- 1Department of Anorectal Surgery, Jiangmen Wuyi Hospital of Traditional Chinese Medicine, Jiangmen, China
- 2Department of Anorectal Surgery, Affiliated Hospital of Jiangxi University of Chinese Medicine, Nanchang, China
Background: Circulating cytokines play a crucial role in the onset and progression of immune skin diseases. However, the causal relationships and the direction of causal effects require further investigation.
Methods: Two-sample Mendelian randomization (MR) analyses were conducted to assess the causal relationships between 41 circulating cytokines and six immune skin diseases including alopecia areata, chloasma, hidradenitis suppurativa (HS), lichen planus (LP), seborrheic dermatitis, and urticaria, using summary statistics from genome-wide association studies. Reverse MR analyses was performed to test for the reverse causation. Pleiotropy and heterogeneity tests were conducted to assess the robustness of the findings.
Results: Twelve unique cytokines showed a suggestive causal relationship with the risk of six immune skin diseases. Among them, the causal effects between 9 unique cytokines and immune skin diseases have strong statistical power. Additionally, the concentrations of six cytokines might be influenced by LP and urticaria. After Bonferroni correction, the following associations remained significant: the causal effect of beta-nerve growth factor on HS (odds ratio [OR] = 1.634, 95% confidence interval [CI] = 1.226-2.177, p = 7.97e-04), interleukin (IL)-6 on LP (OR = 0.615, 95% CI = 0.481-0.786, p = 1.04e-04), IL-4 on LP (OR = 1.099. 95% CI = 1.020-1.184, p = 1.26e-02), and IL-2 on urticaria (OR = 0.712, 95% CI = 0.531-0.955, p = 2.33e-02).
Conclusion: This study provides novel perspectives on the relationship between circulating cytokines and immune skin diseases, potentially providing valuable insights into their etiology, diagnostic approaches, and treatment.
1 Introduction
The skin, being the body’s largest immune organ, serves as a protective barrier against external threats. Numerous skin diseases are associated with the overall homeostasis of the body, including inflammatory response, immune status, metabolic level, and gut flora homeostasis (1, 2). Immune skin diseases, such as alopecia areata (AA), chloasma, hidradenitis suppurativa (HS), lichen planus (LP), seborrheic dermatitis (SD), and urticaria, are highly prevalent and contribute significantly to the global health burden. However, due to their complex and diverse pathogenesis, effective treatments for these conditions are limited (3–8). Genetic research, particularly through genome-wide association studies (GWAS), has identified several single-nucleotide polymorphisms (SNPs) associated with skin diseases (9). However, translating these findings into clinically relevant therapeutic targets and treatments still requires substantial efforts.
Skin diseases often exhibit systemic inflammatory changes (10, 11). Circulating cytokine levels, as crucial inflammatory regulators, exert a significant influence on immune and inflammatory responses in vivo (12). Over 300 cytokines have been identified, primarily including chemokines, interleukins (ILs), interferons (IFNs), and growth factors. These small soluble molecules, secreted by cells, play roles in differentiation, proliferation, and apoptosis promotion (13). Disturbances in cytokine expression are a prominent feature during skin disease episodes (14). During exacerbations, patients with AA show increased levels of serum pro-inflammatory factors such as IFN-γ, IL-2, IL-7, and IL-15 (15). Furthermore, there have been reports on the association between chloasma, HS, LP, SD, and urticaria and cytokines (16–20). However, the existing evidence linking cytokines to these immune skin diseases stems from cross-sectional studies, which are susceptible to confounding factors, such as environment, age, dietary preferences, and lifestyles. Therefore, the causal association between cytokines and these immune skin diseases, as well as the direction of the causal effect remain unknown.
Mendelian randomization (MR) is currently a popular means for exploring causal relationships between exposures and outcomes of interest (21). It leverages the fundamental principles of Mendel’s law, which entail the “random assignment of parental alleles to offspring”, resembling the random assignment of participants in a randomized controlled trial. Thus, MR has emerged as a valuable tool for exploring causal relationships between complex traits, diseases, and phenotypes (22).
In the present study, genetic variants associated with 41 circulating cytokines were obtained from a large GWAS (23). Furthermore, a bidirectional two-sample MR analysis was conducted to investigate the relationship between cytokines and six immune skin diseases. Our findings might help elucidate the association between systemic inflammation and these immune skin diseases, offering new diagnostic biomarkers and revealing new targets for drug therapy.
2 Materials and methods
2.1 Study design
In the MR analysis, SNPs were treated as valid instrumental variables (IVs). These IVs had to fulfil three key assumptions, as shown in Figure 1: (1) a strong correlation between SNPs and exposure; (2) absence of association between SNPs and confounding factors that might influence the association between exposure and outcome; (3) exposure being the only pathway through which SNPs exert an effect on the outcome (24).
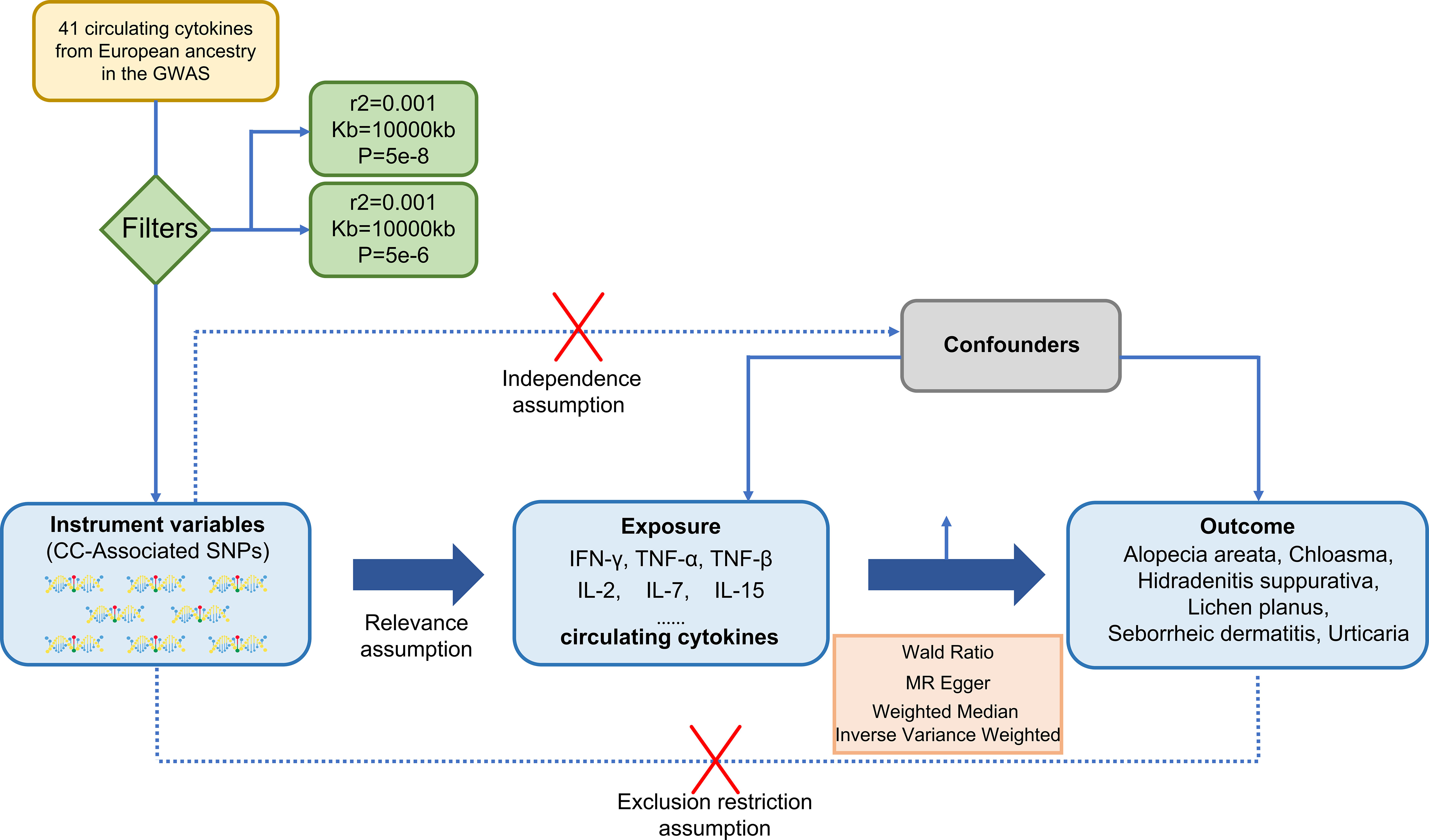
Figure 1 A design of three basic assumptions of Mendelian-randomization analysis: (1) a strong correlation between SNPs and exposure; (2) absence of association between SNPs and confounding factors that might influence the association between exposure and outcome; (3) exposure being the only pathway through which SNPs exert an effect on the outcome. CC, circulating cytokines; IFN-γ, Interferon-gamma; TNF-α, Tumor necrosis factor-alpha; TNFβ, Tumor necrosis factor-beta; IL-2, Interleukin-2; IL-7, Interleukin-7; IL-15, Interleukin-15.
2.2 Data sources
The GWAS conducted by Ahola-Olli et al. provided the genome-wide association summary statistics for 41 systemic inflammatory regulators (23). The study population comprised 8,293 individuals from three different Finnish cohorts, namely the FINRISK1997, FINRISK2002, and the Young Finns Cardiovascular Risk Study (YFS) (Supplementary Table S1). Cytokines and growth factors in plasma were measured for participants in the FINRISK1997 cohort, while heparin plasma and serum cytokine measurements were performed for individuals in the FINRISK2002 and YFS cohorts. The circulating cytokine levels were expressed as standard deviations. Genetic associations were adjusted for the first ten genetic principal components, age, sex, and body mass index. Complete GWAS summary statistics were obtained from https://www.ebi.ac.uk/gwas/downloads/summary-statistics.
The summary statistics for the six immune skin diseases were obtained from FinnGen GWAS available at https://r8.finngen.fi/ (25). The numbers of cases and controls for each phenotype was as follows: 682 patients with AA and 361,140 controls, 95 patients with chloasma and 329,443 controls, 931 patients with HS and 361,140 controls, 3,597 patients with LP and 364,071 controls, 2,688 patients with SD and 336,589 controls, and 3,495 patients with urticaria and 364,583 controls.(More details as) These skin diseases were defined based on the International Classification of Diseases, 10th revision.
2.3 Selection of IVs
A genome-wide statistical significance threshold of <5.0 × 10-8 was established to extract the SNPs robustly associated with the studied exposures. Additionally, the r2 threshold of 0.001 and a clump window size of 10,000 kb were set to eliminate linkage disequilibrium (26). As a result, 27 cytokines were retained based on these criteria. In a secondary analysis, the genome-wide significance threshold was relaxed to 5.0 × 10-6 to allow for the inclusion of all 41 cytokines.
For the reverse direction, the significance threshold employed was p <5 × 10-8, with consistent clumping parameter with the forward-direction analysis. However, due to the lack of sufficient SNPs, four diseases, namely AA, chloasma, HS, and SD, were not subjected to the reverse MR analysis. Consequently, the revere MR analyses were only conducted for LP and urticaria.
2.4 Statistical analysis
The Wald ratio test was used to estimate the association between identified IVs and skin diseases for cases containing only one SNP (27). For multiple SNPs, three MR analysis methods were used, including inverse variance weighted (IVW), MR-Egger and weighted median (WM), with the IVW method being the predominant method. The IVW method, which is the classical approach for MR analysis, incorporates inverse variances of each IV as weights to calculate a weighted average while ensuring the validity of all IVs (28). MR-Egger, on the other hand, employs a form of weighted linear regression analysis. Estimates derived from this method are robust and independent of IV validity, although they might have lower statistical precision and are susceptible to outlying genetic variations (29). The WM method addresses the issue of a large variation in estimation precision. Similar to the IVW method, this approach assigns inverse weights based on the variance of each genetic variant, and it demonstrates reliability even in the presence of violated causal effects (30). When significant causal correlations are evaluated using the IVW method, the MR egger and WM methods serve as supplementary methods and directional validation.
MR-Egger regression was used to assess pleiotropy. Moreover, the presence of pleiotropy and identification of outlying SNPs could be determined through the MR-Pleiotropy RESidual Sum and Outlier (PRESSO) test (31). The Cochran’s Q was used to assess heterogeneity. A p-value of <0.05 indicates the presence of heterogeneity.
The strength of IVs can be evaluated using the F- statistic, which is calculated as follows: F = β2/SE2, where β is the effect size of the allele and SE is the standard error (32). An F statistic greater than 10 suggests the absence of weak IV bias (33). When analyzing multiple outcomes, significance thresholds are adjusted using the Bonferroni method. A p value smaller than the Bonferroni-corrected threshold indicates statistical significance. Additionally, correlations not exceeding the Bonferroni-corrected significance level but <0.05 were considered suggestively significant. All analyses were performed using R version 4.2.2, with the software packages “TwoSampleMR” (34) and “MRPRESSO” (28).
Due to the limited variability of phenotypic characteristics and the small proportion of patients in the outcome trait GWAS data, statistical ability is crucial. In order to calculate the statistical power, an online tool called mRnd (35) was used. In the case of a Type I error of 5%, we determined the expected OR threshold for each circulating cytokine and immune skin disease, which should meet a minimum power requirement of>80%.
3 Results
3.1 Causal effects of circulating cytokines on six immune skin diseases
Figure 2 illustrates the results, with 14 sets of suggestively significant associations (p <0.05) identified at a significance threshold of <5.0 × 10-8. Based on the significant correlation between circulating cytokines and immune skin diseases assessed by Mendelian randomization, further statistical efficacy analysis examined the threshold range of OR that needs to be reached when meeting sufficient statistical efficacy. It is worth noting that six cytokines meet the OR threshold for statistical efficacy analysis (IL-18, IL-17, IL-16, IL2α, TNFβ, Eotaxin). The details of statistic power analyses are presented in Supplementary Table 6. The above results do not meet the threshold range corrected by Bonferroni (p = 0.05/27 = 1.85e-03). The specific SNP profiles, as well as the pleiotropy and heterogeneity results of these 14 groups, are presented in Supplementary Tables S3, 4. No evidence was observed in terms of heterogeneity and pleiotropy (p >0.05). The F-statistics for all groups were >10, indicating the absence of weak IV bias. Supplementary Table S5 presents the MR analyses involving 27 cytokines and six immune skin diseases.
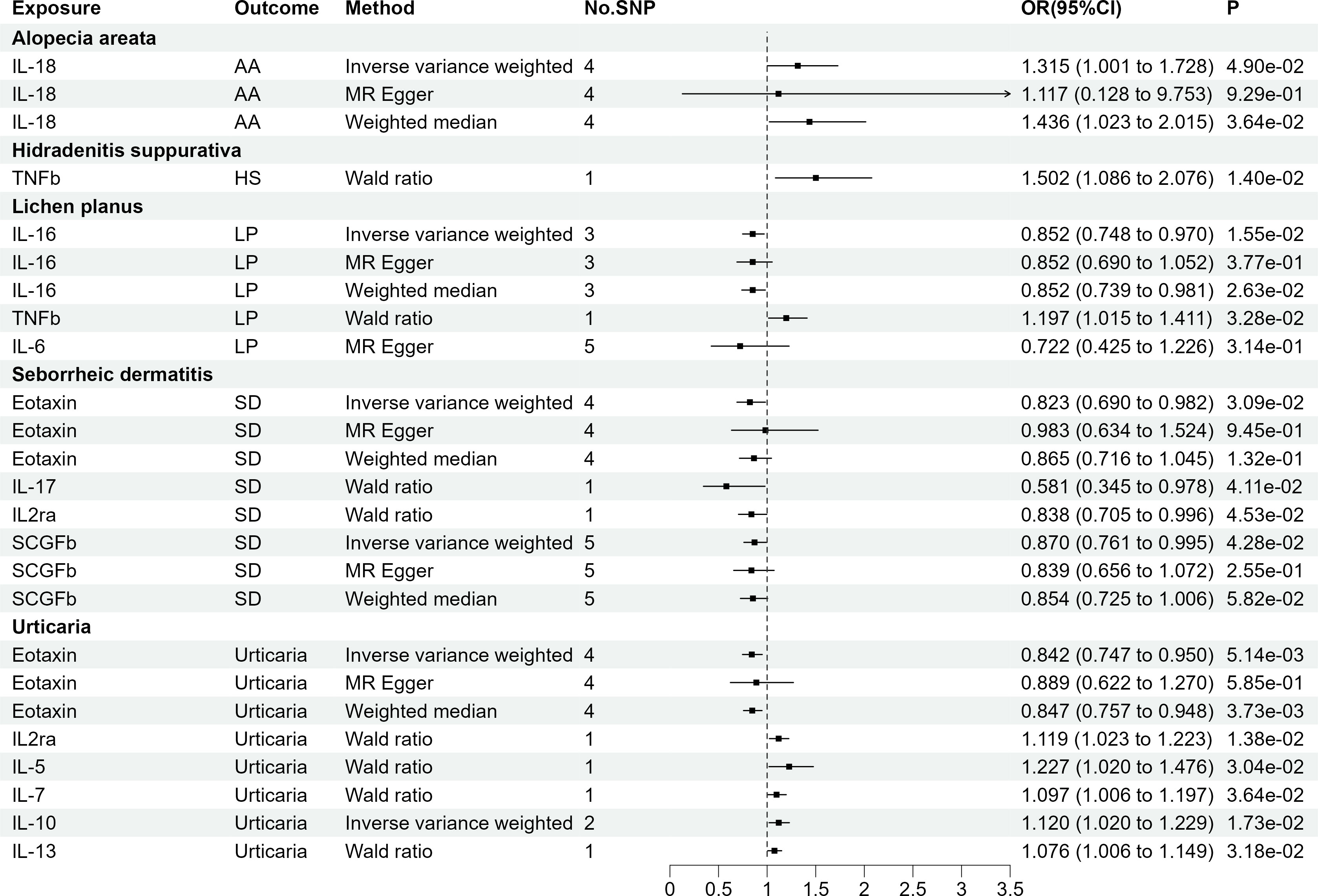
Figure 2 The 14 causal relationships (SNPs reaching P<5×10-8). AA, Alopecia areata; HS, Hidradenitis suppurativa; LP, Lichen planus; SD, Seborrheic dermatitis; IL-18, Interleukin-18; TNFβ, Tumor necrosis factor-beta; IL-16, Interleukin-16; IL-17, Interleukin-17; IL2rα, Interleukin-2 receptor, alpha subunit; SCGFβ, Stem cell growth factor beta.
As presented in Figure 3, employing a significance threshold of <5.0 × 10-6, 14 sets of suggestively significant correlations (p <0.05) were obtained. Based on the significant correlation between circulating cytokines and immune skin diseases assessed by Mendelian randomization, further statistical power analysis examined the threshold range of OR that needs to be reached when meeting sufficient statistical efficacy. Among the 14 pairs of suggestive significantly correlated traits, 4 pairs of suggestive significantly correlated traits that meet sufficient statistical efficacy were obtained through screening. The details of statistic power analyses are presented in Supplementary Table 10. Following Bonferroni correction (p = 0.05/41 = 1.22e-03), bNGF remained statistically significant in HS (OR = 1.634, 95% CI = 1.226-2.177, p = 7.97e-04) and IL6 remained statistically significant in LP (OR = 0.615, 95% CI = 0.481-0.786, p = 1.04e-04). Supplementary Tables S7, 8 provide detailed information on the SNP profiles, as well as the details of pleiotropy and heterogeneity. No heterogeneity or pleiotropy was observed, and there was no evidence of weak IV bias. The details of MR analyses involving 41 cytokines and six immune skin diseases are presented in Supplementary Table S9.
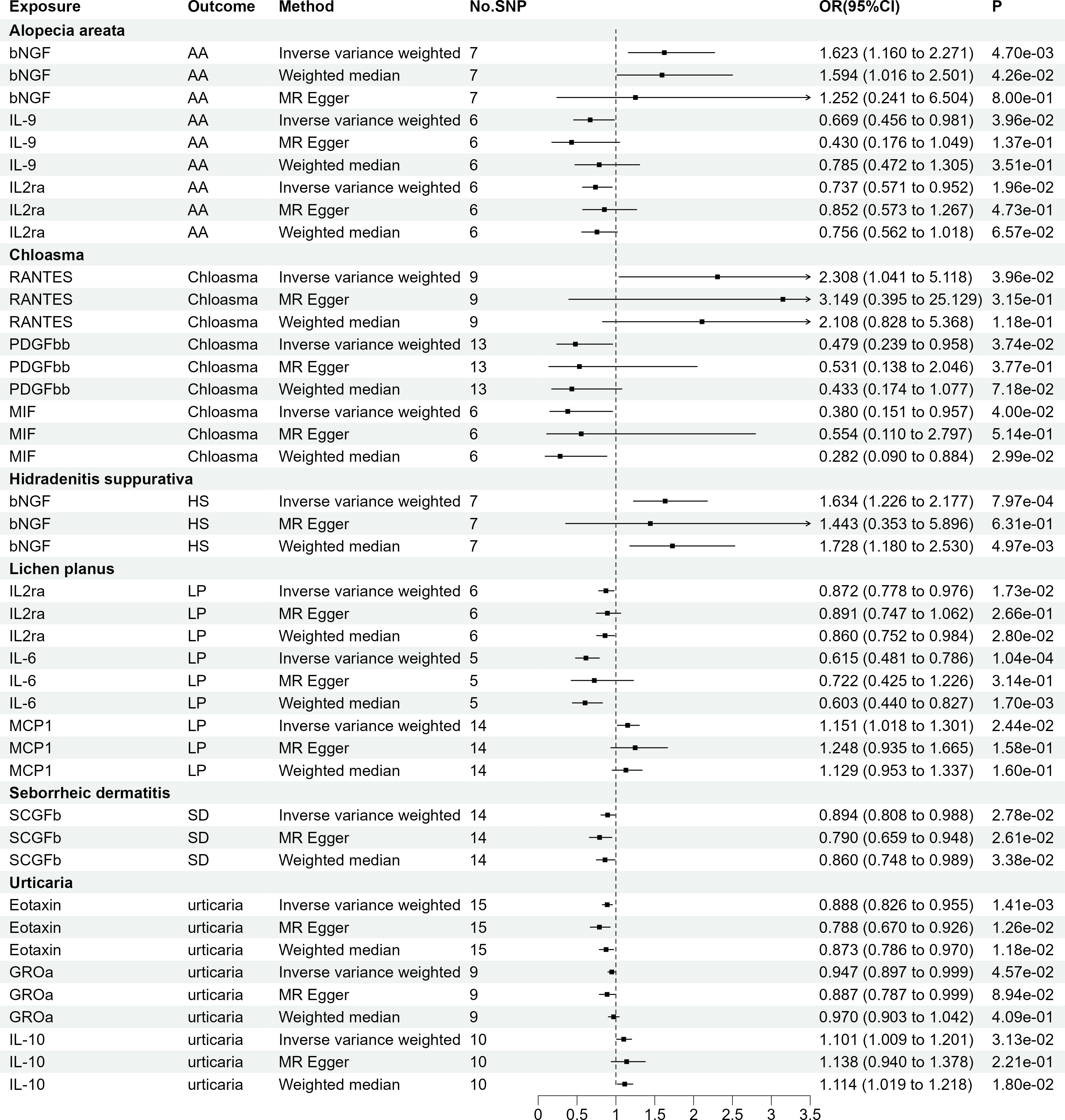
Figure 3 The 14 causal relationships (SNPs reaching P<5×10-6). AA: Alopecia areata, HS, Hidradenitis suppurativa; LP, Lichen planus; SD, Seborrheic dermatitis; βNGF, Beta nerve growth factor; IL-9, Interleukin-9; IL2rα, Interleukin-2 receptor; MIF, Macrophage migration inhibitory factor; PDGFbb, Platelet derived growth factor BB; RANTES, Regulated on Activation, Normal T Cell Expressed and Secreted; IL-6, Interleukin-6; MCP1, Monocyte chemotactic protein-1; SCGFβ, Stem cell growth factor beta; GROα, Growth regulated oncogene-α; IL-10, Interleukin-10.
3.2 Causal effects of six immune skin diseases on circulating cytokines
As AA, chloasma, HS, and SD did not have genome-wide significant SNPs, our assessment was limited to LP and urticaria. Finally, seven sets of suggestively significant correlations (p <0.05) involving six cytokines (eotaxin, IL-12p70, IL-2, IL-4, IL-16, and MCP-1) were obtained. The association of LP with IL-4 (OR = 1.099, 95% CI = 1.020-1.184, p = 1.26e-02) and urticaria with IL-2 (OR = 0.712, 95% CI = 0.531-0.955, p = 2.33e-02) remained significant at the Bonferroni-corrected significance threshold (p = 0.05/2 = 0.025). Figure 4 shows the Mendelian randomization randomized forest map results of circulating cytokines and immune skin diseases with statistically significant (P<0.05) IVW method using multiple methods. Supplementary Tables S9, 10 provide information on SNPs, heterogeneity, and pleiotropy for these groups. Moreover, the F-statistics indicate the absence of weak IV bias. Detailed results of the specific reverse MR analyses involving LP and urticaria are presented in Supplementary Table S11.
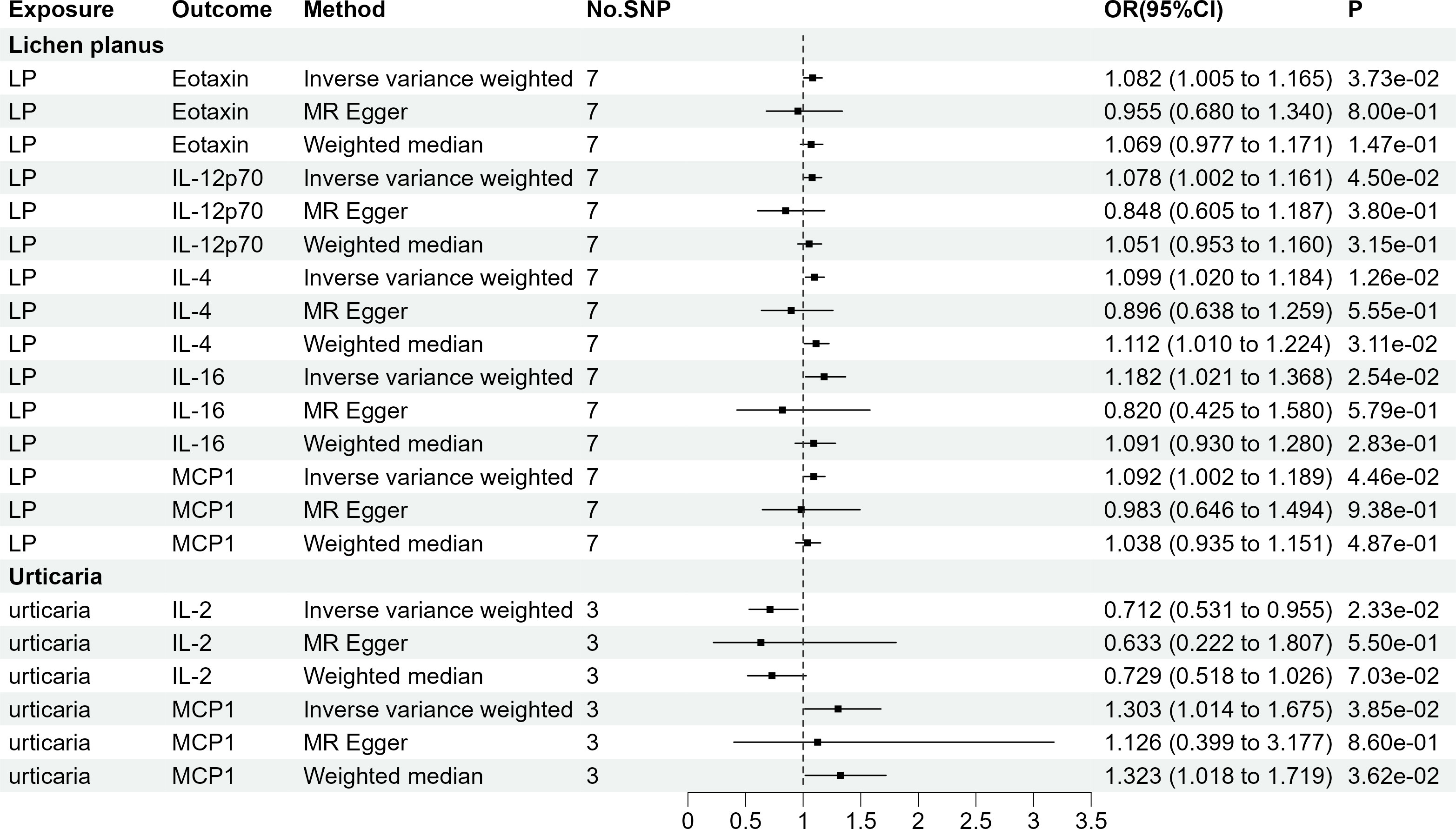
Figure 4 The 7 causal relationships (SNPs reaching P<5×10-8). LP, Lichen planus; IL-12p70, Interleukin-12p70; IL4, Interleukin-4; IL16, Interleukin-16; MCP1, Monocyte chemotactic protein-1; IL2, Interleukin-2.
The four groups that remained statistically significant after Bonferroni correction in this bidirectional MR analysis were selected for scatter plotting, as presented in Figure 5.
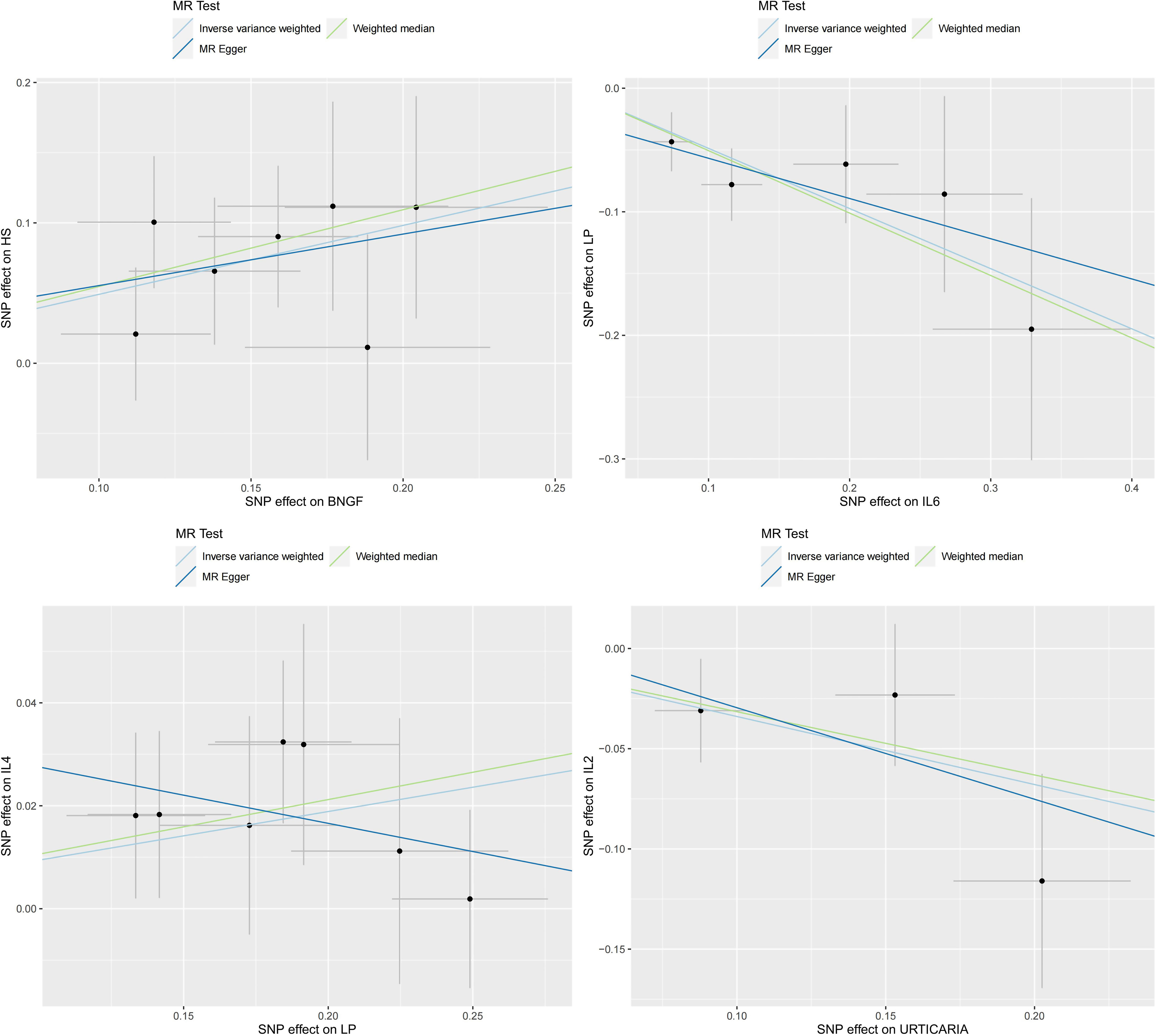
Figure 5 Scatter plots for the 4 significant associations at the Bonferroni corrected significance threshold. β NGF: Beta nerve growth factor, HS, Hidradenitis suppurativa; IL2, Interleukin-2; IL4, Interleukin-4; IL6, Interleukin-6; LP, Lichen planus; SD, Seborrheic dermatitis.
4 Discussion
This is the first study performing a bidirectional MR analysis to explore the relationship between 41 circulating cytokines and six immune skin diseases (AA, chloasma, HS, LP, SD, and urticaria). When cytokines were considered the exposure variable (p <5.0 × 10-6), potential associations were observed between 3 cytokines and the risk of immune skin diseases. Specifically, bNGF was associated with AA and HS, and RANTES was associated with chloasma, IL-6 was associated with LP. On the other hand, when immune skin diseases served as the exposure variable, LP affected eotaxin, IL-12p70, IL-4, IL-16, and MCP-1 and urticaria affected IL-2 and MCP-1. Due to the limited number of cases available in the FinnGen database for the remaining four skin diseases, the genetic variation was insufficient for effective exploration. Therefore, a reanalysis should be performed in the future when an adequate number of patients become available. Although there are some circulating cytokines that have causal associations with immune skin diseases that have not been analyzed through statistical efficacy, some of these cytokines may have predictive value for disease risk and diagnostic purposes. Considering the complexity and complexity of the cytokine network in the body, it is necessary to conduct further clinical and basic experiments to verify these hypotheses.
Healthy skin comprises three layers, namely the epidermis, dermis, and subcutaneous fat. These layers harbor resident cell populations, both immune and non-immune cells, which play crucial roles in maintaining a strong barrier against foreign substances. Moreover, these cells are involved in various inflammatory processes, contributing to an immune response against infections, autoimmunity, tumor immunity, and allergies (10). It can be stated that the pathogenesis of skin diseases is, to a certain extent, linked to inflammatory responses.
AA, a prevalent autoimmune disease, is characterized by non-scarring alopecia. The pathological process of AA involves a shift in the hair follicle microenvironment from a state of immune homeostasis to an active inflammatory state (15). Several GWASs have identified IL2ra as a cytokine gene associated with AA (36–38). IL2ra functions through regulatory T cells, playing a crucial role in maintaining immune homeostasis and suppressing autoantigen-mediated immune responses (39, 40). Notably, the in vivo IL-9 levels are significantly decreased in patients who exhibit effective responses to tetraphenyl cyclopentadienone treatment (41). These findings align with the results obtained in the current study. At present, no research has revealed the biological association and mechanism between bNGF and AA. Our study suggests for the first time that bNGF may have the potential to become an early screening biomarker for AA in the future and provide assistance in exploring the biological mechanisms and therapeutic targets of AA.
Chloasma, a prevalent light-induced pigmentation disorder, results in skin inflammation and the release of melanogenic cytokines and growth factors in response to ultraviolet (UV) radiation. These factors play a crucial role in the hyperpigmentation and reactivation of chloasma lesions (42). MIF is considered a key player in skin biology and wound healing and is negatively correlated with cellular and tissue ageing (43). MIF might serve as a key mediator in photoaging. In vivo and in vitro studies have shown that UVB radiation (UVB) induces the expression of MIF messenger ribonucleic acid in the skin and keratinocyte MIF production in vivo and in vitro (44). Moreover, UVB has been found to inhibit skin inflammation by inhibiting the production of RANTES in epidermal keratinocytes. This provides a new direction for the treatment of skin inflammation (45). PDGFbb production is reduced in terms of quantity and activity by UVB radiation (46). In addition, it promotes wound healing (47). Furthermore, UV irradiation induces RANTES overgrowth, resulting in inflammation (45).
HS, a chronic and inflammatory skin disease, imposes a significant burden on individuals and predominantly affects the axillae, groin, buttocks, and perianal areas. It is characterized by abnormal activation of the innate immune system (48, 49). While specific studies exploring the association between HS and bNGF are currently lacking, the observed relationship between the two cytokines remains significant even after correction. Moreover, a recent study on the relationship between bNGF and chloasma reported higher bNGF levels in affected areas compared with the normal sites (50). Therefore, the relationship between HS and bNGF warrants further investigation.
LP is a T cell-mediated autoimmune disease, specifically CD8+ T cells, which are recruited into the skin and contribute to interface dermatitis (51). Impairment of IL2ra function in patients with oral LP (OLP) results in an increased inflammatory response (52). MCP-1, a chemokine present in LP lesions, plays a crucial role in aggregating monocytes to the skin interface and inducing toxic effects (53). LP and MCP-1 exhibit a bidirectional causal association that warrants further investigation. Reverse MR studies have demonstrated elevated cytokine IL-4 levels in the blood and saliva of patients with OLP (54). The relationship between LP and eotaxin, IL-12p70, and IL163 is novel and requires further investigation.
SD is a chronic, recurrent inflammatory skin disease. SCGFb is a newly discovered protein-secreting sulphated glycoprotein that serves as a growth factor in early hematopoiesis (55). Study have shown that SCGFb may be able to inhibit the pathogenesis of proliferative diabetic retinopathy(PDR) (55). However, the underlying mechanism remains to be elucidated.
Urticaria is a skin disease characterized by clinical symptoms such as wheal and angioedema. The pathogenesis of urticaria involves the activation of mast cells, which in turn release multiple cytokines, leading to sensory nerve activation, vasodilation, and plasma extravasation (56). Chronic urticaria is characterized by increased eotaxin and MCP-1, which are indicative of an inflammatory response (57, 58). Urticaria is mediated by mixed T helper (Th) 1/Th2-reactive lymphocytes, with Th1 cells primarily secreting IL-2 and Th2 cells secreting IL-10. In patients with urticaria, there is an observed increase in IL-10 production and a decrease in IL-2 levels, consistent with the findings of the present study (59).
The present study has certain limitations. Our study elucidates the intricate relationship between circulating cytokines and the risk of six immune skin diseases. However, it’s imperative to recognize its limitations, which warrant attention in subsequent research. Despite our meticulous selection of instrumental variables (IVs) and the deployment of statistical tests, such as MR-PRESSO and MR-Egger intercept, to address potential pleiotropy and confounding, these concerns are not entirely eliminable. Future endeavors should place greater emphasis on understanding and rectifying these challenges, bolstering the validity of conclusions drawn from such data. Moreover, the representativeness of our results is constrained by the limited sample sizes for specific diseases and inflammatory markers. Although F-statistics indicate the potential insignificance of weak IVs, the precision of our inferences, particularly for these diseases and markers, remains ambiguous.
The primary scope of our findings pertains to European populations, given the Finnish origin of our dataset. The mosaic of genetic variations across ethnicities and geographies underscores the need for more comprehensive studies. Genetic disparities have profound implications in clinical scenarios, necessitating the analysis of more heterogeneous GWAS data in the future. Of the 41 cytokines we scrutinized, a mere six demonstrated a pronounced association with skin immune diseases. This underscores their specialized role in disease pathogenesis. However, the magnitude of these associations can be contingent upon factors like sample size, genetic heterogeneity, and disease-specific nuances. The cellular dynamics within skin immune diseases demand further exploration. Deciphering the interactions between various skin cell populations and their associated cytokines will enrich our understanding of disease etiology. Cutting-edge techniques, such as single-cell RNA sequencing and spatial transcriptomics, promise to be revelatory in this domain.
While our Mendelian stochastic approach was instrumental in inferring causality, clinical implications necessitate a more direct approach. This entails measuring cytokine levels in both the affected and healthy populace, juxtaposing them with observed gene polymorphisms. Longitudinal assessments, which monitor the influence of these polymorphisms on cytokine levels across diverse conditions, are paramount.
In conclusion, our study provides a preliminary framework on the cytokine dynamics in immune skin diseases. To fully harness its translational potential, it’s pivotal to address the aforementioned limitations, broaden the scope of datasets, and employ sophisticated methodologies. The potential biomarkers identified warrant rigorous validation in expansive, diverse cohorts and systematic clinical evaluations before they can be integrated into clinical practice guidelines.
5 Conclusion
In summary, this bidirectional MR analysis indicates potential causal associations between specific circulating cytokines and the six immune skin diseases. These findings coffer insights into potential targets and novel biomarkers for their diagnosis and treatment of immune skin diseases in clinical settings.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: Data for 41 circulating cytokines are available at https://www.ebi.ac.uk/gwas/downloads/summary-statistics. Data for the six immune skin diseases can be downloaded from https://r8.finngen.fi.
Ethics statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
QL: Conceptualization, methodology, formal analysis, data curation, writing-original draft preparation; QC: Data curation, writing-original draft preparation, visualization; JG: Writing-original draft preparation, visualization; SC: Data curation, visualization; YW: Supervision, writing-review and editing. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors thank the studies or consortiums referenced and included in the present analysis for providing public datasets. The authors thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1240714/full#supplementary-material
References
1. Karimkhani C, Dellavalle RP, Coffeng LE, Flohr C, Hay RJ, Langan SM, et al. Global skin disease morbidity and mortality: an update from the global burden of disease study 2013. JAMA Dermatol (2017) 153:406–12. doi: 10.1001/jamadermatol.2016.5538
2. Hay RJ, Johns NE, Williams HC, Bolliger IW, Dellavalle RP, Margolis DJ, et al. The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Invest Dermatol (2014) 134:1527–34. doi: 10.1038/jid.2013.446
3. Handel AC, Miot LDB, Miot HA. Melasma: a clinical and epidemiological review. Bras Dermatol (2014) 89:771–82. doi: 10.1590/abd1806-4841.20143063
4. Zhou C, Li X, Wang C, Zhang J. Alopecia areata: an update on etiopathogenesis, diagnosis, and management. Clin Rev Allergy Immunol (2021) 61:403–23. doi: 10.1007/s12016-021-08883-0
5. Saunte DML, Jemec GBE. Hidradenitis suppurativa: advances in diagnosis and treatment. JAMA (2017) 318:2019–32. doi: 10.1001/jama.2017.16691
6. Solimani F, Forchhammer S, Schloegl A, Ghoreschi K, Meier K. Lichen planus - a clinical guide. J Dtsch Dermatol Ges (2021) 19:864–82. doi: 10.1111/ddg.14565
7. Clark GW, Pope SM, Jaboori KA. Diagnosis and treatment of seborrheic dermatitis. Am Fam Phys (2015) 91:185–90.
9. Moitinho-Silva L, Degenhardt F, Rodriguez E, Emmert H, Juzenas S, Möbus L, et al. Host genetic factors related to innate immunity, environmental sensing and cellular functions are associated with human skin microbiota. Nat Commun (2022) 13:6204. doi: 10.1038/s41467-022-33906-5
10. Richmond JM, Harris JE. Immunology and skin in health and disease. Cold Spring Harb Perspect Med (2014) 4:a015339. doi: 10.1101/cshperspect.a015339
11. Pasparakis M, Haase I, Nestle FO. Mechanisms regulating skin immunity and inflammation. Nat Rev Immunol (2014) 14:289–301. doi: 10.1038/nri3646
12. Becher B, Spath S, Goverman J. Cytokine networks in neuroinflammation. Nat Rev Immunol (2017) 17:49–59. doi: 10.1038/nri.2016.123
13. Liszewski W, Gniadecki R. The role of cytokine deficiencies and cytokine autoantibodies in clinical dermatology. J Eur Acad Dermatol Venereol (2016) 30:404–12. doi: 10.1111/jdv.13303
14. Whiteside TL. “Introduction to cytokines as targets for immunomodulation,”. In: House RV, Descotes J, editors. Cytokines in Human Health: Immunotoxicology, Pathology, and Therapeutic Applications Methods in Pharmacology and Toxicology. Totowa, NJ: Humana Press (2007). p. 1–15. doi: 10.1007/978-1-59745-350-9_1
15. Lensing M, Jabbari A. An overview of JAK/STAT pathways and JAK inhibition in alopecia areata. Front Immunol (2022) 13:955035. doi: 10.3389/fimmu.2022.955035
16. Nassar AAE, Ibrahim A-SM, Mahmoud AA. Efficacy and safety of intralesional steroid injection in the treatment of melasma. J Cosmet Dermatol (2021) 20:862–7. doi: 10.1111/jocd.13628
17. Fletcher JM, Moran B, Petrasca A, Smith CM. IL-17 in inflammatory skin diseases psoriasis and hidradenitis suppurativa. Clin Exp Immunol (2020) 201:121–34. doi: 10.1111/cei.13449
18. Shao S, Tsoi LC, Sarkar MK, Xing X, Xue K, Uppala R, et al. IFN-γ enhances cell-mediated cytotoxicity against keratinocytes via JAK2/STAT1 in lichen planus. Sci Transl Med (2019) 11:eaav7561. doi: 10.1126/scitranslmed.aav7561
19. Molinero LL, Gruber M, Leoni J, Woscoff A, Zwirner NW. Up-regulated expression of MICA and proinflammatory cytokines in skin biopsies from patients with seborrhoeic dermatitis. Clin Immunol (2003) 106:50–4. doi: 10.1016/s1521-6616(03)00003-2
20. Hinden S, Klukowska-Rötzler J, Janda J, Marti EI, Gerber V, Roosje PJ. Characterization of the inflammatory infiltrate and cytokine expression in the skin of horses with recurrent urticaria. Vet Dermatol (2012) 23:503–e99. doi: 10.1111/j.1365-3164.2012.01117.x
21. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet (2014) 23:R89–98. doi: 10.1093/hmg/ddu328
22. Long Y, Tang L, Zhou Y, Zhao S, Zhu H. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med (2023) 21:66. doi: 10.1186/s12916-023-02761-6
23. Ahola-Olli AV, Würtz P, Havulinna AS, Aalto K, Pitkänen N, Lehtimäki T, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet (2017) 100:40–50. doi: 10.1016/j.ajhg.2016.11.007
24. Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol (2016) 45:1717–26. doi: 10.1093/ije/dyx028
25. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, et al. FinnGen: Unique genetic insights from combining isolated population and national health register data. medRxiv (2022). doi: 10.1101/2022.03.03.22271360
26. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Võsa U, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet (2019) 51:600–5. doi: 10.1038/s41588-019-0350-x
27. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res (2017) 26:2333–55. doi: 10.1177/0962280215597579
28. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol (2013) 37:658–65. doi: 10.1002/gepi.21758
29. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol (2015) 44:512–25. doi: 10.1093/ije/dyv080
30. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol (2016) 40:304–14. doi: 10.1002/gepi.21965
31. Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
32. Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol (2016) 45:1961–74. doi: 10.1093/ije/dyw220
33. Burgess S, Thompson SG, CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol (2011) 40:755–64. doi: 10.1093/ije/dyr036
34. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PloS Genet (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
35. Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol (2013) 42:1497–501. doi: 10.1093/ije/dyt179
36. Moravvej H, Tabatabaei-Panah P-S, Abgoon R, Khaksar L, Sokhandan M, Tarshaei S, et al. Genetic variant association of PTPN22, CTLA4, IL2RA, as well as HLA frequencies in susceptibility to alopecia areata. Immunol Invest (2018) 47:666–79. doi: 10.1080/08820139.2018.1480032
37. Mosallaei M, Simonian M, Esmaeilzadeh E, Bagheri H, Miraghajani M, Salehi AR, et al. Single nucleotide polymorphism rs10889677 in miRNAs Let-7e and Let-7f binding site of IL23R gene is a strong colorectal cancer determinant: Report and meta-analysis. Cancer Genet (2019) 239:46–53. doi: 10.1016/j.cancergen.2019.09.003
38. Olayinka JJT, Richmond JM. Immunopathogenesis of alopecia areata. Curr Res Immunol (2021) 2:7–11. doi: 10.1016/j.crimmu.2021.02.001
39. Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity (2010) 33:153–65. doi: 10.1016/j.immuni.2010.08.004
40. Redler S, Albert F, Brockschmidt FF, Herold C, Hanneken S, Eigelshoven S, et al. Investigation of selected cytokine genes suggests that IL2RA and the TNF/LTA locus are risk factors for severe alopecia areata. Br J Dermatol (2012) 167:1360–5. doi: 10.1111/bjd.12004
41. Manimaran RP, Ramassamy S, Rajappa M, Chandrashekar L. Therapeutic outcome of diphencyprone and its correlation with serum cytokine profile in alopecia areata. J Dermatolog Treat (2022) 33:324–8. doi: 10.1080/09546634.2020.1752887
42. Rodríguez-Arámbula A, Torres-Álvarez B, Cortés-García D, Fuentes-Ahumada C, Castanedo-Cázares JP. CD4, IL-17, and COX-2 are associated with subclinical inflammation in malar melasma. Am J Dermatopathol (2015) 37:761–6. doi: 10.1097/DAD.0000000000000378
43. Welford SM, Bedogni B, Gradin K, Poellinger L, Broome Powell M, Giaccia AJ. HIF1alpha delays premature senescence through the activation of MIF. Genes Dev (2006) 20:3366–71. doi: 10.1101/gad.1471106
44. Scordi IA, Vincek V. Timecourse study of UVB-induced cytokine induction in whole mouse skin. Photodermatol Photoimmunol Photomed (2000) 16:67–73. doi: 10.1034/j.1600-0781.2000.d01-6.x
45. Arakawa S, Hatano Y, Katagiri K, Terashi H, Fujiwara S. Effects of ultraviolet B irradiation on the production of regulated upon activation normal T-cell expressed and secreted protein in cultured human epidermal keratinocytes. Arch Dermatol Res (2006) 297:377–80. doi: 10.1007/s00403-005-0620-6
46. Evrova O, Kellenberger D, Scalera C, Calcagni M, Giovanoli P, Vogel V, et al. Impact of UV sterilization and short term storage on the in vitro release kinetics and bioactivity of biomolecules from electrospun scaffolds. Sci Rep (2019) 9:15117. doi: 10.1038/s41598-019-51513-1
47. Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regener (2008) 16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x
48. Chopra D, Arens RA, Amornpairoj W, Lowes MA, Tomic-Canic M, Strbo N, et al. Innate immunity and microbial dysbiosis in hidradenitis suppurativa - vicious cycle of chronic inflammation. Front Immunol (2022) 13:960488. doi: 10.3389/fimmu.2022.960488
49. Sabat R, Jemec GBE, Matusiak Ł, Kimball AB, Prens E, Wolk K. Hidradenitis suppurativa. Nat Rev Dis Primers (2020) 6:18. doi: 10.1038/s41572-020-0149-1
50. Byun JW, Park IS, Choi GS, Shin J. Role of fibroblast-derived factors in the pathogenesis of melasma. Clin Exp Dermatol (2016) 41:601–9. doi: 10.1111/ced.12874
51. Boch K, Langan EA, Kridin K, Zillikens D, Ludwig RJ, Bieber K. Lichen planus. Front Med (2021) 8. doi: 10.3389/fmed.2021.737813
52. Zhou L, Cao T, Wang Y, Yao H, Du G, Chen G, et al. Frequently increased but functionally impaired CD4+CD25+ Regulatory T cells in patients with oral lichen planus. Inflammation (2016) 39:1205–15. doi: 10.1007/s10753-016-0356-9
53. Spandau U, Toksoy A, Goebeler M, Bröcker EB, Gillitzer R. MIG is a dominant lymphocyte-attractant chemokine in lichen planus lesions. J Invest Dermatol (1998) 111:1003–9. doi: 10.1046/j.1523-1747.1998.00426.x
54. Mozaffari HR, Zavattaro E, Saeedi M, Lopez-Jornet P, Sadeghi M, Safaei M, et al. Serum and salivary interleukin-4 levels in patients with oral lichen planus: A systematic review and meta-analysis. Oral Surg Oral Med Oral Pathol Oral Radiol (2019) 128:123–31. doi: 10.1016/j.oooo.2019.04.003
55. Sukowati CHC, Patti R, Pascut D, Ladju RB, Tarchi P, Zanotta N, et al. Serum stem cell growth factor beta for the prediction of therapy response in hepatocellular carcinoma. BioMed Res Int (2018) 2018:6435482. doi: 10.1155/2018/6435482
56. Church MK, Kolkhir P, Metz M, Maurer M. The role and relevance of mast cells in urticaria. Immunol Rev (2018) 282:232–47. doi: 10.1111/imr.12632
57. Tedeschi A, Asero R, Lorini M, Marzano AV, Cugno M. Serum eotaxin levels in patients with chronic spontaneous urticaria. Eur Ann Allergy Clin Immunol (2012) 44:188–92.
58. Wang J, Zhao Y, Yan X. Effects of desloratadine citrate disodium on serum immune function indices, inflammatory factors and chemokines in patients with chronic urticaria. J Coll Phys Surg Pak (2019) 29:214–7. doi: 10.29271/jcpsp.2019.03.214
59. Li N, Cao N, Niu Y-D, Bai X-H, Lu J, Sun Y, et al. Effects of the polysaccharide nucleic acid fraction of bacillus Calmette-Guérin on the production of interleukin-2 and interleukin-10 in the peripheral blood lymphocytes of patients with chronic idiopathic urticaria. BioMed Rep (2013) 1:713–8. doi: 10.3892/br.2013.130
Keywords: Mendelian randomization, circulating cytokines, immune skin diseases, genome-wide association studies, causal relationship
Citation: Luo Q, Cao Q, Guo J, Chang S and Wu Y (2023) Genetically predicted levels of circulating cytokines and the risk of six immune skin diseases: a two-sample Mendelian randomization study. Front. Immunol. 14:1240714. doi: 10.3389/fimmu.2023.1240714
Received: 15 June 2023; Accepted: 12 October 2023;
Published: 26 October 2023.
Edited by:
Wang Xiqiao, Shanghai Jiao Tong University, ChinaReviewed by:
Kazufumi Kunimura, Kyushu University, JapanZhang Yong, Shanghai Jiao Tong University, China
Copyright © 2023 Luo, Cao, Guo, Chang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunxiang Wu, V3l4ODQxMTA2QDE2My5jb20=
 Qinghua Luo
Qinghua Luo Qiurui Cao1
Qiurui Cao1 Jinyan Guo
Jinyan Guo