
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 12 September 2023
Sec. Microbial Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1235982
 Ming Yan1,2†
Ming Yan1,2† Yu-Long Ouyang1†
Yu-Long Ouyang1† Li-Yuan Xiao1
Li-Yuan Xiao1 Man Ao1
Man Ao1 Martin Gosau2
Martin Gosau2 Reinhard E. Friedrich2
Reinhard E. Friedrich2 Ralf Smeets2,3
Ralf Smeets2,3 Ling-Ling Fu1,2*
Ling-Ling Fu1,2* Hong-chao Feng1*
Hong-chao Feng1* Simon Burg2
Simon Burg2Purpose: Several existing studies have revealed that the occurrence of lichen planus (LP) is relevant to the gut microbiota, and the causal relationship between gut microbiota and LP was analyzed using the Mendelian randomization (MR) method.
Methods: Through the two-sample MR method, single nucleotide polymorphisms (SNPs) relevant to gut microbiota were selected as instrument variables (IVs) to evaluate the causal association between gut microbiota and the risk of LP.
Results: According to the selection criteria of inverse-variance weighted (IVW), six bacterial genera were found to be significantly linked to the initiation of LP; The IVW results suggested that Oxalobacteraceae, Victivallaceae, and Actinobacteria could restrain the initiation of LP, showing protective effects against LP. Desulfovibrio, Veillonella, and Ruminococcus gauvreauii groups were demonstrated to have casual correlations with the onset of LP.
Conclusion: The relationship between gut microbiota and LP was not a single positive or inverse relationship. Investigation of the causal relationship of these gut microbiota with LP could further provide evidence for the intestine-skin axis theory. However, the specific mechanism of microorganisms affecting the skin remains to be clarified. In this paper, the protective effects and mechanisms of Oxalobacteraceae, Victivallaceae, and Actinobacteria on LP require further exploration.
Lichen planus (LP) is an idiopathic inflammatory skin disorder of unknown etiology, which can involve the skin, mucosa (especially oral mucosa), scalp, fingers, toes, and external genitalia. Among them, oral mucosal diseases are the most common which can ulcerate oral mucosa and Severe cases can cause make it more susceptible to carcinogenesis, especially on the long-term ulcer site. LP is considered a premalignant condition by World Health Organization (WHO) (1), 1%-2% cases malignant transformation to oral squamous cell carcinoma, while the detailed mechanism underlying this process are still obscure (2). When involving the skin, it usually occurs in the inner side of the limbs or throughout the body. The typical skin lesions are manifested with polygonal- or round-shaped purple-red flat papules with clear boundaries and a chronic course (1). Although the etiology and pathogenesis of LP have not been fully elucidated, many scientists believe that LP is mostly related to dysregulation of the immune system (1), and it is a chronic inflammatory autoimmune disease modulated by T cells, with a global incidence of 0.5% - 2.2% (2), most common in middle-aged females.
The gut microbiota is the largest microbial community in the human body. Gut microbiota exerts a significant role in maintaining the balance of the immune and nervous systems in the host, such as the clearance of pathogens, the development of the immune system, and the regulation of the central nervous system (3, 4). Gut microbiota is strongly linked to host immunity, and the activation state of the host and genetic susceptibility can be triggered or motivated by specific microorganisms (3). Disruption of the balance between the host and microbiota can lead to autoimmune diseases such as Sjogren’s syndrome (4), systemic lupus erythematosus (5), and rheumatoid arthritis (6) through different mechanisms.
Microorganisms, which were previously considered pathogenic microorganisms, are generally believed to supply colonization conditions for microorganisms during the initiation and progression of LP but do not result in the initiation and progression of LP. However, dysbiosis of gut microbiota may also play a role in the occurrence of LP (7).
Traditional observational studies are easily affected by reverse causality to produce bias and cannot clarify its causality. Mendelian randomization (MR) is widely applied for etiological inference in the field of genetic epidemiology (8), which can overcome the bias resulting from confounding and reverse causation issues. The exposure files and outcome files in the two-sample MR in this paper are from 2 different cohorts (9). In this study, we intended to reveal the correlation of gut microbiota at the genetic level with the onset of LP through the two-sample MR method based on the genome-wide association study data of gut microbiota and LP, so as to lay the foundation for the formulation of clinical prevention strategies for primary early-stage LP.
Genetic variations in the gut microbiota were derived from the largest genome-wide analysis of gut microbiota composition published to date by the MiBioGen Consortium (10, 11); their analysis enrolled 18,340 individuals from 24 cohorts, the majority of whom had European ancestry (n=13,266). The microbial composition was described targeting the variable regions V4(n=8472), V3-V4(n=5791), and V1-V2(n=4774) of the 16S rRNA gene, and classified using the direct classifying method. Microbiota quantitative trait loci (mbQTL) mapping analysis was performed to identify host genetic variations mapped by gene loci relevant to the abundance level of bacterial taxa in the gut microbiota. In the current study, the genus was the lowest taxonomic level. There are 131 genera with an average abundance higher than 1%, from which 12 unknown genera were excluded. Finally, 119 bacterial genera were identified. The GWAS summary statistics of LP were sourced from FinnGen (8th data release) (12). The GWAS of “lichen planus” adopted in this study contained 334116 individuals, including 3141 cases and 330975 controls.
IVs in MR study must meet three core assumptions: (1) correlation hypothesis: IVs are closely correlated with exposure, with F value > 10 as the standard of a close correlation; (2) Exclusive hypothesis: IVs do not affect outcomes; (3) Independence assumption: IVs are irrelevant to other confounding factors (9, 13). The criteria for the selection of IVs were as follows: (1) the potential IVs were selected with the significance threshold of single nucleotide polymorphisms (SNPs) related to each gut microbial genus within the range of loci (P<5.0×10-6); (2) Linkage disequilibrium (LD) between SNPs was calculated, and only the SNPs with the lowest P-value among SNPs with R2 < 0.001 (Clumping window size=10,000 kb) were retained; (3) SNPs with a minor allele frequency (MAF) ≤ 0.01 were excluded.
Data analysis in this study was performed using R (version 4.2.3) with the TwoSample MR (0.5.6) package and MRPRESSO (1.0). To verify whether there was a causation between exposure of gut microbiota and outcome LP, MR analysis was conducted using five methods such as inverse-variance weighted (IVW) (14), weighted median method (15), MR-Egger regression analysis (16), simple mode (12), and weighted mode (17). In addition, Cochran’s IVW Q was utilized to quantify the heterogeneity of IVs, and the false discovery rate test (FDR-q test) was performed (18), which is a multiple testing correction method used to control the expected proportion of false discoveries among the significant findings. It helps mitigate the risk of finding spurious associations due to testing multiple hypotheses. In the meantime, we performed a “Leave-One-Out” analysis by removing one genetic variant at a time and re-running the MR analysis. MR-egger intercepts to detect polytropy, in addition to which we performed MRPRESSO global tests to detect outliers and polytropy. This helps assess the influence of individual variants on the overall results. To assess the causal relationship between the gut microbiota and LP, we also carried out a reverse MR analysis of bacterial genera (phyla) found to be causally related to LP in the forward MR analysis. We used the GEO database (GSE52130) to retrieve LP-related expression chips, and used R-language “limma” package(3.52.2) for differential genes analysis, ggplot2 was used to visualize the results.
According to the selection criteria of IVs, 2763 SNPs were selected as IVs of 211 genera. As depicted in Table 1, 7 bacterial genera were identified by multiple MR methods, including Oxalobacteraceae, Victivallaceae, and Actinobacteria, which could restrain the onset of LP, showing a protective role, Desulfovibrio, Veillonella, NB1n and Ruminococcus gauvreauii group showed a promoting effect on lichen planus. IVW results showed that three bacterial genera were protective against LP: Actinobacteria (OR=0.64, 95%CI: 0.46-0.89, P=0.0093), Oxalobacteraceae (OR=0.78, 95%CI: 0.65-0.94, P=0.012), Victivallaceae (OR=0.83, 95% CI: 0.70-0.99, P=0.044), and the other four exhibited promoting effect on LP: Desulfovibrio (OR=1.59, 95%CI: 1.12-2.25, P=0.0082), Victivallaceae (OR=1.51. 95%CI: 1.02-2.25, P=0.039), Ruminococcus gauvreauii group (OR=1.52, 95%CI: 1.10-2.10, P=0.011), NB1n (OR=1.23, 95%CI: 1.02-1.49, P=0.034). Scatter plot showing the associations of the SNP effects on each bacterial genera against the SNP effects on LP (Figure 1).
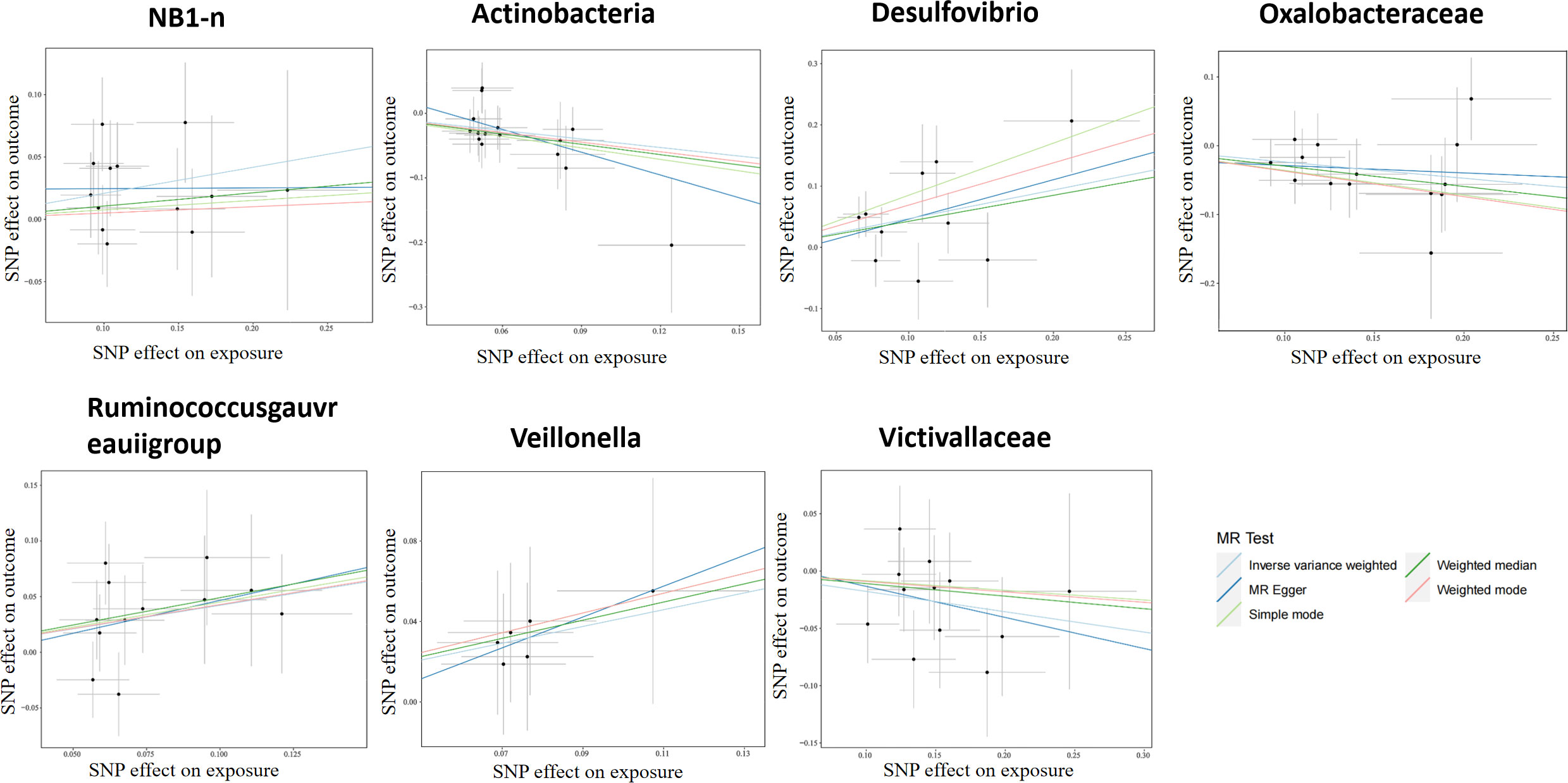
Figure 1 Scatter plot of two-sample MR results. The correlation between gut microbiota and LP was visualized in a scatter plot. In this plot, each black dot represented an SNP. With the correlation between SNP and exposure taken as the X axis, and the correlation between SNP and outcome taken as the Y axis, the slope of the drawn line marked the potential causal correlation of each method.
After the FDR-q test, Actinobacteria (OR=0.64, 95% CI: 0.46-0.89, P=0.0093, q=0.083) still displayed a significant protective role, while the other 6 gut microbiotas were suggested to correlate with LP in the IVW test, however, their correlation was insignificant after FDR-q test: Oxalobacteraceae (OR=0.78, 95%CI: 0.65-0.94, P=0.012, q=0.42), Victivallaceae (OR=0.83, 95%CI: 0.70-0.99, P=0.044, q=0.51), Desulfovibrio (OR=1.59, 95%CI: 1.12-2.25, P=0.0082, q=0.72), Veillonella (OR=1.51, 95%CI:1.02-2.25, P=0.039, q=0.81), and Ruminococcus gauvreauii group (OR=1.52, 95%CI: 1.10-2.10, P=0.011, q=0.72), NB1n (OR=1.23, 95%CI: 1.02-1.49, P=0.034, q=0.40). In addition, it was revealed that bacteria genera (phyla) in gut microbiota related to LP were screened by the forward MR analysis, but no significant correlation was found between these screened bacteria genera (phyla) and LP in the reverse MR analysis (LP and gut microbiota) (Supplementary Table 1).
For all instrumental variables, their F statistics individually range from 14 to 88, and all weak instrumental variables were excluded (Supplementary Table 2). Cochran’s IVW Q test exhibited no heterogeneity in these IVs (Supplementary Table 3). Furthermore, to identify potential heterogeneous SNPs, we performed a “Leave-one-out” analysis by omitting each SNP successively, and did not detect any single SNP that had a strong influence on the results (Figure 2). The q-value program was applied for false discovery rate (FDR) correction, and FDR-q<0.1 was deemed to be statistically significant after the pleiotropy test. However, even if FDR-q>0.1, a value of p<0.05 still suggested a certain correlation between gut microbiota and LP. Besides, based on the MR-Egger regression analysis results of scatter plot intercept, no significant horizontal pleiotropy was noted (P>0.05), and subsequent MR-PRESSO analysis did not find any significant outliers and no horizontal pleiotropy was found using the MR-PRESSO global test (P>0.05) (Supplementary Table 3). The funnel plot displayed a symmetrical and funneled shape, indicating little bias (Supplementary Figure 2).
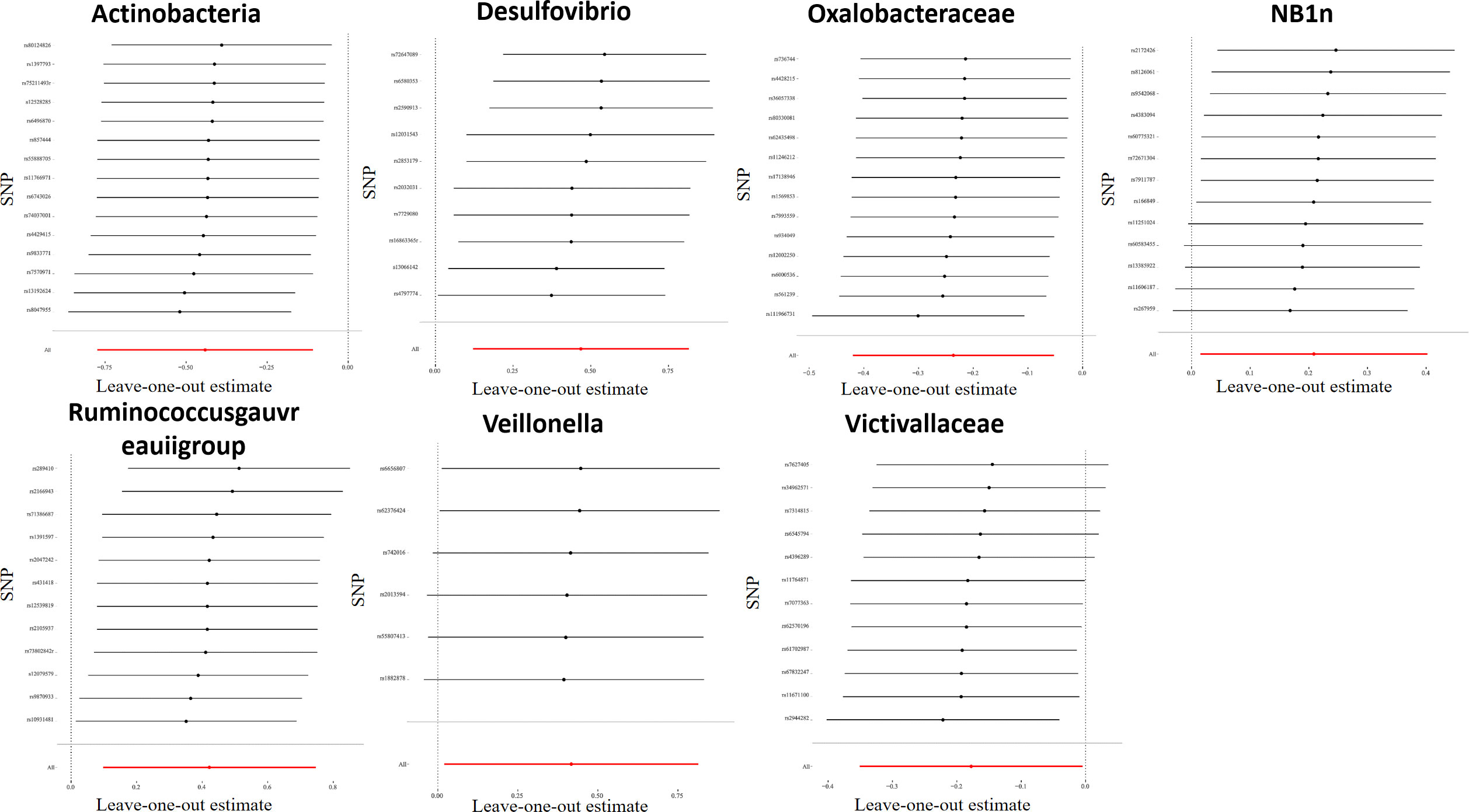
Figure 2 Leave-one-out plots for the causal association between gut microbiota and LP. After eliminating each SNP as an IV one by one, the overall trend did not change significantly, that was, no SNP with a great impact on the outcome among IVs.
It was observed that Oxalobacteraceae, Victivallaceae, and Actinobacteria could inhibit the occurrence of LP, exerting a protective effect, while Desulfovibrio, Veillonella, and Ruminococcus dgauvreauii group are causally related to the initiation of LP.
According to the screening conditions |fold change|>1.5 and P<0.05, a total of 340 genes were screened, including 260 upregulated genes, 90 downregulated genes. A volcano plot showed the distribution of differential genes (Supplementary Figure 1). The results revealed that the upregulated genes were mainly related to inflammation and the downregulated genes were mainly associated with various metabolic processes.
This study conducted a two-sample MR analysis using the summary statistics of gut microbiota in the largest GWAS meta-analysis by the MiBioGen consortium and the summary statistics of LP data published by FinnGen on R8 to further evaluate the causal association between gut microbiota and LP. In this paper, three bacterial genera (Actinobacteria, Oxalobacteraceae, Victivallaceae) were revealed to exhibit protective effects, and four bacterial genera (Desulfovibrio, Veillonella, Ruminococcus gauvreauii group, NB1n) showed promoting impacts on LP. After the FDR-q test of the aforementioned six bacterial genera, Actinobacteria was found to be causally related to LP, while the correlation between the other five bacterial genera and LP was no longer significant.
There are a variety of microbial communities in the human intestine, which play a significant role in maintaining intestine-skin homeostasis. When the relationship between gut microbiota and the immune system is damaged, the skin may be affected, promoting the development of skin diseases. However, the mechanism of gut microbiota affecting skin health remains unclear (19, 20). Mahmud MR et al. systematically evaluated the microecology of healthy skin and intestine, the impact of probiotics on the gut microbiome, and their impact on skin health; The underlying mechanism of the intestine-skin axis and the link between the intestine and skin diseases such as atopic dermatitis were discussed. Dysbiosis of the gut microbiota leads to disruption of the intestinal barrier integrity, resulting in increased intestinal permeability; This allows microorganisms and toxins to enter the circulatory system and reach other target organs, including the skin (21). The consequence is a combination of systemic and local inflammation, thereby triggering skin disease. As an immune disease, LP provides conditions for the colonization of microorganisms in the oral cavity, especially after the occurrence of oral LP, while Actinobacteria may have a protective effect against LP via the intestine-skin axis. In this manuscript, the analysis through Mendelian randomization shown Victivallaceae may be associated with a decreased risk of incident LP. Analysis of the presence of SNPs in 12 presumptive competent-related genes showed rs7627405 associated with IL17, while from the results of differential genes expression analysis in LP, IL1F10, IL1B, IL1F5, IL1F9 are more correlated with occurrence of LP. The above evidence highlight that Victivallaceae may be involved in low incidences of LP through regulating the inflammatory response. But it still needs to be proved by further experiments (22, 23).
In this study, the causal relationship between gut microbiota and LP was determined using MR analysis to exclude the interference of confounding factors and the impact of reversed causality on causal inference. The intensity of IVs in MR analysis was ensured by parameter settings. Horizontal pleiotropy was detected and excluded using MR-PRESSO and MR-Egger regression intercept analysis. In addition, the application of the FDR-q test reduced the incidence of false positives. Meanwhile, there were also some shortcomings. First of all, this study employed summary statistics rather than the original data in the process of analysis. Hence, it was impossible to analyze the subgroups, and the lowest taxonomic level of the exposure data was genus. The current study could only reflect the protective impacts of Actinobacteria on LP. However, the MR analysis of Bifidobacterium, the most common probiotic in Actinobacteria, did not suggest a relationship with LP, and the specific bacteria that inhibited the onset of LP could not be further explored.
Altogether, the two-sample MR study revealed a certain causal association between gut microbiota and LP, and our MR analysis suggested that Actinobacteria in the gut microbiota had a protective effect on the initiation and development of LP (IVW: P<0.05, FDR-q<0.1). According to the intestine-skin axis theory, the dysbiosis of gut microbiota will affect the integrity of the intestinal barrier, leading to changes in intestinal permeability, so that microorganisms in the intestine can reach organs of the whole body including the skin through the circulatory system. Bacteria such as Actinobacteria (the dominant bacteria in the human intestine), Bacteroidetes, Firmicutes, and Proteobacteria account for about 90% of intestinal bacteria (24); In the meantime, Bifidobacterium and Actinobacteria can protect the intestinal barrier (25) Although our MR analysis did not suggest a correlation of Bifidobacterium in the intestine with LP (IVW: P>0.05), a randomized controlled trial (RCT) can be applied to further clarify the inhibitory effect of Actinobacteria on the onset of LP and its specific protective mechanism and further explore whether there is a potential mechanism related to the intestine-skin axis.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
MY, Y-LO: conceived the study and drafted the manuscript. L-YX, MA: data evaluation, manuscript preparation. MG, RF, RS: analyzed the data and revised the manuscript. L-LF, H-CF: conceived the study, designed the data evaluation, and prepared the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by a grant from the Foundation of Guiyang Science and Technology Bureau (No. [2018]1-56). MY was supported by the Merit Scholarship of Hamburg University for International Students (No.7238065).
Authors thank the MiBioGen and FinnGen consortium for sharing the summary-level GWAS data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1235982/full#supplementary-material
1. El-Howati A, Thornhill MH, Colley HE, Murdoch C. Immune mechanisms in oral lichen planus. Oral Dis (2023) 29(4):1400–15. doi: 10.1111/odi.14142
2. Tziotzios C, Lee JYW, Brier T, Saito R, Hsu CK, Bhargava K, et al. Lichen planus and lichenoid dermatoses: Clinical overview and molecular basis. J Am Acad Dermatol (2018) 79(5):789–804. doi: 10.1016/j.jaad.2018.02.010
3. Ansaldo E, Farley TK, Belkaid Y. Control of immunity by the microbiota. Annu Rev Immunol (2021) 39:449–79. doi: 10.1146/annurev-immunol-093019-112348
4. Singh M, Teles F, Uzel NG, Papas A. Characterizing microbiota from sjögren's syndrome patients. JDR Clin Trans Res (2021) 6(3):324–32. doi: 10.1177/2380084420940623
5. Chen Y, Lin J, Xiao L, Zhang X, Zhao L, Wang M, et al. Gut microbiota in systemic lupus erythematosus: A fuse and a solution. J Autoimmun (2022) 132:102867. doi: 10.1016/j.jaut.2022.102867
6. Zhao T, Wei Y, Zhu Y, Xie Z, Hai Q, Li Z, et al. Gut microbiota and rheumatoid arthritis: From pathogenesis to novel therapeutic opportunities. Front Immunol (2022) 13:100716511. doi: 10.3389/fimmu.2022.100716511
7. Jung W, Jang S. Oral microbiome research on oral lichen planus: Current findings and perspectives. Biol (Basel) (2022) 11(5):723. doi: 10.3390/biology11050723
8. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet (2014) 23(R1):R89–98. doi: 10.1093/hmg/ddu328
9. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: The STROBE-MR statement. JAMA (2021) 326(16):1614–21. doi: 10.1001/jama.2021.18236
10. Kurilshikov A, Medina-Gomez C, Bacigalupe R, Radjabzadeh D, Wang J, Demirkan A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet (2021) 53(2):156–65. doi: 10.1038/s41588-020-00763-1
11. MiBioGen consortium. MiBioGen. Available at: https://mibiogen.gcc.rug.nl/.
12. Milne RL, Kuchenbaecker KB, Michailidou K, Beesley J, Kar S, Lindström S, et al. Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat Genet (2017) 49(12):1767–78. doi: 10.1038/ng.3785
13. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife (2018) 7.
14. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol (2013) 37(7):658–65. doi: 10.1002/gepi.21758
15. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol (2016) 40(4):304–14. doi: 10.1002/gepi.21965
16. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method [published correction appears in Eur J Epidemiol. Eur J Epidemiol (2017) 32(5):377–89. doi: 10.1007/s10654-017-0255-x
17. Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol (2017) 46(6):1985–98. doi: 10.1093/ije/dyx102
18. Storey JD, TibshIrani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA (2003) 100(16):9440–5. doi: 10.1073/pnas.1530509100
19. Bonar-Alvarez P, Perez Sayans M, Garcia-Garcia A, Chamorro-Petronacci C, Gandara-Vila P, Luces-Gonzalez R, et al. Correlation between clinical and pathological features of oral lichen planus: A retrospective observational study. Med (Baltimore) (2019) 98(8):e14614.
20. Obermeier KT, Wuersching SN, Liokatis P, Smolka W, Poxleitner P, Kleye C, et al. Metastases of OSCC based on oral lichen ruber planus. Cancers (Basel) (2023) 15(16).
21. Mahmud MR, Akter S, Tamanna SK, Mazumder L, Esti IZ, Banerjee S, et al. Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes (2022) 14(1):2096995. doi: 10.1080/19490976.2022.2096995
22. Wiertsema SP, van Bergenhenegouwen J, Garssen J, Knippels LMJ. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients (2021) 13(3).
23. Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek (2020) 113(12):2019–40.
24. Jethwani P, Grover K. Gut microbiota in health and dis-eases—a review. Int J Curr Microbiol Appl Sci (2019) 8(8):1586–99.
Keywords: Mendelian randomization study, gut microbiota, lichen planus, intestine-skin axis, correlations
Citation: Yan M, Ouyang Y-L, Xiao L-Y, Ao M, Gosau M, Friedrich RE, Smeets R, Fu L-L, Feng H-c and Burg S (2023) Correlations between gut microbiota and lichen planus: a two-sample Mendelian randomization study. Front. Immunol. 14:1235982. doi: 10.3389/fimmu.2023.1235982
Received: 07 June 2023; Accepted: 29 August 2023;
Published: 12 September 2023.
Edited by:
Mario César Salinas-Carmona, Autonomous University of Nuevo León, MexicoReviewed by:
Noé Macias-Segura, Autonomous University of Nuevo León, MexicoCopyright © 2023 Yan, Ouyang, Xiao, Ao, Gosau, Friedrich, Smeets, Fu, Feng and Burg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-chao Feng, aGNmZW5nQGd6dS5lZHUuY24=; Ling-Ling Fu, ZnUubGluZ2xpbmdAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.