- Department of Entomology and Plant Pathology, Oklahoma State University, Stillwater, OK, United States
Insect phenoloxidases (POs) catalyze phenol oxygenation and o-diphenol oxidation to form reactive intermediates that kill invading pathogens and form melanin polymers. To reduce their toxicity to host cells, POs are produced as prophenoloxidases (PPOs) and activated by a serine protease cascade as required. In most insects studied so far, PPO activating proteases (PAPs) generate active POs in the presence of a high Mr cofactor, comprising two serine protease homologs (SPHs) each with a Gly residue replacing the catalytic Ser of an S1A serine protease (SP). These SPHs have a regulatory clip domain at the N-terminus, like most of the SP cascade members including PAPs. In Drosophila, PPO activation and PO-catalyzed melanization have been examined in genetic analyses but it is unclear if a cofactor is required for PPO activation. In this study, we produced the recombinant cSPH35 and cSPH242 precursors, activated them with Manduca sexta PAP3, and confirmed their predicted role as a cofactor for Drosophila PPO1 activation by MP2 (i.e., Sp7). The cleavage sites and mechanisms for complex formation and cofactor function are highly similar to those reported in M. sexta. In the presence of high Mr complexes of the cSPHs, PO at a high specific activity of 260 U/μg was generated in vitro. To complement the in vitro analysis, we measured hemolymph PO activity levels in wild-type flies, cSPH35, and cSPH242 RNAi lines. Compared with the wild-type flies, only 4.4% and 18% of the control PO level (26 U/μl) was detected in the cSPH35 and cSPH242 knockdowns, respectively. Consistently, percentages of adults with a melanin spot at the site of septic pricking were 82% in wild-type, 30% in cSPH35 RNAi, and 53% in cSPH242 RNAi lines; the survival rate of the control (45%) was significantly higher than those (30% and 15%) of the two RNAi lines. These data suggest that Drosophila cSPH35 and cSPH242 are components of a cofactor for MP2-mediated PPO1 activation, which are indispensable for early melanization in adults.
1 Introduction
Proteolytic activation of prophenoloxidase (PPO) and PO-catalyzed pathogen killing and melanin formation have been investigated for more than fifty years in various insects, such as lepidopterans, coleopterans, and dipterans (1–5). Some progress has been made regarding the mechanisms of the activation reaction. In the silkworm Bombyx mori, PPO activating enzyme (a cSP or clip-domain serine protease) alone generated highly active PO (6). Manduca sexta PPO activating protease-1, 2, or 3 (PAP1/2/3) activated the PPOs in the presence of a cofactor containing clip-domain serine protease homologs (cSPHs) (7–11). The high Mr cofactor is composed of a type I SPH (i.e., 1a, 1b, 4, or 101) and a type II SPH (i.e., 2) (12). In another lepidopteran species, Helicoverpa armigera cSPH11 (SPHI) and cSPH50 (SPHII) largely enhanced PPO activation by cSP6 (13). In the Order of Coleoptera, Holotrichia diomphalia PPO activating factor-1 (cSP) and -2 (cSPH) together activated the PPOs (14, 15), and so did Tenebrio molitor Spätzle processing enzyme and serine protease homolog-1 (16, 17). In the mosquito Aedes aegypti, CLIPB9 activated PPO3 along with CLIPA14 (SPHI) to a level higher than CLIPB9 alone did (18). While PPO activation by PAP requires a cofactor in most insects studied, it is unknown if a complex of SPHI and SPHII could lead to a maximal level of PO in B. mori, H. diomphalia, T. molitor, and A. aegypti. Neither is it clear how knocking-down one of the cSPHs by RNAi may affect melanization and survival of pathogenic bacteria in vivo. Beginning to fill this knowledge gap, we extended in vitro studies of the cofactor-assisted PPO activation to Drosophila melanogaster.
D. melanogaster has three PPO genes in the genome, coding for three proenzymes with distinct functions during wound healing, microbial infection, and melanotic encapsulation of parasitoid wasps (19, 20). PPO1 and PPO2 are produced in crystal cells, and PPO3 in lamellocytes may not need a protease for spontaneous activation (21). While PPO1 is made for rapid, early delivery of PO activity upon injury, PPO2 is stored in a crystalline form for deployment in the later phase of infection-induced melanization. PPO3 participates in the encapsulation of wasp eggs, in association with PPO2. Genetic analyses showed that MP1, MP2 (also known as Sp7 or PAE1), and Hayan lead to PPO1 activation (22–26). Defects in these genes or failure to make enough of the proteins differentially affected the survival of the flies after infection with bacteria and fungi. Subsequent experiments using single, double, and triple deletion mutants provided compelling evidence that PPO activation is indispensable for the Drosophila defense response against bacteria, fungi, and parasitoids (19, 20). Furthermore, the authors pointed out that a significant fraction of the Toll-induced killing of bacteria in adults is likely caused by PO activity, which is consistent with the in vitro killing effects (27, 28).
The approach combining the biochemical study of recombinant proteins can be useful for solving problems that are hard to disentangle by genetic analysis alone. For instance, bovine coagulation factor Xa activated the purified proMP2xa expressed in Drosophila S2 cells (29). The active MP2xa cleaved PPO1 from E. coli likely at the classical activation site between Arg52 and Phe53 (30), suggesting that MP2 is a PAP of the PPO1. Despite the fact that half of the correctly folded PPO1 was processed, no PO activity was reported in that paper (29). Based on the cofactor studies in other insects and the low PO activity generated by Drosophila MP2xa alone, we hypothesized that one or two SPHs may act as a cofactor for PPO activation in Drosophila. Therefore, we expressed in insect cells the precursors of cSPH35 and cSPH242, the only SPHI and SPHII orthologs in D. melanogaster (31), and then activated them using M. sexta PAP3 (32). Using MP2xa and PPO1 in the presence of these two cSPHs, we explored the mechanism of the PPO activation and demonstrated the cofactor’s existence and functional importance in Drosophila. To test the roles of these two SPHs in vivo, we knocked down their expression in their RNAi lines and observed the phenotype of significant decreases in PO activity, melanin formation, and survival rate of the flies infected with a strain of Enterococcus faecalis.
2 Materials and methods
2.1 Acquisition, maintenance, and crossing of fly lines for RNAi knockdown of cSPH35 and cSPH242 expression
Wild-type (Oregon-R) and transgenic strains of D. melanogaster were purchased from Carolina Biological and Bloomington Drosophila Stock Center (https://bdsc.indiana.edu/), respectively. BL4414 expresses GAL4 ubiquitously under the control of the Act5C promoter. BL52875 and BL44268 express dsRNA for RNAi of cSPH35 (CG5390) and cSPH242 (CG40160) under the control of GAL4, which binds UAS enhancer (33). The flies were maintained on a standard diet at 25°C (34). Newly emerged male adults (BL4414) with the GAL4 driver were mated with virgin female adults (BL52875 or BL44268) and F1 offspring from these crosses with straight wings were collected 1–2 day-post-eclosion and transferred onto the fresh diet.
2.2 Determination of cSPH35 and cSPH242 mRNA levels by qRT-PCR analysis
For knockdown verification, total RNA samples were extracted from the control (BL4114) and F1 adults (i.e., cSPH35kd and cSPH242kd) five days after emergence. cDNA samples, each equivalent to starting with 50 ng total RNA, were incubated with 1× iTag Universal SYBR Green Supermix (Bio-Rad) and specific primers (0.5 mM each) in triplicate in each reaction (10 μl). The primers were: J1091 (5’-GTGGTCCATTCCACTTCCGT) and J1092 (5’-TCATCATGTCGCGTGGATCA) for ribosomal protein L32 (rpL32) as a reference; J1971 (5’-GAAAGTCCATTCACCCTCCA) and J1972 (5’-TCCTTGCCGAACTTGTTCTT) for cSPH35; J1973 (5’-AAGACGTTGAATCCGACACC) and J1974 (5’-CCGCTTCATTTTGCGATACT) for cSPH242. Each primer pair was tested to ensure amplification of the target gene only. Amplification efficiencies were determined individually and confirmed to be 90−110%. Thermal cycling conditions were 95°C for 2 min and 40 cycles of 95°C 10 s and 60°C 30 s. After PCR was completed on a CFX Connect Real-Time PCR Detection System (Bio-Rad), melting curves of the products in all reactions were examined to ensure proper shape and Tm values. The mRNA levels were first normalized against the internal control of rpL32 using corresponding Ct values for the same cDNA samples and the relative mRNA levels were calculated as 2-ΔCt, where ΔCt = Ct cSPH35/242 - Ct rpL32. The lowest level of cSPH35 or cSPH242 mRNA in a dataset was then used as a reference (set at 1.00) to calculate mRNA fold differences (i.e., 2-ΔΔCt) for target genes in all the samples.
The same method was used to measure relative fold changes of the cSPH35 and cSPH242 transcript levels in wild-type thrid instar larvae, day 2 pupae, and day 2−3 adults for comparison. Similarly, immune inducibility was examined using naïve and infected wild-type adults. At 24 h before total RNA extraction, day 4–5 adult flies in the infection group were pricked with a 0.1 mm stainless needle in the thorax with live Micrococcus luteus (A600 = 100).
2.3 cDNA cloning and baculovirus construction for producing cSPH35, cSPH242, and MP2xa precursors
A cSPH242 cDNA fragment was amplified from FBpp0112536 (https://flybase.org/) using primers J1681 (5’-TCATATGGCTCCTCAGCAGAAC) and J1682 (5’-ACTCGAGTGCGGTGTACACAG). The product was cloned into pGEM-T vector (Promega) and, after sequence validation, the NdeI-XhoI fragment was subcloned into the same sites in pMFFMH6 (Wang et al., unpublished data), a derivative of pMFH6 (35) with FLAG (DYKDDDDK) and c-myc (EQKLISEEDL) tags. cSPH242/pMFFMH6 was used to generate a baculovirus to express pro-cSPH242 (GIHDYKDDDDKHMAPQQN … AVYTALEQKLISEEDLHHHHHH, where the underlined part is encoded by the cDNA, 429 residues, 47,072 Da, pI: 5.52) in insect cells (36). A cSPH35 cDNA fragment was amplified from plasmid FBpp0079653 using J1685 (5’-CATATGCAGGACTCTTCCTTG) and J1686 (5’-TTGCTCGAGGGGTGTATAGTGCCT). After TA cloning and sequence confirmation, the NdeI-XhoI fragment was subcloned into the same sites in pMFFMH6 to generate a baculovirus for producing pro-cSPH35 (GIHDYKDDDDKHMQDSSL … RHYTPLEQKLISEEDLHHHHHH, 418 residues, 46,600 Da, pI: 5.40) in Sf9 cells. An MP2 cDNA fragment was amplified from MP2xa/pFastBac1 (29) using J1551 (5’-CATATGCAAGGAAGTTGTAGG) and J1552 (5’-CTCGAGGGGACGAATGGTCTC). After TA cloning and sequence validation, the NdeI-XhoI region was inserted into pMFFMH6 to generate plasmid, bacmid, and then baculovirus for producing proMP2xa (GIHDYKDDDDKHMQGSCR … ETIRPLEQKLISEEDLHHHHHH, IEGR replacing FSNK, 394 residues, 43,895 kDa, pI: 5.57) in insect cells.
2.4 Expression and purification of the recombinant cSPH35, cSPH242, and MP2xa precursors
Sf9 cells at 2.4×106 cells/ml in 300 ml of insect serum-free medium (Thermo Fisher) were separately infected with the baculovirus stocks at a multiplicity of infection of 10 and grown at 27°C for 96 h with gentle agitation at 100 rpm. After the cells were pelleted by centrifugation at 5,000×g for 10 min, the pH of the conditioned media was adjusted to pH 6.4 with HCl. Cell debris and fine particles were spun down by centrifugation at 22,100×g for 20 min and the supernatant was mixed with an equal volume of 1 mM imidazole in water. The diluted solution was applied to a dextran sulfate-Sepharose column (20 ml bed volume) at a flow rate of 1.5 ml/min. Following a washing step with 100 ml of 0.01% Tween-20, 1 mM benzamidine in 10 mM potassium phosphate, pH 6.4, bound proteins were eluted with a linear gradient of 0−1.0 M NaCl in 80 ml of the same buffer. After SDS-PAGE and immunoblot analysis, the pro-cSPH fractions were combined, adjusted to pH 7.5, loaded onto a 2 ml Ni2+-nitrilotriacetic acid agarose column, and washed with 15 ml of 50 mM sodium phosphate, pH 7.5. Bound proteins were eluted with a gradient of 0−0.3 M imidazole in 20 ml of the same buffer. The pro-cSPH fractions were pooled, dialyzed against 20 mM Tris-HCl, pH 7.5, and concentrated on Amicon Ultra-30 centrifugal filter devices (Millipore). Aliquots of the proteins were flash-frozen in liquid nitrogen prior to long-term storage at -80°C. The MP2xa precursor was produced in Sf9 cells infected by the recombinant virus and purified from the culture medium by following the same procedure.
2.5 Cleavage activation of cSPH242 and cSPH35 precursors by M. sexta PAP3
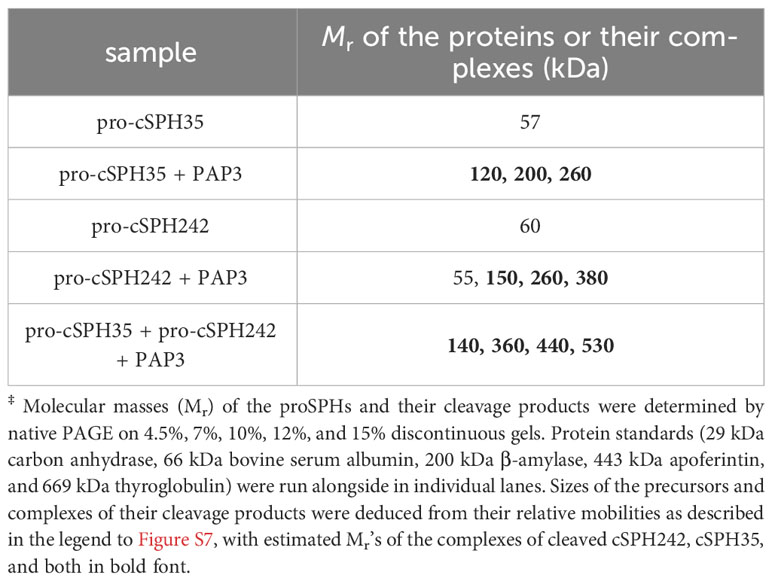
M. sexta PAP3 was isolated from pharate pupal hemolymph (10) and used to activate recombinant proPAP3 (32). Freshly prepared PAP3 was incubated with the purified D. melanogaster pro-cSPH242 and pro-cSPH35. The cleavage reactions, controls, and Mr markers were separated by 10% SDS and native PAGE, followed by electrotransfer and immunoblotting using antibodies against the hexahistidine tag. To better understand the process of PAP3 cleavage and high Mr SPH complex formation, aliquots of the pro-cSPHs were incubated with different amounts of PAP3 for 1 h at 37°C. The mixtures and PAP3 control were resolved by 10% SDS and native PAGE, transferred onto nitrocellulose membranes, and detected using diluted (His)6 antiserum as primary antibody, goat-anti-mouse IgG conjugated to alkaline phosphatase (Bio-Rad) as secondary antibody, and a BCIP-NBT substrate kit (Bio-Rad) for color development. Association states of the precursors and cleaved forms of cSPH35 and cSPH242 were determined by one-dimensional electrophoresis on a series of polyacrylamide gels under non-denaturing conditions (37).
2.6 Identification of the PAP3 cleavage sites in cSPH242 and cSPH35 by LC-MS/MS analysis
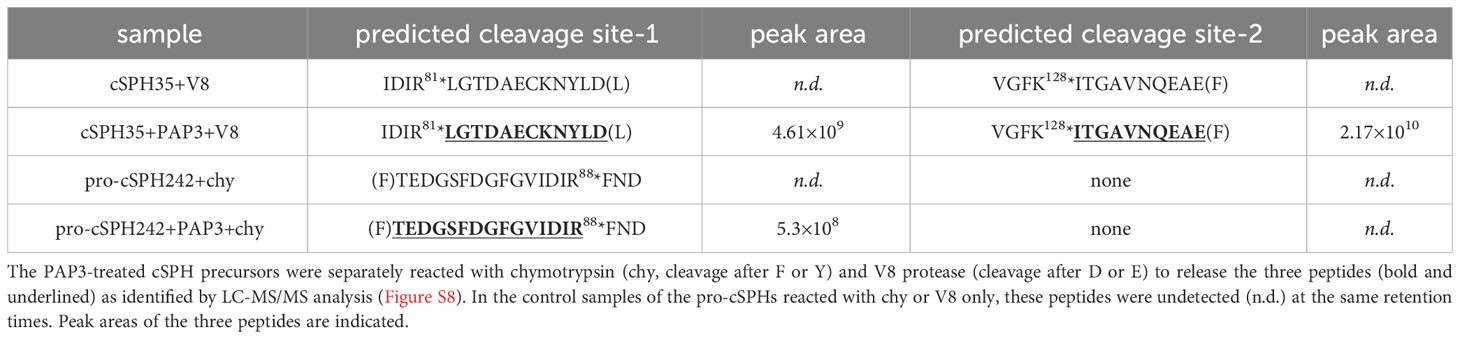
The purified pro-cSPHs (ca. 10 μg) were incubated with PAP3 (1.0 μg) for 1 h at 37 °C. The mixtures and negative controls of the pro-cSPHs were denatured in urea and digested with chymotrypsin or V8 protease overnight at 37 °C (38). The resulting peptides were desalted using Omix C18 affinity media, dried, and dissolved in mobile phase A (0.1% formic acid in H2O). The samples were loaded onto an Acclaim PepMap RSLC C18 column (75 μm × 50 cm, Thermo Fisher) for data-dependent LC-MS/MS analysis as described previously (39). Peptides in each sample were separated via a gradient of 0–35% mobile phase B (0.1% formic acid in 80% AcCN) developed over 120 min (12). The survey scans were followed by both HCD and CID collisional MS/MS events triggered from the hypothetical peptide ions, with scanning of collisional fragments at 15,000 resolutions in the Orbitrap sector.
For identifying the PAP3 cleavage sites in cSPH242 and cSPH35, a database combining D. melanogaster pro-cSPH242 and pro-cSPH35 and background proteins from M. sexta, human, and insect cells were constructed, and peptide spectrum matches were reviewed in Byonic to detect peptides not cut by nonspecific proteases. The details of peak area calculation were described previously to quantify characteristic peptides released by PAP3 and chymotrypsin/V8 protease (12). The original MS imaging data were deposited at a public repository (https://doi.org/10.6084/m9.figshare.23786949).
2.7 Activation of proMP2xa and proPAP3 followed by an amidase assay
The purified poMP2xa (0.44 µg) was incubated with Factor Xa (0.1 µg) at 37 °C for 1 h in 10 μl buffer A. Half of the reaction mixture and controls (proMP2xa and Factor Xa only) were separately incubated with 150 μl 25 µM acetyl-Ile-Glu-Ala-Arg-p-nitroanilide (IEARpNA, Sigma-Aldrich, A0180) in buffer A. Absorbance of the mixtures was monitored at 405 nm on a microplate reader. One unit of the hydrolytic activity is defined as the amount of enzyme causing ΔA405/min of 0.001. To obtain active PAP3, proPAP3 (0.4 µg/µl, 1 µl) was incubated with active PAP3 (40 ng/µl, 1 µl) in 10 μl buffer A at 37°C for 1 h, half of the mixture (0.2 µg) was used to test IEARase activity, as described above for MP2xa.
2.8 Drosophila PPO1 expression, activation, and PO activity assay
D. melanogaster PPO1 was expressed in E. coli BL21 harboring PPO1/pET28a and purified by affinity chromatography on a Ni-NTA agarose column (30). The active MP2xa was produced by preincubating proMP2xa (440 ng/μl, 1 μl) with Factor Xa (100 ng/μl, 1 μl) at 37°C for 1h. The two cSPHs were generated by treating pro-cSPH35 and pro-cSPH242 (each at 200 ng/μl, 1 μl) with M. sexta PAP3 (10 ng/μl, 1 μl) at 25°C for 1h. M. sexta serpin-3 (100 ng/μl, 1 μl) was added to the processed cSPH35 and cSPH242 and incubated at 25°C for 30 min to block PAP3 activity. To activate Drosophila PPO1 under different conditions, PPO1 (1 μl, 270 μg/ml, from E. coli) was processed by pretreated MP2xa (200 ng/μl, 1 μl) with or without cleaved cSPH35 and cSPH242 on ice for 1 h in a total volume of 25 μl buffer A (0.001% Tween-20, pH 7.5, 20 mM Tris-HCl, 5 mM CaCl2). PO activity was determined on a microplate reader immediately after 150 μl of 2.0 mM dopamine in 50 mM sodium phosphate, pH 6.5, was added to each sample well (8). Immunoblot analysis was performed using the antibody against Drosophila PO1 fused with glutathione S-transferase (26), which did not crossreact with PO2 or PO3.
2.9 Hemolymph preparation for DmPPO1 immunoblot analysis, melanization observation, and PO activity measurement
A hole was pierced with a 25-gauge needle in the bottom of a 0.5 ml microfuge tube before placing it in a 1.5 ml tube with its lid removed. Twenty adults were pricked in the thorax with a fine tungsten needle, placed in the small tube, and centrifuged at 5000 rpm for 1 min at 4°C. After discarding the 0.5 ml tube, 1.0 µl of hemolymph in the large tube was transferred to 10 µl buffer A containing 0.1% 1-phenyl-2-thiourea (PTU). After 30 min at room temperature, the mixtures from BL4414 and the two RNAi lines were subjected to 7.5% SDS-PAGE and immunoblot analysis using 1:2000 diluted D. melanogaster PPO1 antiserum (30), a gift from Dr. Erjun Ling at the Shanghai Institute of Plant Physiology and Ecology. Hemolymph samples (1.0 µl, without PTU) from the three groups of adults were also transferred to wells of a microplate for PO activity measurement. After 150 μl of 2.0 mM dopamine solution was added (Section 2.8), PO activities were assayed immediately on a plate reader as described before (8). Spontaneous melanization of the hemolymph was observed at 30 min after hemolymph samples (5 µl each, without PTU) were collected from 50 adults in the three groups, transferred into microplate wells, and incubated at room temperature before imaging.
2.10 Infection of cSPH35 and cSP242 RNAi flies with E. faecalis
An E. faecalis glycerol stock, a gift from Dr. Zhen Zou at the Institute of Zoology, Chinese Academy of Sciences, was revived on an LB agar plate. A single colony was inoculated into 3 mL of LB liquid medium and grown overnight at 37°C with shaking. The bacteria were spun down for 1 min at 15,000×g and the pellet was resuspended in fresh LB to a final OD600 of 1.2 or 0.8 for different tests (40). In the survival test, 4–5-day-old adults in the control (BL4414) and RNAi lines were pricked in the thorax with a needle (0.1-mm stainless steel needle) tinted with E. faecalis at OD600 = 1.2 (41). The number of survivors in each group was counted daily from day 1 to day 10. In the melanization test, an E. faecalis suspension (OD600 = 0.8) was used to infect adults in the three groups under the same conditions. Bacteria at the lower dosage induced melanization but did not kill the flies. Black pigment deposited at the wound site was observed 2 days after infection under an Olympus BH2-RFCA microscope and pictures were taken using a 9MP microscope digital camera (Amscope). Numbers of flies with and without a melanin spot were recorded for three batches of flies in BL4414 and F1 offspring of the two crossed lines (Section 2.1). Besides, the microscope and camera were used to detect and record cuticle decoloring in some F1 offspring of the crossed line with cSPH242 knockdown (Section 2.1). For comparison, a series of images of the 4–5-day-old adult flies in the BL4414 and cSPH35 RNAi lines were also captured and merged into one using ImageJ (https://imagej.nih.gov/ij/).
3 Results
3.1 Rationale for studying D. melanogaster cSPH35 and cSPH242
Among the eighteen clip-domain SPH genes in Drosophila, cSPH35 and cSPH242 are 1:4 and 1:1 orthologous to M. sexta SPH1a, 1b, 4, and 101 (SPHIs) and SPH2 (SPHII) (12, 31), respectively. A new phylogenetic analysis indicated that, except for D. suzukii SPHIa−c and D. elegans SPHIIa, b, all fifteen Drosophila species have a pair of SPHI and SPHII genes in their genomes (Figure S1). In the superfamily Tephritoidea, the olive and Mediterranean fruit flies also possess a single pair of SPHI and SPHII genes. The house fly M. domestica has three SPHs (Ia, Ib, and II). In comparison, expansions of the orthologous genes in mosquitoes and lepidopteran insects are remarkable (12, 42).
Based on the mass spectrometric data on M. sexta PPOs, PAPs, and SPHI-II (12, 43), we indicated that Drosophila cSPH35 and cSPH242 proteins may exist at high levels to act as components of the putative cofactor. Examination of the mRNA profiles (Figure S2A) revealed that, except for the early embryonic stage (0–10 h), their expression patterns were highly similar throughout the life span. We further confirmed by qRT-PCR analysis that levels of the two cSPH transcripts are comparable with each other in larvae, pupae, and adults (Figure 1A). With ΔCt values ranging from 4 to 7, the cSPH mRNA levels are high, in comparison to the vastly abundant rpL32. These data are consistent with their high protein abundances at the whole-body level in embryonic, larval, pupal, and adult stages (Figure S2B) (31, 44). In contrast to the low expression of cSPH35 in early embryos (0–10 h, Figure S1A), moderate cSPH242 transcript and protein levels in the same stage were observed. As shown in Figure 1B, there is an association of Drosophila cSPH35 and cSPH242 with innate immunity, since both cSPH35 and cSPH242 mRNA levels increased significantly after exposure to live M. luteus.
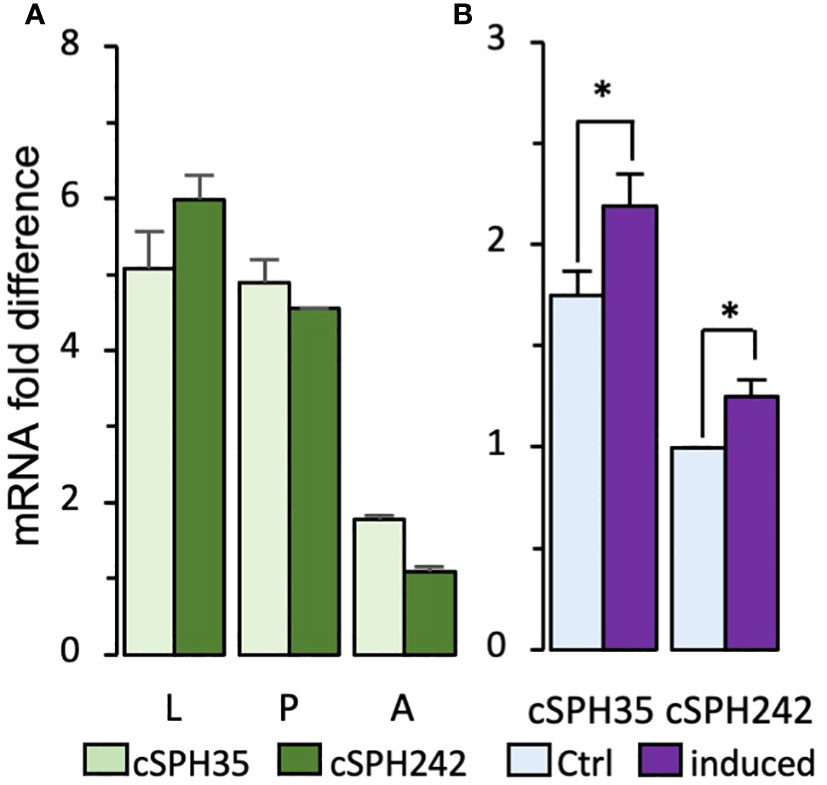
Figure 1 Quantitative RT-PCR analysis of D melanogastor cSPH35 and cSPH242 transcript levels. (A) The mRNA levels in 3rd instar larvae (L), day 2 pupae (P), and day 2–3 adults (A) were determined by qRT-PCR, calculated using ΔΔCt values of cSPH35, cSPH242, and rpL32 (control) for each of the three biological replicates, and plotted in the bar graph, as mean ± SEM (n = 3) (Section 2.2). (B) Immune inducibility. Total RNA samples from naive and immune-challenged adults were analyzed similarly to measure cSPH mRNA fold changes. Statistical significance of the pairwise comparisons was analyzed using Student’s t-test (*, p < 0.05).
3.2 Recombinant expression and characterization of the pro-cSPHs and proMP2xa
To study the functions of cSPH35 and cSPH242 in vitro, we constructed two baculoviruses to infect Sf9 cells for protein production and purification from the cultures. Pro-cSPH35 (0.8 mg) and pro-cSPH242 (1.0 mg) were isolated from the conditioned media (100 ml each), which migrated to the 60 and 63 kDa positions on a 10% SDS polyacrylamide gel (Figure 2). Major deviations from their calculated Mr’s (46.6 and 47.1 kDa) suggested they were modified post-translationally, by glycosylation likely. As shown in native PAGE, most of the precursors existed as a monomer (Table 1) and some pro-cSPH242 were in higher association states (Figure S7). We also expressed and purified proMP2xa (0.5 mg) from the culture medium (150 ml). The protein migrated as a single band at 50 kDa on 10% SDS-PAGE (Figure S3A), larger than its calculated Mr of 43.9 kDa.
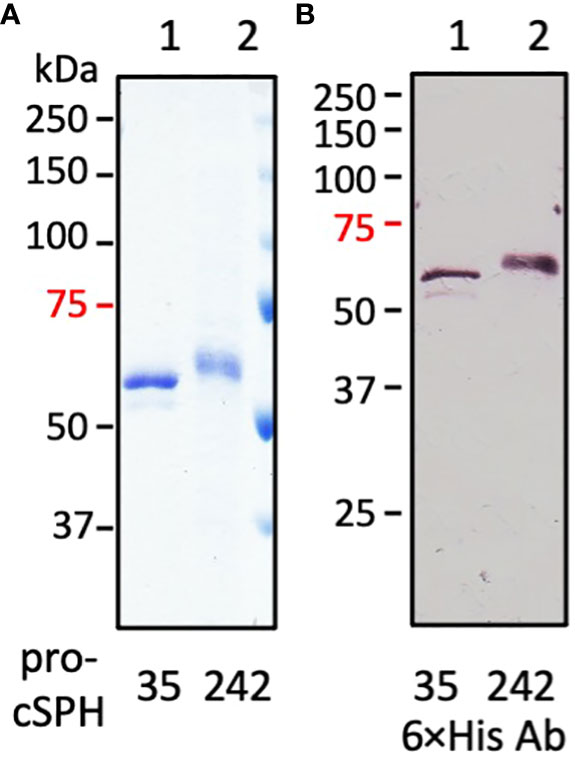
Figure 2 SDS-PAGE and immunoblot analyses of the purified precursors of D. melanogastor cSPH35 and cSPH242. The recombinant pro-cSPHs were resolved by 10% SDS-PAGE under the reducing condition followed with Coomassie brilliant blue staining (panel A, 1 μg per lane) or immunoblotting (panel B, 200 ng per lane) using 1:1000 diluted antiserum against hexahistidine affinity tag. Lane 1: pro-cSPH35; lane 2: pro-cSPH242; Positions and sizes (in kDa) of the prestained Mr standards are marked on the left, with the 75 kDa marker highlighted red.
3.3 Proteolytic processing of the two pro-cSPHs by M. sexta PAP3
Drosophila MP2 acts as a PAP to directly activate PPO1 (29) and some insect PAPs also generate their own cofactors by cleaving SPHI and SPHII precursors (17, 32, 35). Therefore, we activated proMP2xa by bovine factor Xa and detected its catalytic domain at 34 kDa and an increase in IEARpNA hydrolysis from 1.33 U to 2.63 U (Figure S3). However the active MP2xa failed to cleave pro-cSPH35 or pro-cSPH242 and did not cause a remarkable association of the monomers (Figure S4).
We then tested whether Manduca PAP3, an activator of the proSPHs (1a, 1b, 2, 4, and 101) (12), could activate Drosophila pro-cSPH35 and pro-cSPH242. To our surprise, the processing of pro-cSPHs was complete: a 42 kDa fragment was generated in the reaction of pro-cSPH242 and PAP3 and recognized by the hexahistidine antibody, whereas two carboxyl-terminal fragments of cSPH35 were detected at 40 and 30 kDa positions by the same antibody (Figure 3A). Native PAGE and immunoblot analysis indicated that the processed proteins formed a smear of high Mr complexes extending from the stacking gel (Figure 3B).
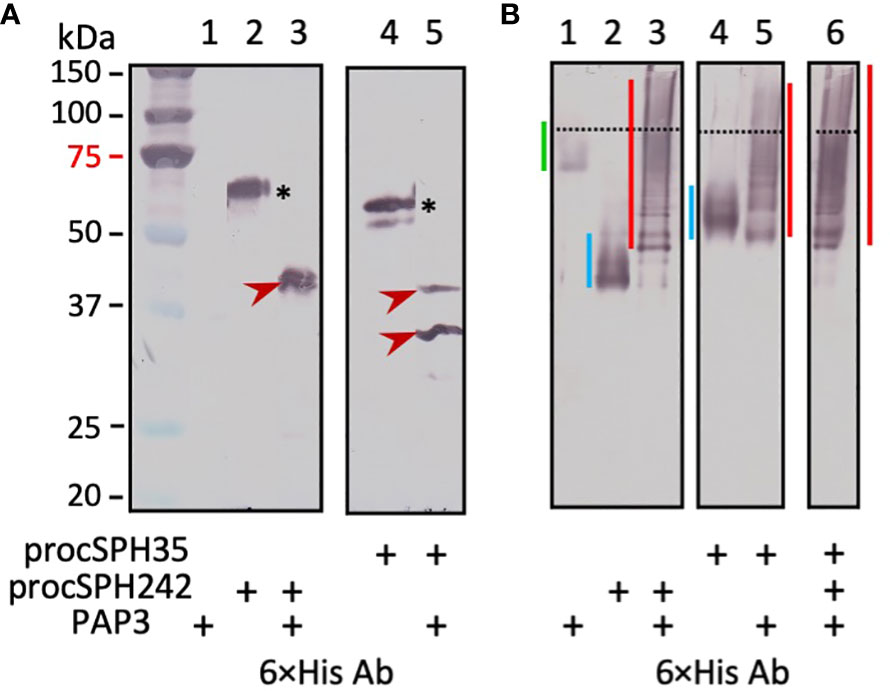
Figure 3 Immunoblot analysis of the PAP3 cleavage products from D melanogastor cSPH35 and cSPH242 precursors after 10% SDS (A) and native (B) PAGE. The pro-cSPHs (200 ng/μl, 1 μl) were separately incubated with M. sexta PAP3 (40 ng/μl, 1 μl) in 10 μl buffer A at 37°C for 1 (h) The reaction mixtures and controls (40 ng PAP3 and 200 ng proSPH alone) were treated with 1×SDS sample buffer at 95°C for 5 min or the same buffer lacking SDS and DTT at 25°C for 5 min prior to 10% SDS-PAGE and 10% native PAGE, respectively. After electrotransfer, immunoblot analysis was performed using 1:1000 diluted antiserum against hexahistidine tag. In panel (A), pro-cSPH242 (lane 2), pro-cSPH35 (lane 4), and major cleavage products (lanes 3 and 5) are marked by asterisks and arrowheads, respectively. Positions and sizes of the prestained Mr standards are indicated, with the 75 kDa marker highlighted red. In panel (B), the dashed line divides the stacking and separating gels. The smeared bands of PAP3, proSPHs, and cleavage products are marked by green, blue, and red vertical bars, respectively.
To examine the dynamic processes of pro-cSPH cleavage, we separately incubated aliquots of the pro-cSPH35 or pro-cSPH242 with decreasing amounts of PAP3, resolved the reaction mixtures by 10% SDS-PAGE, and performed immunoblot analysis using the hexahistidine antibody (Figure 4). At the lowest PAP3 level, the 40 kDa cSPH35 band was major and the 30 kDa band was minor (panel A, lanes 9 and 10), suggesting that cleavage first occurred at site-1. As the PAP3 concentration increased, the intensity of the 30 kDa cSPH35 increased while the 40 kDa band decreased and disappeared due to cleavage at site-2 (panel A, lanes 3−8). On the native PAGE gel, cleavage of the pro-cSPH35 monomer (panel C, lane 2) at the two sites yielded a smear in the stacking gel and a ladder in the separating gel, and the ratios of smear and ladder increased as more PAP3 was added (Figure 4C). In comparison, pro-cSPH242 processing at a single site is much simpler: as PAP3 increased and pro-cSPH242 disappeared, the 42 kDa band reached the highest level (Figure 4B, lanes 3–10). On the native PAGE gel, there was a band migrating slightly faster than pro-cSPH242 (Figure 4D). As the monomer decreased in lanes 10 and 9, the ladder of cSPH242 complexes extended into the stacking gel in lanes 8, 7, 6, and so on. These results clearly indicated that PAP3 efficiently cleaved the pro-cSPHs and readily changed the association states of the cSPHs, orthologs of the Manduca SPHIs, and SPHII.
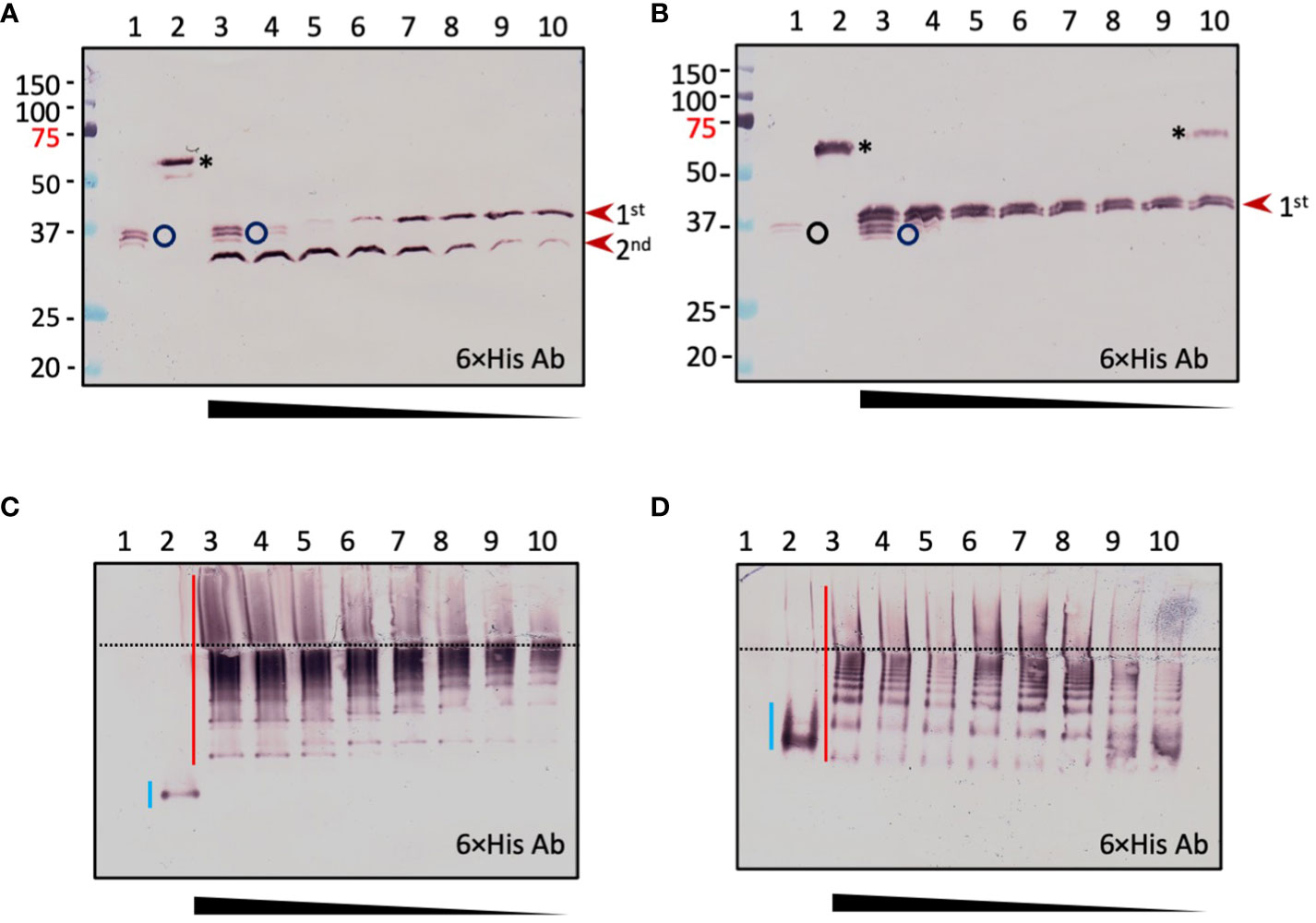
Figure 4 Concentration-dependent PAP3 processing of D melanogaster cSPH35 (A, C) and cSPH242 (B, D) precursors analyzed by 10% SDS and native PAGE followed by immunoblotting. Aliquots of the purified pro-cSPHs (200 ng) were incubated with 80 (lane 3), 40 (lane 4), 20 (lane 5), 10 (lane 6), 5 (lane 7), 2.5 (lane 8), 1.2 (lane 9), 0.6 (lane 10), and 0 (lane 2) ng of PAP3 in 10 μl buffer A at 37°C for 1 (h) The mixtures and PAP3 control (80 ng, lane 1) were subjected to 10% SDS-PAGE (A, B) and 10% native PAGE (C, D), as described in the legend to Figure 3. After electrotransfer, immunoblot analysis was performed using antiserum against hexahistidine tag as primary antibody. In (A, B), PAP3, proSPHs, and major products from the first and second cleavages are marked by circle, asterisk, and arrowheads, respectively. Positions and sizes of the prestained Mr standards are indicated, with the 75 kDa marker highlighted red. In (C, D), the dashed line divides the stacking and separating gels. The smeared bands of pro-cSPHs, and cleavage products are marked by blue and red vertical bars, respectively.
3.4 Identification of the cleavage sites in Drosophila cSPH35 and cSPH242
Similar to Drosophila cSPH35, M. sexta proSPH1b was cleaved by PAP3 first at R82 and then at R133 (9, 11, 12). Like cSPH242, Manduca proSPH2 was activated at a single site (R77). Therefore, we activated pro-cSPH35 and pro-cSPH242 with PAP3, cleaved cSPH35 with V8 protease and cSPH242 with chymotrypsin, and performed LC-MS/MS analysis. Since PAP3 cleaves after R or K, V8 cleaves after D or E, and chymotrypsin cleaves after F, Y, W, M, or L, identification of the peptides released from PAP3-V8 protease and PAP3-chymotrypsin digestion would allow us to deduce the PAP3 cleavage sites in the cSPHs. Indeed, we detected three such peptides and acquired their secondary ion mass spectra (Figure S8) but not in the control pro-cSPH samples treated with chymotrypsin or V8 protease only (Table 2). By mapping these peptides to the mature proteins, we found that pro-cSPH35 was cleaved at R81 and K128 whereas pro-cSPH242 was cleaved at R88. These sites corresponded well with the activation sites in their orthologs from M. sexta.
3.5 PAP3-activated cSPH242 and cSPH35 act as a cofactor for Drosophila PPO1 activation
Drosophila MP2xa cleaved half of the recombinant PPO1 (0.38 μg) but no PO activity was reported (29). Our data showed that MP2xa hardly processed the two pro-cSPHs (Figure S4). Can the PAP3-treated pro-cSPHs greatly enhance PPO1 activation by MP2xa? We found, although there was a clear increase in PPO1 cleavage by MP2xa in the presence of both pro-cSPHs (Figure S6, lane 8 vs. lane 11), a small but significant enhancement in PPO activation (10.5 U) was observed (Figure 5B, bar 7). PAP3 slightly cleaved Drosophila PPO1 but did not increase PO activity (Figure 5A, lane 7; Figure 5B, bar 4). To obtain the active cofactor without interfering with the cleavage of DmPPO1 by MP2xa (especially when the cSPHs were present), we blocked PAP3 with M. sexta serpin-3 (45), which did not inhibit MP2xa’s IEARase activity by forming an inactive complex (Figure S5). After serpin-3 inhibition, the PAP3-generated cSPHs were incubated with Drosophila PPO1 and MP2xa (i.e., Factor Xa-treated proMP2xa). MP2xa cleaved nearly half of the PPO1 and yielded a 38.9 U of PO at a specific activity of 260 U/μg (Figure 5A, lane 13; Figure 5B, bar 11), clearly demonstrating the cofactor role of the SPHI-II complexes versus their precursors (Figure 5A, lane 7). In comparison, PPO1 cleavage at 1/5, 1/5, and 1/10 levels (Figure 5A, lanes 9, 11, and 12) only yielded 4.4, 0.3, and 0.7 U of PO (Figure 5B, bars 3, 8, and 9), respectively. As we reported previously (46), there is no direct correlation between PPO cleavage extent and PO activity, and the presence of a high Mr cofactor of SPHI and SPHII is critically important for generating highly active PO.
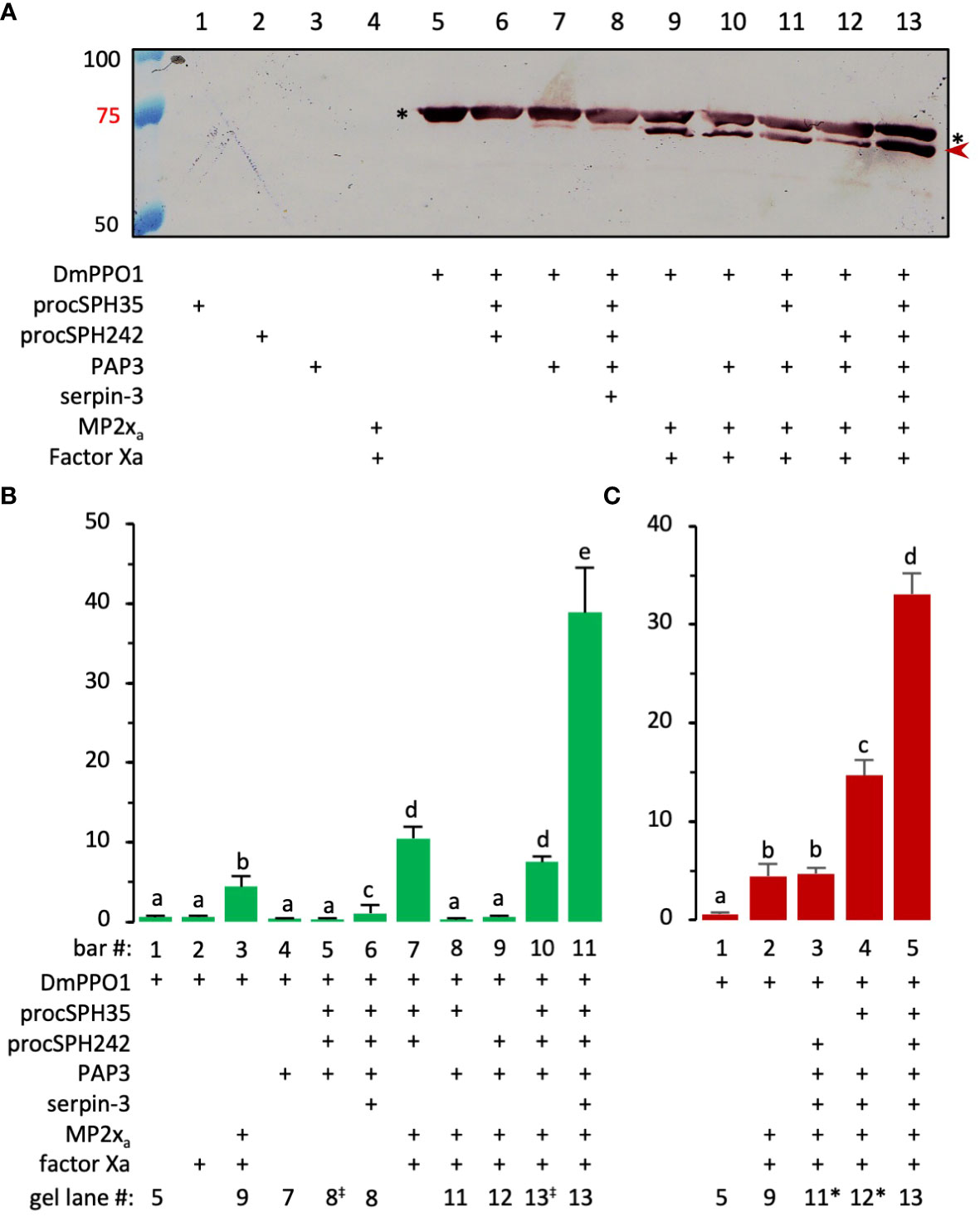
Figure 5 Effect of PAP3-treated pro-cSPH242 and pro-cSPH35 on D melanogaster PPO1 activation by MP2xa. (A) PPO cleavage by MP2xa. ProMP2xa (0.44 μg) was activated by Factor Xa (0.1 μg) in buffer A (2 μl) for 1 h at 37°C. In another reaction, cSPH242 and cSPH35 precursors (400 ng each) were incubated with M. sexta PAP3 (20 ng) in buffer A (8 μl) at 25°C for 1 h, and PAP3 was then inhibited by M. sexta serpin-3 (100 ng/μl, 2 μl) at 25°C for 30 min. D melanogastor PPO1 (2 μl, 0.3 μg/μl) from E coli was incubated with active MP2xa (400 ng) in the presence or absence of active cSPH242 and cSPH35 (400 ng each) on ice for 1 h in 10 μl buffer (A) Half of the mixture (lane 13) and controls (lanes 1−12) were subjected to 7% SDS-PAGE followed by immunoblotting using the specific PPO1 antibody (26). As indicated in panel A, the missing component(s) in the controls were replaced by the same volume of buffer (A, B) PPO1 activation. PO activities in some of the remaining samples were measured in 150 μl of 2.0 mM dopamine in 50 mM sodium phosphate, pH 6.5, using a microplate reader, and the activity data from three biological replicates (mean ± SEM, n = 3) were plotted in the bar graph. (C) Roles of the cSPHs. As described in panel B, a similar experiment was performed to assess the relative contributions of cSPH242 and cSPH35 in D melanogaster PPO1 activation. To facilitate comparison, corresponding lane numbers in panel (A) are indicated below the two bar graphs, with the slight changes indicated as “‡” or “*”. The statistical significance of the differences was determined by one-way ANOVA and indicated by different letters (a−e) for p <0.05.
To assess the relative importance of cSPH35 and cSPH242 to the cofactor activity, we activated one of the pro-cSPHs by PAP3, inhibited PAP3 using serpin-3, and then added PPO1 and MP2xa to the mixture. As shown in Figure 5C, the removal of pro-cSPH242 and pro-cSPH35 caused an activity decrease from 33.1 U to 14.7 U and 4.7 U, respectively. Both cSPHs, cSPH35 in particular, are required for the cofactor function.
3.6 Effects of cSPH35 or cSPH242 knockdown on melanization and survival of infected adults
To examine phenotypes of the single knockdowns, we first measured the mRNA level changes in the two RNAi lines. Compared with the control, 40% and 60% of the cSPH35 and cSPH242 mRNA levels were detected in the adults (Figure 6A). After incubation at 25°C for 10 min with 0.1% PTU (a PO inhibitor that prevents protein crosslinking), nearly one fifth of the PPO1 in the control hemolymph was cleaved whereas no cleavage at the activation site of Arg52 was detected in the samples from cSPH242 or cSPH35 RNAi adults (Figure 6B). In another set of samples lacking PTU, PO activities were detected at 26.0, 4.7, and 1.2 U per μl of hemolymph from the control, cSPH242, and cSPH35 knockdown adults (Figure 6C). Consistently, hemolymph from wild-type flies turned black within 5 min at 25°C while samples from the RNAi lines stayed colorless for 60 min at least (Figure 6E). Some of the cSPH242 knockdowns appeared to have insufficient melanin deposition in the posterior abdominal segments (Figure 6D). This phenotype of light color is not found in the control or cSPH35 RNAi flies, suggesting that cSPH242 participates in cuticle pigmentation and its lower expression (Figure 6A) somehow led to insufficient melanin formation.
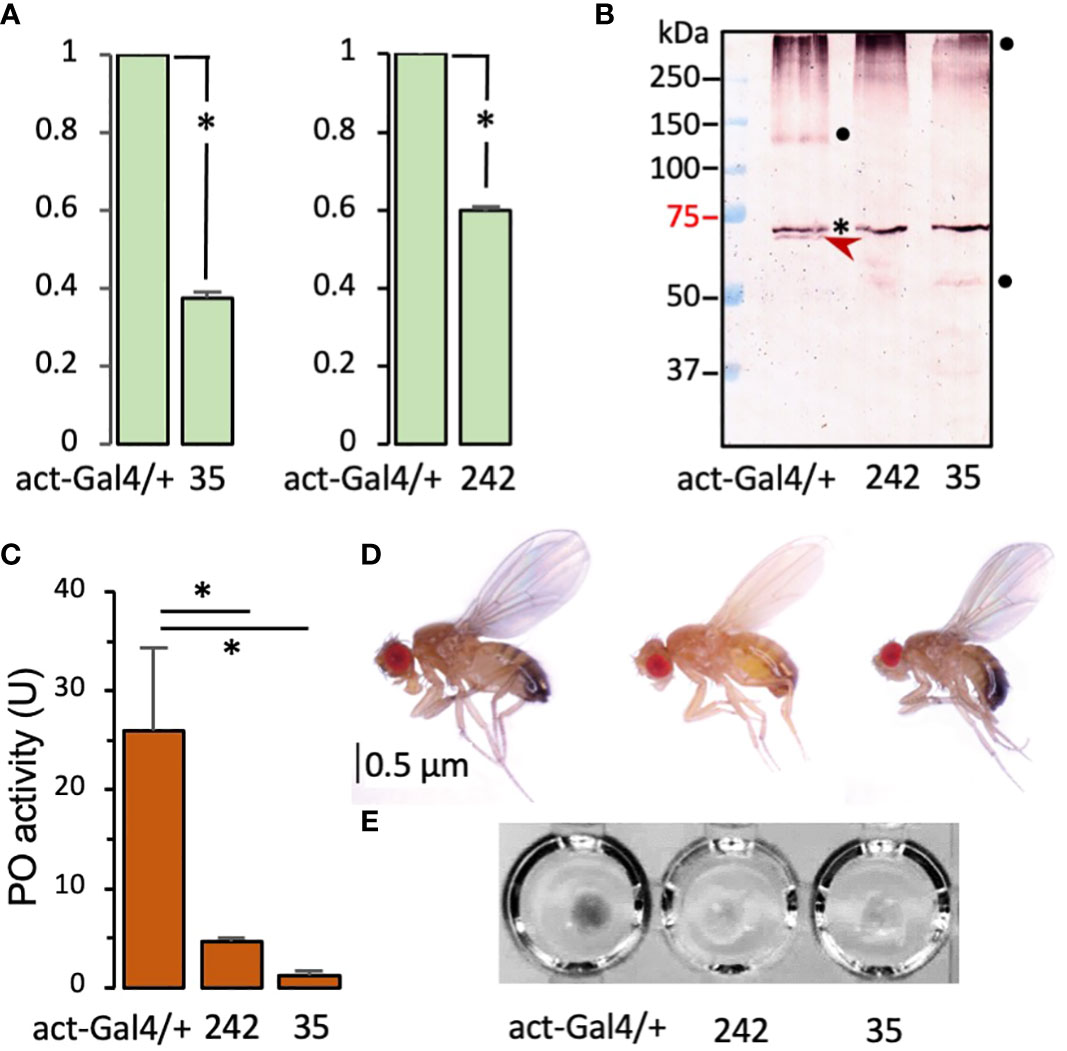
Figure 6 Effects of systemic knockdown of cSPH35 and cSPH242 expression in D melanogastor adults. (A) Decreases in the cSPH mRNA levels. The mRNA levels in adults (5 days after eclosion) were determined by qRT-PCR as described in Section 2.2. In relation to the control (set at 1.00), the cSPH transcript levels (mean ± SEM, n = 3) in the two RNAi lines are shown in the bar graphs. *, p <0.05 in Student’s t-test. (B) Reduction of PPO1 cleavage. Hemolymph samples from ten adults were separately collected from the control, cSPH35, and cSPH242 RNAi lines. The plasma samples (1.0 μl each) were incubated with 0.1% PTU in buffer A (10 μl) for 30 min at room temperature, separated by 7.5% SDS-PAGE, and detected by antibody to D melanogaster PPO1. PPO1 and its 73 kDa cleavage product are marked with asterisk and arrowhead, respectively. The high and low Mr bands (●) may represent cleaved PO in different association states. (C) PO activity decrease. Freshly collected hemolymph samples (1.0 μl from 10 individuals in the control and RNAi groups, three biological replicates) were assayed immediately for PO activity using dopamine as a substrate. The PO activities (mean ± SEM, n = 3) are shown in the bar graph, with * indicating p <0.05 in Student’s t-test. (D) Body color change. Images of adults from the control and RNAi lines, show that 30% of the cSPH242 knockdowns with low pigmentation. (E) Hemolymph melanization. Hemolymph samples (5 μl from 50 individuals) from the three groups of adults were transferred to wells of a microplate, where spontaneous melanization occurred in the control hemolymph within 5 min.
We studied the impacts of the cSPH knockdown on melanization at the site of septic pricking with E. faecalis (OD600 = 0.8). Eighty-two percent of the wild-type flies exhibited a black spot on the thorax two days after inoculation (Figure 7A). The percentage decreased to 53% in the RNAi line of cSPH242. Consistent with the lowest PO activity (Figure 6C), only 30% of the adult flies with cSPH35 expression knocked down had melanin deposited at the wound site. The intergroup differences were highly significant. To further investigate the role of these cSPHs in the immune response, we raised the dosage of E. faecalis (OD600 = 1.2) and monitored the survival of infected adults for ten days. Compared with the wild-type flies, the survival rate decreased from 45% to 30% and 15% in the cSPH35 and cSPH242 RNAi lines (Figure 7B), respectively. The differences between the control and RNAi groups were statistically significant.
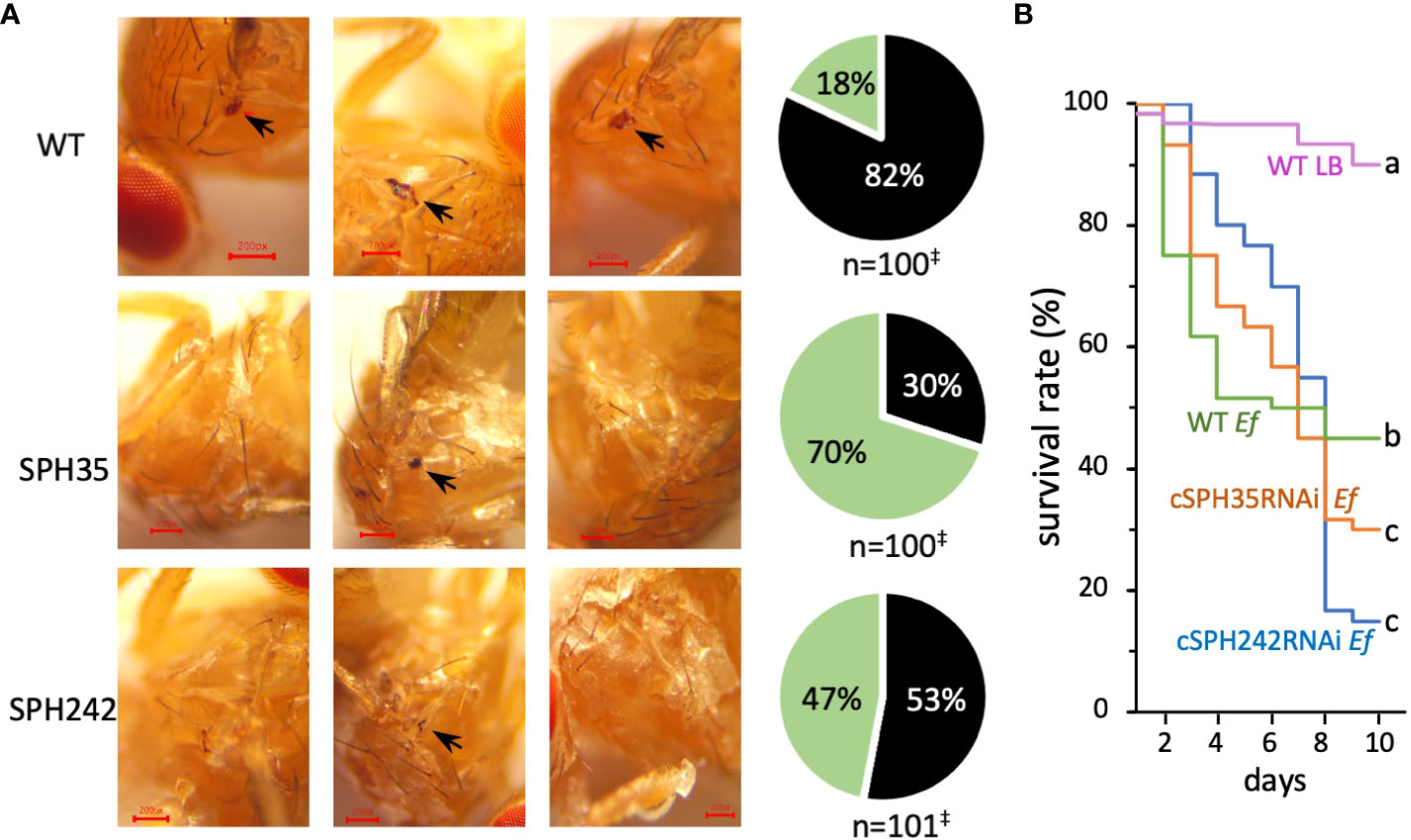
Figure 7 Effects of cSPH242 or cSPH35 knockdown on melanization and survival of infected D melanogastor adults. (A) Melanization at the site of septic pricking. Representative images of the control, cSPH35, and cSPH242 RNAi flies with or without a black spot (black arrow) in the wounded thorax at 24 h after exposure to live E faecalis (OD600 = 0.8). Pie charts on the right show the percentages of flies in the three groups with (black) and without (green) a melanin spot. ‡, data pooled from three biological replicates; intergroup differences: p <0.0005 in Student’s t-test with Boniferoni correction. (B) Survival curves. Twenty-five flies from each group were pricked in the thorax with LB medium or live E faecalis (OD600 = 1.2), transferred to clean vials with a fresh diet, and then incubated at 25°C. Survived flies were counted daily before transfer to new vials. The experiment was repeated at least three times and percentages of survived flies in each pooled group were shown with different time points (days 1–10 after treatment).
4 Discussion
4.1 Requirement of a cofactor for PPO activation in various insects
Despite the apparent discrepancy in B. mori (6), it is now clear that PPO activation is mediated by a clip-domain SP (i.e., PAP) in the presence of one or two cSPHs in all insects studied so far. This seems to be a conserved mechanism that existed before the radiation of holometabolous insects, including beetles, moths, and mosquitoes (9, 12, 13, 15, 17, 18, 47). Guided by the orthologous relationships, we identified the two types of cSPHs in D. melanogaster (Figure S1) and demonstrated by in vitro and in vivo experiments that the mechanism is functional in the fruit fly as well (Figure 8). In most other Drosophila species, single orthologs of cSPH35 and cSPH242 exist to likely play the same role. While their gene orthologs are found in the genomes of B. mori, Tribolium castaneum, and T. molitor (31; data not shown), it is unclear if the corresponding protein products work together to further enhance PPO activation. T. molitor and H. diomphalia PPAF2 (SPHI) alone largely increased PPO activation (14, 16, 17). While the requirement for the SPHI and SPHII co-presence in the beetles and A. aegypti (18) remains unexamined, it is known that Apis mellifera genome has one SPHI but no SPHII gene (31). With a single PPO gene, the honeybee PPO activation may represent the simplest reaction among all holometabolous insects. On the other end of the spectrum, the African malaria mosquito, Anopheles gambiae, has eleven SPHI, one SPHII, and nine PPO genes in its genome (42). Their roles in melanization, a key immune response against pathogens and parasites, are worth exploring. Lineage-specific expansions of the SPHI and SPHII subfamilies in lepidopteran pests (12) and their functional implications and molecular mechanisms also need to be examined in the future to better understand this defense response.
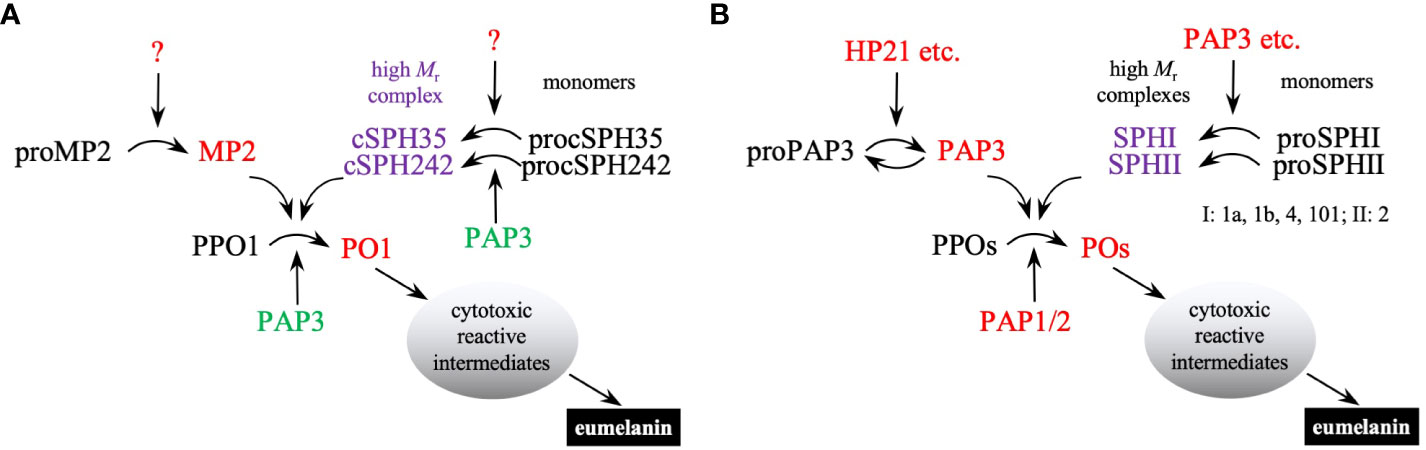
Figure 8 A conserved mechanism of PPO activation in D melanogaster (A) and M. sexta (B). In Drosophila, unknown clip-domain SPs activate MP2, cSPH35, and cSPH242 precursors (black font). Active MP2 (red font for enzyme activity) and its cofactor (purple font for catalytic inactivity but cofactor activity) cleave PPO1 to generate highly active PO1. Under in vitro conditions, M. sexta PAP3 (green font) activates the Drosophila proSPH monomers to form high Mr complexes, which act as a cofactor for PPO1 activation by M. sexta PAP3. In Manduca, HP21 and other hemolymph proteases (including PAP3 itself) activate proPAP3. PAP3 and additional clip-domain SPs cleave proSPHIs and proSPHII to generate active cofactors for PPO activation by PAP1/2/3. Since multiple proteins perform the same function in each step, M. sexta PPO activation appears to be more complicated than in Drosophila. Once active POs are generated, they catalyze the formation of reactive intermediates to kill invading microbes and form melanin sheaths to sequester them.
4.2 Cleavage activation of the pro-cSPHs in D. melanogaster, M. sexta, and H. diomphalia
We have not yet identified any endogenous activating protease of pro-cSPH35 or pro-cSPH242 in the fruit fly. Manduca PAP3 yielded the fly cSPHs to assist the activation of Drosophila PPO1 by MP2xa (Figures 3–5). Cleavage first occurred at the corresponding site between Cys-3 and Cys-4 of their clip domain in the proSPHI and proSPHII, and then at the junction between the linker and protease-like domain of the SPHI intermediate (4), suggesting that their endogenous activating protease(s) act the same way in Drosophila. If proven true, this would be remarkable since Lepidoptera and Diptera separated from each other >250 million years ago and site-1 is located in an exposed loop of the clip domains (48–50). It is likely that cleavage at site-1 leads to the exposure of site-2 in the SPHI to PAP3 for the second cut (Figure 4A). Relative roles of these cleavage events in generating an active cofactor (i.e., SPHI-II complex) remain unclear and need to be determined biochemically.
The formation of high Mr complexes is a vital process for pathogen recognition, melanization, and hemolymph coagulation during insect immune responses (51). In this investigation, cleavage-induced oligomerization of cSPH35 and cSPH242 has been clearly demonstrated (Figures 4C, D, S7). While this agrees well with the positive correlation between native SPHI sizes and cofactor activity in M. sexta (12), we noticed a difference between the SPHIIs in these two insects. Manduca SPH2 had much lower association states and, therefore, may attach to the large scaffolds of SPHIs (e.g., 1b and 101), whereas Drosophila cSPH242 and cSPH35 oligomers had similar Mr’s and could further associate with each other (Figure S7, lanes 7, 9, and 10) for PPO attachment, PAP cleavage, and PO oligomerization (8). Unlike the two-hexameric-toroidal structure of H. diomphalia PPAF2 (48), no such ordered assembly (e.g., hexamer) was detected as a prominent form among the D. melanogaster SPHI or SPHII complexes (Figures 4C, D). Instead, they appeared to grow in size by adding one SPH unit at a time, as suggested in the analysis of association states of cSPH35 and cSPH242 by native gel electrophoresis (Table 1). Oligomers of the two cSPHs are further associated with higher Mr complexes (Figure S7, lanes 7, 9, and 10) and their co-presence is required for displaying the maximal PO activity (Figure 5C). Consistent with the association, the cofactor isolated from hemolymph of M. sexta pharate pupae had an average Mr of 790 kDa, with a continuous distribution from the void volume of the Sephacryl S500 column (11).
4.3 Proteolytic cleavage and PPO activation in various insects
Not all PPO cleavages generate active PO and extra cautions are necessary when a PO activity is assigned to (a) specific cleavage product(s) due to several reasons. First of all, PPO cleavage is essential for in vivo activation (46). Cationic surfactants (e.g., cetylpyridinium chloride) and organic solvents (e.g., 30% ethanol) do activate PPOs in vitro (52), but that is not physiologically relevant. Secondly, there is no direct correlation between cleavage extent and PO activity level (Figure 5, 46). Drosophila MP2xa directly cleaved 50% of the recombinant PPO1 (0.38 μg) but no PO activity was reported (29). On the other hand, since there is a single cleavage in PPO1 probably at the classical activation site between Arg52 and Phe53, the 73 kDa cleavage product and its assemblies are responsible for the high PO-specific activity (Figure 5, lane 13). There are reports for active POs at other sizes, such as 60 kDa PO of H. diomphalia (14), 50 kDa PO3 of A. aegypti (53), and 60 kDa PO1 of D. melanogaster (52). Unfortunately, there is a lack of explicit experimental evidence for these claims. For instance, besides the appearance of the 60 kDa cleavage product, there was a clear intensity increase of the 75 kDa band to account for the boost of PPO activation when H. diomphalia PPAF2 was present (14). In the other cases, only low levels of PO activity were detected, probably due to the absence of a fully functional cofactor of SPHI and SPHII. Thirdly, PPO activation occurs only when PPO, active PAP, and its cofactor coexist (46), suggesting that proper association and maintenance of active conformation is crucial for producing PO with high specific activity (Figure 5). So far, mechanisms for the generation of highly active PO are unclear but, as more cofactors of PPO activation are prepared in vitro from proSPHI and proSPHII, details of the molecular interactions among PPO, PAP, and cofactor are expected to be uncovered in more insects. In D. melanogaster, the knockdown of cSPH35 or cSPH242 expression significantly reduced melanization in adults (Figures 6, 7), similar to the phenotype of PPO1 and PPO2 knockouts (20). It is worth testing if cSPH35 and cSPH242 are also required for PPO2 activation and what specific activity of PO is generated under pathological conditions for comparison with the in vitro data (260 U/μg).
4.4 Conclusions
Proteolytic activation of PPO is required for the survival of D. melanogaster during a microbial infection. This process involves PPO, its activating protease, and a high Mr cofactor (Figure 8). In the reaction of MP2 and PPO1 only, the PAP cleaves the substrate likely between Arg52 and Phe53 to generate active PO1, which seems to quickly lose its activity. The cofactor of cSPH35 and cSPH242 increases the PPO1 cleavage and may maintain PO1 in its active conformation. M. sexta PAP3 cleaves pro-cSPH35 at Arg81 and then at Lys128 to cause cSPH35 association. PAP3 processing of pro-cSPH242 at Arg88 also induces oligomerization of cSPH242. An endogenous activating protease, currently unknown, may play the role of PAP3 to generate the cofactor in Drosophila. In the presence of the SPHI and SPHII complexes formed in vitro, MP2xa-treated PPO1 reached an unprecedentedly high PO-specific activity of 260 U/μg. Coupling active, oligomeric PO near the site of injury or infection through the high Mr cofactor may ensure melanization occurs as a potent, local defense response against pathogens. On the other hand, a rapid activity loss of the diffusing PO could minimize the damage to distant host cells or tissues. These biochemical data are consistent with the in vivo test results. The lower PPO1 cleavage, PO activity, hemolymph melanization, and survival rate highlight the importance of clip-domain SPs and SPHs in this antimicrobial defense response. It is further demonstrated that the combined approach, guided by orthologous relationships and expression patterns, is successful in the elucidation of mechanisms for complex physiological processes in the genetic model species.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.6084/m9.figshare.23786949.
Ethics statement
Ethical review and approval was not required for the study on animals in accordance with the local legislation and institutional requirements.
Author contributions
QJ designed and did most of the experiments, and she also prepared the draft of the manuscript. HJ guided the work, he provided funds and mainly focused on writing the manuscript. YW purified and provided a lot of important proteins. HY helped to do some important in vivo experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Institutes of Health Grants GM58634 and AI139998 to HJ.
Acknowledgments
We would like to thank the reviewers for their critical comments which helped us to improve the manuscript. The plasmids PPO1/pET28a and pMP2xa/pFastBac1 were gifts from Dr. Erjun Ling at the Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences and Dr. Chunju An in the Department of Entomology at China Agricultural University, respectively. Dr. Won-Jae Lee at Seoul National University kindly provided the polyclonal antiserum to Drosophila PPO1. Dr. Steven Hartson at the DNA Protein Core Facility at Oklahoma State University provided support for the determination of PAP3 cleavage sites by mass spectrometry. This article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKL03054.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1244792/full#supplementary-material
Supplementary Figure 1 | Phylogenetic relationships of the SPHI and SPHII subfamily members in Drosophila spp., Bactrocera oleae, Ceratitis capitata, Musca domestica, and Manduca sexta. BLASTP searches of GenBank, hit examination, alignment of entire sequences, and construction of the neighbor-joining tree were performed as described before (12). Species names and their abbreviations in parenthesis are Drosophila albomicans (Dal), D. elegans (Del), D. erecta (Der), D. grimshawi (Dgr), D. innubila (Din), D. mauritiana (Dma), D. melanogastor (Dme), D. persimilis (Dpe), D. pseudoobscura (Dps), D. sechellia (Dse), D. suzukii (Dsu), D. virilis (Dvi), D. willistoni (Dwi), D. yakuba (Dya), B. oleae (Bol), C. capitata (Cca), M. domestica (Mdo), and M. sexta (Mse). Suggested protein names consist of an abbreviated species name and a group name of SPHI or SPHII. The M. sexta SPHIs (i.e., 1a, 1b, 4, and 101) and SPHII (i.e., 2) are in bold font, and so are D. melanogastor cSPH242 and cSPH35. Duplicated genes in a single species are labeled by “‡”. Percentage bootstrap values greater than 50% are marked near branching points of the tree, with values ≥99 simplified as “*”.
Supplementary Figure 2 | Expression profiles of D. melanogastor cSPH35 and cSPH242 genes in different tissue samples and developing stages. (A) The relative mRNA levels, as represented by FPKM values (31), are shown in bar graphs. The stages labeled “*” roughly correspond to the L, P, and F stages in . The 52 cDNA libraries were constructed from various tissues and stages, which are abbreviated using: W, whole insect; E, embryo; L1–3, 1st, 2nd, and 3rd instar larval; PS, puff stage; preP, prepupal; P, pupal; A, adult; M, male; F, female; h, hour; D, day; G, gut; PG, posterior midgut; FB, fat body; ID, imaginal discs, SG, salivary glands; T, testes; O, ovaries of adult mated female; AG-AmM, accessory glands of adult mated male; AvF, adult virgin female; MT, Malpighian tubules. (B) Abundances of cSPH35 and cSPH242 proteins (44) are shown in the bar graphs as log2(LFQ/5×106 + 1) values (mean ± SEM, n = 4). L3c, crawling 3rd instar larvae.
Supplementary Figure 3 | Activation of proMP2xa by Factor Xa and amidase activity of the MP2xa. (A) immunoblot. As described in Section 2.7, the purified proMP2xa was treated with Factor Xa in 10 μl buffer A. Half of the mixture and controls were subjected to 10% SDS-PAGE followed by immunoblot analysis using hexahistidine antibody. The MP2xa precursor (*) and catalytic domain (arrowhead) are labeled. (B) Amidase activity. After IEARpNA was added to the remaining samples, substrate hydrolysis was monitored in a kinetic mode on a plate reader. IEARase activities (mean ± SEM, n = 3) are shown in a bar graph. *, p <0.05 in Student’s t-test.
Supplementary Figure 4 | MP2xa did not significantly activate the cSPH35 and cSPH242 precursors. After proMP2xa (0.44 μg) was preincubated with Factor Xa (0.1 μg) for 1 h at 37 °C in 11 μl buffer A, 1.0 μl of the mixture was added to purified pro-cSPH35 or pro-cSPH242 (200 ng/μl, 1 μl) in 10 μl buffer A. After incubation at 37°C for 1 h, the reaction mixtures and controls were either treated with 1×SDS sample buffer at 95°C for 5 min prior to 10% SDS-PAGE or mixed with the sample buffer lacking SDS and DTT prior to 10% native PAGE. After electrotransfer, immunoblot analysis was performed using 1:1000 diluted antiserum against hexahistidine tag. In panels (A, B), pro-cSPH35 or pro-cSPH242 were marked by “*”. Positions and sizes of the prestained Mr standards are indicated, with the 75 kDa marker highlighted red. In panel (C), the dashed line divides the stacking and separating gels. The bands of MP2xa, pro-cSPH35, and pro-cSPH242 were marked by green, blue, and red vertical bars, respectively.
Supplementary Figure 5 | M. sexta serpin-3 inhibited PAP3 but not Drosophila MP2xa. (A) Active MP2xa (0.44 μg) was first generated as described in Figure S4. M. sexta serpin-3 (1 μl, 0.3 μg/μl) was then added to the mixture and incubated for 30 min at room temperature. The reaction mixture and controls were treated with 1×SDS sample buffer at 95°C for 5 min prior to 10% SDS-PAGE and immunoblot analysis using 1:1000 diluted antiserum against hexahistidine tag. MP2xa precursor and catalytic domain were marked asterisk and arrowhead, respectively. (B) Active PAP3 (0.1 μg) (left) or MP2xa (0.22 μg) (right) was incubated with M. sexta serpin-3 (0.3 μg) at room temperature for 30 minutes. As described in Section 2.7, IEARase activities (mean ± SEM, n = 3) were measured and plotted as bar graphs. Pairwise comparisons were performed between control and treated samples by Student’s t-test. *, p <0.05; ns, not significant (p >0.05).
Supplementary Figure 6 | D. melanogastor PPO1 cleavage by MP2xa in the presence of pro-cSPH35 and pro-cSPH242. ProMP2xa (0.44 μg) was first activated by Factor Xa (0.1 μg) in 2 μl buffer A at 37°C for 1 h. The purified PPO1 (1 μl, 0.3 μg/μl) from E. coli was incubated on ice for 1 h with the active MP2xa (0.2 μg) in the presence of pro-cSPH35 and pro-cSPH242 (0.2 μg each) in 10 μl buffer A on ice for 1h. Half of the mixture (lane 11) and controls (lanes 1–10) were subjected to 7% SDS-PAGE followed by immunoblot analysis using specific antibodies to PPO1 (26). Missing components (s) in the controls were replaced by the same volume of buffer A.
Supplementary Figure 7 | Mobility changes accompanying the cSPH cleavage and association analyzed by native PAGE. (A) An example of the stained gels. The purified pro-cSPH35 and pro-cSPH242 (200 ng/μl, 5 μl for each or 5 + 5 μl for both) were incubated with PAP3 (40 ng/μl, 5 μl) in 50 μl buffer A for 1 h at 37°C. One-fifth of the three mixtures and two controls of pro-cSPH35 and pro-cSPH242 alone were separated along with Mr standards by 4.5%, 7%, 10%, 12%, and 15% native discontinuous PAGE, followed by Coomassie staining. The standard proteins (29 kDa carbonic anhydrase, 67 kDa bovine serum albumin, 200 kDa β-amylase, 443 kDa apoferritin, and 669 kDa thyroglobulin in lanes 1–5), proSPHs (in lanes 6 and 8), and new bands (in lanes 7, 9, and 10) are marked with “●”, “*” and arrowheads, respectively. (B) Native molecular mass (Mr) standard curve. (C) Parameters of the pro-cSPH and cSPH bands. For a specific form or association state, the slope (Kr) of the log10Rm (relative mobility) vs. %T (total monomer concentration) curve from its corresponding bands on the gels was used to derive its unknown mass using the standard curve on Ferguson plot (37). Estimated Mr’s of the complexes of cleaved cSPH35, and cSPH242, and both are in bold font.
Supplementary Figure 8 | Secondary ion mass spectra of the characteristic peptides from the PAP3-treated cSPHs after V8 (A, B) or chymotrypsin (C) proteolytic digestion. Based on the peptide sequences and their locations in the proSPH sequences, the cleavage sites of M. sexta PAP3 were deduced and shown in.
Abbreviations
PO and PPO, phenoloxidase and its precursor; PAP, PPO activating protease; SP and SPH, serine protease and its noncatalytic homolog; CLIP, clip-domain SP or SPH; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
References
1. Nappi AJ, Christensen BM. Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem Mol Biol (2005) 35:443–59. doi: 10.1016/j.ibmb.2005.01.014
2. Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol (2010) 708:181–204. doi: 10.1007/978-1-4419-8059-5_10
3. Park JW, Kim CH, Rui J, Park KH, Ryu KH, Chai JH, et al. Beetle immunity. In: Söderhäll K, editor. Invertebrate Immunity. Adv Exp Med Biol, New York, NY:Springer. vol. 708 (2010). p. 163–80.
4. Kanost MR, Jiang H. Clip-domain serine proteases as immune factors in insect hemolymph. Curr Opin Insect Sci (2015) 11:47–55. doi: 10.1016/j.cois.2015.09.003
5. Marieshwari BN, Bhuvaragavan S, Sruthi K, Mullainadhan P, Janarthanan S. Insect phenoloxidase and its diverse roles: melanogenesis and beyond. J Comp Physiol B (2023) 193:1–23. doi: 10.1007/s00360-022-01468-z
6. Satoh D, Horii A, Ochiai M, Ashida M. Prophenoloxidase activating enzyme of the silkworm, Bombyx mori: purification, characterization, and cDNA cloning. J Biol Chem (1999) 274:7441–53. doi: 10.1074/jbc.274.11.7441
7. Jiang H, Wang Y, Kanost MR. Pro-phenoloxidase activating proteinase from an insect, Manduca sexta: a bacteria-inducible protein similar to Drosophila easter. Proc Natl Acad Sci USA (1998) 95:12220–5.
8. Jiang H, Wang Y, Yu XQ, Kanost MR. Prophenoloxidase-activating proteinase-2 from hemolymph of Manduca sexta: a bacteria-inducible serine proteinase containing two clip domains. J Biol Chem (2003) 278:3552–61. doi: 10.1074/jbc.M205743200
9. Jiang H, Wang Y, Yu XQ, Zhu Y, Kanost MR. Prophenoloxidase-activating proteinase-3 from Manduca sexta hemolymph: a clip-domain serine proteinase regulated by serpin-1J and serine proteinase homologs. Insect Biochem Mol Biol (2003) 33:1049–60. doi: 10.1016/S0965-1748(03)00123-1
10. Yu XQ, Jiang H, Wang Y, Kanost MR. Nonproteolytic serine protease homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem Mol Biol (2003) 33:197–208. doi: 10.1016/S0965-1748(02)00191-1
11. Wang Y, Jiang H. Prophenoloxidase (proPO) activation in Manduca sexta: an analysis of molecular interactions among proPO, proPO-activating proteiase-3, and a cofactor. Insect Biochem Mol Biol (2004) 34:731–42. doi: 10.1016/j.ibmb.2004.03.008
12. Jin Q, Wang Y, Hartson SD, Jiang H. Cleavage activation and functional comparison of Manduca sexta serine protease homologs SPH1a, SPH1b, SPH4, and SPH101 in conjunction with SPH2. Insect Biochem Mol Biol (2022) 144:103762. doi: 10.1016/j.ibmb.2022.103762
13. Wang Q, Yin M, Yuan C, Liu X, Hu Z, Zou Z, et al. Identification of a conserved prophenoloxidase activation pathway in cotton bollworm Helicoverpa armigera. Front Immunol (2020) 11:785. doi: 10.3389/fimmu.2020.00785
14. Lee SY, Kwon TH, Hyun JH, Choi JS, Kawabata SI, Iwanaga S, et al. In vitro activation of prophenoloxidase by two kinds of prophenoloxidase activating factors isolated from hemolymph of coleopteran, Holotrichia diomphalia larvae. Eur J Biochem (1998) 254:50–7. doi: 10.1046/j.1432-1327.1998.2540050.x
15. Kwon TH, Kim MS, Choi HW, Joo CH, Cho MY, Lee BL. A masquerade-like serine proteinase homologue is necessary for phenoloxidase activity in the coleopteran insect, Holotrichia diomphalia larvae. Eur J Biochem (2000) 267:6188–96. doi: 10.1046/j.1432-1327.2000.01695.x
16. Lee KY, Zhang R, Kim MS, Park JW, Park HY, Kawabata S, et al. A zymogen form of masquerade-like serine proteinase homologue is cleaved during pro-phenoloxidase activation by Ca2+ in coleopteran and Tenebrio molitor larvae. Eur J Biochem (2002) 269:4375–83. doi: 10.1046/j.1432-1033.2002.03155.x
17. Kan H, Kim CH, Kwon HM, Park JW, Roh KB, Lee H, et al. Molecular control of phenoloxidase-induced melanin synthesis in an insect. J Biol Chem (2008) 283:25316–23. doi: 10.1074/jbc.M804364200
18. Ji Y, Lu T, Zou Z, Wang Y. Aedes aEgypti CLIPB9 activates prophenoloxidase-3 in the presence of CLIPA14 after fungal infection. Front Immunol (2022) 13:927322. doi: 10.3389/fimmu.2022.927322
19. Binggeli O, Neyen C, Poidevin M, Lemaitre B. Prophenoloxidase activation is required for survival to microbial infections in Drosophila. PLoS Pathog (2014) 10:e1004067. doi: 10.1371/journal.ppat.1004067
20. Dudzic JP, Kondo S, Ueda R, Bergman CM, Lemaitre B. Drosophila innate immunity: regional and functional specialization of prophenoloxidases. BMC Biol (2015) 13:81. doi: 10.1186/s12915-015-0193-6
21. Nam HJ, Jang IH, Asano T, Lee WJ. Involvement of prophenoloxidase 3 in lamellocyte-mediated spontaneous melanization in Drosophila. Mol Cells (2008) 26:606–10.
22. Castillejo-López C, Häcker U. The serine protease Sp7 is expressed in blood cells and regulates the melanization reaction in Drosophila. Biochem Biophys Res Commun (2005) 338:1075–82. doi: 10.1016/j.bbrc.2005.10.042
23. Leclerc V, Pelte N, El Chamy L, Martinelli C, Ligoxygakis P, Hoffmann JA, et al. Prophenoloxidase activation is not required for survival to microbial infections in Drosophila. EMBO Rep (2006) 7:231–5. doi: 10.1038/sj.embor.7400592
24. Tang H, Kambris Z, Lemaitre B, Hashimoto C. Two proteases defining a melanization cascade in the immune system of Drosophila. J Biol Chem (2006) 281:28097–104. doi: 10.1074/jbc.M601642200
25. Ayres JS, Schneider DS. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol (2008) 6:2764–73. doi: 10.1371/journal.pbio.0060305
26. Nam HJ, Jang IH, You H, Lee KA, Lee WJ. Genetic evidence of a redox-dependent systemic wound response via Hayan protease-phenoloxidase system in Drosophila. EMBO J (2012) 31:1253–65. doi: 10.1038/emboj.2011.476
27. Zhao P, Li J, Wang Y, Jiang H. Broad-spectrum antimicrobial activity of the reactive compounds generated in vitro by Manduca sexta phenoloxidase. Insect Biochem Mol Biol (2007) 37:952–9. doi: 10.1016/j.ibmb.2007.05.001
28. Zhao P, Lu Z, Strand MR, Jiang H. Antiviral, anti-parasitic, and cytotoxic effects of 5, 6-dihydroxyindole (DHI), a reactive compound generated by phenoloxidase during insect immune response. Insect Biochem Mol Biol (2011) 41:645–52. doi: 10.1016/j.ibmb.2011.04.006
29. An C, Zhang M, Chu Y, Zhao Z. Serine protease MP2 activates prophenoloxidase in the melanization immune response of Drosophila melanogaster. PLoS One (2013) 8:e79533. doi: 10.1371/journal.pone.0079533
30. Li X, Ma M, Liu F, Chen Y, Lu A, Ling QZ, et al. Properties of Drosophila melanogaster prophenoloxidases expressed in Escherichia coli. Dev Comp Immunol (2012) 36:648–56. doi: 10.1016/j.dci.2011.11.005
31. Cao X, Jiang H. Building a platform for predicting functions of serine protease-related proteins in Drosophila melanogaster and other insects. Insect Biochem Mol Biol (2018) 103:53–69. doi: 10.1016/j.ibmb.2018.10.006
32. Wang Y, Lu Z, Jiang H. Manduca sexta proprophenoloxidase activating proteinase-3 (PAP3) stimulates melanization by activating proPAP3, proSPHs, and proPOs. Insect Biochem Mol Biol (2014) 50:82–91. doi: 10.1016/j.ibmb.2014.04.005
33. Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (1993) 118:401–15. doi: 10.1242/dev.118.2.401
34. Kapila R, Poddar S, Meena N, Prasad NG. Investment in adult reproductive tissues is affected by larval growth conditions but not by evolution under poor larval growth conditions in Drosophila melanogaster. Curr Res Insect Sci (2021) 2:100027. doi: 10.1016/j.cris.2021.100027
35. Lu Z, Jiang H. Expression of Manduca sexta serine proteinase homolog precursors in insect cells and their proteolytic activation. Insect Biochem Mol Biol (2008) 38:89–98. doi: 10.1016/j.ibmb.2007.09.011
36. Sumathipala N, Jiang H. Involvement of Manduca sexta peptidoglycan recognition protein-1 in the recognition of bacteria and activation of prophenoloxidase system. Insect Biochem Mol Biol (2010) 40:487–95. doi: 10.1016/j.ibmb.2010.04.008
37. Gallagher SR. One-dimensional electrophoresis using nondenaturing conditions. In: Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT, editors. Current Protocols in Protein Science. Hoboken, New Jersey:Wiley (1995). p. 10.3.5–10.3.11.
38. Zhang S, Cao X, He H, Hartson S, Jiang H. Semi-quantitative analysis of changes in the plasma peptidome of Manduca sexta larvae and their correlation with the transcriptome variations upon immune challenge. Insect Biochem Mol Biol (2014) 47:46–54. doi: 10.1016/j.ibmb.2014.02.002
39. Cao X, Wang Y, Rogers J, Hartson S, Kanost MK, Jiang H. Changes in composition and levels of hemolymph proteins during metamorphosis of Manduca sexta. Insect Biochem Mol Biol (2020) 127:103489. doi: 10.1016/j.ibmb.2020.103489
40. Chapman JR, Dowell MA, Chan R, Unckless RL. The genetic basis of natural variation in Drosophila melanogaster immune defense against Enterococcus faecalis. Genes (Basel) (2020) 11:234. doi: 10.3390/genes11020234
41. Troha K, Im JH, Revah J, Lazzaro BP, Buchon N. Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster. PLoS Pathog (2018) 14:e1006847. doi: 10.1371/journal.ppat.1006847
42. Cao X, Gulati M, Jiang H. Serine protease-related proteins in the malaria mosquito, Anopheles Gambiae. Insect Biochem Mol Biol (2017) 88:48–62. doi: 10.1016/j.ibmb.2017.07.008
43. He Y, Cao X, Zhang S, Rogers J, Hartson S, Jiang H. Changes in the plasma proteome of Manduca sexta larvae in relation to the transcriptome variations after an immune challenge: evidence for high molecular weight immune complex formation. Mol Cell Proteomics (2016) 15:1176–87.
44. Casas-Vila N, Bluhm A, Sayols S, Dinges N, Dejung M, Altenhein T, et al. The developmental proteome of Drosophila melanogaster. Genome Res (2017) 27:1273–85. doi: 10.1101/gr.213694.116
45. Zhu Y, Wang Y, Gorman M, Jiang H, Kanost MR. Manduca sexta serpin-3 regulates prophenoloxidase activation in response to infection by inhibiting prophenoloxidase activating proteinases. J Biol Chem (2003) 47:46556–64. doi: 10.1074/jbc.M309682200
46. Gupta S, Wang Y, Jiang H. Manduca sexta prophenoloxidase (proPO) activation requires proPO-activating proteinase (PAP) and serine proteinase homologs (SPHs) simultaneously. Insect Biochem Mol Biol (2005) 35:241–8. doi: 10.1016/j.ibmb.2004.12.003
47. Zakhia R, Osta MA. CLIPA7 exhibits pleiotropic roles in the Anopheles Gambiae immune response. J Innate Immun (2022) 24:1–16. doi: 10.1159/000526486
48. Piao S, Song YL, Kim JH, Park SY, Park JW, Lee BL, et al. Crystal structure of a clip-domain serine protease and functional roles of the clip domains. EMBO J (2005) 24:4404–14. doi: 10.1038/sj.emboj.7600891
49. Huang R, Lu Z, Dai H, Velde DV, Prakash O, Jiang H. The solution structure of the clip domains from Manduca sexta prophenoloxidase activating proteinase-2. Biochemistry (2007) 46:11431–9. doi: 10.1021/bi7010724
50. Cao X, He Y, Hu Y, Zhang X, Wang Y, Zou Z, et al. Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta. Insect Biochem Mol Biol (2015) 62:51–63. doi: 10.1016/j.ibmb.2014.10.006
51. Clark KD. Insect hemolymph immune complexes. Subcell Biochem (2020) 94:123–61. doi: 10.1007/978-3-030-41769-7_5
52. Lu A, Li X, Hillyer JF, Beerntsen BT, Söderhäll K, Ling E. Recombinant Drosophila prophenoloxidase 1 is sequentially cleaved by α-chymotrypsin during in vitro activation. Biochimie (2014) 102:154–65. doi: 10.1016/j.biochi.2014.03.007
Keywords: clip domain, insect immunity, melanization, protease cascade, hemolymph protein
Citation: Jin Q, Wang Y, Yin H and Jiang H (2023) Two clip-domain serine protease homologs, cSPH35 and cSPH242, act as a cofactor for prophenoloxidase-1 activation in Drosophila melanogaster. Front. Immunol. 14:1244792. doi: 10.3389/fimmu.2023.1244792
Received: 23 June 2023; Accepted: 16 August 2023;
Published: 15 September 2023.
Edited by:
Uday Kishore, United Arab Emirates University, United Arab EmiratesReviewed by:
Brandon L. Garcia, East Carolina University, United StatesAmr A. Mohamed, Cairo University, Egypt
Kai Wu, Shangrao Normal University, China
Copyright © 2023 Jin, Wang, Yin and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haobo Jiang, aGFvYm8uamlhbmdAb2tzdGF0ZS5lZHU=
 Qiao Jin
Qiao Jin Yang Wang
Yang Wang Haobo Jiang
Haobo Jiang