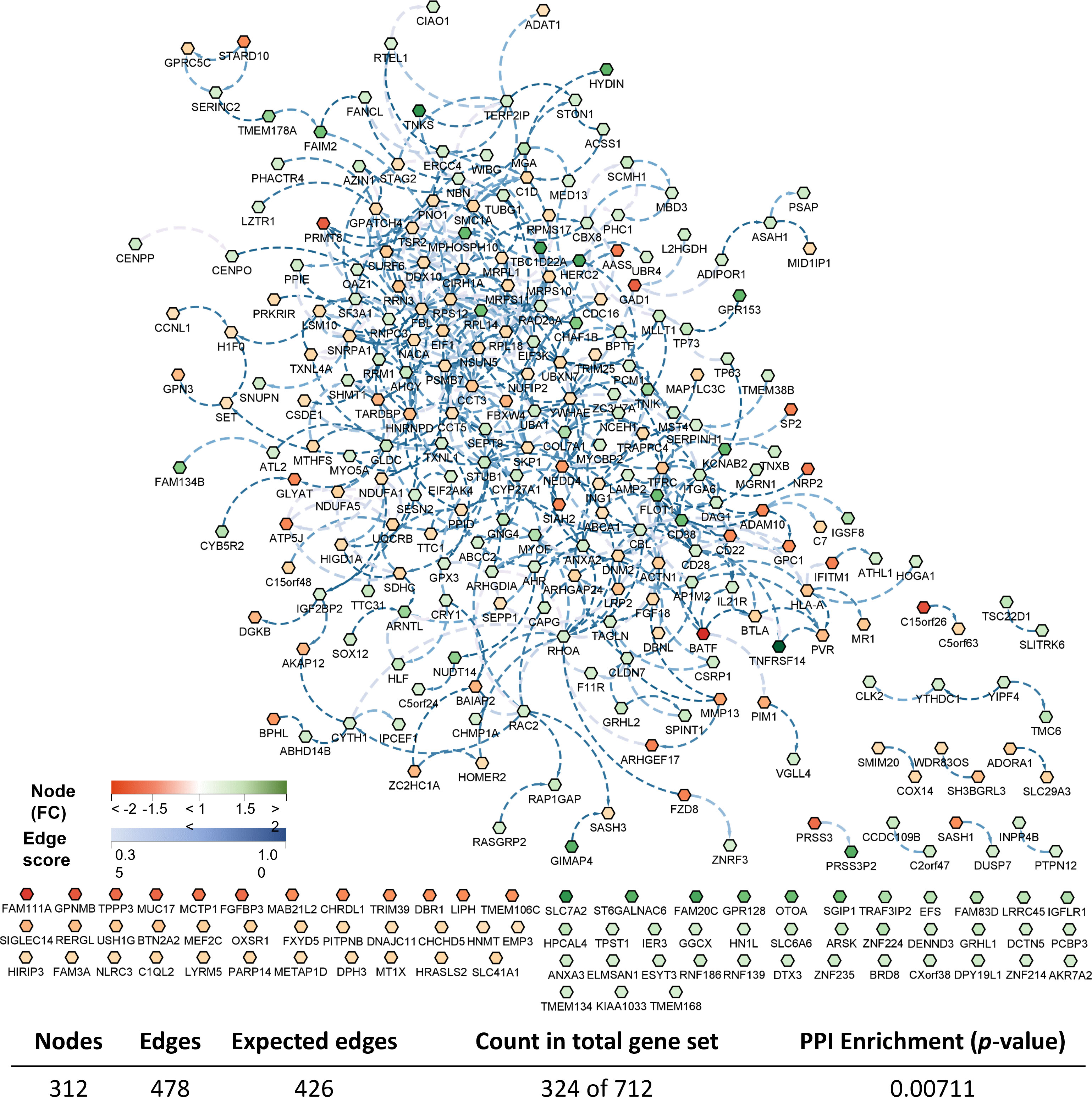
Figure 5 Biological network of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast diet) or devoid of the probiotic (Control diet). Node fills with a continuous mapping color from red (down-regulated DEGs) to green (up-regulated DEGs) represent the fold change (FC) intensity. The edge score was represented by dotted lines with a continuous mapping from 0.3 (light Blue) to 1.00 (dark blue). The transcripteractome was obtained using Cytoscape (v3.9.1) platform with String (v1.7.1). For details, please refer to Supplementary Table 1.
The enrichment analysis identified several biological processes and pathways in the skin of S. aurata when comparing both experimental groups. Thus, most of the Gene Ontology (GO) annotations obtained from the transcripteractome were associated with metabolic, regulatory, and symbiotic interaction-related processes. Selecting the genes associated with the largest subnetwork, which denotes the greatest quantity of interactions described, the enrichment analysis identified 29 GO Biological Processes (GO-BP), 5 GO Molecular Functions (GO-MF), and 13 GO Cellular Components (GO-CC) (Figure 6 and Supplementary Table 2), increasing the protein-protein interaction enrichment p-value up to 1.75 x 1012.
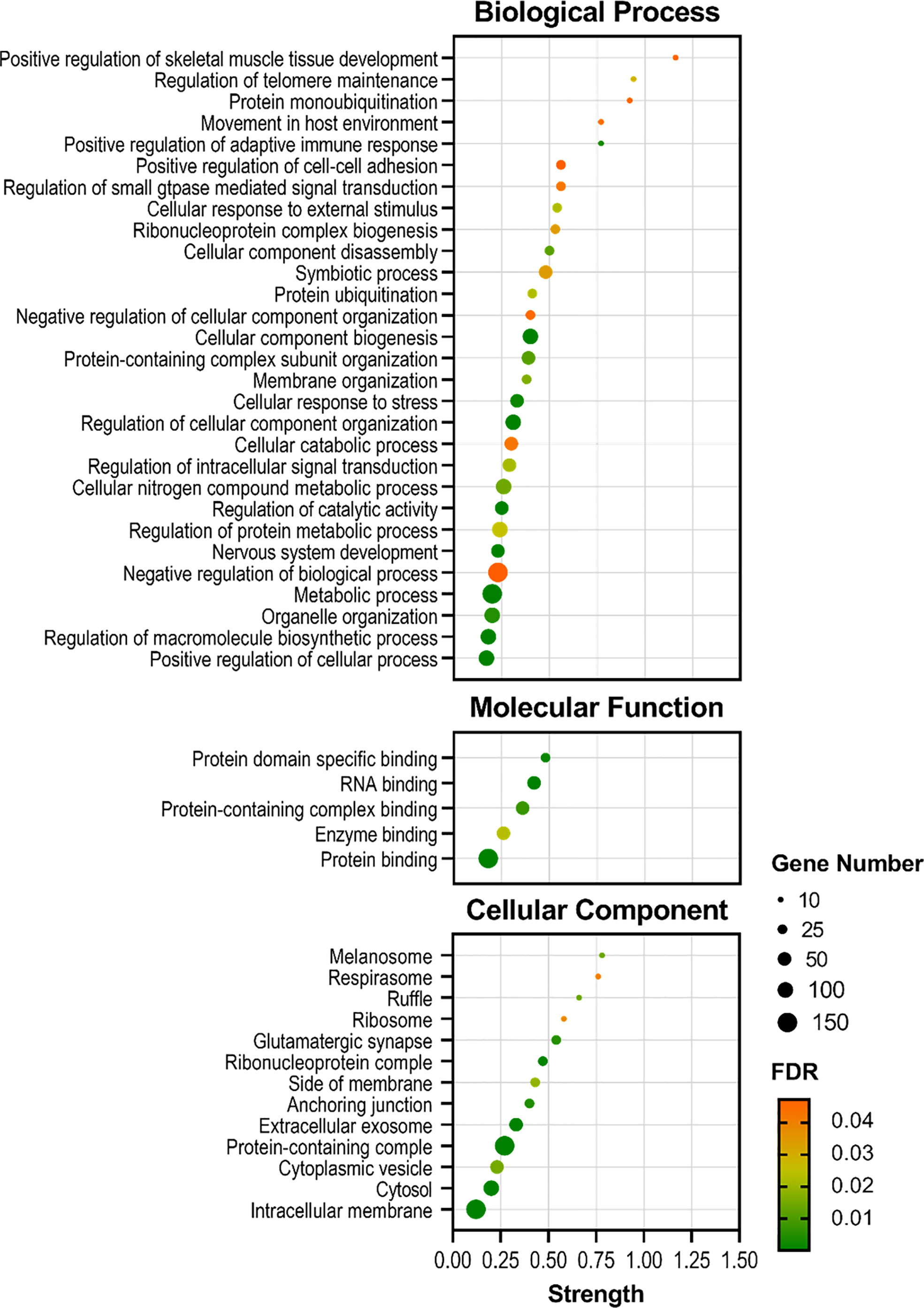
Figure 6 Bubble plots showing the enrichment analysis of the largest DEGs subnetwork (P < 0.05) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The axis variable assignments show: x-axis = strength (Log10(genes observed/genes expected)); y-axis = GO terms (in Biological Process, Molecular Function, and Cellular Component aspects). The size of the black circles associates with the gene number involved in each GO term. The color palette from green (low) to red (high) denotes the False Discovery Rate (FDR; significance of the enrichment p-values) of each GO term. For details, please refer to Supplementary Table 2.
According to the results reported by the enrichment analysis, the GOs were classified into seven representative clusters of Biological Processes as depicted in Figures 7–13 and Supplementary Table 3. The largest cluster was related to metabolic pathways (Figure 7) and contained 171 DEGs (54.8% of known DEGs). The second cluster was related to regulatory mechanisms (Figure 8) and was represented by 140 DEGs (44.9% of the known DEGs). The third cluster encompassed cellular processes and specifically those involved in the cellular component organization (Figure 9), with 104 associated DEGs (33.3% of known DEGs). The following three clusters were related to defense and environmental interactions: a cluster of response to stimulus (72 DEGs; 23.1% of the known DEGs) (Figure 10), a cluster related to immunity (61 DEGs; 19.6% of the known DEGs) (Figure 11), and a cluster related with biological process involved in interspecies interaction between organisms (29 DEGs; 9.3% of the known DEGs) (Figure 12). The last cluster grouped 46 DEGs (14.7% of the known DEGs) and was related to developmental processes (Figure 13).
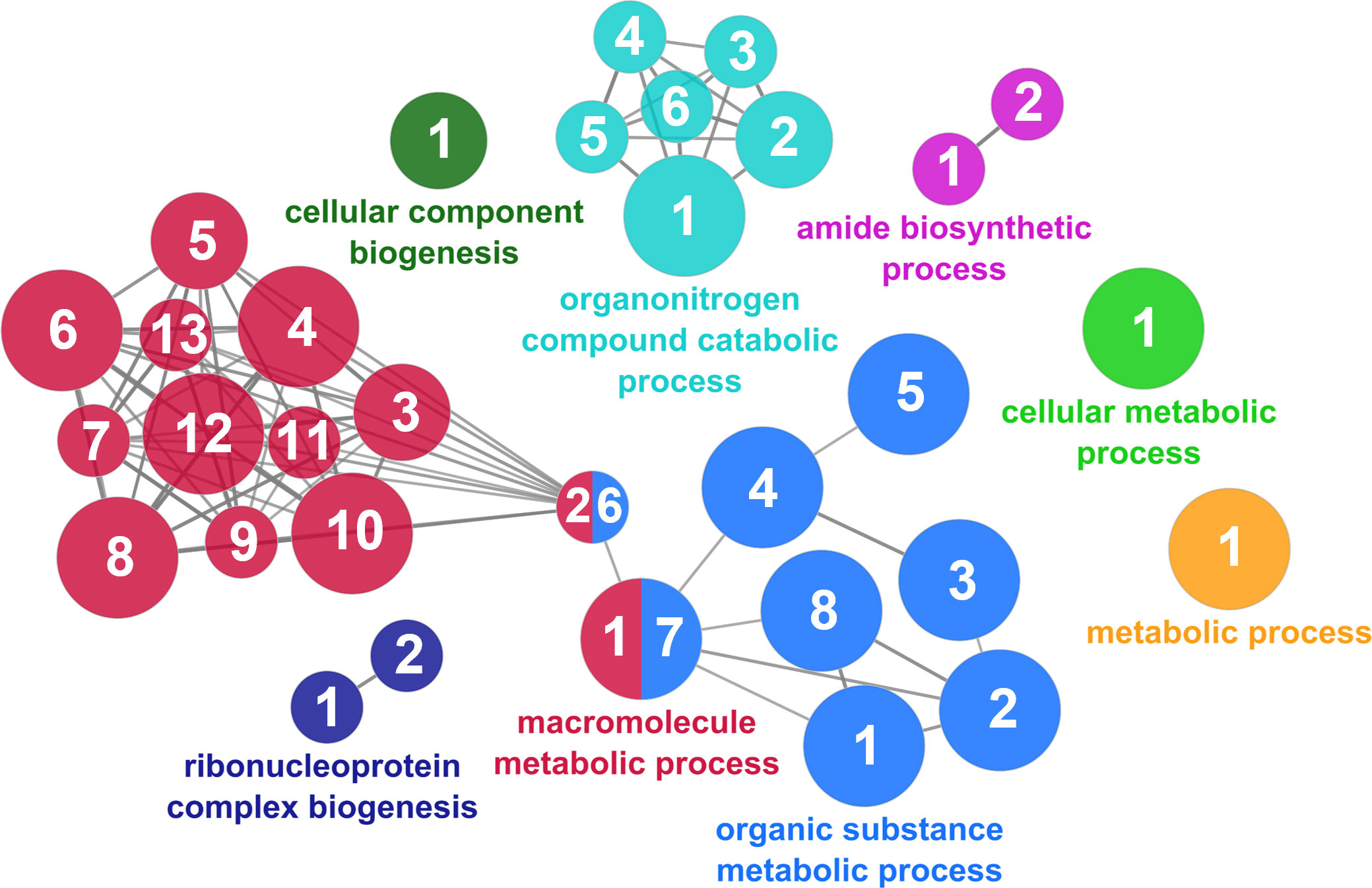
Figure 7 The metabolic cluster of the functional enrichment network for biological processes based on the related functions of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The node size determines the significance (Bonferroni step-down correction; small p < 0.05; medium p < 0.005; large p < 0.0005), the node color determines the principal GO process aggrupation, and the node numbers indicate different GO processes classified in Supplementary Table 3.
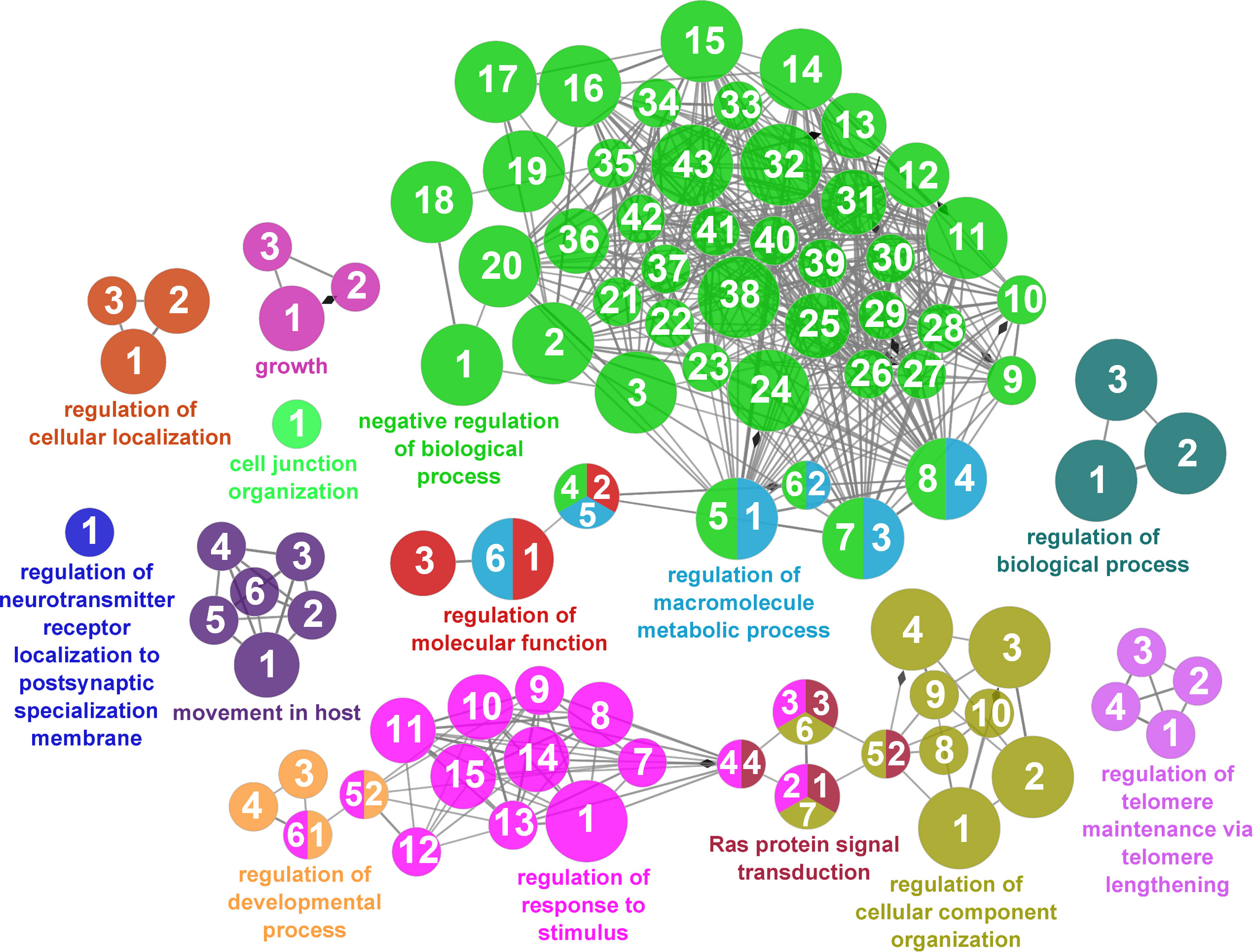
Figure 8 Regulatory mechanisms cluster of the functional enrichment network for biological processes based on the related functions of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The node size determines the significance (Bonferroni step-down correction; small p < 0.05; medium p < 0.005; large p < 0.0005), the node color determines the principal GO process aggrupation, and the node numbers indicate different GO processes classified in Supplementary Table 3.
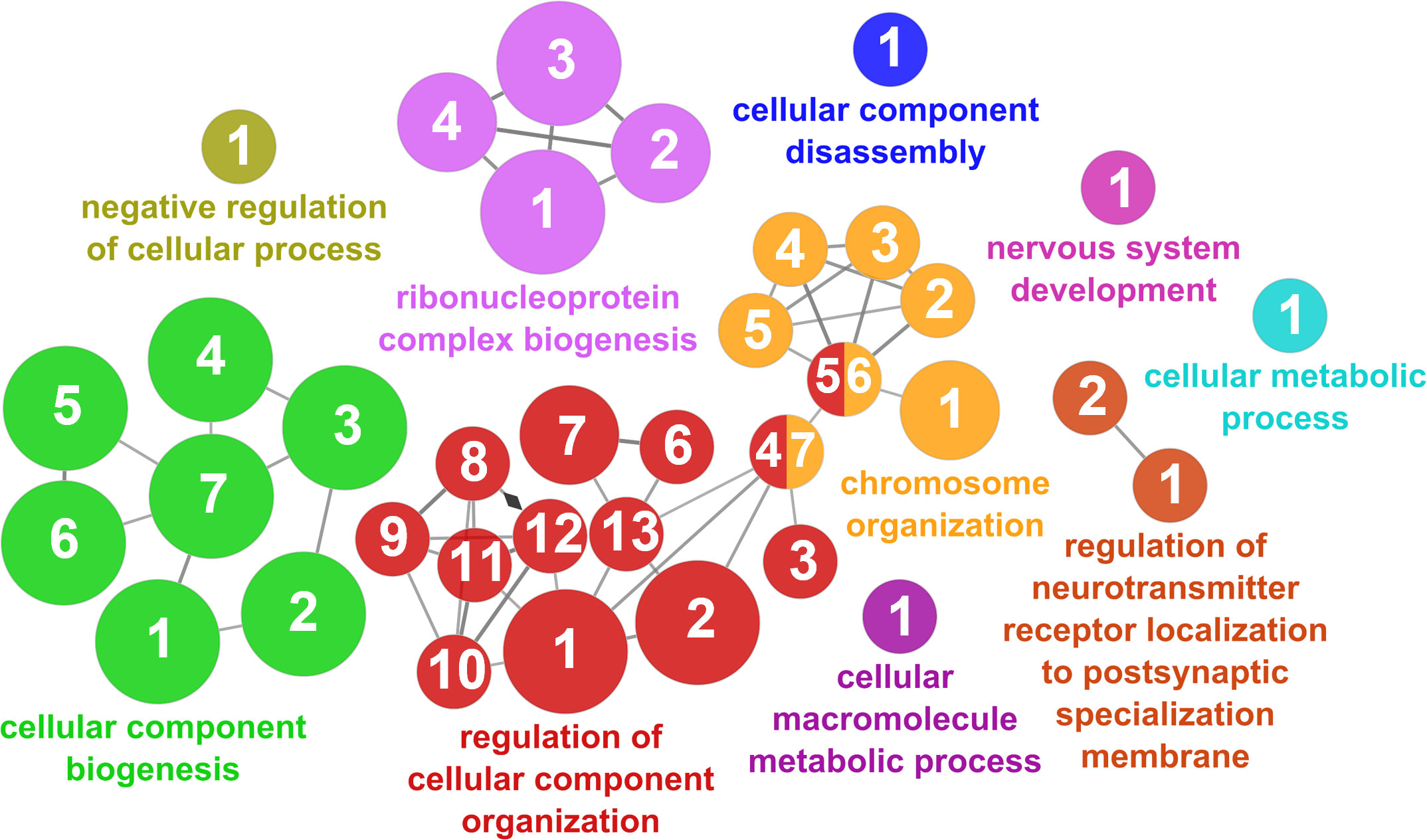
Figure 9 Cellular component organization cluster of the functional enrichment network for biological processes based on the related functions of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The node size determines the significance (Bonferroni step-down correction; small p < 0.05; medium p < 0.005; large p < 0.0005), the node color determines the principal GO process aggrupation, and the node numbers indicate different GO processes classified in Supplementary Table 3.
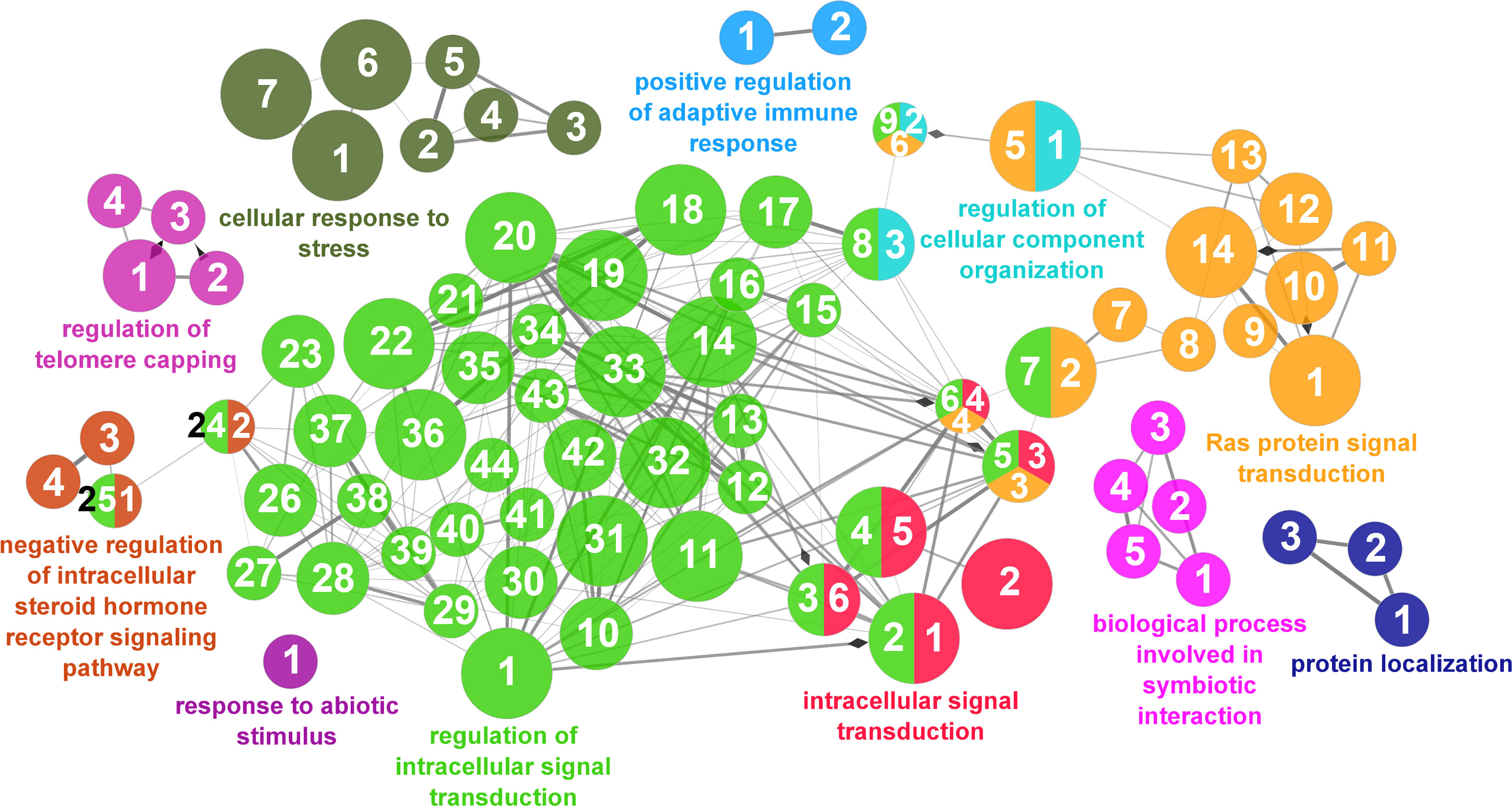
Figure 10 Response to stimulus cluster of the functional enrichment network for biological processes based on the related functions of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The node size determines the significance (Bonferroni step-down correction; small p < 0.05; medium p < 0.005; large p < 0.0005), the node color determines the principal GO process aggrupation, and the node numbers indicate different GO processes classified in Supplementary Table 3.
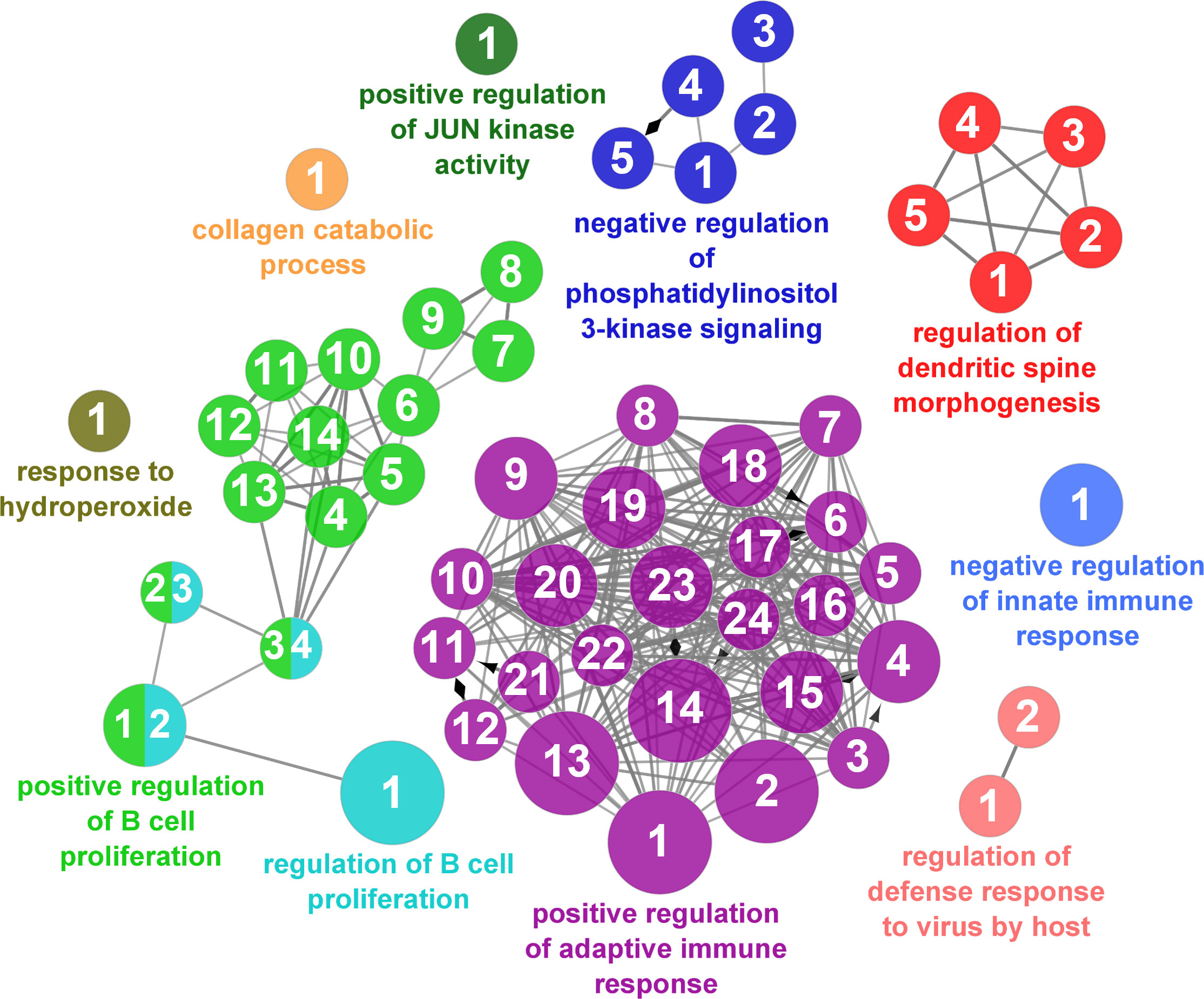
Figure 11 Immune processes cluster of the functional enrichment network for biological processes based on the related functions of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The node size determines the significance (Bonferroni step-down correction; small p < 0.05; medium p < 0.005; large p < 0.0005), the node color determines the principal GO process aggrupation, and the node numbers indicate different GO processes classified in Supplementary Table 3.
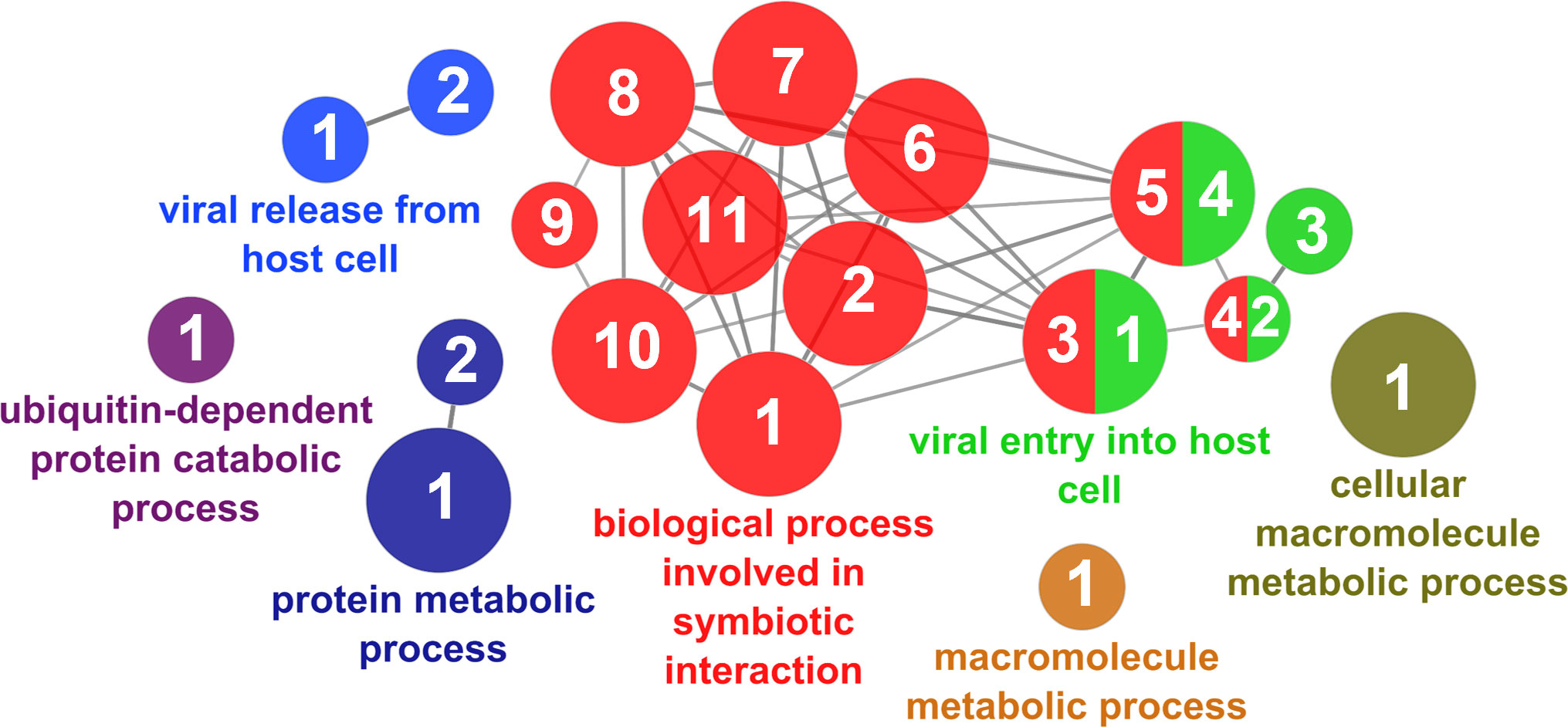
Figure 12 Interspecies interaction between organisms cluster of the functional enrichment network for biological processes based on the related functions of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The node size determines the significance (Bonferroni step-down correction; small p < 0.05; medium p < 0.005; large p < 0.0005), the node color determines the principal GO process aggrupation, and the node numbers indicate different GO processes classified in Supplementary Table 3.
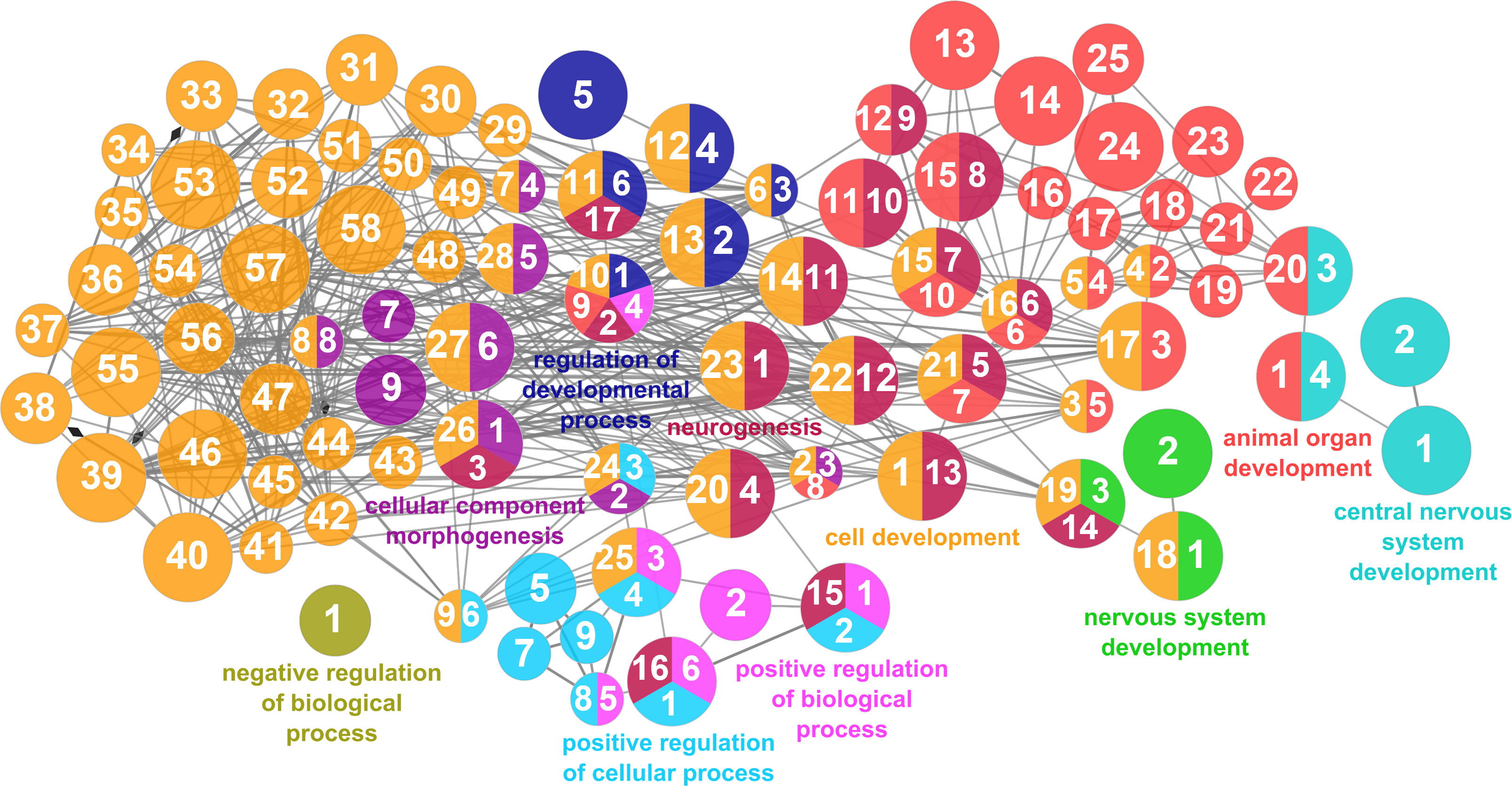
Figure 13 Developmental processes cluster of the functional enrichment network for biological processes based on the related functions of the differentially expressed genes (DEGs) in the skin of gilthead seabream (Sparus aurata) fed experimental low fishmeal diets containing Debaryomyces hansenii (1.1%, 17.2 x 105 cfu; Yeast1.1% diet) or devoid of the probiotic (Control diet). The node size determines the significance (Bonferroni step-down correction; small p < 0.05; medium p < 0.005; large p < 0.0005), the node color determines the principal GO process aggrupation, and the node numbers indicate different GO processes classified in Supplementary Table 3.
In order to elucidate the main activities of DEGs that occur at the molecular level, an enrichment analysis was performed to elucidate different molecular functions related to binding functional terms (Figure 6). Results showed that at the molecular level, the functions of DEGs in fish fed the Yeast1.1% diet were mainly related to protein-containing complex, enzymatic, and RNA-binding associations. Additionally, the locations relative to the cellular structure in which the DEGs performed these biological and molecular functions were mainly structural and at cytosolic spaces. Anchoring, membrane, and vesicle cellular components showed to be the locations of the majority of the DEG products, and to a lesser extent, among some cellular components like the ribosome, respirasome, and melanosome structures. These functions and distributions indicate an increased interaction and response modulations on fish skin cells produced by the dietary supplementation of D. hansenii.
The modulation of health and immunity through dietary strategies is considered a reliable alternative to the use of chemicals and drugs, in addition to being considered a proactive and sustainable practice in farmed animals (40). To this end, probiotic, prebiotic, and symbiotic supplements in diets, together with their constitutive bioactive components, offer viable alternatives to the use of chemical modulators, improving health and fish defense in addition to providing benefits in animal performance and/or improving several physiological processes (41, 42). This is of special interest to the aquaculture industry, a sector vulnerable to different biotic and abiotic challenging episodes, which usually works with shorter margins of error compared to other production industries (43, 44). With this aim, the present study explored the effects of D. hansenii (CBS 8339) supplementation in a low fish meal-based diet on the skin mucosa of S. aurata juveniles. Unlike other mucosal tissues, skin covers the majority of the fish’s body surface. Therefore, it acts as the first frontier between the animal and the surrounding environment, giving it a role as the main defensive barrier.
The holistic approach proposed in this study combined a transcriptomic analysis of the skin tissue and the study of its exuded mucus in terms of SMABs and its bacterial defense capacity. When included in the diet, D. hansenii resulted in an improvement of somatic growth and feed efficiency parameters and supported the immune system in S. aurata. However, the highest supplementation level tested (2.2%) did not provide any significant improvement among the key performance indicators assessed in the current study (same levels as control), which indicated that there was not a dose-response effect of the probiotic administration, suggesting the 1.1% level as the optimal supplementation (45). Therefore, in this study, the Yeast1.1% diet showed enhanced performance indicators, which agree with other studies reporting the improvement of the fish condition in S. aurata at similar levels (46, 47). However, current results differ from other freshwater species presenting no advantages at any supplementation level (48). The significance of these findings was already discussed by Sanahuja et al. (21). In previous reports, it was described that D. hansenii possesses bioactive components with immunomodulatory and digestive promotion effects, like β-glucans and polyamines, whose actions are reported to be indispensable for some biochemical pathways related to cell differentiation and proliferation (19, 22, 49). To test if these effects benefit the skin mucosa, the supplementation level was considered at 1.1% since ultimately, to be considered beneficial for animal health and welfare, the improvement of performance is decisive along with its defensive capacity against biotic and environmental agents.
A detailed transcripteractome was performed from Biological processes, Molecular functions, and Cellular components to unravel the modulatory effects of the yeast-based supplemented diet on the skin mucosal tissue. The enrichment analysis revealed multiple GO annotations corresponding to binding molecular functions, mainly “Protein binding” (GO.0005515), “Enzyme binding” (GO:0019899), and “Protein-containing complex binding” (GO:0044877). In addition, these functions are associated with the presence of regulatory biological processes, which might be correlated with the defensive pluripotency attributed to the fish skin. Specifically, among the main mucosal tissues (i.e., gill, gut, and skin), the skin is considered the principal defensive barrier since it covers almost the entire body of the fish (6, 50). In this line, the analysis of the “Cellular component” GO showed that the DEGs performed their functions, especially in membranes (“Intracellular membrane”, GO:0043231; “Side of membrane”, DO:0098552) in addition to being part of vesicles and exosomes (“Cytoplasmic vesicle”, GO:0031410; “Extracellular exosome”, GO:0070062). In fish, the skin tissue is composed of active cells, with a high turnover ratio to maintain the viability and protective barrier function of this tissue (51). Moreover, the skin epidermis also contains specialized mucus-producing cells which generate and secrete the mucus layer that covers the skin. Therefore, the relation of these biological aspects could represent an improvement in the barrier function of the skin, either by modulating the secretion and/or the composition of the mucus (52) or by enhancing the physical protection capacity at cell-cell junctions or/and cell and extracellular matrix levels (“Anchoring junction”, GO:0070161). These effects are crucial to protecting the organism from environmental fluctuations and biotic stressors (5, 53). The above-mentioned structural and functional characteristics based on the transcriptomic data obtained demonstrate that the supplementation at 1.1% of D. hansenii in a low fish meal-based diet (7%) was capable of modulating and supporting skin functions in S. aurata. These results may be attributed to the contribution of bioactive compounds from D. hansenii since this yeast species is rich in β-glucans, polyamines, and mannan oligosaccharides (19, 20, 54).
Different studies in fish have shown the possibility of modulating skin functions through dietary strategies (27, 30). In our study, we have observed a modulation of some pathways related to amide metabolism (aass, abcc2, acss1, adam10, asah1, eif1, eif3k, glyat, mrpl1, mrps10, mrps11, mthfs, naca, psap, rpl14, rpl18, rpms17, rps12, and shmt1) and amino acid metabolism (shmt1, gldc, glyat, aass, ahcy, serinc2, and arhgef17). Similar findings related to the modulation of amide metabolism were reported in a parallel study in which we evaluated the effect of D. hansenii dietary supplementation at the intestinal level in the same fish species (21); the results may be attributed to the high content of polyamines in this yeast (19). Polyamines are a part of and regulate a large variety of key cellular processes, such as cell growth, differentiation, or proliferation, whose control is crucial to maintain cell homeostasis (55). In the skin, we found two upregulated genes involved in polyamine regulation, oaz1 and azin1. In response to elevated levels of polyamines, ornithine decarboxylase antizyme 1 (OAZ1) and antizyme inhibitor 1 (AZIN1) regulate the entry of polyamine compounds into cells and may inhibit ornithine decarboxylase (ODC). Thereby, these molecules initially regulate polyamine metabolism (55, 56). In addition to this pathway, the slc6a6 gene was upregulated in our study, whose function is the transport of taurine, urea, and biogenic amine (57). This modulation of amide metabolism and its related transport mechanisms are undoubtedly of great interest since D. hansenii dietary supplementation not only has effects at the intestinal level (21) but also at the skin mucosa level as herein observed, which may indicate a similar effect at cell growth, differentiation, or proliferation levels in different MALTs. Moreover, polyamines and their derived compounds participate in protein metabolism, which is highly relevant in carnivorous fish (58). In this sense, the enrichment analysis revealed several biological processes involved in the regulation of protein turnover, suggesting a modulation of the amino acid and protein metabolism in the fish skin by D. hansenii supplementation. Our study also revealed that the slc7a2 gene, an amino acid transporter member of the amino acid-polyamine-organocation (57, 59), was upregulated in the skin cells. The SLC7A2 transporter is responsible for the cellular uptake of arginine, lysine, and ornithine. An increased uptake of arginine and ornithine, which also participates in polyamine metabolism, supports the above-mentioned hypothesis of increased polyamine functions in the skin through dietary uptake. Moreover, the expression and functions of SLC7A2 have been highly related to both innate and adaptive immunity. In particular, SLC7A2 has been reported to control macrophages’ function toward a bacterial infection and under inflammation in mammal peripheral monocytes (60). The up-regulation of these DEGs related to protein metabolism and polyamine turnover might explain a more efficient use of protein sources (61), which would support the improvement of intestinal health status and nutrient digestion (21), altogether with modulation of immune-related processes.
Furthermore, several mediators of the immune response were also observed to be upregulated in the skin of S. aurata fed the Yeast1.1% diet. For instance, the TNF Receptor Superfamily Member 14 (tnfrsf14) gene expression was highly increased. The TNFRSF14 membrane-bound receptor binds several ligands mediating both coinhibitory and costimulatory pathways. Therefore, TNFRSF14 regulates immune cell homeostasis (62–64). In accordance, among its ligands, we found that it down-regulated the B- and T-lymphocyte attenuator (btla) gene, a member of the CD28 family, and its interaction with TNFRSF14 delivers a coinhibitory signal regulating T-cell activation (65). However, due to their wide cellular distribution, their functional analysis is complex. For instance, the balance herein observed in their expressions suggests a naïve or final stage of the T-cell activation phase, and/or an immature phase of dendritic cells (66). In this line, we observed the upregulation of the cd28 gene, a costimulatory receptor that triggers T-cell activating functions. Thus, the lack of modulation of costimulatory T cell receptors (TCR), like their cognate CD80/CD86 ligands, and/or interleukin modulatory expressions, allowed us to discard a proliferative T-cell response associated with the upregulation of CD28, supporting the idea of a naïve/resting stage of T-cells modulation in the skin of S. aurata after feeding with the Yeast1.1% diet for 70 days (67, 68). In addition, T-cell activation requires, among the above-mentioned signals, a complex formed by MCH with T-cell receptors (TCR) (69). Under this scenario, the downregulation of the human leukocyte antigen (hla-a) gene, a major histocompatibility complex class I (MHC-I) molecule present in the cell surface of all nucleated cells, along with genes related to “positive regulation of T cell activation” (btla, tfrc, and sash3; GO:0050870), coupled to the modulation of genes related to “regulation of T cell proliferation” (sox12, and btn2a2; GO:0042129), would also support the above-mentioned hypothesis about the T-cell stage found under the current experimental conditions. Regarding B-cell regulation, one of the co-stimulatory signals for B-cell activation is the CD22 molecule as it has been described in turbot (Scophthalmus maximus) and Japanese flounder (Paralichthys olivaceus) (70). In our study, fish fed the Yeast1.1% diet presented a downregulation of the cd22 gene, which has been suggested as one of the B-cell biomarkers in Atlantic salmon (Salmo salar) as it is consistently expressed in IgM+ or IgT+ lymphoid cells together with cd79a (71). Under the current experimental conditions, the enrichment analysis of DEGs showed several regulatory processes related to B-cell proliferation and adaptative immune responses coupled with the observed upregulation of the cd38 gene. In this sense, CD38 participates in the regulation of intracellular Ca2+, a cell signaling system, which in turn modulates immune functions. In fish, CD38 has been shown to participate in antiviral and antibacterial B-cell responses, and its expression seems to modulate different B-cell stages (72). However, the enrichment analysis herein shows several repressed genes involved in the positive activation and proliferation of B-cells (i.e., mef2c, tfrc, and sash3; GO:0050871 and GO:0030890, respectively). The functionality and modulation of these above-mentioned lymphocyte-related genes reinforced the hypothesis of an immunomodulatory effect of D. hansenii on the skin of fish. With the downregulation of the membrane-attack complex member c7 gene, its function and expression have been observed in response to infection in fish (73), and the modulation of several viral processes and antiviral genes (i.e., trim25, pvr, f11r, among others) suggest that the supplementation of D. hansenii in low fish meal-based diets is capable of modulate and regulate immune functions in the skin of S. aurata. Similar results related to T and B cell development were reported when comparing the effect of D. hansenii on the skin (present study) and the gut in S. aurata fed the Yeast1.1% (21) even though different DEGs were involved in this MALTs, which may be attributed to different tissue-specific response to dietary immunomodulators, being that the gut is more tolerogenic than the skin (3, 50).
Regarding the function of the skin protecting the fish, one of the most distinctive features is the production and secretion of mucus by goblet cells and club cells (74). The mucus layer is mainly composed of glycoprotein molecules called mucins, soluble proteins, several metabolites, and hormones, among other compounds (28, 34). In our study, we found that D. hansenii dietary supplementation could modulate mucin properties through mucin synthesis and secretion. For example, through the upregulation of the GalNAc sialyltransferase (st6galnac6) gene, which adds terminal sialic acids contributing to the mucin negative charges (75). This is of special relevance since sialic acid can act as ligands for receptors on viruses, acting as competitive inhibitors, blocking receptor binding, and limiting infection of epithelial cells (76, 77). Moreover, the upregulation of the myo5a gene, a class V myosin motor capable of moving along actin filaments (78), and the esyt3 gene, a synaptotagmin Ca2+ sensor required for vesicle fusion, might suggest an increased mucin secretion (75, 79). However, this hypothesis could not be supported by an increase in the expression of MUC18, which has been described as the most abundant mucin in S. aurata that plays an important role in adhesion and protection against pathogens (80). In our study, we found a repressed expression of muc17, a membrane-bounded and no secreted mucin, which could indicate a differential modulatory pattern for skin mucins under the present experimental conditions. However, the available information regarding this specific issue is limited, and further studies are needed to discern the complex pattern of mucins, their changes, and their implications in skin mucus homeostasis.
Regarding mucus composition, the SMABs profile analyzed did not show major changes except for a slight increase in skin mucus glucose levels. Glucose, lactate, and protein levels are markers of metabolic and stress factors in fish mucus (34). Increased glucose levels could indicate a stressful condition or a switch in metabolic status. However, the levels of cortisol, the major corticosteroid indicator of stress in fish (81), remained constant through dietary groups. Indeed, when the levels of metabolites were standardized per protein ratios, the Control and Yeast1.1% diets did not show differences in glucose levels, but higher levels on the Yeast2.2% were exhibited. Taking this into consideration, these results also supported that the above-mentioned hypothesis of increased mucus production, as transcriptomic data hinted, is due to the inclusion of yeast in diets. As increased mucus production is shown to be the first defensive mechanism in fish (82), this might be considered a beneficial measure against potential biotic and abiotic stressors due to its incorporated defensive components, as the in vitro skin mucus co-culture with pathogenic bacteria also indicated. Among the above-mentioned metabolites, the skin mucus is also composed of a great variety of innate immune components and defensive molecules that, working together, confer the defensive capacity to the mucus (5, 83). Hernández-Contreras et al. (31) found increased activity of several immune-related enzymatic parameters in the skin mucus (e.g., proteases and lysozyme) when fish were fed a diet supplemented with D. hansenii. In our study, some enzymatic-related genes were modulated, specifically two protease genes (prss2, and prss3), and one phosphatase inhibitor-related gene (set). The upregulation of the serine protease 2 (prss2), which participates in the formation of antimicrobial peptides (AMP) (84, 85), could suggest an increased AMPs level, potentiating the antibacterial defense. The modulation of these defense components has been shown in the grater amberjack fish (Seriola dumerili) when fed a diet supplemented with yeast-derived mannan oligosaccharides (54). This suggests that yeast components might modulate skin defense components potentiating its defensive capacity. Accordingly, the observed reduced growth capacity of V. anguillarum in the skin mucus of S. aurata fed yeast-supplemented diet is especially relevant because Vibrio strains are capable to adhere to surface cells and penetrate host epithelial and vascular tissues (86). Besides, P. anguilliseptica growth was also affected, exhibiting in the final 2 hours of co-culture a decreased growth. Pseudomonas is considered an opportunistic pathogen whose infections in S. aurata occur under environmental stress (87). The results observed herein suggest that feeding the fish with D. hansenii not only could increase mucus production but also could increase its enzymatic and antibacterial defense components, thus protecting the host from potential pathogenic bacteria (88).
In the same line, we found the upregulation of antioxidant-related DEGs (txnl1, gpx3, ipcef1, and sesn2). The antioxidant modulatory defense effects of D. hansenii have been demonstrated in different animal species including fish (46, 89, 90). However, this modulating effect is not reflected in the total antioxidant capacity of the skin mucus, whose values in the Yeast1.1% diet remained close to those from the Control diet. Under certain situations, the skin mucus’s antioxidant capacity is promoted to protect the mucosal surface from oxidative attack, either due to environmental stress (91, 92) or skin disorders (93). Due to the lack of oxidative challenging situations, we hypothesize that the dietary yeast supplementation promoted sentinel antioxidant defensive processes rather than activating response stress by reactive oxygen species (ROS) presence, which is in agreement with other studies focused on functional feeds (30, 32). In addition, skin transcriptomics did not reflect responses linked to oxidative damage but to the downregulation of several genes involved in oxidative phosphorylation, which is the main metabolic ROS producer in cell metabolism (94, 95). This may support the hypotheses that D. hansenii supplementation might result in a better condition of the skin, including a tight regulation of its immune response, enabling a more efficient use of available energy and nutrients for growth (21).
The present holistic study of the skin response to the dietary administration of the probiotic D. hansenii in low fish meal-based diets when administered to S. aurata juveniles provides new insights into its effects on fish skin condition and defensive function. The results observed herein on fish performance indicators demonstrated that D. hansenii, when supplemented at 1.1% (17.2 x 105 cfu), enhances S. aurata juveniles’ growth performance and feed efficiency, whereas when supplemented at higher levels (2.2%; 35.7 x 105 cfu) do not provide any advantages. In addition, at the transcriptomic level, D. hansenii supplementation modulated innate immunity (i.e., T and B cell differentiation), increasing skin barrier and cell trafficking functions, potentiating the physical barrier of this tissue while maintaining its homeostatic immune and metabolic status. Besides, the skin mucus’s defensive capacity was enhanced by increasing its protective function against bacterial fish pathogens, which was associated with immunomodulatory compounds found in yeast. Thus, these results support the dietary administration of D. hansenii as an excellent tool to promote the skin mucosa defensive capacity, which is the first line of fish defense against environmental and biotic challenges, when included in low fish meal-based diets for S. aurata.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GPL13442, https://www.ncbi.nlm.nih.gov/, GSE162504.
The animal study was approved by This study was conducted following the Guiding Principles for Biomedical Research Involving Animals (EU2010/63), the guidelines of the Spanish laws (law 32/2007 and RD 53/2013) and authorized by the Ethical Committee of the Institute for Research and Technology in Food and Agriculture (Spain), and Generalitat de Catalunya government for the use of laboratory animals (FUE-2020–01719167). The study was conducted in accordance with the local legislation and institutional requirements.
EG designed and carried out the experiments. Biological samplings were performed by EG, LF-A, JF, AI, and IS. The transcriptomic data analysis and interpretation were performed by IS, ST, EV-V, and FR-L. LF-A and IS carried out the skin mucus analyses. DT-R reviewed and validated the methodology used in the study. The study was supervised by EG, AI, LT, and IS. IS wrote the original draft. All the authors provided critical feedback, read, and agreed to the published version of the manuscript.
This work has been financed through the DIETAplus project of JACUMAR (Junta de Cultivos Marinos, MAPAMA; Spanish government), which is co-funded with FEMP funds (EU). Furthermore, this research was funded by means of grants from the Spanish Government: PID2019-106878RB-I00, and IS was granted a Postdoctoral fellowship (FJC2020-043933-I). EV-V and FR-L acknowledge the support of Fondecyt iniciación (project number 11221308) and Fondecyt regular (project number 11221308) grants (Agencia Nacional de Investigacioíny Desarrollo de Chile, Government of Chile), respectively. Collaboration between Ibero-American researchers has been done under the framework of the network LARVAplus “Strategies for the development and improvement of fish larvae production in Ibero-America” (117RT0521) funded by the Ibero-American Program of Science and Technology for Development (CYTED, Spain).
We thank the technicians at the IRTA facilities for their valuable help during fish rearing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1247199/full#supplementary-material
1. Schempp C, Emde M, Wölfle U. Dermatology in the Darwin anniversary. Part 1: Evolution of the integument. J Dtsch Dermatol Ges (2009) 7(9):750–7. doi: 10.1111/j.1610-0387.2009.07193.x
2. Lazado CC, Caipang CMA. Mucosal immunity and probiotics in fish. Fish Shellfish Immunol (2014) 39(1):78–89. doi: 10.1016/j.fsi.2014.04.015
3. Gomez D, Sunyer JO, Salinas I. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol (2013) 35(6):1729–39. doi: 10.1016/j.fsi.2013.09.032
4. Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev (2005) 85(1):97–177. doi: 10.1152/physrev.00050.2003
5. Esteban MÁ. An overview of the immunological defenses in fish skin. ISRN Immunol (2012) 2012:853470. doi: 10.5402/2012/853470
6. Beck BH, Peatman E. Why mucosal health? In Mucosal Health in Aquaculture, pp:1-2. Acad Press Elsevier (2015) p:1–395.
7. Oliva-Teles A. Nutrition and health of aquaculture fish. J Fish Dis (2012) 35(2):83–108. doi: 10.1111/j.1365-2761.2011.01333.x
8. Gatlin DM III, Barrows FT, Brown P, Dabrowski K, Gibson Gaylord T, Hardy RW, et al. Expanding the utilization of sustainable plant products in aquafeeds: a review. Aquac Res (2007) 38(6):551–79. doi: 10.1111/j.1365-2109.2007.01704.x
9. Dawood MAO, Koshio S. Application of fermentation strategy in aquafeed for sustainable aquaculture. Rev aquac (2019) 12(2):987–1002. doi: 10.1111/raq.12368
10. Oliva-Teles A, Enes P, Couto A, Peres H. Replacing fish meal and fish oil in industrial fish feeds. In: Davis A, editor. Feed and Feeding Practices in Aquaculture. Woodhead Publishing Elsevier (2022). p. 231–681–454. doi: 10.1016/C2019-0-03580-7
11. Ringø E, Song SK. Application of dietary supplements (synbiotics and probiotics in combination with plant products and β-glucans) in aquaculture. Aquac Nutr (2016) 22(1):4–24. doi: 10.1111/anu.12349
12. Guerreiro I, Oliva-Teles A, Enes P, Alegre C. Prebiotics as functional ingredients: focus on Mediterranean fish aquaculture. Rev aquac (2017) 10(4):800–32. doi: 10.1111/raq.12201
13. Mugwanya M, Dawood MAO, Kimera F, Sewilam H. Updating the role of probiotics, prebiotics, and synbiotics for tilapia aquaculture as leading candidates for food sustainability: a review. Probiotics Antimicrob Proteins (2021) 14(1):130–57. doi: 10.1007/s12602-021-09852-x
14. del Valle JC, Bonadero MC, Fernández-Gimenez AV. Saccharomyces cerevisiae as probiotic, prebiotic, synbiotic, postbiotics and parabiotics in aquaculture: An overview. Aquaculture (2023) 569:739342. doi: 10.1016/j.aquaculture.2023.739342
15. Gatesoupe FJ. Live yeasts in the gut: Natural occurrence, dietary introduction, and their effects on fish health and development. Aquaculture (2007) 267(1–4):20–30. doi: 10.1016/j.aquaculture.2007.01.005
16. Teles A, Alvarez-González CA, Llera-Herrera R, Gisbert E, Salas-Leiva J, del Carmen Rodríguez-Jaramillo M, et al. Debaryomyces hansenii CBS 8339 promotes larval development in. Seriola rivoliana. Aquaculture (2022) 560:738587. doi: 10.1016/j.aquaculture.2022.738587
17. Angulo M, Reyes-Becerril M, Medina-Córdova N, Tovar-Ramírez D, Angulo C. Probiotic and nutritional effects of Debaryomyces hansenii on animals. Appl Microbiol Biotechnol (2020) 104(18):7689–99. doi: 10.1007/s00253-020-10780-z
18. Tovar-Ramírez D, Zambonino IJ, Cahu C, Gatesoupe FJ, Vázquez-Juárez R. Influence of dietary live yeast on European sea bass (Dicentrarchus labrax) larval development. Aquaculture (2004) 234(1–4):415–27. doi: 10.1016/j.aquaculture.2004.01.028
19. Reyes-Becerril M, Ascencio-Valle F, Tovar-Ramírez D, Meseguer J, Esteban MÁ. Effects of polyamines on cellular innate immune response and the expression of immune-relevant genes in gilthead seabream leucocytes. Fish Shellfish Immunol (2011) 30(1):248–54. doi: 10.1016/j.fsi.2010.10.011
20. Morales-Lange B, Djordjevic B, Gaudhaman A, Press CML, Olson J, Mydland LT, et al. Dietary inclusion of hydrolyzed Debaryomyces hansenii yeasts modulates physiological responses in plasma and immune organs of Atlantic salmon (Salmo salar) parr exposed to acute hypoxia stress. Front Physiol (2022) 13:836810. doi: 10.3389/fphys.2022.836810
21. Sanahuja I, Ruiz A, Firmino JP, Reyes-López FE, Ortiz-Delgado JB, Vallejos-Vidal E, et al. Debaryomyces hansenii supplementation in low fish meal diets promotes growth, modulates microbiota and enhances intestinal condition in juvenile marine fish. J Anim Sci Biotechnol (2023) 14:90. doi: 10.1186/s40104-023-00895-4
22. Reyes-Becerril M, Angulo M, Sanchez V, Guluarte C, Angulo C. β-D-glucan from marine yeast Debaryomyces hansenii BCS004 enhanced intestinal health and glucan-expressed receptor genes in Pacific red snapper Lutjanus peru. Microb Pathog (2020) 143:104141. doi: 10.1016/j.micpath.2020.104141
23. Machuca C, Méndez-Martínez Y, Reyes-Becerril M, Angulo C. Yeast β-glucans as fish immunomodulators: a review. Animals (2022) 12(16):2154. doi: 10.3390/ani12162154
24. Vargas O, Gutiérrez MS, Caruffo M, Valderrama B, Medina DA, García K, et al. Probiotic yeasts and Vibrio anguillarum infection modify the microbiome of zebrafish larvae. Front Microbiol (2021) 12:647977. doi: 10.3389/fmicb.2021.647977
25. Raggi P, Lopez P, Diaz A, Carrasco D, Silva A, Velez A, et al. Debaryomyces hansenii and Rhodotorula mucilaginosa comprised the yeast core gut microbiota of wild and reared carnivorous salmonids, croaker and yellowtail. Environ Microbiol (2014) 16(9):2791–803. doi: 10.1111/1462-2920.12397
26. Xu Z, Parra D, Gómez D, Salinas I, Zhang YA, Jørgensen LVG, et al. Teleost skin, an ancient mucosal surface that elicits gut-like immune responses. Proc Natl Acad Sci USA (2013) 110(32):13097–102. doi: 10.1073/pnas.1304319110
27. Reyes-López FE, Ibarz A, Ordóñez-Grande B, Vallejos-Vidal E, Andree KB, Balasch JC, et al. Skin multi-omics-based interactome analysis: integrating the tissue and mucus exuded layer for a comprehensive understanding of the teleost mucosa functionality as model of study. Front Immunol (2021) 11:613824. doi: 10.3389/fimmu.2020.613824
28. Sanahuja I, Ibarz A. Skin mucus proteome of gilthead sea bream: a non-invasive method to screen for welfare indicators. Fish Shellfish Immunol (2015) 46(2):426–35. doi: 10.1016/j.fsi.2015.05.056
29. Sanahuja I, Fernández-Alacid L, Sánchez-Nuño S, Ordóñez-Grande B, Ibarz A. Chronic cold stress alters the skin mucus interactome in a temperate fish model. Front Physiol (2019) 9:1916. doi: 10.3389/fphys.2018.01916
30. Firmino JP, Fernández-Alacid L, Vallejos-Vidal E, Salomón R, Sanahuja I, Tort L, et al. Carvacrol, thymol, and garlic essential oil promote skin innate immunity in gilthead seabream (Sparus aurata) through the multifactorial modulation of the secretory pathway and enhancement of mucus protective capacity. Front Immunol (2021) 12:633621. doi: 10.3389/fimmu.2021.633621
31. Hernández-Contreras Á, Tovar-Ramírez D, Reyes-Becerril M. Modulatory effect of Debaryomyces hansenii and oregano essential oil on the humoral immunity of skin mucus in Longfin yellowtail Seriola rivoliana. Aquac Res (2021) 52(2):749–62. doi: 10.1111/are.14931
32. Fernández-Alacid L, Sanahuja I, Madrid C, Polo J, Firmino JP, Balsalobre C, et al. evaluating the functional properties of spray-dried porcine plasma in gilthead seabream (Sparus aurata) fed low fish meal diets. Animals (2022) 12:3297. doi: 10.3390/ani12233297
33. Gisbert E, Skalli A, Campbell J, Solovyev MM, Rodríguez C, Dias J, et al. Spray-dried plasma promotes growth, modulates the activity of antioxidant defenses, and enhances the immune status of gilthead sea bream (Sparus aurata) fingerlings. J Anim Sci (2015) 93(1):278–86. doi: 10.2527/jas.2014-7491
34. Fernández-Alacid L, Sanahuja I, Ordóñez-Grande B, Sánchez-Nuño S, Viscor G, Gisbert E, et al. Skin mucus metabolites in response to physiological challenges: a valuable non-invasive method to study teleost marine species. Sci Total Environ (2018) 644:1323–35. doi: 10.1016/j.scitotenv.2018.07.083
35. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem (1976) 72(1–2):248–54. doi: 10.1006/abio.1976.9999
36. Fernández-Alacid L, Firmino JP, Sanahuja I, Madrid C, Polo J, de Borba MR, et al. Impact of dietary porcine blood by-products in meagre (Argyrosomus regius) physiology, evaluated by welfare biomarkers and the antibacterial properties of the skin mucus. Fish Shellfish Immunol (2021) 118:241–50. doi: 10.1016/j.fsi.2021.09.011
37. Firmino JP, Vallejos-Vidal E, Sarasquete C, Ortiz-Delgado JB, Carles Balasch J, Tort L, et al. Unveiling the effect of dietary essential oils supplementation in Sparus aurata gills and its efficiency against the infestation by Sparicotyle chrysophrii. Sci Rep (2020) 10:17764. doi: 10.1038/s41598-020-74625-5
38. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res (2018) 47:607–13. doi: 10.1093/nar/gky1131
39. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinf (2016) 54(1). doi: 10.1002/cpbi.5
40. Lieke T, Meinelt T, Hoseinifar SH, Pan B, Straus DL, Steinberg CEW. Sustainable aquaculture requires environmental-friendly treatment strategies for fish diseases. Rev Aquac (2020) 12(2):943–65. doi: 10.1111/raq.12365
41. Loh JY, Chan HK, Yam HC, In LLA, Lim CSY. An overview of the immunomodulatory effects exerted by probiotics and prebiotics in grouper fish. Aquac Inter (2020) 28:729–50. doi: 10.1007/s10499-019-00491-2
42. Rohani MF, Islam SM, Hossain MK, Ferdous Z, Siddik MA, Nuruzzaman M, et al. Probiotics, prebiotics and synbiotics improved the functionality of aquafeed: upgrading growth, reproduction, immunity and disease resistance in fish. Fish Shellfish Immunol (2022) 120:569–89. doi: 10.1016/j.fsi.2021.12.037
43. Lafferty KD, Harvell CD, Conrad JM, Friedman CS, Kent ML, Kuris AM, et al. infectious diseases affect marine fisheries and aquaculture economics. Annu Rev Mar Sci (2015) 7:471–96. doi: 10.1146/annurev-marine-010814-015646
44. Rodger HD. Fish disease causing economic impact in global aquaculture. In: Adams A, editor. Fish Vaccines. Birkhauser Advances in Infectious Diseases. Basel: Springer (2016). doi: 10.1007/978-3-0348-0980-1_1
45. Ouwehand AC. A review of dose-responses of probiotics in human studies. Benef Microbes (2017) 8(2):143–51. doi: 10.3920/BM2016.0140
46. Reyes-Becerril M, Salinas I, Cuesta A, Meseguer J, Tovar-Ramirez D, Ascencio-Valle F, et al. Oral delivery of live yeast Debaryomyces hansenii modulates the main innate immune parameters and the expression of immune-relevant genes in the gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol (2008) 25(6):731–9. doi: 10.1016/j.fsi.2008.02.010
47. Reyes-Becerril M, Ascencio-Valle F, Meseguer J, Tapia-Paniagua ST, Moriñigo MA, Esteban MÁ. Debaryomyces hansenii L2-enriched diet enhances the immunity status, gene expression and intestine functionality in gilthead seabream (Sparus aurata L.). Aquac Res (2012) 43(8):1107–18. doi: 10.1111/j.1365-2109.2011.02914.x
48. Hernández-López IA, Tovar-Ramírez D, de la Rosa-García S, Álvarez-Villagómez CS, Asencio-Alcudia GG, Martínez-Burguete T, et al. Dietary live yeast (Debaryomyces hansenii) provides no advantages in tropical gar, Atractosteus tropicus (Actinopterygii: Lepisosteiformes: Lepisosteidae), juvenile aquaculture. Acta Ichthyol Piscat (2021) 51(3):311–20. doi: 10.3897/aiep.51.67095
49. Vidya SRG, Vijaya C. Efficacy of β-glucan from Debaryomyces hansenii as an immunostimulant in Litopenaeus vannamei culture. Aquac Inter (2021) 29(4):1451–8. doi: 10.1007/s10499-021-00678-6
50. Salinas I, Zhang YA, Sunyer JO. Mucosal immunoglobulins and B cells of teleost fish. Dev Comp Immunol (2011) 35(12):1346–65. doi: 10.1016/j.dci.2011.11.009
51. Sveen L, Karlsen C, Ytteborg E. Mechanical induced wounds in fish – a review on models and healing mechanisms. Rev Aquac (2020) 12(4):2446–65. doi: 10.1111/raq.12443
52. Sanahuja I, Guerreiro PM, Girons A, Fernandez-Alacid L, Ibarz A. Evaluating the repetitive mucus extraction effects on mucus biomarkers, mucous cells, and the skin-barrier status in a marine fish model. Front Mar Sci (2023) 9:1095246. doi: 10.3389/fmars.2022.1095246
53. Reverter M, Tapissier-Bontemps N, Lecchini D, Banaigs B, Sasal P. Biological and ecological roles of external fish mucus: a review. Fishes (2018) 3(4):41. doi: 10.3390/fishes3040041
54. Fernández-Montero Á, Torrecillas S, Izquierdo M, Caballero MJ, Milne DJ, Secombes CJ, et al. Increased parasite resistance of greater amberjack (Seriola dumerili Risso 1810) juveniles fed a cMOS supplemented diet is associated with upregulation of a discrete set of immune genes in mucosal tissues. Fish Shellfish Immunol (2019) 86:35–45. doi: 10.1016/j.fsi.2018.10.034
55. Ramos-Molina B, Lambertos A, Peñafiel R. Antizyme inhibitors in polyamine metabolism and beyond: physiopathological implications. Med Sci (2018) 6(4):89. doi: 10.3390/medsci6040089
56. Abaandou L, Shiloach J. Knocking out ornithine decarboxylase antizyme 1 (OAZ1) improves recombinant protein expression in the HEK293 cell line. Med Sci (2018) 6(2):48. doi: 10.3390/medsci6020048
57. Verri T, Terova G, Romano A, Barca A, Pisani P, Storelli C, et al. The solute carrier (SLC) family series in teleost fish. In: Saroglia, Liu Z, editors. Functional Genomics in Aquaculture. Wiley Online Library (2012). p. 219–320. doi: 10.1002/9781118350041.ch10
58. Wang Q, Xu Z, Ai Q. Arginine metabolism and its functions in growth, nutrient utilization, and immunonutrition of fish. Anim Nutr (2021) 7(3):716–27. doi: 10.1016/j.aninu.2021.03.006
59. Sander M, Bergersen LH, Storm-Mathisen J. Molecular approaches to understanding neural network plasticity and memory: the Kavli Prize Inaugural Symposium on Neuroscience. Neuroscience (2009) 163(4):965–76. doi: 10.1016/j.neuroscience.2009.07.046
60. Sheng L, Luo Q, Chen L. Amino Acid Solute Carrier Transporters in Inflammation and Autoimmunity. Drug Metab Dispos (2022) 50(9):1228–37. doi: 10.1124/dmd.121.000705
61. Andersen SM, Holen E, Aksnes A, Ronnestad I, Zerrahn JE, Espe M. Dietary arginine affects energy metabolism through polyamine turnover in juvenile Atlantic salmon (Salmo salar). Br J Nutr (2013) 110(11):1968–77. doi: 10.1017/S0007114513001402
62. Shui JW, Steinberg MW, Kronenberg M. Regulation of inflammation, autoimmunity, and infection immunity by HVEM-BTLA signaling. J Leukoc Biol (2010) 89(4):517–23. doi: 10.1189/jlb.0910528
63. Tafalla C, Granja AG. Novel insights on the regulation of B cell functionality by members of the tumor necrosis factor superfamily in jawed fish. Front Immunol (2018) 9:1285. doi: 10.3389/fimmu.2018.01285
64. Wu D, Xiang L, Peng L, Gu H, Tang Y, Luo H, et al. Comprehensive analysis of the immune implication of FABP4 in colon adenocarcinoma. PloS One (2022) 17(10):e0276430. doi: 10.1371/journal.pone.0276430
65. Cai G, Freeman GJ. The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation. Immunol Rev (2009) 229(1):244–58. doi: 10.1111/j.1600-065X.2009.00783.x
66. Murphy KM, Nelson CA, Šedý JR. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat Rev Immunol (2006) 6(9):671–81. doi: 10.1038/nri1917
67. Gonzalez LC, Loyet KM, Calemine-Fenaux J, Chauhan V, Wranik B, Ouyang W, et al. A coreceptor interaction between the CD28 and TNF receptor family members B and T lymphocyte attenuator and herpesvirus entry mediator. Proc Natl Acad Sci USA (2005) 102(4):1116–21. doi: 10.1073/pnas.0409071102
68. Bernard D, Hansen JD, Du Pasquier L, Lefranc MP, Benmansour A, Boudinot P. Costimulatory receptors in jawed vertebrates: Conserved CD28, odd CTLA4 and multiple BTLAs. Dev Comp Immunol (2007) 31:255–71. doi: 10.1016/j.dci.2006.06.003
69. Muntjewerff EM, Meesters LD, Van Den Bogaart G, Revelo NH, Van Endert PM, Drake J, et al. Reverse signaling by MHC-I molecules in immune and non-immune cell types. Front Immunol (2020) 11:605958. doi: 10.3389/fimmu.2020.605958
70. Wu L, Qin Z, Liu H, Lin L, Ye J, Li J. Recent advances on phagocytic B cells in teleost fish. Front Immunol (2020) 11:824. doi: 10.3389/fimmu.2020.00824
71. Peñaranda MA, Jensen I, Tollersrud LG, Bruun JA, Jørgensen JB. Profiling the Atlantic salmon IgM + B cell surface proteome: novel information on teleost fish B cell protein repertoire and identification of potential B cell markers. Front Immunol (2019) 10:37. doi: 10.3389/fimmu.2019.00037
72. Martín D, Perdiguero P, Morel E, Soleto I, Herranz-Jusdado JG, Ramón LA, et al. CD38 defines a subset of B cells in rainbow trout kidney with high IgM secreting capacities. Front Immunol (2021) 12:773888. doi: 10.3389/fimmu.2021.773888
73. Shen Y, Zhang J, Xu X, Fu J, Li J. Expression of complement component C7 and involvement in innate immune responses to bacteria in grass carp. Fish Shellfish Immunol (2012) 33(2):448–54. doi: 10.1016/j.fsi.2012.05.016
74. Long Y, Li Q, Zhou B, Song G, Li T, Cui Z. De novo assembly of mud loach (Misgurnus anguillicaudatus) skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion. PloS One (2013) 8(2):e56998. doi: 10.1371/journal.pone.0056998
75. Bansil R, Turner BS. The biology of mucus: Composition, synthesis and organization. Adv Drug Delivery Rev (2017) 124:3–15. doi: 10.1016/j.addr.2017.09.023
76. Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. Mucins in the mucosal barrier to infection. Mucosal Immunol (2008) 1(3):183–97. doi: 10.1038/mi.2008.5
77. Quintana-Hayashi MP, Padra M, Padra JT, Benktander J, Lindén SK. Mucus-pathogen interactions in the gastrointestinal tract of farmed animals. Microorganisms (2018) 6(2):55. doi: 10.3390/microorganisms6020055
78. Yao LL, Cao QJ, Zhang HM, Zhang J, Cao Y, Li XD. Melanophilin stimulates myosin-5a motor function by allosterically inhibiting the interaction between the head and tail of myosin-5a. Sci Rep (2015) 5:10874. doi: 10.1038/srep10874
79. Evans CM, Koo JS. Airway mucus: The good, the bad, the sticky. Pharmacol Ther (2009) 121(3):332–48. doi: 10.1016/j.pharmthera.2008.11.001
80. Pérez-Sánchez J, Estensoro I, Redondo MJ, Calduch-Giner JA, Kaushik S, Sitjà-Bobadilla A. Mucins as diagnostic and prognostic biomarkers in a fish-parasite model: transcriptional and functional analysis. PloS One (2013) 8(6):e65457. doi: 10.1371/journal.pone.0065457
81. Sadoul B, Geffroy B. Measuring cortisol, the major stress hormone in fishes. J Fish Biol (2019) 94(4):540–55. doi: 10.1111/jfb.13904
82. Ibarz A, Ordónez-Grande B, Sanahuja I, Sánchez-Nunõ S, Fernández-Borras J, Blasco J, et al. Using stable isotope analysis to study skin mucus exudation and renewal in fish. J Exp Biol (2019) 222(8):jeb195925. doi: 10.1242/jeb.195925
83. Sanahuja I, Fernández-Alacid L, Ordóñez-Grande B, Sánchez-Nuño S, Ramos A, Araujo RM, et al. Comparison of several non-specific skin mucus immune defences in three piscine species of aquaculture interest. Fish Shellfish Immunol (2019) 89:428–36. doi: 10.1016/j.fsi.2019.04.008
84. Ghosh D, Porter E, Shen B, Lee SK, Wilk D, Drazba J, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol (2002) 3(6):583–90. doi: 10.1038/ni797
85. Stapels DAC, Geisbrecht BV, Rooijakkers SHM. Neutrophil serine proteases in antibacterial defense. Curr Opin Microbiol (2014) 23:42–8. doi: 10.1016/j.mib.2014.11.002
86. Hickey ME, Lee JL. A comprehensive review of Vibrio (Listonella) anguillarum: ecology, pathology and prevention. Rev Aquac (2017) 10(3):585–610. doi: 10.1111/raq.12188
87. Ibarz A, Padrós F, Gallardo MÁ, Fernández-Borràs J, Blasco J, Tort L. Low-temperature challenges to gilthead sea bream culture: review of cold-induced alterations and “Winter Syndrome”. Rev Fish Biol Fish (2010) 20(4):539–56. doi: 10.1007/s11160-010-9159-5
88. Díaz-Puertas R, Adamek M, Mallavia R, Falco A. Fish skin mucus extracts: an underexplored source of antimicrobial agents. Mar Drugs (2023) 21(6):350. doi: 10.3390/md21060350
89. Angulo C, Maldonado M, Delgado K, Reyes-Becerril M. Debaryomyces hansenii up regulates superoxide dismutase gene expression and enhances the immune response and survival in Pacific red snapper (Lutjanus peru) leukocytes after Vibrio parahaemolyticus infection. Dev Comp Immunol (2017) 71:18–27. doi: 10.1016/j.dci.2017.01.020
90. Angulo M, Reyes-Becerril M, Cepeda-Palacios R, Tovar-Ramírez D, Esteban MÁ, Angulo C. Probiotic effects of marine Debaryomyces hansenii CBS 8339 on innate immune and antioxidant parameters in newborn goats. Appl Microbiol Biotechnol (2019) 103(5):2339–52. doi: 10.1007/s00253-019-09621-5
91. De Mercado E, Larrán AM, Pinedo J, Tomás-Almenar C. Skin mucous: a new approach to assess stress in rainbow trout. Aquaculture (2018) 484:90–7. doi: 10.1016/j.aquaculture.2017.10.031
92. Fernández-Alacid L, Sanahuja I, Ordóñez-Grande B, Sánchez-Nuño S, Herrera M, Ibarz A. Comparison between properties of dorsal and ventral skin mucus in Senegalese sole: response to an acute stress. Aquaculture (2019) 513:734410. doi: 10.1016/j.aquaculture.2019.734410
93. Espinosa-Ruíz C, Esteban MÁ. Wound-induced changes in antioxidant enzyme activities in skin mucus and in gene expression in the skin of gilthead seabream (Sparus aurata l.). Fishes (2021) 6(2):15. doi: 10.3390/fishes6020015
94. Banh S, Wiens L, Sotiri E, Treberg JR. Mitochondrial reactive oxygen species production by fish muscle mitochondria: Potential role in acute heat-induced oxidative stress. Comp Biochem Physiol B Biochem Mol Biol (2016) 191:99–107. doi: 10.1016/j.cbpb.2015.10.001
Keywords: Debaryomyces hansenii, skin defense, skin mucus defense, skin mucus associated biomarkers, transcriptomics, yeast probiotic
Citation: Sanahuja I, Fernandez-Alacid L, Torrecillas S, Ruiz A, Vallejos-Vidal E, Firmino JP, Reyes-Lopez FE, Tort L, Tovar-Ramirez D, Ibarz A and Gisbert E (2023) Dietary Debaryomyces hansenii promotes skin and skin mucus defensive capacities in a marine fish model. Front. Immunol. 14:1247199. doi: 10.3389/fimmu.2023.1247199
Received: 25 June 2023; Accepted: 09 August 2023;
Published: 30 August 2023.
Edited by:
Jianmin Ye, South China Normal University, ChinaReviewed by:
Fei Song, South China Normal University, ChinaCopyright © 2023 Sanahuja, Fernandez-Alacid, Torrecillas, Ruiz, Vallejos-Vidal, Firmino, Reyes-Lopez, Tort, Tovar-Ramirez, Ibarz and Gisbert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ignasi Sanahuja, SWduYXNpLnNhbmFodWphQGlydGEuY2F0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.